Abstract
Vesicular stomatitis virus (VSV) is currently being studied as a candidate oncolytic virus for tumor therapies due to its potent tumoricidal activity. Previous studies have demonstrated that VSV selectively infects tumor cells due to defects in their antiviral pathways. These defects make them more susceptible to VSV-induced killing than normal cells. However, some cancer cells display differential sensitivity to VSV. Specifically, LNCaP prostate cancer cells are sensitive to infection with VSV, while PC3 prostate cancer cells are relatively resistant to VSV. This suggests that tumor cells vary in the extent to which they develop defects in antiviral pathways and, thus, permit virus replication. The goal of these studies was to identify the step(s) of the viral replication cycle that is inhibited in PC3 cells. Results showed that although attachment of VSV was not significantly different among cell types, penetration was delayed by 10 to 30 min in PC3 cells relative to LNCaP cells. Primary transcription was delayed by 6 to 8 h in PC3 cells relative to LNCaP cells. Similarly, both secondary transcription and viral protein synthesis rates were delayed by about 6 to 8 h. The progressively increasing delay suggests that more than one step is affected in PC3 cells. Analysis of cellular gene expression showed that in contrast to LNCaP cells, PC3 cells constitutively expressed numerous antiviral gene products, which may enhance their resistance to VSV. These data indicate that the use of VSV for oncolytic virus therapy for prostate tumors may require prescreening of tumors for their level of susceptibility.
Vesicular stomatitis virus (VSV) is one of several viruses currently being developed as oncolytic agents for antitumor therapies (1, 6-8, 17, 20, 24, 25, 27). Oncolytic viruses specifically kill cancer cells while sparing normal cells. One means of achieving this is by exploiting defects in host defense mechanisms that are common in cancer cells. Specifically, many cancer cells have defects in their type I interferon (IFN-I) signaling network and as a result are sensitive to killing by VSV and other oncolytic viruses (24, 25). VSV is an attractive candidate oncolytic virus because it is highly sensitive to the antiviral effects of IFN-I and therefore replicates selectively in cancer cells that have defects in the IFN-I pathway. The specificity of VSV for killing cancer cells rather than normal cells can be further enhanced by pretreatment with IFNs or by using VSV strains that induce IFN-I production in infected cells. Wild type (wt) strains of VSV strongly suppress host antiviral responses due to inhibition of host gene expression by wt M protein (4). Mutations in the M protein abolish the ability of the virus to inhibit host gene expression, and, as a result, infected cells are able to produce IFN-I and other antiviral cytokines in response to viral infection (1, 25). Such M protein mutants have been proposed as strong candidates for oncolytic viral therapy since they are attenuated for replication in normal tissues but replicate as well as wt virus in cancers that have defective antiviral responses.
While VSV has shown promise as a candidate oncolytic virus, it may only be effective in a subset of cancer cells that exhibit defective responses to IFN-I. Ahmed et al. (1) demonstrated that prostate cancer cells display differential sensitivity to VSV. Specifically, LNCaP cells, which have a defective IFN-I response, are highly sensitive to infection and killing by VSV, while PC3 cells have intact IFN-I signaling and are relatively resistant to VSV (1, 11). Although PC3 cells eventually succumb to infection and killing by VSV, their relative resistance was evident by a delay in three parameters: (i) viral gene expression, (ii) production of progeny virus, and (iii) cell death. The goal of the experiments presented here was to determine the mechanism of resistance by identifying which step(s) of the viral replication cycle may be inhibited in PC3 cells to cause the observed delays.
The replication cycle of VSV is typical of that for negative-strand RNA viruses, for which it serves as a widely studied prototype (reviewed in reference 16). Rather than binding to a specific receptor, VSV attaches via nonspecific electrostatic and hydrophobic interactions with the surface of many cells. The virus penetrates by clathrin-dependent endocytosis. Upon acidification of the endosome, the viral G protein mediates fusion of the viral envelope with the endosome membrane and releases the viral nucleocapsid into the cytoplasm of the cell. The nucleocapsid is uncoated, and the parental genomes undergo primary transcription to generate viral messages which are translated using host machinery. After a brief period of primary transcription, the viral polymerase switches from transcription to replication to generate progeny genomes. Progeny genomes then undergo secondary transcription, which is a major step in amplification of viral gene expression. The virus assembles by budding from the host plasma membrane. The early steps of the cycle (adsorption, penetration, and primary transcription) occur within the first 2 h of infection, and these steps are typically sensitive to the amount of input virus, or multiplicity of infection (MOI). The later steps (replication, secondary transcription, and assembly) begin at around 2 h and continue until 12 to 18 h postinfection, when the cell undergoes apoptosis. Because these steps are the results of major amplifications, they are not usually sensitive to the amount of input virus.
In the experiments presented here, we found that the theoretical MOI required to establish a synchronous infection in PC3 cells was high (50 PFU/cell), whereas LNCaP cells were synchronously infected with lower MOIs typical of sensitive cell lines. Attachment of VSV to different cell types did not vary markedly. However, we identified delays in each subsequent step in the virus replication cycle in PC3 cells relative to LNCaP cells. The fact that multiple steps in virus replication contributed to the relative resistance of PC3 cells suggested that these cells were in an antiviral state similar to that observed in cells treated with IFN-I. This idea was supported by analysis of the gene expression profiles of LNCaP and PC3 cells, which showed that numerous antiviral genes were constitutively expressed at higher levels in PC3 cells than in LNCaP cells. These data indicate that prostate tumors may require prescreening for indicators of their level of susceptibility before they can be treated with VSV for oncolytic viral therapy.
MATERIALS AND METHODS
Virus, cells, and infections.
Recombinant wt (rwt) virus was isolated from an infectious cDNA clone, and virus stocks were prepared in BHK cells as previously described (14). PC3 and LNCaP cells were from the American Type Culture Collection and were cultured in RPMI medium containing 10% fetal bovine serum and 2 mM glutamine. HeLa cells were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 7% fetal bovine serum and 2 mM glutamine. Cells were grown in monolayers to about 70 to 90% confluence and were infected in small volumes at the MOIs specified for each experiment.
G protein surface expression.
PC3 and LNCaP cells were seeded in six-well plates and infected at the MOIs and times indicated in the figures. Virus infection and incubation at 4°C frequently caused LNCaP cells to detach from the plates. As a result, both cell types were harvested by scraping cells into the medium with sterile cell lifters, and cells were collected in microcentrifuge tubes. Cells were gently pelleted and washed with ice-cold fluorescence-activated cell sorter (FACS) buffer (100 ml of phosphate-buffered saline [PBS] and 5 ml of fetal bovine serum). The pellet was resuspended in 150 to 200 μl of FACS buffer and transferred to round-bottom 96-well plates. Plates were centrifuged at 1,000 rpm, and the supernatant was gently flicked out of the wells. Cells were washed two more times and were then blocked with a small volume of FACS buffer for 30 min. Surface-expressed G protein was labeled with anti-G protein antibody I1 (15) at a 1:300 dilution (all dilutions in FACS buffer) for 1 h. Cells were washed twice with FACS buffer and incubated with a secondary rabbit anti-mouse antibody conjugated to fluorescein isothiocyanate (ICN Biomedicals, Inc.) at a dilution of 1:200 for at least 30 min. Cells were washed twice with FACS buffer and were resuspended in 100 μl of 2% paraformaldehyde. Samples were stored at 4°C until analyzed by flow cytometry. A Beckman Dickinson FACSCalibur flow cytometer was used to quantify G protein cell surface fluorescence.
Preparation of radioactively labeled VSV.
BHK cells were grown to about 90% confluence in two 100-mm dishes (preparations were made in duplicate). Cells were infected for 6 h with VSV at an MOI of 10 PFU/cell. Cells were washed twice with methionine-free DMEM and were then incubated in methionine-free medium for 25 min to deplete endogenous methionine. Cells were incubated for 1 h with 100 μCi/ml [35S]methionine in methionine-free medium and then washed three times with sterile PBS. [35S]methionine-labeled virus was chased from cells by incubating cells in 3 ml of regular DMEM for 90 min. The supernatant was collected and centrifuged for 20 min at 200 rpm. The supernatant was carefully applied to a 2-ml cushion of 15% sucrose. The sample was centrifuged at 35,000 rpm for 1 h using a Beckman SW 50.1 rotor. The pellet containing the virus was resuspended in 1 ml of DMEM and kept at 4°C until used the following day for the attachment experiment.
Analysis of virus attachment.
The day before preparation of radioactive VSV, HeLa, PC3, and LNCaP cells were seeded (in duplicate) in six-well plates. Culture medium was replaced with 0.5 ml of ice-cold RPMI medium, and cells were harvested by scraping each well with a sterile cell lifter. The harvested cells were transferred to labeled Eppendorf tubes and placed at 4°C for at least 10 min prior to infection. The number of cells per well was determined by trypsinizing an identical well and counting the cells using a hemacytometer (usually, 5 × 105 to 9 × 105 cells). For each experiment, approximately 1.4 × 105 cpm of [35S]methionine-labeled VSV was added to each Eppendorf tube and incubated at 4°C for 30, 60, and 90 min. Cells were washed three times with ice-cold PBS. A total of 200 μl of ice-cold radioimmunoprecipitation assay (RIPA) buffer was added to each tube and incubated at 4°C for 5 min. The radioactivity associated with each sample was determined using a scintillation counter.
Penetration: escape of VSV from the surface of cells.
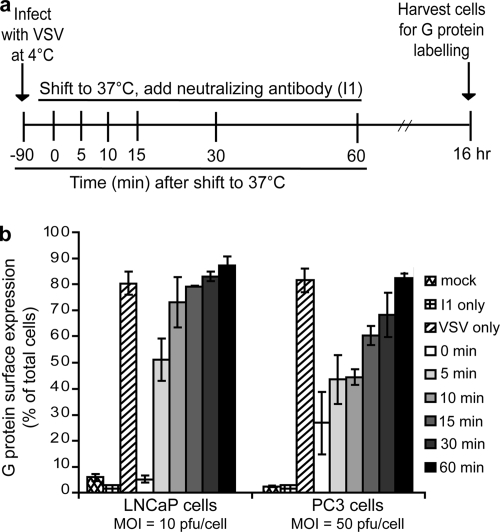
The protocol for the analysis of VSV escape from the cell surface, summarized in Fig. 3a, uses a neutralizing antibody (I1) to bind to the surface virus that has not been internalized. PC3 and LNCaP cells were seeded in 35-mm dishes to be 70% confluent at the time of infection. LNCaP cells were seeded in dishes coated with poly-l-lysine to facilitate continuous attachment throughout the experiment. A total of 700 μl of cold RPMI medium was added to each dish, and cells were placed at 4°C for at least 10 min. Cells were infected with VSV (PC3 cells, MOI of 50 PFU/cell; LNCaP cells, MOI of 10 PFU/cell) for 90 min at 4°C. Cells were washed three times with ice-cold PBS. The mock, rwt-only, I1 antibody-only, and time zero (0 min) samples were kept at 4°C. I1 antibody was added to the 0-min and antibody-only samples at this time. The remaining dishes were placed in a 37°C incubator. I1 antibody was added to the cells after they were warmed at 37°C for 5, 10, 15, 30, and 60 min, at which time cells were immediately moved to 4°C for 30 min to allow antibody binding to virus on the cell surface. All samples were returned to a 37°C incubator for 16 h and then labeled for surface expression of G protein, which was analyzed by flow cytometry.
FIG. 3.
Escape of VSV from the cell surface is delayed in PC3 cells relative to LNCaP cells. Cells were infected with VSV (LNCaP cells at an MOI of 10; PC3 cells at an MOI of 50) for 90 min at 4°C. As diagrammed in panel a, cells were warmed to 37°C for various times to allow endocytosis to proceed. After the warming step, a VSV-neutralizing antibody (I1) was added to the cells, and they were transferred to 4°C for 30 min to allow antibody binding. Cells were then incubated at 37°C for 16 h, labeled for surface expression of G protein, and analyzed by flow cytometry. Data are expressed as the percentage of cells expressing G at their surfaces for each condition (b).
Penetration: escape of VSV from the endosome.
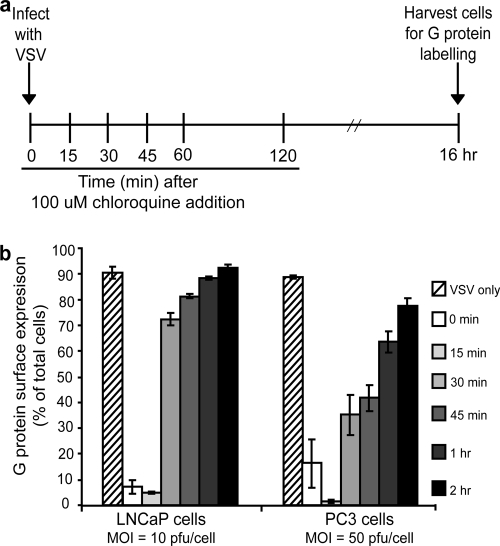
The protocol for the analysis of VSV escape from the endosomes is summarized in Fig. 4a. PC3 and LNCaP cells in six-well plates (70% confluent) were infected with VSV (PC3 cells, MOI 50 PFU/cell; LNCaP cells, MOI 10 PFU/cell). At 0 min, 15 min, 30 min, 45 min, 1 h, and 2 h of infection, 10 μl of 10 mM chloroquine was added to each well (100 μM final concentration). For the 0-min time point, VSV and chloroquine were mixed together before addition to the cells. Cells were incubated for a further 16 h and then labeled for surface expression of G protein.
FIG. 4.
VSV escape from the endosome is delayed in PC3 cells relative to LNCaP cells. As diagrammed in panel a, cells were infected with VSV (LNCaP cells at an MOI of 10; PC3 cells at an MOI of 50) and at various times postinfection were treated with 100 μM chloroquine (to inhibit acidification of the endosome). Sixteen hours later cells were harvested and labeled for surface expression of G protein and analyzed by flow cytometry. Data are expressed as the percentage of cells expressing G at their surfaces for each condition (b).
Real-time RT-PCR.
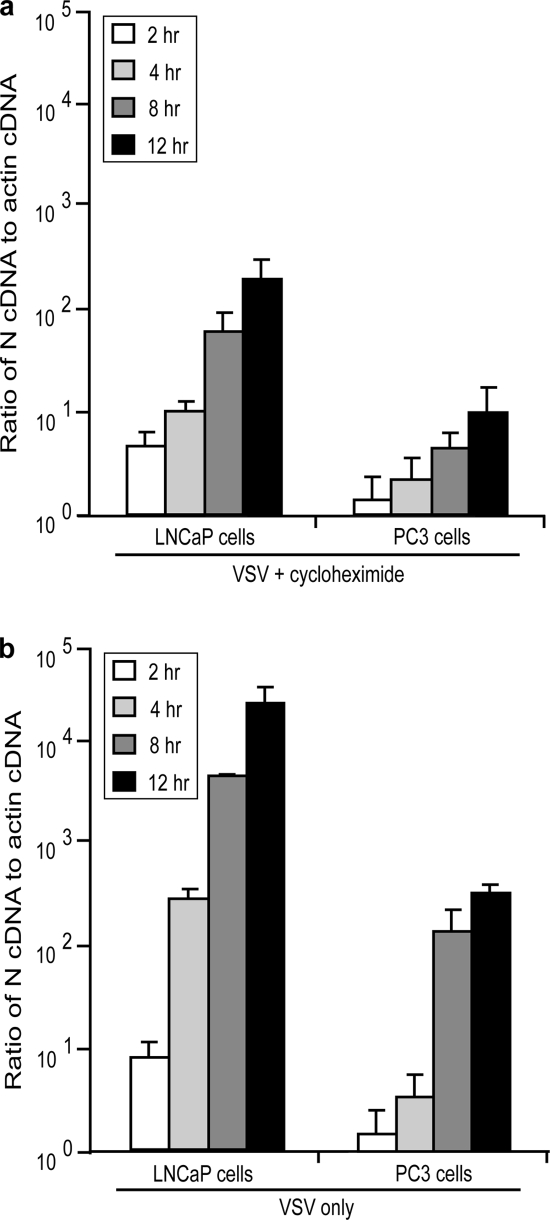
PC3 and LNCaP cells were infected with VSV for 2, 4, 8, and 12 h in either the presence or absence of 100 μg/ml cycloheximide. After infection, total RNA was harvested using TRIzol extraction (Invitrogen). Oligonucleotide primers and probes were designed and purchased from Sigma-Genosys. Primers for VSV nucleocapsid (N) protein were (forward) 5′-GAGTGGGCAGAACACAAATG-3′ and (reverse) 5′-CTTCTGGCACAAGAGGTTCA-3′. The probe for VSV N was 5′-TGCATTGATTTGTCAGCCCATCC-3′, which was labeled at the 5′ end with the reporter dye carboxyfluorescein and at the 3′ with the quencher tetramethylrhodamine. Primers for β-actin were (forward) 5′-CACTCTTCCAGCCTTCCTTC-3′ and (reverse) 5′-GGATGTCCACGTCACACTTC-3′. The probe for β-actin was 5′-TGCCACAGGACTCCATGCCC-3′, which was labeled as described for the VSV N probe. Reverse transcription-PCR analysis (RT-PCR) was performed with a TaqMan One-Step RT-PCR Master Mix Reagents kit (Applied Biosystems) as described by the manufacturer using a 25-μl sample volume with 5 μM concentrations of primers, 2.5 μM concentrations of probes, and 0.5 ng of sample RNA. TaqMan PCR assays were performed using an ABI 7700 instrument (Applied Biosystems, Foster City, CA) using the following amplification profile: 1 cycle at 48°C for 30 min, 1 cycle at 95°C for 10 min, and 40 cycles at 95°C for 15 s and 60°C for 1 min. All samples were tested in triplicate. The critical threshold cycle (CT) is defined as the cycle at which the fluorescence becomes detectable above background and is inversely proportional to the logarithm of the initial number of template molecules. A standard curve was plotted for each primer-probe set with CT values obtained from amplification of known quantities of plasmid DNA coding for either VSV N or β-actin. The standard curves were used to transform CT values of the experimental samples to the relative number of DNA molecules. The quantity of cDNA for each experimental gene was normalized to the quantity of the constitutively transcribed control gene (β-actin) in each sample. For each cell type, the relative change in transcript levels was determined by calculating the ratio of the level of VSV N cDNA in each experimental condition to the level of β-actin cDNA treated under the same conditions.
Quantification of host and viral protein synthesis.
PC3 and LNCaP cells were grown in 35-mm dishes to about 90% confluence and were infected with VSV at MOIs of 10 and 50 PFU/cell. At 4, 8, 12, and 24 h, cells were labeled with a 15-min pulse of [35S]methionine (100 μCi/ml) in a small volume of methionine-free medium. Cells were washed with PBS, and extracts were prepared by harvesting cells in RIPA buffer. Cell extracts were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and the fixed and dried gels were analyzed by phosphorescence imaging. Radioactivity of N protein bands and background host proteins (three sections from each lane excluding viral protein bands) was quantified with ImageQuant software (Molecular Dynamics, Inc.) (14).
Microarrays.
LNCaP and PC3 cells were grown to approximately 80% confluence and were mock infected or infected with rwt virus at an MOI of 10 PFU/cell. At 6 h postinfection, total RNA was isolated using TRIzol reagent (Invitrogen). Each RNA sample was processed according the manufacturer's protocol (Affymetrix) and hybridized to the Affymetrix Human Genome U133A 2.0 Array representing 14,500 well-characterized human genes. Each chip was scaled to a target intensity of 500, normalized to control probe sets present on each chip, and then expressed as a ratio to the nonspecific background on a per-gene basis. Analysis of data was carried out using Affymetrix Data Mining Tool software (Affymetrix). Statistical association of differences between LNCaP and PC3 cells with a data set of 80 different canonical pathways was evaluated using Ingenuity Pathways Analysis (IPA), according to the directions of the software. Parameters used in this analysis are described in the Results section.
Western blot analysis.
LNCaP and PC3 cells were grown to about 80% confluence in six-well plates and were either mock infected or infected with VSV. At the times postinfection indicated in the figures, cells were solubilized in RIPA buffer containing protease and phosphatase inhibitors: 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 1 mM pepstatin, 1 mM aprotinin, 1 mM sodium fluoride, 100 nM okadaic acid, and 100 nM microcystin. Protein concentrations were determined using a Bio-Rad detergent-compatible protein assay, and 10 μg of total protein was loaded onto gels. Proteins were resolved by SDS-PAGE on 8% polyacrylamide gels. Following electrophoresis, proteins were transferred onto polyvinylidene difluoride and blocked in PBS containing 0.2% Tween 20 and 0.1% bovine serum albumin. Immunoblots were then probed with antibodies against total PKR (Abcam), phospho-PKR (Thr451; Abcam), total STAT1 (Abcam), phospho-STAT1 (Tyr701; Abcam), or MxA (supplied by Georg Kochs, University of Frieburg). The positive control was HeLa cells treated for 18 h with IFN-α followed by 15 min with calyculin A. Protein band intensities were quantified by scanning and analysis with Quantity One software (Bio-Rad). For each protein analyzed, the Western blots were exposed together so that expression and/or phosphorylation levels could be compared between LNCaP and PC3 cells.
RESULTS
Synchronous infection in PC3 cells requires more virus than in LNCaP cells.
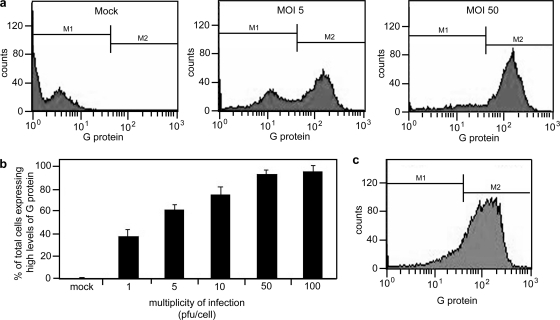
Previous studies have shown that human prostate cancer cells display differential sensitivity to infection with VSV (1). In a single cycle infection, benign prostate cells and LNCaP prostate cancer cells are highly susceptible to VSV infection while PC3 prostate cancer cells appear relatively resistant. In order to determine the steps in the replication cycle that are affected in PC3 cells, we first determined the MOI required for synchronous infection of PC3 cells. PC3 cells were mock infected or infected with VSV at MOIs of 1, 5, 10, 50, and 100 PFU/cell. After 16 h of infection, cells were harvested and labeled for surface expression of the viral G protein using an anti-G antibody, and the levels of G protein expression were determined by flow cytometry.
Figure 1a shows sample histograms of PC3 cells that were mock infected or infected at MOIs of 5 or 50 PFU/cell. Cells infected at an MOI of 5 and 50 PFU/cell expressed levels of G protein above that of mock-infected cells. However, upon infection at an MOI of 5 PFU/cell, there were two distinct populations of cells expressing either a low or a high level of G protein at their surfaces, whereas in cells infected at an MOI of 50, there was a single population of cells expressing a high level of surface G protein. Histograms from a series of experiments were gated as shown in Fig. 1a to analyze the percentage of PC3 cells in the high-expressing population (Fig. 1b). Results indicated that an MOI of 50 PFU/cell was required to synchronously infect PC3 cells such that over 90% of cells were expressing high levels of VSV G protein at their surfaces. LNCaP cells were more typical of cells lines that are susceptible to VSV, in which an MOI of 10 PFU/cell resulted in over 90% of cells expressing high levels of G protein (Fig. 1c).
FIG. 1.
Establishment of synchronous infection of VSV in PC3 cells. PC3 cells were infected at various MOIs (1 to 100 PFU/cell) for 16 h. Cells were then labeled for surface expression of G protein to indicate infected cells and analyzed by flow cytometry. (a) Histograms show different populations of cells expressing low and high levels of G protein in mock-infected PC3 cells and in PC3 cells infected at MOIs of 5 and 50 (b) The histograms were gated to analyze the population expressing high levels of G protein; the data are expressed as the percentage of cells expressing G protein at their surfaces for each condition. PC3 cells require a high MOI (50 PFU/cell) to establish a synchronous infection. (c) LNCaP cells were infected at an MOI of 10 PFU/cell and labeled for surface expression of G protein to show that when infected at this MOI, LNCaP cells are synchronously infected.
Attachment of VSV does not vary markedly among different cell types.
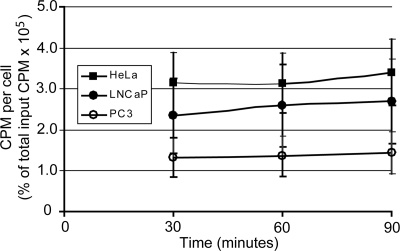
To determine whether the differential sensitivity of LNCaP and PC3 cells to VSV was due to reduced binding of virus to PC3 cells, virus attachment was analyzed in LNCaP and PC3 cells while HeLa cells served as a control for a highly permissive cell line. Attachment was analyzed using VSV radiolabeled with [35S]methionine. Equal amounts of radiolabeled VSV were incubated with HeLa, LNCaP, or PC3 cells for 30, 60, or 90 min at 4°C (a temperature at which virus penetration is inhibited). Cells were washed, and the radioactivity associated with each cell type was analyzed using a scintillation counter. The results are summarized in Fig. 2 which plots the counts per minute (cpm) per cell as a percentage of the total input cpm against time. Attachment of VSV occurred at low levels in all three cell types and did not appear to change significantly over time. There was a small difference in attachment of VSV to the different cell types such that binding to PC3 cells occurred at slightly lower levels than attachment to LNCaP cells, which in turn was slightly lower than attachment to HeLa cells (Fig. 2). However, these differences were not statistically significant, indicating that differences in attachment of VSV are not likely to account for differences in virus infection.
FIG. 2.
VSV attachment is similar among different cell types. VSV particles were labeled with [35S]methionine. HeLa, LNCaP, and PC3 cells were incubated with equal amounts of radiolabeled VSV for 30, 60, and 90 min at 4°C. Radioactivity associated with each cell type was detected using a scintillation counter; data are expressed for each cell type as the cpm per cell as a percentage of the total input cpm over time.
Penetration of VSV is delayed in PC3 cells compared to LNCaP cells.
The penetration step of the VSV replication cycle was analyzed in PC3 and LNCaP cells using two methods. Escape of the virus from the cell surface was measured using a neutralizing antibody to the VSV G protein, and escape from endosomes was measured using the drug chloroquine. The protocol for the antibody escape is diagramed in Fig. 3a. Briefly, cells were infected with VSV (LNCaP at an MOI of 10 PFU/cell and PC3 at an MOI of 50 PFU/cell) for 90 min at 4°C to allow attachment but inhibit endocytosis. Cells were then warmed to 37°C. At the times indicated on the figure, neutralizing antibody was added to the cells, and cultures were transferred to 4°C for 30 min to maximize antibody binding and prevent further endocytosis. Cells were then returned to 37°C for 16 h and were labeled for surface expression of G protein, which was detected by flow cytometry. Figure 3b shows the percentage of cells expressing G protein on their surfaces for each condition.
When the neutralizing antibody was added to LNCaP cells immediately before the cells were warmed to 37°C (0 min), the number of fluorescent cells was similar to that of mock-infected negative controls. However, when the antibody was added only 5 min after cells were warmed, 50% of the cells expressed G protein after 16 h of infection, indicating that infectious virus had escaped the cell surface of half of the cells within 5 min at 37°C. In the case of PC3 cells, the addition of antibody before cells were warmed to 37°C (0 min) reduced the number of cells expressing G protein to approximately 25% of total cells, which was somewhat higher than the negative controls. This likely reflects the larger amount of virus used to establish a synchronous infection. Despite the larger amount of virus, the half-time for virus escape from the cell surface was approximately 15 min, representing a delay of approximately 10 min relative to LNCaP cells.
Escape of virus from endosomes was measured using chloroquine, a drug that inhibits acidification of the endosome. This protocol, as diagramed in Fig. 4a, is similar to the analysis of escape from the cell surface. However, longer time points were analyzed since escape from endosomes is a later step in virus penetration. Briefly, cells were infected with VSV (LNCaP at an MOI of 10 PFU/cell and PC3 at an MOI of 50 PFU/cell), and chloroquine was added at 0, 15, 30, or 45 min or at 1 or 2 h postinfection. Cells were harvested 16 h after infection and were labeled for surface-expressed G protein which was detected by flow cytometry. Figure 4b shows the percentage of cells with surface expression of G protein for each condition.
When chloroquine and VSV were added to LNCaP cells simultaneously (0 min), fewer than 10% of the cells expressed G at their surface after 16 h infection. There was no increase in cells expressing G protein when chloroquine was added 15 min after infection, indicating that it takes longer than 15 min for the virus to escape from the endosome. However, when chloroquine was added 30 min after infection, the majority of LNCaP cells expressed G protein after 16 h of infection. In the case of PC3 cells, when chloroquine was added at the time of infection (0 min) or 15 min after infection, a small percentage of the cells expressed G protein. Similar to results in the previously described experiment, this can likely be attributed to the larger amount of virus used to establish a synchronous infection. When chloroquine was added to PC3 cells 30 or 45 min after infection, only 35 to 45% of cells expressed G protein. Only when chloroquine was added 2 h after infection did most of the PC3 cells express G protein. From these data, the half-times for virus escape from endosomes were estimated to be 23 min in LNCaP cells and 52 min in PC3 cells, representing a delay of almost 30 min.
Primary transcription is delayed in PC3 cells relative to LNCaP cells.
Following penetration, the viral nucleocapsid is released into the cytoplasm, where the parental genome undergoes primary transcription. Once sufficient levels of viral proteins accumulate, the parental genome undergoes replication to generate progeny genomes which then undergo secondary transcription. VSV primary and secondary transcription was analyzed in cells treated or not treated with cycloheximide to inhibit viral protein synthesis. Because genome replication requires new protein synthesis, cells treated with cycloheximide only undergo primary transcription. Cells were infected in the absence or presence of cycloheximide (LNCaP cells at an MOI of 10 PFU/cell and PC3 cells at an MOI of 50 PFU/cell). Total RNA was harvested from cells after 2, 4, 8, and 12 h of infection, and the levels of VSV N RNA were detected using real-time RT-PCR. The VSV N RNA levels were normalized to actin mRNA levels at each time point. The results of primary transcription analysis are shown in Fig. 5a. There was a delay in primary transcription in PC3 cells relative to LNCaP cells such that VSV-infected PC3 cells required 8 h to accumulate levels of VSV N RNA comparable to the levels that LNCaP cells accumulated when infected for only 2 h. Therefore, we conclude that relative to LNCaP cells, VSV primary transcription is delayed by approximately 6 h in PC3 cells.
FIG. 5.
Primary transcription is delayed in PC3 cells relative to LNCaP cells. Cells were infected with VSV (LNCaP cells at an MOI of 10; PC3 cells at an MOI of 50) for 2, 4, 8, and 12 h in the presence (a) and absence (b) of cycloheximide. Total RNA was harvested, and levels of VSV N cDNA were analyzed by reverse transcriptase real time PCR. N cDNA levels were compared to β-actin cDNA at each condition. Data are expressed as the ratio of N cDNA to actin cDNA.
Figure 5b shows analysis of N RNA accumulation in the absence of cycloheximide. The levels of N detected can mostly be attributed to the amplification of viral gene expression during secondary transcription. Similar to results with primary transcription, there was a delay in secondary transcription such that VSV-infected PC3 cells required between 8 to 12 h to accumulate a level of VSV N RNA comparable to the level LNCaP cells accumulated when infected for only 4 h. This result suggests that the delay in secondary transcription in PC3 cells relative to LNCaP cells was mostly due to the delay in primary transcription because there was not much difference in the timing of the delay observed in primary versus secondary transcription.
VSV protein synthesis rates in PC3 cells are reduced at early times and increased at later times relative to LNCaP cells.
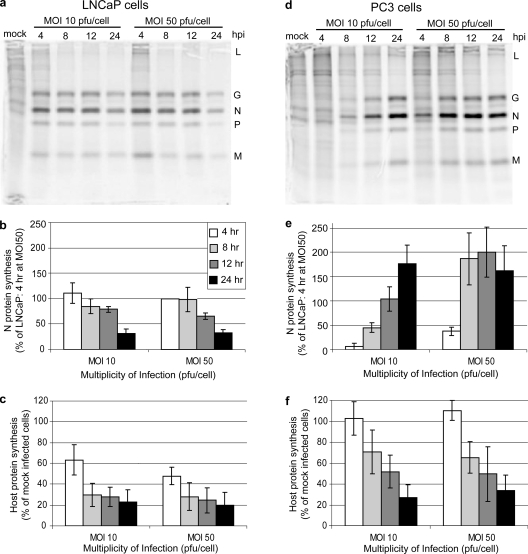
Figure 6 shows the rates of VSV and host protein synthesis in LNCaP and PC3 cells. Cells were mock infected or infected with VSV at an MOI of either 10 or 50 PFU/cell. At 4, 8, 12, and 24 h postinfection, cells were pulsed with [35S]methionine and were harvested for analysis by SDS-PAGE and phosphorescence imaging. Representative images for LNCaP and PC3 cells are shown in Fig. 6a and d, respectively. Figure 6a shows three major results that are typical of VSV infection of sensitive cell lines such as LNCaP cells. First, viral protein synthesis peaked early in infection and declined at late times postinfection. Second, host protein synthesis was rapidly shut off, a result of the ability of the VSV M protein to inhibit host gene expression. Third, the synthesis of viral proteins and the inhibition of host protein synthesis were largely independent of the MOI in this range. These three results were different for VSV infection of PC3 cells, as shown in Fig. 6d. In PC3 cells, viral protein expression was delayed and was barely detectable at early times of infection. Host protein synthesis was inhibited, but the shutoff was more gradual than in LNCaP cells. As expected from the experiment shown in Fig. 1, the expression of viral proteins in PC3 cells was dependent on the MOI since infection at an MOI of 10 PFU/cell was asynchronous.
FIG. 6.
Analysis of protein synthesis in PC3 and LNCaP cells infected with VSV. Both LNCaP (a, b, and c) and PC3 (d, e, and f) cells were infected at MOIs of 10 and 50 PFU/cell. At various times postinfection cells were labeled with [35S]methionine. Lysates were analyzed by SDS-PAGE and phosphorescence imaging (a and d). Viral proteins are indicated to the right of each gel. The radioactivity of the VSV N protein bands was quantified and is shown for LNCaP cells in panel b and for PC3 cells in panel e. The radioactivity of background host protein synthesis (taken from three sections of each lane that exclude viral protein bands) was quantified and is shown for LNCaP cells in panel c and for PC3 cells in panel f. Relative to LNCaP cells, there is a delay in viral protein synthesis in PC3 cells, but it is similar to the delay seen in primary transcription.
Viral and host protein synthesis were quantified, and results of multiple experiments are shown in Fig. 6b, c, e, and f. As a representative of viral protein synthesis (Fig. 6b and e), the density of the VSV N protein band (the most abundant viral protein) was determined and is expressed for both cell types as a percentage of the level of N protein in LNCaP cells infected at an MOI of 50 PFU/cell for 4 h. In LNCaP cells infected at an MOI of either 10 or 50 PFU/cell, N protein expression was highest at 4 h postinfection and declined similarly over the course of infection (Fig. 6b). In contrast, when PC3 cells were infected at an MOI of 10 PFU/cell, there was very little N protein expression at 4 and 8 h postinfection (Fig. 6e). However, by 24 h postinfection, N protein synthesis in PC3 cells was higher than the maximum rate of synthesis in LNCaP cells. When PC3 cells were infected at an MOI of 50 PFU/cell, N protein synthesis was detectable earlier than when cells were infected at an MOI of 10 PFU/cell although N protein synthesis was still delayed relative to LNCaP cells. However, by 8 h postinfection at an MOI of 50 PFU/cell, N protein synthesis was maximal in PC3 cells at a level almost twofold higher than the maximum rate in LNCaP cells. The high level of viral protein synthesis in PC3 cells at later times postinfection was unexpected since the level of production of progeny virus at 24 h postinfection was about 13-fold less in PC3 cells than in LNCaP cells (though by 48 h, viral titers were similar in these cell types) (1). This indicates that there is also a delay in assembly of these proteins into infectious virions.
Host protein synthesis was calculated by analyzing radioactivity in three areas in each lane devoid of viral protein bands. Host protein synthesis in LNCaP and PC3 cells is shown in Fig. 6c and f as a percentage of host protein synthesis in control mock-infected cells. In LNCaP cells, host protein synthesis was reduced to 60% of control by 4 h postinfection and to about 25% by 8 h (Fig. 6c). In contrast, host protein synthesis in PC3 cells was not reduced at 4 h postinfection and was still around 70% of mock infected cells at 8 h postinfection (Fig. 6f). Host protein synthesis declined to about 30% of that of mock-infected cells by 24 h postinfection. These data indicate that host protein synthesis was shut off in PC3 cells infected with VSV, but this shutoff was delayed relative to LNCaP cells.
PC3 cells constitutively overexpress antiviral mRNAs and proteins.
The observation that multiple steps in the VSV replication cycle are delayed in PC3 cells suggests that PC3 cells have some of the properties of cells in an antiviral state since one of the characteristics of antiviral responses is that they affect multiple steps in virus replication. This hypothesis was tested by microarray analysis of mRNAs encoding antiviral proteins expressed in mock- or VSV-infected LNCaP and PC3 cells. Cells were mock infected (three experiments) or VSV infected (two experiments), and RNA was analyzed at 8 h postinfection. Table 1 shows data for IFN-inducible genes whose expression was significantly different (P < 0.05) in comparisons of either mock-infected LNCaP versus PC3 cells or VSV-infected LNCaP versus PC3 cells. The most striking difference between these two cell types is apparent in mock-infected cells. Expression of most of the IFN-inducible genes in LNCaP cells was not detected above the signal from control, nonspecific probe sets (Table 1). In contrast, most of the IFN-inducible genes were expressed at substantially higher levels in mock-infected PC3 cells. The differences between LNCaP and PC3 cells were much less striking following infection with VSV. In fact, levels of expression of most of these genes in PC3 cells were substantially less in VSV-infected cells than in mock-infected cells, consistent with the inhibition of host gene expression by the VSV matrix (M) protein. These data suggest that the resistance of PC3 cells relative to LNCaP cells is related primarily to the constitutive expression of antiviral genes prior to infection.
TABLE 1.
Microarray analysis of IFN-inducible genes whose expression differed between LNCaP and PC3 cells
| Name | Affymetrix probe set | Mean signal intensity in mock-infected cells
|
Mean signal intensity in rwt-infected cells
|
||||
|---|---|---|---|---|---|---|---|
| LNCaP | PC3a | PC3/LNCaP | LNCaP | PC3a | PC3/LNCaP | ||
| LMP7 | 209040_s_at | 8b | 859* | >106 | 1b | 161* | >129 |
| GBP1 | 202270_at | 7b | 681* | >97.7 | 1b | 287* | >410 |
| IFI16b | 208966_x_at | 17b | 1,178* | >67.8 | 7b | 146* | >22.0 |
| MX1 | 202086_at | 103b | 5,658* | >54.8 | 94 | 520 | 5.5 |
| IFIT1 | 203153_at | 61b | 3,218* | >52.8 | 2,487 | 1,442 | 0.6 |
| GBP1 | 202269_x_at | 34 | 1,419* | >42.3 | 3b | 437* | >159 |
| IFITM1 | 214022_s_at | 103 | 4,057* | 39.5 | 27 | 324* | 11.9 |
| IFI27 | 202411_at | 43b | 1,310* | >30.2 | 5b | 233* | >45.7 |
| IFI16 | 206332_s_at | 24b | 727* | >29.8 | 6b | 104 | >16.6 |
| OAS1-E18 | 202869_at | 57b | 1,614* | >28.4 | 55 | 162 | 2.9 |
| OAS1-E16 | 205552_s_at | 35b | 980* | >28.3 | 25b | 147* | >5.9 |
| OAS2 | 204972_at | 45 | 1,261* | 28.1 | 33 | 196 | 6.0 |
| JAK1 | 201648_at | 98 | 2,045* | 20.9 | 34 | 1,121* | 33.2 |
| IFITM1 | 201601_x_at | 232 | 4,684* | 20.2 | 42 | 370* | 8.8 |
| IFIT5 | 203596_s_at | 40b | 676* | >16.7 | 428 | 171 | 0.4 |
| IFI16 | 208965_s_at | 23b | 315* | >13.7 | 2b | 81b | |
| ISG20 | 204698_at | 51b | 693* | >13.7 | 62 | 123 | 2.0 |
| IFI30 | 201422_at | 92b | 928* | >10.1 | 29b | 680 | >23.3 |
| IRF9 | 203882_at | 146b | 1,347* | >9.2 | 25b | 312* | >12.4 |
| OAS3 | 218400_at | 361 | 3,063* | 8.5 | 248 | 671 | 2.7 |
| ISG15 | 205483_s_at | 1,914 | 16,139* | 8.4 | 3,500 | 3,872 | 1.1 |
| IFI6 | 204415_at | 266 | 2,145* | 8.1 | 177 | 631 | 3.6 |
| IRF7 | 208436_s_at | 341 | 2,301* | 6.8 | 27 | 363* | 13.5 |
| STAT1 | AFFX-HUMISGF3A/M97935_3_at | 534 | 2,842* | 5.3 | 78 | 499 | 6.4 |
| IFITM3 | 212203_x_at | 1,809 | 8,014* | 4.4 | 558 | 3,556 | 6.4 |
| IFIT5 | 203595_s_at | 153b | 545* | >3.6 | 200 | 171 | 0.9 |
| IFI41 | 208012_x_at | 389 | 1,310 | 3.4 | 51 | 202* | 4.0 |
| IFITM2 | 201315_x_at | 1,637 | 5,180* | 3.2 | 289 | 1,686 | 5.8 |
| DAP4 | 209323_at | 873 | 1,698* | 1.9 | 34 | 975* | 28.4 |
| IRF3 | 202621_at | 851 | 1,028 | 1.2 | 61 | 620* | 10.2 |
*, P < 0.05 by a Students t test of LNCaP versus PC3 signal intensities.
Signal intensity not significantly different from background intensity of control probe set.
The data in Table 1 were derived by specifically targeting IFN-inducible genes for analysis. In order to generate an analysis that was not biased toward a particular pathway, all mRNAs detected in either LNCaP or PC3 cells were tested for statistical association with a data set of canonical pathways using IPA software. In this analysis, 80 different signaling pathways were evaluated for the fraction of differentially expressed genes associated with each pathway in the database and for the statistical probability that the effect on the pathway would be due to random chance. Data were analyzed as the ratio of expression of each gene (PC3/LNCaP), with the criterion for whether a gene was differentially expressed set as a ratio of either ≥4.0 or ≤−4.0. These criteria included the maximum number of differentially expressed genes recommended by the software (852 out of 5,086 eligible for pathway analysis) and excluded nearly all genes whose expression was not significantly different in repeated experiments.
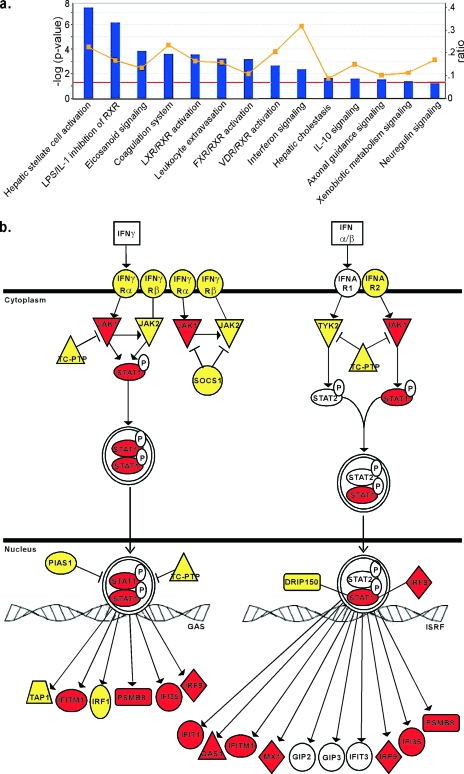
Figure 7a shows the signaling pathways statistically associated with the difference in gene expression between LNCaP and PC3 cells (P < 0.05), sorted according to the statistical significance of the association. Also shown is the fraction of differentially expressed genes associated with each pathway in the database. Several pathways contain gene products that may contribute to resistance/susceptibility to VSV infection, but the most obvious is the IFN signaling pathway, which had the highest fraction of differentially expressed genes (9/29, or 0.31) and a high level of statistical significance (P of 4.41 × 10−3). Increasing the criteria for differentially expressed genes to ratios of ≥10 or ≤−10 had little effect on the statistical significance of the association (P of 4.2 × 10−3) (data not shown). Figure 7b shows the IFN signaling genes used in this analysis, diagrammed according to their product's place in the pathway. The genes used in this analysis that were overexpressed in PC3 cells are a subset of those shown in Table 1. The high level of statistical significance for the association and the large fraction of differentially expressed genes make it unlikely that the results presented in Fig. 7 (and Table 1) are due to chance.
FIG. 7.
IPA analysis of differential mRNA expression in LNCaP and PC3 cells reveals constitutive expression of many antiviral mRNAs in PC3 cells. Microarray analysis of mock-infected LNCaP and PC3 cells was performed, and mRNA levels in the two cell types were further analyzed by IPA software. (a) The genes that were expressed differentially between LNCaP and PC3 cells were analyzed for association with any of 80 canonical pathways and are sorted according to their statistical significance of association (bars; red line indicates a P value of <0.05). The yellow line indicates the fraction associated with each pathway of genes that were expressed differentially between LNCaP and PC3 cells (right y axis). The IFN signaling pathway stands out with the highest fraction of differentially expressed genes. (b) The IFN signaling pathway genes. Red indicates genes that were overexpressed in PC3 cells relative to LNCaP cells. Yellow indicates genes that were present at similar levels in both cells types. Genes shown in white were present in the microarray library, but their signal levels were not detected above background. There were no genes in this pathway that were overexpressed in LNCaP cells relative to PC3 cells.
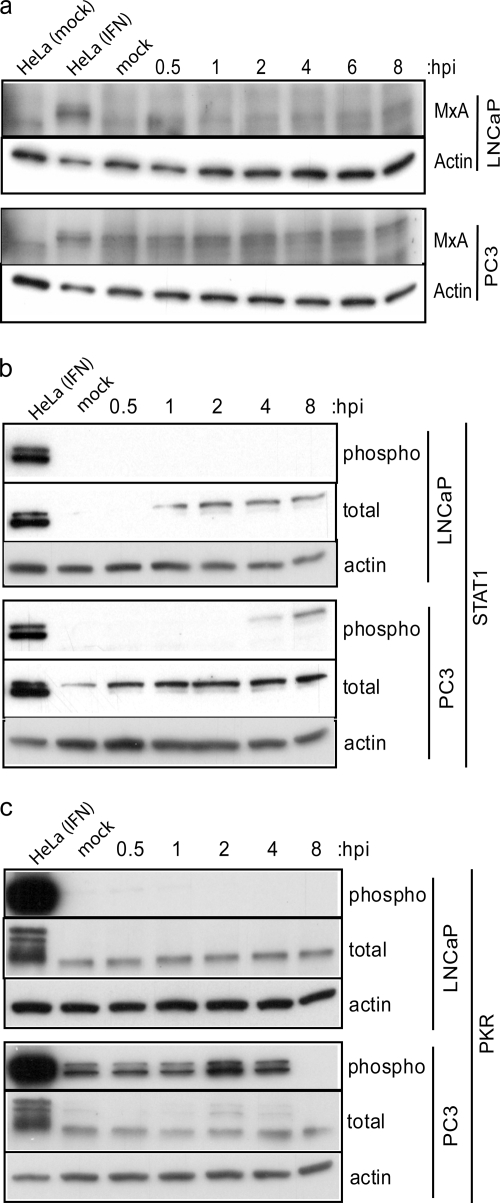
Western blot analyses were used to determine whether expression of antiviral mRNAs in Table 1 and Fig. 7 reflected corresponding proteins levels. Figure 8 shows analysis of MxA, STAT1, and PKR proteins as examples of genes detected only in PC3 cells (Fig. 8a, MxA), detected in both cell types but overexpressed in PC3 cells (Fig. 8b, STAT1), and detected in both cell types at levels that were not significantly different (Fig. 8c, PKR). Expression and/or phosphorylation of these proteins was assessed in both LNCaP and PC3 cells during a time course of VSV infection. IFN-I-treated HeLa cells were used as positive controls.
FIG. 8.
PC3 cells constitutively express antiviral proteins that may provide immediate protection against VSV infection. Western blotting was performed on LNCaP and PC3 cell lysates to detect the expression levels and/or activation status of MxA (a), STAT1 (b), and PKR (c) during a time course of VSV infection. Cells were infected with VSV (LNCaP cells at an MOI of 10; PC3 cells at an MOI of 50), and lysates were made at the indicated times postinfection. The controls for each Western blot were HeLa cells treated for 18 h with IFN-α and 15 min with calyculin A, and actin protein expression levels were included to demonstrate equal loading among lanes. (a) Induction of MxA protein expression was detected with an antibody against total MxA. (b) Total STAT1 protein expression and STAT1 phosphorylation status were detected using total and phospho-specific antibodies. (c) Similarly, total PKR protein expression and PKR phosphorylation status were detected using total and phospho-specific antibodies. The positive signal for phospho-PKR was indicated by a doublet of bands, which was masked in the control lane due to very strong signal strength in this sample. hpi, hours postinfection.
The anti-MxA antibody detected a band corresponding to MxA after IFN-I treatment in HeLa cells (Fig. 8a). The antibody also detected a nonspecific protein that ran slightly below MxA, as seen in the negative control. This nonspecific signal, but not the MxA band, was detected in LNCaP cells, both mock and VSV infected. However, the band representing MxA was detected in both mock- and VSV-infected PC3 cells. Despite the suspected IFN-I signaling in PC3 cells, we did not see an increase in MxA expression after infection with wt VSV, probably because the M protein shuts off host gene expression. The MxA Western blots confirmed the microarray data, in which the Mx1 message was not expressed in LNCaP cells but was detected in PC3 cells.
There was no detectable STAT1 phosphorylation in LNCaP cells at any time tested (Fig. 8b). Despite the lack of functional Jak1 in LNCaP cells (11), which is critical for IFN-I signaling, total STAT1 increased during the time course of infection in LNCaP cells. This is likely due to signals other than IFN-I, produced during virus infection. Although not apparent in the figure, STAT1 was detectable in mock-infected LNCaP cells using longer exposure times. Total STAT1 expression was higher in PC3 cells than LNCaP cells, as expected from the microarray data, and increased slightly following VSV infection. In PC3 cells, phospho-STAT1 was detected at 4 h postinfection and was increased at 8 h postinfection, consistent with the presence of IFN-I signaling in these cells.
Similar to what was observed for STAT1 activation, phospho-PKR was not detected in mock- or VSV-infected LNCaP cells (Fig. 8c). Total PKR expression was similar between the two cell types and remained constant during VSV infection. Surprisingly, phospho-PKR was detected in mock-infected PC3 cells. PKR remained phosphorylated during VSV infection in PC3 cells until 4 h postinfection, after which it was not detected. This result indicates that infection of PC3 cells with VSV induces the dephosphorylation of PKR, and this correlates with the increase in viral protein translation between 4 and 8 h postinfection (Fig. 6e).
DISCUSSION
Several studies have shown that oncolytic vectors based on VSV are promising agents for antitumor therapy (1, 6, 7, 17, 20, 24, 25). Cancer cells that are defective in antiviral signaling are selectively killed by VSV. Numerous cancer cell types, including both primary and immortalized tumor cell lines, have defective IFN signaling pathways (reviewed in reference 30). For example, LNCaP prostate cancer cells are nonresponsive to IFN-I (1). Studies have shown that LNCaP cells have defects in expression of Jak1 (11) and RNase L (21), and, as shown here, PKR phosphorylation was not detected during VSV infection (Fig. 8c). However, some cancer cells retain IFN-I responsiveness. Predictably, these cancers are relatively resistant to oncolytic VSV. In an analysis of VSV infection in the NCI-60 cell panel, 81% of cell lines were found to be unresponsive to IFN-I while the remaining 19% retained IFN-I responsiveness (25). Because these studies were conducted using immortalized cell lines, it is not known whether retention of IFN-I responsiveness is more frequent in naturally occurring cancers.
PC3 prostate cancer cells are an example of a cell line that has retained IFN-I responsiveness, and these cells were observed to be relatively resistant to VSV infection under presumed single-cycle infection conditions (1). Because PC3 cells are more resistant to VSV infection than normal prostate epithelial cells (1), they represent a clear exception to the idea that during tumorigenesis, cells develop defects in their antiviral responses. The goal of the studies reported here was to determine the basis for resistance of PC3 cells to VSV infection.
The experiments presented here demonstrated that multiple steps of the VSV replication cycle, including several early steps (Fig. 2 to 5a), are delayed in PC3 cells. Because we saw progressively longer delays in the early steps of replication and such a large increase in the timing of the delay between penetration (Fig. 3 and 4) and primary transcription (Fig. 5a), we do not think that the overall delay observed can all be attributed to that first delay in escape of the virus from the cell surface but, instead, is an accumulation of delays at several steps. Nevertheless, the delays in virus replication observed in PC3 cells under single-cycle conditions are relatively modest compared to the profound resistance of PC3-derived tumors to VSV infection in mice (1). The delays observed in cultured PC3 cells under single-cycle conditions are amplified during infections at lower MOIs (1). Under the low-MOI conditions intrinsic to the in vivo studies, antiviral signaling in the PC3-derived tumor would be very effective at limiting virus spread. This would be especially true for the inhibition of the virus at early steps of the replication cycle since they are the steps which are most sensitive to MOI.
The observation that multiple steps in virus replication are delayed in PC3 cells suggested that resistance is due to expression of antiviral gene products since inhibition of multiple steps of virus replication is a characteristic of host antiviral responses. For example, in different cell types, IFN-I treatment can affect VSV replication at the level of virus penetration (28, 29), primary transcription (13, 18, 19, 23), or virus assembly (26). While the data presented here correlate the constitutive expression of antiviral gene products in PC3 cells with the anticipated effects of a typical antiviral response (inhibition of the virus at multiple levels), they do not represent a direct link. Our results do not preclude the possibility that there are factors inherent to PC3 cells which mechanistically delay the virus replication cycle, independent of the expression of antiviral proteins. These possibilities are not mutually exclusive; a mechanistic delay could enable PC3 cells to activate an antiviral response, which then further inhibits the virus replication cycle. Future experiments will pursue this distinction.
In addition to constitutive expression of antiviral genes in PC3 cells, we observed constitutive phosphorylation of PKR (Fig. 8c). PKR phosphorylation did not increase further following infection but instead declined after 4 h postinfection, suggesting that VSV infection induces negative regulators of PKR. Phosphorylated PKR is known to inhibit protein synthesis by inactivating eIF2α. However, despite constitutive PKR phosphorylation, PC3 cellular protein synthesis is maintained at high levels. These results are similar to what is observed in transformed mouse embryonic fibroblasts, in which there is a deregulation of signaling downstream of PKR activation (5). Similar observations have been made in other cancer cells (22).
The dephosphorylation of PKR in PC3 cells was correlated with an increase in translation of viral proteins (Fig. 6d and e) in the absence of a corresponding increase in viral mRNA (Fig. 5). Despite the high level of viral protein synthesis in PC3 cells at late times postinfection, there is a pronounced delay in release of infectious virus relative to LNCaP cells (1). This suggests a block in virus assembly in PC3 cells.
The expression of antiviral gene products in PC3 cells was not due to constitutive signaling through STAT1. However, unlike LNCaP cells, PC3 cells were able to signal in response to IFN-I produced during virus infection, as shown by phosphorylation of STAT1 beginning around 4 h postinfection (Fig. 8b). In most cell types, infection with wt VSV results in inhibition of IFN-I production. This is due to the ability of the wt VSV M protein to globally shut off host gene expression (2-4, 9, 10, 31). In PC3 cells, the shutoff of host gene expression is delayed by several hours (Fig. 6f), which is likely a result of the cumulative delays in the virus replication cycle in these cells. During this delay, while virus infection is causing a reduction in gene expression at the mRNA level (Table 1), PC3 cells continue to express and activate antiviral proteins. This likely contributes to inhibition of the virus replication cycle (Fig. 8). IFN-I signaling does play a role during VSV infection of PC3 cells because pretreatment of PC3 cells with neutralizing IFN-I antibody resulted in an increase in cell killing after VSV infection (unpublished data).
The results presented here raise the question of why so many antiviral genes are constitutively expressed in PC3 cells in the absence of constitutive IFN-I signaling. There may be other signaling mechanisms besides IFN-I that activate these genes, possibly involving the constitutive activation of PKR (Fig. 8c) and/or constitutively active NF-κB (12), both of which could lead to the activation of antiviral gene expression through promoter elements other than those used by IFN-I signaling. It is possible that cells express antiviral genes during tumor development as a result of exposure to virus in vivo. Another possibility is that some of these antiviral genes encode survival factors that are important for cancer development. Whatever the cause, if constitutive expression of antiviral genes is common in vivo, such tumors will likely be resistant to many oncolytic viruses. As a result, tumors may need to be screened for resistance to virus to predict whether they will be amenable to treatment with oncolytic VSV. While gene products such as MxA and phospho-PKR may be useful screening tools, future experiments will be needed to identify antiviral gene products in naturally occurring tumors that correlate with resistance to oncolytic VSV.
Acknowledgments
We acknowledge the Microarray Core Laboratory at Wake Forest University. We thank Griffith Parks and Luke Carey for helpful advice and comments on the manuscript. We thank Sean Reid for assistance with real time RT-PCR and Georg Kochs for the anti-MxA antibody.
This work was supported by Public Health Service grants AI 32983 and AI 15892 from the National Institute of Allergy and Infectious Diseases and by grant PC010058 from the U.S. Army Medical Research and Material Command to D. S. Lyles. The Microarray Core Laboratory was supported in part by a core grant for the Comprehensive Cancer Center of Wake Forest University, CA12197, from the National Cancer Institute.
Footnotes
Published ahead of print on 1 October 2008.
REFERENCES
- 1.Ahmed, M., S. D. Cramer, and D. S. Lyles. 2004. Sensitivity of prostate tumors to wild type and M protein mutant vesicular stomatitis viruses. Virology 33034-49. [DOI] [PubMed] [Google Scholar]
- 2.Ahmed, M., and D. S. Lyles. 1998. Effect of vesicular stomatitis virus matrix protein on transcription directed by host RNA polymerases I, II, and III. J. Virol. 728413-8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed, M., and D. S. Lyles. 1997. Identification of a consensus mutation in M protein of vesicular stomatitis virus from persistently infected cells that affects inhibition of host-directed gene expression. Virology 237378-388. [DOI] [PubMed] [Google Scholar]
- 4.Ahmed, M., M. O. McKenzie, S. Puckett, M. Hojnacki, L. Poliquin, and D. S. Lyles. 2003. Ability of the matrix protein of vesicular stomatitis virus to suppress beta interferon gene expression is genetically correlated with the inhibition of host RNA and protein synthesis. J. Virol. 774646-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balachandran, S., and G. N. Barber. 2004. Defective translational control facilitates vesicular stomatitis virus oncolysis. Cancer Cell 551-65. [DOI] [PubMed] [Google Scholar]
- 6.Balachandran, S., and G. N. Barber. 2000. Vesicular stomatitis virus (VSV) therapy of tumors. IUBMB Life 50135-138. [DOI] [PubMed] [Google Scholar]
- 7.Balachandran, S., M. Porosnicu, and G. N. Barber. 2001. Oncolytic activity of vesicular stomatitis virus is effective against tumors exhibiting aberrant p53, Ras, or myc function and involves the induction of apoptosis. J. Virol. 753474-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barber, G. N. 2004. Vesicular stomatitis virus as an oncolytic vector. Viral Immunol. 17516-527. [DOI] [PubMed] [Google Scholar]
- 9.Black, B. L., G. Brewer, and D. S. Lyles. 1994. Effect of vesicular stomatitis virus matrix protein on host-directed translation in vivo. J. Virol. 68555-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Black, B. L., and D. S. Lyles. 1992. Vesicular stomatitis-virus matrix protein inhibits host cell-directed transcription of target genes in vivo. J. Virol. 664058-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn, G. P., K. C. Sheehan, L. J. Old, and R. D. Schreiber. 2005. IFN unresponsiveness in LNCaP cells due to the lack of JAK1 gene expression. Cancer Res. 653447-3453. [DOI] [PubMed] [Google Scholar]
- 12.Gasparian, A. V., Y. J. Yao, D. Kowalczyk, L. A. Lyakh, A. Karseladze, T. J. Slaga, and I. V. Budunova. 2002. The role of IKK in constitutive activation of NF-κB transcription factor in prostate carcinoma cells. J. Cell Sci. 115141-151. [DOI] [PubMed] [Google Scholar]
- 13.Haller, O., P. Staeheli, and G. Kochs. 2007. Interferon-induced Mx proteins in antiviral host defense. Biochimie 89812-818. [DOI] [PubMed] [Google Scholar]
- 14.Kopecky, S. A., M. C. Willingham, and D. S. Lyles. 2001. Matrix protein and another viral component contribute to induction of apoptosis in cells infected with vesicular stomatitis virus. J. Virol. 7512169-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lefrancois, L., and D. S. Lyles. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. II. Monoclonal antibodies of nonneutralizing and cross-reactive epitopes of Indiana and New Jersey serotypes. Virology 121168-174. [DOI] [PubMed] [Google Scholar]
- 16.Lyles, D. S., and C. E. Rupprecht (ed.). 2007. Rhabdoviridae, vol. 1. Lippincott Williams and Wilkins, Philadelphia, PA.
- 17.Obuchi, M., M. Fernandez, and G. N. Barber. 2003. Development of recombinant vesicular stomatitis viruses that exploit defects in host defense to augment specific oncolytic activity. J. Virol. 778843-8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pavlovic, J., and P. Staeheli. 1991. The antiviral potentials of Mx proteins. J. Interferon Res. 11215-219. [DOI] [PubMed] [Google Scholar]
- 19.Pavlovic, J., T. Zurcher, O. Haller, and P. Staeheli. 1990. Resistance to influenza virus and vesicular stomatitis virus conferred by expression of human Mxa protein. J. Virol. 643370-3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Porosnicu, M., A. Mian, and G. N. Barber. 2003. The oncolytic effect of recombinant vesicular stomatitis virus is enhanced by expression of the fusion cytosine deaminase/uracil phosphoribosyltransferase suicide gene. Cancer Res. 638366-8376. [PubMed] [Google Scholar]
- 21.Rennert, H., D. Bercovich, A. Hubert, D. Abeliovich, U. Rozovsky, A. Bar-Shira, S. Soloviov, L. Schreiber, H. Matzkin, G. Rennert, L. Kadouri, T. Peretz, Y. Yaron, and A. Orr-Urtreger. 2002. A novel founder mutation in the RNASEL gene, 471delAAAG, is associated with prostate cancer in Ashkenazi Jews. Am. J. Hum. Genet. 71981-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savinova, O., B. Joshi, and R. Jagus. 1999. Abnormal levels and minimal activity of the dsRNA-activated protein kinase, PKR, in breast carcinoma cells. Int. J. Biochem. Cell. Biol. 31175-189. [DOI] [PubMed] [Google Scholar]
- 23.Staeheli, P., and J. Pavlovic. 1991. Inhibition of vesicular stomatitis virus messenger RNA synthesis by human Mxa protein. J. Virol. 654498- 4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stojdl, D. F., B. Lichty, S. Knowles, R. Marius, H. Atkins, N. Sonenberg, and J. C. Bell. 2000. Exploiting tumor-specific defects in the interferon pathway with a previously unknown oncolytic virus. Nat. Med. 6821-825. [DOI] [PubMed] [Google Scholar]
- 25.Stojdl, D. F., B. D. Lichty, B. R. tenOever, J. M. Paterson, A. T. Power, S. Knowles, R. Marius, J. Reynard, L. Poliquin, H. Atkins, E. G. Brown, R. K. Durbin, J. E. Durbin, J. Hiscott, and J. C. Bell. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4263-275. [DOI] [PubMed] [Google Scholar]
- 26.Trottier, M. D., Jr., B. M. Palian, and C. S. Reiss. 2005. VSV replication in neurons is inhibited by type I IFN at multiple stages of infection. Virology 333215-225. [DOI] [PubMed] [Google Scholar]
- 27.Vaha-Koskela, M. J., J. E. Heikkila, and A. E. Hinkkanen. 2007. Oncolytic viruses in cancer therapy. Cancer Lett. 254178-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whitaker-Dowling, P. A., D. K. Wilcox, C. C. Widnell, and J. S. Youngner. 1983. Interferon-mediated inhibition of virus penetration. Proc. Natl. Acad. Sci. USA 801083-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilcox, D. K., P. A. Whitaker-Dowling, J. S. Youngner, and C. C. Widnell. 1983. Interferon treatment inhibits pinocytosis. Mol. Cell. Biol. 31533-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wong, L. H., K. G. Krauer, I. Hatzinisiriou, M. J. Estcourt, P. Hersey, N. D. Tam, S. Edmondson, R. J. Devenish, and S. J. Ralph. 1997. Interferon-resistant human melanoma cells are deficient in ISGF3 components, STAT1, STAT2, and p48-ISGF3γ. J. Biol. Chem. 27228779-28785. [DOI] [PubMed] [Google Scholar]
- 31.Yuan, H., B. K. Yoza, and S. S. Lyles. 1998. Inhibition of host RNA polymerase II-dependent transcription by vesicular stomatitis virus results form inactivation of TFIID. Virolofy 251383:392. [DOI] [PubMed] [Google Scholar]