Abstract
Human GTPBP3 is an evolutionarily conserved, multidomain protein involved in mitochondrial tRNA modification. Characterization of its biochemical properties and the phenotype conferred by GTPBP3 inactivation is crucial to understanding the role of this protein in tRNA maturation and its effects on mitochondrial respiration. We show that the two most abundant GTPBP3 isoforms exhibit moderate affinity for guanine nucleotides like their bacterial homologue, MnmE, although they hydrolyze GTP at a 100-fold lower rate. This suggests that regulation of the GTPase activity, essential for the tRNA modification function of MnmE, is different in GTPBP3. In fact, potassium-induced dimerization of the G domain leads to stimulation of the GTPase activity in MnmE but not in GTPBP3. The GTPBP3 N-terminal domain mediates a potassium-independent dimerization, which appears as an evolutionarily conserved property of the protein family, probably related to the construction of the binding site for the one-carbon-unit donor in the modification reaction. Partial inactivation of GTPBP3 by small interfering RNA reduces oxygen consumption, ATP production, and mitochondrial protein synthesis, while the degradation of these proteins slightly increases. It also results in mitochondria with defective membrane potential and increased superoxide levels. These phenotypic traits suggest that GTPBP3 defects contribute to the pathogenesis of some oxidative phosphorylation diseases.
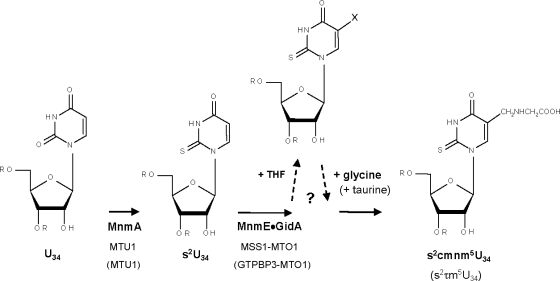
MERRF (myoclonic epilepsy and ragged-red fiber) and MELAS (mitochondrial encephalomyopathy and lactic acidosis with stroke-like episodes) diseases are associated with mutations in the mitochondrial genes for tRNALys and tRNALeu(UUR), respectively (MITOMAP, www.mitomap.org). These mutations cause reduction in oxygen consumption and mitochondrial protein synthesis in the carrier cells (37), which also exhibit reduced ATP production, decreased mitochondrial membrane potential, and increased superoxide levels (1, 16, 33). Interestingly, tRNAs harboring MERRF and MELAS mutations are deficient in normal taurine-containing modifications at the anticodon wobble position [5-taurinomethyluridine (τm5U) in tRNALeu(UUR) and τm5U-2-thiouridine (τm5s2U) in tRNALys] (18, 20, 44, 51-55). Apparently, the pathogenic mutations alter the tRNA determinants recognized by proteins involved in the wobble uridine modification, and the resulting tRNA hypomodification leads to a decoding disorder that appears as the primary cause of MELAS and MERRF at the molecular level (18, 19, 44, 52, 54). Therefore, characterization of the proteins involved in the modification pathway is crucial to understanding the pathogenesis of these diseases.
The biosynthesis of the modified wobble uridine (U34) in mitochondrial tRNALeu(UUR) and tRNALys occurs through a complex pathway involving several genes. The use of the model organisms Escherichia coli and Saccharomyces cerevisiae provides valuable information on this issue because of the high degree of evolutionary conservation of the modification pathway (6, 8-10, 14, 46, 56).
In E. coli, proteins MnmE and GidA assemble in a functional complex involved in formation of the carboxymethylaminomethyl (cmnm) group at position 5 of the wobble uridine of tRNALys, tRNAGlu, tRNA1Gln, tRNA4Leu, tRNA4Arg, and tRNA2Gly (Fig. 1) (4-6, 10, 27, 28, 40, 56, 57). However, the precise role of both proteins in the modification reaction is still unknown, and it is unclear how many steps precede the formation of the 5-carboxymethylaminomethyl (cmnm5) group. In tRNALys, tRNAGlu, and tRNA1Gln, MnmA, together with other proteins, carries out thiolation at position 2 of U34 (Fig. 1) (15). Modifications in positions 2 and 5 occur independently of each other. Modification at position 2, but not at position 5, is important for aminoacylation of tRNAs, whereas modification in both positions 2 and 5 function in the codon recognition process (4).
FIG. 1.
Working model of the U34 modification pathway. Only steps relevant to this work are shown. The first stage in the modification of U34 at position 5 is controlled by MnmE and GidA in bacteria and by their homologous proteins in yeast (MSS1 and MTO1) and human (GTPBP3 and MTO1). MnmE binds 5-formyl-THF, which might be the one-carbon-unit donor in the first step of the modification reaction. MnmE and GidA form a functional complex that would catalyze the production of a still unknown intermediate group (X) at position 5. Taurine (in human) or glycine (in yeast and E. coli) would be subsequently incorporated into tRNA by unidentified transferases to build τm5 or cmnm5, respectively. U34 may undergo thiolation at position 2 mediated by MnmA (in E. coli) or its homologue MTU1 (in eukaryotes), together with other proteins.
In yeast, the protein products of the nuclear genes MSS1, MTO1, and MTU1 (homologues of mnmE, gidA, and mnmA, respectively) localize in mitochondria, and their mutants are defective in modification of mitochondrial tRNA (8, 9, 46, 49). Thus, mitochondrial tRNALys molecules isolated from yeast MTO1 and MSS1 deletion strains contain s2U instead of the cmnm5s2U found in the wild-type tRNALys molecules, whereas disruption of the MTU1 gene eliminates the 2-thio modification of mitochondrial tRNAs (Fig. 1). These results indicate that MTO1 and MSS1 are both involved in the biosynthesis of the cmnm5 group and that MTU1 works as a 2-thiouridylase. Therefore, the function of the GidA, MnmE, and MnmA family proteins seems to be evolutionarily conserved. Human homologues have also been identified and been found to localize in mitochondria (23, 25, 46, 49, 50). It should be noted, however, that in humans taurine is incorporated into mitochondrial tRNALys and tRNALeu(UUR) in place of cmnm (44). Thus, it was proposed that proteins of the MnmE and GidA families jointly catalyze the formation of an unknown intermediate in the modification pathway of U34, whereas the subsequent activity of a taurine or glycine transferase would be responsible for construction of the τm5 group in humans or the cmnm5 group in yeast and bacteria (44, 46, 56) (Fig. 1). However, there is no evidence for this independent, second step. For example, no bacterial or yeast mutants have been isolated in which the putative intermediate produced by the MnmE and GidA proteins accumulates. Therefore, the possibility that the complex formed by these proteins drives all the reaction steps in the cmnm5/τm5 synthesis cannot be discarded.
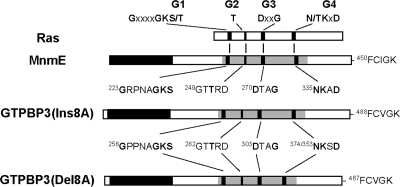
MnmE is a three-domain protein composed of the N-terminal α/β domain (about 120 residues), a central helical domain formed by about 100 residues from the middle region and 70 residues from the C-terminal region, and the G domain formed by approximately 170 residues (40) (Fig. 2). The N-terminal α/β domain induces dimerization. This creates a binding site for tetrahydrofolate(THF), which has been postulated to be the one-carbon-unit donor in the modification reaction. The central helical domain is poorly conserved except for the C-terminal motif, which has been shown to be important for the tRNA modification function of MnmE (57). Finally, the G domain conserves the canonical Ras-like fold and includes the typical G1 to G4 motifs characteristic of all GTPases (Fig. 2).
FIG. 2.
Schematic organization of MnmE, GTPBP3, and Ras proteins. The four motifs (G1, G2, G3, and G4) of the consensus GTP-binding domain are indicated by black bars. In the diagram of MnmE and isoforms of GTPBP3, the shaded area represents the G domain. The sequences of the G1 to G4 and C-terminal motifs of these proteins are also depicted, with the invariant residues of the G motifs indicated in bold. The G motifs are generally involved in binding of guanine nucleotides and Mg2+, hydrolysis of GTP, or controlling the conformational change that regulates the functional state of GTPases. Note that the G4 motif of GTPBP3(Ins8A) and GTPBP3(Del8A) starts at position 374 and 353, respectively, due to the deletion in GTPBP3(Del8A) of a 21-amino-acid sequence (encoded by exon 8A) that extends between the G3 and G4 motifs. In the diagram of MnmE, the black area represents the N-terminal domain involved in dimerization, whereas the empty areas represent the regions that form part of the helical domain. In the diagrams of the GTPBP3 isoforms, the black area represents the N terminal domain, whereas the small empty area at the N terminus represents the mitochondrial targeting signal.
MnmE is a GTPase with peculiar properties (6). Unlike proteins from Ras and translation factor families, MnmE exhibits a high intrinsic GTPase hydrolysis rate and low affinity for GTP and GDP. These properties determine that, in vitro, the GTPase cycle of MnmE proceeds efficiently without auxiliary factors (i.e., GTPase-activating proteins and guanine nucleotide exchange factors). Interestingly, the GTPase activity is essential for the tRNA modifying function of MnmE (27, 57).
The identity of the overall predicted amino acid sequence of MnmE with its human homologue, GTPBP3, is 40%, whereas the similarity is 48% (23). However, such a degree of sequence conservation does not guarantee a perfect match between properties of both homologues. The biochemical properties of GTPBP3 are largely unknown, and conservation of the MnmE properties in the human homologue remains to be clarified.
In E. coli, mnmE and gidA mutations are pleiotropic, affecting diverse phenotypic traits. This may result from a translational control exerted by MnmE and GidA throughout modification of their specific tRNAs (56). Lack of the tRNA modification promoted by the MnmE-GidA complex disturbs codon-anticodon pairing and causes translational frameshifting in particular mRNA sequences (4, 5), which might alter the synthesis of some proteins and contribute to the pleiotropic phenotype. In yeast, ΔMSS1 and ΔMTO1 strains show a growth phenotype similar to that of the wild-type strain and a slightly decreased rate of oxygen consumption (8, 9, 46). This mild phenotype is probably due to the fact that synthesis of the few proteins encoded by the mitochondrial genome is not significantly altered by hypomodification of mitochondrial tRNAs caused by MSS1 and MTO1 defects. Curiously, a combination of mnmE/MSS1 or gidA/MTO1 null mutations with mutations in other genes also involved in the decoding process has a synergistic effect (references 8, 9, 46, and 57 and references therein). The molecular bases of this effect are unknown, but they are probably related to an increase in perturbation of interactions among ribosome, mRNA, and tRNA during decoding on the ribosome. In humans, the phenotype conferred by GTPBP3 and MTO1 mutations is unknown.
In this study, we analyze the transcriptional expression of GTPBP3, determine the biochemical properties of the two most abundant GTPBP3 isoforms, comparing them with those of the MnmE GTPase, and show that the partial inactivation of GTPBP3 by small interfering RNAs (siRNAs) results in mitochondrial dysfunction.
MATERIALS AND METHODS
Bacterial strains, plasmids, oligonucleotides, and microbiological media.
E. coli strains DH5α and BL21(DE3) were used for overproduction and purification of fusion proteins. Plasmids and oligonucleotides used in this study are listed in Tables 1 and 2, respectively. Four predesigned GTPBP3-specific siRNA sequences (Ambion Silencer Predesigned siRNA, annealed; catalog numbers 131736, 34441, 34348, and 34526 for human GTPBP3 GenBank accession nos. NM_032620 and NM_133644) and control nonspecific siRNA oligonucleotides (Silencer Negative Control #1 siRNA; Ambion) were used for GTPBP3 knockdown. For convenience, siRNAs 131736, 34441, 34348, and 34526 are designated siGTPBP3-1, siGTPBP3-2, siGTPBP3-3 and siGTPBP3-4, respectively. LBT (Luria-Bertani broth containing 40 μg/ml thymine) medium and LBT medium containing 20 g of Difco agar per liter were used for routine cultures and plating of E. coli. When required, antibiotics were added as recommended (29).
TABLE 1.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pBAD22 | Expression vector with PB and AraC control (Apr) | 13 |
| pCR-XL-TOPO | Cloning vector (Kanr) | Invitrogen |
| pCR3.1 | Eukaryotic expression vector with Pcmv control (Apr Kanr Neor) | Invitrogen |
| pEGFP-N1 | Eukaryotic expression vector for GFP fusions (Kanr Neor) | Clontech |
| pET15b | His tag fusion vector (Apr) | Novagen |
| pGEX4T1 | GST fusion vector (Apr) | GE Healthcare |
| pIC684 | GST-MnmE fusion | 6 |
| pIC982 | GTPBP3(Del8A) cloned into pCR-XL-TOPO | This work |
| pIC985 | GTPBP3(Ins8A) cloned into pCR-XL-TOPO | This work |
| pIC986 | GTPBP3(Del8A) inserted into pET15b digested with NdeI and BamHI | This work |
| pIC988 | GTPBP3(Ins8A) inserted into pET15b digested with NdeI and BamHI | This work |
| pIC998 | GTPBP3(Ins8A) inserted into EcoRI-cut, filled-in pEGFP-N1 | This work |
| pIC1026 | GTPBP3(Del8A) inserted into EcoRI-cut, filled-in pEGFP-N1 | This work |
| pIC1048 | pIC998 derivative lacking GFP | This work |
| pIC1055 | BglII-XbaI fragment of pIC1048 inserted between the BamHI and XbaI sites on pCR3.1 | This work |
| pIC1089 | pIC1298-related plasmid containing mutation G261A | This work |
| pIC1296 | FLAG-(ΔN)GTPBP3 inserted between sites NcoI and SalI on pBAD22 | This work |
| pIC1298 | GST fusion of GTPBP3(Ins8A) | This work |
| pIC1299 | GST fusion of GTPBP3(Del8A) | This work |
| pIC1318 | GTPBP3 G-domain inserted between sites NcoI and XhoI on pBAD22 | This work |
| pIC1376 | GTPBP3 N-terminal domain inserted between sites NcoI and XhoI on pET15b | This work |
TABLE 2.
Oligonucleotides
| Primer | Sequence (5′ → 3′) | Comment(s) or figure reference |
|---|---|---|
| HUP2 | CGGGTCGCAGGTTGTAAATCCATGTG | Plus-strand primer; start codon in bold |
| HUM2 | TCACTTGCCCACACAGAAGTCCTGGAAG | Minus-strand primer; stop codon in bold |
| LY18 | CGTGGATCCATGTGGCGGGGGCTTTGG | Start codon in bold; BamHI site underlined |
| LY19 | CGCCTCGAGTCACTTGCCCACACAGAAG | Stop codon in bold; XhoI site underlined |
| LY20 | GGAATTCCATATGTGGCGGGGGCTTTGG | Start codon in bold; NdeI site underlined |
| LY21 | CGCGGATCCTCACTTGCCCACACAGAAG | Stop codon in bold; BamHI site underlined |
| LY23 | GGTCGACTGGGGTCTCCAGCACGTCACG | Fig. 3 |
| 50HF1 | CGCCCATGGACTACAAGGACGACGATGACAAGACCATCTTCGCGCTAAGC | Start codon in bold; NcoI site underlined; FLAG-tagged encoding sequence in italics |
| G261HF | CCCAATGCGGCCAAGAGCAGCCTAG | Mutagenic sequence in bold |
| G261HR | CTAGGCTGCTCTTGGCCGCATTGGG | Mutagenic sequence in bold |
| CNT1 | CCGGGAGAGGCTAGAGCAG | Fig. 4 |
| CNT2 | ACTTGTTCAGCACCAGGAGGA | Fig. 4 |
| CNT4 | TATATCGATTTCGGCGAGGATGAC | Fig. 3 |
| CNT5 | TGACCACTACGTGCACCCCT | Fig. 3 |
| CNT33 | CCGGGAGAGTTGCAACTTCCT | Fig. 4 |
| GSP3 | GGCTCTGGGCTCCCCACAGAGGCTACG | Fig. 3 |
| GSP4 | CACTGGACCCCCCAATGCGGGCAAGAG | Fig. 3 |
| E9BR2 | CAGCCCCATTCCAGGGCCTGAATCTCCAGG | Fig. 5 |
| E7BF | TGCTGAGCGACACGGCTGG | Fig. 3 |
| MNT2 | TCCGCCAGCCCCTCCACTTC | Fig. 3 |
| 5IR | AGACAGTGAGGGGAGCCTTTG | Fig. 3 |
| RS3 | GACTTGGCTGAGTTCCCTGTCTTG | Fig. A2 |
| RS5 | AGGGCCCGCCACAGGCTG | Fig. A2 |
| RS6 | CGCAGCCAGCTCGGTCTTCAG | Fig. A2 |
| RS7 | GCCCGCCACAGCTGCAGCT | Fig. A2 |
| P26 | GGACAGGAGCAGGTGAGG | Fig. A2 |
| CYPA-F | CAAATGCTGGACCAAACACAA | Forward primer for cyclophilin A |
| CYPA-R | GCCATCCAGCCACTCAGTCT | Reverse primer for cyclophilin A |
| GAPDH-F | TGGGCTACACTGAGCACCAG | Forward primer for GAPDH |
| GAPDH-R | GGGTGTCGCTGTTGAAGTCA | Reverse primer for GAPDH |
| GFP-BamH1 | GGAATTCGGATCCTGACTTGCCCACACAGAAG | Mutagenic sequence in bold; BamHI site underlined |
| NcoFlagDGH | CGCCCATGGACTACAAGGACGACGATGACAAGAGGCTCCGCTCAGGGGCG | Start codon in bold; NcoI site underlined; FLAG-tagged encoding sequence in italics |
| XhoDGH | GGCCTCGAGTCACACTGCAGCTAGCTCCTTCCTCAGC | Stop codon in bold; XhoI site underlined |
| XhoNTH | GGCCTCGAGTTACGCGTGGATAAGGTCCGCCAGCCCCTC | Stop codon in bold; XhoI site underlined |
RNA and DNA manipulations.
Total cellular RNA was isolated from human lymphocytes and fibroblasts, as well as from different tissues of C57/B6 mice by using a High-Pure RNA isolation kit (Roche), according to the manufacturer's directions. First-strand GTPBP3 cDNA was generated from human fibroblast mRNA with poly(dT) and an Enhanced Avian RT First Strand Synthesis kit (Sigma). The GTPBP3 cDNA coding region was amplified with primers HUP2 and HUM2 and inserted into pCR-XL-TOPO. DNA sequencing of several recombinant plasmids revealed that we had cloned two GTPBP3 cDNAs differing in the insertion/deletion of exon 8A, named GTPBP3(Ins8A) and GTPBP3(Del8A) cDNA, respectively. Two selected plasmids were called pIC985 and pIC982, respectively. These plasmids were then used as templates to amplify the corresponding GTPBP3 cDNA copies with primers LY20 and LY21. The PCR products were digested with NdeI and BamHI and cloned into pET15b digested with the same enzymes, generating plasmids pIC988 and pIC986. In this way, GTPBP3 cDNAs become fused to a His tag at the N terminus and expressed under the control of the PT7 promoter. GTPBP3 cDNA carried by plasmids pIC985 and pIC982 was also amplified by PCR with primers LY18 and LY19. The PCR products were digested with BamHI and XhoI and inserted into pGEX4T1 cut with the same enzymes, producing plasmids pIC1298 and pIC1299. Thus, GTPBP3 cDNAs became fused to glutathione S-transferase (GST) at the N terminus and expressed under the control of the Ptac promoter. It should be pointed out that the GTPBP3 moiety present in pIC1298 and pIC1299 corresponds to GTPBP3 isoforms with accession numbers AAL85493 and AAK39577 in the GenBank database or Q969Y2-1 and Q969Y2-3 the in UniProtKB database, respectively, although pIC1299 carries the A250V change that has been identified as a polymorphism (dbSNP rs3810206, T→C, Val→Ala; heterozygosity index, 0.499). Plasmid pIC985 was also used as template to amplify the GTPBP3(Ins8A) cDNA with primers 50HF1 and LY19. The PCR product was then digested with NcoI and XhoI and inserted into pBAD22 cut with the same enzymes, producing plasmid pIC1296. In addition, plasmids pIC985 and pIC982 were used as templates to amplify the corresponding GTPBP3 cDNA copies with primers LY18 and green fluorescent protein (GFP)-BamHI. Note that primer GFP-BamHI contains a mutagenic sequence that changes the GTPBP3 stop codon (TGA) to a serine codon (TCA). The amplicons were digested with BamHI, filled in with Klenow, and inserted into pEGFP-N1 previously treated with EcoRI and Klenow. The resulting plasmids were named pIC998 and pIC1026, each one expressing the corresponding GTPBP3 isoform fused to GFP. pIC998 was digested with BamHI and NotI, treated with Klenow, and self-ligated to produce pIC1048. Then, the BglII-XbaI fragment of pIC1048 was inserted into pCR3.1, previously cut with BamHI and XbaI, to generate pIC1055, which expresses the GTPBP3(Ins8A) protein. Plasmid pIC1089 is a pIC1298-related plasmid that was used to overproduce protein GST-GTPBP3(Ins8A) G261A. The G261A change was generated by site-directed mutagenesis (QuikChange; Stratagene) with primers G261HF and G261HR. All constructs were verified by DNA sequencing. The expression construct for the GTPBP3 G domain (R245-V418) was made by PCR using pIC985 as a template and primers NcoFlagDGH and XhoDGH. The amplicon was digested with NcoI and XhoI and cloned into pBAD22, previously cut with NcoI and BamHI, to produce plasmid pIC1318. The expression construct for the GTPBP3 N-terminal domain (T35-A167) was made by PCR using pIC985 as a template and primers 50HF1 and XhoNTH. The amplicon was digested with NcoI and XhoI and inserted into pET15b cut with the same enzymes, producing pIC1376.
Cell culture and plasmid transfections.
Human HEK-293 cells were grown in minimum essential medium (Sigma). The FH3349B cell line was grown in the same medium supplemented with 1 mM l-glutamine, 1% vitamins, 2% essential amino acids, and 1% nonessential amino acids (Gibco, Invitrogen Life Technologies). Other cell lines were grown in regular Dulbecco's modified Eagle medium (Sigma) supplemented (SHSY5Y cells) or not (COS-7, HeLa, and HT29 cells) as above. MCF7 and U937 cells were grown in RPMI 1640 medium (Sigma). All culture media were supplemented with 10% heat-inactivated fetal bovine serum (with the exception of FH3349B cells, which used 20% fetal bovine serum) and 1% penicillin-streptomycin antibiotic solution (Gibco, Invitrogen Life Technologies). The HEK-293 cell line was used in transient and permanent transfections. Cells were transfected with plasmid pIC1055 (20 μg) by using 12 μl of Fugene 6 transfection reagent (Roche) and collected using trypsin-EDTA (Sigma) after 36 h (transient transfection experiments) or cultured under Geneticin selection (Gibco, Invitrogen Life Technologies) for permanent GTPBP3 expression.
Protein techniques and production of antisera.
Expression and purification of the GST-MnmE protein was performed as previously described (27). Overproduction of GST-fused GTPBP3 was done in DH5α cells transformed with pIC1298 or pIC1299. The cells were grown to early log phase and induced by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to 10 μM. After 5 h at 30°C, cells were collected, disrupted by sonication, and centrifuged, and the supernatant was applied over glutathione agarose (Sigma). After 2 h of stirring, the resin was loaded into a column. The column was washed with at least 10 column volumes of 50 mM Tris-HCl (pH 7.6), 50 mM KCl, 50 mM NaCl, 2 mM MgCl2, 5% glycerol, 0.15 mM phenylmethylsulfonyl fluoride (PMSF), and 0.5% Triton X-100. Then, the column was further washed with 10 column volumes of the same buffer without PMSF and Triton X-100. GST-GTPBP3 was eluted with 5 ml of a buffer containing 10 mM glutathione, 50 mM Tris-HCl (pH 8), 50 mM KCl, 50 mM NaCl, and 5% glycerol. Overproduction of FLAG-GTPBP3 and the FLAG-tagged GTPBP3 G domain was done in DH5α transformed with pIC1296 and pIC1318, respectively. The cells were grown at 37°C to early log phase, induced by the addition of arabinose (0.1%), and incubated overnight at 25°C. Overproduction of the FLAG-tagged GTPBP3 N-terminal domain was done in BL21 cells transformed with pIC1376. The cells were grown at 37°C to early log phase, induced by the addition of IPTG to 10 μM, and incubated overnight at 25°C. Cells were collected, disrupted by sonication, and centrifuged, and the supernatant was applied over anti-FLAG M2 agarose (Sigma). After the flowthrough was collected, the resin was washed with 10 column volumes of TBS (50 mM Tris-HCl [pH 7.5], 100 mM NaCl, 5 mM dithiothreitol [DTT]) containing 1 mM PMSF and 0.03% Triton X-100. Then, the column was further washed with 10 column volumes of TBS. FLAG-tagged proteins were eluted with 5 ml of TBS containing 0.1 mg/ml of FLAG peptide (Sigma). Proteins were quantified by the Bradford assay using bovine serum albumin as a standard. New Zealand rabbits were inoculated with inclusion bodies produced by overexpression of His-tagged GTPBP3(Ins8A) protein in E. coli after a preimmune serum sample had been taken. The animals were bled and reinoculated at the appropriate times. Serum was aliquoted and stored at −80°C in 0.02% (wt/vol) sodium azide. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting were carried out essentially as described previously (11).
Immunoprecipitation experiments.
For GTPBP3 immunoprecipitation, extracts from HEK-293 cells permanently transfected or not with pIC1055 (about 2 and 5 mg of protein, respectively) in lysis buffer (150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0, containing 0.1 mM leupeptin, and 1 mM PMSF) were precleared by treatment with 100 μl of anti rabbit immunoglobulin G (IgG)-agarose beads (Sigma) for 1 h at 4°C in a rotary wheel and centrifugation at 10,000 × g for 5 min. Then, the supernatants were incubated overnight in a rotary wheel with 40 μl of a rabbit polyclonal antibody toward GTPBP3, washed five times with NET buffer (150 mM NaCl, 0.5% Nonidet P-40, 0.1% sodium deoxycholate, 0.1% SDS, 5 mM EDTA, 0.1% bovine serum albumin, 20 μg/ml chloramphenicol, 50 mM Tris, pH 7.0) and further incubated in the rotary wheel for 2 h at 4°C with 200 μl of anti rabbit IgG-agarose beads. The immunoprecipitate was washed five times with NET buffer, solubilized in sample buffer, separated by SDS-PAGE, and immunoblotted with anti-GTPBP3 and rabbit True-blot (eBioscience, Inc.) in conjunction with a horseradish peroxidase-conjugated detection system.
Gel filtration chromatography and native PAGE.
Gel filtration analysis of the FLAG-(ΔN)GTPBP3(Ins8A) protein, which lacks the first 34 N-terminal residues of GTPBP3, was performed using a Superdex 200 HR 10/30 column (Amersham Biosciences) in 50 mM Tris, pH 7.5, 5 mM MgCl2, 50 mM β-mercaptoethanol, and 100 mM NaCl at a flow rate of 0.4 ml/min. Analysis of the G and N-terminal domains of GTPBP3 was performed using a Superdex 75 10/300 GL column in the appropriate buffers at a flow rate of 0.8 ml/min. Gels and buffers used for native PAGE were made according to the standard Laemmli SDS protocol, omitting the SDS. Native gels (10% polyacrylamide) were run at 15 mA and 4°C for 3.5 to 4 h and stained with Coomassie blue.
GTPase and nucleotide-binding assays.
GTPase assays were done with a malachite green-based kit (602-0120; Innova Biosciences, United Kingdom), following the manufacturer's instructions. Assays were performed in 100 μl of GTPase buffer (50 mM Tris-HCl, pH 7.5, 100 mM KCl, 2 mM MgCl2) containing 2.0 μM protein and 1 mM GTP at 37°C for 10 min. Samples were read within the next 15 min in a Labsystems Multiskan Plus plate reader using a 690-nm filter. To determine values for Vmax and Km, purified GTPBP3(Ins8A) (16 μg) was incubated with increasing concentrations (0 to 1.25 mM) of GTP in 100 μl of GTPase buffer at 37°C for 10 min, and samples were treated as described above. Data were fitted to the Michaelis-Menten equation using nonlinear regression (GraphPad Prism, version 4.0 for Windows; GraphPad Software, Inc). GTPBP3 proteins were titrated against fluorescent mant (N-methyl-3′-O-anthranoyl) nucleotides (Jena Bioscience) until saturation was reached. The mant nucleotides (2 μM) were excited at 360 nm, and the fluorescence was monitored at 440 nm (LS 50 B spectrophotometer; PerkinElmer Life Sciences). All binding assays were performed at 25°C in GTPase buffer (50 mM Tris-HCl, pH 7.5, 50 mM KCl, 2 mM MgCl2, 5% glycerol). The binding constants (Kd values) were calculated by fitting the curves with nonlinear regression, using GraphPad Prism software.
Labeling of double-stranded siRNA and siRNA transfection.
Silencer Negative Control #1 siRNA and GTPBP3 chemically synthesized double-stranded siRNAs were labeled with a Silencer Cy5 Labeling Kit (Ambion). HEK-293 cells (2.5 × 105 cells/well) were inoculated in six-well plates and cultivated for 24 h. Cell confluence was about 50% at the time of transfection. siPORT amine transfection agent (Ambion) was diluted in Opti-MEM1 (Invitrogen) and incubated for 10 min at room temperature. Then, the siRNAs, labeled or not with Cy5, were added to the transfection agent solution and incubated again for 10 min. Finally, transfection mixtures were dropped on the culture dishes containing HEK-293 cells in such a way that the final concentration of siRNA was 100 nM. To avoid cytotoxicity, culture medium was removed and replaced with fresh medium 24 h after transfection. Transfection efficiency was higher than 85% when evaluated by direct fluorescence 72 h after transfection. HEK-293 cells transfected with unlabeled siRNAs were collected by trypsinization after 72 h and used for quantitation of GTPBP3 mRNA by reverse transcription-PCR (RT-PCR), determination of cell viability, mitochondrial membrane potential, oxygen consumption, intracellular ATP, and superoxide levels, as well as for studies of mitochondrial protein synthesis and degradation. It should be pointed out that other concentrations of siRNAs were also used (25, 50, and 75 nM), but quantitation of GTPBP3 mRNA by RT-PCR indicated that GTPBP3 silencing was maximum at 100 nM.
Quantitative real-time PCR.
To investigate the tissue-specific expression of GTPBP3 mRNAs that differ only in the insertion/deletion of exon 8A, we made use of a normalized panel of human cDNAs (Human I MTC panel; Clontech). Primer pairs CNT1/CNT2 and CNT33/CNT2 were used to specifically amplify the insertion and deletion variants, respectively, whereas the CNT4/CNT5 pair was used to amplify total cDNA. Quantitative real-time PCR was performed using Power Sybr Green Master Mix. The efficiency values obtained for the real-time PCR amplification of the cDNA segments were very near to 2. HEK-293 cells transfected with the GTPBP3 siRNAs or Silencer Negative Control #1 siRNA were harvested by trypsinization after 72 h of transfection. Then, cells were centrifuged at 100 × g for 5 min, and total RNA was isolated using a High Pure Total RNA Isolation Kit (Roche). GTPBP3 mRNA levels were measured by quantitative one-step real-time PCR using primers CNT4 (forward primer) and CNT5 (reverse primer), Power Sybr Green Master Mix, and MultiScribe reverse transcriptase (Applied Biosystems). Assays were performed using a Gene Amp 5700 Sequence Detection System (Applied Biosystems). GTPBP3 expression levels were normalized against cyclophilin and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as endogenous controls. Relative expressions were calculated using the comparative ΔΔCT (where CT is threshold cycle) method.
Fluorescence microscopy.
At 72 h posttransfection, cells grown in coverslips were fixed with 3% paraformaldehyde in phosphate-buffered saline (Sigma), pH 7.4, for 30 min. The transfection efficiency was analyzed by direct fluorescence using an Axioscope 2 microscope (Zeiss). Images were taken with a CoolSNAP Fx digital camera (Photometrics), and the RSI image software (Roper Scientific) was used to edit them.
Flow cytometry studies.
Cells were incubated at 37°C with trypsin-EDTA solution (Sigma). After 3 min, the reaction was stopped with 1.5 ml of culture medium. After incubation with the different probes, as described below, cells were analyzed using a Cytomics FC-500 flow cytometer (Beckman Coulter Inc.). Data acquisition and analysis were carried out with the Cytomics RXP analysis software (Beckman Coulter Inc.). For statistical significance, 10,000 cells were analyzed in each sample, and the mean of the emitted fluorescence values was used.
Mitochondrial protein synthesis and degradation.
The effects of GTPBP3 silencing through RNA interference on the incorporation of [3H]leucine into proteins was determined by incubating, 72 h after silencing, exponentially growing cells (∼5 × 105 cells/ml) in culture medium with and without 200 μg/ml cycloheximide, as previously described (21). After 10 min, 50 μCi/ml [3H]leucine (57 Ci/mmol) was added, and the cells were incubated for 60 min at 37°C. Then, after the cells were washed twice with phosphate-buffered saline containing 2 mM leucine plus cycloheximide, they were detached from the flasks and precipitated twice with ice-cold 5% trichloroacetic acid. After the precipitated protein was dissolved in 0.2 N NaOH, the radioactivity was determined by liquid scintillation counting, and protein was assayed by standard procedures. Degradation of proteins synthesized in the presence or in the absence of cycloheximide was measured, after the cells were washed, as the net release, at the various chase times, of trichloroacetic acid-soluble radioactivity from the cells, labeled with [3H]leucine, into fresh culture medium containing 2 mM leucine, as has been previously described (11, 47).
Mitochondrial membrane potential and superoxide determinations.
Cell suspension aliquots (106 cells/ml) were used for the various cytometry analyses. Mitochondrial membrane potential was determined by incubating cells at 37°C with 100 nM MitoTracker Red CMXRos (Invitrogen) for 30 min or 65 nM rhodamine 123 (Calbiochem) for 15 min and analyzing either the red fluorescence emitted by MitoTracker (FL3; 625 nm) or the green fluorescence emitted by rhodamine 123 (FL1; 525 nm). Mitochondrial superoxide was measured by incubating cells at 37°C with 5 μM MitoSOX Red (Molecular Probes) for 30 min and analyzing the red fluorescence emitted by the dye (FL3; 625 nm).
Oxygen consumption.
The endogenous respiration rate was measured as described previously (31). Briefly, at 72 h posttransfection cells were collected by trypsinization and washed twice with culture medium. Cells (3 × 106 cells/ml) were transferred to a chamber equipped with a Clark-type oxygen electrode (Rank Bros., Bottisham, United Kingdom), calibrated with an air-saturated culture medium, assuming an oxygen concentration of 200 μM. Sodium cyanide (1 mM) was used to confirm that oxygen consumption was mainly mitochondrial (approximately 95 to 99%). Measurements were collected using a data acquisition device, Duo.18 (World Precision Instruments, Stevenage, United Kingdom).
Measurement of intracellular ATP.
The amount of ATP was measured with an ATP bioluminescence assay kit (HSII; Roche), according to the manufacturer's instructions. Determinations were made 72 h after transfection with cells collected by trypsinization, resuspended in minimal essential medium (supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin-streptomycin antibiotic solution), centrifuged at 100 × g, and resuspended in the kit dilution buffer at 106 cells/ml. Luminescence was determined using a Wallac Victor V 2 1420 Multilabel HTS Counter (PerkinElmer, Life Sciences).
Statistical analysis.
Statistical comparison among groups was carried out by the Student's test. A P value of <0.05 was considered statistically significant. Values are expressed as the mean ± standard deviation.
RESULTS
Analysis of human GTPBP3 transcripts.
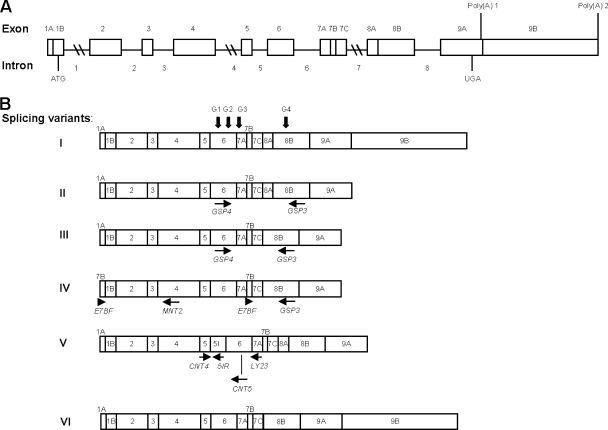
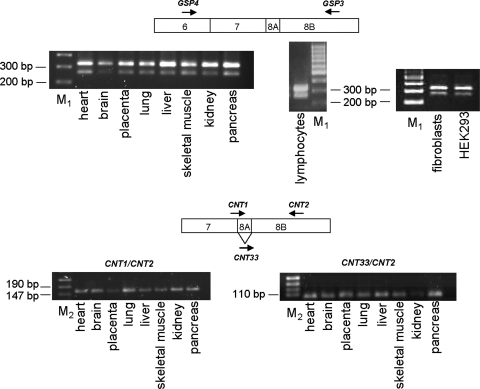
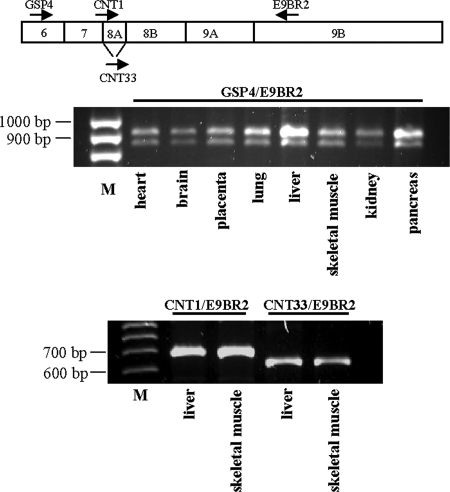
In a previous work, five cDNAs from human GTPBP3 were obtained and proposed to be splicing variants (variants I to V) (Fig. 3); however, their presence in human tissues was not firmly established (23). According to this seminal work, GTPBP3 is composed of nine exons and eight introns, and the five transcripts would result from the alternative splicing of exons 1A, 7B, 8A, and 9B and intron 5 (Fig. 3). Variant I is the longest one and corresponds to the full-length GTPBP3 cDNA. Variants II to V lack the last exon (exon 9B), which affects only the length of the 3′ untranslated region. Variant II differs just in this point from variant I, whereas variant III lacks exon 8A compared to variant II. This is particularly interesting because exon 8A encodes 21 amino acids located between the G3 and G4 motifs of the GTPBP3 G domain. Since the GTPase activity of MnmE is essential for its tRNA modification function (27, 57), the lack of exon 8A could have significant consequences for the functionality of the protein encoded by variant III. Variant IV carries an additional exon 7B at the beginning of cDNA and lacks exons 1A and 8A. Finally, variant V differs from variant II because of the retention of intron 5, which supposes the in-frame insertion of 32 additional amino acids. For confirmation of these splicing variants, a PCR analysis of cDNAs derived from several sources was performed. We used primers GSP3 and GSP4 (Fig. 3B) to detect variants differing in splicing of exon 8A. As shown in Fig. 4 (top panel), two PCR products of about 320 and 260 bp were obtained. The difference in the lengths of the two products fits well with the size of exon 8A (63 bp). These results suggest that partial skipping of exon 8A occurs in all tested tissues. To corroborate the alternative splicing of this exon, we designed primers to specifically amplify each variant (Fig. 4, bottom panel). Thus, primer CNT1 spans the junction region between exons 7 and 8A, whereas CNT33 spans the junction region between exons 7 and 8B. Primer CNT2, located inside exon 8B, was used as reverse primer in each reaction. As shown in Fig. 4, the primer pairs CNT1/CNT2 and CNT33/CNT2 generated single PCR products of around 160 and 100 bp, respectively, confirming the partial skipping of exon 8A. Curiously, we found that splicing of this exon occurs more frequently in primates than in rodents, which may be related to features of the regulatory sequences that precede exon 8A and 8B (see Appendix).
FIG. 3.
Structure of human GTPBP3 gene and its products. (A) Diagram of human GTPBP3 gene structure as proposed by Li and Guan (23). Boxes represent exons, with the positions of initiation and termination codons indicated. Two poly(A) sites are indicated. The nine exons and eight introns are numbered. (B) Schematic representation of six GTPBP3 transcripts. Transcripts I to V were proposed to result from the alternative splicing of exons 1A, 7B, 8A, and 9B and intron 5 (23). Transcript VI is the new variant found in this work. Locations of the G motifs G1 to G4 are indicated by vertical arrows on splicing variant I. Locations of some the primers used in this work are indicated by horizontal arrows below variants II to V.
FIG. 4.
Direct PCRs to detect alternative splicing of human GTPBP3 exon 8A. Primers used are represented by arrows on the diagrams above the gels. PCR analysis was performed on cDNAs from eight different human tissues (Human I MTC panel; Clontech, California) and cDNA synthesized on DNase-treated mRNA isolated from human fibroblasts and lymphocytes and HEK-293 cells. M1, GeneRuler 100-bp DNA Ladder (Fermentas); M2, pUC19 DNA/MspI Marker, 23 (Fermentas).
On the other hand, no PCR product could be synthesized on human cDNA from several sources by using primers E7BF and MNT2, which align with exon 7B and exon 4, respectively (Fig. 3B, variant IV). In contrast, E7BF together with GSP3 (aligning with exon 8B) (Fig. 3B) generated two PCR fragments of about 180 and 120 bp, again revealing the presence of two variants differing in the splicing of exon 8A (data not shown). Thus, it may be concluded that the putative GTPBP3 transcript that carries exon 7B at the 5′ end is not present in the tissues analyzed here and that it probably represents an experimental artifact. Finally, only one band was obtained when primers CNT4 and LY23 (Fig. 3B, variant V) were used to amplify cDNAs synthesized on DNase-treated mRNA derived from lymphocytes, skeletal muscle, kidney, fetal kidney, and HEK-293 cells (data not shown). The size of this band fits well with that expected if intron 5 is not retained (267 bp). Moreover, no direct PCR product was obtained from the aforementioned cDNAs by using primers CNT4 and 5IR (data not shown), the last one pairing with a segment of intron 5 (Fig. 3B, isoform V). These results suggest that variant V is not a true splicing variant or that its expression level is below the detection limit of our PCR technique.
cDNA data indicate that GTPBP3 contains two polyadenylation sites (Fig. 3A) (23, 43) (GenBank accession numbers AY078988, AF360742, BC017207, and BC019261), which generate two sets of transcripts differing in the presence of exon 9B (719 bp). Both sets of transcripts, of about 2.6 and 1.9 kb, were detected by Northern blot analysis, which also indicated that the longer mRNAs were more abundant than the smaller ones (23; also our own unpublished results). It was then suggested that variant I including exon 9B would be responsible for the 2.6-kb transcript, whereas variants II to V lacking exon 9B would contribute to the 1.9-kb mRNA species (23). The 3′ untranslated regions of eukaryotic mRNAs contain regulatory elements affecting mRNA translation, stability, and transport (2, 45). Thus, it was proposed that the presence of exon 9B confers a higher stability to variant I, explaining its prevalence over the shorter ones (23). We wondered whether an mRNA without exon 8A but containing exon 9B could exist and, according to its size, also contribute to the longest mRNA species detected by Northern blot analysis. To test this, we performed a PCR analysis by using primers designed to amplify variants including exon 9B but differing in splicing of exon 8A. As shown in Fig. 5 (top), primers GSP4 (exon 6) and E9BR2 (exon 9B) generated two PCR products with sizes fitting those expected if both variants were present. Moreover, primers CNT1 and CNT33, spanning the junction regions of exons 7/8A and 7/8B, respectively, produced, together with the reverse primer E9BR2 (exon 9B), specific products representative of each splicing variant (Fig. 5, bottom). These results clearly indicate that two transcripts containing exon 9B but differing in the splicing of exon 8A are present in all tested tissues. Therefore, we identified a new mRNA variant (Fig. 3, variant VI) defined by the lack of exon 8A and the presence of exon 9A that may be responsible, together with variant I, for the longest, most stable, mRNA species detected by Northern blotting.
FIG. 5.
Direct PCRs to detect mRNA variants of human GTPBP3 that include exon 9B but differ in the splicing of exon 8A. Primers used are represented by arrows on the diagram and are indicated over each gel. Primer CNT1 spans the junction region between exons 7 and 8A, whereas CNT33 spans that between exons 7 and 8B. PCR analysis was performed on cDNAs from the indicated human tissues (Human I MTC panel; Clontech, California). M, GeneRuler 100-bp DNA Ladder (Fermentas).
Real-time PCR experiments performed on a normalized human tissue cDNA panel (Human I MTC panel; Clontech, California) indicated that amplicons corresponding to the exon 8 insertion/deletion variants, as well as amplicons corresponding to total GTPBP3 mRNA, appeared late during the amplification reaction in relation to the housekeeping genes used as controls (GAPDH and cyclophilin), which suggests that the levels of GTPBP3 transcripts are low (data not shown). These experiments also indicated that the tissue expression pattern of GTPBP3 transcripts differing in the presence of exon 8A is quite similar, with higher levels of both types in skeletal muscle (data not shown). However, we noted some differences in the tissue-specific expression of GTPBP3 among different series of the same panel of cDNAs. This could be due to the procedures used to normalize panels, which could determine that quantification of low-expression genes, such as GTPBP3, becomes imprecise. Therefore, other approaches should be followed in order to exactly quantify the tissue expression of mature mRNA forms of GTPBP3.
In brief, our data support the idea that GTPBP3 has a low transcription level and that the transcript variants I and VI (Fig. 3) are responsible for the most abundant GTPBP3 mRNA species (2.6 kb) previously detected by Northern analysis (23), whereas their 3′-shortened versions (variants II and III, respectively) contribute to the less abundant ones (1.9 kb). Our data also indicate that variants IV and V are not true splicing variants or, alternatively, that they are expressed at extremely low levels. Of note, all these splicing variants contain exons 1B and 2 (Fig. 3), which encode a putative mitochondrial targeting sequence (see below). In fact, Li and Guan (23) found that the full-length GTPBP3 protein (i.e., that encoded by transcript variants I and II), here named GTPBP3(Ins8A), localizes exclusively to mitochondria. Now, we confirm this result and show that the GTPBP3 isoform lacking exon 8A (i.e., that encoded by transcript variants III and VI), here named GTPBP3(Del8A), also localizes to mitochondria (see Fig. S1 in the supplemental material).
Human GTPBP3 protein is expressed at low levels.
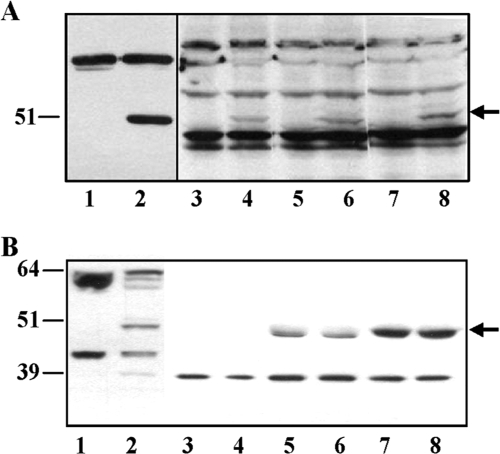
Anti-human GTPBP3 polyclonal antibodies recognized the E. coli-expressed GTPBP3 protein (Fig. 6A, lane 2), but no band that could be clearly assigned to native GTPBP3 was detected in extracts from several human cell lines, including HEK-293 (embryonic kidney) (Fig. 6A, lanes 3, 5, and 7), as well as HeLa (cervix carcinoma), MCF7 (breast adenocarcinoma), U937 (lymphoma), 3349B (fibroblasts), HT29 (colon adenocarcinoma), and SHSY5Y (neuroblastoma) (data not shown). In contrast, the anti-human GTPBP3 antibody recognized one band of about 51 kDa in HEK-293 cells permanently transfected with plasmid pIC1055, which encodes GTPBP3(Ins8A) (Fig. 6A, compare lanes 4, 6, and 8 with lanes 3, 5, and 7). These results suggest that the native expression level of GTPBP3 is very low and that the 51-kDa band detected in pIC1055-transfected cells mostly corresponds to the recombinant GTPBP3(Ins8A) protein. The 3-kDa difference between the 51-kDa band and the calculated molecular mass of GTPBP3(Ins8A), 54 kDa, is probably due to the loss of the mitochondrial targeting signal of GTPBP3(Ins8A) that is present in the GTPBP3(Ins8A) cDNA carried by plasmid pIC1055.
FIG. 6.
Expression of GTPBP3 in HEK-293 cells. (A) Immunoblotting using anti-GTPBP3 of extracts from HEK-293 cells nontransfected (lanes 3, 5, and 7) or permanently transfected with plasmid pIC1055 (lanes 4, 6, and 8) or from E. coli strain DH5α/pIC1296 grown in the absence (lane 1) or in the presence (lane 2) of inducer (arabinose). Samples were separated by 10% SDS-PAGE, and the amounts of bulk protein which were analyzed in each lane were as follows: 100 μg (lanes 1 and 2), 200 μg (lanes 3 and 4), 300 μg (lanes 5 and 6), and 400 μg (lanes 7 and 8). Signals were visualized by ECL (lanes 1 and 2) and ECL Advance (lanes 3 and 8) from GE Healthcare, and during film exposure each pair of lanes (1 and 2, 3 and 4, 5 and 6, and 7 and 8) was independently handled until optimal visualization of GTPBP3 bands was achieved. Nonspecific bands show equal loading in each pair of lanes (paired as above). Position and size (in kDa) of FLAG-(ΔN)GTPBP3 (detected in lane 2) are indicated on the left, and the arrow marks GTPBP3(Ins8A) position (detected in lanes 4, 6, and 8). (B) Extracts from HEK-293 cells permanently transfected or not with pIC1055 (approximately 2 or 5 mg of protein, respectively) were immunoprecipitated using anti-GTPBP3 as described in Materials and Methods. Immunoprecipitates and total extracts were resolved in a NuPAGE 4 to 12% Bis-Tris gel (Invitrogen) and immunoblotted with anti-GTPBP3 and rabbit True-blot (eBioscience, Inc.). Lane 1, total extract of nontransfected HEK-293 cells (100 μg); lane 2, total extract of HEK-293 cells permanently transfected with plasmid pIC1055 (50 μg); lanes 3 and 4, material bound to anti-rabbit IgG agarose beads (no antibody); lanes 5 and 6, immunoprecipitates of nontransfected HEK-293 cells; lanes 7 and 8, immunoprecipitates of HEK-293 cells transfected with plasmid pIC1055. Positions and sizes (in kDa) of mass markers are indicated on the left, and the arrow (on the right) marks GTPBP3 position.
In a new attempt to detect endogenous GTPBP3 protein, immunoprecipitation experiments from extracts of HEK-293 cells, transfected or not with plasmid pIC1055, and Western blot analysis of immunoprecipitates (using rabbit IgG TrueBlot) led to detection of a band of about 51 kDa (Fig. 6B, lanes 5 to 8), which fits well with the expected size for both mature GTPBP3 isoforms, GTPBP3(Ins8A) and GTPBP3(Del8A) (51 and 49 kDa, respectively). Note that this band is more intense in immunoprecipitates obtained from HEK-293 cells permanently transfected with pIC1055 (lanes 7 and 8), which might be due to the joint contribution of the native GTPBP3 isoforms (51 and 49 kDa) and the recombinant GTPBP3(Ins8A) protein (51 kDa) that are being simultaneously expressed in these cells. Considering that mature GTPBP3(Ins8A) and GTPBP3(Del8A) isoforms differ by 2 kDa, we expected them to resolve into two electrophoretically distinguishable species. However, after various attempts, only the ∼51-kDa band was detected in immunoprecipitates of both transfected and nontransfected HEK-293 cells (Fig. 6B, lanes 5 to 8, and data not shown). It is possible that the Ig long chain (55 kDa), which migrates close to the 51-kDa band but is not visible due to the use of rabbit IgG True-Blot, impairs band resolution, making both GTPBP3 isoforms comigrate to the same position. Alternatively, or in addition, the 51-kDa band could mostly correspond to the GTPBP3(Ins8A) protein (native form in lanes 5 and 6; native plus recombinant form in lanes 7 and 8) if the levels of the native GTPBP3(Del8A) are extremely low.
Guanine nucleotide binding capability of GTPBP3 isoforms is similar to that of the bacterial MnmE protein.
The His-tagged GTPBP3 proteins encoded by pIC986 and pIC988 were insoluble when expressed in E. coli. In contrast, the GST-GTPBP3 fusion proteins produced from pIC1298 and pIC1299 were soluble and able to bind nucleotides (see below), but their GST moiety could not be removed by thrombin digestion. Therefore, we decided to overproduce GST-GTPBP3(Ins8A) and GST-GTPBP3(Del8A) and to analyze their biochemical properties in parallel with those of MnmE and GST-MnmE. We reasoned that by comparing MnmE and GST-MnmE, we could estimate the effects of GST on the MnmE properties and, thus, infer its putative effects on GTPBP3. In our analysis, we also used the recombinant protein produced by plasmid pIC1296, in which the FLAG peptide is fused to residue 35 of GTPBP3.
Capability of proteins to bind guanine nucleotides was analyzed by using fluorescent nucleotides as described previously (27). The Kd values of GST-MnmE for mant GPTγS (mGTPγS) and mant GDP (mGDP) were similar to those of MnmE and remained in the low micromolar range, which indicates that the GST moiety does not significantly alter the nucleotide binding properties of MnmE (Table 3). This is not a surprising result considering that the structural independence of the G domain from the remaining MnmE domains is supported not only by crystallographic data but also by the fact that the isolated G domain retains the enzymatic properties of the full-length protein (6, 39, 40). Hence, we can assume that the GST moiety should not significantly change the G-domain properties of the GTPBP3 isoforms either. As shown in Table 3, the calculated Kd values of GST-GTPBP3(Ins8A) and GST-GTPBP3(Del8A) for mGTPγS and mGDP were similar to those obtained for GST-MnmE. Therefore, it may be concluded that both human isoforms have moderate affinity for nucleotides and a slight preference for GTP over GDP. Note that the FLAG-(ΔN)GTPBP3 behavior was similar to that of the GST-GTPBP3 proteins, supporting the idea that the GST moiety does not alter the properties of the GTPBP3 G-domain.
TABLE 3.
Binding parameters of MnmE and GTPBP3 proteins
| Protein |
Kd (μM)
|
|
|---|---|---|
| mGTPγS | mGDP | |
| MnmE | 1.29 ± 0.15 | 3.19 ± 0.23 |
| GST-MnmE | 1.08 ± 0.06 | 2.21 ± 0.22 |
| GST-GTPBP3(Ins8A) | 0.55 ± 0.05 | 1.04 ± 0.05 |
| GST-GTPBP8(Del8A) | 0.41 ± 0.05 | 1.06 ± 0.01 |
| FLAG-(ΔN)GTPBP3 | 0.64 ± 0.06 | 1.71 ± 0.09 |
| GTPBP3 G domain | 0.70 ± 0.07 | 2.17 ± 0.13 |
GTPBP3 isoforms exhibit a very low GTPase activity compared to MnmE.
In the presence of potassium, proteins MnmE from E. coli and Thermatoga maritima show a high intrinsic GTPase activity (kcat, ∼10 min−1) (6, 39, 48) with respect to Ras (kcat, ∼0.03 min−1) (38). Curiously, we have found here that the recombinant human GTPBP3 isoforms, GST-GTPBP3(Ins8A), GST-GTPBP3(Del8), and FLAG-(ΔN)GTPBP3, exhibit a GTPase activity similar to that of the nonactivated Ras proteins, thus differing from MnmE (Table 4).
TABLE 4.
GTPase activities of MnmE and GTPBP3 proteins
| Protein | kcat/min |
|---|---|
| MnmE | 11.645 ± 0.145 |
| GST-MnmE | 10.834 ± 0.262 |
| GST-GTPBP3(Ins8A)a | 0.076 ± 0.008 |
| GST-GTPBP3(Del8A)a | 0.091 ± 0.001 |
| GST-GTPBP3(Ins8A) G261Aa,b | 0.016 ± 0.001 |
| FLAG-(ΔN)GTPBP3a | 0.070 ± 0.007 |
| GTPBP3 G domain | 0.065 ± 0.005 |
The Km for GTP hydrolysis by GTPBP3 proteins (calculated as specified in Materials in Methods) was around 250 μM.
GST-GTPBP3(Ins8A) G261A is a mutant protein impaired for guanine nucleotide binding, which was used as a negative control in these experiments (for location of residue G261, see Fig. 2).
G proteins use different mechanisms to stimulate their intrinsically low GTPase activity. Recent biochemical and structural data support the idea that MnmE uses a new mechanism where the G domains of the protein dimerize in a potassium-dependent manner and induce GTP hydrolysis (39). The MnmE G-domain dimerization may be followed by using fluorescent nucleotides (mGTP or mGDP and aluminum fluoride [aluminum fluoride in solution is represented by an equilibrium between different species; therefore, it is denoted here as AlFx because we do not know which species is predominantly involved in our experiments], which mimics the γ-phosphate of GTP) since it produces a conformational change around the nucleotide that increases the fluorescence signal (39). This change stabilizes the switch regions and reorients the catalytic machinery. Given that dimerization precedes GTP hydrolysis, it is possible to uncouple both steps with appropriate mutations. Thus, mutation E282A, which occurs downstream of motif G3 (Fig. 2), strongly reduces the hydrolysis activity whereas it does not impair dimerization (39).
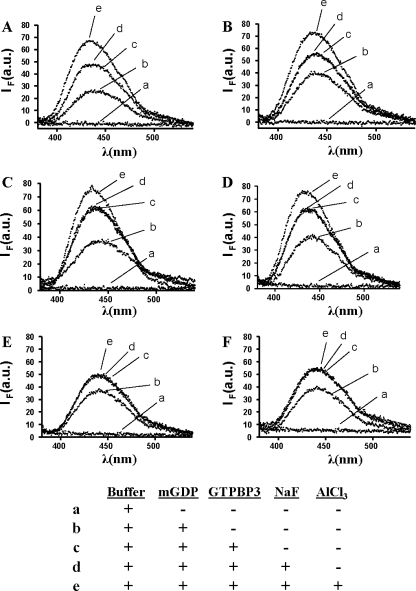
To investigate the origin of the lower GTPase activity of GTPBP3, we explored the capability of its G domain to dimerize by analyzing the interaction of a GTPBP3-mGDP complex with AlFx. As shown in Fig. 7A, formation of the MnmE-mGDP-AlFx complex results in a noticeable increase (28%) of the fluorescence produced by the binding of MnmE to mGDP (compare lines c and e), which reveals the structural changes associated with dimerization of the MnmE G domain and reorientation of the catalytic machinery. Figure 7B indicates that the GST moiety does not affect such reorganization at the G-domain active center, as expected. Figures 7C and D show an ∼19% fluorescence increase when complexes of GST-GTPBP3(Ins8A) and GST-GTPBP3(Del8A) with GDP bind AlFx (compare lines c and e in each figure). Given the extremely low GTPase activity of both GTPBP3 isoforms (Table 4), this fluorescence increase should mostly result from the G-domain dimerization which, in contrast to dimerization of the MnmE G domain, appears unable to reorient the catalytic machinery and drive GTP hydrolysis. If so, the behavior of the GTPBP3 isoforms would be more similar to that of the MnmE E282A mutant which, as described above, is impaired for GTP hydrolysis in spite of the ability of its G domain to dimerize (39). It should be noted that the fluorescence increase resulting from binding of GTPBP3 to mGDP occurs in the absence of potassium (Fig. 7, compare lines b and c in panels E and F), whereas the increase resulting from binding of the GTPBP3-mGDP complex to AlFx (and related to the GTPBP3 G-domain dimerization) was absolutely dependent on this cation since it could not be detected when KCl was replaced by NaCl in the reaction buffer (Fig. 7, compare lines c and e in panels C versus E and D versus F). This indicates that dimerization of the GTPBP3 G domain has the same cation dependence as dimerization of the MnmE G domain. Similar results to those shown in Fig. 7C and E were obtained when protein FLAG-(ΔN)GTPBP3 was used in place of GST-GTPBP3(Ins8A) (data not shown).
FIG. 7.
Binding of GST-GTPBP3 proteins to mGDP and AlFx. Fluorescence spectra (excited at 360 nm) include GTPase buffer (a) and the same solution after subsequent addition of 2 μM mGDP only (b), 10 μM protein (c), 10 mM NaF (d), and 70 μM AlCl3 (e). The table underneath the figure summarizes the conditions under which the spectra were obtained. Note that AlFx is formed by mixing an aluminum salt (AlCl3) with NaF. Fluorescence emission spectra were recorded 15 min after AlCl3 addition. λ, wavelength; IF, fluorescence intensity; au, arbitrary units. (A) MnmE. (B) GST-MnmE. (C) GST-GTPBP3(Ins8A). (D) GST-GTPBP3(Del8A). (E) GST-GTPBP3(Ins8A) (KCl was replaced by NaCl in the assay buffer). (F) GST-GTPBP3(Del8A) (KCl was replaced by NaCl in the assay buffer).
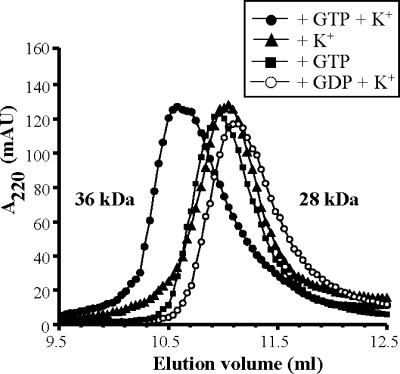
To further explore the properties of the GTPBP3 G domain, we separately cloned the G domain corresponding to isoform GTPBP3(Ins8A) (residues 245 to 418) (Fig. 2). The G domain could be stably expressed and exhibited guanine nucleotide and GTPase activities similar to the full-length protein (Tables 3 and 4). To verify the cation- and nucleotide-dependent oligomerization state of the GTPBP3 G domain, we analyzed its behavior in gel filtration experiments. The GTPBP3 G domain eluted at a position consistent with a dimeric form (36-kDa apparent molecular mass) only in the presence of GTP and potassium (Fig. 8). In the absence of either GTP or potassium, the GTPBP3 G domain eluted as an elongated monomer (28-kDa apparent molecular mass). It should be noted that the monomeric form of the MnmE G domain also eluted at a molecular mass larger than expected (39).
FIG. 8.
Gel filtration analysis of the multimerization behavior of the GTPBP3 G domain. Experiments were performed in 50 mM Tris, pH 7.5, 5 mM MgCl2, 5 mM DTT, and 100 mM KCl or NaCl. This buffer was supplemented with 1 mM GTP or GDP as indicated. In all, a 150 μM concentration of the GTPBP3 G domain was incubated for 30 min on ice, under the conditions being investigated, before application onto the Superdex 75 10/300 GL column. Apparent molecular masses corresponding to elution volumes are indicated. AU, arbitrary units.
In brief, our results support the idea that GTP and potassium induce dimerization of the GTPBP3 G domain. Such dimerization probably produces a conformational change around the nucleotide that may explain the fluorescence increase seen when GTPBP3 binds to mGDP-AlFx (Fig. 7C and D). In contrast to MnmE, dimerization of the G domain does not stimulate the GTPase activity of GTPBP3 (Table 4). Therefore, we propose that structural differences between the G-domain dimer of MnmE and GTPBP3 should be responsible for the lower catalytic activity of the human protein, which probably needs an additional and still unknown factor to efficiently lead the hydrolysis reaction.
Self-association of GTPBP3 is induced by the N-terminal domain.
While the MnmE G domain induces dimerization in the presence of potassium and either GTP or the transition state mimic GDP-AlFx, the N-terminal α/β domain (Fig. 2) induces a nucleotide- and potassium-independent dimerization (39, 40). This creates a binding site for THF, which has been postulated to be the one-carbon-unit donor in the modification reaction.
To investigate whether the N-terminal domain of GTPBP3 also induces a potassium-independent dimerization, we used the protein FLAG-(ΔN)GTPBP3. It should be noted that the GST-GTPBP3 fusion proteins are not appropriate tools in this case because the GST moiety induces dimerization by itself (26). The SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP) predicts a mitochondrial targeting sequence at the 5′ terminal end of GTPBP3 with a cleavage site between positions 44 and 45. However, a sequence of highly conserved residues among the MnmE family proteins starts at residue 35 of human GTPBP3. Some of these residues have counterparts in T. maritima MnmE that have been involved in homodimerization and THF binding (40). Moreover, the GTPBP3(Ins8A) protein expressed in HEK-293 cells permanently transfected with plasmid pIC1055 has an apparent molecular mass of 51 kDa, which is 3 kDa less than expected (Fig. 6). Such a difference corresponds to a mitochondrial signal peptide of about 30 amino acids. For these reasons, we decided to conserve in our construct the 5′ region of GTPBP3 that, according to data from the MnmE structure, might contain residues involved in THF-binding and homodimerization, but to eliminate the first 34 residues of the GTPBP3 N-terminal end since the presence of a putative signal peptide might interfere with dimerization of the GTPBP3 N-terminal domain.
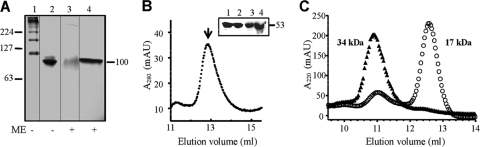
Native PAGE revealed that FLAG-(ΔN)GTPBP3 polymerizes in the absence of guanine nucleotides and potassium, producing several complexes with molecular masses greater than 100 kDa (Fig. 9A, lane 1). All these complexes resolve in one band of about 100 kDa when the protein is mixed with the strong reducing agent β-mercaptoethanol (Fig. 9A, compare lanes 1 and 3). These results suggest that the observed high-molecular-mass bands correspond to self-assemblies of FLAG-(ΔN)GTPBP3 that are maintained in vitro by disulfide bridges (GTPBP3 contains several cysteine residues), whereas the band of about 100 kDa formed in the presence of β-mercaptoethanol corresponds to a dimer stabilized by noncovalent interactions. As a control, the ability of MnmE to dimerize was analyzed in the same way. The E. coli protein has only one cysteine residue, which may contribute to the in vitro dimerization in the absence of β-mercaptoethanol (Fig. 9A, lane 2), but note that the MnmE dimer is also formed in the presence of the reducing agent (Fig. 9A, compare lanes 2 and 4), indicating that MnmE dimerization is mediated by noncovalent interactions, as previously reported (6, 40). The similar behavior of FLAG-(ΔN)GTPBP3 and MnmE in the presence of β-mercaptoethanol suggests that the human protein maintains the dimerization capability of its bacterial homologue that is provided by the N-terminal domain.
FIG. 9.
The GTPBP3 N-terminal domain induces a GTP- and potassium-independent dimerization. (A) Native PAGE analysis of FLAG-(ΔN)GTPBP3(Ins8A). Lanes 1 and 3, FLAG-(ΔN)GTPBP3(Ins8A); lanes 2 and 4, MnmE. Positions and sizes (in kDa) of mass markers are indicated on the left. Position and approximate size (in kDa) of MnmE and FLAG-(ΔN)GTPBP3(Ins8A) dimers are indicated on the right. ME, β-mercaptoethanol. (B) Gel filtration of FLAG-(ΔN)GTPBP3(Ins8A). Experiments were performed as described in Materials and Methods. The peak at an apparent molecular mass of 120 kDa (indicated by an arrow) was collected and analyzed by Western blotting (inset). Lanes 1 and 3, fraction corresponding to the ∼120-kDa peak collected from gel filtration; lanes 2 and 4, input. The Western blot was developed with anti-FLAG (lanes 1 and 2) and anti-GTPBP3 (lanes 3 and 4). The size of a molecular mass marker is indicated in kDa on the right. (C) Gel filtration analysis of the GTPBP3 N-terminal domain. The gel filtration buffer was 50 mM Tris, pH 7.5, 5 mM MgCl2, 5 mM DTT, and 250 mM KCl. Before application onto the column, the protein was incubated overnight at 4°C in the same buffer (filled triangles) or in a buffer containing 50 mM Tris, pH 7.5, 5 mM MgCl2, 5 mM DTT, and 100 mM KCl (open circles). Apparent molecular masses corresponding to elution volumes are indicated. AU, arbitrary units.
FLAG-(ΔN)GTPBP3 was also applied to a Superdex 200 HR gel filtration column. Most of the protein eluted with the void volume peak, suggesting that the protein easily aggregates under the conditions used in the assays. However, a small peak at an apparent molecular mass of 120 kDa could also be detected and collected (Fig. 9B). SDS-PAGE and Western blot analysis with anti-FLAG or anti-GTPBP3 antibody indicated that this peak corresponds to a dimer of FLAG-(ΔN)GTPBP3 (Fig. 9B, inset).
To corroborate the capability of the GTPBP3 N-terminal domain to induce dimerization in the absence of nucleotide, we expressed this domain separately (residues 35 to 157) (Fig. 2) and analyzed its behavior in gel filtration experiments. The N-terminal domain appeared to be subject to a monomer/dimer equilibrium as a function of ionic strength. Thus, when the N-terminal domain was purified in a buffer containing KCl at 100 mM and subsequently analyzed by gel filtration using an elution buffer with KCl at concentrations between 100 and 1,000 mM, two peaks with apparent molecular mass values consistent with the monomeric and dimeric forms of the domain (17 and 34 kDa, respectively) were eluted, with the monomeric form always predominant over the dimeric one (Fig. 9C and data not shown). However, when the purified N-terminal domain was incubated overnight at 4°C in a buffer containing KCl at 250 mM before being analyzed by gel filtration in the same buffer, only a dimeric form with an apparent molecular mass of 34 kDa was eluted (Fig. 9C). A similar result was obtained when the overnight incubation and gel filtration were performed in a buffer containing MgCl2 at 20 mM in place of KCl (data not shown). It seems, therefore, that the exchange between the dimeric and monomeric states of the GTPBP3 N-terminal domain is slower than the characteristic time of chromatography and that the overnight incubation at a given ionic strength is required for shifting the equilibrium toward the dimeric form. This is the first time to our knowledge that the dimerization domain (the ∼120-amino-acid N-terminal domain) of a GTPBP3 family protein has been separately expressed and shown to undergo self-assembly. It is of interest that the crystal structure of the bacterial MnmE protein indicates that the N-terminal domain of one monomer interacts not only with the N-terminal domain of the other monomer but also with its helical domain (40). Such an interaction might be important for stabilization of the dimer formed by the N-terminal domains. Given that our construct contains only the N-terminal domain of GTPBP3, stabilization of the dimer mediated by the buffer components could become more relevant. In any case, it may be concluded that the separately expressed GTPBP3 N-terminal domain, like the entire protein, may form dimers in the absence of guanine nucleotides. Therefore, we propose that the N-terminal domain functions as a dimerization domain of GTPBP3 and may be involved, as occurs in MnmE, in the construction of the THF binding pocket.
Knocking down human GTPBP3 results in mitochondrial dysfunction.
tRNALys and tRNALeu(UUR) harboring MERRF and MELAS mutations are deficient in modifications normally present at the U34 position. Such a deficiency results in defective translation, which seems to be the direct cause of reduction in oxygen consumption and mitochondrial protein synthesis observed in the carrier cells (18, 19, 44, 52, 54). GTPBP3 is thought to control U34 modification of tRNALys and tRNALeu(UUR). Therefore, we wondered whether inactivation of GTPBP3 leads to dysfunction of human mitochondria. To test this hypothesis, we decided to knock down GTPBP3 expression using siRNA. HEK-293 cells were transfected with four different siRNAs that target human GTPBP3 or with Silencer Negative Control #1 siRNA. Total RNA from a sample of transfected cells was subjected to quantitative RT-PCR to estimate the knockdown efficiency of each siRNA. Two GTPBP3 siRNAs (siGTPBP3-1 and siGTPBP3-3, which target GTPBP3 on exon 5) caused the greatest reduction in the GTPBP3 mRNA levels (about 65%) and were chosen for our experiments, whereas the Silencer Negative Control #1 siRNA did not have any silencing effect.
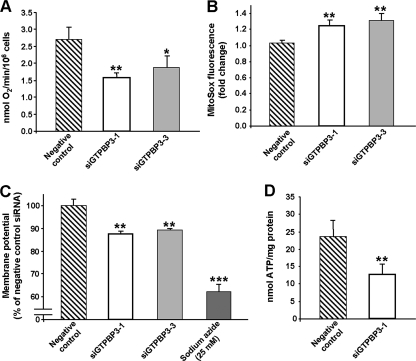
The rate of oxygen consumption of transfected cells was measured with a Clark-type electrode and found clearly decreased in GTPBP3-silenced cells compared with the negative control (Fig. 10A). Moreover, measurement of mitochondrial superoxide generation with MitoSOX Red by flow cytometry showed a 28% ± 3% increase in the GTPBP3-silenced cells (Fig. 10B). We also examined the mitochondrial membrane potential of the knockdown cells by staining with MitoTracker Red, a fluorescent indicator of mitochondrial membrane potential. Flow cytometry revealed a significant decrease of this parameter in the GTPBP3-silenced cells (Fig. 10C). Similar results were obtained using rhodamine 123 (data not shown). Thus, knocking down the expression of GTPBP3 causes a mitochondrial dysfunction that is consistent with phenotypic features exhibited by cells harboring MERRF and MELAS mutations (1, 16, 33, 37).
FIG. 10.
Knockdown of human GTPBP3 results in mitochondrial dysfunction. HEK-293 cells were transfected with GTPBP3 siRNAs or with negative control siRNAs, and oxygen consumption (A), superoxide detection in mitochondria with MitoSOX (B), mitochondrial membrane potential (C), and cellular ATP (D) were determined as specified in Materials and Methods. Bars indicate standard deviations of at least three different experiments (*, P < 0.05; **, P < 0.005; ***, P < 0.0005).
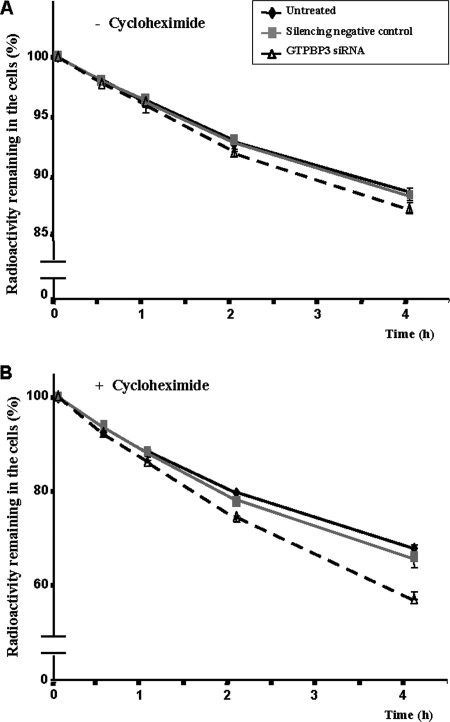
Next, we analyzed mitochondrial protein synthesis by [3H]Leu pulse labeling in the transfected cells and found it to be also reduced in the GTPBP3-knockdown cells. Table 5 shows that in the presence of cycloheximide, an inhibitor of cytosolic protein synthesis, incorporation of [3H]Leu into proteins (which was reduced by about 97% compared to the same cells incubated without cycloheximide) is about 20% lower in GTPBP3-silenced cells than in control cells. Curiously, GTPBP3 inactivation also reduces total protein synthesis, as indicated by the results obtained in the absence of cycloheximide. This could be due to the observed inhibition of mitochondrial protein synthesis and also to inhibition of cytosolic protein synthesis since the impairment of the oxidative phosphorylation produced by GTPBP3 inactivation decreased ATP production. Thus, measurements in GTPBP3- silenced cells revealed a 40% ± 9% decrease in ATP levels (Fig. 10D), whereas there were no changes of ATP content in the negative control. This marked reduction is similar to that found in fibroblasts derived from some patients with nuclear DNA-related disorders (41). Therefore, we conclude that activity of GTPBP3 is important for synthesis of proteins encoded by the mitochondrial genome and that impairment of mitochondrial protein synthesis in GTPBP3-knockdown cells may be the underlying cause of the mitochondrial dysfunction exhibited by these cells.
TABLE 5.
Incorporation of [3H]leucine into proteins by HEK-293 cells subjected to various treatmentsa
| Cell treatment | Cycloheximide | Incorporation of [3H]Leu (cpm/mg of protein)b | % Inhibitionc |
|---|---|---|---|
| None | − | 117,178 ± 16,780 | |
| Negative silencing control | − | 110,734 ± 10,073 | 5.5 |
| GTPBP3 siRNA | − | 93,157 ± 5,739* | 20.5 |
| None | + | 3,398 ± 210 | |
| Negative silencing control | + | 3,109 ± 167 | 8.5 |
| GTPBP3 siRNA | + | 2,464 ± 196** | 27.5 |
Cells, untreated (none) or treated as indicated, were incubated for 1 h with [3H]leucine, in the presence (+) or in the absence (−) of cycloheximide (200 μg/ml), as described in Materials and Methods.
Each value is the mean ± standard deviation from four different experiments with duplicated samples. Differences from the negative silencing control values were considered significant at P < 0.05 (*) and P < 0.005 (**). The values obtained with cells transfected with the negative silencing control were not found to be significantly different from the corresponding values in the cells without treatment (None).
Data are expressed as percent inhibition of [3H]leucine incorporation into proteins with respect to untreated cells (None).
Interestingly, we have also observed a slight increase in protein degradation in the GTPBP3-knockdown cells (Fig. 11), especially of mitochondrially synthesized proteins (Fig. 11B). This can be explained considering that hypomodified tRNAs in GTPBP3- silenced cells may alter the mitochondrial translation process, producing erroneous proteins (including truncated proteins), which could be rapidly degraded by the intramitochondrial degradation systems. Moreover, the impairment of mitochondrial protein synthesis produced by GTPBP3 silencing may affect the biogenesis of oxidative phosphorylation complexes, leading to increased degradation of improperly assembled subunits (17, 22). Finally, the higher oxidative stress detected in the GTPBP3-silenced cells might affect mitochondrial biomolecules. Proteins or protein complexes with abnormal conformation caused by oxidative damage could also be a target for the degradation systems. All these possibilities remain to be investigated.
FIG. 11.
Effect of GTPBP3 silencing on intracellular protein degradation. HEK-293 cells, untreated or treated as indicated, were incubated for 1 h with [3H]leucine in the absence (A) or in the presence (B) of cycloheximide (200 μg/ml), and protein degradation was measured at 0.5, 1, 2, and 4 h as described in Materials and Methods. Results, which are expressed as the percentage of radioactive protein remaining in the cells, are mean ± standard deviation of three different experiments with duplicated samples. Some standard deviation bars are hidden by the symbols. At the 4-h time point, differences between GTPBP3 siRNA and control values were significant at P values of <0.05 (A) and <0.0005 (B).
DISCUSSION
Human GTPBP3 is an evolutionarily conserved, multidomain protein involved in mitochondrial tRNA modification (Fig. 1). Characterization of its biochemical properties and the phenotype conferred by GTPBP3 inactivation is crucial to understanding the role of this protein in tRNA maturation and its effects on mitochondrial respiration.
All members of the GTPBP3 protein family contain a canonical G domain that comprises the typical G1 to G4 motifs involved in binding and hydrolysis of GTP (Fig. 2). In fact, the E. coli GTPBP3 homologue (MnmE) exhibits a GTPase activity that is essential for the tRNA modifying function (27, 57). For this reason, the finding that GTPBP3 may encode two isoforms differing in the length of the G domain is particularly striking. Curiously, both human isoforms show identical biochemical properties in vitro. Thus, they exhibit moderate affinity for guanine nucleotides like their bacterial homologue (Table 3) but hydrolyze GTP at a 100-fold lower rate (Table 4). Our data indicate that potassium induces dimerization of the GTPBP3 G-domain (Fig. 7 and 8), but, in contrast to the bacterial protein, this is not enough to stimulate the GTPase activity (Table 4). Therefore, we conclude that there are some structural differences between MnmE and GTPBP3 that explain their different GTPase properties and that the human protein requires an additional and still unknown factor to promote GTP hydrolysis. Such differences could be related to the formation of a G-domain dimer that in the case of GTPBP3 cannot become activated under our experimental conditions. These results prompt us to suggest a working model for the functional cycle of the MnmE/GTPBP3 family GTPases that can be outlined as follows: G + K+ + GTP ↔ 2[G·K+·GTP] ↔ 2[G*·K+·GTP] → 2[G·K+·GDP·Pi] → G + K+ + GDP + Pi. In this model, G, G*, and Pi represent the G domain, the activated G domain, and inorganic phosphate, respectively. Dimerization is represented by the number 2 preceding the bracket. Considering that the GTPBP3 G-domain is able to dimerize but not to drive GTP hydrolysis, we propose that the second step in the diagram (i.e., that leading to activation of the G-domain dimer) would be the rate-limiting step in the human GTPase cycle. We also propose that the mutation E282A of MnmE, which does not impair the G-domain dimerization but drastically reduces the hydrolysis activity (39), also blocks the step leading to activation of the G-domain dimer. In our model, it is unclear whether the G domain remains as a dimer in the putative state where the reaction products have not yet been released.
GTPBP3 might require the GTPase activity for its tRNA modifying function, as observed with MnmE (27, 57). However, the simultaneous presence in human mitochondria of two GTPBP3 isoforms differing in the G domain makes the situation more complex than in bacteria or yeast mitochondria, unless both isoforms were functionally equivalent. This seems to be the case, given that both GTPBP3 proteins exhibit similar enzymatic properties (Tables 3 and 4 and Fig. 7).
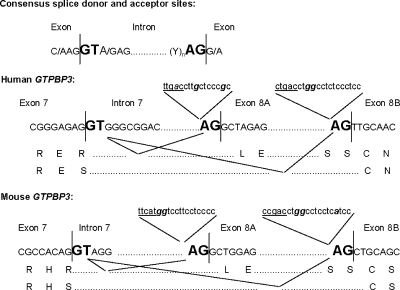
Our results suggest that alternative splicing of exon 8A occurs more frequently in primates than in rodents (Fig. 4; see also Fig. A2) because of the regulatory sequences that precede exon 8A and 8B (see Fig. A1). It seems that mutations that have occurred during evolution in these sequences provide an opportunity for the splicing machinery to skip exon 8A in primates, without compromising the original transcript produced by the gene (that including exon 8A). This is in line with the idea that alternative splicing operates as an evolutionary workshop for tinkering with the protein structure and function (32). In the case of the human GTPBP3 gene, splicing of exon 8A produces a younger isoform that biochemically does not differ from the older one.
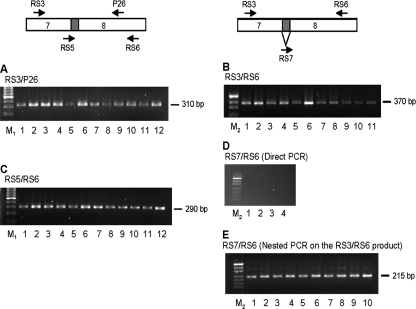
FIG. A2.
PCR analysis to detect alternative splicing of mouse GTPBP3 exon 8A. Primers used are represented by arrows on the diagrams and indicated over each gel. Exon 8A is denoted in gray. Primer RS5 spans the junction region between exons 7 and 8A, whereas RS7 spans that between exons 7 and 8B. PCR analysis was performed on cDNAs from 12 different mouse tissues (Mouse MTC panel I; Clontech, California). For the gels in panels A to E, the lanes are as follows: lanes 1, heart; lanes 2, brain; lanes 3, spleen; lanes 4, lung; lanes 5, liver; lanes 6, skeletal muscle; lanes 7, kidney; lanes 8, testis; lanes 9, 7-day embryo; lanes 10, 11-day embryo; lanes 11, 15-day embryo; lanes 12, 17-day embryo. M1, GeneRuler 100-bp DNA Ladder (Fermentas); M2, pUC19 DNA/MspI Marker, 23 (Fermentas).
FIG. A1.
Alternative splicing events in human and mouse GTPBP3. The consensus splicing sequences in intron 7, the alternative acceptor site in exon 8A, and the polypyrimidine tracks upstream from exon 8A and 8B are shown. The exon/intron boundaries are indicated by vertical sticks. Splice donor and acceptor sites are indicated in bold uppercase letters. Note that the first nucleotide of exon 8B is a pyrimidine and not the more frequent guanine, which is indeed present as the first nucleotide of exon 8A. Sequences including putative branch sites (underlined) and polypyrimidine tracts are denoted in lowercase letters. Purines interrupting polypyrimidine tracts are indicated in bold italics. Amino acid sequences resulting from each splicing variant are denoted by one-letter amino acid code below the corresponding nucleotide sequence.
Interestingly, GTPBP3 forms homodimers in the absence of GTP and potassium (Fig. 9A and B), which suggests that, as occurs in MnmE, dimerization under such conditions is induced by the N-terminal domain of the protein. Supporting this is the finding that the separately expressed GTPBP3 N-terminal domain also forms dimers in the absence of these factors (Fig. 9C). The conservation of this property in the eukaryotic MnmE homologues may have important functional and mechanistic consequences. The yeast proteins MSS1 and MTO1 (the MnmE and GidA homologues, respectively) were proposed to form a heterodimer (8) although some criticisms were raised about the experiments leading to this proposal (56). In contrast, the E. coli MnmE and GidA proteins form homodimers and directly assemble in an α2β2 heterotetrameric complex (6, 28, 40, 56). The finding that the human GTPBP3 protein forms homodimers in the absence of potassium supports the idea that dimerization induced by the N-terminal domain is an evolutionarily conserved property of the MnmE family proteins that is related to the construction of the binding pocket for THF. It remains to be explored whether a GTPBP3 dimer interacts with a dimer of the human MTO1 protein, similarly to their bacterial homologues.
If GTPBP3 is directly involved in the tRNA modification reaction, the protein should contact its substrate in some way. However, we have been unable to demonstrate an interaction between GTPBP3 and in vitro-transcribed tRNA by means of gel shift assays, protease protection experiments, and isothermal titration calorimetry (data not shown). It should be mentioned that previous attempts to biochemically demonstrate an interaction between the bacterial GTPBP3 homologue (MnmE) and in vitro transcribed RNA were reported to be unsuccessful (40). Thus, it is possible that the in vitro transcribed tRNA is not an appropriate substrate for the GTPBP3 family proteins or, alternatively, that the interaction of these proteins with their tRNA substrates is very weak. The recent resolution of the crystal structure of GidA has revealed a large patch of positively charged residues on its surface that has been assigned to the RNA-binding site (28). Given that no such extensive patch of positive charge is apparent on the surface of MnmE, it was suggested that, in the MnmE-GidA complex, GidA is mainly responsible for the tRNA binding. In the case of the mitochondrial proteins, something similar could occur if GTPBP3 also forms a complex with its partner, MTO1.
Several studies support the idea that point mutations in mitochondrial tRNALeu(UUR) and tRNALys associated to MELAS and MERRF diseases exert a great part of their pathogenic effects by hindering modification at the wobble uridine (18, 19, 51-55). If impairment of wobble modification is an important molecular cause of MERRF and MELAS, it is reasonable to think that defects in proteins involved in the modification pathway of the wobble uridine, such as GTPBP3 (Fig. 1), may be a direct underlying cause of some oxidative phosphorylation (OXPHOS) diseases. Here, we show that the partial inactivation of GTPBP3 in HEK-293 cells by siRNA results in phenotypic traits that are observed in MERRF and MELAS. Thus, GTPBP3 silencing reduces oxygen consumption, mitochondrial membrane potential, and ATP production, whereas it causes an increased oxidative stress (Fig. 10). These phenotypic traits are probably caused by defects in mitochondrial protein synthesis, which is reduced by about 20% in GTPBP3-silenced cells (Table 5). Such a reduction may alter biogenesis of oxidative phosphorylation complexes due to an insufficient production of the mitochondrial genome-encoded subunits. Interestingly, we have also found a slight but significant increase in protein degradation (Fig. 11), which might be related to erroneous protein synthesis by mitochondria, generation of unassembled subunits of OXPHOS complexes, and oxidative damage.
The effects of GTPBP3 silencing here observed are prominent, considering that the GTPBP3 inactivation is not total and that the half-life of tRNAs is long (around 24 h or even more) (52, 55), which supposes that a fraction of mitochondrial tRNAs in the silenced cells may be appropriately modified when protein synthesis is measured. Such a fraction may also depend on the GTPBP3 half-life, which remains to be determined. Moreover, the half-life of the inner mitochondrial membrane proteins has been estimated to be about 3.8 days (12) so that a part of the OXPHOS complexes could remain functional at the time of our assays. Curiously, full inactivation of MSS1 in yeast leads to hypomodification of tRNAs, but this has a negligible effect on mitochondrial respiration and protein synthesis (9, 46). The more prominent effects that inactivation of GTPBP3 seems to have in human cells suggest that dynamics of human mitochondrial ribosomes may be somewhat different from that of yeast mitochondrial ribosomes, in such a way that the effects of tRNA hypomodification on mitochondrial translation are more severe in humans than in yeast. One interesting difference between mitochondrial ribosomes from yeast and humans lies in the A-site rRNA. In the yeast 15S rRNA, the 1477C and 1583G bases are in opposition and form a base pair in a region of the small rRNA that is highly conserved from bacteria to mammals since it is an essential part of the ribosome decoding site. If this pairing exists, inactivation of MSS1 has a negligible effect on protein synthesis (9). However, when the C at nucleotide 1477 is mutated to a G, pairing with 1583G cannot be established, and yeast mitochondrial ribosomes become resistant to paromomycin (Parr). Under these conditions, inactivation of MSS1 dramatically impairs mitochondrial protein synthesis (9). In the human mitochondrial 12S rRNA, nucleotides 1494C and 1555A occupy equivalent positions to the yeast 1477C and 1583G (i.e., they are in opposition to each other), but they do not form a base pair (36). In this way, a situation equivalent to that produced by the yeast Parr mutation is natural to the human mitochondrial ribosomes, which might explain why the sole inactivation of GTPBP3 has notorious effects on mitochondrial synthesis. It remains to be established whether GTPBP3 inactivation in humans is sufficient to cause a clinical phenotype.
The phenotype produced by the partial inactivation of GTPBP3 in vitro is reminiscent of MERRF and MELAS cells. Note, however, that several human mitochondrial tRNAs are thought to be substrates for protein GTPBP3. Presence of the τm5 group at the wobble uridine has been unambiguously shown in tRNALys and tRNALeu(UUR) (44), and a modified U34 is known to be present, at least, in tRNAGlu and tRNAGln (3); it is reasonable to think that the modification at this uridine is similar to that found in tRNAs from bacteria (3, 4). Thus, inactivation of GTPBP3 may cause several tRNAs to be inadequately modified, and, accordingly, the effects of such inactivation may not completely correspond to the consequences of MERRF and MELAS mutations, which affect only modification of tRNALys and tRNALeu(UUR), respectively.
In brief, our work describes for the first time some biochemical features of GTPBP3 that may help to elucidate the role of this protein in tRNA modification and reveals its importance for the mitochondrial respiratory functions.
Supplementary Material
Acknowledgments
This work was supported by the Ministerio de Educación y Ciencia (grants PB98-0041, BFU2004-05819, BFU2005-00087, and BFU2007-66509), the Ministerio de Sanidad (grants G03/212, PI051791, and PI070091), and the Generalidad Valenciana (grants GRUPOS04/07 and ACOMP07-187). V.M.V. is a recipient of an FIS contract (CP07/00171).
We thank Adrián Velázquez and Ismaïl Moukadiri for their help with ITC and gel shift experiments, respectively, and Nuria Mas for technical assistance.
APPENDIX
Exon 8A is expressed in a species-specific manner. The mouse GTPBP3 gene contains nine exons and encodes a 486-residue protein with a high degree of identity (81%) to the human homologue (23, 24). Although the full-length of mouse GTPBP3 cDNA was isolated, a careful analysis of its transcription forms has not been performed so far (24). Analysis of the mouse GTPBP3 sequence indicates that the alternative acceptor site of exon 8 is conserved (Fig. A1). To test whether this site is functional, a PCR analysis on cDNAs derived from several sources was performed. In contrast to that observed in humans (Fig. 4, top panel), only one PCR product was detected when different primer pairs were used to analyze the junction region of exons 7 and 8 on cDNAs obtained from tissues of BALB/c, Swiss Webster, and Webster strains (Fig. A2A to C). Identical results were obtained with cDNA synthesized on mRNA from different tissues pooled from five female C57/B6 mice (ages 2 to 24 weeks) (data not shown). Attempts to specifically amplify the putative spliced form of mouse GTPBP3 by a direct PCR were unsuccessful (Fig. A2D), which was also in contrast with the results obtained when a similar approach was used for amplification of the spliced form of the human gene (Fig. 4 bottom right). However, when primers RS6 and RS7 were used to lead a nested PCR on the amplicon obtained with primers RS3 and RS6, detection of the mouse spliced form was possible (Fig. A2E). These results suggest that the skip of exon 8A occurs only in a very small portion of the mouse GTPBP3 transcripts.
The Macaca mulatta GTPBP3 gene is predicted to contain nine exons and to encode a 497-residue protein that shows a very high identity (95%) with the human homologue (http://www.ensembl.org). Interestingly, the study of exon 8A alternative splicing in a monkey cell line (COS cells) revealed a behavior identical to that of the human gene (Fig. 4 and data not shown). These results suggest that there are species-specific mechanisms involved in the regulation of GTPBP3 expression. Exon 8A skipping might be modulated by specific sequences known to take part in the splicing of the primary transcript, such as the branch site and the polypyrimidine tract. The shorter distance separating the branch point and the regular acceptor site of exon 8A in the human gene (Fig. A1) might not be efficiently used by the splicing machinery (7, 42). Moreover, the polypyrimidine tracts preceding exons 8A and 8B in primates and mouse are short and contain a small number of uridine residues (Fig. A1). The introduction of purines during evolution, which makes the polypyrimidine tract preceding exon 8A in human (and monkey) and 8B in mouse even shorter, may affect the 3′ acceptor site selection (30, 34, 35), facilitating skipping of exon 8A in primates and making it more difficult in mouse.
Footnotes
Published ahead of print on 13 October 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Antonická, H., D. Floryk, P. Klement, L. Stratilova, J. Heřmanská, H. Houštková, M. Kalous, Z. Drahota, J. Zeman, and J. Houštěk. 1999. Defective kinetics of cytochrome c oxidase and alteration of mitochondrial potential in fibroblasts and cytoplasmic hybrid cells with the mutation for myoclonus epilepsy with ragged-red fibres (“MERRF”) at position 8344 nt. Biochem. J. 342537-544. [PMC free article] [PubMed] [Google Scholar]
- 2.Beaudoing, E., S. Freier, J. R. Wyatt, J.-M. Claverie, and D. Gautheret. 2000. Patterns of variant polyadenylation signal usage in human genes. Genome Res. 101001-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Björk, G. R. 1998. Modified nucleosides at positions 34 and 37 of tRNAs and their predicted coding capacities, p. 577-581. In H. Grosjean, and R. Benne, R. (ed.), Modification and editing of RNA. ASM Press, Washington, DC.
- 4.Björk, G. R., and T. G. Hagervall. July 2005, posting date. Chapter 4.6.2, Transfer RNA modification. In R. Curtiss III et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org.
- 5.Brégeon, D., V. Colot, M. Radman, and F. Taddei. 2001. Translational misreading: a tRNA modification counteracts a +2 ribosomal frameshift. Genes Dev. 152295-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cabedo, H., F. Macián, M. Villarroya, J. C. Escudero, M. Martínez-Vicente, E. Knecht, and M.-E. Armengod. 1999. The Escherichia coli trmE (mnmE) gene, involved in tRNA modification, codes for an evolutionarily conserved GTPase with unusual biochemical properties. EMBO J. 187063-7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chua, K., and R. Reed. 2001. An upstream AG determines whether a downstream AG is selected during catalytic step II of splicing. Mol. Cell. Biol. 211509-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colby, G., M. Wu, and A. Tzagoloff. 1998. MTO1 codes for a mitochondrial protein required for respiration in paromomycin-resistant mutants of Saccharomyces cerevisiae. J. Biol. Chem. 27327945-27952. [DOI] [PubMed] [Google Scholar]
- 9.Decoster, E., A. Vassal, and G. Faye. 1993. MSS1, a nuclear-encoded mitochondrial GTPase involved in the expression of COX1 subunit of cytochrome c oxidase. J. Mol. Biol. 23279-88. [DOI] [PubMed] [Google Scholar]
- 10.Elseviers, D., L. A. Petrullo, and P. Gallagher. 1984. Novel E. coli mutants deficient in biosynthesis of 5-methylaminomethyl-2-thiouridine. Nucleic Acids Res. 123521-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuertes, G., J. J. Martín de Llano, A. Villarroya, A. J. Rivett, and E. Knecht. 2003. Changes in the proteolytic activities of proteasomes and lysosomes in human fibroblasts produced by serum withdrawal, amino-acid deprivation and confluent conditions. Biochem. J. 37575-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grisolía, S., J. Hernández-Yago, and E. Knecht. 1985. Regulation of mitochondrial protein concentration: a plausible model which may permit assessing protein turnover. Curr. Topics Cell Regul. 27387-396. [DOI] [PubMed] [Google Scholar]
- 13.Guzmán, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1774121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagervall, T. G., C. G. Edmonds, J. A. McCloskey, and G. R. Björk. 1987. Transfer RNA (5-methylaminomethyl-2-thiouridine)-methyltransferase from Escherichia coli K-12 has two enzymatic activities. J. Biol. Chem. 2628488-8495. [PubMed] [Google Scholar]
- 15.Ikeuchi, Y., N. Shigi, J. Kato, A. Nishimura, and T. Suzuki. 2006. Mechanistic insights into sulfur relay by multiple sulfur mediators involved in thiouridine biosynthesis at tRNA wobble positions. Mol. Cell 2197-108. [DOI] [PubMed] [Google Scholar]
- 16.James, A. M., Y.-H. Wei, C.-Y. Pang, and M. P. Murphy. 1996. Altered mitochondrial function in fibroblasts containing MELAS or MERRF mitochondrial DNA mutations. Biochem. J. 318401-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Käser, M., and T. Langer. 2000. Protein degradation in mitochondria. Semin. Cell Dev. Biol. 11181-190. [DOI] [PubMed] [Google Scholar]
- 18.Kirino, Y., T. Yasukawa, S. K. Marjavaara, H. T. Jacobs, I. J. Holt, K. Watanabe, and T. Suzuki. 2006. Acquisition of the wobble modification in mitochondrial tRNALeu(CUN) bearing the G12300A mutation suppresses the MELAS molecular defect. Hum. Mol. Genet. 15897-904. [DOI] [PubMed] [Google Scholar]
- 19.Kirino, Y., T. Yasukawa, S. Ohta, S. Akira, K. Ishihara, K. Watanabe, and T. Suzuki. 2004. Codon-specific translational defect caused by a wobble modification deficiency in mutant tRNA from a human mitochondrial disease. Proc. Natl. Acad. Sci. USA 10115070-15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirino, Y., Y. Goto, Y. Campos, J. Arenas, and T. Suzuki. 2005. Specific correlation between the wobble modification deficiency in mutant tRNAs and the clinical features of a human mitochondrial disease. Proc. Natl. Acad. Sci. USA 1027127-7132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knecht, E., J. Hernández-Yago, A. Martínez-Ramón, and S. Grisolía. 1980. Fate of proteins synthesized in mitochondria of cultured mammalian cells revealed by electron microscope radioautography. Exp. Cell Res. 125191-199. [DOI] [PubMed] [Google Scholar]
- 22.Koopen, M., and T. Langer. 2007. Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Mol. Biol. 42221-242. [DOI] [PubMed] [Google Scholar]
- 23.Li, X., and M.-X. Guan. 2002. A human mitochondrial GTP binding protein related to tRNA modification may modulate phenotypic expression of the deafness-associated mitochondrial 12S rRNA mutation. Mol. Cell. Biol. 227701-7711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, X., and M.-X. Guan. 2003. Identification and characterization of mouse GTPBP3 gene encoding a mitochondrial GTP-binding protein involved in tRNA modification. Biochem. Biophys. Res. Commun. 312747-754. [DOI] [PubMed] [Google Scholar]
- 25.Li, X., R. Li, X. Lin, and M.-X. Guan. 2002. Isolation and characterization of the putative nuclear modifier gene MTO1 involved in the pathogenesis of deafness-associated mitochondrial 12 S rRNA A1555G mutation. J. Biol. Chem. 27727256-27264. [DOI] [PubMed] [Google Scholar]
- 26.Lim, K., J. H. Ho, K. Keeling, G. L. Gilliland, X. Ji, F. Rüker, and D. C. Carter. 1994. Three-dimensional structure of Schistosoma japonicum glutathione S-transferase fused with a six-amino acid conserved neutralizing epitope of gp41 from HIV. Protein Sci. 32233-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martínez-Vicente, M., L. Yim, M. Villarroya, M. Mellado, E. Pérez-Payá, G. R. Björk, and M.-E. Armengod. 2005. Effects of mutagenesis in the switch I region and conserved arginines of Escherichia coli MnmE protein, a GTPase involved in tRNA modification. J. Biol. Chem. 28030660-30670. [DOI] [PubMed] [Google Scholar]
- 28.Meyer, S., A. Scrima, W. Versées, and A. Wittinghofer. 2008. Crystal structures of the conserved tRNA modifying enzyme GidA: implications for its interaction with MnmE and substrate. J. Mol. Biol. 380532-547. [DOI] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Norton, P. A. 1994. Polypyrimidine tract sequences direct selection of alternative branch sites and influence protein binding. Nucleic Acids Res. 223854-3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nuñez, C., V. M. Víctor, R. Tur, A. Alvarez-Barrientos, S. Moncada, J. Esplugues, and P. D'Ocon. 2005. Discrepancies between nitroglycerin and NO-releasing drugs on mitochondrial oxygen consumption, vasoactivity, and the release of NO. Circ. Res. 971063-1069. [DOI] [PubMed] [Google Scholar]
- 32.Nurtdinov, R. M., I. I. Artamonova, A. A. Mironov, and M. S. Gelfand. 2003. Low conservation of the alternative splicing patterns in the human and mouse genomes. Hum. Mol. Genet. 121313-1320. [DOI] [PubMed] [Google Scholar]
- 33.Pang, C.-Y., H.-C. Lee, and Y.-H. Wei. 2001. Enhanced oxidative damage in human cells harbouring A3243G mutation of mitochondrial DNA: implication of oxidative stress in the pathogenesis of mitochondrial diabetes. Diabetes Res. Clin. Pract. 54(Suppl. 2)S45-S56. [DOI] [PubMed] [Google Scholar]
- 34.Reed, R. 1989. The organization of 3′ splice-site sequences in mammalian introns. Genes Dev. 32113-2123. [DOI] [PubMed] [Google Scholar]
- 35.Roscigno, R. F., M. Weiner, and M. A. García-Blanco. 1993. A mutational analysis of the polypyrimidine tract of introns. Effects of sequence differences in pyrimidine tracts on splicing. J. Biol. Chem. 26811222-11229. [PubMed] [Google Scholar]
- 36.Ruiz-Pesini, E., and D. C. Wallace. 2006. Evidence for adaptive selection acting on the tRNA and rRNA genes of human mitochondrial DNA. Hum. Mutat. 271072-1081. [DOI] [PubMed] [Google Scholar]
- 37.Schon, E. A., E. Bonilla, and S. DiMauro. 1997. Mitochondrial DNA mutations and pathogenesis. J. Bioenerg. Biomembr. 29131-149. [DOI] [PubMed] [Google Scholar]
- 38.Schweins, T., M. Geyer, K. Scheffzek, A. Warshel, H. R. Kalbitzer, and A. Wittinghofer. 1995. Substrate-assisted catalysis as a mechanism for GTP hydrolysis of p21ras and other GTP-binding proteins. Nat. Struct. Biol. 236-44. [DOI] [PubMed] [Google Scholar]
- 39.Scrima, A., and A. Wittinghofer. 2006. Dimerisation-dependent GTPase reaction of MnmE: how potassium acts as GTPase-activating element. EMBO J. 252940-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scrima, A., I. R. Vetter, M.-E. Armengod, and A. Wittinghofer. 2005. The structure of the TrmE GTP-binding protein and its implications for tRNA modification. EMBO J. 2423-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shepherd, R. K., N. Checcarelli, A. Naini, D. C. De Vivo, S. DiMauro, and C. M. Sue. 2006. Measurement of ATP production in mitochondrial disorders. J. Inherit. Metab. Dis. 2986-91. [DOI] [PubMed] [Google Scholar]
- 42.Smith, C. W. J., T. T. Chu, and B. Nadal-Ginard. 1993. Scanning and competition between AGs are involved in 3′ splice site selection in mammalian introns. Mol. Cell. Biol. 134939-4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strausberg, R. L., E. A. Feingold, L. H. Grouse, J. G. Derge, R. D. Klausner, F. S. Collins, L. Wagner, C. M. Shenmen, G. D. Schuler, S. F. Altschul, et al. 2002. Generation and initial analysis of more than 15,000 full-length human and mouse cDNA sequences. Proc. Natl. Acad. Sci. USA 9916899-16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki, T., T. Suzuki, T. Wada, K. Saigo, and K. Watanabe. 2002. Taurine as a constituent of mitochondrial tRNA: new insights into the functions of taurine and human mitochondrial diseases. EMBO J. 216581-6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tian, B., J. Hu, H. Zhang, and C. Lutz. 2005. A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33201-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Umeda, N., T. Suzuki, M. Yukawa, Y. Ohya, H. Shindo, K. Watanabe, and T. Suzuki. 2005. Mitochondria-specific RNA-modifying enzymes responsible for the biosynthesis of the wobble base in mitochondrial tRNAs. Implications for the molecular pathogenesis of human mitochondrial diseases. J. Biol. Chem. 2801613-1624. [DOI] [PubMed] [Google Scholar]
- 47.Vargas, J. L., F. Aniento, J. Cervera, and E. Knecht. 1989. Vanadate inhibits degradation of short-lived, but not of long-lived, proteins in L-132 human cells. Biochem. J. 25833-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamanaka, K., J. Hwang, and M. Inouye. 2000. Characterization of GTPase activity of TrmE, a member of a novel GTPase superfamily, from Thermotoga maritime. J. Bacteriol. 1827078-7082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan, Q., X. Li, G. Faye, and M.-X. Guan. 2005. Mutations in MTO2 related to tRNA modification impair mitochondrial gene expression and protein synthesis in the presence of a paromomycin resistance mutation in mitochondrial 15 S rRNA. J. Biol. Chem. 28029151-29157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yan, Q., Y. Bykhovskaya, R. Li, E. Mengesha, M. Shohat, X. Estivill, N. Fischel-Ghodsian, and M.-X. Guan. 2006. Human TRMU encoding the mitochondrial 5-methylaminomethyl-2-thiouridine-methyltransferase is a putative nuclear modifier gene for the phenotypic expression of the deafness-associated 12S rRNA mutations. Biochem. Biophys. Res. Commun. 3421130-1136. [DOI] [PubMed] [Google Scholar]
- 51.Yasukawa, T., Y. Kirino, N. Ishii, I. J. Holt, H. T. Jacobs, T. Makifuchi, N. Fukuhara, S. Ohta, T. Suzuki, and K. Watanabe. 2005. Wobble modification deficiency in mutant tRNAs in patients with mitochondrial diseases. FEBS Lett. 5792948-2952. [DOI] [PubMed] [Google Scholar]
- 52.Yasukawa, T., T. Suzuki, N. Ishii, S. Ohta, and K. Watanabe. 2001. Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J. 204794-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yasukawa, T., T. Suzuki, N. Ishii, Y. Ueda, S. Ohta, and K. Watanabe. 2000. Defect in modification at the anticodon wobble nucleotide of mitochondrial tRNALys with the MERRF encephalomyopathy pathogenic mutation. FEBS Lett. 467175-178. [DOI] [PubMed] [Google Scholar]
- 54.Yasukawa, T., T. Suzuki, S. Ohta, and K. Watanabe. 2002. Wobble modification defect suppresses translational activity of tRNAs with MERRF and MELAS mutations. Mitochondrion 2129-141. [DOI] [PubMed] [Google Scholar]
- 55.Yasukawa, T., T. Suzuki, T. Suzuki, T. Ueda, S. Ohta, and K. Watanabe. 2000. Modification defect at anticodon wobble nucleotide of mitochondrial tRNAsLeu(UUR) with pathogenic mutations of mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes. J. Biol. Chem. 2754251-4257. [DOI] [PubMed] [Google Scholar]
- 56.Yim, L., I. Moukadiri, G. R. Björk, and M.-E. Armengod. 2006. Further insights into the tRNA modification process controlled by proteins MnmE and GidA of Escherichia coli. Nucleic Acids Res. 345892-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yim, L., M. Martínez-Vicente, M. Villarroya, C. Aguado, E. Knecht, and M.-E. Armengod. 2003. The GTPase activity and C-terminal cysteine of the Escherichia coli MnmE protein are essential for its tRNA modifying function. J. Biol. Chem. 27828378-28387. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.