Abstract
A sequencing assay for detection of mutations conferring resistance to human immunodeficiency virus type 1 (HIV-1) integrase inhibitors raltegravir and elvitegravir was developed using the automated TruGene sequencing system. The assay returned clear sequencing results for samples with ≥500 RNA copies/ml for mutation detection and HIV-1 subtyping across a spectrum of HIV-1 subtypes.
Genotypic antiretroviral resistance testing (GART) has become indispensable in the management of patients infected with human immunodeficiency virus type 1 (HIV-1) (2, 12). In clinical laboratories, GART for HIV-1 is mostly performed by reverse transcription (RT)-PCR, followed either by bidirectional sequencing with a fluorochrome dye termination technique or by a CLIP reaction, which uses fluorochrome-labeled forward and reverse primers in conjunction with nucleotide terminators (3-5, 9). In a clinical setting, the latter technique is used with the largely automated TruGene sequencing system (Siemens Medical Solutions Diagnostics, Eschborn, Germany), and a commercially available assay system, the TruGene HIV-1 genotyping kit, allows detection of mutations conferring drug resistance to reverse transcriptase and protease inhibitors (3-5, 9).
The emergence of multiple-drug-resistant HIV particles initiated developments of novel treatment principles. One such development targets the integration of HIV DNA into the host genome by inhibiting HIV-1 integrase (13). Two such compounds (raltegravir and elvitegravir) have been developed and recently introduced into clinical practice (6, 15). Mutations conferring resistance to those two compounds have emerged in treated individuals, requiring expansion of existing GART methods for detection of these compounds in clinical laboratories (11, 16). As the TruGene sequencing system is available in many clinical laboratories, we set out to develop a sequencing assay for detection of HIV-1 integrase inhibitor mutations by using this analysis platform.
Backup EDTA-plasma samples from HIV-1-infected patients were blinded with respect to clinical data and used in an anonymous fashion throughout the study. The clinical data set for each sample specified viral load, subtype, and antiviral drug regimen at the time of sample collection, as all samples had routinely been examined for viral loads by the COBAS TaqMan 48 HIV-1 test (Roche, Vienna, Austria) and for HIV-1 subtype with the TruGene HIV-1 genotyping kit (Siemens), followed by sequence comparison using the Stanford University HIV Drug Resistance Database (http://hivdb.stanford.edu/cgi-bin/IIResiNote.cgi). Plasma from the EDTA-blood samples had been separated within 30 min and stored at −70°C in aliquots of 1 ml.
The AREVIR HIV-1 subtype panel (K. Korn, Institute of Virology, University of Erlangen, Germany), which contains samples from 12 members of group M (members A to H) at about 105 RNA copies/ml each, was used as reference material (1).
RNA was prepared from 1 ml plasma in an automated fashion by using a MagNA Pure LC instrument (Roche Applied Science, Mannheim, Germany) with a MagNA Pure total nucleic acid isolation kit (large volume; Roche). Elution was performed with 50 μl (7).
RT and PCR were performed by using a one-step RT-PCR kit (Qiagen, Hilden, Germany). Prior to the RT, RNA was treated for 2 min at 95°C, placed on ice for 2 min, and incubated at room temperature for 5 min. RT-PCR was carried out with a 50-μl volume containing 7 μl sample RNA, 0.2 μM forward primer (5′-TTAGATGGAATAGATAAGGCCC-3′; GenBank accession no. K03455, nucleotides [nt] 4233 to 4254), 0.4 μM reverse primer (5′-TGCTGTCCCTGTAATAAACCCG-3′; GenBank accession no. K03455, nt 4899 to 4920), 400 μM each deoxynucleoside triphosphate, and 10 U RNase OUT (Invitrogen, Karlsruhe, Germany). Buffer Q solution and the RT-PCR enzyme mixture were added as detailed in the manufacturer's instructions. RT-PCR was run with a Biometra thermocycler (TPersonal; Biometra, Göttingen, Germany) with the following temperature program: 30 min at 50°C and 15 min at 95°C, followed by 42 cycles with 30 s at 94°C, 45 s at 53°C, and 120 s at 72°C and a final extension for 10 min at 72°C. Temperature ramping was set to 1°C/s. The RT-PCR yielded a product of 688 bp of the HIV-1 integrase region, which was initially confirmed by agarose gel electrophoresis.
The CLIP sequencing reaction was performed by using a 7-deaza-dGTP Cy5/Cy5.5 dye primer cycle sequencing kit (Siemens) according to the manufacturer's instructions. The four CLIP reaction mixtures contained CLIP buffer brought to 1×, 10% dimethyl sulfoxide (Sigma, Munich, Germany), 0.4 μM forward primer (5′-Cy5.5-GTAGCCAGCTGTGATAAATGTC-3′; GenBank accession no. K03455, nt 4338 to 4359), 0.2 μM reverse primer (5′-Cy5-CTGCCATTTGTACTGCTGTCT-3′; GenBank accession no. K03455, nt 4747 to 4767), 1.25 μl sample cDNA, one of the four terminator nucleotides, and the CLIP enzyme mixture, according to the manufacturer's instructions. The CLIP-specific cycling program was 5 min at 94°C, followed by 30 cycles of 20 s at 94°C, 20 s at 62°C, and 120 s 70°C and final extension for 5 min at 70°C. Thereafter, stop solution (10 μl) was added. Samples were heated to 94°C for 2 min and incubated for a minimum of 1 h at 4°C. Fragments (1.5 to 2 μl/lane) were separated on a TruGene tower (Siemens) at 60°C, 1,600 V, 50% laser power, and a sampling rate of 0.5 s for 60 min with a size 500 polyacrylamide gel. The sequencing covered a 409-bp stretch of the RT-PCR product, corresponding to codons 41 through 176 of the HIV-1 integrase gene region (13; Stanford University HIV Drug Resistance Database [http://hivdb.stanford.edu/cgi-bin/IIResiNote.cgi]). The forward and reverse sequences were searched and combined using the OpenGene software module GeneObjects (version 4.0; Siemens) (using Macintosh OSX) and read against a newly created HIV-1 integrase sequence-specific reference library. This library was generated with the OpenGene software program (Siemens) by importing the respective sequences into the library file in an aligned fashion, as detailed in the manufacturer's instructions. The consensus sequences of all HIV-1 group M subtypes and circulating recombinant forms were downloaded from the Los Alamos HIV database (http://www.hiv.lanl.gov/). The respective reference sequences were downloaded from the National Center for Biotechnology Information Genotyping resource (http://www.ncbi.nih.gov/projects/genotyping/formpage.cgi) (see Table S1 in the supplemental material) (14). Report generating was set up to classify sequence variations in resistance mutations, silent mutations, polymorphisms (codon changes not at resistance sites), and other mutations at resistance sites, as had been established with the TruGene HIV-1 genotyping kit.
One assay run with four samples was completed after about 2 days when the RT-PCR was run overnight. The total hands-on time required, including sequencing, interpretation, and report generation, was about 2.5 h.
After optimization of RT-PCR and CLIP reaction, the group M members of the AREVIR HIV-1 subtype panel were examined to find out whether the assay allows for sequencing of various HIV-1 group M subtypes. All panel members gave clear, unequivocal sequencing results. Mutations to integrase inhibitors were not detected in any of the panel members. The HIV-1 subtypes of all panel members were correctly identified, and the sequence homologies to the respective reference sequences were found to be between 94.9% and 99.5% (Table 1). Thus, the developed assay did not show any relevant bias against any of the various HIV-1 group M subtypes.
TABLE 1.
Subtype assignment of the AREVIR HIV-1 subtype panel by integrase inhibitor sequencing
| Panel member | Referencea assignment | Assignment by integrase sequencing | Homologyb (%) |
|---|---|---|---|
| A | A | A | 95.35 |
| B | B | B | 97.31 |
| C | C | C | 99.51 |
| D1 | D | D | 98.04 |
| D2 | A | A | 97.07 |
| D3 | D | CRF10_CD | 96.58 |
| D | 96.09 | ||
| E1 | CRF01_AE | CRF01_AE | 99.51 |
| E2 | CRF01_AE | CRF01_AE | 99.27 |
| F1 | B | B | 97.07 |
| F2 | F | CRF12_BF | 96.58 |
| F | 96.33 | ||
| G | G | G | 97.10 |
| H | H | H | 94.87 |
Corresponding AREVIR reference subtype determined by sequencing of the pol region (K. Korn, University of Erlangen).
Sequence homology to the reference sequences in the library of the integrase sequencing assay.
A minimal viral load limit for the sequencing assay was determined by using two high-copy-number samples of subtype B, each spiked into HIV-negative plasma to yield samples with 1,000, 750, 500, and 250 copies/ml. Both sample series were tested twice, each in parallel, on two different days. All samples containing 500 copies/ml or more yielded clear sequencing results (in four runs out of four). With the 250-copy/ml samples, only two out of four runs were successful. Thus, the minimum viral load that allowed consistent sequencing was found to be 500 copies/ml, which is slightly superior or comparable to the level for published GART assays (5, 8).
In order to assess the concordance of the sequencing assay, one high-copy-number subtype B sample was diluted to about 1,000 copies/ml and assayed eight times on eight different days (Table 2). The one sequencing result that showed the greatest similarity to the B consensus sequence in our database (98% homology) was assumed to be correct. Variations at any base within the 409-bp stretch were regarded as inaccuracies, which ranged from 0 to 7 bases per run, corresponding to concordances of 98.3% to 100% (Table 2). Interestingly, the base variations did not represent any false base exchanges, deletions, or insertions. Instead, wobble nucleotides (designated R and Y) were detected, resulting in heterozygous sequences at the positions affected (Table 2). Only one position (321 C→Y) affected an integrase inhibitor mutation codon (encoding S147) and would have been regarded as a silent mutation in a clinical diagnostic setting (Table 2). Thus, we found that the developed assay generated high-quality sequence data comparable to those for published GART assays using the TruGene system (9).
TABLE 2.
Concordance of sequencing for integrase inhibitor mutations of a single subtype B sample
| Sequencing run | No. of variations in 409-bp stretch | Concordance (%) | Homology to subtype B consensus sequence (%) | Sequence variation(s) at indicated nucleotide position(s)a |
|---|---|---|---|---|
| 1 | 0 | 100.0 | 98.0 | |
| 2 | 1 | 99.7 | 97.8 | 395 T→Y |
| 3 | 3 | 99.3 | 97.6 | 126 A→R, 180 T→Y, 321 C→Yb |
| 4 | 3 | 99.3 | 97.6 | 126 A→R, 180 T→Y, 321 C→Yb |
| 5 | 4 | 99.0 | 97.3 | 180 T→Y, 321 C→Y,b 392 T→Y, 395 T→Y |
| 6 | 4 | 99.0 | 96.8 | 126 A→R, 180 T→Y, 321 C→Y,b 392 T→Y |
| 7 | 5 | 98.8 | 96.8 | 126 A→R, 180 T→Y, 213 A→R, 321 C→Y,b 392 T→Y |
| 8 | 7 | 98.3 | 96.6 | 57 A→R, 84 A→R, 1267 A→R, 180 T→Y, 258 T→Y, 321 C→Y,b 372 A→R |
Numbers indicate nucleotide positions on the 409-bp sequencing stretch. Nucleotide exchanges were not detected. At the indicated positions, wobble nucleotides Y and R were observed. A, adenine; C, cytosine; T, thymine; Y, thymine or cytosine; R, adenine or guanine.
T at position 321 represents a silent mutation (corresponding to resistance mutation codon S147G).
The new assay was evaluated by testing 54 backup samples, 15 of which originated from patients on integrase inhibitor (such as raltegravir) treatment. All samples were run only once, and each returned a clear sequencing result. In 11 of those 15 samples, integrase inhibitor resistance mutations were detected (Fig. 1; also see Table S2 in the supplemental material). Eight of those 11 samples exhibited either the Q148H/R or the N155H major raltegravir resistance mutation, sometimes in association with the minor resistance mutations G140A/S and V151I (see Table S2 in the supplemental material) (6). Three samples showed only minor resistance mutations, such as L74M, T97A, Y143R, or E157Q. In 4 of the 15 samples from patients on integrase inhibitor treatment, resistance mutations were not detected. Presumably, treatment duration was too short to have allowed for mutations to occur at detectable levels. The remaining samples were from patients not receiving integrase inhibitor treatment, and the respective mutations were not detected. All samples exhibited several sequence variations classified by the OpenGene mutation report as silent mutations or polymorphisms (coding changes not at resistance sites). This was not surprising, since natural polymorphisms have been described to occur within the HIV-1 integrase gene (10). All samples were assigned to their respective HIV-1 group M subtypes, including some circulating recombinant forms (see Fig. S1 in the supplemental material). The subtype homologies were always ≥93.9% (median, 96.6%).
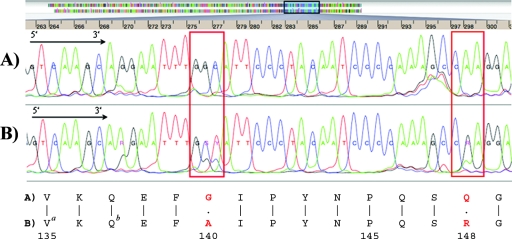
FIG. 1.
Screen shot from the TruGene instrument and single-letter amino acid code panel showing part of a sequence alignment of two HIV-1 patient samples without (A) and with (B) the major and minor integrase inhibitor mutations Q148R and G140S, respectively (red boxes and bold red letters). (A) Drug-naïve patient sample. (B) Sample from a patient on raltegravir for 5 months. a, naturally occurring polymorphism; b, silent mutation. Sequencing was performed in both directions; however, for clarity, only the 5′→3′ sequence is shown.
We have developed a sequencing assay with the TruGene sequencing analysis system to detect mutations in the HIV-1 integrase gene conferring resistance to raltegravir and elvitegravir in HIV-1-infected individuals for the routine clinical laboratory. The sequencing stretch of our assay covers those major resistance mutation codons relevant for raltegravir and elvitegravir, which are reportedly associated with 5- to 10-fold decreases in drug susceptibility (6, 11, 16; Stanford University HIV Drug Resistance Database [http://hivdb.stanford.edu/cgi-bin/IIResiNote.cgi]). However, the C-terminal region and the very N-terminal region of the HIV integrase gene are not covered by our assay. Currently, little is known about the impact of mutations on drug susceptibility in those regions. Recent recommendations suggest that those regions may harbor only minor, accessory resistance mutations (6). If needed, our assay could be extended to also cover those regions. In the current setup, our sequencing primers were selected to anneal at integrase gene consensus regions in order to cover a wide range of HIV-1 group M subtypes.
In conclusion, the developed assay for analysis of HIV-1 integrase mutations conferring resistance to current integrase inhibitors proved robust and accurate and was found suitable for the routine clinical laboratory.
Supplementary Material
Acknowledgments
We are grateful to Klaus Korn, Institute of Virology, University of Erlangen, Erlangen, Germany, for his generous gift of the AREVIR HIV-1 subtype panel. K. Flieger was employed with Siemens until 30 June 2008.
All authors declare that they have no financial interests involving the presented research.
Footnotes
Published ahead of print on 22 October 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Beerenwinkel, N., T. Sing, T. Lengauer, J. Rahnenführer, K. Roomp, I. Savenkov, R. Fischer, D. Hoffmann, J. Selbig, K. Korn, H. Walter, T. Berg, P. Braun, G. Fätkenheuer, M. Oette, J. Rockstroh, B. Kupfer, R. Kaiser, and M. Däumer. 2005. Computational methods for the design of effective therapies against drug resistant HIV strains. Bioinformatics 213943-3950. [DOI] [PubMed] [Google Scholar]
- 2.Department of Health and Human Services. 2008. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf.
- 3.Erali, M., S. Page, L. G. Reimer, and D. R. Hillyard. 2001. Human immunodeficiency virus type 1 drug resistance testing: a comparison of three sequence-based methods. J. Clin. Microbiol. 392157-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gale, H. B., V. L. Kan, And R. C. Shinol. 2006. Performance of the TruGene human immunodeficiency virus type 1 genotyping kit and the OpenGene DNA sequencing system on clinical samples diluted to approximately 100 copies per milliliter. Clin. Vaccine Immunol. 13235-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grant, R. M., D. R. Kuritzkes, V. A. Johnson, J. W. Mellors, J. L. Sulivan, R. Swanstorm, R. T. D'Auila, M. van Gorder, M Holodniy, R. M. Lloyd, Jr., C. Reid, G. F. Morgan, and D. L. Winslow. 2003. Accuracy of the TRUGENE HIV-1 genotyping kit. J. Clin. Microbiol. 411586-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hirsch, M. S., H. F. Günthard, J. M. Schapiro. F. Brun-Vézinet, B. Clotet, S. M. Hammer, V. A. Johnson, D. R. Kuritzkes, J. W. Mellors, D. Pillay, P. G. Yeni, D. M. Jacobson, and D. D. Richman. 2008. Antiretroviral drug resistance testing in adult HIV-1 infection: 2008 recommendations of an international AIDS society-USA panel. Clin. Infect. Dis. 47266-285. [DOI] [PubMed] [Google Scholar]
- 7.Hölzl, G., M. Stöcher, V. Leb, H. Stekel, and J. Berg. 2003. Entirely automated quantification of human immunodeficiency virus type 1 (HIV-1) RNA in plasma by using the ultrasensitive COBAS AMPLICOR HIV-1 monitor test and RNA purification on the MagNA pure LC instrument. J. Clin. Microbiol. 411248-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jagodzinski, L. L., J. D. Cooley, M. Weber, and N. L. Michael. 2003. Performance characteristics of human immunodeficiency virus type 1 (HIV-1) genotyping system in sequence-based analysis of subtypes other than HIV-1 subtype B. J. Clin. Microbiol. 41998-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuritzkes, D. R., R. M. Grant, P. Feorino, M. Griswold, M. Hoover, R. Young, S. Day, R. M. Lloyd, Jr., C. Reid, G. F. Morgan, and D. L. Winslow. 2003. Performance characteristics of the TRUGENE HIV-1 genotyping kit and the OpenGene DNA sequencing system. J. Clin. Microbiol. 411594-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lataillade, M., J. Chiarella, and M. J. Kozal. 2007. Natural polymorphism of the HIV-1 integrase gene and mutations associated with integrase inhibitor resistance. Antivir. Ther. 12563-570. [PubMed] [Google Scholar]
- 11.Malet, I., O. Delelis, M.-A. Valantin, B. Montes, C. Soulie, M. Wirden, L. Tchertanov, G. Peytavin, J. Reynes, J.-F. Mouscadet, C. Katlama, V. Calvez, and A.-G. Marcelin. 2008. Mutations associated with failure of raltegravir treatment affect integrase sensitivity to the inhibitor in vitro. Antimicrob. Agents Chemother. 521351-1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murphy, E. L., A. C. Collier, L. A. Kalish, S. F. Assamnn, M. F. Para, T. P. Flanigan, P. N. Kumar, L. Mintz, F. R. Wallach, and G. J. Nemo. 2001. Highly active antiretroviral therapy decreases mortality and morbidity in patients with advanced HIV disease. Ann. Intern. Med. 13517-26. [DOI] [PubMed] [Google Scholar]
- 13.Pommier, Y., A. A. Johnson, and C. Marchand. 2005. Integrase inhibitors to treat HIV/AIDS. Nat. Rev. Drug Discov. 4236-248. [DOI] [PubMed] [Google Scholar]
- 14.Rozanov, M., U. Pilkat, C. Chappey, A. Kochergin, and T. Tatusova. 2004. A web-based genotyping resource for viral sequences. Nucleic Acids Res. 32W654-W659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Semenova, E. A., C. Marchand, and Y. Pommier. 2008. HIV-1 integrase inhibitors: update and perspectives. Adv. Pharmacol. 56159-228. [DOI] [PubMed] [Google Scholar]
- 16.Shimura, K., E. Kodama, Y. Sakagami, Y. Matsuzaki, W. Watanabe, K. Yamataka, Y. Watanabe, Y. Ohata, S. Doi, M. Sato, M. Kano, S. Ikeda, and M. Matsuoka. 2008. Broad antiretroviral activity and resistance profile of the novel human immunodeficiency virus integrase inhibitor elvitegravir (JTK-303/GS-9137). J. Virol. 82764-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.