Abstract
During lytic infection, the genome of herpes simplex virus 1 (HSV-1) is associated with limited levels of histones but does not form a regular repeating nucleosomal structure. However, the previous observation that chromatin remodeling factors are recruited into viral replication compartments indicates that chromatin remodeling plays a role in HSV-1 gene expression and DNA replication. In this study we demonstrate the presence of histone H3 on HSV-1 DNA early in infection at levels equivalent to those found on a cellular gene. The proportion of viral DNA associated with histone H3 decreases at later times postinfection, independently of either viral DNA replication or transcription. We demonstrate that an immediate-early protein, infected cell protein 0 (ICP0), is required for both a reduction in the proportion of HSV-1 DNA associating with histone H3 and an increase in histone acetylation. This study provides evidence that ICP0 directly alters the chromatin structure of the HSV-1 genome during lytic infection, and this system will serve as a useful model for the reduction of histone load in higher eukaryotes.
Eukaryotic DNA is packaged into a protein-DNA complex known as chromatin. The basic structure of chromatin is the nucleosome, which consists of core histone proteins (H2A, H2B, H3, and H4) around which 147 bp of DNA is wrapped. Modification of the chromatin structure to allow access to proteins involved in DNA replication, recombination and repair, and gene expression is a key mechanism utilized by the cell to regulate these processes. Chromatin structure can be modified by covalent modification of histones and DNA, as well as noncovalent chromatin remodeling. Chromatin remodeling, carried out by ATPase-dependent chromatin remodeling complexes, results in the sliding of nucleosomes along the DNA, unwinding of nucleosomes, and/or complete removal of nucleosomes (65). Histone acetylation is one of the best-characterized covalent histone modifications and is a hallmark of transcriptionally active chromatin. Histone acetylation is thought to result in relaxation of the basic chromatin structure through both increased charge repulsion (20) and by serving as a binding site for chromatin-remodeling complexes (1, 29, 38).
Like cellular DNA, the genomes of DNA viruses that replicate within the nucleus also associate with chromatin, albeit to varied degrees (48). The genome of herpes simplex virus 1 (HSV-1) has been shown to associate with histones during both lytic infection of epithelial cells and latent infection of neurons (13, 32, 39). During latent infection, the viral genome is packaged into nucleosomes and forms a classical laddering pattern following digestion with micrococcal nuclease (13). Consistent with the silencing of lytic genes during a latent infection, lytic gene promoters show markers of heterochromatin, such as H3K9me2, and low levels of histone acetylation (43, 78). In contrast, during lytic infection, regular repeating nucleosome arrays of HSV-1 DNA have not been detected (39, 45, 47). However, at least a portion of the viral DNA associates with histones (32, 39), although one study showed that histone association with immediate-early (IE) gene promoters was low in the presence of functional VP16 (32). A recent study by Oh and Fraser reported that the HSV-1 genome associated with histones at early times postinfection and the proportion of viral DNA associating with histones decreased at late times in the infectious cycle. The authors attributed this decrease to an increase in newly synthesized genomes that were free of histones (55).
The histones present on viral DNA during lytic infection show modifications associated with active gene expression, namely, acetylation of histone H3 and di- and trimethylation of histone H3 lysine 4 (32, 36, 39). Given that regulation of chromatin structure has important consequences for DNA processes, it is likely that both viral and cellular gene products manipulate the HSV-1 chromatin structure to allow gene expression and DNA replication to take place. Consistent with this, proteins involved in both chromatin remodeling and histone modifications are recruited into viral replication compartments (75), which are the sites of late gene transcription and DNA replication (3, 9, 10, 41, 57, 62).
The activation domain of the HSV-1 VP16 virion transactivator has been shown to interact with ATPase-dependent chromatin remodeling proteins and histone acetyltransferases (54, 76). Deletion of the VP16 activation domain results in a decrease in recruitment of ATPase-dependent chromatin remodeling proteins and histone acetyltransferases to IE promoters and an accompanying increase in histone occupancy and decrease in histone acetylation (32). An additional candidate for modifying chromatin on the HSV-1 genome is the immediate-early protein ICP0. ICP0 is able to stimulate the expression of all three classes of viral genes in infected cells (4, 7) and cotransfected genes (14, 21, 53, 56, 61), although it does not bind DNA directly (17). The ability of ICP0 null viruses to replicate in tissue culture cells is dependent upon both the cell type and multiplicity of infection (MOI) (64, 72). ICP0 null viruses show the highest degree of attenuation within primary embryonic or fetal fibroblasts, in which a low multiplicity of infection results in a quiescent infection (15). Infection of a number of other cell types, such as HeLa cells, at a low multiplicity of infection results in attenuation of viral infection, in which gene expression is reduced compared to that in wild-type (WT) virus-infected cells (4, 7). The effects of ICP0 gene deletions on viral gene expression can be overcome in part by treatment with inhibitors of histone deacetylases (HDACs) (34, 59, 60), suggesting that ICP0 may act to increase levels of histone acetylation.
ICP0 has been found to interact with the class II HDACs and reduce their activity in vitro (49). ICP0 also interacts with the RE1 silencing corepressor (REST/CoREST)-HDAC1 complex and causes dissociation of HDAC1 from the complex. Disassociation of HDAC1 from the REST/CoREST complex is able to increase, at least in part, viral yields in the absence of ICP0 (24). Despite the observation that ICP0 interacts with HDACs, there is no direct evidence that ICP0 affects the chromatin structure on the HSV-1 genome during a lytic infection. We therefore employed chromatin immunoprecipitation (ChIP) to determine the effects of ICP0 on the chromatin structure of HSV-1 during lytic infection.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells, Vero cells, and U2OS cells were obtained from the American Type Culture Collection (Manassas, VA). Cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco-BRL) supplemented with 5% heat-inactivated fetal calf serum, 5% heat-inactivated bovine calf serum (BCS), 2 mM l-glutamine, streptomycin, and penicillin.
The WT strain of HSV-1 (KOS) used in this study was grown and titrated as described previously (42, 44). The ICP4− virus (n12) was provided by P. Schaffer (University of Arizona) (12) and grown and titrated on E11 cells (66). The ICP0− (7134) and 7134R rescued viruses, provided by P. Schaffer, were grown and titrated on U2OS cells (5). The DNA polymerase null, HP66, and HP66R rescued viruses, provided by D. Coen (Harvard Medical School), were grown and titrated as described previously (50).
Virus infections.
HeLa cells were plated into 100-mm dishes 24 h prior to infection to obtain 95% confluence at the time of infection. Virus was diluted in cold phosphate-buffered saline (PBS) containing 0.1% glucose and 1% heat-inactivated BCS in the presence or absence of 200 μg/ml sodium phosphonoacetate (PAA). After 1 h of adsorption at 37°C, cells were washed twice with PBS containing 0.1% glucose and 1% heat-inactivated BCS, twice with low-pH wash buffer (40 mM citric acid, 10 mM KCl, 135 mM NaCl, pH 3) (33), and twice with DMEM containing 1% heat-inactivated BCS. Cells were overlaid with DMEM containing 1% heat-inactivated BCS and incubated at 37°C.
Chromatin immunoprecipitation.
ChIP assays were carried out as described previously (78) with a few modifications. At various times postinfection, cells were incubated with 1% formaldehyde for 10 min to cross-link the chromatin. Cells were washed twice with PBS and scraped into PBS containing Complete protease inhibitor tablets (Roche Diagnostics). Cells were resuspended in sodium dodecyl sulfate (SDS) lysis buffer (1% SDS-10 mM EDTA-50 mM Tris, pH 8.1) containing protease inhibitors and incubated on ice for 20 min. The samples were sonicated in 20-s pulses for a total of 4 min, using a Heat Systems XL sonicator, to yield DNA fragments of between 200 and 500 bp in length. The samples were clarified by centrifugation at 13,000 × g at 4°C for 10 min. The supernatant was diluted 10-fold in phosphate-buffered radioimmunoprecipitation assay buffer (0.1% SDS, 1% sodium deoxycholate, 150 mM NaCl, 10 mM Na2PO4, 2 mM EDTA, 1% Nonidet P-40) and precleared for 1 h at 4°C. At this point, 1% of the total volume was removed and reserved as the input. Immunoprecipitation was carried out at 4°C overnight with 5 μg rabbit immunoglobulin G (IgG; Millipore) as the negative control or 1.5 μg anti-histone H3 IgG (Abcam) or 5 μl anti-acetyl histone H3 lysine 9/18 IgG (Millipore).
Immunocomplexes were collected by incubation with a salmon sperm DNA-protein A-agarose slurry for 1 h at 4°C with rotation. Beads were washed for 4 min at 4°C with rotation, two times with low-salt wash buffer (150 mM NaCl, 20 mM Tris·HCl, pH 8.1, 2 mM EDTA, 1% Triton X-100, and 0.1% SDS), one time with high-salt wash buffer (500 mM NaCl, 20 mM Tris·HCl, pH 8.1, 2 mM EDTA, 1% Triton X-100, and 0.1% SDS), one time with lithium chloride wash buffer (0.25 M LiCl, 1% NP-40, 1% sodium deoxycholate, 1 mM EDTA, and 10 mM Tris-HCl, pH 8.1) and two times with Tris-EDTA buffer (10 mM Tris-HCl, pH 8, 1 mM EDTA). The DNA-protein complexes were eluted by adding 200 μl of elution buffer (1% SDS, 0.1 M NaHCO3) preheated to 65°C, rotating for 10 min at room temperature, incubating for 10 min at 65°C, and rotating for 10 min at room temperature. NaCl was added to a final concentration of 0.2 M to both the eluates and the inputs, and the samples were incubated at 65°C for 5 h in the presence of 1 μg RNase A (Ambion). The samples were then digested with proteinase K, and the DNA was purified by using a QIAquick PCR purification kit (Qiagen).
Real-time PCR.
Real-time PCR was performed using the power Sybr green PCR master mix and a Prism 7300 sequence detection system (Applied Biosystems) according to the manufacturer's instructions. The final reaction volume was 25 μl, containing 2.5 μl DNA and 100 nM of each primer. The primers used in this study are shown in Table 1. The specificity of each primer pair was determined by running a dissociation curve of the PCR products. All DNA samples were run in duplicate, and relative copy numbers were determined by comparison with a standard curve generated by 10-fold dilutions of sonicated DNA from HSV-1-infected HeLa cells. The fraction of viral DNA immunoprecipitated compared to the input sample was normalized to the fraction of cellular DNA (glyceraldehyde 3-phosphate dehydrogenase [GAPDH] precipitated in the same reaction.
TABLE 1.
Primers used for real-time PCR analysis
| DNA targeta | Sequence | Locatione |
|---|---|---|
| ICP4 USa | 5′-CGCATGGCATCTCATTACCG-3′ (forward) | −293 to −256 |
| 5′-TAGCATGCGGAACGGAAGC-3′ (reverse) | ||
| ICP4 SSb | 5′-GCGCTCCGTGTGGACGAT-3′ (forward) | −23 to +30 |
| 5′-CGGCCCCTGGGACTATATGA-3′ (reverse) | ||
| ICP4 DS | 5′-GCCCGGGCGCTGCTTGTTCTCC-3′ (forward) | +265 to +332 |
| 5′-CGTCCGCCGTCGCAGCCGTATC-3′ (reverse) | ||
| ICP8 SSb | 5′-CCACGCCCACCGGCTGATGAC-3′ (forward) | −100 to +28 |
| 5′-TGCTTACGGTCAGGTGCTCCG-3′ (reverse) | ||
| ICP8 DSc | 5′-GAGACCGGGGTTGGGGAATGAATC-3′ (forward) | +95 to +144 |
| 5′-CCCCGGGGGTTGTCTGTGAAGG-3′ (reverse) | ||
| UL44 USb | 5′-CCGACCGCCCGCCCGTTGAC-3′ (forward) | −225 to −125 |
| 5′-ACGCTTCGGGGCCTCTTCTTCTCC-3′ (reverse) | ||
| UL44 SSb | 5′-GGGTATAAATTCCGGAAGGGG-3′ (forward) | −30 to +130 |
| 5′-CTGCGAGGGATCGGCTAGCG-3′ (reverse) | ||
| GAPDHd | 5′-TTCGACAGTCAGCCGCATCTTCTT-3′ (forward) | +44 to +153 |
| 5′-CAGGCGCCCAATACGACCAAATC-3′ (reverse) |
US, upstream; DS, downstream; SS, start site.
Primer sequences were taken from the HSV-1 sequence available in GenBank (accession no. NC_001806).
Primer sequences were taken from the KOS sequence kindly provided by Robert Colgrove.
Primer sequences were taken from the GAPDH sequence available in GenBank (accession no. NC_002046).
Relative to the transcriptional start site.
RESULTS
HSV-1 DNA associates with histone H3 to levels comparable to cellular DNA early in infection.
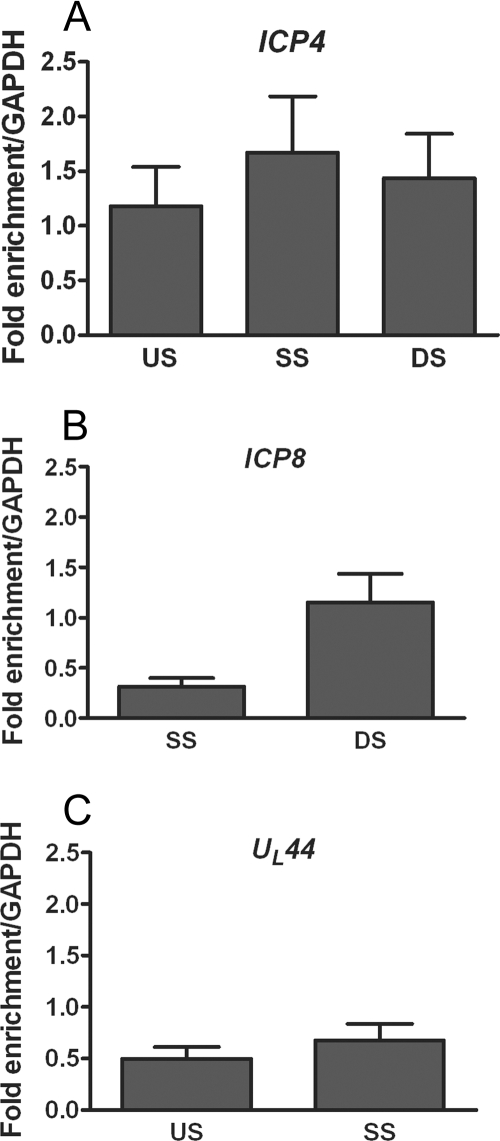
Because replication of ICP0 mutant viruses is attenuated at low MOIs, we first analyzed the relative levels of histone association with the regions of the viral genome following infection at a low MOI. To this end, we carried out ChIP on extracts from HeLa cells infected with WT HSV-1 at 0.1 PFU/cell. After 1 h, cells were subjected to a low-pH buffer wash to inactivate extracellular virus and synchronize infection. At 3 h postinfection (hpi), cell lysates were prepared and ChIP was carried out using an antibody specific for histone H3. The fraction of viral DNA immunoprecipitated was quantified by real-time PCR analysis using the standard curve method and normalized to the fraction of a cellular DNA sequence (GAPDH) immunoprecipitated in the same reaction. Histone H3 associated with all three classes of viral genes, IE (ICP4), E (ICP8), and L (UL44) (Fig. 1). In all regions tested around the ICP4 transcriptional start site, in the promoter, over the transcriptional start site, and downstream of the transcriptional start site in the 5′ untranslated region (UTR), the proportions of viral DNA associated with histone H3 were equivalent to those detected on the GAPDH promoter (Fig. 1A). Levels equivalent to those detected on GAPDH could also be detected within the 5′ UTR downstream of the ICP8 start site (Fig. 1B); although lower levels were detected on the ICP8 start site, the fraction of DNA immunoprecipitated was not significantly different from the fraction of GAPDH DNA immunoprecipitated (P > 0.05). Histones were also detected around the UL44 transcriptional start site but at lower levels than ICP4 and ICP8 (Fig. 1C). These results indicate that multiple regions of the HSV-1 genome associate with histone H3 at levels equivalent to those found on a cellular gene.
FIG. 1.
ChIP analysis of the fraction of HSV-1 DNA associated with histone H3. Total cell extracts were prepared at 3 hpi from HeLa cells infected with HSV-1 at an MOI of 0.1 PFU/cell. ChIP was carried out with an antibody specific to the C terminus of histone H3. The fraction of DNA immunoprecipitated compared to the input value was determined by real-time PCR, with the fraction immunoprecipitated by a nonspecific antibody subtracted, and is expressed as the fold enrichment over the fraction of GAPDH DNA immunoprecipitated. Primers were specific for regions of ICP4 (A), ICP8 (B), and UL44 (C) (see Table 1 for primer locations and sequences). Abbreviations: US, upstream of the transcriptional start site; SS, start site; DS, downstream of the transcriptional start site. The means and standard errors of the means of at least five independent experiments are shown.
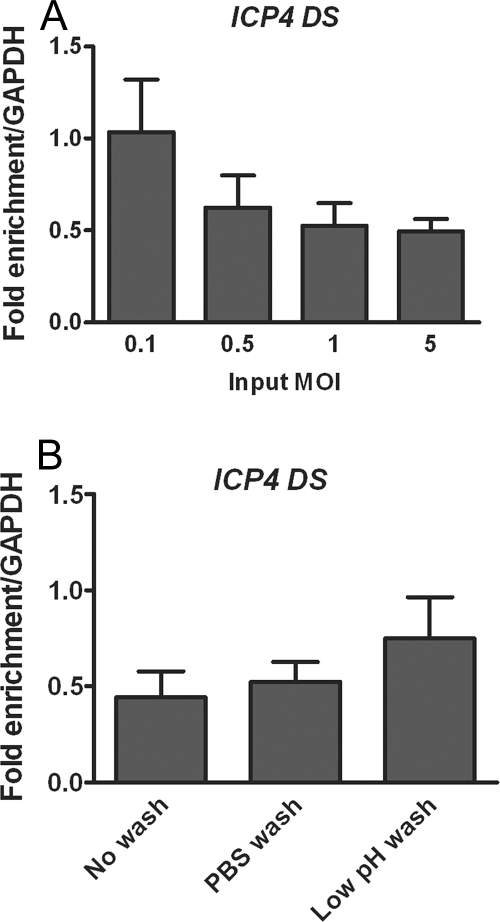
The detection of histone around the ICP4 transcriptional start site at levels equivalent to those of a cellular gene was in contrast with previous observations, in which levels of histone H3 on the ICP4 promoter were found to be only slightly enriched over a no-antibody control and present at much lower levels than on a cellular gene following a high-MOI infection (32). Hence, we examined the effect of MOI on the proportion of viral DNA associated with histone H3. To control for an increasing amount of viral DNA, the amount of viral DNA immunoprecipitated with nonspecific antibody was subtracted from the amount of DNA immunoprecipitated with histone H3 prior to normalization to GAPDH. The highest proportion of viral DNA associated with histone H3 was observed at the lowest MOI tested, 0.1 PFU/cell (Fig. 2A).
FIG. 2.
Effects of MOI and inactivation of nonadsorbed virus on the proportion of viral DNA associated with histone H3. ChIP was carried out at 3 hpi with an antibody for histone H3, and real-time time PCR was undertaken using primers for the region downstream of the ICP4 transcriptional start site and analyzed as for Fig. 1. (A) HeLa cells were infected at MOIs between 0.1 and 5 PFU/cell. (B) HeLa cells were infected at an MOI of 0.1 PFU/cell. After a 1-h adsorption period, the medium was replaced without washing the cells, or the cells were washed with PBS alone or washed with PBS followed by a low-pH wash buffer. The means and standard errors of the means of at least four independent experiments are shown.
To test the effect of synchronizing infection, we conducted different types of washes of infected cells, including PBS, which could remove loosely bound virus, or low-pH buffer, which inactivates the infectivity of virions at the cell surface (33). The addition of the acid wash resulted in an increase in the relative fraction of viral DNA associated with histone H3 at 3 hpi, compared to both no washing at all and washing with only PBS (Fig. 2B). We interpreted this result to mean that low-pH buffer wash reduced the levels of packaged viral DNA present at the cell surface. Given that HSV-1 DNA within virions has been found to be free of histones (22, 55, 58), we conclude that it is important to reduce the amount of encapsidated DNA present prior to carrying out the ChIP assay to establish a more accurate value for the amount of viral DNA within the cell that is associated with histone H3.
The temporal decrease in the association of viral DNA with histone H3 occurs independently of viral DNA replication.
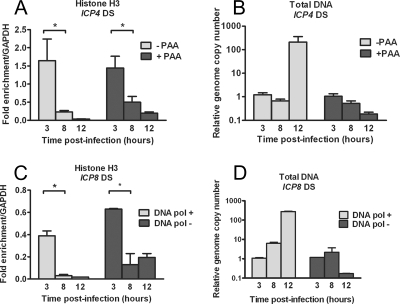
The proportion of viral DNA associated with histone H3 was shown to decrease at late times following infection at a high MOI (55). To determine if the association of viral DNA with histones also decreased in a low-MOI infection, we carried out ChIP assays at various times following infection and analyzed the proportion of the ICP4 gene that was associated with histone H3. ICP4 gene sequences were precipitated with histone H3-specific antibody at approximately the same efficiency as GAPDH at 3 hpi, but the association of viral DNA with histone H3 decreased between 3 and 8 hpi and was further decreased at 12 hpi (Fig. 3A).
FIG. 3.
Kinetics of histone H3 association with the HSV genome and effects of viral DNA replication. HeLa cells were infected at an MOI of 0.1 PFU/cell in the presence or absence of the DNA polymerase inhibitor PAA (A and B) or with either a DNA Pol− virus or the rescued DNA Pol+ virus (C and D). Cell lysates were prepared at 3, 8, and 12 h postinfection. (A and C) ChIP was carried out using anti-histone H3 antibody. The fraction of ICP4 downstream (DS) (A) or ICP8 DS (C) DNA immunoprecipitated as determined by real-time PCR was normalized to the fraction of GAPDH DNA immunoprecipitated. The HSV genome copy number determined by real-time PCR was normalized to the GAPDH copy number and then normalized to the results for the untreated 3-h time point (B and D). The means and standard errors of the means of at least three independent experiments are shown. A significant decrease (*) between the mean fraction of viral DNA immunoprecipitated between 3 and 8 hpi was determined using Student's t test (P < 0.05).
The decreased level of histone H3 could have been due to histones being removed during viral DNA replication, which would be consistent with the observation by Oh and Fraser that newly synthesized viral genomes are free of histones (55). However, DNA accumulation was not seen until after 8 hpi at this low MOI (Fig. 3B), indicating that the decrease in histone association occurred prior to DNA replication. To determine if the decrease in histone association occurred independently of DNA replication, ChIP was carried out with lysates from cells infected in the presence of PAA, an inhibitor of viral DNA replication. Analysis of the genome copy number following infection of the PAA-treated cells confirmed that DNA replication was inhibited (Fig. 3B). In PAA-treated cells, the levels of histone H3 on the ICP4 gene decreased with similar kinetics as in the absence of PAA (Fig. 3A), indicating that a decrease in association of viral DNA with histone H3 occurred independently of viral DNA replication. Similar results were obtained for the ICP8 gene (data not shown).
To rule out any nonspecific effects of PAA on the association of histones with viral DNA, we examined the association of histone H3 with the ICP8 gene in cells infected with a replication-defective, DNA polymerase null virus, HP66 (50), in parallel with the rescued virus, HP66R. Analysis of the genome copy numbers confirmed that viral DNA replication did not occur following infection with HP66 (Fig. 3D). Following infection with both the HP66 DNA polymerase null and the HP66R rescued virus, ICP8 DNA associated with histone H3 to high levels at 3 hpi, but the levels decreased by 8 hpi (Fig. 3C). These results confirmed that viral DNA replication was not a prerequisite for the reduction of histone association with HSV-1 DNA between 3 and 8 hpi.
Total amounts of HSV-1 DNA associated with histone H3 increase as a consequence of viral DNA replication.
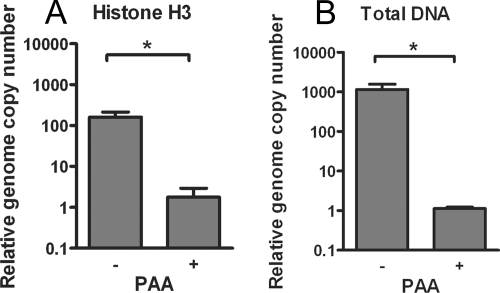
Although levels of histone H3 on viral DNA decreased prior to or in the absence of viral DNA replication, this did not rule out the possibility that histones were also removed from viral DNA during DNA replication, as reported previously (55). At 12 h postinfection, an increase in the proportion of viral DNA associated with histone H3 could be seen in PAA-treated cells compared to the untreated cells (0.033% in the PAA-treated cells compared to 0.0069% in the untreated cells) (Fig. 3A). However, due to the large differences in viral genome copy numbers (approximately 1,000-fold more DNA in the untreated cells) (Fig. 4B), it was difficult to determine whether the slightly decreased proportion of DNA associating with histone H3 in the untreated cells occurred because of an increase in viral DNA load. Therefore, we examined the total amount of viral DNA sequences immunoprecipitated with the H3 antibody normalized to the total amount of GAPDH sequence immunoprecipitated at the time of DNA replication. If histones were removed during replication, the total amount of DNA associated with histone H3 would decrease. Surprisingly, instead of seeing a decrease in the amount of DNA associated with histone H3 following DNA replication, there was a 90-fold increase in the amount of viral DNA associated with histone H3 in the untreated cells in comparison to the PAA-treated cells (Fig. 4A). Taken together, these data suggested that following DNA replication there were increases in the absolute amounts of both histone H3-associated and histone-free viral DNA.
FIG. 4.
Effects of DNA replication on the total amount of viral DNA associated with histone H3. HeLa cells were infected at an MOI of 0.1 PFU/cell in the presence or absence of the DNA polymerase inhibitor PAA. Cell lysates were prepared at 12 hpi. (A) ChIP assays were carried out using anti-histone H3 antibody. The copy number of ICP4 downstream DNA associated with histone H3 was determined by real-time PCR and normalized to the copy number of GAPDH associated with histone H3 and then normalized to the results for PAA-treated cells. (B) Total HSV genome copy number determined by real-time PCR using primers for the ICP4 gene was normalized to the GAPDH copy number and then normalized to results with the PAA-treated cells. The means and standard errors of the means are shown. Samples with mean values that vary significantly (P < 0.05, Student's t test) are indicated (*).
The temporal decrease in association of an early gene promoter with histone H3 occurs without active transcription.
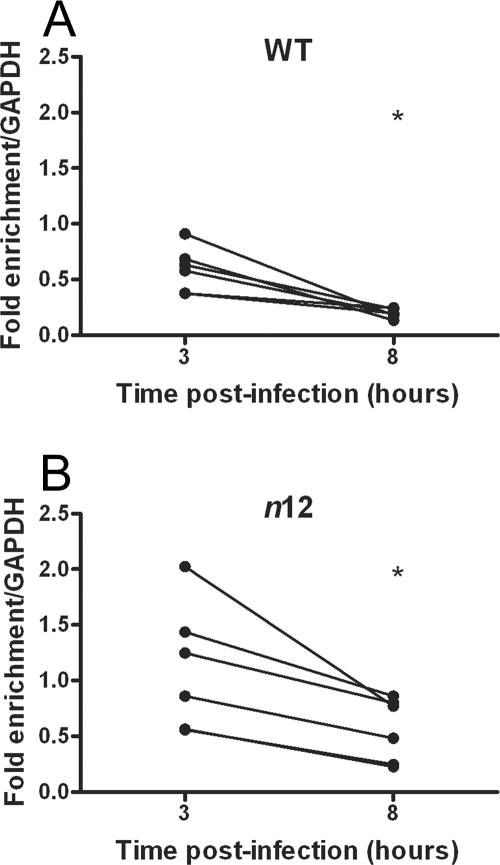
To determine if the decrease in histone H3 association with HSV-1 DNA between 3 and 8 hpi occurred as a consequence of transcription, we carried out ChIP with extracts from cells infected with a virus mutated in the ICP4 gene. ICP4 is an immediate-early protein required for transcription of the early (E) and late (L) genes (11). Work from Sampath and DeLuca has demonstrated that following infection with an ICP4 truncation mutant virus, n12, recruitment of TATA-binding protein and RNA polymerase II (RNA Pol II) to both early and late promoters was greatly reduced (67). Hence, we investigated whether infection with the n12 virus also resulted in the decreased histone association observed for wild-type virus between 3 and 8 hpi. Infection with the n12 virus resulted in slightly increased levels of histone levels at 3 hpi (Fig. 5B) compared to WT virus (Fig. 5A), although this difference was not statistically significant (P = 0.07). Given that at a lower MOI we detected higher levels of histones on the viral genome, it was possible that lower genomic levels may have accounted for the slightly increased histone levels in the absence of ICP4. However, cells infected with the n12 virus had slightly higher viral DNA loads than those infected with KOS at 3 hpi (an average increase of 1.9-fold over KOS). Between 3 and 8 hpi there was a statistically significant decrease in the level of histone H3 on the ICP8 gene following infection with either n12 or WT virus (n12, P = 0.013; WT, P = 0.001), indicating that a decrease in histone association at this promoter did not occur as a result of initiation complex binding and ongoing transcription.
FIG. 5.
Kinetics of histone H3 association with the ICP8 gene following infection with an ICP4 nonsense mutant virus. HeLa cells were infected at an MOI of 0.1 PFU/cell with either WT (A) or ICP4− (B) virus. Cell lysates were prepared at 3 and 8 hpi. The fraction of ICP8 downstream DNA immunoprecipitated, as determined by real-time PCR, was normalized to the fraction of GAPDH DNA immunoprecipitated. Lines connect values for individual experiments. Samples with mean values that varied significantly (P < 0.05, paired Student's t test) are indicated (*).
Role for ICP0 in the decreased association of histone H3 with the HSV-1 genome.
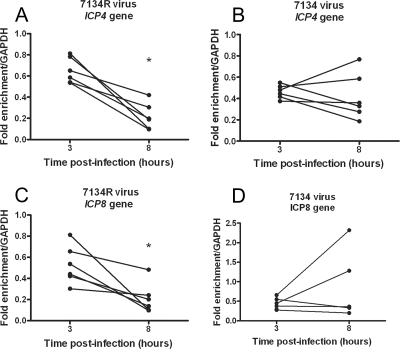
Because the association of E genes with histone H3 decreased between 3 and 8 hpi independently of viral transcription and DNA replication, it was possible that newly expressed viral proteins functioned to reduce the levels of histone H3 on the viral genome. Because of the evidence summarized above, the IE protein, ICP0, is a key candidate for remodeling the chromatin structure on the HSV-1 genome during lytic infection. Hence, we carried out ChIP analysis on HeLa cells following infection with an ICP0 null virus, 7134 (5), in parallel with the rescued virus, 7134R. In 7134R virus-infected cells, the association of histone H3 with both the ICP4 and ICP8 genes decreased significantly (P < 0.05) between 3 and 8 hpi (Fig. 6). However, in 7134 virus-infected cells there were no significant decreases in the levels of histone H3 on the ICP4 or ICP8 genes between 3 and 8 hpi. In addition, on the ICP8 gene there was a small increase in the levels of histone H3 between 3 and 8 hpi, although this increase was not statistically significant (P = 0.29). These results suggested that ICP0 plays a role in the removal of histone H3 from the viral genome between 3 and 8 hpi. The increased histone levels in the absence of ICP0 could not be attributed to a decreased viral genomic DNA load because infection with 7134 resulted in a slightly increased viral DNA load (2.75-fold at 3 hpi) versus infection with 7134R.
FIG. 6.
Kinetics of histone H3 association with the ICP4 and ICP8 genes following infection with a ICP0− virus. HeLa cells were infected at an MOI of 0.1 PFU/cell with either an ICP0− virus (7134) (B and D) or rescued virus (7134R) (A and C). Cell lysates were prepared at 3 and 8 hpi. The fraction of ICP4 downstream (A and B) or ICP8 downstream (C and D) DNA immunoprecipitated determined by real-time PCR was normalized to the fraction of GAPDH DNA immunoprecipitated. Lines connect values for individual experiments. Samples with mean values that varied significantly (P < 0.05, paired Student's t test) are indicated (*).
ICP0 promotes histone H3 acetylation on the ICP8 gene.
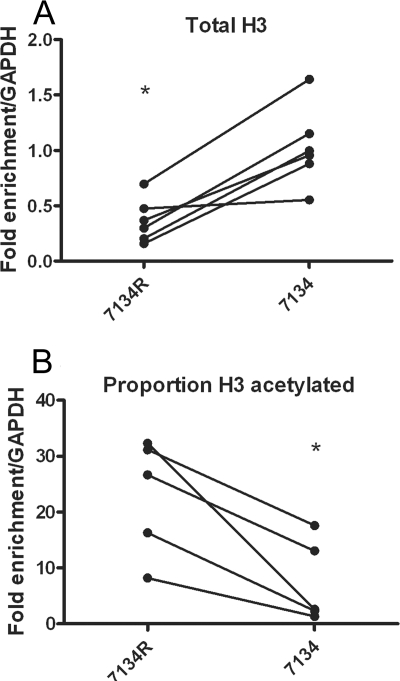
To determine whether ICP0, in addition to promoting the decreased association with total histone H3, was also able to promote the acetylation of histone H3, we conducted ChIP assays using antibodies specific for acetylated histone H3 K9/18 (acH3 K9/18) with extracts from cells infected with 7134 ICP0− virus or 7134R ICP0+ virus at 6 hpi. Because we saw a significantly increased association of histone H3 with the ICP8 gene following infection with 7134 compared to 7134R (Fig. 7A), we expressed the results of the acH3 K9/18 ChIP assay as the proportion of histone H3 that was acetylated. In cells infected with 7134R ICP0+ virus, the proportion of histone H3 that was acetylated was significantly greater than that in cells infected with 7134 ICP0− virus (Fig. 7B). Therefore, ICP0 promotes the acetylation of histone H3 on the ICP8 gene.
FIG. 7.
ChIP for total histone H3 and acetylated histone H3 on the ICP8 gene in the presence and absence of ICP0. HeLa cells were infected an MOI of 0.1 PFU/cell with either 7134 ICP0− virus or 7134R rescued virus (ICP0+). Cell lysates were prepared at 6 hpi, and ChIP was carried out using anti-histone H3 (A and B) and anti-acH3 (K9/18) (B) antibodies. The proportion of acetylated histone H3 associating with viral DNA was determined as a fraction of DNA associating with acH3 K9/14, normalized to the fraction associating with total histone H3. Lines connect values for individual experiments. Samples with mean values that varied significantly (P < 0.05, paired Student's t test) are indicated (*).
DISCUSSION
In this study, ChIP analysis in combination with real-time PCR was used to examine the relative levels of histone H3 on the HSV-1 genome during lytic infection. By normalizing to the histone H3 levels present on a cellular gene, we were able to determine that levels of histone H3 on the HSV-1 genome are equivalent to those on a cellular gene at early times postinfection. This indicates that the chromatin conformation of the viral genome most likely plays a key role in the regulation of gene expression and DNA replication during lytic infection. Consequently, we investigated how the levels of histone H3 change through the course of infection. We found that the proportion of DNA associated with histone H3 decreases as infection progresses, a process that occurs independently of transcription and DNA replication, but is dependent upon ICP0 expression. Later in infection, there is a replication-dependent increase in the amount of viral DNA associated with histone H3, which suggests that histones become loaded onto progeny viral DNA.
HSV-1 DNA associates to higher levels with histone H3 early in infection at a low MOI.
In this study, we extended previous observations that at least a portion of viral DNA associates with histone H3 during lytic infection (32, 39) to conclude that physiologically relevant levels of histone H3 associate with viral DNA early in infection. The association of viral DNA with histones following infection appears to be similar to that seen for transiently transfected DNA, in which the DNA associates with histones but fails to form a regularly spaced nucleosomal structure (31). The lack of a regularly spaced nucleosomal structure on transfected DNA is possibly due to the lower representation of the linker histone, histone H1. It is not known whether histone H1 is found on viral DNA at early time points, but during later time points in infection it has been shown to be excluded from replication compartments (71).
We showed that the level of histones on viral DNA is dependent upon the multiplicity of infection, with an increased proportion of viral DNA associated with histone H3 at a low MOI. Possible reasons for the dependence on MOI are that the cell is unable to assemble histones efficiently onto higher quantities of incoming viral DNA, a limiting pool of available histone H3 for assembly onto increasing amounts of viral DNA, and/or the presence of increased quantities of viral proteins able to reduce the histone association with the viral genome. A study by Yager and Bachenheimer (81) showed that both histone H3 mRNA levels and H3 synthesis rapidly decline following viral infection, which may limit the pool of histones available for loading onto viral DNA at higher MOIs.
Two viral tegument proteins which may limit the histone association with the viral genome are VP16 and VP22. VP16 has been found to interact with members of the SWI/SNF family of ATP-dependent chromatin remodeling proteins (28, 51, 54). In addition, deletion of the VP16 activation domain was found to result in increased levels of histone H3 on IE promoters (32). VP22 has been found to interact with the chromatin remodeling protein TAF-1 (77) and prevent TAF-1-mediated nucleosome deposition onto naked DNA in vitro.
A role for ICP0 in reducing histone H3 occupancy.
Following initial loading of histones onto the viral DNA, we observed a decrease in association of histone H3 with viral DNA. The reduction in histone load is analogous to the loss of histones from promoters following activation of the HS82 genes and the PHO5 genes in Saccharomyces cerevisiae (63, 82). Little is known about eviction of histones from the promoters of higher eukaryotic promoters. However, there is evidence that nucleosome disruption precedes transcriptional activation in the HIV long terminal repeat (reviewed in reference 30) and the human T-cell leukemia virus 1 promoter (46).
Interestingly, histone loss following activation of the HS82, PHO5, and human T-cell leukemia virus 1 promoters occur independently of transcription (46, 68, 69, 82). The loss of histones from an early promoter in the absence of ICP4, and thus the absence of transcription, demonstrated that transcription is also not required for the reduction in histone association on the ICP8 promoter. However, histone loss is dependent upon ICP0. A role for ICP0 in reducing histone association to allow access to the DNA for transcription factor binding is consistent with its role in promoting the expression of genes in the context of both viral infection and transfection (4, 7). Furthermore, the reactivation of viral genomes from at least a subset of latently infected neurons in vivo and quiescently infected cells in vitro can be promoted by ICP0 (25, 26, 83). Given that high levels of histones are present on the HSV-1 genome in latently infected neurons (reviewed in reference 40), it is possible that a function of ICP0 in the reactivation from latency involves the reduction in histone association with the viral genome. A recent study by Coleman et al. found no decrease in histone association with the viral genome following ICP0-mediated derepression in vitro (8). However, this does not rule out the possibility that ICP0 mediates removal of histones from latently infected neuronal cells.
The role of ICP0 in promoting histone acetylation is consistent with previous observations that ICP0 is able to cause the dissociation of HDACs from the REST/CoREST complex (23) and that ICP0 is able to promote histone acetylation during derepression of the quiescent viral genome in vitro (8). Furthermore, previous observations that HDAC inhibition can increase viral gene expression in the absence of ICP0 suggest that ICP0 functions to inhibit HDACs which would otherwise try to silence viral gene expression. However, in some studies inhibition of HDAC administered at the time of infection had no effect on viral replication or gene expression (18, 60). HDAC inhibition has a broad spectrum of effects, including a decrease in the levels and a redistribution of the heterochromatin protein HP1, as well as decreased histone H3 acetylation and lysine 4 methylation at the nuclear periphery (2), which is an early site of HSV-1 genome localization (16, 70). Hence, it is difficult to determine whether the effects of HDAC inhibition are due to direct effects on histone acetylation or indirect effects due to remodeling of the cell nucleus. Further studies are therefore required to determine the effects of HDAC inhibition on the HSV-1 chromatin structure.
A role for ICP0 in reducing total histone load has not been proposed previously. It is possible that through promoting histone acetylation or preventing deacetylation, ICP0 indirectly reduces histone association. Indeed, histone acetylation results in a change in the net charge on the nucleosome, which could loosen histone-DNA interactions (20). Furthermore, acetylated histones have been shown to be easier to displace from reconstituted nucleosomes in vitro (6, 27, 37) in addition to serving as a binding site for chromatin remodeling proteins (1, 29, 38). Conversely, ICP0 perhaps functions directly to reduce histone levels on viral DNA, and the finding that a higher proportion of histones are acetylated is instead a consequence of the reduced histone occupancy or selective removal of nonacetylated histones.
Components of PML bodies, such as PML, Sp100, and hDaxx, play a role in reducing herpesvirus gene expression (18, 19, 79). One protein found within PML bodies, hDaxx, interacts with the core histones and chromatin-remodeling proteins (35, 80). Although hDaxx does not appear to be degraded by ICP0, hDaxx localization to nucleoprotein complexes is inhibited in the presence of ICP0 (19). Hence, a function of PML bodies may be to prevent chromatin remodeling on the viral genome. However, to date there is no direct evidence for a link between PML component repression of HSV-1 gene expression and chromatin structure. It will be of interest to define the roles of cellular proteins and the dependence of histone acetylation on the reduction of histone levels on the HSV-1 genome to determine not only how ICP0 functions to regulate viral gene expression but also the mechanisms underlying nucleosome eviction in higher eukaryotes.
The association of progeny DNA with histone H3.
In a recent study Oh and Fraser reported that viral progeny DNA does not associate with histones (55). In contrast, we observed that the total amount of viral DNA associated with histones increased with viral DNA replication. The different observations for the role of DNA replication could possibly be due to the differing cell types used: Oh and Fraser carried out experiments in a neuroblastoma cell line, whereas in this study HeLa cells were used. A further possibility is that the different observations resulted from the different multiplicities of infection used: Oh and Fraser infected cells at an MOI of 5 PFU/cell, whereas in this study cells were infected at an MOI of 0.1 PFU/cell. Given that the proportion of viral DNA associating with histone H3 at early times was found to decrease at higher MOIs, it is also possible that reduced amounts of histones are loaded onto replicating DNA following high-MOI infections. In addition, the study by Oh and Fraser examined the proportion of progeny DNA associating with histone H3 by performing consecutive ChIPs for histone H3 and bromodeoxyuridine. However, it is possible that the incorporation of bromodeoxyuridine into replicating DNA altered the chromatin structure, as observed previously (52, 73).
It is important to examine the total amount of DNA associating with histones and not just the proportion, which may give misleading results due to increased viral genome copy number. Our data suggest that as DNA replication takes place there is an increase in both histone-free and histone-associated viral DNA. The nature of the two populations of viral DNA following DNA replication possibly reflects the fate of replicated DNA. Given that HSV-1 DNA packaged within capsids is free from histones (22, 55, 58), the histone-free portion of DNA may represent either packaged DNA or DNA destined for packaging. It is tempting to speculate that the histone-associated DNA represents DNA serving as a template for late viral gene expression and that a change in the chromatin structure of viral DNA following DNA replication is involved in the regulation of late gene expression. Therefore, it is possible that a temporal change in histone association throughout the course of infection is a mechanism by which the virus regulates its gene expression throughout the lytic infectious cycle.
Acknowledgments
This work was supported by National Institutes of Health grants PO1 N535133 and AI46006.
We thank Priscilla Schaffer for providing the ICP0 and ICP4 mutant viruses, Don Coen for the DNA polymerase mutant viruses, and Kevin Bryant and Megan Horn for comments on the manuscript.
Footnotes
Published ahead of print on 8 October 2008.
REFERENCES
- 1.Agalioti, T., G. Chen, and D. Thanos. 2002. Deciphering the transcriptional histone acetylation code for a human gene. Cell 111381-392. [DOI] [PubMed] [Google Scholar]
- 2.Bartova, E., J. Pachernik, A. Harnicarova, A. Kovarik, M. Kovarikova, J. Hofmanova, M. Skalnikova, M. Kozubek, and S. Kozubek. 2005. Nuclear levels and patterns of histone H3 modification and HP1 proteins after inhibition of histone deacetylases. J. Cell Sci. 1185035-5046. [DOI] [PubMed] [Google Scholar]
- 3.Bush, M., D. R. Yager, M. Gao, K. Weisshart, A. I. Marcy, D. M. Coen, and D. M. Knipe. 1991. Correct intranuclear localization of herpes simplex virus DNA polymerase requires the viral ICP8 DNA-binding protein. J. Virol. 651082-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai, W., and P. A. Schaffer. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 662904-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai, W. Z., and P. A. Schaffer. 1989. Herpes simplex virus type 1 ICP0 plays a critical role in the de novo synthesis of infectious virus following transfection of viral DNA. J. Virol. 634579-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandy, M., J. L. Gutierrez, P. Prochasson, and J. L. Workman. 2006. SWI/SNF displaces SAGA-acetylated nucleosomes. Eukaryot. Cell 51738-1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J., and S. Silverstein. 1992. Herpes simplex viruses with mutations in the gene encoding ICP0 are defective in gene expression. J. Virol. 662916-2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman, H. M., V. Connor, Z. S. Cheng, F. Grey, C. M. Preston, and S. Efstathiou. 2008. Histone modifications associated with herpes simplex virus type 1 genomes during quiescence and following ICP0-mediated de-repression. J. Gen. Virol. 8968-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Bruyn Kops, A., and D. M. Knipe. 1988. Formation of DNA replication structures in herpes virus-infected cells requires a viral DNA binding protein. Cell 55857-868. [DOI] [PubMed] [Google Scholar]
- 10.de Bruyn Kops, A., S. L. Uprichard, M. Chen, and D. M. Knipe. 1998. Comparison of the intranuclear distributions of herpes simplex virus proteins involved in various viral functions. Virology 252162-178. [DOI] [PubMed] [Google Scholar]
- 11.DeLuca, N. A., A. M. McCarthy, and P. A. Schaffer. 1985. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J. Virol. 56558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLuca, N. A., and P. A. Schaffer. 1988. Physical and functional domains of the herpes simplex virus transcriptional regulatory protein ICP4. J. Virol. 62732-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deshmane, S. L., and N. W. Fraser. 1989. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J. Virol. 63943-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett, R. D. 1984. Trans activation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 33135-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Everett, R. D., C. Boutell, and A. Orr. 2004. Phenotype of a herpes simplex virus type 1 mutant that fails to express immediate-early regulatory protein ICP0. J. Virol. 781763-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Everett, R. D., and J. Murray. 2005. ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 795078-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett, R. D., A. Orr, and M. Elliott. 1991. High level expression and purification of herpes simplex virus type 1 immediate early polypeptide Vmw110. Nucleic Acids Res. 196155-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Everett, R. D., C. Parada, P. Gripon, H. Sirma, and A. Orr. 2008. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J. Virol. 822661-2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Everett, R. D., S. Rechter, P. Papior, N. Tavalai, T. Stamminger, and A. Orr. 2006. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 807995-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia-Ramirez, M., C. Rocchini, and J. Ausio. 1995. Modulation of chromatin folding by histone acetylation. J. Biol. Chem. 27017923-17928. [DOI] [PubMed] [Google Scholar]
- 21.Gelman, I. H., and S. Silverstein. 1985. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc. Natl. Acad. Sci. USA 825265-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson, W., and B. Roizman. 1971. Compartmentalization of spermine and spermidine in the herpes simplex virion. Proc. Natl. Acad. Sci. USA 682818-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gu, H., Y. Liang, G. Mandel, and B. Roizman. 2005. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc. Natl. Acad. Sci. USA 1027571-7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu, H., and B. Roizman. 2007. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc. Natl. Acad. Sci. USA 10417134-17139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halford, W. P., and P. A. Schaffer. 2001. ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J. Virol. 753240-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris, R. A., R. D. Everett, X. X. Zhu, S. Silverstein, and C. M. Preston. 1989. Herpes simplex virus type 1 immediate-early protein Vmw110 reactivates latent herpes simplex virus type 2 in an in vitro latency system. J. Virol. 633513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hassan, A. H., S. Awad, and P. Prochasson. 2006. The Swi2/Snf2 bromodomain is required for the displacement of SAGA and the octamer transfer of SAGA-acetylated nucleosomes. J. Biol. Chem. 28118126-18134. [DOI] [PubMed] [Google Scholar]
- 28.Hassan, A. H., K. E. Neely, and J. L. Workman. 2001. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104817-827. [DOI] [PubMed] [Google Scholar]
- 29.Hassan, A. H., P. Prochasson, K. E. Neely, S. C. Galasinski, M. Chandy, M. J. Carrozza, and J. L. Workman. 2002. Function and selectivity of bromodomains in anchoring chromatin-modifying complexes to promoter nucleosomes. Cell 111369-379. [DOI] [PubMed] [Google Scholar]
- 30.He, G., L. Ylisastigui, and D. M. Margolis. 2002. The regulation of HIV-1 gene expression: the emerging role of chromatin. DNA Cell Biol. 21697-705. [DOI] [PubMed] [Google Scholar]
- 31.Hebbar, P. B., and T. K. Archer. 2008. Altered histone H1 stoichiometry and an absence of nucleosome positioning on transfected DNA. J. Biol. Chem. 2834595-4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herrera, F. J., and S. J. Triezenberg. 2004. VP16-dependent association of chromatin-modifying coactivators and underrepresentation of histones at immediate-early gene promoters during herpes simplex virus infection. J. Virol. 789689-9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Highlander, S. L., W. H. Cai, S. Person, M. Levine, and J. C. Glorioso. 1988. Monoclonal antibodies define a domain on herpes simplex virus glycoprotein B involved in virus penetration. J. Virol. 621881-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hobbs, W. E., II, and N. A. DeLuca. 1999. Perturbation of cell cycle progression and cellular gene expression as a function of herpes simplex virus ICP0. J. Virol. 738245-8255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hollenbach, A. D., C. J. McPherson, E. J. Mientjes, R. Iyengar, and G. Grosveld. 2002. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci. 1153319-3330. [DOI] [PubMed] [Google Scholar]
- 36.Huang, J., J. R. Kent, B. Placek, K. A. Whelan, C. M. Hollow, P. Y. Zeng, N. W. Fraser, and S. L. Berger. 2006. Trimethylation of histone H3 lysine 4 by Set1 in the lytic infection of human herpes simplex virus 1. J. Virol. 805740-5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito, T., T. Ikehara, T. Nakagawa, W. L. Kraus, and M. Muramatsu. 2000. p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 141899-1907. [PMC free article] [PubMed] [Google Scholar]
- 38.Kasten, M., H. Szerlong, H. Erdjument-Bromage, P. Tempst, M. Werner, and B. R. Cairns. 2004. Tandem bromodomains in the chromatin remodeler RSC recognize acetylated histone H3 Lys14. EMBO J. 231348-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kent, J. R., P. Y. Zeng, D. Atanasiu, J. Gardner, N. W. Fraser, and S. L. Berger. 2004. During lytic infection herpes simplex virus type 1 is associated with histones bearing modifications that correlate with active transcription. J. Virol. 7810178-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Knipe, D. M., and A. Cliffe. 2008. Chromatin control of herpes simplex virus lytic and latent infection. Nat. Rev. Microbiol. 6211-221. [DOI] [PubMed] [Google Scholar]
- 41.Knipe, D. M., D. Senechek, S. A. Rice, and J. L. Smith. 1987. Stages in the nuclear association of the herpes simplex virus transcriptional activator protein ICP4. J. Virol. 61276-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knipe, D. M., and A. E. Spang. 1982. Definition of a series of stages in the association of two herpesviral proteins with the cell nucleus. J. Virol. 43314-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kubat, N. J., R. K. Tran, P. McAnany, and D. C. Bloom. 2004. Specific histone tail modification and not DNA methylation is a determinant of herpes simplex virus type 1 latent gene expression. J. Virol. 781139-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee, C. K., and D. M. Knipe. 1983. Thermolabile in vivo DNA-binding activity associated with a protein encoded by mutants of herpes simplex virus type 1. J. Virol. 46909-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leinbach, S. S., and W. C. Summers. 1980. The structure of herpes simplex virus type 1 DNA as probed by micrococcal nuclease digestion. J. Gen. Virol. 5145-59. [DOI] [PubMed] [Google Scholar]
- 46.Lemasson, I., N. J. Polakowski, P. J. Laybourn, and J. K. Nyborg. 2006. Tax-dependent displacement of nucleosomes during transcriptional activation of human T-cell leukemia virus type 1. J. Biol. Chem. 28113075-13082. [DOI] [PubMed] [Google Scholar]
- 47.Lentine, A. F., and S. L. Bachenheimer. 1990. Intracellular organization of herpes simplex virus type 1 DNA assayed by staphylococcal nuclease sensitivity. Virus Res. 16275-292. [DOI] [PubMed] [Google Scholar]
- 48.Lieberman, P. M. 2006. Chromatin regulation of virus infection. Trends Microbiol. 14132-140. [DOI] [PubMed] [Google Scholar]
- 49.Lomonte, P., J. Thomas, P. Texier, C. Caron, S. Khochbin, and A. L. Epstein. 2004. Functional interaction between class II histone deacetylases and ICP0 of herpes simplex virus type 1. J. Virol. 786744-6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marcy, A. I., D. R. Yager, and D. M. Coen. 1990. Isolation and characterization of herpes simplex virus mutants containing engineered mutations at the DNA polymerase locus. J. Virol. 642208-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Memedula, S., and A. S. Belmont. 2003. Sequential recruitment of HAT and SWI/SNF components to condensed chromatin by VP16. Curr. Biol. 13241-246. [DOI] [PubMed] [Google Scholar]
- 52.Miki, K., M. Shimizu, M. Fujii, M. N. Hossain, and D. Ayusawa. 2008. 5-Bromouracil disrupts nucleosome positioning by inducing A-form-like DNA conformation in yeast cells. Biochem. Biophys. Res. Commun. 368662-669. [DOI] [PubMed] [Google Scholar]
- 53.Nabel, G. J., S. A. Rice, D. M. Knipe, and D. Baltimore. 1988. Alternative mechanisms for activation of human immunodeficiency virus enhancer in T cells. Science 2391299-1302. [DOI] [PubMed] [Google Scholar]
- 54.Neely, K. E., A. H. Hassan, A. E. Wallberg, D. J. Steger, B. R. Cairns, A. P. Wright, and J. L. Workman. 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 4649-655. [DOI] [PubMed] [Google Scholar]
- 55.Oh, J., and N. W. Fraser. 2008. Temporal association of the herpes simplex virus genome with histone proteins during a lytic infection. J. Virol. 823530-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Hare, P., and G. S. Hayward. 1985. Three trans-acting regulatory proteins of herpes simplex virus modulate immediate-early gene expression in a pathway involving positive and negative feedback regulation. J. Virol. 56723-733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Phelan, A., J. Dunlop, A. H. Patel, N. D. Stow, and J. B. Clements. 1997. Nuclear sites of herpes simplex virus type 1 DNA replication and transcription colocalize at early times postinfection and are largely distinct from RNA processing factors. J. Virol. 711124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pignatti, P. F., and E. Cassai. 1980. Analysis of herpes simplex virus nucleoprotein complexes extracted from infected cells. J. Virol. 36816-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Poon, A. P., H. Gu, and B. Roizman. 2006. ICP0 and the US3 protein kinase of herpes simplex virus 1 independently block histone deacetylation to enable gene expression. Proc. Natl. Acad. Sci. USA 1039993-9998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Poon, A. P., Y. Liang, and B. Roizman. 2003. Herpes simplex virus 1 gene expression is accelerated by inhibitors of histone deacetylases in rabbit skin cells infected with a mutant carrying a cDNA copy of the infected-cell protein no. 0. J. Virol. 7712671-12678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Quinlan, M. P., and D. M. Knipe. 1985. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol. Cell. Biol. 5957-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Randall, R. E., and N. Dinwoodie. 1986. Intranuclear localization of herpes simplex virus immediate-early and delayed-early proteins: evidence that ICP 4 is associated with progeny virus DNA. J. Gen. Virol. 672163-2177. [DOI] [PubMed] [Google Scholar]
- 63.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 111599-1607. [DOI] [PubMed] [Google Scholar]
- 64.Sacks, W. R., and P. A. Schaffer. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J. Virol. 61829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saha, A., J. Wittmeyer, and B. R. Cairns. 2006. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell Biol. 7437-447. [DOI] [PubMed] [Google Scholar]
- 66.Samaniego, L. A., L. Neiderhiser, and N. A. DeLuca. 1998. Persistence and expression of the herpes simplex virus genome in the absence of immediate-early proteins. J. Virol. 723307-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sampath, P., and N. A. Deluca. 2008. Binding of ICP4, TATA-binding protein, and RNA polymerase II to herpes simplex virus type 1 immediate-early, early, and late promoters in virus-infected cells. J. Virol. 822339-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schwabish, M. A., and K. Struhl. 2007. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol. Cell. Biol. 276987-6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sharma, N., and J. K. Nyborg. 2008. The coactivators CBP/p300 and the histone chaperone NAP1 promote transcription-independent nucleosome eviction at the HTLV-1 promoter. Proc. Natl. Acad. Sci. USA 1057959-7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silva, L., A. Cliffe, L. Chang, and D. M. Knipe. 2008. Role for A-type lamins in herpesviral DNA targeting and heterochromatin modulation. PLoS Pathog. 4e1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Simpson-Holley, M., J. Baines, R. Roller, and D. M. Knipe. 2004. Herpes simplex virus 1 UL31 and UL34 gene products promote the late maturation of viral replication compartments to the nuclear periphery. J. Virol. 785591-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Stow, N. D., and E. C. Stow. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 672571-2585. [DOI] [PubMed] [Google Scholar]
- 73.Suzuki, T., M. Yaginuma, T. Oishi, E. Michishita, H. Ogino, M. Fujii, and D. Ayusawa. 2001. 5-Bromodeoxyuridine suppresses position effect variegation of transgenes in HeLa cells. Exp. Cell. Res. 26653-63. [DOI] [PubMed] [Google Scholar]
- 74.Tagami, H., D. Ray-Gallet, G. Almouzni, and Y. Nakatani. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 11651-61. [DOI] [PubMed] [Google Scholar]
- 75.Taylor, T. J., and D. M. Knipe. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 785856-5866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Utley, R. T., K. Ikeda, P. A. Grant, J. Cote, D. J. Steger, A. Eberharter, S. John, and J. L. Workman. 1998. Transcriptional activators direct histone acetyltransferase complexes to nucleosomes. Nature 394498-502. [DOI] [PubMed] [Google Scholar]
- 77.van Leeuwen, H., M. Okuwaki, R. Hong, D. Chakravarti, K. Nagata, and P. O'Hare. 2003. Herpes simplex virus type 1 tegument protein VP22 interacts with TAF-I proteins and inhibits nucleosome assembly but not regulation of histone acetylation by INHAT. J. Gen. Virol. 842501-2510. [DOI] [PubMed] [Google Scholar]
- 78.Wang, Q. Y., C. Zhou, K. E. Johnson, R. C. Colgrove, D. M. Coen, and D. M. Knipe. 2005. Herpesviral latency-associated transcript gene promotes assembly of heterochromatin on viral lytic-gene promoters in latent infection. Proc. Natl. Acad. Sci. USA 10216055-16059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Woodhall, D. L., I. J. Groves, M. B. Reeves, G. Wilkinson, and J. H. Sinclair. 2006. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J. Biol. Chem. 28137652-37660. [DOI] [PubMed] [Google Scholar]
- 80.Xue, Y., R. Gibbons, Z. Yan, D. Yang, T. L. McDowell, S. Sechi, J. Qin, S. Zhou, D. Higgs, and W. Wang. 2003. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl. Acad. Sci. USA 10010635-10640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yager, D. R., and S. L. Bachenheimer. 1988. Synthesis and metabolism of cellular transcripts in HSV-1 infected cells. Virus Genes 1135-148. [DOI] [PubMed] [Google Scholar]
- 82.Zhao, J., J. Herrera-Diaz, and D. S. Gross. 2005. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol. Cell. Biol. 258985-8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu, X. X., J. X. Chen, C. S. Young, and S. Silverstein. 1990. Reactivation of latent herpes simplex virus by adenovirus recombinants encoding mutant IE-0 gene products. J. Virol. 644489-4498. [DOI] [PMC free article] [PubMed] [Google Scholar]