Abstract
Tissue development and function are exquisitely dependent on proper regulation of gene expression, but it remains controversial whether the genomic signals controlling this process are subject to strong selective constraint. While some studies show that highly constrained noncoding regions act to enhance transcription, other studies show that DNA segments with biochemical signatures of regulatory regions, such as occupancy by a transcription factor, are seemingly unconstrained across mammalian evolution. To test the possible correlation of selective constraint with enhancer activity, we used chromatin immunoprecipitation as an approach unbiased by either evolutionary constraint or prior knowledge of regulatory activity to identify DNA segments within a 66-Mb region of mouse chromosome 7 that are occupied by the erythroid transcription factor GATA1. DNA segments bound by GATA1 were identified by hybridization to high-density tiling arrays, validated by quantitative PCR, and tested for gene regulatory activity in erythroid cells. Whereas almost all of the occupied segments contain canonical WGATAR binding site motifs for GATA1, in only 45% of the cases is the motif deeply preserved (found at the orthologous position in placental mammals or more distant species). However, GATA1-bound segments with high enhancer activity tend to be the ones with an evolutionarily preserved WGATAR motif, and this relationship was confirmed by a loss-of-function assay. Thus, GATA1 binding sites that regulate gene expression during erythroid maturation are under strong selective constraint, while nonconstrained binding may have only a limited or indirect role in regulation.
Most genomic DNA sequences that are conserved (i.e., that align) among two or more mammalian orders do not code for protein, and their functions, if any, are not well understood (Waterston et al. 2002; Miller et al. 2004; Dermitzakis et al. 2005). In particular, the extent of conservation of DNA sequences required for regulation of gene expression, that is, cis-regulatory modules (CRMs) such as enhancers and promoters, is controversial. Some CRMs are conserved across multiple mammalian lineages or beyond (Emorine et al. 1983; Li et al. 1990; Aparicio et al. 1995; Woolfe et al. 2005), and thus deep conservation of noncoding sequences is a common approach to predict CRMs from aligned genomic DNA sequences (Hardison 2000; Pennacchio and Rubin 2001). Indeed, gain-of-function assays show that almost half of the noncoding sequences conserved across diverse vertebrate lineages (from mammals to fish) (Pennacchio et al. 2006) or almost invariant in multiple mammalian orders (Visel et al. 2008) are active as enhancers during early development. Noncoding sequences that are conserved in placental mammals but are under less severe constraint can also be important regulatory regions. For example, noncoding sequences with patterns in multispecies alignments characteristic of known regulatory regions (Taylor et al. 2006), coupled with preservation of a binding site motif for a tissue-specific transcription factor, are validated in gain-of-function assays for over half the cases (Wang et al. 2006).
In contrast, some CRMs are lineage specific (Bodine and Ley 1987; Valverde-Garduno et al. 2004) and others show extensive turnover in binding site motifs (Dermitzakis and Clark 2002) and compensatory evolution (Ludwig et al. 2000). CRMs identified by transcription factor occupancy and chromatin modifications show conservation over more limited phylogenetic spans than those predicted by multispecies alignments (King et al. 2007; Miller et al. 2007), and only about half of the former overlap with highly constrained noncoding sequences (The ENCODE Project Consortium 2007). These contrasting results show that the relationship between evolution and function of regulatory regions is complex, with some occupied DNA segments being deeply conserved and others present only over a limited phylogenetic span.
To clarify the relationship between conservation of binding site motifs in CRMs and their biochemical function, we must distinguish between lineage-specific regulation, in which a CRM regulates a target gene only in a restricted clade such as rodents, and conserved regulation, in which orthologous CRMs regulate orthologous targets in the ancestral and derived species. In all these cases, we accept that within CRMs, transcription factors must bind specific motifs to regulate expression of a target gene. Lineage-specific CRMs are not expected to show evidence of purifying selection when compared with species outside the lineage, either because the DNA is present only in that lineage or because it has been diverging at a sufficiently fast rate in other lineages to make it indistinguishable from neutral.
For conserved CRMs, one can hypothesize that purifying selection (constraint) has rejected changes in the motifs that would affect binding. This motif constraint hypothesis makes two predictions that are tested in this report. First, factor-bound DNA intervals with demonstrable regulatory activity should frequently contain constrained binding site motifs, whereas motifs in occupied intervals lacking enhancer activity should show evidence of constraint only infrequently. Second, regulatory activity should be sensitive to the loss of the constrained binding site motifs.
An alternative hypothesis for conserved CRMs states that selection on the binding site motifs is weak relative to the rate at which other, equivalent binding site motifs arise. In this case, the regulatory activity, while still dependent on occupancy by transcription factors binding to a motif, derives from motifs that vary in location within the occupied intervals in different lineages. Thus, in contrast to the predictions of the motif constraint hypothesis, this motif turnover hypothesis predicts that regulatory activity will not be associated with constraint on the binding site motif, and mutation of poorly preserved motifs will have a substantial effect on activity.
We have tested these hypotheses using a high-quality data set of DNA intervals occupied by the transcription factor GATA1 along 66 Mb of chromosome 7 in mouse erythroid cells. This transcription factor is required for normal development of erythrocytes, megakaryocytes, mast cells, eosinophils, and probably dendritic cells (Pevny et al. 1991, 1995; Simon et al. 1992; Weiss and Orkin 1995b; Shivdasani et al. 1997; Yu et al. 2002; Migliaccio et al. 2003; Gutierrez et al. 2007). GATA1 regulates most of the genes that define the mature erythroid phenotype (Weiss and Orkin 1995a; Blobel and Weiss 2001; Welch et al. 2004) and many genes in megakaryocytes (Orkin et al. 1998; Wang et al. 2002). For this study, we used G1E cells, an immature erythroid line derived from Gata1 null embryonic stem cells (Weiss et al. 1997). A sub-line, G1E–ER4, stably expresses a conditional form of GATA1 in which the full-length protein is fused to the ligand-binding domain of the estrogen receptor. Addition of estradiol to G1E–ER4 cells activates GATA1 and triggers erythroid maturation to a relatively late stage (Weiss et al. 1997; Welch et al. 2004). In this system, chromatin associated with GATA1 can be immunoprecipitated from induced G1E–ER4 cells and compared with background signals in the Gata1 null parental G1E cells (Johnson et al. 2002; Letting et al. 2003). We analyzed global GATA1 chromatin occupancy in a segment of mouse chromosome 7 containing the intensively studied Hbb gene cluster encoding beta-like globin genes expressed in erythroid cells (Bulger et al. 2000, 2003; Forsberg et al. 2000) and an additional 26 other genes whose expression is up- or down-regulated during late erythroid maturation (Welch et al. 2004).
An important aspect to our study is to determine the ability of the GATA1-occupied DNA segments to affect the level of gene expression. To obtain robust, quantitative data on multiple DNA segments, we measured transient expression after transfection of an erythroid cell line, K562, with a reporter construct. This was a recombinant plasmid containing test DNA segments along with a firefly luciferase reporter gene expressed from a gamma-globin gene promoter (Lam and Bresnick 1996). DNA segments that cause a significant increase in expression of luciferase are enhancers. This assay is a well-recognized approach to analyzing the function of CRMs, having been used in experiments ranging from the early definition of enhancers (Banerji et al. 1981; Mellon et al. 1981) to many experiments screening noncoding segments of genomic DNA for effects on expression (e.g., Frazer et al. 2004; Baroukh et al. 2005; Grice et al. 2005; Wang et al. 2006; Petrykowska et al. 2008). This assay provided a tractable method to test a biological activity of the 123 DNA segments examined in this study.
Results
DNA segments bound by GATA1 in vivo
DNA bound by GATA1 in G1E–ER4 cells was isolated by chromatin immunoprecipitation (ChIP), and occupied DNA intervals were deduced by duplicate hybridization to a set of high-density tiling arrays from NimbleGen (ChIP-chip; Ren et al. 2000; Nuwaysir et al. 2002). Hits predicted from the ChIP-chip data (called GHPs, for GATA1 hit positive) were tested using quantitative PCR on an independent ChIP preparation, yielding 63 validated DNA intervals (500–700 bp each) occupied by GATA1 (Fig. 1). Sampling of lower stringency ChIP-chip hits and comparisons with previously identified intervals occupied by GATA1 indicate that this set comprises a substantial majority of the occupied DNA segments in the target region in this cell line. (More information is in the Supplemental material, and results can be viewed on a custom browser at http://bx.psu.edu/~yong/ghp/.) Early work identified WGATAR as the consensus motif bound by GATA1 (Evans et al. 1988; Wall et al. 1988; Orkin 1992), and we found that the motif WGATAR was almost always found in DNA intervals occupied by GATA1, being present in 60 (95%) of the 63 segments. This motif is quite common; it is found in 77% of randomly sampled 500-bp intervals from the 66-Mb target region. However, its frequency in the occupied segments is higher than in randomly sampled, unoccupied 500-bp intervals for 994 trials out of 1000 (empirical P-value = 0.006). This result contrasts with those for other high-throughput studies of occupancy by SP1, MYC, and TP53 (Cawley et al. 2004) and E2F1 (Xu et al. 2007), which show only a minority of occupied segments with the canonical binding site motif. Perhaps the biochemically defined binding site is a stronger contributor to occupancy by some transcription factors than others.
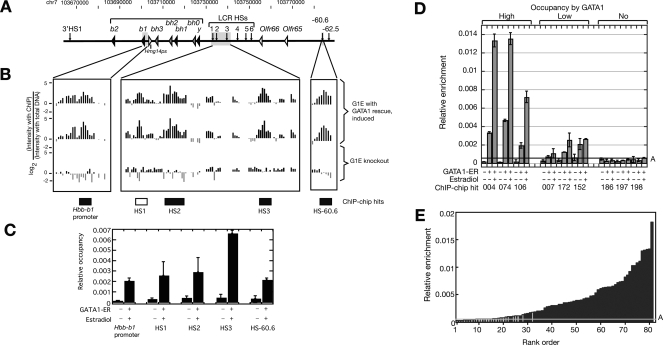
Figure 1.
Occupancy by GATA1: ChIP-chip and qPCR data. (A–C) Sensitivity and specificity of the ChIP-chip data in the Hbb gene cluster. (A) Location of genes and DNase hypersensitive sites in the mouse Hbb gene cluster. (B) ChIP-chip results for GATA1 in the mouse Hbb gene cluster. The first two tracks present the logarithm of the ratio of hybridization intensities between ChIP DNA from G1E ER rescued cell line and the input DNA for two replicates. The third track shows the hybridization signals from the G1E Gata1-null cell. The boxes beneath these tracks show intervals previously identified as bound by GATA1; the boxes are black if they are included in the ChIP-chip peak calls or white if not included. (C) The quantitative PCR results of the previously identified segments occupied by GATA1. The two bars are the qPCR result with ChIP material from G1E cells and from rescued G1E ER4 cells. The mean of two determinations is plotted, and the error bars are half of the range. (D,E) Independent validation of the GATA1 ChIP-chip results by qPCR. (D) DNA segments positive in the ChIP-chip assay shown by qPCR to have high, low, and no occupancy by GATA1. Amplicons for each ChIP-chip hit were assayed in GATA1–ER ChIP material from G1E knockout cells (first bar in each set) and from G1E–ER4 cells before and after induction of the GATA1–ER hybrid with estradiol (second and third bar). The mean of the two determinations is graphed, with half the range shown as error bars. Relative enrichment is the ratio between the amount of the amplicon immunoprecipitated along with GATA1 and the amount of the amplicon in the input material. Line A is drawn at the mean relative enrichment of the negative controls plus three standard deviations. (E) The relative enrichment in ChIP material from induced G1E–ER4 cells for 81 high-stringency ChIP-chip hits tested by qPCR. The black bars are the DNA intervals that not only pass the mean plus three standard deviations of the negative controls set but also show at least a fourfold increase in enrichment compared with the signals from the Gata1-null cells. The gray bars are the ChIP-chip hits that did not pass one or both of the above thresholds. Line A is the same as in D.
Enhancer activity of DNA bound by GATA1
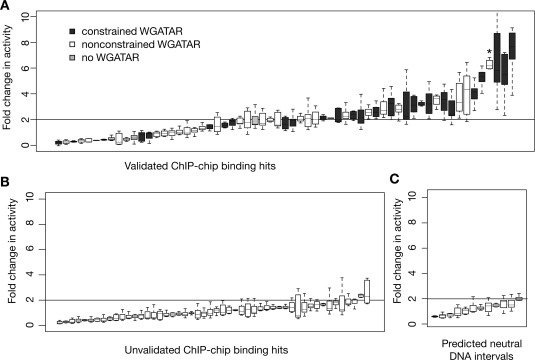
To investigate the extent to which occupancy correlates with measurable regulatory activity, we tested 61 of the 63 occupied segments for their ability to enhance expression of luciferase from an HBG1 promoter in transiently transfected K562 cells. We found that 34 (52%) increase expression at least twofold, and 17 show a high activity of at least a threefold increase (Fig. 2A). In contrast, ChIP-chip hits that are not validated by qPCR are rarely active as enhancers in this assay; only three (6%) of the 50 ChIP-chip hits that did not pass the qPCR validation were active (Fig. 2B). DNA segments that were not identified as ChIP-chip peaks, and thus are predicted to be neutral, show no activity in this assay (Fig. 2C). In addition to the occupied DNA segments that are active as enhancers, another 13 overlap with transcription start sites and hence are likely to be active as promoters (see Supplemental material).
Figure 2.
Activities of DNA segments occupied by GATA1 in enhancer assays. (A) The results of eight to 24 determinations of enhancer activity after transfection for each occupied DNA interval added to a luciferase expression plasmid, rank-ordered by activity. The distribution of results for each occupied segment is represented as a box plot, with the internal line indicating the median, the box extending to the first and third quartiles, and the whiskers extending to the most extreme data point that is no more than 1.5 times the interquartile range. Boxes for occupied segments with at least one constrained GATA1 binding motif (i.e., preserved outside rodents) are shaded dark gray, those with nonconstrained motifs (i.e., found only in rodents) are white, and those with no match to a binding site motif shaded light gray. The asterisk marks the activity for DNA segment GHP304. (B) The results of enhancer activity for DNA intervals that were positive for occupancy by ChIP-chip (many passed only a low stringency threshold, see Supplemental material) but were not validated by qPCR. (C) The transfection results for DNA segments predicted to be neutral because they were not positive for ChIP-chip data. Intervals that generate at least a twofold increase in activity compared with that of the parental reporter gene plasmid (corresponding to the mean of the negative controls plus 3 SDs) in at least two repeats of the assay are considered to be active as enhancers.
Conservation of GATA1-bound segments and preservation of motifs
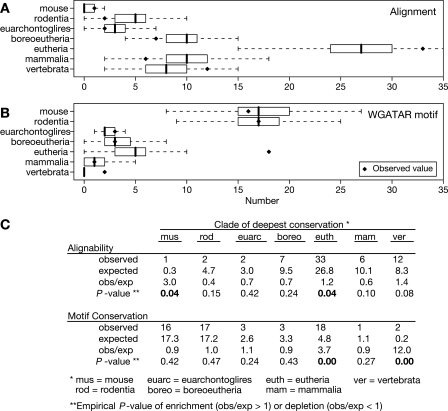
By identifying intervals occupied by GATA1 using an approach that is agnostic to sequence alignments, we can evaluate how frequently these segments are more conserved than the background genomic DNA. Conservation is determined by the presence of an alignment between the sequence of the GATA1-bound segment in mouse and the presumably orthologous sequence in another species in the MULTIZ alignments (Blanchette et al. 2004) for the February 2006 mouse genome assembly (mm8) (Waterston et al. 2002), obtained from the UCSC Genome Browser (Kent et al. 2002). Only 12 of the 63 DNA intervals (19%) show deep conservation across vertebrates (e.g., conserved from mouse to fish), whereas almost all (60 of 63, or 95%) are conserved in multiple mammalian orders or beyond (Fig. 3A). Only three (5%) of the intervals appear to be rodent specific. The frequency of conservation to these distances was then compared with the genomic background by randomly sampling 63 DNA segments 1000 times from the nonrepetitive portion of the 66 Mb represented on the high-density tiling array. The phylogenetic depth of alignment was determined for these random samples. The box plots in Figure 3A summarize the distributions of frequencies that the randomly sampled DNA segments are conserved to a given clade. We used the mean of each distribution to estimate an expected frequency for observing conservation to a particular phylogenetic depth (Fig. 3C). Conservation of the GATA1-bound DNA segments to eutherians or to vertebrates occurs more frequently than expected, with observed to expected ratios of 1.2 and 1.4, respectively. The former is clearly significant, with an empirical P-value of 0.04, and the P-value for the latter is low (0.08, Fig. 3A).
Figure 3.
Conservation of the DNA segments occupied by GATA1 and preservation of the GATA1 binding site motifs within those segments. The 63 GATA1 occupied segments were classified by phylogenetic conservation of sequence (A) and preservation of WGATAR motifs (B), i.e., the motif aligns to the designated clade and still retains a match to WGATAR. Solid diamonds represent the observed number of occupied segments showing conservation of the DNA segments or preservation of the WGATAR motif in the designated clade but no further. The distributions of the same classification found in 1000 iterations of randomly sampling 63 DNA intervals from the nonrepetitive portions of the 66-Mb target region are shown as box plots. The box line is the median, box width is the interquartile range, and whiskers extend to the most extreme data point that is no more than 1.5 times the interquartile range. (C) The observed and expected frequencies of conservation. The mean number of times that a designated clade of deepest conservation is found in the 1000 random samplings is taken as an estimate of the expected value. The ratio of the observed to expected number of times that a given extent of conservation is seen indicates the level of enrichment (ratio > 1) or depletion (ratio < 1). Comparison of the observed value to the distribution of values for the random samplings gives the associated empirical P-values for enrichment or depletion; the P-values < 0.05 are in boldface type.
Given the tendency of the DNA segments occupied by GATA1 to be conserved in multiple eutherian orders or beyond, we then asked whether the WGATAR binding site motifs are also conserved (align to other species) and preserve a match to the motif. This is a more stringent criterion, requiring not only that the sequence in the comparison species aligns but also that it does not change in designated positions. A WGATAR motif aligns with this same motif in nonrodent mammals (or more deeply) for only 27 (45%) of the 60 occupied segments that have the motif (Fig. 3B). The motifs that remain unchanged over this phylogenetic distance appear to be evolutionarily constrained, that is, are subject to purifying selection, based on two observations. First, deep preservation of WGATAR motifs is much less common in randomly sampled, unbound DNA intervals that also contain this motif. For example, the GATA1-occupied DNA segments showed preservation of the motif in eutherians or in vertebrates about four times or 12 times, respectively, more frequently than would be expected from the random sampling (Fig. 3B,C; empirical P-value = 0). Second, estimates of the likelihood of constraint in the binding site motif are significantly higher for the occupied DNA segments with a preserved WGATAR motif than in those in which the motif does not align with WGATAR outside rodents. For each occupied DNA segment, we aggregated the phastCons score, which estimates the posterior probability of a position being in the most slowly changing fraction of DNA (Siepel et al. 2005), over the six positions of the WGATAR motif that aligns with the same motif in the most distant clade. The mean of these aggregated phastCons scores for the 27 occupied segments with preserved WGATAR motifs (0.43) is significantly higher than the mean (0.053) for the other 33 occupied segments (P-value = 6 × 10−5 in a one-tailed Student’s t-test). Thus, a subset of the segments occupied by GATA1 have WGATAR motifs that show significant signs of evolutionary constraint.
Association of enhancement with motif preservation
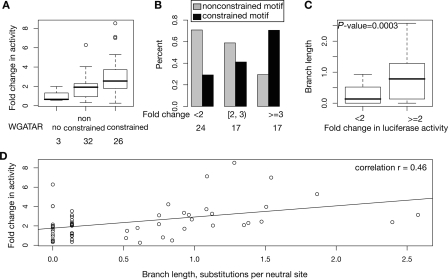
As predicted by the motif-constraint hypothesis, the GATA1-occupied DNA segments that are active as enhancers are strongly associated with preservation of a WGATAR motif beyond rodents. When the occupied DNA segments are ordered by enhancer activity and labeled by constraint on the motif, the ones with more activity tend to be those with constrained WGATAR motifs (Fig. 2A). This relationship was evaluated quantitatively in several ways. When the occupied DNA intervals were partitioned by evidence of constraint on the WGATAR motifs, those with constrained motifs had significantly higher enhancement activity (mean 3.0-fold increase in activity compared with a mean 1.9-fold enhancement, Fig. 4A). The null hypothesis of equal means, evaluated in a one-tailed, two-sample Student’s t-test, has a P-value of 0.008. When the occupied DNA segments were partitioned by level of activity in the enhancer assay, the percentage of occupied segments with constrained motifs increased with the level of activity, while the percentage of occupied segments with nonconstrained motifs showed the opposite trend (Fig. 4B). The phylogenetic depth over which a WGATAR motif is preserved was used to estimate the evolutionary distance over which it has resisted change in each occupied segment, measured as the branch length from mouse in substitutions per neutral site (fourfold degenerate sites in coding sequences). The branch lengths for motif preservation in the active enhancers (mean of 0.8 substitutions per neutral site) are much higher than those for the occupied segments that lack enhancement activity (mean of 0.2 substitutions per neutral site, Fig. 4C). The null hypothesis of equal means has a P-value of 0.0003 (one-tailed, two-sample Student’s t-test). Enhancer activity of the GATA1-occupied segments correlates positively with branch length for preservation of the motif (Pearson’s correlation r = 0.46, Fig. 4D). Furthermore, the mean of phastCons scores for the most deeply preserved WGATAR motifs in the 34 occupied segments with enhancer activity (0.314) is significantly higher than in the 24 occupied segments with a WGATAR but not active as enhancers (0.092, P-value = 0.005 for a test of the null hypothesis of equal means by Student’s one-tailed t-test). All these analyses support the motif-constraint hypothesis for DNA segments occupied by the transcription factor GATA1 in erythroid cells. These results also show that the motif-turnover hypothesis does not hold for all the conserved enhancers dependent on GATA1.
Figure 4.
Positive correlation of enhancer activity with constraint on GATA1 binding site motifs. (A) The range of enhancer activities as box plots for occupied segments partitioned by constraint on the GATA1 binding site motif. The total numbers of occupied DNA segments in each category are given at the bottom of A and B. (B) The fraction of occupied DNA segments with constrained (black bars) or nonconstrained (gray bars) binding site motifs in classes of increasing enhancer activity. (C) The distribution of branch lengths over which the binding site motif is preserved for intervals without or with enhancer activity. (D) The mean enhancer activity (fold change compared with that of the parental plasmid) of each occupied segment as a function of branch length over which the motif resists alteration to remain a match to WGATAR. The P-values evaluate the null hypothesis of equal means in a one-tailed, two-sample Student’s t-test.
Sensitivity of constrained motifs to mutagenesis
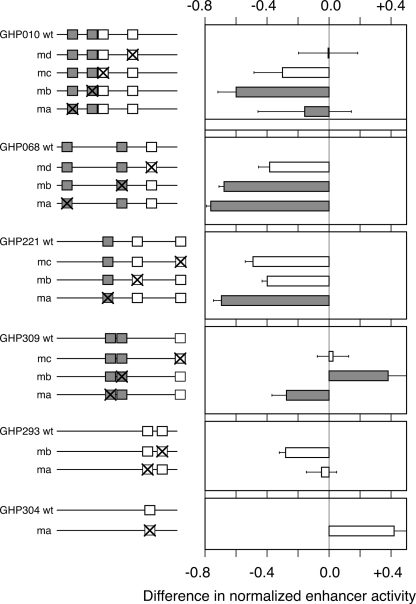
If enhancement activity dependent on binding of a transcription factor to a particular motif is the function under purifying selection, then mutation of constrained motifs should have a larger effect than mutation of other motifs. This prediction of the motif-constraint hypothesis was tested in several GATA1-occupied DNA intervals that are active as enhancers in the transfection assay. All but one have multiple WGATAR motifs, which were classified as nonconstrained (rodent-only) or constrained (preserved in multiple eutherian lineages). After mutation of each WGATAR motif, changes in activity of the altered DNA fragments were measured by the luciferase enhancer assay (Fig. 5; Supplemental material). Four enhancers contain motifs in both constraint classes (GHPs 010, 068, 221, and 309), and in each case mutation of constrained motifs had a larger effect than mutation of nonconstrained motifs; this was seen for six constrained motifs. The sole exception is mutation of constrained motif “a” in GHP010, but in this same enhancer, mutation of constrained motif “b” has a much larger effect than the nonconstrained motifs. Overall, the differences in enhancer activity are significantly higher for the constrained motifs than for the nonconstrained ones (P = 0.008 for a one-tailed Student’s t-test; box plots are in Supplemental material). These results fit the predictions of the motif-constraint hypothesis.
Figure 5.
Mutation of constrained GATA1 binding site motifs reduces enhancement more than alteration of lineage-specific ones. The left panel shows the positions of WGATAR motifs that are present in mouse plus multiple mammalian lineages (constrained, gray boxes) or present only in rodents (nonconstrained, white boxes) in six wild-type (wt) DNA segments occupied by GATA1. Mutations in each motif (labeled “ma” for mutation in motif a, etc.) are indicated by an X in the box. The enhancer activity of each wild-type DNA fragment was normalized to 1, and the difference in normalized activity (mutant − wild type) is presented in the graph. The bar (shaded to match the constraint status of the mutated motif) gives the mean of four independent transfection experiments, each assayed in duplicate to give eight measurements, with error bars showing the standard deviation.
It is also clear that some of the nonconstrained motifs do affect enhancement activity, albeit less than is observed for the constrained motifs (Fig. 5). GHP304 is an interesting case of acquisition of a WGATAR motif in mouse that reduces enhancer activity. This DNA segment has the fourth highest enhancer activity of the GATA1-occupied DNA fragments tested in this study (Fig. 2A), but the WGATAR motif in the mouse sequence is not preserved in other species, and no WGATAR is found in the sequences orthologous to GHP304 in most mammals. (More details on motifs in the alignments of this and all GHPs are at http://www.bx.psu.edu/~dcking/ghp/img2/.) The enhancer activity actually increases when this motif is mutated (Fig. 5), and thus the activity of this enhancer is not dependent on the WGATAR motif. Other portions of GHP304 do appear to be under purifying selection, and investigation of non-WGATAR motifs in that part of the enhancer may reveal motifs needed for activity that are under constraint. Mutational analyses of WGATAR motifs with more complex evolutionary interpretations (present in coding exons or with an unusual phylogenetic pattern) in two other enhancers are presented in Supplemental materials.
Discussion
This study integrates results from three types of analyses to show that constraint on enhancement activity leads to preservation of binding site motifs, at least for the protein GATA1. We identified DNA segments occupied by the transcription factor GATA1 comprehensively over a large chromosomal region (66 Mb), using a combination of ChIP-chip and independent validation by quantitative PCR. These techniques are agnostic to evolutionary conservation or prior knowledge about regulatory regions, and thus they provide a relatively unbiased set of in vivo occupied DNA intervals. The binding data do not reveal specific biological functions, and thus we also assayed the occupied DNA segments for one activity critical to gene regulation, that is, enhancement of expression. By partitioning the bound DNA intervals into those active or inactive as enhancers, we found a strong association with the third line of evidence, namely, the phylogenetic extent of preservation of binding site motifs. GATA1-bound DNA segments with binding site motifs preserved in multiple mammalian lineages (or beyond) tend to be active as enhancers, and constrained binding motifs contribute more to the enhancer activity than nonconstrained ones (as shown by mutagenesis studies).
Our study employing data on occupancy, activity, and conservation provides important insights for the effective use of multiple sequence alignments to identify gene regulatory sequences. First, examining conservation at the motif level is more informative than evaluating overall conservation of a bound DNA segment. Recent studies reveal only a limited overlap between DNA segments occupied by transcription factors and segments under strong evolutionary constraint (The ENCODE Project Consortium 2007). One explanation discussed in that paper is that only smaller subregions within the larger intervals identified as occupied may be under purifying selection. Our results support this explanation. While we do observe a small but significant enrichment for alignability to eutherians and vertebrates for the bound DNA segments (size of 500–700 bp), the enrichment for preservation of binding site motifs (size of 6 bp) is much greater (Fig. 3, cf. A and B). Second, it is important to differentiate between in vivo occupancy and specific biological function when evaluating conservation of motifs in CRMs. For the set of DNA segments occupied in vivo by GATA1 identified in our study, we find that the phylogenetic depth of preservation of binding site motifs varies over a wide range, with the majority not preserved beyond rodents. However, the subset of GATA1-occupied sites active as enhancers shows a pronounced association with evolutionary preservation of the binding site motif, that is, they are constrained across mammalian orders. This observation confirms one of the two predictions of the motif-constraint hypothesis. The other prediction, that constrained motifs would show a stronger phenotype upon mutation than nonconstrained motifs, was also confirmed. Thus, we conclude that purifying selection acting on enhancer activity does frequently result in preservation of a binding site motif in a particular position in the enhancer.
This conclusion is consistent with a recent analysis of enhancer activity and evolution for muscle genes in Ciona species (Brown et al. 2007). In this study, individual binding site motifs within known enhancers were mutated and evaluated for effects on expression. Motifs that contribute significantly to the activity of an enhancer are almost invariably constrained, as shown by clear orthologs that have sustained many fewer substitutions than expected between distant Ciona species. Motifs that contribute weakly to enhancer activity sustain substantially more substitutions, such that motif activity is correlated with percent identity.
These evolutionary insights provide guidance in deciding which DNA segments occupied by transcription factors are likely to be involved in enhancement. For a set of DNA segments occupied by a sequence-specific binding protein, the relevant binding site motifs should be identified and the phylogenetic depth of preservation determined for each instance of the motifs. Those DNA segments containing binding site motifs preserved over a large phylogenetic distance (equivalent to the time of separation of multiple eutherian orders, estimated as 60 to 100 million years ago) are good candidates for enhancer activity.
While that is likely to be a good rule for finding biological activity within a set of DNA segments occupied by a protein, we should consider several caveats and limitations to our study. Our assay for enhancement, transient transfection of cultured mammalian cells, provides meaningful activity data, as shown by the striking difference in effects for the occupied DNA segments compared with those for the unoccupied segments (Fig. 2, cf. A and B). However, this assay can reveal an activity only in the cell line used, and thus some of the occupied DNA segments with no activity could be false negatives. Furthermore, analysis of the occupied DNA segments after gene transfer in whole animals would allow many additional aspects of regulation to be examined, including tissue specificity and developmental timing of expression. In future studies it will be useful to examine a few of these GATA1-bound DNA segments in greater detail through transgenic or knock-in strategies. Those bound segments with deep preservation of the binding site motif are particularly interesting in this regard. We note that our biological assay has been confined to GATA1-dependent enhancers in erythroid cells. It will be important to study the phylogenetic extent of motif preservation in other types of CRMs (such as promoters and silencers), using other transcription factors and examining additional developmental systems, to see how consistent the results are with the present study.
Our studies, comparing the level of enhancer activity between occupied sites with and without constraint on the motifs, and those of Brown et al. (2007) both show that high enhancer activity is associated with strong purifying selection on the binding site motifs. Other studies emphasize turnover (Dermitzakis and Clark 2002; Moses et al. 2006) and compensatory changes (Ludwig and Kreitman 1995; Ludwig et al. 2000) in binding site motifs when orthologous enhancers are compared. Both conservation and turnover of binding sites motifs have been observed in multiple comparative studies in a range of species (Dermitzakis et al. 2003; Moses et al. 2003). We also observe many DNA segments occupied by GATA1 whose binding site motifs are not deeply preserved but rather the position of the motifs change over evolutionary time. The motif turnover hypothesis can explain the evolution of motifs in these bound segments. They tend to have low or no activity in enhancer assays, and thus the selection against changes in the binding site motif is not expected to be as severe as for enhancers with high activity. Mutations in the binding site motif are more likely to persist in a population, especially if mutations at other nucleotides in the vicinity generate a motif closer to the preferred binding site. A few events like this could lead to a motif “moving” from one position to another in the vicinity, but only if the effects of the alterations were nearly neutral. One possibility is that occupied segments with nonconserved motifs could function to modulate the activity of enhancers but have little activity on their own.
Several GATA1-occupied DNA segments show evidence of constraint on some motifs and turnover for others within the same segment. Mutation of constrained motifs in enhancers gave the strongest phenotypes, but mutation of the nonconstrained motifs also gave significant effects in several cases (GHPs 010, 068, and 221 in Fig. 5). One interpretation is that selective constraint on the enhancer activity preserves certain critical binding site motifs, leaving other instances of the motif in the enhancer free to change over evolutionary time. The deeply preserved motifs may anchor the enhancement activity, while other motifs that can turn over may fine-tune the activity.
In other cases, the lack of constraint on motifs could reflect lineage-specific differences in activity. The changes in motif patterns in eve stripe 2 enhancer relate to important differences in activity between Drosophila species (Ludwig et al. 2005). Some erythroid enhancers are found in primates but not mice (Bodine and Ley 1987; Valverde-Garduno et al. 2004), presumably playing a role in lineage-specific differences in expression. Some of the newly identified segments occupied by GATA1 may fall into this category. For example, the mouse-specific WGATAR in GHP304 appears to be reducing the activity of an enhancer that is deeply conserved but not dependent on that motif.
Another interpretation of the occupied DNA segments with no activity in the transient transfection assay is that they have no biological activity. While it is possible that they have functions that we have not assayed, it is also possible that they play a static role, such as storing unused proteins. Our results show that partitioning protein-occupied DNA segments by evolutionary preservation of binding site motifs is biologically meaningful; in particular, those with constrained motifs are enriched for strong enhancer activity. However, we expect occupied DNA segments lacking constrained motifs to be a mix of weak enhancers (perhaps modulating activities of strong enhancers), lineage-specific CRMs, and sequences with a static role. Additional experiments are needed to distinguish among those possibilities.
An underlying assumption of our study is that DNA segments are occupied by GATA1 through direct binding to a motif in the DNA sequence. This assumption is supported by the very high frequency with which the consensus motif occurs in the occupied DNA segments and the sensitivity of the enhancement activity to mutation of the motif. However, proteins can tightly associate with DNA segments through interactions with other proteins. Such cases will greatly complicate the analysis of functional significance of conservation on binding site motifs. Ideally, the studies would be done with sets of occupied DNA segments in which the effect of each motif on occupancy has been measured in vivo. Accomplishing this in a high-throughput manner is a technical challenge, but it is a worthwhile goal.
Much new high-throughput data on biochemical features of chromosomes, such as occupancy by transcription factors, histone modifications, and chromatin alterations, is becoming available (The ENCODE Project Consortium 2007). Connecting these biochemical features with biological activities and physiological consequences will be a challenge. Combining the high-throughput data with assays for biological activity and evolutionary analysis will lead to clearer insights about the history of the functional regions and their current roles.
Methods
Cell culture
Cells were cultured as described previously (Welch et al. 2004). In brief, G1E and G1E–ER4 cells were grown in Iscove's modified Dulbecco's media (IMDM) with 15% fetal calf serum, 2 U/mL erythropoietin, and 50 ng/mL kit ligand. To activate the conditional GATA1–ER, cells were cultured in the presence of 10−7 mol/L beta-estradiol for 24 h.
Chromatin immunoprecipitation (ChIP)
The ChIP assay was conducted as described previously (Welch et al. 2004). Three different cells were used in this assay: G1E knockout cells and the G1E–ER4 cell before and after induction of the GATA1–ER hybrid with estradiol.
For the ChIP-chip assay, ChIP DNA from induced G1E–ER4 cells after cross-linking and immunoprecipitation with antibody against the ER domain of GATA1-ER was amplified by ligation-mediated PCR according to the protocols provided by NimbleGen. It was hybridized to the NimbleGen array 16 from the mm6 version of the high-density tiling array. This array covers mouse chromosome 7 from position 69,577,286 to 133,051,535 in the mm6 mouse genome assembly. The tiling array consists of oligonucleotide probes of 50 nucleotides, spaced every 100 bp in the nonrepetitive DNA.
ChIP-chip positives were validated using a conventional quantitative PCR (qPCR) assay for GATA1 occupancy and unamplified ChIP material. Primers were designed using primer quest (www.idtdna.com) to amplify DNA intervals (amplicons) between 120 and 150 bp in size and located within a GATA1 binding hits identified by ChIP-chip. A thermal disassociation curve was examined to ensure that all the primer pairs generated single amplicon. The PCR products were measured by SYBR green fluorescence in 20-μL reactions on an ABI 7300 machine. The amounts of products were determined relative to the standard curve generated from a serial dilution of the input DNA. The number of cycles required to generate a PCR product signal of a given magnitude was compared with the cycle number required to generate that signal in a serial dilution of the relevant input. These comparisons for each DNA segment give a value for the relative enrichment.
Peak finding in ChIP-chip data
Both Mpeak (Zheng et al. 2007) and TAMALPAIS (Bieda et al. 2006) programs were applied to identify the GATA1 binding hits. For Mpeak, the two replicate ChIP-chip results from the induced G1E–ER4 cells were used separately as input. Only the peaks identified in the outputs of analysis of both duplicates were selected as the GATA1 binding hits. Different prefiltering thresholds (mean + 1 SD, mean + 2 SD, mean + 2.5 SD, and mean + 3 SD) were chosen, and the default was chosen for all the other parameters. The “peak only” output returns DNA segments of 50 bp, which were then extended 250 bp on both sides to get the final GATA1 binding hits. For the program TAMALPAIS, the two replicate ChIP-chip data sets from the induced G1E–ER4 cells were used as input, and only the peaks identified in both data sets were selected as binding hits. We then constructed a nonredundant union (merging common hits). The original coordinates are in mouse genome assembly mm6, and the chromosomal coordinates of the binding segment hits were lifted over to mouse assembly mm8.
Coordinates of ChIP-chip hits and results of quantitative PCR binding assays and enhancer assays may be browsed as custom tracks on the UCSC Genome Browser and downloaded via the Table Browser at http://bx.psu.edu/~yong/ghp/.
Phylogenetic analysis of conservation of occupied DNA and binding site motifs
MULTIZ alignments (Blanchette et al. 2004) for the February 2006 mouse genome assembly (mm8) (Waterston et al. 2002) were downloaded from the UCSC Genome Browser (Kent et al. 2002). These alignments contain 17 species: mouse, rat, rabbit, human, chimp, macaque, dog, cow, armadillo, elephant, tenrec, Monodelphis, chicken, frog, zebrafish, Tetraodon, and Fugu. The unaligned regions of mouse were supplemented by the mouse sequence. The criteria for inclusion in the alignment are determined by the parameters of the MULTIZ alignment; we did not filter the alignments any further for percent identity or length.
Sequences that match WGATAR in mouse were compared with the aligned sequences of 16 other species in the alignment. The phylogenetic preservation of any given motif was described as the set of species that also contain a WGATAR match at the aligned position. Analyses in this paper focused on high-quality mammalian sequences from that alignment: mouse, rat, human, chimp, dog, cow, armadillo, elephant, and Monodelphis. Diagrams of the positions of WGATAR motifs in the aligned sequences of all DNA segments in this study are at http://www.bx.psu.edu/~dcking/ghp/img2/.
In general, the broadest phylogenetic term was applied to the pattern of species matches to a motif; conservation patterns need not include all intermediate species to yield a given term. The following phylogenetic groups were used to characterize the depth of conservation with mouse: mouse-only, rodent-conserved, euarchontoglires-conserved (extends to primates), boreoeutheria-conserved (extends to cow or dog), eutheria-conserved (extends to tenrec, armadillo, or elephant), mammalia-conserved (extends to the marsupial Monodelphis), amniota-conserved (extends to chicken), tetrapod (extends to frog), and lastly, vertebrate-conserved (extends to the fishes).
For conservation measures using the neutral substitution rate, the branch lengths of the phylogenetic tree were estimated on fourfold degenerate sites. For each conserved motif, the subtree was taken for only the matching species, and the total branch length of the subtree was used to measure the conservation of that motif.
Enhancer assays by transient transfection
The enhancer assays were similar to those described previously (Wang et al. 2006) with the following modifications. DNA segments of ∼1 kb were amplified from mouse DNA for intervals occupied by GATA1 (inferred from ChIP-chip analysis) and negative controls (not positive by ChIP-chip). These were inserted into the luciferase reporter genes driven by the HBG1 gene promoter.
The plasmid DNAs were transiently transfected into K562 cells in a 48-well format using (per well) 0.4 μg of plasmid containing firefly luciferase reporter and 0.0001 μg of cotransfection control plasmid expressing Renilla luciferase in OptiMEM medium (Invitrogen), adding 0.4 μL of PLUS Reagent (Invitrogen) and 0.6 μL Lipofectamine LTX per well. The K562 cells were plated at 8 × 104 cells per well. For each experiment, each plasmid was transfected in quadruplicate. Each plasmid was tested in at least two separate experiments.
Two days after the transfection, cell extracts were harvested and luciferase activity measured in a dual luciferase assay following the manufacturer’s protocol (Promega). For each of the quadruplicate transfections, duplicate measurements were made on the cell extracts for a total of eight measurements of both luciferases for each plasmid in each experiment. The firefly luciferase activity of the test plasmid (divided by the Renilla luciferase activity of the cotransfection control) was normalized by the firefly luciferase activity from the parental MCSluc (divided by the Renilla luciferase activity of the cotransfection control) to obtain a fold change.
Tested DNA segments that caused at least a twofold increase in activity compared with that of the parental reporter gene plasmid in at least two separate transfection experiments are considered to be active as enhancers. The twofold increase is greater than the mean of the negative controls plus 3 standard deviations.
Acknowledgments
This work was supported by NIH grants from NIDDK (DK65806, R.C.H.) and NHGRI (HG002238, W.M.), by Tobacco Settlement Funds from the Pennsylvania Department of Health, and by the Huck Institutes of Life Sciences, Pennsylvania State University. M.J.W. is a Leukemia and Lymphoma Society Scholar.
Footnotes
[Supplemental material is available online at www.genome.org. The ChIP-chip and quantitative PCR data, enhancer results, and analysis of phylogenetic depth of conservation presented in this paper are available at http://bx.psu.edu/~yong/supplementary/GR2008.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.083089.108.
References
- Aparicio S., Morrison A., Gould A., Gilthorpe J., Chaudhuri C., Rigby P., Krumlauf R., Brenner S. Detecting conserved regulatory elements with the model genome of the Japanese puffer fish, Fugu rubripes. Proc. Natl. Acad. Sci. 1995;92:1684–1688. doi: 10.1073/pnas.92.5.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji J., Rusconi S., Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299–308. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- Baroukh N., Ahituv N., Chang J., Shoukry M., Afzal V., Rubin E.M., Pennacchio L.A. Comparative genomic analysis reveals a distant liver enhancer upstream of the COUP-TFII gene. Mamm. Genome. 2005;16:91–95. doi: 10.1007/s00335-004-2442-9. [DOI] [PubMed] [Google Scholar]
- Bieda M., Xu X., Singer M.A., Green R., Farnham P.J. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchette M., Kent W.J., Riemer C., Elnitski L., Smit A.F.A., Roskin K.M., Baertsch R., Rosenbloom K., Clawson H., Green E.D., et al. Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res. 2004;14:708–715. doi: 10.1101/gr.1933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blobel G.A., Weiss M.J. Disorders of hemoglobin: Genetics, pathophysiology, and clinical management. Cambridge University Press; Cambridge, UK: 2001. Nuclear factors that regulate erythropoiesis; pp. 72–94. [Google Scholar]
- Bodine D., Ley T. An enhancer element lies 3′ to the human Aγ globin gene. EMBO J. 1987;6:2997–3004. doi: 10.1002/j.1460-2075.1987.tb02605.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.D., Johnson D.S., Sidow A. Functional architecture and evolution of transcriptional elements that drive gene coexpression. Science. 2007;317:1557–1560. doi: 10.1126/science.1145893. [DOI] [PubMed] [Google Scholar]
- Bulger M., Bender M.A., von Doorninck J.H., Wertman B., Farrell C., Felsenfeld G., Groudine M., Hardison R. Comparative structural and functional analysis of the olfactory receptor genes flanking the human and mouse β-globin gene clusters. Proc. Natl. Acad. Sci. 2000;97:14560–14565. doi: 10.1073/pnas.97.26.14560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M., Schubeler D., Bender M.A., Hamilton J., Farrell C.M., Hardison R.C., Groudine M. A complex chromatin “landscape” revealed by patterns of nuclease sensitivity and histone modification within the mouse β-globin locus. Mol. Cell. Biol. 2003;23:5234–5244. doi: 10.1128/MCB.23.15.5234-5244.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. doi: 10.1016/s0092-8674(04)00127-8. [DOI] [PubMed] [Google Scholar]
- Dermitzakis E., Clark A. Evolution of transcription factor binding sites in mammalian gene regulatory regions: Conservation and turnover. Mol. Biol. Evol. 2002;19:1114–1121. doi: 10.1093/oxfordjournals.molbev.a004169. [DOI] [PubMed] [Google Scholar]
- Dermitzakis E.T., Bergman C.M., Clark A.G. Tracing the evolutionary history of Drosophilaregulatory regions with models that identify transcription factor binding sites. Mol. Biol. Evol. 2003;20:703–714. doi: 10.1093/molbev/msg077. [DOI] [PubMed] [Google Scholar]
- Dermitzakis E.T., Reymond A., Antonarakis S.E. Conserved non-genic sequences—An unexpected feature of mammalian genomes. Nat. Rev. Genet. 2005;6:151–157. doi: 10.1038/nrg1527. [DOI] [PubMed] [Google Scholar]
- Emorine L., Kuehl M., Weir L., Leder P., Max E.E. A conserved sequence in the immunoglobulin Jκ-Cκ intron: Possible enhancer element. Nature. 1983;304:447–449. doi: 10.1038/304447a0. [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T., Reitman M., Felsenfeld G. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc. Natl. Acad. Sci. 1988;85:5976–5980. doi: 10.1073/pnas.85.16.5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg E.C., Downs K.M., Christensen H.M., Im H., Nuzzi P.A., Bresnick E.H. Developmentally dynamic histone acetylation pattern of a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. 2000;97:14494–14499. doi: 10.1073/pnas.97.26.14494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer K.A., Tao H., Osoegawa K., de Jong P.J., Chen X., Doherty M.F., Cox D.R. Noncoding sequences conserved in a limited number of mammals in the SIM2 interval are frequently functional. Genome Res. 2004;14:367–372. doi: 10.1101/gr.1961204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grice E.A., Rochelle E.S., Green E.D., Chakravarti A., McCallion A.S. Evaluation of the RET regulatory landscape reveals the biological relevance of a HSCR-implicated enhancer. Hum. Mol. Genet. 2005;14:3837–3845. doi: 10.1093/hmg/ddi408. [DOI] [PubMed] [Google Scholar]
- Gutierrez L., Nikolic T., van Dijk T.B., Hammad H., Vos N., Willart M., Grosveld F., Philipsen S., Lambrecht B.N. Gata1 regulates dendritic-cell development and survival. Blood. 2007;110:1933–1941. doi: 10.1182/blood-2007-05-091397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardison R.C. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- Johnson K.D., Grass J.A., Boyer M.E., Keikhaefer C.M., Blobel G.A., Weiss M.J., Bresnick E.H. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. 2002;99:11760–11765. doi: 10.1073/pnas.192285999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King D.C., Taylor J., Zhang Y., Cheng Y., Lawson H.A., Martin J., Chiaromonte F., Miller W., Hardison R.C., ENCODE groups for Transcriptional Regulation and Multispecies Sequence Analysis Finding cis-regulatory elements using comparative genomics: Some lessons from ENCODE data. Genome Res. 2007;17:775–786. doi: 10.1101/gr.5592107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam L., Bresnick E.H. A novel DNA binding protein, HS2NF5, interacts with a functionally important sequence of the human beta-globin locus control region. J. Biol. Chem. 1996;271:32421–32429. doi: 10.1074/jbc.271.50.32421. [DOI] [PubMed] [Google Scholar]
- Letting D.L., Rakowski C., Weiss M.J., Blobel G.A. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA1. Mol. Cell. Biol. 2003;23:1334–1340. doi: 10.1128/MCB.23.4.1334-1340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Zhou B., Powers P., Enver T., Stamatoyannopoulos G. Beta-globin locus activation regions: Conservation of organization, structure and function. Proc. Natl. Acad. Sci. 1990;87:8207–8211. doi: 10.1073/pnas.87.21.8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig M.Z., Kreitman M. Evolutionary dynamics of the enhancer region of even-skipped in Drosophila. Mol. Biol. Evol. 1995;12:1002–1011. doi: 10.1093/oxfordjournals.molbev.a040277. [DOI] [PubMed] [Google Scholar]
- Ludwig M.Z., Bergman C., Patel N.H., Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–567. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- Ludwig M.Z., Palsson A., Alekseeva E., Bergman C.M., Nathan J., Kreitman M. Functional evolution of a cis-regulatory module. PLoS Biol. 2005;3:e93. doi: 10.1371/journal.pbio.0030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellon P., Parker V., Gluzman Y., Maniatis T. Identification of DNA sequences required for transcription of the human α1-globin gene in a new SV40 host-vector system. Cell. 1981;27:279–288. doi: 10.1016/0092-8674(81)90411-6. [DOI] [PubMed] [Google Scholar]
- Migliaccio A.R., Rana R.A., Sanchez M., Lorenzini R., Centurione L., Bianchi L., Vannucchi A.M., Migliaccio G., Orkin S.H. GATA1 as a regulator of mast cell differentiation revealed by the phenotype of the GATA1low mouse mutant. J. Exp. Med. 2003;197:281–296. doi: 10.1084/jem.20021149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller W., Makova K.D., Nekrutenko A., Hardison R.C. Comparative genomics. Annu. Rev. Genomics Hum. Genet. 2004;5:15–56. doi: 10.1146/annurev.genom.5.061903.180057. [DOI] [PubMed] [Google Scholar]
- Miller W., Rosenbloom K., Hardison R.C., Hou M., Taylor J., Raney B., Burhans R., King D.C., Baertsch R., Blankenberg D., et al. 28-Way vertebrate alignment and conservation track in the UCSC Genome Browser. Genome Res. 2007;17:1797–1808. doi: 10.1101/gr.6761107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses A.M., Chiang D.Y., Kellis M., Lander E.S., Eisen M.B. Position specific variation in the rate of evolution in transcription factor binding sites. BMC Evol. Biol. 2003;3:19. doi: 10.1186/1471-2148-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses A.M., Pollard D.A., Nix D.A., Iyer V.N., Li X.Y., Biggin M.D., Eisen M.B. Large-scale turnover of functional transcription factor binding sites in Drosophila. PLoS Comput. Biol. 2006;2:e130. doi: 10.1371/journal.pcbi.0020130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuwaysir E.F., Huang W., Albert T.J., Singh J., Nuwaysir K., Pitas A., Richmond T., Gorski T., Berg J.P., Ballin J., et al. Gene expression analysis using oligonucleotide arrays produced by maskless photolithography. Genome Res. 2002;12:1749–1755. doi: 10.1101/gr.362402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin S.H. GATA-binding transcription factors in hematopoietic cells. Blood. 1992;80:575–581. [PubMed] [Google Scholar]
- Orkin S.H., Shivdasani R.A., Fujiwara Y., McDevitt M.A. Transcription factor GATA1 in megakaryocyte development. Stem Cells. 1998;16(Suppl 2):79–83. doi: 10.1002/stem.5530160710. [DOI] [PubMed] [Google Scholar]
- Pennacchio L.A., Rubin E.M. Genomic strategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- Pennacchio L.A., Ahituv N., Moses A.M., Prabhakar S., Nobrega M.A., Shoukry M., Minovitsky S., Dubchak I., Holt A., Lewis K.D., et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Petrykowska H., Vockley C., Elnitski L. Detection and characterization of silencers and enhancer-blockers in the greater CFTR locus. Genome Res. 2008;18:1238–1246. doi: 10.1101/gr.073817.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny L., Simon M.C., Robertson E., Klein W.H., Tsai S.-F., D'Agati V., Orkin S.H., Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- Pevny L., Lin C.S., D'Agati V., Simon M.C., Orkin S.H., Costantini F. Development of hematopoietic cells lacking transcription factor GATA1. Development. 1995;121:163–172. doi: 10.1242/dev.121.1.163. [DOI] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Shivdasani R.A., Fujiwara Y., McDevitt M.A., Orkin S.H. A lineage-selective knockout establishes the critical role of transcription factor GATA1 in megakaryocyte growth and platelet development. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon M.C., Pevny L., Wiles M.V., Keller G., Costantini F., Orkin S.H. Rescue of erythroid development in gene targeted GATA1− mouse embryonic stem cells. Nat. Genet. 1992;1:92–98. doi: 10.1038/ng0592-92. [DOI] [PubMed] [Google Scholar]
- Taylor J., Tyekucheva S., King D.C., Hardison R.C., Miller W., Chiaromonte F. ESPERR: Learning strong and weak signals in genomic sequence alignments to identify functional elements. Genome Res. 2006;16:1596–1604. doi: 10.1101/gr.4537706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde-Garduno V., Guyot B., Anguita E., Hamlett I., Porcher C., Vyas P. Differences in the chromatin structure and cis-element organization of the human and mouse GATA1 loci: Implications for cis-element identification. Blood. 2004;104:3106–3116. doi: 10.1182/blood-2004-04-1333. [DOI] [PubMed] [Google Scholar]
- Visel A., Prabhakar S., Akiyama J.A., Shoukry M., Lewis K.D., Holt A., Plajzer-Frick I., Afzal V., Rubin E.M., Pennacchio L.A. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat. Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall L., deBoer E., Grosveld F. The human beta-globin gene 3′ enhancer contains multiple binding sites for an erythroid-specific protein. Genes & Dev. 1988;2:1089–1100. doi: 10.1101/gad.2.9.1089. [DOI] [PubMed] [Google Scholar]
- Wang X., Crispino J., Letting D., Nakazawa M., Poncz M., Blobel G. Control of megakaryocyte-specific gene expression by GATA1 and FOG-1: Role of Ets transcription factors. EMBO J. 2002;21:5225–5234. doi: 10.1093/emboj/cdf527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang Y., Cheng Y., Zhou Y., King D.C., Taylor J., Chiaromonte F., Kasturi J., Petrykowska H., Gibb B., et al. Experimental validation of predicted mammalian erythroid cis-regulatory modules. Genome Res. 2006;16:1480–1492. doi: 10.1101/gr.5353806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Weiss M.J., Orkin S.H. GATA transcription factors: Key regulators of hematopoiesis. Exp. Hematol. 1995a;23:99–107. [PubMed] [Google Scholar]
- Weiss M.J., Orkin S.H. Transcription factor GATA1 permits survival and maturation of erythroid precursors by preventing apoptosis. Proc. Natl. Acad. Sci. 1995b;92:9623–9627. doi: 10.1073/pnas.92.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M.J., Yu C., Orkin S.H. Erythroid-cell-specific properties of transcription factor GATA1 revealed by phenotypic rescue of a gene-targeted cell line. Mol. Cell. Biol. 1997;17:1642–1651. doi: 10.1128/mcb.17.3.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch J.J., Watts J.A., Vakoc C.R., Yao Y., Wang H., Hardison R.C., Blobel G.A., Chodosh L.A., Weiss M.J. Global regulation of erythroid gene expression by transcription factor GATA1. Blood. 2004;104:3136–3147. doi: 10.1182/blood-2004-04-1603. [DOI] [PubMed] [Google Scholar]
- Woolfe A., Goodson M., Goode D.K., Snell P., McEwen G.K., Vavouri T., Smith S.F., North P., Callaway H., Kelly K., et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 2005;3:e7. doi: 10.1371/journal.pbio.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Bieda M., Jin V.X., Rabinovich A., Oberley M.J., Green R., Farnham P.J. A comprehensive ChIP-chip analysis of E2F1, E2F4, and E2F6 in normal and tumor cells reveals interchangeable roles of E2F family members. Genome Res. 2007;17:1550–1561. doi: 10.1101/gr.6783507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Cantor A.B., Yang H., Browne C., Wells R.A., Fujiwara Y., Orkin S.H. Targeted deletion of a high-affinity GATA-binding site in the GATA1 promoter leads to selective loss of the eosinophil lineage in vivo. J. Exp. Med. 2002;195:1387–1395. doi: 10.1084/jem.20020656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M., Barrera L.O., Ren B., Wu Y.N. ChIP-chip: Data, model, and analysis. Biometrics. 2007;63:787–796. doi: 10.1111/j.1541-0420.2007.00768.x. [DOI] [PubMed] [Google Scholar]