Abstract
GADD34, the product of a growth arrest and DNA damage-inducible gene, is expressed at low levels in unstressed cells. In response to stress, the cellular content of GADD34 protein increases and, on termination of stress, rapidly declines. We investigated the mechanisms that control GADD34 levels in human cells. GADD34 proteins containing either an internal FLAG or a C-terminal green fluorescent protein epitope were degraded at rates similar to endogenous GADD34. However, the addition of epitopes at the N terminus or deletion of N-terminal sequences stabilized GADD34. N-terminal peptides of GADD34, either alone or fused to heterologous proteins, exhibited rapid degradation similar to wild-type GADD34, thereby identifying an N-terminal degron. Deletion of internal PEST repeats had no impact on GADD34 stability but modulated the binding and activity of protein phosphatase 1. Proteasomal but not lysosomal inhibitors enhanced GADD34 stability and eukaryotic initiation factor 2α (eIF-2α) dephosphorylation, a finding consistent with GADD34's role in assembling an eIF-2α phosphatase. GADD34 was polyubiquitinated, and this modification enhanced its turnover in cells. A stabilized form of GADD34 promoted the accumulation and aggregation of the mutant cystic fibrosis transmembrane conductance regulator (CFTRΔF508), highlighting the physiological importance of GADD34 turnover in protein processing in the endoplasmic reticulum and the potential impact of prolonged GADD34 expression in human disease.
Protein translation in higher eukaryotes is regulated by the activation of multiple protein kinases and the reversible phosphorylation of numerous components of the protein translation machinery. Among these, the phosphorylation of eukaryotic translation initiation factor 2α (eIF-2α) (at serine-51) inhibits the assembly of the preinitiation complex containing the 40S ribosome, mRNA, and the initiating methionyl-tRNA, and is the most evolutionarily conserved (7). Thus, it is remarkable that eIF-2α phosphorylation, which controls protein translation in all eukaryotic cells, appears dispensable for mammalian cell growth, differentiation, and embryogenesis such that the substitution of a single amino acid (S51A) in the mouse eIF-2α gene abolished eIF-2α phosphorylation but yielded mutant mice with the expected Mendelian ratio and normal tissue development. However, the homozygous eIF-2α (S51A) animals died within the first day after birth with severe hypoglycemia and low plasma insulin. Although basal protein synthesis was increased in the eIF-2α (S51A) mutant mice, they failed to induce hepatic gluconeogenic enzymes, were unable to store liver glycogen, and displayed severe pancreatic β-cell deficiency (32). This suggested that eIF-2α phosphorylation, while inhibiting global protein synthesis, is required for the translation of some proteins, particularly those involved in the execution of an integrated stress response (ISR) that enables recovery of cells from stress and commits the irreversibly damaged cells to programmed cell death. Consistent with this idea, mice lacking PERK, the major pancreatic eIF-2α kinase, also developed early onset type-1 diabetes with significant loss of β-cells and displayed skeletal abnormalities associated with the defective synthesis of bone matrix proteins by osteoblasts, thus resembling individuals with Wolcott-Rallison syndrome associated with mutations in the human PERK gene (39). Finally, the heterozygous eIF-2α (S51A) mice, while phenotypically normal on a low-fat diet, rapidly gained weight and became diabetic on high-fat diet (33), demonstrating the delayed processing of proinsulin and reduced glucose-stimulated insulin secretion. In contrast, protein translation required for long-term memory (19) was basally elevated in the heterozygous eIF-2α (S51A) mice enhancing their capacity to convert short-term training to long-lasting spatial memory (6). These studies highlighted that metabolic and other physiological stress placed distinct demands on the cellular reserve for eIF-2α phosphorylation and suggested that aberrant eIF-2α phosphorylation, resulting from either the malfunction or misregulation of eIF-2α kinases and phosphatases, may play a role in human disease.
The four mammalian kinases, GCN2, PKR, HRI, and PERK, which catalyze the phosphorylation of eIF-2α at serine-51, are differentially expressed in mammalian tissues and activated by a diverse array of cell stress. By comparison, GADD34, first identified as the product of a gene activated by growth arrest and DNA damage (12), is also expressed in response to many forms of cell stress and binds protein phosphatase-1α to assemble an eIF-2α phosphatase (5, 26) that antagonizes the actions of the eIF-2α kinases. Interestingly, mice that lack the major reticulocyte eIF-2α kinase, HRI, show a significantly reduction in reticulocytes and defective hemoglobin synthesis (10), a phenotype similar to mice lacking a functional GADD34 gene (28). These data suggested that cells such as β-cells, osteoblasts, and reticulocytes, which are dedicated to high levels of protein synthesis, are uniquely susceptible to aberrant eIF-2α phosphorylation-dephosphorylation, requiring a careful coordination of the eIF-2α kinases and phosphatases to ensure a level and duration of eIF-2α phosphorylation that maintains normal cell function and viability.
Analysis of homozygous eIF-2α (S51A) mutant mouse embryo fibroblasts (MEFs) in culture highlighted their remarkable resistance to apoptosis induced by double-stranded RNA, tumor necrosis factor alpha, and serum deprivation (34). In contrast, the viability of these cells to other insults, including glucose deprivation and inhibition of N-linked glycosylation by tunicamycin, was significantly reduced (32). This hinted at significant differences in the programmed activation of ISR genes downstream of eIF-2α phosphorylation that determined cell survival or death. Interestingly, eIF-2α phosphorylation is required for the expression of GADD34 (21, 27), itself the product of an ISR gene. Thus, GADD34-null mouse embryonic fibroblasts (MEFs) showed a delayed recovery of protein synthesis and diminished activation of several stress-induced genes (27). These studies suggested that GADD34 functioned in a feedback loop that promoted cell recovery from translational inhibition, and thus the precise control of GADD34 expression was critical for determining the cellular fate following various forms of stress.
The present study noted significant differences in the steady-state levels of GADD34 protein in human cell lines subjected to various physiological and environmental stress. This was in part attributable to the remarkable instability of the GADD34 protein, which was rapidly eliminated upon removal of cell stress. Our studies established for the first time that GADD34 was polyubiquitinated in cultured human cells, and inhibiting ubiquitination or proteasome function stabilized GADD34. These studies identified an N-terminal degron in GADD34, which when fused to heterologous proteins, promoted their degradation by the proteasome. Finally, our data showed that a stabilized form of GADD34 enhanced the accumulation and aggregation of CFTR(ΔF508), the mutant protein associated with human cystic fibrosis and processed in the endoplasmic reticulum (ER). Together, the data suggested that proteasome-mediated turnover of GADD34 is critical for the orderly synthesis and folding of proteins in the ER and highlighted the potential role of prolonged GADD34 function in the proteotoxicity associated with human disease.
MATERIALS AND METHODS
Fugene 6 was obtained from Roche Diagnostics. Anti-FLAG M2 affinity gel, thapsigargin, sodium arsenite, MG132, chloroquine (CQ), and cycloheximide (CHX) were from Sigma-Aldrich. Okadaic acid (OA), MG262, and protease inhibitor cocktail set III were from Calbiochem. Calyculin A (CA) was from Alexis Biochemicals, and leucine-free Dulbecco modified Eagle medium (DMEM) from MP Biomedicals.
The following antibodies were utilized at the dilutions in parentheses: polyclonal anti-GADD34 (1:400) was from Calbiochem, polyclonal anti-eIF-2α (phosphoserine-51) was from Biosource International (1:500) or Cell Signaling Technologies (1:200), polyclonal Living Colors anti-green fluorescent protein (anti-GFP, 1:4,000) was from Clontech, monoclonal anti-FLAG M2 (1:4,000) and monoclonal anti-α-tubulin (1:8,000) were from Sigma-Aldrich, monoclonal antiubiquitin (1:1,000) and monoclonal antihemagglutinin (anti-HA; 1:1,000) were from Santa Cruz Biotechnology, and monoclonal anti-GM130 (1:100) was from BD Transduction Laboratories. Horseradish peroxidase-linked rabbit anti-goat immunoglobulin G (Zymed Laboratories) and sheep anti-mouse IgG and sheep anti-rabbit IgG (GE Healthcare) were used as secondary antibodies.
GADD34 expression plasmids.
The cDNA encoding full-length GADD34 was amplified from pSG5-FLAG-GADD34 (3) by using PCR primers containing BglII and EcoRI restriction sites. GFP was fused to either the N or the C terminus of GADD34 by ligating the PCR-amplified cDNA into pEGFP-C1 or pEGFP-N1 (Clontech), respectively. An internal FLAG epitope was introduced into FLAG(109)GADD34 by ligating the PCR-amplified GADD34 cDNA into pGRE5-2 vector (David Pallas, Emory University), followed by site-directed mutagenesis that replaced amino acids 109 to 116 (GLLDDDDG) from wild-type GADD34 with the FLAG epitope (DYKDDDDK). FLAG-GADD34, which contained an N-terminal FLAG epitope (FLAG2), was amplified from pSG5-FLAG-GADD34, and ligated into BglII and EcoRI sites in pGRE5-2.
GADD34(Δ1-10)-GFP, GADD34(Δ1-60)-GFP, and GADD34(1-60)-GFP were generated by amplifying appropriate sequences from pSG5-FLAG-GADD34 and ligating them into pEGFP-N3 (Clontech). The internal deletion, GADD34(ΔPESTs)-GFP, eliminating residues 323 to 512 encompassing the four internal PEST repeats, was generated by two-step PCR using pSG5-FLAG-GADD34 as a template. The product of the PCR was cloned into pEGFP-N1.
GADD34(1-155)-HA was generated by PCR amplification of a cDNA encoding N-terminal 155 residues of wild-type GADD34 from pSG5-FLAG-GADD34, using 5′ primer containing BglII cleavage site and 3′ primer encoding the HA epitope followed by an EcoRI restriction site. The product of the PCR was digested with BamHI and EcoRI and ligated into pcDNA3.1. Lysine-51 and lysine-99 were substituted with arginines in GADD34(1-155, 0K)-HA by site-directed mutagenesis. All cDNAs were sequenced by Duke University Comprehensive Cancer Center DNA Sequencing Facility.
GFP-CFTRΔF508 and GFP-250 expression vectors were provided by Tso-Pang Yao (Duke University). GFP-GRP94 (lacking the C-terminal ER-retention sequence, KDEL) was from Christopher Nicchitta (Duke University). Expression vectors for wild-type and K48R myc-ubiquitin were provided by Michael Ehlers (Duke University). Monomeric red fluorescent protein (mRFP) was provided by Roger Tsien (University of California, San Diego). N-terminally FLAG-tagged monomeric RFP was generated by PCR amplifying mRFP cDNA from the vector described above and subcloning it into pFLAG-CMV-2 (Sigma). The expression vector for ER-marker, pDsRed2-ER, was obtained from Clontech.
GADD34 expression in cultured cells.
Human embryonic kidney (HEK293T), cervical adenocarcinoma (HeLa), lung epithelial carcinoma (A549), and colorectal adenocarcinoma (SW480) cells were obtained from the American Type Culture Collection and maintained in DMEM with 10% (vol/vol) fetal bovine serum. All cells were grown at 37°C in an atmosphere of 5% CO2 and 95% air.
Transfections of plasmid DNA were undertaken in 12- or 6-well plates (Falcon) using Fugene 6 as described in the manufacturer's instructions. Transfections of pGRE5-2 FLAG-GADD34 plasmid DNA were undertaken in the presence of 0.5 to 2 nM dexamethasone, which activated the glucocorticoid response element to drive FLAG-GADD34 transcription.
To induce endogenous GADD34 levels, cells grown to 75 to 90% confluence in six-well plates were treated with thapsigargin (1 μM), sodium arsenite (25 μM), and MG132 (5 μM) or maintained in leucine-free DMEM. In recovery experiments, cells were first incubated in leucine-free medium or treated with the proteasome inhibitor, MG132, for 8 h to induce GADD34. Cells were then washed three times in DMEM containing 10% (vol/vol) FBS and maintained in fresh medium prior to homogenization in sodium dodecyl sulfate (SDS) sample buffer, followed by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting with anti-GADD34, anti-GFP, anti-FLAG, or anti-HA antibody.
Analysis of eIF-2α dephosphorylation.
HeLa or HEK293T cells treated with 5 μM MG132 for 8 h to induce GADD34 were washed with drug-free medium. At 0, 4, and 8 h, a phosphatase inhibitor, either 1 μM OA, which preferentially inhibited cellular PP2A to activate eIF-2α kinases, or 100 nM CA, which inhibited both PP2A and the eIF-2α phosphatase containing PP1 (3), was added to the culture medium, and cells were incubated for 30 min at 37°C prior to homogenization in SDS sample buffer and then analysis by SDS-PAGE and immunoblotting with anti-phospho-eIF-2α antibody.
Analysis of GADD34 degradation.
To investigate GADD34 degradation in cells following their recovery from cell stress, we utilized reversible stressors (leucine deprivation and MG132) to increase cellular GADD34 levels. Cell stress was terminated by transferring cells into drug-free complete medium. At selected intervals, cells were lysed in SDS sample buffer, and proteins were analyzed by SDS-PAGE and immunoblotting.
To analyze the turnover of ectopically expressed GADD34, cells were transfected with plasmids expressing GFP- or FLAG-tagged GADD34. After 20 to 24 h, GADD34-expressing cells were treated with CHX (30 μg/ml) to terminate protein synthesis, and cells were lysed at selected time intervals to analyze the cellular content of GFP- or FLAG-GADD34 using SDS-PAGE and immunoblotting. To elucidate the cellular mechanism that mediated GADD34 degradation, the proteasome inhibitor MG262 (1 μM) or the lysosomal inhibitor CQ (200 μM) was included in medium containing CHX. For time course studies, cells were scraped in culture medium followed by the addition of dimethyl sulfoxide (10% [vol/vol]) and flash-freezing in dry ice-ethanol bath prior to storage at −80°C. This allowed subsequent simultaneous analysis of all timed samples from any given experiment. Quantitation of GADD34 was undertaken by laser scanning of Western immunoblots and analysis using Quantity One 4.1.1 software (Bio-Rad). The results of three to five independent experiments are presented with standard deviations.
GADD34 immunoassays.
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.5% [wt/vol] deoxycholic acid, 0.1% [wt/vol] SDS, 1% [wt/vol] NP-40) and analyzed by SDS-PAGE. In the ubiquitin immunoprecipitation experiments, HEK293T cells expressing GADD34 proteins were grown in 35-mm plates and, at 20 to 24 h after transfection, the cells were treated with either 1 μM MG262 or vehicle for 8 h. Cells were washed with phosphate-buffered saline (PBS) and lysed in ice-cold RIPA buffer (500 μl) containing a protease inhibitor cocktail, 20 μM MG132, and 5 mM N-ethylmaleimide to prevent deubiquitination. Lysates clarified by centrifugation at 10,000 × g were incubated with anti-FLAG M2 beads (25-μl bed volume) for 2 h at 4°C. The beads were washed four times with NETN250 buffer (10 mM Tris-HCl [pH 7.5] containing 250 mM NaCl, 1 mM EDTA, and 0.5% [wt/vol] NP-40) at 4°C, and the bound proteins were eluted by boiling in 50 μl of SDS sample buffer. After SDS-PAGE, the proteins were electrophoretically transferred to polyvinylidene difluoride membranes, which were blocked for 1 to 2 h at room temperature with 2% (wt/vol) bovine serum albumin in TBS (50 mM Tris-HCl [pH 7.5] containing 150 mM NaCl) before immunoblotting with selected antibodies.
Fluorescence microscopy of cultured cells.
HEK293T cells were cotransfected with plasmids encoding GFP-CFTRΔF508 and either mRFP (a cytosolic marker) or DsRed2-ER (an ER luminal marker) and, in some cases, pSG5-GADD34-FLAG. Where indicated, cells were treated with 1 μM MG262 for 8 h prior to imaging with a Nikon Eclipse TE200 fluorescence microscope.
Analysis of protein aggregation.
Cells were grown to 50% confluence in six-well dishes prior to transfection with plasmids that expressed either GFP-CFTRΔF508 or GFP-GRP94, alone or in combination with pSG5-GADD34-FLAG. After 24 h, the cells were treated with 1 μM MG262 for 8 h and harvested in 150 μl of RIPA buffer containing protease inhibitors and DNase at 40 μg/ml. An aliquot (20 μl) of cell lysate was retained as “total lysate” (T), and the remainder was subjected to centrifugation at 20,000 × g for 15 min to yield an insoluble pellet and the “soluble fraction” (S). The pellet was resuspended in 25 μl of RIPA buffer containing 1% (wt/vol) SDS, 10% (vol/vol) glycerol, protease inhibitors, and DNase and heated at 65°C for 10 min prior to dilution and sonication in a total volume of 150 μl of RIPA buffer. After centrifugation at 20,000 × g for 15 min, the supernatant, termed the “insoluble fraction” (I), was analyzed with T and S samples by using SDS-PAGE and immunoblotting.
RESULTS
GADD34 induction by cell stress.
Prior studies showed that GADD34 mRNA levels were increased by many forms of cell stress (13, 14, 24, 25). We also noted significant increases in GADD34 protein in human cell lines subjected to diverse cell stress (Fig. 1). Interestingly, the ER stressors tunicamycin and thapsigargin, amino acid (leucine) deprivation, and the DNA-damaging agent methyl methanesulfonate all stimulated much higher levels of GADD34 expression in A549 cells than in either HeLa or HEK293T cells. By comparison, oxidative stress induced by arsenite and treatment with the phosphatase inhibitor, OA, induced higher levels of GADD34 in HeLa and HEK293T than in A549 cells. Inhibition of proteasome by MG262 (and other inhibitors [data not shown]) resulted in high levels of GADD34 in all cells analyzed. While prior work has emphasized the transcriptional activation of the single human GADD34 gene (13, 14, 24, 25) by many forms of cell stress, these new findings hinted at possibly other factors, such as protein translation and stability, that may also account for the steady-state levels of GADD34 protein in different human cell lines.
FIG. 1.
GADD34 expression in human cell lines subjected to various forms of stress. A549, HeLa, and HEK293T cells were exposed to various stress, including ER stress, nutrient deprivation, DNA damage, apoptosis, oxidative stress, and inhibition of protein phosphatase or the proteasome. Specifically, cells were treated with either the vehicle (0.1% [vol/vol] dimethyl sulfoxide [DMSO]), 1 μM thapsigargin (TG), 10 μg/ml (wt/vol) tunicamycin (TN), leucine-free DMEM (-Leu); 125 μg/ml (wt/vol) methyl methanesulfonate (MMS), 6 μM etoposide (Etop), 0.5 μg/ml (wt/vol) staurosporine (Staur), 50 μM C2-ceramide (C2), 1 mM hydrogen peroxide (H2O2), 75 μM sodium arsenite (Ars), 100 nM OA, or 1 μM Z-Leu-Leu-Leu-B(OH)2 (MG262) for 5 h. Cells were lysed, and the lysates were analyzed by immunoblotting with an anti-GADD34 antibody. Actin levels were used to equalize for protein loading.
Time course analysis of GADD34 accumulation in SW480 human colorectal adenocarcinoma cells after treatment with thapsigargin, arsenite, leucine deprivation, and MG132, another proteasome inhibitor, showed remarkably similar increases in GADD34 between 12 and 24 h (Fig. 2A), although some differences in GADD34 levels were noted at earlier time points with individual stresses. The fate of GADD34 following the reversal of stress was also examined utilizing the experimental scheme shown in Fig. 2B. After 8 h in medium lacking leucine or glucose or in medium containing MG132, a significant elevation of GADD34 protein was noted. On restoration in complete media, the cellular levels of GADD34 rapidly decreased, returning to basal levels in 2 to 4 h after nutrient restoration and 4 to 8 h after MG132 removal (Fig. 2C). Similar results were obtained in A549, HEK293T, and HeLa cells (data not shown). The slightly slower decrease in GADD34 in MG132-treated cells may reflect a delayed proteasome reactivation and suggests a role for the 26S proteasome in regulating GADD34 turnover.
FIG. 2.
Cellular GADD34 levels following various stress. (A) Time-dependent accumulation of endogenous GADD34 was analyzed in SW480 cells during continued exposure to 1 μM thapsigargin (Tg), 50 μM arsenite (Ars), leucine-deficient DMEM (-Leu), or 5 μM MG132 by immunoblotting with anti-GADD34 antibody as described in Materials and Methods. (B) The schematic shows the experimental design in which cells were subjected to various stress stimuli for 8 h to induce GADD34 (stage I). Then, replacement with complete medium was used to promote the recovery of cells from the stress stimuli (stage II). (C) A549 cells were incubated in media lacking either leucine (Leu) or glucose (Glc) or containing 5 μM MG132 to elicit GADD34 induction. At the times indicated (0, 2, 4, and 8 h) following their transfer into complete media, cells were lysed and GADD34 levels analyzed by immunoblotting as described in Materials and Methods. Immunoblotting with antitubulin antibody was used to assess protein loading.
GADD34 levels regulate eIF-2α phosphorylation.
In our previous studies, a brief treatment with the cell-permeable phosphatase inhibitor OA at concentrations that preferentially inhibit PP2A resulted in increased eIF-2α phosphorylation (13), reflecting the activation of one or more eIF-2α kinases that are antagonized by PP2A (see schematic in Fig. 3A). In contrast, CA inhibits both PP1 and PP2A and is thus predicted to also inhibit the cellular PP1-containing eIF-2α phosphatases, further enhancing eIF-2α phosphorylation. Accordingly, the exposure of HeLa cells to 1 μM OA for 30 min increased eIF-2α phosphorylation compared to untreated cells. By comparison, treatment with 100 nM CA resulted in an even higher increase in eIF-2α phosphorylation, a finding consistent with the added inhibition of one or more eIF-2α phosphatases (Fig. 3A, lanes 1 to 3). Since neither the OA nor the CA treatment resulted in any detectable elevation of endogenous GADD34 (Fig. 3A, lanes 2 and 3), the eIF-2α phosphatase inhibited by CA most likely represented PP1 complexed with the constitutive GADD34 homolog, CReP (18).
FIG. 3.
GADD34 modulates eIF-2α phosphorylation. (A) HeLa cells either uninduced (lanes 1 to 3) or exposed to 5 μM MG132 for 8 h to induce GADD34 expression (lanes 4 to 6) were subjected to brief (0.5 h) treatment with either OA or CA at the times indicated. Lysates were subjected to immunoblotting with anti-GADD34 and anti-phospho-eIF-2α antibodies. The schematic (inset) shows the kinase and phosphatase that control eIF-2α phosphorylation and the proposed mode of action of OA and CA, which target PP2A and/or PP1 to modulate eIF-2α phosphorylation. (B) The schematic shows the experimental design by which MG132 treatment for 8 h was used to induce GADD34 (stage I), followed by the washout of drug with fresh media lacking MG132 and cell recovery (stage II). At the times indicated, cells were treated with OA or CA for 30 min (stage III) to elicit eIF-2α phosphorylation and monitor the activity of the residual GADD34. (C) Cells at stages I, II, and III were subjected to immunoblotting to monitor GADD34 expression and eIF-2α phosphorylation. Parallel immunoblots for tubulin established equivalent protein loading.
After the prior induction of GADD34 by 8 h of exposure of cells to MG132, OA treatment no longer promoted eIF-2α phosphorylation (Fig. 3A, lanes 2 and 5). This was consistent with the ability of the stress-induced GADD34 to assemble a highly active eIF-2α phosphatase that overrides the actions of the eIF-2α kinase(s) activated by OA. In contrast, despite the presence of high levels of GADD34 in MG132-treated cells, CA, which inhibits both CReP/PP1 and GADD34/PP1 complexes, increased eIF-2α phosphorylation to levels similar to those seen in uninduced cells (Fig. 3A, lanes 3 and 6). It is noteworthy that OA and particularly CA elicited a noticeable shift in the electrophoretic mobility of GADD34 (Fig. 3A), hinting at the hyperphosphorylation of GADD34 in cells treated with phosphatase inhibitors.
The washout of MG132 from cells following several hours of exposure to the proteasome inhibitor resulted in rapid loss of the GADD34 protein (Fig. 3C). To establish the quantitative relationship between cellular GADD34 levels and eIF-2α phosphorylation, cells treated with MG132 for 8 h to first induce GADD34 were transferred into fresh medium lacking MG132 and, at various times, cells were treated with either OA or CA for 30 min, using the experimental scheme shown in Fig. 3B. GADD34 levels and eIF-2α phosphorylation were monitored by parallel immunoblotting with anti-GADD34 and phospho-eIF-2α antibody (Fig. 3C). As noted in Fig. 2C, the removal of MG132 caused a rapid decay in cellular GADD34 levels such that at 4 h, less than 20% of that seen at time zero was observed. Moreover, GADD34 was virtually undetectable by 8 h. OA failed to induce eIF-2α phosphorylation at either 0 or 4 h after MG132 washout (Fig. 3C, lanes 1 and 2), suggesting that even the much-reduced GADD34 levels (at 4 h) could override the actions of OA-activated eIF-2α kinase(s). OA only elicited eIF-2α phosphorylation at 8 h, when GADD34 had been essentially eliminated (Fig. 3C, lane 3). In contrast, CA induced robust eIF-2α phosphorylation at all times, regardless of the presence or absence GADD34, a finding consistent with its ability to inhibit eIF-2α phosphatase. These data highlighted the remarkable ability of the GADD34-assembled eIF-2α phosphatase to suppress eIF-2α phosphorylation and suggested that reduction of GADD34 to very low or near undetectable levels was required to restore cellular signaling by eIF-2α kinases in mammalian cells.
GADD34 degradation requires 26S proteasome.
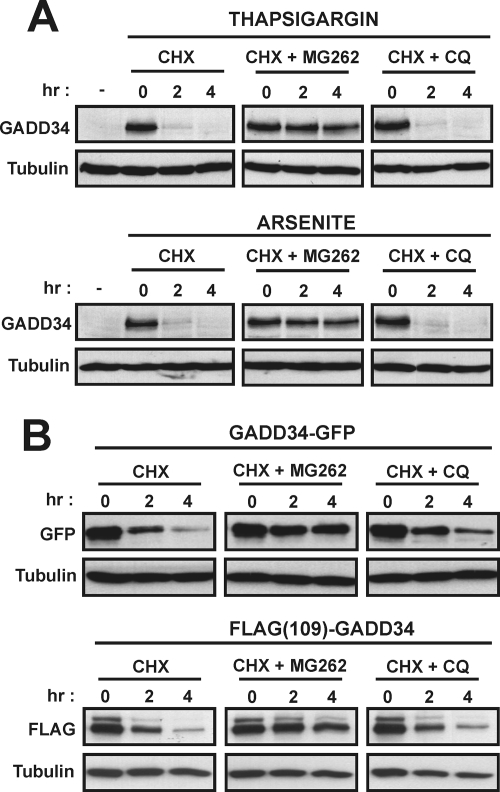
To identify the cellular mechanisms that degrade GADD34, SW480 cells were treated with thapsigargin or arsenite for 5 h to induce GADD34 expression. After the addition of a protein synthesis inhibitor, CHX, GADD34 levels decayed to barely detectable levels, as seen in untreated cells within 2 to 4 h (Fig. 4A). The presence of a proteasome inhibitor, MG262, stabilized cellular GADD34, with little reduction in the protein in the 4-h period. In contrast, a lysosomal inhibitor, CQ, had no discernible effect on GADD34 turnover. These data strongly implicated the 26S proteasome in the cellular degradation of GADD34. Similar results were obtained in HeLa, HEK293T, and A549 cells (data not shown).
FIG. 4.
Proteasomal degradation of GADD34. (A) SW480 cells exposed to 1 μM thapsigargin or 25 μM arsenite for 5 h to induce GADD34 were treated with CHX (30 μg/ml) either alone or in combination with MG262 (5 μM) or CQ (200 μM), and GADD34 levels were analyzed at selected times by immunoblotting. (B) GADD34 fused at its C terminus to GFP (GADD34-GFP) or containing an internal FLAG epitope, FLAG(109)GADD34, were expressed in HEK293T cells. After CHX treatment to inhibit protein synthesis, degradation of the expressed GADD34 proteins was analyzed by immunoblotting with anti-GFP or anti-FLAG antibodies. Cells were treated with either MG132 or CQ to investigate the role of proteasome or lysosome in GADD34 degradation. Tubulin levels monitored in all immunoblots ensured equal protein loading.
To further characterize the mechanism that degraded GADD34, we expressed GADD34 fused at its C terminus with GFP (GADD34-GFP) or containing an internal FLAG epitope, FLAG(109)GADD34, in HEK293T cells. In the presence of CHX, both GADD34 proteins decayed at similar rates to endogenous GADD34 (Fig. 4B). As seen for the endogenous GADD34, MG262 but not CQ stabilized the cellular levels of the ectopically expressed GADD34 proteins. These data established GADD34-GFP and FLAG(109)GADD34 as proteasome substrates that were degraded similarly to endogenous GADD34.
GADD34 is polyubiquitinated.
Covalent addition of branched ubiquitin chains to lysines in the substrate proteins represents the primary mechanism by which proteins are targeted for destruction by 26S proteasome. To determine whether GADD34 was subject to ubiquitin conjugation, FLAG(109)GADD34 was expressed in HeLa cells treated with MG262 to inhibit proteasome activity and accumulate polyubiquitinated proteins. Anti-FLAG immunoprecipitates from cell lysates treated with or without MG262 were subjected to SDS-PAGE and immunoblotting with an anti-ubiquitin antibody. The immunoprecipitated FLAG(109)GADD34 migrated on SDS-PAGE as a smear characteristic of polyubiquitination (Fig. 5A, lanes 1 and 2). This occurred only in the presence of MG262, and little or no immunoreactivity was seen in the absence of the proteasome inhibitor. This hinted at a very rapid turnover of the ubiquitinated GADD34. No ubiquitin immunoreactivity was observed in anti-FLAG immunoprecipitates from cells expressing control FLAG-RFP (Fig. 5A, lane 3). To alleviate any concern that the GADD34-mediated enhancement of protein may have overloaded the proteasome resulting in aberrant or nonspecific polyubiquitination of proteins, we performed additional controls. As seen in Fig. 5A, lanes 4 and 5, overexpression of GADD34-GFP either alone or in combination with FLAG-RFP failed to result in the sedimentation of ubiquitin conjugates by the FLAG-resin, thereby excluding the possibility that GADD34 overexpression promoted nonspecific protein ubiquitination. These data provided the first direct experimental evidence that cellular GADD34 was polyubiquitinated.
FIG. 5.
GADD34 is polyubiquitinated. (A) HeLa cells expressing either FLAG(109)GADD34 (lanes 1 and 2), FLAG-RFP (lane 3), GADD34-GFP (lane 4), or both FLAG-RFP and GFP-GADD34 (lane 5) were treated with or without MG262 (5 μM, 8 h). Cell lysates were subjected to immunoprecipitation using agarose-conjugated anti-FLAG antibody. Lysates and immunoprecipitates were subjected to SDS-PAGE and immunoblotted with antiubiquitin, anti-FLAG, and anti-GFP antibodies. Asterisks indicate the location of heavy and light chains of anti-FLAG IgG in all immunoprecipitates. Molecular mass markers (in kilodaltons) migrated as indicated on each blot. (B) GADD34-GFP was expressed in HEK293T cells in combination with empty vector, with vector encoding wild-type myc-tagged ubiquitin (myc-Ub), or with a mutant myc-ubiquitin (myc-Ub K48R). After 24 h, protein synthesis was inhibited with CHX, and GADD34 levels analyzed at the indicated times by immunoblotting. Tubulin levels established equivalent protein loading. (C) The results from three independent experiments (shown in panel B) were quantified by laser scanning and are shown with standard errors.
To establish the physiological significance of GADD34 polyubiquitination, we coexpressed GADD34-GFP with either empty vector, myc-tagged ubiquitin (Myc-Ub), or a mutant Myc-Ub (K48R) that inhibits polyubiquitin chain elongation and the delivery of polyubiquitinylated proteins to the 26S proteasome. GADD34-GFP turnover was monitored in the presence of CHX as described above. As anticipated, the presence of wild-type Myc-Ub had no effect on the decay rate of GADD34-GFP, which was comparable to that seen in control cells transfected with the empty vector. By comparison, the coexpression of the Myc-Ub(K48R) mutant increased basal GADD34-GFP levels and significantly slowed its degradation (Fig. 5B). Quantitation of data from several independent experiments revealed that Myc-Ub(K48R) coexpression doubled the half-life of GADD34-GFP (Fig. 5C), demonstrating a key role for polyubiquitination in the degradation of GADD34 by the 26S proteasome.
Identification of N-terminal degron in GADD34.
Human GADD34 contains four PEST repeats, motifs that have previously been implicated in the degradation of many cellular proteins (30). To investigate the contribution of these repeats to GADD34 instability, we analyzed several PEST-deleted GADD34-GFP proteins expressed in HEK293T cells. To our surprise, GADD34-GFP lacking individual (data not shown) or all four PEST repeats were all rapidly degraded, essentially at the same rate as wild-type GADD34-GFP (Fig. 5B). It is noteworthy that like wild-type GADD34-GFP, GADD34 proteins with one or more PEST deletions were all targeted to ER (data not shown), most bound PP1 and catalyzed eIF-2α dephosphorylation (see Fig. S1 in the supplemental material). These data suggested that the evolutionarily conserved PEST sequences played at best a minor role in GADD34 instability.
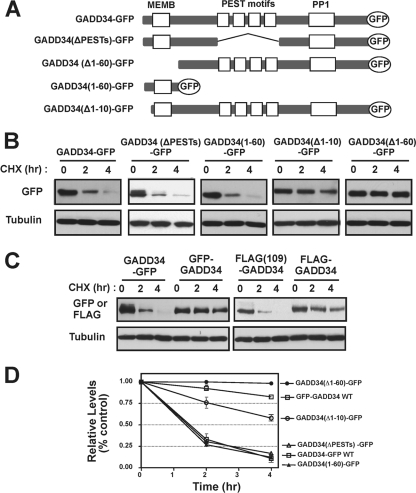
In studies aimed at identifying destabilizing elements (degrons) in GADD34, polypeptides representing N-terminal regions of the protein showed increased expression in cells in the presence of proteasome inhibitors (data not shown). This pointed to the presence of a destabilizing element or degron at or near the N terminus of GADD34. Consistent with this hypothesis, the deletion of the N-terminal 60 amino acids significantly stabilized GADD34(Δ1-60)-GFP with little reduction in protein observed over 4 h in the presence of CHX (Fig. 6B). Prior studies established that GFP displayed a half-life of >10 h in mammalian cells (2). However, after the fusion of N-terminal 60 amino acids from GADD34, the turnover of GADD34(1-60)-GFP was dramatically enhanced, with a half-life of <2 h (Fig. 6B). These data confirmed that that the N-terminal 60 residues of GADD34 encompassed a transportable degron, which was both necessary and sufficient for rapid proteasomal degradation of GADD34. Interestingly, the deletion of 10 residues from the N terminus of GADD34 also increased the stability of GADD34(Δ1-10)-GFP, albeit to a lesser extent than the deletion of N-terminal 60 residues (Fig. 6B). Finally, the fusion of GFP or FLAG at the N terminus of wild-type GADD34 produced GFP-GADD34 or FLAG-GADD34), respectively. Both proteins were significantly more stable than either GADD34-GFP containing a C-terminal fusion of GFP or FLAG(109)GADD34 containing an internal FLAG epitope (Fig. 6C). Quantification of three independent experiments highlighted the stability of GFP-GADD34 and GADD34(Δ1-60)-GFP, which showed half-lives 10- to 20-fold longer than either wild-type GADD34-GFP or GADD34(1-60)-GFP (Fig. 6D).
FIG. 6.
N-terminal degron in GADD34. (A) Schematic represents all GADD34 proteins analyzed in turnover studies. The positions of a putative ER membrane-association domain (MEMB), PEST, and PP1-binding domains are shown in boxes. (B) C-terminally GFP-tagged GADD34 proteins were expressed in HEK293T cells and subjected to turnover studies. (C) GADD34 proteins fused at the N terminus (GFP-GADD34) or the C terminus (GADD34-GFP) with GFP were expressed in HEK293T cells, and their rate of degradation analyzed as described in Materials and Methods. Similarly, turnover of an N-terminally tagged FLAG-GADD34 was compared to that of the internally tagged FLAG(109)GADD34. (D) Quantification of degradation of all GFP-fusion proteins analyzed in the present study is shown and represents the summary of three independent experiments. All data are shown with standard errors. In panels B and C, tubulin immunoblots served as loading controls.
The two established modes of proteasome-mediated degradation that utilized N-terminal sequences in the substrate proteins are the N-end rule pathway (9) and direct N-terminal α-NH2 ubiquitination (4). Although the fusion of N-terminal epitopes impairs both modes of protein degradation, a clear distinction between these two pathways is the requirement for one or more internal lysines to execute ɛ-NH2 ubiquitination required for the N-end rule pathway. Human GADD34 contains 27 lysines that are potentially subject to polyubiquitination. However, we previously observed that N-terminal fragments of GADD34, specifically one containing the amino acids 1 to 155, were degraded by the proteasome at rates similar to the full-length protein and thus likely contained the critical elements directing GADD34 degradation in human cells. The wild-type polypeptide (1-155 WT) and the mutant protein (1-155 0K), with its two internal lysines substituted with arginines, were fused with a C-terminal HA epitope, which was itself devoid of lysines. In cells treated with CHX, the turnover of 1-155 WT and 1-155 0K was essentially identical (Fig. 7). Moreover, the degradation of 1-155 0K was inhibited by MG262, albeit to a slightly lesser extent than 1-155 WT. These data suggested that ɛ-NH2 ubiquitination of internal lysines was not essential for the degradation of GADD34(1-155) and favored α-NH2 ubiquitination as the likely requirement for the degradation of GADD34 by the proteasome.
FIG. 7.
Internal lysines and GADD34 degradation. (A) C-terminal HA-tagged wild-type (WT) and lysineless (0K) GADD34 polypeptides, residues 1 to 155, were expressed in HEK293T cells and subjected to turnover analyses in the absence or presence of 5 μM MG262. Lysates were generated at 0, 1.5, and 3 h following the addition of CHX; subjected to SDS-PAGE; and immunoblotted for HA and tubulin.
Sustained GADD34 expression impairs ER protein processing.
Premature translational recovery induced by GADD34 during ER stress can promote cell death, resulting from a backlog of protein processing and excessive oxidative burden in the ER (25). Moreover, a number of human diseases result from mutations in proteins that impair their processing in the ER. For example, human cystic fibrosis results from mutations in the cystic fibrosis transmembrane conductance regulator (CFTR), most commonly the deletion of phenylalanine-508 (ΔF508), which impairs its synthesis and trafficking through the ER (1). At low levels of expression, CFTR(ΔF508) can be processed normally and delivered to the plasma membrane (17). However, the errors on CFTR(ΔF508) processing become much more noticeable when the mutant protein was expressed at higher levels and result in its prolonged retention and/or accumulation in the ER, with the subsequent formation of protein aggregates (2). This provided a very sensitive assay for evaluating the impact of sustained GADD34 expression on protein processing in the ER. We expressed GFP-CFTRΔ508 and the N-terminally tagged FLAG-GADD34, which showed significantly greater stability compared to either endogenous or GADD34-GFP in HEK293T cells. Analysis of cells expressing GFP-CFTRΔF508 using fluorescence microscopy showed relatively low levels of this protein when coexpressed with a control cytosolic marker (mRFP) or ER marker (DsRed2-ER) (Fig. 8A, control). Treating these cells with MG262 resulted in a modest increase in CFTRΔF508 expression (Fig. 8A, +MG262), a finding consistent with the known degradation of CFTR by the 26S proteasome (8, 37). Remarkably, the cotransfection of FLAG-GADD34 dramatically increased GFP-CFTRΔF508 expression even in the absence of a proteasome inhibitor (Fig. 8A). Since neither mRFP nor DsRed2-ER levels were increased, despite being driven by the same promoter as GFP-CFTRΔ508, the preferential increase in GFP-CFTRΔF508 likely reflected its distinct translational and posttranslational processing in the ER. These data highlighted the potential impact of prolonged GADD34 activity, impairing ER function by enhancing the accumulation of folding-impaired proteins such as CFTRΔF508.
FIG. 8.
Stabilized GADD34 facilitates protein aggregation. (A) GFP-CFTRΔF508 was coexpressed in HEK293T cells with either cytosolic (RFP) or ER (DsRed2-RFP) proteins. Cells were untreated (control), exposed to 5 μM MG262 for 8 h, or cotransfected with FLAG-GADD34. Cells were analyzed by fluorescence microscopy. (B) GFP-CFTRΔF508 or GFP-GRP94 were expressed in HEK293T cells that were either untreated (control), exposed to 5 μM MG262 for 8 h, or cotransfected with FLAG-GADD34. At 24 h posttransfection, cells were lysed in RIPA buffer. After removing an aliquot (T), the lysates were subjected to centrifugation to separate soluble (S) and insoluble (I) fractions. Proteins were solubilized by heating and sonication in RIPA containing 1% SDS. T, S, and I fractions were subjected to SDS-PAGE and analyzed by immunoblotting. Double asterisks (**) indicate faster-migrating band glycosylation intermediates of CFTR, and a single asterisk (*) indicates HMW aggregated forms of CFTR. (C) Distribution of FLAG-GADD34 in HEK293T cells cotransfected with GFP-GRP94, CFTRΔF508, or GFP-250 was analyzed in the T, S, and I fractions by immunoblotting. Tubulin and GM130 immunoblots served as controls.
High expression of CFTRΔF508 prompts the formation of protein aggregates or aggresomes that are thought to redirect the mutant protein for degradation by the proteasome (2). Thus, we analyzed the formation of protein aggregates containing the mutant CFTR in HEK293T cells. The distribution of GFP-CFTRΔF508 in detergent-soluble and insoluble pools was compared to a control folding-competent protein, GFP-GRP94. In untreated cells, GFP-CFTRΔF508 was expressed at lower levels and largely present in the detergent-soluble fraction, where a series of bands representing immature or incompletely glycosylated intermediates of CFTR (indicated by “**” in the figure) are seen (Fig. 8B, left panel). Very little insoluble high-molecular-weight (HMW) species (indicated by “*” in the figure), indicative of aggregated CFTRΔF508, was observed in these cells. Treatment with MG262 resulted in the increased expression of mutant CFTR and notable reduction in the soluble species with majority of the protein being seen in the insoluble fraction or HMW aggregates (Fig. 8B, middle panel). The most striking observation was that coexpression of FLAG-GADD34 resulted in the accumulation of even higher levels of mutant CFTR, with significant accumulation of both immature soluble and aggregated insoluble species (Fig. 8B, right panel). Similar accumulation of GFP-CFTRΔ508 aggregates was not observed when coexpressed with a mutant FLAG-GADD34 that lacked PP1-binding (data not shown), suggesting that CFTR accumulation and aggregation resulted from the ability of stabilized GADD34 to assemble an eIF-2α phosphatase and promote protein translation. Interestingly, the expression and subcellular distribution of GFP-GRP94, or endogenous tubulin, was not affected by either proteasome inhibition or FLAG-GADD34 coexpression (Fig. 8B and C). This demonstrated that the elevated GADD34 function more severely impacted the accumulation and solubility of folding-compromised proteins such as GFP-CFTRΔ508. This notion was also supported by the observation that another folding-deficient model protein, the cytosolic GFP-250 protein (20), behaved similarly to CFTRΔF508 and was aggregated in the presence of FLAG-GADD34 (data not shown). To our surprise, FLAG-GADD34 was also incorporated into the protein aggregates found in cells that expressed folding compromised proteins such as GFP-CFTRΔF508 or GFP-250 (Fig. 8C). In contrast, other proteins, such as GM130 and tubulin, were excluded from these insoluble aggregates. This raised the intriguing possibility that incorporation of excess GADD34 in aggregates containing misfolded or partially processed proteins represented a novel mechanism for lowering cellular GADD34 activity and limiting the detrimental effects of prolonged or elevated protein translation. Together, our data showed that sustained GADD34 expression and the resulting increase in protein translation can overwhelm ER quality control mechanisms, resulting in defective protein maturation, folding, and degradation, which eventually triggers the apoptosis of irreversibly damaged cells.
DISCUSSION
Transient expression of GADD34.
Cellular insults, such as DNA damage, nutrient deprivation, UV irradiation, viral infection, and ischemia, trigger the activation of one or more kinases that catalyze eIF-2α phosphorylation to inhibit general protein translation and activate a complex transcriptional program known as the ISR. A genetic suppressor element of ISR, A1 GSE, encoded a fragment of hamster GADD34 (Myd116) and established GADD34 as a potent suppressor, as well as a target of ISR (21, 27). This is consistent with the observation GADD34 binds PP1 to assemble an eIF-2α phosphatase complex that attenuates eIF-2α phosphorylation (5, 26) and the expression of ISR genes (21, 27). Subsequent studies showed that ISR genes, such as CHOP, ATF3, and ATF4, activate GADD34 transcription (14, 24, 25), suggesting that GADD34 functions in a negative-feedback loop that enables a transient and orderly expression of stress response genes and tempers the potentially negative physiological effect of prolonged eIF-2α phosphorylation. Since prolonged eIF-2α phosphatase activity can exacerbate cell stress, by elevating protein synthesis and overtaxing the cell's protein quality control machinery, the rapid degradation of GADD34 following the resolution of stress is also critical to restore homeostasis and allow the cell to respond effectively to subsequent insults.
Rutkowski et al. (31) analyzed the temporal relationship for the induction of several stress response genes, including GADD34, by both mild/reversible and chronic or terminal ER stress. These studies suggested a higher threshold for the induction of proapoptotic effectors, such as CHOP/GADD153 and GADD34, compared to survival proteins, like the ER chaperone BiP. The intrinsic instability of mRNAs and possibly also the proteins representing the proapoptotic components, such as CHOP and GADD34 (31), made it more difficult to activate this pathway and required more severe and/or persistent stress signals. This was particularly true for distal components such as GADD34, which required the prior upregulation of ATF4 and CHOP. Thus, cells seemed to adapt to mild forms of stress by the rapid and preferential activation of the prosurvival genes, which elevated their protein folding capacity and avoided the need for ISR activation by subsequent mild disruptions in protein folding. Interestingly, GADD34 levels correlated strongly with cell fate, suggesting that persistent GADD34 expression triggered cell death. This raised the intriguing possibility that under severe or irreparable cell stress, the sustained GADD34 expression committed the damaged cell to abandoning adaptive measures and embracing apoptosis. Recent findings support this paradigm, since premature or prolonged GADD34 activity exacerbates the unfolded protein response, increases the oxidative burden on the protein processing machinery, and promotes cell death (25, 31). The present report established the dependence of GADD34 turnover on proteasome function. This hinted that under severe stress, when proteasome function is partially compromised, cellular GADD34 levels might be severely elevated. By assembling an eIF-2α phosphatase, the elevated GADD34 may enhance the translation of GADD34 and other proteins to create a vicious feed-forward cycle and further impair proteasome function in the irreversibly damaged cells. It should be noted that significant induction of GADD34 by environmental insults occurs in MEFs that lack individual ISR genes (14, 24, 25), thereby hinting at a second pathway, independent of eIF-2α phosphorylation, for the transcriptional activation of GADD34. Thus, under conditions of severe stress, the transcriptional activation of GADD34 combined with its increased stability may maintain high levels of GADD34 in the stressed cells despite significant attenuation of eIF-2α phosphorylation by the GADD34-containing eIF-2α phosphatase.
Proteasomal degradation of GADD34.
The present study noted significant differences in GADD34 accumulation in several human cell lines subjected to various forms of cell stress. These differences in GADD34 content were largely eliminated when cells were treated with proteasome inhibitors. Our studies further established that on the reversal of cell stress, endogenous GADD34 was rapidly degraded. Moreover, near-basal levels of GADD34 were required before stress-induced eIF-2α phosphorylation could be fully restored. This emphasized that the active degradation of GADD34 following the resolution of cell stress is necessary to restore the cell's ability to respond to subsequent stress.
Both endogenous and ectopically expressed GADD34 displayed half-lives of <2 h in cells treated with the protein synthesis inhibitor, CHX. Conversely, proteasome inhibitors but not lysosome inhibitors significantly prolonged the half-life of GADD34. We also demonstrated that cellular GADD34 underwent polyubiquitination, and its stabilization in cells expressing the chain-terminating ubiquitin mutant (Ub-K48R) established the physiological role of polyubiquitination in the proteasome-mediated degradation of GADD34. Together, the data suggested that GADD34 levels reflect not only changes in gene transcription but also protein turnover and raise the possibility that stress signals that inhibit proteasome function achieve higher levels of GADD34 and trigger ISR-induced programmed cell death.
ER stress induced by arsenite, in contrast to thapsigargin or amino acid deprivation, was shown to inhibit proteasome activity in MEFs, resulting in significant accumulation of GADD45, the product of another stress response gene (16). However, arsenite elicited weak induction of GADD34 in A549 cells, although more robust GADD34 induction was observed in HeLa and HEK293T cells subjected to the same insult (Fig. 1). By comparison, direct proteasome inhibition results in high GADD34 accumulation in all cells, possibly illustrating differences in stress signaling and the role of the 26S proteasome in regulating GADD34 and other ISR proteins in human cells. Moreover, prior induction of GADD34 by the proteasome inhibitors strongly attenuated eIF-2α phosphorylation in many human cell lines (Fig. 3). Consistent with this observation, proteasome inhibition attenuated the cellular response to ER stress (22). In contrast, MG132 enhanced eIF-2α phosphorylation in MEFs despite significant induction of GADD34 (15). This may hint at additional and as-yet-unidentified posttranslational mechanisms that regulate the assembly and disassembly of the cellular GADD34-PP1 complex (5).
N-terminal degron in GADD34.
Investigating proteasome-mediated GADD34 degradation, we noted that the fusion of epitopes at its C terminus had no discernible effect on GADD34 degradation in cells. With the same epitopes fused at its N terminus, GADD34's half-life was prolonged >4-fold. Deletion of N-terminal 10 amino acids, and even more so, 60 amino acids, also stabilized GADD34. Conversely, the fusion of N-terminal 60 amino acids from GADD34 shortened the half-life of GFP, from more than 10 h as reported in the literature (2) to less than 2 h, similar to that of wild-type GADD34. These studies established the presence of an N-terminal degron in GADD34. Proteins that contain an N-terminal degron are degraded by either the N-end rule or by direct N-terminal ubiquitination. In eukaryotes, the N-end rule is the more common mechanism, which utilizes two key structural elements, an “N-degron” or more specifically a destabilizing N-terminal residue and ubiquitination of one or more internal lysines. The binding of an N-recognin complex to the destabilizing N-terminal residue enables an E3-ubiquitin ligase to target internal lysines for ɛ-NH2 ubiquitination. As eukaryotic proteins are synthesized with N-terminal methionine, a “stabilizing” residue that is bound weakly by N-recognin, a covalent modification to generate a new N terminus either through proteolysis or addition of destabilizing amino acids activates the N-end rule. The second mode of protein degradation is the more recently identified N-terminal ubiquitination (4). Here, a distinct E3-ligase catalyzes α-NH2 ubiquitination of the amino-terminal residue in the target protein. Although recent studies linked protein degradation by the N-end rule to the induction of ER stress (38), this is unlikely to mediate the proteasome-mediated degradation of GADD34. Comparing the decay rates for 1-155, an N-terminal fragment of GADD34 to that of 1-155 0K, containing the substitution of all internal lysines, we established that both polypeptides were similarly degraded by the proteasome. In this regard, GADD34 degradation fulfills all criteria previously established for the N-terminal α-NH2 ubiquitination and proteasomal degradation of the CDK inhibitor, p21, MyoD, and other proteins (4). Future studies will investigate the direct N-terminal ubiquitination of GADD34 and its role in rapid protein degradation following cell stress.
Cellular studies showed that A1 GSE encoding a fragment of hamster GADD34/Myd116, amino acids 292 to 590, generated a more active eIF-2α phosphatase than wild-type GADD34 (26). The authors of that study postulated that the more restricted localization of wild-type GADD34 at the ER, a potentially “inactive” site, accounted for its reduced eIF-2α phosphatase activity compared to the A1 protein that was widely distributed in the cytoplasm. Current studies suggest that the absence of an N-degron may also increase or prolong the actions of the A1 peptide, generating a more active eIF-2α phosphatase than the rapidly degraded wild-type GADD34.
Role of PEST repeats.
GADD34 contains four PEST repeats that are highly conserved across species. Rich in prolines, acidic residues, serines, and threonines, PEST sequences are frequently found in unstable proteins and play a key role in protein turnover (30). However, deletion of all four PEST repeats had no impact of GADD34 turnover in human cells. Thus, alternate roles for the PEST sequences were investigated. Our prior studies showed that the PKA-activated PP1 inhibitor, inhibitor-1, bound GADD34 in a region that encompassed the PEST repeats (5). The current studies analyzed both eIF-2α phosphatase activity (see Fig. S1B in the supplemental material) and PP1 binding (see Fig. S1C in the supplemental material) by GADD34-GFP polypeptides lacking one or more PEST repeats and showed that deletion of the fourth or most C-terminal PEST repeat reduced PP1 binding and eIF-2α phosphatase activity. The more surprising finding was that GADD34-GFP proteins lacking the first PEST repeat bound PP1 like wild-type GADD34-GFP but were severely impaired in catalyzing eIF-2α dephosphorylation. Most notably, OA-induced eIF-2α phosphorylation was indistinguishable in cells expressing Δ1,2P, Δ1,2,3P, and KARA, a mutant GADD34 that lacked the core PP1-binding motif and thus could not assemble an eIF-2α phosphatase (3). This provided new evidence for collaboration between the GADD34 PEST repeats and the C-terminal PP1-binding domain (5), conserved in the HSV-1 γ34.5 protein, in assembling a functional eIF-2α phosphatase.
Treatment of cells with OA or CA resulted in an upward shift in GADD34 mobility on SDS-PAGE (see Fig. S2A in the supplemental material), suggesting the increased phosphorylation of GADD34. Metabolic labeling of cells expressing GADD34 proteins with internal PEST deletions (see Fig. S2A in the supplemental material) or systematic N- and C-terminal deletions (see Fig. S2C to E in the supplemental material) established that the majority of the protein-bound phosphates resided in the second PEST repeat. While the functional role of GADD34 phosphorylation remains to be defined, prior studies suggested a cross talk between PEST phosphorylation and protein ubiquitination during the proteasomal degradation of some proteins (23). Reversible phosphorylation may also regulate the assembly of the cellular GADD34 complex, modulating the recruitment of PP1, inhibitor-1, or other proteins, to control eIF-2α phosphorylation.
GADD34 and human disease.
Protein overload and impaired processing/folding of mutant proteins in ER contributes to many human diseases (11). The most common mutation associated with human cystic fibrosis, ΔF508, generates a CFTR protein that is poorly processed, resulting in its ER retention and destruction by ER-associated degradation pathway (8, 36). The delayed ER processing/trafficking of CFTRΔ508 also elicits ER stress in the lungs of cystic fibrosis patients (35) and would be predicted to activate ISR genes, including GADD34. The dephosphorylation of eIF-2α by the GADD34/PP1 complex may increase the translation of CFTRΔ508 and account for the buildup of misfolded protein aggregates in the ER. Protein aggregates are thought to represent a fail-safe mechanism that reduces the demand on the already overburdened ER protein processing machinery and redirects misfolded proteins for polyubiquination and degradation by the 26S proteasome (17). However, aggregates formed by CFTRΔ508 also impair proteasome function (2). Other studies implicated PKR, PERK, and eIF-2α phosphorylation in promoting ubiquitin-dependent proteasome-mediated degradation of cyclin D1 (29). This would predict that by antagonizing eIF-2α phosphorylation, elevated GADD34 levels inhibit the degradation of some proteasome substrates, possibly including the ISR proteins. The present studies that showed that proteasome inhibition increased cellular content of GADD34 and enhanced CFTRΔ508 expression and aggregation suggest a key role for GADD34 in the tissue damage and cell death seen in diseases such as cystic fibrosis.
In conclusion, our studies suggest that the short half-life of GADD34 protects normal cells from transient delays in protein processing or milder forms of stress. In contrast, the prolonged expression or activity of stabilized forms of GADD34 result in much higher accumulation of CFTRΔ508 and protein aggregation in the ER than seen with pharmacological inhibition of the proteasome. Interestingly, FLAG-GADD34 was also incorporated in the protein aggregates, suggesting a novel cellular mechanism clearing excess GADD34. Together, the studies emphasized the deleterious effects of prolonged GADD34 function in human health and hinted at a role for GADD34 in the pathophysiology of cystic fibrosis and other protein-folding diseases.
Supplementary Material
Acknowledgments
We thank Christopher V. Nicchitta (Duke University) for advice and support during the course of these studies.
This study was supported by NIH grant RO1-DK52054 (to S.S.).
Footnotes
Published ahead of print on 15 September 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Akabas, M. H. 2000. Cystic fibrosis transmembrane conductance regulator: structure and function of an epithelial chloride channel. J. Biol. Chem. 2753729-3732. [DOI] [PubMed] [Google Scholar]
- 2.Bence, N. F., R. M. Sampat, and R. R. Kopito. 2001. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 2921552-1555. [DOI] [PubMed] [Google Scholar]
- 3.Brush, M. H., D. C. Weiser, and S. Shenolikar. 2003. Growth arrest and DNA damage-inducible protein, GADD34, targets protein phosphatase 1 to the endoplasmic reticulum and promotes dephosphorylation of α-subunit of eukaryotic translation initiation factor 2. Mol. Cell. Biol. 231292-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciechanover, A., and R. Ben-Saadon. 2004. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 14103-106. [DOI] [PubMed] [Google Scholar]
- 5.Connor, J. H., D. C. Weiser, S. Li, J. M. Hallenbeck, and S. Shenolikar. 2001. Growth arrest and DNA damage-inducible protein GADD34 assembles a novel signaling complex containing protein phosphatase 1 and inhibitor 1. Mol. Cell. Biol. 216841-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Costa-Mattioli, M., D. Gobert, E. Stern, K. Gamache, R. Colina, C. Cuello, W. Sossin, R. Kaufman, J. Pelletier, K. Rosenblum, K. Krnjevic, J. C. Lacaille, K. Nader, and N. Sonenberg. 2007. eIF2α phosphorylation bidirectionally regulates the switch from short- to long-term synaptic plasticity and memory. Cell 129195-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganoza, M. C., M. C. Kiel, and H. Aoki. 2002. Evolutionary conservation of reactions in translation. Microbiol. Mol. Biol. Rev. 66460-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gelman, M. S., E. S. Kannegaard, and R. R. Kopito. 2002. A principal role for the proteasome in endoplasmic reticulum-associated degradation of misfolded intracellular cystic fibrosis transmembrane conductance regulator. J. Biol. Chem. 27711709-11714. [DOI] [PubMed] [Google Scholar]
- 9.Gonda, D. K., A. Bachmair, I. Wunning, J. W. Tobias, W. S. Lane, and A. Varshavsky. 1989. Universality and structure of the N-end rule. J. Biol. Chem. 26416700-16712. [PubMed] [Google Scholar]
- 10.Han, A. P., C. Yu, L. Lu, Y. Fujiwara, C. Browne, G. Chin, M. Fleming, P. Leboulch, S. H. Orkin, and J. J. Chen. 2001. Heme-regulated eIF2α kinase (HRI) is required for translational regulation and survival of erythroid precursors in iron deficiency. EMBO J. 206909-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hebert, D. N., and M. Molinari. 2007. In and o of the ER: protein folding, quality control, degradation, and related human diseases. Physiol. Rev. 871377-1408. [DOI] [PubMed] [Google Scholar]
- 12.Hollander, M. C., Q. Zhan, I. Bae, and A. J. Fornace. 1997. Mammalian GADD34, an apoptosis- and DNA damage-inducible gene. J. Biol. Chem. 27213731-13737. [DOI] [PubMed] [Google Scholar]
- 13.Hollander, M. C., M. S. Sheikh, K. Yu, Q. Zhan, M. Iglesias, C. Woodworth, and A. J. Fornace. 2001. Activation of GADD34 by diverse apoptotic signals and suppression of its growth inhibitory effects by apoptotic inhibitors. Int. J. Cancer 9622-31. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, H. Y., S. A. Wek, B. C. McGrath, D. Lu, T. Hai, H. P. Harding, X. Wang, D. Ron, D. R. Cavener, and R. C. Wek. 2004. Activating transcription factor 3 is integral to the eukaryotic initiation factor 2 kinase stress response. Mol. Cell. Biol. 241365-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang, H. Y., and R. C. Wek. 2005. Phosphorylation of the α-subunit of the eukaryotic initiation factor-2 (eIF2) reduces protein synthesis and enhances apoptosis in response to proteasome inhibition. J. Biol. Chem. 28014189-14202. [DOI] [PubMed] [Google Scholar]
- 16.Jiang, H. Y., L. Jiang, and R. C. Wek. 2007. The eukaryotic initiation factor-2 kinase pathway facilitates differential GADD45α expression in response to environmental stress. J. Biol. Chem. 2823755-3765. [DOI] [PubMed] [Google Scholar]
- 17.Johnston, J. R., C. L. Ward, and R. R. Kopito. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 1431883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jousse, C., S. Oyadomari, I. Novoa, P. Lu, Y. Zhang, H. P. Harding, and D. Ron. 2003. Inhibition of a constitutive translation initiation factor 2α phosphatase, CReP, promotes survival of stressed cells. J. Cell Biol. 163767-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kandel, E. R. 2001. The molecular biology of memory storage: a dialogue between genes and synapses. Science 2941030-1038. [DOI] [PubMed] [Google Scholar]
- 20.Kawaguchi, Y., J. J. Kovacs, A. McLaurin, J. M. Vance, A. Ito, and T. P. Yao. 2003. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115727-738. [DOI] [PubMed] [Google Scholar]
- 21.Kojima, E., A. Takeuchi, M. Haneda, F. Yagi, T. Hasegawa, K. Yamaki, K. Takeda, S. Akira, K. Shimokata, and K. Isobe. 2003. The function of GADD34 is a recovery from a shutoff of protein synthesis induced by ER stress: elucidation by GADD34-deficient mice. FASEB J. 171573-1575. [DOI] [PubMed] [Google Scholar]
- 22.Lee, A. H., N. N. Iwakoshi, K. C. Anderson, and L. H. Glimcher. 2003. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc. Natl. Acad. Sci. USA 1009946-9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, R., P. Beauparlant, C. Makris, S. Meloche, and J. Hiscott. 1996. Phosphorylation of IκBα in the C-terminal PEST domain by casein kinase II affects intrinsic protein stability. Mol. Cell. Biol. 161401-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma, Y., and L. M. Hendershot. 2003. Delineation of a negative feedback regulatory loop that controls protein translation during endoplasmic reticulum stress. J. Biol. Chem. 27834864-34873. [DOI] [PubMed] [Google Scholar]
- 25.Marciniak, S. J., C. Y. Yun, S. Oyadomari, I. Novoa, Y. Zhang, R. Jungreis, K. Nagata, H. P. Harding, and D. Ron. 2004. CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Gene Dev. 183066-3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novoa, I., H. Zeng, H. P. Harding, and D. Ron. 2001. Feedback inhibition of the unfolded protein response by GADD34-mediated dephosphorylation of eIF2. J. Cell Biol. 1531011-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novoa, I., Y. Zhang, H. Zeng, R. Jungreis, H. P. Harding, and D. Ron. 2003. Stress-induced gene expression requires programmed recovery from translational repression. EMBO J. 221180-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patterson, A. D., M. C. Hollander, G. F. Miller, and A. J. Fornace. 2006. GADD34 requirement for normal hemoglobin synthesis. Mol. Cell. Biol. 261644-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raven, J. F., D. Baltzis, S. Wang, Z. Mounir, A. I. Papadakis, H. Q. Gao, and A. E. Koromilas. 2008. PKR and PKR-like endoplasmic reticulum kinase induce the proteasome-dependent degradation of cyclin D1 via a mechanism requiring eukaryotic initiation factor 2α phosphorylation. J. Biol. Chem. 2833097-3108. [DOI] [PubMed] [Google Scholar]
- 30.Rechsteiner, M. 1990. PEST sequences are signals for rapid intracellular proteolysis. Semin. Cell Biol. 1433-440. [PubMed] [Google Scholar]
- 31.Rutkowski, D. T., S. M. Arnold, C. N. Miller, J. Wu, J. Li, K. M. Gunnison, K. Mori, A. A. S. Akha, D. Raden, and R. J. Kaufman. 2006. Adaptation to ER stress is mediated by differential stabilities of pro-survival and proapoptotic mRNAs and proteins. PLoS Biol. 42024-2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheuner, D., B. Song, E. McEwen, C. Liu, R. Laybutt, P. Gillespie, T. Saunders, S. Bonner-Weir, and R. J. Kaufman. 2001. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol. Cell 71165-1176. [DOI] [PubMed] [Google Scholar]
- 33.Scheuner, D., D. Vander Mierde, B. Song, D. Flamez, J. W. M. Creemers, K. Tsukamoto, M. Ribick, F. C. Schuit, and R. J. Kaufman. 2005. Control of mRNA translation preserves endoplasmic reticulum function in beta cells and maintains glucose homeostasis. Nat. Med. 11757-764. [DOI] [PubMed] [Google Scholar]
- 34.Scheuner, D., R. Patel, F. Wang, K. Lee, K. Kumar, J. Wu, A. Nilsson, M. Karin, and R. J. Kaufman. 2006. Double-stranded RNA-dependent protein kinase phosphorylation of the α-subunit of eukaryotic translation initiation factor 2 mediates apoptosis. J. Biol. Chem. 28121458-21468. [DOI] [PubMed] [Google Scholar]
- 35.Turnbull, E. L., M. F. N. Rosser, and D. M. Cyr. 2007. The role of the UPS in cystic fibrosis. BMC Biochem. 8S11-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vij, N., S. Fang, and P. L. Zeitlin. 2006. Selective inhibition of endoplasmic reticulum-associated degradation rescues ΔF508-cystic fibrosis transmembrane regulator and suppresses interleukin-8 levels: therapeutic implications. J. Biol. Chem. 28117369-17378. [DOI] [PubMed] [Google Scholar]
- 37.Ward, C. L., S. Omura, and R. R. Kopito. 1995. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83121-127. [DOI] [PubMed] [Google Scholar]
- 38.Wojcik, C., M. Rowicka, A. Kudlicki, D. Nowis, E. McConnell, M. Kujawa, and G. N. DeMartino. 2006. Valosin-containing protein (p97) is a regulator of endoplasmic reticulum stress and of the degradation of N-End rule and ubiquitin-fusion degradation pathway substrates in mammalian cells. Mol. Biol. Cell 174606-4618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, P., B. McGrath, S. Li, A. Frank, F. Zambito, J. Reinert, M. Gannon, K. Ma, K. McNaughton, and D. R. Cavener. 2002. The PERK eukaryotic initiation factor 2α kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol. Cell. Biol. 223864-3874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.