Abstract
Here, we report the sequence characterization of the bovine pseudoautosomal boundary (PAB) and its neighborhood. We demonstrate that it maps to the 5′ end of the GPR143 gene, which has concomitantly lost upstream noncoding exons on the Y chromosome. We show that the bovine PAB was created ∼20.7 million years ago by illegitimate intrachromatid recombination between inverted, ruminant-specific Bov-tA repeats. Accordingly, we demonstrate that cattle share their PAB with all other examined ruminants including sheep, but not with cetaceans or more distantly related mammals. We provide evidence that, since its creation, the ancestral ruminant PAB has been displaced by attrition, which occurs at variable rates in different species, and that it is capable of retreat by attrition erasure. We have estimated the ratio of male to female mutation rates in the Bovidae family as ∼1.7, and we provide evidence that the mutation rate is higher in the recombining pseudoautosomal region than in the adjacent, nonrecombining gonosome-specific sequences.
Maleness in placental mammals and marsupials is determined by the SRY gene located on the Y chromosome. This major sex determinant arose ∼166 million years ago (Mya) on an ancestral autosome as an allele of the SOX3 gene (Veyrunes et al. 2008). As is commonly observed for chromosomes carrying sex-determining genes (Ohno 1967), the Y has since undergone progressive degeneration, being reduced in present-day man to a mere 25 Mb of euchromatin harboring no more than 27 distinct protein-coding genes or gene families, appended with an approximately equal amount of dispensable heterochromatin (Skaletsky et al. 2003). These numbers are to be compared with the ∼155 Mb and 1100 genes of its ancestral partner, the X chromosome (Ross et al. 2005).
The decay of the Y is thought to result from the successive selection of male-beneficial/female-deleterious alleles embedded in haplotypes that lost the ability to recombine with the X and are hence confined to males (Charlesworth 1991). Absence of recombination causes rapid degeneration by mutation, deletion, and transposon invasion accumulating as a result of a higher mutation rate in the male versus the female germline (due to the larger number of cell divisions required to produce male vs. female gametes), inefficient repair (e.g., Muller’s ratchet), and inefficient selection (e.g., shielding of deleterious recessives and Hill–Robertson interference) (e.g., Charlesworth et al. 2005; Bachtrog 2006; Graves 2006).
The most commonly invoked recombination-blocking mechanism is chromosomal inversion. The observation of a stepwise increase in sequence similarity between genes ordered on the human X with their gametologs on the Y (“evolutionary strata”) suggests that five such recombination-blocking inversions have occurred in the human lineage (Lahn and Page 1999; Ross et al. 2005). These have isolated an increasing proportion of the Y from its X partner, progressively reducing the region of X–Y homology to the ∼2.7 Mb pseudoautosomal region 1 (PAR1). The five inversions in the human lineage were initially dated to 240–320 Mya, 130–170 Mya, 80–130 Mya, 38–44 Mya, and 29–32 Mya, respectively, yet recent reexamination of the age of the therian sex chromosomes (Veyrunes et al. 2008) forces reevaluation of these estimates.
Loss of genes from the Y causes male hemizygosity and thus a different gonosome-to-autosome balance in the two sexes. This is thought to drive progression of dosage compensation involving (in mammals) doubling of expression levels from the X (Nguyen and Disteche 2006) and compensatory XIST-dependent inactivation of one X chromosome in females (Lyon 1961; Heard and Disteche 2006). Notably, while virtually all genes located in the older strata undergo X inactivation, their proportion decreases in the younger layers (Carrel and Willard 2005). Concomitantly, the enrichment in L1 interspersed repeats, which may operate as way stations spreading the inactivation process (Lyon 1998; Carrel et al. 2006), increases with stratum age (Ross et al. 2005).
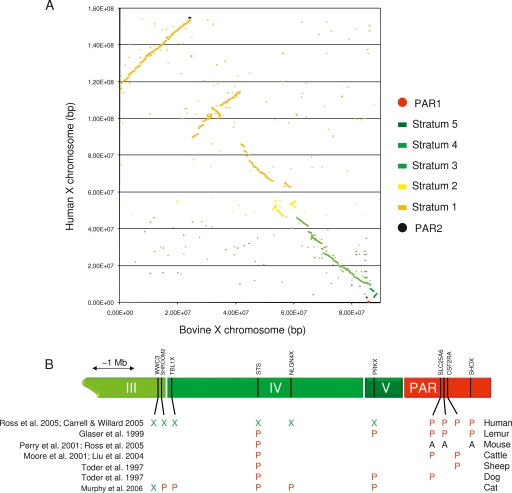
The generalization of dosage compensation across most of the X chromosome is thought to underlie its “frozen” gene content in mammals (Ohno 1967). The human and dog X chromosome sequences, for instance, are essentially colinear, while the human and mouse X chromosomes are nearly perfectly syntenic despite multiple intrachromosomal rearrangements (Ross et al. 2005). Figure 1A illustrates the equally remarkable conservation of gene content and order between the human and bovine X chromosomes.
Figure 1.
(A) Graphical representation of unique BLAST hits (E-value < 10−2) between the human and bovine X chromosomes. Sequences mapping to human PAR1-2 or strata 1–5 are color-labeled as indicated. (B) Schematic representation of distal Xp in human indicating the limits between strata III, IV, and V (green) and PAR1 (red), and approximate positions of 10 genes. Known pseudoautosomal (P), X-specific (X), or autosomal (A) status of orthologs in six mammals with corresponding references.
Despite the largely frozen gene content of the X, the evolution of mammalian sex chromosomes has been punctuated by interchromosomal exchanges. An autosome to proto-gonosome translocation occurring after placental mammals diverged from marsupials has increased the size of the eutherian neo-gonosome by addition of the “X added region” (XAR) (Graves 2006). Autosome to Y transposition has augmented the content of the Y chromosome in male-beneficial genes, including retrotransposition of CDY before the divergence of marsupials and eutherians (Lahn and Page 1999; Skaletsky et al. 2003), transposition of DAZ during primate evolution (Saxena et al. 1996), and transposition of FLJ36031 prior to carnivore radiation and TETY1 following the divergence of cat and dog lineages (Murphy et al. 2006). Moreover, the human Y euchromosome has acquired an X transposed region (XTR) after its divergence from chimpanzees (Skaletsky et al. 2003). In addition, X-linked genes have generated pseudogenes by retrotransposition to autosomes, presumably to compensate for their silencing during male meiotic sex chromosome inactivation (MSCI) (e.g., Potrzebowski et al. 2008).
Pseudoautosomal regions (PARs)
Despite their growing divergence, the mammalian X and Y maintain a short region of homology, allowing pairing and recombination in males that is required for faithful segregation of the sex chromosomes. Obligatory crossing-over in the PAR in males erases sex linkage for the more distal markers, justifying the “pseudoautosomal” designation.
The exceptionally high centimorgan to megabase ratio in the PAR (∼20-fold higher than the genome average in humans [Brown 1988; Petit et al. 1988; Lien et al. 2000]) is thought to account for its higher GC content, as a result of recombination-associated gene conversion biased toward GC. Notably, a gradual increase in GC content is observed on the human X when moving from old to young strata and finally to the present-day PAR (e.g., Ross et al. 2005). Enhanced recombination is also thought to account for the accelerated rate of evolution noted for genes in the PAR (particularly in rodents and to a lesser extent in primates), as recombination is accompanied by DNA repair relying on low-fidelity DNA polymerases (Perry and Ashworth 1999; Filatov and Gerrard 2003; Galtier 2004; Yi et al. 2004).
As stated above, the human Xp PAR1 measures ∼2.7 Mb and harbors 24 genes. In other mammals studied, the PAR is thought to be larger as it encompasses genes that are pseudoautosomal in human (notably SHOX, IL3RA, CSF2RA, and SLC25A6 [ANT3]), plus genes that have become X-specific in the human lineage (notably PRKX and STS mapping to human stratum 4). This assumption applies to lemurs (Gläser et al. 1999), sheep (Toder et al. 1997), cattle (Moore et al. 2001), dog (Toder et al. 1997), and cat (Murphy et al. 2006). The PAR of Mus musculus domesticus includes Sts, but none of the genes that are pseudoautosomal in human. This is apparently due to the loss of ∼9 Mb from its distal end, reducing the size of the PAR to a mere ∼700 kb (Perry et al. 2001; Ross et al. 2005).
Taken together, these results suggest that the human pseudoautosomal boundary (PAB) has advanced further in the ancestral eutherian PAR when compared with most other mammals (Fig. 1B). The human sex chromosomes are also unique in that they possess two PARs. PAR2, on distal Xq, measures 330 kb and contains five genes. It is thought to be a piece of the X chromosome recently acquired by the Y by means of LINE-mediated illegitimate recombination (Charchar et al. 2003). The recombination rate in PAR2 is reportedly much lower than in PAR1 (Lien et al. 2000).
Extant and ancestral pseudoautosomal boundaries (PABs)
To the best of our knowledge, extant PABs have only been characterized only at the sequence level for humans, Great Apes, Old World Monkeys and the domestic mouse. In Catarrhini, the PAB maps within the gene coding for the XG blood group antigen, also called PBDX (pseudoautosomal boundary divided on the X) (Ellis et al. 1990, 1994). As a result, XG is disrupted on the Y, missing nine exons on the 3′ side. This is compatible either with a pericentric inversion (Ellis et al. 1994) or with the intrachromosomal transposition of a chromosome fragment including SRY (Gläser et al. 1999). Since its creation, the PAB of Catarrhini has shifted ∼240 bp into the PAR by “attrition,” accounting for the fact that the present-day PAB is flanked by a 240-bp segment of reduced homology (∼77%) on its proximal side. It is thought that an Alu element has subsequently been inserted at the exact location of the PAB in the common ancestor of humans and Great Apes, without perturbing its position, and separating the pseudoautosomal “Alu-distal region” from the sex-specific “Alu-proximal region” (Ellis et al. 1990).
The PAB of M. musculus domesticus is located in the third intron of the Mid1 (also known as Fxy) gene and truncates the 5′ end of the Y copy. The pseudoautosomal 3′ end of the gene starts with a variable number of tandem (intron 3–exon 4)n copies (Palmer et al. 1997). The history of the PAB in rodents remains somewhat confusing. Mid1 is X-specific in M. spretus and rat, indicating that the likely position of the ancestral rodent PAB is distal from Mid1 (Perry and Ashworth 1999). As the M. musculus domesticus PAB coincides with Mid1, this means either that the PAB moved backward to adopt a more proximal position in the domesticus lineage, or that Mid1 was translocated to a more distal position, as proposed by Galtier (2004). The completion of the sequences of the murine X and Y chromosomes should clarify this issue.
The feline PAB has not been defined at the sequence level but has been tentatively positioned between SHROOM2 and WWC3 based on an abrupt drop in retention frequency in radiation hybrids obtained from male cells (Murphy et al. 2007).
Ancestral PABs in the human lineage have been tentatively mapped to gene intervals corresponding with abrupt changes in Ks between gametologs. Hence, the boundaries between strata 1–2, 2–3, 3–4, and 4–5 have been positioned in the CXORF39–ZXDA, RGN–PHF16, WWC3–GPR143 (=OA1), and NLGN4X–AA971220 intervals, respectively (Skaletsky et al. 2003; Carrel and Willard 2005; Ross et al. 2005). Note that, especially for strata 3 and 4, the boundary is blurry, with some confidence intervals of Ks values being nonoverlapping within strata, while overlapping between strata (Skaletsky et al. 2003; Ross et al. 2005). As an example, while TBL1X, GPR143 (OA1), SHROOM2 (APXL), and AMELX map in that order on the human X, the Ks for TBL1X and SHROOM2 matches that of stratum 3 best, while that of GPR143 and AMELX places these in stratum 4. However, the investigators considered that it was premature to conclude that suppression of X–Y crossing over evolved in more than five steps, as alternative explanations including local changes in gene order and/or gene conversion might account for these findings (Skaltesky et al. 2003).
Intriguingly, the observation of an abrupt increase in GC content and decrease in divergence between gametologs when moving from the 5′ to the 3′ end of the AMELX/Y genes in six mammalian species, plus the fact that in phylogenetic analyses orthologs cluster at the 5′ end of the gene while gametologs cluster at the 3′ end of the gene, suggest that the AMEL locus may span the ancestral PAB separating human strata 3 and 4 (Iwase et al. 2003; Marais and Galtier 2003). As the AMELX/Y genes are not interrupted, this suggests that recombination-blocking mechanisms other than chromosomal rearrangements may be involved in isolating the X and Y chromosomes.
In this work, we report the identification and sequence characterization of the PAB and its neighborhood in ruminants.
Results
Identifying and sequencing Y- and X-specific BACs spanning the bovine PAB
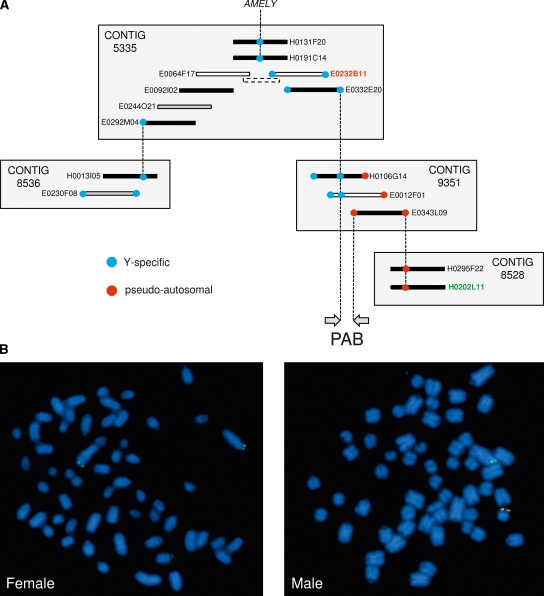
To identify Y-specific bacterial artificial chromosomes (BACs) spanning the bovine PAB, we initiated a bidirectional chromosome walk starting from AMELY, the Y-specific locus mapping closest to the PAB (Liu et al. 2002). A male BAC library (Warren et al. 2000) was screened with AMELY probes. A BAC contig (no. 5335) containing PCR-confirmed positive clones was retrieved from the BAC-based fingerprint map of the bovine genome (http://www.bcgsc.ca/platform/mapping/bovine) using iCE (Fjell et al. 2003). New probes were designed from the end sequences of the outer BACs of this contig. PCR on male and female DNA was used to discriminate between pseudoautosomal or Y-specific probes before a new library screening was carried out. Several rounds of hybridization were carried out until a probe designed on a Y-specific contig (no. 9351) turned out to be pseudoautosomal. This implied that contig 9351 spanned the PAB. BAC end-derived probes covering the entire contig length were used to refine the position of the PAB within the contig by PCR. Two BAC clones were hence shown to lie across the PAB on the Y chromosome: E0012F01 and H0106G14 (Fig. 2A). E0012F01 was completely sequenced as described in the Supplemental Materials. The predicted restriction pattern of the resulting 180,781 kb of finished sequence was compared with the experimental pattern obtained by restriction digestion (AsuII, BamHI, XbaI) and subsequent pulsed-field gel electrophoresis (data not shown).
Figure 2.
(A) Chromosome walk from the Y-specific AMELY gene across the bovine PAB. BACs are sorted by contigs as retrieved from the BAC-based fingerprint map of the bovine genome (http://www.bcgsc.ca/platform/mapping/bovine). Only BACs relevant in the context of this study are shown. (Blue dots) Y-specific BAC end sequences, (red dots) pseudoautosomal BAC end sequences. Dotted lines connect BAC ends used as probes in filter hybridization and resulting positive BAC clones. Sequenced BACs are shown as finished (white bars) or draft (gray bars). (White bar with dotted contour) Sequenced PCR product and plasmids connecting E0064F17 and E0232B11. (Arrows) Interval encompassing the bovine PAB. The identifiers of the BACs used as probes in the fluorescence in situ hybridization (FISH) experiment are labeled (red and green, respectively). (B) Representative FISH results obtained with the Y-specific E0232B11 (red) and pseudoautosomal H0202L11 (green) BACs on female (left panel) and male (right panel) metaphases, respectively.
Alignment of the E0012F01 sequence with the BTAX sequences available in the public domain (Btau_3.1), indicated that BAC E0383I16 must span the PAB on the X chromosome. However, only 6 kb of contiguous X-specific sequences bordering the PAB were reported at the time. To better characterize the X-specific PAB neighborhood, we completed the corresponding sequence as described in the Supplemental materials. This led to a contiguous 156,628-kb long sequence, including 129,705 kb of X-specific sequence proximal to the PAB.
To obtain additional Y-specific sequences, we sequenced BACs E0232B11 and E0064F17, as well as a 12-kb bridging fragment, to yield a total of 425,809 kb of contiguous finished sequence adjacent to the PAB (Fig. 2A; Supplemental materials).
To confirm the Y-specific and pseudoautosomal origin of the identified BACs, we performed fluorescence in situ hybridization (FISH) on male and female metaphase chromosomes using H0202L11 and E0232B11 as probes. As expected for a pseudoautosomal sequence, H0202L11 labeled the extremity of the two Xq arms in the female, while hybridizing to Xq and distal Yp in the male. E0232B11, on the other hand, hybridized exclusively to distal Yp, in the immediate vicinity of the H0202L11 signal, as expected for a Y-specific probe adjacent to the PAB (Fig. 2B).
Pinpointing the bovine PAB and annotating genes in its neighborhood
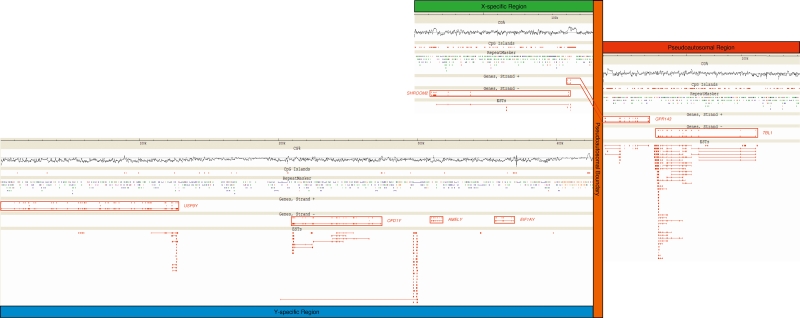
The obtained sequences were annotated as follows: (1) Genes were predicted by BLASTing the masked BAC sequences against human cDNA at Ensembl (http://www.ensembl.org) and bovine EST at NCBI (http://www.ncbi.nlm.nih.gov), (2) the moving average [G+C] content was determined using a 200-bp sliding window, (3) CpG islands were identified following Gardiner-Garden and Frommer (1987), and (4) repetitive elements were identified using Repeat Masker (A.F.A Smit and P. Green, http://repeatmasker.genome.washington.edu). The resulting genomic landscape is shown in Figure 3 (see gatefold).
Figure 3.
Genomic landscape surrounding the bovine pseudoautosomal boundary: pseudoautosomal region (red), X-specific sequences (green), Y-specific sequences (blue), and pseudoautsomal boundary (orange). (CG%) [G+C] content calculated in a 200-bp sliding window; the horizontal line corresponds to 50% [G+C]. (CpG Islands) CpG islands defined according to Gardiner-Garden and Frommer (1987). (RepeatMasker) Repetitive elements identified with RepeatMasker (A.F.A Smit and P. Green, http://repeatmasker.genome.washington.edu): LTRs (orange), SINEs (blue), LINEs (green), simple repeats (red), tRNAs and snRNAs (black). (Genes) Protein-encoding genes are transcribed from right to left (strand +) or left to right (strand −); gene names are given adjacent to the corresponding boxed gene. (ESTs) ESTs BLASTing to the region with an identity >95% and an E-value <10−50. Most ESTs correspond to identified exons with the notable exception of a cluster of ESTs located between OFD1Y and AMELY. Although very similar, these ESTs show some differences with the genomic sequence and with each other. They probably originate from paralogous loci.
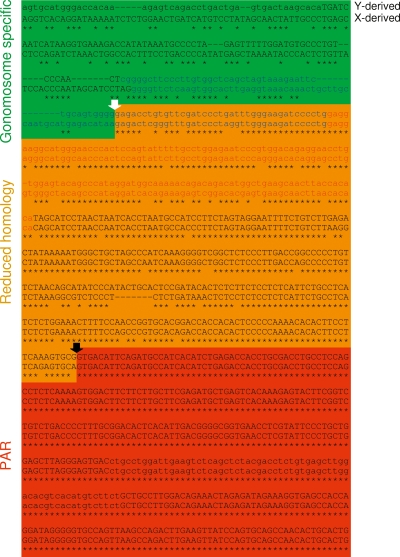
Alignment of the PAB-spanning E0012F01 (Y chromosome) and H0025A18 (X chromosome) sequences identified segments of near perfect homology (99.97%) corresponding to the PAR, diverging respectively into Y- and X-specific sequences, hence defining the bovine PAB. Detailed examination of the gonosome-specific sequences adjoining the PAB revealed a 413-bp segment of reduced homology (86.20%) that separates the PAR sequences from the clearly nonhomologous X- and Y-specific sequences. This segment of reduced homology is reminiscent of the Alu-proximal region of the human PAB, which is supposed to reflect progressive displacement of the PAB by attrition (Ellis et al. 1990, 1994). The boundary between the segment of reduced homology and the gonosome-specific sequences coincides with the tRNA portion of a Bov-tA1 SINE element on the X and a closely related Bov-tA2 element on the Y (Fig. 4). This strongly suggests that the original PAB (i.e., before it shifted by attrition) was created by intrachromatid recombination between inverted Bov-tA repeats. As Bov-tA elements are reportedly ruminant-specific (Shimamura et al. 1999), this finding implied that the bovine PAB had to be younger than the time of divergence of the ruminant lineage from the other mammals.
Figure 4.
Bovine pseudoautosomal boundary (PAB). Corresponding Y- and X-derived sequences were aligned using ClustalW, revealing (1) a region of near perfect homology (red) corresponding to the PAR, (2) a region of reduced homology (orange) resulting from the displacement of the PAB by attrition, and (3) nonhomologous gonosome-specific segments (green). (Black arrow) Present-day bovine PAB, (white arrow) ancestral PAB. (Lowercase letters) Repetitive sequences, (uppercase letters) unique sequences. (Red) Monomer domain of the Bov-tA repeat overlapping the ancestral PAB, (blue) tRNA-like domains differentiating the Bov-tA1 repeat of the X and the Bov-tA2 repeat of the Y.
According to the Ensembl annotation of the X chromosome, the bovine PAB is located just upstream of the GPR143 gene. This places the bovine PAB in the intergenic region separating SHROOM2 and GPR143. However, a detailed examination of bovine EST sequences (e.g., DV913014 and EH378090) identified a putative, noncoding, upstream exon of GPR143 lying across the PAB on the X chromosome. We performed 5′ RACE experiments (Fig. 3; Supplemental Fig. 1) and confirmed that at least some of the X-derived GPR143 transcripts are indeed initiated proximally from the PAB. The bovine PAB thus truncates at least two 5′ exons from the GPR143 gene on the Y chromosome. The GPR143 gene on the Y chromosome seems nevertheless transcriptionally competent. Indeed, using a SNP in exon 5, we demonstrated the existence of Y-derived GPR143 transcripts, initiated distally from the PAB (Supplemental Fig. 1; Supplemental materials).
We identified four genes within the available bovine Y-specific sequences, in the order USP9Y–OFD1Y–AMELY–EIF1AY–PAB (Fig. 3). USP9Y and EIF1AY have all the hallmarks of functional housekeeping genes encoding widely expressed ubiquitin-specific protease 9 and eukaryotic translation initiation factor 1A, respectively. They show minor idiosyncrasies when compared with their X-linked gametologs, which are unlikely to be inactivating, yet may provide some functional specificity. USP9Y is thought to be functional in humans and mice as well (albeit with testis-specific expression in the mouse), while EIF1AY is functional in humans but has not yet been described in the mouse. The structure of the bovine AMELY also suggests that it is functional, as it is in other species. However, the absence of an AMELY EST in the bovine databases suggests that it is expressed exclusively in the developing tooth in cattle as well. EST data indicate that OFD1Y is transcribed, yet analysis of the corresponding sequence reveals numerous differences with OFD1X, which, more than likely, preclude the production of a functional protein. In cattle, OFD1Y is therefore likely to be a transcribed pseudogene. On the human Y chromosome, OFD1 has evolved in a multicopy pseudogene family, while no Y-linked Ofd1 copy has yet been reported in the mouse. In addition, we identified five of the 20 exons of the SHROOM2 gene, which encodes the human homolog of the Xenopus laevis apx gene, on the bovine X chromosome, and 15 of the 17 exons of the TBL1 gene, encoding transducin β-like 1, in the PAR. A more detailed description of each of these seven genes is provided as Supplemental material.
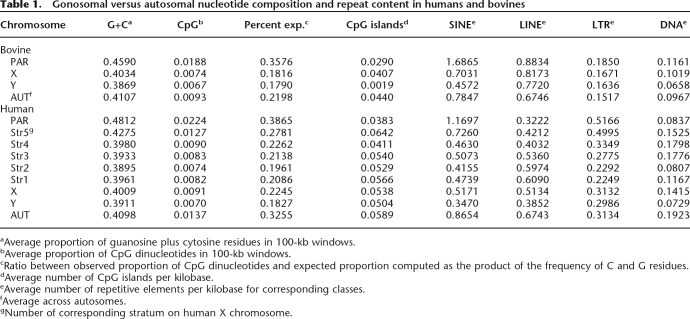
Base-pair composition and repeat content of the bovine PAR and sex chromosomes
Strikingly, the bovine PAB is flanked, on both the X and Y chromosomes, by ∼1- to 5-kb segments of very high G+C content (>70%) that correspond to unusually long CpG islands. Moreover, it is flanked on the Y chromosome by an unusual, ∼17-kb long segment, composed primarily of MaLRs (Mammalian apparent LTR-retrotransposons) elements. Neither of these features is shared by the human PAB. Further examination of the BAC sequences points toward a high G+C and CpG island content for the PAR, intermediate values for the X-specific sequences, and the lowest values for the Y-specific sequences (Fig. 3). To obtain a more complete view of the base-pair composition of the bovine sex chromosomes, we extracted all available PAR, X-specific, and autosomal sequences from build Btau4.0 of the bovine genome sequence and compared the corresponding base-pair compositions with that of the available Y-derived sequences. We extracted the equivalent human sequences (build NCBI 36) for comparison (Table 1).
Table 1.
Gonosomal versus autosomal nucleotide composition and repeat content in humans and bovines
aAverage proportion of guanosine plus cytosine residues in 100-kb windows.
bAverage proportion of CpG dinucleotides in 100-kb windows.
cRatio between observed proportion of CpG dinucleotides and expected proportion computed as the product of the frequency of C and G residues.
dAverage number of CpG islands per kilobase.
eAverage number of repetitive elements per kilobase for corresponding classes.
fAverage across autosomes.
gNumber of corresponding stratum on human X chromosome.
From this, it appears that the G+C and CpG dinucleotide content is well correlated with recombinational activity, being highest for the PAR, followed by the autosomes, X-specific, and Y-specific sequences. It is higher for the human than for the bovine PAR, which is compatible with the smaller size and hence higher recombination rate per base pair in the human PAR. It is worthwhile noting, however, that when compared with the ends of several autosomes, the rise in G+C and CpG content in the PAR is not unusual in magnitude (Supplemental Fig. 2).
In humans, the density of CpG islands is highest on the autosomes, followed by the X chromosome, Y chromosome, and PAR. The same ranking is observed in bovines except for the available Y-specific sequences, which were remarkably depleted of CpG islands. It remains to be determined whether this feature will extend to the rest of the bovine Y.
Y chromosome decay is predicted to cause accumulation of transposable elements (e.g., Bachtrog 2006). However, this was not observed either on the bovine or the human Y. The bovine PAR, however, was enriched in the four classes of repeats when compared with the rest of the genome (Table 1; Supplemental Fig. 2). This was particularly striking for SINEs, with the PAR having a density in SINEs more than twice that of the X-specific region or of the autosomal average. This enrichment of interspersed repeats on the PAR was not observed in humans. The increase in LINE density on the X when compared with autosomes, first noticed by Lyon (1998), was also apparent in bovines (Supplemental Fig. 2). However, in bovines, we found a higher LINE density in the PAR than for the remainder of the X.
The bovine PAB is ruminant-specific
To verify whether Bos taurus shares its PAB with other ruminants, we used bovine primers to amplify the orthologuous sequences of four Bovinae (Bison, Yak, Banteng, and Zebu) and one Caprinae (sheep). Y- and X-specific products spanning the PAB could be amplified and sequenced for all species (Supplemental Fig. 3), demonstrating that this boundary predates the divergence of Bovinae and Caprinae and is thus at least ∼18 million years (Myr) old (Hassanin and Ropiquet 2004).
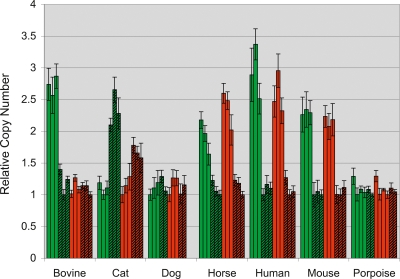
To verify whether the identified PAB is indeed ruminant-specific (as suggested by the occurrence of a Bov-tA element bridging the limit of homology), we compared the number of SHROOM2 and GPR143 copies in cattle, porpoises, horses, cats, dogs, mice, and humans of both sexes. Porpoises are cetaceans, which are assumed (with hippopotamus) to be the closest relatives of ruminants, having diverged an estimated ∼50 Mya (e.g., Graur and Higgins 1994; Gatesy et al. 1996; Shimamura et al. 1997). Note that the genome of cetaceans does not contain Bov-tA repeats (Shimamura et al. 1999).
We reasoned that the female-to-male copy ratio should be one for genes in the PAR, and two for X-specific genes. To ensure that our species-specific SHROOM2 and GPR143 PCR primers would amplify only the X-specific copy of non-PAR genes, we designed at least one of the primers in intronic sequences (except for porpoise) and verified the homogeneity of the amplified sequences by monitoring their melting behavior (dissociation curve) and, for some of them, by sequencing. One to three species-specific autosomal amplicons were used to control for varying amounts of template DNA, and relative copy numbers of SHROOM2 and GPR143 were estimated in males and females using qBase (Fig. 5; Hellemans et al. 2007). For the species with known PAB, we obtained the expected results: In humans and mice, the female-to-male copy-number ratio was about two for both SHROOM2 and GPR143, while in cattle the ratio was about two for SHROOM2 but only about one for GPR143. Most interestingly, in the examined cetacean, the ratio was approximately one for both genes. This indicates that both genes are located in the PAR, and thus that the PAB is more proximal in this species. This finding suggests that the chromosomal event that created the PAB of ruminants occurred after the divergence of cetaceans.
Figure 5.
Relative copy numbers of the SHROOM2 (green) and GPR143 (red) genes in females (plain) and males (hatched) of different mammalian species determined by quantitative PCR. Each bar corresponds to one individual. Error bars correspond to the standard deviation over three replicates.
In the horse, the female-to-male copy ratio was about two for both genes (as in human and mice), thus indicating their X-specific location and hence a more distal position of the equine PAB. In dogs, the ratio was about one for both genes, implying a pseudoautosomal location as in porpoises. Unexpectedly, in cat the ratio was about one-half for both genes. This suggests that the feline Y chromosome harbors SHROOM2 and GPR143 sequences that are very closely related to the X-specific gametolog sequences, possibly pointing toward recent X to Y transposition. It precludes conclusions regarding their location with respect to the PAB. However, it indicates that the higher retention rate of SHROOM2 relative to X-specific sequences that was observed in male radiation hybrids (Murphy et al. 2006) may not be due to its location on the PAR.
Lineage-specific PAB attrition and retreat
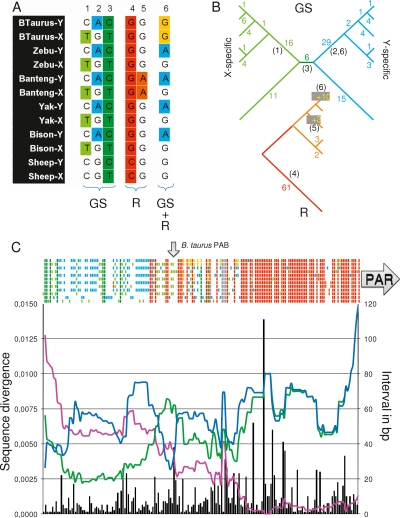
To further characterize the ruminant-specific PAB, we aligned the available homologous X- and Y-derived sequences (i.e., the proximal gonosome-specific 413-bp segment of reduced homology and 1233 bp of PAR sequence distal from the PAB) across the six studied species using ClustalW (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form=clustalw-multialign). 181 residues were found to be variable. 173 of these were characterized by two states and eight by three states. The genotypes of the studied species at a given site (“gonotypes”) resulted from one or more mutations, and—for some of the sites—one or more recombination events between the X and Y. Mutations and recombination events can be parsimoniously (i.e., trying to minimize the number of events needed to explain the observed genotype vector) mapped on the species tree (assumed to be known; Hassanin and Ropiquet 2004). Unless recombination blurred their gonosomal origin, mutations can also be assigned to either the X or Y chromosome. The 181 gonotype vectors could be interpreted in terms of 178 mutations and 82 recombinations. Gonosomal origin could be inferred for 110 out of the 178 mutations. These will be referred to as gonosome-specifying (GS) events, while recombinations will be referred to as R events (Fig. 6A,B).
Figure 6.
Comparative sequence analysis of the PAB in sheep and Bovinae (Bovine, Zebu, Banteng, Yak, and Bison), focusing on the 413 bp of reduced homology and 1233 bp in the PAR. X- and Y-derived sequences were aligned using ClustalW, and 181 variant sites were identified. (A) Examples of variant sites with color-labeled gonosome-specifying (GS) (green or blue) or recombinational (R) events (yellow, orange, or red). (1–3) GS events corresponding to: (1) a C to T transition on the X of the ancestor of Bovinae, (2) a G to A transition on the Y of the ancestor of Bovinae, and (3) a C to T transition on the X or Y of the ancestor of Bovinae and Caprinae. (4,5) R events corresponding to: (4) a C to G transversion and proximal X↔Y recombination having occurred on the red segment in the R-tree, (5) a G to A transition and proximal X↔Y recombination having occurred on the Banteng-specific branch in the R-tree. (6) Composite GS/R sites having undergone a G to A transition on the Y of the Bovinae ancestor and a X↔Y recombination on the cattle branch. (B) Tallying of the identified GS and R events on the known species tree (Hassanin and Ropiquet 2004). GS events are mapped on a tree distinguishing the X and Y chromosomes. R events are mapped on the species tree. The numbers of events mapped to a given branch are given. (Red and orange R numbers) Mutations that could not be assigned to the X or Y, (yellow R numbers) mutations with known gonosomal origin mapped on the GS tree (e.g., example 6). The numbers in parentheses correspond to the examples in A. (C) (Colored bars above the graph) Status of the six species (ordered vertically as in A) for the 181 variant sites. (Black vertical bars) Number of base pairs between adjacent sites with corresponding y-axis on the right. (Pink curve) Average within-species sequence divergence (average of moving 100-bp window) between the X- and Y-derived sequences. (Blue curve) Average sequence divergence for the Y-derived sequence of sheep versus the five Bovinae (average of moving 100-bp window). (Green curve) Average sequence divergence for the X-derived sequence of sheep versus the five Bovinae (average of moving 100-bp window). (Arrows) Positions of the B. taurus PAB and the PAR.
As shown in Figure 6C, the sequence alignement is characterized by two opposite gradients of GS and R events, respectively. The proximal part is dominated by GS events, hence with very little evidence for ancestral recombinations. The distal part, on the other hand, is dominated by R events, indicative of numerous intergonosomal recombinations. The intermediate part is characterized by intermingled GS and R events, supporting the progressive displacement of the recombination barrier over time, with the most proximal R sites marking the oldest exchanges between the sex chromosomes.
The Bos taurus PAB, i.e., the boundary between the segment of reduced homology and the PAR, maps in the middle of this intermediate part. Only one cattle-specific GS event is observed distally from this point in Bos taurus, which is compatible with regular interallelic nucleotide diversity in domestic cattle (∼1/2000) (e.g., Steele and Georges 1991). Detailed examination of the aligned Y- and X-derived sequences in the other species (Fig. 6C; Supplemental Fig. 3) indicates that the segment of reduced homology (characterized by levels of sequence divergence between Y and X that are well above normal levels of allelic nucleotide diversity [>1/500]) is considerably larger in these species than in cattle. This is particularly striking for Zebu and Banteng for which the segment of reduced homology extends over virtually the entire length of the sequenced amplicon. This suggests that attrition may proceed at different rates in different lineages.
Several of the GS events on the distal side of the Bos taurus PAB predate the Bovinae or even ruminant radiation, as several species share the same gonotype. The fact that the X and Y of Bos taurus have the same residues at the corresponding sites (the Y-specifying state for some, the X-specifying state for others), implies that the distal progression of the PAB by attrition can occasionally be reversed by recombination events between diverging X- and Y-specific sequences. This phenomenon of “PAB retreat” or “attrition erasure” has not been described before.
Lineage-, sex-, and region-specific mutation rates
It is noteworthy that the number of GS events mapped to a lineage from the common ancestor of ruminants to any one of the Bovinae is higher than for the equivalent lineage to sheep. This is observed for the Y (33.2 vs. 15) and the X chromosome (19.8 vs. 11). It suggests (P < 0.06) that the mutation rate (both male and female) is lower in Caprinae than in Bovinae.
The ratio of GS events assigned to the Y versus X chromosome corresponds to 60/44 (∼1.36) and is identical in Bovinae (45/33) and Caprinae (15/11). Knowing that the Y chromosome traverses only the male germline, while the X traverses the female germline twice versus once in the male germline, the ratio of male versus female mutation rates (α) in this region can be estimated at ∼1.67 (95% confidence interval: 0.7–8.5) (Miyata et al. 1987; Makova and Li 2002).
We quantified the average sequence divergence between sheep and bovinae, separately for the Y- and X-derived sequences, using a moving 100-bp window (Fig. 6A). As expected, on the proximal side (where GS sites dominate), values for the Y chromosome are in general superior to values for the X chromosome, reflecting the abovementioned higher mutation rate in the male versus the female germline. Y values and X values tend to merge when moving toward the distal end (where R sites dominate), as sequences increasingly behave pseudoautosomally. Interestingly, interspecies sequence divergence (for both Y- and X-derived sequences) tends to become larger on the distal side than even the Y values on the proximal side. Assuming identical mutation rates on either side of the PAB in both the male and female germline, the sequence divergence between sheep and bovinae on the PAR side should be intermediate between the Y and X divergence on the PAB-proximal side. The fact that it tends to become higher suggests a higher mutation rate in this region, possibly the direct result of the higher recombination rate in this region (Perry and Ashworth 1999; Filatov and Gerrard 2003; Galtier 2004; Yi et al. 2004).
Probing the age of the bovine PAB
Y-specific sequences have accumulated on average ∼24.1 GS mutations after the divergence between Bovinae and Caprinae (i.e., ∼18 Mya), while the corresponding figure for X-specific sequences is ∼15.4. Before the corresponding speciation event, Y- and X-specific sequences have accumulated six such differences (Fig. 6B). Assuming that the male and female mutation rates before the Caprinae–Bovinae split corresponded to the average of the mutation rates after the split, the creation of the corresponding PAB can be dated at ∼2.7 Myr before the Caprinae–Bovinae divergence, i.e., ∼20.7 Mya. Note that this is probably a lower bound as recombination events would have erased early traces of X from Y divergence.
Discussion
We herein report the identification and sequence characterization of the bovine PAB. It is located in the SHROOM2–GPR143 interval, coinciding exactly with the presumed limit between strata 3 and 4 of the human X chromosome (Lahn and Page 1999; Carrel and Willard 2005; Ross et al. 2005). This suggested that, in the ruminant lineage, the position of the PAB might not have changed since the occurrence of the chromosomal inversion that created the limit between strata 3 and 4 in a common ancestor of human and cattle. However, we found that the breakpoint of homology between the bovine sex chromosomes coincides with a Bov-tA1 SINE element on the X and a Bov-tA2 element on the Y. As Bov-tA elements are ruminant-specific, this implied that the bovine PAB could not predate the divergence of ruminants from other mammals, which is estimated at ∼50 Mya. Accordingly, we found that all analyzed ruminants (five Bovinae and one Caprinae) share their PAB with cattle, but that both SHROOM2 and GPR143 are pseudoautosomal in porpoise. This is in agreement with a more proximal PAB in the common ancestor of cetaceans and ruminants, and the occurrence of at least one subsequent chromosomal inversion in the ruminant lineage, displacing the PAB more distally. Comparative sequence analysis dates the last of these inversions to ∼20.7 Mya. The PAB of porpoise may still be the ancestral boundary between strata 3 and 4, although this remains to be proven. SHROOM2 and GPR143 were both shown to be pseudoautosomal in dogs, pointing toward a more proximal position of the PAB in this species as well. It will be interesting to determine whether porpoises and dogs share the same PAB.
104 GS events could be assigned either to the Y or to the X chromosome. From the corresponding proportions, we estimated the ratio of male to female mutation rates (α) at 1.7, thus supporting weak “male-driven evolution” (Li et al. 2002) in ruminants. Indeed, this figure is slightly lower than previous estimates of approximately two in murids (e.g., Chang et al. 1994; Chang and Li 1995; Rat Genome Sequencing Project Consortium 2002), and is 1.5- to threefold lower than corresponding figures in hominids (Shimmin et al. 1993; Chang et al. 1996; Huang et al. 1997; Makova and Li 2002; The Chimpanzee Sequencing and Analysis Consortium 2005), carnivores (Slattery and O’Brien 1998; Lindblad-Toh et al. 2005), and birds (Ellegren and Fridolfsson 1997; Kahn and Quinn 1999; Carmichael et al. 2000). It is noteworthy that α has been previously estimated at about four in Caprinae (Lawson and Hewitt 2002). As the confidence intervals in both studies are large, additional data will have to be collected to obtain a conclusive picture for ruminants. If a value of about two were to be confirmed in Bovinae, it would certainly argue against a simple correlation between α and generation time (Li et al. 2002).
Available bovine Y-specific sequences were found to be characterized by a very pronounced depletion in CpG dinucleotides (Table 1). The same feature was observed in humans, indicating that this might be a genuine characteristic of the Y chromosome. In addition to the absence of recombination (recombination would promote de novo CpG creation), we propose that this might be due to the fact that the Y chromosome transits only through the male germline, in which DNA sequences remain methylated for a much longer period than in the female germline, thereby providing more opportunities for methyl-C to T transition by oxidative deamination (Bourc’his and Bestor 2006; Schaefer et al. 2007). We acknowledge that this hypothesis predicts an enrichment of CpG dinucleotides on the X chromosome relative to autosomes (as the X chromosome spends twice as much time in the female germline than in the male germline while autosomes share their time equally between both germlines), while the opposite tendency is observed. However, this might be due to the unique biology of the X chromosome, such as constraints on sequence composition imposed by the mechanisms underlying X inactivation in females and doubling of expression levels from the only active X in both sexes (Heard and Disteche 2006; Nguyen and Disteche 2006).
We observed the same enrichment of LINE repeats on the bovine X chromosome when compared with autosomes, which has been reported in other species and is thought to reflect the involvement of LINE elements in the spreading of the X-inactivation process (Lyon 1998; Carrel et al. 2006). Intriguingly, however, the bovine PAR proved to be even richer in LINE elements than the remainder of the X chromosome. A high density in LINE sequences alone is thus certainly not a sufficient condition for the spreading of X inactivation. It will be interesting to compare the word composition of inactivated portions of the X chromosome with the PAR in bovine, as was recently performed for inactivated segments versus segment escaping inactivation on the human X chromosome (e.g., Carrel et al. 2006; McNeil et al. 2006). Note that the bovine PAR is also enriched in SINE elements, which were shown to be enriched in regions of the X chromosome escaping X inactivation in human (Carrel et al. 2006). They might thus operate in a compensatory way to protect the bovine PAR from spreading of X inactivation.
In conclusion, the comparative sequence analysis of the ruminant PAB has allowed us to document new facets of the evolutionary history of sex chromosomes in mammals, to provide independent evidence supporting previously established hypotheses including male-driven evolution and the effect of recombination on mutation rate and nucleotide composition, as well as to reveal novel phenomena including the reversibility of PAB progression by attrition.
Methods
DNA purification
Genomic DNA was purified by phenol–chloroform extraction following standard procedures. BAC DNA, plasmid DNA, and PCR products were purified with a QIAGEN Large-Construct Kit, QIAprep Spin Miniprep Kit, and QIAquick Gel Extraction Kit (QIAGEN), respectively.
Library screening
Filters one to six from the RPCI-42 bovine BAC library (Warren et al. 2000) were hybridized with 100 ng of 32P-labeled PCR products (Hexalabel DNA Labeling Kit, Fermentas) in Church buffer (1% BSA, 1 mM EDTA, 0.5 M NaPO4 at pH 7.2, 7% SDS) and washed twice with wash 1 (0.5% BSA, 0.5 mM EDTA, 40 mM NaPO4 at pH 7.2, 5% SDS) and twice with wash 2 (1 mM EDTA, 40 mM NaPO4 at pH 7.2, 1% SDS). Filters were exposed on Hyperfilm (Amersham Biosciences) for ∼20 h at −80°C. Contigs of BACs containing the positive clones were identified with Internet Contig Explorer (iCE) (Fjell et al. 2003).
Sequencing
Sequencing reactions were performed with the Big Dye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems), ethanol purified, and analyzed on a 3730 DNA Analyser (Applied Biosystems).
Pulsed-field gel electrophoresis (PFGE) and subcloning
Two to three micrograms of BAC DNA were digested overnight with 15 U of one of the following enzymes: Acc65I, BamHI, EcoRI, HindIII, KpnI, SphI, or XbaI. Digestion products were ethanol purified and run on a CHEF-DR II Pulsed Field Electrophoresis System (Bio-Rad). Electrophoresis was carried out at 14°C in 0.5× TBE at 4 V/cm, with pulse times ramping from 0.5–5 sec for 16 h. Gels were subsequently stained with ethidium bromide and visualized by UV light. A subset of restriction fragments was gel purified and subcloned into pUC19.
In silico sequence annotation
Assembly of BAC sequences was carried out with Sequencher 4.5 software (Gene Codes Corporation). Alignment of sequences surrounding the PAB was performed with ClustalW (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form=clustalw-multialign). Repetitive elements were detected with RepeatMasker (version open-3.1.9) on the RepeatMasker Web Server (Institute for Systems Biology, http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker). Genes were identified by BLASTing the masked sequences against human cDNAs at Ensembl (http://www.ensembl.org/index.html) and bovine ESTs at NCBI (http://www.ncbi.nlm.nih.gov/). Results of the bioinformatic analyses of the BAC sequences were displayed with the purpose-build DNA Viewer software (A. Kvasz and W. Coppieters, unpubl.).
Fluorescence in situ hybridization (FISH)
BACs E0232B11 and H0202L11 were labeled with Spectrum Orange (Vysis, Abbott Molecular, catalog no. 30-803000) and Spectrum Green (Vysis, catalog no. 30-803200), respectively, using the Nick Translation Kit from Vysis (32-801300). Probes were added to male and female blood cell metaphase spreads that were sealed under glass with rubber cement, denatured for 5 min at 75°C and incubated overnight at 37°C in a humidified chamber. The slides were washed for 2 min in 0.4× SSC 0.3% Tween-20 at 72°C and for 2 min in 2× SSC 0.1% Tween-20 at room temperature. They were placed in 2× SSC for 3 min before being counterstained in a DAPI bath for 5 min. They were briefly rinsed in 2× SSC, dehydrated through graded alcohols, air-dried in the dark, and mounted with Vectashield H100 (Vecta Laboratories).
5′ RACE
Total RNA was extracted from the cerebellum of a male calf with TRIzol (Invitrogen) according to the manufacturer’s protocol and was used as starting material for the 5′ rapid amplification of cDNA ends (RACE). This experiment was performed with the GeneRacer kit (Invitrogen) according to the manufacturer’s protocol. The 5′ end of GPR143 was amplified by nested PCR with the following primer pairs: GeneRacer 5′ (CGACTGGAGCACGA GGACACTGA) + GPR143_race3 (CGTGGTGATGTAGTGGGGG ATGG) followed by GeneRacer 5′Nested (GGACACTGACATG GACTGAAGGAGTA) + GPR143_race2 (CAGAACCACCACC AGAAGCAGGC). PCRs were performed in a volume of 50 μL with 1 μL of the cDNA (or of the first PCR product), 0.4 μM of both primers, 200 μM of each dNTP, 1.625 mM MgCl2, 2.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems) and 1× PCR buffer supplied with the polymerase. The following cycling conditions were applied: 10 min at 95°C; 5 cycles of 30 sec at 95°C and 1.5 min at 72°C; 5 cycles of 30 sec at 95°C, 30 sec at 71°C, and 1 min at 72°C; 5 cycles of 30 sec at 95°C, 30 sec at 70°C, and 1 min at 72°C; 25 cycles of 30 sec at 95°C, 30 sec at 69°C, and 1 min at 72°C; and a final extension of 10 min at 72°C. The PCR products were separated by electrophoresis on a 1.5% agarose gel and purified with the QIAquick Gel Extraction Kit (QIAGEN) and either directly sequenced with primers GeneRacer 5′Nested and GPR143_race2 or cloned with the TA cloning kit (Invitrogen) following the manufacturer’s protocol and sequenced with M13F and M13R.
qPCR
PCRs were performed in a volume of 15 μL with 1× Absolute Blue SYBR Green ROX Mix (AB-4163/D) (Applied Biosystems), 70 nM of each primer, and 20–30 ng of genomic DNA. Cycling was performed on an AbiPrism 7900 HT (Applied Biosystems) with the following parameters: 15 min at 95°C, 40 cycles of 15 sec at 95°C and 1 min at 60°C. Following the amplification, a dissociation curve was generated under the following conditions: 15 min at 95°C, 15 min at 60°C, and 15 min at 95°C. Samples were run in triplicate. Results were analyzed with qBase 1.3.5 (Center for Medical Genetics, Ghent University Hospital, Belgium).
Estimating α, the ratio between male and female mutation rates
α was estimated from the ratio between the number of mutational events assigned to the Y chromosome (Y) and the X chromosome (X) as (2Y/X)/[3 − (Y/X)] (Miyata et al. 1987). 95% confidence intervals for α were obtained from 10,000 pairs of 1646 (sequence length) simulated Bernouilli trials with respective probability of success of 60/1,646 (pY) and 44/1,646 (pX). The corresponding α-values were computed using the abovementioned equation. The limits of the 95% confidence interval for α were determined as the 2.5% and 97.5% percentiles of the simulated series.
Acknowledgments
This work was funded by a grant from the Walloon Ministry of Agriculture and was partly supported by EADGENE (European Animal Disease Genomics Network of Excellence for Animal Health and Food Safety). A.-S.V.L. is a fellow from the Belgian Fonds National de la Recherche Scienctifique. We thank Mauricette Jamar and Carine Deusings for their assistance in the FISH analysis, and Alex Kvasz for his help in developing tools for bioinformatic analysis. We thank Michel Milinkovitch for providing us with DNA samples from cetaceans.
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data from this study have been submitted to GenBank under accession nos. FJ195351–FJ195356 and FJ195359–FJ195366.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.082487.108.
References
- Bachtrog D. A dynamic view of sex chromosome evolution. Curr. Opin. Genet. Dev. 2006;16:578–585. doi: 10.1016/j.gde.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Bourc’his D., Bestor T.H. Origins of extreme sexual dimorphism in genomic imprinting. Cytogenet. Genome Res. 2006;113:36–40. doi: 10.1159/000090813. [DOI] [PubMed] [Google Scholar]
- Brown W.R. A physical map of the human pseudoautosomal region. EMBO J. 1988;7:2377–2385. doi: 10.1002/j.1460-2075.1988.tb03082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael A.N., Fridolfsson A.K., Halverson J., Ellegren H. Male-biased mutation rates revealed from Z and W chromosome-linked ATP synthase α-subunit (ATP5A1) sequences in birds. J. Mol. Evol. 2000;50:443–447. doi: 10.1007/s002390010046. [DOI] [PubMed] [Google Scholar]
- Carrel L., Willard H.F. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–404. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Carrel L., Park C., Tyekucheva S., Dunn J., Chiaromonte F., Makova K.D. Genomic environment predicts expression patterns on the human inactive X chromosome. PLoS Genet. 2006;2:1477–1486. doi: 10.1371/journal.pgen.0020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B.H., Li W.H. Estimating the intensity of male-driven evolution in rodents by using X-linked and Y-linked Ube 1 genes and pseudogenes. J. Mol. Evol. 1995;40:70–77. doi: 10.1007/BF00166597. [DOI] [PubMed] [Google Scholar]
- Chang B.H., Shimmin L.C., Shyue S.K., Hewett-Emmett D., Li W.H. Weak male-driven molecular evolution in rodents. Proc. Natl. Acad. Sci. 1994;91:827–831. doi: 10.1073/pnas.91.2.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B.H., Hewett-Emmett D., Li W.H. Male-to-female ratios of mutation rate in higher primates estimated from intron sequences. Zool. Stud. 1996;35:36–48. doi: 10.1007/pl00006166. [DOI] [PubMed] [Google Scholar]
- Charchar F.J., Svartman M., El-Mogharbel N., Ventura M., Kirby P., Matarazzo M.R., Ciccodicola A., Rocchi M., D'Esposito M., Graves J.A. Complex events in the evolution of the human pseudoautosomal region 2 (PAR2) Genome Res. 2003;13:281–286. doi: 10.1101/gr.390503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of sex chromosomes. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- Charlesworth D., Charlesworth B., Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- The Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- Ellegren H., Fridolfsson A.K. Male-driven evolution of DNA sequences in birds. Nat. Genet. 1997;17:182–184. doi: 10.1038/ng1097-182. [DOI] [PubMed] [Google Scholar]
- Ellis N., Yen P., Neiswanger K., Shapiro L.J., Goodfellow P.N. Evolution of the pseudoautosomal boundary in Old World monkeys and great apes. Cell. 1990;63:977–986. doi: 10.1016/0092-8674(90)90501-5. [DOI] [PubMed] [Google Scholar]
- Ellis N.A., Ye T.Z., Patton S., German J., Goodfellow P.N., Weller P. Cloning of PBDX, an MIC2-related gene that spans the pseudoautosomal boundary on chromosome Xp. Nat. Genet. 1994;6:394–400. doi: 10.1038/ng0494-394. [DOI] [PubMed] [Google Scholar]
- Filatov D.A., Gerrard D.T. High mutation rates in human and ape pseudoautosomal genes. Gene. 2003;317:67–77. doi: 10.1016/s0378-1119(03)00697-8. [DOI] [PubMed] [Google Scholar]
- Fjell C.D., Bosdet I., Schein J.E., Jones S.J., Marra M.A. Internet Contig Explorer (iCE)—A tool for visualizing clone fingerprint maps. Genome Res. 2003;13:1244–1249. doi: 10.1101/gr.819303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N. Recombination, GC-content and the human pseudoautosomal boundary paradox. Trends Genet. 2004;20:347–349. doi: 10.1016/j.tig.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M., Frommer M. CpG islands in vertebrate genomes. J. Mol. Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gatesy J., Hayashi C., Cronin M.A., Arctander P. Evidence from milk casein genes that cetaceans are close relatives of hippopotamid artiodactyls. Mol. Biol. Evol. 1996;13:954–963. doi: 10.1093/oxfordjournals.molbev.a025663. [DOI] [PubMed] [Google Scholar]
- Gläser B., Myrtek D., Rumpler Y., Schiebel K., Hauwy M., Rappold G.A., Schempp W. Transposition of SRY into the ancestral pseudoautosomal region creates a new pseudoautosomal boundary in a progenitor of simian primates. Hum. Mol. Genet. 1999;8:2071–2078. doi: 10.1093/hmg/8.11.2071. [DOI] [PubMed] [Google Scholar]
- Graur D., Higgins D.G. Molecular evidence for the inclusion of cetaceans within the order Artiodactyla. Mol. Biol. Evol. 1994;11:357–364. doi: 10.1093/oxfordjournals.molbev.a040118. [DOI] [PubMed] [Google Scholar]
- Graves J.A. Sex chromosome specialization and degeneration in mammals. Cell. 2006;124:901–914. doi: 10.1016/j.cell.2006.02.024. [DOI] [PubMed] [Google Scholar]
- Hassanin A., Ropiquet A. Molecular phylogeny of the tribe Bovini (Bovidae, Bovinae) and the taxonomic status of the Kouprey, Bos sauveli Urbain. Mol. Phylogenet. Evol. 2004;33:896–907. doi: 10.1016/j.ympev.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Heard E., Disteche C.M. Dosage compensation in mammals: Fine-tuning the expression of the X chromosome. Genes & Dev. 2006;20:1848–1867. doi: 10.1101/gad.1422906. [DOI] [PubMed] [Google Scholar]
- Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Chang B.H., Gu X., Hewett-Emmett D., Li W. Sex differences in mutation rate in higher primates estimated from AMG intron sequences. J. Mol. Evol. 1997;44:463–465. doi: 10.1007/pl00006166. [DOI] [PubMed] [Google Scholar]
- Iwase M., Satta Y., Hirai Y., Hirai H., Imai H., Takahata N. The amelogenin loci span an ancient pseudoautosomal boundary in diverse mammalian species. Proc. Natl. Acad. Sci. 2003;100:5258–5263. doi: 10.1073/pnas.0635848100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn N.W., Quinn T.W. Male-driven evolution among Eoaves? A test of the replicative division hypothesis in a heterogametic female (ZW) system. J. Mol. Evol. 1999;49:750–759. doi: 10.1007/pl00006597. [DOI] [PubMed] [Google Scholar]
- Lahn B.T., Page D.C. Retroposition of autosomal mRNA yielded testis-specific gene family on human Y chromosome. Nat. Genet. 1999;21:429–433. doi: 10.1038/7771. [DOI] [PubMed] [Google Scholar]
- Lawson L.J., Hewitt G.M. Comparison of substitution rates in ZFX and ZFY introns of sheep and goat related species supports the hypothesis of male-biased mutation rates. J. Mol. Evol. 2002;54:54–61. doi: 10.1007/s00239-001-0017-x. [DOI] [PubMed] [Google Scholar]
- Li W.-H., Yi S., Makova K. Male-driven evolution. Curr. Opin. Genet. Dev. 2002;12:650–656. doi: 10.1016/s0959-437x(02)00354-4. [DOI] [PubMed] [Google Scholar]
- Lien S., Szyda J., Schechinger B., Rappold G., Arnheim N. Evidence for heterogeneity in recombination in the human pseudoautosomal region: High resolution analysis by sperm typing and radiation-hybrid mapping. Am. J. Hum. Genet. 2000;66:557–566. doi: 10.1086/302754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Chang J.L., Kulbokas E.J., Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Liu W.S., de Ponce León F.A. Assignment of SRY, ANT3, and CSF2RA to the bovine Y chromosome by FISH and RH mapping. Anim. Biotechnol. 2004;15:103–109. doi: 10.1081/LABT-200032337. [DOI] [PubMed] [Google Scholar]
- Liu W.S., Mariani P., Beattie C.W., Alexander L.J., De Ponce Leon F.A. A radiation hybrid map for the bovine Y chromosome. Mamm. Genome. 2002;13:320–326. doi: 10.1007/s00335-001-2133-8. [DOI] [PubMed] [Google Scholar]
- Lyon M.F. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–373. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Lyon M.F. X-chromosome inactivation: A repeat hypothesis. Cytogenet. Cell Genet. 1998;80:133–137. doi: 10.1159/000014969. [DOI] [PubMed] [Google Scholar]
- Makova K.D., Li W.-H. Strong male-driven evolution of DNA sequences in humans and apes. Nature. 2002;416:624–626. doi: 10.1038/416624a. [DOI] [PubMed] [Google Scholar]
- Marais G., Galtier N. Sex chromosomes: How X-Y recombination stops. Curr. Biol. 2003;13:R641–R643. doi: 10.1016/s0960-9822(03)00570-0. [DOI] [PubMed] [Google Scholar]
- McNeil J.A., Smith K.P., Hall L.L., Lawrence J.B. Word frequency analysis reveals enrichment of dinucleotide repeats on the human X chromosome and [GATA]n in the X escape region. Genome Res. 2006;16:477–484. doi: 10.1101/gr.4627606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata T., Hayashida H., Kuma K., Mitsuyasu K., Yasunaga T. Male-driven molecular evolution: A model and nucleotide sequence analysis. Cold Spring Harb. Symp. Quant. Biol. 1987;52:863–867. doi: 10.1101/sqb.1987.052.01.094. [DOI] [PubMed] [Google Scholar]
- Moore S.S., Byrne K., Johnson S.E., Kata S., Womack J.E. Physical mapping of CSF2RA, ANT3 and STS on the pseudoautosomal region of bovine chromosome X. Anim. Genet. 2001;32:102–104. doi: 10.1046/j.1365-2052.2001.00740.x. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Pearks Wilkerson A.J., Raudsepp T., Agarwala R., Schaffer A.A., Stanyon R., Chowdhary B.P. Novel gene acquisition on carnivore Y chromosomes. PLoS Genet. 2006;2:e43. doi: 10.1371/journal.pgen.0020043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy W.J., Davis B., David V.A., Agarwala R., Schaffer A.A., Wilkerson A.J., Neelam B., O’Brien S.J., Menotti-Raymond M. A 1.5-Mb-resolution radiation hybrid map of the cat genome and comparative analysis with the canine and human genomes. Genomics. 2007;89:189–196. doi: 10.1016/j.ygeno.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen D.K., Disteche C.M. Dosage compensation of the active X chromosome in mammals. Nat. Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- Ohno S. Sex chromosomes and sex-linked genes. In: Labhart A., et al., editors. Monographs on endocrinology. Vol. 1. Springer Verlag; Berlin/Heidelberg/New York: 1967. pp. 5–24. [Google Scholar]
- Palmer S., Perry J., Kipling D., Ashworth A. A gene spans the pseudoautosomal boundary in mice. Proc. Natl. Acad. Sci. 1997;94:12030–12035. doi: 10.1073/pnas.94.22.12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J., Ashworth A. Evolutionary rate of a gene affected by chromosomal position. Curr. Biol. 1999;9:987–989. doi: 10.1016/s0960-9822(99)80430-8. [DOI] [PubMed] [Google Scholar]
- Perry J., Palmer S., Gabriel A., Ashworth A. A short pseudoautosomal region in laboratory mice. Genome Res. 2001;11:1826–1832. doi: 10.1101/gr.203001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C., Levilliers J., Weissenbach J. Physical mapping of the human pseudoautosomal region; comparison with genetic linkage map. EMBO J. 1988;7:2369–2376. doi: 10.1002/j.1460-2075.1988.tb03081.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrzebowski L., Vinckenbosch N., Marques A.C., Chalmel F., Jégou B., Kaessmann H. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 2008;6:e80. doi: 10.1371/journal.pbio.0060080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rat Genome Sequencing Project Consortium Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2005;428:493–521. doi: 10.1038/nature02426. [DOI] [PubMed] [Google Scholar]
- Ross M.T., Grafham D.V., Coffey A.J., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Bird C.P., et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena R., Brown L.G., Hawkins T., Alagappan R.K., Skaletsky H., Reeve M.P., Reijo R., Rozen S., Dinulos M.B., Disteche C.M., et al. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat. Genet. 1996;14:292–299. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- Schaefer C.B., Ooi S.K., Bestor T.H., Bourc'his D. Epigenetic decisions in mammalian germ cells. Science. 2007;316:398–399. doi: 10.1126/science.1137544. [DOI] [PubMed] [Google Scholar]
- Shimamura M., Yasue H., Ohshima K., Abe H., Kato H., Kishiro T., Goto M., Munechika I., Okada N. Molecular evidence from retroposons that whales form a clade within even-toed ungulates. Nature. 1997;388:666–670. doi: 10.1038/41759. [DOI] [PubMed] [Google Scholar]
- Shimamura M., Abe H., Nikaido M., Ohshima K., Okada N. Genealogy of families of SINEs in cetaceans and artiodactyls: The presence of a huge superfamily of tRNA(Glu)-derived families of SINEs. Mol. Biol. Evol. 1999;16:1046–1060. doi: 10.1093/oxfordjournals.molbev.a026194. [DOI] [PubMed] [Google Scholar]
- Shimmin L.C., Chang B.H., Li W.H. Male-driven evolution of DNA sequences. Nature. 1993;362:745–747. doi: 10.1038/362745a0. [DOI] [PubMed] [Google Scholar]
- Skaletsky H., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Slattery J., O’Brien S.J. Patterns of Y and X chromosome DNA sequence divergence during the Felidae radiation. Genetics. 1998;148:1245–1255. doi: 10.1093/genetics/148.3.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele M.R., Georges M. Generation of bovine multisite haplotypes using random cosmid clones. Genomics. 1991;10:889–904. doi: 10.1016/0888-7543(91)90177-g. [DOI] [PubMed] [Google Scholar]
- Toder R., Gläser B., Schiebel K., Wilcox S.A., Rappold G., Graves J.A., Schempp W. Genes located in and near the human pseudoautosomal region are located in the X-Y pairing region in dog and sheep. Chromosome Res. 1997;5:301–306. doi: 10.1023/B:CHRO.0000038760.84605.0d. [DOI] [PubMed] [Google Scholar]
- Veyrunes F., Waters P.D., Miethke P., Rens W., McMillan D., Alsop A.E., Grützner F., Deakin J.E., Whittington C.M., Schatzkamer K., et al. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008;18:965–973. doi: 10.1101/gr.7101908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren W., Smith T.P., Rexroad C.E., Fahrenkrug S.C., Allison T., Shu C.L., Catanese J., de Jong P.J. Construction and characterization of a new bovine bacterial artificial chromosome library with 10 genome-equivalent coverage. Mamm. Genome. 2000;11:662–663. doi: 10.1007/s003350010126. [DOI] [PubMed] [Google Scholar]
- Yi S., Summers T.J., Pearson N.M., Li W.H. Recombination has little effect on the rate of sequence divergence in pseudoautosomal boundary 1 among humans and great apes. Genome Res. 2004;14:37–43. doi: 10.1101/gr.1777204. [DOI] [PMC free article] [PubMed] [Google Scholar]