Abstract
The Caenorhabditis elegans genome encodes more than 100 microRNAs (miRNAs). Genetic analyses of miRNA deletion mutants have only provided limited insights into miRNA function. To gain insight into the function of miRNAs, it is important to determine their spatiotemporal expression pattern. Here, we use miRNA promoters driving the expression of GFP as a proxy for miRNA expression. We describe a set of 73 transgenic C. elegans strains, each expressing GFP under the control of a miRNA promoter. Together, these promoters control the expression of 89 miRNAs (66% of all predicted miRNAs). We find that miRNA promoters drive GFP expression in a variety of tissues and that, overall, their activity is similar to that of protein-coding gene promoters. However, miRNAs are expressed later in development, which is consistent with functions after initial body-plan specification. We find that miRNA members belonging to families are more likely to be expressed in overlapping tissues than miRNAs that do not belong to the same family, and provide evidence that intronic miRNAs may be controlled by their own, rather than a host gene promoter. Finally, our data suggest that post-transcriptional mechanisms contribute to differential miRNA expression. The data and strains described here will provide a valuable guide and resource for the functional analysis of C. elegans miRNAs.
Differential gene expression can be regulated at many levels and by various trans-acting factors. MicroRNAs (miRNAs) and transcription factors (TFs) are pivotal regulators of metazoans gene expression. While TFs physically interact with cis-regulatory DNA elements to activate or repress gene expression, miRNAs mainly repress gene expression post-transcriptionally by imperfect base-pairing to sequences located in the 3′UTR of their target mRNAs (for review, see Bartel 2004; Walhout 2006). Like TFs, many miRNAs are highly conserved between related species and even across phyla. Typically, miRNAs are transcribed by RNA polymerase II into a primary transcript (pri-miRNA) that is further processed by RNASEN (also known as DROSHA) into an ∼60-nt-long precursor (pre-miRNA), and subsequently by DICER1 (also known as Dicer) into a mature ∼23-nt-long miRNA (for review, see Bartel 2004). The two founding miRNAs, lin-4 and let-7, were identified genetically as temporal regulators of development in the nematode Caenorhabditis elegans (Lee et al. 1993; Reinhart et al. 2000). MiRNAs regulate a broad range of biological processes in animals and plants, including patterning of the nervous system, cell death, cell proliferation, and development (Ambros 2004; Stefani and Slack 2008). In addition, as for TFs, there is increasing evidence that mammalian miRNA expression may also be regulated at the post-transcriptional level (Obernosterer et al. 2006; Thomson et al. 2006; Wulczyn et al. 2007; Viswanathan et al. 2008;).
Genome-wide genetic analyses in many organisms have demonstrated a myriad of critical roles that TFs play in controlling gene expression during development, homeostasis, and disease. For instance, more than 30% of C. elegans TFs confer a detectable phenotype when knocked down by RNAi (291 out of 940 predicted TFs tested; data obtained from WormBase WS180) (Vermeirssen et al. 2007b). In contrast, with the exceptions of lin-4 (Lee et al. 1993), let-7 (Reinhart et al. 2000), lsy-6 (Johnston and Hobert 2003), and mir-1 (Simon et al. 2008), a single null mutation does not result in an easily detectable phenotype for most C. elegans miRNA genes (Miska et al. 2007). The observation that most C. elegans miRNAs appear to be individually dispensable may reflect roles in processes that have not yet been readily assayable. Alternatively, there may be considerable genetic redundancy between miRNAs and other regulators such as TFs, other miRNAs, and, in some cases, members of the same miRNA family. For example, the three related miRNAs lin-58 (hereafter referred to as mir-48), mir-84, and mir-241 function redundantly in the control of developmental timing in C. elegans (Abbott et al. 2005). One approach that will help to delineate the biological function of miRNAs is by determining when and where they are expressed. The particular pattern of expression of each miRNA gene should help to identify potential genetic interactors that exhibit similar expression patterns, and to design experiments for the delineation of phenotypes of miRNA mutants.
A simple anatomy, invariant cell lineage, transparent body, and high-quality complete genome sequence make C. elegans a highly suitable model to study spatiotemporal miRNA expression. In addition, many biological processes are conserved between nematodes and higher organisms, so the analysis of miRNA function in C. elegans may potentially be applicable to other animals. For instance, it has been demonstrated that mir-1, a highly conserved miRNA, is expressed and functions in muscle in diverse organisms such as mice, zebrafish, fruit flies, and nematodes (Sokol and Ambros 2005; Wienholds et al. 2005; Zhao et al. 2007; Simon et al. 2008). Therefore, the spatiotemporal expression pattern and, perhaps, function of many other miRNAs may also be also conserved.
Previous studies in various organisms have examined miRNA expression by in situ hybridization (Aboobaker et al. 2005; Wienholds et al. 2005), Northern blotting (Lau et al. 2001; Lee and Ambros 2001), or small RNA library sequencing from enriched tissues (Landgraf et al. 2007; Ruby et al. 2007). Although powerful, such studies can be limited by a relatively low sensitivity. In addition, these methods do not enable the analysis of spatiotemporal expression patterns in living animals as they depend on animal fixation (in situ hybridization) or RNA purification (Northern blotting, sequencing). Reporter genes such as that encoding the green fluorescent protein (GFP) have provided powerful tools for the analysis of gene expression in vivo. Indeed, promoter∷gfp fusions in C. elegans have already been used to analyze more than 350 TFs and ∼1800 other protein-coding genes (referred to here as “all genes”) (Hunt-Newbury et al. 2007; Reece-Hoyes et al. 2007). Importantly, this approach faithfully recapitulates known gene expression in the majority of cases examined (Reece-Hoyes et al. 2007).
Here, we present a collection of 73 transgenic C. elegans strains (see Note Added in Proof), each containing a miRNA promoter∷gfp fusion construct. We used promoter activity as a proxy for miRNA expression in vivo. We examined miRNA promoter activity across all developmental stages, and, frequently, to the level of individual cells. We compared miRNA promoter activity to that of the TF and “all gene” data sets introduced above. We find that miRNA promoters are active in all major tissues and cell types. However, miRNA promoters are active later in development than protein-coding gene promoters, which is consistent with roles for miRNAs after the initial specification of body plan, organs, and tissues (Wienholds et al. 2005; Schier and Giraldez 2006). We correlate promoter activity with previously reported Northern blotting data and examine two endogenous pri-miRNAs by RT-PCR. Our data suggest that post-transcriptional regulation of pri-miRNAs provides an additional layer of differential miRNA expression in nematodes. The data and transgenic lines that we present provide a platform for functional miRNA studies to delineate their roles in the development of the animal and to understand their function in gene regulatory networks.
Results
Generation of transgenic PmiRNA∷gfp C. elegans strains
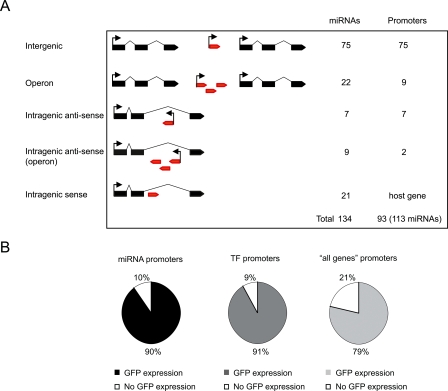
Of the 134 C. elegans miRNA genes currently available in miRBase V9.0, 75 reside in intergenic regions, i.e., between protein-coding genes, and can be assigned to their own promoter (Fig. 1A). An additional 22 intergenic miRNAs are transcribed in a total of nine intergenic operons, with a single promoter regulating each operon. Sixteen miRNAs are embedded within the intron of protein-coding genes in the antisense orientation either as single genes (seven miRNAs), or as operons (nine miRNAs into two operons) (Fig. 1A; Supplemental Table S1). Twenty-one miRNAs are embedded within the intron of a protein-coding gene in the sense orientation. It has been hypothesized that such miRNAs are under the control of the host gene promoter (Baskerville and Bartel 2005), and therefore, we largely focus on the set of 113 miRNAs with presumed independent promoters.
Figure 1.
A) Number of miRNA genes and promoters considered according to genome annotation. Black boxes indicate protein-coding gene exons; red boxes, miRNA genes. (B) Expression rate of Pmir∷gfp constructs compared with TFs and “all genes.”
We generated miRNA promoter∷gfp (PmiRNA∷gfp) fusions by Gateway cloning (Walhout et al. 2000). We used the PmiRNA Entry clones we generated previously (Martinez et al. 2008) and a Gateway-compatible GFP destination vector (Dupuy et al. 2004). Promoter sequences were defined as the intergenic genomic sequence upstream of annotated miRNA genes with a minimum length of 300 bp and a maximum length of 2 kb. It has been shown previously that upstream sequences defined by these criteria are often sufficient for rescue of miRNA mutant phenotypes (Lee et al. 1993; Johnson et al. 2003; Chang et al. 2004) and/or to recapitulate miRNA expression (Johnson et al. 2005; Li et al. 2005; Yoo and Greenwald 2005). PmiRNA∷gfp constructs were then used to transform unc-119(ed3) worms by microparticle bombardment as described (Berezikov et al. 2004; Reece-Hoyes et al. 2007).
In total, we generated a collection of transgenic lines for 70 PmiRNA∷gfp constructs (we will introduce another three below). These 70 constructs together include upstream sequences for 61 single gene miRNAs and nine miRNA operons, corresponding to a total of 86 miRNAs (out of 113 considered, or 76%). On average, we obtained four independent lines per construct. We observed a high transmission rate of the PmiRNA∷gfp transgene for most of the lines (data not shown). With only one exception, all independent lines for a given construct show similar expression patterns. The exception is the promoter of mir-227-80. While one line shows mosaic expression in excretory cells, vulva, body wall muscle, and head neurons, two other independent lines show expression in the pharynx and head neurons. All strains were genotyped to verify the presence of Pmir-227-80, and both expression patterns are provided in our EDGEdb database (Barrasa et al. 2007).
Characterization of miRNA expression patterns
We examined the activity of miRNA promoters throughout the whole organism and across all developmental stages in living animals, and, when feasible, to the level of individual cells. Specifically, for each transgenic line we examined GFP expression in a mixed stage population of hermaphrodites. We only recorded the expression pattern of a given PmiRNA∷gfp reporter strain that was observed consistently in each of the independent PmiRNA∷gfp transgenic lines (data not shown). Detailed descriptions and representative images can be found in Supplemental Table S2 and in our publicly available EDGEdb database.
In total, 90% of the miRNA promoters confer GFP expression (63 out of 70) (Supplemental Table S3). The expression rate of PmiRNA∷gfp fusions is comparable to that of TFs (91%) (Reece-Hoyes et al. 2007) and “all genes” (79%) (Fig. 1B; Hunt-Newbury et al. 2007). This demonstrates that the chosen genomic sequences upstream of miRNAs indeed function as promoters. The promoters of seven miRNAs did not drive detectable GFP expression in vivo. Two of these miRNAs are conserved in the related nematode Caenorhabditis briggsae: lsy-6, a well-characterized miRNA involved in neuronal specification (Johnston and Hobert 2003), and mir-77, for which a phenotype has not been described but which has been detected in large-scale sequencing analyses (Ruby et al. 2006). The fact that we did not observe GFP expression for these promoters may be because they lack elements required for expression or because the transgene is present at a low copy number, which may not suffice for the detection of GFP expression. The other five miRNAs for which we did not detect promoter activity, mir-257, mir-258, mir-261, mir-267, and mir-271, are not conserved in C. briggsae and have not been detected by large-scale sequencing (Ruby et al. 2006). We found a significant correlation between GFP expression and conservation (Fisher exact test, P-value < 0.05) and between GFP expression and detection by sequencing (Fisher exact test, P-value < 0.05). Based on these observations, it is possible that some or all of mir-257, mir-258, mir-261, mir-267, and mir-271 are not genuine miRNAs and/or are not transcribed under normal culture conditions.
Temporal PmiRNA∷gfp activity correlates with Northern blot analysis
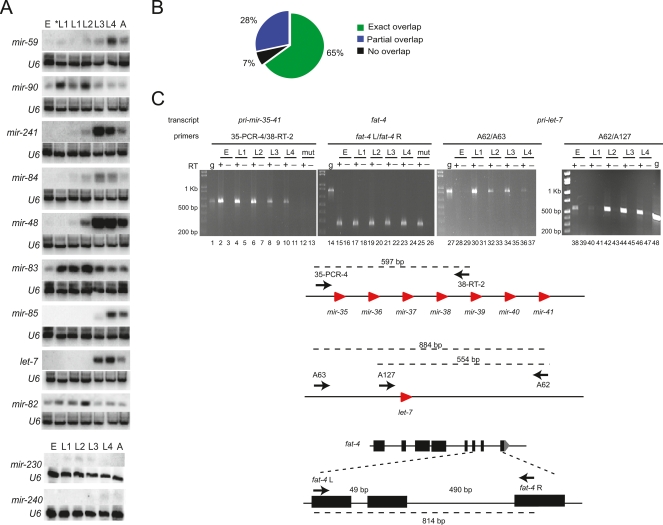
Northern blots have been extensively used to determine the temporal expression of miRNAs in C. elegans (Lau et al. 2001; Lee and Ambros 2001; Ambros et al. 2003; Lim et al. 2003). We searched WormBase (WS180) for information regarding temporal miRNA expression. Of the 81 miRNAs for which we had temporal information based on PmiRNA∷gfp transgenic lines, equivalent information was available for 58 (Supplemental Table S4). We found that the observed temporal GFP expression pattern agrees with the pattern detected by developmental Northern blots in most of the cases. Four PmiRNA∷gfp strains did not match the temporal expression pattern. For instance, we only detected mir-82 promoter activity in the L4 and adult stage, while Northern blotting detected mature miRNA in all developmental stages (Fig. 2A, see also below). These discrepancies may be due to a lack of regulatory elements in the chosen genomic DNA fragment. In other cases, the temporal pattern partially agrees with previously reported patterns (Supplemental Table S4). Twelve PmiRNA∷gfp strains exhibited earlier expression than reported previously by Northern blotting. There are several explanations for this difference. For instance, the DNA fragments used as promoters may lack transcriptional elements that are required for repression of miRNA expression in early developmental stages. Also, GFP transgenics may be more sensitive for detecting spatially restricted miRNA expression in early stages of development. For instance, mature mir-237 was detected from L3 to adult stages; however, we observed GFP expression in Pmir-237∷gfp animals as early as the first larval stage (see also Esquela-Kerscher et al. 2005). We performed additional Northern blotting using StarFire probes to detect the temporal expression of nine miRNAs: mir-241, mir-84, mir-48, let-7, mir-83, mir-230, mir-240, mir-82, and mir-85. This allowed for the more sensitive detection of low levels of mature miRNAs than traditional oligonucleotide probes (Behlke et al. 2000; Abbott et al. 2005; Ow et al. 2008). For four of these miRNAs (mir-241, mir-84, mir-48, and mir-83), we detected a weak miRNA signal at earlier stages than previously reported, consistent with our PmiRNA∷gfp expression data (Supplemental Table S4; Fig. 2A). We also detected the temporal expression of two additional miRNAs (mir-59 and mir-90) for which there was no information in WormBase, and found that it was consistent with promoter activity (Supplemental Table S4; Fig. 2A).
Figure 2.
Temporal PmiRNA∷gfp activity correlates with Northern blot analysis and uncovers possible post-transcriptional mechanisms that control miRNA expression. (A) Northern blot analyses using StarFire probes detect temporal expression of mature miRNAs. E indicates embryo; L, larvae; *L1, starved L1; A, adult. Probe against the U6 snRNA was used as control. (B) Comparison between miRNA expression determined by Northern blotting and promoter∷gfp reporters. (C) Detection of mir-35-41 and let-7 primary transcripts by RT-PCR. As control, we used primers to amplify a protein-coding mRNA (fat-4). Total RNA from N2 embryos, L1, L2, L3, and L4 stages and total RNA from VC514 mir-35-41 mutant embryos (mut) were subjected to reverse transcription (+RT, lanes 2,4,6,8,10,12,15,17,19,21,23,25,28,30,32,34,36,38,40,42,44,46) or mock reactions (−RT, lanes 3,5,7,9,11,13,16,18,20,22,24,26,29,31,33,35,37,39,41,43,45,47). Genomic DNA was used as size marker (g, lanes 1,14,27,48). Cartoons indicate the predicted size of PCR amplicons from mir-35-41 primary transcript, let-7 primary transcript, and fat-4 mRNA and indicate the primers that were used. Note that fat-4 L and R primers amplify a product of different size when genomic DNA (lane 14) or cDNA (lanes 15–26) was used as template.
We have also observed cases in which mature miRNAs were only detected early by Northern blotting, while we detected continuous PmiRNA∷gfp activity in later stages. This was the case for two miRNA operons: mir-42-44 and mir-35-41 (a total of nine miRNAs). In addition to the aforementioned reasons, the differences observed between mature miRNA expression and miRNA promoter activity may be due to post-transcriptional mechanisms that may regulate transcript stability or processing of either the pri-miRNA, the pre-miRNA, or the mature miRNA (see below).
Taken together, the temporal expression in Pmir∷gfp animals was consistent with the expression determined by Northern blotting for 65% (39/60) of the miRNAs in our data set. For only 7% (4/60) of the miRNAs, the expression determined by Northern blotting does not agree with promoter activity, while the remaining 28% (17/60) partially agrees (Fig. 2B).
Post-transcriptional mechanisms contribute to differential miRNA expression
Our data suggest that post-transcriptional mechanisms affect transcript processing or stability of several miRNAs. For instance, while mir-61 is detected by Northern blotting in all developmental stages, mir-250 is only detected starting from the L1 stage (Lee and Ambros 2001; Lim et al. 2003), even though both miRNAs are likely transcribed from the same promoter (Pmir-61-250). This suggests that post-transcriptional mechanisms may either prevent processing of the pre-mir-250 transcript or affect mature miR-250 stability. Similarly, three other miRNAs that are expressed from a single operon and are thus controlled by one promoter, mir-42, mir-43, and mir-44, are differentially expressed: Mature mir-42 and mir-43 are only detected in embryos, while mir-44 is detected not only in embryos but also in larval and adult stages (Lau et al. 2001). Consistent with the expression of mature mir-44, we observed promoter activity from Pmir-42-44 in all developmental stages, suggesting that mir-42 and mir-43 might be subject to post-transcriptional regulation. Lau et al. (2001) only detected mature miRNAs from the mir-35-41 operon in the embryo by Northern blotting. However, they detected the precursor of miR-35 (pre-mir-35) both in the embryo and at the L4 stage, suggesting that it is down-regulated between those stages. We detected Pmir-35-41 activity (by GFP fluorescence) not only in embryos and L4 stages but also in the other larval stages and in adults (EDGEdb) (Supplemental Table S3). To test whether this may be the result of GFP stability rather than promoter activity, we used RT-PCR to detect the endogenous mir-35-41 primary transcript (pri-mir-35-41). We observed pri-mir-35-41 in embryos and L4, where mature and pre-miRNAs are detected, as well as in L1, L2, and L3 stages, where neither mature nor pre-miRNAs from this cluster were detected. This suggests that post-transcriptional mechanisms regulate the processing or stability of the mir-35-41 primary transcript, pre-miRNAs, or mature miRNAs during L1 to L4 stages (Fig. 2C). We also compared the expression of the let-7 primary transcript to the expression of mature let-7 as described previously (Bracht et al. 2004). We detected mature let-7 by Northern blotting starting at the L3 stage, which is in agreement with previous observations (Fig. 2A; Reinhart et al. 2000). However, we detected pri-let-7 by RT-PCR as early as the embryonic stage, consistent with the GFP expression observed in Plet-7∷gfp strains (EDGEdb) (Fig. 2C). Similar observations have been made for let-7 in mammalian systems, where let-7 processing is selectively blocked in embryonic stem cells (Wulczyn et al. 2007; Viswanathan et al. 2008). Our results suggest that post-transcriptional mechanisms likely regulate pri-let-7 processing at early stages (Fig. 2C).
Taken together, our results show that miRNA promoter activity largely overlaps with mature miRNA expression and that post-transcriptional mechanisms likely contribute to differences in primary and mature miRNA expression.
The promoters of miRNAs are active later in development
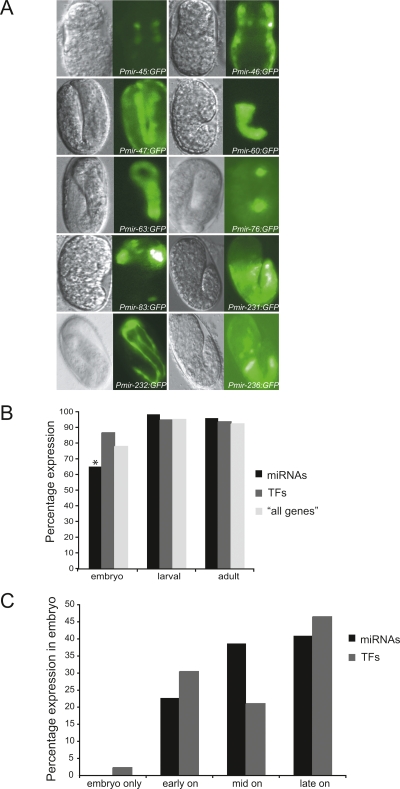
We detected GFP expression conferred by miRNA promoters in all developmental stages. Representative examples of embryonic promoter activity are shown in Figure 3A. We compared the temporal expression conferred by promoter of miRNAs to that of TFs and “all genes” and noticed that miRNA promoters overall tend to be less active in embryos (P-value < 0.05) (Fig. 3B). In addition, the majority of miRNA promoters that confer embryonic expression tend to do so at later embryonic stages, on average, than do promoters of TFs (this analysis could not be done for the “all genes” data set as analogous temporal expression information was not available) (Fig. 3C). This observation is in agreement with previous studies in other organisms that suggest that miRNAs are involved in tissue differentiation and maintenance rather than the establishment of body plan, organs, and tissues (Wienholds et al. 2005; Schier and Giraldez 2006).
Figure 3.
Temporal miRNA promoter activity. (A) Representative images of miRNA promoters that drive GFP expression in the embryo. (Left) DIC images; (right) GFP fluorescence. Additional images can be found in the EDGEdb database (Barrasa et al. 2007). (B) miRNAs tend to be expressed later in development compared to TFs and “all genes.” The asterisk indicates a significant difference (P < 0.05). (C) Percentage of miRNA and TF promoters that drive expression in the embryonic stage: embryo only, early, mid, and late embryonic stages.
Most miRNA promoters drive GFP expression in a tissue-specific manner
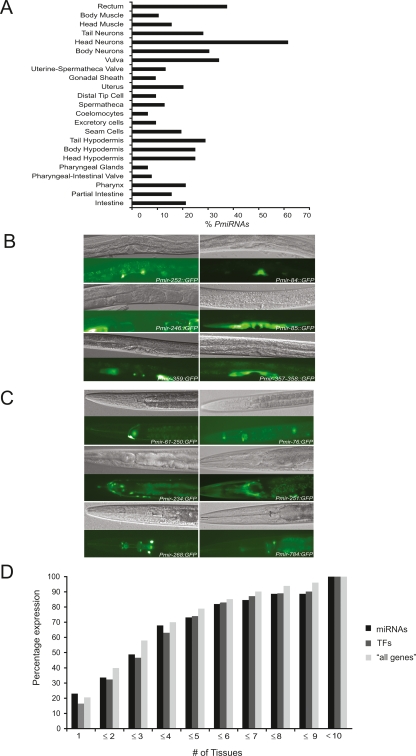
We found that miRNA promoters drive expression in all major tissues and cell types, except the germline (Fig. 4A; Supplemental Table S3). Representative examples of miRNA promoter activity in various parts of the somatic gonad and neuronal cells are shown in Figure 4B and C. Microparticle bombardment has been reported to be the method of choice when germline expression is desired (Praitis et al. 2001). However, none of the miRNA promoters are able to direct GFP expression in the germinal gonad. Thus, it is possible that the miRNAs assayed here are exclusively expressed in somatic tissues. However, promoters of protein-coding genes also generally fail to drive GFP reporter expression in the germline (Hunt-Newbury et al. 2007; Reece-Hoyes et al. 2007). Thus, we think that it is more likely that the GFP transgene is silenced in the germinal gonad. Future studies that use germline-specific deep sequencing of miRNAs will reveal whether any of the miRNAs are expressed in this tissue.
Figure 4.
Spatial miRNA promoter activity. (A) Percentage of miRNA promoters that drive expression in a survey of tissues. (B) Multiple miRNA promoters drive expression in various parts of the somatic gonad, including gonadal sheath, vulva, and uterus. (Top) DIC images; (bottom) GFP fluorescence. (C) Multiple miRNA promoters drive GFP expression in the nervous system. (Top) DIC images; (bottom) GFP fluorescence. Additional images can be found in the EDGEdb database (Barrasa et al. 2007). (D) Most miRNAs as well as TFs and “all genes” promoters confer GFP expression in a tissue-specific manner.
To enable the comparison at a tissue level between miRNAs and protein-coding genes, we reannotated the TF and “all genes” data sets according to a systematic spatiotemporal expression scheme that we devised. We defined 23 categories (hereafter referred to as “tissues”), including intestine, vulva, head neurons, etc. (for precise definitions, see Methods and Supplemental Table S5). Some of these are highly specific (e.g., distal tip cells), and others are broader (e.g., head neurons). We observed that most miRNA promoters confer GFP expression in only a few tissues or cell types. For instance, ∼50% of the promoters confer expression in three or fewer tissues, while only less than 5% of promoters confer ubiquitous somatic expression (lin-4, let-7, and mir-53). A high degree of tissue specificity has also been observed for miRNAs in other organisms, including chicken and zebrafish (Wienholds et al. 2005; Xu et al. 2006). The promoters of TFs and “all genes” drive GFP expression with a similar degree of tissue specificity (Fig. 4D). We recently obtained a genome-scale miRNA transcriptional network (Martinez et al. 2008) that reveals a similar overall network architecture as protein-coding gene networks (Deplancke et al. 2006; Vermeirssen et al. 2007a; Martinez et al. 2008). Together, these observations indicate that the regulation of miRNA gene promoters is not fundamentally different from that of protein-coding gene promoters.
Members of miRNA families can be expressed in distinct or overlapping patterns
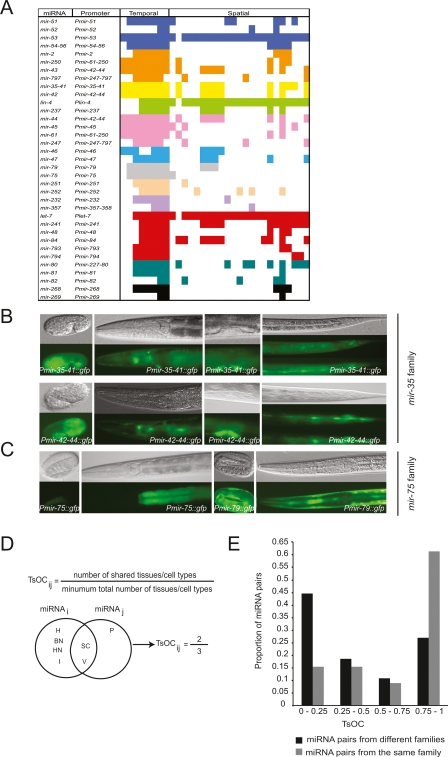
MiRNAs can be classified into families according to sequence similarities (Bartel 2004). Sixty percent of C. elegans miRNAs (78 out of 134) can be classified into 24 families, each containing between two and eight members (Ruby et al. 2006). Members of a given family are predicted to share target mRNAs and may function redundantly (Abbott et al. 2005; Miska et al. 2007). For instance, the let-7 family members mir-48, mir-84, and mir-241 function together to regulate the L2 to L3 cell fate transitions in the hypodermis (Abbott et al. 2005). Redundancy among miRNAs from the same family can occur if miRNA family members are (partially) coexpressed. We examined the extent to which spatiotemporal promoter activity of members of a miRNA family overlap. We were able to compare the expression patterns for 10 complete miRNA families, as well as for two families for which we have expression patterns for most, but not all of the members (Fig. 5A). Interestingly, we observed that some families are expressed with a high degree of overlap, whereas other families exhibit largely nonoverlapping spatiotemporal expression. For instance, miRNAs from the mir-35 family (mir-35-36-37-38-39-40-41 cluster and mir-42) are expressed throughout all stages and in overlapping tissues, including the vulva, seam cells, head neurons, and the rectum (Fig. 5B). Similarly, members of the lin-4 (lin-4, mir-237) and mir-46 (mir-46, mir-47) families are expressed in overlapping tissues (Supplemental Table S3). In contrast, miRNAs from the mir-75 family (mir-75 and mir-79) are expressed in different tissues. While Pmir-75 confers GFP expression exclusively in the intestine, Pmir-79 drives expression in the hypodermis (Fig. 5C). Similarly, members of the mir-232 (mir-232, mir-357) and mir-251 (mir-251, mir-252) families exhibit distinct expression patterns (Supplemental Table S3).
Figure 5.
miRNAs from a given family can have overlapping as well as different spatiotemporal expression patterns. (A) Cartoon depicting expression patterns of 10 complete and two incomplete (let-7 and mir-80 families shown at the bottom) miRNA families. Each color represents a family. Spatiotemporal expression is as in Supplemental Table S3. (B) miRNAs from the mir-35 family are expressed in overlapping tissues/cell types. Pmir35-41∷gfp and Pmir-42-44∷gfp are shown. (C) miRNAs from the mir-75 family are expressed in different tissues/cell types. (Top) DIC images; (bottom) GFP fluorescence. (D) Definition and example of TsOC between any two miRNAs. (E) Distribution of TsOC among miRNA pairs from the same or different family.
We introduce a “tissue overlap coefficient” (TsOC) as the number of tissues shared between two miRNAs divided by the smallest of the total number of tissues where either miRNA is expressed (Fig. 5D). This coefficient is similar to the topological overlap coefficient (TOC) that is used for network modularity analysis (Vermeirssen et al. 2007a). We used TsOC as a measure to determine if the overlap in expression between miRNAs from the same family is different than the overlap in expression between miRNAs from distinct families. We calculated TsOCs for all pairs of miRNAs from distinct families, as well as pairs of miRNAs from the same family (see Methods). We found that the distribution of TsOCs for pairs of miRNAs from the same family is significantly different than the distribution of TsOCs for pairs of miRNAs from distinct families (Fisher Freeman Halton test P-value < 0.001) (Fig. 5E). Pairs of miRNAs from the same family tend to have a higher TsOC compared with pairs of miRNAs from distinct families. Taken together, the degree of overlapping expression varies per miRNA family; however, miRNAs from the same family do tend to exhibit overlapping expression patterns. Thus, it is likely that the lack of phenotypes for individual miRNAs can be explained (at least partly) by familial redundancy and that, in addition, many miRNAs may have a synthetic genetic interaction with other miRNAs, or perhaps with protein-coding genes.
Intragenic miRNAs
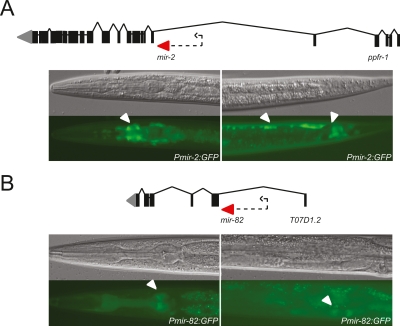
MiRNA genes that are located within the intron of a protein-coding gene in the sense orientation are thought to be under the control of the host gene promoter (Baskerville and Bartel 2005). We generated PmiRNA∷gfp constructs using the immediate upstream sequence of three of these intragenic miRNAs: mir-58, mir-2, and mir-82, which are embedded in the intron of Y67D8A.1, ppfr-1, and T07D1.2, respectively (Fig. 6; also see Note Added in Proof). We found that the region upstream of mir-58 does not confer GFP expression (data not shown). Surprisingly, however, sequences upstream of both mir-82 and mir-2 drive tissue-specific GFP expression (Fig. 6). In addition, the annotation of lin-4 has recently changed; rather than being located in an intergenic region (WS140), it is now annotated to be located in an intron of F59G1.4 (WS180). We and others have shown that the genomic fragment immediately upstream of lin-4 does function as a promoter (Esquela-Kerscher et al. 2005; Ow et al. 2008; this study). It has been previously shown that internal promoters in operons are a common feature in the C. elegans genome (Huang et al. 2007). It is tempting to speculate that internal miRNA promoters located in the introns of protein-coding genes might be common as well. In contrast to C. elegans miRNAs, most human miRNAs are located within introns (Rodriguez et al. 2004). It will be interesting to see if the genomic sequences upstream of these miRNAs can function as promoters in mammals or whether this is specific to nematodes.
Figure 6.
Upstream sequences of two intragenic miRNAs can drive GFP expression in vivo. (A) Pmir-2 drives expression in the nerve ring (left) and ventral nerve cord and tail neurons (right). (B) Pmir-82 drives expression in pharyngeal muscle and head neurons (left), and developing spermatheca (right). (Top) DIC images; (bottom) GFP fluorescence. Arrows indicate expression. Dotted arrows indicate sequence used as promoter.
Discussion
We present here the generation and analysis of transgenic animals for 73 PmiRNA∷gfp constructs that represent the expression of 89 C. elegans miRNAs. Several lines of evidence indicate that the majority of these transgenic animals likely recapitulate endogenous miRNA transcription. First, it has been demonstrated previously that a 2-kb fragment upstream of the translational start site of protein-coding genes accurately drives gene expression in the majority of cases examined (Reece-Hoyes et al. 2007). Second, the majority of PmiRNA∷gfp lines completely or partially recapitulate previously reported temporal expression of miRNAs detected by Northern blotting (Lau et al. 2001; Lee and Ambros 2001; Ambros et al. 2003; Lim et al. 2003). Third, for a handful of miRNAs it has been shown that such a fragment is sufficient for miRNA rescue and in other expression experiments (Lee et al. 1993; Johnston and Hobert 2003; Chang et al. 2004; Johnston Jr. et al. 2005; Li et al. 2005; Yoo and Greenwald 2005).
We compared miRNA promoter activity to mature miRNA expression determined by Northern blotting. While in the majority of cases promoter activity exactly agrees with mature miRNA expression, there are cases in which they only partially agree. We have shown in the case of let-7 and the mir-35-41 operon that this partial agreement is likely due to post-transcriptional mechanisms that contribute to differential miRNA expression. Such mechanisms can in principle control pri-miRNA, pre-miRNA, or mature miRNA stability and/or processing. It has been shown previously that mammalian miRNAs can be regulated post-transcriptionally (Obernosterer et al. 2006). Viswanathan et al. (2008) have identified LIN-28 as a developmentally regulated RNA-binding protein that selectively blocks the processing of pri-let-7 in embryos. In the future, it will be important to dissect the factors that play a role in post-transcriptional regulation of C. elegans miRNAs.
We found that miRNAs are expressed in a variety of tissues. In zebrafish and fruit flies, previous studies have also shown a broad expression for many miRNAs (Aboobaker et al. 2005; Wienholds et al. 2005). We also found that miRNAs are expressed relatively late in development, which is in agreement with results obtained in zebrafish and likely reflects a function of miRNAs in tissue differentiation and maintenance, rather than in tissue establishment (Wienholds et al. 2005).
Most miRNAs do not confer a detectable phenotype when deleted (Miska et al. 2007). It is likely that the lack of phenotypes for individual miRNAs can be explained not only by familial redundancy but also by genetic interactions with miRNAs from other families, or perhaps by interactions with protein-coding genes, such as TFs. The spatiotemporal miRNA expression patterns will provide an important tool for the identification of genes with which they may act redundantly and, hence, will be an important tool that can be used toward understanding the cellular functions of each miRNA.
Our study provides some important advantages over other studies of miRNA expression. First, our method is noninvasive, which means that expression can be studied in living animals. Second, in contrast to methods such as Northern blotting or sequencing, we can frequently annotate miRNA expression to the single cell level. Third, we will provide all the strains to the C. elegans community, which should help to delineate the expression patterns at greater levels of resolution. Fourth, the transgenic lines will enable the study of miRNA expression under different (experimental) conditions, including dauer, stress, etc., and in males. And finally, the transgenic lines will be available for other studies. For example, they can be used to identify or validate upstream regulators of miRNA expression. We recently mapped a genome-scale miRNA regulatory network by high-throughput yeast one-hybrid assays (Deplancke et al. 2004, 2006) and used several of the transgenic lines described here for in vivo validation of the interactions obtained (Martinez et al. 2008; Ow et al. 2008).
Methods
Generation of Pmir∷gfp constructs
For our network study (Martinez et al. 2008) we used the 115 miRNA predictions available in WormBase WS130 (http://www.wormbase.org) and miRNA registry V4.0 (http://microrna.sanger.ac.uk) (Ambros et al. 2003; Lim et al. 2003; Griffiths-Jones et al. 2006). We completed these with 19 recently discovered miRNAs (WormBase WS175 and miRBase V9.0). A miRNA promoter is defined as the intergenic region upstream of the predicted stem-loop sequence or from the mature miRNA as annotated in miRBase V4.0 (Supplemental Table S1). We used a minimal length of 300 bp and a maximal length of 2 kb. In total, 93 promoters (that control 113 miRNAs) were selected. Seventy-three promoters (controlling 89 miRNAs) were successfully cloned into pDEST-DD04 by Gateway cloning as described (Walhout et al. 2000; Dupuy et al. 2004). Constructs were verified by DNA sequencing using either GFP Fw (5′-TTCTACTTCTTTTAC TGAACG) or GFP Rv (5′-CTCCACTGACAGAAAATTTG) primers.
The following PmiRNA∷gfp constructs were generated by conventional restriction enzyme-based cloning into the pPD97.75 vector (for information on restriction sites used, see Supplemental Table S1): Pmir-257, Pmir-51, Pmir-2, Pmir-228, Pmir-54, Pmir-81, Pmir-235, Pmir-227-80 and Pmir-234, Plet-7, Plin-4, Pmir-48, Pmir-237, Pmir-241, Pmir-84.
C. elegans strains
Routine C. elegans maintenance and culture were done as described (Brenner 1974). The DP38 strain (unc-119(ed3)) was cultured in liquid media for microparticle bombardment as described (Reece-Hoyes et al. 2007) or in egg plates (Wood 1988).
Transformation of C. elegans by microparticle bombardment
Transgenic PmiRNA∷gfp animals were generated as described previously (Berezikov et al. 2004; Reece-Hoyes et al. 2007).
Genotyping
The genotype of each transgenic line was confirmed by single animal PCR (Williams et al. 1992) using GFP Fw and GFP Rv primers (see above) as described, followed by DNA sequencing to confirm the identity of the miRNA promoter in the PmiRNA∷gfp transgene.
Characterization of GFP expression patterns
Mixed populations of hermaphrodites were examined by fluorescence microscopy using a Zeiss Axioskop 2 plus microscope equipped with a FITC filter. We recorded the expression pattern conferred by each miRNA promoter that was consistent in each of the independently derived transgenic lines (except for Pmir-227-80, see main text). Fluorescence photographs representative of each expression pattern were taken using a Hamamatsu Orca-ER/1394 video camera and Axiovision Rel. 4.5 software and stored in the EDGEdb database (Barrasa et al. 2007). For each genotype, we stored up to three independent lines into frozen stocks. These lines were chosen based on highest transmission level and/or GFP expression (data not shown). These lines will be made available through the CGC.
PmiRNA∷gfp expression pattern annotation
We devised a standarized temporal and spatial annotation to record the expression pattern of each PmiRNA∷gfp. Temporal expression patterns were classified into eight stages: early, mid, and late embryo; all four larval stages; and adult stage. We defined early embryo as the pre-comma stage, mid-embryo as comma stage, and late embryo as two and threefold embryos. Spatial expression patterns were classified into 23 categories that correspond to tissues, cell types, organs, and, when feasible, to individual cells (i.e., coelomocytes and distal tip cells) (Supplemental Table S5). For GFP expression analysis purposes, temporal and spatial expression was standardized into a binary code, where 1 represents expression detected and 0 represents no expression detected (Supplemental Table S3).
Other data sets
GFP expression patterns driven by “TFs” and other protein-coding gene (“all genes”) promoters where obtained from Reece-Hoyes et al. (2007) and Hunt-Newbury et al. (2007), and converted into our binary annotation scheme. Specifically, “all genes” patterns were classified as follows: BM (body wall muscle); BN (body neurons, lateral nerve cords/commissures, ventral nerve cord); C (coelomocytes); DTC (distal tip cell); GS (gonad sheath cells); HH (hypodermis); HN (amphids, dorsal nerve cord, head neurons, labial sensilla, nerve ring, pharyngeal neurons); I (intestinal, intestinal muscle); O (other: amphid socket cells, developing gonad, head mesodermal cell, mechanosensory neurons, pvt interneuron, unidentified body, unidentified cells, unidentified tail, unidentified head, uterine-seam cell, other); P (arcade cells, pharynx); PG (pharyngeal gland cells); PIV (pharyngeal-intestinal valve); R (anal depressor muscle, anal sphincter, rectal epithelium, rectal gland cells); S (developing spermatheca, spermatheca); SC (seam cells); TN (phasmids, tail neurons); U (developing uterus, uterine muscle, uterus); USV (spermatheca-uterine valve); V (developing vulva, vulva other, vulval muscle); and X (excretory cells, excretory gland cells). For comparison analyses, several tissues/systems were fused in one or more of the data sets to allow the same category types in all three data sets: HH and BH categories were fused into one category, H (hypodermis); HM and BM categories were fused into one M (muscle); PG and P were fused into P (pharynx); and I and PI were fused into I (intestinal).
Northern blot analyses
Total RNA was extracted using TRIzol reagent (Invitrogen) and analyzed by Northern blotting using 5 μg of RNA from each stage as described before (Ow et al. 2008).
RT-PCR analyses
Total RNA was extracted as above and digested with RNase-Free DNase Set (Qiagen) following the manufacturer’s recommendations. First strand cDNA synthesis was performed using 2.5 μg of total RNA, random primers, and SuperScript II (Invitrogen) following manufacturer’s recommendations.
Primer sequences used in the PCR reactions were as follows:
38-RT-2: 5′-GGGCTCTCGGTATATCAGG-3′
35-PCR-4: 5′-GGAAATGGTCCATTCAGTCATC-3′
fat-4 L: 5′-TGTTTCTATCTTGTTGGAGG
fat-4 R: 5′-GGTAAACCATTTGCTGCTGC
Primers used to detect the let-7 primary transcript are A62, A127, and A63 (Bracht et al. 2004).
TsOC analysis
The TsOC between any two miRNAs was defined as the number of tissues where both miRNAs are expressed divided by the smallest of the total number of tissues in which either miRNA is expressed (see Fig. 5D). In case of operons, where several miRNAs are expressed from a single promoter, the same expression pattern was assigned to all miRNAs in the operon. We calculated a TsOC for all individual pairs of miRNAs from different families (3160 total pairs) and all pairs of miRNAs from the same families (80 total pairs). We grouped the TsOCs into four bins (0 < TsOC ≥ 0.25, 0.25 < TsOC ≥ 0.5, 0.5 < TsOC ≥ 0.75, and 0.75 > TsOC ≥ 1) and calculated if the distribution of TsOCs within families was significantly different from the distribution between families using a Fisher Freeman Halton test for 4-rows × 2-columns tables (Freeman and Halton 1951).
Acknowledgments
We thank the members of the Walhout and Ambros laboratories and Job Dekker for advice and critical reading of the manuscript. We thank the Caenorhabditis Genetics Center (CGC), which is funded by the NIH National Center for Research Resources (NCRR), for providing the unc-119(ed3) strain. We thank A. Fire for providing pPD95.75. M.C.O. was supported in part by NIH postdoctoral fellowship GM070118-02. This work was supported by NIH grants DK068429 to A.J.M.W. and GM348642 to V.R.A.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.083055.108.
Note added in proof
During the review of this paper, we have generated an additional C. elegans transgenic strain containing Pmir-71∷gfp. This strain was not included in the analysis, but information regarding GFP expression is available in Supplemental Tables and EDGEdb. mir-71 is an intragenic miRNA, annotated in the intron of ppfr-1, the same intron where mir-2 is annotated. Sequence upstream of mir-71 drives GFP expression in vivo.
References
- Abbott A.L., Alvarez-Saavedra E., Miska E.A., Lau N.C., Bartel D.P., Horvitz H.R., Ambros V. The let-7 microRNA family members mir-48, mir-84 and mir-241 function together to regulate developmental timing in Caenorhabditis elegans. Dev. Cell. 2005;9:403–414. doi: 10.1016/j.devcel.2005.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aboobaker A.A., Tomancak P., Patel N.H., Rubin G.M., Lai E.C. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc. Natl. Acad. Sci. 2005;102:18017–18022. doi: 10.1073/pnas.0508823102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Ambros V., Lee R.C., Lavanway A., Williams P.T., Jewell D. MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr. Biol. 2003;13:807–818. doi: 10.1016/s0960-9822(03)00287-2. [DOI] [PubMed] [Google Scholar]
- Barrasa M.I., Vaglio P., Cavasino F., Jacotot L., Walhout A.J.M. EDGEdb: A transcription factor-DNA interaction database for the analysis of C. elegans differential gene expression. BMC Genomics. 2007;8:21. doi: 10.1186/1471-2164-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baskerville S., Bartel D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlke M., Dames S.A., McDonald W.H., Gould K.L., Devor E.J., Walder J.A. Use of high specific activity StarFire oligonucleotide probes to visualize low-abundance pre-mRNA splicing intermediates in S. pombe. Biotechniques. 2000;29:892–897. doi: 10.2144/00294pf01. [DOI] [PubMed] [Google Scholar]
- Berezikov E., Bargmann C.I., Plasterk R.H. Homologous gene targeting in Caenorhabditis elegans by biolistic transformation. Nucleic Acids Res. 2004;32:e40. doi: 10.1093/nar/gnh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracht J., Hunter S., Eachus R., Weeks P., Pasquinelli A.E. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Johnston R.J., Frokjaer-Jensen C., Lockery S., Hobert O. MicroRNAs act sequentially and asymmetrically to control chemosensory laterality in the nematode. Nature. 2004;430:785–789. doi: 10.1038/nature02752. [DOI] [PubMed] [Google Scholar]
- Deplancke B., Dupuy D., Vidal M., Walhout A.J.M. A Gateway-compatible yeast one-hybrid system. Genome Res. 2004;14:2093–2101. doi: 10.1101/gr.2445504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplancke B., Mukhopadhyay A., Ao W., Elewa A.M., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stam L., Reece-Hoyes J.S., Hope I.A., et al. A gene-centered C. elegans protein–DNA interaction network. Cell. 2006;125:1193–1205. doi: 10.1016/j.cell.2006.04.038. [DOI] [PubMed] [Google Scholar]
- Dupuy D., Li Q., Deplancke B., Boxem M., Hao T., Lamesch P., Sequerra R., Bosak S., Doucette-Stam L., Hope I.A., et al. A first version of the Caenorhabditis elegans promoterome. Genome Res. 2004;14:2169–2175. doi: 10.1101/gr.2497604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Johnson S.M., Bai L., Saito K., Partridge J., Reinert K.L., Slack F.J. Post-embryonic expression of C. elegans microRNAs beloning to the lin-4 and let-7 families in the hypodermis and reproductive system. Dev. Dyn. 2005;234:868–877. doi: 10.1002/dvdy.20572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman G.H., Halton J.H. Note on an exact treatment of contingency, goodness of fit and other problems of significance. Biometrika. 1951;38:141–149. [PubMed] [Google Scholar]
- Griffiths-Jones S., Grocock R.J., van Dongen S., Bateman A., Enright A.J. miRBase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Pleasance E.D., Maydan J.S., Hunt-Newbury R., O’Neil N.J., Mah A., Baillie D.L., Marra M.A., Moerman D.G., Jones S.J. Identification and analysis of internal promoters in Caenorhabditis elegans operons. Genome Res. 2007;17:1478–1485. doi: 10.1101/gr.6824707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt-Newbury R., Viveiros R., Johnsen R., Mah A., Anastas D., Fang L., Halfnight E., Lee D., Lin J., Lorch A., et al. High-throughput in vivo analysis of gene expression in Caenorhabditis elegans. PLoS Biol. 2007;5:e237. doi: 10.1371/journal.pbio.0050237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S.M., Lin S.-Y., Slack F.J. The time of appearance of the C. elegans let-7 microRNA is transcriptionally controlled utilizing a temporal regulatory element in its promoter. Dev. Biol. 2003;259:364–379. doi: 10.1016/s0012-1606(03)00202-1. [DOI] [PubMed] [Google Scholar]
- Johnson S.M., Grosshans H., Shingara J., Byrom M., Jarvis R., Cheng A., Labourier E., Reinert K.L., Brown D., Slack F. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Johnston R.J., Hobert O. A microRNA controlling left/right neuronal asymmetry in Caenorhabditis elegans. Nature. 2003;426:845–849. doi: 10.1038/nature02255. [DOI] [PubMed] [Google Scholar]
- Johnston R.J., Jr., Chang S., Etchberger J.F., Ortiz C.O., Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc. Natl. Acad. Sci. 2005;102:12449–12454. doi: 10.1073/pnas.0505530102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Li M., Jones-Rhoades M.W., Lau N.C., Bartel D.P., Rougvie A.E. Regulatory mutations of mir-48, a C. elegans let-7 family microRNA, cause developmental timing defects. Dev. Cell. 2005;9:415–422. doi: 10.1016/j.devcel.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Weinstein E.G., Abdelhakim A., Yekta S., Rhoades M.W., Burge C.B., Bartel D.P. The microRNAs of Caenorhabditis elegans. Genes & Dev. 2003;17:991–1008. doi: 10.1101/gad.1074403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez N.J., Ow M.C., Barrasa M.I., Hammell M., Sequerra R., Doucette-Stamm L., Roth F.P., Ambros V., Walhout A.J.M. A genome-scale microRNA regulatory network in C. elegans reveals composite feedback motifs that provide high information flow. Genes & Dev. 2008;22:2535–2549. doi: 10.1101/gad.1678608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miska E.A., Alvarez-Saavedra E., Abbott A.L., Lau N.C., Hellman A.B., McGonagle S.M., Bartel D.P., Ambros V.R., Horvitz H.R. Most Caenorhabditis elegans microRNAs are individually not essential for development or viability. PLoS Genet. 2007;3:e215. doi: 10.1371/journal.pgen.0030215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obernosterer G., Leuschner P.J., Alenius M., Martinez J. Post-transcriptional regulation of microRNA expression. RNA. 2006;12:1161–1167. doi: 10.1261/rna.2322506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ow M.C., Martinez N.J., Olsen P., Silverman S., Barrasa M.I., Conradt B., Walhout A.J.M., Ambros V.R. The FLYWCH transcription factors FLH-1, FLH-2 and FLH-3 repress embryonic expression of microRNA genes in C. elegans. Genes & Dev. 2008;22:2520–2534. doi: 10.1101/gad.1678808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praitis V., Casey E., Collar D., Austin J. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics. 2001;157:1217–1226. doi: 10.1093/genetics/157.3.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reece-Hoyes J.S., Shingles J., Dupuy D., Grove C.A., Walhout A.J., Vidal M., Hope I.A. Insight into transcription factor gene duplication from Caenorhabditis elegans Promoterome-driven expression patterns. BMC Genomics. 2007;8:27. doi: 10.1186/1471-2164-8-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Rodriguez A., Griffith-Jones S., Ashurst J.L., Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruby J.G., Jan C., Player C., Axtell M.J., Lee W., Nusbaum C., Ge H., Bartel D.P. Large-scale sequencing reveals 21U-RNAs and additional microRNAs and endogenous siRNAs in C. elegans. Cell. 2006;127:1193–1207. doi: 10.1016/j.cell.2006.10.040. [DOI] [PubMed] [Google Scholar]
- Ruby J.G., Stark A., Johnston W.K., Kellis M., Bartel D.P., Lai E.C. Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res. 2007;17:1850–1864. doi: 10.1101/gr.6597907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schier A.F., Giraldez A.J. MicroRNA function and mechanism: Insights from zebrafish. Cold Spring Harb. Symp. Quant. Biol. 2006;71:195–203. doi: 10.1101/sqb.2006.71.055. [DOI] [PubMed] [Google Scholar]
- Simon D.J., Madison J.M., Conery A.L., Thompson-Peer K.L., Soskis M., Ruvkun G.B., Kaplan J.M., Kim J.K. The microRNA miR-1 regulates a MEF-2-dependent retrograde signal at neuromuscular junctions. Cell. 2008;133:891–902. doi: 10.1016/j.cell.2008.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol N.S., Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes & Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefani G., Slack F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008;9:219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- Thomson J.M., Newman M., Parker J.S., Morin-Kensicki E.M., Wright T., Hammond S.M. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes & Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V., Barrasa M.I., Hidalgo C., Babon J.A.B., Sequerra R., Doucette-Stam L., Barabasi A.L., Walhout A.J.M. Transcription factor modularity in a gene-centered C. elegans core neuronal protein-DNA interaction network. Genome Res. 2007a;17:1061–1071. doi: 10.1101/gr.6148107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeirssen V., Deplancke B., Barrasa M.I., Reece-Hoyes J.S., Arda H.E., Grove C.A., Martinez N.J., Sequerra R., Doucette-Stamm L., Brent M., et al. Matrix and Steiner-triple-system smart pooling assays for high-performance transcription regulatory network mapping. Nat. Methods. 2007b;4:659–664. doi: 10.1038/nmeth1063. [DOI] [PubMed] [Google Scholar]
- Viswanathan S.R., Daley G.Q., Gregory R.I. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walhout A.J.M. Unraveling transcription regulatory networks by protein-DNA and protein–protein interaction mapping. Genome Res. 2006;16:1445–1454. doi: 10.1101/gr.5321506. [DOI] [PubMed] [Google Scholar]
- Walhout A.J.M., Temple G.F., Brasch M.A., Hartley J.L., Lorson M.A., van den Heuvel S., Vidal M. GATEWAY recombinational cloning: Application to the cloning of large numbers of open reading frames or ORFeomes. Methods Enzymol. 2000;328:575–592. doi: 10.1016/s0076-6879(00)28419-x. [DOI] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Williams B.D., Schrank B., Huynh C., Shownkeen R., Waterston R.H. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics. 1992;131:609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood W.B. The nematode Caenorhabditis elegans. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1988. [Google Scholar]
- Wulczyn F.G., Smirnova L., Rybak A., Brandt C., Kwidzinski E., Ninnemann O., Strehle M., Seiler A., Schumacher S., Nitsch R. Post-transcriptional regulation of the let-7 microRNA during neural cell specification. FASEB J. 2007;21:415–426. doi: 10.1096/fj.06-6130com. [DOI] [PubMed] [Google Scholar]
- Xu H., Wang X., Du Z., Li N. Identification of microRNAs from different tissues of chicken embryo and adult chicken. FEBS Lett. 2006;508:3610–3616. doi: 10.1016/j.febslet.2006.05.044. [DOI] [PubMed] [Google Scholar]
- Yoo A.S., Greenwald I. LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans. Science. 2005;310:1330–1333. doi: 10.1126/science.1119481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Ransom J.F., Li A., Vedantham V., von Drehle M., Muth A.N., Tsuchihashi T., McManus M.T., Schwartz R.J., Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]