Abstract
Transition metals require exquisite handling within cells to ensure that cells are not harmed by an excess of free metal species. In gram-negative bacteria, copper is only required in low amounts in the periplasm, not in the cytoplasm, so a key aspect of protection under excess metal conditions is to export copper from the periplasm. Additional protection could be conferred by a periplasmic chaperone in order to limit the free metal species prior to export. Using isothermal titration calorimetry, we have demonstrated that two periplasmic proteins, CusF and CusB, of the E. coli Cu(I)/Ag(I) efflux system undergo a metal dependent interaction. Through the development of a novel X-ray absorption spectroscopy approach using selenomethionine labeling to distinguish the metal sites of the two proteins, we have demonstrated transfer of Cu(I) occurs between CusF and CusB. The interaction between these proteins is highly specific, as a homolog of CusF with 51% sequence identity and similar affinity for metal, did not function in metal transfer. These experiments establish a metallochaperone activity for CusF in the periplasm of gram-negative bacteria, serving to protect the periplasm from metal-mediated damage.
Transition metals such as copper are double-edged swords for living cells. Their redox capability is essential in numerous enzymatic processes (1). However, this ability to undergo redox transitions makes them commensurably toxic when present in excess of the required concentration. Additionally, the strong bonds that transition metals form with functional groups such as thiolates and imidazolium nitrogens in proteins contributes to their toxicity (2). Thus, the dual nature of transition metals continually challenges cells to maintain a delicate concentration balance within the cellular milieu. An effective means of preventing the cytotoxic effects of copper is to keep it complexed (3). In other cells, most notably studied in yeast, cytoplasmic copper chaperone systems are useful for both sequestering and delivering copper to various proteins within the cell (4–10).
E. coli cells, in common with the vast majority of bacterial cells, do not require copper for cytoplasmic enzymes, but instead only need copper for periplasmic or inner membrane enzymes such as multicopper oxidases, Cu, Zn-superoxide dismutases or for the final enzyme of the respiratory chain, cytochrome c oxidase. Thus, while there is a need for some copper in the periplasm, the periplasm also appears to be the primary target of copper-mediated damage making protective mechanisms necessary (11).
Efflux systems which remove excess copper are a common protective measure for the cells. The E. coli CusCFBA system is a Cu(I)/Ag(I) efflux system that pumps Cu(I) and Ag(I) to the extracellular space using the proton gradient across the inner membrane as an energy source (12). Three proteins in this system, CusA, CusB, and CusC, are members of the RND-(Resistance, Nodulation, Division) or CBA-type of efflux systems (13). The most-well studied members of this family have been those involved in multidrug resistance (e.g. AcrAB-TolC), and the metal resistance systems are thought to have many similarities in structure and function to these systems. CusA, CusB and CusC are expected to form a protein complex that spans the inner and outer membranes. Pathways for substrate efflux have not been directly determined in these kinds of systems, but studies of multidrug resistance systems suggest substrate uptake is likely from the periplasmic space or potentially from within the inner membrane in the case of hydrophobic compounds (14, 15). In the Cus system, metal binding by the periplasmic protein CusB serves an essential role in Cus-mediated resistance (16), and thus CusB could potentially provide a direct entry point for metal substrates into the efflux complex. In monovalent metal resistance systems there is a fourth component, CusF, which is a small periplasmic metal binding protein of unknown function. CusF could act as a metallochaperone to deliver metal to the Cus system for removal from the periplasm, or alternately it could serve as a metal dependent regulator of the CusCBA complex (17).
A chaperone for an efflux system could play a dual protective role through both sequestration of metal and through enhancing delivery of metal for subsequent export. In this work we demonstrate specific and direct metal transfer between CusF and CusB, providing evidence for CusF as a metallochaperone. CusF could thus play a role in protecting the periplasmic compartment from metal-induced damage by both limiting the free copper in the periplasmic space and aiding in removal of copper from the cell.
MATERIALS AND METHODS
Protein Expression and Purification
The expression and purification of CusF and CusB in Escherichia coli was performed as described previously (16, 17) except as noted below. For X-ray absorption spectroscopy (XAS) studies, selenomethionine-labeled CusF (SeM-CusF) was produced from E. coli BL21-(λDE3) cells containing the pASKIBA3/cusF plasmid grown in M9 minimal medium supplemented with L-selenomethionine, leucine, isoleucine and valine at 50 mg/L and lysine, phenylalanine and threonine at 100 mg/L (18). All the buffers used for the purification of SeM-CusF contained 10 mM dithiothreitol.
Plasmid pMG101 from S. typhimurium was used to amplify the silF gene. The primers used for the PCR reaction contained unique restriction sites at the 5′ end (EcoRI) and 3′ end (XhoI). After restriction enzyme digestion of the PCR product, it was ligated into the pASK-IBA3 (IBA, Germany) vector. The result was a construct, pASK-silF8–94, encoding the SilF protein lacking the signal peptide sequence and the first 7 residues at the N-terminus. The protein produced from this construct contains an additional 7 residues at the N-terminus encoded by the plasmid.
The pASK-silF8–94 plasmid was transformed into E. coli BL21-(λDE3). Cells were grown in LB media containing 100 μg/mL ampicillin at 37 °C until an O.D.600 of 0.6–1.0 was reached. Cells were induced with 200 μg/L of anhydrotetracycline (AHT) and grown at 30 °C for another 6–8 hours. Cells were harvested by centrifugation and frozen at −20 °C. The cell pellet was thawed and resuspended in approximately 75 mL of 100 mM Tris (pH 8.0), 150 mM NaCl buffer per liter of cell culture. The procedure used for processing of the cell pellet, including lysis and centrifugation was the same as that used for the CusF and CusB purifications. The supernatant obtained from centrifugation of the cell lysate was dialyzed against 60 mM lactate (pH 3.5), 50 mM NaCl buffer. After 3 changes of dialysis buffer, the precipitated proteins were removed by centrifugation at 12000Xg and the resulting supernatant was loaded onto a HighPrep 16/10 Sepharose Fast Flow cation exchange column (Amersham) equilibrated with the lactate buffer used for dialysis. SilF was eluted using a linear gradient of 50–500 mM NaCl in 60 mM lactate buffer with a pH gradient of 3.5–5.0. Fractions containing SilF were identified using SDS-PAGE, combined, and concentrated to approximately 0.5 mL using Amicon concentrators with a 5 kDa molecular weight cut-off. The concentrated protein was applied to a S100 size exclusion column (Amersham) equilibrated with 100 mM phosphate (pH 8.0), 100 mM NaCl buffer. Aliquots of the fractions were run on SDS polyacrylamide gels and fractions >95% pure, as determined using Coomassie staining, were pooled and dialyzed in 50 mM cacodylate (pH 7.0) buffer. Proteins were concentrated as described above. Protein concentrations were determined using the BCA assay (Pierce Biotechnology).
Isothermal Titration Calorimetry (ITC)
ITC measurements were performed on a Microcal VP-ITC Microcalorimeter (Northampton, MA, USA), typically at 25 °C. To load proteins with Ag(I), AgNO3 solution in H2O from a 100X stock was added at approximately twice the concentration of the protein, followed by dialysis in 50 mM cacodylate (pH 7.0) to remove excess Ag(I). Protein in either the apo- or Ag(I)-bound state was concentrated using Amicon concentrators with a 5 kDa cutoff to the desired concentrations for the ITC experiments. The titrand and the titrant were thoroughly degassed in a ThermoVac apparatus (Microcal). For a titration experiment, approximately 1.5 mL of titrand protein was placed in a reaction cell and injected with titrant protein over 20 seconds. The first injection was 2 μL, and all subsequent injections were 10 μL. A total of 25 injections were made with 5-minute intervals between each injection. Titrations were performed of apo-CusF (375 μM) into apo-CusB (20 μM), Ag(I)-CusF (375 μM) into apo-CusB (24 μM), Ag(I)-CusF (375 μM) into Ag(I)-CusB (23 μM) and Ag(I)-CusB (142 μM) into apo-CusF (17 μM), where the concentrations given are the protein concentrations of the starting material. In order to ensure adequate mixing of the two proteins, the reaction cell was continuously stirred at 300 rpm. The heat due to dilution, mechanical effects and other non-specific effects was accounted for by averaging the last three points of the titration and subtracting that value from all data points (27). No attempt was made at fitting these data to obtain binding affinities or stoichiometries, since the experiments are expected to be reflective of both protein-protein interactions and metal binding/release.
For ITC experiments with SilF, Ag(I)-bound protein where needed was prepared as described above for CusF and CusB, and the following titrations were performed: AgNO3 (300 μM) into apo-SilF (36 μM), Ag(I)-SilF (170 μM) into apo-CusB (21 μM), Ag(I)-SilF (170 μM) into apo-CusF (29 μM), and Ag(I)-CusF (275 μM) into apo-SilF (30 μM). In each case, the buffer was 50 mM cacodylate, pH 7.0. As controls for all the ITC experiments, each protein was titrated into buffer, or buffer was titrated into each protein to determine the heat changes due to protein dilution. For the titration of apo-SilF with AgNO3, a single-site binding model was fitted to the data using the Origin software package (MicroCal). The software uses a non-linear least-squares algorithm and the concentrations of the titrant and the titrand to fit the enthalpy change per injection to an equilibrium binding equation. The binding enthalpy change ΔH, association constant Ka, and the binding stoichiometry n were permitted to float during the least-squares minimization process and taken as the best-fit values.
Preparation of Samples for X-Ray Absorption Spectroscopy (XAS)
CusB and SeM-CusF in 20 mM N-(2-acetamido)-2-aminoethanesulfonic acid (ACES), pH 7.0 were argon purged, then transferred to the anaerobic chamber. Ascorbate solution buffered at pH 7.0, which was prepared inside the anaerobic chamber, was added to the argon-purged proteins to a final concentration of 50 mM. CuCl2 was then added such that the final copper concentration was 25% in excess of the protein concentration. The proteins were dialyzed against 20 mM ACES, 10 mM ascorbate, pH 7.0, to remove unbound copper. The final concentration of protein was determined using the Bradford assay (19) (Biorad). For extended X-ray absorption fine structure (EXAFS) sample preparation, proteins were mixed with 20–30% ethylene glycol, transferred to EXAFS cuvettes and flash frozen in liquid nitrogen. The final EXAFS samples were SeM-CusF in apo- and Cu(I)-bound forms, at concentrations of 380 μM and 200 μM, respectively, Cu(I)-SeM-CusF mixed with apo-CusB and Cu(I)-CusB mixed with the apo-SeM-CusF. In the samples containing Cu(I)-SeM-CusF and apo-CusB, the final concentrations of both proteins were 110 μM. To measure different timepoints, three identical samples of CusF and CusB were prepared, which only differed in the amount of time they were mixed before adding ethylene glycol and flash freezing after either 4, 14 or 34 minutes. For the sample containing Cu(I)-bound CusB and apo-SeM-CusF, the final concentration of apo-SeM-CusF was 85 μM and that of Cu(I)-CusB was 125 μM.
XAS Data Collection and Analysis
Cu K-edge (8.980 keV) and Se K-edge (12.658 keV) EXAFS data for CusF and CusB were collected at the Stanford Synchrotron Radiation Laboratory operating at 3 GeV with currents between 100 and 50 mA. All samples were measured on beam line 9–3 by use of a Si[220] monochromator and a Rh-coated mirror upstream of the monochromator with a 13 keV (Cu) or 15 keV (Se) energy cutoff to reject harmonics. A second Rh mirror downstream of the monochromator was used to focus the beam. Data were collected in fluorescence mode on a high-count-rate Canberra 30-element Ge array detector with maximum count rates below 120 kHz. A 6 μ Z-1 metal oxide (Ni, As) filter and Soller slit assembly were placed in front of the detector to reduce the elastic scatter peak. Four to six scans of a sample containing only sample buffer (50 mM sodium phosphate, pH 7.2) were collected at each absorption edge, averaged, and subtracted from the averaged data for the protein samples to remove Z-1 Kβ fluorescence and produce a flat pre-edge baseline. This procedure allowed data with an excellent signal-to-noise ratio to be collected down to 100 μM total copper in the sample. The samples (80 μL) were measured as aqueous glasses (>20% ethylene glycol) at 10–15 K. Energy calibration was achieved by reference to the first inflection point of a copper metal foil (8980.3 eV) for Cu K-edges and a selenium metal foil (12658.0 eV) for Se K-edges, placed between the second and third ionization chamber. Data reduction and background subtraction were performed with the program modules of EXAFSPAK (8). Data from each detector channel were inspected for glitches or drop-outs before inclusion in the final average. Spectral simulation was carried out with the program EXCURVE 9.2 (20–23) as previously described using 8985 and 12663 eV for the start of the EXAFS in k space (k=0) for Cu and Se EXAFS respectively (24).
For Se EXAFS analysis of the sample of apo-CusB added to Cu(I)-SeM-CusF, the Se-Cu coordination number was allowed to vary, while its DW factor was held constant at 0.005 Å2. Small variations in metrical parameters (RSe-C, RSe-Cu, RSe-Se) were permitted. The analysis led to excellent fits with 50 percent reduction in Se-Cu shell occupancy and a small increase (~0.02 Å) in the Se-Cu bond length, but no difference in Se-C or Se-Se interactions.
RESULTS
Metal dependent interaction of CusF and CusB
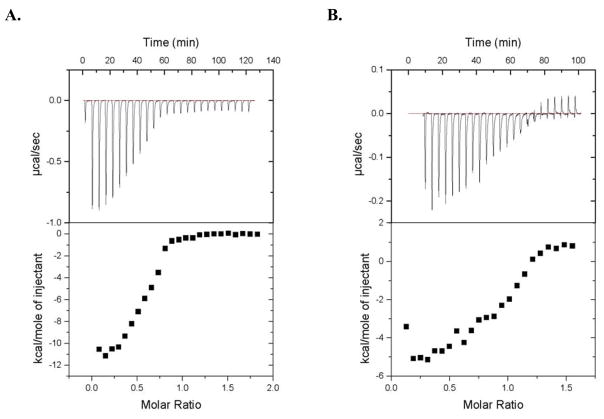
To establish whether CusF could play a role as a metallochaperone, we first tested for evidence of an interaction between the two proteins in vitro using isothermal titration calorimetry, a technique that detects the heat absorbed or released during a binding event (i.e. the binding enthalpy change). The titration of Ag(I)-CusF into the solution of apo-CusB (Fig. 1a) and the reverse titration of Ag(I)-CusB into apo-CusF (Fig. 1b) both showed significant changes in enthalpy. Because the observed enthalpy change could include contributions from both metal binding and protein-protein interactions, no attempt was made to extract binding affinities from the ITC data. An enthalpy change was not observed in the control titrations of Ag(I)-CusF into buffer or buffer into apo-CusB (data not shown). Additionally, no change in the heat absorbed or released was detected upon titration of apo-CusF into apo-CusB (Supplementary Fig. 1a) demonstrating that the observed interaction depended on the presence of metal ion. Furthermore, titration of Ag(I)-CusF into Ag(I)-CusB also showed no change in enthalpy (Supplementary Fig. 1b), indicating that an enthalpy change for this system requires one of the two proteins to exist in the apo-form.
Figure 1.
Isothermal titration calorimetry of CusF and CusB. (A) Ag(I)-CusF (375 μM) titrated into apo-CusB (24 μM); (B) Ag(I)-CusB (142 μM) titrated into apo-CusF (17 μM).
Transfer of copper between CusF and CusB
If CusF functions as a metallochaperone, it should be able to transfer metal to CusB. To observe direct transfer of metal between the two proteins, a novel X-ray absorption spectroscopy (XAS) technique was developed to follow the copper environment in samples of CusF and CusB. XAS is a spectroscopic technique that provides both electronic structure and atomic resolution molecular structure information, acting as a bridge between molecular and electronic structure techniques. CusF and CusB both have 3-coordinate metal sites, with CusF having 2 sulfur ligands from methionines and 1 nitrogen ligand from a histidine (25, 26) and CusB having 3 sulfur ligands from methionines (16). To distinguish the ligands arising from the methionines of each protein, selenomethionine was incorporated in CusF in place of methionine (SeM-CusF). Extended X-Ray Absorption Fine Structure (EXAFS) data at the Se edge were collected for apo-SeM-CusF and Cu(I)-SeM-CusF (Table 1 and Supplementary Figs. 2 and 3). The Fourier transform (FT) spectra show the expected features for the apo and Cu(I)-bound proteins as anticipated from the previous EXAFS characterization of CusF without SeM incorporation (17). The similarity of the Se-Se distance in the apo and Cu(I) bound proteins implies that the relative positions of the Se atoms in the metal binding site does not change when Cu(I) binds and that the site is therefore pre-formed for metal binding. The Cu EXAFS data similarly support the structural description of the metal binding site with Cu(I) bound by 1 His and 2 SeM ligands (Table 1 and Supplementary Fig. 4).
Table 1.
Fits obtained to the EXAFS of CusF and CusB by curve-fitting using the program EXCURVE 9.2
| Fa | Noc | R (Å)d | DW (Å2) | Noc | R (Å)d | DW (Å2) | Noc | R (Å)d | DW (Å2) | −E0 | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Cu-edge | C-N(His)b | Cu-Se | Cu-S | ||||||||
| Cu(I)-CusF | 0.364 | 1 | 2.01 | 0.003 | 2.0 | 2.41 | 0.009 | 4.75 | |||
|
| |||||||||||
| Cu(I)-CusF+ apo-CusB | |||||||||||
| 4 mins | 0.336 | 0.48 | 2.05 | 0.003 | 0.96 | 2.43 | 0.005 | 1.57 | 2.29 | 0.009 | 5.07 |
| 14 mins | 0.374 | 0.62 | 2.06 | 0.002 | 1.24 | 2.43 | 0.007 | 1.14 | 2.29 | 0.006 | 6.98 |
| 34 mins | 0.444 | 0.49 | 2.08 | 0.003 | 0.99 | 2.42 | 0.005 | 1.52 | 2.28 | 0.010 | 5.32 |
|
| |||||||||||
| Se-edge | Se-C(Met) | Se-Cu | Se-Se | ||||||||
|
| |||||||||||
| apo-CusF | 0.518 | 2.0 | 1.96 | 0.004 | 1.0 | 2.84 | 0.036 | 5.58 | |||
|
| |||||||||||
| Cu(I)-CusF | 0.545 | 2.0 | 1.96 | 0.005 | 0.5 | 2.41 | 0.005 | 1.0 | 2.85 | 0.030 | 5.98 |
|
| |||||||||||
| Cu(I)-CusF+ apo-CusB | |||||||||||
| 4 mins | 0.522 | 2.0 | 1.97 | 0.005 | 0.30 | 2.42 | 0.005 | 1.0 | 2.84 | 0.030 | 6.13 |
| 14 mins | 0.548 | 2.0 | 1.96 | 0.006 | 0.26 | 2.43 | 0.004 | 1.0 | 2.84 | 0.029 | 5.62 |
| 34 mins | 0.675 | 2.0 | 1.96 | 0.004 | 0.29 | 2.43 | 0.005 | 1.0 | 2.83 | 0.032 | 5.62 |
|
| |||||||||||
| Cu(I)-CusB+ apo-CusF | 0.880 | 2 | 1.96 | 0.005 | 0.3 | 2.43 | 0.005 | 1.0 | 2.85 | 0.029 | 5.98 |
F is a least-squares fitting parameter defined as
Fits modeled histidine coordination by an imidazole ring, which included single and multiple scattering contributions from the second shell (C2/C5) and third shell (C3/N4) atoms respectively. The Cu-N-Cx angles were as follows: Cu-N-C2 126°, Cu-N-C3 −126°, Cu-N-N4 163°, Cu-N-C5 −163°.
Coordination numbers are generally considered accurate to ± 25%. The fractional coordination numbers listed in the Table represent the results obtained from allowing the coordination numbers to vary using the constraints NCu-N = 0.5*NCu-Se, NCu-S = 3 – 1.5*NCu-Se (see text). While this approach overestimates the accuracy obtainable from the simulations, we have retained the two significant digit format to ensure consistency with the formula for the imposed constraints.
In any one fit, the statistical error in bond-lengths is ±0.005 Å. However, when errors due to imperfect background subtraction, phase-shift calculations, and noise in the data are compounded, the actual error is probably closer to ±0.02 Å.
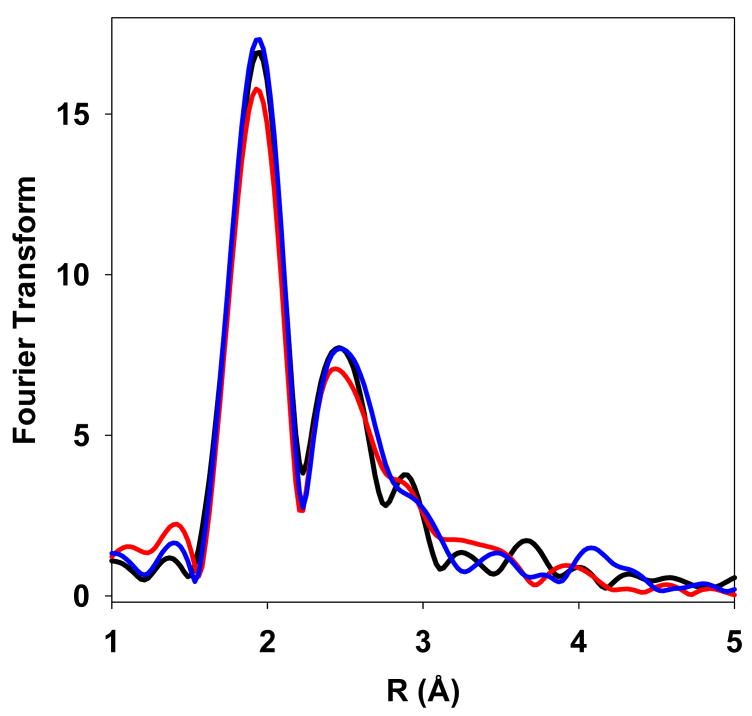
The addition of apo-CusB to a Cu(I)-SeM-CusF sample caused significant changes in the Se and Cu EXAFS spectra (Supplementary Fig. 5). The most dramatic effects are seen in the FT of the Se EXAFS where a significant decrease in the intensity of the Se-Cu peak is observed, while the intensity of the Se-C peak (Fig. 2a) remains unchanged. The Se-C interaction derives from the methyl and methylene carbons from the methionine side chain and thus acts as an internal standard for comparing intensities of Fourier transforms. Since CusF, but not CusB, is Se-labeled, the decrease in Se-Cu measured by Se EXAFS is the result of loss of Cu(I) specifically from the CusF binding site. Spectral fitting showed transfer of ~50 % of the Cu(I) out of the Se environment. This distribution likely reflects the similar affinities of CusF and CusB for metal (16, 27). These results demonstrate the unique strength of SeM substitution coupled to Se-XAS analysis as a ligand-directed monitor of metal transfer reactions.
Figure 2.
EXAFS data of SeM-CusF and CusB mixtures. (A) Comparison of the Fourier transforms of EXAFS data measured at the Se edge of Cu(I)-SeM-CusF (red), Cu(I)-SeM-CusF mixed with apo-CusB for 34 minutes before freezing (blue) and apo-SeM-CusF (black). (B) Comparison of the Fourier transforms of EXAFS data measured at the Cu edge of Cu(I)-SeM-CusF (black), and Cu(I)-SeM-CusF mixed with apo-CusB for 34 minutes before freezing (red).
Analysis of the Cu EXAFS provided an additional probe of metal transfer. In previous work we have reported that CusB binds Cu(I) in an all-S environment, with coordination to 3 methionine residues with Cu-S bond lengths of 2.28 –2.30 Å (16). Copper transfer from CusF to CusB should therefore be accompanied by loss of Cu-N and Cu-Se (the ligands to CusF) and replacement by the CuS (Met) environment of CusB. Knowledge of the binding site environment of both donor and acceptor protein allowed us to set constraints on the coordination numbers NCu-N and NCu-S as follows:
The data were analyzed by imposing these constraints and allowing only the Cu-Se coordination number, NCu-Se, to vary, with small variations in distances permitted. The result of these simulations is that the Cu-Se coordination number dropped by 50 percent, exactly equivalent to the change observed at the Se edge, while an appropriate increase in Cu-S shell occupancy at R = 2.29 Å was observed (Table 1). The FT of the Cu EXAFS data is shown in Fig. 2b. While not as easily detectable as the changes observed at the Se edge, transfer from CusF to CusB is evident from the shift in the main transform peak to lower R value, as the Cu transfers out of the Cu-Se (R=2.4 Å) environment of CusF into the Cu-S (R=2.3 Å) environment of CusB. The Cu EXAFS data are thus consistent with 50 percent Cu(I) transfer into the all-S environment of the CusB protein, with a Cu-S distance equal to that obtained previously from studies on the fully reconstituted Cu(I)-CusB protein (16).
In addition to the sample described above, which was incubated for 34 minutes before freezing, two additional samples, incubated for 4 and 14 minutes before freezing, were made. In both cases, the extent of transfer was close to 50 percent and the Se data were identical to the 34 minute sample (Fig. 3, Table 1, and Supplementary Fig. 5). This result indicates that metal transfer is relatively rapid and proceeds to an end point of equal Cu(I) distribution between donor and acceptor in less than 4 minutes (the time taken to mix and freeze the solution).
Figure 3.
Comparison the Fourier transforms of EXAFS data measured at the Se edge for samples of Cu(I)-SeM-CusF mixed with apo-CusB for different times before freezing: 4 minutes (black), 14 minutes (red), and 34 minutes (blue)
To examine whether metal transfer can directly occur in the opposite direction from CusB to CusF, we monitored the Cu and Se EXAFS of a sample of Cu(I)-CusB mixed with apo-CusF. The spectra show the loss of the Cu(I)-S environment of CusB and replacement by the Cu(I)-Se environment of SeM-CusF (Fig. 4). Again, Cu(I) shows an approximately 50 percent distribution between the CusB and CusF proteins. Therefore, Cu(I) transfer between CusF and CusB is reversible and proceeds to an equivalent end-point regardless of which protein is pre-loaded with Cu(I).
Figure 4.
EXAFS data of Cu(I)-CusB mixed with apo-SeM-CusF measured at the Se edge. Experimental (black traces) versus simulated (red traces) FT and EXAFS (inset) using metrical parameters listed in Table 1.
Interaction and metal transfer between CusF and CusB is specific
To confirm that metal is directly transferred between CusF and CusB, not released into solution and re-bound by the proteins, we performed ITC experiments with a homolog of CusF, SilF, which has 51% sequence identity to CusF, and binds Ag(I) with similar affinity (Kd = 35 nM) (Supplementary Fig. 6a). The titrations of both Ag(I)-CusF into apo-SilF and Ag(I)-SilF into apo-CusF show no significant enthalpy changes (Supplementary Fig. 6b,c), indicating no metal transfer between these proteins.
Importantly, we determined that the homolog of CusF, SilF, is unable to productively interact with CusB. Using ITC, no enthalpy changes are observed during the course of a titration of Ag(I)-SilF into apo-CusB (Supplementary Fig. 6d). This result implies that specific recognition is required for the metal transfer event and that metal transfer is direct between proteins of the Cus system. This high degree of specificity in the transfer process may serve as a protective mechanism to ensure that metals remain sequestered among the Cus proteins and help prevent the toxic effects of free copper or silver.
DISCUSSION
Our results demonstrate a role for CusF as a periplasmic metallochaperone for a copper efflux system. Through isothermal titration calorimetry experiments we have demonstrated that interactions only occur between CusF and CusB in the presence of metal. A close homolog of CusF is unable to interact with or transfer metal to CusB, thus demonstrating a high degree of specificity in the interaction. Using selenomethionine-labeled protein to distinguish the metals sites of the two proteins in XAS experiments, we were able to unequivocally show the direct transfer of Cu(I) from one protein to the other. Periplasmic metallochaperones, in analogy to eukaryotic metallochaperones, play a dual protective role within the cell. By sequestering metal within the confines of a protein, coupled with specific transfer to a target protein, the cytotoxic effects of the free metal can be avoided.
CusF may provide specificity for monovalent metal resistance systems that is lacking in other RND- or CBA-type exporters. The homologous multidrug resistance systems, which lack an equivalent of CusF, have very broad substrate specificity, leading to their description as “periplasmic vacuum cleaners” (28). In addition to the sequestration and delivery function implied by the identification of CusF as a metallochaperone, the CusF protein may play a role in specific substrate selection, so as to not remove needed metals from the cell.
We have shown that metal transfer occurs in both directions between CusF and CusB and proceeds to a 50% equilibrium distribution in vitro. In light of these findings, how could CusF effectively function as a metallochaperone? Because the affinities of the individual proteins are similar, in a system where only the two purified components are present at equilibrium the metal is expected to partition equally between the two proteins, as we have experimentally observed. This shallow thermodynamic gradient is similar to what has been previously reported for a yeast copper chaperone and the regulatory domain of a copper-transporting ATPase (29). In the case of the Cus system, in the cell where the complete efflux complex is present and where energy from the proton gradient can be utilized to drive transport, our model is that metal will be continually removed from CusB and therefore unidirectional transfer of metal from CusF to CusB will occur, even in the case of a shallow thermodynamic gradient. Consistent with this, in another system where the transport site is present, transfer from the chaperone to an ATPase is essentially irreversible (30). In this model, CusF could efficiently function as a metallochaperone though both proteins have similar binding affinities for metal and metal transfer can occur in both directions.
The site of metal entry into the efflux complex is not yet known, however, metal transfer from CusF to CusB provides evidence for entry of substrates from the periplasm to the RND-efflux system. In homologous multidrug resistance systems, evidence suggests that the substrates are bound by the periplasmic domains of inner membrane proteins (14, 15, 31), with no substrate binding function by the periplasmic component equivalent to CusB. For hydrophobic substrates dissolved in the inner membrane, this pathway would make sense. Though at this point a regulatory role for metal binding by CusB cannot be ruled out, in the metal efflux system the substrates are likely to be entering the complex from the aqueous periplasmic space, and thus the transfer of metal from CusF to CusB may be the direct point of entry for the transport system.
Supplementary Material
Supplementary Figs. 1, 2, 3, 4, 5, and 6
This material is available free of charge on the Internet at http://pubs.acs.org
Abbreviations
- AHT
anhydrotetracycline
- EXAFS
extended X-ray absorption fine structure
- ITC
isothermal titration calorimetry
- RND
resistance, nodulation, division
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- SeM-CusF
selenomethionine-labeled CusF
- XAS
x-ray absorption spectroscopy
Footnotes
We gratefully acknowledge the use of facilities at the Stanford Synchrotron Radiation Laboratory, which is supported by the National Institutes of Health Biomedical Research Technology Program, Division of Research Resources, and by the U.S. Department of Energy, Basic Energy Sciences (BES) and Office of Biological and Environmental Research (OBER). This work was supported by grants from the National Institutes of Health GM54803 and PO1 GM067166 to N.J.B. and GM079192 to M.M.M.
References
- 1.Nelson N. Metal ion transporters and homeostasis. EMBO J. 1999;18:4361–4371. doi: 10.1093/emboj/18.16.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong MD, Fan B, Rosen BP. Handbook of ATPases: Biochemistry, Cell Biology, Pathophysiology. Weinheim: Wiley-VCH; 2004. [Google Scholar]
- 3.Singleton C, Le Brun NE. Atx1-like chaperones and their cognate P-type ATPases: copper-binding and transfer. Biometals. 2007;20:275–289. doi: 10.1007/s10534-006-9068-1. [DOI] [PubMed] [Google Scholar]
- 4.Poulos TL. Helping copper find a home. Nat Struc Biol. 1999;6:709–711. doi: 10.1038/11464. [DOI] [PubMed] [Google Scholar]
- 5.Culotta VC, Lin SJ, Schmidt P, Klomp LWJ, Casareno RLB, Gitlin J. Intracellular pathways of copper trafficking in yeast and humans. Adv Exp Med Biol. 1999;448:247–254. doi: 10.1007/978-1-4615-4859-1_22. [DOI] [PubMed] [Google Scholar]
- 6.O’Halloran TV, Culotta VC. Metallochaperones, an intracellular shuttle service for metal ions. J Biol Chem. 2000;275:25057–25060. doi: 10.1074/jbc.R000006200. [DOI] [PubMed] [Google Scholar]
- 7.Rosenzweig AC. Copper delivery by metallochaperone proteins. Acc Chem Res. 2001;34:119–128. doi: 10.1021/ar000012p. [DOI] [PubMed] [Google Scholar]
- 8.Finney LA, O’Halloran TV. Transition metal speciation in the cell: insights from the chemistry of metal ion receptors. Science. 2003;300:931–6. doi: 10.1126/science.1085049. [DOI] [PubMed] [Google Scholar]
- 9.Culotta VC, Yang M, O’Halloran TV. Activation of superoxide dismutases: Putting the metal to the pedal. Biochim Biophys Acta-Mol Cell Res. 2006;1763:747–758. doi: 10.1016/j.bbamcr.2006.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cobine PA, Pieffel F, Winge DR. Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim Biophys Acta-Mol Cell Res. 2006;1763:759–772. doi: 10.1016/j.bbamcr.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 11.Macomber L, Rensing C, Imlay JA. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol. 2007;189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franke S, Grass G, Rensing C, Nies DH. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J Bacteriol. 2003;185:3804–3812. doi: 10.1128/JB.185.13.3804-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tseng TT, Gratwick KS, Kollman J, Park D, Nies DH, Goffeau A, Saier MH., Jr The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J Mol Microbiol Biotechnol. 1999;1:107–25. [PubMed] [Google Scholar]
- 14.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443:173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 15.Seeger MA, Schiefner A, Eicher T, Verrey F, Diederichs K, Pos KM. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313:1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 16.Bagai I, Liu W, Rensing C, Blackburn NJ, McEvoy MM. Substrate-linked conformational change in the periplasmic component of a Cu(I)/Ag(I) efflux system. J Biol Chem. 2007;282:35695–35702. doi: 10.1074/jbc.M703937200. [DOI] [PubMed] [Google Scholar]
- 17.Loftin IR, Franke S, Roberts SA, Weichsel A, Heroux A, Montfort WR, Rensing C, McEvoy MM. A novel copper-binding fold for the periplasmic copper resistance protein CusF. Biochemistry. 2005;44:10533–40. doi: 10.1021/bi050827b. [DOI] [PubMed] [Google Scholar]
- 18.Doublie S. Preparation of selenomethionyl proteins for phase determination. Macromol Cryst, Pt A. 1997;276:523–530. [PubMed] [Google Scholar]
- 19.Bradford MM. Rapid and Sensitive Method for Quantitation of Microgram Quantities of Protein Utilizing Principle of Protein-Dye Binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Gurman SJ, Binsted N, Ross I. A rapid, exact, curved-wave theory for EXAFS calculations. J Phys Chem. 1984;17:143–151. [Google Scholar]
- 21.Gurman SJ, Binsted N, Ross I. A rapid, exact, curved-wave theory for EXAFS calculations, II. The multiple-scattering contributions. J Phys Chem. 1986;19:1845–1861. [Google Scholar]
- 22.Binsted N, Gurman SJ, Campbell JW. Daresbury Laboratory. Warrington; England: 1998. [Google Scholar]
- 23.Binsted N, Hasnain SS. State-of-the-art analysis of whole X-ray absorption spectra. J Synch Rad. 1996;3:185–196. doi: 10.1107/S0909049596005651. [DOI] [PubMed] [Google Scholar]
- 24.Blackburn NJ, Ralle M, Hassett R, Kosman DJ. Spectroscopic analysis of the trinuclear cluster in the Fet3 protein from yeast, a multinuclear copper oxidase. Biochemistry. 2000;39:2316–24. doi: 10.1021/bi992334a. [DOI] [PubMed] [Google Scholar]
- 25.Loftin IR, Franke S, Blackburn NJ, McEvoy MM. Unusual Cu(I)/Ag(I) coordination of Escherichia coli CusF as revealed by atomic resolution crystallography and X-ray absorption spectroscopy. Prot Sci. 2007;16:2287–2293. doi: 10.1110/ps.073021307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xue Y, Davis AV, Balakrishnan G, Stasser JP, Staehlin BM, Focia P, Spiro TG, Penner-Hahn JE, O’Halloran TV. Cu(I) recognition via cation-pi and methionine interactions in CusF. Nat Chem Biol. 2008;4:107–9. doi: 10.1038/nchembio.2007.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kittleson JT, Loftin IR, Hausrath AC, Engelhardt KP, Rensing C, McEvoy MM. Periplasmic metal-resistance protein CusF exhibits high affinity and specificity for both Cu-I and Ag-I. Biochemistry. 2006;45:11096–11102. doi: 10.1021/bi0612622. [DOI] [PubMed] [Google Scholar]
- 28.Lomovskaya O, Zgurskaya HI, Totrov M, Watkins WJ. Waltzing transporters and ‘the dance macabre’ between humans and bacteria. Nat Rev Drug Disc. 2007;6:56–65. doi: 10.1038/nrd2200. [DOI] [PubMed] [Google Scholar]
- 29.Huffman DL, O’Halloran TV. Energetics of copper trafficking between the Atx1 metallochaperone and the intracellular copper transporter, Ccc2. J Biol Chem. 2000;275:18611–4. doi: 10.1074/jbc.C000172200. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez-Guerrero M, Arguello JM. Mechanism of Cu+-transporting ATPases: soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc Natl Acad Sci U S A. 2008;105:5992–7. doi: 10.1073/pnas.0711446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu EW, Aires JR, McDermott G, Nikaido H. A periplasmic drug-binding site of the AcrB multidrug efflux pump: a crystallographic and site-directed mutagenesis study. J Bacteriol. 2005;187:6804–6815. doi: 10.1128/JB.187.19.6804-6815.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figs. 1, 2, 3, 4, 5, and 6
This material is available free of charge on the Internet at http://pubs.acs.org