Abstract
Paramecium bursaria chlorella virus 1 (PBCV-1) is the prototype of a family of large, double-stranded DNA, plaque-forming viruses that infect certain eukaryotic chlorella-like green algae from the genus Chlorovirus. PBCV-1 infection results in rapid host membrane depolarization and potassium ion release. One interesting feature of certain chloroviruses is that they code for functional potassium ion-selective channel proteins (Kcv) that are considered responsible for the host membrane depolarization and, as a consequence, the efflux of potassium ions. This report examines the relationship between cellular depolarization and solute uptake. Annotation of the virus host Chlorella strain NC64A genome revealed 482 putative transporter-encoding genes; 224 are secondary active transporters. Solute uptake experiments using seven radioactive compounds revealed that virus infection alters the transport of all the solutes. However, the degree of inhibition varied depending on the solute. Experiments with nystatin, a drug known to depolarize cell membranes, produced changes in solute uptake that are similar but not identical to those that occurred during virus infection. Therefore, these studies indicate that chlorovirus infection causes a rapid and sustained depolarization of the host plasma membrane and that this depolarization leads to the inhibition of secondary active transporters that changes solute uptake.
Chloroviruses belong to the family Phycodnaviridae. They are large (190 nm in diameter), icosahedral, plaque-forming viruses with linear double-stranded DNA genomes (41, 45). The type member of the genus Chlorovirus is Paramecium bursaria chlorella virus 1 (PBCV-1); it has a genome of 331 kb that encodes ∼366 proteins and 11 tRNAs. The PBCV-1 virion has a glycoprotein shell that surrounds a lipid bilayered membrane. Proteomic experiments established that the PBCV-1 virion contains more than 100 different virus-encoded proteins (D. Dunigan et al., unpublished data).
Chloroviruses infect certain freshwater, unicellular, eukaryotic chlorella-like green algae, which normally exist as endosymbionts in protists. The addition of PBCV-1 to its host, Chlorella strain NC64A, leads to the following program of events: (i) virus attachment to the cell wall is host specific and occurs at a unique virus vertex (45), followed by wall degradation at the point of attachment (25); (ii) rapid host membrane depolarization (11) and potassium ion release (28) occur within minutes of virus attachment; (iii) host nuclear DNA degradation begins at 3 to 5 min postinfection (p.i.) (1); (iv) early viral transcripts begin to appear at 5 to 10 min p.i. (17, 36); (v) virus DNA replication begins at 60 to 90 min p.i. (42); (vi) late virus transcription begins at 60 to 90 min p.i. (36); and (vii) viral-induced lysis and particle release occur at 6 to 8 h p.i.
Chlorella cells typically have a negative resting membrane potential of −120 to −150 mV (19) (Fig. 1). As in other plant cells, this negative voltage is achieved by a H+ ATPase, which pumps H+ out of the cytoplasm, and a low conductance of all passive transporters. This resulting proton motive force across the plasma membrane provides the principal driving force for solute fluxes across the plasma membrane via secondary active transporters (also referred to as secondary transporters) (22).
FIG. 1.
Electrochemical potential and transport mechanisms of a plant cell (adapted from reference 22). (A) Plant cell electrochemical potential is usually based on a proton (H+) gradient. A cell establishes the H+ gradient across the cell plasma membrane by using ATP energy to pump protons out of the cell. (B and C) Active transporters can be classified as primary (ATP powered) or secondary (driven by the electrochemical gradient potential). Secondary transporters translocate a solute across the cell membrane into the cell using the energy of the proton moving down its concentration gradient. Some solutes have very complex transport pathways. A cell can have several transporters for a solute, it can be an active transport system (ATP powered; e.g., ABC transporters), and it can have secondary active transporters working at the same time. All of these transporters may be controlled differently; some of them are constitutively expressed, some need induction for expression, and many of them are regulated by a feedback, depending on solute concentration or other factors (22).
Many chloroviruses encode a channel protein (called Kcv) that forms a functional K+-selective channel. PBCV-1 Kcv was the first virus-encoded K+ channel protein to be discovered, and it is also the smallest protein (94 amino acids) known to form a functional K+ channel when expressed in Xenopus oocytes (33) and mammalian HEK293 (27) and CHO (12) cells. Chloroviruses expressing Kcv homologs have selective sensitivity to known K+ channel blockers, such as Ba2+ and Cs+, resulting in diminished virus replication (24). Kcv is postulated to be located in the internal ∼40-Å-thick membrane of the virus particles and to play the following role during virus infection: after PBCV-1 infects its host by degrading the cell wall, presumably there is contact between the virus and host membrane, resulting in a fusion of the viral and the host cell membranes. Because of the high conductance of the Kcv channel in the viral membrane, this fusion of membranes causes a rapid elevation of the K+ conductance and a consequent depolarization of the entire host plasma membrane/virus membrane continuum. The combination of membrane depolarization and elevated K+ conductance leads to K+ release from the host cell. This release reduces the host cell osmotic pressure, which can reach 0.7 MPa in plant cells (44), and facilitates viral DNA ejection into the host (24, 28).
The present report further examines the effect of chloroviruses on membrane transport features of the host cell. The data show that virus infection results in a drastic decrease in the secondary active transport of solutes across the plasma membrane into the host cell. This decrease provides additional support for the hypothesis that chlorovirus infection causes a depolarization of the host cell membrane; as a consequence, the uptake of solutes by secondary active transporters rapidly decreases. This means that all of the material which is required for the synthesis of virus progeny must be recruited from within the host cell. In addition, the data also imply that infected host cells are impaired in taking up radioactive adenine so that results from previous experiments on RNA synthesis during virus infection must be reinterpreted.
MATERIALS AND METHODS
Strains and culture conditions.
The growth of the host Chlorella strain NC64A on bold basal medium (BBM) modified by the addition of 0.5% sucrose and 0.1% peptone (MBBM) and the production and purification of viruses PBCV-1 and NY-2A have been previously described elsewhere (1, 43).
Solute uptake analysis.
Chlorella cells (5 × 107 to 6 × 107 per ml) were infected with virus at a multiplicity of infection (MOI) of 10 PFU per cell. Mock-infected cells (using Tris buffer; 50 mM Tris-HCl, pH 7.8) served as a control. The radioactively labeled solutes (Table 1) were added for 10 min, starting at 10 min p.i.; radioactive solutes not taken up by the cells were removed by diluting the samples 20-fold with ice-cold growth medium, followed by centrifugation at 3,000 × g for 5 min at 4°C. The cell pellets were resuspended in fresh growth medium, and three samples (500 μl) were taken from each treatment, mixed with 5 ml of Opti-Fluor (PerkinElmer, Boston, MA), and counted in a liquid scintillation counter.
TABLE 1.
Characteristics of the radioactive solutes testeda
| Solute | Properties and type of transporter/class | Isotope | Specific activity (MBq/ml) | Solute concn (nM) |
|---|---|---|---|---|
| Adenine | Nucleobase | 3H | 37 | 215 |
| Glucose | Monosaccharide | 3H | 37 | 80 |
| Proline | Hydrophobic amino acid | 3H | 37 | 50 |
| Histidine | Basic amino acid | 14C | 1.85 | 0.8 |
| Methionine | Hydrophobic amino acid | 3H | 37 | 40 |
| Ornithine | Basic amino acid | 14C | 1.85 | 4.8 |
| Putrescine | Polyamine | 14C | 1.85 | 2.34 |
All tested solutes were obtained from Amersham/GE Healthcare, Piscataway, NJ.
Nystatin treatment.
Prior to infection, chlorella cells (5 × 107 to 6 × 107 per ml) were incubated for 10 min with nystatin (Sigma, St. Louis, MO) at a final concentration of 5 μg/ml (nystatin was dissolved in methanol at 5 mg/ml). An equal amount of methanol (1 μl/ml) was added to control cells. The radioactively labeled solutes were added for 10 min from 10 to 20 min p.i., and the samples were then processed as described above.
Sodium azide treatment.
Sodium azide (100 μM) was added to chlorella cells (1.2 × 107 to 1.5 × 107 cells/ml) for 3 min before virus infection or the addition of nystatin (5 μg/ml). The radioactively labeled solutes were added for 10 min from 10 to 20 min p.i. Reactions were stopped by adding ice-cold growth medium, and the samples were processed for radiometric analysis as described above. Mock-infected cells (Tris buffer) and chlorella cells with methanol (1 μl/ml) served as controls.
Growth medium pH adjustments.
Chlorella cells grown in MBBM to 1.2 × 107 to 1.5 × 107 cells per ml were collected by centrifugation; the cells were resuspended in either MBBM at pH 5.0 (adjusted with H3PO4), MBBM at pH 8.0 (adjusted with NaOH) or regular MBBM (pH 7.0) to 5 × 107 to 6 × 107 cells per ml. The cells were equilibrated in the modified media for 3 h prior to initiating experiments. Cells were infected with PBCV-1 at an MOI of 10. Mock-infected cells (Tris buffer) in MBBM with adjusted pH values served as the controls. Radioactively labeled solutes were added for 10 min, starting at 10 min p.i., and then the radioactive tracer was removed by dilution and the samples were processed as described above.
Exogenous digestion of the chlorella cell wall.
The cell walls of Chlorella NC64A were disrupted using a crude mixture of cell wall-digesting enzymes (called “lysin”) prepared from Acanthocystis turfacea chlorellavirus 1 (ATCV-1) (5). ATCV-1 has a different host (Chlorella SAG 3.83), and the virus does not attach to Chlorella NC64A cells. Chlorella NC64A cells (4 × 108 to 5 × 108 cells/ml) in BBM were treated for 1 h with the ATCV-1 lysin. Control cells had an equal amount of Tris buffer. Radioactive [3H]adenine was added for 10 min from 10 to 20 min p.i., and the samples were processed as described above.
Lysin preparation and activity assay.
An ATCV-1 virus preparation (3 mg/ml) (5) in Tris buffer was sonicated (Tekmar ultrasonic processor; 100 W model) in an ice bath for 2 min (5-s pulses for 24 pulses) and centrifuged at 4°C for 15 min at 18,000 × g. The supernatant fraction (200 μl) containing the soluble virion-associated proteins (ATCV-1 lysin) was mixed with 800 μl Chlorella NC64A cells (4 × 108 to 5 × 108 cells/ml) and incubated for 1 h at 20°C. Chlorella NC64A wall digestion by ATCV-1 lysin was assayed by measuring sodium dodecyl sulfate (SDS)-mediated chlorophyll release (18). After ATCV-1 lysin treatment, SDS was added to the samples to a final concentration of 2% and incubated for 30 min at 20°C before centrifuging for 10 min at 18,000 × g. The optical densities of the supernatant fractions were determined at a λ of 435 nm to measure chlorophyll release, indicating the extent of cell wall damage.
Treatment with potassium ion channel blockers.
Chlorella cells in MBBM (1.2 × 107 to 1.5 × 107cells/ml) were infected with viruses at an MOI of 10. The K+ channel blockers at 10 mM Ba2+ (as BaCl2) and Cs+ (as CsCl) were added to the cells 3 min before virus infection. Mock-infected cells (Tris buffer) with the corresponding K+ channel inhibitor served as controls. The radioactively labeled solute was added for 10 min, from 10 to 20 min p.i., and then the samples were processed for radiometric analysis as described above.
RNA synthesis analysis.
Exponentially growing chlorella cells at 1.2 × 107 to 2.0 × 107 cells/ml were collected by centrifugation and then resuspended in fresh growth medium at a concentration of 5 × 107 to 6 × 107 cells/ml. Virus was added at an MOI of 10. Mock-infected control cells had an equal amount of Tris buffer.
Pulse-labeling cells with adenine.
Infected cells were pulse-labeled for 5 min with [3H]adenine at 5, 15, 25, 40, 55, and 85 min p.i. Infections were stopped by adding an equal volume of 10% trichloroacetic acid (TCA) to the reaction mixtures. Cells were washed three times with 5% TCA, resuspended in 0.5 M NaOH, incubated overnight at 37°C, and centrifuged for 10 min at 18,000 × g, and three samples (250 μl) of the supernatant fraction were neutralized with an equal volume of 10% TCA, mixed with 5 ml of Opti-Fluor, and counted with a liquid scintillation analyzer as an indicator of RNA synthesis.
Preloading cells with adenine.
Chlorella cells were incubated with [3H]adenine for 5 min, and then the radioactive tracer was removed by centrifugation and the cells were washed with growth medium. A sample taken at the time of infection served as a baseline. Samples were collected at 10, 20, 30, 45, 60, and 90 min p.i., and infection was stopped by adding an equal volume of 10% TCA. Samples were processed for radiometric analysis as described above.
Membrane potential recordings.
Changes in membrane voltage of Chlorella NC64A cells were recorded with the voltage-sensitive dye bisoxonol as described previously (11). To account for photobleaching during the long measurements, each recording was accompanied by monitoring the fluorescence under the same conditions but without viruses. The data were normalized to the value at the beginning of the recording, and the data that were obtained without virus infection were subtracted from those measured in the presence of viruses.
Cytosolic pH.
The cytosolic pH of chlorella cells was measured with the ratiometric pH-sensitive dye 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein (BCECF) (Mobitec, Göttingen, Germany). Chlorella cells (3.5 × 106 cells/ml) were incubated for 90 min in the dark in BBM containing 20 μM dye in its acetoxymethyl ester form (BCECF-AM). This molecule is membrane permeable; the ester is cleaved by esterases inside the cell, and the pH-sensitive dye is then trapped inside the cells and fluoresces. Ratiometric measurements were performed in a fluorescence spectrometer (Jasco FP-6200; Tokyo, Japan), switching between the two excitation wavelengths 490 ± 5 nm and 436 ± 5 nm. The emitted fluorescent light was collected at 530 ± 10 nm. Cells with a density of 3.5 × 106 cells/ml were incubated in a 3.5-ml quartz cuvette; after obtaining a stable baseline, the virus was added at an MOI of 10. In vivo calibrations were performed according to Dixon at el. (9) as follows: chlorella cells were first loaded with dye as described above and were then transferred into MBBM with defined pH values of between pH 5.0 and pH 9.0. Buffering was achieved with either 10 mM MES (morpholineethanesulfonic acid) or HEPES buffers. The permeability of the plasma membrane for protons was increased by adding 20 μg/ml of nigericin (Sigma, Deisenhoven, Germany) to the incubation buffer. The fluorescence ratio F490/F436 was recorded after reaching a new steady state, assuming an equilibration of the cytoplasmic pH with that of the bath medium. The calibration curve was fitted as previously described by Plieth et al. (32). From the calibration, we estimated a resting pH of about pH 6.5 for the Chlorella NC64A cells. This value is low for a cytoplasmic pH and may indicate that the measurement is influenced by some dye uptake into the chloroplast.
Statistical analysis.
All experiments were conducted at least twice with three subsamplings in each treatment. The effect of a particular treatment on the transport of different solutes was calculated as the percentage of control for each subsampling in the experiment. The results are expressed as the mean ± the standard deviation (SD). Statistical analysis between different solutes and treatments was carried out using the least squares means method with SAS software (SAS Institute, Inc., Cary, NC). The differences between mean values were considered significant if the P value was less than 0.05.
RESULTS
Effect of virus infection on adenine uptake.
Adenine is commonly used as a precursor to monitor RNA and DNA synthesis in vivo. Adenine uptake typically occurs by a secondary transporter named purine permease (PUP) (8, 13). PUP is an integral membrane protein that transports adenine and cytosine with high affinity. The protein is a secondary cotransporter that uses the proton motive force across the plasma membrane to take up the nucleosides. Adenine uptake occurs against a concentration gradient and is sensitive to protonophores (13).
An experiment to monitor the effect of virus PBCV-1 infection on adenine uptake indicates that adenine transport was inhibited about 77% in virus-infected cells from 10 to 20 min p.i. (Fig. 2A). The cells were also monitored for adenine uptake from 170 min to 180 min p.i. to determine if the initial inhibition of uptake was transitory (Fig. 2A). Adenine transport did not return to the mock-infected level during the first 3 h p.i., which is well into the virus DNA synthesis phase of infection.
FIG. 2.
Effect of virus PBCV-1 infection and nystatin treatment on Chlorella NC64A adenine uptake. (A) Effect of PBCV-1 infection on adenine uptake. Chlorella cells were infected with PBCV-1 at an MOI of 10. Cells were pulse labeled with [3H]adenine 10 min before the indicated time (a 10-min pulse). Mock-infected chlorella cells served as a control. Results are plotted as the mean ± SD (n = 6). Asterisk, P ≥ 0.05. (B) Comparison of PBCV-1 infection to nystatin treatment. Chlorella cells were treated with nystatin 5 μg/ml for 10 min before labeling with [3H]adenine for 10 min. Untreated cells served as a control. Results are plotted as the mean ± SD (n = 9). Two asterisks, P ≥ 0.05.
The reduced import of adenine could in principle be due to depolarization of the host membrane and/or acidification of the cytosol. Both processes would result in a decrease in the proton motive force, which facilitates adenine import. Therefore, we explored both possibilities. First, we monitored chlorella membrane voltage over the entire 3 h p.i. The results indicate that infection of Chlorella NC64A rapidly depolarized the cell membrane (Fig. 3). After passing a peak depolarization of about 700 s p.i., the membrane only partially repolarized; during the entire period, the membrane remained more positive than the resting voltage prior to infection. Collectively, this means that virus infection is associated with a sustained lowering of the electrical driving force for secondary transport.
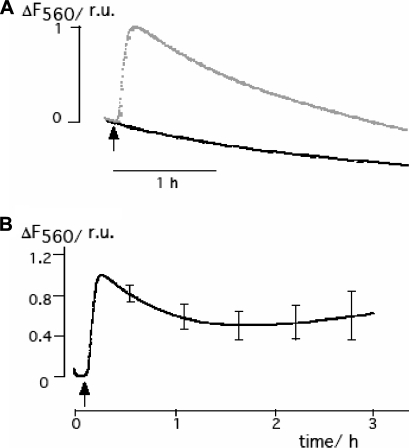
FIG. 3.
Long-term effect of PBCV-1 on the membrane potential of Chlorella NC64A cells as reported from the fluorescence of bisoxonol at 560 nm (F560). Approximately 3.5 × 106 chlorella cells/ml were maintained in MBBM containing 1 μM bisoxonol. Viruses were added at an MOI of 10 at the time indicated by the arrow. (A) Change in bisoxonol fluorescence in solution with Chlorella NC64A cells with (gray line) and without (black line) the addition of viruses. The black curve reports the photobleaching of the fluorescent dye; the virus-induced increase in bisoxonol fluorescence, which results from a depolarization of the cell, is given by the difference between the two curves. (B) Mean difference (± SD; n = 4) in PBCV-1-induced bisoxonal fluorescence. To compare different measurements, the data were normalized to the peak of the fluorescence increase.
Second, we measured the cytoplasmic pH of Chlorella NC64A cells during virus infection. Chlorella NC64A cells were loaded with the fluorescent pH-sensitive dye BCECF. The ratio of fluorescence obtained at an excitation of F490 and F436 is pH sensitive (32); an increase in the ratio indicates alkalinization of the cell, while a decrease indicates acidification. As expected, loading of the cells with the weak acid by adding 10 mM acetate to the extracellular medium resulted in a decrease in the fluorescence ratio (Fig. 4). The trajectory of the fluorescence ratio in Fig. 4 shows that virus infection of Chlorella NC64A cells (an MOI of 10) resulted in an increase in the ratio, indicating alkalinization of the cytoplasm. Similar results were obtained in eight independent experiments, giving a mean increase in the fluorescence ratio of 0.25 ± 0.06. From in vivo calibrations, this increase in ratio translates into a pH difference of 0.4 to 0.5 pH units. The effect of PBCV-1 infection on the cytosolic pH was specific for the host. Similar experiments with Chlorella vulgaris, a nonhost for the virus, produced no appreciable change in pH (Fig. 4). Collectively, these data indicate that infection of Chlorella NC64A cells is associated with an increase in the cytosolic pH of the host cells. However, this alkalinization does not explain the inhibition of adenine import; in contrast, for thermodynamic reasons, alkalinization should favor adenine import via secondary transport. Hence, the primary reason for the reduced uptake of adenine after virus infection must be due to the rapid and sustained depolarization of the plasma membrane.
FIG. 4.
Virus PBCV-1 infection evokes an alkalinization of the internal pH of Chlorella NC64A cells. Fluorescence of Chlorella NC64A cells was monitored with the preloaded BCECF dye (inset). The continuous recording indicates the trajectory of the ratiometric fluorescence recording of cells before and after (at arrow) infection with PBCV-1 at an MOI of 10. The bar graph shows the mean difference (± SD; n > 6) in fluorescence ratio ΔF490/F436 over a period of 3,000 s in response to virus infection of Chlorella NC64A (solid bar) or Chlorella vulgaris (gray bar). The hatched bar reports the decrease in fluorescence ratio ΔF490/F436 upon the addition of 10 mM acetate to the incubation buffer.
Effect of nystatin on Chlorella NC64A adenine transport.
Cells were treated with different concentrations of nystatin (0.5 to 200 μg/ml), a drug known to reduce chlorella cell membrane potential (24, 30), in order to evaluate the secondary active nature of adenine transport in Chlorella NC64A cells. Nystatin reduced adenine uptake about 80% at 5 μg/ml (∼5 μM) (Fig. 2B); higher concentrations did not increase the inhibition of adenine uptake significantly. Apparently, 5 μg/ml nystatin is a saturating dose for Chlorella NC64A cells. Separate experiments established that 5 μg/ml nystatin inhibits Chlorella NC64A growth and prevents PBCV-1 replication (results not shown).
The inhibition of adenine uptake by nystatin mimicked the inhibition by PBCV-1 infection, indicating that plasma membrane potential is required for adenine transport into Chlorella NC64A cells. This similarity between virus infection and nystatin treatment also supports the concept that the change in adenine uptake in virus-infected cells is due to virus-mediated membrane depolarization, probably leading to an inhibition of PUP.
Transporter proteins encoded by Chlorella NC64A.
The Chlorella NC64A genome was recently sequenced by the Joint Genome Institute (U.S. Department of Energy), and ninefold coverage data are available for annotation (http://www.jgi.doe.gov/NC64A/). The Chlorella NC64A genome was analyzed for genes encoding cytoplasmic membrane transport proteins. This analysis also led to the identification of the types of solutes that might be altered by depolarization of the plasma membrane.
The sequence analysis revealed that Chlorella NC64A encodes at least 482 putative transporters and ion channels (Table 2). This number of transport-associated proteins is similar to that of other algae whose genomes have been sequenced (35), e.g., Chlamydomonas reinhardtii (486 transporter proteins), Thalassiosira pseudonana (429 transporters), and Ostreococcus tauri (323 transporters). The density of genes encoding transporters for Chlorella NC64A is about 11.3 transporter proteins per 1 Mb of genome. Other algae range between 4.9 transporters per 1 Mb of genome for C. reinhardtii and 28 transporters per 1 Mb for O. tauri.
TABLE 2.
Putative solute transporters encoded by Chlorella NC64A
| Characteristic | Valuea |
|---|---|
| Genome size | 46.2 |
| Transporters per Mb genome | 11.3 |
| ATP dependent | 194 |
| Ion channels | 64 |
| Secondary active transporter | 224 |
| Amino acid, polyamine, and oligopeptide transporter | 73 |
| Nucleobases and nucleoside transporter | 17 |
| Hexose and carbohydrate transporter | 37 |
| Inorganic ion transporter | 60 |
| Other | 37 |
| Total | 482 |
All values are number of transporters except for genome size (Mb).
Sixty-four of the Chlorella NC64A genes are predicted to encode ion channel proteins; such channel proteins transport solutes in a passive fashion. The transporter group driven by ATP hydrolysis consists of 194 proteins; transporters in this class are often referred to as primary active transporters. These proteins couple energy released during ATP hydrolysis to translocate a substance against its electrochemical potential. Transmembrane transporters of the other major class are referred to as secondary active transporters. In plants, this type of transport utilizes energy stored in the electrochemical gradient of protons across the cell membrane (22). Chlorella NC64A encodes 224 such genes, as follows: 73 amino acid, polyamine, and oligopeptide transporters; 17 nucleobase and nucleoside transporters, including PUP; 37 hexose and sugar transporters; 60 inorganic ion transporters; and 37 other secondary transporters.
Chlorella NC64A annotation is preliminary, and the final number of transporters encoded by the alga may differ slightly from these estimates. Nevertheless, these results indicate that Chlorella NC64A shares many aspects of solute transport with other eukaryotic organisms.
Effect of virus infection on solute uptake.
Members of three of the five families of secondary transporters were evaluated for their response to virus infection as follows: (i) the amino acid, polyamine, and oligopeptide family; (ii) the nucleobase and nucleoside family; and (iii) the hexose and carbohydrate family. The amino acid, polyamine, and oligopeptide family was evaluated using four amino acids (proline, methionine, histidine, and ornithinine) and one polyamine precursor (putrescine). Adenine was used to evaluate the nucleobase and nucleoside family, and glucose was used to evaluate the hexose and carbohydrate family. These compounds are commercially available as radioactive tracers (their properties are listed in Table 1).
All experiments on the effect of virus infection on solute uptake were accompanied by experiments with nystatin to evaluate the secondary nature of the particular solute transport system. Experiments with different solutes revealed that virus infection caused changes in the uptake of all the solutes tested, but the degree of effect depended on the solute (Table 3). The uptake changes caused by virus infection and nystatin treatment were similar but not identical for most of the solutes. Both virus infection and nystatin had the highest inhibitory effect on proline uptake, which was nearly abolished, about 3% to 6% of the control in both cases.
TABLE 3.
Effect of PBCV-1 infection on solute transport in Chlorella NC64A cells
| Solute | Transporter class | Solute uptake as a % of that of the mock-infected controla
|
||
|---|---|---|---|---|
| Virus | Nystatin | P value | ||
| Adenine | Nucleobase | 23 ± 7.9 (37) | 21 ± 4.9 (9) | 0.476 |
| Glucose | Monosaccharide | 66 ± 15.1 (9) | 30 ± 9.2 (6) | 0.014 |
| Proline | Hydrophobic amino acid | 3 ± 3.4 (28) | 6 ± 4.7 (9) | 0.8521 |
| Methionine | Hydrophobic amino acid | 19 ± 8.3 (9) | 25 ± 7.2 (9) | 0.657 |
| Histidine | Basic amino acid | 66 ± 6.8 (9) | 82 ± 8.2 (6) | 0.273 |
| Ornithine | Basic amino acid | 63 ± 9.3 (18) | 100 ± 13.0 (9) | 0.016 |
| Putrescine | Polyamine | 35 ± 13.0 (12) | 10 ± 2.7 (9) | 0.096 |
Values are means ± SD (number of experimental replicates).
Glucose and putrescine uptake was suppressed by both PBCV-1 infection and nystatin, but it was significantly less for both solutes in virus-infected cells than in nystatin-treated cells. Glucose transport in nystatin-treated cells was about 30% of that of the control, whereas in PBCV-1-infected cells it was 66% of that of mock-infected cells (Table 3). Putrescine uptake was 10% compared to that of the control in nystatin-treated cells versus 35% of that in virus-infected cells.
Histidine and ornithine (both basic amino acids) transport were more affected by virus infection than by nystatin treatment (Table 3). In PBCV-1-infected cells, ornithine uptake was 63% of that of control cells. Nystatin treatment did not alter ornithine uptake, indicating that this solute is likely not transported by secondary transporters. Histidine transport was 66% in PBCV-1-infected cells compared to that in mock-infected cells, and in nystatin-treated cells it was 82% of that of the control cells.
Effect of sodium azide on Chlorella NC64A glucose and putrescine transport.
The inhibition of both glucose and putrescine uptake was less in virus-infected cells than in nystatin-treated cells. To determine the impact of ATP on these transport processes, cells were treated with sodium azide (NaN3), an uncoupler of oxidative phosphorylation. Studies of putrescine and glucose uptake were conducted in the presence of 100 μM NaN3 added 3 min before virus infection or nystatin treatment. NaN3 diminished the differences in glucose and putrescine uptake between virus infection and nystatin treatment and reduced the uptake of both solutes to ∼10% of that of the control (results not shown). These results are consistent with the view that glucose and putrescine uptake into Chlorella NC64A depends on energy from ATP hydrolysis. The results of these experiments further imply that the effect of virus-induced depolarization on the proton motive force for solute uptake is less severe than that obtained by the ionophore nystatin or by depletion of the cellular ATP concentration in response to azide.
Effect of K+ channel blockers on solute transport in virus-infected Chlorella NC64A.
As mentioned in the introduction, the virus-encoded K+ channel protein (Kcv) is predicted to be responsible for cell membrane depolarization during virus infection. Chlorovirus-encoded Kcv channels have selective sensitivity to known K+ channel blockers (24, 28). PBCV-1 Kcv is blocked by Ba2+ and is less sensitive to Cs+. The virus NY-2A Kcv ion channel is blocked by both Ba2+ and Cs+. Adenine and proline uptake were measured in PBCV-1- and NY-2A-infected cells in the presence of either Ba2+ or Cs+ to test the hypothesis that virus infection induced changes in the membrane potential and, consequently, that the inhibition of secondary transporter proteins is dependent on the Kcv channels.
Adding Ba2+ decreased the inhibition of adenine transport by PBCV-1 infection. Adenine uptake went from 23% of that of the control to 84% of that of the control in Ba2+-treated cells. Ba2+ had a similar effect on proline uptake, reducing the inhibitory affect of virus infection from 3% of the mock-infected control to 42% (Table 4). Cs+ did not have a significant effect on the uptake of either adenine or proline by PBCV-1-infected cells, consistent with the linkage to Kcv and membrane depolarization sensitivity.
TABLE 4.
Effect of K+ channel inhibitors on adenine and proline transport in virus-infected Chlorella NC64A cellsa
| Solute | Treatment | Solute uptake as a % of that of the mock-infected control
|
|||
|---|---|---|---|---|---|
| PBCV-1
|
NY-2A
|
||||
| Mean ± SD | P value | Mean ± SD | P value | ||
| Adenine | None | 23 ± 7.9 (37) | 12 ± 6.8 (6) | ||
| Ba2+ | 84 ± 6.8 (21) | 0.0001 | 74 ± 12.0 (6) | 0.025 | |
| Cs+ | 38 ± 12.7 (6) | 0.177 | 72 ± 13.7 (6) | 0.034 | |
| Proline | None | 3 ± 3.4 (28) | 2 ± 0.2 (6) | ||
| Ba2+ | 42 ± 10.4 (15) | <0.0001 | 4 ± 1.6 (6) | 0.021 | |
| Cs+ | 9 ± 1.5 (6) | 0.300 | 7 ± 4.7 (6) | 0.049 | |
Chlorella cells were treated with 10 mM Ba2+ or Cs+ for 3 min and infected with PBCV-1 at an MOI of 10. Radioactively labeled solutes were added for 10 min starting at 10 min p.i. Control cells had an equal amount of inhibitor. Results are presented as means ± SD (number of experimental replicates). The differences between mean values were considered significant if the P value was less than 0.05. PBCV-1 Kcv is blocked by Ba2+ and is less sensitive to Cs+. The NY-2A Kcv ion channel is blocked by both Ba2+ and Cs+.
In NY-2A infection, the inhibition of adenine uptake was significantly altered by Ba2+ treatment (from 12% of that of the mock-infected control to 74% for adenine) and Cs+ treatment (from 2% to 72% for adenine). However, the effect of Ba2+ and Cs+ on proline uptake is difficult to explain. The addition of both of these cations led to increased proline uptake, which was statistically significant (Table 4); however, the increase from 2% to 4% for Ba2+ and 2% to 7% for Cs+ in absolute values does not represent a major reversal of virus-mediated inhibition.
One possible explanation for these observations is that different transporters have different dependencies on membrane potential. Such differences are expected, depending on the mode of operation of the respective secondary transporters. An electrogenic symporter, for example, exhibits a much stronger dependency on the membrane potential than an electroneutral transporter.
Effect of medium pH on adenine and proline transport in Chlorella NC64A.
Most secondary transporters in plants utilize protons to cotransport solutes against their gradient. For thermodynamic reasons, it can be expected that external acidification increases while alkalinization decreases secondary active transporters, because the cell has to invest less or more energy, respectively, to maintain the proton gradient across the plasma membrane. The full impact of extracellular pH on transport, however, is more complex because the activity of transport enzymes usually shows an inherent pH dependency, and the protonation of the transporter could be sensitive to the prevailing membrane voltage (37). As a result of these concerted actions, the activity of extracellular H+ on secondary transporters usually shows a sigmoidal decrease in activity, with progressive alkaline pH values (37).
To examine pH dependency, adenine and proline uptake were measured in PBCV-1-infected Chlorella NC64A cells at different medium pHs (Fig. 5A and B). Transport of adenine into Chlorella NC64A cells did not differ significantly between pH 5.0 and pH 7.0. Consistent with the hypothesis of H+-coupled transport, adenine uptake was lower at pH 8.0 than at both pH 5.0 and 7.0 (Fig. 5A). The amount of adenine taken up in virus-infected cells was about the same at all three pH values, suggesting that H+-coupled import is no longer relevant in infected chlorella cells.
FIG. 5.
Effect of pH on solute uptake in PBCV-1-infected Chlorella NC64A cells. Cells in media with different pHs were infected with PBCV-1 at an MOI of 10. Radioactively labeled solutes were added for 10 min, starting at 10 min p.i. Mock-infected cells in medium with a corresponding pH served as a control. Results are plotted as the mean ± SD (n = 6). (A) Effect of pH on adenine uptake. (B) Effect of pH on proline uptake.
The proline results support the view that uptake of this amino acid into cells is coupled to proton transport (Fig. 5B). In mock-infected cells, proline uptake is about 37% higher when the pH of the medium is 5.0 than when it is pH 7.0 (the normal pH of MBBM), and uptake is about 70% lower when the pH of the medium is 8.0 than when it is pH 7.0. At all three pH values, PBCV-1 infection resulted in about 95% inhibition of proline uptake. Thus, the degree of inhibition of proline uptake by PBCV-1 infection was not altered by the pH of the medium. This result can be explained by the fact that virus infection inhibits proline transport to nearly background levels, but the uninfected cell is hypersensitive to changes in the membrane potential.
These results, along with those shown in Table 4, indicate that nucleobases and amino acid transporters in Chlorella NC64A may have very different mechanisms. While proline transport seems to use the H+ concentration gradient, the transport of adenine makes more use of the electrical gradient. But in spite of the different mechanisms of secondary transport, both uptake systems are strongly inhibited by virus infection.
Effect of cell wall degradation on adenine uptake in Chlorella NC64A.
The cell wall is the first barrier that chloroviruses have to overcome when infecting their hosts. Chloroviruses encode and package several enzymes in the virion that are probably involved in degrading the host cell wall during infection (6, 38-40, 45). One possibility is that this enzymatic “tampering” with the host cell wall triggers a defense process in the host, which indirectly causes membrane depolarization and hence a drop in the proton motive force of the host. Such phenomena are known in higher plant cells (4). To evaluate this possibility, a crude cell wall-degrading enzyme preparation, called lysin, was isolated from virus ATCV-1 virions. Although virus ATCV-1 does not attach to Chlorella NC64A, the ATCV-1 lysin degrades Chlorella NC64A walls sufficiently so that the cells become sensitive to SDS lysis, resulting in the release of measurable chlorophyll. That is, a 1-h treatment of Chlorella NC64A cells with ATCV-1 lysin caused a ninefold increase in released chlorophyll from the treated cells (Table 5). However, the lysin treatment did not alter adenine uptake by Chlorella NC64A (Table 5). We conclude that the change in solute transport is not the result of cell wall digestion. The change probably results from the fusion of the virus membrane with the host membrane rather than a secondary effect due to a perturbed cell wall.
TABLE 5.
Effect of cell wall damage on adenine uptake in Chlorella NC64A cells
| Treatment | Chlorophyll released (A435) (n = 3) | Relative chlorophyll released (A435) | Adenine uptake (CPM, 103) (n = 3) |
|---|---|---|---|
| NC64A cells, untreated (control) | 0.043 ± 0.002 | 1.0 | 254 ± 4.13 |
| NC64A cells treated with ATCV-1 solubilized lysin | 0.365 ± 0.062 | 9.3 | 242 ± 12.05 |
Effect of PBCV-1 infection on adenine incorporation into RNA.
Several experiments have been conducted in the past 25 years on the relationship between PBCV-1 infection and the apparent rates of macromolecular syntheses (36, 42). These experiments have produced incongruent results, depending on the labeling technique. In the context of the current findings, we reevaluated the role of transport in the radiolabeling of nucleic acids during virus infection.
Nucleic acid synthetic rates (RNA and DNA) are commonly measured by pulse-labeling techniques, where radioactive tracers are used to track the incorporation of substrate molecules, including nucleobases and the corresponding nucleosides, into a polymer. Nucleotides are used less frequently as a synthesis tracer because these anionic molecules are usually not transported through cell membranes. The rates of RNA synthesis in certain Chlorella species can be estimated by incubating cells with an exogenous source of [3H]adenine for short time periods (e.g., 5 to 10 min); the alkaline soluble fraction of total cellular nucleic acid is monitored by scintillation counting. In light of the data presented here, we now believe that chlorovirus infection induces changes in chlorella membrane potential (i.e., depolarization) that alters the intracellular pool of the nucleotides by disrupting the proton motive force and consequently inhibiting PUP. This inhibition disrupts adenine uptake and thus alters the intracellular pool of radiolabeled ATP relative to that of the mock-infected control. Consequently, the net effect is an apparent suppression of transcription rate, when in fact the suppression occurs because of reduced substrate transport, resulting in an altered intracellular nucleotide pool size.
This phenomenon is illustrated with a 5-min pulse-labeling experiment with [3H]adenine during PBCV-1 infection; 94% inhibition of adenine incorporation into RNA occurred at 20 min p.i. compared to that of mock-infected cells (Fig. 6). In a companion experiment, [3H]adenine was preloaded into Chlorella NC64A cells for 5 min, and the exogenous radioactive adenine was removed from the medium before virus infection. In this experiment, the results indicated only a 60% decrease in RNA synthesis at 20 min p.i. (Fig. 6). A key feature of both labeling methods is the uptake of adenine from the exogenous environment, which is subsequently converted to ATP via a series of steps known as the salvage pathway (26). This discrepancy in the data produced by different labeling techniques can be explained by the fact that adenine uptake changes after virus infection.
FIG. 6.
Effect of PBCV-1 infection on Chlorella NC64A RNA synthesis. Chlorella NC64A cells were infected with PBCV-1 at an MOI of 10. Mock-infected chlorella cells served as a control. For “Pulse labeling,” chlorella cells were pulse labeled with [3H]adenine (a 5-min pulse) after virus infection, starting at 5 min p.i.; for “Preloaded adenine,” chlorella cells were labeled before virus infection with [3H]adenine for 5 min, and then the unincorporated radioactive tracer was removed by centrifugation. Results are plotted as the mean ± SD.
DISCUSSION
All viruses must pass through the host cell plasma membrane to establish an infection de novo. Many plant and animal viruses enter a cell intact through penetration or endocytic pathways. In contrast, most bacteriophages and certain animal and algal viruses deposit the genome and associated proteins in the cell, leaving structural or envelope elements of the virion on the exterior of the cell (14). This transgression of the plasma membrane has physiological consequences.
The immediate-early events of chlorovirus infection are preprogrammed into a complex virion. PBCV-1 contains over 100 unique virus-encoded proteins (Dunigan, unpublished data). In addition to structural functions, these proteins direct several important events leading up to the initial viral gene expression. As noted earlier, virus PBCV-1 has a gene encoding a protein that forms a functional K+ channel (named Kcv) in heterologous cells (12, 27, 33). We hypothesize that Kcv is located in the inner membrane of the virus particle and plays the following role during infection (24). PBCV-1 infects its host by attaching rapidly, specifically, and irreversibly to the external surface of the algal cell wall (25). Attachment occurs at a unique virus vertex (29; C. Xiao, V. Kostyucherko, J. L. Van Etten, and M. Rossmann, unpublished data), followed by digestion of the cell wall at the point of attachment by virion-packaged, cell wall-degrading enzymes. Presumably, this leads to fusion of the virion membrane with the host plasma membrane and activation of Kcv, which leads to depolarization of the host membrane, followed by ejection of the virus DNA plus accompanying virus-packaged proteins into the cell.
Several experiments support the following hypothesis: (i) host membrane depolarization occurs almost immediately after mixing PBCV-1 with its host chlorella (11); (ii) PBCV-1 infection results in a rapid efflux of K+ from the host cells (28); (iii) K+ channel blockers that inhibit Kcv activity in heterologous cells also block host membrane depolarization, K+ release from the host cells, and virus infection (11, 28); and (iv) the kcv gene is expressed late in infection (16) and typically virus-packaged proteins are expressed late. However, direct evidence that the Kcv protein is packaged in the virus particle is lacking. To test this Kcv hypothesis, one would like to delete the kcv gene from PBCV-1 and determine if the mutated virion is infectious, and if it is, if virus infection results in host membrane depolarization. Unfortunately, these experiments are not possible at the present time because procedures for manipulating the chlorella virus genomes are lacking.
In the present manuscript, we demonstrate that one of the consequences of host cell membrane depolarization is a rapid and sustained effect on the secondary transport of seven solutes across the plasma membrane into the host cell. Agents that depolarize the plasma membrane inhibit the uptake of solutes dependent on secondary transporters. Virus infection results in the altered function of all tested transporters in the same time period as depolarization. The fact that PBCV-1 infection does not affect the uptake of all solutes equally argues against the possibility that the agent simply creates holes in the cell, such as what occurs with certain animal viruses and results in transient increased cell permeability (14).
Nystatin, a compound known to depolarize chlorella cells (24, 30), caused the inhibition of secondary solute uptake in a manner that was similar, but not identical, to virus infection. The inhibition of virus-mediated depolarization by Ba2+ for PBCV-1 and virus NY-2A, or Cs+ for NY-2A, also reversed the inhibition of the associated transport dysfunction. Thus, we conclude that chlorella virus infection results in cell depolarization, which leads to the dysfunction of secondary solute uptake. Additionally, these experiments reveal the differential nature of certain solute transporters when the cells were treated with both virus and K+ channel blockers.
In light of the fact that virus infection results in cell depolarization (11, 24), we evaluated the uptake of adenine solute into Chlorella NC64A cells, particularly in the context of RNA synthesis. The uptake of adenine, a nucleobase precursor of ATP, was inhibited ∼75% within minutes of infection compared to what occurred with a mock-infected control. Uptake remained inhibited for at least 3 h p.i. (Fig. 2A), well past the transition point from early to late infection. Similar results were obtained with nystatin (Fig. 2B); thus, the transport of adenine is due primarily to a secondary transporter, and virus infection inhibits this transport function, consistent with the depolarization-sensitive transport model. This depolarization causes the dysfunction of other secondary transporters, which alters the uptake of many solutes into the cell, including adenine, glucose, proline, methionine, histidine, ornithine, and putrescine. Consequently, one has to use caution when studying changes in metabolic synthetic rates during virus infection of Chlorella NC64A using pulse-labeling procedures.
Virus-mediated cell permeabilization is a commonly observed event, especially with animal viruses. The virus agents responsible for permeabilization are called viroporins, and several viruses encode these small hydrophobic proteins that interact with the host membranes (14). The permeabilization observed is transient and often biphasic, where there is a period shortly after infection when the cells become permeable to both large and small molecules that are otherwise prevented from entering the cells. Later in infection, cells often become leaky to a wide range of molecules as the virus is about to be released. In conjunction with these events, ionic homeostasis is disrupted, and this can be a precursor to induced cell death (23). Mitochondrial membranes are known to depolarize with virus infection, leading to apoptosis (2). Certain mitochondrial-targeted regulators of apoptosis of viral origin are known to modify mitochondrial membrane permeabilization (MMP) (10). For example, human immunodeficiency virus type 1 Vpr protein, influenza virus p13II protein, and the hepatitis B virus HVB-X protein are proapoptotic factors that appear to be linked to MMP (3), whereas Kaposi's sarcoma-associated herpesvirus K7 protein, cytomegalovirus vMIA protein, and myxoma virus M11L protein act as antiapoptotic factors that tend to suppress MMP. However, in none of these cases have cell membrane dysfunctions been linked to inhibition of secondary transporters.
Bacteriophage infections are also known to cause membrane depolarization and in certain cases are known to be transient, MOI dependent, and biphasic (21). Indeed, certain types of virus-encoded holins depolarize bacterial cells, typically as a late event (31). In the cases of the double-stranded RNA phages φ6 and φ13, virus entry is associated with a limited depolarization of the plasma membrane along with a transient increase in permeability of the gram-negative Pseudomonas syringae outer membrane (7). Indeed, nucleocapsid binding of φ6 to the plasma membrane is independent of the membrane potential (ΔΨ), yet internalization requires a high ΔΨ (34). The subsequent transient depolarization is thought to preclude secondary infections. Likewise, bacteriophage T4 and T5 infections of Escherichia coli cause a reversible depolarization of the cell membrane, and the infection requires a high ΔΨ (less than −100 mV) (15, 20). But again, the membrane permeabilization is nonspecific, unlike the alteration of the algal cell secondary transporters observed with chlorovirus infection.
In summary, PBCV-1 infection of Chlorella NC64A leads to rapid alterations in the physiology of the host cell. These alterations include a depolarization of the host plasma membrane and an alkalinization of the cytoplasm. In particular, the depolarization causes the dysfunction of secondary transporters, which alters the uptake of many solutes into the cell, including adenine, glucose, proline, methionine, histidine, ornithine, and putrescine. The observations presented here indicate that virus infections may alter cell physiology significantly; however, little is known about how widespread virus-mediated depolarization is in the virosphere, the consequences of these events to productive infections, or how they relate to host-virus interactions in general. Nevertheless, in the case of Chlorovirus infection of Chlorella NC64A, it appears that these physiological manifestations lead to a productive and robust infection with a large burst size (approximately 1,000 particles per cell [41]), indicating that this altered cellular environment is conducive to acute lytic infections.
Acknowledgments
We thank Martin Ehle (Darmstadt) for the in vivo calibration of BCECF. We thank the U.S. Department of Energy Joint Genome Institute for sequencing and assembling the Chlorella NC64A genome (http://genome.jgi-psf.org/ChlNC64A_1/ChlNC64A_1.info.html). We also thank Michael Graves and Xiao Li, Department of Biological Sciences at the University of Massachusetts-Lowell for the tools and Chlorella NC64A genome sequence, available at the GreenGene website (http://greengene.uml.edu).
This research was supported in part by Public Health Service grant GM32441 (J.L.V.E.) and NIH grant P21RR15635 from the COBRE program of the National Center for Research Resources (J.L.V.E. and D.D.) and by a grant from the Deutsche Forschungsgemeinschaft (G.T.).
Footnotes
Published ahead of print on 8 October 2008.
REFERENCES
- 1.Agarkova, I. V., D. D. Dunigan, and J. L. Van Etten. 2006. Virion-associated restriction endonucleases of chloroviruses. J. Virol. 808114-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boya, P., A. L. Pauleau, D. Poncet, R. A. Gonzalez-Polo, N. Zamzami, and G. Kroemer. 2004. Viral proteins targeting mitochondria: controlling cell death. Biochim. Biophys. Acta 1659178-189. [DOI] [PubMed] [Google Scholar]
- 3.Boya, P., T. Roumier, K. Andreau, R. A. Gonzalez-Polo, N. Zamzami, M. Castedo, and G. Kroemer. 2003. Mitochondrion-targeted apoptosis regulators of viral origin. Biochem. Biophys. Res. Commun. 304575-581. [DOI] [PubMed] [Google Scholar]
- 4.Brüdern, A., and G. Thiel. 1999. Effect of cell wall-digesting enzymes on physiological state and competence of maize coleoptile cells. Protoplasma 209246-255. [Google Scholar]
- 5.Bubeck, J. A., and A. J. Pfitzner. 2005. Isolation and characterization of a new type of chlorovirus that infects an endosymbiotic Chlorella strain of the heliozoon Acanthocystis turfacea. J. Gen. Virol. 862871-2877. [DOI] [PubMed] [Google Scholar]
- 6.Chuchird, N., S. Hiramatsu, I. Sugimoto, M. Fujie, S. Usami, and T. Yamada. 2001. Digestion of chlorella cells by chlorovirus-encoded polysaccharide degrading enzymes. Microbes Environ. 16206-212. [Google Scholar]
- 7.Daugelavicius, R., V. Cvirkaite, A. Gaidelyte, E. Bakiene, R. Gabrenaite-Verkhovskaya, and D. H. Bamford. 2005. Penetration of enveloped double-stranded RNA bacteriophages φ13 and φ6 into Pseudomonas syringae cells. J. Virol. 795017-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Koning, H., and G. Diallinas. 2000. Nucleobase transporters. Mol. Membr. Biol. 7575-94. [DOI] [PubMed] [Google Scholar]
- 9.Dixon, G. K., C. Brownlee, and M. J. Merrett. 1989. Measurement of internal pH in the coccolithophore Emiliana huxleyi using 2′,7′-bis-(2-carboxyethyl)-5(and-6) carboxyfluorescein acetoxymethyl ester and digital imaging microscopy. Planta 178443-449. [DOI] [PubMed] [Google Scholar]
- 10.Ferri, K. F., E. Jacotot, J. Blanco, J. A. Esté, and G. Kroemer. 2000. Mitochondrial control of cell death induced by HIV-1-encoded proteins. Ann. N. Y. Acad. Sci. 926149-164. [DOI] [PubMed] [Google Scholar]
- 11.Frohns, F., A. Käsmann, D. Kramer, B. Schäfer, M. Mehmel, M. Kang, J. L. Van Etten, S. Gazzarrini, A. Moroni, and G. Thiel. 2006. Potassium ion channels of chlorella viruses cause rapid depolarization of host cells during infection. J. Virol. 802437-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gazzarrini, S., M. Severino, M. Lombardi, M. Morandi, D. DiFrancesco, J. L. Van Etten, G. Thiel, and A. Moroni. 2003. The viral potassium channel Kcv: structural and functional features. FEBS Lett. 55212-16. [DOI] [PubMed] [Google Scholar]
- 13.Gillissen, B., L. Burkle, B. Andre, C. Kuhn, D. Rentsch, B. Brandl, and W. B. Frommer. 2000. A new family of high-affinity transporters for adenine, cytosine, and purine derivatives in Arabidopsis. Plant Cell 12291-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez, M. E., and Luis Carrasco. 2003. Viroporins. FEBS Lett. 55228-34. [DOI] [PubMed] [Google Scholar]
- 15.Kalasauskaite, E. V., D. L. Kadisaite, R. J. Daugelavicius, L. L. Grinius, and A. A. Jasaitis. 1983. Requirement for membrane potential in Escherichia coli infection by phage T4. Eur. J. Biochem. 130123-130. [PubMed] [Google Scholar]
- 16.Kang, M., A. Moroni, S. Gazzarrini, D. DiFrancesco, G. Thiel, M. Severino, and J. L. Van Etten. 2004. Small potassium ion channel proteins encoded by chlorella viruses. Proc. Natl. Acad. Sci. USA 1015318-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawasaki, T., M. Tanaka, M. Fujie, S. Usami, and T. Yamada. 2004. Immediate early genes expressed in chlorovirus infections. Virology 318214-223. [DOI] [PubMed] [Google Scholar]
- 18.Kennedy, J. E. 1987. Purification and characterization of a lytic enzyme from viral infection of Chlorella-like green algae. M.S. thesis. University of Nebraska, Lincoln.
- 19.Komor, E., and W. Tanner. 1976. The determination of the membrane potential of Chlorella vulgaris. Evidence for electrogenic sugar transport. Eur. J. Biochem. 70197-204. [DOI] [PubMed] [Google Scholar]
- 20.Letellier, L., and B. Labedan. 1984. Involvement of envelope-bound calcium in the transient depolarization of the Escherichia coli cytoplasmic membrane induced by bacteriophage T4 and T5 adsorption. J. Bacteriol. 157789-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letellier, L., L. Plançon, M. Bonhivers, and P. Boulanger. 1999. Phage DNA transport across membranes. Res. Microbiol. 150499-505. [DOI] [PubMed] [Google Scholar]
- 22.Ludewig, U., and W. Frommer. 2002. Genes and proteins for solute transport and sensing. In C. R. Somerville and E. M. Meyerowitz (ed.), The Arabidopsis book. American Society of Plant Biologists, Rockville, MD. [DOI] [PMC free article] [PubMed]
- 23.Madan, V., A. Castelló, and L. Carrasco. 2008. Viroporins from RNA viruses induce caspase-dependent apoptosis. Cell. Microbiol. 10437-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mehmel, M., M. Rothermel, T. Meckel, J. L. Van Etten, A. Moroni, and G. Thiel. 2003. Possible function for virus encoded K+ channel Kcv in the replication of chlorella virus PBCV-1. FEBS Lett. 5527-11. [DOI] [PubMed] [Google Scholar]
- 25.Meints, R. H., K. Lee, D. E. Burbank, and J. L. Van Etten. 1984. Infection of a chlorella-like alga with the virus, PBCV-1: ultrastructural studies. Virology 13841-346. [DOI] [PubMed] [Google Scholar]
- 26.Moffatt, B. A., and H. Ashihara. 2002. Purine and pyrimidine nucleotide synthesis and metabolism. In C. R. Somerville and E. M. Meyerowitz (ed.), The Arabidopsis book. American Society of Plant Biologists, Rockville, MD. [DOI] [PMC free article] [PubMed]
- 27.Moroni, A., C. Viscomi, V. Sangiorgio, C. Pagliuca, T. Meckel, F. Horvath, S. Gazzarrini, P. Valbuzzi, J. L. Van Etten, D. DiFrancesco, and G. Thiel. 2002. The short N-terminus is required for functional expression of the virus-encoded miniature K+ channel Kcv. FEBS Lett. 53065-69. [DOI] [PubMed] [Google Scholar]
- 28.Neupärtl, M., C. Meyer, I. Woll, F. Frohns, M. Kang, J. L. Van Etten, D. Kramer, B. Hertel, A. Moroni, and G. Thiel. 2008. Chlorella viruses evoke a rapid release of K+ from host cells during early phase of infection. Virology 372340-348. [DOI] [PubMed] [Google Scholar]
- 29.Onimatsu, H., K. Suganuma, S. Uenoyama, and T. Yamada. 2006. C-terminal repetitive motifs in Vp130 present at the unique vertex of the chlorovirus capsid are essential for binding to the host chlorella cell wall. Virology 353432-442. [DOI] [PubMed] [Google Scholar]
- 30.Opekarová, M., and W. Tanner. 1994. Nystatin changes the properties of transporters for arginine and sugars. FEBS Lett. 35046-50. [DOI] [PubMed] [Google Scholar]
- 31.Park, T., D. K. Struck, J. F. Deaton, and R. Young. 2006. Topological dynamics of holins in programmed bacterial lysis. Proc. Natl. Acad. Sci. USA 10319713-19718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Plieth, C., B. Sattelmacher, and U.-P. Hansen. 1997. Cytoplasmic Ca2+-H+-exchange buffers in green algae. Protoplasma 198107-124. [Google Scholar]
- 33.Plugge, B., S. Gazzarrini, M. Nelson, R. Cerana, J. L. Van Etten, C. Derst, D. DiFrancesco, A. Moroni, and G. Thiel. 2000. A potassium channel protein encoded by chlorella virus PBCV-1. Science 2871641-1644. [DOI] [PubMed] [Google Scholar]
- 34.Poranen, M. M., R. Daugelavicius, P. M. Ojala, M. W. Hess, and D. H. Bamford. 1999. A novel virus-host cell membrane interaction: membrane voltage-dependent endocytic-like entry of bacteriophage straight φ6 nucleocapsid. J. Cell Biol. 147671-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ren, Q., K. Chen, and I. Paulsen. 2007. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 35D274-D279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuster, A. M., L. Girton, D. E. Burbank, and J. L. Van Etten. 1986. Infection of a Chlorella-like alga with the virus PBCV-1: transcriptional studies. Virology 148181-189. [DOI] [PubMed] [Google Scholar]
- 37.Schwab, W. G. W., and E. Komor. 1978. A possible mechanistic role of the membrane potential in proton-sugar cotransport of Chlorella. FEBS Lett. 87157-160. [DOI] [PubMed] [Google Scholar]
- 38.Sugimoto, I., S. Hiramatsu, D. Murakami, M. Fujie, S. Usami, and T. Yamada. 2000. Algal-lytic activities encoded by chlorella virus CVK2. Virology 277119-126. [DOI] [PubMed] [Google Scholar]
- 39.Sun, L., B. Adams, J. Gurnon, Y. Ye, and J. L. Van Etten. 1999. Characterization of two chitinase genes and one chitosanase gene encoded by chlorella virus PBCV-1. Virology 263376-387. [DOI] [PubMed] [Google Scholar]
- 40.Sun, L., J. R. Gurnon, B. J. Adams, M. V. Graves, and J. L. Van Etten. 2000. Characterization of a β-1,3-glucanase encoded by chlorella virus PBCV-1. Virology 27627-36. [DOI] [PubMed] [Google Scholar]
- 41.Van Etten, J. L. 2003. Unusual life style of giant chlorella viruses. Annu. Rev. Genet. 37153-195. [DOI] [PubMed] [Google Scholar]
- 42.Van Etten, J. L., D. E. Burbank, J. Joshi, and R. H. Meints. 1984. DNA synthesis in a Chlorella-like alga following infection with the virus, PBCV-1. Virology 134443-449. [DOI] [PubMed] [Google Scholar]
- 43.Van Etten, J. L., D. E. Burbank, Y. Xia, and R. H. Meints. 1983. Growth cycle of a virus, PBCV-1, that infects chlorella-like algae. Virology 126117-125. [DOI] [PubMed] [Google Scholar]
- 44.Wendler, S., and U. Zimmermann. 1982. A new method for the determination of hydraulic conductivity and cell volume of plant cells by pressure clamp. Plant Physiol. 69998-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamada, T., H. Onimatsu, and J. L. Van Etten. 2006. Chlorella viruses Adv. Virus Res. 66 293-366. [DOI] [PMC free article] [PubMed] [Google Scholar]