Abstract
Nucleophosmin (NPM) (B23) is an essential protein in mouse development and cell growth; however, it has been assigned numerous roles in very diverse cellular processes. Here, we present a unified mechanism for NPM's role in cell growth; NPM directs the nuclear export of both 40S and 60S ribosomal subunits. NPM interacts with rRNA and large and small ribosomal subunit proteins and also colocalizes with large and small ribosomal subunit proteins in the nucleolus, nucleus, and cytoplasm. The transduction of NPM shuttling-defective mutants or the loss of Npm1 inhibited the nuclear export of both the 40S and 60S ribosomal subunits, reduced the available pool of cytoplasmic polysomes, and diminished overall protein synthesis without affecting rRNA processing or ribosome assembly. While the inhibition of NPM shuttling can block cellular proliferation, the dramatic effects on ribosome export occur prior to cell cycle inhibition. Modest increases in NPM expression amplified the export of newly synthesized rRNAs, resulting in increased rates of protein synthesis and indicating that NPM is rate limiting in this pathway. These results support the idea that NPM-regulated ribosome export is a fundamental process in cell growth.
The nucleolus is a dynamic subnuclear organelle that contains proteins involved in not only ribosome production but also oncogenesis and tumor suppression (28). As the site for ribosome biogenesis, a cell's nucleoli are centered around ribosomal DNA (rDNA) genes, which are transcribed to produce rRNAs (49). A host of proteins are involved in the intricate processing and assembly steps required to produce the small, 40S, and large, 60S, ribosomal subunits (9, 30, 49). Once exported from the nucleus, the 40S and 60S subunits are combined in the cytoplasm to form functional 80S subunits (20, 30). While the steps involved in the transcription and processing of rRNAs within the nucleolus are beginning to be understood, little is known about how ribosomal subunits are exported to the cytosol in mammalian cells. Studies of Saccharomyces cerevisiae and Xenopus laevis oocytes have shown that the process is dependent on the CRM1-RanGTP export receptor pathway (20). Additionally, NMD3, a highly conserved adapter protein, assists in the nuclear export of the 60S subunit in yeast (18). However, experiments with mammalian cells have provided evidence that a complex export network exists. The nuclear export of some mammalian rRNAs is dependent on NMD3, while the export of others is not (47). Thus, additional nucleolar proteins must be involved in the nuclear export of ribosomes.
Nucleophosmin (NPM) is an abundant nucleolar phosphoprotein that is highly expressed in proliferating cells (10). NPM is involved in numerous diverse cellular processes such as ribosome biogenesis (33), centrosome duplication (31), protein chaperoning (32), and transcriptional control (7). We have shown that NPM can shuttle from the nucleolus/nucleus to the cytoplasm (4), with its shuttling requiring a conserved CRM1 binding site (51). The mutation of two critical leucine residues (Leu-42 and Leu-44 to Ala-42 and Ala-44) in NPM's conserved CRM1 binding domain blocks NPM's ability to transit from the nucleus, resulting in nuclear and nucleolar retention (51). Additionally, NPM is an essential nucleolar protein with an acute loss of expression, resulting in a severe attenuation of cellular proliferation and increased apoptosis (3, 4, 19). In agreement with these in vitro findings, two reports have shown that Npm1 is required for embryonic development and that fibroblasts derived from Npm1-deficient embryos have a reduced capacity to grow and proliferate (6, 15).
In an effort to define NPM's role in the cell, we first characterized endogenous NPM protein complexes. A majority of NPM binding proteins were ribosomal subunit proteins or were known players in ribosome biogenesis. In vivo, NPM associated with ribosomes in the nucleolus, nucleus, and cytosol. We found that NPM directed ribosome nuclear export in a rate-limiting mechanism and that the export of ribosomes was dependent on NPM expression. By setting the rate at which ribosomes are exported to the cytosol, we discovered that NPM was a potent inducer of enhanced polysome formation and protein synthesis. Thus, we now describe a role for NPM in driving ribosome nuclear export, providing a consensus function for NPM in regulating cell growth.
MATERIALS AND METHODS
Cell culture, virus production, and nucleofection.
HeLa cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM glutamine, 0.1 mM nonessential amino acids, and 100 U penicillin and streptomycin. Npm+/+ and Npmhy/hy litter-matched mouse embryo fibroblasts (MEFs), Arf−/− MEFs, p53−/− MEFs, double-knockout (DKO) MEFs (p53−/− Mdm2−/−), and triple-knockout (TKO) MEFs (Arf−/− p53−/− Mdm2−/−) were maintained in identical media with gentamicin. Virus production and infection of MEFs were carried out using retroviral helper and vector plasmids provided by Charles Sawyers (UCLA). HeLa cells (2 × 106 cells) were transduced with 2 μg of the indicated plasmids according to the manufacturer's instructions (Amaxa Inc.).
Plasmid constructs.
Retroviral vectors encoding full-length His-tagged murine NPM and NPMdL were described elsewhere previously (4, 16, 51), and pBabe-GFP-NPM and pBabe-GFP-NPMdL were made by subcloning the Nhe-PstI green fluorescent protein (GFP)-NPM or GFP-NPMdL fragment from pEGFP-NPM and pEGFP-NPMdL (51) into the SnaBI site of pBabe-puro for retroviral production. His epitope-tagged NPM and His-NPMdL in pcDNA3.1 expression vectors were previously described (51). Short interfering RNAs (siRNAs) against NPM and a scrambled control siRNA were also previously described (51).
Fluid-phase liquid chromatography.
HeLa cells were lysed in Tween 20 lysis buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.1% Tween 20, 1 μM NaVO4, 10 μM NaF, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μg/ml aprotinin) by sonication. Lysates (600 μg) were injected onto a HiPrep 16/60 Sephacryl S-300 column (Amersham). Proteins were eluted with 150 mM NaCl-50 mM NaH2PO4 (pH 7.2) using BioLogic fluid-phase liquid chromatography and HR software (Bio-Rad). Fractions were precipitated with trichloroacetic acid (TCA), resuspended in 1 M Tris-HCl (pH 7.4), separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, transferred onto polyvinylidene difluoride membranes, and immunoblotted with antibodies recognizing NPM (Zymed). For affinity chromatography, a custom rabbit polyclonal antibody recognizing the N terminus of NPM (Sigma) was bound to NHS-activated Sepharose (Amersham). HeLa cells were lysed in 20 mM Tris (pH 7.4)-0.1% Tween 20 and sonicated. Lysates (600 μg) were first injected over the sizing column, and subsequent NPM pools were loaded onto the NPM affinity column, washed with 20 mM Tris, and eluted with an increasing NaCl gradient (0.1 to 1 M). Collected fractions were pooled, precipitated, and subjected to proteomic analysis.
Proteomic analysis.
Proteins from fluid-phase liquid chromatography fractions were precipitated with TCA and resuspended in rehydration buffer (Bio-Rad). Five hundred micrograms of proteins and 12.5 μl of 200 mM tributylphosphine (Bio-Rad) were mixed and loaded by passive in-gel rehydration. IPG strips (pH 3 to 10; Bio-Rad) were focused, and separated proteins were stained with Sypro ruby (Bio-Rad). Tryptic peptides were calibrated with a Sequazyme peptide mass standard kit (PE Biosystems) and analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry (Voyager DE Pro; Applied Biosystems). The identification of proteins was performed using MS-Fit software (http://prospector.ucsf.edu/ucsfhtml4.0/msfit.htm) and verified using tandem mass spectrometry techniques.
rRNA immunoprecipitation.
Arf−/− MEFs were starved in methionine-free medium for 15 min prior to labeling with 50 μCi/ml [methyl-3H]methionine (GE Healthcare) for 3 h. Cells were harvested and resuspended in NET2 lysis buffer (50 mM Tris HCl [pH 7.4], 150 mM NaCl, 0.05% NP-40, 1 mM PMSF, protease inhibitors, and 5 U/ml RNasin [Promega]). Five hundred micrograms or 1 mg of total protein was immunoprecipitated with mouse anti-NPM (Zymed) or nonimmune mouse serum overnight at 4°C. The beads were washed in NET2 lysis buffer and incubated in 100 μl proteinase K buffer (7 mM Tris-HCl [pH 7.5], 0.7 mM EDTA, 20 mM NaCl, 0.7% SDS, 5 U/ml RNasin, and 30 μg/ml proteinase K) at 37°C for 30 min. Immunoprecipitated RNAs were isolated using RNA-solv (Omega Bio-tek), separated on 1% agarose-formaldehyde gels, transferred onto a Hybond XL membrane (GE Healthcare), sprayed with En3Hance (Perkin-Elmer), and subjected to autoradiography.
Western blot analysis and immunoprecipitations.
Cells were lysed by sonication in EBC buffer (25 mM Tris-HCl [pH 8], 150 mM NaCl, 1 mM EDTA, and 0.1% NP-40 with PMSF and protease inhibitors) at the indicated times, or cells were fractionated into the cytoplasm and nuclei as described previously (51). Lysates used for immunoprecipitation were incubated with NPM, L7, DHX9, or S3 antibody, and immunoprecipitates were probed by immunoblotting. NPM, rpL7, DHX9, rpS3, lamin A/C, SOD, and GFP-tagged and His-tagged proteins were visualized by direct immunoblotting with NPM (Zymed), rpL7, DHX9 (Bethyl Laboratories), rpS3 (Cell Signaling) lamin A/C (Santa Cruz), SOD (Santa Cruz), GFP (Santa Cruz), and His (Santa Cruz) antibodies, respectively. Blots were also probed with γ-tubulin antibody (Santa Cruz) as indicated.
Cell growth curves.
HeLa cells (2 × 106 cells) were transduced with the indicated constructs, or TKO MEFs (5 × 105 MEFs) were infected with the indicated retroviral constructs for 48 h, and 2 × 104 cells/condition were plated in triplicate in 35-mm dishes. Every 24 h thereafter, cells were harvested and counted.
BrdU assay.
HeLa cells (2 × 106 cells) transduced with the indicated plasmids or TKO MEFs infected with the indicated retrovirus for 48 h were plated onto glass coverslips at 7.5 × 104 cells/3 ml culture medium, and 24 h later, they were labeled with 10 mM bromodeoxyuridine (BrdU) for 2 h to label cells in S phase. Cells were fixed, stained, and counted for BrdU and NPM constructs (His for HeLa and GFP for TKO) as previously described (4). At least 100 cells per condition were counted in triplicate using a Nikon epifluorescent compound microscope (magnification, ×40).
Apoptosis assay.
TKO MEFs were infected with pBabe (control), pBabe-GFP-NPM, or pBabe-GFP-NPMdL retrovirus for 48 h prior to harvesting. Cells were stained with annexin V and propidium iodide using Vybrant apoptosis assay kit 3 (Molecular Probes) according to the manufacturer's specifications. Cells were analyzed by flow cytometry using a Becton Dickinson FACSCalibur cell sorter with CELLQuest Pro (v5.2) analytical software.
Cell volume.
HeLa cells were transduced with the indicated plasmids for 24 h or siRNAs for 72 h, or TKO MEFs were infected with the indicated retrovirus for 48 h. Cells were harvested, and the cellular volume was calculated based on the average diameter of the cells measured (at least 1,200 cells per condition) with a Coulter Vi-Cell apparatus.
[35S]methionine labeling of newly synthesized proteins.
HeLa cells were transduced with the indicated plasmids for 24 h or siRNAs for 72 h, or TKO MEFs were infected with the indicated retrovirus for 48 h before labeling with 150 μCi [35S]methionine in 5 ml methionine-free medium for the indicated times. Cells were harvested and counted, and equal numbers of cells were lysed. Total protein was precipitated with TCA, and 35S cpm were measured for each time point.
rRNA [methyl-3H]methionine labeling.
HeLa cells (2 × 106 cells) were transduced with pcDNA3.1-His, His-tagged NPM, or His-NPMdL for 24 h or with control and NPM siRNAs for 72 h. TKO MEFs were infected with the indicated retroviruses for 48 h. Cells were methionine starved for 15 min and labeled with 50 μCi/ml l-[methyl-3H]methionine (GE Healthcare) for 30 min, followed by a 10-fold excess of cold methionine chase for the indicated times. For rRNA export assays, cells were harvested and counted, and equal numbers were fractionated into the cytoplasm and nuclei (51). Total RNA was isolated from the cytosolic and nuclear fractions with Trizol (Invitrogen), fractionated by gel electrophoresis, transferred onto a Hybond NX membrane (GE Healthcare), sprayed with En3Hance (Perkin-Elmer), and subjected to autoradiography. For rRNA processing assays, cells were harvested, total RNA was isolated, and equal cpm were separated on agarose gels and processed as described above.
Ribosome fractionation.
HeLa cells (2 × 106 cells) were transduced with pcDNA3.1-His, His-tagged NPM, or His-NPMdL for 24 h or with siRNA against NPM or control siRNA for 72 h. TKO MEFs were infected with the indicated retroviruses for 48 h. Cells were treated with 50 μg/ml cycloheximide prior to harvesting and counting. Cells were subjected to cytoplasmic (3 × 106 cells/condition) or nuclear (2.5 × 106 cells/condition) ribosome fractionation essentially as described previously (43) using a density gradient system (Teledyne ISCO). For radiolabeling of newly synthesized rRNAs in polysomes, cells were labeled with [methyl-3H]methionine and fractionated as described above. Fractions were collected, RNA was extracted with Trizol, and 3H cpm were measured for each fraction and graphed for the 40S, 60S, 80S, and polysome peaks.
47S pre-rRNA quantitative PCR.
Quantitative PCR for human 47S pre-rRNA transcripts was performed in real time essentially as described previously by Cui and Tseng (8) and modified for mouse sequences for murine 47S pre-rRNA transcripts. Briefly, total RNA was isolated from cells (wild-type, Npmhy/hy, Arf−/−, p53−/−, DKO, and TKO MEFs or HeLa cells) with Trizol (Invitrogen) according to the manufacturer's instructions. First-strand cDNA was prepared using the SuperScript III first-strand synthesis system (Invitrogen) with the following rDNA-specific primers: mouse primer 5′-CGTGGCATGAACACTTGG-3′ or human primer 5′-CGACGTCACCACATCGATCG-3′. Real-time PCR, measuring the copy number of mouse and human 47S pre-rRNA, was performed using iQ Sybr green supermix (Bio-Rad) on an iCycler apparatus (Bio-Rad) at 95°C for 3 min and 45 cycles of 95°C for 45s, 60°C for 45s, and 72°C for 75s, and the plate was read (forward mouse primer 5′CTGACACGCTGTCCTTTCCC-3′ and reverse mouse primer 5′-GTGAGCCGAAATAAGGTGGC-3′ [324-bp product] and human forward primer 5′-GCTCACACGCTGTCCTCTGG-3′ and reverse primer 5′-GAGAACGCCTGACACGCACG-3′ [320-bp product]). Melting curve analysis was from 55°C to 95°C at 0.5°C/read and with a 10-s hold. The experimental data were plotted against a standard curve generated from serial dilutions of a known quantity of subcloned cDNA of the 47S pre-rRNA product to obtain copy numbers.
Immunogold labeling and electron microscopy.
Npm+/+ and Npmhy/hy MEFs were fixed with 4% paraformaldehyde-0.05% glutaraldehyde in phosphate-buffered saline (pH 7.2) at room temperature for 2 h, rinsed three times in phosphate-buffered saline, and embedded in 10% gelatin. After pelleting the cells, the gelatin was solidified on ice and infused in 2 M sucrose-polyvinylpyrrolidone. Ultrathin sections were prepared and immunolabeled as described previously by Slot et al. (42), with the following modifications: 10% goat serum was used in the blocking buffer in place of 1% bovine serum albumin. Immunolabeling was carried out with primary antibodies (rabbit anti-L7 [1:250] and mouse anti-NPM [1:250]) for 1 h, followed by secondary antibodies (12-nm-gold-labeled goat anti-rabbit antibodies and 18-nm-gold-labeled goat anti-mouse antibodies) for 30 min. After washing, sections were stained with uranyl acetate and embedded in methyl cellulose according to a modification of the method described previously by Tokuyasu (46), which was introduced by Griffiths et al. (14). Npm+/+ and Npmhy/hy MEFs were embedded in plastic and processed for electron microscopy as described previously (48). Specimens were viewed on a Zeiss 902 electron microscope, and photographs were recorded with Kodak EM film. At least 10 fields over eight cells were examined and scored for NPM-L7 or NPM-S16 colocalization and nuclear membrane and rough endoplasmic reticulum (ER) morphology.
5S rRNA in situ hybridization.
Sections from embryonic day 10.5 (E10.5) Npm+/+ and Npm−/− embryos were mounted onto glass slides as previously described (15). In situ RNA fluorescent in situ hybridization analysis was performed using a biotin-labeled probe recognizing 5S rRNA (GeneDetect) according to the manufacturer's instructions. Biotin was detected using an in situ hybridization biotin detection kit from DakoCytomation (purple). Nuclear Fast Red (DakoCytomation) counterstaining was used to demarcate nuclei (pink). Whole embryos were visualized at magnifications of ×2 (Npm+/+) and ×4 (Npm−/−) and insets were visualized at a magnification of ×60 with a Nikon CoolSnap ES camera attached to a Nikon Eclipse E800 microscope and processed with Metamorph V6 software.
Densitometry, image, and statistical analysis.
Autoradiograms were scanned using an ImageScanner III apparatus (GE), and densities were determined using ImageQuant V. 2005 (GE). Statistical analyses were performed using a Student's t test.
RESULTS
Endogenous NPM protein complexes contain ribosomal proteins.
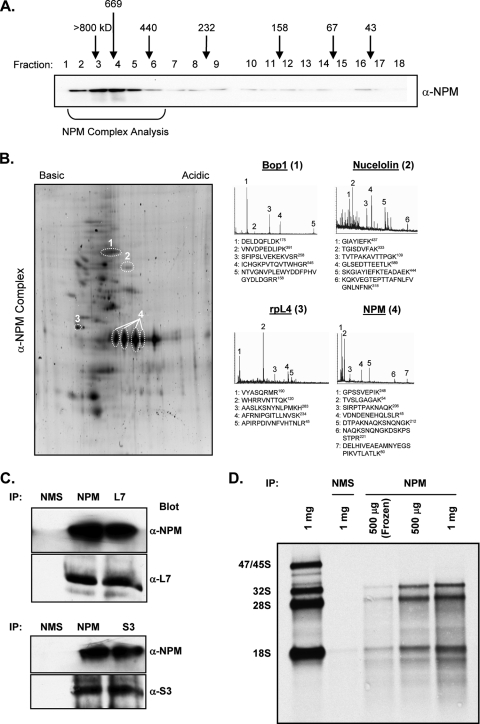
Given NPM's nuclear localization (10) and implied role in ribosome biogenesis (33), we wanted to examine the composition of in vivo NPM protein complexes isolated from HeLa cells. Gel filtration chromatography of HeLa cell lysates revealed numerous distinct endogenous NPM complexes (Fig. 1A). Due to the oligomeric structure of NPM, we chose to focus on protein complexes greater than 370 kDa in size (Fig. 1A). In an attempt to identify the protein components of the NPM complex, we generated an NPM polyclonal antibody affinity column. Pooled NPM complexes eluted from the sizing column were cleared over a control immunoglobulin G column prior to passage over the NPM affinity column and eluted with increasing salt concentrations. Further separation of NPM protein complexes by two-dimensional electrophoresis aided in the positive identification of various NPM-associated proteins via mass spectrometry analysis (Fig. 1B). Among those proteins bound to NPM, a cluster of proteins associated with ribosome biogenesis as well as the nuclear pore complex proteins Nup50, Nup62, and Crm1 were identified (Fig. 1B and Table 1). A list of identified NPM complex proteins is shown in Table 1. Notably, the rpL5, nucleolin, and Brca1 proteins identified from our isolated NPM complexes were previously shown to interact with NPM, demonstrating the effectiveness of our approach in identifying NPM complex components (27, 40, 51). Of the 23 known proteins identified in our NPM complex, 10 constitute integral components of the 40S and 60S ribosomal subunits. Additionally, 9 of the remaining 13 NPM binding proteins are involved in translational regulation. Although we could not determine the exact stoichiometry of these complexes to one another, it was apparent that NPM protein complexes involved in some facet of ribosome biogenesis predominated.
FIG. 1.
Isolation of endogenous NPM protein complexes. (A) HeLa cell lysates were injected onto a Sephacryl 300 gel filtration column, and fractions were collected. Proteins were separated by SDS-polyacrylamide gel electrophoresis and immunoblotted with antibodies recognizing NPM. Gel filtration sizes are indicated above the fractions, and the presence of NPM complexes is indicated below. (B) Pooled sizing column fractions (fractions 2 to 6) containing NPM, precleared over an immunoglobulin G column, were injected onto an NPM polyclonal antibody affinity column and eluted with an increasing NaCl gradient (0.1 to1.0 M). Eluted proteins were separated by two-dimensional electrophoresis and visualized with Sypro ruby dye (2D). Tryptic peptides were analyzed by mass spectrometry. Representative MALDI-TOF spectra of labeled spots from above are presented. (C) HeLa cell lysates were immunoprecipitated (IP) with mouse anti-NPM, anti-rpL7, and anti-rpS3 antibodies or normal mouse serum (NMS) as a control. Immunoprecipitated proteins were subjected to Western blot analysis for NPM, rpL7, and rpS3. (D) Arf−/− MEFs labeled with [methyl-3H]methionine were lysed, and the indicated amount of total protein was immunoprecipitated with mouse anti-NPM antibodies or normal mouse serum. Total RNA was isolated from the immunoprecipitates, separated, transferred onto membranes, and subjected to autoradiography.
TABLE 1.
Components of the NPM protein complexa
| Protein | Function (reference) |
|---|---|
| rpL4 | 60S ribosome subunit |
| rpL5b | Molecular chaperone for 5S rRNA |
| rpL7 | 60S ribosome subunit |
| rpL13a | 60S ribosome subunit |
| rpL22 | 60S ribosome subunit |
| rpL27 | 60S ribosome subunit |
| rpS3a | 40S ribosome subunit |
| rpS5 | 40S ribosome subunit |
| rpS6 | 40S ribosome subunit |
| rpS7 | 40S ribosome subunit |
| Nucleolinb | Represses p53 translation |
| Bopl | Required for rRNA processing (43) |
| CPSF6 | mRNA processing |
| hnRNP Al | mRNA processing and transport |
| hnRNP H2 | mRNA processing |
| EF-2 | Translational regulation |
| DDX1 | RNA helicase |
| DDX5 | RNA helicase |
| DHX9 | RNA helicase |
| Ebpl | Nuclear export factor |
| Nup50 | Nuclear pore complex |
| Nup62 | Nuclear pore complex |
| Crml | Nuclear export receptor |
| Brcalb | E3 ubiquitin ligase for NPM (40) |
Isolated by NPM affinity chromatography and identified by MALDI-TOF or tandem mass spectrometry.
Known NPM binding proteins.
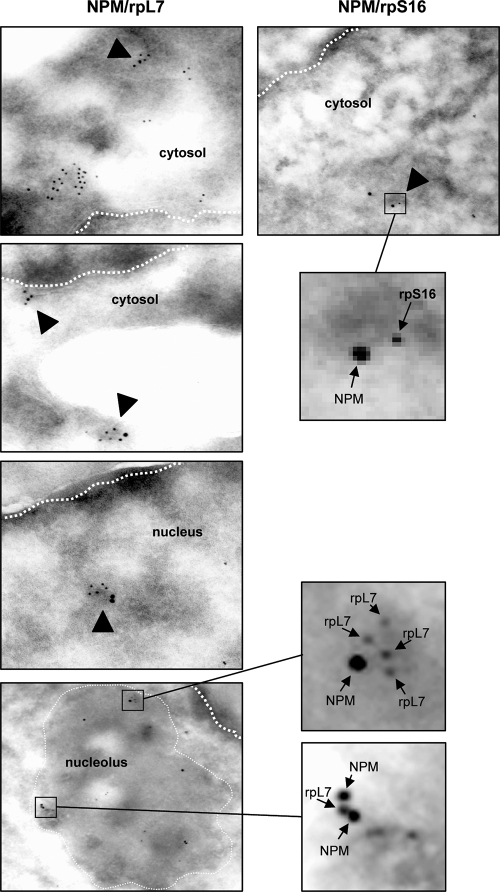
We verified the interactions between NPM and ribosomal subunit proteins with coimmunoprecipitation assays in HeLa cell lysates. Endogenous NPM was immunoprecipitated, and NPM-bound proteins were analyzed by Western blotting. We confirmed NPM interactions with both large (rpL7) and small (rpS3) ribosomal subunit proteins in our pull-down assay, and these two proteins pulled down NPM in reciprocal immunoprecipitations (Fig. 1C). In addition, we were able to show that the immunoprecipitation of endogenous NPM from whole-cell lysates of [methyl-3H]methionine-labeled Arf−/− MEFs was able to pull down newly synthesized and processed rRNAs (Fig. 1D). Finally, we performed electron microscopy on wild-type MEFs with dual immunogold labeling of NPM (oversized gold) and large (rpL7) and small (rpS16) subunit proteins (undersized gold). NPM colocalized with both rpL7 and rpS16 in the cytosol of wild-type MEFs in over 50% of the fields examined (Fig. 2). Further colocalization was found between NPM and rpL7 in the nucleoplasm as well as in nucleoli in over 50% of the fields examined (Fig. 2). Taken together, the data indicate that NPM exists in complex with both large and small ribosomal subunits throughout the cell.
FIG. 2.
Endogenous NPM localizes with large and small ribosomal subunit proteins. Wild-type MEFs were fixed, sectioned, and stained with mouse anti-NPM and rabbit anti-rpL7 (left) or rabbit anti-rpS16 (right) and with 18-mm gold anti-mouse and 12-mm gold anti-rabbit secondary antibodies prior to visualization by electron microscopy (magnification, ×20,000). Arrows indicate colocalization of NPM and ribosomal proteins in the cytosol, nucleus, and nucleolus. Thick dashed lines mark the nuclear membrane, and thin dashed lines demarcate the nucleolus.
NPM nuclear export is required for protein synthesis.
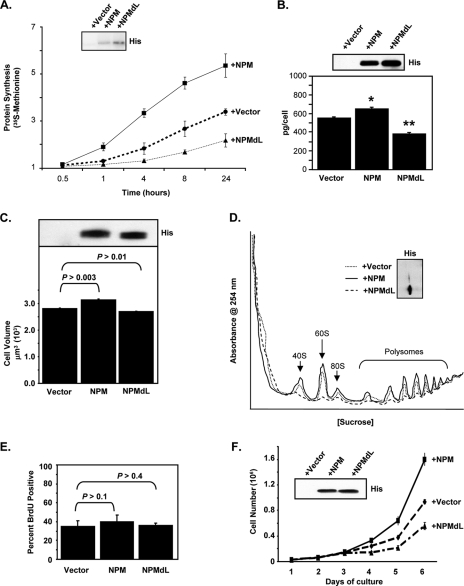
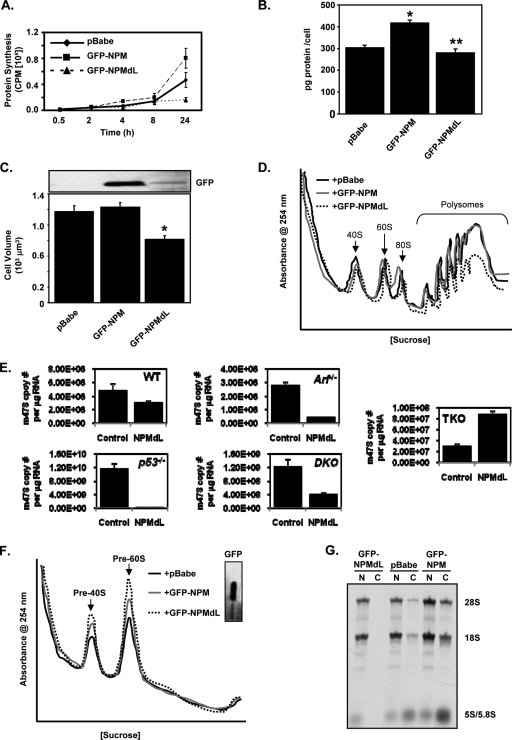
Given that NPM-ribosomal protein complexes exist in cells, we wanted to determine if NPM might play a role in regulating protein synthesis. HeLa cells transduced with empty vector, wild-type NPM, or NPMdL (a double-leucine mutant that is unable to be exported from the nucleus) (51) for 24 h were labeled with [35S]methionine for the indicated times, and total TCA-precipitable proteins were counted for label incorporation. The addition of the shuttling-defective mutant NPMdL significantly attenuated the amount of total protein synthesis. Conversely, a slight overexpression of wild-type NPM (Fig. 3A, inset) resulted in a significant increase in the amount of protein synthesis. In agreement with the 35S labeling study, the amount of protein per cell was also dramatically increased by NPM overexpression. The shuttling-defective NPMdL mutant significantly attenuated the amount of protein produced on a per-cell basis (Fig. 3B). This also correlated with alterations in cellular volume. As shown in Fig. 3C, the volume of cells increased with NPM overexpression, and the loss of NPM shuttling significantly reduced cellular volume.
FIG. 3.
NPM nuclear export is required for new protein production and polysome formation. HeLa cells were transduced with pcDNA3.1-His (dotted line), His-NPM (solid line), or His-NPMdL (dashed line) and incubated for 24 h. (A) Cells were labeled with [35S]methionine for the indicated times. Equal numbers of cells (3 × 106 cells) were lysed, and TCA-precipitated counts were measured. His-tagged NPM protein expression was measured by anti-His immunoblot (inset). (B) Cells were harvested, counted, and lysed, and total protein was measured by a Bradford assay (3a). The results are the averages ± standard deviations (SD) for three independent experiments performed in triplicate (*, P > 0.0001 for vector versus NPM; **, P > 0.0001 for vector versus NPMdL). His-tagged NPM protein expression was measured by anti-His immunoblot (inset). (C) Cells were harvested, and live-cell diameter was measured using a Coulter Vi-Cell apparatus. Results are the averages ± SD for at least 1,200 cells/condition from three independent experiments. P values are indicated. His-tagged NPM protein expression was measured by anti-His immunoblot (inset). (D) Cytosolic extracts from equal numbers of transduced cells (3 × 106 cells) were separated on sucrose gradients with constant UV (254 nm) monitoring. His-tagged NPM protein expression was measured by anti-His immunoblot (inset). (E) Cells were labeled with BrdU for 2 h, fixed, and subjected to immunofluorescence for BrdU and His. Results are the averages of data from three independent experiments performed in triplicate (100 cells/condition counted) ± SD. P values are indicated. (F) Cells (2 × 104) were plated in triplicate for each of the indicated time points. Cells were harvested and counted every 24 h after transduction. His-tagged NPM protein expression levels were measured by anti-His immunoblotting (inset). Results are averages ± SD.
We next examined NPM's ability to influence cytosolic ribosome profiles. Cytosolic ribosomes were isolated via sucrose gradient centrifugation from equal numbers of HeLa cells transduced with empty vector, wild-type NPM, or NPMdL for 24 h. Continuous evaluation of the gradient by UV absorbance for the presence of rRNAs indicated that in the absence of NPM shuttling (NPMdL), the formation of cytosolic 40S and 60S ribosome subunits was dramatically impaired (Fig. 3D, dashed line), while a mild overexpression of NPM (Fig. 3D, inset) resulted in the enhanced formation of cytosolic 40S and 60S subunits (Fig. 3D, solid line). Notably, NPM overexpression amplified (∼30%) the amount of cytosolic ribosomes engaged in active mRNA translation (polysomes in Fig. 3D, solid line) in a mechanism that was dependent on the nuclear export activities of NPM molecules (Fig. 3D, dashed line).
This was not a result of a simple acceleration of cell cycle progression. HeLa cells expressing exogenous NPM or NPMdL proliferated at a rate similar to that of control cells 24 h after expression (Fig. 3E). However, over a prolonged growth curve, the overexpression of wild-type NPM resulted in an increase in cell number, while the overexpression of the shuttling mutant NPMdL resulted in a decrease in cell number (Fig. 3F). These effects on cell proliferation were not observed until 72 h posttransduction, well after the 24-h time point of our experiments.
NPM is a rate-limiting factor for rRNA nuclear export and cytosolic ribosome accumulation.
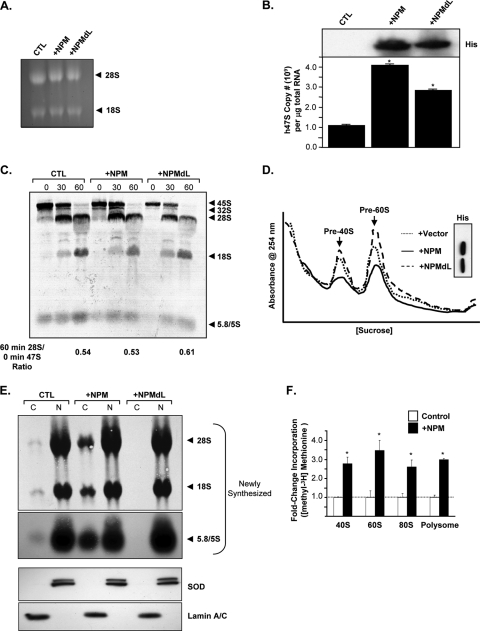
We have presented data that NPM nuclear export is required for the enhanced accumulation of cytosolic ribosomal subunits and polysomes (Fig. 3). To understand the mechanism behind this observed increase in cytosolic ribosomes, we examined the effects of NPM on several steps in the process of ribosome biogenesis. The overexpression of NPM and the shuttling mutant NPMdL in HeLa cells for 24 h did not alter total cellular rRNA compared to vector control levels as shown by ethidium bromide staining (Fig. 4A). However, NPM and NPMdL overexpression increased the amounts of the 47S pre-rRNA transcript produced in HeLa cells four- and twofold, respectively, as measured by quantitative real-time PCR (Fig. 4B). While there appeared to be more 47S pre-rRNA transcription, the expression of wild-type NPM and NPMdL did not affect newly synthesized rRNA processing as measured by [methyl-3H]methionine incorporation (Fig. 4C) (44), demonstrating that even though rDNA transcription is elevated, the processing of the resultant rRNA transcripts was not performed at an increased rate. This also indicates that rRNA processing is limiting and not altered by NPM expression (either the wild type or shuttling mutants).
FIG. 4.
NPM is a rating-limiting factor for rRNA export. HeLa cells were transduced with pcDNA3.1-His (control [CTL]), His-NPM (+NPM), or His-NPMdL (+NPMdL) for 24 h. (A) Cells were harvested, and total RNA extracted from each condition was separated on a 1% agarose gel and stained for total 28S and 18S rRNA with ethidium bromide. (B) Total RNA from cells was analyzed by Sybr green quantitative PCR for 47S pre-rRNA transcript copy number. Results are means ± SD (P > 0.001 versus control). His-tagged NPM protein expression was measured by anti-His immunoblotting (inset). (C) Cells were labeled with [methyl-3H]methionine for 15 min and chased for the indicated times. Total RNA extracted from each sample was separated, transferred onto membranes, and subjected to autoradiography. Newly synthesized and processed rRNAs are indicated by arrows. (D) Nuclear extracts from equal numbers of transduced cells (2.5 × 106 cells) were separated on sucrose gradients with constant UV (254 nm) monitoring. His-tagged NPM protein expression was measured by anti-His immunoblotting (inset). (E) Cells were labeled with [methyl-3H]methionine for 30 min and chased for 2 h. Equal numbers of labeled cells (2 × 106 cells) were subjected to fractionation into cytoplasmic (C) and nuclear (N) extracts. Total RNA extracted from each fraction was separated, transferred onto membranes, and subjected to autoradiography. Newly synthesized and labeled 28S, 18S, and 5.8S/5S rRNAs are indicated by arrows. The bottom 5.8S/5S panel is a longer exposure of the same autoradiograph shown above. Parallel samples were harvested prior to labeling and subjected to fractionation in cytosolic (C) and nuclear (N) extracts. Proteins were subjected to Western blot analysis for lamin A/C (cytosol control) and SOD (nuclear control). (F) Cells were labeled with [methyl-3H]methionine for 30 min and chased for 2 h, and 3 × 106 cells were subjected to polysome fractionation. Fractions from the sucrose gradient were isolated and counted for [methyl-3H]methionine incorporation into newly synthesized rRNA-assembled cytosolic ribosomes. 3H counts were normalized for the efficiency of label incorporation into total RNA for each condition (*, P > 0.005).
Once transcribed and processed, rRNAs are assembled with ribosomal proteins to form the pre-40S and pre-60S ribosomal subunits in the nucleolus/nucleus. A block in the assembly process can be monitored by nuclear ribosome profiles, with a shift of the pre-40S and/or pre-60S peaks indicating a block in proper ribosome assembly (11). We assessed the effects of NPM and the NPMdL shuttling mutant on ribosome assembly by performing nuclear ribosome profiling. As shown in Fig. 4D, the overexpression of NPM or NPMdL did not alter the assembly of the pre-40S and pre-60S ribosomal complexes, as the pre-40S and pre-60S peaks matched the vector control. However, the overexpression of NPMdL (Fig. 4D, dashed line) for 24 h in HeLa cells resulted in a significant increase in pre-40S and pre-60S ribosomal complexes over vector control levels (Fig. 4D, dotted line). In contrast, the overexpression of wild-type NPM (Fig. 4D, solid line) resulted in a significant attenuation in levels of both preribosomal complexes in the nucleus compared to that of the vector control, suggesting that rRNAs were being accumulated in the nucleus of NPMdL-transduced cells and rapidly exported in NPM-expressing cells.
We previously showed that NPM shuttles from the nucleus to the cytoplasm (4, 51) and, thus, next asked if NPM's shuttling activity was required for the export of assembled ribosomal subunits from the nucleus to the cytoplasm. HeLa cells transduced with control vector, wild-type NPM, or the NPMdL shuttling mutant for 24 h were pulsed with [methyl-3H]methionine to label newly synthesized and processed rRNAs (44). Consistent with our earlier findings for protein synthesis and cytoplasmic and nuclear ribosome profiles, NPMdL significantly attenuated (>12-fold) the cytoplasmic accumulation of all rRNAs to nearly undetectable levels (Fig. 4E). In contrast, cells overexpressing wild-type NPM exhibited a significant increase (more than sevenfold) in the cytoplasmic accrual of rRNAs (Fig. 4E). Western blot analysis of lamin A/C (nuclear) and SOD (cytoplasmic) showed no contamination in the fractionation procedure (Fig. 4E). Taken together, these data indicate that rRNA export was dependent on the NPM shuttling activity and could not be attributed to a block in ribosome biogenesis, as transcription, processing, and assembly are not attenuated in NPMdL-expressing cells.
To determine the distribution of newly exported rRNAs into cytosolic ribosomes and polysomes, we pulse-labeled cells with [methyl-3H]methionine and measured the amount of incorporated label in each sucrose gradient fraction containing 40S, 60S, 80S, and polysomes. Importantly, absolute amounts of newly processed rRNAs were identical regardless of the level of NPM expression (Fig. 4C). While NPM overexpression appeared to affect the steady-state levels of polysome production (∼30%) (Fig. 3D), the transduction of cells with wild-type NPM greatly increased (by about threefold) the amounts of newly exported cytosolic ribosomal subunits, including 40S, 60S, 80S, and polysomes available for protein synthesis (Fig. 4F). This underscores the ability of NPM not only to enhance the export of ribosomal subunits but also to provide more ribosomes for polysome-directed protein synthesis.
NPM-mediated changes in polysome production do not rely on p19ARF or p53.
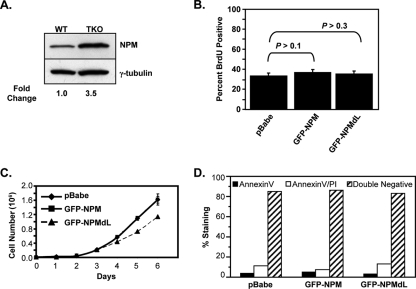
Given the interactions of NPM with p19ARF and p53 (3, 4, 7, 19), we wanted to know if the NPM-induced changes in rRNA export and cytosolic polysome formation that we had observed were influenced by these tumor suppressors. We utilized TKO (Arf−/− p53−/− Mdm2−/−) MEFs deficient in the ARF-Mdm2-p53 axis. Retroviral infection of TKO MEFs with pBabe (control), GFP-tagged NPM, or GFP-tagged NPMdL for 48 h showed results similar to those previously observed in HeLa cells (Fig. 3 and 4). TKO MEFs overexpressing GFP-NPM or GFP-NPMdL (Fig. 5C, inset) for 48 h were labeled with [35S]methionine over 24 h, and TCA-precipitable counts were measured at the indicated times. As shown in Fig. 5A, NPM overexpression increased protein synthesis rates in TKO MEFs, while the inhibition of NPM shuttling with the NPMdL mutant reduced protein output. In addition, TKO MEFs overexpressing NPM for 48 h appreciably increased their total cellular protein levels, while the overexpression of the NPM shuttling mutant NPMdL reduced them (Fig. 5B). Finally, while TKO MEFs overexpressing NPM for 48 h did not greatly increase cell size, the loss of NPM shuttling activity through the overexpression of NPMdL significantly reduced the total cellular volume.
FIG. 5.
NPM-mediated rRNA export occurs in the absence of Arf and p53. TKO (Arf−/− p53−/− Mdm2−/−) (A to G) and wild-type (WT), Arf−/−, p53−/−, and DKO (Mdm2−/− p53−/−) (E only) MEFs were infected with pBabe, GFP-NPM, or GFP-NPMdL retroviruses for 48 h. (A) Cells (5 × 104) were labeled with [35S]methionine for the indicated times and lysed, and TCA-precipitated counts were measured. Results are averages ± SD. (B) Cells were harvested, counted, and lysed, and total protein was measured by Bradford assay (3a). The results are the averages ± SD for two independent experiments performed in triplicate. (*, P > 0.0001 for pBabe versus NPM; **, P > 0.01 for pBabe versus NPMdL). (C) Cells were harvested, and live-cell diameter was measured using a Coulter Vi-Cell apparatus. Results are the averages ± SD for at least 1,200 cells/condition from three independent experiments (*, P > 0.001 for pBabe versus NPMdL). (D) Equal numbers of cells (3 × 106 cells) transduced with pBabe (dotted line), NPM (solid line), or NPMdL (dashed line) were lysed and separated on sucrose gradients with constant UV (254 nm) monitoring. (E) Total RNA from cells was analyzed by Sybr green quantitative PCR for 47S pre-rRNA transcript copy number. Results are means ± SD. (F) Nuclear extracts from equal numbers of transduced cells (2.5 × 106 cells) were separated on sucrose gradients with constant UV (254 nm) monitoring. GFP-tagged NPM protein expression levels were measured by anti-GFP immunoblotting (inset). (G) Cells were labeled with [methyl-3H]methionine for 30 min and chased for 2 h. Equal numbers of labeled cells (2 × 106 cells) were subjected to fractionation into cytoplasmic (C) and nuclear (N) extracts. Total RNA extracted from each fraction was separated, transferred onto membranes, and subjected to autoradiography. Newly synthesized and labeled 28S, 18S, and 5.8S/5S rRNAs are indicated. GFP-tagged NPM and NPMdL protein expression levels were measured by anti-GFP immunoblotting.
We next examined the effects of the loss of Arf and p53 on NPM-mediated cytosolic polysome formation. Equal numbers of TKO MEFs transduced with empty pBabe, GFP-NPM, or GFP-NPMdL for 48 h were lysed, and cytosolic ribosomes were isolated via sucrose gradient centrifugation. Continuous monitoring of RNA levels in gradients by UV absorbance indicated that in the absence of NPM shuttling (NPMdL), the buildup of cytosolic 40S and 60S ribosome subunits was mildly impaired (Fig. 5D, dashed line), while the overexpression of NPM (Fig. 5C, inset) resulted in a modestly enhanced formation of cytosolic 40S and 60S subunits (Fig. 5D, solid line). Importantly, the overexpression of NPMdL resulted in dramatic decreases in actively translating polysomes (Fig. 5D, dashed line). Conversely, the overexpression of NPM had little effect on actively translating polysome levels (Fig. 5D). This may be due to the high levels of endogenous NPM protein expression already present in TKO MEFs (>3.5-fold versus WT MEFs) (Fig. 6A).
FIG. 6.
NPMdL does not alter cell proliferation or death in the absence of Arf and p53. (A) NPM protein levels were determined by anti-NPM immunoblotting using wild-type (WT) and TKO MEF lysates. NPM levels were normalized to γ-tubulin, and changes are indicated. (B to D) TKO (Arf−/− p53−/− Mdm2−/−) MEFs were infected with pBabe, GFP-NPM, or GFP-NPMdL retroviruses for 48 h. (B) Cells were labeled with BrdU for 2 h, fixed, and subjected to immunofluorescence for BrdU and GFP. Results are the averages of two independent experiments performed in triplicate (100 cells/condition counted) ± SD. P values are indicated. (C) Cells (2 × 104) were plated in triplicate for each of the indicated time points. Cells were harvested and counted every 24 h after transduction. Results are averages ± SD. (D) Cells were harvested and stained with annexin V and propidium iodide (PI) prior to flow cytometric analysis. Apoptotic cells are annexin V positive, dead and/or necrotic cells are annexin V/propidium iodide positive, and living cells are double negative.
Given the effects of the inhibition of NPM shuttling on protein synthesis levels and cytosolic ribosome formation in the absence of the Arf and p53 tumor suppressors, we wanted to examine the effects of NPM on ribosome biogenesis in this setting. The overexpression of both NPM and NPMdL in TKO MEFs for 48 h did not alter total 28S and 18S levels, as determined by ethidium bromide staining (data not shown), similar to our data for HeLa cells, which lack functional ARF and p53 (Fig. 4A). We next wanted to determine the effects of NPM shuttling on 47S pre-rRNA transcription. The overexpression of NPM and NPMdL for 48 h in TKO MEFs (Fig. 5E, right, and data not shown) resulted in a significant increase in the levels of the 47S pre-rRNA transcripts similar to that seen in HeLa cells lacking functional ARF and p53 (Fig. 4B). Interestingly, the presence of either p53 or ARF resulted in an inhibition of 47S pre-rRNA transcription in the absence of functional NPM. As shown in Fig. 5E, wild-type, Arf−/−, p53−/−, or DKO (Mdm2−/− p53−/−) MEFs which express either ARF or p53 are able to decrease 47S pre-rRNA transcription when NPM shuttling was inhibited. Taken together, these data indicate that in the absence of functional NPM, either ARF or p53 is required to properly adjust 47S pre-rRNA transcription; cells lacking both ARF and p53 (TKO MEFs and HeLa cells) fail to properly regulate rRNA transcription.
No block in ribosome assembly was detected upon NPM or NPMdL expression in TKO MEFs (Fig. 5F). However, the overexpression of both NPM (gray line) and NPMdL (dotted line) resulted in an increase of the amount of nuclear pre-40S and pre-60S ribosomal complexes over control levels (Fig. 5F, black line), suggesting that in TKO cells, NPM function is critical in ribosome subunit export, but factors other than NPM might also be rate limiting.
Finally, we wanted to examine the effects of NPM shuttling on rRNA export in the absence of ARF and p53. TKO MEFs infected with pBabe, GFP-tagged NPM, and GFP-tagged NPMdL for 48 h (Fig. 5C inset) were labeled with [methyl-3H]methionine and chased for 2 h, and RNA was isolated from nuclear and cytosolic fractions to monitor the export of newly synthesized and processed rRNAs. The overexpression of wild-type NPM resulted in an increase in all forms of rRNA exported into the cytoplasm (more than twofold each for 28S, 18S, and 5S/5.8S) (Fig. 5G), while the inhibition of NPM shuttling with the NPMdL mutant attenuated the export of all four mature rRNAs (>90% inhibition for 28S and 18S and 73% inhibition for 5S/5.8S) (Fig. 5G). These data indicate that in the absence of the ARF and p53 tumor suppressors, NPM can still exert its effects on rRNA export, polysome formation, protein synthesis, and, ultimately, cellular growth.
Notably, the overexpression of NPM or NPMdL in TKO MEFs for 48 h did not alter S-phase progression in these cells, as measured by a 2-h pulse of BrdU incorporation into newly synthesized DNA (Fig. 6B); however, prolonged (>120 h) expression of NPMdL did block cell cycle progression (Fig. 6C) (51), while NPM overexpression had no effect on long-term growth. In addition, the overexpression of NPM and NPMdL for 48 h in TKO MEFs did not increase apoptotic (black bars) or necrotic (white bars) cell death over control levels (Fig. 6D). These data indicate that the effects of NPM and NPMdL on rRNA biogenesis, export, polysome formation, and protein production in the absence of Arf and p53 are not due to alterations in cell cycle progression or cell death.
Loss of NPM blocks rRNA export in vitro and in vivo.
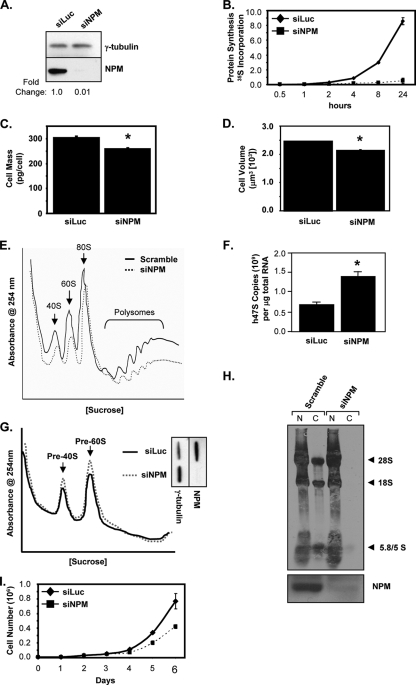
The inhibition of NPM shuttling activity using the dominant negative NPMdL mutant indicated that NPM nuclear export is required for rRNA export to the cytoplasm. However, we cannot rule out the possibility that the NPMdL mutant prevents ribosome export in an indirect and passive manner. Thus, we directly examined the effect of the loss of NPM expression on protein synthesis. As shown in Fig. 7B, the loss of NPM protein expression through the siRNA-mediated knockdown of NPM (99% decrease compared to the siRNAs directed against luciferase control) (Fig. 7A) in HeLa cells for 72 h resulted in a dramatic attenuation of 35S-precipitable counts compared to the control. In addition, the total cellular protein level was significantly reduced with the loss of NPM (Fig. 7C). The knockdown of NPM resulted in a significant decrease in cell size compared to that of control siRNAs (Fig. 7D). We next examined the effects of the loss of NPM on cytosolic ribosome levels. The transduction of HeLa cells with NPM siRNAs significantly decreased the steady-state levels of all cytosolic ribosomal subunits (40S, 60S, 80S, and polysomes) compared to that of the scrambled control (Fig. 7E), providing strong evidence that NPM is an essential component in the nuclear export of ribosomal subunits and the maintenance of cytosolic polysomes.
FIG. 7.
Endogenous NPM is an essential mediator of ribosome nuclear export. HeLa cells were transduced with scrambled or NPM siRNAs and incubated for 72 h. (A) Western blot analysis using antibodies recognizing NPM was used to demonstrate knockdown. Knockdown was normalized to γ-tubulin, and changes are indicated. (B) Cells (5 × 104) were labeled with [35S]methionine for the indicated times and lysed, and TCA-precipitated counts were measured. Results are averages ± SD. (C) Cells were harvested, counted, and lysed, and total protein was measured by Bradford assay (3a). The results are averages ± SD for two independent experiments performed in triplicate. (*, P > 0.003). (D) Cells were harvested, and live-cell diameter was measured using a Coulter Vi-Cell apparatus. Results are averages ± SD for at least 1,200 cells/condition from three independent experiments (*, P > 0.003). (E) Cytosolic extracts from equal numbers of cells (3 × 106 cells) were separated on sucrose gradients with constant UV (254 nm) monitoring. (F) Total RNA from cells was analyzed by Sybr green quantitative PCR for 47S pre-rRNA transcript copy number. Results are averages ± SD (*, P > 0.005). (G) Nuclear extracts from equal numbers of cells (2.5 × 106 cells) were separated on sucrose gradients with constant UV (254 nm) monitoring. (H) Cells were labeled with [methyl-3H]methionine for 30 min and chased for 2 h, and equal numbers of cells (2 × 106 cells) were subjected to fractionation into cytoplasmic (C) and nuclear (N) extracts. Total RNA extracted from each fraction was separated, transferred onto membranes, and subjected to autoradiography. Newly synthesized and labeled 28S, 18S, and 5.8S/5S rRNAs are indicated by arrows. Western blot analysis using antibodies recognizing NPM was used to visualize efficient knockdown (bottom). (I) Cells (2 × 104) were plated in triplicate for each of the indicated time points. Cells were harvested and counted every 24 h after transduction. Results are averages ± SD.
As observed with the NPMdL shuttling mutant, the loss of NPM expression did not alter ribosome biogenesis. The knockdown of NPM with siRNAs in HeLa cells for 72 h resulted in an increase in 47S pre-rRNA transcript levels (>2.5-fold) (Fig. 7F). In addition, siRNA-mediated NPM knockdown did not inhibit ribosome assembly as measured by nuclear ribosome profiling and resulted in the nuclear accumulation of pre-40S and pre-60S ribosome complexes (Fig. 7G) similar to that observed with the NPMdL shuttling mutant in HeLa cells and TKO MEFs (Fig. 4D and 5F, respectively).
To confirm that NPM was required for the nuclear export of rRNAs, we examined the effects of the loss of NPM expression on the export of newly synthesized rRNAs. HeLa cells transduced with scrambled control or NPM-specific siRNAs (51) were pulsed with [methyl-3H]methionine to label newly synthesized and processed rRNAs. The specific knockdown of endogenous NPM was confirmed (Fig. 7H, bottom). The dramatic reduction of NPM protein expression significantly attenuated the cytoplasmic accumulation of all newly processed rRNAs to nearly undetectable levels (Fig. 7H, top). Moreover, cells lacking NPM displayed normal processing of rRNAs (Fig. 7H, lanes 1 and 3). The reduction in protein synthesis, cytosolic ribosome accumulation, and ribosome export were not due to an inhibition in cellular proliferation. As shown in Fig. 7I, HeLa cells were transduced with siRNAs directed against luciferase and NPM for 48 h (day 0) prior to plating for the cell growth assay. The loss of NPM protein expression resulted in a significant reduction in cell growth only past 120 h (day 4) after siRNA transduction. Our experiments were performed at 72 h after siRNA transduction (day 1), when no difference in cell proliferation was observed.
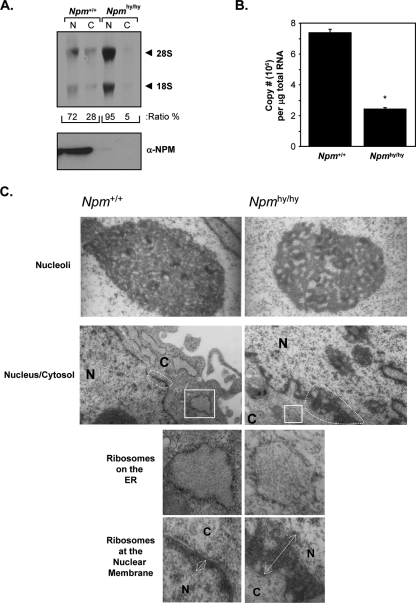
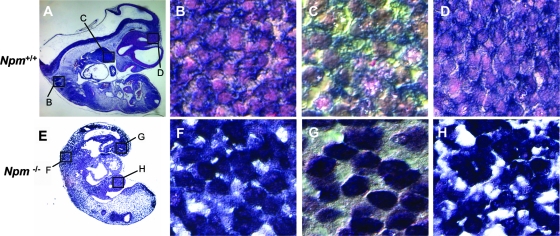
Similarly, MEFs carrying hypomorphic NPM alleles (Npmhy/hy), which express nearly undetectable levels of NPM (15) (Fig. 8A, bottom), showed a striking decrease in the nuclear export of 28S and 18S rRNA compared to that of littermate-matched wild-type MEFs (Fig. 8A). Again, we observed normal processing of rRNAs in the absence of Npm1 (Fig. 8A, lanes 1 and 3). The chronic loss of Npm1 in the presence of ARF and p53 resulted in an inhibition of 47S pre-rRNA transcription (Fig. 8B), as opposed to the acute loss of NPM in the absence of ARF and p53 (seen with siRNA treatment), resulting in increased levels of 47S pre-rRNA transcription (Fig. 7D). However, the 60% decrease in transcription did not result in lower levels of processed rRNA as seen in Fig. 8A, indicating that Npm1 is not absolutely required for rRNA processing but, rather, appears to be necessary for the nuclear export of newly processed rRNAs. These data also suggest that ARF and/or p53 is required to dampen 47S rRNA transcription in the absence of NPM expression given that HeLa and TKO cells are incapable of lowering 47S rRNAs in the absence of NPM (Fig. 4, 5, and 7). These findings are further supported by electron microscopy of Npm+/+ and Npmhy/hy MEFs. As shown in Fig. 8C, the loss of Npm1 resulted in a severe loss of electron density in nucleoli (100% of cells examined), similar to that observed when rDNA transcription is inhibited (35). In addition, Npmhy/hy MEFs exhibited an accumulation of ribosome particles at the nuclear membrane (100% of cells examined) (Fig. 8C, right) and a paucity of rough ER in the cytoplasm (100% of cells examined) (Fig. 8C, bottom), supporting a requirement of NPM in ribosome export.
FIG. 8.
Loss of Npm results in inhibition of rRNA export and 47S pre-rRNA transcription. (A) Npm+/+ and Npmhy/hy MEFs were labeled with [methyl-3H]methionine for 30 min and chased for 2 h, and equal numbers of cells (3 × 106 cells) were subjected to fractionation into cytoplasmic (C) and nuclear (N) extracts. Total RNA extracted from each fraction was separated, transferred onto membranes, and subjected to autoradiography. Newly synthesized and labeled 28S and 18S rRNAs are indicated by arrows. The lack of NPM protein expression in Npmhy/hy cells was monitored via Western blot analysis using antibodies recognizing NPM. (B) Total RNA from Npm+/+ and Npmhy/hy MEFs was analyzed by Sybr green quantitative PCR for 47S pre-rRNA transcript copy number. Results are averages ± SD (*, P > 0.001). (C) Npm+/+ and Npmhy/hy MEFs were fixed, sectioned, and visualized by electron microscopy. Top panels showing nucleoli were taken at a magnification of ×30,000, and bottom panels showing the nucleus (N) and cytosol (C) were taken at a magnification of ×12,000. Bottom panels are enlargements of the nuclear membrane and rough ER from the indicated areas of the panels with a magnification of ×12,000.
Embryos nullizygous for Npm1 fail to develop properly and die in utero between days 11.5 and 16.5 (15). To determine whether a defect in rRNA nuclear export might be visible in these Npm1-null embryos, we examined in vivo 5S rRNA export. Utilizing a biotinylated 5S rRNA probe, we performed RNA in situ hybridization on wild-type and Npm−/− embryos at E10.5. Littermate-matched wild-type embryos readily exported 5S rRNA (purple staining) out of the nucleus (pink staining) and into the cytoplasm in numerous embryonic cell compartments (Fig. 9A to D). In contrast, Npm−/− embryos did not export 5S rRNA in any of the embryonic cells as evidenced by the accumulation (over 10-fold) of dark nuclear 5S rRNA staining (purple) and the lack of cytoplasmic 5S rRNA (Fig. 9E to H). Taken together, these data indicate that NPM is required for the export of rRNAs in vivo.
FIG. 9.
Loss of Npm prevents 5S rRNA export in vivo. In situ RNA fluorescent in situ hybridization analysis was performed on Npm+/+ (A to D) and Npm−/− (E to H) E10.5 embryos using a probe recognizing 5S rRNA (purple). Nuclear fast red counterstaining was used to demarcate nuclei (pink). Whole embryos were visualized at magnifications of ×2 (wild type) and ×4 (Npm−/−). Cell images were observed at a magnification of ×60.
DISCUSSION
The nucleolus is a dynamic subnuclear organelle tasked with producing the core of the protein translation machinery of a cell, the ribosome (30, 49). Historic models of nucleolar function have depicted ribosome biogenesis as more of a passive, reactionary process in the cell, where growth signals are relayed to the nucleolus to increase rRNA synthesis, resulting in higher rates of ribosome production to meet the demands of increased proliferation rates. However, a more current view of the nucleolus as a sensor of both stress and growth has emerged. In challenge of this “static ribosome factory” model, numerous proteins originally thought to reside solely in the nucleolus have recently been shown to continuously shuttle from the nucleolus to various subcellular compartments in a regulated manner, providing evidence that the nucleolus is a dynamic site of numerous cellular events (2, 4, 21, 22, 51, 52). One of these nucleolar proteins, NPM, has been suggested to be involved in a variety of important cellular processes in and out of the nucleolus, including ribosome processing, molecular chaperoning, centrosome duplication, and transcriptional regulation in order to regulate cell growth and proliferation (5, 10, 23, 31-33).
One of the challenges surrounding NPM is its apparent ability to perform functions that are in stark contrast to one another. Specifically, NPM has been reported to interact with all three components of the ARF-p53-Mdm2 pathway to both activate and suppress this cascade (3, 4, 7, 19, 24-26, 45). However, the overarching theme of this pathway is the regulation of growth and proliferation. Taken as single observations, numerous conflicting views of NPM function can be readily made. However, when placed together with our current findings, a more unified outlook on NPM function emerges, as we will discuss below. Our current study offers data that integrate many of these seemingly disconnected functions of NPM. We show that NPM interacts with components of the ribosome, acting as a chaperone to direct the nuclear export of ribosomes. The loss of functional NPM (either through a loss of expression or shuttling-defective mutants) results in a complete loss of ribosome nuclear export and subsequent attenuation in cell growth. Moreover, the transcription of the rDNA locus is severely reduced in the absence of NPM, most likely through an indirect mechanism controlled by ARF and p53. Thus, it is possible that NPM's apparent indirect control of numerous cellular functions stems from its ability to regulate ribosome production and nuclear export using the ARF and p53 tumor suppressors to serve as feedback sensors set to prevent these cell growth processes (e.g., rDNA synthesis, rRNA processing, and centrosome duplication) in the absence of NPM. This is consistent with the notion that the loss of Npm1 is tumorigenic (15). We predict that the loss of Npm1, which induces a p53 response (15), places genetic pressure on the ARF/p53 axis through the cell's ultimate need to produce functional cytosolic ribosomes, eventually selecting for the loss of ARF/p53 pathway components and leading to cellular transformation. In agreement with this notion, the concomitant loss of p53 rescues the growth defects observed in Npm1−/− MEFs (15).
The mechanism of NPM's growth-promoting properties is thought to be mandated through alterations in rRNA processing (41). We have provided evidence that overexpressed NPM does not alter rRNA processing or ribosome assembly, nor is Npm1 obligatory for rRNA processing or ribosome assembly to occur normally. We now show that NPM serves as a nuclear export chaperone for the ribosome, drawing similarities to previous studies that showed an in vitro propensity for NPM to serve as a protein chaperone (32, 45). Even modest increases in NPM protein expression amplified ribosome export, resulting in increased rates of protein synthesis, demonstrating that NPM is rate limiting in its capacity to shuttle ribosomes to the cytosol. Under this model, NPM would serve as an adaptor, analogous to NMD3, bridging the ribosome with CRM1 prior to export through the nuclear pore (47). In fact, we were able to copurify the nuclear pore proteins Nup50 and Nup62 with NPM, demonstrating that these export complexes exist in vivo.
The similarities between NPM and NMD3 are intriguing, but there are enough measurable differences to suggest that they do not perform identical functions and rather, most likely, complement one another in their abilities to act as nuclear export chaperones. Unlike NMD3, we found NPM in complexes with ribosomal proteins and rRNAs from both the small and large ribosome subunits. Additionally, the mutation of the NMD3 nuclear export signal resulted in only an attenuation of 28S rRNA export (47), whereas NPMdL readily inhibits the nuclear export of all newly synthesized 28S, 18S, and 5S rRNAs, suggesting that NPM may reside upstream of NMD3 in regulating the nuclear export of all ribosomal subunits. Moreover, the overexpression of NPM is capable of dramatically increasing ribosome nuclear export. The striking sequestration of the entire ribosome in the nucleus by mutant NPM molecules is underscored further by the rapid depletion of newly assembled polysomes in cells unable to shuttle NPM protein complexes. Thus, we propose that NPM serves as a “growth thermostat” in the nucleolus, ready to respond to changes in the cell that might require more or less ribosome output for protein synthesis. The loss of Npm1 confirms this hypothesis, with Npmhy/hy and Npm−/− cells displaying a severe attenuation of all rRNA export. Moreover, cells displaying hyperactivated mTOR signaling exhibit elevated NPM protein levels and produce dramatic increases in levels of rRNA export and polysome formation, all of which require functional NPM (36).
Given NPM's role in the nucleolar growth-promoting pathway, through ribosome export regulation, it is not surprising that several groups including our own have shown NPM to be a p53-independent target of the nucleolar ARF tumor suppressor (3, 4, 19). In order to maintain proper nucleolar function, ARF should be sensitive to abrupt changes in ribosome output, constantly monitoring the rate of ribosome synthesis and export by NPM. ARF is often referred to as a sensor of hyperproliferative signals, such as those emanating from Myc and Ras (16, 34, 54), but both of these oncoproteins are quite capable of eliciting hypergrowth signals as well. Indeed, recent evidence has demonstrated that oncogenic myc translocates to the nucleolus in order to greatly increase rDNA synthesis (1, 12), and under some settings, ARF can act to antagonize nucleolar myc oncoproteins (13, 38). In order to maintain nucleolar integrity in the face of myc-induced increases in rRNAs, the cell might also require NPM to enhance the rate of ribosome nuclear export. In support of this notion, Npm1 is a direct transcriptional target of myc (17, 29, 50, 53). Accordingly, nucleolar ARF may have evolved to monitor this process and, as such, is also a target of myc induction (54). In cancers, it then becomes advantageous to disrupt the normal regulation of this pathway. Under normal conditions, NPM is devoted to ribosome nuclear export to maintain protein synthesis at a constant rate. We predict that the interruption of this nucleolar homeostasis occurs through two different signals. First, stress signals that alter nucleolar structures (39) and delocalize NPM to the nucleoplasm, where it binds to p53 or Mdm2 (7, 25), would serve to not only activate the p53 pathway but also divert NPM from its normal role in rRNA export. Dramatic decreases in protein synthesis rates are hallmarks of these cellular stress outcomes (37). Second, oncogenic growth signals that upregulate NPM protein expression (36) might also trigger the ARF checkpoint, with accumulated nucleolar ARF proteins preventing the nuclear export of NPM-ribosome complexes (4, 51). However, in the absence of functional ARF (such as in HeLa or TKO cells), NPM function would be unopposed. Thus, endogenous NPM proteins are capable of amplifying the rate of rRNA export, resulting in dramatic increases in polysome formation and protein translation, a hallmark of tumorigenesis.
Acknowledgments
We are grateful to Myla Ashfaq and Mike Benjamin for excellent technical assistance and Alan Diehl, Joe Baldassare, Helen Piwnica-Worms, Sheila Stewart, and other members of the laboratory of J.D.W. for discussion, critical reading of the manuscript, and support.
M.K. is supported by the Cancer Biology Pathway through the Washington University Siteman Cancer Center, and D.Y.A.D. is supported by the BioMed RAP Program. This work was funded by grants from the National Institutes of Health to P.P.P. and J.D.W. and the Susan G. Komen Breast Cancer Race for the Cure and the Pew Scholars Program in Biomedical Sciences to J.D.W. This research was supported by the National Center for Research Resources of the National Institutes of Health grant P41RR000954 and the W. M. Keck Foundation.
Footnotes
Published ahead of print on 22 September 2008.
REFERENCES
- 1.Arabi, A., S. Wu, K. Ridderstrale, H. Bierhoff, C. Shiue, K. Fatyol, S. Fahlen, P. Hydbring, O. Soderberg, I. Grummt, L. G. Larsson, and A. P. Wright. 2005. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 7303-310. [DOI] [PubMed] [Google Scholar]
- 2.Azzam, R., S. L. Chen, W. Shou, A. S. Mah, G. Alexandru, K. Nasmyth, R. S. Annan, S. A. Carr, and R. J. Deshaies. 2004. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science 305516-519. [DOI] [PubMed] [Google Scholar]
- 3.Bertwistle, D., M. Sugimoto, and C. J. Sherr. 2004. Physical and functional interactions of the Arf tumor suppressor protein with nucleophosmin/B23. Mol. Cell. Biol. 24985-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72248-254. [DOI] [PubMed] [Google Scholar]
- 4.Brady, S. N., Y. Yu, L. B. Maggi, Jr., and J. D. Weber. 2004. ARF impedes NPM/B23 shuttling in an Mdm2-sensitive tumor suppressor pathway. Mol. Cell. Biol. 249327-9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, W. Y., Q. R. Liu, J. Borjigin, H. Busch, O. M. Rennert, L. A. Tease, and P. K. Chan. 1989. Characterization of the cDNA encoding human nucleophosmin and studies of its role in normal and abnormal growth. Biochemistry 281033-1039. [DOI] [PubMed] [Google Scholar]
- 6.Colombo, E., P. Bonetti, E. L. Denchi, P. Martinelli, R. Zamponi, J. C. Marine, K. Helin, B. Falini, and P. G. Pelicci. 2005. Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol. Cell. Biol. 258874-8886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Colombo, E., J. C. Marine, D. Danovi, B. Falini, and P. G. Pelicci. 2002. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 4529-533. [DOI] [PubMed] [Google Scholar]
- 8.Cui, C., and H. Tseng. 2004. Estimation of ribosomal RNA transcription rate in situ. BioTechniques 36134-138. [DOI] [PubMed] [Google Scholar]
- 9.Eichler, D. C., and N. Craig. 1994. Processing of eukaryotic ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 49197-239. [DOI] [PubMed] [Google Scholar]
- 10.Feuerstein, N., S. Spiegel, and J. J. Mond. 1988. The nuclear matrix protein, numatrin (B23), is associated with growth factor-induced mitogenesis in Swiss 3T3 fibroblasts and with T lymphocyte proliferation stimulated by lectins and anti-T cell antigen receptor antibody. J. Cell Biol. 1071629-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feunteun, J., R. Monier, C. Vola, and R. Rosset. 1974. Ribosomal assembly defective mutants of Escherichia coli. Nucleic Acids Res. 1149-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grandori, C., N. Gomez-Roman, Z. A. Felton-Edkins, C. Ngouenet, D. A. Galloway, R. N. Eisenman, and R. J. White. 2005. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat. Cell Biol. 7311-318. [DOI] [PubMed] [Google Scholar]
- 13.Gregory, M. A., Y. Qi, and S. R. Hann. 2005. The ARF tumor suppressor: keeping Myc on a leash. Cell Cycle 4249-252. [PubMed] [Google Scholar]
- 14.Griffiths, G., A. McDowall, R. Back, and J. Dubochet. 1984. On the preparation of cryosections for immunocytochemistry. J. Ultrastruct. Res. 8965-78. [DOI] [PubMed] [Google Scholar]
- 15.Grisendi, S., R. Bernardi, M. Rossi, K. Cheng, L. Khandker, K. Manova, and P. P. Pandolfi. 2005. Role of nucleophosmin in embryonic development and tumorigenesis. Nature 437147-153. [DOI] [PubMed] [Google Scholar]
- 16.Groth, A., J. D. Weber, B. M. Willumsen, C. J. Sherr, and M. F. Roussel. 2000. Oncogenic Ras induces p19ARF and growth arrest in mouse embryo fibroblasts lacking p21Cip1 and p27Kip1 without activating cyclin D-dependent kinases. J. Biol. Chem. 27527473-27480. [DOI] [PubMed] [Google Scholar]
- 17.Guo, Q. M., R. L. Malek, S. Kim, C. Chiao, M. He, M. Ruffy, K. Sanka, N. H. Lee, C. V. Dang, and E. T. Liu. 2000. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 605922-5928. [PubMed] [Google Scholar]
- 18.Ho, J. H.-N., and A. W. Johnson. 1999. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol. Cell. Biol. 192389-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Itahana, K., K. P. Bhat, A. Jin, Y. Itahana, D. Hawke, R. Kobayashi, and Y. Zhang. 2003. Tumor suppressor ARF degrades B23, a nucleolar protein involved in ribosome biogenesis and cell proliferation. Mol. Cell 121151-1164. [DOI] [PubMed] [Google Scholar]
- 20.Johnson, A. W., E. Lund, and J. Dahlberg. 2002. Nuclear export of ribosomal subunits. Trends Biochem. Sci. 27580-585. [DOI] [PubMed] [Google Scholar]
- 21.Juan, G., and C. Cordon-Cardo. 2001. Intranuclear compartmentalization of cyclin E during the cell cycle: disruption of the nucleoplasm-nucleolar shuttling of cyclin E in bladder cancer. Cancer Res. 611220-1226. [PubMed] [Google Scholar]
- 22.Keough, R. A., E. M. Macmillan, J. K. Lutwyche, J. M. Gardner, F. J. Tavner, D. A. Jans, B. R. Henderson, and T. J. Gonda. 2003. Myb-binding protein 1a is a nucleocytoplasmic shuttling protein that utilizes CRM1-dependent and independent nuclear export pathways. Exp. Cell Res. 289108-123. [DOI] [PubMed] [Google Scholar]
- 23.Kondo, T., N. Minamino, T. Nagamura-Inoue, M. Matsumoto, T. Taniguchi, and N. Tanaka. 1997. Identification and characterization of nucleophosmin/B23/numatrin which binds the anti-oncogenic transcription factor IRF-1 and manifests oncogenic activity. Oncogene 151275-1281. [DOI] [PubMed] [Google Scholar]
- 24.Kuo, M. L., W. den Besten, D. Bertwistle, M. F. Roussel, and C. J. Sherr. 2004. N-terminal polyubiquitination and degradation of the Arf tumor suppressor. Genes Dev. 181862-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurki, S., K. Peltonen, L. Latonen, T. M. Kiviharju, P. M. Ojala, D. Meek, and M. Laiho. 2004. Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell 5465-475. [DOI] [PubMed] [Google Scholar]
- 26.Li, J., X. Zhang, D. P. Sejas, G. C. Bagby, and Q. Pang. 2004. Hypoxia-induced nucleophosmin protects cell death through inhibition of p53. J. Biol. Chem. 27941275-41279. [DOI] [PubMed] [Google Scholar]
- 27.Liu, H. T., and B. Y. Yung. 1999. In vivo interaction of nucleophosmin/B23 and protein C23 during cell cycle progression in HeLa cells. Cancer Lett. 14445-54. [DOI] [PubMed] [Google Scholar]
- 28.Maggi, L. B., Jr., and J. D. Weber. 2005. Nucleolar adaptation in human cancer. Cancer Investig. 23599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Menssen, A., and H. Hermeking. 2002. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc. Natl. Acad. Sci. USA 996274-6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moss, T., and V. Y. Stefanovsky. 2002. At the center of eukaryotic life. Cell 109545-548. [DOI] [PubMed] [Google Scholar]
- 31.Okuda, M., H. F. Horn, P. Tarapore, Y. Tokuyama, A. G. Smulian, P. K. Chan, E. S. Knudsen, I. A. Hofmann, J. D. Snyder, K. E. Bove, and K. Fukasawa. 2000. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell 103127-140. [DOI] [PubMed] [Google Scholar]
- 32.Okuwaki, M., K. Matsumoto, M. Tsujimoto, and K. Nagata. 2001. Function of nucleophosmin/B23, a nucleolar acidic protein, as a histone chaperone. FEBS Lett. 506272-276. [DOI] [PubMed] [Google Scholar]
- 33.Okuwaki, M., M. Tsujimoto, and K. Nagata. 2002. The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol. Biol. Cell 132016-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmero, I., C. Pantoja, and M. Serrano. 1998. p19ARF links the tumour suppressor p53 to Ras. Nature 395125-126. [DOI] [PubMed] [Google Scholar]
- 35.Panse, S. L., C. Masson, L. Heliot, J. M. Chassery, H. R. Junera, and D. Hernandez-Verdun. 1999. 3-D organization of ribosomal transcription units after DRB inhibition of RNA polymerase II transcription. J. Cell Sci. 1122145-2154. [DOI] [PubMed] [Google Scholar]
- 36.Pelletier, C. L., L. B. Maggi, Jr., S. N. Brady, D. K. Scheidenhelm, D. H. Gutmann, and J. D. Weber. 2007. TSC1 sets the rate of ribosome export and protein synthesis through nucleophosmin translation. Cancer Res. 671609-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pyronnet, S., and N. Sonenberg. 2001. Cell-cycle-dependent translational control. Curr. Opin. Genet. Dev. 1113-18. [DOI] [PubMed] [Google Scholar]
- 38.Qi, Y., M. A. Gregory, Z. Li, J. P. Brousal, K. West, and S. R. Hann. 2004. p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature 431712-717. [DOI] [PubMed] [Google Scholar]
- 39.Rubbi, C. P., and J. Milner. 2003. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 226068-6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato, K., R. Hayami, W. Wu, T. Nishikawa, H. Nishikawa, Y. Okuda, H. Ogata, M. Fukuda, and T. Ohta. 2004. Nucleophosmin/B23 is a candidate substrate for the BRCA1-BARD1 ubiquitin ligase. J. Biol. Chem. 27930919-30922. [DOI] [PubMed] [Google Scholar]
- 41.Savkur, R. S., and M. O. Olson. 1998. Preferential cleavage in pre-ribosomal RNA byprotein B23 endoribonuclease. Nucleic Acids Res. 264508-4515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slot, J. W., H. J. Geuze, S. Gigengack, G. E. Lienhard, and D. E. James. 1991. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J. Cell Biol. 113123-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strezoska, Z., D. G. Pestov, and L. F. Lau. 2000. Bop1 is a mouse WD40 repeat nucleolar protein involved in 28S and 5.8S rRNA processing and 60S ribosome biogenesis. Mol. Cell. Biol. 205516-5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugimoto, M., M. L. Kuo, M. F. Roussel, and C. J. Sherr. 2003. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol. Cell 11415-424. [DOI] [PubMed] [Google Scholar]
- 45.Szebeni, A., and M. O. Olson. 1999. Nucleolar protein B23 has molecular chaperone activities. Protein Sci. 8905-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tokuyasu, K. T. 1980. Immunochemistry on ultrathin frozen sections. Histochem. J. 12381-403. [DOI] [PubMed] [Google Scholar]
- 47.Trotta, C. R., E. Lund, L. Kahan, A. W. Johnson, and J. E. Dahlberg. 2003. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 222841-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wainszelbaum, M. J., B. M. Proctor, S. E. Pontow, P. D. Stahl, and M. A. Barbieri. 2006. IL4/PGE2 induction of an enlarged early endosomal compartment in mouse macrophages is Rab5-dependent. Exp. Cell Res. 3122238-2251. [DOI] [PubMed] [Google Scholar]
- 49.Warner, J. R. 1990. The nucleolus and ribosome formation. Curr. Opin. Cell Biol. 2521-527. [DOI] [PubMed] [Google Scholar]
- 50.Watson, J. D., S. K. Oster, M. Shago, F. Khosravi, and L. Z. Penn. 2002. Identifying genes regulated in a Myc-dependent manner. J. Biol. Chem. 27736921-36930. [DOI] [PubMed] [Google Scholar]
- 51.Yu, Y., L. B. Maggi, Jr., S. N. Brady, A. J. Apicelli, M. S. Dai, H. Lu, and J. D. Weber. 2006. Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol. Cell. Biol. 263798-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yung, B. Y., and P. K. Chan. 1987. Identification and characterization of a hexameric form of nucleolar phosphoprotein B23. Biochim. Biophys. Acta 92574-82. [DOI] [PubMed] [Google Scholar]
- 53.Zeller, K. I., T. J. Haggerty, J. F. Barrett, Q. Guo, D. R. Wonsey, and C. V. Dang. 2001. Characterization of nucleophosmin (B23) as a Myc target by scanning chromatin immunoprecipitation. J. Biol. Chem. 27648285-48291. [DOI] [PubMed] [Google Scholar]
- 54.Zindy, F., C. M. Eischen, D. H. Randle, T. Kamijo, J. L. Cleveland, C. J. Sherr, and M. F. Roussel. 1998. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 122424-2433. [DOI] [PMC free article] [PubMed] [Google Scholar]