Abstract
Bovine herpesvirus 1 (BHV-1) infected cell protein 0 (bICP0) stimulates productive infection, in part by activating viral gene expression. The C3HC4 zinc RING finger of bICP0 is crucial for activating viral transcription and productive infection. In this study, we used a bacterial artificial chromosome containing a wild-type (wt) virulent BHV-1 strain to generate a single amino acid mutation in the C3HC4 zinc RING finger of bICP0. This virus (the 51g mutant) contains a cysteine-to-glycine mutation (51st amino acid) in the C3HC4 zinc RING finger of bICP0. A plasmid expressing the 51g mutant protein did not transactivate viral promoter activity as efficiently as wt bICP0. The 51g mutant virus expressed higher levels of the bICP0 protein than did the 51g rescued virus (51gR) but yielded reduced virus titers following infection of permissive bovine cells. The 51g mutant virus, but not the 51gR virus, grew poorly in bovine cells pretreated with imiquimod to stimulate interferon production. During acute infection of calves, levels of infectious virus were 2 to 3 logs lower in ocular or nasal swabs with 51g than with 51gR. Calves latently infected with the 51g mutant did not reactivate from latency because virus shedding did not occur in ocular or nasal cavities. As expected, calves latently infected with 51gR reactivated from latency following dexamethasone treatment. These studies demonstrate that mutation of a single well-conserved cysteine residue in the C3HC4 zinc RING finger of bICP0 has dramatic effects on the growth properties of BHV-1.
Infection of cattle with bovine herpesvirus 1 (BHV-1) can cause conjunctivitis, pneumonia, genital disorders, abortions, and an upper respiratory infection known as bovine respiratory disease complex (BRDC), or “shipping fever” (67). BHV-1 initiates BRDC by immunosuppressing cattle (9, 22-24), which leads to secondary bacterial infections and pneumonia. For example, CD8+ T-cell recognition is impaired by BHV-1 infection because antigen presentation is repressed (27, 30, 53). CD4+ T-cell function is also impaired because BHV-1 infects these cells and induces apoptosis (71). The virus-encoded protein BHV-1 infected cell protein 0 (bICP0) also inhibits interferon (IFN)-dependent transcription, in part by degrading a transcription factor (IFN regulatory factor 3 [IRF3]) that activates the IFN-β promoter (28, 63). Modified live vaccines are available, and in general they prevent clinical disease in adults. However, the same vaccine strains can be immunosuppressive and cause serious disease in young calves or abortions in pregnant cows. BRDC and BHV-1 infections cost the cattle industry at least $3 billion/year in the United States (35, 41, 59).
Like other Alphaherpesvirinae subfamily members, BHV-1 establishes lifelong latency in ganglionic neurons of the peripheral nervous system following acute replication in the mucosal epithelium (36, 37). Virus reactivation and spread to other susceptible cattle occur after natural or corticosteroid-induced stress (61, 65). Viral gene expression is temporally regulated in three distinct phases during productive infection: immediate early (IE), early (E), and late (L) (36, 37). IE gene expression is stimulated by a virion component, bTIF, that interacts with a cellular transcription factor (Oct-1), and this complex subsequently interacts with and transactivates IE promoters (36, 37). Two IE transcription units exist: IEtu1 and IEtu2. IEtu1 encodes homologues of two herpes simplex virus type 1 (HSV-1) proteins, ICP0 and ICP4. IEtu2 encodes a protein similar to the HSV ICP22 protein (75). IE proteins activate E gene expression, and viral DNA replication occurs. L gene expression then occurs, culminating in virion assembly and release. The bICP0 protein is crucial for productive infection because it activates all viral promoters and is constitutively expressed throughout productive infection (19, 74, 75).
bICP0 (34), HSV-1 ICP0 (12-15, 45), and equine herpesvirus 1 ICP0 (4, 5) contain a C3HC4 zinc RING finger (referred to as RING finger herein) near their N terminus that activates productive infection. ICP0 (16-18, 46, 47) and bICP0 (34, 54) localize to and disrupt promyelocytic leukemia (PML) protein-containing nuclear domains. bICP0 (11) and ICP0 (1, 3, 69) contain RING fingers that possess intrinsic E3 ubiquitin ligase activity. Consequently, bICP0 and ICP0 can promote ubiquitin-dependent proteolysis of certain proteins (16, 18, 42, 55). Mutagenesis of the bICP0 RING finger impairs its ability to activate transcription in all cells tested (34, 76), and in certain cell types IFN-dependent transcription is inhibited (28, 63).
In this study, we generated a single point mutation in a conserved cysteine residue at position 51 within the zinc RING finger of bICP0 by using a virulent BHV-1 bacterial artificial chromosome (BAC) clone, resulting in the 51g mutant. The 51g mutant virus grew less efficiently than the 51g rescued virus (51gR) or wild-type (wt) BHV-1 in cultured bovine cells. Following infection of calves, the 51g mutant grew poorly, induced few clinical symptoms, and did not reactivate from latency following dexamethasone (DEX) treatment. Conversely, the 51gR virus grew to high titers and reactivated from latency after DEX treatment. In summary, this study provides evidence that the wt zinc RING finger of bICP0 is crucial for efficient growth in cultured bovine cells and virulence in calves.
MATERIALS AND METHODS
Cells and virus.
Bovine kidney cells (CRIB cells) were grown in Earle's modified Eagle's medium supplemented with 5% fetal calf serum. CRIB cells, derived from MDBK cells, are resistant to infection with bovine viral diarrhea virus and were obtained from Ruben Donis (Centers for Disease Control and Prevention, Atlanta, GA). All media contained penicillin (10 U/ml) and streptomycin (100 μg/ml). The wt BHV-1 Cooper strain was obtained from the National Veterinary Services Laboratory, Animal and Plant Health Inspection Services (Ames, IA). BHV-1 strains were propagated and titrated in CRIB cells.
Construction of the BHV-1 bICP0 zinc RING finger domain mutant (51g).
For mutation of the bICP0 zinc RING finger domain, a two-step red-mediated recombination technique described by Tischer et al. (68) was used, with slight modifications that we recently described (44). The primers used for mutagenesis are shown below. To mutate the zinc RING finger domain of bICP0, codon TGC (nucleotides shown in bold below) encoding a cysteine residue (bICP0 residue 51) was mutated to glycine (GGC). The 5′ 60 bp of the forward and reverse primer sequences are homologous to the bICP0 gene sequence and contain the intended mutated base. While the 5′ 20 bp of the forward and reverse primers are not complementary, the remaining 40 bp of the bICP0-specific primer sequences are complementary to each other, including the mutated base (G instead of T in the case of the forward primer and C instead of A for the reverse primer). At the 3′ terminus of each of the respective primers, 20 bp of the forward and reverse primers are homologous to the pEPkan-S plasmid sequence (underlined nucleotides). When these primers are used as a template for PCR (44), the product overlaps with an I-SceI site located upstream of AphI, which confers kanamycin resistance to the plasmid. Furthermore, the 3′ end of the reverse primer is complementary to sequences downstream of AphAI within pEPKanS (68). The primers were custom synthesized and purified by polyacrylamide gel electrophoresis (PAGE) (Integrated DNA Technologies, Coralville, IA). For the forward primer (TGC → GGC) used for the 51g mutation, the original sequence is 5′ATCCGCCGGTGGCTGGAGGGGCGCCCGACCTGCCCGCTGTGCAAGGCGCCCGTGCAGTCTAGGATGACGACGATAAGTAGGG3′, and the mutated sequence is 5′ATCCGCCGGTGGCTGGAGGGGCGCCCGACCTGCCCGCTGGGCAAGGCGCCCGTGCAGTCTAGGATGACGACGATAAGTAGGG3′. For the reverse primer (ACG → CCG) used for the 51g mutation, the original sequence is 5′AGGCGACGCTGTGGATGAGAGACTGCACGGGCGCCTTGCACAGCGGGCAGGTCGGGCGCCCCAACCAATTAACCAATTCTGATTAG3′, and the mutated sequence is 5′AGGCGACGCTGTGGATGAGAGACTGCACGGGCGCCTTGCCCAGCGGGCAGGTCGGGCGCCCCAACCAATTAACCAATTCTGATTAG3′. The PCR products (44) were used for the electroporation of electrocompetent SW105 cells harboring pBHV-1BAC (44). Following mutagenesis, pBHV-1BAC plasmids containing the mutation were verified by sequencing after PCR amplification of the region of interest. Primers used for PCR are as follows: forward bICP0 primer, 5′ GCC TTT CGC CCG CCC G 3′; reverse bICP0 primer, 5′ CAA CGC GCC GTC CGC CCC 3′. The positive pBHV-1 BAC mutant plasmids were maintained in Escherichia coli DH10B (68). The 51g mutant virus without the BAC sequence was then reconstituted by cotransfection of pBHV-1 mutated BAC plasmid and pCre DNA into CRIB cells, as described earlier (44).
To generate 51gR, CRIB cells were transfected with 2.5 μg plasmid phDK/bICP0 (contains a 5-kb fragment of BHV-1 that spans the bICP0 gene) by use of Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. At 24 h after transfection, cells were infected with the 51g mutant virus at a multiplicity of infection (MOI) of 0.1. The supernatant was collected 72 h after infection, and virus was harvested by three freeze-thaw cycles. Plaque assays were then performed. Since BHV-1 containing a wt bICP0 rescued virus was anticipated to grow faster and more efficiently than the original 51g mutant virus, we predicted that it would be easy to identify wt plaques. 51gR was readily observed in a plaque assay and was subsequently purified by three rounds of plaque purification. Three plaques were selected after the final round for viral genomic DNA isolation. PCR using bICP0-specific primers was performed with the viral genomic DNA as the template. PCR products were gel purified and sequenced. In each case, the 51gR virus was present.
Extraction of viral genomic DNA.
CRIB cells were infected with the indicated viruses at an MOI of 1.0, and viral genomic DNA was isolated as described previously (33). At 24 h after infection, the supernatant was collected and centrifuged at 7,000 rpm at 4°C for 20 min. Virus particles were collected as a pellet by centrifugation (25,000 rpm for 2 h in a Beckman LT-65 instrument) using an SW28 rotor at 4°C in a 30% sucrose-Tris-EDTA cushion. The pellet was suspended in Tris-EDTA buffer and extracted with phenol:chloroform:isoamyl alcohol (25:24:1) three times, followed by three ether extractions. Viral genomic DNA was then precipitated overnight at −80°C in 100% ethanol. The quality and quantity of viral DNA were determined by agarose gel electrophoresis (1%) and spectrophotometry (optical density at 260 nm). For sequencing of the bICP0 region, PCR was performed using viral genomic DNA as the template and the bICP0 primers described above. The PCR product was gel purified and sequenced.
One-step growth kinetics.
CRIB cells were plated onto 100-mm2 dishes 24 h prior to virus infection. Cells were infected at different MOIs (0.1, 1.0, and 5 PFU/cell). After 1 h of adsorption at 37°C, cells were rinsed with phosphate-buffered saline (PBS) and overlaid with Earle's modified Eagle's medium containing 5% fetal calf serum. Virus was collected at 8, 16, 24, and 48 h after infection. Subjecting cells to three freeze-thaw cycles resulted in obtaining intracellular virus. The virus-containing supernatant was clarified by centrifugation at 3,500 rpm for 30 min at 4°C and frozen at −80°C. Virus titers were determined by standard plaque assays on CRIB cells.
Cell lysis and Western blotting.
Cells were infected with the wt, 51g, or 51gR at an MOI of 1.0. Cells were harvested at the indicated times after infection. Cells were washed in PBS and suspended in cell lysis buffer (100 mM Tris, pH 8.0, 1 mM EDTA, 100 mM NaCl, 1% NP-40, 1 mM phenylmethylsulfonyl fluoride) and one tablet of complete protease inhibitor (Roche Molecular Biochemicals) per 10 ml of buffer. The cell suspension was sonicated three times, incubated at 4°C for 15 min, and then centrifuged at 12,000 rpm at 4°C for 15 min. Protein concentrations were then determined by the Bradford assay (Bio-Rad), and the lysate was boiled in sodium dodecyl sulfate-PAGE (SDS-PAGE) buffer. Western blot analyses were performed as described previously (63). The anti-BHV-1 antibody was purchased from Veterinary Medical Research and Development (VMRD, Pullman, WA). This antiserum recognizes virion proteins (210-70-IBR). The anti-bICP0 antibodies were prepared against the C-terminal half of the protein or a peptide antibody. Both were produced in rabbits, and they yielded similar results.
Animal experiments.
BHV-1-free crossbred calves (∼200 kg) were randomly assigned and housed in isolation rooms to prevent cross-contamination. Calves were inoculated with 106 PFU of the indicated virus into each nostril and eye, without scarification, for a total of 4 × 106 PFU per animal, as described previously (32, 33, 64, 70, 71, 73). Experiments with animals were performed in accordance with the American Association of Laboratory Animal Care guidelines. Calves were housed under strict isolation containment and given antibiotics before and after BHV-1 infection to prevent secondary bacterial infection. Nasal swabs, ocular swabs, and serum samples were taken at the designated times. Swabs were stored at −80°C in 2 ml of medium containing 2% serum. Samples were thawed in a 37°C water bath, vortexed, and centrifuged (1,500 × g for 10 min). Virus titrations were performed using 10-fold serial dilutions, and samples were plated in triplicate.
To initiate reactivation from latency, 100 mg of water-soluble DEX (Sigma) in 1.5 ml of medium was injected into the jugular vein of calves at 45 days after infection. Additional intramuscular injections of DEX (25 mg) were given at 2 and 4 days after the initial DEX injection. This protocol consistently leads to reactivation from latency (32).
RESULTS
Two-step red-mediated mutagenesis of bICP0 RING finger domain.
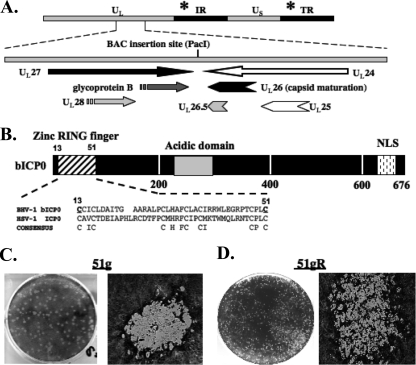
A BHV-1 BAC clone was constructed by inserting the BAC into the intergenic region between UL27 and UL24 (Fig. 1A) (44). Following transfection of the wt BHV-1 BAC plasmid into bovine cells, infectious virus containing only 34 bp of loxP sequences in the intergenic region between UL27 and UL24 was obtained. Recent studies demonstrated that the wt BHV-1 BAC, in a fashion similarl to wt BHV-1, grows efficiently in bovine cells, is virulent in calves, and reactivates from latency following treatment with DEX (44). The 51st amino acid (aa) of bICP0 was chosen for mutagenesis because this cysteine residue is important for the transactivation potential of bICP0, inhibition of IFN-dependent transcription, and reduced toxicity in cultured cells (28, 29, 34, 63, 76) and is conserved in the zinc RING finger of HSV-1-encoded ICP0 (Fig. 1B). Two-step red-mediated recombination was used to introduce mutations in the 51st aa of the zinc RING finger (see Materials and Methods for the primers used to make the mutations) (68). DNA sequencing confirmed that this mutation was present in both repeats (data not shown).
FIG. 1.
BHV-1 BAC organization and analysis of wt BHV-1 BAC. (A) BHV-1 genomic organization, showing the intergenic region between the UL27/UL28 and UL26/UL26.5/UL25/UL24 genes and the relative transcription directions/orientations of these genes. The site of the BAC insertion (PacI site) is shown. The position of the bICP0 gene located in the inverted repeat (IR) and the terminal repeat (TR) is denoted by the asterisk. (B) The bICP0 protein is 676 aa long and contains several important functional domains. The bICP0 zinc RING finger (aa 13 to 51) is similar to the zinc RING finger present in the HSV-1 ICP0 protein. Positions of conserved C residues in a consensus C3HC4 zinc RING finger are shown. The locations of the acidic domain and the nuclear localization signal (NLS) within bICP0 are also denoted. The numbers below the diagram denote amino acid sequence numbers of bICP0. Mutagenesis of aa 13 (cysteine to glycine) and aa 51 (cysteine to alanine) (underlined) impairs the ability of bICP0 to activate transcription in transient-transfection assays (34), and plasmids containing these mutated cysteines do not inhibit IFN-dependent transcription (28, 63). (C and D) CRIB cells were infected with 51g (C) or 51gR (D) at an MOI of 0.1. After incubation at 37°C for 1 h, cells were rinsed three times with calcium magnesium-free PBS and replaced with fresh media. Plaques were stained with crystal violet at 72 h after infection for 51g or at 36 h after infection for 51gR. Images were taken at a ×10 (left panels) or a ×100 (right panels) magnification using a Leica DM IRB microscope.
The mutant BAC DNA was transfected in MDBK cells, and 4 to 5 days later, plaques were observed. Virus stocks were prepared by infecting CRIB cells with the initial virus stock obtained after transfection. In general, large, well-defined, circular plaques that failed to spread were observed following infection of bovine cells with the 51g mutant virus (Fig. 1C). Conversely, plaques generated by 51gR (Fig. 1D) or the wt BHV-1 BAC (data not shown) were smaller and irregular, and these plaques spread and fused with other plaques as a function of time after infection. Although plaques produced by the 51g mutant were larger than those produced by the 51gR strain, we consistently observed fewer plaques, suggesting that the 51g mutant was growth restricted. Furthermore, the plaques induced by the 51g mutant took 72 h to appear, whereas by 24 to 36 h after infection plaques were readily detected when cells were infected with the 51gR strain or wt BHV-1. DNA sequencing confirmed that the 51g mutation was present after several passages in CRIB cells, suggesting that the 51g mutant was stable. 51gR was prepared by transfecting CRIB cells with a plasmid (phDK) that contains wt bICP0 sequences. Plaques resembling those of wt BHV-1 were selected and amplified (Fig. 1D shows representative plaques of 51gR). The plaque morphologies of wt BHV-1 and 51gR are indistinguishable. DNA sequencing of a PCR fragment spanning the zinc RING finger of 51gR demonstrated that the 51g mutation was rescued back to the wt. In addition, a similar HindIII digestion pattern was observed when the 51g mutant and the 51gR strain were compared to wt BHV-1 (data not shown).
Expression of bICP0 and viral proteins during productive infection.
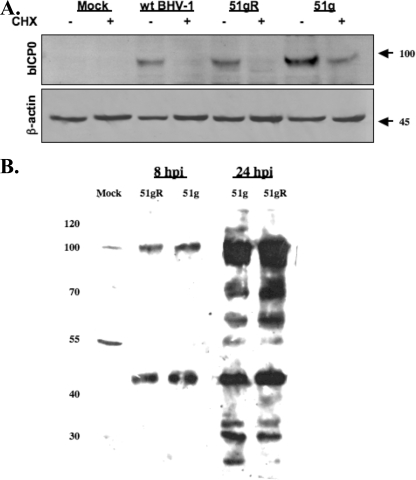
bICP0 expression was examined following infection of CRIB cells with the 51g mutant or 51gR at an MOI of 1.0. Cell lysate was collected at different times after infection, and Western analysis was performed with an anti-bICP0 peptide antibody. Higher levels of bICP0 protein expression were consistently observed following infection of bovine cells with the 51g mutant than following infection with 51gR or wt BHV-1 (Fig. 2A and data not shown). Previous studies demonstrated that HSV-1 ICP0 mutants with reduced E3 ubiquitin ligase activity have increased stability as a result of decreased autoubiquitination (2, 8). We predicted that the cysteine-to-glycine mutation in the 51st aa of bICP0 disrupted the putative E3 ubiquitin ligase activity of bICP0 and, consequently, that the 51g mutant version of bICP0 would have a longer half-life. To test this prediction, CRIB cells were infected with wt BHV-1, the 51gR strain, or the 51g mutant, and at 6 h after infection, cells were treated with 100 μg/ml cycloheximide. bICP0 protein levels were examined 2 h after cycloheximide treatment. The bICP0 protein was readily detected after treatment with cycloheximide when cells were infected with the 51g mutant. Conversely, bICP0 was not readily detected in cells infected with wt BHV-1 or the 51gR strain after treatment with cycloheximide. As expected, the bICP0-specific antiserum did not recognize a protein in mock-infected cells. Similar levels of β-actin were present in each lane, confirming that similar protein levels were loaded for each sample (Fig. 2A, bottom). In summary, these studies suggested that the 51g mutant bICP0 protein had increased stability during productive infection and was consequently expressed at higher levels.
FIG. 2.
Western blot analysis of bICP0 and viral proteins in productively infected cells. (A) CRIB cells were mock infected or infected with the 51g mutant, 51gR, or wt BHV-1 at an MOI of 1.0. At 6 h after infection, some cultures were treated with 100 μg/ml cycloheximide (Sigma) (CHX; + lanes) to inhibit protein synthesis. Two hours later (8 h after infection), cells were lysed by treatment with NP-40 and sonication and then processed for Western blot analysis as described in Materials and Methods. Following SDS-PAGE (8%), immunoblotting was performed with a polyclonal antibody generated against the N-terminal 361 aa of bICP0 (1:500). The bICP0 protein migrates at approximately 97 kDa, and β-actin migrates at approximately 45 kDa (marked at right). For each lane, 50 μg protein was added. (B) CRIB cells were infected with the designated strains of BHV-1 for 8 or 24 h postinfection (hpi). After cells were lysed, proteins (50 μg/lane) were electrophoresed by 8% SDS-PAGE. Immunoblotting was performed using an anti-BHV-1 virion antibody that was commercially available from VMRD (Pullman, WA). Markers (in kilodaltons) are shown at left.
A commercially available antiserum that recognizes BHV-1 structural proteins was also used to examine the accumulation of viral proteins following infection of bovine cells with the 51g mutant or 51gR (Fig. 2B). Similar levels of proteins were detected at 8 and 24 h after infection regardless of which virus was used for infection. Since this antibody recognizes viral structural proteins and many of these proteins are glycosylated, it was not surprising to find that this antibody recognized several bands that appeared as broad, diffuse bands.
Analysis of the 51g mutant virus in CRIB cells.
CRIB cells were infected with 51gR or the 51g mutant strain at an MOI of 1.0 or 5.0. Infectious virus was collected and virus titers were measured as described in Materials and Methods. At an MOI of 1, cultures infected with the 51g mutant did not contain detectable virus at 16 h after infection whereas cultures infected with the wt or 51gR contained approximately 1 × 105 PFU/ml (Fig. 3A). By 48 h after infection, there were 2 to 3 logs difference in the titer of the 51g mutant versus that of 51gR or wt BHV-1. At an MOI of 5, there was approximately 1 log difference in virus titers at 24 h after infection (Fig. 3B). The 51g mutant grew better at early times after infection at an MOI of 5 than it did at an MOI of 1.0.
FIG. 3.
Growth of the 51g mutant, 51gR, or wt BHV-1 in CRIB cells. CRIB cells were infected with the 51g mutant, 51gR, or wt BHV-1 at an MOI of 1.0 (A) or 5.0 (B). Error bars show the standard errors of the results of three independent studies. (C) Viral DNA was prepared from 107 PFU of virus stocks that were obtained from CRIB cells infected with wt BHV-1, the 51g mutant virus, or 51gR. Tenfold dilutions of viral DNA were amplified using gB-specific primers (forward, 5′-GTGGTGGCCTTTGACCGCGAC-3′; reverse, 5′-GCTCCGGCGAGTAGCTGGTGT-3′). Amplified products were electrophoresed on a 1% agarose gel, and the ethidium bromide gel was photographed. The values above the lanes refer to the log of the original MOI of the respective virus stocks. The arrow denotes the position of the gB-specific amplified product. The “+” lane denotes a positive control. The lowest dilution of virus stocks that yielded a visible band was quantified using an RX biomolecular imager (Bio-Rad), and the arbitrary DNA levels are shown (D). These results are representative of comparing two different stocks of the respective viruses.
It is known that HSV-1 ICP0 mutants have a high particle/PFU ratio following infection of certain cultured cells (7). To test whether the 51g mutant had higher particle/PFU ratios than wt BHV-1, the viral DNA levels present in the same MOI of virus derived from stocks of the 51g mutant, 51gR, or wt BHV-1 were measured. Tenfold dilutions of DNA derived from these virus stocks were subjected to PCR using gB-specific primers. At an MOI of the 51g mutant virus of 10−1, a gB-specific amplified product was readily detected (Fig. 3C). Conversely, the lowest dilution of wt BHV-1 or the 51gR mutant that yielded a gB-specific band was 1. When the intensity of the lowest dilution of virus stock that yielded a gB-specific band was quantified, we concluded that the 51g mutant virus contained approximately fourfold-higher levels of viral DNA than did 51gR or wt BHV-1 at the same MOI (Fig. 3D). When the growth curves were repeated using an MOI of 1 PFU/cell for wt BHV-1 and 51gR as well as similar amounts of viral genomes of the 51g mutant virus, there were no dramatic differences compared to the results shown in Fig. 3A (data not shown).
The 51g mutant grows inefficiently in MDBK cells pretreated with imiquimod.
HSV-1-encoded ICP0 has been reported to play a crucial role in allowing virus replication to grow in the face of an IFN response (50-52). The ICP0 homologue encoded by pseudorabies virus (EP0) is also important for counteracting the antiviral state in swine cells but not cells from nonnatural hosts (6). Thus, it was of interest to determine whether the growth of the 51g mutant was reduced more by IFN than was the growth of 51gR.
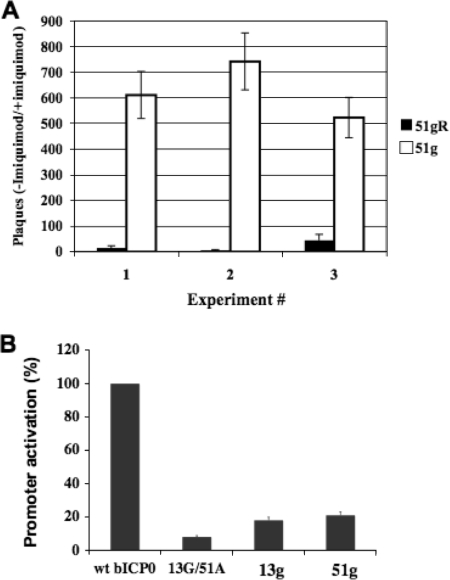
To test whether the 51g mutant was capable of growing in cells actively producing IFN, bovine kidney cells (MDBK) were treated with imiquimod for 12 h and then infected with the 51g mutant or 51gR. Imiquimod stimulates IFN as well as cytokine production (48) and IFN-β promoter activity in MDBK cells (56). The effects of imiquimod treatment on virus replication were calculated by dividing the number of plaques in control cells not treated with imiquimod by the number of plaques in cultures treated with imiquimod (Fig. 4A). The plaquing efficiency of 51g was more than 500 in three independent experiments. 51gR had a plaquing efficiency of approximately 5, indicating that it grew better after imiquimod treatment than did the 51g mutant. The ability of 51gR to grow efficiently after imiquimod treatment was consistent with results from a previous study, which concluded that the growth properties of BHV-1 are not dramatically reduced in the presence of IFN (31). Since bICP0 has the potential to inhibit IRF3- and IRF7-dependent activation of the IFN-β promoter (63), bICP0, directly or indirectly, may play a role in overcoming the effects of imiquimod. Conversely, the inability of the 51g mutant to grow in CRIB cells after imiquimod treatment may merely mean that a disabled virus does not grow efficiently under suboptimal growth conditions.
FIG. 4.
Analysis of effects of the zinc RING finger mutants on virus growth in cells treated with imiquimod and on transactivation. (A) Bovine cells (CRIB cells) were treated with 20 μg/ml imiquimod (InvivoGen, San Diego, CA) for 12 h and then infected with the 51g mutant virus or 51gR at an MOI of 1. As a negative control, some cultures were not treated with imiquimod. Plaque assays were then performed. The number of plaques in cultures not treated with imiquimod was divided by the number of plaques in cultures that were treated with imiquimod. The results of three independent experiments are shown. For each experiment, duplicate samples were used (the mean from each experiment is given). (B) Bovine cells (9.1.3 cells) were transfected with a TK promoter construct (pBLcat4; 1 μg DNA), which has a chloramphenicol acetyltransferase (CAT) gene at the 3′ end of the TK promoter. Results for human cytomegalovirus expression plasmids containing wt bICP0, a bICP0 mutant containing a cysteine-to-glycine mutation in the 13th aa (13G), a bICP0 mutant containing a cysteine-to-alanine mutation in the 51st aa (51A), and the double mutant (13G/51A) (1 μg DNA) are shown. The ratio between pBLcat4 and wt bCP0 is optimal for transactivation of the TK promoter. DNA concentrations in the transfection mixtures were kept at a constant level by adding pcDNA3.1, an empty expression vector. At 48 h posttransfection, cell lysate was collected and assayed for CAT activity. The CAT activity of cells transfected with the TK promoter and wt bICP0 was set to 100%. All other values are expressed as the level of activation (n-fold) with respect to that of the control. The TK promoter construct and the plasmids expressing wt bICP0 and the 13G/51A mutant were previously described (34, 76). The means of three independent studies are shown.
Transactivation of a simple promoter by zinc RING finger mutants.
Although previous studies demonstrated that mutations in both the 13th and 51st aa of the zinc RING finger reduced the ability of bICP0 to transactivate a simple promoter (34, 76), single point mutations in these amino acids were not examined. To this end, transient-transfection assays were performed to determine whether a single cysteine-to-glycine or cysteine-to-alanine mutation affected bICP0 transcriptional activation potential. Cytomegalovirus expression plasmids containing the 13th-aa (pbICP013G; designated 13G) or 51st-aa (pbICP051A; designated 51A) mutation in bICP0 were unable to transactivate a simple HSV-1 thymidine kinase (TK) promoter with the same efficiency as wt bICP0 (Fig. 4B). A plasmid with both the 13th and 51st cysteine residues mutated in the zinc RING finger (pbICP013G/51A; designated 13G/51A) also did not transactivate the TK promoter, which was expected (34). In general, the 13G/51A mutant transactivated the TK promoter less efficiently than the 13g or 51g mutant construct.
Growth of the 51g mutant virus in calves.
Calves were infected with a total of 4 × 106 PFU of 51gR or the 51g mutant virus via intraocular and intranasal instillation, as described previously (32, 33, 57, 58, 64, 70-73). Loss of appetite, increased breathing, inflammation, persistent nasal discharge, and conjunctivitis were observed as early as 3 days after infection of calves with 51gR. These symptoms were similar to those observed following infection of calves with wt BHV-1. In general, clinical symptoms during acute infection of calves were most severe between 4 and 8 days after infection, which is also similar to results for calves acutely infected with wt BHV-1. Clinical symptoms were not evident after infection of calves with the 51g mutant.
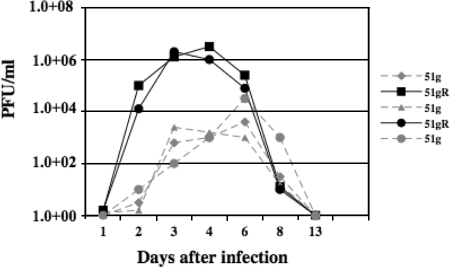
To quantify infectious-virus shedding from ocular or nasal cavities, swabs were obtained following infection and viral growth was measured by limiting dilution assays. In general, at the peak of acute infection, ocular shedding of the 51g mutant was approximately 2 to 3 logs less than that for calves infected with 51gR (Fig. 5). Ocular swabs from two calves infected with the 51g mutant contained less than 1 × 102 PFU/ml infectious virus at 2 or 3 days after infection. The peak of virus infection for the 51g mutant was also delayed compared to that for the 51gR strain, because the most infectious virus detected in ocular swabs was at 6 days after infection. There was also 1 to 2 logs less virus in swabs collected from calves infected with the 51g mutant at 6 days after infection. Infectious virus obtained from ocular swabs of calves infected with the 51g mutant retained the expected mutation regardless of whether virus was obtained from ocular or nasal swabs. After 8 days of infection, infectious virus was not detected in ocular swabs regardless of which virus was used to infect calves.
FIG. 5.
Ocular shedding of virus during acute infection. Calves were infected with the designated virus as described in Materials and Methods. At the indicated days after infection, ocular swabs were collected from calves infected with the 51g mutant or 51gR. Infectious virus present in swabs was quantified by infecting CRIB cells.
Nasal swabs from one calf infected with the 51g mutant contained approximately 4 × 104 PFU/ml infectious virus at 6 days after infection (Fig. 6), which was approximately 2 logs less than the level in calves infected with 51gR at the peak of shedding. However, the other three calves infected with the 51g mutant virus shed less than 104 PFU/ml in nasal swabs during the course of acute infection. In general, the levels of infectious virus shed from the nasal cavity were higher than the levels from ocular shedding in calves infected with 51gR, which was consistent with previous studies (32, 33, 57, 58, 64, 70-73). In summary, virus titers in nasal or ocular swabs collected from calves infected with the 51g mutant virus were reduced at least 2 logs compared to those for calves infected with 51gR.
FIG. 6.
Virus shedding from the nasal cavity during acute infection. Calves were infected with the designated virus as described in Materials and Methods. At the indicated days after infection, nasal swabs were collected from calves infected with the 51g mutant or 51gR. Infectious virus present in swabs was quantified by infecting CRIB cells.
DEX induces reactivation from latency of 51gR but not 51g.
BHV-1 can consistently be reactivated from latently infected calves or rabbits following treatment with DEX (36, 37, 39, 40). To test whether calves latently infected with the 51g mutant or 51gR reactivate from latency, calves were given three injections of DEX, one intravenous injection and two intramuscular injections, to initiate reactivation from latency. For these studies, the calves had been infected for 45 days, they exhibited no signs of clinical disease, they were not shedding infectious virus, and thus they were operationally defined as being latently infected. Infectious virus was detected in nasal and ocular swabs of 1/3 calves latently infected with 51gR 2 days after DEX treatment (Table 1). From 5 to 14 days after the initial DEX injection, infectious virus was detected in ocular or nasal swabs of 3/3 calves latently infected with 51gR (Table 1). No infectious virus was detected in ocular or nasal swabs after DEX treatment of calves latently infected with the 51g mutant virus (Table 1).
TABLE 1.
Virus shedding from ocular and nasal cavities after DEX-induced reactivationa
| Virus (no. of calves) | No. of calves with infectious virus detected/total no. of calves tested
|
||||
|---|---|---|---|---|---|
| Day 0 | Day 2 | Day 5 | Day 7 | Day 14 | |
| 51gR (3) | 0/3 | 1/3 | 3/3 | 3/3 | 3/3 |
| 51g (4) | 0/4 | 0/4 | 0/4 | 0/4 | 0/4 |
As described in Materials and Methods, ocular or nasal swabs were collected from each calf at the designated times after the initial intravenous injection of DEX. Infectious virus in swabs was detected by inoculating CRIB cells with a 1:4 dilution of the swab medium solution. The results shown are from two different experiments. When the cultures were positive for nasal swabs, they were also positive for ocular swabs, and thus the results were combined.
DISCUSSION
In this study, we used a virulent BAC clone containing wt BHV-1 to construct a zinc RING finger mutant that contains a glycine instead of a cysteine at the 51st residue (the 51g mutant). Relative to the rescued virus (51gR), the 51g mutant virus did not grow efficiently in calves or cultured cells, had reduced clinical symptoms in calves, and did not reactivate from latency. However, the 51g mutant, unlike a bICP0 null mutant previously characterized (21), formed plaques in cultured bovine cells. Although the plaques produced by the 51g mutant appeared to be larger than those produced by wt BHV-1 or 51gR, they took longer to appear and they did not spread and fuse with other plaques. A previous study demonstrated that a bICP0 null mutant virus was unable to inhibit UV-induced apoptosis following infection of bovine kidney cells, whereas the rescued virus or wt BHV-1 efficiently inhibited UV light-induced apoptosis (20). Furthermore, the bICP0 null mutant appeared to induce autophagy in bovine kidney cells. These observations suggested that bICP0 inhibits cell death during productive infection, perhaps because bICP0 transactivates viral or cellular promoters that interfere with virus-induced cell death. We suggest that the 51g mutant induced atypical plaques in CRIB cells in part because the mutant prematurely induced cell death or because additional cell death pathways were activated.
When HSV-1-encoded ICP0 is compared to bICP0, the zinc RING finger is the only well-conserved domain (74). ICP0 RING finger mutants generally exhibit impaired replication, delays in early or late gene expression, and reduced plaque-forming efficiency at a low MOI (reviewed in reference 14). Previous studies indicated that mutations in the 13th and 51st aa of bICP0, which includes two cysteine residues within the RING finger, eliminated the transactivation potential of bICP0 (34, 76) and its ability to inhibit IFN-dependent transcription (28, 63). The ability of the 51g mutant virus to grow in cells pretreated with imiquimod was greatly reduced compared to that of 51gR, which is consistent with studies demonstrating that HSV-1 ICP0 mutants are hypersensitive to the effects of IFN (50). The reduced growth of the 51g mutant after imiquimod treatment could also be due to (i) the overall reduced growth potential of the 51g mutant or (ii) the wt bICP0 (but not the 51g mutant protein) transactivated expression of a viral or cellular gene that promotes growth following imiquimod treatment. In conclusion, these studies demonstrate that the ability of bICP0 to directly or indirectly stimulate growth of BHV-1 in cultured bovine cells correlates with the virulence and growth of BHV-1 in infected calves.
Viral DNA was detected in tonsils and trigeminal ganglia of calves latently infected with the 51g mutant (data not shown), suggesting that the 51g mutant virus established latency. Additional studies are in progress to determine whether there are differences in viral gene expression in trigeminal ganglia of calves infected with the 51g mutant versus 51gR or whether the 51g mutant establishes latency at a reduced frequency. Calves latently infected with the 51g mutant did not reactivate from latency following DEX treatment (Table 1), as judged by virus shedding from ocular or nasal cavities. Although one could argue that we missed shedding of the 51g mutant virus during the course of reactivation, we were unable to detect an increase in virus-specific neutralizing antibodies after calves latently infected with the 51g mutant were treated with DEX three times to stimulate reactivation from latency (62). A marked increase in virus-specific neutralizing antibodies is a sensitive method to monitor BHV-1 shedding during reactivation from latency (32, 36-38). After DEX treatment, calves latently infected with 51gR shed virus in ocular or nasal swabs (Table 1), and titers of virus-specific antibodies increased. Several studies have concluded that HSV-1 ICP0 is important for reactivation from latency in mouse models of infection (25, 26, 43). In the context of an in vitro model for latency, reactivation of gene expression from quiescent HSV-1 genomes can occur in the absence of ICP0 if the cells are highly stressed and if the proper MOI is used (49, 60, 66). By use of an in vivo mouse model in which reactivation from latency is stimulated by hyperthermic stress (66), it was shown that ICP0 is not required for initiating lytic gene expression but that ICP0 is important for detecting infectious virus. At this point, our studies do not allow us to discriminate whether bICP0 was necessary for initiation of reactivation from latency or for production of infectious virus during reactivation. Regardless of the mechanism, wt bICP0 expression was important for viral replication during acute infection and for virus shedding in latently infected calves following DEX-induced reactivation from latency.
During transient transfection (34, 76) or productive infection (Fig. 2A), high levels of bICP0 were detected in cells expressing bICP0 proteins containing zinc RING finger mutants. It is well established that HSV-1 ICP0 zinc RING finger mutants have increased half-lives because they cannot induce their own ubiquitination (3, 8, 12). Consequently, we suggest that bICP0 regulates its own half-life by self-ubiquitination. In support of this hypothesis, transient-transfection assays and cell-free assays indicated that the bICP0 protein possesses E3 ubiquitin ligase activity and that the zinc RING finger is crucial for this activity (11). The 51g mutation is predicted to disrupt the structure of the zinc RING finger and the putative E3 ubiquitin ligase activity of bICP0 because mutation of a single amino acid within HSV-1 ICP0 disrupts the secondary and tertiary structures of the zinc RING finger (12). Studies designed to test whether the 51g mutant has impaired E3 ubiquitin ligase activity are under way.
Acknowledgments
This work was supported primarily by two USDA grants (08-00891 and 06-01627) to C.J. and two USDA grants to S.C. (04-35204-14657 and 0735204-17358). In addition, a NIAID grant to C.J. (R21AI069176) and a COBRE grant to the NE Center for Virology (1P20RR15635) supported certain aspects of these studies. N.G. was partially supported by an NIH Ruth L. Kirschstein fellowship (T32 AI060547).
Footnotes
Published ahead of print on 8 October 2008.
REFERENCES
- 1.Boutell, C., and R. D. Everett. 2003. The herpes simplex virus type 1 (HSV-1) regulatory protein ICP0 interacts with and ubiquitinates p53. J. Biol. Chem. 27836596-36602. [DOI] [PubMed] [Google Scholar]
- 2.Boutell, C., M. Canning, A. Orr, and R. D. Everett. 2005. Reciprocal activities between herpes simplex virus type 1 regulatory protein ICP0, a ubiquitin E3 ligase, and ubiquitin-specific protease USP7. J. Virol. 7912342-12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowles, D. E., V. R. Holden, Y. Zhao, and D. J. O'Callaghan. 1997. The ICP0 protein of equine herpesvirus 1 is an early protein that independently transactivates expression of all classes of viral promoters. J. Virol. 714904-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowles, D. E., S. K. Kim, and D. J. O'Callaghan. 2000. Characterization of the trans-activation properties of equine herpesvirus 1 EICP0 protein. J. Virol. 741200-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brukman, A., and L. W. Enquist. 2006. Pseudorabies virus EP0 protein counteracts an interferon-induced antiviral state in a species-specific manner. J. Virol. 8010871-10873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai, W., and P. A. Schaffer. 1992. Herpes simplex virus type 1 ICP0 regulates expression of immediate-early, early, and late genes in productively infected cells. J. Virol. 662904-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Canning, M., C. Boutell, J. Parkinson, and R. D. Everett. 2004. A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. J. Biol. Chem. 27938160-38168. [DOI] [PubMed] [Google Scholar]
- 9.Carter, J. J., A. D. Weinberg, A. Pollard, R. Reeves, J. A. Magnuson, and N. S. Magnuson. 1989. Inhibition of T-lymphocyte mitogenic responses and effects on cell functions by bovine herpesvirus 1. J. Virol. 631525-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Devireddy, L. R., and C. Jones. 1999. Activation of caspases and p53 by bovine herpesvirus 1 infection results in programmed cell death and efficient virus release. J. Virol. 733778-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diao, L., B. Zhang, J. Fan, X. Gao, S. Sun, K. Yang, D. Xin, N. Jin, Y. Geng, and C. Wang. 2005. Herpes virus proteins ICP0 and BICP0 can activate NF-kB by catalyzing IkBa ubiquitination. Cell. Signal. 17217-229. [DOI] [PubMed] [Google Scholar]
- 12.Everett, R., P. O'Hare, D. O'Rourke, P. Barlow, and A. Orr. 1995. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J. Virol. 697339-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett, R. D. 1988. Analysis of the functional domains of herpes simplex virus type 1 immediate-early polypeptide Vmw110. J. Mol. Biol. 20287-96. [DOI] [PubMed] [Google Scholar]
- 14.Everett, R. D. 2000. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays 22761-770. [DOI] [PubMed] [Google Scholar]
- 15.Everett, R. D., P. Barlow, A. Milner, B. Luisi, A. Orr, G. Hope, and D. Lyon. 1993. A novel arrangement of zinc-binding residues and secondary structure in the C3HC4 motif of an alpha herpes virus protein family. J. Mol. Biol. 2341038-1047. [DOI] [PubMed] [Google Scholar]
- 16.Everett, R. D., W. C. Earnshaw, J. Findlay, and P. Lomonte. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 181526-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everett, R. D., P. Lomonte, T. Sternsdorf, R. van Driel, and A. Orr. 1999. Cell cycle regulation of PML modification and ND10 composition. J. Cell Sci. 1124581-4588. [DOI] [PubMed] [Google Scholar]
- 18.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 161519-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fraefel, C., J. Zeng, Y. Choffat, M. Engels, M. Schwyzer, and M. Ackermann. 1994. Identification and zinc dependence of the bovine herpesvirus 1 transactivator protein BICP0. J. Virol. 683154-3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geiser, V., S. Rose, and C. Jones. 2008. The bovine herpes virus 1 bICP0 protein regulates toxicity in a cell type dependent fashion. Mol. Pathog. 44459-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geiser, V., Y. Zhang, and C. Jones. 2005. Characterization of a BHV-1 strain that does not express the major regulatory protein, bICP0. J. Gen. Virol. 861987-1996. [DOI] [PubMed] [Google Scholar]
- 22.Griebel, P., H. B. Ohmann, M. J. Lawman, and L. A. Babiuk. 1990. The interaction between bovine herpesvirus type 1 and activated bovine T lymphocytes. J. Gen. Virol. 71369-377. [DOI] [PubMed] [Google Scholar]
- 23.Griebel, P., L. Qualtiere, W. C. Davis, A. Gee, H. Bielefeldt Ohmann, M. J. Lawman, and L. A. Babiuk. 1987. T lymphocyte population dynamics and function following a primary bovine herpesvirus type-1 infection. Viral Immunol. 1287-304. [DOI] [PubMed] [Google Scholar]
- 24.Griebel, P. J., L. Qualtiere, W. C. Davis, M. J. Lawman, and L. A. Babiuk. 1987. Bovine peripheral blood leukocyte subpopulation dynamics following a primary bovine herpesvirus-1 infection. Viral Immunol. 1267-286. [DOI] [PubMed] [Google Scholar]
- 25.Halford, W. P., C. D. Kemp, J. A. Isler, D. J. Davido, and P. A. Schaffer. 2001. ICP0, ICP4, or VP16 expressed from adenovirus vectors induces reactivation of latent herpes simplex virus type 1 in primary cultures of latently infected trigeminal ganglion cells. J. Virol. 756143-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halford, W. P., and P. A. Schaffer. 2001. ICP0 is required for efficient reactivation of herpes simplex virus type 1 from neuronal latency. J. Virol. 753240-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hariharan, M. J., C. Nataraj, and S. Srikumaran. 1993. Down regulation of murine MHC class I expression by bovine herpesvirus 1. Viral Immunol. 6273-284. [DOI] [PubMed] [Google Scholar]
- 28.Henderson, G., Y. Zhang, and C. Jones. 2005. The bovine herpesvirus 1 gene encoding infected cell protein 0 (bICP0) can inhibit interferon-dependent transcription in the absence of other viral genes. J. Gen. Virol. 862697-2702. [DOI] [PubMed] [Google Scholar]
- 29.Henderson, G., Y. Zhang, M. Inman, D. Jones, and C. Jones. 2004. The infected cell protein 0 encoded by bovine herpes virus 1 (bICP0) can activate caspase 3 when over-expressed in transfected cells. J. Gen. Virol. 853511-3516. [DOI] [PubMed] [Google Scholar]
- 30.Hinkley, S., A. B. Hill, and S. Srikumaran. 1998. Bovine herpesvirus-1 infection affects the peptide transport activity in bovine cells. Virus Res. 5391-96. [DOI] [PubMed] [Google Scholar]
- 31.Hohle, C., A. Karger, P. Konig, K. Glesow, and G. M. Keil. 2005. High-level expression of biologically active bovine alpha interferon by bovine herpesvirus 1 interferes only marginally with recombinant virus replication in vitro. J. Gen. Virol. 862685-2695. [DOI] [PubMed] [Google Scholar]
- 32.Inman, M., L. Lovato, A. Doster, and C. Jones. 2002. A mutation in the latency-related gene of bovine herpesvirus 1 interferes with the latency reactivation cycle of latency in calves. J. Virol. 766771-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Inman, M., L. Lovato, A. Doster, and C. Jones. 2001. A mutation in the latency-related gene of bovine herpesvirus 1 leads to impaired ocular shedding in acutely infected calves. J. Virol. 758507-8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Inman, M., Y. Zhang, V. Geiser, and C. Jones. 2001. The zinc ring finger in the bICP0 protein encoded by bovine herpes virus-1 mediates toxicity and activates productive infection. J. Gen. Virol. 82483-492. [DOI] [PubMed] [Google Scholar]
- 35.Ishmael, W. 2001. Gasping for dollars. Angus Beef Bulletin. www.mycattle.com/health/updates/gaspingfordollars.
- 36.Jones, C. 1998. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv. Virus Res. 5181-133. [DOI] [PubMed] [Google Scholar]
- 37.Jones, C. 2003. Herpes simplex virus type 1 and bovine herpesvirus 1 latency. Clin. Microbiol. Rev. 1679-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones, C., T. J. Newby, T. Holt, A. Doster, M. Stone, J. Ciacci-Zanella, C. J. Webster, and M. W. Jackwood. 2000. Analysis of latency in cattle after inoculation with a temperature sensitive mutant of bovine herpesvirus 1 (RLB106). Vaccine 183185-3195. [DOI] [PubMed] [Google Scholar]
- 39.Jones, C., V. Geiser, G. Henderson, Y. Jiang, F. Meyer, S. Perez, and Y. Zhang. 2006. Functional analysis of bovine herpesvirus 1 (BHV-1) genes expressed during latency. Vet. Microbiol. 113199-210. [DOI] [PubMed] [Google Scholar]
- 40.Jones, C., and S. Chowdhury. 2007. A review of the biology of bovine herpesvirus type 1 (BHV-1), its role as a cofactor in the bovine respiratory disease complex, and development of improved vaccines. Adv. Anim. Health 8187-205. [DOI] [PubMed] [Google Scholar]
- 41.Kapil, S., and R. J. Basaraba. 1997. Infectious bovine rhinotracheitis, parainfluenza-3, and respiratory coronavirus. Bovine respiratory disease update. Vet. Clin. North Am. Food Anim. Pract. 13455-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lees-Miller, S. P., M. C. Long, M. A. Kilvert, V. Lam, S. A. Rice, and C. A. Spencer. 1996. Attenuation of DNA-dependent protein kinase activity and its catalytic subunit by the herpes simplex virus type 1 transactivator ICP0. J. Virol. 707471-7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leib, D. A., D. M. Coen, C. L. Bogard, K. A. Hicks, D. R. Yager, D. M. Knipe, K. L. Tyler, and P. A. Schaffer. 1989. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J. Virol. 63759-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu, S. F., M. C. S. Brum, A. Doster, C. Jones, and S. I. Chowdhury. 2008. A bovine herpesvirus type 1 mutant virus specifying a carboxyl-terminal truncation of glycoprotein E is defective in anterograde neuronal transport in rabbits and calves. J. Virol. 827432-7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lium, E. K., and S. Silverstein. 1997. Mutational analysis of the herpes simplex virus type 1 ICP0 C3HC4 zinc ring finger reveals a requirement for ICP0 in the expression of the essential α27 gene. J. Virol. 718602-8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 751223-1233. [DOI] [PubMed] [Google Scholar]
- 47.Maul, G. G., H. H. Guldner, and J. G. Spivack. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0). J. Gen. Virol. 742679-2690. [DOI] [PubMed] [Google Scholar]
- 48.Megyeri, K., W.-C. Au, I. Rosztoczy, N. B. K. Raj, R. L. Miller, M. A. Tomai, and P. M. Pitha. 1995. Stimulation of interferon and cytokine gene expression by imiquimod and stimulation by Sendai virus utilize similar signal transduction pathways. Mol. Cell. Biol. 152207-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller, C. S., R. J. Danaher, and R. J. Jacob. 2006. ICP0 is not required for efficient stress-induced reactivation of herpes simplex virus type 1 from cultured quiescently infected neuronal cells. J. Virol. 803360-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mossman, K. L., H. A. Saffran, and J. R. Smiley. 2000. Herpes simplex virus ICP0 mutants are hypersensitive to interferon. J. Virol. 742052-2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mossman, K. L., P. F. Macgregor, J. J. Rozmus, A. B. Goryachev, A. M. Edwards, and J. R. Smiley. 2001. Herpes simplex virus triggers and then disarms a host antiviral response. J. Virol. 75750-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mossman, K. L., and J. R. Smiley. 2002. Herpes simplex virus ICP0 and ICP34.5 counteract distinct interferon-induced barriers to virus replication. J. Virol. 761995-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nataraj, C., S. Eidmann, M. J. Hariharan, J. H. Sur, G. A. Perry, and S. Srikumaran. 1997. Bovine herpesvirus 1 downregulates the expression of bovine MHC class I molecules. Viral Immunol. 1021-34. [DOI] [PubMed] [Google Scholar]
- 54.Parkinson, J., and R. D. Everett. 2000. Alphaherpesvirus proteins related to herpes simplex virus type 1 ICP0 affect cellular structures and proteins. J. Virol. 7410006-10017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parkinson, J., S. P. Lees-Miller, and R. D. Everett. 1999. Herpes simplex virus type 1 immediate-early protein Vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J. Virol. 73650-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perez, S., F. Meyer, K. Saira, A. Doster, and C. Jones. 2008. Premature expression of the latency-related RNA encoded by bovine herpesvirus 1 correlates with higher levels of beta interferon RNA expression in productively infected cells. J. Gen. Virol. 891338-1345. [DOI] [PubMed] [Google Scholar]
- 57.Perez, S., L. Lovato, J. Zhou, A. Doster, and C. Jones. 2006. Comparison of inflammatory infiltrates in trigeminal ganglia of cattle infected with wild type BHV-1 versus a virus strain containing a mutation in the LR (latency-related) gene. J. Neurovirol. 12392-397. [DOI] [PubMed] [Google Scholar]
- 58.Perez, S., M. Inman, A. Doster, and C. Jones. 2005. Latency-related gene encoded by bovine herpesvirus 1 promotes virus growth and reactivation from latency in tonsils of infected calves. J. Clin. Microbiol. 43393-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Powell, J. 2005. Bovine respiratory disease. FSA 3082. University of Arkansas Division of Agriculture Cooperative Extension Service, Fayetteville, AR.
- 60.Preston, C. M. 2007. Reactivation of expression from quiescent herpes simplex virus type 1 genomes in the absence of immediate-early protein ICP0. J. Virol. 8111781-11789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rock, D., J. Lokensgard, T. Lewis, and G. Kutish. 1992. Characterization of dexamethasone-induced reactivation of latent bovine herpesvirus 1. J. Virol. 662484-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saira, K. 2008. Functional analysis of the bovine herpesvirus-1 gene encoding bICP0, a promiscuous trans-activator, that stimulates productive infection and interferon signaling pathways. Ph.D. dissertation. University of Nebraska, Lincoln.
- 63.Saira, K., Y. Zhou, and C. Jones. 2007. The infected cell protein 0 encoded by bovine herpesvirus 1 (bICP0) induces degradation of interferon response factor 3 and, consequently, inhibits beta interferon promoter activity. J. Virol. 813077-3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schang, L., and C. Jones. 1997. Analysis of bovine herpesvirus 1 transcripts during a primary infection of trigeminal ganglia of cattle. J. Virol. 716786-6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheffy, B. E., and D. H. Davies. 1972. Reactivation of a bovine herpesvirus after corticosteroid treatment. Proc. Soc. Exp. Biol. Med. 140974-976. [DOI] [PubMed] [Google Scholar]
- 66.Thompson, R. L., and N. M. Sawtell. 2006. Evidence that the herpes simplex virus type 1 ICP0 protein does not initiate reactivation from latency in vivo. J. Virol. 8010919-10930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tikoo, S. K., M. Campos, and L. A. Babiuk. 1995. Bovine herpesvirus 1 (BHV-1): biology, pathogenesis, and control. Adv. Virus Res. 45191-223. [DOI] [PubMed] [Google Scholar]
- 68.Tischer, B. K., J. Von Einem, B. Kaufer, and N. Osterrieder. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. BioTechniques 40191-197. [DOI] [PubMed] [Google Scholar]
- 69.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin-conjugating enzyme, and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 988815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winkler, M. T., A. Doster, J. H. Sur, and C. Jones. 2002. Analysis of bovine trigeminal ganglia following infection with bovine herpesvirus 1. Vet. Microbiol. 86139-155. [DOI] [PubMed] [Google Scholar]
- 71.Winkler, M. T., A. Doster, and C. Jones. 1999. Bovine herpesvirus 1 can infect CD4+ T lymphocytes and induce programmed cell death during acute infection of cattle. J. Virol. 738657-8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winkler, M. T., L. S. Schang, A. Doster, T. Holt, and C. Jones. 2000. Analysis of cyclins in trigeminal ganglia of calves infected with bovine herpesvirus-1. J. Gen. Virol. 812993-2998. [DOI] [PubMed] [Google Scholar]
- 73.Winkler, M. T. C., A. Doster, and C. Jones. 2000. Persistence and reactivation of bovine herpesvirus 1 in the tonsils of latently infected calves. J. Virol. 745337-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wirth, U. V., C. Fraefel, B. Vogt, C. Vlcek, V. Paces, and M. Schwyzer. 1992. Immediate-early RNA 2.9 and early RNA 2.6 of bovine herpesvirus 1 are 3′ coterminal and encode a putative zinc finger transactivator protein. J. Virol. 662763-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wirth, U. V., B. Vogt, and M. Schwyzer. 1991. The three major immediate-early transcripts of bovine herpesvirus 1 arise from two divergent and spliced transcription units. J. Virol. 65195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang, Y., and C. Jones. 2005. Identification of functional domains within the bICP0 protein encoded by BHV-1. J. Gen. Virol. 86879-886. [DOI] [PubMed] [Google Scholar]