Abstract
Bronchial epithelial cells play a pivotal role in airway inflammation, but little is known about posttranscriptional regulation of mediator gene expression during the inflammatory response in these cells. Here, we show that activation of human bronchial epithelial BEAS-2B cells by proinflammatory cytokines interleukin-4 (IL-4) and tumor necrosis factor alpha (TNF-α) leads to an increase in the mRNA stability of the key chemokines monocyte chemotactic protein 1 and IL-8, an elevation of the global translation rate, an increase in the levels of several proteins critical for translation, and a reduction of microRNA-mediated translational repression. Moreover, using the BEAS-2B cell system and a mouse model, we found that RNA processing bodies (P bodies), cytoplasmic domains linked to storage and/or degradation of translationally silenced mRNAs, are significantly reduced in activated bronchial epithelial cells, suggesting a physiological role for P bodies in airway inflammation. Our study reveals an orchestrated change among posttranscriptional mechanisms, which help sustain high levels of inflammatory mediator production in bronchial epithelium during the pathogenesis of inflammatory airway diseases.
Persistent inflammation of the respiratory tract in chronic pulmonary disease is mediated by increased expression of multiple inflammatory mediators (reviewed, e.g., in references 3 and 4). During allergic airway inflammation, the Th2 cell-associated cytokines (e.g., interleukin-4 [IL-4] and IL-13) strongly synergize with other proinflammatory cytokines, such as tumor necrosis factor alpha (TNF-α), to stimulate production of eosinophil- and Th2-specific chemokines in epithelial cells, endothelial cells, and tissue macrophages (2, 3, 67). As chemokines help shape the manner by which the immune system responds to inflammatory stimuli (59), increased chemokine expression could sustain an inflammatory reaction well beyond its original purpose, thereby leading to chronic inflammation (reviewed in, e.g., references 23 and 59).
The bronchial airway epithelium is directly exposed to the environment and is the first line of defense against airborne particulate matter, allergens, and infectious agents; it plays a critical role not only in the maintenance of physicochemical homoeostasis of the airways but also in the pathogenesis of airway diseases. During allergic airway inflammation the epithelium is both a source of mediator production as well as a target of remodeling processes (31, 61). Recent findings further support the view that the epithelium also plays a central role in the Th2 cell sensitization process by influencing the function of antigen-presenting dendritic cells in the airways (for recent reviews, see references 3 and 61). Thus, understanding the biochemical and molecular basis on which the expression of inflammatory mediators is regulated in the bronchial epithelium will provide important new insight into inflammatory airway diseases and identify novel therapeutic targets.
Previous studies have revealed a wealth of knowledge about the complex transcriptional programs that regulate the expression of genes coding for various mediators of inflammation (13, 28, 30, 56). However, relatively little is known about the mechanisms controlling the expression of these genes at posttranscriptional levels in inflammation. In this study we used the human bronchial epithelial BEAS-2B cell line stimulated with proinflammatory cytokines IL-4 and TNF-α as a cellular model to investigate gene regulation at the level of mRNA turnover and translation in activated bronchial epithelial cells. Previously, it was shown that the stability of a chemokine mRNA coding for eotaxin-1 (or CCL11) in BEAS-2B cells increased when the cells were treated with both IL-4 and TNF-α (2), suggesting the feasibility of using this in vitro cellular model system to study gene regulation relevant to airway inflammation. This approach is further substantiated by our current genome-wide microarray analysis. Using this system, we found that both mRNA and protein levels of key chemokines such as monocyte chemotactic protein 1 (MCP-1) and IL-8 increased upon stimulation of BEAS-2B cells and that their mRNAs were more stable in the activated cells. In addition, an enhancement of the global translation rate in activated bronchial epithelial cells was observed. Moreover, our data show that IL-4/TNF-α stimulation of BEAS-2B cells led to an elevation of the level of translation factors critical for enhancing translation efficiency accompanied by a reduction of the translational silencing mediated by microRNAs (miRNA). This group of evolutionarily conserved and abundant small noncoding RNAs downregulate gene expression across species and in multiple biological processes through base-pairing to the 3′ untranslated regions (UTRs) of target mRNAs (reviewed in, e.g., references 21 and 34).
Recently, a newly identified cytoplasmic domain, termed RNA processing body (P body), was found to be a site for storage and/or degradation of translationally silenced mRNAs including miRNA-silenced mRNPs (17, 37, 49). Using both the BEAS-2B cell model and an in vivo mouse model of allergic airway inflammation, we show that P-body formation is significantly reduced in activated bronchial epithelial cells. These observations are consistent with our data showing that mRNA degradation and translational repression are compromised during allergic airway inflammation. To our knowledge, this finding represents the first demonstration of a physiologically relevant regulation of mammalian P bodies observed in vivo. Collectively, this study reveals a dynamic linkage among posttranscriptional mechanisms that facilitate and fine-tune a cell's response to external influences.
MATERIALS AND METHODS
Plasmids.
To construct the plasmids pTet-BBB+ARE(IL-8) (where Tet is tetracycline and BBB is β-globin) and pTet-BBB+ARE(MCP-1), AU-rich element (ARE) sequences of IL-8 and MCP-1 were amplified by PCR using genomic DNA extracted from BEAS-2B cells as a template and then cloned into the unique BglII site of plasmid pTet-BBB (11). The IL-8 ARE sequence is from positions 998 to 1199 in human IL-8 cDNA (NM_000584), and the MCP-1 (CCL2) ARE sequence is from positions 441 to 653 in human CCL2 (NM_002982). Construction of plasmid pSVα1/GAPDH (where GAPDH is glyceraldehyde-3-phosphate dehydrogenase) has been described previously (8). Plasmids expressing Renilla luciferase mRNA and its derivatives carrying three let-7 miRNA target sites, three mutated nonfunctional let-7 target sites, or a single site that forms a perfect match with let-7 miRNAs (54) were kindly provided by W. Filipowicz and are referred to here as RL, RL(let-7wt), RL(let-7mut), and RL(let-7per), respectively. To create RL+miR-29a and RL+miR-155, sequences encoding three copies of the miR-155 or miR-29a target site (searched by miRBase and TargetScan) were synthesized and inserted between the XbaI and NotI sites of the Renilla luciferase reporter gene.
Cell culture and transfection.
The human bronchial epithelial BEAS-2B cell line was purchased from ATCC and cultured in F12-Dulbecco's modified Eagle's medium containing heat-inactivated fetal calf serum (5%), l-glutamine (2 mM), penicillin (100 μg/ml), and streptomycin (100 mg/ml). For IL-4/TNF-α stimulation, cells were cultured for 18 h in medium containing 50 ng/ml of each cytokine.
For transient transfection experiments, a tTA-expressing BEAS-2B-19 stable cell line (12) that supports transcription of genes driven by the Tet-off or Tet-on promoter was first seeded in six-well plates at a density of 0.25 × 106 to 0.3 × 106 cells per well for 1 day and then transfected with pTet-BBB or pTet-BBB+ARE plasmids with a control plasmid, pSVα1/GAPDH (8), using FuGENE 6 (Roche). Time course experiments using the Tet-off system for transcriptional pulsing and Northern blot analysis to determine mRNA stability were performed as described previously (12).
RNA extraction and analysis.
Total RNA was extracted using Trizol (Invitrogen) or RNeasy Mini Kit (Qiagen) following the manufacturer's protocols. For real-time quantitative reverse transcription-PCR (RT-PCR), 250 to 500 ng of RNA was used in a 10-μl RT reaction mixture containing 1 unit of SuperScript II reverse transcriptase (Invitrogen). The reaction was incubated at 50°C for 30 min, followed by heat inactivation at 72°C for 5 min. After RT, PCRs containing a 1:20,000 dilution of Sybr Green (Cambrex Bio Science Rockland, Inc.) and 0.1 U/μl JumpStart Taq DNA polymerase (Sigma-Aldrich) were performed according to the manufacturer's protocol (Eppendorf) using an Eppendorf Mastercycler ep realplex quantitative PCR machine. The reactions were performed as follows: 94°C for 2 min; 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; followed by 72°C for 5 min and a default melting curve programmed by the instrument.
Analyses of polysome profiles by sucrose gradient fractionation were performed as described previously (9). Briefly, cytoplasmic lysates prepared from approximately 5 × 107 nonstimulated or IL-4/TNF-α stimulated BEAS-2B cells were overlaid on a continuous sucrose gradient (15% to 40%). The gradient was centrifuged using a Beckman SW41 rotor at 38,000 rpm for 2 h with medium acceleration and no brake. Immediately after centrifugation, 500-μl aliquots were taken from top to bottom of the gradient, with 20 aliquots prepared in total. RNA was extracted from each fraction and used for electrophoresis in 1% agarose to visualize the distributions of 5.5S, 18S, and 28S rRNA bands.
Illumina microarrays.
Triplicate samples of total cytoplasmic mRNA were prepared from nonstimulated or IL-4/TNF-α-stimulated BEAS-2B cells. RNA amplification and microarray hybridization were carried out in the Microarray Core Laboratory at the University of Texas Health Science Center (Houston, TX) using Illumina Genome-Wide Expression BeadChips (Human Ref-6 v1; Illumina Inc.), representing ∼43,000 human transcripts (40). Two hundred nanograms of total RNA was reverse transcribed and amplified overnight with T7 RNA polymerase and labeled with biotin following the manufacturer's protocol. Biotin-labeled amplified RNA (1.5μg) was hybridized to the microarrays at 58°C overnight. Arrays were incubated with Cy3-streptavidin and washed according to the manufacturer's instructions. Initial analysis of the microarray data was done using Illumina's Beadstudio, version 3. After background subtraction, arrays were normalized to each other by rank-invariant normalization. Changes in gene expression were tested using a modified t test, which employs estimates of variation that include sequence-specific biological variation (σbio), nonspecific biological variation (σneg), and technical error (σtech) as described in the user's guide (Illumina Inc.).
Western blot analysis.
Cytoplasmic or total lysates were prepared as described previously (52). Cell lysates (5 to 40 μg) were resolved on an 8% or 15% sodium dodecyl sulfate-polyacrylamide gel and analyzed using an ECL Western blotting kit (Bio-Rad). The polyvinylidene difluoride blots were probed with specific antibodies as indicated in each figure and detected with SuperSignal West Pico Chemiluminescent Substrate (Pierce). Membranes were incubated with one of the primary antibodies at the following dilutions: anti-eukaryotic initiation factor 4E (eIF4E) (Cell Signaling Technology) at 1:500, anti-4E binding protein 1 (4E-BP1) phosphorylated at Ser 65 (Cell Signaling Technology) at 1: 2,000, anti-termination release factor 1 (eRF1; Abcam) at 1:500, anti-poly(A)-binding protein (PABP; a gift from R. Lloyd) at 1:3,000, anti-Dcp1a (a gift from S. Ohno) at 1:4,000, anti-Rck/p54 (Bethyl) at 1:3,000, and anti-α-tubulin monoclonal antibody (Sigma) at 1:10,000. The fluorescence signals and intensity of detected bands were captured and quantified by an imaging system (Syngene) and the software GeneTools (Syngene), respectively.
Global translational rate measurement.
To pulse label proteins with [35S]methionine in BEAS-2B cells, 0.43 × 106 cells were first seeded in each 6-cm plate. Twenty-four hours later, cells were either left untreated or stimulated by IL-4/TNF-α for 18 h. Both nonstimulated and stimulated cells were incubated for 1 h with 2 ml of Gibco's methionine- and cysteine-free Dulbecco's modified Eagle's medium containing fetal bovine serum (also containing IL-4/TNF-α for stimulated cells). The medium was then replaced with fresh medium containing 100 μCi of 35S labeling mix (Perkin Elmer). After 0, 30, and 60 min of labeling, cells were harvested and lysed for trichloroacetic acid precipitation to quantitate incorporation of [35S]methionine and cysteine. The experiment was performed in triplicate.
Quantification of cytokine and chemokine proteins.
Equivalent numbers of BEAS-2B cells without or with IL-4/TNF-α stimulation were lysed in cytoplasmic lysis buffer (10 mM HEPES [pH 7.6], 3 mM MgCl2, 40 mM KCl, 5% glycerol, 0.5% NP-40, 1× protease inhibitor cocktail, 1 mM dithiothreitol), and nuclei were removed by centrifugation. The protein concentration of the supernatant (cytoplasmic lysate) was adjusted to 0.5 mg/ml. Fifty microliters of each cytoplasmic lysate was used for cytokine and chemokine detection and quantification by a Quantibody Human Inflammation Array I kit (RayBiotech, Inc.) according to the manufacturer's protocol. The signals (green fluorescence, Cy3 channel, 555-nm excitation, and 565-nm emission) were captured using an Axon GenePix laser scanner (the Microarray Core Laboratory at the University of Texas Health Science Center, Houston, TX). Quantitative data analysis was performed using RayBio Q Analyzer software (RayBiotech, Inc.).
Renilla and firefly dual luciferase assay.
BEAS-2B cells were seeded (0.3 × 106 cells/6-cm plate) 24 h before transfection. For each plate, 0.02 ng of Renilla luciferase plasmid, 4 ng of firefly luciferase plasmid (pUHC13), and 2.2 μg of pT18 plasmid were transfected using FuGENE 6 transfection reagent (Roche) following the manufacturer's instructions. The next day, half of the plates were stimulated by 50 ng/ml TNF-α/IL-4 for 18 h. Both nonstimulated and stimulated samples were harvested 42 h after transfection. The pellet was prepared for luciferase assay using a Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's protocol.
Immunofluorescence microscopy.
BEAS-2B cells in slide chambers were fixed with 3.7% paraformaldehyde and permeabilized with methanol and 0.5% Triton X-100. Following incubation with rabbit anti-human Dcp1a antibody (1:500 dilution), fluorescent donkey anti-rabbit antibody, coupled to Alexa 555 (Invitrogen) was used for secondary detection at 1:1,000 dilution. Fluorescent mounting medium containing DAPI (4′,6′-diamidino-2-phenylindole) was applied to the slides before the placement of coverslip and microscopy. Images were obtained at room temperature by optical z-sectioning (20 sections in total with a 0.2-μm space between sections) using an Olympus objective lens (100× with a numerical aperture of 1.35) of a DeltaVision deconvolution microscope system containing an Olympus IX70 inverted microscope.
Ovalbumin sensitization and challenge.
Six-week old BALB/c female mice were sensitized by an intraperitoneal injection of 500 μl of solution containing 8 mg/ml ovalbumin (Sigma-Aldrich) and 8 mg/ml alum (Sigma-Aldrich) on days 0, 7, and 14 (75). Mice were challenged intranasally with 30 ml of saline or ovalbumin (8 mg/ml) on days 14, 21, 22, and 23 and were sacrificed 24 h after the last installation. The lungs were processed for analysis of lavage cellularity and then infused with 10% buffered formalin and processed for paraffin embedding.
Dcp1a and MCP-1 immunodetection in mouse bronchial epithelium.
Lung sections (5 μm) were deparaffinized and processed for Dcp1a immunofluorescence. Sections were blocked in 1% bovine serum albumin and incubated for 1 h at room temperature with a 1:500 dilution of rabbit anti-human Dcp1a antibody. This was followed by incubation with a 1:1,000 dilution of donkey anti-rabbit secondary antibody coupled to Alexa 555 (Invitrogen). Coverslips were mounted using mounting medium containing DAPI to visualize nuclei. Quantitation of P bodies was determined by counting the number of bronchial epithelial cells per millimeter of airway basement membrane that contained 20 or more P bodies (P-body index) using ImagePro Plus software (Media Cybernetics). At least three airways from each of four different mice per group were analyzed.
For MCP-1 immunoperoxidase detection, 5-μm lung sections were deparaffinized and quenched with 0.5% hydrogen peroxide, and antigen retrieval was performed (Dako). Slides were then incubated overnight at 4°C with a 1:600 dilution of goat anti-mouse MCP-1 antibody (R&D Systems). A rabbit anti-goat Elite detection system (Vector Laboratories) was used in conjunction with 3,3′-diaminobenzidine peroxidase detection (Sigma) and counterstaining with methyl green.
Microarray data accession number.
The raw microarray data without normalization has been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/projects/geo/) under accession number GSE13119.
RESULTS
BEAS-2B cell model for the study of posttranscriptional regulation of genes related to airway inflammation.
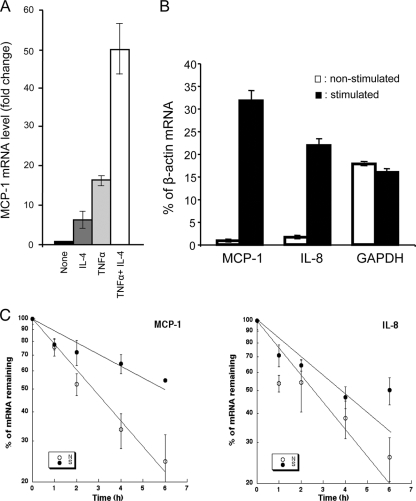
We first tested whether IL-4, a Th2 cell-associated cytokine, and TNF-α, a key proinflammatory cytokine, act synergistically in BEAS-2B cells to upregulate genes preferentially expressed during Th2 responses. We examined the level of the mRNA encoding a key chemokine, MCP-1 (42, 47), in cells treated with either IL-4 or TNF-α alone or with both. The results of real-time RT-PCR quantification show that treatment with either IL-4 or TNF-α alone increases MCP-1 mRNA to a level only 35% of that produced by treatment with both TNF-α and IL-4 together (Fig. 1A). Importantly, while the 18-h treatment with both IL-4 and TNF-α activates BEAS-2B cells effectively, it does not produce any significant apoptosis of the cells (data not shown).
FIG. 1.
Characterizing the response of BEAS-2B cells to IL-4 and TNF-α stimulation at the mRNA level and the effect of IL-4/TNF-α stimulation on mRNA stability. (A) Synergistic effect of IL-4 and TNF-α on induction of MCP-1 mRNA expression in BEAS-2B cells. mRNA levels were quantified by real-time RT-PCR. The levels of MCP-1 mRNA in cells treated with IL-4, TNF-α, or both in comparison to the MCP-1 mRNA levels in cells without cytokine treatment (None; assigned a value of 1) are shown. (B) Bar graph showing that the levels of MCP-1, IL-8 mRNAs, but not the control GAPDH message are increased in IL-4/TNF-α stimulated BEAS-2B cells. The levels of indicated cytoplasmic mRNAs in either nonstimulated or IL-4/TNF-α-stimulated BEAS-2B cells were quantified by real-time RT-PCR and normalized to β-actin mRNA (percentage of β-actin). (C) Stabilization effect of IL-4/TNF-α stimulation on chemokine mRNA decay. BEAS-2B cells, either nonstimulated (N) or stimulated (S) with IL-4/TNF-α (50 ng/ml each) for 18 h, were treated with ActD (5 mg/ml) to block transcription. Cells were harvested immediately (time zero) or after 1, 2, 4, or 6 h of ActD treatment. The levels of IL-8 and MCP-1 mRNAs were quantified by real-time RT-PCR and normalized by the amount of GAPDH mRNA. Decay curves shown in the semilog plots were obtained by least squares analysis of the percentage of mRNA remaining as a function of time. All data represent the mean of three experiments ± standard errors.
The changes in mRNA level in BEAS-2B cells upon 18-h IL-4/TNF-α stimulation were examined by microarray mRNA profiling with high-density oligonucleotide arrays using mRNA samples prepared from control and stimulated cells. We identified 112 genes whose mRNA levels increased at least fourfold (P < 0.01; n = 3) following stimulation, and 41 of them are directly related to airway inflammation and remodeling (Table 1). In addition to these 41 genes, the mRNA levels of another 9 genes directly related to airway inflammation and remodeling were increased two- to threefold in the stimulated cells (Table 1). These include genes coding for many key cytokines and chemokines and genes encoding factors involved in dendritic cell recruitment and activation, recruitment of innate immune cells, promotion of Th2 cell polarization, or perpetuation of epithelium changes related to asthma, fibrosis, and tissue remodeling. Importantly, many of these genes are also induced in bronchial epithelium following various physiological or pathological stimuli that occur during allergic inflammatory responses characteristic of asthma (29, 32). Taken together, these observations indicate that BEAS-2B cells not only maintain expression of many genes directly related to airway inflammation and remodeling but also respond to IL-4/TNF-α treatment by upregulating these genes, thereby supporting the use of this cellular model system to study the regulation of gene expression relevant to airway inflammation.
TABLE 1.
mRNAs upregulated in activated BEAS-2B cells
| Gene symbol(s) or product(s)a | Fold increase (P < 0.01)b | Effect(s) related to airway allergic inflammation |
|---|---|---|
| CCL2 (MCP-1) | 97 | Chemotactic effects; recruitment of dendritic cells and other innate immune cells; eosinophilia |
| CCL26 (Eotaxin-3) | 67 | |
| CXCL1 (Gro-α) | 23 | |
| CXCL6 (GCP-2) | 13 | |
| CXCL8 (IL-8) | 242 | |
| IL-1α | 282 | Proinflammatory and proallergic cytokines; dendritic cell recruitment and activation |
| IL-1β | 18 | |
| IL-6 | 22 | |
| IL-32 | 44 | |
| CSF2 (GM-CSF) | 35 | |
| VEGFC | 3 | |
| C3 | 46 | Recruitment of eosinophils and other; innate immune cells |
| ICAM1 | 23 | |
| CFB | 7 | |
| OLR1 | 6 | |
| VACM1 | 3 | |
| IL-4Rα | 2 | Promote Th2 cell polarization and perpetuate epithelium changes related to asthma; neutrophil recruitment and lung fibrosis |
| IL-17Rα | 4 | |
| CD82 | 6 | |
| SLCO2A1 | 9 | |
| ALOX5AP | 4 | |
| CD83 | 4 | |
| CLDN1 | 12 | Epithelium permeability and dendritic cell activation |
| Claudin11 | 3 | |
| HBEGF | 4 | Tissue remodeling |
| COL6A1 | 16 | |
| COL6A2 | 9 | |
| Serpins (E2, E1, B8, A3, B2) | 2-37 | |
| SOD2 | 18 | |
| PAPPA | 9 | |
| TFPI2 | 8 | |
| ADORA2A | 8 | |
| LOX | 5 | |
| CITED4 | 3 | |
| NFKBIA | 10 | Inflammation/immune response-related transcription factors |
| NFKBIE | 4 | |
| STAT5A | 4 | |
| TNFAIP3 | 27 | TNF-α-inducible proteins or signaling proteins that contribute to TNF-α effects |
| BIRC3 | 26 | |
| PTX3 (TNFAIP5) | 21 | |
| IRAK2 | 11 | |
| NUAK2 | 8 | |
| TNFAIP2 | 7 | |
| TNFAIP4 (Ephrin A1) | 6 | |
| TRAF1 | 3 |
Abbreviations: GM-CSF, granulocyte-macrophage colony-stimulating factor; VEGFC, vascular endothelial growth factor C; IL-4Rα, IL-4 receptor α; IL-17Rα, IL-17 receptor α; SOD2, superoxide dismutase 2; HBEFG, heparin-binding epidermal growth factor; COL6A1 and COL6A2, collagen genes; NFKBIA, NF-κB inhibitor α; NFKBIE, NF-κB inhibitor ε.
Relative to cells not stimulated with IL-4/TNF-α.
Stability of MCP-1 and IL-8 mRNAs is increased in activated BEAS-2B cells.
To determine whether mRNA stability may contribute to the elevation of the mRNA levels of the mediator genes that are upregulated in activated bronchial epithelial cells, we examined the mRNA decay of two key chemokines in airway inflammation, MCP-1 and IL-8, in nonstimulated and IL-4/TNF-α stimulated BEAS-2B cells. We first performed real-time RT-PCR analysis and validated that the mRNA levels of MCP-1 and IL-8 are increased in IL-4/TNF-α-stimulated BEAS-2B cells (Fig. 1B). In contrast, the level of the control GAPDH transcript remains similar in the cells with or without stimulation (Fig. 1B). To determine the stability of MCP-1 and IL-8 mRNAs, cells were treated with the transcription inhibitor actinomycin D (ActD), and RNA samples were extracted and analyzed at various time points. The results (Fig. 1C) show that IL-4/TNF-α treatment slowed the decay of these mRNAs. The half-lives (i.e., the time when 50% of mRNA remained) of MCP-1 and IL-8 mRNAs increased from ∼3 h to ∼6 h and from ∼2.5 h to ∼4 h, respectively. The twofold increase in the mRNA stability is physiologically significant because even a moderate change in mRNA half-life can greatly affect message levels (58). These findings, together with the observation that eotaxin-1 mRNA is also stabilized in activated BEAS-2B cells (2), support a critical role of mRNA turnover in the upregulation of chemokine gene expression during airway inflammation and remodeling.
IL-4/TNF-α stimulation has little effect on the mRNA destabilizing function of MCP-1 and IL-8 AREs in BEAS-2B cells.
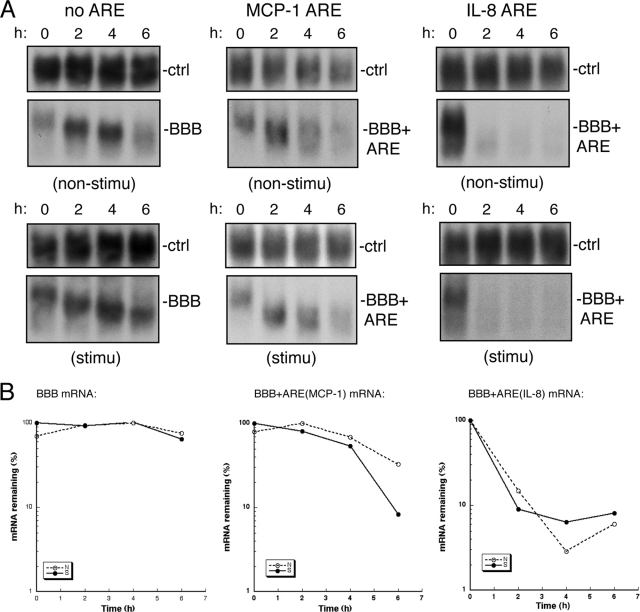
Most cytokine and chemokine mRNAs carry AU-rich sequences in their 3′ UTRs, and decay of some of these ARE-containing mRNAs was shown to be compromised under various immune responses; therefore, ARE-mediated mRNA decay has been ascribed a key role in regulating the mRNA level of inflammatory mediators (e.g., see reviews in references 19, 39 and 65). Since the stability of MCP-1 and IL-8 mRNAs in BEAS-2B cells increased upon TNF-α/IL-4 stimulation (Fig. 1C), we examined whether the stabilization was due to a compromise of the destabilizing function of their AREs. For this, MCP-1 and IL-8 AREs (see ARE sequences in Fig. S1 in the supplemental material) were inserted into a stable BBB mRNA, and the decay of the reporter messages was analyzed in BEAS-2B cells with or without IL-4/TNF-α treatment. The Tet-off promoter-driven transcriptional pulsing approach (12, 74) was used to monitor decay kinetics of BBB mRNAs carrying an MCP-1 or IL-8 ARE (BBB+ARE). A tTA-expressing BEAS-2B-19 stable cell line that supports transcription of genes driven by the Tet-off or Tet-on promoter (12) was transfected with plasmids encoding BBB+ARE mRNAs, which were expressed under the control of an inducible Tet-off promoter (26). This well-established promoter-reporter system allows a transcriptional pulsing for monitoring the nearly synchronous deadenylation of a uniformly sized population of mRNA molecules, permitting evaluation of mRNA deadenylation and decay kinetics (12, 74). The results (Fig. 2) show that both MCP-1 and IL-8 AREs have the ability to promote rapid decay of the corresponding BBB+ARE mRNA in BEAS-2B cells. However, the decay of BBB+ARE mRNAs did not slow down when the cells were stimulated with IL-4/TNF-α (Fig. 2), indicating that the mRNA-destabilizing function of MCP-1 and IL-8 AREs remained active after stimulation.
FIG. 2.
IL-4/TNF-α stimulation has little effect on the mRNA destabilizing function of MCP-1 and IL-8 AREs in BEAS-2B cells. (A) Northern blots showing decay of BBB mRNA (left panels), BBB+ARE (MCP-1) mRNA (middle panels), and BBB+ARE (IL-8) mRNA (right panels) in nonstimulated (non-stimu) or in IL-4/TNF-αstimulated (stimu) BEAS-2B cells. BEAS-2B-19 cells were transiently cotransfected with a Tet-off promoter-regulated plasmid encoding a reporter mRNA as indicated and a control plasmid encoding constitutively expressed α-globin/GAPDH hybrid mRNA that serves as an internal standard for transfection efficiency and sample handling (ctrl). After 2 h of transcriptional pulse by culturing the transfected cells in the absence of Tet, Tet (500 ng/ml) was added to block further transcription. Times correspond to hours after transcription blockage by Tet addition. Cytoplasmic RNAs were extracted at the indicated times for Northern blot analysis. The experiments were repeated at least once, and the results were reproducible. (B) Semilog plots showing the decay kinetics for BBB, BBB+ARE (MCP-1), and BBB+ARE (IL-8) mRNAs. Quantification of RNA was obtained by scanning radioactive blots, as described for panel A, with an imager (Bio-Rad).
Protein synthesis is upregulated in activated BEAS-2B cells.
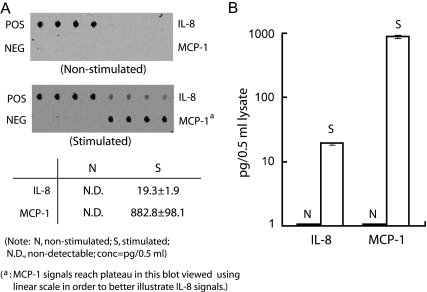
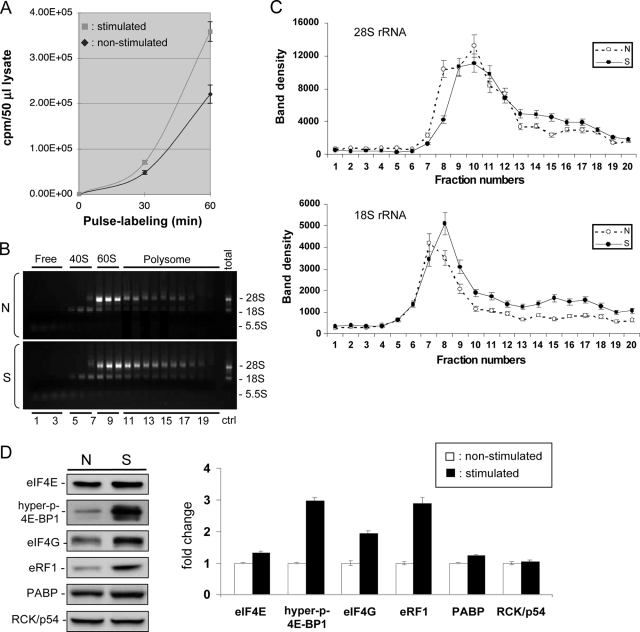
A sandwich enzyme-linked immunosorbent assay was used to quantify the levels of MCP-1 and IL-8 proteins in cytoplasmic lysates prepared from both nonstimulated and stimulated BEAS-2B cells (Fig. 3). The data show that IL-4/TNF-α treatment elevated the production of IL-8 and MCP-1 proteins from a nondetectable level to about 20 and 880 pg/0.5 ml of cytoplasmic lysate, respectively, indicating that these chemokines are robustly produced in activated cells. We then measured the protein synthesis rate in both nonstimulated and stimulated BEAS-2B cells by pulse labeling nascent proteins with [35S]methionine and cysteine. We observed a significant increase in the global translation rate when cells were stimulated with TNF-α/IL-4 (Fig. 4A). To further substantiate this result, changes in ribosome loading onto mRNAs (polysome profile) were determined by performing sucrose gradient fractionation of cytoplasmic lysates. Comparison of the polysome profiles from nonstimulated and stimulated cells revealed a significant increase in the polysome fraction (Fig. 4B and C, fractions 11 to 19) in stimulated cells, indicating that more ribosomes are loaded on mRNAs for translation in activated cells than in nonstimulated cells.
FIG. 3.
Increased MCP-1 and IL-8 protein production in stimulated BEAS-2B cells. BEAS-2B cells were left untreated (nonstimulated) or treated with IL-4/TNF-α for 18 h (stimulated). Cytoplasmic lysates, prepared from equal numbers of cells, were incubated with a Human Quantibody Inflammation array (RayBiotech, Inc.). (A) Representative images showing the results obtained using an Axon GenePix laser scanner. POS, positive control (a biotinylated immunoglobulin G); NEG, negative control (bovine serum albumin). (B) Quantification (pg/0.5 ml) of the data obtained from the combination of three biological and four technical repeats. Note that the y axis is on a logarithmic scale. Quantitative data analysis was done using RayBio Q Analyzer software (RayBiotech, Inc.).
FIG. 4.
Global translation enhancement in stimulated BEAS-2B cells. (A) BEAS-2B cells with or without IL-4/TNF-α stimulation were pulse labeled with [35S]methionine and cysteine for the indicated periods of time. Equal numbers of cells were lysed in the same volume of lysis buffer, and 50 μl was used for trichloroacetic acid precipitation and scintillation counting. Experiments were done in triplicate. (B) Polysome profiles of nonstimulated (N) and stimulated (S) BEAS-2B cells. RNA was extracted from each of the 20 fractions, and an aliquot was used for the electrophoresis in 1% agarose to visualize the distributions of 5.5S, 18S, and 28S rRNA bands. A representative is shown in this panel. Ctr, control. (C) The patterns showing the shift in 28S or 18S rRNA distribution following IL-4/TNF-α stimulation were separately illustrated graphically with the error bars based on the average of three independent experiments, including the one shown in panel B. The amounts of 18S and 28S rRNAs from nonstimulated (N) and stimulated (S) cells were quantified by GeneTools (Syngene), and band intensities were all normalized to the same range based on the band intensities exhibited by the representative profile shown in panel B. (D) Western blot (left) showing that the levels of eIF4E, hyperphosphorylated 4E-BP1, eIF4G, eRF1, and PABP in BEAS-2B cells increased upon stimulation, whereas the level of Rck/p54, a translation repressor, did not change. Cytoplasmic cell lysates from the same number of nonstimulated (N) and stimulated (S) cells were used. Chemiluminescence of target bands on Western blots was captured with an imaging system (Syngene) and quantified using GeneTools (Syngene). The increase in expression of each protein from stimulated cells in comparison to the level of the corresponding protein from nonstimulated cells (assigned a value of 11) is shown in the bar-graph. All data represent the mean of three experiments ± standard errors.
A major way to control translation in eukaryotes is through altering the level of the eIF4E, a 5′ cap-binding protein which has been found to be a major target for translational control by extracellular stimuli (25). eIF4E is the least abundant eIF and limits formation of the translation initiation complex (16). Thus, availability of eIF4E greatly affects the translation profile of the cytoplasmic mRNA pool and accordingly is tightly regulated (44). We therefore checked the levels of eIF4E and other translation regulatory factors in both nonstimulated and stimulated cells. As shown in Fig. 4D, the level of eIF4E was 30% higher in BEAS-2B cells after stimulation with IL-4/TNF-α, whereas the level of Rck/p54, a translation repressor (14, 71), did not change.
The availability of eIF4E for translation initiation is tightly controlled by 4E-BPs, particularly by 4E-BP1, the most abundant member of the 4E-BP family of eIF4E repressor proteins (55). Hypophosphorylated 4E-BP1 binds and prevents eIF4E from forming a translation initiation complex. However, eIF4E is released from hyperphosphorylated 4E-BP1 as a result of consecutive phosphorylation of 4E-BP1, with Ser 65 being phosphorylated last via the phosphatidylinositol-3 kinase/Akt/mTOR pathway (24, 44). Using antibody that specifically recognizes human 4E-BP1 proteins phosphorylated at Ser 65 (24), we found that the level of hyperphosphorylated 4E-BP1 increased about threefold upon IL-4/TNF-α stimulation (Fig. 4D). This observation is fully consistent with the notion that activation of BEAS-2B cells triggers release of eIF4E proteins from the eIF4E/4E-BP1 complexes, making them available to elevate translation initiation efficiency.
The levels of several other translation regulatory factors such as eIF4G, termination release factor 1 (eRF1), and PABP also increased in activated BEAS-2B cells (Fig. 4D). eIF4G is a large scaffolding protein in the translation initiation complex and is crucial for translation initiation by recruiting the 40S ribosomal subunit, by providing a platform for 5′ cap-3′ poly(A) interactions on mRNA and by helping select AUG initiation codon (53). eRF1 has been known to play a role in enhancing efficient translation termination and also has been suggested to play a role in promoting translation reinitiation, thereby increasing the translation rate (33, 68). PABP enhances translation initiation through its interaction with eIF4G (35, 45) and promotes translation termination through its interaction with the termination complex involving eRF1 (33, 68). Thus, by coordinately upregulating the levels of key factors necessary for translation, activated BEAS-2B cells can accommodate the elevated levels of mRNAs induced by IL-4/TNF-α treatment.
miRNA-mediated translational silencing is compromised in activated BEAS-2B cells.
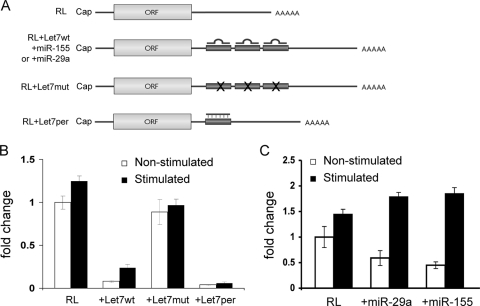
The above findings indicate that posttranscriptional regulation at the translation level plays an important role in sustaining high levels of production of inflammatory mediator proteins during inflammatory responses. Protein production may also be elevated through mechanisms that release translational silencing, e.g., by miRNAs, which have emerged as important posttranscriptional regulators of gene expression (reviewed in references 21 and 34). We therefore examined whether the miRNA-mediated translational silencing is reduced in activated BEAS-2B cells using a let-7 miRNA reporter system (54) because the let-7 miRNA family is one of the most abundant groups of miRNAs ubiquitously expressed in mammalian cells (63), including BEAS-2B cells. The reporter system consists of a gene coding for the Renilla luciferase (RL) or its derivatives RL(let-7wt), RL(let-7mut), and RL(let-7per) in the 3′ UTR (Fig. 5A). The ability of let-7 miRNA to repress translation in our cellular model was demonstrated by observations that overall luciferase activity from the RL(let-7wt) gene was dramatically lower than that from the control RL gene in both stimulated and nonstimulated BEAS-2B cells, whereas there was only a modest difference when RL(let-7mut) was tested (Fig. 5B). Importantly, after IL-4/TNF-α stimulation, there was a significant increase (∼3.5-fold) in luciferase activity from RL(let-7wt) (Fig. 5B), indicating a reduction in translation repression of mRNA carrying let-7 target sites. In contrast, there was only a modest increase (∼1.3-fold) in the expression level of the RL, RL(let-7mut), or RL(let-7per)gene(Fig. 5B). This modest increase is consistent with the data showing a global increase in the translation initiation rate in activated BEAS-2B cells (Fig. 4). Thus, our data demonstrate that let-7 miRNA-mediated translational repression is downregulated in activated airway epithelial cells.
FIG. 5.
miRNA-mediated translational repression is downregulated in activated BEAS-2B cells. (A) Schematic diagram showing the basic Renilla luciferase reporter mRNA (RL), containing three let-7, miR-155, or miR-29a target sites (RL+Let7wt, +miR-155, or +miR-29a), three mutated nonfunctional let-7 target sites (RL+Let7mut), or a single site that forms a perfect match with let-7 miRNAs (RL+Let7per). (B and C) Histograms representing relative changes of Renilla luciferase activity detected in extracts from nonstimulated or stimulated BEAS-2B cells expressing the indicated RL reporter mRNA. Cells were cotransfected with a control construct expressing the firefly luciferase (FL). The RL/FL activity detected in nonstimulated extract expressing RL mRNA was set as 1. All data represent the normalized mean of three repeats ± standard errors. ORF, open reading frame.
To test the generality of the observed decrease in miRNA-mediated mRNA silencing, we then examined the ability of two additional miRNAs that are also found in BEAS-2B cells, miR-155 and miR-29a, to repress translation in both IL-4/TNF-α stimulated and nonstimulated BEAS-2B cells. miR-155 regulates various immune responses in B and T cells (reviewed in reference 57), and miR-29a is predicted to target ∼18 collagen mRNAs (unpublished observation). Alteration of collagenous matrix has an important role in airway tissue remodeling and fibrosis (e.g., see references 7 and 36). Three copies of the target site corresponding to each miRNA were added to the RL reporter gene to create RL+miR-155 and RL+miR-29a. The results (Fig. 5C) show that both miRNAs repressed Renilla luciferase expression in nonstimulated BEAS-2B cells, and a consistent derepression of translation was observed in both cases in activated BEAS-2B cells. Together, we showed that translation silencing mediated by three different miRNAs (let-7, miR-155, and miR-29a) is compromised in BEAS-2B cells stimulated with IL-4 and TNF-α, thereby supporting a general reduction of miRNA-mediated translation silencing.
The number of P bodies is reduced in activated bronchial epithelial cells, both in vitro and in vivo.
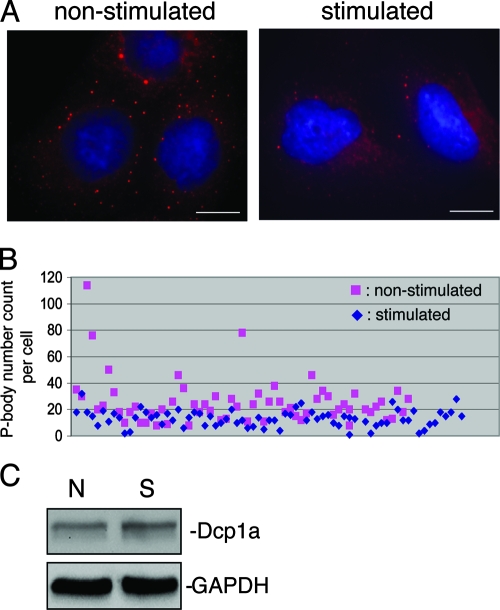
P bodies are known to be a site for storing nontranslatable mRNAs and many mRNA decay factors in mammalian tissue culture cells. Observations of MCP-1 and IL-8 mRNA stabilization, increased translation rate, elevated levels of factors necessary for translation, and reduced miRNA-mediated translational repression in activated BEAS-2B cells suggest a reduction of the nontranslatable mRNA pool, which in turn may decrease the number of P bodies. To test this hypothesis, we examined P bodies in stimulated and nonstimulated BEAS-2B cells by immunofluorescence microscopy. Dcp1a, a well-characterized P-body component required for eukaryotic mRNA decay at the decapping step (6, 15, 38, 43), was used as a marker to visualize P bodies. The results show a pronounced reduction in P-body numbers in cells treated with IL-4/TNF-α (Fig. 6A). Only 10% of stimulated cells contained 20 or more P bodies, compared with over 54% of nonstimulated cells (Fig. 6B). Western blot analysis showed that cytoplasmic levels of Dcp1a did not change following IL-4/TNF-α stimulation (Fig. 6C), indicating that the loss of P bodies in activated BEAS-2B cells was not due to a reduction of cytoplasmic Dcp1a protein.
FIG. 6.
Change in P-body numbers in BEAS-2B cells. (A) Immunofluorescence microscopy showing a reduction in P-body numbers in BEAS-2B cells treated with IL-4/TNF-α. Rabbit anti-Dcp1a antibody was used to detect P bodies (red signals). Nuclei were visualized by DAPI staining (blue signals). The scale bar in each panel corresponds to 15 μm. (B) P-body counts of individual cells from nonstimulated or stimulated cells (60 cells were counted for each treatment). The plot is a merged result of at least two independent experiments. (C) Western blot showing that the cytoplasmic level of Dcp1a did not change following IL-4/TNF-α stimulation. GAPDH served as a loading control. N, nonstimulated; S, stimulated.
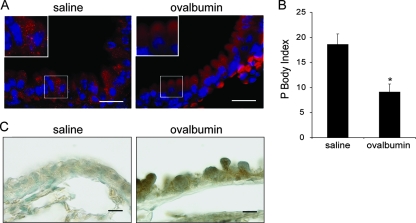
We then analyzed P-body numbers in bronchial epithelium in vivo using an allergen (ovalbumin)-challenged mouse model of pulmonary inflammation (60, 76). P bodies are strikingly abundant throughout the bronchial airway epithelium of control mice challenged with saline (Fig. 7A, left). Consistent with our findings in BEAS-2B cells (Fig. 6), significantly fewer P bodies were observed in the bronchial epithelium of mice with ovalbumin-induced airway inflammation (Fig. 7A, right, and B). The inflammatory response in ovalbumin-challenged mice was confirmed by demonstration of increased MCP-1 levels in the bronchial epithelium (Fig. 7C) and by the elevated numbers of eosinophils in the bronchial alveolar lavage fluid (75).
FIG. 7.
Change in P-body number in mouse activated bronchial airway epithelium. BALB/c mice were sensitized with ovalbumin, and then the airways were challenged with either saline or ovalbumin. (A) High magnification of bronchial airways from mice challenged with saline or ovalbumin and then subjected to immunofluorescence using rabbit anti-Dcp1a antibody to detect P bodies (red signal). Blue signal, DAPI-stained nuclei. Scale bar, 100 μm. (B) Quantification of P bodies in bronchial epithelial cells was determined by counting the number of bronchial epithelial cells per millimeter of airway basement membrane that contained 20 or more P bodies (P-body index) using ImagePro Plus software (Media Cybernetics). Data are presented as the mean P-body index ± standard error of the mean (n = 4 mice per group). The asterisk denotes a significant difference. (C) Sections from the same mice described in panel A were stained with an antibody against MCP-1 followed by immunoperoxidase detection (shown in brown). Scale bar, 100 μm.
Taken together, the observations that P bodies are abundant both in nonstimulated human bronchial epithelial BEAS-2B cells (Fig. 6A, left) and in normal mouse bronchial epithelium (Fig. 7A, left) and that the number of P bodies is reduced in activated bronchial epithelial cells, both in vitro and in vivo (Fig. 6 and 7), suggest that decreased mRNA turnover, upregulation of translation, and the loss of P bodies are general features of activated bronchial epithelial cells during allergic airway inflammation.
DISCUSSION
The results of the present study shed new light on the posttranscriptional regulation of inflammatory mediator production at the levels of mRNA turnover and translation in activated bronchial epithelial cells. Using a comprehensive microarray analysis of changes in mRNA expression between nonactivated and IL-4/TNF-α-activated human bronchial epithelial BEAS-2B cells, we found (Table 1) the following: (i) that activated epithelial cells express high levels of mRNAs for several cytokines and chemokines, such as IL-1-β, IL-6, granulocyte-macrophage colony-stimulating factor, MCP-1, IL-8, and exotaxin-3; (ii) expression of adhesion molecules (such as C3, ICAM-1, and VCAM-1) that are important for recruitment of innate immune cells is also greatly upregulated in activated BEAS-2B cells; and (iii) several genes related to airway remodeling and fibrosis are significantly upregulated in activated cells, including genes for collagens, serine protease inhibitors, and superoxide dismutase 2 (Table 1). It is worth noting that mRNAs encoding five different collagens (COL1A1, COL5A1, COL6A1, COL7A1, and COL8A1) are rather abundant in BEAS-2B cells, suggesting a direct and active role of bronchial epithelium in airway remodeling. Collectively, these observations demonstrate that BEAS-2B cells stimulated with IL-4/TNF-α represent a useful in vitro cellular model for investigating molecular and cellular mechanisms of mRNA decay and translational control in airway inflammation. Moreover, these findings support the view that bronchial epithelium is a major source of many key inflammatory and remodeling molecules (3, 29, 31, 61) and thereby play a pivotal role in the pathogenesis of airway diseases.
Kinetic experiments conducted in this study measuring MCP-1 and IL-8 mRNA decay and in a previous study measuring exotaxin-1 mRNA decay (2) support a critical role of mRNA turnover in the upregulation of chemokine gene expression during airway inflammation and remodeling. Intriguingly, while the stability of mRNAs encoding two key chemokines, MCP-1 and IL-8, increases in BEAS-2B cells activated by IL-4 and TNF-α (Fig. 1C), little change in the destabilization function of their AREs was observed (Fig. 2). This finding is of particular interest because downregulation of ARE-mediated mRNA decay was often considered to account for the increased mRNA stability of an ARE-containing mediator mRNA in response to the proinflammatory stimulation (27, 41, 66, 72, 73). Our findings that alteration of ARE-mediated decay is not always responsible for changes in stability of an ARE-containing message support the notion that an ARE plays different roles in different cell types or under different physiological conditions (5). Given the fact that RNA-destabilizing elements can be anywhere in a message (e.g., 5′ UTR, protein coding region, or 3′ UTRs) (10, 50), it would be interesting to identify mRNA decay pathways that are involved in the observed mRNA stabilization in activated BEAS-2B cells.
Several lines of evidence from this study demonstrate an enhancement of global translation rate in activated bronchial epithelial cells. These include the following: (i) increased [35S]methionine and cysteine incorporation into nascent polypeptides (Fig. 4A); (ii) increased ribosomes in the polysome fractions (Fig. 4B and C); and (iii) increased levels of several proteins critical for elevating translation efficiency (e.g., eIF4E, eRF1, and PABP) (Fig. 4D). It is worth noting that eIF4E, which is also a 5′ cap-binding protein, is the least abundant eIF and limits formation of the translation initiation complex (16). Moreover, eIF4E has been found to be a major target for translational control by extracellular stimuli, such as stress, cytokines, growth factors, and mitogens (25). Thus, availability of eIF4E greatly affects the translation profile of the cytoplasmic mRNA pool and is tightly regulated. As chronic airway inflammation is often associated with wound healing, tissue remodeling, and fibrosis, it is also worth noting that administration of eIF4E mRNA has been shown to augment wound healing in a rat model (62). The observation that the level of hyperphosphorylated 4E-BP1 significantly increased upon IL-4/TNF-α stimulation (Fig. 4D) is fully consistent with the notion that activation of BEAS-2B cells triggers release of eIF4E proteins from the eIF4E/4E-BP1 complexes, making them available to elevate translation initiation efficiency. These data reveal a new example of translational control via altering eIF4E/4E-BP1 interaction and thus provide new insight into the mechanism by which global translation is enhanced in activated BEAS-2B cells.
eRF1 and PABP levels were also found to increase in activated BEAS-2B cells (Fig. 4D). eRF1 associates with eRF3, a PABP-interacting protein, to form a complex that is required for efficient translation termination (33, 68). Recently, it was proposed that interaction between the eRF1-eRF3 complex and PABP/poly(A) not only enhances efficient translation termination but also promotes translation reinitiation, thereby increasing the translation rate (33, 68). Thus, by upregulating the expression of factors necessary for translation, activated BEAS-2B cells can accommodate the elevated levels of mRNAs induced by IL-4/TNF-α treatment.
Recently, miRNAs, a group of evolutionarily conserved and abundant class of small silencing RNAs, have emerged as important posttranscriptional regulators of gene expression across species and in multiple biological processes (21, 48, 64). In this study, we tested three different miRNAs exhibiting moderate to high abundance in BEAS-2B cells (data not shown) and found that miRNA-mediated mRNA silencing is compromised in human bronchial epithelial BEAS-2B cells following IL-4/TNF-α treatment, leading to an increase in protein levels of their targets (Fig. 5). These findings open up a new avenue for studying mechanisms by which the modulation of miRNA function is accomplished to help cells reset their protein profile in response to external stimuli. One possibility is that the increased number of translation initiation complexes (see above) overcomes the translation initiation repression by miRNAs (46). This interpretation is consistent with an in vitro study showing that the addition of purified initiation complex eIF4F, consisting of eIF4E, eIF4G, and eIF4A, to an ascites extract rescued mRNA from miRNA-mediated translation repression in vitro (46). Another intriguing possibility is that under the physiological condition of activated BEAS-2B cells, Ago2-miRNA complex may somehow turn into a translation activator that promotes translation of mRNA targets. This notion is consistent with the findings of two recent reports showing that under specific serum-starvation conditions of some mammalian tissue culture cells, miRNAs including endogenous let-7 and miR-396-3 miRNAs and a synthetic miRNA positively promote translation of reporter mRNAs bearing the cognate miRNA target sites (69, 70).
Another important and novel finding in this study is that P bodies, which are closely linked to mRNA decay and translation repression, are abundant both in nonstimulated human bronchial epithelial BEAS-2B cells (Fig. 6) and in the normal mouse bronchial epithelium (Fig. 7). Prolonged treatment of IL-4 and TNF-α caused a significant decrease of P bodies in BEAS-2B cells (Fig. 6). A similar reduction in the P-body population was observed in bronchial epithelium from mice exhibiting allergic airway inflammation (Fig. 7). These findings suggest that decreased mRNA turnover, upregulation of translation, and the loss of P bodies are general features of activated bronchial epithelial cells during allergic airway inflammation. It is also worth noting that the bronchial epithelium from normal mice typically has 30 to 50 P bodies per epithelial cell (Fig. 7A), which is much more than many cultured mammalian cells (e.g., see references 15, 18, 22, and 51) (Fig. 6A). These observations suggest that bronchial epithelium is primed to elicit a response to airborne allergens or other insults by releasing factors and mRNPs from P bodies to the cytoplasm, e.g., for a boost of translation. Along this line, it is worth noting that eIF4E, the translation initiation factor that plays a rate-limiting role in formation of the translation initiation complex (see above), can be stored in P bodies (1, 20). The observation of a reduced number of P bodies in activated bronchial epithelial cells raises a possibility that translation initiation is boosted by releasing eIF4E from P bodies to the cytoplasm.
While the exact function of P bodies in mRNA turnover and translational control remains controversial and a subject of intensive experimentation (17, 49), our findings support a physiological role of P bodies as a cellular reservoir that allows storage and release of translation factors and dormant mRNPs in the cytoplasm of bronchial airway epithelium. General mRNA decay activity and translational silencing pathways may well be active in normal bronchial epithelium to keep certain mRNAs (e.g., those coding for inflammatory and remodeling mediators) from being effectively translated. Upon activation of the epithelium with inflammatory cytokines such as IL-4 and TNF-α, a concerted reduction of P bodies along with a downregulation of pathways that produce nontranslatable mRNPs promotes production of mediators. To our knowledge, this study represents the first report of a physiologically relevant modulation of mammalian P bodies in vivo. It will be important to determine the mechanism governing the loss of P bodies following the stimulation of bronchial epithelial cells.
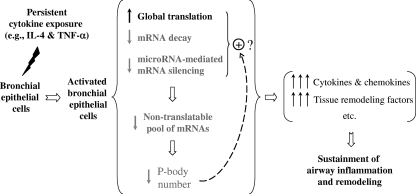
In summary, our data suggest that a combination of decreasing mRNA decay, upregulating global translation, and decreasing P-body abundance results in persistently elevated production of inflammatory mediators in bronchial epithelium (Fig. 8). This may represent an important amplification pathway in the regulation of inflammatory and remodeling genes in the airway epithelium. Thus, the present study reveals a potential new pathogenic mechanism operating at the levels of mRNA decay and translation in bronchial epithelium to sustain the chronic allergic inflammation and remodeling seen in allergic airway diseases. To the best of our knowledge, this study is among the first to demonstrate coordinate regulation of multiple posttranscriptional pathways following stimulation by factors largely thought to be activators of transcriptional pathways.
FIG. 8.
Coordinated changes in mRNA turnover, translation, and RNA P bodies in bronchial epithelial cells following inflammatory stimulation. Coordinate regulation of multiple posttranscriptional pathways following stimulation by proinflammatory cytokines represents a novel mechanism that allows amplification and sustainment of protein production in bronchial epithelium during airway inflammation. See Discussion for details.
Supplementary Material
Acknowledgments
We thank R. Kulmacz, J. Lever, and J. Garcia-Sanz for critical reading of the manuscript and valuable comments; W. Filipowicz for let-7-luciferase reporter constructs; R. Lloyd for rabbit anti-PABP antibody; S. Ohno for rabbit anti-Dcp1a antibody; and W. Zhu for technical assistance.
This work was supported by the Sandler Foundation for Asthma Research to A.-B. Shyu and in part by National Institutes of Health (GM 46454 to A.-B. Shyu and AI43572 and HL70952 to M. Blackburn) and the Houston Endowment, Inc., to A.-B. Shyu.
Footnotes
Published ahead of print on 20 October 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Andrei, M. A., D. IIngelfinger, R. Heintzmann, T. Achsel, R. Rivera-Pomar, and R. Luhrmann. 2005. A role for eIF4E and eIF4E-transporter in targeting mRNPs to mammalian processing bodies. RNA 11717-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atasoy, U., S. L. Curry, I. Lopez de Silanes, A.-B. Shyu, V. Casolaro, M. Gorospe, and C. Stellato. 2003. Regulation of eotaxin gene expression by TNF-α and IL-4 through mRNA stabilization: involvement of the RNA-binding protein HuR. J. Immunol. 1714369-4378. [DOI] [PubMed] [Google Scholar]
- 3.Barnes, P. J. 2008. Immunology of asthma and chronic obstructive pulmonary disease. Nat. Rev. Immunol. 8183-192. [DOI] [PubMed] [Google Scholar]
- 4.Barnes, P. J., K. F. Chung, and C. P. Page. 1998. Inflammatory mediators of asthma: an update. Pharmacol. Rev. 50515-596. [PubMed] [Google Scholar]
- 5.Barreau, C., L. Paillard, and H. B. Osborne. 2005. AU-rich elements and associated factors: are there unifying principles? Nucleic Acids Res. 337138-7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beelman, C. A., A. Stevens, G. Caponigro, T. E. LaGrandeur, L. Hatfield, D. M. Fortner, and R. Parker. 1996. An essential component of the decapping enzyme required for normal rates of mRNA turnover. Nature 382642-646. [DOI] [PubMed] [Google Scholar]
- 7.Chapman, H. A. 2004. Disorders of lung matrix remodeling. J. Clin. Investig. 113148-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, C. Y., T. M. Chen, and A. B. Shyu. 1994. Interplay of two functionally and structurally distinct domains of the c-fos AU-rich element specifies its mRNA-destabilizing function. Mol. Cell. Biol. 14416-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, C. Y., N. Xu, and A. B. Shyu. 1995. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol. Cell. Biol. 155777-5788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, C.-Y., R. Gherzi, J. S. Andersen, G. Gaietta, K. Jurchott, H.-D. Royer, M. Mann, and M. Karin. 2000. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 141236-1248. [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, C.-Y. A., and A.-B. Shyu. 2003. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol. Cell. Biol. 234805-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, C.-Y. A., Y. Yamashita, T.-C. Chang, A. Yamashita, W. Zhu, Z. Zhong, and A.-B. Shyu. 2007. Versatile applications of transcriptional pulsing to study mRNA turnover in mammalian cells. RNA 131775-1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Christman, J. W., R. T. Sadikot, and T. S. Blackwell. 2000. The role of nuclear factor-κB in pulmonary diseases. Chest 1171482-1487. [DOI] [PubMed] [Google Scholar]
- 14.Chu, C.-Y., and T. M. Rana. 2006. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biology 4e210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cougot, N., S. Babajko, and B. Seraphin. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 16531-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duncan, R., S. C. Milburn, and J. W. Hershey. 1987. Regulated phosphorylation and low abundance of HeLa cell initiation factor eIF-4F suggest a role in translational control. Heat shock effects on eIF-4F. J. Biol. Chem. 262380-388. [PubMed] [Google Scholar]
- 17.Eulalio, A., I. Behm-Ansmant, and E. Izaurralde. 2007. P bodies: at the crossroads of post-transcriptional pathways. Nat. Rev. Mol. Cell Biol. 89-22. [DOI] [PubMed] [Google Scholar]
- 18.Eulalio, A., I. Behm-Ansmant, D. Schweizer, and E. Izaurralde. 2007. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell. Biol. 273970-3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fan, J., N. M. Heller, M. Gorospe, U. Atasoy, and C. Stellato. 2005. The role of post-transcriptional regulation in chemokine gene expression in inflammation and allergy. Eur. Respir. J. 26933-947. [DOI] [PubMed] [Google Scholar]
- 20.Ferraiuolo, M. A., S. Basak, J. Dostie, E. L. Murray, D. R. Schoenberg, and N. Sonenberg. 2005. A role for the eIF4E-binding protein 4E-T in P-body formation and mRNA decay. J. Cell Biol. 170913-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Filipowicz, W., S. N. Bhattacharyya, and N. Sonenberg. 2008. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9102-114. [DOI] [PubMed] [Google Scholar]
- 22.Franks, T. M., and J. Lykke-Andersen. 2007. TTP and BRF proteins nucleate processing body formation to silence mRNAs with AU-rich elements. Genes Dev. 21719-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerard, C., and B. J. Rollins. 2001. Chemokines and disease. Nat. Immunol. 2108-115. [DOI] [PubMed] [Google Scholar]
- 24.Gingras, A.-C., B. Raught, S. P. Gygi, A. Niedzwiecka, M. Miron, S. K. Burley, R. D. Polakiewicz, A. Wyslouch-Cieszynska, R. Aebersold, and N. Sonenberg. 2001. Hierarchical phosphorylation of the translation inhibitor 4E-BP1. Genes Dev. 152852-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gingras, A.-C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Ann. Rev. Biochem. 68913-963. [DOI] [PubMed] [Google Scholar]
- 26.Gossen, M., and H. Bujard. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 895547-5551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gueydan, C., L. Droogmans, P. Chalon, G. Huez, D. Caput, and V. Kruys. 1999. Identification of TIAR as a protein binding to the translational regulatory AU-rich element of tumor necrosis factor alpha mRNA. J. Biol. Chem. 2742322-2326. [DOI] [PubMed] [Google Scholar]
- 28.Haegens, A., T. F. Barrett, J. Gell, A. Shukla, M. MacPherson, P. Vacek, M. E. Poynter, K. J. Butnor, Y. M. Janssen-Heininger, C. Steele, and B. T. Mossman. 2007. Airway epithelial NF-κB activation modulates asbestos-induced inflammation and mucin production in vivo. J. Immunol. 1781800-1808. [DOI] [PubMed] [Google Scholar]
- 29.Hammad, H., and B. N. Lambrecht. 2008. Dendritic cells and epithelial cells: linking innate and adaptive immunity in asthma. Nat. Rev. Immunol. 8193-204. [DOI] [PubMed] [Google Scholar]
- 30.Hart, L. A., V. L. Krishnan, I. M. Adcock, P. J. Barnes, and K. F. Chung. 1998. Activation and localization of transcription factor, nuclear factor-κB, in asthma. Am. J. Respir. Crit. Care Med. 1581585-1592. [DOI] [PubMed] [Google Scholar]
- 31.Holgate, S. T. 2007. The epithelium takes centre stage in asthma and atopic dermatitis. Trends Immunol. 28248-251. [DOI] [PubMed] [Google Scholar]
- 32.Holgate, S. T., P. M. Lackie, D. E. Davies, W. R. Roche, and A. F. Walls. 1999. The bronchial epithelium as a key regulator of airway inflammation and remodeling in asthma. Clin. Exp. Allergy 29(Suppl. 2)90-95. [DOI] [PubMed] [Google Scholar]
- 33.Hoshino, S., M. Imai, T. Kobayashi, N. Uchida, and T. Katada. 1999. The eukaryotic polypeptide chain releasing factor (eRF3/GSPT) carrying the translation termination signal to the 3′-poly(A) tail of mRNA. Direct association of erf3/GSPT with polyadenylate-binding protein. J. Biol. Chem. 27416677-16680. [DOI] [PubMed] [Google Scholar]
- 34.Jackson, R. J., and N. Standart. 2007. How do microRNAs regulate gene expression? Sci. STKE 2007re1. [DOI] [PubMed] [Google Scholar]
- 35.Kahvejian, A., Y. V. Svitkin, R. Sukarieh, M.-N. M'Boutchou, and N. Sonenberg. 2005. Mammalian poly(A)-binding protein is a eukaryotic translation initiation factor, which acts via multiple mechanisms. Genes Dev. 19104-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Keane, M. P., S. C. Donnelly, J. A. Belperio, R. B. Goodman, M. Dy, M. D. Burdick, M. C. Fishbein, and R. M. Strieter. 2002. Imbalance in the expression of CXC chemokines correlates with bronchoalveolar lavage fluid angiogenic activity and procollagen levels in acute respiratory distress syndrome. J. Immunol. 1696515-6521. [DOI] [PubMed] [Google Scholar]
- 37.Kedersha, N., and P. Anderson. 2007. Mammalian stress granules and processing bodies. Methods Enzymol. 43161-81. [DOI] [PubMed] [Google Scholar]
- 38.Kedersha, N., G. Stoecklin, M. Ayodele, P. Yacono, J. Lykke-Andersen, M. J. Fritzler, D. Scheuner, R. J. Kaufman, D. E. Golan, and P. Anderson. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169871-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kracht, M., and J. Saklatvala. 2002. Transcriptional and post-transcriptional control of gene expression in inflammation. Cytokine 2091-106. [DOI] [PubMed] [Google Scholar]
- 40.Kuhn, K., S. C. Baker, E. Chudin, M.-H. Lieu, S. Oeser, H. Bennett, P. Rigault, D. Barker, T. K. McDaniel, and M. S. Chee. 2004. A novel, high-performance random array platform for quantitative gene expression profiling. Genome Res. 142347-2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis, T., C. Gueydan, G. Huez, J. J. Toulme, and V. Kruys. 1998. Mapping of a minimal AU-rich sequence required for lipopolysaccharide-induced binding of a 55-kDa protein on tumor necrosis factor-alpha mRNA. J. Biol. Chem. 27313781-13786. [DOI] [PubMed] [Google Scholar]
- 42.Lukacs, N. W. 2001. Role of chemokines in the pathogenesis of asthma. Nat. Rev. Immunol. 1108-116. [DOI] [PubMed] [Google Scholar]
- 43.Lykke-Andersen, J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 228114-8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mamane, Y., E. Petroulakis, O. LeBacquer, and N. Sonenberg. 2006. mTOR, translation initiation and cancer. Oncogene 256416-6422. [DOI] [PubMed] [Google Scholar]
- 45.Mangus, D. A., M. C. Evans, and A. Jacobson. 2003. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4233:http://genomebiology.com/2003/4/7/223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mathonnet, G., M. R. Fabian, Y. V. Svitkin, A. Parsyan, L. Huck, T. Murata, S. Biffo, W. C. Merrick, E. Darzynkiewicz, R. S. Pillai, W. Filipowicz, T. F. Duchaine, and N. Sonenberg. 2007. MicroRNA inhibition of translation initiation in vitro by targeting the cap-binding complex eIF4F. Science 3171764-1767. [DOI] [PubMed] [Google Scholar]
- 47.Nickel, R., L. A. Beck, C. Stellato, and R. P. Schleimer. 1999. Chemokines and allergic disease. J. Allergy Clin. Immunol. 104723-742. [DOI] [PubMed] [Google Scholar]
- 48.Nilsen, T. W. 2007. Mechanisms of microRNA-mediated gene regulation in animal cells. Trends Genet. 23243-249. [DOI] [PubMed] [Google Scholar]
- 49.Parker, R., and U. Sheth. 2007. P bodies and the control of mRNA translation and degradation. Mol. Cell 25635-646. [DOI] [PubMed] [Google Scholar]
- 50.Paste, M., G. Huez, and V. Kruys. 2003. Deadenylation of interferon-beta mRNA is mediated by both the AU-rich element in the 3′-untranslated region and an instability sequence in the coding region. Eur. J. Biochem. 2701590-1597. [DOI] [PubMed] [Google Scholar]
- 51.Pauley, K. M., T. Eystathioy, A. Jakymiw, J. C. Hamel, M. J. Fritzler, and E. K. Chan. 2006. Formation of GW bodies is a consequence of microRNA genesis. EMBO Rep. 7904-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Peng, S.-S., C.-Y. A. Chen, N. Xu, and A.-B. Shyu. 1998. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 173461-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pestova, T. V., V. G. Kolupaeva, I. B. Lomakin, E. V. Pilipenko, I. N. Shatsky, V. I. Agol, and C. U. T. Hellen. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Nat. Acad. Sci. USA 987029-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pillai, R. S., S. N. Bhattacharyya, C. G. Artus, T. Zoller, N. Cougot, E. Basyuk, E. Bertrand, and W. Filipowicz. 2005. Inhibition of translational initiation by let-7 micro-RNA in human cells. Science 3091573-1576. [DOI] [PubMed] [Google Scholar]
- 55.Poulin, F., A.-C. Gingras, H. Olsen, S. Chevalier, and N. Sonenberg. 1998. 4E-BP3, a new member of the eukaryotic initiation factor 4E-binding protein family. J. Biol. Chem. 27314002-14007. [DOI] [PubMed] [Google Scholar]
- 56.Poynter, M. E., C. G. Irvin, and Y. M. W. Janssen-Heininger. 2003. A prominent role for airway epithelial NF-κB activation in lipopolysaccharide-induced airway inflammation. J. Immunol. 1706257-6265. [DOI] [PubMed] [Google Scholar]
- 57.Rodriguez, A., E. Vigorito, S. Clare, M. V. Warren, P. Couttet, D. R. Soond, S. van Dongen, R. J. Grocock, P. P. Das, E. A. Miska, D. Vetrie, K. Okkenhaug, A. J. Enright, G. Dougan, M. Turner, and A. Bradley. 2007. Requirement of bic/microRNA-155 for normal immune function. Science 316608-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ross, J. 1995. mRNA stability in mammalian cells. Microbiol. Rev. 59423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rot, A., and U. H. von Andrian. 2004. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Ann. Rev. Immunol. 22891-928. [DOI] [PubMed] [Google Scholar]
- 60.Sakai, K., A. Yokoyama, N. Kohno, and K. Hiwada. 1999. Effect of different sensitizing doses of antigen in a murine model of atopic asthma. Clin. Exp. Immunol. 1189-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schleimer, R. P., A. Kato, R. Kern, D. Kuperman, and P. C. Avila. 2007. Epithelium: at the interface of innate and adaptive immune responses. J. Allergy Clin. Immunol. 1201279-1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwarz, K. W., M. T. Murray, R. Sylora, R. L. Sohn, and S. A. Dulchavsky. 2002. Augmentation of wound healing with translation initiation factor eIF4E mRNA. J. Surg. Res. 103175-182. [DOI] [PubMed] [Google Scholar]
- 63.Sempere, L. F., S. Freemantle, I. Pitha-Rowe, E. Moss, E. Dmitrovsky, and V. Ambros. 2004. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shyu, A. B., M. F. Wilkinson, and A. van Hoof. 2008. Messenger RNA regulation: to translate or to degrade. EMBO J. 27471-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stoecklin, G., and P. Anderson. 2006. Posttranscriptional mechanisms regulating the inflammatory response. Adv. Immunol. 891-37. [DOI] [PubMed] [Google Scholar]
- 66.Suk, K., and K. L. Erickson. 1996. Differential regulation of tumour necrosis factor-alpha mRNA degradation in macrophages by interleukin-4 and interferon-gamma. Immunology 87551-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas, P. S. 2001. Tumour necrosis factor-alpha: the role of this multifunctional cytokine in asthma. Immunol. Cell Biol. 79132-140. [DOI] [PubMed] [Google Scholar]
- 68.Uchida, N., S.-i. Hoshino, H. Imataka, N. Sonenberg, and T. Katada. 2002. A novel role of the mammalian GSPT/eRF3 associating with poly(A)-binding protein in Cap/poly(A)-dependent translation. J. Biol. Chem. 27750286-50292. [DOI] [PubMed] [Google Scholar]
- 69.Vasudevan, S., and J. A. Steitz. 2007. AU-rich-element-mediated upregulation of translation by FXR1 and Argonaute 2. Cell 1281105-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vasudevan, S., Y. Tong, and J. A. Steitz. 2007. Switching from repression to activation: microRNAs can up-regulate translation. Science 3181931-1934. [DOI] [PubMed] [Google Scholar]
- 71.Weston, A., and J. Sommerville. 2006. Xp54 and related (DDX6-like) RNA helicases: roles in messenger RNP assembly, translation regulation and RNA degradation. Nucleic Acids Res. 343082-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Winzen, R., M. Kracht, B. Ritter, A. Wilhelm, C.-Y. A. Chen, A.-B. Shyu, M. Muller, M. Gaestel, K. Resch, and H. Holtmann. 1999. The p38 MAP kinase pathway signals for cytokine-induced mRNA stabilization via MAP kinase-activated protein kinase 2 and an AU-rich region-targeted mechanism. EMBO J. 184969-4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Winzen, R., B. K. Thakur, O. Dittrich-Breiholz, M. Shah, N. Redich, S. Dhamija, M. Kracht, and H. Holtmann. 2007. Functional analysis of KSRP interaction with the AU-rich element of interleukin-8 and identification of inflammatory mRNA targets. Mol. Cell. Biol. 278388-8400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu, N., P. Loflin, C.-Y. A. Chen, and A.-B. Shyu. 1998. A broader role for AU-rich element-mediated mRNA turnover revealed by a new transcriptional pulse strategy. Nucleic Acids Res. 26558-565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Young, H. W. J., J. G. Molina, D. Dimina, H. Zhong, M. Jacobson, L.-N. L. Chan, T.-S. Chan, J. J. Lee, and M. R. Blackburn. 2004. A3 adenosine receptor signaling contributes to airway inflammation and mucus production in adenosine deaminase-deficient mice. J. Immunol. 1731380-1389. [DOI] [PubMed] [Google Scholar]
- 76.Young, H. W. J., C.-X. Sun, C. M. Evans, B. F. Dickey, and M. R. Blackburn. 2006. A3 adenosine receptor signaling contributes to airway mucin secretion after allergen challenge. Am. J. Respir. Cell Mol. Biol. 35549-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.