Abstract
Thirteen extensively drug-resistant tuberculosis isolates which were highly resistant to a broad spectrum of antituberculosis drugs were identified from 1,926 clinical isolates in China. They had highly diverse mycobacterial interspersed repetitive-unit-variable-number tandem-repeat patterns. Most, but not all, of the drug target genes had mutations contributing to resistance to the corresponding drug.
Extensively drug-resistant tuberculosis (XDR-TB) has emerged worldwide as one of the biggest threats to public health and TB control programs. XDR-TB is defined as TB caused by a Mycobacterium tuberculosis strain that is resistant to at least rifampin (RIF) and isoniazid (INH) among the first-line anti-TB drugs (the definition of multidrug-resistant TB [MDR-TB] strains), resistant to a fluoroquinolone (FQ), and resistant to at least one of the three injectable second-line drugs (11). XDR-TB is difficult and expensive to treat: an extremely high mortality (98%) was reported in patients with human immunodeficiency virus (HIV) coinfection in South Africa, and a 33% mortality rate was reported in those without HIV coinfection (1, 3, 4, 7). So far, XDR-TB has been documented in 41 countries in all continents (12). Although China has a high annual risk for TB cases and an increasing prevalence of MDR-TB cases, XDR-TB has not yet been reported in China, except in Hong Kong (12). In the present study, 13 XDR-TB isolates were identified from patients in mainland China and were further characterized as to drug resistance phenotype and genotype. To our best knowledge, this is the first report of XDR-TB in mainland China.
From January 2002 to December 2005, 1,926 clinical Mycobacterium tuberculosis isolates were collected from 1,926 HIV-negative TB patients in the Beijing Chest Hospital. These isolates were screened initially by the absolute two-concentration method, and then the MICs of nine TB drugs for them were determined by the agar dilution method. The nine drugs included both first-line and second-line anti-TB drugs. Specifically, they were INH, RIF, amikacin (AMK), ofloxacin (OFX), streptomycin (STR), ethambutol (EMB), pyrazinamide (PZA), ethionamide (ETH), and sodium p-aminosalicylic acid (PAS). The MIC results showed that 18.8% (362/1,926) and 20.2% (389/1,926) of the isolates were resistant to INH and RIF, respectively. Moreover, 9.2% (177/1,926), 5.9% (113/1,926), and 8.7% (167/1,926) of the isolates were resistant to FQs, AMK, and other anti-TB drugs, respectively. Furthermore, 10.75% (207/1,926) of the isolates showed resistance to both INH and RIF (i.e., MDR-TB), and of the 207 MDR-TB isolates, 13 (6.28%) were resistant to INH, RIF, OFX, and AMK, which met the revised definition of XDR-TB (11) (Table 1). Of the 13 XDR-TB isolates identified, 12 isolates (92.3%) were resistant to more than four anti-TB drugs and six isolates (46.1%) were resistant to at least seven drugs. Besides the four drugs described in the definition of XDR-TB, resistance to STR and PZA was found in nine isolates.
TABLE 1.
Results of MIC testing of 13 XDR-TB strains isolated from January 2002 to December 2005
| Strain | Patient's sex (age in yr) | MIC (μg/ml) ofa:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RIF (50) | INH (1) | OFX (5) | AMK (5) | STR (10) | EMB (5) | PZA (50) | PAS (1) | ETH (25) | ||
| H37Rv | 6.25 | 0.125 | 0.625 | 0.625 | 1.25 | 0.625 | 6.25 | 0.125 | 3.125 | |
| A88 | Female (51) | 4,000 | 10 | 20 | 20 | 20 | 10 | 100 | 0.25 | 12.5 |
| B42 | Female (37) | 500 | 20 | 10 | 80 | 1,600 | 20 | 800 | 4 | 50 |
| C10 | Male (40) | 125 | 5 | 20 | 10 | 200 | 1.25 | 12.5 | 1 | 12.5 |
| C28 | Male (41) | 125 | 40 | 10 | 10 | 5 | 10 | 12.5 | 0.25 | 6.25 |
| C29 | Male (43) | 2,000 | 20 | 10 | 160 | 2.5 | 0.625 | 100 | 16 | 100 |
| C52 | Male (28) | 2,000 | 10 | 20 | 10 | 800 | 40 | 3,200 | 32 | 25 |
| C57 | Male (45) | 2,000 | 160 | 20 | 10 | 20 | 1.25 | 1,600 | 0.5 | 6.25 |
| C80 | Male (44) | 1,000 | 160 | 10 | 10 | 2.5 | 2.5 | 400 | 0.25 | 12.5 |
| C98 | Male (40) | 4,000 | 5 | 10 | 40 | 1,600 | 1.25 | 12.5 | 1 | 12.5 |
| D17 | Male (45) | 4,000 | 1.25 | 20 | 20 | 5 | 0.625 | 25 | 1 | 25 |
| D22 | Male (50) | 2,000 | 2.5 | 10 | 10 | 200 | 1.25 | 1,600 | 0.5 | 25 |
| E19 | Male (54) | 2,000 | 20 | 40 | 160 | 20 | 1 | 3,200 | 32 | 50 |
| E42 | Male (42) | 2,000 | 10 | 10 | 160 | 20 | 10 | 400 | 0.5 | 6.25 |
The cutoff MICs used in this study are given at the top in parentheses. MICs lower than the cutoff MICs are shown in italic.
The extreme drug resistance of these XDR-TB strains was reflected not only in the broad spectrum of anti-TB drugs that they were resistant to, as described above, but also in the high degree of drug resistance. If high resistance is defined as having a MIC higher than 10 times the cutoff value, of the 13 XDR-TB strains, 11, 9, 5, 5, 4, and 3 strains, respectively, were highly resistant to RIF, INH, PZA, STR, AMK, and PAS (Table 1). Strikingly, the MICs for INH and STR were as high as 160 times the cutoff value in several isolates, and the MIC for RIF reached 80 times the cutoff value in some isolates. The extreme resistance to INH, STR, and RIF may be attributed to the drugs being the first choice in treatment and to the longtime use in both the old standard anti-TB drug regimens (INH plus STR plus PAS) and current regimens in China, which focus on INH, RIF, PZA and EMB (see the patient history section of Table S1 in the supplemental material). On the other hand, the extreme resistance to INH, STR, and RIF might indicate that XDR-TB isolates had a very long in vivo evolution and selection history. Patient documents indicated that five patients were retreatment TB patients with disease history of 3 to 12 years (see Table S1 in the supplemental material). Presumably drug overuse and noncompliance in these patients contributed to their XDR-TB since most patients were treated at hospitals for only a short period. Most patients used INH, RIF, PZA, and EMB, as recommended by the WHO. Fluoroquinolones were also widely used, and the second-line injectable drugs (such as AMK) were less frequently used. Unfortunately, case documents available to our institution were limited, as most of those patients were transferred to Beijing Chest Hospital from local hospitals, stayed for less than 3 months, and then checked out with their medicines.
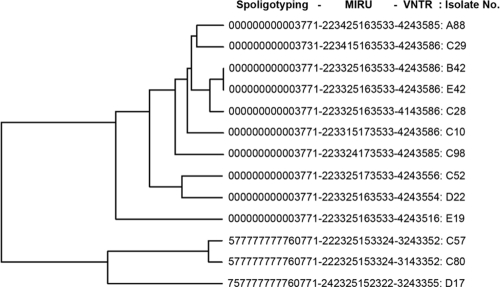
The genetic background of the 13 XDR-TB isolates was analyzed by spoligotyping and mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) analysis. Spoligotyping was performed with a commercially available kit (Isogen Bioscience BV, Maarssen, The Netherlands) according to the manufacturer's protocols. PCR-based MIRU-VNTR analysis was performed using the 12-MIRU-locus method (5, 6) and 7 VNTR loci (ETR-A, ETR-B, ETR-C, ETR-D, ETR-E, QUB11a, and QUB11b) as described by Frothingham and Meeker-O'Connell (2, 10). Spoligotyping results showed 10 of 13 XDR-TB strains had the Beijing genotype pattern (000000000003771 and 000000000003731) (Fig. 1). MIRU and VNTR analysis identified nine MIRU patterns and nine VNTR patterns (Fig. 1). Five strains shared the MIRU pattern 2233-2516-3533, 2 strains (C57 and C80) shared the MIRU pattern 2223-2515-3324, and the other six XDR-TB strains had different MIRU patterns. By VNTR analysis, five strains shared one pattern (4243586) and two strains (A88 and C98) shared another pattern (4243585), while the others had different patterns. Taken together, these results indicated the heterogeneous genetic background of these 13 XDR-TB strains. Only B42 and E42 strains had the exactly same genotype, while the other 11 XDR-TB isolates were of different genotypes. These supported the current understanding that XDR-TB strains emerged independently in many different places and on multiple occasions (8, 9). In addition, although 10 of 13 XDR-TB strains (76.9%) belonged to the Beijing family, which is the dominant TB genotype in China, no evidence showed a significant correlation between the Beijing genotype and drug resistance in China.
FIG. 1.
Dendrogram showing clustering of XDR-TB genotypes. Strain numbers A88, B42, C10, C28, C29, C52, C98, D22, E19, and E42 represent XDR-TB strains with the typical pattern of the Beijing family genotype. Strain numbers C57, C80, and D17 represent the genotype of the T1 family. M. tuberculosis H37Rv and Mycobacterium bovis BCG were used as controls.
In order to uncover whether and to what degree the major important known mutations in drug target genes contributed to the extreme drug resistance, nine genes (katG, inhA, rpoB, pncA, embB, rpsL, gyrA, and rrs and its promoter), which included most important known drug target genes, except for the PAS resistance-related thyA gene, were amplified from all 13 XDR-TB strains and sequenced. The 10 sets of primers specific to these genes were based on the genome of M. tuberculosis H37Rv and amplified the hot spots or the whole genes (see Table S2 in the supplemental material) (13). Sequencing results revealed that most of these genes had mutations contributing to resistance to the corresponding drug, and the detailed mutation patterns are presented in Table 2. However, several strains had no sense mutations in a drug target gene but were resistant to the corresponding drug, and examples of such could be found with the B42, C29, and E19 strains (inhA [ETH]); C98 and D22 strains (gyrA [OFX]); C29, C57, and D22 strains (pncA [PZA]); and C10 and E42 strains (rrs [AMK]) (Table 2). These data suggested that the extensive drug resistance can be largely attributed to, but could not be completely explained by, the accumulation of known mutations in drug target genes.
TABLE 2.
Mutation patterns of the different drug target genes
| Strain | Mutation pattern of target gene (corresponding drug)a:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| rpoB (RIF) | katG (INH) | gyrA (FQ) | rrs (AMK, STR) | rpsL (STR) | embB (EMB) | pncA (PZA) | inhA (ETH, INH) | |
| A88 | S531L (R) | T180C, S315T (R) | D94N (R) | C-1401→T (R, R) | K43R (R) | M306V (R) | D12A (S) | None (S, R) |
| B42 | H526Y (R) | S315T (R) | D94G (R) | A-1400→G (R, R) | 47T,b,d K43R (R) | M306I, D328G (R) | None (R) | None (R, R) |
| C10 | L533P (R) | W477 stop (R) | D94Y (R) | None (R, R) | K43R (R) | None (S) | None (S) | None (S, R) |
| C28 | S531W (R) | H108Q (R) | D94A (R) | Multiple (R, S) | None (S) | M306I (R) | None (S) | None (S, R) |
| C29 | S531L (R) | W328L (R) | D94H (R) | A-1400→G (R, S) | None (S) | None (S) | None (R) | None (R, R) |
| C52 | S531L (R) | N138S, S315T (R) | D94G (R) | C-1296→Gd (R, R) | K43R, G92,b K88R (R) | M306V, G331R,d E327G,d D328G (R) | W68Rd (R) | None (S, R) |
| C57 | S531L (R) | T262R, F252L (R) | A90V (R) | C-1296→Gd (R, R) | K88R (R) | None (S) | None (R) | None (S, R) |
| C80 | None (R) | T262R (R) | A90V (R) | C-1296→G,d G-1483→A (R, S) | None (S) | None (S) | K48Ed (R) | None (S, R) |
| C98 | Q517H, S531L, A532Gd (R) | S315T (R) | None (R) | C-1293→G,d C-1296→G,d A-1400→G (R, S) | K43R (S) | None (S) | None (R) | None (S, R) |
| D17 | S531L (R) | None (R) | D94G (R) | C-1293→G,d C-1296→Gd (R, S) | None (S) | None (S) | None (S) | None (S, R) |
| D22 | S531L (R) | H108E, W477 stop (R) | None (R) | C-1296→G,d C-1351→Td (R, R) | K88R (R) | None (S) | None (R) | None (S, R) |
| E19 | S531L (R) | S315T (R) | D94Y (R) | A-1400→G (R, R) | K43R (R) | M306V (S) | V180Fd (R) | None (R, R) |
| E42 | S531L (R) | S140N (R) | D94A (R) | None (R, R) | K43R (R) | M306V (R) | −11A-→Gc (R) | None (S, R) |
Resistance (R) or sensitivity (S) of strains to the corresponding anti-TB drugs is noted in parentheses. Strains without any sense mutations in the drug target genes but resistant to the corresponding drugs are noted with “None” in boldface.
Base lost in this gene position.
Mutation in the promoter of the drug target gene.
New mutation.
In conclusion, 13 XDR-TB strains were identified by screening 1,926 clinical isolates in China. These strains were highly resistant to a broad spectrum of anti-TB drugs. Ten strains belonged to the Beijing family, but high genotypic diversity was found by MIRU-VNTR analysis. Mutations in the corresponding drug target genes could be found for most but not all of the drugs the strains were resistant to, indicating other drug resistance mechanisms contributed to the drug resistance. To our knowledge, this is the first report of XDR-TB strains in China.
Supplementary Material
Acknowledgments
We thank the National High-Tech R&D Program (863) of China (2007AA02Z405), the National Natural Science Foundation of China (30770030), and the China National Basic Research Program (973) (2006CB504401) for financial support.
Footnotes
Published ahead of print on 22 October 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Cohen, J. 2006. Infectious disease. Extensively drug-resistant TB gets foothold in South Africa. Science 3131554. [DOI] [PubMed] [Google Scholar]
- 2.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 1441189-1196. [DOI] [PubMed] [Google Scholar]
- 3.Gandhi, N. R., A. Moll, and R. Pawinski. 2006. High prevalence and mortality from extensively-drug resistant (XDR) TB in TB/HIV coinfected patients in rural South Africa, p. 13-18. In Program of the XVI International AIDS Conference, Toronto, Canada.
- 4.Gandhi, N. R., A. Moll, A. W. Sturm, R. Pawinski, T. Govender, U. Lalloo, K. Zeller, J. Andrews, and G. Friedland. 2006. Extensively drug-resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 3681575-1580. [DOI] [PubMed] [Google Scholar]
- 5.Han, H., F. Wang, Y. Xiao, Y. Ren, Y. Chao, A. Guo, and L. Ye. 2007. Utility of mycobacterial interspersed repetitive unit typing for differentiating Mycobacterium tuberculosis isolates in Wuhan, China. J. Med. Microbiol. 561219-1223. [DOI] [PubMed] [Google Scholar]
- 6.Kwara, A., R. Schiro, L. S. Cowan, N. E. Hyslop, M. F. Wiser, S. Roahen Harrison, P. Kissinger, L. Diem, and J. T. Crawford. 2003. Evaluation of the epidemiologic utility of secondary typing methods for differentiation of Mycobacterium tuberculosis isolates. J. Clin. Microbiol. 412683-2685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwoon, Y. S., Y. H. Kim, G. Y. Suh, M. P. Chung, H. Kim, O. J. Kwon, Y. S. Choi, K. Kim, J. Kim, Y. M. Shim, and W. J. Koh. 2008. Treatment outcomes of HIV-uninfected patients with multidrug-resistant and extensively drug-resistant tuberculosis. Clin. Infect. Dis. 47496-502. [DOI] [PubMed] [Google Scholar]
- 8.Lawn, S. D., and R. Wilkinson. 2006. Extensively drug resistant tuberculosis. BMJ 333559-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mlambo, C. K., R. M. Warren, X. Poswa, T. C. Victor, A. G. Duse, and E. Marais. 2008. Genotypic diversity of extensively drug-resistant tuberculosis (XDR-TB) in South Africa. Int. J. Tuberc. Lung Dis. 1299-104. [PubMed] [Google Scholar]
- 10.Skuce, R. A., T. P. McCorry, J. F. McCarroll, S. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148519-528. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization. 2006. Case definition for extensively drug-resistant tuberculosis. Wkly. Epidemiol. Rec 81408. [PubMed] [Google Scholar]
- 12.World Health Organization. November 2007, posting date. XDR-TB facts (November 2007). World Health Organization, Geneva, Switzerland. http://www.who.int/tb/challenges/xdr/facts_nov2007_en.pdf.
- 13.Zhang, Y., C. Vilchèze, and W. R. Jacobs, Jr. 2005. Mechanisms of drug resistance in Mycobacterium tuberculosis, p. 115-140. In S. T. Cole, K. D. Eisenach, D. N. McMurray, and W. R. Jacobs, Jr. (ed.), Tuberculosis and the tubercle bacillus. ASM Press, Washington, DC.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.