Abstract
The loss of E-cadherin gene expression can cause the dysfunction of the cell-cell junction to trigger tumor metastasis. Members of the Snail family of transcription factors are repressors of the expression of the E-cadherin gene. In this study, we showed that the activated androgen receptor (AR) is a novel repressor of E-cadherin gene expression and can promote metastasis. Our results demonstrated that the activated AR could bind to the E-cadherin promoter in vitro and in vivo. The activated AR and HDAC1 had synergistic effects in downregulating E-cadherin gene expression. Treating cells with the AR ligand, dihydrotestosterone (DHT), triggered the reduction of E-cadherin expression and induced changes in cell morphology from an epithelial-like to a mesenchymal-like appearance. When nonmetastatic breast cancer cells expressing cytoplasmic AR were transplanted into mice and the mice were treated with DHT, tumors were detected at metastatic sites, whereas no tumors were detected in transplanted mice without DHT treatment. Furthermore, clinical data from breast cancer patients with invasive ductal carcinomas showed high levels of AR expression in the nuclei and low levels of E-cadherin expression. These results suggest that, similarly to Snail and Twist, the activated AR can downregulate E-cadherin expression to promote the activation of epithelial-mesenchymal transition and tumor metastasis.
The efforts of cancer research from the past several decades have led to an understanding of the mechanisms of tumorigenesis (23, 24, 47). However, cancer death remains one of the top killers in annual mortality reports from public health agencies. More than 95% of cancer deaths are due to cancer metastasis. The metastatic progression is a complex multistep process. Malignant tumors invade and break out of the confinement of adjacent tissues and travel to distant sites, where they establish new cancer colonies (14, 36). At the mechanistic level, for cancer cells to develop into metastatic cancer cells at least four interrelated processes are involved: (i) the activation of epithelial-mesenchymal transition (EMT) (51), (ii) the remodeling of the extracellular matrix (44), (iii) neoangiogenesis (8), and (iv) migration to specific secondary sites (38).
EMT is a vital process that controls morphogenic changes in multicellular organisms during embryonic development (50). EMT leads epithelial cell layers to lose polarity and cell-cell contacts and triggers the remodeling of the cytoskeleton. Activated EMT is also a necessary process for the development of invasion (51). It allows many epithelial tumor cells to increase cell motility and become metastatic cancer cells. E-cadherin is regarded as a main indicator of the epithelial/mesenchymal phenotype switch (29, 39). It plays a critical role in establishing cell polarity, cellular differentiation, and maintaining cell structure. The downregulation of E-cadherin has been implicated in the activation of EMT (15, 25, 45). Therefore, E-cadherin has been suggested as a tumor suppressor in various carcinomas.
We still do not know the genetic or epigenetic basis of tumor metastasis. Thus, the study of regulatory mechanisms of genes related to metastasis is essential for a better understanding of the molecular changes that turn cancer cells into metastatic cancer cells. E-cadherin has been used as an indicator in the study of the repression of tumor metastasis, especially in the mouse E-cadherin gene (4, 11, 49). Several mechanisms have been suggested for the loss of E-cadherin expression during tumor metastasis (11, 49). The hypermethylation of the E-cadherin gene promoter, the deacetylation of chromatin, and a host of repressive factors binding to elements on the E-cadherin regulatory sequence have emerged as the main mechanisms in most carcinomas (3, 5, 7, 18, 19, 33, 41-43, 48, 54). Snail, the transcription factor of the zinc finger class, is a strong repressor of E-cadherin transcription and a well-known inducer of EMT (18, 19). Other repressors of the E-cadherin gene that have been implicated in EMT are the Ebox binding proteins, such as Snail family member Slug (5), the basic helix-loop-helix factors E47 (43) and Twist (54), and the two-handed zinc factors ZEBl and SIPl (5). Most studies on the repression of the E-cadherin gene have focused on the Eboxes that are proximal to the transcription start site of the E-cadherin gene. The functions of regulatory elements distal to the transcription start site of the human E-cadherin gene still are poorly understood.
Our previous study that focused on the regulation of human E-cadherin gene expression in metastatic and nonmetastatic cancer cells showed that both methylation states and chromatin constraints played important roles in the downregulation of E-cadherin gene expression (33). We also identified an additional Snail binding site that could repress E-cadherin gene activity. In addition, we showed that HNF3, a member of the Fork Head domain-containing transcription factor (28), could bind the enhancer elements on the regulatory sequence of the E-cadherin gene and could reverse EMT activity to reduce cell mobility. In that study, we also had detected a binding element for a putative repressor located between −357 and −195 on the human E-cadherin regulatory sequence that appeared to be different from the Snail and Twist binding sites. There may be a novel transcription factor that binds to this negative regulatory element to suppress the human E-cadherin gene.
In the present report, we extend that study to characterize this element and identify factors interacting with this element. Our results show that the novel repressor for the E-cadherin gene is the androgen receptor (AR). We show that the AR is expressed in most breast cancer cell lines and some colon and lung cancer cell lines. In nonmetastatic cancer cells, the AR is sequestered in the cytoplasm. In the presence of adequate concentrations of the steroid hormone dihydrotestosterone (DHT), the AR is activated and translocated into the nuclei to repress the E-cadherin gene. When nonmetastatic breast cancer cells expressing cytoplasmic AR are transplanted into mice and the mice are treated with DHT, the cancer cells survive and proliferate at distant sites to form new cancer colonies. Our results show that the activated AR is a repressor of E-cadherin gene expression, and it can increase the metastatic potential of cancer cells. These results also are corroborated by pathological observations showing an increase in nuclear AR and a decrease in E-cadherin staining in invasive breast carcinomas.
MATERIALS AND METHODS
Generation of plasmids and stable cell lines.
The E-cadherin regulatory sequence from 357 bp upstream to 135 bp downstream of the E-cadherin transcription start site was cloned by genomic PCR, using human genomic DNA as a template. Variously sized deletions of the promoter also were generated by PCR with various 5′ primers and a fixed 3′ primer (data not shown). The PCR program was 95°C for 5 min, followed by 35 cycles of 95°C for 1 min, 56°C for 1 min, and 72°C for 1 min. At the end of the PCR, the reaction was extended for 8 min at 72°C and then the mixture was cooled to 4°C. Aliquots of the PCR products were analyzed by electrophoresis in 1% agarose. The expected sizes of the PCR fragments were 510, 451, 401, 380, 355, and 330 bp. All serially deleted E-cadherin promoter fragments were cloned into luciferase reporter vector pGL3 (Promega). The HDAC1 and HDAC3 cDNA expression vectors were obtained from H. Y. Kao, Case Western Reserve University, Cleveland, OH. All E-cadherin regulatory sequence mutants were generated by site-directed mutagenesis PCR with various primers (data not shown). The PCR products were incubated with 1 μl DpnI (New England BioLabs). The DpnI-treated PCR products were transformed into Escherichia coli strain XL1-blue. All mutant constructs were verified by sequencing.
The T47D/AR and MCF7/AR transfectants were generated by stable transfection with 10 μg of plasmid AR/pcDNA3 into T47D and MCF7 cells, respectively, and were selected with 400 μg/ml of G418 for 1 month. The TARS (T47D/AR-DHT-selected) and MARS (MCF7/AR-DHT-selected) cells were cloned from T47D/AR and MCF7/AR stable transfectants, respectively, that were infiltrated from 8.0-μm-pore-size transwell inserts (Nunc) after DHT treatment.
Cell culture and treatment.
The following cell lines were used in this study: breast cancer cell lines T47D, MCF7, MDA-MD-435, and MDA-MD-231; melanoma cell line A2058; colon cancer cell lines LoVo and DLD1; and hepatocarcinoma cell line Huh7. All cell lines were grown in Dulbecco's modified Eagle's medium (DMEM) (Sigma) supplemented with 10% fetal bovine serum. Cells were grown at 37°C in a 5% CO2 atmosphere. DHT and methyltrienolone (R1881; Sigma) were dissolved in methanol and added to the DMEM at 10 nM. A corresponding volume of methanol was added to the control untreated cells. Trichostatin A (TSA) (Sigma) was dissolved in ethanol and added to the DMEM at 300 nM. A corresponding volume of ethanol was added to the control untreated cells.
Luciferase reporter assay.
Cells (1 × 105 per well) were seeded in 24-well plates with 0.5 ml of an appropriate DMEM. One microgram of various DNA constructs and 0.1 μg of Renilla construct (Promega) were mixed with 10 μl of Superfect (Qiagen). The mixture was incubated at room temperature for 10 min. After the cells were washed with 1× phosphate-buffered saline (PBS), the DNA/Superfect mixtures were transferred to the cells and incubated at 37°C in a CO2 incubator for 24 h. During the cotransfection with constructs expressing various transcription factors, appropriate control plasmids such as pcDNA3 and pGL3 also were transfected into separate cultured cells as controls, and DNA was maintained in equal amounts. Subsequently, the transfected cells were washed with 1× PBS and cultured for an additional 48 h in DMEM. Seventy-two hours after transfection, the transfected cells were lysed with reporter lysis buffer (Promega). The enzymatic activity was measured for firefly and Renilla luciferase using the dual-luciferase reporter assay system (Promega) with a luminometer (MGM). The activities were the averages from three experiments. The luciferase activities were expressed as the change in activity (n-fold) over that of the control vector.
RNA isolation and reverse transcription-PCR (RT-PCR).
When cells reached 80% confluence they were harvested, and the RNA was extracted using the acid quanidinium method (10). The RNA (4 μg) from each cell line was converted into single-strand cDNA primed by oligo(dT) using the SuperScript preamplification system for first-strand cDNA synthesis kit (Promega) by following the vender's protocol. One microliter of each single-strand cDNA was used as a template to generate the E-cadherin, AR, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) partial sequences of cDNA (data not shown). The PCR program was 95°C for 5 min followed by 35 cycles of 95°C for 1 min, 56°C for 1 min, and 72°C for 1 min. At the end of the PCR, the reaction was extended for 8 min at 72°C, and then the mixture was cooled to 4°C. Aliquots of the PCR products were analyzed by electrophoresis in 1% agarose.
Cell extraction and Western blotting.
After washing cells with 1× PBS twice, cells were scraped with 0.5 ml 1× PBS and collected in a microcentrifuge tube. The cells were spun at 3,000 rpm for 5 min, and the supernatants were discarded. Pellets were resuspended with 1 ml radioimmunoprecipitation assay buffer (50 mM Tris, 0.5% NP-40, 150 mM NaCl, 20% glycerol, and 2 mM dithiothreitol [DTT]) containing protease inhibitor (Roche), and the samples were incubated on ice for 30 min. These samples then were spun at 10,000 rpm for 10 min at 4°C, and the supernatants were collected. The methods for the preparation of the nuclear and cytoplasmic extracts were the same as those for the chromatin immunoprecipitation (ChIP) assay described below. The protein concentrations of cell extracts were measured using a protein assay (Bio-Rad). One hundred micrograms of the total cell extracts from each cell line was electrophoresed in a sodium dodecyl sulfate (SDS)-6% polyarcrylamide gel. The separated proteins were transferred to polyvinyl difluoride transfer membranes (Perkin-Elmer) for Western blot analysis. Membranes were blocked in 5% skim milk diluted with 1× PBST (1× PBS containing 0.1% Tween 20) for 1 h. The membranes then were incubated in diluted (1:2,000 diluted in 1× PBST) primary antibody (anti-E-cadherin, antivimentin, anti-AR, anti-ERK2, or antiactin antibody) (Santa Cruz) for 1 h and then washed three times (15 min each time) with 1× PBST. Membranes were incubated with diluted (1:5,000 diluted in 1× PBST) secondary antibody (anti-mouse, anti-rabbit, or anti-goat antibody) (Molecular Probes) at room temperature for 1 h. After being washed with 1× PBST, membranes were incubated in Western Lightning chemiluminescence reagent (PerkinElmer) for 1 min and then exposed to Kodak BioMax light film or UVP bioimaging systems (Biospectrum-AC w/Bio Chemi camera; UVP) for 1 to 5 min with repeated exposures for an optimal signal.
EMSA.
To prepare oligonucleotides for the electrophoresis mobility shift assay (EMSA), 500 ng of each sense and antisense oligonucleotide was mixed in 100 μl water, heated at 60°C for 15 min, and then slowly cooled to 4°C. Double-stranded oligonucleotides (100 ng) were end labeled with 32P (specific activity, 1 × 107/μg to 5 × 107/μg). For the EMSA reactions, 1 ng of labeled probe (10,000 to 50,000 cpm/sample) was mixed with nuclear extracts (2 μg/μl) in 50 μl EMSA buffer containing 10 mM HEPES, pH 7.9, 10 mM KCl, 5 mM MgCl2, 1 mM DTT, 1 μg poly(dI-dC), and 10% glycerol. The mixtures were kept on ice for 1 h and then loaded onto a 6% polyacrylamide gel and electrophoresed at 150 V in 1× TBE buffer (22.5 mM Tris-borate, 0.5 mM EDTA, pH 8.0). The sequence of the AR consensus binding site oligonucleotides is 5′-AGAACACCCTGTACC-3′.
For the supershift EMSA, 1 ng 32P-labeled probe (10,000 to 50,000 cpm/sample) was incubated with 2 μg of nuclear extract derived from MDA-MD-435 cells on ice for 30 min. The anti-immunoglobulin G (IgG) (0.5 and 1 μg/sample) (Sigma) or anti-AR (0.5 and 1 μg/sample) antibody (Santa Cruz) was added and incubated with mixtures at room temperature for 30 min and then loaded onto 6% polyacrylamide and electrophoresed at 150 V in 1× TBE buffer.
ChIP.
Cells used for the ChIP assay were fixed with formaldehyde directly in tissue culture medium at a final concentration of 1%. Cells were incubated on a shaking platform for 10 min at room temperature. The cross-linking reaction was stopped by adding glycine to a final concentration of 0.125 M at room temperature for 5 min. The medium was poured off, and the plate was rinsed twice with cold 1× PBS. Cells were scraped from the dishes after adding 5 ml of 1× PBS. The cells were pelleted by centrifugation at 1,500 rpm and then washed once with 1× PBS plus phenylmethylsulfonyl fluoride. The cell pellet was resuspended in cell lysis buffer [5 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES), pH 8.0, 85 mM KCl, 0.5% NP-40] plus the protease inhibitors and then incubated on ice for 10 min. The lysate was centrifuged at 5,000 rpm for 5 min at 4°C to pellet the nuclei. The supernatant was collected as the cytoplasmic extract for Western blotting. The nuclear pellet was resuspended in 1 ml nuclei lysis buffer (50 mM Tris-Cl, pH 8.0, 10 mM EDTA, 1% SDS) plus protease inhibitors and incubated on ice for 10 min. The lysate was collected as the nuclear extract for Western blotting. The chromatin was sonicated to yield a DNA size with an average length of approximately 0.5 to 1.0 kb. After sonication, samples were centrifuged at 14,000 rpm for 10 min at 4°C. The supernatant was carefully removed and transferred to a new tube. The chromatin mixture was incubated on a rotating platform at 4°C for 15 min, and then the mixture was centrifuged at 14,000 rpm for 4 min. The supernatant was transferred to a new tube and divided equally among immunoprecipitation (IP) samples. We generally added 1 μg of the specific antibody (anti-AR and anti-Flag; Santa Cruz and Sigma, respectively) to the appropriate samples. The chromatin samples were incubated with the antibodies on a rotating platform at 4°C overnight and then centrifuged at 14,000 rpm for 4 min. The supernatant was discarded, and the pellet was washed twice with 1.4 ml of 1× dialysis buffer (2 mM EDTA, 50 mM Tris-Cl, pH 8.0, 0.2% Sarkosyl) and then centrifuged at 14,000 rpm for 4 min, and all traces of the wash buffer were removed. One hundred fifty microliters of IP elution buffer (5 mM NaHCO3, 1% SDS) then was added, and the mixture was vortexed on setting 3 for 15 min at room temperature. The tubes were spun for 4 min, and the elution buffer was transferred to a fresh tube. After elution, the samples were centrifuged at 14,000 rpm for 4 min. The supernatants then were transferred to new tubes, and 12 μl of 5 M NaCl was added to a final concentration of 0.3 M. The samples were incubated at 67°C for 4 to 5 h to reverse the formaldehyde cross-links. After the incubation, 2.5 volumes of ethanol were added to each sample and precipitated at −20°C overnight. The samples then were centrifuged at 14,000 rpm for 20 min at 4°C. The supernatant was discarded and respun to remove any residual ethanol. The pellets were air dried and dissolved in 100 μl of Tris-EDTA buffer. Twenty-five microliters of 5× digestion buffer (50 mM Tris-Cl, pH 7.5, 25 mM EDTA, 1.25% SDS) and 1.5 μl of proteinase K (25 mg/ml) were added to each sample, and the samples were incubated at 45°C for 1 to 2 h. To remove all proteins and contaminants from the immunoprecipitated chromatin, the chromatin was extracted once with 300 μl of phenol-chloroform-isoamyl alcohol and once with 300 μl chloroform-isoamyl alcohol. After collecting the aqueous phase, 30 μl of 5 M NaCl and 5 μg of glycogen were added to each sample. Ethanol (750 μl) was added to the samples and mixed well. The DNA was precipitated in a −20°C freezer overnight and collected by centrifuging the samples at 14,000 rpm for 20 min at 4°C. The DNA was dissolved in water and analyzed by PCR. The program of the PCR was the same as that for the generation of cDNA with the primers mentioned above (data not shown). The PCR product was analyzed with a 1% agarose gel. The expected ChIP product was 322 bp.
siRNA transfection.
Cells (1 × 105 per well) were seeded on a 24-well plate in 0.5 ml of an appropriate culture medium containing serum and antibiotics before transfection. The AR siRNA or nonspecific siRNA was diluted in 100 μl serum-free medium to a final short interfering RNA (siRNA) concentration of 5, 12.5, or 25 nM. The sequence of the sense AR siRNA-1 was r(GGA ACU CGA UCG UAU CAU U)dTdT (r indicates RNA sequence). The diluted siRNAs were mixed with 3 μl HiPerFect transfection reagent (Qiagen) by vortexing. The samples were incubated at room temperature for 10 min to allow the formation of transfection complexes. The cells were incubated with the transfection complexes under their normal growth conditions, and the gene silencing was monitored after 72 h.
Migration assay.
Cell migration was performed using 8.0-μm-pore-size transwell inserts (Nunc). The six-well dish was prepared by adding 1.5 ml DMEM or 10 nM DHT containing DMEM to each well. An insert was placed into each prepared well with the membrane toward the well bottom. The cell suspension (2 × 105 cells in 1.5 ml DMEM or 10 nM DHT containing DMEM) was added to the interior of each insert. After 72 h of incubation, each well was trypsinized and the transwell cells were stained with trypan blue. The numbers of migratory cells were calculated by dividing by the total number of cells. Each data point represented the averages from three or four individual experiments.
IF staining.
For immunofluorescence (IF) staining, stably transfected cells were grown on coverslips and fixed in 1% formaldehyde (200 ml of formaldehyde mixed with 7.2 ml PHEM; PHEM contains 60 mM PIPES, 25 mM HEPES, pH 6.9, 10 mM EGTA, and 4 mM MgCl2) for 10 min, and 1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate was added to perforate cells at room temperature for 4 min. Samples then were blocked three times with skim milk at room temperature for 10 min and stained with mouse anti-E-cadherin or rabbit anti-AR (Santa Cruz) antibody (1:100) at 37°C for 1 h. The cells then were washed with MBST (10 mM morpholinepropanesulfonic acid, pH 7.4, 150 mM NaCl, 0.05% Tween-20) three times at room temperature for 10 min and then stained with secondary anti-mouse or anti-rabbit antibody (1:300; Molecular Probes) at 37°C for 1 h. Samples then were washed with MBST three times at room temperature for 10 min and then counterstained with 4′,6′-diamidino-2-phenylindole (DAPI) (1:500; Molecular Probes) at room temperature for 10 min. The cells were mounted on p-phenylenediamine, and the preparations were visualized with an immunofluorescence microscope.
Transplantation.
To mark cells with luciferase, TARS, T47D, and MDA-MD-435 (metastatic-positive control) cells were stably transfected with Luc/pcDNA3 vector. Cells were injected into the tail veins of NOD/SCID mice. NOD/SCID female mice, age matched between 5 and 7 weeks old, were used for intravenous injection. For DHT (Sigma) treatment studies, one group of TARS cell-transplanted mice (n = 8) was treated with 100 μg/ml DHT (dissolved in methanol and mixed 1:1 with gingili) every 3 days intramuscularly to maintain the plasma DHT concentration (2). A corresponding volume of methanol/gingili mixture was injected into the untreated mice (n = 6). The MDA-MD-435 transplantation mice (n = 4) were not treated with DHT.
IHC staining.
For immunohistochemistry (IHC) staining, 1× peroxide blocking buffer (Thermo) was applied to cover the specimen, and the samples were incubated at room temperature for 15 min. The sections were drained and blotted gently, and an appropriate volume of diluted primary anti-AR (1:500 diluted in 0.5× peroxide blocking buffer), anti-E-cadherin (1:500 diluted in 0.5× peroxide blocking buffer) (Santa Cruz), and antiluciferase (1:100 diluted in 0.5× peroxide blocking buffer) (Sigma) antibodies was added to cover the specimen according to tissue size. Likewise, negative control serum was added to the negative control slides. The slides were incubated at 4°C overnight. The samples were washed three times with 1× PBST for 10 min. An appropriate volume of Super Enhancer reagent (Biogenex) was added to cover the specimen, and the specimens were incubated for 30 min at room temperature. Samples were rinsed with 1× PBST, and an appropriate volume of poly-horseradish peroxidase reagent (Biogenex) was added to cover the specimen. The samples were incubated for 40 min at room temperature and then washed three times with 1× PBST for 10 min each. An appropriate volume of DAB substrate (Biogenex) solution was added to cover the specimen, and the samples were incubated for 2 min at room temperature and then washed three times with 1× PBST for 10 min each. The slides were immersed in a bath of Mayer's hematoxylin for 10 s and washed with water for 5 min. While the slides were still wet, coverslips were mounted with 1 to 2 drops of aqueous immu-mounting medium (Thermo).
Statistical analysis.
Data are shown as averages and standard deviations. We used the Student's t test for luciferase reporter analyses and migration analyses. All statistical analyses were done with Excel 2003 (Microsoft) with the Statcel2 add on (OMS). Comparisons between nuclear AR expression levels and E-cadherin expression patterns with clinical pathological parameters were evaluated using Student's t or the Welch test. A P value of <0.05 was considered to have statistical significance.
RESULTS
The novel repressor binding element is the binding site for the AR.
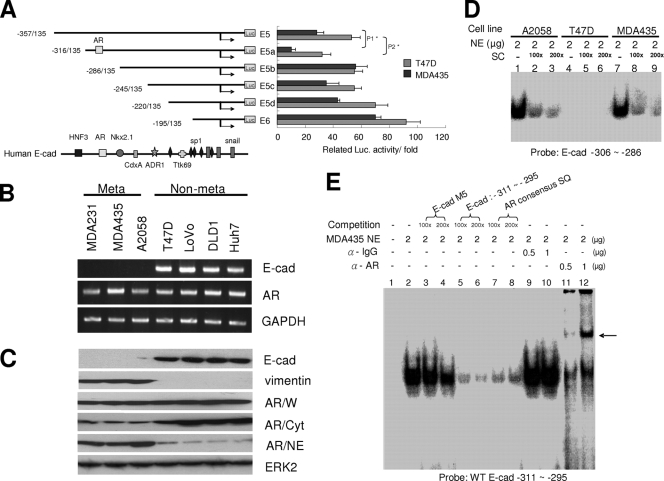
To identify the novel repressor binding element, we serially deleted the human E-cadherin regulatory sequence from −357 to −195 into four fragments and cloned them into the luciferase reporter vector. Several transcription factor binding sites were identified using the TFsearch software program (Fig. 1A). Our results showed that the construct E5a had greatly reduced reporter activity. When the 30 bp of nucleotides from −318 to −286 were deleted the reporter activity was restored, suggesting that the 30-bp DNA sequence might contain a potent repressive element. The 30-bp DNA contained the AR binding element, suggesting that the AR could be a negative regulator for human E-cadherin expression. To determine the expression levels of the AR and the E-cadherin gene in metastatic and nonmetastatic breast cancer cell lines, we carried out RT-PCR (Fig. 1B) and Western blotting (Fig. 1C). Although the AR was detected in all of the cancer cell lines, most of the AR was present in the nuclear extracts of metastatic cancer cells. EMSA also showed that the binding complexes were detectable in nuclear extracts that were derived from metastatic cancer cells (A2058 and MDA-MD-435) but not from that of nonmetastatic cancer cells (T47D) (Fig. 1D). To characterize the AR binding site further, we used increasing concentrations of oligonucleotides representing an AR consensus binding element to compete with the probe that is the AR binding element of the E-cadherin gene. A decrease in probe/nuclear extract complexes was indicated by EMSA (Fig. 1E, lanes 7 and 8). Furthermore, when anti-AR antibody was used in EMSA, supershifts of the complex were observed (Fig. 1E, lanes 11 and 12). In addition, the ChIP assay showed that the AR could interact with the E-cadherin regulatory sequence in vivo in DHT-treated T47D cells (Fig. 2A, left, lane 4) but not in T47D cells without DHT treatment (Fig. 2A, left, lane 1). However, the AR could interact with the E-cadherin gene in metastatic cancer cells (MDA-MB-435) when treated either with or without DHT (Fig. 2A, right, lanes 9 and 12). These results suggest that the AR is sequestered in the cytoplasm of nonmetastatic breast cancer cells and needs to be activated to translocate into the nucleus to interact with the E-cadherin regulatory sequence.
FIG. 1.
Possible AR binding site is located between −306 and −286 of the human E-cadherin gene regulatory sequence. (A) E-cadherin promoter activity was analyzed in E-cadherin-positive (T47D) and -negative (MDA-MD-435) cells. E-cadherin reporter constructs of various lengths were transfected into T47D and MDA-MD-435 cells. Forty-eight hours after transfection, the cell lysates were analyzed for luciferase activities. The luciferase (Luc.) activities were expressed as the change in activity compared to that of the control vector. P values: P1 = 0.00933; P2 = 0.02487 (Student's t test). (B) The AR, E-cadherin (E-cad), and GAPDH transcripts in various tumor cells. Equal amounts (4 μg) of total RNA from various cell lines were converted to cDNAs by reverse transcriptase. These cDNAs were used as the templates for PCR using primers designed from E-cadherin, AR, and GAPDH genes that covered partial coding sequences. Meta, metastatic; Non-meta, nonmetastatic. (C) The expression of E-cadherin, vimentin, AR, and ERK2 proteins in various cancer cells. Equal amounts (100 μg) of whole cell (W), cytoplasmic (Cyt), and nuclear (NE) extracts were analyzed by Western blotting with anti-E-cadherin, anti-AR, and anti-ERK2 antibodies. (D) Interaction between the E-cadherin regulatory sequence and various nuclear extracts. The 32P-labeled probe of wild-type E-cadherin oligonucleotides (E-cad −306 ∼ −286) was mixed with nuclear extracts derived from various cancer cell lines (A2058, lanes 1 to 3; T47D, lanes 4 to 6; and MDA-MD-435, lanes 7 to 9). The reaction products were analyzed by EMSA. The competing oligonucleotides are different concentrations of homologous probe sequences (SC). (E) Interaction between AR and the E-cadherin regulatory sequence by EMSA. The 32P-labeled wild-type E-cadherin oligonucleotides (WT E-cad −306 ∼ −286) were mixed with the MDA-MD-435 nuclear extract. The competing oligonucleotides represent the mutant of the probe (M3) (lanes 3 and 4), homologous probe sequences (lanes 5 and 6), and the consensus binding site of the AR (lanes 7 and 8). The arrow points to the supershift with anti-AR antibody (α-AR) (lanes 11 and 12). α-IgG, anti-IgG antibody. SQ, sequence.
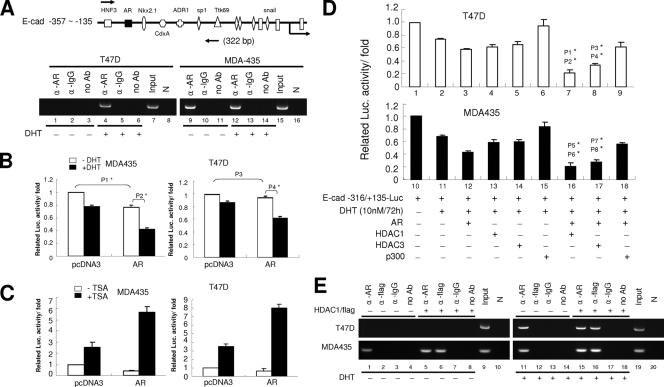
FIG. 2.
Activated AR cooperates with HDAC1 and recruits onto E-cadherin regulatory sequences. (A) Binding of the ligand-activated AR on the E-cadherin gene analyzed by ChIP assay. (Top) Sketch of the E-cadherin regulatory sequence and various transcription factor binding motifs. Arrows indicate the size of the PCR product. (Bottom) Nonmetastatic (T47D) and metastatic (MDA-MD-435) cells were treated with 10 nM DHT for 72 h (lanes 4 to 6 and lanes 12 to 14) or were left untreated (lanes 1 to 3 and lanes 9 to 11). Chromatin extracts were immunoprecipitated with anti-AR (α-AR) antibody, anti-IgG (α-IgG) antibody (Ab), or without antibodies and were analyzed by the ChIP assay as described in Materials and Methods. (B) The activity of the E-cadherin promoter was analyzed in MDA-MD-435 and T47D cells in the presence of DHT. E-cadherin reporter vector was cotransfected with the AR expression vector or empty vector into various cells, and the cells were further treated with 10 nM DHT (black) or were left untreated (white). Seventy-two hours after DHT treatment, luciferase (Luc.) activities were determined. P values: P1 = 0.03377; P2 = 0.01043; P3 = 0.08933; P4 = 0.01877 (Student's t test). (C) The E-cadherin promoter activity was analyzed in MDA-MD-435 and T47D cells in the presence of TSA. The E-cadherin reporter vector was cotransfected with the AR expression vector or empty vector into various cells, and cells were treated with 10 nM DHT. Forty-eight hours after DHT treatment, cells were treated with 300 nM TSA (black) or were left untreated (white). Twenty-four hours after TSA treatment, luciferase activities were determined. (D) The E-cadherin promoter activity was analyzed in T47D (white) and MDA-MD-435 (black) cells in the presence of AR, HDAC1, HDAC3, p300, and pcDNA3 expression vectors. Various expression vectors were cotransfected with E-cadherin reporter vector, and then cells were treated with 10 nM DHT or were left untreated. Seventy-two hours after DHT treatment, luciferase activities were analyzed. P values: P1 = 0.00734 (lane 3 versus lane 7); P2 = 0.00603 (lane 4 versus lane 7); P3 = 0.01087 (lane 3 versus lane 8); P4 = 0.01166 (lane 5 versus lane 8); P5 = 0.01732 (lane 3 versus lane 7); P6 = 0.00912 (lane 4 versus lane 7); P7 = 0.01848 (lane 3 versus lane 8); P8 = 0.01498 (lane 5 versus lane 8) (Student's t test). (E) Recruitment of the activated AR and HDAC1 onto the E-cadherin gene, as analyzed by ChIP assay. T47D and MDA-MD-435 cells were transiently transfected with Flag-tagged HDAC1 expression vector (lanes 5 to 8 and lanes 15 to 18) or were left untransfected (lanes 1 to 4 and lanes 11 to 14) and then were treated with 10 nM DHT for 72 h (lanes 1 to 8) or were left untreated (lanes 11 to 18). Chromatin extracts were immunoprecipitated with anti-AR, anti-Flag, or anti-IgG antibody or without antibodies and were analyzed by ChIP assay as described in Materials and Methods. N, negative control.
Activated AR cooperates with HDAC1 to repress E-cadherin expression.
To test whether the AR is a negative regulator of the human E-cadherin gene in metastatic and nonmetastatic breast cancer cells, we cotransfected the AR expression vector and E-cadherin regulatory sequence reporter construct into MDA-MD-435 and T47D cells and treated cells with the AR ligand DHT or left them untreated. In the presence of DHT, the AR expression vector could reduce E-cadherin reporter activity in both metastatic and nonmetastatic breast cancer cells (Fig. 2B). In the absence of DHT, the overexpressed AR could not reduce the E-cadherin reporter activity in T47D cells, but the AR could reduce E-cadherin reporter activity in MDA-MD-435 cells (Fig. 2B). These results suggest that the repressive ability of the AR is ligand dependent in nonmetastatic breast cancer cells. To test the possibility that the repressive effect of the activated AR is due to the remodeling of the chromatin structure, the AR expression vector and E-cadherin reporter constructs were cotransfected into MDA-MD-435 and T47D cells. The DHT-treated cells were further treated with the histone deacetylase (HDAC) inhibitor TSA or were left untreated. The results showed that when cells were treated with TSA, E-cadherin reporter activities were increased in the presence of DHT (Fig. 2C). This suggests that the repressive effect of the activated AR on E-cadherin expression involves histone deacetylation and requires the cooperation of HDACs.
To further identify the cooperative effects of the AR with chromatin-remodeling factors (e.g., HDACs) in metastatic and nonmetastatic breast cancer cells, HDAC1 (or HDAC3) and AR expression vectors were cotransfected into MDA-MD-435 and T47D cells. The cotransfection of AR and HDAC1/3 expression vectors reduced more luciferase activity than AR or HDAC1/3 transfection alone in the presence of DHT (Fig. 2D). These results suggest that the cooperation between the activated AR and HDAC1/3 can downregulate E-cadherin gene expression. In addition, the ChIP assay also showed that the AR could cooperate with HDAC1 to interact with the E-cadherin gene in both nonmetastatic and metastatic cancer cells in the presence of DHT (Fig. 2E, lanes 15 and 16). Taken together, these results suggest that the activated AR cooperates with HDAC1 to interact with the E-cadherin promoter and to repress E-cadherin gene expression.
Activated AR changes the cell morphology and expression of E-cadherin and vimentin in nonmetastatic breast carcinomas.
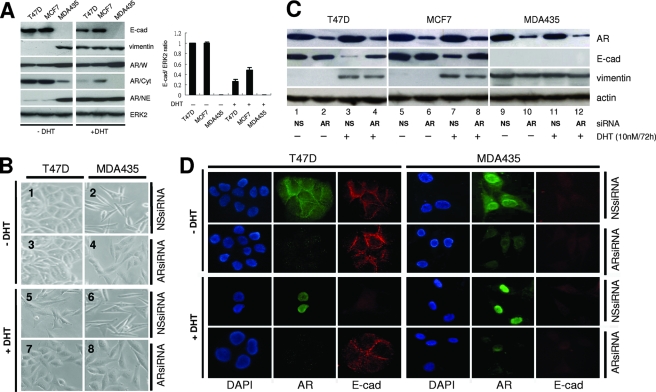
When cells were treated with 10 nM DHT, the E-cadherin expression was reduced and the vimentin expression was increased in MCF7 and T47D cells (Fig. 3A). In order to see whether the activated AR could induce changes in cell morphology, cells were treated with DHT and examined for changes in cell morphology. Seventy-two hours after DHT treatment, T47D cells had changed from epithelial-like to mesenchymal-like (Fig. 3B, image 5). In contrast, cells that were not treated with DHT remained epithelial-like (Fig. 3B, image 1). Furthermore, when AR siRNA was introduced into cells and the cells were treated with DHT, the cells retained their epithelial morphology (Fig. 3B, image 7). Western blotting also showed the reduction of E-cadherin expression and the induction of vimentin expression in MCF7 and T47D cells (Fig. 3C, lanes 3 and 7). AR siRNA could reverse the activated AR-mediated suppression of the E-cadherin gene and could increase E-cadherin expression in DHT-treated cells (Fig. 3C, lanes 4 and 8). An IF staining assay was used to determine whether the activated AR was translocated from the cytoplasm into the nucleus to suppress E-cadherin expression in nonmetastatic breast cancer cells. In the presence of DHT, the AR was translocated into the nucleus from the cytoplasm in T47D cells (Fig. 3D, left). At the same time, the E-cadherin expression that should have been located in cell-cell adhesion was reduced. However, the above phenomena were not observed in AR siRNA-treated cells. Thus, the endogenous AR can be activated by DHT and is translocated into the nucleus, where it represses E-cadherin gene expression in nonmetastatic breast cancer cells and changes the morphology of these cells.
FIG. 3.
Ligand induces the translocation of AR and changes the protein expression of E-cadherin and vimentin. (A, left) The expression of E-cadherin, vimentin, AR, and ERK2 proteins in the presence of DHT in various breast cancer cells. Different extracts were analyzed by Western blotting as described in the legend to Fig. 1C. (Right) Quantification of the intensity of the bands of immunoblot products (represented as the ratio of E-cadherin to ERK2 immunoblot product). Results represent the averages ± standard deviations from at least three experiments. (B) The morphology changes in the presence of DHT or AR siRNA in T47D cells. T47D and MDA-MD-435 cells were treated with 12 nM AR siRNA or nonspecific (NS) siRNA and then treated with 10 nM DHT or were left untreated. Seventy-two hours after DHT treatment, cells were examined by microscopy. (C) The expression of E-cadherin, vimentin, AR, and actin proteins in the presence of DHT or AR siRNA in various breast cancer cells. Cells were treated with DHT or siRNA as described for panel B and analyzed by Western blotting as described in the legend to Fig. 1C. (D) The expression of DAPI, AR, and E-cadherin proteins in the presence of DHT or AR siRNA in T47D and MDA-MD-435 cells by an IF assay. T47D and MDA-MD-435 cells were treated with DHT or siRNA or were left untreated as described for panel B and then stained with DAPI (blue), anti-AR (green), and anti-E-cadherin (red) antibodies.
Regulation of the ARE-containing gene by AR.
The results of our studies so far showed that the AR can downregulate the E-cadherin gene. To determine whether other AR response element (ARE)-containing genes can be either down- or upregulated by the AR in breast cancer cells, we used the FKBP5 intro-5 enhancer report construct, which contains the strongest ARE (ARE-7) (34), to carry out reporter assays. Results showed that the AR could activate the FKBP5 gene in both breast (MDA-MB-435) and prostate (PC-3 and LNCaP) cancer cells (data not shown). In addition, RT-PCR analysis showed that FKBP5 mRNAs were constitutively expressed in breast and prostate cancer cells (data not shown). These results indicate that the AR plays positive roles in the activation of the FKBP5 gene in these cells. This is in contrast to results showing the activated AR could downregulate the E-cadherin gene in both breast and prostate cancer cells (data not shown). Thus, the regulation of the ARE-containing gene by the AR is cell and gene specific. The AR may cooperate with other transcription regulators whose binding motifs are adjacent to the AR binding site.
Activated AR induces cell motility in epithelial type breast and colon cancer cells.
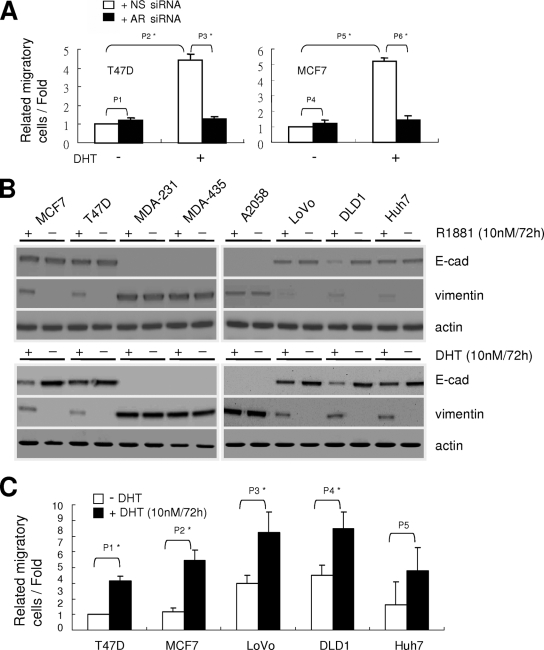
To determine if the activation of the AR could affect the migratory ability in nonmetastatic breast cancer cells, T47D and MCF7 cells were treated with DHT or were left untreated, and transwell assays were carried out. Seventy-two hours after DHT treatment, T47D and MCF7 cells showed increasing cell motility (Fig. 4A). This effect could be attenuated when AR siRNA was transfected into these cells. This phenomenon was not restricted to breast cancer cells. When nonmetastatic colon cancer cells (LoVo and DLD1) and liver cancer cells (Huh7) were treated with DHT or R1881, a synthetic AR ligand, reduced E-cadherin and increased vimentin expression were observed by Western blotting (Fig. 4B). However, the increased cell motility was observed only in breast and colon cancer cells and not in liver cancer cells (Fig. 4C). Thus, the activated AR can induce cell motility not only in breast cancer cells but also in colon cancer cells.
FIG. 4.
Ligand induces changes in cell type markers and enhances the migration of various epithelial type cancer cell lines. (A) Changes in the migratory ability of MCF7, T47D, and MDA-MD-435 cells in the presence of DHT, AR siRNA (white), or nonspecific (NS) siRNA (black). Cells were treated with DHT or siRNA as described in the legend to Fig. 3B and then analyzed by a transwell assay. The migratory ability is presented as the percentage of migrating cells over the total number of cells in the chamber. P1 = 0.07993; P2 = 0.00231; P3 = 0.00433; P4 = 0.06649; P5 = 0.00123; P6 = 0.00272 (Student's t test). (B) The expression of E-cadherin, vimentin, and actin proteins in the presence of AR ligand in various cancer cells. Cells were treated with 10 nM R1881 or 10 nM DHT for 72 h or were left untreated and were analyzed by Western blotting as described in the legend to Fig. 1C. (C) Changes in the migratory ability of various epithelial-type cancer cells in the presence of DHT. Various cancer cell lines (breast cancer, MCF7 and T47D; colon cancer, LoVo and DLD1; and liver carcinoma, Huh7) were treated with 10 nM DHT for 72 h (black) or were left untreated (white) and then were analyzed by a transwell assay as described for panel A. P1 = 0.01041; P2 = 0.00939; P3 = 0.03542; P4 = 0.03133; P5 = 0.07827 (Student's t test).
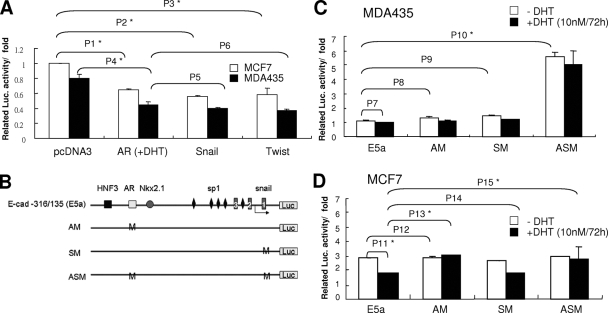
Activated AR has repressive effects similar to those of Snail and Twist in E-cadherin gene expression.
Snail and Twist have been shown to play important roles in the suppression of E-cadherin (16, 17, 49). The repressive effects of the activated AR on the E-cadherin gene were compared to those of Snail and Twist and proved to be similar (Fig. 5A). It could significantly downregulate E-cadherin reporter activities in metastatic (MDA-MD-435) and nonmetastatic (MCF7) breast cells. To further compare the repressive effect of the activated AR to that of Snail and Twist, the AR binding site and Ebox on the E-cadherin reporter constructs were mutated and reporter activities were measured (Fig. 5B). In MDA-MD-435 cells, AM (AR binding site mutant) or SM (Snail binding site mutant) did not significantly increase reporter activity. ASM (AR/Snail binding site double mutant) had significantly increased reporter activity (Fig. 5C). Nuclear AR and Snail both were expressed in MDA-MD-435 cells. A single mutation on either the AR or Snail binding site could not affect E-cadherin reporter activity. Only double mutations on these two binding sites could totally attenuate the repressive effect and increase reporter activity. In MCF7 cells, in which Snail or Twist was not expressed, the reporter activity of wild-type and SM constructs could be repressed by the activated AR when cells were treated with DHT (Fig. 5D). In addition, the AR binding site mutants (AM and ASM) attenuated the activated AR-mediated repression and increased reporter activity. However, without DHT there were no changes in reporter activity. Thus, in addition to Snail and Twist, the activated AR may be a transcription factor that can downregulate E-cadherin gene expression.
FIG. 5.
Activated AR has effects similar to those of Snail and Twist in suppressing human E-cadherin gene expression. (A) E-cadherin promoter activity was analyzed in MCF7 (white) and MDA-MD-435 (black) cells in the presence of pcDNA3, AR, Snail, or Twist vector. Various expression vectors were cotransfected with E-cadherin reporter vector into different cells. The AR-transfected cells were further treated with 10 nM DHT. Seventy-two hours after DHT treatment, luciferase (Luc.) activities were analyzed as described in the legend to Fig. 1A. P1 = 0.01143; P2 = 0.01121; P3 = 0.00866; P4 = 0.01443; P5 = 0.08822; P6 = 0.05669 (Student's t test). (B) Sketch of the wild-type E-cadherin reporter construct (E5a) and various mutated constructs. (C and D) The promoter activity of various E-cadherin reporter constructs was analyzed in MDA-MD-435 (C) and MCF7 (D) cells. Various cells were transfected with different E-cadherin reporter constructs and treated with 10 nM DHT (black) or were left untreated (white). Seventy-two hours after DHT treatment, luciferase activities were analyzed as described in the legend to Fig. 1A. P7 = 0.09944; P8 = 0.08586; P9 = 0.06763; P10 = 0.00076; P11 = 0.01833; P12 = 0.06294; P13 = 0.01173; P14 = 0.07723; P15 = 0.01238 (Student's t test).
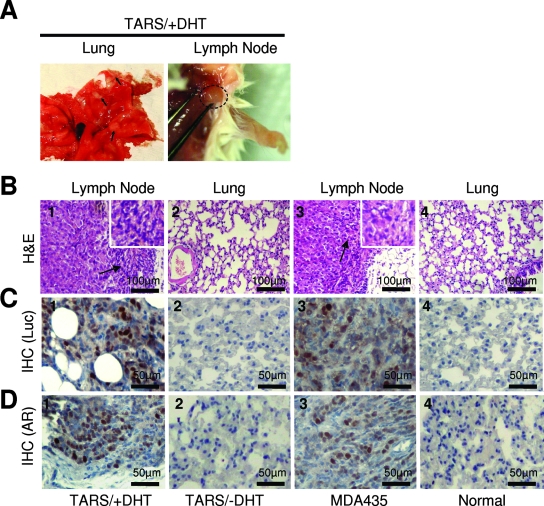
Activated AR can increase the invasiveness of transplanted cancer cells in mice.
To explore whether the activated AR can increase the metastatic potential of nonmetastatic cancer cells, TARS-Luc cells (Luc/pcDNA3 stable clone in TARS cells as described in Materials and Methods) were transplanted into NOD/SCID mice through their tail veins, and the mice then were treated with DHT. Seven weeks after treatment, the mice were sacrificed and the bilateral lungs and visible lymph nodes were removed for pathological examination. Seven mice in the DHT-treated group (7/8) and four mice in the positive control group (4/4) had multiple tumor nodules in the lungs or lymphadenopathy in the neck (Fig. 6A). Sections of the tumor samples stained with hematoxylin and eosin showed that the lymphandenopathy tissues were filled with cancer cells and had lost their original germinal center (Table 1 and Fig. 6B, images 1 and 3). In the untreated group, we did not observe these pathological findings. To test if these cancer cells were derived from transplanted cells, IHC staining to detect luciferase-positive cells was carried out. The results showed that the seven mice in the DHT-treated group (7/8) and the four mice in the positive control group (4/4) showed positive nuclear luciferase staining in the metastatic sites (Table 1 and Fig. 6C, images 1 and 3). These results indicated that these cancer cells were derived from transplanted cells. Furthermore, the metastatic lesions showed positive nuclear AR expression in DHT-treated mice (7/8) and in the positive control group (4/4) (Table 1 and Fig. 6D, images 1 and 3). Thus, the activated AR may increase cancer cell survival, allowing proliferation at distant sites with the formation of new cancer colonies.
FIG. 6.
Activated AR promotes the formation of metastatic carcinoma in mice. (A) DHT induces TARS cell metastasis formation in the lungs and lymph nodes of NOD/SCID mice. Representative photos of the lungs and lymph nodes from mice with TARS cell injection and DHT treatment carrying mammary tumors 7 weeks after DHT treatment. The arrows indicate metastatic nodules in the lung. The circle indicates metastatic nodules in the lymph node. (B) Metastatic tumors were detected in the lungs and lymph nodes of mice with transplanted TARS cells in the presence of DHT. Various cells were transplanted into NOD/SCID mice through the tail vain. Various mice were treated with DHT or were left untreated. The DHT treatment is described in Materials and Methods. Seven weeks after DHT treatment, hematoxylin and eosin staining (H&E) was done on the lungs and lymph nodes from various mice to determine tumor formation. (C and D) Nuclear luciferase (C) and the AR (D) were detected in the metastatic tumors of mice transplanted with TARS cells in the presence of DHT. The lungs and lymph nodes from various mice were analyzed for nuclear luciferase (Luc) and AR expression by IHC staining with anti-Luc and anti-AR antibodies.
TABLE 1.
Expression of metastatic carcinoma, nuclear luciferase, and nuclear AR in NOD/SCID mice
| Cell line | DHT treatment | No. of mice | No. (%) of mice positive for:
|
||
|---|---|---|---|---|---|
| Metastatic tumor | Nuclear luciferase | Nuclear AR | |||
| TARS | No | 6 | 0 | 0 | 0 |
| TARS | Yes | 8 | 7 (87.5) | 7 (87.5) | 7 (87.5) |
| MDA435 | No | 4 | 4 (100) | 4 (100) | 4 (100) |
| Medium | No | 2 | 0 | 0 | 0 |
Increased nuclear AR and decreased E-cadherin expression observed in human breast carcinomas.
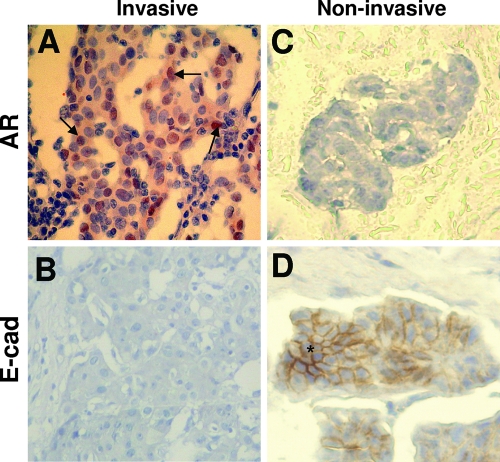
The results from the molecular and cell biology studies and the transplantation experiment strongly suggest that the activated AR represses E-cadherin gene expression and promotes tumor metastasis. To see whether the activated AR could increase the invasiveness of human tumor samples, two different types of breast cancer samples (invasive ductal and noninvasive mucinous carcinoma) from patients were analyzed for nuclear AR and E-cadherin by IHC staining. In 76 human invasive (invasive ductal carcinoma) breast cancer samples, 59 showed positive nuclear AR (Table 2 and Fig. 7A and B). Forty-five of the 59 nuclear AR-positive tumors were negative for E-cadherin (Table 2). Twelve of the 17 nuclear AR-negative invasive tumors were positive for E-cadherin (Table 2). Five of the 21 human noninvasive (mutinous carcinoma) breast cancer samples had positive nuclear AR, and 16 were negative (Table 2 and Fig. 7C and D). Four of the five nuclear AR-positive noninvasive tumors were E-cadherin negative, and 14 of the 16 nuclear AR-negative noninvasive tumors were E-cadherin positive (Table 2). Taken together, these results showed that in invasive breast cancer samples, the more positive the nuclear AR, the less the E-cadherin staining was observed. In contrast, in noninvasive breast cancer samples, the lower the positive nuclear AR, the more the E-cadherin staining was detected. In conclusion, these results of nuclear AR and E-cadherin staining are consistent with our hypothesis that the activated AR is a repressor of E-cadherin gene expression and can downregulate E-cadherin expression in human breast cancer cells to promote tumor metastasis.
TABLE 2.
Comparison of nuclear AR and E-cadherin expression in invasive ductal and noninvasive mucinous samples of human breast carcinomas
| Sample type | % of samples showing expression (no. of samples/total no. of samples) |
|---|---|
| Invasive ductal breast carcinoma (n = 76) | |
| Nuclear AR positive | 77.6 (59/76)a |
| E-cadherin positive | 23.7 (14/59)b |
| E-cadherin negative | 76.3 (45/59)b |
| Nuclear AR negative | 22.4 (17/76)a |
| E-cadherin positive | 70.6 (12/17)c |
| E-cadherin negative | 29.4 (5/17)c |
| Noninvasive mucinous breast carcinomas (n = 21) | |
| Nuclear AR positive | 23.8 (5/21)d |
| E-cadherin positive | 20.0 (1/5)e |
| E-cadherin negative | 80.0 (4/5)e |
| Nuclear AR negative | 76.2 (16/21)d |
| E-cadherin positive | 87.5 (14/16)f |
| E-cadherin negative | 12.5 (2/16)f |
P = 0.000287.
P = 1.5E-5.
P = 6.22E-5.
P = 3.88E-5.
P = 6.61E-5.
P = 6.57E-5.
FIG. 7.
Inversed expression pattern of nuclear AR and E-cadherin in human invasive ductal and noninvasive mucinous carcinomas. Carcinoma cells show positive nuclear AR expression (A) and negative E-cadherin expression (B) in invasive breast ductal carcinomas, as well as negative nuclear AR expression (C) and positive E-cadherin expression (D) in noninvasive breast mucinous carcinomas on IHC staining with anti-E-cadherin (E-cad) and anti-AR antibodies. AR staining is primarily nuclear (arrow) within invasive cells; E-cadherin staining is mainly on the cell boundary, like a beehive in epithelial cells (star).
DISCUSSION
The genetic, cell biological, and biochemical mechanisms of the spreading of cancer cells or metastasis still are elusive. Accumulated evidence suggests that the loss of E-cadherin strongly enhances progression from nonmetastatic to metastatic carcinoma (15, 25, 28). E-cadherin is an important tumor suppressor gene, and it is an indicator of metastasis (11, 29, 39, 49). Changes in the expression of E-cadherin can lead to the loss of the cell-cell junction and consequently increase cell migratory ability. However, the regulatory mechanisms of the human E-cadherin gene in metastatic and nonmetastatic cancer cells still are poorly understood. Promoter hypermethylation, histone deacetylation, and transcriptional repression are known to result in the downregulation of many genes, including the E-cadherin gene (3, 5, 7, 18, 19, 33, 41-43, 48, 54). Previous studies have focused on the repressive functions of Snail family members and Twist on the E-cadherin gene (5, 18, 19, 41, 42, 48). The results of our study show that the activated AR has equal repressive effects on E-cadherin expression.
The AR is essential for male sexual differentiation and prostatic epithelial cell proliferation (9, 27, 55). It also plays a critical role in the growth of androgen-dependent prostate cancer. Recent studies showed that the AR is significantly upregulated in metastatic prostate carcinoma (8, 27, 37). Some reports indicated that the activated AR could promote prostate cancer cell migration (26, 40), but others showed that androgen derivatives could inhibit this phenomenon (1, 22). Whether the activated AR can promote prostate cancer cell migration remains unsettled. There are limited investigations on the role of the AR in regulating the expression of cell-cell adhesion molecules in breast cancer. The AR is expressed in most breast cancer cells (32, 37, 46). However, the role of the AR in invasion and metastasis in breast cancer is not clear. Our data suggest that the activated AR is another repressor, in addition to Snail and Twist, of E-cadherin gene expression in nonmetastatic breast cancer cells.
In the presence of TSA, E-cadherin reporter activity was increased, suggesting that the downregulation of the E-cadherin gene by the activated AR may require the cooperation of HDACs. The induction of E-cadherin expression in TSA-treated cells could be due to the chromatin decondensation effect of TSA (41). Our previous study showed that the TSA treatment of various E-cadherin-negative cells (e.g., MDA-MB-435 and MDA-MB-231) for 24 h could induce the reexpression of E-cadherin (33). It is clear that TSA is an HDAC inhibitor and can strongly increase the acetylation of the N-terminal tails of histone H3 (16, 41). This can explain the results of our pervious study, which showed that TSA-induced E-cadherin reexpression in E-cadherin-negative cells was due to the inhibition of histone deacetylase or the increased histone acetylation of E-cadherin gene chromatin. The AR also was shown to serve as a target of acetylation by the effects of TSA (16). That report showed that the acetylation of AR by TSA treatment could increase ARE-containing reporter activity. Therefore, it is possible that the TSA treatment of AR expression cells acetylates the AR, increasing its affinity for the regulatory sequence of ARE-containing genes. In the meantime, TSA inhibits the HDACs that may turn the AR into an activator for the ARE-containing gene. Whether this is true remains to be determined. The present study showed that the activated AR could cooperate with HDAC1 or HDAC3 to downregulate the expression of E-cadherin and promote the cell migration of nonmetastatic breast cancer cells. The results support a previous observation, which showed that the activated AR could interact with HDAC1 in vitro and in vivo (17, 52).
In our study, we found that the AR was sequestered in the cytoplasm and remained inactive in nonmetastatic cancer cells. This could be due to the AR-interacting protein that usually is associated with the AR and inhibits the translocation of the AR to the nucleus to regulate its target genes. For example, selective androgen receptor modulators or protein inhibitors of activated STAT (PIASs), the coregulators of the AR, display distinct effects on AR-mediated gene activation in prostate cancer cells (20, 21, 31, 35). Our results showed more PIAS1 expression in nonmetastatic breast cancer MCF7 cells than in MDA-MD-231 and MDA-MD-435 metastatic breast cancer cells (data not shown). This implies that PIAS1 helps in sequestering the AR in the cytoplasm of nonmetastatic breast cancer cells. When these cells are treated with adequate amounts of DHT, the AR is activated and translocated into the nucleus to repress E-cadherin gene expression. However, a Western blotting assay of AR siRNA-treated cells showed that the reduced AR did not restore E-cadherin expression in MDA-MB-435 cells (Fig. 3C). This suggested that there are additional factors other than the AR that repress the E-cadherin gene in MDA-MB-435 cells. Our previous study had shown that different cells used more than one mechanism to repress E-cadherin gene expression (33). In that report, we showed that the inhibition of E-cadherin in MDA-MB-435 cells was due to promoter hypermethylation and the histone deacetylation of the E-cadherin gene. In the present study, reporter assays of the mutated ARE and the Snail binding site containing E-cadherin reporter constructs in MDA-MB-435 cells showed that only the double mutant in AR and Snail binding sites could significantly increase E-cadherin reporter activity. This suggested that Snail also participated in the repression of the E-cadherin gene in MDA-MB-435 cells. These results indicate that the inhibition of E-cadherin in MDA-MB-435 cells has multiple mechanisms.
The downregulation of E-cadherin has been shown to be associated with tumor metastasis. Previous studies have focused on the repressive functions of Snail and Twist on the E-cadherin gene (18, 19, 54). The results of our study showed that the activated AR had an equally repressive effect on E-cadherin expression. We generated several site-directed mutagenesis constructs to compare the repressive effect of the AR to that of Snail. The data indicated that AM did not increase the E-cadherin reporter construct activity in MDA-MD-435 metastatic cancer cells. This may be due to the repressive effect of the binding of Snail or Twist to the wild-type Eboxes on the regulatory sequence of the E-cadherin gene. Conversely, SM did not increase E-cadherin reporter construct activity, indicating that the AR could exert repressive effects on E-cadherin gene expression in the absence of Snail function. Only ASM showed increases in E-cadherin reporter construct activity. In the Snail- and Twist-negative MCF7 cells, the AM and ASM mutants had increased E-cadherin reporter construct activity when treated with DHT. Thus, the activated AR is a new repressive transcription factor that downregulates E-cadherin gene expression similarly to Snail and Twist. There are three Eboxes on this E-cadherin reporter construct. Previous studies demonstrated that Eboxes 2 and 3 were nonfunctional (3, 33), whereas Ebox 1 had the most repression ability on human E-cadherin gene expression. Even though we did not create mutant sites for Eboxes 2 and 3, this would not affect the results.
Our data showed that the activated AR could downregulate the E-cadherin gene in both metastatic (MDA-MB-435) and nonmetastatic (T47D) breast cancer cells. In addition, the E-cadherin gene also was downregulated by the AR in metastatic prostate cancer cells (e.g., PC-3 or LNCaP) (data not shown). In contrast, AR appears to play a positive regulatory role in other consensus ARE-containing genes (e.g., FKBP5) in most of the cells we tested (data not shown). Thus, the regulation of ARE-containing genes by AR is gene and cell type specific. The AR differentially regulates ARE-containing genes in the same cell and may depend on other transcription factor binding motifs adjacent to ARE. This possibility has been reported recently (6).
In our transplantation experiment, we introduced nonmetastatic breast cancer cells into NOD/SCID mice and then treated these mice with DHT or left them untreated. In the DHT-treated groups, tumor nodules were detected in the lung and lymph nodes. Without DHT treatment, no tumors were detected on gross or histological examination. Our animal studies were carried out by tail vein injection to mimic the migratory process in vivo. Nevertheless, these results indicate that the activated AR can enhance cancer cell survival and proliferation at distant sites to form new cancer colonies and may enhance the migratory ability of nonmetastatic cancer cells in vivo. Steroid hormone could activate cancer cell growth and invasiveness, as frequently reported in prostate cancer studies (12, 13, 27, 30, 53). In our in vitro experiments, DHT also could increase the migratory ability of breast and colon cancer cells and decrease E-cadherin expression. Thus, the possibility exists that the misuse of steroid hormone may trigger dormant cancer cells into proliferating developing metastatic tumors. Our in vitro experiments showed that R1881 treatment did not significantly repress the E-cadherin gene in MCF7, T47D, LoVo, and Huh7 cells. R1881 is a synthetic AR agonist, whereas DHT is a natural AR ligand. Both natural and synthetic steroids can interact with AR and play different agonist effects. This could be due to differences in binding affinities between R1881 and AR and those of DHT and AR.
The transplanted breast cancer cells we used were derived from nonmetastatic breast cancer cells (T47D). The T47D cells were stably transfected with the AR expression vector and were treated with DHT to generate an individual subline (TARS). Comparisons of gene expression patterns, migratory ability, and morphology changes between TARS and nonmetastatic breast cancer cells (T47D or MCF7) indicated that TARS-like T47D cells have characteristics similar to those of TARS in the absence of DHT (data not shown). However, in the presence of DHT, TARS cells had more repressed E-cadherin gene expression and more increased cell motility than T47D cells (data not shown). We also carried out similar transplant experiments using T47D with results similar to those with TARS (data not shown). More metastatic tumor colonies were observed in TARS-transplanted/DHT-treated mice than with T47D-transplanted/DHT-treated mice. These data suggest that the dosage of AR expression affects tumor progression in breast cancer cells.
In summary, we have shown that the activated AR can be translocated into the nucleus from the cytoplasm to cooperate with HDAC1 and bind to the human E-cadherin regulatory sequence. The activated AR downregulates the expression of the E-cadherin gene and changes cell morphology from epithelial-like to mesenchymal-like. We also have shown that DHT can increase cell motility by reducing E-cadherin expression in breast and colon cancer cells. The capacity of the activated AR to downregulate E-cadherin expression is equal to that of Snail and Twist. The activated AR could promote cell survival and proliferation at distant sites in our animal model. Finally, an analysis of clinical tumor samples showed that the more positive the nuclear AR, the less positive the E-cadherin staining detected on the cell-cell boundary of human breast invasive ductal carcinoma. These results corroborate our results in the cell biological and animal studies. Our study supports our argument that the AR is a repressor of the E-cadherin gene and can promote metastasis.
Acknowledgments
We thank Y. L. Juang and Z. I. Yeh for comments on the manuscript and M. H. Wang and Y. Y. Chiang for assistance in the transplantation study. We also thank H. Y. Kao for providing HDAC1 and HDAC3 cDNA expression vectors and members of the Chen laboratory for technical assistance, discussion, and suggestions.
This work was supported by grants from the National Science Council, Taiwan (NSC 91-23320-B-320-012).
Footnotes
Published ahead of print on 15 September 2008.
REFERENCES
- 1.Baek, S. H., K. A. Ohgi, C. A. Nelson, D. Welsbie, C. Chen, C. L. Sawyers, D. W. Rose, and M. G. Rosenfeld. 2006. Ligand-specific allosteric regulation of coactivator functions of androgen receptor in prostate cancer cells. Proc. Natl. Acad. Sci. USA 1033100-3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baisch, H., U. Otto, and H. Fack. 1998. Growth of human prostate carcinomas with and without hormone alpha-dehydrotestosterone in nude mice. Euro. Urol. 34505-511. [DOI] [PubMed] [Google Scholar]
- 3.Batlle, E., E. Sancho, C. Franci, D. Dominguez, M. Monfar, J. Baulida, and A. G. de Herreros. 2000. The transcription factor Snail is a repressor of E-cadherin gene expression in epithelial tumor cells. Nat. Cell Biol. 284-89. [DOI] [PubMed] [Google Scholar]
- 4.Birchmeier, W., and J. Behrens. 2003. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. BBA/Rev. Cancer 119811-26. [DOI] [PubMed] [Google Scholar]
- 5.Bolós, V., H. Peinado, M. A. Perez-Moreno, M. F. Fraga, M. Esteller, and A. Cano. 2003. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressers. J. Cell Sci. 116499-511. [DOI] [PubMed] [Google Scholar]
- 6.Bolton, E. C., A. Y. So, C. Chaivorapol, C. M. Haqq, H. Li, and K. R. Yamamoto. 2007. Cell- and gene-specific regulation of primary target genes by the androgen receptor. Genes Dev. 212005-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cano, A., M. A. Perez-Moreno, I. Rodrigo, A. Locascio, M. J. Blanco, M. G. DelBarrio, F. Portillo, and M. A. Nieto. 2000. The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 276-83. [DOI] [PubMed] [Google Scholar]
- 8.Chambers, A. F., A. C. Groom, and L. C. MacDonald. 2002. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2563-572. [DOI] [PubMed] [Google Scholar]
- 9.Chesire, D. R., and W. B. Isaacs. 2003. Beta-catenin signaling in prostate cancer: an early perspective. Endocr. Relat. Cancer 10537-560. [DOI] [PubMed] [Google Scholar]
- 10.Chirgwin, J. M., A. E. Przybyla, R. J. MacDonald, and W. J. Rutter. 1979. Isolation of biologically active ribonucleic acid from sources enriched in ribonuclease. Biochemistry 185294-5299. [DOI] [PubMed] [Google Scholar]
- 11.Christofori, G., and H. Semb. 1999. The role of the cell-adhesion molecule E-cadherin as a tumour-suppressor gene. Trends Biochem. Sci. 2473-76. [DOI] [PubMed] [Google Scholar]
- 12.Cunha, G. R., W. Ricke, A. Thomson, P. C. Marker, G. Risbridger, S. W. Hayward, Y. Z. Wang, A. A. Donjacoura, and T. Kurita. 2004. Hormonal, cellular, and molecular regulation of normal andneoplastic prostatic development. J. Steroid Biochem. Mol. Biol. 92221-236. [DOI] [PubMed] [Google Scholar]
- 13.Dürnberger, H., and K. Kratochwii. 1980. Specificity of tissue interaction and origin of mesenchymal cells in the androgen response of the embryonic mammary gland. Cell 19465-471. [DOI] [PubMed] [Google Scholar]
- 14.Fidler, I. J. 2003. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer 3453-458. [DOI] [PubMed] [Google Scholar]
- 15.Frixen, U. H., J. Behrens, M. Sachs, G. Eberle, B. Voss, A. Warda, D. Löchner, and W. Birchmeier. 1991. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J. Cell Biol. 113173-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fu, M., M. Rao, K. Wu, C. Wang, X. Zhang, M. Hessien, Y. G. Yeung, D. Gioeli, M. J. Weber, and R. G. Pestell. 2004. The androgen receptor acetylation site regulates cAMP and AKT but not ERK-induced activity. J. Biol. Chem. 27929436-29449. [DOI] [PubMed] [Google Scholar]
- 17.Gaughan, L., I. R. Logan, D. E. Neal, and C. N. Robson. 2005. Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic Acids Res. 3313-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grady, W. M., J. Willis, P. J. Guilford, A. K. Dunbier, T. T. Toro, H. Lynch, G. Wiesner, K. Ferguson, C. Eng, J. G. Park, S. J. Kirn, and S. Markowitz. 2000. Methylation of the CDHI promoter as the second genetic hit in hereditary diffuse gastric cancer. Nat. Genet. 2616-17. [DOI] [PubMed] [Google Scholar]
- 19.Graff, J. R., J. G. Herman, R. G. Lapidus, H. Chopra, R. Xu, D. F. Jarrard, W. B. Isaacs, P. M. Pitha, N. E. Davidson, and S. B. Baylin. 1995. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 555195-5199. [PubMed] [Google Scholar]
- 20.Gross, M., B. Liu, J. Tan, F. S. French, M. Carey, and K. Shuai. 2001. Distinct effects of PIAS proteins on androgen-mediated gene activation in prostate cancer cells. Oncogene 203880-3887. [DOI] [PubMed] [Google Scholar]
- 21.Gross, M., R. Yang, I. Top, C. Gasper, and K. Shuai. 2004. PIASy-mediated repression of the androgen receptor is independent of sumoylation. Oncogene 233059-3066. [DOI] [PubMed] [Google Scholar]
- 22.Guerini, V., D. Sau, E. Scaccianoce, P. Rusmini, P. Ciana, A. Maggi, P. G. V. Martini, B. S. Katzenellenbogen, L. Martini, M. Motta, and A. Poletti. 2005. The androgen derivative 5α-androstane-3β, 17β-diol inhibits prostate cancer cell migration through activation of the estrogen receptor β subtype. Cancer Res. 655445-5453. [DOI] [PubMed] [Google Scholar]
- 23.Gupta, G. P., and J. Massague. 2006. Cancer metastasis: building a framework. Cell 127679-695. [DOI] [PubMed] [Google Scholar]
- 24.Hanahan, D., and R. A. Weinberg. 2000. The hallmarks of cancer. Cell 10057-70. [DOI] [PubMed] [Google Scholar]
- 25.Handschuh, G., S. Candidus, B. Luber, U. Reich, C. Schott, S. Oswald, H. Becke, P. Hutzler, W. Birchmeier, H. Hofler, and K. F. Becker. 1999. Tumour-associated E-cadherin mutations alter cellular morphology, decrease cellular adhesion and increase cellular motility. Oncogene 184301-4312. [DOI] [PubMed] [Google Scholar]
- 26.Hara, T., H. Miyazaki, A. Lee, C. P. Tran, and R. E. Reiter. 2008. Androgen receptor and invasion in prostate cancer. Cancer Res. 681128-1135. [DOI] [PubMed] [Google Scholar]
- 27.Heuberger, B., I. Fitzka, G. Wasner, and K. Kratochwil. 1982. Induction of androgen receptor formation by epithelium-mesenchymel interaction in embryonic mouse mammary gland. Proc. Natl. Acad. Sci. USA 792957-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ishizaka, S., A. Shiroi, S. Kanda, M. Yoshikawa, H. Tsujinoue, S. Kuriyama, T. Hasuma, K. Nakatani, and K. Takahashi. 2002. Development of hepatocytes from embryonic stem cells after transfection with the HNF-3β gene. FASEB J. 161444-1446. [DOI] [PubMed] [Google Scholar]
- 29.Kemler, R. 1993. From cadherins to catenins: Cytoplasmatic protein interactions and regulation of cell adhesion. Trends Genet. 9317-321. [DOI] [PubMed] [Google Scholar]
- 30.Labaree, D. C., T. J. Brown, R. M. Hoyte, and R. B. Hochberg. 1997. 7α-Iodine-125-iodo-5α-dihydrotestosterone: a radiolabeled ligand for the androgen receptor. J. Nuclear Med. 38402-409. [PubMed] [Google Scholar]
- 31.Lee, H., J. C. Quinn, K. V. Prasanth, V. A. Swiss, K. D. Economides, M. M. Camacho, D. L. Spector, and C. Abate-Shen. 2006. PIAS1 confers DNA-binding specificity on the Msx1 homeoprotein. Genes Dev. 20784-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lillie, E. O., L. Bernstein, and G. Ursin. 2003. The role of androgens and polymorphisms in the androgen receptor in the epidemiology of breast cancer. Breast Cancer Res. 5164-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu, Y. N., W. W. Lee, C. Y. Wang, T. H. Chao, Y. V. Chen, and J. H. Chen. 2005. Regulatory mechanisms controlling human E-cadherin gene expression. Oncogene 248277-8290. [DOI] [PubMed] [Google Scholar]
- 34.Magee, J. A., L. W. Chang, G. D. Stormo, and J. Milbrandt. 2006. Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology 147590-598. [DOI] [PubMed] [Google Scholar]
- 35.Mantovani, A. 2007. Cancer: an infernal triangle. Nature 448547-548. [DOI] [PubMed] [Google Scholar]
- 36.Mareel, M., and A. Leroy. 2003. Clinical, cellular, and molecular aspects of cancer invasion. Physiol. Rev. 83337-376. [DOI] [PubMed] [Google Scholar]
- 37.Moinfar, F., M. Okcu, O. Tsybrovskyy, P. Regitnig, S. F. Lax, W. Weybora, M. Ratschek, F. A. Tavassoli, and H. Denk. 2003. Androgen receptors frequently are expressed in breast carcinomas—potential relevance to new therapeutic strategies. Cancer 98703-711. [DOI] [PubMed] [Google Scholar]
- 38.Müller, A., B. Homey, H. Soto, N. Ge, D. Catron, M. E. Buchanan, T. McClanahan, E. Murphy, W. Yuan, S. N. Wagner, J. L. Barrera, A. Mohar, E. Verástegui, and A. Zlotnik. 2001. Involvement of chemokine receptors in breast cancer metastasis. Nature 41050-56. [DOI] [PubMed] [Google Scholar]
- 39.Nagafuchi, A., and M. Takeichi. 1988. Cell binding function of E-cadherin is regulated by the cytoplasmic domain. EMBO J. 73679-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nightingale, J., K. S. Chaudhary, P. D. Abely, A. P. Stubbs, H. M. Romanska, S. E. Mitchell, G. W. H. Stamp, and E. N. Lalani. 2003. Ligand activation of the androgen receptor downregulates E-cadherin mediated cell adhesion and promotes apoptosis of prostatic cancer cells. Neoplasia 5347-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peinado, H., E. Ballestar, M. Esteller, and A. Cano. 2004. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase I (HDACI)/HDAC2 complex. Mol. Cell. Biol. 24306-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peinado, H., F. Portillo, and A. Cano. 2004. Transcriptional regulation of cadherins during development and carcinogenesis. Int. J. Dev. Biol. 48365-375. [DOI] [PubMed] [Google Scholar]
- 43.Perez-Moreno, M. A., A. Locascio, I. Rodrigo, G. Dhondt, F. Portillo, M. A. Nieto, and A. Cano. 2001. A new role for E12/E47 in the repression of E-cadherin expression and epithelial-mesenchymal transitions. J. Biol. Chem. 27627424-27431. [DOI] [PubMed] [Google Scholar]
- 44.Sasisekharan, R., Z. Shriver, G. Venkataraman, and U. Narayanasami. 2002. Roles of heparan-sulphate glycosaminoglycans in cancer. Nat. Rev. Cancer 2521-528. [DOI] [PubMed] [Google Scholar]
- 45.Shiozaki, H., H. Oka, M. Inoue, S. Tamura, and M. Monden. 1996. E-cadherin mediated adhesion system in cancer cells. Cancer 771605-1613. [DOI] [PubMed] [Google Scholar]
- 46.Spady, T. J., D. M. E. Harvell, M. C. Snyder, K. L. Pennington, R. D. McComb, and J. D. Shull. 1998. Estrogen-induced tumorigenesis in the Copenhagen ratdisparate susceptibilities to development of prolactin-producing pituitary tumors and mammary carcinomas. Cancer Lett. 12495-103. [DOI] [PubMed] [Google Scholar]
- 47.Sporn, M. B. 1996. The war on cancer. Lancet 3471377-1381. [DOI] [PubMed] [Google Scholar]
- 48.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 40341-45. [DOI] [PubMed] [Google Scholar]
- 49.Takeichi, M. 1993. Cadherins in cancer: implications for invasion and metastasis. Curr. Opin. Cell Biol. 5806-811. [DOI] [PubMed] [Google Scholar]
- 50.Tepass, U., K. Truong, D. Godt, M. Ikura, and M. Peifer. 2000. Cadherins in embryonic and neural morphogenesis. Nat. Rev. Mol. Cell Biol. 191-100. [DOI] [PubMed] [Google Scholar]
- 51.Thiery, J. P. 2002. Epithelial-mesenchymal transitions in tumour progression. Nat. Rev. Cancer 2442-454. [DOI] [PubMed] [Google Scholar]
- 52.Tóth, K. F., T. A. Knoch, M. Wachsmuth, M. Frank-Stöhr, M. Stöhr, C. P. Bacher, G. Müller, and K. Rippe. 2004. Trichostatin A-induced histone acetylation causes decondensation of interphase chromatin. J. Cell Sci. 1174277-4287. [DOI] [PubMed] [Google Scholar]
- 53.Visakorpi, T. 2003. The molecular genetics of prostate cancer. Urology 62 (Suppl. 1)3-10. [DOI] [PubMed] [Google Scholar]
- 54.Yang, J., S. A. Mani, J. L. Donaher, S. Ramaswamy, R. A. Ltzykson, C. Come, P. Savagner, I. Gitelman, A. Richardson, and R. A. Weinberg. 2004. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell 117927-939. [DOI] [PubMed] [Google Scholar]
- 55.Zhu, P., S. H. Baek, E. Bourk, K. A. Ohgi, I. Garcia-Bassets, H. Sanjo, S. Akira, P. Kotol, C. Glass, and M. G. Rosenfeld. 2006. Macrophage/cancer cell interactions mediate hormone resistance through a conserved nuclear receptor derepression pathway. Cell 124615-629. [DOI] [PubMed] [Google Scholar]