Abstract
Viral infection of the liver can lead to severe tissue damage when high levels of viral replication and spread in the organ are coupled with strong induction of inflammatory responses. Here we report an unexpected correlation between the expression of a functional X domain encoded by the hepatotropic mouse hepatitis virus strain A59 (MHV-A59), the high-level production of inflammatory cytokines, and the induction of acute viral hepatitis in mice. X-domain (also called macro domain) proteins possess poly-ADP-ribose binding and/or ADP-ribose-1′′-phosphatase (ADRP) activity. They are conserved in coronaviruses and in members of the “alpha-like supergroup” of phylogenetically related positive-strand RNA viruses that includes viruses of medical importance, such as rubella virus and hepatitis E virus. By using reverse genetics, we constructed a recombinant murine coronavirus MHV-A59 mutant encoding a single-amino-acid substitution of a strictly conserved residue that is essential for coronaviral ADRP activity. We found that the mutant virus replicated to slightly reduced titers in livers but, strikingly, did not induce liver disease. In vitro, the mutant virus induced only low levels of the inflammatory cytokines tumor necrosis factor alpha and interleukin-6 (IL-6). In vivo, we found that IL-6 production, in particular, was reduced in the spleens and livers of mutant virus-infected mice. Collectively, our data demonstrate that the MHV X domain exacerbates MHV-induced liver pathology, most likely through the induction of excessive inflammatory cytokine expression.
Acute viral hepatitis is accompanied by strong host innate immune responses that include the expression of type I interferons (IFNs) and the release of proinflammatory cytokines (31, 37). These responses are considered essential to control early virus replication in the liver (14, 30). Type I IFNs induce the expression of antiviral effector molecules (12, 16) but also shape the upcoming adaptive immune response (29). Proinflammatory cytokines and chemokines mediate the migration of cells with antiviral effector functions into the liver and thereby promote viral clearance (23). This process can, when excessive activation occurs, also result in severe tissue damage (4). Although the main producer cells of type I IFNs and proinflammatory cytokines have been identified as plasmacytoid dendritic cells (pDCs) and activated macrophages, respectively (25, 31), very little is known about if and how viruses may affect their expression during acute viral hepatitis and how these virus-host interactions may impact the course of the infection and disease.
Mouse hepatitis virus (MHV) is a positive-strand RNA virus of the Coronaviridae family. Its natural host is the mouse, and MHV has been extensively studied in the context of various disease models and host innate and adoptive immune responses (3, 38). MHV strain A59 (MHV-A59) is both hepatotropic and neurotropic and can infect hepatocytes, macrophages, conventional DCs (cDCs), and pDCs. Although virus replication is controlled by pDC-mediated alpha IFN (IFN-α) production during the early phase of infection (5), acute viral hepatitis and markedly elevated serum alanine aminotransferase (ALT) levels become apparent at day 5 postinfection (p.i.). Clearly, the coronavirus spike protein, as the major determinant of virus target cell tropism (39), is responsible for the pronounced MHV-A59 liver tropism. However, our knowledge of other coronaviral factors that may be involved in the induction of acute hepatitis is limited.
There is accumulating evidence that coronavirus replicase gene products impact virus pathogenicity (7, 28, 33, 41). Replicase gene expression involves the translation of large polyproteins that undergo extensive proteolytic processing by viral proteinases to give rise to 15 to 16 nonstructural proteins (nsps) (40). They assemble to form the viral replicase-transcriptase complex at endoplasmic reticulum-derived double-membrane vesicles (17). Interestingly, the ADP-ribose-1′′-phosphatase (ADRP) activity (26) encoded in nsp3 appears to be dispensable for virus RNA synthesis (20), suggesting a possible role in vivo. The ADRP domain, also called the X domain, is strictly conserved among coronaviruses, and moreover, a homologous domain can be found in viruses belonging to the “alpha-like supergroup” of positive-strand RNA viruses (10, 11). This group of phylogenetically related viruses includes alphaviruses such as Semliki Forest virus (SFV), a number of plant viruses, and viruses of medical importance, such as rubella virus and hepatitis E virus (HEV). Viral X domains are related to a large family of macro domain proteins found in many cellular organisms (15, 26). It is generally accepted that macro domains are associated with ADP-ribose binding or with the processing of ADP-ribose derivatives such as ADP-ribose-1′′-phosphate (Appr-1′′-p) (9, 15). The X domains of human coronavirus 229E (HCoV-229E), severe acute respiratory syndrome coronavirus (SARS-CoV), transmissible gastroenteritis virus, and HEV possess highly specific ADRP activity that converts Appr-1′′-p to ADP-ribose and inorganic phosphate (9, 20, 22). However, since the X domains of SARS-CoV, SFV, and HEV also possess poly-ADP-ribose binding activity, it has been questioned whether the enzymatic ADRP activity represents the sole relevant biological function (9). Structural data from cellular and viral X-domain proteins have revealed a common macro domain fold, including four conserved stretches of amino acids that line the ADP-ribose binding pocket and make up the catalytic center of viral ADRP enzymes (Fig. 1a) (9, 15). The first stretch contains two asparagines, of which the second one is absolutely essential for the catalytic ADRP activity of HCoV-229E and SARS-CoV (9, 20).
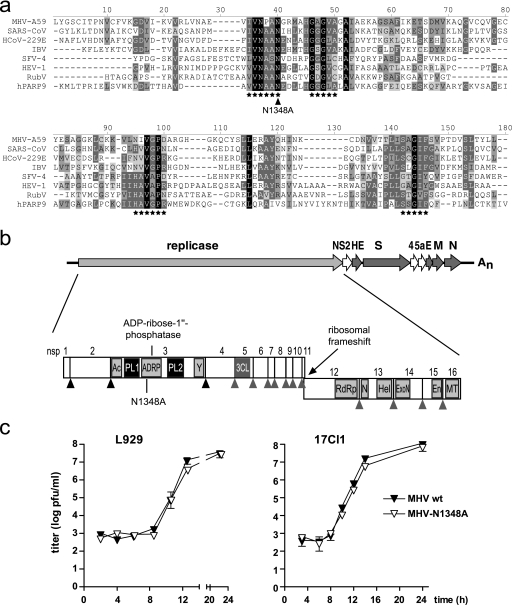
FIG. 1.
Generation of MHV-N1348. (a) Alignment of macro domain sequences. The presented alignment was produced by using AlignX of Vector-NTI-9 with manual adjustment and cross verification with previously published data (9, 10, 20, 22). Highlighted are amino acid stretches that line the substrate binding pocket (asterisks) (22) and MHV-A59 asparagine1348 (N1348A, arrowhead). MHV-A59, AY700211; SARS-CoV Frankfurt-1, AY291315; HCoV-229E, AF304460; infectious bronchitis virus Beaudette (IBV), NC_001451; SFV clone 4 (SFV-4), NC_003215; HEV genotype 1 (HEV-1), D10330; human poly-ADP-ribose-polymerase-9 (hPARP9), NM_031458; rubella virus (RubV), Q6X2V4. (b) Graphic representation of MHV-A59 genome organization. Replicase open reading frames and nsps 1 to 16 are depicted with papain-like proteinase (black triangles) and 3C-like proteinase (gray triangles) cleavage sites. Domains shown include ADRP; acidic domain (Ac); papain-like proteinase 1 (PL1); papain-like proteinase 2 (PL2); a domain of unknown function (Y) (10); 3C-like proteinase (3CL); RNA-dependent RNA polymerase (RdRp); NTPase (N); RNA helicase (Hel); 3′-to-5′ exoribonuclease (ExoN); endoribonuclease (En); and 2′-O-methyltransferase (MT). (c) Growth kinetics of MHV-A59 (MHV wt) and MHV-N1348A in L929 and 17Cl1 cells (MOI = 1). Data shown are mean values ± standard errors of the means (SEMs) from representative experiments performed in quadruplicate (L929) or duplicate (17Cl1).
Here we addressed the functional role of viral ADRP activity in a murine model of coronavirus infection. We generated and phenotypically analyzed an MHV ADRP catalytic core mutant that did not induce acute hepatitis in the natural host. The attenuated phenotype was associated with reduced interleukin 6 (IL-6)-expression in mice. Our data suggest a biological role for viral ADRPs in modulating the expression of proinflammatory immune mediators that impact virus pathogenicity.
MATERIALS AND METHODS
Mice and cells.
C57BL/6 (B6) mice were obtained from Charles River Laboratories (Sulzfeld, Germany). 129Sv and A129 mice (type I IFN receptor-deficient [IFNAR−/−] mice on the 129Sv background) (19) were obtained from the Institut für Labortierkunde (University of Zürich). IFNAR−/− mice on the B6 strain background (B6-IFNAR−/−) were a kind gift from M. Bachmann (Cytos Biotechnology, Schlieren, Switzerland). Mice were maintained in individually ventilated cages and were used at between 6 and 9 weeks of age. All animal experiments were performed in accordance with Swiss federal legislation on animal protection. BHK-21, L929, 293, and CV-1 cells were purchased from the European Collection of Cell Cultures. 17Clone1 (17Cl1), D980R, and BHK-MHV-N cells were described previously (6). All cell lines were maintained in minimal essential medium supplemented with fetal bovine serum (5% to 10%) and antibiotics. Murine peritoneal macrophages and bone marrow-derived cDCs and pDCs were generated and purified as described previously (41). The murine periportal Kupffer cell line KC-13 was a kind gift from R. Landmann, University Hospital, Basel, Switzerland, and was cultured as described previously (8).
Recombinant DNA and viruses.
Wild-type MHV-A59 was generated from a molecularly cloned cDNA (vMHV-inf-1) (6). To obtain vMHV-GFP, accessory gene 4 in vMHV-inf-1 was replaced by a gene encoding a fusion protein of enhanced green fluorescent protein (EGFP) and the lymphocytic choriomeningitis virus-derived cytotoxic T lymphocyte epitope KAVYNFATC (GP33-GFP), as described previously (41). Coronaviruses and recombinant vaccinia viruses were propagated and purified, and the titers of the viruses were determined as described previously (6, 13, 34). To generate the recombinant MHV mutants MHV-N1348A and MHV-N1348A-GFP, vMHV-inf-1 and vMHV-GFP were modified in two steps, using vaccinia virus-mediated homologous recombination and Escherichia coli guanine phosphoribosyltransferase (gpt) as a selection marker (13). Briefly, a plasmid DNA encoding the MHV-A59 nucleotides (nts) 3751 to 4161, the gpt gene, and MHV-A59 nts 5044 to 5460 was transfected into vMHV-inf-1- or vMHV-GFP-infected CV-1 cells, and recombined vaccinia viruses were obtained after gpt-positive selection (13). These viruses were used to recombine with a plasmid encoding MHV-A59 nts 3751 to 5460, which contained the N1348A-encoding mutation (4252AAT4254 to GCT). gpt-negative selection yielded the recombinant vaccinia virus clones vMHV-N1348A and vMHV-N1348A-GFP. Recombinant CoV MHV-N1348A and MHV-N1348A-GFP were obtained from supernatants of BHK-MHV-N cells that had been transfected with in vitro-transcribed RNA derived from vMHV-N1348A or vMHV-N1348A-GFP DNA templates (6).
Virus infections and determination of virus titers.
Peritoneal macrophages, Kupffer cells, pDCs, or cDCs (5 × 105 to 1 × 106) were infected with MHV at a multiplicity of infection (MOI) of 1 for 2 h at 37°C. IFN-α treatment of 1 × 105 peritoneal macrophages or cDCs prior to MHV infection at an MOI of 0.01 or 1 was performed, using universal type I IFN (IFN-αA/D; Sigma). Mice were injected intraperitoneally (i.p.) with 5, 500, or 50,000 PFU of MHV and analyzed at different time points as indicated in Results. Virus titers were assessed from frozen organs after they were weighed and homogenized or from the tissue culture supernatants by standard plaque assay on L929 cells.
Histology, ELISA, and ALT measurements.
Organs were fixed in 4% formalin and embedded in paraffin. Sections were stained with hematoxylin and eosin and analyzed by microscopy with a Leica DMRA microscope, a Leica DC300 FX camera, and IM2000 software (Leica, Heerbrugg, Switzerland). Images were processed using Adobe Photoshop (Adobe Systems, San Jose, CA). Commercial enzyme-linked immunosorbent assays (ELISAs) were used to determine the concentrations of IFN-α (PBL Biomedical Laboratories, Piscataway, NJ), tumor necrosis factor alpha (TNF-α), and IL-6 (BD Biosciences, Basel, Switzerland) according to the manufacturer's instructions. Serum ALT levels were measured by using a Hitachi 747 autoanalyzer (Tokyo, Japan).
RESULTS
Generation of a recombinant MHV encoding a mutated ADRP.
To assess the biological consequences of viral ADRP expression, we generated a MHV-A59 mutant lacking ADRP activity. Asparagine1348 encoded by the MHV-A59 replicase gene was replaced with alanine, and the resulting recombinant MHV mutant was designated MHV-N1348A (Fig. 1b). The corresponding asparagine residue is highly conserved in macro domain proteins (Fig. 1a), and structural data derived from crystallized SARS-CoV ADRP further suggests that this residue is directly involved in Appr-1′′-p processing, since it is located in the catalytic center of the enzyme (9, 22). In addition, biochemical analyses revealed that an asparagine-to-alanine substitution at this position completely abrogated the enzymatic activity of the SARS-CoV and HCoV-229E ADRPs, confirming that this residue is essential for the catalytic activity of coronaviral ADRPs (9, 20).
To assess the growth kinetics of MHV-N1348A, murine L929 and 17Cl1 cells were infected (MOI = 1) with MHV-N1348A or wild-type MHV-A59 (Fig. 1c). Analyses of virus production revealed that MHV-N1348A and wild-type MHV-A59 grew with similar kinetics and reached equal final titers. Furthermore, the plaque sizes and morphologies were indistinguishable (data not shown). To determine if there was a selective pressure for the generation of revertant viruses in tissue culture, we grew MHV-N1348A over 12 passages in L929 cells. Sequence analysis of the ADRP-coding region confirmed that the N1348A substitution remained stably encoded and that no revertant viruses had emerged. Thus, in accordance with a report on a corresponding HCoV-229E ADRP mutant (20), the MHV-A59 replicase-encoded ADRP activity is dispensable for viral RNA synthesis and does not impact virus growth kinetics in tissue culture.
MHV-N1348A does not cause acute viral hepatitis.
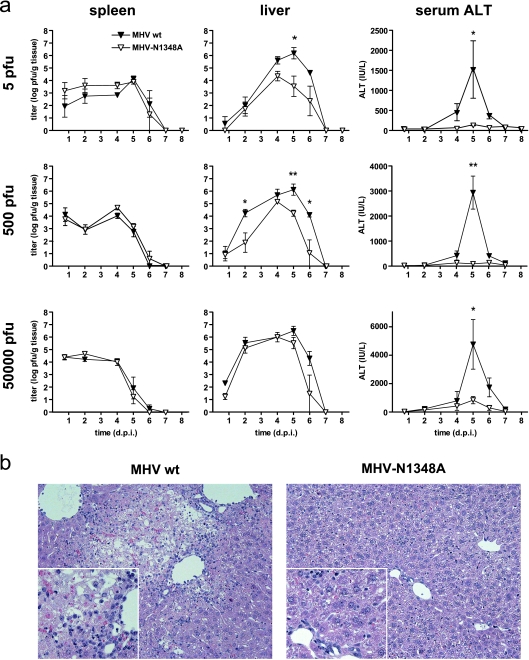
To investigate if the virally encoded ADRP activity interferes with viral replication in the natural host, we infected B6 mice i.p. with low, intermediate, and high doses (5, 500, and 50,000 PFU, respectively) of wild-type MHV-A59 or MHV-N1348A (Fig. 2a). The viral titers in spleens for the two viruses were comparable at all virus doses used until virus clearance at days 6 to 7 p.i.. In livers, we detected significantly reduced titers of MHV-N1348A at low and intermediate doses, whereas at high virus doses no significant differences in growth kinetics were observed. In contrast to the rather moderate growth differences, serum ALT levels, which serve as a marker for MHV-induced acute hepatitis (5, 41), were dramatically increased in wild-type MHV-A59- infected mice but not in MHV-N1348A-infected mice, irrespective of which virus dose was used for infection (Fig. 2a). These observations correlated well with the absence of hepatocyte necrosis and parenchymal inflammation following MHV-N1348A infection (Fig. 2b). Collectively, these data demonstrate that the asparagine-to-alanine substitution in the MHV-A59 ADRP domain results in attenuation in the natural host and, in particular, abolishes the ability of MHV to cause severe liver damage.
FIG. 2.
MHV-N1348A does not cause acute viral hepatitis. (a) B6 mice (n = 3 to 10) were infected with 5, 500, or 50,000 PFU of wild-type MHV-A59 (MHV wt) or MHV-N1348A. Viral titers in spleens and livers and serum ALT values were determined at the indicated time points p.i. Statistical analyses of titers and ALT values were performed, using Student's t test and the Mann-Whitney nonparametric test, respectively (*, P < 0.05; **, P < 0.01). d.p.i., day p.i. (b) Hematoxylin and eosin staining of 4% formaldehyde-fixed liver sections at day 5 p.i of B6 mice infected with 500 PFU (magnification, ×200; inset, ×400) of wild-type MHV-A59 or MHV-N1348A.
MHV-N1348 replication in macrophages and DCs.
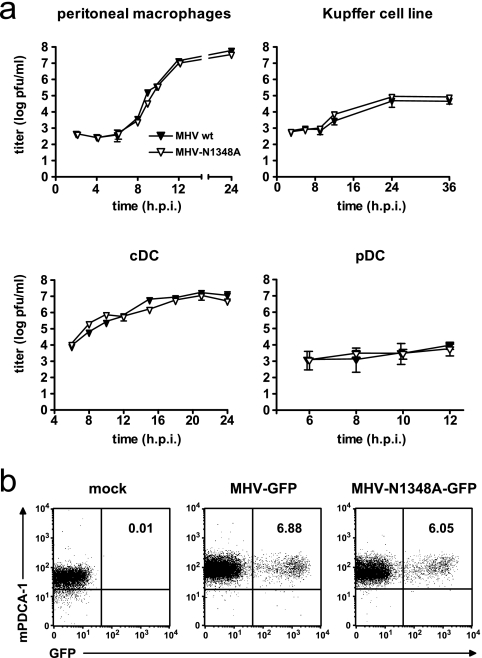
In order to assess if MHV-N1348 replication is impaired in important MHV-A59 target cells, we infected primary peritoneal macrophages, bone marrow-derived cDCs and pDCs, and a Kupffer cell line (KC-13) (7) with MHV-N1348A or wild-type MHV-A59 (MOI = 1). As shown in Fig. 3a, both viruses displayed similar growth kinetics in each cell type. Notably, both viruses replicated to only low titers in pDCs, suggesting that they are both controlled in this cell type. It has been proposed previously that MHV replication in pDCs is controlled through pDC-derived IFN-α (4). However, to confirm that both wild-type MHV-A59 and MHV-N1348A indeed replicate in pDCs, we used a corresponding virus pair expressing GFP (MHV-GFP and MHV-N1348-GFP; see Materials and Methods) for pDC infection. As shown in Fig. 3b, fluorescence-activated cell sorter analysis revealed that MHV-GFP- and MHV-N1348-GFP-infected pDCs displayed green fluorescence indicative of viral replication. Thus, the growth kinetics of wild-type MHV-A59 and MHV-N1348A in macrophages and DCs were indistinguishable.
FIG. 3.
MHV-N1348A replication in macrophages and DCs. (a) Peritoneal macrophages, a Kupffer cell line, and bone marrow-derived cDCs and pDCs were infected with wild-type MHV-A59 or MHV-N1348A (MOI = 1). Viral titers in supernatants were measured at the indicated time points. The data shown represent mean values ± SEMs from two independent experiments. (b) Flow cytometric analysis of frequency of GFP-expressing bone marrow-derived pDCs (B220+ CD11chigh mPDCA+) after infection with MHV-GFP or MHV-N1348A-GFP (MOI = 10). The values indicated in the upper right quadrants are the percentages of GFP-positive cells.
MHV-N1348 infection results in elevated IFN-α expression in pDCs.
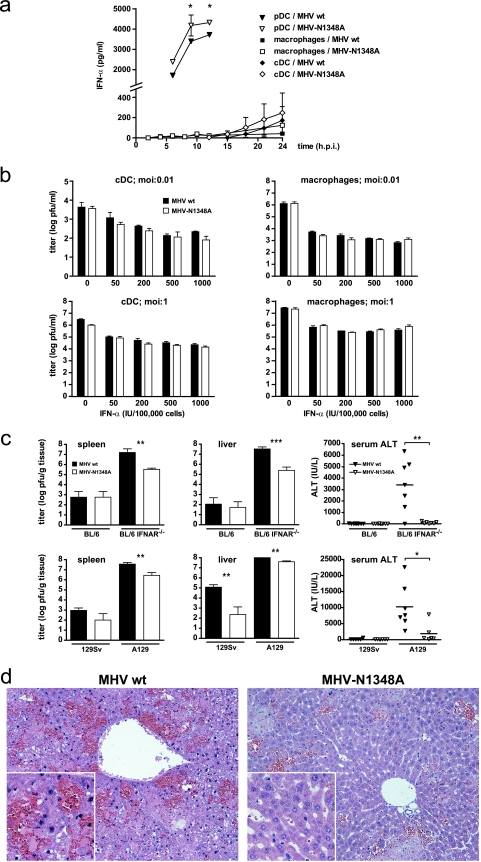
The control of MHV-A59 in mice is strongly dependent on swift IFN-α expression by pDCs during the early phase of infection (5). We found no difference in the ability of MHV-N1348A to replicate in pDCs (Fig. 3); however, compared to that for wild-type MHV-A59 infection, IFN-α production was significantly increased following MHV-N1348A infection (Fig. 4a). Notably, neither macrophages nor cDCs produced elevated IFN-α levels upon MHV-N1348A infection, suggesting that, similar to that for wild-type MHV-A59, immediate IFN-α expression upon MHV-N1348A infection is still restricted to pDCs. We also assessed if MHV-N1348A replication is more vulnerable to IFN-α pretreatment than wild-type MHV-A59 replication. Macrophages and cDCs were pretreated with different dosages of IFN-α and, 4 h later, were infected with wild-type MHV-A59 or MHV-N1348A. As shown in Fig. 4b, the viral titers determined at 18 h p.i. were indistinguishable between wild-type MHV-A59 and MHV-N1348A infections, irrespective of whether the infections were done at low (0.01) or high (1) MOIs.
FIG. 4.
MHV-N1348A and type I IFNs. (a) Bone marrow-derived pDCs and cDCs, as well as peritoneal macrophages, were infected with wild-type MHV-A59 (MHV wt) or MHV-N1348A (MOI = 1). The levels of IFN-α in the supernatants were measured at the indicated time points. For macrophages and cDCs, the data represent the mean values ± SEMs from two independent experiments. For pDCs, the data represent the mean values ± SEMs from one experiment representative of three. (b) Bone marrow-derived cDCs and peritoneal macrophages were pretreated with the indicated amounts of IFN-α for 4 h and then infected with wild-type MHV-A59 or MHV-N1348A (MOI = 0.01 and 1). The titers in the supernatants were determined at 18 h p.i. The data represent the mean values ± SEMs from a quadruplicate experiment. (c) B6, B6-IFNAR−/−, 129Sv, and A129 mice (n = 6 to 9) were infected with 5 PFU of wild-type MHV-A59 or MHV-N1348A. The viral titers in the spleen and liver and serum ALT levels were measured at 48 h p.i. Statistical analyses of titers and ALT levels were performed, using Student's t test and the Mann-Whitney nonparametric test, respectively (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (d) Hematoxylin and eosin staining of 4% formaldehyde-fixed liver sections at 48 h p.i of A129 mice infected with 5 PFU of wild-type MHV-A59 or MHV-N1348A (×200; inset, ×400).
MHV-N1348A is attenuated in livers of IFNAR−/− mice.
The phenotypic analysis of MHV-N1348A revealed a profound impact of this mutant virus on the induction of viral hepatitis in mice, and we observed elevated IFN-α expression in MHV-N1348A-infected pDCs. To clarify if the lack of acute viral hepatitis is related to elevated IFN-α expression, we infected type I IFN receptor-deficient mice (IFNAR−/− mice) that cannot respond to type I IFNs. MHV-A59 can replicate essentially uncontrolled in IFNAR−/− mice, and these mice succumb to the infection at day 2 p.i. (4). Therefore, at day 2 p.i. we assessed the viral titers in the spleens and livers and the serum ALT values of IFNAR−/− mice (on the B6 or 129Sv background) that were infected with 5 PFU (i.p.) of wild-type MHV-A59 or MHV-N1348A. As shown in Fig. 4c, wild-type MHV-A59 and MHV-N1348A titers were increased by several orders of magnitude in mice lacking the IFNAR compared to those for wild-type mice. However, MHV-N1348A titers did not fully reach those of wild-type MHV-A59 in B6-IFNAR−/− and A129 (IFNAR−/− on the 129Sv background) mice. Consistent with the observation of comparable reductions of mutant and wild-type virus replication in IFN-α-pretreated cells in vitro (Fig. 4b), these data suggest that elevated IFN-α expression also may not lead to gross differences in viral growth kinetics between wild-type MHV-A59 and MHV-N1348A in vivo. Remarkably, however, despite efficient growth of MHV-N1348A in IFNAR−/− mice, serum ALT values were significantly lower than in IFNAR−/− mice infected with wild-type MHV-A59 (Fig. 4c). Histological analysis revealed that wild-type MHV-A59 induced acute hemorrhagic liver disease with massive hepatocyte necrosis within 48 h, whereas the livers of MHV-N1348A-infected IFNAR−/− mice remained healthy, with only minor leukocyte infiltration (Fig. 4c). Collectively, we concluded that the host type I IFN response has only a minor effect on MHV-N1348A growth in vivo and that the observed reduction of liver pathogenicity of MHV-N1348A is still apparent in type I IFN receptor-deficient mice.
MHV-N1348A infection results in diminished expression of inflammatory cytokines.
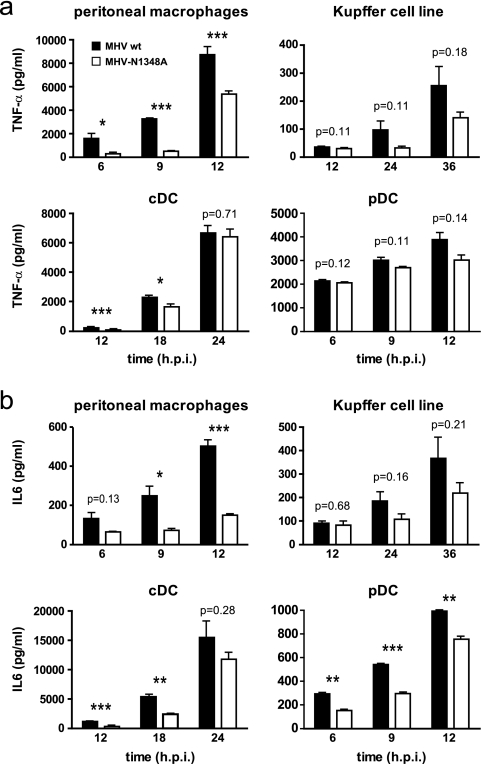
Since the impact of IFN-α appeared of minor importance in relation to the observed reduced liver pathology, we assessed the expression of the proinflammatory cytokines TNF-α and IL-6 in MHV-infected cells. To this end, primary peritoneal macrophages, a Kupffer cell line, and bone-marrow derived cDC and pDCs were investigated, as they represent both major cytokine producer cells and MHV target cells. As shown in Fig. 3, wild-type MHV-A59 and MHV-N1348A grew with similar kinetics in each of these cell types. However, we detected reduced amounts of TNF-α and IL-6 in the supernatants of mutant virus-infected cells compared to those for wild-type virus infections (Fig. 5). Particularly in peritoneal macrophages, MHV-N1348A elicited significantly lower TNF-α and IL-6 responses than wild-type MHV-A59. Likewise, in a Kupffer cell line we consistently observed lower TNF-α and IL-6 expression levels following MHV-N1348A infection. In cDCs and pDCs, IL-6 expression was significantly lower following MHV-N1348A infection, whereas TNF-α expression was induced to comparable levels in both wild-type and mutant virus infections.
FIG. 5.
MHV-N1348A-induced inflammatory cytokine expression in vitro. (a and b) Peritoneal macrophages, a Kupffer cell line, and bone marrow-derived cDCs and pDCs were infected with wild-type MHV-A59 (MHV wt) or MHV-N1348A (MOI = 1). TNF-α and IL-6 production in supernatants was measured by ELISA at the indicated time points. The data show mean values ± SEMs for representative experiments performed in triplicate (macrophages, Kupffer cell line, and cDC) or duplicate (pDC). Statistical analyses were performed using Student's t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Assessment of IFN-α, TNF-α, and IL-6 expression in MHV-infected mice.
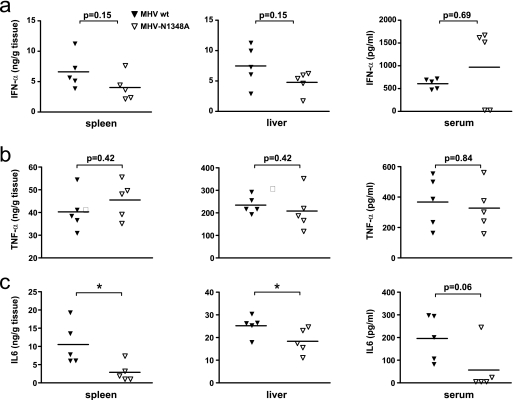
Finally, we compared IFN-α, TNF-α, and IL-6 production in wild-type MHV-A59- and MHV-N1348A-infected mice. In order to choose an appropriate virus dose for this experiment, we took into account that virus titers greatly influence IFN-α and inflammatory cytokine expression levels. For example, infection with low or intermediate viral doses (e.g., 5 or 500 PFU) resulted in approximately 100-fold-higher wild-type MHV-A59 than MHV-N1348A titers in livers at day 5 p.i. (Fig. 2a). We therefore decided to measure IFN-α, TNF-α, and IL-6 after infection with a high viral dose at day 4 p.i., since under these conditions viral titers in the livers and spleens of mutant virus-infected mice were indistinguishable from those of wild-type virus-infected mice (Fig. 2a, bottom panel). The assessment of IFN-α revealed no significant differences in IFN-α expression in the spleen, liver, and serum (Fig. 6a), further suggesting that a potentially elevated level of IFN-α production by MHV-N1348A-infected pDCs may not play a major role in the apparent lack of liver pathology in MHV-N1348A-infected mice. We next assessed TNF-α expression in mice and observed comparable expression levels in wild-type MHV-A59 and MHV-N1348A infections (Fig. 6b). In contrast, IL-6 expression was significantly reduced in the spleens and livers of MHV-N1348A-infected mice compared to that in the spleens and livers of wild-type virus-infected mice (Fig. 6c), indicating that, in vivo, MHV-N1348A infection rather selectively impacts the production of this inflammatory cytokine.
FIG. 6.
MHV-N1348A-induced IFN-α and inflammatory cytokine expression in vivo. 129Sv mice (n = 5) were infected with 5 × 104 PFU of wild-type MHV-A59 (MHV wt) or MHV-N1348A. The levels of IFN-α (a), TNF-α (b), and IL-6 (c) in spleens, livers, and sera were determined 4 days p.i. by ELISA. Statistical analyses were performed by using the Mann-Whitney nonparametric test (*, P < 0.05).
DISCUSSION
Despite the long-recognized conservation of a viral X domain in coronaviruses and RNA viruses of the alpha-like supergroup, its biological role remained elusive. Here we provide conclusive evidence that the X domain of a murine CoV impacts viral pathogenicity. Specifically, a single-amino-acid substitution in the ADRP catalytic core of the hepatotropic MHV-A59 prevented induction of acute viral hepatitis in mice. Mechanistically, our data support the idea that expression of the coronaviral X domain interferes with host inflammatory and possibly innate responses, since compared to those for wild-type virus infections, we observed increased IFN-α expression in pDCs and diminished proinflammatory cytokine expression in DCs and macrophages following MHV ADRP mutant infection in vitro.
Type I IFN and proinflammatory cytokine production is one of the earliest events in acute hepatitis. These mediators initiate the production of other cytokines and chemokines, resulting in the recruitment of inflammatory cells and the generation of an inflammatory microenvironment that can lead to extensive hepatocyte death and liver damage. The phenotype of MHV-N1348A indicates that the MHV X domain is likely to be involved in inflammatory processes that exacerbate MHV-induced liver pathology. We have thoroughly assessed MHV-N1348A replication in mice, using a variety of experimental settings. Most strikingly, the mutant virus did not induce acute viral hepatitis irrespective of whether infections were done with low, intermediate, or high viral doses. In contrast to those of wild-type MHV-A59-infected mice, the livers of MHV-N1348A-infected mice remained healthy and displayed neither marked hepatocyte necrosis nor characteristic parenchymal infiltration of leukocytes, and accordingly, serum ALT values remained low. Interestingly, mutant virus titers were only slightly reduced in mice after low- and intermediate-dose infections, and with high-dose infections there were no detectable growth differences observed. It is conceivable that the slightly reduced growth of MHV-N1348A after low- or intermediate-dose infection resulted from increased pDC-mediated type I IFN expression, as the mutant virus was shown to be equally as sensitive to IFN-α pretreatment as wild-type MHV-A59. However, under conditions of rapid growth to high titers (i.e., after a high-dose infection) the potential effect of type I IFN in reducing MHV-N1348A replication may be overridden. Collectively, our analyses revealed that increased IFN-α expression by MHV-N1348A-infected pDCs may not have a major impact on the greatly reduced liver pathology in MHV-N1348A-infected mice. We could also demonstrate that the MHV X-domain mutant affected host cell inflammatory cytokine expression in DCs and macrophages, resulting in reduced levels compared to those for wild-type MHV-A59 infection. Notably, reduced IL-6 production was also observed in MHV-N1348A-infected mice, providing a reasonable explanation for the absence of liver pathology. However, delineation of the extent to which decreased inflammatory cytokine expression may prevent severe liver pathology in MHV-N1348A infections and which cytokines are important in that respect requires further studies. Thus, it is likely that a number of cytokines and chemokines are differentially expressed in MHV-N1348A infections, and we have already commenced comparative transcriptional profiling, using microarrays to gain more insight into cellular pathways that may be affected by the MHV X domain. Our data also show that the magnitude of inflammatory cytokine expression greatly varies depending on the cell type infected. For example we have observed that IL-6 production in cDCs exceeds that in macrophages by at least one order of magnitude after MHV infection. Therefore, in order to delineate the impact of individual cell types on the induction of viral hepatitis, future studies have to carefully take into account which and how many target cells are infected at which time points p.i.
The coronaviral ADRP may also impact other coronavirus-induced diseases, such as SARS, feline infectious peritonitis, and avian infectious bronchitis. It has been reported that inflammatory cytokines play pivotal roles in these infections and that their expression levels may be decisive for the severity of disease (1, 24, 32). Moreover, single-amino-acid substitutions within the SFV X domain, or combinations thereof, were reported to significantly alter neurovirulence and survival in a mouse model (35). Therefore, it is tempting to speculate that the X domains of other viruses of the alpha-like supergroup, such as the HEV X domain, may modulate host innate and inflammatory responses. The availability of reverse genetic techniques for the generation of recombinant HEV will certainly help to address this issue. However, future studies should also aim at extending our knowledge of HEV target cell tropism and clarify (i) if HEV can infect macrophages or DCs and (ii) if these cell types represent major type I IFN and/or cytokine producer cells during HEV infection.
The apathogenic phenotype of MHV-N1348A should encourage further studies to identify molecular targets of viral X domains. The ADRP activity of the coronaviral X domain was proposed because of its homology to cellular macro domain proteins that process Appr-1′′-p in cellular pre-tRNA splicing (26). However, it is unlikely that viral X domains are involved in pre-tRNA splicing, since they are replicase encoded and, at least in coronavirus infections, colocalize with other replicase proteins at perinuclear cytoplasmic membranes where RNA synthesis takes place (17, 27, 36). Notably, Appr-1′′-p is also produced during cytoplasmic splicing of the mRNA encoding XBP1, a mediator of the unfolded-protein response of the endoplasmic reticulum (21). Alternatively or in addition to an ADRP activity, viral X domains may also function as ADP-ribose binding modules, as has been proposed for cellular macro domain proteins (15). Viral X domains are connected to enzymatic functions within the replicase polyprotein and may direct these functions to their molecular substrates or targets. For example, the coronaviral X domain is expressed on nsp3 together with papain-like proteinase domains that have been shown to possess deubiquitinating activity and to function as an IFN antagonist (2, 7, 18).
In conclusion, we demonstrated that the MHV ADRP domain is a pathogenicity factor affecting inflammatory processes associated with the induction of acute viral hepatitis. The evolutionary conservation of viral X domains among positive-strand RNA viruses of the alpha-like supergroup suggests that this domain may also have an impact on inflammatory processes in other virus infections and may, therefore, represent a decisive factor in the clinical outcomes of a number of RNA virus-induced animal and human diseases.
Acknowledgments
This work was supported by the Swiss National Science Foundation and the European Commission (SARS-DTV SP22-CT-2004-511064, AIDSCoVAC LSHP-CT-2006-037416, and TOLERAGE HEALTH-F4-2008-202156).
We thank Regine Landmann, University Hospital, Basel, Switzerland, and Martin Bachmann, Cytos Biotechnology, Schlieren, Switzerland, for valuable cell lines and mice and Reinhard Maier for critically reading the manuscript.
Footnotes
Published ahead of print on 15 October 2008.
REFERENCES
- 1.Asif, M., J. W. Lowenthal, M. E. Ford, K. A. Schat, W. G. Kimpton, and A. G. Bean. 2007. Interleukin-6 expression after infectious bronchitis virus infection in chickens. Viral Immunol. 20479-486. [DOI] [PubMed] [Google Scholar]
- 2.Barretto, N., D. Jukneliene, K. Ratia, Z. Chen, A. D. Mesecar, and S. C. Baker. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 7915189-15198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann, C. C., T. E. Lane, and S. A. Stohlman. 2006. Coronavirus infection of the central nervous system: host-virus stand-off. Nat. Rev. Microbiol. 4121-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bertoletti, A., and M. K. Maini. 2000. Protection or damage: a dual role for the virus-specific cytotoxic T lymphocyte response in hepatitis B and C infection? Curr. Opin. Microbiol. 3387-392. [DOI] [PubMed] [Google Scholar]
- 5.Cervantes-Barragan, L., R. Zust, F. Weber, M. Spiegel, K. S. Lang, S. Akira, V. Thiel, and B. Ludewig. 2007. Control of coronavirus infection through plasmacytoid dendritic-cell-derived type I interferon. Blood 1091131-1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coley, S. E., E. Lavi, S. G. Sawicki, L. Fu, B. Schelle, N. Karl, S. G. Siddell, and V. Thiel. 2005. Recombinant mouse hepatitis virus strain A59 from cloned, full-length cDNA replicates to high titers in vitro and is fully pathogenic in vivo. J. Virol. 793097-3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devaraj, S. G., N. Wang, Z. Chen, M. Tseng, N. Barretto, R. Lin, C. J. Peters, C. T. Tseng, S. C. Baker, and K. Li. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 28232208-32221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dory, D., H. Echchannaoui, M. Letiembre, F. Ferracin, J. Pieters, Y. Adachi, S. Akashi, W. Zimmerli, and R. Landmann. 2003. Generation and functional characterization of a clonal murine periportal Kupffer cell line from H-2Kb-tsA58 mice. J. Leukoc. Biol. 7449-59. [DOI] [PubMed] [Google Scholar]
- 9.Egloff, M. P., H. Malet, A. Putics, M. Heinonen, H. Dutartre, A. Frangeul, A. Gruez, V. Campanacci, C. Cambillau, J. Ziebuhr, T. Ahola, and B. Canard. 2006. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J. Virol. 808493-8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko, and V. M. Blinov. 1989. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 174847-4861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorbalenya, A. E., E. V. Koonin, and M. M. Lai. 1991. Putative papain-related thiol proteases of positive-strand RNA viruses. Identification of rubi- and aphthovirus proteases and delineation of a novel conserved domain associated with proteases of rubi-, alpha- and coronaviruses. FEBS Lett. 288201-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haller, O., G. Kochs, and F. Weber. 2006. The interferon response circuit: induction and suppression by pathogenic viruses. Virology 344119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hertzig, T., E. Scandella, B. Schelle, J. Ziebuhr, S. G. Siddell, B. Ludewig, and V. Thiel. 2004. Rapid identification of coronavirus replicase inhibitors using a selectable replicon RNA. J. Gen. Virol. 851717-1725. [DOI] [PubMed] [Google Scholar]
- 14.Heydtmann, M., P. Shields, G. McCaughan, and D. Adams. 2001. Cytokines and chemokines in the immune response to hepatitis C infection. Curr. Opin. Infect. Dis. 14279-287. [DOI] [PubMed] [Google Scholar]
- 15.Karras, G. I., G. Kustatscher, H. R. Buhecha, M. D. Allen, C. Pugieux, F. Sait, M. Bycroft, and A. G. Ladurner. 2005. The macro domain is an ADP-ribose binding module. EMBO J. 241911-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Katze, M. G., Y. He, and M. Gale, Jr. 2002. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2675-687. [DOI] [PubMed] [Google Scholar]
- 17.Knoops, K., M. Kikkert, S. H. Worm, J. C. Zevenhoven-Dobbe, Y. van der Meer, A. J. Koster, A. M. Mommaas, and E. J. Snijder. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindner, H. A., N. Fotouhi-Ardakani, V. Lytvyn, P. Lachance, T. Sulea, and R. Menard. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 7915199-15208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Müller, U., U. Steinhoff, L. F. Reis, S. Hemmi, J. Pavlovic, R. M. Zinkernagel, and M. Aguet. 1994. Functional role of type I and type II interferons in antiviral defense. Science 2641918-1921. [DOI] [PubMed] [Google Scholar]
- 20.Putics, Á., W. Filipowicz, J. Hall, A. E. Gorbalenya, and J. Ziebuhr. 2005. ADP-ribose-1′′-monophosphatase: a conserved coronavirus enzyme that is dispensable for viral replication in tissue culture. J. Virol. 7912721-12731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ron, D., and P. Walter. 2007. Signal integration in the endoplasmic reticulum unfolded protein response. Nat. Rev. Mol. Cell Biol. 8519-529. [DOI] [PubMed] [Google Scholar]
- 22.Saikatendu, K. S., J. S. Joseph, V. Subramanian, T. Clayton, M. Griffith, K. Moy, J. Velasquez, B. W. Neuman, M. J. Buchmeier, R. C. Stevens, and P. Kuhn. 2005. Structural basis of severe acute respiratory syndrome coronavirus ADP-ribose-1′′-phosphate dephosphorylation by a conserved domain of nsP3. Structure 131665-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salazar-Mather, T. P., T. A. Hamilton, and C. A. Biron. 2000. A chemokine-to-cytokine-to-chemokine cascade critical in antiviral defense. J. Clin. Investig. 105985-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pang, B. S., Z. Wang, L. M. Zhang, Z. H. Tong, L. L. Xu, X. X. Huang, W. J. Guo, M. Zhu, C. Wang, X. W. Li, Z. P. He, H. X. Li, and F. J. Zhao. 2003. Dynamic changes in blood cytokine levels as clinical indicators in severe acute respiratory syndrome. Chinese Med. J. 1161283-1287. [PubMed] [Google Scholar]
- 25.Siegal, F. P., N. Kadowaki, M. Shodell, P. A. Fitzgerald-Bocarsly, K. Shah, S. Ho, S. Antonenko, and Y. J. Liu. 1999. The nature of the principal type 1 interferon-producing cells in human blood. Science 2841835-1837. [DOI] [PubMed] [Google Scholar]
- 26.Snijder, E. J., P. J. Bredenbeek, J. C. Dobbe, V. Thiel, J. Ziebuhr, L. L. Poon, Y. Guan, M. Rozanov, W. J. Spaan, and A. E. Gorbalenya. 2003. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 331991-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snijder, E. J., Y. van der Meer, J. Zevenhoven-Dobbe, J. J. Onderwater, J. van der Meulen, H. K. Koerten, and A. M. Mommaas. 2006. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 805927-5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sperry, S. M., L. Kazi, R. L. Graham, R. S. Baric, S. R. Weiss, and M. R. Denison. 2005. Single-amino-acid substitutions in open reading frame (ORF) 1b-nsp14 and ORF 2a proteins of the coronavirus mouse hepatitis virus are attenuating in mice. J. Virol. 793391-3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinman, R. M., and H. Hemmi. 2006. Dendritic cells: translating innate to adaptive immunity. Curr. Top. Microbiol. Immunol. 31117-58. [DOI] [PubMed] [Google Scholar]
- 30.Szabo, G., and A. Dolganiuc. 2005. Subversion of plasmacytoid and myeloid dendritic cell functions in chronic HCV infection. Immunobiology 210237-247. [DOI] [PubMed] [Google Scholar]
- 31.Szabo, G., P. Mandrekar, and A. Dolganiuc. 2007. Innate immune response and hepatic inflammation. Semin. Liver Dis. 27339-350. [DOI] [PubMed] [Google Scholar]
- 32.Takano, T., T. Hohdatsu, A. Toda, M. Tanabe, and H. Koyama. 2007. TNF-alpha, produced by feline infectious peritonitis virus (FIPV)-infected macrophages, upregulates expression of type II FIPV receptor feline aminopeptidase N in feline macrophages. Virology 36464-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thiel, V. 2007. Reverse genetic analysis of coronavirus replication, p. 109-132. In V. Thiel (ed.), Coronaviruses: molecular and cellular biology. Caister Academic Press, Norfolk, United Kingdom.
- 34.Thiel, V., J. Herold, B. Schelle, and S. G. Siddell. 2001. Infectious RNA transcribed in vitro from a cDNA copy of the human coronavirus genome cloned in vaccinia virus. J. Gen. Virol. 821273-1281. [DOI] [PubMed] [Google Scholar]
- 35.Tuittila, M., and A. E. Hinkkanen. 2003. Amino acid mutations in the replicase protein nsP3 of Semliki Forest virus cumulatively affect neurovirulence. J. Gen. Virol. 841525-1533. [DOI] [PubMed] [Google Scholar]
- 36.van der Meer, Y., E. J. Snijder, J. C. Dobbe, S. Schleich, M. R. Denison, W. J. Spaan, and J. K. Locker. 1999. Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 737641-7657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Visvanathan, K., and S. R. Lewin. 2006. Immunopathogenesis: role of innate and adaptive immune responses. Semin. Liver Dis. 26104-115. [DOI] [PubMed] [Google Scholar]
- 38.Weiss, S. R., and S. Navas-Martin. 2005. Coronavirus pathogenesis and the emerging pathogen severe acute respiratory syndrome coronavirus. Microbiol. Mol. Biol. Rev. 69635-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wentworth, D. E., and K. V. Holmes. 2007. Coronavirus binding and entry, p. 3-32. In V. Thiel (ed.), Coronaviruses: molecular and cellular biology. Caister Academic Press, Norfolk, United Kingdom.
- 40.Ziebuhr, J., B. Schelle, N. Karl, E. Minskaia, S. Bayer, S. G. Siddell, A. E. Gorbalenya, and V. Thiel. 2007. Human coronavirus 229E papain-like proteases have overlapping specificities but distinct functions in viral replication. J. Virol. 813922-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Züst, R., L. Cervantes-Barragan, T. Kuri, G. Blakqori, F. Weber, B. Ludewig, and V. Thiel. 2007. Coronavirus non-structural protein 1 is a major pathogenicity factor: implications for the rational design of coronavirus vaccines. PLoS Pathog. 3e109. [DOI] [PMC free article] [PubMed] [Google Scholar]