Abstract
To learn how well ungapped sequence comparisons of multiple species can predict cis-regulatory elements in Caenorhabditis elegans, we made such predictions across the large, complex ceh-13/lin-39 locus and tested them transgenically. We also examined how prediction quality varied with different genomes and parameters in our comparisons. Specifically, we sequenced ∼0.5% of the C. brenneri and C. sp. 3 PS1010 genomes, and compared five Caenorhabditis genomes (C. elegans, C. briggsae, C. brenneri, C. remanei, and C. sp. 3 PS1010) to find regulatory elements in 22.8 kb of noncoding sequence from the ceh-13/lin-39 Hox subcluster. We developed the MUSSA program to find ungapped DNA sequences with N-way transitive conservation, applied it to the ceh-13/lin-39 locus, and transgenically assayed 21 regions with both high and low degrees of conservation. This identified 10 functional regulatory elements whose activities matched known ceh-13/lin-39 expression, with 100% specificity and a 77% recovery rate. One element was so well conserved that a similar mouse Hox cluster sequence recapitulated the native nematode expression pattern when tested in worms. Our findings suggest that ungapped sequence comparisons can predict regulatory elements genome-wide.
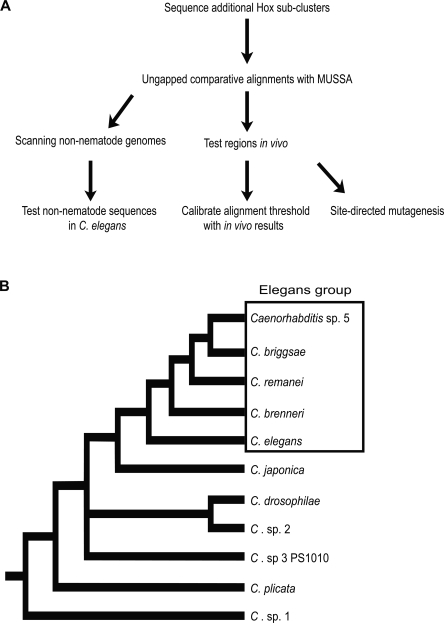
Despite knowledge of entire genome sequences, discovering cis-regulatory DNA elements remains surprisingly inefficient. In animal genomes, cis-regulatory elements are located unpredictably around or within the genes they regulate (Woolfe et al. 2005; Davidson 2006; Pennacchio et al. 2006; Engström et al. 2007). These elements, when dissected further, often prove to be composed of individual transcription factor binding sites that are often very loosely defined (Sandelin et al. 2004). Transgenic analysis in vivo is the most definitive way to show that a sequence is regulatory, but it is also the most time consuming and expensive. It is therefore desirable to use other criteria, such as preferential sequence conservation, to identify regions most likely to be functional. To evaluate a strategy for phylogenetic footprinting using four other Caenorhabditis species, we dissected the cis-regulatory structure of a Hox cluster in the nematode Caenorhabditis elegans (Fig. 1A).
Figure 1.
Experimental flow and Caenorhabditis phylogeny. (A) The experimental rationale of the project is shown. (B) Phylogeny of nematodes within the Caenorhabditis genus from Kiontke et al. (2007). The Elegans group and C. sp. 3 PS1010 are dealt with in this study.
If two or more species are evolutionarily close enough to show common development and physiology, their genomes are expected to share an underlying gene regulatory network driven by cis-regulatory elements with conserved sequences of several hundred base pairs (Tagle et al. 1988; Davidson 2006; Brown et al. 2007; Li et al. 2007). Within a functional cis-regulatory element, individual transcription-factor binding sites are generally short (∼6–20 bp) with statistical preferences, not strict requirements, for specific bases (Sandelin et al. 2004). Statistical over-representation of such motifs has been useful for identifying transcription-factor binding sites common to coregulated genes in C. elegans (Ao et al. 2004; Gaudet et al. 2004; Wenick and Hobert 2004; Pauli et al. 2006; Etchberger et al. 2007; McGhee et al. 2007; Zhao et al. 2007). However, this approach requires a known set of coregulated genes, a limitation that cross-species genomic comparison methods do not have. The simplest genomic comparison method is all-against-all matching of ungapped sequence windows, which is well suited for finding cis-regulatory elements under selective pressure against insertions and deletions (Brown et al. 2002; Cameron et al. 2005). This kind of comparison reveals orientation-independent, one-to-many, and many-to-many relationships, all of which are possible for conserved cis-regulatory sequences, yet invisible in standard global alignments. While ungapped comparisons can highlight regulatory regions, they are not expected to resolve individual transcription-factor binding sites within them. However, different prediction biases from sequence conservation versus statistical over-representation can complement one another (Wang and Stormo 2003; Bigelow et al. 2004; Tompa et al. 2005; Chen et al. 2006).
Since purely random pairing of unrelated 100-bp DNA segments typically yields two perfect 6-bp matches (Dickinson 1991), comparing three or more species should identify sequences under selective pressure with greater accuracy than comparing only two (Boffelli et al. 2004; Sinha et al. 2004; Eddy 2005; Stone et al. 2005). This has recently been done for budding yeasts (Cliften et al. 2003; Kellis et al. 2003), Drosophila (Stark et al. 2007), and vertebrates (Krek et al. 2005; Xie et al. 2005, 2007; Pennacchio et al. 2006; McGaughey et al. 2008). Vertebrates have many conserved sequences that may be regulatory, but most have unknown functions (Bejerano et al. 2004; Boffelli et al. 2004; Ovcharenko et al. 2005; Ahituv et al. 2007) that are difficult to test in all cell types throughout the life cycle, especially in mammals.
The nematode Caenorhabditis elegans has a compact genome (100 Mb, ∼27,000 genes) and body (∼1000 somatic cells in adults), which should allow candidate regulatory elements to be tested for function throughout development and across all cell types (Sulston and Horvitz 1977; Kimble and Hirsh 1979; Hillier et al. 2005). Although C. elegans is the most familiar Caenorhabditis species, others are available for multispecies genomic comparisons (Fig. 1B) (Sudhaus and Kiontke 1996, 2007; Baldwin et al. 1997; Stothard and Pilgrim 2006). Sibling species (the Elegans group, including C. brenneri) are difficult to distinguish from C. elegans morphologically, save for sex differences (Sudhaus and Kiontke 1996; Kiontke et al. 2004). C. japonica, the closest outgroup, shows some morphological differences, but they are relatively minor (Kiontke et al. 2002), while the more distant C. sp. 3 PS1010 has distinct morphology and behavior (Sudhaus and Kiontke 1996; Cho et al. 2004; Kiontke et al. 2004). Since C. brenneri subdivides an evolutionary branch between C. elegans and the siblings C. briggsae and C. remanei, comparisons of its genome with the others might help weed out nonfunctional DNA sequences that had failed to diverge in the sibling species. Comparisons with the more remote C. sp. 3 PS1010 might define more highly conserved sequences invariant within the Caenorhabditis genus and not simply within the Elegans group. We therefore undertook a pilot project to sequence and analyze ∼0.5% of the genomes of C. brenneri and C. sp. 3 PS1010, including the Hox subcluster ceh-13/lin-39 (Streit et al. 2002; Stoyanov et al. 2003; Sternberg 2005; Wagmaister et al. 2006).
ceh-13 and lin-39 are a linked pair of Hox genes, orthologous to labial/HOXA1 and Sex combs reduced/HOXA5. Hox genes, an ancient class of developmental control genes, pose a special challenge to cis-regulatory analysis because they are not regulated as isolated loci. Instead, they are found throughout bilateria as conserved multigene clusters encoding paralogous transcription factors that are crucial for development, and that are expressed in complex spatiotemporal patterns requiring intricate transcriptional regulation (Garcia-Fernandez 2005; Lemons and McGinnis 2006). Hox genes not only function similarly in disparate animal phyla, but may also be regulated similarly (Malicki et al. 1992; Frasch et al. 1995; Popperl et al. 1995; Haerry and Gehring 1997; Streit et al. 2002; Garcia-Fernandez 2005), although few cis-regulatory elements shared by Hox clusters of different phyla have actually been found (Haerry and Gehring 1997; Streit et al. 2002).
Nematodes have only a single set of Hox genes. Several megabases of DNA and numerous non-Hox genes separate the C. elegans Hox cluster into three subclusters of two genes each: ceh-13/lin-39, mab-5/egl-5, and nob-1/php-3 (Supplemental Fig. S1) (Aboobaker and Blaxter 2003). This differs from most vertebrate genomes, which have four or five versions of a single large, unfragmented Hox gene cluster (Lemons and McGinnis 2006). Some Hox genes have been lost in the C. elegans lineage, but all those present have vertebrate and arthropod orthologs (Clark et al. 1993; Maloof and Kenyon 1998; Aboobaker and Blaxter 2003; Stoyanov et al. 2003; Wagmaister et al. 2006). Cis-regulation is almost certainly confined within each C. elegans subcluster: The ceh-13/lin-39 subcluster is thus a natural experiment, in which two genes represent a cluster of vertebrate orthologs (Lemons and McGinnis 2006).
The ceh-13/lin-39 subcluster is vital for much anterior and mid-body development in C. elegans, but deciphering its cis-regulation has been difficult and remains incomplete. It is large by C. elegans standards, with almost 20 kb of intergenic DNA encoding only a single microRNA gene. ceh-13 is required for both embryonic and postembryonic development; null ceh-13 mutations are lethal (Brunschwig et al. 1999). In the embryo, ceh-13 is expressed in the A, D, E, and MS lineages and is required for normal gastrulation (Wittmann et al. 1997). Two upstream regulatory sites have been reported to drive expression in the embryo, one of which also acts in the male tail (Streit et al. 2002; Stoyanov et al. 2003). Cis-regulation of post-embryonic ceh-13 expression, which includes the anterior dorsal hypodermis, anterior bodywall muscle, and ventral nerve cord (Brunschwig et al. 1999), is not yet well understood, especially in tissues where it is coexpressed with lin-39. While lin-39 is dispensable for viability, it is required for normal vulval development, migration of the QR and QL neuroblasts, muscle formation, and specification of VC neurons (Burglin and Ruvkun 1993; Clark et al. 1993; Wang et al. 1993; Clandinin et al. 1997; Grant et al. 2000; McKay et al. 2003). A recent study of the lin-39 promoter delimited several elements to ∼300 bp by generating many transgenic reporter strains without using comparative genomics information; one of these elements was critical for vulval expression (Wagmaister et al. 2006). Our working hypothesis is that the complex expression of the ceh-13/lin-39 locus arises from the summed actions of independent conserved cis-regulatory elements.
We have dissected ceh-13/lin-39 cis-regulation through comparative genomics, and thus defined parameters likely to be useful for genome-wide analyses. This revealed several known and new regulatory elements, including one with functional similarity in mammalian Hox clusters.
Results
DNA sequencing
To enable comparisons to C. elegans, 1.1 Mb of genomic sequences from C. brenneri and C. sp. 3 PS1010 were sequenced and assembled (Supplemental Tables S1, S2). This comprised ∼0.5% of each genome, assuming genome sizes roughly equal to C. elegans. The primary DNA sequence data were generally well assembled; the exception was a set of C. brenneri clones covering the mab-5/egl-5 intergenic region, which may have suffered from high polymorphism found in gonochoristic Caenorhabditis species (Graustein et al. 2002).
Sequence comparison
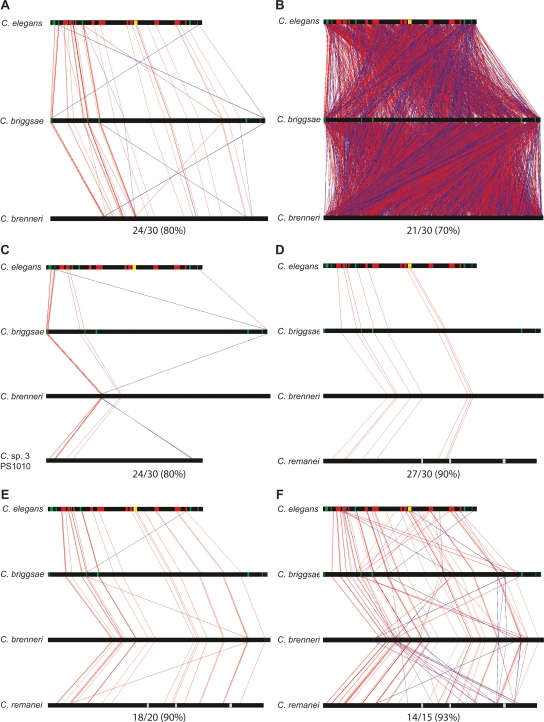
We used MUSSA (multi-species sequence analysis; http://mussa.caltech.edu) to find preferentially conserved sequences. MUSSA is a N-way sequence comparison algorithm, generalized from Family Relations (Brown et al. 2002), which integrates similarities among three or more genomes (see Methods). It compares, via sliding window, every frame in each participating sequence with every frame in all other sequences, allowing users to choose a window size and threshold of conservation for ungapped sequence matches (here called “MUSSA matches”). MUSSA produces an orientation-independent map of all one-to-one, one-to-many, and many-to-many transitive matches (Fig. 2). MUSSA matches highlight regions intolerant of insertions and deletions that may contain regulatory elements when found outside coding sequences (Cameron et al. 2005).
Figure 2.
MUSSA comparisons highlighted ungapped sequence matches. Horizontal black bars represent the nematode sequences. The top sequence, C. elegans, has green sections for exons (with lin-39 on the left and ceh-13 on the right), red sections for each of the N regions, and a yellow section for region N8, which encompasses mir-231 and its promoter. The vertical lines highlight ungapped sequence MUSSA matches, with red lines for matches facing the same direction and blue lines for reverse-complement matches. The MUSSA matches represent transitive alignments, meaning they match across all sequences compared. (A) At high thresholds the vertical red lines are largely parallel, reflecting predominant colinearity of conserved sequence identified with 80% (24/30) sequence identity for a 30-bp window. As the threshold (identity/window length) decreases, more matches are identified by MUSSA but the noise also increases. (B) At a lower threshold, 70% (21/30), the graph is packed with many lines that cross each other, producing a cluttered, cross-hatched pattern. The number of species being compared may also be varied, giving a range of matches. Comparisons, using a 30-bp window, are shown between C. elegans, C. briggsae, and C. brenneri at 80% (24/30) (A) and C. elegans, C. briggsae, C. brenneri, and C. sp. 3 PS1010 at 80% (24/30) (C). The window size can also be varied at a constant threshold, as between 27/30 (90%) (D), 18/20 (90%) (E), and 14/15 (93%) (F).
A number of parallel lines from visualizing MUSSA matches (at a given threshold of conservation) identified domains of similarity between the sequences, indicating the uniqueness and colinearity of potential regulatory elements (Fig. 2). Noise from repeats and low-complexity DNA sequence tended to create a cross-hatched pattern, reflecting many-to-many alignments that could be eliminated by raising similarity thresholds (Fig. 2A).
We initially performed two-way comparisons using a 30-bp window size, which minimized cross-hatched noise and had been useful in comparing mammalian genomes (T. De Buysscher, unpubl.). In principle, the threshold which gives P ≤ 0.05 for spurious matches in a 30-bp window should be 19/30 identities in 1 kb of completely random sequence (Brown 2006). Since nonconserved sequence is not actually random, the real P-value must be larger. For thresholds of ≤21/30, we found that cross-hatched connections marred the readout (Fig. 2B), while higher thresholds of ≥24/30 revealed a much sparser set of nearly parallel connections (Supplemental Fig. S2A). As expected, comparisons of three or more genomic sequences allowed clean results at lower thresholds than pairwise comparisons, improving the signal-to-noise ratio (Fig. 2A,C; Supplemental Fig. S2A,B).
Three-way comparison of ceh-13/lin-39 sequences from C. elegans, C. briggsae, and C. brenneri with 30-bp windows identified several conserved regions (Fig. 2A). In C. elegans, the ceh-13/lin-39 locus includes 19 kb of intergenic sequence and 8 kb of intronic sequence, of which only ∼2% was highlighted in MUSSA matches at a threshold of 24/30 (80%). This 50-fold enrichment was the basis for experimental dissection of the locus. In contrast, comparison of C. elegans, C. briggsae, and C. sp. 3 PS1010 revealed substantially fewer MUSSA matches and gained no new alignments across the range of parameters (Fig. 2C; Supplemental Fig. S2C–F). After experimentally testing predicted elements, as reported below, we could re-evaluate the effects of window size and genome numbers, as well as determine the effects of using the C. remanei ceh-13/lin-39 locus (which was unavailable during the earlier part of our work).
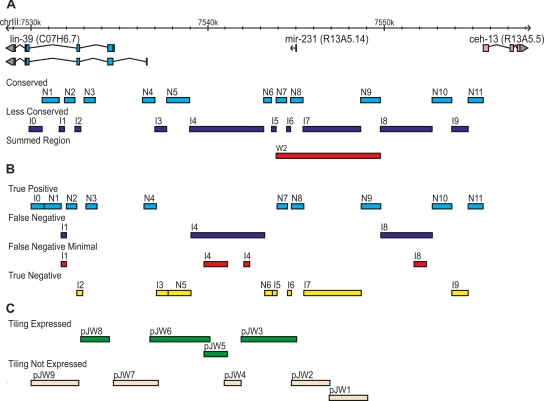
Cis-regulatory elements operating during development are typically composed of multiple binding sites arrayed over several hundred base pairs (Davidson 2006; Li et al. 2007). We expected that not all of these binding sites would be preserved as ungapped sequence blocks. To ensure that our comparison parameters did not omit functional sequences from transgenic assays, we buffered each MUSSA match with 200 bp of flanking DNA on each side. Aligned features located close to each other were grouped into single regions for testing. In this manner, 11 different regions (N1–N11) were predicted to be functional (Fig. 3A). The intervening noncoding regions selected for study (I0–I9), being less conserved, were deemed less likely to be functional (Fig. 3A; Supplemental Table S3) but were also tested transgenically.
Figure 3.
ceh-13/lin-39 Hox subcluster dissection based on sequence conservation. The ceh-13/lin-39 Hox locus was dissected into 21 sections for in vivo expression analysis based on the presence of MUSSA matches in a three-way alignment between C. elegans, C. briggsae, and C. brenneri. (A) MUSSA matches were used to identify similar, presumably conserved regions (N regions), which include the sequence match windows, 200 bp of 5′ and 3′ flanking sequence, and additional sequence for primer selection. The intervening, less-similar regions (I regions) located between the N regions were also tested. A “summed” region (W2) encompassing several component regions is shown as well. (B) With revised parameters of 100% match of 15-bp windows, the regions were repartitioned and true positives, true negatives, and false negatives were identified. The minimal region to recover the observed expression in the false negatives is identified (Streit et al. 2002; Wagmaister et al. 2006). (C) The regions assayed in the tiling analysis from Wagmaister et al. (2006) are shown for comparison, noting which drove expression (green) and which did not (beige).
Four of the 11 conserved regions corresponded to sequences previously shown to have some function. Region N8 corresponds precisely to the microRNA mir-231 and its upstream promoter. mir-231 is expressed from embryonic through adult stages, but its biological role is unknown (Lim et al. 2003). Region N3 drives larval ventral nerve cord expression (pJW8) (Wagmaister et al. 2006); region N9 drives embryonic expression (enh450) (Streit et al. 2002); and a region including element N10 drives larval and male tail expression (271-bp enhancer) (Stoyanov et al. 2003). Because our comparison rediscovered elements of the ceh-13/lin-39 subcluster previously shown to be important, it seemed likely that the newly defined blocks of similarity would also have biological activities.
Expression in C. elegans
We tested nine of the 11 strongly conserved regions, and all 10 intervening weakly conserved regions, for their ability to positively regulate expression; their repressor activity (if any) was not assayed. We did not retest the previously characterized N8 and N10, but did retest N3 and N9 to show that our assays reproduced published expression patterns in our reporter system (a Δpes-10 basal promoter driving nuclear-localized GFP with an unc-54 3′ untranslated region [UTR]). Background expression from the reporter is described in the Supplemental material, as are experiments showing that different basal promoters gave identical expression patterns in elements that were retested.
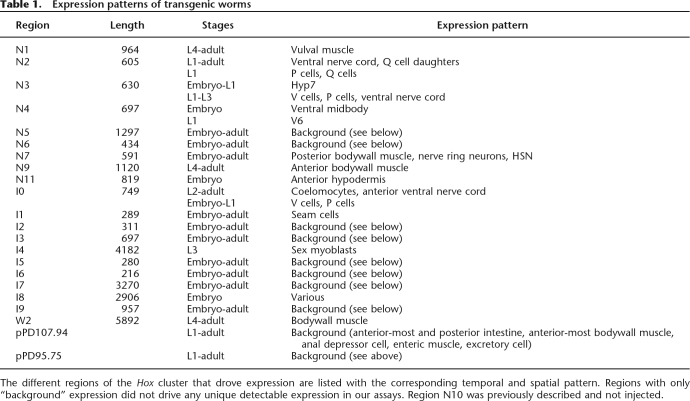
Most conserved regions drove expression in specific cell types (Table 1). In all cases, the described expression pattern was reproducible in multiple independent lines. Despite some spatial and temporal overlap, the expression patterns for each region were unique.
Table 1.
Expression patterns of transgenic worms
The different regions of the Hox cluster that drove expression are listed with the corresponding temporal and spatial pattern. Regions with only “background” expression did not drive any unique detectable expression in our assays. Region N10 was previously described and not injected.
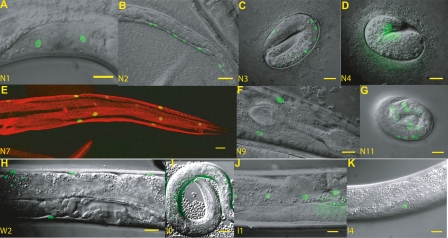
The intronic element N1 drove expression in vulval muscle, starting during the L4 larval stage and continuing through the adult (Fig. 4A). This element was well conserved with two MUSSA matches. Region N2 was expressed in the ventral nerve cord during the L1 larval stage (Fig. 4B). Expression of region N2 was also seen in some P cells and in the neural precursor Q cells, which are known to require lin-39 to regulate proper migration. N2 was also highly conserved: It consisted of two intronic MUSSA matches next to one another in all species except for C. sp. 3 PS1010, in which one match was inverted and moved 5′ with respect to lin-39. N2 occupies the same intron as N1, but is sufficiently separated (by 500 bp in C. elegans) to designate N1 and N2 as separate elements. Region N3, identified by one very well-conserved MUSSA match in the first intron of lin-39, was expressed in the hypodermal hyp7 cells in the late embryo and early L1 larvae (Fig. 4C) as well as in the V cells, P cells, and ventral nerve cord of the early L1 through L3 larvae. This expression pattern matched and expanded on that previously observed for this region (Wagmaister et al. 2006). Region N4 is in the proximal promoter region of lin-39; it drove expression in the ventral mid-body of the early embryo shortly after gastrulation (Fig. 4D). During early larval development N4 also drove expression in V6. Region N7 drove expression in the posterior bodywall muscle cells (Fig. 4E), starting in the late embryo and continuing through adulthood, and in the diagonal and longitudinal muscles of the male tail. Region N9 drove previously reported embryonic expression, along with previously unreported anterior bodywall muscle expression in L4 larvae and adults (Fig. 4F) (Streit et al. 2002). Region N11 was in the proximal promoter region of ceh-13 and drove expression in the anterior hypodermis of late embryos (Fig. 4G). Neither N5 nor N6 drove expression; this could be due to the limited conditions (e.g., non-dauer, noninfected, etc.) in which we scored the worms.
Figure 4.
In vivo expression patterns. Many well-conserved and some poorly conserved regions drive independent and reproducible expression. Expression is observed in a variety of tissues that largely agree with published antibody staining for ceh-13 and lin-39. (A) Element N1 directs expression in the L4 to adult vulval muscles. (B) Element N2 directs expression in the late embryo through L2 in the ventral nerve cord and P cells. (C) Element N3 directs expression in late embryonic through L3 hyp7, and in the V cells and P cells soon after hatching. (D) N4 directs expression in cells of the AB lineage in the dorsal mid-body during the comma stage. (E) N7 directs expression in the posterior bodywall muscle in the late embryo through the adult. N8 contains mir-231 and was not assayed. (F) N9 directs expression in the anterior bodywall muscle in the adult. (G) N11 directs expression in anterior late embryos. (H) W2, a large region spanning N7, N8, and N9, directs expression in both the anterior and posterior bodywall muscles, demonstrating additive coexpression of N7 and N9. (I) I0 directs expression in the posterior ventral coelomocyte. (J) I1 directs expression in the seam cells. (K) I4 directs expression in the SM cells. All scale bars are equal to 10 microns. For background expression from the reporter, see Supplemental material and Supplemental Figure S4.
Potential regulatory sequences were found for both ceh-13 and lin-39. For conserved regions closer to ceh-13 (N9 and N11), observed patterns agreed well with expected ones (Wittmann et al. 1997; Brunschwig et al. 1999; Streit et al. 2002). Expression of lin-39 in the bodywall muscles, intestine, and central body region have all been described and were reproduced, for the most part, by conserved regions closer to lin-39: N1–N4 and N7 (Clark et al. 1993; Wang et al. 1993; Maloof and Kenyon 1998; McKay et al. 2003). Furthermore, expression in the anterior midbody is predicted for both transcription factors, meaning that regions N2–N4 could be acting on both genes. Published patterns for both ceh-13 and lin-39 may be incomplete, which would account for observed activities beyond those expected.
Each region drove a different expression pattern. The fusion of a large region (W2) that included both N7 and N9 drove expression in both anterior and posterior bodywall muscle, a simple summation of N7 (strictly posterior) and N9 (strictly anterior) expression patterns (Figs. 3A, 4H). It is unknown whether these regions regulate ceh-13, lin-39, mir-231, or all three genes.
We then asked what regulatory activities, if any, resided in the less-conserved regions between our conserved elements. Four of the 10 less-conserved regions (I0, I1, I4, and I8) yielded expression apart from the expected background. Region I0 drove expression in the ventral posterior coelomocytes (Fig. 4I) and the two anterior inner longitudinal muscles of the male tail. This element had one MUSSA match that was strongly identified only when the window size was reduced to 15 or 20 bp. Region I1 drove expression in seam cells, starting with the embryo and continuing through to young adults (Fig. 4J). This element had no components strongly identified by MUSSA, with alignments appearing only at relatively low and noisy thresholds. Region I4 drove expression in the sex myoblasts through two cell divisions (Fig. 4K), as previously described by Wagmaister et al. (2006). Although expression was also reported in the Pn.p cells, we did not observe this, perhaps because I4 was not identical to the pJW5 region assayed by Wagmaister et al. (2006). I4 showed no MUSSA matches until a lower threshold of 22/30 bp or a 20-bp window was used, at which point the regions necessary for sex myoblast and ventral hypodermal Pn.p cell expression described by Wagmaister et al. (2006) were identified. Region I8 drove early embryonic expression, as previously reported (Streit et al. 2002). This region had a number of MUSSA matches that appeared as the threshold or window size was lowered.
Testing for sequence necessity
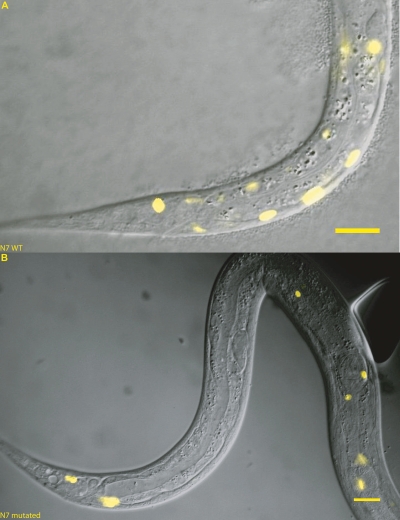
Our DNA regions from the ceh-13/lin-39 Hox subcluster contained not only blocks of ungapped sequence similarity, but also nonconserved sequences in which they were embedded. While these regions clearly drove expression in transgenic worms, our initial survey did not test whether the small conserved matches within them were crucial for regulatory activity. We therefore assayed in vivo constructs derived from some of the most highly conserved regions (N1, N2, N3, and N7; Supplemental Tables S3, S4), in which we mutated the MUSSA match in C. elegans. For N7, mutating the MUSSA match completely eliminated expression in the posterior bodywall muscle, showing the match to be needed for regulation (Fig. 5). In contrast, the remaining mutated regions from N1–N3 had the same expression patterns as their respective wild-type constructs. The conserved matches in N1–N3 were themselves dispensable for regulatory activity, yet were closely associated with active regulatory sequences. Our data paralleled previous negative results of Wagmaister et al. (2006) for a point mutation in the N3 region (HP2), which was a possible Hox or Pbx binding site.
Figure 5.
Mutating a conserved window in N7 knocked out expression. Element N7 (592 bp) normally drives expression in the posterior bodywall muscle (A). (B) When the 20-bp MUSSA match was reversed, all expression in the posterior bodywall muscle was abolished. Scale bars,10 microns.
Ultraconserved elements
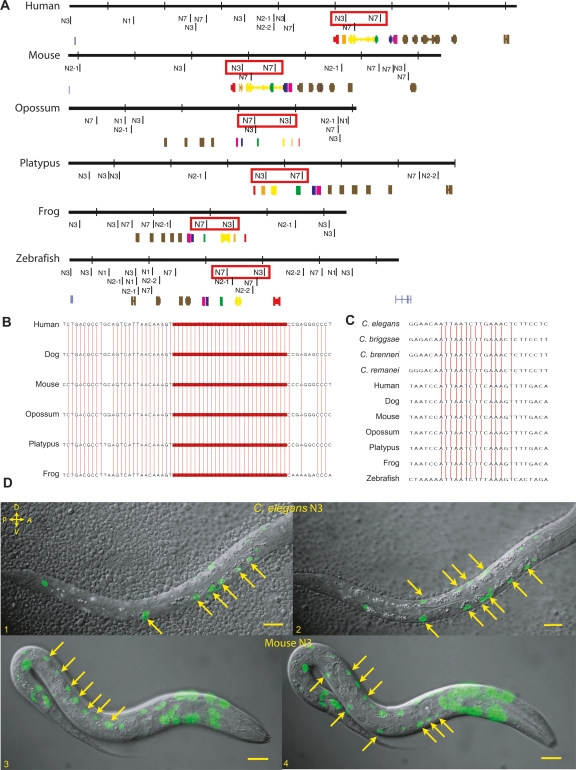
Hox clusters are evolutionarily ancient, sharing a common origin for all bilaterians (Garcia-Fernandez 2005; Lemons and McGinnis 2006), meaning that some cis-regulatory elements in C. elegans ceh-13/lin-39 might be conserved in other bilaterian phyla (Haerry and Gehring 1997; Streit et al. 2002). The following Hoxclusters were searched for any possible MUSSA matches to our conserved elements: the single Hox clusters of Drosophila melanogaster, Aedes aegypti (mosquito), Anopheles gambiae (mosquito), Apis mellifera (honey bee), Branchiostoma floridae (lancelet), Capitella sp. I (polychaete worm), Helobdella robusta (leech), Lottia gigantea (snail), Schistosoma mansoni (trematode), Schmidtea mediterranea (flatworm), and Tribolium castaneum (beetle); the four Hox clusters of mouse and human; and the seven Hox clusters of zebrafish. In each of these genomes we found several matches of uncertain significance. We therefore searched orthologous Hox regions for recurrent patterns of MUSSA matches (Fig. 6A). In newly characterized phyla, for which several related genomes had not yet been sequenced, this approach did not help to evaluate hits; but it was useful in vertebrates and insects, for which many related genomes were available.
Figure 6.
N3 cis-regulatory elements from either nematodes or vertebrates drove expression equivalently. (A) MUSSA analysis was used to identify any ungapped matches between nematodes and various vertebrates. Synteny of two elements, N3 and N7 highlighted by red boxes, suggested the match was not noise. All figures are to the same scale (hash marks represent 50-kb distances), with the regions examined in each case bounded by the next 5′ or 3′ curated genes on the chromosome. The Hox genes are color coded: (red) HOXA1, (orange) HOXA2, (yellow) HOXA3, (green) HOXA4, (blue) HOXA5, (purple) HOXA6. (B) Apparent conservation of N3 among vertebrates was very high, with similarity still at 100% in a 30-bp window. Vertical red lines represent base conservation between all six species. (C) N3 sequences shared 75% identity, using a 20-bp window, across 11 vertebrate and nematode species. (D) A mouse N3-like region drove expression in C. elegans that was almost identical to that driven by the C. elegans N3 region. Expression is seen in L1 larvae in the V cells on the left (D1, D3), and P cells and hypodermal syncytium on the right (D2, D4). Additional expression in observed in the head with the mouse construct. Scale bars, 10 microns.
In both mouse and human, N3 and N7-like MUSSA matches were paired with each other in the HOXA cluster near the ceh-13 and lin-39 orthologs, HOXA1 and HOXA5, respectively. Scans of the HOXA clusters in dog, opossum, platypus, and frog also revealed this pairing (Fig. 6A). Among the vertebrates alone, sequence conservation was high, indicating that these hits were located in functionally important DNA (Fig. 6B), although these sites had not been previously described. Using a low threshold, the matches showed similarity through nematodes and vertebrates, with the N3-like MUSSA match just 3′ of HOXA1 being more similar (86%) than the N7-like MUSSA match just 5′ of HOXA5 (73%) (Fig. 6C; Supplemental S3A). Similar searches within 11 Drosophila species yielded matches highly conserved among insects, but with only low levels of similarity to either nematodes or vertebrates.
To test whether the interphylum similarities revealed functional sequences, we cloned a 700-bp region of mouse Hox genomic DNA centered on the mouse N3-like MUSSA match and a 650-bp region centered on the N7-like MUSSA match, each containing local sequence conserved among mammals. We assayed both regions in C. elegans transgenes. The mouse N3-like region drove almost the same expression pattern as the C. elegans N3 region (Fig. 6D) in hyp7, P cells, V cells, and the ventral nerve cord, with discordant activity in only a few extra anterior hypodermal cells. Whereas C. elegans N3 was previously predicted to include a Hox/Pbx autoregulatory site for lin-39 (Wagmaister et al. 2006), the mouse N3-like MUSSA match is found closer to Hoxa1 (a ceh-13 ortholog) than to Hoxa4 (a lin-39 ortholog). N3 could be a general Hox binding site, or its role may have changed over time. In contrast, the mouse N7-like region failed to drive the posterior bodywall muscle expression as the C. elegans N7 region did, though its background expression level was noticeably increased (Supplemental Fig. S4A).
If N3′s similarities between nematodes and vertebrates result from common descent, N3-like matches should exist in other animal phyla. We found co-occurrence of two top-scoring MEME motifs and a MUSSA match in the nematodes, vertebrates, B. floridae, Capitella sp. I, H. robusta, and S. mansoni (Supplemental Fig. S3B; Supplemental material). MUSSA comparison of N3-like sequences in nematodes, vertebrates, and B. floridae yielded a 70% match, while a comparison of nematodes, vertebrates, S. mansoni, and H. robusta yielded a 65% match (Supplemental Fig. S3B). These matches encompass deuterostomes, ecdysozoa, and lophotrochozoa—all of the major divisions of bilateria. Thus, we interpret the N3 site to be evolutionarily conserved rather than convergent.
Threshold revision
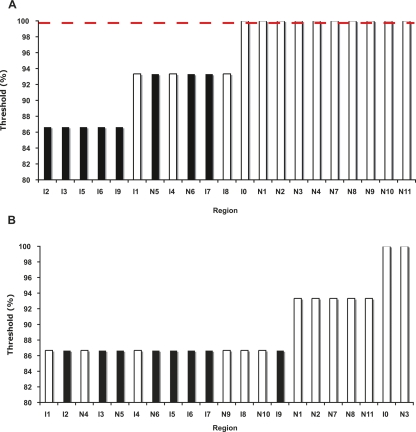
Having had some success with our initial parameters for ungapped sequence comparison, we then adjusted them empirically and retested them computationally against well-characterized genes in the hope of optimizing our parameters for genome-wide analysis. Initially, nine of the 11 regions (82%) identified by conservation gave expression, while three of the 10 less conserved regions (30%) gave expression; this was promising, but left room for possible improvement. When we tried lower thresholds or smaller windows, MUSSA found matches in some regions that had previously given no hits despite having regulatory activity (and that we had originally classified as false negatives). We therefore optimized the parameter settings and genome combination to achieve the best yield of functional elements while keeping false positives to a minimum (Fig. 2D–F; Supplemental Figs. S2G–L, S5, and S6). A 15-bp window and perfect conservation between C. elegans, C. briggsae, C. remanei, and C. brenneri identified MUSSA matches in 77% of all expressing regions with no false positives (Fig. 7A). Using a different window size (14 or 16–30 bp) decreased the resolution and efficiency (see Supplemental material; Supplemental Figs. S5, S6A,B). Including C. sp. 3 PS1010 sequences adequately selected the top hits, but only at the expense of eliminating many other hits and considerably reducing predictive power (Fig. 7B). Though the four Elegans group species together gave the best analysis, inclusion of C. remanei masked matches in the I4 region (Supplemental Fig. S2E; see Discussion).
Figure 7.
Revising MUSSA parameters for well-conserved regions. (A) A 15-bp window and four-way comparison among C. elegans, C. briggsae, C. brenneri, and C. remanei identified the thresholds at which MUSSA matches are observed within a region. Regions capable of driving expression are shown in white and those not capable of driving expression are shown in black. With a threshold of 100%, there is a 77% recovery of expressing regions with perfect specificity. (B) Using five-way comparisons and a 15-bp window among the four above species and C. sp. 3 PS1010, the thresholds where conservation was still observed were identified for each element. The predictive power for identifying functional regions is considerably reduced from the four-way comparison.
The intervening regions were often much larger than any conserved region. For instance, region I4 was 4.2 kb; however, the subsection of I4 sufficient to drive expression was 1.6 kb (38% of I4) (Wagmaister et al. 2006). Likewise, region I8 was 2.9 kb, but expression could be recapitulated with only 0.7 kb within it (24% of I8) (Streit et al. 2002). Thus, the density of regulatory regions within nonconserved sequences is probably even lower than our data indicate (Fig. 3B). When compared with tiling, as performed by Wagmaister et al. (2006), conservation-based analysis confers an efficiency advantage, with 100% instead of 40% specificity (Fig. 3C; Wagmaister et al. 2006).
To test whether the revised parameters are useful outside the Hox cluster, we analyzed the previously described C. elegans genes hlh-1, myo-2, myo-3, and unc-54 (Okkema et al. 1993; Krause et al. 1994). These were chosen for analysis because their promoter dissections had been screened for expression across all tissues, unlike most studies that identify positive expression in a specific tissue but did not screen for negative activity across other tissues. Using our strict 15-bp threshold and technique of including 200 bp of flanking DNA, all known regulatory elements of the myosin genes myo-2, myo-3, and unc-54 (Okkema et al. 1993) were identified with no false positives (Supplemental Fig. S7). For the hlh-1 locus, two of four regulatory sites (Krause et al. 1994) were recovered at a lower threshold. Therefore, MUSSA predictions were accurate at some non-Hox loci, but as in the Hox locus itself, some functional elements could not be identified this way.
Discussion
This study found four known and seven new cis-regulatory elements in the ceh-13/lin-39 Hox subcluster of C. elegans, using ungapped sequence conservation across four genomes and verification by transgenic analyses. Remarkably, one conserved element’s mouse counterpart recapitulated the native nematode expression pattern. The observed expression patterns generally paralleled those found by prior antibody staining and expression from the parental undissected promoters, suggesting that the union of these cis-regulatory elements drives the entire endogenous expression pattern, and that we have identified most cis-regulatory regions of ceh-13/lin-39 (Clark et al. 1993; Wang et al. 1993; Wittmann et al. 1997; Maloof and Kenyon 1998; Brunschwig et al. 1999; Streit et al. 2002; McKay et al. 2003).
For ceh-13/lin-39, our first parameters for sequence conservation worked well, even though we later improved them empirically. They identified 11 possible elements, of which nine showed function experimentally, leaving two false positives—a threefold enrichment for functional regulatory elements compared with simple, unselected tiling. With revised parameters, 100% of the computationally identified elements were functional. For these nematode sequences, we found that MUSSA predicted function with highest reliability and resolution when we used windows of 15 bp. Smaller windows gave noisier alignments with poor resolution, while larger windows tended to miss shorter conserved sequences with regulatory activities. These parameters correctly rediscovered regulatory regions in other well-characterized genes, but made some errors, suggesting additional possible refinements as functional data becomes available at other loci. However, we do not expect that this method, used on its own, will discover all elements. We also expect parameters to change when the set of compared genomes is changed, as we have already found. For instance, the conserved regions for vertebrate Hox sequences (e.g., the N3-like mouse region) were much longer than in nematodes, and could be detected at a lower MUSSA threshold with a larger window size. Such differences in sequence conservation might arise from different rates and types of mutations, or from altered selection pressures.
Our aim was to efficiently predict new elements with bona fide biological activity, accepting that this runs the risk of missing some regulatory regions. Nevertheless, correctly identifying even two-thirds of all C. elegans regulatory elements with a low false-positive rate, as we did prior to refinement, could significantly advance our knowledge of the worm regulatory genome. Recent uses of sequence constraint in vertebrates have been less sensitive in finding regulatory elements, perhaps because vertebrates undergo qualitatively different regulation (Pennacchio et al. 2006; McGaughey et al. 2008), although there are many differences, both biological and methodological, between their studies and this one. Only a representative subset of regulatory sites are needed to derive refined, genome-wide motifs in C. elegans, as we did with N2-1 (Supplemental material), which can then be statistically correlated with traits of their neighboring genes (Wenick and Hobert 2004; Mortazavi et al. 2006; Etchberger et al. 2007).
If a given regulatory element is mutated or fragmented in some species, comparing it with different sets of related species can still allow detection of that element. Such regulatory mutations are known to be responsible for subtle evolutionary changes in the salt resistance and excretory canal phenotypes of C. elegans, which have diverged from the ancestral phenotypes retained in C. briggsae and C. brenneri (Wang and Chamberlin 2004). The most striking difference in conservation we observed was between Elegans group species and the outlying C. sp. 3 PS1010. Four-way comparison of C. elegans, C. briggsae, C. brenneri, and C. remanei predicted the most regulatory elements, many of which could only be detected in C. sp. 3 PS1010 with much lower and noisier thresholds. Although all regions identified with C. sp. 3 PS1010 drove expression, there was no added benefit from this comparison; rather, it increased the false-negative rate. Similarly, neither lin-3 nor lin-11 in C. sp. 3 PS1010 had the organization or the sequence motifs of the genes in the Elegans group species (Supplemental Fig. S8; Supplemental Table S5). Additional Caenorhabditis genomic sequences should clarify which parts of the C. elegans genome encode species- or group-specific traits.
The regulatory organization of the ceh-13/lin-39 locus appears to be modular, with each regulatory element functioning independently in transgenes: The expression output of two elements on a single DNA fragment (N7 and N9 on W2) or of four coinjected elements (N1, N2, N3, and N7) matched the sum of their individual activities. Nevertheless, the linear order of conserved elements across the ceh-13/lin-39 locus has been conserved between the different Caenorhabditis species, including the relatively distant C. sp. 3 PS1010, suggesting that element order is under selective pressure. Among the elements, there is also potential for some functional redundancy, as has been noted in mammals (e.g., Ahituv et al. 2007). ceh-13, for example, is expressed in the larval ventral nerve cord (Brunschwig et al. 1999) and three different elements drive expression there.
Multiple regulatory elements distributed throughout large introns and flanking sequences control many metazoan genes expressed in complex spatiotemporal patterns (Woolfe et al. 2005; Davidson 2006; Pennacchio et al. 2006) and ceh-13/lin-39 follows this trend. Only two of the nine expressing regions were located within the proximal 2-kb promoter sequences of ceh-13 or lin-39, and four were in lin-39 introns. We did not assay for the effect that these regions had on ceh-13, lin-39, or mir-231 expression. Other examples of distal elements in C. elegans include remote regulation of ceh-10 and osm-9 (Colbert et al. 1997; Wenick and Hobert 2004).
Conservation analysis helped define elements without inadvertently splitting them, a hazard in blind deletion analysis. Moreover, it may have freed elements from inhibitory sequences, as we found that some large segments were less active when assayed than their subdomains. The entire second intron of lin-39 yielded no expression in a prior study (Wagmaister et al. 2006), but we identified four different active cis-regulatory elements (N1, N2, I0, and I1) by subdividing the region. One possibility is that poorly conserved DNA separating ceh-13/lin-39 elements harbors hidden regulatory functions that our assay misses, such as repression. The basal promoter construct we used to screen for in vivo enhancer activity is not expected to detect isolated transcriptional silencers or insulators. This could explain moderately conserved but inactive regions, as might enhancers dependent on untested culture conditions or promoter-specific interactions with regulatory elements (Wenick and Hobert 2004; Etchberger et al. 2007).
Although large regions can be split into smaller functional components (such as the W2 region dividing into N7, N8, and N9, and the lin-39 intron dividing into N1, N2, I0, and I1), further dissection of functional elements might simply disrupt them, yielding weak and variable expression. This has been observed for ceh-13 male tail expression when multiple sites within N10 were mutated (V. Wegewitz and A. Streit, pers. comm.).
Biologically relevant sequence motifs often appear in or near the best-conserved regions, even if the MUSSA matches themselves are not essential for regulatory activity. For instance, two conserved MUSSA matches <200-bp apart identify the element N9; but a known motif that is not part of either conserved window is located next to them, and is necessary for proper regulatory function (Supplemental Fig. S9A). In four of five mutageneses, changing just one conserved feature had little effect, which is consistent with functional redundancy often seen in multi-site regulatory elements. Our assays used injected transgenes, for which multiple copies generally exist of a cloned reporter (Mello and Fire 1995); this might have provided a relaxed context for gene expression, tolerating the loss of “redundant” sites actually required in vivo. A site that subtly controls the quantity or spatiotemporal pattern of gene activity could easily lack an observable impact on GFP expression. Thus, it is important to test not only conserved sequences for regulatory activity, but the sequences near them.
The apparent conservation of N3 and N7 regions across phyla suggests that they predate the divergence of bilateria. Although mouse N7 was not active in the cross-phylum assay, the mouse N3-like region was strikingly positive and contains a potentially autoregulatory Hox/Pbx binding site. To test regulatory elements for functional conservation between different animal phyla, Drosophila enhancers and promoters have been compared with those of C. elegans and mammals: This generally involved isolating an enhancer or promoter with a known expression pattern in a donor organism, and testing it transgenically for similar expression in a second, distantly related organism (Malicki et al. 1992; Frasch et al. 1995; Popperl et al. 1995; Haerry and Gehring 1997; Streit et al. 2002; Ruvinsky and Ruvkun 2003). With nematode and mouse N3 regions, we instead tested the donor enhancer for activity equivalent to that already defined for its ortholog in the recipient species. This provides an alternative for comparisons over very long evolutionary distances, across which anatomical similarities may not be obvious. Moreover, additional MEME motifs, one of which may have been independently identified in mammals (as LM115 and LM171 of Xie et al. [2007]) (Supplemental Results), are shared by the vertebrate and nematode sequences. Based on these in vivo data and computational analyses, we consider N3 a pan-phyletic regulatory sequence. Such sequences may be rare, and only present in the most ancient regulatory loci, such as the ParaHox or NK clusters (Garcia-Fernandez 2005).
Methods
General methods and strains
We obtained Caenorhabditis elegans, C. brenneri CB5161, and C. sp. 3 PS1010 from the CGC strain collection and cultured them on OP50 at 20°C, using methods standard for C. elegans (Sulston and Hodgkin 1988). unc-119(ed4) hermaphrodites were microinjected with a mixture of 60 ng/μL unc-119 vector, 12 ng/μL unpurified fusion product, and either 100 ng/μL pBluescript or 100 ng/μL digested genomic DNA to generate transgenic animals (Mello and Fire 1995; Kelly et al. 1997). All noted expression patterns were observed in two or more independent transgenic lines. In nonexpressing lines, at least 16 hermaphrodites from three independent lines (each line driving background GFP to guarantee GFP’s functionality) were observed at each stage (early embryos, late embryos, L1–L4 larvae, young adults, and mature adults) with 100× magnification; males and dauers were observed for some, but not all, reporter lines.
DNA preparation
DNA was prepared by standard methods (Sulston and Hodgkin 1988). pEpiFos-5 (Epicentre), based on pBeloBAC11 (Birren et al. 1999), was used as the fosmid library vector. Fosmid sequences were shotgun sequenced and assembled into contigs by the Department of Energy’s Joint Genome Institute at Walnut Creek (http://www.jgi.doe.gov/sequencing/protocols).
Sequence analysis
Sequence contigs from JGI were initially linked by BLASTN (Korf et al. 2003) and then merged with the revseq and megamerger functions of EMBOSS (Olson 2002). Our C. brenneri data had 22 genomic contigs, totaling 680,633 nucleotides (Supplemental Table S1). Our C. sp. 3 PS1010 data had seven genomic contigs, totaling 417,129 nucleotides (Supplemental Table S2). Gene predictions were made with Twinscan 3.5 running in single-species mode with C. elegans parameters (Wei et al. 2005); predicted protein sequences were extracted with BioPerl (Stajich et al. 2002). C. brenneri and C. sp 3 PS1010 protein sequences were tested for orthology against one another and against the protein-coding gene sets of C. elegans, C. briggsae, and C. remanei (from the WS170 release of WormBase) with OrthoMCL 1.3 (Li et al. 2003). Inferred ortholog groups were considered specific (i.e., unique) if they contained only one C. elegans gene, and only one gene from either C. briggsae or C. remanei. Our C. brenneri contigs encode 141 predicted proteins of ≥100 residues in length, of which 88 have unique C. elegans orthologs (Supplemental Table S1). Our C. sp. 3 PS1010 contigs encode 86 predicted ≥100-residue proteins, 68 with C. elegans orthologs (Supplemental Table S2). SVG genomic sequence images were generated by GBrowse for nematodes and vertebrates at the Wormbase (http://www.wormbase.org) and UCSC Genome Browser (http://genome.ucsc.edu) websites.
MUSSA (mulitple species sequence analysis) (http://mussa.caltech.edu), a program written in C++ with a Python controlled user interface, was used to identify evolutionarily conserved sequences. MUSSA uses N-way transitivity (all-against-all) so that only windows passing the selected similarity threshold across all species are reported as alignments. No sequences were repeat-masked in the comparisons performed here, though use of MUSSA in other phyla may benefit from masking as a preprocessing step (T. De Buysscher, D. Trout, and B.J. Wold, unpubl.).
For regulatory element dissection in the ceh-13/lin-39 cluster, published sequences from C. elegans, C. briggsae, and C. remanei (http://www.wormbase.org) were used with novel sequences from C. brenneri and C. sp. 3 PS1010. The mab-5/egl-5 Hox cluster comparisons used sequences from C. elegans, C. briggsae, and C. remanei. Additional comparisons with non-nematodes used sequences from all of each organism’s available Hox clusters (http://www.ensembl.org; http://genome.ucsc.edu; http://www.genedb.org/genedb/smansoni; http://racerx00.tamu.edu; and http://genome.jgi-psf.org). Known regulatory regions of non-Hox genes were linked from C. elegans to other species using MUSSA.
MEME
Multiple EM for Motif Elicitation (MEME) v3.5.4 was used to identify nonaligned motifs shared by different animal phyla (http://meme.sdsc.edu/meme) (Bailey and Elkan 1994). MEME motifs from the N3 element were tested for similarities to previously published genomic motifs by examining two 14-nt human sequences with up to two mismatches against JASPAR CNE (Bryne et al. 2007; Xie et al. 2007).
Transgene design and construction
PCR fusions were generated using standard protocols, essentially as in Hobert (2002). Genomic DNA and the cosmids R13A5 and C07H6 (from A. Fraser and R. Shownkeen at the Sanger Institute) were used as sequence templates. The Fire Lab Vector pPD107.94 was used as the template for the Δpes-10∷4X-NLS∷eGFP∷LacZ∷unc-54 sequence (Mello and Fire 1995). The Fire Lab Vector pPD95.75 was used as the template for the “promoterless” eGFP∷unc-54 sequence (Etchberger and Hobert 2008), used as a control in four constructs to demonstrate identical expression patterns under different basal promoters. Mutation primers were used to mutate target sites in plasmids. The mutated and sequenced enhancers were fused to Fire Lab Vector pPD122.53, where GFP was replaced with YFP, to give a Δpes-10∷4X-NLS∷YFP∷unc-54. GFP was replaced with CFP for unmutated controls. We mutated conserved sequences by reversal, not reverse complementation; such reversal maintained the base content, but was expected to destroy any sequence-specific binding of transcription factors. Complete methods are described in the Supplemental material.
Acknowledgments
We dedicate this study to the memory of E.B. Lewis, who pioneered the analysis of Hox clusters at Caltech. We thank C.T. Brown for discussions, N. Mullaney for work on an early version of MUSSA, E. Moon for aid in fosmid library construction, and E. Rubin and his colleagues at the DOE JGI for fosmid sequencing and assembly. We thank L.R. Baugh, C.T. Brown, C. Dalal, J. Green, M. Kato, K. Kiontke, A. Mortazavi, A. Seah, and B. Williams for comments on the manuscript. Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the NIH National Center for Research Resources (NCRR). Unpublished metazoan genomic sequences were generously provided by the DOE JGI and GeneDB. This work was supported by grants from DOE to B.J.W. and P.W.S., from NASA to B.J.W., from NIH to B.J.W., and from the HHMI, with which P.W.S. is an Investigator.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) under accession nos. FJ362353–FJ36238.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.085472.108.
References
- Aboobaker A., Blaxter M. Hox gene evolution in nematodes: Novelty conserved. Curr. Opin. Genet. Dev. 2003;13:593–598. doi: 10.1016/j.gde.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Ahituv N., Zhu Y., Visel A., Holt A., Afzal V., Pennacchio L.A., Rubin E.M. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao W., Gaudet J., Kent W.J., Muttumu S., Mango S.E. Environmentally induced foregut remodeling by PHA-4/FoxA and DAF-12/NHR. Science. 2004;305:1743–1746. doi: 10.1126/science.1102216. [DOI] [PubMed] [Google Scholar]
- Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- Baldwin J.G., Frisse L.M., Vida J.T., Eddleman C.D., Thomas W.K. An evolutionary framework for the study of developmental evolution in a set of nematodes related to Caenorhabditis elegans. Mol. Phylogenet. Evol. 1997;8:249–259. doi: 10.1006/mpev.1997.0433. [DOI] [PubMed] [Google Scholar]
- Bejerano G., Pheasant M., Makunin I., Stephen S., Kent W.J., Mattick J.S., Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- Bigelow H.R., Wenick A.S., Wong A., Hobert O. CisOrtho: A program pipeline for genome-wide identification of transcription factor target genes using phylogenetic footprinting. BMC Bioinformatics. 2004;5:27. doi: 10.1186/1471-2105-5-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birren B., Mancino V., Shizuya H. Genome analysis: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. Bacterial artificial chromosomes; pp. 241–295. [Google Scholar]
- Boffelli D., Nobrega M.A., Rubin E.M. Comparative genomics at the vertebrate extremes. Nat. Rev. Genet. 2004;5:456–465. doi: 10.1038/nrg1350. [DOI] [PubMed] [Google Scholar]
- Brown C.T. “Tackling the regulatory genome.” Ph.D. thesis. California Institute of Technology; Pasadena: 2006. [Google Scholar]
- Brown C.T., Rust A.G., Clarke P.J., Pan Z., Schilstra M.J., De Buysscher T., Griffin G., Wold B.J., Cameron R.A., Davidson E.H., et al. New computational approaches for analysis of cis-regulatory networks. Dev. Biol. 2002;246:86–102. doi: 10.1006/dbio.2002.0619. [DOI] [PubMed] [Google Scholar]
- Brown C.D., Johnson D.S., Sidow A. Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science. 2007;317:1557–1560. doi: 10.1126/science.1145893. [DOI] [PubMed] [Google Scholar]
- Brunschwig K., Wittmann C., Schnabel R., Burglin T.R., Tobler H., Muller F. Anterior organization of the Caenorhabditis elegans embryo by the labial-like Hox gene ceh-13. Development. 1999;126:1537–1546. doi: 10.1242/dev.126.7.1537. [DOI] [PubMed] [Google Scholar]
- Bryne J.C., Valen E., Tang M.H., Marstrand T., Winther O., da Piedade I., Krogh A., Lenhard B., Sandelin A. JASPAR, the open access database of transcription factor-binding profiles: New content and tools in the 2008 update. Nucleic Acids Res. 2007;36:D102–D106. doi: 10.1093/nar/gkm955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burglin T.R., Ruvkun G. The Caenorhabditis elegans homeobox gene cluster. Curr. Opin. Genet. Dev. 1993;3:615–620. doi: 10.1016/0959-437x(93)90097-9. [DOI] [PubMed] [Google Scholar]
- Cameron R.A., Chow S.H., Berney K., Chiu T.Y., Yuan Q.A., Kramer A., Helguero A., Ransick A., Yun M., Davidson E.H. An evolutionary constraint: Strongly disfavored class of change in DNA sequence during divergence of cis-regulatory modules. Proc. Natl. Acad. Sci. 2005;102:11769–11774. doi: 10.1073/pnas.0505291102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Mah A., Blacque O.E., Chu J., Phgora K., Bakhoum M.W., Newbury C.R., Khattra J., Chan S., Go A., et al. Identification of ciliary and ciliopathy genes in Caenorhabditis elegans through comparative genomics. Genome Biol. 2006;7:R126. doi: 10.1186/gb-2006-7-12-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S., Jin S.W., Cohen A., Ellis R.E. A phylogeny of Caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res. 2004;14:1207–1220. doi: 10.1101/gr.2639304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clandinin T.R., Katz W.S., Sternberg P.W. Caenorhabditis elegans HOM-C genes regulate the response of vulval precursor cells to inductive signal. Dev. Biol. 1997;182:150–161. doi: 10.1006/dbio.1996.8471. [DOI] [PubMed] [Google Scholar]
- Clark S.G., Chisholm A.D., Horvitz H.R. Control of cell fates in the central body region of C. elegans by the homeobox gene lin-39. Cell. 1993;74:43–55. doi: 10.1016/0092-8674(93)90293-y. [DOI] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Colbert H.A., Smith T.L., Bargmann C.I. OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J. Neurosci. 1997;17:8259–8269. doi: 10.1523/JNEUROSCI.17-21-08259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson E.H. The regulatory genome: Gene regulatory networks in development and evolution. Academic Press; San Diego, CA: 2006. [Google Scholar]
- Dickinson W. The evolution of regulatory genes and patterns in Drosophila. Evol. Biol. 1991;25:127–173. [Google Scholar]
- Eddy S.R. A model of the statistical power of comparative genome sequence analysis. PLoS Biol. 2005;3:e10. doi: 10.1371/journal.pbio.0030010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström P.G., Ho Sui S.J., Drivenes O., Becker T.S., Lenhard B. Genomic regulatory blocks underlie extensive microsynteny conservation in insects. Genome Res. 2007;17:1898–1908. doi: 10.1101/gr.6669607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etchberger J.F., Hobert O. Vector-free DNA constructs improve transgene expression in C. elegans. Nat. Methods. 2008;5:3. doi: 10.1038/nmeth0108-3. [DOI] [PubMed] [Google Scholar]
- Etchberger J.F., Lorch A., Sleumer M.C., Zapf R., Jones S.J., Marra M.A., Holt R.A., Moerman D.G., Hobert O. The molecular signature and cis-regulatory architecture of a C. elegans gustatory neuron. Genes & Dev. 2007;21:1653–1674. doi: 10.1101/gad.1560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frasch M., Chen X., Lufkin T. Evolutionary-conserved enhancers direct region-specific expression of the murine Hoxa-1 and Hoxa-2 loci in both mice and Drosophila. Development. 1995;121:957–974. doi: 10.1242/dev.121.4.957. [DOI] [PubMed] [Google Scholar]
- Garcia-Fernandez J. The genesis and evolution of homeobox gene clusters. Nat. Rev. Genet. 2005;6:881–892. doi: 10.1038/nrg1723. [DOI] [PubMed] [Google Scholar]
- Gaudet J., Muttumu S., Horner M., Mango S.E. Whole-genome analysis of temporal gene expression during foregut development. PLoS Biol. 2004;2:e352. doi: 10.1371/journal.pbio.0020352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant K., Hanna-Rose W., Han M. sem-4 promotes vulval cell-fate determination in Caenorhabditis elegans through regulation of lin-39 Hox. Dev. Biol. 2000;224:496–506. doi: 10.1006/dbio.2000.9774. [DOI] [PubMed] [Google Scholar]
- Graustein A., Gaspar J.M., Walters J.R., Palopoli M.F. Levels of DNA polymorphism vary with mating system in the nematode genus Caenorhabditis. Genetics. 2002;161:99–107. doi: 10.1093/genetics/161.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haerry T.E., Gehring W.J. A conserved cluster of homeodomain binding sites in the mouse Hoxa-4 intron functions in Drosophila embryos as an enhancer that is directly regulated by Ultrabithorax. Dev. Biol. 1997;186:1–15. doi: 10.1006/dbio.1997.8582. [DOI] [PubMed] [Google Scholar]
- Hillier L.W., Coulson A., Murray J.I., Bao Z., Sulston J.E., Waterston R.H. Genomics in C. elegans: So many genes, such a little worm. Genome Res. 2005;15:1651–1660. doi: 10.1101/gr.3729105. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kelly W.G., Xu S., Montgomery M.K., Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caenorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J., Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Kiontke K., Hironaka M., Sudhaus W. Description of Caenorhabditis japonica n. sp. (Nematoda: Rhabditida) associated with the burrower bug Parastrachia japonensis (Heteroptera: Cydnidae) in Japan. Nematology. 2002;4:933–941. [Google Scholar]
- Kiontke K., Gavin N.P., Raynes Y., Roehrig C., Piano F., Fitch D.H. Caenorhabditis phylogeny predicts convergence of hermaphroditism and extensive intron loss. Proc. Natl. Acad. Sci. 2004;101:9003–9008. doi: 10.1073/pnas.0403094101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiontke K., Barriere A., Kolotuev I., Podbilewicz B., Sommer R., Fitch D.H., Felix M.A. Trends, stasis, and drift in the evolution of nematode vulva development. Curr. Biol. 2007;17:1925–1937. doi: 10.1016/j.cub.2007.10.061. [DOI] [PubMed] [Google Scholar]
- Korf I., Yandell M., Bedell J. BLAST. O'Reilly; Sebastopol, CA: 2003. [Google Scholar]
- Krause M., Harrison S.W., Xu S.Q., Chen L., Fire A. Elements regulating cell- and stage-specific expression of the C. elegans MyoD family homolog hlh-1. Dev. Biol. 1994;166:133–148. doi: 10.1006/dbio.1994.1302. [DOI] [PubMed] [Google Scholar]
- Krek A., Grün D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Lemons D., McGinnis W. Genomic evolution of Hox gene clusters. Science. 2006;313:1918–1922. doi: 10.1126/science.1132040. [DOI] [PubMed] [Google Scholar]
- Li L., Stoeckert C.J., Jr., Roos D.S. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Zhu Q., He X., Sinha S., Halfon M.S. Large-scale analysis of transcriptional cis-regulatory modules reveals both common features and distinct subclasses. Genome Biol. 2007;8:R101. doi: 10.1186/gb-2007-8-6-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P. The microRNAs of Caenorhabditis elegans. Genes & Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malicki J., Cianetti L.C., Peschle C., McGinnis W. A human HOX4B regulatory element provides head-specific expression in Drosophila embryos. Nature. 1992;358:345–347. doi: 10.1038/358345a0. [DOI] [PubMed] [Google Scholar]
- Maloof J.N., Kenyon C. The Hox gene lin-39 is required during C. elegans vulval induction to select the outcome of Ras signaling. Development. 1998;125:181–190. doi: 10.1242/dev.125.2.181. [DOI] [PubMed] [Google Scholar]
- McGaughey D.M., Vinton R.M., Huynh J., Al-Saif A., Beer M.A., McCallion A.S. Metrics of sequence constraint overlook regulatory sequences in an exhaustive analysis at phox2b. Genome Res. 2008;18:252–260. doi: 10.1101/gr.6929408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J.D., Sleumer M.C., Bilenky M., Wong K., McKay S.J., Goszczynski B., Tian H., Krich N.D., Khattra J., Holt R.A., et al. The ELT-2 GATA-factor and the global regulation of transcription in the C. elegans intestine. Dev. Biol. 2007;302:627–645. doi: 10.1016/j.ydbio.2006.10.024. [DOI] [PubMed] [Google Scholar]
- McKay S.J., Johnsen R., Khattra J., Asano J., Baillie D.L., Chan S., Dube N., Fang L., Goszczynski B., Ha E., et al. Gene expression profiling of cells, tissues, and developmental stages of the nematode C. elegans. Cold Spring Harb. Symp. Quant. Biol. 2003;68:159–169. doi: 10.1101/sqb.2003.68.159. [DOI] [PubMed] [Google Scholar]
- Mello C., Fire A. DNA transformation. Methods Cell Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- Mortazavi A., Leeper Thompson E.C., Garcia S.T., Myers R.M., Wold B. Comparative genomics modeling of the NRSF/REST repressor network: From single conserved sites to genome-wide repertoire. Genome Res. 2006;16:1208–1221. doi: 10.1101/gr.4997306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okkema P.G., Harrison S.W., Plunger V., Aryana A., Fire A. Sequence requirements for myosin gene expression and regulation in Caenorhabditis elegans. Genetics. 1993;135:385–404. doi: 10.1093/genetics/135.2.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson S. EMBOSS opens up sequence analysis. European Molecular Biology Open Software Suite. Bioinformatics. 2002;3:87–91. doi: 10.1093/bib/3.1.87. [DOI] [PubMed] [Google Scholar]
- Ovcharenko I., Loots G.G., Nobrega M.A., Hardison R.C., Miller W., Stubbs L. Evolution and functional classification of vertebrate gene deserts. Genome Res. 2005;15:137–145. doi: 10.1101/gr.3015505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli F., Liu Y., Kim Y.A., Chen P.J., Kim S.K. Chromosomal clustering and GATA transcriptional regulation of intestine-expressed genes in C. elegans. Development. 2006;133:287–295. doi: 10.1242/dev.02185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Popperl H., Bienz M., Studer M., Chan S.K., Aparicio S., Brenner S., Mann R.S., Krumlauf R. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell. 1995;81:1031–1042. doi: 10.1016/s0092-8674(05)80008-x. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I., Ruvkun G. Functional tests of enhancer conservation between distantly related species. Development. 2003;130:5133–5142. doi: 10.1242/dev.00711. [DOI] [PubMed] [Google Scholar]
- Sandelin A., Alkema W., Engstrom P., Wasserman W.W., Lenhard B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. doi: 10.1093/nar/gkh012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha S., Schroeder M.D., Unnerstall U., Gaul U., Siggia E.D. Cross-species comparison significantly improves genome-wide prediction of cis-regulatory modules in Drosophila. BMC Bioinformatics. 2004;5:129. doi: 10.1186/1471-2105-5-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stajich J.E., Block D., Boulez K., Brenner S.E., Chervitz S.A., Dagdigian C., Fuellen G., Gilbert J.G., Korf I., Lapp H., et al. The Bioperl toolkit: Perl modules for the life sciences. Genome Res. 2002;12:1611–1618. doi: 10.1101/gr.361602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Lin M.F., Kheradpour P., Pedersen J.S., Parts L., Carlson J.W., Crosby M.A., Rasmussen M.D., Roy S., Deoras A.N., et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–232. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg P.W. (TheC. elegans Research Community) WormBook. 2005. Vulval development. http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E.A., Cooper G.M., Sidow A. Trade-offs in detecting evolutionarily constrained sequence by comparative genomics. Annu. Rev. Genomics Hum. Genet. 2005;6:143–164. doi: 10.1146/annurev.genom.6.080604.162146. [DOI] [PubMed] [Google Scholar]
- Stothard P., Pilgrim D. Conspecific and interspecific interactions between the FEM-2 and the FEM-3 sex-determining proteins despite rapid sequence divergence. J. Mol. Evol. 2006;62:281–291. doi: 10.1007/s00239-005-0084-5. [DOI] [PubMed] [Google Scholar]
- Stoyanov C.N., Fleischmann M., Suzuki Y., Tapparel N., Gautron F., Streit A., Wood W.B., Muller F. Expression of the C. elegans labial orthologue ceh-13 during male tail morphogenesis. Dev. Biol. 2003;259:137–149. doi: 10.1016/s0012-1606(03)00138-6. [DOI] [PubMed] [Google Scholar]
- Streit A., Kohler R., Marty T., Belfiore M., Takacs-Vellai K., Vigano M.A., Schnabel R., Affolter M., Muller F. Conserved regulation of the Caenorhabditis elegans labial/Hox1 gene ceh-13. Dev. Biol. 2002;242:96–108. doi: 10.1006/dbio.2001.0544. [DOI] [PubMed] [Google Scholar]
- Sudhaus W., Kiontke K. Phylogeny of Rhabditis subgenus Caenorhabditis (Rhabditidae, Nematoda) J. Zoo. Syst. Evol. 1996;34:217–233. [Google Scholar]
- Sudhaus W., Kiontke K. Comparison of the cryptic nematode species Caenorhabditis brenneri sp. n. and C. remanei (Nematoda: Rhabditidae) with the stem species pattern of the Caenorhabditis elegans group. Zootaxa. 2007;1456:45–62. [Google Scholar]
- Sulston J., Hodgkin J. Methods In The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1988. pp. 587–606. [Google Scholar]
- Sulston J.E., Horvitz H.R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Tagle D.A., Koop B.F., Goodman M., Slightom J.L., Hess D.L., Jones R.T. Embryonic and globin genes of a prosimian primate (Galago crassicaudatus). Nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J. Mol. Biol. 1988;203:439–455. doi: 10.1016/0022-2836(88)90011-3. [DOI] [PubMed] [Google Scholar]
- Tompa M., Li N., Bailey T.L., Church G.M., De Moor B., Eskin E., Favorov A.V., Frith M.C., Fu Y., Kent W.J., et al. Assessing computational tools for the discovery of transcription factor binding sites. Nat. Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- Wagmaister J.A., Miley G.R., Morris C.A., Gleason J.E., Miller L.M., Kornfeld K., Eisenmann D.M. Identification of cis-regulatory elements from the C. elegans Hox gene lin-39 required for embryonic expression and for regulation by the transcription factors LIN-1, LIN-31 and LIN-39. Dev. Biol. 2006;297:550–565. doi: 10.1016/j.ydbio.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Wang X., Chamberlin H.M. Evolutionary innovation of the excretory system in Caenorhabditis elegans. Nat. Genet. 2004;36:231–232. doi: 10.1038/ng1301. [DOI] [PubMed] [Google Scholar]
- Wang T., Stormo G.D. Combining phylogenetic data with co-regulated genes to identify regulatory motifs. Bioinformatics. 2003;19:2369–2380. doi: 10.1093/bioinformatics/btg329. [DOI] [PubMed] [Google Scholar]
- Wang B.B., Muller-Immergluck M.M., Austin J., Robinson N.T., Chisholm A., Kenyon C. A homeotic gene cluster patterns the anteroposterior body axis of C. elegans. Cell. 1993;74:29–42. doi: 10.1016/0092-8674(93)90292-x. [DOI] [PubMed] [Google Scholar]
- Wei C., Lamesch P., Arumugam M., Rosenberg J., Hu P., Vidal M., Brent M.R. Closing in on the C. elegans ORFeome by cloning TWINSCAN predictions. GenomeRes. 2005;15:577–582. doi: 10.1101/gr.3329005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenick A.S., Hobert O. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev. Cell. 2004;6:757–770. doi: 10.1016/j.devcel.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Wittmann C., Bossinger O., Goldstein B., Fleischmann M., Kohler R., Brunschwig K., Tobler H., Muller F. The expression of the C. elegans labial-like Hox gene ceh-13 during early embryogenesis relies on cell fate and on anteroposterior cell polarity. Development. 1997;124:4193–4200. doi: 10.1242/dev.124.21.4193. [DOI] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Mikkelsen T.S., Gnirke A., Lindblad-Toh K., Kellis M., Lander E.S. Systematic discovery of regulatory motifs in conserved regions of the human genome, including thousands of CTCF insulator sites. Proc. Natl. Acad. Sci. 2007;104:7145–7150. doi: 10.1073/pnas.0701811104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G., Schriefer L.A., Stormo G.D. Identification of muscle-specific regulatory modules in Caenorhabditis elegans. Genome Res. 2007;17:348–357. doi: 10.1101/gr.5989907. [DOI] [PMC free article] [PubMed] [Google Scholar]