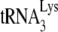Abstract
Attempts to use the mouse as a model system for studying AIDS are stymied by the multiple blocks to human immunodeficiency virus type 1 (HIV-1) replication that exist in mouse cells at the levels of viral entry, transcription, and Gag assembly and processing. In this report, we describe an additional block in the selective packaging of 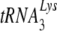 into HIV-1 produced in murine cells. HIV-1 and murine leukemia virus (MuLV) use
into HIV-1 produced in murine cells. HIV-1 and murine leukemia virus (MuLV) use 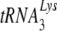 and tRNAPro, respectively, as primers for reverse transcription. Selective packaging of
and tRNAPro, respectively, as primers for reverse transcription. Selective packaging of 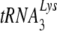 into HIV-1 produced in human cells is much stronger than that for tRNAPro incorporation into MuLV produced in murine cells, and different packaging mechanisms are used. Thus, both lysyl-tRNA synthetase and GagPol are required for
into HIV-1 produced in human cells is much stronger than that for tRNAPro incorporation into MuLV produced in murine cells, and different packaging mechanisms are used. Thus, both lysyl-tRNA synthetase and GagPol are required for 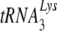 packaging into HIV-1, but neither prolyl-tRNA synthetase nor GagPol is required for tRNAPro packaging into MuLV. In this report, we show that when HIV-1 is produced in murine cells, the virus switches from an HIV-1-like incorporation of
packaging into HIV-1, but neither prolyl-tRNA synthetase nor GagPol is required for tRNAPro packaging into MuLV. In this report, we show that when HIV-1 is produced in murine cells, the virus switches from an HIV-1-like incorporation of 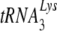 to an MuLV-like packaging of tRNAPro. The primer binding site in viral RNA remains complementary to
to an MuLV-like packaging of tRNAPro. The primer binding site in viral RNA remains complementary to 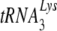 , resulting in a significant decrease in reverse transcription and infectivity. Reduction in
, resulting in a significant decrease in reverse transcription and infectivity. Reduction in 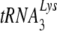 incorporation occurs even though both murine lysyl-tRNA synthetase and HIV-1 GagPol are packaged into the HIV-1 produced in murine cells. Nevertheless, the murine cell is able to support the select incorporation of
incorporation occurs even though both murine lysyl-tRNA synthetase and HIV-1 GagPol are packaged into the HIV-1 produced in murine cells. Nevertheless, the murine cell is able to support the select incorporation of 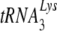 into another retrovirus that uses
into another retrovirus that uses 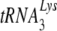 as a primer, the mouse mammary tumor virus.
as a primer, the mouse mammary tumor virus.
In retroviruses, tRNA is used as a primer to initiate the reverse transcriptase (RT)-catalyzed synthesis of minus-strand strong-stop DNA (−SS DNA). Different retroviruses use different tRNAs as a primer. In lentiviruses, including human immunodeficiency virus type 1 (HIV-1), 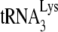 serves as the primer tRNA (36, 40). In avian retroviruses, the primer is tRNATrp (17, 26, 44, 45, 54, 55), whereas tRNAPro is the common primer for murine leukemia virus (MuLV) (25, 43, 51). In avian retroviruses and HIV-1, the primer tRNAs are selectively packaged, i.e., the percentage of the tRNA population representing primer tRNA increases in going from the cytoplasm to the virus. For example, in avian myeloblastosis virus, the relative concentration of tRNATrp changes from 1.4% to 32% (54). In HIV-1, the relative concentration of tRNALys (which includes both primer
serves as the primer tRNA (36, 40). In avian retroviruses, the primer is tRNATrp (17, 26, 44, 45, 54, 55), whereas tRNAPro is the common primer for murine leukemia virus (MuLV) (25, 43, 51). In avian retroviruses and HIV-1, the primer tRNAs are selectively packaged, i.e., the percentage of the tRNA population representing primer tRNA increases in going from the cytoplasm to the virus. For example, in avian myeloblastosis virus, the relative concentration of tRNATrp changes from 1.4% to 32% (54). In HIV-1, the relative concentration of tRNALys (which includes both primer 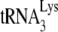 and the other major tRNALys isoacceptors,
and the other major tRNALys isoacceptors, 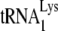 and
and 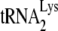 , which differ by only one base pair in the anticodon stem and are referred to together as
, which differ by only one base pair in the anticodon stem and are referred to together as 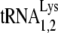 ) changes from 5 to 6% in the cytoplasm to 50 to 60% in virus produced from transfected COS7 cells (39). In AKR MuLV, selective packaging of primer tRNAPro is less dramatic, going from a relative cytoplasmic concentration of 5 to 6% to 12 to 24% of low-molecular-weight RNA (54).
) changes from 5 to 6% in the cytoplasm to 50 to 60% in virus produced from transfected COS7 cells (39). In AKR MuLV, selective packaging of primer tRNAPro is less dramatic, going from a relative cytoplasmic concentration of 5 to 6% to 12 to 24% of low-molecular-weight RNA (54).
In HIV-1, an increase in the concentration of primer 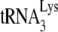 in the viral population is correlated with an increase in both
in the viral population is correlated with an increase in both 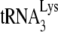 annealing and viral infectivity (19). Within a complex formed by Gag and GagPol, GagPol is required for packaging tRNALys into Gag virus-like particles (VLPs) or into HIV-1 (39), and more specifically, the thumb domain in RT sequences plays an important role in the interaction between GagPol and tRNALys (32). Nevertheless, Gag plays an important role in selecting tRNALys isoacceptors for incorporation into Gag VLPs. Lysyl-tRNA synthetase (LysRS), the protein that aminoacylates tRNALys, is also selectively packaged into HIV-1 (10). An important role of LysRS in the viral life cycle appears to be to target tRNALys for incorporation into the virion through a specific interaction between LysRS and Gag (10, 24, 30, 34, 35). Gag alone is sufficient for incorporation of LysRS into Gag VLPs (10), but GagPol is required for tRNALys incorporation as well (39).
annealing and viral infectivity (19). Within a complex formed by Gag and GagPol, GagPol is required for packaging tRNALys into Gag virus-like particles (VLPs) or into HIV-1 (39), and more specifically, the thumb domain in RT sequences plays an important role in the interaction between GagPol and tRNALys (32). Nevertheless, Gag plays an important role in selecting tRNALys isoacceptors for incorporation into Gag VLPs. Lysyl-tRNA synthetase (LysRS), the protein that aminoacylates tRNALys, is also selectively packaged into HIV-1 (10). An important role of LysRS in the viral life cycle appears to be to target tRNALys for incorporation into the virion through a specific interaction between LysRS and Gag (10, 24, 30, 34, 35). Gag alone is sufficient for incorporation of LysRS into Gag VLPs (10), but GagPol is required for tRNALys incorporation as well (39).
In contrast to HIV-1, the process responsible for the weaker select packaging of tRNAPro in MuLV appears to be different, i.e., the incorporation of tRNAPro occurs independently of both prolyl-tRNA synthetase (ProRS), which is not found in this virus (8, 24), and GagPol (18, 37). We have investigated whether these differences in the mechanism of primer tRNA incorporation into HIV and MuLV are due to differences in viral and/or cellular factors by examining the incorporation of tRNAs into HIV-1 produced in murine cells.
There are multiple HIV replication blocks in murine cells that occur at the levels of viral entry, transcriptional elongation, splicing, and Gag assembly and processing (3, 5, 38, 41, 42, 52, 57, 61). While blocks in viral entry and transcription have been partially overcome through engineering mouse cells to synthesize HIV-1 receptors CD4 and CCR5 (chemokine [C-C motif] receptor 5), and the TAT cofactor, cyclin T1 (20, 57), such cells release only trace amounts of HIV-1 capsid and infectious virus (5, 20, 42). Further problems are encountered with Gag assembly at membranes, and this has been partially remedied through altering the nuclear export pathway of HIV-1 RNA from a Rev-dependent to a Rev-independent one (49), or by replacing HIV-1 MA with either MuLV or simian immunodeficiency virus MA (13, 29). It has also been found that murine cells containing human chromosome 2 were more efficient at facilitating both HIV-1 Gag assembly at membranes and the budding of infectious virus, presumably due to one or more cell factors expressed from chromosome 2 (15).
In this work, we demonstrate that another block to HIV-1 replication in murine cells is the inability to selectively package 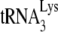 . We have examined the incorporation of tRNA into HIV-1 produced in transfected murine A9 cells, where viral production was facilitated either by using A9 cells containing human chromosome 2 and human cyclin T1, or through transfection with HIV-1 DNA that produces mRNA utilizing a Rev-independent mRNA nuclear export pathway. The HIV-1 produced in murine cells shows a strong reduction in the incorporation of tRNALys, but instead, like MuLV, selectively packages tRNAPro, using a mechanism that is independent of either ProRS or GagPol incorporation.
. We have examined the incorporation of tRNA into HIV-1 produced in transfected murine A9 cells, where viral production was facilitated either by using A9 cells containing human chromosome 2 and human cyclin T1, or through transfection with HIV-1 DNA that produces mRNA utilizing a Rev-independent mRNA nuclear export pathway. The HIV-1 produced in murine cells shows a strong reduction in the incorporation of tRNALys, but instead, like MuLV, selectively packages tRNAPro, using a mechanism that is independent of either ProRS or GagPol incorporation.
MATERIALS AND METHODS
Plasmids and cell lines.
SVC21.BH10 is a simian virus 40-based vector that contains full-length wild-type HIV-1 proviral DNA, and it was a gift from E. Cohen, University of Montreal. Human Gag (hGag) and hGag/GagPol were gifts from Y. Huang and G. Nabel, Vaccine Research Center, National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH), Bethesda, MD (27). The Gag and GagPol proteins coded by these plasmids have amino acid sequences that are identical to the sequences of their viral counterparts, but the mRNAs coding for them have had their codons optimized for mammalian cell codon usage, which results in more efficient translation and protein production and also makes nuclear export of these mRNAs Rev independent through modification of the multiple inhibitory sequences (27). HIV-1 Gag-Rev response element (Gag-RRE) is a gift from David Rekosh (University of Virginia at Charlotte), while HIV-1 Gag/GagPol-RRE is a gift from Chen Liang (McGill University, Montreal, Quebec, Canada). HIV-1 Gag-4CTE, Gag/GagPol-4CTE plasmids were constructed by PCR. PCR products of Gag or Gag/GagPol from HIV-1 BH10 were cloned into pcDNA3.1/V5-His vector (pcDNA3.1/V5-His TOPO TA expression kit; Invitrogen). Four copies of the Mason-Pfizer monkey virus constitutive transport element (4CTE), obtained from Hans-Georg Kräusslich, University Hamburg, Germany (59), were cloned into the unique sites of NotI and BstBI of the above vector. The plasmid coding for MuLV was a gift from Alan Rein (National Cancer Institute, Bethesda, MD).
HEK-293T cells (CRL-11268), A9 mouse connective tissue cells (CCL-1.4), and MM5MT, a mouse mammary gland cell line stably producing mouse mammary tumor virus (MMTV) (CRL-1637) were all obtained from the American Type Culture Collection (ATCC). The somatic cell hybrid GM11686 (A9 plus human chromosome 2) was obtained from the Coriell Cell Repositories, Camden, NJ. GM11686-cycT1 and A9-cycT1 are the GM11686 and A9 cell lines transduced with human cycT1 genes and are gifts from Richard Sutton (Baylor College of Medicine). The indicator cell line TZM-bl (a HeLa cell line containing CD4, CCR5, and a reporter luciferase gene associated with the HIV-1 long terminal repeat) was used for measuring HIV-1 infectivity and was obtained from NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH (catalog no. 8129) (58). HEK-293T and A9 cells were grown in complete Dulbecco's modified Eagle's medium plus 10% fetal bovine serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. GM11686 cells were cultured in complete Dulbecco's modified Eagle's medium-F12 (1:1) (catalog no. 11330; Invitrogen) plus 10% fetal bovine serum, 100 U of penicillin/ml, 100 μg streptomycin/ml, 500 μg/ml G418, and 1× minimal essential medium nonessential amino acid solution (catalog no. 11140; Invitrogen). For GM11686-cycT1 cells, 5 μg puromycin/ml was also added to the above medium.
Cell transfection and virus purification.
In addition to problems with assembly, which can be partially countered by the presence of human chromosome 2, production of HIV-1 from murine cells is also inhibited by low viral RNA transcription, and it was found that this can be improved by constitutively expressing human cyclin T1 in the GM11686 cells, i.e., GM11686-cycT1 cells (15). Human or murine cells were plated at 3.0 × 106 to 3.4 × 106 cells in 100-mm-diameter plates for 24 h and then transfected with 2 μg BH10 or other plasmids indicated using Lipofectamine 2000 (Invitrogen), according to the manufacturer's instructions. Another problem encountered is the low transfection efficiency of murine cells, which was improved through the use of spin transfection, i.e., immediately after transfection by Lipofectamine 2000, the plates containing murine cells were centrifuged for 10 min at 1,200 × g (MicroPlus Carrier; Beckman Coulter). Spin transfection had little or no effect on the number of human 293T cells transfected, but the number of GM11686-cycT1 cells transfected increased from 2 to 8% without spin to 10 to 30% with spin (data not shown).
Transfected human and murine cell lines, as well as the stably transfected murine MM5MT cell line, were incubated for 48 h at 37°C, and VLPs were collected from the cell culture supernatant by centrifugation in a Beckman type 45Ti rotor at 35,000 rpm for 1 h. The viral pellets were then purified by centrifugation in a Beckman SW41 rotor at 26,500 rpm for 1 h through 15% sucrose onto a 65% sucrose cushion. The band of purified virus was removed and repelleted at 35,000 rpm for 1 h in a Beckman SW41 rotor.
Viral RNA isolation and quantification.
Total viral RNA was extracted from these pellets using the guanidinium isothiocyanate procedure as previously described (14). The RNA pellets were dissolved in 5 mM Tris-HCl (pH 7.5) and stored at −80°C. Hybridization to dot blots of total viral RNA was carried out as previously described (9), with 5′-32P-end-labeled DNA probes complementary to either the 3′ terminal 18 nucleotides of 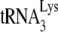 (5′-TGGCGCCCGAACAGGGAC-3′), tRNAPro (5′-TGGGGGCTCGTCCGGGAT-3′), or tRNAHis (5′-TGGTGCCGTGACTCGGAT-3′) or to the 5′ end of the HIV-1 genomic RNA, upstream of the primer binding site (PBS) (5′-CTGACGCTCTCGCACCC-3′).
(5′-TGGCGCCCGAACAGGGAC-3′), tRNAPro (5′-TGGGGGCTCGTCCGGGAT-3′), or tRNAHis (5′-TGGTGCCGTGACTCGGAT-3′) or to the 5′ end of the HIV-1 genomic RNA, upstream of the primer binding site (PBS) (5′-CTGACGCTCTCGCACCC-3′).
2D PAGE.
Total viral RNA was 3′ end labeled with 32pCp by T4 RNA ligase and resolved by two-dimensional polyacrylamide gel electrophoresis (2D PAGE) as previously described (31). To identify tRNAPro, spots resolved by 2D PAGE were excised, ground, dissolved in buffer (20 mM Tris-HCl [pH 7.5], 5 mM EDTA, 400 mM sodium acetate) for 2 h, and precipitated with ethanol (1). The pellet was dissolved in RNase-free water and used as a template for reverse transcription-PCR (RT-PCR) (ThermoScript reverse transcriptase [Invitrogen]; Hot-star Taq polymerase [Qiagen]). The primers for tRNAPro are as follows: mouse-tRNAPro-F1 (forward primer) (5′-GGCTCGTTGGTCTAGGGGTA-3′) and mouse-tRNAPro-R1 (reverse primer) (5′-CTCGTCCGGGATTTGAAC-3′). Nested primers were also used: mouse-tRNAPro-F2 (5′-GCTCGTTGGTCTAGGGGTAT-3′) and mouse-tRNAPro-R2 (5′-GATTTGAACCCGGGACCT-3′). The primers for both 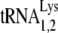 and
and 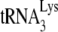 are mouse-tRNALys-F1 (5′-CCGGMTAGCTCAGTCGGTA-3′), mouse-tRNALys-R1 (5′-CCGAACAGGGRCTYGAAC-3′), mouse-tRNALys-F2 (5′-CGGMTAGCTCAGTCGGTAG-3′), and mouse-tRNALys-R2 (5′-CGAACAGGGRCTYGAACC-3′), where M is C+A, Y is C+T, and R is A+G. The PCR products were cloned into the TOPO vector (TOPO TA cloning kit; Invitrogen) and sequenced to identify the tRNA.
are mouse-tRNALys-F1 (5′-CCGGMTAGCTCAGTCGGTA-3′), mouse-tRNALys-R1 (5′-CCGAACAGGGRCTYGAAC-3′), mouse-tRNALys-F2 (5′-CGGMTAGCTCAGTCGGTAG-3′), and mouse-tRNALys-R2 (5′-CGAACAGGGRCTYGAACC-3′), where M is C+A, Y is C+T, and R is A+G. The PCR products were cloned into the TOPO vector (TOPO TA cloning kit; Invitrogen) and sequenced to identify the tRNA.
Real-time PCR quantitation of synthesis of minus-strand strong-stop HIV-1 DNA.
Equal amounts of DNase-treated HIV-1 produced in either 293T cells or GM11686-cycT1 cells (1 ng CAp24) were used to spin infect 1 × 106 SupT1 cells of each well on the 24-well plates at 1,200 × g for 1 hour. At different times postinfection, aliquots of cells were collected and washed with phosphate-buffered saline, and cellular DNA was extracted using the DNeasy tissue kit (Qiagen). Using equal amounts of cellular genomic DNA (determined spectrophotometrically at an optical density of 260 nm), −SS DNA synthesis (R-U5) was quantitated by the Light Cycler instrument (Roche Diagnostics GmbH) using the following primers: RT forward (5′-TTAGACCAGATCTGAGCCTGGGAG-3′) and RT reverse (5′-GGGTCTGAGGGATCTCTAGTTACC-3′) (23).
The −SS DNA containing the tRNA primer was also used to identify the tRNA primer as previously described (56). Equal amounts of cellular genomic DNA were amplified with PCR. The forward primer is complementary to the −SS DNA synthesized (5′-GCTCTAGACCAGATCTGAGCCTGGGAGCTC-3′). The reverse primer is complementary to either the 5′ end of tRNAPro (5′-GGCTCGTTGGTCTAGGGGTA-3′) or the 5′ end of 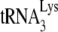 (5′-CCGGMTAGCTCAGTCGGTA-3′, where M is C+A).
(5′-CCGGMTAGCTCAGTCGGTA-3′, where M is C+A).
Protein analysis.
Cellular and viral proteins were extracted with CytoBuster protein extraction reagent (catalog no. 71009; Novagen) with protease inhibitor cocktail tablets (Roche). The cell and viral lysates were analyzed by sodium dodecyl sulfate-PAGE (10% acrylamide), followed by blotting onto nitrocellulose membranes (Amersham Pharmacia). Western blots were probed with monoclonal antibodies that are specifically reactive with HIV-1 capsid (Zeptometrix Inc.), HIV-1 RT (catalog no. 7372; NIH AIDS Research and Reference Reagent Program), V5 (Invitrogen), and β-actin (Sigma), and a rabbit polyclonal antibody for ProRS, a gift from S. Kim (Seoul National University, Seoul, South Korea) (33). Detection of proteins was performed by enhanced chemiluminescence (PerkinElmer Life Sciences, Inc.), using anti-mouse (for capsid, V5, and β-actin) and anti-rabbit antibodies (for RT and ProRS) as secondary antibodies, both obtained from Amersham Life Sciences. Bands in Western blots were quantified using ImageJ software (NIH).
Expression and purification of human and murine LysRS.
Human LysRS cDNA was a gift from K. Shiba (Japanese Foundation for Cancer Research, Tokyo, Japan) (47). Murine LysRS cDNA was constructed by first extracting mRNA from A9 cells with TRIzol (Invitrogen), followed by reverse transcription (SuperScript II; Invitrogen) and amplification by PCR (Expand high-fidelity PCR system, Roche). The following PCR primers were used: forward, 5′-GACTGAATTCGTGTTCCGCCATGTTGATGC-3′; reverse, 5′-GAACTCGAGGACAGAGGGGCCGGCTGTTGT-3′. The PCR products were digested with EcoRI and XhoI and cloned into the pcDNA3.1/V5-His vector (Invitrogen). The human LysRS and murine LysRS expressed were C terminally tagged with V5.
Human and murine LysRS were purified by a previously published procedure involving expression of His-tagged LysRS in Escherichia coli and purification on Ni+ affinity columns (47).
Infectivity assay.
Viral infectivity was determined by challenging 1 × 105 TZM-bl indicator cells with equal amounts of viruses (1 ng of CAp24), and measuring the induction of luciferase activity as previously described (6, 60).
Fluorescence anisotropy measurements.
Equilibrium dissociation constants for the CAp24/LysRS interaction were determined using fluorescein isothiocyanate-labeled HIV-1 CAp24 (FITC-CAp24) prepared as previously described (34). The fluorescence anisotropy of FITC-CAp24 (50 nM) was measured as a function of increasing concentrations of unlabeled LysRS, and the data were analyzed as previously described (34).
RESULTS
HIV-1 produced in murine cells does not selectively package tRNALys isoacceptors and shows both reduced synthesis of minus-strand strong-stop DNA and reduced infectivity.
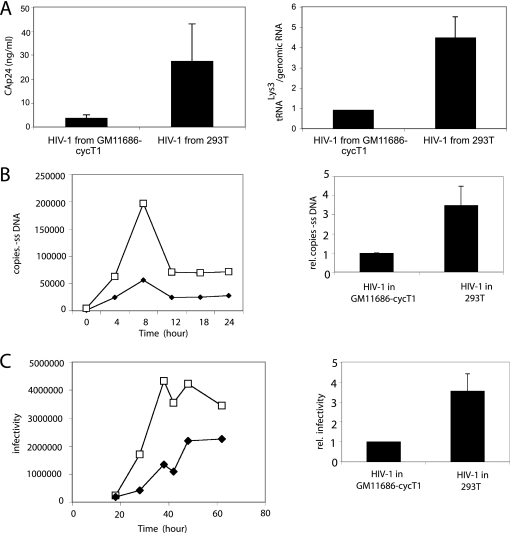
HIV-1 particles were produced by spin transfecting HIV-1 DNA into either 293T cells or into A9 mouse cells containing human chromosome 2 and cyclin T1 (GM11686-cycT1). As shown in the left panel in Fig. 1A, production of extracellular HIV-1 (CAp24) from GM11686-cycT1 cells is <25% of that produced from a similar number of cultured 293T cells. The viruses produced were tested for their ability to incorporate 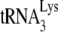 . The difference in the viral concentration of
. The difference in the viral concentration of 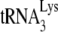 found in HIV-1 in 293T cells or GM11686-cycT1 cells was examined by measuring the
found in HIV-1 in 293T cells or GM11686-cycT1 cells was examined by measuring the 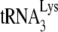 /genomic RNA in each viral type, using dot blots of viral RNA hybridized with DNA probes specific for either HIV-1 genomic RNA or
/genomic RNA in each viral type, using dot blots of viral RNA hybridized with DNA probes specific for either HIV-1 genomic RNA or 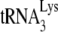 . The viral genomic RNA/CAp24 ratios for HIV-1 produced from each cell type were similar (data not listed), and equal amounts of viral genomic RNA for each cell type were used in the dot blots. Normalizing to HIV-1 (GM11686-cycT1), the graph in Fig. 1A shows that the ratio of
. The viral genomic RNA/CAp24 ratios for HIV-1 produced from each cell type were similar (data not listed), and equal amounts of viral genomic RNA for each cell type were used in the dot blots. Normalizing to HIV-1 (GM11686-cycT1), the graph in Fig. 1A shows that the ratio of 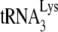 to genomic RNA in HIV-1 in 293T cells is approximately 4.5 times higher than that found in HIV-1 in GM11686-cycT1 cells.
to genomic RNA in HIV-1 in 293T cells is approximately 4.5 times higher than that found in HIV-1 in GM11686-cycT1 cells.
FIG. 1.
HIV-1 produced in murine cells show reduced 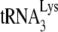 (tRNALys3) packaging, −SS DNA synthesis, and infectivity. 293T and GM11686-cycT1 cells were spin transfected with HIV-1, and the resulting virions were analyzed. (A) Viral production and
(tRNALys3) packaging, −SS DNA synthesis, and infectivity. 293T and GM11686-cycT1 cells were spin transfected with HIV-1, and the resulting virions were analyzed. (A) Viral production and 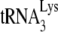 packaging. (Left) Extracellular amount of CAp24 per milliliter of culture medium. (Right) Total viral RNA was isolated and analyzed by dot blot hybridization using DNA probes specific for
packaging. (Left) Extracellular amount of CAp24 per milliliter of culture medium. (Right) Total viral RNA was isolated and analyzed by dot blot hybridization using DNA probes specific for 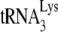 and viral genomic RNA. (B) −SS DNA synthesis. SupT1 cells were infected with equal amounts of HIV-1 produced from either 293T cells or GM11686-cyc-T1 cells. DNA was extracted at different times postinfection. Early (R-U5) minus-strand cDNA production was monitored by real-time PCR as described in Materials and Methods. (Left) Time course of DNA production. Symbols: □, HIV-1 produced in 293T cells; ♦, HIV-1 produced in GM11686-cycT1 cells. (Right) Relative (rel.) production of −SS DNA at 8 h. (C) Viral infectivity. Equal amounts of viral CAp24 were used to challenge TZM-bl indicator cells, and productive infection was measured as the induction of luciferase activity. (Left) Time course of infection. Symbols: □, HIV-1 produced in 293T cells; ♦, HIV-1 produced in GM11686-cycT1 cells. (Right) Relative (rel.) infectivity at 28 h postinfection, normalized to HIV-1 produced in GM11686-cycT1 cells.
and viral genomic RNA. (B) −SS DNA synthesis. SupT1 cells were infected with equal amounts of HIV-1 produced from either 293T cells or GM11686-cyc-T1 cells. DNA was extracted at different times postinfection. Early (R-U5) minus-strand cDNA production was monitored by real-time PCR as described in Materials and Methods. (Left) Time course of DNA production. Symbols: □, HIV-1 produced in 293T cells; ♦, HIV-1 produced in GM11686-cycT1 cells. (Right) Relative (rel.) production of −SS DNA at 8 h. (C) Viral infectivity. Equal amounts of viral CAp24 were used to challenge TZM-bl indicator cells, and productive infection was measured as the induction of luciferase activity. (Left) Time course of infection. Symbols: □, HIV-1 produced in 293T cells; ♦, HIV-1 produced in GM11686-cycT1 cells. (Right) Relative (rel.) infectivity at 28 h postinfection, normalized to HIV-1 produced in GM11686-cycT1 cells.
Figure 1B shows the ability of these viruses to synthesize −SS DNA upon infection of SupT1 cells. The T-lymphocyte cell line SupT1 was infected with equal amounts of HIV-1 (CAp24) produced from either human or mouse cells. −SS DNA (R-U5) synthesis was monitored over a 24-h postinfection time period using real-time fluorescence-monitored PCR with equal amounts of cellular DNA, and the results are graphed in Fig. 1B. −SS PCR products reached a maximum concentration at 8 h postinfection (Fig. 1B, left panel), and the relative values at the 8-h time point are graphed on the right side of Fig. 1B. The production of −SS DNA synthesis in cells infected with HIV-1 produced in 293T cells is 3.5 to 4 times greater than that obtained for HIV-1 infection in murine cells. After 8 h, the abundance of −SS DNA declined, a phenomenon that has been previously reported (7), and probably the result of degradation of viral DNA not converted into integrated proviral DNA (7, 22).
To measure infectivity of the viruses, TZM-b1 cells, containing a luciferase reporter gene, were infected with equal amounts of HIV-1 (CAp24) produced from either human or mouse cells, and at various times postinfection, cells were lysed, and luciferase activity was measured. The results are graphed in Fig. 1C. Luciferase activity at various times postinfection are plotted in the left graph of Fig. 1C, and the relative infectivity at 28 h is graphed in the right panel. These results indicate that HIV-1 produced in murine cells have a 3.5- to 4-fold decrease in infectivity compared to HIV-1 produced in 293T cells.
In spite of the numerous defects in HIV-1 replication encountered during replication in murine cells, the data in Fig. 1 show a correlation between reduced 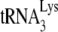 packaging, reduced −SS DNA synthesis, and reduced infectivity. This is because entry or assembly defects that exist for HIV-1 production in murine cells should not play a role in our measurements of either −SS DNA production or viral infectivity. We are using equal amounts of viral p24 produced in human or murine cells to infect human cells with the proper HIV-1 receptors for cell entry, i.e., SupT1 cells for measuring −SS DNA and TZM-b1 cells for measuring infectivity, and the parameters being measured occur prior to viral assembly in these cells. Furthermore, when wild-type HIV-1 is replaced with envelope-negative HIV-1 pseudotyped with vesicular stomatitis virus G, as we have previously described (23), the reduction in infectivity of virions produced in the GM11686-cycT1 cells is similar to that of wild-type HIV-1 produced in the same cells (data not shown).
packaging, reduced −SS DNA synthesis, and reduced infectivity. This is because entry or assembly defects that exist for HIV-1 production in murine cells should not play a role in our measurements of either −SS DNA production or viral infectivity. We are using equal amounts of viral p24 produced in human or murine cells to infect human cells with the proper HIV-1 receptors for cell entry, i.e., SupT1 cells for measuring −SS DNA and TZM-b1 cells for measuring infectivity, and the parameters being measured occur prior to viral assembly in these cells. Furthermore, when wild-type HIV-1 is replaced with envelope-negative HIV-1 pseudotyped with vesicular stomatitis virus G, as we have previously described (23), the reduction in infectivity of virions produced in the GM11686-cycT1 cells is similar to that of wild-type HIV-1 produced in the same cells (data not shown).
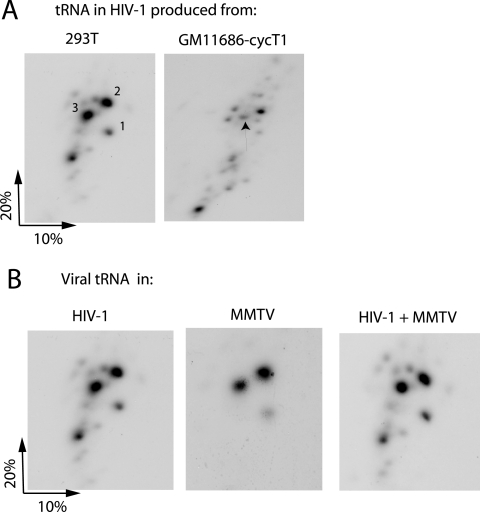
Figure 2A shows the two-dimensional polyacrylamide gel electrophoresis pattern of low-molecular-weight RNA extracted from HIV-1 produced in either 293T or GM11686-cycT1 cells. The pattern from HIV-1 produced in COS7 cells has been characterized (31), and it is known that spot 3 is 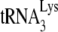 , while spots 2 and 1 correspond to
, while spots 2 and 1 correspond to 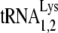 . The lower-mobility spot seen in this gel represents tRNAAsn (unpublished), and its appearance in the HIV-1 tRNA population varies between different viral samples. The electrophoretic pattern for tRNA extracted from HIV-1 produced in murine cells is quite different and does not indicate selective packaging of tRNALys isoacceptors. The spot marked with an arrow has been identified as
. The lower-mobility spot seen in this gel represents tRNAAsn (unpublished), and its appearance in the HIV-1 tRNA population varies between different viral samples. The electrophoretic pattern for tRNA extracted from HIV-1 produced in murine cells is quite different and does not indicate selective packaging of tRNALys isoacceptors. The spot marked with an arrow has been identified as 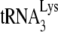 by RT-PCR, using primers specific for
by RT-PCR, using primers specific for 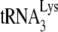 (see Materials and Methods). As described later in the article (Fig. 3B), the major high-mobility spot is identified by RT-PCR as tRNAPro.
(see Materials and Methods). As described later in the article (Fig. 3B), the major high-mobility spot is identified by RT-PCR as tRNAPro.
FIG. 2.
2D PAGE patterns of viral tRNA. (A) Viral tRNA patterns in HIV-1 produced from transfected 293T cells or GM11686-cycT1 cells. (B) Viral tRNA patterns from HIV-1 produced in 293T cells (left panel) and from mouse mammary tumor virus produced in the stably transfected murine cell line, MM5MT (middle panel). The right panel shows the 2D PAGE pattern when RNA from both viruses are mixed.
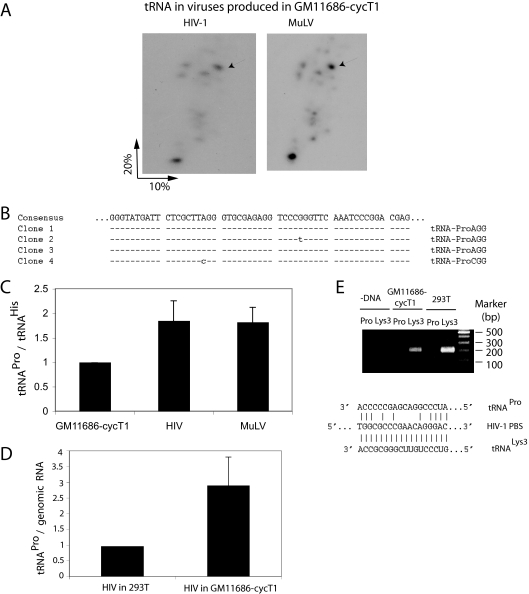
FIG. 3.
HIV-1 produced in murine cells packages tRNAPro. (A) 2D PAGE patterns of tRNAs in HIV-1 and MuLV, both produced in transiently transfected GM11686-cycT1 cells. (B) The numbered spots marked by arrows in Fig. 2A were extracted, amplified by RT-PCR using tRNAPro-specific primers, and sequenced. Clones 1 and 2 are MuLV, while clones 3 and 4 are HIV-1. (C) tRNAPro/tRNAHis ratios in GM11686-cycT1 cells and in the HIV-1 and MuLV produced in these cells. The tRNAPro/tRNAHis ratios were determined by hybridizing dot blots of cytoplasmic or viral RNA with probes specific for tRNAPro or tRNAHis. (D) tRNAPro/HIV-1 genomic RNA in HIV-1 produced in 293T cells or GM11686cyc-T1 cells, as determined by hybridizing dot blots of viral RNA with probes specific for tRNAPro and HIV-1 genomic RNA. (E) Identification of tRNA used to prime the synthesis of −SS DNA. SupT1 cells were infected with equal amounts of HIV-1 produced from either 293T cells or GM11686-cyc-T1 cells. DNA was extracted at different times postinfection, and equal amounts of DNA were amplified by PCR, using primer pairs complementary to −SS DNA (−DNA) and sequences near the 5′ terminus of either 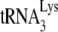 (tRNALys3) or tRNAPro. (Top) PCR products were resolved by agarose gel electrophoresis. (Bottom) HIV-1 PBS sequence and sequences of the 3′ termini of tRNALys3 and tRNAPro.
(tRNALys3) or tRNAPro. (Top) PCR products were resolved by agarose gel electrophoresis. (Bottom) HIV-1 PBS sequence and sequences of the 3′ termini of tRNALys3 and tRNAPro.
Figure 2B shows that the different tRNA pattern in HIV-1 produced in murine cells is not due to a difference in electrophoretic mobility between human and murine tRNALys isoacceptors. It is known that mouse mammary tumor virus also uses 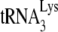 as the primer for reverse transcriptase, and in Fig. 2B, we have compared the 2D PAGE patterns of tRNA extracted from either HIV-1 produced in 293T cells or MMTV produced in the chronically infected MM5MT cell line. It can be seen that the tRNALys isoacceptors found in human or murine cells have identical electrophoretic mobility. Genes for human or murine
as the primer for reverse transcriptase, and in Fig. 2B, we have compared the 2D PAGE patterns of tRNA extracted from either HIV-1 produced in 293T cells or MMTV produced in the chronically infected MM5MT cell line. It can be seen that the tRNALys isoacceptors found in human or murine cells have identical electrophoretic mobility. Genes for human or murine 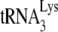 and
and 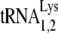 are distributed over many chromosomes, and their sequences were downloaded from the Genomic tRNA Database website (http://lowelab.ucsc.edu/GtRNAdb/) and aligned using Clustal multiple-sequence alignment software (http://www.clustal.org). The results are shown in Fig. S1 in the supplemental material and indicate that at some chromosomal loci, murine and human tRNALy3 or
are distributed over many chromosomes, and their sequences were downloaded from the Genomic tRNA Database website (http://lowelab.ucsc.edu/GtRNAdb/) and aligned using Clustal multiple-sequence alignment software (http://www.clustal.org). The results are shown in Fig. S1 in the supplemental material and indicate that at some chromosomal loci, murine and human tRNALy3 or 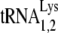 genes are identical, while at others, polymorphisms occur. The contribution of each loci to viral tRNALys is not known. The sensitivity of our
genes are identical, while at others, polymorphisms occur. The contribution of each loci to viral tRNALys is not known. The sensitivity of our 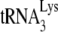 probe for detecting murine and human
probe for detecting murine and human 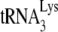 , however, seems to be similar. Thus, we have measured the changes in the
, however, seems to be similar. Thus, we have measured the changes in the 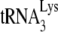 /tRNAHis ratio that occur between cytoplasm and virus for both HIV-1 (a five- to sixfold increase [see Fig. 4B]) and for MMTV (an approximately sevenfold increase [data not listed]).
/tRNAHis ratio that occur between cytoplasm and virus for both HIV-1 (a five- to sixfold increase [see Fig. 4B]) and for MMTV (an approximately sevenfold increase [data not listed]).
FIG. 4.
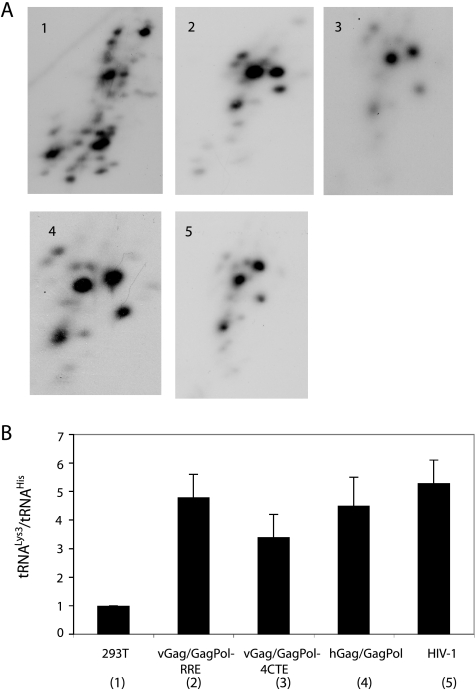
Viral tRNA in HIV-1 and Gag VLPs. 293T cells were transfected with plasmids coding for HIV-1 or HIV-1 Gag/GagPol VLPs. The RNA from the resulting virus or Gag VLPs was extracted and analyzed by 2D PAGE. (A) 2D PAGE patterns of viral tRNA extracted from VLPs produced from viral Gag/GagPol-RRE (panel 2), viral Gag/GagPol-4CTE (panel 3), or hGag/hGagPol (panel 4). The viral tRNA patterns in HIV-1 (panel 5) and in the cytoplasm of 293T cells (panel 1) are also shown. (B) 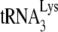 (tRNALys3)/tRNAHis ratios in HIV-1 and VLPs, normalized to that of the cell cytoplasm.
(tRNALys3)/tRNAHis ratios in HIV-1 and VLPs, normalized to that of the cell cytoplasm.
tRNA incorporation into HIV-1 produced in GM11686-cycT1 cells is qualitatively similar to tRNA incorporation into MuLV.
We next transfected GM11686-cycT1 cells with either HIV-1 or MuLV DNA, and Fig. 3A shows that the 2D PAGE patterns of the major tRNA species in HIV-1 and MuLV are identical. Since MuLV is known to selectively package tRNAPro, the spot in both patterns labeled by an arrow was extracted from the gels, amplified by RT-PCR using primers specific for tRNAPro, cloned, and sequenced (Fig. 3B). Clones 1 and 2 represent the high-mobility spot in MuLV (arrow) and were identified as tRNAPro with the AGG anticodon. Clones 3 and 4 represent the high-mobility spot in HIV-1 (arrow) and were identified as tRNAPro with either the AGG or CGG anticodon. The major low-mobility spot in either virus was not identified as tRNAPro by this procedure and has not yet been identified.
These results indicate that HIV-1 produced in murine cells selectively packages tRNAPro and another unidentified species. In Fig. 3C, the tRNAPro/tRNAHis ratios were determined for GM11686-cycT1 cells and for HIV-1 or MuLV produced in these cells by hybridizing dot blots of cytoplasmic or viral RNA with probes specific for tRNAPro or tRNAHis. It can be seen that for both HIV-1 and MuLV, the tRNAPro/tRNAHis ratios increase on average 1.8-fold over the cytoplasmic ratio. As mentioned earlier, selection of tRNAPro in MuLV is not as strong as selection of tRNALys in HIV-1. In Fig. 3D, the tRNAPro/viral genomic RNA ratio is compared using HIV-1 produced in either human (293T) or murine (GM11686-cycT1) cells, and it can be seen that the viral content of tRNAPro in HIV-1 produced in murine cells is on average three times higher than that found for HIV-1 produced in human cells.
Figure 3E shows that the −SS DNA produced upon infection of GM11686-cycT1 cells by HIV-1 is primed only by 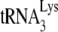 , and not by tRNAPro. Equal amounts of the total DNA extracted from SupT1 cells infected with HIV-1 produced from either 293T cells or GM11686-cycT1 cells (Fig. 1B) were amplified by PCR, using primer pairs complementary to −SS DNA and to the primer binding site for either
, and not by tRNAPro. Equal amounts of the total DNA extracted from SupT1 cells infected with HIV-1 produced from either 293T cells or GM11686-cycT1 cells (Fig. 1B) were amplified by PCR, using primer pairs complementary to −SS DNA and to the primer binding site for either 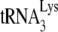 or tRNAPro (shown at the bottom of Fig. 1E). The top panel of Fig. 1E shows the electrophoretic resolution of the PCR products on agarose gels and shows that while the −SS DNA contains the
or tRNAPro (shown at the bottom of Fig. 1E). The top panel of Fig. 1E shows the electrophoretic resolution of the PCR products on agarose gels and shows that while the −SS DNA contains the 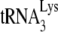 PBS, it does not contain the tRNAPro PBS, indicating that tRNAPro is not used as a primer. Thus, although tRNAPro is packaged into HIV-1, its inability to serve as a primer for reverse transcription is not surprising if one assumes that the HIV-1 genome does not contain a functional PBS complementary to tRNAPro. There is no reason to assume that the incorporation of tRNAPro into the virus is dependent upon a PBS complementary to tRNAPro, since the selective packaging of
PBS, it does not contain the tRNAPro PBS, indicating that tRNAPro is not used as a primer. Thus, although tRNAPro is packaged into HIV-1, its inability to serve as a primer for reverse transcription is not surprising if one assumes that the HIV-1 genome does not contain a functional PBS complementary to tRNAPro. There is no reason to assume that the incorporation of tRNAPro into the virus is dependent upon a PBS complementary to tRNAPro, since the selective packaging of 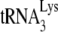 in HIV-1 has been shown to occur independently of the presence of the
in HIV-1 has been shown to occur independently of the presence of the 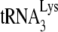 PBS (28).
PBS (28).
The selective incorporation of tRNALys into HIV-1 is unaffected by the mRNA nuclear export pathway used by Gag and GagPol.
Our studies have thus far used a murine cell line containing human chromosome 2 (GM11686-cycT1), which is required for the production of extracellular HIV-1 particles from murine cells. It is possible that the alteration in tRNALys packaging into HIV-1 is due to a product of human chromosome 2 interacting with murine factors. However, HIV-1 or HIV-1 VLPs may also be produced in murine cells not containing chromosome 2 if the nuclear export pathway is changed from Crm-1 dependence (utilizing Rev and RRE) to a TAP-dependent one. This can be achieved either by replacing the Rev response element with four copies of the constitutive transport element of Mason-Pfizer monkey virus (i.e., the viral Gag-4CTE [vGag-4CTE] or vGag/Gagpol-4CTE vector) or by optimizing codon usage of the viral mRNAs so as to use mammalian rather than viral codon usage (i.e., the hGag or hGag/GagPol vector) (49, 59). In Fig. 4, we show, using 293T cells, that the production of Gag/GagPol VLPs using different nuclear export pathways does not alter the ability of these VLPs to selectively incorporate tRNALys. Fig. 4A shows the 2D PAGE patterns of viral tRNAs produced by these different constructs, and it can be seen that the selective incorporation of tRNALys occurs regardless of which nuclear export pathway is used. Fig. 4B shows the results of hybridizing labeled DNA probes specific for 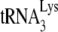 and tRNAHis to dot blots of RNA extracted from each type of VLP. These results show similar quantitative selection of
and tRNAHis to dot blots of RNA extracted from each type of VLP. These results show similar quantitative selection of 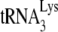 into HIV-1 (BH10) and the Gag/GagPol VLPs.
into HIV-1 (BH10) and the Gag/GagPol VLPs.
tRNAPro incorporation into HIV-1 VLPs produced in murine cells occurs independently of both GagPol and human chromosome 2.
We next used the Rev-independent constructs to show that tRNAPro incorporation into HIV-1 VLPs occurs independently of GagPol and independently of human chromosome 2. While GagPol is required for the incorporation of tRNALys into HIV-1 (32, 39), tRNAPro packaging in MuLV occurs independently of GagPol (18, 37). We therefore examined whether tRNAPro incorporation into HIV-1 in murine cells also occurred independently of GagPol. To examine this, GM11686 cells were transfected with Rev-independent plasmid constructs coding for Gag alone or for Gag and GagPol. 2D PAGE patterns of RNAs extracted from the different Gag or Gag/GagPol VLPs are shown in Fig. 5A, and it is clear that tRNAPro is packaged into these particles independently of the presence of GagPol, as has been found for MuLV.
FIG. 5.
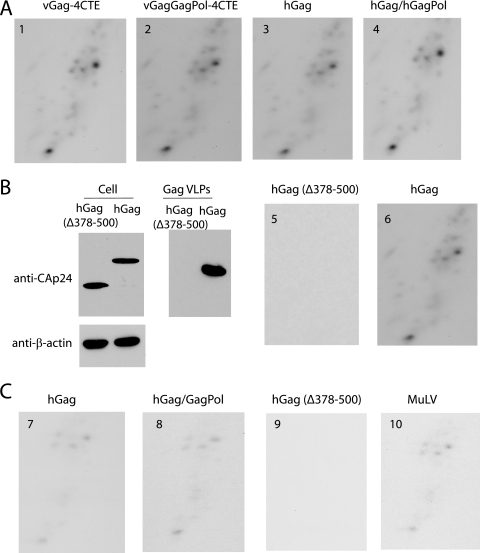
tRNAPro incorporation into HIV-1 VLPs produced in murine cells. (A) 2D PAGE resolution of tRNAs in VLPs produced from GM11686 cells. VLPs are produced from vGag-4CTE (panel 1), vGag/GagPol-4CTE (panel 2), hGag (panel 3), and hGag/hGagPol (panel 4). (B, left) Western blots of the expression in GM11686 cells and the extracellular production of hGag and C-terminally deleted hGag (Δ378-500). (Right) 2D PAGE of tRNA extracted from hGag and hGag (Δ378-500) VLPs produced from GM11686 cells. (C) 2D PAGE resolution of tRNAs in HIV-1 hGag/hGagPol VLPs and in MuLV produced in transiently transfected murine A9 cells, the parent cell of GM11686, and lacking human chromosome 2. VLPs are produced from hGag (panel 7), hGag/hGagPol (panel 8), and hGag (Δ378-500) (panel 9), which is expressed in the cell but does not form extracellular VLPs (panel 7). The 2D PAGE pattern of tRNAs in MuLV (panel 10) is also shown.
The data in Fig. 5B show that the incorporation of tRNAs into extracellular particles is dependent upon the expression of extracellular HIV-1 Gag VLPs. GM11686 cells were transfected with plasmids coding for either hGag or hGag Δ378-500, which produces C-terminally-deleted Gag missing nucleocapsid, SP2, and p6 sequences. As shown in the leftmost panel in Fig. 5B, this mutant Gag is expressed in the cell but does not produce extracellular Gag VLPs. The two right panels in Fig. 5B shows that when RNA is extracted from Gag VLPs and resolved by 2D PAGE, no RNA-containing particles are produced from cells expressing Gag Δ378-500.
In Fig. 5C, we show that tRNAPro incorporation into HIV-1 VLPs occurs even when the murine cells producing the VLPs do not contain human chromosome 2 or human cyclin T1. Figure 5C shows the tRNA incorporation pattern in VLPs produced in A9 cells, the parent cell of GM11868, and which does not contain human chromosome 2 or human cyclin T1. Virions are produced by transfecting cells with codon-optimized genes for Gag (hGag) or Gag and GagPol (hGag/ hGagPol). A comparison with the 2D PAGE pattern of low-molecular-weight RNA from virus with that of MuLV produced in these cells indicates similar patterns, i.e., VLPs produced in A9 cells selectively package tRNAPro, not tRNALys. In A9 cells, as in GM11686 cells, when Gag Δ378-500 was expressed, no RNA-containing extracellular VLPs were detected (Fig. 5, panel 9).
Altered tRNA packaging in HIV-1 produced in murine cells is not associated with alterations in the viral incorporation of LysRS, ProRS, or GagPol.
As described above, tRNALys packaging in HIV-1 produced in 293T cells requires both a specific interaction of Gag with LysRS, which targets the tRNALys isoacceptors for incorporation, and the presence of GagPol, which binds to tRNALys and stabilizes the tRNA in the packaging complex (10, 39). Therefore, we have examined whether the inability of LysRS or GagPol to be packaged into HIV-1 produced in murine cells could be the cause of the lack of selective packaging of tRNALys.
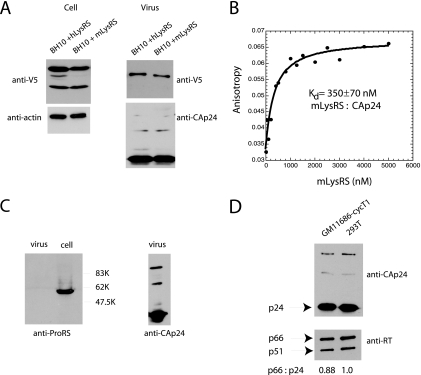
Because our antibody to human LysRS was relatively insensitive for detection of endogenous mouse LysRS, GM11686-cycT1 cells were cotransfected with BH10 and C-terminally tagged V5-human or mouse LysRS, and antibody to V5 was used to detect either human or murine LysRS in Western blots of cell or viral lysates. The results shown in Fig. 6A show that both human LysRS and murine LysRS are readily incorporated into HIV-1 made in GM11686-cycT1 cells.
FIG. 6.
Incorporation of LysRS, ProRS, and GagPol into HIV-1 made in murine cells. (A) Viral incorporation of human or mouse LysRS. GM11686-cycT1 cells were cotransfected with BH10 DNA and DNA coding for either human LysRS (hLysRS) or murine LysRS (mLysRS), both of which were tagged with V5. Forty-eight hours posttransfection, viruses were collected from the supernatant, and cells and viruses were lysed. Western blots of cell and viral lysates were probed with anti-V5 and anti-β-actin or with anti-V5 and anti-CAp24, respectively. (B) Fluorescence anisotropy experiment, showing binding of mouse LysRS (mLysRS) to FITC-CAp24. Only a representative data set is shown, but measurements were carried out at least three times. The equilibrium dissociation constant (Kd) is shown. (C) Incorporation of ProRS. GM11686-cycT1 cells were transfected with BH10, and 48 h posttransfection, viruses were collected from the supernatant, and cells and viruses were lysed. Western blots of cell lysate were probed with anti-ProRS, and Western blots of viral lysates were probed with anti-ProRS and anti-CAp24. (D) Incorporation of GagPol. GM11686-cycT1 cells and 293T cells were transfected with BH10. Forty-eight hours posttransfection, viruses were collected from the supernatant and lysed. Western blots of viral lysates were probed with anti-CAp24 and anti-RT.
We previously reported that human LysRS interacts with the C-terminal domain of HIV-1 CAp24 (30). Using fluorescence anisotropy, an apparent equilibrium dissociation constant of 420 nM was measured for this interaction in vitro (34, 35). Using this technique, we now demonstrate that murine LysRS binds to HIV-1 CAp24 with a similar affinity (equilibrium dissociation constant of 350 nM; Fig. 6B). Figure S2 in the supplemental material compares the amino acid sequences of murine (A9) LysRS and human LysRS. Helix 7, which contains the putative CAp24 binding site (30, 34, 35) differs by only one amino acid between the two species (I247V), consistent with the similar measured binding affinities.
We have also previously shown that the packaging of tRNAPro into MuLV occurs without a corresponding incorporation of ProRS into this virus (8), and in Fig. 6C, we demonstrate the absence of ProRS in HIV-1 produced in murine cells as well.
The incorporation of GagPol into Gag VLPs is required for the viral packaging of tRNALys isoacceptors. The ability of GagPol to be incorporated into HIV-1 produced in either GM11686-cycT1 or 293T cells is demonstrated in Fig. 6D. Western blots of lysates of virus produced from either type of cell transfected with HIV-1 (BH10) DNA are probed with either anti-CAp24 or anti-RT. It can be seen that there is no significant change in the RTp66/CAp24 ratio.
DISCUSSION
This study shows that HIV-1 produced in murine cells is unable to selectively incorporate tRNALys isoacceptors. As shown in Fig. 1, this contributes to a reduction in both the synthesis of −SS DNA and viral infectivity. We have previously shown using 293T cells that HIV-1 Gag particles alone can incorporate LysRS but that GagPol is also required for the packaging of tRNALys (10, 39). A scenario for the formation of a 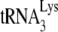 packaging/annealing complex may involve a Gag/GagPol/viral RNA complex interacting with a LysRS/
packaging/annealing complex may involve a Gag/GagPol/viral RNA complex interacting with a LysRS/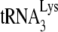 complex, with Gag specifically interacting with LysRS and GagPol interacting with both Gag and
complex, with Gag specifically interacting with LysRS and GagPol interacting with both Gag and 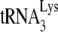 . In fact, in vitro evidence indicates that
. In fact, in vitro evidence indicates that 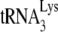 interacts with the thumb domain in RT (2, 16), and we have shown that
interacts with the thumb domain in RT (2, 16), and we have shown that 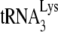 is packaged into mutant HIV-1 containing C-terminal deletions of GagPol, as long as the thumb domain is not included in the deletions (32). The inability to package
is packaged into mutant HIV-1 containing C-terminal deletions of GagPol, as long as the thumb domain is not included in the deletions (32). The inability to package 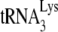 into HIV-1 produced in murine cells could therefore be due to an inability to incorporate either LysRS or GagPol, but in fact, Fig. 6 shows that HIV-1 produced in murine cells can incorporate LysRS and GagPol (RT). Therefore, there may be additional factors required for packaging that are missing.
into HIV-1 produced in murine cells could therefore be due to an inability to incorporate either LysRS or GagPol, but in fact, Fig. 6 shows that HIV-1 produced in murine cells can incorporate LysRS and GagPol (RT). Therefore, there may be additional factors required for packaging that are missing.
It has been shown that HIV-1 Pol alone (missing cis-Gag sequences) will be incorporated into HIV-1 Gag particles and that HIV-1 Pol can replace GagPol in facilitating the selective incorporation of tRNALys (12). A similar situation exists in human foamy viruses, which use 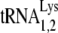 as a primer, and where Gag and Pol are made from separate mRNAs (48). Figure 7 presents a model, based on biochemical evidence, showing the possible relationships between the components of a
as a primer, and where Gag and Pol are made from separate mRNAs (48). Figure 7 presents a model, based on biochemical evidence, showing the possible relationships between the components of a 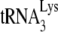 packaging/annealing complex. Because of the demonstrated interaction (direct or indirect) of Pol with Gag (12), we predict that GagPol may have a conformation that results in its folding back due to a Pol/Gag interaction. In this model,
packaging/annealing complex. Because of the demonstrated interaction (direct or indirect) of Pol with Gag (12), we predict that GagPol may have a conformation that results in its folding back due to a Pol/Gag interaction. In this model, 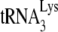 is shown bound to the RT thumb domain, which is based both on in vitro (2, 16) and in vivo (11, 32) studies. The 5′ region of viral RNA is also shown, and the nucleocapsid (NC) sequences within Gag are shown bound to the RNA region containing the RNA packaging sequence of viral RNA that includes stem-loop 3 (4, 21). This stem-loop is only 112 nucleotides downstream of the viral RNA PBS to which
is shown bound to the RT thumb domain, which is based both on in vitro (2, 16) and in vivo (11, 32) studies. The 5′ region of viral RNA is also shown, and the nucleocapsid (NC) sequences within Gag are shown bound to the RNA region containing the RNA packaging sequence of viral RNA that includes stem-loop 3 (4, 21). This stem-loop is only 112 nucleotides downstream of the viral RNA PBS to which  anneals. Several advantages to the virus can arise from a folding back of GagPol that would bring Pol sequences closer to both LysRS and the PBS. This conformation could facilitate transfer of
anneals. Several advantages to the virus can arise from a folding back of GagPol that would bring Pol sequences closer to both LysRS and the PBS. This conformation could facilitate transfer of 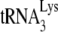 from the LysRS to the RT thumb domain and/or facilitate transfer of
from the LysRS to the RT thumb domain and/or facilitate transfer of 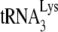 from RT to the PBS. Thus, even though both LysRS and GagPol are incorporated into HIV-1 produced in murine cells, an inhibition of a Pol/Gag interaction might inhibit
from RT to the PBS. Thus, even though both LysRS and GagPol are incorporated into HIV-1 produced in murine cells, an inhibition of a Pol/Gag interaction might inhibit 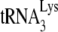 incorporation.
incorporation.
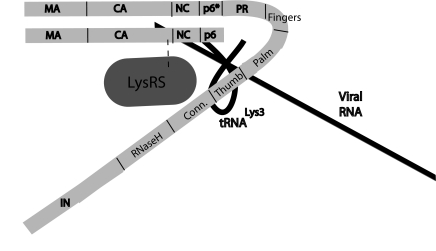
FIG. 7.
Model illustrating proposed relationships between components of the 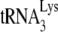 (tRNALys3) packaging/annealing complex. In this figure, Gag domains shown include the matrix (MA), capsid (CA), nucleocapsid (NC), and p6 domains. Pol domains within GagPol include p6* and integrase (IN), as well as the reverse transcriptase subdomains that include fingers, palm, thumb, connection (Conn.), and RNaseH.
(tRNALys3) packaging/annealing complex. In this figure, Gag domains shown include the matrix (MA), capsid (CA), nucleocapsid (NC), and p6 domains. Pol domains within GagPol include p6* and integrase (IN), as well as the reverse transcriptase subdomains that include fingers, palm, thumb, connection (Conn.), and RNaseH.
Murine cells are, however, capable of facilitating the selective packaging of tRNALys into another retrovirus, MMTV. This is seen clearly in Fig. 2, which shows that MMTV, which also uses 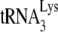 as a primer (53), selectively packages tRNALys when produced from the MM5MT murine cell line. One difference between MMTV and HIV-1 is the cell location for capsid assembly. While lentiviruses and the C-type MuLV assemble into immature virions at the membrane, MMTV, a B-type virus, assembles its capsid in the cytoplasm prior to moving to the membrane for budding (50). Thus, the cell location of assembly in murine cells might play a role in determining which tRNAs are incorporated into the virion as a result of the specific cellular locations of required factors, some of which might be required for the Gag/Pol interaction.
as a primer (53), selectively packages tRNALys when produced from the MM5MT murine cell line. One difference between MMTV and HIV-1 is the cell location for capsid assembly. While lentiviruses and the C-type MuLV assemble into immature virions at the membrane, MMTV, a B-type virus, assembles its capsid in the cytoplasm prior to moving to the membrane for budding (50). Thus, the cell location of assembly in murine cells might play a role in determining which tRNAs are incorporated into the virion as a result of the specific cellular locations of required factors, some of which might be required for the Gag/Pol interaction.
It also appears that in the absence of select packaging of tRNALys into HIV-1, the virus selectively incorporates tRNAPro. The 2D PAGE pattern of viral tRNAs looks identical for either MuLV or HIV-1 produced in murine cells. Also, like MuLV, incorporation of tRNAPro occurs independently of the incorporation of both GagPol and ProRS. As shown in Fig. 5, extracellular particles containing tRNA were obtained only when HIV-1 Gag VLPs were produced, i.e., the cytoplasmic expression of a truncated HIV-1 Gag unable to assemble and form extracellular VLPs produced no such tRNA-containing particles. The selective packaging of primer tRNA is much weaker in MuLV than in avian retroviruses (54) or HIV-1 (39), and while the incorporation of GagPol is required for primer tRNA incorporation in avian sarcoma virus (44), Rous sarcoma virus (46), and HIV-1 (32, 39), it is not required for MuLV (18). Also, while tRNALys incorporation into HIV-1 requires LysRS, tRNAPro incorporation into MuLV does not require ProRS (8). Therefore, the possibility that tRNAPro packaging into retrovirus uses a more primitive mechanism that will occur by default when more efficient mechanisms for incorporation of other tRNA primers do not function exists.
Supplementary Material
Acknowledgments
This work was supported by Canadian Institutes of Health Research operating grant MOP-81389 (to L.K.), a Canadian Institutes of Health Research postdoctoral fellowship award (to M.W.), National Institutes of Health grant AI054145 (to L.K. and K. M.-F.), and Ruth L. Kirschstein National Service Award GM069339 (to R.K.).
Footnotes
Published ahead of print on 8 October 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ambros, V., and R. C. Lee. 2004. Identification of microRNAs and other tiny noncoding RNAs by cDNA cloning. Methods Mol. Biol. 265131-158. [DOI] [PubMed] [Google Scholar]
- 2.Arts, E. J., J. T. Miller, B. Ehresmann, and S. F. Le Grice. 1998. Mutating a region of HIV-1 reverse transcriptase implicated in tRNALys-3 binding and the consequences for (−)-strand DNA synthesis. J. Biol. Chem. 27314523-14532. [DOI] [PubMed] [Google Scholar]
- 3.Atchison, R. E., J. Gosling, F. S. Monteclaro, C. Franci, L. Digilio, I. F. Charo, and M. A. Goldsmith. 1996. Multiple extracellular elements of CCR5 and HIV-1 entry: dissociation from response to chemokines. Science 2741924-1926. [DOI] [PubMed] [Google Scholar]
- 4.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214177-218. [DOI] [PubMed] [Google Scholar]
- 5.Bieniasz, P. D., and B. R. Cullen. 2000. Multiple blocks to human immunodeficiency virus type 1 replication in rodent cells. J. Virol. 749868-9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop, K. N., R. K. Holmes, and M. H. Malim. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 808450-8458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Butler, S. L., M. S. Hansen, and F. D. Bushman. 2001. A quantitative assay for HIV DNA integration in vivo. Nat. Med. 7631-634. [DOI] [PubMed] [Google Scholar]
- 8.Cen, S., H. Javanbakht, S. Kim, K. Shiba, R. Craven, A. Rein, K. Ewalt, P. Schimmel, K. Musier-Forsyth, and L. Kleiman. 2002. Retrovirus-specific packaging of aminoacyl-tRNA synthetases with cognate primer tRNAs. J. Virol. 7613111-13115. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
9.Cen, S., A. Khorchid, J. Gabor, L. Rong, M. A. Wainberg, and L. Kleiman. 2000. Roles of Pr55gag and NCp7 in
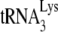 genomic placement and the initiation step of reverse transcription in human immunodeficiency virus type 1. J. Virol. 7410796-10800. [DOI] [PMC free article] [PubMed] [Google Scholar]
genomic placement and the initiation step of reverse transcription in human immunodeficiency virus type 1. J. Virol. 7410796-10800. [DOI] [PMC free article] [PubMed] [Google Scholar] - 10.Cen, S., A. Khorchid, H. Javanbakht, J. Gabor, T. Stello, K. Shiba, K. Musier-Forsyth, and L. Kleiman. 2001. Incorporation of lysyl-tRNA synthetase into human immunodeficiency virus type 1. J. Virol. 755043-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cen, S., M. Niu, and L. Kleiman. 2004. The connection domain in reverse transcriptase facilitates the in vivo annealing of tRNALys3 to HIV-1 genomic RNA. Retrovirology 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cen, S., M. Niu, J. Saadatmand, F. Guo, Y. Huang, G. J. Nabel, and L. Kleiman. 2004. Incorporation of Pol into human immunodeficiency virus type 1 Gag virus-like particles occurs independently of the upstream Gag domain in Gag-Pol. J. Virol. 781042-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen, P., W. Hubner, K. Riviere, Y. X. Liu, and B. K. Chen. 2006. Chimeric HIV-1 containing SIV matrix exhibit enhanced assembly in murine cells and replicate in a cell-type-dependent manner in human T cells. Virology 3491-12. [DOI] [PubMed] [Google Scholar]
- 14.Chomczynski, P., and N. Sacchi. 1987. RNA isolation from cultured cells. Anal. Biochem. 162156-159. [DOI] [PubMed] [Google Scholar]
- 15.Coskun, A. K., M. van Maanen, V. Nguyen, and R. E. Sutton. 2006. Human chromosome 2 carries a gene required for production of infectious human immunodeficiency virus type 1. J. Virol. 803406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dufour, E., J. Reinbolt, M. Castroviejo, B. Ehresmann, S. Litvak, L. Tarrago-Litvak, and M. L. Andreola. 1999. Cross-linking localization of a HIV-1 reverse transcriptase peptide involved in the binding of primer tRNALys3. J. Mol. Biol. 2851339-1346. [DOI] [PubMed] [Google Scholar]
- 17.Faras, A. J., and N. A. Dibble. 1975. RNA-directed DNA synthesis by the DNA polymerase of Rous sarcoma virus: structural and functional identification of 4S primer RNA in uninfected cells. Proc. Natl. Acad. Sci. USA 72859-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fu, W., A. Ortiz-Conde, R. J. Gorelick, S. H. Hughes, and A. Rein. 1997. Placement of tRNA primer on the primer binding site requires pol gene expression in avian but not murine retroviruses. J. Virol. 716940-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
19.Gabor, J., S. Cen, H. Javanbakht, M. Niu, and L. Kleiman. 2002. Effect of altering the
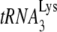 concentration in human immunodeficiency virus type 1 upon its annealing to viral RNA, GagPol incorporation, and viral infectivity. J. Virol. 769096-9102. [DOI] [PMC free article] [PubMed] [Google Scholar]
concentration in human immunodeficiency virus type 1 upon its annealing to viral RNA, GagPol incorporation, and viral infectivity. J. Virol. 769096-9102. [DOI] [PMC free article] [PubMed] [Google Scholar] - 20.Garber, M. E., P. Wei, V. N. KewalRamani, T. P. Mayall, C. H. Herrmann, A. P. Rice, D. R. Littman, and K. A. Jones. 1998. The interaction between HIV-1 Tat and human cyclin T1 requires zinc and a critical cysteine residue that is not conserved in the murine CycT1 protein. Genes Dev. 123512-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geigenmüller, U., and M. L. Linial. 1996. Specific binding of human immunodeficiency virus type 1 (HIV-1) Gag-derived proteins to a 5′ HIV-1 genomic RNA sequence. J. Virol. 70667-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goncalves, J., Y. Korin, J. Zack, and D. Gabuzda. 1996. Role of Vif in human immunodeficiency virus type 1 reverse transcription. J. Virol. 708701-8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
23.Guo, F., S. Cen, M. Niu, J. Saadatmand, and L. Kleiman. 2006. The inhibition of
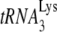 -primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 8011710-11722. [DOI] [PMC free article] [PubMed] [Google Scholar]
-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 8011710-11722. [DOI] [PMC free article] [PubMed] [Google Scholar] - 24.Halwani, R., S. Cen, H. Javanbakht, J. Saadatmand, S. Kim, K. Shiba, and L. Kleiman. 2004. Cellular distribution of lysyl-tRNA synthetase and its interaction with Gag during human immunodeficiency virus type 1 assembly. J. Virol. 787553-7564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harada, F., G. G. Peters, and J. E. Dahlberg. 1979. The primer tRNA for Moloney murine leukemia virus DNA synthesis: nucleotide sequence and aminoacylation of tRNAPro. J. Biol. Chem. 25410979-10985. [PubMed] [Google Scholar]
- 26.Harada, F., R. C. Sawyer, and J. E. Dahlberg. 1975. A primer RNA for initiation of in vitro Rous sarcoma virus DNA synthesis: nucleotide sequence and amino acid acceptor activity. J. Biol. Chem. 2503487-3497. [PubMed] [Google Scholar]
- 27.Huang, Y., W.-P. Kong, and G. J. Nabel. 2001. Human immunodeficiency virus type 1-specific immunity after genetic immunization is enhanced by modification of Gag and Pol expression. J. Virol. 754947-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
28.Huang, Y., J. Mak, Q. Cao, Z. Li, M. A. Wainberg, and L. Kleiman. 1994. Incorporation of excess wild-type and mutant
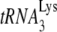 into human immunodeficiency virus type 1. J. Virol. 687676-7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
into human immunodeficiency virus type 1. J. Virol. 687676-7683. [DOI] [PMC free article] [PubMed] [Google Scholar] - 29.Hubner, W., and B. K. Chen. 2006. Inhibition of viral assembly in murine cells by HIV-1 matrix. Virology 35227-38. [DOI] [PubMed] [Google Scholar]
- 30.Javanbakht, H., R. Halwani, S. Cen, J. Saadatmand, K. Musier-Forsyth, H. G. Gottlinger, and L. Kleiman. 2003. The interaction between HIV-1 Gag and human lysyl-tRNA synthetase during viral assembly. J. Biol. Chem. 27827644-27651. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, M., J. Mak, A. Ladha, E. Cohen, M. Klein, B. Rovinski, and L. Kleiman. 1993. Identification of tRNAs incorporated into wild-type and mutant human immunodeficiency virus type 1. J. Virol. 673246-3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khorchid, A., H. Javanbakht, M. A. Parniak, M. A. Wainberg, and L. Kleiman. 2000. Sequences within Pr160gag-pol affecting the selective packaging of tRNALys into HIV-1. J. Mol. Biol. 29917-26. [DOI] [PubMed] [Google Scholar]
- 33.Kim, T., S. G. Park, J. E. Kim, W. Seol, Y.-G. Ko, and S. Kim. 2000. Catalytic peptide of human glutaminyl-tRNA synthetase is essential for its assembly to the aminoacyl-tRNA synthetase complex. J. Biol. Chem. 27521768-21772. [DOI] [PubMed] [Google Scholar]
- 34.Kovaleski, B. J., R. Kennedy, M. K. Hong, S. A. Datta, L. Kleiman, A. Rein, and K. Musier-Forsyth. 2006. In vitro characterization of the interaction between HIV-1 Gag and human lysyl-tRNA synthetase. J. Biol. Chem. 28119449-19456. [DOI] [PubMed] [Google Scholar]
- 35.Kovaleski, B. J., R. Kennedy, A. Khorchid, L. Kleiman, H. Matsuo, and K. Musier-Forsyth. 2007. Critical role of helix 4 of HIV-1 capsid C-terminal domain in interactions with human lysyl-tRNA synthetase. J. Biol. Chem. 28232274-32279. [DOI] [PubMed] [Google Scholar]
- 36.Leis, J., A. Aiyar, and D. Cobrinik. 1993. Regulation of initiation of reverse transcription of retroviruses, p. 33-47. In A. M. Skalka and S. P. Goff (ed.), Reverse transcriptase, vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 37.Levin, J. G., and J. G. Seidman. 1981. Effect of polymerase mutations on packaging of primer tRNAPro during murine leukemia virus assembly. J. Virol. 38403-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maddon, P. J., A. G. Dalgleish, J. S. McDougal, P. R. Clapham, R. A. Weiss, and R. Axel. 1986. The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 47333-348. [DOI] [PubMed] [Google Scholar]
- 39.Mak, J., M. Jiang, M. A. Wainberg, M.-L. Hammarskjöld, D. Rekosh, and L. Kleiman. 1994. Role of Pr160gag-pol in mediating the selective incorporation of tRNALys into human immunodeficiency virus type 1 particles. J. Virol. 682065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mak, J., and L. Kleiman. 1997. Primer tRNAs for reverse transcription. J. Virol. 718087-8095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malim, M. H., D. F. McCarn, L. S. Tiley, and B. R. Cullen. 1991. Mutational definition of the human immunodeficiency virus type 1 Rev activation domain. J. Virol. 654248-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mariani, R., G. Rutter, M. E. Harris, T. J. Hope, H.-G. Kräusslich, and N. R. Landau. 2000. A block to human immunodeficiency virus type 1 assembly in murine cells. J. Virol. 743859-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peters, G., F. Harada, J. E. Dahlberg, A. Panet, W. A. Haseltine, and D. Baltimore. 1977. Low-molecular-weight RNAs of Moloney murine leukemia virus: identification of the primer for RNA-directed DNA synthesis. J. Virol. 211031-1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peters, G. G., and J. Hu. 1980. Reverse transcriptase as the major determinant for selective packaging of tRNAs into avian sarcoma virus particles. J. Virol. 36692-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawyer, R. C., and J. E. Dahlberg. 1973. Small RNAs of Rous sarcoma virus: characterization by two-dimensional polyacrylamide gel electrophoresis and fingerprint analysis. J. Virol. 121226-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sawyer, R. C., and H. Hanafusa. 1979. Comparison of the small RNAs of polymerase-deficient and polymerase-positive Rous sarcoma virus and another species of avian retrovirus. J. Virol. 29863-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiba, K., T. Stello, H. Motegi, T. Noda, K. Musier-Forsyth, and P. Schimmel. 1997. Human lysyl-tRNA synthetase accepts nucleotide 73 variants and rescues E. coli double-defective mutant. J. Biol. Chem. 27222809-22816. [DOI] [PubMed] [Google Scholar]
- 48.Stenbak, C. R., and M. L. Linial. 2004. Role of the C terminus of foamy virus Gag in RNA packaging and Pol expression. J. Virol. 789423-9430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanson, C. M., B. A. Puffer, K. M. Ahmad, R. W. Doms, and M. H. Malim. 2004. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 232632-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. Coffin, S. Hughes, and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed]
- 51.Taylor, J. M. 1977. An analysis of the role of tRNA species as primers for transcription into DNA of RNA tumor virus genomes. Biochim. Biophys. Acta 4757-71. [DOI] [PubMed] [Google Scholar]
- 52.Trono, D., and D. Baltimore. 1990. A human cell factor is essential for HIV-1 Rev action. EMBO J. 94155-4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waters, L. C. 1978. Lysine tRNA is the predominant tRNA in murine mammary tumor virus. Biochem. Biophys. Res. Commun. 81822-827. [DOI] [PubMed] [Google Scholar]
- 54.Waters, L. C., and B. C. Mullin. 1977. Transfer RNA in RNA tumor viruses. Prog. Nucleic Acid Res. Mol. Biol. 20131-160. [DOI] [PubMed] [Google Scholar]
- 55.Waters, L. C., B. C. Mullin, T. Ho, and W. K. Yang. 1975. Ability of tryptophan tRNA to hybridize with 35S RNA of avian myeloblastosis virus and prime reverse transcription in vitro. Proc. Natl. Acad. Sci. USA 722155-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
-
56.Wei, M., S. Cen, M. Niu, F. Guo, and L. Kleiman. 2005. Defective replication in human immunodeficiency virus type 1 when non-
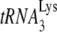 primers are used for reverse transcription. J. Virol. 799081-9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
primers are used for reverse transcription. J. Virol. 799081-9087. [DOI] [PMC free article] [PubMed] [Google Scholar] - 57.Wei, P., M. E. Garber, S. M. Fang, W. H. Fischer, and K. A. Jones. 1998. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell 92451-462. [DOI] [PubMed] [Google Scholar]
- 58.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 461896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wodrich, H., A. Schambach, and H. G. Krausslich. 2000. Multiple copies of the Mason-Pfizer monkey virus constitutive RNA transport element lead to enhanced HIV-1 Gag expression in a context-dependent manner. Nucleic Acids Res. 28901-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang, Y., F. Guo, S. Cen, and L. Kleiman. 2007. Inhibition of initiation of reverse transcription in HIV-1 by human APOBEC3F. Virology 36592-100. [DOI] [PubMed] [Google Scholar]
- 61.Zheng, Y. H., H. F. Yu, and B. M. Peterlin. 2003. Human p32 protein relieves a posttranscriptional block to HIV replication in murine cells. Nat. Cell Biol. 5611-618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.