Abstract
Coronaviruses express two very large replicase polyproteins, the 16 autoproteolytic cleavage products of which collectively form the membrane-anchored replication complexes. How these structures are assembled is still largely unknown, but it is likely that the membrane-spanning members of these nonstructural proteins (nsps) are responsible for the induction of the double-membrane vesicles and for anchoring the replication complexes to these membranes. For 3 of the 16 coronavirus nsps—nsp3, nsp4, and nsp6—multiple transmembrane domains are predicted. Previously we showed that, consistent with predictions, nsp4 occurs in membranes with both of its termini exposed in the cytoplasm (M. Oostra et al., J. Virol. 81:12323-12336, 2007). Strikingly, however, for both nsp3 and nsp6, predictions based on a multiple alignment of 27 coronavirus genome sequences indicate an uneven number of transmembrane domains. As a consequence, the proteinase domains present in nsp3 and nsp5 would be separated from their target sequences by the lipid bilayer. To look into this incongruity, we studied the membrane disposition of nsp3 and nsp6 of the severe acute respiratory syndrome coronavirus and murine hepatitis virus by analyzing tagged forms of the proteins expressed in cultured cells. Contrary to the predictions, in both viruses, both proteins had their amino terminus, as well as their carboxy terminus, exposed in the cytoplasm. We established that two of the three hydrophobic domains in nsp3 and six of the seven in nsp6 are membrane spanning. Subsequently, we verified that in nsp4, all four hydrophobic domains span the lipid bilayer. The occurrence of conserved non-membrane-spanning hydrophobic domains in nsp3 and nsp6 suggests an important function for these domains in coronavirus replication.
Positive-strand RNA viruses induce the formation of cytoplasmic membrane structures in their host cells to accomplish the efficient replication of their genomes. These structures probably facilitate the orchestration of the replication process and the recruitment of the components required for RNA synthesis and may shield the RNA intermediates from recognition by the host cell's defense mechanisms. The membranes of these structures can be acquired from different cellular compartments. In many virus families, such as Picorna-, Flavi-, and Bromoviridae, the RNA replication complex is associated with membranes derived from the endoplasmic reticulum (ER). However, endosomes and lysosomes (Togaviridae), peroxisomes and chloroplasts (Tombusviridae), and mitochondria (Nodaviridae) are also used as membrane donors (for a review, see reference 44). In general, viral nonstructural proteins (nsps) are responsible for the assembly of the replication complex at these specific cellular organelles and for the observed membrane rearrangements.
Coronaviruses are enveloped, plus-strand RNA viruses belonging to the family Coronaviridae that, together with the Arteri- and Roniviridae, belong to the order Nidovirales. With sizes ranging between 27 and 32 kb, coronaviruses possess the largest genomes among all known RNA viruses. The 5′ two-thirds of the coronaviral genome is occupied by open reading frames (ORFs) that encode the viral replicase. The remaining part of the genome codes for the structural proteins, which invariably comprise at least the spike (S), envelope (E), membrane (M), and nucleocapsid (N) proteins, and for a variable number of accessory proteins. Except for the replicase ORFs, all genes are translated from subgenomic mRNAs, which are generated by a process of discontinuous transcription (for a recent review, see reference 45). The viral replicase is encoded by the two most 5′ ORFs on the genomic RNA, orf1a and orf1b, which are translated into two very large precursor polyproteins, pp1a and pp1ab, comprising approximately 4,000 and 7,000 amino acids, respectively. pp1ab is translated only after a −1 frameshift induced by a slippery sequence at the end of orf1a, which only occurs in a fraction of translational events (5).
The replicase polyproteins are processed by virus-encoded proteinases to produce 16 mature nsps (19, 63). nsp1, -2, and -3 are released through cleavage by a papain-like proteinase, one or two functional copies of which are contained within nsp3; all other cleavages are performed by the 3C-like main proteinase located in nsp5 (2, 11, 17, 55, 64). pp1a, which contains the first 11 nsps, including the proteinases, is three to five times more abundantly produced than pp1ab, which additionally gives rise to nsp12 to nsp16 (13). Functions involving the actual replication and transcription of the viral genome have been assigned to several of the orf1b-encoded nsps, like RNA-dependent RNA polymerase (RdRp) activity in nsp12, helicase activity in nsp13, exonuclease activity in nsp14, endoribonuclease activity in nsp15, and methyltransferase activity in nsp16 (3, 7, 9, 25-27, 43, 63). Except for the proteases, the functions of the orf1a-encoded nsps are less clearly defined. ADP-ribose-1″-monophosphatase activity has been identified in nsp3, whereas in nsp8 a second RdRp activity was discovered (24, 42). nsp7 and nsp8 together were shown to form a hexadecameric complex able to bind nucleic acids and suggested to function as a processivity factor for the RdRp (62). nsp9 and nsp10 were also found to bind nucleic acids, while nsp10, which contains two zinc finger domains, might additionally be somehow involved in the processing of the polyproteins (12, 31, 52).
The nsps collectively assemble into membrane-associated complexes that constitute the sites of de novo viral RNA synthesis (47, 58). The virus-encoded N protein and possibly several cellular proteins are also recruited to these sites (4, 46). The replication complexes are found in the perinuclear region of the host cell anchored to double-membrane vesicles (DMVs) (6, 20, 48). The origin of the membranes in these structures has not unambiguously been established. Several cellular pathways and organelles, such as the ER, Golgi complex, endosomal/lysosomal system, and autophagic pathway, have been implicated in the formation of the replication complexes (40, 47, 48, 57). However, recent studies indicate the ER to be the most likely lipid donor compartment (37, 48).
Essentially nothing is known about how the membrane-anchored replication complexes are induced and assembled and how the individual nsps and other necessary constituents are recruited to these sites. Coronavirus orf1a encodes three nsps—nsp3, nsp4, and nsp6—that are predicted to contain transmembrane domains. It is likely that these proteins not only function in the membrane anchoring of the multisubunit replication complex but also induce the formation of the membrane structures. The membrane association of mouse hepatitis virus (MHV) and severe acute respiratory syndrome-associated coronavirus (SARS-CoV) nsp3 has been demonstrated previously (22, 28). In addition, the membrane association, as well as the topology, of nsp4 of these two viruses has been resolved (37). The involvement of these nsps in the formation of replication complexes is supported by the fact that for the related arterivirus equine arteritis virus (EAV), coexpression of the counterparts of nsp3 and nsp4, i.e., nsp2 and nsp3, is sufficient for the induction of DMVs (49). Furthermore, mutations in MHV nsp4 or EAV nsp3 impaired DMV formation (8, 39).
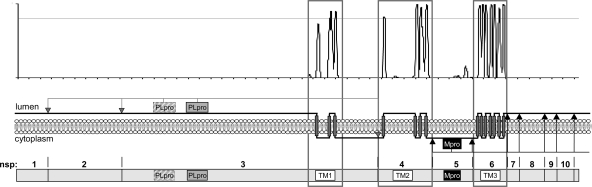
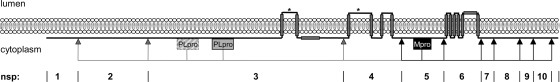
In this study, we wanted to fill a gap in the existing knowledge of coronaviruses by establishing an experimentally verified topology model of the complete coronavirus replicase polyprotein. When transmembrane predictions (51) were performed on the entire orf1a region based on the multiple alignment of 27 coronavirus sequences, with representatives from each of the different coronavirus groups (present as reference sequences [RefSeq] in the NCBI CoreNucleotide database), some discrepancies were observed (Fig. 1). For nsp4, four transmembrane domains were predicted, which is in agreement with our previous experimental data that showed that both termini are located on the cytoplasmic face of the membrane (37). However, for both nsp3 and nsp6, an uneven number of transmembrane domains was predicted, three and seven, respectively, resulting in a model in which the proteinases, present in nsp3 and nsp5, would be separated from some of their target sequences by the lipid bilayer. Furthermore, this prediction places nsp1, nsp2, and most of nsp3 on the lumenal side of the membrane, which is unlikely as pp1a lacks an amino-terminal signal sequence. In addition, several studies have shown that nsp1 and nsp2, as well as a number of nsps downstream of nsp6, localize to the cytosol (6, 21, 22, 57). To solve these discrepancies, we focused on the topology and membrane integration of the hydrophobic nsps, particularly nsp3 and nsp6. To strengthen our conclusions, the nsps of both MHV and SARS-CoV were studied. Contrary to the predictions, in each virus both nsp3 and nsp6 appeared to have the amino terminus as well as the carboxy terminus exposed on the cytoplasmic side of the membrane. We show that in both nsp3 and nsp6, not all of the predicted transmembrane domains are used as such. In nsp3, only two, and in nsp6, only six of the predicted transmembrane domains actually span the lipid bilayer. These results raise the question of why coronaviruses have conserved hydrophobic domains in their nsps that do not function as membrane-spanning domains.
FIG. 1.
pp1a transmembrane domain prediction based on multiple alignment. The presence of transmembrane domains in the pp1a precursor was predicted (51) on the basis of a multiple alignment of 27 different coronavirus sequences with representatives from each of the different groups (RefSeq in the NCBI CoreNucleotide database). The resulting hydrophobicity plot is shown in the upper panel, with peaks reaching the threshold (dotted line) representing predicted membrane-spanning domains. The black line in the middle panel represents the protein and shows, based on the known topology of nsp4 (37), its predicted localization on the lumenal or cytoplasmic side of the membrane, which is symbolized by the gray bar. At the bottom, a schematic representation of pp1a is shown in which the regions containing the putative transmembrane (TM) domains, the papain-like protease (PLpro, two in MHV and one in SARS-CoV), and the main protease (Mpro) are highlighted. The protease cleavage sites are indicated by arrowheads, with the nsp3-encoded PLpro cleavage sites in gray and the nsp5-encoded Mpro cleavage sites in black.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
OST7-1 cells, obtained from B. Moss (16), were maintained as monolayer cultures in Dulbecco's modified Eagle's medium (DMEM) (Cambrex Bio Science Verviers) containing 10% fetal calf serum (FCS) (Bodinco B.V.), 100 IU of penicillin per ml, and 100 μg of streptomycin per ml (referred to as culture medium). Recombinant vaccinia virus encoding the bacteriophage T7 RNA polymerase (vTF7-3) was also obtained from B. Moss (18).
Rabbit polyclonal antisera directed against the enhanced green fluorescent protein (EGFP) or the influenza virus hemagglutinin (HA) tag were obtained from ICL. Rabbit antiserum recognizing the C-terminal domain of the MHV membrane (M) protein (anti-MC) has been described previously (30), while mouse monoclonal antibody against the amino terminus of MHV M (J1.3 or anti-MN) was provided by J. Fleming (53).
Plasmid constructions.
First, a plasmid was created in which all gene fragments could be cloned behind a T7 promoter in frame with the sequence encoding the EGFP tag. To this end, the pEGFP-N3 vector (Clontech) was digested with EcoRI and NotI, of which the latter restriction site was filled in with Klenow polymerase (Invitrogen) and this fragment was cloned into the EcoRI- and BamHI-digested pTUG31 vector (59), of which the BamHI restriction site was also filled in with Klenow polymerase (Invitrogen), thereby creating pTug-EGFP. An N glycosylation site was created in the EGFP gene by performing site-directed mutagenesis with the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions on pEGFP-N3 with primers 3212 and 3213. This mutated EGFP gene was cloned into the pTUG31 vector similar to the wild-type EGFP gene, thereby creating pTug-EGFPglyc. (For the sequences and locations on the viral genomes of all of the primers used, see Table S1 in the supplemental material.)
The SARS-CoV nsp gene fragments were obtained by reverse transcriptase-PCR amplification of viral RNA isolated from SARS-CoV isolate 5688 (29) with primers 3072 and 3073 for nsp3 (nsp3s) and primers 3070 and 3071 for nsp6 (nsp6s). The MHV nsp gene fragments were obtained by reverse transcription-PCR amplification of viral genomic RNA isolated from MHV strain A59 with primers 3632 and 2933 for nsp3 (nsp3m) and primers 2974 and 2975 for nsp6 (nsp6m). The PCR products were cloned into the pGEM-T Easy vector (Promega), and their sequences were confirmed by sequence analysis. Site-directed mutagenesis to mutate the N glycosylation sites was performed on the pGem-T Easy constructs containing the nsp3 gene fragments with primers 3354 and 3355 for SARS-CoV nsp3 and primers 3630 and 3631 for MHV nsp3.
The nsp gene fragments were cloned into the pTug-EGFP or pTug-EGFPglyc vector by digesting the pGem-T Easy constructs with EcoRI and BamHI and cloning the fragments obtained into the EcoRI-BamHI-digested pTug-EGFP and pTug-EGFPglyc vectors. The plasmids created encode the different nsps fused C terminally to the wild-type or mutant EGFP tag. The same EcoRI-BamHI nsp3 and nsp6 fragments were also cloned into the EcoRI-BamHI-digested pTUG31 vector together with a primer dimer of primers 3050 and 3051, resulting in plasmids encoding the nsps C terminally fused to a HA tag. In these latter constructs, as well as in the pTug construct encoding MHV nsp6 fused to EGFPglyc (pTug-nsp6m-EGFPglyc), an MHV M (MN) tag-encoding sequence was inserted in front of the nsps by cloning a primer dimer of primers 3019 and 3020, coding for the 10-residue amino-terminal sequence of the MHV M protein (MSSTTQAPEP), into the XhoI-EcoRV-restricted plasmids, thereby creating constructs that encode nsps tagged at both termini.
MHV nsp6 lacking the first hydrophobic domain (nsp6mΔHD1′) was amplified by PCR with primers 3566 and 2975. The PCR product was cloned into the pGEM-T Easy vector (Promega), and the sequence was confirmed by sequence analysis. A fragment was obtained by digestion with EcoRV and BamHI and cloned into the EcoRV-BamHI-digested pTugMN-nsp6m-EGFPglyc plasmid, thereby creating a construct containing MHV nsp6 without the first hydrophobic domain fused N terminally to the MHV M tag and C terminally to the EGFP tag containing the N glycosylation site.
Progressive C-terminal deletion mutant forms lacking one or more hydrophobic domains were made for both nsp3 and -6. PCRs were performed with the same forward primers as described before; for the reverse primers used, see Table S1 in the supplemental material. For nsp3, the PCRs were performed on the constructs with the mutated N glycosylation sites. The PCR products were cloned into the pGEM-T Easy vector (Promega), and the sequences were confirmed by sequence analysis. Fragments were obtained by digestion with EcoRI and BamHI and cloned into the EcoRI-BamHI-digested pTug-EGFPglyc vector.
By using combinations of the primers used to create the C-terminal deletions and primers for N-terminal deletions, the sequences encoding each of the hydrophobic domains of MHV nsp6 were also amplified separately. These sequences were cloned into the pGEM-T Easy vector (Promega) and confirmed by sequence analysis. Fragments were obtained by digestion with EcoRV and BamHI and cloned into the EcoRV-BamHI-digested pTugMN-nsp6m-EGFPglyc plasmid, thereby creating constructs encoding the MHV nsp6 fragments fused N terminally to the MHV M tag and C terminally to the EGFP tag containing the N glycosylation site.
The construct containing SARS-CoV nsp4 in which the N glycosylation site had been removed by mutation has been described previously (37). This construct was used to create C-terminal deletion mutants lacking one to three of the hydrophobic domains. PCRs were performed with primer 3648 and primer 3848, 3650, or 3651; the products were cloned into the pGEM-T Easy vector (Promega); and the sequences were confirmed by sequence analysis. Fragments were obtained by digestion with EcoRI and BamHI and cloned into the EcoRI-BamHI-digested pTug-EGFPglyc vector.
The construct containing MHV nsp4 has also been described previously (37). The glycosylation sites in MHV nsp4 were mutated by performing sequential site-directed mutagenesis reactions with primers 3758 and 3759 and primers 3760 and 3761. The construct with the mutated glycosylation sites was used to create C-terminal deletion mutants by performing PCRs with primer 2890 and primer 3847, 3756, or 3757. The PCR products were cloned into the pGEM-T Easy vector (Promega), and the sequences were confirmed by sequence analysis. Fragments were obtained by digestion with EcoRI and BamHI and cloned into the EcoRI-BamHI-digested pTug-EGFPglyc vector.
The constructs encoding the equine arterivirus (EAV) membrane protein N terminally extended with the MN tag (EAV M + 9A) (10) and the 8a protein C terminally tagged with the EGFP tag with or without the N glycosylation site (36) have been described previously.
Infection and transfection.
Subconfluent monolayers of OST7-1 cells grown in 10-cm2 tissue culture dishes were inoculated with vTF7-3 at a multiplicity of infection of 10 for 1 h, after which the medium was replaced with a transfection mixture consisting of 0.5 ml of DMEM without FCS but containing 10 μl Lipofectin (Invitrogen) and 5 μg of each selected construct. After a 5-min incubation at room temperature, 0.5 ml of DMEM was added and incubation was continued at 37°C. At 3 h postinfection (p.i.), the medium was replaced with culture medium. Where indicated, tunicamycin (5 μg/ml) or brefeldin A (6 μg/ml) was added to the culture medium at 3 h p.i.
Metabolic labeling and immunoprecipitation.
Prior to labeling, the cells were starved for 30 min in cysteine- and methionine-free modified Eagle's medium containing 10 mM HEPES (pH 7.2) and 5% dialyzed FCS. This medium was replaced with 1 ml of a similar medium containing 100 μCi of 35S in vitro cell-labeling mixture (Amersham), after which the cells were further incubated for the indicated time periods. After labeling, the cells were washed once with phosphate-buffered saline (PBS) containing 50 mM Ca2+ and 50 mM Mg2+ and then lysed on ice in 1 ml of lysis buffer (0.5 mM Tris [pH 7.3], 1 mM EDTA, 0.1 M NaCl, 1% Triton X-100) per 10-cm2 dish. The lysates were cleared by centrifugation for 5 min at 15,000 rpm and 4°C.
Coupled in vitro transcription and translation reactions were performed with the TNT coupled reticulocyte lysate system from Promega, according to manufacturer's instructions, in the presence of 35S in vitro labeling mixture (Amersham) but without the use of microsomal membranes.
Radioimmunoprecipitations were performed essentially as described previously (35); 200-μl aliquots of the cell lysates or 5-μl volumes of in vitro translation reaction mixtures were diluted in 1 ml detergent buffer (50 mM Tris [pH 8.0], 62.5 mM EDTA, 1% NP-40, 0.4% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) containing antibodies (2 μl rabbit anti-EGFP or rabbit anti-HA serum or 25 μl of J1.3 monoclonal anti-MHV M serum). The immunoprecipitation mixtures were incubated overnight at 4°C. The immune complexes were adsorbed to Pansorbin cells (Calbiochem) for 60 min at 4°C and subsequently collected by centrifugation. The pellets were washed three times by resuspension and centrifugation with RIPA buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 0.1% SDS, 1% NP-40, 1% sodium deoxycholate). The final pellets were suspended in Laemmli sample buffer (LSB) and heated at 95°C for 1 min before analysis by SDS-polyacrylamide gel electrophoresis (PAGE) with 10 to 15% polyacrylamide gels.
Where indicated, immunoprecipitates were treated with peptide-N-glycosidase F (PNGaseF; New England BioLabs). To this end, the final immunoprecipitation pellets were suspended in PBS instead of LSB, 2 μl PNGaseF was added, and the samples were incubated at 37°C for 2 h. Before analysis by SDS-PAGE, 0.5 volume of a three-times-concentrated solution of LSB was added to the samples, which were then heated at 95°C for 1 min.
Immunofluorescence microscopy.
OST7-1 cells grown on glass coverslips were fixed at the indicated times postinfection with 3% paraformaldehyde for 1 h at room temperature. The fixed cells were washed with PBS and permeabilized with either 0.1% Triton X-100 for 10 min at room temperature or 0.5 μg/ml digitonin (diluted in 0.3 M sucrose-25 mM MgCl2+-0.1 M KCl-1 mM EDTA-10 mM PIPES [pH 6.8]) for 5 min at 4°C. Next, the permeabilized cells were washed with PBS and incubated for 15 min in blocking buffer (PBS-10% normal goat serum), followed by a 45-min incubation with antibodies directed against HA or against the C- or N-terminal domain of MHV M. After four washes with PBS, the cells were incubated for 45 min with either fluorescein isothiocyanate-conjugated goat anti-rabbit immunoglobulin G antibodies (ICN) or Cy5-conjugated donkey anti-mouse immunoglobulin G antibodies (Jackson Laboratories). After four washes with PBS, the samples were mounted on glass slides in FluorSave (Calbiochem). The samples were examined with a confocal fluorescence microscope (Leica TCS SP2).
RESULTS
Processing of nsp3 and nsp6.
We started our studies of nsp3 and nsp6 by analyzing the co- and posttranslational modifications of the two proteins from both SARS-CoV and MHV. To this end, gene fragments encoding nsp3 and nsp6 of both viruses were cloned into the pTUG31 vector behind a T7 promoter and fused to an EGFP tag, since no antibodies to the proteins themselves were available. As the full-length nsp3-encoding genome segments are very large (approximately 6 kb) and difficult to clone, only the 3′-terminal 2-kb fragments, which encode all of the hydrophobic domains, were cloned. nsp3 of both SARS-CoV and MHV contains potential N glycosylation sites (NXS/T) in front of the first hydrophobic domain, between the first and second hydrophobic domains, and behind the third hydrophobic domain (Fig. 1 shows the localization of the hydrophobic domains). The glycosylation sites between the first and second hydrophobic domains, two for SARS-CoV and one for MHV, have previously been shown to be functional (22, 28). No potential N glycosylation sites were identified in the sequence of MHV nsp6, while for SARS-CoV nsp6, an N glycosylation site was predicted between the fifth and sixth hydrophobic domains.
We studied the expression and processing of the nsps by in vitro translation and by using the recombinant vaccinia virus bacteriophage T7 RNA polymerase (vTF7-3) expression system. OST7-1 cells were infected with vTF7-3, transfected with plasmids containing the nsp3-EGFP or nsp6-EGFP gene, and labeled with [35S]methionine from 5 to 6 h p.i. The cells were lysed and processed for immunoprecipitation with a rabbit polyclonal antiserum directed to the EGFP tag. In parallel, in vitro translations were performed with the TNT coupled reticulocyte lysate system from Promega in the absence of membranes to analyze the electrophoretic mobility of the nonprocessed proteins. To demonstrate the presence of the N-linked sugars on the nsps, the proteins were expressed in the presence and absence of tunicamycin, which is an inhibitor of N-linked glycosylation, and/or the N-linked glycans were enzymatically removed with PNGaseF.
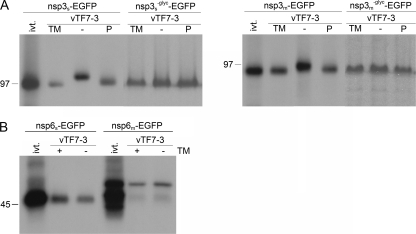
As shown in Fig. 2A, the electrophoretic mobility of nsp3 expressed in OST7-1 cells in the presence of tunicamycin was similar to that of the in vitro-translated product, whereas the protein expressed in the absence of tunicamycin migrated slower. Treatment of this latter protein with PNGaseF shifted its electrophoretic mobility to that of the in vitro-translated product and of the protein expressed in the presence of tunicamycin. This result confirms the addition of N-linked glycans to nsp3, as has been demonstrated previously (22, 28). Next, the N glycosylation sites between the first and second hydrophobic domains were mutated and these proteins, in fusion with EGFP (nsp3−glyc-EGFP), were also expressed in the presence and absence of tunicamycin and/or treated with PNGaseF. The presence of tunicamycin or treatment with PNGaseF did not influence the electrophoretic mobility of these mutant proteins (Fig. 2A). This clearly demonstrates that the N glycosylation sites between the first and second hydrophobic domains are the only N-glycan attachment sites. The other potential sites are likely inaccessible, probably because they are located on the cytoplasmic side of the membrane.
FIG. 2.
Processing of SARS-CoV and MHV nsp3 and nsp6. vTF7-3-infected OST7-1 cells were transfected with the indicated constructs. The cells were labeled with [35S]methionine from 5 to 6 h p.i., lysed, and processed for immunoprecipitation with antiserum directed to the EGFP tag, followed by SDS-PAGE. (A) Cells were transfected with SARS-CoV or MHV nsp3-EGFP (nsp3s-EGFP or nsp3m-EGFP, respectively)-encoding constructs without or with mutation (−glyc) of the N glycosylation sites in the presence (TM) or absence (− and P) of tunicamycin. The constructs with intact glycosylation sites were also transcribed and translated in vitro with the TNT coupled reticulocyte lysate system from Promega (ivt.). After immunoprecipitations, the samples were mock (TM, −) or PNGaseF (P) treated. (B) Cells were transfected with SARS-CoV or MHV nsp6-EGFP (nsp6s-EGFP and nsp6m-EGFP, respectively)-encoding constructs in the presence (+) or absence (−) of tunicamycin (TM). The same constructs were also transcribed and translated in vitro with the TNT coupled reticulocyte lysate system from Promega (ivt.). The positions and masses (in kilodaltons) of protein size markers are indicated at the left. Only the relevant portion of the gels is shown.
vTF7-3-expressed nsp6 of both viruses comigrated in the gel with the corresponding in vitro-translated products, both in the presence and in the absence of tunicamycin (Fig. 2B). Also, some lower-molecular-weight products were observed after the in vitro translation of MHV nsp6, which probably resulted from translation initiation at more downstream start codons. Apparently, nsp6 of SARS-CoV or MHV is not N glycosylated, indicating that the putative glycosylation site in the region between the fifth and sixth hydrophobic domains of SARS-CoV nsp6 is either located on the cytoplasmic side of the membrane or not accessible for glycosylation for other reasons. Furthermore, it appeared that both nsp6 fusion proteins migrated with lower mobility in the gel than predicted on the basis of their amino acid sequences. Similar results have been obtained before for other highly hydrophobic proteins (37, 41).
Membrane topology of nsp3 and nsp6.
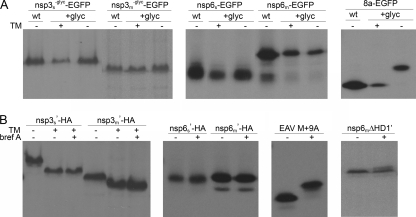
In order to elucidate the membrane topology of SARS-CoV and MHV nsp3 and nsp6, we studied the disposition of their amino and carboxy termini. Therefore, the proteins were C or N terminally extended with tags containing potential glycosylation sites. The C termini of the proteins were fused to an EGFP tag in which an N glycosylation site had been created (EGFPglyc). As a control, we used a fusion protein generated earlier which essentially consists of a cleavable signal sequence fused to the same tag (8a-EGFP) (36). For nsp3, the tag was fused to either the SARS-CoV or the MHV gene fragment in which the N glycosylation sites between the first and second hydrophobic domains had been disabled by mutation (nsp3−glyc) to allow discrimination between glycosylation of the tag and that of nsp3 itself. The fusion proteins were expressed with the vTF7-3 expression system in the presence and absence of tunicamycin. N glycosylation of the C-terminal EGFP tag would demonstrate that the carboxy terminus of the protein is located on the lumenal side of the membrane.
For each of the proteins (nsp3 or nsp6), a similar electrophoretic mobility was observed regardless of the EGFP tag used (i.e., with or without the N glycosylation site) or the presence of tunicamycin (Fig. 3A). The control protein, 8a, behaved as expected. In the presence of tunicamycin, the protein with the EGFPglyc tag migrated with the same mobility as the protein with the wild-type tag in the absence of tunicamycin, whereas in the absence of tunicamycin the protein with the EGFPglyc tag migrated slower (Fig. 3A). The results demonstrate that the carboxy termini of nsp3 and nsp6 of both SARS-CoV and MHV are located on the cytoplasmic side of the membrane.
FIG. 3.
Glycosylation of tagged SARS-CoV and MHV nsp3 and nsp6. vTF7-3-infected OST7-1 cells were transfected with the indicated constructs. The cells were labeled with [35S]methionine from 5 to 6 h p.i., lysed, and processed for immunoprecipitation, followed by SDS-PAGE. (A) Cells were transfected with constructs encoding SARS-CoV or MHV nsp3 or nsp6 or encoding the SARS-CoV orf8a protein (8a), each fused either to a wild-type EGFP tag (wt) or to an EGFP tag with an N glycosylation site (+glyc). The proteins were expressed in the presence (+) or absence (−) of tunicamycin (TM). Immunoprecipitations were performed with rabbit antiserum directed to the EGFP tag. (B) Cells were transfected with constructs encoding SARS-CoV or MHV nsp3 or nsp6 with a C-terminal HA tag and an N-terminal MN tag (') or with a construct encoding the EAV M protein with the same MN tag (EAV M + 9A). MHV nsp6 lacking the first hydrophobic domain (nsp6mΔHD1′) contains the N-terminal MN tag in combination with a C-terminal EGFP tag. The cells were incubated in the presence (+) or absence (−) of brefeldin A (bref A) and or tunicamycin (TM). Immunoprecipitations were performed with rabbit antiserum directed to the HA tag or, for EAV M, with monoclonal antibody J1.3 (directed against the MN tag). Only the relevant portions of the gels are shown.
To examine the disposition of the nsp3 and nsp6 amino termini, the N-terminal 10-residue sequence of the MHV M protein (MN), which contains a well-defined O glycosylation motif, was fused to the amino terminus of each nsp. The functionality of this tag was previously demonstrated after fusion to the EAV type III M protein, resulting in EAV M + 9A (10). This protein has a Nexo/Cendo topology and is retained in the ER. Yet it became O glycosylated upon the addition of brefeldin A, a drug which causes the redistribution of Golgi enzymes, including the ones involved in O glycosylation, to the ER. By a similar approach, the location of the N termini of nsp3 and nsp6 was assessed. The nsp fusion proteins, containing the N-terminal MN and a C-terminal HA tag, were expressed with the vTF7-3 expression system in OST7-1 cells in the presence or absence of brefeldin A and/or tunicamycin. Tunicamycin was added to the cells expressing the nsp3 fusion protein to prevent its N glycosylation, as this could obscure the detection of its O glycosylation.
As shown in Fig. 3B, the EAV M + 9A control protein showed a shift in electrophoretic mobility when it was expressed in the presence of brefeldin A. In contrast, the electrophoretic mobilities of the nsp3 and nsp6 fusion proteins were unaffected by the addition of brefeldin A. As expected, the presence of tunicamycin did prevent the addition of N-linked sugars to nsp3. These results indicate that the amino termini of SARS-CoV and MHV nsp3 and nsp6 are not accessible to enzymes that mediate the addition of O-linked sugars, which is most likely explained by the cytoplasmic exposure of these termini, although misfolding of the amino-terminal tag as the cause can also not be ruled out completely. We do not consider the latter explanation very likely, since the presence of two proline residues in the tag has previously been demonstrated to induce a conformation favorable for glycosylation (10). Indeed, when the first hydrophobic domain of MHV nsp6 was removed, the resulting protein carrying the amino-terminal tag (now in combination with a C-terminal EGFP tag) did become modified by O-linked sugars in the presence of brefeldin A, as shown by the appearance of an extra band which runs at a slightly higher position in the gel. The difference in electrophoretic mobility between the glycosylated and unglycosylated protein species is smaller for the nsp6 mutant than for the EAV M protein because of the much higher molecular weight of EGFP-tagged nsp6 (Fig. 3B).
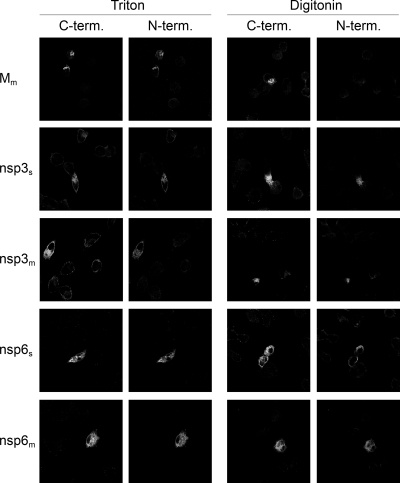
The localization of the amino and carboxy termini of each nsp was also determined by immunofluorescence assays. In these experiments, nsps were used that were tagged at both ends, containing the N-terminal MN and a C-terminal HA extension. OST7-1 cells were infected with vTF7-3 and transfected with plasmids encoding the fusion proteins. The cells were fixed at 6 h p.i. with 3% paraformaldehyde and permeabilized under strictly controlled conditions with either Triton X-100, which permeabilizes all cellular membranes, or digitonin, which selectively permeabilizes the plasma membrane. The type III MHV M protein, which has a known Nexo/Cendo topology and localizes to the Golgi compartment, was used as a control. A rabbit polyclonal antibody directed to the C terminus and a mouse monoclonal antibody directed to the N terminus were used to detect this protein after the use of each of the permeabilization methods. As expected, the antibody directed to the C terminus detected the protein after Triton X-100 permeabilization, as well as after digitonin permeabilization, whereas the antibody directed to the N terminus only detected the protein after permeabilization with Triton X-100 and not after treatment with digitonin, thereby validating the assay conditions (Fig. 4).
FIG. 4.
Membrane topology of SARS-CoV and MHV nsp3 and nsp6. vTF7-3-infected OST7-1 cells were transfected with constructs encoding the proteins indicated at the left. The cells were fixed at 6 h p.i. and permeabilized with Triton X-100 (left two columns) or digitonin (right two columns). Immunofluorescence analysis was performed with antibodies against the C-terminal (C-term.) tag, anti-Mc for Mm and anti-HA for the nsps (first and third columns), or against the N-terminal (N-term.) tag, J1.3 (anti-MN, second and fourth columns).
Similar experiments were performed for SARS-CoV and MHV nsp3 and nsp6, with the exception that a different rabbit antiserum, directed against the C-terminal HA tag, was used. Both nsp3 and nsp6 appeared to localize in a reticular pattern reminiscent of the ER, as observed at higher magnification (data not shown). After permeabilization with Triton X-100 or digitonin, cells were stained with the rabbit antibody directed to the C terminus and the mouse antibody directed to the N terminus. The results, shown in Fig. 4, demonstrate that the amino and carboxy termini of both nsp3 and nsp6 are located on the cytoplasmic face of the membrane and confirm the results obtained with the biochemical experiments shown in Fig. 3. Furthermore, the localization of the nsp3 amino terminus on the cytoplasmic face of the membrane is in agreement with the observed N glycosylation in the region between the first and second transmembrane domains.
Membrane integration of nsp3.
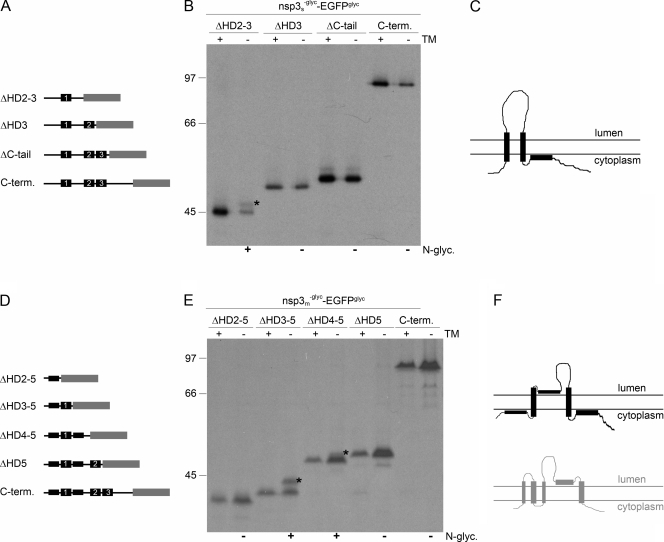
The above results show that both nsp3 and nsp6 have a Nendo/Cendo membrane topology, indicating that both nsps have an even number of transmembrane domains. This is not in agreement with the three and seven transmembrane domains that are predicted for nsp3 and nsp6, respectively. Therefore, we examined which of the predicted transmembrane domains are indeed used as such. To this end, constructs were made that encode progressive C-terminal deletion mutant forms of the nsps, lacking one or more hydrophobic domains, fused to the EGFPglyc tag.
As SARS-CoV nsp3 appeared to be N glycosylated between the first two of its three predicted transmembrane domains, it seems most likely that either the second or the third hydrophobic domain does not function as a membrane-spanning domain. Mutant forms were made in which the C-terminal hydrophilic tail was deleted or in which the C-terminal deletion was extended to include either the third hydrophobic domain or both the third and second hydrophobic domains (Fig. 5A). These deletion mutant forms, which were fused to the EGFPglyc tag, additionally carried the mutations that disrupt the N glycosylation motifs between the first and second hydrophobic domains.
FIG. 5.
Membrane integration of deletion mutant forms of SARS-CoV and MHV nsp3. (A, D) Schematic representations of the C-terminal (C-term.) deletion mutant forms of SARS-CoV (A) and MHV (D) nsp3, with the hydrophobic domains presented as black rectangles and the EGFPglyc tag in gray. The corresponding hydrophobic domains in the two proteins are indicated by numbers. (B, E) vTF7-3-infected OST7-1 cells were transfected with the indicated constructs and expressed in the presence (+) or absence (−) of tunicamycin (TM). The cells were labeled with [35S]methionine from 5 to 6 h p.i., lysed, and processed for immunoprecipitation with anti-EGFP antiserum followed by SDS-PAGE. The positions and masses (in kilodaltons) of the protein size markers are indicated at the left. The asterisks indicate the position of the glycosylated protein species. Below the panels, the observed presence or absence of N glycosylation is indicated by a plus or a minus sign, respectively. (C, F) Models of the membrane structures of SARS-CoV (C) and MHV (F) nsp3, with the hydrophobic domains presented as black rectangles. For comparison, the MHV nsp3 model proposed by Baker and coworkers is shown below in gray.
vTF7-3-infected OST7-1 cells were transfected with constructs encoding the different fusion proteins and labeled from 5 to 6 h p.i. in the presence or absence of tunicamycin. The cells were lysed and processed for immunoprecipitation with the anti-EGFP serum. As shown in Fig. 5B, complete removal of the hydrophilic C terminus of nsp3 (ΔC-tail) does not affect the glycosylation of the fusion protein, as expected. When the third hydrophobic domain was additionally deleted, the fusion protein still remained indifferent to tunicamycin and was thus not glycosylated, indicating that this third hydrophobic domain did not function as a transmembrane domain (Fig. 5B). However, when the second hydrophobic domain was deleted as well, an extra protein species with a slightly lower electrophoretic mobility appeared after expression in the absence of tunicamycin compared to in its presence (Fig. 5B). Although the protein was only partially glycosylated, this result demonstrates that the carboxy-terminal EGFP tag fused to this mutant form of nsp3 is translocated to the lumenal side of the membrane. As nsp3ΔHD2-3 was the only mutant form that became modified by N-linked sugars, we conclude that SARS-CoV nsp3 spans the lipid bilayer only twice and that the third hydrophobic domain does not function as a transmembrane domain (Fig. 5C).
The transmembrane domain predictions for MHV nsp3 are somewhat different from those for SARS-CoV nsp3. Several programs predict the presence of five rather than three transmembrane domains. Three of these correspond to those in SARS-CoV nsp3, but additional hydrophobic domains are located immediately up- and downstream of the first domain (Fig. 5D). However, not all programs predict these additional hydrophobic domains to be membrane spanning (28). Earlier work by Baker and coworkers, with a combination of in vitro translations, carbonate extraction assays, and proteinase K treatments, suggested that four of the five predicted hydrophobic domains functioned as transmembrane domains, with only the fourth domain (i.e., the one corresponding to the second hydrophobic domain in SARS-CoV nsp3) not spanning the lipid bilayer (28). As these results are in conflict with the results we obtained for SARS-CoV nsp3, we prepared a similar set of progressive C-terminal deletion mutant forms of MHV nsp3 lacking one, two, three, or four of the putative transmembrane domains and again fused to the EGFPglyc tag (Fig. 5D).
The different constructs were expressed with the vTF7-3 system in the presence or absence of tunicamycin. The EGFP tag was again not glycosylated when fused to the unmodified carboxy terminus of nsp3 or when the last hydrophobic domain (HD5) was deleted, consistent with the results obtained for SARS-CoV nsp3 (Fig. 5E). However, when the fourth hydrophobic domain was deleted as well, the fusion protein became partially glycosylated, as shown by the presence of a protein species the appearance of which was inhibited by tunicamycin (Fig. 5E), indicating that the EGFP tag had become translocated. The same result, though with more efficient glycosylation, was obtained when also the third hydrophobic domain was removed. Finally, additional deletion of the second hydrophobic domain, which corresponds to the first hydrophobic domain of SARS-CoV nsp3, resulted in an unglycosylated protein as its electrophoretic mobility was not affected by the presence of tunicamycin (Fig. 5E). As nsp3ΔHD4-5 and nsp3ΔHD3-5 were the only MHV nsp3 mutant constructs that became modified by N-linked sugars, while the other mutants showed no trace of glycosylation, we conclude that MHV nsp3 integrates into the membrane similarly to SARS-CoV nsp3, with only two membrane-spanning domains at approximately the same positions in the protein (Fig. 5F).
Membrane integration of nsp6.
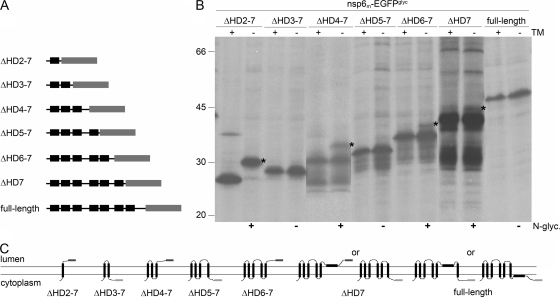
Transmembrane domain predictions for nsp6 yielded similar results for the MHV and SARS-CoV proteins, as well as for other coronaviruses shown in the multiple alignment of the orf1a sequence. Because SARS-CoV nsp6 consistently appeared as a fuzzy band when analyzed by SDS-PAGE, which will complicate the interpretation of our assay, we limited our focus to MHV nsp6. The observed cytoplasmic localization of both the amino and the carboxy termini of nsp6 implies the presence of an even number of transmembrane domains, although invariably seven such domains are predicted. Thus, as for nsp3, one of the predicted transmembrane domains is probably not used as such. To confirm this conclusion and identify this particular domain, we made progressive C-terminal deletion mutant constructs lacking one to six of the potential transmembrane domains and tagged once again with the EGFPglyc tag (Fig. 6A) and evaluated these mutant constructs as before.
FIG. 6.
Membrane integration of deletion mutant forms of MHV nsp6. (A) Schematic representation of the MHV nsp6 C-terminal deletion mutant forms, with the hydrophobic domains presented in black and the EGFPglyc tag in gray. (B) vTF7-3-infected OST7-1 cells were transfected with the constructs presented in panel A and expressed in the presence (+) or absence (−) of tunicamycin (TM). The cells were labeled with [35S]methionine from 5 to 6 h p.i., lysed, and processed for immunoprecipitation with anti-EGFP antiserum, followed by SDS-PAGE. The positions and masses (in kilodaltons) of the protein size markers are indicated at the left. The asterisks indicate the positions of the glycosylated protein species. Below the gel, the observed presence or absence of N glycosylation is indicated by a plus or a minus sign, respectively. (C) Models of the membrane structures of the C-terminal deletion mutant forms of MHV nsp6. The hydrophobic domains are presented as black rectangles, and the EGFP tag is presented as a dark gray rectangle when glycosylated and in light gray when unglycosylated.
As shown in Fig. 6B, when only the first hydrophobic domain was present (ΔHD2-7), the clear electrophoretic mobility difference caused by tunicamycin indicated that the EGFP tag was glycosylated and thus present on the lumenal side of the membrane, implying that the first hydrophobic domain functions both as a signal sequence and as a transmembrane domain. The mutant protein containing the first two hydrophobic domains (ΔHD3-7) remained unglycosylated, indicating that also the second hydrophobic domain spans the membrane. Mutant proteins with additional transmembrane domains continued to reveal alternatingly the appearance and disappearance of N-linked modifications, indicating that the predicted transmembrane domains are integrated in the lipid bilayer. However, when reaching the sixth hydrophobic domain, the regular succession was interrupted as this protein, ΔHD7, appeared to become glycosylated, though only marginally. The wild-type protein showed no sign of glycosylation. These observations lead to a model in which only six hydrophobic domains in MHV nsp6 are actually used as transmembrane domains. Our data appear to indicate that the sixth hydrophobic domain does not span the lipid bilayer; however, in view of the very inefficient glycosylation of the ΔHD7 mutant construct, we cannot fully exclude the possibility that the seventh hydrophobic domain is not membrane spanning (Fig. 6C).
The efficiency of glycosylation that we observed in this deletion assay appeared to decrease with increasing protein length. We have no clear explanation for this. Perhaps the accessibility of the glycosylation site differs between the different proteins, though this does not seem very likely, assuming that the EGFP moiety folds independently and similarly in all cases its glycosylation site is always presented in the same way. Alternatively, the longer proteins might adopt multiple alternative membrane topologies, though this is again not a very likely possibility since such behavior is not observed for ΔHD3-7, ΔHD5-7, and full-length nsp6, as judged by their complete lack of glycosylation. It seems more plausible that the functioning of the third, and particularly the fifth, hydrophobic domain as an internal signal sequence for membrane insertion is somehow hampered by the EGFP extension, resulting in decreased efficiency of tag translocation.
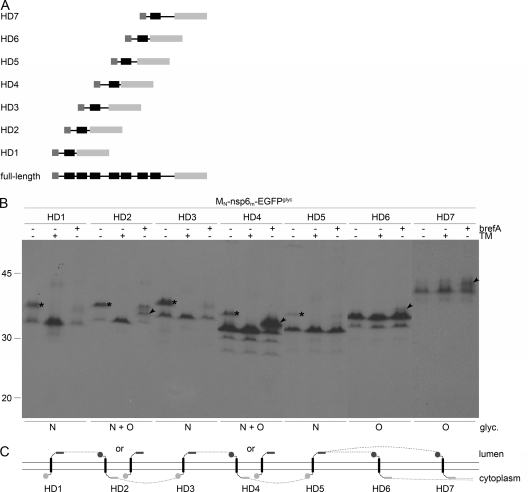
In view of these uncertainties and to obtain further evidence for our model, an additional set of mutant proteins was created in which each one of the MHV nsp6 hydrophobic domains was individually positioned between a C-terminal EGFPglyc tag and an N-terminal MN tag containing an O glycosylation site (Fig. 7A). The HD1 protein is, in fact, identical to nsp6ΔHD2-7 (Fig. 6), except for the amino-terminal tag containing the O glycosylation site. The proteins were expressed with the vTF7-3 system in the presence and absence of brefeldin A or tunicamycin and analyzed as before (Fig. 7B). The fusion proteins containing hydrophobic domain 1, 2, 3, 4, or 5 appeared in two electrophoretic forms when expressed in the absence of any drug. The slower-migrating form of these was not observed when tunicamycin had been applied, indicating that these proteins became N glycosylated. The proteins that contained either hydrophobic domain 2, 4, 6, or 7 exhibited a partial shift in electrophoretic mobility upon expression in the presence of brefeldin A, indicating that these proteins were modified by O-linked sugars. The addition of O-linked sugars was less efficient for the proteins containing either hydrophobic domain 6 or 7. Strikingly, the proteins that contained hydrophobic domain 2 or 4 were apparently able to adopt two alternative topologies, as both types of glycosylation were detected. These proteins are either N or O glycosylated, but not both, since in that case expression in the presence of brefeldin A would have resulted in an additional reduction in electrophoretic mobility relative to the N-glycosylated protein species. Thus, these proteins are still membrane associated. These results indicate that, in principle, all hydrophobic domains are able to individually mediate insertion into the membrane and to function as transmembrane domains, although with different efficiencies, where especially domains 6 and 7 were inserted with lower efficiency (Fig. 7C). All together, the results are consistent with our model in which nsp6 has both of its termini exposed on the cytoplasmic face of the membrane while spanning the lipid bilayer six times. Of the seven predicted transmembrane domains, the sixth or seventh is least likely to function as such.
FIG. 7.
Membrane integration of MHV nsp6 hydrophobic domains. (A) Schematic representation of the MHV nsp6 constructs. (B) vTF7-3-infected OST7-1 cells were transfected with the constructs presented in panel A and expressed in the presence (+) or absence (−) of tunicamycin (TM) or brefeldin A (brefA). The cells were labeled with [35S]methionine from 5 to 6 h p.i., lysed, and processed for immunoprecipitation with anti-EGFP antiserum, followed by SDS-PAGE. The positions and masses (in kilodaltons) of the protein size markers are indicated at the left. Only the relevant portion of the gel is shown. The asterisks indicate the position of the N-glycosylated protein species, and the arrowhead indicates that of the O-glycosylated protein species. Below the panels, the observed presence or absence of N- and or O-linked oligosaccharides is indicated. (C) Model of the membrane topologies of the nsp6 hydrophobic domains. The hydrophobic domains are presented as black rectangles, the EGFPglyc tag is presented as a gray rectangle, and the MN tag is presented as a gray circle. Dark gray rectangles or circles represent glycosylated tags, and light gray rectangles or circles represent unglycosylated tags. The dotted line connects the transmembrane domains in accordance with the full-length nsp6 structure, indicating that either the sixth or the seventh hydrophobic domain does not traverse the lipid bilayer.
Membrane integration of nsp4.
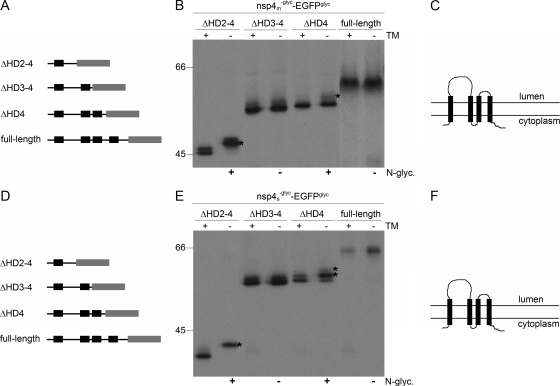
In view of the observations with nsp3 and nsp6, we wanted to complete these evaluations by similarly analyzing the membrane structure of the third viral membrane protein involved in anchoring the replication complex, nsp4. Our earlier study already revealed that both termini of this protein are oriented cytoplasmically and that the protein, both for SARS-CoV and for MHV, is N glycosylated in a region between the first two of its four hydrophobic domains, which led us to propose a tetraspanning structure (37). Yet, we could not exclude the possibility that actually two of the three carboxy-terminal hydrophobic domains are not membrane spanning. Hence, we constructed progressive C-terminal deletion mutant constructs of both MHV and SARS-CoV nsp4 in which the natural glycosylation sites had additionally been removed by mutagenesis and which were again fused to the EGFPglyc tag (Fig. 8A and D). The proteins were expressed in the presence and absence of tunicamycin and analyzed as before.
FIG. 8.
Membrane integration of deletion mutant forms of MHV and SARS-CoV nsp4. (A, D) Schematic representations of the MHV (A) and SARS-CoV (D) nsp4 C-terminal deletion mutant forms, with the hydrophobic domains presented as black rectangles and the EGFPglyc tag shown in gray. (B, E) vTF7-3-infected OST7-1 cells were transfected with the indicated constructs and expressed in the presence (+) or absence (−) of tunicamycin (TM). The cells were labeled with [35S]methionine from 5 to 6 h p.i., lysed, and processed for immunoprecipitation with anti-EGFP antiserum, followed by SDS-PAGE. The positions and masses (in kilodaltons) of the protein size markers are indicated at the left. Only the relevant portions of the gels are shown. The asterisks indicate the position of the glycosylated protein species. Below the panels, the observed presence or absence of N glycosylation is indicated by a plus or a minus sign, respectively (C, F). Models of the membrane structure of nsp4 of MHV (C) and SARS-CoV (F), with the hydrophobic domains presented as black rectangles.
The nsp4 mutant forms of both viruses gave similar results (Fig. 8B and E). Full-length nsp4-EGFPglyc remained unglycosylated, as expected (37). When only the first hydrophobic domain was present (ΔHD2-4), the fusion proteins became efficiently N glycosylated, as shown by the complete shift in electrophoretic mobility due to the presence of tunicamycin. This is again as expected and confirms that the first hydrophobic domain spans the lipid bilayer. When the first two hydrophobic domains were present (ΔHD3-4), the electrophoretic mobility was not influenced by the presence of tunicamycin. Since this fusion protein was not N glycosylated, its C terminus was located on the cytoplasmic side of the membrane, implying that also the second hydrophobic domain spans the lipid bilayer. N glycosylation was again observed, though especially for MHV nsp4 not very efficiently, when only the fourth hydrophobic domain was deleted (ΔHD4), as demonstrated by the higher-molecular-weight band seen in the absence of tunicamycin but not in its presence, indicating that also the third and fourth hydrophobic domains span the membrane. For several nsp4 deletion mutant constructs, two rather than one tunicamycin-resistant protein species could be detected, which is explained by the partial removal of the first transmembrane domain by signal peptidase (37; data not shown). It appeared that signal peptidase cleavage of nsp4 was more efficient for the smaller deletion mutant proteins. N glycosylation of the SARS-CoV nsp4ΔHD4 mutant form combined with signal peptidase cleavage resulted in an electrophoretic mobility similar to that of the unglycosylated and uncleaved protein (hence the two asterisks in Fig. 8E, which indicate the position of the N-glycosylated protein species), similar to what has been observed previously (37). The results show that, in contrast to nsp3 and nsp6, all of the predicted transmembrane domains of nsp4 span the lipid bilayer (Fig. 8C and F).
DISCUSSION
Coronaviruses have exceptionally large RNA genomes and exhibit complex replication and transcription strategies. These processes take place at DMVs located in the perinuclear region of the host cell. Although the membranes of these structures are most likely derived from the ER, the way they are organized and modified to function as the scaffolds of the replication complexes is still largely unknown. Presumably, as has been demonstrated for other RNA viruses, the hydrophobic nsps are key organizers of the observed membrane rearrangements. All known members of the families Coronaviridae and Arteriviridae encode three nsps containing hydrophobic domains. Invariably, hydrophobic domains are present in the two nsps surrounding the 3C-like main proteinase encoded by nsp5 of coronaviruses and by nsp4 of arteriviruses, while additional hydrophobic domains are found in the large nsp immediately upstream of this cluster, which further contains the papain-like proteinase domains (19). For EAV, the coexpression of nsp2 and nsp3, which are the homologues of coronavirus nsp3 and nsp4, is sufficient to induce the formation of DMVs (49). Similar results, however, have so far not been reported for coronaviruses. In order to get more insight into the function of the coronavirus hydrophobic nsps, we studied the topology and membrane integration of these proteins. The results demonstrate that, similar to nsp4 (37), both nsp3 and nsp6 have a Nendo/Cendo topology with an even number of transmembrane domains, although for both proteins an uneven number of transmembrane domains was predicted. The conservation of hydrophobic domains not actually serving as transmembrane domains suggests that these domains may play important roles in the formation, stabilization, or functioning of the replication complexes.
Membrane structures of complex multispanning proteins are notoriously difficult to establish. Ideally, one would like to probe the disposition of particular domains in such proteins by introducing or removing glycosylation sites and by using specific antisera raised to peptides corresponding to these domains. Removal of the glycosylation sites in the large lumenal loops of nsp3 and nsp4 was relatively straightforward. Unfortunately, however, no glycosylation motifs occurred in the other, much smaller loops, and motifs introduced into such loops appeared to be nonfunctional (data not shown), probably because they are not accessible to the enzymes. As domain-specific antibodies are not available—and are probably hard to generate due to the small size of most loops—we decided to use other frequently applied approaches, which involved tagging of the nsps with epitopes or EGFP carrying O or N glycosylation sites in combination with progressive deletions of hydrophobic domains. A potential drawback of these techniques is that the manipulation may affect the conformational behavior of the protein. Thus, deletion mutant proteins often appeared to be only partially glycosylated. While this might be taken as an indication that the protein can adopt more than one membrane topology, we do not consider this interpretation very likely. First of all, a large number of our mutant proteins did not become glycosylated at all and apparently do not adopt a dual topology. Furthermore, glycosylation was invariably observed for those deletion mutant proteins in which the glycosylatable tag replaced a lumenal loop with an authentic glycosylation site. It seems more likely that the partial glycosylation of the proteins is inherent to the type of approach and relates to their very hydrophobic nature and their often drastically modified state, which makes them prone to aggregation when expressed out of their natural context. Furthermore, we confirmed our results by performing the experiments with nsps of both SARS-CoV and MHV, which yielded essentially identical results.
We studied the topology and membrane integration of SARS-CoV and MHV hydrophobic nsps by expressing the proteins independently, i.e., out of their polyprotein context. Similar strategies have been employed before to study the membrane integration of viral nsps derived from large precursor proteins (33, 34, 37). Unlike for nsp4 and nsp6, we expressed only the C-terminal approximately 700 amino acids of nsp3, the part that contains all of the putative transmembrane domains. The full-size, approximately 2,000-amino-acid polypeptide would have been too large to detect the often subtle size differences inherent to our analytic approach. Moreover, it appeared difficult to express complete nsp3, presumably due to the presence of toxic sequences, as have also been reported by others to occur in approximately the same region of other coronavirus genomes (1, 60, 61). The N-terminal region of nsp3 lacking in our expression constructs does not contain any appreciable hydrophobic domains, and prediction programs do not identify any transmembrane domains in this region. Hence, we believe that the models obtained for the partial proteins apply as well to complete MHV and SARS-CoV nsp3.
By using biochemical and immunofluorescence assays, we demonstrated that in both SARS-CoV and MHV, nsp3 has a Nendo/Cendo topology. While this is inconsistent with the predicted number of transmembrane domains (Fig. 1), it is in agreement with a previous study on the topology of MHV nsp3 (28). A Nendo/Cendo topology obviously makes more sense than the predicted Nexo/Cendo orientation with respect to the localization of the PLpro domain and its substrates (Fig. 9). However, whereas previously four transmembrane domains were identified for MHV nsp3 (28), we found evidence for the presence of only two such domains in both of the nsps studied. We could not confirm the presence of additional membrane-spanning domains located immediately up- and downstream of the first transmembrane domain, nor were these additional domains predicted on the basis of the multiple alignment. Our study did confirm, however, the use of the previously identified N-linked glycosylation sites between the first and second transmembrane domains (22, 28). It further demonstrated that in both SARS-CoV and MHV, the second, but not the third predicted transmembrane domain of nsp3 is used. This result is in conflict with findings of Baker and coworkers, who identified the third rather than the second predicted transmembrane domain of MHV nsp3 as spanning the lipid bilayer (28). Their conclusion was mainly based on electrophoretic mobility measurements of hydrophobic nsp3 fragments obtained after proteinase K digestions. Hydrophobic proteins, including coronavirus nsp4 and nsp6, often exhibit anomalous migration in gels, and this might explain the different interpretations. Regardless of the number of transmembrane domains, it is striking that the first transmembrane domain in coronavirus pp1a and pp1ab is found only after more than 2,000 amino acids, while the polyproteins and mature nsp3 lack an identifiable signal sequence at the amino-terminal end. Apparently, the first hydrophobic domain of nsp3 is able to function as an internal signal sequence for membrane insertion, as we confirmed in our study.
FIG. 9.
Model of the membrane disposition of coronavirus pp1a. The black line represents the protein, with the hydrophobic domains depicted as gray rectangles, and shows its localization with respect to the membrane. The proteases, papain-like protease (PLpro) and main protease (Mpro), and their cleavage sites (arrowheads) are indicated, with the PLpro cleavage sites in gray and the Mpro cleavage sites in black. The confirmed N glycosylation sites of the proteins are indicated by asterisks. In this model, the sixth hydrophobic domain of nsp6 does not span the lipid bilayer; however, we cannot exclude the possibility that the seventh rather than the sixth predicted transmembrane domain is not used as such.
Like nsp4 and nsp3, nsp6 of SARS-CoV and MHV appeared to have a Nendo/Cendo membrane topology. This is again inconsistent with the prediction, which identified seven transmembrane domains, compatible with a Nendo/Cexo topology. The experimentally established topology obviously makes more sense, as it positions all of the main proteinase cleavage sites on the same side of the membrane as the proteinase itself (Fig. 9). Demonstrating which of the hydrophobic domains in nsp6 actually served as transmembrane domains was not as straightforward as it was for nsp3. It appeared that, in principle, all seven domains have the capacity to function as signal sequences and membrane anchors, irrespective of their fate in the full-length protein. The combined observations appear to fit best a model in which nsp6 contains six transmembrane domains, with the sixth or seventh hydrophobic domain not spanning the lipid bilayer. The transmembrane prediction program SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/sosuimenu0.html) also predicts seven transmembrane-spanning helices but additionally calculates that the hydrophobicity of the sixth domain, but not that of the others, is similar to the hydrophobicity of helices found in soluble proteins (23). This prediction hence supports the model in which the sixth putative transmembrane domain does not span the lipid bilayer.
Our experience with nsp3 and nsp6 prompted us to revisit also the membrane structure of nsp4. With four hydrophobic domains, the known Nendo/Cendo topology of this protein could, in principle, be achieved with either two or all of those as transmembrane domains. Our deletion assay led us to conclude that all four domains are membrane spanning. Remarkably, in MHV, the removal of the fourth domain was found by Sparks and coworkers (50) to yield a virus that grew in cultured cells with kinetics and to titers similar to those of the wild-type virus, although the kinetics of its RNA replication were significantly delayed. Given the topological consequences of this deletion, one has to assume that in this mutant virus either the third hydrophobic membrane is not functioning as a transmembrane domain or, as the authors suggest, the effect of the deletion is compensated by the region between the third and fourth transmembrane domains, which is also somewhat hydrophobic. Mutant viruses lacking any of the other three hydrophobic domains of nsp4 could not be recovered.
Combining all of the information obtained for nsp3, -4, and -6 into one model, we arrive at the membrane structure of the pp1a precursor that is depicted in Fig. 9. As a result of the Nendo/Cendo topology of all three membrane-anchored nsps, the major part of the polyproteins, including all known functional domains, is located on the cytoplasmic side of the membrane. Of the entire approximately 4,400-amino-acid pp1a precursor, only six relatively short stretches are exposed lumenally, exceptions being the glycosylated polypeptides occurring between the first and second transmembrane domains of nsp3 and nsp4, which comprise some 80 and 250 residues, respectively. Interestingly, both of these polypeptide loops contain several highly conserved cysteine residues. Mutations in the lumenally exposed part of MHV nsp4 (and its EAV counterpart) were shown to affect DMV formation (8, 39).
Also, the pp1b precursor, which lacks membrane-spanning domains but carries cleavage sites processed by the nsp5-encoded main protease, and its cleavage products nsp12 to -16 end up cytoplasmically. This topological arrangement fits into a picture of the biogenesis of the replication complexes in which membrane proteins nsp3, -4 and -6, while still contained in the precursor, direct the formation of the membranous structures on which RNA replication is to take place. These proteins probably shape the platform onto which the subsequently appearing nonanchored cleavage products derived from pp1a and pp1b remain associated and to which cellular, as well as viral (e.g., N protein), components may additionally attach to generate the functional replication complex.
What role(s) the conserved non-membrane-spanning hydrophobic domains in nsp3 and -6 play in the formation and functioning of the complex remains to be established. A cytoplasmically exposed hydrophobic domain might serve as an interaction partner for other components of the complex, while the hydrophobic non-membrane-spanning domains might also contribute to the double-membrane configuration, for instance, by dipping into the hydrophobic phase of the membrane. The latter has been observed earlier for proteins containing amphipathic helices, showing that these domains can act as a wedge in the membrane and thereby induce curvature (32, 65).
The induction of membrane rearrangements to create sites for RNA replication and transcription in infected cells is not unique to coronaviruses but is common among plus strand RNA viruses. These viruses generally encode nsps that induce membrane rearrangements. These nsps can be integral membrane proteins, but this is not always the case. The occurrence of hydrophobic/amphipathic regions that do not span the bilayer but are peripherally associated with membranes is not an exception; it rather is a common theme. Interestingly, dengue virus, which also replicates on DMVs (56), encodes two nonstructural membrane proteins that, in addition to transmembrane domains, also contain hydrophobic domains that do not span the lipid bilayer (33, 34). The NS4A and NS4B proteins of this virus contain one and two hydrophobic domains, respectively, that do not function as transmembrane domains in the full-length protein. Yet, the non-membrane-spanning hydrophobic domain of NS4A is integrated into the membrane when expressed individually (33, 34), comparable to our observations with the hydrophobic domains of MHV nsp6. In the case of hepatitis C virus, which, like dengue virus, also belongs to the flavivirus family, the amphipathic helix contained in NS4B was shown to be required for the formation of replication sites (15). Likewise, the poliovirus 2C protein, which is able to induce the membrane rearrangements associated with RNA replication, also contains an essential amphipathic domain (14, 38, 54). In view of these observations, it is tempting to speculate that these similarities among the viral nonstructural membrane-associated proteins relate to their common ability to induce membrane rearrangements.
Supplementary Material
Acknowledgments
We thank Matthijs Raaben and Mijke Vogels for stimulating discussions.
This work was supported by grants from The Netherlands Organization for Scientific Research (NWO-VIDI-700.54.421) and Utrecht University (High Potential) to C. A. M. de Haan and from the European Community (Frame VI, DISSECT PROJECT, SP22-CT-2004-511060) to P. J. M. Rottier.
Footnotes
Published ahead of print on 8 October 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Almazán, F., J. M. Gonzalez, Z. Penzes, A. Izeta, E. Calvo, J. Plana-Duran, and L. Enjuanes. 2000. Engineering the largest RNA virus genome as an infectious bacterial artificial chromosome. Proc. Natl. Acad. Sci. USA 975516-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker, S. C., K. Yokomori, S. Dong, R. Carlisle, A. E. Gorbalenya, E. V. Koonin, and M. M. Lai. 1993. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J. Virol. 676056-6063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhardwaj, K., L. Guarino, and C. C. Kao. 2004. The severe acute respiratory syndrome coronavirus Nsp15 protein is an endoribonuclease that prefers manganese as a cofactor. J. Virol. 7812218-12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bost, A. G., R. H. Carnahan, X. T. Lu, and M. R. Denison. 2000. Four proteins processed from the replicase gene polyprotein of mouse hepatitis virus colocalize in the cell periphery and adjacent to sites of virion assembly. J. Virol. 743379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bredenbeek, P. J., C. J. Pachuk, A. F. Noten, J. Charite, W. Luytjes, S. R. Weiss, and W. J. Spaan. 1990. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 181825-1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brockway, S. M., C. T. Clay, X. T. Lu, and M. R. Denison. 2003. Characterization of the expression, intracellular localization, and replication complex association of the putative mouse hepatitis virus RNA-dependent RNA polymerase. J. Virol. 7710515-10527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng, A., W. Zhang, Y. Xie, W. Jiang, E. Arnold, S. G. Sarafianos, and J. Ding. 2005. Expression, purification, and characterization of SARS coronavirus RNA polymerase. Virology 335165-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clementz, M. A., A. Kanjanahaluethai, T. E. O'Brien, and S. C. Baker. 2008. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology 375118-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decroly, E., I. Imbert, B. Coutard, M. Bouvet, B. Selisko, K. Alvarez, A. E. Gorbalenya, E. J. Snijder, and B. Canard. 2008. Coronavirus nonstructural protein 16 is a Cap-0 binding enzyme possessing (nucleoside-2′O)-methyltransferase activity. J. Virol. 828071-8084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Haan, C. A., P. Roestenberg, M. de Wit, A. A. de Vries, T. Nilsson, H. Vennema, and P. J. Rottier. 1998. Structural requirements for O glycosylation of the mouse hepatitis virus membrane protein. J. Biol. Chem. 27329905-29914. [DOI] [PubMed] [Google Scholar]
- 11.Denison, M. R., A. C. Sims, C. A. Gibson, and X. T. Lu. 1998. Processing of the MHV-A59 gene 1 polyprotein by the 3C-like proteinase. Adv. Exp. Med. Biol. 440121-127. [DOI] [PubMed] [Google Scholar]
- 12.Donaldson, E. F., R. L. Graham, A. C. Sims, M. R. Denison, and R. S. Baric. 2007. Analysis of murine hepatitis virus strain A59 temperature-sensitive mutant TS-LA6 suggests that nsp10 plays a critical role in polyprotein processing. J. Virol. 817086-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dos Ramos, F., M. Carrasco, T. Doyle, and I. Brierley. 2004. Programmed −1 ribosomal frameshifting in the SARS coronavirus. Biochem. Soc. Trans. 321081-1083. [DOI] [PubMed] [Google Scholar]
- 14.Echeverri, A. C., and A. Dasgupta. 1995. Amino terminal regions of poliovirus 2C protein mediate membrane binding. Virology 208540-553. [DOI] [PubMed] [Google Scholar]
- 15.Elazar, M., P. Liu, C. M. Rice, and J. S. Glenn. 2004. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J. Virol. 7811393-11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elroy-Stein, O., and B. Moss. 1990. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc. Natl. Acad. Sci. USA 876743-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan, K., L. Ma, X. Han, H. Liang, P. Wei, Y. Liu, and L. Lai. 2005. The substrate specificity of SARS coronavirus 3C-like proteinase. Biochem. Biophys. Res. Commun. 329934-940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 838122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorbalenya, A. E., L. Enjuanes, J. Ziebuhr, and E. J. Snijder. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res. 11717-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 763697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graham, R. L., A. C. Sims, S. M. Brockway, R. S. Baric, and M. R. Denison. 2005. The nsp2 replicase proteins of murine hepatitis virus and severe acute respiratory syndrome coronavirus are dispensable for viral replication. J. Virol. 7913399-13411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harcourt, B. H., D. Jukneliene, A. Kanjanahaluethai, J. Bechill, K. M. Severson, C. M. Smith, P. A. Rota, and S. C. Baker. 2004. Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity. J. Virol. 7813600-13612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14378-379. [DOI] [PubMed] [Google Scholar]
- 24.Imbert, I., J. C. Guillemot, J. M. Bourhis, C. Bussetta, B. Coutard, M. P. Egloff, F. Ferron, A. E. Gorbalenya, and B. Canard. 2006. A second, non-canonical RNA-dependent RNA polymerase in SARS coronavirus. EMBO J. 254933-4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivanov, K. A., T. Hertzig, M. Rozanov, S. Bayer, V. Thiel, A. E. Gorbalenya, and J. Ziebuhr. 2004. Major genetic marker of nidoviruses encodes a replicative endoribonuclease. Proc. Natl. Acad. Sci. USA 10112694-12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ivanov, K. A., V. Thiel, J. C. Dobbe, Y. van der Meer, E. J. Snijder, and J. Ziebuhr. 2004. Multiple enzymatic activities associated with severe acute respiratory syndrome coronavirus helicase. J. Virol. 785619-5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivanov, K. A., and J. Ziebuhr. 2004. Human coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 787833-7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanjanahaluethai, A., Z. Chen, D. Jukneliene, and S. C. Baker. 2007. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology 361391-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuiken, T., R. A. Fouchier, M. Schutten, G. F. Rimmelzwaan, G. van Amerongen, D. van Riel, J. D. Laman, T. de Jong, G. van Doornum, W. Lim, A. E. Ling, P. K. Chan, J. S. Tam, M. C. Zambon, R. Gopal, C. Drosten, S. van der Werf, N. Escriou, J. C. Manuguerra, K. Stohr, J. S. Peiris, and A. D. Osterhaus. 2003. Newly discovered coronavirus as the primary cause of severe acute respiratory syndrome. Lancet 362263-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Locker, J. K., J. K. Rose, M. C. Horzinek, and P. J. Rottier. 1992. Membrane assembly of the triple-spanning coronavirus M protein. Individual transmembrane domains show preferred orientation. J. Biol. Chem. 26721911-21918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthes, N., J. R. Mesters, B. Coutard, B. Canard, E. J. Snijder, R. Moll, and R. Hilgenfeld. 2006. The non-structural protein Nsp10 of mouse hepatitis virus binds zinc ions and nucleic acids. FEBS Lett. 5804143-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMahon, H. T., and J. L. Gallop. 2005. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438590-596. [DOI] [PubMed] [Google Scholar]
- 33.Miller, S., S. Kastner, J. Krijnse-Locker, S. Buhler, and R. Bartenschlager. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 2828873-8882. [DOI] [PubMed] [Google Scholar]
- 34.Miller, S., S. Sparacio, and R. Bartenschlager. 2006. Subcellular localization and membrane topology of the Dengue virus type 2 non-structural protein 4B. J. Biol. Chem. 2818854-8863. [DOI] [PubMed] [Google Scholar]
- 35.Oostra, M., C. A. de Haan, R. J. de Groot, and P. J. Rottier. 2006. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. J. Virol. 802326-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oostra, M., C. A. de Haan, and P. J. Rottier. 2007. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J. Virol. 8113876-13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oostra, M., E. G. te Lintelo, M. Deijs, M. H. Verheije, P. J. Rottier, and C. A. de Haan. 2007. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 8112323-12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paul, A. V., A. Molla, and E. Wimmer. 1994. Studies of a putative amphipathic helix in the N-terminus of poliovirus protein 2C. Virology 199188-199. [DOI] [PubMed] [Google Scholar]
- 39.Posthuma, C. C., K. W. Pedersen, Z. Lu, R. G. Joosten, N. Roos, J. C. Zevenhoven-Dobbe, and E. J. Snijder. 2008. Formation of the arterivirus replication/transcription complex: a key role for nonstructural protein 3 in the remodeling of intracellular membranes. J. Virol. 824480-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prentice, E., W. G. Jerome, T. Yoshimori, N. Mizushima, and M. R. Denison. 2004. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 27910136-10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Prentice, E., J. McAuliffe, X. Lu, K. Subbarao, and M. R. Denison. 2004. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 789977-9986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Putics, A., W. Filipowicz, J. Hall, A. E. Gorbalenya, and J. Ziebuhr. 2005. ADP-ribose-1″-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 7912721-12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ricagno, S., M. P. Egloff, R. Ulferts, B. Coutard, D. Nurizzo, V. Campanacci, C. Cambillau, J. Ziebuhr, and B. Canard. 2006. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc. Natl. Acad. Sci. USA 10311892-11897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salonen, A., T. Ahola, and L. Kaariainen. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285139-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sawicki, S. G., D. L. Sawicki, and S. G. Siddell. 2007. A contemporary view of coronavirus transcription. J. Virol. 8120-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi, S. T., and M. M. Lai. 2005. Viral and cellular proteins involved in coronavirus replication. Curr. Top. Microbiol. Immunol. 28795-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi, S. T., J. J. Schiller, A. Kanjanahaluethai, S. C. Baker, J. W. Oh, and M. M. Lai. 1999. Colocalization and membrane association of murine hepatitis virus gene 1 products and de novo-synthesized viral RNA in infected cells. J. Virol. 735957-5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snijder, E. J., Y. van der Meer, J. Zevenhoven-Dobbe, J. J. Onderwater, J. van der Meulen, H. K. Koerten, and A. M. Mommaas. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 805927-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Snijder, E. J., H. van Tol, N. Roos, and K. W. Pedersen. 2001. Non-structural proteins 2 and 3 interact to modify host cell membranes during the formation of the arterivirus replication complex. J. Gen. Virol. 82985-994. [DOI] [PubMed] [Google Scholar]
- 50.Sparks, J. S., X. Lu, and M. R. Denison. 2007. Genetic analysis of Murine hepatitis virus nsp4 in virus replication. J. Virol. 8112554-12563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sutormin, R. A., and A. A. Mironov. 2006. Membrane probability profile construction based on amino acids sequences multiple alignment. Mol. Biol. [Engl. Transl. Mol. Biol. (Moscow)] 40541-545. [PubMed] [Google Scholar]
- 52.Sutton, G., E. Fry, L. Carter, S. Sainsbury, T. Walter, J. Nettleship, N. Berrow, R. Owens, R. Gilbert, A. Davidson, S. Siddell, L. L. Poon, J. Diprose, D. Alderton, M. Walsh, J. M. Grimes, and D. I. Stuart. 2004. The nsp9 replicase protein of SARS-coronavirus, structure and functional insights. Structure 12341-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Taguchi, F., and J. O. Fleming. 1989. Comparison of six different murine coronavirus JHM variants by monoclonal antibodies against the E2 glycoprotein. Virology 169233-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Teterina, N. L., A. E. Gorbalenya, D. Egger, K. Bienz, and E. Ehrenfeld. 1997. Poliovirus 2C protein determinants of membrane binding and rearrangements in mammalian cells. J. Virol. 718962-8972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thiel, V., K. A. Ivanov, A. Putics, T. Hertzig, B. Schelle, S. Bayer, B. Weissbrich, E. J. Snijder, H. Rabenau, H. W. Doerr, A. E. Gorbalenya, and J. Ziebuhr. 2003. Mechanisms and enzymes involved in SARS coronavirus genome expression. J. Gen. Virol. 842305-2315. [DOI] [PubMed] [Google Scholar]
- 56.Uchil, P. D., and V. Satchidanandam. 2003. Architecture of the flaviviral replication complex. Protease, nuclease, and detergents reveal encasement within double-layered membrane compartments. J. Biol. Chem. 27824388-24398. [DOI] [PubMed] [Google Scholar]
- 57.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 737641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Meer, Y., H. van Tol, J. K. Locker, and E. J. Snijder. 1998. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J. Virol. 726689-6698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vennema, H., R. Rijnbrand, L. Heijnen, M. C. Horzinek, and W. J. Spaan. 1991. Enhancement of the vaccinia virus/phage T7 RNA polymerase expression system using encephalomyocarditis virus 5′-untranslated region sequences. Gene 108201-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yount, B., K. M. Curtis, and R. S. Baric. 2000. Strategy for systematic assembly of large RNA and DNA genomes: transmissible gastroenteritis virus model. J. Virol. 7410600-10611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yount, B., M. R. Denison, S. R. Weiss, and R. S. Baric. 2002. Systematic assembly of a full-length infectious cDNA of mouse hepatitis virus strain A59. J. Virol. 7611065-11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhai, Y., F. Sun, X. Li, H. Pang, X. Xu, M. Bartlam, and Z. Rao. 2005. Insights into SARS-CoV transcription and replication from the structure of the nsp7-nsp8 hexadecamer. Nat. Struct. Mol. Biol. 12980-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ziebuhr, J. 2005. The coronavirus replicase. Curr. Top. Microbiol. Immunol. 28757-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ziebuhr, J., E. J. Snijder, and A. E. Gorbalenya. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J. Gen. Virol. 81853-879. [DOI] [PubMed] [Google Scholar]
- 65.Zimmerberg, J., and M. M. Kozlov. 2006. How proteins produce cellular membrane curvature. Nat. Rev. Mol. Cell Biol. 79-19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.