Abstract
Summary: As random shotgun metagenomic projects proliferate and become the dominant source of publicly available sequence data, procedures for the best practices in their execution and analysis become increasingly important. Based on our experience at the Joint Genome Institute, we describe the chain of decisions accompanying a metagenomic project from the viewpoint of the bioinformatic analysis step by step. We guide the reader through a standard workflow for a metagenomic project beginning with presequencing considerations such as community composition and sequence data type that will greatly influence downstream analyses. We proceed with recommendations for sampling and data generation including sample and metadata collection, community profiling, construction of shotgun libraries, and sequencing strategies. We then discuss the application of generic sequence processing steps (read preprocessing, assembly, and gene prediction and annotation) to metagenomic data sets in contrast to genome projects. Different types of data analyses particular to metagenomes are then presented, including binning, dominant population analysis, and gene-centric analysis. Finally, data management issues are presented and discussed. We hope that this review will assist bioinformaticians and biologists in making better-informed decisions on their journey during a metagenomic project.
INTRODUCTION
For the purposes of this review, we define metagenomics as the application of shotgun sequencing to DNA obtained directly from an environmental sample or series of related samples, producing at least 50 Mbp of randomly sampled sequence data. This distinguishes it from functional metagenomics, as reviewed elsewhere previously (58), whereby environmental DNA is cloned and screened for specific functional activities of interest. Metagenomics is a derivation of conventional microbial genomics, with the key difference being that it bypasses the requirement for obtaining pure cultures for sequencing. Therefore, metagenomics holds the promise of revealing the genomes of the majority of microorganisms that cannot be readily obtained in pure culture (62). In addition, since the samples are obtained from communities rather than isolated populations, metagenomics may serve to establish hypotheses concerning interactions between community members.
Indeed, metagenomics is increasingly being viewed as a baseline technology for understanding the ecology and evolution of microbial ecosystems, upon which hypotheses and experimental strategies are built (145), and with new sequencing technologies producing hundreds of megabases of data for well under $20,000 (see “Sequencing”), metagenomics is within the reach of many laboratories.
In this review, we address the bioinformatic aspects of analyzing metagenomic data sets, stressing the differences with standard genomic analyses. Although our focus is on bioinformatics, we will begin by considering experimental planning and implementation of metagenomic projects, as these aspects can have major impacts on subsequent bioinformatic analyses.
Throughout the review, we will follow the workflow of a typical metagenomic project at the Joint Genome Institute (JGI) (summarized in Fig. 1). This process begins with sample and metadata collection and proceeds with DNA extraction, library construction, sequencing, read preprocessing, and assembly. Genes are then called on either reads, contigs, or both, and binning is applied. Community composition analysis is employed at several stages of this workflow, and databases are used to facilitate the analysis. All of these stages will be discussed in detail below. We expect that some details of the workflow will be different in other sequencing facilities, and some aspects may be difficult to reproduce in a small research laboratory embarking alone on a metagenomic project without the support of a dedicated facility. Moreover, the rapid advancement of sequencing technologies will change the suite of tools available for metagenomic analysis. Therefore, rather than focusing on available tools, we emphasize the considerations and pitfalls of a typical metagenomic project. We hope that most considerations that we highlight will be useful even when current tools become obsolete.
FIG. 1.
Typical workflow for Sanger-based metagenomic projects of bacterial and archaeal communities at the JGI. Oval boxes indicate processes, and half-circles indicate data. See the text for discussion.
PRESEQUENCING CONSIDERATIONS
Community Composition
Community composition has a deciding influence on the types of analyses that can be performed on a metagenomic data set. Microbial communities comprise combinations of bacteria, archaea, microbial eukaryotes, and viruses, often with all four groups co-occurring in a single habitat. Historically, however, microbiologists are trained to think of themselves as either bacteriologists, virologists, or protistologists, and ecological studies investigating more than one of these taxonomic groups are still remarkably uncommon (74). To be frank, the authors are no exception; therefore, when we talk about community composition in the following sections, we are referring primarily to bacterial and archaeal species that have been the focus of most of our metagenomic studies.
At the current sequencing capacity, metagenomic sequencing of communities containing eukaryotes, in particular protists, is mostly cost-prohibitive because of their enormous genome sizes and low gene coding densities (133). Therefore, selection of a community that does not contain eukaryotes, or from which eukaryotes or their DNA can be excluded, is an important consideration prior to embarking on a metagenomic analysis. For example, one of the main reasons that the hindgut of a higher rather than lower termite was sequenced (146) is because the former lacks protist symbionts. When sequencing microbial communities that are found in tight symbiotic relationships with eukaryotic hosts, the removal of host cells or extracted host DNA is important to avoid eukaryotic contamination. For example, in the analysis of a gutless worm microbial symbiont community, host cells were physically separated from bacterial endosymbiont populations using a Nycodenz gradient (150).
Simply excluding eukaryotes from a metagenomic analysis is not ideal from an ecological perspective, as it compromises our ability to assess a microbial community in its entirety. An alternative or complementary strategy could be to obtain molecular data at the RNA (metatranscriptomics) or protein (metaproteomics) level, thus bypassing the problem of large amounts of noncoding eukaryotic sequence data. Emerging sequencing technologies such as pyrosequencing (89) may ultimately allow metagenomic sequencing of communities comprising eukaryotes, but the data are likely to present numerous challenges for many downstream bioinformatic analyses (see “Selection of Sequencing Technology”).
Within the sequence-tractable bacterial, archaeal, and viral components of a community, community complexity should be assessed prior to shotgun sequencing (see “Premetagenome Community Composition Profiling” for a description of assessment methods). Community complexity is a function of the number of species in the community (richness) and their relative abundance (evenness). A community with more species that are closer to equal abundance is more complex than a community with less species that have unequal abundance. As a consequence, for a constant sequencing effort, sequence data from a less complex community will tend to assemble into larger contigs (contiguous genomic stretches comprised of overlapping reads). However, in our experience, the key variable affecting the type of downstream analyses that can be performed on a metagenomic data set is the presence or absence of dominant populations regardless of the total number of species. Dominant populations that comprise more than a few percent of the total number of cells or virions in a community will have a higher representation in a metagenomic data set, resulting in a greater likelihood of assembly and recovery of contigs. Note that we define assembled contigs arising from a population as composite genomic fragments because each component read likely comes from a different individual within the population in which individuals are usually not clonal.
We therefore distinguish between two basic types of communities throughout this review: those comprising dominant populations and those that do not. Broadly speaking, communities of the first type produce contigs of >10 kbp up to several hundred kbp, depending on the degree of dominance and amount of sequence obtained. Examples include simple communities that are comprised mostly of a few dominant species, such as acid mine drainage biofilms (138) and a gutless worm symbiont community (150). However, species-rich communities can also fall into this category, such as enhanced biological phosphorus-removing (EBPR) sludge (47) and an anaerobic ammonia-oxidizing reactor (129), which have one dominant population flanked by a long tail of low-abundance species. Communities of the second type lack populations abundant enough to result in assembled contigs of >10 kbp using on the order of 100 Mbp of Sanger data (Fig. 2). Such communities also tend to be species rich.
FIG. 2.
Contig size distribution for assemblies of around 100 Mbp of Sanger data obtained from each of seven microbial communities. The gray area indicates small contigs with a higher likelihood of chimeric assemblies (see “Assembly”). Communities with contigs found mostly in this zone (termite hindgut [146], soil, and whale fall [135]) lack dominant populations, whereas communities with larger contigs outside this zone have dominant populations: gutless worm (150), phosphorus-removing sludges from U.S. and Australian (OZ) laboratory-scale bioreactors (47), and an acid mine drainage (AMD) biofilm (138). Note that the gutless worm scaffolds (end-pair-linked contigs) are shown, explaining the larger size.
Sequencing of a community with dominant species is likely to reproduce a significant part of the genomes of the dominant organisms and, in some cases, near-complete genomes (47, 138). Therefore, analysis of large genomic fragments is similar to conventional comparative genomics. In contrast, sequences obtained from a complex system without dominating species will not contain large genomic fragments of any component population using current technologies (135, 140). The analysis will therefore normally be focused on averaged properties of the community, such as gene content and abundance, since information on any given component species will be sparse.
Selection of Sequencing Technology
The number of sequencing technologies is currently expanding, drawn by demand to bring down the cost of sequencing. At the time of writing of this review, Sanger (dye terminator) sequencing (118, 119) remains the major source of metagenomic sequence data. Alternative strategies have also been used, namely, pyrosequencing (89), which has been applied to viral (9) and bacterial (36) communities. Advantages of pyrosequencing over Sanger sequencing include a much lower per-base cost and no requirement for cloning (114). The latter is useful for both bacterial and virion communities because of the demonstrated cloning bias of bacterial genes (127) and promoters (48) in Escherichia coli and difficulties with cloning viral nucleic acids (17). However, the major disadvantage of pyrosequencing has been its average read length, initially ∼100 bp on the GS20 platform and ∼200 bp on the GS FLX platform. Reads of this length present additional challenges for assembly and gene calling. Indeed, most studies that have used pyrosequencing for metagenomic analysis did not attempt assembly or gene calling, instead relying on similarity searches of the short reads against a reference database as the basis of the analysis (9, 36) (see also Table 1). Therefore, the sections on bioinformatics processing below refer mostly to Sanger data. Notably, however, 454 Life Sciences is currently evaluating 400- to 500-bp (titanium) pyroreads (http://www.454.com/). If, in conjunction with longer read length, technical problems such as reagent dilution and maintaining nucleotide extension synchronization (114) can be adequately addressed to produce read quality comparable to that of Sanger data, then pyrosequencing will be able to supplant Sanger sequencing as the preferred data type for metagenomic analysis.
TABLE 1.
Gene prediction methods used in metagenomic projects
| Project | Institution(s) | Gene prediction method | Reference |
|---|---|---|---|
| Acid mine drainage biofilm communities from Richmond mine | University of California, Berkeley, JGI | fgenesb | 138 |
| Aquatic microbial communities from Drinking water networks | University of Goettingen | BLAST | 120 |
| Aquatic microbial communities from Soudan Mine in Minnesota | San Diego State University | BLAST | 36 |
| Fossil microbial community from Whale Fall at Santa Cruz Basin of the Pacific Ocean | JGI | fgenesb | 135 |
| Gut microbiome of healthy human adults | J. Craig Venter Institute, Washington University, Stanford University | BLAST | 50 |
| Gut microbiome of healthy human Japanese infants and adults | University of Tokyo | Metagene | 77 |
| Gut microbiome of lean and obese mice | Washington University | BLAST | 136 |
| Gut viriome of healthy human adults | Genome Institute of Singapore | BLAST | 152 |
| Marine archaeal anaerobic methane oxidation communities from Eel River sediments | JGI, MBARI | fgenesB | 57 |
| Marine microbial communities from Bras del Port saltern in Santa Pola, Spain, crystallizer pond | Universitas Miguel Hernandez | GLIMMER | 80 |
| Marine microbial communities from Global Ocean Sampling | J. Craig Venter Institute | Similarity searches and filtering of ORFs | 115 |
| Marine microbial communities from Sargasso Sea | J. Craig Venter Institute | BLAST | 140 |
| Marine plankton communities from deep Mediterranean Sea Ionian station Km3 | Universitas Miguel Hernandez | BLAST | 93 |
| Marine planktonic communities from Hawaii Ocean Times Series Station | JGI | BLAST | 33 |
| Marine RNA viral communities from coastal samples | University of British Columbia | BLAST | 27 |
| Marine viral communities from ocean environments | SDSU | BLAST | 9 |
| Olavius algarvensis (gutless worm) microbiome from Mediterranean Sea | Max Planck Institute, JGI | mORFind | 150 |
| Oral TM7 microbial communities of healthy human adults | JGI, Stanford University | fgenesb | 88 |
| Soil microbial communities from Minnesota farm | JGI | fgenesb | 134 |
| Wastewater EBPR microbial communities from bioreactor | JGI | fgenesb | 47 |
Combinations of different sequencing technologies have been evaluated for producing high-quality draft assemblies of microbial isolates (51) that could be applied to metagenomes containing one or more dominant populations. The Illumina (http://www.illumina.com) and ABI SOLiD (http://www.appliedbiosystems.com) sequencing technologies have not yet been applied to environmental samples, but their application is likely to be limited to the resequencing of dominant populations since reads are currently too short (<50 bp) to be used for de novo assembly or gene calling. One next-generation sequencing technology worth keeping an eye on is real-time single-molecule sequence determination that aims to produce multikilobase-length reads at throughputs comparable to those of short-read technologies (71; http://visigenbio.com). If such an ambitious goal can be achieved with acceptable sequence quality and cost, this platform will become the choice for metagenomic studies, since even single reads will contain contextual data of one or more neighboring genes, and assembly will be simplified.
How Much Sequence Data?
A common question asked by researchers embarking on their first metagenomic analysis is how much sequence data they should request or allocate for their project. Unlike genome projects, metagenomes have no fixed end point, i.e., a completed genome. Therefore, decisions on how much sequence data to generate for an environmental sample have been based on pragmatic reasons, chiefly sequencing budget. For example, 100 Mbp is a typical Sanger sequencing request for a metagenomic project through the JGI community sequencing program (http://www.jgi.doe.gov/CSP/index.html). However, with the per-base cost of sequencing continuing to drop, other more objective criteria can be brought to the fore, such as estimates of sequence coverage (number of reads covering each base in a contig) of the community. Since species do not have uniform abundance in a community, it is simpler to address the coverage of individual populations for which an approximate average genome size is known. For example, if a dominant population represents 10% of the total community and 100 Mbp is obtained, then this population is expected to be represented by 10 Mbp, assuming completely random sampling of the community. If the average genome size of individuals in this population is 2 Mbp, then an average of 5× coverage of the composite population genome will be expected. To place this in perspective, 6× to 8× coverage of microbial isolates is a common target to obtain a draft genome suitable for finishing. In the most extreme example to date, the complete genome of a low-abundance uncultured species, “Candidatus Cloacamonas acidaminovorans,” was elucidated from a complex anaerobic digester community at the cost of the end sequencing of more than a million fosmids, generating 1.7 million reads (1.12 Gbp) (4). Ultimately, the objectives of the study should guide sequence allocation. For example, if the aim is to determine the single-nucleotide polymorphism (SNP) frequency profile of a dominant population as part of a population genetic analysis (67), then ideally, a coverage of 20× or greater will be needed for the dominant population. If the aim is to identify overrepresented gene functions in the community as a whole (see “Gene-Centric Analysis”), then much less sequence data will be needed. Indeed, we recently found that an extremely low coverage of a highly complex and stratified hypersaline mat community (estimated dominant population coverage of <0.01×) was still sufficient to detect genetic gradients in the mat community using 10 Mbp per layer (76).
SAMPLING AND DATA GENERATION
Sample Collection for Metagenomes and Other Molecular Analyses
Metagenomes are sequence inventories of genomic DNAs from environmental samples. Extraction and purification of high-quality DNA are still some of the main bottlenecks in metagenomics, compounded by the fact that there is not a “one-size-fits-all” extraction method for all environmental samples. Low-biomass samples yield small quantities of DNA that may be insufficient for library construction. In general, microgram quantities of genomic DNA are required for cloning (see below) and pyrosequencing. Whole-genome amplification has been used on small yields of environmental DNAs to provide microgram quantities for sequencing (9). One major advantage of this technique is that it can process and retain single-stranded DNA, which is invaluable for viral samples. However, the relative representation of genomic DNAs may be compromised, particularly if the amount of starting material is small (10, 12, 110, 111). This is important to keep in mind for downstream comparative analyses, particularly between samples that used whole-genome amplification and those that did not.
In many cases, it may be beneficial to collect additional sample material for complementary analyses. Examples of additional molecular analyses that will leverage and enhance metagenomic data from cellular microbial communities include metatranscriptomics (54, 74, 139), metaproteomics (81), viral metagenomics (37), and imaging methods such as fluorescence in situ hybridization (FISH) using group-specific oligonucleotide probes (8, 62, 144). For example, colocalization studies by combining FISH with digital image analysis can provide spatial information in structured ecosystems to support metabolic interactions between community members inferred from metagenomic data.
While it is sometimes possible to resample many habitats, two temporally separated samples may not be directly comparable. For example, habitats that have seasonal patterns such as the marine water column (32) cannot be considered equivalent at different times of the year. Even in habitats that do not show seasonal variation, such as controlled laboratory-scale bioreactors, community composition may be influenced by predators, parasites, or other variables that confound comparisons of metagenomic data. For example, from an initial metagenomic analysis of two laboratory-scale sequencing batch reactors, we implicated bacteriophages as being important determinants in driving bacterial community composition (74). Unfortunately, we did not have appropriately stored material from the original sampling and characterized the virion community in a reactor sample taken 7 months after the initial metagenomic sampling. During this time, both the bacterial and viral communities had changed, complicating the comparative analysis. It is of course impossible to store sample material in the appropriate manner for every conceivable downstream molecular analysis, but as a number of techniques become more routine, such as metatranscriptomics, metaproteomics, metabolomics, and viral metagenomics, subsamples can be inexpensively stored in standardized ways to provide researchers with the potential to perform these analyses if needed.
Sample Metadata Collection
Collecting collateral nonsequence data associated with an environmental sample greatly enhances the ability to interpret the sequence data, particularly for a comparative analysis of temporal or spatial series (33, 140). Such “metadata” include biochemical data such as pH, temperature, and salinity; geographical data such as global positioning system coordinates; and depth, height, and sample-processing data such as collection date, DNA extraction method, and clone library details (42). The type of metadata can vary considerably depending on the sample type; for instance, environmental and clinical samples historically have very different metadata. Databases housing metagenomic data already include various degrees of metadata (91, 123), but cross-referencing such data is problematic due to a lack of consistency and standards. Initiatives are under way to standardize metadata collection, e.g., by use of a controlled vocabulary, where possible (42). Such data are expected to prove invaluable once enough data are generated to compare communities along environmental, spatial, or longitudinal gradients (140).
Premetagenome Community Composition Profiling
To facilitate decisions on sequence allocation and processing, the community composition of the environmental sample under study should be assessed prior or at least in parallel to the metagenomic analysis using a conserved marker gene survey, ideally conducted on the same sample. Indeed, several samples could be prescreened using marker genes to aid in the selection of a subset for metagenomic analysis. The small-subunit rRNA (16S rRNA) gene is usually the marker gene of choice for bacterial and archaeal communities owing to its widespread use and consequent large reference database (25, 34). One drawback of the 16S rRNA gene is that copy number can vary by an order of magnitude between bacterial species, which, along with PCR-induced biases (130, 143), can skew estimates of community composition. PCR products are normally cloned and sequenced to provide a semiquantitative phylogenetic profile of a community. At the JGI, we typically sequence one 384-well plate containing 16S clones (called a ribosomal panel) to provide a baseline estimate of community structure.
For most microbial communities, however, 384 clones are a gross undersampling of diversity and highlight only relatively dominant taxa. Other approaches that have higher resolution include microarrays to which fluorescently labeled 16S PCR amplicons or rRNAs are applied (19, 105, 108). For example, the Phylochip comprises 500,000 probes redundantly targeting ∼9,000 phylogenetic groups (operational taxonomic units) and has a sensitivity that is 1 to 2 orders of magnitude higher than that of a PCR clone library sequenced to ∼102 (19). On the downside, species that are not represented by probes on the microarray will be missed, and the relative abundance of sequence types cannot be easily estimated. This means that dominant populations are currently difficult to detect from Phylochip data alone.
Pyrosequencing has recently been applied to PCR-amplified 16S rRNA genes, providing 100- or 200-bp 16S “pyrotags” to evaluate community composition (61, 63, 126). This approach has the benefits of high resolution (due to the large number of pyrotags, ∼500,000 per bulk 454-FLX run) comparable to that of a 16S microarray while retaining relative amplicon abundance like a clone library. The main limitation of this approach is the reduced phylogenetic resolution afforded by 100 to 200 bp, so the method is dependent on a high-quality reference 16S database for the accurate classification of pyrotags.
FISH using group-specific 16S rRNA gene-targeted oligonucleotide probes (8, 144) can also be used to profile community composition. Fluorescently labeled cells can be quantified by microscopy either manually or with the aid of image analysis software (28) or in combination with flow cytometry (121). In principle, FISH-based counting is the most accurate method for determining relative and absolute abundances of populations since it is not affected by 16S copy number variation. In practice, only a few phylogenetic groups can be targeted per sample due to logistical considerations (e.g., number of fluorochromes that can be visualized simultaneously, availability, and cost of suitable probes), and for gross community composition estimates, these tend to target broader groups such as domains or phyla. Therefore, the complete or even widespread population-level characterization of communities using FISH has not been feasible to date.
Since no universally conserved marker genes exist for viruses, none of the methods described above can be used to profile viral communities, and direct metagenomic investigations are the only option at this point.
Shotgun Library Preparation
Shotgun clone libraries for genome sequencing are typically prepared using three different average sizes of cloned DNA: 3, 8, and 40 kbp (fosmids). This facilitates primarily assembly and finishing since longer clones will have a greater likelihood of spanning gaps and repeats in the genome assembly. The JGI uses a ratio of 4:4:1 for 3, 8, and 40 kbp end-sequence data to produce high-quality draft assemblies (largest correctly assembled contigs) economically. We have more or less adopted the same insert-size libraries and sequencing ratios for metagenomic projects even though the end product may be vastly different from that of a genomic project. In the case of microbial communities with one or more dominant populations, the ratio of insert-size sequencing will serve the same function of improving assembly (and occasionally finishing) of composite population genomes. For microbial communities lacking dominant populations, the main purpose of the larger-size inserts is to provide gene neighborhood context, usually through the complete sequencing of selected fosmids (40, 146). Bacterial artificial chromosomes allow access to even larger pieces of contiguous genomic DNA from environmental samples (14); however, they are technically more demanding to prepare than are fosmids and small-insert libraries.
Occasionally, the environmental sample will dictate which libraries can be created. For example, despite repeated attempts, DNA extracted from acid mine drainage biofilm samples could not be obtained with a purity and a molecular weight high enough to create an 8-kbp or fosmid clone library, limiting the study to data from a 3-kbp library only (138). The preparation of clone libraries requires between 5 μg (for a 3-kb library) and 20 μg (for a fosmid library) of DNA, which often cannot be obtained directly from low-biomass communities. Whole-genome amplification via multiple-displacement amplification can circumvent this problem, but the typical length of the amplified DNA, ∼15 kbp, is too short in general to allow large-insert-library construction, although fosmid libraries from amplified environmental DNA have been reported (99).
Sequencing
At the JGI, metagenomic projects are sequenced in at least two stages for quality control (QC). The first stage is a 20-plate QC of a 3-kbp insert (pUC) library generating approximately 10 Mbp of Sanger sequence data followed by a preliminary informatic analysis to guide the allocation of the remainder (majority) of the sequence allotment. First and foremost, the QC sequencing confirms that the shotgun clone libraries produce sequence data of sufficient quality to warrant further sequencing. For genome projects, sufficient quality typically means that 95% of clones produce reads with at least 650 Q20 bases (see “Sequence Read Preprocessing”), i.e., a 95% pass rate. For metagenomic projects, this bar is dropped sometimes to as low as an 85% pass rate because of the greater difficulty in making high-quality libraries from environmental DNAs and the often precious nature of difficult-to-collect environmental samples. The preliminary analysis usually involves assembly but not gene prediction primarily to confirm initial community composition estimates but also to determine if populations can be easily discriminated in the data. For example, similarity searches against public nucleotide and protein databases will identify populations via conserved marker genes and provide some indication of relative abundance according to the size and read depth of the contig that the marker genes were found on. A histogram of contig read depth will alert the researcher to the presence of one or more dominant populations, since 10 Mbp is sufficient to result in the assembly of genomic fragments from dominant populations. Plotting contig depth against another variable, such as GC content, often helps to discriminate populations. If a dominant population was expected based on community composition profiling and not noted by contig read depth, this indicates greater-than-expected microheterogeneity in the population hindering assembly (see “Finishing”) or a technical error in the experimental workup. For example, QC sequencing of EBPR sludge from a laboratory-scale bioreactor revealed that the primary target organism “Candidatus Accumulibacter phosphatis” type I was grossly underrepresented relative to the initial community composition estimate (4% versus 60%). The discrepancy arose because this organism was poorly lysed in the DNA extraction, a fact that was missed because the community was profiled using a type I-specific FISH probe (S. He and K. McMahon, personal communication). At this point, it was not too late to reextract DNA from the EBPR sludge using a different method.
SEQUENCE PROCESSING
Processing of genomic sequence data and processing of metagenomic sequence data have many features in common, namely, read preprocessing, assembly including selected instances of finishing (dominant populations), and gene prediction and annotation. As mentioned above, the key difference between genomes and metagenomes is that the latter, with the exception of finishable dominant populations, do not have a fixed end point, i.e., one or more completed chromosomes as for microbial isolate genomes. This means that metagenomes rarely progress beyond draft assemblies and lack many of the quality assurance procedures associated with producing finished genomes. Therefore, greater care needs to be taken when processing sequences of metagenomic data sets than when processing genomic data sets.
Sequence Read Preprocessing
Preprocessing of sequence reads prior to assembly, gene prediction, and annotation is a critical and largely overlooked aspect of metagenomic analysis. Preprocessing comprises the base calling of raw data coming off the sequencing machines, vector screening to remove cloning vector sequence, quality trimming to remove low-quality bases (as determined by base calling), and contaminant screening to remove verifiable sequence contaminants. Errors in each of these steps can have greater downstream consequences in metagenomes than in genomes and will be discussed in turn.
Base calling is the procedure of identifying DNA bases from the readout of a sequencing machine. There are surprisingly few choices for base callers, and the differences between them for the purposes of metagenomics are small; therefore, we have no specific recommendation from the ones described below. By far, the dominant base caller used today is phred (41). phred initiated the widespread use of probabilistic-based quality scores, which all later base callers adopted. phred quality scores are estimates of per-base error probabilities. The quality score, q, assigned to a base is related to the estimated probability, p, of erroneously calling the base by the following formula: q = −10 × log10(p). Thus, a phred quality score of 20 corresponds to an error probability of 1%. Other frequently used base callers are Paracel's TraceTuner (www.paracel.com) and ABI's KB (www.appliedbiosystems.com), which behave very similarly to phred, converting raw data into accuracy probability base calls. In general, however, metagenomic assemblies have lower coverage than do genomes, and therefore, errors are more likely to propagate to the consensus. For complex communities, the majority of reads will not assemble into contigs, and base-calling errors in these unassembled reads will appear directly in the final data set.
Vector screening is the process of removing cloning vector sequences from base-called sequence reads. The complete and accurate removal of cloning vector sequence is especially important in metagenomic data sets since these data sets often have large regions of very low coverage in which each read uniquely represents a part of a genome. The assembly of these data without vector trimming can produce chimeric contigs in which the vector sequence, being common to most reads, acts to draw together unrelated sequences (Fig. 3). Also, genes may be predicted on the vector sequence introducing phantom gene families into downstream analyses (see “Gene-Centric Analysis”).
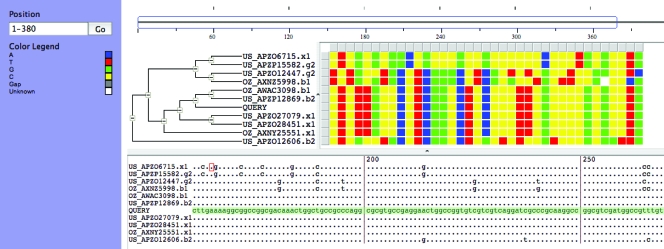
FIG. 3.
Phrap assemblies visualized with the Consed (53) program. The consensus sequence is shown at the top of the display and is derived from aligned reads shown below the consensus. Note that the Phrap assembler uses the highest-quality base for the consensus regardless of base frequency at each position. Read identifiers and orientation (arrowheads) are shown on the left of the display. Low-quality bases and masked regions are grayed out. Green bars indicate sequence fragments found elsewhere in the assembly. (A) Example of a good-quality assembly with high read depth. Note the consistent alignment of all residues. (B) Example of a misassembled contig drawn together by a common repeat sequence (indicated by purple bars at left). Note the misaligned residues in red and the meaningless “consensus” sequence that does not correspond to any single read below it. (C) Chimeric contig produced by coassembly of closely related strains (haplotypes) in a metagenomic data set. Note that the consensus sequence is a chimera of the two haplotypes (based on the highest-quality base at each position) and likely does not represent an extant organism. (Screen shots are printed with permission of the software publisher.)
A number of different tools are available for vector screening, including cross_match (www.phrap.org), LUCY (22), and vector_clip (128). Also, some assemblers include vector trimming as part of a preprocessing pipeline, including PGA (http://www.paracel.com) and Arachne (13, 65). The most commonly used tool is cross_match, which uses a modified Smith-Waterman algorithm to identify matches to vectors that are extended to produce optimal alignments. However, cross_match requires exact matches to vector sequences and has no expectation for the location of the vector sequence in a read. In our experience, this program frequently fails to remove vector sequences because of frequent base-calling errors on the edges of reads where the vector sequence is found. Another vector-trimming tool, LUCY, avoids this problem by specifying error rates as a function of sequence position. In every case that we have tested to date, LUCY results are substantially better than those achieved with cross_match. The downstream effects of improved vector screening are fewer spurious protein predictions and fewer errors in predictions of real protein-coding sequences, particularly open reading frames (ORFs) at the ends of reads (see “Gene Prediction and Annotation”).
Most postprocessing pipelines appear to ignore base quality scores associated with reads and contigs, and few take positional sequence depth into account as a weighting factor for consensus reliability. Therefore, low-quality data will be indistinguishable to the average user from the rest of the data set and should be removed. An extreme example of a poor-quality read that inadvertently passed through to gene prediction is shown in Fig. 4. In the worst-case scenario, such phantom genes called on a low-quality sequence may pass unchecked into public repositories. We recommend quality trimming to be performed after vector screening, as described above. The reason is that the trimming of low-quality bases might truncate the vector sequence and impede the ability of vector-screening programs to recognize the remainder of the vector. In such cases, significant parts of the vector might still remain for the next stages of the pipeline. LUCY combines vector and quality trimming into one tool.
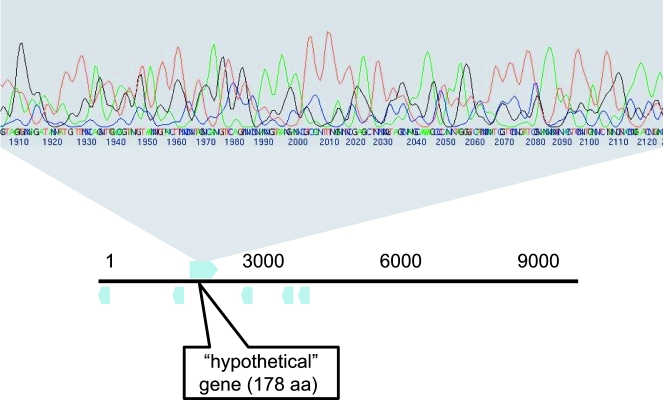
FIG. 4.
Part of the chromatogram of a low-quality read without quality trimming on which multiple nonexistent genes were predicted (bottom).
Recognition of sequence contamination of metagenomic data sets, other than vector sequence, is nontrivial. Sanger data sets from clonal organisms are routinely screened for E. coli genomic sequence because E. coli is the cloning vector host, and small amounts of its genome may get through plasmid purification. Pyrosequencing, which does not rely on cloning of DNA into E. coli, will not have this problem; however, other types of contamination cannot be excluded. For metagenomic data sets, host contamination screening should be considered carefully because the environment under study may have E. coli or close relatives as bona fide members of the community, and screening would therefore bias the representation of these species in the data set. Occasionally, the mislabeling of sequence plates occurs in production pipelines. These types of cross-contamination between two data sets can usually be detected if one of the data sets is from an isolate by differences in GC content or BLAST. If plates from two metagenomic projects are mixed up, the contamination may be harder to detect, since neither data set is likely to be homogeneous. It is quite common that reads and even contigs are not incorporated into finished microbial genomes, and these are usually dismissed as being either low-quality or contaminant sequences. In contrast, metagenomic projects will keep high-quality contaminating reads and contigs, as they will probably not be easily distinguishable from the rest of the data set and may therefore skew downstream analyses such as gene-centric analysis, depending on the degree of contamination. Presently, there is no solution to this quandary, and suspected contaminant sequences would need to be investigated on a case-by-case basis.
Assembly
Assembly is the process of combining sequence reads into contiguous stretches of DNA called contigs, based on sequence similarity between reads. The consensus sequence for a contig is either based on the highest-quality nucleotide in any given read at each position or based on majority rule, i.e., the most frequently encountered nucleotide at each position. The number of reads underlying each consensus base is called depth or coverage. Sequencing is typically performed from both sides of an insert in a vector plasmid, and such pairs are called paired reads or mate pairs. Knowledge of the approximate insert size of the library facilitates the production of a more accurate assembly since mate pairs provide an external constraint to guide assembly. The presence of paired reads in two different contigs allows those contigs to be linked into a larger noncontiguous DNA sequence called a scaffold, whose intercontig gap size can be estimated based on the insert size of the read pairs. For this reason, large-insert clones such as fosmids are particularly useful for improving assemblies.
The major cause of misassembly in genomic projects is repetitive regions that can be resolved in the finishing process (79). The assembly of metagenomic projects will also be confounded by repeats but pose additional assembly challenges in the form of nonuniform read depth due to nonuniform species abundance distribution and the potential for the coassembly of reads originating from different species. Therefore, not only can misassembled reads be retained in the final published data set due to the absence of finishing, but reads from more than one species can also be assembled together, producing chimeric contigs. Coassembly is more likely to happen with reads from closely related genomes where the sequence similarity is higher (we routinely observe homologous regions of two or more strains with up to 4% nucleotide sequence divergence coassembling) but has been found between reads originating from phylogenetically distant taxa, with conserved genes serving as the focal point for misassembly. For example, a contig from a surface seawater metagenome comprised reads originating from bacteria and archaea, as evidenced by gene calls, with the 16S rRNA gene serving as the focal point in this instance (32). A recent simulation study found that chimeras are particularly prevalent among contigs lower than 10 kbp in size (94). High-complexity microbial communities lacking dominant populations rarely produced contigs larger than 10 kbp (Fig. 2), prompting the recommendation that such data sets should not be assembled at all (94).
A variety of assembly programs are publicly available, including Phrap (www.phrap.org), Arachne (13, 65), the Celera Assembler (97), PGA (http://www.paracel.com/), and CAP3 (60; for a description and history of these assemblers, we refer the reader to reference 79). Most currently available assemblers were designed to assemble individual genomes or, in some cases, genomes of polyploid eukaryotes; however, they were not designed to assemble metagenomes comprising multiple species with nonuniform sequence coverage, and therefore, their performance with metagenomic data sets varies significantly (94). For example, the Celera assembler does not assemble contigs with atypically high read depths (based on an expected Poisson distribution) because it interprets them as potential assembly artifacts due to the coassembly of repeats, whereas in metagenomic data, they may be bona fide contigs arising from dominant populations (140). A second example is that Phrap is optimized for making maximal use of its input data using a “greedy” algorithm and will extend contigs as far as possible. This is a good approach for assembling low-coverage nonrepetitive regions from low-quality reads, as it makes the most of the available data, particularly if the assembly will be verified by finishing but is not desirable for metagenomes since it is more likely to produce chimeras when data include reads from multiple strains and species. More conservative assembly programs such as Arachne have been shown to produce smaller but more reliable contigs than Phrap (94).
A useful auxiliary approach to de novo assembly is comparative assembly, that is, aligning reads and/or contigs to a reference genome of a closely related organism. The AMOS comparative assembler has been developed specifically for this purpose (112). For metagenomic data sets, this can improve the assembly of dominant populations since it provides a mechanism to span hypervariable regions in a composite population genome and is computationally much less expensive than de novo assembly (4). A major caveat of the approach, however, is that it will be useful for only a small subset of the average metagenomic data set since reference genomes cover only a fraction, and a highly biased fraction at that, of microbial diversity (see “Postsequencing Community Composition Estimates”).
One thing is clear: there is no magic bullet for assembling metagenomic data sets, and all assemblers will make numerous errors. Ideally, therefore, every metagenomic assembly should be manually inspected for errors before public release. Assembly errors can be easily identified with visualization tools, such as Consed (Fig. 3) (53), which are used to facilitate genome finishing; however, the sheer scale of most metagenomic data sets precludes manual inspection let alone the correction of all identified assembly errors. One approach that we have taken to address this limitation is to make two or more assemblies of the same data using different assemblers (47) to facilitate the identification of misassemblies during the downstream analysis phase following gene calling. It is, however, feasible and worthwhile to resolve misassemblies of the largest contigs in a metagenomic assembly, especially contigs that are greater in length than or equal in length to fosmids, using standard initial steps in the finishing process (79).
The final products of assembly, contigs and scaffolds, are submitted to public databases as flat text files, meaning that all information about the underlying reads is lost, including sequencing depth and quality scores of each base, length and overlaps between reads, and quality of vector trimming. This is not ideal for two reasons. Firstly, the quality of the contigs cannot be assessed and is also not taken into consideration by tools such as BLAST. Secondly, meaningful polymorphisms in the data due to coassembled strains (haplotypes) (see “Analyzing Dominant Populations”) are lost because a single consensus sequence is submitted. Methods for weighting consensus accuracy and preserving polymorphism information for subsequent analyses are needed. A first step in this direction has been taken by public databases with the establishment of the Trace and Assembly archives, which archive raw read files and assemblies associated with submitted genomic and metagenomic data sets, respectively (147). In practice, however, most users will work only with the flat text consensus data and ignore read and consensus quality unless it is presented to them in a more convenient user interface. Such interfaces are beginning to be provided by dedicated comparative genome and metagenome platforms (see Data Management).
Finishing
Genome closure and finishing are commonplace for microbial isolate projects and part of the standard processing pipeline at sequence facilities such as JGI. For most metagenomes, finishing is not possible. However, for dominant populations within metagenome data sets that have draft-level coverage, finishing may be an option. This is dependent largely on the degree of microheterogeneity within the population. Genome rearrangements such as insertions, deletions, and inversions will break assemblies, whereas point mutations usually will not. Even in instances where chromosomal walking along large-insert clones is used instead of shotgun sequencing, microheterogeneity can still complicate assembly (56). However, there are now several examples in the literature of complete or near-complete composite population genomes of uncultivated organisms derived from environmental sources including Cenarchaeaum symbiosum, the sole archaeal symbiont of a marine sponge (56); Kuenenia stuttgartiensis, an anaerobic ammonia-oxidizing planctomycete sequenced from a laboratory-scale bioreactor enrichment (129); a Rice Cluster 1 methanogen from an enrichment culture (40); “Candidatus Cloacamonas acidaminovorans,” the first sequenced representative of the candidate phylum WWE1, from an anaerobic digester (107); and Ferroplasma acidarmanus, one of a few dominant populations in an acid mine drainage biofilm (5). In the last case, the assembly was facilitated by the availability of an isolate genome (fer1) obtained from the same habitat. The Kuenenia, Rice Cluster 1 methanogen, and “Candidatus Cloacamonas” genomes, however, could be assembled without reference to an isolate genome because the populations were near clonal. We make the general observation that sequence microheterogeneity within populations often seems to reflect spatial heterogeneity within the ecosystem from which the populations were derived. Homogenized systems such as bioreactors or enrichment cultures have produced composite population genomes with very low levels of polymorphism (40, 107, 129), perhaps due to the higher likelihood of selective sweeps through the population curtailing genomic divergence (24). Therefore, if the goal is to assemble a complete population genome from an environmental sample, we recommend the use of ecosystems with low spatial heterogeneity if at all possible or finer-scale sampling to reduce the effect of spatial heterogeneity.
Gene Prediction and Annotation
Gene prediction (or gene calling) is the procedure of identifying protein and RNA sequences coded on the sample DNA. Depending on the applicability and success of the assembly, gene prediction can be done on postassembly contigs, on reads from the unassembled metagenome, and, finally, for a mixture of contigs and individual unassembled reads.
There are two main approaches for gene prediction. The “evidence-based” gene-calling methods use homology searches to identify genes similar to those observed previously. Simple BLAST comparisons against protein databases as well as tools like CRITICA (11) and Orpheus (46) use such an approach. Conversely, the second approach, “ab initio” gene calling, relies on intrinsic features of the DNA sequence to discriminate between coding and noncoding regions, allowing the identification of genes without homologs in the available databases. The use of gene training sets, i.e., sets of parameters derived from known genes of the same or related organisms, can enhance the quality of the predicted genes for some of those programs (e.g., fgenesB [http://www.softberry.com]), while others are self-trained on the target sequence (Genemark [16], GLIMMER [31], and MetaGene [100]).
Pipelines that use a combination of evidence-based and “ab initio” gene calling are frequently used for complete genomes. In the first step, genes are identified based on homology searches of the sequence of interest versus public databases. Hits to genes in databases are considered to be real genes and can be used as a training set for the ab initio gene-calling programs. Subsequently, an “ab initio” method fine-tuned for a particular genome is used to identify more genes that were missed in the previous step. One such pipeline, called mORFind, uses a combination of Orpheus, CRITICA, and GLIMMER.
In metagenomic sequences, genes can originate from many, frequently diverse organisms. When dominant populations exist, their sequences can be separated from the rest of the data set (see “Binning”) and the pipeline generally used for complete genomes applied to this subset of the data. For communities or their parts that defy assembly or assemble poorly, no training is possible. In these cases, “generic” gene prediction models or models fine-tuned to the closest phylogenetic group can be used. For example, MetaGene (100) is a gene prediction program developed specifically for metagenomic data sets using two generic models, one for archaea and one for bacteria. Due to the fragmented nature of such data sets and the quality of the sequencing, gene prediction is further complicated by the fact that many genes are represented only by fragments, contain frameshifts, or are chimeras due to errors in assembly. Recently, a tool that allows gene prediction despite these problems, even on short 454 reads, has been reported (73), although its performance has yet to be evaluated in real applications. The method is based on similarity comparisons of the metagenomic nucleotide sequences either to the same metagenome or to other external sequences and the subsequent discrimination of conserved coding sequences from conserved noncoding sequences by synonymous substitution rates. BLAST searches are conducted at the amino acid level to provide higher resolution than nucleotide searches.
Both evidence-based and “ab initio” methods have been used for the prediction and analysis of metagenomic data sets (Table 1). Evidence-based gene calling has been used as the sole method of gene calling in at least one metagenomic study using Sanger reads (140) and all metagenomic studies using unassembled pyrosequence data due to short read lengths (Table 1). Since this approach relies entirely on comparisons to existing databases, it has two major drawbacks. Low values of similarity to known sequences either due to evolutionary distance or due to the short length of metagenomic coding sequences and the presence of sequence errors prevent the identification of homologs. Moreover, novel genes without similarities are completely ignored. Despite these drawbacks, this approach has been used in several studies and can be useful for gene-centric comparisons of metagenomes, especially in cases where the size of the sequence fragments is not adequate for the ab initio gene prediction, such as high-complexity metagenomes and metagenomes sequenced by high-throughput parallel pyrosequencing.
Treating all ORFs as putative genes usually produces prohibitive amounts of data, contains too much noise, and is therefore very hard to use. Methods based on features of the sequences, the size of the predicted ORFs, and the similarity to known sequences have been used to lower the total number of candidate coding sequences from a population of ORFs (151).
At the JGI, we are using two “ab initio” gene prediction pipelines for the analysis of metagenomic data sets. The first gene prediction pipeline uses fgenesB with specific training models for sequences that can be assigned to phylogenetic groups and generic models for the unassigned sequences (Table 1). The second uses Genemark, which allows gene prediction without the need for training sets and classification of sequences. Both pipelines have proved to be quite accurate when used on simulated data sets (http://fames.jgi-psf.org). Other studies have employed GLIMMER, MetaGene, and the mORFind pipeline (Table 1).
RNA genes (tRNA and rRNA) are predicted using tools such as tRNAscan (82) for tRNAs and similarity searches for rRNAs. While tRNA predictions are quite reliable, it is not uncommon for rRNA genes to be incompletely identified (incorrect gene boundary coordinates) or even entirely missed. In these instances, it is also not uncommon to see nonexistent hypothetical protein-coding genes called in the place of rRNA genes. Other types of noncoding RNA (ncRNA) genes can be detected by comparison to covariance models (55) and sequence-structure motifs (84). However, searching of covariance models and motifs is computationally expensive, and it is prohibitively long for large metagenomic data sets. Overall, the identification of other ncRNA genes is difficult, since their sequences are not conserved and reliable “ab initio” methods are lacking even for isolate genomes. High-throughput transcript (cDNA) sequencing holds great promise for improving the accuracy of RNA gene prediction. Currently, genes encoding ncRNAs are largely excluded from downstream analyses; however, we may expect this situation to change in the coming years as transcriptomic data enrich our inventories of these genes.
There are several types of errors that can be made by a gene-calling pipeline. A gene can be missed completely or called on the wrong strand. A less severe mistake would call part of the gene correctly but fail in estimating gene boundaries or call genes that are partly correct and partly wrong due to chimeric assemblies or frameshifts (94). The quality of the gene prediction relies on the quality of read preprocessing and assembly. Gene-calling methods used on accurately assembled sequences predict more than 90% of the genes that are included in the data set correctly, as evidenced from studies of simulated data sets (http://fames.jgi-psf.org). This large number was achieved with training on generic models or self-trained algorithms. Gene prediction on unassembled reads exhibits lower accuracy than that on contigs (∼70% versus >80%, respectively) (94), a result attributed to the small size and greater chance of sequencing errors for individual reads.
Often, even in low-complexity communities, a large number of reads belonging to less abundant organisms remain unassembled. Although the genes predicted on the assembled sequences allow the metabolic reconstruction of the abundant organisms, a better representation of the metabolic capacity of the community is gained when genes from both contigs and reads are included in the subsequent analyses as a majority of the functionality may in fact be encoded in the unassembled reads (94). Therefore, it is advisable to perform gene calling on both reads and contigs. For high-complexity communities, where assembly is minimal, gene calling on unassembled reads is the only possibility.
Gene prediction is usually followed by functional annotation. Functional annotation of metagenomic data sets is very similar to genomic annotation and relies on comparisons of predicted genes to existing, previously annotated sequences. The goal is to propagate accurate annotations to correctly identified othologs. However, there are additional complications in metagenomic data where predicted proteins are often fragmented and lack neighborhood context. The annotation of metagenomic data created by short-read methods, such as 454, is even more complicated since most reads contain only fractions of proteins.
At the JGI, we use profile-to-sequence searches to identify functions. Protein sequences are compared to sequence alignment profiles from the protein families TIGRFAM (122), PFAM (43), and COGs (131) using RPS-BLAST (91). PFAMs allow the identification and annotation of protein domains. TIGRFAMs include models for both domain and full-length proteins. COGs also allow the annotation of the full-length proteins. Unfortunately, although PFAMs and TIGRFAMs are updated regularly, allowing the annotation of new protein families, COGs are still lacking such updates. As a rule, the assignment of protein function solely based on BLAST results should be avoided, mainly because of the potential for error propagation through databases (49, 75, 78).
In addition to annotation by homology, several methods for context annotation are available. These include genomic neighborhood (30, 103), gene fusion (38, 86), phylogenetic profiles (106), and coexpression (87). We are aware of one study that performed adapted neighborhood analysis on metagenomic data, which, combined with homology searches, inferred specific functions for 76% of the metagenomic data sets (83% when nonspecific functions are considered) (59). It is possible that more context information will be used to predict protein function in metagenomic data in the future.
It is common practice that all gene predictions and annotations for microbial genomes are manually checked as part of informatic QC pipelines. Such manual curation is not feasible for metagenomic projects, although, as for the assembly, we recommend manual curation of larger contigs. Therefore, the quality of gene calling and annotation for the majority of metagenomic data rests solely on automated procedures. A recent benchmarking study using simulated metagenomic data sets suggests that there is significant room for improvement in existing gene prediction and annotation tools (94). One final note of caution: some vector-screening and -trimming programs only mask out rather than remove vector and low-quality sequences, resulting in runs of N′s at the ends of reads and contigs. When sequences are submitted to public databases, terminal runs of N′s are removed as part of the submission process, which can introduce systematic errors in the start-stop coordinates of any genes predicted on the untrimmed reads and contigs. Therefore, all reads and contigs should be trimmed of terminal N runs prior to gene prediction and annotation.
DATA ANALYSIS
Gene prediction and annotation complete the list of procedures that are routinely applied to both genomic and metagenomic data. While there is still great room for improvement in applying a number of these steps to metagenomic data, they constitute part of the standard data-processing pipeline at sequencing centers such as the JGI. Beyond this point, the data analysis methods apply specifically to metagenomes.
Postsequencing Community Composition Estimates
One of the first analyses that can be performed on metagenomic data according to standard processing steps is a reevaluation of the community composition estimate, this time directly from the metagenomic data itself. This is important for interpretations of the data since biases in the initial estimates, such as PCR skewing (130, 143), are different from biases introduced during metagenomic data generation (described below). Mapping of conserved phylogenetically informative marker genes such as 16S and 23S rRNA (rRNAs), RecA (DNA repair protein), EF-Tu, EF-G (elongation factors), HSP70 (heat shock protein), and RpoB (RNA polymerase subunit) onto their reference trees has been used to assess both organism identity and relative abundance (140). Single-copy, mostly ribosomal, genes have been applied for the same purpose (23, 47, 141). Ubiquitous single-copy genes have the advantage of being present once in all microbial genomes and are therefore thought to provide more accurate estimates of community composition than markers such as 16S rRNA genes with a variable copy number (141).
Marker gene analyses are performed as follows. An alignment of each gene is prepared from a reference data set, usually from all available complete genomes. The marker genes are identified in the metagenomic data set of interest and included in the reference alignment. For the quantification of populations, the depth of contigs containing the marker genes should be taken into account (135, 142). Trees are calculated, and the relative positions of metagenomic genes are identified in the tree. There are several limitations to community composition estimates based on the phylogenetic inference of single-copy genes identified in metagenomic data sets.
First, the reference genome database is currently incomplete and highly biased toward just three bacterial phyla (Proteobacteria, Firmicutes, and Actinobacteria) out of at least 50 phyla (62). This means that the accurate placement of metagenomic genes is compromised if they originate from organisms not belonging to the three well-represented phyla, with the exception of the 16S rRNA gene, which is broadly used to define taxonomic groups (34). Initiatives to improve the genome sequence representation of the tree of life should help to rectify this problem, such as the Genomic Encyclopedia of Bacteria and Archaea pilot project at the JGI (http://www.jgi.doe.gov/programs/GEBA/). Even so, the majority of microbial lineages still lack cultured representatives (3, 62), complicating our ability to obtain representative genome sequences. Another strategy to improve the reference database for comparative analyses is to obtain genome sequences of isolates in parallel with metagenome sequences from the same habitat, as is being done, for example, in the Human Microbiome Project (http://nihroadmap.nih.gov/hmp/).
Second, genes derived from metagenomic data sets, particularly those with minimal assembly, are often fragmented and produce incomplete alignments. Indeed, it is often the case that metagenomic gene fragments from the same protein family are entirely nonoverlapping. This precludes the use of evolutionary distance methods, as infinite distances are created in the pairwise distance matrix, severely compromising the resulting tree (18). Discrete character inference methods, particularly maximum likelihood, can tolerate incomplete alignments to a certain extent. Alternative approaches to address the problem include making separate trees for each metagenomic gene only in the context of the reference data set, subdividing the alignment into smaller parts to produce more complete subalignments that can still contain multiple metagenome-derived genes, or inserting partial sequences into a reference tree of full-length sequences using, for example, probabilistic maximum likelihood placement (142) or the ARB parsimony insertion tool (83).
Third, erroneous gene calls, particularly ribosomal proteins, are sometimes missed by automatic gene callers because of their small size (94).
Finally, and perhaps most importantly, conserved phylogenetically informative genes represent only a small fraction of the total metagenomic data set. For example, 100 Mbp of Sanger sequence will typically yield about a dozen mostly partial-length sequences of any given marker gene. In addition, it has recently come to light that single-copy genes are particularly prone to underrepresentation in shotgun libraries due to their toxicity to the E. coli host (127). Furthermore, since the toxicity is due to the expression of the introduced gene, it varies between organisms depending on the ability of E. coli to transcribe and translate the introduced gene (127). Therefore, small numbers of incompletely overlapping marker sequences, together with the toxicity effect, compromise the ability to reliably infer community composition from single-copy genes.
Sequence similarity tools such as BLAST (7) can be used to identify homologs in reference sequences (64). Such an analysis results in a much greater fraction of the data set being involved in the composition estimate but suffers from other effects. Potentially, larger genomes are expected to generate more matches than smaller genomes (125), and therefore, the assessment is of gene rather than organism abundance. The closest BLAST hit is not necessarily the nearest phylogenetic neighbor (72), and therefore, classifying by BLAST hits can be misleading, particularly if only distantly related homologs are available in the reference database. Additionally, the potential for horizontal gene transfer between sympatric populations can cause the recipient organism to be identified as the donor organism. Presently, the biggest problem for BLAST-based composition estimation is the poor representation of microbial diversity by sequenced isolates (62, 66), often resulting in remote matches to phylogenetically distant organisms or the absence of any hits. In our experience, BLAST-based methods overestimate the abundance of highly covered taxa such as the Proteobacteria and Firmicutes, especially if only the top hit is taken into consideration. One recent implementation of BLAST-based community composition profiling, MEGAN (64), addresses this problem by assigning sequence fragments to the lowest common ancestor of the set of taxa that it hit in the comparison, thereby reducing false matches. Unfortunately, this often results in the bulk of a data set being assigned to very-high-level groupings, such as Bacteria, or being unclassified altogether. Again, the problem lies with the reference genome database rather than the tool and can be expected to improve as the bias in the database is addressed.
Finally, given that fundamental upstream processes such as DNA extraction can produce an equal or greater skewing of community representation as any bioinformatic analysis, researchers should, if possible, calibrate their data against the original intact community using methods such as 16S rRNA-targeted FISH.
Binning
A metagenomic sequence pipeline produces a collection of reads, contigs, and genes. Associating these data with the organisms from which they were derived is highly desirable for the interpretation of the ecosystem. This process of association between sequence data and contributing species (or higher-level taxonomic groups) is called binning or classification. The most reliable binning is assembly; that is, in a good assembly, all reads in a contig are derived from the same species, with the optimal binning being a closed chromosome. As described above, this is often not the case, and some level of coassembly is usually encountered in metagenomic data sets, particularly between strains (see “Assembly”). However, binning methods rarely have the resolution to discriminate between strains of the same species, so strain coassembly is not a practical concern when it comes to binning. In fact, a much coarser level assignment of sequences can be useful for interpreting microbial communities, such as the classification of fragments from a termite hindgut analysis into two dominant class-level groups, the treponeme spirochetes and fibrobacter-like bacteria, with each group comprising numerous but functionally related species (146). In this regard, less stringent “extreme” assemblies (115), which certainly produce chimeric and misassembled contigs, may be a useful binning approach.
In many ways, binning and community composition estimates share a common goal, the classification of sequence data into taxonomic groups, and so there is overlap in the methods to achieve this goal. Phylogenetic marker genes can be used to bin sequence fragments, but this approach suffers from the same problems as those in community profiling, namely, an incomplete and biased reference database, difficulties with tree building, and a low overall incidence of marker genes (∼1%) in the metagenomic data set. Similarly, sequence comparison and visualization tools such as BLAST and MEGAN (64) can also be used to bin a larger cross section of sequence fragments to phylogenetic groups, with the associated problems described above.
An entirely different binning approach is based on genome sequence composition. Cellular processes such as codon usage, restriction-modification systems, and DNA repair mechanisms produce sequence composition signatures, primarily oligonucleotide (word) frequencies, that are distinct in different genomes (35, 69, 70). This property of genomes has been exploited by a variety of methods to identify groups of sequences with similar composition features and to determine their phylogenetic origins (2, 29, 98, 117, 132), which can be used not only to bin metagenomic data but also to identify atypical regions within genomes, such as laterally transferred genes. The words can be of any length, usually from 1 (GC content) to 4 nucleotides and usually no longer than 8 nucleotides. Typically, longer words give better resolution but also require longer sequences and are more computationally expensive, with the best results being provided by words between 3 and 6 nucleotides long.
Composition-based methods can be divided into supervised and unsupervised (clustering) procedures. Unsupervised procedures cluster metagenomic fragments in composition signature space without the need to train models on reference sequences and include self-organizing maps (1) and the program TETRA (132). An advantage of unsupervised classification is that phylogenetically novel populations lacking close or even distantly related sequenced taxa can potentially be binned by shared sequence composition features, although the identification of the clustered fragments still relies on sequence similarity to reference organisms. Such populations, even when well represented in metagenomes, cannot be binned directly by homology-based methods. A drawback of unsupervised methods is that they tend to focus on major classes in a data set and will not perform well on low-abundance populations. Supervised methods classify metagenomic fragments against models trained on classified reference sequences and, in principle, can assign fragments from low-abundance populations if there is a model learned from reference data. Examples of supervised approaches include Bayesian classifiers (29) and the support vector machine-based phylogenetic classifier Phylopythia (95). As they are able to learn the relevant features that distinguish a particular population from others using the labeled reference sequences, supervised methods usually achieve higher classification accuracy (sensitivity and specificity) than unsupervised methods and, therefore, are preferable if training data are available. Further details on the underlying principles and relative merits of different binning methods can be found in a recent opinion article on metagenomic binning (96).
At the JGI, we have had most experience with the supervised classifier Phylopythia (95). This program uses generic or sample-specific models, with the former usually being derived from reference genomes and the latter usually being derived from the metagenomic data set itself. Perhaps not surprisingly, sample-specific models based on training data from the metagenome under study produced the most specific and sensitive binning of the available approaches as determined by simulated data sets (94) or the subsequent assembly of the targeted population (95), often increasing the amount of classified sample data by an order of magnitude over the training set. Ideally, at least 100 kbp of training data is required to make a sample-specific model (96). For dominant populations, this amount of target population data can often be found using a single phylogenetic marker gene identified on a large contig that can be extended to other contigs by mate pair information. For low-abundance populations, identifying 100 kbp of training data may not be possible based on marker genes, particularly if the population is not closely related to sequenced reference genomes. However, higher-level taxonomic models may still be feasible in which multiple species contribute to the training set. This approach was used successfully for the sample-specific binning of treponeme spirochete species that were collectively the dominant group in a termite hindgut symbiont community (146).
Sequence length is a critical parameter for all composition-based classifiers, with no method convincingly classifying sequences of less than 1 kb long due to the limited number of words that are contained in short sequences (96). This precludes the classification of individual Sanger and pyrosequence reads, meaning that largely or completely unassembled complex communities cannot be binned at all by composition-based methods.
Finally, simulations of fractionating a community into even-course subsets of component species prior to sequencing suggest that the overall proportion of assembled sequences will be greater (15), thereby simplifying the binning process.
Analyzing Dominant Populations
In several aspects, the analysis of low-complexity communities resembles the analysis of isolate genomes. As with isolate genomes, draft-level composite genomes of dominant populations have sufficient coverage and gene context to allow a reasonably comprehensive metabolic reconstruction in which most major pathways can be elucidated. If more than one dominant population is sequenced, the potential metabolic interplay of those populations may also become apparent. For example, a metagenomic study of an acid mine drainage biofilm revealed that while all dominant bacterial and archaeal populations were potentially capable of iron oxidation (the main energy-generating reaction in this habitat), only Leptospirillum group III had genes for nitrogen fixation, suggesting a keystone function for this species since the habitat is limited in externally derived fixed nitrogen (138). Similarly, a metabolic reconstruction of the dominant bacterial symbiont populations in a gutless worm suggested a model for how these organisms together satisfy the nutritional requirements of their host (150). As with draft genomes of isolates, caution needs to be exercised in inferring the absence of metabolic traits since the relevant genes may be present in sequencing gaps, particularly if the trait is encoded by only one or two genes. For example, respiratory nitrate reductase necessary for denitrification was not found in the draft composite population genome of “Candidatus Accumulibacter phosphatis” type II despite circumstantial experimental evidence, suggesting that this organism is capable of denitrification (47).
The major difference between isolate genomes and composite dominant population genomes is that the latter are usually not clonal due to genetic variation inherent in natural populations (148). Genomic differences between individuals and strains within a population can take the form of SNPs and rearrangements (insertions, deletions, inversions, transitions, and duplications). The coassembly of genetically distinct strains (haplotypes) will produce high-quality discrepancies (SNPs) in the consensus that finishing would normally try to resolve. However, in metagenomic data sets, SNPs can be mined in a number of ways to provide insights into population structure and evolution. For example, total SNP frequency provides a quantitative estimate of the degree of genetic variation within a species population, which has been found to range from virtually clonal in enrichment cultures (129) and an anaerobic digester (107) to highly polymorphic in acid mine drainage archaeal populations (138). The ratio of nonsynonymous to synonymous SNPs in protein-coding genes within a population provides an estimate of the fraction of genes under selective pressure. Furthermore, the ratio of haplotypes for individual SNPs (site frequency spectra) can be used to estimate important parameters in population genetics such as the scaled mutation rate and scaled exponential growth rate (68). SNPs also highlight junctions of homologous recombination between strains, allowing the degree of sexuality within a population to be estimated (148). In all cases, the clear advantage of using environmental shotgun sequence data for these analyses over isolate sequence data is a broader and less biased sampling of genetic variation within a population (4, 148).
A complication associated with interpreting these data is sequencing error. Setting base quality thresholds too low will introduce noise into the analysis, while setting it too high will discard potentially useful information. The latter may be an important consideration when read depth is low. A conservative approach to avoid mistaking errors as polymorphisms is to score only SNPs with haplotypes represented by at least two different reads requiring a minimum read depth of 4. A second complication is the inability to easily distinguish between orthologous and paralogous regions. Unless repeats occur on the same (manually verified) contig or scaffold, such as in the case of a neighboring gene duplication, it is difficult to distinguish repeats from orthologous regions in different organisms. This problem is alleviated if the composite population is finished.
Several tools are available for the visualization and analysis of polymorphisms in composite population assemblies. Consed, developed to assist in the finishing process, is a generically useful graphical tool for viewing assemblies at the nucleotide level (53). A note of caution, however, is that Consed sometimes masks stretches of nucleotide sequence with X's, and when SNP analysis is performed, it identifies these X characters as being SNPs. Therefore, manual postprocessing is required for Consed results. SNP-VISTA (124) is an adaptation of the comparative genomics tool VISTA (44), developed specifically to visualize SNPs in alignments. The input for this program is the BLASTn output for user friendliness. Reads are ordered by haplotype using a clustering algorithm calculated for sliding windows. Putative recombination sites are detected by sudden changes in cluster composition between adjacent windows (Fig. 5). Strainer is also dedicated software for the analysis of genetic variation in populations (39). As the name suggests, it facilitates the reconstruction of individual strains from coassembled sequences, clusters reads by haplotype from which it predicts gene and protein variants, identifies conserved regulatory sequences, and quantifies and displays homologous recombination sites along contigs.
FIG. 5.
Screenshot of SNP-VISTA showing SNPs in individual reads relative (and aligned) to a reference contig belonging to “Candidatus Accumulibacter phosphatis” (74) (labeled query at the bottom and highlighted in pale green). (Top) Alignment condensed to show only polymorphic columns color coded by base (see left for color coding). (Bottom) Expanded alignment. Note that reads are ordered dynamically by similarity for the window under investigation to facilitate SNP pattern recognition.
As for fine-scale genetic variation, methods for visualizing and analyzing gross within-population variation caused by rearrangements are beginning to emerge. For example, recruitment plots display alignments of environmental reads against a reference sequence such as an isolate genome, with one axis showing read location along the reference and the other axis showing sequence identity to the reference. The depth of alignment at each point is a measure of the frequency of occurrence of the particular genomic region. Genomic regions that are present in all members of the species will be covered by multiple reads, while strain-specific regions will have shallow or no coverage (Fig. 6), effectively highlighting hypervariable regions in a population. A number of important biological insights have been made using this type of analysis, including the discovery of genomic islands encoding ecologically important genes (26), and phage defense mechanisms, notably CRISPRs, are among the fastest-evolving elements in the genome (137).
FIG. 6.
Screenshot of JCVI's Advanced Reference Viewer (http://gos.jcvi.org/openAccess/advancedReferenceviewer.html). A reference contig or genome, in this case, Prochlorococcus marinus strain AS9601, shown on the x axis, against which metagenomic reads, in this case, from the Global Ocean Survey (115), is aligned and arrayed by similarity to the reference sequence on the y axis. Reads have been color coded according to sampling site to highlight site-to-site variations in Prochlorococcus populations but can be color coded by any type of metadata or other features such as the consistency of read mate pairs. Genomic islands peculiar to strain AS9601 are easily identified as gaps in the read coverage (between 60 and 70 kb). This viewer also allows users to zoom into regions of interest for higher resolution. (Image courtesy of Doug Rusch.)
Recruitment plots can be enhanced by displaying data from multiple metagenomes against a reference sequence distinguished by color coding. This is particularly effective for spatial series where differences between allopatric populations can be highlighted and correlated with metadata (115). Rearrangements such as inversions or indels can be specifically visualized using a variant of recruitment plotting. Instead of plotting all reads, only reads with inconsistently distanced end pairs are shown, which draws attention to rearrangements (115). Similarly, individual reads that do not map 1:1 onto the reference genome can be plotted to highlight inversion, insertion, or deletion boundaries. As has been discussed in the context of several other analyses, recruitment plots can be limited by the availability of reference genomes unless reference sequences are forthcoming from the metagenomic data set itself.
Gene-Centric Analysis
Metagenomic sequencing of high-complexity microbial communities results in little or no assembly of reads (135), which precludes the use of microheterogeneity analyses described above for dominant populations. The high coding density of bacterial and archaeal genomes and average gene size do, however, mean that most reads will capture a coding sequence. This allows a gene-centric analysis of the data that treats the community as an aggregate, largely ignoring the contribution of individual species. Genes and gene fragments from a given metagenomic data set are mapped to gene families, providing an estimate of relative representation (Fig. 7). The power of the method lies in comparing relative gene family or subsystem abundances between metagenomes to highlight functional differences. Since determining relative gene family frequencies within and between metagenomic data sets is a key aspect of the method, it is important that the frequencies are not masked by assembly. Either the analysis should be conducted on unassembled reads or the read depth of contigs should be taken into account (94). The approach was first described by Tringe et al. (134, 135), in which they coined the term environmental gene tags because of the fragmentary nature of the data, akin to expressed sequence tags. Other groups published similar but distinct approaches in quick succession (50, 33, 113).
FIG. 7.
Screenshot (at left) from the IMG/M database (91) showing one implementation of gene-centric analysis available through this system. Four PFAM families involved in cellulose hydrolysis are shown in columns color coded to match the pathway schematic to the right. The relative representation of these families in 12 metagenomic data sets (rows) is shown as fractions normalized for data set size. Overrepresented families are further highlighted by color: bisque, moderately overrepresented; yellow, highly overrepresented. This figure shows that termite hindgut followed by human gut samples have the greatest overrepresentation of genes involved in cellulose hydrolysis and, indeed, are the only communities of the compared data sets that appear to have the enzymatic potential to break down cellulose. It also shows that one whale fall sample, a soil sample from the drainage path of a silage storage bunker, and one laboratory-scale phosphorus-removing sludge sample are moderately overrepresented in genes for processing the dimer cellobiose. (Image courtesy of Falk Warnecke.)
The implicit assumption of gene-centric analysis is that high relative abundance equates to metabolic and ecological significance. Knowledge of the ecosystem is required for simple sanity checks. For example, one of the most overrepresented gene families in ocean surface waters relative to soil and whale fall (deep ocean) samples is the proteorhodopsin family, which function as light-driven proton pumps (134), a function that is receiving great attention as a major missed energy flux in surface waters (116). A recent RNA-based study of a picoplankton community in the photic zone confirmed that proteorhodopsins are indeed highly expressed; however, other overrepresented gene families, such as DNA repair photolyase, were not highly expressed, bringing into question the metabolic or ecological significance of their high copy numbers in the community (45). Conversely, other gene families that were poorly represented in the metagenomic data, such as pufB, encoding a subunit of a light-harvesting protein, were highly expressed (45), indicating that potentially important functions will be overlooked or underestimated by DNA-based gene-centric analysis. In addition to expression levels, other factors such as the stability of mRNAs and proteins are likely important determinants of ecological significance.
In addition to violations of the implicit assumption, the method has a number of technical limitations. Chen and Pachter estimated that 6 Gbp of sequence data would be required to sample half the genes in a simulated soil community (21), whereas a typical metagenome project is on the order of 100 Mbp. Therefore, only genes present in high copy numbers in higher-abundance organisms will be sampled, meaning that the method is actually very low resolution. Environmental gene tag data are also noisy due to the uneven cloning efficiency of different genes (127), differences in gene length (longer genes will be detected more often on reads than short genes), and errors in gene calling and annotation. A more pervasive problem may be the inability to normalize gene prediction between data sets. For example, read length will affect the ability to call genes: the shorter the read, the lower the gene prediction resolution. Therefore, Sanger (∼750-bp reads) and pyrosequence (100- to 200-bp reads) data sets cannot be directly compared using gene-centric analysis because of the differences in gene-calling sensitivities between the two data types (149). A final word of caution on technical considerations: whole-genome amplification of environmental DNAs is becoming a more common method, particularly for low-biomass microbial communities (9, 36). Several studies have shown that although some degree of bias is introduced by multiple-strand-displacement whole-genome amplification using Phi29 DNA polymerase, it has sufficient fidelity to allow meaningful comparative analyses in most instances (10, 12, 110). However, the amplification step should be kept in mind when interpreting gene-centric analyses, particularly between amplified and nonamplified data sets.
To differentiate between signal and noise, statistical tests to estimate the confidence of over- and underrepresentations of gene families have been reported (50, 113). Despite these statistical reassurances, simulated metagenomic data sets show that up to 20% of COGs may have incorrect frequency calls and should be interpreted with caution (94). However, the error rate is reduced when gene family frequencies are grouped by metabolic pathway, because error in any given gene family will be averaged out in a multigene pathway. One important potential source of error when gene family frequencies are mapped onto pathways is an uneven coverage of the pathway. For example, broad gene families such as oxidoreductases can be nonspecifically mapped to a pathway via incomplete EC numbers and give the false appearance that the pathway is overrepresented. In the extreme case, the pathway may be entirely absent from the community, and only the nonspecific gene family is mapped to the pathway. This type of error can be overcome by weighting pathways for gene coverage or excluding incomplete EC numbers from the analysis. In addition, to avoid spurious prediction, there is no substitution for manual inspection by experts of all results obtained by automatic data mining.
DATA MANAGEMENT
Shotgun sequencing of environmental samples produces massive amounts of data that already dwarf the data for existing genomic sequences in public databases. This trend will not only continue but accelerate as the cost of sequencing continues to fall and more researchers enter into the field, drawn by the promise of metagenomics and greater access to high-throughput sequencing via new sequencing technologies. For the average researcher to make sense of this mountain of data, dedicated data management resources are required. There is a variety of Web-based and standalone computational resources available for comparative genomic analyses including ACT (20), MicrobesOnLine (6), CMR (109), ERGO (104), PUMA2 (85), COGENT++ (52), and IMG (92), but data management systems have only recently been developed specifically for metagenomic analysis, notably CAMERA (123), IMG/M (91), and SEED (102).
These systems allow the comparison of a metagenome of interest to other genomes and metagenomes on multiple levels, including at the gene, protein family, pathway, scaffold, or complete genome level, and all systems include variants of the metagenome-specific tools described in the preceding sections (90). Most systems also allow some degree of curation by users to improve annotation. Although the same type of analyses can be performed without the aid of such systems, prepackaged tools with transparent user interfaces can save considerable amounts of time even for expert users. Custom analyses need to be performed externally, and the main use of dedicated metagenomic databases in these cases is improved curation over generic databases.
It is fair to say that all developers of metagenomic data management and analysis systems are struggling to keep pace with new data. This acute problem is manifest at two levels. The first level is data volume. Genomic data are more compressed than metagenomic data by virtue of assembly, and underlying read data are typically not incorporated into comparative genome systems. In contrast, some metagenomic systems keep not only read information but also quality data associated with reads for population analysis and QC. The problem is expected to accelerate in the future as new sequencing technologies produce much larger volumes of data than traditional Sanger sequencing. For example, a single Illumina run produces ∼1 Gbp of sequence data (albeit short reads of <50 bp), compared to only 0.7 Mbp of ∼750-bp reads for a standard Sanger run. While trace quality information may be important for quality assessment, their storage together with the sequence and incorporation of quality information into sequence search methods might not be feasible. The second level is pairwise comparisons. The cornerstone of comparative analysis is all-against-all comparisons. Ideally, these should be precomputed to prevent lengthy on-the-fly calculations for users. Unfortunately, all-against-all comparisons scale poorly (quadratically) and can become extremely computationally expensive for metagenomic data. For example, 28.6 million protein sequences were compared using all-against-all BLAST searches in the Global Ocean Survey study, which required more than 1 million computing hours (151). The sheer size of the computational effort needed for this metagenomic data set was unprecedented in sequence analysis. A parallelized implementation of BLAST, ScalaBLAST (101), is used to precompute all pairwise gene similarities at the amino acid level for IMG/M, reducing the computation time by ∼30-fold (90). ScalaBLAST uses a combination of database sharing and task scheduling to achieve high computational performance (101). Computationally intensive tasks can also be bypassed by profile scans using profile databases such as TIGRFAM, PFAM, COGS, and InterProScan. Because the number of profiles is constant, computational complexity scales linearly with the growth of the data, as opposed to quadratically in the case of all-against-all comparisons. One drawback of profile searches is that new families will not be identified, but such novel families will have unknown functions (hypothetical families) and will not contribute to metabolic reconstruction efforts in the first instance.
It remains to be seen if any data management system will be capable of incorporating all metagenomic data and present the data in a precomputed format for comparative analyses. More likely, subsets of the data united by common phylogenetic or functional themes will be made into separate databases for analyses.
The final stage of any sequencing project is the submission of the data to public repositories such as GenBank. Metagenomic data submission is more problematic than isolate genome submission because it is usually not discrete. For example, should a metagenomic data set be described as a single entry or as multiple entries? On one hand, the data are a collection of sequence fragments from multiple species, which argues for multiple entries. On the other hand, there is often a single sampling site and a single study performed on the sequence, although this too is changing as single studies incorporate spatial or temporal sampling. At the JGI, we submit the data as one entry, and, whenever possible, subdivide it into bins of organisms. For example, the metagenome of the Olavius algarvensis symbionts was submitted under accession number AASZ00000000, with scaffolds ranging between accession numbers AASZ01000001 and AASZ01005597. The scaffolds assigned to particular genome bins were then assigned to subaccession numbers, such as subaccession numbers DS021107 to DS021197 for the O. algarvensis gamma 1 symbiont.
CONCLUDING REMARKS
We hope that this review will serve as a useful primer for researchers embarking on their first metagenomic project. The field is moving forward rapidly, driven by enormous improvements in sequencing technology and the availability of many complementary technologies (145). We therefore anticipate that methodological details presented in this review will change markedly in the coming years or even months, particularly when Sanger sequencing is no longer the main source of metagenomic data. The discussed methodological considerations and approaches for analyzing communities and populations, however, will no doubt persist for much longer, enabling interpretations of metagenomic data sets and likely contributing many more profound insights into the microbial world.
Acknowledgments
We thank Alice McHardy, Susannah Tringe, Tanja Woyke, and Gene Tyson for useful input and feedback during the course of preparing this review. We also thank three anonymous reviewers for constructive criticism and overall bravery in agreeing to review such a long article.
This work was performed under the auspices of the Biological and Environmental Research Program of the U.S. Department of Energy's Office of Science and by the University of California Lawrence Berkeley National Laboratory under contract no. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under contract no. DE-AC52-07NA27344, and Los Alamos National Laboratory under contract no. DE-AC02-06NA25396.
REFERENCES
- 1.Abe, T., S. Kanaya, M. Kinouchi, Y. Ichiba, T. Kozuki, and T. Ikemura. 2003. Informatics for unveiling hidden genome signatures. Genome Res. 13693-702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abe, T., H. Sugawara, M. Kinouchi, S. Kanaya, and T. Ikemura. 2005. Novel phylogenetic studies of genomic sequence fragments derived from uncultured microbe mixtures in environmental and clinical samples. DNA Res. 12281-290. [DOI] [PubMed] [Google Scholar]
- 3.Achtman, M., and M. Wagner. 2008. Microbial diversity and the genetic nature of microbial species. Nat. Rev. Microbiol. 6431-440. [DOI] [PubMed] [Google Scholar]
- 4.Allen, E. E., and J. F. Banfield. 2005. Community genomics in microbial ecology and evolution. Nat. Rev. Microbiol. 3489-498. [DOI] [PubMed] [Google Scholar]
- 5.Allen, E. E., G. W. Tyson, R. J. Whitaker, J. C. Detter, P. M. Richardson, and J. F. Banfield. 2007. Genome dynamics in a natural archaeal population. Proc. Natl. Acad. Sci. USA 1041883-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alm, E. J., K. H. Huang, M. N. Price, R. P. Koche, K. Keller, I. L. Dubchak, and A. P. Arkin. 2005. The MicrobesOnline Web site for comparative genomics. Genome Res. 151015-1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amann, R., B. M. Fuchs, and S. Behrens. 2001. The identification of microorganisms by fluorescence in situ hybridisation. Curr. Opin. Biotechnol. 12231-236. [DOI] [PubMed] [Google Scholar]
- 9.Angly, F. E., B. Felts, M. Breitbart, P. Salamon, R. A. Edwards, C. Carlson, A. M. Chan, M. Haynes, S. Kelley, H. Liu, J. M. Mahaffy, J. E. Mueller, J. Nulton, R. Olson, R. Parsons, S. Rayhawk, C. A. Suttle, and F. Rohwer. 2006. The marine viromes of four oceanic regions. PLoS Biol. 4e368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arriola, E., M. B. Lambros, C. Jones, T. Dexter, A. Mackay, D. S. Tan, N. Tamber, K. Fenwick, A. Ashworth, M. Dowsett, and J. S. Reis-Filho. 2007. Evaluation of Phi29-based whole-genome amplification for microarray-based comparative genomic hybridisation. Lab. Investig. 8775-83. [DOI] [PubMed] [Google Scholar]
- 11.Badger, J. H., and G. J. Olsen. 1999. CRITICA: coding region identification tool invoking comparative analysis. Mol. Biol. Evol. 16512-524. [DOI] [PubMed] [Google Scholar]
- 12.Ballantyne, K. N., R. A. van Oorschot, I. Muharam, A. van Daal, and R. J. Mitchell. 2007. Decreasing amplification bias associated with multiple displacement amplification and short tandem repeat genotyping. Anal. Biochem. 368222-229. [DOI] [PubMed] [Google Scholar]
- 13.Batzoglou, S., D. B. Jaffe, K. Stanley, J. Butler, S. Gnerre, E. Mauceli, B. Berger, J. P. Mesirov, and E. S. Lander. 2002. ARACHNE: a whole-genome shotgun assembler. Genome Res. 12177-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beja, O., M. T. Suzuki, E. V. Koonin, L. Aravind, A. Hadd, L. P. Nguyen, R. Villacorta, M. Amjadi, C. Garrigues, S. B. Jovanovich, R. A. Feldman, and E. F. DeLong. 2000. Construction and analysis of bacterial artificial chromosome libraries from a marine microbial assemblage. Environ. Microbiol. 2516-529. [DOI] [PubMed] [Google Scholar]
- 15.Bergeron, A., M. Belcaid, G. F. Steward, and G. Poisson. 2007. Divide and conquer: enriching environmental sequencing data. PLoS ONE 2e830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Besemer, J., and M. Borodovsky. 1999. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 273911-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breitbart, M., P. Salamon, B. Andresen, J. M. Mahaffy, A. M. Segall, D. Mead, F. Azam, and F. Rohwer. 2002. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. USA 9914250-14255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brocchieri, L. 2001. Phylogenetic inferences from molecular sequences: review and critique. Theor. Popul. Biol. 5927-40. [DOI] [PubMed] [Google Scholar]
- 19.Brodie, E. L., T. Z. Desantis, D. C. Joyner, S. M. Baek, J. T. Larsen, G. L. Andersen, T. C. Hazen, P. M. Richardson, D. J. Herman, T. K. Tokunaga, J. M. Wan, and M. K. Firestone. 2006. Application of a high-density oligonucleotide microarray approach to study bacterial population dynamics during uranium reduction and reoxidation. Appl. Environ. Microbiol. 726288-6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carver, T. J., K. M. Rutherford, M. Berriman, M. A. Rajandream, B. G. Barrell, and J. Parkhill. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 213422-3423. [DOI] [PubMed] [Google Scholar]
- 21.Chen, K., and L. Pachter. 2005. Bioinformatics for whole-genome shotgun sequencing of microbial communities. PLoS Comput. Biol. 1106-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chou, H. H., and M. H. Holmes. 2001. DNA sequence quality trimming and vector removal. Bioinformatics 171093-1104. [DOI] [PubMed] [Google Scholar]
- 23.Ciccarelli, F. D., T. Doerks, C. von Mering, C. J. Creevey, B. Snel, and P. Bork. 2006. Toward automatic reconstruction of a highly resolved tree of life. Science 3111283-1287. [DOI] [PubMed] [Google Scholar]
- 24.Cohan, F. M. 2002. What are bacterial species? Annu. Rev. Microbiol. 56457-487. [DOI] [PubMed] [Google Scholar]
- 25.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, A. S. Kulam-Syed-Mohideen, D. M. McGarrell, A. M. Bandela, E. Cardenas, G. M. Garrity, and J. M. Tiedje. 2007. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 35D169-D172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coleman, M. L., M. B. Sullivan, A. C. Martiny, C. Steglich, K. Barry, E. F. Delong, and S. W. Chisholm. 2006. Genomic islands and the ecology and evolution of Prochlorococcus. Science 3111768-1770. [DOI] [PubMed] [Google Scholar]
- 27.Culley, A. I., A. S. Lang, and C. A. Suttle. 2006. Metagenomic analysis of coastal RNA virus communities. Science 3121795-1798. [DOI] [PubMed] [Google Scholar]
- 28.Daims, H., S. Lucker, and M. Wagner. 2006. daime, a novel image analysis program for microbial ecology and biofilm research. Environ. Microbiol. 8200-213. [DOI] [PubMed] [Google Scholar]
- 29.Dalevi, D., D. Dubhashi, and M. Hermansson. 2006. Bayesian classifiers for detecting HGT using fixed and variable order Markov models of genomic signatures. Bioinformatics 22517-522. [DOI] [PubMed] [Google Scholar]
- 30.Dandekar, T., B. Snel, M. Huynen, and P. Bork. 1998. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. 23324-328. [DOI] [PubMed] [Google Scholar]
- 31.Delcher, A. L., D. Harmon, S. Kasif, O. White, and S. L. Salzberg. 1999. Improved microbial gene identification with GLIMMER. Nucleic Acids Res. 274636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeLong, E. F. 2005. Microbial community genomics in the ocean. Nat. Rev. Microbiol. 3459-469. [DOI] [PubMed] [Google Scholar]
- 33.DeLong, E. F., C. M. Preston, T. Mincer, V. Rich, S. J. Hallam, N. U. Frigaard, A. Martinez, M. B. Sullivan, R. Edwards, B. R. Brito, S. W. Chisholm, and D. M. Karl. 2006. Community genomics among stratified microbial assemblages in the ocean's interior. Science 311496-503. [DOI] [PubMed] [Google Scholar]
- 34.DeSantis, T. Z., P. Hugenholtz, N. Larsen, M. Rojas, E. L. Brodie, K. Keller, T. Huber, D. Dalevi, P. Hu, and G. L. Andersen. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 725069-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Deschavanne, P. J., A. Giron, J. Vilain, G. Fagot, and B. Fertil. 1999. Genomic signature: characterization and classification of species assessed by chaos game representation of sequences. Mol. Biol. Evol. 161391-1399. [DOI] [PubMed] [Google Scholar]
- 36.Edwards, R. A., B. Rodriguez-Brito, L. Wegley, M. Haynes, M. Breitbart, D. M. Peterson, M. O. Saar, S. Alexander, E. C. Alexander, Jr., and F. Rohwer. 2006. Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edwards, R. A., and F. Rohwer. 2005. Viral metagenomics. Nat. Rev. Microbiol. 3504-510. [DOI] [PubMed] [Google Scholar]
- 38.Enright, A. J., I. Iliopoulos, N. C. Kyrpides, and C. A. Ouzounis. 1999. Protein interaction maps for complete genomes based on gene fusion events. Nature 40286-90. [DOI] [PubMed] [Google Scholar]
- 39.Eppley, J. M., G. W. Tyson, W. M. Getz, and J. F. Banfield. 2007. Strainer: software for analysis of population variation in community genomic datasets. BMC Bioinformatics 8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Erkel, C., M. Kube, R. Reinhardt, and W. Liesack. 2006. Genome of Rice Cluster I archaea—the key methane producers in the rice rhizosphere. Science 313370-372. [DOI] [PubMed] [Google Scholar]
- 41.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8175-185. [DOI] [PubMed] [Google Scholar]
- 42.Field, D., G. Garrity, T. Gray, N. Morrison, J. Selengut, P. Sterk, T. Tatusova, N. Thomson, M. J. Allen, S. V. Angiuoli, M. Ashburner, N. Axelrod, S. Baldauf, S. Ballard, J. Boore, G. Cochrane, J. Cole, P. Dawyndt, P. De Vos, C. DePamphilis, R. Edwards, N. Faruque, R. Feldman, J. Gilbert, P. Gilna, F. O. Glockner, P. Goldstein, R. Guralnick, D. Haft, D. Hancock, H. Hermjakob, C. Hertz-Fowler, P. Hugenholtz, I. Joint, L. Kagan, M. Kane, J. Kennedy, G. Kowalchuk, R. Kottmann, E. Kolker, S. Kravitz, N. Kyrpides, J. Leebens-Mack, S. E. Lewis, K. Li, A. L. Lister, P. Lord, N. Maltsev, V. Markowitz, J. Martiny, B. Methe, I. Mizrachi, R. Moxon, K. Nelson, J. Parkhill, L. Proctor, O. White, S. A. Sansone, A. Spiers, R. Stevens, P. Swift, C. Taylor, Y. Tateno, A. Tett, S. Turner, D. Ussery, B. Vaughan, N. Ward, T. Whetzel, I. San Gil, G. Wilson, and A. Wipat. 2008. The minimum information about a genome sequence (MIGS) specification. Nat. Biotechnol. 26541-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frazer, K. A., L. Pachter, A. Poliakov, E. M. Rubin, and I. Dubchak. 2004. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 32W273-W279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Frias-Lopez, J., Y. Shi, G. W. Tyson, M. L. Coleman, S. C. Schuster, S. W. Chisholm, and E. F. Delong. 2008. Microbial community gene expression in ocean surface waters. Proc. Natl. Acad. Sci. USA 1053805-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Frishman, D., A. Mironov, H. W. Mewes, and M. Gelfand. 1998. Combining diverse evidence for gene recognition in completely sequenced bacterial genomes. Nucleic Acids Res. 262941-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia Martin, H., N. Ivanova, V. Kunin, F. Warnecke, K. W. Barry, A. C. McHardy, C. Yeates, S. He, A. A. Salamov, E. Szeto, E. Dalin, N. H. Putnam, H. J. Shapiro, J. L. Pangilinan, I. Rigoutsos, N. C. Kyrpides, L. L. Blackall, K. D. McMahon, and P. Hugenholtz. 2006. Metagenomic analysis of two enhanced biological phosphorus removal (EBPR) sludge communities. Nat. Biotechnol. 241263-1269. [DOI] [PubMed] [Google Scholar]
- 48.Gase, K., J. J. Ferretti, C. Primeaux, and W. M. McShan. 1999. Identification, cloning, and expression of the CAMP factor gene (cfa) of group A streptococci. Infect. Immun. 674725-4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gilks, W. R., B. Audit, D. De Angelis, S. Tsoka, and C. A. Ouzounis. 2002. Modeling the percolation of annotation errors in a database of protein sequences. Bioinformatics 181641-1649. [DOI] [PubMed] [Google Scholar]
- 50.Gill, S. R., M. Pop, R. T. Deboy, P. B. Eckburg, P. J. Turnbaugh, B. S. Samuel, J. I. Gordon, D. A. Relman, C. M. Fraser-Liggett, and K. E. Nelson. 2006. Metagenomic analysis of the human distal gut microbiome. Science 3121355-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldberg, S. M., J. Johnson, D. Busam, T. Feldblyum, S. Ferriera, R. Friedman, A. Halpern, H. Khouri, S. A. Kravitz, F. M. Lauro, K. Li, Y. H. Rogers, R. Strausberg, G. Sutton, L. Tallon, T. Thomas, E. Venter, M. Frazier, and J. C. Venter. 2006. A Sanger/pyrosequencing hybrid approach for the generation of high-quality draft assemblies of marine microbial genomes. Proc. Natl. Acad. Sci. USA 10311240-11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goldovsky, L., P. Janssen, D. Ahren, B. Audit, I. Cases, N. Darzentas, A. J. Enright, N. Lopez-Bigas, J. M. Peregrin-Alvarez, M. Smith, S. Tsoka, V. Kunin, and C. A. Ouzounis. 2005. CoGenT++: an extensive and extensible data environment for computational genomics. Bioinformatics 213806-3810. [DOI] [PubMed] [Google Scholar]
- 53.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8195-202. [DOI] [PubMed] [Google Scholar]
- 54.Grant, S., W. D. Grant, D. A. Cowan, B. E. Jones, Y. Ma, A. Ventosa, and S. Heaphy. 2006. Identification of eukaryotic open reading frames in metagenomic cDNA libraries made from environmental samples. Appl. Environ. Microbiol. 72135-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Griffiths-Jones, S., S. Moxon, M. Marshall, A. Khanna, S. R. Eddy, and A. Bateman. 2005. Rfam: annotating non-coding RNAs in complete genomes. Nucleic Acids Res. 33D121-D124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hallam, S. J., K. T. Konstantinidis, N. Putnam, C. Schleper, Y. Watanabe, J. Sugahara, C. Preston, J. de la Torre, P. M. Richardson, and E. F. DeLong. 2006. Genomic analysis of the uncultivated marine crenarchaeote Cenarchaeum symbiosum. Proc. Natl. Acad. Sci. USA 10318296-18301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hallam, S. J., N. Putnam, C. M. Preston, J. C. Detter, D. Rokhsar, P. M. Richardson, and E. F. DeLong. 2004. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 3051457-1462. [DOI] [PubMed] [Google Scholar]
- 58.Handelsman, J., M. R. Rondon, S. F. Brady, J. Clardy, and R. M. Goodman. 1998. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5R245-R249. [DOI] [PubMed] [Google Scholar]
- 59.Harrington, E. D., A. H. Singh, T. Doerks, I. Letunic, C. von Mering, L. J. Jensen, J. Raes, and P. Bork. 2007. Quantitative assessment of protein function prediction from metagenomics shotgun sequences. Proc. Natl. Acad. Sci. USA 10413913-13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huang, X., and A. Madan. 1999. CAP3: a DNA sequence assembly program. Genome Res. 9868-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huber, J. A., D. B. M. Welch, H. G. Morrison, S. M. Huse, P. R. Neal, D. A. Butterfield, and M. L. Sogin. 2007. Microbial population structures in the deep marine biosphere. Science 31897-100. [DOI] [PubMed] [Google Scholar]
- 62.Hugenholtz, P. 2002. Exploring prokaryotic diversity in the genomic era. Genome Biol. 3REVIEWS0003. [DOI] [PMC free article] [PubMed]
- 63.Huse, S. M., J. A. Huber, H. G. Morrison, M. L. Sogin, and D. M. Welch. 2007. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol. 8R143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huson, D. H., A. F. Auch, J. Qi, and S. C. Schuster. 2007. MEGAN analysis of metagenomic data. Genome Res. 17377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jaffe, D. B., J. Butler, S. Gnerre, E. Mauceli, K. Lindblad-Toh, J. P. Mesirov, M. C. Zody, and E. S. Lander. 2003. Whole-genome sequence assembly for mammalian genomes: Arachne 2. Genome Res. 1391-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Janssen, P., B. Audit, I. Cases, N. Darzentas, L. Goldovsky, V. Kunin, N. Lopez-Bigas, J. M. Peregrin-Alvarez, J. B. Pereira-Leal, S. Tsoka, and C. A. Ouzounis. 2003. Beyond 100 genomes. Genome Biol. 4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Johnson, P. L., and M. Slatkin. 2008. Accounting for bias from sequencing error in population genetic estimates. Mol. Biol. Evol. 25199-206. [DOI] [PubMed] [Google Scholar]
- 68.Johnson, P. L., and M. Slatkin. 2006. Inference of population genetic parameters in metagenomics: a clean look at messy data. Genome Res. 161320-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Karlin, S., and C. Burge. 1995. Dinucleotide relative abundance extremes: a genomic signature. Trends Genet. 11283-290. [DOI] [PubMed] [Google Scholar]
- 70.Karlin, S., J. Mrazek, and A. M. Campbell. 1997. Compositional biases of bacterial genomes and evolutionary implications. J. Bacteriol. 1793899-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Korlach, J., P. J. Marks, R. L. Cicero, J. J. Gray, D. L. Murphy, D. B. Roitman, T. T. Pham, G. A. Otto, M. Foquet, and S. W. Turner. 2008. Selective aluminum passivation for targeted immobilization of single DNA polymerase molecules in zero-mode waveguide nanostructures. Proc. Natl. Acad. Sci. USA 1051176-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koski, L. B., and G. B. Golding. 2001. The closest BLAST hit is often not the nearest neighbor. J. Mol. Evol. 52540-542. [DOI] [PubMed] [Google Scholar]
- 73.Krause, L., N. N. Diaz, D. Bartels, R. A. Edwards, A. Puhler, F. Rohwer, F. Meyer, and J. Stoye. 2006. Finding novel genes in bacterial communities isolated from the environment. Bioinformatics 22e281-e289. [DOI] [PubMed] [Google Scholar]
- 74.Kunin, V., S. He, F. Warnecke, S. B. Peterson, H. Garcia Martin, M. Haynes, N. Ivanova, L. L. Blackall, M. Breitbart, F. Rohwer, K. D. McMahon, and P. Hugenholtz. 2008. A bacterial metapopulation adapts locally to phage predation despite global dispersal. Genome Res. 18293-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kunin, V., and C. A. Ouzounis. 2005. Clustering the annotation space of proteins. BMC Bioinformatics 624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kunin, V., J. Raes, J. K. Harris, J. R. Spear, J. J. Walker, N. Ivanova, C. von Mering, B. M. Bebout, N. R. Pace, P. Bork, and P. Hugenholtz. 2008. Millimeter-scale genetic gradients and community-level molecular convergence in a hypersaline microbial mat. Mol. Syst. Biol. 4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kurokawa, K., T. Itoh, T. Kuwahara, K. Oshima, H. Toh, A. Toyoda, H. Takami, H. Morita, V. K. Sharma, T. P. Srivastava, T. D. Taylor, H. Noguchi, H. Mori, Y. Ogura, D. S. Ehrlich, K. Itoh, T. Takagi, Y. Sakaki, T. Hayashi, and M. Hattori. 2007. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 14169-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kyrpides, N. C., and C. A. Ouzounis. 1999. Whole-genome sequence annotation: ‘going wrong with confidence.’ Mol. Microbiol. 32886-887. [DOI] [PubMed] [Google Scholar]
- 79.Lapidus, A. Genome sequence databases (overview): sequencing and assembly. In M. Schaechter (ed.), The encyclopedia of microbiology, in press. Elsevier, New York, NY.
- 80.Legault, B. A., A. Lopez-Lopez, J. C. Alba-Casado, W. F. Doolittle, H. Bolhuis, F. Rodriguez-Valera, and R. T. Papke. 2006. Environmental genomics of “Haloquadratum walsbyi” in a saltern crystallizer indicates a large pool of accessory genes in an otherwise coherent species. BMC Genomics 7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lo, I., V. J. Denef, N. C. Verberkmoes, M. B. Shah, D. Goltsman, G. DiBartolo, G. W. Tyson, E. E. Allen, R. J. Ram, J. C. Detter, P. Richardson, M. P. Thelen, R. L. Hettich, and J. F. Banfield. 2007. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature 446537-541. [DOI] [PubMed] [Google Scholar]
- 82.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 321363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Macke, T. J., D. J. Ecker, R. R. Gutell, D. Gautheret, D. A. Case, and R. Sampath. 2001. RNAMotif, an RNA secondary structure definition and search algorithm. Nucleic Acids Res. 294724-4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Maltsev, N., E. Glass, D. Sulakhe, A. Rodriguez, M. H. Syed, T. Bompada, Y. Zhang, and M. D'Souza. 2006. PUMA2—grid-based high-throughput analysis of genomes and metabolic pathways. Nucleic Acids Res. 34D369-D372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marcotte, E. M., M. Pellegrini, H. L. Ng, D. W. Rice, T. O. Yeates, and D. Eisenberg. 1999. Detecting protein function and protein-protein interactions from genome sequences. Science 285751-753. [DOI] [PubMed] [Google Scholar]
- 87.Marcotte, E. M., M. Pellegrini, M. J. Thompson, T. O. Yeates, and D. Eisenberg. 1999. A combined algorithm for genome-wide prediction of protein function. Nature 40283-86. [DOI] [PubMed] [Google Scholar]
- 88.Marcy, Y., C. Ouverney, E. M. Bik, T. Losekann, N. Ivanova, H. G. Martin, E. Szeto, D. Platt, P. Hugenholtz, D. A. Relman, and S. R. Quake. 2007. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc. Natl. Acad. Sci. USA 10411889-11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Margulies, M., M. Egholm, W. E. Altman, S. Attiya, J. S. Bader, L. A. Bemben, J. Berka, M. S. Braverman, Y. J. Chen, Z. Chen, S. B. Dewell, L. Du, J. M. Fierro, X. V. Gomes, B. C. Godwin, W. He, S. Helgesen, C. H. Ho, G. P. Irzyk, S. C. Jando, M. L. Alenquer, T. P. Jarvie, K. B. Jirage, J. B. Kim, J. R. Knight, J. R. Lanza, J. H. Leamon, S. M. Lefkowitz, M. Lei, J. Li, K. L. Lohman, H. Lu, V. B. Makhijani, K. E. McDade, M. P. McKenna, E. W. Myers, E. Nickerson, J. R. Nobile, R. Plant, B. P. Puc, M. T. Ronan, G. T. Roth, G. J. Sarkis, J. F. Simons, J. W. Simpson, M. Srinivasan, K. R. Tartaro, A. Tomasz, K. A. Vogt, G. A. Volkmer, S. H. Wang, Y. Wang, M. P. Weiner, P. Yu, R. F. Begley, and J. M. Rothberg. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Markowitz, V. M. 2007. Microbial genome data resources. Curr. Opin. Biotechnol. 18267-272. [DOI] [PubMed] [Google Scholar]
- 91.Markowitz, V. M., N. Ivanova, K. Palaniappan, E. Szeto, F. Korzeniewski, A. Lykidis, I. Anderson, K. Mavromatis, V. Kunin, H. Garcia Martin, I. Dubchak, P. Hugenholtz, and N. C. Kyrpides. 2006. An experimental metagenome data management and analysis system. Bioinformatics 22e359-e3567. [DOI] [PubMed] [Google Scholar]
- 92.Markowitz, V. M., F. Korzeniewski, K. Palaniappan, E. Szeto, G. Werner, A. Padki, X. Zhao, I. Dubchak, P. Hugenholtz, I. Anderson, A. Lykidis, K. Mavromatis, N. Ivanova, and N. C. Kyrpides. 2006. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 34D344-D348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Martin-Cuadrado, A. B., P. Lopez-Garcia, J. C. Alba, D. Moreira, L. Monticelli, A. Strittmatter, G. Gottschalk, and F. Rodriguez-Valera. 2007. Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS ONE 2e914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mavromatis, K., N. Ivanova, K. Barry, H. Shapiro, E. Goltsman, A. C. McHardy, I. Rigoutsos, A. Salamov, F. Korzeniewski, M. Land, A. Lapidus, I. Grigoriev, P. Richardson, P. Hugenholtz, and N. C. Kyrpides. 2007. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Nat. Methods 4495-500. [DOI] [PubMed] [Google Scholar]
- 95.McHardy, A. C., H. G. Martin, A. Tsirigos, P. Hugenholtz, and I. Rigoutsos. 2007. Accurate phylogenetic classification of variable-length DNA fragments. Nat. Methods 463-72. [DOI] [PubMed] [Google Scholar]
- 96.McHardy, A. C., and I. Rigoutsos. 2007. What's in the mix: phylogenetic classification of metagenome sequence samples. Curr. Opin. Microbiol. 10499-503. [DOI] [PubMed] [Google Scholar]
- 97.Myers, E. W., G. G. Sutton, A. L. Delcher, I. M. Dew, D. P. Fasulo, M. J. Flanigan, S. A. Kravitz, C. M. Mobarry, K. H. Reinert, K. A. Remington, E. L. Anson, R. A. Bolanos, H. H. Chou, C. M. Jordan, A. L. Halpern, S. Lonardi, E. M. Beasley, R. C. Brandon, L. Chen, P. J. Dunn, Z. Lai, Y. Liang, D. R. Nusskern, M. Zhan, Q. Zhang, X. Zheng, G. M. Rubin, M. D. Adams, and J. C. Venter. 2000. A whole-genome assembly of Drosophila. Science 2872196-2204. [DOI] [PubMed] [Google Scholar]
- 98.Nakashima, H., M. Ota, K. Nishikawa, and T. Ooi. 1998. Genes from nine genomes are separated into their organisms in the dinucleotide composition space. DNA Res. 5251-259. [DOI] [PubMed] [Google Scholar]
- 99.Neufeld, J. D., Y. Chen, M. G. Dumont, and J. C. Murrell. 2008. Marine methylotrophs revealed by stable-isotope probing, multiple displacement amplification and metagenomics. Environ. Microbiol. 101526-1535. [DOI] [PubMed] [Google Scholar]
- 100.Noguchi, H., J. Park, and T. Takagi. 2006. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res. 345623-5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Oehmen, C., and J. Nieplocha. 2006. ScalaBLAST: a scalable implementation of BLAST for high-performance data-intensive bioinformatics analysis. IEEE Trans. Parallel Distrib. Syst. 17740-749. [Google Scholar]
- 102.Overbeek, R., T. Begley, R. M. Butler, J. V. Choudhuri, H. Y. Chuang, M. Cohoon, V. de Crecy-Lagard, N. Diaz, T. Disz, R. Edwards, M. Fonstein, E. D. Frank, S. Gerdes, E. M. Glass, A. Goesmann, A. Hanson, D. Iwata-Reuyl, R. Jensen, N. Jamshidi, L. Krause, M. Kubal, N. Larsen, B. Linke, A. C. McHardy, F. Meyer, H. Neuweger, G. Olsen, R. Olson, A. Osterman, V. Portnoy, G. D. Pusch, D. A. Rodionov, C. Ruckert, J. Steiner, R. Stevens, I. Thiele, O. Vassieva, Y. Ye, O. Zagnitko, and V. Vonstein. 2005. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 335691-5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Overbeek, R., M. Fonstein, M. D'Souza, G. D. Pusch, and N. Maltsev. 1999. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. USA 962896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Overbeek, R., N. Larsen, T. Walunas, M. D'Souza, G. Pusch, E. Selkov, Jr., K. Liolios, V. Joukov, D. Kaznadzey, I. Anderson, A. Bhattacharyya, H. Burd, W. Gardner, P. Hanke, V. Kapatral, N. Mikhailova, O. Vasieva, A. Osterman, V. Vonstein, M. Fonstein, N. Ivanova, and N. Kyrpides. 2003. The ERGO genome analysis and discovery system. Nucleic Acids Res. 31164-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Palmer, C., E. M. Bik, M. B. Eisen, P. B. Eckburg, T. R. Sana, P. K. Wolber, D. A. Relman, and P. O. Brown. 2006. Rapid quantitative profiling of complex microbial populations. Nucleic Acids Res. 34e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pellegrini, M., E. M. Marcotte, M. J. Thompson, D. Eisenberg, and T. O. Yeates. 1999. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl. Acad. Sci. USA 964285-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pelletier, E., A. Kreimeyer, S. Bocs, Z. Rouy, G. Gyapay, R. Chouari, D. Riviere, A. Ganesan, P. Daegelen, A. Sghir, G. N. Cohen, C. Medigue, J. Weissenbach, and D. Le Paslier. 2008. “Candidatus Cloacamonas acidaminovorans”: genome sequence reconstruction provides a first glimpse of a new bacterial division. J. Bacteriol. 1902572-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Peplies, J., S. C. Lau, J. Pernthaler, R. Amann, and F. O. Glockner. 2004. Application and validation of DNA microarrays for the 16S rRNA-based analysis of marine bacterioplankton. Environ. Microbiol. 6638-645. [DOI] [PubMed] [Google Scholar]
- 109.Peterson, J. D., L. A. Umayam, T. Dickinson, E. K. Hickey, and O. White. 2001. The Comprehensive Microbial Resource. Nucleic Acids Res. 29123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Pinard, R., A. de Winter, G. J. Sarkis, M. B. Gerstein, K. R. Tartaro, R. N. Plant, M. Egholm, J. M. Rothberg, and J. H. Leamon. 2006. Assessment of whole genome amplification-induced bias through high-throughput, massively parallel whole genome sequencing. BMC Genomics 7216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Podar, M., C. B. Abulencia, M. Walcher, D. Hutchison, K. Zengler, J. A. Garcia, T. Holland, D. Cotton, L. Hauser, and M. Keller. 2007. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl. Environ. Microbiol. 733205-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Pop, M., A. Phillippy, A. L. Delcher, and S. L. Salzberg. 2004. Comparative genome assembly. Brief. Bioinformatics 5237-248. [DOI] [PubMed] [Google Scholar]
- 113.Rodriguez-Brito, B., F. Rohwer, and R. A. Edwards. 2006. An application of statistics to comparative metagenomics. BMC Bioinformatics 7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ronaghi, M. 2001. Pyrosequencing sheds light on DNA sequencing. Genome Res. 113-11. [DOI] [PubMed] [Google Scholar]
- 115.Rusch, D. B., A. L. Halpern, G. Sutton, K. B. Heidelberg, S. Williamson, S. Yooseph, D. Wu, J. A. Eisen, J. M. Hoffman, K. Remington, K. Beeson, B. Tran, H. Smith, H. Baden-Tillson, C. Stewart, J. Thorpe, J. Freeman, C. Andrews-Pfannkoch, J. E. Venter, K. Li, S. Kravitz, J. F. Heidelberg, T. Utterback, Y. H. Rogers, L. I. Falcon, V. Souza, G. Bonilla-Rosso, L. E. Eguiarte, D. M. Karl, S. Sathyendranath, T. Platt, E. Bermingham, V. Gallardo, G. Tamayo-Castillo, M. R. Ferrari, R. L. Strausberg, K. Nealson, R. Friedman, M. Frazier, and J. C. Venter. 2007. The Sorcerer II Global Ocean Sampling Expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 5e77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sabehi, G., A. Loy, K. H. Jung, R. Partha, J. L. Spudich, T. Isaacson, J. Hirschberg, M. Wagner, and O. Beja. 2005. New insights into metabolic properties of marine bacteria encoding proteorhodopsins. PLoS Biol. 3e273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sandberg, R., G. Winberg, C. I. Branden, A. Kaske, I. Ernberg, and J. Coster. 2001. Capturing whole-genome characteristics in short sequences using a naive Bayesian classifier. Genome Res. 111404-1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Sanger, F., and A. R. Coulson. 1975. A rapid method for determining sequences in DNA by primed synthesis with DNA polymerase. J. Mol. Biol. 94441-448. [DOI] [PubMed] [Google Scholar]
- 119.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 745463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Schmeisser, C., C. Stockigt, C. Raasch, J. Wingender, K. N. Timmis, D. F. Wenderoth, H. C. Flemming, H. Liesegang, R. A. Schmitz, K. E. Jaeger, and W. R. Streit. 2003. Metagenome survey of biofilms in drinking-water networks. Appl. Environ. Microbiol. 697298-7309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sekar, R., B. M. Fuchs, R. Amann, and J. Pernthaler. 2004. Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl. Environ. Microbiol. 706210-6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Selengut, J. D., D. H. Haft, T. Davidsen, A. Ganapathy, M. Gwinn-Giglio, W. C. Nelson, A. R. Richter, and O. White. 2007. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 35D260-D264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Seshadri, R., S. A. Kravitz, L. Smarr, P. Gilna, and M. Frazier. 2007. CAMERA: a community resource for metagenomics. PLoS Biol. 5e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Shah, N., M. V. Teplitsky, S. Minovitsky, L. A. Pennacchio, P. Hugenholtz, B. Hamann, and I. L. Dubchak. 2005. SNP-VISTA: an interactive SNP visualization tool. BMC Bioinformatics 6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Snel, B., P. Bork, and M. A. Huynen. 1999. Genome phylogeny based on gene content. Nat. Genet. 21108-110. [DOI] [PubMed] [Google Scholar]
- 126.Sogin, M. L., H. G. Morrison, J. A. Huber, D. M. Welch, S. M. Huse, P. R. Neal, J. M. Arrieta, and G. J. Herndl. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. USA 10312115-12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sorek, R., Y. Zhu, C. J. Creevey, M. P. Francino, P. Bork, and E. M. Rubin. 2007. Genome-wide experimental determination of barriers to horizontal gene transfer. Science 3181449-1452. [DOI] [PubMed] [Google Scholar]
- 128.Staden, R., K. F. Beal, and J. K. Bonfield. 2000. The Staden package, 1998. Methods Mol. Biol. 132115-130. [DOI] [PubMed] [Google Scholar]
- 129.Strous, M., E. Pelletier, S. Mangenot, T. Rattei, A. Lehner, M. W. Taylor, M. Horn, H. Daims, D. Bartol-Mavel, P. Wincker, V. Barbe, N. Fonknechten, D. Vallenet, B. Segurens, C. Schenowitz-Truong, C. Medigue, A. Collingro, B. Snel, B. E. Dutilh, H. J. Op den Camp, C. van der Drift, I. Cirpus, K. T. van de Pas-Schoonen, H. R. Harhangi, L. van Niftrik, M. Schmid, J. Keltjens, J. van de Vossenberg, B. Kartal, H. Meier, D. Frishman, M. A. Huynen, H. W. Mewes, J. Weissenbach, M. S. Jetten, M. Wagner, and D. Le Paslier. 2006. Deciphering the evolution and metabolism of an anammox bacterium from a community genome. Nature 440790-794. [DOI] [PubMed] [Google Scholar]
- 130.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278631-637. [DOI] [PubMed] [Google Scholar]
- 132.Teeling, H., A. Meyerdierks, M. Bauer, R. Amann, and F. O. Glockner. 2004. Application of tetranucleotide frequencies for the assignment of genomic fragments. Environ. Microbiol. 6938-947. [DOI] [PubMed] [Google Scholar]
- 133.Thomas, C. A., Jr. 1971. The genetic organization of chromosomes. Annu. Rev. Genet. 5237-256. [DOI] [PubMed] [Google Scholar]
- 134.Tringe, S. G., and E. M. Rubin. 2005. Metagenomics: DNA sequencing of environmental samples. Nat. Rev. Genet. 6805-814. [DOI] [PubMed] [Google Scholar]
- 135.Tringe, S. G., C. von Mering, A. Kobayashi, A. A. Salamov, K. Chen, H. W. Chang, M. Podar, J. M. Short, E. J. Mathur, J. C. Detter, P. Bork, P. Hugenholtz, and E. M. Rubin. 2005. Comparative metagenomics of microbial communities. Science 308554-557. [DOI] [PubMed] [Google Scholar]
- 136.Turnbaugh, P. J., R. E. Ley, M. A. Mahowald, V. Magrini, E. R. Mardis, and J. I. Gordon. 2006. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 4441027-1031. [DOI] [PubMed] [Google Scholar]
- 137.Tyson, G. W., and J. F. Banfield. 2008. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ. Microbiol. 10200-207. [DOI] [PubMed] [Google Scholar]
- 138.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 42837-43. [DOI] [PubMed] [Google Scholar]
- 139.Urich, T., A. Lanzen, J. Qi, D. H. Huson, C. Schleper, and S. C. Schuster. 2008. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS ONE 3e2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 30466-74. [DOI] [PubMed] [Google Scholar]
- 141.von Mering, C., P. Hugenholtz, J. Raes, S. G. Tringe, T. Doerks, L. J. Jensen, N. Ward, and P. Bork. 2007. Quantitative phylogenetic assessment of microbial communities in diverse environments. Science 3151126-1130. [DOI] [PubMed] [Google Scholar]
- 142.von Mering, C., L. J. Jensen, M. Kuhn, S. Chaffron, T. Doerks, B. Kruger, B. Snel, and P. Bork. 2007. STRING 7—recent developments in the integration and prediction of protein interactions. Nucleic Acids Res. 35D358-D362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.von Wintzingerode, F., U. B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21213-229. [DOI] [PubMed] [Google Scholar]
- 144.Wagner, M., M. Horn, and H. Daims. 2003. Fluorescence in situ hybridisation for the identification and characterisation of prokaryotes. Curr. Opin. Microbiol. 6302-309. [DOI] [PubMed] [Google Scholar]
- 145.Warnecke, F., and P. Hugenholtz. 2007. Building on basic metagenomics with complementary technologies. Genome Biol. 8231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Warnecke, F., P. Luginbuhl, N. Ivanova, M. Ghassemian, T. H. Richardson, J. T. Stege, M. Cayouette, A. C. McHardy, G. Djordjevic, N. Aboushadi, R. Sorek, S. G. Tringe, M. Podar, H. G. Martin, V. Kunin, D. Dalevi, J. Madejska, E. Kirton, D. Platt, E. Szeto, A. Salamov, K. Barry, N. Mikhailova, N. C. Kyrpides, E. G. Matson, E. A. Ottesen, X. Zhang, M. Hernandez, C. Murillo, L. G. Acosta, I. Rigoutsos, G. Tamayo, B. D. Green, C. Chang, E. M. Rubin, E. J. Mathur, D. E. Robertson, P. Hugenholtz, and J. R. Leadbetter. 2007. Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite. Nature 450560-565. [DOI] [PubMed] [Google Scholar]
- 147.Wheeler, D. L., T. Barrett, D. A. Benson, S. H. Bryant, K. Canese, V. Chetvernin, D. M. Church, M. Dicuccio, R. Edgar, S. Federhen, M. Feolo, L. Y. Geer, W. Helmberg, Y. Kapustin, O. Khovayko, D. Landsman, D. J. Lipman, T. L. Madden, D. R. Maglott, V. Miller, J. Ostell, K. D. Pruitt, G. D. Schuler, M. Shumway, E. Sequeira, S. T. Sherry, K. Sirotkin, A. Souvorov, G. Starchenko, R. L. Tatusov, T. A. Tatusova, L. Wagner, and E. Yaschenko. 2008. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 36D13-D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Whitaker, R. J., and J. F. Banfield. 2006. Population genomics in natural microbial communities. Trends Ecol. Evol. 21508-516. [DOI] [PubMed] [Google Scholar]
- 149.Wommack, K. E., J. Bhavsar, and J. Ravel. 2008. Metagenomics: read length matters. Appl. Environ. Microbiol. 741453-1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Woyke, T., H. Teeling, N. N. Ivanova, M. Huntemann, M. Richter, F. O. Gloeckner, D. Boffelli, I. J. Anderson, K. W. Barry, H. J. Shapiro, E. Szeto, N. C. Kyrpides, M. Mussmann, R. Amann, C. Bergin, C. Ruehland, E. M. Rubin, and N. Dubilier. 2006. Symbiosis insights through metagenomic analysis of a microbial consortium. Nature 443950-955. [DOI] [PubMed] [Google Scholar]
- 151.Yooseph, S., G. Sutton, D. B. Rusch, A. L. Halpern, S. J. Williamson, K. Remington, J. A. Eisen, K. B. Heidelberg, G. Manning, W. Li, L. Jaroszewski, P. Cieplak, C. S. Miller, H. Li, S. T. Mashiyama, M. P. Joachimiak, C. van Belle, J. M. Chandonia, D. A. Soergel, Y. Zhai, K. Natarajan, S. Lee, B. J. Raphael, V. Bafna, R. Friedman, S. E. Brenner, A. Godzik, D. Eisenberg, J. E. Dixon, S. S. Taylor, R. L. Strausberg, M. Frazier, and J. C. Venter. 2007. The Sorcerer II Global Ocean Sampling Expedition: expanding the universe of protein families. PLoS Biol. 5e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zhang, T., M. Breitbart, W. H. Lee, J. Q. Run, C. L. Wei, S. W. Soh, M. L. Hibberd, E. T. Liu, F. Rohwer, and Y. Ruan. 2006. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 4e3. [DOI] [PMC free article] [PubMed] [Google Scholar]