Abstract
Single-stranded positive-sense RNA viruses induce the biogenesis of cytoplasmic membranous vesicles, where viral replication takes place. However, the mechanism underlying this characteristic vesicular proliferation remains poorly understood. Previously, a 6-kDa potyvirus membrane protein (6K) was shown to interact with the endoplasmic reticulum (ER) and to induce the formation of the membranous vesicles. In this study, the involvement of the early secretory pathway in the formation of the 6K-induced vesicles was investigated in planta. By means of live-cell imaging, it was found that the 6K protein was predominantly colocalized with Sar1, Sec23, and Sec24, which are known markers of ER exit sites (ERES). The localization of 6K at ERES was prevented by the coexpression of a dominant-negative mutant of Sar1 that disables the COPII activity or by the coexpression of a mutant of Arf1 that disrupts the COPI complex. The secretion of a soluble secretory marker targeting the apoplast was arrested at the level of the ER in cells overexpressing 6K or infected by a potyvirus. This blockage of protein trafficking out of the ER by 6K and the distribution of 6K toward the ERES may account for the aggregation of the 6K-bound vesicles. Finally, virus infection was reduced when the accumulation of 6K at ERES was inhibited by impairing either the COPI or COPII complex. Taken together, these results imply that the cellular COPI and COPII coating machineries are involved in the biogenesis of the potyvirus 6K vesicles at the ERES for viral-genome replication.
Upon entry into host cells, single-stranded positive-sense RNA viruses outcompete cellular mRNAs for translation (11, 42, 70). Subsequently, by an as yet poorly understood mechanism, the biogenesis of unique intracellular membranous structures that are committed to housing the viral replication complex for viral-genome replication occurs (1, 2, 39, 43, 44, 58, 59). The membranous origins and natures of these cytoplasmic vesicles are diverse and often depend on the type of invading virus (36, 61). For instance, endosomes/lysosomes, chloroplasts, and peroxisomes/mitochondria have been shown to be the replication sites for togaviruses, tymoviruses, and tombusviruses, respectively (22, 23, 55). Remarkably, many plant and animal viruses, e.g., potyviruses, comoviruses, nepoviruses, picornaviruses, flaviviruses, arteriviruses, bromoviruses, and tobamoviruses, assemble their viral replication complexes in association with the endoplasmic reticulum (ER) (39, 58, 59, 65, 81).
The family Potyviridae, which includes Tobacco etch virus (TEV), Potato virus Y, Plum pox virus, Soybean mosaic virus, and Turnip mosaic virus, is the largest and most agriculturally important virus family (10, 50, 72, 73). Based on the similarity of the replicases, genomic organizations, and gene expression strategies, potyviruses are classified into the picornavirus supergroup of positive-strand RNA viruses (35). The genomic RNA of members of the genus Poytvirus is about 10 kb in length and has a viral protein covalently linked to its 5′ terminus and a poly(A) tail at its 3′ end (50, 72). It contains a single long open reading frame encoding a large polyprotein of about 360 kDa that is ultimately cleaved into 10 mature proteins (72). Recently, an additional 25-kDa protein, termed P3-PIPO, has been discovered (17). This protein is derived from a frameshift on the P3 cistron. Of these 11 mature viral proteins, the 6-kDa protein (6K or 6K2) contains a central hydrophobic domain and is an integral membrane protein (49, 60). TEV is one of the best-characterized potyviruses. In seminal work, Schaad and colleagues determined that the TEV 6K protein induces the formation of the ER-derived vesicles and that virus replication occurs in association with these vesicles (60). Since then, many viral membrane proteins from different viruses have been shown to induce the production of ER-derived vesicles for virus replication (39, 58, 59). The mechanism by which the vesicular structures proliferate and develop from the ER remains to be characterized.
The ER is the cytoplasmic endomembrane network where cellular proteins are produced. The ER, the Golgi apparatus, endosomes, prevacuolar compartments, and lytic compartments constitute the endomembrane system of plant cells (16, 53). Membrane trafficking between endomembrane compartments is essential for transport of proteins to various destinations (16, 53). The early secretory pathway originates from the ER. The export of protein from the ER to the Golgi apparatus occurs at specialized subdomains of the ER called ER exit sites (ERES) (68). The coat protein complex II (COPII) vesicles bud from the plant ERES (53, 63). COPII, generally thought to mediate anterograde traffic out of the ER, is composed of three cytosolic components, the small GTPase Sar1 and two heterodimeric complexes, Sec23/24 and Sec13/31, which are recruited sequentially to the ER membrane at ERES (29, 46, 53). The plant Golgi apparatus receives exported proteins from the ER via ERES. ERES and Golgi bodies seem to have a continuous association in tobacco leaf epidermal cells (18, 28, 29, 64) or, more likely, a transient (stop-and-go) association in Arabidopsis root cells or cultured BY-2 cells (63, 80). The association of ERES and Golgi bodies is assumed to be mediated by the scaffold layer of the COPII vesicles, which captures and pulls passing Golgi stacks off the actin track (63). The retrograde traffic out of the Golgi apparatus back to the ER is modulated by COPI vesicles, which consist of the small GTPase Arf1 and a heptameric complex of structural coat components (29, 53). Inhibition of COPI function by the dominant-negative mutant of Arf1 results in impaired ER export and disruption of the ERES, indicating a possible role for Arf1 in anterograde transport, in addition to its role in retrograde transport (64).
Accumulated evidence suggests that early secretory pathways, i.e., COPI and COPII, are involved in the infection and replication of animal picornaviruses (6, 7, 8, 9, 24, 76, 77, 78). However, very little is known about plant viruses. As mentioned above, plant potyviruses, nepoviruses, and comoviruses belong to a picornavirus-like superfamily and bear strong resemblance to animal picornaviruses in the amino acid sequence of replication proteins, as well as the arrangement of genes encoding these proteins. As reviewed previously by Koonin and Dolja (35), the core replication proteins, including, in sequential order in the genome, helicase (2C), small membrane protein (3A), viral genome-linked protein (VPg or 3B), protease (3C), and RNA-dependent RNA polymerase (RdRp or 3D), of enteroviruses, rhinoviruses, and cardioviruses are very similar to CI, 6K2, NIa-VPg, NIa-Pro, and NIb of potyviruses and to NTB (containing helicase and membrane domains), VPg, protease, and RdRp of nepoviruses and comoviruses, respectively. Thus, it is possible that cellular early secretory pathways may also play a role in the replication of plant picorna-like viruses (12, 52).
In this study, we investigated the membrane dynamics of the early secretory pathway in the process of the formation of potyvirus 6K vesicles using live-cell imaging. Our results suggest that the 6K-induced ER proliferations take place at the ERES in Nicotiana benthamiana leaves. Moreover, we demonstrate that both COPI and COPII machineries are involved in the biogenesis of the 6K-containing membranous vesicles.
MATERIALS AND METHODS
Gene cloning and plasmid construction.
Gateway technology (Invitrogen, Burlington, Ontario, Canada) was used to generate all the clones reported in this study. RNA extraction from Arabidopsis thaliana leaves was performed using an RNeasy Plant Mini Kit (Qiagen, Mississauga, Ontario, Canada). Reverse transcription was catalyzed by Superscript III reverse transcriptase (Invitrogen) with oligonucleotide dT20. The primer sequences used for cloning and mutagenesis in this study are listed in Table S1 in the supplemental material. Gene sequences were amplified by PCR using Phusion DNA polymerase (NEB, Pickering, Ontario, Canada). The resulting DNA fragments were purified and transferred by recombination into the entry vector pDONR201 (Invitrogen) using BP clonase II (Invitrogen) following the manufacturer's protocol. The insert of the resulting pDONR clone was verified by sequencing. The insert was then transferred by recombination to the indicated binary destination vector using LR clonase II (Invitrogen) following the standard conditions and procedure recommended by the supplier. ERD2 (AT1G29330), Sar1 (AT1G56330), Sec23 (AT3G23660), and Sec24 (AT3G07100) coding sequences were amplified from cDNA derived from Arabidopsis RNA and recombined into pDONR201. The inserts of the resulting intermediate clones containing ERD2 and Sar1 were further transferred by recombination into the binary destination vectors pEarleygate101 and pEarleygate102 (20) to generate plasmids ERD2-yellow fluorescent protein (YFP), ERD2-cyan fluorescent protein (CFP), Sar1-YFP, and Sar1-CFP, respectively. The entry vector pDONR201 containing Sec23 or Sec24 was transferred by recombination into the binary destination vectors pEarleygate104 (20) and pH7WGC2 (34) to generate plasmids YFP-Sec23, CFP-Sec23, YFP-Sec24, and CFP-Sec24. The plasmid pMT121-AtSAR1 H74L containing the Sar1 mutant [Sar1(H74L)] was a kind gift from Akihiko Nakano (Riken, Japan). The DNA fragment for Sar1(H74L) was amplified and recombined into pDONR201 and then into the binary destination vector pEarleygate101 to give plasmid Sar1(H74L)-YFP. The plasmid pYSC14 containing the mouse talin (mTalin) DNA sequence was a generous gift from Nam-Hai Chua (Rockefeller University, New York, NY) (37, 48). A similar strategy was used to generate plasmid mTalin-YFP with plasmids pDONR201 and pEarleygate101. The pTEV7DA-GFP infectious clone was kindly provided by James C. Carrington (Oregon State University, Corvallis). The 6K and VPg cistrons of TEV were amplified by PCR. Gateway technology with the entry vector pDONR201 and destination vectors pEarleygate101 and pEarleygate102 was again used to produce plasmids 6K-YFP, 6K-CFP, VPg-YFP, and VPg-CFP. The constructs for expressing Arf1-YFP, Arf1-CFP, and Arf1(T31N)-YFP fusion proteins were kindly provided by Ben Scheres (Utrecht University, The Netherlands) (79). A similar strategy was used to generate expression vectors containing Arf1 and Arf1(T31N). Constructs for expressing YFP-HDEL and SecYFP fusion proteins were kindly provided by Ian Moore (Oxford Brookes University, United Kingdom) (3, 69). The entry vector pDONR201 containing Sar1, Sar1(H74L), Arf1, and Arf1(T31N) was transferred by recombination into the binary destination vector pEarleygate100 to generate plasmids that code for untagged proteins.
Transient expression in N. benthamiana.
Four-week-old N. benthamiana plants grown in a greenhouse at 22°C to 24°C were used for Agrobacterium tumefaciens (strain GV3101)-mediated transient expression as described previously (62). The relevant binary vectors were transformed into GV3101 using electroporation and infiltrated into leaf tissues using a 1-ml syringe without a needle by gentle pressure through the stomata on the lower epidermal surface. For leaf infiltration, agrobacteria harboring the relevant plasmids were grown overnight in LB plus the appropriate antibiotics, collected by centrifugation, and then resuspended in 1 ml of 10 mM MgCl2 containing 100 μM acetosyringone. The bacterial optical density at 600 nm (OD600) used for plant transformation was 0.02 to 0.04 for SecYFP and HDEL-YFP and 0.1 to 0.3 for other constructs. Agroinfiltrated plants were incubated under normal growth conditions for 2 to 4 days at 22°C to 24°C.
Drug treatments.
Chemical treatment was carried out following transient coexpression of 6K-CFP and mTalin-YFP and in 4-week-old N. benthamiana plants. For the disruption of actin, leaf tissues were treated with 25 μM latrunculin B (Lat B) (Sigma, Toronto, Canada) for 2 h.
Virus infection.
The pTEV7DA-green fluorescent protein (GFP) infectious clone was propagated in Escherichia coli HB101 (Invitrogen), and DNA was purified using a Qiagen maxikit. Approximately 20 μg of plasmids was linearized by BglII (Invitrogen). After purification, transcription of 5′-capped RNAs was done using an SP6 mMessage mMachine kit (Ambion Inc., Austin, TX) following instructions provided by the manufacturer. The transcripts were precipitated with 1.5 volumes of diethyl pyrocarbonate (DEPC)-treated ultrapure water and 1.5 volumes of 7.5 M LiCl-50 mM EDTA for at least 30 min at −20°C, collected by centrifugation at 16,000 × g for 15 min at 4°C, washed with 70% ethanol, and suspended in DEPC-treated ultrapure water. The purified transcripts were mixed with a 1:10 volume of inoculation buffer (100 mg/ml Carborundum, 0.5 M K2HPO4). Four-week-old N. benthamiana plants were inoculated by abrasion with 4 μl of inoculum applied to the third true leaf. The plants were maintained in a greenhouse at 24°C with 16 h of light. Symptoms appeared 4 to 5 days postinfection in all the inoculated plants. The presence of TEV was monitored by an enzyme-linked immunosorbent assay (ELISA) with TEV-specific ELISA reagents (Agdia, Indiana). ELISA was performed according to the standard protocol provided by the supplier.
Confocal microscopy.
Plant tissues were imaged at room temperature using a Leica TCS SP2 inverted confocal microscope with a 63× oil immersion objective. For confocal microscopy, a UV laser and a krypton/argon laser were used to examine fluorescence. GFP was excited at 488 nm, and the emitted light was captured at 497 to 510 nm; CFP was excited at 405 nm, and the emitted light was captured at 440 to 470 nm; YFP was excited at 514 nm, and the emitted light was captured at 525 to 650 nm. Images were captured digitally and handled using the Leica LCS software. Time-lapse scanning was performed with the Leica TCS SP2 imaging system software. Postacquisition image processing was done with Adobe Photoshop 5.0 software.
The fluorescence intensities of SecYFP in control, 6K-expressing, or TEV-infected leaves were quantified as described previously (3, 82).
RESULTS
The TEV 6K fusion protein localizes at the ERES and at additional ringlike structures on the ER membrane in N. benthamiana leaves.
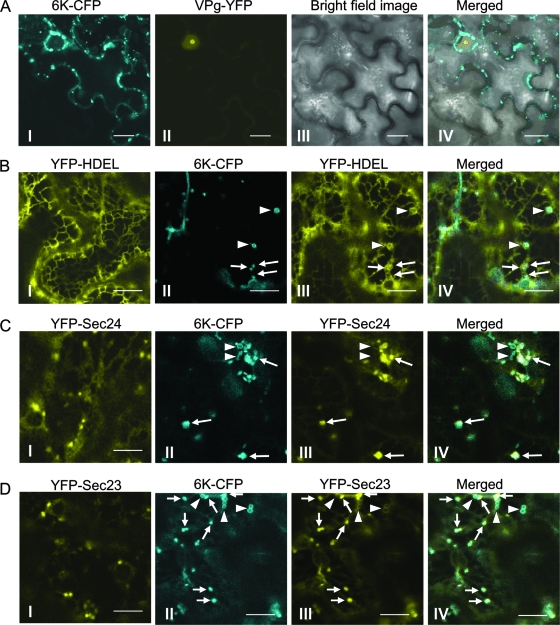
Previously, it was shown that the TEV 6K protein associates with ER membranes as an integral protein and induces the formation of the ER-derived vesicles for virus replication (49, 60). To further investigate the subcellular distribution of the 6K-induced vesicles in planta, transient expression vectors encoding a TEV 6K-CFP fusion and a TEV VPg-YFP fusion were constructed. Transient expression of these chimeric genes was achieved through agroinfiltration. The expression of 6K-CFP in N. benthamiana led to the appearance of a population of vesicular structures with a size range of 0.5 to 4 μm in the cytoplasm, in comparison with cells expressing VPg-YFP, which localized at the nucleus (Fig. 1A), as reported previously (5, 14). Many of the 6K-CFP vesicles appeared to move along actin microfilaments (see below). To visualize the ER network, a soluble ER marker, e.g., the YFP fused with the HDEL motif (YFP-HDEL), was constructed. In the epidermal cells expressing only YFP-HDEL, fluorescence signals were associated with a characteristic reticular network, which is typical of the cortical ER (Fig. 1B). Several small cortical 6K-CFP aggregates showing ringlike structures formed at the vortexes of ER networks (Fig. 1B), in agreement with the previous finding that the TEV 6K protein induces vesiculation on the ER membrane and is associated with these ER-derived vesicles (60).
FIG. 1.
6K is localized at the ERES and at additional ringlike structures on the ER membrane in N. benthamiana leaves. (A) Confocal images of cells expressing 6K-CFP (I) and VPg-YFP (II). Panels III and IV show a bright-field image and merged images, respectively. (B) Confocal images of cells expressing YFP-HDEL alone (I) or in combination with 6K-CFP (II to IV). The arrows indicate colocalization of punctate structures of 6K with cortical aggregates of the ER membrane. The arrowheads indicate colocalization of ringlike structures of 6K with cortical aggregates of the ER membrane. (C) Confocal images of cells expressing YFP-Sec24 alone (I) or in combination with 6K-CFP (II to IV). The arrows indicate colocalization of punctate structures of 6K-CFP with YFP-Sec24-labeled ERES. The arrowheads indicate that the ringlike structures did not colocalize with the ERES labeled by YFP-Sec24. (D) Confocal images of cells expressing YFP-Sec23 alone (I) or in combination with 6K-CFP (II to IV). The arrows indicate colocalization of punctate structures of 6K-CFP with YFP-Sec23-labeled ERES. The arrowheads indicate the ringlike structure that did not colocalize with the ERES staining by YFP-Sec23. Bars, 9 μm.
Recent studies have shown that the plant ER contains functionally distinct subdomains, e.g., ERES, where cargo molecules are packed into transport carriers for delivery (68). Thus, it is possible that the potyvirus 6K-induced vesicles associate with ERES. To examine this possibility, an ERES marker, Sec24, was fused with YFP and cloned into a transient expression vector to generate the YFP-Sec24 expression vector. Sec24 is an essential coat component of the COPII complex that is responsible for anterograde traffic out of the ER and that localizes to the ERES (28, 29, 64). The YFP-Sec24 expression vector was either agroinfiltrated alone or coinfiltrated with the vector containing 6K-CFP into N. benthamiana epidermal cells. When expressed alone, the YFP-Sec24 fusion protein strongly labeled punctate structures and, to a much lesser extent, the cytosol (Fig. 1C). It has been shown that the punctate structures correspond to the ERES (28, 64). Furthermore, most of the 6K-induced punctate structures (vesicles) coincided with YFP-Sec24 fluorescence at ERES, but the ringlike 6K structures did not localize to the ERES (Fig. 1C). To further verify the colocalization of the 6K protein and the ERES, another ERES-labeling protein, Sec23, was cloned and fused with YFP to generate a YFP-Sec23 expression vector for transient expression and localization analyses. Sec23 is also an essential coatomer of the COPII complex (64). When expressed alone or coexpressed with 6K-CFP, the distribution pattern of YFP-Sec23 was evident and was very similar to that of YFP-Sec24 (Fig. 1C and D). Taken together, these results suggest that 6K-induced vesicles are predominantly located at ER export domains, which are labeled by the ERES markers.
The TEV 6K protein-induced vesicles coalign with and traffic along microfilaments.
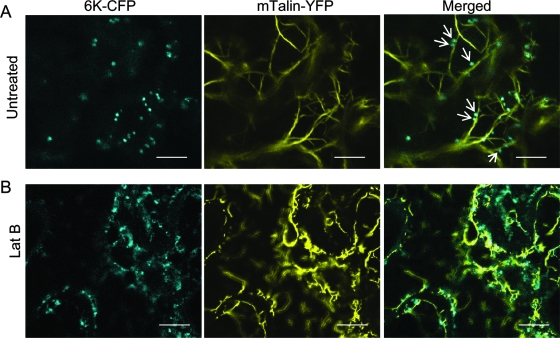
As mentioned above, a characteristic feature of the expression of the 6K-CFP fusion in N. benthamiana leaves was the appearance of many punctate vesicles that appeared to move along actin microfilaments. To confirm this, the association of the 6K vesicles with microfilaments was analyzed by dual-labeling experiments. The actin binding region of mTalin was fused with YFP to generate an mTalin-YFP fusion as a reporter for microfilaments. Coexpression of mTalin-YFP with 6K-CFP in the leaf epidermal cells of N. benthamiana showed that many 6K vesicles coaligned with microfilaments (Fig. 2A). Time-lapse imaging experiments were carried out to determine the dynamic status of the 6K vesicles on microfilaments. It was found that the 6K vesicles trafficked along the microfilaments with an average velocity of approximately 2 μm/s (see Movies S1 and S2 in the supplemental material). Lat B, an inhibitor of actin polymerization, was applied during coinfiltration of N. benthamiana with Agrobacterium strains harboring 6K-CFP and mTalin-YFP vectors. The drug treatment disrupted microfilaments and induced a slight diffusion of the 6K-derived vesicles (Fig. 2B). Further time-lapse imaging analysis showed that Lat B treatment blocked intracellular trafficking of the 6K vesicles (see Movie S3 in the supplemental material). Together, these results demonstrate that the 6K vesicles move intracellularly along the microfilaments.
FIG. 2.
Coalignment of 6K vesicles with microfilaments. (A) Expression of 6K-CFP and mTalin-YFP in an N. benthamiana leaf epidermal cell after agroinfiltration. The arrows indicate coalignment of microfilament markers with 6K-CFP. (B) LatB at 25 μM concentration was infiltrated into N. benthamiana leaves coexpressing 6K-CFP and mTalin-YFP. At 2 h postinfiltration, the infiltrated area was observed by confocal laser scanning microscopy. The 6K-CFP vesicles still formed after treatment with Lat B but did not always coalign with collapsed microfilaments (B). Bars, 9 μm.
The 6K-induced structures and Golgi bodies are intimately associated and highly mobile in N. benthamiana.
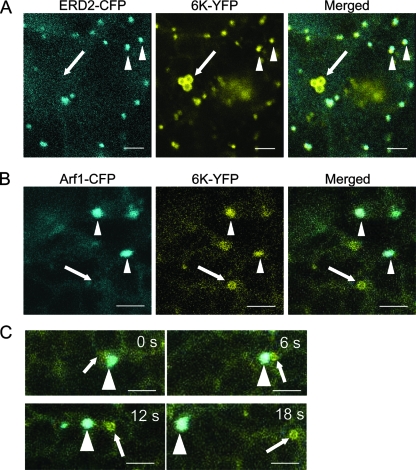
ERES and Golgi bodies in plants have been suggested to be single mobile secretory units in plant cells (18, 28, 29, 64). To examine if 6K-containing ERES also colocalize with Golgi stacks, the distribution of 6K with respect to the Golgi apparatus was monitored in epidermal cells coexpressing 6K-YFP and two Golgi markers tagged with CFP (ERD2-CFP and Arf1-CFP). ERD2 is a well-described secretory marker whose translocation to the Golgi apparatus depends on the functional COPII secretory pathway (57). Arf1, a known marker for Golgi bodies, regulates the recruitment of the COPI coat complex to membranes (38, 64). Coexpression of 6K-YFP with ERD2-CFP or Arf1-CFP in the leaf epidermal cells of N. benthamiana showed that a 6K-YFP punctate structure always colocalized with a Golgi stack labeled by ERD2-CFP or by Arf1-CFP (Fig. 3A and B). However, the ringlike structures of 6K did not localize to the Golgi apparatus (Fig. 3A and B). Interestingly, some ringlike structures labeled by 6K-YFP were transiently associated with and eventually detached from mobile punctate structures containing 6K-YFP and Arf1-CFP (Fig. 3C). When treated with 25 μM Lat B to depolymerize the actin microfilaments, the movement of the punctate structures labeled by 6K-YFP was inhibited, but they remained colocalized with the Golgi stack (data not shown). These data suggest that the 6K-induced structures and Golgi bodies are intimately associated and highly mobile in N. benthamiana.
FIG. 3.
Golgi stacks and 6K-YFP punctate structures move together. (A) Confocal images of N. benthamiana epidermal cells coexpressing ERD2-CFP and 6K-YFP. (B) Confocal images of N. benthamiana epidermal cells coexpressing Arf1-CFP and 6K-YFP. (C) High-magnification time-lapse imaging of a cell coexpressing Arf1-CFP and 6K-YFP. Note that Golgi stacks and 6K-YFP punctate structures moved together in the cell at all times. The times of acquisition of frames are indicated in the upper corners. The arrowheads indicate the localization of 6K-YFP punctate structures with the Golgi area. The arrows point to the ringlike structure of 6K-YFP that did not overlap with the Golgi area. Bars, 15 μm.
The inhibition of COPII transport via the coexpression of dominant-negative Sar1 GTP blocks 6K-CFP accumulation at ERES.
Since the 6K-induced vesicles colocalized with two COPII markers at the ERES, experiments were designed to test whether the COPII complex is required for the biogenesis of the 6K vesicles. To manipulate COPII transport in the N. benthamiana system, two expression vectors were constructed for the transient expression of wild-type or mutated (H74L) Sar1 tagged with YFP [i.e., Sar1-YFP and Sar1(H74L)-YFP]. Sar1, also known as the small GTPase Sar1, is one of three essential cytosolic components of the COPII complex (18, 27). Sar1(H74L) is the GTP-locked mutant that exerts the known dominant-negative effect on COPII vesicle transport. This mutant has previously been shown to be less sensitive to the GTPase-activating activity of Sec23 and to trap vesicles in a coated configuration so that they are unable to fuse with the target membrane (18, 27, 53, 66).
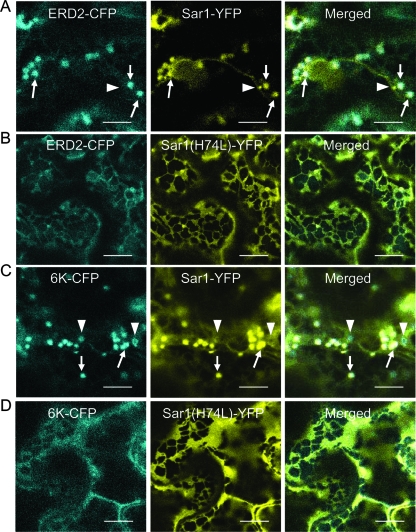
When wild-type Sar1-YFP and ERD2-CFP were transiently coexpressed in N. benthamiana leaf epidermal cells, the ERD2-CFP fluorescence was detected in punctate structures likely representing Golgi stacks (Fig. 4A), as reported previously (28). Sar1-YFP highlighted the peri-Golgi stacks labeled by ERD2-CFP. Moreover, some strong Sar1-YFP punctate labeling did not overlap with the Golgi stack (Fig. 4A), consistent with previous observations (27). These structures may represent embryonic ERES for the biogenesis of new Golgi bodies (64). The coexpression of ERD2-CFP with the Sar1(H74L) mutant led to the retention of the Golgi body-resident protein ERD2-CFP within the ER (Fig. 4B), in agreement with results published previously (18, 66). In addition, Sar1(H74L)-YFP was uniformly distributed along the ER network, colocalizing with ERD2-CFP (Fig. 4B). These experiments demonstrate that Sar1(H74L) can be used to effectively arrest COPII transport out of the ER in N. benthamiana.
FIG. 4.
Coexpression of dominant-negative Sar1 GTP blocks 6K-CFP accumulation on ERES. (A) Cell coexpressing ERD2-CFP and Sar1-YFP. The arrows indicate colocalization of Golgi bodies with ERES stained with Sar1-YFP. The arrowheads show rare Sar1-YFP punctate labeling that did not overlap with the Golgi stack. (B) Coexpression of Sar1(H74L) led to the retention within the ER of ERD2-CFP. (C) Cell coexpressing 6K-CFP and Sar1-YFP. The arrows indicate colocalization of 6K-CFP punctate structures with ERES stained with Sar1-YFP. The arrowheads show ringlike structures of 6K-CFP that did not colocalize with Sar1-YFP punctate labeling. (D) Expression of Sar1(H74L)-YFP resulted in the retention of 6K-CFP in the ER. Bars, 12 μm.
The same system was employed to test whether the inhibition of COPII transport by Sar1(H74L) also affects the accumulation of the TEV 6K protein at the ERES. When 6K-CFP was coexpressed with the wild-type Sar1-YFP in planta, the 6K-CFP punctate structures were closely associated with Sar1-YFP (Fig. 4C). However, in about 90% of cells coexpressing Sar1(H74L)-YFP and 6K-CFP, the distribution pattern of 6K-CFP was completely altered. As shown in Fig. 4D, the 6K punctate structures were not evident. Instead, the 6K-CFP fluorescence was distributed within the reticulate network (Fig. 4D). Taken together, these results indicate that the accumulation of 6K-CFP at the ERES occurs in a COPII-dependent manner.
The accumulation of the 6K protein at ERES depends on active retrograde protein export.
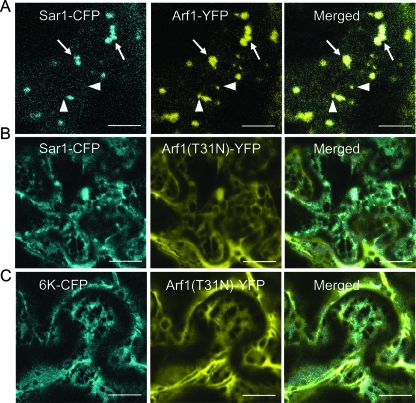
A recent study has suggested that the integrity of protein export from the ER and ERES depends on active COPI involved in retrograde protein transport (64). To test the influence of active retrograde protein export on the biogenesis of the 6K vesicles, transient expression vectors were generated for the expression of Sar1 fused to CFP (Sar1-CFP) in N. benthamiana. The wild-type Arf1 protein or a GDP-restricted mutant of Arf1 [Arf1(T31N)] was fused with YFP. Arf1(T31N) is a mutant of Arf1 that impairs the normal COPI function and inhibits the formation of ERES labeled by YFP-Sec24 and YFP-Sec23 in tobacco leaf epidermal cells (38, 64, 67, 79). In this study, Arf1-YFP was found to localize to the Golgi apparatus and also to additional small structures in the epidermal cells of N. benthamiana (Fig. 5A), consistent with published results (64, 79). In the presence of Arf1-YFP, Sar1-CFP was distributed in the Golgi apparatus area and coincided with the majority of the Arf1-YFP-derived fluorescence. However, some small punctate structures were labeled by Arf1-YFP but not by Sar1-CFP (Fig. 5A). These small structures labeled by the Arf1 fluorescence but not by the COPII coatomer have also been observed in Nicotiana tabacum and likely represent the trans-Golgi network or trans-Golgi network-derived structures that detach from the Golgi bodies (64, 79). Coexpression of Arf1(T31N)-YFP with Sar1-CFP caused the redistribution of this ERES marker into the ER (Fig. 5B). We also detected a cytosolic distribution of Arf1(T31N)-YFP in those cells that were overexpressing the mutant (Fig. 5B), as reported previously (64). All these results confirm that the expression of Arf1(T31N) impairs COPI activity in N. benthamiana.
FIG. 5.
Coexpression of dominant-negative Arf1 blocks the accumulation of 6K-CFP at the ERES. (A) Cell coexpressing Sar1-CFP and Arf1-YFP. Sar1-CFP localized in the Golgi area (arrows) but not at the additional small Arf1-YFP structures (arrowheads). (B) Coexpression of Arf1(T31N) led to the retention of Sar1-CFP within the ER. (C) Cell coexpression of Arf1(T31N) causes the retention of 6K-CFP within the ER. Bars, 12 μm.
The above-mentioned system was employed to test whether the inhibition of COPI by expressing Arf1(T31N) also affects the accumulation of the TEV 6K protein at the ERES. The expression of the Arf1 mutant Arf1(T31N)-YFP resulted in the redistribution of 6K-CFP into the ER network in about 85% of epidermal cells coexpressing 6K-CFP and Arf1(T31N)-YFP (Fig. 5C). The ER membranes appeared enlarged in those cells where 6K-CFP was redistributed into the ER in comparison to control cells (Fig. 5C). The fluorescence of Arf1(T31N)-YFP was also cytosolic in the cells coexpressing the Arf1 mutant and 6K-CFP (Fig. 5C). These data demonstrate that the collapse of the Golgi apparatus-associated COPI machinery perturbs the integrity of the ERES, leading to the containment of the TEV 6K protein within the ER.
Expression of 6K or TEV infection partially suppresses the secretion of a soluble marker, secYFP.
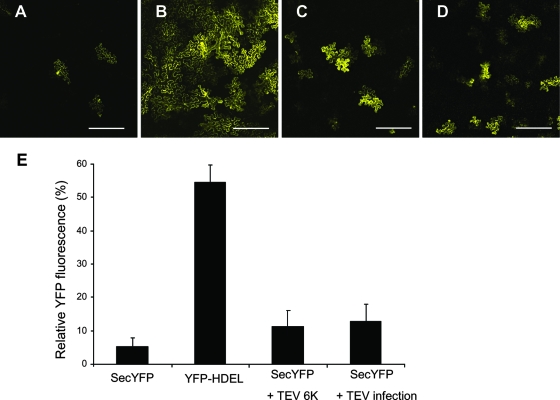
To determine whether the expression of 6K or TEV infection has an effect on the anterograde ER-Golgi apparatus pathway, a transport assay using a secreted YFP marker, secYFP, was carried out as described previously (3, 82). After expression, secYFP is transported from the ER to the apoplast, where it accumulates poorly and exhibits weak fluorescence. When the secretion traffic from the ER to the apoplast is inhibited, the fluorescent secYFP protein accumulates intracellularly and can be readily visualized. This assay has been widely used to test diverse inhibitors and mutants to perturb membrane traffic between the ER and the plasma membrane in plant cells (3, 15, 54, 69, 71, 82). The N. benthamiana leaves infected by TEV or expressing 6K were infiltrated with Agrobacterium species strain GV3101 containing plasmid SecYFP with an OD600 of 0.03. Forty-eight hours after infiltration, secYFP fluorescence intensity was examined by confocal laser scanning microscopy at low magnification. At this time, control cells expressing only secYFP exhibited very dim fluorescence (Fig. 6A) in comparison with cells expressing an ER-resident YFP marker, YFP-HDEL (OD600 = 0.03) (Fig. 6B). In the majority of cells (approximately 80%), coexpression of secYFP with 6K or in the presence of viral infection resulted in an increase in YFP fluorescence relative to the expression of secYFP alone (Fig. 6C and D). Quantitative analysis of confocal images confirmed that leaves cotransfected with TEV 6K had approximately two to four times the fluorescence intensity of leaves expressing secYFP alone but similar to the intensity measured in the presence of viral infection (Fig. 6E). These data suggest that TEV 6K or viral infection partially inhibited secYFP traffic, albeit to a low degree.
FIG. 6.
TEV infection or expression of 6K-CFP reduces the secretion of secYFP. (A to D) Confocal images at low magnification of N. benthamiana epidermal cells expressing secYFP (A), YFP-HDEL (B), secYFP and 6K (C), and secYFP (D) in the presence of viral infection. (E) Relative YFP fluorescence intensities of N. benthamiana epidermal cells expressing secYFP, YFP-HDEL, secYFP plus 6K, and secYFP in the presence of viral infection. The data are means plus standard errors of three independent experiments. Bars, 150 μm.
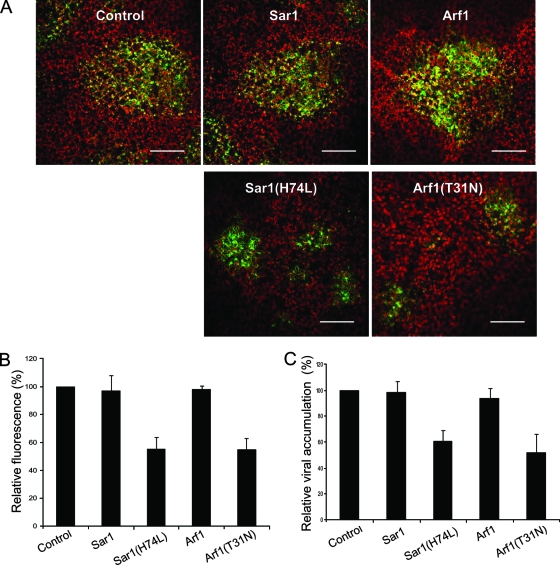
Ectopic expression of dominant-negative mutants of Sar1 and Arf1 reduces TEV infection.
As demonstrated above, both COPII and COPI mutants, i.e., Sar1(H74L) and Arf1(T31N), abolished the formation of the 6K vesicles in cells coexpressing 6K-CFP and the mutants. To test if these dominant-negative mutants affect viral replication, wild-type Sar1 or Sar1(H74L) and wild-type Arf1 or Arf1(T31N) were expressed in the epidermal cells of N. benthamiana, followed by the inoculation of TEV tagged with GFP (from a GFP-tagged TEV infectious clone). The initial infection foci were visualized 3 days postinoculation using the GFP fluorescence. The sizes of these initial infection foci were similar in leaves expressing either wild-type Sar1 or wild-type Arf1 or in buffer-treated control leaves (Fig. 7A). This suggests that the expression of these wild-type COP1 and COPII components did not significantly affect viral infection. In contrast, TEV infection was reduced in the leaves where mutant Sar1(H74L) or Arf1(T31N) was expressed (Fig. 7). The size of the GFP foci was half that of foci observed in control leaves or in the leaves expressing wild-type Sar1 and Arf1 (Fig. 7A and 7B). This was confirmed by ELISA (Fig. 7C). Taken together, these data suggest that both COPII and COPI complex machineries, which are required for the maintenance of the functional ERES and for the biogenesis of the 6K vesicles, may be involved in potyvirus infection.
FIG. 7.
Ectopic expression of dominant-negative mutants of Sar1 and Arf1 reduces TEV infection. (A) Confocal microscopy micrographs of N. benthamiana leaves that were infiltrated with Agrobacterium strains carrying plasmids encoding either Sar1, Sar1(H74L), Arf1, or Arf1(T31N) 24 h before TEV-GFP infection. The photographs show the typical conditions of TEV-GFP replication expressed as the different sizes of GFP foci 3 days postinfection in the absence or presence of the expression of Sar1, Sar1(H74L), Arf1, or Arf1(T31N), respectively. GFP foci are shown in green, and chloroplast autofluorescence is shown in red. Bars, 300 μm. (B) Quantification of the effect of either Sar1, Sar1(H74L), Arf1, or Arf1(T31N) expression upon TEV-GFP replication expressed as the relative average areas of GFP foci with standard errors (SE). The data are means plus SE of three independent experiments. (C) Effect of transient expression of Sar1, Sar1(H74L), Arf1, or Arf1(T31N) on TEV-GFP infection in N. benthamiana. Leaves were infected with TEV 24 h after agroinfiltration and assayed for accumulation of TEV by ELISA. The values represent means plus SE and are given as the percentage of the control.
DISCUSSION
The ER subdomain for the COPII-dependent biogenesis of potyvirus 6K-induced vesicles.
The replication of many positive-strand RNA viruses is closely associated with ER-derived punctate bodies or vesicles (4, 7, 13, 21, 25, 26, 32, 33, 40, 47, 52, 56, 60, 65). Specific viral proteins, e.g., nsp3 of arterivuses, NTB of nepoviruses, 3A (possibly in conjunction with 2BC) of picornaviruses, and 6K of potyviruses, are responsible for the induction of these membranous vesicles. To study how the potyvirus 6K membranous vesicles form in planta, we used live-cell imaging technology. In the epidermal cells of N. benthamiana expressing the 6K-CFP fusion protein, we observed punctate and ringlike structures in the cytoplasm (Fig. 1). We have shown colocalization of the punctate structures induced and labeled by the TEV 6K-CFP protein with Sar1, Sec23, and Sec24 (Fig. 1C and D and 4C), which are three established markers for ERES in tobacco leaf epidermal cells. Moreover, we have presented evidence that the accumulation of the 6K-CFP at the ERES no longer occurred in the cells expressing a dominant-negative mutant of Sar1 (Fig. 4D). Thus, the Sar1-restricted mutant that inhibited the COPII transport system interfered with the ability of 6K to accumulate at the ERES. These results imply that potyvirus 6K vesicles originate and develop in association with the ERES and that the COPII machinery plays an important role in the biogenesis of the 6K vesicles at these sites.
ERES is the term used to define the distinct subdomains or discrete spots of the ER where COPII-coated buds are formed and cargo molecules are loaded into transport carriers for anterograde traffic out of the ER (28, 53, 68). Sar1 modulates the recruitment of the COPII complex to the ER and facilitates the formation of transport carriers (vesicles) for delivery (18). It is possible that the cytosolic domains of 6K interact directly or indirectly with Sar1 to stimulate the assembly of the COPII complex at the ERES. Interestingly, despite striking differences between plant and animal early secretory pathways, the 3A protein of animal coxsackievirus (an enterovirus of the family Picornaviridae) shares many common properties with the potyvirus 6K protein. The 3A protein is a small membrane protein with multiple functions in the viral life cycle and was shown to localize to the ERES in Buffalo green monkey kidney cells (77). Similarly, the replication of a coronovirus is inhibited in murine LR7 cells expressing a dominant mutant of Sar1 that disables the COPII-associated ER export machinery (45). However, in both cases, it is not clear if the COPII complex is involved in the biogenesis of the induced membranous vesicles.
We found that the 6K-induced ringlike structures did not colocalize with the ERES marker but probably corresponded to the small cortical network of the ER membrane (Fig. 1B). It seems that these ringlike structures occur when the 6K protein directly induces vesiculation on the ER membrane independent of the secretory pathway. In fact, when 6K-CFP-expressing cells were treated with FM4-64 dye, which is often used to trace membranes from the endocytic pathway, the red fluorescence observed in endocytic vesicles never overlapped with the 6K-CFP-expressing ringlike structures (data not shown). These data support the notion that the 6K-induced ringlike structures are derived from the ER membrane and are unrelated to endosomes. They may result from defects in vesicle formation triggered by 6K, as suggested for similar structures induced by the TGBp2 membrane protein of Potato virus X (32).
In this study, we reported that virus infection or the expression of 6K partially blocked the export of a secretory marker from the ER in tobacco leaf epidermal cells (Fig. 6). Since 6K is an integral membrane protein, it is possible that the 6K protein stabilizes the COPII coat components on the vesicles and prevents a subsequent fusion process into the Golgi apparatus. This would lead to the proliferation of membranes and blockage of the normal trafficking or recycling of transport carriers. Interestingly, transient expression of NTB (the protein functionally equivalent to CI and 6K of potyviruses) of Grapevine fanleaf virus (GFLV) (a nepovirus) in cultured tobacco cells prevents the export of the resident Golgi apparatus protein α-1,2 mannosidase I (51). The coxsackievirus 3A (equivalent to 6K) and the poliovirus 2B and 3A proteins have also been shown to inhibit ER-to-Golgi apparatus transport (19, 78). Wessels and colleagues have elegantly unraveled the molecular mechanism underlying the 3A-associated inhibition of the secretory pathway (75, 76, 77). They have shown that 3A specifically interacts with the N terminus of GBF1, a guanine nucleotide exchange factor for Arf1. This interaction inhibits the function of GBF1, which deactivates Arf1. As a result, the normal secretion from the ER to the Golgi apparatus is blocked. It would be interesting to determine if the 6K protein inhibits anterograde traffic by a similar mechanism in plant cells.
The association of the potyvirus 6K-induced vesicles with the Golgi apparatus and the COPI machinery.
In this work, we have shown that the 6K-YFP punctate structures coincided with cis-Golgi stacks labeled by Arf1-CFP (Fig. 3). This finding provides support for the model proposed by daSilva et al., Hanton et al., and Stefano and colleagues (18, 28, 64) in which ERES and their associated Golgi bodies function as mobile secretory units in plant leaf epidermal cells. It is possible that the association of 6K-containing ERES with the Golgi stack promotes the COPI-mediated retrograde traffic of biomolecules from the Golgi apparatus to the ER for virus replication. Indeed, the fact that both anterograde and retrograde membrane traffic flows are rerouted toward the formation of the viral replication complexes may account for the decrease in the average number of Golgi stacks in GFLV-infected cultured tobacco cells (52) and the disassembly of the Golgi complex in poliovirus-infected human cells (6).
Brefeldin A (BFA) inhibits the replication of many positive-strand viruses, including plant viruses, such as GFLV (52), and human viruses, such as poliovirus (30, 41). BFA interferes with vesicular transport for the intracellular trafficking of proteins, membrane materials, and soluble cargo between the ER and the Golgi apparatus (7, 64, 74). We also found that the replication of two potyviruses, TEV and Turnip mosaic virus, was suppressed by this fungal metabolite (data not shown). BFA interacts with specific guanine nucleotide exchange factor that recycle Arf1 between an active (GTP-bound) membrane-bound state and an inactive (GDP-bound) cytosolic state. Through this interaction, BFA inhibits the activation and function of the small-GTPase family and directly suppresses the COPI pathway. Thus, the active COPI machinery participates in the replication of these BFA-sensitive viruses either directly or indirectly. In poliovirus-infected HeLa cells, it has been shown that Arf1 is redistributed to the new structures, where viral replication takes place (9). Differential requirements for COPI coats in the formation of replication complexes have been found among three genera of Picornaviridae (24). In this study, we have shown that an Arf1 mutant that impairs the activation of Arf1 disrupted the distribution of membrane markers in the Golgi apparatus and simultaneously led to the inhibition of 6K accumulation at ERES (Fig. 5), suggesting that the biogenesis of the 6K vesicles is also dependent on the active COPI machinery. It is believed that the integrity of ERES is maintained by the balanced action of the COPI and COPII systems and that Arf1 coatomer is indirectly required for forward trafficking out of the ER due to its role in recycling components essential for differentiation of the ERES (64).
Involvement of the early secretory pathway in virus infection.
To test if the early secretory pathway is required for potyvirus replication, we disabled the COPI and COPII functions in N. benthamiana by expressing the Arf1 mutant, i.e., Arf1(T31N), and the Sar1 mutant, i.e., Sar1(H74L), respectively. We found that the expression of either the Arf1 mutant or the Sar1 mutant disrupted ERES and that this disruption was coupled with an ∼50% reduction in TEV accumulation (Fig. 7). This reduction was less than expected, possibly due to the fact that the incorporation of 6K vesicles into ERES was not abolished in some cells expressing Sar1(H74L) and Arf1(T31N). In this study, we estimated that 6K distribution was not affected in 10 to 15% of cells coexpressing the COP mutants and 6K-CFP. Since virus gene expression outcompetes mRNA expression, the percentage of these cells with unaffected 6K distribution probably is higher in coexpression of TEV and the COP mutants. It is also possible that potyviruses may have an alternative, less favorable replication site when the ER is not available. We cannot exclude the possibility that the partial reduction was due to the reduced viability of cells resulting from the expression of the COPI or COPII mutant, but this possibility is not favored by the following recent observations. First, Jiang and colleagues have screened the 800 essential genes present in the yeast Tet promoter Hughes Collection and found that downregulation of COP1 suppresses the replication of a tombusvirus (31). Second, BFA-mediated deactivation of Arf1 that disables the COPI activity inhibits the replication of BFA-sensitive viruses (30, 41, 52). Finally, the expression of the Sar1 mutant blocks the replication of a coronovirus (45). In this study, the inhibition of virus accumulation was paralleled by the disruption of the 6K vesicles, and the ERES was the putative site where the 6K vesicles originated and developed. Therefore, it is reasonable to suggest that the early secretory pathway is likely involved in the infection of potyviruses.
In summary, our data provide evidence to suggest that the potyvirus 6K-induced membranous vesicles localize at the ERES and associate with the cis-Golgi stack. Moreover, we have shown that the formation of the 6K vesicles at the ERES is COPI and COPII dependent and that the deactivation of either the COPI or COPII machinery disrupts the formation of the 6K vesicles at the ERES and suppresses virus accumulation.
Supplementary Material
Acknowledgments
We thank Richard Nelson and Phil Harris (Samuel Roberts Noble Foundation), Jean-François Laliberté (INRS-Institut Armand Frappier, Canada) and Hélène Sanfaçon (AAFC, Canada) for critical reading of the manuscript. We are grateful to Jim Carrington (Oregon State University) for kindly providing the GFP-tagged TEV infectious clone pTEV7DA-GFP; to Akihiko Nakano (Riken, Japan) for kindly providing plasmid pMT121-AtSAR1 H74L; to Nam-Hai Chua (Rockefeller University) for kindly providing plasmid pYSC14; to Ben Scheres (Utrecht University, The Netherlands) for plasmids containing Arf1-YFP, Arf1-CFP, and Arf1(T31N)-YFP; and to Ian Moore (Oxford Brookes University, United Kingdom) for plasmids containing YFP-HDEL and SecYFP. We are grateful to Hongyan Chen for ELISA and Jamie McNeil for technical support. We also thank the anonymous reviewers for their very helpful comments in improving this paper.
This work was supported by Agriculture and Agri-Food Canada and the Natural Sciences and Engineering Research Council of Canada.
Footnotes
Published ahead of print on 8 October 2008.
Supplemental material for this article may be found at http://jvi.asm.org/.
REFERENCES
- 1.Ahlquist, P. 2006. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 4371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahlquist, P., A. O. Noueiry, W. M. Lee, D. B. Kushner, and B. T. Dye. 2003. Host factors in positive-strand RNA virus genome replication. J. Virol. 778181-8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Batoko, H., H. Q. Zheng, C. Hawes, and I. Moore. 2000. A rab1 GTPase is required for transport between the endoplasmic reticulum and Golgi apparatus and for normal Golgi movement in plants. Plant Cell 122201-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beauchemin, C., and J.-F. Laliberté. 2007. The poly(A) binding protein is internalized in virus-induced vesicles or redistributed to the nucleolus during turnip mosaic virus infection. J. Virol. 8110905-10913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beauchemin, C., N. Boutet, and J. F. Laliberté. 2007. Visualization of the interaction between the precursors of VPg, the viral protein linked to the genome of turnip mosaic virus, and the translation eukaryotic initiation factor iso 4E in planta. J. Virol. 81775-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belov, G. A., N. Altan-Bonnet, G. Kovtunovych, C. L. Jackson, J. Lippincott-Schwartz, and E. Ehrenfeld. 2007. Hijacking components of the cellular secretory pathway for replication of poliovirus RNA. J. Virol. 81558-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belov, G. A., and E. Ehrenfeld. 2007. Involvement of cellular membrane traffic proteins in poliovirus replication. Cell Cycle 636-38. [DOI] [PubMed] [Google Scholar]
- 8.Belov, G. A., M. H. Fogg, and E. Ehrenfeld. 2005. Poliovirus proteins induce membrane association of GTPase ADP-ribosylation factor. J. Virol. 797207-7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belov, G. A., C. Habbersett, D. Franco, and E. Ehrenfeld. 2007. Activation of cellular Arf GTPases by poliovirus protein 3CD correlates with virus replication. J. Virol. 819259-9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger, P. H., M. J. Adams, O. W. Barnett, A. A. Brunt, J. Hammond, J. H. Hill, R. L. Jordan, S. Kashiwazaki, E. Rybicki, N. Spence, D. C. Stenger, S. T. Ohki, I. Uyeda, A. van Zaayen, J. Valkonen, and H. J. Vetten. 2005. Potyvirida, p. 819e-841e. In C. M. Fauquet, M. A. Mayo, J. Maniloff, U. Desselbergar, and L. A. Ball (ed.), Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, San Diego, CA.
- 11.Bushell, M., and P. Sarnow. 2002. Hijacking the translation apparatus by RNA viruses. J. Cell Biol. 158395-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carette, J. E., J. Verver, J. Martens, T. van Kampen, J. Wellink, and A. van Kammen. 2002. Characterization of plant proteins that interact with cowpea mosaic virus ‘60K’ protein in the yeast two-hybrid system. J. Gen. Virol. 83885-893. [DOI] [PubMed] [Google Scholar]
- 13.Carette, J. E., J. van Lent, S. A. MacFarlane, J. Wellink, and A. van Kammen. 2002. Cowpea mosaic virus 32- and 60-kilodalton replication proteins target and change the morphology of endoplasmic reticulum membranes. J. Virol. 766293-6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carrington, J. C., D. D. Freed, and A. J. Leinicke. 1991. Bipartite signal sequence mediates nuclear translocation of the plant potyviral NIa protein. Plant Cell 3953-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chatre, L., F. Brandizzi, A. Hocquellet, C. Hawes, and P. Moreau. 2005. Sec22 and Memb11 are v-SNAREs of the anterograde endoplasmic reticulum-Golgi pathway in tobacco leaf epidermal cells. Plant Physiol. 1391244-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cheung, A. Y., and S. C. de Vries. 2008. Membrane trafficking: intracellular highways and country roads. Plant Physiol. 1471451-1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung, B. Y.-W., W. A. Miller, J. F. Atkins, and A. E. Firth. 2008. An overlapping essential gene in the Potyviridae. Proc. Natl. Acad. Sci. USA 1055897-5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.daSilva, L. L., E. L. Snapp, J. Denecke, J. Lippincott-Schwartz, C. Hawes, and F. Brandizzi. 2004. Endoplasmic reticulum export sites and Golgi bodies behave as single mobile secretory units in plant cells. Plant Cell 161753-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doedens, J. R., and K. Kirkegaard. 1995. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 14894-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Earley, K. W., J. R. Haag, O. Pontes, K. Opper, T. Juehne, K. Song, and C. S. Pikaard. 2006. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45616-629. [DOI] [PubMed] [Google Scholar]
- 21.Egger, D., N. Teterina, E. Ehrenfeld, and K. Bienz. 2000. Formation of the poliovirus replication complex requires coupled viral translation, vesicle production, and viral RNA synthesis. J. Virol. 746570-6580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Froshauer, S., J. Kartenbeck, and A. Helenius. 1988. Alphavirus RNA replicase is located on the cytoplasmic surface of endosomes and lysosomes. J. Cell Biol. 1072075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garnier, M., T. Candresse, and J. M. Bové. 1986. Immunocytochemical localization of TYMV-coded structural and non-structural proteins by the protein A-gold technique. Virology 151100-109. [DOI] [PubMed] [Google Scholar]
- 24.Gazina, E. V., J. M. Mackenzie, R. J. Gorrell, and D. A. Anderson. 2002. Differential requirements for COPI coats in formation of replication complexes among three genera of Picornaviridae. J. Virol. 7611113-11122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gosert, R., A. Kanjanahaluethai, D. Egger, K. Bienz, and S. C. Baker. 2002. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J. Virol. 763697-3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han, S., and H. Sanfaçon. 2003. Tomato ringspot virus proteins containing the nucleoside triphosphate binding domain are transmembrane proteins that associate with the endoplasmic reticulum and cofractionate with replication complexes. J. Virol. 70523-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanton, S. L., L. Chatre, L. A. Matheson, M. Rossi, M. A. Held, and F. Brandizzi. 2008. Plant Sar1 isoforms with near-identical protein sequences exhibit different localisations and effects on secretion. Plant Mol. Biol. 67283-294. [DOI] [PubMed] [Google Scholar]
- 28.Hanton, S. L., L. Chatre, L. Renna, L. A. Matheson, and F. Brandizzi. 2007. De novo formation of plant endoplasmic reticulum export sites is membrane cargo induced and signal mediated. Plant Physiol. 1431640-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanton, S. L., L. A. Matheson, and F. Brandizzi. 2006. Seeking a way out: export of proteins from the plant endoplasmic reticulum. Trends Plant Sci. 11335-343. [DOI] [PubMed] [Google Scholar]
- 30.Iruzun, A., L. Perez, and L. Carasco. 1992. Involvement of membrane traffic in the replication of poliovirus genomes: effects of brefeldin A. Virology 191166-175. [DOI] [PubMed] [Google Scholar]
- 31.Jiang, Y., E. Serviene, J. Gal, T. Panavas, and P. D. Nagy. 2006. Identification of essential host factors affecting tombusvirus RNA replication based on the yeast Tet promoters Hughes Collection. J. Virol. 807394-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ju, H. J., J. E. Brown, C. M. Ye, and J. Verchot-Lubicz. 2007. Mutations in the central domain of potato virus X TGBp2 eliminate granular vesicles and virus cell-to-cell trafficking. J. Virol. 811899-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ju, H. J., T. D. Samuls, Y. S. Wang, E. Blancaflor, M. Payton, R. Mitra, K. Krishnamurthy, R. S. Nelson, and J. Verchot-Lubicz. 2005. The potato virus X TGBp2 movement protein associates with endoplasmic reticulum-derived vesicles during virus infection. Plant Physiol. 1381877-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karimi, M., D. Inzé, and A. Depicker. 2002. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7193-195. [DOI] [PubMed] [Google Scholar]
- 35.Koonin, E., and V. V. Dolja. 1993. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit. Rev. Biochem. Mol. Biol. 28375-430. [DOI] [PubMed] [Google Scholar]
- 36.Kopek, B. G., G. Perkins, D. J. Miller, M. H. Ellisman, and P. Ahlquist. 2007. Three-dimensional analysis of a viral RNA replication complex reveals a virus-induced mini-organelle. PLoS Biol. 5e220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kost, B., P. Spielhofer, and N. H. Chua. 1998. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualizes the actin cytoskeleton in growing pollen tubes. Plant J. 16393-401. [DOI] [PubMed] [Google Scholar]
- 38.Lee, M. H., M. H. Min, Y. J. Lee, J. B. Jin, D. H. Shin, D. H. Kim, K. H. Lee, and I. Hwang. 2002. ADP-ribosylation factor 1 of Arabidopsis plays a critical role in intracellular trafficking and maintenance of endoplasmic reticulum morphology in Arabidopsis. Plant Physiol. 1291507-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mackenzie, J. 2005. Wrapping things up about virus RNA replication. Traffic 6967-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Más, P., and R. N. Beachy. 2000. Role of microtubules in the intracellular distribution of tobacco mosaic virus movement proteins. Proc. Natl. Acad. Sci. USA 9712345-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maynell, L. A., K. Kirkegaard, and M. W. Klymkowsky. 1992. Inhibition of poliovirus RNA synthesis by brefeldin A. J. Virol. 661985-1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nelson, R. S., and V. Citovsky. 2005. Plant viruses. Invaders of cells and pirates of cellular pathways. Plant Physiol. 1381809-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Netherton, C. K., K. Moffat, E. Brooks, and T. Wileman. 2007. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv. Virus Res. 70101-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Novoa, R. R., G. Calderita, R. Arranz, J. Fontana, H. Granzow, and C. Risco. 2005. Virus factories: associations of cell organelles for viral replication and morphogenesis. Biol. Cell 97147-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oostra, M., E. G. te Lintelo, M. Deijs, M. H. Verheije, P. J. Rottier, and C. A. de Haan. 2008. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 8112323-12336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Phillipson, B. A., P. Pimpl, L. L. daSilva, A. J. Crofts, J. P. Taylor, A. Movafeghi, D. G. Robinson, and J. Denecke. 2001. Secretory bulk flow of soluble proteins is efficient and COPII dependent. Plant Cell 132005-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Posthuma, C. C., K. W. Pedersen, Z. Lu, R. G. Joosten, N. Roos, J. C. Zevenhoven-Dobbe, and E. J. Snijder. 2008. Formation of the aterivirus replication/transcription complex: a key role for non-structural protein 3 in the remodeling of intracellular membranes. 2008. J. Virol. 824480-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rees, D. J., S. E. Ades, S. J. Singer, and R. O. Hynes. 1990. Sequence and domain structure of talin. Nature 347685-689. [DOI] [PubMed] [Google Scholar]
- 49.Restrepo-Hartwig, M. A., and J. C. Carrington. 1994. The tobacco etch potyvirus 6-kilodalton protein is membrane associated and involved in viral replication. J. Virol. 682388-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Riechmann, J. L., S. Lain, and J. A. Garcia. 1992. Highlights and prospects of potyvirus molecular biology. J. Gen. Virol. 731-16. [DOI] [PubMed] [Google Scholar]
- 51.Ritzenthaler, C., and R. Elamawi. 2006. The ER in replication of positive-strand RNA viruses, p. 309-330. In D. G. Robinson (ed.). Plant cell monographs, vol. 4. Springer Publishing, Berlin, Germany. [Google Scholar]
- 52.Ritzenthaler, C., C. Laporte, F. Gaire, P. Dunoyer, C. Schmitt, S. Duval, A. Piéquet, A. M. Loudes, O. Rohfritsch, C. Stussi-Garaud, and P. Pfeiffer. 2002. Grapevine fanleaf virus replication occurs on endoplasmic reticulum-derived membranes. J. Virol. 768808-8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson, D. G., M.-C. Herranz, J. Bubeck, R. Pepperkok, and C. Ritzenthaler. 2007. Membrane dynamics in the early secretory pathway. Crit. Rev. Plant Sci. 26199-225. [Google Scholar]
- 54.Rojo, E., and J. Denecke. 2008. What is moving in the secretory pathway of plants? Plant Physiol. 1471493-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rubino, L., F. Weber-Lotfi, A. Dietrich, C. Stussi-Garaud, and M. Russo. 2001. The open reading frame 1-encoded (′36K') protein of Carnation Italian ringspot virus localizes to mitochondria. J. Gen. Virol. 8229-34. [DOI] [PubMed] [Google Scholar]
- 56.Rust, R. C., L. Landmann, R. Gosert, B. L. Tang, W. Hong, H.-P. Hauri, D. Egger, and K. Bienz. 2001. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J. Virol. 759808-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saint-Jore, C. M., J. Evins, H. Batoko, F. Brandizzi, I. Moore, and C. Hawes. 2002. Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 29661-678. [DOI] [PubMed] [Google Scholar]
- 58.Salonen, A., T. Ahola, and L. Kääriäinen. 2005. Viral RNA replication in association with cellular membranes. Curr. Top. Microbiol. Immunol. 285139-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sanfaçon, H. 2005. Replication of positive-strand RNA viruses in plants: contact points between plant and virus components. Can. J. Bot. 831529-1549. [Google Scholar]
- 60.Schaad, M. C., P. E. Jensen, and J. C. Carrington. 1997. Formation of plant RNA virus replication complexes on membranes: role of an endoplasmic reticulum-targeted viral protein. EMBO J. 164049-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schwartz, M., J. Chen, W. M. Lee, M. Janda, and P. Ahlquist. 2004. Alternate, virus-induced membrane rearrangements support positive-strand RNA virus genome replication. Proc. Natl. Acad. USA 10111263-11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sparkes, I. A., J. Runions, A. Kearns, and C. Hawes. 2006. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 12019-2025. [DOI] [PubMed] [Google Scholar]
- 63.Staehelin, L. A., and B. H. Kang. 2008. Nanoscale architecture of endoplasmic reticulum export sites and of Golgi membranes as determined by electron tomography. Plant Physiol. 1471454-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stefano, G., L. Renna, L. Chatre, S. L. Hanton, P. Moreau, C. Hawes, and F. Brandizzi. 2006. In tobacco leaf epidermal cells, the integrity of protein export from the endoplasmic reticulum and of ER export sites depends on active COPI machinery. Plant J. 4695-110. [DOI] [PubMed] [Google Scholar]
- 65.Suhy, D. A., T. H. Giddings, Jr., and K. Kirkegaard. 2000. Remodeling the endoplasmic reticulum by poliovirus infection and by individual viral proteins: an autophagy-like origin for virus-induced vesicles. J. Virol. 748953-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Takeuchi, M., T. Ueda, K. Sato, H. Abe, T. Nagata, and A. Nakano. 2000. A dominant negative mutant of sar1 GTPase inhibits protein transport from the endoplasmic reticulum to the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 23517-525. [DOI] [PubMed] [Google Scholar]
- 67.Takeuchi, M., T. Ueda, N. Yahara, and A. Nakano. 2002. Arf1 GTPase plays roles in the protein traffic between the endoplasmic reticulum and the Golgi apparatus in tobacco and Arabidopsis cultured cells. Plant J. 31499-515. [DOI] [PubMed] [Google Scholar]
- 68.Tang, B. L., Y. Wang, Y. S. Ong, and W. Hong. 2005. COPII and exit from the endoplasmic reticulum. Biochim. Biophys. Acta 1744293-303. [DOI] [PubMed] [Google Scholar]
- 69.Teh, O. K., and I. Moore. 2007. An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448493-496. [DOI] [PubMed] [Google Scholar]
- 70.Thivierge, K., V. Nicaise, P. J. Dufresne, S. Cotton, J.-F. Laliberté, O. Le Gall, and M. Fortin. 2005. Plant virus RNAs: coordinated recruitment of conserved host functions by (+) ssRNA viruses during early infection events. Plant Physiol. 1381822-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tyrrell, M., P. Campanoni, J. U. Sutter, R. Pratelli, M. Paneque, S. Sokolovski, and M. R. Blatt. 2007. Selective targeting of plasma membrane and tonoplast traffic by inhibitory (dominant-negative) SNARE fragments. Plant J. 511099-1115. [DOI] [PubMed] [Google Scholar]
- 72.Urcuqui-Inchima, S., A. L. Haenni, and F. Bernardi. 2001. Potyvirus proteins: a wealth of functions. Virus Res. 74157-175. [DOI] [PubMed] [Google Scholar]
- 73.Wang, A., H. Sanfaçon, L. W. Stobbs, D. James, D. Thompson, A. M. Svircev, and D. C. W. Brown. 2006. Plum pox virus in Canada: progress in research and future prospects for disease control. Can. J. Plant Pathol. 28182-196. [Google Scholar]
- 74.Wei, T., T. Shimizu, and T. Omura. 2008. Endomembranes and myosin mediate assembly into tubules of Pns10 of Rice dwarf virus and intercellular spreading of the virus in cultured insect vector cells. Virology 372349-356. [DOI] [PubMed] [Google Scholar]
- 75.Wessels, E., D. Duijsings, K. H. W. Lanke, W. J. G. Melchers, C. L. Jackson, and F. J. M. van Kuppeveld. 2007. Molecular determinants of the interaction between cosackievirus protein 3A and guanine nucleotide exchange factor GBF1. J. Virol. 815238-5245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wessels, E., D. Duijsings, T. K. Niu, K. H. W. Lanke, S. H. J. van Dooren, C. L. Jackson, W. J. G. Melchers, and F. J. M. van Kuppeveld. 2006. Effects of picornavirus 3A proteins on protein transport and GBF1-dependent COPI recruitment. J. Virol. 8011852-11860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wessels, E., D. Duijsings, T. K. Niu, S. Neumann, V. M. Oorschot, F. de Lange, K. H. W. Lanke, J. Klumperman, A. Henke, C. L. Jackson, W. J. G. Melchers, and F. J. M. van Kuppeveld. 2006. A viral protein that blocks Af1-mediated COPI assembly by inhibiting the guanine nucleotide exchange factor GBF1. Dev. Cell 11191-201. [DOI] [PubMed] [Google Scholar]
- 78.Wessels, E., D. Duijsings, R. A. Notebaart, W. J. G. Melchers, and F. J. M. van Kuppeveld. 2005. A proline-rich region in the coxsackievirus 3A protein is required for the protein to inhibit endoplasmic reticulum-to-Golgi transport. J. Virol. 795163-5173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xu, J., and B. Scheres. 2005. Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17525-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang, Y. D., R. Elamawi, J. Bubeck, R. Pepperkok, C. Ritzenthaler, and D. G. Robinson. 2005. Dynamics of COPII vesicles and the Golgi apparatus in cultured Nicotiana tabacum BY-2 cells provides evidence for transient association of Golgi stacks with endoplasmic reticulum exit sites. Plant Cell 171513-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang, S. C., G. Zhang, L. Yang, J. Chisholm, and H. Sanfacon. 2005. Evidence that insertion of tomato ringspot nepovirus NTB-VPg protein in endoplasmic reticulum membranes is directed by two domains: a C-terminal transmembrane helix and an N-terminal amphipathic helix. J. Virol. 7911752-11765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zheng, H., L. Camacho, E. Wee, H. Batoko, J. Legen, C. J. Leaver, R. Malhó, P. J. Hussey, and I. Moore. 2005. A Rab-E GTPase mutant acts downstream of the Rab-D subclass in biosynthetic membrane traffic to the plasma membrane in tobacco leaf epidermis. Plant Cell 172020-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.