Abstract
There are many unique aspects of vesicular stomatitis virus (VSV) transcription. In addition to its unusual mRNA capping and methyltransferase mechanisms, the addition of S-adenosyl homocysteine (SAH), which is the by-product and competitive inhibitor of S-adenosyl methionine (SAM)-mediated methyltransferase reactions, leads to synthesis of poly(A) tails on the 3′ end of VSV mRNAs that are 10- or 20-fold longer than normal. The mechanism by which this occurs is not understood, since it has been shown that productive transcription is not dependent on 5′ cap methylation and full-length VSV mRNAs can be synthesized in the absence of SAM. To investigate this unusual phenotype, we assayed the effects of SAH on transcription using a panel of recombinant viruses that contained mutations in domain VI of the VSV L protein. The L proteins we investigated displayed a range of 5′ cap methyltransferase activities. In the present study, we show that the ability of the VSV L protein to catalyze methyl transfer correlates with its sensitivity to SAH with respect to polyadenylation, thereby indicating an intriguing connection between 5′ and 3′ end mRNA modifications. We also identified an L protein mutant that hyperpolyadenylates mRNA irrespective of the presence or absence of exogenous SAH. Further, the data presented here show that the wild-type L protein hyperpolyadenylates a percentage of VSV mRNAs in infected cells as well as in vitro.
Vesicular stomatitis virus (VSV) is a member of the Mononegavirales. Viruses within this order have a single strand of nonsegmented negative-sense genomic RNA. For VSV, there are five genes, and the order is 3′-(leader)-N-P-M-G-L-(trailer)-5′ (3). The nucleocapsid (N) protein encapsidates the viral RNA genome and the N:RNA complex comprises the active template for all viral RNA synthesis. The phosphoprotein (P) is a cofactor for the RNA-dependent RNA polymerase (RdRp) and a solubility factor for the N protein to prevent N-protein aggregation (15, 19). The matrix (M) is the most abundant structural protein and is responsible for the bullet-shaped morphology of the virion (30). The glycoprotein (G) provides both attachment and fusion functions for entry of the virus into the host cell (31). The large (L) protein is the catalytic component of the viral RdRp (11). Expression of the genes of VSV is controlled primarily at the level of transcription (reviewed in reference 7). The general mechanism for transcription of the Mononegavirales is that the RdRp enters the genome at a single 3′ entry site and transcribes each gene in an obligatorily sequential manner (1-3). A central feature of this mode of transcription is that RdRp access to a downstream gene for transcription is dependent upon prior termination of the upstream mRNA. Due to a poorly understood process, known as attenuation, which has been localized to each gene junction, there is a gradient of mRNA synthesis, such that the levels of each mRNA decrease with each successive RdRp initiation event: genes closer to the 3′ entry site are transcribed more abundantly than 3′ distal genes (27).
Each mRNA is capped and methylated at the 5′ end and polyadenylated at the 3′ end. These activities are all carried out by the L-protein component of the RdRp in response to conserved sequences located at the beginning and end of each gene (16, 23, 32, 36, 40, 41; see reference 7 for a review). Modifications of the 5′ and 3′ ends of the mRNAs are tightly coupled to transcription. As such, a thorough understanding of the coordination of these modification events with respect to transcription is important for understanding the regulation of VSV transcription.
The length of poly(A) tails on VSV mRNAs is affected by the presence of S-adenosyl homocysteine (SAH), the by-product and competitive inhibitor of S-adenosyl methionine (SAM)-mediated methyltransferase (20, 21, 23, 37). When in vitro transcription reactions are performed in the presence of SAM, the RdRp synthesizes mRNA products with mobility similar to those synthesized in VSV-infected cells. However, VSV mRNAs synthesized in vitro in the presence of 1 mM SAH are unmethylated and heterogeneous in length due to having extremely long poly(A) tails, from 700 to 2,400 nucleotides (nt) in length, rather than the normal 100 to 200 AMP nt (21, 37). There are no data available regarding the length of VSV poly(A) tails in vivo in the presence of elevated levels of SAH. To our knowledge, it is not known how the presence of extremely long poly(A) tails affects the translation or stability of mRNAs. Shortening the poly(A) tail in eukaryotic cells is associated with reduced translatability and mRNA degradation (8). Alternatively, lengthening the poly(A) tails on cellular mRNAs leads to increased translatability and decreased mRNA turnover (8, 34, 35). However, the studies examining the effects of extending the poly(A) tails were conducted on cellular mRNAs containing poly(A) tails of approximately 20 nt that were extended to approximately 150 nt, which is far below the 700 to 2,400 nt observed for VSV in the presence of SAH.
Hyperpolyadenylation of VSV mRNAs has been reported also in the absence of SAH through studies of a VSV mutant, ts(G)16 (21, 22, 24, 25). ts(G)16 was isolated in 1970 based on its ability to grow at 31°C but not at 39°C (39). The mRNAs synthesized by the ts(G)16 polymerase were excessively polyadenylated at the 3′ end (21). The characterization of ts(G)16 revertants and subsequent sequence analysis showed two amino acid changes, C1291Y and F1488S, in the L protein of ts(G)16 compared to the wild type. The defect in polyadenylation reverted with replacement of S1488F in the ts(G)16 L protein, as found in the wild-type L protein (22, 24, 25).
The effect of SAH on polyadenylation is an intriguing aspect of VSV transcription that is not understood. It is not known why the polymerase synthesizes long polyadenylate tails when a compound is added that interferes with the ability to methylate the 5′ cap. The polymerase can synthesize full-length mRNAs in vitro in the absence of SAM, indicating that transcription is not dependent on cap methylation (21). Therefore, it seems that it is the presence of SAH, rather than the absence of SAM, that causes the polymerase to hyperpolyadenylate (21).
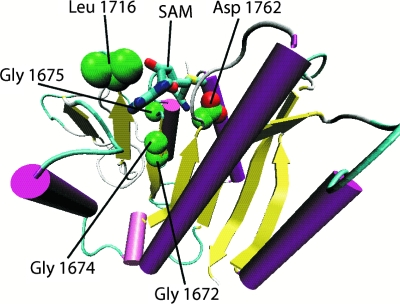
In a separate study, we described a structural homology model of the 2′-O-ribose methyltransferase domain within the VSV L protein (12). The model encompassed L-protein amino acid residues 1644 to 1842. Based on the model, we identified residues in domain VI of the VSV L protein for mutational analysis that were putatively involved in SAM binding (G1672, G1674, and G1675) or methyltransferase catalytic activity (D1762). An additional residue, L1716, was identified that appears to be important in the formation of the putative SAM binding pocket (Fig. 1). We generated a panel of mutants with various substitutions of each of these residues in the background of infectious virus and analyzed these mutants for effects on viral transcription and 5′ cap methylation. Our results showed that all substitutions to the invariantly conserved D1762 reduced transcription slightly, with the exception of the D1762E substitution, which synthesized greater levels of mRNA relative to the wild type, and that all D1762 mutants abrogated 5′ methyltransferase activity. Mutation of the glycines in the SAM binding motif at 1672, 1674, and 1675 gave a spectrum of phenotypes; all reduced viral transcription levels compared to wild-type and abrogated or reduced 5′ cap methyltransferase activity. These results agree with mutational analyses done on these residues by others (28, 29). The mutations introduced at position 1716 yielded L proteins that were temperature sensitive for RNA synthesis, and overall viral production had marked reductions in viral mRNA synthesis at the permissive temperature, but only a slight inhibition of 5′ cap methyltransferase activity (12). In addition, in the course of these studies we noted aberrant migration of mRNAs synthesized by the D1762E L protein and found it was associated with alterations in poly(A) tail length.
FIG. 1.
Location of domain VI L-protein residues examined. Spatial arrangement of the domain VI residues examined previously for their role in methyltransferase activity (12) and in the present study for SAH sensitivity. The model is shown as a ribbon diagram except for the highlighted residues, which are shown in space-fill arrangement. Alpha helices are colored purple. Beta strands are colored yellow. Random coils are colored light blue. Carbon atoms are green, and oxygen atoms are red. The SAM molecule is shown as a stick model: carbon atoms are colored light blue, nitrogen atoms are colored dark blue, oxygen atoms are colored red, and sulfur atoms are colored yellow.
In the present study, we examined this panel of mutants to investigate the effects of SAH on polyadenylation in the context of the L proteins shown to be deficient in methyltransferase activity. We assayed the effects of these mutations on mRNA transcription and polyadenylation in vitro. The results presented here show that mRNA hyperpolyadenylation by the VSV L protein in the presence of exogenous SAH is dependent upon the methyltransferase activity of the L protein.
MATERIALS AND METHODS
Cells and viruses.
BHK-21 cells were used for analysis of viral RNA synthesis. Vero-76 cells were used for titrating virus stocks. A new cell-line, termed AlphaV-T7 (kindly provided by Ilya Frolov, University of Texas Medical Branch), was used for the recovery of recombinant viruses containing the F1488S and F1488A mutations in the L gene. The AlphaV-T7 cell line is a BHK-derived cell line that expresses an alphavirus replicon, which encodes four nonstructural proteins that are required for replication of the RNA replicon, as well as T7 RNA polymerase and puromycin acetyltransferase, each of which are expressed under the control of separate subgenomic promoters. The recombinant viruses used in the present study (L1716T, L1716Y, D1762E, D1762G, D1762N, G1672A, G1672P, G1674P, and G1675P) were described previously (12).
Plasmid construction and mutagenesis.
To introduce the F1488S and F1488A mutations into the VSV full-length cDNA, we used a subclone that was described previously (12). The mutations were introduced into the subclone via site-directed mutagenesis using the QuikChange methodology (Stratagene, La Jolla, CA). Sequence analysis of a 3.5-kb fragment that spanned the AvaI and AflII restriction sites verified the presence of the desired mutation. The VSV subclone and pVSV1(+) were digested with AvaI and AflII, and the 3.5-kb subclone fragment was cloned into the pVSV1(+) backbone. The presence of the desired mutation was further verified via sequence analysis of the full-length cDNA plasmid.
Recovery and purification of recombinant VSV.
For recovery of recombinant VSV using AlphaV-T7 cells, it was necessary to clone the VSV N, P, and L genes into a T7 expression vector that contained an internal ribosomal entry site from encephalomyocarditis virus to allow for cap-independent translation of the VSV N, P, and L messages. These support plasmids have been described previously (12). Recombinant VSV was recovered from cDNA by transfection of AlphaV-T7 cells as described previously (12). Transfected cell supernatants were harvested at between 48 and 72 h, and one-fourth of the harvested supernatant was passaged onto BHK-21 cells. Infected cells were incubated at 31°C. Supernatants from infected BHK-21 cells were harvested at between 30 and 72 h postinfection (hpi) and clarified to remove cell debris at 3,000 × g for 10 min. Concentrated stocks of each virus were prepared as previously described (12). Virus titers were determined via plaque assay on Vero-76 cells.
Analysis of viral RNA synthesis in infected cells.
To analyze viral RNA synthesis, BHK-21 cells were infected at a multiplicity of infection (MOI) of 5 in Dulbecco modified Eagle medium plus 2% newborn calf serum at 37°C. At 2 hpi, infected cells were either treated with 5 μM adenosine dialdehyde (AdOX) or left untreated. At 4 hpi, infected cells were treated with 10 μg of actinomycin D/ml for 30 min and subsequently labeled with 33 μCi of [3H]uridine or 33 μCi of [3H]adenine/ml in the presence of an additional 10 μg of actinomycin D/ml for 1 h at 37°C. RNA was harvested in lysis buffer and then phenol-chloroform extracted and ethanol precipitated. RNAs were either digested with RNase A or RNase A followed by digestion with RNase H after annealing with oligonucleotide (dT), as described below. Undigested and digested RNAs were visualized by 1.5% acid agarose-urea gel electrophoresis and fluorography.
Analysis of in vitro-transcribed RNA.
A total of 2 μg of recombinant VSV was activated with 0.05% Triton N-101 in 0.4 M NaCl for 5 min at room temperature. Transcription reactions consisted of 1× in vitro transcription buffer (30 mM Tris-HCl [pH 8.1]; 33 mM NH4Cl; 4.5 mM magnesium acetate; 7 mM KCl; 10 mM dithiothreitol; 0.2 mM spermidine; 0.5 mM concentrations [each] of ATP, CTP, GTP, and UTP), 30% (vol/vol) nuclease-treated rabbit reticulocyte lysate, 5 μg of actinomycin D/ml, 0 or 1 mM SAH, and 15 μCi of [α-33P]ATP or 15 μCi of [α-33P]UTP. The 1× in vitro transcription buffer was modified to account for the addition of [α-33P]ATP; normal reactions contained 1 mM ATP, but here cold ATP was decreased to 0.5 mM. Transcription reactions were incubated at 30°C for 5 h and labeled RNA was harvested by using an RNeasy kit (Qiagen).
RNA size markers were generated from RNA Century Plus marker templates (Ambion) by using a MAXIscript T7 in vitro transcription kit (Ambion) according to the manufacturer's recommendations. In vitro-transcribed RNAs were labeled with [α-33P]-ATP and were phenol extracted and ethanol precipitated. For acid agarose-urea gel analysis, 1 μl of a 1:100 dilution was resolved alongside the digested RNAs described below.
RNase digestions.
For dT/RNase H digestions, one-fifteenth of the RNA isolated from in vitro transcription reactions was incubated with an excess (1 μg) of oligo(dT)15. RNA/DNA oligomers were denatured at 98°C for 1 min and chilled on ice for 2 min. To the RNA/DNA hybrids, 2× RNase H buffer (100 mM Tris, 20 mM MgCl2, 2 mM EDTA, 20 mM dithiothreitol, 20% glycerol) and 2 U of Escherichia coli RNase H (Invitrogen) were added, followed by incubation at 37°C for 20 min. Digested RNAs were ethanol precipitated prior to electrophoresis on 1.5% acid agarose-urea gels.
For RNase A (Epicentre) digestions, one-fifth of each RNA sample isolated from in vitro transcription reactions or one-third of each RNA sample isolated from infected cells was digested in a final volume of 10 μl that consisted of 1× TNE buffer (100 mM Tris, 2 M NaCl, 100 mM EDTA [pH 7.4]) and 5 μg of RNase A. Reactions were incubated at 37°C for 30 min and ethanol precipitated prior to gel analysis. A portion of the RNase A-digested RNAs was subsequently digested with dT/RNase H as described above. RNase 1 (Epicentre) digestions were conducted in a total volume of 10 μl that consisted of 1× TNE buffer and 2 U of RNase 1, followed by incubation at 37°C for 30 min. For analysis of RNase A-digested RNA, we used ultrapure agarose 1000 (Invitrogen) instead of ultrapure agarose (Invitrogen) for acid agarose-urea gel analysis, since the former resulted in better resolution of the smaller RNase A-resistant RNAs.
Analysis of cap structures from in vitro-transcribed RNA.
One-fourth of the RNA purified from in vitro transcription reactions labeled with [α-33P]GTP was digested with tobacco acid pyrophosphatase (TAP; Invitrogen), and one-fourth of the [3H]SAM-labeled RNA was digested with TAP and/or nuclease P1 (U.S. Biological) and calf intestinal alkaline phosphatase (New England Biolabs). All digestions were done in 1× TAP buffer at 37°C for 30 min. One-fifth of the [α-33P]GTP-digested RNA was resolved on polyethyleneimine-F plates (EM Biosciences) using 1.2 M LiCl as a solvent. Plates were dried and exposed to film. For reactions labeled with [3H]SAM, digested RNAs were resolved on polyethyleneimine-F plates using a sodium phosphate solvent. Plates were dried and treated with 5% 2,5-diphenyloxazole-acetone prior to exposure to film at −80°C.
RESULTS
Analysis of RNAs transcribed in vitro in the presence of exogenous SAH.
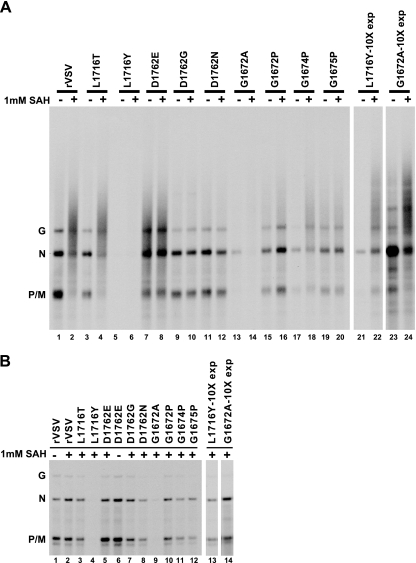
SAH has been shown to cause hyperpolyadenylation of VSV mRNAs when added at 1 mM to in vitro transcription reactions (20, 23, 37). To investigate whether the mutations in domain VI of the VSV L protein affected the sensitivity to SAH with respect to polyadenylation, we examined the effects of SAH on poly(A) tail addition to mRNA transcribed by the VSV L protein during transcription in vitro. In vitro transcription reactions were carried out in the presence of [α-33P]ATP and were supplemented with 1 mM SAH, or no additional substrate, to the usual transcription buffer (as described in Materials and Methods). Labeled RNAs were purified by RNeasy columns and resolved on acid agarose-urea gels (Fig. 2A). When wild-type VSV in vitro transcription reactions were performed in the absence of exogenous SAH, the characteristic VSV mRNA pattern was observed (Fig. 2A, lane 1). However, when wild-type in vitro transcription reactions were supplemented with 1 mM SAH, the mRNAs resolved as a heterogeneous population (Fig. 2A, lane 2). We observed a similar disparity in mRNA migration in the absence (−) or presence (+) of exogenous SAH for the mRNAs synthesized by the L-protein mutants that carried out 5′ cap methylation, L1716T, L1716Y, G1672A, and G1674P, (Fig. 2A, lanes 3 to 6, 13, 14, 17, and 18). RNAs synthesized by the L1716Y and G1672A L proteins were only visualized after a 10-fold longer exposure of film (Fig. 2A, lanes 21 to 24). Alternatively, the RNAs synthesized by the D1762G, D1762N, G1672P, and G1675P L proteins, all of which were unable to carry out 5′ cap methylation, did not resolve as heterogeneous populations in the presence of 1 mM SAH, as indicated by the indistinguishable mobility of the mRNA transcribed (−) and (+) 1 mM SAH (Fig. 2A, lanes 9 to 12, 15, 16, 19, and 20). The RNAs synthesized by the D1762E L protein resolved as a heterogeneous population even in the absence of exogenous SAH (Fig. 2A, lanes 7 and 8).
FIG. 2.
Sensitivity of domain VI L protein mutants to SAH in vitro. (A) Viral RNAs were transcribed in vitro in the presence of 1 mM SAH or no additional substrate. Transcription reactions were labeled with [33P]ATP. Labeled RNAs were purified and resolved on 1.5% acid agarose-urea gels. The gels were fluorogrammed, and radiolabeled RNAs were detected by autoradiography. In order to visualize RNAs transcribed by L1716Y and G1672A rVSVs, the films were exposed for 10 times longer (lanes labeled “10X exp”). The identities of VSV mRNAs are denoted on the left. (B) The RNAs shown in panel A were digested with RNase H following annealing with oligo(dT). Digested RNAs were ethanol precipitated and resolved on 1.5% acid agarose-urea gels. The gels were fluorogrammed, and radiolabeled RNAs were detected via autoradiography. The identities of labeled RNAs are denoted on the left.
Effect of poly(A) tail removal.
To determine whether the heterogeneity observed for the RNAs synthesized in the presence of 1 mM SAH was the result of hyperpolyadenylation, the RNAs were digested with RNase H following annealing with oligonucleotide dT [oligo(dT)/RNase H], which will cleave off the poly(A) tails. RNAs transcribed (+) and (−) exogenous SAH by the D1762E L protein were digested with oligo(dT)/RNase H because this mutation appeared to display the hyper-polyadenylating phenotype regardless of the presence of SAH. Upon oligo(dT)/RNase H digestion, (+)SAH RNAs and D1762E (+) and (−)SAH RNAs resolved as discrete VSV mRNAs, indicating that the heterogeneous populations of RNA observed in Fig. 2A were the result of hyperpolyadenylation (Fig. 2B). As noted above, due to their low transcription levels, the L1716Y and G1672A digested RNAs were visualized only upon a 10-fold longer exposure of film (Fig. 2B, lanes 13 and 14).
Analysis of poly(A) tail length.
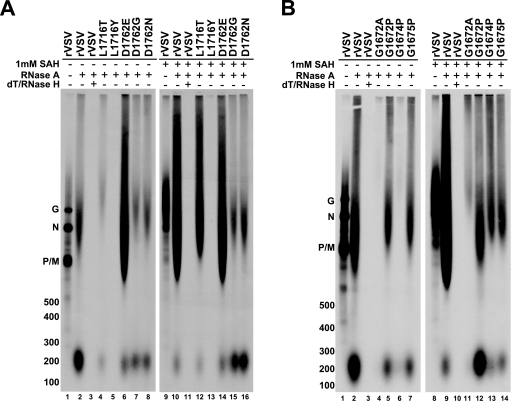
To directly analyze the poly(A) tails generated by each mutant L protein (+) or (−) 1 mM SAH, the in vitro-transcribed [α-33P]ATP-labeled RNAs shown in Fig. 2A were digested with RNase A, which cleaves single-stranded RNA at the 3′ end of pyrimidine residues, leaving the poly(A) tail intact. When RNA was synthesized in the absence of exogenous SAH by wild-type L protein and digested with RNase A, the predominant product resolved as a relatively discrete product on acid agarose-urea gels, sized at ∼200 nt, as determined by comparison to in vitro transcripts (Fig. 3A, lane 2; the migrations of size markers are noted on the left). However, when RNA was synthesized in the presence of 1 mM exogenous SAH by wild-type polymerase and digested with RNase A, the predominant product migrated as a heterogeneous population with a mobility indicating a size range from 800 to 2000 nt and even up to the origin of the gel (Fig. 3A, lane 10). Interestingly, the poly(A) tails synthesized by the wild-type L protein in the absence of exogenous SAH (Fig. 3A, lane 2) had a minor population of RNase A-resistant RNA that migrated with the large heterogeneous RNAs synthesized in the presence of exogenous SAH, although their abundance was small compared to the poly(A) synthesized in the presence of exogenous SAH (Fig. 3A, compare lanes 2 and 10). These data suggest that the wild-type polymerase also synthesized a small population of extended poly(A) tails even in the absence of exogenous SAH.
FIG. 3.
Direct analysis of the poly(A) tails synthesized by domain VI L protein mutants in the presence or absence of exogenous SAH. (A) The L1716T/Y and D1762E/G/N RNAs shown in Fig. 2A were digested with RNase A and resolved on 1.5% acid agarose-urea gels. Representative digests for rVSV (+/−) SAH are shown in each panel: rVSV samples were digested with RNase A or RNase A, followed by dT/RNase H digestion. The gels were fluorogrammed, and radiolabeled RNAs were detected by autoradiography. [33P]ATP-labeled in vitro-transcribed size markers were generated from RNA Century Plus marker templates (Ambion) and resolved alongside undigested and digested RNAs. The location of each size marker is denoted on the left. The identities of undigested VSV mRNAs are also denoted on the left. The sizes of each undigested VSV mRNA minus their respective poly(A) tails are 1,332 nt (N), 821 nt (P), 837 nt (M), and 1,672 nt (G). (B) Same as in panel A, except the RNAs analyzed were synthesized by G1672A/P, G1674P, and G1675P L proteins.
Confirmation of the RNase A-resistant RNA as poly(A) was done by RNase H digestion after annealing with oligo(dT). The undigested, RNase A, and RNase A/oligo(dT)/RNase H digestions for rVSV (+/−) exogenous SAH are shown as representative control samples (Fig. 3A, lanes 1 to 3 and lanes 9 to 11; Fig. 3B, lanes 1 to 3 and lanes 8 to 10). Digestion of the RNase A-resistant RNA synthesized by the wild-type L protein with oligo(dT)/RNase H resulted in the digestion of all labeled RNA, which indicated that the RNase A-resistant RNAs were poly(A) (Fig. 3A, lanes 3 and 11, and Fig. 3B, lanes 3 and 10). Similarly, when the RNase A-resistant RNA synthesized by each mutant polymerase was digested with oligo(dT)/RNase H, all labeled RNA was digested, also indicating that the labeled RNA was poly(A) (data not shown). RNase A-resistant RNAs were also characterized by RNase 1 digestion, which resulted in complete digestion of all radiolabeled products (data not shown). As an additional control for the specificity of RNase A, RNA synthesized by rVSV in the presence (+) or absence (−) of exogenous SAH was labeled with [α-33P]UTP and digested with RNase A, which resulted in the digestion of all labeled RNA (data not shown).
In the absence of exogenous SAH, RNase A digestion showed that all mutant polymerases, except D1762E, synthesized mRNAs whose poly(A) tails exhibited sizes of ∼200 nt, as found for RNase A-digested rVSV (Fig. 3A, lanes 2, 4, 7, and 8; Fig. 3B, lane 2 and lanes 4 to 7). The L1716Y L protein transcribed so poorly that RNase A-resistant RNAs were not detected. Interestingly, in the absence of exogenous SAH, the D1762E L protein had a poly(A) phenotype more similar to the wild-type L protein in the presence of exogenous SAH (Fig. 3A, compare lanes, 2, 6, and 10). These data again confirmed that the heterogeneous population observed in Fig. 2A for the D1762E L protein was the result of hyperpolyadenylation.
The mutants that showed a large disparity in the size of the poly(A) tails synthesized in the absence or presence of 1 mM SAH were L1716T, L1716Y, G1672A, and G1674P, indicating that these L proteins were sensitive to SAH with respect to polyadenylation, as is the wild-type polymerase (Fig. 3A, compare lanes 4 to 12; Fig. 3B, compare lanes 4 to 11 and lanes 6 to 13). As shown in Fig. 2, the L1716Y L protein transcribed so poorly that RNase A-resistant RNAs were not detected. The L proteins with mutations—D1762G, D1762N, G1672P, and G1675P—did not synthesize excessive amounts of aberrantly long poly(A) tails in reactions containing 1 mM SAH, as determined by their similarity to RNAs isolated from reactions carried out in the absence of SAH (Fig. 3A, compare lanes 7 and 15 and lanes 8 and 16; Fig. 3B, compare lanes 5 and 12 and lanes 7 and 14). These mutant L proteins were shown in previous work to be defective in 5′ cap methylation (12). Although the D1762E L protein synthesized long poly(A) tails in the absence or presence of 1 mM SAH, the lengths of the poly(A) tails generated in the presence of SAH did not appear to be larger overall than those generated in the absence of SAH (Fig. 3A, lanes 6 and 14). These data indicated that the D1762E L protein was not sensitive to the presence of exogenous SAH. The insensitivity to SAH with regard to poly(A) tail length in the in vitro transcription reactions correlated with the inability of polymerase to methylate the 5′ mRNA cap. In our previous study, we showed that substitutions to the 2′-O-ribose methyltransferase catalytic residue, D1762, completely abrogated the ability of the L protein to carry out 5′ cap methyltransferase activity. Mutations to residues in the putative SAM binding motif, GxG1672xG1674G1675, disparately affected 5′ cap methyltransferase activity; G1672A and G1674P retained activity, and L proteins with mutations of G1672P and G1675P significantly reduced methyltransferase activity. Lastly, mutation of L1716, a residue that is hypothesized to be structurally important for proper formation of the SAM binding pocket, to T or Y both retained cap methyltransferase activity (12). (These data are summarized in Table 2.)
TABLE 2.
Summary of cap methylation and SAH sensitivity
| Recombinant virus | G-N-7 (% of G cap released)a | Hyperpoly(A)
|
|
|---|---|---|---|
| (-)SAH | (+)SAH | ||
| rVSV | 92 | - | + |
| L1716T | 91 | - | + |
| L1716Y | 64 | - | + |
| D1762E | 8 | + | + |
| D1762G | 9 | - | - |
| D1762N | 7 | - | - |
| G1672A | 28 | - | + |
| G1672P | 8 | - | - |
| G1674P | 69 | - | + |
| G1675P | 9 | - | - |
| F1488S | 90 | + | + |
That is, the percent G-N-7 methylation catalyzed by each L-protein mutant. Data are summarized from a separate study (12).
Analysis of SAH sensitivity of a previously identified hyperpolyadenylating L protein.
Due to the phenotypes associated with the D1762E L protein, which we identified as hyperpolyadenylation in the absence of exogenous SAH, defective 5′ cap methyltransferase activity, and insensitivity to the presence of exogenous SAH, we wanted to test the hypothesis that L-protein sensitivity to SAH is dependent upon the ability of the L protein to catalyze 5′ cap methyltransferase activity. We introduced the F1488S mutation previously found in ts(G)16 (22, 25) into the L gene of a functional cDNA clone encoding the VSV genome and recovered the recombinant virus using a new cell line that constitutively expresses T7 RNA polymerase from an alphavirus replicon (described in Materials and Methods). Sequence analysis of the entire L gene confirmed the presence of the F1488S mutation, as well as the absence of second site mutations. We characterized this virus for growth, RNA synthesis, and 5′ cap methylation. These data are summarized in Table 1. We assayed this virus at 31 and 39°C due to the original temperature-sensitive phenotype described for ts(G)16 (33, 39). We found that the F1488S rVSV had a slight reduction in RNA transcription levels at 39°C and that the plaque size was reduced at 39°C compared to 31°C. However, virus growth, as determined by titers at 24 hpi, was not significantly reduced at 39°C compared to wild-type or compared to the F1488S rVSV at 31°C. We examined the methyltransferase activity of the F1488S L protein and found that it catalyzed methyltransferase activity with near wild-type levels.
TABLE 1.
Summary of analysis of recombinant F1488S VSV
| rVSV | Mean ± SEM
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Transcription (% wild type)
|
Plaque sizea (mm)
|
Titer at 24 h (log10 PFU/ml)
|
G-N-7b (% total G cap) | 2′-Oc (% of total methylation) | ||||
| 31°C | 39°C | 31°C | 39°C | 31°C | 39°C | |||
| Wild type | 100 | 100 | 2.7 ± 0.3 | 3.2 ± 0.3 | 9.4 ± 0.5 | 8.3 ± 0.6 | 92.8 ± 2.7 | 52 ± 2.3 |
| F1488S | 118 ± 6.9 | 84.1 ± 8.7 | 2.1 ± 0.3 | 1.1 ± 0.2 | 8.6 ± 0.4 | 8.4 ± 0.5 | 90.7 ± 1.9 | 47 ± 1.3 |
Plaque assays were incubated at 31°C for 45 h and at 39°C for 30 h.
G-N-7, methylation at the G-N-7 position; amounts determined from TAP-digested [α-33P]GTP in vitro-labeled RNA. Values are expressed as a percentage 7mGp of the total G cap released upon TAP digestion.
2′-O, methylation at the 2′-O-ribose position. Amounts were determined from TAP/nuclease P1/CIP-digested [3H]SAM in vitro-labeled RNA. Values are expressed as the percentage of 2′-O methylation of the total amount of methylation at the G-N-7 and 2′-O positions.
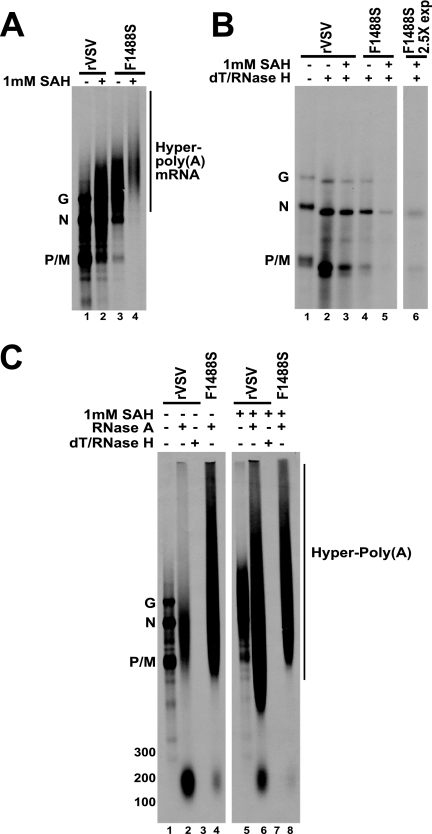
Having established that the F1488S rVSV retained significant methyltransferase activity, we examined its sensitivity to the presence of exogenous SAH with respect to polyadenylation in vitro. To assay RNA transcription in vitro in the absence or presence of exogenous SAH, we utilized the VSV in vitro transcription system described above. RNAs were labeled in the presence of [α-33P]ATP and were resolved on 1.5% acid agarose-urea gels (Fig. 4A). In the absence of exogenous SAH, the RNAs synthesized by the F1488S L protein were heterogeneous in length, confirming the hyperpolyadenylating phenotype previously described (Fig. 4A, lane 3). In the presence of 1 mM SAH, no discrete VSV mRNAs were observed, and the mRNAs migrated as a diffuse population larger than the G mRNA, as determined by comparison to the RNAs synthesized by the wild-type L protein in the absence of exogenous SAH, suggesting that the poly(A) effect on the F1488S L protein was further enhanced (Fig. 4A, lane 4). When the mRNAs shown in Fig. 4A were digested with RNase H following annealing with oligo(dT), each VSV mRNA was detected (N, P, M, and G), and all had indistinguishable migration compared to the wild type, indicating that the heterogeneity observed in Fig. 4A for the F1488S L protein in the absence or presence of exogenous SAH and for the wild-type L protein in the presence of 1 mM SAH was the result of hyperpolyadenylation (Fig. 4B). Notably, the level of RNA synthesized by the F1488S L protein in the presence of 1 mM SAH was greatly reduced compared to the synthesis in the absence of 1 mM SAH, which we suggest is likely due to polymerase pausing for increased lengths of time due to excessive polyadenylation at each gene junction (Fig. 4B, compare lanes 4 and 5). The dT/RNase H-digested RNA synthesized by the F1488S L protein after a 2.5-fold longer period of exposure of film is shown in Fig. 4B (lane 6).
FIG. 4.
Analysis of poly(A) tail synthesis by the F1488S rVSV. (A) Viral RNAs were transcribed in vitro in the presence of 1 mM SAH or no additional substrate. Transcription reactions were labeled with [33P]ATP. Labeled RNAs were purified and resolved on 1.5% acid agarose-urea gels. The gels were fluorogrammed, and radiolabeled RNAs were detected by autoradiography. The identities of VSV mRNAs are denoted on the left. The sizes of each undigested VSV mRNA minus their respective poly(A) tails are 1,332 nt (N), 821 nt (P), 837 nt (M), and 1,672 nt (G). (B) The RNAs shown in panel A were digested with RNase H in the presence of oligo(dT). Digested RNAs were ethanol precipitated and resolved on 1.5% acid agarose-urea gels. The gels were fluorogrammed, and radiolabeled RNAs were detected via autoradiography. The dT/RNase H-digested RNA synthesized by the F1488S L protein in the presence of 1 mM SAH after a 2.5-fold exposure of film is shown in lane 6. The identities of labeled RNAs are denoted on the left. (C) The RNAs shown in panel A were digested with RNase A and resolved on 1.5% acid agarose-urea gels. Representative digests for rVSV (+/−) SAH are shown in each panel: rVSV samples were digested with RNase A or RNase A, followed by dT/RNase H digestion. The gels were fluorogrammed, and radiolabeled RNAs were detected by autoradiography. [33P]ATP-labeled in vitro-transcribed size markers were generated from RNA Century Plus marker templates (Ambion) and resolved alongside undigested and digested RNAs. The location of each size marker is denoted on the left. The identities of undigested VSV mRNAs are also denoted on the left. The sizes of each undigested VSV mRNA minus their respective poly(A) tails are 1,332 nt (N), 821 nt (P), 837 nt (M), and 1,672 nt (G).
To examine the lengths of the poly(A) tails synthesized by the F1488S L protein, we utilized the RNase A assay described above. Briefly, the RNAs shown in Fig. 4A were digested with RNase A to separate the poly(A) tails from the nucleotides of the coding portion of the mRNA. We performed the same controls as described above, which included oligo(dT)/RNase H digestion of the RNase A-resistant RNAs (Fig. 4C, lanes 3 and 7), RNase 1 digestion of the RNase A-resistant RNAs (data not shown), and RNase A digestion of [α-33P]UTP-labeled RNAs (data not shown), all of which resulted in the digestion of all labeled RNA. As shown in Fig. 4C, when RNAs synthesized by wild-type polymerase were digested with RNase A, the resultant RNAs resolved as a relatively discrete product that migrated around 200 nt, as determined by comparison to size markers generated from in vitro transcription reactions (Fig. 4C, lane 2; the migration of size markers is noted on the left of the panel). Alternatively, when RNAs synthesized by wild-type rVSV in the presence of 1 mM SAH were digested with RNase A, the majority of resistant RNAs resolved as a heterogeneous population of 800 nt and larger (Fig. 4C, lane 6). When RNAs synthesized by the F1488S L protein in the absence of exogenous SAH were digested with RNase A, the resultant RNAs migrated as a large heterogeneous population similar to those synthesized by the wild-type L protein in the presence of exogenous SAH. Likewise, when the RNAs synthesized by the F1488S L protein in the presence of exogenous SAH were digested with RNase A, a similar heterogeneous profile was observed that appeared to migrate slightly slower, suggesting that these poly(A) tails were even longer than those synthesized in the absence of 1 mM SAH (Fig. 4A, lanes 3 and 4; Fig. 4C, lanes 4 and 8). These data further confirm the correlation between hyperpolyadenylation in the presence of exogenous SAH and the ability of the L protein to provide methyltransferase activity, which the F1488S L protein does (Table 1).
Analysis of poly(A) tail length in vivo.
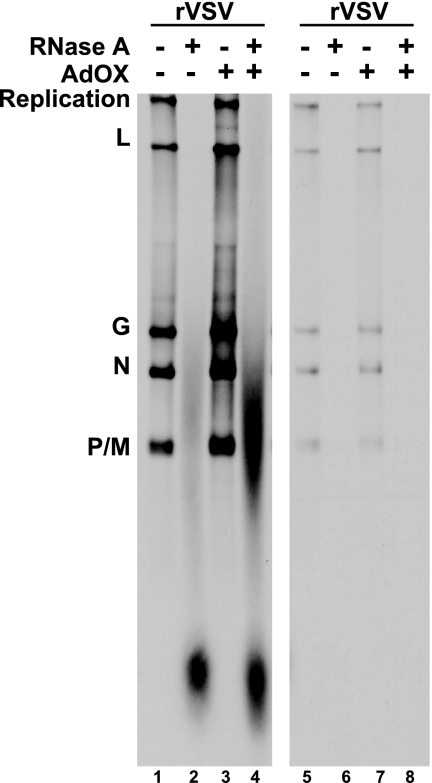
To date, all analyses of the effects of SAH on poly(A) tail synthesis by the VSV L protein have been done in vitro and not in cells. To investigate whether SAH-induced hyperpolyadenylation also occurred in VSV-infected cells, we examined transcription and poly(A) tail synthesis in the presence of elevated intracellular levels of SAH during a wild-type VSV infection. Elevating SAH levels in cells is more complicated than in vitro, which only requires the direct addition of the compound to the transcription reaction. To achieve elevated SAH levels in cells, a compound was used that inhibits the activity of SAH hydrolase, which is the enzyme responsible for metabolizing SAH into adenosine and homocysteine to maintain the appropriate balance of SAM to SAH within the cell, AdOX (10). Briefly, BHK cells were infected at an MOI of 5 at 37°C, and at 2 hpi infections were either treated with AdOX or left untreated. At 4.5 hpi, RNAs were labeled metabolically with [3H]adenine in the presence of actinomycin D. RNAs were isolated from cytoplasmic extracts and digested with either RNase A or RNase A, followed by RNase H digestion after annealing of oligo(dT). As a control, we labeled VSV-infected cells both in the absence and in the presence of AdOX with [3H]uridine and performed the same digestions mentioned above. Undigested and digested RNAs were visualized by acid agarose-urea gel electrophoresis and fluorography (Fig. 5). As shown in Fig. 5, when RNAs synthesized during a wild-type VSV infection in the absence of AdOX are digested with RNase A, the major population of poly(A) tails migrate at approximately 200 nt (Fig. 5, lane 2). Interestingly, similar to our observations in vitro in the absence of exogenous SAH, there is a minor population of poly(A) that migrated more slowly and was greater than 800 nt in size (Fig. 5, lane 2). When a wild-type VSV infection was treated with AdOX, not only was the overall incorporation of [3H]AMP greater than in the absence of AdOX, but when these RNAs were digested with RNase A, there was a significant increase in the levels of the slower migrating poly(A). These data indicated that the wild-type L protein was sensitive to elevated levels of SAH, which was achieved through the treatment of infected cells with AdOX, and showed that the hyperpolyadenylating effect in the presence of elevated levels of SAH occurs during mRNA synthesis in cells. These data further indicate that the wild-type L protein also synthesizes a small population of hyperpolyadenylated mRNAs during synthesis in infected cells, similar to that shown in vitro (Fig. 3A, lane 2; Fig. 5, lane 2).
FIG. 5.
Effect of AdOX on VSV transcription in cells. BHK cells were infected with wild-type VSV at an MOI of 5 at 37°C. At 2 hpi, cells were treated with 5 μM AdOX and incubated at 37°C. Viral RNAs were labeled with [3H]adenine (lanes 1 to 4) or [3H]uridine (lanes 5 to 8) in the presence of actinomycin D for 1 h. RNAs were harvested, and two-thirds of each sample was digested with RNase A as described in Materials and Methods. Radiolabeled RNAs were visualized by acid agarose-urea gel electrophoresis and fluorography.
DISCUSSION
In a recent study, we developed a structural homology model of the 2′-O-ribose methyltransferase domain of the VSV L protein. Based on that model, we identified residues in the putative 2′-O-ribose methyltransferase catalytic tetrad and the SAM binding domain for mutational analysis: G1672, G1674, G1675, L1716, and D1762 (Fig. 1) (12). We engineered a range of substitutions at these positions to test the structural and functional constraints of each selected residue. Mutation of residues G1672, G1674, and G1675 in the SAM binding motif and D1762 of the putative catalytic tetrad all diminished or reduced methyltransferase activity depending on the substitution made (see Table 2 for a summary). Alteration of L1716 reduced methyltransferase activity only if changed to tyrosine (Table 2). In the present study, we examined mRNA synthesis and polyadenylation by recombinant VSVs with mutations in domain VI of the VSV L protein at positions L1716, D1762, G1672, G1674, and G1675. In particular, we investigated their sensitivity to the presence of exogenous SAH with respect to polyadenylation. We found that polymerases that were defective in 5′ cap methyltransferase activity did not synthesize mRNAs with excessively long poly(A) tails in the presence of exogenous SAH, with the exception of the D1762E L protein, which synthesized excessively long poly(A) tails in the presence or absence of SAH, as shown in Fig. 2 and 3 (see Table 2 for summary). However, L proteins that retained the ability to methylate the 5′ cap were sensitive to SAH, as was the wild type. This was shown by the large poly(A) tails on mRNAs synthesized by L1716T, L1716Y, G1674P, and G1672A L proteins in the presence of exogenous SAH (Fig. 3 and Table 2). Further, the work presented here demonstrated that SAH-induced hyperpolyadenylation also occurs in cells, as evidenced by the excessively long poly(A) tails on mRNAs synthesized during a wild-type VSV infection in the presence of AdOX (Fig. 5).
The connection between SAH sensitivity and methyltransferase activity is unknown. One possibility is that if the residues analyzed by mutational analysis are involved in SAM binding, either directly or by formation of the binding pocket, then the mutations introduced may alter accessibility not only of SAM, but SAH also, thereby reducing the potential for SAH to affect polyadenylation. However, if SAM/SAH binding is unaffected by the presence of the mutation, the ability to render polymerase sensitive to SAH might be dependent on the ability to catalyze the cap methyltransferase reaction.
The mechanism by which SAH-induced hyperpolyadenylation occurs is also not understood. It has been shown that full-length mRNAs are synthesized in the absence of SAM and, likewise, SAH (21). The data indicate that it is the presence of SAH rather than the absence of SAM that causes this phenotype. It is possible that, in the presence of SAH, the polymerase adopts an altered conformation that favors increased pausing (decreased processivity relative to the template), which might allow polymerase to stutter and repeatedly add AMP residues to the 3′ end of an mRNA until SAM outcompetes SAH for binding, the poly(A) tail reaches a critical length, or an unknown factor signals polymerase to terminate polyadenylation. The altered conformation hypothesis might also explain why SAH sensitivity is not observed with L proteins that inherently lack the ability to provide cap methyltransferase activity. If the reason for the methyl transfer defect were the result of the inability of SAM or SAH to bind within the catalytic binding pocket of the L protein, the L protein would not have the opportunity to adopt this paused/less-processive conformation. However, this hypothesis requires further investigation.
The details of the mechanism of polyadenylation are not known, nor has the location of the active site on the polymerase that carries out polyadenylation been unambiguously established. The available evidence indicates that the U7 tract at each VSV gene end acts as the template for synthesis of the 3′ poly(A) tail by the polymerase via reiterative transcription or “slippage.” Shortening the U tract by a single nucleotide eliminates termination and slippage (6). In addition, it has been shown for VSV that the A/U-rich composition of the gene end tetra-nucleotide preceding the U-tract controls the ability of the polymerase to slip on the U7 tract. This suggests that polymerase slippage is controlled, at least in part, by the strength of base-pairing between the template and nascent strand (5). There have been other observations of polymerase slippage in polycistronic RNAs synthesized in vitro and in vivo, which contained large intervening poly(A) tracts or poly(U) tracts synthesized from an A7 template of a VSV subgenomic replicon (6, 17, 18, 26, 38), and during VSV G mRNA synthesis when the sequence 5′-UUUUUAA-3′ is transcribed (9). Taken together, these data have led to a general view that polyadenylation is carried out by the polymerase domain (domain III) of the VSV L protein. Extrapolation from eukaryotic systems, which utilize separate proteins for transcription and polyadenylation, led us to ask whether perhaps the poly(A) polymerase activity of the VSV L protein was carried out by another domain. However, BLAST searches using the L protein as the query failed to return any homology to known poly(A) polymerases. There are, however, three putative ATP binding sites located in the VSV L protein, two of which match the ATP binding site consensus sequence (GxGxxG[x]16-23K) and the other being a permutation of the consensus. The locations of the putative ATP binding sites are domain III (G754xGxxG[x]18K778), the variable region between domains V and VI (G1332xGxxG[x]14K1351), and domain VI (K1651X18GxGxxG1675), the last of the three being the noncanonical ATP binding site. Importantly, the site located within domain VI contains the SAM-binding motif (G1670xGxG1674) and the conserved catalytic tetrad (K1651DKE) that are important for methyltransferase activity (12, 13, 29). The site located within the variable region between domains V and VI, G1332xGxxG[x]14K1351, is an attractive candidate for the site of polyadenylation. This site is located in proximity to the phenylalanine at 1488 in the VSV L protein that was previously shown to hyperpolyadenylate VSV mRNAs when mutated to serine (22), which we have also confirmed here. This residue was also recently examined in a study that identified a region, VSV L protein residues 1450 to 1481, that was necessary for 5′ cap methylation (14). In light of the poly(A) phenotype associated with the F1488S L protein and the proximity of the ATP binding site to this newly identified methyltransferase region, it would be interesting if the VSV L protein utilized this putative ATP binding site to synthesize poly(A) tails. Although there are no data available suggesting that the polymerase utilizes a separate domain to carry out 3′ polyadenylation, it is conceivable that polymerase slippage can occur by reiterative transcription using an active site other than the polymerase active site located in domain III if the transfer of the nascent strand to the poly(A) site occurred without disruption of the template-nascent strand interaction.
The L-protein mutant, D1762E, which affects one of the residues of the putative catalytic tetrad involved in methyltransferase activity, synthesized hyperpolyadenylated mRNAs irrespective of the presence of exogenous SAH (Fig. 2 and 3). The hyper-poly(A) phenotype associated with the D1762E L protein was intriguing since this was the second L protein identified with such activity. A mutation was previously identified in the L protein of a temperature-sensitive VSV, ts(G)16, which displayed thermosensitive transcriptase activity, aberrantly long poly(A) tails, an increased response to SAH, and increased synthesis of polycistronic viral mRNAs (21, 22, 24-26). Analysis of revertants of ts(G)16, identified F1488S as the mutation responsible for the hyperpolyadenylating phenotype (22, 24, 25). This residue was recently investigated as part of a larger study that proposed a previously unidentified region of the L protein, located in proximity to this residue, was involved in 5′ cap methylation: amino acids 1450 to 1481 (14). In a previously mentioned study, the authors showed that mutation of the phenylalanine at position 1488 to alanine did not result in a methyltransferase defective L protein. The poly(A) activity of the F1488A L protein was not examined, however. During analysis of the recombinant F1488S virus, we introduced the F1488A mutation into the L gene of the VSV full-length clone and found that this mutation also resulted in hyperpolyadenylation of VSV mRNAs (data not shown). Similar to the analysis of the F1488A L protein, we analyzed the ability of a recombinant virus containing the F1488S mutation in the L gene to carry out methyltransferase activities and examined its sensitivity to exogenous SAH with respect to polyadenylation. The F1488S L protein synthesized 5′ methylated cap structures that were indistinguishable from wild-type, similar to the findings of Grdzelishvili et al. (14) for the F1488A rVSV and of Hunt (21) for ts(G)16 (14, 21). Due to the ability of the F1488S L protein to synthesize fully methylated cap structures, we expected that this L protein would be sensitive to the presence of exogenous SAH. We found that while the F1488S L protein synthesized large heterogeneous poly(A) tails in the absence of exogenous SAH, transcription in the presence of SAH resulted in poly(A) tails that migrated as if they were even longer (Fig. 4A and C).
Considering the structural homology model of the 2′-O-ribose methyltransferase domain of the L protein that we recently presented, if residue 1762, when mutated to glutamic acid, results in an L protein that hyperpolyadenylates even in the absence of exogenous SAH, is positioned with respect to SAM as depicted in our model, it is reasonable to suggest that the D1762E polymerase may favor binding to SAH over SAM. The difference in size between aspartic acid and glutamic acid is roughly equivalent to the methyl group of SAM, with the exception of a single hydrogen atom and thus may reasonably occupy similar three-dimensional space. The presence of glutamic acid at position 1762 may be large enough to interfere with SAM binding, but not SAH binding. Our viral in vitro transcription reactions contain SAM and SAH, as provided by the rabbit reticulocyte lysate; therefore, these experiments do not conclusively determine whether the hyperpolyadenylation observed for the D1762E polymerase is due to polymerase that favors SAH binding over SAM binding, or whether it is due to an intrinsic component of the mutant polymerase. Additional studies aimed at investigating the substrate binding ability of this L protein will be important in addressing this phenotype further.
Another interesting finding brought to light by the poly(A) analyses presented here, both in vitro and in infected cells, was that with wild-type VSV L protein in the absence of SAH, there was a minor population of poly(A) tails that were excessively long (>800 nt), similar to those observed with wild-type VSV in the presence of SAH (Fig. 3A, compare lanes 2 and 10; Fig. 5, compare lanes 2 and 4). Despite extensive studies delineating the cis-acting regulatory sequences involved in VSV transcription, there are many aspects of transcription that are poorly understood. In particular, the mechanism of transcriptional attenuation that results in the polar gradient of mRNA synthesis is not known. To date, we know that extending the gene end U-tract results in increased attenuation that becomes greater with each inserted UMP residue (6). Also, it was shown recently that extending the distance between the gene end and the gene start sequence, either by increasing the intergenic region or by overlapping the gene end and gene start sequences by various lengths, resulted in increased transcriptional attenuation (4). It has been postulated that hyperpolyadenylation may also affect downstream initiation (21). It was originally suggested that a population of polymerases disengage from the template during the events that occur at each gene junction (27). However, the facts are that under normal conditions, the VSV polymerase synthesizes ca. 30% fewer transcripts after crossing each gene junction. Our data suggest that a possible reason for transcriptional attenuation with wild-type VSV might be due, in part, to a low level of hyperpolyadenylation that occurs naturally, which may result in conditions where polymerase is more susceptible to disengagement from the template. However, more extensive investigation is needed to test this hypothesis. For example, are the long poly(A) tails synthesized in the absence of SAH equally distributed among all mRNAs, or is a particular mRNA more susceptible to hyperpolyadenylation?
In summary, the data presented here indicate that hyperpolyadenylation in the presence of SAH is dependent upon the methyltransferase activity of the L protein, suggesting a connection between 5′- and 3′-end mRNA modification. L proteins with mutations that rendered them unable to methylate the viral 5′ cap were not sensitive to the presence of SAH with respect to polyadenylation, whereas those that retained cap methyltransferase activity synthesized an excessively long poly(A) tail in the presence of 1 mM SAH. The data presented also show for the first time that elevated levels of SAH result in synthesis of mRNAs with excessively long poly(A) tails in VSV-infected cells, as well as in vitro. We have also identified an L-protein mutation, D1762E, that hyperpolyadenylates mRNA without regard to the presence or absence of exogenous SAH. Interestingly, this mutant does not possess methyltransferase activity, which may be due to preferential binding of SAH versus SAM.
Acknowledgments
We thank Andy Ball, John Barr, and fellow members of the Wertz lab for critical review of the manuscript and unwavering enthusiasm for this project. We also thank Ilya Frolov for the kind gift of the AlphaV-T7 cells and Paul Richardson for creating the image in Fig. 1.
This research was supported by NIH grant R37AI12464 and RO1AI12464 to G.W.W. and the Freedom to Discover Award from Bristol Myers Squibb to G.W.W.
Footnotes
Published ahead of print on 1 October 2008.
REFERENCES
- 1.Abraham, G., and A. K. Banerjee. 1976. Sequential transcription of the genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 731504-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball, L. A. 1977. Transcriptional mapping of vesicular stomatitis virus in vivo. J. Virol. 21411-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball, L. A., and C. N. White. 1976. Order of transcription of genes of vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 73442-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barr, J. N., X. Tang, E. Hinzman, R. Shen, and G. W. Wertz. 2008. The VSV polymerase can initiate at mRNA start sites located either up or downstream of a transcription termination signal but size of the intervening intergenic region affects efficiency of initiation. Virology 374361-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barr, J. N., and G. W. Wertz. 2001. Polymerase slippage at vesicular stomatitis virus gene junctions to generate poly(A) is regulated by the upstream 3′-AUAC-5′ tetranucleotide: implications for the mechanism of transcription termination. J. Virol. 756901-6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barr, J. N., S. P. Whelan, and G. W. Wertz. 1997. cis-Acting signals involved in termination of vesicular stomatitis virus mRNA synthesis include the conserved AUAC and the U7 signal for polyadenylation. J. Virol. 718718-8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barr, J. N., S. P. Whelan, and G. W. Wertz. 2002. Transcriptional control of the RNA-dependent RNA polymerase of vesicular stomatitis virus. Biochim. Biophys. Acta 1577337-353. [DOI] [PubMed] [Google Scholar]
- 8.Beilharz, T. H., and T. Preiss. 2007. Widespread use of poly(A) tail length control to accentuate expression of the yeast transcriptome. RNA 13982-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bilsel, P. A., and S. T. Nichol. 1990. Polymerase errors accumulating during natural evolution of the glycoprotein gene of vesicular stomatitis virus Indiana serotype isolates. J. Virol. 644873-4883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cools, M., and E. De Clercq. 1990. Influence of S-adenosylhomocysteine hydrolase inhibitors on S-adenosylhomocysteine and S-adenosylmethionine pool levels in L929 cells. Biochem. Pharmacol. 402259-2264. [DOI] [PubMed] [Google Scholar]
- 11.Emerson, S. U., and R. R. Wagner. 1973. L protein requirement for in vitro RNA synthesis by vesicular stomatitis virus. J. Virol. 121325-1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galloway, S. E., P. E. Richardson, and G. W. Wertz. 2008. Analysis of a structural homology model of the 2′-O-ribose methyltransferase domain within the vesicular stomatitis virus L protein. Virology [Epub ahead of print.] doi: 10.1016/j.virol.2008.08.041. [DOI] [PMC free article] [PubMed]
- 13.Grdzelishvili, V. Z., S. Smallwood, D. Tower, R. L. Hall, D. M. Hunt, and S. A. Moyer. 2005. A single amino acid change in the l-polymerase protein of vesicular stomatitis virus completely abolishes viral mRNA cap methylation. J. Virol. 797327-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grdzelishvili, V. Z., S. Smallwood, D. Tower, R. L. Hall, D. M. Hunt, and S. A. Moyer. 2006. Identification of a new region in the vesicular stomatitis virus L polymerase protein which is essential for mRNA cap methylation. Virology 350394-405. [DOI] [PubMed] [Google Scholar]
- 15.Green, T. J., S. Macpherson, S. Qiu, J. Lebowitz, G. W. Wertz, and M. Luo. 2000. Study of the assembly of vesicular stomatitis virus N protein: role of the P protein. J. Virol. 749515-9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hercyk, N., S. M. Horikami, and S. A. Moyer. 1988. The vesicular stomatitis virus L protein possesses the mRNA methyltransferase activities. Virology 163222-225. [DOI] [PubMed] [Google Scholar]
- 17.Herman, R. C., S. Adler, R. A. Lazzarini, R. J. Colonno, A. K. Banerjee, and H. Westphal. 1978. Intervening polyadenylate sequences in RNA transcripts of vesicular stomatitis virus. Cell 15587-596. [DOI] [PubMed] [Google Scholar]
- 18.Herman, R. C., M. Schubert, J. D. Keene, and R. A. Lazzarini. 1980. Polycistronic vesicular stomatitis virus RNA transcripts. Proc. Natl. Acad. Sci. USA 774662-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard, M., and G. Wertz. 1989. Vesicular stomatitis virus RNA replication: a role for the NS protein. J. Gen. Virol. 70(Pt. 10)2683-2694. [DOI] [PubMed] [Google Scholar]
- 20.Hunt, D. M. 1989. Effect of analogues of S-adenosylmethionine on in vitro polyadenylation by vesicular stomatitis virus. J. Gen. Virol. 70(Pt. 3)535-542. [DOI] [PubMed] [Google Scholar]
- 21.Hunt, D. M. 1983. Vesicular stomatitis virus mutant with altered polyadenylic acid polymerase activity in vitro. J. Virol. 46788-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunt, D. M., and K. L. Hutchinson. 1993. Amino acid changes in the L polymerase protein of vesicular stomatitis virus which confer aberrant polyadenylation and temperature-sensitive phenotypes. Virology 193786-793. [DOI] [PubMed] [Google Scholar]
- 23.Hunt, D. M., R. Mehta, and K. L. Hutchinson. 1988. The L protein of vesicular stomatitis virus modulates the response of the polyadenylic acid polymerase to S-adenosylhomocysteine. J. Gen. Virol. 69(Pt. 10)2555-2561. [DOI] [PubMed] [Google Scholar]
- 24.Hunt, D. M., E. F. Smith, and D. W. Buckley. 1984. Aberrant polyadenylation by a vesicular stomatitis virus mutant is due to an altered L protein. J. Virol. 52515-521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hutchinson, K. L., D. P. Bouknight, W. Fan, and D. M. Hunt. 1990. Revertants of a mutant of vesicular stomatitis virus which has an aberrant polyadenylation activity and a temperature-sensitive transcriptase. Virology 174444-449. [DOI] [PubMed] [Google Scholar]
- 26.Hutchinson, K. L., R. C. Herman, and D. M. Hunt. 1992. Increased synthesis of polycistronic mRNA associated with increased polyadenylation by vesicular stomatitis virus. Virology 18967-78. [DOI] [PubMed] [Google Scholar]
- 27.Iverson, L. E., and J. K. Rose. 1981. Localized attenuation and discontinuous synthesis during vesicular stomatitis virus transcription. Cell 23477-484. [DOI] [PubMed] [Google Scholar]
- 28.Li, J., E. C. Fontaine-Rodriguez, and S. P. Whelan. 2005. Amino acid residues within conserved domain VI of the vesicular stomatitis virus large polymerase protein essential for mRNA cap methyltransferase activity. J. Virol. 7913373-13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li, J., J. T. Wang, and S. P. Whelan. 2006. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA 1038493-8498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyles, D. S., M. O. McKenzie, P. E. Kaptur, K. W. Grant, and W. G. Jerome. 1996. Complementation of M gene mutants of vesicular stomatitis virus by plasmid-derived M protein converts spherical extracellular particles into native bullet shapes. Virology 21776-87. [DOI] [PubMed] [Google Scholar]
- 31.Matlin, K. S., H. Reggio, A. Helenius, and K. Simons. 1982. Pathway of vesicular stomatitis virus entry leading to infection. J. Mol. Biol. 156609-631. [DOI] [PubMed] [Google Scholar]
- 32.Ogino, T., and A. K. Banerjee. 2007. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell 2585-97. [DOI] [PubMed] [Google Scholar]
- 33.Pringle, C. R. 1970. Genetic characteristics of conditional lethal mutants of vesicular stomatitis virus induced by 5-fluorouracil, 5-azacytidine, and ethyl methane sulfonate. J. Virol. 5559-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Read, R. L., R. G. Martinho, S. W. Wang, A. M. Carr, and C. J. Norbury. 2002. Cytoplasmic poly(A) polymerases mediate cellular responses to S-phase arrest. Proc. Natl. Acad. Sci. USA 9912079-12084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Read, R. L., and C. J. Norbury. 2002. Roles for cytoplasmic polyadenylation in cell cycle regulation. J. Cell Biochem. 87258-265. [DOI] [PubMed] [Google Scholar]
- 36.Rhodes, D. P., S. A. Moyer, and A. K. Banerjee. 1974. In vitro synthesis of methylated messenger RNA by the virion-associated RNA polymerase of vesicular stomatitis virus. Cell 3327-333. [DOI] [PubMed] [Google Scholar]
- 37.Rose, J. K., H. F. Lodish, and M. L. Brock. 1977. Giant heterogeneous polyadenylic acid on vesicular stomatitis virus mRNA synthesized in vitro in the presence of S-adenosylhomocysteine. J. Virol. 21683-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schubert, M., and R. A. Lazzarini. 1981. In vivo transcription of the 5′-terminal extracistronic region of vesicular stomatitis virus RNA. J. Virol. 38256-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szilagyi, J. F., and C. R. Pringle. 1972. Effect of temperature-sensitive mutations on the virion-associated RNA transcriptase of vesicular stomatitis virus. J. Mol. Biol. 72281-292. [DOI] [PubMed] [Google Scholar]
- 40.Testa, D., and A. K. Banerjee. 1977. Two methyltransferase activities in the purified virions of vesicular stomatitis virus. J. Virol. 24786-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, J. T., L. E. McElvain, and S. P. Whelan. 2007. Vesicular stomatitis virus mRNA capping machinery requires specific cis-acting signals in the RNA. J. Virol. 8111499-11506. [DOI] [PMC free article] [PubMed] [Google Scholar]