Abstract
Summary: Lactobacilli have been crucial for the production of fermented products for centuries. They are also members of the mutualistic microbiota present in the human gastrointestinal and urogenital tract. Recently, increasing attention has been given to their probiotic, health-promoting capacities. Many human intervention studies demonstrating health effects have been published. However, as not all studies resulted in positive outcomes, scientific interest arose regarding the precise mechanisms of action of probiotics. Many reported mechanistic studies have addressed mainly the host responses, with less attention being focused on the specificities of the bacterial partners, notwithstanding the completion of Lactobacillus genome sequencing projects, and increasing possibilities of genomics-based and dedicated mutant analyses. In this emerging and highly interdisciplinary field, microbiologists are facing the challenge of molecular characterization of probiotic traits. This review addresses the advances in the understanding of the probiotic-host interaction with a focus on the molecular microbiology of lactobacilli. Insight into the molecules and genes involved should contribute to a more judicious application of probiotic lactobacilli and to improved screening of novel potential probiotics.
INTRODUCTION
During the last decades, it became clear that the human body lives in close harmony with a complex ecosystem that is composed of more than 1,000 different bacterial species inhabiting the oral cavity, upper respiratory tract, gastrointestinal tract (GIT), vagina, and skin. This collection, known as the microbiota, is acquired soon after birth and persists throughout life. Together, these microbes play an important role in the physiology of their host, including the digestion and assimilation of nutrients, protection against pathogen colonization, modulation of immune responses, regulation of fat storage, and stimulation of intestinal angiogenesis (12). However, understanding how these different species contribute to human health remains a major challenge. One main difficulty is correlating the health status of the host with the presence or absence of certain bacterial species, bearing in mind that the microbiota varies extensively among individuals. Obviously, the complexity of the microbiota makes genetic approaches to define microbe-microbe and microbe-host interactions a challenging task.
Within this complex research area on the microbiota, the deliberate administration of probiotic bacteria can contribute substantially to gain better knowledge of beneficial microbe-host interactions whereby fundamental, medical, nutritional, and commercial aspects are taken into account. As implicated in the definition of a probiotic bacterium, i.e., “a live microorganism that, when administered in adequate amounts, confers a health benefit on the host” (81), this field investigates mainly the health effects of specific strains after oral intake, for example, in functional food products. Although the term “probiotic” cannot be used merely as a synonym for putatively beneficial members of the microbiota, members of the human microbiota are often sources from which probiotics are isolated based on recommended properties such as specific health benefits, survival and persistence in the host, proven safety, and stability (250). While bifidobacteria and other genera are also increasingly being applied as probiotics, this review will focus on lactobacilli, given their long history of traditional use in food fermentations of products derived from animals (milk and meat, etc.) or plants (vegetables and olives, etc.).
The lactobacilli belong to the lactic acid bacteria (LAB), since their main end product of carbohydrate metabolism is lactic acid. The genus Lactobacillus comprises a large heterogeneous group of low-G+C gram-positive, nonsporulating, and anaerobic bacteria (51). Taxonomically, the genus Lactobacillus belongs to the phylum Firmicutes, class Bacilli, order Lactobacillales, family Lactobacillaceae. They are nutritionally fastidious, requiring rich media to grow (carbohydrates, amino acids, peptides, fatty acid esters, salts, nucleic acid derivatives, and vitamins) (119).
Besides their key role in food fermentations, lactobacilli are found in the GIT of humans and animals in variable amounts depending on the animal species, age of the host, or location within the gut. However, it is difficult to distinguish true autochthonous lactobacilli from allochthonous or transiently passing lactobacilli, for instance, originating from fermented foods or from the oral cavity, which is home to a considerable amount of lactobacilli (267). Lactobacilli seem to form only a minor proportion of the human adult fecal microbiota, i.e., around 0.01% to 0.6% of total bacterial counts (58, 125, 226, 247). L. gasseri, L. reuteri, L. crispatus, L. salivarius, and L. ruminis appear to be predominant autochthonous Lactobacillus species (267). L. acidophilus, L. fermentum, L. casei, L. rhamnosus, L. johnsonii, L. plantarum, L. brevis, L. delbrueckii, L. curvatus, and L. sakei can also be found in the human GIT at fluctuating levels (104, 267). Although less accessible, lactobacilli are commonly identified in biopsy samples from stomach, small intestine, and colon but in variable and usually rather low numbers (267).
In comparison to the adult microbiota, the infant microbiota is highly unstable but also contains lactobacilli in variable amounts (153). The number of Lactobacillus cells in neonates was found to be in the range of 105 CFU/g of feces, while in infants 1 month of age and older, the counts ranged from 106 to 108 CFU/g of feces (153). Sequences retrieved from infant feces grouped with L. salivarius, L. rhamnosus, and L. paracasei (104).
In contrast to the GIT, the presence of lactobacilli is more pronounced in the female urogenital tract, where they often dominate the healthy microbiota (202, 284). The most frequently occurring species found in the vagina are L. crispatus, L. gasseri, L. iners, and L. jensenii (7, 36, 37, 259). Moreover, a healthy, stable Lactobacillus population seems to protect against urogenital infections and bacterial vaginosis (80).
HEALTH EFFECTS OF LACTOBACILLI
Reported Beneficial Effects of Lactobacilli on the Host
Lactobacilli have been shown to exert health benefits when applied under various conditions. The best evidence exists for the treatment and prevention of enteric infections and postantibiotic syndromes. Several meta-analyses have established the efficacy of some lactobacilli in acute infectious diarrhea and the prevention of antibiotic-associated diarrhea (216). Certain lactobacilli may reduce the recurrence of Clostridium difficile-associated diarrhea (192) and prevent necrotizing enterocolitis in preterm neonates (69). Some promising results have also been obtained for the prevention and treatment of inflammatory bowel disease (IBD) (103), prevention of colorectal cancer (199), and treatment of irritable bowel syndrome (41). Although the GIT is the site where probiotic lactobacilli are believed to exert most health-modulating activities, probiotic applications with some Lactobacillus strains at other sites of the body have shown promise, such as the prevention and treatment of urogenital diseases and bacterial vaginosis in women (80), the prevention of atopic disease and food hypersensitivity (28), and the prevention of dental caries (168). Probiotic lactobacilli have a high safety profile, recognized by a “generally-regarded-as-safe” status, and the tolerance is usually excellent. However, in rare cases, reports of infections presumably caused by probiotic lactobacilli in immunocompromised patients or patients with severe underlying disease have been published (21, 146).
The levels of evidence for these health effects vary greatly between different clinical trials, different clinical conditions, and different probiotic strains used. Notably, it is also fair to state that many clinical studies did not result in positive outcomes. On the other hand, studies that did report health benefits of lactobacilli often lacked control groups, blinding, validated outcomes, or standards for reporting adverse events (158). It is clear that for a more judicious and scientifically supported application of probiotic lactobacilli, their efficacy should be addressed in double-blind, placebo-controlled studies of the appropriate sample size. Preferentially, different samples (blood, feces, urine, and biopsies) of individuals should be analyzed. In this way, information on biomarkers (microbes, cytokines, and metabolites, etc.) and putative modes of action can be obtained (see below).
General Mechanisms of Action of Probiotic Lactobacilli
The application of probiotic lactobacilli starts with the general assumption that the mechanisms underlying the health-promoting capacities of lactobacilli belong to one of the following, sometimes overlapping, categories (Fig. 1) (26, 82, 158): (i) pathogen inhibition and restoration of microbial homeostasis through microbe-microbe interactions, (ii) enhancement of epithelial barrier function, and (iii) modulation of immune responses. The capacity of lactobacilli to inhibit pathogens is well known since they have been used for centuries in food preservation to prevent microbial spoilage. Gradually, lactobacilli have been investigated for their capacities to exert immunostimulatory (adjuvant) and immunoregulatory (e.g., in IBD) properties. As probiotics are applied to the GIT mostly via beverages, food, or pills, their capacity to enhance the barrier function of the gut wall epithelium against pathogens and toxins is also increasingly gaining attention.
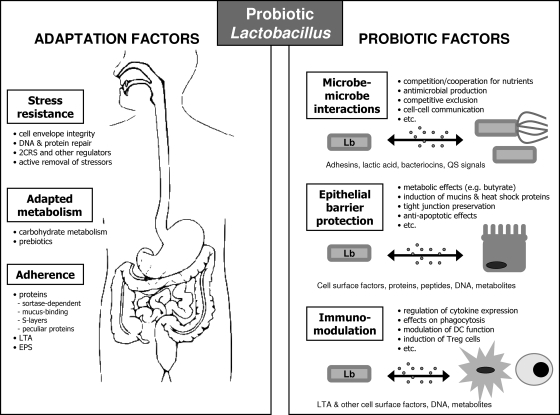
FIG. 1.
Mechanistic view of probiotic actions by lactobacilli. Molecular studies of probiotics with focus on lactobacilli (Lb) aim to identify factors that promote survival in, adaptation to, and colonization of the host (adaptation factors) and factors that directly contribute to health-promoting effects (probiotic factors). As probiotic lactobacilli are generally studied in their relation with the GIT, this niche is depicted. The health-promoting effects are thought to be mediated by three main mechanisms of probiotic actions, which include pathogen inhibition, restoration of the microbial balance, enhancement of epithelial barrier function, and immunomodulatory effects via interactions with immune cells such as DCs. The figure reflects the outline of this review.
Given the complexity of these three main functions, it can be understood that different strains evoke different responses in the host. Therefore, results with one specific Lactobacillus strain cannot be generalized. Molecular research on lactobacilli should carefully pay attention to these strain-specific properties. Different probiotic Lactobacillus strains have been associated with different effects related to their specific capacities to express particular surface molecules or to secrete proteins and metabolites directly interacting with host cells. The specific aspects of the three main mechanisms of action will be discussed in detail at the molecular level below (see Mechanisms of the Health-Promoting Effects of Lactobacilli: Probiotic Factors). First, we will discuss some general aspects (concept of adaptation and probiotic factors, specific cell surface architecture, and genetic tools) that are related to molecular research on the microbial partners in these probiotic interactions.
Adaptation and Probiotic Factors of Lactobacilli
It is believed that many mechanistic studies of probiotic lactobacilli are somehow biased to the characterization of the host response. Considering the significant strain differences and the sometimes disappointing results from clinical trials with lactobacilli while strains showed great promise in vitro, molecular characterization of probiotic strains is of utmost importance. Molecular studies that focus on the probiotic microbes themselves have two important goals: defining the best conditions driving the best performance of the probiotic strains and allowing the selection of novel probiotic strains with well-defined molecular criteria. There are two main categories of factors that contribute to the optimal functioning of probiotic lactobacilli: factors that allow optimal adaptation to the new niches that they temporarily encounter in the host (adaptation factors) and factors that directly contribute to the health-promoting effects (probiotic factors) (Fig. 1). These two categories are also implicitly integrated in the definition of probiotic strains, highlighting live and active microbes with health-promoting capacities (81). Probiotic factors involve the three main mechanisms of probiotic action: maintaining microbial balance, epithelial protection, and immunomodulation (as indicated above). The term “adaptation factors” refers to the factors that contribute to these probiotic effects, without themselves being health promoting, although the distinction between these two categories is sometimes difficult to make. Adaptation factors include stress resistance, active metabolism adapted to the host environment, and adherence to the intestinal mucosa and mucus (see below).
In this aspect, there are many parallels that can be drawn between the unwanted consumption of food-borne gastrointestinal pathogens like Salmonella enterica serovar Typhimurium and enteropathogenic Escherichia coli and the dedicated consumption of probiotics. From the bacterial point of view, they both need to survive the harsh conditions of the stomach and bile, and both need to be able to interact with the host (as elaborated below). For pathogens, this interaction is characterized by mechanisms of invasion and pathogenesis. For probiotics, this interaction is purported to be health promoting, a more symbiotic (or mutualistic) interaction from which both partners, microbes and the host, gain advantage. Thus, these adaptation and probiotic factors are proposed in analogy with virulence factors of pathogens where some factors contribute to the survival and adhesion of these strains and other factors are directly disease causing, such as toxin production (167).
CELL SURFACE STRUCTURES OF LACTOBACILLI
Specific metabolic and physiological characteristics of lactobacilli that play a key role in the adaptation to the host and the production and availability of probiotic factors will be discussed below. In this section, we describe the typical cell surface structures of lactobacilli, since these structures are in direct contact with the environment and can function as both key adaptation and probiotic factors. We focus on some aspects of the enormous diversity in cell surface structures and secreted factors of lactobacilli. Functional analyses of these structures related to adaptation and probiotic effects will be dealt with below.
In gram-positive bacteria, the cell wall consists of several characteristic structures: a thick, multilayered peptidoglycan (PG) sacculus decorated with proteins, teichoic acids, and polysaccharides and, in some species, surrounded by an outer shell of proteins packed in a paracrystalline layer (S layer) (Fig. 2). These molecular structures provide the bacteria with species- and strain-specific properties. For a more extensive overview on the cell wall biology of lactobacilli, see other reviews (65, 219, 256), although specific biochemical or genetic data on the cell wall biosynthesis pathways are rather scarce for lactobacilli (65).
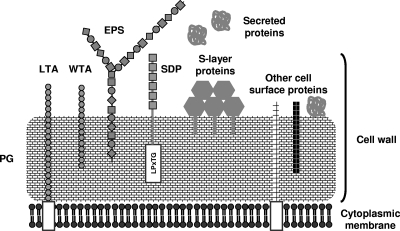
FIG. 2.
Cell surface architecture of lactobacilli. The cell wall of lactobacilli is composed of different macromolecules together determining the strain-specific properties that include adaptation to the changing host environment and interaction with host immune receptors and epithelial cells. A thick multilayered PG layer is decorated with teichoic acids (WTA and/or LTA), proteins, and EPSs (65). SDPs (24, 258) and S-layer proteins (9) are best studied in lactobacilli, but many other types of cell surface proteins and protein anchors exist. In contrast to coccoid bacteria, PG and WTA biosynthesis and protein secretion via the general secretion machinery appear to occur in helical patterns around the cell surface of rod-shaped bacteria such as B. subtilis (42, 59, 85, 219). Such a helical pattern of cell wall biosynthesis, although not yet documented, can also be postulated for Lactobacillus rods.
Peptidoglycan
Like most gram-positive species, the cell wall of lactobacilli is characterized by a thick PG layer. This 20- to 100-nm-thick PG multilayer, sometimes referred to as the murein sacculus, plays a key role in structural integrity and protects the cell against lysis. Cell wall PG is further covalently and noncovalently decorated with teichoic acids, polysaccharides, and proteins (65) (Fig. 2).
PG is composed of glycan chains of repeating β-1,4-linked N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) residues extensively cross-linked by two pentapeptide side chains. The chemistries of the glycan chains vary only slightly among different bacteria, but there is considerable variation in the compositions of stem peptides, which are linked to the carboxyl group of MurNAc (219, 256). In LAB, the consensus sequence for the pentapeptide is l-alanine-d-glutamate-meso-diaminopimelic acid or l-lysine-d-alanine-d-alanine, with a preference for l-lysine (65). In several lactobacilli, e.g., L. casei and L. plantarum, the C-terminal d-alanine residue of the muramyl pentapeptide is replaced by d-lactate, referred to as a pentadepsipeptide. This pentadepsipeptide confers resistance to the glycopeptide antibiotic vancomycin (65).
The biosynthesis of PG involves the synthesis of the nucleotide sugar-linked precursors UDP-MurNAc-pentapeptide and UDP-GlcNAc in the cytoplasm, followed by the assembly of the PG subunits at the cytoplasmic membrane, lipid anchored to undecaprenol (lipids I and II) (219, 256). The building blocks are translocated across the cytoplasmic membrane. Polymerization and cross-linking occur on the external face of the cytoplasmic membrane. This is achieved mainly through the action of the so-called penicillin-binding proteins, which catalyze the transglycosylation and transpeptidation reactions responsible for the formation of the glycosidic and peptide bonds of the PG, respectively (65, 219). Although this overall scheme of PG biosynthesis is quite similar in all LAB, many variations in the PG structure that may affect interactions of lactobacilli with the environment and the host can be introduced (65). Following polymerization and incorporation into the cell wall PG, GlcNAc and/or MurNAc may undergo different modifications, such as the O-acetylation of the cell wall MurNAc residues in L. casei (23). Additionally, the peptide cross-links can differ considerably. Frequently, d-asparagine is used as a cross-bridge between d-alanine and l-lysine in LAB (23), and this residue may also be amidated (65).
Teichoic Acids
Teichoic acids (TAs) are the second major component of the cell walls of most gram-positive bacteria. These anionic cell wall polymers are generally made of polyglycerol phosphate or polyribitol phosphate repeating units covalently anchored to either PG (wall TAs [WTAs]) or attached to the cytoplasmic membrane (lipoteichoic acids [LTAs]) (179). Both WTA and LTA are often substituted with glycosyl or d-alanyl (d-Ala) esters. The d-Ala ester substitution of LTA (and WTAs) requires four proteins that are encoded by the dlt operon. Two of these proteins are the d-alanyl:d-alanyl carrier protein ligase (Dcl, encoded by dltA), which activates d-alanine by use of ATP, and the d-alanyl carrier protein (Dcp), which is encoded by dltC. DltB is a transport protein predicted to be involved in the passage of the activated d-alanyl-Dcp complex across the glycerol phosphate backbone of LTA, while the DltD membrane protein facilitates the binding of Dcp for ligation with d-Ala and has thioesterase activity for removing mischarged d-alanyl carrier proteins (65, 179). Substitution contributes greatly to the functionality of these anionic polymers, as will be discussed below.
There are considerable differences between the WTA and LTA molecules of different Lactobacillus strains, especially in the degree of substitution and chain length (97, 184) and the quantity, which may account for 20 to 50% of the dry weight of the cell wall of lactobacilli, depending on the species or strain, growth stage, pH of the medium, carbon source, and availability of phosphate, etc. (65). Moreover, although all lactobacilli have TAs in their cell walls, not all Lactobacillus cell walls seem to contain WTA. For example, the cell walls of many L. rhamnosus and L. casei strains appear to contain only LTA (188), in contrast to most L. plantarum strains, which contain the two types of TAs (184). Additionally, a fraction of LTA may be released through deacylation (removal of the lipid anchor) or the inside-to-outside expansion of PG without the removal of the lipid anchor (65), indicating that LTAs of certain Lactobacillus strains can sometimes act as soluble factors, which is important for the interpretation of some functional studies discussed below.
Exopolysaccharides
Polysaccharides are also ubiquitous components of the cell envelope of lactobacilli, in addition to PG and TA (65). Conceptually, a distinction is made between the capsular polysaccharides, which form the thick outermost shell intimately associated with, and often covalently bound to, the cell wall, and cell-bound polysaccharides, which are loosely associated with it. Some extracellular polysaccharides are also released into the medium (206, 242). For some members of the Bacillaceae, a different class of “nonclassical” secondary cell wall polymers has been identified, which is involved in the anchoring of S-layer proteins to the bacterial cell surface (218). In the case of Lactobacillus buchneri, it was shown that hydroxyl groups of a neutral cell wall polysaccharide are responsible for the attachment of the S-layer protein to the cell wall (162). However, differentiation between these various classes of cell wall polysaccharides is often difficult. For lactobacilli, the term exopolysaccharides (EPSs) is preferred and relates to extracellular polysaccharides that can be attached to the cell wall or be secreted into the surroundings. Like polysaccharides in general, EPSs of lactobacilli are complex structures that differ not only in the nature of sugar monomers but also in their modes of linkage, branching, and substitution, greatly contributing to the structural variety of the Lactobacillus cell wall (71). EPSs of lactobacilli are generally heteropolysaccharides (HePSs) composed of different sugar moieties (glucose, galactose, rhamnose, GlcNAc, and N-acetylgalactosamine) (70, 71). Residues such as glucuronic acid and glycerol-3-phosphate can also be present, as can phosphate groups, acetyl, and pyruvyl groups although only in a subset of Lactobacillus strains (70). Information about the structures and biosynthesis pathways of these HePSs in lactobacilli is fragmented. The genes encoding HePS biosynthesis are typically located in a gene cluster of 12 to 25 kb depending on the complexity of the HePS (65, 71, 185, 276). The gene organization of these clusters seems to be highly conserved: a central region with similarity to glycosyltransferases is flanked by regions with similarity to genes involved in polymerization, export, and regulation. Based on homology, it is assumed that in lactobacilli, the repeating units of EPSs are synthesized in a stepwise manner that involves the intracellular formation of sugar nucleotides and the assembly of the subunit on undecaprenol at the cytoplasmic membrane by specific glycosyltransferases (65, 71, 185, 276). This final subunit is translocated across the membrane, followed by the linkage of the repeat units into long-chain polysaccharides. In lactobacilli, most EPSs are secreted or remain weakly bound to the cell wall by electrostatic interactions (ionic, hydrogen bonds, or hydrophobic interactions) (65, 71, 185, 245, 276). EPS production is also highly dependent on the environmental conditions, and the regulation of EPS production and chain length occurs, at least in part, through the action of a phosphoregulatory system that includes an autophosphorylating tyrosine kinase (18, 170, 175, 176). Nevertheless, much remains to be learned about the specific aspects of the biosynthetic pathway to EPSs in lactobacilli. Additionally, more knowledge is needed about the exact locations, compositions, polymer sizes, and conformation properties of surface polysaccharides on lactobacilli. It was recently shown by the application of single-molecule force spectroscopy that the probiotic strain L. rhamnosus GG has two major classes of surface polysaccharides: long galactose-rich EPS molecules and shorter glucose-rich (and/or mannose-rich) polysaccharides of unknown nature (87). Preliminary experiments indicate that some polysaccharide chains could be present as glycoproteins, adding an extra level of complexity to the Lactobacillus cell wall architecture.
In contrast to typical heteropolymeric EPS molecules, some Lactobacillus strains can also synthesize homopolysaccharides (HoPSs), glucans or fructans, from sucrose by the single action of extracellular glycosyltransferases, termed glucosyltransferases or fructosyltransferases, respectively. These glycosyltransferases use the energy of the osidic bond of sucrose to catalyze the transfer of a glycosyl moiety of sucrose to H2O, an acceptor carbohydrate, or a growing polymer chain (91). As for HePSs, these HoPSs can show a high degree of diversity in polymer length, linkages, and branching, etc. Because the biosynthesis of these HoPSs does not involve transport processes or the use of activated carbohydrate precursors, there is no energy requirement for the producer organisms other than the biosynthesis of the glycosyltransferase enzymes.
Cell Surface Proteins
Finally, the bacterial envelope of lactobacilli may comprise different cell wall-associated proteins, which are often large proteins consisting of repeating modules or particular domains. Cell surface proteins can be anchored to the cell wall by different mechanisms (178): by single N- or C-terminal transmembrane anchors, lipoprotein anchors (lipobox), LPxTG-type anchors (see below), or other cell wall-binding (repeated) domains such as LysM domains or glycine-tryptophan (GW) dipeptide motifs (16, 258). Other proteins are secreted in the surroundings, mediating interactions with the environment independent from direct cell contact. Some secreted proteins have also been shown to reassociate to the cell wall by electrostatic interactions (9, 16). Sortase-dependent proteins (SDPs) and S-layer proteins are best characterized in lactobacilli and will be further discussed below.
SDPs are an important group of cell surface proteins suggested to play a crucial role in Lactobacillus-host interactions (24). These SDPs are characterized by the presence of a cell wall-sorting signal at the C terminus that comprises a short pentapeptide motif (LPxTG) followed by a stretch of hydrophobic side chains and a positively charged tail. After transfer of the surface protein precursor to the plasma membrane and cleavage of the signal peptide, the SDP is retained within the cell wall by the cleavage of the sorting signal at the pentapeptide motif by a membrane-associated sortase. Sortase A (SrtA) cleaves between the threonine and glycine residues and then covalently links the threonine residue to an amino group of PG cross-bridges (177). Interestingly, gram-positive bacteria also utilize sortase-dependent mechanisms to assemble pili or fimbriae where sortase cross-links individual pilin monomers and ultimately joins the resulting covalent polymer to the cell wall PG (156). The occurrence of pili in lactobacilli does not seem to be a general rule, but they have been suggested to occur, for instance, in L. johnsonii (196).
Some specific lactobacillus strains are surrounded by a superimposed surface layer, the S layer, made of protein subunits packed into a paracrystalline hexagonal or tetragonal monolayer (9). Examples of S-layer-containing lactobacilli are L. acidophilus (27), L. gasseri, L. johnsonii (261), L. brevis (263), L. helveticus (40), and L. crispatus (228). S-layer proteins are usually small and highly basic proteins of 40 to 60 kDa with generally highly stable tertiary structures. These proteins are noncovalently bound to the cell wall, mostly to secondary cell wall polymers (LTA, WTA, and neutral polysaccharides), and assemble into surface layers with high degrees of positional order, often completely covering the cell wall. Although glycan structures have been identified in the S-layer proteins of several gram-positive bacteria, most appear to be nonglycosylated in lactobacilli (9). To date, a detailed glycan structure on S-layer proteins has been reported only for L. buchneri (9). Nevertheless, as mentioned above, some lactobacilli such as L. rhamnosus GG appear to have glycoproteins on their cell surface (87; data not shown).
GENETIC TOOLS TO STUDY LACTOBACILLI
Recent advances in genome sequencing projects, molecular tools, and genomic-based strategies for functional studies of lactobacilli contribute greatly to the identification of adaptation and probiotic factors (127). The number of genetic tools that have been developed has increased tremendously during the last 20 years. Genetic analysis is made possible for several strains of known probiotic lactobacilli, such as L. plantarum WCFS1 (128), L. acidophilus NCFM (4), L. johnsonii NCC533 (67, 196), L. salivarius UCC118 (50), L. reuteri ATCC 55730 (266, 277), and L. rhamnosus GG (62, 136). For these organisms, the first dedicated mutant analyses have confirmed some postulated roles of genes and molecules of lactobacilli involved in probiotic action, as will be discussed below (see Tables 2 to 5). Mutant studies are of the utmost importance in the unraveling of modes of action of lactobacilli as they can often directly relate genotype to phenotype. Nevertheless, this is still a technical challenge for many lactobacilli, and the number of currently identified genetic loci hypothesized to encode features supporting probiotic action confirmed by mutant analysis is still limited (see Table 5).
TABLE 2.
Genes of lactobacilli putatively involved in stress resistance studied by mutant analysis
| Functional category | Gene or locus tag(s)a | Identification approachb | Putative function | Organism | Mutant phenotype | Reference(s) |
|---|---|---|---|---|---|---|
| Cell envelope | lr1516 | Microarray expression analysis | Putative esterase involved in PG biosynthesis and reorganization | L. reuteri ATCC 55730 | Increased sensitivity to acid and bile | 266, 277 |
| LBA1272 | Dedicated approach | Cyclopropane fatty acid synthase | L. acidophilus NCFM | Increased sensitivity to acid | 127 | |
| dltD | Dedicated approach | d-Alanylation of LTA | L. rhamnosus GG | Increased sensitivity to simulated gastric juice (pH 2), increased sensitivity to defensins | 188 | |
| dltA | Dedicated approach | d-Alanylation of LTA | L. reuteri 100-23 | Decreased adaptation to acid, increased sensitivity to defensins | 270 | |
| slpA | Dedicated approach | S-layer protein | L. acidophilus NCFM | Increased resistance to bile, decreased resistance to osmotic stress | 127 | |
| cdpA | Comp. gen. (weak similarity to S-layer proteins) | Cell division and separation protein | L. acidophilus NCFM | Increased resistance to bile, decreased resistance to osmotic stress | 3, 127 | |
| Protection and repair DNA and proteins | dps | Microarray expression analysis | DNA protection during starvation and other stresses | L. reuteri ATCC 55730 | No significantly increased sensitivity to bile | 277 |
| clpL | Microarray expression analysis | Clp ATPase (chaperone) | L. reuteri ATCC 55730 | Increased sensitivity to acid and bile | 6, 277 | |
| clpE | Microarray expression analysis | Clp ATPase (chaperone) | L. reuteri ATCC 55730 | No significantly increased sensitivity to bile | 277 | |
| clpC | R-IVET | Clp ATPase (chaperone) | L. plantarum WCFS1 | Reduced persistence capacity in vivo | 31, 33 | |
| msrB | IVET | Methionine sulfoxide reductase | L. reuteri 100-23 | Reduced persistence capacity in vivo | 268, 269 | |
| luxS | Dedicated approach | Activated methyl cycle | L. rhamnosus GG | Reduced persistence capacity in vivo | 135 | |
| 2CRS regulators | LBA1524 | Comp.gen. (in silico) | Histidine protein kinase | L. acidophilus NCFM | Increased sensitivity to acid | 11 |
| LBA1430* | Microarray expression analysis | Histidine protein kinase | L. acidophilus NCFM | Increased sensitivity to bile | 191 | |
| LBA1431* | Microarray expression analysis | Response regulator | L. acidophilus NCFM | Increased sensitivity to bile | 191 | |
| LBA1432* | Microarray expression analysis | Hypothetical protein with similarity to RelA/SpoT | L. acidophilus NCFM | Increased sensitivity to bile | 191 | |
| rrp-1 | Dedicated approach | Response regulator | L. sakei 23K | Increased sensitivity to acid | 173 | |
| rrp-48 | Dedicated approach | Response regulator | L. sakei 23K | Increased sensitivity to acid | 173 | |
| Active removal of stressors | copA | R-IVET | Copper-transporting ATPase | L. plantarum WCFS1 | Reduced competitive ability in mouse intestine | 31, 33 |
| gadC (LBA0057) | Comp.gen. (genome)c | Glutamate/γ-aminobutyrate antiporter | L. acidophilus NCFM | Increased sensitivity to acid | 10 | |
| LBA0867 | Comp.gen. (genome)c | Transcriptional regulator | L. acidophilus NCFM | Increased sensitivity to acid and bile | 10 | |
| LBA0995 | Comp.gen. (genome)c | Amino acid permease | L. acidophilus NCFM | Increased sensitivity to acid and bile | 10 | |
| LBA0996 | Comp.gen. (genome)c | Ornithine decarboxylase | L. acidophilus NCFM | Increased sensitivity to acid but increased resistance to bile | 10 | |
| Lr1265 | Microarray expression analysis | Multidrug resistance protein (ABC transporter family) | L. reuteri ATCC 55730 | Decreased adaptation to bile | 277 | |
| Lr1584 | Microarray expression analysis | MDR protein (major facilitator family) | L. reuteri ATCC 55730 | Decreased adaptation to bile | 277 | |
| LBA1427* | Microarray expression analysis | Putative oxidoreductase, role in bile salt modification? | L. acidophilus NCFM | Increased tolerance to bile | 191 | |
| LBA1428* | Microarray expression analysis | Hypothetical protein with similarity to redox proteins, role in bile salt modification? | L. acidophilus NCFM | Increased tolerance to bile | 191 | |
| LBA1429* | Microarray expression analysis | MDR protein (major facilitator family) | L. acidophilus NCFM | Increased sensitivity to bile | 191 | |
| bshA | Comp.gen. (genome) | Bile salt hydrolase | L. acidophilus NCFM | Inability to hydrolyze bile salts conjugated to chenodeoxycholic acid; no altered bile tolerance | 165 | |
| bshB | Comp.gen. (genome) | Bile salt hydrolase | L. acidophilus NCFM | Inability to hydrolyze bile salts conjugated to taurine; no altered bile tolerance | 165 | |
| LJ0056, LJ1147, LJ1413 | Comp.gen. (genome) | Bile salt hydrolases | L. johnsonii NCC533 | A triple mutant showed gut persistence similar to that of the wild type | 67 | |
| bsh1 | Comp.gen. (genome) | Bile salt hydrolase | L. plantarum WCFS1 | Reduced tolerance to glycodeoxycholic acid but not to taurodeoxycholic acid | 132 |
*, these genes are located in an operon in L. acidophilus NCFM (191).
Comp.gen., comparative genomics.
Genome, information based on genome sequence.
TABLE 5.
Genes of lactobacilli encoding putative probiotic functions studied by mutant analysis
| Functional category | Gene | Identification approacha | Predicted function | Lactobacillus strain | Mutant phenotype | Reference(s) |
|---|---|---|---|---|---|---|
| Antipathogenic effects | labT | Comp.gen. (genome) | Putative ABC exporter for lactacin B | L. acidophilus NCFM | Reduced bacteriocin activity against closely related strains | 74 |
| abpT | Comp.gen. (genome) | Putative ABC exporter for bacteriocin Abp118 | L. salivarius UCC118 | Reduced effectiveness against L. monocytogenes infections in mice | 53 | |
| luxS | Dedicated approach | Direct role in the production of AI-2; indirect in the production of AI-3-like agonist molecules | L. reuteri 100-23 | Reduced capacity to induce virulence genes of pathogenic E. coli (EHEC) | 246 | |
| luxS | Dedicated approach | Direct role in production of AI-2; indirect role in production of AI-3-like antagonist molecules | L. reuteri ATCC 55730 | Reduced capacity to repress virulence genes of pathogenic E. coli (EHEC) | 115 | |
| Immunomodulation | dltB | Dedicated approach | d-Alanylation of LTA | L. plantarum NCIMB8826 | Increased anti-inflammatory potential in vitro in PBMCs and in vivo in a murine model of colitis or in a rat model for visceral pain perception | 76, 97 |
| dltD | Dedicated approach | d-Alanylation of LTA | L. rhamnosus GG | No significant difference in immunomodulation in PBMCs or HT-29 cells; increased sensitivity to human β-defensin-2 | 188 | |
| cps1A-J | Comp.gen. (genome) | Cell wall-associated high-molecular-mass polysaccharide biosynthesis | L. casei Shirota | Reduced proinflammatory potential in mouse macrophages and spleen cells | 283 | |
| LJ1680 | Microarray and CGH | 30% identity to IgA proteases | L. johnsonii NCC533 | Reduced persistence in murine GIT | 67 |
Comp.gen., comparative genomics.
With the increasing availability of genome sequences of lactobacilli, holistic approaches that complement traditional genetic and biochemical approaches are possible (51). Genome-scale comparisons of lactobacilli (comparative genomics) can reveal common as well as unique sequences that may shed light on the evolution of these strains and the genetic basis of particular traits, including differences between strains inhabiting the GIT or strains used in dairy industries. For instance, in silico analysis of the genome sequence of the cheese isolate L. helveticum DPC4571 (39) reflects considerable differences with the closely related L. acidophilus NCFM, a probiotic strain isolated from infant feces (4). L. helveticum DPC4571 is particularly marked by the loss of genes reported to be important for the adaptation to the gut environment. Half of the phosphoenol-pyruvate-dependent sugar phosphotransferases (PTS) (PEP-PTS), cell wall-anchoring proteins, and all the mucus-binding proteins predicted for L. acidophilus NCFM are absent or classified as being pseudogenes in DPC4571 (39). Genomes can also be compared by using DNA microarrays of a reference strain (comparative genome hybridization [CGH]), as exemplified for L. plantarum by a study that investigated the level of diversity in this species by a comparison of 20 strains (172). The main differences were found in transferable regions like prophages and insertion sequence elements but also in other regions that are predicted to encode the production of the bacteriocin plantaricin (see below), nonribosomal peptides, or EPSs. High levels of strain-specific variation were encountered in a 600-kb region containing genes involved mainly in sugar metabolism and which represents a lifestyle adaptation island (128, 172).
Although the availability of genome sequences will certainly advance the field, they need to be complemented with functional studies. Different methodologies have been developed for large-scale comparisons of differential gene expression, for instance, by comparing expression profiles of a strain grown in vitro in standard laboratory medium versus those of strains grown in vivo or in GIT-related in vitro simulations. Among the genes differently expressed in the GIT environment, candidate genes contributing to the adaptation and fitness of the microbes in the host environment are likely present. Examples of the methods that are yet applied for differential gene expression analysis for lactobacilli under relevant conditions are genome-wide comparisons of RNA profiles using microarrays (see, e.g., reference 66), comparison of protein profiles with two-dimensional (2D) difference gel electrophoresis (see, e.g., reference 141, 149), in vivo expression technology (IVET) using a promoter probe library (see, e.g., reference 31), and differential-display PCR (DD-PCR) (see, e.g., reference 130). Table 1 summarizes some advantages and disadvantages of these genetic tools that need to be taken into account when results are interpreted. Therefore, these molecular techniques can be considered as complementary, as will become apparent in the next sections, where we describe the functions identified by these studies.
TABLE 1.
Genetic tools used so far in the identification of adaptation and probiotic factors of lactobacilli
| Molecular tool | Description | Potential use | Pitfalls | Key reference(s) |
|---|---|---|---|---|
| Dedicated mutant analysis | Targeted gene mutation by gene deletion or insertional inactivation | Identification of cause-consequence relationships linking genes and their functions; allows in vivo confirmation of proposed functions; allows studies on the role of surface components in their native conformation | Requires a hypothesis from which to start (“bottom-up” approach); requires that a strain is genetically accessible; pleiotropic effects of a certain mutation can occur, complicating cause-consequence relationships | 53, 97a |
| In silico analysis | Algorithm-based analysis of sequences | Mining for yet-undocumented genetic elements and interactions; predictions on evolution | Requires the availability of the genome sequence; assigned functions are putative and need to be confirmed experimentally; erroneously annotated sequences can easily be spread | 4, 39, 50, 128, 196 |
| CGH | DNA-DNA hybridization-based comparison of sequences | Enables identification of unique sequences in the reference strain | Gives information only about genes present in the reference strain; cross-hybridization of similar sequences is possible, complicating interpretation | 67, 172 |
| DNA microarray for transcription profiling | DNA-cDNA hybridization-based method for analyzing transcription | Provides a global view of transcription under specific conditions | Requires genome sequence; depends on the annotation of open reading frames for a classical microarray (not for a tiling array); only a “snapshot” view of transcription; difficult to obtain sufficient RNA from in vivo samples; good analytical tools are needed; role of identified genes needs to be confirmed by downstream analyses | 34, 66, 67, 266, 277 |
| Proteomics | Large-scale analysis of protein profiles (gel or non-gel based) | Proteins are most directly related to functions of the cell; can reveal posttranslational modifications | Identification of proteins requires mass spectrometry facility; coverage is mostly only partial; difficult to obtain sufficient protein from in vivo samples; downstream analyses are needed | 141, 149 |
| IVET and R-IVET | Promoter-trap technique that allows selection of active promoters in vivo | Allows in vivo identification of putative adaptation factors; with R-IVET, weak and transiently expressed genes can be identified | Requires a genetically accessible strain; only promoter activities are detected; downstream analyses are needed by, e.g., construction of knockout mutants | 31, 269 |
| DD-PCR | Based on PCRs of reverse transcribed RNA (cDNA) with random primers | Availability of genome sequence or special genetic tools is not required | Overrepresentation of structural RNA in total RNA can result in false positives; isolation of bacterial RNA from in vivo samples is difficult | 130 |
MECHANISMS OF SURVIVAL AND PERSISTENCE OF LACTOBACILLI IN THE HOST: ADAPTATION FACTORS
Active Stress Resistance Mechanisms in the Host
Probiotic lactobacilli encounter various environmental conditions upon ingestion by the host and during transit in the GIT (Fig. 1). Firstly, they need to survive the harsh conditions of the stomach. Humans secrete approximately 2.5 liters of gastric juice each day, generating a fasting pH of 1.5, increasing to pH 3 to 5 during food intake (55). The precise effects of acid stress on bacterial physiology are not fully known. Lowering the intracellular pH reduces the transmembrane pH difference, which determines the proton motive force used as an energy source in numerous transmembrane transport processes. Internal acidification also reduces the activity of acid-sensitive enzymes and results in damage to proteins and DNA (255).
Given that the liver excretes approximately 1 liter of bile each day into the small intestine, exposure to bile represents another challenge for bacteria entering the GIT (17). Bile acids are synthesized from cholesterol and are conjugated to either glycine or taurine. Although the toxicity of bile acids for bacterial cells is not well understood, bile acids are surface-active, amphipatic molecules with potent antimicrobial activity and act as detergents, disrupting biological membranes. Moreover, bile salts also seem to induce an intracellular acidification so that many resistance mechanisms are common for bile and acid stress (17). Indeed, the protonated form of bile salts is thought to exhibit toxicity through intracellular acidification in a manner similar to those of organic acids like the lactic acid produced by the lactobacilli themselves (17). These acids can passively diffuse in the undissociated form through the cell membrane (or via a transporter) and, after entry into the cytoplasm, rapidly dissociate into protons and charged derivatives to which the cell membrane is impermeable (255).
In addition to coping with acid and bile, the contribution of other stress responses to the survival capacity of lactobacilli in the GIT should not be overlooked, although they remain rather unexplored (22). In analogy to the stresses encountered by intestinal pathogens (212), these certainly include oxidative and osmotic stress. Moreover, it can be anticipated that interactions with other microbes and with cells of the immune system and the various antimicrobial products that they produce can also impose a serious threat for the probiotic microbes, two aspects that will be covered below. Interestingly, the phenomenon of cross-adaptation is often observed, i.e., that adaptation to one stress condition also protects against another stress factor, implying some common mechanisms (17, 61, 255). In this respect, non-actively-growing stationary-phase cells are generally more resistant to various stressors than early-log-phase cells.
Multiple genome-wide studies and downstream functional analyses have now been performed to characterize various stress responses in lactobacilli (Table 2). Although different culture conditions, types of stresses, strains, exposure times, and growth phases were used for these studies, some common themes have emerged, which are summarized below. Some factors contribute rather aspecifically to stress resistance, such as maintaining the cell envelope integrity and protecting and repairing macromolecules, while other mechanisms contribute in a specific way, such as dedicated stress-sensing and export systems. For a detailed overview of acid, bile, and other stress resistance mechanisms of lactobacilli, the reader is referred to more extensive reviews (17, 55, 61, 255).
Maintaining integrity of the cell envelope.
The different macromolecules constituting the cell membranes and cell walls of lactobacilli have been shown to contribute to maintaining cell integrity during stress to various degrees. For example, low pH caused a shift in the fatty acid composition of the cell membrane of an oral strain of L. casei (86). Similarly, bile salts have been shown to induce changes in the lipid cell membrane of L. reuteri CRL1098 (249). For some lactobacilli, the genetic factors involved have been identified, for example, by using a microarray analysis; a gene encoding a putative phosphatidyl glycerophosphatase was shown to be two- to threefold induced in L. reuteri ATCC 55730 after an acid shock to pH 2.7 (266). Klaenhammer and colleagues also reported that the inactivation of LBA1272 in L. acidophilus NCFM, encoding a cyclopropane fatty acid synthase, resulted in increased acid sensitivity (127).
In addition, genes involved in PG biosynthesis and organization have been identified in various screens for acid responses in lactobacilli. A putative esterase gene, lr1516, belonging to the family of penicillin-binding proteins, was shown to be induced in L. reuteri ATCC 55730 under acidic conditions by the same microarray study mentioned above (266). The disruption of lr1516 significantly increased the sensitivity to acid shock. Interestingly, Whitehead et al. identified lr1516 as being induced by bile exposure as well (277). They also confirmed the importance of this gene for the survival of bile by mutant analysis. In addition, by use of microarray analysis, Pfeiler et al. identified a relatively high number of L. acidophilus NCFM genes involved in cell membrane, PG, and surface protein (e.g., srtA) biosynthesis as being differentially expressed after exposure to bile (191). Furthermore, genetic characterization of the bile salt response in L. plantarum WCFS1 by screening of a promoter probe library also resulted in the identification of several genes involved in cell envelope functions, including genes encoding muramidases (32). Interestingly, in L. acidophilus NCFM, the disruption of cdpA, a putative cell wall-modifying enzyme that promotes cell division and separation, resulted in an increased level of resistance to bile salts compared to that of the wild type, while the cdpA mutant showed reduced resistance to osmotic stress (3, 127). According to those authors, these effects might result from a presumed immature structure of the cell wall in the mutant, where certain components remained cross-linked or uncleaved. Similarly, an slpA mutant of L. acidophilus NCFM was reported to be more bile resistant while being more sensitive to osmotic stress (127).
Additionally, d-Ala esters of LTA and WTA have been suggested to be necessary for the proper functioning of cell integrity at low pH and in the presence of bile (179). For instance, DNA microarray studies identified the L. plantarum WCFS1 dlt operon as being induced by bile (34). Additionally, a dltD mutant of L. rhamnosus GG was shown to be severely affected in its capacity to survive in simulated gastric juice at pH 2 (188). In contrast, the inactivation of dltA in L. reuteri 100-23, a rodent commensal, did not affect the viability of this strain at low pH in vitro. However, the dltA mutation had a pronounced effect on in vitro growth at low pH (acid adaptation) and the colonization capacity of the murine forestomach (270).
The role of EPS in acid and bile resistance is less clear. Microarray expression analyses of L. acidophilus (191) and L. reuteri (277) identified genes of EPS biosynthesis as being reduced after bile exposure, including epsB, epsC, and epsE in L. acidophilus and lr0957 in L. reuteri, respectively. EpsB, EpsC, and lr0957 show homology to proteins involved in the phosphoregulatory system that regulates EPS biosynthesis and chain length in streptococci (18, 174). epsE encodes the putative priming glycosyltransferase, which catalyzes the transfer of the first sugar in EPS polymer biosynthesis (191). However, EPS production has not been studied in detail after exposure to bile. In fact, to our knowledge, phenotypic analyses of dedicated Lactobacillus mutants affected in EPS biosynthesis genes have not yet been performed. HoPSs from L. reuteri have been reported to have a more established role in stress resistance by the maintenance of the cell membrane in the physiological liquid crystalline phase under adverse conditions (91). However, although both an inu (inulose sucrase) and gftA (glucosyltransferase) mutant of L. reuteri TMW1.106 showed a lower resistance to lactic acid, the resistance of the mutant strains to low pH was not affected (223).
Repair and protection of DNA and proteins.
A number of proteins that play a role in the protection or repair of macromolecules such as DNA and proteins also seem to be essential for acid and bile resistance. Intracellular acidification can result in a loss of purines and pyrimidines from DNA. For example, using Northern analysis and reverse transcription (RT)-PCR, Cappa et al. observed an increased level of expression of uvrA, coding for subunit A of the excinuclease ATP-binding cassette (ABC) complex, involved in nucleotide excision repair, at low pH and suggested a role for this system in acid adaptation in L. helveticus CNBL 1156 (43). Bile acids have also been shown to induce DNA damage and the activation of enzymes involved in DNA repair (17). In L. reuteri, it was observed by microarray analysis that the level of expression of dps (DNA protection during starvation) increased after bile exposure (277). Dps is involved in several types of stress adaptation in E. coli, including oxidative stress, irradiation, metal toxicity, heat stress, and pH stress (160). However, the disruption of dps in L. reuteri did not significantly affect the organism's ability to survive bile shock or adapt to bile (277), probably due to the redundancy of DNA protection and repair enzymes.
Perhaps even more vital in the general stress response are chaperones that intervene in numerous stresses for important tasks such as protein folding, renaturation, protection of denatured proteins, and removal of damaged proteins. Important molecular chaperones include DnaK, GroEL, and GroES, the well-known heat shock proteins. A 2D proteomic approach to study acid adaptation in L. delbrueckii subsp. bulgaricus identified three highly induced proteins, i.e., GroES, GroEL, and DnaK (144). Similar approaches were used to demonstrate DnaK, DnaJ, GrpE, GroES, and GroEL production in L. acidophilus as a response to acid adaptation (149); GrpE upregulation in acid-tolerant mutants of L. sanfranciscensis (60); and increased levels of expression of GrpE and DnaK in L. reuteri ATCC 23272 after 1 h of incubation at pH 4 (141). Similarly, in a microarray analysis of L. acidophilus NCFM after bile exposure, groES, groEL, dnaK, htrA, and grpE were found to be upregulated (191). These heat shock proteins seem to be especially pivotal for long-term acid stress resistance.
Clp ATPases perform a similar role by targeting misfolded proteins for degradation by the ClpP peptidase, in addition to protein reactivation and remodeling activities (88). In contrast to the heat shock proteins described above, these Clp proteins seem to be particularly important for the fast response of lactobacilli when they encounter adverse conditions in the GIT. The microarray approach to the study of acid shock in L. reuteri ATCC 55730 revealed clpL as being one of the induced genes (266). Interestingly, bile shock induced the same clpL chaperone gene, while the clp chaperone genes were not overexpressed during bile adaptation (277). Mutant analysis confirmed the importance of this ATPase with chaperone activity. The clpL knockout mutant of L. reuteri showed a significantly decreased rate of survival after incubation at pH 2.7 (266) or incubation in 0.3% bile (277). The vital role of these chaperones for GIT survival is further highlighted by the identification of clpC as being one of the genes of L. plantarum WCFS1 being induced in the murine GIT through in vivo expression technology (recombinase-based IVET [R-IVET]) (31). A follow-up mutant analysis showed that the persistence capacity of a WCFS1 clpC mutant in the murine GIT was 10- to 100-fold decreased compared to that of the wild-type control (33).
As mentioned above, other stressful situations such as oxidative stress can be encountered in the GIT. An IVET study of L. reuteri 100-23 identified a gene encoding a methionine sulfoxide reductase (Msr) to be induced in the murine GIT (269). Msr reverses the loss of the biological activity of proteins caused by the oxidation of methionine to methionine sulfoxide and therefore contributes to the protection of bacteria against oxidative damage caused by, e.g., reactive nitrogen and oxygen intermediates. The exact role of Msr in the in vivo survival of L. reuteri remains to be determined. Nevertheless, the ecological performance of an msrB mutant was significantly impaired in mice in further downstream mutant analysis experiments (268). Bile stress has also been shown to induce oxidative stress (17). Bron and coworkers identified increased levels of expression of glutathione reductase and the metC-cysK operon upon bile treatment in L. plantarum (34). Glutathione is an important biomarker for oxidative stress and might play an important role in the in vivo survival of bacteria. In addition to its key role in maintaining the proper oxidation state of protein thiol groups, glutathione also serves a key function in protecting the bacterial cell from the action of low pH, chlorine compounds, and osmotic stresses (161). The metabolism of glutathione and cysteine is tightly linked to the activated methyl cycle and the metabolism of S-adenosylmethionine (SAM) (278). This SAM cycle plays a central metabolic role and is involved in rRNA nucleotide modification, polyamine synthesis, and methylation processes. All these processes can be implicated in promoting the stability of macromolecules under stresses (147). Interestingly, the two IVET studies of lactobacilli identified one common gene encoding a putative vitamin B12-independent methionine synthase belonging to this activated methyl cycle (31, 269). The inactivation of this gene in L. reuteri, designated met, did not affect the ecological performance, probably as L. reuteri carries redundant functions. In contrast, the LuxS enzyme, catalyzing the conversion of S-ribosylhomocysteine to homocysteine in the same pathway, seems to be crucial for the survival of the probiotic strain L. rhamnosus GG in the murine GIT (135). In a competition assay with the wild type, the number of luxS mutant cells that survived passage through the GIT gradually decreased to less than 1% compared to the wild type (135). Recently, Lee et al. showed by proteomic analysis that SAM synthetase is upregulated in bile-stressed L. reuteri cells (140). Those authors also linked the upregulation of this enzyme to the central metabolic role of the SAM cycle in conferring stability to bacterial components.
Two-component and other regulatory systems.
Mechanisms to specifically sense the presence of certain stress factors and regulate gene expression in response to these stimuli are also crucial for bacterial survival under adverse conditions. Although these mechanisms are not well characterized for lactobacilli, they often involve two-component regulatory systems (2CRSs). 2CRSs allow bacteria to sense and respond to changes in their environment after receiving an environmental signal through transmembrane sensing domains of the histidine protein kinase (HPK). Once it receives a signal input, the HPK is activated to autophosphorylate a specific histidine residue. The phosphoryl group is then transferred to the regulatory domain of the response regulator (RR), which in turn induces a transcriptional response through its DNA-binding domain (237).
Various studies have shown a role for 2CRSs in stress responses of lactobacilli. In L. sakei 23K, the disruption of the rrp-1 and rrp-48 genes, encoding RRs, resulted in an increased susceptibility to low pH (173). Klaenhammer and coworkers identified a 2CRS (LBA1524-LBA1525) in L. acidophilus NCFM that was similar to the acid-related system LisRK from Listeria monocytogenes (11). The insertional inactivation of the HPK resulted in decreased rates of survival of log-phase cells after exposure to pH 3.5. Moreover, microarray analysis identified approximately 80 genes in L. acidophilus NCFM for which the expression was changed by the HPK mutation (11). The most dramatic changes in expression in the HPK mutant were observed for genes predicted to encode components of the proteolytic enzyme system, including two oligopeptide transport systems. One major function of oligopeptide transport (Opp) systems for bacterial cells is to internalize peptides to be used as carbon and nitrogen sources. These transport systems are also involved in the recycling of the cell wall peptides, which are likely the first targets of physiochemical stress, but this role is not well established in gram-positive bacteria (134). Similarly, in L. reuteri, the microarray analysis mentioned above also identified an RR (lr1804) as being induced after acid shock (266). This lr1804 gene is part of an operon homologous to the yycFG operon in Bacillus subtilis, where this RR regulates genes involved in cell wall metabolism, such as components of TA biosynthesis (110). However, no further analysis of the signaling events by this 2CRS has yet been performed.
The microarray study by Pfeiler and coworkers to characterize the bile response of L. acidophilus NCFM also identified, among many other genes, a 7-kb eight-gene operon encoding a 2CRS, a transporter, an oxidoreductase, and four hypothetical proteins (191). Mutations in the transporter, the HPK, the RR, and a hypothetical protein that shows similarity to RelA (SpoT) (see below) each resulted in a loss of tolerance to bile. Mutations in other genes of the 7-kb operon encoding another hypothetical protein and a putative oxidoreductase resulted in significant increases in bile tolerance, showing the importance of this operon in both bile tolerance and bile sensitivity. These data suggest that this 2CRS could have a complex bile-sensing role, but the details of the regulatory network still need to be defined (191).
Other common themes for important regulators in the stress responses of lactobacilli are less easy to delineate. For instance, RelA is involved in the synthesis and hydrolysis of (p)ppGpp, a signal known to be involved in the stringent response and induction of tolerance against different types of stresses (235). In L. lactis, the inactivation of genes (guaA, encoding the GMP synthetase, and relA) involved in guanine nucleotide metabolism resulted in increased acid tolerance (201). In L. reuteri ATCC 55730, microarray analysis showed that the level of expression of relA was decreased after acid shock (266). However, further functional analyses are needed to characterize the role of this system in lactobacilli.
Active removal of acid- and bile-related stress factors.
Bacteria have also evolved many direct and rather specific strategies to actively remove different stress factors.
(i) ATPases.
The multisubunit FoF1 ATPase, which facilitates the extrusion of protons from the cytoplasm by proton motive force, is one of the main proton pumps utilized by gram-positive bacteria (55). DD-PCR experiments showed that exposure to low pH in L. acidophilus causes an increase in mRNA levels of a pH-inducible, proton-translocating F1Fo ATPase (130), but dedicated mutant analyses have not yet been reported. Corcoran et al. used spontaneous neomycin-resistant mutants of the probiotic strain L. rhamnosus GG with reduced FoF1 ATPase activity to highlight the importance of the presence of fermentable sugars and ATP generation via glycolysis in proton exclusion by the FoF1 ATPase (52). Lee et al. also observed by 2D analysis a significant overexpression of some glycolytic proteins in response to acid stress, including glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate mutase, and pyruvate kinase, stressing the importance of the generation of energy-rich intermediates (ATP and NADH) at a low pH (141). Furthermore, the genes encoding the FoF1 ATPase were also upregulated in L. plantarum when exposed to bile (34), consistent with the fact that bile exposure results in a mild acidification of the cytoplasm, as mentioned above.
Recently, a role for heavy-metal-transporting ATPases and copper homeostasis in the acid tolerance of L. bulgaricus ATCC 11842 has been suggested (186). The exact function of these transporters is not known, but it is remarkable that Kleerelezem and coworkers also identified a copA gene coding for a putative copper-transporting ATPase (lp_3055) as being induced in the murine GIT (31, 157). A competition experiment between the wild type and the copA mutant showed that the relative abundance of the copA mutant was 100- to 1,000-fold decreased after passage through the murine GIT (33). Interestingly, Denou and coworkers also identified the stomach-specific expression of a copper-transporting ATPase in L. johnsonii NCC533 (66), indicating that it must have some important, but not yet completely understood, in vivo role for lactobacilli.
(ii) Amino acid decarboxylation-antiporter reactions.
In amino acid decarboxylation-antiporter reactions, an amino acid is transported into the cell, where decarboxylation occurs. A proton is consumed in the reaction, and the product is exported from the cell via an antiporter. The net result is an increase in the intracellular pH. An example of this system is glutamate decarboxylase (GAD), which was biochemically characterized for L. brevis (253). It has been proposed that ATP could be generated by glutamate conversion to γ-aminobutyrate in lactobacilli, thereby coupling the GAD system to ATP synthesis (108). In L. acidophilus NCFM, insertional inactivation showed the importance of an ornithine decarboxylase, an adjacent amino acid permease, a glutamate γ-aminobutyrate antiporter, and a transcriptional regulator with weak similarity to the regulator of the GAD system in L. lactis for acid tolerance (10). Moreover, that study confirmed that stationary-phase cells are generally more tolerant to low pH than are log-phase cells, as elaborated above.
(iii) ADI pathway.
An additional mechanism that has been implicated in acid and bile tolerance is the production of alkaline compounds, more specifically, ammonia, by the arginine deiminase (ADI) pathway, which catalyzes the conversion of arginine to ornithine, ammonia, and CO2. This also results in the generation of ATP, enabling the extrusion of cytoplasmic protons by the FoF1 ATPase (57). The system has three main enzymes, ADI, ornithine transcarbamylase, and carbamate kinase, encoded by arcA, arcB, and arcC, respectively. Additionally, ArcD, an arginine-ornithine transporter, is present in many organisms and allows the exchange of these two molecules at no energy cost. Whitehead et al. also found the genes of the ADI pathway to be specifically induced during bile adaptation in L. reuteri ATCC 55730 (277), further highlighting that exposure to bile can result in a mild acidification of the cytoplasm.
(iv) Transport and hydrolysis of bile.
Some bacteria use efflux systems belonging to the family of multidrug resistance (MDR) transporters to export bile (17). The role of an MDR transporter gene of L. acidophilus NCFM, as one of the genes of the identified 2CRS operon important for bile tolerance (191), is mentioned above in the paragraph on 2CRS systems. In their screening for bile-responsive genes in L. plantarum WCFS1, Bron et al. identified three possible exporter proteins, including a putative MDR transporter gene (lp_3160) (32). Microarray analysis of L. reuteri ATCC 55730 also identified two putative MDR genes, lr1265 and lr1584, as being induced after bile treatment (277). Mutational analysis showed that the disruption of lr1265 and lr1584 did not decrease the survival capacity in bile. However, these mutants could not grow in the presence of bile, suggesting that these MDR transporters play an important role in bile adaptation (277).
Several bacteria are also known to enzymatically modify bile salts (17). Bile salt hydrolases (BSHs) are generally intracellular enzymes that catalyze the hydrolysis of the amide bond between the steroid moiety and the amino acid side chain of the bile acids. BSH activity is found primarily in organisms isolated from the GIT of mammals (Bifidobacterium species, L. acidophilus, L. gasseri, L. johnsonii, and some strains of L. plantarum), while organisms isolated from environments that do not contain bile acids, such as fermented milk products and vegetables (L. lactis, L. delbrueckii, L. helveticus, and Streptococcus thermophilus), do not exhibit BSH activity (17). Nevertheless, the role of BSH in the GIT survival capacity of these lactobacilli remains elusive. A bsh-1 mutant of L. plantarum WCFS1 was recently reported to be affected in tolerance to glycine-conjugated bile salts (131). However, the inactivation of two genes, bshA and bshB, coding for BSH enzymes with different catalytic activities in L. acidophilus NCFM, did not affect bile tolerance (165). Moreover, a triple-knockout mutant for all three BSH proteins of L. johnsonii NCC533 was not affected in its capacity to persist in the murine GIT (67).
Adaptation to the Host Nutritional Environment
In addition to the survival capacity of lactobacilli under various stress conditions, the capacity to adapt to the special nutritional conditions is of utmost importance for their residence time and survival in the various microhabitats of the GIT. Pioneering studies with the important human symbiont Bacteroides thetaiotaomicron have nicely shown the importance of a flexible carbohydrate-foraging behavior in the lower parts of the intestine (109, 231). Additionally, mutations in sugar acquisition pathways were found to affect the colonization of commensal E. coli of the murine GIT (46). Important sources of carbon and energy for bacteria growing in the gut are simple sugars that are readily utilized in the upper GIT and nondigestible complex carbohydrates that remain abundant in the lower part of the GIT and originate from the diet or from host mucins. Thus, bacteria such as Bifidobacterium longum, which contain numerous genes predicted to be involved in polysaccharide degradation, are probably better suited to reside in the colon, whereas species containing various sugar transporters, such as most intestinal lactobacilli, seem better adapted to the proximal compartments of the GIT (83).
Importance of carbohydrate metabolism in the host.
For a description of the metabolic activities of lactobacilli that are important for adaptation in the host, we integrated the available genomic information with functional studies (see Table 3 for the studies that included mutant analyses).
TABLE 3.
Genes of lactobacilli putatively involved in active metabolism in the host studied by mutant analysis
| Functional category | Gene | Identification approacha | Predicted function | Organism | Mutant phenotype | Reference(s) |
|---|---|---|---|---|---|---|
| Carbohydrate metabolism | LJ1654-LJ1656 | Microarray expression and CGH | PTS sugar transporter | L. johnsonii NCC533 | Reduced persistence time in mouse intestine | 67 |
| pts14C | R-IVET | Cellobiose PTS, EIIC | L. plantarum WCFS1 | Reduced competitive ability in the murine GIT | 31, 33 | |
| xylA | IVET | Xylose isomerase | L. reuteri 100-23 | No reduced competitive ability in the murine GIT | 268, 269 | |
| inu | Dedicated approach | Inulosucrase | L. reuteri TMW1.106 | Reduced competitive ability in the murine GIT | 271 | |
| gtfA | Dedicated approach | Glucosyltransferase | L. reuteri TMW1.106 | Reduced competitive ability in the murine GIT with L. johnsonii 21 but not with the wild type | 271 | |
| bfrA | Comp.gen. | Intracellular β-fructosidase (FOS metabolism) | L. acidophilus NCFM | Reduced ability to grow on FOS; no in vivo studies reported yet | 13 | |
| msmE | Comp.gen. | ABC transporter substrate binding protein (FOS metabolism) | L. acidophilus NCFM | Reduced ability to grow on FOS; no in vivo studies reported yet | 13 | |
| fosE | Shotgun microarray approach | Extracellular β-fructosidase (FOS metabolism | L. paracasei 1195 | Reduced ability to grow on FOS; no in vivo studies performed yet | 93 | |
| treC | Comp.gen. (genome) | Trehalase, osmoprotection? | L. acidophilus NCFM | Reduced ability to grow on trehalose, cryosensitive; no in vivo studies reported yet | 77 | |
| Protein metabolism | prtP (LJ1840) | Comp.gen. | Cell wall-bound proteinase | L. johnsonii NCC533 | No altered competitive ability in murine GIT | 67 |
| met | IVET | Putative vitamin B12-independent methionine synthase | L. reuteri 100-23 | No altered competitive ability in murine GIT | 268, 269 |
Comp.gen., comparative genomics.
(i) L. johnsonii.
The genome sequence of L. johnsonii NCC533 (formerly L. acidophilus La1) revealed that this organism lacks genes encoding biosynthetic pathways for amino acids, purine nucleotides, and most cofactors, explaining its complex nutritional requirements (196). In contrast, the L. johnsonii genome is predicted to code for highly expressed sugar and amino acid transporters and a large number of peptidases and peptide transporters, illustrating how this bacterium may have evolved efficient mechanisms to acquire such nutrients from its environment. Functional analysis of genes associated with the long gut persistence of this L. johnsonii NCC533 strain confirmed the importance of sugar metabolism (67). Denou and coworkers used a combination of comparative genome hybridization with the type strain L. johnsonii ATCC 33200 and microarray analysis of genes of NCC533 expressed in the jejunum of NCC533-monoassociated mice (67). The interest in this comparison arose from the fact that L. johnsonii NCC533 is able to reside significantly longer in the murine GIT than type strain ATCC 33200. The combination of the microarray data sets of both experiments identified three gene loci that were both specific to NCC533 and expressed in vivo. Among these loci was LJ1654-LJ1656, encoding a PEP-PTS transporter annotated as mannose PTS. Mutational analysis showed a distinct in vitro sugar growth pattern compared to that of the wild type. Moreover, the deletion of LJ1654-LJ1656 resulted in a significantly reduced persistence time in the murine GIT (67). As indicated above, these sugar transporters are especially important in the small intestine. Indeed, L. johnsonii NCC533 was recently shown to be rather metabolically inactive in the colon, with only 26 genes being expressed, as detected by microarray analysis (66). In contrast, L. johnsonii isolated from the jejunum showed transcripts for 297 genes, including the jejunum-specific expression of three sugar PTS transporters (annotated with fructose, glucose, and cellobiose specificity) and high-level expression of sugar-digesting enzymes. Additionally, the specific expression pattern of NCC533 in the cecum revealed several PTS transporters, including a cecum-specific galactosamine PTS transporter, highlighting the importance of sugar import. On the contrary, an L. johnsonii NCC533 mutant of the gene encoding the Prp protease, for degradation and growth on milk proteins, was not affected in its gut persistence capacity, suggesting that this protein acquisition system is less important in vivo (67).
(ii) L. acidophilus.
Similarly to L. johnsonii NCC533, L. acidophilus NCFM appears to be unable to synthesize most amino acids, vitamins, and cofactors but compensates with a large number of transport systems for amino acids and peptides and genes encoding peptidases and proteases (4). In addition, the genome of L. acidophilus NCFM encodes a large number of proteins related to carbohydrate transport and metabolism, reflecting its ability to use a variety of sugars including mono-, di-, and polysaccharides such as raffinose and fructooligosaccharides (FOS). Recently, the transport and catabolic machinery involved in carbohydrate utilization by L. acidophilus NCFM was characterized by using microarrays. Specific transporters were identified for different sugars, including PTS transporters for monosaccharides and ABC transporters for the uptake of oligosaccharides. Examples are the previously identified locus for trehalose utilization putatively involved in osmo- and cryoprotection (77) and the operon for FOS utilization (13). Genes central to glycolysis were found to be among the most highly expressed in the genome (14). According to those authors, this flexible carbohydrate metabolism of L. acidophilus NCFM likely contributes to its competitive ability for limited carbohydrate sources available in the human GIT (14). Moreover, the capacity of strain NCFM to degrade a variety of complex carbohydrates, next to simple sugars, suggests that it is adapted to survive in the lower parts of the intestine, more so than L. johnsonii NCC533 (see above). Additional evidence for the importance of this flexible carbohydrate utilization for the competitive ability of L. acidophilus NCFM in the GIT results from the comparison with the closely related strain L. helveticus DPC4571, which has adapted to milk and dairy environments (39). This cheese isolate has lost all the ABC transporters for FOS and raffinose and most of the glucosidase enzymes and contains only 9 out of the 20 PEP-PTS transporters described for L. acidophilus NCFM, as mentioned above. However, to our knowledge, the confirmation of the in vivo role of this versatile carbohydrate utilization capacity in the persistence capacity of L. acidophilus NCFM has not yet been reported.
(iii) L. plantarum.
The L. plantarum WCFS1 genome has also yielded information on how this bacterium may have adapted to growth in diverse environments such as fermented foods, plants, and the human GIT (128). For example, the flexible and adaptive behavior of WCFS1 is reflected by the relatively large number of regulatory and transport functions, including 25 complete PTS sugar transport systems. Consistent with the classification of L. plantarum as a facultative heterofermentative lactic acid bacterium, the genome also encodes all enzymes required for the glycolysis and phosphoketolase pathways, all of which appear to belong to the class of potentially highly expressed genes in this organism. Moreover, L. plantarum WCFS1 encodes a large pyruvate-dissipating potential, emphasizing its fermentative capacity. Many genes encoding sugar transport and utilization, as well as genes encoding extracellular functions, appear to be clustered in the 600-kb lifestyle adaptation island mentioned above. This genome information on L. plantarum WCFS1 has been complemented with some functional analyses. Several genes encoding proteins involved in metabolizing various carbon sources were indeed identified by R-IVET analysis in L. plantarum, including five PTS systems (for GlcNAc, sorbitol, sucrose, and cellobiose), a ribose permease, a ribokinase, and two di- and polysaccharide-hydrolyzing enzymes (31). Follow-up real-time quantitative RT-PCR confirmed that the L. plantarum ribokinase is expressed along the murine GIT (157). Moreover, the transcription of an important indicator for active fermentation, ldhL, was induced more than twofold in the cecum and colon, probably because of the longer residence time in these distal compartments (157). Mutational analysis of the cellobiose PTS (lp_1164), ribose transport protein, and ribokinase (lp_3559-lp_3660) confirmed the key role of the cellobiose PTS in the functionality of L. plantarum WCFS1 in the murine GIT (33). In a competition experiment, the relative abundance of the Δlp1164 mutant was 100- to 1,000-fold lower than that of the wild-type control strain after passage through the GIT.
Although carbohydrate metabolism is of primary importance in the metabolism of lactobacilli in the GIT (and other niches), other nutrients also need to be acquired, as shown by the R-IVET study of L. plantarum WCFS1. Bron and coworkers identified nine genes that are involved in the acquisition of nonsugars, including factors involved in the uptake and synthesis of amino acids, nucleotides, cofactors, and vitamins (31). Examples are genes involved in arginine, proline, and biotin metabolism and metal ion acquisition. In L. monocytogenes, proline metabolism is induced under high-osmolarity conditions (229), such as those present in the small intestine and colon. Under conditions of osmotic stress, lactobacilli have systems for accumulating specific solutes that do not interfere with the physiology of the cell, such as the accumulation of glycine betaine, carnitine, proline, and glutamate (61).
(iv) L. salivarius.
In L. salivarius UCC118, the genome comprises a 1.8-Mb chromosome and a 242-kb megaplasmid (pMP118) (50). The annotation of the pMP118 sequence suggests that it confers a range of additional metabolic capabilities upon L. salivarius UCC118, such as rhamnose, ribose, and sorbitol utilization, and two key enzymes of the pentose phosphate pathway (transketolase and transaldolase). According to those authors, this megaplasmid could provide a selective advantage for this L. salivarius strain to adapt to environments known for frequent dietary fluctuations, such as the GIT. However, to our knowledge, no in vivo functional studies have yet been performed.
(v) L. reuteri.
Although no genome sequence is publicly available for an L. reuteri strain, an IVET study was performed with L. reuteri 100-23 (269). One of the genes identified was a gene predicted to encode a xylose isomerase, indicating that xylose is available in the GIT for lactobacilli either from direct feeding or from the breakdown of dietary xylan and pectin by other intestinal microorganisms. In a follow-up study, the ecological performance of the xylA mutant in the murine GIT was not significantly impaired (268). This might be due to a flexible carbohydrate-foraging behavior for L. reuteri and the presence of redundant functions.
As mentioned above, some L. reuteri strains also synthesize HoPSs (glucans or fructans) from sucrose by extracellular glycosyltransferases. These HoPSs are suggested to have a function as a carbohydrate reserve for conditions of carbohydrate depletion (91). Some of these HoPSs might have prebiotic characteristics (see below). In many strains of L. reuteri, sucrose hydrolysis is mediated exclusively or predominantly by these glycosyltransferases, and fructose liberated by Gtfs or the hydrolase activity of Ftfs can be reduced to mannitol with the concomitant oxidation of NADH to NAD+, enabling the formation of acetate from acetyl phosphate and the gain of an additional molecule of ATP in the metabolic flux through the pentose phosphate pathway (91). Such an energy-conserving system may turn out to be beneficial in the GIT, where substrate availability is subject to sudden changes. To test this hypothesis, the in vivo performances of two HoPS mutants in the murine GIT were investigated (271). The inactivation of gtfA in L. reuteri TMW1.106 abolished glucan synthesis, while the inactivation of inu abolished the production of FOS. Both mutations affected sucrose turnover, mannitol formation, and acetate formation (223). Although it is difficult to discriminate between the roles of these HoPSs in metabolism and adherence (see below), experiments using ex-Lactobacillus-free mice revealed that the ecological performance of the inu mutant of L. reuteri TMW1.106 was reduced in the GIT when in competition with the parental strain. The competitive ability of the gtfA mutant was impaired only when it was administered together with L. johnsonii 21, but not with wild-type L. reuteri, indicating extracellular complementation by wild-type glycan (271).
(vi) L. casei.
A promoter study of L. casei DN-114 001 compared the promoter activities for four genes in the GIT: the lactose operon promoter, the promoter of lactate dehydrogenase, the ccpA promoter involved in catabolite repression, and the dlt operon promoter for LTA biosynthesis (181). The lacT and ldh promoters were induced in the GIT, indicating that L. casei is metabolically active during GIT transit. However, the fact that the dlt promoter was not induced indicated that the majority of the administered L. casei cells did not multiply in the GIT, since LTA biosynthesis occurs mainly during cell division (181).
Application of prebiotics to improve persistence capacity.
As elaborated above, the capacity to ferment sugars plays a key role in the competitive ability of lactobacilli and gut commensals to survive and persist in the GIT. This concept is exploited by the application of prebiotics, which are administered mainly to fortify the resident beneficial microbiota. They are also applied in combination with probiotics (as synbiotics) to improve their ecological performance in the gut. A prebiotic is defined as “a nondigestible food ingredient that beneficially affects the host by selectively stimulating the growth and/or activity of one or a limited number of bacteria in the colon and thus improves host health” (92). Although bifidobacteria are the main targets, some lactobacilli are also known to ferment different prebiotics such as FOS. However, this capacity seems to be highly strain specific (121), and the mechanisms by which FOS metabolism occurs in Lactobacillus species are only beginning to be elucidated.
Based on an in silico analysis of the L. acidophilus NCFM genome sequence, a gene cluster that encodes an operon involved in FOS utilization that resembles the msm operon of Streptococcus mutans was identified (13). Genes encoding the ABC transport system (msmEFGK) as well as a putative intracellular β-fructosidase (bfrA) were located in this multiple-sugar metabolism (msm) operon. All of the genes in this operon are coexpressed in the presence of sucrose and FOS but not glucose or fructose, suggesting specificity for non-readily-fermentable sugars and regulation by catabolite repression. The insertional inactivation of the genes encoding the ABC transporter substrate-binding protein and the fructosidase confirmed the function of these genes, as the mutants were unable to grow on FOS. Another system for FOS utilization was detected in L. paracasei 1195 (94). Microarray expression analyses of this strain grown on FOS led to the identification of a putative fosABCDXE operon that encodes a putative fructose/mannose PTS transporter (FosABCDX) and a β-fructofuranosidase precursor (FosE). The latter contains an N-terminal signal peptide sequence and a sortase-dependent cell wall sorting signal at the C terminus, suggesting that FOS is hydrolyzed extracellularly by FosE, with the subsequent uptake of the hydrolysis products mediated by the FosABCDX PTS. The inactivation of fosE disrupted the ability to grow on FOS, and further functional studies confirmed that FOS hydrolysis occurs exclusively at the cell surface (94). Moreover, this system was also shown to be subject to catabolite regulation by glucose (93, 94). In L. plantarum WCFS1, the β-fructofuranosidase is apparently intracellular (215). It is part of a sucrose transport and metabolic system, as identified after microarray analysis of L. plantarum grown on FOS (215). This locus is predicted to encode a sucrose PTS (possibly also for the uptake of short-chain FOS), a β-fructofuranosidase, a fructokinase, an α-glucosidase, and a sucrose operon repressor. In contrast to L. paracasei 1195 (94) and L. acidophilus NCFM (13), L. plantarum showed a preference for short-chain FOS (215).
The functional importance of these FOS utilization systems and the application of FOS to improve the survival of these probiotic strains in vivo remain to be validated. However, a recent study showed that FOS can increase the retention time of certain exogenously applied Lactobacillus strains in the murine GIT. In contrast to glucose, FOS and inulin prolonged the retention period of L. acidophilus LAFTIL10 from 24 to 30 h and that of L. casei LAFTIL26 from 2 to ≥6 days after a single administration (240). Additionally, FOS has also been described to increase the number of endogenous luminal and mucosa-associated lactobacilli in humans (133).
Adherence Mechanisms in the Host
It is generally assumed that a good adherence capacity is a desirable trait for probiotic lactobacilli, as it can promote the gut residence time, pathogen exclusion, and interaction with host cells for the protection of epithelial cells or immune modulation (224) (also see below). The adhesive capacity of probiotic lactobacilli is usually studied in vitro. Mostly, adherence to epithelial cell lines (e.g., Caco-2 or HT-29 human-derived adenocarcinoma cells) or immobilized intestinal mucus or extracellular matrix molecules (e.g., collagen and fibronectin) is investigated in short-term assays (187). However, in most natural niches, adherent bacteria can form microcolonies and multicellular structures, recognized as biofilm-like communities (30). This more dynamic adherence process can also be simulated in vitro for lactobacilli (137, 270). In the human gut of healthy individuals, isolated bacteria and microcolonies seem to be the predominant colonization form (151). Thick biofilms appear to be observed only in the human gut under disease conditions such as in IBD (243). Additionally, biofilms are frequently found on nonglandular, squamous, stratified epithelial cells in the forestomach of rodents or the crop of chicken (267).
It is of crucial importance that in vitro results for adherence capacities of lactobacilli are difficult to extrapolate to the GIT situation in vivo, where the host defense systems, competition with the resident microbiota for nutrients and space, mucosal shedding, and peristaltic flow that continuously washes the GIT epithelium are likely to modify adhesion (224). A few studies have already investigated the adhesion and colonization dynamics of lactobacilli in vivo in animal models (270, 271) or humans (254). However, exogenously applied lactobacilli are generally able to only temporarily colonize the GIT. This phenomenon was linked to colonization resistance or the niche exclusion principle, where each niche in the GIT is colonized by well-adapted species (267). A healthy human gut microbiota is quite stable and seems to show some resistance to colonization with new species. For example, Tannock et al. showed that the establishment of L. rhamnosus DR20 in the intestinal microbiota of human subjects was inversely related to the presence of a stable indigenous population of lactobacilli (247). Other researchers also reported that in contrast to human adults, a more permanent colonization by probiotic lactobacilli such as L. rhamnosus GG could be achieved in the gut of newborn infants, whose microbiota still needs to develop (222).
Even though the in vivo adherence capacity of lactobacilli depends on many external factors, in the next paragraphs, we will summarize the genetic studies that have been performed so far on the cell surface factors of lactobacilli that can mediate adherence (recently reviewed in reference 187). Although the information on these Lactobacillus surface molecules is still fragmentary, there are some common themes that have emerged. We have classified these cell surface factors as mediating either specific adhesin-receptor interactions with molecules of the host or mediating nonspecific interactions, e.g., by electrostatic interactions, or interactions based on hydrophobicity. However, these specific and aspecific adherence mechanisms are sometimes intertwined (see below). For an overview of studies including Lactobacillus mutants affected in adhesion, see Table 4.
TABLE 4.
Genes of lactobacilli putatively involved in adhesion studied by mutant analysis
| Functional category | Gene | Identification approacha | Predicted function | Lactobacillus strain | Mutant phenotype | Reference(s) |
|---|---|---|---|---|---|---|
| Cell surface proteins | srtA | Comp.gen. (genome) | Sortase | L. plantarum WCFS1 | Reduced mannose-specific binding (yeast agglutination); no altered competitive ability in the murine GIT | 195 |
| msa | CGH and gene-trait matching | Mannose-specific adhesin | L. plantarum WCFS1 | Reduced mannose-specific binding (yeast agglutination) | 195 | |
| lp_2940 | R-IVET | Sortase-dependent cell wall protein | L. plantarum WCFS1 | Reduced persistence in the murine GIT | 31, 33 | |
| lp_1403 | R-IVET | Putative secreted cell surface protein | L. plantarum WCFS1 | No altered competitive ability in the murine GIT | 31, 33 | |
| fbpA | Comp.gen. (genome) | Putative fibronectin-binding protein | L. acidophilus NCFM | Reduced adherence to Caco-2 cells | 35 | |
| mub | Comp. gen. (genome) | Putative mucus-binding protein | L. acidophilus NCFM | Reduced adherence to Caco-2 cells | 35 | |
| slpA | Dedicated approach | S-layer protein | L. acidophilus NCFM | Reduced adherence to Caco-2 cells | 35 | |
| cdpA | Comp.gen. (genome) | Cell wall-modifying enzyme involved in cell division and separation | L. acidophilus NCFM | Reduced adherence to Caco-2 cells | 3 | |
| LBA1663-LBA1664 | Comp.gen. (genome) | R28 homologs (adhesins in S. pyogenes) | L. acidophilus NCFM | No significantly altered adherence to Caco-2 cells | 35 | |
| srtA | Comp.gen. (genome) | Sortase | L. salivarius UCC118 | Reduced adherence to HT-29 and Caco-2 cells | 258 | |
| lspA | Comp.gen. (genome) | Large surface protein, putative mucus-binding protein | L. salivarius UCC118 | Reduced adherence to HT-29 and Caco-2 cells | 258 | |
| lspB | Comp.gen. (genome) | Large surface protein, putative mucus-binding protein | L. salivarius UCC118 | No altered adherence to HT-29 and Caco-2 cells | 258 | |
| lspD | Comp.gen. (genome) | Large surface protein, putative mucus-binding protein | L. salivarius UCC118 | No altered adherence to HT-29 and Caco-2 cells | 258 | |
| lsp | Screening of a phage library with an antiserum | Large surface protein | L. reuteri 100-23 | Reduced persistence in murine GIT | 268 | |
| LJ1476 | Comp.gen. (genome) | Transpeptidase sortase | L. johnsonii NCC533 | Gut persistence pattern similar to that of the wild-type strain | 67 | |
| LTA | dltD | Dedicated approach | d-Alanylation of LTA | L. rhamnosus GG | No significant difference in adherence to Caco-2 and HT-29 cells; increased biofilm formation on polystyrene under certain conditions | 137, 188 |
| dltA | Dedicated approach | d-Alanylation of LTA | L. reuteri 100-23 | Decreased in vivo biofilm formation in the murine forestomach | 270 | |
| EPS | LJ1021-LJ1035 | Microarray expression and CGH | Entire EPS cluster | L. johnsonii NCC533 | Slightly prolonged persistence in the murine GIT | 67 |
| lamA | Comp.gen. (genome) | Response regulator of a 2CRS involved in regulation of EPSs and membrane proteins | L. plantarum WCFS1 | Decreased biofilm formation of glass substrates | 239 | |
| wzb | Dedicated approach | EPS biosynthesis, phosphotyrosine protein phosphatase | L. rhamnosus GG | Altered biofilm formation on polystyrene | 137; our unpublished observations | |
| inu | Dedicated approach | Inulosucrase | L. reuteri TMW1.106 | Reduced in vitro biofilm formation; no reduced biofilm formation in vivo on the forestomach | 271 | |
| gtfA | Dedicated approach | Glucosyltransferase | L. reuteri TMW1.106 | Reduced in vitro biofilm formation; no reduced biofilm formation in vivo on the forestomach | 271 |
Comp.gen., comparative genomics.
SDPs and specific adherence mechanisms.
Many specific interactions between lactobacilli and host cells seem to be mediated by SDPs with a typical modular structure that includes an N-terminal signal peptide and a C-terminal LPxTG motif (177). Additionally, other surface proteins can be involved in specific adherence mechanisms, as will be exemplified below for several Lactobacillus species.
(i) L. plantarum.
An elegant approach has been followed to identify the mannose-specific adhesin in L. plantarum WCFS1 (195). First, a genome-wide microarray-based genotyping of L. plantarum strains was performed to select WCFS1 genes whose presence or absence in the different strains was significantly correlated with the presence or absence, respectively, of the yeast agglutination phenotype observed for the same strains (gene-trait matching). This yeast agglutination capacity was previously shown to be related to the mannose-specific adherence of L. plantarum to human intestinal epithelial cells (1). This resulted in four candidate genes that showed a 100% gene-trait match, including two SDPs (lp_0373 and lp_1229). To validate the role of SDPs for mannose-specific adhesion, a sortase-lacking mutant (srtA) was constructed. This srtA mutant was not able to agglutinate yeast cells. Subsequently, knockout mutants were constructed for lp_0373 and lp_1229. The deletion of lp_1229 completely abolished the agglutination capacity. Moreover, the overexpression of lp_1229 resulted in a slight but significant enhancement of this capacity. Overall, those researchers concluded that lp_1229 encodes the mannose-specific adhesin of L. plantarum WCFS1 and renamed the gene msa. It encodes a large surface protein (>1,000 residues) with characteristic multidomain structure, including the mucus-binding domains described below.
The R-IVET study in L. plantarum WCFS1 identified two SDPs (lp_0800 and lp_2940) as being induced in the murine GIT (31). The follow-up quantitative RT-PCR study showed that they are induced primarily in the small intestine (157). lp_0800 was induced along the length of the small intestine, while lp_2940 was most active at the distal end of the small intestine and in the cecum. Subsequent mutational analyses showed that the persistence capacity of the lp_2940 knockout mutant was 100- to 1,000-fold decreased in the murine GIT, while the colonization dynamics of an lp_1403 knockout were not significantly different from those of the wild type (33). This lp_1403 protein is a putative secreted cell surface protein of L. plantarum, for which the expression has also been shown to be induced in the murine GIT by R-IVET (31) and, more specifically, in the small intestine (157). However, the in vivo persistence capacity of an srtA mutant of L. plantarum was not significantly affected (195), which might be due to the fact that the analysis of fecal samples reflects mainly the survival of the majority of administered bacteria during GIT transit.
(ii) L. reuteri.
Roos and Jonsson also applied a rewarding method to identify the mub gene, encoding mucus-binding protein in L. reuteri ATCC 53608 (strain 1023) (209). Using the immunoglobulin G (IgG) fraction of an antiserum raised against cell surface proteins of L. reuteri 1023, they screened a λ phage library and identified a number of clones that were reactive with the antiserum and adhered to mucus. Subcloning resulted in the identification of the mub gene, encoding a very large SDP with a highly repetitive structure (>3,000 residues). Domains with the two main types of repeats (designated Mub1 and Mub2) were shown to adhere to mucus after recombinant expression in E. coli. Competition experiments indicated that Mub binds to carbohydrates on the mucus components. In another L. reuteri strain, 100-23, a similar approach using an antiserum against the surface proteins was used to identify the lsp gene, which encodes a high-molecular-mass cell wall protein, Lsp (268). Mutational analysis showed a reduced ecological performance of the lsp mutant in the murine GIT. When the lsp mutant was administered to mice in a 1:1 ratio with the wild type, it was recovered in the feces at rates of <1.5% of the total Lactobacillus population. Moreover, when the lsp mutant was administered as pure cultures to mice, scanning electron microscopic observations of forestomachs revealed less biofilm formation by the mutant.
(iii) L. acidophilus.
The complete genome sequence of L. acidophilus NCFM was in silico analyzed for the presence of genes encoding proteins that were previously implicated in the adherence of other bacteria. Some of these genes were inactivated, including two streptococcal R28 homologs, a fibronectin-binding protein (FbpA), a mucin-binding protein (Mub), and a surface layer protein (SlpA) (also see below), of which the R28 homologs and the Mub protein (LBA1392) are SDPs (35). That study showed that the genes encoding FbpA, Mub, and SlpA all contribute to the ability of L. acidophilus NCFM to adhere to Caco-2 cells in vitro, confirming that adhesion is determined by multiple factors. mub and fbpA mutations resulted in 65% and 76% decreases in adherence, respectively.
(iv) L. salivarius.
van Pijkeren and coworkers mined the genome of L. salivarius UCC118 for the presence of sortase gene homologs and genes encoding SDPs (258). The sortase gene srtA was deleted, three genes encoding SPDs (lspA, lspB, and lspD) were disrupted, and the capacity of adherence of these mutants to HT-29 and Caco-2 cells was investigated. Both the srtA and the lspA mutant showed a significant decrease in adherence. While the adherence of the srtA mutant was on average 50% of wild-type levels, the lspA mutant adhered at around 65%, only slightly better than the srtA mutant, indicating that LspA plays a key role in adherence to these intestinal cells. LspA is a large protein (>1,000 residues), which contains seven repeats similar to mucus-binding domains previously identified in L. reuteri (209).
(v) L. johnsonii.
Genome analysis of L. johnsonii NCC533 also predicted an abundance of SDPs (>16) (196, 258). LJ0047, LJ0484, and LJ1839 are of special interest because they share significant similarity to the Mub protein of L. reuteri described above (209), but no functional analyses of the role of these Mub homologs in the reported protease-sensitive ability of NCC533 to bind intestinal mucins in vitro (251) have yet to be reported. The knockout of the sortase gene LJ1476 resulted in a gut persistence pattern for the sortase mutant similar to that for the wild type (67). Nevertheless, as mentioned above, the analysis of fecal samples, which is primarily a measure for survival during GIT transit, generally does not reflect subtle differences in adherence capacities. Further mechanistic studies for this sortase, LJ1476, of NCC533 will have to delineate its role in adherence. Moreover, L. johnsonii NCC533 also encodes a second sortase (196), which could be involved in the assembly of the putative pili or fimbriae (156). The genome of NCC533 contains a nine-gene predicted fimbrial operon (196). According to those authors, one predicted cell surface protein of this operon, LJ0391, and another putative surface protein, LJ1711, both with extensive sequence repeats, may encode glycosylated cell surface-adhesive or fimbrial proteins analogous to Fap1 of Streptococcus parasanguis (236). However, this glycoprotein or fimbria-mediated adhesion is not proven yet.
On the contrary, the adhesive capacities for two peculiar proteins of L. johnsonii NCC533 have been documented. The first corresponds to elongation factor Tu (EF-Tu), normally involved in protein synthesis in the cytoplasm. However, EF-Tu of NCC533 has been located at the cell surface, although no secretion or cell wall-binding motifs are present to explain this observation (95). Competition experiments with the isolated protein suggested that EF-Tu has an important role in the binding of NCC533 to Caco-2 and HT-29 intestinal epithelial cells and mucins. Another usually cytoplasmic protein has been found to be located at the cell surface of NCC533, namely, the heat shock protein GroEL (20). This protein was also shown to mediate adhesion to epithelial cells and mucins. Interestingly, the presence of normally cytoplasmic proteins at the cell surface and a role for these proteins in adhesion have also been described for other lactobacilli. In L. plantarum LA 318, a surface plasmon resonance BIAcore-based assay demonstrated that cell surface glyceraldehyde-3-phosphate dehydrogenase (GAPDH) adheres to human colonic mucin (126). In L. crispatus, the glycolytic enzymes enolase and GAPDH were found to bind and activate plasminogen (112). They were localized on the cell surface at pH 5, bound to LTA but released into the medium at an alkaline pH (6). Importantly, these results indicate that lactobacilli rapidly modify their surface properties in response to changes in pH and that the pH used in buffers is a crucial factor in in vitro adhesion experiments (6).
(vi) Mucus-binding proteins.
A common theme that has emerged from the analysis of these specific adherence mechanisms is the frequent identification of mucus-binding proteins. Based on the above-described Mub protein of L. reuteri 1063 (209) and the Msa protein of L. plantarum (195), Boekhorst et al. performed an in silico search for potential mucus-binding proteins present in several publicly available databases (25). As a result, a total of 48 proteins containing at least one MUB domain were identified in 10 lactic acid bacterial species. The identified MUB domains varied in size, ranging from ca. 100 to more than 200 amino acid residues per domain, and appeared most abundant, although not exclusively, in lactobacilli that are found mainly in the GIT, i.e., 13 in L. gasseri ATCC 33323, 12 in L. acidophilus NCFM, and 9 in L. johnsonii NCC533. Recent evidence that these mucus-binding proteins are involved mainly in GIT colonization also results from the genome sequence of the dairy isolate L. helveticus DPC4571 (39). In comparison to the closely related gut isolate L. acidophilus NCFM, with which it has 75% of genes in common, strain DPC4571 has lost all mucus-binding proteins. However, experimental confirmation of the specific binding of these putative mucus-binding proteins to specific mucus components is still needed. A striking difference between the various mucus-binding proteins is the number of repeats of the MUB domain. This high variability suggests that the MUB domain is often duplicated (or deleted) in evolution, and it might be interesting to investigate whether the number of repeats correlates with the capacity of binding to mucus (25).
Other cell surface factors determining adherence. (i) S-layer proteins.
S-layer proteins of lactobacilli have been commonly suggested to be involved in the adherence of lactobacilli, although not all lactobacilli have an S layer (9). We mentioned above that an slpA mutant of L. acidophilus NCFM was severely affected in its capacity to adhere to Caco-2 cells, although it is likely that multiple surface-associated proteins are disrupted with the removal of the S layer (35). Interestingly, the cell morphology of this slpA mutant was significantly altered (small, curved bacilli), indicating a role for SlpA in cell shape determination. That same group identified another cell surface protein, designated CdpA, as being involved in adherence to Caco-2 cells (3). The cdpA mutant adhered only ca. 17% of wild-type levels. According to those authors, this significant drop in adhesion is caused by pleiotropic effects generated by the mutation of this cell wall-modifying enzyme that promotes cell division and adherence. The lack of CdpA is suggested to result in a loss of anchoring or translocation or a loss of integrity of important adherence-determining proteins (3).
In L. crispatus JCM 5810, the S layer has been suggested to act as a specific adhesin, as it was reported to promote binding to components of the extracellular matrix of target cells in vitro, particularly collagen (5, 228). The domain structure of this collagen-binding protein, CbsA, was also analyzed after heterologous expression in E. coli and L. casei (5, 228). These analyses revealed that CbsA has an N-terminal assembly domain exhibiting affinity for pericellular tissue components (collagens and laminin) and a cationic C-terminal domain binding to negatively charged LTA. Flagellar display experiments also showed that SlpA of L. brevis ATCC 8287 has an N-terminal adhesive domain but with affinity for epithelial cells and fibronectin (113).
(ii) LTA.
LTA is suggested to provide the main component of the hydrophobicity of the Lactobacillus cell envelope, although this depends on the d-alanine ester substitutions (65). Therefore, LTA seems to contribute mainly to adhesiveness in a nonspecific way. For instance, it was shown that the adhesion of L. johnsonii strain La1 to Caco-2 human intestinal cells could be inhibited by LTA purified from that strain in a concentration-dependent manner (96). The inactivation of dltD in the probiotic strain L. rhamnosus GG revealed that the d-alanylation of LTA is not required for short-term adherence to Caco-2 cells (188), while the dltD mutation increased the biofilm formation capacity of L. rhamnosus GG after 72 h of growth on polystyrene (137). In L. reuteri 100-23, a dltA mutant, also deficient in d-Ala residues on LTA, was shown to be severely impaired in colonizing the GIT of ex-Lactobacillus-free mice (270). This mutant was especially affected in its capacity to form in vivo biofilms in the forestomachs of these mice. However, when the ex vivo adherence of the wild type and the dltA mutant to tissue from the forestomach was investigated, no difference in adherence capacities was observed, indicating that d-alanyl esters on LTA do not contribute to initial adhesion events of L. reuteri. It should be noted that the modulation of the LTA structure and charge by mutation can have a significant impact on binding and conformational properties of attached cell surface proteins. For instance, for the dltD mutant of L. rhamnosus GG, we observed a marked difference in the secretion of proteins compared to that of the wild type (unpublished observations). Moreover, the formation of biofilms in the GIT is a more complex process than mere adherence to a substrate, as was also apparent from in vitro simulations of lactobacilli (137). For L. reuteri 100-23, d-alanylation might be important for later events in biofilm formation. Increased repulsive electrostatic forces in the mutant due to increased negative charges might contribute to the disruption of biofilm structure. Moreover, the reported impaired growth of the dltA mutant under acidic culture conditions and increased susceptibility to cationic peptides might further impair the growth of mature biofilms under normal biofilm-permissive conditions.
(iii) EPS.
As for LTA, EPSs generally play a role in nonspecific interactions of lactobacilli with abiotic and biotic surfaces by contributing to the cell surface physicochemical properties. EPSs have also been shown to have an indirect effect on adhesion by shielding other cell surface adhesins. For example, in L. johnsonii NCC533, the deletion of the entire EPS cluster slightly prolonged the gut persistence of the knockout mutant compared to the persistence of the parental control strains (67). In L. acidophilus CRL639, EPSs have also been shown to negatively mediate adhesion to extracellular matrices (148). Ruas-Madiedo and coworkers found both positive and negative effects of EPS on the adhesion of probiotics and enteropathogens to human intestinal mucus (211). Generally, aspecific mechanisms seem to dominate, but these EPS molecules could also act as ligands for host or pathogenic lectins, mediating specific adhesion and coaggregation (211). Much remains to be learned about the specific interactions of EPSs with lectin adhesin receptors.
In contrast to these assays that investigate relative short-time adherence events, EPSs seem to play a more specific role in the formation of microcolonies and biofilms (30). EPSs can promote intercellular interactions and the formation of microcolonies, although this step also depends on many other intrinsic and environmental factors. In L. plantarum WCFS1, an agr-like 2CRS regulatory system, encoded by lamBDCA, which regulates EPS production and adherence, was recently identified (239) (see also below). An RR-deficient lamA mutant showed reduced biofilm formation on glass substrates. Global transcription analysis of the wild type and the RR lamA mutant showed that LamA is involved in the regulation of the expression of genes encoding surface polysaccharides, cell membrane proteins, and sugar utilization proteins, indicating that this system has a key role in the regulation of cell surface properties. For L. rhamnosus GG, cell surface polysaccharides have also been shown to play an important, but condition-dependent, role in in vitro biofilm formation (135, 137). A wzb knockdown mutant in a gene encoding a putative EPS chain-length modulator and a spontaneous polysaccharide mutant, CMPG5413, of L. rhamnosus GG were significantly affected in their biofilm formation capacities (135, 137). Recently, the role of two HoPSs of L. reuteri TMW1.106 was also investigated in biofilm formation (271). In vitro assays showed that both the gftA and inu mutant were impaired in biofilm formation under acidic conditions. This was related to a role for glucan in autoaggregation and a role for Inu as a glucan-binding protein mediating coaggregation. However, experiments using ex-Lactobacillus-free mice could not show a significant difference in biofilm formation in vivo on forestomach epithelial cells (271).
MECHANISMS OF HEALTH-PROMOTING EFFECTS OF LACTOBACILLI: PROBIOTIC FACTORS
Microbe-Microbe Interactions
Although adaptation factors contribute to the survival of lactobacilli in the host and are relatively easy to study mechanistically, the ultimate goal of molecular research on probiotics is the characterization of the key probiotic factors that result in direct health-promoting effects (see Table 5 for an overview of the studies that have yet included mutant analyses of lactobacilli).
An important mechanism of probiotic action is the capacity of probiotics to beneficially affect the host by direct effects on the microbiota. Traditionally, most attention is given to the antipathogenic properties of probiotics by competition for nutrients, the production of antimicrobials, and/or competitive exclusion. However, synergistic interactions could also occur between probiotics and endogenous beneficial members of the microbiota.
Competition and cooperation for nutrients.
Interactions at the metabolome level are very complex to decipher, and it is almost impossible to investigate the impact of probiotic lactobacilli. However, Sonnenburg and coworkers used a simplified mouse model to investigate the metabolic impact of probiotics such as L. casei DN-114001 on the human symbiont Bacteroides thetaiotaomicron (233). They showed that this Bacteroides strain expands its capacity to utilize polysaccharides upon cocolonization with certain probiotic strains. This expansion is characterized by a general shift from mainly host mucus-derived to also dietary plant-derived glycans, such as xylose- and arabinose-containing glycans. This clearly shows the importance of competition for carbohydrates, as is expected from the importance of carbohydrate metabolism for microbes in the GIT (see above). Additionally, synergistic metabolic interactions could occur, particularly in the lower part of the GIT. For instance, primary degraders of complex carbohydrates such as Bacteroides may release a wider range of polysaccharides than they can utilize. The oligosaccharides generated by the numerous glycosylhydrolases of these species can be fermented by, e.g., LAB to lactic acid, which can then be metabolized by lactic acid-utilizing bacteria (279). The removal of substrates from the vicinity of the producer cells may also be advantageous for thermodynamic reasons, driving reactions that are otherwise energetically unfavorable. The net result is the promotion of symbiotic and syntrophic relationships. Moreover, nutrient harvest and metabolic interactions seem to be stimulated when the bacteria are present in multispecies biofilm-like communities (232).
Although the contribution of endogenous and exogenously applied lactobacilli to this metabolic cross-feeding has not been investigated, some studies have shown that ldh (encoding lactate hydrogenase) of exogenously applied lactobacilli is induced upon delivery into the GIT (157, 181), indicating that lactobacilli do produce lactate in situ. This lactic acid could be converted to butyric acid by lactate-utilizing butyrate-producing colon bacteria such as Eubacterium hallii (75). This metabolic conversion is especially interesting, as butyrate has been shown to have beneficial effects on the GIT epithelium (220). This interaction can also explain why lactic acid generally does not accumulate in healthy adults and why many researchers have described an increase in butyrate levels after prebiotic consumption of, e.g., FOS that target mainly the LAB bifidobacteria and lactobacilli (see, e.g., reference 138). Additionally, some L. reuteri strains were shown to produce fructans and FOS, which could act as prebiotics in vivo and stimulate certain beneficial members of the GIT microbiota (91). Although this concept is attractive, this hypothesis remains to be verified in vivo.
Production of antimicrobial compounds.
Besides competition for nutrients, lactobacilli are known to produce a variety of compounds that exert a direct antimicrobial action toward competing bacteria and viruses, although effects on viruses are not yet well documented at the molecular level.
(i) Lactic acid.
Lactic acid can be considered to be a key antimicrobial compound produced by lactobacilli (224). For example, the strong antimicrobial activity of L. rhamnosus GG against S. enterica serovar Typhimurium was shown to be due to the accumulation of lactic acid (64). The exact mode of action underlying this observed antimicrobial effect of lactic acid has not yet been completely clarified, although it is clear that both Salmonella growth and the expression of virulence factors are affected by lactic acid (78). Besides exerting its activity through lowering the pH and through its undissociated form, lactic acid is also known to function as a permeabilizer of the gram-negative bacterial outer membrane (2), allowing other compounds to act synergistically with lactic acid. In addition, organic acids such as lactic acid can capture elements essential for growth, such as iron, by their chelating properties (194).
(ii) Bacteriocins.
Many lactobacilli are reported to secrete antimicrobial peptides called bacteriocins. These bacteriocins are usually active against closely related bacteria that are likely to reside in the same ecological niche. Most Lactobacillus bacteriocins are small, heat-stable proteins with a high isoelectric point (class II bacteriocins) that act generally by inducing membrane permeabilization and the subsequent leakage of molecules from target bacteria (79). Bacteriocin production is controlled in many strains in a population density-dependent manner using a secreted peptide pheromone for quorum sensing (QS). The sensing of its own growth, which is likely to be comparable to that of related species, is suggested to enable the producing organism to switch on bacteriocin production at times when competition for nutrients is likely to become more severe (79). Although this proposed role for bacteriocin production has not been validated in vivo for lactobacilli, the R-IVET study of L. plantarum WCFS1 already gave some indications, as a plnI gene, encoding the plantaricin immunity protein, was found to be induced in the murine GIT (31). This gene belongs to a bacteriocin locus in L. plantarum WCFS1 that includes plnABCD, encoding a plantaricin 2CRS and an autoinducing peptide, PlnA, and several genes encoding class II bacteriocins (plnE-plnF, plnJ-plnK, and plnN) (128, 238). Marco et al. further confirmed that plnI is induced in the small intestine, cecum, and colon of mice (157), suggesting that the production of this bacteriocin is important for L. plantarum in the highly competitive environment of the GIT. However, no mutant analysis has yet been performed to confirm this hypothesis.
Nevertheless, some functional studies of lactobacillus mutants affected in bacteriocin production have been reported. In L. acidophilus NCFM, an operon involved in lactacin B production that includes genes encoding a 2CRS and an ABC transporter was identified (74). The inactivation of a predicted ABC transporter (encoded by labT) completely abolished bacteriocin activity against closely related strains such as L. delbrueckii ATCC 4797. Additionally, a pheromone-like peptide encoded by this operon (by LBA1800 or labIP) that was able to induce lactacin B production was identified. However, no in vivo competition experiment or experiments with pathogens have yet been reported for these mutants. This is in contrast to a recent study by Corr et al. demonstrating the in vivo functionality of a bacteriocin of L. salivarius UCC118 as an effective antimicrobial strategy (53). UCC118 produces a potent broad-spectrum class II bacteriocin, Abp118, and its production is also regulated by an induction peptide, AbpIP. To investigate the role of this bacteriocin in the antimicrobial activity of UCC118 against L. monocytogenes, the abpT gene, encoding the AbpT transporter, was inactivated, resulting in abolished bacteriocin production. In contrast to the wild type, this mutant was not able to reduce Listeria infection in mice. To confirm that this was due to a direct antimicrobial action of the bacteriocin Abp118, a strain of L. monocytogenes immune to Abp118 was created by the heterologous expression of the associated immunity determinant, abpIM, which conferred specific resistance to Abp118. This way, the protective effect of UCC118 against L. monocytogenes was lost when the Abp118-resistent variant was used as the infectious strain.
(iii) H2O2.
H2O2 production by lactobacilli has also been suggested to be an important antimicrobial mechanism, especially in the vagina of healthy women (224). Recently, Pridmore et al. (197) described an in vitro role for H2O2 in the anti-Salmonella activity of L. johnsonii NCC533. This strain produces up to millimolar quantities when resting cells are incubated in the presence of oxygen. The genetic basis for this H2O2 production is not clear. The roles of different enzymes were investigated by mutant analysis: LJ1853, encoding a putative pyruvate oxidase enzyme; LJ1826, encoding a putative lactate oxidase; LJ1254-LJ1255, encoding a predicted NADH oxidase gene interrupted by a perfect 13-bp direct repeat; and LJ1810-LJ1814, encoding a putative cytochrome d ubiquinol oxidase operon. Single mutants still produced wild-type levels of H2O2, but the combination of all four mutations resulted in a non-H2O2-producing mutant. However, this mutant rapidly reversed to the production of H2O2, complicating further functional analyses.
Competitive exclusion.
Intestinal pathogens such as type 1-fimbriated E. coli utilize oligosaccharide receptor sites in the gut (139). There is some evidence that probiotics could use the same attachment site so that the pathogen is in competition for binding to the host mucosal interface and thereby could be inhibited from invading the mucosal layer. This antipathogenic mechanism is known as competitive exclusion and generally requires that the probiotic lactobacilli are administered in a preventive setup, as the displacement of a pathogen by a Lactobacillus strain is usually not observed. The various specific adhesins described above probably contribute to this mechanism of probiotic action, although aspecific mechanisms based on steric hindrance are also possible.
One putative competitive exclusion factor is the mannose-specific adhesin Msa of L. plantarum (195). Its identification allows detailed studies with deletion and overproduction strains to identify strains that effectively exclude pathogens containing type 1 fimbriae. Mack et al. demonstrated that a spontaneous and noncharacterized adh mutant of the probiotic strain L. plantarum 299v, but which is suggested to be affected in the msa gene, showed 10-times-less adherence to HT-29 epithelial cells and was not able to inhibit the adherence of enteropathogenic E. coli, in contrast to the wild type (152).
Surface layer extracts from L. helveticus R0052 were recently shown to inhibit the adhesion of E. coli O157:H7 to epithelial cells (118). This process seems to be mediated partly by the high hydrophobicity of the S layers, and it is not yet known whether it involves interactions with specific receptors. Similar results were obtained for the S layers of L. crispatus ZJ001, which were shown to play a role in the competitive exclusion against enterohemorrhagic E. coli (EHEC) and S. enterica serovar Typhimurium (47). Another example is the putative collagen-binding protein of L. fermentum RC-14, which was reported to inhibit the adhesion of Enterococcus faecalis 1131 (105). An additional mechanism to decrease the pathogenic load during infections is coaggregation, which is thought to facilitate the clearance of pathogens during mucus flushing. For example, a cpf gene encoding a surface protein that mediates coaggregation with E. coli K88, Campylobacter coli, and Campylobacter jejuni was identified in L. coryniformis DSM20001T (217). Bergonzelli et al. also described a role for the cell surface-located GroEL of L. johnsonii NCC 533 in the specific aggregation of Helicobacter pylori (20). Nevertheless, whether the preventive administration of probiotics is really able to reduce infections in vivo by competitive exclusion remains to be determined.
Bacterial cell-cell communication.
Given the high number and level of diversity of bacteria that comprise the GIT, it was postulated that the members of this community somehow communicate to coordinate various adaptive processes that include competition and cooperation for nutrients and adhesion sites (120). QS is a cell-to-cell signaling mechanism through which bacteria produce and/or respond to chemical signals called autoinducers.
(i) Intraspecies communication.
As described above, QS is best studied in lactobacilli in relation to bacteriocin production. Intraspecies bacterial communication in gram-positive bacteria is generally mediated by specific autoinducing signaling peptides that are often posttranslationally modified and exported by dedicated transport systems (238). These signals are sensed by responsive cells via dedicated 2CRSs. In silico analysis predicted the presence of five QS 2CRSs in L. plantarum WCFS1, two in the intestinal species L. acidophilus NCFM and L. johnsonii NCC533, one in the intestinal species L. salivarius UCC118 and the food species L. delbrueckii subsp. bulgaricus ATCC BAA-365, and none in the intestinal species L. gasseri ATCC 33323 (238). The high number identified in L. plantarum WCFS1 could reflect the ecological flexibility of this species, which can be found on plants, in fermented foods, and in the GIT (128). In comparison, the other lactobacilli described seem to be more restricted to specific environments, possibly resulting in fewer peptide-based QS 2CRSs (238). The functionality of abpIPKR in L. salivarius UCC118 (53) and LBA1798-LBA1800 in L. acidophilus NCFM (74) in QS-regulated bacteriocin production is described above. Additionally, a Lactobacillus agr-like module was functionally characterized in L. plantarum WCFS1 (239). Analogous with the staphylococcal agr system, the L. plantarum lamBDCA locus is involved in the production of a cyclic thiolactone peptide. Analysis of a lamA mutant revealed a role for the lam operon in the L. plantarum biofilm-forming capacity and EPS production, as we described above in relation to adhesion. Nevertheless, an in vivo role for these QS systems in the competitive ability of lactobacilli in the GIT remains to be elucidated.
(ii) Interspecies communication.
In the late 1990s, a new family of signal molecules, autoinducer-2 (AI-2), and its cognate synthase LuxS, which are present in both gram-negative and gram-positive bacteria, were described (241). In Vibrio harveyi, AI-2, a furanosyl borate diester, is one of the signals that regulate bioluminescence through a complex phosphorelay system (106). The binding of AI-2 to the periplasmic receptor LuxP modulates the activity of the inner membrane sensor kinase LuxQ, transducing AI-2 information into the cytoplasm. V. harveyi can produce light in response to the AI-2 produced by many other bacterial species. These observations of V. harveyi resulted in the development of a bioassay to detect AI-2 production and led to the suggestion that AI-2 acts as a universal signal molecule that fosters interspecies cell-cell communication (15). Using the Vibrio bioluminescence assay, De Keersmaecker and Vanderleyden tested the spent culture supernatant of various lactobacilli for the presence of AI-2-type molecules (63). Strains such as L. rhamnosus GG, L. plantarum NCIMB8826, L. johnsonii VPI1088, and L. casei ATCC 393 were shown to produce AI-2-like molecules (63).
In the gastrointestinal pathogen Vibrio cholerae, the accumulation of two QS signals, cholerae AI-1 (CAI-1) and AI-2, represses the expression of virulence factors at high population density. AI-2 functions synergistically with CAI-1 to control virulence gene expression, although CAI-1 is the stronger of the two signals (107). However, apart from bioluminescence in V. harveyi and a role in virulence of V. cholerae, no obvious phenotype has been associated with the extracellular accumulation of this molecule in most bacteria (260). The functional studies of AI-2-mediated QS are complicated by the fact that the AI-2 synthase LuxS forms an integral part of the activated methyl cycle, as mentioned above in relation to stress resistance. In this cycle, LuxS catalyzes the conversion of S-ribosylhomocysteine, yielding (S)-4,5-dihydroxy-2,3-pentanedione and homocysteine (260). (S)-4,5-Dihydroxy-2,3-pentanedione undergoes spontaneous rearrangements to form AI-2, while homocysteine is recycled to methionine and SAM. The disruption of luxS also disrupts the activated methyl cycle, resulting in many metabolic defects that cannot be easily discriminated from QS defects. However, since only the LuxS AI-2 synthase is widespread in bacteria, as opposed to the LuxPQ AI-2 receptor, most functional studies have been performed with luxS knockout mutants. This is also the case for lactobacilli, and only two functional analyses of luxS knockout mutants have been described in detail. In L. reuteri 100-23, the disruption of luxS resulted in increased biofilm formation in vitro in a bioreactor and on the epithelial surface of the murine forestomach (246). Whether this was due to disrupted QS control of the biofilm thickness or disturbed metabolism is difficult to discriminate, as the addition of purified AI-2 to the biofilm culture medium could not restore the phenotype to the wild-type level, and the luxS mutant showed a reduced ATP level in exponentially growing cells. Moreover, the ecological performance of the luxS mutant, when in competition with L. reuteri strain 100-93, was significantly reduced in the highly competitive conditions of the murine cecum but not in the stomach or jejunum (246). In L. rhamnosus GG, the disruption of the luxS gene resulted in pleiotropic effects on in vitro growth, biofilm formation, and in vivo persistence in the murine GIT, and these effects were shown to be caused merely by metabolic defects (135, 136). Therefore, although AI-2 is an attractive candidate for multispecies communication in the GIT, it is very difficult to verify this hypothesis in vivo. Clearly, the strategy to investigate this hypothesis has to include, in addition to the characterization of luxS mutants, the identification of putative AI-2 receptors and signaling pathways between different species, both pathogenic and probiotic bacteria.
(iii) Interkingdom communication.
Recently, it was discovered that bacteria can also exploit QS signals to communicate with the host in a process referred to as cross-kingdom cell-to-cell signaling (111). This cross-kingdom signaling involves small molecules, such as hormones that are produced by eukaryotes and hormone-like chemicals that are produced by bacteria. For instance, Sperandio and colleagues could show a role for (nor)epinephrine produced by the host, similar to the aromatic signal AI-3 produced by commensal microbiota: both signaling molecules could activate virulence genes of EHEC mainly by inducing ler expression (234). ler (locus of enterocyte effacement [LEE]-encoded regulator) encodes the principal transcriptional activator of the LEE genes present on a pathogenicity island referred to as LEE of EHEC (234). The identity and exact structure of this AI-3 signal is currently unknown, but its production in EHEC seems to be indirectly dependent on the LuxS enzyme described above. AI-3 appears to be an aromatic compound derived from tyrosine, whose biosynthesis is linked to the activated methyl cycle and metabolism of methionine (272). Interestingly, lactobacilli were recently shown to interfere with this signaling. The supernatant of L. reuteri ATCC 55730 exhibited a negative effect on ler expression by EHEC, in contrast to the one of the corresponding ATCC 55730 luxS mutant and growth medium alone (115). Similarly, L. acidophilus La-5 also reduced the expression of virulence genes of EHEC (166). On the contrary, the stationary-phase supernatant of L. reuteri 100-23C induced ler expression, and this induction was abolished in the isogenic luxS mutant (246). Therefore, Medellin-Pena and coworkers proposed a LuxS-dependent production of as-yet-unknown competing antagonistic molecules in L. reuteri ATCC 55730 and L. acidophilus La-5 versus AI-3-like agonistic molecules in L. reuteri 100-23C (166). As LuxS plays a central role in metabolism, and mutation has pleiotropic effects on many compounds in the supernatant, it will be difficult to identify these AI-3-like molecules.
Beneficial Interactions with Gut Epithelium
The intestinal epithelial cells are presumed to be the first and most important target cells of probiotic action. In this section, we discuss Lactobacillus molecules that can beneficially influence the optimal functioning of the gut epithelium, related to its nutritive and its barrier function.
Metabolic interactions.
Metabolic interactions between lactobacilli and the host could modulate the primarily nutritive function of the epithelium, but these interactions are difficult to delineate. A holistic approach was recently used by Martin and coworkers (159). They investigated the metabolic effects of exposure to either L. paracasei NCC2461 or L. rhamnosus NCC4007 in a humanized microbiome mouse model (germ-free mice colonized with human baby microbiota) by an impressive top-down systems biology strategy of samples taken from plasma, urine, liver, ileum, cecum, and feces. Although it concerned a simplified model with only seven different strains making up the microbiota, they could show that probiotic exposure can modify the microbiota and alter hepatic lipid metabolism, coupled with lowered plasma lipoprotein, increased triglyceride levels, and apparent stimulated glycolysis. They also observed changes in a diverse range of other pathways, including amino acid metabolism, methylamines, and short-chain fatty acids such as butyrate (159). As mentioned above, butyrate is a particularly important source of energy for the colonic mucosal cells, and it was suggested to be necessary for the maintenance of the colonic epithelium (100, 220). It was also suggested that part of these observed effects is due to the capacity of these lactobacilli to metabolize bile acids, the primary function of which is to emulsify fats, so that these probiotics seem to affect how much fat that the body can absorb. However, the relationship with the presence of, for instance, bsh genes in these lactobacilli has not yet been made. This holistic approach is of particular interest to further investigate the potential of probiotic lactobacilli in modulating mainly metabolic disease risks for, e.g., diabetes (244), obesity (143), and colon cancer (68).
Other metabolic effects of lactobacilli on the host are less easy to delineate. Some lactobacilli are able to catalyze the isomerization of the double bond at the C-9 position in linoleic acid (c9,c12; 18:2) to form t10,c12-conjugated linoleic acid (18:2), which is hypothesized to be involved in additional metabolic effects of lactobacilli (142). Also, although lactobacilli are traditionally known as being auxotrophic for B vitamins, some strains of lactobacilli such as L. reuteri JCM1112 can synthesize the B vitamins folate (vitamin B9) and/or vitamin B12, which could be interesting in cases of vitamin deficiency (213, 214). Additionally, five genes essential for folate biosynthesis from Lactococcus lactis have successfully been cloned in the folate auxotroph L. gasseri, changing this strain from a folate consumer to a folate producer (274), but the potential of such engineering in relation to probiotic effects remains to be studied.
Preservation of epithelial barrier function.
Alterations in epithelial barrier function are implicated in a variety of intestine-related disorders including enteric infections, IBD, food allergy, autism, and stress (252). Probiotic lactobacilli are suggested to strengthen the epithelial barrier by various mechanisms such as the induction of mucin secretion (152), enhancement of tight-junction functioning (117, 183, 225), upregulation of cytoprotective heat shock proteins (190, 248) and prevention of apoptosis of epithelial cells (280). In some cases, parts of the signaling pathways have been identified (Fig. 3), but the Lactobacillus effector molecules and their corresponding receptors mediating these effects are largely unknown.
FIG. 3.
Modulation of epithelial barrier function by lactobacilli. Several in vitro studies have identified signaling pathways that are involved in the interaction between lactobacilli and epithelial cells. The MAPKs p38, ERK1/2, and JNK have an important function in the dynamic regulation of the cell cytoskeleton, tight junctions (TJ), and other effectors of epithelium barrier function, and these MAPKs are often influenced by lactobacilli (see, e.g., reference 205). Given the critical role of EGF signaling in many aspects of gastrointestinal physiology and epithelial repair, some beneficial effects of probiotics are also related to interference with this signaling (204, 282). Akt (or protein kinase B) plays a central role in promoting epithelial cell survival by lactobacilli by the inactivation of several proapoptotic pathways, including caspase 9 and caspase 3, and stimulation of cell proliferation by the activation of cell cycle regulators (see, e.g., reference 280). Akt is generally activated in a phosphatidylinositol 3-kinase (PI3-K)-dependent manner by, e.g., EGF receptor (EGFr) signaling or TLR signaling. Additionally, inhibition of the activation of the NF-κB pathway that plays a key role in inflammatory responses seems to be a primary target of lactobacilli by inhibiting the ubiquitination and proteasome degradation of IκB and thus preventing the nuclear translocation of the NF-κB transcription factor (see, e.g., reference 190). Clearly, a complex network of interacting signaling pathways can be influenced by lactobacilli. However, there is a general lack of knowledge of the Lactobacillus effector molecules and their corresponding host receptors mediating these effects (89, 116, 190, 198, 204, 205, 225, 248, 280) (double arrows indicate cross-signaling events; dotted lines indicate ligand-receptor interactions that are not yet well defined). PKC, protein kinase C.
(i) Cell surface factors.
Many studies have demonstrated that direct cell contact between lactobacilli and epithelial cells is needed for certain beneficial effects on the epithelium, and some studies identified important cell surface factors of lactobacilli involved. For instance, LTA molecules from L. johnsonii La1 and L. acidophilus La10 inhibited E. coli- and lipopolysaccharide (LPS)-induced interleukin-8 (IL-8) release by HT-29 epithelial cells, and the lipid moiety of the LTA molecules proved to be important (262). Mack et al. also showed that the induction of mucin expression by probiotic lactobacilli was dependent on direct cell contact between lactobacilli and epithelial cells (152). To show this, they used a spontaneous adh mutant of the probiotic strain L. plantarum 299v, resulting in 10-times-less adherence to HT-29 epithelial cells. This adh mutant had lost the ability to induce mucin expression and was not able to inhibit the adherence of enteropathogenic E. coli, in contrast to the wild type. However, the exact molecular details of the adh mutation remain to be determined (as elaborated above).
Also, other studies have shown that some host pathways are modulated by direct contact with lactobacilli without identifying the Lactobacillus elicitors. For instance, L. acidophilus ATCC 4356 was reported to mediate anti-inflammatory and antiapoptotic effects in epithelial cells dependent on direct cell contact by the activation of mitogen-activated protein kinases (MAPK) and Akt, the prevention of IκB degradation, and the prevention of inactivation of the epidermal growth factor (EGF) receptor (204, 205, 280). Yan and Polk showed that L. rhamnosus GG exerts a specific antiapoptotic effect through the inhibition of tumor necrosis factor alpha (TNF-α)-stimulated activation of the proapoptotic MAPK p38, for which direct contact is needed (281), in addition to the activation of Akt, which is induced by soluble proteins (see below). As-yet-unknown cell surface factors were also found to be responsible for the inhibition of IκB degradation and induction of nerve growth factor by L. reuteri in epithelial cells, resulting in an anti-inflammatory effect (150). Interestingly, a quite unexpected receptor that mediated beneficial effects of a specific Lactobacillus strain was recently identified. L. acidophilus NCFM was shown to induce the expression of opioid and cannabinoid receptors in intestinal epithelial cells and mediate analgesic functions in the gut, similar to the effects of morphine (210). These effects seemed to be mediated by direct cell contact, but the probiotic effectors are yet unknown.
(ii) Secreted proteins.
Other beneficial effects of lactobacilli on epithelial cells do not require direct cell contact. For instance, two secreted proteins of L. rhamnosus GG, designated p40 and p75, were recently identified to promote in vitro intestinal epithelial homeostasis through specific signaling pathways (280, 281). p40 and p75 show low similarity with putative cell wall-associated hydrolases or cell wall-modifying enzymes and are abundantly present in spent L. rhamnosus GG supernatant. Similar proteins were identified in the supernatant of L. casei ATCC 334 and ATCC 393 but not in the supernatant of L. acidophilus ATCC 4356 (280). These proteins stimulated the activation of Akt, promoted epithelial cell growth, and inhibited TNF-α-induced epithelial cell apoptosis. Akt was activated by p40 and p75 in a phosphatidylinositol 3-kinase-dependent manner, probably mediated by the activation of the EGF receptor (280, 282). In another model system, p40 and p75 of L. rhamnosus GG appeared to protect the intestinal epithelial tight junctions and attenuate the H2O2-induced epithelial barrier disruption in Caco-2 cells. These effects of p40 and p75 were shown to be mediated by protein kinase C and the MAPK extracellular signal-regulated kinase 1 (ERK1)/ERK2 (225), indicating that these proteins can affect various pathways. The probiotic mixture VSL#3, which includes L. casei, L plantarum, L acidophilus, and L. delbrueckii subsp. bulgaricus, was also reported to stabilize tight junctions and induce mucins in intestinal epithelial cells by large but unidentified proteinaceous soluble factors (>50 kDa) (183). Also, more research is needed to identify the unknown factor(s) in spent culture medium of various probiotic lactobacilli and VSL#3 that were reported to induce expression of enterocyte β-defensin 2, an antimicrobial peptide that is important for gut barrier function, via the induction of pathways including NF-κB and activator protein 1 (AP-1) as well as MAPKs (221).
(iii) Soluble peptides.
Low-molecular-weight, heat- and acid-stable peptides of L. rhamnosus GG were shown to activate the MAPK p38 and JNK and to induce cytoprotective heat shock proteins in intestinal epithelial cells (248). Similarly, unknown peptide-soluble factors of the probiotic mixture VSL#3 were shown to inhibit the degradation of the NF-κB inhibitor IκB and induce heat shock proteins through specific proteasome inhibition (190). In contrast, the well-characterized pentapeptide QS molecule colony-stimulating factor (CSF) of B. subtilis was recently reported to exert a protective effect against the loss of epithelial barrier function in gut epithelial cells by the induction of cytoprotective heat shock proteins (89). Moreover, Fujiya et al. identified a mammalian apical membrane cationic oligopeptide transporter (OCTN2) for CSF uptake that is typically present on epithelial cells. Whether peptides of lactobacilli could serve a similar function remains to be evaluated, but apparently, L. rhamnosus GG and L. plantarum exerted similar effects on the induction of heat shock proteins and interactions with the OCTN2 receptor (89).
(iv) Unmethylated CpG DNA.
Finally, in the presence of proinflammatory stimuli, bacterial DNA of the VSL#3 mixture was shown to inhibit IL-8 secretion, reduce p38 MAPK activation, delay NF-κB activation, stabilize levels of IκB, and inhibit proteasome function (116). Rachmilewitz et al. reported that Toll-like receptor 9 (TLR9) signaling mediates the anti-inflammatory effects of VSL#3 DNA in murine experimental colitis (198).
(v) Multiple probiotic factors and multiple signaling pathways.
Taken together, multiple cell surface and secreted factors of lactobacilli seem to exert a protective effect on intestinal epithelial cells mediated by multiple signaling pathways (Fig. 3). Some of these effects are related to immune responses (see below). However, the exact effector-receptor interactions and downstream signaling events still need to be characterized in most cases. Moreover, many of these in vitro studies need to be complemented with in vivo data. For instance, Di Caro et al. investigated the influence of L. rhamnosus GG administration on gene expression in the small bowel mucosa by using human gene arrays (72). L. rhamnosus GG affected the expression of genes involved mainly in the immune response and inflammation (transforming growth factor β [TGF-β] and TNF family members, cytokines, nitric oxide synthase 1, and α-defensin), apoptosis, cell growth, cell differentiation (cyclins, caspases, and oncogenes), cell-cell signaling (ICAMs and integrins), cell adhesion (cadherins), and signal transcription and transduction (72). This is in agreement with some of the in vitro effects of L. rhamnosus GG on epithelial cells that are described above.
Immunomodulatory Interactions
Different cell types involved.
In addition to their nutritive function and their role as a physical barrier that separates luminal contents from the internal milieu, intestinal epithelium cells (IECs) actively participate in immune reactions. Together with IECs, dendritic cells (DCs) and macrophages continuously sense the environment and coordinate various defenses for the protection of mucosal tissues (Fig. 4). Innate defenses include the production of antimicrobial compounds (defensins and nitric oxide, etc.) and the secretion of chemokines such as IL-8 that recruit neutrophils, i.e., phagocytes that are capable of ingesting microorganisms or particles. In addition, many adaptive immune responses against commensal, probiotic, and pathogenic bacteria are mediated in mucosal lymphoid follicles that are distributed throughout the GIT and are referred to as the gut-associated lymphoid tissue (GALT) (8) (Fig. 4). DCs are the most important antigen-presenting cells in the mucosa. Particularly, the modulation of DC function and induction of regulatory T cells are increasingly gaining attention as important mechanisms of probiotic action (26). Related to this, the “hygiene hypothesis” suggests that the lack of exposure to certain organisms, referred to as “old friends microbes,” could be partially responsible for the increased rates of some chronic diseases such as allergies and IBD (99). Lactobacilli are suggested to be among these “old friends microbes” that are thought to be especially needed to prime immunoregulatory responses and to induce regulatory DCs and T cells (208).
FIG. 4.
Interaction of lactobacilli with the GALT. Together with IECs, DCs and macrophages continuously sense the environment and coordinate defenses for the protection of mucosal tissues. DCs are the most important antigen-presenting cells of the mucosa. It has been demonstrated that immature DCs in the lamina propria can even extend their appendices between epithelial cells, like periscopes, into the intestinal lumen to take up bacteria (203). DCs are also involved in the sampling of antigens and bacteria, including lactobacilli that are transported through microfold epithelial cells (M-cells) to the dome region of the GALT. M cells are specialized epithelial cells for antigen sampling, which are located in the follicle-associated epithelium (FAE) overlying the GALT such as Peyer's patches (PP) in isolated lymphoid follicles and are individually present in the gut epithelium. Once released into the dome region, the antigens are captured by immature DCs, which become activated when they encounter microbial products through these different pathways. This triggers a switch in cytokine and chemokine production and an upregulation in costimulatory molecules. This activation is mediated by PRRs such as TLRs, DC-SIGN on DCs, and mannose receptor (CD206) on macrophages, and these PRRs recognize pathogen-associated molecular patterns (PAMP). Activation allows the DCs to migrate to the draining lymph nodes such as the mesenteric lymph nodes (MLN) or subepithelial dome of the GALT, were the DCs orchestrate the conversion of naïve T cells into a mature, balanced response of T-helper cells or regulatory T cells, depending on the microbial products which they have encountered (8). The DCs can also activate naïve plasma cells into becoming protective sIgA-producing B cells, especially in the Peyer's patches (155). Tolerance and homeostasis in the intestine are also maintained by specialized subsets of T lymphocytes, which can all be influenced by lactobacilli (Lb). T-helper 1 (Th1) responses are usually associated with inflammatory reactions, and Th2 cells are usually associated with allergic responses. Some cytokines are released by both cell types, e.g., IL-3 and TNF-α, whereas Th1 cells secrete cytokines such as gamma interferon (IFN-γ) and IL-12 and Th2 cells secrete IL-4 and IL-5, etc. Treg cells are essential in modulating immune responses and preventing overreaction and are thought to be a key target of probiotics. At least three different Treg cells have been identified: CD4+ CD25+ Treg cells, Tr1 cells mediating bystander suppressor function by secreting IL-10, and Th3 cells that produce TGF-β and are believed to play a role in oral tolerance (54).
First, we will give examples of various effects of lactobacilli on immune cells. Subsequently, the Lactobacillus factors and their putative receptors in the host will be summarized, although knowledge of these Lactobacillus elicitors is limited, given the focus on host responses by most studies reported recently.
(i) Specific effects of lactobacilli on macrophages.
It was shown that macrophages exposed to L. rhamnosus strain GG or GR-1 produce large amounts of granulocyte CSF, resulting in reduced levels of TNF-α production, related to the ability of these probiotics to activate Stat3 and the subsequent inhibition of JNK (124). Lin and coworkers also reported that unknown small soluble factors of L. reuteri strain ATCC PTA6475 suppressed TNF-α in primary monocyte-derived macrophages from children with Crohn's disease by inhibiting the activation of the MAPK JNK and the transcription factor AP-1 and not by affecting NF-κB activation (145). Others reported that live L. rhamnosus GG induces NF-κB with subsequent STAT1- and STAT3 DNA-binding activity in human macrophages (169). Roessler et al. showed that probiotic lactobacilli can even increase the phagocytic activity of monocytes and granulocytes in healthy subjects (207). Interactions by certain lactobacilli and macrophages seem to be, at least in part, mediated by the mannose receptor (CD206) (155a).
(ii) Specific effects of lactobacilli on DCs.
As DCs play a key role in mucosal immunity, many studies have investigated the effect of lactobacilli on DCs. Most studies have shown that lactobacilli can modulate DC function by differentially inducing their maturation and the expression of MHC class II, costimulatory, adhesion, and activation molecules for antigen presentation to T cells or regulatory cytokines such as IL-10. However, lactobacilli seem to differ significantly in their capacities to modulate DC responses and T-cell balances. For instance, Christensen et al. showed that mouse bone marrow DCs were differentially stimulated by various Lactobacillus species (49). In particular, those authors showed that some strains were strong inducers of IL-12 and TNF-α, while others were considerably less potent inducers. Similarly, the various strains exhibited different capacities to induce IL-10 and IL-6. Mohamadzadeh et al. demonstrated that L. gasseri ATCC 19992, L. johnsonii ATCC 33200, and L. reuteri ATCC 23272 were able to induce the secretion of IL-12 and not of IL-10 and hence skew T-cell polarization toward Th1 and Tc1 CD8+ cytotoxic T cells, although differences among the three strains in this capacity were observed (171). Additionally, they also suggested that TLR2 is the principal TLR involved, as the Lactobacillus species used in their study upregulated the expression of TLR2 transcripts (for TLRs, see below). Karlsson et al. showed that monocytes produced higher levels of IL-12 and TNF-α in response to L. plantarum than in response to E. coli (122). In contrast, DCs secreted large amounts of IL-12, TNF-α, IL-6, and IL-10 in response to E. coli but were practically unresponsive to L. plantarum. On the other hand, Hart et al. observed that VSL#3 organisms induce high levels of IL-10 and low levels of IL-12 compared to levels of LPS in DCs from either the lamina propria or the circulation (101). In addition, these organisms inhibited LPS-induced IL-12 while maintaining IL-10 production.
(iii) Specific effects of lactobacilli on regulatory T cells.
Some studies have focused on the induction of regulatory T (Treg) cells by lactobacilli in more in detail. The probiotic mixture VSL#3 was reported to ameliorate Th1-mediated colitis by inducing TGF-β-bearing regulatory cells (73). Interactions between DCs and L. rhamnosus were also shown to induce hyporesponsive T-helper cells (29). Smits and coworkers reported that certain lactobacilli (L. casei NIZO B255 and L. reuteri ASM20016) but not L. plantarum NIZO B253 or commensal E. coli induced T cells to produce IL-10 when cultured with DCs and that such T cells exert mild to moderate suppressive effects on peripheral CD4+ T cells (230). Of interest, these Lactobacillus strains did not activate TLRs, whereas E. coli (an organism without the capacity to induce regulatory cells) did induce TLR signaling. On the other hand, L. casei NIZO B255 and L. reuteri ASM20016 were found to bind to cells transfected with the C-type lectin DC-SIGN (DC-specific intercellular adhesion molecule 3-grabbing nonintegrin) (see below), and this binding mediated by DC-SIGN was necessary to induce regulatory T cells (230). Foligne and coworkers reported that L. salivarius Ls33 and L. rhamnosus Lr32, but not L. acidophilus NCFM, induce regulatory or tolerogenic DCs (only partially matured) and CD4+ CD25+ regulatory cells in an IL-10-independent but TLR2- and NOD2-dependent way (84).
Overall, the results suggest that various Lactobacillus strains can direct immunological responses toward pro- or anti-inflammatory responses, depending on the specific bacterial strains applied, the specific immune cells used, and the specific experimental setup. This is also nicely illustrated by a study by O'Mahony and coworkers that showed that DCs from different lymphoid compartments exhibit divergent cytokine responses to probiotic and pathogenic bacteria (180). In particular, DCs from mesenteric lymph nodes produced IL-10 and TGF-β in response to L. salivarius UCC118, while those from peripheral blood produced IL-12. On the other hand, S. enterica serovar Typhimurium stimulated IL-12 in both populations, seemingly maintaining the optimal responsiveness of these cells to pathogens.
PRRs.
How are some lactobacilli able to exert these immunomodulatory effects, and which ligand-receptor interactions are involved? Many immune responses against the GIT microbiota are mediated by pattern recognition receptors (PRRs) such as TLRs that are present on IECs, DCs, and macrophages and intracellular nucleotide binding oligomerization domain (NOD)-like receptors (NLRs) present in the cytosols of many immune and epithelial cells. These PRRs play a crucial role in the innate immune system and have broad specificities for conserved, invariant, and generally repetitive features of microorganisms, in contrast to the specific antigen receptors of the adaptive immune system (167). The targets of these PRRs are often components of the bacterial cell wall such as LPS, PG, LTA, and cell wall lipoproteins. For example, triacylated bacterial lipoproteins interact with TLR2, LPS interacts with TLR4, flagellin interacts with TLR5, DNA interacts with TLR9, and muropeptides derived from PG interact with NOD1 or NOD2. These microbial ligands are present in pathogens and nonpathogenic organisms, including lactobacilli. To discriminate between microbes, it seems that the information for the different ligand-PRR interactions is integrated and converged to determine a final response. These signaling events need to be delicately balanced toward tolerance against commensals and reactivity against pathogens, and imbalance might result in the uncontrolled upregulation of inflammatory responses toward commensal bacteria, as seen in IBD (167).
(i) TLR and NLR signaling.
TLRs are transmembrane proteins with an extracellular domain made of leucine-rich repeats involved in ligand recognition and an intracytoplasmic domain containing the highly conserved Toll/IL-1 receptor domain. These Toll/IL-1 receptor domains are homologous to the IL-1β receptor-like intracellular domain and utilize some of the same signaling components involved in the response to IL-1β, including the cytoplasmic adapter molecule MyD88, the protein kinase IL-1 receptor-associated kinase (IRAK), and the adapter protein TNF receptor-associated factor 6 (TRAF6) as well as Tollip (Toll-interacting protein). Upon stimulation, IRAK is recruited to the TLR through MyD88. IRAK subsequently undergoes phosphorylation and relays the signal downstream by interacting with TRAF6 (Fig. 3). The cytosolic NLR proteins also have a leucine-rich repeat at the C terminus for ligand recognition, in addition to a Nod domain, and caspase activation and recruitment domains (CARDs) at the N terminus. TLR and NLR expressions and responsivenesses are highly localized and vary extensively with cell type (DCs versus IECs), location in the body (e.g., spleen versus mucosa), and disease status, etc. The interaction of these PRRs with their specific ligand induces NF-κB signaling and MAPK pathways, with the subsequent secretion of proinflammatory cytokines, chemokines, costimulatory molecules, and antimicrobial peptides (44). NOD and TLR signaling can also modulate each other's pathways, e.g., Watanabe et al. showed that NOD2 can be a negative regulator of TLR2-dependent NF-κB signaling (273). Moreover, innate immune recognition of microorganisms by TLRs not only is involved in induction of proinflammatory responses but also has been shown to have an important role in intestinal homeostasis and protection against intestinal injury (200). Otte et al. also reported that repeated contact with bacterial components including LTA inhibited intracellular signaling through TLRs by upregulating Tollip, an inhibitor of TLR-mediated cell activation (182). Of note, this dampening of the innate immunity in IECs and other cells does not totally mute these cells. Immune responses can still be observed upon exposure to proinflammatory cytokines and newly encountered potentially dangerous microbial products, all of which presumably activate alternative TLRs or pathways (182).
(ii) DC-SIGN signaling.
The calcium-dependent C-type lectin DC-SIGN present on DCs is an interesting PRR as it relates to the induction of Treg cells by lactobacilli (230). How lactobacilli interact with DC-SIGN is currently unknown. DC-SIGN binds to mainly mannose-containing glycoepitopes, triggering the internalization of microbes for processing and antigen presentation. Certain pathogens also use DC-SIGN as an escape mechanism for immune surveillance and the induction of immunosuppressive effects (257). For instance, it was shown that some pathogens trigger DC-SIGN on human DCs to activate the serine and threonine kinase Raf-1, which subsequently leads to the acetylation of the NF-κB subunit p65 but only after the TLR-induced activation of NF-κB, implying cross talk between TLR and DC-SIGN (98). The acetylation of p65 both prolonged and increased IL-10 transcription to enhance anti-inflammatory cytokine responses (98).
Lactobacillus ligands for PRRs.
As mentioned above, ligands for PRRs are generally cell surface components, although some ligands can be released upon lysis (e.g., DNA fragments). Additionally, as interactions between ligands and PRRs are not as specific as those between antigens and antibodies, the ligands for PRRs, such as TLR and DC-SIGN, are generally present in repetitive structures to increase avidity.
(i) LTA.
Since TAs are abundantly present on the cell surface of lactobacilli and are present only on gram-positive bacteria, they have until now gained most attention as probiotic effector molecules interacting with PRRs. It was shown that LTA molecules from L. casei YIT9029 and L. fermentum YIT0159 have the capacity to induce TNF-α in murine macrophages (163). Experiments by using cells from TLR2−/− mice or using expression plasmids for TLR2 or TLR4 and CD14 (a coreceptor for TLR2 and TLR4), along with a luciferase reporter plasmid for NF-κB in HEK293T cells, showed that TLR2, and not TLR4, is involved in this activity.
Grangette and colleagues used a dedicated dltB knockout mutant of L. plantarum NCIMB8826 to investigate the relevance of d-alanine substituents of LTA on the modulation of specific immune responses (97). A dltB mutant showed a substantial reduction in d-alanine content with a marked increase in glucose substitutions on its LTA molecules. This dltB mutant of L. plantarum induced an increased level of secretion of the anti-inflammatory cytokine IL-10 in peripheral blood mononuclear cells (PBMCs). The use of the dltB mutant in a murine colitis model was also found to be more protective against TNBS (2,4,6-trinitrobenzene sulfonic acid)-induced colitis than was the use of the wild type. Moreover, the importance of TLR2 was investigated by using cells from TLR2−/− mice. Highly purified LTA isolated from wild-type L. plantarum was able to induce TNF-α in bone marrow cells from TLR2+/+ mice but not from TLR2−/− mice. The TNF-α-inducing capacity of isolated LTA from the dltB mutant was significantly reduced in cells from TLR2+/+ mice and lost from TLR2−/− mice. However, cytokine induction by whole L. plantarum cells was not completely lost in cells from TLR2−/− mice, indicating that other surface molecules mediated by other PRRs play a role.
Interestingly, the role of d-alanylation of LTA in immunomodulation by lactobacilli seems to be species specific (188). A dltD mutant of L. rhamnosus GG, which completely lacks d-alanine residues on its LTA molecules, was also tested in the same assay using PBMCs. In contrast to L. plantarum NCIMB8826, no major differences in the levels of induction of cytokines in PBMCs for the dltD mutant and wild-type strains of L. rhamnosus GG were observed. Interestingly, the cytokine levels induced by wild-type L. plantarum NCIMB8826 and those induced by wild-type L. rhamnosus GG in PBMCs also seem to differ considerably. It will be very interesting to investigate the various differences in cell wall composition between these strains at the molecular level. Nevertheless, it is already clear that the LTA compositions of the respective wild-type and dlt mutant strains differ significantly with respect to d-alanine contents, presence of glucose residues, presence of WTA, lengths of the glycerol phosphate chain, and lipid anchors (97, 188). Moreover, since mutations affecting LTA and its charge properties as described here can have pleiotropic consequences affecting other cell surface structures such as cell surface proteins, EPS, and/or PG, only a detailed molecular characterization of different mutant strains can help to elucidate the exact role of LTA and its specific characteristics in the immunomodulatory capacity of lactobacilli. Interestingly, certain LTA molecules are also suggested to interact with the DC-SIGN receptor of DCs (38). However, the exact cell surface structures interacting with DC-SIGN have not yet been determined (also see below).
(ii) Cell surface proteins.
The putative key immunostimulatory activity of LTA is sometimes debated. It was recently shown that contaminants in LTA purifications can significantly affect immune interactions and cytokine induction. Not LTA but lipoproteins appeared to be the dominant immunologically active compounds in some LTA isolations from gram-positive bacteria interacting with TLR2 (102). The role of lipoproteins in Lactobacillus immune interactions is quite unexplored. In the L. johnsonii La1 genome sequence, sequences encoding two putative lipoproteins, one with similarity to the CD4+ T-cell-stimulating antigen of L. monocytogenes and the second with similarity to the saliva-binding protein of Streptococcus sanguis, have been identified (196), but to our knowledge, functional analyses have not yet been reported. Additionally, glycoproteins are also attractive candidates for interactions with lectin receptors such as DC-SIGN or TLR receptors such as TLR2, but their role in bacteria in general is quite unexplored (19).
On the other hand, immunomodulation studies have shown that cell surface EF-Tu and GroEL from L. johnsonii La1 stimulate IL-8 secretion in HT-29 cells and isolated blood macrophages in a soluble-CD14-dependent mechanism (20, 95). CD14 is known as a coreceptor for TLR2 and TLR4, but its exact function as a possible PRR for lactobacilli is currently not known. Similarly, experiments with a spontaneous nonaggregation mutant, MU5, of L. crispatus M247 also suggest a role for surface proteins and EF-Tu as interesting PRR ligands (265). Although the exact mutation causing the nonaggregation phenotype in MU5 is currently not known, it is suggested to be related to the differential expression of cell surface proteins, with a possible important role for EF-Tu (227). L. crispatus M247 was shown to increase TLR2 mRNA levels both in the colonic mucosa and in epithelial cells, while it reduced TLR4 expression levels in epithelial cells, in a pathway possibly involving ERK1 signaling. On the contrary, the mutant MU5 did not modulate TLR2 and TLR4 levels in the colonic mucosa (265).
The role of other surface proteins in immune reactions remains elusive. Secretory IgA (sIgA) is abundantly present in the GIT and plays an important role in the modulation of gut immune responses against commensal bacteria (189) and the modulation of bacterial adherence to the gut mucosa (232). Although sIgA is not a low-affinity PRR of the innate immune system, it has been shown that IgA responses against commensal bacteria, in contrast to responses against toxins and pathogens, are mediated by a primitive T-cell-independent mechanism, resulting in the control of the microbiota by a broad spectrum of reduced-affinity sIgA (154, 155). How lactobacilli interact with sIgA responses is currently unknown, but Denou et al. identified LJ1680, encoding a putative sortase-dependent IgA protease, as being specifically induced in L. johnsonii NCC533 cells in the small intestine (67). The mutation of LJ1680 significantly decreased the gut persistence time of NCC533. LJ1680 contains a PF07580 peptidase domain and shows 30% identity to the IgA1 protease of S. pneumoniae, which cleaves the antibody in the hinge region so that Fab fragments, which bind to the bacterial cell surface and increase the binding to epithelial cells, are released (275). However, the role of LJ1680 in L. johnsonii NCC533 and its possible interaction with IgA remain unclear. Peptidase domains are also present in proteins involved in the metabolism of the bacterial cell wall, and the specific induction of LJ1680 in the GIT might also be involved in general stress resistance, as most genes identified by IVET or related strategies such as those followed by Denou et al. (67) belong to this functional class (see above).
(iii) EPS.
EPSs and, particularly, the heteropolysaccharides are also attractive candidates as probiotic effector molecules interacting with PRRs, as they display a high level of diversity and complexity among lactobacilli, but they have not yet been studied in detail by molecular analysis. One obvious question would be why many Lactobacillus strains invest biosynthetic energy in producing such complex structures. A possible answer is that they do so to circumvent enzymatic breakdown by competing microbiota or host enzymes, implying an important role in the competitive environment of the GIT. Another but complementary explanation would be to evade the host immune system and induce tolerance. Support for this explanation comes from studies with the gut symbiont Bacteroides fragilis (see, e.g., reference 56) and studies with pathogenic streptococci where capsular polysaccharides have been shown to impede phagocytosis by macrophages and neutrophils (123). In this way, the role of EPS in immune responses seems to be rather indirect, by protecting or shielding other surface molecules such as LTA from their cognate host receptors.
It thus remains to be determined whether Lactobacillus EPS molecules can interact with specific lectin receptors of the immune system, such as TLR2, DC-SIGN, or CD14. Nevertheless, isolated EPS molecules of lactobacilli have been shown to exert some immune responses. For instance, it was reported that the oral administration of EPS (in rather high concentrations of 2 mg per day) produced by L. kefiranofaciens ATCC 43761, consisting of equal amounts of glucose and galactose, induced a mucosal immune response in mice that included the induction of IgA-positive cells (264). High-rhamnose-EPS molecules from L. rhamnosus RW-9595 M, composed of heptasaccharide subunits of rhamnose-, glucose-, and pyruvate-substituted galactose in a molar ratio of 4:2:1, were reported to stimulate TNF-α, IL-6, and IL-12 in human PBMCs and macrophages and gamma interferon in mouse splenocytes in significantly higher concentrations than LPS from E. coli (45). Others also reported that the purified polysaccharide-PG fraction of L. casei Shirota is active in a concentration of 10 μg/ml in suppressing the cytokine production by macrophages after induction by LPS and that the polysaccharide moiety is responsible for this activity (164). Recently, knockout mutants defective in the major cell wall-associated EPS molecule of L. casei Shirota were shown to induce more proinflammatory cytokine secretion in the mouse macrophages and spleen cells than the corresponding wild type (283). These results indicate that EPS can function as an immune modulator to reduce excessive immune reactions during the activation of macrophages by L. casei Shirota. However, those authors used heat-killed wild-type and mutant bacteria for these assays, demanding some caution with the interpretation of the results.
(iv) PG.
NOD1 and NOD2 are the principal intracellular PRRs for PG. NOD1 senses γ-d-glutamyl-meso-diaminopimelic acid (44), present mostly in gram-negative bacteria but also present in some lactobacilli such as some L. plantarum strains (65). NOD2 recognizes muramyl peptide (44), which is present in all lactobacilli (65). For PG fragments to be released, this requires autolysis or an exogenous lysozyme attack, which is common in the gut. Muramylpeptides derived from L. plantarum ATCC 8014, for instance, are known to display immunoadjuvant activity (129). It was also shown that L. salivarius Ls33 and L. rhamnosus Lr32, but not L. acidophilus NCFM, induce regulatory DCs and T cells in a TLR2- and NOD2-dependent way (84), but the role of PG fragments in the immunomodulatory effects of lactobacilli remains rather unexplored. This is particularly of interest for IBD, as a genetic link between NOD2 mutations and susceptibility to Crohn's disease exists, mainly in European and North American patients (48). Most polymorphisms in the NOD2 (or Card15) gene that are linked to Crohn's disease are located within or near the leucine-rich repeat of the NOD2 protein, which is the domain that senses the bacterial products, indicating an altered capacity to recognize microbial ligands. This loss of NOD2 function in certain Crohn's disease patients needs to be taken into account when probiotics are applied. It is important that most intervention studies reported to date with probiotic lactobacilli in this class of IBD patients failed to demonstrate any benefit (26, 103, 193).
(v) Unmethylated CpG DNA.
DNA of the probiotic cocktail VSL#3 has been shown to exert a systemically induced protective effect against dextran sulfate sodium-induced colitis mediated by TLR9 (198). Those authors suggested that DNA-TLR9 signaling in a subset of mucosal plasmacytoid DCs probably resulted in the differentiation of naïve CD4+ T lymphocytes into Treg cells, mediating the protective effect. Another example of the strong immunomodulatory capacity of probiotic DNA is shown by chromosomal DNA of L. rhamnosus GG (including a derived TTTCGTTT oligonucleotide) that was able to induce murine B-cell proliferation and activate DCs (114). However, further functional studies are required to investigate the relative contribution of DNA to the overall immunomodulatory activity of lactobacilli.
CONCLUSIONS
In this review, we have summarized the current knowledge of genes and molecules that contribute to the presently best-documented probiotic and health-promoting actions of lactobacilli. Important Lactobacillus factors can be grouped into two categories: adaptation and probiotic factors directly mediating health effects. Adaptation factors include determinants of stress resistance, metabolism in the host, and adherence to the gut mucosa. Knowledge of these adaptation factors will help to determine the optimal frequency, dose, and mode of administration for potential probiotic Lactobacillus strains. Probiotic factors include antipathogenic, epithelium barrier-preserving, and immunomodulatory molecules. A literature survey reveals that in contrast to adaptation factors, knowledge of the probiotic factors of lactobacilli is still limited but is starting to be built. In order to advance the field, future molecular studies should continue to address these factors, taking into account that various cell surface structures or metabolites of lactobacilli also influence the adaptation and adherence capacity in the host.
Related to adaptation to the host, molecular studies have shown that a combination of cell surface structures, chaperones, specific regulatory systems, and exporters or enzymes determines the survival capacity of particular Lactobacillus strains encountering acid and bile stress. However, in vitro screening for acid and bile tolerance does not always predict the in vivo survival capacity. This indicates that other stresses such as osmotic and oxidative stress, nutrient limitation, and the presence of antimicrobial products produced by competing microbes and host cells also need to be considered when potential probiotic strains are selected. Nevertheless, the fact that most probiotic lactobacilli were originally isolated and selected from the GIT is meant to overcome possible problems of surviving the harsh environments after oral administration.
The need to include a selection criterion based on adhesion and colonization of the human intestinal mucosa by lactobacilli to obtain probiotic effects is currently being debated. Several in vitro mechanistic studies have shown a role for adhesion (or direct cell contact) in pathogen exclusion, the enhancement of epithelial barrier function, and immunomodulation. Moreover, these studies showed that the best effects are observed when the lactobacilli are administered in a preventive setup. However, others have shown that short contact times or the release of various soluble factors is sometimes sufficient to modulate signaling pathways in the host. Extrapolation of these findings to clinical applications requires caution. Permanent colonization by exogenously applied lactobacilli seems difficult to achieve due to the niche exclusion principle and colonization of the GIT by thousands of well-adapted species. However, the available data suggest that probiotic effects can be obtained when lactobacilli are administered regularly. Although the criteria for live probiotics still remain valid, there is currently no ground to state that introduced probiotic lactobacilli need to persist and rapidly multiply in the GIT. Detailed mechanistic studies, including adhesion mutants and in vivo experiments, should further substantiate the importance of adhesion as a key selection criterion.
The antipathogenic effect of lactobacilli is one of the hallmarks of probiotic action. However, mechanistic data on their in vivo efficacy are currently scarce, notwithstanding that a relatively high number of publications reported in vitro antipathogenic effects of certain molecules of lactobacilli. In this respect, the study showing an in vivo role for bacteriocin production by L. salivarius against L. monocytogenes infection (53) is an important contribution to the field. Additionally, although the concept of competitive exclusion of pathogens by adherent lactobacilli is attractive, there is currently almost no evidence that this concept is applicable in vivo to limit pathogen colonization in the GIT.
Related to the beneficial effects of lactobacilli on the host epithelium and immune cells, much remains to be learned about the specific Lactobacillus ligand-host receptor interactions. Interpretation of some of the currently available data related to Lactobacillus ligands is blurred by the fact that isolated cell wall components may be contaminated or that mutations in genes determining a given cell surface factor may significantly affect the presence or presentation of other surface appendages. Nevertheless, many mechanistic in vitro studies have convincingly shown that lactobacilli or their products can modulate signaling pathways, resulting in beneficial effects. Interestingly, both anti-inflammatory and (transiently induced) mild proinflammatory responses have been reported for lactobacilli. Although this seems to be contradictory at first sight, it indicates that probiotic lactobacilli have a principally balance-controlling effect on gut epithelium homeostasis whereby strain specificity is observed. For example, a limited activation of NF-κB and MAPKs may be favorable, as activation below the inflammation threshold might render the host defense system more alert against hostile confrontations. This implies that the health conditions in which specific Lactobacillus strains are administered need to be carefully defined, as it can be anticipated that different immune-stimulating effects are intended for, e.g., IBD patients than for subjects with constipation. Additionally, most mechanistic studies of immunological and epithelial responses are currently performed in vitro only. They need to be confirmed in complex in vivo environments where the information of many intracellular and extracellular signals is integrated. Hereby, researchers studying Lactobacillus-host interactions must consider that the impact of probiotics on the host physiology is usually much smaller and more subtle than that of pharmaceutical products, which complicates the delineation of clear cause-consequence relationships between Lactobacillus factors and host responses.
In conclusion, the closer the molecular machinery is studied, the more similarities are observed between ingested probiotic and pathogenic bacteria. It is perhaps not surprising that efficient probiotic Lactobacillus strains resemble pathogens in many aspects, such as in survival and adherence (i.e., adaptation factors). It can be imagined that for efficient competition with pathogens, lactobacilli must utilize similar nutrients and adhesion sites on host cells. Additionally, the available data indicate that probiotic lactobacilli interact with various receptors of immune cells and modulate epithelial cell functions. Some of these interactions are similar for pathogens and absent for resident and commensal microbiota. In this respect, it is interesting to look at the mechanisms that some pathogenic bacteria use for immune evasion and the downregulation of inflammatory reactions. A remarkable example is that, similarly to some pathogens using DC-SIGN as an escape mechanism, some lactobacilli interact with DC-SIGN and induce regulatory T cells. As lactobacilli lack tissue-destructive functions and genuine virulence factors, the overall result of their interactions with the host is generally beneficial for the host.
A continuing challenge for the future is to reveal more effector-receptor relationships in Lactobacillus-host interactions that contribute to the beneficial effects of lactobacilli on mucosal homeostasis. These fundamental studies involving many disciplines will substantiate translational research attempting to directly relate insights from the bench to the manufacturer, the consumer, and the clinic.
Acknowledgments
S.L. and S.C.J.D.K. are a research assistant and a postdoctoral fellow of the FWO-Vlaanderen, respectively.
We are also grateful for the financial support of the FWO-Vlaanderen through project G.0236.07.
REFERENCES
- 1.Adlerberth, I., S. Ahrne, M. L. Johansson, G. Molin, L. A. Hanson, and A. E. Wold. 1996. A mannose-specific adherence mechanism in Lactobacillus plantarum conferring binding to the human colonic cell line HT-29. Appl. Environ. Microbiol. 622244-2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alakomi, H. L., E. Skytta, M. Saarela, T. Mattila-Sandholm, K. Latva-Kala, and I. M. Helander. 2000. Lactic acid permeabilizes gram-negative bacteria by disrupting the outer membrane. Appl. Environ. Microbiol. 662001-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altermann, E., B. L. Buck, R. Cano, and T. R. Klaenhammer. 2004. Identification and phenotypic characterization of the cell division protein CdpA. Gene 342189-197. [DOI] [PubMed] [Google Scholar]
- 4.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 1023906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antikainen, J., L. Anton, J. Sillanpaa, and T. K. Korhonen. 2002. Domains in the S-layer protein CbsA of Lactobacillus crispatus involved in adherence to collagens, laminin and lipoteichoic acids and in self-assembly. Mol. Microbiol. 46381-394. [DOI] [PubMed] [Google Scholar]
- 6.Antikainen, J., V. Kupannen, K. Lahteenmaki, and T. K. Korhonen. 2007. pH-dependent association of enolase and glyceraldehyde-3-phosphate dehydrogenase of Lactobacillus crispatus with the cell wall and lipoteichoic acids. J. Bacteriol. 1894539-4543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Anukam, K. C., E. O. Osazuwa, I. Ahonkhai, and G. Reid. 2006. Assessment of Lactobacillus species colonizing the vagina of apparently healthy Nigerian women, using PCR-DGGE and 16S rRNA gene sequencing. World J. Microbiol. Biotechnol. 221055-1060. [Google Scholar]
- 8.Artis, D. 2008. Epithelial-cell recognition of commensal bacteria and maintenance of immune homeostasis in the gut. Nat. Rev. Immunol. 8411-420. [DOI] [PubMed] [Google Scholar]
- 9.Avall-Jaaskelainen, S., and A. Palva. 2005. Lactobacillus surface layers and their applications. FEMS Microbiol. Rev. 29511-529. [DOI] [PubMed] [Google Scholar]
- 10.Azcarate-Peril, M. A., E. Altermann, R. L. Hoover-Fitzula, R. J. Cano, and T. R. Klaenhammer. 2004. Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl. Environ. Microbiol. 705315-5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Azcarate-Peril, M. A., O. McAuliffe, E. Altermann, S. Lick, W. M. Russell, and T. R. Klaenhammer. 2005. Microarray analysis of a two-component regulatory system involved in acid resistance and proteolytic activity in Lactobacillus acidophilus. Appl. Environ. Microbiol. 715794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 3071915-1920. [DOI] [PubMed] [Google Scholar]
- 13.Barrangou, R., E. Altermann, R. Hutkins, R. Cano, and T. R. Klaenhammer. 2003. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc. Natl. Acad. Sci. USA 1008957-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barrangou, R., M. A. Azcarate-Peril, T. Duong, S. B. Conners, R. M. Kelly, and T. R. Klaenhammer. 2006. Global analysis of carbohydrate utilization by Lactobacillus acidophilus using cDNA microarrays. Proc. Natl. Acad. Sci. USA 1033816-3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 1794043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bath, K., S. Roos, T. Wall, and H. Jonsson. 2005. The cell surface of Lactobacillus reuteri ATCC 55730 highlighted by identification of 126 extracellular proteins from the genome sequence. FEMS Microbiol. Lett. 25375-82. [DOI] [PubMed] [Google Scholar]
- 17.Begley, M., C. G. M. Gahan, and C. Hill. 2005. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29625-651. [DOI] [PubMed] [Google Scholar]
- 18.Bender, M. H., R. T. Cartee, and J. Yother. 2003. Positive correlation between tyrosine phosphorylation of CpsD and capsular polysaccharide production in Streptococcus pneumoniae. J. Bacteriol. 1856057-6066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benz, I., and M. A. Schmidt. 2002. Never say never again: protein glycosylation in pathogenic bacteria. Mol. Microbiol. 45267-276. [DOI] [PubMed] [Google Scholar]
- 20.Bergonzelli, G. E., D. Granato, R. D. Pridmore, L. F. Marvin-Guy, D. Donnicola, and I. E. Corthesy-Theulaz. 2006. GroEL of Lactobacillus johnsonii La1 (NCC 533) is cell surface associated: potential role in interactions with the host and the gastric pathogen Helicobacter pylori. Infect. Immun. 74425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Besselink, M. G. H., H. C. van Santvoort, E. Buskens, M. A. Boermeester, et al. 2008. Probiotic prophylaxis in predicted severe acute pancreatitis: a randomised, double-blind, placebo-controlled trial. Lancet 371651-659. (Erratum, 371:1246.) [DOI] [PubMed] [Google Scholar]
- 22.Bezkorovainy, A. 2001. Probiotics: determinants of survival and growth in the gut. Am. J. Clin. Nutr. 73399S-405S. [DOI] [PubMed] [Google Scholar]
- 23.Billot-Klein, D., R. Legrand, B. Schoot, J. vanHeijenoort, and L. Gutmann. 1997. Peptidoglycan structure of Lactobacillus casei, a species highly resistant to glycopeptide antibiotics. J. Bacteriol. 1796208-6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boekhorst, J., M. W. H. J. De Been, M. Kleerebezem, and R. J. Siezen. 2005. Genome-wide detection and analysis of cell wall-bound proteins with LPxTG-like sorting motifs. J. Bacteriol. 1874928-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boekhorst, J., Q. Helmer, M. Kleerebezem, and R. J. Siezen. 2006. Comparative analysis of proteins with a mucus-binding domain found exclusively in lactic acid bacteria. Microbiology 152273-280. [DOI] [PubMed] [Google Scholar]
- 26.Boirivant, M., and W. Strober. 2007. The mechanism of action of probiotics. Curr. Opin. Gastroenterol. 23679-692. [DOI] [PubMed] [Google Scholar]
- 27.Boot, H. J., C. P. A. M. Kolen, J. M. Vannoort, and P. H. Pouwels. 1993. S-layer protein of Lactobacillus acidophilus ATCC 4356: purification, expression in Escherichia coli, and nucleotide sequence of the corresponding gene. J. Bacteriol. 1756089-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyle, R. J., and M. L. K. Tang. 2006. The role of probiotics in the management of allergic disease. Clin. Exp. Allergy 36568-576. [DOI] [PubMed] [Google Scholar]
- 29.Braat, H., J. van den Brande, E. van Tol, D. Hommes, M. Peppelenbosch, and S. van Deventer. 2004. Lactobacillus rhamnosus induces peripheral hyporesponsiveness in stimulated CD4(+) T cells via modulation of dendritic cell function. Am. J. Clin. Nutr. 801618-1625. [DOI] [PubMed] [Google Scholar]
- 30.Branda, S. S., A. Vik, L. Friedman, and R. Kolter. 2005. Biofilms: the matrix revisited. Trends Microbiol. 1320-26. [DOI] [PubMed] [Google Scholar]
- 31.Bron, P. A., C. Grangette, A. Mercenier, W. M. de Vos, and M. Kleerebezem. 2004. Identification of Lactobacillus plantarum genes that are induced in the gastrointestinal tract of mice. J. Bacteriol. 1865721-5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bron, P. A., M. Marco, S. M. Hoffer, E. Van Mullekom, W. M. de Vos, and M. Kleerebezem. 2004. Genetic characterization of the bile salt response in Lactobacillus plantarum and analysis of responsive promoters in vitro and in situ in the gastrointestinal tract. J. Bacteriol. 1867829-7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bron, P. A., M. Meijer, R. S. Bongers, W. M. de Vos, and M. Kleerebezem. 2007. Dynamics of competitive population abundance of Lactobacillus plantarum ivi gene mutants in faecal samples after passage through the gastrointestinal tract of mice. J. Appl. Microbiol. 1031424-1434. [DOI] [PubMed] [Google Scholar]
- 34.Bron, P. A., D. Molenaar, W. M. Vos, and M. Kleerebezem. 2006. DNA micro-array-based identification of bile-responsive genes in Lactobacillus plantarum. J. Appl. Microbiol. 100728-738. [DOI] [PubMed] [Google Scholar]
- 35.Buck, B. L., E. Altermann, T. Svingerud, and T. R. Klaenhammer. 2005. Functional analysis of putative adhesion factors in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 718344-8351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burton, J. P., P. A. Cadieux, and G. Reid. 2003. Improved understanding of the bacterial vaginal microbiota of women before and after probiotic instillation. Appl. Environ. Microbiol. 6997-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burton, J. P., and G. Reid. 2002. Evaluation of the bacterial vaginal flora of 20 postmenopausal women by direct (Nugent score) and molecular (polymerase chain reaction and denaturing gradient gel electrophoresis) techniques. J. Infect. Dis. 1861770-1780. [DOI] [PubMed] [Google Scholar]
- 38.Calder, P. C., S. Krauss-Etschmann, E. C. de Jong, C. Dupont, J. S. Frick, H. Frokiaer, J. Heinrich, H. Garn, S. Koletzko, G. Lack, G. Mattelio, H. Renz, P. T. Sangild, J. Schrezenmeir, T. M. Stulnig, T. Thymann, A. E. Wold, and B. Koletzko. 2006. Early nutrition and immunity—progress and perspectives. Br. J. Nutr. 96774-790. [PubMed] [Google Scholar]
- 39.Callanan, M., P. Kaleta, J. O'Callaghan, O. O'Sullivan, K. Jordan, O. McAuliffe, A. Sangrador-Vegas, L. Slattery, G. F. Fitzgerald, T. Beresford, and R. P. Ross. 2008. Genome sequence of Lactobacillus helveticus, an organism distinguished by selective gene loss and insertion sequence element expansion. J. Bacteriol. 190727-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Callegari, M. L., B. Riboli, J. W. Sanders, P. S. Cocconcelli, J. Kok, G. Venema, and L. Morelli. 1998. The S-layer gene of Lactobacillus helveticus CNRZ 892: cloning, sequence and heterologous expression. Microbiology 144719-726. [DOI] [PubMed] [Google Scholar]
- 41.Camilleri, M. 2006. Probiotics and irritable bowel syndrome: rationale, putative mechanisms, and evidence of clinical efficacy. J. Clin. Gastroenterol. 40264-269. [DOI] [PubMed] [Google Scholar]
- 42.Campo, N., H. Tjalsma, G. Buist, D. Stepniak, M. Meijer, M. Veenhuis, M. Westermann, J. P. Muller, S. Bron, J. Kok, O. P. Kuipers, and J. D. H. Jongbloed. 2004. Subcellular sites for bacterial protein export. Mol. Microbiol. 531583-1599. [DOI] [PubMed] [Google Scholar]
- 43.Cappa, F., D. Cattivelli, and P. S. Cocconcelli. 2005. The uvrA gene is involved in oxidative and acid stress responses in Lactobacillus helveticus CNBL1156. Res. Microbiol. 1561039-1047. [DOI] [PubMed] [Google Scholar]
- 44.Cario, E. 2005. Bacterial interactions with cells of the intestinal mucosa: Toll-like receptors and NOD2. Gut 541182-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chabot, S., H. L. Yu, L. De Leseleuc, D. Cloutier, M. R. Van Calsteren, M. Lessard, D. Roy, M. Lacroix, and D. Oth. 2001. Exopolysaccharides from Lactobacillus rhamnosus RW-9595M stimulate TNF, IL-6 and IL-12 in human and mouse cultured immunocompetent cells, and IFN-gamma mouse splenocytes. Lait 81683-697. [Google Scholar]
- 46.Chang, D. E., D. J. Smalley, D. L. Tucker, M. P. Leatham, W. E. Norris, S. J. Stevenson, A. B. Anderson, J. E. Grissom, D. C. Laux, P. S. Cohen, and T. Conway. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA 1017427-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen, X. Y., J. J. Xu, J. B. Shuai, J. S. Chen, Z. F. Zhang, and W. H. Fang. 2007. The S-layer proteins of Lactobacillus crispatus strain ZJ001 is responsible for competitive exclusion against Escherichia coli O157: H7 and Salmonella typhimurium. Int. J. Food Microbiol. 115307-312. [DOI] [PubMed] [Google Scholar]
- 48.Cho, J. H. 2008. The genetics and immunopathogenesis of inflammatory bowel disease. Nat. Rev. Immunol. 8458-466. [DOI] [PubMed] [Google Scholar]
- 49.Christensen, H. R., H. Frokiaer, and J. J. Pestka. 2002. Lactobacilli differentially modulate expression of cytokines and maturation surface markers in murine dendritic cells. J. Immunol. 168171-178. [DOI] [PubMed] [Google Scholar]
- 50.Claesson, M. J., Y. Li, S. Leahy, C. Canchaya, J. P. van Pijkeren, A. M. Cerdeno-Tarraga, J. Parkhill, S. Flynn, G. C. O'Sullivan, J. K. Collins, D. Higgins, F. Shanahan, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Multireplicon genome architecture of Lactobacillus salivarius. Proc. Natl. Acad. Sci. USA 1036718-6723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Claesson, M. J., D. van Sinderen, and P. W. O'Toole. 2007. The genus Lactobacillus—a genomic basis for understanding its diversity. FEMS Microbiol. Lett. 26922-28. [DOI] [PubMed] [Google Scholar]
- 52.Corcoran, B. M., C. Stanton, G. F. Fitzgerald, and R. P. Ross. 2005. Survival of probiotic lactobacilli in acidic environments is enhanced in the presence of metabolizable sugars. Appl. Environ. Microbiol. 713060-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corr, S. C., Y. Li, C. U. Riedel, P. W. O'Toole, C. Hill, and C. G. M. Gahan. 2007. Bacteriocin production as a mechanism for the antfinfective activity of Lactobacillus salivarius UCC118. Proc. Natl. Acad. Sci. USA 1047617-7621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corthesy, B., H. R. Gaskins, and A. Mercenier. 2007. Cross-talk between probiotic host immune system. J. Nutr. 137781S-790S. [DOI] [PubMed] [Google Scholar]
- 55.Cotter, P. D., and C. Hill. 2003. Surviving the acid test: responses of gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67429-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Coyne, M. J., B. Reinap, M. M. Lee, and L. E. Comstock. 2005. Human symbionts use a host-like pathway for surface fucosylation. Science 3071778-1781. [DOI] [PubMed] [Google Scholar]
- 57.Cunin, R., N. Glansdorff, A. Pierard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dal Bello, F., J. Walter, W. P. Hammes, and C. Hertel. 2003. Increased complexity of the species composition of lactic acid bacteria in human feces revealed by alternative incubation condition. Microb. Ecol. 45455-463. [DOI] [PubMed] [Google Scholar]
- 59.Daniel, R. A., and J. Errington. 2003. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113767-776. [DOI] [PubMed] [Google Scholar]
- 60.De Angelis, M., L. Bini, V. Pallini, P. S. Cocconcelli, and M. Gobbetti. 2001. The acid-stress response in Lactobacillus sanfranciscensis CB1. Microbiology 1471863-1873. [DOI] [PubMed] [Google Scholar]
- 61.De Angelis, M., and M. Gobbetti. 2004. Environmental stress responses in Lactobacillus: a review. Proteomics 4106-122. [DOI] [PubMed] [Google Scholar]
- 62.De Keersmaecker, S. C. J., K. Braeken, T. L. A. Verhoeven, M. Perea Vélez, S. Lebeer, J. Vanderleyden, and P. Hols. 2006. Flow cytometric testing of green fluorescent protein-tagged Lactobacillus rhamnosus GG for response to defensins. Appl. Environ. Microbiol. 724923-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.De Keersmaecker, S. C. J., and J. Vanderleyden. 2003. Constraints on detection of autoinducer-2 (Al-2) signalling molecules using Vibrio harveyi as a reporter. Microbiology 1491953-1956. [DOI] [PubMed] [Google Scholar]
- 64.De Keersmaecker, S. C. J., T. L. A. Verhoeven, J. Desair, K. Marchal, J. Vanderleyden, and I. Nagy. 2006. Strong antimicrobial activity of Lactobacillus rhamnosus GG against Salmonella typhimurium is due to accumulation of lactic acid. FEMS Microbiol. Lett. 25989-96. [DOI] [PubMed] [Google Scholar]
- 65.Delcour, J., T. Ferain, M. Deghorain, E. Palumbo, and P. Hols. 1999. The biosynthesis and functionality of the cell-wall of lactic acid bacteria. Antonie van Leeuwenhoek 76159-184. [PubMed] [Google Scholar]
- 66.Denou, E., B. Berger, C. Barretto, J. M. Panoff, F. Arigoni, and H. Brussow. 2007. Gene expression of commensal Lactobacillus johnsonii strain NCC533 during in vitro growth and in the murine gut. J. Bacteriol. 1898109-8119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Denou, E., R. D. Pridmore, B. Berger, J. M. Panoff, F. Arigoni, and H. Brussow. 2008. Identification of genes associated with the long gut persistence phenotype of the probiotic Lactobacillus johnsonii strain NCC533 using a combination of genomics and transcriptome analysis. J. Bacteriol. 1903161-3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De Preter, V., T. Vanhoutte, G. Huys, J. Swings, L. De Vuyst, P. Rutgeerts, and K. Verbeke. 2007. Effects of Lactobacillus casei Shirota, Bifidobacterium breve, and oligofructose-enriched inulin on colonic nitrogen-protein metabolism in healthy humans. Am. J. Physiol. Gastrointest. Liver Physiol. 292G358-G368. [DOI] [PubMed] [Google Scholar]
- 69.Deshpande, G., S. Rao, and S. Patole. 2007. Probiotics for prevention of necrotising enterocolitis in preterm neonates with very low birthweight: a systematic review of randomised controlled trials. Lancet 3691614-1620. [DOI] [PubMed] [Google Scholar]
- 70.De Vuyst, L., F. De Vin, F. Vaningelgem, and B. Degeest. 2001. Recent developments in the biosynthesis and applications of heteropolysaccharides from lactic acid bacteria. Int. Dairy J. 11687-707. [Google Scholar]
- 71.De Vuyst, L., and B. Degeest. 1999. Heteropolysaccharides from lactic acid bacteria. FEMS Microbiol. Rev. 23153-177. [DOI] [PubMed] [Google Scholar]
- 72.Di Caro, S., H. Tab, A. Grillo, C. Elia, G. Gasbarrini, A. R. Sepulveda, and A. Gasbarrini. 2005. Effects of Lactobacillus GG on genes expression pattern in small bowel mucosa. Dig. Liver Dis. 37320-329. [DOI] [PubMed] [Google Scholar]
- 73.Di Giacinto, C., M. Marinaro, M. Sanchez, W. Strober, and M. Boirivant. 2005. Probiotics ameliorate recurrent Th1-mediated murine colitis by inducing IL-10 and IL-10-dependent TGF-beta-bearing regulatory cells. J. Immunol. 1743237-3246. [DOI] [PubMed] [Google Scholar]
- 74.Dobson, A. E., R. B. Sanozky-Dawes, and T. R. Klaenhammer. 2007. Identification of an operon and inducing peptide involved in the production of lactacin B by Lactobacillus acidophilus. J. Appl. Microbiol. 1031766-1778. [DOI] [PubMed] [Google Scholar]
- 75.Duncan, S. H., P. Louis, and H. J. Flint. 2004. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 705810-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Duncker, S. C., L. Wang, P. Hols, and J. Bienenstock. 2008. The D-alanine content of lipoteichoic acid is crucial for Lactobacillus plantarum-mediated protection from visceral pain perception in a rat colorectal distension model. Neurogastroenterol. Motil. 20843-850. [DOI] [PubMed] [Google Scholar]
- 77.Duong, T., R. Barrangou, W. M. Russell, and T. R. Klaenhammer. 2006. Characterization of the tre locus and analysis of trehalose cryoprotection in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 721218-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Durant, J. A., D. E. Corrier, and S. C. Ricke. 2000. Short-chain volatile fatty acids modulate the expression of the hilA and invF genes of Salmonella Typhimurium. J. Food Prot. 63573-578. [DOI] [PubMed] [Google Scholar]
- 79.Eijsink, V. G. H., L. Axelsson, D. B. Diep, L. S. Havarstein, H. Holo, and I. F. Nes. 2002. Production of class II bacteriocins by lactic acid bacteria; an example of biological warfare and communication. Antonie van Leeuwenhoek 81639-654. [DOI] [PubMed] [Google Scholar]
- 80.Falagas, M. E., G. I. Betsi, and S. Athanasiou. 2007. Probiotics for the treatment of women with bacterial vaginosis. Clin. Microbiol. Infect. 13657-664. [DOI] [PubMed] [Google Scholar]
- 81.FAO/WHO. 2001. Evaluation of health and nutritional properties of powder milk and live lactic acid bacteria. Food and Agriculture Organization of the United Nations and World Health Organization expert consultation report. FAO, Rome, Italy.
- 82.Fedorak, R. N., and K. L. Madsen. 2004. Probiotics and the management of inflammatory bowel disease. Inflamm. Bowel Dis. 10286-299. [DOI] [PubMed] [Google Scholar]
- 83.Fisseha, M., and F. Arigoni. 2005. Beyond genome sequences: approaches to genome-wide analysis of gut bacteria, p. 97-128. In G. W. Tannock (ed.), Probiotics & prebiotics: scientific aspects. Caister Academic Press, Norfolk, United Kingdom.
- 84.Foligne, B., G. Zoumpopoulou, J. Dewulf, A. Ben Younes, F. Chareyre, J. C. Sirard, B. Pot, and C. Grangette. 2007. A key role of dendritic cells in probiotic functionality. PLOS One 2e313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Formstone, A., R. Carballido-Lopez, P. Noirot, J. Errington, and D. J. Scheffers. 2008. Localization and interactions of teichoic acid synthetic enzymes in Bacillus subtilis. J. Bacteriol. 1901812-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fozo, E. M., J. K. Kajfasz, and R. G. Quivey. 2004. Low pH-induced membrane fatty acid alterations in oral bacteria. FEMS Microbiol. Lett. 238291-295. [DOI] [PubMed] [Google Scholar]
- 87.Francius, G., S. Lebeer, D. Alsteens, L. Wildling, H. J. Gruber, P. Hols, S. C. J. De Keersmaecker, J. Vanderleyden, and Y. F. Dufrene. 2008. Detection, localization and conformational analysis of single polysaccharide molecules on live bacteria. ACS Nano 21921-1929. [DOI] [PubMed] [Google Scholar]
- 88.Frees, D., K. Savijoki, P. Varmanen, and H. Ingmer. 2007. Clp ATPases and ClpP proteolytic complexes regulate vital biological processes in low GC, gram-positive bacteria. Mol. Microbiol. 631285-1295. [DOI] [PubMed] [Google Scholar]
- 89.Fujiya, M., M. W. Musch, Y. Nakagawa, S. Hu, J. Alverdy, Y. Kohgo, O. Schneewind, B. Jabri, and E. B. Chang. 2007. The Bacillus subtilis quorum-sensing molecule CSF contributes to intestinal homeostasis via OCTN2, a host cell membrane transporter. Cell Host Microbe 1299-308. [DOI] [PubMed] [Google Scholar]
- 90.Reference deleted.
- 91.Ganzle, M. G., and C. Schwab. 2005. Exopolysaccharide production by intestinal lactobacilli, p. 83-96. In G. W. Tannock (ed.), Probiotics & prebiotics: scientific aspects. Caister Academic Press, Norfolk, United Kingdom.
- 92.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota—introducing the concept of prebiotics. J. Nutr. 1251401-1412. [DOI] [PubMed] [Google Scholar]
- 93.Goh, Y. J., J. H. Lee, and R. W. Hutkins. 2007. Functional analysis of the fructooligosaccharide utilization operon in Lactobacillus paracasei 1195. Appl. Environ. Microbiol. 735716-5724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Goh, Y. J., C. M. Zhang, A. K. Benson, V. Schlegel, J. H. Lee, and R. W. Hutkins. 2006. Identification of a putative operon involved in fructooligosaccharide utilization by Lactobacillus paracasei. Appl. Environ. Microbiol. 727518-7530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Granato, D., G. E. Bergonzelli, R. D. Pridmore, L. Marvin, M. Rouvet, and I. E. Corthesy-Theulaz. 2004. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect. Immun. 722160-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Granato, D., F. Perotti, I. Masserey, M. Rouvet, M. Golliard, A. Servin, and D. Brassart. 1999. Cell surface-associated lipoteichoic acid acts as an adhesion factor for attachment of Lactobacillus johnsonii La1 to human enterocyte-like Caco-2 cells. Appl. Environ. Microbiol. 651071-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Grangette, C., S. Nutten, E. Palumbo, S. Morath, C. Hermann, J. Dewulf, B. Pot, T. Hartung, P. Hols, and A. Mercenier. 2005. Enhanced antiinflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc. Natl. Acad. Sci. USA 10210321-10326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gringhuis, S. I., J. den Dunnen, M. Litjens, B. V. Hof, Y. van Kooyk, and T. B. H. Geijtenbeek. 2007. C-type lectin DC-SIGN modulates Toll-like receptor signaling via Raf-1 kinase-dependent acetylation of transcription factor NF-kappa B. Immunity 26605-616. [DOI] [PubMed] [Google Scholar]
- 99.Guarner, F., R. Bourdet-Sicard, P. Brandtzaeg, H. S. Gill, P. McGuirk, W. van Eden, J. Versalovic, J. V. Weinstock, and G. A. W. Rook. 2006. Mechanisms of disease: the hygiene hypothesis revisited. Nat. Clin. Pract. Gastroenterol. Hepatol. 3275-284. [DOI] [PubMed] [Google Scholar]
- 100.Hamer, H. M., D. Jonkers, K. Venema, S. Vanhoutvin, F. J. Troost, and R. J. Brummer. 2008. Review article: the role of butyrate on colonic function. Aliment. Pharmacol. Ther. 27104-119. [DOI] [PubMed] [Google Scholar]
- 101.Hart, A. L., K. Lammers, P. Brigidi, B. Vitali, F. Rizzello, P. Gionchetti, M. Campieri, M. A. Kamm, S. C. Knight, and A. J. Stagg. 2004. Modulation of human dendritic cell phenotype and function by probiotic bacteria. Gut 531602-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hashimoto, M., K. Tawaratsumida, H. Kariya, A. Kiyohara, Y. Suda, F. Krikae, T. Kirikae, and F. Gotz. 2006. Not lipoteichoic acid but lipoproteins appear to be the dominant immunobiologically active compounds in Staphylococcus aureus. J. Immunol. 1773162-3169. [DOI] [PubMed] [Google Scholar]
- 103.Hedin, C., K. Whelani, and J. O. Lindsay. 2007. Evidence for the use of probiotics and prebiotics in inflammatory bowel disease: a review of clinical trials. Proc. Nutr. Soc. 66307-315. [DOI] [PubMed] [Google Scholar]
- 104.Heilig, H. G. H. J., E. G. Zoetendal, E. E. Vaughan, P. Marteau, A. D. L. Akkermans, and W. M. de Vos. 2002. Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Environ. Microbiol. 68114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Heinemann, C., J. E. T. V. Vlieg, D. B. Janssen, H. J. Busscher, H. C. van der Mei, and G. Reid. 2000. Purification and characterization of a surface-binding protein from Lactobacillus fermentum RC-14 that inhibits adhesion of Enterococcus faecalis 1131. FEMS Microbiol. Lett. 190177-180. [DOI] [PubMed] [Google Scholar]
- 106.Henke, J. M., and B. L. Bassler. 2004. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 1866902-6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Higgins, D. A., M. E. Pomianek, C. M. Kraml, R. K. Taylor, M. F. Semmelhack, and B. L. Bassler. 2007. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature 450883-886. [DOI] [PubMed] [Google Scholar]
- 108.Higuchi, T., H. Hayashi, and K. Abe. 1997. Exchange of glutamate and γ-aminobutyrate in a Lactobacillus strain. J. Bacteriol. 1793362-3364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hooper, L. V., J. Xu, P. G. Falk, T. Midtvedt, and J. I. Gordon. 1999. A molecular sensor that allows a gut commensal to control its nutrient foundation in a competitive ecosystem. Proc. Natl. Acad. Sci. USA 969833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Howell, A., S. Dubrac, K. K. Andersen, D. Noone, J. Fert, T. Msadek, and K. Devine. 2003. Genes controlled by the essential YycG/YycF two-component system of Bacillus subtilis revealed through a novel hybrid regulator approach. Mol. Microbiol. 491639-1655. [DOI] [PubMed] [Google Scholar]
- 111.Hughes, D. T., and V. Sperandio. 2008. Inter-kingdom signalling: communication between bacteria and their hosts. Nat. Rev. Microbiol. 6111-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hurmalainen, V., S. Edelman, J. Antikainen, M. Baumann, K. Lahteenmaki, and T. K. Korhonen. 2007. Extracellular proteins of Lactobacillus crispatus enhance activation of human plasminogen. Microbiology 1531112-1122. [DOI] [PubMed] [Google Scholar]
- 113.Hynonen, U., B. Westerlund-Wikstrom, A. Palva, and T. K. Korhonen. 2002. Identification by flagellum display of an epithelial cell- and fibronectin-binding function in the SlpA surface protein of Lactobacillus brevis. J. Bacteriol. 1843360-3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Iliev, I. D., H. Kitazawa, T. Shimosato, S. Katoh, H. Morita, F. He, M. Hosoda, and T. Saito. 2005. Strong immunostimulation in murine immune cells by Lactobacillus rhamnosus GG DNA containing novel oligodeoxynucleotide pattern. Cell. Microbiol. 7403-414. [DOI] [PubMed] [Google Scholar]
- 115.Jelcic, I., E. Hufner, H. Schmidt, and C. Hertel. 2008. Repression of the locus of the enterocyte effacement-encoded regulator of gene transcription of Escherichia coli O157:H7 by Lactobacillus reuteri culture supernatants is LuxS and strain dependent. Appl. Environ. Microbiol. 743310-3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jijon, H., J. Backer, H. Diaz, H. Yeung, D. Thiel, C. McKaigney, C. De Simone, and K. Madsen. 2004. DNA from probiotic bacteria modulates murine and human epithelial and immune function. Gastroenterology 1261358-1373. [DOI] [PubMed] [Google Scholar]
- 117.Johnson-Henry, K. C., K. A. Donato, G. Shen-Tu, A. Gordanpour, and P. A. Sherman. 2008. Lactobacillus rhamnosus strain GG prevents enterohemorrhagic Escherichia coli O157:H7-induced changes in epithelial barrier function. Infect. Immun. 761340-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Johnson-Henry, K. C., K. E. Hagen, M. Gordonpour, T. A. Tompkins, and P. M. Sherman. 2007. Surface-layer protein extracts from Lactobacillus helveticus inhibit enterohaemorrhagic Escherichia coli O157:H7 adhesion to epithelial cells. Cell. Microbiol. 9356-367. [DOI] [PubMed] [Google Scholar]
- 119.Kandler, O., and N. Weiss. 1986. Regular, nonsporing gram-positive rods, p. 1208-1234. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Williams & Wilkins, Baltimore, MD.
- 120.Kaper, J. B., and V. Sperandio. 2005. Bacterial cell-to-cell signaling in the gastrointestinal tract. Infect. Immun. 733197-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kaplan, H., and R. W. Hutkins. 2000. Fermentation of fructooligosaccharides by lactic acid bacteria and bifidobacteria. Appl. Environ. Microbiol. 662682-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Karlsson, H., P. Larsson, A. E. Wold, and A. Rudin. 2004. Pattern of cytokine responses to gram-positive and gram-negative commensal bacteria is profoundly changed when monocytes differentiate into dendritic cells. Infect. Immun. 722671-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kasper, D. L. 1986. Bacterial capsule—old dogmas and new tricks. J. Infect. Dis. 153407-415. [DOI] [PubMed] [Google Scholar]
- 124.Kim, S. O., H. I. Sheikh, S. D. Ha, A. Martins, and G. Reid. 2006. G-CSF-mediated inhibition of JNK is a key mechanism for Lactobacillus rhamnosus-induced suppression of TNF production in macrophages. Cell. Microbiol. 81958-1971. [DOI] [PubMed] [Google Scholar]
- 125.Kimura, K., A. L. McCartney, M. A. McConnell, and G. W. Tannock. 1997. Analysis of fecal populations of bifidobacteria and lactobacilli and investigation of the immunological responses of their human hosts to the predominant strains. Appl. Environ. Microbiol. 633394-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kinoshita, H., H. Uchida, T. Kawasaki, N. Wakahara, H. Matuo, Y. Kawai, H. Kitazawa, S. Ohmura, K. Miura, K. Shiiba, A. Horii, and T. Saito. 2007. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expressed on the cell surface of Lactobacillus plantarum LA 318 mediates adhesion to human colonic mucin. J. Dairy Sci. 90427.17183111 [Google Scholar]
- 127.Klaenhammer, T. R., R. Barrangou, B. L. Buck, M. A. Azcarate-Peril, and E. Altermann. 2005. Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiol. Rev. 29393-409. [DOI] [PubMed] [Google Scholar]
- 128.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. E. J. Fiers, W. Stiekema, R. M. K. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 1001990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kotani, S., Y. Watanabe, T. Shimono, F. Kinoshita, T. Narita, K. Kato, D. E. S. Stewarttull, I. Morisaki, K. Yokogawa, and S. Kawata. 1975. Immunoadjuvant activities of peptidoglycan subunits from cell walls of Staphylococcus aureus and Lactobacillus plantarum. Biken J. 1893-103. [PubMed] [Google Scholar]
- 130.Kullen, M. J., and T. R. Klaenhammer. 1999. Identification of the pH-inducible, proton-translocating F1FO-ATPase (atpBEFHAGDC) operon of Lactobacillus acidophilus by differential display: gene structure, cloning and characterization. Mol. Microbiol. 331152-1161. [DOI] [PubMed] [Google Scholar]
- 131.Lambert, J., R. Bongers, W. de Vos, and M. Kleerebezem. 2008. Functional analysis of four bile salt hydrolase and penicillin acylase family members in Lactobacillus plantarum WCFS1. Appl. Environ. Microbiol. 744719-4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lambert, J. M., R. S. Bongers, and M. Kjeerebezem. 2007. Cre-lox-based system for multiple gene deletions and selectable-marker removal in Lactobacillus plantarum. Appl. Environ. Microbiol. 731126-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Langlands, S. J., M. J. Hopkins, N. Coleman, and J. H. Cummings. 2004. Prebiotic carbohydrates modify the mucosa associated microflora of the human large bowel. Gut 531610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lazazzera, B. A. 2001. The intracellular function of extracellular signaling peptides. Peptides 221519-1527. [DOI] [PubMed] [Google Scholar]
- 135.Lebeer, S., I. J. J. Claes, T. L. A. Verhoeven, C. Shen, I. Lambrichts, J. L. Ceuppens, J. Vanderleyden, and S. C. J. De Keersmaecker. 2008. Impact of luxS and suppressor mutations on the gastrointestinal transit of Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 744711-4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lebeer, S., S. C. J. De Keersmaecker, T. L. A. Verhoeven, A. A. Fadda, K. Marchal, and J. Vanderleyden. 2007. Functional analysis of luxS in the probiotic strain Lactobacillus rhamnosus GG reveals a central metabolic role important for growth and biofilm formation. J. Bacteriol. 189860-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lebeer, S., T. L. A. Verhoeven, M. Perea Vélez, J. Vanderleyden, and S. C. J. De Keersmaecker. 2007. Impact of environmental and genetic factors on biofilm formation by the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 736768-6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Le Blay, G., C. Michel, H. M. Blottiere, and C. Cherbut. 1999. Prolonged intake of fructo-oligosaccharides induces a short-term elevation of lactic acid-producing bacteria and a persistent increase in cecal butyrate in rats. J. Nutr. 1292231-2235. [DOI] [PubMed] [Google Scholar]
- 139.Le Bouguenec, C. 2005. Adhesins and invasins of pathogenic Escherichia coli. Int. J. Med. Microbiol. 295471-478. [DOI] [PubMed] [Google Scholar]
- 140.Lee, K., H. G. Lee, and Y. J. Choi. 2008. Proteomic analysis of the effect of bile salts on the intestinal and probiotic bacterium Lactobacillus reuteri. J. Biotechnol. 13714-19. [DOI] [PubMed] [Google Scholar]
- 141.Lee, K., H. G. Lee, K. Pi, and Y. J. Choi. 2008. Effect of low pH on protein expression by the probiotic bacterium Lactobacillus reuteri. Proteomics 81624-1630. [DOI] [PubMed] [Google Scholar]
- 142.Lee, K., K. Paek, H. Y. Lee, J. H. Park, and Y. Lee. 2007. Antiobesity effect of trans-10, cis-12-conjugated linoleic acid-producing Lactobacillus plantarum PL62 on diet-induced obese mice. J. Appl. Microbiol. 1031140-1146. [DOI] [PubMed] [Google Scholar]
- 143.Ley, R. E., P. J. Turnbaugh, S. Klein, and J. I. Gordon. 2006. Microbial ecology—human gut microbes associated with obesity. Nature 4441022-1023. [DOI] [PubMed] [Google Scholar]
- 144.Lim, E. M., S. D. Ehrlich, and E. Maguin. 2000. Identification of stress-inducible proteins in Lactobacillus delbrueckii subsp bulgaricus. Electrophoresis 212557-2561. [DOI] [PubMed] [Google Scholar]
- 145.Lin, Y. P., C. H. Thibodeaux, J. A. Pena, G. D. Ferry, and J. Versalovic. 2008. Probiotic Lactobacillus reuteri suppress proinflammatory cytokines via c-Jun. Inflamm. Bowel Dis. 141068-1083. [DOI] [PubMed] [Google Scholar]
- 146.Liong, M. T. 2008. Safety of probiotics: translocation and infection. Nutr. Rev. 66192-202. [DOI] [PubMed] [Google Scholar]
- 147.Loenen, W. A. M. 2006. S-Adenosylmethionine: jack of all trades and master of everything? Biochem. Soc. Trans. 34330-333. [DOI] [PubMed] [Google Scholar]
- 148.Lorca, G., M. I. Torino, G. F. de Valdez, and A. Ljungh. 2002. Lactobacilli express cell surface proteins which mediate binding of immobilized collagen and fibronectin. FEMS Microbiol. Lett. 20631-37. [DOI] [PubMed] [Google Scholar]
- 149.Lorca, G. L., G. F. de Valdez, and A. Ljungh. 2002. Characterization of the protein synthesis dependent adaptive acid tolerance response in Lactobacillus acidophilus. J. Mol. Microbiol. Biotechnol. 4525-532. [PubMed] [Google Scholar]
- 150.Ma, D. L., P. Forsythe, and J. Bienenstock. 2004. Live Lactobacillus reuteri is essential for the inhibitory effect on tumor necrosis factor alpha-induced interleukin-8 expression. Infect. Immun. 725308-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Macfarlane, S., E. Furrie, J. H. Cummings, and G. T. Macfarlane. 2004. Chemotaxonomic analysis of bacterial populations colonizing the rectal mucosa in patients with ulcerative colitis. Clin. Infect. Dis. 381690-1699. [DOI] [PubMed] [Google Scholar]
- 152.Mack, D. R., S. Ahrne, L. Hyde, S. Wei, and M. A. Hollingsworth. 2003. Extracellular MUC3 mucin secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut 52827-833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mackie, R. I., A. Sghir, and H. R. Gaskins. 1999.Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 691035S-1045S. [DOI] [PubMed] [Google Scholar]
- 154.MacPherson, A. J., D. Gatto, E. Sainsbury, G. R. Harriman, H. Hengartner, and R. M. Zinkernagel. 2000. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 2882222-2226. [DOI] [PubMed] [Google Scholar]
- 155.MacPherson, A. J., and T. Uhr. 2004. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science 3031662-1665. [DOI] [PubMed] [Google Scholar]
- 155a.Maldonado Galdeano, C. M., and G. Perdigón. 2006. The probiotic bacterium Lactobacillus casei induces activation of the gut mucosal immune system through innate immunity. Clin. Vaccine Immunol. 13219-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mandlik, A., A. Swierczynski, A. Das, and H. Ton-That. 2008. Pili in gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 1633-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Marco, M. L., R. S. Bongers, W. M. de Vos, and M. Kleerebezem. 2007. Spatial and temporal expression of Lactobacillus plantarum genes in the gastrointestinal tracts of mice. Appl. Environ. Microbiol. 73124-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Marco, M. L., S. Pavan, and M. Kleerebezem. 2006. Towards understanding molecular modes of probiotic action. Curr. Opin. Biotechnol. 17204-210. [DOI] [PubMed] [Google Scholar]
- 159.Martin, F. P. J., Y. Wang, N. Sprenger, I. K. S. Yap, T. Lundstedt, P. Lek, S. Rezzi, Z. Ramadan, P. van Bladeren, L. B. Fay, S. Kochhar, J. C. Lindon, E. Holmes, and J. K. Nicholson. 2008. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol. Syst. Biol. 4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Martinez, A., and R. Kolter. 1997. Protection of DNA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 1795188-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Masip, L., K. Veeravalli, and G. Georgioui. 2006. The many faces of glutathione in bacteria. Antioxid. Redox Signal. 8753-762. [DOI] [PubMed] [Google Scholar]
- 162.Masuda, K., and T. Kawata. 1985. Reassembly of a regularly arranged protein in the cell wall of Lactobacillus buchneri and its reattachment to cell walls—chemical modification studies. Microbiol. Immunol. 29927-938. [PubMed] [Google Scholar]
- 163.Matsuguchi, T., A. Takagi, T. Matsuzaki, M. Nagaoka, K. Ishikawa, T. Yokokura, and Y. Yoshikai. 2003. Lipoteichoic acids from Lactobacillus strains elicit strong tumor necrosis factor alpha-inducing activitives in macrophages through Toll-like receptor 2. Clin. Diagn. Lab. Immunol. 10259-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Matsumoto, S., T. Hara, T. Hori, K. Mitsuyama, M. Nagaoka, N. Tomiyasu, A. Suzuki, and M. Sata. 2005. Probiotic Lactobacillus-induced improvement in murine chronic inflammatory bowel disease is associated with the down-regulation of pro-inflammatory cytokines in lamina propria mononuclear cells. Clin. Exp. Immunol. 140417-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.McAuliffe, O., R. J. Cano, and T. R. Klaenhammer. 2005. Genetic analysis of two bile salt hydrolase activities in Lactobacillus acidophilus NCFM. Appl. Environ. Microbiol. 714925-4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Medellin-Pena, M. J., H. F. Wang, R. Johnson, S. Anand, and M. W. Griffiths. 2007. Probiotics affect virulence-related gene expression in Escherichia coli O157:H7. Appl. Environ. Microbiol. 734259-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Medzhitov, R. 2007. Recognition of microorganisms and activation of the immune response. Nature 449819-826. [DOI] [PubMed] [Google Scholar]
- 168.Meurman, J. H., and I. Stamatova. 2007. Probiotics: contributions to oral health. Oral Dis. 13443-451. [DOI] [PubMed] [Google Scholar]
- 169.Miettinen, M., A. Lehtonen, I. Julkunen, and S. Matikainen. 2000. Lactobacilli and streptococci activate NF-kappa B and STAT signaling pathways in human macrophages. J. Immunol. 1643733-3740. [DOI] [PubMed] [Google Scholar]
- 170.Minic, Z., C. Marie, C. Delorme, J. M. Faurie, G. Mercier, D. Ehrlich, and P. Renault. 2007. Control of EpsE, the phosphoglycosyltransferase initiating exopolysaccharide synthesis in Streptococcus thermophilus, by EpsD tyrosine kinase. J. Bacteriol. 1891351-1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mohamadzadeh, M., S. Olson, W. V. Kalina, G. Ruthel, G. L. Demmin, K. L. Warfield, S. Bavari, and T. R. Klaenhammer. 2005. Lactobacilli activate human dendritic cells that skew T cells toward T helper 1 polarization. Proc. Natl. Acad. Sci. USA 1022880-2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Molenaar, D., F. Bringel, F. H. Schuren, W. M. de Vos, R. J. Siezen, and M. Kleerebezem. 2005. Exploring Lactobacillus plantarum genome diversity by using microarrays. J. Bacteriol. 1876119-6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Morel-Deville, F., F. Fauvel, and P. Morel. 1998. Two-component signal-transducing systems involved in stress responses and vancomycin susceptibility in Lactobacillus sakei. Microbiology 1442873-2883. [DOI] [PubMed] [Google Scholar]
- 174.Morona, J. K., D. C. Miller, R. Morona, and J. C. Paton. 2004. The effect that mutations in the conserved capsular polysaccharide biosynthesis genes cpsA, cpsB, and cpsD have on virulence of Streptococcus pneumoniae. J. Infect. Dis. 1891905-1913. [DOI] [PubMed] [Google Scholar]
- 175.Morona, J. K., R. Morona, D. C. Miller, and J. C. Paton. 2002. Streptococcus pneumoniae capsule biosynthesis protein CpsB is a novel manganese-dependent phosphotyrosine-protein phosphatase. J. Bacteriol. 184577-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Morona, J. K., J. C. Paton, D. C. Miller, and R. Morona. 2000. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol. Microbiol. 351431-1442. [DOI] [PubMed] [Google Scholar]
- 177.Navarre, W. W., and O. Schneewind. 1994. Proteolytic cleavage and cell-wall anchoring at the LPXTG motif of surface proteins in gram-positive bacteria. Mol. Microbiol. 14115-121. [DOI] [PubMed] [Google Scholar]
- 178.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Neuhaus, F. C., and J. Baddiley. 2003. A continuum of anionic charge: structures and functions of d-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67686-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.O'Mahony, L., L. O'Callaghan, J. McCarthy, D. Shilling, P. Scully, S. Sibartie, E. Kavanagh, W. O. Kirwan, H. P. Redmond, J. K. Collins, and F. Shanahan. 2006. Differential cytokine response from dendritic cells to commensal and pathogenic bacteria in different lymphoid compartments in humans. Am. J. Physiol. Gastrointest. Liver Physiol. 290G839-G845. [DOI] [PubMed] [Google Scholar]
- 181.Oozeer, R., J. P. Furet, N. Goupil-Feuillerat, J. Anba, J. Mengaud, and G. Corthier. 2005. Differential activities of four Lactobacillus casei promoters during bacterial transit through the gastrointestinal tracts of human-microbiota-associated mice. Appl. Environ. Microbiol. 711356-1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Otte, J. M., E. Cario, and D. K. Podolsky. 2004. Mechanisms of cross hyporesponsiveness to Toll-like receptor bacterial ligands in intestinal epithelial cells. Gastroenterology 1261054-1070. [DOI] [PubMed] [Google Scholar]
- 183.Otte, J. M., and D. K. Podolsky. 2004. Functional modulation of enterocytes by gram-positive and gram-negative microorganisms. Am. J. Physiol. Gastrointest. Liver Physiol. 286G613-G626. [DOI] [PubMed] [Google Scholar]
- 184.Palumbo, E., M. Deghorain, P. S. Cocconcelli, M. Kleerebezem, A. Geyer, T. Hartung, S. Morath, and P. Hols. 2006. d-Alanyl ester depletion of teichoic acids in Lactobacillus plantarum results in a major modification of lipoteichoic acid composition and cell wall perforations at the septum mediated by the Acm2 autolysin. J. Bacteriol. 1883709-3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Peant, B., G. LaPointe, C. Gilbert, D. Atlan, P. Ward, and D. Roy. 2005. Comparative analysis of the exopolysaccharide biosynthesis gene clusters from four strains of Lactobacillus rhamnosus. Microbiology 1511839-1851. [DOI] [PubMed] [Google Scholar]
- 186.Penaud, S., A. Fernandez, S. Boudebbouze, S. D. Ehrlich, E. Maguin, and M. van de Guchte. 2006. Induction of heavy-metal-transporting CPX-type ATPases during acid adaptation in Lactobacillus bulgaricus. Appl. Environ. Microbiol. 727445-7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Perea Vélez, M., S. C. J. De Keersmaecker, and J. Vanderleyden. 2007. Adherence factors of Lactobacillus in the human gastrointestinal tract. FEMS Microbiol. Lett. 276140-148. [DOI] [PubMed] [Google Scholar]
- 188.Perea Vélez, M., T. L. A. Verhoeven, C. Draing, S. Von Aulock, M. Pfitzenmaier, A. Geyer, I. Lambrichts, C. Grangette, B. Pot, J. Vanderleyden, and S. C. J. De Keersmaecker. 2007. Functional analysis of D-alanylation of lipoteichoic acid in the probiotic strain Lactobacillus rhamnosus GG. Appl. Environ. Microbiol. 733595-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Peterson, D. A., N. P. McNulty, J. L. Guruge, and J. I. Gordon. 2007. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe 2328-339. [DOI] [PubMed] [Google Scholar]
- 190.Petrof, E. O., K. Kojima, M. J. Ropeleski, M. W. Musch, Y. Tao, C. De Simone, and E. B. Chang. 2004. Probiotics inhibit nuclear factor-kappa B and induce heat shock proteins in colonic epithelial cells through proteasome inhibition. Gastroenterology 1271474-1487. [DOI] [PubMed] [Google Scholar]
- 191.Pfeiler, E. A., M. A. Azcarate-Peril, and T. R. Klaenhammer. 2007. Characterization of a novel bile-inducible operon encoding a two-component regulatory system in Lactobacillus acidophilus. J. Bacteriol. 1894624-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Pillai, A., and R. Nelson. 2008. Probiotics for treatment of Clostridium difficile-associated colitis in adults. Cochrane Database Syst. Rev. 2008CD004611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Prantera, C., M. L. Scribano, G. Falasco, A. Andreoli, and C. Luzi. 2002. Ineffectiveness of probiotics in preventing recurrence after curative resection for Crohn's disease: a randomised controlled trial with Lactobacillus GG. Gut 51405-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Presser, K. A., D. A. Ratkowsky, and T. Ross. 1997. Modelling the growth rate of Escherichia coli as a function of pH and lactic acid concentration. Appl. Environ. Microbiol. 632355-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Pretzer, G., J. Snel, D. Molenaar, A. Wiersma, P. A. Bron, J. Lambert, W. M. de Vos, R. van der Meer, M. A. Smits, and M. Kleerebezem. 2005. Biodiversity-based identification and functional characterization of the mannose-specific adhesin of Lactobacillus plantarum. J. Bacteriol. 1876128-6136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A. C. Pittet, M. C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 1012512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Pridmore, R. D., A. C. Pittet, F. Praplan, and C. Cavadini. 2008. Hydrogen peroxide production by Lactobacillus johnsonii NCC 533 and its role in anti-Salmonella activity. FEMS Microbiol. Lett. 283210-215. [DOI] [PubMed] [Google Scholar]
- 198.Rachmilewitz, D., K. Katakura, F. Karmeli, T. Hayashi, C. Reinus, B. Rudensky, S. Akira, K. Takeda, J. Lee, K. Takabayashi, and E. Raz. 2004. Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology 126520-528. [DOI] [PubMed] [Google Scholar]
- 199.Rafter, J., M. Bennett, G. Caderni, Y. Clune, R. Hughes, P. C. Karlsson, A. Klinder, M. O'Riordan, G. C. O'Sullivan, B. Pool-Zobel, G. Rechkemmer, M. Roller, I. Rowland, M. Salvadori, H. Thijs, J. Van Loo, B. Watzl, and J. K. Collins. 2007. Dietary synbiotics reduce cancer risk factors in polypectomized and colon cancer patients. Am. J. Clin. Nutr. 85488-496. [DOI] [PubMed] [Google Scholar]
- 200.Rakoff-Nahoum, S., J. Paglino, F. Eslami-Varzaneh, S. Edberg, and R. Medzhitov. 2004. Recognition of commensal microflora by Toll-like receptors is required for intestinal homeostasis. Cell 118229-241. [DOI] [PubMed] [Google Scholar]
- 201.Rallu, F., A. Gruss, S. D. Ehrlich, and E. Maguin. 2000. Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol. Microbiol. 35517-528. [DOI] [PubMed] [Google Scholar]
- 202.Redondo-Lopez, V., R. L. Cook, and J. D. Sobel. 1990. Emerging role of lactobacilli in the control and maintenance of the vaginal bacterial microflora. Rev. Infect. Dis. 12856-872. [DOI] [PubMed] [Google Scholar]
- 203.Rescigno, M., M. Urbano, B. Valzasina, M. Francolini, G. Rotta, R. Bonasio, F. Granucci, J. P. Kraehenbuhl, and P. Ricciardi-Castagnoli. 2001. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat. Immunol. 2361-367. [DOI] [PubMed] [Google Scholar]
- 204.Resta-Lenert, S., and K. E. Barrett. 2003. Live probiotics protect intestinal epithelial cells from the effects of infection with enteroinvasive Escherichia coli (EIEC). Gut 52988-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Resta-Lenert, S., and K. E. Barrett. 2006. Probiotics and commensals reverse TNF-alpha- and IFN-gamma-induced dysfunction in human intestinal epithelial cells. Gastroenterology 130731-746. [DOI] [PubMed] [Google Scholar]
- 206.Roberts, I. S. 1996. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 50285-315. [DOI] [PubMed] [Google Scholar]
- 207.Roessler, A., U. Friedrich, H. Vogelsang, A. Bauer, M. Kaatz, U. C. Hipler, I. Schmidt, and G. Jahreis. 2008. The immune system in healthy adults and patients with atopic dermatitis seems to be affected differently by a probiotic intervention. Clin. Exp. Allergy 3893-102. [DOI] [PubMed] [Google Scholar]
- 208.Rook, G. A. W., and L. R. Brunet. 2005. Microbes, immunoregulation, and the gut. Gut 54317-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Roos, S., and H. Jonsson. 2002. A high-molecular-mass cell-surface protein from Lactobacillus reuteri 1063 adheres to mucus components. Microbiology 148433-442. [DOI] [PubMed] [Google Scholar]
- 210.Rousseaux, C., X. Thuru, A. Gelot, N. Barnich, C. Neut, L. Dubuquoy, C. Dubuquoy, E. Merour, K. Geboes, M. Chamaillard, A. Ouwehand, G. Leyer, D. Carcano, J. F. Colombel, D. Ardid, and P. Desreumaux. 2007. Lactobacillus acidophilus modulates intestinal pain and induces opioid and cannabinoid receptors. Nat. Med. 1335-37. [DOI] [PubMed] [Google Scholar]
- 211.Ruas-Madiedo, P., M. Gueimonde, A. Margolles, C. G. D. L. Reyes-Gavilan, and S. Salminen. 2006. Exopolysaccharides produced by probiotic strains modify the adhesion of problotics and enteropathogens to human intestinal mucus. J. Food Prot. 692011-2015. [DOI] [PubMed] [Google Scholar]
- 212.Rychlik, I., and P. A. Barrow. 2005. Salmonella stress management and its relevance to behaviour during intestinal colonisation and infection. FEMS Microbiol. Rev. 291021-1040. [DOI] [PubMed] [Google Scholar]
- 213.Santos, F., J. L. Vera, R. van der Heijden, G. Valdez, W. M. de Vos, F. Sesma, and J. Hugenholtz. 2008. The complete coenzyme B-12 biosynthesis gene cluster of Lactobacillus reuteri CRL1098. Microbiology 15481-93. [DOI] [PubMed] [Google Scholar]
- 214.Santos, F., A. Wegkamp, W. M. de Vos, E. J. Smid, and J. Hugenholtz. 2008. High-level folate production in fermented foods by the B12 producer Lactobacillus reuteri JCM1112. Appl. Environ. Microbiol. 743291-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Saulnier, D. A. A., D. Molenaar, W. A. de Vos, G. R. Gibson, and S. Kolida. 2007. Identification of prebiotic fructooligosaccharide metabolism in Lactobacillus plantarum WCFS1 through microarrays. Appl. Environ. Microbiol. 731753-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Sazawal, S., G. Hiremath, U. Dhingra, P. Malik, S. Deb, and R. E. Black. 2006. Efficacy of probiotics in prevention of acute diarrhoea: a meta-analysis of masked, randomised, placebo-controlled trials. Lancet Infect. Dis. 6374-382. [DOI] [PubMed] [Google Scholar]
- 217.Schachtsiek, M., W. P. Hammes, and C. Hertel. 2004. Characterization of Lactobacillus coryniformis DSM 20001T surface protein Cpf mediating coaggregation with and aggregation among pathogens. Appl. Environ. Microbiol. 707078-7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Schaffer, C., and P. Messner. 2005. The structure of secondary cell wall polymers: how gram-positive bacteria stick their cell walls together. Microbiology 151643-651. [DOI] [PubMed] [Google Scholar]
- 219.Scheffers, D. J., and M. G. Pinho. 2005. Bacterial cell wall synthesis: new insights from localization studies. Microbiol. Mol. Biol. Rev. 69585-607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Scheppach, W. 1994. Effects of short chain fatty acids on gut morphology and function. Gut 35S35-S38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Schlee, M., J. Harder, B. Koten, E. F. Stange, J. Wehkamp, and K. Fellermann. 2008. Probiotic lactobacilli and VSL#3 induce enterocyte beta-defensin 2. Clin. Exp. Immunol. 151528-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Schultz, M., C. Gottl, T. J. Young, T. Iwen, and T. A. Vanderhoof. 2004. Administration of oral probiotic bacteria to pregnant women causes temporary infantile colonization. J. Pediatr. Gastroenterol. Nutr. 38293-297. [DOI] [PubMed] [Google Scholar]
- 223.Schwab, C., J. Walter, G. W. Tannock, R. F. Vogel, and M. G. Ganzle. 2007. Sucrose utilization and impact of sucrose on glycosyltransferase expression in Lactobacillus reuteri. Syst. Appl. Microbiol. 30433-443. [DOI] [PubMed] [Google Scholar]
- 224.Servin, A. L. 2004. Antagonistic activities of lactobacilli and bifidobacteria against microbial pathogens. FEMS Microbiol. Rev. 28405-440. [DOI] [PubMed] [Google Scholar]
- 225.Seth, A., F. Yan, D. B. Polk, and R. K. Rao. 2008. Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. Am. J. Physiol. Gastrointest. Liver Physiol. 294G1060-G1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Sghir, A., G. Gramet, A. Suau, V. Rochet, P. Pochart, and J. Dore. 2000. Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 662263-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Siciliano, R. A., G. Cacace, M. F. Mazzeo, L. Morelli, M. Elli, M. Rossi, and A. Malorni. 2008. Proteomic investigation of the aggregation phenomenon in Lactobacillus crispatus. Biochim. Biophys. Acta 1784335-342. [DOI] [PubMed] [Google Scholar]
- 228.Sillanpaa, J., B. Martinez, J. Antikainen, T. Toba, N. Kalkkinen, S. Tankka, K. Lounatmaa, J. Keranen, M. Hook, B. Westerlund-Wikstrom, P. H. Pouwels, and T. K. Korhonen. 2000. Characterization of the collagen-binding S-layer protein CbsA of Lactobacillus crispatus. J. Bacteriol. 1826440-6450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sleator, R. D., T. Clifford, and C. Hill. 2007. Gut osmolarity: a key environmental cue initiating the gastrointestinal phase of Listeria monocytogenes infection? Med. Hypotheses 691090-1092. [DOI] [PubMed] [Google Scholar]
- 230.Smits, H. H., A. Engering, D. van der Kleij, E. C. de Jong, K. Schipper, T. M. M. van Capel, B. A. J. Zaat, M. Yazdanbakhsh, E. A. Wierenga, Y. van Kooyk, and M. L. Kapsenberg. 2005. Selective probiotic bacteria induce IL-10-producing regulatory T cells in vitro by modulating dendritic cell function through dendritic cell-specific intercellular adhesion molecule 3-grabbing nonintegrin. J. Allergy Clin. Immunol. 1151260-1267. [DOI] [PubMed] [Google Scholar]
- 231.Sonnenburg, E. D., J. L. Sonnenburg, J. K. Manchester, E. E. Hansen, H. C. Chiang, and J. I. Gordon. 2006. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc. Natl. Acad. Sci. USA 1038834-8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sonnenburg, J. L., L. T. Angenent, and J. I. Gordon. 2004. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 5569-573. [DOI] [PubMed] [Google Scholar]
- 233.Sonnenburg, J. L., C. T. L. Chen, and J. I. Gordon. 2006. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 42213-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Sperandio, V., A. G. Torres, B. Jarvis, J. P. Nataro, and J. B. Kaper. 2003. Bacteria-host communication: the language of hormones. Proc. Natl. Acad. Sci. USA 1008951-8956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Srivatsan, A., and J. D. Wang. 2008. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 11100-105. [DOI] [PubMed] [Google Scholar]
- 236.Stephenson, A. E., H. Wu, J. Novak, M. Tomana, K. Mintz, and P. Fives-Taylor. 2002. The Fap1 fimbrial adhesin is a glycoprotein: antibodies specific for the glycan moiety block the adhesion of Streptococcus parasanguis in an in vitro tooth model. Mol. Microbiol. 43147-157. [DOI] [PubMed] [Google Scholar]
- 237.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69183-215. [DOI] [PubMed] [Google Scholar]
- 238.Sturme, M. H. J., C. Francke, R. J. Siezen, W. M. de Vos, and M. Kleerebezem. 2007. Making sense of quorum sensing in lactobacilli: a special focus on Lactobacillus plantarum WCFS1. Microbiology 1533939-3947. [DOI] [PubMed] [Google Scholar]
- 239.Sturme, M. H. J., J. Nakayama, D. Molenaar, Y. Murakami, R. Kunugi, T. Fujii, E. E. Vaughan, M. Kleerebezem, and W. M. de Vos. 2005. An agr-like two-component regulatory system in Lactobacillus plantarum is involved in production of a novel cyclic peptide and regulation of adherence. J. Bacteriol. 1875224-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Su, P., A. Henriksson, and H. Mitchell. 2007. Prebiotics enhance survival and prolong the retention period of specific probiotic inocula in an in vivo murine model. J. Appl. Microbiol. 1032392-2400. [DOI] [PubMed] [Google Scholar]
- 241.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 961639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Sutherland, I. W. 1972. Bacterial exopolysaccharides. Adv. Microb. Physiol. 8143-213. [DOI] [PubMed] [Google Scholar]
- 243.Swidsinski, A., J. Weber, V. Loening-Baucke, L. P. Hale, and H. Lochs. 2005. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 433380-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Tabuchi, M., M. Ozaki, A. Tamura, N. Yamada, T. Ishida, M. Hosoda, and A. Hosono. 2003. Antidiabetic effect of Lactobacillus GG in streptozotocin-induced diabetic rats. Biosci. Biotechnol. Biochem. 671421-1424. [DOI] [PubMed] [Google Scholar]
- 245.Tallon, R., P. Bressollier, and M. C. Urdaci. 2003. Isolation and characterization of two exopolysaccharides produced by Lactobacillus plantarum EP56. Res. Microbiol. 154705-712. [DOI] [PubMed] [Google Scholar]
- 246.Tannock, G. W., S. Ghazally, J. Walter, D. Loach, H. Brooks, G. Cook, M. Surette, C. Simmers, P. Bremer, F. Dal Bello, and C. Hertel. 2005. Ecological behavior of Lactobacillus reuteri 100-23 is affected by mutation of the luxS gene. Appl. Environ. Microbiol. 718419-8425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Tannock, G. W., K. Munro, H. J. M. Harmsen, G. W. Welling, J. Smart, and P. K. Gopal. 2000. Analysis of the fecal microflora of human subjects consuming a probiotic product containing Lactobacillus rhamnosus DR20. Appl. Environ. Microbiol. 662578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Tao, Y., K. A. Drabik, T. S. Waypa, M. W. Musch, J. C. Alverdy, O. Schneewind, E. B. Chang, and E. O. Petrof. 2006. Soluble factors from Lactobacillus GG activate MAPKs and induce cytoprotective heat shock proteins in intestinal epithelial cells. Am. J. Physiol. Cell Physiol. 290C1018-C1030. [DOI] [PubMed] [Google Scholar]
- 249.Taranto, M. P., M. L. F. Murga, G. Lorca, and G. F. de Valdez. 2003. Bile salts and cholesterol induce changes in the lipid cell membrane of Lactobacillus reuteri. J. Appl. Microbiol. 9586-91. [DOI] [PubMed] [Google Scholar]
- 250.Tuomola, E., R. Crittenden, M. Playne, E. Isolauri, and S. Salminen. 2001. Quality assurance criteria for probiotic bacteria. Am. J. Clin. Nutr. 73393S-398S. [DOI] [PubMed] [Google Scholar]
- 251.Tuomola, E. M., A. C. Ouwehand, and S. J. Salminen. 2000. Chemical, physical and enzymatic pre-treatments of probiotic lactobacilli alter their adhesion to human intestinal mucus glycoproteins. Int. J. Food Microbiol. 6075-81. [DOI] [PubMed] [Google Scholar]
- 252.Turner, J. R. 2006. Molecular basis of epithelial barrier regulation—from basic mechanisms to clinical application. Am. J. Pathol. 1691901-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Ueno, Y., K. Hayakawa, S. Takahashi, and K. Oda. 1997. Purification and characterization of glutamate decarboxylase from Lactobacillus brevis IFO 12005. Biosci. Biotechnol. Biochem. 611168-1171. [DOI] [PubMed] [Google Scholar]
- 254.Valeur, N., P. Engel, N. Carbajal, E. Connolly, and K. Ladefoged. 2004. Colonization and immunomodulation by Lactobacillus reuteri ATCC 55730 in the human gastrointestinal tract. Appl. Environ. Microbiol. 701176-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.van de Guchte, M., P. Serror, C. Chervaux, T. Smokvina, S. D. Ehrlich, and E. Maguin. 2002. Stress responses in lactic acid bacteria. Antonie van Leeuwenhoek 82187-216. [PubMed] [Google Scholar]
- 256.van Heijenoort, J. 2007. Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol. Mol. Biol. Rev. 71620-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.van Kooyk, Y., and T. B. H. Geijtenbeek. 2003. DC-sign: escape mechanism for pathogens. Nat. Rev. Immunol. 3697-709. [DOI] [PubMed] [Google Scholar]
- 258.van Pijkeren, J. P., C. Canchaya, K. A. Ryan, Y. Li, M. J. Claesson, B. Sheil, L. Steidler, L. O'Mahony, G. F. Fitzgerald, D. van Sinderen, and P. W. O'Toole. 2006. Comparative and functional analysis of sortase-dependent proteins in the predicted secretome of Lactobacillus salivarius UCC118. Appl. Environ. Microbiol. 724143-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Vasquez, A., T. Jakobsson, S. Ahrne, U. Forsum, and G. Molin. 2002. Vaginal Lactobacillus flora of healthy Swedish women. J. Clin. Microbiol. 402746-2749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Vendeville, A., K. Winzer, K. Heurlier, C. M. Tang, and K. R. Hardie. 2005. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3383-396. [DOI] [PubMed] [Google Scholar]
- 261.Ventura, M., I. Jankovic, D. C. Walker, R. D. Pridmore, and R. Zink. 2002. Identification and characterization of novel surface proteins in Lactobacillus johnsonii and Lactobacillus gasseri. Appl. Environ. Microbiol. 686172-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Vidal, K., A. Donnet-Hughes, and D. Granato. 2002. Lipoteichoic acids from Lactobacillus johnsonii strain La1 and Lactobacillus acidophilus strain La10 antagonize the responsiveness of human intestinal epithelial HT29 cells to lipopolysaccharide and gram-negative bacteria. Infect. Immun. 702057-2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Vidgren, G., I. Palva, R. Pakkanen, K. Lounatmaa, and A. Palva. 1992. S-layer protein gene of Lactobacillus brevis—cloning by polymerase chain reaction and determination of the nucleotide sequence. J. Bacteriol. 1747419-7427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Vinderola, G., G. Perdigon, J. Duarte, E. Farnworth, and C. Matar. 2006. Effects of the oral administration of the exopolysaccharide produced by Lactobacillus kefiranofaciens on the gut mucosal immunity. Cytokine 36254-260. [DOI] [PubMed] [Google Scholar]
- 265.Voltan, S., I. Castagliuolo, M. Elli, S. Longo, P. Brun, R. D'Inca, A. Porzionato, V. Macchi, G. Palu, G. C. Sturniolo, L. Morelli, and D. Martines. 2007. Aggregating phenotype in Lactobacillus crispatus determines intestinal colonization and TLR2 and TLR4 modulation in murine colonic mucosa. Clin. Vaccine Immunol. 141138-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Wall, T., M. Bath, R. A. Britton, H. Jonsson, J. Versalovic, and S. Roos. 2007. The early response to acid shock in Lactobacillus reuteri involves the ClpL chaperone and a putative cell wall-altering esterase. Appl. Environ. Microbiol. 733924-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Walter, J. 2005. The microecology of lactobacilli in the gastrointestinal tract, p. 51-82. In G. W. Tannock (ed.), Probiotics & prebiotics: scientific aspects. Caister Academic Press, Norfolk, United Kingdom.
- 268.Walter, J., P. Chagnaud, G. W. Tannock, D. M. Loach, F. Dal Bello, H. F. Jenkinson, W. P. Hammes, and C. Hertel. 2005. A high-molecular-mass surface protein (Lsp) and methionine sulfoxide reductase B (MsrB) contribute to the ecological performance of Lactobacillus reuteri in the murine gut. Appl. Environ. Microbiol. 71979-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Walter, J., N. C. K. Heng, W. P. Hammes, D. M. Loach, G. W. Tannock, and C. Hertel. 2003. Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 692044-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Walter, J., D. M. Loach, M. Alqumber, C. Rockel, C. Hermann, M. Pfitzenmaier, and G. W. Tannock. 2007. d-Alanyl ester depletion of teichoic acids in Lactobacillus reuteri 100-23 results in impaired colonization of the mouse gastrointestinal tract. Environ. Microbiol. 91750-1760. [DOI] [PubMed] [Google Scholar]
- 271.Walter, J., C. Schwab, D. M. Loach, M. G. Ganzle, and G. W. Tannock. 2008. Glucosyltransferase A (GtfA) and inulosucrase (Inu) of Lactobacillus reuteri TMW1.106 contribute to cell aggregation, in vitro biofilm formation, and colonization of the mouse gastrointestinal tract. Microbiology 15472-80. [DOI] [PubMed] [Google Scholar]
- 272.Walters, M., M. P. Sircili, and V. Sperandio. 2006. AI-3 synthesis is not dependent on luxS in Escherichia coli. J. Bacteriol. 1885668-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Watanabe, T., A. Kitani, P. J. Murray, and W. Strober. 2004. NOD2 is a negative regulator of Toll-like receptor 2-mediated T helper type 1 responses. Nat. Immunol. 5800-808. [DOI] [PubMed] [Google Scholar]
- 274.Wegkamp, A., M. Starrenburg, W. M. de Vos, J. Hugenholtz, and W. Sybesma. 2004. Transformation of folate-consuming Lactobacillus gasseri into a folate producer. Appl. Environ. Microbiol. 703146-3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Weiser, J. N., D. Bae, C. Fasching, R. W. Scamurra, A. J. Ratner, and E. N. Janoff. 2003. Antibody-enhanced pneumococcal adherence requires IgA1 protease. Proc. Natl. Acad. Sci. USA 1004215-4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Welman, A. D., and I. S. Maddox. 2003. Exopolysaccharides from lactic acid bacteria: perspectives and challenges. Trends Biotechnol. 21269-274. [DOI] [PubMed] [Google Scholar]
- 277.Whitehead, K., J. Versalovic, S. Roos, and R. A. Britton. 2008. Genomic and genetic characterization of the bile stress response of probiotic Lactobacillus reuteri ATCC 55730. Appl. Environ. Microbiol. 741812-1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Winzer, K., K. R. Hardie, and P. Williams. 2003. LuxS and autoinducer-2: their contribution to quorum sensing and metabolism in bacteria. Adv. Appl. Microbiol. 53291-396. [DOI] [PubMed] [Google Scholar]
- 279.Xu, J., and J. I. Gordon. 2003. Honor thy symbionts. Proc. Natl. Acad. Sci. USA 10010452-10459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Yan, F., H. W. Cao, T. L. Cover, R. Whitehead, M. K. Washington, and D. B. Polk. 2007. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology 132562-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Yan, F., and D. B. Polk. 2002. Probiotic bacterium prevents cytokine-induced apoptosis in intestinal epithelial cells. J. Biol. Chem. 27750959-50965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Yan, F., C. Vanderpool, H. Cao, and D. B. Polk. 2007. Soluble proteins produced by Lactobacillus rhamnosus GG (LGG) activate EGF receptor to regulate the anti-apoptotic response in intestinal epithelial cells. Gastroenterology 132A102-A103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Yasuda, E., M. Serata, and T. Sako. 2008. Lactobacillus casei strain Shirota genes determining the synthesis of cell wall-associated polysaccharides: their suppressive effect on the activation of macrophages. Appl. Environ. Microbiol. 744746-4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Zhou, X., C. J. Brown, Z. Abdo, C. C. Davis, M. A. Hansmann, P. Joyce, J. A. Foster, and L. J. Forney. 2007. Differences in the composition of vaginal microbial communities found in healthy Caucasian and black women. ISME J. 1121-133. [DOI] [PubMed] [Google Scholar]