Abstract
Hepatitis C virus (HCV) RNA detection and quantification are the key diagnostic tools for the management of hepatitis C. Commercially available HCV RNA assays are calibrated to the HCV genotype 1 (gt1)-based WHO standard. Significant differences between assays have been reported. However, it is unknown which assay matches the WHO standard best, and little is known about the sensitivity and linear quantification of the assays for non-gt1 specimens. Two real-time reverse transcriptase PCR-based assays (RealTime HCV and Cobas Ampliprep/Cobas TaqMan HCV [CAP/CTM]) and one signal amplification-based assay (the Versant HCV RNA, version 3.0, branched DNA [bDNA] assay) were compared for their abilities to quantify HCV RNA in clinical specimens (n = 65) harboring HCV isolates of gt1 to g5. The mean differences in the amounts detected by RealTime HCV in comparison to those detected by the bDNA assay and CAP/CTM were −0.02 and 0.72 log10 IU/ml HCV RNA, respectively, for gt1; −0.22 and 0.03 log10 IU/ml HCV RNA, respectively, for gt2; −0.27 and −0.22 log10 IU/ml HCV RNA, respectively, for gt3; −0.19 and −1.27 log10 IU/ml HCV RNA, respectively, for gt4; and −0.03 and 0.09 log10 IU/ml HCV RNA, respectively, for gt5. The lower limits of detection for RealTime HCV and CAP/CTM were 16.8 and 10.3 IU/ml, respectively, for the WHO standard and in the range of 4.7 to 9.0 and 3.4 to 44.4 IU/ml, respectively, for clinical specimens harboring gt1 to gt6. Direct comparison of the two assays with samples of the WHO standard (code 96/798) with high titers yielded slightly smaller amounts by RealTime HCV (−0.2 log10 at 1,500 IU/ml and −0.3 log10 at 25,000 IU/ml) and larger amounts by CAP/CTM (0.3 log10 at 1,500 IU/ml and 0.2 log10 at 25,000 IU/ml). Finally, all three tests were linear between 4.0 × 103 and 1.0 × 106 IU/ml (correlation coefficient, ≥0.99). In conclusion, the real-time PCR based assays sensitively detected all genotypes and showed comparable linearities for the quantification of HCV RNA, with the exception of gt1 and gt4. The previously reported differences in the absolute quantification of samples harboring gt1 were confirmed and may be explained by different calibrations to the WHO standard.
Hepatitis C virus (HCV) infection is a common cause of chronic liver disease that can lead to end-stage liver disease, including hepatocellular carcinoma.
At present, combination therapy with pegylated interferon and ribavirin is the standard of care. However, this treatment regimen appears to be effective in only 40% to 50% of patients infected with genotype 1 and approximately 80% of patients infected with genotypes 2 and 3 (12, 13, 16).
A sustained virologic response, defined as undetectable HCV RNA at least 6 months after the completion of therapy, may be predicted by a number of host and viral factors, including age, race, liver fibrosis, the HCV genotype, and the baseline viral load.
Recent reports suggest that decisions on the optimal treatment duration may be made on the basis of the baseline viral load and the virologic response at weeks 4, 12, and 24 of therapy (2, 11, 15, 28). In addition, stopping algorithms have been established on the basis of a less than 2-log10 drop in the viral load at week 12 of therapy, due to a very low rate (no greater than 3%) of a sustained response (9, 12). Moreover, the detection of HCV RNA at week 24 is associated with a low sustained virologic response rate, and treatment discontinuation is recommended due to nonresponse (9). Measurement of HCV RNA has therefore become the key parameter for the tailoring of individualized treatment.
For HCV RNA measurement, different standardized quantification assays based on signal amplification techniques (the branched DNA [bDNA] assay) and target amplification techniques (reverse transcriptase PCR [RT-PCR]) with different sensitivities and different ranges of quantification are commercially available.
Recently, real-time RT-PCR-based assays have been introduced in routine diagnostics and are rapidly replacing standard RT-PCR- and signal amplification-based assays.
These quantification assays offer the advantage of amplification over a broad dynamic range, thus improving limits of detection (LODs) to ≤10 IU/ml and linear quantification up to 107 to 108 IU/ml without the need for predilution. Previous qualitative and quantitative RT-PCR-based tests may be combined in a single assay.
The development of a WHO HCV international unit (IU) standard has contributed to better accuracy and the better comparability of the results obtained by different assays. However, the standard is based on the consensus value attained from the quantitative analysis only of samples harboring HCV RNA of genotype 1 (22, 23). Although all current commercially available assays are calibrated to the WHO standard, little is known about the sensitivity and the linear quantification of these assays when they are directly compared with each other by the use of both clinical and WHO standard specimens. In addition, it has been shown that the results obtained by assays performed with different HCV genotypes may vary significantly, despite standardization to IU, and it remains unclear which assay produces the correct results (7, 24).
The aim of our study was to evaluate and compare the performance characteristics of three currently available HCV RNA quantification methods, including two real-time PCR-based assays (RealTime HCV [Abbott Molecular Inc., Abbott Park, IL] and the Roche Cobas Ampliprep/Cobas TaqMan HCV assay [CAP/CTM; Roche Molecular Systems, Pleasanton, CA]) and a signal amplification-based assay (Siemens Versant HCV RNA, version 3.0 [bDNA], assay; Siemens Healthcare Diagnostics, Tarrytown, NY) by testing serum samples from patients infected with HCV genotypes 1 to 5 (6) and the HCV WHO standard.
(Part of this study was presented at the Annual Meeting of the American Association for the Study of Liver Diseases, Boston, MA, 2 to 6 November 2007.)
MATERIALS AND METHODS
HCV genotyping.
The HCV genotypes were determined by using the Versant HCV genotype (version 2.0) assay (a line probe assay; Siemens Healthcare Diagnostics, Tarrytown, NY), in accordance with the manufacturer's instructions. While with version 1 of the line probe assay HCV subtypes 1a and 1b were correctly identified in only <90% of the patient samples, a >95% accuracy for correct subtyping is achievable with version 2 (1, 4, 6). In addition, version 2 correctly identifies samples harboring genotype 6 (4, 20).
HCV RNA assays. (i) Abbott RealTime HCV.
RealTime HCV is based on the reverse transcription-PCR technology combined with real-time fluorescent detection for the quantification of HCV RNA.
HCV RNA was isolated by the Abbott m2000sp instrument from 1,000-μl aliquots (sample volume required, 500 μl), as described previously (17). Extracted samples and controls were then amplified and detected with the Abbott m2000rt instrument, according to the manufacturer's instructions.
In short, an RNA sequence that is unrelated to the HCV target sequence is added as an internal control (IC). Specimens and ICs are extracted highly efficiently by binding to magnetic particles.
After sample preparation, reverse transcription and amplification are carried out with the thermostable recombinant Tth DNA polymerase. Both, HCV and IC have their respective primers and probes. The HCV-specific primers and probes bind within the highly conserved 5′ nontranslated region of HCV, and the IC primers and probes bind to a target sequence of an armored RNA particle derived from the pumpkin plant (Cucurbita pepo).
For detection of the amplification products, the assay utilizes real-time PCR technology with two different dually labeled fluorescent oligonucleotide probes that bind to the HCV or the IC target sequence. The probes carry a fluorescent moiety that is covalently linked to the 5′ end and a quenching moiety that is covalently linked to the 3′ end. When the HCV or the IC target sequence is absent, the fluorescent moiety is quenched and there is no fluorescent emission (randomly coiled probes). In the presence of the target sequences, probe hybridization to complementary sequences separates the two moieties and allows fluorescence emission and detection.
The amplification cycle at which the fluorescent signal is detected by the m2000rt instrument is proportional to the log of the amount of HCV RNA present in the original sample; thus, the HCV RNA concentration in the specimen can be calculated.
The assay has adopted the second international WHO standard for HCV RNA (code 96/798) for calibration, and HCV RNA concentration is reported in IU per ml.
Generally, four different modes of results are possible: undetectable (below the LOD of the assay), positive but below 12 IU/ml, positive above 12 IU/ml with an exact HCV RNA concentration, or positive above 8.0 log10 IU/ml (which represents the upper limit of quantification).
RealTime HCV has a reported linear quantification range of between 12 and 107 IU/ml. The assay's 95% detection rate or LOD is 10.5 IU/ml, as stated in the package insert. Similar results have been reported in an analysis conducted by researchers who were independent of the manufacturer (27).
(ii) Roche CAP/CTM.
CAP/CTM is also based on the real-time reverse transcription-PCR technology.
HCV RNA extraction from 1,000-μl aliquots (sample volume required, 850 μl) of clinical samples and controls was performed with the automated Cobas Ampliprep instrument, according to the manufacturer's instructions. The extracted samples and controls were then processed for amplification and detection with the Cobas TaqMan 48 analyzer, as described previously (24). CAP/CTM is standardized against the first WHO international standard for HCV RNA (code 96/790), and titers are reported in IU per ml. As for RealTime HCV, the results of CAP/CTM are reported in four different ways: undetectable (below or above the assays’ LOD), positive but below 15 IU/ml, positive above 15 IU/ml with an exact HCV RNA concentration, or positive and above 6.9 × 107 IU/ml (upper limit of quantification).
The assay's linear quantification range is between 43 and 6.9 × 107 IU/ml, as indicated in the package insert. The assay's 95% detection rate is 12.6 IU/ml (package insert) and has ranged from 7.4 to 10.5 IU/ml in recently published studies (26, 29).
(iii) Versant bDNA assay.
The Versant bDNA assay is a signal amplification nucleic acid probe test that uses a sandwich nucleic acid hybridization procedure.
The test has been standardized to the first WHO international standard (code 96/790) and has a reported dynamic range between the lower LOD of 615 IU/ml and 7.7 × 106 IU/ml, as stated in the package insert.
The testing procedure has been described elsewhere and was performed by following the manufacturer's instructions (21).
The bDNA assay is currently the only commercially available assay that has gained FDA approval, but all three tests (RealTime HCV, CAP/CTM, and the bDNA assay) have received the Communauté Européenne (CE) mark.
Clinical correlation of HCV RNA assays.
The performance characteristics of the three HCV RNA assays (RealTime HCV, CAP/CTM, and the bDNA assay) were evaluated by the comparative quantification of HCV in clinical specimens sorted by genotype.
Sixty-five undiluted clinical serum samples from patients infected with HCV genotype 1 (n = 30), 2 (n = 12), 3 (n = 16), 4 (n = 4), or 5 (n = 3) were obtained from the outpatient clinic at Saarland University Hospital. Written informed consent was obtained from each patient, and the studies were approved by the Ethics Committee of Medical Research in Homburg, Germany, in accordance with the 1975 Declaration of Helsinki.
All samples were stored at −80°C. To allow identical conditions for all assay procedures, the samples were thawed to generate appropriate aliquots and were subsequently stored again at −80°C prior to parallel testing by all three assays. To determine the intra-assay variability, aliquots were analyzed in triplicate in a single run. Samples with invalid, nondetectable, and outlying results were excluded from further analysis.
WHO and genotype-specific assay sensitivity.
To measure the analytical sensitivity of the two real-time PCR-based assays, panels of the second international WHO standard HCV RNA (code 96/798) were serially diluted to the following concentrations: 50, 25, 15, 10, 7.5, 5, and 2.5 IU/ml. The dilutions were stored at −80°C and were subsequently tested in parallel by both assays. Fifteen replicates of each concentration of the panel were tested in a single run.
In addition, the LOD was also evaluated with serial dilutions of six clinical specimens infected with HCV genotypes 1 to 6. For this purpose, serum samples were collected from patients with chronic HCV infection in the outpatient clinic at the Saarland University Hospital in the same fashion as described above. Panels with the following concentrations were produced: 50, 25, 12.5, 6.25, and 3.125 IU/ml. The original HCV RNA levels used for calculation of the dilutions were determined by RealTime HCV. Aliquots were again tested in parallel by both assays (RealTime HCV, CAP/CTM). Twelve replicates of each concentration were tested in a single run.
Quantification of HCV WHO standard RNA.
Even though all commercially available assays have adopted the HCV WHO standard for calibration, differences between assays have been reported (7, 24), and it remains unclear which assay's results correspond best to the standard material.
The assigned unitage of the second international WHO standard HCV RNA is 50,000 IU/ml. For direct comparison of the two real-time PCR-based assays, high-titer dilutions of the standard were prepared by using HCV antibody-negative human serum. Four aliquots of 25,000 IU/ml and three aliquots of 1,500 IU/ml were each tested in a single run by RealTime HCV and CAP/CTM, respectively.
Genotype-specific assay linearities.
The linearities of all three assays were determined on the basis of results of tests with high-titer clinical specimens from patients infected with HCV genotypes 1 through 5. For each genotype, one high-titer sample (>105 IU/ml) was selected. Determination of the original HCV RNA level was performed by RealTime HCV.
Six dilutions (1.0 × 106, 3.3 × 105, 1.1 × 105, 3.7 × 104, 1.2 × 104, and 4.1 × 103) were prepared, and HCV antibody-negative and/or HCV RNA-negative plasma obtained from healthy volunteers was used as a dilution matrix. Each dilution was tested in triplicate by all three assays in a single run.
Data analysis.
The results are expressed as the mean, median, and standard deviation (SD), as appropriate. Correlation coefficients (R values) and the rates of agreement between RealTime HCV, the bDNA assay, and CAP/CTM were determined from the mean differences in quantification for averaged log values by Bland-Altman plot analysis (Bias, version 8.4.6; Epsilon Verlag, Frankfurt, Germany) (3). Intra-assay variability was expressed as the SD and the coefficient of variation (CV), based on the mean log10-transformed HCV RNA concentrations. Probit analysis was performed to determine the LOD, with the results for standard reference samples used as independent variables and the numbers of positive results used as dichotomous variables (SPSS program, version 16.0; SPSS Inc., Chicago, IL). The LOD was determined as the 95% probability of obtaining a positive result for HCV RNA. R values were calculated for linearity data.
RESULTS
Agreement of HCV RNA assays.
Sixty-five undiluted clinical samples infected with HCV genotypes 1 to 5 were available for parallel testing by RealTime HCV, CAP/CTM, and the bDNA assay.
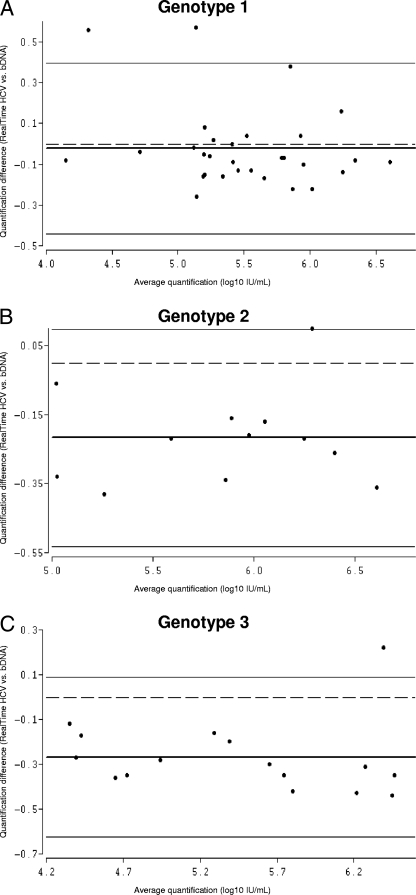
Bland-Altman plots of the results for HCV genotypes 1 to 3 were used to determine the agreement between RealTime HCV and the bDNA assay. The differences between the two assays were plotted against the averaged log10 results. For RealTime HCV versus the bDNA assay, the mean difference between the values of the assays for genotype 1 was −0.02 log10 IU/ml HCV RNA, with limits of agreement (mean difference ± 2 SDs) of −0.44 and 0.40 log10 IU/ml, and more than 95% of the differences fell within these limits. The mean difference between the values for genotype 2 was −0.22 log10 IU/ml, with limits of agreement of −0.53 and 0.10 log10 IU/ml. The mean difference in the values for genotype 3 was −0.27 log10 IU/ml, with limits of agreement of −0.63 and 0.09 log10 IU/ml. The mean differences between the values of the two assays for genotypes 4 and 5 were −0.19 and −0.03 log10 IU/ml, respectively (Fig. 1A and 2A to C; Table 1). R values for genotypes 1 to 5 were also calculated and ranged from 0.94 to 0.99.
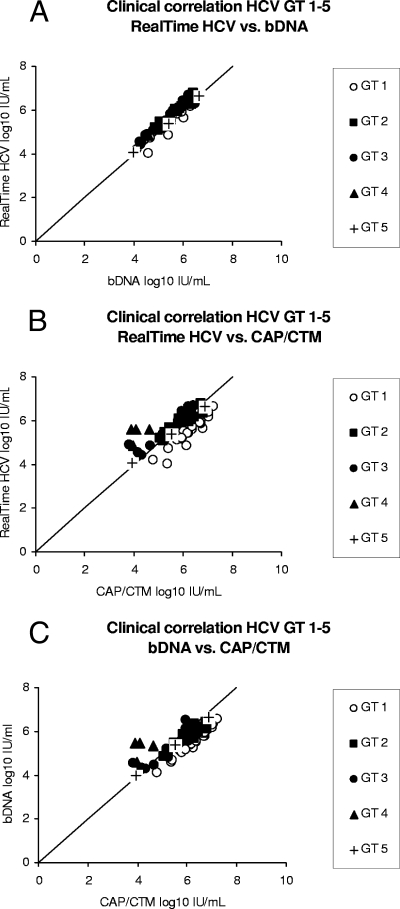
FIG. 1.
Correlation of the results for clinical specimens (n = 65) from patients with chronic HCV infection, as assessed by direct comparison of the results of RealTime HCV versus those of the bDNA assay (A), the results of RealTime HCV versus those of CAP/CTM (B), and the results of CAP/CTM versus those of the bDNA assay (C). The different genotypes (GTs) are represented by different symbols.
FIG. 2.
Bland-Altman analysis of genotype-specific mean differences in HCV RNA quantification by RealTime HCV versus that by the bDNA assay. Due to the relatively low number of samples harboring genotypes 4 and 5, Bland-Altman analysis was performed only for genotypes 1 (A), 2 (B), and 3 (C). The bold lines represent the mean differences for the samples, the thin lines represent the 95% limits of agreement, and the dashed lines are the reference lines.
TABLE 1.
Correlation of RealTime HCV with bDNA assay and CAP/CTM for determination of HCV RNA concentrations in patients infected with genotypes 1 to 5
| Genotype | No. of samples | Mean (range) concna (log10 IU/ml) by RealTime HCV | bDNA assay
|
CAP/CTM
|
||
|---|---|---|---|---|---|---|
| Mean (range) concn (log10 IU/ml) | Difference in concn (log10 IU/ml) from that by RealTime HCV | Mean (range) concn (log10 IU/ml) | Difference in concn (log10 IU/ml) from that by RealTime HCV | |||
| 1 | 30 | 5.50 (4.0-6.7) | 5.48 (4.1-6.6) | −0.02 | 6.22 (5.4-7.2) | 0.72 |
| 2 | 12 | 5.96 (5.1-6.8) | 5.74 (4.9-6.4) | −0.22 | 5.99 (5.1-6.8) | 0.03 |
| 3 | 16 | 5.58 (4.4-6.7) | 5.31 (4.3-6.5) | −0.27 | 5.36 (3.8-6.4) | −0.22 |
| 4 | 4 | 5.41 (4.9-5.6) | 5.22 (4.6-5.5) | −0.19 | 4.14 (3.9-4.6) | −1.27 |
| 5 | 3 | 5.36 (4.1-6.7) | 5.33 (4.0-6.6) | −0.03 | 5.45 (3.9-6.9) | 0.09 |
Mean (range) concentration, mean (range) HCV RNA concentration.
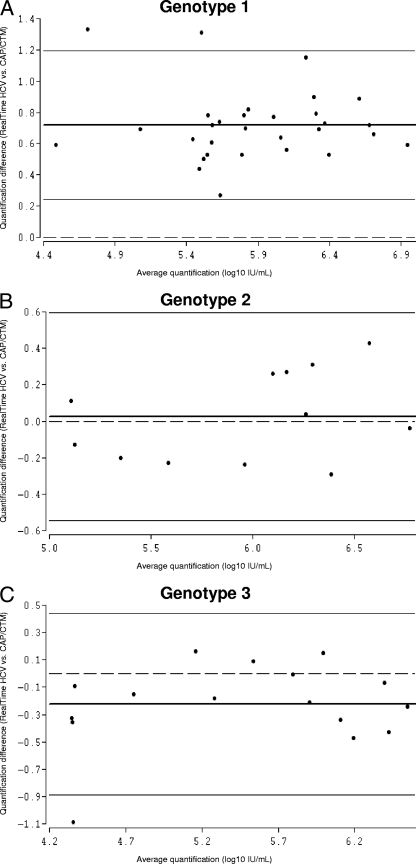
For genotype 1, the agreement of the values between RealTime HCV and CAP/CTM was poor, with a mean difference of 0.72 log10 IU/ml (limit of agreement, 0.24 and 1.20 log10 IU/ml). The agreement for genotypes 2 and 3 was good, with mean differences of 0.03 log10 IU/ml (limit of agreement, −0.55 and 0.60 log10 IU/ml) and −0.22 log10 IU/ml (limit of agreement, −0.89 and 0.44 log10 IU/ml), respectively. The mean differences for genotypes 4 and 5 were −1.27 and 0.09 log10 IU/ml, respectively (Fig. 1B and 3A to C; Table 1). R values for genotypes 1 to 5 ranged from 0.36 to 0.99.
FIG. 3.
Bland-Altman analysis of genotype-specific mean differences in HCV RNA quantification by RealTime HCV versus that by CAP/CTM. Due to the relatively low number of samples harboring genotypes 4 and 5, Bland-Altman analysis was performed only for genotypes 1 (A), 2 (B), and 3 (C). The bold lines represent the mean differences for the samples, the thin lines represent the 95% limits of agreement, and the dashed lines are the reference lines.
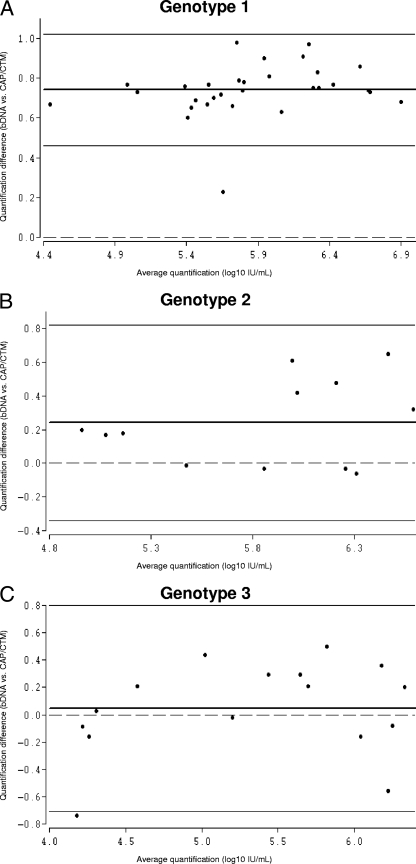
Comparison of the bDNA assay with CAP/CTM demonstrated poor agreement of the results for samples harboring genotype 1. The mean difference was 0.74 log10 IU/ml, with limits of agreement of 1.02 and 0.46 log10 IU/ml.
The mean differences for genotypes 2 and 3 were 0.24 log10 IU/ml (limit of agreement, 0.82 and −0.34 log10 IU/ml) and 0.05 log10 IU/ml (limit of agreement, 0.80 and −0.70 log10 IU/ml), respectively. The mean differences for genotypes 4 and 5 were −1.08 and 0.12 log10 IU/ml, respectively (Fig. 1C and 4A to C). R values for genotypes 1 to 5 were in the range of 0.21 to 0.99.
FIG. 4.
Bland-Altman analysis of genotype-specific mean differences in HCV RNA quantification by CAP/CTM versus that the bDNA assay. Due to the relatively low number of samples harboring genotypes 4 and 5, Bland-Altman analysis was performed only for genotypes 1 (A), 2 (B), and 3 (C). The bold lines represent the mean differences for the samples, the thin lines represent the 95% limits of agreement, and the dashed lines are the reference lines.
Intra-assay variability was calculated on the basis of the three replicates measured for each patient sample. The CVs varied from 0.72% to 1.3% for RealTime HCV, 0.55% to 1.35% for the bDNA assay, and 1.4% to 3.02% for CAP/CTM (Table 2). The CVs for specimens with low viral loads (<400,000 IU/ml HCV RNA, as assessed by RealTime HCV) were between 1.01% and 1.69% for RealTime HCV, 0.5% and 1.54% for the bDNA assay, and 1.66% and 3.24% for CAP/CTM (Table 2).
TABLE 2.
Intra-assay variability by SD based on variance analyses and estimation of CVs for all samples and samples with low viral loads
| Sample type and genotype | No. of samples | RealTime HCV
|
bDNA assay
|
CAP/CTM
|
|||
|---|---|---|---|---|---|---|---|
| Mean CV (%) | SD (log10) | Mean CV (%) | SD (log10) | Mean CV (%) | SD (log10) | ||
| All samples | |||||||
| 1 | 30 | 1.17 | 0.06 | 0.67 | 0.04 | 1.40 | 0.08 |
| 2 | 12 | 0.96 | 0.06 | 0.55 | 0.03 | 2.38 | 0.14 |
| 3 | 16 | 0.83 | 0.04 | 1.35 | 0.08 | 2.04 | 0.10 |
| 4 | 4 | 1.30 | 0.07 | 0.64 | 0.03 | 2.58 | 0.11 |
| 5 | 3 | 0.72 | 0.04 | 1.11 | 0.05 | 3.02 | 0.16 |
| LVLa | |||||||
| 1 | 17 | 1.38 | 0.07 | 0.73 | 0.04 | 1.66 | 0.09 |
| 2 | 3 | 1.20 | 0.06 | 0.92 | 0.05 | 2.64 | 0.13 |
| 3 | 8 | 1.12 | 0.05 | 0.84 | 0.04 | 2.86 | 0.13 |
| 4 | 3 | 1.69 | 0.09 | 0.50 | 0.02 | 2.54 | 0.11 |
| 5 | 2 | 1.01 | 0.05 | 1.54 | 0.07 | 3.24 | 0.16 |
LVL, low viral load (<400,000 IU/ml), as assessed by RealTime HCV.
Assay sensitivity for WHO standard and genotype-specific clinical samples.
The positive hit rates (positive values of <12 IU/ml or ≥12 IU/ml) for the WHO HCV RNA standard dilution series for RealTime HCV were 73% (11 of 15 samples) at 10 IU/ml, 87% (13 of 15 samples) at 15 IU/ml, and 100% at 25 IU/ml. The positive hit rates (positive values of <15 IU/ml or ≥15 IU/ml) for CAP/CTM were 87% (13 of 15 samples) at 10 IU/ml and 100% at 15 and 25 IU/ml.
The lower LOD determined by probit analysis yielded sensitivities of 16.8 (95% confidence interval, 13.1 to 27.9) for RealTime HCV and 10.3 (95% confidence interval, 8.4 to 15.1) for CAP/CTM (Table 3 and Table 4).
TABLE 3.
LOD of RealTime HCVa
| Nominal input HCV RNA concn (IU/ml) | No. of samples | No. of samples with positive measurements detectable at:
|
Hit rate (%) | |
|---|---|---|---|---|
| ≥12 IU/ml | ≥12 and <12 IU/ml | |||
| 50 | 15 | 15 | 15 | 100 |
| 25 | 15 | 7 | 15 | 100 |
| 15 | 15 | 5 | 13 | 87 |
| 10 | 15 | 1 | 11 | 73 |
| 7.5 | 15 | 1 | 11 | 73 |
| 5 | 15 | 10 | 67 | |
| 2.5 | 15 | 3 | 20 | |
Panels of the second international WHO standard HCV RNA (code 96/798) were used. The LOD determined by probit analysis was 16.8 IU/ml (95% confidence interval, 13.1 to 27.9 IU/ml).
TABLE 4.
LOD of CAP/CTMa
| Nominal input HCV RNA concn (IU/ml) | No. of samples | No. of samples with positive measurements detectable at:
|
Hit rate (%) | |
|---|---|---|---|---|
| ≥12 IU/ml | ≥12 and <12 IU/ml | |||
| 50 | 14 | 14 | 14 | 100 |
| 25 | 15 | 5 | 15 | 100 |
| 15 | 15 | 1 | 15 | 100 |
| 10 | 15 | 1 | 13 | 87 |
| 7.5 | 15 | 1 | 14 | 93 |
| 5 | 15 | 9 | 60 | |
| 2.5 | 15 | 4 | 27 | |
Panels of the second international WHO standard HCV RNA (code 96/798) were used. The LOD determined by probit analysis was 10.3 IU/ml (95% confidence interval, 8.4 to 15.1 IU/ml).
RealTime HCV achieved positive hit rates (positive values of <12 IU/ml or ≥12 IU/ml) of ≥95% at a concentration of 6.25 IU/ml for clinical samples harboring genotypes 1, 2, 3, 5, and 6. However, a ≥95% hit rate for genotype 4 was achieved only at 12.5 IU/ml (Table 5).
TABLE 5.
LOD of the RealTime HCV assay based on clinical specimens representing genotypes 1 to 6a
| Sample dilution (IU/ml) | GT 1
|
GT 2
|
GT 3
|
GT 4
|
GT 5
|
GT 6
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of samples | No. of samples with positive measurements detectable at:
|
Hit rate (%) | No. of samples | No. of samples with positive measurements detectable at:
|
Hit rate (%) | No. of samples | No. of samples with positive measurements detectable at:
|
Hit rate (%) | No. of samples | No. of samples with positive measurements detectable at:
|
Hit rate (%) | No. of samples | No. of samples with positive measurements detectable at:
|
Hit rate (%) | No. of samples | No. of samples with positive measurements detectable at:
|
Hit rate (%) | |||||||
| ≥12 IU/ml | <12 and ≥12 IU/ml | ≥12 IU/ml | <12 and ≥12 IU/ml | ≥12 IU/ml | <12 and ≥12 IU/ml | ≥12 IU/ml | <12 and ≥12 IU/ml | ≥12 IU/ml | <12 and ≥12 IU/ml | ≥12 IU/ml | <12 and ≥12 IU/ml | |||||||||||||
| 50 | 12 | 12 | 12 | 100 | 12 | 12 | 12 | 100 | 12 | 12 | 12 | 100 | 12 | 12 | 12 | 100 | 12 | 12 | 12 | 100 | 11 | 11 | 11 | 100 |
| 25 | 12 | 11 | 12 | 100 | 12 | 11 | 12 | 100 | 12 | 12 | 12 | 100 | 12 | 12 | 12 | 100 | 12 | 12 | 12 | 100 | 12 | 12 | 12 | 100 |
| 12.5 | 12 | 12 | 12 | 100 | 12 | 8 | 12 | 100 | 12 | 7 | 12 | 100 | 12 | 12 | 12 | 100 | 12 | 8 | 12 | 100 | 12 | 8 | 12 | 100 |
| 6.25 | 12 | 3 | 12 | 100 | 12 | 1 | 12 | 100 | 12 | 1 | 12 | 100 | 12 | 10 | 83 | 12 | 12 | 100 | 12 | 3 | 12 | 100 | ||
| 3.125 | 12 | 8 | 67 | 12 | 9 | 75 | 12 | 1 | 8 | 67 | 12 | 10 | 83 | 12 | 1 | 10 | 83 | 12 | 10 | 83 | ||||
For genotypes (GTs) 1 to 6, the LODs determined by probit analysis were 5.4 IU/ml, 5.2 IU/ml, 5.4 IU/ml, 9.0 IU/ml, 4.7 IU/ml, and 4.7 IU/ml, respectively.
For CAP/CTM, positive hit rates (positive values of <15 IU/ml or ≥15 IU/ml) of ≥95% at a concentration of 6.25 IU/ml were achieved only for samples harboring genotypes 1 and 5. A ≥95% positive hit rate was achieved for genotype 3 at 12.5 IU/ml. Positive hit rates of ≥95% were achieved for genotypes 2 and 4 at 25 IU/ml. In addition, 92% positive hit rates were achieved for genotypes 1, 2, 4, and 5 at higher concentrations (Table 6).
TABLE 6.
LOD of the CAP/CTM assay based on clinical specimens representing genotypes 1 to 6a
| Sample dilution (IU/ml) | GT 1
|
GT 2
|
GT 3
|
GT 4
|
GT 5
|
GT 6
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. of samples | No. of samples with positive measurements detectable at:
|
Hit rate (%) | No. of samples | No. of samples with positive measurements detectable at:
|
Hit rate (%) | No. of samples | No. of samples with positive measurements detectable at:
|
Hit rate (%) | No. of samples | No. of samples with positive measurements detectable at:
|
Hit rate (%) | No. of samples | No. of samples with positive measurements detectable at:
|
Hit rate (%) | No. of samples | No. of samples with positive measurements detectable at:
|
Hit rate (%) | |||||||
| ≥15 IU/ml | <15 and ≥15 IU/ml | ≥15 IU/ml | <15 and ≥15 IU/ml | ≥15 IU/ml | <15 and ≥15 IU/ml | ≥15 IU/ml | <15 and ≥15 IU/ml | ≥15 IU/ml | <15 and ≥15 IU/ml | ≥15 IU/ml | <15 and ≥15 IU/ml | |||||||||||||
| 50 | 12 | 12 | 12 | 100 | 12 | 11 | 11 | 92 | 12 | 11 | 12 | 100 | 12 | 1 | 11 | 92 | 12 | 12 | 12 | 100 | 12 | 12 | 12 | 100 |
| 25 | 12 | 11 | 11 | 92 | 12 | 7 | 12 | 100 | 12 | 2 | 12 | 100 | 12 | 12 | 100 | 12 | 4 | 12 | 100 | 12 | 12 | 12 | 100 | |
| 12.5 | 12 | 10 | 12 | 100 | 12 | 10 | 83 | 11 | 1 | 11 | 100 | 12 | 8 | 67 | 12 | 11 | 92 | 12 | 10 | 12 | 100 | |||
| 6.25 | 12 | 2 | 12 | 100 | 12 | 11 | 92 | 12 | 10 | 83 | 12 | 5 | 42 | 12 | 12 | 100 | 12 | 11 | 92 | |||||
| 3.125 | 12 | 1 | 11 | 92 | 12 | 8 | 67 | 12 | 9 | 75 | 12 | 4 | 33 | 12 | 5 | 42 | 12 | 1 | 10 | 83 | ||||
For genotypes (GTs) 1 to 6, the LODs determined by probit analysis were 3.4 IU/ml, 44.4 IU/ml, 14.1 IU/ml, 40.5 IU/ml, 11.1 IU/ml, and 7.0 IU/ml, respectively.
For clinical specimens harboring genotypes 1 to 6, the sensitivities obtained by probit analysis were in the range of 4.7 to 9.0 IU/ml for RealTime HCV. For CAP/CTM, the lower LOD ranged from 3.4 to 14.1 IU/ml for samples harboring genotypes 1, 3, 5, and 6. For samples harboring genotypes 2 and 4, significantly higher values were observed at 44.4 and 40.5 IU/ml, respectively (Tables 5 and 6).
Quantification of HCV WHO standard RNA.
For estimation of the correct quantification of high concentrations of the WHO standard by the two real-time PCR-based assays, repeat testing at concentrations of 1,500 and 25,000 IU/ml was performed.
RealTime HCV quantification results for the WHO standard samples yielded mean deviations of −0.2 log10 IU/ml (range, −0.4 to −0.2 log10 IU/ml) and −0.3 log10 IU/ml (range, −0.4 to −0.2 log10 IU/ml) at 3.2 log10 (1,500 IU/ml) and 4.4 log10 (25,000 IU/ml), respectively. For CAP/CTM, the quantification difference was 0.3 log10 IU/ml at 3.2 log10 and 0.2 log10 IU/ml (range, −0.1 to 0.4 log10 IU/ml) at 4.4 log10 (Table 7).
TABLE 7.
Quantification of the second international WHO standard HCV RNAa
| Nominal input concn (IU/ml [log10]) of WHO standard | RealTime HCV
|
CAP/CTM
|
||||
|---|---|---|---|---|---|---|
| Mean (range) concn
|
Mean difference from WHO standard | Mean (range) concn
|
Mean difference from WHO standard | |||
| IU/ml | log10 transformed | IU/ml | log10 transformed | |||
| 1,500 (3.2) | 920 (674-1,102) | 3.0 (2.8-3.0) | −0.2 | 3,064 (2,857-3,096) | 3.5 (3.5-3.5) | 0.3 |
| 25,000 (4.4) | 13,558 (8,997-16,102) | 4.1 (4.0-4.2) | −0.3 | 43,489 (22,085-69,852) | 4.6 (4.3-4.8) | 0.2 |
The second international WHO standard HCV RNA is code 96/798.
Genotype-specific assay linearity.
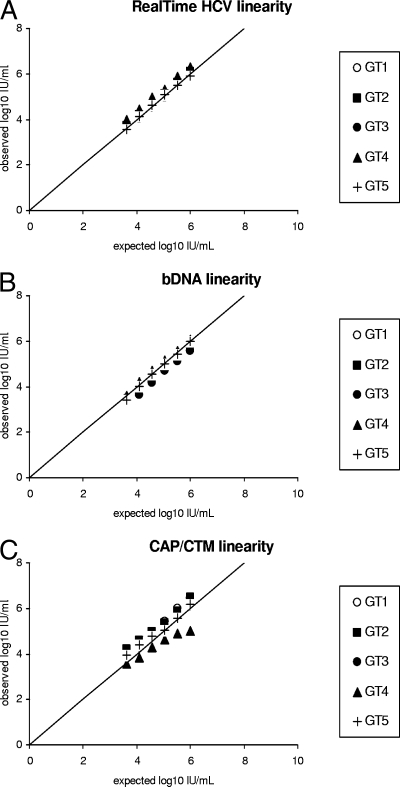
For RealTime HCV, CAP/CTM, and the bDNA assay, the quantification of HCV RNA of five different HCV genotypes (1 to 5) was mostly linear between 4.0 × 103 and 1.0 × 106 IU/ml. The exception was the CAP/CTM assay, which showed results lower than expected for the sample harboring genotype 4 at concentrations above 1.0 × 104 IU/ml (Fig. 5A to C).
FIG. 5.
Linearities of RealTime HCV (A), the bDNA assay (B), and CAP/CTM (C), as assessed with five threefold serial dilutions of clinical specimens representing genotypes (GTs) 1 to 5. The expected and the observed HCV RNA concentrations are shown on the basis of known HCV RNA concentrations, as assessed by RealTime HCV.
The overall R values between the expected and the observed results were >0.99, >0.99, and 0.99 for RealTime HCV, the bDNA assay, and CAP/CTM, respectively, with mean differences in the expected and the observed values of −0.06, 0.14, and −0.14 log10 IU/ml for the three assays, respectively.
DISCUSSION
The clinical utility of HCV RNA quantification is well established (7, 24). Indeed, viral load monitoring before, during, and after antiviral therapy is crucial for the management of hepatitis C. Among other aspects, HCV RNA assays ideally should be sensitive, offer precise and reproducible quantification results, and be reliable across all the different HCV genotypes.
The lack of standardization among HCV quantification assays has been overcome, in part, by the development of an IU standard (22, 23). However, the results differ significantly between assays, despite the standardization to IU (5, 14, 24), and it remains unknown which assay calibration best matches the standard. In addition, relative quantification results and LODs for different genotypes may vary among assays, since standardization to IU and the calibration of assay sensitivity are based on genotype 1a (22, 23).
In this study, we evaluated and compared the performance characteristics of two quantitative real-time reverse transcription-PCR-based assays (RealTime HCV and CAP/CTM) and one signal amplification-based assay (the bDNA assay).
The quantification of undiluted clinical specimens by RealTime HCV in comparison to that by the bDNA assay displayed a high correlation and good agreement among all genotypes tested. Mean differences were below ±0.3 log10 IU/ml.
Comparison of RealTime HCV and CAP/CTM showed a good correlation only for samples harboring genotypes 2, 3, and 5. The mean difference between RealTime HCV and CAP/CTM for specimens harboring genotype 1 was 0.72 log10 IU/ml; i.e., the quantification results obtained by the CAP/CTM assay were found to be higher than those obtained by RealTime HCV. In contrast, the quantification results for samples harboring genotype 4 obtained by the CAP/CTM assay were lower than those obtained by RealTime HCV (−1.27 log10 IU/ml).
Comparison of CAP/CTM and the bDNA assay again showed a high concordance for samples harboring genotypes 2, 3, and 5, whereas a discrepancy for samples harboring genotypes 1 and 4 similar to that found for RealTime HCV and CAP/CTM was observed (higher levels of quantification for genotype 1 and lower levels of quantification for genotype 4 by CAP/CTM; 0.74 and −1.08 log10 IU/ml, respectively).
Taken together, our results demonstrated an overall good correlation of the results of all assays. However, when the HCV RNA levels in samples harboring genotype 1 were measured by CAP/CTM, they were always greater than the corresponding levels obtained by both RealTime HCV and the bDNA assay. In addition, a lower level of quantification by CAP/CTM was observed for samples harboring genotype 4. Despite the small number of samples available, these results are in line with those of a previous comparative study of Cobas TaqMan assays, the Cobas Amplicor Monitor assay, and the bDNA assay (24). More recently, Chevaliez et al. have described the overestimation of HCV RNA levels by CAP/CTM (7). However, in that study the global overquantification of HCV RNA of approximately 0.6 log10 IU/ml was claimed for all genotypes. In the present study, as well as in other previous studies, major differences in HCV RNA quantification between CAP/CTM, the bDNA assay, and RealTime HCV were restricted only to HCV genotype 1 and/or 4 (5, 17, 24, 30).
To date, the underquantification obtained for samples harboring genotype 4 has not been fully understood (7, 24, 29). The mismatch of primers and/or the TaqMan probe and the target viral sequence is unlikely to be the only reason, since the recent introduction of a revised version of the manual High Pure system for specimen extraction, which is used together with the CTM amplification and detection system (version 2), has overcome the prior underestimation of the amounts of genotypes 2 through 5 without changing the set of primers and/or probes used. In fact, only changes in the ethanol concentration of the wash buffer used for sample preparation and the temperature of the reverse transcription step were introduced (8). However, it has been suggested that suboptimal binding of oligonucleotides due to the secondary structure of the internal ribosome entry site and/or genotype-specific polymorphisms within the highly conserved 5′ nontranslated region may be responsible for underestimation of the quantity of genotype 4 by CAP/CTM (24, 29).
The clinical impact of genotype 4 underestimation by CAP/CTM may be the greatest in Egypt and the Middle East, where this genotype is prevalent. However, this technical issue may have minor implications (i.e., the use of different cutoff values for low versus high viral loads at the baseline) for patient management if viral load testing before, during, and after antiviral therapy is always performed by the same assay and in the same laboratory.
Since the bDNA assay has a reported lower LOD of 615 IU/ml, it was not included in our sensitivity experiments. In the present study, RealTime HCV had a sensitivity of 16.8 IU/ml for the second international WHO standard HCV RNA (code 96/798) that was comparable to that of the CAP/CTM assay (10.3 IU/ml) and that correlated with previous results (17, 27, 29).
The sensitivity of RealTime HCV was even lower for clinical specimens harboring genotypes 1 to 6 (4.7 to 9.0 IU/ml). Interestingly, CAP/CTM had a limit of detection between 3.4 and 14.1 IU/ml for samples harboring genotypes 1, 3, 5, and 6, whereas samples harboring genotype 2 and 4 yielded higher probit values of 44.4 and 40.5 IU/ml, respectively. However, the results for the lower LODs of clinical specimens may vary between different samples of different HCV subtypes and origins. In a recent publication by Sizmann et al., equal lower LODs between 6.5 and 15.8 IU/ml for HCV genotypes 1 to 6 were reported (29). In that study, however, no comparison with other real-time PCR-based assays was performed, and the results may vary depending on the method used for assessment of the HCV RNA concentration in the original undiluted sample.
Generally, patients who may have tested HCV RNA negative during or after antiviral therapy by older assays with lower LODs of ≥50 IU/ml may test HCV RNA positive by highly sensitive HCV RNA assays. Highly sensitive HCV RNA assays are now used in clinical practice to define a virologic nonresponse to antiviral therapy and to predict relapses after antiviral therapy (10, 18, 19, 25). In addition, the large dynamic range of the real-time PCR-based assays allows precise HCV RNA quantification without predilution, which was frequently required with previous standard PCR-based assays.
As discussed above, previous studies have shown differences of 0.5 to 0.7 log10 IU/ml for clinical specimens harboring genotype 1 between different assays (7, 17, 24). Although the bDNA assay, RealTime HCV, and CAP/CTM are calibrated to the WHO standard, it remains unclear which of the assays corresponds best to the WHO standard on direct comparison. In order to investigate whether one of the assays over- or underestimates the true HCV RNA concentration, we performed for the first time a direct comparison of the two real-time PCR-based assays with the WHO standard.
The standard, produced by the National Institute for Biological Standards and Control (South Mimms, United Kingdom), is available only at an assigned unitage of 50,000 IU/ml. Two concentrations (25,000 and 1,500 IU/ml) were chosen and tested in multiple aliquots in a single run each.
The present analysis revealed a consistently lower level of quantification of the WHO standard by RealTime HCV (−0.2 to −0.3 log10 IU/ml) and a consistently higher level of quantification of the WHO standard by CAP/CTM (+0.2 to +0.3 log10 IU/ml). Thus, the total difference adds up to approximately 0.5 log10 IU/ml, and this corresponds to the findings of previous clinical studies that evaluated CAP/CTM versus the bDNA assay and RealTime HCV (7, 17, 24).
In conclusion, the real-time PCR-based HCV RNA assays showed comparable, linear HCV RNA quantification abilities and a comparable sensitive detection of all HCV genotypes, with the exception of genotypes 1 and 4. The previously reported differences in the absolute quantification of samples harboring HCV genotype 1, which showed higher quantification results by CAP/CTM and lower quantification results by the bDNA assay and/or RealTime HCV, resulting in a total difference of approximately 0.5 to 0.7 log10 IU/ml, were confirmed (7, 17, 24). Comparative analysis with the current WHO standard suggests that the differences may be explained by the different calibration methods used and/or the different WHO standards used (RealTime HCV is standardized against the second international WHO standard HCV RNA [code 96/798], whereas CAP/CTM is standardized against the first international WHO standard HCV RNA [code 96/790]).
Footnotes
Published ahead of print on 17 September 2008.
REFERENCES
- 1.Andonov, A., and R. K. Chaudhary. 1995. Subtyping of hepatitis C virus isolates by a line probe assay using hybridization. J. Clin. Microbiol. 33254-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berg, T., V. Weich, G. Teuber, H. Klinker, B. Moeller, J. Rasenack, H. Hinrichsen, T. Gerlach, U. Spengler, P. Buggisch, H. Balk, M. Zankel, C. Sarrazin, S. Zeuzem, et al. 2007. Time to HCV RNA negativation in hepatitis C virus (HCV) type 1-infection during PEG-interferon-alpha-2B plus ribavirin therapy. Differences in relation to the assay sensitivity. Hepatology 46360A. [Google Scholar]
- 3.Bland, J. M., and D. G. Altman. 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet i307-310. [PubMed] [Google Scholar]
- 4.Bouchardeau, F., J. F. Cantaloube, S. Chevaliez, C. Portal, A. Razer, J. J. Lefrere, J. M. Pawlotsky, P. De Micco, and S. Laperche. 2007. Improvement of hepatitis C virus (HCV) genotype determination with the new version of the Inno-LiPA HCV assay. J. Clin. Microbiol. 451140-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caliendo, A. M., A. Valsamakis, Y. Zhou, B. Yen-Lieberman, J. Andersen, S. Young, A. Ferreira-Gonzalez, G. J. Tsongalis, R. Pyles, J. W. Bremer, and N. S. Lurain. 2006. Multilaboratory comparison of hepatitis C virus viral load assays. J. Clin. Microbiol. 441726-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Z., and K. E. Weck. 2002. Hepatitis C virus genotyping: interrogation of the 5′ untranslated region cannot accurately distinguish genotypes 1a and 1b. J. Clin. Microbiol. 403127-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chevaliez, S., M. Bouvier-Alias, R. Brillet, and J. M. Pawlotsky. 2007. Overestimation and underestimation of hepatitis C virus RNA levels in a widely used real-time polymerase chain reaction-based method. Hepatology 4622-31. [DOI] [PubMed] [Google Scholar]
- 8.Colucci, G., J. Ferguson, C. Harkleroad, S. Lee, D. Romo, S. Soviero, J. Thompson, M. Velez, A. Wang, Y. Miyahara, S. Young, and C. Sarrazin. 2007. Improved COBAS TaqMan hepatitis C virus test (version 2.0) for use with the High Pure system: enhanced genotype inclusivity and performance characteristics in a multisite study. J. Clin. Microbiol. 453595-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davis, G. L., J. B. Wong, J. G. McHutchison, M. P. Manns, J. Harvey, and J. Albrecht. 2003. Early virologic response to treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C. Hepatology 38645-652. [DOI] [PubMed] [Google Scholar]
- 10.Ferenci, P., H. Laferl, T. M. Scherzer, A. Maieron, M. Gschwantler, H. Brunner, R. Hubmann, M. Bischof, K. Staufer, C. Datz, P. Steindl-Munda, and H. Kessler. 2006. Customizing treatment with peginterferon alfa-2a (40kd) (PegasysR) plus ribavirin (CopegusR) in patients with HCV genotype 1 or 4 infection. Interim results of a prospective randomized trial. Hepatology 44336A. [Google Scholar]
- 11.Ferenci, P., M. W. Fried, M. L. Shiffman, C. I. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, M. Chaneac, and K. R. Reddy. 2005. Predicting sustained virological responses in chronic hepatitis C patients treated with peginterferon alfa-2a (40 KD)/ribavirin. J. Hepatol. 43425-433. [DOI] [PubMed] [Google Scholar]
- 12.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Goncales, Jr., D. Haussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffman, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347975-982. [DOI] [PubMed] [Google Scholar]
- 13.Hadziyannis, S. J., H. Sette, Jr., T. R. Morgan, V. Balan, M. Diago, P. Marcellin, G. Ramadori, H. Bodenheimer, Jr., D. Bernstein, M. Rizzetto, S. Zeuzem, P. J. Pockros, A. Lin, and A. M. Ackrill. 2004. Peginterferon-alpha2a and ribavirin combination therapy in chronic hepatitis C: a randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 140346-355. [DOI] [PubMed] [Google Scholar]
- 14.Halfon, P., M. Bourliere, G. Penaranda, H. Khiri, and D. Ouzan. 2006. Real-time PCR assays for hepatitis C virus (HCV) RNA quantitation are adequate for clinical management of patients with chronic HCV infection. J. Clin. Microbiol. 442507-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen, D. M., T. R. Morgan, P. Marcellin, P. J. Pockros, K. R. Reddy, S. J. Hadziyannis, P. Ferenci, A. M. Ackrill, and B. Willems. 2006. Early identification of HCV genotype 1 patients responding to 24 weeks peginterferon alpha-2a (40 kd)/ribavirin therapy. Hepatology 43954-960. [DOI] [PubMed] [Google Scholar]
- 16.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358958-965. [DOI] [PubMed] [Google Scholar]
- 17.Michelin, B. D., Z. Muller, E. Stelzl, E. Marth, and H. H. Kessler. 2007. Evaluation of the Abbott RealTime HCV assay for quantitative detection of hepatitis C virus RNA. J. Clin. Virol. 3896-100. [DOI] [PubMed] [Google Scholar]
- 18.Mihm, U., W. P. Hofmann, B. Kronenberger, M. Wagner, S. Zeuzem, and C. Sarrazin. 2005. Highly sensitive hepatitis C virus RNA detection assays for decision of treatment (dis)continuation in patients with chronic hepatitis C. J. Hepatol. 42605-606. [DOI] [PubMed] [Google Scholar]
- 19.Morishima, C., T. R. Morgan, J. E. Everhart, E. C. Wright, M. L. Shiffman, G. T. Everson, K. L. Lindsay, A. S. Lok, H. L. Bonkovsky, A. M. Di Bisceglie, W. M. Lee, J. L. Dienstag, M. G. Ghany, and D. R. Gretch. 2006. HCV RNA detection by TMA during the hepatitis C antiviral long-term treatment against cirrhosis (Halt-C) trial. Hepatology 44360-367. [DOI] [PubMed] [Google Scholar]
- 20.Nadarajah, R., G. Y. Khan, S. A. Miller, and G. F. Brooks. 2007. Evaluation of a new-generation line-probe assay that detects 5′ untranslated and core regions to genotype and subtype hepatitis C virus. Am. J. Clin. Pathol. 128300-304. [DOI] [PubMed] [Google Scholar]
- 21.Ross, R. S., S. Viazov, S. Sarr, S. Hoffmann, A. Kramer, and M. Roggendorf. 2002. Quantitation of hepatitis C virus RNA by third generation branched DNA-based signal amplification assay. J. Virol. Methods 101159-168. [DOI] [PubMed] [Google Scholar]
- 22.Saldanha, J., A. Heath, C. Aberham, J. Albrecht, G. Gentili, M. Gessner, and G. Pisani. 2005. World Health Organization collaborative study to establish a replacement WHO international standard for hepatitis C virus RNA nucleic acid amplification technology assays. Vox Sang. 88202-204. [DOI] [PubMed] [Google Scholar]
- 23.Saldanha, J., N. Lelie, M. W. Yu, and A. Heath. 2002. Establishment of the first World Health Organization international standard for human parvovirus B19 DNA nucleic acid amplification techniques. Vox Sang. 8224-31. [DOI] [PubMed] [Google Scholar]
- 24.Sarrazin, C., B. C. Gartner, D. Sizmann, R. Babiel, U. Mihm, W. P. Hofmann, M. von Wagner, and S. Zeuzem. 2006. Comparison of conventional PCR with real-time PCR and branched DNA-based assays for hepatitis C virus RNA quantification and clinical significance for genotypes 1 to 5. J. Clin. Microbiol. 44729-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sarrazin, C., G. Teuber, R. Kokka, H. Rabenau, and S. Zeuzem. 2000. Detection of residual hepatitis C virus RNA by transcription-mediated amplification in patients with complete virologic response according to polymerase chain reaction-based assays. Hepatology 32818-823. [DOI] [PubMed] [Google Scholar]
- 26.Sarrazin, C., A. Dragan, B. C. Gärtner, M. S. Forman, S. Traver, S. Zeuzem, and A. Valsamakis. 2008. Evaluation of an automated, highly sensitive, real-time PCR based assay (Cobas Ampliprep/Cobas TaqMan) for quantification of HCV RNA. J. Clin. Virol. 43162-168. [DOI] [PubMed] [Google Scholar]
- 27.Schutten, M., E. Fries, C. Burghoorn-Maas, and H. G. Niesters. 2007. Evaluation of the analytical performance of the new Abbott RealTime RT-PCRs for the quantitative detection of HCV and HIV-1 RNA. J. Clin. Virol. 4099-104. [DOI] [PubMed] [Google Scholar]
- 28.Shiffman, M. L., S. Pappas, B. Bacon, E. Godofsky, D. Nelson, H. Harley, M. Diago, A. Lin, G. Hooper, and S. Zeuzem. 2006. Utility of virological response at weeks 4 and 12 in the prediction of SVR rates in genotype 2/3 patients treated with peginterferon-alfa-2a (40KD) plus ribavirin: findings from ACCELERATE. Hepatology 44316A-317A. [Google Scholar]
- 29.Sizmann, D., C. Boeck, J. Boelter, D. Fischer, M. Miethke, S. Nicolaus, M. Zadak, and R. Babiel. 2007. Fully automated quantification of hepatitis C virus (HCV) RNA in human plasma and human serum by the Cobas AmpliPrep/Cobas TaqMan system. J. Clin. Virol. 38326-333. [DOI] [PubMed] [Google Scholar]
- 30.Tuaillon, E., A.-M. Mondain, L. Ottomani, L. Roudière, P. Perney, M.-C. Picot, F. Séguret, F. Blanc, D. Larrey, P. Van de Perre, and J. Ducos. 2007. Impact of hepatitis C virus (HCV) genotypes on quantification of HCV RNA in serum by Cobas AmpliPrep/Cobas TaqMan HCV test, Abbott HCV RealTime assay [corrected], and Versant HCV RNA assay. J. Clin. Microbiol. 453077-3081. [DOI] [PMC free article] [PubMed] [Google Scholar]