Abstract
During T helper cell differentiation, distinct programs of gene expression play a key role in defining the immune response to an environmental challenge. How chromatin remodeling events at the associated cytokine loci control differentiation is not known. We found that the ATP-dependent remodeling enzyme subunit BRG1 was required for T helper 2 (Th2) differentiation and Th2 cytokine transcription. BRG1 binding to cytokine genes was regulated by the extent of differentiation, the extent of activation, and cell fate. BRG1 was required for some features of the chromatin structure in target genes (DNase I hypersensitivity and histone acetylation), suggesting that BRG1 remodeling activity was directly responsible for changes in gene expression. NFAT and STAT6 activity were required for BRG1 recruitment to the Th2 locus control region, and STAT6 associated with BRG1 in a differentiation-inducible manner, suggesting direct recruitment of BRG1 to the bound loci. Together, these findings suggest BRG1 interprets differentiation signals and plays a causal role in gene regulation, chromatin structure, and cell fate.
Changes in chromatin structure are catalyzed by chromatin remodeling enzymes. ATP-dependent remodeling enzymes remodel or reposition nucleosomes, while other classes of remodeling enzymes covalently modify histone proteins or DNA. ATP-dependent remodeling can directly alter gene expression in cell free systems and also in cells (49). Remodeling may alter transcription factor binding, transcription factor function, or RNA polymerase binding. ATP-dependent remodeling enzymes are often multiprotein complexes, classified by their ATPase subunit into subfamilies such as SWI/SNF, ISWI, and Mi2 (49). BRG1 is an ATPase in the SWI/SNF subfamily and is essential for embryonic development (9). In cell-free systems SWI/SNF enzymes can displace, unfold, and slide nucleosomes (36, 50, 58). Remodeling enzymes are thought to bind DNA nonspecifically and be recruited by interactions with transcription factors and modified histones (21, 43, 59, 62). The role of BRG1 as an important regulator of T-cell development has been recently reviewed (13); BRG1 also plays an important role in macrophages (47) and T helper 1 (Th1) gene expression (34, 64). Although BRG1 is known to bind the T helper 2 (Th2) locus in T-cell clones (11), little is known about the role of ATP-dependent remodeling enzymes in the chromatin reorganization and gene expression changes that take place during Th2 development.
When peripheral T helper (Th) cells first encounter antigen they have the potential to differentiate into one of several different Th effector lineages characterized by specific cytokine profiles (19, 41, 56, 57). One effector population, Th2 cells, produces the cytokines interleukin-4 (IL-4), IL-5, IL-13, and IL-10; activates mast cells and eosinophils; and directs B-cell responses against extracellular pathogens. In contrast, Th1 cells produce gamma interferon (IFN-γ) and promote cell-mediated immunity directed toward intracellular pathogens, while Th17 cells produce IL-17 and target bacterial pathogens. Th lineage commitment is largely influenced by the local cytokine environment where the naive precursor cells first encounter antigen. Antigenic signals through the T-cell receptor (TCR) in the presence of IL-4 silence IFN-γ expression and program Th2 differentiation, ultimately leading to the expression of IL-4, IL-5, IL-13, and IL-10. IL-4 binding to the IL-4 receptor activates the transcription factor STAT6 through Jak kinases, leading to elevated expression of the transcription factor GATA-3. In contrast, IFN-γ-induced STAT1 activation induces the expression of the Th1-specific transcription factor, T-bet, which collaborates with IL-12-induced STAT4 to drive Th1 differentiation and expression of the cytokines IFN-γ and IL-2. Inappropriate Th responses can be pathological by either preventing efficient clearing of pathogens or triggering allergy, asthma, atopy, or autoimmunity.
Th2 cell differentiation is one of the best-studied mammalian systems for cell fate determination and gene regulation. The Th2 cytokine locus, consisting of the IL-4, IL-5, and IL-13 genes and the RAD50 housekeeping gene in a compact (120 kb) region on mouse chromosome 11, has been examined extensively to identify the changes in chromatin structure and gene regulation during Th lineage determination. During Th2 differentiation, the entire locus undergoes a number of chromatin modifications including the creation of nuclease accessible sites (2, 30), histone acetylation (6, 16, 20), and histone methylation (61), as well as DNA demethylation (2, 20), as summarized in recent reviews (3, 32). It is not known what remodeling enzymes mediate these changes. These modifications at specific regulatory regions are frequently associated with conserved, noncoding sequences; occur rapidly after the induction of Th2 differentiation; and may control the accessibility of DNA elements to transcription factors such as NFATs, STATs, GATA3, and c-Maf. It is not known how the sites of remodeling are targeted. Ectopic expression of the Th2-specific transcription factor GATA3 has been shown to program many of the chromatin changes in the Th2 locus, but the mechanism and remodeling enzymes responsible are not known (46, 55). Ectopic expression of STAT6 can induce GATA3, c-Maf, IL-4, IL-13, IL-5, and IL-10 (27).
Many of the Th2 locus regulatory elements have been functionally defined genetically and shown to contribute both positively and negatively to Th2 cytokine gene expression and lineage commitment (4, 31, 33, 40, 53). Included in these sites and embedded in the RAD50 gene is a Th2-specific locus control region (LCR) that participates in intrachromosomal interactions with the Th2 promoter elements, as well as interchromosomal interactions with the IFN-γ promoter in Th precursor (Thp) cells (54). (RAD50 is a constitutively expressed, essential DNA repair gene [38] that is largely independent of the regulatory signals for the cytokine genes.) At the Th2 cluster the three genes are often coordinately regulated by an LCR that is 16 to 70 kb away. In contrast, the best-studied mammalian gene cluster sharing an LCR is probably the β-globin cluster, where promoter competition results in a single gene being active. STAT6 has been found to play a key role in the chromatin structure of the Th2 LCR (29). Although much has been learned about Th2 cytokine regulation and Th2 differentiation, little is known about the key issues of how chromatin structure is established and maintained at the Th2 cytokine cluster and GATA3 gene, the enzyme(s) responsible for chromatin remodeling, and whether the observed changes in chromatin structure are important for regulation.
Here, we sought to determine whether the SWI/SNF subunit BRG1 is required for Th2 differentiation or gene expression. We found that knockdown of BRG1 expression during Th2 differentiation impaired differentiation and Th2 gene transcription. BRG1 knockdown after differentiation was complete also impaired Th2 gene expression. BRG1 knockdown during differentiation reduced GATA3 and c-Maf mRNA expression; in contrast, BRG1 knockdown after differentiation did not alter GATA3 and c-Maf expression. BRG1 bound to multiple enhancers in the Th2 cytokine cluster in an inducible manner; little if any binding was found in naive cells. BRG1 bound to the GATA3 gene in naive and effector cells. We detected changes in chromatin structure at some BRG1 binding sites when BRG1 expression was reduced. We found that endogenous BRG1 and STAT6 associated, specifically in response to IL-4, and BRG1 binding to part of the Th2 LCR was dependent on the activity of STAT6 and NFAT transcription factors. Together, these results suggest BRG1 directly regulates chromatin structure in Th cells, causing changes in gene expression and cell fate.
MATERIALS AND METHODS
Lymphocyte preparation and culture.
CD4+ T cells were purified from the lymph nodes of 4 to 6-week-old BALB/c mice (Taconic) by CD4 MACS according to the manufacturer's instructions (Miltenyi). Naive Thp cells were purified from lymph node and spleens by using a CD4+ CD62+ T-cell isolation kit (Miltenyi) to 95% purity (data not shown). Lymphocytes were cultured in RPMI 1640 supplemented with 10% fetal calf serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, 1 mM sodium pyruvate, 2 mM l-glutamine, 25 mM HEPES, and 50 μM β-mercaptoethanol. Stat6−/− (BALB/c background) mice were from Jackson Laboratories (24). Cyclosporine was from Sigma.
Institutional and national guidelines and regulations.
Animal approval was from the NIA ACUC (protocol ASP-365-MJP-Mi), and all experiments conform to the relevant regulatory standards.
In vitro Th cell differentiation.
Purified CD4+ LN T cells or naive Thp cells were plated onto anti-CD3 (1 μg/ml)- and anti-CD28 (2 μg/ml)-coated plates at 1 × 106 to 2 × 106/ml in the presence of 10 ng of IL-4 and 10 μg of anti-IFN-γ/ml (Th2 conditions) or 1 ng of IL-12 and 10 μg of anti-IL-4/ml (Th1 conditions). IL-2 (100 U/ml) was added 24 h later. Cultures were expanded in IL-2 (100 U/ml) 3 days after initial culture.
Nucleofection of Th cells.
A total of 2.5 × 106 to 4 × 106 purified Th cells was resuspended in 100 μl of mouse T-cell Nucleofector solution (Amaxa) and mixed with the indicated small interfering (siRNA) oligonucleotide (500 nM). Electroporation was performed as described by the manufacturer's protocol using the program x-001, and the cells were transferred into prewarmed mouse T-cell growth medium (Amaxa) supplemented with hIL-2 (100 U/ml). After transfection of naive Thps, the cells were allowed to rest in media for 3 h before transfer to anti-CD3/anti-CD28-coated plates and Th skewing conditions. The efficiency of transfection of naive Thps as determined by fluorescence-activated cell sorting analysis of fluorescently labeled oligonucleotides (Qiagen catalog no. 1022079) was ∼75%. The efficiencies with green fluorescent protein plasmid were ∼30% for naive Thps and ∼75% in Th2 effector cells (see Fig. S1C in the supplemental material and data not shown). We did not purify transduced cells. siRNA oligonucleotides (BRGm sc-29830) and negative control siRNA (sc-37007) used in the present study were obtained from Santa Cruz Biotechnology. Similar results were obtained using independent BRG1-specific siRNA pools obtained from Santa Cruz Biotechnology (sc-44289) and Dharmacon (m-041135-00) or three individual oligonucleotides (BRGm sc-29830). Based on sequence comparisons using BLASTn, all of these sequences are predicted to target BRG1, and none are predicted to target BRM.
Cytokines and antibodies.
The antibodies to CD3, CD28, IL-4, and IFN-γ used in Th cell differentiation cultures were obtained from Pharmingen. Recombinant hIL2, mIL-4, and mIL-12 were obtained from Peprotech. Antibodies used in immunoblots were specific to BRG1 (Upstate/Millipore), BRM (Pharmingen), phospho-Stat6 (Cell Signaling Technology), and Stat6 and SNF2H (Santa Cruz Biotechnology). Antibodies used in chromatin immunoprecipitation (ChIP) experiments were specific to BRG1 (J1; Weidong Wang), H3K9,14 acetylated histones (catalog no. 06-599; Upstate/Millipore), NFAT1 (catalog no. 07-136; Upstate/Millipore), and Stat6 (M-200; Santa Cruz Biotechnology).
mRNA quantitation.
Total RNA was purified by using RNeasy columns (Qiagen). cDNA was made by using iScript (Bio-Rad) according to the manufacturer's instructions. The mRNA levels of chromatin remodeling factors, cytokines, and transcription factors were determined by real-time PCR using Sybr green (Qiagen) on an ABI 7500 Fast. Expression levels were normalized to mTBP or m-actin as indicated. The oligonucleotide sequences are in Fig. S14 in the supplemental material.
Immunoblot analysis.
Whole-cell extracts were prepared by lysing cells in 50 mM Tris 7.4, 1% NP-40, 150 mM NaCl, 0.5% deoxycholate, and 0.1% sodium dodecyl sulfate (SDS) and clearing the lysates by centrifugation. Protein extracts were separated on a 6% polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Bio-Rad). The immunoblots were blocked for 1 h at room temperature in 5% milk in TBST (50 mM Tris [pH 7.5], 100 mM NaCl, 0.03% Tween 20) and incubated with the indicated antibody overnight at 4°C. The blots were washed with TBST and incubated with anti-rabbit horseradish peroxidase-conjugated antibody (Zymed) at room temperature. After the blots were washed with TBST, detection was carried out by using enhanced chemiluminescence (Amersham) according to the manufacturer's instructions.
ChIP assay.
ChIP was performed using methods similar to those described previously (37); the details are available on request. Approximately 20 million cells (for three to five immunoprecipitations) were cross-linked with 1% formaldehyde and quenched with glycine. Cells were lysed with buffer containing 1% SDS, treated with micrococcal nuclease, sonicated until the average DNA size was approximately 500 bp, and adjusted to 0.1% SDS-1% Triton X-100-150 mM NaCl at 5 ml. Sonicates were precleared with protein A-Sepharose (Upstate), and immunoprecipitation was performed with the following antibodies: H3K9,14 (Upstate catalog no. 06-599), BRG1 (J1), NFAT1 (Upstate catalog no. 07136) STAT6 (Santa Cruz sc-1698), or rabbit immunoglobulin G (IgG; Santa Cruz sc-2027). Chromatin was collected with protein A, washed, and eluted with sodium bicarbonate-SDS, and the cross-links were reversed, followed by protease treatment. Chromatin was quantified by real-time PCR using an Applied Biosystems 7500 with Sybr green detection (Qiagen). For each primer set for each experiment, a six-point standard curve of genomic DNA (0 to 40 ng) was compared to input and immunoprecipitated samples, thus correcting for the efficiency of the individual primer sets. Graphs indicate immunoprecipitated chromatin amounts relative to input DNA (percent input). For C-ChIP (45), we modified the above procedure by combining 1 to 5 million mouse T cells with 20 million carrier cells (E6-1 Jurkat cells) and then repeating the procedure described above beginning with the formaldehyde cross-linking. The oligonucleotide sequences are presented in Fig. S14 in the supplemental material.
DNase I hypersensitivity analysis.
Nuclei were purified from differentiated or effector Th2 cells as described previously (2). Briefly, nuclei were released by hypotonic lysis in the presence of 0.5% NP-40 and digested with the indicated amounts of DNase 1 (Worthington) for 3 min at room temperature. The samples were then treated with proteinase K and RNase A, and the DNA was recovered after phenol-chloroform extraction and ethanol precipitation. Stimulated effector Th2 cells were enriched for viable cells prior to nuclear isolation by Ficoll-Hypaque separation. DNase I accessibility was assessed by real-time PCR of the DNA samples (39) with the following modifications. Briefly, DNase-treated DNA was subjected to real-time PCR using primers to the indicated regulatory elements in the Th2 cytokine cluster and the GATA-3 locus. PCRs were performed by using a QuantiTect Sybr green PCR kit (Qiagen) according to manufacturer's instructions on an ABI 7500 apparatus. The DNase I sensitivity is indicated by the percent DNA remaining compared to the undigested sample, and DNA content was normalized to a known DNase I-resistant locus (Nfm). The oligonucleotide sequences are presented in Fig. S14 in the supplemental material.
Coimmunoprecipitation of BRG and STAT6.
On day 6, Th2 effector cells (107) were washed and replated in fresh medium for 1 h. IL-4 was added at 10 ng/ml for 2 h. The cells were washed with phosphate-buffered saline, and whole-cell extracts were made in 100 mM NaCl-20 mM HEPES (pH 7.5)-1 mM EDTA-10% glycerol-0.1% NP-40 in the presence of protease inhibitors and sodium vanadate. Extracts were cleared and immunoprecipitated with either BRG (Upstate 07-478) or Stat6 (Santa Cruz sc-1698). Similar results were obtained if the intact cells were first cross-linked with dithiobis(succinimidyl proprionate) (data not shown).
RESULTS
Remodeling ATPase expression in primary T cells.
Chromatin structure and transcription factor binding change dramatically during Th2 differentiation; however, the remodeling enzymes directly have not been identified. We found that a variety of chromatin remodeling ATPases were present in primary mouse Th cells (see Fig. S1A in the supplemental material). Given the high mRNA levels of the SWI/SNF remodeling ATPase BRG1 and the strong evidence for a role of BRG1 in T-cell development (13), we tested the role of BRG1 in the differentiation of Th2 cells.
BRG1 is required during Th2 differentiation for Th2 cytokine expression.
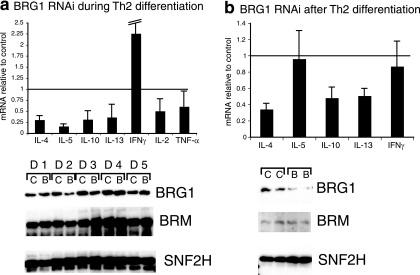
We sought to determine whether BRG1 expression was necessary for Th differentiation and cytokine expression. BRG1 was transiently knocked down in Thp cells (CD4+ cells from mouse lymph nodes), Th2 differentiation was initiated, and after 1 week the cells were restimulated and the cytokine expression was assessed (see the scheme in Fig. S1B in the supplemental material). We observed a decrease (typically 3- to 10-fold) in the amount of mRNA encoding the Th2 cytokines IL-4, IL-5, IL-13, and IL-10, which are markers for Th2 differentiation (Fig. 1a). This decrease is at the level of transcription, since primers specific for newly synthesized (unspliced) IL-4, IL-5, IL-13, and IL-10 RNA detect similar BRG1-dependent decreases in expression (data not shown). Similar results were obtained using two other siRNA reagents directed against BRG1, suggesting this is a specific effect of targeting BRG1 rather than an off-target or nonspecific effect (data not shown). We sought to determine whether the reduced Th2 cytokine expression was the result of the cells adopting alternative fates. The Th1 markers IL-2 and tumor necrosis factor alpha were not increased; the expression of IFN-γ increased in some experiments, although not to typical Th1 levels (Fig. 1a), and there was little if any change in the Treg marker FoxP3 and the Th17 marker IL-17A (data not shown). Decreased IL-4 protein and increased IFN-γ protein levels were also observed after BRG1 knockdown by enzyme-linked immunosorbent assay and intracellular cytokine staining, reflecting the mRNA changes (see Fig. S2A and B in the supplemental material). Intracellular cytokine staining revealed a decrease in the number of IL-4-producing cells in BRG1 knockdown cells, suggesting a decrease in the efficiency of differentiation to the Th2 lineage and a decrease in expression per cell (see Fig. S2B in the supplemental material). Others have reported that the inhibition of BRG1 decreases the number of IFN-γ-producing cells under Th1 conditions (64). Thus, BRG1 regulates Th2 cytokines; importantly, the cells do not appear to adopt the alternative Th1, Th17, or Treg fates. Since our conditions reduce, but do not eliminate, BRG1 expression during differentiation, the residual Th2 differentiation may also be BRG1 dependent or BRM dependent.
FIG. 1.
BRG1 RNAi impairs expression of Th2 cytokines. RNA was quantified by real-time reverse transcription-PCR, and each bar indicates the average and standard deviation of at least three independent experiments. Cytokine mRNA in BRG1 knockdown cells is normalized to mRNA in control knockdown cells; the horizontal line at y = 1 indicates no effect. Protein was detected with by immunoblotting. (a) Th2 cytokine expression is impaired by BRG1 knockdown during Th2 differentiation. Similar results were obtained using either CD4+ CD62L+ or CD4+ lymph node cells as starting material. The IFN-γ error bar extends to 3.9. D1 indicates day 1 of differentiation, etc.; C indicates control RNAi, B indicates BRG1 RNAi. BRM and SNF2H are reprobes of the same immunoblot as BRG1. BRM shows the effect of the knockdown on the closest homolog of BRG1, while SNF2H is another remodeling ATPase used here as a loading control. (b) BRG1 knockdown after Th2 differentiation impairs Th2 cytokine expression. C indicates control RNAi, B indicates BRG1 RNAi. BRM and SNF2H are reprobes of the same immunoblot as BRG1. BRM shows the effect of the knockdown on the closest homolog of BRG1, while SNF2H is another remodeling ATPase used here as a loading control.
BRG1 protein was transiently reduced on day 2 and day 3 after knockdown (Fig. 1a). The closely related BRM protein, the other ATPase that can be present in SWI/SNF complexes, was not affected as strongly. Consequently, we primarily attribute the effects we see to BRG1; however, although our experiments suggest a role for BRG1, they do not imply that BRM has no function. BRG1 knockdown during Th2 cell differentiation had little or no effect on the expression of cell surface markers (including CD4, CD3, and CD69), the proliferative response of cells to anti-CD3/CD28 during the first 48 to 72 h of culture, Stat6 expression and phosphorylation, and cell viability (see Fig. S3 in the supplemental material; also data not shown). However, we reproducibly observed that, after 1 week in culture, there was a decrease in cell number after BRG1 knockdown, suggesting that the loss of BRG1 expression during Th2 differentiation affected the potential outgrowth of the cells. Perhaps this limits the decrease in BRG1 expression obtained with siRNA.
BRG1 is also required after Th2 differentiation for expression of Th2 cytokine genes.
These experiments suggest a role for BRG1 in programming Thp cells to initiate Th2 cytokine gene expression. However, it was unclear whether BRG1 acted directly on Th2 cytokine genes or alternatively blocked differentiation upstream of cytokine expression. We sought to determine whether BRG1 was required for Th2 cytokine expression after differentiation. We knocked down BRG1 expression after Th2 differentiation was complete (day 5 [see Fig. S1B in the supplemental material]) and assessed the cytokine expression from restimulated cells 2 days after knockdown (day 7). We observed decreases in IL-4, IL-13, and IL-10 expression, similar to the reduction when BRG1 was reduced during differentiation. In contrast, IL-5 expression was not consistently affected (Fig. 1b). Again, the effect of BRG1 was at the level of transcription (data not shown). BRG1 protein was decreased, whereas BRM and SNF2H, another remodeling protein, were unaffected (Fig. 1b). We reproducibly observed stronger BRG1 knockdown after Th2 differentiation than during Th2 differentiation (compare Fig. 1a and b). IL-4 protein expression was reduced (see Fig. S2A in the supplemental material). The pattern of impaired IL-4 and IL-13 production with typical IL-5 production resembles the phenotype of the deletion of the Th2 LCR (33). Again, since the knockdown is partial and probably does not affect all cells, our results likely underestimate the importance of BRG1. These results suggest that BRG1 is important for Th2 cytokine expression (i) during differentiation of Thp cells and (ii) after Th2 differentiation is complete.
BRG1 regulates Th2 transcription factors during differentiation but not after.
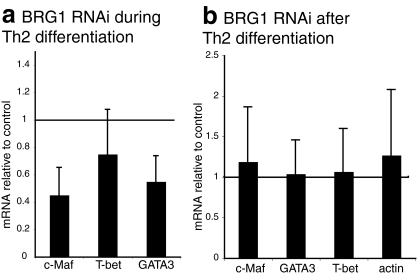
STAT6, GATA3, and c-Maf have been identified as transcription factors that control Th2 differentiation and expression of Th2-specific cytokines. When BRG1 expression was decreased after Th2 differentiation was complete, GATA3 and c-Maf expression were not affected (Fig. 2b), in contrast to the Th2 cytokine genes. Thus, the role of BRG1 in regulating cytokine expression after Th2 differentiation is probably direct, rather than an indirect effect of GATA3 or c-Maf regulation.
FIG. 2.
BRG1 knockdown after Th2 differentiation does not alter expression of Th2 transcription factor mRNA. Each bar is the average and standard deviation of at least three independent experiments; similar results were obtained with either CD4+ CD62L+ or CD4+ lymph node cells as starting material. (a) BRG1 knockdown during Th2 differentiation alters expression of GATA-3 and c-Maf in restimulated Th2 cells. (b) BRG1 knockdown after Th2 differentiation does not change transcription factor expression in restimulated Th2 cells.
When BRG1 was knocked down during differentiation, we observed a decrease of GATA-3 and c-Maf mRNA in restimulated cells, a finding consistent with a defect in Th2 differentiation (Fig. 2a). However, at the protein level, neither GATA3, c-Maf, nor STAT6 expression were strongly altered by BRG1 knockdown during or after differentiation (see Fig. S4 and S3A in the supplemental material).
BRG1 binding to the Th2 cluster varies with extent of differentiation, activation, and cell fate.
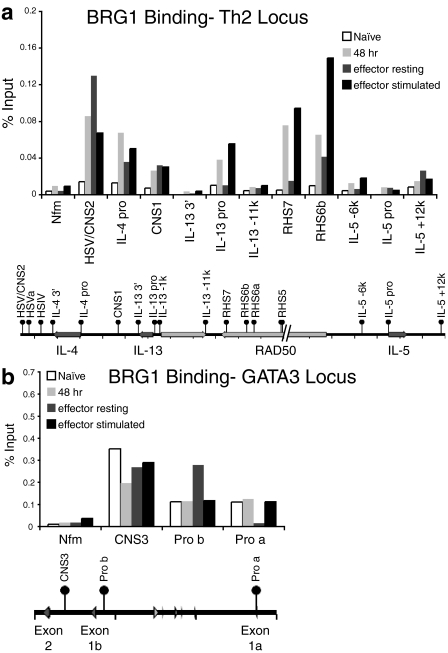
We hypothesized that if BRG1 were directly regulating the clustered Th2 cytokine genes, BRG1 would bind these genes in cells, perhaps at regions where chromatin remodeling occurs. Previous reports demonstrated a number of dramatic changes in chromatin structure at the Th2 cytokine locus during the first 72 h of Th2 differentiation (2, 7, 16). We performed ChIP for BRG1 at four stages of differentiation: Thp cells (naive), Th cells during Th2 differentiation (48 h), Th2 cells after differentiation was complete (effector resting), and Th2 effector cells restimulated 1.5 h (effector stimulated). BRG1 binding was readily detected at a number of known regulatory regions and DNase I hypersensitive sites (HS) during Th2 differentiation (48 h) (Fig. 3a; also see Fig. S5A in the supplemental material). In contrast, there was little if any BRG1 binding in naive cells at the tested sites within the Th2 locus (Fig. 3a; also see Fig. S5A in the supplemental material). Binding in naive cells was often similar to a brain-specific control locus (Nfm/Nef3) or control IgG immunoprecipitation, which may reflect little or no BRG1 occupancy (see Fig. S6A in the supplemental material). Binding was strongest (10- to 20-fold greater than the background value) at the Th2 LCR (RHS7 and RHS6b) and other distal regulatory elements (HSV/CNS2) and weaker at proximal promoter regions (IL-4 pro and IL-13 pro). BRG1 binding was often, although not always, associated with regions that were enriched for H3 acetylation at the Th2 locus (see Fig. S5B and S6B in the supplemental material; also data not shown). BRG1 binding was not detected in Th2 cells at the IFN-γ promoter or at a nonconserved region 11 kb upstream of the IL-13 promoter (Fig. 3a; see also Fig. S5A in the supplemental material; also data not shown). In our knockdown experiments during differentiation, this is the same time that BRG1 protein expression is most strongly reduced.
FIG. 3.
BRG1 binds to Th2 cytokine loci in differentiated, but not undifferentiated (CD4+ lymph node) cells. All of the Th2 cytokine locus samples are from one experiment (a), while all of the GATA3 locus samples are from another experiment (b). These panels are plotted at the same scale as in Fig. S6A in the supplemental material for direct comparison with the IgG control immunoprecipitation. These results are typical of at least three independent experiments (see also Fig. S5 in the supplemental material). The tick marks indicate 10 kb on the Th2 locus and 5 kb on the GATA3 locus.
Importantly, inducible BRG1 binding was found at loci that have been previously determined to be functional regulatory elements by deletion of these elements in mice (RHS7, CNS1, HSV/CNS2, HSVa, HSIV) (Fig. 3a; also see Fig. S5A in the supplemental material, and data not shown). Thus, BRG1 is specifically recruited to the Th2 cytokine cluster during early Th2 differentiation and may directly initiate subsequent Th2 cytokine gene expression.
In resting Th2 effector cells, BRG1 binding was elevated compared to naive cells at some loci; however, at several loci, including the Th2 LCR, the binding was reduced compared to differentiating Th2 cells (Fig. 3a; also see Fig. S5A in the supplemental material). BRG1 binding in restimulated Th2 effector cells was rapidly induced at some loci (RHS7, RHS6b, IL-4 pro, and IL-13 pro), although other loci were unchanged (CNS1) (Fig. 3a; also see Fig. S5A in the supplemental material). Again, little or no BRG1 binding was detected at the IFN-γ promoter, Nfm, or a nonconserved region 11 kb upstream of the IL-13 promoter (Fig. 3a; also see Fig. S5A in the supplemental material; also data not shown). In contrast, in mouse brain the brain-specific control locus Nfm exon 1 binds BRG1 and is highly acetylated, whereas RHS7 has little if any BRG1 or acetylation (M. J. Pazin, unpublished data). These results suggest activation changes BRG1 binding in Th2 effector cells, which is consistent with a direct role for BRG1 in regulating Th2 gene expression after Th2 differentiation.
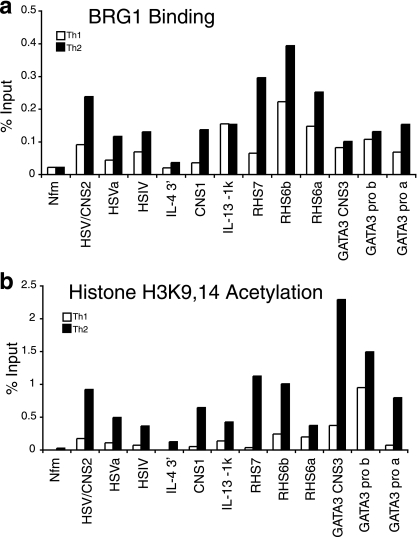
To examine the specificity of BRG1 recruitment by cell fate, we compared BRG1 binding between restimulated Th1 and Th2 effector cells. We found that BRG1 binding was enriched in Th2 cells at the Th2 cytokine cluster, although detectable at many loci in both Th1 and Th2 cells (Fig. 4a; also see Fig. S7 in the supplemental material). The greatest Th2 specificity was found at regions such as RHS7, HSVa, HSV/CNS2, and CNS1, which are DNase I HS that are preferentially or exclusively Th2 specific. In contrast, BRG1 binding was only slightly enriched at RHS6a, RHS6b, and HSIV, which are DNase I HS in both Th1 and Th2 cells. BRG1 is also recruited at low levels to Th2 sites in Th1 cells (HSV/CNS2); perhaps BRG1 plays a role in establishing repressive marks in the Th1 lineage (25). As reported by others (64), we found that BRG1 binding to the IFN-γ promoter was Th1 specific (see Fig. S7 in the supplemental material). The histone acetylation patterns were consistent with previous reports of Th1 and Th2 cells (Fig. 4b; also see Fig. S5B and S6B in the supplemental material). The finding that BRG1 binding is enriched at some Th2 cytokine loci in Th2 cells relative to Th1 cells, especially at regions with a Th2-specific chromatin structure, is consistent with a direct role for BRG1 in Th2-specific gene expression; the data also suggest BRG1 may play a role in chromatin structure that is similar between Th1 and Th2 cells.
FIG. 4.
More BRG1 is bound to Th2 loci in restimulated Th2 cells than in restimulated Th1 cells. (a) BRG1 binding. (See also Fig. S7 in the supplemental material.) (b) H3K9,14 acetylation. Similar results were obtained with either CD4+ CD62L+ or CD4+ lymph node cells as starting material. The control immunoprecipitation average for this experiment is 0.006 ± 0.005. The same chromatin was used for both panels a and b.
BRG1 binds to the GATA3 locus.
We hypothesized that the effect of BRG1 on GATA3 mRNA expression might be direct, and we began testing this by measuring BRG1 binding to the GATA3 locus. GATA3 regulation is complex, and GATA3 is required for embryogenesis, neuronal development, T-cell development, and Th2 differentiation (22, 28, 63, 66). GATA3 is expressed in naive Th cells; expression is increased during Th2 differentiation and decreased during Th1 expression. However, little is known about the Th2-specific chromatin structure and transcriptional regulation of GATA3, in contrast to the regulation of the Th2 cytokines. An alternative GATA3 promoter (promoter a) was identified whose activity increases during Th2 differentiation, while the activity of the previously identified promoter (promoter b) remains unchanged (5). This promoter appears to be located at a previously mapped DNase I HS (35).
BRG1 was bound to GATA3 (both known promoters and a conserved intronic noncoding sequence, CNS3) in naive CD4+ cells (Fig. 3b), in contrast to the lack of BRG1 binding at this time to the cytokine loci. Only modest changes in BRG1 binding were observed at the GATA3 locus during and after Th2 differentiation (Fig. 3b). One notable exception is the loss of BRG1 binding at the GATA3 promoter a in resting effector cells that is rapidly restored after cell stimulation. BRG1 binding and histone acetylation are enriched at promoter a in Th2 cells relative to Th1 cells (Fig. 4). BRG1 binding to the tested GATA3 elements did not directly correlate with the progressive histone acetylation (Fig. 3b). These results are consistent with a direct role for BRG1 in the regulation of GATA3 mRNA during Th2 differentiation.
BRG1 is required for remodeling the Th2 cytokine and GATA3 loci.
Regions of chromatin that demonstrate enhanced sensitivity to nucleases are frequently associated with transcriptional regulatory elements. The Th2 cytokine cluster has been well mapped for DNase I HS before, during and at the completion of Th2 differentiation (reviewed in references 3 and 32). Frequently, these DNase HS are located in conserved, noncoding sequences (17, 42), and subsequent genetic studies have demonstrated the importance of some of these regulatory elements in Th2 cytokine expression (4, 33, 40, 53). We hypothesized that if BRG1 were directly regulating cytokine and transcription factor genes, then BRG1 should be required to establish and/or maintain the chromatin structure of these target genes.
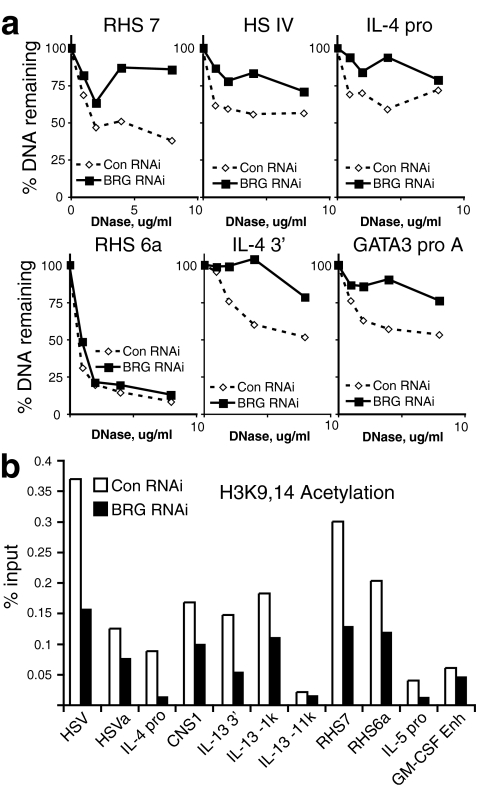
We sought to determine whether BRG1 was required after differentiation for the DNase I HS in effector Th2 cells. Under these conditions, BRG1 RNA interference (RNAi) does not appear to alter GATA3 or c-Maf RNA or protein. BRG1 was targeted after Th2 differentiation was complete, followed by restimulation of these effector cells. We investigated the DNase I HS at the time of maximal cytokine transcription to determine whether the structure and function were correlated. In the Th2 locus, BRG1 was required at the Th2-specific RHS7 (part of the Th2 LCR) for the DNase I HS (Fig. 5a and see Fig. S8A in the supplemental material). In addition, BRG1 was required at HSIV, the IL-4 promoter, the IL-4 3′ end, and GATA3 promoter a (Fig. 5a; also see Fig. S8A in the supplemental material; also data not shown). In contrast, BRG1 was not required at other BRG1 binding sites, such as RHS6a and GATA3 promoter b; it is possible our knockdown is not complete enough to reveal an effect, that BRG1 may be redundant with other remodeling enzymes, or that BRG1 may be important in Mast cells or at other times (Fig. 5a; also see Fig. S8A, S12, and S13 in the supplemental material). The loci affected by BRG1 knockdown are all binding sites for BRG1, and known or putative regulatory regions. Using another probe of chromatin structure, MNase, we find that BRG1 is required for accessibility (see Fig. S8B in the supplemental material). We also found that RHS7 is an inducible site in Th2 effector cells; this is in agreement with one report (29) but differs from another 16a, perhaps because of subtle differences in culture conditions (3). These findings indicate that BRG1 is required after Th2 differentiation to maintain or modify the typical chromatin structure at the Th2 and GATA3 loci.
FIG. 5.
BRG1 is required for typical chromatin structure after Th2 differentiation is complete. CD4+ lymph node cells were differentiated into Th2 cells in culture. Control or BRG1 RNAi was performed after Th2 differentiation was complete, as indicated. (a) BRG1 is required for the DNase I HS at RHS7, HSIV, IL-4 promoter, IL-4 3′ end, and GATA3 promoter a. (See also Fig. S8 in the supplemental material.) (b) BRG1 is required for histone acetylation in the Th2 locus. (See also Fig. S9 in the supplemental material.)
We continued to examine chromatin structure by measuring histone acetylation when BRG1 was knocked down after Th2 differentiation was complete, since BRG1 has been found to cooperate with histone acetyltransferases (8, 10, 14, 34, 47). Under these conditions, BRG1 RNAi does not appear to alter GATA3 or c-Maf RNA or protein. We found that in restimulated Th2 effector cells, BRG1 was required for H3K9,14 acetylation at several locations, especially at the IL-4 promoter and RHS7, which are preferentially Th2-specific DNase I HS and require BRG1 for typical chromatin structure (Fig. 5b; also see Fig. S9 in the supplemental material). In contrast, the granulocyte-macrophage colony-stimulating factor enhancer, the β2-microglobulin promoter, and a nonconserved region 11 kb upstream of the IL-13 promoter that fails to bind BRG1 and is not DNase I hypersensitive showed little if any effect, whereas acetylation increased at the promoter for CD4, a gene regulated by BRG1 during early T cell development (13a), suggesting that BRG1 is not required for all H3K9,14 acetylation. This is consistent with a causal role for BRG1 in maintenance or establishment of the histone acetylation pattern.
Transcription factors recruit BRG1 to the Th2 LCR.
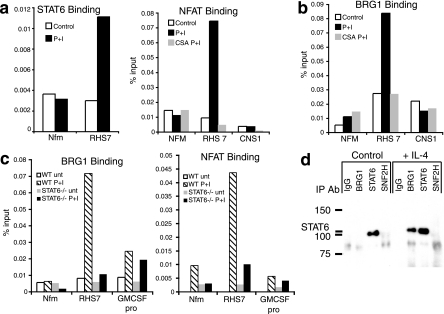
We decided to focus our studies on the Th2 LCR, located 16 to 70 kb from the Th2 promoters, for a number of reasons. Reduced expression of BRG1 after Th2 differentiation altered cytokine expression in a manner that resembled the deletion of the Th2 LCR element RHS7 (Fig. 2), BRG1 binding to RHS7 occurred preferentially in Th2 cells and was inducible upon activation (Fig. 3 and 4), and RHS7 chromatin structure was dependent upon BRG1 activity (Fig. 5). Since IL-4 and STAT6 are thought to play key roles in the formation of the HS at RHS7, we sought to determine whether STAT6 was involved in recruiting BRG1 to RHS7. In Th2 effector cells, endogenous STAT6 was bound to RHS7 in a stimulation-dependent manner (Fig. 6a). PMA plus ionomycin were sufficient to promote STAT6 binding and BRG1 binding under these conditions, whereas IL-4 was able to induce STAT6 binding but not BRG1 binding, suggesting that another transcription factor might also be required (data not shown). NFAT, a transcription factor family rapidly activated after TCR activation and known to be involved in cytokine transcription, also bound RHS7 after stimulation (Fig. 6a). NFAT binding required stimulation and was blocked by cyclosporine, a known inhibitor of the NFAT pathway. Activation-induced BRG1 binding to RHS7 was cyclosporine sensitive, suggesting that NFAT might play a role in recruiting BRG1 (Fig. 6b), which is consistent with a report of NFAT-BRG1 interaction in T cells (64). BRG1 did not bind to RHS7 in cells from STAT6-deficient mice differentiated under Th2 conditions, although binding to a STAT6-independent locus was unaffected (Fig. 6C). As reported by others (29), we found that RHS7 DNase I HS were reduced in cells from STAT6−/− mice differentiated under Th2 conditions (data not shown). NFAT binding to RHS7 was also impaired in cells from STAT6-deficient mice; again, binding to a STAT6-independent locus was unaffected (Fig. 6C). We performed sequential ChIP and found that BRG1 was present at the same chromatin fragments as the STAT6 and NFAT transcription factors (see Fig. S10 in the supplemental material). Thus, it was unlikely that BRG1 and transcription factors were binding to RHS7 in different populations of cells or at different alleles. Finally, we detected association of endogenous STAT6 and BRG1 in primary Th2 cells (Fig. 6d). The association required the stimulation of cells with exogenous IL-4, the same cytokine used to program Th2 differentiation, which may indicate that SWI/SNF interacts with phosphorylated STAT6. Together, these findings suggest a role for Th2 transcription factors in targeting the SWI/SNF complex to the Th2 LCR.
FIG. 6.
Transcription factor activity is required for BRG1 binding to the Th2 LCR after Th2 differentiation. (a) STAT6 and NFAT bind to RHS7 in the Th2 LCR in restimulated Th2 effector cells. NFAT binding is suppressed by cyclosporine. Cells were stimulated with phorbol myristate acetate and ionomycin where indicated, in the absence of exogenous IL-4. Binding was determined by ChIP. (b) Cyclosporine blocks BRG1 binding to RHS7. Binding was determined by ChIP. (c) STAT6 expression is required for BRG1 and NFAT binding to RHS7. Binding was determined by ChIP. Cells were differentiated under Th2 conditions. (d) Endogenous BRG1 and STAT6 associate in primary Th2 cells. IL-4 was added as indicated.
The BAF complex regulates Th2 cytokine expression.
BRG1 is known to exist in at least two different SWI/SNF complexes, BAF and P-BAF. Using ChIP, we further characterized the SWI/SNF complex at the Th2 LCR, finding two BAF-specific components at RHS7: BRM and BAF250a/Arid1a (see Fig. S11 in the supplemental material). Knockdown of BAF250a/Arid1a resulted in reduced IL-4 expression, similar to BRG1 knockdown, suggesting that the BAF complex plays a functional role in regulating the Th2 cytokine cluster (see Fig. S11 in the supplemental material). We did not detect a role for the P-BAF complex (BAF180/polybromo-1 and BAF200/Arid2); however, there is no known P-BAF target in these cells for comparison (data not shown). First, these findings confirm the role of SWI/SNF complexes in regulating Th2 cytokine expression, and, second, they suggest that the BAF version of the SWI/SNF complex is responsible for at least part of the observed SWI/SNF activity.
DISCUSSION
We tested the role of ATP-dependent remodeling in differentiation using primary T cells and BRG1 as a model system. We found BRG1 was required for Th2 differentiation. BRG1 was required during and after differentiation for Th2 cytokine transcription and during differentiation (but not after) for expression of the c-Maf and GATA3 mRNAs. BRG1 bound to the Th2 cytokine cluster, including the LCR, and also the GATA3 gene. Reduction in BRG1 expression affected IL-4 and IL-13 more strongly than IL-5, a finding reminiscent of the previously described phenotype of deletion of part of the LCR. BRG1 binding was regulated by the extent of differentiation and the choice of cell fate and cell activation. BRG1 was acting at least in part through the BAF complex. BRG1 was required for the Th2 cytokine cluster and GATA3 genes to adopt their typical chromatin structure. STAT6 bound the Th2 LCR, was required for NFAT and BRG1 binding to the LCR, and was associated with BRG1. NFAT activity was also required for BRG1 binding to the Th2 LCR.
We infer that at least part of the observed changes in gene expression are the result of BRG1 directly regulating the target genes rather than indirect (i.e., regulating a regulator of the target genes). First, we found that BRG1 bound to target loci with kinetics such that it is competent to regulate the cytokines and transcription factors. Second, reduction in BRG1 expression triggered both changes in gene expression and changes in chromatin structure at BRG1 binding sites, which is consistent with a direct, causal role for BRG1 remodeling in transcriptional regulation. Third, in mature effector cells BRG1 knockdown does not appear to regulate known regulators of Th2 cytokines. Fourth, BRG1 binding was observed specifically at known and putative regulatory regions. In differentiating cells BRG1 knockdown decreases mRNA encoding GATA3 and c-Maf, two transcription factors known to positively regulate Th2 cytokines. However, it does not strongly change the protein expression. Thus, in differentiating cells, GATA3 (and perhaps c-Maf) is a direct target of BRG1, whereas the Th2 cluster appears to be regulated by a combination of direct and perhaps indirect BRG1 effects. We propose the following simple model: BRG1 changes the chromatin structure of target loci, which allows enhancers and perhaps silencers to change their activity, which directly regulates gene expression.
We found that BRG1 plays a direct role in changing chromatin structure at the Th2 cytokine and transcription factor genes, which provides insight into the mechanism of a remodeling enzyme during choice of developmental fates. The change in DNase I hypersensitivity in the Th2 cytokine cluster have been carefully documented by several labs under various conditions. To our knowledge, this is the first report identifying an ATP-dependent chromatin remodeling enzyme that plays a role in their formation. Given the well-documented activity of BRG1 and SWI/SNF complexes in cell-free systems, we hypothesize that BRG1 is unfolding, sliding, or displacing nucleosomes to create DNase I HS; our experiments do not distinguish among these possibilities. DNase I hypersensitivity can reflect nucleosome-free regions, transcription factor binding, histone modifications, or nucleosome unfolding. At least part of this BRG1 activity appears to be through the BAF complex.
It is perhaps surprising that BRG1 is required in effector cells, since some of the Th2 cytokine locus DNase I HS, such as HSIV, are present in resting Th2 cells (1). One might have predicted that the chromatin structure was mostly programmed during differentiation, and after it was set there would be little if any need for remodeling. However, we found that BRG1 is required for Th2 effector cell chromatin structure and that BRG1 is functionally important in these cells. We found that nuclease sensitivity at RHS7 is inducible after Th2 differentiation is complete, consistent with a prior report (29); thus, the function of BRG1 may be to alter chromatin structure in effector cells. Our finding is also consistent with the recently described role of SWI/SNF in mediating the recall of transcriptional regulation memory in budding yeast (26).
We found that BRG1 binding to the Th2 LCR is dependent on STAT6 and, furthermore, that BRG1 directly interacts with this transcription factor. Interestingly, a number of other STAT proteins have also been suggested to activate transcription and remodel chromatin, either directly or indirectly, through BRG1, suggesting that the BRG/STAT pathway is general (18, 23, 44). In our system, we found that BRG1 recruitment to the Th2 LCR depends both on cytokine signals through STAT6, as well as signaling through the TCR via NFAT1. Since the differentiation of a Thp to a Th2 cell depends on these same signals, we suggest that BRG1 uses the mechanism of chromatin remodeling to integrate these signals. This is reminiscent of results reported for the IFN-γ promoter, where BRG1 binding in Th1 cells is dependent on both NFAT and STAT4 (64). It is tempting to speculate that the recruitment of chromatin remodeling factors to STAT binding sites via STAT proteins is a general feature of cytokine-induced transcription, providing a direct connection between extracellular signals and epigenetic changes at the level of nucleosome remodeling.
Although our data suggest that the developmentally regulated and inducible recruitment of BRG1 to the Th2 locus is dependent on the activation of Th2 transcription factors, we cannot rule out a role for direct modification of BRG1-containing complexes or Th2 transcription factors by T-cell activation. Antigen receptor signaling has been shown to induce the rapid association of BRG complexes to chromatin, an activity that is dependent on phosphatidylinositol 4,5-bisphosphate (PIP2) (65). In addition, p38 mitogen-activated protein (MAP) kinase has been demonstrated to directly phosphorylate the BAF60 subunit of SWI-SNF resulting in the recruitment of SWI-SNF to a target promoter (52). BRG1 itself is also capable of being phosphorylated by ERK1 in a cell cycle-dependent manner, resulting in the inactivation of BRG1 activity (51). GATA3 has been found to be a target of MAP kinase cascades (12). Given the established role for MAP kinases in Th differentiation (15, 48, 60), future experiments will be needed to determine whether this signaling pathway plays a role in directing BRG1 recruitment to cytokine loci.
We do not yet know how BRG1 is activating transcription in this system. We found that BRG1 binds strongly to RHS7 in a Th2-specific manner, before DNase I hypersensitivity is established, and that BRG1 is required for DNase I HS at RHS7 in Th2 effector cells. Our finding that IL-4, IL-13, and IL-10 require BRG1 in Th2 effector cells, whereas IL-5 does not, is reminiscent of a previous finding that RHS7 is required in vitro under Th2 conditions for IL-4 and IL-13 mRNA production, but not for IL-5 (33). It is unclear why IL-5 is strongly affected by BRG1 knockdown during differentiation but is unaffected after differentiation. We note that BRG1 binding is weak or absent at the IL-5 proximal regions that we have tested to date; perhaps another remodeling enzyme is regulating IL-5. It may be significant that we measure the strongest BRG1 binding at sites that are distant from the transcriptional start sites rather than the more frequently studied promoters. BRG1-mediated remodeling might be affording access to transcriptional activators or RNA polymerase, facilitating the action of other remodeling enzymes such as histone-modifying enzymes, or mediating changes in higher-order chromatin structure that permit long-range interactions in the locus.
We hypothesized that BRG1 would be important for the chromatin structure of at least some of the previously mapped DNase I HS. Unexpectedly, BRG1 binds to nearly every regulatory element we tested. Our assays are specific, since nonconserved regions in the Th2 locus (IL-13, kb -11; IL-5, kb -6) and a neuron-specific gene (NfM/Nef3, exon 1) lack BRG1 binding, histone acetylation, and DNase I HS. Moreover, BRG1 bound the IFN-γ promoter in Th1 cells but not Th2 cells. Although we favor a model wherein BRG1 is binding to specific regulatory elements, further experiments are in progress to determine whether BRG1 binds discrete elements or broad regions. We also detected BRG1 binding at sites that do not appear to have BRG1-dependent chromatin structure; we do not yet know whether BRG1 has no effect at these sites or is redundant with other remodeling enzymes, or whether the residual BRG1 following knockdown is sufficient to program the chromatin structure at these sites.
We have preferentially reduced BRG1 expression and revealed a role for the SWI/SNF component in Th2 differentiation, cytokine gene expression, and chromatin structure. The remaining Th2 function may be dependent on the residual BRG1 expression, BRM, or other remodeling enzymes; our experiments do not exclude a role for BRM. Our observation that reduced expression of BAF250a, another SWI/SNF component, has a similar effect as BRG1 provides independent confirmation of the role of SWI/SNF in these processes. This finding also detects a function for the BAF version of SWI/SNF; however, this does not exclude a role for P-BAF versions of SWI/SNF.
In a recent study examining the role of SATB1 in the Th2 locus, it was reported that BRG1 was present throughout the locus in a Th2 cell clone and that binding increased upon activation (11). We extended these findings here by using primary cells to determine that BRG1 is not bound in naive cells and that binding occurs during Th2 differentiation. We found high BRG1 binding to the LCR and low binding to the IL-4 promoter and CNS1, whereas this pattern is reversed in the Th2 cell clone; this might reflect a difference between primary cells versus a T-cell clone maintained in vitro.
BRG1 is essential for embryonic development and is ubiquitously expressed. BRG1 has demonstrated roles in T-cell development, T-cell lineage choice, Th differentiation and function, and macrophage function. However, much remains to be discovered about how BRG1 helps to control these processes. We have found that in Th2 differentiation, BRG1 directly regulates targets at the transcriptional level by binding to proximal and distal regulatory regions and programming chromatin structure. We speculate this mechanism is likely to be recapitulated in other cell types throughout the body during development rather than being specific to T cells.
Supplementary Material
Acknowledgments
We thank Weidong Wang for BRG1 (J1) antibody; Bryan Turner and Laura P. O'Neill for providing a detailed C-ChIP protocol and advice prior to publication; Sung-Yun Pai for the IL-13 promoter primer sequences and helpful advice; and Sebastian Fugmann, Dan Longo, Nan-Ping Weng, Weidong Wang, Kerri Mowen, Ranjan Sen, Rebecca Potts, Patricia Precht, and Bob Wersto for helpful discussions.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging.
Footnotes
Published ahead of print on 13 October 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Agarwal, S., O. Avni, and A. Rao. 2000. Cell-type-restricted binding of the transcription factor NFAT to a distal IL-4 enhancer in vivo. Immunity 12643-652. [DOI] [PubMed] [Google Scholar]
- 2.Agarwal, S., and A. Rao. 1998. Modulation of chromatin structure regulates cytokine gene expression during T-cell differentiation. Immunity 9765-775. [DOI] [PubMed] [Google Scholar]
- 3.Ansel, K. M., I. Djuretic, B. Tanasa, and A. Rao. 2006. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 24607-656. [DOI] [PubMed] [Google Scholar]
- 4.Ansel, K. M., R. J. Greenwald, S. Agarwal, C. H. Bassing, S. Monticelli, J. Interlandi, I. M. Djuretic, D. U. Lee, A. H. Sharpe, F. W. Alt, and A. Rao. 2004. Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nat. Immunol. 51251-1259. [DOI] [PubMed] [Google Scholar]
- 5.Asnagli, H., M. Afkarian, and K. M. Murphy. 2002. Cutting edge: identification of an alternative GATA-3 promoter directing tissue-specific gene expression in mouse and human. J. Immunol. 1684268-4271. [DOI] [PubMed] [Google Scholar]
- 6.Avni, O., D. Lee, F. Macian, S. J. Szabo, L. H. Glimcher, and A. Rao. 2002. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat. Immunol. 3643-651. [DOI] [PubMed] [Google Scholar]
- 7.Baguet, A., and M. Bix. 2004. Chromatin landscape dynamics of the Il4-Il13 locus during T helper 1 and 2 development. Proc. Natl. Acad. Sci. USA 10111410-11415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brockmann, D., O. Lehmkuhler, U. Schmucker, and H. Esche. 2001. The histone acetyltransferase activity of PCAF cooperates with the brahma/SWI2-related protein BRG-1 in the activation of the enhancer A of the MHC class I promoter. Gene 277111-120. [DOI] [PubMed] [Google Scholar]
- 9.Bultman, S., T. Gebuhr, D. Yee, C. La Mantia, J. Nicholson, A. Gilliam, F. Randazzo, D. Metzger, P. Chambon, G. Crabtree, and T. Magnuson. 2000. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 61287-1295. [DOI] [PubMed] [Google Scholar]
- 10.Bultman, S. J., T. C. Gebuhr, and T. Magnuson. 2005. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in beta-globin expression and erythroid development. Genes Dev. 192849-2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai, S., C. C. Lee, and T. Kohwi-Shigematsu. 2006. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat. Genet. 381278-1288. [DOI] [PubMed] [Google Scholar]
- 12.Chen, C. H., D. H. Zhang, J. M. LaPorte, and A. Ray. 2000. Cyclic AMP activates p38 mitogen-activated protein kinase in Th2 cells: phosphorylation of GATA-3 and stimulation of Th2 cytokine gene expression. J. Immunol. 1655597-5605. [DOI] [PubMed] [Google Scholar]
- 13.Chi, T. 2004. A BAF-centered view of the immune system. Nat. Rev. Immunol. 4965-977. [DOI] [PubMed] [Google Scholar]
- 13a.Chi, T. H., M. Wan, K. Zhao, I. Taniuchi, L. Chen, D. R. Littman, and G. R. Crabtree. 2002. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418195-199. [DOI] [PubMed] [Google Scholar]
- 14.DiRenzo, J., Y. Shang, M. Phelan, S. Sif, M. Myers, R. Kingston, and M. Brown. 2000. BRG-1 is recruited to estrogen-responsive promoters and cooperates with factors involved in histone acetylation. Mol. Cell. Biol. 207541-7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong, C., D. D. Yang, C. Tournier, A. J. Whitmarsh, J. Xu, R. J. Davis, and R. A. Flavell. 2000. JNK is required for effector T-cell function but not for T-cell activation. Nature 40591-94. [DOI] [PubMed] [Google Scholar]
- 16.Fields, P. E., S. T. Kim, and R. A. Flavell. 2002. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J. Immunol. 169647-650. [DOI] [PubMed] [Google Scholar]
- 16a.Fields, P. E., G. R. Lee, S. T. Kim, V. V. Bartsevich, and R. A. Flavell. 2004. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity 21865-876. [DOI] [PubMed] [Google Scholar]
- 17.Frazer, K. A., L. Pachter, A. Poliakov, E. M. Rubin, and I. Dubchak. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32W273-W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Giraud, S., A. Hurlstone, S. Avril, and O. Coqueret. 2004. Implication of BRG1 and cdk9 in the STAT3-mediated activation of the p21waf1 gene. Oncogene 237391-7398. [DOI] [PubMed] [Google Scholar]
- 19.Grogan, J. L., and R. M. Locksley. 2002. T helper cell differentiation: on again, off again. Curr. Opin. Immunol. 14366-372. [DOI] [PubMed] [Google Scholar]
- 20.Guo, L., J. Hu-Li, J. Zhu, C. J. Watson, M. J. Difilippantonio, C. Pannetier, and W. E. Paul. 2002. In TH2 cells the Il4 gene has a series of accessibility states associated with distinctive probabilities of IL-4 production. Proc. Natl. Acad. Sci. USA 9910623-10628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hassan, A. H., K. E. Neely, and J. L. Workman. 2001. Histone acetyltransferase complexes stabilize swi/snf binding to promoter nucleosomes. Cell 104817-827. [DOI] [PubMed] [Google Scholar]
- 22.Hendriks, R. W., M. C. Nawijn, J. D. Engel, H. van Doorninck, F. Grosveld, and A. Karis. 1999. Expression of the transcription factor GATA-3 is required for the development of the earliest T-cell progenitors and correlates with stages of cellular proliferation in the thymus. Eur. J. Immunol. 291912-1918. [DOI] [PubMed] [Google Scholar]
- 23.Huang, M., F. Qian, Y. Hu, C. Ang, Z. Li, and Z. Wen. 2002. Chromatin-remodeling factor BRG1 selectively activates a subset of interferon-alpha-inducible genes. Nat. Cell Biol. 4774-781. [DOI] [PubMed] [Google Scholar]
- 24.Kaplan, M. H., U. Schindler, S. T. Smiley, and M. J. Grusby. 1996. Stat6 is required for mediating responses to IL-4 and for development of Th2 cells. Immunity 4313-319. [DOI] [PubMed] [Google Scholar]
- 25.Koyanagi, M., A. Baguet, J. Martens, R. Margueron, T. Jenuwein, and M. Bix. 2005. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J. Biol. Chem. 28031470-31477. [DOI] [PubMed] [Google Scholar]
- 26.Kundu, S., P. J. Horn, and C. L. Peterson. 2007. SWI/SNF is required for transcriptional memory at the yeast GAL gene cluster. Genes Dev. 21997-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurata, H., H. J. Lee, A. O'Garra, and N. Arai. 1999. Ectopic expression of activated Stat6 induces the expression of Th2-specific cytokines and transcription factors in developing Th1 cells. Immunity 11677-688. [DOI] [PubMed] [Google Scholar]
- 28.Lakshmanan, G., K. H. Lieuw, K. C. Lim, Y. Gu, F. Grosveld, J. D. Engel, and A. Karis. 1999. Localization of distant urogenital system-, central nervous system-, and endocardium-specific transcriptional regulatory elements in the GATA-3 locus. Mol. Cell. Biol. 191558-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee, D. U., and A. Rao. 2004. Molecular analysis of a locus control region in the T helper 2 cytokine gene cluster: a target for STAT6 but not GATA3. Proc. Natl. Acad. Sci. USA 10116010-16015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, G. R., P. E. Fields, and R. A. Flavell. 2001. Regulation of IL-4 gene expression by distal regulatory elements and GATA-3 at the chromatin level. Immunity 14447-459. [DOI] [PubMed] [Google Scholar]
- 31.Lee, G. R., P. E. Fields, T. J. Griffin, and R. A. Flavell. 2003. Regulation of the Th2 cytokine locus by a locus control region. Immunity 19145-153. [DOI] [PubMed] [Google Scholar]
- 32.Lee, G. R., S. T. Kim, C. G. Spilianakis, P. E. Fields, and R. A. Flavell. 2006. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity 24369-379. [DOI] [PubMed] [Google Scholar]
- 33.Lee, G. R., C. G. Spilianakis, and R. A. Flavell. 2005. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nat. Immunol. 642-48. [DOI] [PubMed] [Google Scholar]
- 34.Letimier, F. A., N. Passini, S. Gasparian, E. Bianchi, and L. Rogge. 2007. Chromatin remodeling by the SWI/SNF-like BAF complex and STAT4 activation synergistically induce IL-12Rβ2 expression during human Th1 cell differentiation. EMBO J. 261292-1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lieuw, K. H., G. Li, Y. Zhou, F. Grosveld, and J. D. Engel. 1997. Temporal and spatial control of murine GATA-3 transcription by promoter-proximal regulatory elements. Dev. Biol. 1881-16. [DOI] [PubMed] [Google Scholar]
- 36.Lorch, Y., B. Maier-Davis, and R. D. Kornberg. 2006. Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl. Acad. Sci. USA 1033090-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu, J., M. J. Pazin, and K. Ravid. 2004. Properties of ets-1 binding to chromatin and its effect on platelet factor 4 gene expression. Mol. Cell. Biol. 24428-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo, G., M. S. Yao, C. F. Bender, M. Mills, A. R. Bladl, A. Bradley, and J. H. Petrini. 1999. Disruption of mRad50 causes embryonic stem cell lethality, abnormal embryonic development, and sensitivity to ionizing radiation. Proc. Natl. Acad. Sci. USA 967376-7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McArthur, M., S. Gerum, and G. Stamatoyannopoulos. 2001. Quantification of DNase 1 sensitivity by real-time PCR: quantitative analysis of DNase 1 hypersensitivity of the mouse beta-globin LCR. J. Mol. Biol. 31327-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohrs, M., C. M. Blankespoor, Z. E. Wang, G. G. Loots, V. Afzal, H. Hadeiba, K. Shinkai, E. M. Rubin, and R. M. Locksley. 2001. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat. Immunol. 2842-847. [DOI] [PubMed] [Google Scholar]
- 41.Murphy, K. M., and S. L. Reiner. 2002. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2933-944. [DOI] [PubMed] [Google Scholar]
- 42.Nardone, J., D. U. Lee, K. M. Ansel, and A. Rao. 2004. Bioinformatics for the ‘bench biologist’: how to find regulatory regions in genomic DNA. Nat. Immunol. 5768-774. [DOI] [PubMed] [Google Scholar]
- 43.Neely, K. E., A. H. Hassan, A. E. Wallberg, D. J. Steger, B. R. Cairns, A. P. Wright, and J. L. Workman. 1999. Activation domain-mediated targeting of the SWI/SNF complex to promoters stimulates transcription from nucleosome arrays. Mol. Cell 4649-655. [DOI] [PubMed] [Google Scholar]
- 44.Ni, Z., E. Karaskov, T. Yu, S. M. Callaghan, S. Der, D. S. Park, Z. Xu, S. G. Pattenden, and R. Bremner. 2005. Apical role for BRG1 in cytokine-induced promoter assembly. Proc. Natl. Acad. Sci. USA 10214611-14616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Neill, L. P., M. D. Vermilyea, and B. M. Turner. 2006. Epigenetic characterization of the early embryo with a chromatin immunoprecipitation protocol applicable to small cell populations. Nat. Genet. 38835-841. [DOI] [PubMed] [Google Scholar]
- 46.Ouyang, W., M. Lohning, Z. Gao, M. Assenmacher, S. Ranganath, A. Radbruch, and K. M. Murphy. 2000. Stat6-independent GATA-3 autoactivation directs IL-4-independent Th2 development and commitment. Immunity 1227-37. [DOI] [PubMed] [Google Scholar]
- 47.Ramirez-Carrozzi, V. R., A. A. Nazarian, C. C. Li, S. L. Gore, R. Sridharan, A. N. Imbalzano, and S. T. Smale. 2006. Selective and antagonistic functions of SWI/SNF and Mi-2β nucleosome remodeling complexes during an inflammatory response. Genes Dev. 20282-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rincon, M., H. Enslen, J. Raingeaud, M. Recht, T. Zapton, M. S. Su, L. A. Penix, R. J. Davis, and R. A. Flavell. 1998. Interferon-gamma expression by Th1 effector T cells mediated by the p38 MAP kinase signaling pathway. EMBO J. 172817-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saha, A., J. Wittmeyer, and B. R. Cairns. 2006. Chromatin remodelling: the industrial revolution of DNA around histones. Nat. Rev. Mol. Cell. Biol. 7437-447. [DOI] [PubMed] [Google Scholar]
- 50.Schnitzler, G., S. Sif, and R. E. Kingston. 1998. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell 9417-27. [DOI] [PubMed] [Google Scholar]
- 51.Sif, S., P. T. Stukenberg, M. W. Kirschner, and R. E. Kingston. 1998. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 122842-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simone, C., S. V. Forcales, D. A. Hill, A. N. Imbalzano, L. Latella, and P. L. Puri. 2004. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat. Genet. 36738-743. [DOI] [PubMed] [Google Scholar]
- 53.Solymar, D. C., S. Agarwal, C. H. Bassing, F. W. Alt, and A. Rao. 2002. A 3′ enhancer in the IL-4 gene regulates cytokine production by Th2 cells and mast cells. Immunity 1741-50. [DOI] [PubMed] [Google Scholar]
- 54.Spilianakis, C. G., M. D. Lalioti, T. Town, G. R. Lee, and R. A. Flavell. 2005. Interchromosomal associations between alternatively expressed loci. Nature 435637-645. [DOI] [PubMed] [Google Scholar]
- 55.Takemoto, N., Y. Kamogawa, H. Jun Lee, H. Kurata, K. I. Arai, A. O'Garra, N. Arai, and S. Miyatake. 2000. Cutting edge: chromatin remodeling at the IL-4/IL-13 intergenic regulatory region for Th2-specific cytokine gene cluster. J. Immunol. 1656687-6691. [DOI] [PubMed] [Google Scholar]
- 56.Tato, C. M., A. Laurence, and J. J. O'Shea. 2006. Helper T-cell differentiation enters a new era: le roi est mort; vive le roi! J. Exp. Med. 203809-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tato, C. M., and J. J. O'Shea. 2006. Immunology: what does it mean to be just 17? Nature 441166-168. [DOI] [PubMed] [Google Scholar]
- 58.Whitehouse, I., A. Flaus, B. R. Cairns, M. F. White, J. L. Workman, and T. Owen-Hughes. 1999. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400784-787. [DOI] [PubMed] [Google Scholar]
- 59.Wysocka, J., T. Swigut, H. Xiao, T. A. Milne, S. Y. Kwon, J. Landry, M. Kauer, A. J. Tackett, B. T. Chait, P. Badenhorst, C. Wu, and C. D. Allis. 2006. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature 44286-90. [DOI] [PubMed] [Google Scholar]
- 60.Yamashita, M., M. Kimura, M. Kubo, C. Shimizu, T. Tada, R. M. Perlmutter, and T. Nakayama. 1999. T-cell antigen receptor-mediated activation of the Ras/mitogen-activated protein kinase pathway controls interleukin 4 receptor function and type-2 helper T-cell differentiation. Proc. Natl. Acad. Sci. USA 961024-1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamashita, M., R. Shinnakasu, Y. Nigo, M. Kimura, A. Hasegawa, M. Taniguchi, and T. Nakayama. 2004. Interleukin (IL)-4-independent maintenance of histone modification of the IL-4 gene loci in memory Th2 cells. J. Biol. Chem. 27939454-39464. [DOI] [PubMed] [Google Scholar]
- 62.Yudkovsky, N., C. Logie, S. Hahn, and C. L. Peterson. 1999. Recruitment of the SWI/SNF chromatin remodeling complex by transcriptional activators. Genes Dev. 132369-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, D. H., L. Cohn, P. Ray, K. Bottomly, and A. Ray. 1997. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the interleukin-5 gene. J. Biol. Chem. 27221597-21603. [DOI] [PubMed] [Google Scholar]
- 64.Zhang, F., and M. Boothby. 2006. T helper type 1-specific Brg1 recruitment and remodeling of nucleosomes positioned at the IFN-gamma promoter are Stat4 dependent. J. Exp. Med. 2031493-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao, K., W. Wang, O. J. Rando, Y. Xue, K. Swiderek, A. Kuo, and G. R. Crabtree. 1998. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell 95625-636. [DOI] [PubMed] [Google Scholar]
- 66.Zheng, W., and R. A. Flavell. 1997. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89587-596. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.