Abstract
Hepatitis C virus (HCV) is one of the major causative agents of virus-related hepatitis, liver cirrhosis, and hepatocellular carcinoma in humans. Translation of the HCV polyprotein is mediated by an internal ribosomal entry site (IRES) in the 5′ nontranslated region of the genome. Here, we report that a cellular protein, hnRNP D, interacts with the 5′ border of HCV IRES (stem-loop II) and promotes translation of HCV mRNA. Overexpression of hnRNP D in mammalian cells enhances HCV IRES-dependent translation, whereas knockdown of hnRNP D with small interfering RNAs (siRNAs) inhibits translation. In addition, sequestration of hnRNP D with an interacting DNA oligomer inhibits the translation of HCV mRNA in an in vitro system. Ribosome profiling experiments reveal that HCV RNA is redistributed from heavy to light polysome fractions upon suppression of the hnRNP D level using specific siRNA. These results collectively suggest that hnRNP D plays an important role in the translation of HCV mRNA through interactions with the IRES. Moreover, knockdown of hnRNP D with siRNA significantly hampers infection by HCV. A potential role of hnRNP D in HCV proliferation is discussed.
Hepatitis C virus (HCV), one of the major causative agents of virus-related liver cirrhosis and hepatocellular carcinoma in humans, contains a single-stranded RNA genome of positive polarity. HCV RNA contains nontranslated regions (NTRs) at the 5′ and 3′ ends (18, 23) and a long open reading frame encoding polyprotein that is synthesized through a single translational initiation event directed by an RNA element designated the internal ribosomal entry site (IRES) (21, 38) at the 5′ NTR (46). The polyprotein is proteolytically processed into 10 or more viral proteins.
In general, IRES elements need several canonical translation factors (except eukaryotic initiation factor 4E [eIF4E]) for their activities (40, 42). However, HCV IRES-dependent translation requires only a few canonical factors (eIF2, eIF3, eIF5, and eIF5B) for function (30, 39, 41). Additionally, cellular proteins known as IRES-specific cellular transacting factors (ITAFs) are required for the efficient translation of HCV mRNA. For instance, polypyrimidine tract-binding protein (PTB) interacting with HCV IRES is required for IRES function (2, 3). La antigen interacting with the GCAC site near the initiator AUG is necessary for the optimal function of the HCV IRES (1, 10, 43). Recent studies have shown that NSAP1 interacting with the adenosine-rich core-coding region of HCV mRNA augments HCV IRES-dependent translation (27).
Heterogeneous ribonucleoprotein D (hnRNP D), also known as AU-rich element RNA-binding protein 1 (AUF1), is an hnRNP family member that shuttles between the nucleus and cytoplasm (44, 48). hnRNP D was initially identified owing to its ability to bind and destabilize c-myc mRNA in a crude in vitro decay system (6). The protein has four isoforms of different molecular weights (p37, p40, p42, and p45), all of which are produced by alternate splicing of a single transcript (11, 47). The hnRNP D protein has various functions, including mRNA decay (6), telomere maintenance (12), translation initiation (31, 36), and mCRD-mediated mRNA turnover (16). Another recent report showed that hnRNP D enhances the translation of c-myc mRNA (33).
Here, we show that hnRNP D binds HCV IRES and promotes translation of the viral protein. Specifically, hnRNP D interacts with the stem-loop II of HCV 5′ NTR, and its overexpression enhances HCV IRES-dependent translation. Conversely, knockdown of hnRNP D inhibits the translation process. Analysis of the ribosomal profile with or without hnRNP D depletion also indicates that hnRNP D functions as an ITAF of HCV IRES. Interestingly, knockdown of hnRNP D results in increased HCV RNA replication inside cells, indicating that the protein represses HCV RNA replication. Based on the results, we conclude that hnRNP D functions in modulating HCV proliferation by balancing replication and translation of RNA.
MATERIALS AND METHODS
Plasmid construction and small interfering RNA (siRNA).
Dual reporters harboring the HCV IRES, encephalomyocarditis virus (EMCV) IRES, and c-myc IRES were constructed as described previously (27). The monocistronic reporter containing the HCV IRES and EMCV IRES, followed by firefly luciferase used for in vitro translation, were prepared according to methods described in a previous report (28). Plasmids expressing hnRNP D, pFLAG-CMV2 p37, pFLAG-CMV2 p40, pFLAG-CMV2 p42, and pFLAG-CMV2 p45 were kindly provided by R. J. Schneider at New York University School of Medicine (44). To generate pRSET A-hnRNP D for recombinant hnRNP D purification, pFLAG-CMV2 p45 was treated with HindIII-Klenow-EcoRI, and the resulting DNA fragment was cloned into NcoI-Klenow-EcoRI-treated pRSET A (Invitrogen).
To construct pH(130-228)CAT and pH(229-402)CAT used for generating an RNA probe (see Fig. 2B, below), HCV IRES corresponding to positions 130 to 228 and 229 to 402 were amplified from pH(18-402)CAT (27) using the following primer pairs: 5′-CTAGGTACCGGGAGAGCCATAG-3′ and 5′-CGGGATCCAAATCTCCAGGC-3′ for pH(130-228)CAT and 5′-CTAGGTACCGGGCGTGCCCCC-3′ and 5′-CGGGATCCCCTGTGGGCGGC-3′ for pH(229-402)CAT. PCR fragments were treated with Asp718-BamHI and cloned into the corresponding sites of pH(18-402)CAT.
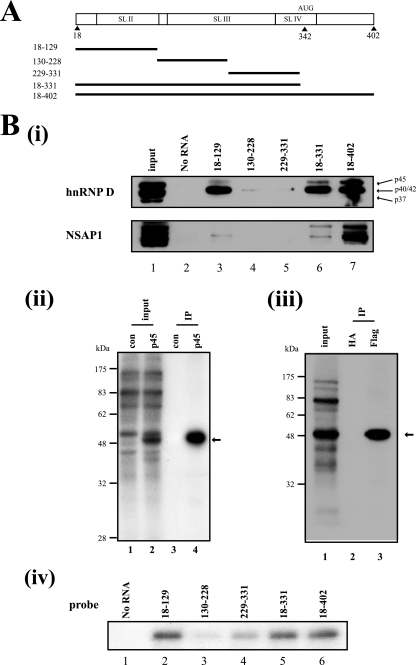
FIG. 2.
hnRNP D interacts with SL II of the HCV IRES. (A) Schematic diagrams of HCV RNA used in UV cross-linking experiments and affinity chromatography. AUG denotes the initiator codon. (B, panel i) Cytoplasmic extracts of Huh7 cells were subjected to an in vitro RNA-binding assay with biotinylated RNA probes corresponding to different regions of the HCV IRES (positions 18 to 129, 130 to 228, 229 to 331, 18 to 331, and 18 to 402 in lanes 3 to 7, respectively). Immunoblot analysis was performed with anti-hnRNP D (upper panel) and anti-NSAP1 antibodies (lower panel). Each hnRNP D isoform is denoted with arrows. (ii) Immunoprecipitation of UV cross-linked Flag-tagged p45 isoform of hnRNP D with a 32P-labeled HCV RNA probe (nt 18 to 402). After UV cross-linking with cytoplasmic extracts of empty vectors and hnRNP D expression plasmid-transfected 293T cells, samples were precleared with protein G-agarose resin and immunoprecipitated with 2 μg of anti-Flag antibodies. Resin-bound proteins were resolved by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis (SDS-PAGE; lanes 3 and 4). Lanes 1 and 2 depict 32P-labeled proteins before immunoprecipitation in empty vector-transfected and hnRNP D-coding plasmid-transfected cells, respectively. The arrow signifies immunoprecipitated protein. (iii) UV cross-linked cytoplasmic extracts of Flag-hnRNP D overexpressing 293T cells were immunoprecipitated with 2 μg of anti-Flag antibodies or anti-HA antibodies as a negative control. After immunoprecipitation, resin-bound proteins were subjected to SDS-PAGE (lanes 2 and 3). The arrow indicates immunoprecipitated protein. (iv) UV cross-linking experiments were performed with 200 ng of purified recombinant p45 isoform of hnRNP D protein and 32P-labeled RNA corresponding to probes 18-129 (lane 2), 130-228 (lane 3), 229-331 (lane 4), 18-331 (lane 5), and 18-402 (lane 6).
To construct a plasmid expressing short hairpin RNA (shRNA) against hnRNP D, we selected the target site on hnRNP D mRNA corresponding to nucleotides (nt) 867 to 895 (exon 7) downstream of the initiation codon. The DNA oligomers 5′-CTAGTCGACCTGGAACCAGGGATATAGTAACTATTGGATTCAAGAGATCCAATAGTTACTATATCCCTGGTTCCAGTTTTTCTAGAGA-3′ and 5′-CTAGTCGACCTGGAACCAGGGATATAGTAACTATTGGACTTCCTGTCATCCAATAGTTACTATATCCCTGGTTCCAGTTTTTCTAGAGA-3′ were annealed and ligated into plasmid pEBV-U6+27 (8) treated with SalI and XbaI.
Duplex siRNAs targeting exon 1 of hnRNP D (nt 201 to 219) and control siRNA were purchased from Bioneer Inc. (Korea). The siRNA sequence targeting hnRNP D and the control RNA sequence were 5′-GAUUGACGCCAGUAAGAAC(dTdT)-3′ and 5′-CCUACGCCACCAAUUUCGU(dTdT)-3′, respectively.
Cell culture.
293T, Huh7, and Huh 7.5.1 cells were cultured in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (Clontech). FK cells containing the full-length HCV genome (32) and Huh-luc/neo-ET cells containing the subgenomic replicon carrying both the neomycin phosphotransferase and firefly luciferase genes (25) were grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal bovine serum (Clontech) and the antibiotic G418 (600 μg/ml; Calbiochem). Huh 5-15 cells containing the subgenomic replicon I389hyg-ubi/NS3-3′ were provided by R. Bartenschlager at the University of Heidelberg (13). Cells were grown in Dulbecco's modified Eagle's medium (Gibco) with 10% fetal bovine serum (Clontech) and hygromycin (300 μg/ml; Calbiochem).
Antibodies.
Anti-Flag and anti-NSAP1 antibodies were purchased from Sigma, anti-actin antibody from ICN, and anti-hnRNP D antibody from Upstate Biotechnology. Anti-PTB and anti-La (7), anti-NS5B and anti-core (32), and anti-NS5A (24) antibodies have been described elsewhere.
UV cross-linking and immunoprecipitation of cross-linked proteins.
All experiments were performed according to previous reports (26), except that 32P-labeled RNA corresponding to fragments of HCV IRES and purified hnRNP D were used as the interacting RNA and protein, respectively.
RNA affinity chromatography.
RNA affinity chromatography experiments were performed as described previously (27), except that Huh7 cytoplasmic extracts and biotinylated RNA corresponding to HCV IRES fragments were used.
Establishment of cell clones expressing hnRNP D shRNAs.
HeLa cells were transfected with control plasmids or those expressing shRNA (pEBV-U6+27 and pEBV-U6+27/hnRNP D; 1 μg each) by electroporation. From 48 h posttransfection, cells were maintained in Dulbecco's modified Eagle's medium containing hygromycin (300 μg/ml; Calbiochem). After 1 month of selection, hygromycin-resistant colonies were pooled and cultivated for further analysis.
Preparation of HeLa S10 cell extracts and in vitro translation.
Cytoplasmic S10 extracts of HeLa S3 cells were prepared as specified in an earlier report (9). In vitro translation reactions in HeLa S10 were performed for 1 h in a 12.5-μl reaction mixture (130 mM potassium acetate and 1.5 mM magnesium acetate) containing 10 nM mRNA at 30°C.
RNA purification and analysis.
Total RNA was extracted using the TRI reagent (Invitrogen). Total RNA (20 μg) from transfected 293T cells was analyzed by Northern blot analysis (8). Total RNA (2 μg) was reverse transcribed using Improm II reverse transcriptase (Promega), and the cDNA was subjected to real-time PCR analysis for quantification using Sybr premix Ex Taq (Takara). Primer sequences for reverse transcription-PCR and real-time PCR were as follows: HCV, 5′-GTCTAGCCATGGCGTTAGTA-3′ and 5′-CTCCCGGGGCACTCGCAAGC-3′ (29); glyceraldehyde-3-phosphate dehydrogenase (GAPDH), 5′-TGCACCACCAACTGCTTAG-3′ and 5′-GAGGCAGGGATGATGTTC-3′ (45).
Ribosomal profiling.
Huh 5-15 cells were subjected to sucrose gradient analysis, as described elsewhere (8). Fractionated samples were classified as shown below in Fig. 6. Total RNA of each fraction was purified using TRI reagent (Invitrogen) and subjected to real-time PCR analysis for quantification.
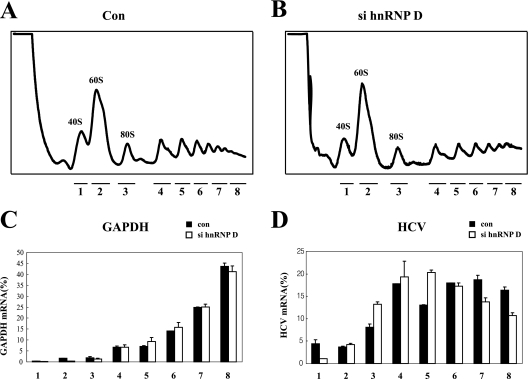
FIG. 6.
Knockdown of hnRNP D results in redistribution of HCV RNAs in a ribosomal profile. (A and B) Ribosomal profiles in sucrose gradients. Huh 5-15 cells were transfected with control or si hnRNP D by using oligofectamine. Ribosomal distribution in sucrose density gradients was analyzed 48 h posttransfection in cell extracts. RNA samples were purified from eight fractions in the sucrose gradient, as follows: fraction 1, 40S ribosomal fraction; 2, 60S ribosomal fraction; 3, monosomal fraction; 4 to 8, disome and polysomal fractions. (C and D) Distribution of mRNA in sucrose gradients. The amounts of GAPDH mRNA (C) and HCV RNA (D) across the gradient were analyzed by real-time PCR, and the relative amounts of RNA in each fraction are depicted by black bars (RNA in cells treated with control siRNA) or white bars (RNA in cells treated with si hnRNP D). The numbers under the graphs depict the fraction number.
Virus infection.
Preparation of infectious JFH 5A-RLuc and infection experiments were performed as described in a previous report (24).
RESULTS
hnRNP D enhances HCV IRES-dependent translation.
Since hnRNP D is cofractionated and coimmunoprecipitated with NSAP1 (16), which enhances HCV IRES activity (27), we examined its effects on HCV IRES-dependent translation. The effects of hnRNP D isoforms were monitored by cotransfection of 293T cells with effector plasmids expressing these proteins (Fig. 1A, panel ii) and a reporter plasmid producing dicistronic mRNA consisting of Renilla luciferase (RLuc), which is translated in a cap-dependent manner, followed by firefly luciferase (FLuc) under the translational control of HCV IRES (nt 18 to 402) (Fig. 1A, panel i, top). Among the hnRNP D isoforms, p37 and p45 enhanced HCV IRES-dependent translation, as measured from the relative ratio of firefly luciferase to Renilla luciferase activity (Fig. 1B, compare lanes 2 and 5 with lane 1). The p40 and p42 isoforms had no significant effect on the translation of HCV mRNA in 293T cells (Fig. 1B, panel i, compare lanes 3 and 4 with lane 2), despite similar expression levels of the isoforms on a Western blot (Fig. 1B, panel 1, α-Flag). The p45 isoform showed the strongest translational enhancing activity in Huh7 cells (hepatocellular carcinoma cells), the same as in 293T cells. Interestingly, p42 but not p37 enhanced translation through the HCV IRES in Huh7 cells (Fig. 1B, panel i, compare lanes 2 and 4 with the corresponding lanes in C, panel i). This discrepancy may be attributed, at least in part, to the varied amounts of hnRNP D isoforms in different cells and to the varied strength of binding affinity of hnRNP D isoforms to RNA (19, 22). Since p45 isoform of hnRNP D enhanced HCV IRES-dependent translation to a maximal extent in both cells (Fig. 1B, panel i, and C, panel i, lane 5), we focused on the p45 isoform of hnRNP D (designated p45′) in subsequent experiments.
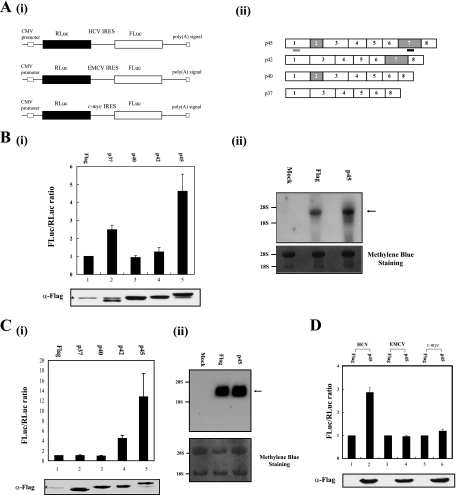
FIG. 1.
hnRNP D augments HCV IRES-dependent translation in vivo. (A, panel i) Schematic diagrams of dicistronic reporter plasmids used to monitor the efficiency of cap- and IRES-dependent translation in vivo. The vectors contain the cytomegalovirus (CMV) immediate-early enhancer-promoter (CMV promoter) to direct transcription in cells. (ii) Schematic diagrams of mRNAs encoding different hnRNP D isoforms. The shRNA and siRNA target sites are underlined in black and gray, respectively. (B and C, panel i) 293T cells (B) or Hun7 cells (C) were cotransfected with a dicistronic reporter plasmid harboring the HCV IRES and an effector plasmid, pFLAG-CMV2 (lane 1), and its derivatives expressing hnRNP D isoforms (lanes 2 to 5). At 48 h posttransfection, the relative luciferase activities in transfected cells were measured. The ratio of firefly luciferase to Renilla luciferase activity in cells transfected with plasmid pFLAG-CMV2 was set to 1 (upper panels). Experiments were performed at least three times for each set, and standard deviations are presented as error bars. Lysates were analyzed by Western blotting with a monoclonal anti-Flag antibody (lower panels). The asterisk indicates a nonspecific band observed in all cells. (ii) Integrity of dicistronic mRNA. Northern blot analysis was performed with 20 μg of total RNA isolated from transfected 293T cells (B) or Huh7 cells (C). The RNA content was visualized by methylene blue staining (lower panels). The positions of the human 28S and 18S bands are indicated. The arrow signifies the position of reporter mRNA. (D) Translation activities of IRES-containing dicistronic mRNA in 293T cells. Dicistronic reporter constructs (HCV IRES in lanes 1 and 2, EMCV IRES in lanes 3 and 4, c-myc IRES in lanes 5 and 6), an effector plasmid expressing p45 (lanes 2, 4, and 6), and a negative control effector pFLAG-CMV2 (lanes 1, 3, and 5) were cotransfected. At 48 h after transfection, cells were harvested and their luciferase activities measured. Luciferase activities in cells transfected with the pFLAG-CMV2 plasmid were set to 1 (upper panel). Error bars represent standard deviation values. Lysates were subjected to immunoblot analysis with antibodies against Flag (lower panel).
To exclude the possibility that the increase in firefly luciferase activity by p45 protein is due to monocistronic reporter mRNA, which may be generated by cryptic promoter activity, aberrant splicing, or cleavage of dicistronic mRNA by a putative endonuclease, we monitored the integrity of the dicistronic reporter mRNA containing the HCV IRES element by Northern blot analysis with a 32P-labeled probe corresponding to the firefly luciferase gene (Fig. 1B, panel ii, and C, panel ii). Overexpression of p45 did not affect the integrity of reporter mRNA, and putative RNA of monocistronic mRNA size was not detected (Fig. 1B, panel ii, and C, panel ii). Together, these results indicate that some hnRNP D isoforms enhance HCV IRES-dependent translation in vivo, and the p45 isoform displays the strongest ITAF activity.
We investigated the effects of p45 on several IRES elements through cotransfection of dual reporter plasmids containing HCV, EMCV, and c-myc IRES (Fig. 1A, panel i, and D). Overexpression of p45 enhanced the IRES activity of HCV mRNA, as expected, but did not affect EMCV or c-myc IRES-dependent translation in 293T cells (Fig. 1D, upper panel), despite similar levels of the effector protein (p45) (Fig. 1D, lower panel).
We monitored the effects of cotransfection of plasmids expressing NSAP1 and hnRNP D, since hnRNP D interacts with NSAP1. The overproduction of NSAP1 and hnRNP D together showed an additive effect rather than synergistic effect, which might be observed if they functioned in a cooperative manner (data not shown). This suggests that NSAP1 and hnRNP D function independently.
hnRNP D interacts with the HCV IRES element.
Several known ITAFs function by interacting with IRES RNA. To validate whether hnRNP D binds the HCV IRES element, we performed in vitro RNA-protein binding assays using various segments of HCV RNA corresponding to the 5′ NTR and the coding region of the N terminus of the core protein (Fig. 2A). Cytoplasmic extracts from Huh7 cells were incubated with biotinylated RNA probes corresponding to regions shown in Fig. 2A, and the RNA-protein complexes were precipitated with streptavidin-agarose beads, followed by detection of hnRNP D protein by Western blotting. Consistent with the riboproteomics data with HCV IRES (35), hnRNP D protein interacted with the full-length HCV IRES element (Fig. 2B, panel i, lane 7). Interestingly, binding strength to the HCV IRES varied among the hnRNP D isoforms (Fig. 2B, panel i, lane 7). The region between nt 18 and 129, including stem-loop II (SL II) of HCV IRES, was sufficient for binding to hnRNP D (Fig. 2B, panel i, lanes 3, 6, and 7). On the other hand, hnRNP D did not bind RNA corresponding to positions 130 to 228 or 229 to 331 (Fig. 2B, panel i, lanes 4 and 5).
Direct interactions of hnRNP D with HCV IRES were confirmed using UV cross-linking and immunoprecipitation. Cytoplasmic extracts of 293T cells transfected with a plasmid expressing the Flag-tagged p45 isoform of hnRNP D or a negative control vector were immunoprecipitated using an anti-Flag antibody after UV cross-linking reactions with 32P-labeled HCV RNA (nt 18 to 402) and RNase digestion (Fig. 2B, panel ii). The p45 protein was clearly detected by UV cross-linking and immunoprecipitation of cell extracts transfected with p45-expressing cells (Fig. 2B, panel ii, lane 4, and panel iii, lane 3). No band was detected with the extracts of cells transfected with the control vector (Fig. 2B, panel ii, lane 3) or when antihemagglutinin (anti-HA) antibody was used in the immunoprecipitation (Fig. 2B, panel iii, lane 2). These data strongly suggest that p45 directly interacts with HCV IRES in the cell.
The hnRNP D and HCV IRES interactions were further confirmed using purified p45 protein and radiolabeled RNA corresponding to different portions of the HCV IRES (Fig. 2A). Purified hnRNP D protein bound efficiently to RNA containing nt 18 to 129 (Fig. 2B, panel iv, lanes 2, 5, and 6) and weakly to those lacking nt 18 to 129 (Fig. 2B, panel iv, lanes 3 and 4). These data are consistent with the results of protein precipitation with biotinylated RNA and Western blotting shown in Fig. 2B, panel i. These results collectively indicate that hnRNP D interacts directly with HCV IRES through nt 18 to 129, including SL2 of the IRES.
Knockdown of hnRNP D by shRNA reduces HCV IRES-dependent translation.
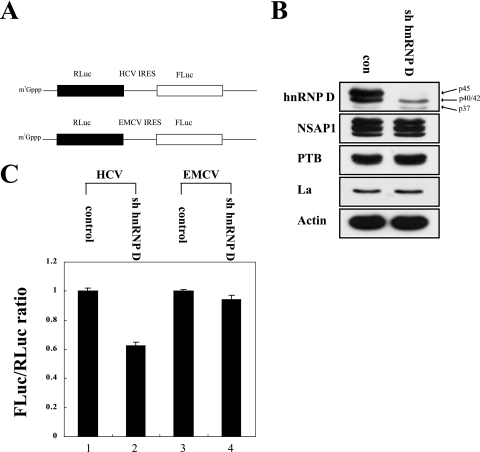
To further analyze the effects of hnRNP D on HCV IRES-dependent translation, we monitored the effects of hnRNP D knockdown with shRNA. We generated a plasmid expressing hnRNP D-specific shRNA (sh hnRNP D) targeting exon 7, which is only found in the p42 and p45 isoforms of hnRNP D mRNA (Fig. 1A, panel ii), because p45 showed the strongest ITAF activity (Fig. 1B). Cell lines expressing shRNA against hnRNP D and negative controls were generated by transfecting HeLa cells with a plasmid expressing hnRNP D-specific shRNA or empty vector and then collecting hygromycin-resistant cells. Cells expressing shRNA against exon 7 of hnRNP D displayed a markedly reduced level of p45 and reduced level of p40/p42 isoforms of hnRNP D, which was most likely due to knockdown of p42, but retained p37 isoform (Fig. 3B, hnRNP D). On the other hand, the levels of actin and other ITAFs that affect HCV IRES-dependent translation, such as NSAP1, PTB, and La, remained unchanged (Fig. 3B, NSAP1, PTB, La, and actin), indicating the specificity of this shRNA. The effects of hnRNP D knockdown on HCV IRES-dependent translation were monitored via transfection of dicistronic reporter mRNAs containing HCV IRES or EMCV IRES in the intercistronic regions (Fig. 3A) into control and hnRNP-D knockdown cells. The RNA transfection method was used to eliminate the possibility of aberrant mRNA production through a putative cryptic promoter or cryptic splicing accepter in HCV IRES that might be activated when dicistronic mRNAs are generated by DNA transfection. HCV IRES-dependent translation was reduced upon transfection of dicistronic RNA into hnRNP D knockdown cells compared with control cells (Fig. 3C, lanes 1 and 2). Notably, about 60% of HCV IRES activity remained, even after knockdown of the p42/p45 isoforms of hnRNP D. Residual translation activity through HCV IRES is possibly attributed, at least in part, to the other isoforms of hnRNP D (p37/p40), because p37 augments HCV IRES-dependent translation (Fig. 1B) and siRNA that suppressed all isoforms of hnRNP D induced much stronger inhibition of HCV IRES activity (see below). On the other hand, translation of EMCV IRES was not affected by knockdown of hnRNP D (Fig. 3C, lanes 3 and 4). These results, along with p45 expression data (Fig. 1B), strongly suggest that the p45 isoform of hnRNP D protein augments HCV IRES-dependent translation.
FIG. 3.
Effects of an hnRNP D-specific shRNA on HCV IRES activity. (A) Schematic diagrams of dicistronic mRNA used in RNA transfection experiments. The intercistronic regions of dicistronic mRNA contain the HCV IRES (nt 18 to 402) and EMCV IRES, respectively, are shown. Dicistronic mRNAs were synthesized with T7 RNA polymerase in the presence of 7-methyl-GpppG to add cap structures at the 5′ end. (B) Western blot analysis of HeLa cells expressing hnRNP D-specific shRNA (sh hnRNP D). Stably transformed HeLa cells were generated by transfection of the control vector and that encoding sh hnRNP D. Cells were harvested, and protein levels were analyzed by immunoblotting with anti-hnRNP D, -NSAP1, -PTB, -La, and -actin antibodies. (C) Translation activities of dicistronic mRNA in shRNA-expressing cells. Capped dicistronic mRNAs were transfected into control cells (lanes 1 and 3) and sh hnRNP D-expressing cells (lanes 2 and 4). After 3 h of incubation, luciferase activities in cells were measured. The ratio of FLuc to RLuc activity in control cells was set to 1 (lanes 1 and 3). Experiments were performed at least three times for each set, and standard deviations are presented as error bars.
Sequestration of hnRNP D with an oligodeoxynucleotide impedes HCV IRES-dependent translation in vitro.
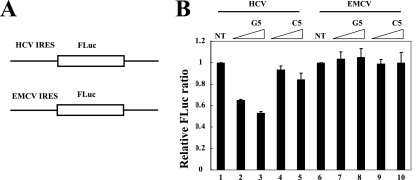
Previously, we developed a useful method to identify the cellular proteins augmenting IRES activities (9). The underlying principle of this method is that several RNA-binding proteins bind to specific DNA sequences. Thus, addition of an oligodeoxynucleotide that specifically interacts with an RNA-binding protein, such as ITAF, inactivates protein function through sequestration from its own target RNA (9). hnRNP D interacts strongly with the G-rich strand of telomeric DNA repeats (TTAGGG)4 (12). A G-to-C substitution of the fifth nucleotide, (TTAGCG)4, abolishes the hnRNP D-binding capacity of telomeric DNA repeats. To determine the function of hnRNP D in HCV IRES-dependent translation, we monitored the effects of (TTAGGG)4 and (TTAGCG)4 on HCV IRES-dependent translation using an in vitro translation system. In vitro translation reactions of monocistronic reporter mRNA containing HCV IRES or EMCV IRES, followed by firefly luciferase (Fig. 4A), were performed in HeLa S10 extracts in the presence of (TTAGGG)4 or (TTAGCG)4 (hereafter referred to as G5 and C5, respectively). Addition of G5 DNA oligomers, which bind hnRNP D with high affinity, inhibited HCV IRES-dependent translation in a dose-dependent manner (Fig. 4B, lanes 2 and 3). On the other hand, C5 DNA oligomers that bind weakly to hnRNP D inhibited HCV IRES-dependent translation only slightly, even at high concentrations (Fig. 4B, lanes 4 and 5). Neither G5 nor C5 oligomers inhibited EMCV IRES-dependent translation, indicating that the inhibitory effect of the G5 oligomer on HCV IRES-dependent translation is not attributed to nonspecific inactivation of the translation system (Fig. 4B, lanes 6 to 10). These data also indicate that hnRNP D plays a positive role in HCV IRES-dependent, but not in EMCV IRES-dependent, translation.
FIG. 4.
hnRNP D-interacting DNA oligomers inhibit HCV IRES-dependent translation in vitro. (A) Schematic diagrams of monocistronic reporter mRNA used in an in vitro translation assay. The HCV IRES (nt 18 to 402) and EMCV IRES sequences are followed by firefly luciferase. (B) Effects of G5 and C5 oligomers on IRES-dependent translation in HeLa S10 extracts. Monocistronic reporters (10 nM, final concentration) were used in translation reactions in the presence of G5 (lanes 2, 3, 7, and 8) and C5 (lanes 4, 5, 9, and 10) oligomers or in the absence of oligonucleotides (lanes 1 and 6). The final concentrations of oligomers were 2 μM (lanes 2, 4, 7, and 9) or 4 μM (lanes 3, 5, 8, and 10). The firefly luciferase activity in extracts without oligomers was set to 1. Experiments were performed at least three times for each experimental set, and standard deviations are presented as error bars. NT, not treated.
Knockdown of hnRNP D with siRNA inhibits HCV IRES-dependent translation in HCV replicon-containing cells.
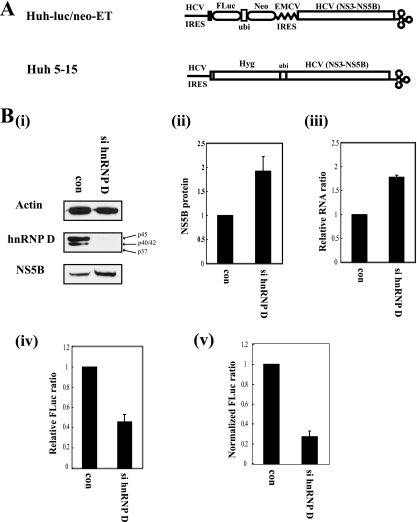
HCV replicons mimic viral proliferation occurring in infected cells (5) and are therefore good model systems to investigate the replication process and the translation event of HCV. We used a dicistronic HCV replicon, denoted Huh-luc/neo-ET, which contains two cistrons composed of a fusion protein (firefly luciferase [a reporter]-ubiquitin-neomycin phosphotransferase II [a selection marker] consecutively) under the control of HCV IRES and HCV nonstructural proteins (from NS3 to NS5B, required for replication of HCV RNA) under the control of the EMCV IRES (Fig. 5A, upper panel) (25) for investigating the effects of siRNA against hnRNP D on HCV IRES activity. In this case, we used siRNA targeting all different isoforms of hnRNP D (Fig. 1A, panel ii). Compared with cells transfected with control siRNA, those transfected with siRNA against hnRNP D displayed markedly reduced levels of all isoforms of hnRNP D (Fig. 5B, panel i, hnRNP D). Interestingly, the level of NS5B protein under the control of the EMCV IRES was increased about twofold in cells depleted of hnRNP D (Fig. 5B, panel i, NS5B, and panel ii). This effect is possibly attributable to the twofold increase in the level of HCV RNA in hnRNP D-depleted cells, as measured by real-time PCR in cells transfected with siRNA against hnRNP D (Fig. 5B, panel iii; also see below). Depletion of hnRNP D did not affect EMCV IRES-dependent translation (Fig. 3C and 4B). It is plausible that hnRNP D blocks replication of HCV to avoid the collision between translational and replication machineries. An analogous situation is reported for the interplay between translation and replication of polioviral RNA (4, 15), where PCBP and/or PTB have key roles in the switch from translation to replication (see Discussion, below). Further investigation is required to clarify the molecular basis of the increase in the HCV RNA level in hnRNP D-depleted cells. Importantly, the level of firefly luciferase under the control of the HCV IRES was reduced by about 60% in cells transfected with siRNA against hnRNP D (Fig. 5B, panel iv). The actual effect of hnRNP D knockdown on HCV IRES function is even more dramatic (about 80%), considering the increased HCV RNA level (Fig. 5B, panel v; relative luciferase activities in panel iv were normalized with RNA levels in panel iii).
FIG. 5.
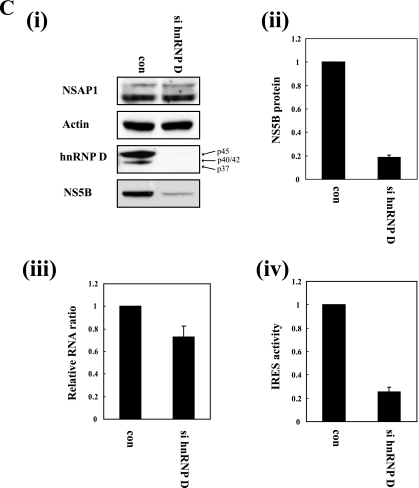
hnRNP D is required for HCV IRES-dependent translation in HCV replicon-containing cells. (A) Schematic diagrams of subgenomic HCV replicons. Huh-luc/neo-ET cell lines contain a bicistronic selectable HCV replicon including both neomycin phosphotransferase II (Neo) and FLuc genes under translational control of the HCV IRES. Huh 5-15 cell lines contain a monomeric selectable HCV replicon harboring a hygromycin-resistant gene, followed by HCV nonstructural proteins. (B, panel i) Western blot analysis of Huh-luc/neo-ET cells transfected with control (con) or hnRNP D siRNA (si hnRNP D). Control siRNA and si hnRNP D were transfected into Huh-luc/neo-ET cells using Lipofectamine 2000. At 48 h after transfection, cells were harvested and protein levels analyzed by Western blotting with anti-actin, -hnRNP D, and -NS5B antibodies. (ii) The NS5B bands in panel i were quantified and normalized to actin bands. (iii) Levels of HCV mRNA (relative to GAPDH mRNA) in Huh-luc/neo-ET cells transiently transfected with control or hnRNP D-targeting siRNA were determined using real-time PCR analysis. The amount of HCV mRNAs in control siRNA-transfected cells was set to 1. (iv) The effects of si hnRNP D on HCV IRES-dependent translation in Huh-luc/neo-ET cells were monitored by measuring firefly luciferase activity. Firefly luciferase activity was normalized to the amounts of protein in cell extracts, and the luciferase activity in cells transfected with control siRNA was set to 1. (v) The firefly luciferase activities in panel iv were normalized by mRNA levels in panel iii, and the ratio is depicted in the graph. (C, panel i) Western blot analysis of Huh 5-15 cells transfected with control siRNA (con) or hnRNP D-specific siRNA (si hnRNP D). Huh 5-15 cells were transfected with siRNA by using oligofectamine, and protein levels were analyzed by Western blotting with anti-NSAP1, -actin, -hnRNP D,and -NS5B antibodies. (ii) NS5B bands in panel i were quantified and normalized to actin bands. (iii) Levels of replicon RNA (relative to GAPDH mRNA) in Huh 5-15 cells transfected with control or si hnRNP D were determined by real-time PCR analysis. The amount of replicon RNA in control siRNA-transfected cells was set to 1. (iv) The NS5B level in panel ii was normalized to RNA levels in panel iii to obtain relative IRES activity, as shown in panel iv.
The necessity of hnRNP D in proliferation of HCV, which requires both translation and replication of RNA, was monitored using a monocistronic HCV replicon, Huh 5-15, containing the hygromycin resistance gene and HCV NS3 to NS5B in a single open reading frame (Fig. 5A, lower panel) (13). This artificial polyprotein is translated through the HCV IRES and processed by ubiquitinase and the viral proteinase NS3, resulting in functional proteins. Under these conditions, expression of NS5B controlled by HCV IRES was reduced by 80% upon suppression of hnRNP D with siRNA (Fig. 5C, panel i, NS5B, and panel ii). On the other hand, the HCV RNA level was reduced by 20% (Fig. 5C, panel iii). The reduction in the RNA level might be due to the suboptimal levels of viral proteins, such as NS5B polymerase, which are under the control of the HCV IRES. Considering the HCV RNA concentration, HCV IRES activity was reduced by about 70% in hnRNP D knockdown cells. This effect of hnRNP D on HCV IRES-dependent translation in a monocistronic context is consistent with that in a dicistronic context (Fig. 5B and C).
The importance of hnRNP D in HCV mRNA translation was further analyzed by monitoring the ribosomal profiles of HCV mRNA with or without treatment with siRNAs against hnRNP D (Fig. 6). The overall profiles of ribosomes in sucrose gradient analyses were not altered by treatment with siRNA against hnRNP D. This is reflected in the levels of a control GAPDH mRNA (Fig. 6C). On the other hand, a shift of HCV mRNA from heavy polysomes (fractions 7 and 8) to light polysomes (fractions 3 to 6) was observed in cells treated with siRNA against hnRNP D (Fig. 6D, lanes 3 to 8). The effects of hnRNP D on HCV IRES activity in replicons collectively suggest an important role of the protein in translation of HCV mRNA.
Effect of hnRNP D on translation of HCV mRNA during infection.
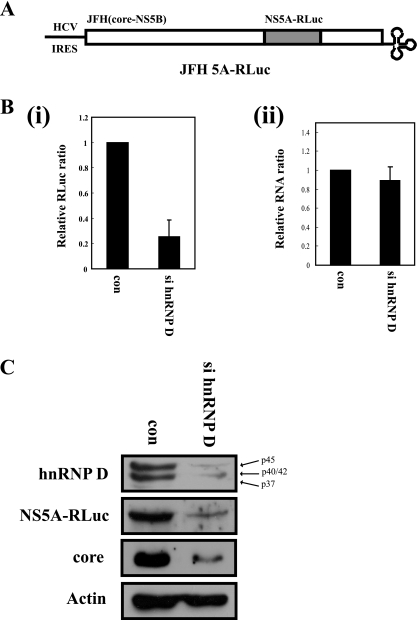
Finally, we monitored the effects of hnRNP D suppression on viral RNA proliferation, using an infectious HCV clone containing Renilla luciferase fused with NS5A viral protein (24). The luciferase activity reflects the level of viral protein translated in HCV-infected cells. Upon treatment with specific siRNA, the hnRNP D protein level was markedly reduced, with no effects on the amount of the negative control protein, actin (Fig. 7C). Expression of viral proteins in hnRNP D siRNA-treated cells was reduced to about 70%, as measured by luciferase activity (Fig. 7B, panel i) or Western blotting of NS5A-Rluc fusion protein and core protein (Fig. 7C, NS5A-Rluc and core). Similar to the replicon system, the viral RNA level in infected cells was marginally reduced in hnRNP D-depleted cells (Fig. 7B, panel ii). These data strongly suggest that hnRNP D participates in HCV proliferation by augmenting IRES activity.
FIG. 7.
Knockdown of hnRNP D blocks translation of HCV RNA in HCV-infected cells. (A) Schematic diagram of an infectious JFH derivative containing the Renilla luciferase gene within NS5A. (B) Huh 7.5.1 cells were transfected twice with either control or hnRNP D-specific siRNA using oligofectamine. At 24 h after the final transfection, cells were infected with JFH 5A-Rluc and further cultivated for 48 h. (i) Virus infectivity was monitored by Renilla luciferase activity in cell extracts. Renilla luciferase activity was normalized, based on the total protein levels. Relative luciferase activity in cells is presented. (ii) Levels of HCV RNA (relative to GAPDH mRNA) in cells transfected with control or si hnRNP D were determined by real-time PCR analysis. The amount of JFH RNA in control siRNA-transfected cells was set to 1 for comparing RNA levels. (C) The hnRNP D, NS5A-RLuc, core, and actin levels in virus-infected cells were analyzed by Western blotting using anti-hnRNP D, -NS5A, -core, and -actin antibodies, respectively.
DISCUSSION
Translational initiation through the HCV IRES is unique for eukaryotic mRNA, because only a few canonical translation factors (eIF2, eIF3, eIF5, and eIF5B, but not eIF4E, eIF4A, or eIF4G) are used for activity (40-42). However, a number of noncanonical translation factors (PTB, La, and NSAP1) augment HCV IRES-dependent translation. Here, we report that hnRNP D, an RNA-binding protein, augments HCV IRES activity through binding to the SL II region of the IRES element. Several lines of evidence confirm that hnRNP D is a positive regulator of HCV IRES-dependent translation. First, overexpression of hnRNP D enhanced HCV IRES-dependent translation (Fig. 1B and C). The p45 isoform of hnRNP D displayed the strongest IRES-enhancing activity (Fig. 1B and C). Additionally, hnRNP D specifically activated HCV IRES-dependent translation among the IRES elements examined (Fig. 1D). Second, hnRNP D interacted with SL II of the HCV IRES element (Fig. 2). Third, knockdown of hnRNP D with siRNAs inhibited HCV IRES activity monitored using a dicistronic reporter (Fig. 3), HCV replicons (Fig. 5), and an infectious HCV clone (Fig. 7). Fourth, the G5 oligomer that binds to hnRNP D specifically inhibits HCV IRES-dependent translation in vitro (Fig. 4). Finally, a significant amount of HCV replicon RNA was shifted from heavy to light polysomal fractions in hnRNP D knockdown cells (Fig. 6).
We speculate on the mechanism by which hnRNP D augments IRES-dependent translation, based on the molecular properties of hnRNP D. SL II, the binding site of hnRNP D (Fig. 2B, panels i and iv), is related with IRES-dependent translation. Mutations in this region inhibit transition from the 48S to 80S complex (37). This effect may be related with the SL II function mediating eIF2 release by promoting eIF5-induced GTP hydrolysis, which facilitates 80S complex formation (34, 39). Therefore, it is possible that hnRNP D facilitates 60S joining by inducing the critical bent structure in SL II (34) through its intrinsic property of rearranging nucleic acid chains (12). Alternatively, hnRNP D may enhance HCV IRES-dependent translation through interactions with another protein(s) that promote translational initiation. In this respect, it is worth noting that hnRNP D interacts with NSAP1 (16), enhancing HCV IRES-dependent translation (27). However, synergistic activation of the HCV IRES was not observed when these two proteins were overexpressed together in the cells (data not shown), indicating that these proteins enhance HCV IRES activity independently.
Intriguingly, knockdown of ITAF hnRNP D facilitates the replication of HCV RNA. This finding suggests that hnRNP D is involved in the functional switch from the translational mode of incoming HCV RNA to the replication mode. After the synthesis of viral proteins, including RNA-dependent RNA polymerase (NS5B), genomic viral RNA should serve as a template for the synthesis of negative-strand RNA. The switch from translation to replication should occur properly, because continuous translation of viral mRNA blocks the replication of viral RNA (15). RNA-binding proteins play key roles in the functional switch from translation to replication of poliovirus, which displays a similar life cycle to HCV. Specifically, poliovirus is a positive-sense RNA virus, and its translation is directed by an IRES element at the 5′ NTR of viral RNA, such as HCV (38). Poly(rC)-binding protein (PCBP) interactions with domain IV in the poliovirus IRES element enhance IRES activity. Functional switching from translation to replication of viral RNA possibly occurs by changes in PCBP2-binding sites from domain IV to the 5′-terminal cloverleaf of the polioviral 5′ NTR (14, 15). More recent results suggest that proteolytic cleavage of PTB (which binds to the IRES element of polioviral RNA and enhances IRES function) by the virus-encoded proteinase 3Cpro induces functional switching from translation to replication (4). An analogous functional switch may occur during infection of HCV. One possible hypothesis is that hnRNP A1, which displays similar binding specificity to hnRNP D (17, 20) and is required for HCV RNA replication (25), competes with hnRNP D for the same binding site in SL II. In this respect, it is interesting that hnRNP A1 in the nucleus in noninfected cells relocalizes to the cytoplasm in HCV-infected cells, where translation and replication occur (25). Therefore, the shift from hnRNP D-bound RNA to hnRNP A1 may result in a switch from translation to replication of RNA.
Further investigation of the functions of hnRNP D should improve our understanding of the mechanisms of IRES-dependent translation and replication. Moreover, this improved knowledge may lead to the development of novel therapeutic approaches for the treatment of life-threatening HCV infections.
Acknowledgments
We are grateful to R. J. Schneider at the New York University School of Medicine for providing pFLAG-CMV2 p37, pFLAG-CMV2 p40, pFLAG-CMV2 p42, and pFLAG-CMV2 p45 and R. Bartenschlarger at the University of Heidelberg for Huh-luc/neo-ET and Huh 5-15 cell lines. We also thank S. J. Kum for technical assistance and lab members for helpful discussion.
This work is supported in part by the Brain Korea 21 Project, Acceleration Research (Center for Translational Mechanism) of MEST/KOSEF, grant FPR08B1-220 of the 21C Frontier Functional Proteomics Project from MEST, the Korea Science and Engineering Foundation (KOSEF) grant funded by the Korean government (MEST) (no. R15-2004-033-05001-0), and a grant from POSCO.
Footnotes
Published ahead of print on 8 October 2008.
REFERENCES
- 1.Ali, N., G. J. Pruijn, D. J. Kenan, J. D. Keene, and A. Siddiqui. 2000. Human La antigen is required for the hepatitis C virus internal ribosome entry site-mediated translation. J. Biol. Chem. 27527531-27540. [DOI] [PubMed] [Google Scholar]
- 2.Ali, N., and A. Siddiqui. 1995. Interaction of polypyrimidine tract-binding protein with the 5′ noncoding region of the hepatitis C virus RNA genome and its functional requirement in internal initiation of translation. J. Virol. 696367-6375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anwar, A., N. Ali, R. Tanveer, and A. Siddiqui. 2000. Demonstration of functional requirement of polypyrimidine tract-binding protein by SELEX RNA during hepatitis C virus internal ribosome entry site-mediated translation initiation. J. Biol. Chem. 27534231-34235. [DOI] [PubMed] [Google Scholar]
- 4.Back, S. H., Y. K. Kim, W. J. Kim, S. Cho, H. R. Oh, J. E. Kim, and S. K. Jang. 2002. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3Cpro. J. Virol. 762529-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartenschlager, R. 2005. The hepatitis C virus replicon system: from basic research to clinical application. J. Hepatol. 43210-216. [DOI] [PubMed] [Google Scholar]
- 6.Brewer, G. 1991. An A + U-rich element RNA-binding factor regulates c-myc mRNA stability in vitro. Mol. Cell. Biol. 112460-2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho, S., J. H. Kim, S. H. Back, and S. K. Jang. 2005. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1 mRNA and modulates transition from G1 to S phase. Mol. Cell. Biol. 251283-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, S., S. M. Park, T. D. Kim, J. H. Kim, K. T. Kim, and S. K. Jang. 2007. BiP internal ribosomal entry site activity is controlled by heat-induced interaction of NSAP1. Mol. Cell. Biol. 27368-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi, K., J. H. Kim, X. Li, K. Y. Paek, S. H. Ha, S. H. Ryu, E. Wimmer, and S. K. Jang. 2004. Identification of cellular proteins enhancing activities of internal ribosomal entry sites by competition with oligodeoxynucleotides. Nucleic Acids Res. 321308-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa-Mattioli, M., Y. Svitkin, and N. Sonenberg. 2004. La autoantigen is necessary for optimal function of the poliovirus and hepatitis C virus internal ribosome entry site in vivo and in vitro. Mol. Cell. Biol. 246861-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dempsey, L. A., M. J. Li, A. DePace, P. Bray-Ward, and N. Maizels. 1998. The human HNRPD locus maps to 4q21 and encodes a highly conserved protein. Genomics 49378-384. [DOI] [PubMed] [Google Scholar]
- 12.Eversole, A., and N. Maizels. 2000. In vitro properties of the conserved mammalian protein hnRNP D suggest a role in telomere maintenance. Mol. Cell. Biol. 205425-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frese, M., K. Barth, A. Kaul, V. Lohmann, V. Schwarzle, and R. Bartenschlager. 2003. Hepatitis C virus RNA replication is resistant to tumour necrosis factor-alpha. J. Gen. Virol. 841253-1259. [DOI] [PubMed] [Google Scholar]
- 14.Gamarnik, A. V., and R. Andino. 2000. Interactions of viral protein 3CD and poly(rC) binding protein with the 5′ untranslated region of the poliovirus genome. J. Virol. 742219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamarnik, A. V., and R. Andino. 1998. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 122293-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosset, C., C. Y. Chen, N. Xu, N. Sonenberg, H. Jacquemin-Sablon, and A. B. Shyu. 2000. A mechanism for translationally coupled mRNA turnover: interaction between the poly(A) tail and a c-fos RNA coding determinant via a protein complex. Cell 10329-40. [DOI] [PubMed] [Google Scholar]
- 17.Hamilton, B. J., E. Nagy, J. S. Malter, B. A. Arrick, and W. F. Rigby. 1993. Association of heterogeneous nuclear ribonucleoprotein A1 and C proteins with reiterated AUUUA sequences. J. Biol. Chem. 2688881-8887. [PubMed] [Google Scholar]
- 18.Han, J. H., V. Shyamala, K. H. Richman, M. J. Brauer, B. Irvine, M. S. Urdea, P. Tekamp-Olson, G. Kuo, Q. L. Choo, and M. Houghton. 1991. Characterization of the terminal regions of hepatitis C viral RNA: identification of conserved sequences in the 5′ untranslated region and poly(A) tails at the 3′ end. Proc. Natl. Acad. Sci. USA 881711-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He, C., and R. Schneider. 2006. 14-3-3sigma is a p37 AUF1-binding protein that facilitates AUF1 transport and AU-rich mRNA decay. EMBO J. 253823-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henics, T., A. Sanfridson, B. J. Hamilton, E. Nagy, and W. F. Rigby. 1994. Enhanced stability of interleukin-2 mRNA in MLA 144 cells. Possible role of cytoplasmic AU-rich sequence-binding proteins. J. Biol. Chem. 2695377-5383. [PubMed] [Google Scholar]
- 21.Jang, S. K., H. G. Krausslich, M. J. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 622636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kajita, Y., J. Nakayama, M. Aizawa, and F. Ishikawa. 1995. The UUAG-specific RNA binding protein, heterogeneous nuclear ribonucleoprotein D0. Common modular structure and binding properties of the 2xRBD-Gly family. J. Biol. Chem. 27022167-22175. [DOI] [PubMed] [Google Scholar]
- 23.Kato, N., M. Hijikata, Y. Ootsuyama, M. Nakagawa, S. Ohkoshi, T. Sugimura, and K. Shimotohno. 1990. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc. Natl. Acad. Sci. USA 879524-9528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, C. S., J. H. Jung, T. Wakita, S. K. Yoon, and S. K. Jang. 2007. Monitoring the antiviral effect of alpha interferon on individual cells. J. Virol. 818814-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim, C. S., S. K. Seol, O. K. Song, J. H. Park, and S. K. Jang. 2007. An RNA-binding protein, hnRNP A1, and a scaffold protein, septin 6, facilitate hepatitis C virus replication. J. Virol. 813852-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim, J. H., K. Y. Paek, K. Choi, T. D. Kim, B. Hahm, K. T. Kim, and S. K. Jang. 2003. Heterogeneous nuclear ribonucleoprotein C modulates translation of c-myc mRNA in a cell cycle phase-dependent manner. Mol. Cell. Biol. 23708-720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, J. H., K. Y. Paek, S. H. Ha, S. Cho, K. Choi, C. S. Kim, S. H. Ryu, and S. K. Jang. 2004. A cellular RNA-binding protein enhances internal ribosomal entry site-dependent translation through an interaction downstream of the hepatitis C virus polyprotein initiation codon. Mol. Cell. Biol. 247878-7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim, W. J., J. H. Kim, and S. K. Jang. 2007. Anti-inflammatory lipid mediator 15d-PGJ2 inhibits translation through inactivation of eIF4A. EMBO J. 265020-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Komurian-Pradel, F., M. Perret, B. Deiman, M. Sodoyer, V. Lotteau, G. Paranhos-Baccala, and P. Andre. 2004. Strand specific quantitative real-time PCR to study replication of hepatitis C virus genome. J. Virol. Methods 116103-106. [DOI] [PubMed] [Google Scholar]
- 30.Lancaster, A. M., E. Jan, and P. Sarnow. 2006. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA 12894-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laroia, G., R. Cuesta, G. Brewer, and R. J. Schneider. 1999. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284499-502. [DOI] [PubMed] [Google Scholar]
- 32.Lee, S. H., Y. K. Kim, C. S. Kim, S. K. Seol, J. Kim, S. Cho, Y. L. Song, R. Bartenschlager, and S. K. Jang. 2005. E2 of hepatitis C virus inhibits apoptosis. J. Immunol. 1758226-8235. [DOI] [PubMed] [Google Scholar]
- 33.Liao, B., Y. Hu, and G. Brewer. 2007. Competitive binding of AUF1 and TIAR to MYC mRNA controls its translation. Nat. Struct. Mol. Biol. 14511-518. [DOI] [PubMed] [Google Scholar]
- 34.Locker, N., L. E. Easton, and P. J. Lukavsky. 2007. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 26795-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu, H., W. Li, W. S. Noble, D. Payan, and D. C. Anderson. 2004. Riboproteomics of the hepatitis C virus internal ribosomal entry site. J. Proteome Res. 3949-957. [DOI] [PubMed] [Google Scholar]
- 36.Lu, J. Y., N. Bergman, N. Sadri, and R. J. Schneider. 2006. Assembly of AUF1 with eIF4G-poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. RNA 12883-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Otto, G. A., and J. D. Puglisi. 2004. The pathway of HCV IRES-mediated translation initiation. Cell 119369-380. [DOI] [PubMed] [Google Scholar]
- 38.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334320-325. [DOI] [PubMed] [Google Scholar]
- 39.Pestova, T. V., S. de Breyne, A. V. Pisarev, I. S. Abaeva, and C. U. Hellen. 2008. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 271060-1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pestova, T. V., C. U. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 166859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1267-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pestova, T. V., I. N. Shatsky, and C. U. Hellen. 1996. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell. Biol. 166870-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pudi, R., P. Srinivasan, and S. Das. 2004. La protein binding at the GCAC site near the initiator AUG facilitates the ribosomal assembly on the hepatitis C virus RNA to influence internal ribosome entry site-mediated translation. J. Biol. Chem. 27929879-29888. [DOI] [PubMed] [Google Scholar]
- 44.Sarkar, B., J. Y. Lu, and R. J. Schneider. 2003. Nuclear import and export functions in the different isoforms of the AUF1/heterogeneous nuclear ribonucleoprotein protein family. J. Biol. Chem. 27820700-20707. [DOI] [PubMed] [Google Scholar]
- 45.Schmittgen, T. D., and B. A. Zakrajsek. 2000. Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods 4669-81. [DOI] [PubMed] [Google Scholar]
- 46.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 661476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wagner, B. J., C. T. DeMaria, Y. Sun, G. M. Wilson, and G. Brewer. 1998. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics 48195-202. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, W., B. J. Wagner, K. Ehrenman, A. W. Schaefer, C. T. DeMaria, D. Crater, K. DeHaven, L. Long, and G. Brewer. 1993. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol. Cell. Biol. 137652-7665. [DOI] [PMC free article] [PubMed] [Google Scholar]