Abstract
The neurotropic virus Borna disease virus (BDV) persists in the central nervous systems of a wide variety of vertebrates and causes behavioral disorders. BDV represents an intriguing example of a virus whose persistence in neurons leads to altered brain function in the absence of overt cytolysis and inflammation. The bases of BDV-induced behavioral impairment remain largely unknown. To better characterize the neuronal response to BDV infection, we compared the proteomes of primary cultures of cortical neurons with and without BDV infection. We used two-dimensional liquid chromatography fractionation, followed by protein identification by nanoliquid chromatography-tandem mass spectrometry. This analysis revealed distinct changes in proteins implicated in neurotransmission, neurogenesis, cytoskeleton dynamics, and the regulation of gene expression and chromatin remodeling. We also demonstrated the selective interference of BDV with processes related to the adaptative response of neurons, i.e., defects in proteins regulating synaptic function, global rigidification of the cytoskeleton network, and altered expression of transcriptional and translational repressors. Thus, this work provides a global view of the neuronal changes induced by BDV infection together with new clues to understand the mechanisms underlying the selective interference with neuronal plasticity and remodeling that characterizes BDV persistence.
The analysis of the response of a host cell to a pathogenic microorganism represents a daunting task, as it often results in complex and numerous changes in gene expression (37). Generally, these changes strongly depend on the nature of the pathogen interaction with its host. In the case of the central nervous system (CNS), the deleterious consequences of viral infection are often due to the cytopathic nature of viral replication (25, 38), or, alternatively, they can result from the immune response to the virus (31). However, some viruses can also persist in the CNS and cause diseases without an overt cytopathic effect or inflammation (1). These viral models provide a unique opportunity to unravel the molecular mechanisms underlying virus-induced neuronal dysfunction. A better understanding of the pathological consequences of viral persistence in the CNS may help to shed light on the pathogenesis of many neurological diseases of unclear etiology where viruses are thought to play a role (34, 47).
Infection with Borna disease virus (BDV) represents an ideal paradigm for the investigation of the neuronal consequences due to the persistence of a noncytolytic virus. BDV is an enveloped virus with a nonsegmented, negative-strand RNA genome (13, 44). BDV infects a wide variety of mammals (35), possibly including humans (6, 29). Infected hosts develop a large spectrum of neurological disorders, ranging from immune-mediated diseases to behavioral alterations without inflammation (35, 41), reminiscent of symptoms observed in certain human neuropsychiatric diseases (28). These neurobehavioral manifestations reflect the selective localization of BDV in the CNS. The virus targets mainly neurons of the cortex and hippocampus (20, 23), which governs many cognitive and behavioral functions (8).
One striking feature of BDV infection is its noncytolytic strategy of replication (20) in vivo and in vitro. Indeed, many studies using cells infected with BDV, either primary neuronal cells or established cell lines, have repeatedly shown that infection proceeds without any overt phenotype or impaired survival (reviewed in reference 21). However, when appropriately stimulated, BDV-infected neurons exhibit selective impairment in signaling pathways that are important for proper neuronal functioning and neuronal communication (21, 22, 26, 34, 50, 51). Together, these results imply that some biochemical pathways in neurons must actually be targeted by the infection, even at steady state, but have not been detected with the resolution of the phenotypic analyses performed so far. To address this question more thoroughly, an unbiased and comprehensive analysis of BDV-infected neurons was needed.
The recent development of improved proteomic methods has greatly enhanced our ability to assess cellular changes at a global scale, and these methods are very well suited for the characterization of the diversity of cellular responses to a virus (37, 45). Here, we fractionated protein extracts from uninfected and BDV-infected primary cultures of neurons using two-dimensional liquid chromatography (2D-LC). Thereafter, the identity of the proteins present in fractions differing in profile between samples was determined by nano-liquid chromatography (nanoLC)-tandem mass spectrometry (MS/MS). Even using such a proteomic approach, we did not detect any change in the expression of markers for neuronal stress, apoptosis, or neurodegeneration, further confirming the remarkable noncytolytic replication of BDV. Nevertheless, we identified changes at different levels that provide new clues to an understanding of the mechanisms underlying the selective interference with neuronal plasticity and remodeling, which characterizes BDV persistence.
MATERIALS AND METHODS
Primary cultures and viral infection.
Cerebral cortical neurons were prepared from embryonic Sprague-Dawley rats at gestational day 18 and maintained in serum-free Neurobasal medium (Invitrogen, Cergy-Pontoise, France) supplemented with 0.5 mM glutamine and 2% B-27 supplement (Invitrogen), as described previously (3). One day after the plating of neurons, half of the culture was infected with cell-free BDV (strain He/80) (105 focus-forming units/ml), while the other half was maintained as a mock control sample. By 13 days postinfection, the efficacy of BDV infection was verified by immunofluorescence for each experiment using an anti-BDV nucleoprotein serum. Neuronal cultures contained more than 90% neurons, as assessed by staining with neuron-specific markers, and all neurons were infected at the time when experiments were performed (3).
Preparation of the protein samples.
Proteins were extracted from neurons according to the ProteomeLab PF2D protocol (Beckman Coulter, Fullerton, CA). Samples were first desalted on a PD-10 Sephadex G-25 gel filtration column with a 5-kDa cutoff (Amersham Biosciences, Piscataway, NJ) and eluted using chromatofocusing start buffer. Protein concentrations were determined in duplicate using the BCA protein assay (Thermo Scientific, Pierce, Rockford, IL).
Protein fractionation by 2D-LC.
Control and infected neuronal protein samples were fractionated using the ProteomeLab PF2D protein fractionation system (Beckman Coulter), which consists of two high-performance liquid chromatographs, two UV detectors, an autosampler, and a fraction collector. The first-dimension fractionation of PF2D, which consists of chromatofocusing, was performed on a high-performance chromatofocusing first-dimension column (250 by 2.1 mm; Beckman Coulter). The pH gradient was generated using Start buffer (pH 8.5) and eluent buffer (pH 4), both included in the ProteomeLab PF2D kit. The chromatofocusing column was first equilibrated with start buffer at pH 8.5 before being loaded with 2 ml (3 mg) of the protein extract. The flowthrough was collected, and after a stable baseline was established, a pH gradient was generated by infusing the eluent buffer with a constant flow rate of 0.2 ml/min, allowing the elution of proteins according to their pIs. The proteins with a pI of <4 were finally eluted by washing the column with 1 M NaCl. The second dimension of PF2D used reverse-phase high-performance liquid chromatography (HPLC) on a C18 column (4.6 by 33 mm; Beckman Coulter) packed with 1.5-μm nonporous silica and kept at 50°C in a heated column jacket. The fractions from the first dimension were injected in the column and eluted using a water-acetonitrile gradient at 0.75 ml/min. The second-dimension fractions were collected in 96-well plates and stored at −80°C until use. The second-dimension fractions differing in profile between control and infected samples were selected for MS analysis. This selection was carried out by performing a peak-to-peak analysis of the UV chromatograms. After subtraction of the baseline, a peak was selected if it was present only in one sample (with a minimal value of 0.02 arbitrary units [AU]) or if its difference in intensity between samples was at least twofold.
Treatment of fractions for proteomic analysis.
Selected fractions were adjusted with variable volumes of HPLC-grade water (depending on the gradient of pH and the water-to-acetonitrile ratio) to ensure the same volume of water in all samples before evaporation. Fractions were concentrated using a SpeedVac concentrator (Jouan, Saint-Herblain, France) at 40°C until there was 50 μl left. One M NH4HCO3 and 10 mM dithiothreitol were added to the concentrate (to final concentrations of 100 mM and 1 mM, respectively) to neutralize the concentrate to pH 7.8, and samples were reduced for 10 min at 60°C. Sequencing-grade modified trypsin (Promega, Charbonnières, France) was added (100 ng for peaks of <0.05 AU and 200 ng for the others) and incubated for 24 h at 37°C. Next, fractions were completely evaporated in a SpeedVac concentrator at room temperature, and plates were stored at −80°C prior to MS analysis.
nanoLC-MS/MS analysis.
The resulting peptides were analyzed by nanoLC-MS/MS using an LC Packings system (Dionex, Amsterdam, The Netherlands) coupled to a QStar XL mass spectrometer (Applied Biosystems, Foster City, CA). Dried peptides were reconstituted in 14 μl of solvent A′ (5% acetonitrile [ACN] and 0.05% trifluoroacetic acid in HPLC-grade water), and 5 μl was loaded onto a precolumn (300-μm internal diameter [ID] by 5 mm) using the Switchos unit of the LC Packings system, delivering a flow rate of 20 μl/min of solvent A′. After desalting for 7 min, the precolumn was switched online with the analytical column (75-μm-ID by 15-cm PepMap C18 column) equilibrated in 95% solvent A (5% ACN and 0.1% formic acid in HPLC-grade water) and 5% solvent B (95% ACN and 0.1% formic acid in HPLC-grade water). Peptides were eluted from the precolumn to the analytical column and then to the mass spectrometer with a gradient from 5 to 50% solvent B for 60 min. The QStar XL mass spectrometer was operated in information-dependent acquisition mode with Analyst QS 1.1 software. MS and MS/MS data were recorded continuously with a 7-s cycle time. Within each cycle, MS data were accumulated for 1 s over the mass range m/z 300 to 2,000, followed by two MS/MS acquisitions of 3 s each on the two most abundant ions over the mass range m/z 80 to 2,000. Dynamic exclusion was employed within 60 s to prevent the repetitive selection of the same ions. Collision energies were automatically adjusted according to the charge state and mass value of the precursor ions. The MS-to-MS/MS switch threshold was set to 10 counts.
Database search and data analysis.
Mascot server software (version 2.2.0; Matrix Science, London, United Kingdom) was used to perform database searches in batch mode (Mascot Daemon) with all the wiff files acquired for each plate well. A peak list was created for each fraction analyzed, and individual Mascot searches were performed for each fraction. Data were searched against a homemade database containing mammal entries of the Sprot-Trembl_20071010 database (240,989 sequences) in which the six protein sequences from BDV were added manually. Oxidation of methionines was set as a variable modification for all Mascot searches. The specificity of trypsin digestion was set for cleavage after K or R, and two missed trypsin cleavage sites were allowed. Mascot results were parsed, and protein hits were automatically validated with the in-house-developed software Mascot File Parsing and Quantification (MFPaQ), version 3.0.4 (7). From all the validated result files corresponding to the wells of one specific PF2D plate, MFPaQ was used to generate a unique, nonredundant list of proteins found in different wells by creating clusters of protein groups (composed of all the protein sequences matching the same set of peptides). The lists obtained for each plate can be found in the supplemental material. A limited search was performed on specific fractions to study the impact of BDV on posttranslational modifications of histone 2B (H2B). Methylation, dimethylation, trimethylation, and acetylation of lysines and arginines were set as new variable modifications for this Mascot search.
Bioinformatic pathway analysis.
All identified proteins were evaluated by Ingenuity Pathways Analysis (IPA; Ingenuity Systems, Mountain View, CA). IPA constructs hypothetical protein interaction clusters on the basis of a regularly updated and a very large curated database that consists of millions of individual relationships between proteins collected from the literature. This database also integrates a broad range of systems biology including protein function, cellular localization, and small-molecule and disease interrelationships. For each identified network, IPA computes a p score (defined as −log10(P value) determined by Fischer's exact test) according to the fit of that network to the inputted proteins.
Antibodies and reagents.
We used mouse monoclonal antibodies to acetylated tubulin (clone 6-11B1), beta-tubulin (clone SDL.3D10; Sigma-Aldrich), growth-associated protein 43 (GAP-43) (clone GAP-7B10; Sigma-Aldrich), synapsin 1 (clone 46.1; Synaptic Systems), and tyrosinated tubulin (clone TUB-1A2; Sigma-Aldrich). We also used rabbit polyclonal antibodies to methyl CpG-binding protein 2 (MeCP2) (kindly provided by Z. Zhou and M. E. Greenberg, Harvard Medical School, Boston, MA); rabbit antibodies specific for synapsin 1 phosphorylated by CaMK II at site 3 (clone RU19) or by PKA and CaMK I at site 1 (clone G257), provided by P. Greengard (The Rockefeller University, New York, NY); stathmin (kindly provided by A. Sobel, Paris, France); Tau (Sigma-Aldrich); and Y-box protein 1 (YB1) (Cell Signaling Technology). All other antibodies were described previously (26). For F-actin staining, we used fluorescein isothiocyanate-coupled phalloidin (Sigma-Aldrich). For cytoskeleton depolymerization assays, we used nocodazole (diluted at 10 μg/ml in dimethyl sulfoxide; Sigma-Aldrich). Paclitaxel (Taxol; Sigma-Aldrich) was diluted at 10 μM in dimethyl sulfoxide.
Western blot analysis.
Cell extracts and Western blots were performed as described previously (26). Briefly, equivalent amounts of cell lysates were separated by electrophoresis using 10% Bis-Tris Nu-PAGE gels (Invitrogen) and then transferred onto nitrocellulose membranes (Hybond-C Extra; Amersham Biosciences). After blocking (Li-Cor blocking buffer or Tris-buffered saline containing 5% nonfat dry milk), membranes were incubated with primary antibodies. Secondary fluorescent antibodies used in experiments were as follows: IRDye 800CW goat anti-mouse immunoglobulin G (Li-Cor, ScienceTec, Les Ulis, France) and Alexa Fluor 680 goat anti-rabbit immunoglobulin G (Invitrogen). Laser scanning and quantitative analyses of the blots were performed using the Odyssey infrared imaging system (Li-Cor).
Recovery assays after cytoskeleton depolymerization and immunofluorescence analysis.
In order to reversibly depolymerize the neuronal cytoskeleton network, neuronal cultures were incubated for 1 h in medium containing 10 μg/ml nocodazole. After being rinsed three times in fresh medium, cultures were again incubated at 37°C for the indicated time intervals to allow the recovery of the cytoskeleton network. Thereafter, cells were incubated for 5 min in PHEM buffer [60 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2 (pH 6.9)] containing 10 mM paclitaxel at 37°C to remove unpolymerized tubulin and rinsed three times in warm PHEM buffer before fixation. Cultures were fixed by incubation for 20 min in warm 4% paraformaldehyde and 5 mM paclitaxel diluted in PHEM. Thereafter, samples were processed for immunofluorescence analysis as described previously (26).
Image acquisition and analysis.
Images were acquired using a confocal microscope (Axiovert 200 M; Zeiss, Le Pecq, France) equipped with a Leica DC 200 camera and either a 20× or a Plan-Apochromat 63×/1.40 oil objective (Zeiss). Acquisition and analysis were carried out using Zeiss-LSM 3.2 software.
RNA isolation and cDNA synthesis.
Total RNA was isolated from primary infected or control neuronal cultures using the RNeasy minikit (Qiagen, Courtaboeuf, France). Three to 10 μg of total RNA was primed with 2 μg of oligo(dT) (GE Healthcare, Vélizy, France) in the presence of 40 U of RNaseOUT recombinant RNase inhibitor (Invitrogen) and reverse transcribed with Moloney murine leukemia virus reverse transcriptase RNase H minus (300 U; Promega), according to the manufacturer's instructions. cDNAs were stored at −20°C until use.
Quantitative PCR.
Transcript levels of brain-derived neurotrophic factor (BDNF), phospholemman (FXYD1), and the housekeeping gene HPRT were quantified using real-time quantitative PCR (ABI Prism 7000; Perkin-Elmer Applied Biosystems) and Sybr green DNA dye from a Sybr green I reaction system (Eurogentec, Angers, France). Primer sequences were as follows: forward primer 5′-CCCAGTCTCTGCCTAGATCAAATGG-3′ and reverse primer 5′-ACTCGCACGCCTTCAGTGAGAA-3′ for BDNF (32), forward primer 5′-AGTGCAGAAGCTCCGCAGGAA-3′ and reverse primer 5′-TACCGCCTGCGGGTGGACAGA-3′ (14) for FXYD1, and forward primer 5′-TGTTGGATACAGGCCAGACTTTGT-3′ and reverse primer 5′-TCCACTTTCGCTGATGACACA-3′ (53) for HPRT.
In each case, sense and antisense primers (300 nM) were used in a final reaction mixture volume of 25 μl. Real-time quantitative PCR was performed according to the manufacturer's protocol, except for the hybridization of BDNF primers, for which a two-step hybridization was done at 55°C for 30 s and 60°C for 30 s. Quantitative analysis of the data was performed using the relative quantification (ΔΔCT) model, where the ΔΔCT is the difference between the ΔCT of the BDV-infected sample and that of the corresponding control sample. ΔCT is the difference between the threshold cycle (CT) of the target gene and the CT of the HPRT gene.
RESULTS
Experimental setup.
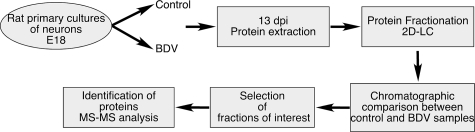
To analyze the impact of BDV persistence on the neuronal proteome, we applied 2D-LC fractionation to cellular extracts prepared from primary cultures of cortical neurons infected or not with BDV. These cells were chosen because they represent one of the main targets for BDV in vivo and because sufficient numbers of cortical neurons (70 million cells per experiment) can be obtained for a proteomic study. We used neurons that had been infected with BDV the day after plating and subsequently cultured for 13 days. At that time point, the neurons had established a mature network, and the virus had spread to all of them, in agreement with our previous findings (3, 26, 50). There was no detectable effect of the infection on the morphology or viability of the cultures, consistent with the noncytolytic replication strategy of BDV (3, 26, 50). After the fractionation step, we selected the fractions differing in profiles between control and BDV-infected samples, which were subsequently digested with trypsin prior to nanoLC-MS/MS analysis. The whole experimental process is schematically represented in Fig. 1.
FIG. 1.
Experimental outline. Whole-cell extracts were prepared from control and BDV-infected primary cultures of rat cortical neurons. Protein extracts were fractionated using automated 2D-LC, and chromatographic profiles of each fraction were compared between samples. Fractions differing in profile were selected and digested by trypsin before being subjected to identification by nanoLC-MS/MS. dpi, days postinfection.
PF2D-based fractionation of control and BDV-infected cortical neurons.
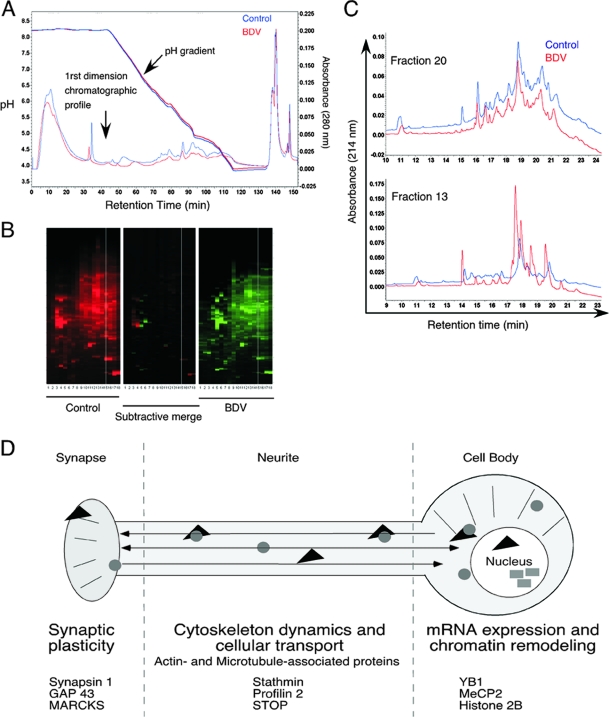
The control of the initial step of chromatofocusing fractionation is particularly important, as any variability in the pH gradient may lead to shifts in fractionation. Importantly, the pH gradients generated for the fractionation of control and BDV-infected samples were remarkably superposable (Fig. 2A), thereby ensuring that the observed differences in our study would not be drastically affected by shifts of pH during fractionation. Moreover, the pH gradients were also superposable when we compared the fractionations of BDV-infected neurons prepared in two independent experiments performed at a 3-month interval (data not shown). The maps summarizing the differences observed between control and BDV-infected neurons after the 2D-LC procedure are displayed in Fig. 2B and revealed clear differences between control and BDV-infected neurons. Two representative examples of comparative analyses are displayed in Fig. 2C. These two fractions were chosen within the pH gradient. As can be seen from these two examples, the differences between control and BDV-infected neurons were not evenly distributed along the fractions, with some fractions, such as fraction 20 (Fig. 2C, top) being almost identical in both cases. However, marked differences in other fractions, such as fraction 13, were noted (Fig. 2C, bottom). As stated above, we selected all peaks with an intensity of >0.02 AU for which at least a twofold difference was noted between samples. However, it should be stressed that the generation of quantitative data cannot be directly inferred by the analysis of the 2D-LC chromatograms, and further assessment and validation by other methods are necessary. Among the 600 fractions resulting from the separation, we selected 90 fractions from control and BDV-infected neurons (i.e., 180 in total), which were subjected to MS/MS identification.
FIG. 2.
Comparative analysis of the chromatograms between control and BDV-infected neuronal extracts. (A) Reproducibility of the pH gradients during the first dimension of automated 2D-LC. The pH gradients of the chromatofocusing steps for control (blue curves) and BDV-infected (red curves) samples as well as their chromatographic profiles are displayed on the same graph to illustrate the overlap between the two fractionation experiments. (B) Overview of the protein maps obtained after fractionation. The x axis of the 2D-LC map displays fractionation according to pI values, and the y axis displays increasing hydrophobicity. Each UV peak is represented by a horizontal band, whose intensity is proportional to the height of the peak. The center map is a differential map corresponding to a point-to-point subtraction of corresponding lanes from the control (red) and BDV-infected (green) maps, revealing fractions presenting differently expressed proteins between samples. (C) Representative examples of the comparison of 2D-LC chromatograms. Two chromatograms obtained after second-dimension fractionation using first-dimension fractions 20 (top) and 13 (bottom) are shown. Fraction 20 is representative of a chromatogram profile that was very similar between BDV and control samples and for which no fraction was selected for subsequent protein identification by MS. Fraction 13 is representative of a profile for which significant differences were noted. In this case, fractions collected between retention times of 17 and 21 min were selected for MS/MS analysis. (D) Schematic overview of the main neuronal proteome changes identified following infection of primary neurons with BDV. Changes were arbitrarily regrouped into three main functional categories, for which several examples of proteins are given.
Identification of differently expressed proteins by MS.
A total of 5 to 40 proteins were identified in each fraction subjected to nanoLC-MS/MS analysis. Most of the identified proteins were expressed by neurons and belonged to all subcellular compartments; i.e., some were cytosolic, membrane bound, enclosed, nuclear, or cytoskeletal proteins (Table 1). This finding suggests that the cell lysis and fractionation procedures had resulted in homogeneous protein extracts, representing all subcellular structures. We focused on the proteins with the highest scores, either in control or in BDV-infected samples, leading to the selection of a total of 87 nonredundant proteins (Table 1) (the full list of the identified proteins can be found in the supplemental material). Several proteins were identified in more than two nonsequential fractions, suggesting that these proteins may have potential posttranslational modifications (60). As an internal control, we verified the presence of BDV proteins among fractions from the infected samples. Indeed, four out of the six known BDV proteins (namely, the phosphoprotein, the X protein, the glycoprotein, and the matrix protein) were found in fractions originating from the BDV-infected neuronal extract (Table 1). In addition, a fifth viral protein, the nucleoprotein, was also identified in a fraction that had not been selected for MS/MS analysis by using Western blot analysis of all fractions from the first-dimension separation. We did not detect the expression of BDV polymerase either because it was not included in the fractions analyzed by MS/MS or because its large size and strong basic properties, together with its low level of expression, may have led to its hindrance by highly expressed neuronal proteins. The majority of identified neuronal proteins could be arbitrarily grouped into three categories (Fig. 2D): (i) proteins implicated in neurotransmission and neurogenesis, (ii) proteins involved in cytoskeleton dynamics, and (iii) proteins involved in the regulation of gene expression and modification in neurons, including factors regulating the local transport or modification of mRNA and nuclear proteins involved in transcriptional regulation and chromatin remodeling.
TABLE 1.
Database of identified proteins
| Protein | GenBank accession no. | Identification code | No. of peptidesb | Sequence coverage (%)b | No. of MS/MSb | Plate | Fraction(s)a | Scoreb |
|---|---|---|---|---|---|---|---|---|
| BVD proteins | ||||||||
| Envelope protein p57 precursor (G) | P52638 | VGLG_BDV | 1 | 1.6 | 2 | 3 | 17 (13-14) | 52.63 |
| Matrix protein (M) | P52637 | MATRX_BDV | 5 | 35.9 | 8 | 1 | 10 (19); 11 (19) | 111.54 |
| Matrix protein (M) | P52637 | MATRX_BDV | 2 | 17.6 | 8 | 2 | 13 (19-20); 14 (18-19); 15 (19) | 89.69 |
| Matrix protein (M) | P52637 | MATRX_BDV | 3 | 18.3 | 4 | 3 | 17 (19-20); 18 (18) | 73.15 |
| Matrix protein (M) | P52637 | MATRX_BDV | 4 | 26.8 | 5 | 5 | 25 (18) | 92.29 |
| Nucleoprotein (N) | Q01552 | NCAP_BDV | WBd | WBd | WBd | 2 | 16 | WBd |
| Phosphoprotein (P) | P26668 | PHOSP_BDV | 3 | 13.9 | 3 | 4 | 24 (21) | 30.31 |
| X protein | Q912Z9 | X_BDV | 1 | 9.2 | 1 | 1 | 9 (12); 11 (12) | 44.01 |
| Proteins involved in neurotransmission and neurogenesis | ||||||||
| AH receptor-interacting protein (AIP) | Q5FWY5 | AIP_RAT | 4 | 10.3 | 4 | 3 | 17 (16-17) | 56.76 |
| Alpha-2-macroglobulin receptor-associated precursor (Alpha-2-MRAP) | Q99068 | AMRP_RAT | 41 | 70.3 | 114 | 3 | 18 (14-15) | 939.8 |
| Alpha-2-MRAP | Q99068 | AMRP_RAT | 18 | 52.2 | 27 | 1 | 9 (15-16) | 201.64 |
| Amyloid-like protein 1 precursor (APLP) | Q03157 | APLP1_MOUSE | 6 | 10.9 | 6 | 5 | 31 (15) | 64.66 |
| BASP-1 (NAP22) | Q05175 | BASP_RAT | 3 | 15.9 | 3 | 4 | 23 (9-10) | 60.54 |
| Brain FABP | O15540 | FABPB_HUMANc | 1 | 6.8 | 1 | 4 | 22 (14) | 49.41 |
| Brain FABP | P55051 | FABPB_RAT | 7 | 63.6 | 26 | 1 | 9 (14); 10 (18) | 214.75 |
| Cholecystokinin precursor | P01355 | CCKN_RAT | 4 | 28.7 | 15 | 1 | 9 (11) | 58.58 |
| Complexin 1 | P63040 | CPLX1_MOUSEc | 13 | 70.1 | 41 | 5 | 25 (13-14) | 281.14 |
| Dihydropyrimidinase-related protein 5 (DRP-5) (ULIP6 protein) (CRMP-5) | Q9JHU0 | DPYL5_RAT | 3 | 6.9 | 7 | 1 | 11 (11-12) | 108.14 |
| Dihydropyrimidinase-related protein 5 (DRP-5) (ULIP6 protein) (CRMP-5) | Q9JHU0 | DPYL5_RAT | 33 | 51.2 | 146 | 3 | 17 (17-18-19-20) | 572.26 |
| DRP-2 | O02675 | DPYL2_BOVINc | 5 | 7.9 | 25 | 1 | 10 (13) | 172.44 |
| Glutamate dehydrogenase 1 mitochondrial precursor (GDH) | P26443 | DHE3_MOUSEc | 15 | 21.3 | 15 | 1 | 9 (18) | 175.53 |
| Glutamate dehydrogenase 1 mitochondrial precursor (GDH) | P26443 | DHE3_MOUSEc | 18 | 24.7 | 18 | 2 | 16 (18) | 321.84 |
| Glutamine synthetase | P09606 | GLNA_RAT | 16 | 29.2 | 17 | 3 | 17 (16) | 152.79 |
| MARCKS | P30009 | MARCS_RAT | 5 | 18.8 | 5 | 4 | 24 (9-10-11) | 89.83 |
| MARCKS | P30009 | MARCS_RAT | 12 | 55.7 | 24 | 5 | 25 (9-10) | 494.6 |
| MARCKS-related protein 1 | Q9EPH2 | MRP_RAT | 3 | 28.1 | 3 | 4 | 23 (9-10) | 86.02 |
| MSAP (MIR-interacting saposin-like protein) | A0JN30 | A0JN30_RAT | 5 | 28.6 | 5 | 5 | 25 (17) | 124.64 |
| mUNC18 | P61763 | STXB1_BOVINc | 7 | 10.1 | 14 | 4 | 24 (21) | 207.11 |
| Neuromodulin (GAP43) | P07936 | NEUM_RAT | 25 | 89.8 | 156 | 5 | 31 (15) | 2178.44 |
| Neuronal pentraxin receptor | O35764 | NPTXR_RAT | 7 | 13 | 7 | 3 | 17 (17-18) | 113.51 |
| Paralemmin (Palm protein) | Q920Q0 | Q920Q0_RAT | 6 | 15.9 | 6 | 5 | 31 (15) | 132.61 |
| Phosphatidylethanolamine-binding protein 1 | P31044 | PEBP1_RAT | 1 | 7 | 1 | 1 | 9 (13) | 70.45 |
| Proenkephalin A precursor | P22005 | PENK_MOUSEc | 1 | 3.3 | 1 | 5 | 25 (9-10) | 62.61 |
| Secretogranin 2 precursor | P10362 | SCG2_RAT | 5 | 8.4 | 8 | 5 | 25 (13-14) | 75.79 |
| Secretogranin-3 precursor (SgIII) | P47868 | SCG3_RAT | 5 | 12.5 | 5 | 5 | 25 (18) | 87.84 |
| Synapsin 1 | P09951 | SYN1_RAT | 23 | 48.9 | 78 | 1 | 9 (17) | 421.08 |
| Synapsin 1 | P09951 | SYN1_RAT | 6 | 12.2 | 6 | 2 | 16 (17) | 111.25 |
| Synapsin 1 | P09951 | SYN1_RAT | 3 | 4.5 | 3 | 3 | 17 (17-18) | 90.87 |
| VGF nerve growth factor inducible | Q0VGU4 | Q0VGU4_MOUSEc | 4 | 4.9 | 5 | 3 | 20 (11) | 126.26 |
| VGF nerve growth factor inducible | Q0VGU4 | Q0VGU4_MOUSEc | 2 | 5 | 3 | 5 | 31 (15); 25 (9-10) | 88.33 |
| Proteins involved in cytoskeleton dynamics | ||||||||
| Actin-binding LIM protein 1 | Q8K4G5 | ABLM1_MOUSEc | 18 | 26.1 | 63 | 2 | 15 (15); 16(16) | 398.36 |
| Actin-binding LIM protein 1 | Q8K4G5 | Q3KR72_RAT | 5 | 12.9 | 5 | 3 | 17 (16) | 110.85 |
| Arp2/3 complex | Q2LE71 | Q2LE71_HUMAN | 5 | 22.5 | 12 | 2 | 16 (18) | 113.59 |
| Cofilin | P45592 | COF1_RAT | 6 | 46.4 | 12 | 1 | 9 (14) | 154.31 |
| Ezrin-radixin-moesin binding phosphoprotein 50 (EBP50) | Q9JJ19 | NHERF_RAT | 5 | 16 | 5 | 3 | 18 (14-15) | 80.29 |
| EBP50 | P70441 | NHERF_MOUSEc | 4 | 9.3 | 4 | 5 | 25 (13-14) | 41.37 |
| Growth arrest-specific protein 7 (GAS-7) | O55148 | GAS7_RAT | 22 | 37 | 39 | 3 | 17 (16) | 340.59 |
| MAP1B (neuraxin) | P15205 | MAP1B_RAT | 8 | 4.3 | 33 | 3 | 17 (14-15); 18 (18-19) | 260.41 |
| Neuronal migration protein doublecortin (lissencephalin-X) (Lis-X) (doublin) | O43602 | DCX_HUMANc | 4 | 10.2 | 13 | 1 | 11 (15-16-17); 12 (18-19) | 94.64 |
| Platelet-activating factor acetylhydrolase 1B gamma | Q61205 | PA1B3_MOUSEc | 3 | 10.8 | 3 | 3 | 18 (18-19) | 65.26 |
| Platelet-activating factor acetylhydrolase 1B alpha | P43034 | LIS1_HUMANc | 6 | 14.1 | 11 | 2 | 15 (16) | 124.82 |
| Profilin 2 | Q09430 | PROF2_BOVINc | 4 | 23.6 | 6 | 3 | 17 (14-15);17 (19-20) | 78.77 |
| Septin 11 | Q8C1B7 | SEP11_MOUSE | 11 | 25.3 | 28 | 1 | 9 (15-16) | 174.36 |
| Septin 11 | Q8C1B7 | SEP11_MOUSE | 23 | 33.4 | 34 | 2 | 16 (17) | 348.68 |
| Septin 11 | Q8C1B7 | SEP11_MOUSE | 28 | 43.6 | 63 | 3 | 17 (17-18) | 495.88 |
| Septin 8 | Q5SX81 | SEPT8_BOVIN | 10 | 13.6 | 10 | 3 | 17 (17-18) | 44.97 |
| Septin 9 | Q9ZQZR6 | SEPT9_RAT | 6 | 11.3 | 7 | 4 | 21 (16) | 86.04 |
| Stathmin | P13668 | STMN1_RAT | 7 | 40.3 | 65 | 1 | 9 (15-16) | 151.23 |
| Stathmin | P13668 | STMN1_RAT | 21 | 67.8 | 82 | 4 | 24 (9-10-11) | 450.18 |
| STOP protein | Q63560 | Q63560_RAT | 26 | 33.7 | 145 | 1 | 9 (14); 12 (12-13) | 685.15 |
| Proteins involved in regulation of mRNA localization | ||||||||
| ELAV-like protein 3 (Hu-antigen C) (HuC) | Q60900 | ELAV3_MOUSEc | 3 | 7.1 | 7 | 1 | 10 (17-18-19) | 59.5 |
| Lupus La protein homolog | P38656 | LA_RAT | 16 | 32.5 | 28 | 1 | 11 (18) | 255.12 |
| Matrin 3 | P43244 | MATR3_RAT | 4 | 3.6 | 5 | 3 | 17 (16) | 44.45 |
| NoNo | Q5FVM4 | NONO_RAT | 35 | 43.3 | 52 | 1 | 9 (18) | 364.94 |
| NoNo | Q15233 | NONO_HUMANc | 13 | 21.7 | 20 | 3 | 17 (17-18) | 54.78 |
| Treacle | O08784 | TCOF_MOUSE | 2 | 1.5 | 2 | 2 | 14 (13-14) | 35.63 |
| Wiskott-Aldrich syndrome protein-interacting protein (WASP-interacting protein) | Q8K1I7 | WIPF1_MOUSE | 2 | 3.9 | 2 | 1 | 12 (10-11) | 43.39 |
| Transcriptional and translational repressors | ||||||||
| MeCP2 | Q00566 | MECP2_RAT | 12 | 25.8 | 12 | 1 | 12 (14; 16) | 245.22 |
| MeCP2 | Q00566 | MECP2_RAT | 25 | 42.1 | 25 | 2 | 13 (14); 14 (13-14) | 410.03 |
| Nuclease-sensitive element-binding protein 1 (YB1) | P62960 | YBOX1_MOUSEc | 23 | 67.1 | 186 | 1 | 12 (10-11) | 848.98 |
| Nuclease-sensitive element-binding protein 1 (YB1) | Q3ZAV2 | Q3ZAV2_RAT | 34 | 73 | 100 | 2 | 13 (14); 14 (10-14); 16 (11) | 897.77 |
| Nuclease-sensitive element-binding protein 1 (YB1) | P67808 | YBOX1_BOVINc | 8 | 29.3 | 9 | 4 | 23 (9-10) | 61.37 |
| Histones | ||||||||
| H2A histone family; J member | Q3ZBX9 | Q3ZBX9_BOVIN | 6 | 44.2 | 151 | 2 | 16 (17) | 228.09 |
| Histone cluster 1.H2aj | A2RUU6 | A2RUU6_HUMAN | 8 | 66.4 | 384 | 2 | 13 (17) | 801.24 |
| Histone H2A.x (H2a/x) | P16104 | H2AX_HUMAN | 8 | 63.6 | 91 | 2 | 13 (17) | 183.97 |
| Histone H2A.x (H2a/x) | P27661 | H2AX_MOUSE | 8 | 63.6 | 90 | 2 | 13 (17) | 157.01 |
| Histone H2B (fragment) | Q29574 | Q29574_PIGc | 17 | 66.7 | 277 | 1 | 11 (15-16) | 1149.24 |
| Histone H2B type 1-N | Q32L48 | H2B1N_BOVIN | 13 | 67.5 | 1643 | 1 | 11 (16-17) | 2405.83 |
| Histone H2B | Q3KP43 | Q3KP43_HUMAN | 7 | 63.8 | 92 | 1 | 11 (15) | 449.79 |
| Histone H2B type 1-N | Q32L48 | H2B1N_BOVIN | 12 | 52.4 | 1271 | 2 | 13 (19-20); 16 (11-16) | 1419.12 |
| Histone cluster 1.H2bh | A2RTP6 | A2RTP6_MOUSE | 18 | 73 | 2155 | 2 | 13 (14-16); 14 (15); 15 (16) | 1637.31 |
| Histone H2B (fragment) | Q29574 | Q29574_PIGc | 21 | 79.7 | 187 | 2 | 13 (15); 14 (14-15-16) | 1392.22 |
| Histone H4 (fragment) | A0AUM5 | A0AUM5_MOUSEc | 13 | 62 | 349 | 1 | 11 (18); 12 (16) | 3459.32 |
| Histone H4 | Q9D0C9 | Q9D0C9_MOUSEc | 8 | 60.2 | 28 | 2 | 13 (16) | 509.01 |
| Histone H4 (fragment) | A0AUM5 | A0AUM5_MOUSEc | 17 | 67 | 276 | 2 | 13 (16); 14 (16); 16 (17) | 1404.42 |
| Other proteins | ||||||||
| 60S ribosomal protein L6 (fragment) | Q6P790 | Q6P790_RAT | 20 | 44.1 | 25 | 3 | 18 (14) | 193.28 |
| Aconitate hydratase. mitochondrial | Q9ER34 | ACON_RAT | 39 | 45.1 | 126 | 3 | 17 (17-18) | 775.81 |
| Ataxin-2-like protein | Q8WWM7 | ATX2L_HUMAN | 13 | 12.5 | 13 | 5 | 31 (15) | 113.41 |
| ATP synthase e chain; mitochondrial | P29419 | ATP5I_RAT | 7 | 71.8 | 26 | 2 | 13 (15) | 232.03 |
| ATP synthase e chain; mitochondrial | P29419 | ATP5I_RAT | 5 | 69 | 11 | 3 | 18 (14) | 127.14 |
| ATP synthase f chain; mitochondrial | P56135 | ATPK_MOUSE | 2 | 20.5 | 4 | 2 | 14 (16); 15 (16) | 59.07 |
| ATP synthase alpha chain; mitochondrial precursor | P15999 | ATPA_RAT | 15 | 25.9 | 15 | 3 | 17 (20) | 309.34 |
| ATP synthase subunit O; mitochondrial precursor | Q06647 | ATPO_RAT | 9 | 40.4 | 16 | 2 | 13 (20); 15 (20) | 216.85 |
| BolA-like 2 | Q0P601 | Q0P601_MOUSE | 5 | 51.2 | 11 | 3 | 17 (14) | 130.83 |
| Cyclic AMP-dependent protein kinase inhibitor gamma (PKI-gamma) | Q7YQJ3 | IPKG_BOVINc | 1 | 21.1 | 1 | 5 | 31 (9) | 60.74 |
| Catalase (EC 1.11.1.6) | P04762 | CATA_RAT | 25 | 46.5 | 74 | 2 | 15 (17-18); 16 (17) | 480.78 |
| Chaperonin subunit 6a (zeta) | Q3MHS9 | Q3MHS9_RAT | 19 | 32.8 | 19 | 3 | 17 (21) | 114.72 |
| Cold-inducible RNA-binding protein (CIRP) | P60826 | CIRBP_CRIGRc | 6 | 41.9 | 19 | 1 | 9 (13) | 173.67 |
| Cytochrome c oxidase polypeptide VIIa-liver/heart; mitochondrial precursor | P35171 | CX7A2_RAT | 1 | 12 | 4 | 2 | 13 (16); 14 (16) | 75.48 |
| Eukaryotic translation initiation factor 2beta | Q6P685 | Q6P685_RAT | 16 | 41.1 | 18 | 5 | 31 (9) | 463.48 |
| Heterogeneous nuclear ribonucleoprotein A1 (helix-destabilizing protein) | P09867 | ROA1_BOVINc | 7 | 32.8 | 7 | 3 | 17 (13) | 84.1 |
| Heterogeneous nuclear ribonucleoprotein A1 | P09651 | ROA1_HUMANc | 23 | 41.4 | 116 | 1 | 9 (11-13) | 641.89 |
| Heterogeneous nuclear ribonucleoprotein D0 | Q14103 | HNRPD_HUMANc | 9 | 21.4 | 20 | 4 | 22 (14) | 186.16 |
| Heterogeneous nuclear ribonucleoprotein H | P31943 | HNRH1_HUMAN | 6 | 16 | 15 | 3 | 17 (15) | 125.09 |
| Heterogeneous nuclear ribonucleoprotein Q | O60506 | HNRPQ_HUMANc | 5 | 10 | 7 | 2 | 13 (18) | 115.12 |
| Inorganic pyrophosphatase 2; mitochondrial precursor | Q91VM9 | IPYR2_MOUSE | 8 | 22.4 | 8 | 3 | 18 (15) | 115.17 |
| p21 Ras GTPase-activating protein-associated p62 (Sam68) | Q91V33 | SAM68_RAT | 10 | 29.8 | 10 | 5 | 25 (17) | 116.12 |
| Low-molecular-wt phosphotyrosine protein phosphatase (LMW-PTP) | P41498 | PPAC_RAT | 6 | 41.1 | 15 | 3 | 18 (14-15) | 155.75 |
| Mitochondrial aconitase 2 (fragment) | Q1P9Q3 | Q1P9Q3_BOVINc | 19 | 21.3 | 20 | 2 | 16 (17) | 424.43 |
| NADH dehydrogenase (ubiquinone) Fe-S protein 7 | Q5RJN0 | Q5RJN0_RAT | 3 | 16.5 | 8 | 2 | 13 (18) | 104.15 |
| NADH-ubiquinone oxidoreductase MLRQ subunit | Q62425 | NDUA4_MOUSE | 3 | 36.6 | 7 | 2 | 13 (16) | 75.72 |
| Prohibitin-2 (B-cell receptor-associated protein BAP37) | Q99623 | PHB2_HUMANc | 5 | 16.7 | 5 | 3 | 17 (20) | 78.89 |
| Ribosomal protein L13a | Q5M9M0 | Q5M9M0_MOUSEc | 18 | 53.7 | 66 | 3 | 17 (15) | 200.01 |
| RRS1 ribosome biogenesis regulator homolog | A1A5P2 | A1A5P2_RAT | 10 | 26.8 | 10 | 3 | 17 (15) | 128.07 |
| Stress-induced-phosphoprotein 1 (STI1) (Hsc70/Hsp90-organizing protein) (Hop) | O35814 | STIP1_RAT | 28 | 46.6 | 28 | 4 | 21 (16) | 227.95 |
| Transcription elongation factor B polypeptide 2 (elongin B) | P62869 | ELOB_MOUSEc | 6 | 39 | 10 | 4 | 22 (14) | 140.23 |
| Ubiquitin-conjugating enzyme E2 variant 2 | A0M8W4 | A0M8W4_HUMANc | 1 | 6.9 | 1 | 5 | 25 (14) | 56.44 |
Fraction numbering is indicated as follows: fraction number after the first-dimension separation (corresponding fraction number after the second dimension separation).
MS/MS data are indicated for the selected fraction with highest score. See Fig. S1 and S2 in the supplemental material for the full list.
The corresponding rat protein can be found in Fig. S3 in the supplemental material.
Protein identified by Western blot (WB) analysis of the indicated fraction.
BDV infection alters proteins that are key modulators of synaptic activity.
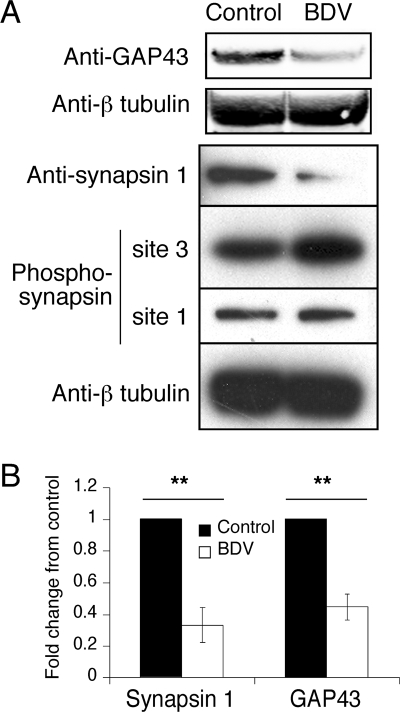
Many proteins identified by MS/MS are localized at the synapse, where they play key roles in regulating neuronal growth or communication. These proteins included axonal guidance factors, synaptic proteins, and neurotransmitter-producing enzymes (Table 1). A surprisingly high proportion of the identified proteins were found in different nonadjacent fractions, probably as a result of posttranslational modifications. For example, this was the case for key enzymes in the synthesis of neurotransmitters as well as for proteins that regulate synaptic vesicle recycling and mobilization for neurotransmitter release, such as myristoylated alanine-rich C kinase substrate (MARCKS), mUNC18, or synapsin 1. Several of the identified changes at the synapse were indeed in agreement with data reported in previous studies, such as cDNA array analyses that demonstrated changes in B-FABP (30) or functional assays that showed differences for MARCKS and mUNC18 (50). Using quantitative Western blot analysis, we demonstrated a significant downregulation of GAP-43, a presynaptic membrane protein that accumulates in neuronal growth cones and plays key roles in synaptic density and plasticity (Fig. 3) (39). We also observed a significant decrease in the levels of synapsin 1 in BDV-infected neurons (Fig. 3). Moreover, the availability of different phosphospecific synapsin 1 antibodies allowed us to demonstrate that the multiple fractions in which synapsin 1 was found likely resulted from various posttranslational modifications. Indeed, we observed strong signals in BDV-infected neurons with an antibody specific for synapsin 1 phosphorylated by CaMK II at site 3 (clone RU19). Combined with its reduced total levels, our findings suggest that BDV infection leads to a hyperphosphorylation of synapsin 1. Given the key roles of synapsin 1 in the compartmentalization and mobilization of vesicular pools at the synapse (19, 24), this result may underlie the defects in presynaptic-synaptic vesicle organization (as revealed by electron microscopy analysis) described previously upon BDV infection (26).
FIG. 3.
Expression of GAP-43 and synapsin 1 is impaired following BDV infection. (A) Western blot analysis of neuronal extracts from control and BDV-infected cultures using antibodies specific for GAP-43, total synapsin 1, phosphosynapsin 1 (site 3, specific for CaMK II, and site 1, specific for PKA and CaMK I), and beta-tubulin for normalization. Results are representative of four independent experiments. (B) Quantification of the Western blot signals using the Odyssey imager. Due to the intrinsic variability between neuronal cultures, results were expressed as their change compared to control neurons that were arbitrarily set to 1 in all cases. **, P < 0.05 by paired t test.
BDV interferes with cytoskeleton dynamics.
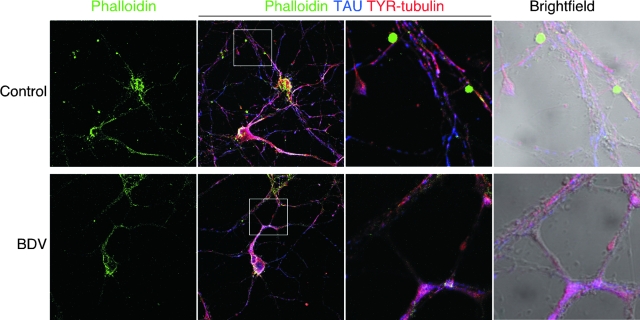
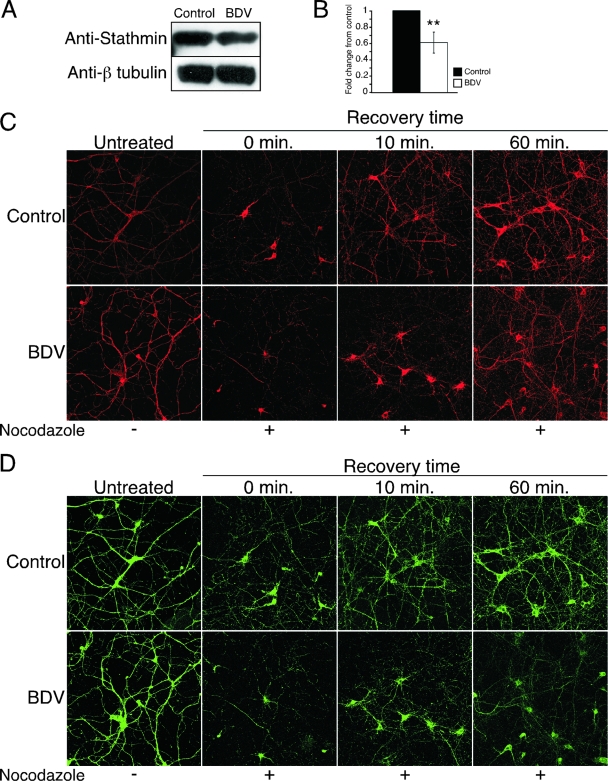
Accumulating evidence indicates that many signaling cascades converge at the actin and microtubule cytoskeletons and their associated proteins, thereby regulating neuronal growth and remodeling (8). Indeed, the cytoskeleton is a highly dynamic structure that not only controls cell morphology but also plays a major role in neuronal plasticity, transport, and signaling. The dynamics of actin filaments and microtubules are controlled by a complex set of actin- and microtubule-binding proteins. To date, up to 28 actin-binding proteins and 13 microtubule-binding proteins in the neuronal growth cone have been described (15). Interestingly, we identified differences in 10 of 28 and in 5 of 13 of these previously described actin- and microtubule-binding proteins (Table 1). Importantly, we did not identify changes in the structural proteins that constitute the cytoskeleton, such as actin or tubulin, in agreement with the fact that BDV infection does not drastically affect neuronal morphology. Rather, differences were detected in proteins such as cofilin, profilin II, and the Arp2/3 complex, which regulate actin polymerization upon stimulation (15). Likewise, other identified proteins such as STOP, doublecortin, or stathmin are essential regulators of microtubule polymerization. These results led us to reexamine the impact of BDV infection on the neuronal cytoskeleton more thoroughly. We first assessed the pattern of F-actin expression in control and BDV-infected neurons using fluorescently labeled phalloidin (Fig. 4). By high-magnification confocal microscopy, we observed a loss of the characteristic punctuate staining for F actin in BDV-infected neurons. In control neurons, this staining was concentrated at the contact nodes between neurites. It has been suggested that these actin-rich “ruffles” may correspond to enforced synapses (10, 57). Given the important roles of F actin in synapse maintenance or elimination (18), these changes may contribute to the defects in synaptic plasticity linked to BDV persistence. Next, we examined the phenotypic consequences of changes detected in microtubule-binding proteins. Using quantitative Western blot analysis, we first showed that levels of one these proteins, stathmin, were reduced in BDV-infected neurons (Fig. 5A).
FIG. 4.
BDV infection interferes with localization of F actin at contact nodes between neurites. Shown is confocal microscopic imaging of control (top) and BDV-infected (bottom) cortical neurons triply stained for F actin using phalloidin-fluorescein isothiocyanate (FITC) (green), the neuronal marker Tau (blue), and the tyrosinated form of polymerized tubulin (TYR) to stain microtubule structures (red). Original magnification, ×630. An enlarged view of the highlighted white square of the triple-merge image is displayed on the two right pictures (fluorescence and bright field). At this higher magnification, note that the punctuated phalloidin staining colocalizes with contact nodes between neurites in control neurons and is lost following BDV infection.
FIG. 5.
Infection with BDV delays recovery of the microtubule network following depolymerization with nocodazole. (A) BDV impairs stathmin expression. Shown is a Western blot analysis of neuronal extracts from control and BDV-infected cultures using antibodies specific for the microtubule-associated protein stathmin and beta-tubulin for normalization. Data are representative of two independent experiments. (B) Quantification of the Western blot signals using the Odyssey imager. **, P < 0.05 by paired t test. (C and D) Analysis of the microtubule network recovery kinetics following nocodazole-induced depolymerization. Neurons were treated (+) or not (−) with nocodazole and processed for immunofluorescence analysis at different times following washout of nocodazole. (C) Confocal microscopy imaging of staining using an antibody specific for the stable acetylated form of tubulin (ACE) (red). (D) Confocal microscopy imaging of staining using an antibody specific for the microtubule-associated protein Tau (green). Note the incomplete recolonization of the microtubule network by Tau even after 1 h of recovery time. Original magnification, ×400.
To demonstrate the functional relevance of the observed changes in microtubule-binding proteins, we treated neuronal cultures with the microtubule-depolymerizing agent nocodazole (5) and monitored the kinetics of recovery after washout of the drug using immunofluorescence analysis for the detection of acetylated tubulin and Tau proteins. The repolymerization of microtubules, which could be visualized using acetylated tubulin staining (16), was very rapid and had returned to normal levels 60 min after the washout of nocodazole. However, the recovery kinetics were significantly delayed in BDV-infected neurons (Fig. 5C). Likewise, the recolonization of the microtubule network by the Tau protein was also significantly delayed in BDV-infected neurons, even 1 h after nocodazole washout (Fig. 5D).
BDV impacts the regulation of mRNA expression in neurons.
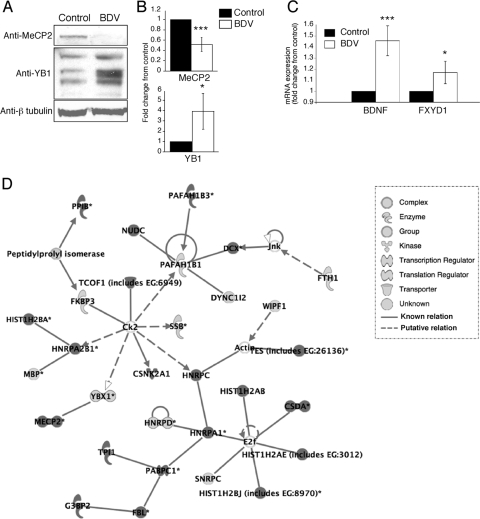
We were particularly interested in the identification of changes in the transcriptional repressor MeCP2. MeCP2 is a DNA-binding protein that selectively binds methylated DNA CpG dinucleotides and plays important roles in neuronal physiology (4). MeCP2 is expressed during postnatal mammalian brain development and is a marker for neuronal maturity. MeCP2 mutations have been shown to be responsible for Rett syndrome, a severe mental retardation disease associated with defects in synaptic plasticity. MeCP2 interacts with YB1, a principal component of messenger ribonucleoprotein particles that controls multiple steps of mRNA processing, including the selection of alternative splice sites (55). Western blot and immunofluorescence analyses (Fig. 6A and data not shown) revealed that MeCP2 levels were significantly reduced upon infection with BDV. Conversely, levels of the different YB1 isoforms were increased in BDV-infected neurons (Fig. 6B), a result consistent with the proposed model of interaction between MeCP2 and YB1 (4). MeCP2 regulation of neuronal differentiation is mediated through chromatin remodeling and its action on multiple target genes, most of which are still unknown. BDNF and FXYD1 are two genes repressed by MeCP2 repression (14, 32, 59). Consistent with the reduced MeCP2 levels detected in BDV-infected neurons, we demonstrated that mRNA levels for both BDNF and FXYD1 were significantly increased upon infection (Fig. 6C).
FIG. 6.
Infection with BDV modifies the expression pattern of the MeCP2 and YB1 proteins as well as that of MeCP2 target genes. (A) Western blot analysis of neuronal extracts from control and BDV-infected cultures using antibodies specific for MeCP2, YB1, and beta-tubulin for normalization. Results are representative of four to six independent experiments. (B) Quantification of the signals using the Odyssey imager. Due to the intrinsic variability between neuronal cultures, results were expressed as their change compared to control neurons that were arbitrarily set to 1 in all cases. ***, P < 0.0; *, P < 0.1 (by paired t test). (C) Real-time quantitative reverse transcription-PCR analysis of the expression of MeCP2 target genes. Levels of BDNF and phospholemman (FXYD1) transcripts were determined using total RNA extracted from control and BDV-infected neurons. Transcript levels were normalized to HPRT. Results are displayed as means ± standard errors of the means for six independent experiments. ***, P < 0.01; *, P < 0.1 (by paired t test). (D) Representative example of the signaling network/function analyses performed using IPA. The list of identified proteins was analyzed using IPA tools as described in the text. The network shown in the figure had the highest score (score of 49) and included 30 focus proteins. The legend for each node shape and arrow is indicated at the bottom. Focus proteins were identified mainly in BDV-infected neuronal extracts, whereas others were found mainly in control neurons.
In order to gain further insight into the repercussions of the detected changes between control and BDV-infected neurons on neuronal physiology, we applied the 87 identified proteins as a group for network analysis. We used the Ingenuity Pathway Knowledge Base criteria and uploaded this set of proteins with their Swiss-Prot numbers into IPA. IPA created 19 biological networks with highly significant scores (>11). Among these networks, four networks had a score of >30 and included more than 20 focus proteins. Interestingly, the highest-scoring network (score of 49) predicted by IPA included 30 focus proteins and included both MeCP2 and YB1 (Fig. 6D).
Histone modifications upon BDV infection.
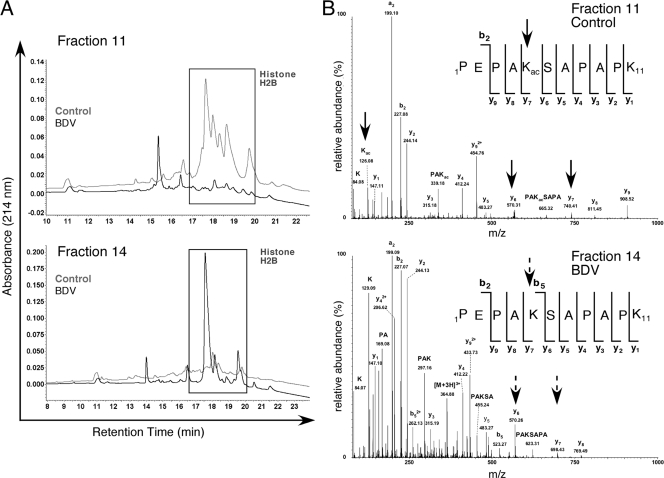
The analysis of the 2D-LC chromatograms revealed important changes for a group of peaks identified in two nonadjacent fractions of the control and BDV-infected protein extracts (Fig. 7A). MS identified this differently expressed set of peaks as corresponding to H2B. Moreover, using specific fragment ions as previously described (56), MS/MS analysis of the H2B peptide spectra revealed unambiguously that H2B acetylations on Lys-5 (Fig. 7B) and Lys-20 (data not shown) were not observed following BDV infection. As a matter of fact, a trypsin miscleavage in Lys-5 allowed us to identify both the modified and unmodified peptides in control and BDV-infected samples, clearly showing that Lys-5 acetylation was absent in this selected fraction (Fig. 7B).
FIG. 7.
BDV infection affects H2B posttranslational modifications. (A) Chromatographic profiles of BDV (black line) and control (gray line) samples for 2D-LC fractions 11 and 14. Differences in profiles between samples (square) were identified by nanoLC-MS/MS as being due to H2B. (B) Analysis of MS/MS spectra from a specific peptide sequence of histone H2B reveals differences in posttranslational modifications. The identified peptide sequences are indicated in the upper corner of the spectra. (Top) MS/MS spectrum of the peptide with m/z 567.81 corresponding to [M + 2H]2+ (control). The peptide was determined to be 1PEPAKacSAPAPK11, where lysine 5 is acetylated. (Bottom) MS/MS spectrum of the peptide with m/z at 364.85 corresponding to [M + 3H]3+ (BDV). The peptide was determined to be 1PEPAKSAPAPK11 from histone H2B, where lysine 5 is unmodified. The immonium ion at m/z 126.08 indicated the presence of ɛ-acetyllysine and was observed only on the control peptide (top). Acetylation was assigned on Lys-5 because ion y7 at m/z 740.41 was shifted by 42 Da in a mass spectrum (top) (control), whereas y7 at m/z 698.43 (bottom) (BDV) showed no mass shift.
DISCUSSION
The goal of this study was to analyze the effects of BDV infection on the neuronal proteome in order to gain further insight into BDV pathogenesis. To date, only a limited number of studies have addressed the virus-induced changes on the cellular proteome (reviewed in reference 37), and even fewer have used primary cultures of neurons as a cellular target (11, 17). Moreover, the proteomic profiling of virus-infected cells has led mostly to the identification of proteins whose altered expression results from the cytopathic consequences of viral replication. These include modifications in heat shock proteins or markers of apoptosis and/or neurodegeneration, as illustrated in studies using West Nile virus-infected neurons or cells lines infected with Nipah virus, respiratory syncytial virus, or African swine fever virus (2, 9, 11, 17). A striking feature is that our study using BDV-infected neurons did not reveal any change in proteins falling into the above-mentioned categories, further stressing the exquisite noncytolytic adaptation of BDV to its neuronal target cell.
Previous studies aimed at analyzing global cellular changes upon BDV infection used microarrays and detected cellular changes at the transcriptional level (30, 52). Although we confirmed the impact of BDV infection on B-FABP expression, there was little overlap with data from those previous transcriptome reports. Indeed, changes observed at the mRNA level do not always correspond to changes at the protein level (46). In addition, previous studies were based on data from total extracts prepared from infected brains and could not discriminate between the impact of BDV on neurons and that on glial cells that also become infected at later time points. Here, the use of neuron-rich cultures allowed us to gain further insight into the impact of BDV persistence on neurons. Nevertheless, it is likely that many of the events taking place in the brain of BDV-infected rats are the result of a complex interplay between the virus and different cell types, including glial cells such as astrocytes and microglial cells, which may not be recreated in the primary cortical neuron cultures.
Although this study provides considerable new information about the neuronal proteome changes upon BDV infection, it is clear that we may have not yet uncovered the totality of these changes. Indeed, we selected peaks for which a difference could be detected upon analysis of the 2D-LC chromatograms. Therefore, we may have missed the protein changes that did not result in any strong modification of the chromatogram either because they were below the limit of detection or because they were hidden by other highly represented and unchanged proteins present in the same fraction.
Damage to the CNS cytoskeleton is a hallmark of different viral infections and is also observed in many neurodegenerative diseases (36, 58). Although there was no alteration of the neuronal cytoskeleton following BDV infection, in sharp contrast to other neurotropic viral systems, we identified changes in many actin- and microtubule-binding proteins, suggesting a global “rigidity” of the network, i.e., the inability to respond or adapt when requested. Indeed, the depolymerization and recovery assays described herein clearly showed that BDV leads to impaired cytoskeleton dynamics. The most classical way to demonstrate such a phenotype is to use growing neurons during the first days after plating and to monitor the process of axonal sprouting, as has been done to demonstrate the key role of proteins such as stathmins in the neuronal growth cone (43). However, this kind of approach is not feasible with BDV, as infected cells produce extremely low levels of infectious viral particles. Thus, BDV stocks have usually low titers, and a significant number of infected cells is observed only after 6 to 8 days, after the process of axonal growth in vitro is nearly completed. In any event, given the complexity of the functions controlled by cytoskeleton-binding proteins and their possible link with other identified proteins, such as synapsin or MARCKS, it is plausible that the identified changes may impact neuronal function significantly and lead to long-term plasticity impairment.
BDV has the property to replicate and transcribe in the nucleus of infected cells, a feature that distinguishes BDV from other members of the order Mononegavirales (12). Consequently, BDV has established a strategy for persistence that allows the long-term retention of its genome packed in viral ribonucleoprotein complexes within the nucleus. Moreover, BDV must also ensure an efficient nuclear export and transfer of viral ribonucleoproteins along neuronal processes to permit viral transmission to neighboring cells. Most of the mechanisms underlying these events are still largely unknown (48). In this study, we identified many changes in nuclear proteins and proteins involved in RNA transport in neurons that may provide clues for future studies. Indeed, proteins such as YB1, HuC, La, or NoNo play important roles in mRNA processing and/or the transport of RNA granules in neuronal processes (40). In addition, the identified changes in histone proteins may reflect the consequences of BDV persistence in the nucleus on the cellular chromatin. Indeed, viruses that persist in the cell nucleus have evolved diverse mechanisms to manipulate cellular chromatin to ensure efficient viral genome survival and propagation (33). Thus, it is likely that the observed modifications in the histone acetylation pattern may induce important changes in the pattern of expression of the infected neurons. Histone proteins and their accompanying posttranslational modifications have received much attention for their ability to affect chromatin structure and consequently to regulate gene expression. The acetylation and methylation of histones are usually related to transcriptionally active chromatin (27) and are best described for H3 and H4. Interestingly, the activation of cellular histone deacetylases and the concomitant deacetylation of histones H3 and H4 have been demonstrated for many persistent viruses, including the retroviruses human immunodeficiency virus and human T-cell leukemia virus or the herpesvirus Epstein-Barr virus (42, 49, 54). BDV infection may use a similar mechanism but focused on H2B, a hypothesis that will require further investigation. Such changes in histones and their consequences on chromatin structure may also explain the decreased levels of expression of transcriptional repressors such as MeCP2 (4).
One of the major challenges resulting from the proteomic profiling of the virus-cell interaction lies in trying to extrapolate from a global analysis to gain new insight into viral pathogenesis. This issue is complicated by the complexity of the identified proteins and the difficulty in organizing them in a somehow hierarchical manner. The use of IPA allowed us to gain clues about possible interrelationships and revealed that MeCP2 and YB1 could play an important role. Given the drastic consequences of any qualitative or quantitative change in MeCP2 expression (4), we consider the reported changes in MeCP2 following BDV infection, together with the accompanying modification in the mRNA expression of target genes, to be highly relevant. We hypothesize that BDV replication could lead to the genetic reprogramming of infected neurons through its action on cellular chromatin. This, in turn, would affect key transcriptional repressors and impair the ability of the neuron to respond to certain stimuli. Further studies will be required to formally test this hypothesis.
Supplementary Material
Acknowledgments
This work was supported financially by INSERM and an ANR Agence Nationale de la Recherche grant to D.G.-D. (ANR-06-MIME-005-01). E.S. is a recipient of a poste d'accueil from INSERM. D.G.-D. is supported by the CNRS. B.M. is supported by the CNRS, the Génopôle Toulouse Midi-Pyrénées, the Région Midi-Pyrénées, and grants from Institut National du Cancer, Agence Nationale de la Recherche, and Fondation pour la Recherche Médicale.
We thank Z. Zhou and M. E. Greenberg (Harvard Medical School, Boston, MA), P. Greengard (The Rockefeller University, New York, NY), and A. Sobel, (INSERM U839, Paris, France) for their gifts of antibodies; the Genopôle and Canceropôle Grand-Sud-Ouest for providing access to the proteomic platforms; S. Allart for expert assistance at the IFR30 cellular imaging platform; and R. Liblau, A. Saoudi, C. Prat, C. Duval, S. Boullier, L. Mars, and M. Lazarczyk for critical reading of the manuscript.
Footnotes
Published ahead of print on 1 October 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ahmed, R., L. A. Morrison, and D. M. Knipe. 1996. Persistence of viruses, p. 219-249. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed., vol. 1. Lippincott-Raven, Philadelphia, PA. [Google Scholar]
- 2.Alfonso, P., J. Rivera, B. Hernaez, C. Alonso, and J. M. Escribano. 2004. Identification of cellular proteins modified in response to African swine fever virus infection by proteomics. Proteomics 42037-2046. [DOI] [PubMed] [Google Scholar]
- 3.Bajramovic, J. J., S. Münter, S. Syan, U. Nerhbass, M. Brahic, and D. Gonzalez-Dunia. 2003. Borna disease virus glycoprotein is required for viral dissemination in neurons. J. Virol. 7712222-12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bienvenu, T., and J. Chelly. 2006. Molecular genetics of Rett syndrome: when DNA methylation goes unrecognized. Nat. Rev. Genet. 7415-426. [DOI] [PubMed] [Google Scholar]
- 5.Bigot, D., A. Matus, and S. P. Hunt. 1991. Reorganization of the cytoskeleton in rat neurons following stimulation with excitatory amino acids in vitro. Eur. J. Neurosci. 3551-558. [DOI] [PubMed] [Google Scholar]
- 6.Bode, L., and H. Ludwig. 2003. Borna disease virus infection, a human mental-health risk. Clin. Microbiol. Rev. 16534-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bouyssie, D., A. Gonzalez de Peredo, E. Mouton, R. Albigot, L. Roussel, N. Ortega, C. Cayrol, O. Burlet-Schiltz, J. P. Girard, and B. Monsarrat. 2007. Mascot File Parsing and Quantification (MFPaQ), a new software to parse, validate, and quantify proteomics data generated by ICAT and SILAC mass spectrometric analyses: application to the proteomics study of membrane proteins from primary human endothelial cells. Mol. Cell. Proteomics 61621-1637. [DOI] [PubMed] [Google Scholar]
- 8.Brady, S., D. R. Colman, and P. Brophy. 1999. Subcellular organization of the nervous system, p. 71-106. In M. J. Zigmond, F. E. Bloom, S. C. Landis, J. L. Roberts, and L. R. Squire (ed.), Fundamental neuroscience. Academic Press, San Diego, CA.
- 9.Brasier, A. R., H. Spratt, Z. Wu, I. Boldogh, Y. Zhang, R. P. Garofalo, A. Casola, J. Pashmi, A. Haag, B. Luxon, and A. Kurosky. 2004. Nuclear heat shock response and novel nuclear domain 10 reorganization in respiratory syncytial virus-infected A549 cells identified by high-resolution two-dimensional gel electrophoresis. J. Virol. 7811461-11476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Capani, F., M. E. Martone, T. J. Deerinck, and M. H. Ellisman. 2001. Selective localization of high concentrations of F-actin in subpopulations of dendritic spines in rat central nervous system: a three-dimensional electron microscopic study. J. Comp. Neurol. 435156-170. [DOI] [PubMed] [Google Scholar]
- 11.Chang, L. Y., A. R. Ali, S. S. Hassan, and S. AbuBakar. 2007. Human neuronal cell protein responses to Nipah virus infection. Virol. J. 454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cubitt, B., and J. C. de la Torre. 1994. Borna disease virus (BDV), a nonsegmented RNA virus, replicates in the nuclei of infected cells where infectious BDV ribonucleoproteins are present. J. Virol. 681371-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de la Torre, J. C. 1994. Molecular biology of Borna disease virus: prototype of a new group of animal viruses. J. Virol. 687669-7675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng, V., V. Matagne, F. Banine, M. Frerking, P. Ohliger, S. Budden, J. Pevsner, G. A. Dissen, L. S. Sherman, and S. R. Ojeda. 2007. FXYD1 is an MeCP2 target gene overexpressed in the brains of Rett syndrome patients and Mecp2-null mice. Hum. Mol. Genet. 16640-650. [DOI] [PubMed] [Google Scholar]
- 15.Dent, E. W., and F. B. Gertler. 2003. Cytoskeletal dynamics and transport in growth cone motility and axon guidance. Neuron 40209-227. [DOI] [PubMed] [Google Scholar]
- 16.Dent, E. W., and K. Kalil. 2001. Axon branching requires interactions between dynamic microtubules and actin filaments. J. Neurosci. 219757-9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dhingra, V., Q. Li, A. B. Allison, D. E. Stallknecht, and Z. F. Fu. 2005. Proteomic profiling and neurodegeneration in west-Nile-virus-infected neurons. J. Biomed. Biotechnol. 2005271-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dillon, C., and Y. Goda. 2005. The actin cytoskeleton: integrating form and function at the synapse. Annu. Rev. Neurosci. 2825-55. [DOI] [PubMed] [Google Scholar]
- 19.Evergren, E., F. Benfenati, and O. Shupliakov. 2007. The synapsin cycle: a view from the synaptic endocytic zone. J. Neurosci. Res. 852648-2656. [DOI] [PubMed] [Google Scholar]
- 20.Gonzalez-Dunia, D., C. Sauder, and J. C. de la Torre. 1997. Borna disease virus and the brain. Brain Res. Bull. 44647-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gonzalez-Dunia, D., R. Volmer, D. Mayer, and M. Schwemmle. 2005. Borna disease virus interference with neuronal plasticity. Virus Res. 111224-234. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez-Dunia, D., M. Watanabe, S. Syan, M. Mallory, E. Masliah, and J. C. de la Torre. 2000. Synaptic pathology in Borna disease virus persistent infection. J. Virol. 743441-3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gosztonyi, G., and H. Ludwig. 1984. Neurotransmitter receptors and viral neurotropism. Neuropsychiatr. Clin. 3107-114. [Google Scholar]
- 24.Greengard, P., F. Valtorta, A. J. Czernik, and F. Benfenati. 1993. Synaptic vesicle phosphoproteins and regulation of synaptic function. Science 259780-785. [DOI] [PubMed] [Google Scholar]
- 25.Griffin, D. E. 2005. Neuronal cell death in alphavirus encephalomyelitis. Curr. Top. Microbiol. Immunol. 28957-77. [DOI] [PubMed] [Google Scholar]
- 26.Hans, A., J. J. Bajramovic, S. Syan, E. Perret, I. Dunia, M. Brahic, and D. Gonzalez-Dunia. 2004. Persistent, noncytolytic infection of neurons by Borna disease virus interferes with ERK 1/2 signaling and abrogates BDNF-induced synaptogenesis. FASEB J. 18863-865. [DOI] [PubMed] [Google Scholar]
- 27.He, H., and N. Lehming. 2003. Global effects of histone modifications. Brief. Funct. Genomics Proteomics 2234-243. [DOI] [PubMed] [Google Scholar]
- 28.Hornig, M., M. Solbrig, N. Horscroft, H. Weissenbock, and W. I. Lipkin. 2001. Borna disease virus infection of adult and neonatal rats: models for neuropsychiatric disease. Curr. Top. Microbiol. Immunol. 253157-177. [DOI] [PubMed] [Google Scholar]
- 29.Ikuta, K., M. S. Ibrahim, T. Kobayashi, and K. Tomonaga. 2002. Borna disease virus and infection in humans. Front. Biosci. 7470-495. [DOI] [PubMed] [Google Scholar]
- 30.Jehle, C., I. Herpfer, M. Rauer, M. Schwemmle, and C. Sauder. 2003. Identification of differentially expressed genes in brains of newborn Borna disease virus-infected rats in the absence of inflammation. Arch. Virol. 14845-63. [DOI] [PubMed] [Google Scholar]
- 31.Kaul, M., G. A. Garden, and S. A. Lipton. 2001. Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature 410988-994. [DOI] [PubMed] [Google Scholar]
- 32.Klein, M. E., D. T. Lioy, L. Ma, S. Impey, G. Mandel, and R. H. Goodman. 2007. Homeostatic regulation of MeCP2 expression by a CREB-induced microRNA. Nat. Neurosci. 101513-1514. [DOI] [PubMed] [Google Scholar]
- 33.Lieberman, P. M. 2006. Chromatin regulation of virus infection. Trends Microbiol. 14132-140. [DOI] [PubMed] [Google Scholar]
- 34.Lipkin, W. I., and M. Hornig. 2004. Psychotropic viruses. Curr. Opin. Microbiol. 7420-425. [DOI] [PubMed] [Google Scholar]
- 35.Ludwig, H., and L. Bode. 2000. Borna disease virus: new aspects on infection, disease, diagnosis and epidemiology. Rev. Sci. Tech. 19259-288. [DOI] [PubMed] [Google Scholar]
- 36.Maloney, M. T., and J. R. Bamburg. 2007. Cofilin-mediated neurodegeneration in Alzheimer's disease and other amyloidopathies. Mol. Neurobiol. 3521-44. [DOI] [PubMed] [Google Scholar]
- 37.Maxwell, K. L., and L. Frappier. 2007. Viral proteomics. Microbiol. Mol. Biol. Rev. 71398-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morimoto, K., D. C. Hooper, S. Spitsin, H. Koprowski, and B. Dietzschold. 1999. Pathogenicity of different rabies virus variants inversely correlates with apoptosis and rabies virus glycoprotein expression in infected primary neuron cultures. J. Virol. 73510-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mosevitsky, M. I. 2005. Nerve ending “signal” proteins GAP-43, MARCKS, and BASP1. Int. Rev. Cytol. 245245-325. [DOI] [PubMed] [Google Scholar]
- 40.Perrone-Bizzozero, N., and F. Bolognani. 2002. Role of HuD and other RNA-binding proteins in neural development and plasticity. J. Neurosci. Res. 68121-126. [DOI] [PubMed] [Google Scholar]
- 41.Pletnikov, M., D. Gonzalez-Dunia, and L. Stitz. 2002. Experimental infection: pathogenesis of neurobehavioral disease, p. 125-178. In K. Carbone (ed.), Borna disease virus and its role in neurobehavioral disease. ASM Press, Washington, DC.
- 42.Poleshko, A., I. Palagin, R. Zhang, P. Boimel, C. Castagna, P. D. Adams, A. M. Skalka, and R. A. Katz. 2008. Identification of cellular proteins that maintain retroviral epigenetic silencing: evidence for an antiviral response. J. Virol. 822313-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Poulain, F. E., and A. Sobel. 2007. The “SCG10-like protein” SCLIP is a novel regulator of axonal branching in hippocampal neurons, unlike SCG10. Mol. Cell. Neurosci. 34137-146. [DOI] [PubMed] [Google Scholar]
- 44.Schneemann, A., P. A. Schneider, R. A. Lamb, and W. I. Lipkin. 1995. The remarkable coding strategy of Borna disease virus: a new member of the nonsegmented negative strand RNA viruses. Virology 2101-8. [DOI] [PubMed] [Google Scholar]
- 45.Tian, Q. 2006. Proteomic exploration of the Wnt/beta-catenin pathway. Curr. Opin. Mol. Ther. 8191-197. [PubMed] [Google Scholar]
- 46.Tian, Q., S. B. Stepaniants, M. Mao, L. Weng, M. C. Feetham, M. J. Doyle, E. C. Yi, H. Dai, V. Thorsson, J. Eng, D. Goodlett, J. P. Berger, B. Gunter, P. S. Linseley, R. B. Stoughton, R. Aebersold, S. J. Collins, W. A. Hanlon, and L. E. Hood. 2004. Integrated genomic and proteomic analyses of gene expression in mammalian cells. Mol. Cell. Proteomics 3960-969. [DOI] [PubMed] [Google Scholar]
- 47.Tomonaga, K. 2004. Virus-induced neurobehavioral disorders: mechanisms and implications. Trends Mol. Med. 1071-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tomonaga, K., T. Kobayashi, and K. Ikuta. 2002. Molecular and cellular biology of Borna disease virus infection. Microbes Infect. 4491-500. [DOI] [PubMed] [Google Scholar]
- 49.Villanueva, R., A. H. Iglesias, S. Camelo, L. C. Sanin, S. G. Gray, and F. Dangond. 2006. Histone deacetylase 3 represses HTLV-1 tax transcription. Oncol. Rep. 16581-585. [PubMed] [Google Scholar]
- 50.Volmer, R., C. Monnet, and D. Gonzalez-Dunia. 2006. Borna disease virus blocks potentiation of synaptic activity through inhibition of protein kinase C signaling. PLoS Pathog. 2e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Volmer, R., C. M. Prat, G. Le Masson, A. Garenne, and D. Gonzalez-Dunia. 2007. Borna disease virus infection impairs synaptic plasticity. J. Virol. 818833-8837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams, B. L., and W. I. Lipkin. 2006. Endoplasmic reticulum stress and neurodegeneration in rats neonatally infected with Borna disease virus. J. Virol. 808613-8626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xystrakis, E., A. S. Dejean, I. Bernard, P. Druet, R. Liblau, D. Gonzalez-Dunia, and A. Saoudi. 2004. Identification of a novel natural regulatory CD8 T-cell subset and analysis of its mechanism of regulation. Blood 1043294-3301. [DOI] [PubMed] [Google Scholar]
- 54.Ye, J., L. Gradoville, D. Daigle, and G. Miller. 2007. De novo protein synthesis is required for lytic cycle reactivation of Epstein-Barr virus, but not Kaposi's sarcoma-associated herpesvirus, in response to histone deacetylase inhibitors and protein kinase C agonists. J. Virol. 819279-9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Young, J. I., E. P. Hong, J. C. Castle, J. Crespo-Barreto, A. B. Bowman, M. F. Rose, D. Kang, R. Richman, J. M. Johnson, S. Berget, and H. Y. Zoghbi. 2005. Regulation of RNA splicing by the methylation-dependent transcriptional repressor methyl-CpG binding protein 2. Proc. Natl. Acad. Sci. USA 10217551-17558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang, K., K. E. Williams, L. Huang, P. Yau, J. S. Siino, E. M. Bradbury, P. R. Jones, M. J. Minch, and A. L. Burlingame. 2002. Histone acetylation and deacetylation: identification of acetylation and methylation sites of HeLa histone H4 by mass spectrometry. Mol. Cell. Proteomics 1500-508. [DOI] [PubMed] [Google Scholar]
- 57.Zhang, W., and D. L. Benson. 2002. Developmentally regulated changes in cellular compartmentation and synaptic distribution of actin in hippocampal neurons. J. Neurosci. Res. 69427-436. [DOI] [PubMed] [Google Scholar]
- 58.Zheng, X., L. Hong, L. Shi, J. Guo, Z. Sun, and J. Zhou. 2008. Proteomics analysis of host cells infected with infectious bursal disease virus. Mol. Cell. Proteomics 7612-625. [DOI] [PubMed] [Google Scholar]
- 59.Zhou, Z., E. J. Hong, S. Cohen, W. N. Zhao, H. Y. Ho, L. Schmidt, W. G. Chen, Y. Lin, E. Savner, E. C. Griffith, L. Hu, J. A. Steen, C. J. Weitz, and M. E. Greenberg. 2006. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron 52255-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu, K., J. Zhao, D. M. Lubman, F. R. Miller, and T. J. Barder. 2005. Protein pI shifts due to posttranslational modifications in the separation and characterization of proteins. Anal. Chem. 772745-2755. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.