Abstract
The proliferation and differentiation of neural precursor cells are mutually exclusive during brain development. Despite its importance for precursor cell self renewal, the molecular linkage between these two events has remained unclear. Fibroblast growth factor 2 (FGF2) promotes neural precursor cell proliferation and concurrently inhibits their differentiation, suggesting a cross talk between proliferation and differentiation signaling pathways downstream of the FGF receptor. We demonstrate that FGF2 signaling through phosphatidylinositol 3 kinase activation inactivates glycogen synthase kinase 3β (GSK3β) and leads to the accumulation of β-catenin in a manner different from that in the Wnt canonical pathway. The nuclear accumulated β-catenin leads to cell proliferation by activating LEF/TCF transcription factors and concurrently inhibits neuronal differentiation by potentiating the Notch1-RBP-Jκ signaling pathway. β-Catenin and the Notch1 intracellular domain form a molecular complex with the promoter region of the antineurogenic hes1 gene, allowing its expression. This signaling interplay is especially essential for neural stem cell maintenance, since the misexpression of dominant-active GSK3β completely inhibits the self renewal of neurosphere-forming stem cells and prompts their neuronal differentiation. Thus, the GSK3β/β-catenin signaling axis regulated by FGF and Wnt signals plays a pivotal role in the maintenance of neural stem/precursor cells by linking the cell proliferation to the inhibition of differentiation.
The proliferation of neural precursor cells and their neuronal differentiation are mutually exclusive; once the cells start to differentiate into neurons, they exit from the cell cycle. Conversely, the withdrawal of cells from the cell cycle is critical for their subsequent neuronal differentiation. However, the molecular basis for the transition from proliferation to differentiation, or for the mutual inhibition between proliferation- and differentiation-inducing machineries, largely has remained elusive. Environmental factors present in the neuroepithelium may play an important role in cell cycle progression. Among such factors, fibroblast growth factor 2 (FGF2) is well known to promote the proliferation of neural precursor cells and to maintain them in an undifferentiated state. For this reason, FGF2 has been used widely to generate neurospheres and expand neural precursor cells in culture. Nevertheless, the intracellular components of FGF2 signaling pathways in neural precursor cells that are involved in the link between the promotion of proliferation and the inhibition of differentiation have not been elucidated.
The Wnt family of proteins also is known to promote the proliferation of neural precursor cells. The canonical Wnt signaling pathway has been well studied in Drosophila and Xenopus embryos as well as in several mammalian cell lines (8). Wnt ligands initiate signaling by binding to their receptors, Frizzled and LRP5/6. The formation of this ternary complex reduces the activity of glycogen synthase kinase 3β (GSK3β), a serine/threonine kinase. In the absence of Wnt ligands, GSK3β phosphorylates β-catenin and directs it to the proteasome degradation pathway, whereas β-catenin is not phosphorylated and remains intact in the presence of Wnt ligands. Unphosphorylated β-catenin subsequently accumulates in the nucleus and trans-activates its target genes. Chenn and Walsh generated transgenic mice expressing stabilized β-catenin in which the NH2 terminus was truncated in neural precursors and reported an expansion of the precursor population (9). Recent studies have further demonstrated that the activation of the canonical Wnt pathway promotes the self renewal of stem cells prepared from various organs, including hematopoietic stem cells and embryonic stem (ES) cells (40, 42).
Although both FGF2 and canonical Wnts act as mitogens for neural precursor cells, the correlation between FGF2 and Wnt signaling pathways in neural precursor cells is not yet well understood. In the present study, we show that suboptimal doses of FGF2 and Wnt-3a displayed an additive effect on neural precursor cell proliferation, suggesting that FGF2 and Wnt-3a signaling share a common pathway for promoting neural precursor cell proliferation. The activation of FGF2 signaling through phosphatidylinositol 3 kinase (PI3K) activation induces the accumulation of β-catenin in the cell nucleus. Our data further reveal that the nuclear accumulated β-catenin prompts two independent transcriptional machineries that lead to the proliferation of neural precursor cells and the inhibition of their neuronal differentiation: (i) the FGF2-induced accumulation of β-catenin promotes neural precursor cell proliferation through the activation of LEF/TCF transcription factors in a manner different from that involved in the activation of the Wnt canonical pathway; and (ii) the nuclear accumulated β-catenin also induces antineurogenic hes1 gene expression through the enhancement of Notch1- and RBP-Jκ-mediated transcription. Noteworthy findings in this study are that β-catenin can associate with the Notch1 intracellular domain (N1IC), and it is present in a nuclear protein-DNA complex containing the hes1 gene promoter. The β-catenin-N1IC complex is efficiently formed when transcriptional coactivators p300 and P/CAF both are present. The activation of the canonical Notch signaling pathway is more significant in neural stem cells than intermediate neuronal progenitors. Mizutani et al. (33) have reported that the canonical Notch signaling through RBP-Jκ in neural stem cells induces the expressions of hes genes that maintain stem cell character and inhibit neurogenesis, but this pathway is attenuated in the intermediate progenitors. Therefore, the signaling interaction between Notch and β-catenin that we show here may be more significant in neural stem cells than in intermediate neuronal progenitors. Consistently, the misexpression of dominant-active GSK3β in our hands completely inhibits secondary neurosphere formation and promotes neuronal differentiation, suggesting that GSK3β inactivation and β-catenin stabilization are essential for the self renewal of neurosphere-forming stem cells. Thus, our results provide a new framework for understanding neural stem cell maintenance in view of the molecular link between proliferation and differentiation.
MATERIALS AND METHODS
Antibodies and recombinant proteins.
Mouse monoclonal anti-bromodeoxyuridine (BrdU) antibody was purchased from Sigma (St. Louis, MO) and used according to the manufacturer's instructions. Rabbit polyclonal anti-active caspase 3 and mouse monoclonal anti-Ki67 antibodies were from BD PharMingen (San Diego, CA). Mouse monoclonal anti-β-catenin and anti-GSK3β antibodies were from Transduction Laboratories (Lexington, KY). Rabbit polyclonal anti-phospho-GSK3β-Ser9 antibody was from Cell Signaling Technology (Beverly, MA). Mouse monoclonal anti-Notch1 antibody was from Chemicon (Temecula, CA). The rabbit polyclonal anti-hemagglutinin (HA) probe, goat polyclonal anti-Notch1, and rabbit polyclonal anti-β-catenin (H-102) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse monoclonal anti-myc (9E10) antibody was from Upstate (Charlottesville, VA). Rabbit anti-green fluorescent protein (GFP) antibody was from MBL (Nagoya, Japan). Immunocytochemistry and immunoblotting were performed as described previously (46). Recombinant mouse Wnt-3a (rmWnt-3a; catalog no. 1324-WN), recombinant mouse frizzled-8/Fc chimera (catalog no. 112-FZ), and recombinant human immunoglobulin G1 (IgG1) Fc (catalog no. 110-HG) were purchased from R&D Systems (Minneapolis, MN). LY294002 and wortmannin were purchased from Sigma. L685458 was purchased from Calbiochem (San Diego, CA).
BrdU incorporation assay.
Neural precursor cells were grown for 4 days, replated on an 8-well chamber slide (Nunc, Naperville, IL), and incubated in N2-supplemented Dulbecco's modified Eagle's medium (DMEM)-F-12 medium containing 10 ng/ml FGF2. On the next day, the medium was switched to fresh medium containing rmWnt-3a (0 or 200 ng/ml) and FGF2 (0, 2, or 10 ng/ml) for 12 h. BrdU was added to media for a further 6 h; cells were fixed with 4% paraformaldehyde (PFA) and immunostained with anti-BrdU antibody. To examine the contribution of the Akt/PI3K pathway, neural precursor cells on 8-well chamber slides were cultured in the absence of FGF2 for 10 h. The cells then were treated with FGF2 (0 or 10 ng/ml) and LY294002 (0 or 50 μM) together with 10 μM BrdU for 6 h. Cells were fixed with 4% PFA and immunostained with anti-BrdU antibody.
Retroviruses.
For recombinant retrovirus construction, GSK3βS9A cDNA (43) was inserted into pMY-IRES-GFP (36), and the plasmid was introduced into Plat-E cells (33a) using TransIT-293 (Mirus). After 48 h of incubation, culture medium containing retroviruses was centrifuged, and the virus pellets were resuspended in N2-supplemented DMEM-F-12 containing 10 ng/ml FGF2.
Reverse transcription-PCR (RT-PCR).
First-strand cDNA was synthesized from total RNA prepared from embryonic day 14.5 (E14.5) neural precursor cells using Superscript II (Invitrogen) as described previously (47). The PCR for detecting hes1 and hes5 consisted of 35 cycles of denaturation at 95°C for 1 min, annealing at 58°C for 45 s, and extension at 72°C for 1.5 min. Specific primers were the following: Hes1 sense primer, 5′-CAGCCAGTGTCAACACGACAC-3′; antisense primer, 5′-TCGTTCATGCACTCGCTGAG-3′; Hes5 sense primer, 5′-CGCATCAACAGCAGCATAGAG-3′; antisense primer, 5′-TGGAAGTGGTAAAGCAGCTTC-3′; and glyceraldehyde-3-phosphate dehydrogenase (G3PDH) sense primer, 5′-GTCATCATCTCCGCCCCTTCTGC-3′; antisense primer, 5′-GATGCCTGCTTCACCACCTTCTTG-3′. The PCR for detecting cyclin D1 consisted of 35 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 2 min, and extension at 72°C for 3 min. The cyclin D1-specific primers were the following: sense primer, 5′-CTGGCCATGAACTACCTGGA-3′; antisense primer, 5′-GTCACACTTGATCACTCTGG-3′. The PCR for detecting Notch1, Notch2, Notch3, and Delta1 consisted of 33 cycles of denaturation at 94°C for 1 min, annealing at 55°C for 1 min, and extension at 72°C for 2 min. The specific primers were the following: mouse Notch1 sense primer, 5′-TTACAGCCACCATCACAGCCACACC-3′; antisense primer, 5′-ATGCCCTCGGACCAATCAGA-3′; mouse Notch2 sense primer, 5′-GAGGCGCTCTTCTGCTGTTGAAGA-3′; antisense primer, 5′-ATAGAGTCACTGAGCTCTCGGACAG-3′; mouse Notch3 sense primer, 5′-ACACTGGGAGTTCTCTGT-3′; antisense primer, 5′-GTCTGCTGGCATGGGATA-3′; and mouse Delta1 sense primer, 5′-TGTGACGAGCACTACTACGGAGAAG-3′; antisense primer, 5′-AGTAGTTCAGGTCTTGGTTGCAGAA-3′. The PCR products were run on 1.2% agarose gels and visualized by ethidium bromide staining.
Immunoprecipitation.
HEK293 cells were transfected with combinations of pMY-β-catenin-HA, pEF-BOS-Myc-N1IC, pEF-BOS-HA-p300, pcDEF3-Flag-P/CAF, pcDEF3-Flag-p300, pcDEF3-Flag-p300ΔC(1-1736), and pcDEF3-Flag-p300ΔN(1736-2414) (34, 47) using TransIT-LT1 (Mirus). Forty-eight hours after transfection, cells were washed twice with chilled phosphate-buffered saline (PBS) and lysed on ice with 0.5 ml of NP-40 lysis buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5% NP-40, 5 mM EDTA, 0.4 mM sodium orthovanadate, 1 mM phenylmethylsulfonyl fluoride, and 10 μg/ml aprotinin). Whole-cell lysates were sonicated in a closed-type sonicator (Cosmo Bio) and centrifuged at 15,000 rpm for 15 min at 4°C. Primary antibody was incubated with the supernatant overnight at 4°C with gentle agitation. Ten microliters of protein A-Sepharose beads were added to each lysate/antibody mixture and incubated for 1 h at 4°C with gentle agitation. The immune complex was precipitated by a brief centrifugation at 5,000 rpm at 4°C. The complex was washed five times with NP-40 lysis buffer, and the proteins were eluted by boiling the beads with 20 μl sodium dodecyl sulfate sample buffer for 5 min. To detect the interaction between endogenous β-catenin and Notch1 proteins, neural precursor cells expanded for 4 days were plated on 60-mm dishes and cultured for another 2 days in the presence of FGF2. Anti-Notch1 antibody was used for coimmunoprecipitation, and anti-β-catenin antibody was used for Western blot analysis. For the blocking experiment, anti-Notch1 antibody was premixed with a fivefold (by weight) excess of its blocking peptide in PBS and incubated overnight at 4°C.
RNA interference.
A synthetic double-stranded short interfering RNA (siRNA) for β-catenin (5′-AUUACAAUCCGGUUGUGAACGUCCC-3′) was purchased from Invitrogen (catalog no. 1320003). The expression level of β-catenin protein in NIH 3T3 cells that were cotransfected with the siRNA and pMY-HA-β-catenin-IRES-GFP was examined by Western blot analysis with anti-HA antibody. For the luciferase reporter assay, the siRNA was introduced into neural precursor cells using Lipofectamine 2000 (Invitrogen, CA) together with the reporter plasmid (pHes1-Luc), an internal control plasmid (pRL-tk), and the expression vector (pEF-BOS-Myc-N1IC). The dual luciferase reporter assay was performed as described previously (34).
ChIP assay.
A chromatin immunoprecipitation (ChIP) assay was performed according to the protocol reported by Schwartz et al. (42a). Neural precursor cells were transfected with stabilized β-catenin cDNA using the Nucleofector system (Amaxa). The cells were seeded at a high density (3.2 × 105 cells/cm2) or a low density (5.4 × 104 cells/cm2) to control the involvement of Notch signaling through cell-cell contacts. Cells prepared from each culture were equalized in number at the onset of the ChIP assay. The PCR template was amplified with the following primers: 5′ primer, 5′-TGTCTCTTCCTCCCATTGG-3′; 3′ primer, 5′-AACTACTGAGCAGTTGAAGG-3′.
Cell preparation.
Monolayer cultures of neural precursor cells were prepared from E14.5 mouse telencephalon as described previously (34). In the neurosphere assay, monolayers of cultured neural precursor cells were infected with GSK3βS9A (43) or control GFP retroviruses on day 3 of the culture. After 24 h of incubation with the retroviruses, cells were detached and cultured at a density of 1 × 105 cells per poly-HEME-coated 90-mm dish with N2-supplemented DMEM-F-12 medium (8 ml) containing 10 ng/ml FGF2 for 8 days. The size of 8-day-old GFP-positive neurospheres was measured. For the secondary neurosphere assay, 8-day-old primary spheres were dissociated with trypsin and replated at a density of 0.5 × 105 cells on poly-HEME-coated 90-mm dishes in medium (8 ml) containing both FGF2 and epidermal growth factor (10 ng/ml each; Peprotech, Rocky Hill, NJ). The cells were cultured for another 8 days.
Luciferase reporter assay.
Neural precursor cells were transfected with the reporter plasmids pHes1-Luc (a gift from Ryuichiro Kageyama, Kyoto University), H5-Luc, RMH5-Luc (47), 7×TCF/siman virus 40 (SV40)-Luc and SV40-Luc (pGL3-Promoter Vector purchased from Promega), and an internal control plasmid, pRL-tk (Promega), together with the expression plasmids pEF-BOS-Myc-N1IC (47) and pMY-gsk3βS9A, by using TransIT-LT1 (Mirus). On the following day, cells were cultured in medium without FGF2 and insulin for 6 h and retreated with 20 ng/ml FGF2 for 7 h. Luciferase activity was measured using the Pikkagene dual luciferase assay system (Tokyo Ink Inc.). For the reporter assay with stabilized β-catenin (a gift from A. Nagafuchi at IMEG, Kumamoto University), cells were transfected with pHes1-Luc together with the expression plasmids pMY-β-catenin-HA, pEF-BOS-Myc-N1IC, pcDNA3-Myc-P/CAF (47), and pcDEF3-Flag-p300 (34).
RESULTS
FGF2-induced β-catenin accumulation and cyclin D1 gene expression through PI3K activation.
FGF2 and Wnts act as mitogens for neural precursor cells. Suboptimal doses of FGF2 and Wnt-3a displayed an additive effect on neural precursor cell proliferation (data not shown), suggesting that FGF2 and Wnt-3a signaling share a common pathway for promoting neural precursor cell proliferation. Previous reports have shown the FGF2-triggered activation of the PI3K-Akt pathway in breast cancer cells (3, 49). Akt phosphorylates GSK3β at Ser9 in insulin-stimulated L6 myotubes (11), and this Ser9 phosphorylation reduces the kinase activity of GSK3β, a critical component of the canonical Wnt signaling pathway (45). Thus, we investigated whether FGF2 stimulation increased the Ser9 phosphorylation of GSK3β through PI3K activation in neural precursor cells. Insulin that had been used for neural precursor cell preparation could affect the PI3K-Akt pathway. Therefore, both FGF2 and insulin were depleted from the culture medium for 6 h, and cells were retreated with 20 ng/ml FGF2 in the absence of insulin for 3 h. The phosphorylation of GSK3β-Ser9 was examined by Western blot analysis using an anti-phospho-GSK3β-Ser9-specific antibody. FGF2 indeed induced GSK3β-Ser9 phosphorylation, as did Wnt-3a (Fig. 1A). Simultaneous stimulation by FGF2 and Wnt-3a resulted in slightly greater GSK3β-Ser9 phosphorylation. LY294002 and wortmannin are well-known compounds that inhibit PI3K (4, 50). LY294002 and wortmannin reduced the effect of FGF2 signaling on GSK3β-Ser9 phosphorylation (Fig. 1B; wortmannin data not shown), suggesting that GSK3β-Ser9 was phosphorylated at least partly through PI3K activation. In addition, wortmannin treatment slightly reduces the GSK3β protein level with an unknown mechanism, since wortmannin inhibits PI3K activity but may affect some other signaling as well. The Wnt-induced phosphorylation of GSK3β was only slightly reduced by LY294002 (Fig. 1B). Although insulin has been reported to activate the PI3K-Akt pathway, it should be noted that FGF2 treatment induced GSK3β-Ser9 phosphorylation in neural precursor cells regardless of the presence or absence of insulin in the culture medium (data not shown).
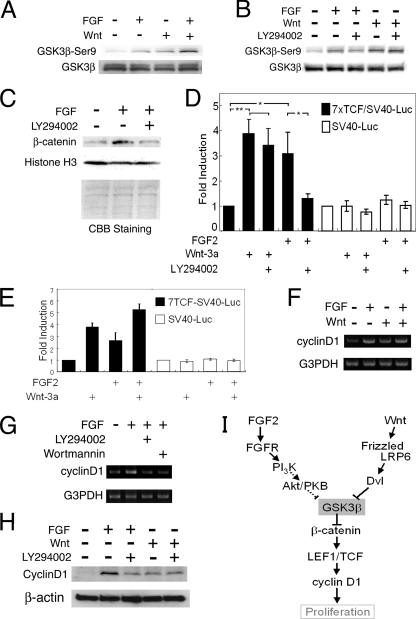
FIG. 1.
Nuclear β-catenin accumulation and induction of cyclin D1 expression through FGF2-mediated PI3K activation and GSK3β inactivation in neural precursor cells. (A) The upper panel shows the Western blot analysis with anti-phospho-GSK3β-Ser9 antibody. The lower panel shows the total amount of GSK3β examined with anti-GSK3β antibody. The upper of the two bands appeared to be phosphorylated GSK3β. Neither FGF2 nor Wnt3a affected the β-actin protein level (data not shown). (B) The PI3K inhibitor LY294002 (50 μM) impaired the GSK3β Ser9 phosphorylation induced by FGF2 treatment (20 ng/ml; n = 6). LY294002 slightly reduced GSK3β Ser9 phosphorylation induced by Wnt3a treatment (150 ng/ml; n = 3). (C) Accumulation of β-catenin was detected in the nuclear fraction of neural precursor cells treated with 20 ng/ml FGF2 for 1 h. This accumulation was blocked by 50 μM LY294002. The amounts and purities of the nuclear fraction were confirmed by Coomassie brilliant blue staining, the expression level of histone H3, and leukemia inhibitory factor receptor (LIFR; data not shown). (D) 7×TCF-SV40 promoter activity (black bars) and SV40 promoter activity (white bars) were investigated by a dual luciferase reporter assay. The height of each bar indicates the increase of luciferase activity compared to that of unstimulated cells. rmWnt-3a (100 ng/ml) was applied as a positive control for 7×TCF-Luc reporter activity (n = 3; *, P < 0.05; **, P < 0.001; Student's t test). (E) Coapplication of 20 ng/ml FGF2 and 100 ng/ml rmWnt-3a displayed an additive effect on 7×TCF promoter activation (n = 3). The height of each bar indicates the increase of luciferase activity compared to that of unstimulated cells. (F) FGF2 (20 ng/ml) and rmWnt-3a (200 ng/ml) increased the expression of cyclin D1 mRNA, as detected by RT-PCR. (G) LY294002 (50 μM) and wortmannin (100 nM) impaired FGF2-induced cyclin D1 mRNA expression (n = 3). (H) Western blot analysis with anti-cyclin D1 antibody. LY294002 (50 μM) abolished the induction of cyclin D1 by FGF2 (20 ng/ml; n = 3) but did not affect that by rmWnt-3a (150 ng/ml; n = 2). All experiments in this figure were done with cells seeded at a high density (3.2 × 105 cells/cm2). (I) A schematic drawing summarizing the signaling pathways analyzed in this figure.
In the canonical Wnt signaling pathway, the inactivation of GSK3β leads to the nuclear accumulation of β-catenin, which promotes LEF/TCF-dependent transcription and functions as a transcriptional coactivator (8). Similarly to the Wnt signaling pathway, neural precursor cells treated with FGF2 for 1 h showed an increase of β-catenin protein in the 0.5% NP-40-insoluble nuclear fraction, but LY294002 impaired the effect of FGF treatment (Fig. 1C). FGF2 likely induced β-catenin stabilization rather than its translocation into the nucleus, since the protein level of β-catenin was increased in both the nucleus and cytoplasm (data not shown). To explore whether β-catenin accumulation by FGF2 increases LEF/TCF-mediated transcription, we prepared, according to Ueda et al. (48), a luciferase reporter construct containing seven repeats of a TCF binding motif located upstream of an SV40 promoter of a pGL3 promoter vector (Promega). FGF2 treatment increased reporter activity, but this increase was blocked by LY294002 treatment (Fig. 1D), suggesting that FGF2 activates LEF/TCF-mediated transcription through PI3K signaling. In contrast, LY294002 did not affect Wnt3a-induced LEF/TCF transcription (Fig. 1D). The activation of the FGF2 and Wnt3a signaling pathways showed additive effects on the induction of TCF-dependent transcription, suggesting that they share a common pathway after the inactivation of GSK3β (Fig. 1E).
The cyclin D1 gene has LEF/TCF binding sites in its promoter region, and thus it is a representative Wnt target. Cyclin D1 plays a critical role at the G1/S transition in the cell cycle. Cyclin D1 mRNA and protein levels both were increased in cells treated with FGF2 for 3 h (Fig. 1F, G, and H), but LY294002 and wortmannin abolished FGF2-induced cyclin D1 expression (Fig. 1G and H). Thus, the induction of cyclin D1 expression by FGF2 depended upon PI3K activation. There still remained the possibility that FGF2-induced cyclin D1 gene expression was due to the autocrine secretion of Wnt ligands. However, this possibility was excluded by the fact that recombinant mouse Frizzled-8/Fc chimera protein, a soluble Wnt antagonist, showed no effect on FGF2-induced cyclin D1 expression, whereas it blocked rmWnt-3a-induced cyclin D1 expression (data not shown). These results suggest that FGF2 signaling directly induces cyclin D1 expression regardless of Wnt signal initiation. It should be noted that FGF2 treatment did not alter the STAT3 phosphorylation status that affected cell proliferation (data not shown). The schematic drawing in Fig. 1I summarizes the signaling pathway shown in Fig. 1.
Cell cycle progression by FGF2 signaling through PI3K activation and GSK3β inactivation.
To investigate the functional consequences of the FGF2-induced activation of the PI3K-Akt-GSK3β pathway in the proliferation of neural precursor cells, we tested a dominant-negative Akt (dnAkt) to see if it blocked the FGF2-induced cell proliferation. Many cells expressing dnAkt underwent apoptosis within a day after gene transfer (data not shown). As Akt was reported to be involved in the survival of various cells, the long-term blocking of Akt activity by dnAkt overexpression might cause neural precursor cell death. Thus, we next treated the cells with a pharmacological PI3K inhibitor, LY294002, for a short time period. After 10 h of FGF2 depletion, cells were treated with 10 μM BrdU, 10 ng/ml FGF2, and 0, 10, or 50 μM of LY294002 for 6 h. The number of BrdU+ cells was increased by FGF2 retreatment, but this increase was abolished by LY294002 (Fig. 2A, B, and C), suggesting a contribution of PI3K to FGF2-induced cell proliferation. Active caspase 3-positive apoptotic cells were not increased in number by LY294002 under this culture condition (Fig. 2D). To further test the involvement of GSK3β Ser9 phosphorylation in neural precursor cell proliferation, an Akt-insensitive GSK3β, in which serine9 was replaced with alanine (GSK3βS9A), was used as a dominant-active protein (43). GSK3βS9A cDNA was inserted into the retroviral pMY-IRES-GFP vector (36). GSK3βS9A or control retrovirus was introduced into neural precursor cells cultured in the presence of FGF2. The size of the population of Ki67+ proliferating cells became smaller when GSK3βS9A was expressed (Fig. 2E, F, and G). These results indicate that the inactivation of GSK3β plays a significant role in neural precursor cell proliferation induced by FGF2.
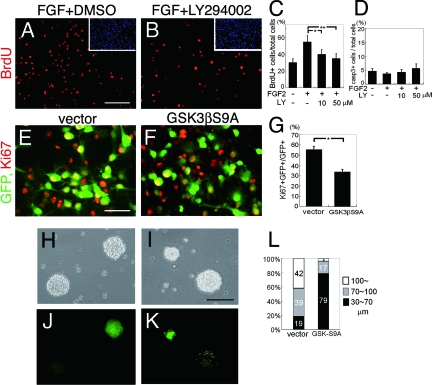
FIG. 2.
Involvement of PI3K activation and GSK3β Ser9 phosphorylation in the proliferation of neural precursor cells in response to FGF2. (A to D) FGF2 (10 ng/ml) increased the number of BrdU+ cells (red in panel A), and LY294002 abolished this effect in a dose-dependent manner (B). DMSO, dimethylsulfoxide. (C) Ratio of BrdU+ cells/total cells (n = 4; *, P < 0.01; **, P < 0.001; Student's t test). (D) Ratio of active caspase 3+ (casp3+) cells/total cells (n = 4). (E to G) Ki67+ proliferative cells (red in panels E and F) were less frequent among GSK3βS9A-expressing cells (green in panel F) than among control GFP virus-infected cells (green in panel E). (G) The ratio of Ki67+ cells/GFP+ cells (n = 5; *, P < 0.01 compared to values for the control; Student's t test). All of these experiments were done with cells seeded at a high density (3.2 × 105 cells/cm2). (H to L) Misexpression of GSK3βS9A significantly decreased the diameter of primary neurospheres (green sphere in panel K) compared to that of the control (green sphere in panel J; also see the corresponding uncolored spheres in panels H and I). Yellow dots seen in panel K are artifacts caused by the reflection of the lights from clean benches. (L) Summary of results (n = 4). Scale bar for panels A and B, 100 μm. Scale bar for panels E and F, 50 μm. Scale bar for panels H to K, 100 μm.
Neural stem cells in vitro can generate neurospheres. To assess the involvement of GSK3β inactivation in neurosphere formation, GSK3βS9A or control virus-infected neural precursor cells were replated in floating conditions on nonadhesive dishes at a clonal density and cultured for 8 days in the presence of FGF2 to form neurospheres. GSK3βS9A-expressing neurospheres were much smaller in size than control spheres expressing GFP alone (Fig. 2H, I, J, K, and L), suggesting that GSK3βS9A inhibited the proliferation of sphere-forming cells. We further examined the ability of GSK3βS9A-expressing cells to form secondary neurospheres, an ability that is a characteristic feature of self-renewing neural precursor cells. Primary sphere-forming cells were dissociated and cultured at a clonal density on nonadhesive dishes for a further 8 days. GSK3βS9A-expressing cells generated only negligible numbers of secondary spheres compared to the level generated by control GFP-expressing cells and uninfected cells (Fig. 3A). Instead, the misexpression of GSK3βS9A promoted neuronal differentiation within the primary neurosphere even in the presence of FGF2 (Fig. 3B). Thus, the self-renewal ability of neural precursor cells as indicated by secondary neurosphere formation, which is maintained by FGF2, involves the inactivation of GSK3β.
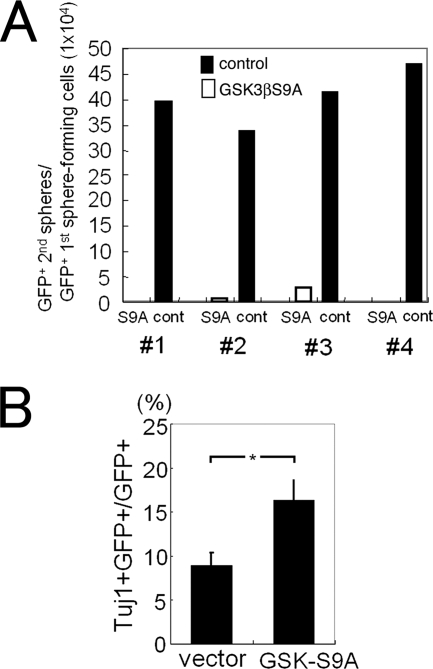
FIG. 3.
Defect in secondary sphere formation by dominant-active GSK3β-expressing cells. (A) GSK3βS9A-GFP- or GFP-retrovirus-infected cells in primary neurospheres were dissociated and used for a secondary sphere assay. GSK3βS9A-expressing cells (S9A) formed no or very few secondary spheres compared to control GFP-expressing cells (cont). The numbers at the bottom indicate each of the four independent experiments. (B) Neural precursor cells were infected with GSK3βS9A-GFP or GFP virus, and primary neurospheres formed after 8 days were dissociated and replated on 8-well chamber slides. Six hours later, the number of Tuj1+ cells among GSK3βS9A-GFP-expressing cells and that among GFP-expressing cells were counted (n = 3; *, P < 0.05 compared to control values; Student's t test).
Induction of hes1 gene expression by FGF2.
Since neural precursor cell proliferation and neuronal differentiation are observed in a mutually exclusive manner from worms to mammals, these two events may have a molecular link involving an evolutionarily conserved classical factor(s). Meanwhile, Temple and colleagues reported that neural stem cells cultured at clonal densities exhibit trends toward cell cycle exit and differentiation into neurons and glial cells, even in the presence of FGF2 (38), suggesting that the FGF2-induced self renewal of neural stem cells requires cell-cell contact-mediated signal transduction. Notch signaling plays a central role in inhibiting the neuronal differentiation of neural precursor cells, and mutations in key components of the Notch signaling pathway cause premature neuronal differentiation. For these reasons, we hypothesized that FGF2 signaling contributes to the maintenance of neural precursor cells, in part via an interaction with the Notch signaling pathway (Fig. 4A).
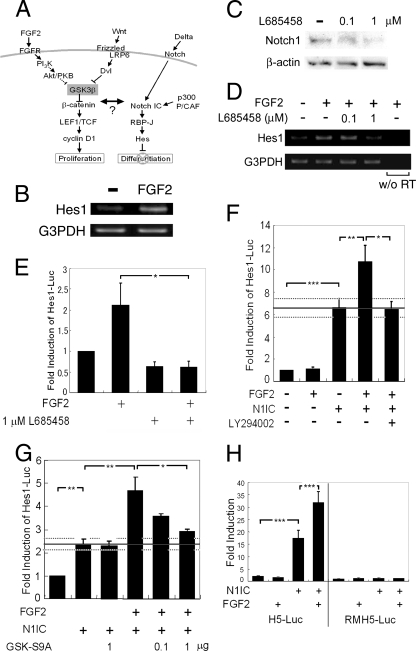
FIG. 4.
Potentiation of Notch-mediated hes promoter activation by FGF2 signaling via PI3K activation and GSK3β inactivation. (A) Our working hypothesis. FGF signaling and Notch signaling cooperate to regulate cell fate decisions. (B) FGF2 treatment (20 ng/ml) of neural precursor cells for 3 h increased the level of Hes1 mRNA, as detected by RT-PCR. PCR products were not amplified from samples without reverse transcriptase treatment (data not shown). (C) γ-Secretase inhibitor L685458 decreased the amount of N1IC in a high density of neural precursor cells cultured for 3 days. (D) L685458 abolished FGF2-induced hes1 gene expression in a high-cell-density culture of neural precursor cells examined by RT-PCR. The hes1 mRNA level was increased by a 3-h FGF2 treatment; however, the FGF2 effect was abolished by the addition of 1 μM L685458. No band was detected without reverse transcriptase (w/o RT). (E) L685458 (1 μM) abolished FGF2-induced hes1 gene promoter activation. Neural precursor cells cultured at a high cell density were transfected with a plasmid containing pHes1-Luc. hes1 promoter activity was investigated by a dual luciferase reporter assay. Each bar indicates the increase of luciferase activity compared to that of untreated cells (n = 3; *, P < 0.001; Student's t test). (F) Neural precursor cells cultured at a low cell density were cotransfected with plasmid containing pHes1-Luc with or without the cDNA encoding N1IC. FGF2 (20 ng/ml) potentiated hes1 promoter activity only when the cells expressed N1IC. LY294002 treatment (50 μM) abolished FGF2-induced potentiation of hes1 promoter activity (n = 5; *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student's t test). (G) Misexpression of GSK3βS9A blocked the ability of FGF2 to potentiate N1IC-induced hes1 promoter activation (n = 4; *, P < 0.05; **, P < 0.01; Student's t test). (H) Neural precursor cells were transfected with N1IC cDNA together with H5-Luc or RMH5-Luc with mutations in the RBP-Jκ binding site. (n = 3; ***, P < 0.001; Student's t test).
The expression of Notch1, Notch2, Notch3, and Delta1 mRNAs was detected by RT-PCR in the E14.5 mouse telencephalon and a neural precursor cell-enriched culture (data not shown). Hes1, a representative critical effector molecule of Notch signaling, is a basic helix-loop-helix (bHLH) transcription factor that inhibits neurogenic bHLH factors. According to the hypothesis described above, we focused on hes1 gene expression to assess whether FGF2 stimulation influences Notch signaling. As shown in Fig. 4B, the hes1 mRNA level was elevated by FGF2 treatment for 3 h when cells were seeded at a high density (3.2 × 105 cells/cm2). The γ-secretase-mediated cleavage and nuclear translocation of the Notch intracellular domain are critical for Notch target/effector gene expression. L685458, a commercially available γ-secretase inhibitor, significantly decreased the amount of cleaved Notch1 in a high-density culture of neural precursor cells (Fig. 4C). L685458 at a dose of 1 μM abolished FGF2-induced hes1 gene expression in a high-density culture of neural precursor cells (Fig. 4D). We further analyzed the effect of FGF2 signaling on hes1 promoter (−2500 to +46) activation in neural precursor cells using a luciferase reporter assay. FGF2 caused a significant induction of hes1 promoter activity in neural precursor cells cultured at a high density (Fig. 4E, left two bars), and this was completely inhibited by L685458 (Fig. 4E, fourth bar) or was never observed when cells were cultured at a low density (5.4 × 104 cells/cm2) (Fig. 4F, left two bars). These results suggested that FGF2-induced hes1 expression requires Notch signal activation through cell-cell contacts. This idea was supported by the result that, when an expression vector for N1IC (a constitutively active form of Notch1) was introduced into low-density neural precursor cell cultures, the hes1 promoter was activated, and this activation was enhanced by FGF2 stimulation (Fig. 4F). Thus, FGF2, although having no effect by itself, evidently enhanced N1IC-induced hes1 promoter activation.
Since dominant-active GSK3β inhibited the self renewal of neurosphere-forming cells in the presence of FGF2 (Fig. 3), we further hypothesized that GSK3β activity should influence the FGF2-mediated potentiation of hes1 promoter activity. As expected, the PI3K inhibitor LY294002 abolished the potentiation of hes1 promoter activity by FGF2 (Fig. 4F, right three bars), as did another PI3K inhibitor, wortmannin (data not shown), suggesting that PI3K activity plays a major role in this process. The potentiation of hes1 promoter activity by FGF2 also was prevented by the misexpression of a dominant-active form of GSK3β, namely, GSK3βS9A, in a dose-dependent manner (Fig. 4G). Importantly, GSK3βS9A showed no inhibitory effect on hes1 promoter activation induced by N1IC alone (Fig. 4G). These results suggested that the inactivation of GSK3β is essential for the FGF2-mediated potentiation of Notch-induced hes1 promoter activation. To further confirm the involvement of GSK3β in hes1 promoter activation, we examined whether Wnt-3a stimulation affects hes1 promoter activation. Wnt-3a treatment elevated hes1 promoter activity only when N1IC is expressed, although this elevation was less pronounced than that observed with FGF2 (data not shown). These findings provided us with hints into molecular mechanisms underlying the observation shown in Fig. 3 that dominant-active GSK3β-expressing cells lose their self-renewal activity and generate very few secondary neurospheres.
β-Catenin-mediated potentiation of N1IC-induced promoter activation is RBP-Jκ dependent.
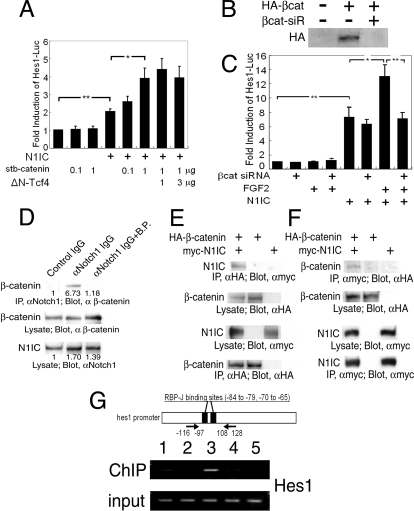
Similarly to the hes1 gene promoter, the hes5 gene promoter (−179 to +72) was potentiated by FGF2 in neural precursor cells only when Notch signaling was activated (Fig. 4H, left four bars). The RBP-Jκ binding sites in the hes5 promoter have been well characterized. We previously demonstrated that a hes5 reporter construct with a mutated RBP-Jκ binding site (RMH5-Luc, in which the nucleotides at −79 to −72, TGTGGAA, were replaced by TGTGCTGA) has no responsiveness to N1IC in cultured neural precursor cells (47). As shown in Fig. 4H (right four bars), neither N1IC alone nor N1IC plus FGF2 led to any activation of this mutant hes5 promoter. These results suggested that the binding of RBP-Jκ protein to the hes5 promoter is essential for the potentiation of its gene expression by FGF2 in the presence of Notch signaling. The involvement of RBP-Jκ binding to promoter DNA in the FGF2-mediated potentiation of N1IC-induced hes1 gene expression was indicated in further experiments (Fig. 5), as described later.
FIG. 5.
Novel function of β-catenin as a transcriptional coactivator for Notch signaling. (A) Expression of stabilized (stb) β-catenin potentiated N1IC-induced hes1 promoter activation in neural precursor cells in a dose-dependent manner. The misexpression of dominant-negative human TCF4 (ΔN-hTCF4) did not affect hes1 promoter activity (n = 4; *, P < 0.01; **, P < 0.001; Student's t test). (B) The ectopic expression of HA-tagged β-catenin (βcat) was potently inhibited by the cotransfection of NIH 3T3 cells with β-catenin siRNA (siR). (C) β-Catenin siRNA abolished the effect of FGF2 on hes1 promoter activation, whereas it did not affect N1IC-mediated hes1 promoter activation (n = 4; *, P < 0.01; **, P < 0.001; Student's t test). (D) Endogenous β-catenin and Notch1 proteins in neural precursor cells were coimmunoprecipitated. Preincubation of anti-Notch1 antibody (αNotch1) with its blocking peptide (B.P.) prevented the coprecipitation of β-catenin and N1IC. The β-catenin signal density was quantified with densitometry, and the result is indicated under each band. IP, immunoprecipitation. (E and F) The molecular interaction between N1IC and β-catenin was analyzed by coimmunoprecipitation assays in HEK293 cells that overexpressed the cDNAs for HA-tagged β-catenin and myc-tagged N1IC. Anti-HA (αHA) antibody was used for immunoprecipitation, and anti-myc antibody was used for the Western blotting analysis shown in panel E. Anti-myc antibody (αmyc) was used for immunoprecipitation, and anti-HA antibody was used for the Western blotting analysis shown in panel F. (G) ChIP assay demonstrated that β-catenin is associated with the hes1 promoter region in neural precursor cells. Chromosomal DNA was precipitated with normal rabbit IgG from Upstate (lane 1), normal rabbit IgG from Santa Cruz Biotechnology (lane 2), or anti-β-catenin antibody (α β-catenin) (lanes 3, 4, and 5). Chromosomal DNA was prepared from high-density neural precursor cells (lanes 1, 2, 3, and 4), from the cells treated with 1 μM L685458 (lane 4), or from low-density cultures of neural precursor cells (lane 5). Arrows in the drawing indicate PCR primer positions.
GSK3β was reported to phosphorylate the Notch2 serine/threonine-rich domain and to be involved in the Notch signaling pathway in HEK293 cells (14). Therefore, we hypothesized that GSK3β also could phosphorylate N1IC and downregulate Notch signaling in neural precursor cells. However, the structural equivalent of the Notch2 serine/threonine-rich domain was not found in Notch1. FGF2 treatment did not change the phosphorylation status of N1IC, as examined by a band shift phosphorylation assay (data not shown). Calf intestine alkaline phosphatase treatment did not modify the electrophoretic mobility of N1IC that was extracted from either FGF2-treated or nontreated neural precursors (data not shown). These data do not support the hypothesis that GSK3β phosphorylates N1IC and regulates the Notch signaling activity.
As previously mentioned, FGF2 treatment led to the nuclear accumulation of β-catenin in neural precursor cells (Fig. 1). As shown in Fig. 5A, the forced expression of stabilized β-catenin, in which all four GSK3β target residues were mutated, mimicked the effect of FGF2 on hes1 promoter activation in the sense that neither FGF2 nor stabilized β-catenin activated it by themselves, but they did potentiate N1IC-induced hes1 promoter activation. Even though no putative binding site for LEF/TCF was found in the hes1 promoter region, it is possible that the LEF/TCF-β-catenin transcription factor complex binds to and transactivates the hes1 promoter in cooperation with N1IC, which binds to the promoter via RBP-Jκ. To test this possibility, we used dominant-negative human TCF4, which lacks the β-catenin binding domain in its amino-terminal region (ΔN-TCF4) (41). In the control experiment, ΔN-TCF4 totally prevented the 7×TCF-Luc reporter transactivation that was induced by stabilized β-catenin (data not shown). ΔN-TCF4 showed no detectable effect on the hes1 promoter activation that was induced by N1IC and stabilized β-catenin (Fig. 5A, right three bars). These data suggest that hes1 promoter activation mediated by N1IC together with stabilized β-catenin does not involve LEF/TCF transcription factors.
To confirm whether endogenous β-catenin is involved in FGF2-induced hes1 promoter activation, we performed a β-catenin knockdown analysis using RNA interference (siRNA). The expression of HA-tagged β-catenin protein in NIH 3T3 cells by a plasmid vector was potently inhibited by the cotransfection of cells with β-catenin siRNA (Fig. 5B). Neural precursor cells were transfected with combinations of the β-catenin siRNA, Hes1-Luc reporter, and N1IC expression vector. β-Catenin siRNA abolished FGF2-induced hes1 promoter activation, whereas it did not affect N1IC-mediated hes1 promoter activation (Fig. 5C), demonstrating that endogenous β-catenin is required for the effect of FGF2 on hes1 promoter activation. We looked for a DNA binding protein that could anchor FGF2-stabilized β-catenin on the hes1 gene promoter. As described above, RBP-Jκ is suggested to be important for the effect of FGF2 on hes1 promoter activation. In Drosophila embryo lysates, Armadillo, a Drosophila homolog of β-catenin, was detected in a protein complex containing Notch (21), raising the possibility of an interaction between β-catenin and the N1IC-RBP-Jκ complex in the mouse neural precursor cells used in our experiments. In fact, a coimmunoprecipitation assay detected an interaction between endogenous β-catenin and N1IC in neural precursor cells (Fig. 5D). The preabsorption of anti-Notch1 antibody with its blocking peptide inhibited the coprecipitation of β-catenin with N1IC (Fig. 5D). An interaction between β-catenin and N1IC was further confirmed by coimmunoprecipitation experiments with epitope-tagged β-catenin and N1IC expressed in HEK293 cells (Fig. 5E and F), while β-catenin and RBP-Jκ by themselves were not coimmunoprecipitated (data not shown). We next examined whether β-catenin could associate with the hes1 gene promoter at RBP-Jκ binding sites. Neural precursor cells were transfected with stabilized β-catenin cDNA. A ChIP assay suggested that the stabilized β-catenin associates with the 5′-flanking region of the hes1 gene containing two putative RBP-Jκ binding sites (Fig. 5G). The association of β-catenin with the hes1 gene promoter was reduced when cells were cultured at a low density or cultured in the presence of L685458 (Fig. 5G). These results suggest that the interaction between β-catenin and the hes1 gene promoter requires Notch signal activation.
Physical and functional association of transcriptional coactivators with the β-catenin-N1IC complex.
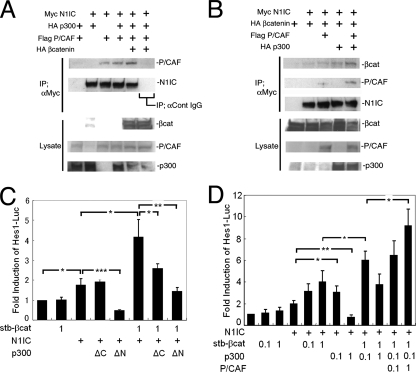
Since the misexpression of stabilized β-catenin enhanced the N1IC-stimulated hes1 promoter activity, there might be an interaction between the β-catenin-N1IC complex and transcriptional coactivator(s). The CBP/p300 family is a widely known family of transcriptional coactivator proteins. These proteins are expressed and also function in neural precursor cells (34). Previous studies with several mammalian cell lines demonstrated that β-catenin could bind to CBP/p300 (37), and N1IC could interact with CBP/p300 and p300/CBP-associated factor (P/CAF) (28, 44). Consistently with these early studies, our in vitro analysis with tagged proteins expressed in HEK293 cells confirmed that N1IC could interact with P/CAF (Fig. 6A, lane 3). Interestingly, the interaction of N1IC and P/CAF was enhanced by the coexpression of p300 and, to a greater extent, by the additional the coexpression of β-catenin (Fig. 6A, lanes 4 and 5). Furthermore, β-catenin and N1IC were more efficiently coimmunoprecipitated in the presence of both p300 and P/CAF (Fig. 6B), suggesting the cooperative binding of p300, N1IC, β-catenin, and P/CAF. To further verify the involvement of endogenous p300 in hes1 promoter activation in neural precursor cells, p300 deletion mutants (34) were employed. The p300ΔN (comprised of amino acids 1736 to 2414) mutant retained the N1IC binding domain but lacked the β-catenin-interacting, CREB binding, and HAT domains. Therefore, the misexpression of p300ΔN was expected to antagonize endogenous p300 binding to N1IC and act as a dominant-negative form of p300 (44). Indeed, it decreased N1IC-induced hes1 promoter activation (Fig. 6C, lanes 5 and 8). The p300ΔC (comprised of amino acids 1 to 1736) mutant, which lacked the N1IC binding domain, bound to and eliminated β-catenin from the N1IC/RBP-Jκ complex (37). Indeed, the misexpression of p300ΔC had no effect on N1IC-induced hes1 promoter activation, whereas it suppressed the hes1 promoter activity induced by N1IC plus stabilized β-catenin (Fig. 6C, lane 4 and 7). Supporting this result, p300ΔC reduced, to some extent, the interaction between β-catenin and N1IC in the coimmunoprecipitation assay (data not shown). These results suggest that endogenous p300 is involved in the transcriptional activation of the hes1 promoter mediated by N1IC and β-catenin in neural precursor cells. This idea was further supported by a gain-of-function experiment in which the overexpression of full-length p300 cDNA at a moderate dose (0.1 μg/well) in neural precursor cells activated the hes1 promoter in the presence of β-catenin and N1IC (Fig. 6D). A high dose of p300 cDNA of, for example, 1 μg/well reduced the hes1 promoter activity. Such a nonlinear bell-shaped dose-response function via the quenching of positive factor activity is not unusual for transcriptional modulators in many biological processes. The overexpression of P/CAF in neural precursor cells led to a dose-dependent increase in hes1 promoter activity when N1IC, β-catenin, and p300 were coexpressed (Fig. 6D), suggesting that P/CAF is involved in the transactivation of the hes1 promoter together with N1IC, β-catenin, and p300.
FIG. 6.
Physical and functional interactions among N1IC, β-catenin, p300, and P/CAF in hes1 promoter activation. (A and B) HEK293 cells were transfected with expression vectors containing tagged cDNAs. The molecular interaction between N1IC and P/CAF was facilitated in the presence of both p300 and β-catenin (βcat). Furthermore, β-catenin was efficiently immunoprecipitated with N1IC in the presence of both p300 and P/CAF (B). IP; αCont IgG, immunoprecipitation with anti-control IgG. (C) The contribution of endogenous p300 to hes1 promoter activation in neural precursor cells was examined using truncated forms of dominant-negative p300 (n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.001; Student's t test). stb, stabilized. (D) Neural precursor cells were transfected with combinations of pHes1-Luc, N1IC, stabilized β-catenin, and P/CAF and p300 cDNAs (n = 4; *, P < 0.05; **, P < 0.01; Student's t test). αmyc, anti-myc antibody.
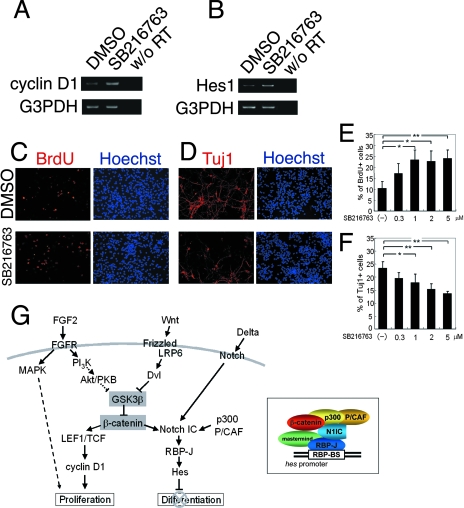
Lastly, we assessed whether the inactivation of endogenous GSK3β is sufficient to promote cell proliferation and inhibit neuronal differentiation. This idea was examined primarily by gain-of-function experiments, in which the misexpression of GSK3βS9A inhibited cell proliferation and the self-renewal of sphere-forming cells (Fig. 2 and 3) and promoted neuronal differentiation (Fig. 3B). SB216763 is a potent, selective, and cell-permeable inhibitor of GSK3β (10). After SB216763 treatment for 3 h, both cyclin D1 and Hes1 mRNA levels were elevated in neural precursor cells, similarly to the results obtained by FGF2 treatment (Fig. 7A and B). Moreover, SB216763 treatment increased the number of BrdU-positive proliferating cells and decreased the number of Tuj1-positive neuronal cells in a dose-dependent manner (Fig. 7C to F). Thus, the inactivation of endogenous GSK3β is involved in both the promotion of neural precursor cell proliferation and the inhibition of their differentiation.
FIG. 7.
Correlation of GSK3β activity with cell proliferation and neuronal differentiation. (A and B) SB216763 (5 μM) treatment of neural precursor cells for 3 h increased the amount of cyclin D1 that had not been amplified from samples without reverse transcriptase (w/o RT) in the reaction mixture. (C to F) SB216763 treatment of neural precursor cells increased the number of BrdU+ cells (red in panel C) and decreased the number of Tuj1+ cells (red in panel D). The ratios of BrdU+ cells/total cells and Tuj1+ cells/total cells are shown in panels E and F, respectively (n = 3; *, P < 0.05, **; P < 0.01; Student's t test). (G) Schematic drawing summarizing our model. β-Catenin plays an important role in linking the promotion of proliferation and the inhibition of differentiation in neural precursor cells by coordinating signals initiated by FGF2, canonical Wnts, and Notch ligands. The inset represents the protein complex governing hes1 promoter activation. BS, binding site; DMSO, dimethylsulfoxide.
DISCUSSION
Involvement of GSK3β and β-catenin in neural precursor cell proliferation.
FGF2 is well known to promote the proliferation of neural stem/precursor cells; however, the molecular mechanism underlying its action has not been fully elucidated. Israsena et al. reported that FGF2 upregulated β-catenin mRNA. They also observed the FGF2-induced rapid translocation of β-catenin protein into the nucleus through an unknown molecular mechanism (24). Jin et al. reported that FGF2 activated the PI3K-Akt pathway, leading to the phosphorylation of GSK3β in postnatal hippocampal progenitor cells (25). These data fit well with the results of our current study. We independently hypothesized and showed evidence that FGF2 inactivates GSK3β through PI3K signaling, leads to the nuclear accumulation of β-catenin, activates LEF/TCF transcription factors, and induces cyclin D1 expression. The application of a PI3K inhibitor or dominant-active GSK3β decreased the number of Ki67+ cells and BrdU-incorporating cells and reduced the size of neurospheres formed in the presence of FGF2, suggesting that this signaling pathway plays an essential role in the proliferation of neural precursor cells and neurosphere-forming cells.
Contribution of GSK3β inactivation and β-catenin stabilization to the inhibition of neuronal differentiation.
It is noteworthy that even when canonical Notch signaling is supposed to be activated through cell-cell contacts in neurospheres, FGF2 removal causes massive neuronal differentiation. Thus, the activation of FGF2 signaling is critical for boosting the Notch signal intensity up to a level sufficient to inhibit neurogenesis. Even in the presence of FGF2, the misexpression of dominant-active GSK3β inhibited secondary neurosphere formation and led to an increase in the proportion of differentiated neurons. As Mizutani et al. (33) reported that Notch signaling is more significant in the maintenance of neural stem cell character than the inhibition of neuronal differentiation from the intermediate progenitor, the inactivation of GSK3β in response to FGF2 is required for the self renewal of neural stem cells. The inactivation of GSK3β by SB216763 mimicked, at least partly, the promotion of cell proliferation and the inhibition of differentiation induced by FGF2 (Fig. 7E and F). GSK3β inactivation and β-catenin stabilization potentiated the promoter activity of hes1 and hes5 genes only when Notch signaling was activated. We demonstrated that stabilized β-catenin associates with N1IC and enhances its RBP-Jκ-dependent transcription activity to induce hes1 and hes5 gene expression. This is the first report to show an interaction between β-catenin and N1IC in mammalian cells. Based on the results that (i) β-catenin was present in a nuclear protein complex associating with the 5′-flanking region of the hes1 gene, which contains RBP-Jκ binding sites, (ii) a stabilized form of β-catenin induced hes1 gene expression in cooperation with N1IC by the recruitment of the transcriptional coactivators p300 and P/CAF, and (iii) the coimmunoprecipitation of β-catenin and N1IC is enhanced by the presence of both p300 and P/CAF, a model is proposed in Fig. 7G for the cooperation between β-catenin and N1IC. This complex may contain Mastermind, which associates with N1IC/RBP-Jk and recruits p300 into the core complex to activate N1IC-mediated transcription (27, 35, 52). Interestingly, Alves-Guerra et al. reported that Mastermind-like 1, a member of the Mastermind-like family, also participated in the Wnt signaling as a specific coactivator of β-catenin/TCF transcription in colon carcinoma cells (1). Thus, Mastermind may be another key molecule, in addition to β-catenin, to link the proliferation and differentiation of neural stem cells. Further studies of Mastermind are necessary to understand its precise roles in the maintenance of neural stem cell character.
We have demonstrated by the use of β-catenin siRNA that β-catenin plays a critical role in the FGF2-mediated potentiation of N1IC-induced hes1 promoter activation (Fig. 5C). However, β-catenin siRNA could not be adequately applied to the cell fate analysis of neural precursor cells due to another function of β-catenin as a member of scaffold proteins. Therefore, the overall reduction of β-catenin by siRNA disturbs cell viability and, thus, may affect the outcomes of cell populations after a culture period necessary for cell differentiation assays.
A molecular link between promotion of proliferation and inhibition of differentiation involving β-catenin stabilization.
Proliferation and differentiation are observed as mutually exclusive events during neural development. How the transition from proliferation to differentiation is managed has been a major unanswered question. We propose that the transcriptional coactivator β-catenin exerts a dual function: β-catenin promotes neural precursor cell proliferation when it binds to LEF/TCF transcription factors, and it also contributes to the maintenance of neural precursor cells in an undifferentiated state when it associates with N1IC and RBP-Jκ under the condition of Notch activation (Fig. 7G). Although the induction of cyclin D1 expression largely involves the activation of LEF/TCF transcription factors, some other downstream effectors of the FGF2-PI3K pathway also may contribute to neural precursor cell proliferation, as discussed in the following paragraphs.
An increasing number of genes have been implicated as targets of the canonical Wnt pathway. By analogy to the Wnt pathway, the FGF2-triggered LEF/TCF activation may induce neural precursor cell proliferation not only through cyclin D1 but also many other LEF/TCF target gene expressions, for example, other cyclin D family members such as c-myc, c-jun, etc. Several other reports have identified Wnt targets through microarray expression profiling. Interestingly, the activation of the Wnt signaling pathway induces the expressions of ID2, REST/NRSF, and Jagged 1, which inhibit neural differentiation in human colon cancer, human embryonic carcinoma cells, and developing mouse hair follicles (15, 51). Although it is still unclear whether FGF2-triggered GSK3β inactivation induces the expression of these genes in neural precursor cells, if so, they also may contribute to the maintenance of neural precursor cells in addition to cyclin D1 and Hes1. There is an interesting report that the misexpression of dominant-negative TCF4 in cortical precursor cells using in utero electroporation-mediated gene transfer causes increased cell cycle exit and precocious neuronal differentiation (53), suggesting that LEF/TCF target genes have some roles in the maintenance of neural precursor cells. However, the hes1 gene may not be the direct target of LEF/TCF-mediated transcription, since dominant-negative TCF4 does not affect hes1 promoter activity (Fig. 5A), which predominantly inhibits neuronal differentiation in our experimental paradigm. Therefore, it will be interesting to examine whether the misexpression of dominant-negative TCF4 affects ID2, REST/NRSF, and Jagged 1 gene expression.
Functional redundancy among Hes family members was reported previously (20, 26). Since FGF2 and N1IC activate the hes5 promoter (Fig. 4H), Hes5 is an obvious candidate for the β-catenin/NICD target. Hes gene products regulate the maintenance of precursor cells in an undifferentiated state and the normal timing of their differentiation, while recent studies suggest that they do not directly regulate neural cell proliferation but do so in a context-dependent manner (26).
Differences between the FGF2 and Wnt signaling pathways.
Similarly to the effect of FGF2, Wnt-3a treatment elevated hes1 promoter activity in the presence of N1IC, but the elevation induced by Wnt-3a was less than that induced by FGF2 (data not shown). This was probably because the molecular mechanisms for inactivating GSK3β are different between the FGF2 and Wnt-3a pathways. FGF receptor signaling activates Akt to phosphorylate GSK3β at Ser9, and the phosphorylated GSK3β diminishes the interaction between GSK3β and Axin in NIH 3T3 cells (55). Wnts initiate downstream signal transduction by binding to their receptors, Frizzled and LRP5/6, leading to the segregation of β-catenin from Axin and GSK3β (22). Although we detected a weak phosphorylation of GSK3β Ser9 induced by rmWnt-3a stimulation, GSK3 phosphorylation itself may not be critical for the Wnt-mediated inactivation of GSK3 (see below). In addition to the signaling pathway common to FGF2 and Wnts, FGF2 also activates the ERK pathway in neural progenitor cells in the fetal subventricular zone and postnatal hippocampus (25, 29). Indeed, we detected the phosphorylation of ERK in neural precursor cells by a 3-h treatment with 20 ng/ml FGF2 (data not shown). Ten micromolars of U0126, a MEK inhibitor, completely abolished the FGF2-induced phosphorylation of ERK and partially suppressed the FGF2-induced proliferation of neural precursor cells (data not shown). Thus, the activation of the MEK-ERK signaling pathway contributes at least in part to FGF2-induced neural precursor cell proliferation. Taken together, our results imply that the PI3K-GSK3β and MEK-ERK signaling pathways both are necessary for FGF2-induced neural precursor cell proliferation.
Inactivation of GSK3β.
GSK3β-Ser9 is a well-known Akt phosphorylation site, and the Ser9 phosphorylation reduces the kinase activity of GSK3β. We demonstrated that FGF2 signaling though PI3K induced the phosphorylation of GSK3β at Ser9. The overexpression of dominant-active GSK3β that lacked the Akt phosphorylation site disrupted the effects of FGF2 on neural precursor cell proliferation and hes1 promoter activation, suggesting the involvement of Akt or equivalent kinases in the FGF2 signaling pathway. However, we could not conclude yet that Akt actually phosphorylated and inactivated GSK3β in neural precursor cells. Although the overexpression of dnAkt was supposed to show some evidence for linkage between Akt and GSK3β activities, dnAkt disturbed cell viability before the end of the culture period, which was long enough for cell proliferation or differentiation assays. LY294002 and wortmannin antagonize PI3K activity; however, the inactivation of PI3K affects not only Akt activity but also various PI3K downstream targets. For example, the PI3K-PDK1 pathway phosphorylates and activates a number of kinases, including protein kinase C, serum and glucocorticoid-regulated kinase-1, and p70S6-kinase (p70S6K) independently of Akt (5), and one of these may act downstream of PI3K to inactivate GSK3β.
The inactivation of GSK3 is essential for normal development. GSK3α/β double-knockout ES cells display hyperactivated Wnt/β-catenin signaling and are severely compromised in their differentiation ability, confirming the importance of GSK3α/β inactivation in maintaining pluripotent stem cell properties (12). In contrast, the biological significance of GSK3 phosphorylation remains unclear. Some studies have shown the Akt-mediated phosphorylation of GSK3α-Ser21 and GSK3β-Ser9 and its importance in biological events (17, 45). Therefore, it is surprising that the knock-in mice missing phosphorylation sites in both GSK3α/β genes (Gsk3αS21A/S21A/βS9A/S9A) are viable and fertile (31). One notable report was published by Gärtner et al., in which an in vivo analysis of Gsk3αS21A/S21A/βS9A/S9A knock-in neurons showed axonal and dendritic projections similar to that in the wild-type control. However, their figures also displayed distinguishable distribution patterns of MAP2 and TauA immunoreactivities between the knock-in and wild-type brains (19). The detailed analysis of the Gsk3αS21A/S21A/βS9A/S9A knock-in phenotype will reveal their importance in the course of brain development.
Multiple functions of β-catenin.
β-Catenin has been implicated to increase neural precursor cell proliferation and suppress neuronal differentiation. β-Catenin-ablated embryos showed a reduced tissue mass of spinal cords and exhibited partial but significant loss of the nestin-positive neural precursor population (56). Conversely, mice that expressed stabilized β-catenin showed an enlarged mass of neural tissue, and the sizes of their neural precursor populations were increased (9, 56). Thus, these studies suggest that β-catenin activation allows neural precursor cells to reenter the cell cycle but not to differentiate. Another noteworthy finding of these β-catenin mutant experiments was that the above abnormalities were observed in various regions of the nervous system, indicating that β-catenin signals can, in general, control the size of the precursor pool. In these previous studies, molecular mechanisms that firmly link and finely modulate the promotion of cell cycle reentry and the inhibition of differentiation were not proposed.
In contrast, at some stages and in some regions of the developing brain, Wnt signal activation was reported to inhibit the self-renewal capacity of neural progenitor cells and promote their neuronal differentiation. Hirabayashi et al. prepared neural precursor cells from neurospheres derived from E11.5 mouse cortex. They demonstrated that the overexpression of Wnt-7a or β-catenin in neural precursors induced neuronal differentiation and that a β-catenin-TCF complex appeared to directly regulate the promoter activity of neurogenin 1, a gene encoding a neurogenic bHLH transcription factor, in cortical neural precursor cells (23). Israsena et al. demonstrated that the overexpression of β-catenin in the presence of FGF2, a condition similar to that of our experiment, helped to maintain neural progenitor cells in a proliferative state, consistently with our present study (24). However, they further showed that the overexpression of β-catenin in the absence of FGF2 enhanced neuronal differentiation accompanied by the activation of the neurogenin 1 promoter by β-catenin-Lef1 binding. Besides these studies with fetal telencephalic neural precursor cells, Lie et al. reported that the in vivo blockade of endogenous Wnt signaling by a dominant-negative Wnt-1 abolished neurogenesis in the adult hippocampus (30). They further suggested that Wnt signaling is involved in both the control of neuronal fate commitment and the proliferation of neuronally committed precursor cells during adult hippocampal neurogenesis. Future experiments should be designed to address the precise contribution of Wnt signaling to proliferation and differentiation in a developmental stage-dependent and region-specific manner.
Maintenance of hESCs and cancer cells.
Pluripotent human ES cells (hESCs) provide potential applications in regenerative medicine and the study of early embryonic development. Although hESCs are capable of unlimited proliferation and maintain pluripotency in vitro, little is known about the regulatory mechanisms that support their undifferentiated proliferation. FGF2 and Wnts both were reported to promote the self renewal of hESCs (2, 54). Furthermore, a pharmacological GSK3-specific inhibitor by itself sustained the pluripotency of hESCs (42), suggesting the importance of GSK3 targets in the maintenance of hESCs. The importance of FGF2 in the maintenance of hESCs is obvious, but we think its direct cross-talk with Wnt should be investigated in more detail, since FGF2 seems to be important in establishing the regulatory niche in the hESC-maintaining culture system (6). GSK3 also plays important roles in cell cycle exit and differentiation in mouse ES cells (mESCs), since GSK3α/β double-knockout mESCs display hyperactivated Wnt/β-catenin signaling and lose their differentiation ability (12). It was recently reported that Notch receptor activation induces hESCs proliferation but does not support their self renewal (18). Therefore, hESCs require some signaling other than that of Notch to maintain their stem cell properties.
Mutations in β-catenin are linked to tumorigenesis. The activation of the FGF receptor signaling pathway and the accumulation of β-catenin in the nuclei often are observed in malignant tumor cells, suggesting their involvement in tumor cell proliferation and tumor maintenance (13, 39). Many brain tumors contain stem-like cells, also called tumor-initiating cells or cancer-initiating cells, which can undergo self renewal and multilineage cell differentiation similarly to normal neural stem cells (7, 32). Interestingly, the pharmacological inhibition of Notch signaling depletes these stem-like cells and blocks the growth of embryonal brain tumors both in vitro and in xenografts (16). Thus, the maintenance and amplification of certain cancer stem-like cells may involve the interplay of the FGF2, Wnt, and Notch pathways through the complex formations of β-catenin and LEF/TCF as well as β-catenin and N1IC. Our present study provides new insights into the molecular basis for the maintenance of stem cell properties, including neural stem cells, ES cells, and cancer stem-like cells, in view of the FGF-Wnt-Notch signaling connection by β-catenin.
Acknowledgments
We greatly thank Morris J. Birnbaum (University of Pennsylvania) for the gift of GSK3β-S9A cDNA, Hans Clevers (The Netherlands Institute of Developmental Biology) for the gift of pcDNA3.1-ΔNTCF4, Makoto Hijikata (Kyoto University) for the gift of the original 7×TCF-Luc plasmid, Akira Nagafuchi (Kumamoto University) for the gift of stabilized β-catenin and pEF-mβH cDNA, Brian A. Hemmings (Friedrich Miescher Institute, Basel, Switzerland) for the gift of dnAkt cDNA (pCMV5-AktAAA), and Atsuko Sakurai (National Cardio Vascular Center) for useful information on the anti-β-catenin antibody. We also thank Kinichi Nakashima (Nara Institute of Science and Technology) and Yutaka Yoshinaga (Kumamoto University) for helpful discussions. We thank Yuko Saiki, Kaori Kaneko, and Sayomi Iwaki for their technical assistance. We also thank Michiko Teramoto for her secretarial assistance.
This work was supported by a grant-in-aid for 21st Century COE research from the Ministry of Education, Culture, Sports, Science and Technology Cell Fate Regulation Research and Education Unit, the Naito Foundation (T.S.); grants-in-aid 16047223 and 18053018 for Scientific Research on Priority Areas on the elucidation of glia-neuron network mediated information processing systems (T.K.), grants-in-aid for scientific research on priority areas on molecular brain science (T.T.), and grants-in-aid for scientific research on priority areas on self-renewal and pluripotency of the stem cells (T.T.) from the Ministry of Education, Culture, Sports, Science and Technology; grant-in-aid 18700365 for Scientific Research for Young Scientist (T.S.), grants-in-aid 17500255 for scientific research (C) (T.K.), and grants-in-aid for scientific research (B) (T.T.) from the Japan Society for the Promotion of Sciences, NCNP, and CREST.
Footnotes
Published ahead of print on 13 October 2008.
REFERENCES
- 1.Alves-Guerra, M. C., C. Ronchini, and A. J. Capobianco. 2007. Mastermind-like 1 is a specific coactivator of beta-catenin transcription activation and is essential for colon carcinoma cell survival. Cancer Res. 678690-8698. [DOI] [PubMed] [Google Scholar]
- 2.Amit, M., M. K. Carpenter, M. S. Inokuma, C. P. Chiu, C. P. Harris, M. A. Waknitz, J. Itskovitz-Eldor, and J. A. Thomson. 2000. Clonally derived human embryonic stem cell lines maintain pluripotency and proliferative potential for prolonged periods of culture. Dev. Biol. 227271-278. [DOI] [PubMed] [Google Scholar]
- 3.Andjelkoviæ, M., D. R. Alessi, R. Meier, A. Fernandez, N. J. Lamb, M. Frech, P. Cron, P. Cohen, J. M. Lucocq, and B. A. Hemmings. 1997. Role of translocation in the activation and function of protein kinase B. J. Biol. Chem. 27231515-31524. [DOI] [PubMed] [Google Scholar]
- 4.Arcaro, A., and M. P. Wymann. 1993. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: the role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem. J. 296297-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belham, C., S. Wu, and J. Avruch. 1999. Intracellular signalling: PDK1-a kinase at the hub of things. Curr. Biol. 9R93-R96. [DOI] [PubMed] [Google Scholar]
- 6.Bendall, S. C., M. H. Stewart, P. Menendez, D. George, K. Vijayaragavan, T. Werbowetski-Ogilvie, V. Ramos-Mejia, A. Rouleau, J. Yang, M. Bossé, G. Lajoie, and M. Bhatia. 2007. IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature 4481015-1021. [DOI] [PubMed] [Google Scholar]
- 7.Bjerkvig, R., B. B. Tysnes, K. S. Aboody, J. Najbauer, and A. J. Terzis. 2005. Opinion: the origin of the cancer stem cell: current controversies and new insights. Nat. Rev. Cancer 5899-904. [DOI] [PubMed] [Google Scholar]
- 8.Cadigan, K. M., and R. Nusse. 1997. Wnt signaling: a common theme in animal development. Genes Dev. 113286-3305. [DOI] [PubMed] [Google Scholar]
- 9.Chenn, A., and C. A. Walsh. 2002. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science 297365-369. [DOI] [PubMed] [Google Scholar]
- 10.Coghlan, M. P., A. A. Culbert, D. A. Cross, S. L. Corcoran, J. W. Yates, N. J. Pearce, O. L. Rausch, G. J. Murphy, P. S. Carter, L. Roxbee Cox, D. Mills, M. J. Brown, D. Haigh, R. W. Ward, D. G. Smith, K. J. Murray, A. D. Reith, and J. C. Holder. 2000. Selective small molecule inhibitors of glycogen synthase kinase-3 modulate glycogen metabolism and gene transcription. Chem. Biol. 7793-803. [DOI] [PubMed] [Google Scholar]
- 11.Cross, D. A., D. R. Alessi, P. Cohen, M. Andjelkovich, and B. A. Hemmings. 1995. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 378785-789. [DOI] [PubMed] [Google Scholar]
- 12.Doble, B. W., S. Patel, G. A. Wood, L. K. Kockeritz, and J. R. Woodgett. 2007. Functional redundancy of GSK-3alpha and GSK-3beta in Wnt/beta-catenin signaling shown by using an allelic series of embryonic stem cell lines. Dev. Cell 12957-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dvorak, P., D. Dvorakova, and A. Hampl. 2006. Fibroblast growth factor signaling in embryonic and cancer stem cells. FEBS Lett. 5802869-2874. [DOI] [PubMed] [Google Scholar]
- 14.Espinosa, L., J. Ingles-Esteve, C. Aguilera, and A. Bigas. 2003. Phosphorylation by glycogen synthase kinase-3 beta down-regulates Notch activity, a link for Notch and Wnt pathways. J. Biol. Chem. 27832227-32235. [DOI] [PubMed] [Google Scholar]
- 15.Estrach, S., C. A. Ambler, C. Lo Celso, K. Hozumi, and F. M. Watt. 2006. Jagged 1 is a beta-catenin target gene required for ectopic hair follicle formation in adult epidermis. Development 1334427-4438. [DOI] [PubMed] [Google Scholar]
- 16.Fan, X., W. Matsui, L. Khaki, D. Stearns, J. Chun, Y. M. Li, and C. G. Eberhart. 2006. Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 667445-7452. [DOI] [PubMed] [Google Scholar]
- 17.Forde, J. E., and T. C. Dale. 2007. Glycogen synthase kinase 3: a key regulator of cellular fate. Cell Mol. Life Sci. 641930-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox, V., P. J. Gokhale, J. R. Walsh, M. Matin, M. Jones, and P. W. Andrews. 2008. Cell-cell signaling through NOTCH regulates human embryonic stem cell proliferation. Stem Cells 26715-723. [DOI] [PubMed] [Google Scholar]
- 19.Gärtner, A., X. Huang, and A. Hall. 2006. Neuronal polarity is regulated by glycogen synthase kinase-3 (GSK-3beta) independently of Akt/PKB serine phosphorylation. J. Cell Sci. 1193927-3934. [DOI] [PubMed] [Google Scholar]
- 20.Hatakeyama, J., Y. Bessho, K. Katoh, S. Ookawara, M. Fujioka, F. Guillemot, and R. Kageyama. 2004. Hes genes regulate size, shape and histogenesis of the nervous system by control of the timing of neural stem cell differentiation. Development 1315539-5550. [DOI] [PubMed] [Google Scholar]
- 21.Hayward, P., K. Brennan, P. Sanders, T. Balayo, R. DasGupta, N. Perrimon, and A. Martinez Arias. 2005. Notch modulates Wnt signalling by associating with Armadillo/beta-catenin and regulating its transcriptional activity. Development 1321819-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, X. 2003. A Wnt-Wnt situation. Dev. Cell 4791-797. [DOI] [PubMed] [Google Scholar]
- 23.Hirabayashi, Y., Y. Itoh, H. Tabata, K. Nakajima, T. Akiyama, N. Masuyama, and Y. Gotoh. 2004. The Wnt/β-catenin pathway directs neuronal differentiation of cortical neural precursor cells. Development 1312791-2801. [DOI] [PubMed] [Google Scholar]
- 24.Israsena, N., M. Hu, W. Fu, L. Kan, and J. A. Kessler. 2004. The presence of FGF2 signaling determines whether beta-catenin exerts effects on proliferation or neuronal differentiation of neural stem cells. Dev. Biol. 268220-231. [DOI] [PubMed] [Google Scholar]
- 25.Jin, L., X. Hu, and L. Feng. 2005. NT3 inhibits FGF2-induced neural progenitor cell proliferation via the PI3K/GSK3 pathway. J. Neurochem. 931251-1261. [DOI] [PubMed] [Google Scholar]
- 26.Kageyama, R., T. Ohtsuka, and T. Kobayashi. 2007. The Hes gene family: repressors and oscillators that orchestrate embryogenesis. Development 1341243-1251. [DOI] [PubMed] [Google Scholar]
- 27.Kovall, R. A. 2007. Structures of CSL, Notch and Mastermind proteins: piecing together an active transcription complex. Curr. Opin. Struct. Biol. 17117-127. [DOI] [PubMed] [Google Scholar]
- 28.Kurooka, H., and T. Honjo. 2000. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 27517211-17220. [DOI] [PubMed] [Google Scholar]
- 29.Learish, R. D., M. D. Bruss, and M. Haak-Frendscho. 2000. Inhibition of mitogen-activated protein kinase kinase blocks proliferation of neural progenitor cells. Brain Res. Dev. Brain Res. 12297-109. [DOI] [PubMed] [Google Scholar]
- 30.Lie, S., S. A. Colamarino, H. J. Song, L. Desire, H. Mira, A. Consiglio, E. S. Lein, S. Jessberger, H. Lansford, A. R. Dearie, and F. H. Gage. 2005. Wnt signalling regulates adult hippocampal neurogenesis. Nature 4371370-1375. [DOI] [PubMed] [Google Scholar]
- 31.McManus, E. J., K. Sakamoto, L. J. Armit, L. Ronaldson, N. Shapiro, R. Marquez, and D. R. Alessi. 2005. Role that phosphorylation of GSK3 plays in insulin and Wnt signalling defined by knockin analysis. EMBO J. 241571-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mimeault, M., R. Hauke, P. P. Mehta, and S. K. Batra. 2007. Recent advances in cancer stem/progenitor cell research: therapeutic implications for overcoming resistance to the most aggressive cancers. J. Cell Mol. Med. 11981-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizutani, K., K. Yoon, L. Dang, A. Tokunaga, and N. Gaiano. 2007. Differential Notch signalling distinguishes neural stem cells from intermediate progenitors. Nature 449351-355. [DOI] [PubMed] [Google Scholar]
- 33a.Morita, S., T. Kojima, and T. Kitamura. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 71063-1066. [DOI] [PubMed] [Google Scholar]
- 34.Nakashima, K., M. Yanagisawa, H. Arakawa, N. Kimura, T. Hisatsune, M. Kawabata, K. Miyazono, and T. Taga. 1999. Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science 284479-482. [DOI] [PubMed] [Google Scholar]
- 35.Nam, Y., P. Sliz, L. Song, J. C. Aster, and S. C. Blacklow. 2006. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 124973-983. [DOI] [PubMed] [Google Scholar]
- 36.Nosaka, T., T. Kawashima, K. Misawa, K. Ikuta, A. L. Mui, and T. Kitamura. 1999. STAT5 as a molecular regulator of proliferation, differentiation and apoptosis in hematopoietic cells. EMBO J. 184754-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oswald, F., B. Tauber, T. Dobner, S. Bourteele, U. Kostezka, G. Adler, S. Liptay, and R. M. Schmid. 2001. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol. 217761-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Qian, X., A. A. Davis, S. K. Goderie, and S. Temple. 1997. FGF2 concentration regulates the generation of neurons and glia from multipotent cortical stem cells. Neuron 1881-93. [DOI] [PubMed] [Google Scholar]
- 39.Reya, T., and H. Clevers. 2005. Wnt signalling in stem cells and cancer. Nature 434843-850. [DOI] [PubMed] [Google Scholar]
- 40.Reya, T., A. W. Duncan, L. Ailles, J. Domen, D. C. Scherer, K. Willert, L. Hintz, R. Nusse, and I. L. Weissman. 2003. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature 423409-414. [DOI] [PubMed] [Google Scholar]
- 41.Roose, J., G. Huls, M. van Beest, P. Moerer, K. van der Horn, R. Goldschmeding, T. Logtenberg, and H. Clevers. 1999. Synergy between tumor suppressor APC and the beta-catenin-Tcf4 target Tcf1. Science 2851923-1926. [DOI] [PubMed] [Google Scholar]
- 42.Sato, N., L. Meijer, L. Skaltsounis, P. Greengard, and A. H. Brivanlou. 2004. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat. Med. 1055-63. [DOI] [PubMed] [Google Scholar]
- 42a.Schwartz, D. R., R. Wu, S. L. Kardia, A. M. Levin, C. C. Huang, K. A. Shedden, R. Kuick, D. E. Misek, S. M. Hanash, J. M. Taylor, H. Reed, N. Hendrix, Y. Zhai, E. R. Fearon, and K. R. Cho. 2003. Novel candidate targets of beta-catenin/T-cell factor signaling identified by gene expression profiling of ovarian endometrioid adenocarcinomas. Cancer Res. 632913-2922. [PubMed] [Google Scholar]
- 43.Summers, S. A., A. W. Kao, A. D. Kohn, G. S. Backus, R. A. Roth, J. E. Pessin, and M. J. Birnbaum. 1999. The role of glycogen synthase kinase 3beta in insulin-stimulated glucose metabolism. J. Biol. Chem. 27417934-17940. [DOI] [PubMed] [Google Scholar]
- 44.Sun, Y., F. T. Kolligs, M. O. Hottiger, R. Mosavin, E. R. Fearon, and G. J. Nabel. 2000. Regulation of beta-catenin transformation by the p300 transcriptional coactivator. Proc. Natl. Acad. Sci. USA 9712613-12618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sutherland, C., I. A. Leighton, and P. Cohen. 1993. Inactivation of glycogen synthase kinase-3 beta by phosphorylation: new kinase connections in insulin and growth-factor signalling. Biochem. J. 29615-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takizawa, T., K. Nakashima, M. Namihira, W. Ochiai, A. Uemura, M. Yanagisawa, N. Fujita, M. Nakao, and T. Taga. 2001. DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev. Cell 1749-758. [DOI] [PubMed] [Google Scholar]
- 47.Takizawa, T., W. Ochiai, K. Nakashima, and T. Taga. 2003. Enhanced gene activation by Notch and BMP signaling cross-talk. Nucleic Acids Res. 315723-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ueda, Y., M. Hijikata, S. Takagi, R. Takada, S. Takada, T. Chiba, and K. Shimotohno. 2002. Wnt/beta-catenin signaling suppresses apoptosis in low serum medium and induces morphologic change in rodent fibroblasts. Int. J. Cancer 99681-688. [DOI] [PubMed] [Google Scholar]
- 49.Vandermoere, F., I. El Yazidi-Belkoura, E. Adriaenssens, J. Lemoine, and H. Hondermarck. 2005. The antiapoptotic effect of fibroblast growth factor-2 is mediated through nuclear factor-kappaB activation induced via interaction between Akt and IkappaB kinase-beta in breast cancer cells. Oncogene 245482-5491. [DOI] [PubMed] [Google Scholar]
- 50.Vlahos, C. J., W. F. Matter, K. Y. Hui, and R. F. Brown. 1994. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002). J. Biol. Chem. 2695241-5248. [PubMed] [Google Scholar]
- 51.Willert, J., M. Epping, J. R. Pollack, P. O. Brown, and R. Nusse. 2002. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev. Biol. 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wilson, J. J., and R. A. Kovall. 2006. Crystal structure of the CSL-Notch-Mastermind ternary complex bound to DNA. Cell 124985-996. [DOI] [PubMed] [Google Scholar]
- 53.Woodhead, G. J., C. A. Mutch, E. C. Olson, and A. Chenn. 2006. Cell-autonomous beta-catenin signaling regulates cortical precursor proliferation. J. Neurosci. 2612620-12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, C., M. S. Inokuma, J. Denham, K. Golds, P. Kundu, J. D. Gold, and M. K. Carpenter. 2001. Feeder-free growth of undifferentiated human embryonic stem cells. Nat. Biotechnol. 19971-974. [DOI] [PubMed] [Google Scholar]
- 55.Yuan, H., J. Mao, L. Li, and D. Wu. 1999. Suppression of glycogen synthase kinase activity is not sufficient for leukemia enhancer factor-1 activation. J. Biol. Chem. 27430419-30423. [DOI] [PubMed] [Google Scholar]
- 56.Zechner, D., Y. Fujita, J. Hulsken, T. Muller, I. Walther, M. M. Taketo, E. B. Crenshaw III, W. Birchmeier, and C. Birchmeier. 2003. β-Catenin signals regulate cell growth and the balance between progenitor cell expansion and differentiation in the nervous system. Dev. Biol. 258406-418. [DOI] [PubMed] [Google Scholar]