Abstract
Nucleotide excision repair (NER) is the principal pathway for counteracting cytotoxic and mutagenic effects of UV irradiation. To provide insight into the in vivo regulation of the DNA damage recognition step of global genome NER (GG-NER), we constructed cell lines expressing fluorescently tagged damaged DNA binding protein 1 (DDB1). DDB1 is a core subunit of a number of cullin 4-RING ubiquitin ligase complexes. UV-activated DDB1-DDB2-CUL4A-ROC1 ubiquitin ligase participates in the initiation of GG-NER and triggers the UV-dependent degradation of its subunit DDB2. We found that DDB1 rapidly accumulates on DNA damage sites. However, its binding to damaged DNA is not static, since DDB1 constantly dissociates from and binds to DNA lesions. DDB2, but not CUL4A, was indispensable for binding of DDB1 to DNA damage sites. The residence time of DDB1 on the damage site is independent of the main damage-recognizing protein of GG-NER, XPC, as well as of UV-induced proteolysis of DDB2. The amount of DDB1 that is temporally immobilized on damaged DNA critically depends on DDB2 levels in the cell. We propose a model in which UV-dependent degradation of DDB2 is important for the release of DDB1 from continuous association to unrepaired DNA and makes DDB1 available for its other DNA damage response functions.
Nucleotide excision repair (NER) removes a wide spectrum of bulky DNA lesions induced by UV irradiation and chemical mutagens (21, 29). Inherited mutations in NER genes result in the UV-sensitive and cancer-prone syndrome xeroderma pigmentosum (11). NER is divided into two subpathways: transcription-coupled NER (TCR) and global genome NER (GG-NER). TCR repairs DNA lesions in the transcribed strand of active genes (16), whereas GG-NER removes damage throughout the genome (24). The two subpathways differ only in the first step, damage recognition. In TCR, blocking of RNA polymerase II transcription elongation at the DNA lesion serves as a damage recognition signal and stimulates accumulation of downstream repair factors (17, 60). In GG-NER, the principal damage recognition factor is the XPC-HR23B-CEN2 complex (3, 58, 64), which is essential for GG-NER (1, 6).
Several studies demonstrated that the DDB1-DDB2 heterodimer (UV-DDB protein) is an auxiliary damage-recognizing factor of GG-NER stimulating the binding of XPC to UV damage sites. Unlike other NER factors (TFIIH, XPG, XPA-RPA, and ERCC1-XPF), which require functional XPC to bind DNA damage sites (64), DDB2 binds to DNA lesions in XPC-deficient cells (65). Recruitment of XPC to UV lesions in human cells was significantly decreased in the absence of functional DDB2 (43, 68), whereas overexpression of DDB2 results in enhanced recruitment of XPC to cyclobutane pyrimidine dimers (CPD) (15).
The UV-DDB protein has affinity for the two major cytotoxic/mutagenic types of lesions introduced in DNA by UV irradiation [pyrimidine (6-4) pyrimidone photoproducts (6-4PP) and CPD] as well as for other bulky DNA lesions (18, 70). DDB2 mutations were found in the mildest form of xeroderma pigmentosum syndrome, complementation group E (XP-E) (10, 46). Cells of XP-E patients are deficient in GG-NER of CPD (33) and show delayed repair of 6-4PP (35, 43). Knockdown of DDB1 in human fibroblasts by small interfering RNA (siRNA) also resulted in deficiency in GG-NER of CPD (39). Overexpression of DDB2 in murine cells, which are usually expressing a very small amount of DDB2 (2, 62) and are deficient in GG-NER of CPD (9, 55), increases the rates of repair of both CPD and 6-4PP (2). Together, these studies show the significance of UV-DDB in GG-NER.
DDB1 and DDB2 are subunits of a larger protein complex, which also contains cullin 4A (CUL4) and Roc1 and possesses ubiquitin ligase (E3) activity (23). The DDB1-DDB2-CUL4A-Roc1 E3 ligase complex is inactive in nonirradiated cells but becomes active in response to UV irradiation (23) and polyubiquitylates XPC (59). This ubiquitylation positively regulates the NER function of XPC and links the ubiquitin ligase and NER functions of UV-DDB (59, 67). Other targets for ubiquitylation by DDB2-contaning ubiquitin ligase are histones H2A, H3, and H4 (36, 66). These findings suggest a possible role for this ubiquitin ligase in post-UV irradiation chromatin remodeling in order to make it accessible for the downstream factors.
DDB2 itself is also a substrate for ubiquitylation, which results in its proteosomal degradation by the 26S proteasome (8, 45, 53). This degradation of DDB2 is independent of downstream NER factors (53). The biological significance of this paradoxical UV-dependent degradation of a factor functionally involved in the repair of UV-induced lesions is still not understood. It was suggested that this degradation is possibly required to provide access to the damage for the downstream repair factors (13).
The DDB2-containing DDB1-CUL4A-Roc1 E3 ligase complex is one in a series of ubiquitin ligase complexes, each of which contains DDB1, cullin 4A (CUL4), Roc1, and one of the many different WD40 repeat proteins (e.g., DDB2) responsible for substrate specificity (25). So far, 49 such WD40-repeat proteins have been identified (28). Another NER-specific DDB1-CUL4A-Roc1 ubiquitin ligase contains the WD40 repeat protein CSA, one of the key factors of TCR (23). CSA-mediated proteosomal degradation of CSB by CSA-DDB1-CUL4A-Roc1 is essential for post-TCR recovery of transcription (22). DDB1 is also essential for UV-dependent proteolysis of Cdt1, a DNA replication-licensing factor (26, 32, 47). Binding of Cdt1 to proliferating cell nuclear antigen (PCNA) is essential for this degradation (4), demonstrating a role for DDB1-CUL4A in controlling the cell cycle in response to DNA damage in proliferating cells. Therefore, DDB1 can be regarded as a multifunctional regulator of the cellular response to genotoxic stress.
Other functions of DDB1-CUL4 ligases include regulation of transcription via degradation of c-Jun and STAT proteins (51, 69), control of the cell cycle via regulation of cyclin E and cyclin-dependent kinase inhibitor p27Kip1 (known targets of SCF ubiquitin ligases) (27), and regulation of cellular levels of tumor suppressor p53 (5, 44). Some of these functions of DDB1 might also be linked to the regulation of the cellular response to DNA damage; however, their implication in response to genotoxic stress needs to be elucidated. Characteristically for a cell cycle-regulating protein, DDB1 is essential for proliferating but not for postmitotic cells (7). Mutations in DDB1 are lethal early in development in Drosophila flies (61) and mice (7).
In the present study, we analyzed the dynamical behavior of DDB1 in live mammalian cells in response to UV irradiation to further investigate the role of the DDB1-DDB2 complex in cellular response to UV damage.
MATERIALS AND METHODS
Cell lines.
The cell lines used were simian virus 40 (SV40)-immortalized human fibroblasts: VH10 (wild type), MRC5 (wild type), XP4PA (XPC mutant), and XP12RO (XPA mutant). Cell lines were cultured in a 1:1 mixture of Ham's F10 and Dulbecco's modified Eagle's medium (Gibco), supplemented with antibiotics and 10% fetal calf serum, at 37°C, 20% O2, and 5% CO2.
Expression of fusion proteins.
To generate the plasmid pmCherry-C1, mCherry was amplified by PCR on plasmid pRSET-B-mCherry (56) (a kind gift from Roger Tsien) and inserted into peGFP-C1 (Clontech). Full-length murine DDB1 cDNA was excised from the DDB1-eCFP plasmid produced by Moser et al. (43) and inserted into the pmCherry-C1 plasmid. The resulting vector (pmCherry-DDB1), expressing the DDB1 protein tagged on the N terminus with mCherry fluorescent protein (mCherry-DDB1), was transfected into human wild-type fibroblast cells (VH10). Stably expressing cells were isolated after selection using G418, followed by single-cell sorting using FACSVantage (Becton Dickinson). The resulting line was designated A634.
XP-C and XP-A lines expressing mCherry-DDB1 were produced by transducing the cell lines XP4PA-SV40 and XP12RO-SV40 (respectively) with a lentivirus (48) containing mCherry-DDB1. Clones expressing mCherry-DDB1 were isolated by fluorescence-activated cell sorting (FACSVantage; Becton Dickinson). Cells used for fluorescence recovery after photobleaching on local damage (FRAP-LD) and fluorescence loss in photobleaching (FLIP) were produced by simultaneous transduction of VH10-SV40 by lentiviruses expressing mCherry-DDB1 and YFP-DDB2, recloned from an enhanced yellow fluorescent protein (eYFP)-DDB2-expressing construct (43). Clones expressing both mCherry-DDB1 and eYFP-DDB2 were selected by fluorescence-activated cell sorting.
For overexpression of eYFP-DDB2 in cells used for FRAP experiments, A634 cells were transiently transfected with the eYFP-DDB2-expressing construct (43), using FuGENE transfection reagent (Roche) according to the producer's instructions. Expression of green fluorescent protein (GFP)-CUL4A was achieved by transient transfection of cells, using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions.
We also used the previously described eYFP-DDB2-expressing MRC5 (wild-type) (43) and XPC-enhanced GFP (eGFP)-expressing XP4PASV40 (XPC-deficient) fibroblast cell lines (31).
Electrophoretic mobility shift assay.
The electrophoretic mobility shift assay was performed as described in reference 2. Briefly, a 32P-labeled 50-bp DNA fragment was irradiated with 5 kJ/m2 (or mock irradiated) and incubated with nuclear extracts in the presence of specific (nonlabeled, nonirradiated probe) and nonspecific (deoxyinosinic-deoxycytidylic acid) competitors. After incubation, samples were separated on a native polyacrylamide gel.
RNA interference.
siRNA against human DDB2 was purchased from Qiagen (Hs-DDB2-1 HP, a kind gift from R. Nishi and K. Sugasawa, Kobe University). Small nuclear RNA against cullin 4A was produced by Dharmacon, using target sequence GAACAGCGATCGTAATCAA (13) with 3′-UU overhangs. Nontargeting siRNA (no. 5) was purchased from Dharmacon. Transfection of siRNA into cells was performed using Lipofectamine 2000 reagent (Invitrogen) and Opti-MEM medium (Invitrogen) according to the manufacturer's instructions. Before transfections, siRNA was mixed with siGLO Green transfection indicator (Dharmacon) at a ratio of 4 (siRNA):1 (siGLO). The transfected cells were incubated at 37°C for 72 h before experiments. Transfected cells were identified by the green fluorescence of the siGLO Green transfection indicator.
UV irradiation and special treatments.
For induction of global UV DNA damage, cells cultured on 24-mm coverslips were rinsed with phosphate-buffered saline (PBS) (warmed to 37°C) and irradiated with a Philips TUV lamp (254 nm) at a dose rate of 0.8 J/m2/s. To induce local UV damage, cells were irradiated with UV-C with a dose of 100 J/m2 through a polycarbonate filter (Millipore Billerica) with 5-μm or 8-μm pores as described previously (42). For determining the assembly kinetics of mCherry-DDB1, GFP-CUL4A, and eYFP-DDB2 on DNA lesions, the cells were grown to confluence in glass-bottomed dishes (MatTek, Ashland, MA) and irradiated with a UV-C dose of 100 J/m2 using a homemade box containing four UV lamps (Philips TUV 9W PL-S) above the microscope stage as described elsewhere (71).
To investigate the influence of proteasome activity on the mobility of mCherry-DDB1, cells were exposed to the proteasome inhibitor MG-132 (Calbiochem) at a 20-μmol concentration, starting at 3 h before UV irradiation.
Immunoblot analysis and immunofluorescence.
Immunoblot analysis was performed as described previously (17). DDB1 was detected with goat polyclonal anti-DDB1 antibodies (1:1,000; Abcam), and CUL4A was detected with rabbit polyclonal CUL4A antibodies (1:1,000; Abcam). Protein bands were visualized via chemiluminescence (ECL; GE Healthcare) using horseradish peroxidase-conjugated secondary antibodies and exposure to Amersham Hyperfilm ECL (GE Healthcare).
Immunofluorescent staining on fixed cells was carried out as described previously (43). Rabbit polyclonal anti-DDB1 antibodies (1:1,000; Abcam) and Alexa Fluor 594 goat anti-rabbit immunoglobulin G (H+L) (Molecular Probes, Invitrogen) were used as primary and secondary antibodies. All antibodies were diluted in PBS containing 0.15% glycine and 0.5% bovine serum albumin.
FRAP, calculation of immobile fractions of DDB1, and Monte Carlo simulations.
Live-cell imaging experiments were performed using a Zeiss LSM 510 meta confocal laser scanning microscope (Zeiss). The assembly kinetics of mCherry-DDB1, YFP-DDB2, and GFP-CUL4A at the site of local irradiation were measured as detailed before (41).
FRAP experiments were performed as described previously (30, 71), with modifications specific for the use of mCherry fluorescent protein. Briefly, a strip spanning the nucleus was bleached for 80 ms at 100% power with a digital shape sampling and processing laser (561 nm, 10 mW) (Zeiss). Recovery of fluorescence in the strip was monitored every 20 ms for 20 s at 0.5% laser intensity. All FRAP curves are normalized to the average prebleaching fluorescence (based on 200 measurements before photobleaching).
To estimate the relative immobile fractions (IF) from the FRAP measurements, we first subtracted the lowest value of the FRAP curve (i.e., Fab, the first data point after bleaching) from the data set and then renormalized it to the average fluorescence before photobleaching. The immobile fraction is then given by IF = 1 − Fr, where Fr is the average relative fluorescence once recovery is complete (between 15 and 23 s upon the start of the FRAP experiment). We corrected for the photobleached fraction (i.e., incomplete recovery of fluorescence due to irreversible mCherry bleaching during the FRAP experiment) as follows: a whole nucleus of a cell expressing fluorescent protein was imaged, subsequently strip bleached (80 ms at 100% laser intensity), and then imaged 30 s after the bleach pulse. The bleached fraction (BF) was estimated as the relative fluorescence intensity loss between the average fluorescence of the whole nucleus before photobleaching (Fnbb) and the average fluorescence of the whole nucleus after bleaching (Fnab): BF = (1 − Fnab/Fnbb)/(1 − Fab). The corrected immobile fraction (IFcorr) is then given by IFcorr = (IF − BF)/(1 − BF).
To compensate for the effect of fluorescence blinking (19, 41), the corrected immobile fraction was normalized to the size of the corrected immobile fraction of mCherry-DDB1 in fixed A634 cells (treated with 2% paraformaldehyde in PBS for 24 h at room temperature), where mCherry-DDB1 protein was fully immobile (34).
Each estimation of the immobile fraction was based on 12 to 20 cells. Statistical significance was checked by using Student's t test (two samples, two tailed).
Monte Carlo simulations for analysis of FRAP data were performed as described elsewhere (71).
FRAP-LD and FLIP.
To determine the residence times of DDB1 and DDB2 on the local damage, FRAP-LD experiments were performed as described previously (20), with modifications specific for simultaneous bleaching of mCherry and eYFP signals. Briefly, cells were irradiated with 100 J/m2 of UV-C through a polycarbonate filter with 8-μm pores. At 10 to 15 min post-UV irradiation, when accumulation of the fluorescent proteins reached equilibrium (with the dissociation constant [koff] equal to the association constant [kon]), locally damaged areas were photobleached for 2 s at 100% intensities with the DSSP laser (561 nm, 10 mW) and an argon laser (514 nm, 60 mW) (both from Zeiss, Germany). Monitoring of the mCherry and eYFP fluorescence recovery was followed by imaging of the cell every 15 s for 300 s. A single image was taken prior to the photobleaching. The results are expressed at the ratio between the damaged area and the undamaged area. The first data point after photobleaching is set to 0, while later time points are normalized to the plateau value. Time zero corresponds to the first measure after the bleaching. All values were corrected for the background fluorescence. Error bars represent the standard errors of the means.
FLIP was performed as previously described (30). Briefly, cells were irradiated with UV-C through the filter to introduce local damage. One-third of a locally damaged nucleus (in the part of the nucleus distant from the damage) was simultaneously bleached every 5 s at 100% intensities with the DSSP laser (561 nm, 10 mW) and the argon laser (514 nm, 60 mW) (both from Zeiss, Germany). The fluorescence of the locally damaged area was monitored at low intensities with the lasers. The fluorescence in the bleached area was plotted as normalized to the fluorescence of the damaged area before photobleaching. All values were corrected for the background fluorescence.
RESULTS
Production of the mCherry-DDB1-expressing cell line and subcellular localization of mCherry-DDB1.
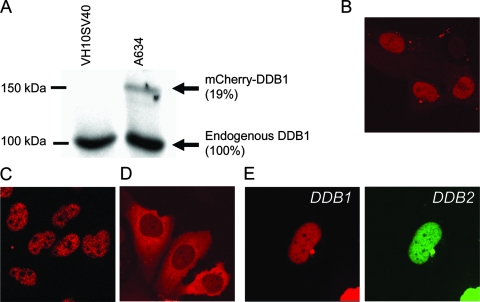
In order to study the response of DDB1 to genotoxic stress in living cells, we fused the cDNA of murine DDB1 at its N terminus with the fluorescent protein mCherry (56). This construct was stably expressed in immortalized human fibroblasts (VH10SV40), and the cell line was designated A634. Immunoblot analysis of A634 cells by use of anti-DDB1 antibody showed a band corresponding to the full-length fusion protein (∼150 kDa), which was expressed at a significantly lower level than the endogenous protein (Fig. 1A).
FIG. 1.
Expression and subcellular localization of mCherry-tagged DDB1. (A) Immunoblot analysis of parental VH10SV40 cells and VH10SV40 cells stably expressing mCherry-DDB1 (A634), using antibodies against DDB1. (B) Subcellular localization of mCherry-DDB1 in A634 cells. (C) Subcellular localization of endogenous DDB1 in cells detected by immunostaining of VH10SV40 cells by use of antibodies against DDB1. (D) Subcellular localization of mCherry-DDB1 in VH10SV40 cells transiently transfected with the mCherry-DDB1-expressing construct (48 h after transfection). (E) Nuclear localization of mCherry-DDB1 in VH10SV40 cells transiently cotransfected with mCherry-DDB1- and eYFP-DDB2-expressing constructs (48 h after transfection).
Fluorescent DDB1 was localized predominantly in the nucleus (Fig. 1B) and was excluded from nucleoli, identical to the localization of endogenous DDB1 (Fig. 1C). In VH10SV40 cells transiently transfected with the mCherry-DDB1 plasmid, the fluorescent signal was localized mostly in cytoplasm (Fig. 1D), suggesting a tight regulation of the nuclear level of DDB1. When cells were transiently cotransfected with plasmids expressing mCherry-DDB1 and eYFP-DDB2, both fusion proteins showed nuclear localizations (Fig. 1E), which correlates with an earlier reported role for DDB2 in regulation of nuclear concentration of DDB1 (40, 57). Apparently, expression of fluorescent mCherry-DDB1 protein in A634 cells did not significantly change the cellular concentration and localization of DDB1.
mCherry-DDB1 accumulates at the site of UV damage with the same kinetics as DDB2 and cullin 4A proteins.
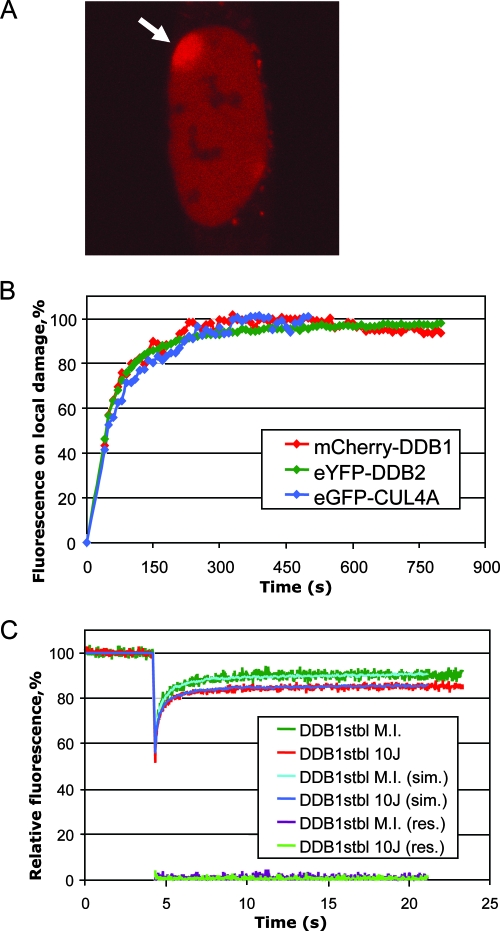
To assess the kinetics of recruitment of DDB1 to the sites of DNA damage, we locally inflicted damage in cell nuclei by UV-C (100 J/m2) through a polycarbonate filter with 5-μm pores. We observed a clear accumulation of mCherry-DDB1 at UV-irradiated areas immediately after UV irradiation (Fig. 2A). The binding kinetics of mCherry-DDB1 (half-life [t1/2] of 44 s) (Fig. 2B) is similar to earlier measured kinetics of accumulation of DDB2 (t1/2 of 40 s) and of cullin 4A (CUL-4A), a component of DDB1-containing ubiquitin ligases (t1/2 of 47 s) (41), suggesting that DDB1 is recruited to the site of UV damage as a part of the DDB1-DDB2-CUL4A-ROC1 protein complex. This result shows that the mCherry-tagged DDB1 protein is able to bind damaged DNA and demonstrates that this protein is incorporated into the E3 complex that was described for endogenous DDB1 protein.
FIG. 2.
Recruitment of mCherry-DDB1 to UV-damaged DNA. (A) Accumulation of mCherry-DDB1 at the site of local damage in A634 cells (indicated with an arrow) irradiated with 100 J/m2 of UV-C through a polycarbonate filter (5 min after UV irradiation). (B) Quantification of the accumulation kinetics of mCherry-DDB1, eYFP-DDB2, and GFP-CUL4A at the site of local damage. Curves were normalized to the plateau value. Time point 0 corresponds to the beginning of the UV irradiation. (C) FRAP analysis of mCherry-DDB1 mobility in mock-irradiated (M.I.) and UV-irradiated (10 J/m2) A634 cells. Experimental curves, curves obtained by Monte Carlo simulations (sim.), and residuals (res.) are shown.
To estimate how genotoxic stress modulates the dynamics of DDB1, we applied FRAP to A634 cells irradiated with 10 J/m2 of UV-C (5 to 20 min post-UV irradiation) and mock-irradiated A634 cells (Fig. 2C). UV-irradiated cells showed an incomplete recovery of fluorescence in comparison with mock-irradiated cells (Fig. 2C), which demonstrates UV-dependent immobilization of mCherry-DDB1. Using Monte Carlo simulations (14), we calculated an increase of mCherry-DDB1 immobilization from 5% to 15% upon UV irradiation yielded the best fit to the experimental FRAP curves. For the diffusing part of the mCherry-DDB1 pool, Monte Carlo simulations yielded similar effective diffusion constants for mCherry-DDB1 in mock-irradiated (3.2 ± 0.3 μm2/s) and irradiated (3.8 ± 0.6 μm2/s) cells, indicating that most DDB1-containing complexes are not rearranged upon UV irradiation.
UV-induced immobilization of DDB1 is dose dependent and requires DDB2.
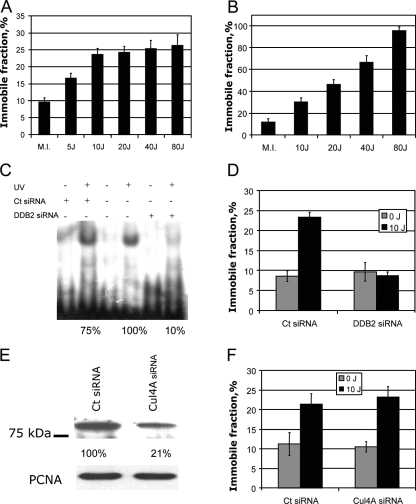
To assess the dose dependency of UV-induced DDB1 immobilization, we applied FRAP to cells exposed to different doses of UV. The immobile fraction of mCherry-DDB1 (calculated using FRAP data as described in Materials and Methods) increased in a dose-dependent manner, with approximately 16% immobilization after a relatively low dose of 5 J/m2 and a further increase to 23% immobilization after 10 J/m2 (Fig. 3A). These results were in line with the immobile fractions calculated by Monte Carlo simulations (see previous paragraph). Further increase of the UV dose, however, did not result in a statistically significant increase of the immobile fraction of mCherry-DDB1 (Fig. 3A). Thus, UV-induced immobilization of DDB1 is a dose-dependent process at low doses of UV-C, already showing saturation with a moderate dose of around 10 J/m2 of UV.
FIG. 3.
Factors influencing UV-induced immobilization of DDB1. (A) Immobilization of mCherry-DDB1 in A634 cells after mock irradiation (M.I.) or different doses of UV. (B) Immobilization of mCherry-DDB1 in A634 cells transiently overexpressing eYFP-DDB2 after different doses of UV-C. (C) Electrophoretic mobility shift assay showing reduction of damaged DNA binding activity in A634 cells transfected with siRNA against DDB2 (DDB2 siRNA) or control nontargeting siRNA (Ct siRNA). (D) Immobile fraction of mCherry-DDB1 in A634 cells transfected with siRNA against DDB2 or control nontargeting siRNA after mock irradiation or irradiation with 10 J/m2 of UV-C. (E) Western blot analysis of CUL4A depletion by siRNA against CUL4A. (F) Immobile fraction of mCherry-DDB1 in A634 cells transfected with siRNA against CUL4A or control (nontargeting) siRNA (mock irradiated or irradiated with 10 J/m2 of UV-C).
We hypothesized that the amount of UV-induced DDB1 immobilization might be determined by the level of intracellular DDB2. To test this hypothesis, we transiently overexpressed eYFP-tagged DDB2 in A634 cells. Transfected cells demonstrated a dose-dependent increase of immobilization of mCherry-DDB1, and at the dose of 80 J/m2, virtually all DDB1 was immobile (Fig. 3B).
Since DDB1 protein is believed to be involved in multiple pathways of cellular response to genotoxic stress besides NER, it is important to know whether all UV-induced immobilization of DDB1 requires DDB2. To address this question, we transfected A634 cells with siRNA against DDB2. Since no reliable antibody against DDB2 was available to us, we used reduction of the damaged DNA binding activity in nuclear extracts as a measure of DDB2 depletion (Fig. 3C). We found that no UV-induced immobilization of mCherry-DDB1 was observed in the DDB2-depleted cells (Fig. 3D), showing that DDB2 is indispensable for UV-induced immobilization of DDB1.
UV-induced immobilization of DDB1 does not require cullin 4A.
To test if the functional ubiquitin ligase complex is required for UV-induced DDB1 immobilization, we transfected A634 cells with interfering RNA against CUL4A, another component of DDB1-containing E3 ligases. Transfection of the cells with CUL4A siRNA resulted in a fivefold depletion of CUL4A, as shown by immunoblotting (Fig. 3E). However, transfection efficiency as estimated by the fluorescence of the siGLO Green transfection indicator (Dharmacon) reached only 70 to 80% (data not shown). Therefore, depletion of cullin 4A in the cells used for FRAP experiments (which were selected based on transfection indicator fluorescence) was probably even higher than 80%. Nevertheless, such depletion of CUL4A did not result in a significant change of the amount of mCherry-DDB1 immobilized upon UV irradiation (Fig. 3F). This result strongly suggests that CUL4A is not required for UV-induced immobilization of DDB1, which is in line with known damaged DNA binding activity of purified UV-DDB in vitro (18, 37, 70).
Immobilization of mCherry-DDB1 depends on the presence of DNA damage and DDB2 degradation.
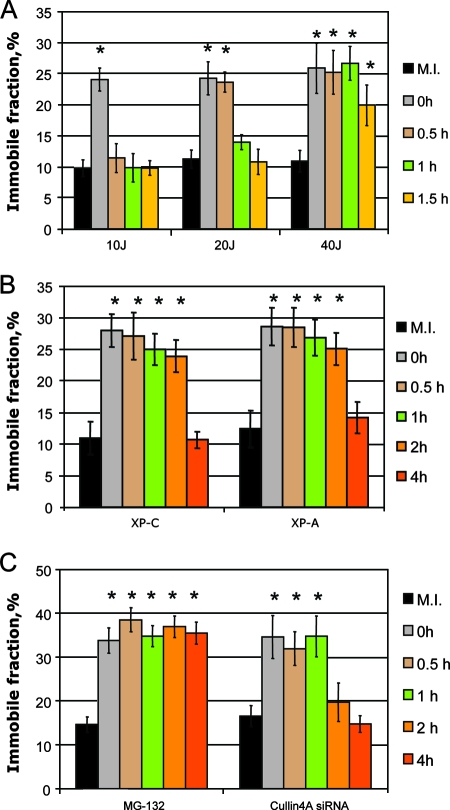
To estimate how long the UV-dependent immobile fraction of mCherryDDB1 is retained upon UV irradiation, we performed FRAP analysis of A634 cells at different time points post-UV irradiation. UV-dependent immobilization of mCherryDDB1 after a saturating UV-C dose of 10 J/m2 lasted less than half an hour (Fig. 4A). However, after irradiation of the A634 cells with 20 J/m2, UV-dependent immobilization was detected beyond 30 min after UV irradiation, and at 40 J/m2, it was retained longer than 1 h (Fig. 4A). Since the maximum UV-induced immobilization was already reached at a dose of 10 J/m2, we assumed that DDB1 immobilization at higher UV doses is retained longer due to the longer persistence of its binding substrate (nonrepaired UV damage) after higher doses of UV. To check this hypothesis, we assessed the time of persistence of the UV-induced immobilization of DDB1 in NER-deficient cell lines expressing mCherry DDB1.
FIG. 4.
Decrease of UV-dependent DDB1 immobilization with time. (A) UV-dependent mCherry-DDB1 immobilization after 10, 20, and 40 J/m2 of UV-C at different time points after UV irradiation or mock irradiation (M.I.). (B) UV-dependent mCherry-DDB1 immobilization in XP-C and XP-A cells after 10 J/m2 of UV-C at different time points after UV irradiation or mock irradiation. (C) UV-dependent mCherry-DDB1 immobilization at different time points after 10 J/m2 of UV-C or mock irradiation in A634 cells treated with the proteasome inhibitor MG-132 and in A634 cells transfected with siRNA against CUL4A. Asterisks mark the immobile fractions of mCherry-DDB1, which differed from those of mock-irradiated cells (P < 0.05).
In both XP-C (deficient in GG-NER) and XP-A (totally deficient in NER) cells, UV-induced immobilization of mCherry-DDB1 was observed as late as 2 h after UV irradiation. However, at 4 h post-UV irradiation, the immobile fraction of mCherry-DDB1 was reduced to that of nonirradiated cells (Fig. 4B). Since in repair-deficient XP-C and XP-A cells there is no reduction of binding substrate (i.e., DNA damage) for DDB1 even after longer incubation times, the decrease of the immobile fraction of DDB1 might be a result of degradation of DDB2 by the 26S proteasome. Indeed, in cells exposed to proteasome inhibitor MG-132, UV-dependent immobilization of DDB1 was retained beyond 4 h post-UV irradiation (Fig. 4C). Therefore, DDB2 degradation might cause reduction of the immobile fraction of DDB1 in XP-C and XP-A cells.
CUL4A, a component of DDB1-containing E3 ligase, was shown to be required for DDB2 degradation (13). We found that UV-induced immobilization of mCherry-DDB1 in cells transfected with interfering RNA against Cul-4A was observed for longer than that in cells transfected with control siRNA and was detectable beyond 1 h post-UV irradiation but disappeared at later time points (Fig. 4C), which was in agreement with earlier observations that CUL4A is involved in but not essential for repair of UV-induced damage (13), similar to DDB2, which is also not absolutely required but significantly enhanced the repair rate at low doses.
The kinetics of DDB1 on DNA lesions is independent of XPC and degradation of DDB2.
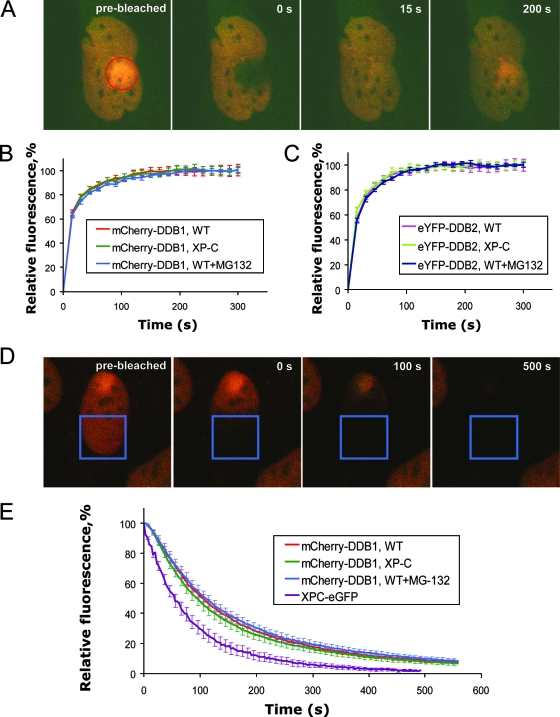
Recent studies showed that DDB2, the binding partner of DDB1, constantly binds and leaves UV damage in DNA and that this process is independent of XPC and uncoupled from DDB2 degradation (41). To investigate whether DDB1 exhibits the same properties of dynamic binding to damaged DNA or shows a more stable binding to UV damage sites, we measured the binding times of DDB1 and DDB2 at the site of UV damage. For this purpose, we produced cell lines simultaneously expressing mCherry-DDB1 and eYFP-DDB2 in VH10-SV40 (wild-type) and XP4PA-SV40 (XPC mutant) cells by using lentiviral vectors expressing the respective fluorescent proteins. To assess the binding kinetics of DDB1 and DDB2 proteins, we applied FRAP-LD (20, 30) (Fig. 5A). In less than 15 s after simultaneous bleaching of eYFP and mCherry fluorescence in the locally UV-irradiated area in the nucleus, the fluorescence levels of both eYFP-DDB2 and mCherry-DDB1 in the bleached area were restored to the levels in the nonbleached part of the nucleus (Fig. 5B and data not shown). This part of the recovery was considered as the free diffusing fractions of DDB1 and DDB2 were unbound from DNA damage sites. However, further increase of the fluorescence in the bleached area (Fig. 5B and C) was due to a specific accumulation of fluorescent DDB1 and DDB2 proteins at the DNA damage sites. This swift association of nonbleached mCherry-DDB1 and eYFP-DDB2 proteins to the damage site shows that these proteins constantly leave and associate to DNA damage sites, as was earlier shown for TFIIH (30). The recovery of fluorescence in the bleached area reaches its saturation between 150 and 200 s after UV irradiation (Fig. 5B), which was less time than that required for such saturation in a similar experiment with TFIIH (∼250 s) (30), showing that the association of DDB1 with the DNA damage site is faster than that in the TFIIH-containing preincision complex. The kinetics of fluorescence recovery of eYFP-DDB2 was very similar to that for mCherry-DDB1 (Fig. 5C), which shows that both dissociation of DDB1 from the UV lesions in DNA and its consequent rebinding to UV-damaged DNA occur within the context of a DDB2-containing complex. The FRAP curves for DDB1 and DDB2 in XP-C cells and wild-type cells exposed to the proteasome inhibitor MG-132 did not differ from those obtained with nontreated wild-type cells (Fig. 5B and C), showing that dissociation and rebinding of DDB1 and DDB2 to the DNA damage site are independent of XPC and uncoupled from DDB2 degradation.
FIG. 5.
Residence time and dissociation kinetics of DDB1 and DDB2 proteins on local DNA damage. (A) Illustration of FRAP-LD in wild-type cells expressing mCherry-DDB1 and eYFP-DDB2. A cell is shown before the bleaching and at different time points after the bleaching. (B) FRAP-LD analysis of mCherry-DDB1 on local DNA damage in the nuclei of wild-type (WT) cells, XP-C cells, and wild-type cells treated with the proteasome inhibitor MG-132. (C) FRAP-LD analysis of eYFP-DDB2 on local DNA damage in the nuclei of wild-type cells, XP-C cells, and wild-type cells treated with the proteasome inhibitor MG-132. (D) Illustration of FLIP in wild-type cells expressing mCherry-DDB1 and eYFP-DDB2. A cell is shown before the bleaching and at different time points after the beginning of the repeated bleachings. (E) Dissociation kinetics of mCherry-DDB1 on local DNA damage in the nuclei of wild-type cells, XP-C cells, and wild-type cells treated with the proteasome inhibitor MG-132, as measured by FLIP. Also shown is a FLIP curve for XPC-eGFP. Error bars represent standard errors of the means. FRAP-LD and FLIP curves are based on 9 to 12 cells.
To independently verify the results of FRAP-LD, we performed FLIP experiments (52). A region of the nucleus distant from the local damage (approximately half of the nucleus) was continuously bleached with appropriate lasers, and the rate of the loss of fluorescence at the local damage site was measured (Fig. 5D). The t1/2 of the FLIP curve can be used as a measure for the dissociation constant (koff) of DDB1 at sites of DNA damage. The t1/2 of release of DDB1 at sites of UV damage was ∼105 s, which was similar to the t1/2 of DDB2 (110 s) obtained earlier (41). In consistence with FRAP-LD experiments, FLIP experiments also showed that the release of DDB1 from the sites of the damage happened independently of XPC and DDB2 degradation (Fig. 5E). Binding of XPC-eGFP to the damage site was more dynamic than that of mCherry-DDB1, which is in agreement with earlier findings indicating less-stable association for XPC than for DDB2 (41).
DISCUSSION
DDB1 was first identified as a subunit of a heterodimeric DNA damage-binding protein (UV-DDB) that it forms together with DDB2. UV-DDB plays a role in the damage recognition step of GG-NER, but its exact function in DNA repair is still unclear. More recently, it was found that DDB1 via its interactions with CUL4A and ROC1 is involved in regulation of multiple cellular processes by ubiquitylation of numerous substrates, many of which undergo proteosomal degradation as a result of ubiquitylation. DDB2 is one of a long series of WD40 repeat proteins that are responsible for the substrate specificity of DDB1-CUL4A-ROC1 ligases (reviewed in references 28 and 38). A recent study of dynamic behavior of DDB2 in live cells showed that virtually all DDB2 is bound to damaged DNA at relatively low doses of UV-C and that DDB2 accumulation is independent of the principal damage-recognizing complex, XPC-HR23B-Centrin2 (41). Since DDB1, apart from its interaction with DDB2, is also included in other protein complexes involved in DNA damage response, we aimed to (i) assess its participation in this process by determining the dynamic properties of DDB1 in relation to accumulation/dissociation of DDB1 and DDB2 at the DNA damage site, (ii) determine which factors regulate association of DDB1 with DNA lesions, and (iii) find a possible interplay between functions of DDB1 in GG-NER and its other DNA damage-reacting functions.
To obtain a tool to monitor the dynamic behavior of DDB1, we produced a cell line that expresses mCherry-DDB1 at a significantly lower level than endogenous DDB1 to avoid possible artifacts coming from overexpression of DDB1. Since the total cellular concentration of DDB1 was not significantly changed, the measured mobility of fluorescently tagged protein reflects the mobility of the total pool of endogenous DDB1. Fluorescent DDB1 (similar to endogenous DDB1) showed predominantly nuclear localization, which correlates with most of its known functions, such as GG-NER, transcription regulation, and licensing of replication (26, 32, 47). Earlier reported data on predominantly cytoplasmic localization of DDB1 (40, 57) were based on transient transfection with fluorescently tagged DDB1 expressing constructs likely causing overexpression and imbalance of subcellular localization of DDB1. However, an important role for DDB2 in regulation of nuclear concentration of DDB1 reported by these authors was further supported by our finding of predominantly nuclear localization of DDB1 even in cells overexpressing DDB1 when transiently cotransfected with plasmids expressing both DDB1 and DDB2.
Accumulation of DDB1 at locally UV-damaged areas in the nuclei indicated that the functionality of mCherry-DDB1 was not compromised by the fluorescent tag. The accumulation of CFP-tagged DDB1 at the locally damaged area in the nucleus was previously shown under conditions of overexpression of both DDB1 and DDB2 (43). Here, we show that DDB1 at a physiological concentration is indeed able to translocate to DNA damage sites. This accumulation occurred with kinetics similar to those for accumulation of DDB2 and CUL4A, suggesting that DDB1 is recruited to the site of UV damage as a part of the DDB1-DDB2-CUL4A-ROC1 ubiquitin ligase complex. The effective diffusion constant of mCherry-DDB1 of 3.1 ± 0.3 μm2/s, estimated by fitting to Monte Carlo-simulated fluorescence recovery curves, was somewhat higher than the effective diffusion constant for DDB2 (2.4 ± 0.4 μm2/s) (41) previously measured by this method. This difference might be explained by the compositions of different DDB1-CUL4A-ROC1 ubiquitin ligase complexes. DDB1-DDB2-CUL4A-ROC1 in non-UV-irradiated cells is bound by COP9 signalosome, which dissociates from the complex upon UV irradiation (23). Unlike DDB2-containing E3, CSA-DDB1-CUL4A-ROC1 is not bound to COP9 signalosome in unchallenged cells (23) (this is possibly also the case for some of the other DDB1-CUL4A ubiquitin ligases), which can account for the faster overall mobility of DDB1-containing complexes.
The immobile fraction of mCherry-DDB1 found in nonirradiated cells showed that a certain amount of DDB1 is involved in long-term interactions with immobile structures in the nucleus, presumably chromatin. This UV-independent immobilization of DDB1 was DDB2 independent (Fig. 3D). In contrast, UV-induced immobilization of mCherry-DDB1 observed in our experiments critically depends on the cellular concentration of DDB2 (compare Fig. 3A and B).
Surprisingly, despite the multiple DNA damage response functions of DDB1, UV-induced immobilization of DDB1 was completely abolished in DDB2 knockdown cells (Fig. 3). This finding shows that other activities of DDB1 in response to genotoxic stress do not contribute to measurable immobilization on the damaged chromatin.
UV-induced immobilization of DDB1 decreased to undetectable levels in less than 0.5 h at a relatively high dose of 10 J/m2, which was faster than the actual repair of 6-4PP (3 h) (54) and CPD (24 h) (63) after this dose of UV. This phenomenon might be explained by a scenario in which the initial processing of damage or accumulation of downstream repair factors makes the lesion inaccessible for UV-DDB binding. A prolonged conservation of the unprocessed/unrepaired DNA lesions after high doses of UV-C (20 and 40 J/m2) was obviously the reason for the dose-dependent increase of the time during which the UV-dependent immobilization of mCherry-DDB1 was detectable.
In GG-NER-deficient XPC and totally NER-deficient XPA cells, UV-dependent immobilization of DDB1 can be detected for much longer period upon UV irradiation than in wild-type cells. This longer period of detectable immobilization cannot be explained by a difference in DDB2 degradation, since in XP-C and XP-A cells DDB2 degradation occurs as in wild-type cells (53). However, the strong reduction of the immobile fraction of DDB1 in XP-C and XP-A cells at 4 h after UV irradiation, despite the presence of the unrepaired damage, is likely due to the DDB2 degradation. Indeed, a previous study (41) showed that approximately 75% of DDB2 in human fibroblasts is degraded at 4 h after irradiation with 8 J/m2 of UV-C (whereas only about 30% of DDB2 was degraded at 2 h post-UV irradiation). When proteasome activity was blocked by adding the proteasome inhibitor MG-132, the UV-dependent immobile fraction of DDB1 remained unchanged for at least 4 h. Proteasome inhibition by MG-132 not only blocks UV-dependent degradation of DDB2 (2) but also (even at concentrations lower than those used in our experiments) considerably inhibits repair of both 6-4PP and CPD (67). Depletion of cullin 4, which was shown to completely inhibit DDB2 degradation and moderately delayed CPD (but not 6-4PP) repair (13), also resulted in prolonged UV-dependent immobilization. Thus, repair of UV damage in DNA and degradation of DDB2 are two processes responsible for reduction of UV-dependent DDB1 immobilization in time. In the absence of these processes, a fraction of DDB1 is constantly associated to damaged DNA.
Binding of DDB1 and DDB2 to the damaged DNA is not static. These proteins are constantly dissociating and associating to the damaged DNA, as demonstrated by the relatively fast redistribution of fluorescent DDB1 and DDB2 in FRAP-LD experiments. The dissociation kinetics of DDB1 are equal in both wild-type and XPC mutant cells (Fig. 5), indicating that binding of DNA damage sites by DDB1 is not regulated by the XPC complex or by any other known NER factors, since their recruitment to DNA damage sites requires XPC (64). This is in line with the XPC independence previously found for dynamic association of DDB2 to damaged DNA (41). Since the dynamic behavior patterns of DDB1 and DDB2 were identical, it might be concluded that dynamic binding of the UV damage sites is performed by the DDB1-DDB2 complex as a whole.
Degradation of DDB2 is triggered by activation of the DDB2-containing ubiquitin ligase after binding to the damaged DNA (23), which suggests that DDB2 degradation is a part of the cellular response to UV. The current view on the role of UV-dependent DDB2 degradation is that degradation is required to provide access to the damage for the downstream repair factors (13). However, the association of the DDB1-DDB2-containing complex to the damaged DNA is very dynamic (this study and reference 41); thus, degradation of DDB2 is not needed for access of the NER factors to the damage. The degradation of DDB2 is also not triggered by recruitment of the downstream repair factors, since various cells with mutations in NER genes are fully capable in DDB2 degradation (41, 53). Furthermore, DDB2 was shown to perform multiple binding cycles prior to its degradation; thus, degradation of DDB2 is uncoupled from binding of DDB2 to the damage site (41). It was also found that siRNA-mediated knockdown of a replication checkpoint protein, Claspin, inhibits UV-dependent degradation of DDB2 but does not interfere with GG-NER of UV lesions (50). Finally, inhibition of DDB2 degradation by the proteasome inhibitor MG-132 did not influence the exchange kinetics of DDB1 at the site of the damage (this study) (Fig. 5B and E). Taken together, these data suggest that degradation of DDB2 does not play a role in NER.
Inhibition of GG-NER by proteasome inhibitor (67) is therefore not connected with inhibition of DDB2 degradation. The effect of proteasome inhibition of NER by MG-132 (67) was much more severe than NER deficiency caused by total absence of DDB2 (43) and is most likely due to depletion of the pool of free ubiquitin by proteasome inhibition (12). Inhibition of ubiquitylation processes might result in blocking of GG-NER-stimulating ubiquitylation of XPC (59), absence of UV-dependent chromatin remodeling via histone ubiquitylation (36, 66), and, possibly, absence of some other NER-specific ubiquitylation processes.
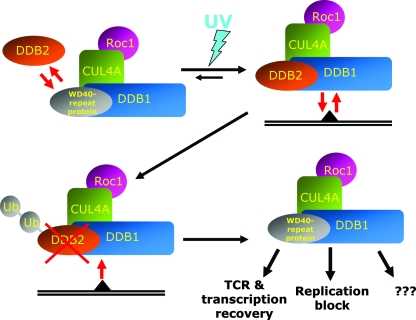
We propose the following hypothesis for the role of UV dose-dependent degradation of DDB2 (Fig. 6). We showed that UV-dependent immobilization of DDB1 is regulated by cellular concentrations of DDB2 (Fig. 3A to D). Binding of DDB2-contaning complexes to damaged DNA depletes cells of DDB1 by titrating this factor away from its other interactions, which is demonstrated by the virtually complete immobilization of DDB1 in DDB2-overexpressing cells irradiated with high doses of UV (Fig. 3B). Degradation of DDB2 is dose dependent (53) and, paradoxically, reduces the concentrations of functional damage recognition complexes when needed most. However, when too much lesion is encountered, priority must be given to other functions of DDB1 (e.g., replication arrest and transcription regulation, etc.) over DNA repair. Apparently, dose-dependent DDB2 degradation regulates implication of DDB1 in UV-damaged DNA damage response. Degradation of DDB2 releases DDB1 (and other shared components of ubiquitin ligases) from continuous association to unrepaired DNA damage sites and makes it available for its other damage response functions.
FIG. 6.
Hypothesis proposing the role of UV-induced DDB2 degradation in release of DDB1 for the interactions outside NER. Without UV irradiation, different WD40 repeat proteins associate with DDB1-ubiquitine ligase complexes. In the presence of the unrepaired DNA lesion, equilibrium is shifted toward assembly of the DDB2-DDB1-CUL4A-ROC1 complex, which constantly rebinds to the damaged DNA. At the same time, binding of the DDB2-containing ubiquitin ligase complex to UV-damaged DNA triggers ubiquitylation and proteasomic degradation of DDB2, which makes DDB1 available for inclusion in other E3 complexes, providing cellular response to the genotoxic stress.
In the mCherry-DDB1-expressing A634 cell line, which we used in our experiments, the maximal range of UV-induced immobilization was about 13% of the total pool of DDB1, which, considering the multiple functions of DDB1, could already have significant physiological consequences for other DDB1-regulated processes. It should be noted, however, that the cells used in our study were SV40-immortalized fibroblasts that express a lower level of DDB2 than primary cells due to functional inhibition of p53 by the SV40 large T antigen, while expression of DDB2 was found to be partially dependent on p53 (33). In addition, it was found that keratinocytes, the cells encountering most of the UV irradiation in vivo, express DDB2 at a much higher level than fibroblasts (49). Thus, in human keratinocytes the concentrations of DDB2 are much higher than in A634 cells, and the impact of the UV-induced, DDB2-dependent immobilization of DDB1 is expected to be high and probably closer to that of DDB2-overexpressing cells in our experiments. Therefore, the role of DDB2 degradation as a UV dose-dependent regulator at the interplay of the different activities of DDB1 in the cellular response to DNA damage appears to be extremely important.
Acknowledgments
S.A. was financed by the European Science Foundation, EuroDYNA program 03-DYNA-F-18 (Spatio-temporal Organization of Genome Surveillance in Live Cells). A.P. was financed by an FP6-EU grant (DNA Repair; LSHG-CT-2005-512113). M.S.L., A.B.H., and W.V. were supported by grants from The Netherlands Organization for Health Research and Development (ZonMW; grants 912-03-012, 917-46-371, and 917-46-364), P.-O.M. and H.L. were supported by grants from the Association for International Cancer Research (AICR 07-0129 and AICR 08-045), and H.L. was also supported by the Human Frontiers Science Program Organization (RGP0007/2004). G.G.-M. was supported by an Erasmus University Fellowship.
Footnotes
Published ahead of print on 20 October 2008.
REFERENCES
- 1.Aboussekhra, A., M. Biggerstaff, M. K. Shivji, J. A. Vilpo, V. Moncollin, V. N. Podust, M. Protic, U. Hubscher, J. M. Egly, and R. D. Wood. 1995. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell 80859-868. [DOI] [PubMed] [Google Scholar]
- 2.Alekseev, S., H. Kool, H. Rebel, M. Fousteri, J. Moser, C. Backendorf, F. R. de Gruijl, H. Vrieling, and L. H. Mullenders. 2005. Enhanced DDB2 expression protects mice from carcinogenic effects of chronic UV-B irradiation. Cancer Res. 6510298-10306. [DOI] [PubMed] [Google Scholar]
- 3.Araki, M., C. Masutani, M. Takemura, A. Uchida, K. Sugasawa, J. Kondoh, Y. Ohkuma, and F. Hanaoka. 2001. Centrosome protein centrin 2/caltractin 1 is part of the xeroderma pigmentosum group C complex that initiates global genome nucleotide excision repair. J. Biol. Chem. 27618665-18672. [DOI] [PubMed] [Google Scholar]
- 4.Arias, E. E., and J. C. Walter. 2006. PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 884-90. [DOI] [PubMed] [Google Scholar]
- 5.Banks, D., M. Wu, L. A. Higa, N. Gavrilova, J. Quan, T. Ye, R. Kobayashi, H. Sun, and H. Zhang. 2006. L2DTL/CDT2 and PCNA interact with p53 and regulate p53 polyubiquitination and protein stability through MDM2 and CUL4A/DDB1 complexes. Cell Cycle 51719-1729. [DOI] [PubMed] [Google Scholar]
- 6.Bohr, V. A., D. S. Okumoto, and P. C. Hanawalt. 1986. Survival of UV-irradiated mammalian cells correlates with efficient DNA repair in an essential gene. Proc. Natl. Acad. Sci. USA 833830-3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cang, Y., J. Zhang, S. A. Nicholas, J. Bastien, B. Li, P. Zhou, and S. P. Goff. 2006. Deletion of DDB1 in mouse brain and lens leads to p53-dependent elimination of proliferating cells. Cell 127929-940. [DOI] [PubMed] [Google Scholar]
- 8.Chen, X., Y. Zhang, L. Douglas, and P. Zhou. 2001. UV-damaged DNA-binding proteins are targets of CUL-4A-mediated ubiquitination and degradation. J. Biol. Chem. 27648175-48182. [DOI] [PubMed] [Google Scholar]
- 9.Cheo, D. L., H. J. Ruven, L. B. Meira, R. E. Hammer, D. K. Burns, N. J. Tappe, A. A. van Zeeland, L. H. Mullenders, and E. C. Friedberg. 1997. Characterization of defective nucleotide excision repair in XPC mutant mice. Mutat. Res. 3741-9. [DOI] [PubMed] [Google Scholar]
- 10.Chu, G., and E. Chang. 1988. Xeroderma pigmentosum group E cells lack a nuclear factor that binds to damaged DNA. Science 242564-567. [DOI] [PubMed] [Google Scholar]
- 11.Cleaver, J. E. 2005. Cancer in xeroderma pigmentosum and related disorders of DNA repair. Nat. Rev. Cancer 5564-573. [DOI] [PubMed] [Google Scholar]
- 12.Dantuma, N. P., T. A. Groothuis, F. A. Salomons, and J. Neefjes. 2006. A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J. Cell Biol. 17319-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El-Mahdy, M. A., Q. Zhu, Q. E. Wang, G. Wani, M. Praetorius-Ibba, and A. A. Wani. 2006. Cullin 4A-mediated proteolysis of DDB2 protein at DNA damage sites regulates in vivo lesion recognition by XPC. J. Biol. Chem. 28113404-13411. [DOI] [PubMed] [Google Scholar]
- 14.Farla, P., R. Hersmus, B. Geverts, P. O. Mari, A. L. Nigg, H. J. Dubbink, J. Trapman, and A. B. Houtsmuller. 2004. The androgen receptor ligand-binding domain stabilizes DNA binding in living cells. J. Struct. Biol. 14750-61. [DOI] [PubMed] [Google Scholar]
- 15.Fitch, M. E., S. Nakajima, A. Yasui, and J. M. Ford. 2003. In vivo recruitment of XPC to UV-induced cyclobutane pyrimidine dimers by the DDB2 gene product. J. Biol. Chem. 27846906-46910. [DOI] [PubMed] [Google Scholar]
- 16.Fousteri, M., and L. H. Mullenders. 2008. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 1873-84. [DOI] [PubMed] [Google Scholar]
- 17.Fousteri, M., W. Vermeulen, A. A. van Zeeland, and L. H. Mullenders. 2006. Cockayne syndrome A and B proteins differentially regulate recruitment of chromatin remodeling and repair factors to stalled RNA polymerase II in vivo. Mol. Cell 23471-482. [DOI] [PubMed] [Google Scholar]
- 18.Fujiwara, Y., C. Masutani, T. Mizukoshi, J. Kondo, F. Hanaoka, and S. Iwai. 1999. Characterization of DNA recognition by the human UV-damaged DNA-binding protein. J. Biol. Chem. 27420027-20033. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Parajo, M. F., G. M. Segers-Nolten, J. A. Veerman, J. Greve, and N. F. van Hulst. 2000. Real-time light-driven dynamics of the fluorescence emission in single green fluorescent protein molecules. Proc. Natl. Acad. Sci. USA 977237-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giglia-Mari, G., C. Miquel, A. F. Theil, P. O. Mari, D. Hoogstraten, J. M. Ng, C. Dinant, J. H. Hoeijmakers, and W. Vermeulen. 2006. Dynamic interaction of TTDA with TFIIH is stabilized by nucleotide excision repair in living cells. PLoS Biol. 4e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gillet, L. C., and O. D. Scharer. 2006. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem. Rev. 106253-276. [DOI] [PubMed] [Google Scholar]
- 22.Groisman, R., I. Kuraoka, O. Chevallier, N. Gaye, T. Magnaldo, K. Tanaka, A. F. Kisselev, A. Harel-Bellan, and Y. Nakatani. 2006. CSA-dependent degradation of CSB by the ubiquitin-proteasome pathway establishes a link between complementation factors of the Cockayne syndrome. Genes Dev. 201429-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Groisman, R., J. Polanowska, I. Kuraoka, J. Sawada, M. Saijo, R. Drapkin, A. F. Kisselev, K. Tanaka, and Y. Nakatani. 2003. The ubiquitin ligase activity in the DDB2 and CSA complexes is differentially regulated by the COP9 signalosome in response to DNA damage. Cell 113357-367. [DOI] [PubMed] [Google Scholar]
- 24.Hanawalt, P. C., J. M. Ford, and D. R. Lloyd. 2003. Functional characterization of global genomic DNA repair and its implications for cancer. Mutat. Res. 544107-114. [DOI] [PubMed] [Google Scholar]
- 25.He, Y. J., C. M. McCall, J. Hu, Y. Zeng, and Y. Xiong. 2006. DDB1 functions as a linker to recruit receptor WD40 proteins to CUL4-ROC1 ubiquitin ligases. Genes Dev. 202949-2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Higa, L. A., I. S. Mihaylov, D. P. Banks, J. Zheng, and H. Zhang. 2003. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 51008-1015. [DOI] [PubMed] [Google Scholar]
- 27.Higa, L. A., X. Yang, J. Zheng, D. Banks, M. Wu, P. Ghosh, H. Sun, and H. Zhang. 2006. Involvement of CUL4 ubiquitin E3 ligases in regulating CDK inhibitors Dacapo/p27Kip1 and cyclin E degradation. Cell Cycle 571-77. [DOI] [PubMed] [Google Scholar]
- 28.Higa, L. A., and H. Zhang. 2007. Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div. 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoeijmakers, J. H. 2001. Genome maintenance mechanisms for preventing cancer. Nature 411366-374. [DOI] [PubMed] [Google Scholar]
- 30.Hoogstraten, D., A. L. Nigg, H. Heath, L. H. Mullenders, R. van Driel, J. H. Hoeijmakers, W. Vermeulen, and A. B. Houtsmuller. 2002. Rapid switching of TFIIH between RNA polymerase I and II transcription and DNA repair in vivo. Mol. Cell 101163-1174. [DOI] [PubMed] [Google Scholar]
- 31.Hoogstraten, D., S. Bergink, V. H. M. Verbiest, M. S. Luijsterburg, B. Geverts, A. Raams, C. Dinant, J. H. J. Hoeijmakers, W. Vermeulen, and A. B. Houtsmuller. 2008. Versatile DNA damage probing by the global genome nucleotide excision repair protein XPC. J. Cell Sci. 1212850-2859. [DOI] [PubMed] [Google Scholar]
- 32.Hu, J., C. M. McCall, T. Ohta, and Y. Xiong. 2004. Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 61003-1009. [DOI] [PubMed] [Google Scholar]
- 33.Hwang, B. J., J. M. Ford, P. C. Hanawalt, and G. Chu. 1999. Expression of the p48 xeroderma pigmentosum gene is p53-dependent and is involved in global genomic repair. Proc. Natl. Acad. Sci. USA 96424-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ibrahim, S. M. 2006. Quantitative fluorescence microscopy of protein dynamics in living cells. Ph.D. thesis. Erasmus University, Rotterdam, The Netherlands.
- 35.Itoh, T., T. Mori, H. Ohkubo, and M. Yamaizumi. 1999. A newly identified patient with clinical xeroderma pigmentosum phenotype has a non-sense mutation in the DDB2 gene and incomplete repair in (6-4) photoproducts. J. Investig. Dermatol. 113251-257. [DOI] [PubMed] [Google Scholar]
- 36.Kapetanaki, M. G., J. Guerrero-Santoro, D. C. Bisi, C. L. Hsieh, V. Rapic-Otrin, and A. S. Levine. 2006. The DDB1-CUL4ADDB2 ubiquitin ligase is deficient in xeroderma pigmentosum group E and targets histone H2A at UV-damaged DNA sites. Proc. Natl. Acad. Sci. USA 1032588-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kulaksiz, G., J. T. Reardon, and A. Sancar. 2005. Xeroderma pigmentosum complementation group E protein (XPE/DDB2): purification of various complexes of XPE and analyses of their damaged DNA binding and putative DNA repair properties. Mol. Cell. Biol. 259784-9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee, J., and P. Zhou. 2007. DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol. Cell 26775-780. [DOI] [PubMed] [Google Scholar]
- 39.Li, J., Q. E. Wang, Q. Zhu, M. A. El-Mahdy, G. Wani, M. Praetorius-Ibba, and A. A. Wani. 2006. DNA damage binding protein component DDB1 participates in nucleotide excision repair through DDB2 DNA-binding and cullin 4A ubiquitin ligase activity. Cancer Res. 668590-8597. [DOI] [PubMed] [Google Scholar]
- 40.Liu, W., A. F. Nichols, J. A. Graham, R. Dualan, A. Abbas, and S. Linn. 2000. Nuclear transport of human DDB protein induced by ultraviolet light. J. Biol. Chem. 27521429-21434. [DOI] [PubMed] [Google Scholar]
- 41.Luijsterburg, M. S., J. Goedhart, J. Moser, H. Kool, B. Geverts, A. B. Houtsmuller, L. H. Mullenders, W. Vermeulen, and R. van Driel. 2007. Dynamic in vivo interaction of DDB2 E3 ubiquitin ligase with UV-damaged DNA is independent of damage-recognition protein XPC. J. Cell Sci. 1202706-2716. [DOI] [PubMed] [Google Scholar]
- 42.Mone, M. J., M. Volker, O. Nikaido, L. H. Mullenders, A. A. van Zeeland, P. J. Verschure, E. M. Manders, and R. van Driel. 2001. Local UV-induced DNA damage in cell nuclei results in local transcription inhibition. EMBO Rep. 21013-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moser, J., M. Volker, H. Kool, S. Alekseev, H. Vrieling, A. Yasui, A. A. van Zeeland, and L. H. Mullenders. 2005. The UV-damaged DNA binding protein mediates efficient targeting of the nucleotide excision repair complex to UV-induced photo lesions. DNA Repair (Amsterdam) 4571-582. [DOI] [PubMed] [Google Scholar]
- 44.Nag, A., S. Bagchi, and P. Raychaudhuri. 2004. Cul4A physically associates with MDM2 and participates in the proteolysis of p53. Cancer Res. 648152-8155. [DOI] [PubMed] [Google Scholar]
- 45.Nag, A., T. Bondar, S. Shiv, and P. Raychaudhuri. 2001. The xeroderma pigmentosum group E gene product DDB2 is a specific target of cullin 4A in mammalian cells. Mol. Cell. Biol. 216738-6747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nichols, A. F., P. Ong, and S. Linn. 1996. Mutations specific to the xeroderma pigmentosum group E Ddb- phenotype. J. Biol. Chem. 27124317-24320. [DOI] [PubMed] [Google Scholar]
- 47.Nishitani, H., N. Sugimoto, V. Roukos, Y. Nakanishi, M. Saijo, C. Obuse, T. Tsurimoto, K. I. Nakayama, K. Nakayama, M. Fujita, Z. Lygerou, and T. Nishimoto. 2006. Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 251126-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oklejewicz, M., E. Destici, F. Tamanini, R. A. Hut, R. Janssens, and G. T. van der Horst. 2008. Phase resetting of the mammalian circadian clock by DNA damage. Curr. Biol. 18286-291. [DOI] [PubMed] [Google Scholar]
- 49.Pines, A., C. Backendorf, S. Alekseev, M. Fousteri, F. R. de Gruijl, H. Vrieling, and L. H. F. Mullenders. Endogenous levels of DDB2 expression strongly influence the susceptibility of the hairless mice to UV-induced skin cancer. DNA Repair (Amsterdam), in press.
- 50.Praetorius-Ibba, M., Q. E. Wang, G. Wani, M. A. El-Mahdy, Q. Zhu, S. Qin, and A. A. Wani. 2007. Role of Claspin in regulation of nucleotide excision repair factor DDB2. DNA Repair (Amsterdam) 6578-587. [DOI] [PubMed] [Google Scholar]
- 51.Precious, B., K. Childs, V. Fitzpatrick-Swallow, S. Goodbourn, and R. E. Randall. 2005. Simian virus 5 V protein acts as an adaptor, linking DDB1 to STAT2, to facilitate the ubiquitination of STAT1. J. Virol. 7913434-13441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rademakers, S., M. Volker, D. Hoogstraten, A. L. Nigg, M. J. Mone, A. A. Van Zeeland, J. H. Hoeijmakers, A. B. Houtsmuller, and W. Vermeulen. 2003. Xeroderma pigmentosum group A protein loads as a separate factor onto DNA lesions. Mol. Cell. Biol. 235755-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rapic-Otrin, V., M. P. McLenigan, D. C. Bisi, M. Gonzalez, and A. S. Levine. 2002. Sequential binding of UV DNA damage binding factor and degradation of the p48 subunit as early events after UV irradiation. Nucleic Acids Res. 302588-2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Riou, L., E. Eveno, A. van Hoffen, A. A. van Zeeland, A. Sarasin, and L. H. Mullenders. 2004. Differential repair of the two major UV-induced photolesions in trichothiodystrophy fibroblasts. Cancer Res. 64889-894. [DOI] [PubMed] [Google Scholar]
- 55.Ruven, H. J., R. J. Berg, C. M. Seelen, J. A. Dekkers, P. H. Lohman, L. H. Mullenders, and A. A. van Zeeland. 1993. Ultraviolet-induced cyclobutane pyrimidine dimers are selectively removed from transcriptionally active genes in the epidermis of the hairless mouse. Cancer Res. 531642-1645. [PubMed] [Google Scholar]
- 56.Shaner, N. C., R. E. Campbell, P. A. Steinbach, B. N. Giepmans, A. E. Palmer, and R. Y. Tsien. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 221567-1572. [DOI] [PubMed] [Google Scholar]
- 57.Shiyanov, P., S. A. Hayes, M. Donepudi, A. F. Nichols, S. Linn, B. L. Slagle, and P. Raychaudhuri. 1999. The naturally occurring mutants of DDB are impaired in stimulating nuclear import of the p125 subunit and E2F1-activated transcription. Mol. Cell. Biol. 194935-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sugasawa, K., J. M. Ng, C. Masutani, S. Iwai, P. J. van der Spek, A. P. Eker, F. Hanaoka, D. Bootsma, and J. H. Hoeijmakers. 1998. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol. Cell 2223-232. [DOI] [PubMed] [Google Scholar]
- 59.Sugasawa, K., Y. Okuda, M. Saijo, R. Nishi, N. Matsuda, G. Chu, T. Mori, S. Iwai, K. Tanaka, K. Tanaka, and F. Hanaoka. 2005. UV-induced ubiquitylation of XPC protein mediated by UV-DDB-ubiquitin ligase complex. Cell 121387-400. [DOI] [PubMed] [Google Scholar]
- 60.Svejstrup, J. Q. 2002. Mechanisms of transcription-coupled DNA repair. Nat. Rev. Mol. Cell Biol. 321-29. [DOI] [PubMed] [Google Scholar]
- 61.Takata, K., H. Yoshida, M. Yamaguchi, and K. Sakaguchi. 2004. Drosophila damaged DNA-binding protein 1 is an essential factor for development. Genetics 168855-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan, T., and G. Chu. 2002. p53 binds and activates the xeroderma pigmentosum DDB2 gene in humans but not mice. Mol. Cell. Biol. 223247-3254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Venema, J., A. van Hoffen, V. Karcagi, A. T. Natarajan, A. A. van Zeeland, and L. H. Mullenders. 1991. Xeroderma pigmentosum complementation group C cells remove pyrimidine dimers selectively from the transcribed strand of active genes. Mol. Cell. Biol. 114128-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Volker, M., M. J. Mone, P. Karmakar, A. van Hoffen, W. Schul, W. Vermeulen, J. H. Hoeijmakers, R. van Driel, A. A. van Zeeland, and L. H. Mullenders. 2001. Sequential assembly of the nucleotide excision repair factors in vivo. Mol. Cell 8213-224. [DOI] [PubMed] [Google Scholar]
- 65.Wakasugi, M., A. Kawashima, H. Morioka, S. Linn, A. Sancar, T. Mori, O. Nikaido, and T. Matsunaga. 2002. DDB accumulates at DNA damage sites immediately after UV irradiation and directly stimulates nucleotide excision repair. J. Biol. Chem. 2771637-1640. [DOI] [PubMed] [Google Scholar]
- 66.Wang, H., L. Zhai, J. Xu, H. Y. Joo, S. Jackson, H. Erdjument-Bromage, P. Tempst, Y. Xiong, and Y. Zhang. 2006. Histone H3 and H4 ubiquitylation by the CUL4-DDB-ROC1 ubiquitin ligase facilitates cellular response to DNA damage. Mol. Cell 22383-394. [DOI] [PubMed] [Google Scholar]
- 67.Wang, Q. E., M. A. Wani, J. Chen, Q. Zhu, G. Wani, M. A. El-Mahdy, and A. A. Wani. 2005. Cellular ubiquitination and proteasomal functions positively modulate mammalian nucleotide excision repair. Mol. Carcinog. 4253-64. [DOI] [PubMed] [Google Scholar]
- 68.Wang, Q. E., Q. Zhu, G. Wani, J. Chen, and A. A. Wani. 2004. UV radiation-induced XPC translocation within chromatin is mediated by damaged-DNA binding protein, DDB2. Carcinogenesis 251033-1043. [DOI] [PubMed] [Google Scholar]
- 69.Wertz, I. E., K. M. O'Rourke, Z. Zhang, D. Dornan, D. Arnott, R. J. Deshaies, and V. M. Dixit. 2004. Human De-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 3031371-1374. [DOI] [PubMed] [Google Scholar]
- 70.Wittschieben, B. O., S. Iwai, and R. D. Wood. 2005. DDB1-DDB2 (xeroderma pigmentosum group E) protein complex recognizes a cyclobutane pyrimidine dimer, mismatches, apurinic/apyrimidinic sites, and compound lesions in DNA. J. Biol. Chem. 28039982-39989. [DOI] [PubMed] [Google Scholar]
- 71.Zotter, A., M. S. Luijsterburg, D. O. Warmerdam, S. Ibrahim, A. Nigg, W. A. van Cappellen, J. H. Hoeijmakers, R. van Driel, W. Vermeulen, and A. B. Houtsmuller. 2006. Recruitment of the nucleotide excision repair endonuclease XPG to sites of UV-induced DNA damage depends on functional TFIIH. Mol. Cell. Biol. 268868-8879. [DOI] [PMC free article] [PubMed] [Google Scholar]