Abstract
Mutations in components of the Wnt signaling pathway initiate colorectal carcinogenesis by deregulating the β-catenin transcriptional coactivator. β-Catenin activation of one target in particular, the c-Myc proto-oncogene, is required for colon cancer pathogenesis. β-Catenin is known to regulate c-Myc expression via sequences upstream of the transcription start site. Here, we report that a more robust β-catenin binding region localizes 1.4 kb downstream from the c-Myc transcriptional stop site. This site was discovered using a genome-wide method for identifying transcription factor binding sites termed serial analysis of chromatin occupancy. Chromatin immunoprecipitation-scanning assays demonstrate that the 5′ enhancer and the 3′ binding element are the only β-catenin and TCF4 binding regions across the c-Myc locus. When placed downstream of a simian virus 40-driven promoter-luciferase construct, the 3′ element activated luciferase transcription when introduced into HCT116 cells. c-Myc transcription is negligible in quiescent HCT116 cells but is induced when cells reenter the cell cycle after the addition of mitogens. Using these cells, we found that β-catenin and TCF4 occupancy at the 3′ enhancer precede occupancy at the 5′ enhancer. Association of c-Jun, β-catenin, and TCF4 specifically with the downstream enhancer underlies mitogen stimulation of c-Myc transcription. Our findings indicate that a downstream enhancer element provides the principal regulation of c-Myc expression.
The Wnt signaling pathway is essential for normal intestinal growth and development (37). Inappropriate activation of this pathway, most commonly caused by mutations in the adenomatous polyposis coli (APC) gene, is associated with colorectal cancer (11, 16, 19, 33). Activation of Wnt signaling or mutations in APC cause nuclear accumulation of the transcriptional coactivator, β-catenin. Nuclear β-catenin associates with members of the T-cell factor/lymphoid enhancer factor (TCF/Lef) family of sequence-specific transcription factors and activates expression of target genes involved in cell proliferation and growth. Proper β-catenin/TCF regulation of one important target gene, the c-Myc proto-oncogene, is required to maintain cellular homeostasis in response to Wnt signaling within the intestinal crypt microenvironment (21, 38).
c-Myc was among the first β-catenin target genes identified in mammalian cells (21). In a seminal report, He et al. used serial analysis of gene expression to identify genes that were differentially expressed in colorectal carcinoma cells in response to induced Wnt/β-catenin signaling (21). c-Myc was one of the most responsive genes detected. c-Myc is a basic helix-loop-helix-zipper protein that predominantly functions as a transcriptional activator (18). c-Myc exerts positive effects on cell cycle progression and cell growth by activating an array of target genes involved in DNA replication and ribosome biogenesis (12, 26). In the study by He et al., portions of the c-Myc promoter were fused to a reporter, and the ability of these fragments to be activated by β-catenin was evaluated (21). Two TCF consensus motifs were identified in the proximal 5′ c-Myc promoter, and mutation of both largely abrogated β-catenin-dependent reporter activity. Sierra et al. recently expanded upon these findings by using chromatin immunoprecipitation (ChIP) to monitor transcription factor occupancy at the c-Myc promoter during activation and cessation of Wnt/β-catenin signaling (43). This study showed that Wnt signaling induced β-catenin binding and recruitment of transcriptional coactivator complexes to the 5′ c-Myc promoter prior to c-Myc gene expression (43). When the Wnt signal was blocked, c-Myc transcription was repressed, and β-catenin and its coactivators vacated the c-Myc promoter. Thus, it was proposed that β-catenin (and associated coactivator complexes) bind to 5′ promoter elements to activate c-Myc gene expression in response to Wnt signaling.
A recent study confirmed the importance of c-Myc in intestinal epithelial cell transformation (38). APC−/− intestinal crypts contain high levels of nuclear β-catenin and display abnormal morphology, cell proliferation, and apoptosis—all hallmarks of colorectal carcinoma (38, 39). Surprisingly, specific deletion of c-Myc within APC−/− intestinal cells rescued all morphological abnormalities, suggesting that c-Myc was required for generation of these abnormalities (38). Moreover, c-Myc deletion reduced expression of the majority of β-catenin target genes in APC−/− intestinal cells. These findings suggest that c-Myc is critically involved in the cellular transformation that occurs in the colonic crypt epithelium as a result of deregulated Wnt/β-catenin signaling. Thus, deciphering the mechanisms involved in controlling Wnt/β-catenin regulation of c-Myc expression is paramount to understanding the molecular events that initiate colorectal carcinogenesis.
Enhancers are DNA elements that bind transcription factors to activate gene expression (34). Traditionally, enhancers have been identified in mammalian systems as sites that are sensitive to cleavage by DNase I or through reporter assays in transfected cells. Recently, ChIP has been used to identify and characterize these elements (13). Early ChIP studies focused on localizing potential enhancer elements in regions surrounding the transcription start sites which include the proximal 5′ promoter and first intron (6). This bias stemmed from studies in prokaryotes and lower eukaryotes where transcription is primarily controlled through sequences surrounding the transcription start site. To identify novel transcription factor binding elements, our laboratory developed an unbiased, genome-wide methodology, termed serial analysis of chromatin occupancy (SACO) (23). SACO combines ChIP with long serial analysis of gene expression, a technique originally designed to quantify mixtures of RNA (36). In SACO, the immunoprecipitated DNA, corresponding to transcription factor binding regions, is processed such that large numbers of genomic signature tags (GSTs) can be sequenced and positioned onto the genome. In a recent study, we used SACO to identify more than 400 β-catenin targets in the HCT116 colorectal cancer cell line (54). Our SACO screen discovered not only new gene targets of Wnt/β-catenin signaling but also potential novel enhancer elements.
In the present study, we identify and characterize a novel enhancer element that is localized 1.4 kb downstream from the c-Myc transcriptional stop site. Interestingly, this region coincided with a DNase I hypersensitivity site identified by Polack and colleagues more than 10 years ago (28). We find that the 3′ c-Myc enhancer mediated higher levels of β-catenin and TCF4 occupancy in ChIP assays and functions as a stronger enhancer of luciferase activity in reporter assays than the well-characterized 5′ enhancer. Moreover, upon interrogation of the mitogen response of c-Myc expression in cell cycle-synchronized HCT116 cells, we find that β-catenin and TCF4 binding to the 3′ enhancer precedes occupancy at the 5′ enhancer. Finally, we demonstrate that cooperation between β-catenin, TCF4, and c-Jun is required for c-Myc 3′ enhancer activity. Together, these results demonstrate the utility of unbiased genome-wide screens for transcription factor binding sites in identifying novel regulatory elements.
MATERIALS AND METHODS
Cells.
Human HCT116 colorectal cancer cells (ATCC CCL-247) were cultured in McCoy's 5A modified medium (ATCC). HEK293 cells were grown in Dulbecco's modified Eagle's medium (Invitrogen). HT29-APC cells were grown in RPMI medium and 600 μg/ml hygromycin (29). The medium was supplemented with 10% fetal bovine serum (HyClone), 100 units/ml penicillin, 100 units/ml streptomycin, and 5 mM l-glutamine. Cells were maintained at 37°C and 5% CO2.
Plasmids.
Construction of plasmids encoding Lef-1 and β-catenin S45F cDNAs was described previously (53). For luciferase reporter constructs, the pGL3-promoter (Promega) was used as the vector backbone. The c-Myc 5′ enhancer-pGL3-luciferase construct was made by PCR amplification of the 5′ enhancer of c-Myc from HCT116 genomic DNA (prepared using a DNeasy kit; Qiagen) with 5′-CAT GGT ACC CTA GCA CCT TTG ATT TCT CCC-3′ and 5′-CAT GCT AGC CGC TTT GAT CAA GAG TCC CAG-3′. The PCR products were subcloned as NheI-KpnI fragments into pGL3-promoter. The c-Myc 5′ enhancer-pGL3-luciferase construct containing mutations in the TCF consensus motifs was made by amplifying HCT116 genomic DNA with 5′-CAT GGT ACC CTA GCA CTG GTG CAT CTC CCA AAC CCG GCA GCC CG-3′ and 5′-CAT GCT AGC GCC GCG TGG TGC ACA AGA GTC CCA GGG AGA GTG-3′. The c-Myc 5′ enhancer contains the same sequence described by Vogelstein and colleagues (21). The c-Myc 3′ enhancer-pGL3-luciferase construct was made by PCR amplification of HCT116 genomic DNA with 5′-CAT GGA TCC GCT ATT GCT GTT CTA ATT ACC-3′ and 5′-CAT GGA TCC CAA GAT CAG CCT GAC TTT CGG-3′ and subcloning the PCR products as BamHI fragments into the pGL3-promoter. The c-Myc 3′ enhancer reverse-pGL3-luciferase construct was made by PCR amplification of HCT116 genomic DNA with 5′-CAT GCT AGC GCT ATT GCT GTT CTA ATT ACC-3′ and 5′-CAT GGT ACC CAA GAT CAG CCT GAC TTT CGG-3′ and subcloning the PCR products as NheI-KpnI fragments into pGL3-promoter. A 1-kb fragment from pBluescript SK+ (Stratagene) was PCR amplified using 5′-CAT GGC TAG CTA GGC CCG CTT TCC AGT C-3′ and 5′-CAT GCT CGA GGG TAA CTG TCA GAC C-3′, and the PCR product was subcloned as an NheI-Xho fragment into the c-Myc 3′ enhancer reverse-pGL3-luciferase plasmid to generate the c-Myc 3′ enhancer reverse-DNA spacer-pGL3-luciferase construct. The TCF consensus motifs within the c-Myc 3′ enhancer were mutated using a QuickChange site-directed mutagenesis kit (Stratagene). In QuickChange PCRs, 5′-TGG CAC GTC ATA TAG GCG AAT TTC AGC GGG AGA TGC AAT C-3′ and 5′-GAT TGC ATC TCC CGC TGA AAT TCG CCT ATA TGA CGT GCC A-3′ were used to mutate the first TCF motif, and 5′-GAG ATG CAA TCC ACA GAA GTA TAG TAG TTC AGC GGG TTA CAA AAG CAA G-3′ and 5′-CTT GCT TTT GTA ACC CGC TGA ACT ACT ATA CTT CTG TGG ATT GCA TCT C-3′ were used to mutate the second TCF motif. The AP-1 motif within the c-Myc 3′ enhancer was mutated with 5′-GAG TGC AGC TCT GGG GGT ACT CAC TTG GGA ATC G-3′ and 5′-CGA TTC CCA AGT GAG TAC CCC CAG AGC TGC ACT C-3′. All constructs and mutations were verified by DNA sequencing.
ChIP assays.
Antibodies used for ChIP included the following: 3 μg of anti-β-catenin (no. 610154; BD Transduction), anti-TCF4 (no. 05-0511; Upstate Biotechnology), anti-β-galactosidase (no. Z378B; Promega), anti-TATA-binding protein ([TBP] no. ab818-100; Abcam), anti-RNA polymerase II (RNAPII) C-terminal domain repeat YSPTSPS (phospho-S5) (no. ab5131-50; Abcam), anti-RNAPII (no. ab62655; Abcam), 5 μg of anti-acetyl-histone H3 (no. 06-599; Upstate Biotechnology), 5 μl of anti-trimethyl-histone H3 (Lys4) (no. 07-473; Upstate Biotechnology), and 4 μg of anti-c-Jun (no. SC44X; Santa Cruz). ChIP and quantitative real-time PCR were performed as described previously (54, 55). Sequences of oligonucleotides used in real-time PCRs are available upon request.
SACO.
For the detailed protocol used to construct the β-catenin SACO library, see the work of Impey et al. (23).
Western blot analysis.
Preparation of protein extracts and Western blotting were conducted as described previously (53). The following antibodies were used at the indicated dilutions: 1:1,000 anti-β-catenin (no. 610154; BD Transduction), 1:200 anti-TCF4 (no. 05-511; Upstate Biotechnology), 1:10,000 antitubulin (no. T3526; Sigma), and 1:500 anti-c-Myc (no. 05-419; Upstate Biotechnology).
Quantitative reverse transcription-PCR.
RNA was isolated from 5 × 106 HCT116 or HT29-APC cells, and cDNA was synthesized as described previously (53). APC expression was induced in HT29-APC cells with 100 μM ZnCl2 (29). tubulin was amplified from random-primed cDNA with 5′-GGG GCT GGG TAA ATG GCA AA-3′ and 5′-TGG CAC TGG CTC TGG GTT CG-3′. c-Myc expression was detected using 5′-GCA AAC CTC CTC ACA GCC CAC T-3′ and 5′-AAC TTG ACC CTC TTG GCA GCA-3′. APC expression was detected using 5′-CCC AAG CAA CAG AAG CAG AGA GGT-3′ and 5′-TTG AAC CCT GAC CAT TAC CAG AAG T-3′. Reactions lacking reverse transcriptase were run in parallel as a control, and real-time PCR was conducted as for ChIP except 10 ng of cDNA was used as template.
Luciferase reporter assays.
For each sample, examined in quadruplicate, 1 × 105 HEK293 or HCT116 cells were seeded into a single well of a 24-well plate. The following amounts of plasmids were transfected using Lipofectamine (Invitrogen) according to the manufacturer's instructions: pGL3-luciferase reporter construct, 125 ng; pcDNA3.1-β-catenin S45F, 250 ng; pME18-Lef-1, 250 ng; and pRL-simian virus 40 ([SV40] Renilla luciferase), 25 ng. β-Catenin and Lef-1 plasmids were excluded in transfection mixtures for assays in HCT116 cells. Total DNA was adjusted to 2 μg with pBluescript SK+ (Stratagene). Medium containing transfection complexes was removed after 6 h and replaced with fetal bovine serum containing culture medium. Luciferase assays were conducted using a dual luciferase kit (no. E1960; Promega), and firefly and Renilla luciferase activities were measured using an LMax II384 luminometer (Molecular Devices).
HCT116 cell synchronization.
Cells were synchronized as reported by Toualbi et al.(46). For ChIP assays, cells were grown to confluence on a 10-cm dish. The cells were then cultured for 48 h in medium lacking serum. Serum-containing medium was then added for 1, 2, 4, or 8 h, and ChIP assays were conducted as described above. Prior to fixation of serum-starved cells (zero time point), serum-containing medium was replaced to ensure equal exposure to formaldehyde. For luciferase assays, cells were transfected at 80% confluence and, the following day, serum starved as outlined above.
Cell cycle analysis.
A total of 1 × 107 HCT116 cells were harvested in suspension buffer (1× phosphate buffered saline [PBS]-0.1% bovine serum albumin) and collected by centrifugation at 3,000 × g. The cell pellet was washed twice in suspension buffer, centrifuged, and resuspended in 10 ml of suspension buffer. Three milliliters of ice-cold 100% ethanol was added to a 1-ml aliquot containing 1 × 106 cells. The cells were fixed for 1 h at 4°C, collected by centrifugation at 3,000 × g for 5 min, and washed twice in 1× PBS. The cell pellets were resuspended in 1 ml of propidium iodide staining solution (3.8 mM sodium citrate, 50 μg/ml propidium iodide [no. P4170; Sigma], 1× PBS). Before cells were counted with a fluorescence-activated cell sorter (FACS), they were incubated with RNase (0.5 μg) for 3 h at 4°C.
RESULTS
β-Catenin and TCF4 bind a region downstream from the c-Myc transcription stop site in HCT116 human colorectal carcinoma cells.
One surprising finding from our SACO analysis of β-catenin binding sites was that 12% of the protein-coding genes in the library contained a binding site downstream from the transcriptional stop site (54). c-Myc was one such target, containing multiple GSTs in a region that is 1.4 kb downstream from the transcription termination signal (Fig. 1A). Importantly, this region is as highly conserved across mammalian species as the previously characterized upstream β-catenin/TCF enhancer, suggesting that it may play an important role in c-Myc gene regulation.
FIG. 1.
β-Catenin and TCF4 bind a region downstream from the c-Myc transcription stop site in HCT116 cells. (A) Diagram of the human c-Myc gene locus. The c-Myc gene is depicted in blue with exons as rectangles, introns as horizontal lines, and the 5′ and 3′ untranslated regions as thin rectangles. An arrow marks the transcription start site. Coordinates of the region of chromosome 8 (chr 8) that contains c-Myc are shown at the top. Below, a red rectangle identifies the characterized 5′ β-catenin/TCF enhancer (21), and the green vertical lines indicate the positions of the β-catenin GSTs identified in the SACO screen (54). Clustered vertical lines below indicate the degree of conservation across mammalian species. This representation was downloaded from the UCSC Genome Browser (http://genome.ucsc.edu/). (B) Real-time PCR analysis of DNA fragments precipitated in a ChIP assay by using an anti-β-catenin antibody (blue bars) or an anti-TCF4 antibody (red bars). Primers were designed to the 5′ promoters of cycD1 and c-Myc to detect specific β-catenin and TCF4 binding and to an internal region of tubulin, an internal region of β-actin, and a region downstream of cycD1 to monitor nonspecific interactions. The primers used to detect binding to the region downstream of c-Myc were designed adjacent to the β-catenin SACO GSTs. As a control, ChIP assays were conducted in TIG fibroblasts which lack appreciable levels of nuclear β-catenin. Data are presented as percent input signal, and error bars indicate standard errors of the means.
Binding sites represented by two GSTs in the SACO screen (as was the case for the region downstream from c-Myc) had an 85% confirmation rate when tested for β-catenin binding using repeat ChIP assays (54). To confirm binding to this element, we used β-catenin and TCF4 antibodies in ChIP assays. β-Catenin and TCF4 bound the 5′ promoter elements of c-Myc and cyclin D1 (cycD1) in HCT116 colorectal carcinoma cells, while little binding was detected at tubulin and β-actin genes, which are not known to be β-catenin targets (Fig. 1B). β-Catenin and TCF4 also bound the 3′ region downstream from c-Myc, and, interestingly, binding of both factors was enhanced relative to that at the known upstream c-Myc 5′ enhancer. No significant β-catenin or TCF4 occupancy was detected at a region downstream from cycD1. As an additional control for nonspecific binding of β-catenin and TCF4 antibodies, ChIP assays were conducted in a TIG fibroblast cell line. TIG fibroblasts contain cytoplasmic and membrane-localized β-catenin but lack detectable nuclear β-catenin (30). Low levels of β-catenin and TCF4 binding, comparable to background levels, were detected using all primer sets, attesting to the specificity of the antibodies used in ChIP assays. Thus, we concluded that the β-catenin SACO screen uncovered a novel β-catenin and TCF4 binding region downstream of c-Myc that was more robust than the element in the 5′ promoter.
We next searched for additional TCF consensus elements across the 16-kb chromosomal sequence surrounding the genomic c-Myc locus. In addition to the two 5′ TCF sequences reported previously, we found five more core consensus motifs containing the sequence CTTTG(A/T)(A/T) [or the complementary sequence (A/T)(A/T)CAAAG], upstream from the transcription start site (Fig. 2A). Nine consensus sequences localized downstream from the c-Myc transcriptional stop site, two of which overlapped the region identified in the β-catenin SACO screen. No consensus motifs were found within the coding region. We then utilized ChIP-scanning to detect β-catenin and TCF4 binding across the c-Myc genomic locus (50). For this assay, we designed 16 oligonucleotide pairs at approximately 1-kb intervals across the c-Myc region (Fig. 2A). These oligonucleotides were used to detect precipitated DNA in real-time PCRs. Both β-catenin and TCF4 binding occurred only at the 5′ promoter and the downstream site identified by SACO (Fig. 2B). Again, binding to the 3′ element was significantly greater than to the 5′ region. Notably, not all consensus TCF motifs were occupied by TCF4.
FIG. 2.
ChIP scanning analysis of the c-Myc genomic locus in HCT116 cells. (A) Red boxes correspond to the amplicons produced in PCRs using oligonucleotide primer sets designed across the c-Myc genomic locus. These amplicons are arbitrarily designated 1 to 16. Purple and green vertical lines indicate the positions of consensus TCF motifs and β-catenin SACO GSTs, respectively. c-Myc is depicted at the top of the diagram with an arrow marking the transcription start site. (B) Real-time PCR analysis of ChIP assays performed in HCT116 cells using antibodies directed against β-catenin (red bars), TCF4 (blue bars), or control β-galactosidase (gray bars). The amplicons produced in the real-time PCRs are labeled 1 to 16 along the x axis. Real-time PCR analysis of ChIP assays in HCT116 cells using anti-TBP (C), anti-RNAPII S5 (D), anti-RNAPII (E), or anti-H3K9/K14ac and anti-H3K4me3 (F) antibodies. Error bars indicate standard errors of the means.
Earlier studies had shown that an antisense transcript was expressed from the N-Myc locus in neuroblastomas (1, 25). Additionally, we found that β-catenin induced expression of an antisense E2F4 transcript (53). These findings raised the possibility that the 3′ β-catenin/TCF4 binding region in c-Myc could also drive expression of an antisense transcript. To test this possibility, we analyzed TBP and serine-5 phosphorylated RNAPII (RNAPII S5) occupancy across the c-Myc locus. Because these proteins associate specifically with promoter elements, binding of TBP and RNAPII S5 downstream from c-Myc would suggest that the downstream β-catenin/TCF4 binding site regulates the activity of an antisense promoter. However, TBP and RNAPII S5 binding localized only to regions surrounding the 5′ promoter (Fig. 2C and D). Total RNAPII, in contrast, localized throughout the c-Myc gene (Fig. 2E). Furthermore, we were unable to detect an antisense c-Myc transcript using a c-Myc strand-specific reverse transcription-PCR assay (data not shown). Together, this analysis suggests that the only promoter within this chromosomal region lies at the 5′ end of the c-Myc gene.
To further characterize the c-Myc genomic locus, we localized histone modifications that correlate with gene activation. One such modification is trimethylated lysine 4 on histone H3 (H3K4me3), which is deposited by SET domain-containing proteins (4). Sierra et al. recently demonstrated that this modification was indeed found at the c-Myc promoter when the Wnt signaling pathway was induced (43). We found that H3K4me3 localized to a region encompassing the promoter and first intron of c-Myc (Fig. 2F). β-Catenin has also been shown to recruit the histone acetyltransferase CBP/p300 (22, 45). CBP/p300 acetylates several amino-terminal histone tail residues on histone H3, including lysines 9 and 14 (H3K9/K14ac) (41). These modifications also localized to the 5′ promoter region, consistent with the involvement of CBP/p300 in β-catenin activation of c-Myc (Fig. 2F). Together, this ChIP-scanning analysis suggests that the new β-catenin binding site does not identify a 3′ promoter; rather, the upstream site is the only promoter involved in c-Myc expression. We suggest that the downstream region identifies a c-Myc enhancer.
The downstream β-catenin and TCF4 binding region identifies a transcriptional enhancer.
Classically, enhancers are believed to bind transcriptional activators and function in an orientation-independent manner (2). To test whether the downstream element could function in this fashion, we used luciferase reporter assays. We initiated these studies by engineering a series of plasmids designed to assess the ability of the 5′ and 3′ c-Myc elements to regulate expression of a luciferase gene driven by the minimal SV40 viral promoter (Fig. 3A). These plasmids were first tested in HEK293 cells, which contain little nuclear β-catenin. To activate the reporter constructs, cDNAs encoding Lef-1 and stabilized β-catenin (containing an S45F mutation, which impairs proteasomal degradation) were cotransfected. The combination of β-catenin and Lef-1 activated a reporter containing the c-Myc 5′ enhancer, as previously described (Fig. 3B) (21). Mutation of the two TCF consensus motifs within the c-Myc 5′ enhancer impaired activation, also as reported previously (21). Cells were then transfected with a reporter construct containing the 615-bp region flanking the c-Myc 3′ GSTs inserted downstream from luciferase. β-Catenin/Lef-1 activated this construct considerably more robustly than the construct containing the 5′ enhancer alone. Two consensus TCF motifs, both consisting of the sequence TTCAAAG and separated by 28 nucleotides, are localized within the 3′ enhancer region. These sequences were mutated to TTCAGCG, which has been demonstrated to disrupt TCF binding (9). Mutation of either TCF consensus motif alone impaired β-catenin/Lef-1 transactivation while the double mutation was slightly repressive. Inclusion of the c-Myc 3′ enhancer with the 5′ elements augmented luciferase expression. This activation was also largely impaired by mutations in TCF elements within the c-Myc 3′ enhancer. Luciferase assays were then conducted in HCT116 cells, which have high levels of endogenous nuclear β-catenin. The luciferase activities of reporters in HCT116 cells largely paralleled the activities in the HEK293 cells (Fig. 3C). Together, these experiments indicate that the c-Myc 3′ element requires two coupled TCF consensus motifs to activate transcription of a heterologous reporter plasmid.
FIG. 3.
The c-Myc 3′ β-catenin/TCF4 binding region enhances SV40 promoter-driven luciferase gene expression. (A) Diagram of reporter constructs with the SV40 promoter represented as green rectangles, and the firefly luciferase gene is shown as yellow rectangles. The rectangles labeled c-Myc 5′ EN correspond to the 588 bp β-catenin/TCF enhancer (21). The rectangles labeled c-Myc 3′ EN correspond to a 615-bp fragment that begins 1,410 bp downstream from the c-Myc transcription stop site and binds β-catenin and TCF4 in ChIP assays. An “X” indicates that a TCF consensus motif was mutated. (B) The firefly luciferase reporter plasmids, a control plasmid expressing Renilla luciferase, and plasmids encoding β-catenin S45F and Lef-1 were transfected into HEK293 cells. After 24 h, firefly luciferase levels were assayed and normalized to Renilla luciferase levels. The activities of the respective reporters are aligned with their schematic representations. Activity is represented as relative change in the presence of β-catenin and Lef-1 and is normalized to background β-catenin/Lef-1 activation of the pGL3-promoter vector. (C) HCT116 cells were transfected with the indicated firefly luciferase reporters and a control Renilla luciferase plasmid. Luciferase activities were measured as in HEK293 cells, and data are represented as the change in expression relative to levels obtained with the pGL3-promoter vector alone. Error bars indicate standard deviations.
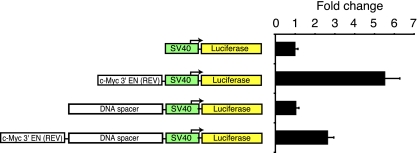
We then designed additional reporter constructs to determine whether the c-Myc 3′ element could activate transcription irrespective of position or orientation relative to the SV40 promoter. These reporters were tested in HCT116 cells. The luciferase level from a plasmid containing the c-Myc 3′ element inserted in reverse orientation and upstream from the SV40 promoter was approximately fivefold greater than the control when assayed in HCT116 cells (Fig. 4). In a second plasmid, we placed the c-Myc 3′ element in the reverse orientation and 1 kb upstream from the SV40 promoter. The luciferase level from this plasmid was also enhanced relative to the control. Thus, by using standard luciferase assays, we find that the c-Myc 3′ element activates transcription in a position- and orientation-independent manner and thereby satisfies the criteria for its designation as a transcriptional enhancer.
FIG. 4.
The c-Myc 3′ enhancer activates transcription in a position- and orientation-independent manner. (Left) Diagram of reporter constructs with the SV40 promoter represented as green rectangles and the firefly luciferase gene shown as yellow rectangles. The rectangles labeled c-Myc 3′ EN (REV) correspond to the 615 bp c-Myc 3′ element inserted in reverse orientation relative to the SV40 promoter. DNA spacers corresponding to 1-kb fragments from the pBluescript plasmid are indicated. The firefly luciferase reporter plasmids and a control plasmid expressing Renilla luciferase were transfected into HCT116 cells. Luciferase levels were measured 24 h following transfection. Firefly luciferase values were normalized to Renilla luciferase values, and the data are represented as the change in expression relative to levels in cells transfected with the pGL3-promoter vector and control Renilla vector. Error bars indicate standard deviations.
The 3′ enhancer is required for activation of c-Myc expression by serum mitogens.
The response of quiescent cells to growth factors has long been used as a model system to study regulation of c-Myc gene expression (24). Previous studies have shown that c-Myc expression is negligible in serum-starved quiescent cells but is then dramatically induced upon serum addition (42, 49). Once the cells reach the G1/S boundary, or restriction point, c-Myc expression subsides. We first confirmed an earlier report that HCT116 cells entered quiescence 48 h after serum withdrawal and then reentered the cell cycle following addition of serum (46). FACS analysis of propidium iodide-stained serum-deprived cells indicated that most are in the G0/G1 stage (Fig. 5A). Treatment of quiescent cells with serum over a time course of 1 to 8 h indicated that synchronized HCT116 cells progressed through G1 and entered S phase. To evaluate c-Myc expression in HCT116 cells cultured under these conditions, we collected RNAs from quiescent cells and cells treated with serum for 1, 2, 4, and 8 h. c-Myc expression was monitored by real-time PCR. Consistent with other reports, we found very low levels of c-Myc RNA in quiescent cells (Fig. 5B) (42, 49). A 10-fold increase in c-Myc transcript was apparent after exposure to serum for 2 h, and a 70-fold increase occurred after 4 h, compared to quiescent cells. After 8 h in the presence of serum, c-Myc RNA levels substantially decreased. At this time, the synchronized cells have entered S phase, where c-Myc expression is repressed. Protein extracts were prepared under the same conditions, and Western blot analysis was performed with a c-Myc-specific antibody. c-Myc protein levels were not detectable in quiescent cells, and peak levels of c-Myc protein were detected at 4 h after the addition of serum (Fig. 5C). c-Myc protein levels then decreased at 8 h. Using the same protein preparations, we evaluated the expression profiles of β-catenin and TCF4. While total β-catenin levels remained constant, TCF4 levels, surprisingly, decreased after exposure of cells to serum for 2 h. This low level persisted as the cells transited into S phase. To evaluate nuclear levels of β-catenin, we isolated nuclei from synchronized cells, and protein extracts were subjected to Western blot analysis. Nuclear β-catenin levels were elevated after serum addition for 1 and 2 h compared to levels seen in quiescent cells. At 4 and 8 h after serum addition, nuclear β-catenin was reduced to levels seen in quiescent cells (Fig. 5C). We then used this system to assess whether β-catenin binding to the 5′ promoter and/or 3′ enhancer accompanied c-Myc gene expression.
FIG. 5.
c-Myc expression is tightly regulated in synchronized HCT116 cells. (A) FACS profiles of propidium iodide-stained HCT116 cells that were serum deprived for 48 h and then cultured in the presence of serum-containing medium for the number of hours indicated at the top of each panel. FL2-H is peak emission values of propidium iodide-stained DNA fluorescence. (B) RNAs were isolated from HCT116 cells treated as described for panel A, and cDNA was synthesized using a random primer and avian myeloblastosis virus reverse transcriptase. Real-time quantitative PCR was conducted using primers designed against the third exon of c-Myc or the fourth exon of tubulin as an internal control. The data are presented as relative c-Myc mRNA levels normalized to tubulin. Error bars indicate standard errors of the means. (C) Protein extracts prepared from HCT116 cells synchronized as in panel A were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and probed with anti-c-Myc, anti-β-catenin, anti-TCF4, or antitubulin antibodies in a Western blot.
We along with others have shown that β-catenin and TCF4 binding to the 5′ c-Myc promoter accompanied c-Myc gene expression (30, 43, 54). However, it is unknown whether β-catenin/TCF4 binding occurs in response to serum. We used ChIP coupled with real-time PCR to monitor binding of β-catenin and TCF4 as HCT116 cells exited quiescence and entered the cell cycle. The specific oligonucleotides used to monitor occupancy at the c-Myc promoter amplify a region approximately 500 bases upstream from the transcription start site that is adjacent to the two characterized TCF motifs. Consequently, these primers also interrogate occupancy at the c-Myc 5′ enhancer. β-Catenin binding to the c-Myc promoter was minimal in serum-starved cells but was dramatically increased following addition of serum for 2 h (Fig. 6A). This finding indicates that β-catenin was present at the c-Myc promoter prior to maximal c-Myc transcript induction (Fig. 5B, 4-h time point) and suggests that β-catenin participated in mitogen-dependent c-Myc activation in HCT116 cells. Consistent with this idea, β-catenin binding was absent at 8 h, a time when c-Myc transcript was reduced. TCF4 binding to the c-Myc promoter, unlike β-catenin, was only marginally induced as cells were released into the cell cycle (Fig. 6A). This finding is in agreement with earlier reports indicating that TCF4 binding is constitutive at the c-Myc promoter (43).
FIG. 6.
β-Catenin, TCF4, and transcriptional regulatory factors assemble at the c-Myc 3′ enhancer prior to binding to the 5′ c-Myc promoter following exposure of arrested HCT116 cells to serum. (A) Real-time PCR analysis of ChIP assays using anti-β-catenin or anti-TCF4 antibodies to precipitate chromatin isolated from serum-starved cells (0) and starved cells released into serum for 1, 2, 4, or 8 h. Primers specific to the 5′ c-Myc promoter and the c-Myc 3′ enhancer (primer set 5 and set 13, respectively) (Fig. 2A) were used in the real-time PCRs. (B to D) Real-time PCR analysis as in panel A except that anti-TFIIB (B), anti-H3K4me3 (C), or anti-H3K9/K14ac (D) antibodies were used in the ChIP assays. Error bars indicate standard errors of the means.
Having confirmed β-catenin/TCF4 binding at the 5′ promoter, we next determined occupancy of these factors at the 3′ enhancer. Surprisingly, maximal β-catenin binding to this site occurred after serum treatment for only 1 h (Fig. 6A), a full hour before occupancy at the 5′ promoter. Additionally, when we interrogated TCF4 occupancy at the 3′ enhancer, we found a dramatic (∼60-fold) increase in binding in cells treated with serum for 1 h compared to serum-deprived cells. As with binding to the 5′ promoter, β-catenin and TCF4 occupancy decreased as cells transited into S phase (i.e., 8 h after serum addition). These findings indicate that β-catenin binding to the c-Myc 3′ enhancer preceded binding to the 5′ promoter. Furthermore, these results suggest that mitogen-stimulated activation of c-Myc expression involves recruitment of TCF4 to the 3′ element.
The general transcription factor TFIIB positions the coding strand of DNA into the catalytic center of RNAPII to determine the first nucleotide of the nascent mRNA (7) and is required for RNAPII-dependent transcription (35, 40). TFIIB occupancy was coincident with that of β-catenin at the 5′ promoter element, peaking at 2 h in the presence of serum and then decreasing as cells transited G1 and entered into S phase (Fig. 6B). The binding kinetics of TFIIB is consistent with assembly of the RNAPII holoenzyme prior to and during c-Myc expression and then disassembly of this complex during the period when gene expression is repressed.
High levels of H3K4me3 at the promoter region accompany c-Myc expression. Sierra et al. demonstrated that when the Wnt pathway was blocked, c-Myc transcript levels decreased as well as levels of H3K4me3 (43). Consistent with that study, we found that H3K4me3 was absent at the c-Myc promoter during repression (i.e., serum starvation) and then increased as c-Myc was activated (i.e., 2 h after serum addition) (Fig. 6C). As was the case for β-catenin, induction of H3K4me3 binding at the 3′ enhancer preceded the increase in H3K4me3 at the promoter. H3K9/K14ac histone modifications paralleled the H3K4me3 changes (Fig. 6D). The finding that increases in H3K4me3- and H3K9/K14ac-modified histones accompanied β-catenin and TCF4 binding to the 3′ enhancer suggests that β-catenin/TCF4 directly recruited chromatin-remodeling complexes to this site. These data further suggest that β-catenin/TCF4 and coactivator complexes assembled first at the 3′ enhancer and then at the 5′ enhancer during mitogen stimulation.
β-Catenin and the TCF motifs in the c-Myc 3′ enhancer are required for mitogen-dependent activation of c-Myc gene expression.
The human colorectal cancer cell line, HT29, contains a mutant APC allele (44). APC protein derived from this allele no longer coordinates the destruction of β-catenin by the proteasome. As a result, Wnt/β-catenin signaling is constitutive. To block Wnt signaling in these cells, Vogelstein and colleagues inserted a metallothionein promoter-driven transgene encoding full-length APC (29). In the presence of ZnCl2, APC is synthesized, and β-catenin is targeted for proteasomal degradation. We used the HT29-APC cells to determine whether β-catenin is required for mitogen activation of c-Myc expression. Cells were synchronized by serum deprivation for 2 days. Serum-containing medium, with or without ZnCl2, was then added, and RNA was collected at subsequent time points. APC and c-Myc transcript levels were measured by quantitative real-time PCR. In the absence of zinc, APC transcripts were not detected, and c-Myc expression was induced by serum (Fig. 7A). In the presence of zinc, mitogen activation of c-Myc was completely blocked by the induced APC. Western blot analysis indicated that APC induction also blocked serum-dependent accumulation of c-Myc protein and nuclear β-catenin (Fig. 7B). Importantly, a ChIP assay demonstrated that the enhanced β-catenin binding to the c-Myc 3′ enhancer seen after 1 h of serum addition was also blocked by APC induction (Fig. 7C). These findings suggest that nuclear β-catenin and β-catenin binding to the c-Myc 3′ enhancer were required for mitogen activation of c-Myc expression.
FIG. 7.
β-Catenin and the TCF-responsive sites in the c-Myc 3′ enhancer are required for mitogen-dependent activation of c-Myc gene expression. (A) HT29-APC cells were synchronized in the cell cycle by serum withdrawal for 48 h followed by addition of serum-containing medium for 1, 2, 4, or 8 h. Where indicated, 100 μM ZnCl2 was included in the medium. ZnCl2 induces expression of a transgene encoding full-length APC in the HT29-APC cells (29). At each time point, RNAs were isolated and reverse transcribed using avian myeloblastosis virus reverse transcriptase, and the synthesized cDNA was amplified in real-time PCRs using APC-, c-Myc-, or tubulin-specific primer sets. Each point is normalized to signal generated with tubulin primers as an internal control. Error bars indicate standard errors of the means. (B) Protein extracts prepared from HT29-APC cells treated as in panel A were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis and probed with anti-c-Myc- or anti-β-catenin-specific antibodies in Western blotting. Whole-cell extracts were used in the c-Myc experiment while nuclear extracts were used in the β-catenin experiment. (C) Real-time PCR analysis of the c-Myc 3′ enhancer precipitated from HT29-APC cells with anti-β-catenin antibodies in a ChIP assay. Prior to the ChIP, HT29-APC cells were serum starved for 48 h (0), and then serum was added for 1 or 2 h in the presence or absence of ZnCl2 as indicated. Error bars indicate standard errors of the means. (D) The luciferase constructs diagrammed above the graph were transfected into HCT116 cells and, the following day, medium lacking serum was added. After 48 h, serum was added for 1, 2, or 4 h, and then levels of luciferase were quantified. Both TCF4 consensus sequences within the 3′ enhancer are mutated in the construct labeled 5′EN/3′EN (mut TBE3/4). Error bars indicate standard deviations. EN, enhancer; X, mutation in a TCF consensus motif.
As shown in Fig. 6A, mitogens stimulated β-catenin and TCF4 binding to the c-Myc 3′ enhancer. We used luciferase assays to test whether the consensus TCF motifs within the 3′ enhancer were required for activation in response to serum. HCT116 cells were transfected with a luciferase construct containing both the 5′ and 3′ c-Myc enhancers and were then starved for 2 days. Serum-containing medium was then added, and levels of luciferase were monitored. Serum activated the reporter construct containing the 5′ and 3′ enhancers while activities of reporters containing mutations in the 3′ TCF motifs were attenuated upon serum addition (Fig. 7C). This suggests that assembly of a transcriptional coactivator complex at the 3′ TCF sequences was required for serum-stimulated activation. Furthermore, the 5′ c-Myc enhancer alone was insufficient for mitogen-dependent transcriptional activity.
c-Jun binds to an AP-1 site within the c-Myc 3′ enhancer.
The c-Myc 5′ enhancer and 3′ enhancer both contain two consensus TCF motifs. Why did mitogen stimulation specifically induce β-catenin/TCF4 binding to the 3′ enhancer? We entertained the possibility that binding of an additional transcription factor(s) may account for our observations. Using the TRANSFAC database, we searched for transcription factor motifs embedded in the c-Myc 3′ enhancer that were highly conserved in rat, mouse, and human. We reasoned that if the motif was functionally relevant for c-Myc transcription, it was likely to be conserved among the three species. This search revealed the sequence TGACTCA, an AP-1 consensus motif (10), immediately downstream from the two TCF consensus sequences (Fig. 8A). AP-1 is a heterodimeric transcription factor composed of members from the c-Jun and c-Fos families of transcription factors (15). AP-1 is induced by mitogens and is thus classified as an immediate-early factor. Moreover, AP-1 is known to interact with TCF4 and β-catenin to activate gene transcription in HCT116 cells (46). We used ChIP analysis to determine whether c-Jun binding occurred at the c-Myc 5′ and 3′ enhancers during mitogen activation of serum-starved HCT116 cells. c-Jun bound the 3′ enhancer in serum-starved cells, and this binding increased when the cells were exposed to serum for 1 h (Fig. 8B). c-Jun binding diminished as cells transited into S phase (i.e., at 8 h). Levels of c-Jun binding to the c-Myc 5′ enhancer were similar to those seen at a control sequence (approximately 6 kb downstream from the c-Myc transcriptional stop site) and therefore likely represented background binding. The profile of c-Jun binding to the 3′ enhancer paralleled β-catenin and TCF4 binding, suggesting that a β-catenin/TCF4/c-Jun complex may account for specific binding of coactivator complexes to the downstream c-Myc enhancer.
FIG. 8.
AP-1 is involved in β-catenin/TCF-dependent activation of c-Myc expression in response to mitogens. (A) Diagram of the c-Myc gene (blue) with the 3′ enhancer represented by a black rectangle. The sequence of a 158-bp segment from the enhancer containing two TCF motifs (bold) and an AP-1 motif (underlined) is shown. (B) Real-time PCR analysis of ChIP assays using an anti-c-Jun-specific antibody was conducted in starved (0) HCT116 cells and starved cells cultured in serum-containing medium for 1, 2, 4, or 8 h. Error bars indicate standard errors of the means. (C) Diagram of luciferase constructs where X indicates mutation in the TCF motif, and a black oval indicates a mutation in the AP-1 motif. HCT116 cells were transfected with the indicated luciferase reporters. The next day, cells were starved in serum-deprived medium for 48 h. Serum-containing medium was added to half of the samples for 4 h, and luciferase activities were measured in cell lysates. Data are presented as the relative change in luciferase activity (in relative light units [RLU]) in response to serum and are normalized to levels obtained in cells transfected with pGL3-promoter. Error bars indicate standard deviations. EN, enhancer.
We then used luciferase assays to test whether the consensus AP-1 motif within the 3′ enhancer was required for activation in response to serum. For these experiments, luciferase reporter constructs were transfected into HCT116 cells that were then cultured in serum-deprived medium for 2 days. On the next day, serum was added to half of the samples for 4 h. As previously shown (Fig. 7D), mitogen activation of the reporter containing the c-Myc 5′ and 3′ enhancers depended on the integrity of the two TCF motifs in the 3′ enhancer (Fig. 8C). To test whether AP-1 binding was also required for the mitogen response, the consensus AP-1 motif within the 3′ enhancer was mutated to GTACTCA (shown previously to block this interaction with AP-1), and this reporter was assayed as above (27). Mutation of the AP-1 motif abolished mitogen activation. Furthermore, mutation of the AP-1 motif in the context of the TCF double mutant did not cause an additive impairment of 3′-dependent activation. Together, these results demonstrate that the TCF motifs and the AP-1 motif within the 3′ enhancer are required for the maximal transcriptional response to mitogen stimulation.
DISCUSSION
Genome-wide screens are a powerful approach to discover new transcription factor targets. Once the binding sites have been identified, determining which of these sites are functional can be challenging. One characteristic of transcription factor binding sites revealed by genome-wide approaches is that they are commonly located in regions other than the 5′ flanking elements of protein-coding genes (8, 20, 23, 31, 54, 55). In a previous report, we used SACO to identify β-catenin binding sites in human colorectal carcinoma cells and found that, of those associated with protein-coding genes, 12% localized downstream from the transcriptional stop site (54). In a subsequent study, we described a β-catenin/TCF4-regulated enhancer that localized within the 3′ untranslated region of the gene encoding the E2F4 cell cycle transcription factor (53). Activation of Wnt/β-catenin signaling induced expression of an antisense E2F4 transcript which, in turn, diminished E2F4 protein levels. In this study, we describe a β-catenin binding site that localized downstream from the c-Myc proto-oncogene. This site identified an enhancer that controlled the mitogen-dependent activation of c-Myc expression in colon cancer cells. However, unlike the downstream enhancer regulating E2F4, the c-Myc 3′ enhancer did not regulate an antisense transcript but rather cooperated with 5′ elements to activate c-Myc expression.
Several recent reports have described functional intersections between Wnt/β-catenin and AP-1 signaling (17, 30, 46). Nateri et al. demonstrated that phosphorylated c-Jun interacted with TCF4 and, together with β-catenin, activated c-Jun gene expression (30). Toulabi et al. showed using reporter assays that c-Fos and c-Jun activated c-Myc expression and demonstrated that a complex containing c-Jun family members, β-catenin, and TCF4 assembled on the cycD1 promoter in serum-synchronized HCT116 cells (46). Whether this complex associated with the endogenous c-Myc promoter in a mitogen-regulated manner was not addressed. Finally, Gan et al. demonstrated that binding of c-Jun, TCF4, β-catenin, and disheveled was required for full c-Myc expression (17). Our results complement and extend these findings. We suggest that c-Jun binding underlies the specific association of β-catenin and TCF4 at the 3′ c-Myc enhancer following mitogen stimulation. The fact that the AP-1 consensus motif within the 3′ enhancer is required for activation suggests that c-Jun binds directly to this site. We failed to detect c-Fos binding, however, suggesting that another Fos family member may be involved. Furthermore, we detected little c-Jun interaction with the c-Myc 5′ promoter. This finding contrasts that reported by Gan et al.; however, differences in experimental systems may account for this observation (17). Gan et al. examined β-catenin and c-Jun interactions in cells stimulated by Wnt3A while we focused on cell cycle-regulated associations. Our data support a model wherein c-Jun acts as a mitogen sensor and, when phosphorylated in response mitogen signaling, cooperates with β-catenin and TCF4 to activate c-Myc expression via the 3′ enhancer.
Members of the TCF/Lef family of sequence-specific DNA binding factors were thought to associate constitutively with target sites irrespective of Wnt/β-catenin signaling activity (48). According to this model, TCF/Lef binds to their cognate DNA sequence motifs and provides docking sites for associations with corepressor or coactivator complexes. Wnt signaling then induces an exchange of coactivators for corepressors, and transcription of the target genes ensues. While this model may apply for Wnt/β-catenin regulation of some target genes, recent studies indicate that TCF binding to other targets is more dynamic (32, 51). Wöhrle et al. demonstrated that Wnt signaling induced TCF binding to promoters of the developmentally regulated target genes, Axin2, Cdx1, and Tbra (51). However, in cell types that are not Wnt responsive, these target gene promoters contain methylated cytosines and lack TCF occupancy. In addition, Parker et al. reported that TCF binding is induced at the naked-cuticle and notum loci in response to Wg (the Wnt homolog) signaling in Drosophila (32). Our ChIP analysis using β-catenin and TCF4 antibodies suggests that Wnt/β-catenin regulation of c-Myc expression involves both inducible and constitutive TCF binding. We confirmed the findings of Sierra et al. that TCF binding to the 5′ enhancer is largely constitutive (43). However, we find that TCF4 occupancy at the 3′ enhancer is dynamic, as binding this element was clearly induced as cells entered G1 phase from quiescence. What is not clear at this time is whether c-Myc regulation through the downstream enhancer occurs only in cells that contain high levels of nuclear β-catenin such as the colon cancer cells used in this study. It is tempting to speculate that deregulated β-catenin signaling, as occurs in colon cancer, activates a normally quiescent enhancer.
H3K4me3 correlates with gene transcriptional activity. With respect to protein-coding genes (including targets of Wnt/β-catenin signaling), this modification typically localizes to the 5′ promoters and first introns (3, 56). In agreement with Sierra et al., we find that levels of promoter-localized H3K4me3 correlate with c-Myc activity; i.e., there are high levels of H3K4me3 when c-Myc is expressed in early G1 and low levels when c-Myc expression is repressed in S phase and in serum-deprived cells (43). Surprisingly, we found that high levels of H3K4me3 accompanied β-catenin/TCF4/c-Jun binding to the c-Myc 3′ enhancer as synchronized cells entered early G1. Interestingly, this 3′ localization of H3K4me3-modified histones was seen in synchronized cells only and not in logarithmically growing cultures (Fig. 2F and 6C). We hypothesize that cell cycle-dependent epigenetic modifications may underlie the ability of a β-catenin/TCF4/c-Jun complex to bind to the 3′ enhancer. Alternatively, nucleosomes that block access to the 3′ enhancer may require repositioning to allow binding by the β-catenin/TCF4/c-Jun complex. Once these epigenetic barriers are removed, c-Myc expression during subsequent cycles would not require chromatin modifications at the 3′ enhancer.
Our findings suggest that the 5′ and 3′ enhancers cooperate to regulate c-Myc expression. We envision two mechanisms that may account for this cooperation. First, complexes assembled at the 3′ enhancer may physically associate with complexes at the 5′ enhancer via a chromatin loop. The resultant enhanceosome would then recruit RNAPII and other transcriptional machinery to activate c-Myc expression. There are many examples of such regulatory chromatin loops involving transcription factor complexes separated by several kilobases or more (14). Alternatively, chromatin-modifying coactivator complexes recruited by β-catenin, TCF4, and c-Jun to the 3′ enhancer may initiate spreading of histones modifications to the 5′ promoter region. Indeed, such a process has been ascribed to TCF binding in Drosophila, where induced histone changes correlate with remodeling of 5′ promoters to a transcriptionally facilitative state (32).
One surprising observation from our cell cycle analysis was that the level of TCF4 protein decreased after exposure of cells to serum. This decrease suggests that TCF4 is regulated at the posttranslational level. Yamada et al. recently reported that TCF/Lef proteins phosphorylated by the Nemo-like kinase are ubiquitinated and targeted for degradation by the proteasome (52). Furthermore, Tran et al. found that TCF proteins are stabilized when proteasome activity is inhibited in HEK293 cells (47). It will be of interest to determine which of the 6,868 recently identified TCF4-binding sites display differential binding throughout the cell cycle (20). Coupling analysis of TCF4 binding with large-scale transcript profiling in synchronized cells may elucidate targets that are coregulated by the cell cycle and Wnt/β-catenin signaling.
The idea that c-Myc expression could be controlled by a 3′ regulatory element first was proposed on the basis of DNase I hypersensitivity mapping (28). Interestingly, the hypersensitivity site 1.5 kb downstream from c-Myc was detected in some, but not all, cell types. Notably, the downstream hypersensitivity site was prominent in Colo320 colorectal cancer cells. Our results suggest that this downstream enhancer is the primary controlling element for c-Myc expression in colorectal cancer cells. The importance of c-Myc in intestinal carcinogenesis has been firmly established using conditional knockout approaches in mouse models (38). However, the contribution of Wnt/β-catenin signaling to pathogenic c-Myc expression was not addressed. It was subsequently suggested that deletion of the 5′ β-catenin-responsive elements would ascertain the role of c-Myc in Wnt/β-catenin signaling (5). Our data suggest that the 3′ β-catenin-responsive element might be equally important for c-Myc expression.
Acknowledgments
This work was supported by grants from the National Institutes of Health.
We thank Bert Vogelstein and colleagues (Johns Hopkins School of Medicine) for the generous gift of the HT29-APC cell line. We are grateful to Gail Mandel, members of the Goodman and Mandel laboratories, and colleagues who coattended the 2008 Wnt/β-catenin Signaling in Development and Disease Keystone Symposium for helpful comments.
Footnotes
Published ahead of print on 13 October 2008.
REFERENCES
- 1.Armstrong, B. C., and G. W. Krystal. 1992. Isolation and characterization of complementary DNA for N-cym, a gene encoded by the DNA strand opposite to N-myc. Cell Growth Differ. 3385-390. [PubMed] [Google Scholar]
- 2.Banerji, J., S. Rusconi, and W. Schaffner. 1981. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 27299-308. [DOI] [PubMed] [Google Scholar]
- 3.Barski, A., S. Cuddapah, K. Cui, T. Y. Roh, D. E. Schones, Z. Wang, G. Wei, I. Chepelev, and K. Zhao. 2007. High-resolution profiling of histone methylations in the human genome. Cell 129823-837. [DOI] [PubMed] [Google Scholar]
- 4.Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. [DOI] [PubMed] [Google Scholar]
- 5.Bommer, G. T., and E. R. Fearon. 2007. Role of c-Myc in Apc mutant intestinal phenotype: case closed or time for a new beginning? Cancer Cell 11391-394. [DOI] [PubMed] [Google Scholar]
- 6.Buck, M. J., and J. D. Lieb. 2004. ChIP-chip: considerations for the design, analysis, and application of genome-wide chromatin immunoprecipitation experiments. Genomics 83349-360. [DOI] [PubMed] [Google Scholar]
- 7.Bushnell, D. A., K. D. Westover, R. E. Davis, and R. D. Kornberg. 2004. Structural basis of transcription: an RNA polymerase II-TFIIB cocrystal at 4.5 angstroms. Science 303983-988. [DOI] [PubMed] [Google Scholar]
- 8.Cawley, S., S. Bekiranov, H. H. Ng, P. Kapranov, E. A. Sekinger, D. Kampa, A. Piccolboni, V. Sementchenko, J. Cheng, A. J. Williams, R. Wheeler, B. Wong, J. Drenkow, M. Yamanaka, S. Patel, S. Brubaker, H. Tammana, G. Helt, K. Struhl, and T. R. Gingeras. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116499-509. [DOI] [PubMed] [Google Scholar]
- 9.Chamorro, M. N., D. R. Schwartz, A. Vonica, A. H. Brivanlou, K. R. Cho, and H. E. Varmus. 2005. FGF-20 and DKK1 are transcriptional targets of beta-catenin and FGF-20 is implicated in cancer and development. EMBO J. 2473-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chinenov, Y., and T. K. Kerppola. 2001. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 202438-2452. [DOI] [PubMed] [Google Scholar]
- 11.Clevers, H. 2006. Wnt/beta-catenin signaling in development and disease. Cell 127469-480. [DOI] [PubMed] [Google Scholar]
- 12.Dang, C. V., K. A. O'Donnell, K. I. Zeller, T. Nguyen, R. C. Osthus, and F. Li. 2006. The c-Myc target gene network. Semin. Cancer Biol. 16253-264. [DOI] [PubMed] [Google Scholar]
- 13.Das, P. M., K. Ramachandran, J. vanWert, and R. Singal. 2004. Chromatin immunoprecipitation assay. BioTechniques 37961-969. [DOI] [PubMed] [Google Scholar]
- 14.Dekker, J. 2008. Gene regulation in the third dimension. Science 3191793-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eferl, R., and E. F. Wagner. 2003. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3859-868. [DOI] [PubMed] [Google Scholar]
- 16.Fodde, R., R. Smits, and H. Clevers. 2001. APC, signal transduction and genetic instability in colorectal cancer. Nat. Rev. Cancer. 155-67. [DOI] [PubMed] [Google Scholar]
- 17.Gan, X. Q., J. Y. Wang, Y. Xi, Z. L. Wu, Y. P. Li, and L. Li. 2008. Nuclear Dvl, c-Jun, beta-catenin, and TCF form a complex leading to stabilization of beta-catenin-TCF interaction. J. Cell Biol. 1801087-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grandori, C., S. M. Cowley, L. P. James, and R. N. Eisenman. 2000. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu. Rev. Cell Dev. Biol. 16653-699. [DOI] [PubMed] [Google Scholar]
- 19.Gregorieff, A., and H. Clevers. 2005. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 19877-890. [DOI] [PubMed] [Google Scholar]
- 20.Hatzis, P., L. G. van der Flier, M. A. van Driel, V. Guryev, F. Nielsen, S. Denissov, I. J. Nijman, J. Koster, E. E. Santo, W. Welboren, R. Versteeg, E. Cuppen, M. van de Wetering, H. Clevers, and H. G. Stunnenberg. 2008. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol. Cell. Biol. 282732-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He, T. C., A. B. Sparks, C. Rago, H. Hermeking, L. Zawel, L. T. da Costa, P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Identification of c-MYC as a target of the APC pathway. Science 2811509-1512. [DOI] [PubMed] [Google Scholar]
- 22.Hecht, A., K. Vleminckx, M. P. Stemmler, F. van Roy, and R. Kemler. 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 19:1839-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Impey, S., S. R. McCorkle, H. Cha-Molstad, J. M. Dwyer, G. S. Yochum, J. M. Boss, S. McWeeney, J. J. Dunn, G. Mandel, and R. H. Goodman. 2004. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell 1191041-1054. [DOI] [PubMed] [Google Scholar]
- 24.Kelly, K., and U. Siebenlist. 1985. The role of c-myc in the proliferation of normal and neoplastic cells. J. Clin. Immunol. 565-77. [DOI] [PubMed] [Google Scholar]
- 25.Krystal, G. W., B. C. Armstrong, and J. F. Battey. 1990. N-myc mRNA forms an RNA-RNA duplex with endogenous antisense transcripts. Mol. Cell. Biol. 104180-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee, L. A., and C. V. Dang. 2006. Myc target transcriptomes. Curr. Top. Microbiol. Immunol. 302145-167. [DOI] [PubMed] [Google Scholar]
- 27.Lee, M. E., M. S. Dhadly, D. H. Temizer, J. A. Clifford, M. Yoshizumi, and T. Quertermous. 1991. Regulation of endothelin-1 gene expression by Fos and Jun. J. Biol. Chem. 26619034-19039. [PubMed] [Google Scholar]
- 28.Mautner, J., S. Joos, T. Werner, D. Eick, G. W. Bornkamm, and A. Polack. 1995. Identification of two enhancer elements downstream of the human c-myc gene. Nucleic Acids Res. 2372-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morin, P. J., B. Vogelstein, and K. W. Kinzler. 1996. Apoptosis and APC in colorectal tumorigenesis. Proc. Natl. Acad. Sci. USA 937950-7954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nateri, A. S., B. Spencer-Dene, and A. Behrens. 2005. Interaction of phosphorylated c-Jun with TCF4 regulates intestinal cancer development. Nature 437281-285. [DOI] [PubMed] [Google Scholar]
- 31.Otto, S. J., S. R. McCorkle, J. Hover, C. Conaco, J. J. Han, S. Impey, G. S. Yochum, J. J. Dunn, R. H. Goodman, and G. Mandel. 2007. A new binding motif for the transcriptional repressor REST uncovers large gene networks devoted to neuronal functions. J. Neurosci. 276729-6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parker, D. S., Y. Y. Ni, J. L. Chang, J. Li, and K. M. Cadigan. 2008. Wingless signaling induces widespread chromatin remodeling of target loci. Mol. Cell. Biol. 281815-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polakis, P. 2000. Wnt signaling and cancer. Genes Dev. 141837-1851. [PubMed] [Google Scholar]
- 34.Ptashne, M. 1988. How eukaryotic transcriptional activators work. Nature 335683-689. [DOI] [PubMed] [Google Scholar]
- 35.Reinberg, D., and R. G. Roeder. 1987. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and functional analysis of initiation factors IIB and IIE. J. Biol. Chem. 2623310-3321. [PubMed] [Google Scholar]
- 36.Saha, S., A. B. Sparks, C. Rago, V. Akmaev, C. J. Wang, B. Vogelstein, K. W. Kinzler, and V. E. Velculescu. 2002. Using the transcriptome to annotate the genome. Nat. Biotechnol. 20508-512. [DOI] [PubMed] [Google Scholar]
- 37.Sancho, E., E. Batlle, and H. Clevers. 2004. Signaling pathways in intestinal development and cancer. Annu. Rev. Cell Dev. Biol. 20695-723. [DOI] [PubMed] [Google Scholar]
- 38.Sansom, O. J., V. S. Meniel, V. Muncan, T. J. Phesse, J. A. Wilkins, K. R. Reed, J. K. Vass, D. Athineos, H. Clevers, and A. R. Clarke. 2007. Myc deletion rescues Apc deficiency in the small intestine. Nature 446676-679. [DOI] [PubMed] [Google Scholar]
- 39.Sansom, O. J., K. R. Reed, A. J. Hayes, H. Ireland, H. Brinkmann, I. P. Newton, E. Batlle, P. Simon-Assmann, H. Clevers, I. S. Nathke, A. R. Clarke, and D. J. Winton. 2004. Loss of Apc in vivo immediately perturbs Wnt signaling, differentiation, and migration. Genes Dev. 181385-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawadogo, M., and R. G. Roeder. 1985. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc. Natl. Acad. Sci. USA 824394-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schiltz, R. L., C. A. Mizzen, A. Vassilev, R. G. Cook, C. D. Allis, and Y. Nakatani. 1999. Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem. 2741189-1192. [DOI] [PubMed] [Google Scholar]
- 42.Schorl, C., and J. M. Sedivy. 2003. Loss of protooncogene c-Myc function impedes G1 phase progression both before and after the restriction point. Mol. Biol. Cell 14823-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sierra, J., T. Yoshida, C. A. Joazeiro, and K. A. Jones. 2006. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 20586-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sparks, A. B., P. J. Morin, B. Vogelstein, and K. W. Kinzler. 1998. Mutational analysis of the APC/beta-catenin/Tcf pathway in colorectal cancer. Cancer Res. 581130-1134. [PubMed] [Google Scholar]
- 45.Takemaru, K.-I., and R. T. Moon. 2000. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression J. Cell Biol. 149249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Toualbi, K., M. C. Guller, J. L. Mauriz, C. Labalette, M. A. Buendia, A. Mauviel, and D. Bernuau. 2007. Physical and functional cooperation between AP-1 and beta-catenin for the regulation of TCF-dependent genes. Oncogene 263492-3502. [DOI] [PubMed] [Google Scholar]
- 47.Tran, H., F. Hamada, T. Schwarz-Romond, and M. Bienz. 2008. Trabid, a new positive regulator of Wnt-induced transcription with preference for binding and cleaving K63-linked ubiquitin chains. Genes Dev. 22528-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van Es, J. H., N. Barker, and H. Clevers. 2003. You Wnt some, you lose some: oncogenes in the Wnt signaling pathway. Curr. Opin. Genet. Dev. 1328-33. [DOI] [PubMed] [Google Scholar]
- 49.Walker, W., Z. Q. Zhou, S. Ota, A. Wynshaw-Boris, and P. J. Hurlin. 2005. Mnt-Max to Myc-Max complex switching regulates cell cycle entry. J. Cell Biol. 169405-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang, J. C., M. K. Derynck, D. F. Nonaka, D. B. Khodabakhsh, C. Haqq, and K. R. Yamamoto. 2004. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc. Natl. Acad. Sci. USA 10115603-15608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wohrle, S., B. Wallmen, and A. Hecht. 2007. Differential control of Wnt target genes involves epigenetic mechanisms and selective promoter occupancy by T-cell factors. Mol. Cell. Biol. 278164-8177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamada, M., J. Ohnishi, B. Ohkawara, S. Iemura, K. Satoh, J. Hyodo-Miura, K. Kawachi, T. Natsume, and H. Shibuya. 2006. NARF, a Nemo-like kinase (NLK)-associated ring finger protein regulates the ubiquitylation and degradation of T cell factor/lymphoid enhancer factor (TCF/LEF). J. Biol. Chem. 28120749-20760. [DOI] [PubMed] [Google Scholar]
- 53.Yochum, G. S., R. Cleland, S. McWeeney, and R. H. Goodman. 2007. An antisense transcript induced by Wnt/beta-catenin signaling decreases E2F4. J. Biol. Chem. 282871-878. [DOI] [PubMed] [Google Scholar]
- 54.Yochum, G. S., S. McWeeney, V. Rajaraman, R. Cleland, S. Peters, and R. H. Goodman. 2007. Serial analysis of chromatin occupancy identifies beta-catenin targets in colorectal carcinoma cells. Proc. Natl. Acad. Sci. USA 1043324-3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yochum, G. S., V. Rajaraman, R. Cleland, and S. McWeeney. 2007. Localization of TFIIB binding regions using serial analysis of chromatin occupancy. BMC Mol. Biol. 8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao, X. D., X. Han, J. L. Chew, J. Liu, K. P. Chiu, A. Choo, Y. L. Orlov, W. K. Sung, A. Shahab, V. A. Kuznetsov, G. Bourque, S. Oh, Y. Ruan, H. H. Ng, and C. L. Wei. 2007. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 1286-298. [DOI] [PubMed] [Google Scholar]