Abstract
The commercial PremiTest Salmonella kit uses a multiplexed DNA typing test aimed at identifying common serovars of Salmonella enterica. It was used in assays over a 9-month period in the Belgian reference laboratory that performs the routine identification of Salmonella strains of animal origin. A blind analysis of 754 strains was conducted in parallel by classical serotyping and the PremiTest assay. Full results were available for 685 strains (90.8%) by serotyping, while the remaining 69 strains were found to be nontypeable due to either a lack of surface antigen expression or autoagglutination properties. When the PremiTest assay (version 4.2) was performed with crude bacterial extracts, it identified 658 strains (87.3%), including most strains found to be nontypeable by serotyping. In contrast, it gave no, wrong, dual, or noninterpretable results for 96 strains, for which 23 were caused by assay failures. When purified DNA instead of crude extracts were tested, the number of strains successfully identified to the serovar level increased to 714 (94.7%), while all assay failures were cleared. Our conclusion is that, in its actual development stage, the application of the investigated kit to purified DNA samples offers a valuable alternative to classical serotyping for laboratories performing the routine identification of Salmonella strains belonging to commonly encountered serovars and isolated from a given geographical area, assuming that the system has been validated beforehand with a significant number of strains originating from that particular area.
The scoring of antigenic formulae for Salmonella enterica subsp. enterica strain typing uses the Kaufmann-White scheme, which is based on the reactivities of specific antisera with the Salmonella surface antigens and which is considered the reference method (3, 16). More than 150 different somatic O and flagellar H antigens are used for the characterization of over 2,500 serovars (3, 15). To compensate for the methodological constraints associated with antigen profiling, alternative strategies aimed at replacing or complementing the reference method have been proposed over the last decade. These include PCR and real-time PCR specific identification and typing methods (6, 8, 9, 14, 20, 22, 23), multiplex PCR (1, 10), DNA sequencing (13), and DNA microarray analysis (5, 11, 12, 17, 18, 27). Antibody-based arrays have been developed as well (4, 7, 21, 26).
The PremiTest Salmonella assay (PT; DSM Nutritional Products) was previously evaluated in our laboratory (25). It uses a DNA-based methodology called the multiplex ligation detection reaction (LDR) to generate a collection of ligated probes from the template genome that are further amplified by PCR with a single pair of amplimers (19, 24, 25). The amplified molecules are subsequently hybridized to a low-density DNA microarray spotted with probe-specific complementary oligonucleotides. A biotin label is incorporated into one of the PCR primers, generating biotinylated amplification products that are detected on the microarray by colorimetric detection. Array images are analyzed with a photometric detector and are computed online through the use of customized software. The target genetic markers were selected to give, as far as possible, a unique microarray hybridization profile (i.e., a PT signature) for each S. enterica serovar considered (2). The system was initially evaluated with strains isolated from Belgium and The Netherlands, and the strains were selected to fairly represent the local diversity of Salmonella serovars associated with food-producing animals (25). In the present work, a blind comparison of PT with the classical serotyping method was conducted over a 9-month period (from April to December 2007) with every strain that arrived at the Belgian reference laboratory for routine testing of Salmonella strains of animal origin. A total of 754 strains belonging to 58 different serovars were tested. Each strain was typed in parallel by slide agglutination by the classical serotyping scheme with antisera purchased from the Statens Serum Institute (Copenhagen, Denmark) and Bio-Rad (La Jolla, CA). The antisera were used according to the manufacturers' instructions.
PT is supplied as a kit containing premixed LDR buffers and probes, customized DNA microarrays in a microtube format (Array Tubes; ClonDiag, Jena, Germany), and reagents and buffers for microarray hybridization and staining. The assay requires standard laboratory equipment, such as a minicentrifuge, a PCR thermocycler, and a rotating heating block for hybridization and staining of the array tubes (Eppendorf, Hamburg, Germany). The array images are generated with a dedicated microarray reader (Array Tube reader; ClonDiag) connected to a standard computer running software customized for PT data analysis (version 4.2). The cost of the assay (without DNA extraction and any other laboratory cost allowance) was €31.5 per sample at the time of testing. The analysis time was about 1.5 working days for a 36-sample batch.
According to the instructions of the manufacturer (Check-Points, Wageningen, The Netherlands), the assays were conducted with crude DNA preparations obtained by boiling a small amount of colony material. In our hands, however, such crude extracts yielded correct results for only 87.3% of the samples tested. Testing of the remaining 12.7% of the samples led to assay failures or to wrong, partial, or noninterpretable results. An increase or decrease in the amount of sample and the use of a lysis buffer prior to boiling of the sample improved the results in some instances but did not do so systematically. Using the commercial kit DNeasy blood and tissue kit (Qiagen Inc., Valencia, CA), we therefore extracted the total genomic DNA from those strains with assay failures or suspect results. By the use of purified DNA, PTs were 100% successful and allowed the results to be updated to 714 strains (94.7%) with correct identifications, 16 strains (2.1%) with a PT signature corresponding to two possible serovars, 19 strains (2.5%) with unknown PT signatures (i.e., the strains were not recognized as defined serovars), and 5 strains (0.6%) with incorrect identifications. The serovars determined by classical slide agglutination and PT were found to be significantly associated, as indicated by independency χ2 statistical assessment (P < 0.01). The probabilities of obtaining a successful result by PT performed with crude bacterial extracts and DNA were 0.894 and 0.968, respectively. The results are summarized in Table 1. See Table S1 in the supplemental material for detailed strain typing data.
TABLE 1.
Comparison of PT with classical serotyping for serovar identification of Salmonella enterica subsp. enterica
| Assay | No. (%) of samples with:
|
|||||
|---|---|---|---|---|---|---|
| Correct serovar identification | No result | Noninterpretable result | Dual result | Wrong result | Total | |
| Classical serotyping | 685 (90.8) | 69 (9.1)b | 0 | 0 | —f | 754 |
| PT | ||||||
| Crude extract | 658 (87.3) | 23 (3.0)c | 50 (6.6)d | 16 (2.1)e | 7 (0.9) | 754 |
| Purified genomic DNAa | 714 (94.7) | 0c | 19 (2.5)d | 16 (2.1)e | 5 (0.6) | 754 |
Purified DNA was assayed only if the crude extract yielded an assay failure or a wrong or noninterpretable result.
No serotyping possible due to strain autoagglutination or a lack of antigen expression.
Assay failure.
Unknown serovar code.
Numeric code associated with two possible serovars.
—, classical serotyping was considered the reference method. Strains yielding discrepant results by both classical serotyping and PT were retested in a third reference laboratory to ascertain their correct serovar designations.
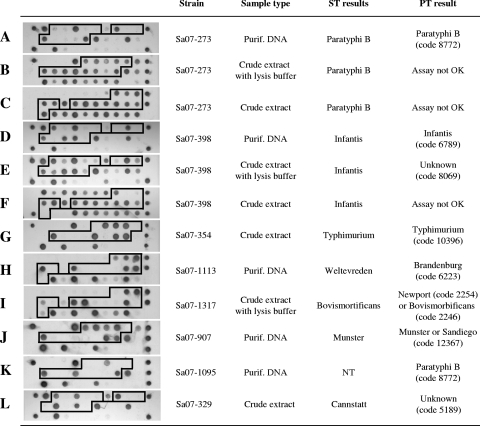
Salmonella strains leading to recurrent assay failures with crude material often belonged to serovar Typhimurium, Infantis, or Paratyphi B var. Java (Fig. 1A to F). Such crude strain extracts are thought to contain an unidentified bacterial compound that interferes with the assay procedure. The relatively high incidence of these serovars during the assessment period probably accounts for the observed frequency of assay failures.
FIG. 1.
Typical microarray results obtained by PT. The boxed microarray spots are used by the PT software (version 4.2) to calculate a PT signature that is used as serovar identifier. (A to F) Strains identified correctly only if purified DNA was assayed; (G) a strain yielding the correct result when crude material was assayed; (H) incorrect identification by PT; (I and J) a single PT signature matching two possible serovars; (K) a strain found to be nontypeable by classical serotyping and identified as Salmonella serovar Paratyphi B by PT; (L) uncommon serovar yielding a PT signature not recognized by the system. ST, serotyping; NT, nontypeable; Purif., purified.
PT results yielding a PT signature but no serovar name typically belonged to uncommon serovars, as estimated by classical serotyping, suggesting that their associated PT signature either was never encountered during system validation or was rarely so (Fig. 1L). We also noticed that more than two-thirds of the strains (37/50) yielding unknown PT signatures when crude extracts were tested were, in fact, assigned a wrong PT signature by the software. When purified DNA from these strains was tested, these strains displayed a different PT signature, suggesting that the software had misinterpreted the corresponding poor-quality profiles obtained with the crude extracts (Fig. 1E).
Incorrect serovar identifications issued from strains that up to now displayed PT signatures found to be exclusively associated with another serovar. Therefore, the interpretation of some PT signatures as defined in version 4.2 of the software should be revised (Fig. 1H). Dual results occurred for strains with PT signatures associated with two serovars (Fig. 1I and J).
In summary, we found that PT performs significantly better when purified DNA rather than crude extracts are used. The proportion of Salmonella strains successfully identified to the serovar level was 4% greater than that successfully identified by the classical serotyping method. This value can be increased by another 2% if strains assigned two possible serovars are considered, which leads to a maximum success rate of 96.8%. Our assessment of the method also pointed to some incomplete or questionable PT signatures, and these should be regularly updated on the basis of present and future results. The current version of PT makes use of a microarray profile made up of 13 DNA markers for serovar discrimination. Given the relatively low level of complexity of the microarray profiles generated, on the one hand, and the limited number of serovars recognized by the system, on the other, it is expected that additional DNA markers will be required to identify unequivocally any of the 2,500 Salmonella serovars defined to date. Therefore, future versions of the system should be extended to generate PT signatures that do not overlap different serovars. In our opinion, the present version of PT already offers a valuable alternative to the classical serotyping method for the routine identification of common Salmonella serovars. Future versions that increase the robustness of the test and the number of DNA markers for serovar discrimination will further enhance the value of the system.
Supplementary Material
Acknowledgments
We thank Sophie Bertrand and Bernard China (Belgian Institute of Public Health) for their support with the classical serotyping of uncommon Salmonella serovars and with data processing, respectively.
This work was sponsored through a governmental grant from the Dutch Agency for Sustainability and Innovation (Senternovem).
We declare that none of the participating individuals (authors, laboratory staff, and acknowledged people and bodies) has any commercial interest (direct or indirect) in the results presented here.
Footnotes
Published ahead of print on 8 October 2008.
Supplemental material for this article may be found at http://jcm.asm.org/.
REFERENCES
- 1.Alvarez, J., M. Sota, A. B. Vivanco, I. Perales, R. Cisterna, A. Rementeria, and J. Garaizar. 2004. Development of a multiplex PCR technique for detection and epidemiological typing of Salmonella in human clinical samples. J. Clin. Microbiol. 421734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreoli, P., J. Thijssen, R. Anthony, P. Vos, and W. De Levita. December 2004. Fast method for detecting micro-organisms in food samples. Patent WO2004/106547.
- 3.Bopp, C. A., F. W. Brenner, P. I. Fields, J. G. Wells, and N. A. Stockbine. 2003. Escherichia, Shigella, and Salmonella, p. 654-671. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, vol. 1, 8th ed. ASM Press, Washington, DC. [Google Scholar]
- 4.Cai, H. Y., L. Lu, C. A. Muckle, J. F. Prescott, and S. Chen. 2005. Development of a novel protein microarray method for serotyping Salmonella enterica strains. J. Clin. Microbiol. 433427-3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, K., S. Baker, C. C. Kim, C. S. Detweiler, G. Dougan, and S. Falkow. 2003. Genomic comparison of Salmonella enterica serovars and Salmonella bongori by use of an S. enterica serovar Typhimurium DNA microarray. J. Bacteriol. 185553-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esteban, E., K. Snipes, D. Hird, R. Kasten, and H. Kinde. 1993. Use of ribotyping for characterization of Salmonella serotypes. J. Clin. Microbiol. 31233-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fitzgerald, C., M. Collins, S. van Duyne, M. Mikoleit, T. Brown, and P. Fields. 2007. Multiplex, bead-based suspension array for molecular determination of common Salmonella serogroups. J. Clin. Microbiol. 453323-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoorfar, J., P. Ahrens, and P. Radstrom. 2000. Automated 5′ nuclease PCR assay for identification of Salmonella enterica. J. Clin. Microbiol. 383429-3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen, M. A., and R. J. Hubner. 1996. Use of homoduplex ribosomal DNA spacer amplification products and heteroduplex cross-hybridization products in the identification of Salmonella serovars. Appl. Environ. Microbiol. 622741-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim, S., J. G. Frye, J. Hu, P. J. Fedorka-Cray, R. Gautom, and D. S. Boyle. 2006. Multiplex PCR-based method for identification of common clinical serotypes of Salmonella enterica subsp. enterica. J. Clin. Microbiol. 443608-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majtan, T., L. Majtanova, J. Timko, and V. Majtan. 2007. Oligonucleotide microarray for molecular characterization and genotyping of Salmonella spp. strains. J. Antimicrob. Chemother. 60937-946. [DOI] [PubMed] [Google Scholar]
- 12.Malorny, B., C. Bunge, B. Guerra, S. Prietz, and R. Helmuth. 2007. Molecular characterisation of Salmonella strains by an oligonucleotide multiprobe microarray. Mol. Cell. Probes 2156-65. [DOI] [PubMed] [Google Scholar]
- 13.Mortimer, C. K., T. M. Peters, S. E. Gharbia, J. M. Logan, and C. Arnold. 2004. Towards the development of a DNA-sequence based approach to sero-typing of Salmonella enterica. BMC Microbiol. 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nair, S., T. K. Lin, T. Pang, and M. Altwegg. 2002. Characterization of Salmonella serovars by PCR-single-strand conformation polymorphism analysis. J. Clin. Microbiol. 402346-2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perch, M., C. R. Braden, R. Bishop, P. Fields, R. Plikaytis, and R. V. Tauxe. 2003. Salmonella surveillance summary, 2003. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Atlanta, GA.
- 16.Popoff, M. Y., J. Bockemuhl, and L. L. Gheesling. 2004. Supplement 2002 (no. 46) to the Kauffmann-White scheme. Res. Microbiol. 155568-570. [DOI] [PubMed] [Google Scholar]
- 17.Porwollik, S., E. F. Boyd, C. Choy, P. Cheng, L. Florea, E. Proctor, and M. McClelland. 2004. Characterization of Salmonella enterica subspecies I genovars by use of microarrays. J. Bacteriol. 1865883-5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scaria, J., R. U. Palaniappan, D. Chiu, J. A. Phan, L. Ponnala, P. McDonough, Y. T. Grohn, S. Porwollik, M. McClelland, C. S. Chiou, C. Chu, and Y. F. Chang. 2008. Microarray for molecular typing of Salmonella enterica serovars. Mol. Cell. Probes 22238-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schouten, J. P., C. J. McElgunn, R. Waaijer, D. Zwijnenburg, F. Diepvens, and G. Pals. 2002. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 30e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shangkuan, Y. H., and H. C. Lin. 1998. Application of random amplified polymorphic DNA analysis to differentiate strains of Salmonella typhi and other Salmonella species. J. Appl. Microbiol. 85693-702. [DOI] [PubMed] [Google Scholar]
- 21.Taitt, C. R., Y. S. Shubin, R. Angel, and F. S. Ligler. 2004. Detection of Salmonella enterica serovar Typhimurium by using a rapid, array-based immunosensor. Appl. Environ. Microbiol. 70152-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torpdahl, M., and P. Ahrens. 2004. Population structure of Salmonella investigated by amplified fragment length polymorphism. J. Appl. Microbiol. 97566-573. [DOI] [PubMed] [Google Scholar]
- 23.Uzzau, S., M. Hovi, and B. A. Stocker. 1999. Application of ribotyping and IS200 fingerprinting to distinguish the five Salmonella serotype O6,7:c:1,5 groups: Choleraesuis sensu stricto, Choleraesuis var. Kunzendorf, Choleraesuis var. Decatur, Paratyphi C, and Typhisuis. Epidemiol. Infect. 12337-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Eijk, M. J., J. L. Broekhof, H. J. van der Poel, R. C. Hogers, H. Schneiders, J. Kamerbeek, E. Verstege, J. W. van Aart, H. Geerlings, J. B. Buntjer, A. J. van Oeveren, and P. Vos. 2004. SNPWave: a flexible multiplexed SNP genotyping technology. Nucleic Acids Res. 32e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wattiau, P., T. Weijers, P. Andreoli, C. Schlicker, H. Vanderveken, H. M. Maas, A. J. Verbruggen, M. E. Heck, W. J. Wannet, H. Imberechts, and P. Vos. 2008. Evaluation of the Premi®Test Salmonella, a commercial low-density DNA microarray system intended for routine identification and typing of Salmonella enterica. Int. J. Food Microbiol. 123293-298. [DOI] [PubMed] [Google Scholar]
- 26.Wilkes, J. G., L. Rushing, R. Nayak, D. A. Buzatu, and J. B. Sutherland. 2005. Rapid phenotypic characterization of Salmonella enterica strains by pyrolysis metastable atom bombardment mass spectrometry with multivariate statistical and artificial neural network pattern recognition. J. Microbiol. Methods 61321-334. [DOI] [PubMed] [Google Scholar]
- 27.Yoshida, C., K. Franklin, P. Konczy, J. R. McQuiston, P. I. Fields, J. H. Nash, E. N. Taboada, and K. Rahn. 2007. Methodologies towards the development of an oligonucleotide microarray for determination of Salmonella serotypes. J. Microbiol. Methods 70261-271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.