Abstract
Exchange proteins activated by cAMP (cyclic AMP) 2 (Epac2) is a guanine nucleotide exchange factor for Rap1, a small G protein involved in many cellular functions, including cell adhesion, differentiation, and exocytosis. Epac2 interacts with Ras-GTP via a Ras association (RA) domain. Previous studies have suggested that the RA domain was dispensable for Epac2 function. Here we show for the first time that Ras and cAMP regulate Epac2 function in a parallel fashion and the Ras-Epac2 interaction is required for the cAMP-dependent activation of endogenous Rap1 by Epac2. The mechanism for this requirement is not allosteric activation of Epac2 by Ras but the compartmentalization of Epac2 on the Ras-containing membranes. A computational modeling is consistent with this compartmentalization being a function of both the level of Ras activation and the affinity between Ras and Epac2. In PC12 cells, a well-established model for sympathetic neurons, the Epac2 signaling is coupled to activation of mitogen-activated protein kinases and contributes to neurite outgrowth. Taken together, the evidence shows that Epac2 is not only a cAMP sensor but also a bona fide Ras effector. Coincident detection of both cAMP and Ras signals is essential for Epac2 to activate Rap1 in a temporally and spatially controlled manner.
Rap1 is a small GTPase involved in the regulation of multiple cellular functions such as adhesion, differentiation, and exocytosis. Like all small G proteins, Rap1 cycles between a GTP-loaded active state and a GDP-loaded inactive state, which is mediated via the opposing actions of G protein activation proteins that promote hydrolysis of bound GTP to GDP and guanine nucleotide exchange factors (GEFs) that catalyze the exchange of bound GDP for GTP.
Exchange proteins activated by cAMP (Epac1 and Epac2) are unique Rap1 GEFs that link cAMP elevation to Rap1 activation (5). This is achieved via the direct binding of cAMP to the Epac protein itself, thereby defining a novel cAMP signaling pathway that is independent of protein kinase A (PKA). Both Epac1 and Epac2 contain a catalytic region and a regulatory region. The catalytic region consists of a CDC25 homology domain that catalyzes Rap1 activation, a REM domain, and a Ras association (RA) domain that lies in between. The regulatory region consists of one or two cAMP binding domains (cNBDs) and a DEP (disheveled, Egl-10, and pleckstrin homology) domain. Under resting conditions, Epac proteins are inactive due to the inhibitory interaction between the regulatory and catalytic regions. The binding of cAMP to the cNBD relieves the intramolecular inhibition by exposing the catalytic site to Rap1 (36-39). Previous studies have been largely focused on the mechanism of cAMP regulation of Epacs. Whether other molecular interactions play a role in Epac regulation is unknown.
A number of features of Epac2 distinguish it from Epac1, which suggests distinctive regulatory mechanisms for Epac2. While Epac1 is expressed ubiquitously, Epac2 is highly enriched in neuronal tissues (22). Moreover, Epac1 is localized to the perinuclear mitochondria through a specific N-terminal sequence (34) or interaction with specific AKAPs (6), while Epac2 is largely cytosolic. Importantly, although both proteins contain potential RA domains, only Epac2 interacts with Ras-GTP (26). In these studies, Quilliam and colleagues have demonstrated the ability of Ras-GTP to recruit Epac2 to the plasma membrane (26). However, the necessity of the RA domain for Epac2-mediated Rap1 activation has not been firmly established. Our study shows that Ras-Epac2 interaction is required for Epac2 activity, suggesting that Epac2 is a bona fide Ras effector, as well as a cAMP sensor, thereby acting as a coincidence detector for signals emanating from Ras and cAMP.
Several models could explain the mechanism for the regulation of Epac2 by Ras-GTP. First, Ras-GTP could facilitate the cAMP-mediated relief of autoinhibition. Second, Ras-GTP could enhance the enzymatic activity of Epac2 by inducing allosteric changes within the catalytic domain through its interaction with the RA domain. This model is analogous to the model of allosteric activation of Son of sevenless (SOS) by Ras-GTP (28). A third model considers the compartmentalization of Epac2. As both Ras and Rap1 are lipid modified at their carboxy termini and tethered to the lipid membrane, the interaction between Ras and Epac2 may greatly increase the Epac2 concentration at the membrane, therefore accelerating the rate of Rap1 activation. We examined each of the three models and demonstrate here that Ras regulates Epac2 function independently of cAMP and that enrichment of Epac2 on the membrane through Ras binding is crucial for Epac2-mediated Rap1 activation.
It is possible that this Ras-dependent mode of Rap1 activation could favor the activation of selective targets of Rap1. As we have shown recently, relocation of Epac1 from the perinuclear region to the plasma membrane allows Rap1 activation to be coupled to the phosphorylation of extracellular signal-regulated kinases (ERKs) (51). While this relocation of Epac1 was artificially achieved, we hypothesized that the Ras-dependent membrane recruitment of Epac2 could also activate Rap1 on Ras-containing membranes to trigger ERK activation. Our results show that Epac2 potentiates ERK activation induced by Ras activation and cAMP elevation and this pathway contributes to neurite outgrowth in PC12 cells as a model of sympathetic neurons.
MATERIALS AND METHODS
Reagents.
The following antibodies were used for Western blotting or immunoprecipitation. Anti-Rap1 A/B and unconjugated and agarose-coupled anti-Flag (M2) antibodies were from Sigma-Aldrich (St. Louis, MO). Anti-phospho-ERK (threonine 202 and tyrosine 204) was from Cell Signaling Technology (Beverly, MA). Anti-Epac2 (rabbit) and green fluorescent protein (GFP) antiserum (rabbit) were from Abcam (Cambridge, MA). Anti-Ras (RAS10) antibody was from Upstate Biotechnology. Antihemagglutinin (anti-HA) antibody was from Covance (Princeton, NJ). Anti-ERK2, anti-H-Ras, and anti-glutathione S-transferase (anti-GST) antibodies were from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). Secondary antibodies against mouse and rabbit immunoglobulins G were from GE Healthcare. Forskolin, 3-isobutyl-1-methylxanthine (IBMX), 8-Br-cAMP, and H89 were from Calbiochem (Riverside, CA). Epidermal growth factor (EGF), isoproterenol (ISO), GTP, GTPγS, glutathione peptide, and glutathione-agarose beads were from Sigma-Aldrich (St. Louis, MO). Nerve growth factor (NGF) was from Axxora (San Diego, CA). 3′-(N-Methylanthraniloyl)-2′-deoxyguanosine-5′-diphosphate, triethylammonium salt (mant-dGDP), was from Jena Bioscience (Jena, Germany).
Plasmids.
Mouse Epac2 was subcloned downstream of GFP in the pEGFP-C1 vector (Clontech, Mountain View, CA) between a PstI site and a blunted NotI/SmaI fusion site or downstream of Flag in the pcDNA3 vector (Invitrogen) between the EcoRI and NotI sites. The K684E and R667E mutations were introduced into Flag-Epac2 with a QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA) according to the manufacturer's instructions. Epac2-684E was also subcloned downstream of GFP. GFP-Epac2Δ430 and GFP-Epac2Δ430-684E were generated by deletion of the sequences corresponding to the 430 amino acids at the amino terminus from the parent plasmids by using the available EcoRI and SalI sites, filling in with T4 polymerase, and self-ligation. Human H-RasV12 and H-RasN17 were subcloned into the mCherry-C1 vector (Clontech) by using the available EcoRI and ApaI sites. For the mCherry-RasV12-SAAX construct, the C186S mutation was introduced by PCR through the antisense primer and then the mutant was subcloned back into the mCherry-C1 vector by using the available EcoRI and XbaI sites. The Flag-Epac2-CAAX and Flag-Epac2-684E-CAAX mutant forms were created by inserting annealed double-stranded oligonucleotides corresponding to the carboxy-terminal 24 amino acids of human H-Ras containing the H-CAAX motif (amino acids 166 to 189 of H-Ras) into Flag-Epac2 and Flag-Epac2 684E, respectively. HA-SOScat, comprising the catalytic domain of the Ras exchanger SOS, was a gift from Dafna Bar-Sagi, NYU School of Medicine. HA-Epac2 was a gift from Lawrence Quilliam, Indiana University. Epac1 was a gift from Johannes Bos, Utrecht University.
Cell culture conditions and treatments.
COS and HEK293 cells were cultured in Dulbecco modified Eagle medium plus 10% fetal calf serum, penicillin-streptomycin, and l-glutamine at 37°C and 5% CO2. PC12 cells were kindly provided by Pat Casey (Duke University) and cultured in Dulbecco modified Eagle medium with 10% horse serum and 5% fetal calf serum plus penicillin, streptomycin, and l-glutamine at 37°C and 5% CO2. To generate stable cell populations, HEK293 cells were transfected with GFP-Epac2 and selected with 0.5 mg/ml G418 (Invitrogen, Carlsbad, CA) for 4 weeks. Cells were starved in serum-free medium for 16 h prior to treatments. Cells were treated with 5 μM forskolin and 50 μM IBMX for 15 min unless otherwise indicated. H89 was used at 10 μM and was added 15 min prior to forskolin treatment. EGF and NGF were used at 10 and 50 ng/ml, respectively, unless otherwise indicated. 8-Br-cAMP was used at 100 μM for the times indicated. Transient transfections were performed with Lipofectamine 2000 according to the manufacturer's instructions. The control vector, pcDNA3, pEGFP-C1, or mCherry-C1, was included in each set of transfections to ensure that each plate received the same amount of transfected DNA.
RNA interference.
The RNA interference target within Epac2 was identified and validated by testing three double-stranded RNA oligoribonucleotides from Ambion Inc. (Austin, TX). The most effective target site, which corresponds to positions 1144 to 1162 of mouse Epac2 (NM_019688) and positions 1238 to 1256 of rat Epac2 (XM_001060956), was chosen. The sequences used for Rap1a efficiently eliminate rat Rap1a, the major isoform of Rap1 in PC12 cells (15). The Epac2, Rap1a, and scrambled pairs of oligonucleotides were synthesized by Integrated DNA Technologies Inc. (Coralville, IA), annealed, and cloned into the pTER vector (49) by using the available BglII and HindIII sites. The sequences of the oligonucleotides used for Epac2, Rap1a, and scrambled short hairpin RNA (shRNA) were as follows: for Epac2, 5′-ATTATTAGATCTGGATCCGTGAATGTAGTCATTCAAGAGATGACTACATTCACGGATCCTTTTTGGAAAAAGCTTATTATT-3′; for Rap1a, 5′-ATTATTAGATCTCAGAATTTAGCAAGACAGTGGTGTTCAAGAGACACCACTGTCTTGCTAAAT TCTGTTTTTGGAAAAAGCTTATTATT-3′; for scrambled shRNA, 5′-AATAATAAGCTTTTTCCAAAAAGCGCGCTTTGTAGGATTCGTCTCTTGAACGAATCCTACAAAGCGCGCAGATCTAATAAT-3′.
Western blotting, immunoprecipitation, and GST pull-down assay.
Western blotting and immunoprecipitation were performed as described previously (51). For the GST pull-down assay, 10 μg GST-RasV12 loaded with GTPγS and 20 μl of 25% agarose beads were incubated with the amounts of cell lysates indicated at 4°C for 3 h, followed by three washes. The proteins were eluted from the beads via 2× Laemmli buffer and subjected to 7.5 to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Rap1 activation assay.
Active GTP-bound Rap1 was assayed with the GST-tagged Ras binding domain (RBD) of RalGDS (gift from J. L. Bos, Utrecht University, Utrecht, The Netherlands) in an in vitro pull-down assay as previously described (9).
Reverse transcription (RT)-PCR.
Total RNAs were extracted from PC12 cells left untreated or treated as indicated with TRIzol from Invitrogen in accordance with the manufacturer's instructions. Equal amounts of total RNA were used for first-strand cDNA synthesis with oligo(dT) and Superscript II reverse transcriptase (Invitrogen), and the cDNA obtained was used as the template for PCR. The Epac2 primers (sense, 5′-CATTACCACGCACAGCCTTC-3′; antisense, 5′-TTGTACTCCTTGCAGTGAGC-3′) generated an 881-bp fragment corresponding to nucleotides 1620 to 2500 of the Epac2 cDNA. 9.5/ubiquitin carboxyl-terminal hydrolase (PGP9.5), was used as an internal control (25). The PGP primers (sense, 5′-TAATGTGGACGGCCACCTCT-3′; antisense, 5′-GCTCGCGCTCAGTGAATTCT-3′; gift from B. Habecker, Oregon Health & Science University, Portland) generated a 109-bp fragment corresponding to nucleotides 531 to 639 of the PGP9.5 cDNA. Amplification of the Epac2 cDNA was analyzed at 28 and 30 cycles, both of which yielded similar results.
Cell fractionation.
Cell fractionation was performed as previously described (51). Briefly, cells were scraped into ice-cold hypotonic buffer (10 mM KCl, 1.5 mM MgCl2, 1 mM Na-EDTA, 1 mM dithiothreitol [DTT], 1 μM leupeptin, 10 μg/ml soybean trypsin inhibitor, 0.1 μM aprotinin, 1 mM sodium orthovanadate, 1 mM β-glycerophosphate, 10 mM Tris-HCl, pH 7.4). Cells were homogenized with 50 strokes in a Dounce-type homogenizer. Lysates were then spun at 800 × g for 5 min at 4°C to pellet the nuclear fraction and to isolate the supernatant. The supernatant was transferred to a new tube and spun at 20,000 × g for 30 min at 4°C to pellet the membrane fraction (M), which was washed twice in the above-described hypotonic buffer and then resuspended in 50 μl lysis buffer. The remaining supernatant (400 μl) represented the cytosolic fraction (C). Forty percent (20 μl) and 5% (20 μl) of the total volumes of the M and C fractions were supplemented with 6× Laemmli buffer and loaded onto the gel for Western blotting, and from the percentages of the total input and densities of the Western blotting bands, the percentage of membrane recruitment could be estimated. The presence of endogenous Ras and β-actin was also examined as membrane and cytosol markers, respectively.
Protein expression and purification.
Human H-RasV12 (referred to as RasV12) and bovine Rap1b (referred to as Rap1) were subcloned into the pGEX-4T3 vector downstream of the GST tag between the EcoRI and fused XbaI/SmaI sites. The construct was transformed into bacterial strain BL21(DE3) from Invitrogen and kept as a glycerol stock. One hundred milliliters of standard LB medium was inoculated, and the bacteria were grown overnight at 37°C as a preculture, which was used to inoculate 2 liters of medium and rocked at 37°C until it reaching an optical density at 600 nm of 0.8. Protein expression was induced by isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 100 μM, and the culture was grown at 37°C for 10 h at 200 rpm. The cells were harvested and frozen at −80°C overnight. The pellets were then resuspended in ice-cold phosphate-buffered saline supplemented with 5 mM DTT, 5 mM EDTA, and protease inhibitors and lysed with a French press at 15,000 lb/in2 for 3 cycles. The debris was removed from the lysates by centrifugation, and the supernatants were incubated with 2 ml of 50% glutathione-agarose for 3 h at 4°C. The beads were washed three times with 10 ml phosphate-buffered saline. For GST-Rap1, the fusion protein was eluted with 10 mM glutathione (50 mM Tris-HCl, pH 8), followed by dialysis to remove the glutathione. For RasV12, the GST tag was cleaved by incubation with thrombin (Sigma-Aldrich), followed by elution. Epac2Δ430 and Epac2Δ430-684E were subcloned into the pGEX-4T3 vector (GE Healthcare) downstream of the GST tag by using the available EcoRI and NotI sites. The transfection and expression processes were similar to those used for RasV12, except that the cells were induced and rocked at 25°C for 15 h. The purification procedure was similar to that described previously for Epac1 (35), except that the cells were lysed with a French press instead of by sonication. The concentrations of the purified proteins were determined by both the bicinchoninic acid assay (Bio-Rad protein assay reagent) and A280 measurement, and the values generated by the two methods agreed with each other.
Nucleotide exchange assay.
We used mant-dGDP for the exchange assay to avoid artifacts caused by isomerization of the fluorescent label (11). RasV12 and GST-Rap1 were loaded with GTPγS and mant-dGDP, respectively, in loading buffer (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 5 mM EDTA, 5 mM DTT, 5% glycerol) by using a 10-fold molar excess of the respective nucleotides, and the free nucleotides were removed by gel filtration with a NAP-5 column (GE Healthcare) equilibrated in exchange buffer (50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 5 mM MgCl2, 5 mM DTT, 5% glycerol). The exchange assays were performed by a method similar to that described previously (10, 28, 35). Briefly, dissociation rates were measured on a Photon Technology International spectrophotometer with a DeltaRam excitation source. Fluorescence was excited at 360 nm, and emission was monitored at 435 nm. In a quartz cuvette, 100 nM Rap1-mant-dGDP was mixed with the exchange buffer supplemented with 100 μM GTP and incubated at 25°C in a final volume of 250 μl. When indicated, reactions were supplemented with 1 μM Epac2Δ430 or Epac2Δ430-684E and/or 1 μM RasV12-GTPγS. Dissociation was measured for 1,000 s. The data were fitted to a single-exponential function (Y = A0 + A1e−kt), and the decay due to photobleaching was minimal and ignored. After fitting, the raw data for each reaction were normalized independently between 0 and 1 by using the formula Ynormalized = (Yraw − A0)/(M − A0), where A0 represents the offset value from the exponential fit and M is the initial, maximum, fluorescence.
Fluorescence microscopy.
COS cells were cultured in a 12-well plate and transfected with the GFP- or mCherry-tagged plasmids as indicated. The cells were imaged after 24 h alive at room temperature. Wide-field microscopy images were obtained with a Leica DMIRE2 inverted fluorescence microscope. The TIFF images were acquired with MagnaFIRE 2.1 software and processed with Adobe Photoshop. Intensity profiling data obtained along the indicated line across the cell with Scion Image (Scion Corp., Frederick, MD) and plotted with Prism 3 (GraphPad Software, La Jolla, CA) are provided.
Neurite outgrowth assay.
PC12 cells were grown on poly-d-lysine-coated plates (Sigma-Aldrich) and serum-starved for 16 h prior to treatment with EGF (50 ng/ml) or forskolin (0.5 μM) and pretreatment with H89 (10 μM), which was added 15 min prior to forskolin treatment. After 24 h, the cells were examined microscopically for evidence of neurite outgrowth (32). Processes greater than two cell body lengths were scored as neurites. Representative photomicrographs of more than 200 cells examined for each condition in five independent experiments are shown. For experiments utilizing transfection, a vector expressing GFP was cotransfected as a reporter for transfected cells and morphological assessment was restricted to GFP-positive cells. For pretreatment experiments with NGF, NGF (50 ng/ml) was applied for 8 h and washed out prior to subsequent treatment with EGF (50 ng/ml) and/or forskolin (0.5 μM) plus H89 (10 μM).
Data processing and statistics.
The densities of the bands from Western blotting or RT-PCR assays were quantified with Scion Image (Scion Corp., Frederick, MD). The densities of the bands of interest were adjusted according to the input or loading controls. All of the experiments were repeated at least three times, and data from multiple data sets were normalized on a scale of 0 to 100% and analyzed with Prism 3 (GraphPad Software, La Jolla, CA). Unpaired t tests were performed between two groups of data as indicated, and P < 0.05 was regarded as statistically significant.
RESULTS
Epac2 interacts with Ras-GTP in a cAMP-independent manner.
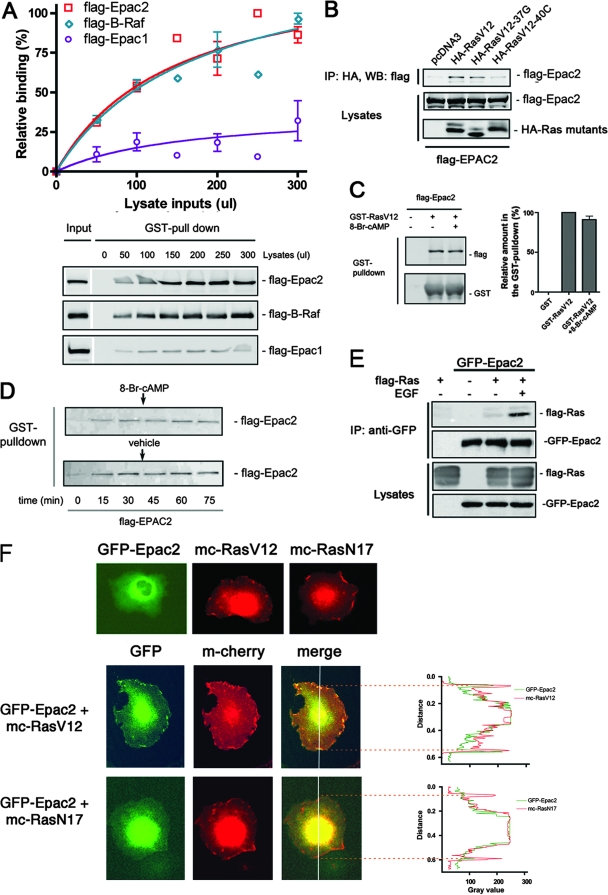
In the GTP-loaded active state, Ras binds the classical effectors like B-Raf and C-Raf via their RBDs. Epac2, but not Epac1, was also reported to interact with Ras-GTP (26). To estimate the strength of Ras-Epac2 binding, the profiles of Epac2 and B-Raf binding to Ras were compared in vitro. We expressed Flag-tagged Epac2, B-Raf, and Epac1 in COS cells and performed GST pull-down assays with increasing amounts of lysates in the presence of purified GST-RasV12 (a constitutively active Ras mutant) that was loaded with GTPγS. Epac1 interacted poorly with GST-RasV12, whereas both Epac2 and B-Raf bound to RasV12 at much higher and comparable levels (Fig. 1A). When transfected into mammalian cells, RasV12 became stably GTP loaded under basal conditions (data not shown) and associated with Flag-Epac2 in the absence of extracellular stimuli (Fig. 1B). Ras effectors interact with Ras mainly through the effector loop of Ras, and specific mutations along the effector loop can selectively interfere with a subset of effectors. For example, RasV12-40C interacts poorly with B-Raf and C-Raf but interacts normally with phosphoinositide 3-kinase (52). In contrast, 37G interacts poorly with C-Raf but interacts well with another Ras effector, RalGDS (29). As shown in Fig. 1B, Epac2 associated with RasV12 at levels similar to that seen with RasV12-37G, whereas its interaction with RasV12-40C was almost completely lost (Fig. 1B). Therefore, the Ras-Epac2 interaction requires an intact effector loop, as shown by Quilliam and colleagues (26).
FIG. 1.
Epac2 interacts with Ras-GTP independently of cAMP. (A) Epac2 and B-Raf, but not Epac1, bind to Ras-GTP. COS cells were transfected with Flag-Epac1, Epac2, or B-Raf as indicated, and increasing amounts of lysates were incubated with purified GST-RasV12 loaded with GTPγS, followed by a GST pull-down assay and Western blotting. The upper panel shows data from three experiments (mean ± standard error) and curve fitting. The lower panel shows a representative result from one experiment. Input and protein levels recovered after pull-down are shown for Epac2 (top), B-Raf (middle), and Epac1 (bottom). (B) Epac2 interacts with Ras at its effector loop. Flag-Epac2 was cotransfected along with either HA-RasV12 or its effector loop mutant form (37G or 40C) into COS cells, followed by immunoprecipitation (IP) with anti-HA antibody and Western blotting (WB, top). The expression levels of Flag-Epac2 (middle) and HA-Ras V12 and mutant forms (bottom) are shown. (C) Ras-Epac2 interaction is not enhanced by the cAMP analog 8-Br-cAMP in vitro. Purified GST-RasV12 loaded with GTPγS was incubated with cell lysates containing Flag-Epac2 in the presence or absence of 8-Br-cAMP, followed by GST pull-down assay. The left panel shows Western blotting with anti-Flag antibody (top) and anti-GST antibody (bottom). The right panel shows the quantification of three experiments (mean ± standard error). (D) The time course of Ras-Epac2 binding is not affected by 8-Br-cAMP. Purified GST-RasV12 loaded with GTPγS was incubated with cell lysates containing Flag-Epac2. 8-Br-cAMP or vehicle was added to the incubation mixture after 30 min. GST pull-down assays were performed at the time points indicated. (E) Ras-Epac2 association was induced by EGF treatment. Flag-Ras was transiently expressed in HEK293 cells stably transfected with GFP-Epac2 or in wild-type cells as indicated, and immunoprecipitation was performed with anti-GFP antibody after EGF (+) or mock (−) treatment, followed by Western blotting. The amounts of Flag-Ras and GFP-Epac2 recovered by immunoprecipitation and the expression of transfected proteins are shown. (F) Epac2 and Ras-GTP colocalized on the plasma membrane. (Upper row) GFP-Epac2 (left), mCherry-RasV12 (middle), and mCherry-RasN17 (right) were transfected into COS cells, respectively, and imaged by epifluorescence microscopy. (Middle row) GFP-Epac2 (left) and mCherry-RasV12 (middle) were cotransfected into COS cells. The merged fluorescent images are shown in the right panel. (Lower row) GFP-Epac2 (left) and mCherry-RasN17 (middle) were cotransfected. The merged images are shown in the right panel. In both the middle and bottom rows, the far right panel shows intensity profiles across the cell at the white line indicated in the right panel. The broken lines denote the positions of the plasma membranes.
cAMP is a well-studied regulator of Epac2 and was reported to be necessary for the interaction between Ras and Epac2 in cells (26). However, treatment with 8-Br-cAMP, an analog of cAMP that is capable of activating Epac2 (data not shown), neither enhanced nor inhibited the binding of Epac2 and RasV12 in vitro (Fig. 1C and D), suggesting that the conformational change in the regulatory region triggered by cAMP was not required for Ras binding. Importantly, the cAMP-independent nature of the Ras-Epac2 interaction is consistent with the crystal structure of Epac2 in its autoinhibited state, in which its RA domain is well exposed in the solvent and free of steric hindrance from its regulatory region (37).
To examine whether the Ras-Epac2 interaction could be induced by physiological stimuli, Flag-Ras was expressed in HEK293 cells stably transfected with GFP-Epac2. Low levels of interaction between Epac2 and Ras were detected under basal conditions; this interaction was significantly enhanced upon EGF stimulation (Fig. 1E). Next, we used epifluorescence microscopy to investigate the distribution of Epac2 in the presence of either RasV12 or RasN17, an inactive mutant form of Ras that is constitutively in the GDP-bound state. When transfected alone, mCherry-RasV12 and mCherry-RasN17 were both detected at the plasma membrane while GFP-Epac2 was not seen at the plasma membrane (Fig. 1F). When cotransfected with mCherry-RasV12, GFP-Epac2 colocalized with RasV12 at the plasma membrane. In contrast, when cotransfected with mCherry-RasN17, GFP-Epac2 remained largely in the cytosol (Fig. 1F). In summary, Epac2 binds to the effector loop of Ras-GTP in a cAMP-independent manner and this interaction recruits Epac2 to the plasma membrane.
Identification of Epac2 mutants with loss of Ras binding.
The Epac2 RA domain is located between the REM domain and the CDC25 domain. Therefore, it is possible that deletion of the RA domain may affect the interaction between REM and CDC25 domains, as well as the overall structure of Epac2. Indeed, an Epac2 deletion mutant lacking the entire RA domain appeared to be unstable when expressed in COS cells (data not shown). Therefore, to further our understanding of the Epac2-Ras interaction, we looked for specific point mutations within the Epac2 RA domain that could disrupt the Ras-Epac2 interaction without affecting the overall structure of the Epac2 protein.
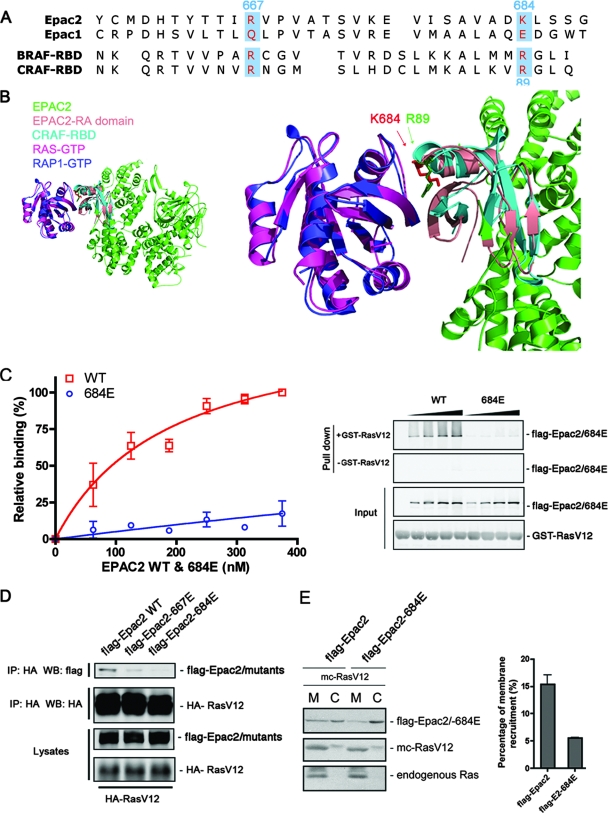
Comparison of the sequences of RA domains from Epac2 and Epac1 revealed two positively charged residues in Epac2, K684 and R667, that are conserved across species in Epac2 but are replaced by glutamate and glutamine, respectively, in Epac1 (blue boxes in Fig. 2A). To predict the role of these residues in RA domain function, we modeled the interaction between Epac2 and Ras-GTP, taking advantage of the existing structure of Epac2 (37), as well as the structure of the complex of C-Raf RBD and Rap1, which has served as a structural model of RBD-Ras interaction (31) (Fig. 2B). A high degree of similarity between the RA domain of Epac2 and the RBD of C-Raf was observed. Importantly, the position of the side chain of K684 of Epac2 partially overlapped that of R89 of the C-Raf RBD. R89 of C-Raf has previously been identified as a critical residue for Ras interaction (1, 8) (Fig. 2B). Therefore, we predict that mutation of K684 could similarly abolish the Ras-Epac2 interaction. The above model also highlighted the lack of steric restriction from the regulatory region of Epac2 on the Ras-Epac2 interaction.
FIG. 2.
Disruption of Ras-Epac2 interaction by a single point mutation within the RA domain. (A) Sequence alignment of the RA domains from Epac1 and Epac2, as well as the RBDs from B-Raf (BRAF-RBD) and C-Raf (CRAF-RBD). The residues within Epac2 that were targeted for mutational analysis are highlighted. (B) Structural modeling demonstrating the position of K684 at the Ras-Epac2 binding interface. The crystal structure of Rap1 in a complex with the C-Raf RBD was used as the template, and the structures of Ras and Epac2 were superimposed onto Rap1 and the C-Raf RBD, respectively, with the program Chimera (University of California at San Francisco). The left panel shows an overview of the predicted Ras-Epac2 complex; the right panel shows a close-up of the binding interface. The side chains of K684 (Epac2) and R89 (C-Raf) are highlighted as red and green sticks, respectively (arrows). (C) Epac2-684E was incapable of Ras binding. Increasing amounts of wild-type (WT) Flag-Epac2 or Epac2-684E were incubated with GTPγS-loaded GST-RasV12, followed by a GST pull-down assay and Western blotting. The concentration of Flag-tagged protein was quantified by comparison to purified protein standards. The graph shows data from three experiments (mean ± standard error) with curve fitting. The right panel shows the amounts of Epac proteins pulled down in the presence or absence of GST-RasV12, as well as the input levels of all of the proteins. (D) Ras association with wild-type Epac2 and Epac2 mutant forms. Wild-type Flag-Epac2 and mutant Epac2-667E or Epac2-684E were cotransfected with HA-RasV12 into COS cells, and immunoprecipitation (IP) was performed with anti-HA agarose antibody, followed by Western blotting (WB). The first two rows show the levels of Epac2 proteins and HA-RasV12 immunoprecipitated. The third and fourth rows show the levels of transfected proteins within the lysates. (E) The recruitment of Epac2-684E to the membrane is reduced compared to that of wild-type Epac2. Wild-type Flag-Epac2 or Epac2-684E was cotransfected with mCherry (mc)-RasV12 into COS cells, followed by cell fractionation. The membrane fraction (M) and the cytosolic fraction (C) were isolated and examined by Western blotting with Flag antibody (top). Both transfected RasV12 (mc-RasV12; middle) and endogenous Ras (bottom) were also monitored as markers for the membrane fraction with Ras antibody. The percentages of wild-type Epac2 and mutant Epac2-684E that were recruited to the membrane were calculated as described in Materials and Methods, and the right panel summarizes data from three experiments (mean ± standard error).
As predicted by the sequence alignment and structural modeling, mutation of K684 to glutamate (Epac2-684E) dramatically decreased the Epac2 and Ras binding as measured by GST pull-down assay (Fig. 2C), and the dissociation constant (Kd) increased from 189.8 ± 9.6 to 2,898 ± 369.4 nM. The loss of the association of this mutant with Ras-GTP was also demonstrated by immunoprecipitation in cells cotransfected with RasV12 and either wild-type Epac2 or Epac2-684E (Fig. 2D). Mutant Epac2-667E was also included in this experiment, but the loss of Ras binding by this mutant was less pronounced than that by Epac2-684E. Because Ras-Epac2 interaction recruits Epac2 to the plasma membrane (Fig. 1F), we asked if this effect was reduced when examining the RA domain mutant. We demonstrated that compared to wild-type Epac2, the amount of the RA domain mutant within the membrane fraction in the presence of RasV12 was reduced (Fig. 2E). Taken together, the mutational analysis indicates that an intact RA domain is required for Ras-Epac2 interaction and that the 684E mutant can be used to investigate how loss of Ras binding affects Epac2 function.
Effect of Ras binding on Rap1 activation via Epac2.
In general, Ras effectors are selective Ras-GTP binding proteins whose functions are modified by that association (40). To examine whether the interaction of Epac2 with Ras modulated Epac2 function, we examined Epac2-dependent activation of Rap1. Previous studies have suggested that the RA domain is dispensable for Epac2 function and that Ras binding to Epac2 does not change its overall ability to activate Rap1 (26). However, those studies examined transfected Rap1, which was basally activated by cotransfected Epac2 in the absence of cAMP. To avoid any potential pitfall of Rap1 overexpression, we focused on the activation of endogenous Rap1 by Epac2. To elevate intracellular cAMP levels, we used a combination of forskolin (an adenylyl cyclase activator) and IBMX (a phosphodiesterase inhibitor). To eliminate cAMP-dependent activation of PKA, we pretreated the cells with H89 (a PKA inhibitor). As shown in Fig. 3A, this cocktail (referred to throughout this report as F/H) modestly activated endogenous Rap1 following the transfection of wild-type Epac2 but not Epac2-684E (Fig. 3A). Similar results were seen with ISO (Fig. 3B). That the RA domain mutant exhibited diminished activity in response to cAMP was surprising since the mutation is distant from the catalytic core of Epac2 and is not expected to have any influence on its catalytic activity. Thus, the interaction between Epac2 and Ras appears to be necessary for efficient activation of Rap1 by Epac2 following cAMP elevation. However, F/H is a poor activator of Ras in these cells (data not shown). Therefore, the different activities of Epac2 versus Epac2-684E may reflect their abilities to associate with low levels of basally active Ras.
FIG. 3.
Ras-Epac2 interaction is required for efficient Rap1 activation by Epac2. (A) Time course of activation of endogenous Rap1 triggered by F/H (H89 plus forskolin and IBMX) in cells expressing wild-type Epac2 and mutant Epac2-684E. COS cells were transfected with pcDNA3, Flag-Epac2, or Flag-Epac2-684E for 24 h. Serum-starved cells were treated with F/H for the times indicated and harvested. Rap1 activation assay was performed with GST-RalGDS RBD, followed by Western blotting for endogenous Rap1. The left panel shows the quantification of data from three independent experiments (mean ± standard error). The right panel shows representative gels from one experiment. In this and all of the following Rap1 activation assays, Rap1 activation and Rap1 protein levels are shown in the top two rows, respectively, and the levels of transfected proteins are shown in the bottom rows. At least three independents experiments were performed. (B) Time course of activation of endogenous Rap1 triggered by ISO in cells expressing wild-type Epac2 and mutant Epac2-684E. COS cells were transfected with pcDNA3, Flag-Epac2, or Flag-Epac2-684E for 24 h. Serum-starved cells were treated with ISO for the times indicated, harvested, and subjected to a Rap1 activation assay. The left panel shows the quantification of data from three independent experiments (mean ± standard error); the right panel shows representative gels from one experiment. (C) Coexpression of RasV12 enhances Rap1 activation by Epac2. COS cells were transfected with mCherry (mc)-RasV12, Flag-Epac2, or both, as indicated, and treated with F/H (+) for 15 min or left untreated (−). The lysates were subjected to a Rap1 activation assay. Epac2-684E was included for comparison. (D) Time course of RasV12-enhanced Rap1 activation by Epac2. COS cells were transfected as for panel C and treated with F/H for the indicated times, and lysates were subjected to a Rap1 activation assay. (E) RasV12-dependent enhancement of Rap1 activation is specific for Epac2 but not Epac1. Flag-tagged Epac1 or Epac2 was transfected alone or together with mc-RasV12 (V12). After F/H treatment for 10 min, cells were harvested for a Rap1 activation assay. (F) Specific activation of endogenous Ras enhances Epac2-mediated Rap1 activation. COS cells were transfected with the HA-tagged catalytic domain of SOS (SOScat) and/or HA-Epac2 as indicated and treated with F/H for 10 min or left untreated (Untr.). Lysates were subjected to a Rap1 activation assay. The left panel shows representative gels from one experiment. The right panel shows the quantification of data from three experiments (mean ± standard error).
We next examined whether an increased level of Ras-GTP can potentiate Rap1 activation by Epac2. Indeed, RasV12 dramatically augmented the Epac2-mediated Rap1 activation triggered by cAMP, while Rap1 activation by Epac2-684E was slightly enhanced by RasV12, probably due to residual Ras binding of the mutant (Fig. 3C). Furthermore, RasV12 boosted the cAMP-triggered Rap1 activation at all of the time points examined (Fig. 3D). This enhancement was specific for Epac2 but not Epac1 (Fig. 3E). SOS is a well-studied Ras exchanger whose catalytic domain shows constitutive and specific activity toward Ras (12). When cotransfected into cells, the catalytic domain of SOS also dramatically enhanced the ability of Epac2 to activate Rap1 (Fig. 3F), demonstrating the ability of endogenous Ras-GTP to couple Epac2 to Rap1 activation.
Mechanism of Ras-dependent Epac2 activation.
cAMP binding to the cNBD within the regulatory region of Epac2 has been proposed to reorient the regulatory region away from the catalytic domain, allowing access of Rap1 to the catalytic site (36-38). To understand the mechanism of the requirement of Ras-GTP in Epac2-mediated Rap1 activation, we first tested whether Ras-Epac2 interaction facilitated this action of cAMP. Deletion of the regulatory region of Epac2, including both cNBDs and the DEP domain, produced a constitutively active truncation (Epac2Δ430) whose function was no longer regulated by cAMP. Epac2Δ430 activated Rap1 in a dose-dependent manner when expressed in COS cells. In contrast, the Epac2Δ430 mutant containing the additional K684E mutation (Epac2Δ430-684E) exhibited a significantly reduced ability to activate Rap1 (Fig. 4A). This suggests that an intact RA domain is still required for Rap1 activation by the catalytic region even in the absence of intramolecular inhibition. Importantly, the Epac2Δ430-mediated Rap1 activation could be further enhanced by RasV12, which was ineffective in enhancing Epac2Δ430-684E activity (Fig. 4B). Together, these results indicate that Ras-mediated regulation of Epac2 is independent of the mechanism by which cAMP activates Epac2.
FIG. 4.
Ras-GTP potentiates Rap1 activation by the catalytic region of Epac2 in vivo but not in vitro. (A) Comparison of Rap1 activation by Epac2Δ430 and Epac2Δ430-684E. Increasing amounts of Epac2Δ430 or Epac2Δ430-684E were expressed in COS cells, and lysates were subjected to a Rap1 activation assay. The left panel shows linear regression of data from one representative experiment. Levels of Rap1 activation were plotted against the expression levels of constructs as indicated. The right panel shows representative gels from one experiment. Three independent experiments were performed. AU, artificial units; Ctrl, control. (B) RasV12 enhancement of Rap1 activation by Epac2Δ430 requires an intact RA domain. GFP-Epac2Δ430 and Epac2Δ430-684E were transfected alone or cotransfected with mCherry (mc)-RasV12, and lysates were subjected to a Rap1 activation assay. The left panel shows the quantification of data from three experiments (mean ± standard error); the right panel displays gels from one representative experiment. (C) Coomassie staining of purified Epac2Δ430, Epac2Δ430-684E, GST-Rap1, and RasV12. Protein molecular size markers are shown on the left. (D) Both Epac2Δ430 and Epac2Δ430-684E catalyzed nucleotide exchange reactions on Rap1 at identical rates in vitro. The upper panel shows a comparison of the intrinsic (red) exchange reaction and that catalyzed by Epac2Δ430 (cyan) or Epac2Δ430-684E (pink). Rap1-mant-dGDP (100 nM) was incubated in buffer containing 100 μM unlabeled GTP in the absence or presence of 1 μM Epac2Δ430 or Epac2Δ430-684E. Dissociation of mant-dGDP was monitored by the decrease in fluorescence emission at 435 nm over time. The bottom panel shows reaction rates fitted to single exponentials, and three to six independent measurements for each condition were pooled in the bar graph (mean ± standard error). (E) RasV12 does not enhance the exchange activity of Epac2Δ430 in vitro. The upper panel shows a comparison of the intrinsic (red) exchange reaction and that catalyzed by Epac2Δ430 in the presence (orange) or absence (cyan) of Ras-GTP. Rap1-mant-dGDP (100 nM) was incubated in buffer containing 100 μM unlabeled GTP in the absence or presence of 1 μM Epac2Δ430 alone or in addition to GTPγS-loaded RasV12. Dissociation of mant-dGDP was monitored. The bottom panel shows reaction rates fitted to single exponentials, and three independent measurements are summarized in the bar graph (mean ± standard error).
To test the possibility of allosteric activation of Epac2 by Ras-GTP, we examined the kinetics of nucleotide exchange in vitro with purified proteins (Fig. 4C). The rate of nucleotide release from Rap1 in the presence of Epac2Δ430 ([5 ± 0.2] × 10−3 s−1for 1 μM exchange factor) is comparable to the rate in the presence of Epac2Δ430-684E ([5 ± 0.7] × 10−3 s−1 for 1 μM exchange factor) and is significantly higher than the intrinsic rate of nucleotide release by isolated Rap1 (0.3 × 10−3 s−1; Fig. 4D). These data confirm that the Epac2-684E mutant is fully functional and the loss of Rap1 activation by this RA mutant in cell-based Rap1 activation assays cannot be explained by the loss of its intrinsic catalytic activity (Fig. 3A and 4A). In addition, in the presence of saturating concentrations of RasV12 preloaded with GTPγS, the rate of nucleotide release from Rap1 catalyzed by Epac2Δ430 was not accelerated (Fig. 4E). These results suggest that Ras does not enhance Epac2-mediated Rap1 activation in vitro, despite activating it within cells.
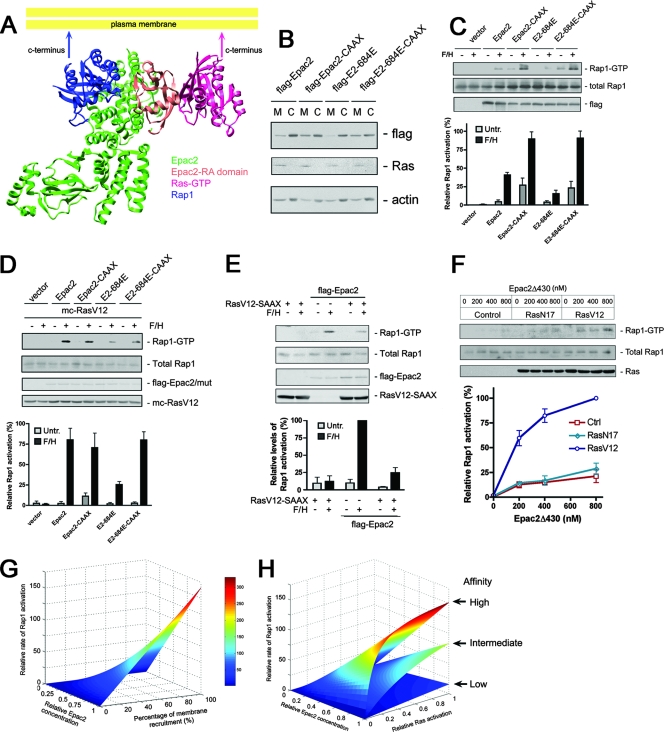
One possible explanation for these findings is that the compartmentalization of Ras and Rap1 is critical for Ras-dependent Rap1 activation by Epac2 in intact cells. To test this hypothesis, we first performed structural modeling to visualize the Ras-Epac2Δ430-Rap1 ternary complex in the context of the plasma membrane. The ternary complex was assembled based on the crystal structures of Epac2 in complex with Rap1 and a cAMP analog (36) and the complex of Rap1-C-Raf RBD (for detailed methods, see the section on structural modeling in the supplemental material). From this model (Fig. 5A), we observed that Ras, Rap1, and the RA and cdc25 domains of Epac2 were aligned on a single plane parallel to the membrane plane. Importantly, the carboxy-terminal ends of both Ras and Rap1 were oriented toward the plasma membrane in this model. This is a necessary constraint, since both Ras and Rap1 are lipid modified at the carboxy terminals and tethered to the lipid membrane.
FIG. 5.
Mechanism of Ras-facilitated Rap1 activation by Epac2. (A) Structural modeling of the Ras-Epac2-Rap1 ternary complex in relation to the plane of the lipid membrane. The ternary complex demonstrates that Rap1 engages the CDC25 homology domain of Epac2 to the left; the RA domain of Epac2 interacts with Ras to the right. The carboxy-terminal ends of Rap1 and Ras point toward the membrane plane. This accommodates the lipid modifications of both proteins that tether both carboxy-terminal ends to the membrane. For additional details, see the section on structural modeling in the supplemental material. (B) Addition of the H-Ras CAAX motif increased the amounts of Epac2 and Epac2-684E localized to the membrane. Flag-tagged Epac2, Epac2-CAAX, Epac2-684E (E2-684E) and Epac2-684E-CAAX (E2-684E-CAAX) were transfected into COS cells for 24 h, and cell fractionation was performed. The membrane (M) and cytosolic (C) fractions were subjected to Western blotting. Representative gels from one experiment are shown. The upper row shows Flag-tagged proteins within the two fractions. The middle and lower rows are endogenous Ras and β-actin, which were used as markers for membrane and cytosol, respectively. (C) Anchoring Epac2 to the plasma membrane by the CAAX motif from H-Ras results in an enhanced level of Rap1 activation and rescues the defect of Epac2-684E (E2-684E). COS cells were transfected with Epac2, Epac2-CAAX, Epac2-684E, and Epac2-684E-CAAX as indicated, starved, and treated with F/H or left untreated (Untr.). Lysates were subjected to a Rap1 activation assay. The upper panel shows representative gels from one experiment. The bottom panel shows the quantification of three experiments (mean ± standard error). (D) Membrane targeting of Epac2 (Epac2-CAAX) occluded the effect of RasV12 on Epac2-mediated Rap1 activation. mCherry RasV12 was cotransfected with Epac2, Epac2-CAAX, Epac2-684E, or Epac2-684E-CAAX into COS cells as indicated, starved, and treated with F/H for 10 min or left untreated. Lysates were subjected to a Rap1 activation assay. The upper panels show representative gels from one experiment. The bottom panel shows the quantification of three experiments (mean ± standard error). (E) Sequestration of Epac2 in the cytosol by RasV12-SAAX prevents Epac2 from activating endogenous Rap1. COS cells were transfected with Flag-Epac2 and/or Ras-SAAX as indicated, and starved cells were treated with F/H for 10 min or left untreated and then subjected to a Rap1 activation assay. The upper panel shows gels from one representative experiment. The bottom panel shows the quantification of three experiments (mean ± standard error). (F) Purified Epac2Δ430 activates Rap1 within membrane preparations containing RasV12 in vitro. Increasing amounts of Epac2Δ430 were incubated with membrane fractions isolated from COS cells that were transfected with pcDNA3, mCherry-RasN17 (RasN17), or mCherry-RasV12 (RasV12) in 100 μl of exchange buffer and the presence of 100 μM GTP for 15 min at room temperature. The membrane fractions were recovered by centrifuge, and endogenous Rap1 activity within the membranes was assayed. The upper panel shows a representative result from one experiment. The bottom panel shows the quantification of three experiments (mean ± standard error). Ctrl, control. (G and H) Computational model of Epac2-mediated Rap1 activation. For details, see the section on computational modeling in the supplemental material. (G) The relative rate of Rap1 activation is shown as a function of the relative intracellular concentration of Epac2 (ranging from 0 to 1) and the percentage of its membrane recruitment (ranging from 0 to 100%). (H) The relative rate of Rap1 activation is shown as a function of the relative levels of Ras activation (varying from 0 to 1) and the affinity between Ras and Epac2 or its mutant forms. The upper, middle, and lower surfaces represent high, intermediate, and low Epac2-Ras interaction affinities, respectively.
Based on this structural model, it is possible that Ras-mediated membrane recruitment may play a crucial role in Rap1 activation by Epac2. We predict that tethering Epac2 to the membrane may mimic the effect of Ras-Epac2 binding. To address this possibility, we targeted Epac2 to the membrane by introducing a CAAX motif at its carboxy-terminal end. This increased the amount of Epac2 that could be recovered from membrane fractions (Fig. 5B) and dramatically enhanced the ability of Epac2 to activate Rap1 (Fig. 5C). Importantly, membrane targeting rescued the defect of Epac2-684E (Fig. 5C) and occluded the enhancing effect of RasV12 on Epac2-mediated Rap1 activation (Fig. 5D). Next, we tested whether sequestration of Epac2 in the cytosol could prevent it from activating Rap1. We achieved this by cotransfection of RasV12-SAAX, which was constitutively GTP loaded but mislocalized in the cytosol due to the loss of lipid modification (data not shown). RasV12-SAAX dramatically reduced Epac2-mediated Rap1 activation in response to cAMP (Fig. 5E), suggesting that RasV12-SAAX was acting as an interfering mutant. This finding further supports the role of endogenous Ras-GTP in the Epac2-mediated activation of Rap1. We next validated the above-described membrane recruitment model in vitro by incubating purified Epac2Δ430 and membrane fractions from cells expressing RasV12 or control cells, and RasV12 significantly enhanced the activation of endogenous Rap1 within the membrane fraction by Epac2Δ430 (Fig. 5F).
We propose that compartmentalization of Epac2 by Ras-GTP increases the concentration of Epac2 in the submembrane space, thus greatly enhancing the subsequent interaction between Epac2 and Rap1. This was demonstrated by using computational simulation (Fig. 5G). In the absence of active recruitment of Epac2 to the membrane, the rate of Rap1 activation is very slow and solely dependent on the expression level of Epac2 (see the section on computational modeling in the supplemental material for details). However, active membrane recruitment by Ras binding provided an additional level of regulation, which could dramatically accelerate the reaction rate even in the presence of a small increase in membrane recruitment (Fig. 5G).
In this model, membrane recruitment of Epac2 is influenced by two factors: the level of Ras activation and the affinity of Epac2 for Ras-GTP. As demonstrated in Fig. 5H, the rate of Epac2-mediated Rap1 activation is predicted to be very sensitive to initial changes in Ras-GTP levels and reaches a plateau at higher Ras-GTP levels. This may explain the apparent dependence of Epac2 activation on interaction with low levels of Ras-GTP that may be present in cells stimulated with cAMP alone (Fig. 3A). The model also explains the decreased level of Rap1 activation by Epac2-684E by showing that it can be fully accounted for as a result of decreased Ras association (Fig. 5H). Taken together, the data show that Ras association converts Epac2 into an efficient, membrane-based Rap1 activator and that this recruitment is required for Epac2 function.
Epac2-mediated Rap1 activation at the membrane is coupled to ERK activation and neurite outgrowth.
We have recently shown that activation of Rap1 at the plasma membrane is coupled to ERK activation via B-Raf (51). Therefore, we tested whether recruitment of Epac2 to the plasma membrane by Ras could also couple Rap1 to ERK activation in B-Raf-expressing cells. PC12 cells express abundant levels of B-Raf, and Rap1 activation of B-Raf/ERKs in these cells has been well documented (15, 27, 53). However, these cells express low levels of Epac1 or Epac2 and have shown little PKA-independent activation of Rap1(51). Accordingly, F/H treatment triggered little activation of ERKs, even in the presence of EGF (Fig. 6A and B). However, upon the expression of Epac2, F/H and EGF were able to synergistically and robustly activate ERKs in a PKA-independent manner (Fig. 6A and B). Importantly, Rap1a contributed to the robust ERK activation by F/H and EGF in the presence of Epac2, as knockdown of Rap1a with shRNA (Fig. 6C) significantly reduced this effect (Fig. 6D). The modest ERK activation via Epac2 triggered by F/H alone was enhanced when Epac2 was targeted to the membrane via the CAAX motif and was absent in cells expressing the Epac2-684E mutant protein (Fig. 6E). Moreover, the enhanced activity of Epac2-CAAX no longer required Ras binding, as similar levels of ERK activation were seen with the Epac2-684E-CAAX mutant protein (Fig. 6E).
FIG. 6.
Enhancement of ERK activation and neurite outgrowth by Epac2. (A) Expression of Epac2 enhances the activation of ERKs by EGF and F/H. PC12 cells were transfected with vector (pcDNA3) or Flag-ERK2 and treated for 15 min with F/H, EGF, or EGF plus F/H or left untreated. For this and all subsequent ERK activation assays, the levels of phosphorylated Flag-ERK2 are shown with a phospho-ERK antibody following Flag immunoprecipitation (first row). Total levels of transfected Flag-ERK2 (second row) and Flag-Epac2 (third row) are also shown. (B) Expression of Epac2 enhances the activation of ERKs by EGF and F/H. PC12 cells were transfected with Flag-ERK2 along with GFP or Epac2 and treated with EGF or EGF plus F/H. Representative gels from one experiment are shown on the left. The right panel shows the quantification of data from three independent experiments (mean ± standard error). (C) Rap1a shRNA reduces the expression of cotransfected Rap1a. PC12 cells were cotransfected with Flag-Rap1a (rat) and Flag-ERK2 cDNA with either Rap1a shRNA or a scrambled control (scrm.) for 24 h, followed by Flag immunoprecipitation (IP). The left panel shows levels of Flag-Rap1a (upper row) and Flag-ERK2 (lower row), used as a transfection control, determined by Western blotting. The right panel shows the quantification of data from three independent experiments (mean ± standard error). (D) Rap1a knockdown blocks ERK activation via Epac2 triggered by EGF plus F/H. PC12 cells were cotransfected with Flag-Epac2 and Flag-ERK2 along with scrambled (scrm.) shRNA or Rap1a shRNA and treated as indicated. The upper panel shows the quantification of data from three experiments, and the lower panel shows representative gels from one experiment. (E) Membrane targeting of Epac2 overcomes mutation within the RA domain. PC12 cells were transfected with Flag-ERK2 along with wild-type or mutant Epac2 (E2) as indicated and treated with F/H. An ERK activation assay was performed, and the upper panel shows gels from a representative experiment. The bottom panel shows the quantification of data from three independent experiments (mean ± standard error). Untr., untreated. (F) Expression of Epac2 increases neurite outgrowth induced by EGF and F/H. For the left panel, PC12 cells were transfected with GFP and either vector, Epac2, Epac2-684E (E2-684E), or Epac2-CAAX and treated with EGF and/or F/H as indicated. Twenty-four hours later, neurite outgrowth was assessed by epifluorescence microscopy. In the right panel, representative photomicrographs are shown. The left panel shows the quantification of data from five independent experiments (mean ± standard error). The bar in the lower left panel represents 20 μm.
Augmented ERK signaling has been associated with neurite outgrowth in PC12 cells (4, 15, 20). To provide a physiological correlation for the Epac2-mediated augmentation of ERK signaling, we measured neurite outgrowth in PC12 cells with or without exogenous Epac2. In the presence of overexpressed Epac2, EGF and F/H induced neurite outgrowth in about 10 and 40% of the transfected cells, respectively. However, the combination of EGF and F/H had a synergistic action, promoting neurite outgrowth in 80% of the transfected cells (Fig. 6F). These data correlated with the pattern of ERK activations observed with these treatments (Fig. 6A and B). Interestingly, in the absence of transfected Epac2, the combination of EGF and F/H also synergistically induced neurite outgrowth in about 40% of the cells (Fig. 6F). This may reflect the presence of endogenous Epac2 in these cells (see below). The ability of F/H to induce neurite outgrowth to a level greater than that induced by EGF alone may depend on a low level of basal Ras activation in PC12 cells. Moreover, the inability of EGF to promote neurite outgrowth is consistent with the inability of EGF to stimulate cAMP levels.
The levels of neurite outgrowth via Epac2 mutant proteins also correlated with the levels of ERK activation (Fig. 6E). Unlike Epac2, Epac2-684E did not enhance neurite outgrowth above the levels seen in cells transfected with GFP alone, suggesting that the action of Epac2 required an intact RA domain (Fig. 6E). Targeting Epac2 to the plasma membrane via the CAAX motif resulted in equally high levels of neurite outgrowth by F/H in the presence or absence of EGF. This suggests that the EGF potentiated the F/H effect primarily through recruitment of Epac2 to the membrane. Taken together, these data suggest that Epac2 activation at the membrane is coupled to ERK activation and neurite outgrowth.
Endogenous Epac2 is increased by NGF and contributes to ERK activation and neurite outgrowth.
Epac2 is enriched in the neuronal tissues (22). Therefore, we hypothesized that NGF-induced differentiation of PC12 cells might be accompanied by increased expression of Epac2. Indeed, although basal levels of Epac2 mRNA and protein were detected by RT-PCR and Western blotting, respectively, both were significantly enhanced after 8 h of NGF pretreatment (Fig. 7A and B). Therefore, we could increase endogenous levels of Epac2 by brief pretreatment with NGF and examine the role of Epac2 in neurite outgrowth without elevating Epac2 levels by transfection. Importantly, the 8-h NGF pretreatment dramatically augmented the synergism between EGF and F/H in the activation of endogenous ERKs (Fig. 7C). Pretreatment with NGF can also enhance the basal level of Ras-GTP. This may account for the ability of F/H to induce significantly higher levels of ERK activation following NGF pretreatment, compared to F/H treatment alone in the absence of NGF pretreatment (Fig. 7C). Interestingly, EGF was also capable of inducing a modest increase in Epac2 expression (Fig. 7A and B).
FIG. 7.
Endogenous Epac2 is increased by NGF and contributes to ERK activation and neurite outgrowth. (A) Epac2 mRNA is increased by NGF or EGF treatment. PC12 cells were treated with NGF or EGF (50 ng/ml) for 8 h or left untreated. Total RNAs extracted from the cells were subjected to RT-PCR for Epac2 (upper row). PGP (lower row) was used as a control (Ctrl). The lower panel shows the quantification of data from five experiments (mean ± standard error). (B) Epac2 protein is increased by NGF or EGF treatment. PC12 cells were treated with NGF or EGF for 8 h or left untreated. Total cell lysates were subjected to Western blotting. In the upper panel, Western blotting shows endogenous Epac2 protein (upper row). ERK2 was used as the loading control (lower row). The bottom panel shows the quantification of data from four experiments (mean ± standard error). (C) NGF pretreatment enhances the activation of ERKs by EGF plus F/H. PC12 cells were pretreated with NGF for 8 h or left without pretreatment and subsequently treated with EGF and/or F/H as indicated. The top panel shows the average of data from three independent experiments. In the bottom panel, the levels of phosphorylation of endogenous ERKs and total levels of ERKs are shown. (D) Epac2 shRNA reduces the expression of cotransfected Epac2. PC12 cells were cotransfected with Flag-Epac2 and Flag-ERK2 cDNAs with either Epac2 shRNA or a scrambled (scrm.) control for 24 h, followed by Flag immunoprecipitation (IP). The left panel shows levels of Flag-Epac2 (upper row) and Flag-ERK2 (lower row), used as a transfection control, determined by Western blotting. The right panel shows the quantification of data from three independent experiments (mean ± standard error). (E) Knockdown of endogenous Epac2 by shRNA decreases the activation of ERKs by EGF plus F/H after NGF pretreatment. PC12 cells were cotransfected with Flag-ERK2 along with scrambled shRNA or Epac2 shRNA and treated as indicated. The upper panel shows gels from one representative ERK activation assay. The lower panel shows the quantification of data from three independent experiments (mean ± standard error). (F) Epac2 knockdown decreases neurite outgrowth induced by F/H after NGF pretreatment. PC12 cells were transfected with GFP, scrambled shRNA, or shRNA for Epac2. Cells were pretreated with NGF for 8 h and subsequently treated with F/H. Twenty-four hours later, neurite outgrowth was assessed by epifluorescence microscopy. In the top panel, representative photomicrographs are shown. In the bottom panel, the percentage of cells bearing neurites is shown with the standard error from at least three independent experiments. (G) Epac2 knockdown decreases neurite outgrowth induced by treatment with EGF plus F/H in the presence or absence of NGF pretreatment (pretreat.). PC12 cells were transfected with GFP, scrambled shRNA, or shRNA for Epac2. Cells were pretreated with NGF for 8 h or left without pretreatment and subsequently treated with EGF and F/H. Twenty-four hours later, neurite outgrowth was assessed by epifluorescence microscopy. In the right panel, representative photomicrographs are shown. The bar in the lower left photomicrograph represents 20 μm. In the left panel, the percentage of cells bearing neurites is shown with the standard error from five independent experiments.
The enhanced ERK activation was significantly diminished in cells expressing shRNA targeting Epac2 but not scrambled shRNA (Fig. 7E). The efficacy of shRNA knockdown of Epac2 is shown in Fig. 7D. Knockdown of Epac2 by shRNA also inhibited neurite outgrowth (Fig. 7F and G). In the presence of Epac2 shRNA (but not scrambled shRNA or the GFP control), the level of neurite outgrowth triggered by EGF and F/H was reduced from 40 to 20% (Fig. 7G), suggesting that the basal level of Epac2 mediates the synergistic effect of EGF and F/H on neurite outgrowth in PC12 cells. Importantly, short-term NGF pretreatment itself did not trigger neurite outgrowth by itself if NGF was washed out after 8 h. However, this pretreatment had an enhancing effect on the outcome of subsequent treatment with F/H alone or F/H plus EGF (Fig. 7F and G). This priming effect was largely dependent on the expression of Epac2, as the enhancement was blocked by Epac2 shRNA but not scrambled shRNA (Fig. 7F and G). Taken together, the findings show that endogenous Epac2, either basal or induced by NGF, integrates signals from cAMP and Ras and contributes to ERK activation and neurite outgrowth in PC12 cells.
DISCUSSION
Recently, Quilliam and colleagues showed that Ras recruits Epac2 to the plasma membrane. Here we extend their model by demonstrating that this recruitment is essential for the maximal action of Epac2 as a Rap1 exchanger. The classical definition of Ras effectors requires that effector binding be dependent on the GTP-bound state of Ras and that binding modify effector function (30). Epac2 has met this definition and is a bona fide Ras effector. While Epac proteins need cAMP to relieve their autoinhibition, our study shows for the first time that Epac2 also requires the interaction with Ras-GTP via the RA domain for its ability to activate Rap1 efficiently. This is based on a number of findings. (i) A specific mutation that disrupts the association of Epac2 with Ras-GTP diminishes the Epac2-dependent activation of Rap1 without affecting intrinsic exchange activity or cAMP-mediated regulation of Epac2. (ii) Interference with endogenous Ras by using the mutant protein RasV12-SAAX, which cannot associate with the membrane, blocks cAMP-dependent activation of Epac2. (iii) Activation of endogenous Ras by a Ras-specific exchanger, as well as expression of a constitutively active Ras mutant protein, enhances Epac2-dependent activation of Rap1. These data provide strong support for the necessity of Ras interaction for Epac2 function.
Thus, Epac2 is both a cAMP sensor and a novel Ras effector that couples coincident cAMP elevation and Ras activation to the activation of Rap1. This Rap1 activation is significantly higher than that seen following stimulation via either Ras or cAMP alone. While the cross talk between the cAMP and Ras signaling pathways has been shown to occur at multiple levels (7, 48), Epac2 provides a unique example of a single molecule integrating the two pathways through the independent actions of discrete domains.
Regarding the mechanism for Ras-dependent regulation of Epac2, Ras neither facilitates the relief of Epac2 autoinhibition nor induces allosteric changes to activate the Epac2 catalytic domain. Instead, Ras recruits Epac2 to the membrane, dramatically increases its local concentration, and accelerates the rate of Rap1 activation. This action may be similar to what was recently proposed for Ras activation of SOS. However, in that model, both membrane recruitment and allosteric modulation of SOS by Ras contributed to SOS activation (4, 12). As supported by the structural modeling and our experimental data, cAMP binding to the cNBD and the interaction between Ras and the RA domain are two independent events, which allows the relief of autoinhibition and membrane localization of Epac2 to be independently regulated by cAMP and Ras, respectively.
The requirement of Ras in the Epac2-dependent activation of Rap1 has many potential consequences for cell signaling. Temporally, Rap1 activated by Epac2 would accompany both cAMP elevation and Ras activation. Spatially, Epac2-mediated Rap1 activation would be restricted to Ras-containing membranes. Rap1 signaling from Ras-containing membranes has previously been shown to couple to ERKs through the Raf isoform B-Raf (51). This may reflect the properties of B-Raf itself, as the Raf family of kinases has been shown to be much more efficient at coupling to ERKs from the plasma membranes compared to intracellular locales (14). The participation of Rap1 in Ras-dependent activation of ERKs may also provide amplification, since many Rap1 molecules can be activated following the recruitment of a single Epac2 molecule to Ras. Moreover, the inactivation of Rap1 by Rap1-selective G protein activation proteins may be slower than that of Ras (44, 47), permitting Rap1 activation to be sustained. Distinct Ras isoforms are selectively activated in different subcellular compartments, resulting in the activation of distinct intracellular signaling pathways (13). Epac2 has been reported to interact with all Ras isoforms via its RA domain (26). Therefore, it is possible that recruitment of Epac2, and resultant Rap1 activation, differentially modulates the signals emanating from each of these Ras isoforms.
Neurite outgrowth of PC12 cells has been a well-studied physiological reporter of the neuronal differentiation that accompanies activation of the ERK pathway, with increased activation of ERK correlating with the extension of neuritic processes (4, 15, 44, 53). Importantly, transient activation of ERKs by EGF is not capable of triggering neurite outgrowth in the absence of additional signals. Using PC12 cells, we show that the inclusion of Epac2/Rap1 signaling with that of EGF can enhance EGF-dependent signaling to ERKs and can induce robust neurite outgrowth. Of course, the Epac2-mediated Rap1 activation is also likely to regulate other actions of Rap1 that occur at or near the plasma membrane, including exocytosis (16, 33) and cell adhesion (2, 3).
Indirect activation of Rit by Epac has also been reported to mediate PACAP-triggered p38 phosphorylation and neurite outgrowth in related PC6 cells (46) and ERK activation in other cell types (24, 43). These reports and our findings together reveal the complexity of Epac signaling upstream of mitogen-activated protein kinase cascades. It may be that Rap1 and Rit, working through ERKs and p38, act in concert to promote neurite outgrowth in these cells. Rap1 activation of p38 has been reported (17, 45), and it is possible that this is mediated by the coupling of Rap1 to Rit activation. Since Epac is not the direct GEF for Rit (46), it will be important to identify the Rit exchanger and to determine whether there is direct interaction between Rap1 and this exchanger that might mediate the coupling between these two small G proteins. Conversely, the possibility that Rit itself can bind to the RA domain of Epac and potentiate its actions remains to be tested.
Epac2 is a neuron-expressed isoform of the Epac family of exchangers. It is possible that Epac2 is involved in neuronal functions such as neuronal differentiation and synaptic plasticity that can be potentiated by both Ras-dependent and cAMP-dependent signals (19, 50). Because Ras can be activated via multiple signals, including intracellular calcium (23), Epac2 might also function as a coincidence detector for cAMP and calcium. In pancreatic beta cells, exocytosis of insulin stimulated by cAMP has a PKA-independent component that requires Epac2 (21). It will be important to determine whether Ras-dependent recruitment of Epac2 by glucose, growth factors, or hormones potentiates this action, as well as other PKA-independent actions of cAMP.
We propose that Epac2 levels will also be induced by NGF in neuronal cells. The Epac2 promoter contains cAMP-responsive element sites (18, 54) that are well established targets of NGF signaling (41). The induction of Epac2 by NGF may serve to enhance signaling through Rap1 as part of a positive feedback loop to potentiate neurite outgrowth following its initiation. It is also possible that NGF also induces posttranslational modifications of Epac2 to further enhance its function. We do not believe that the induction of Epac2 by NGF is unique to NGF, but its induction by EGF does not, by itself, contribute to neurite outgrowth in the absence of additional stimulators of cAMP. However, it does provide a mechanism by which EGF and cAMP can act together to promote neurite outgrowth.
In summary, we show that Epac2 is a novel Ras effector that can act as a coincidence detector for Ras and cAMP. The mechanism by which Epac2 permits Ras to activate Rap1 may be shared by other GEFs that contain functional RA domains (42). Therefore, activation of small G proteins by recruitment of their cognate GEFs to the plasma membrane via RA domains may be a fundamental mechanism to couple distinct G proteins to each other.
Supplementary Material
Acknowledgments
We thank David Farrens, Satinder Singh, Peter Rotwein, Mike Forte, and Zhiping Wang for scientific discussion and Quentin Low, Mark DeWitt, Jon Fay, and Srilatha Tavisala for technical assistance. We also thank Dafna Bar-Sagi, NYU School of Medicine; Lawrence Quilliam (Indiana University), Johannes Bos (Utrecht University), and Daniel Altschuler (University of Pittsburgh) for generous sharing of reagents.
This work was funded by NIH NCI grant CA72971.
Footnotes
Published ahead of print on 29 September 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Block, C., R. Janknecht, C. Herrmann, N. Nassar, and A. Wittinghofer. 1996. Quantitative structure-activity analysis correlating Ras/Raf interaction in vitro to Raf activation in vivo. Nat. Struct. Biol. 3244-251. [DOI] [PubMed] [Google Scholar]
- 2.Bos, J. L. 2005. Linking Rap to cell adhesion. Curr. Opin. Cell Biol. 17123-128. [DOI] [PubMed] [Google Scholar]
- 3.Bos, J. L., J. de Rooij, and K. A. Reedquist. 2001. Rap1 signalling: adhering to new models. Nat. Rev. Mol. Cell Biol. 2369-377. [DOI] [PubMed] [Google Scholar]
- 4.Boykevisch, S., C. Zhao, H. Sondermann, P. Philippidou, S. Halegoua, J. Kuriyan, and D. Bar-Sagi. 2006. Regulation of ras signaling dynamics by Sos-mediated positive feedback. Curr. Biol. 162173-2179. [DOI] [PubMed] [Google Scholar]
- 5.de Rooij, J., F. J. Zwartkruis, M. H. Verheijen, R. H. Cool, S. M. Nijman, A. Wittinghofer, and J. L. Bos. 1998. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature 396474-477. [DOI] [PubMed] [Google Scholar]
- 6.Dodge-Kafka, K. L., J. Soughayer, G. C. Pare, J. J. Carlisle Michel, L. K. Langeberg, M. S. Kapiloff, and J. D. Scott. 2005. The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature 437574-578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dumaz, N., and R. Marais. 2005. Integrating signals between cAMP and the RAS/RAF/MEK/ERK signalling pathways. Based on the anniversary prize of the Gesellschaft fur Biochemie und Molekularbiologie Lecture delivered on 5 July 2003 at the Special FEBS Meeting in Brussels. FEBS J. 2723491-3504. [DOI] [PubMed] [Google Scholar]
- 8.Fabian, J. R., A. B. Vojtek, J. A. Cooper, and D. K. Morrison. 1994. A single amino acid change in Raf-1 inhibits Ras binding and alters Raf-1 function. Proc. Natl. Acad. Sci. USA 915982-5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Franke, B., J.-W. Akkerman, and J. L. Bos. 1997. Rapid Ca2+-mediated activation of rap1 in human platelets. EMBO J. 16252-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freedman, T. S., H. Sondermann, G. D. Friedland, T. Kortemme, D. Bar-Sagi, S. Marqusee, and J. Kuriyan. 2006. A Ras-induced conformational switch in the Ras activator Son of sevenless. Proc. Natl. Acad. Sci. USA 10316692-16697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo, Z., M. R. Ahmadian, and R. S. Goody. 2005. Guanine nucleotide exchange factors operate by a simple allosteric competitive mechanism. Biochemistry 4415423-15429. [DOI] [PubMed] [Google Scholar]
- 12.Gureasko, J., W. J. Galush, S. Boykevisch, H. Sondermann, D. Bar-Sagi, J. T. Groves, and J. Kuriyan. 2008. Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat. Struct. Mol. Biol. 15452-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock, J. F. 2003. Ras proteins: different signals from different locations. Nat. Rev. Mol. Cell Biol. 4373-384. [DOI] [PubMed] [Google Scholar]
- 14.Harding, A., T. Tian, E. Westbury, E. Frische, and J. F. Hancock. 2005. Subcellular localization determines MAP kinase signal output. Curr. Biol. 15869-873. [DOI] [PubMed] [Google Scholar]
- 15.Hisata, S., T. Sakisaka, T. Baba, T. Yamada, K. Aoki, M. Matsuda, and Y. Takai. 2007. Rap1-PDZ-GEF1 interacts with a neurotrophin receptor at late endosomes, leading to sustained activation of Rap1 and ERK and neurite outgrowth. J. Cell Biol. 178843-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holz, G. G., G. Kang, M. Harbeck, M. W. Roe, and O. G. Chepurny. 2006. Cell physiology of cAMP sensor Epac. J. Physiol. 5775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, C.-C., J.-L. You, M.-Y. Wu, and K.-S. Hsu. 2004. Rap1-induced p38 mitogen-activated protein kinase activation facilitates AMPA receptor trafficking via the GDI.Rab5 complex. Potential role in (S)-3,5-dihydroxyphenylglycene-induced long term depression. J. Biol. Chem. 27912286-12292. [DOI] [PubMed] [Google Scholar]
- 18.Impey, S., S. R. McCorkle, H. Cha-Molstad, J. M. Dwyer, G. S. Yochum, J. M. Boss, S. McWeeney, J. J. Dunn, G. Mandel, and R. H. Goodman. 2004. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 1191041-1054. [DOI] [PubMed] [Google Scholar]
- 19.Je, H. S., F. Yang, J. Zhou, and B. Lu. 2006. Neurotrophin 3 induces structural and functional modification of synapses through distinct molecular mechanisms. J. Cell Biol. 1751029-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kao, S., R. K. Jaiswal, W. Kolch, and G. E. Landreth. 2001. Identification of the mechanisms regulating the differential activation of the MAPK cascade by epidermal growth factor and nerve growth factor in PC12 cells. J. Biol. Chem. 27618169-18177. [DOI] [PubMed] [Google Scholar]
- 21.Kashima, Y., T. Miki, T. Shibasaki, N. Ozaki, M. Miyazaki, H. Yano, and S. Seino. 2001. Critical role of cAMP-GEFII-Rim2 complex in incretin-potentiated insulin secretion. J. Biol. Chem. 27646046-46053. [DOI] [PubMed] [Google Scholar]
- 22.Kawasaki, H., G. M. Springett, N. Mochizuki, S. Toki, M. Nakaya, M. Matsuda, D. E. Housman, and A. M. Graybiel. 1998. A family of cAMP-binding proteins that directly activate Rap1. Science 2822275-2279. [DOI] [PubMed] [Google Scholar]
- 23.Krapivinsky, G., L. Krapivinsky, Y. Manasian, A. Ivanov, R. Tyzio, C. Pellegrino, Y. Ben-Ari, D. E. Clapham, and I. Medina. 2003. The NMDA receptor is coupled to the ERK pathway by a direct interaction between NR2B and RasGRF1. Neuron 40775-784. [DOI] [PubMed] [Google Scholar]
- 24.Lein, P. J., X. Guo, G. X. Shi, M. Moholt-Siebert, D. Bruun, and D. A. Andres. 2007. The novel GTPase Rit differentially regulates axonal and dendritic growth. J. Neurosci. 274725-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, W., D. Knowlton, W. R. Woodward, and B. A. Habecker. 2003. Regulation of noradrenergic function by inflammatory cytokines and depolarization. J. Neurochem. 86774-783. [DOI] [PubMed] [Google Scholar]
- 26.Li, Y., S. Asuri, J. F. Rebhun, A. F. Castro, N. C. Paranavitana, and L. A. Quilliam. 2006. The RAP1 guanine nucleotide exchange factor Epac2 couples cyclic AMP and Ras signals at the plasma membrane. J. Biol. Chem. 2812506-2514. [DOI] [PubMed] [Google Scholar]
- 27.Limpert, A. S., J. C. Karlo, and G. E. Landreth. 2007. Nerve growth factor stimulates the concentration of TrkA within lipid rafts and extracellular signal-regulated kinase activation through c-Cbl-associated protein. Mol. Cell. Biol. 275686-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Margarit, S. M., H. Sondermann, B. E. Hall, B. Nagar, A. Hoelz, M. Pirruccello, D. Bar-Sagi, and J. Kuriyan. 2003. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell 112685-695. [DOI] [PubMed] [Google Scholar]
- 29.McFall, A., A. Ulku, Q. T. Lambert, A. Kusa, K. Rogers-Graham, and C. J. Der. 2001. Oncogenic Ras blocks anoikis by activation of a novel effector pathway independent of phosphatidylinositol 3-kinase. Mol. Cell. Biol. 215488-5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mitin, N., K. L. Rossman, and C. J. Der. 2005. Signaling interplay in Ras superfamily function. Curr. Biol. 15R563-R574. [DOI] [PubMed] [Google Scholar]
- 31.Nassar, N., G. Horn, C. Herrmann, A. Scherer, F. McCormick, and A. Wittinghofer. 1995. The 2.2 Å crystal structure of the Ras-binding domain of the serine/threonine kinase cRaf1 in complex with Rap1A and a GTP analogue. Nature 375554-560. [DOI] [PubMed] [Google Scholar]
- 32.Obara, Y., K. Labudda, T. J. Dillon, and P. J. Stork. 2004. PKA phosphorylation of Src mediates Rap1 activation in NGF and cAMP signaling in PC12 cells. J. Cell Sci. 1176085-6094. [DOI] [PubMed] [Google Scholar]
- 33.Ozaki, N., T. Shibasaki, Y. Kashima, T. Miki, K. Takahashi, H. Ueno, Y. Sunaga, H. Yano, Y. Matsuura, T. Iwanaga, Y. Takai, and S. Seino. 2000. cAMP-GEFII is a direct target of cAMP in regulated exocytosis. Nat. Cell Biol. 2805-811. [DOI] [PubMed] [Google Scholar]
- 34.Qiao, J., F. C. Mei, V. L. Popov, L. A. Vergara, and X. Cheng. 2002. Cell cycle-dependent subcellular localization of exchange factor directly activated by cAMP. J. Biol. Chem. 27726581-26586. [DOI] [PubMed] [Google Scholar]
- 35.Rehmann, H. 2006. Characterization of the activation of the Rap-specific exchange factor Epac by cyclic nucleotides. Methods Enzymol. 407159-173. [DOI] [PubMed] [Google Scholar]
- 36.Rehmann, H., E. Arias-Palomo, M. A. Hadders, F. Schwede, O. Llorca, and J. L. Bos. 2008. Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B. Nature 455124-127. [DOI] [PubMed] [Google Scholar]
- 37.Rehmann, H., J. Das, P. Knipscheer, A. Wittinghofer, and J. L. Bos. 2006. Structure of the cyclic-AMP-responsive exchange factor Epac2 in its auto-inhibited state. Nature 439625-628. [DOI] [PubMed] [Google Scholar]
- 38.Rehmann, H., B. Prakash, E. Wolf, A. Rueppel, J. De Rooij, J. L. Bos, and A. Wittinghofer. 2003. Structure and regulation of the cAMP-binding domains of Epac2. Nat. Struct. Biol. 1026-32. [DOI] [PubMed] [Google Scholar]
- 39.Rehmann, H., A. Rueppel, J. L. Bos, and A. Wittinghofer. 2003. Communication between the regulatory and the catalytic region of the cAMP-responsive guanine nucleotide exchange factor Epac. J. Biol. Chem. 27823508-23514. [DOI] [PubMed] [Google Scholar]
- 40.Repasky, G. A., E. J. Chenette, and C. J. Der. 2004. Renewing the conspiracy theory debate: does Raf function alone to mediate Ras oncogenesis? Trends Cell Biol. 14639-647. [DOI] [PubMed] [Google Scholar]
- 41.Riccio, A., B. A. Pierchala, C. L. Ciarallo, and D. D. Ginty. 1997. An NGF-TrkA-mediated retrograde signal to transcription factor CREB in sympathetic neurons. Science 2771097-1100. [DOI] [PubMed] [Google Scholar]
- 42.Rodriguez-Viciana, P., C. Sabatier, and F. McCormick. 2004. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol. Cell. Biol. 244943-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudolph, J. L., G. X. Shi, E. Erdogan, A. P. Fields, and D. A. Andres. 2007. Rit mutants confirm role of MEK/ERK signaling in neuronal differentiation and reveal novel Par6 interaction. Biochim. Biophys. Acta 17731793-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasagawa, S., Y. Ozaki, K. Fujita, and S. Kuroda. 2005. Prediction and validation of the distinct dynamics of transient and sustained ERK activation. Nat. Cell Biol. 7365-373. [DOI] [PubMed] [Google Scholar]
- 45.Sawada, Y., K. Nakamura, K. Doi, K. Takeda, K. Tobiume, M. Saitoh, K. Morita, I. I. Komuro, K. De Vos, M. Sheetz, and H. Ichijo. 2001. Rap1 is involved in cell stretching modulation of p38 but not ERK or JNK MAP kinase. J. Cell Sci. 1141221-1227. [DOI] [PubMed] [Google Scholar]
- 46.Shi, G. X., H. Rehmann, and D. A. Andres. 2006. A novel cyclic AMP-dependent Epac-Rit signaling pathway contributes to PACAP38-mediated neuronal differentiation. Mol. Cell. Biol. 269136-9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stork, P. J. 2005. Directing NGF's actions: it's a Rap. Nat. Cell Biol. 7338-339. [DOI] [PubMed] [Google Scholar]
- 48.Stork, P. J. S., and J. M. Schmitt. 2002. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 12258-266. [DOI] [PubMed] [Google Scholar]
- 49.van de Wetering, M., I. Oving, V. Muncan, M. T. Pon Fong, H. Brantjes, D. van Leenen, F. C. Holstege, T. R. Brummelkamp, R. Agami, and H. Clevers. 2003. Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep. 4609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waltereit, R., and M. Weller. 2003. Signaling from cAMP/PKA to MAPK and synaptic plasticity. Mol. Neurobiol. 2799-106. [DOI] [PubMed] [Google Scholar]
- 51.Wang, Z., T. J. Dillon, V. Pokala, S. Mishra, K. Labudda, B. Hunter, and P. J. Stork. 2006. Rap1-mediated activation of extracellular signal-regulated kinases by cyclic AMP is dependent on the mode of Rap1 activation. Mol. Cell. Biol. 262130-2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.White, M. A., C. Nicolette, A. Minden, A. Polverino, L. Van Aelst, M. Karin, and M. H. Wigler. 1995. Multiple Ras functions can contribute to mammalian cell transformation. Cell 80533-541. [DOI] [PubMed] [Google Scholar]
- 53.York, R. D., H. Yao, T. Dillon, C. L. Ellig, S. P. Eckert, E. W. McCleskey, and P. J. S. Stork. 1998. Rap1 mediates sustained MAP kinase activation induced by nerve growth factor. Nature 392622-625. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, X., D. T. Odom, S. H. Koo, M. D. Conkright, G. Canettieri, J. Best, H. Chen, R. Jenner, E. Herbolsheimer, E. Jacobsen, S. Kadam, J. R. Ecker, B. Emerson, J. B. Hogenesch, T. Unterman, R. A. Young, and M. Montminy. 2005. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. USA 1024459-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.