Abstract
The cell division cycle 25A (Cdc25A) phosphatase is a critical regulator of cell cycle progression under normal conditions and after stress. Stress-induced degradation of Cdc25A has been proposed as a major way of delaying cell cycle progression. In vitro studies pointed toward serine 123 as a key site in regulation of Cdc25A stability after exposure to ionizing radiation (IR). To address the role of this phosphorylation site in vivo, we generated a knock-in mouse in which alanine was substituted for serine 123. The Cdc25 S123A knock-in mice appeared normal, and, unexpectedly, cells derived from them exhibited unperturbed cell cycle and DNA damage responses. In turn, we found that Cdc25A was present in centrosomes and that Cdc25A levels were not reduced after IR in knock-in cells. This resulted in centrosome amplification due to lack of induction of Cdk2 inhibitory phosphorylation after IR specifically in centrosomes. Further, Cdc25A knock-in animals appeared sensitive to IR-induced carcinogenesis. Our findings indicate that Cdc25A S123 phosphorylation is crucial for coupling centrosome duplication to DNA replication cycles after DNA damage and therefore is likely to play a role in the regulation of tumorigenesis.
Cyclin-dependent kinases (CDKs) regulate progression through phases of the cell division cycle. The activation of CDKs, in turn, is dependent on their phosphorylation at Thr14 and Tyr15 by the Cdc25 family of protein phosphatases. Three Cdc25-related proteins have been identified in mammalian cells (10, 25). Cdc25B and Cdc25C appear to regulate progression from G2 to M phase (9, 21), while Cdc25A is active in all stages of the cell cycle (4, 19, 22, 30). Cdc25A is capable of binding to different cyclins and dephosphorylating CDKs in vitro (10, 26, 29). Cdc25 is a transcriptional target of c-Myc (11) and E2F (28) and can cooperate with activated Ras in inducing mouse cell transformation and in vivo tumor growth (12). In addition, a subset of aggressive human cancers shows increased expression of Cdc25A (12, 13).
The activity and level of Cdc25A are regulated by reversible phosphorylation, protein-protein interactions, and ubiquitin-mediated proteolysis (3, 6, 18, 22, 24). Regulation of Cdc25A is crucial for unperturbed cell cycle progression and for eliciting arrest in response to checkpoint activation (30). Cdc25A is rapidly degraded in a proteasome-dependent manner in cells exposed to UV light, hydroxyurea, or ionizing radiation (IR) (3, 8, 22, 24, 30). In addition, overexpression of Cdc25A causes bypass of the IR-induced S and G2 phase checkpoints, as well as the DNA replication checkpoint (8, 22, 24). Several residues have been identified as critical for mediating Cdc25A degradation depending on the type of DNA damage incurred, including serines 76 and 123 (8, 14, 15, 24).
Phosphorylation of serine 123 has been reported to regulate Cdc25A stability after IR exposure (8, 14). In vitro studies have shown that phosphorylation of serine 123 is induced by IR and is mediated by the Chk1 and Chk2 kinases (8, 14, 30). To demonstrate a role for serine 123 phosphorylation under in vivo conditions, we generated knock-in mice in which this site was mutated to alanine. Cdc25 S123A knock-in mice appeared normal, and, in contrast to expectations, cells derived from them exhibited unperturbed cell cycle and DNA damage responses. In turn, we found that Cdc25A was present in centrosomes and that Cdc25A levels were not reduced after IR in knock-in cells. This resulted in centrosome amplification due to lack of induction of Cdk2 inhibitory phosphorylation after IR specifically in centrosomes. Further, Cdc25A knock-in animals appeared sensitive to IR-induced carcinogenesis. Our findings indicate that Cdc25A S123 is crucial for maintaining genomic integrity through the coupling of centrosome duplication and DNA replication cycles after DNA damage.
MATERIALS AND METHODS
Generation of Cdc25A S123A knock-in mice.
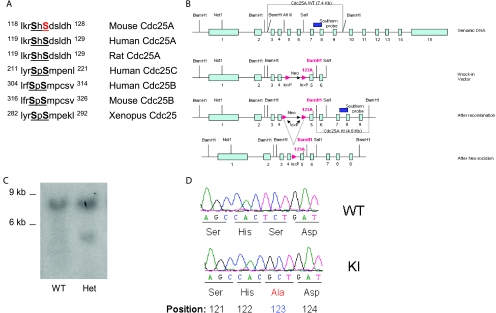
Targeting vectors were constructed that contained the equivalent of 6.5 kb of mouse DNA (from NotI to SalI sites) (Fig. 1B), including the serine 123 site. A phosphoglycerate kinase promoter neo cassette (in either of two orientations) was introduced into a SacI site, and an additional BamHI site was introduced by site-directed mutagenesis ∼1.3 kb from the SalI site. The final cassette was flanked by two thymidine kinase genes. The two targeting vectors were electroporated into embryonic stem (ES) cells (129/SvJ), and colonies were selected in G418 and ganciclovir. For genotyping by genomic Southern blot analysis, we probed BamHI-digested DNA with a 600-bp fragment corresponding to a region in genomic DNA 5′ to that in the targeting vector. All animal protocols used in this study were approved by the Institute of Molecular and Cell Biology, Animal Use and Safety Committee.
FIG. 1.
Targeting of Cdc25A serine at position 123. (A) Alignment of the Cdc25 conserved motif among various species. (B) Design of Cdc25A S123A targeting constructs. (C) Southern blot analysis of targeted ES cells showing the presence of the wild-type (WT; 7.4 kb) and knock-in (KI; 4.6 kb) band in wild-type and targeted ES cells. (D) Sequencing analysis of cDNAs from Cdc25A S123A wild-type and knock-in mice to confirm the presence of the alanine substitution at position 123. Het, heterozygous.
Generation of mouse dermal fibroblasts.
Mouse dermal fibroblasts (DFs) were derived from newly born pups. Skin was removed and placed in Dulbecco's modified Eagle's medium (DMEM) containing 0.1% collagenase A. After overnight incubation at 4°C, the epidermal cell layer was carefully removed. The remaining dermis was suspended by pipetting in normal medium containing 10% serum and cultured until cells reached confluence. Early-passage cells (passages 2 [P2] to P4) were analyzed for cell cycle progression and for cellular responses to DNA damage.
3T3 proliferation assay.
Dermal fibroblasts were plated at 105 in triplicates onto six-well tissue culture plates. Cells were trypsinized, counted, and replated every third day until senescence was reached.
S to G1 phase progression.
A total of 106 DFs were seeded onto 10-cm tissue culture dishes 24 h prior to bromodeoxyuridine (BrdU) labeling. Cells were incubated in 5 ml of DMEM (10% fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin G/ml, and 100 mg of streptomycin/ml) containing 20 μM BrdU (Zymed Laboratories) at 37°C for 1 h. Medium was removed and replaced with 10 ml of culture medium, and cells were incubated for the indicated times. Cells were harvested by trypsinization and collected by centrifugation. After the removal of the supernatant by aspiration, cells were washed once in phosphate-buffered saline (PBS) and were then suspended in 0.5 ml of PBS. Cells were fixed by the addition of 5 ml of 70% ethanol at 4°C. Pelleted cells were washed twice in PBS and resuspended in 1 ml of 2 M HCl for 30 min. Cells were then washed twice in PBS and once in PBS-T (PBS supplemented with 0.1% bovine serum albumin [BSA] and 0.2% Tween 20, pH 7.4). Cell pellets were then incubated in 200 μl of anti-BrdU antibody (Becton Dickinson) diluted 1:50 in PBS-T for 1 h at room temperature in the dark. Following two washes in PBS-T, cells were stained with chicken anti-mouse antibody (AlexaFluor) for 30 min in the dark. Cells were then washed with PBS and incubated with 100 μg of RNase A and 25 μg of propidium iodide (PI) for 30 min in the dark. Cells were analyzed for DNA content by flow cytometry using a FACSCalibur instrument (Becton Dickinson Instruments). The data were analyzed using CELLQUEST analysis software (Becton Dickinson).
G0 to S phase progression.
A total of 106 DFs were seeded onto 10-cm-diameter tissue culture dishes. The following day, cells were incubated in 10 ml of DMEM containing 0.25% fetal bovine serum for 96 h. Cells were then pulsed with 20 μM BrdU prior to harvest. Harvesting, fixing, staining, and analysis of cells by flow cytometry were carried out as described above.
IR-induced G1 phase checkpoint in DFs.
Asynchronously growing cells were exposed to 5 Gy of IR. After 14 h of irradiation, 20 μM BrdU was added for 2 h, and cells were harvested. Cells were fixed, stained, and analyzed by flow cytometry as described above.
IR-induced S-phase checkpoint.
DFs were labeled with 10 nCi of [14C]thymidine (Amersham) per ml for 24 h to control for total DNA content between samples. Cells were either mock irradiated or exposed to 5, 10, or 20 Gy of IR; incubated for 30 min; and then pulse labeled with 2.5 μCi of [3H]thymidine (Amersham) per ml for 15 min. Cells were harvested, washed twice in PBS, and then incubated in 0.5 ml of 0.1 M NaCl containing 0.01 M EDTA (pH 8) per plate. An aliquot (200 μl) was added to a separate tube containing 200 μl of lysis buffer (1 M NaOH, 0.02 M EDTA), alkali-lysed DNA was collected on a glass microfiber (GFC) filter and air dried, and the amount of radioactivity was assayed in a liquid scintillation counter. The resulting ratios of 3H counts per minute to 14C counts per minute, corrected for those counts per minute that were the result of channel crossover, were a measure of DNA synthesis.
IR-induced G2 phase checkpoint.
DFs were either mock irradiated or exposed to 5 Gy of IR. After 40 min of incubation, 100 ng of nocodazole/ml was added to trap cells in mitosis, and samples were harvested by trypsinization 1 h 20 min later. Cells were fixed by the addition of 5 ml of 70% ethanol at 4°C for as long as 24 h. After fixation, cells were washed twice with PBS, suspended in 1 ml of 0.25% Triton X-100 in PBS, and incubated at 4°C with rocking for 15 min. After centrifugation, cell pellets were suspended in 100 μl of PBS containing 1% BSA and 0.75 μg of anti-phospho-histone H3 antibody (Upstate) and incubated for 1.5 h at room temperature. Cells were rinsed with PBS containing 1% BSA and incubated with fluorescein isothiocyanate-conjugated chicken anti-rabbit immunoglobulin G (AlexaFluor) antibody diluted at a ratio of 1:500 in PBS containing 1% BSA. After a 30-min incubation at room temperature in the dark, cells were washed again and incubated with 500 μl of PBS with 1% BSA containing 25 μg of PI/ml and 100 μg of RNase A/ml for 30 min, also in the dark. Cells were analyzed by flow cytometry as described above.
Exit from G2/M checkpoint.
DFs were either mock irradiated or exposed to 5 Gy of IR. Nocodazole was added at 100 ng/ml to trap cells in mitosis, and samples were harvested by trypsinization 18 h later. Cells were fixed and stained for phospho-histone H3 by immunofluorescence.
Immunofluorescence.
DFs were either mock irradiated or exposed to 5 Gy of IR. Nocodazole was added at 100 ng/ml to trap cells in mitosis, and samples were fixed in 100% methanol for 20 min at −20°C. Cells were incubated in 1% BSA and PBS-T for 30 min and then in pericentrin antibody (Covance Research Products, Denver, PA) diluted at 1:500 for 1 h. Cells were washed three times in PBS and then incubated with AlexaFluor 594 chicken anti-rabbit antibody diluted at 1:500 for 30 min. Coverslips were mounted with a drop of mounting medium containing DAPI (4′,6′-diamidino-2-phenylindole).
Immunoprecipitation and Western blotting.
Cultures were washed twice in ice-cold PBS, lysed in ice-cold radioimmunoprecipitation assay buffer (50 mM Tris-HCl, pH 8.0, 0.14 M NaCl, 1% NP-40, 20 mM glycerophosphate, 10 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM phenylmethylsulfonyl fluoride), and centrifuged at 14 000 × g for 15 min. For immunoprecipitation, lysates were precleared with protein A/G beads (Santa Cruz), and 2 μg of antibody specific to Cdc25A (Santa Cruz) was added and incubated in a rolling shaker overnight at 4°C. Twenty-five microliters of protein A/G beads was added, and lysates were incubated for another 2 h. The beads were then washed and boiled after the addition of 50 μl of 2× Laemmli sample buffer. For Western blotting, proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to Immobilon-P membranes (Millipore). Immunoblots were probed with antibodies specific to Cdc25A (kind gift from H. Piwnica-Worms), total p38 (C-20), cyclin A (H-432), cyclin B (H-433), phospho-Cdk2 (at Thr14/Tyr15) (all from Santa Cruz), Cdk2 (BD Transduction Laboratories), and γ-tubulin (GTU-88; Sigma).
Cdc25A steady-state and cycloheximide half-life experiments.
For Cdc25A protein half-life experiments, mock- or IR-treated DFs were incubated in the presence of 25 μg/ml of cycloheximide. Cells were harvested at different time points, and the level of Cdc25A was analyzed using Western blotting.
Centrosome isolation.
Exponentially growing cells were incubated with culture medium containing 1 μg of cytochalasin D/ml and 0.2 μM nocodazole for 1 h at 37°C to depolymerize the actin and microtubule filaments. Then, 1 × 107 to 3 × 107 cells were harvested by trypsinization and lysed in a solution containing 1 mM HEPES (pH 7.2), 0.5% NP-40, 0.5 mM MgCl2, and 0.1% 2-mercaptoethanol with proteinase and phosphatase inhibitors. Swollen nuclei and chromatin aggregates were removed by centrifugation at 2,500 × g for 10 min, and the supernatant was filtered through a 37-μm-pore-size nylon mesh. The supernatant was treated with 2 U of DNase I/ml in 10 mM HEPES (pH 7.2) for 30 min on ice. The lysate was then underlaid with a 60% sucrose solution (60% [wt/wt] sucrose in 10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 7.2, with 0.1% Triton X-100 and 0.1% 2-mercaptoethanol). Centrosomes were sedimented into the sucrose cushion by centrifugation at 10,000 × g for 30 min at 4°C in a Beckman ultracentrifuge equipped with an SW41 Ti rotor. After centrifugation the crude centrosomes were purified further by using a discontinuous gradient consisting of 1,500 μl of 70% sucrose, 900 μl of 50% sucrose, and 900 μl of 40% sucrose solution at 120,000 × g for 1 h at 4°C in an SW60 Ti rotor. Aliquots of 350 μl were collected as fractions, and the first six fractions were used for analysis of centrosomes. Each fraction was diluted with 1 ml of 10 mM PIPES (pH 7.2) and centrifuged for 10 min at 15,000 rpm, and the pellet was resuspended in sodium dodecyl sulfate sample buffer.
RESULTS
Generation and phenotype of Cdc25 S123A knock-in mice.
We introduced a serine 123-to-alanine (S123A) substitution in the mouse Cdc25A gene (Fig. 1A and B). Genomic Southern blot analysis of ES cell clones exhibited an additional band of predicted size for the mutated allele (Fig. 1C). The neo cassette was removed after the F1 Cdc25A heterozygous mice were crossed with transgenic mice expressing a Cre recombinase under the control of the beta-actin promoter. To facilitate genotyping, PCR was used to detect the presence of endogenous and mutant genes in mice after confirmation by genomic Southern blot analysis. The presence of the alanine substitution was further confirmed by the sequencing of cDNA from wild-type and Cdc25A S123A knock-in (hereafter S123AKI/KI) mice (Fig. 1 D).
Matings between Cdc25A S123A heterozygotes yielded the expected frequency of wild-type, knock-in (Cdc25A S123AKI/KI), and heterozygous (Cdc25A S123AKI/+) offspring, indicating that phosphorylation of Cdc25A at S123 is not required for normal mouse development. Cdc25A S123AKI/KI mice grew to adulthood and displayed no obvious differences from their wild-type littermates.
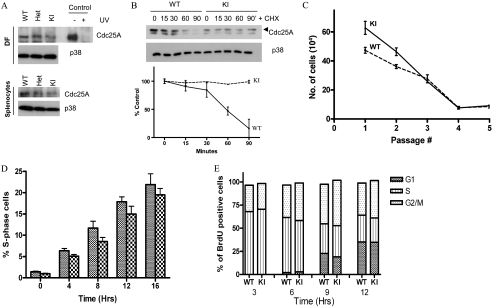
Cdc25A levels were analyzed in DFs, splenocytes, and mouse embryonic fibroblasts (MEFs) derived from wild-type, Cdc25A S123AKI/KI, and Cdc25A S123AKI/+ mice (Fig. 2A; see also Fig. S5A in the supplemental material). We further reconfirmed specificity of Cdc25A antibody using small interfering RNA depletion experiments (see Fig. S6 in the supplemental material). No difference in levels was observed between the genotypes, indicating that Cdc25A phosphorylation at serine 123 does not alter basal levels of Cdc25A phosphatase.
FIG. 2.
Cell cycle analysis of Cdc25A S123A mutant cells. (A) Asynchronously growing DFs or splenocytes from wild-type, heterozygous, or Cdc25 S123A knock-in S123A mice were harvested, and Cdc25A expression was analyzed by Western blotting. Extracts prepared from simian virus 40-transformed MEFs that were either mock-irradiated or irradiated with 40J/m2 UVC are shown as a control. p38 levels were used as a loading control. (B) DFs were treated with cycloheximide (CHX) (25 μg/ml) and harvested at the indicated time points. Cdc25A protein levels were analyzed. Statistical analysis is shown as a graph below. (C) DFs were established from either wild-type or S123A knock-in mice, and the number of cells per dish was determined using a 3T3 passaging protocol. (D) Wild-type and knock-in DFs were serum starved for 96 h. Cells were then incubated in DMEM with serum and BrdU and harvested at the indicated time points. Cells were stained with PI and for BrdU and analyzed by flow cytometry. (E) DFs were pulse labeled with BrdU for 1 h. Cells were harvested at the indicated times and stained with PI and for BrdU and analyzed by flow cytometry WT, wild-type; KI, knock-in; Het, heterozygous.
To determine whether the stability of Cdc25A protein is regulated differently in wild-type versus S123AKI/KI cells, the half-life of Cdc25A protein was measured in DFs (Fig. 2B). Wild-type Cdc25A had a half-life of approximately 60 min whereas Cdc25A S123A exhibited a prolonged half-life of greater than 90 min. This indicates that phosphorylation of S123 is important for turnover of Cdc25A.
Normal cycle progression in Cdc25A S123AKI/KI DFs.
To determine the proliferation capacity of DFs, a 3T3 proliferation assay was carried out (Fig. 2C). Cdc25A S123AKI/KI fibroblasts grew more rapidly than wild-type cells at early passages. However, both wild-type and Cdc25A S123AKI/KI DFs attained similar growth rates by P3, and both cell lines entered replicative senescence by P6. Thus, phosphorylation of Cdc25A S123 is not required for unperturbed cell proliferation.
Next, we analyzed the ability of fibroblasts to progress from G0 into S phase (Fig. 2D). DFs were synchronized by serum starvation. Cells were harvested at various times after the addition of complete medium containing BrdU, stained with PI and anti-BrdU antibody, and analyzed by flow cytometry. BrdU-positive cells were gated to monitor the timing of S phase entry from G0. As shown in Fig. 2D, no significant difference was observed, indicating that the progression from G0 into S phase is not perturbed in the absence of the serine 123 phosphorylation.
We then analyzed the ability of DFs to traverse from S phase through G2/M and into G1 (Fig. 2E). Asynchronously growing DFs were pulsed with BrdU, and cells were harvested at various times after incubation and processed for flow cytometry analysis. At all time points analyzed, no difference was observed between wild-type and Cdc25A S123AKI/KI fibroblasts, indicating that the time required for cells to progress through the cell cycle phases is unaltered.
Normal response to IR in Cdc25A S123AKI/KI cells.
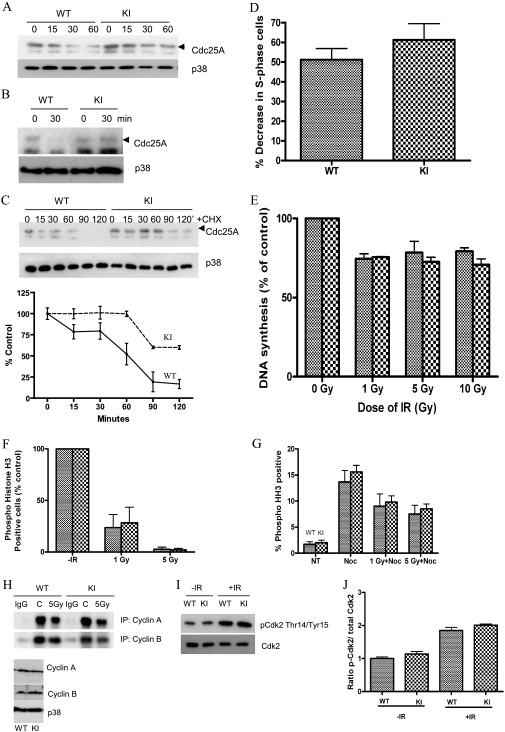
Cells respond to IR by activating checkpoint pathways that prevent cell cycle progression. Cdc25A is a key target of negative regulation by various checkpoint and stress pathways. To investigate whether the S123A mutation stabilizes Cdc25A, DFs were irradiated, and levels of Cdc25A were analyzed either by direct Western blotting or after immunoprecipitation. As shown in Fig. 3A and B, Cdc25A was stabilized in Cdc25A S123AKI/KI fibroblasts compared to wild-type cells, consistent with a previous report that IR-induced destruction of Cdc25A requires phosphorylation of Cdc25A at serine 123 (8, 14).
FIG. 3.
DNA damage-induced checkpoints in Cdc25A S123A mutant cells. (A) DFs were irradiated with 5 Gy of IR, and the Cdc25A level was analyzed by Western blotting at the indicated time points. p38 levels were used as a loading control. (B) DFs were treated as described in panel A, and Cdc25A levels were analyzed after immunoprecipitation with appropriate antibody. (C) DFs were irradiated with 5 Gy of IR followed immediately by the addition of cycloheximide (CHX; 25 μg/ml), and the Cdc25A level was analyzed by Western blotting at the indicated time points. p38 levels were used as a loading control. Statistical analysis is shown as a graph below. (D) Early-passage DFs were serum starved for 96 h. Arrested cells were mock irradiated or gamma irradiated and then incubated in DMEM containing BrdU. Cells were stained for DNA and BrdU and analyzed by flow cytometry. The decrease in the percentage of S phase cells is indicated. (E) DFs were exposed to various doses of IR, and radioresistant DNA synthesis was assessed after 1 h. (F) DFs were mock treated or gamma irradiated, and nocodazole was added 40 min later. Cells were costained for DNA and phospho-histone H3 and analyzed by flow cytometry. The percentages of mitotic cells are shown. (G) DFs were either mock treated or exposed to various doses of IR and incubated in medium containing nocodazole (Noc) for 18 h. Cells were costained for PI and phospho-histone H3 and analyzed by immunofluorescence protocol. NT, no treatment. (H) DFs were either mock treated or irradiated, and cell extracts were prepared after 1 h. Kinase activity associated with cyclin A/B was immunoprecipitated (IP) and assayed using histone H1 as a substrate. Total levels of cyclin A/B are shown in the lower panel. p38 was used as a loading control. (I) Mock-treated (−IR) and IR-treated (+IR) whole protein extracts obtained from DFs were analyzed for phospho-Cdk2/total Cdk2 levels. (J) The intensity ratio of phospho-Cdk2 to total Cdk2 from panel I was quantified and plotted. WT, wild-type; KI, knock-in; p-Cdk2, phospho-Cdk2.
To determine how IR influences the half-life of Cdc25A, DFs were treated with cycloheximide immediately after 5 Gy of irradiation. As shown in Fig. 3C, Cdc25A in wild-type cells had a half-life of approximately 15 min whereas Cdc25A S123A protein exhibited a prolonged half-life of around 90 min. This indicates that serine 123 phosphorylation is required for IR-induced destruction of Cdc25A (8, 14).
Cells arrest in response to IR at various stages of the cell cycle. To monitor the IR-induced G1 phase checkpoint, wild-type and Cdc25A S123AKI/KI DFs were irradiated and 14 h later were pulsed with BrdU for 2 h before flow cytometry analysis. As shown in Fig. 3D, wild-type and Cdc25A S123AKI/KI DFs displayed 51% and 61% reduction, respectively, in the number of S phase cells present relative to nonirradiated control cells. The difference was not statistically significant (P = 0.091), indicating that the G1 phase checkpoint is intact in Cdc25A S123AKI/KI cells.
To determine whether the S phase checkpoint was functional in Cdc25A S123AKI/KI cells, [3H]thymidine incorporation into DNA was monitored 1 h after exposure of cells to various doses of IR (Fig. 3E). No significant difference was observed in the dose-dependent decrease of [3H]thymidine incorporation into newly synthesized DNA, indicating a functional intra-S phase checkpoint in knock-in cells. We next investigated whether the IR-induced G2 phase checkpoint was intact in Cdc25A S123AKI/KI fibroblasts (Fig. 3F). The percentage of cells positive for phospho-histone H3 (a mitotic marker) following exposure to IR was determined. Both wild-type and Cdc25A S123AKI/KI DFs showed a dose-dependent decrease of phospho-histone H3-positive cells, indicative of an intact G2/M checkpoint. Next, in order to check for precocious recovery from a checkpoint, we analyzed the ability of wild-type and Cdc25A S123AKI/KI DFs to exit from an IR-induced G2 arrest. As shown in Fig. 3G, wild-type and knock-in cells exited G2/M arrest with no significant difference between them.
Progression through different phases of the cell cycle is regulated by CDKs, and, in turn, their activity is reduced after DNA damage in a Cdc25A phosphatase-dependent manner (1). To measure CDK activity levels, we next treated wild-type and knock-in cells with 5 Gy of IR, and cyclin A- and B-associated kinases were immunoprecipitated and analyzed in in vitro kinase reactions. As shown in Fig. 3H and Fig. S1 in the supplemental material, wild-type and knock-in cells showed similar decreases in kinase activities after IR. Further, we observed a similar increase in Cdk2 inhibitory phosphorylation in IR-treated knock-in cells compared to wild-type DFs (Fig. 3I and J). Thus, regulation of Cdc25A protein stability is not the only mechanism contributing to regulation of CDK phosphorylation and activity after DNA damage.
Centrosome amplification in Cdc25A S123AKI/KI cells.
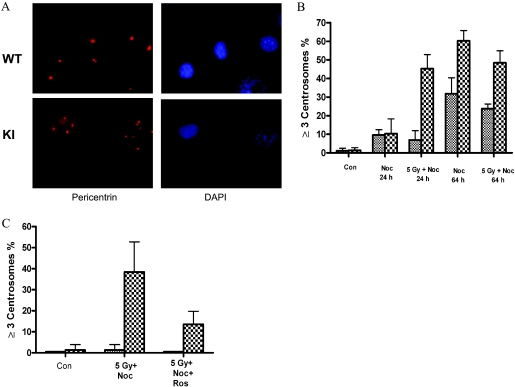
An important observation emerged from our preliminary experiments that mitotic morphology for some nocodazole-treated Cdc25A S123AKI/KI cells appeared normal, but many cells showed a variety of abnormalities in all phases of mitosis (data not shown). As unbalanced chromosomal segregation could be a result of centrosome abnormalities, we next stained cells for the centrosome marker pericentrin. Representative photomicrographs depict centrosome staining in IR-treated wild-type and Cdc25A S123AKI/KI cells (Fig. 4A). As shown in Fig. 4B (see also Fig. S2 in the supplemental material), knock-in DFs display a significant increase in the number of cells with three or more centrosomes after treatment with nocodazole for 64 h. The difference between wild-type and Cdc25A S123AKI/KI cells was even more dramatic when cells were treated with 5 Gy of IR and maintained in the presence of nocodazole for either 24 or 64 h. No difference was observed in fluorescence-activated cell sorter profiles between different genotypes (see Fig. S3 in the supplemental material) except for the appearance of aneuploid cells with nearly 4N DNA content in Cdc25A S123AKI/KI cells (see Fig. S3 in the supplemental material). These data suggest that regulation of Cdc25A stability through S123 phosphorylation is critical to prevent centrosome amplification after IR.
FIG. 4.
Centrosome amplification in Cdc25A S123A cells. (A) Immunofluorescent staining of centrosomes was performed using pericentrin antibody on DFs exposed to 5 Gy of IR followed by incubation in DMEM containing nocodazole for 24 h. DNA was stained with DAPI. (B and C) Cells with three or more centrosome numbers were quantified for different samples as shown in the figure. The panels are representative results of the number of cells counted from four experiments. WT, wild-type; KI, knock-in; Con, control; Noc, nocodazole; Ros, roscovitine.
To understand whether centrosome amplification in knock-in cells requires CDK activity, we next treated both wild-type and knock-in cells with the selective CDK inhibitor roscovitine. While we observed no difference in DNA distribution between different genotypes (see Fig. S4 in the supplemental material), a substantial reduction of Cdc25A S123AKI/KI cells with increased numbers of centrosomes and aneuploid (nearly 4N) DNA were detected in knock-in cells treated with CDK inhibitors (Fig. 4C; also see Fig. S4 in the supplemental material). Thus, CDK activity is required for centrosome amplification in Cdc25A S123AKI/KI cells. Similar results were obtained with another CDK inhibitor (see Fig. S2 in the supplemental material).
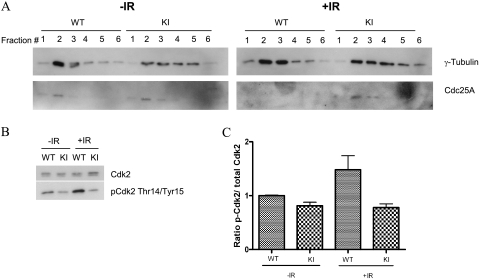
Because we had observed IR-induced centrosome amplification in S123AKI/KI DFs that appeared to be sensitive to CDK inhibitors (Fig. 4; also see Fig. S2 in the supplemental material), we speculated that CDK activities in these cells might be altered specifically at centrosomes. First, we checked whether Cdc25A is present in centrosomes. Centrosomes were isolated from either mock- or IR-treated DFs, and different fractions obtained from a gradient centrifugation were blotted for Cdc25A and the centrosome marker γ-tubulin. We found that Cdc25A colocalized with γ-tubulin and observed maximum expression in fraction 2 (Fig. 5A, left panel). Following IR, Cdc25A levels were reduced and not detectable in centrosome fractions obtained from wild-type cells. However, Cdc25A remained at high levels in centrosomes from Cdc25A S123AKI/KI cells (Fig. 5A, right panel). We then analyzed the inhibitory phosphorylation of Cdk2 in centrosomes as a measure of Cdk2 activity. As shown in Fig. 5B and C, the levels of inhibitory Cdk2 phosphorylation at Thr14 and Tyr15 in centrosome fraction 2 increased approximately twofold in IR-treated wild-type cells while it remained unchanged in Cdc25A S123AKI/KI cells. These data argue that regulation of Cdk2 inhibitory phosphorylation and, thus, activity is fully dependent on the levels of Cdc25A in centrosomes but not in whole-cell extracts (compare Fig. 3I and 5B).
FIG. 5.
Colocalization of Cdc25A on centrosomes. (A) DFs were mock (−IR) or IR (+IR) treated, and centrosomes were purified as described in Materials and Methods. The first six fractions were used for staining of γ-tubulin (a centrosome marker) and Cdc25A. (B) The γ-tubulin-enriched fraction (fraction 2) was further analyzed for phospho-Cdk2/total Cdk2 levels. (C) The intensity ratio of phospho-Cdk2 to total Cdk2 from panel B was quantified and plotted. WT, wild-type; KI, knock-in; p-Cdk2, phospho-Cdk2.
Increased radiation-induced carcinogenesis in Cdc25A S123AKI/KI mice.
Previous studies showed that Cdc25A overexpression can efficiently overcome the growth suppressor activity of various oncogenes and may complement them in the transformation of primary cells (12). Next, we investigated whether primary cells established from wild-type and S123AKI/KI mice would continue to proliferate in the presence of oncogenes and undergo a single oncogene transformation. Early-passage MEFs were infected with retroviruses carrying either E1A or Ras oncogenes, selected in the presence of puromycin for 5 days, and seeded into methylcellulose. We found no colonies in oncogene-expressing MEFs established from either wild-type or S123AKI/KI mice, which was in contrast to p53-deficient cells expressing similar oncogenes (data not shown). Thus, phosphorylation of Cdc25A S123 is not critical in regulating oncogene-induced transformation of rodent primary cells.
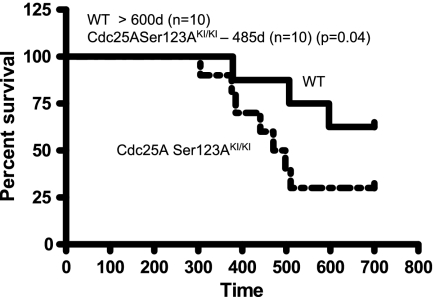
To understand whether the differences in Cdc25A level after IR (Fig. 3A and B) and the number of centrosomes (Fig. 4) in primary knock-in cells could have physiological significance in vivo, Cdc25A S123AKI/+ mice were intercrossed with each other, and wild-type and Cdc25A S123AKI/KI littermates were irradiated once with 4 Gy, a sublethal schedule. Eighteen months after treatment, there was a threefold increase in the frequency of Cdc25A S123AKI/KI mice that succumbed to tumors compared to wild-type mice (Fig. 6). Although some wild-type mice did develop tumors, latency was substantially longer than for Cdc25A S123AKI/KI mice. By 600 days of age, 30% of wild-type mice developed tumors, compared to 80% for Cdc25A S123AKI/KI mice. Histological examination demonstrated that most tumors were lymphomas, and necropsy often revealed large thymic masses, consistent with thymic tumor origin.
FIG. 6.
Mutation of Cdc25A S123 to alanine accelerates IR-induced carcinogenesis. Survival of Cdc25A wild type (WT) and Cdc25A S123AKI/KI mice after irradiation with 4 Gy. The median life span for the different genotypes is shown on top of the panel. d, days.
DISCUSSION
Serine 123 was proposed as a key phosphorylation site in regulation of Cdc25A stability after IR and, thus, cell cycle progression (8). Here, we found that while S123 was critical for regulating Cdc25A stability (Fig. 3A to C), the cell cycle profiles and responses to IR were similar between wild-type and S123A knock-in cells (Fig. 3D to G). Depletion of Cdc25A levels is sufficient for cell cycle arrest after DNA damage, which is supported by Cdc25A RNA interference experiments (30). However, under conditions in which Cdc25A levels remain unchanged after stress, as in S123A knock-in cells or after treatment with low doses of UVC light (16), additional mechanisms are essential for checkpoint activation. Such mechanisms may include the inhibition of Cdc25A's interaction with various CDK-cyclin complexes, which was shown to be required for full checkpoint activation (5, 14). Several phosphorylation sites have been identified on Cdc25A, including threonine 507, which has been shown to regulate Cdc25A binding with Cdk1 complex and play an important role in controlling cell cycle progression (5).
Although normal cell cycle progression and DNA damage-induced checkpoints are not perturbed in Cdc25A S123AKI/KI cells, S123 phosphorylation and Cdc25A stability appear critical for centrosome duplication under certain conditions (Fig. 4). By regulating Cdc25 activity, cells regulate CDK activity, and, in turn, CDK-associated kinase activity is required for initiation of centrosome duplication and ensures that centrosomes are duplicated once within the normal cell cycle (17, 20, 23). Coupling of centrosome duplication and DNA replication cycles is crucial to prevent multiple rounds of centrosome duplication within a single DNA replication cycle. Uncoupling of these two processes, as in Cdc25A S123AKI/KI cells (Fig. 4), is one mechanism by which generation of supernumerary centrosomes may arise (7, 27), ultimately leading to increased tumorigenesis (Fig. 6). As CDK activity is required for centrosome duplication in IR-treated Cdc25A S123AKI/KI cells (Fig. 4C and 5B; see also Fig. S4 in the supplemental material), the question remaining to be answered is why the regulation of Cdc25A stability is sufficient to control CDK activity at centrosomes but not in the nucleus. One potential explanation comes from the fact that alternative pathways (through either Myt1 or Wee1 kinases), which are activated by Chk1/2 kinases after DNA damage, may not be operational at centrosomes. As a result, DNA damage-induced inactivation of CDKs in the nucleus is achieved through targeting both Cdc25 and Wee1/Myt1 but can only take place via inactivation of Cdc25 phosphatases at centrosomes. This idea is further supported by our findings that the level of inhibitory phosphorylation of Cdk2 in centrosomes is not increased after IR in Cdc25A S123AKI/KI cells (Fig. 5B and C). While we cannot completely rule out other mechanisms by which Cdc25A phosphatase regulates the centrosome multiplication observed in our experiments, it is possible that regulation of CDK activity at centrosomes fully relies on Cdc25, which is in turn dependent on the protein stability provided by regulation of Cdc25A S123 phosphorylation after IR.
Increasing evidence supports a role for centrosome amplification in cancer. A recent report in Drosophila demonstrated a direct link between centrosome amplification and tumorigenesis (2). It was revealed that in the presence of extra centrosomes, the asymmetric division of the larval neural stem cells was compromised, which led to metastatic tumors when cells were transplanted into the abdomens of wild-type hosts. While the connection between centrosome amplification and tumorigenesis in mammals requires further experimental support, our results with increased IR-induced tumorigenesis in Cdc25A S123AKI/KI mice (Fig. 6) support this connection and provide a new in vivo genetic mouse model for analysis of the role of centrosome amplification in tumorigenesis.
Supplementary Material
Acknowledgments
This work was supported by the Agency for Science, Technology and Research, Singapore.
We thank Helen Piwnica-Worms for providing Cdc25A antibody and Oleg Demidov for his help with Cdc25A analysis.
Footnotes
Published ahead of print on 20 October 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aleem, E., H. Kiyokawa, and P. Kaldis. 2005. Cdc2-cyclin E complexes regulate the G1/S phase transition. Nat. Cell Biol. 7831-836. [DOI] [PubMed] [Google Scholar]
- 2.Basto, R., K. Brunk, T. Vinadogrova, N. Peel, A. Franz, A. Khodjakov, and J. W. Raff. 2008. Centrosome amplification can initiate tumorigenesis in flies. Cell 1331032-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernardi, R., D. A. Liebermann, and B. Hoffman. 2000. Cdc25A stability is controlled by the ubiquitin-proteasome pathway during cell cycle progression and terminal differentiation. Oncogene 192447-2454. [DOI] [PubMed] [Google Scholar]
- 4.Blomberg, I., and I. Hoffmann. 1999. Ectopic expression of Cdc25A accelerates the G1/S transition and leads to premature activation of cyclin E- and cyclin A-dependent kinases. Mol. Cell. Biol. 196183-6194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, M. S., C. E. Ryan, and H. Piwnica-Worms. 2003. Chk1 kinase negatively regulates mitotic function of Cdc25A phosphatase through 14-3-3 binding. Mol. Cell. Biol. 237488-7497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conklin, D. S., K. Galaktionov, and D. Beach. 1995. 14-3-3 proteins associate with cdc25 phosphatases. Proc. Natl. Acad. Sci. USA 927892-7896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doxsey, S. 2002. Duplicating dangerously: linking centrosome duplication and aneuploidy. Mol. Cell 10439-440. [DOI] [PubMed] [Google Scholar]
- 8.Falck, J., N. Mailand, R. G. Syljuasen, J. Bartek, and J. Lukas. 2001. The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature 410842-847. [DOI] [PubMed] [Google Scholar]
- 9.Gabrielli, B. G., C. P. De Souza, I. D. Tonks, J. M. Clark, N. K. Hayward, and K. A. Ellem. 1996. Cytoplasmic accumulation of cdc25B phosphatase in mitosis triggers centrosomal microtubule nucleation in HeLa cells. J. Cell Sci. 1091081-1093. [DOI] [PubMed] [Google Scholar]
- 10.Galaktionov, K., and D. Beach. 1991. Specific activation of cdc25 tyrosine phosphatases by B-type cyclins: evidence for multiple roles of mitotic cyclins. Cell 671181-1194. [DOI] [PubMed] [Google Scholar]
- 11.Galaktionov, K., X. Chen, and D. Beach. 1996. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature 382511-517. [DOI] [PubMed] [Google Scholar]
- 12.Galaktionov, K., A. K. Lee, J. Eckstein, G. Draetta, J. Meckler, M. Loda, and D. Beach. 1995. CDC25 phosphatases as potential human oncogenes. Science 2691575-1577. [DOI] [PubMed] [Google Scholar]
- 13.Gasparotto, D., R. Maestro, S. Piccinin, T. Vukosavljevic, L. Barzan, S. Sulfaro, and M. Boiocchi. 1997. Overexpression of CDC25A and CDC25B in head and neck cancers. Cancer Res. 572366-2368. [PubMed] [Google Scholar]
- 14.Goloudina, A., H. Yamaguchi, D. B. Chervyakova, E. Appella, A. J. Fornace, Jr., and D. V. Bulavin. 2003. Regulation of human Cdc25A stability by Serine 75 phosphorylation is not sufficient to activate a S phase checkpoint. Cell Cycle 2473-478. [PubMed] [Google Scholar]
- 15.Hassepass, I., R. Voit, and I. Hoffmann. 2003. Phosphorylation at serine 75 is required for UV-mediated degradation of human Cdc25A phosphatase at the S-phase checkpoint. J. Biol. Chem. 27829824-29829. [DOI] [PubMed] [Google Scholar]
- 16.Heffernan, T. P., K. Unsal-Kaçmaz, A. N. Heinloth, D. A. Simpson, R. S. Paules, A. Sancar, M. Cordeiro-Stone, and W. K. Kaufmann. 2007. Cdc7-Dbf4 and the human S checkpoint response to UVC. J. Biol. Chem. 2829458-9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hinchcliffe, E. H., C. Li, E. A. Thompson, J. L. Maller, and G. Sluder. 1999. Requirement of Cdk2-cyclin E activity for repeated centrosome reproduction in Xenopus egg extracts. Science 283851-854. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann, I., G. Draetta, and E. Karsenti. 1994. Activation of the phosphatase activity of human cdc25A by a cdk2-cyclin E dependent phosphorylation at the G1/S transition. EMBO J. 134302-4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jinno, S., K. Suto, A. Nagata, M. Igarashi, Y. Kanaoka, H. Nojima, and H. Okayama. 1994. Cdc25A is a novel phosphatase functioning early in the cell cycle. EMBO J. 131549-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lacey, K. R., P. K. Jackson, and T. Stearns. 1999. Cyclin-dependent kinase control of centrosome duplication. Proc. Natl. Acad. Sci. USA 962817-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lammer, C., S. Wagerer, R. Saffrich, D. Mertens, W. Ansorge, and I. Hoffmann. 1998. The cdc25B phosphatase is essential for the G2/M phase transition in human cells. J. Cell Sci. 1112445-2453. [DOI] [PubMed] [Google Scholar]
- 22.Mailand, N., J. Falck, C. Lukas, R. G. Syljuasen, M. Welcker, J. Bartek, and J. Lukas. 2000. Rapid destruction of human Cdc25A in response to DNA damage. Science 2881425-1429. [DOI] [PubMed] [Google Scholar]
- 23.Meraldi, P., J. Lukas, A. M. Fry, J. Bartek, and E. A. Nigg. 1999. Centrosome duplication in mammalian somatic cells requires E2F and Cdk2-cyclin A. Nat. Cell Biol. 188-93. [DOI] [PubMed] [Google Scholar]
- 24.Molinari, M., C. Mercurio, J. Dominguez, F. Goubin, and G. F. Draetta. 2000. Human Cdc25A inactivation in response to S phase inhibition and its role in preventing premature mitosis. EMBO Rep. 171-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadhu, K., S. I. Reed, H. Richardson, and P. Russell. 1990. Human homolog of fission yeast cdc25 mitotic inducer is predominantly expressed in G2. Proc. Natl. Acad. Sci. USA 875139-5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saha, P., Q. Eichbaum, E. D. Silberman, B. J. Mayer, and A. Dutta. 1997. p21CIP1 and Cdc25A: competition between an inhibitor and an activator of cyclin-dependent kinases. Mol. Cell. Biol. 174338-4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sluder, G., and J. J. Nordberg. 2004. The good, the bad and the ugly: the practical consequences of centrosome amplification. Curr. Opin. Cell Biol. 1649-54. [DOI] [PubMed] [Google Scholar]
- 28.Vigo, E., H. Muller, E. Prosperini, G. Hateboer, P. Cartwright, M. C. Moroni, and K. Helin. 1999. CDC25A phosphatase is a target of E2F and is required for efficient E2F-induced S phase. Mol. Cell. Biol. 196379-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu, X., and S. P. Burke. 1996. Roles of active site residues and the NH2-terminal domain in the catalysis and substrate binding of human Cdc25. J. Biol. Chem. 2715118-5124. [DOI] [PubMed] [Google Scholar]
- 30.Zhao, H., J. L. Watkins, and H. Piwnica-Worms. 2002. Disruption of the checkpoint kinase 1/cell division cycle 25A pathway abrogates ionizing radiation-induced S and G2 checkpoints. Proc. Natl. Acad. Sci. USA 9914795-14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.