Abstract
Heterochromatin protein 1 (HP1) is a conserved chromosomal protein with important roles in chromatin packaging and gene silencing. In fission yeast, two HP1 family proteins, Swi6 and Chp2, are involved in transcriptional silencing at heterochromatic regions, but how they function and whether they act cooperatively or differentially in heterochromatin assembly remain elusive. Here, we show that both Swi6 and Chp2 are required for the assembly of fully repressive heterochromatin, in which they play distinct, nonoverlapping roles. Swi6 is expressed abundantly and plays a dose-dependent role in forming a repressive structure through its self-association property. In contrast, Chp2, expressed at a lower level, does not show a simple dose-dependent repressive activity. However, it contributes to the recruitment of chromatin-modulating factors Clr3 and Epe1 and possesses a novel ability to bind the chromatin-enriched nuclear subfraction that is closely linked with its silencing function. Finally, we demonstrate that a proper balance between Swi6 and Chp2 is critical for heterochromatin assembly. Our findings provide novel insight into the distinct and cooperative functions of multiple HP1 family proteins in the formation of higher-order chromatin structure.
In eukaryotic cells, heterochromatin is a distinct chromatin structure that plays pivotal roles in chromosomal function and epigenetic gene regulation. Heterochromatin is generally transcriptionally silent, and its higher-order chromatin structure can spread, causing the heritable inactivation of euchromatic genes adjacent to heterochromatin and leading to varied patterns of gene expression (12, 13, 37). The establishment of heterochromatin is intimately correlated with changes in the posttranslational modifications of histone tails. The methylation of lysine 9 of histone H3 (H3K9me) is a hallmark of heterochromatin and is catalyzed by SUV39H, a SET-domain-containing histone methyltransferase. The H3K9me serves as a specific binding site for heterochromatin protein 1 (HP1), an evolutionarily conserved chromosomal protein (4, 23, 30). HP1 family proteins share a basic structure, consisting of an amino-terminal chromodomain (CD) and a carboxyl-terminal chromoshadow domain (CSD) linked by a hinge (H) region (17). The HP1 CD functions as a binding module that targets H3K9me (19, 33) while the CSD functions as a dimerization module, which is thought to provide a recognition surface for the binding of other chromatin proteins (5, 7, 17, 43). Although dimerization is a conserved property of HP1 family proteins and is thought to facilitate their dose-dependent effects on heterochromatin silencing (8, 10), the exact mechanisms by which these proteins contribute to the spreading of repressive chromatin structure remain elusive.
Several HP1 isoforms have been identified in many organisms, including mammals (HP1α, HP1β, and HP1γ), Xenopus (HP1α and HP1γ), Drosophila (HP1, HP1b, and HP1c), Caenorhabditis elegans (HPL-1 and HPL-2), and fission yeast (Swi6 and Chp2) (25). These HP1 isoforms exhibit isoform-specific characteristics regarding their binding partners (3, 31, 32, 47), cellular localization (16, 27, 42), posttranslational modifications (24, 51), biochemical properties (26, 28), and genetic redundancy (20, 40). In particular, human HP1γ is involved in the transcriptional elongation of euchromatic genes rather than the formation of repressive heterochromatin (46). Although these isoform-specific functions are thought to explain the versatile roles of HP1 family proteins, the distinct or overlapping functions of HP1 isoforms, especially in forming large heterochromatin domains, known as constitutive heterochromatin, are unknown.
In the fission yeast Schizosaccharomyces pombe, the higher-order chromatin structure is also essential for the functional organization of heterochromatic domains, such as the centromeres, mating-type region, and telomeres (2, 11). Two HP1 family proteins, Swi6 and Chp2, are involved in transcriptional silencing at these heterochromatic regions (Fig. 1A) (9, 15, 29, 45). Clr4 is a mammalian SUV39H homolog and provides the H3K9me mark on heterochromatin, which then serves as a docking site for Swi6 and Chp2 (4, 30, 38). While H3K9me and the binding of HP1 family proteins are a conserved structural property of repressive heterochromatin, it is still unclear how the two HP1 family proteins Swi6 and Chp2 function in the maintenance of the repressive chromatin structure or whether they act cooperatively or differentially. Several lines of evidence from genetic studies suggest that these proteins have distinct functions in heterochromatin silencing (38, 45) although the underlying molecular mechanisms are not clear. In addition to their role in forming the higher-order chromatin structure, these proteins may facilitate the recruitment of chromatin-modulating enzymes, such as Clr3 histone deacetylase and a JmjC-domain-containing protein, Epe1 (18, 50, 52). Although the balance between Clr3 and Epe1 is likely to be critical for heterochromatin maintenance, exactly how the HP1 homologs Swi6 and Chp2 participate in this dynamic process remains elusive.
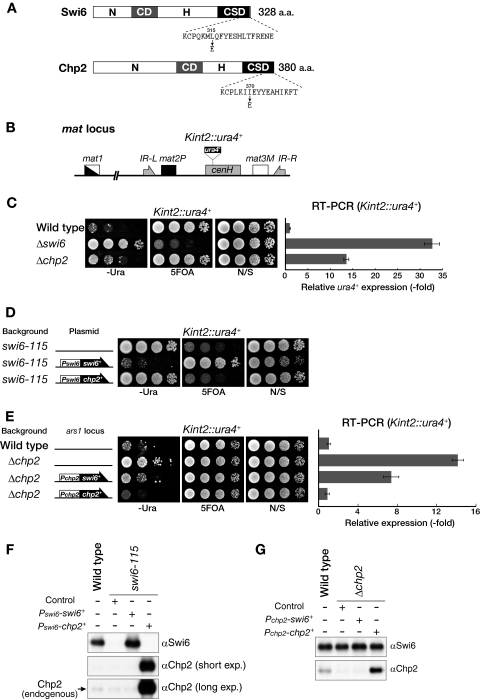
FIG. 1.
Swi6 and Chp2 play distinct roles in heterochromatin assembly. (A) Schematic drawing of Swi6 and Chp2. Gray and black boxes represent the conserved CD and CSD, respectively. N-terminal (N) and hinge (H) regions are also indicated. The amino acid sequence of C-terminal Swi6 or Chp2 is shown beneath each drawing, and the positions of amino acids mutated in this study (L315E and I370E) are indicated by an arrow. (B) Schematic diagram of the mating-type (mat) locus. The position of the silencing reporter gene (Kint2::ura4+) is shown. The mat locus is not drawn to scale. (C) Both Swi6 and Chp2 are required for heterochromatin silencing. A 10-fold serially diluted culture of the indicated strain was spotted onto nonselective medium (N/S), −Ura medium, or medium containing 5-FOA (left). The expression level of the ura4+ transcript of the wild-type, Δswi6, or Δchp2 strain was evaluated by real-time PCR analysis and compared with the level in wild-type cells (right). Error bars represent the standard error of the mean. (D) Chp2 production fails to rescue the defective silencing in swi6-115 mutants. Swi6 or Chp2 protein was produced from the swi6 promoter (Pswi6) on an episomal plasmid (pRE), and silencing of the Kint2::ura4+ reporter in swi6-115 mutant cells was assayed by the plating efficiency on selective medium. Medium lacking leucine was used to select the cells harboring episomal plasmids. Empty pRE vector was used as the control. (E) Swi6 production does not fully rescue the silencing defect in Δchp2 cells. Each gene was introduced into the ars1 locus with the LEU2 gene and ectopically expressed from the chp2 promoter (Pchp2). Recovery of the Kint2::ura4+ silencing defect was evaluated by spotting assay (left) and RT-PCR analysis (right). (F and G) Western blot analysis of ectopically expressed Swi6 and Chp2. The level of Swi6 or Chp2 produced from Pswi6 on an episomal plasmid pRE on a swi6-115 background was compared with that of wild-type cells (F). The level of Swi6 or Chp2 expressed from Pchp2 at the ars1 locus on a Δchp2 background was compared with that of wild-type cells (G). α, anti; exp, exposure; N/S, nonselective medium.
In this study, we show that Swi6 and Chp2 play distinct roles in the formation of higher-order chromatin structure. We demonstrate that Swi6 has a dose-dependent function in forming repressive chromatin but that a specific Chp2 function is also required to ensure a fully repressed chromatin state. We further show that a proper balance between Swi6 and Chp2 is critical for heterochromatin assembly. Our findings indicate that heterochromatin organization is dependent on the balance between the distinct functions of different HP1 isoforms.
MATERIALS AND METHODS
Strains and plasmids.
The strains used in this study are listed in Table S1 in the supplemental material. The deletion and tagging of endogenous genes were conducted using a PCR-based gene-targeting protocol (22). To construct plasmids for producing recombinant Swi6 or Chp2 proteins in Escherichia coli, the coding sequence for swi6+ or chp2+ was amplified by PCR and cloned into the pRSET (Invitrogen), pET15b (Novagen), pGEX4T (GE Healthcare), or pMALc2 (NEB) vector. To express CSD-mutated recombinant Swi6 proteins (Swi6 with the mutation L315E [Swi6L315E]), these plasmids were subjected to site-directed mutagenesis as described previously (39). The internal CD regions (amino acids [aa] 80 to 136 of Swi6 and aa 175 to 231 of Chp2) were PCR amplified and cloned into pGEX4T to express glutathione S-transferase (GST)-CD proteins. To express Swi6, Chp2, or Clr3 from the inducible nmt1+ promoter, each coding sequence was amplified by PCR and cloned into the pREP-1, -41, or -81 vector. To express Swi6, Chp2, or chimeric proteins from a multicopy episomal plasmid using the native swi6+ promoter, the swi6+ coding sequence with its potential promoter and terminator regions was first cloned into pBluescript (pAL2pBK), and then two restriction enzyme sites (BamHI and PacI sites immediately after the ATG and stop codons, respectively) were introduced by site-directed mutagenesis. The Swi6 and Chp2 proteins were divided into four domains: the N terminus (N; aa 1 to 66 of Swi6 and aa 1 to 160 of Chp2), chromodomain (CD; aa 66 to 133 of Swi6 and aa 159 to 230 of Chp2), hinge region (H; aa 134 to 261 of Swi6 and aa 229 to 316 of Chp2), and chromoshadow domain (CSD; aa 262 to 328 of Swi6 and aa 315 to 380 of Chp2). Chimeric Swi6 proteins containing Chp2N, Chp2CD, Chp2H, or Chp2CSD were designated Swi6-ch1, -ch2, -ch3, and -ch4, respectively. Each coding sequence (Swi6, Chp2, or chimeric) with the swi6+ promoter and terminator regions was then introduced into a pRE vector, which was created from the pREP vector by eliminating the nmt1+ promoter region (pRE-Pswi6-swi6+, -chp2+, or -swi6-ch1∼4).
To obtain a strain expressing chp2+ or swi6+ from the endogenous chp2+ promoter, the chp2+ genomic region was amplified by PCR and cloned into the pRE vector. Two restriction sites, BamHI and PacI, were introduced immediately after the ATG and stop codons, respectively, using site-directed mutagenesis (pRE-Pchp2-chp2+), and the chp2+coding region was replaced with swi6+ or other chp2 chimeric genes using these sites (pRE-Pchp2-swi6+ and pRE-Pchp2-chp2-ch1∼4). pRE-Pchp2-chp2I370E, which carries the chp2 gene with the mutation I370E, was also created by site-directed mutagenesis. These plasmids were cleaved with MluI for introduction into the ars1 locus, and transformed cells were isolated using the LEU2 marker gene.
Ectopic silencing constructs were generated and introduced as previously described (38). Transformed strains were confirmed by genomic PCR or Southern blotting. All other strains were constructed using standard genetic crosses.
Antibodies.
The following antibodies were used: anti-Swi6 (38), anti-Chp2 (this study), anti-H3K9me2 (38), anti-FLAG horseradish peroxidase-conjugated (A8592; Sigma); anti-His6 (Qiagen), anti-FLAG M2 affinity gel (Sigma), and anti-histone H3 acetylated at K14 (H3K14Ac) (07-353; Upstate).
RNA preparation and reverse transcription-PCR (RT-PCR) analysis.
Total RNA was extracted from cells as described previously (38). Total RNA prepared from each strain was preincubated with RNase-free DNase I (0.4 U/μg RNA; TaKaRa) to digest genomic DNA. cDNA samples were synthesized using Superscript III reverse transcriptase (Invitrogen) and an oligo(dT) primer and subjected to quantitative PCR analyses using qPCR MasterMix Plus for Sybr Green (Eurogentec) and a 7300 Real-Time PCR System (ABI). The primers used in these analyses were ura4-RT-Fw1 (5′-GGCCTCAAAGAAGTTGGTTTACC-3′) and ura4-RT-Rv1 (5′-GAAGACATTTCAGCCAAAAGCA-3′) for the Kint2::ura4+ locus.
Silencing assay.
Silencing assays were performed using unsaturated cultures grown in YEA (yeast extract with adenine) medium. Serial dilutions (10-fold) were prepared (1 × 105 to 1 × 103 cells) and spotted on plates with minimal (amino acids [AA]) medium, AA medium lacking uracil (−Ura), or AA medium containing 5-fluoroorotic acid (5-FOA). The plates were then incubated at 30°C for 2.5 to 4 days. For cells harboring pRE plasmids, AA medium lacking leucine was used for the culture and spotting.
BIAcore surface plasmon resonance analysis.
Interactions between each CD and the histone H3 peptides were examined using a Biacore 3000 instrument (GE Healthcare). H3 peptide that was unmodified, K9-dimethylated (H3K9me2), or K9-trimethylated (H3K9me3) was immobilized on different flow cells of a CM5 sensor chip using an amine-coupling kit (GE Healthcare). The interaction assays were performed at a constant flow rate (15 ml/min) at 20°C. Distinct concentrations ranging from 250 nM to 900 nM of purified recombinant GST alone or each GST-CD protein were injected as analytes. The sensor chip surface was regenerated with 10 ml of 50 mM NaOH. Sensorgrams were analyzed using BIA evaluation software, version 3.0. The bivalent analyte model was employed to fit the data, which was based on the observation that the GST moiety forms a stable dimer that binds two ligands on the sensor chip.
Gel filtration analyses.
Recombinant His6-tagged proteins (Swi6, Swi6L315E, Chp2, and Chp2I370E) or Swi6 fused with maltose binding protein (MBP-Swi6) were further purified by anion exchange chromatography (Source 15Q; GE Healthcare). Two milligrams of each purified protein was loaded onto Superdex 200 pg (GE Healthcare) equilibrated with TSG buffer (50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1 mM dithiothreitol, 5% glycerol). Eluate fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and proteins were visualized by Coomassie brilliant blue staining.
ChIP.
Chromatin immunoprecipitation (ChIP) was performed as described previously (38). To immunoprecipitate FLAG-tagged proteins, anti-FLAG M2 affinity gel (Sigma) was used. Antibodies used in this study were anti-Swi6 polyclonal antibody, anti-Chp2 polyclonal antibody, anti-H3K9me monoclonal antibody (38), or anti-H3K14Ac (Upstate 07-353). PCR products were separated and analyzed on a 15% polyacrylamide gel (ATTO). Chip-on-chip analysis was carried out using in vitro transcription amplification method as described previously (49).
GST pull-down assays.
Recombinant His6-tagged proteins (Swi6, Swi6L315E, and Chp2) were incubated with GST-Swi6 or GST-Chp2 protein in binding buffer (25 mM Tris-HCl, pH 7.5, 250 mM NaCl, 1 mM EDTA, 10% glycerol, 0.2% NP-40) at 4°C for 4 h. GST fusion proteins and associated proteins were pulled down by adding 10 μl of glutathione-Sepharose 4B. After proteins were washed with the binding buffer, the bound proteins were eluted with elution buffer (50 mM Tris-HCl, pH 8.0, 10 mM reduced glutathione). The eluted proteins were resolved by 10% SDS-PAGE, and the pulled-down His-tagged proteins were detected by Western blotting using an anti-His6 antibody (Qiagen).
Chromatin fractionation assay.
The chromatin fractionation assay was performed as described previously (34) with some modifications. Cells (2.5 × 108) were harvested, washed once with stop buffer (150 mM NaCl, 50 mM NaF, 10 mM EDTA, 1 mM NaN3), and placed on ice for 5 min. The cells were resuspended in PEMS (100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 10 mM EGTA, 10 mM MgSO4, 1.2 M sorbitol) containing 1 mg/ml Novozyme and 1 mg/ml Zymolyase 100T, and incubated at 37°C for 20 min. The cell suspension was spun at 400 × g at 4°C for 5 min, and the resulting cell pellet was washed twice with 1.2 M sorbitol and then lysed with HBS buffer (25 mM MOPS, 60 mM β-glycerolphosphate, 15 mM MgCl2, 15 mM EGTA, 15 mM p-nitrophenylphosphate, 0.1 mM sodium vanadate, pH 7.2) containing 1 mM phenylmethylsulfonyl fluoride, 1× Complete (Roche), and 1.5% Triton X-100. The resulting lysate was the whole-cell extract, which was spun at 22,000 × g at 4°C for 15 min to obtain supernatant and pellet fractions. Proteins from each fraction were separated by SDS-PAGE, and either Swi6 or Chp2 was detected by Western blotting using anti-Swi6, anti-Chp2, or anti-FLAG antibodies.
RESULTS
Both Swi6 and Chp2 are required for fully repressive heterochromatin.
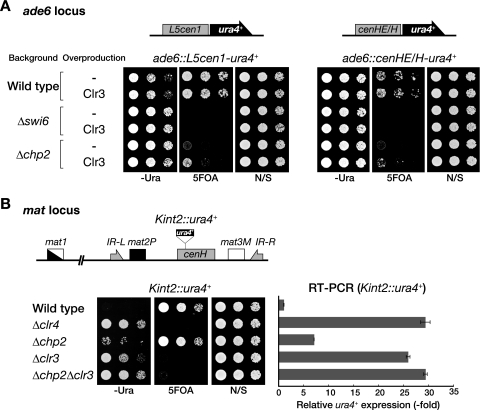
A previous report showed that the deletion of swi6+ or chp2+ alleviates silencing at the mat locus (45). To confirm the roles of these proteins, we examined the effect of swi6+ or chp2+ deletion (Δswi6 or Δchp2) on the silencing of a ura4+ marker gene inserted within the mating-type K region (Kint2::ura4+) (Fig. 1B), combined with real-time RT-PCR analyses (Fig. 1C). Serially diluted wild-type or mutant cells were spotted onto nonselective control medium or onto selective medium lacking uracil (−Ura) or containing 5-FOA to evaluate ura4+ expression. As previously observed, Δswi6 clearly abolished heterochromatic silencing at the mat locus (Fig. 1C). Δchp2 also alleviated silencing at the mat locus, but its effects were milder than those of Δswi6 (Fig. 1C) (45). Real-time RT-PCR analysis supported the results of the spotting assay and clearly showed that the silencing defect of Δswi6 was stronger than that of Δchp2 (Fig. 1C), thus confirming the previous results and demonstrating that both Swi6 and Chp2 are required, albeit to different degrees, for the assembly of fully repressive heterochromatin.
Swi6 and Chp2 play distinct roles in heterochromatin assembly.
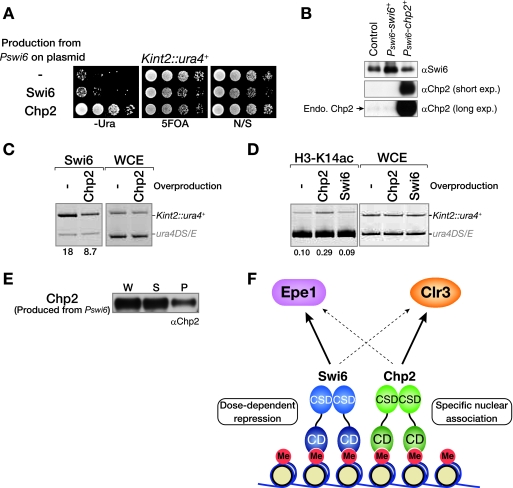
The above results raised the testable question of whether these proteins simply have an overlapping function in heterochromatin assembly. To address this possibility, we first examined whether the complementary expression of Chp2 suppressed the silencing defect in swi6 mutant cells (swi6-115). In a control experiment, the expression of swi6+ from its native promoter clearly suppressed the silencing defect of swi6 mutant cells (Fig. 1D, Pswi6-swi6+). However, complementary expression of Chp2 from the swi6+ promoter showed no suppressive effect on the silencing defect of swi6 mutant cells (Fig. 1D, Pswi6-chp2+), suggesting that Chp2 has little or no overlapping function with Swi6. To confirm the distinct functions of these proteins, we also performed the reverse experiment. While the ectopic expression of chp2+ from its native promoter successfully suppressed the silencing defect of Δchp2 (Fig. 1E, Pchp2-chp2+), complementary expression of Swi6 from the chp2+ promoter failed to restore the Δchp2 silencing defect and only partially suppressed the Δchp2 phenotype (Fig. 1E, Pchp2-swi6+). Collectively, these results suggest that Swi6 and Chp2 play distinct roles in heterochromatin assembly. Furthermore, the partial suppression of the Δchp2 phenotype by Swi6 might be attributable to Swi6's having a dose-dependent function in forming repressive chromatin. Alternatively, the silencing derepression in Δchp2 cells might reflect a combined phenotype caused by the scarcity of Swi6 at this locus and the deficiency of a Chp2-specific function; in this case, Swi6 expression from the chp2+ promoter can alleviate the Swi6 scarcity but cannot rescue the Chp2-specific function.
Expression levels of Swi6 and Chp2 are differentially regulated.
When analyzing the protein levels of ectopically expressed Swi6 and Chp2, we noticed that the level of Chp2 produced from the Swi6 promoter (Pswi6) was clearly higher than that of endogenous Chp2 (Fig. 1F). To confirm this difference in expression levels, we analyzed the relative abundance of the proteins. Using polyclonal antibodies raised against each of the recombinant proteins, the amounts of Swi6 and Chp2 in a single fission yeast cell were determined by Western blot analyses (Fig. 2A and B). The number of Swi6 molecules in a single cell was estimated to be 19,400 (the amount of Swi6 in 2.5 × 105 cells corresponded to approximately 0.318 ng of the recombinant protein). Given that a single Swi6 recognizes one histone H3 tail methylated at lysine 9, this amount of Swi6 appears more than sufficient; in fact, there was nearly twice as much as would be required to cover all the heterochromatic regions in a cell (Fig. 2A). The estimated amount of Chp2 was only ∼240 molecules in a single cell (36.6 pg of Chp2 in 2.5 × 106 cells), which is nearly 1/80 the amount of Swi6 (Fig. 2B). These results demonstrate that the levels of these HP1 proteins are differentially regulated in fission yeast cells.
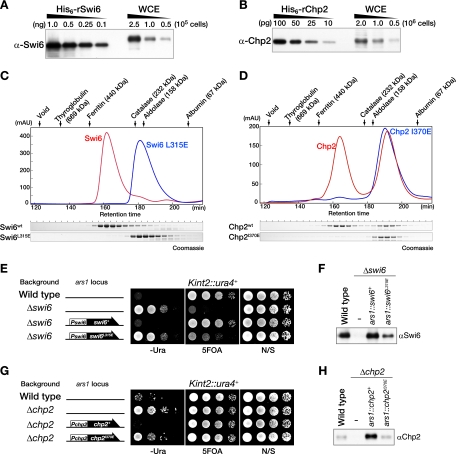
FIG. 2.
Cellular abundance and dimer formation of Swi6 and Chp2. (A and B) Cellular abundance of Swi6 and Chp2 proteins. The indicated amounts of recombinant His6-Swi6 or His6-Chp2 proteins and of whole-cell extracts (WCE) prepared from the indicated number of wild-type cells were subjected to Western blot analyses using anti-Swi6 (A) and anti-Chp2 (B) antibodies. The total heterochromatic region of a G2 phase cell is estimated to be ∼950 kb, consisting of ∼430 kb of centromeres, ∼40 kb of silent mating type loci, and ∼520 kb of telomeres. Given that every ∼200 bp of heterochromatic DNA is organized into a nucleosome with two H3 tails, the heterochromatin region contains ∼9,500 H3 tails. (C and D) The Chp2 dimer is less stable than the Swi6 dimer. (C) Chromatogram and elution profile of the Swi6 protein by size exclusion chromatography. Recombinant His6-tagged Swi6 and Swi6L315E proteins were loaded onto a Sephadex 200 pg column. Elution profiles for Swi6 (red) and Swi6L315E (blue) are shown (top), and the eluted proteins were resolved by 10% SDS-PAGE and visualized by Coomassie staining (bottom). (D) Chromatogram and elution profile of Chp2 protein by size exclusion chromatography. Recombinant His6-tagged Chp2 and Chp2I370E proteins were resolved by a Sephadex 200 pg column. Elution profiles are shown as chromatograms (top) and 10% SDS-PAGE visualized by Coomassie staining (bottom). The positions of eluted marker proteins are indicated above each chromatogram. (E to H) The dimer configuration is critical for the in vivo function of Swi6 and Chp2. (E) The swi6+ or swi6L315E gene was introduced into the ars1 locus of Δswi6 cells with the LEU2 gene and ectopically expressed from the swi6 promoter. Silencing of the Kint2::ura4+ reporter in control wild-type, Δswi6, and Δswi6-derivative cells was evaluated by spotting assay. (F) Western blot analysis of Swi6 in wild-type cells or cells expressing Swi6 or Swi6L315E from the swi6 promoter at the ars1 locus on a Δswi6 background. (G) The chp2+ or chp2I370E gene was introduced into the ars1 locus of Δchp2 cells with the LEU2 gene and ectopically expressed from the chp2 promoter. Silencing of the Kint2::ura4+ reporter in control wild-type, Δchp2, and Δchp2-derivative cells was evaluated by spotting assay. (H) Western blot analysis of Chp2 in wild-type cells or cells expressing Chp2 or Chp2I370E from the chp2 promoter at the ars1 locus on a Δchp2 background. α, anti; AU, arbitrary units; N/S, nonselective medium.
Swi6 and Chp2 possess distinct biochemical properties.
The functions of HP1 family proteins are tightly linked to their biochemical properties, which include their ability to bind H3K9me and to form multimeric complexes (17). To gain insight into the distinct functions of Swi6 and Chp2, we analyzed their biochemical properties. We first determined the binding affinity of Swi6 and Chp2 for H3K9me. Surface plasmon resonance analyses revealed that a GST fusion with the CD of Swi6 (GST-Swi6CD) and GST-Chp2CD bound to H3K9me2 and H3K9me3 with similar affinities, although GST-Chp2CD displayed slightly higher affinities for both (Table 1). These results suggest that Swi6 and Chp2 have similar binding abilities for the H3K9-methylated chromosomal regions.
TABLE 1.
Binding affinities of Swi6 and Chp2 for H3K9me peptidesa
| CD | H3K9 peptide | ka (M−1s−1) | kd (s−1) | KD (μM) |
|---|---|---|---|---|
| Swi6 | me2 | (1.2 ± 0.15) × 103 | (8.3 ± 0.91) × 10−3 | 7.1 ± 0.40 |
| me3 | (1.8 ± 0.23) × 103 | (1.7 ± 0.33) × 10−2 | 10 ± 2.6 | |
| Chp2 | me2 | (2.6 ± 0.06) × 103 | (9.6 ± 0.60) × 10−3 | 3.7 ± 0.33 |
| me3 | (2.8 ± 0.11) × 103 | (1.5 ± 0.27) × 10−2 | 5.5 ± 0.96 |
The bivalent analyte model was employed to fit the data. ka, association rate constant; kd, dissociation rate constant; KD, equilibrium dissociation rate constant. KD was calculated as kd/ka.
Swi6 was shown to form a multimeric complex (48), but it is not known whether Chp2 shares this property. To address this issue, we examined the structural configuration of the recombinant proteins (rSwi6 and rChp2) using gel filtration chromatography. Wild-type recombinant Swi6 (rSwi6) eluted as a single peak corresponding to an apparent molecular mass of 300 to 350 kDa (Fig. 2C, Swi6). Although the estimated size was clearly larger than the calculated molecular mass (39.5 kDa) or the size previously reported (∼164 kDa) (48), sedimentation velocity analyses revealed that the molecular weight of rSwi6 was 72,260 (see Fig. S2 in the supplemental material), and its frictional ratio was 1.59 (data not shown), which is a deviation from the average ratio of globular proteins (∼1.2). These results indicate that rSwi6 predominantly forms stable dimers with an elongated shape under physiological buffer conditions. Its dimer configuration was also confirmed by introducing an amino acid substitution in the CSD of rSwi6 (Fig. 2C, Swi6L315E; see also Fig. S2 in the supplemental material), which caused rSwi6 to elute as a monomer. The mutation of the corresponding isoleucine (Fig. 1A; see also Fig. S1B in the supplemental material) in mammalian HP1 is also reported to abolish the dimer formation (5).
When recombinant Chp2 (rChp2) was subjected to the same chromatographic analysis, it eluted as two distinct peaks (Fig. 2D, Chp2). The slowly eluting peak clearly overlapped with that of a CSD mutant (I370E) of rChp2 (Fig. 2D, Chp2I370E), suggesting that the rChp2 dimers are less stable than those of rSwi6. We also tested whether rSwi6 and rChp2 could form heterodimers by producing these proteins simultaneously in E. coli. However, we were unable to obtain any Swi6-Chp2 heterodimers (data not shown), indicating that Swi6 and Chp2 predominantly exist as homodimers.
To examine the importance of the dimer configuration in vivo, we performed silencing complementation analyses. While silencing defects in Δswi6 and Δchp2 mutant cells were clearly rescued by the complementary expression of wild-type swi6+ and chp2+, respectively (Fig. 2E, Pswi6-swi6+, and G, Pchp2-chp2+), the non-dimer-forming Swi6 (Swi6L315E) and Chp2 (Chp2I370E) mutant proteins could not rescue the silencing defects efficiently in their respective mutant strains (Fig. 2E, Pswi6-swi6L315E, and G, Pchp2-chp2I370E). Western blot analysis further revealed that, although the wild-type and mutant proteins were similarly expressed from an ectopic ars1 locus, the in vivo protein level of the non-dimer-forming mutants was less than that of the wild-type protein (Fig. 2F and H). These results demonstrated that a dimer configuration through the CSD, regardless of the difference in stability, is critical for the in vivo functions of both proteins, and they suggest that dimer formation is correlated with the in vivo stability of these proteins. It is thus likely that the instability of Chp2 dimer formation is linked with its in vivo protein level (Fig. 2B).
Swi6, but not Chp2, undergoes intermolecular interactions.
Although our analysis clearly showed the presence of homodimeric Swi6 and Chp2, other modes of intermolecular association are proposed to be involved in the spreading of the heterochromatin structure (17). To investigate whether Swi6 or Chp2 has such properties, we conducted a GST pull-down experiment. Recombinant GST-fusion proteins (see Fig. S3A in the supplemental material) were immobilized on glutathione beads and assayed for their interactions with His-tagged proteins. We found that His-Swi6 was successfully pulled down with GST-Swi6 (see Fig. S3B in the supplemental material). In contrast, no association was observed between His-Chp2 and GST-Chp2. We also failed to detect interactions between Swi6 and Chp2 in either combination. These results suggested that Swi6, but not Chp2, has an intrinsic ability to interact with itself.
One possible explanation for this interaction is a direct association between GST-Swi6 and His-Swi6 dimers, and another is that they exchange dimer partners. To address these possibilities, we tested whether the behavior of His-Swi6 in gel filtration chromatography was altered by its interaction with MBP-fused Swi6 (MBP-Swi6), which has nearly twice the molecular mass of His-Swi6 (80.5 kDa). When His-Swi6 or MBP-Swi6 was loaded separately, the proteins eluted as a single peak corresponding to an apparent molecular mass of ∼300 kDa and ∼450 kDa, respectively (see Fig. S3D in the supplemental material). However, after His-Swi6 and MBP-Swi6 were incubated together (10 h at 4°C), the elution profile was shifted from the original peak positions to include an intermediate one. This result suggests that the intermolecular interactions between Swi6 proteins primarily involved the exchange of dimer-forming partners. This conclusion was further supported by the observation that GST-Swi6 failed to pull down the non-dimer-forming mutant Swi6 (see Fig. S3C in the supplemental material). Together, these results suggest that while the majority of Swi6 forms stable dimers, it undergoes dynamic partner exchange, which presumably facilitates the ability of repressive heterochromatin structure to undergo dynamic changes.
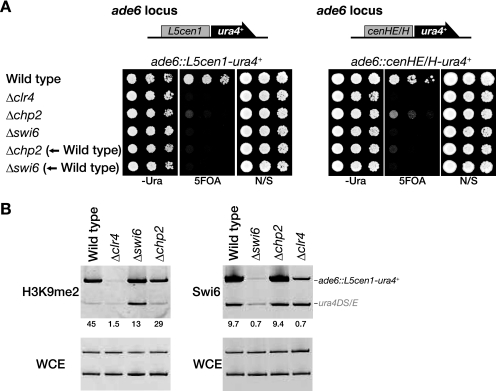
Chp2 functions are required to maintain ectopically induced heterochromatin.
The biochemical properties and cellular abundance of Swi6 are consistent with the general features of HP1 family proteins, but it was not clear how Chp2 plays a specific role at a lower protein level. Our previous studies showed that another chromodomain protein, Chp1, is specifically required for the establishment step of heterochromatin assembly (38). To investigate whether Chp2 has a specific function in the establishment of heterochromatin, we analyzed its effect on ectopically induced heterochromatin formation, in which a DNA fragment from a centromere (L5cen1) or the mating-type region (cenHE/H) is introduced into the euchromatic ade6+ locus with a ura4+ reporter gene (35, 38). In wild-type cells, silencing of the ura4+ gene was induced, and the cells grew on 5-FOA medium (Fig. 3A). As previously demonstrated, the same DNA construct introduced into Δclr4, Δswi6, or Δchp2 cells failed to induce ectopic silencing (Fig. 3A) (38). If Chp2 functions specifically in the establishment step, the ectopic silencing in wild-type cells would be maintained after the chp2+ deletion. We found, however, that the chp2+ deletion clearly abolished the preexisting ectopic silencing (Fig. 3A), indicating that Chp2's function is not exclusively associated with the establishment step and that its continued activity is critical for the maintenance of heterochromatin. Remarkably, a ChIP assay revealed that the levels of Swi6 and H3K9me at the ectopic locus (ade6::L5cen1-ura4+) in the Δchp2 cells were almost the same as in wild-type cells (Fig. 3B). These results clearly demonstrated that H3K9me and the subsequent binding of Swi6 are not sufficient to form the repressive heterochromatin structure and that a Chp2-specific function is required for higher-order chromatin assembly.
FIG. 3.
Chp2 functions are required to maintain ectopically induced heterochromatin. (A) Chp2 is required for L5cen1- or cenHE/H-mediated ectopic silencing. Schematic representations of the L5cen1-ura4+ and cenHE/H-ura4+ constructs inserted into the ade6 locus are shown (top). Ectopically induced silencing was evaluated by spotting assay (bottom). Δclr4, Δchp2, and Δswi6 indicate that the ectopic silencing constructs were introduced in the cells of each mutant background. Δchp2 (←Wild type) and Δswi6 (←Wild type) indicate that the deletion mutants were constructed from the cells in which ectopic silencing was already established in the wild-type background (wild type). (B) Association of Swi6 and H3K9me2 with the ectopic locus in Δswi6, Δchp2, or Δclr4 cells. ChIP assays were performed with DNA isolated from anti-Swi6 or anti-H3K9me2 immunoprecipitates. Purified DNA from the indicated strains was used as the template for PCR amplification of the ade6::L5cen1-ura4+ and control ura4 mini gene (ura4DS/E). The increase in the ade6::L5cen1-ura4+ signal relative to the ura4DS/E signal in the ChIP results was normalized to the whole-cell extract (WCE) signal and is shown beneath each lane. N/S, nonselective medium.
Swi6 and Chp2 contribute differentially to the recruitment of Epe1 and Clr3.
To gain further insight into the functional differences between Swi6 and Chp2, we next focused on their ability to recruit chromatin-modulating factors. In previous studies, Epe1 and Clr3 were shown to associate with heterochromatic regions in an Swi6-dependent manner (18, 50, 52). Although Chp2 is also required for the recruitment of Clr3 to the Kint2::ura4+ locus (50), exactly how it contributes to Epe1 or Clr3 recruitment to heterochromatic regions is not completely understood. To address this question, we examined the localization of Epe1 (Epe1-FLAG) and Clr3 (Clr3-FLAG) in Δswi6 or Δchp2 cells by ChIP analyses (Fig. 4).
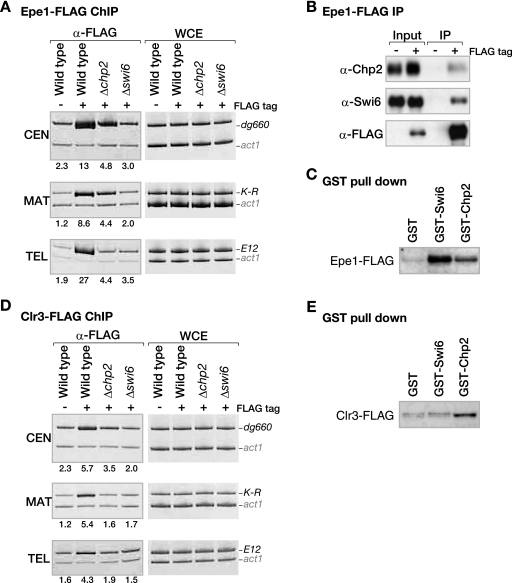
FIG. 4.
Distinct contributions of Swi6 and Chp2 to the heterochromatin localization of Epe1 or Clr3. (A) Epe1 localization to heterochromatic loci is differentially regulated by Swi6 or Chp2. DNA isolated from an anti-FLAG-immunoprecipitated chromatin fraction (α-FLAG) or whole-cell extract (WCE) was used as a template for PCR amplifying centromeric dg660, mat locus K-R, or telomeric E12. The samples were prepared from control nontagged (−FLAG tag) cells or strains expressing Epe1-FLAG (+FLAG tag) from its native promoter. The enrichment ratios of the dg660, K-R, or E12 signals to the act1 signals in the ChIP results are shown beneath each lane. (B) Epe1 efficiently interacts with both Swi6 and Chp2. Anti-FLAG antibodies were used to immunoprecipitate Epe1-FLAG from a strain expressing it (IP). Chp2, Swi6, or Epe1-FLAG was detected by Western blotting with anti-Chp2, anti-Swi6, or anti-FLAG antibodies, respectively. (C) Both Swi6 and Chp2 physically interact with Epe1. Control GST, GST-Swi6, or GST-Chp2 was incubated with whole-cell lysate prepared from cells expressing Epe1-FLAG. Pulled-down Epe1-FLAG was detected by Western blotting using the anti-FLAG antibody. (D) Clr3 localization at heterochromatic loci depends on both Swi6 and Chp2. Association of Clr3-FLAG with the indicated heterochromatic regions was assayed by ChIP as in panel A, using the anti-FLAG antibody. (E) Chp2 has a higher affinity than Swi6 for Clr3. The association of Clr3-FLAG with either Swi6 or Chp2 was assayed as in panel C. α, anti.
In wild-type cells, Epe1 localized to the three heterochromatic loci, centromeres (CEN), the mat locus (MAT), and telomeres (TEL), and as shown previously, this localization was almost completely lost in Δswi6 cells (Fig. 4A) (18, 52). Interestingly, although the degree of Epe1 delocalization was less severe and varied for different loci, Δchp2 also caused a reduction in Epe1 at all three heterochromatic loci, indicating that both Swi6 and Chp2 are required for Epe1 recruitment to native heterochromatic loci. Next, we asked whether Chp2 interacts with Epe1, as previously reported for Swi6 (52). We found that Chp2 as well as Swi6 was efficiently coimmunoprecipitated with Epe1 (Fig. 4B). Physical interactions between these proteins were also confirmed by GST pull-down experiments (Fig. 4C). Together, these results suggested that both Swi6 and Chp2 are involved in recruiting Epe1 to the heterochromatic regions. Our ChIP experiments showed that Swi6 has a more dominant role in the recruitment of Epe1. We think this is simply attributable to the difference in their relative abundances and/or abilitities to form intermolecular associations.
We next sought to analyze the effect of swi6+ or chp2+ deletion on the recruitment of Clr3 to heterochromatin. As previously observed, the Clr3 ChIP experiment showed that Clr3's localization to CEN, MAT, and TEL was abolished in Δswi6 cells (Fig. 4D) (50). Interestingly, chp2 deletion also led to a clear reduction of Clr3 at these heterochromatic regions (Fig. 4D), suggesting that both Swi6 and Chp2 are required for Clr3 recruitment to native heterochromatic loci. Their cooperative function to recruit Clr3 histone deacetylase was further confirmed by the observation of increased levels of H3K14Ac at the Kint2::ura4+ locus (see Fig. S4 in the supplemental material). Although we failed to obtain clear evidence of physical associations between Clr3 and either Swi6 or Chp2 by coimmunoprecipitation experiments (data not shown), the GST pull-down experiment showed that GST-Chp2 efficiently pulled down Clr3-FLAG (Fig. 4E). In contrast, only a negligible amount of Clr3-FLAG was detected in the control GST or GST-Swi6 fraction. Although we could not determine whether this interaction was direct or indirect, these results suggest that Chp2 has a higher affinity for Clr3 than Swi6 does, and thus it is likely that Chp2 plays a major part in the recruitment of Clr3 to heterochromatic regions.
Chp2 is functionally linked with Clr3.
If silencing defects in Δchp2 cells are caused, at least in part, by the delocalization of Clr3, it is possible that an increased amount of Clr3 would suppress the chp2+ deletion phenotype. To test this possibility, we overproduced Clr3 and analyzed the effect on ectopic silencing (Fig. 5A). Although Clr3 overproduction did not affect the overall silencing state at the ectopic loci in wild-type or Δswi6 mutant cells, it clearly did so in the Δchp2 mutant cells (Fig. 5A). These results suggested that Chp2 is functionally linked with Clr3. As described above, H3K9me and Swi6 were present at the ectopic locus in the Δchp2 cells at almost the same levels as in wild-type cells (Fig. 3B). Therefore, it is likely that Clr3 recruitment is a key regulatory step for forming the repressive heterochromatin structure from intermediate chromatin states.
FIG. 5.
Chp2 is functionally linked with Clr3. (A) Overexpression of Clr3 suppresses the Δchp2 mutant phenotype. A control pREP41 (-) or pREP41-clr3+ (Clr3) plasmid was introduced into cells harboring the L5cen1-ura4+ or cenHE/H-ura4+ construct in the ade6 locus (top), and the ectopically induced silencing of the ura4+ gene was evaluated by spotting assay (bottom). A wild-type, Δswi6, or Δchp2 strains was used. (B) Δchp2 Δclr3 mutant cells showed an additive effect for silencing derepression. The silencing of Kint2::ura4+ in the indicated strains was evaluated by spotting assay (bottom left) and RT-PCR analyses (bottom right). A schematic representation of Kint2::ura4+ is shown at the top. IR-L, left inverted repeat; IR-R, right inverted repeat; N/S, nonselective medium.
To clarify the relationship between Chp2 and Clr3, we asked whether chp2+ genetically interacts with clr3+. As previously described, the deletion of clr3+ led to silencing derepression at Kint2::ura4+ (50), and we noticed that the Δclr3 phenotype was more severe than the Δchp2 phenotype (Fig. 5B). Intriguingly, a Δchp2 Δclr3 double mutant showed an additive silencing derepression effect. This result suggested that Chp2 plays some other specific roles in forming repressive heterochromatin in addition to the recruitment of Clr3. This might explain why Clr3 overproduction only partially suppressed the Δchp2 mutant phenotype (Fig. 5A).
Chp2 is tightly associated with a chromatin-enriched nuclear subfraction.
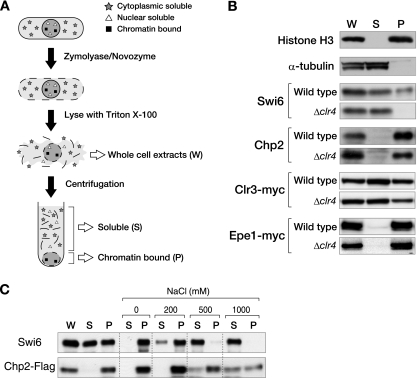
Although the K9-methylated histone tail is thought to be critical for HP1 to associate with chromatin, previous studies suggest that HP1 uses multiple mechanisms for this association (26, 28, 31). To investigate whether the association of Swi6 and Chp2 with chromatin is correlated with their specific functions, we performed a chromatin fractionation assay. In this assay, whole-cell lysates were separated by centrifugation into soluble and pellet fractions, and the proteins from each fraction were subjected to Western blot analyses (Fig. 6A). The enrichment of chromatin in the pellet fraction was verified by a control experiment using antibodies against total histone H3 and α-tubulin (Fig. 6B).
FIG. 6.
Chp2 and Swi6 bind the chromatin-enriched nuclear subfraction in distinct manners. (A) Schematic representation of the chromatin fractionation assay used in the present study. Whole-cell extracts (W) were spun and separated into soluble supernatant (S) and chromatin-enriched pellet (P) fractions. (B) Chromatin fractionation assay in the presence or absence of the histone H3K9 methyltransferase gene, clr4+. Proteins from each fraction were separated by SDS-PAGE, and Swi6, Chp2, Clr3-myc, or Epe1-myc was detected by Western blot analysis. Histone H3 and α-tubulin were examined as controls. (C) Chromatin fractionation assay with various concentrations of salt. The pellet fraction was treated with 0 to 1,000 mM NaCl followed by centrifugation to divide the extract into supernatant and pellet fractions. Swi6 and Chp2-FLAG were detected by Western blotting.
In wild-type cells, Swi6 was present in both the soluble and pellet fractions, with roughly 30 to 40% of the total Swi6 protein reproducibly detected in the chromatin-enriched pellet fraction (Fig. 6B). Interestingly, in Δclr4 cells, Swi6 was redistributed to the soluble fraction, suggesting that a limited fraction of the Swi6 is stably associated with chromatin, and this association is exclusively dependent on Clr4. Under the same experimental conditions, Chp2 was preferentially found in the pellet fraction. Surprisingly, the Chp2 in this fraction was not affected by the clr4+ deletion (Fig. 6B). These results suggested that Chp2 uses some other mode for associating with the chromatin-enriched nuclear subfraction. Consistent with this idea, a large amount of Chp2 persisted in the pellet fraction even after extraction with high-salt buffer (1 M NaCl) (Fig. 6C), while Swi6 was efficiently eliminated from the pellet with buffer containing 0.5 M NaCl. These results demonstrated that Chp2 is tightly associated with the chromatin-enriched nuclear fraction in a Clr4-independent manner.
Heterochromatic localization of Chp2 and Swi6 is dependent on Clr4.
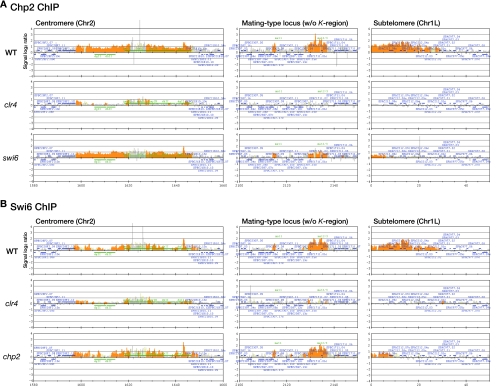
Our previous studies showed that myc-tagged Chp2 associates with three heterochromatic domains, CEN, MAT, and TEL, through clr4+-mediated H3K9 methylation (38). However, our above results showing the Clr4-independent association of Chp2 with the chromatin-enriched nuclear fraction left open the possibility that Chp2 could localize to heterochromatic regions in the absence of H3K9me. Alternatively, Chp2 could bind other euchromatic regions without H3K9me. To address these possibilities, we performed a ChIP assay using an anti-Chp2 antibody, and analyzed Chp2's localization with a tiling array that covers almost the entire genome of fission yeast at 250-bp resolution, except for the telomeres and rDNA.
Chp2 was located exclusively at the three heterochromatic regions (Fig. 7A), and its localization pattern was largely superimposable with that of Swi6 (Fig. 7B) (6). We further observed that Chp2's association with the mating-type locus and subtelomere was reduced in Δswi6 cells, and chp2+ deletion caused a decrease in the subtelomeric association of Swi6. This is consistent with our previous observation (38) and underscores the interdependent relationship between these HP1 family proteins. Importantly, Chp2's association with these heterochromatic regions was nearly completely abolished in Δclr4 cells (Fig. 7A), suggesting that the heterochromatic localization of Chp2 is dependent on Clr4. In agreement with the notion that Chp2 and Swi6 play a major role in the recruitment of Clr3, the dominant role of Clr4 in maintaining the hypo H3K14Ac state was confirmed by ChIP assay (see Fig. S4 in the supplemental material). Although we observed that Chp2 associated with several euchromatic regions and its binding persisted even in the Δclr4 background (see Fig. S5 in the supplemental material), these signals were relatively weak, and it is thus unlikely that these euchromatic binding sites contributed to the tight chromatin association of Chp2 in the fractionation assay. Since neither RNase nor DNase treatment released Chp2 from this nuclear fraction (data not shown), other nuclear components are probably involved in the novel Chp2 association.
FIG. 7.
Heterochromatic localization of Chp2 and Swi6 is dependent on Clr4. (A) Location of Chp2 in three heterochromatic regions: centromere 2, the mating-type locus, and the subtelomere (Chr1L). Wild-type, Δclr4, or Δswi6 cells were subjected to ChIP using an anti-Chp2 antibody. Enrichment in the immunoprecipitated fractions relative to a sample of whole-genome DNA is shown along the length of the chromosome. The orange shading represents the binding ratio of loci that showed significant binding, as described previously (21). The blue bars above and below the graph represent, respectively, genes transcribed from left to right, and from right to left. The green bars represent inverted repeats of centromere 2. A mat2-mat3 interval of the mating-type region was omitted because the tiling arrays used in this study are designed according to the genome sequence of the heterothallic h− strain. (B) Locations of Swi6 in three heterochromatic regions. Wild-type, Δclr4, or Δchp2 cells were subjected to ChIP using an anti-Swi6 antibody. Enrichment in the immunoprecipitated fractions relative to a sample of whole-genome DNA is shown along the length of the chromosome as in panel A. w/o, without; WT, wild type.
The chromatin/nuclear association property is linked with the dosage and silencing functions of Swi6 and Chp2.
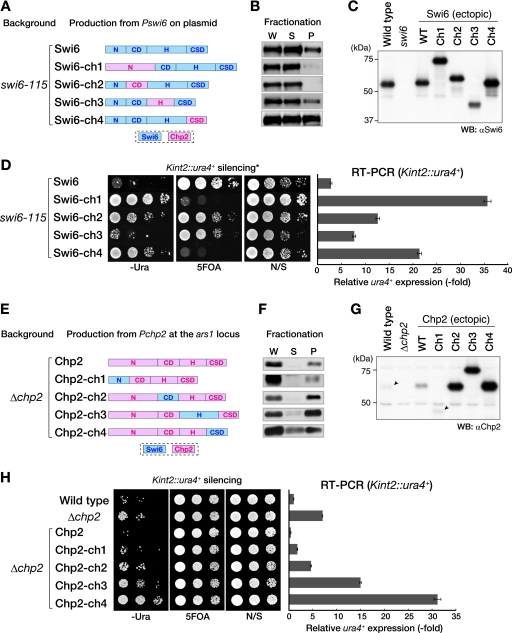
We next determined whether the property of associating with the chromatin-enriched nuclear subfraction (chromatin/nuclear association hereafter) is linked with the silencing function of these proteins. Swi6 and Chp2 were divided into four parts (N terminus, CD, H, and CSD), and chimeric proteins expressed from the native promoter were tested for chromatin binding and the ability to suppress the silencing defect of their corresponding mutant cells (Fig. 8). Chimeric Swi6 proteins containing either the N terminus of Chp2 (Chp2N) or Chp2CD (Swi6-ch1 and -ch2) showed weak chromatin binding and failed to suppress the swi6 mutant phenotype (Fig. 8B and D), suggesting that Swi6N and Swi6CD are important for Swi6 to associate with chromatin via H3K9me. Chimeric Swi6 containing Chp2H (Swi6-ch3) showed a level of chromatin binding comparable to that of wild-type Swi6 and greater suppressive activity. Interestingly, although replacing the Swi6CSD with the Chp2CSD (Swi6-ch4) increased the chromatin-bound fraction (to approximately 50%), this chimeric protein failed to suppress the silencing defect of the swi6 mutant cells (Fig. 8D). These results indicated that the mode of chromatin/nuclear association mediated by Chp2CSD is distinct from that of Swi6CSD and that the proper chromatin binding of Swi6 is critical for its function in heterochromatin assembly.
FIG. 8.
Silencing functions of Swi6 and Chp2 are linked with the property of chromatin/nuclear association. (A to D) Swi6 function is correlated with its chromatin association state. (A) Using the alignment of conserved regions (see Fig. S1 in the supplemental material), the Swi6 and Chp2 proteins were divided into four domains: the N terminus (aa 1 to 66 of Swi6; aa 1 to 160 of Chp2), CD (aa 66 to 133 of Swi6; aa 159 to 230 of Chp2), H region (aa 134 to 261 of Swi6; aa 229 to 316 of Chp2), and CSD (aa 262 to 328 of Swi6; aa 315 to 380 of Chp2). Chimeric Swi6 proteins containing Chp2N, Chp2CD, Chp2H, or Chp2CSD were produced from the swi6 promoter on an episomal plasmid (pRE) in a swi6-115 mutant background. Expressed chimeric proteins were detected by Western blotting using anti-Swi6 antibody (C) and were subjected to the chromatin fractionation assay (B). The silencing of Kint2::ura4+ in swi6-115 mutant cells was evaluated by spotting assay and RT-PCR (D). Domains derived from Swi6 and Chp2 are indicated by blue and pink boxes, respectively. (E to H) Chp2 function is correlated with its chromatin-associated state. Chimeric Chp2 protein containing Swi6N, Swi6CD, Swi6H, or Swi6CSD was produced from the chp2+ promoter at the ars1 locus (E). Expressed chimeric proteins were detected by Western blotting using an anti-Chp2 antibody (G) and were subjected to the chromatin fractionation assay (F). W, whole-cell extract; S, supernatant fraction; P, pellet fraction. Arrowheads indicate the positions of endogenous Chp2 in wild-type cells and ectopically expressed Chp2-ch1, respectively (G). The silencing of Kint2::ura4+ in Δchp2 cells was also evaluated by spotting assay and RT-PCR (H). α, anti; WT, wild type; N/S, nonselective medium.
To confirm this conclusion, chimeric Chp2 proteins were also examined using these assays (Fig. 8E to H). Chimeric Chp2 containing Swi6N suppressed the Δchp2 phenotype, but the other chimeric proteins failed to do so, suggesting that the other domains are important for Chp2 function. Interestingly, replacing these domains also caused an increase in protein levels and enhanced silencing derepression. In particular, replacing the Chp2CSD with the Swi6CSD increased the level of the protein in the soluble fraction, and its expression clearly enhanced the silencing derepression in Δchp2 cells (Fig. 8H, Chp2-ch4). These results suggest that the chromatin/nuclear-associated state of Chp2 is linked with its in vivo protein level and silencing function and that the Chp2CSD plays a major role in the tight chromatin/nuclear association of Chp2. The enhanced silencing derepression is presumably caused by the soluble form of Chp2 interfering with other silencing components, such as Swi6. The detailed molecular mechanisms and biological significance of the distinct chromatin/nuclear associations of Chp2 are currently unclear. We noticed, however, that about half the Clr3 and most of the Epe1 were stably associated with the chromatin-enriched fraction, and these associations were independent of Clr4 (Fig. 6B). These results suggest a possible link between Chp2 function and a nuclear subfraction that is enriched in chromatin-modifying enzymes (see Discussion).
The balance between Swi6 and Chp2 is critical for repressive heterochromatin assembly.
Our analysis showed that Swi6 is more abundant than Chp2 in wild-type cells (Fig. 2A and B) and that the expression of chimeric Chp2 proteins enhanced the silencing derepression (Fig. 8H). We next examined whether the balance between the cellular levels of these two HP1 isoforms is critical for heterochromatin assembly. To this end, we altered the balance of these proteins by overexpressing Chp2. When Chp2 was expressed in wild-type cells from the native swi6+ promoter, the Kint2::ura4+ silencing was clearly derepressed (Fig. 9A). A similar silencing derepression was observed for centromeric otr1R::ura4+ silencing (see Fig. S6A in the supplemental material). This derepression appeared consistent with a previous observation that overexpressing Chp2 causes an increased rate of the mitotic loss of a mini chromosome (15). Together these results suggest that Chp2 does not simply serve as a dose-dependent structural component of heterochromatin and that it plays a specific role that is associated with its lower expression level. Even more importantly, these data also indicated that heterochromatin is sensitive to the balance between Swi6 and Chp2.
FIG. 9.
Proper balance between Swi6 and Chp2 is critical for repressive heterochromatin assembly. (A and B) Overproduction of Chp2 causes a silencing defect at the mat locus in wild-type cells. Swi6 or Chp2 protein was overproduced from the swi6+ promoter on an episomal plasmid (pRE). Silencing of the Kint2::ura4+ reporter in wild-type cells was evaluated by spotting assay (A), and the expressed proteins were detected by Western blot analysis (B). Empty pRE vector was used as the control. (C and D) Chp2 overproduction causes a reduction of Swi6 association (C) and the upregulation of H3-K14 acetylation (D) at Kint2::ura4+. WCE, whole-cell extract. The level of Swi6 and H3-K14Ac at Kint2::ura4+ was assayed by ChIP using anti-Swi6 or anti-acetylated H3-K14 antibodies, respectively. The increase in Kint2::ura4+ signal relative to the ura4DS/E signal is shown beneath each lane. (E) Chromatin fractionation assay for overproduced Chp2. Chp2 expressed from the swi6+ promoter on an episomal plasmid (pRE) in swi6-115 mutant cells was subjected to the chromatin fractionation assay. Fractionated Chp2 was detected by Western blot analysis using an anti-Chp2 antibody. W, whole-cell extract; S, supernatant fraction; P, pellet fraction. (F) A model showing the interaction network among Swi6, Chp2, Epe1, and Clr3. α, anti; exp, exposure; Endo, endogenous.
To understand the mechanisms underlying this silencing derepression, we performed ChIP experiments. We found that overexpressing Chp2led to a decrease in the level of Swi6 at the Kint2::ura4+ locus (Fig. 9C), suggesting that overproduced Chp2 competes with Swi6 for binding to the H3K9me and thus impairs the dose-dependent function of Swi6. We also observed that the H3K14 acetylation levels were increased in cells overexpressing Chp2 (Fig. 9D). This change was probably not caused by a defect in the recruitment of Clr3 and Epe1, since their levels at the Kint2::ura4+ locus were not noticeably altered in the cells overexpressing Chp2(data not shown). A chromatin fractionation assay revealed that most of the overproduced Chp2 was in the soluble fraction (Fig. 9E), suggesting that the expression level and chromatin-binding state of Chp2 are critical for its specific function in heterochromatin assembly. It is highly likely that the soluble form of Chp2 can disturb the dose-dependent Swi6 function and/or impair the association of Chp2 with the aforementioned nuclear subfractions.
Recent studies have suggested that HP1 has a role not only in heterochromatin formation and gene silencing but also in the transcriptional regulation of euchromatic genes (14, 17). While our results demonstrated that the proper balance between the two HP1 proteins is critical for heterochromatin assembly, it is possible that the HP1 protein level affects global cellular processes. To assess this possibility, we produced Swi6 or Chp2 at different expression levels and analyzed the effect on cellular growth. Interestingly, the overexpression of Swi6 from an episomal nmt1-promoter caused severe growth defects (see Fig. S6C in the supplemental material) (44). The microscopic analysis of cells overexpressing Swi6 revealed a variety of abnormal nuclear and cellular morphologies, such as unequal chromosome segregation and anucleate cells (see Fig. S6F in the supplemental material), suggesting that excessive Swi6 affects chromosome segregation or nuclear integrity, which causes growth defects.
The overexpression of Chp2 also led to severe growth defects (see Fig. S6C in the supplemental material). Although similar, abnormal nuclear morphologies were observed in the cells overexpressing Chp2, we noticed that many cells were elongated (see Fig. S6F in the supplemental material), which was not a common phenotype of the cells overexpressing Swi6, implying a possible link between Chp2 function and cell cycle checkpoints. Although the detailed molecular mechanisms underlying these growth defects remain to be elucidated, these results suggest that the expression levels of Swi6 and Chp2 differentially affect cellular growth, thus underscoring how important it is that HP1 proteins be expressed in the proper amounts for heterochromatin assembly and global cellular processes.
DISCUSSION
HP1 family proteins are highly conserved and play critical roles in establishing and maintaining heterochromatic domains. The genetic and biochemical experiments presented in this study reveal novel functional relationships between the two HP1 family proteins in fission yeast, Swi6 and Chp2, and suggest that heterochromatin organization requires the proper balance between the two HP1 isoforms, which have distinct functions.
Swi6 and Chp2 play distinct roles in heterochromatin assembly.
Consistent with previous observations, we showed that Chp2, like Swi6, is required for the formation of fully repressive heterochromatin (45). In this study, we further showed by complementary expression experiments that neither protein could rescue the other's deletion phenotype (Fig. 1D and E). Thus, Swi6 and Chp2 do not merely play overlapping roles but have distinct functions in the formation of the repressive heterochromatin structure.
Swi6 is an abundant protein, and its biochemical properties are consistent with the general features of HP1 family proteins. Therefore, it seems clear that Swi6 plays a dose-dependent role as a building block and promotes the repressive heterochromatin structure through its ability to self-associate. In contrast, Chp2, which is present in lesser amounts, does not play a dose-dependent role in heterochromatin assembly, and its biochemical properties do not fit well with the general characteristics of HP1 family proteins. However, it is also evident that a Chp2-specific function is required for higher-order chromatin assembly (Fig. 3).
What are the specific functions of Chp2? We show here that Chp2 makes a distinct contribution to the recruitment of Epe1 and Clr3 (Fig. 4) and that its function is closely associated with Clr3 (Fig. 5A and 9F). These findings, however, do not fully explain the Chp2-specific functions, because Δchp2 Δclr3 mutant cells showed an additive phenotype (Fig. 5B). Considering that the Chp2 function is tied to its lower abundance, our finding that Chp2 has a distinct mode for associating with the chromatin-enriched nuclear fraction is quite interesting. Our chromatin fractionation assay showed that substantial amounts of Clr3 and Epe1 were associated with the same chromatin fraction as Chp2 in a Clr4-independent manner. Therefore, it is possible that Chp2 plays a role in anchoring or attracting H3K9me-marked chromosome regions to a specific nuclear compartment that is enriched in chromatin-modulating factors. Although we showed that Chp2CSD is important for Chp2's chromatin association (Fig. 8), the underlying molecular mechanisms and target factor(s) are currently unclear. It will be critical to identify a factor or factors that interact differentially with Swi6 and Chp2. Since the nuclear periphery is generally regarded as a zone for transcriptionally repressed genes (1, 41), it will also be interesting to test whether Chp2's function is linked with nuclear components associated with the nuclear envelope or nuclear pore complex.
Distinct functions of HP1 isoforms and heterochromatic domains.
We found that the overproduction of Chp2 caused silencing derepression, Swi6 delocalization, and increased H3K14Ac (Fig. 9). These results show that Chp2 competes with Swi6 for H3K9me binding and interferes with Swi6's heterochromatin spreading and dose-dependent functions. Thus, the lower expression level of Chp2 or the molar balance between Swi6 and Chp2 appears to be critical for ensuring their heterochromatin functions. Although HP1 isoform-specific properties have been described in other systems, to our knowledge this is the first report showing that a balance between the levels of HP1 isoforms affects their functions and is critical for heterochromatin assembly. Therefore, it will be interesting to determine whether the balance among HP1 isoforms is important for heterochromatin function in other organisms.
Previously, we demonstrated that each chromodomain protein makes distinct contributions to the formation of heterochromatin (38). In the present study, we mainly focused on the silent mating type locus although distinct functions of Swi6 and Chp2 were also observed in the centromeres and telomeres (Fig. 7 and our unpublished observations). It remains unclear whether the balance between Swi6 and Chp2 at specific heterochromatic domains is determined simply by their cellular abundance or if unidentified mechanisms actively regulate this balance by preferentially recruiting one of these isoforms. Our previous study revealed that centromeric heterochromatin possesses dynamic properties that require frequent establishment steps compared with other heterochromatic regions (38). It would therefore be interesting to examine whether the balance between different HP1 isoforms determines the dynamic or silencing properties of each heterochromatic domain.
Several lines of evidence support a possible role for HP1 in transcriptional activation. In Drosophila, HP1 is localized to heat shock-induced transcriptionally active loci (36). In humans, HP1γ, one of three HP1 isoforms, is preferentially associated with transcriptional elongation (46). Although it is not clear whether the recruitment of HP1 isoforms is a prerequisite for transcriptional activity, these observations suggest that HP1 family proteins have functions besides their roles in the formation of repressive heterochromatin. In this sense, it is of interest to examine how Chp2 expression leads to the enhancement of heterochromatic transcription.
Another unresolved question is how different HP1 isoforms are recruited to specific chromosomal locations. If the transcriptionally active loci contain the same H3K9me marks as have been observed in pericentromeric regions, it is likely that the recruitment of HP1 isoforms is not simply determined by the presence of H3K9me but is regulated by other mechanisms. Further studies are necessary to elucidate the molecular mechanisms by which different HP1 isoforms are cooperatively or differentially targeted to heterochromatin and contribute to the formation of higher-order chromatin structures.
Supplementary Material
Acknowledgments
We thank F. Ishikawa, Y. Murakami, and T. Matsumoto for strains and plasmids. We thank A. Hayashi, K. Hamada, T. Hayakawa, and H. Hiraga for critical reading; S. Seno for excellent secretarial work; and other laboratory members in the RIKEN Center for Developmental Biology for discussion and support.
None of the authors has a financial interest related to this work.
This research was supported by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. M.S. was supported by a Special Postdoctoral Research Program of RIKEN.
Footnotes
Published ahead of print on 22 September 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alfredsson-Timmins, J., F. Henningson, and P. Bjerling. 2007. The Clr4 methyltransferase determines the subnuclear localization of the mating-type region in fission yeast. J. Cell Sci. 1201935-1943. [DOI] [PubMed] [Google Scholar]
- 2.Allshire, R. C. 1996. Transcriptional silencing in the fission yeast: a manifestation of higher order chromosome structure and functions, p. 443-466. In V. E. A Russo, R. A. Martienssen, and A. D. Riggs (ed.), Epigenetic mechanisms of gene regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 3.Badugu, R., M. M. Shareef, and R. Kellum. 2003. Novel Drosophila heterochromatin protein 1 (HP1)/origin recognition complex-associated protein (HOAP) repeat motif in HP1/HOAP interactions and chromocenter associations. J. Biol. Chem. 27834491-34498. [DOI] [PubMed] [Google Scholar]
- 4.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410120-124. [DOI] [PubMed] [Google Scholar]
- 5.Brasher, S. V., B. O. Smith, R. H. Fogh, D. Nietlispach, A. Thiru, P. R. Nielsen, R. W. Broadhurst, L. J. Ball, N. V. Murzina, and E. D. Laue. 2000. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 191587-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cam, H. P., T. Sugiyama, E. S. Chen, X. Chen, P. C. FitzGerald, and S. I. Grewal. 2005. Comprehensive analysis of heterochromatin- and RNAi-mediated epigenetic control of the fission yeast genome. Nat. Genet. 37809-819. [DOI] [PubMed] [Google Scholar]
- 7.Cowieson, N. P., J. F. Partridge, R. C. Allshire, and P. J. McLaughlin. 2000. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr. Biol. 10517-525. [DOI] [PubMed] [Google Scholar]
- 8.Eissenberg, J. C., G. D. Morris, G. Reuter, and T. Hartnett. 1992. The heterochromatin-associated protein HP-1 is an essential protein in Drosophila with dosage-dependent effects on position-effect variegation. Genetics 131345-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ekwall, K., J. P. Javerzat, A. Lorentz, H. Schmidt, G. Cranston, and R. Allshire. 1995. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 2691429-1431. [DOI] [PubMed] [Google Scholar]
- 10.Festenstein, R., S. Sharghi-Namini, M. Fox, K. Roderick, M. Tolaini, T. Norton, A. Saveliev, D. Kioussis, and P. Singh. 1999. Heterochromatin protein 1 modifies mammalian PEV in a dose- and chromosomal-context-dependent manner. Nat. Genet. 23457-461. [DOI] [PubMed] [Google Scholar]
- 11.Grewal, S. I. 2000. Transcriptional silencing in fission yeast. J. Cell Physiol. 184311-318. [DOI] [PubMed] [Google Scholar]
- 12.Grewal, S. I., and S. C. Elgin. 2002. Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12178-187. [DOI] [PubMed] [Google Scholar]
- 13.Grewal, S. I., and D. Moazed. 2003. Heterochromatin and epigenetic control of gene expression. Science 301798-802. [DOI] [PubMed] [Google Scholar]
- 14.Gullerova, M., and N. J. Proudfoot. 2008. Cohesin complex promotes transcriptional termination between convergent genes in S. pombe. Cell 132983-995. [DOI] [PubMed] [Google Scholar]
- 15.Halverson, D., G. Gutkin, and L. Clarke. 2000. A novel member of the Swi6p family of fission yeast chromo domain-containing proteins associates with the centromere in vivo and affects chromosome segregation. Mol. Gen. Genet. 264492-505. [DOI] [PubMed] [Google Scholar]
- 16.Hayakawa, T., T. Haraguchi, H. Masumoto, and Y. Hiraoka. 2003. Cell cycle behavior of human HP1 subtypes: distinct molecular domains of HP1 are required for their centromeric localization during interphase and metaphase. J. Cell Sci. 1163327-3338. [DOI] [PubMed] [Google Scholar]
- 17.Hiragami, K., and R. Festenstein. 2005. Heterochromatin protein 1: a pervasive controlling influence. Cell Mol. Life Sci. 622711-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Isaac, S., J. Walfridsson, T. Zohar, D. Lazar, T. Kahan, K. Ekwall, and A. Cohen. 2007. Interaction of Epe1 with the heterochromatin assembly pathway in Schizosaccharomyces pombe. Genetics 1751549-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs, S. A., and S. Khorasanizadeh. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 2952080-2083. [DOI] [PubMed] [Google Scholar]
- 20.Kaller, M., U. Euteneuer, and W. Nellen. 2006. Differential effects of heterochromatin protein 1 isoforms on mitotic chromosome distribution and growth in Dictyostelium discoideum. Eukaryot. Cell 5530-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katou, Y., Y. Kanoh, M. Bando, H. Noguchi, H. Tanaka, T. Ashikari, K. Sugimoto, and K. Shirahige. 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 4241078-1083. [DOI] [PubMed] [Google Scholar]
- 22.Krawchuk, M. D., and W. P. Wahls. 1999. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast 151419-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410116-120. [DOI] [PubMed] [Google Scholar]
- 24.Lomberk, G., D. Bensi, M. E. Fernandez-Zapico, and R. Urrutia. 2006. Evidence for the existence of an HP1-mediated subcode within the histone code. Nat. Cell Biol. 8407-415. [DOI] [PubMed] [Google Scholar]
- 25.Lomberk, G., L. Wallrath, and R. Urrutia. 2006. The heterochromatin protein 1 family. Genome Biol. 7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meehan, R. R., C. F. Kao, and S. Pennings. 2003. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. EMBO J. 223164-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Minc, E., Y. Allory, H. J. Worman, J. C. Courvalin, and B. Buendia. 1999. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 108220-234. [DOI] [PubMed] [Google Scholar]
- 28.Muchardt, C., M. Guilleme, J. S. Seeler, D. Trouche, A. Dejean, and M. Yaniv. 2002. Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1alpha. EMBO Rep. 3975-981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakayama, J., A. J. Klar, and S. I. Grewal. 2000. A chromodomain protein, Swi6, performs imprinting functions in fission yeast during mitosis and meiosis. Cell 101307-317. [DOI] [PubMed] [Google Scholar]
- 30.Nakayama, J., J. C. Rice, B. D. Strahl, C. D. Allis, and S. I. Grewal. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292110-113. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen, A. L., M. Oulad-Abdelghani, J. A. Ortiz, E. Remboutsika, P. Chambon, and R. Losson. 2001. Heterochromatin formation in mammalian cells: interaction between histones and HP1 proteins. Mol. Cell 7729-739. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen, A. L., C. Sanchez, H. Ichinose, M. Cervino, T. Lerouge, P. Chambon, and R. Losson. 2002. Selective interaction between the chromatin-remodeling factor BRG1 and the heterochromatin-associated protein HP1α. EMBO J. 215797-5806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nielsen, P. R., D. Nietlispach, H. R. Mott, J. Callaghan, A. Bannister, T. Kouzarides, A. G. Murzin, N. V. Murzina, and E. D. Laue. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416103-107. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa, Y., T. Takahashi, and H. Masukata. 1999. Association of fission yeast Orp1 and Mcm6 proteins with chromosomal replication origins. Mol. Cell. Biol. 197228-7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Partridge, J. F., K. S. Scott, A. J. Bannister, T. Kouzarides, and R. C. Allshire. 2002. cis-Acting DNA from fission yeast centromeres mediates histone H3 methylation and recruitment of silencing factors and cohesin to an ectopic site. Curr. Biol. 121652-1660. [DOI] [PubMed] [Google Scholar]
- 36.Piacentini, L., L. Fanti, M. Berloco, B. Perrini, and S. Pimpinelli. 2003. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 161707-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richards, E. J., and S. C. Elgin. 2002. Epigenetic codes for heterochromatin formation and silencing: rounding up the usual suspects. Cell 108489-500. [DOI] [PubMed] [Google Scholar]
- 38.Sadaie, M., T. Iida, T. Urano, and J. Nakayama. 2004. A chromodomain protein, Chp1, is required for the establishment of heterochromatin in fission yeast. EMBO J. 233825-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawano, A., and A. Miyawaki. 2000. Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schott, S., V. Coustham, T. Simonet, C. Bedet, and F. Palladino. 2006. Unique and redundant functions of C. elegans HP1 proteins in post-embryonic development. Dev. Biol. 298176-187. [DOI] [PubMed] [Google Scholar]
- 41.Sexton, T., H. Schober, P. Fraser, and S. M. Gasser. 2007. Gene regulation through nuclear organization. Nat. Struct. Mol. Biol. 141049-1055. [DOI] [PubMed] [Google Scholar]
- 42.Smothers, J. F., and S. Henikoff. 2001. The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol. Cell. Biol. 212555-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thiru, A., D. Nietlispach, H. R. Mott, M. Okuwaki, D. Lyon, P. R. Nielsen, M. Hirshberg, A. Verreault, N. V. Murzina, and E. D. Laue. 2004. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 23489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thon, G., P. Bjerling, C. M. Bunner, and J. Verhein-Hansen. 2002. Expression-state boundaries in the mating-type region of fission yeast. Genetics 161611-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thon, G., and J. Verhein-Hansen. 2000. Four chromo-domain proteins of Schizosaccharomyces pombe differentially repress transcription at various chromosomal locations. Genetics 155551-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vakoc, C. R., S. A. Mandat, B. A. Olenchock, and G. A. Blobel. 2005. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell 19381-391. [DOI] [PubMed] [Google Scholar]
- 47.Vassallo, M. F., and N. Tanese. 2002. Isoform-specific interaction of HP1 with human TAFII130. Proc. Natl. Acad. Sci. USA 995919-5924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, G., A. Ma, C. M. Chow, D. Horsley, N. R. Brown, I. G. Cowell, and P. B. Singh. 2000. Conservation of heterochromatin protein 1 function. Mol. Cell. Biol. 206970-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wendt, K. S., K. Yoshida, T. Itoh, M. Bando, B. Koch, E. Schirghuber, S. Tsutsumi, G. Nagae, K. Ishihara, T. Mishiro, K. Yahata, F. Imamoto, H. Aburatani, M. Nakao, N. Imamoto, K. Maeshima, K. Shirahige, and J. M. Peters. 2008. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451796-801. [DOI] [PubMed] [Google Scholar]
- 50.Yamada, T., W. Fischle, T. Sugiyama, C. D. Allis, and S. I. Grewal. 2005. The nucleation and maintenance of heterochromatin by a histone deacetylase in fission yeast. Mol. Cell 20173-185. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, R., W. Chen, and P. D. Adams. 2007. Molecular dissection of formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 272343-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zofall, M., and S. I. Grewal. 2006. Swi6/HP1 recruits a JmjC domain protein to facilitate transcription of heterochromatic repeats. Mol. Cell 22681-692. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.