Abstract
Among retroviruses, lentiviruses are unusual in their ability to efficiently infect both dividing and nondividing cells, such as activated T cells and macrophages, respectively. Recent studies implicate the viral capsid protein (CA) as a key determinant of cell-cycle-independent infection by human immunodeficiency virus type 1 (HIV-1). We investigated the effects of the host cell protein cyclophilin A (CypA), which binds to HIV-1 CA, on HIV-1 infection of nondividing cells. The HIV-1 CA mutants A92E, T54A, and R132K were impaired for infection of aphidicolin-arrested HeLa cells, but not HOS cells. The mutants synthesized normal quantities of two-long-terminal-repeat circles in arrested HeLa cells, indicating that the mutant preintegration complexes can enter the nuclei of both dividing and nondividing cells. The impaired infectivity of the CA mutants on both dividing and nondividing HeLa cells was relieved by either pharmacological or genetic disruption of the CypA-CA interaction or by RNA interference-mediated depletion of CypA expression in target cells. A second-site suppressor of the CypA-restricted phenotype also restored the ability of CypA-restricted HIV-1 mutants to infect growth-arrested HeLa cells. These results indicate that CypA-restricted mutants are specifically impaired at a step between nuclear import and integration in nondividing HeLa cells. This study reveals a novel target cell-specific restriction of HIV-1 CA mutants in nondividing cells that is dependent on CypA-CA interactions.
Following fusion of the viral envelope with a susceptible target cell, retroviruses undergo a poorly defined process of uncoating of the viral core. Uncoating involves dissociation of the viral capsid and is required for efficient reverse transcription in target cells (18). Reverse transcription occurs in the cytoplasm and results in the formation of a high-molecular-weight complex of proviral DNA bound by viral proteins, including NC and IN. This “preintegration complex” (PIC) is competent for integration, but it must first enter the cell nucleus.
A defining characteristic of lentiviruses is their ability to efficiently infect nondividing cells (22, 27, 52). Such cells include terminally differentiated macrophages, a relevant target of human immunodeficiency virus type 1 (HIV-1) in vivo. In contrast, gammaretroviruses, such as murine leukemia virus (MLV), enter nondividing cells but are impaired for entry into the nucleus. Efficient infection by these viruses depends on active cell cycle progression, specifically the mitotic stage (28). Despite intensive study, the mechanism employed by lentiviruses to infect nondividing cells is poorly understood. Previous studies have focused on the nuclear location signals present within the HIV-1 proteins, including matrix (MA), integrase (IN), and viral protein R (Vpr) (8, 14, 20, 25, 52). Such studies revealed roles of these proteins in nuclear import and/or infection of arrested cells, but these conclusions have not endured. Subsequent studies of the central polypurine tract and the DNA flap, a structure synthesized during reverse transcription, implicated these cis-acting elements in viral-DNA docking and nuclear entry (2, 3, 56), but the central polypurine tract requirement for infection of nondividing cells has been inconsistently observed (13, 29). More recently, studies of chimeric MLV/HIV-1 have revealed a role of the capsid protein (CA) in infection of aphidicolin-arrested HeLa cells and monocyte-derived macrophages (51). Specific amino acid substitutions in CA were then shown to result in selective impairment for infection of nondividing cells (53). These findings have led to the notion that CA protein is a key determinant for the ability of HIV-1 to infect nondividing cells, suggesting a potential link between uncoating and nuclear import.
HIV-1 and some other lentiviruses encode Gag proteins that bind specifically to the host protein cyclophilin A (CypA) (6, 30, 33). CypA is specifically incorporated into HIV-1 particles via binding to a flexible loop in the N-terminal domain of CA between helices IV and V (7, 21). An initial model postulated that incorporation of CypA into HIV-1 particles facilitates uncoating of the viral core in target cells (31). More recent studies have demonstrated that incorporation of CypA into virions appears to be biologically irrelevant; rather, it is the binding of CypA to the viral core in the target cell that promotes infection (23, 42, 47). Although the CA-CypA interaction is well characterized in vitro, the precise role of CypA in promoting HIV-1 infection is unclear despite intensive study (reviewed in references 32 and 46).
Cyclosporine (CsA) is an immunosuppressive drug that binds CypA, promoting its interaction with calcineurin, an inhibitor of the host protein phosphatase 2A. CsA thus competitively inhibits the interaction of CypA with HIV-1 CA. When added at the time of virus inoculation, CsA reduces the infectivity of most strains of HIV-1. In CsA-treated T-cell cultures, HIV-1 spread is also inhibited. Several HIV-1 CA mutations have been identified that enable HIV-1 replication in the presence of CsA and some of its nonimmunosuppressive analogues (1, 6, 10). The mutant viruses can replicate in the presence of CsA and actually require the drug for efficient infection of some cell types; they are therefore referred to as CsA-resistant/dependent mutants. Two such mutations, A92E and G94D, map to the CypA binding loop within CA. These mutants infect nonpermissive cell types, such as HeLa and CEM, only in the presence of the drug, while infection of permissive cell lines (e.g., HOS and Jurkat) is efficient and is not significantly affected by the drug. Additional CA mutations conferring a CsA-resistant/dependent phenotype have been identified outside the CypA binding loop, including T54A and R132K (40, 54). The mechanism by which these substitutions render HIV-1 dependent on CsA is unclear, but heterokaryons generated between permissive and nonpermissive cells exhibit the nonpermissive cell phenotype, suggestive of a dominant restriction that depends on binding of CypA to the viral capsid (43).
Recently, Yamashita and coworkers reported that an HIV-1 mutant encoding substitutions of alanine for Thr54 and Asn57 is selectively impaired for infection of nondividing cells (53). Because this mutant contained the T54A substitution, which alone confers CsA resistance/dependence, we asked whether CsA-dependent infection of HeLa cells is related to the impaired infection of nondividing cells by some HIV-1 CA mutants. In this study, we show that CsA-resistant/dependent HIV-1 mutants are selectively impaired for infection of mitotically arrested HeLa cells at a step following nuclear entry.
MATERIALS AND METHODS
Cells and viruses.
293T, HeLa-CD4/LTR-lacZ (HeLa-P4), and HOS cells were cultured in Dulbecco's modified Eagle's medium (Cellgro) supplemented with 10% fetal bovine serum, penicillin (50 IU/ml), and streptomycin (50 μg/ml) at 37°C with 5% CO2. The wild-type HIV-1 proviral DNA construct R9 (20), encoding full-length open reading frames for all HIV-1 structural and accessory genes, was used for these studies. The point mutations T54A and T54A/N57A in the CA region of R9 were previously described; mutants carrying these mutations were the generous gift of Wes Sundquist (49). Mutants A92E and R132K were generated by PCR and cloned into R9. All mutations were confirmed by sequencing. The T54A/A105T, A92E/A105T, and R132K/A105T mutants were constructed by PCR-based mutagenesis. In some experiments, HIV-GFP, an envelope-defective pNL4-3-based HIV-1 reporter virus clone encoding green fluorescent protein (GFP) in place of Nef (24), was employed. Viruses were produced by calcium phosphate transfection of 293T cells (20 μg of plasmid DNA per 2 × 106 cells) (11). Vesicular stomatitis virus glycoprotein (VSV-G)-pseudotyped reporter virus particles were produced by cotransfection of HIV-GFP plasmid DNA with the VSV-G expression plasmid pHCMV-G (11). Two days after transfection, the culture supernatants were harvested and clarified by filtration through 0.45-μm-pore-size filters, and aliquots were frozen at −80°C. The CA concentrations of the virus stocks were quantified by p24 enzyme-linked immunosorbent assay, as previously described (50).
Highly purified, activated primary CD4+ T lymphocytes were prepared as described previously (34). Three days following stimulation, cultures were expanded at a ratio of 1:2 in medium containing interleukin-2 (NIH AIDS Research and Reference Reagent Program; 200-U/ml final concentration). The cultures were further expanded 1:2 every 2 days in interleukin-2-containing medium and inoculated with HIV-1 at 7 days poststimulation.
We used a Monocyte Negative Isolation kit from Dynal Biotech to purify monocytes from peripheral blood mononuclear cells according to the manufacturer's protocol. Purified monocytes were differentiated by culturing them in T-cell medium supplemented with recombinant human granulocyte-macrophage colony-stimulating factor (50 ng/ml; R&D Systems). The cells were cultured in a 96-well plate in 0.2 ml at a concentration of 5 × 106 cells/well. Medium containing granulocyte-macrophage colony-stimulating factor was replenished after 3 days. We inoculated T cells and monocyte-derived macrophages with VSV-G-pseudotyped viruses (20 ng p24) in 0.1-ml volumes of T-cell medium. The next day, we added fresh medium to the infected cultures. Infectivity was quantified by flow cytometric detection of GFP expression 2 days after inoculation.
Viral-infectivity assay.
The HeLa-P4 cell line, a HeLa cell clone engineered to express CD4 and an integrated long terminal repeat (LTR)-lacZ reporter cassette, was used to quantify HIV-1 infectivity as previously described (9), with the following modifications. HIV-1 stocks were serially diluted in culture medium, and samples (0.125 ml) were used to inoculate HeLa-P4 target cells seeded the previous day (20,000 cells per well in 48-well plates). Two hours after inoculation, the cultures were supplemented with additional medium (0.5 ml) and cultured for another 48 h prior to being stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) to detect infected cells. To quantify infected cells, individual wells were visualized using a Scion CFW 1312 M camera equipped with a Navitar Macro Zoom (18- to 108-mm) lens, and images were captured with a Dell computer. Blue cells were quantified using ImageJ software in the particle-counting mode. Infections were performed in triplicate, and only values within the linear range of the infection assay (up to 1,000 blue cells per well) were used to calculate infectivity. Infections with HIV-GFP reporter viruses were analyzed by flow cytometric analysis of GFP expression as previously described (43).
Cell cycle arrest.
To generate arrested target cells for infectivity assays, HeLa-P4 or HOS cells (4 × 104 cells/well) were plated in 48-well plates. Aphidicolin was added to a final concentration of 2 μg/ml upon seeding (22, 51, 53); 24 h after seeding, cell cycle arrest was confirmed by flow cytometric analysis after the cells were stained with propidium iodide or inoculated with HIV-1. The cultures were exposed to viruses overnight; aphidicolin was also present during this period. The wells were washed thoroughly using PBS, and cells were cultured in 0.5 ml fresh medium for another 24 h prior to staining or cell cycle analysis. In some experiments, gamma irradiation was used to arrest the cells. Cells (1 × 105) were seeded 1 day before irradiation. The cells were irradiated with a dose of 50 Gy from a 137Cs source and were inoculated with VSV-G-pseudotyped viruses 24 h after irradiation (28).
Knockdown of CypA expression in HeLa-P4 cells.
We used Open Biosystems pGIPZ-based short hairpin RNA (shRNA) lentiviral vectors to deplete CypA expression. The vectors were obtained from the Vanderbilt Shared Microarray Resource (clone numbers 66713 and 66716). The vectors were packaged by cotransfection of 293T cells with CMV-ΔR8.2 (36), an HIV Gag-Pol expression plasmid, and pHCMV-G. Stable populations of lentivector-transduced cells were selected by culture in puromycin (3 μg/ml) for 2 weeks prior to use in HIV-1 infection assays. CypA depletion in the cell lines was confirmed by immunoblotting and functionally by enhanced permissiveness to infection by the HIV-1 A92E CA mutant virus.
Quantitative analyses of HIV-1 reverse transcription in HeLa-P4 cells.
One day prior to infection, HeLa-P4 cells (100,000) were plated into each well of 12-well plates. Viruses were treated with 20 μg/ml of DNase I and 10 mM MgCl2 at 37°C for 1 h to remove contaminating plasmid DNA. HeLa-P4 cells were inoculated with VSV-G-pseudotyped viruses (30 ng p24/well) in the presence of 8 μg/ml Polybrene (Sigma). As a control for determining the presence of contaminating plasmid DNA, wild-type HIV-1 was inoculated in the presence of a nonnucleoside reverse transcriptase inhibitor (efavirenz; 5 μM; obtained from the NIH AIDS Research and Reference Reagent Program). At 12 h postinfection, infected cells were washed with 1 ml of phosphate-buffered saline (PBS) and then treated with 500 μl of trypsin. The trypsin was inactivated by addition of 750 μl of Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, and cells were collected and washed once with 500 μl of PBS. The cell pellets were resuspended in 200 μl of PBS, and DNA was isolated using a DNeasy kit (Qiagen) following the manufacturer's instructions. Viral DNA was quantified by real-time PCR using an MX-3000p thermocycler (Stratagene) utilizing TaqMan chemistry. Early reverse transcription products (minus-strand strong-stop DNA) were amplified with the primers ERT-SS-F (5′-GCTAACTAGGGAACCCACTGCTT-3′) and ERT-SS-R (5′-ACAACAGACGGGCACACACTAC-3′) and were detected with the probe ERT-SS (5′-6-carboxyfluorescein-AGCCTCAATAAAGCTTGCCTTGAGTGCTTC-6-carboxytetramethylrhodamine-3′). Late reverse transcription products (U5-Gag) were amplified with primers MH531 (5′-TGTGTGCCCGTCTGTTGTGT-3′) and MH532 (5′-GAGTCCTGCGTCGAGAGAGC-3′) and were detected with the probe LTR-P (5′-6-carboxyfluorescein-CAGTGGCGCCCGAACAGGGA-6-carboxytetramethylrhodamine-3′) as previously described (12, 43).
Quantification of two-LTR circular DNA was performed essentially as previously described (12), with the following modifications. DNA from acutely infected cells was isolated at 18 h postinfection by using the DNeasy kit (Qiagen). Two-LTR circular DNA was detected by quantitative real-time PCR utilizing primers and a TaqMan probe specific for the LTR-LTR junction. The forward primer MH535 (5′-ACTAGGGAACCCACTGCTTAAG-3′), the reverse primer MH536 (5′-TCCACAGATCAAGGATATCTTGTC-3′), and the two-LTR probe MH603 (5′-[6-carboxyfluorescein]-ACACTACTTGAAGCACTCAAGGCAAGCTTT-[6-carboxytetramethylrhodamine]-3′) were used. The amplification conditions were as follows: 2 min at 50°C, 10 min at 95°C, and 40 cycles of 95°C for 15 s and 60°C for 90 s (12).
RESULTS
CsA-dependent HIV-1 mutants are impaired for infecting nondividing cells.
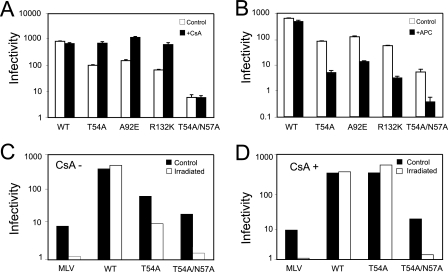
To test the hypothesis that CypA restricts infection of nondividing cells by CsA-dependent CA mutants, we generated virus stocks by transfecting 293T cells and titrated the viruses on HeLa-P4 indicator cells. The mutations had no deleterious effects on HIV-1 particle production, as the virus production was comparable as quantified by p24 enzyme-linked immunosorbent assay (data not shown). We sought first to verify the CsA-dependent phenotype of the HIV-1 CA mutants T54A, A92E, and R132K in dividing cells. We inoculated cultures with dilutions of viral supernatants and treated the cultures with CsA during the inoculation. Twenty-four hours after incubation, the cultures were washed with PBS and the medium was refreshed. Two days after inoculation, the cells were fixed, and infected cells were identified by staining them with X-Gal. CsA treatment increased the infectivity of each mutant by approximately 10-fold but had minimal effects on infection by wild-type HIV-1 and T54A/N57A (Fig. 1A). To determine if the mutant viruses were further impaired for infection of nondividing cells, we titrated the virus stocks on aphidicolin-arrested HeLa cells. Relative to wild-type HIV-1, which infected both dividing and nondividing cells efficiently, the infectivity of each mutant was reduced by 100-fold (Fig. 1B). The T54A/N57A mutant is selectively impaired for infection of arrested cells (53) and was therefore employed as a positive control for cell arrest. However, the mutant was unaffected by CsA, thus indicating that the N57A substitution renders T54A CsA independent. To test the possibility that the cell-cycle-dependent phenotype is caused by a side effect of aphidicolin, we tested infection of HeLa-P4 cells arrested by irradiation. As expected, infection by the cell-cycle-dependent MLV was blocked in the irradiated cells (Fig. 1C). Infection by T54A and T54A/N57A was also restricted in these cells. We conclude from these data that CsA-dependent CA mutants were selectively impaired for infection of arrested HeLa-P4 cells. However, the results obtained with the T54A/N57A mutant demonstrate that cell cycle dependence of HIV-1 CA mutants does not necessarily require CsA dependence.
FIG. 1.
CsA-dependent HIV-1 mutants are impaired for infection of arrested HeLa-P4 cells. (A) Infectivities of CA viruses on dividing HeLa-P4 cells. CsA was used in these assays at 10 μM. (B) HIV-1 infectivity on aphidicolin (APC)-arrested HeLa-P4 cells. Shown are the mean values of triplicate infections, with error bars representing 1 standard deviation. The results shown are from one representative of three independent experiments. WT, wild type. (C and D) HeLa-P4 cells were arrested by gamma irradiation and challenged with the indicated VSV-G-pseudotyped viruses. For HIV-1, infectivity was determined as the number of infected cells per nanogram of p24 in the inoculum. For MLV, the infectivity was determined as the number of GFP-positive cells per microliter of viral supernatant. The results shown are the mean values of three independent experiments, with error bars representing 1 standard deviation. (D) Assays were performed in cultures containing CsA (5 μM). The results shown in panels C and D are representative of two independent experiments.
CypA-CA interaction is necessary for the cell-cycle-dependent infectivity of CsA-dependent CA mutants.
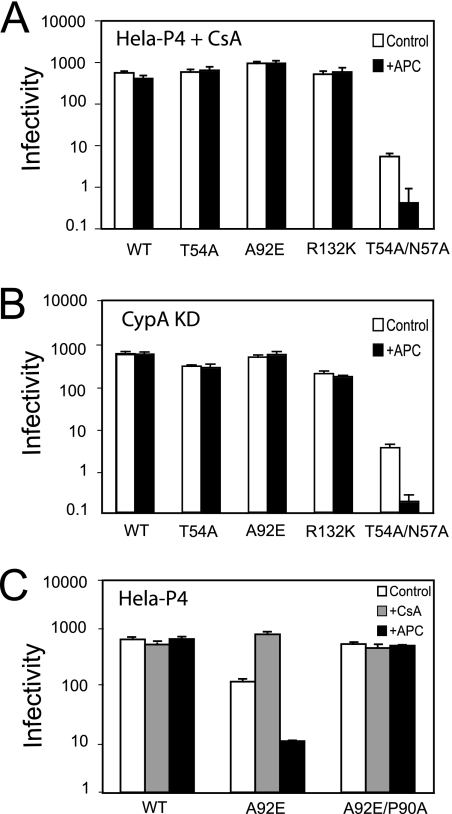
We next asked whether the impaired infection of arrested HeLa cells by the mutants was related to their CsA-dependent infectivity. We first tested if CsA treatment could relieve the cell cycle dependence of these mutants. For this purpose, aphidicolin-arrested HeLa cells were infected with wild-type and mutant viruses in the presence and absence CsA. All of the mutants exhibited similar infectivities in dividing and nondividing cells if CsA was added upon infection (Fig. 2A). This result indicates that CypA plays an important role in the cell cycle dependence. We also observed that CsA relieves the specific infection impairment by the T54A mutant induced upon cell arrest by irradiation (Fig. 1D). CsA is an immunosuppressive drug (19); thus, to eliminate other effects caused by the drug and to determine if target cell CypA is essential for this rescue, we assayed the viruses for infection of cells in which CypA was depleted by expression of a specific shRNA. All of the CsA-dependent mutants exhibited similar infectivities in dividing and nondividing CypA-depleted cells (Fig. 2B). These data confirmed that target cell CypA is necessary for cell cycle dependence of the CsA-dependent mutants.
FIG. 2.
The CypA-CA interaction is necessary for the cell-cycle-dependent infectivity of CsA-resistant/dependent HIV-1 mutants. (A) Infectivities of the CsA-dependent HIV-1 mutants in arrested HeLa-P4 cells. CsA (10 μM) was added during inoculation and maintained throughout the 2-day assay period. WT, wild type. APC, aphidicolin. (B) Infectivities of HIV-1 mutants on aphidicolin-arrested HeLa-P4 cells depleted for CypA expression. (C) Infectivities of the indicated viruses as determined on CsA-treated or aphidicolin-arrested cells. The results shown are the mean values of triplicate determinations, with error bars representing 1 standard deviation. The data are from one representative of three independent experiments.
To test the role of CypA-CA interactions in cell-cycle-dependent infection by the CsA-dependent mutants, we generated a double mutant, A92E/P90A, which exhibits CsA-resistant replication in T-cell lines and has been shown to exhibit impaired interaction with CypA (55). This virus was equally infectious in both arrested and proliferating HeLa cells and was unaffected by CsA (Fig. 2C). Collectively, these results indicated that CsA-dependent HIV-1 CA mutants are impaired for infection of arrested HeLa cells by a mechanism involving binding of CypA to the incoming viral capsid.
A second-site mutation relieves the CsA dependence and the cell-cycle dependence of all the mutants.
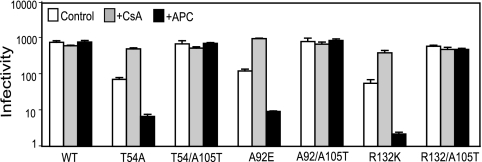
In a previous study, we identified the CA mutation A105T as a suppressor of the CsA-dependent phenotype of T54A and A92E mutants (54). We therefore asked whether introduction of the A105T mutation in CA would relieve the CypA-dependent restriction of infection of arrested HeLa cells by these mutants. The double mutant T54A/A105T exhibited similar infectivity in control cells, CsA treated cells, and aphidicolin-arrested cells (Fig. 3). Thus, the A105T second-site mutation relieved both the CsA dependence and the cell cycle dependence of T54A. We also observed that A92E and R132K mutants were rescued by addition of the A105T mutation (Fig. 3). We conclude that A105T is a general suppressor of the CsA-dependent phenotype in both dividing and nondividing cells.
FIG. 3.
The second-site suppressor mutation A105T relieves the impaired infectivity of CsA-resistant/dependent mutants on dividing and nondividing cells. Control cells were inoculated in the presence and absence of CsA (10 μM). Shown are the mean values of triplicate infections, with error bars representing 1 standard deviation. The results shown are from one representative of three independent experiments. WT, wild type; APC, aphidicolin.
Infection of nondividing cells by CsA-dependent CA mutants is blocked at a step following nuclear entry.
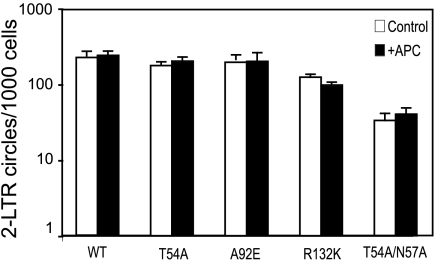
To determine at which step in the HIV-1 life cycle infection by the CA mutants is blocked, we performed quantitative PCR analysis for early and late products of reverse transcription. Consistent with a previous report describing other CA mutants (53), we observed no significant differences in the levels of early and late reverse transcription products in dividing and nondividing cells (data not shown). We then asked whether the mutations affect the nuclear import and integration steps in infection. Two-LTR circles are a convenient indicator of nuclear import for HIV-1, so we quantified two-LTR circle accumulation for these CA mutants by quantitative PCR in control and aphidicolin-arrested cells. Surprisingly, all the CA mutants produced comparable levels of two-LTR circles in arrested and control HeLa cells (Fig. 4). These results indicate that the PICs of all the CA mutants enter the nuclei of nondividing cells efficiently, and thus, there is no apparent defect in nuclear import. Because the HeLa-P4 infection assay scores the early steps in the life cycle, from entry to the expression of Tat protein, these results indicate that the mutant viruses are impaired for a step between nuclear import and integration in nondividing cells.
FIG. 4.
CsA-dependent mutants are not impaired for entry into the nuclei of nondividing cells. Viral two-LTR circles were assayed by quantitative PCR. Shown are the mean values of triplicate infections, with error bars representing 1 standard deviation. The results shown are from one representative of three independent experiments. WT, wild type; APC, aphidicolin.
Reversibility of the CypA-imposed block to integration by CsA-dependent mutants in arrested cells.
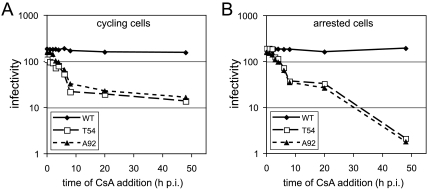
Our results with the A92E/P90A double mutant indicated that CypA-CA interactions inhibit infection by the A92E mutant in nondividing cells. CsA has been shown to act rapidly to penetrate cells and prevent CypA binding to the viral core (38). We exploited this to analyze the reversibility of CypA-restricted viral cores in target cells. Normal and aphidicolin-arrested cells were pulsed with wild-type and mutant viruses; then, CsA was added at various times postinoculation and maintained throughout the culture period. Two days later, the extent of infection was determined. In both normal and arrested cells, the infection was rescued to the unrestricted level (i.e., CsA added at time zero) with a half-life of approximately 4 h (Fig. 5A and B). In nonarrested cultures, addition of CsA at 8 h resulted in a level of infection similar to that when no drug was added (i.e., the 48-h time point) (Fig. 5A). Similar results were observed for the aphidicolin-treated cells (Fig. 5B). However, when CsA was added to the arrested cell cultures at 20 h postinoculation, the extent of infection was restored to that observed in the control nonarrested cultures (compare the 20-h time point to the 48-h time points in Fig. 5A and B). These results indicate that there are two distinct impairments to infection of arrested cells by CsA-dependent mutants, which differ from one another in the kinetic window of reversibility by CsA. The results imply that, in arrested cells, the mutant PIC forms a stable, reversibly restricted complex with CypA.
FIG. 5.
The infectivity impairment of CsA-dependent mutants in arrested HeLa cells can be rescued 20 h after inoculation (p.i.). Viral stocks were inoculated in control (A) and aphidicolin-arrested (B) cells. Two hours later, the inocula were removed, the cells were washed, and the media were replenished. At various times, CsA (10 μM) was added to the cultures and maintained for another 24 h. Infected cells were quantified 2 days after inoculation. Shown are the mean infectivity values of triplicate infections, with error bars representing 1 standard deviation. The results shown are from one representative of three independent experiments. WT, wild type.
Cell-cycle-dependent infection by CsA-dependent HIV-1 mutants is observed in HeLa, but not HOS, cells.
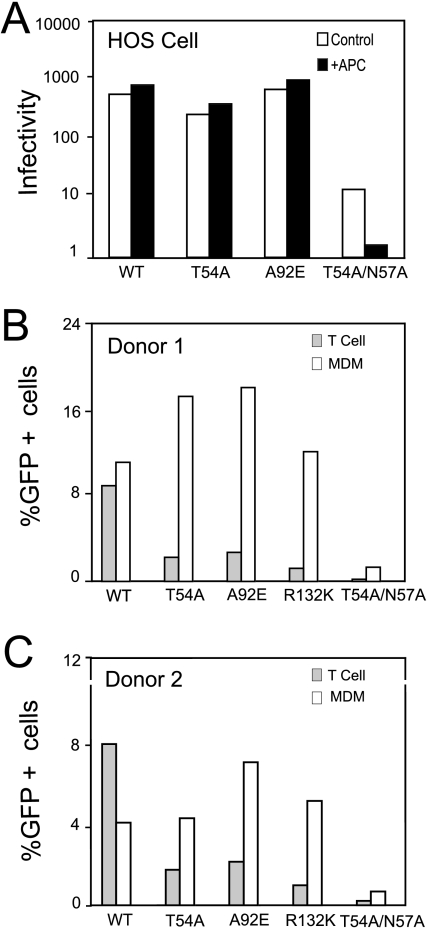
Previous studies have shown that infection by the CsA-dependent HIV-1 mutants is restricted in HeLa cells, whereas other cell lines, including HOS and 293T, are permissive to these viruses (23, 42). Heterokaryon studies revealed that the HeLa cell restriction is dominant, since heterokaryons between permissive and nonpermissive cells exhibited the nonpermissive phenotype (43). To ask whether the apparent restriction to infection by CsA-dependent CA mutants is cell type dependent, we infected aphidicolin-arrested HOS cells with wild-type HIV-1 and the T54A and A92E mutant viruses. Flow cytometric analysis of the cell cycle following staining with propidium iodide confirmed that both HeLa and HOS cell types were effectively arrested by aphidicolin (data not shown). In contrast to HeLa cells, we observed that aphidicolin-arrested HOS cells were as permissive for the CA mutants T54A and A92E as the corresponding nonarrested cells (Fig. 6). As previously reported (53), the control virus T54A/N57A exhibited reduced infectivity in arrested versus nonarrested HOS cells. These results demonstrated that infection by CsA-dependent mutants is inhibited in aphidicolin-arrested HeLa cells but not in HOS cells.
FIG. 6.
The cell-cycle dependence of CsA-dependent mutants varies with the cell type. VSV-G-pseudotyped viruses were used to infect HOS cells (A), as well as activated CD4+ T cells and monocyte-derived macrophages (MDM), from two individual donors (B and C). Two days later, the extent of infection was quantified by flow cytometry and was calculated as the percentage of GFP-expressing cells per nanogram of input p24 antigen in the inoculum. WT, wild type; APC, aphidicolin.
To determine whether cell-cycle-dependent infection by the CsA-dependent CA mutants occurs in natural targets of HIV infection, we inoculated primary monocyte-derived macrophages and activated primary CD4+ T cells. Macrophages are terminally differentiated and incapable of undergoing mitosis, whereas activated T cells are proliferating. Cells prepared from two donors were challenged with equivalent quantities of VSV-G-pseudotyped HIV-GFP. The overall levels of infection by the wild-type virus were similar in macrophages and T cells, likely reflecting the efficient entry of the pseudotyped viruses. Surprisingly, the T54A, A92E, and R132K mutant viruses infected macrophages efficiently (Fig. 6B and C). In contrast, these mutants were impaired for infection of primary T cells. Addition of CsA had no enhancing effect on any of the viruses in macrophages, consistent with the apparent lack of restriction of the mutants in the cells (data not shown). We have not yet tested whether CsA relieves the apparent restriction of the mutants in primary T cells. Collectively, these results demonstrate that the cell-cycle-dependent infection by CA mutants is a cell-type-specific phenomenon.
DISCUSSION
Recently, several cellular restrictions of HIV-1 infection have been identified, including TRIM5α, TRIM-Cyp, APOBEC3G, and CD317/tetherin (37, 39, 41, 44, 48). Our data suggest that there is an additional cell-dependent restriction that inhibits infection by specific HIV-1 CA mutants in nondividing cells. We observed that CsA-dependent mutants are impaired for infection of aphidicolin-arrested HeLa cells. In several respects, this impairment is similar to the CsA-dependent block in dividing cells. First, both defects are critically dependent on CypA-CA interactions and are manifested as a failure to complete the early postentry steps in infection. Secondly, the CA mutation A105T rescues the CypA-imposed block to infection by the HIV-1 mutants in both dividing and nondividing cells. Thirdly, both blocks are present in HeLa, but not HOS, cells. Despite these similarities, the block to infection of nondividing cells appears to be mechanistically different from the CsA-dependent infectivity defect observed in dividing cells. Specifically, the impaired infection of dividing cells was reversible by CsA addition only during the first few hours after inoculation, while the selective impairment for infection of nondividing cells could be relieved by addition of CsA at up to 20 h after virus entry. Our observation that the HIV-1 mutants form normal quantities of two-LTR circles in nondividing cells further indicates that the viruses form a stable, CypA-restricted PIC in the nuclei of nondividing cells that fails to integrate unless CypA is dissociated from CA.
Relative to MLV, which poorly infects nondividing cells, purified HIV-1 PICs contain only small amounts of CA (5, 15-17, 35). Previous findings from our laboratory and Emerman's group suggest that the rate and/or extent of HIV-1 uncoating may be a controlling factor in HIV-1 infection of nondividing cells (12, 51, 53). Our study provides further evidence that impaired infection of nondividing cells can result after the PIC enters the nucleus. Like the previously characterized T54A/N57A mutant, the CsA-dependent CA mutants synthesized equivalent quantities of two-LTR circles in dividing and nondividing cells, arguing against a specific effect on nuclear import. In other experiments, we have examined the intrinsic stability of the CsA-resistant/dependent mutant cores and obtained paradoxical results. While we observed that purified T54A cores have a decreased level of CA, suggestive of unstable capsids (54), the level of CA protein associated with A92E cores was comparable to the wild type (our unpublished observations), suggesting that an intrinsic defect in capsid stability is not responsible for the CsA-dependent phenotype of these mutants. Yamashita and coworkers reported that the T54A/N57A mutant escapes inhibition by CsA less rapidly than wild-type HIV-1, which was interpreted as possible evidence for a delay in uncoating (53). However, analysis of the CA association by imaging of intracellular HIV-1 cores revealed a more rapid dissociation of CA from the mutant core (53). In the present study, we observed that the specific block to infection in nondividing cells by T54A and A92E viruses can be rescued by CsA as late as 20 h postentry (Fig. 5). Because CypA exerts its effects on HIV-1 infection via interaction with the viral capsid, it is likely that a persistent association of CA with the viral core underlies the cell-cycle-dependent phenotype of these mutants. Based on quantitation of viral two-LTR circular DNA, the CsA-dependent mutant PICs appear to access the nucleus as efficiently as wild-type HIV-1 (Fig. 4), thus suggesting that the mutants are impaired for intranuclear trafficking or integration. Our results provide further evidence that efficient dissociation of CA from the PICs is required for efficient HIV-1 integration to occur, particularly in nondividing cells.
In this study, we observed cell-cycle-dependent restriction of CsA-dependent mutants in HeLa cells, but not in the HOS cell line or in monocyte-derived macrophages. Why are these HIV-1 mutants restricted in a cell-dependent manner? One plausible explanation is that a specific host factor restricts these mutants only after modification of the CA protein by CypA. Attractive candidate proteins would be TRIM family members or a factor that is specifically induced upon cell cycle arrest. Rhesus macaque TRIM5α has been reported to restrict cells in a CypA-dependent manner, providing a precedent for CypA-dependent inhibition of infection by a specific host factor (4, 26, 45). Identification of a host factor responsible for CsA-dependent infection by HIV-1 CA mutants would lead to improved understanding of early postentry events in HIV-1 infection.
Acknowledgments
We thank Jing Zhou for purified T cells and macrophages and Greg Towers for sharing results prior to publication. We are also grateful to Hailun Wang and the Vanderbilt University Flow Cytometry Core for assistance with cell cycle analysis. Efavirenz was obtained from the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH. Clones used in the shRNA experiments were obtained from Open Biosystems and were maintained and distributed by the Vanderbilt Microarray Shared Resource.
The Vanderbilt Microarray Shared Resource was supported by the Vanderbilt Ingram Cancer Center (P30 CA68485), the Vanderbilt Diabetes Research and Training Center (P60 DK20593), the Vanderbilt Digestive Disease Center (P30 DK58404), and the Vanderbilt Vision Center (P30 EY08126). This work was supported by NIH grant AI073137.
Footnotes
Published ahead of print on 1 October 2008.
REFERENCES
- 1.Aberham, C., S. Weber, and W. Phares. 1996. Spontaneous mutations in the human immunodeficiency virus type 1 gag gene that affect viral replication in the presence of cyclosporins. J. Virol. 703536-3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ao, Z., X. Yao, and E. A. Cohen. 2004. Assessment of the role of the central DNA flap in human immunodeficiency virus type 1 replication by using a single-cycle replication system. J. Virol. 783170-3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arhel, N., S. Munier, P. Souque, K. Mollier, and P. Charneau. 2006. Nuclear import defect of human immunodeficiency virus type 1 DNA flap mutants is not dependent on the viral strain or target cell type. J. Virol. 8010262-10269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berthoux, L., S. Sebastian, E. Sokolskaja, and J. Luban. 2005. Cyclophilin A is required for TRIM5α-mediated resistance to HIV-1 in Old World monkey cells. Proc. Natl. Acad. Sci. USA 10214849-14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowerman, B., P. O. Brown, J. M. Bishop, and H. E. Varmus. 1989. A nucleoprotein complex mediates the integration of retroviral DNA. Genes Dev. 3469-478. [DOI] [PubMed] [Google Scholar]
- 6.Braaten, D., C. Aberham, E. K. Franke, L. Yin, W. Phares, and J. Luban. 1996. Cyclosporine A-resistant human immunodeficiency virus type 1 mutants demonstrate that Gag encodes the functional target of cyclophilin A. J. Virol. 705170-5176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukovsky, A. A., A. Weimann, M. A. Accola, and H. G. Gottlinger. 1997. Transfer of the HIV-1 cyclophilin-binding site to simian immunodeficiency virus from Macaca mulatta can confer both cyclosporin sensitivity and cyclosporin dependence. Proc. Natl. Acad. Sci. USA 9410943-10948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubel, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charneau, P., M. Alizon, and F. Clavel. 1992. A second origin of DNA plus-strand synthesis is required for optimal human immunodeficiency virus replication. J. Virol. 662814-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chatterji, U., M. D. Bobardt, R. Stanfield, R. G. Ptak, L. A. Pallansch, P. A. Ward, M. J. Jones, C. A. Stoddart, P. Scalfaro, J. M. Dumont, K. Besseghir, B. Rosenwirth, and P. A. Gallay. 2005. Naturally occurring capsid substitutions render HIV-1 cyclophilin A independent in human cells and TRIM-cyclophilin-resistant in Owl monkey cells. J. Biol. Chem. 28040293-40300. [DOI] [PubMed] [Google Scholar]
- 11.Chen, C., and H. Okayama. 1987. High-efficiency tranformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 72745-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dismuke, D. J., and C. Aiken. 2006. Evidence for a functional link between uncoating of the human immunodeficiency virus type 1 core and nuclear import of the viral preintegration complex. J. Virol. 803712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dvorin, J. D., P. Bell, G. G. Maul, M. Yamashita, M. Emerman, and M. H. Malim. 2002. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 7612087-12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emerman, M. 1996. HIV-1, Vpr and the cell cycle. Curr. Biol. 61096-1103. [DOI] [PubMed] [Google Scholar]
- 15.Farnet, C. M., and W. A. Haseltine. 1991. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol. 651910-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fassati, A., and S. P. Goff. 2001. Characterization of intracellular reverse transcription complexes of human immunodeficiency virus type 1. J. Virol. 753626-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fassati, A., and S. P. Goff. 1999. Characterization of intracellular reverse transcription complexes of Moloney murine leukemia virus. J. Virol. 738919-8925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Forshey, B. M., U. von Schwedler, W. I. Sundquist, and C. Aiken. 2002. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J. Virol. 765667-5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Franke, E. K., and J. Luban. 1996. Inhibition of HIV-1 replication by cyclosporine A or related compounds correlates with the ability to disrupt the Gag-cyclophilin A interaction. Virology 222279-282. [DOI] [PubMed] [Google Scholar]
- 20.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 949825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 871285-1294. [DOI] [PubMed] [Google Scholar]
- 22.Hatziioannou, T., and S. P. Goff. 2001. Infection of nondividing cells by Rous sarcoma virus. J. Virol. 759526-9531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hatziioannou, T., D. Perez-Caballero, S. Cowan, and P. D. Bieniasz. 2005. Cyclophilin interactions with incoming human immunodeficiency virus type 1 capsids with opposing effects on infectivity in human cells. J. Virol. 79176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He, J., Y. Chen, M. Farzan, H. Choe, A. Ohagen, S. Gartner, J. Busciglio, X. Yang, W. Hofmann, W. Newman, C. R. Mackay, J. Sodroski, and D. Gabuzda. 1997. CCR3 and CCR5 are co-receptors for HIV-1 infection of microglia. Nature 385645-649. [DOI] [PubMed] [Google Scholar]
- 25.Heinzinger, N. K., M. I. Bukrinsky, S. A. Haggerty, A. M. Ragland, V. Kewalramani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 917311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keckesova, Z., L. M. Ylinen, and G. J. Towers. 2006. Cyclophilin A renders human immunodeficiency virus type 1 sensitive to Old World monkey but not human TRIM5 alpha antiviral activity. J. Virol. 804683-4690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis, P., M. Hensel, and M. Emerman. 1992. Human immunodeficiency virus infection of cells arrested in the cell cycle. EMBO J. 113053-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lewis, P. F., and M. Emerman. 1994. Passage through mitosis is required for oncoretroviruses but not for the human immunodeficiency virus. J. Virol. 68510-516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Limon, A., N. Nakajima, R. Lu, H. Z. Ghory, and A. Engelman. 2002. Wild-type levels of nuclear localization and human immunodeficiency virus type 1 replication in the absence of the central DNA flap. J. Virol. 7612078-12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, T. Y., and M. Emerman. 2006. Cyclophilin A interacts with diverse lentiviral capsids. Retrovirology 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luban, J. 1996. Absconding with the chaperone: essential cyclophilin-Gag interactions in HIV-1 virions. Cell 871157-1159. [DOI] [PubMed] [Google Scholar]
- 32.Luban, J. 2007. Cyclophilin A, TRIM5, and resistance to human immunodeficiency virus type 1 infection. J. Virol. 811054-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 731067-1078. [DOI] [PubMed] [Google Scholar]
- 34.Lundquist, C. A., M. Tobiume, J. Zhou, D. Unutmaz, and C. Aiken. 2002. Nef-mediated downregulation of CD4 enhances human immunodeficiency virus type 1 replication in primary T lymphocytes. J. Virol. 764625-4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 715382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naldini, L., U. Blomer, P. Gallay, D. Ory, R. Mulligan, F. H. Gage, I. M. Verma, and D. Trono. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science 272263-267. [DOI] [PubMed] [Google Scholar]
- 37.Neil, S. J., T. Zang, and P. D. Bieniasz. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451425-430. [DOI] [PubMed] [Google Scholar]
- 38.Perez-Caballero, D., T. Hatziioannou, F. Zhang, S. Cowan, and P. D. Bieniasz. 2005. Restriction of human immunodeficiency virus type 1 by TRIM-CypA occurs with rapid kinetics and independently of cytoplasmic bodies, ubiquitin, and proteasome activity. J. Virol. 7915567-15572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sayah, D. M., E. Sokolskaja, L. Berthoux, and J. Luban. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430569-573. [DOI] [PubMed] [Google Scholar]
- 40.Schneidewind, A., M. A. Brockman, R. Yang, R. I. Adam, B. Li, S. Le Gall, C. R. Rinaldo, S. L. Craggs, R. L. Allgaier, K. A. Power, T. Kuntzen, C. S. Tung, M. X. LaBute, S. M. Mueller, T. Harrer, A. J. McMichael, P. J. Goulder, C. Aiken, C. Brander, A. D. Kelleher, and T. M. Allen. 2007. Escape from the dominant HLA-B27-restricted cytotoxic T-lymphocyte response in Gag is associated with a dramatic reduction in human immunodeficiency virus type 1 replication. J. Virol. 8112382-12393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheehy, A. M., N. C. Gaddis, J. D. Choi, and M. H. Malim. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418646-650. [DOI] [PubMed] [Google Scholar]
- 42.Sokolskaja, E., D. M. Sayah, and J. Luban. 2004. Target cell cyclophilin A modulates human immunodeficiency virus type 1 infectivity. J. Virol. 7812800-12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song, C., and C. Aiken. 2007. Analysis of human cell heterokaryons demonstrates that target cell restriction of cyclosporine-resistant human immunodeficiency virus type 1 mutants is genetically dominant. J. Virol. 8111946-11956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stremlau, M., C. M. Owens, M. J. Perron, M. Kiessling, P. Autissier, and J. Sodroski. 2004. The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427848-853. [DOI] [PubMed] [Google Scholar]
- 45.Stremlau, M., B. Song, H. Javanbakht, M. Perron, and J. Sodroski. 2006. Cyclophilin A: an auxiliary but not necessary cofactor for TRIM5α restriction of HIV-1. Virology 351112-120. [DOI] [PubMed] [Google Scholar]
- 46.Towers, G. J. 2007. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Towers, G. J., T. Hatziioannou, S. Cowan, S. P. Goff, J. Luban, and P. D. Bieniasz. 2003. Cyclophilin A modulates the sensitivity of HIV-1 to host restriction factors. Nat. Med. 91138-1143. [DOI] [PubMed] [Google Scholar]
- 48.Van Damme, N., D. Goff, C. Katsura, R. L. Jorgenson, R. Mitchell, M. C. Johnson, E. B. Stephens, and J. Guatelli. 2008. The interferon-induced protein BST-2 restricts HIV-1 release and is downregulated from the cell surface by the viral Vpu protein. Cell Host Microbe 3245-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Schwedler, U. K., K. M. Stray, J. E. Garrus, and W. I. Sundquist. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J. Virol. 775439-5450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wehrly, K., and B. Chesebro. 1997. p24 antigen capture assay for quantification of human immunodeficiency virus using readily available inexpensive reagents. Methods 12288-293. [DOI] [PubMed] [Google Scholar]
- 51.Yamashita, M., and M. Emerman. 2004. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J. Virol. 785670-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamashita, M., and M. Emerman. 2006. Retroviral infection of non-dividing cells: old and new perspectives. Virology 34488-93. [DOI] [PubMed] [Google Scholar]
- 53.Yamashita, M., O. Perez, T. J. Hope, and M. Emerman. 2007. Evidence for direct involvement of the capsid protein in HIV infection of nondividing cells. PLoS Pathog. 31502-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, R., and C. Aiken. 2007. A mutation in alpha helix 3 of CA renders human immunodeficiency virus type 1 cyclosporine A resistant and dependent: rescue by a second-site substitution in a distal region of CA. J. Virol. 813749-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yin, L., D. Braaten, and J. Luban. 1998. Human immunodeficiency virus type 1 replication is modulated by host cyclophilin A expression levels. J. Virol. 726430-6436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101173-185. [DOI] [PubMed] [Google Scholar]