Abstract
Summary: There has been a great expansion in the number of small regulatory RNAs identified in bacteria. Some of these small RNAs repress the synthesis of potentially toxic proteins. Generally the toxin proteins are hydrophobic and less than 60 amino acids in length, and the corresponding antitoxin small RNA genes are antisense to the toxin genes or share long stretches of complementarity with the target mRNAs. Given their short length, only a limited number of these type I toxin-antitoxin loci have been identified, but it is predicted that many remain to be found. Already their characterization has given insights into regulation by small RNAs, has suggested functions for the small toxic proteins at the cell membrane, and has led to practical applications for some of the type I toxin-antitoxin loci.
INTRODUCTION
It has become clear that RNA molecules act as regulators in all organisms in which they have been characterized. In bacteria, these regulatory RNAs are generally referred to as small RNAs (sRNAs), given that most are between 50 and 200 nucleotides in length. The first regulatory sRNAs to be discovered were plasmid encoded, where they are required for plasmid replication or maintenance. Chromosomally encoded sRNAs were initially detected due to their abundance or found by serendipity, though in recent years there have been an increasing number of systematic screens for these molecules (reviewed in references 3 and 33).
The sRNAs that have been characterized act by two general mechanisms (reviewed in reference 50). A small number of sRNAs bind proteins and modify their activities. The other sRNAs function by base pairing with target mRNAs. Base pairing can lead to changes in gene expression by altering the stability and/or translation of the target. The majority of the characterized chromosomally encoded sRNAs act by base pairing with targets that have limited complementarity (generally 6 to 12 contiguous nucleotides). In contrast, most of the sRNAs carried on plasmids are encoded on the antisense strand relative to their targets and have extensive complementarity with the mRNA. Until recently only a limited number of chromosomally encoded sRNAs with potential for extensive base pairing with their target mRNAs were known, but an increasing number are being discovered. Intriguingly, most of these sRNAs repress the expression of proteins that are under 60 amino acids in length, highly hydrophobic, and toxic at high levels. These mRNA-sRNA pairs, which have been classified as type I toxin-antitoxins (27), are the focus of this review.
Before examining type I toxin-antitoxins in depth, it is worth summarizing what is known about type II toxin-antitoxins (reviewed in references 8, 17, and 27). While the toxin is a protein in both cases, the antitoxin for type II systems is also a protein, in contrast to the RNA antitoxin associated with type I systems. In general, much more is known about the type II toxin-antitoxin modules. The protein antitoxin, which is labile, binds to the more stable toxin and inhibits its activity. For cases in which the toxin-antitoxin pairs are encoded on plasmids, these modules prevent the growth of plasmid-free cells, conferring what has been denoted plasmid addiction or postsegregational killing. If a plasmid is lost, the antitoxin is degraded, and with no new synthesis of the antitoxin, the toxin is released from inhibition, leading to killing of the plasmid-free cells. The roles of the chromosomally encoded type II toxin-antitoxin modules are less well defined, although the genes are surprisingly abundant. Mycobacterium tuberculosis, for example, has more than 30 pairs. In some cases, induction of the toxin genes has been shown to lead to growth arrest, which might allow quality control or persistence under times of stress. Others have reported that induction of the toxins leads to cell death, possibly providing nutrients to neighboring cells. The toxins potentially could also contribute toward stabilizing adjacent regions of the bacterial chromosome, analogous to their role in plasmids. The biochemical activities of several of the type II toxin proteins, which are generally around 100 amino acids in length, have been characterized and fall into several general classes, including inhibition of DNA gyrase (for example, CcdB and ParE) and RNA cleavage (for example, MazF).
While type I toxin-antitoxin loci have been studied less extensively than the type II loci, recent studies summarized below indicate that further identification and characterization of both the antitoxin sRNAs and the small toxin proteins promise to be exciting directions for future research.
DISCOVERY OF TYPE I TOXIN-ANTITOXIN LOCI
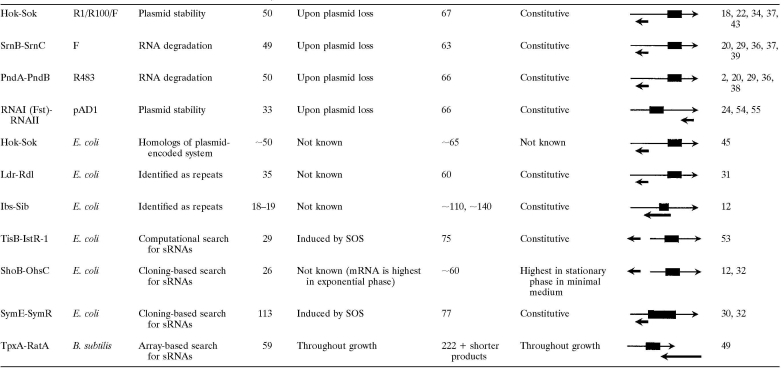
The properties of the identified type I toxin-antitoxin pairs are listed in Table 1. Some pairs are found on both plasmids and chromosomes, while other are exclusively plasmid or chromosomally encoded. In most cases, the protein toxin and RNA antitoxin are encoded on opposite strands, with the overlap occurring at either the 5′ end or the 3′ end of the mRNA transcript. For two pairs, the mRNA and sRNA are encoded divergently in the same intergenic region but share 19 and 23 nucleotides of contiguous complementarity.
TABLE 1.
Characteristics of type I toxin-antitoxin systems
The mRNA is shown as a thin arrow and the sRNA as a thick arrow. The toxin open reading frame is indicated by a box. The sizes are approximate.
Plasmid-Encoded Type I Toxin-Antitoxin
The Hok-Sok system of R1 plasmids was the first type I toxin-antitoxin pair to be discovered through the characterization of a locus that stabilized various plasmids in gram-negative bacteria (18). Due to this role in plasmid partitioning, the locus was first denoted “par” (18), but it was renamed when further characterization showed that one gene encoded a small protein capable of host killing (Hok) and a second gene was capable of suppression of host killing (Sok) (21). Homologs of Hok-Sok are found on the F plasmid, where the locus was denoted flm (F leading maintenance) (34). Two other loci encoding Hok-Sok-like systems, SrnB-SrnC (stable RNA negative) of the F plasmid (37, 39) and PndA-PndB (promotion of nucleic acid degradation) of plasmid R483 (2, 38), were noted when it was observed that the addition of rifampin led to membrane damage, RNase I influx from the periplasm, degradation of stable RNA, and cell killing (29). It was subsequently shown that the arrangement of genes was very similar to that for Hok-Sok (20), with srnB and pndA encoding toxin proteins containing single transmembrane domains and srnC and pndB encoding small antisense RNAs (36). RNAI-RNAII encoded on the pAD1 plasmid of Enterococcus faecalis, the first type I toxin-antitoxin pair found for a gram-positive bacterium, was identified on the basis of a postsegregational killing phenotype (54, 55). For this pair, RNAI encodes the small Fst (E. faecalis plasmid-stabilizing toxin), protein while RNAII functions as the regulatory sRNA (24).
Chromosomally Encoded Type I Toxin-Antitoxin
Homologs of the R1 plasmid-carried Hok-Sok locus were noted on the Escherichia coli chromosome (45) and subsequently on the chromosomes of other enteric species (11). Interestingly, all of the hok-sok genes appear to have degenerated with mutations and transposon insertions in E. coli K-12 but are intact in other E. coli strains (45).
Two families of chromosomally encoded type 1 toxin-antitoxin pairs were initially identified as genomic repeat sequences. The long direct repeat (LDR) sequences are approximately 530 nucleotides in length, and each encodes an Ldr toxin and an Rdl antitoxin RNA (31). The LDR sequence is repeated four times with slight variation on the E. coli K-12 chromosome; there are three tandem repeats (LDR-A, LDR-B, and LDR-C) at one locus at 27.4 min and a single repeat (LDR-D) on the opposite side of the chromosome at 79.7 min. Various numbers of LDR repeat sequences are found in other closely related enteric bacteria; for example, three are present in E. coli O157:H7, and two are detected in Salmonella enterica serovar Typhimurium (11). Genomic repeat sequences of approximately 165 nucleotides were initially denoted the QUAD repeats in E. coli K-12 (48) but were subsequently renamed SIB (short, intergenic, abundant) sequences when it be became clear that a fifth repeat was present (12). As for the LDR sequences, the number of SIB repeats varies between strains; for example, seven SIB repeats are detected for E. coli O157:H7 EDL933, four repeats are predicted for E. coli CFT073, and three repeats are found in Salmonella enterica serovar Typhimurium. In all cases, the SIB repeats are in three locations on the chromosome, with expansion or contraction in the number of repeats at each locus. The toxin genes encoded by each of the repeat sequences have been named ibs (induction brings stasis), and the antitoxin genes have been named sib.
Four toxin-antitoxin pairs were discovered more recently in global searches for sRNAs. The gene encoding the IstR-1 RNA (inhibitor of SOS-induced toxicity by RNA) was noted in a computational search for promoter and terminator sequences in the intergenic regions of E. coli (4, 53). It is located upstream of and divergent from the ysdAB operon, whose expression is induced during the SOS response. When the ysdAB transcript was found to confer toxicity upon overexpression, the operon was renamed tisAB (toxicity-induced by SOS). The OhsC (oppression of hydrophobic open reading frame by sRNA) RNA was identified in a cloning-based screen for E. coli sRNAs, and the divergently located shoB (short hydrophobic open reading frame) gene was noted because this region had a 19-nucleotide region complementary to ohsC (12, 32). An sRNA denoted SymR (symbiotic RNA) and encoded opposite yjiW was cloned in the same screen. The yjiW transcript, which is also induced during the SOS response, was subsequently renamed symE (SOS-induced yjiW gene with similarity to MazE) (30). The final toxin-antitoxin pair, TxpA (toxic peptide)-RatA (RNA antitoxin) of Bacillus subtilis, was discovered in a microarray-based expression screen for sRNAs (49).
DISTRIBUTION OF TYPE I TOXIN-ANTITOXIN LOCI
While all chromosomally encoded type I toxin-antitoxin pairs have been characterized in the model organisms E. coli and B. subtilis, homologs of all of the type I toxins can be found in related bacteria (Table 2). The toxin with the broadest distribution is the Hok protein, possibly because many plasmids carry hok-sok genes that could serve as sources for copies incorporated into genomes. Interestingly, the Ibs proteins, which are conserved in some enteric species, also can be found in two Haemophilus species and in Mannheimia haemolytica (12).
TABLE 2.
Distribution of type I toxins found in enteric bacteria
| Genus | No. of toxic protein homologs identified in sequenced species
|
||||
|---|---|---|---|---|---|
| HokA | IbsA | TisB | ShoB | LdrD | |
| Escherichia | 4-15 | 3-7 | 1 | 1 | 2-7 |
| Shigella | 7-12 | 3-6 | 1 | 1 | 5-10 |
| Citrobacter | 0 | 1 | 1 | 0 | 0 |
| Salmonella | 0 | 3 | 1 | 0 | 2 |
| Enterobacter | 6 | 0 | 1 | 0 | 0 |
| Klebsiella | 2 | 0 | 1 | 0 | 0 |
| Serratia | 5 | 0 | 0 | 0 | 0 |
| Vibrio | 1 | 0 | 0 | 0 | 0 |
| Yersiniaa | 1 | 0 | 0 | 0 | 0 |
| Photobacterium | 1-2 | 0 | 0 | 0 | 0 |
| Photorhabdus | 3 | 0 | 0 | 0 | 0 |
| Shewanellaa | 1-2 | 0 | 0 | 0 | 0 |
| Haemophilus | 0 | 1 | 0 | 0 | 0 |
| Mannheimiaa | 0 | 1 | 0 | 0 | 0 |
Homologs were identified only in genome shotgun sequences in this genus. Thus, the total number of repeat sequences remains to be determined.
No global searches for type I toxin-antitoxins have been reported, but we predict that they will be found to be as broadly distributed as type II toxin-antitoxins. A possible reason for the paucity of identified type I loci is that type I toxins are smaller than the type II toxins and in some cases consist of only a short transmembrane helix, making reliable prediction difficult. In addition, the hydrophobic properties of the toxins could be maintained even with substantial sequence divergence.
It is worth considering how one might systematically search for these genes. Many of the chromosomally encoded pairs were discovered during systematic searches for sRNAs. As these searches are carried out in a wide range of bacteria, additional candidates for type I toxin-antitoxins will likely be found. Any putative sRNA locus which is present as a repeated sequence, for which there is extensive complementarity with a neighboring gene, or for which transcripts can be detected for both strands should be viewed as a possible component of a type I toxin-antitoxin system. In fact, it is probably worthwhile to scan the opposite strands of all sRNA genes for the hallmarks of the small hydrophobic open reading frames.
Both the Ldr and Ibs genes are repeated multiple times on the E. coli chromosome and are found in various numbers in closely related species. What evolutionary pressures could force the cell to maintain so many copies of potentially toxic genes? Possibly the different copies are required under slightly different growth conditions. Alternatively, the cell may require a “threshold” of protein activity to be reached. If the regulation is particularly tight, induction or derepression of multiple genes could rapidly increase the amount of the Ldr or Ibs proteins. In this case, what would be important is the sum of the different proteins, not the individual amounts.
REGULATION BY ANTITOXIN sRNAs
Thus far, many of the studies of the type I toxin-antitoxin loci have focused on the regulation by the antitoxin sRNA. In addition to providing insights into when the type I toxins are expressed, these studies have contributed to the understanding of the mechanisms of regulation by base pairing sRNAs. The simplest model for how antisense RNAs repress the synthesis of proteins is that base pairing across the ribosome binding site blocks translation and/or leads to mRNA degradation. However, as illustrated by some of the well-studied examples described below and shown in Fig. 1, the regulation can also be more intricate.
FIG. 1.
Models for three types of regulation of type I toxin-antitoxin loci. SD, Shine-Dalgarno sequences. Red denotes SDs that are blocked, green denotes SDs that are accessible, and black denotes SDs that are accessible upon ribosome binding to an upstream SD. The full-length hok and tisB mRNAs need to be processed (indicated by the broken circle) at the 3′ and 5′ ends, respectively, before they can be translated. This processing brings about a change in secondary structure required to make the critical SD accessible. In addition to blocking ribosome binding, the sRNAs promote cleavage (indicated by the scissors) of the mRNA, thus irreversibly inactivating the target.
Block in Ribosome Binding
A block in ribosome binding together with mRNA cleavage appears to be the mechanism by which SymR regulates the synthesis of the SOS-induced protein SymE (Fig. 1). SymR is encoded opposite the 5′ untranslated region (UTR) of SymE, and base pairing can extend over the ribosome binding site as well as the symE ATG start codon (30). A mutation that abolishes the symR promoter led to a sevenfold increase in the levels of the tagged protein and a threefold increase in the symE transcript, supporting the hypothesis that SymR blocks translation and probably also leads to symE mRNA degradation. A similar scenario is likely for Sib RNA regulation of Ibs protein synthesis. The Sib RNAs are complementary to the entire ibs coding sequences as well as the predicted ribosome binding sites (12). Synthesis of the conserved Ibs proteins has not been examined, but the ibsC mRNA could be detected only if the sibC promoter sequence was deleted, suggesting that base pairing normally leads to degradation of the target mRNA.
Processing and Block in Translation of an Overlapping Open Reading Frame
The type I toxin-antitoxin system for which the regulatory mechanism has been studied most extensively is the Hok-Sok pair of plasmid R1 (Fig. 1). The simple model that the Sok RNA blocks translation of Hok was not applicable, since the region of complementarity does not overlap the hok Shine-Dalgarno sequence. However, the 398-nucleotide hok mRNA also encodes a second, 70-amino-acid open reading frame, mok (mediation of killing), that encompasses hok (51). Translation of the hok open reading frame is dependent upon translation of the mok open reading frame, and it is the mok ribosome binding site where the Sok RNA and ribosomes compete for binding (51). An additional complication is that the full-length mRNA (here denoted hok rather than mok-hok) is highly structured and is not accessible to binding by either ribosomes or the Sok RNA (13, 14, 35). It was found that the hok mRNA is processed at the 3′ end (22) and upon truncation folds into a new structure in which the binding sites for the Sok RNA and the ribosome are more accessible (14, 35). Once formed, the hok mRNA-Sok RNA duplex is subject to cleavage by RNase III, thus ensuring that hok cannot be translated (19). The relative stabilities of both the hok mRNA and Sok RNA are another important aspect of the regulation. The Sok RNA has a half-life of less than 30 s, while the Hok mRNA is much more stable (22). If the R1 plasmid is lost, the inherently unstable Sok RNA is quickly degraded, and with no new synthesis of the Sok RNA, the hok gene is translated and cell death ensues.
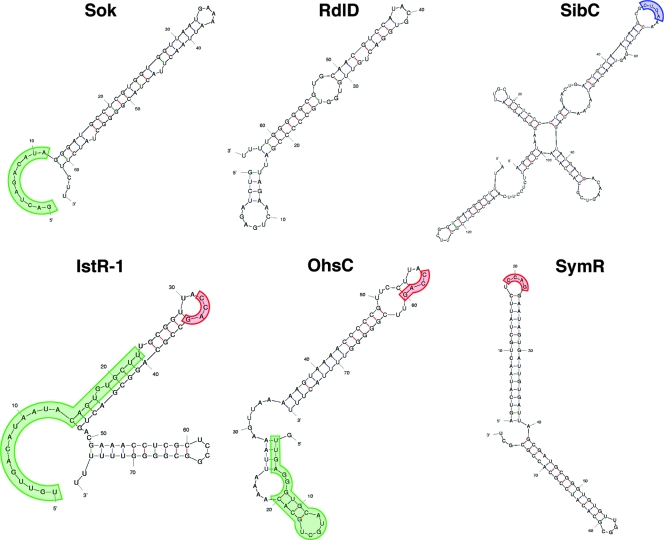
The Hok-Sok pair has been a model system for studies of RNA-RNA base pairing (15). These studies showed that the initial interaction between the two RNAs involves only limited base pairing and is facilitated by accessible bases in a U-turn structure in the target loop of the hok mRNA. In this light, it is worth noting that U-turn RNA structural motifs (YUNR, where Y is a pyrimidine, N is any nucleotide, and R is a purine), in which a uridine is involved in a bend in the direction of the RNA backbone, are also found in the ldr mRNAs and Sib RNAs (Fig. 2).
FIG. 2.
Predicted structures of antitoxin sRNAs. The structures of the plasmid R1 Sok RNA and the E. coli RdlD, SibC, IstR-1, OhsC, and SymR sRNAs were predicted by the program M-fold (http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/rna-form1.cgi). The region of the Sok RNA involved in the initial base-pairing interaction with the hok mRNA and the regions of complementarity between IstR-1 and OhsC and their targets are highlighted in green. The U-turn motif found in SibC is highlighted in blue, and a motif shared by IstR-1, OhsC, and SymR is indicated in red.
The main characteristics of Hok-Sok regulation, i.e., RNA processing and the presence of an overlapping open reading frame, appear to be true for all of the other type I toxin-antitoxin loci carried on plasmids found in the gram-negative bacteria (36). Gerdes and Wagner also have suggested that synthesis of the chromosomally encoded Ldr toxins is regulated in a manner similar to that for Hok (23). The Rdl RNAs are encoded opposite the long 5′ UTR of the ldr genes, and overexpression of RdlD can repress ldrD translational reporter gene fusions. However, the overlap between RdlD and ldrD does not encompass the ribosome binding site (31). Gerdes and Wagner (23) noted the presence of an open reading frame, referred to as ldrX, that begins upstream of and encompasses part of ldrD. The RdlD RNA base-pairing region overlaps the ribosome binding site as well as the start codon for ldrX. This overlap is reminiscent of the case for Hok/Sok/Mok, though experimental evidence is needed to confirm the hypothesis that Ldr synthesis is regulated by the same mechanism.
Processing and Block of a Standby Ribosome
The regulation of TisB synthesis shares many similarities with the regulation of Hok synthesis, though instead of blocking the translation of an overlapping open reading frame, the sRNA obstructs the binding of a standby ribosome (Fig. 1). Like the hok transcript, the tisAB mRNA has multiple forms. In vitro experiments demonstrated that a truncated form (+42) of the tisAB mRNA, which is processed at the 5′ end, is translated, whereas the full-length transcript is not (9). Structure mapping of the +42 form revealed that the ribosome binding site for tisB is in a tight secondary structure (9). The mRNA was also annotated to carry tisA, although this 37-amino-acid open reading frame upstream of tisB is not conserved and not likely to be translated (53). However, the predicted tisA ribosome binding site appears to be a loading site for ribosomes and is where ribosomes and the IstR-1 RNA compete for binding. If the ribosome out-competes IstR-1, the ribosome is loaded and stands by until the secondary structure opens and allows the ribosome to move and begin translating TisB. If IstR-1 binds, the RNA duplex is cleaved by RNase III, eliminating all further translation, as was seen for Hok-Sok (53).
Similar to the case for TisB-IstR-1, the mRNA encoding the ShoB toxin is encoded divergently from the OhsC RNA but nevertheless shares 19 nucleotides of perfect complementarity. It is intriguing that the 5′ UTR of ShoB is very long and contains a small open reading frame of five amino acids, an arrangement reminiscent of that for TisB-IstR-1. In addition, there are two shoB transcripts with 5′ ends that map 40 nucleotides apart (32), and only fusions to the shorter 5′ UTR were expressed (12). Further experimentation is needed to determine whether the regulation of ShoB synthesis is in fact similar to that of TisB synthesis, as well as to understand additional aspects of the regulation such as how the shorter shoB and tisB transcripts are generated.
Overlapping 3′ Ends
The RNAI-RNAII locus of the pAD1 plasmid of E. faecalis and the TpxA and RatA locus of B. subtilis, both in gram-positive bacteria, differ from the loci described above in that the toxin-antitoxin RNAs are transcribed convergently and have overlapping 3′ ends (Table 1). RNAI and RNAII overlap by 35 nucleotides (54), and the stem-loops of the two terminators form an initial complex (25). However, other interactions are required to prevent translation of fst encoded by RNAI. The RNAs have two complementary direct repeats in their 5′ ends, and base pairing between these direct repeats is required to block ribosome access and translation both in vivo and in vitro (24, 26). The 3′ ends of the TpxA mRNA and RatA RNA overlap by approximately 75 nucleotides (49). It was proposed that upon base pairing between the two RNAs, degradation of the RNA duplex leads to the formation of a truncated txpA message, which is not translated. Future experiments to test whether the 3′ end of the RatA RNA alone can prevent TxpA toxicity will confirm whether this 3′ interaction is sufficient for repression or if, as seen for RNAII regulation of RNAI, additional sequences are necessary.
OUTSTANDING ISSUES REGARDING REGULATION
Even with a simple block in translation, there are several parameters, such as RNA levels and structure, which are not well understood but are likely to affect the repression of toxin protein synthesis by the antitoxin sRNAs in particular and the regulation by base-pairing sRNAs in general.
RNA Levels
Clearly, the relative abundances of the mRNA and the corresponding sRNA are a key factor in determining whether repression takes place. Transcription of some toxin genes, such as the SOS-induced tisB and symE genes, occurs under very specific conditions, suggesting a need for derepression under these conditions. In some cases, the transcription of the antitoxin genes also appears to be regulated. For example, the levels of the OhsC RNA are high in stationary-phase cells but low in exponentially growing cells (32). In these cases, a decrease in the amount of the sRNA relative to the mRNA can explain how the toxin synthesis might be derepressed. However, in other cases, the sRNAs appear to be transcribed constitutively. The Sib and SymR RNAs are detected under all conditions tested, raising the question of when the corresponding toxins are synthesized. Another factor affecting regulation is the stability of the RNAs. As summarized above, the plasmid-encoded hok mRNA has a much longer half-life than the Sok RNA. Similarly, the ldrD mRNA has a half-life of 30 min, in contrast to the RdlD RNA half-life of 2 min (31). Thus, rapid turnover of RdlD could lead to increased LdrD synthesis. A further compounding factor is that little is known about the actual levels of the antitoxin sRNA and toxin mRNA. In one case where it has been measured, the SymR RNA levels are always at least 10-fold higher than the levels of the symE mRNA (30).
It is possible that the ability of the antisense RNAs to base pair with their targets is regulated. As shown in Fig. 2, the sRNAs discussed here are predicted to have extensive secondary structure with very long stems and few unpaired nucleotides. This is in contrast to sRNAs that possess only limited complementarity with their targets and are more loosely structured with shorter stems and more unpaired nucleotides. It may be that specific conditions or proteins are required to unfold portions of the sRNA such that productive base pairing to the toxin mRNA occurs only under these circumstances. However, the region of the Sok RNA involved in the initial base-pairing interaction with the hok mRNA and the regions in IstR-1 and OhsC complementary to tisB and shoB, respectively, are predicted to be primarily single stranded (Fig. 2). Conceivably, the sRNAs have evolved additional cellular roles, aside from regulating the synthesis of toxins. For example, the sRNAs could base pair with other mRNA targets or even bind specific proteins. These additional roles could titrate the regulatory sRNA away from toxin mRNA. For RatA regulation of TxpA and SibC regulation of IbsC, it was observed that the sRNAs had to be induced prior to the toxin in order to repress the toxic effects of overexpression of the small proteins (12, 49), suggesting that a preexisting pool of the sRNA was necessary to repress toxin synthesis.
Specificity
In many cases, mRNA-sRNA pairs have extensive complementarity (greater than 60 nucleotides), though it is not known how much complementarity is needed for regulation. An extensive base-pairing interaction could help to ensure that there is minimal dissociation of the RNAs. For the toxin-antitoxin loci present in multiple copies, such as the Ldr-Rdl and Sib-Ibs loci, the different copies are extremely homologous, and one could imagine that there might be cross-regulation. However, genetic evidence indicates that each Sib RNA uniquely represses the synthesis of the corresponding Ibs protein (12). Further exploration of this specificity and how it occurs will add to our understanding of base-pairing rules.
The Hfq protein serves as an RNA chaperone to facilitate the specific interaction between sRNA-target mRNA pairs which share only limited complementarity and is required for the regulatory activity of this class of sRNAs (reviewed in references 1 and 7). SymR repression of SymE synthesis does not require the E. coli Hfq protein (30). Similarly, the B. subtilis Hfq homolog YmaH is not required for RatA regulation of TxpA (49). The potential for extensive base pairing between the type I toxin mRNA and antitoxin sRNA may bypass the need for the chaperone; however, it is unknown whether additional protein factors are involved in facilitating the effects of the antitoxin sRNAs or in conferring specificity. Intriguingly, the IstR-1, OhsC, and SymR RNAs are all predicted to contain the sequence CCAG in a loop at the end of a long stem (Fig. 2); such a sequence could be a protein binding site.
In all of the type I toxin-antitoxin systems, the sRNA and toxin protein are encoded either adjacent to or opposite each other. This close proximity may increase the efficiency of regulation, though the advantages of cis- versus trans-encoded sRNAs have not been examined. For the chromosomal pairs described here, the sRNAs encoded in trans on plasmids still act as repressors, although in these cases the sRNAs are highly overexpressed (12, 30, 31, 49, 53). It is possible that cis-encoded RNAs might pair with their targets more rapidly, which could be an advantage when the sRNA is unstable.
Additional Levels of Regulation
As has been observed for the Hok and TisB proteins, the control of the synthesis of most, if not all, toxic proteins is likely to be multilayered, with repression by sRNAs being only one component. The 5′ UTRs of the shoB and ldrD transcripts are long (180 and 185 nucleotides, respectively), and structural predictions suggest that these leader regions form extensive secondary structures. These secondary structures are likely to affect translation and may also be subjected to processing, as has been shown for the hok and tisB mRNAs.
Proteolysis is another potential mechanism for maintaining low levels of the toxic proteins. For the type II toxin-antitoxin systems, the Lon protease degrades some of the protein antitoxins (reviewed in reference 17). The SymR-regulated SymE protein also has been found to be subject to Lon protease degradation (30). Based on these observations, it could be interesting to examine the levels of the type I toxin proteins in protease-deficient strains.
ADVANTAGES OF ANTITOXIN RNA REGULATORS
The emergence of both sRNA and protein antitoxin regulators suggests that each may have particular advantages. It is worth considering possible advantages of sRNA-based repression, which occurs before the translation of the toxin. If even small amounts of the protein can cause damage to the cell, it might be critical to block translation. By preventing translation, the cell also does not expend energy in synthesizing the toxin. In addition, as the toxic proteins described here are likely to be integral membrane proteins, it may be difficult to efficiently neutralize the small proteins through protein-protein interactions before the toxin becomes integrated into the membrane. Another difference between sRNA and protein antitoxins is that the sRNA-mRNA interaction usually promotes cleavage of the mRNA, rendering the effects of the sRNA irreversible, while antitoxin proteins can dissociate from their corresponding toxin proteins, making their inhibitory effects reversible.
ROLES OF SMALL TOXIN PROTEINS
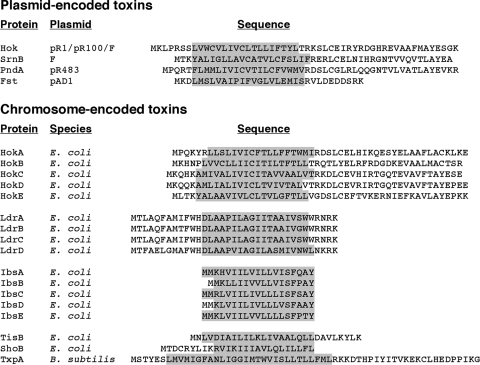
Although the exact functions of the type I toxins is not known, the small proteins are intriguing. Some general properties of the type I toxin proteins can be noted by simply examining their amino acid sequences (Fig. 3). With the exception of SymE, all are very hydrophobic and are predicted to contain an α-helical transmembrane domain. Several type I proteins, such as Fst, PndA, SrnB, and TxpA as well as the Hok and Ldr family members, also are likely to have cytosolic or periplasmic domains. Others, such as TisB, ShoB, and the Ibs family members, are predicted to consist of little more than the transmembrane helix. It is also worth noting that a cysteine, located within the predicted transmembrane domain and proposed to be involved in Hok dimerization (47), is conserved in all Hok proteins. Below we discuss what is known about the small, hydrophobic type I toxins. The one exclusion is SymE protein, which is 113 amino acids in length and is not hydrophobic. This protein shows homology to the type II antitoxin proteins but shares properties of the type II toxins that cleave RNA (30).
FIG. 3.
Protein sequences of characterized type I toxins. Transmembrane domains were predicted using the TMPred program (http://www.ch.embnet.org/software/TMPRED_form.html) and are shaded in gray for each protein.
Toxicity upon Overexpression
The available information on the functions of the small, hydrophobic proteins is based largely on overexpression experiments. The proteins are classified as toxic because high levels lead to cell death as measured by decreased optical density and colony-forming ability (12, 16, 49). Overexpression of Hok, SrnB, PndA, Fst, IbsC, TisB, and ShoB also has been shown to lead to membrane depolarization or membrane disruption (12, 16, 40, 52, 56). These effects on the membrane are not surprising given the hydrophobic nature of these proteins and are similar to those observed for some bacteriophage holin proteins (5), as well as some antimicrobial peptides (28), which multimerize and form pores that disrupt the membrane potential and ultimately lead to cell lysis. Other small, hydrophobic phage proteins lyse cells by interacting with membrane proteins, so there may be multiple mechanisms for membrane disruption by the type I toxin proteins. Subcellular localization experiments with Gef, a chromosomally encoded Hok protein, showed that the majority of the protein is in the membrane, consistent with this site of action (47). In addition, mutations that abolished the toxicity of Gef overexpression localize primarily to the transmembrane domain of the protein (47). TisB has also recently been found to fractionate with the inner membrane (52). It is tempting to speculate that the type I toxin proteins might act extracellularly, similar to toxic microcin peptides (10). However, there are no data to support this hypothesis, and exogenously applied Hok did not significantly kill the six bacterial strains tested, including E. coli (43).
Studies for at least a subset of the type I toxin proteins have shown that there are physiological effects aside from membrane damage. Overexpression of the LdrD protein leads to nucleoid condensation in E. coli (31), and Fst overexpression disrupts nucleoid structure, chromosome segregation, and cell division in E. faecalis (41). For Fst, these physiological effects have been observed at lower levels of expression than those required for membrane disruption (41). High levels of TisB lead to decreased DNA, RNA, and protein synthesis in vivo, but these effects are likely to be indirect consequences of membrane damage, since TisB had no effects on transcription and translation in vitro (52). Whole-genome expression analysis has shown that overexpression of LdrD, IbsC, ShoB, or TisB results in the induction and repression of specific genes (12, 31). While some genes, such as soxS, which encodes a regulator of the superoxide stress response, are induced by overexpression of each of the four toxins, other genes are affected by only a subset of the toxins, suggesting that these small hydrophobic proteins do not all act on the same targets. It is noteworthy, though, that many of the induced genes encode membrane proteins or are members of the heat shock or envelope stress regulons.
Given the strong toxic phenotype associated with overexpression of type I proteins, it is interesting that very few resistant strains have been reported (46). This is in contrast to the large number of resistant mutants that can be isolated after exposure to antibiotics. One mutation that confers resistance to the Hok family Gef protein was mapped to tadA, a gene encoding a tRNA-specific adenosine deaminase involved in the processing of tRNAArg2 (46), but this did not help to elucidate the function of the Gef protein. If the proteins are acting to permeabilize the membrane, the toxicity phenotype might be hard to suppress given the pleiotropic effects of disrupting the cell membrane. On the other hand, if the proteins are killing cells in other ways, additional genetic screens that exploit the toxic phenotype, possibly at barely toxic levels of the proteins, might provide insights into interacting proteins, key amino acids, or the functions of these hydrophobic proteins.
Roles at Endogenous Levels
For plasmid-encoded type I toxin proteins, the physiological role of the protein seems clear. The protein induces stasis or death in cells that lack the plasmid after cell division. In this capacity the toxic protein can be extremely efficient, leading to a 200- to 300-fold-increased stabilization of the plasmid (18, 36). Another proposed function of the Hok-Sok loci on plasmid R1 is to protect transformed E. coli cells from infection with T4 phage (44). When carried on a high-copy plasmid, the Hok-Sok cassette reduced the efficiency of T4 plating and decreased the plaque size.
For type I toxins encoded on bacterial chromosomes, the functions of endogenous levels of the proteins are less obvious. It may be that these proteins have some of the same functions described for the type II toxin proteins, such as persistence, quality control, chromosome stabilization, or, alternatively, cell death. As discussed above, the transcription of certain toxin genes is induced under very specific conditions, suggesting that the toxin might play a role under these conditions. For the SOS-induced TisB and SymR proteins, TisB has been suggested to promote a reversible decrease in growth rate, allowing cells to recover and carry out DNA repair (9), while SymR has been suggested to play a role in recycling damaged RNAs (30). The characterization of toxin proteins expressed at cellular levels has been difficult because, in almost all cases tested, no obvious phenotype has been observed upon reduction or elimination of the antitoxin RNA. One exception is the IbsC-SibC locus of E. coli, where a reduction in SibC RNA levels through a promoter deletion was associated with an increase in the levels of the pspABDCE mRNA encoding phage shock proteins (12). An even more dramatic phenotype of cell lysis was observed for mutants lacking the RatA RNA of the TxpA-RatA locus of B. subtilis (49). To fully elucidate the physiological roles of the type I toxin proteins, more will need to be learned about the conditions under which these proteins are synthesized.
It is worth mentioning the possibility that the type I toxin-antitoxin modules may not have any function in the cell but may be examples of “selfish DNA,” especially since several of the type I toxin-antitoxin pairs have been deleted from bacterial chromosomes without observable consequences to the bacteria (12, 31). Like transposons and other mobile DNA elements, these RNA-regulated toxins could be collected by bacterial species as a result of horizontal transfer or duplication. In this light, it is intriguing that the TpxA-RatA pair is encoded on a 48-kb phage-like element denoted skin (49). At least some of the type I toxin-antitoxin pairs may confer a selective advantage to cells, since no point mutations are present in either the coding sequences or the ribosome binding sites of all copies of the ldr or ibs genes identified to date.
Perspectives
The small size, potent toxicity, and broad range of susceptible bacterial and eukaryotic cell types make the small, hydrophobic toxin proteins potential tools for a variety of practical applications. One of the best-studied uses for toxin-antitoxin systems is plasmid stabilization in large-scale bacterial cell cultures (57). The advantages of using toxin-antitoxin cassettes as plasmid stabilizers include not needing the constant presence of antibiotics, a wide range of effectiveness in gram-negative (and some gram-positive) bacteria, and no requirement to modify the host chromosome. In addition, the fact that the type I toxin-antitoxin cassettes are small facilitates their incorporation into plasmids. The plasmid R1 Hok-Sok locus has been shown to increase stability in large-scale cultures, and combining the locus with the plasmid RP4 parD-parE type II antitoxin-toxin locus increases stability even further (42). More recently, there has been an interest in using the small, hydrophobic proteins as cell-killing agents in eukaryotic cells. Studies with breast cancer-derived cells transfected with the E. coli gef toxin gene under the control of a mouse mammary tumor virus promoter showed that Gef expression leads to apoptosis and decreased proliferation of the cell line (6). One can also imagine antimicrobial therapies based on derepressed toxin expression by interference with the antitoxin sRNAs. Undoubtedly the continued identification and characterization of type I toxin-antitoxin modules will lead to more applications, as well as give further insights into the mechanisms of sRNA regulation and small protein function.
ADDENDUM IN PROOF
R. Weel-Sneve, M. Bjørås, and K. I. Kristiansen (Nucleic Acids Res., in press) have recently proposed another function for the tisAB mRNA. They report that base pairing between the tisAB mRNA and the SOS-inducible dinD mRNA as well as the uxaA mRNA dampens the SOS response.
Acknowledgments
We thank E. Koonin and K. Makarova for helpful discussions and S. Durand, F. Fontaine, E. Hobbs, S. Gottesman, M. Kawano, and L. Waters for comments on the manuscript.
This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development as well as a Research Associateship from the National Research Council (to E.M.F.) and a postdoctoral fellowship from the Life Sciences Foundation (to M.R.H).
REFERENCES
- 1.Aiba, H. 2007. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 10134-139. [DOI] [PubMed] [Google Scholar]
- 2.Akimoto, S., and Y. Ohnishi. 1982. R483 and F plasmid genes promoting RNA degradation: comparative restriction mapping. Microbiol. Immunol. 26779-793. [DOI] [PubMed] [Google Scholar]
- 3.Altuvia, S. 2007. Identification of bacterial small non-coding RNAs: experimental approaches. Curr. Opin. Microbiol. 10257-261. [DOI] [PubMed] [Google Scholar]
- 4.Argaman, L., R. Hershberg, J. Vogel, G. Bejerano, E. G. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11941-950. [DOI] [PubMed] [Google Scholar]
- 5.Bernhardt, T. G., I. N. Wang, D. K. Struck, and R. Young. 2002. Breaking free: “protein antibiotics” and phage lysis. Res. Microbiol. 153493-501. [DOI] [PubMed] [Google Scholar]
- 6.Boulaiz, H., J. Prados, C. Melguizo, A. M. García, J. A. Marchal, J. L. Ramos, E. Carrillo, C. Vélez, and A. Aranega. 2003. Inhibition of growth and induction of apoptosis in human breast cancer by transfection of gef gene. Br. J. Cancer 89192-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brennan, R. G., and T. M. Link. 2007. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 10125-133. [DOI] [PubMed] [Google Scholar]
- 8.Buts, L., J. Lah, M. H. Dao-Thi, L. Wyns, and R. Loris. 2005. Toxin-antitoxin modules as bacterial metabolic stress managers. Trends Biochem. Sci. 30672-679. [DOI] [PubMed] [Google Scholar]
- 9.Darfeuille, F., C. Unoson, J. Vogel, and E. G. H. Wagner. 2007. An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell 26381-392. [DOI] [PubMed] [Google Scholar]
- 10.Duquesne, S., V. Petit, J. Peduzzi, and S. Rebuffat. 2007. Structural and functional diversity of microcins, gene-encoded antibacterial peptides from enterobacteria. J. Mol. Microbiol. Biotechnol. 13200-209. [DOI] [PubMed] [Google Scholar]
- 11.Faridani, O. R., A. Nikravesh, D. P. Pandey, K. Gerdes, and L. Good. 2006. Competitive inhibition of natural antisense Sok-RNA interactions activates Hok-mediated cell killing in Escherichia coli. Nucleic Acids Res. 345912-5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fozo, E. M., M. Kawano, F. Fontaine, Y. Kaya, K. S. Mendieta, K. L. Jones, A. Ocampo, K. E. Rudd, and G. Storz. 2008. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol. Microbiol. 701076-1093. [DOI] [PMC free article] [PubMed]
- 13.Franch, T., and K. Gerdes. 1996. Programmed cell death in bacteria: translational repression by mRNA end-pairing. Mol. Microbiol. 211049-1060. [DOI] [PubMed] [Google Scholar]
- 14.Franch, T., A. P. Gultyaev, and K. Gerdes. 1997. Programmed cell death by hok/sok of plasmid R1: processing at the hok mRNA 3′-end triggers structural rearrangements that allow translation and antisense RNA binding. J. Mol. Biol. 27338-51. [DOI] [PubMed] [Google Scholar]
- 15.Franch, T., M. Petersen, E. G. H. Wagner, J. P. Jacobsen, and K. Gerdes. 1999. Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J. Mol. Biol. 2941115-1125. [DOI] [PubMed] [Google Scholar]
- 16.Gerdes, K., F. W. Bech, S. T. Jørgensen, A. Løbner-Olesen, P. B. Rasmussen, T. Atlung, L. Boe, O. Karlstrom, S. Molin, and K. von Meyenburg. 1986. Mechanism of postsegregational killing by the hok gene product of the parB system of plasmid R1 and its homology with the relF gene product of the E. coli relB operon. EMBO J. 52023-2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerdes, K., S. K. Christensen, and A. Løbner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3371-382. [DOI] [PubMed] [Google Scholar]
- 18.Gerdes, K., J. E. Larsen, and S. Molin. 1985. Stable inheritance of plasmid R1 requires two different loci. J. Bacteriol. 161292-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerdes, K., A. Nielsen, P. Thorsted, and E. G. H. Wagner. 1992. Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable Hok, SrnB and PndA effector messenger RNAs. J. Mol. Biol. 226637-649. [DOI] [PubMed] [Google Scholar]
- 20.Gerdes, K., L. K. Poulsen, T. Thisted, A. K. Nielsen, J. Martinussen, and P. H. Andreasen. 1990. The hok killer gene family in gram-negative bacteria. New Biol. 2946-956. [PubMed] [Google Scholar]
- 21.Gerdes, K., P. B. Rasmussen, and S. Molin. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 833116-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerdes, K., T. Thisted, and J. Martinussen. 1990. Mechanism of post-segregational killing by the hok/sok system of plasmid R1: sok antisense RNA regulates formation of a hok mRNA species correlated with killing of plasmid-free cells. Mol. Microbiol. 41807-1818. [DOI] [PubMed] [Google Scholar]
- 23.Gerdes, K., and E. G. H. Wagner. 2007. RNA antitoxins. Curr. Opin. Microbiol. 10117-124. [DOI] [PubMed] [Google Scholar]
- 24.Greenfield, T. J., E. Ehli, T. Kirshenmann, T. Franch, K. Gerdes, and K. E. Weaver. 2000. The antisense RNA of the par locus of pAD1 regulates the expression of a 33-amino-acid toxic peptide by an unusual mechanism. Mol. Microbiol. 37652-660. [DOI] [PubMed] [Google Scholar]
- 25.Greenfield, T. J., T. Franch, K. Gerdes, and K. E. Weaver. 2001. Antisense RNA regulation of the par post-segregational killing system: structural analysis and mechanism of binding of the antisense RNA, RNAII and its target, RNAI. Mol. Microbiol. 42527-537. [DOI] [PubMed] [Google Scholar]
- 26.Greenfield, T. J., and K. E. Weaver. 2000. Antisense RNA regulation of the pAD1 par post-segregational killing system requires interaction at the 5′ and 3′ ends of the RNAs. Mol. Microbiol. 37661-670. [DOI] [PubMed] [Google Scholar]
- 27.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 3011496-1499. [DOI] [PubMed] [Google Scholar]
- 28.Henriques, S. T., M. N. Melo, and M. A. Castanho. 2006. Cell-penetrating peptides and antimicrobial peptides: how different are they? Biochem. J. 3991-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito, R., and Y. Ohnishi. 1983. The roles of RNA polymerase and RNAase I in stable RNA degradation in Escherichia coli carrying the srnB+ gene. Biochim. Biophys. Acta 73927-34. [DOI] [PubMed] [Google Scholar]
- 30.Kawano, M., L. Aravind, and G. Storz. 2007. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 64738-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawano, M., T. Oshima, H. Kasai, and H. Mori. 2002. Molecular characterization of long direct repeat (LDR) sequences expressing a stable mRNA encoding for a 35-amino-acid cell-killing peptide and a cis-encoded small antisense RNA in Escherichia coli. Mol. Microbiol. 45333-349. [DOI] [PubMed] [Google Scholar]
- 32.Kawano, M., A. A. Reynolds, J. Miranda-Rios, and G. Storz. 2005. Detection of 5′ and 3′ UTR-derived small RNAs and cis-encoded antisense RNAs in Escherichia coli. Nucleic Acid Res. 331040-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livny, J., and M. K. Waldor. 2007. Identification of small RNAs in diverse bacterial species. Curr. Opin. Microbiol. 1096-101. [DOI] [PubMed] [Google Scholar]
- 34.Loh, S. M., D. S. Cram, and R. A. Skurray. 1988. Nucleotide sequence and transcriptional analysis of a third function (Flm) involved in F-plasmid maintenance. Gene 66259-268. [DOI] [PubMed] [Google Scholar]
- 35.Møller-Jensen, J., T. Franch, and K. Gerdes. 2001. Temporal translational control by a metastable RNA structure. J. Biol. Chem. 27635707-35713. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen, A. K., P. Thorsted, T. Thisted, E. G. H. Wagner, and K. Gerdes. 1991. The rifampicin-inducible genes srnB from F and pnd from R483 are regulated by antisense RNAs and mediate plasmid maintenance by killing of plasmid-free segregants. Mol. Microbiol. 51961-1973. [DOI] [PubMed] [Google Scholar]
- 37.Ohnishi, Y. 1975. F factor promotes turnover of stable RNA in Escherichia coli. Science 187257-258. [DOI] [PubMed] [Google Scholar]
- 38.Ohnishi, Y., and S. Akimoto. 1980. I-like R plasmids promote degradation of stable ribonucleic acid in Escherichia coli. J. Bacteriol. 144833-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ohnishi, Y., H. Iguma, T. Ono, H. Nagaishi, and A. J. Clark. 1977. Genetic mapping of the F plasmid gene that promotes degradation of stable ribonucleic acid in Escherichia coli. J. Bacteriol. 132784-789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ono, T., S. Akimoto, K. Ono, and Y. Ohnishi. 1986. Plasmid genes increase membrane permeability in Escherichia coli. Biochim. Biophys. Acta 86781-88. [DOI] [PubMed] [Google Scholar]
- 41.Patel, S., and K. E. Weaver. 2006. Addiction toxin Fst has unique effects on chromosome segregation and cell division in Enterococcus faecalis and Bacillus subtilis. J. Bacteriol. 1885374-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pecota, D. C., C. S. Kim, K. Wu, K. Gerdes, and T. K. Wood. 1997. Combining the hok/sok, parDE, and pnd postsegregational killer loci to enhance plasmid stability. Appl. Environ. Microbiol. 631917-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pecota, D. C., G. Osapay, M. E. Selsted, and T. K. Wood. 2003. Antimicrobial properties of the Escherichia coli R1 plasmid host killing peptide. J. Biotechnol. 1001-12. [DOI] [PubMed] [Google Scholar]
- 44.Pecota, D. C., and T. K. Wood. 1996. Exclusion of T4 phage by the hok/sok killer locus from plasmid R1. J. Bacteriol. 1782044-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pedersen, K., and K. Gerdes. 1999. Multiple hok genes on the chromosome of Escherichia coli. Mol. Microbiol. 321090-1102. [DOI] [PubMed] [Google Scholar]
- 46.Poulsen, L. K., N. W. Larsen, S. Molin, and P. Andersson. 1992. Analysis of an Escherichia coli mutant strain resistant to the cell-killing function encoded by the gef gene family. Mol. Microbiol. 6895-905. [DOI] [PubMed] [Google Scholar]
- 47.Poulsen, L. K., A. Refn, S. Molin, and P. Andersson. 1991. Topographic analysis of the toxic Gef protein from Escherichia coli. Mol. Microbiol. 51627-1637. [DOI] [PubMed] [Google Scholar]
- 48.Rudd, K. E. 1999. Novel intergenic repeats of Escherichia coli K-12. Res. Microbiol. 150653-664. [DOI] [PubMed] [Google Scholar]
- 49.Silvaggi, J. M., J. B. Perkins, and R. Losick. 2005. Small untranslated RNA antitoxin in Bacillus subtilis. J. Bacteriol. 1876641-6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storz, G., and S. Gottesman. 2006. Versatile roles of small RNA regulators in bacteria, p. 567-594. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 51.Thisted, T., and K. Gerdes. 1992. Mechanism of post-segregational killing by the hok/sok system of plasmid R1. Sok antisense RNA regulates hok gene expression indirectly through the overlapping mok gene. J. Mol. Biol. 22341-54. [DOI] [PubMed] [Google Scholar]
- 52.Unoson, C., and E. G. H. Wagner. 2008. A small SOS-induced toxin is targeted against the inner membrane in Escherichia coli. Mol. Microbiol. 70258-270. [DOI] [PubMed] [Google Scholar]
- 53.Vogel, J., L. Argaman, E. G. H. Wagner, and S. Altuvia. 2004. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 142271-2276. [DOI] [PubMed] [Google Scholar]
- 54.Weaver, K. E., K. D. Jensen, A. Colwell, and S. I. Sriram. 1996. Functional analysis of the Enterococcus faecalis plasmid pAD1-encoded stability determinant par. Mol. Microbiol. 2053-63. [DOI] [PubMed] [Google Scholar]
- 55.Weaver, K. E., and D. J. Tritle. 1994. Identification and characterization of an Enterococcus faecalis plasmid pAD1-encoded stability determinant which produces two small RNA molecules necessary for its function. Plasmid 32168-181. [DOI] [PubMed] [Google Scholar]
- 56.Weaver, K. E., D. M. Weaver, C. L. Wells, C. M. Waters, M. E. Gardner, and E. A. Ehli. 2003. Enterococcus faecalis plasmid pAD1-encoded Fst toxin affects membrane permeability and alters cellular responses to lantibiotics. J. Bacteriol. 1852169-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu, K., D. Jahng, and T. K. Wood. 1994. Temperature and growth rate effects on the hok/sok killer locus for enhanced plasmid stability. Biotechnol. Prog. 10621-629. [DOI] [PubMed] [Google Scholar]