Abstract
Transforming growth factor β (TGF-β) superfamily members are critical in maintaining cell growth and differentiation in the ovary. Although signaling of activins, TGF-βs, growth differentiation factor 9, and nodal converge preferentially to SMAD2 and SMAD3, the in vivo functions and redundancy of these SMADs in the ovary and female reproduction remain largely unidentified. To circumvent the deleterious phenotypic aspects of ubiquitous deletion of Smad2 and Smad3, a conditional knockout strategy was formulated to selectively inactivate Smad2, Smad3, or both Smad2 and Smad3 in ovarian granulosa cells. While granulosa cell ablation of individual Smad2 or Smad3 caused insignificant changes in female fertility, deletion of both Smad2 and Smad3 led to dramatically reduced female fertility and fecundity. These defects were associated with the disruption of multiple ovarian processes, including follicular development, ovulation, and cumulus cell expansion. Furthermore, the impaired expansion of cumulus cells may be partially associated with altered cumulus expansion-related transcripts that are regulated by SMAD2/3 signaling. Our results indicate that SMAD2 and SMAD3 function redundantly in vivo to maintain normal female fertility and further support the involvement of an intraovarian SMAD2/3 pathway in mediating oocyte-produced signals essential for coordinating key events of the ovulatory process.
Ligands of the transforming growth factor β (TGF-β) superfamily (TGF-βs, activins/inhibins, growth differentiation factor 9 [GDF9], bone morphogenetic proteins [BMPs], nodal, etc.) are multifunctional proteins that are critically involved in various physiological and developmental processes (9, 42, 43). Transduction of TGF-β-related signals is initiated when the ligands bind their serine/theonine kinase type 2 and type 1 (activin receptor-like kinase) receptors to form an oligomeric receptor complex (31, 33, 42, 65). Subsequent phosphorylation and activation of receptor-regulated SMADs (R-SMADs) by the type I receptor are accomplished in concert with SMAD anchor for receptor activation. The R-SMADs then form heteromeric complexes with a common SMAD (Co-SMAD; SMAD4) and accumulate in the nucleus to regulate ligand-specific gene expression via recruitment of distinct transcription factors, coactivators, and corepressors (41-43). Activation of SMADs can be modulated at multiple levels by numerous molecules including the ligand-induced inhibitory SMADs, SMAD6 and SMAD7, which function as negative regulators of SMAD activity (9, 43, 44, 48, 50, 66).
Although accumulating evidence highlights the intraovarian TGF-β superfamily signaling cascades as key regulatory pathways of fundamental female reproductive events, including folliculogenesis and ovulation (22, 30-33, 45, 54), knowledge of the functional identity of the individual components of this pathway in reproductive physiology is far from complete due to the complex and promiscuous nature of the ligands, receptors, and SMAD signaling proteins. For example, activins, TGF-βs, GDF9, and nodal can all transduce their signals in vitro through SMAD2 and SMAD3, which share more than 90% identity in their amino acid sequences (7), whereas BMPs activate SMAD1, SMAD5, and SMAD8 (7, 10). Despite the fact that signals of the aforementioned ligands converge to SMAD2 and SMAD3, the in vivo functions of these SMADs in the ovary remain to be illustrated. An essential role of SMAD2 in embryonic development was initially revealed by genetic studies on Smad2 mutant mice, which are lethal during early development (embryonic days 7.5 to 12.5) with defects in egg cylinder elongation, mesoderm formation, and left-right patterning (26, 27, 51, 70, 73). The lethal phenotype of Smad2-null mice precludes direct examination of the reproductive roles of SMAD2. In contrast, functional inactivation of mouse Smad3 gene results in viable animals with reduced body size (11, 76, 79). Smad3-null mice develop colorectal cancer (79) or have immune defects (76) with accelerated wound healing (3). However, discrepancies in female fertility were reported for the two lines of Smad3 knockout mice that have targeted deletions of distinct exons (exon 2 versus exon 8) (67, 68, 79). The complex functions of SMAD2 and SMAD3 were further suggested by studies on the potential downstream targets of SMAD2 and SMAD3 using mouse embryonic fibroblasts (11, 61, 77), as well as experiments performed in vitro to silence (34) or overexpress (49) these Smads.
The spatiotemporal expression patterns of SMAD2 and SMAD3 in the ovary (4, 74) provide additional rationale to perform further functional studies. To understand the SMAD2/3 signaling cascade in the ovary, a conditional knockout (cKO) strategy was undertaken to circumvent the lethal phenotype associated with Smad2 deletion and the deleterious effects of colorectal cancer resulting from loss of SMAD3. We herein generated three independent mouse lines with granulosa cell deletions of Smad2, Smad3, or both Smad2 and Smad3 genes by using the Cre-loxP recombination system that has been validated in our previous reports (1, 2, 29, 56-58). The present study identified redundant in vivo functions of ovarian SMAD2 and SMAD3 in the maintenance of female fertility.
MATERIALS AND METHODS
Construction of Smad2 and Smad3 conditional alleles and generation of Smad2, Smad3, and Smad2/3 cKO mice.
All mouse lines used in the present study were maintained on a mixed C57BL/6/129S6/SvEv genetic background and manipulated according to the NIH Guide for the Care and Use of Laboratory Animals. The Smad2-null allele and conditional allele with exons 9 and 10 flanked by loxP sites were generated in previous studies (37, 38). For construction of the Smad3 conditional allele, the 5′ and 3′ arms for homologous recombination were generated as previously described (79). The targeting vector was constructed to contain two loxP sites flanking Smad3 exons 2 and 3, which were deleted in the Smad3-null mice (79). The targeting vector was electroporated into embryonic stem cells, and positive clones were selected and injected into blastocysts. The chimeric males were bred to wild-type (WT) females to obtain germ line transmission. The Smad3flox/+ mice were then derived and intercrossed to obtain the homozygous Smad3flox/flox mice, which were viable and fertile.
Generation of Smad2, Smad3, or Smad2/3 cKO mice was conducted using mice carrying the Smad2 and Smad3 conditional alleles and anti-Müllerian hormone receptor type 2 (Amhr2)-cre knock-in allele (insertion of a Cre-Neo cassette into the fifth exon of Amhr2 gene) (28). Three independent cKO mouse lines, including Smad2 cKO (Smad2flox/−; Amhr2cre/+), Smad3 cKO (Smad3flox/−; Amhr2cre/+), and Smad2 and Smad3 double cKO (Smad2/3 cKO, Smad2flox/−; Smad3flox/−; Amhr2cre/+) were produced. Mice were genotyped by PCR analyses of genomic tail DNA using specific primers (Table 1) . Analyses of DNA recombination in the granulosa cell compartment were performed using DNA isolated from preovulatory stage granulosa cells as described previously (57, 58).
TABLE 1.
Primers for PCR genotyping and quantitative real-time PCR using Sybr green
| Gene | Primer sequence (5′-3′)
|
|
|---|---|---|
| Forward | Reverse | |
| Amhr2cre | CGC ATT GTC TGA GTA GGT GT | GAA ACG CAG CTC GGC CAG C |
| Smad2 flox | TAC TTG GGG CAA TCT TTT CG | GTC ACT CCC TGA ACC TGA AG |
| Smad3 flox | CTC CAG ATC GTG GGC ATA CAG C | GGT CAC AGG GTC CTC TGT GCC |
| Smad2 null (set 1)a | ACT TCG CTA GTT GCT CAT GG | CCA GAC TGC CTT GGG AAA AGC |
| Smad2 null (set 2)a | GCT GAG TGC CTA AGT GAT AGT GCA | TCT TCT TTT TCC CCG CTG G |
| Smad3 WT | TGG ACT TAG GAG ACG GCA GTC C | CTT CTG AGA CCC TCC TGA GTA GG |
| Smad3 null | TGG ACT TAG GAG ACG GCA GTC C | CTC TAG AGC GGC CTA CGT TTG G |
| Smad2 Rec | GAG CTG CGC AGA CCT TGT TAC | GTC ACT CCC TGA ACC TGA AG |
| Smad3 Rec | TCG TCG ATC GAC CTC GAA TAA C | GGT CAC AGG GTC CTC TGT GCC |
| Smad3 exon1-2 | ACC AAG TGC ATT ACC ATC C | CAG TAG ATA ACG TGA GGG AGC CC |
| Smad2 exon10 | GCT CTT CTG GCT CAG TCT GTC A | GGT GCA CAT TCG GGT TAG CT |
The first and the second sets of primers detect the Smad2-null allele with the Neo cassette or without the Neo cassette, respectively.
Histological analysis and quantification of growing follicles.
Mouse ovaries were collected and fixed in 10% (vol/vol) neutral buffered formalin. The specimens were then washed with 70% ethanol before being embedded in paraffin. The ovaries were sectioned and stained with periodic acid-Schiff-hematoxylin using standard procedures in the Pathology Core Services Facility at Baylor College of Medicine. Follicles were classified according to the morphological criteria described by Pedersen and Peters (60). Follicle counting (n = 3 to 4 per genotype) was performed as reported elsewhere (57), and the number of follicles was normalized to the area of the section. The measurements were obtained by using AxioVision 4.0 software (Carl Zeiss), and the results are reported as average number of follicles/mm2.
Hormone analyses.
Mice were anesthetized with isoflurane inhalation, and cardiac puncture was performed to collect the blood samples into serum separator tubes (BD). The serum was separated by centrifugation and stored at −20°C until assayed for hormone levels. Serum follicle-stimulating hormone (FSH), luteinizing hormone (LH), progesterone (P4), and estradiol (E2) were measured by the ligand assay and analysis core at the Center for Research in Reproduction, University of Virginia. The limit of detection of the assays is as follows: FSH (2 ng/ml), E2 (10 pg/ml), LH (0.07 ng/ml), and P4 (0.1 ng/ml). Samples that are below the assay threshold were assigned with the threshold value. Detailed information on the hormone analyses is available at http://www.healthsystem.virginia.edu/internet/crr/ligand.cfm.
Superovulation.
Superovulation experiments were carried out as described previously (56). Briefly, immature female mice (WT, Smad2flox/−; Amhr2cre/+, Smad3flox/−; Amhr2cre/+, and Smad2flox/−; Smad3flox/−; Amhr2cre/+) were injected intraperitoneally (i.p.) with 5 IU of pregnant mare serum gonadotropin (PMSG; Calbiochem), followed by administration of 5 IU of hCG (i.p.) 44 to 46 h later. After 18 h of hCG injection, the ovaries and oviducts were surgically removed, and the cumulus oocyte complexes (COCs) mass was recovered from the oviduct and collected into M2 medium (Sigma) containing 1 mg of hyluronase/ml (Sigma) to dissociate the cumulus cells from oocytes. The numbers of oocytes were then counted and recorded.
Cumulus expansion analyses.
To study the in vivo cumulus expansion, immature (21- to-23 day-old) mice, including both control and experimental groups, were treated with PMSG, followed by injection of hCG 44 h later. The ovaries were collected 6 h after hCG injection and fixed in 10% neutral buffered formalin. The ovaries were then processed for periodic acid-Schiff-hematoxylin staining and histologically examined. In vitro cumulus expansion assay was performed according to a previously described protocol with slight modifications (57). In brief, immature WT and Smad2/3 cKO female mice were injected with 5 IU of PMSG (i.p.), and 44 h later, COCs were surgically dissected from large antral follicles. Intact COCs were cultured in droplets of DME-F12 (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Sigma), 0.25 mM sodium pyruvate (Invitrogen), 3 mM l-glutamine (Invitrogen), and 100 U of penicillin-streptomycin (Invitrogen)/ml in the presence or absence of 10 ng of epidermal growth factor (EGF; BD Bioscience)/ml.
Production of recombinant mouse GDF9.
Stably transfected Chinese hamster ovary (CHO) cells carrying the full-length mouse Gdf9 cDNA or parent vector were used to produce recombinant mouse GDF9 conditioned medium and control medium, respectively. Procedures for the GDF9 production have been detailed elsewhere (18, 55). The GDF9 conditioned medium containing 0.4 μg of GDF9/ml and 2% FBS was stored at 4°C.
Granulosa cell isolation and culture.
Isolation of granulosa cells was performed as described previously (55-57). Briefly, immature female mice (21- to 23-day-old) were injected i.p. with 5 IU of PMSG, and 44 h later, large antral follicles were punctured with needles, and granulosa cells were collected into DME-F12 medium containing 0.3% bovine serum albumin, 100 U of penicillin-streptomycin/ml, and 10 mM HEPES (Invitrogen). After filtration through a 40-μm-pore-size nylon mesh (Nalgene), the granulosa cells were recovered and subjected to RNA isolation for studies on Smad2 and Smad3 transcript levels by using an RNeasy minikit (Qiagen) or further cultured to test the effect of recombinant GDF9 on the expression of cumulus expansion-related transcripts (pentraxin 3 [Ptx3], hyaluronan synthase 2 [Has2], prostaglandin synthase 2 [Ptgs2], and tumor necrosis factor alpha-induced protein 6 [Tnfaip6]).
For the culture experiment, granulosa cells were isolated from WT and Smad2/3 cKO mice and plated in 24-well plates in DME-F12 supplemented with 1× insulin-transferrin-selenite (Sigma), 2% heat-inactivated FBS, and 100 U of penicillin-streptomycin/ml. The culture was maintained at 37°C in a humidified incubator with 5% CO2 overnight. The granulosa cells were then further cultured in the presence or absence of recombinant mouse GDF9 conditioned medium containing 100 ng of recombinant mouse GDF9/ml or conditioned control medium supplemented with 0.5% heat-inactivated FBS for 5 h. The granulosa cell cultures were then harvested, and the cells were washed with phosphate-buffered saline and lysed in RLT buffer (Qiagen) for RNA extraction by using the RNAeasy microkit (Qiagen) according to the manufacturer's instructions.
RT.
Briefly, appropriate amount of total granulosa cell RNA (50 ng for granulosa cell culture experiment or 200 ng for nonculture experiment) was reverse transcribed by using Superscript III reverse transcriptase (Invitrogen) and oligo(dT)12-18 primers (Invitrogen). When 50 ng of total RNA was used, RNaseOUT (Invitrogen) was included in the reverse transcription (RT) reaction, and the incubation time for RT was increased to 2 h. The resultant RT products were kept at −20°C until assayed.
Quantitative real-time PCR.
Quantitative real-time PCR was conducted to measure the relative mRNA levels of Smad2, Smad3, and the effect of GDF9 on gene expression in granulosa cells using an ABI Prism 7500 sequence detection system (Applied Biosystems). The real-time PCR program consists of 40 cycles of 95°C for 15 s and 60°C for 1 min (35, 57). The assay was performed in a 20-μl reaction volume in a 96-well plate (Applied Biosystems) with mouse Gapdh as an internal control. Quantification of Smad2 and Smad3 transcripts was performed using Sybr PCR master mix (Applied Biosystems) and gene specific primers designed by Primer Express (Applied Biosystems) (Table 1). Analyses of cumulus expansion-related genes were performed using TaqMan gene expression assays (Ptx3, Mm00477267_g1; Has2, Mm00515089_m1; Tnfaip6, Mm00493736_m1; and Ptgs2, Mm00478374_m1) and TaqMan Universal PCR Master Mix (Applied Biosystems). Expression of mRNA for target genes was normalized relative to that of the endogenous control (Gapdh) mRNA using the ΔΔCT method (39). Negative controls using RT products from reactions where reverse transcriptase was omitted were included in the analysis to monitor potential genomic DNA contamination.
Western blot.
Granulosa cells collected from WT, Smad2flox/−, Smad3flox/−, Smad2 cKO, and Smad3 cKO mice (two to four animals were pooled for each genotype) were lysed in radioimmunoprecipitation assay buffer on ice (50 mM Tris-HCl [pH 7.5], 250 mM NaCl, 0.5% NP-40, and 50 mM NaF) in the presence of proteinase inhibitors. The cells were sonicated and the lysates centrifuged before quantification (Pierce BCA protein assay kit). Approximately 35 μg of total proteins from each sample were separated on a NuPAGE 4 to 12% Bis-Tris gel (Invitrogen) and then electroblotted to nitrocellulose membranes at 30 V for 70 min. Membranes were blocked and incubated with primary antibodies in 3% milk (SMAD2 [L16D3] mouse monoclonal antibody [#3103], 1:1,000 dilution; Cell Signaling Technology) or 1% bovine serum albumin (SMAD3 [C67H9] rabbit monoclonal antibody [#9523], 1:500 dilution; Cell Signaling Technology) overnight at 4°C. Membranes were then probed with peroxidase-conjugated goat anti-mouse or anti-rabbit antibodies (1:10,000 dilution in 3% milk; Jackson Immunoresearch) for 1 h at room temperature. Protein bands were visualized by using a SuperSignal West Pico detection kit (Pierce). To verify equal loading of proteins, membranes were stripped by using stripping buffer (Pierce) and reprobed with a mouse monoclonal antibody against actin (1:8,000 dilution in 3% milk; Sigma).
Statistical analysis.
Differences among groups were assessed by analysis of variance, and the mean between individual groups was further compared by using Tukey's HSD test (fertility data, hormone data, and real-time PCR results from the recombinant GDF9 experiments) or Dunnett's test (Smad2 and Smad3 transcript level and superovulation data). The hormone data were log transformed before statistical analysis. The data are shown as means ± the standard errors of the mean (SEM), and a P value of <0.05 was considered statistically significant.
RESULTS
Generation of Smad2, Smad3, and Smad2/3 cKO mice.
A conditional allele of the Smad2 gene with exons 9 and 10 flanked by two loxP sites was generated (38) to overcome the embryonic lethal phenotype caused by the Smad2 deletion (Fig. 1A). The Smad3 floxed allele was constructed to conditionally delete exons 2 and 3 (Fig. 1B). Since the Amhr2cre knock-in mouse has been previously validated in our laboratory (1, 29, 56-58), we chose this Cre line in the present study to conditionally delete Smad2 and Smad3 in the granulosa cell compartment. The Smad2 cKO mice (Smad2flox/−; Amhr2cre/+) were generated by crossing the Smad2+/−; Amhr2cre/+ mice to Smad2flox/flox homozygous mice, while Smad3 cKO mice (Smad3flox/−; Amhr2cre/+) were produced by crossing the Smad3+/−; Amhr2cre/+ mice to Smad3flox/flox mice. To generate the Smad2/3 cKO mouse model, the Smad2+/− mice were first crossed with Smad3+/− mice to produce Smad2/3 double heterozygous mice, and the mice were obtained at a lower ratio (1/10) than expected (1/4). However, this ratio is higher than that observed in other studies when a different Smad3-null allele was used and defects in embryonic development observed (72). The Smad2 and Smad3 double heterozygous mice were then bred to Amhr2cre/+ mice to obtain mice with the genotype of Smad2+/−; Smad3+/−; Amhr2cre/+, which were subsequently crossed with Smad2flox/flox; Smad3flox/flox double homozygous mice. Representative genotype analyses of Smad2 and Smad3 cKO mice are depicted in Fig. 1I.
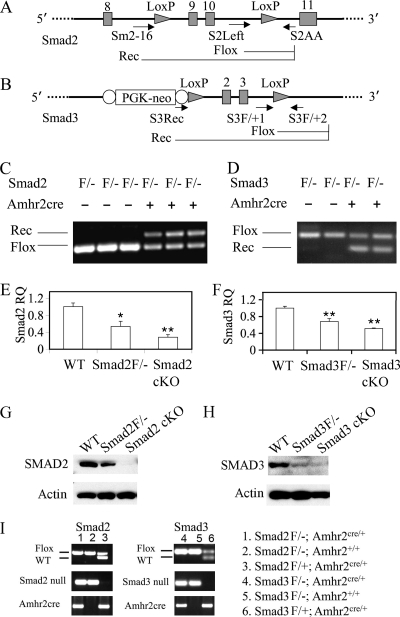
FIG. 1.
Construction and recombination of Smad2 and Smad3 floxed alleles. (A) Schematic representation of Smad2 conditional allele with exons 9 and 10 (shaded vertical bars) flanked by two loxP sites (shaded triangles). Primers S2Left and S2AA detect the Smad2 floxed allele, while Sm2-16 and S2AA identify the recombined allele of Smad2. Real-time PCR primers were designed based on the nucleotide sequence of exon 10. (B) Schematic representation of Smad3 conditional allele with exons 2 and 3 (shaded vertical bars) flanked by two loxP sites (shaded triangles). A PGK promoter driven neomycin (PGK-neo) cassette flanked by flip recombinase consensus sequences (open ovals) was illustrated. Primers S3F/+1 and S3F/+2 detect the Smad3 floxed allele, while S3Rec and S3F/+2 detect the recombined allele. Quantitative PCR primers were designed based on the nucleotide sequence of exon 1 (forward primer) and exon 2 (reverse primer; deleted exon) of Smad3. (C and D) Recombination of Smad2 and Smad3 floxed alleles in the genomic DNA of granulosa cells. Note that the recombined Smad2 or Smad3 allele was only detectable in mice that express Amhr2cre recombinase. (E and F) Relative Smad2 and Smad3 mRNA level in granulosa cells of control and Smad2 or Smad3 cKO mice. The relative mRNA abundance of genes relative to WT controls was determined by using the ΔΔCT method. The data are shown as means ± the SEM. *, P < 0.05l **, P < 0.01 (versus WT controls). (G and H) Representative Western blots of total SMAD2 and SMAD3 protein levels in the granulosa cells of control and cKO mice. Compared to WT mice, SMAD2 (∼58 kDa) was markedly reduced (data not shown) or became undetectable in the independent pools of granulosa cells from Smad2 cKO mice (G). SMAD3 protein (50 kDa) was reduced to background levels in the Smad3 cKO mice versus WT controls (H). (I) Genotyping of Smad2 or Smad3 cKO mice using genomic PCR. Representative PCR images are shown on the left, and the corresponding genotypes are denoted on the right. Flox/− and Flox/+ are abbreviated as F/− and F/+, respectively.
To determine whether the conditional alleles of Smad2 and Smad3 can undergo recombination in the mouse ovarian granulosa cells, genomic PCR analysis was performed using primers that can detect and distinguish the flox and recombined alleles (Fig. 1A to D and Table 1). As expected, the Smad2 or Smad3 recombined alleles can be detected in the cKO mice (Smad2flox/−; Amhr2cre/+ and Smad3flox/−; Amhr2cre/+) that express Amhr2cre, but not in control mice lacking the Amhr2cre knock-in alleles (Fig. 1C and D). The effect of Smad2 and Smad3 recombination on the transcription of these genes in granulosa cells was also evaluated. We analyzed the mRNA abundance of Smad2 and Smad3 in the granulosa cells from exogenous hormone-primed mice using quantitative real-time PCR with specific primers for the exons of Smad2 (both primers on exon 10 [deleted exon]) and Smad3 (forward primer on exon 1 and reverse primer on exon 2 [deleted exon]) (Table 1). A significant reduction of Smad2 and Smad3 mRNA abundance in the cKO mice was detected compared to the WT controls (Fig. 1E and F). However, incomplete and considerably variable ablation of mRNA transcripts in both Smad2 and Smad3 cKO mice at 21 days of age was observed, potentially due to the variable efficiency of Cre recombinase, which has been observed in studies using the Amhr2cre (5, 56, 58). Moreover, a correlation of Cre recombination with age has been previously reported in mice expressing Amhr2cre (5), although the percentage of recombination is likely allele specific. We further performed Western blot to examine the protein levels of total SMAD2 or SMAD3 in the granulosa cells of cKO mice. While SMAD2 protein was detected as a single band of approximately 58 kDa in the granulosa cells of both WT and Smad2flox/− mice, it was markedly reduced (data not shown) or essentially undetectable in the independent pools of granulosa cells from Smad2 cKO mice (Fig. 1G). A 50-kDa immunoreactive band corresponding to SMAD3 protein was readily detectable in the granulosa cells of WT mice, but it was almost reduced to background levels in the Smad3 cKO mice (Fig. 1H). A cross-reactive band of ∼60 kDa was also visualized in the granulosa cell samples using the SMAD3 antibody (data not shown). The 50-kDa band was confirmed to be present in the thymus tissues of WT mice but not Smad3−/− mice (data not shown).
Double cKO of Smad2 and Smad3 leads to severe fertility and fecundity defects.
To begin to analyze the ovarian functions of SMAD2 and SMAD3, the fertility and fecundity of Smad2 and Smad3 cKO mice were tested over a 6-month period. The Smad2 cKO mice exhibited normal breeding activity and demonstrated a continuous accumulation of pups during the testing period. The litter size (6.7 ± 0.5 versus 7.5 ± 0.5 pups/litter; P > 0.05; Fig. 2A) and litter/month (0.9 ± 0.0 versus 1.1 ± 0.1; P > 0.05; Fig. 2B) of the Smad2 cKO mice were not statistically different from the controls. To overcome the adverse effects of Smad3 deletion, especially those resulting from the colon cancer development, and to achieve a valid interpretation of the ovarian roles of SMAD3 because it is also expressed in the anterior pituitary, we conditionally deleted Smad3 in the ovarian granulosa cells. As expected, Smad3 cKO mice were viable and had body sizes and weights similar to those of their littermates that did not have the Amhr2cre knock-in allele (data not shown). Fertility tests showed that the Smad3flox/−; Amhr2cre/+ mice had a minor and insignificant reduction in the litter size (5.9 ± 0.3 versus 7.3 ± 0.3 pups/litter; P > 0.05; Fig. 2A). The average litter/month obtained in the Smad3 cKO mice was not significantly different from that of controls during the examined time course (Fig. 2B).
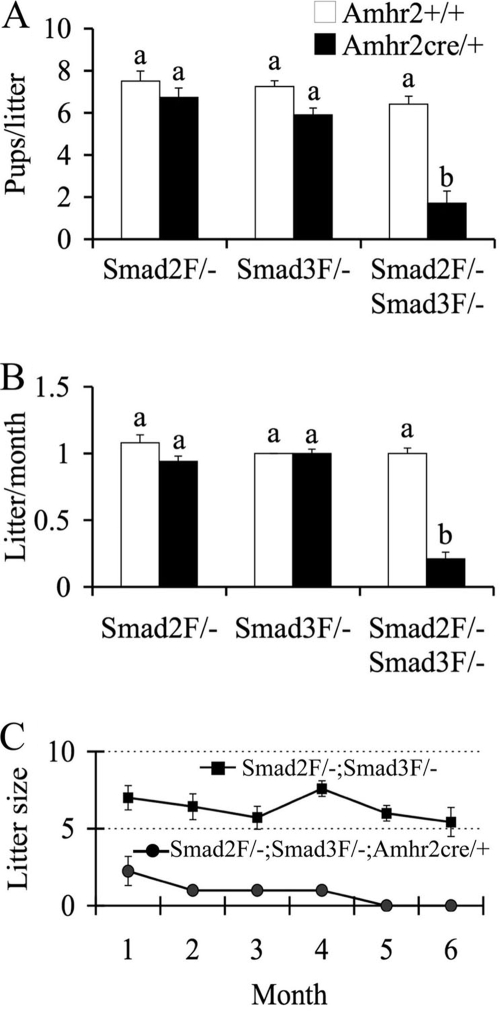
FIG. 2.
Fertility changes in Smad2, Smad3, and Smad2/3 cKO mice during a 6-month testing period. (A) Litter size of Smad2, Smad3, and Smad2/3 cKO mice versus controls. Smad2 cKO (n = 6) and Smad3 cKO (n = 8) did not show significant differences in the litter size versus Smad2flox/−; Amhr2+/+ (n = 6) and Smad3flox/−; Amhr2+/+ (n = 7) controls. The litter size of the Smad2/3 cKO mice (n = 7) was dramatically reduced compared to Smad2flox/−; Smad3flox/−; Amhr2+/+ controls (n = 8). (B) Litters/month in Smad2, Smad3, and Smad2/3 cKO mice versus controls. The litter/month of Smad2flox/−; Amhr2cre/+ and Smad3flox/−; Amhr2cre/+ mice did not significantly differ from the controls. However, the litters/month were dramatically decreased in Smad2/3 cKO mice compared to the controls. (C) Time course of litter sizes in Smad2/3 cKO mice during a 6-month testing period. The Smad2/3 cKO mice became infertile after 4 months of breeding. The data are shown as means ± the SEM, and bars without a common superscript are significantly different at P < 0.01. Flox/− is abbreviated as F/−.
Based on the observations of minimal fertility changes in the single Smad2 or Smad3 cKO females, we hypothesized that SMAD2 and SMAD3 function redundantly in the ovary. To test this hypothesis, we generated Smad2 and Smad3 double cKO mice and examined the effects of simultaneous deletion of Smad2 and Smad3 in ovarian granulosa cells. In contrast to the single Smad2 or Smad3 cKO mice, the Smad2/3 cKO (Smad2flox/−; Smad3flox/−; Amhr2cre/+) females demonstrated dramatically reduced fertility and fecundity during the testing period compared to age-matched controls (Fig. 2). The control mice showed an increase in the accumulated pups per month, whereas no obvious increase in the pup numbers of the Smad2flox/−; Smad3flox/−; Amhr2cre/+ mice was observed. The litter size (1.7 ± 0.6 versus 6.4 ± 0.4 pups/litter; P < 0.01; Fig. 2A) and litter/month (0.2 ± 0.0 versus 1.0 ± 0.0; P < 0.01; Fig. 2B) of the Smad2/3 cKO mice were substantially reduced. The litter size was further examined on a monthly basis to depict a timeline of the fertility changes. Interestingly, the Smad2/3 cKO mice demonstrated premature ovarian failure and became infertile after 4 months of breeding (Fig. 2C). These findings indicated important redundant roles of SMAD2 and SMAD3 in the ovary.
To evaluate the hormone profiles of Smad2/3 cKO mice, we measured the hormone levels (FSH, LH, E2, and P4) in these cKO mice at 3 months of age. However, no significant alterations of the hormone levels were detected. Since the Smad2/3 cKO mice become infertile after 4 months of breeding, we further analyzed the hormone levels in the infertile Smad2/3 cKO mice at 8 months of age. The results showed that the double cKO mice had elevated FSH and LH levels compared to the age-matched controls (Table 2).
TABLE 2.
Serum hormone levels of Smad2, Smad3, and Smad2/3 3-month-old cKO mice
| Mouse group and genotype | Mean hormone levela ± SEM (n)
|
|||
|---|---|---|---|---|
| FSH (ng/ml) | LH (ng/ml) | E2 (pg/ml) | P4 (ng/ml) | |
| Three-month-old mice | ||||
| Smad2flox/− | 5.83 ± 0.63 (7) | 0.23 ± 0.10 (7) | 30.48 ± 4.70 (7) | ND |
| Smad2flox/−; Amhr2cre/+ | 7.20 ± 1.38 (12) | 0.57 ± 0.14 (11) | 25.78 ± 3.83 (9) | ND |
| Smad3flox/− | 5.93 ± 0.70 (9) | 0.27 ± 0.05 (8) | 27.96 ± 4.42 (9) | ND |
| Smad3flox/−; Amhr2cre/+ | 7.02 ± 1.09 (13) | 0.35 ± 0.06 (13) | 35.84 ± 8.28 (10) | ND |
| Smad2flox/−; Smad3flox/− | 14.33 ± 5.28 (6) | 0.25 ± 0.08 (6) | 26.16 ± 2.50 (6) | 8.35 ± 3.17 (7) |
| Smad2flox/−; Smad3flox/−; Amhr2cre/+ | 10.36 ± 5.70 (5) | 0.22 ± 0.07 (5) | 24.96 ± 6.76 (5) | 4.26 ± 0.77 (3) |
| Eight-month-old mice | ||||
| Smad2flox/−; Smad3flox/− | 5.22 ± 0.36 (7) | 0.09 ± 0.02 (7) | 14.89 ± 1.86 (7) | 18.12 ± 2.56 (6) |
| Smad2flox/−; Smad3flox/−; Amhr2cre/+ | 13.23 ± 3.35* (7) | 0.21 ± 0.04* (7) | 13.61 ± 2.40 (7) | 14.12 ± 3.47 (7) |
*, P < 0.05 versus controls; ND, not determined; n, number of animals.
Smad2/3 cKO mice demonstrate disrupted follicular development and reduced ovulation efficacy.
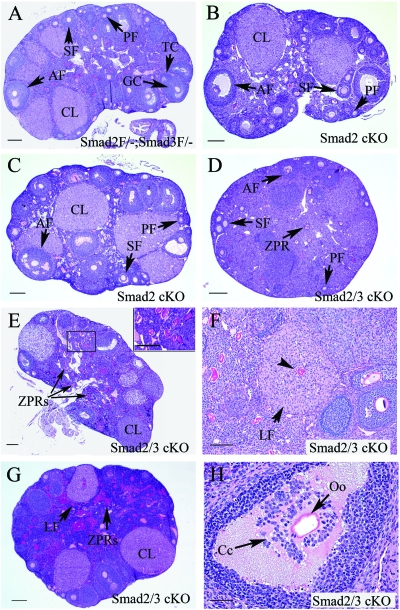
To explore the potential ovarian defects contributing to the compromised female fertility in Smad2/3 cKO mice, the ovarian histology was examined in the cKO mice at 3 and/or 8 months of age. Conditional deletion of Smad2 did not cause discernible alterations in the ovarian histology (Fig. 3B and C). Likewise, Smad3 cKO mice demonstrated minimal changes in ovarian histology (data not shown). However, histological examination of the ovaries of Smad2/3 cKO mice identified several histological abnormalities. At 3 months of age, control ovaries contained follicles at all developmental stages (primordial, primary, secondary and antral follicles) and corpora lutea (Fig. 3A). The majority of the Smad2/3 cKO mice contained fewer identifiable antral follicles (Fig. 3D and E and Table 3) compared to the controls (Fig. 3A). To determine whether there are alterations of the expanding follicles in the absence of SMAD2 and SMAD3, we compared the counts of secondary and preantral follicles between the Smad2/3 cKO mice and controls. Significant changes of secondary and preantral follicles were not observed although there was a trend toward reduction in the preantral follicles in the Smad2/3 cKO mice (P = 0.07; Table 3). Moreover, zona pellucida remnants (ZPRs), a marker of follicular atresia due to the loss of oocytes (75), accumulated in the ovaries of Smad2/3 cKO mice (Fig. 3E). The presence of luteinizing follicles encompassing trapped oocytes was another feature observed in Smad2/3 cKO mice (Fig. 3F). Although the histological observations among individual cKO mice were variable, potentially due to the differential recombination efficacy of the conditional alleles, the histological aberrations in the 8-month-old Smad2/3 cKO mice represents a more severe version of that observed in the 3-month-old mice with increased accumulation of ZPRs (Fig. 3G). As illustrated in Fig. 3H, aberrant cumulus cell-oocyte histology could also be observed in the Smad2/3 cKO mice.
FIG. 3.
Histological analyses of ovaries from Smad2/3 cKO mice. (A) Ovarian histology of a 3-month-old Smad2flox/−; Smad3flox/− mouse. The ovary contained follicles at all developmental stages (primordial, primary, secondary, and antral follicles) and corpora lutea. Flox/− is abbreviated as F/−. (B) Ovarian histology of a 3-month-old Smad2 cKO mouse demonstrating normal follicular development. (C) Ovarian histology of an 8-month-old Smad2 cKO mouse demonstrating indiscernible abnormalities. (D, E, and F) Ovarian histology of 3-month-old Smad2/3 cKO mice. Note that the ovaries of Smad2/3 cKO mice contained fewer antral follicles (D and E) compared to the controls (A). Other histological features of Smad2/3 cKO ovaries were the accumulation of ZPRs (D to F) and the presence of luteinizing follicles (F; arrow) with trapped oocytes (F; arrowhead). (G) Ovarian histology of an 8-month-old Smad2/3 cKO mouse. Note the lack of large antral follicles, the accumulation of ZPRs, and the presence of a luteinizing follicle. (H) Ovarian histology of an 8-month-old Smad2/3 cKO mouse demonstrating aberrant cumulus histology (arrow). PF, primordial and primary follicle; SF, secondary follicle; AF, antral follicle; CL, corpora lutea; GC, granulosa cell; Cc, cumulus cell; TC, theca cell; Oo, oocyte; LF, luteinizing follicle. Scale bars: A to E and G, 200 μm; F, 100 μm; H, 50 μm.
TABLE 3.
Quantification of growing follicles in Smad2/3 cKO mice
| Genotype | Mean no. of follicles/mm2 ± SEM
|
||
|---|---|---|---|
| Secondary | Preantral | Antral | |
| Smad2flox/−; Smad3flox/− | 1.07 ± 0.22 | 1.82 ± 0.14 | 1.29 ± 0.13 |
| Smad2flox/; Smad3flox/−; Amhr2cre/+ | 0.63 ± 0.14 | 1.15 ± 0.28 | 0.73 ± 0.10a |
P < 0.05 versus control.
To address potential changes in ovulation efficiency, pharmacologic superovulation experiments were performed on these cKO mice using exogenous hormones (PMSG-hCG). The numbers of oocytes recovered from Smad2 cKO (36.0 ± 2.2) and Smad3 cKO mice (31.8 ± 3.8) were not significantly different from that of controls (42.4 ± 2.9; P > 0.05; Table 4). However, a dramatic reduction in the number of ovulated oocytes was found in the Smad2/3 cKO mice, which ovulated about half of the oocytes of the controls (23.3 ± 4.6 versus 42.4 ± 2.9; P < 0.01; Table 4).
TABLE 4.
Superovulation data of Smad2, Smad3, and Smad2/3 cKO
| Genotype | No. of mice | Mean no. of oocytes/female ± SEM |
|---|---|---|
| WT | 7 | 42.4 ± 2.9 |
| Smad2flox/−; Amhr2cre/+ | 3 | 36.0 ± 2.2 |
| Smad3flox/−; Amhr2cre/+ | 5 | 31.8 ± 3.8 |
| Smad2flox/−; Smad3flox/−; Amhr2cre/+ | 3 | 23.3 ± 4.6a |
P < 0.01 versus WT controls.
Smad2/3 cKO mice have impaired cumulus expansion.
Since the histological examination of the Smad2/3 cKO ovaries provides an indication of potential cumulus cell defects in the Smad2/3 cKO mice, we next performed both in vivo and in vitro cumulus cell expansion analyses in the Smad2/3 cKO mice to define whether there is impaired cumulus expansion during the development of preovulatory follicles. PMSG induced follicular development in the control mice with typical cumulus cell expansion present in the preovulatory follicles 6 h after hCG administration (Fig. 4A and B). In contrast, the Smad2/3 cKO mice demonstrated minimal cumulus expansion activity after the same hormonal treatment (Fig. 4C and D). In the majority of preovulatory follicles examined in the Smad2/3 cKO mice, fewer cumulus cells were attached to the oocytes with minimal expansion of cumulus cells (Fig. 4C and D). Even in cases where cumulus expansion did occur, the extension of expansion seldom matched that of the WT controls (data not shown).
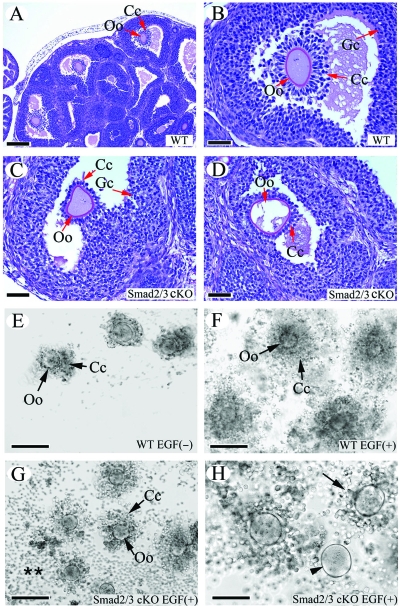
FIG. 4.
In vivo and in vitro cumulus expansion defects in Smad2/3 cKO mice. (A and B) Cumulus expansion in WT mice treated with PMSG-hCG. Note the outward radiation pattern of the expanded cumulus cells from the oocytes illustrated in panel B. (C and D) The cumulus cells failed to undergo expansion in response to PMSG-hCG treatment in Smad2/3 cKO mice. Note that fewer cumulus cells were attached to the oocyte, and these cells did not undergo the typical expansion observed in control mice (A and B). Even though expansion of cumulus cells was observed occasionally in some large antral follicles, the expansion pattern observed in WT mice was lacking (data not shown). (E) COCs from WT mice cultured in the absence of EGF. (F) In vitro expansion of cumulus cells from WT mice stimulated by EGF (10 ng/ml). Note the expansion pattern of cumulus cells outward from the oocytes in the COC culture. (G and H) Cultured COCs from Smad2/3 cKO mice stimulated by EGF (10 ng/ml). Note the impaired expansion of cumulus cells (H; arrow) and the denuded oocyte (H; arrowhead) in the COC culture. The detached cumulus cells from COCs are indicated (**). Oo, oocyte; Cc, cumulus cell; Gc, granulosa cell. Scale bars: A, 200 μm; B to D and H, 50 μm; E to G, 100 μm.
Complementary to the in vivo analysis, we examined the in vitro cumulus expansion in the Smad2/3 cKO mice by culturing COCs from both controls and the Smad2/3 cKO mutants and comparing the EGF-induced cumulus expansion. COCs from the WT controls (Fig. 4E) and Smad2/3 cKO mice (data not shown) remained compact and did not expand in the absence of EGF. EGF (10 ng/ml) induced cumulus cell expansion in the control mice 18 h after culture (Fig. 4F). In contrast to the typical cumulus expansion observed in the controls (Fig. 4F), the majority of COCs (71%) from Smad2/3 cKO mice underwent disorganized and limited expansion in response to EGF stimulation (Fig. 4G and H). Some of the culmulus cells from the mutant mice were readily detached from the COCs and attached to the culture plate (Fig. 4G and H). In the most severe case, the oocytes were denuded (18%) during the culture period (Fig. 4H). Thus, redundant signaling through both SMAD2 and SMAD3 is required for normal cumulus expansion.
Effect of recombinant mouse GDF9 on the expression of cumulus expansion-related transcripts in granulosa cells deficient in SMAD2 and SMAD3.
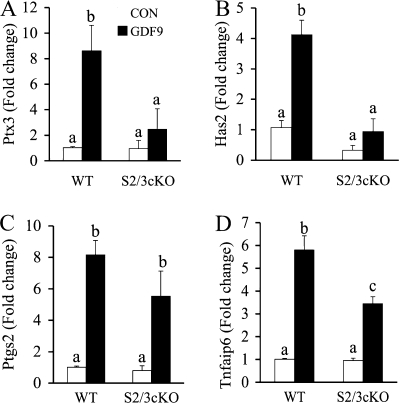
The expansion of cumulus cells requires cumulus expansion-enabling factors (CEEFs), and our previous studies identified GDF9 as a potent stimulator of cumulus expansion-related transcripts Ptx3, Has2, Ptgs2, and Tnfaip6 in mouse granulosa cells (18, 19, 69). To test whether SMAD2/3 is required for GDF9 induction of these cumulus expansion-related transcripts in the ovarian granulosa cells, we cultured the granulosa cells from control and Smad2/3 cKO mice in the absence or presence of recombinant mouse GDF9 (100 ng/ml). The cumulus expansion-related transcript levels in the granulosa cells were determined by real-time PCR assays. The transcript levels of expansion-related transcripts in untreated granulosa cells of Smad2/3 cKO mice are not significantly altered compared to WT controls, which can be possibly explained by the incomplete deletion of SMAD2 and SMAD3 in ovarian granulosa cells (i.e., attenuated SMAD2/3 signaling may be still sufficient for the baseline expression of these transcripts) and/or the existence of an unidentified mechanism contributing to basal level expression of the expansion-related transcripts. Consistent with the defective cumulus cell expansion phenotype, the pattern of GDF9 stimulated-cumulus expansion related-transcripts in ovarian granulosa cells was altered in the Smad2/3 cKO mice (Fig. 5A to D). GDF9 induction of Ptx3 and Has2 is substantially suppressed in the granulosa cells of Smad2/3 cKO mice (Fig. 5A and B), whereas stimulation of Ptgs2 and Tnfaip6 by GDF9 still occurred (Fig. 5C) or was partially affected (Fig. 5D) in the granulosa cells of Smad2/3 cKO mice.
FIG. 5.
Effect of conditional deletion of Smad2 and Smad3 on GDF9-stimulated cumulus expansion-related transcript expression in granulosa cells. Immature female mice (WT, n = 5; Smad2/3 cKO [S2/3 cKO], n = 3) were primed with PMSG, and granulosa cells were collected and cultured in the absence or presence of 100 ng of recombinant mouse GDF9/ml. GDF9 significantly induced expression of Ptx3 (A), Has2 (B), Ptgs2 (C), and Tnfaip6 (D) (4- to 8-fold increases) in the granulosa cells of WT mice. However, in the granulosa cells of Smad2/3 cKO mice, GDF9-stimulated Ptx3 (A) and Has2 (B) increases were suppressed. GDF9-stimulated Ptgs2 (C) and Tnfaip6 (D) increases still occurred in the Smad2/3 cKO granulosa cells but in an attenuated pattern. Changes in the relative mRNA expression of genes relative to WT control were determined by using the ΔΔCT method. The data are shown as means ± the SEM, and bars without a common superscript are significantly different.
DISCUSSION
Redundancy of SMAD2 and SMAD3 in female fertility.
The incomplete knowledge of the functional properties of SMAD2 and SMAD3 in reproduction is partially attributable to the fact that Smad2-null mice are embryonically lethal while Smad3 homozygous mutants are predisposed to colorectal cancer (79) or demonstrate immune defects (76) with reduced life span. With the aim to define the in vivo roles of SMAD2 and SMAD3 in the ovary, we undertook a cKO strategy to produce mouse models that lack SMAD2, SMAD3, and both SMAD2 and SMAD3 in ovarian granulosa cells. Three different mouse lines of Smad3 knockouts have been generated with targeted disruption of the respective exon 1, exon 2, or exon 8 (11, 76, 79). Interestingly, distinct phenotypic consequences of Smad3 deletion are reported in the three mouse lines except the consistent observations of body size reduction by all authors (11, 76, 79). Severe fertility defects were reported in mice with exon 8 knockout (67, 68) but not in mice with exon 2 inactivation (79). Moreover, the exon 2 knockout mice are able to respond to exogenous hormone stimulation, although with reduced responsiveness (Q. Li and M. M. Matzuk, unpublished observations). The mechanisms underlying the distinct phenotype of the different Smad3 knockout mouse lines are unknown. However, the deletion of exon 8 retains the linker region of SMAD3, which can be potentially targeted by non-TGF-β signals (12). In addition, the truncated protein resulting from exon 8 deletion demonstrates a dominant-negative effect in an in vitro assay system when expressed at a high level (76). Therefore, we used the Smad3 exon 2 knockout mice for the generation of cKO models in the present study.
The Smad2 cKO mouse model provided evidence that SMAD2 per se is not essential in transducing TGF-β/activin/nodal/GDF9 signals in granulosa cells, since ablation of SMAD2 in this cell lineage does not lead to discernible ovarian phenotype. Consistent with the previous observation (79), the Smad3−/− females were subfertile in our colony. However, our attempt to establish a comprehensive fertility record was hampered by the progressive lethality of these mice secondary to the development of colorectal cancer. Likewise, the observed fertility deficiency in Smad3−/− mice might partially reflect the systematic effect of Smad3 deletion (i.e., hormonal imbalance due the pituitary involvement of SMAD3, propensity of tumor development and tumor burden, growth retardation, or immune dysfunction, etc.). As expected, mice with granulosa cell ablation of SMAD3 did not suffer from colorectal cancer and have a normal life span. Thus, the Smad3 cKO mice represent a more appropriate model to interpret the ovary specific roles of SMAD3, particularly since FSH is not altered in the cKO mice but remains elevated in the Smad3-null mice. Interestingly, a dramatic fertility reduction was found in the Smad2 and Smad3 double cKO mice, although minimal fertility changes were observed in Smad2 or Smad3 single cKO mice. The current results support the concept that SMAD3 is capable of activating essential target genes downstream of TGF-β-related ligands (17). Furthermore, loss of SMAD2/3 signaling in granulosa cells led to disrupted follicular development and reduced ovulation efficacy. Therefore, SMAD2 and SMAD3 function redundantly in the mouse ovary to maintain female fertility. Of note, mice expressing a Smad2 dominant-negative transgene (Smad2-dn) directed by the Amh promoter (6) exhibit subfertility and ovarian epithelial cyst formation as early as 3 months of age (8), which was not observed in the Smad2/3 cKO mice. The phenotypic differences between the Smad2 cKO or Smad2/3 cKO and the Smad2-dn mouse models await further investigation.
Interpretation of the global phenotypic outcome of Smad2/3 cKO mice.
Despite the pivotal roles of SMAD2/3 signaling in female fertility revealed by the present study, the TGF-β superfamily ligands in addition to GDF9 required for the activation of ovarian SMAD2/3 pathway are unknown, because multiple ligands including TGF-βs, activins, and nodal impinge on these SMADs in vitro and, along with GDF9, might signal through both SMAD-dependent and -independent pathways (12). As predicted from the above fact, absence of SMAD2/3 signaling may therefore not phenocopy the loss of any individual SMAD2/3-related ligand. In fact, conditional deletion of ovarian activins (both activin βA and βB subunits) results in a distinct ovarian phenotype from that demonstrated in the present study; the activin β subunit-deficient ovaries have increased numbers of corpora lutea (56). Moreover, ablation of all TGF-β signaling in the ovarian granulosa cells by conditionally inactivating Smad4 results in premature luteinization of granulosa cells, leading to the predisposition of premature ovarian failure (57). The Smad4 cKO mice do not phenocopy the loss of any individual TGF-β family ligands reported (e.g., loss of GDF9, activins, etc.), likely due to the loss of both BMP and activin/TGF-β signaling activity. Therefore, the phenotype associated with deletion of Smad2 and Smad3 is a converged and profound manifestation of the loss of interactions/balance among ligands that signal through SMAD2/3. Recently, Smad1/5/8 cKO mice have been generated by our group, and these mice develop metastatic tumors with increased signaling of SMAD2/3 in granulosa cells, leading to the hypothesis that SMAD1/5/8 acts as a tumor suppressor pathway that potentially antagonizes the SMAD2/3 signaling (58). Based on this hypothesis, loss of SMAD2/3 signaling in the normal ovary should not result in a tumor phenotype but may attenuate tumor development/progression in ovaries that are tumorigenic. This notion is in line with our recent findings (35) and a report from another laboratory (40) that loss of SMAD3 results in reduced gonadal tumor formation and progression in mice deficient in inhibin. Although the cross talk between the SMAD2/3 and SMAD1/5/8 pathways is not well defined, one recent study demonstrated that BMP signaling is potentiated in SMAD3-deficient chondrocytes (36), further supporting the existence of a counterregulatory mechanism between SMAD2/3 and SMAD1/5/8 pathways. Thus, the balance between the SMAD2/3 and SMAD1/5/8 pathways and the complex cross talk/interactions at multiple levels may define the normal cellular responses to the TGF-β family ligands. This may also help to explain why loss of SMAD2/3 signaling leads to a distinct phenotype from that of genetic deletion of any individual SMAD2/3-related ligand. Identification of the potential interactions between these two signaling pathways remains our further aim.
SMAD2/3 signaling is indispensable for normal cumulus expansion.
Bidirectional communication between oocytes and the surrounding somatic cells has been recognized as an essential regulatory loop for modulating/coordinating various fundamental processes of both cell types (20, 21, 24, 46). The gonadotropin surge induces the resumption of oocyte meiosis and expansion of the cumulus cells characterized by the formation of the hyaluronan-rich matrix by the COCs within the preovulatory follicles (62, 63). Expansion of cumulus cells protects the oocyte from the subsequent mechanic and enzymatic stresses during ovulation, and the initiation of cumulus expansion requires CEEFs, some of which belong to the TGF-β superfamily (e.g., GDF9 and BMP15) and can signal through SMAD2/3 (13, 16, 23). Although the argument regarding CEEFs remains unsettled (59), a combination of GDF9, BMP15, and other unidentified oocyte-secreted factors likely function as CEEFs (15, 18, 25, 78). Importantly, CEEFs can induce the expression of cumulus expansion-related transcripts (Ptx3, Has2, Ptgs2, Tnfaip6, etc.) required for cumulus expansion (14, 16, 25), and mice lacking Ptx3 (64, 69), Ptgs2 (71), and Tnfaip6 (52, 53) have defects in cumulus expansion. In support of the involvement of SMAD2/3 signaling in cumulus expansion, our studies herein show that deletion of Smad2/3 in the granulosa cell compartment causes defective expansion of the cumulus cells. Moreover, loss of SMAD2 and SMAD3 does not seem to affect the integrity of the GDF9 receptor machinery since the gene expression of GDF9 receptors, TGF-β receptor type 1 (Tgfbr1) and BMP receptor type 2 (Bmpr2) (47), was not altered in COCs from Smad2/3 cKO mice (Li and Matzuk, unpublished). Therefore, the defective cumulus expansion observed in the Smad2/3 cKO mice is the result of loss of SMAD2 and SMAD3 signaling proteins. However, our results cannot distinguish whether the impaired cumulus expansion in the Smad2/3 cKO mice is attributable to a defective response to EGF or CEEFs because of the absence of SMAD2 and SMAD3 at the time of treatment or due to prior defective differentiation of cumulus cells resulting from loss of SMAD2/3 signaling during follicular development.
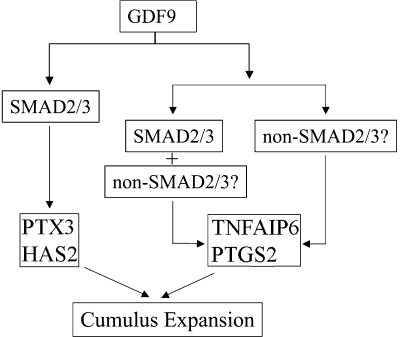
To further understand the mechanisms underlying the defective cumulus expansion in Smad2/3 cKO mice, we utilized a granulosa cell culture system to examine the effect of GDF9 on the expansion-related transcripts for the following reasons. (i) Granulosa cells are the most abundant source of cells in which Smad2 and Smad3 are deleted in our cKO model. (ii) GDF9 induction of cumulus expansion-related genes in the mouse granulosa cell culture system has been characterized, and a correlation between GDF9-induced gene expression (e.g., Has2 and Ptgs2) in granulosa cell culture and GDF9-promoted cumulus expansion in oocytectomized COCs (OOX) has been suggested in our previous study (18). As indirect evidence corroborating the cumulus expansion defects, the induction of cumulus expansion-related transcripts by GDF9 in the granulosa cells of Smad2/3 cKO mice was altered. However, variable effects were found on the transcripts examined. Loss of SMAD2/3 signaling had a dramatic impact on GDF9-induced Ptx3 and Has2 expression in the ovarian granulosa cells, whereas stimulation of Ptgs2 and Tnfaip6 transcripts by GDF9 still occurred, although somewhat intermediate between treated and untreated WT controls. One concern is that the incomplete recombination of Smad2/3 in the cKO mice could as well contribute to the differential effects of GDF9 on cumulus expansion-related transcripts in the granulosa cells. However, this seems less likely in view of the dramatic loss of GDF9 induction of SMAD2/3-dependent regulation of Ptx3 and Has2 in the Smad2/3 cKO mice. Interestingly, some cumulus cells from the Smad2/3 cKO mice detached from the COCs during the in vitro culture, which is a milder version of that observed in PTX3-deficient cumulus cells that readily detach from the cumulus oophorus, resulting in denuded oocytes (64, 69). Therefore, alteration of GDF9-induced cumulus expansion-related transcripts expression in Smad2/3 cKO mice may partially account for the defective phenotype of cumulus expansion (see Fig. 6 for a hypothetical model delineating the GDF9 regulation of cumulus expansion-related transcripts through SMAD2/3-dependent and/or -independent pathways).
FIG. 6.
Hypothetical working model for GDF9 induction of cumulus expansion-related transcripts through SMAD2/3-dependent and/or SMAD2/3-independent pathways. GDF9 can signal through SMAD2/3 to regulate PTX3 and HAS2, since conditional deletion of Smad2 and Smad3 substantially suppressed GDF9 induced Ptx3 and Has2 expression. In contrast, GDF9-induced Ptgs2 and Tnfaip6 expression still occurred in Smad2/3 conditionally deleted cells, suggesting that GDF9 might signal through both SMAD2/3-dependent and/or SMAD2/3-independent pathways to confer the regulation of these genes. The nature of the SMAD-independent pathways remains to be identified.
Recent studies from Diaz et al. (15) elegantly elaborated the complex involvement of SMAD2/3 signaling in the regulation of expansion-related transcripts in cumulus cells using SMAD3 and SMAD2/3 inhibitors (SIS3 and SB431542). Interestingly, inhibition of both SMAD2 and SMAD3 has a marked effect on EGF-induced increases in Ptx3 and Has2, while the regulation of Ptgs2 and Tnfaip6 mRNAs is not acutely dependent on the SMAD2/3 activation (13), leading to the hypothesis that one or more CEEFs function during cumulus expansion. Indeed, studies from Dragovic et al. (15, 16) also support the involvement of more than one CEEF in cumulus expansion. The present study further supports GDF9 as an oocyte-secreted CEEF whose function is mediated at least partially through SMAD2/3 signaling. In combination with studies using the SMAD2 and SMAD3 inhibitors (13, 16), our results indicate that both SMAD2 and SMAD3 are required for the full cumulus expansion response and provide relevant albeit indirect evidence supporting the mechanistic implication of altered induction of cumulus expansion-related transcripts by oocyte secreted factors in the defective cumulus expansion of Smad2/3 cKO mice.
Collectively, the present study demonstrates that SMAD2 and SMAD3 are functionally redundant in mouse ovary in the maintenance of female fertility. Our results provide genetic evidence that the intraovarian SMAD2/3 signaling machinery is implicated in mediating oocyte-produced signals that are essential for coordinating key events of the ovulatory process including cumulus expansion.
Acknowledgments
We thank John Eppig for critical reading of the manuscript and for insightful comments. We thank Richard Behringer for providing the Amhr2cre mice, Maria Festing for helpful information on the Smad2 genotyping, and Claudia Andreu-Vieyra for suggestions on follicle quantification.
This project was supported by National Institutes of Health grant HD32067 (to M.M.M.) and a Burroughs Wellcome Career Award in the Biomedical Sciences grant (to S.A.P.). Serum hormone analyses, performed at the University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core, were supported by NICHD (SCCPRR) grant U54-HD28934.
Footnotes
Published ahead of print on 22 September 2008.
REFERENCES
- 1.Andreu-Vieyra, C., R. Chen, and M. M. Matzuk. 2007. Effects of granulosa cell-specific deletion of Rb in Inha-alpha null female mice. Endocrinology. 1483837-3849. [DOI] [PubMed] [Google Scholar]
- 2.Andreu-Vieyra, C., R. Chen, and M. M. Matzuk. 2008. Conditional deletion of the retinoblastoma (Rb) gene in ovarian granulosa cells leads to premature ovarian failure. Mol. Endocrinol. 222141-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashcroft, G. S., S. J. Mills, K. C. Flanders, L. A. Lyakh, M. A. Anzano, S. C. Gilliver, and A. B. Roberts. 2003. Role of Smad3 in the hormonal modulation of in vivo wound healing responses. Wound Repair Regen. 11468-473. [DOI] [PubMed] [Google Scholar]
- 4.Billiar, R. B., J. B. St Clair, N. C. Zachos, M. G. Burch, E. D. Albrecht, and G. J. Pepe. 2004. Localization and developmental expression of the activin signal transduction proteins Smads 2, 3, and 4 in the baboon fetal ovary. Biol. Reprod. 70586-592. [DOI] [PubMed] [Google Scholar]
- 5.Boerboom, D., M. Paquet, M. Hsieh, J. Liu, S. P. Jamin, R. R. Behringer, J. Sirois, M. M. Taketo, and J. S. Richards. 2005. Misregulated Wnt/β-catenin signaling leads to ovarian granulosa cell tumor development. Cancer Res. 659206-9215. [DOI] [PubMed] [Google Scholar]
- 6.Bristol-Gould, S. K., C. G. Hutten, C. Sturgis, S. M. Kilen, K. E. Mayo, and T. K. Woodruff. 2005. The development of a mouse model of ovarian endosalpingiosis. Endocrinology 1465228-5236. [DOI] [PubMed] [Google Scholar]
- 7.Brown, K. A., J. A. Pietenpol, and H. L. Moses. 2007. A tale of two proteins: differential roles and regulation of Smad2 and Smad3 in TGF-β signaling. J. Cell Biochem. 1019-33. [DOI] [PubMed] [Google Scholar]
- 8.Burdette, J. E., R. M. Oliver, V. Ulyanov, S. M. Kilen, K. E. Mayo, and T. K. Woodruff. 2007. Ovarian epithelial inclusion cysts in chronically superovulated CD1 and Smad2 dominant-negative mice. Endocrinology 1483595-3604. [DOI] [PubMed] [Google Scholar]
- 9.Chang, H., C. W. Brown, and M. M. Matzuk. 2002. Genetic analysis of the mammalian transforming growth factor-beta superfamily. Endocrinol. Rev. 23787-823. [DOI] [PubMed] [Google Scholar]
- 10.Chang, H., A. L. Lau, and M. M. Matzuk. 2001. Studying TGF-beta superfamily signaling by knockouts and knockins. Mol. Cell Endocrinol. 18039-46. [DOI] [PubMed] [Google Scholar]
- 11.Datto, M. B., J. P. Frederick, L. Pan, A. J. Borton, Y. Zhuang, and X. F. Wang. 1999. Targeted disruption of Smad3 reveals an essential role in transforming growth factor beta-mediated signal transduction. Mol. Cell. Biol. 192495-2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derynck, R., and Y. E. Zhang. 2003. Smad-dependent and Smad-independent pathways in TGF-β family signalling. Nature 425577-584. [DOI] [PubMed] [Google Scholar]
- 13.Diaz, F. J., K. Wigglesworth, and J. J. Eppig. 2007. Oocytes determine cumulus cell lineage in mouse ovarian follicles. J. Cell Sci. 1201330-1340. [DOI] [PubMed] [Google Scholar]
- 14.Diaz, F. J., M. J. O'Brien, K. Wigglesworth, and J. J. Eppig. 2006. The preantral granulosa cell to cumulus cell transition in the mouse ovary: development of competence to undergo expansion. Dev. Biol. 29991-104. [DOI] [PubMed] [Google Scholar]
- 15.Dragovic, R. A., L. J. Ritter, S. J. Schulz, F. Amato, D. T. Armstrong, and R. B. Gilchrist. 2005. Role of oocyte-secreted growth differentiation factor 9 in the regulation of mouse cumulus expansion. Endocrinology 1462798-2806. [DOI] [PubMed] [Google Scholar]
- 16.Dragovic, R. A., L. J. Ritter, S. J. Schulz, F. Amato, J. G. Thompson, D. T. Armstrong, and R. B. Gilchrist. 2007. Oocyte-secreted factor activation of SMAD2/3 signaling enables initiation of mouse cumulus cell expansion. Biol. Reprod. 76848-857. [DOI] [PubMed] [Google Scholar]
- 17.Dunn, N. R., C. H. Koonce, D. C. Anderson, A. Islam, E. K. Bikoff, and E. J. Robertson. 2005. Mice exclusively expressing the short isoform of Smad2 develop normally and are viable and fertile. Genes Dev. 19152-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elvin, J. A., A. T. Clark, P. Wang, N. M. Wolfman, and M. M. Matzuk. 1999. Paracrine actions of growth differentiation factor-9 in the mammalian ovary. Mol. Endocrinol. 131035-1048. [DOI] [PubMed] [Google Scholar]
- 19.Elvin, J. A., C. Yan, and M. M. Matzuk. 2000. Growth differentiation factor-9 stimulates progesterone synthesis in granulosa cells via a prostaglandin E2/EP2 receptor pathway. Proc. Natl. Acad. Sci. USA 9710288-10293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eppig, J. J. 2001. Oocyte control of ovarian follicular development and function in mammals. Reproduction 122829-838. [DOI] [PubMed] [Google Scholar]
- 21.Eppig, J. J., F. Chesnel, Y. Hirao, M. J. O'Brien, F. L. Pendola, S. Watanabe, and K. Wigglesworth. 1997. Oocyte control of granulosa cell development: how and why. Hum. Reprod. 12127-132. [PubMed] [Google Scholar]
- 22.Findlay, J. K., A. E. Drummond, M. L. Dyson, A. J. Baillie, D. M. Robertson, and J. F. Ethier. 2002. Recruitment and development of the follicle; the roles of the transforming growth factor-beta superfamily. Mol. Cell Endocrinol. 19135-43. [DOI] [PubMed] [Google Scholar]
- 23.Gilchrist, R. B., L. J. Ritter, and D. T. Armstrong. 2004. Oocyte-somatic cell interactions during follicle development in mammals. Anim. Reprod. Sci. 82-83431-446. [DOI] [PubMed] [Google Scholar]
- 24.Gilchrist, R. B., L. J. Ritter, S. Myllymaa, N. Kaivo-Oja, R. A. Dragovic, T. E. Hickey, O. Ritvos, and D. G. Mottershead. 2006. Molecular basis of oocyte-paracrine signalling that promotes granulosa cell proliferation. J. Cell Sci. 1193811-3821. [DOI] [PubMed] [Google Scholar]
- 25.Gui, L.-M., and I. M. Joyce. 2005. RNA interference evidence that growth differentiation factor-9 mediates oocyte regulation of cumulus expansion in Mice. Biol. Reprod. 72195-199. [DOI] [PubMed] [Google Scholar]
- 26.Hamamoto, T., H. Beppu, H. Okada, M. Kawabata, T. Kitamura, K. Miyazono, and M. Kato. 2002. Compound disruption of smad2 accelerates malignant progression of intestinal tumors in apc knockout mice. Cancer Res. 625955-5961. [PubMed] [Google Scholar]
- 27.Heyer, J., D. Escalante-Alcalde, M. Lia, E. Boettinger, W. Edelmann, C. L. Stewart, and R. Kucherlapati. 1999. Postgastrulation Smad2-deficient embryos show defects in embryo turning and anterior morphogenesis. Proc. Natl. Acad. Sci. USA 9612595-12600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jamin, S. P., N. A. Arango, Y. Mishina, M. C. Hanks, and R. R. Behringer. 2002. Requirement of Bmpr1a for Mullerian duct regression during male sexual development. Nat. Genet. 32408-410. [DOI] [PubMed] [Google Scholar]
- 29.Jorgez, C. J., M. Klysik, S. P. Jamin, R. R. Behringer, and M. M. Matzuk. 2004. Granulosa cell-specific inactivation of follistatin causes female fertility defects. Mol. Endocrinol. 18953-967. [DOI] [PubMed] [Google Scholar]
- 30.Juengel, J. L., and K. P. McNatty. 2005. The role of proteins of the transforming growth factor-beta superfamily in the intraovarian regulation of follicular development. Hum. Reprod. Update 11143-160. [DOI] [PubMed] [Google Scholar]
- 31.Kaivo-oja, N., L. A. Jeffery, O. Ritvos, and D. G. Mottershead. 2006. Smad signalling in the ovary. Reprod. Biol. Endocrinol. 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knight, P. G., and C. Glister. 2003. Local roles of TGF-beta superfamily members in the control of ovarian follicle development. Anim. Reprod. Sci. 78165-183. [DOI] [PubMed] [Google Scholar]
- 33.Knight, P. G., and C. Glister. 2006. TGF-beta superfamily members and ovarian follicle development. Reproduction 132191-206. [DOI] [PubMed] [Google Scholar]
- 34.Kretschmer, A., K. Moepert, S. Dames, M. Sternberger, J. Kaufmann, and A. Klippel. 2003. Differential regulation of TGF-beta signaling through Smad2, Smad3, and Smad4. Oncogene 226748-6763. [DOI] [PubMed] [Google Scholar]
- 35.Li, Q., J. M. Graff, A. E. O'Connor, K. L. Loveland, and M. M. Matzuk. 2007. SMAD3 regulates gonadal tumorigenesis. Mol. Endocrinol. 212472-2486. [DOI] [PubMed] [Google Scholar]
- 36.Li, T.-F., M. Darowish, M. J. Zuscik, D. Chen, E. M. Schwarz, R. N. Rosier, H. Drissi, and R. J. O'Keefe. 2006. Smad3-deficient chondrocytes have enhanced BMP signaling and accelerated differentiation. J. Bone Miner. Res. 214-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu, Y., M. Festing, J. C. Thompson, M. Hester, S. Rankin, H. M. El-Hodiri, A. M. Zorn, and M. Weinstein. 2004. Smad2 and Smad3 coordinately regulate craniofacial and endodermal development. Dev. Biol. 270411-426. [DOI] [PubMed] [Google Scholar]
- 38.Liu, Y., M. H. Festing, M. Hester, J. C. Thompson, and M. Weinstein. 2004. Generation of novel conditional and hypomorphic alleles of the Smad2 gene. Genesis 40118-123. [DOI] [PubMed] [Google Scholar]
- 39.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 40.Looyenga, B. D., and G. D. Hammer. 2007. Genetic removal of Smad3 from inhibin-null mice attenuates tumor progression by uncoupling extracellular mitogenic signals from the cell cycle machinery. Mol. Endocrinol. 212440-2457. [DOI] [PubMed] [Google Scholar]
- 41.Massague, J. 1992. Receptors for the TGF-beta family. Cell 691067-1070. [DOI] [PubMed] [Google Scholar]
- 42.Massague, J. 1998. TGF-beta signal transduction. Annu. Rev. Biochem. 67753-791. [DOI] [PubMed] [Google Scholar]
- 43.Massague, J. 2000. How cells read TGF-beta signals. Nat. Rev. Mol. Cell Biol. 1169-178. [DOI] [PubMed] [Google Scholar]
- 44.Massague, J., S. W. Blain, and R. S. Lo. 2000. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell 103295-309. [DOI] [PubMed] [Google Scholar]
- 45.Matzuk, M. M. 2000. Revelations of ovarian follicle biology from gene knockout mice. Mol. Cell Endocrinol. 16361-66. [DOI] [PubMed] [Google Scholar]
- 46.Matzuk, M. M., K. H. Burns, M. M. Viveiros, and J. J. Eppig. 2002. Intercellular communication in the mammalian ovary: oocytes carry the conversation. Science 2962178-2180. [DOI] [PubMed] [Google Scholar]
- 47.Mazerbourg, S., and A. J. W. Hsueh. 2006. Genomic analyses facilitate identification of receptors and signalling pathways for growth differentiation factor 9 and related orphan bone morphogenetic protein/growth differentiation factor ligands. Hum. Reprod. Update 12373-383. [DOI] [PubMed] [Google Scholar]
- 48.Mehra, A., and J. L. Wrana. 2002. TGF-beta and the Smad signal transduction pathway. Biochem. Cell Biol. 80605-622. [DOI] [PubMed] [Google Scholar]
- 49.Moustakas, A., and D. Kardassis. 1998. Regulation of the human p21/WAF1/Cip1 promoter in hepatic cells by functional interactions between Sp1 and Smad family members. Proc. Natl. Acad. Sci. USA 956733-6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moustakas, A., S. Souchelnytskyi, and C. H. Heldin. 2001. Smad regulation in TGF-beta signal transduction. J. Cell Sci. 1144359-4369. [DOI] [PubMed] [Google Scholar]
- 51.Nomura, M., and E. Li. 1998. Smad2 role in mesoderm formation, left-right patterning and craniofacial development. Nature 393786-790. [DOI] [PubMed] [Google Scholar]
- 52.Ochsner, S. A., A. J. Day, M. S. Rugg, R. M. Breyer, R. H. Gomer, and J. S. Richards. 2003. Disrupted function of tumor necrosis factor-α-stimulated gene 6 blocks cumulus cell-oocyte complex expansion. Endocrinology 1444376-4384. [DOI] [PubMed] [Google Scholar]
- 53.Ochsner, S. A., D. L. Russell, A. J. Day, R. M. Breyer, and J. S. Richards. 2003. Decreased expression of tumor necrosis factor-α-stimulated gene 6 in cumulus cells of the cyclooxygenase-2 and EP2 null mice. Endocrinology 1441008-1019. [DOI] [PubMed] [Google Scholar]
- 54.Pangas, S. A. 2007. Growth factors in ovarian development. Semin. Reprod. Med. 25225-234. [DOI] [PubMed] [Google Scholar]
- 55.Pangas, S. A., C. J. Jorgez, and M. M. Matzuk. 2004. Growth differentiation factor 9 regulates expression of the bone morphogenetic protein antagonist gremlin. J. Biol. Chem. 27932281-32286. [DOI] [PubMed] [Google Scholar]
- 56.Pangas, S. A., C. J. Jorgez, M. Tran, J. Agno, X. Li, C. W. Brown, T. R. Kumar, and M. M. Matzuk. 2007. Intraovarian activins are required for female fertility. Mol. Endocrinol. 212458-2471. [DOI] [PubMed] [Google Scholar]
- 57.Pangas, S. A., X. Li, E. J. Robertson, and M. M. Matzuk. 2006. Premature luteinization and cumulus cell defects in ovarian-specific Smad4 knockout mice. Mol. Endocrinol. 201406-1422. [DOI] [PubMed] [Google Scholar]
- 58.Pangas, S. A., X. Li, L. Umans, A. Zwijsen, D. Huylebroeck, C. Gutierrez, D. Wang, J. F. Martin, S. P. Jamin, R. R. Behringer, E. J. Robertson, and M. M. Matzuk. 2008. Conditional deletion of Smad1 and Smad5 in somatic cells of male and female gonads leads to metastatic tumor development in mice. Mol. Cell. Biol. 28248-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pangas, S. A., and M. M. Matzuk. 2005. The art and artifact of GDF9 activity: cumulus expansion and the cumulus expansion-enabling factor. Biol. Reprod. 73582-585. [DOI] [PubMed] [Google Scholar]
- 60.Pedersen, T., and H. Peters. 1968. Proposal for a classification of oocytes and follicles in the mouse ovary. J. Reprod. Fertil. 17555-557. [DOI] [PubMed] [Google Scholar]
- 61.Piek, E., W. J. Ju, J. Heyer, D. Escalante-Alcalde, C. L. Stewart, M. Weinstein, C. Deng, R. Kucherlapati, E. P. Bottinger, and A. B. Roberts. 2001. Functional characterization of transforming growth factor beta signaling in Smad2- and Smad3-deficient fibroblasts. J. Biol. Chem. 27619945-19953. [DOI] [PubMed] [Google Scholar]
- 62.Richards, J. S. 2005. Ovulation: new factors that prepare the oocyte for fertilization. Mol. Cell Endocrinol. 23475-79. [DOI] [PubMed] [Google Scholar]
- 63.Richards, J. S., D. L. Russell, S. Ochsner, M. Hsieh, K. H. Doyle, A. E. Falender, Y. K. Lo, and S. C. Sharma. 2002. Novel signaling pathways that control ovarian follicular development, ovulation, and luteinization. Recent Prog. Horm. Res. 57195-220. [DOI] [PubMed] [Google Scholar]
- 64.Salustri, A., C. Garlanda, E. Hirsch, M. De Acetis, A. Maccagno, B. Bottazzi, A. Doni, A. Bastone, G. Mantovani, P. B. Peccoz, G. Salvatori, D. J. Mahoney, A. J. Day, G. Siracusa, L. Romani, and A. Mantovani. 2004. PTX3 plays a key role in the organization of the cumulus oophorus extracellular matrix and in in vivo fertilization. Development 1311577-1586. [DOI] [PubMed] [Google Scholar]
- 65.Schmierer, B., and C. S. Hill. 2007. TGFbeta-SMAD signal transduction: molecular specificity and functional flexibility. Nat. Rev. Mol. Cell Biol. 8970-982. [DOI] [PubMed] [Google Scholar]
- 66.ten Dijke, P., and C. S. Hill. 2004. New insights into TGF-beta-Smad signalling. Trends Biochem. Sci. 29265-273. [DOI] [PubMed] [Google Scholar]
- 67.Tomic, D., K. P. Miller, H. A. Kenny, T. K. Woodruff, P. Hoyer, and J. A. Flaws. 2004. Ovarian follicle development requires Smad3. Mol. Endocrinol. 182224-2240. [DOI] [PubMed] [Google Scholar]
- 68.Tomic, D., S. G. Brodie, C. Deng, R. J. Hickey, J. K. Babus, L. H. Malkas, and J. A. Flaws. 2002. Smad3 may regulate follicular growth in the mouse ovary. Biol. Reprod. 66917-923. [DOI] [PubMed] [Google Scholar]
- 69.Varani, S., J. A. Elvin, C. Yan, J. DeMayo, F. J. DeMayo, H. F. Horton, M. C. Byrne, and M. M. Matzuk. 2002. Knockout of pentraxin 3, a downstream target of growth differentiation factor-9, causes female subfertility. Mol. Endocrinol. 161154-1167. [DOI] [PubMed] [Google Scholar]
- 70.Waldrip, W. R., E. K. Bikoff, P. A. Hoodless, J. L. Wrana, and E. J. Robertson. 1998. Smad2 signaling in extraembryonic tissues determines anterior-posterior polarity of the early mouse embryo. Cell 92797-808. [DOI] [PubMed] [Google Scholar]
- 71.Wang, H., W.-G. Ma, L. Tejada, H. Zhang, J. D. Morrow, S. K. Das, and S. K. Dey. 2004. Rescue of female infertility from the loss of cyclooxygenase-2 by compensatory up-regulation of cyclooxygenase-1 is a function of genetic makeup. J. Biol. Chem. 27910649-10658. [DOI] [PubMed] [Google Scholar]
- 72.Weinstein, M., S. P. Monga, Y. Liu, S. G. Brodie, Y. Tang, C. Li, L. Mishra, and C. X. Deng. 2001. Smad proteins and hepatocyte growth factor control parallel regulatory pathways that converge on beta1-integrin to promote normal liver development. Mol. Cell. Biol. 215122-5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weinstein, M., X. Yang, C. Li, X. Xu, J. Gotay, and C. X. Deng. 1998. Failure of egg cylinder elongation and mesoderm induction in mouse embryos lacking the tumor suppressor Smad2. Proc. Natl. Acad. Sci. USA 959378-9383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu, J., J. Oakley, and E. A. McGee. 2002. Stage-specific expression of Smad2 and Smad3 during folliculogenesis. Biol. Reprod. 661571-1578. [DOI] [PubMed] [Google Scholar]
- 75.Yan, C., P. Wang, J. DeMayo, F. J. DeMayo, J. A. Elvin, C. Carino, S. V. Prasad, S. S. Skinner, B. S. Dunbar, J. L. Dube, A. J. Celeste, and M. M. Matzuk. 2001. Synergistic roles of bone morphogenetic protein 15 and growth differentiation factor 9 in ovarian function. Mol. Endocrinol. 15854-866. [DOI] [PubMed] [Google Scholar]
- 76.Yang, X., J. J. Letterio, R. J. Lechleider, L. Chen, R. Hayman, H. Gu, A. B. Roberts, and C. Deng. 1999. Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T-cell responsiveness to TGF-β. EMBO J. 181280-1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang, Y. C., E. Piek, J. Zavadil, D. Liang, D. Xie, J. Heyer, P. Pavlidis, R. Kucherlapati, A. B. Roberts, and E. P. Bottinger. 2003. Hierarchical model of gene regulation by transforming growth factor beta. Proc. Natl. Acad. Sci. USA 10010269-10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yoshino, O., H. E. McMahon, S. Sharma, and S. Shimasaki. 2006. A unique preovulatory expression pattern plays a key role in the physiological functions of BMP-15 in the mouse. Proc. Natl. Acad. Sci. USA 10310678-10683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhu, Y., J. A. Richardson, L. F. Parada, and J. M. Graff. 1998. Smad3 mutant mice develop metastatic colorectal cancer. Cell 94703-714. [DOI] [PubMed] [Google Scholar]