Abstract
To examine the role of chromatin in transcriptional regulation of the major histocompatibility complex (MHC) class I gene, we determined nucleosome occupancy and positioning, histone modifications, and H2A.Z occupancy across its regulatory region in murine tissues that have widely different expression levels. Surprisingly, nucleosome occupancy and positioning were indistinguishable between the spleen, kidney, and brain. In all three tissues, the 200 bp upstream of the transcription start site had low nucleosome occupancy. In contrast, nuclease hypersensitivity, histone modifications, and H2A.Z occupancy showed tissue-specific differences. Thus, tissue-specific differences in MHC class I transcription correlate with histone modifications and not nucleosomal organization. Further, activation of class I transcription by gamma interferon or its inhibition by α-amanitin did not alter nucleosome occupancy, positioning, nuclease hypersensitivity, histone modifications, or H2A.Z occupancy in any of the tissues examined. Thus, chromatin remodeling was not required to dynamically modulate transcriptional levels. These findings suggest that the MHC class I promoter remains poised and accessible to rapidly respond to infection and environmental cues.
Accurate gene expression is the result of diverse transcriptional responses to both tissue-specific “intrinsic” and dynamic “extrinsic” stimuli. In particular, widely expressed genes, including members of the major histocompatibility complex (MHC) class I family, are regulated by complex and overlapping developmental, tissue-specific, and inducible stimuli. Although MHC class I genes are ubiquitously expressed, distinct tissue-specific regulatory mechanisms lead to dramatically different levels of expression (55). For example, MHC class I levels in neural tissues and germ line cells are approximately 2 orders of magnitude lower than in lymphoid tissues. Further, numerous extracellular stimuli, in particular hormones and cytokines, are capable of dynamically modulating intrinsic tissue-specific MHC class I expression patterns (55). For example, gamma interferon (IFN-γ) increases class I transcription in nearly all tissues, whereas thyroid-stimulating hormone decreases it. The mechanisms that integrate these intrinsic and extrinsic regulatory signals are only partially understood.
MHC class I expression is primarily regulated at the transcriptional level, and many of the DNA sequence elements that mediate both tissue-specific and hormonal/cytokine regulation have been identified. Tissue-specific expression is achieved through the combined effects of a promoter-distal complex regulatory element and a series of promoter-proximal elements. The promoter-distal element, located between bp −700 and −800, consists of overlapping enhancer and silencer elements (63). Hormone/cytokine signaling is mediated by a series of promoter-proximal elements, located between bp −68 and −500. These elements include enhancer A (6, 17, 18, 26, 59), IFN-γ-stimulated response element (2, 16, 19, 20, 28, 62), and a composite RFX/cyclic AMP response element (CRE) (19, 47). A variety of DNA-binding transcription factors have been identified that interact with these promoter-proximal DNA sequence elements. For example, an enhanceosome consisting of the IFN-γ-inducible coactivator CIITA and DNA-bound transcription factors RFX, CREB/ATF, and NF-Y mediates constitutive cell surface class I expression in B lymphocytes (27, 38, 39, 56) and in increased levels in response to IFN-γ in other tissues. In addition, differences in the rates of transcription correlate with differences in polymerase II (Pol II) occupancy at the promoter (24). Despite the detailed understanding of the role of DNA sequence elements and transcription factors in establishing tissue-specific levels of MHC class I expression and hormone/cytokine-mediated responses, the role of chromatin structure in the regulation of this gene family has not been examined.
Chromatin structure regulates transcription at multiple levels: nucleosome positioning and occupancy, higher-order packaging, and histone modifications. In general, chromosomal packaging functions to reduce transcription such that active remodeling or changes in histone modifications are required to allow transcription to proceed. Nucleosomes, the basic units packaging DNA into chromatin, repress transcription if they occlude regulatory DNA target sites. The importance of nucleosome positioning is highlighted by the fact that gene activation or repression is often associated with discrete changes within regulatory regions (45). Chromatin remodeling enzymes can alter chromatin configuration by histone eviction or sliding and thus result in better access to transcription factors. In one of the best-characterized examples of this phenomenon, positioned nucleosomes at the yeast PHO5 gene promoter, are lost as a result of gene activation, while repression accompanies de novo chromatin assembly (see, for example, references 8 and 51). Nucleosome eviction associated with transcriptional activation has been demonstrated by global analyses in yeasts, flies, and humans where promoters of transcriptionally active genes are depleted of nucleosomes relative to inactive genes (5, 23, 31, 32, 41, 44). A variety of posttranslational modifications of the histone components of nucleosomes have been shown to correlate with positive or negative effects on transcription and hence are presumed to be important in regulating access of the transcription machinery to chromatin (reviewed in references 4, 30, and 34). These modifications serve to change the structure of chromatin by altering histone-DNA interactions, are binding sites for other proteins, and help in the recruitment of ATP-dependent chromatin remodeling enzymes. In addition, several histone variants have been found to affect transcription. One of these variants, H2A.Z, has been shown to flank nucleosome-depleted regions at promoters (reviewed in reference 22) and is often associated with the promoters of poised genes. It is thought to facilitate their rapid induction, presumably because it is more easily dissociated from nucleosomes than is H2A.
Most of the information that has contributed to our understanding of the role of chromatin in regulating gene expression has been obtained from studies of inducible genes. There have been few studies on chromatin in primary mammalian cells or on the regulation of constitutively expressed genes, such as MHC class I. In the present study, we have examined the role of chromatin in regulating tissue-specific and dynamic transcription of a ubiquitously expressed MHC class I gene.
Defining the role of chromatin organization in regulating MHC class I gene expression is hampered by the fact that the endogenous MHC class I gene family consists of a large number of closely related but functionally distinct genes in both humans and mice. To overcome this limitation, we have examined the chromatin structure of a heterologous swine MHC class I gene, PD1, in transgenic mice since it is sufficiently divergent in sequence from all murine class I genes to be readily distinguishable (12, 14). Importantly, the regulation of PD1 transgene expression parallels that of the endogenous classical MHC class I genes and its own native in situ pattern of expression (12). Furthermore, the PD1 transgene product functions as a classical transplantation antigen and mediates graft rejection (14). Therefore, this transgene provides an excellent model system in which to characterize the role of chromatin structure in regulating MHC class I gene expression.
We report here the surprising finding that nucleosome occupancy and positioning around the MHC class I core promoter are virtually indistinguishable among different tissues, irrespective of the expression level under basal, induced, or repressed conditions. Although nuclease hypersensitivity at the core promoter, histone modifications, and H2A.Z occupancy differ constitutively among tissues expressing different levels of class I, these patterns are largely unaltered by cytokine stimulation. The results of the present study show that in the constitutively active MHC class I gene, nucleosomal occupancy and positioning do not correlate with levels of expression, nor is chromatin remodeling necessary to achieve different rates of transcription. Although histone modifications correlate with constitutive transcription rates, they are not altered when transcription is dynamically modulated.
MATERIALS AND METHODS
Mice and tissues used.
C57BL/10 mice homozygous for the PD1 transgene (B10.PD1 mice) were used for all of the studies, which were carried out in accordance with the National Institutes of Health Animal Care and Use committee (protocol EIB-076). C57BL/6 or C57BL/10 mice were used as controls. Mice were euthanized by using CO2, and whole-body perfusion was carried out using phosphate-buffered saline so that tissues were free of contamination from circulating lymphocytes, which express high levels of MHC class I. Spleens, kidneys, brains, and livers were harvested; tissues from at least two to six mice were pooled for each experiment. Livers were frozen immediately on dry ice and stored at −80°C and used for genomic DNA extraction (58). Spleens and kidneys were harvested into separate tubes containing 25 ml of complete RPMI (10% fetal calf serum) or Dulbecco modified Eagle medium (10% fetal calf serum), while the brains were harvested into 25 ml of complete Hibernate (BrainBits, Springfield, IL) with B27 supplement (Gibco/Invitrogen, Grand Island, NY) or Neurobasal (Gibco/Invitrogen) medium (with B27 supplement).
IFN-γ treatment of B10.PD1 mice.
Mice were injected intraperitoneally with 50 kU of mouse IFN-γ (Calbiochem, Gibbstown, NJ) or an equal volume of saline. Tissues were harvested 24 h postinjection. Tissues from B10.PD1 obtained before and after treatment were analyzed by Northern blotting or real-time reverse transcription-PCR (RT-PCR) to verify IFN-γ stimulation of MHC class I expression.
Northern blotting and real-time RT-PCR.
RNA was prepared by using TRIzol (Invitrogen). Northern blotting was done as described before (9). The membrane was probed with the XbaI/BamHI fragment of the PD1 gene that extends from intron 3 to exon 7. The probe was labeled by using nick translation (GE Healthcare, Piscataway, NJ). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) probe (Ambion/Applied Biosystems, Austin, TX) was used as the internal control. For real-time RT-PCR, randomly primed cDNA (Stratascript; Stratagene/Agilent Technologies, Santa Clara, CA) was amplified by using Sybr green PCR master mix (Applied Biosystems, Foster City, CA) and PD1 primer sets (forward, 5′-GGA ATG TCA AGG AAA CCG CAC-3′; reverse, 5′-ATG CTC TGG AGG GTG TGA GAC C-3′). Primers for 18S RNA (AM1716, Ambion/Applied Biosystems) were used as an internal control.
Splenocyte culture and α-amanitin treatment.
Spleens from three to six mice were disaggregated in 1.5 to 3 ml of complete RPMI medium. Single cell suspensions of splenocytes were treated with 10 ml of ACK at room temperature for 1 min. The cells were then washed and resuspended in complete RPMI (medium alone) or in complete RPMI containing 50 μg of α-amanitin (Calbiochem)/ml to a final concentration of 10 million cells/ml. After incubation in a humidified CO2 incubator for 5 h at 37°C, the cells were washed three times with PBS to wash off the α-amanitin and then lysed by using NP-40 lysis buffer (11). The nuclei thus obtained were used for all further experiments as described below.
Nuclear run-on assay.
Thirty million cells were used to make nuclei that were then resuspended in 150 μl of nucleus storage buffer. A total of 50 μl of the thawed nuclei was used for a nuclear run-on assay carried out as described previously (3). The exposures from the nuclear run-on assays were obtained on a phosphorimager and quantitated by using TotalLab (Nonlinear USA, Inc., Durham, NC).
Isolation of nuclei from tissues.
Spleen, kidney, and brain tissues were minced in 10 ml of ice-cold nuclear buffer (15 mM HEPES [pH 7.5], 60 mM KCl, 15 mM NaCl, 2 mM EDTA, 0.5 mM EGTA, and 0.34 M sucrose, to which 0.15 mM 2-mercaptoethanol, 0.15 mM spermine, and 0.5 mM spermidine were added immediately prior to use) (65). The suspension was homogenized with 10 to 12 strokes in a chilled Dounce homogenizer, and the homogenate was filtered through a 70-μm-pore-size nylon cell strainer into a 50-ml conical tube on ice. The recovery and purity of the nuclei were determined microscopically after trypan blue staining of an aliquot.
Low-resolution MNase hypersensitivity assay.
This was done as previously described (11) with minor variations. Briefly, nuclei from the spleens, kidneys, or brains of B10.PD1 mice were washed with ice-cold micrococcal nuclease (MNase) digestion buffer (10 mM Tris-HCl [pH 7.4], 15 mM NaCl, and 60 mM KCl, to which 0.2 mM phenylmethylsulfonyl fluoride, 0.15 mM spermine, and 0.5 mM spermidine were added immediately prior to use) and then suspended in MNase digestion buffer containing 1 mM CaCl2 to obtain ∼10 million nuclei per 100-μl reaction. Each 100-μl aliquot of nuclear suspension was digested with 0, 1, 2, or 3 U of MNase (Worthington Biochemical Corp., Lakewood, NJ) at room temperature for 5 min. The reaction was stopped by adding 240 μl of MNase digestion buffer, 60 μl of MNase stop buffer (100 mM EDTA, 10 mM EGTA [pH 7.5]), 6 μl of 25 mg of proteinase K/ml, and 40 μl of 10% sodium dodecyl sulfate. Samples were incubated overnight at 37°C and extracted by using multiple rounds of phenol and isoamyl alcohol-chloroform. The final aqueous phase was subjected to RNase A digestion (100 μg/ml) at 37°C for 2 h, reextracted, and precipitated. The resulting DNA pellets were resuspended in 10 mM Tris-HCl (pH 7.5)-1 mM EDTA. In control MNase digestions of genomic DNA, 100 μg of genomic DNA was digested in a 300-μl reaction volume with 0, 0.1, 0.2, or 0.3 U of MNase (65).
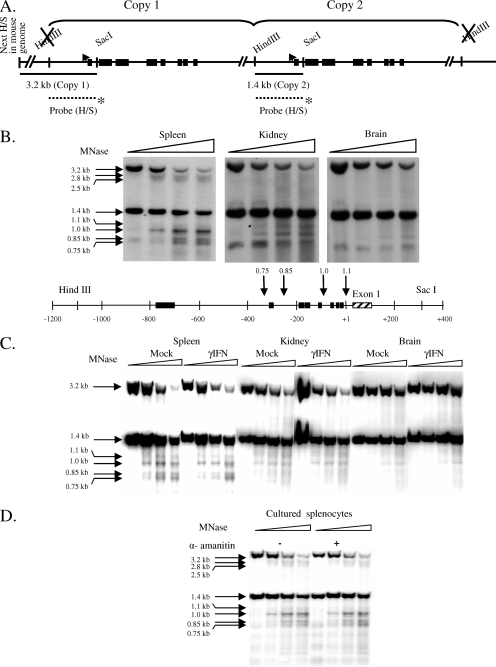
The MNase hypersensitive sites were assessed by Southern blotting using a probe corresponding to the 1.4-kb region between the HindIII and SacI sites (shown in Fig. 5A). This probe hybridizes to and reveals both copies of the gene, as shown in the figure. Then, 20 μg of DNA purified from MNase-treated nuclei and MNase-treated genomic DNA samples from B10.PD1 mice was digested to completion using 120 U each of HindIII and SacI. Digested samples were electrophoresed on 1% agarose gels in 1× Tris-borate-EDTA, transferred onto nitrocellulose membranes, and cross-linked using a UV cross-linker. The membranes were hybridized with the indicated probe labeled by nick translation and washed using high-stringency conditions. The results shown are representative of at least two independent experiments.
FIG. 5.
MNase hypersensitivity correlates with expression level in tissues but does not change with induction or depend on active transcription. (A) Map of the PD1 gene in B10.PD1 transgenic mice. The regions examined for nuclease-hypersensitive regions are indicated by horizontal lines. The labeled HindIII/SacI probe is represented by a dotted line with an asterisk at the bottom of the figure. Its location shows where it hybridizes to the PD1 gene. (B to D) Southern blots with MNase-treated and HindIII/SacI-digested samples from untreated (B) and mock- and IFN-γ-treated spleen, kidney, and brain (C) or from splenocytes cultured in medium alone or medium with α-amanitin (D) probed with a fragment corresponding to the 1.4-kb PD1 5′ region are presented. Fragment sizes corresponding to the band are marked on the left. At the bottom of panel B, arrows indicate the positions of MNase-hypersensitive sites at the 5′ end of the PD1 gene, and the numbers on top of the arrows represent sizes of the bands (in kilobases) generated as a result of MNase hypersensitivity. Note that the MNase concentration was standardized to optimize the cleavage of the 1.4-kb fragment and the generation of its derivative 1.0-kb hypersensitive band. These conditions of MNase result in greater digestion of the 3.2-kb fragment (upstream copy of the gene). Due to the decreased MNase hypersensitivity in the kidney and brain, the weaker hypersensitive bands derived from the 3.2-kb fragment are not always visible.
High-resolution nucleosome positioning by LMPCR.
Nuclei from the spleens, kidneys, and brains of B10.PD1 mice (mock treated or IFN-γ treated) and nuclei from splenocytes cultured in the presence or absence of α-amanitin were extensively digested with 100 U of MNase per 100-μl reaction at room temperature for 5, 10, or 15 min. DNA was purified as described before except that RNase digestion and reextraction were done twice to ensure that any residual RNA did not pose problems for subsequent steps. The purified DNA from the time point showing almost complete digestion of chromatin into mononucleosome-sized fragments was used for all further manipulations. Ligation-mediated PCR (LMPCR) was done as described previously (12) with the following modifications. Approximately 15 μg of purified mononucleosome-sized DNA was phosphorylated using 10 U of T4 DNA polynucleotide kinase (New England Biolabs, Ipswich, MA) in a 50-μl reaction volume with 70 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 5 mM dithiothreitol, 1 mM ATP, and 2 mM spermidine. The reactions were incubated at 37°C for 1 h, heat inactivated at 65°C for 20 min, precipitated, and resuspended in 50 μl of water. The DNA was ligated to a 20-fold excess of a unidirectional linker at 15°C overnight. The reaction was stopped by incubation at 70°C for 10 min, after which the entire reaction was loaded (in 5% glycerol) on a 6% polyacrylamide gel. The samples were electrophoresed in 1× Tris-borate-EDTA, along with a pBR322 MspI digest marker. Bands corresponding to mononucleosome-sized fragments ligated to linker were excised from the gel, crushed in 200 μl of 10:0.1 Tris-EDTA, and vortexed. After overnight incubation at 4°C in a shaker-incubator, the samples were vortex mixed and spun down, and the supernatant was collected. DNA was quantitated by running it on a 2% agarose gel. A total of 100 ng of this DNA was used in LMPCRs.
As controls, genomic DNA from B10.PD1 and control mice were digested with MNase, phosphorylated, and ligated to linker and then used in the LMPCRs. The linker primers used were 25-mer (5′-GCG GTG ACC CGG GAG ATC TGA ATT C-3′) and 11-mer (5′-GAA TTC AGA TC-3′). The primer sets used for the LMPCRs were as follows: set I (maps the downstream edge of nucleosomes roughly between positions −110 and −25), 5′-CCC GTG TCC CCA GTT TCA CTT CTC CG-3′ and 5′-GCA ACC TGT GTG GGA CCG TCC TGC CC-3′; set II (maps the downstream edge of nucleosomes roughly between positions −230 and −80), 5′-GAG TCC CAG CTC CGC AGC C-3′ and 5′-CTC CGC AGC CAG GCG TGG CTC-3′; set III (maps the upstream edge of nucleosomes roughly between positions −300 and −200), 5′-GTT GCG AGA CGG AGA AGT GAA ACT G-3′ and 5′-CGG AGA AGT GAA ACT GGG GAC ACG G-3′; set IV (maps the upstream edge of nucleosomes roughly between positions −175 and −105), 5′-GAA ACC GGA CAC GCA ATA GGA G-3′ and 5′-GGA CAC GCA ATA GGA GAG AGA AG-3′; and set V (maps the upstream edge of nucleosomes roughly between positions +45 and +125), 5′-GGT GAG GTG GCG GGT CTG-3′ and 5′-CAG CAG AGT CGC ACC TTC CCC-3′.
The results shown are representative of at least two independent experiments. Densitometric analysis was done by using TotalLab.
Nucleosome quantitation.
The samples purified for LMPCRs were also used for nucleosome quantitation. Each sample (2.5 ng) was amplified by real-time PCR using the Sybr green PCR master mix and primer sets across the upstream regulatory region and further down the gene in exon 5. The primer sets used are given below, and their positions are illustrated in the bottom panel of Fig. 2. From 5′ to 3′ across the gene, the sets were as follows: set 1, 5′-TAC ATA TGA AAC ACT CCT GCT ACC TTC C-3′ and 5′-CCA GTA AAG GTT GTA TTC CAT GA-3′; set 2, 5′-AAT CAT TCT GTG AGC TGC ACT AGC C-3′ and 5′-GTA AGA GTT TTA AGA CCG AAT ACA TTG-3′; set 3, 5′-ACT GAT TCA GGT CCA CAT TCA-3′ and 5′-GAG TCC TTT TGG TGG CTG ACA TC-3′; set 4, 5′-TGT GCG GGG CTT TTA CAT TTC-3′ and 5′-CAC TGG AGG TTT ATG TCT GCT TCT G-3; set 5, 5′-CTC ACT AAA AGG TTT GGA AAT CGC-3′ and 5′-CCC TGC TGC TCT TCA GAA AGC-3′; set 6, 5′-TCA GGG TCT CAG GCT CCA-3′ and 5′-GGA CAC GCA ATA GGA GAG AGA AG-3′; set 7, 5′-CCC GTG TCC CCA GTT TCA CTT CTC CG-3′ and 5′-GGG AAC CGC AGT GGC-3′; set 8, 5′-CGC AAC CTG TGT GGG AC-3′ and 5′-GGG TGG GTG GAG AGT TT-3′; set 9, 5′-CTT CTC TCT CCT ATT GCG TGT CC-3′ and 5′-TCA GCC TCG GAG TCT GG-3′; set 10, 5′-CCA GAC TCC GAG GCT GA-3′ and 5′-ATC CCG CAC TCA CCC G-3′; set 11, 5′-GAA CAA GGC CGC TGC G-3′ and 5′-CAG CAG AGT CGC ACC TTC-3′; set 12, 5′-GAA GGT GCG ACT CTG CTG-3′ and 5′-TGG AGG TCG GAG ACG G-3′; and set 13, 5′-CAG ACC CTG CTC AGC CC-3′ and 5′-GCG TCT TCC TCC AGA T-3′.
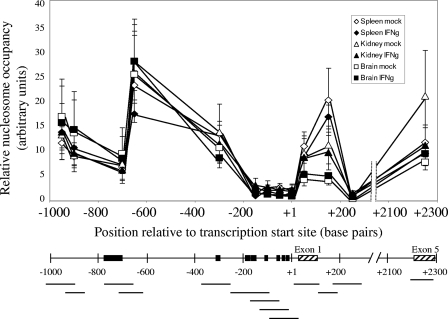
FIG. 2.
Nucleosome occupancy across the MHC class I regulatory region does not correlate with either tissue-specific or induced levels of expression, except immediately downstream of transcription initiation. The top panel panel shows nucleosome occupancy in mock-treated spleen (⋄), IFN-γ-treated spleen (♦), mock-treated kidney (▵), IFN-γ-treated kidney (▴), mock-treated brain (□), and IFN-γ-treated brain (▪). The bottom panel presents the MHC class I upstream regulatory region, showing amplified regions (horizontal lines), important regulatory elements (▪), and exons (▒). The results are an average of two independent experiments. Error bars represent the standard errors of the mean. The lines drawn connecting data points do not necessarily imply levels between points.
Although the efficiencies of the different primer sets were not identical, the following calculations internally corrected for any differences. The CT value obtained with each primer set was plotted on a standard curve derived from amplifying genomic DNA from B10.PD1 mice. Amplification from one sample was arbitrarily assigned a value of 1, and the values of all of the other samples were adjusted accordingly. The results shown are from at least two independent experiments.
Native ChIP.
Native chromatin immunoprecipitation (ChIP) assays were performed as described previously (43), with the following modifications. All solutions used for this protocol included 5 mM sodium butyrate. Tissues pooled from two to four mice were Dounce homogenized in 10 ml of nuclear buffer A (recipe described above; with 0.34 M sucrose and 5 mM soidum butyrate) and strained through a 70-μm-pore-size cell strainer. Then, 4.5 ml of nuclear suspension in nuclear buffer A (with 0.34 M sucrose) was layered onto 2.5 ml of nuclear buffer A (with 1.43 M or 50% sucrose), followed by centrifugation at 700 × g at 4°C for 20 min. Mononucleosomes were prepared as described above. Prior to immunoprecipitation, the suspension was precleared with 75 μl of protein A-Sepharose slurry per ml of chromatin. The antibodies used for the native ChIP were anti-acetyl H3K9/K14 (catalog no. 06-599; Upstate/Millipore, Billerica, MA), anti-trimethyl H3K4 (catalog no. 07-473; Upstate), anti-trimethyl H3K9 (catalog no. 07-442; Upstate), anti-H2A.Z (ab4174; Abcam, Cambridge, MA), and nonspecific rabbit immunoglobulin G (Jackson Immunoresearch Laboratories, Westgrove, PA). For 50 μg of chromatin, 12.5 μg of each antibody was used. In the case of H3K4 trimethyl antibody, which is an antiserum, 12.5 μl was used.
The immunoprecipitated material was quantitated by real-time PCR using the primer sets shown in the bottom of panels of Fig. 6. Primer sets 4, 5, 8, 11, and 13 described in the nucleosome quantitation section above were used. An additional primer set in the 3′ region was also used: set 14, 5′-TCT GTG TTC CTA TGA GCA TC-3′ and 5′-GAA CAC AGG TCA GGG TGA G-3′.
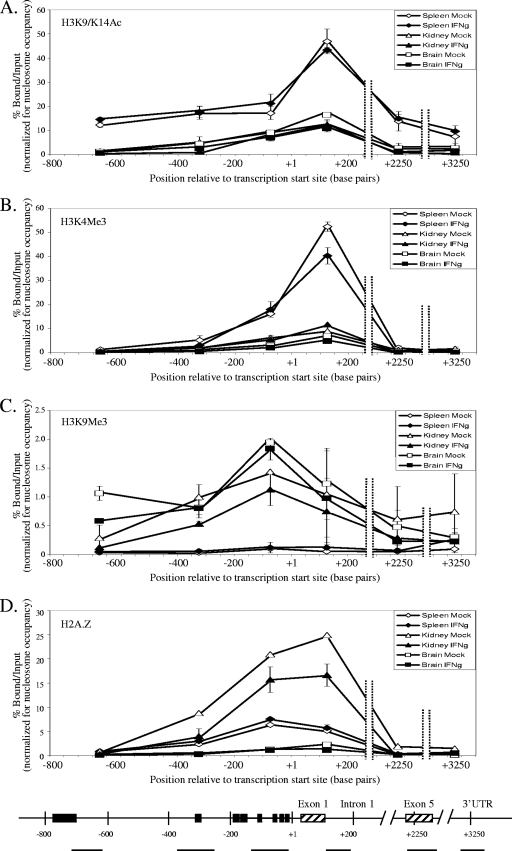
FIG. 6.
Histone modifications positively associated with transcription (H3K9/K14 acetylation [A] and H3K4 trimethylation [B]) are higher at the PD1 gene in highly expressing spleen cells than in lower expressing kidney or brain cells. On the other hand, kidney and brain have higher levels of a histone modification negatively associated with transcription (H3K9 trimethylation [C]). In contrast, H2A.Z occupancy (D) showed a unique tissue-specific pattern, where the kidney had the highest levels of H2A.Z, followed by the spleen, whereas the brain had very low levels of H2A.Z. Panels A to D show the percentage of bound versus input in mock-treated spleen (⋄), IFN-γ-treated spleen (⧫), mock-treated kidney (▵), IFN-γ-treated kidney (▴), mock-treated brain (□), and IFN-γ-treated brain (▪). The bottom panel shows the MHC class I gene with amplified regions (horizontal lines), important regulatory elements (▪), and exons (▒). The results are an average of two independent experiments. Error bars represent the standard errors of mean. Input is the entire mononucleosomal fraction before immunoprecipitation, and hence the results obtained are normalized to nucleosome occupancy. The lines drawn connecting data points do not necessarily imply levels between points.
The results were plotted as the “percent bound versus input.” Each graph was compiled from at least two independent experiments done with mock-treated (these were combined with untreated since the values obtained were identical) and IFN-γ-treated mice. The bars represent the average value, while the error bars represent the standard error of mean.
RESULTS
The patterns of nucleosome occupancy across a class I gene are indistinguishable among tissues with various levels of expression.
The levels of MHC class I transgene expression in vivo differ by almost 2 orders of magnitude among tissues (Fig. 1A) (55). Therefore, we sought to determine whether the chromatin structure contributes to its regulation by comparing the nucleosomal organization of the PD1 transgene in three tissues expressing widely different levels of class I. The extent of nucleosome occupancy across the transgene in the spleen (high expresser), kidney (intermediate expresser), and brain (very low expresser) was quantitated by comparing the abundance of the transgene in total mononucleosomal DNA relative to genomic DNA by real-time PCR. This measurement provides an assessment of the extent of DNA protection via packaging into nucleosomes. Using a series of PCR primers spanning the upstream regulatory region and coding sequences (Fig. 2, bottom panel), we observed that the overall pattern of nucleosomal occupancy was indistinguishable among the three tissues (Fig. 2). In all three tissues, the nucleosomal occupancy was reduced around the core promoter, between approximately bp −200 upstream and the transcription start site (TSS) at position +1, as assessed by four overlapping primer sets within the 200-bp segment (Fig. 2). However, it is important to note that the region was not completely devoid of nucleosomes. The paucity of nucleosomes around the core promoter, unrelated to the level of class I transcription, is consistent with the interpretation that the region is relatively open and available for the assembly of transcription machinery, independent of the frequency of transcription.
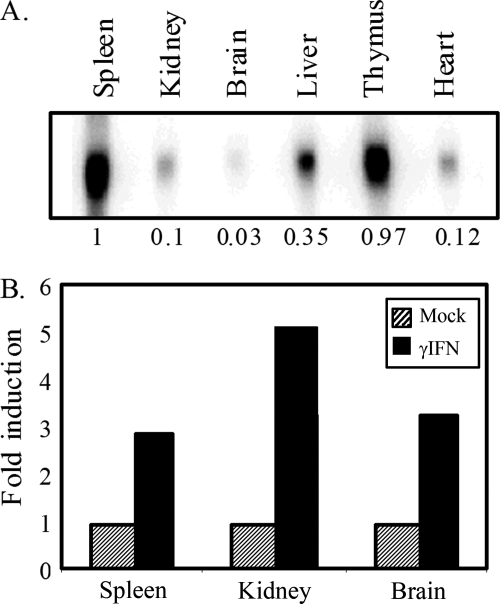
FIG. 1.
PD1 expression varies widely in different tissues and can be induced by IFN-γ in the spleens, kidneys, and brains of transgenic mice. (A) Northern blot of RNA from tissues from B10.PD1 transgenic mice. The numbers at the bottom represent relative amounts. (B) Induction of PD1 transcript levels in the spleen, kidney, and brain by in vivo treatment with IFN-γ as assessed by Northern blotting. GAPDH RNA levels served as the internal control.
We considered the possibility that the nucleosome-depleted region upstream of the TSS was a result of remodeled nucleosomes being lost during MNase treatment. However, earlier genome-wide reports found no differences in nucleosome occupancy between cross-linked and un-cross-linked chromatin (44, 52). Therefore, we conclude that the region upstream of the TSS is relatively depleted of nucleosomes.
In two other regions of the gene, nucleosomal occupancy was also consistently much lower than elsewhere in the gene in all three tissues. Nucleosomal occupancy was decreased in the upstream regulatory region between bp −690 and −770 that encompasses a complex regulatory element involved in establishing tissue-specific levels of transcription (63). Another region of reduced nucleosomal occupancy occurs within intron 1, where a potential regulatory element resides (H. Cohen et al., unpublished data).
Although the overall pattern of nucleosome occupancy was the same in all three tissues, a modest relationship was observed between nucleosomal occupancy and tissue-specific expression levels immediately downstream of the TSS at exon 1/intron 1. Surprisingly, high-expressing spleen cells had a higher occupancy of nucleosomes compared to the lower-expressing brain.
The nucleosomal patterns across the class I gene in the tissues reflect the “steady-state” chromatin organization, in the absence of either hormonal or cytokine stimuli. Therefore, we next considered the possibility that induction of class I expression by the inflammatory cytokine IFN-γ might trigger chromatin remodeling and changes in nucleosomal occupancy. To test this possibility, class I expression was induced in transgenic mice by IFN-γ treatment. Under these conditions, class I RNA levels were increased in all three tissues, by two- to fivefold (Fig. 1B). Surprisingly, neither the overall pattern nor the relative level of nucleosomal occupancy across the gene was affected by in vivo induction of class I expression with IFN-γ (Fig. 2). Thus, the pattern of nucleosome occupancy is independent of either tissue-specific or cytokine-modulated levels of expression. Furthermore, these results indicate that activation of class I transcription does not require extensive chromatin remodeling. In support of this latter conclusion, we observed activation of MHC class I transcription by IFN-γ in SW13 cells that are devoid of the remodeling factor, BRG1 (J. Weissman, Z. Sercan, and D. Singer, unpublished observations).
Nucleosomes occupy multiple, discrete positions around the MHC class I TSS.
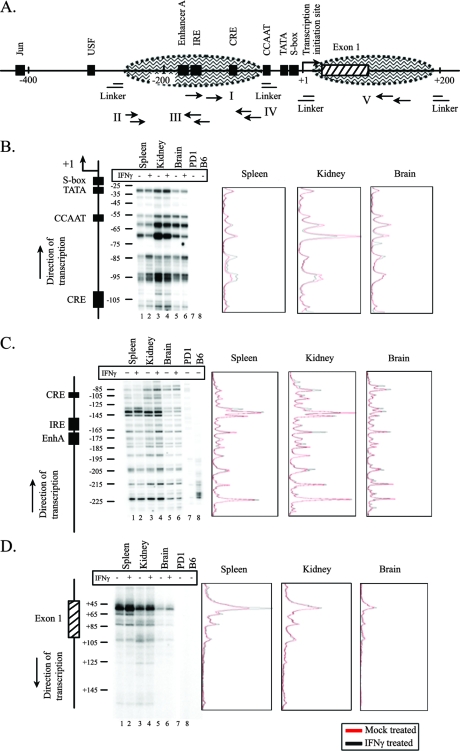
The lack of correlation between the level of expression in tissues and nucleosomal occupancy across the extended regulatory region of the MHC class I gene prompted us to examine more closely the organization of nucleosomes around the core promoter. Although this segment has a low nucleosomal occupancy relative to the surrounding regions, it is not completely devoid of nucleosomes. This raised the possibility that the precise positioning (i.e., differences in even a few base pairs) of nucleosomes at core promoter sequences correlated with class I expression levels. To this end, nucleosome positions within approximately 200 bp on either side of the TSS were mapped to single nucleotide resolution. Mononucleosomal DNA fragments isolated from the spleen, kidney, and brain nuclei of either IFN-γ-treated or mock-treated mice were probed by LMPCR with five different primer sets (Fig. 3A). Nucleosomes associated with core promoter sequences upstream of the TSS occupy seven to eight preferred positions on the underlying DNA sequence, as evidenced by the multiplicity of bands, where each band reflects a discrete nucleosome position (Fig. 3B). (The multiple positions mapped in the present study are unlikely to be a result of “trimming” of the nucleosome at the edges since the bands did not differ by 10 bp or a multiple thereof.) The same set of major nucleosomal positions was occupied in each of the three tissues (Fig. 3B, see lanes 1, 3, and 5). (It should be noted that we cannot exclude the possibility that on a subset of promoters, the nucleosomes are randomly arrayed, which would not be detected in this analysis.) Induction of class I gene transcription with IFN-γ did not elicit any reproducible differences in nucleosome positioning in the tissues (Fig. 3B and C, compare lanes 1 to 6). Analysis of other regions upstream of the TSS also showed no differences in nucleosomal positioning among the tissues in either the presence or absence of IFN-γ (see Fig. S1A and B in the supplemental material). Taken together, these results demonstrate that nucleosomes occupy multiple, discrete sites upstream of transcription initiation and that the pattern and relative occupancy of these sites do not vary significantly after in vivo induction with IFN-γ.
FIG. 3.
Nucleosomes occupy multiple positions at the 5′ end of the MHC class I gene in all tissues studied, and these did not change on induction of the gene with IFN-γ. (A) Locations of primer sets used for fine-mapping nucleosome positions at the MHC class I promoter using LMPCR. The MHC class I upstream region is shown with important regulatory elements (▪), exon 1 (▒), and hypothetical nucleosome positions (oval areas bordered by dashed lines). (B) Lanes 1, 3, and 5, LMPCR products from mock-treated spleen, kidney, and brain, respectively, using primer set I that maps the downstream edge of nucleosome(s) roughly from positions −110 to −25. Lanes 2, 4, and 6, LMPCR products from IFN-γ-treated spleen, kidney, and brain, respectively. Lanes 7 and 8, LMPCR products from MNase-digested genomic DNA from B10.PD1 and B6 mice, respectively. The pattern of bands obtained has been reproduced in four independent experiments. A map of the PD1 5′ end and the position with respect to TSS are shown on the left. The densitometric scans of the LMPCR results from mock-treated (red line) and IFN-γ-treated (black line) spleen, kidney, and brain are shown on the right. (C) Same as panel B, except using primer set II that maps the downstream edge of nucleosome(s) roughly from positions −230 to −80. (D) Same as panel B, except using primer set V that maps the upstream edge of nucleosome(s) roughly from positions +45 to +125. Note that because it was not possible to internally control for loading differences on the gels, no quantitative comparison between lanes could be made.
We next mapped nucleosome positioning downstream of the TSS in spleen, kidney, and brain tissues isolated from IFN-γ-treated or control mice. In contrast to nucleosomes located upstream of the TSS, nucleosomes located downstream of the core promoter predominantly occupied a single site, with a small number of other sites occupied infrequently (Fig. 3D). This is most evident in the spleen, where nearly all nucleosomes mapped to a single site at the beginning of exon 1. Nucleosomes in the kidney and brain displayed the same pattern of occupancy. However, the level of amplification at each position was lower in the kidney and even lower in the brain, a finding consistent with the relative pattern of nucleosomal occupancy at ca. +100 bp in the three tissues (Fig. 2).
Active transcription is not required to maintain either nucleosome occupancy or positioning.
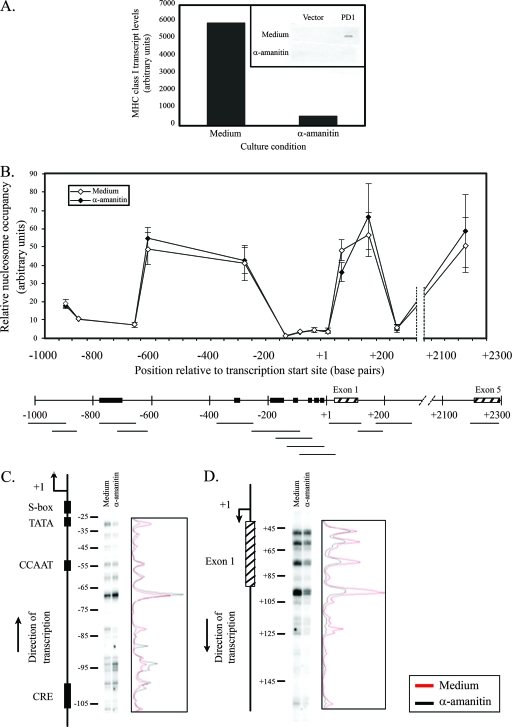
Although promoter activity differs enormously in the brain, kidney, and spleen, the class I gene is transcribed in all three. Therefore, we considered the possibility that the pattern of nucleosomal occupancy is a reflection of ongoing transcription and might be affected by complete abrogation of transcription. To address this possibility, we cultured freshly isolated splenocytes in the presence or absence of α-amanitin and then assessed nucleosome occupancy across the class I gene. Under conditions where transcription was completely inhibited by α-amanitin (Fig. 4A), there was no change in the pattern of nucleosomal occupancy across the gene relative to mock-treated controls (Fig. 4B). In particular, the nucleosome occupancy around the core promoter was indistinguishable between α-amanitin-treated and control cells. Thus, overall nucleosomal occupancy did not depend on ongoing transcription.
FIG. 4.
The pattern of nucleosome occupancy and positioning across the MHC class I gene does not depend on active transcription. (A) MHC class I transcription is abrogated in splenocytes cultured for 5 h in medium containing 50 μg of α-amanitin/ml. The inset shows MHC class I transcript levels obtained using a nuclear run-on assay. (B) The top panel shows nucleosome occupancy in splenocytes incubated in medium alone (⋄) or in medium with 50 μg of α-amanitin/ml (⧫). The bottom panel shows the MHC class I upstream regulatory region indicating amplified regions (horizontal lines), important regulatory elements (▪), and exons (▒). The results are an average of two independent experiments. Error bars represent the standard errors of mean. The lines drawn connecting data points do not necessarily imply levels between points. (C) Lanes 1 and 2, LMPCR products from splenocytes incubated in medium alone or in medium containing 50 μg of α-amanitin/ml, respectively, using primer set I that maps the downstream edge of nucleosome(s) roughly from positions −110 to −25. The pattern of bands obtained has been reproduced in two independent experiments. A map of the PD1 5′ end and the position with respect to TSS are shown on the left. Densitometric scans of LMPCR results are shown on the right (medium, red line; α-amanitin, black line). (D) Same as panel C, except using primer set V that maps the upstream edge of nucleosome(s) roughly from positions +45 to +125.
We next examined the possibility that active transcription determines precise nucleosome positioning. As shown in Fig. 4C and D, inhibition of transcription by α-amanitin did not alter the profile of nucleosomal positioning either upstream or downstream of the TSS (see also Fig. S2A to C in the supplemental material). These results indicate that maintenance of the pattern of nucleosome occupancy along the MHC class I promoter does not depend on the transcriptional activity of the gene.
Nuclease hypersensitivity in the promoter of the MHC class I transgene correlates with expression levels.
Since functional regulatory elements within chromatin are often characterized by hypersensitivity to nuclease digestion, we probed for nuclease-hypersensitive regions in the promoter of the transgene. Nuclei of cells from the spleens, kidneys, or brains of B10.PD1 transgenic mice were subjected to digestion with low concentrations of MNase, and hypersensitive regions were identified with a probe spanning the promoter. Previous characterization of the B10.PD1 mouse determined that it carries two copies of the PD1 transgene in a head-to-tail configuration. Digestion with HindIII/SacI releases two promoter fragments of 3.2 and 1.4 Kb. (See the map of the PD1 transgene and the scheme of the experiment in Fig. 5A.) Hypersensitive regions would appear as subfragments of greater electrophoretic mobility.
All three tissues displayed hypersensitive sites within the upstream regulatory region of the MHC class I gene. However, in contrast to their similar chromatin organization, the relative intensities of the hypersensitive sites differed sharply among the tissues and correlated with their levels of expression (Fig. 5B and see Fig. S3A in the supplemental material). In spleen cells, a prominent hypersensitive site mapped to a region located ca. 100 bp upstream of the TSS (1-kb band, Fig. 5B, top left panel). Other minor sites were also observed (see the map showing the location of the hypersensitive sites in Fig. 5B, bottom panel). The positions of the hypersensitive sites were confirmed by using a smaller probe (data not shown). Both of the tandemly arrayed copies of the transgene displayed the same pattern of hypersensitivity (Fig. 5B and see Fig. S3B in the supplemental material), providing an internal confirmation of the findings. The hypersensitive sites map within the region of low nucleosomal occupancy. The same hypersensitive sites were observed in nuclei from kidney and brain tissues. However, in contrast to the spleen, the hypersensitive sites in the kidney and brain were weaker (see quantitation in Fig. S3A in the supplemental material). Similar results were obtained when DNase was used instead of MNase (data not shown). Thus, there is a direct correlation between nuclease hypersensitivity of the class I core promoter and tissue-specific expression levels. This is a striking and surprising finding, given that the nucleosomal occupancies were indistinguishable among the brain, kidney, and spleen. It indicates that the generation of a nuclease-hypersensitive site is not directly related to nucleosomal occupancy.
The finding that hypersensitivity did not directly correlate with nucleosome occupancy suggested that it might be established by factors associated with transcription. To test this possibility, the effect on the hypersensitive sites of increasing class I transcription by in vivo treatment with IFN-γ was examined. Unexpectedly, induction of expression did not alter the levels of hypersensitivity in any of the tissues (Fig. 5C). Thus, the intensity of the hypersensitive site is not simply a reflection of the transcription rate. To pursue this finding, the effect of inhibiting transcription on MNase hypersensitivity was assessed. Inhibition of transcription by α-amanitin also did not result in any change in MNase hypersensitivity (Fig. 5D). Therefore, ongoing transcription is not necessary to maintain the hypersensitive site. Taken together, these results suggest that the presence of a nuclease-hypersensitive site is not directly correlated with the rate of transcription but may reflect the stable association of tissue-specific factors.
Differences in histone modifications and variants correlate with class I expression levels.
Distinct histone modifications and histone variants have been associated with actively transcribed and silent genes (reviewed in reference 34). To further characterize the chromatin structure of the three tissues expressing different levels of class I, we compared the extent of their histone modifications, specifically for marks associated with active (H3K9/K14 acetylation marks both actively transcribed genes and those poised for transcription; H3K4 trimethylation marks actively transcribed genes) and inactive (H3K9 trimethylation) chromatin by ChIP analysis of native mononucleosomes. Since the histone variant, H2A.Z, has been reported to be present at inactive promoters that are poised for expression (reviewed in reference 22), we also measured its association with the class I promoter. Regions of the gene probed by real-time PCR are shown schematically in Fig. 6, at the bottom of each panel. It is important to note that all of the results have been normalized for nucleosome occupancy.
Consistent with the high level of class I expression in the spleen, splenic nucleosomes were more highly acetylated throughout the class I gene than nucleosomes derived from the kidney and brain (Fig. 6A), peaking around intron 1. In the kidney and brain, acetylation levels were uniformly low throughout the gene, with a small increase around intron 1.
High levels of H3K4 trimethylation, another mark associated with actively transcribed genes, were also observed in spleen cells (Fig. 6B). Unlike the acetylation pattern, the H3K4 trimethylation was observed only at the 5′ end of the gene. This modification, like acetylation, peaked at intron 1. A similar pattern of H3K4 trimethylation, but at much lower levels, was observed in kidney and brain cells. Thus, increased H3K9/K14 acetylation and H3K4 trimethylation of nucleosomes immediately downstream of transcription initiation correlates with increased expression of the class I gene. However, neither of these marks was increased as a result of IFN-γ-mediated increases in gene expression (Fig. 6A and B).
H3K9 trimethylation, which is associated with inactive regions of the genome, was found only in kidney and brain cells and not in spleen cells (Fig. 6C). This modification was also unaffected by IFN-γ induction of the gene.
The presence of the histone variant, H2A.Z, has been proposed to contribute to maintaining promoters poised for expression. On the PD1 gene, H2A.Z occupancy was restricted to around 200 bp on either side of the TSS (Fig. 6D) in all three tissues. H2A.Z was not detected at any significant level elsewhere along the gene. The highest levels of H2A.Z on the class I promoter were found in kidney cells, followed by intermediate levels in spleen cells, and were almost nonexistent on brain cells. H2A.Z levels were unaffected by the induction of the gene with IFN-γ.
Thus, histone modifications are directly correlated with tissue-specific levels of expression, while H2A.Z occupancy of nucleosomes differs among the tissues. Surprisingly, activation of transcription by IFN-γ does not markedly alter these histone modifications or H2A.Z occupancy.
DISCUSSION
The MHC class I genes encode cell surface molecules that provide immune surveillance, triggering cellular immunity against intracellular pathogens. Consistent with this role, class I genes are ubiquitously expressed, although their expression is highly regulated, varying over 2 orders of magnitude among different tissues. Superimposed on the tissue-specific patterns of expression, class I gene transcription is induced by inflammatory cytokines, such as IFN, in all tissues. In the present study, we have examined the role of chromatin structure in regulating both the tissue-specific and the cytokine-mediated expression of the class I gene, PD1. We report that, in sharp contrast to the remodeling of chromatin structure that accompanies the regulation of many other inducible genes, the chromatin structure of the MHC class I gene is remarkably constant in vivo. Thus, the patterns of nucleosomal occupancy across the gene and nucleosomal positioning around the promoter are virtually indistinguishable in tissues expressing widely different levels of class I in both the presence and the absence of IFN-γ. Tissue-specific differences occur in histone modifications and H2A.Z occupancy, but these are unaltered in response to IFN-γ-mediated activation of transcription. Our findings indicate that chromatin organization of the class I gene is uniform in different tissues, maintaining a conformation poised for transcription, which is consistent with the need to rapidly induce transcription in response to intracellular pathogens.
Although numerous studies have addressed the role of chromatin structure in transcriptional regulation in inducible genes, most of those have been done in yeast or in cultured mammalian cells, which may behave very differently than in their in vivo environment. In contrast, we have examined the role played by chromatin in transcriptional regulation of constitutively active genes in vivo. We find that although the ubiquitously expressed MHC class I gene is transcribed at widely different levels in the spleen, kidney, and brain, the overall pattern of nucleosomal occupancy across the gene was indistinguishable in the three tissues. Three regions of relative nucleosomal depletion were observed, two of which correspond to known regulatory regions. One region is located between bp −700 and −800 and encompasses a complex tissue-specific regulatory element. The second region spans approximately 200 bp upstream of the TSS and encompasses many of the important sequence elements associated with the transcriptional regulation of the gene. The third region that is relatively depleted of nucleosomes occurs around intron 1, suggesting that it also may be the site of a regulatory element. These results extend to tissues the results of large-scale studies on nucleosome occupancy from yeast (5, 31, 32, 64) and human (44) cell lines that have shown that promoters and TSSs of actively transcribed genes are demarcated by nucleosome-depleted regions. We speculate that the regions of relative nucleosomal depletion are mediated either by binding of ubiquitous transcription factors or by a promoter DNA structure refractory to nucleosome formation. Several studies have revealed that DNA structural parameters contribute to nucleosome occupancy and positioning in vivo (5, 25, 32, 36, 53, 54). Indeed, computational analysis (33) of the class I promoter region predicts very low nucleosome-forming potential (data not shown), suggesting that accessibility to the MHC class I promoter may be established by DNA sequences that are intrinsically poor at positioning nucleosomes.
Several studies have suggested that nucleosomal occupancy is inversely proportional to transcriptional rates (5, 8, 13, 31, 32, 35, 46, 48, 49). However, this relationship does not hold for the MHC class I gene. Nucleosomal occupancy upstream of the TSS does not correlate with transcription rate, since no differences were observed among the three tissues that vary dramatically in expression. The only exception occurs immediately downstream of the TSS, within intron 1, where nucleosome occupancy does correlate with promoter activity. However, it is a direct correlation, not an inverse one: nucleosomal occupancy was higher in the spleen than in the kidney or brain. Similar results have been reported in yeast (32) and human (52) cells. The latter report includes an analysis of human MHC class I genes that is consistent with our results. We speculate that the higher occupancy downstream of the TSS in spleen cells may be established by the higher occupancy of Pol II at the class I promoter (24; Weissman et al., unpublished). It has been previously suggested that higher transcriptional rates result in increased occupancy of Pol II and recruitment of HDAC Rpd3S complex, leading to greater nucleosomal occupancy (32). However, neither transcription activation by IFN-γ treatment nor transcription aborted by α-amanitin alters the pattern of nucleosomal occupancy of the class I gene in any of the tissues. Taken together, we conclude that neither the overall nucleosomal organization across the MHC class I gene nor the tissue-specific occupancy at the promoter, once established, is actively altered in response to the dynamic modulation of transcription rates.
The only direct correlation between chromatin structure and variations in transcription rate was at a nuclease-hypersensitive site around the core promoter. The site is strong in the spleen and successively weaker in the kidney and brain. Since nucleosome occupancy at the core promoter is indistinguishable among the three tissues, these findings indicate that the hypersensitive site does not reflect the extent of nucleosome occupancy. Rather, we speculate that the extent of hypersensitivity may reflect the relative abundance of transcription factors, such as Pol II, on actively transcribed promoters. In support of this interpretation, we have observed much higher levels of Pol II at the promoter in spleen cells versus kidney or brain cells (24; Weissman et al., unpublished). The low occupancy of nucleosomes around the core promoter may keep it accessible and poised for transcription; binding of specific transcription factors establish the hypersensitive site. Significantly, the tissue-specific nuclease hypersensitivity did not change on induction of the gene (the present study; L. Satz and D. Singer, unpublished data). Thus, the class I gene differs markedly from other genes, such as the human interleukin-12 (p35) gene where induction by lipopolysaccharide and IFN-γ results in increased nuclease hypersensitivity (21). Furthermore, abrogation of class I transcription did not result in a loss of the hypersensitive site, indicating that a hypersensitive site once formed does not change with transcription status. This is in contrast to what has been observed for hepatic genes, where hypersensitivity to nucleases disappears on α-amanitin treatment (29). It is possible that these differences are due to differences in Pol II occupancy. Whereas Pol II is lost from hepatic genes after α-amanitin treatment (29), we did not observe loss of Pol II from the class I promoter after α-amanitin treatment (Weissman et al., unpublished).
Nucleosomes occupied multiple discrete positions with preferences for certain positions. Although we could not rule out the possibility that relative occupancy at any given position may vary between different tissues, occupancy did not correlate with differences in expression. Interestingly, these preferred positions remained the same across tissues in the presence or absence of IFN-γ or when transcription was abrogated by α-amanitin. These results suggest that various transcriptional rates did not change positioning. Since nucleosomes can occupy seven to eight discrete positions around the class I promoter region, it is unlikely that the precise nucleosome position influences class I expression. This is in contrast to the yeast PHO5 gene, where the precise positioning of a single nucleosome affects expression (37); even a 2- to 3-bp shift in nucleosome positioning over the TATA box abrogated expression. Multiple nucleosome positions, such as those observed here, have been reported at the glucocorticoid responsive unit of the tyrosine aminotransferase (Tat) gene in rat hepatoma cells (13). However, in contrast to the class I gene, the nucleosomes at the Tat gene were remodeled and lost on activation.
As opposed to nucleosome occupancy and positioning, histone modifications and H2A.Z occupancy displayed a clear correlation with the level of class I expression in tissues. The histone marks associated with active transcription, H3K9/14 acetylation and H3K4 trimethylation, were much higher in the spleen relative to the kidney or brain. Acetylation of H3K9/K14 peaked downstream of the promoter, around intron 1, and was observed even in brain. These results are consistent with earlier observations in yeast that H3 acetylation levels peak in regions downstream of the TSS (46, 50) and that many inducible genes are marked by histone acetylation even when they are not active (50, 61). Surprisingly, although these modifications correlated with tissue-specific levels of transcription, neither H3K9/14 acetylation nor H3K4 trimethylation changed after activation of transcription by IFN-γ. This finding suggests that these marks may be established during development when tissue-specific transcription levels are established; once established, these marks remain unchanged. Further support for this conclusion derives from a very recent genome-wide analysis that revealed few changes in chromatin modifications following T-cell activation (A. Barski and K. Zhao, unpublished observations).
A histone mark negatively associated with transcription, H3K9 trimethylation, was detected across the extended promoter and through intron 1 of the class I gene in both kidney and brain cells but was barely detectable in spleen cells. Although H3K9 trimethylation has primarily been observed in heterochromatin (34), recent studies indicate that it is also associated with the transcribed region of expressed genes (57, 60). Thus, these marks may serve to ensure low levels of transcription, while keeping the gene poised for expression.
H2A.Z has been shown to occur immediately upstream and downstream of the TSS and the nucleosome-depleted region at promoters (1; reviewed in reference 22). Here we find that more H2A.Z is associated with the MHC class I promoter in kidney cells than in spleen cells, although the total amount of H2A.Z protein is equivalent in both cell types (A. S. Kotekar, unpublished observations). This argues for a tissue-specific role for H2A.Z in transcriptional regulation. It has been suggested that H2A.Z is preferentially associated with inactive genes to enable their rapid induction (reviewed in reference 22). These findings are consistent with the interpretation that although the promoter is only weakly active in kidney, it is maintained in a poised configuration for a rapid response to inductive stimuli. Accordingly, the promoter in the spleen, which is constitutively active, presumably has a high turnover of H2A.Z and hence appears to have relatively lower levels of H2A.Z. Its absence in brain cells may be related to the distinct mechanism of transcriptional control known to exist in these cells (42). Thus, in contrast to observations using whole-genome scanning in Drosophila (40), H2A.Z occupancy did not directly correlate with nucleosome organization at the MHC class I promoter.
Overall, our results indicate that chromatin structure does not actively regulate class I transcription but rather functions to keep the core promoter poised and accessible for transcription, to allow rapid activation of the gene without changes in nucleosome occupancy, positioning, or histone modifications. Supporting this model, MHC class I transcription can be induced by IFN-γ in the absence of BRG1, the ATPase subunit of the chromatin remodeling complex SWI/SNF (Weissman et al., unpublished). It is the tissue-specific differences in histone modifications and H2A.Z occupancy that may play a role in recruiting different proteins/complexes to effect proper tissue-specific expression of the gene in different tissues. The present study of a constitutively active gene suggests that genes that need to be “on” at all times have evolved mechanisms (e.g., constitutive binding of transcription factors, a DNA structure refractive to nucleosome assembly) that ensure promoter access. These conclusions are supported by reports that housekeeping and/or widely expressed genes have lower nucleosome-forming potential at their promoters than do tissue-specific genes (15, 25, 33).
A recent analysis of the yeast Pho5 promoter has found that transcriptional activation is accompanied by assembly and disassembly of nucleosomes at the promoter, possibly by a sliding-mediated nucleosome disassembly process (7, 10). Similar nucleosomal cycling may be responsible for the nucleosomal depletion and multiplicity of positions that occur upstream of the TSS of the class I gene. Such cycling would be independent of active transcription, induction, or tissue-specific transcription, and this process may be a prerequisite for transcription “readiness.”
The surprising finding in the present study is that chromatin organization does not directly correlate with transcription rates in vivo. Rather, in the case of the MHC class I gene, nucleosomal organization is constant among different tissues with widely different rates of transcription. Although histone modifications largely correlate with the homeostatic tissue specific levels of transcription, they are not altered by cytokine-mediated changes in transcription. Unlike other genes that have been examined, regulation of MHC class I gene expression is largely independent of changes in chromatin organization. The differences that we have described between the ubiquitously expressed MHC class I gene and many other genes highlight the complexity and diversity in chromatin organization that are needed to achieve proper transcriptional regulation.
Supplementary Material
Acknowledgments
We thank Artem Barski (NHLBI), John Brady (NCI/LCO), Arkady Celeste (NCI/EIB), Michael Kruhlak (NCI/EIB), Dave Levens (NCI/LP), and past and present members of our lab for helpful discussions. We thank Ranjan Sen (NIA) and Keji Zhao (NHLBI) for helpful discussions and critical review of the manuscript. We acknowledge the help of Terry Guinter (NCI/EIB), Karen Hathcock (NCI/EIB), and Steve Jay (NCI/LASP) with splenocyte cultures and animal procedures.
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Published ahead of print on 22 September 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Albert, I., T. N. Mavrich, L. P. Tomsho, J. Qi, S. J. Zanton, S. C. Schuster, and B. F. Pugh. 2007. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446572-576. [DOI] [PubMed] [Google Scholar]
- 2.Baldwin, A. S., Jr., and P. A. Sharp. 1987. Binding of a nuclear factor to a regulatory sequence in the promoter of the mouse H-2Kb class I major histocompatibility gene. Mol. Cell. Biol. 7305-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banerji, S. S., N. G. Theodorakis, and R. I. Morimoto. 1984. Heat shock-induced translational control of HSP70 and globin synthesis in chicken reticulocytes. Mol. Cell. Biol. 42437-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berger, S. L. 2007. The complex language of chromatin regulation during transcription. Nature 447407-412. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, B. E., C. L. Liu, E. L. Humphrey, E. O. Perlstein, and S. L. Schreiber. 2004. Global nucleosome occupancy in yeast. Genome Biol. 5R62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanar, M. A., L. C. Burkly, and R. A. Flavell. 1989. NF-κB binds within a region required for B-cell-specific expression of major histocompatibility complex class II gene E alpha d. Mol. Cell. Biol. 9844-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boeger, H., J. Griesenbeck, and R. D. Kornberg. 2008. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell 133716-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boeger, H., J. Griesenbeck, J. S. Strattan, and R. D. Kornberg. 2003. Nucleosomes unfold completely at a transcriptionally active promoter. Mol. Cell 111587-1598. [DOI] [PubMed] [Google Scholar]
- 9.Brown, T., K. Mackey, and T. Du. 2004. Analysis of RNA by Northern and slot blot hybridization, p. 4.9.2-4.9.8. In Current protocols in molecular biology. Wiley Interscience, New York, NY. [DOI] [PubMed]
- 10.Cairns, B. R. 2007. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat. Struct. Mol. Biol. 14989-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carey, M., and S. T. Smale. 2000. In vivo analysis of an endogenous control region, p. 338-364. In Transcriptional regulation in eukaryotes: concepts, strategies, and techniques. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Ehrlich, R., S. O. Sharrow, J. E. Maguire, and D. S. Singer. 1989. Expression of a class I MHC transgene: effects of in vivo alpha/beta-interferon treatment. Immunogenetics 3018-26. [DOI] [PubMed] [Google Scholar]
- 13.Flavin, M., L. Cappabianca, C. Kress, H. Thomassin, and T. Grange. 2004. Nature of the accessible chromatin at a glucocorticoid-responsive enhancer. Mol. Cell. Biol. 247891-7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frels, W. I., J. A. Bluestone, R. J. Hodes, M. R. Capecchi, and D. S. Singer. 1985. Expression of a microinjected porcine class I major histocompatibility complex gene in transgenic mice. Science 228577-580. [DOI] [PubMed] [Google Scholar]
- 15.Ganapathi, M., P. Srivastava, S. K. Das Sutar, K. Kumar, D. Dasgupta, G. Pal Singh, V. Brahmachari, and S. K. Brahmachari. 2005. Comparative analysis of chromatin landscape in regulatory regions of human housekeeping and tissue specific genes. BMC Bioinform. 6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Girdlestone, J. 1995. Regulation of HLA class I loci by interferons. Immunobiology 193229-237. [DOI] [PubMed] [Google Scholar]
- 17.Giuliani, C., M. Saji, G. Napolitano, L. A. Palmer, S. I. Taniguchi, M. Shong, D. S. Singer, and L. D. Kohn. 1995. Hormonal modulation of major histocompatibility complex class I gene expression involves an enhancer A-binding complex consisting of Fra-2 and the p50 subunit of NF-κB. J. Biol. Chem. 27011453-11462. [DOI] [PubMed] [Google Scholar]
- 18.Gobin, S. J., V. Keijsers, M. van Zutphen, and P. J. van den Elsen. 1998. The role of enhancer A in the locus-specific transactivation of classical and nonclassical HLA class I genes by nuclear factor κB. J. Immunol. 1612276-2283. [PubMed] [Google Scholar]
- 19.Gobin, S. J., A. Peijnenburg, V. Keijsers, and P. J. van den Elsen. 1997. Site alpha is crucial for two routes of IFN gamma-induced MHC class I transactivation: the ISRE-mediated route and a novel pathway involving CIITA. Immunity 6601-611. [DOI] [PubMed] [Google Scholar]
- 20.Gobin, S. J., M. van Zutphen, A. M. Woltman, and P. J. van den Elsen. 1999. Transactivation of classical and nonclassical HLA class I genes through the IFN-stimulated response element. J. Immunol. 1631428-1434. [PubMed] [Google Scholar]
- 21.Goriely, S., D. Demonte, S. Nizet, D. De Wit, F. Willems, M. Goldman, and C. Van Lint. 2003. Human IL-12(p35) gene activation involves selective remodeling of a single nucleosome within a region of the promoter containing critical Sp1-binding sites. Blood 1014894-4902. [DOI] [PubMed] [Google Scholar]
- 22.Guillemette, B., and L. Gaudreau. 2006. Reuniting the contrasting functions of H2A.Z. Biochem. Cell Biol. 84528-535. [DOI] [PubMed] [Google Scholar]
- 23.Heintzman, N. D., R. K. Stuart, G. Hon, Y. Fu, C. W. Ching, R. D. Hawkins, L. O. Barrera, S. Van Calcar, C. Qu, K. A. Ching, W. Wang, Z. Weng, R. D. Green, G. E. Crawford, and B. Ren. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39311-318. [DOI] [PubMed] [Google Scholar]
- 24.Howcroft, T. K., J. D. Weissman, A. Gegonne, and D. S. Singer. 2005. A T lymphocyte-specific transcription complex containing RUNX1 activates MHC class I expression. J. Immunol. 1742106-2115. [DOI] [PubMed] [Google Scholar]
- 25.Ioshikhes, I. P., I. Albert, S. J. Zanton, and B. F. Pugh. 2006. Nucleosome positions predicted through comparative genomics. Nat. Genet. 381210-1215. [DOI] [PubMed] [Google Scholar]
- 26.Israel, A., O. Le Bail, D. Hatat, J. Piette, M. Kieran, F. Logeat, D. Wallach, M. Fellous, and P. Kourilsky. 1989. TNF stimulates expression of mouse MHC class I genes by inducing an NF κB-like enhancer binding activity which displaces constitutive factors. EMBO J. 83793-3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jabrane-Ferrat, N., N. Nekrep, G. Tosi, L. Esserman, and B. M. Peterlin. 2003. MHC class II enhanceosome: how is the class II transactivator recruited to DNA-bound activators? Int. Immunol. 15467-475. [DOI] [PubMed] [Google Scholar]
- 28.Kimura, A., A. Israel, O. Le Bail, and P. Kourilsky. 1986. Detailed analysis of the mouse H-2Kb promoter: enhancer-like sequences and their role in the regulation of class I gene expression. Cell 44261-272. [DOI] [PubMed] [Google Scholar]
- 29.Kouskouti, A., and I. Talianidis. 2005. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J. 24347-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kouzarides, T. 2007. Chromatin modifications and their function. Cell 128693-705. [DOI] [PubMed] [Google Scholar]
- 31.Lee, C. K., Y. Shibata, B. Rao, B. D. Strahl, and J. D. Lieb. 2004. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nat. Genet. 36900-905. [DOI] [PubMed] [Google Scholar]
- 32.Lee, W., D. Tillo, N. Bray, R. H. Morse, R. W. Davis, T. R. Hughes, and C. Nislow. 2007. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 391235-1244. [DOI] [PubMed] [Google Scholar]
- 33.Levitsky, V. G. 2004. RECON: a program for prediction of nucleosome formation potential. Nucleic Acids Res. 32W346-W349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 35.Lomvardas, S., and D. Thanos. 2002. Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110261-271. [DOI] [PubMed] [Google Scholar]
- 36.Mai, X., S. Chou, and K. Struhl. 2000. Preferential accessibility of the yeast his3 promoter is determined by a general property of the DNA sequence, not by specific elements. Mol. Cell. Biol. 206668-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez-Campa, C., P. Politis, J. L. Moreau, N. Kent, J. Goodall, J. Mellor, and C. R. Goding. 2004. Precise nucleosome positioning and the TATA box dictate requirements for the histone H4 tail and the bromodomain factor Bdf1. Mol. Cell 1569-81. [DOI] [PubMed] [Google Scholar]
- 38.Masternak, K., A. Muhlethaler-Mottet, J. Villard, M. Zufferey, V. Steimle, and W. Reith. 2000. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 141156-1166. [PMC free article] [PubMed] [Google Scholar]
- 39.Masternak, K., and W. Reith. 2002. Promoter-specific functions of CIITA and the MHC class II enhanceosome in transcriptional activation. EMBO J. 211379-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mavrich, T. N., C. Jiang, I. P. Ioshikhes, X. Li, B. J. Venters, S. J. Zanton, L. P. Tomsho, J. Qi, R. L. Glaser, S. C. Schuster, D. S. Gilmour, I. Albert, and B. F. Pugh. 2008. Nucleosome organization in the Drosophila genome. Nature 453358-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mito, Y., J. G. Henikoff, and S. Henikoff. 2005. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 371090-1097. [DOI] [PubMed] [Google Scholar]
- 42.Murphy, C., D. Nikodem, K. Howcroft, J. D. Weissman, and D. S. Singer. 1996. Active repression of major histocompatibility complex class I genes in a human neuroblastoma cell line. J. Biol. Chem. 27130992-30999. [DOI] [PubMed] [Google Scholar]
- 43.O'Neill, L. P., and B. M. Turner. 2003. Immunoprecipitation of native chromatin: NChIP. Methods 3176-82. [DOI] [PubMed] [Google Scholar]
- 44.Ozsolak, F., J. S. Song, X. S. Liu, and D. E. Fisher. 2007. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 25244-248. [DOI] [PubMed] [Google Scholar]
- 45.Pazin, M. J., P. Bhargava, E. P. Geiduschek, and J. T. Kadonaga. 1997. Nucleosome mobility and the maintenance of nucleosome positioning. Science 276809-812. [DOI] [PubMed] [Google Scholar]
- 46.Pokholok, D. K., C. T. Harbison, S. Levine, M. Cole, N. M. Hannett, T. I. Lee, G. W. Bell, K. Walker, P. A. Rolfe, E. Herbolsheimer, J. Zeitlinger, F. Lewitter, D. K. Gifford, and R. A. Young. 2005. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell 122517-527. [DOI] [PubMed] [Google Scholar]
- 47.Raval, A., T. K. Howcroft, J. D. Weissman, S. Kirshner, X. S. Zhu, K. Yokoyama, J. Ting, and D. S. Singer. 2001. Transcriptional coactivator, CIITA, is an acetyltransferase that bypasses a promoter requirement for TAF(II)250. Mol. Cell 7105-115. [DOI] [PubMed] [Google Scholar]
- 48.Reinke, H., and W. Horz. 2003. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol. Cell 111599-1607. [DOI] [PubMed] [Google Scholar]
- 49.Richard-Foy, H., and G. L. Hager. 1987. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 62321-2328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roh, T. Y., W. C. Ngau, K. Cui, D. Landsman, and K. Zhao. 2004. High-resolution genome-wide mapping of histone modifications. Nat. Biotechnol. 221013-1016. [DOI] [PubMed] [Google Scholar]
- 51.Schermer, U. J., P. Korber, and W. Horz. 2005. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol. Cell 19279-285. [DOI] [PubMed] [Google Scholar]
- 52.Schones, D. E., K. Cui, S. Cuddapah, T. Y. Roh, A. Barski, Z. Wang, G. Wei, and K. Zhao. 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132887-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Segal, E., Y. Fondufe-Mittendorf, L. Chen, A. Thastrom, Y. Field, I. K. Moore, J. P. Wang, and J. Widom. 2006. A genomic code for nucleosome positioning. Nature 442772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sekinger, E. A., Z. Moqtaderi, and K. Struhl. 2005. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol. Cell 18735-748. [DOI] [PubMed] [Google Scholar]
- 55.Singer, D. S., E. Mozes, S. Kirshner, and L. D. Kohn. 1997. Role of MHC class I molecules in autoimmune disease. Crit. Rev. Immunol. 17463-468. [PubMed] [Google Scholar]
- 56.Spilianakis, C., A. Kretsovali, T. Agalioti, T. Makatounakis, D. Thanos, and J. Papamatheakis. 2003. CIITA regulates transcription onset viaSer5-phosphorylation of RNA Pol II. EMBO J. 225125-5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Squazzo, S. L., H. O'Geen, V. M. Komashko, S. R. Krig, V. X. Jin, S. W. Jang, R. Margueron, D. Reinberg, R. Green, and P. J. Farnham. 2006. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res. 16890-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strauss, W. M. 1998. Preparation of genomic DNA from mammalian tissue, p. 2.2.1-2.2.3. In Current protocols in molecular biology. Wiley Interscience, New York, NY. [DOI] [PubMed]
- 59.Taniguchi, S. I., M. Shong, C. Giuliani, G. Napolitano, M. Saji, V. Montani, K. Suzuki, D. S. Singer, and L. D. Kohn. 1998. Iodide suppression of major histocompatibility class I gene expression in thyroid cells involves enhancer A and the transcription factor NF-κB. Mol. Endocrinol. 1219-33. [DOI] [PubMed] [Google Scholar]
- 60.Vakoc, C. R., S. A. Mandat, B. A. Olenchock, and G. A. Blobel. 2005. Histone H3 lysine 9 methylation and HP1γ are associated with transcription elongation through mammalian chromatin. Mol. Cell 19381-391. [DOI] [PubMed] [Google Scholar]
- 61.Vogelauer, M., J. Wu, N. Suka, and M. Grunstein. 2000. Global histone acetylation and deacetylation in yeast. Nature 408495-498. [DOI] [PubMed] [Google Scholar]
- 62.Waring, J. F., J. E. Radford, L. J. Burns, and G. D. Ginder. 1995. The human leukocyte antigen A2 interferon-stimulated response element consensus sequence binds a nuclear factor required for constitutive expression. J. Biol. Chem. 27012276-12285. [DOI] [PubMed] [Google Scholar]
- 63.Weissman, J. D., and D. S. Singer. 1991. A complex regulatory DNA element associated with a major histocompatibility complex class I gene consists of both a silencer and an enhancer. Mol. Cell. Biol. 114217-4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan, G. C., Y. J. Liu, M. F. Dion, M. D. Slack, L. F. Wu, S. J. Altschuler, and O. J. Rando. 2005. Genome-scale identification of nucleosome positions in Saccharomyces cerevisiae. Science 309626-630. [DOI] [PubMed] [Google Scholar]
- 65.Zaret, K. 2005. Micrococcal nuclease analysis of chromatin structure, p. 21.1.6-21.1.13. In F. M. Ausubel et al. (ed.), Current protocols in molecular biology. Wiley Interscience, New York, NY. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.