Abstract
Previous to this work, typical genes were thought to move from one position to another infrequently. On the contrary, we now estimate that between one-fourth and three-fourths of the genes in Arabidopsis transposed in the Brassicales. We used the CoGe comparative genomics system to perform and visualize multiple orthologous chromosomal alignments. Using this tool, we found large differences between different categories of genes. Ten of the gene families examined, including genes in most transcription factor families, exhibited a median frequency of 5% transposed genes. In contrast, other gene families were composed largely of transposed genes: NB-LRR disease-resistance genes, genes encoding MADS-box and B3 transcription factors, and genes encoding F-box proteins. A unique method involving transposition-rich regions of genome allowed us to obtain an indirect estimate of the positional stability of the average gene. The observed differences between gene families raise important questions concerning the causes and consequences of gene transposition.
One of the most striking results that comes from comparing related genomes is the prevalence of collinear runs of genes. Broadly speaking, even distantly related species within the same family have roughly the same gene content in roughly the same order (Gale and Devos 1998; Bennetzen 2007). However, order is readily detected and it is easy to overlook exceptions to that order. These exceptions are the subject of this work.
It is important here to distinguish between collinearity, which is a direct and empirical comparison of gene order, and synteny, which is an inference about a common ancestral gene order shared between two or more chromosomal regions. In the absence of collinearity, synteny can be difficult to infer. There are several reasons for this. Most importantly, plant lineages have often undergone repeated tetraploidies and/or large segmental duplications. Such large-scale duplications are eventually reduced back to near that of the pre-tetraploid in terms of gene content and chromosome number by a mutational process called fractionation. However, the resulting genome is scrambled due to deletions, translocations, and inversions (Bowers et al. 2003; Yogeeswaran et al. 2005; Thomas et al. 2006). These rearrangements and fractionations disrupt or even eliminate collinearity, but synteny can usually be deduced by comparison to outgroup genomes.
The second reason that synteny can be difficult to measure involves gene detectability. In some cases, genes or families of genes may evolve by base substitution so rapidly that they cannot be detected in outgroups. We call such genes or gene families “rapidly diverged,” but the term “lineage-specific genes” has also been used (Lespinet et al. 2002). Also undetectable are newly originated genes (Bosch et al. 2007; Zhou et al. 2008). Neither of these classes of genes can be measured for synteny or a lack thereof. Thus, if a gene seems to have moved from an ancestral chromosomal position to a new position, we require that the newly positioned gene must be detectable somewhere in the outgroup genome. Only detectable genes are designated as transposed in our analysis.
Finally, there are genes and families of genes whose distribution among related species is patchy. These gene families can be detected in some outgroup genomes but not others; a given family may have gone extinct in particular lineages but is nonetheless ancient. This behavior has been explained by combinations of high gene birth-and-death coupled with strong purifying selection (Nei 1992; Nei et al. 2000). The most dramatic examples of this are transposons; they only survive to the extent that active elements can move to new positions within a genome and individual transposon lineages are often lost in particular clades. This results in very high birth-and-death and a near absence of synteny (Brookfield 1986; Petrov et al. 1996; Marino-Ramirez et al. 2005).
Our purpose here is to test genes and gene families for movement from an ancestral chromosomal position to a new position beginning with the origin of the order Brassicales and along the lineage leading to modern Arabidopsis thaliana (At). We measure movement by finding genes in At that are flanked closely by neighbor genes that are adjacent in the Brassicales outgroup genome. We call this the “flanking gene method.”
The flanking gene method does not work for all chromosomal positions and it is important to recognize its limitations. First, the methodology depends on adequate sequences from outgroup genomes. If sequences flanking a given gene are absent or incomplete in an outgroup, or if an inversion could have inserted into unsequenced DNA in the region, the movement of that gene cannot be evaluated for transposition. Second, we depend on local synteny between At and orthologous outgroup chromosomes. If the gene in question is in a region showing no synteny (Fortna et al. 2004; Gordon et al. 2007; Kurahashi et al. 2007) the gene cannot be evaluated using the flanking gene method.
At has the best-annotated plant genome and is the preferred species with which to begin an analysis of the evolution of gene chromosomal position. The most recent analyses of intragenomic collinearity within At inferred two sequential tetraploidies (designated α and β), although the timing and exact gene contents of these two events differed markedly when estimated by two independent research groups (Bowers et al. 2003; Maere et al. 2005). Most of the genes that had been duplicated as a consequence of tetraploidy were subsequently removed by fractionation. Those duplicates that were not fractionated enriched the At genome for genes involved in complex regulatory interactions (Blanc and Wolfe 2004; Seoighe and Gehring 2004; Birchler et al. 2005; Maere et al. 2005; Freeling and Thomas 2006). According to the Gene Balance Hypothesis (for review, see Birchler et al. 2005), such genes are retained following tetraploidy events because they are dose sensitive. The net effect of this process is to drive regulatory, and probably morphological, complexity upward (Freeling and Thomas 2006). Biases in gene family expansion or contraction—for any reason—have profound evolutionary consequences involving drives and directions (Freeling 2008). We suggest that the same may be true for biases in a gene’s propensity to transpose.
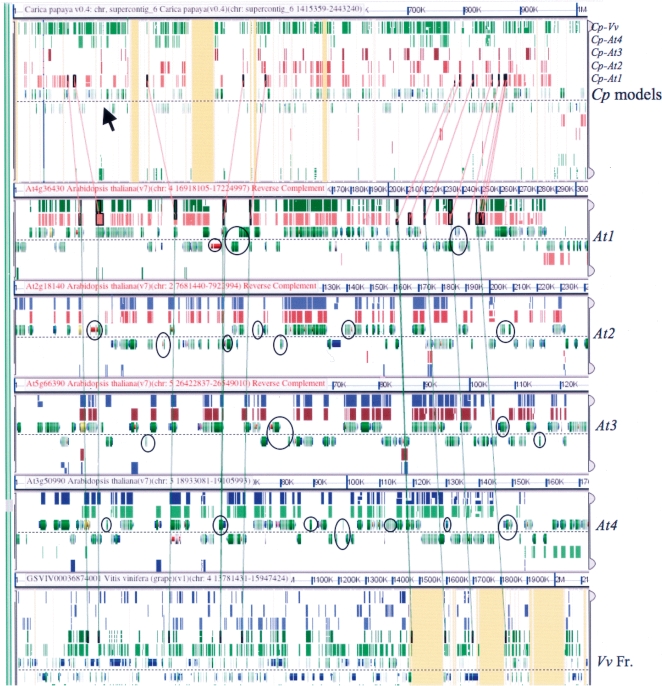
The power of comparative genomics is enhanced by using proper outgroups. Fortunately, two excellent comparators for Arabidopsis research have been recently sequenced. A 3× papaya (Carica papaya, or Cp) genome was released recently (Ming et al. 2008). Although only 75% of the sequence is represented in this assembly, the authors estimate that >90% of the gene content is represented. This genome is a particularly important outgroup for At because it has not undergone a tetraploidy in its lineage for over 100 MY and is a basal Brassicales, the order that also includes At. It is clear that Cp diverged from At before either of the two most recent At lineage tetraploidies occurred, so any one Cp chromosomal segment is often represented as four different segments in At, each one of which, because of fractionation, contains only a subset of the genes in the Cp segment. In order to distinguish between gain versus loss, we use a second outgroup. The grape (Vitis vinifera, or Vv) genome (Jaillon et al. 2007) is also recently sequenced and, like Cp, is devoid of obvious whole-genome duplications subsequent to the radiation of the rosids. Figure 1 illustrates this 1Cp:1Vv:4At relationship. It portrays a GEvo (Lyons and Freeling 2008) graphic representation of a BLASTZ alignment output, where all sequences are compared with Cp. The CoGe platform for organizing whole genomic data and its GEvo tool for comparison of genomic regions (Lyons and Freeling 2008) has been tailored specifically to support comparisons among rosid genomes (Lyons et al. 2008). High-scoring pairs are represented as colored rectangular BLAST “hits.” Note that nearly every gene present in this region of Cp is present on at least one of the At segments. Genes present in these At segments that are not present in a syntenous region in Cp have been either gained at this position in At, lost in Cp, or are not authentic genes. Returning to Figure 1, At genes that are flanked by syntenic genes in Cp and Vv (identified by the lines) but do not appear to be present in either of these outgroup species at this position, are the subject of this study. Examples of these potentially interesting genes are enclosed in ovals.
Figure 1.
A GEvo graphic of a BLASTZ six-way multiple alignments including one syntenous region from Cp, the four homeologous (orthologous) At regions, and the orthologous Vv segment. Both Cp (top) and Vv (bottom) are references, so a maximum of five high-scoring pairs (HSPs, colored boxes) could be piled above, or, for inversion, below the gene models. If syntenic lines connecting HSPs could be drawn without obscuring the graphic, essential collinearity of all HSPs would be demonstrated. The arrow in the Cp panel marks a Cp gene that is present in the expected syntenic position in Vv, and is present in all four At homeologs, meaning that the gene was retained following both α and β tetraploidies in the At-specific lineage. The other Cp genes tend to be on one, two, or three of the four possible At homeologs, reflecting various patterns of fractionation. At genes that are flanked by ancestral genes but are not hit by BLASTZ in either outgroup in this region are circled. These are candidate transpositions. Unsequenced nucleotides, n’s, are marked orange. Continue research at http://tinyurl.com/23lybd.
There are a variety of ways that single genes are known to have moved. One way is via transposon-mediated transduplication, a process in which portions of genes are captured by transposons such as MULEs (Jiang et al. 2004; Juretic et al. 2005; Lisch 2005) or helitrons (Morgante et al. 2005). There are thousands of examples of transduplication in rice and maize. In addition to transduplication, there are three other ways that genes are known experimentally to have transposed singly or in small groups: (1) Excision and reinsertion, mediated by two flanking transposons (Tonzetich et al. 1990); (2) reverse transcription of a pre-existing mRNA and retro-transposition (retroposition) of the intronless copy to a new location (Neufeld et al. 1991); or (3) intrachromosomal recombination among locally duplicated genes or genes flanking repetitive sequences (Yi and Charlesworth 2000).
Although there is little evidence against the common occurrence of single gene transposition in plants, the possibility is rarely mentioned. There is an exception (Fischer et al. 1995): Studies mapping MADS-box genes in maize concluded that many of them acted like transposons. With this exception, the possibility of transposition can come up when disease-resistance (especially NB-LRR) genes (Jones and Dangl 2006) are involved, but the word “transposition” is avoided. Leister (2004) called transpositions “ectopic duplications,” and did not exclude them from explanations of the positions of singlets and clusters of NB-LRR genes in all plants, as had others (Baumgarten et al. 2003). A recent study (Ameline-Torregrosa et al. 2008) called transpositions “ectopic translocations” and inferred a significant number of them among the disease-resistance genes of the legume Medicago truncatula. NB-LRR genes are certainly diverse within Arabidopsis thaliana, Columbia (Meyers et al. 1998), and some of them are particularly polymorphic in their LRR regions (Bakker et al. 2006; Shen et al. 2006; Borevitz et al. 2007), as judged from resequencing in wild accessions. There is at least one suggestion that plant disease-resistance gene clusters might generate diversity under extreme stress (Friedman and Baker 2007). Here we present evidence that NB-LRR genes, and many others, are particularly prone to have become transposed. In contrast, genes in other gene families, like those encoding most sorts of transcription factors, tend to stay in an ancestral chromosomal position.
Results
Our goal was to determine the frequency with which genes were relocated or transposed into the At lineage genome subsequent to its divergence from Cp. In order to do this, it was necessary to identify genes in At that are flanked by syntenic genes in Cp, but that are not themselves present at that syntenic position in Cp. Ultimately, we examine flanked genes using BLASTN or TBLASTX, and display the results, as illustrated in Figures 1 and 2. We then calculate the resulting “not ancestral” frequency for each of several gene families. If the gene is detectable in the outgroups (not rapidly diverging or high birth-and-death), then we infer that the gene transposed into Arabidopsis. By “transposition,” we do not imply a specific mechanism, only that these genes were apparently mobilized and inserted at some point subsequent to the divergence of At and Cp without disrupting ancestral flanking markers.
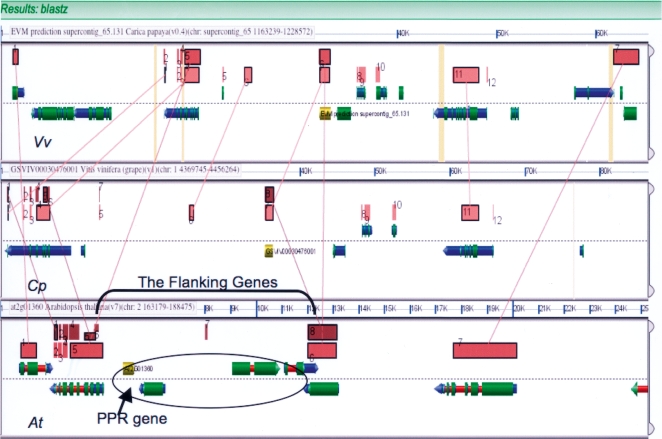
Figure 2.
A GEvo screenshot of a complete flanking gene analysis of a cluster of four potentially nonsyntenic Arabidopsis genes (enclosed in the oval) using both Cp (top) and Vv (middle) outgroups. In GEvo, when an HSP is clicked with the cursor, a line-of-synteny is drawn. For example, the orange HSPs and lines show clearly the one-to-one collinearity of the Cp and Vv outgroups, the most direct evidence for synteny. Note how the two flanking genes (indicated) in At each have hits to orthologous (syntenic) genes in both Cp (light brown) and Vv (dark brown). There are no runs of n’s between the flanking genes in either outgroup that could account for a missing gene. These four genes, including the PPR gene, were each transposed into this position in the At lineage.
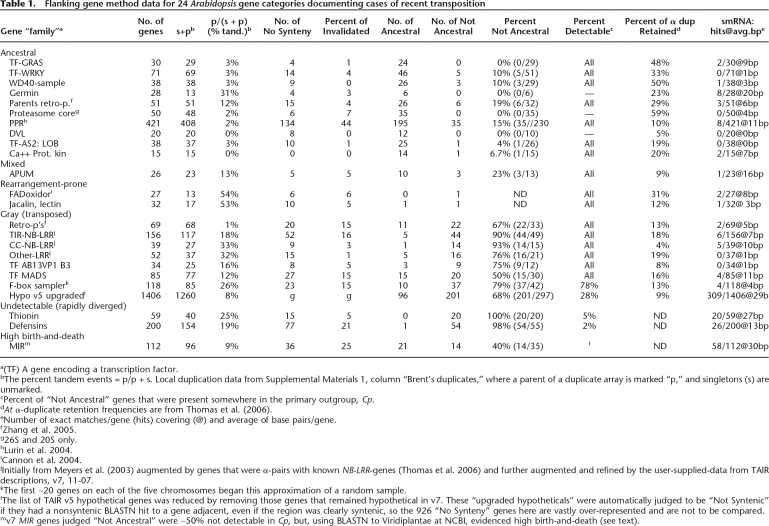
In order to accurately determine the frequency of transposition for each family examined, we used a series of protocols and controls, detailed below. Results are summarized in numbers in Table 1, words in Table 2, and cataloged in Supplemental Information 1.
Table 1.
Flanking gene method data for 24 Arabidopsis gene categories documenting cases of recent transposition
a(TF) A gene encoding a transcription factor.
bThe percent tandem events = p/p + s. Local duplication data from Supplemental Materials 1, column “Brent’s duplicates,” where a parent of a duplicate array is marked “p,” and singletons (s) are unmarked.
cPercent of “Not Ancestral” genes that were present somewhere in the primary outgroup, Cp.
dAt α-duplicate retention frequencies are from Thomas et al. (2006).
eNumber of exact matches/gene (hits) covering (@) and average of base pairs/gene.
g26S and 20S only.
jInitially from Meyers et al. (2003) augmented by genes that were α-pairs with known NB-LRR-genes (Thomas et al. 2006) and further augmented and refined by the user-supplied-data from TAIR descriptions, v7, 11-07.
kThe first ∼20 genes on each of the five chromosomes began this approximation of a random sample.
lThe list of TAIR v5 hypothetical genes was reduced by removing those genes that remained hypothetical in v7. These “upgraded hypotheticals” were automatically judged to be “Not Syntenic” if they had a nonsyntenic BLASTN hit to a gene adjacent, even if the region was clearly syntenic, so the 926 “No Synteny” genes here are vastly over-represented and are not to be compared.
mv7 MIR genes judged “Not Ancestral” were ∼50% not detectable in Cp, but, using BLASTN to Viridiplantae at NCBI, evidenced high birth-and-death (see text).
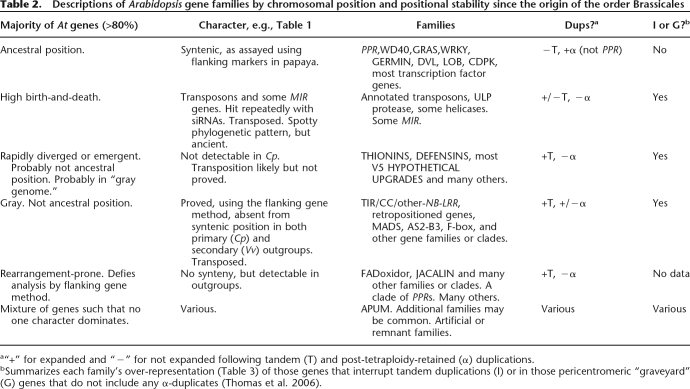
Table 2.
Descriptions of Arabidopsis gene families by chromosomal position and positional stability since the origin of the order Brassicales
a“+” for expanded and “−” for not expanded following tandem (T) and post-tetraploidy-retained (α) duplications.
bSummarizes each family’s over-representation (Table 3) of those genes that interrupt tandem duplications (I) or in those pericentromeric “graveyard” (G) genes that do not include any α-duplicates (Thomas et al. 2006).
Protocol 1: Minimizing the Atv7 genome
There are 31,762 annotated genes in the v7 TAIR Arabidopsis genome. For the purposes of counting positions (loci), we removed annotated transposons as well as ULP protease genes that are hitchhiking within transposons, and we condensed local duplications to one arbitrary gene-space identifier (Methods; details of our gene list explained in header of column S, Supplementary Information 1). The minimized Atv7 genome comprises 26,646 loci.
Protocol 2: Defining the “graveyard,” abbreviated “G”
There are 6326 At loci that have been noted as members of “α-pairs,” defined as pairs of loci that are retained from the most recent paleotetraploidy (Bowers et al. 2003; Thomas et al. 2006). Over 80% of the Arabidopsis genome can be homeologously paired using these α-pairs. Following this tetraploidy, most duplicate (homeologous) genes were fractionated from one or the other, but not both, homeologs, leaving ∼25% of the genome as α-pairs. It is interesting to note that both transposable elements and pseudogenes are relatively rare in these α-regions of the At genome, suggesting that, particularly in relatively small genomes such as At, neutral, redundant, and/or deleterious genes are actively removed from these regions. In contrast, non-α parts of the genome are rich in both transposons and pseudogenes and correlate roughly with cytologically and biochemically defined heterochromatin. We call these unpaired regions “the graveyard,” or G, because they house a significant excess of pseudogenes (715, or 37.5% of G-genes), defined as genes that do not make typical proteins and tend to be covered by small RNA exact matches (121-bp/average G-pseudogene; Supplemental Information 1). It is interesting to note that while plant transposons preferentially transpose into low-copy regions (Dietrich et al. 2002; Pan et al. 2005; Piffanelli et al. 2007), it’s the graveyards that end up enriched for these elements. The excess of pseudogenes and transposons in the graveyard suggest that this portion of the genome exhibits a reduced rate of DNA deletion, perhaps as a consequence of reduced recombination, a characteristic feature of pericentromeric regions (Tanksley et al. 1992). Instead, it appears that genes in the graveyard are disappearing by sequence randomization, which is often called the “pseudogene pathway.” This makes the graveyard particularly valuable with respect to the detection of transposed genes. If transposed typical genes, as with transposons and pseudogenes, were less likely to be lost in the graveyard via deletion mechansims, then we would expect to find a significant enrichment of all transposed genes in the graveyard without much regard to selection.
A complete graveyard would include the five centromeric regions (Meinke et al. 2003) as well as several smaller regions (Thomas et al. 2006). There were 4039 TAIRv7 genes in the five pericentromeric (Meinke et al. 2003) graveyards, as defined by the absence of α-pairs. This number drops to 2194 when the many transposons are removed and when local gene duplicates are condensed. The total gene compliment in this pericentromeric graveyard constitutes 8.4% of the minimized genome.
Protocol 3: Defining “interrupter gene,” I
A second potentially useful region within the At genome lies within local (usually tandem) arrays of duplicated genes. If a gene interrupts a tandem array of genes, we designate it an “interrupter” (or I) gene. I-gene content is particularly informative because it is reasonable to hypothesize that most I-genes were inserted within a local array after the array existed. If an I gene came from outside the array on a short inversion, for example, one of the flanks would invert. However, interrupter genes are not duplicates of nearby genes, strongly suggesting that they are legitimate insertions.
There are 3088 locally duplicated At genes in 1953 arrays (see Methods). Removing transposons, there are 1773 duplicated genes, yielding a tandem duplication frequency of 6.8% in the minimized genome. Within these arrays, we detected 937 I-genes. These genes were rarely duplicates themselves (Supplemental Information I). As expected, and as is the case for G-genes, a significant fraction (24.7%) of I-genes are transposons. After transposons were removed, there were 704 I-genes remaining, or 2.65% of the minimized genome. Therefore, a gene in an “average or typical” gene family would be positioned as an I-gene 2.6% of the time if they were randomly distributed. I-gene pseudogenes were targeted by small RNAs to about the same extent as G-gene pseudogenes (136 bp/average pseudogene, Supplemental Information 1); this is about half of that expected of annotated transposons.
Protocol 4: Defining nonsyntenic regions
Some gene families included genes that were preferentially positioned in “rearrangement-prone” regions of chromosomes, genes that were not flanked by syntenic markers. This lack of synteny was determined by scanning columns of most-homologous chromosomal/supercontig positions in a spreadsheet (Supplemental Information 2) and confirmed using SynMap (Methods). Genes located in rearrangement-prone regions could not be subjected to the flanking gene method, so we could not test for transposition directly. Interestingly, some gene families were significantly over-represented in these nonsyntenic regions. Genes encoding JACALIN lectins and FAD oxidoreductases (FADoxidor) are exemplary, with 46% (6/13) and 59% (10/17) of the genes, respectively, located in regions without synteny (Table 1). Almost all of the rest were invalidated for technical reasons, which can be caused by rearrangements as well. Only 10% of jacalin and FADoxidors could be analyzed, even though all were detectable in Cp. These two gene families—and probably hundreds of other, similar families that we have not analyzed—also have very high local (tandem) duplication frequencies. Genes positioned in regions we found to be in rearrangement-prone regions were denoted “No Synteny” in Table 1 (and “No” in Supplemental Information 1). Even within essentially ancestral families, both individuals and clades of “No Synteny” genes can occur, comprising meaningful data.
Control 1: Genes that are known to have transposed
In addition to regions of the genome that are more likely to contain transposed copies of genes, we wanted to examine classes of genes that are known to have transposed in order to compare them with gene families that we hypothesized to have transposed.
Zhang et al. (2005) annotated 69 retropositioned genes in the Arabidopsis genome, with one being a tandem duplicate. All are intronless compared with their 51 probable genes of origin. The “parents” of retroposons, a small but “normal” set of genes, are not present at their ancestral position (“not ancestral” in Table 1) 19% of the time (6/32), while the retropositioned genes themselves are not present at their ancestral position 67% of the time (22/33). Each of these presumptively transposed genes were detectable in other regions of papaya, with TBLASTN scores >45 and almost always >100. As with all families or groups of genes analyzed, genes in rearrangement-prone regions (“No Synteny” in Table 1), and genes invalidated for technical reasons (“Invalidated” in Table 1) were noted, and these were not subjected to the flanking gene test. The ancestral retropositions must have occurred before At and Cp lineages branched; those few that were retained as α-duplicates must have transposed before the α-tetraploidy. So, the flanking gene method detects known transposed genes.
Most of the 92 At genes annotated “ULP Proteases” are actually MULE transposons carrying an ULP protease gene somewhere between the transposons’ inverted repeats (Hoen et al. 2006). Data from this special gene transposition class provides an additional control. Not surprisingly, ULP proteases are repositioned preferentially as single insertions within tandem arrays (I) and near centromeres (G) at 8% (7/92) and 35% (38/92), respectively. Since only 2.65% of the minimized genome is I, 8% is an over-representation (P < 0.001 by χ2). Similarly, since 8% of v7 minimized genes are in these graveyards, ULP proteases are clearly over-represented in this part of the genome as well (P < 0.001 by χ2). As with other transposons, At ULP protease genes tend to be hit multiply and covered by exact matches to small RNA sequences. For ULP genes, the mean is 10 unique hits/gene covering an average of 184 bp of ULP-protease CDS; this is more than 40-fold greater than coverage of the average ancestral gene. This “hitchhiking” mechanism for single gene transposition is explicable, and, for that reason only, trivial to this discussion, but the over-representation in I and G regions is an additional control.
Control 2: Rapidly diverged genes are undetectable
Some individual Arabidopsis genes and some families of genes have no believable BLAST hits anywhere in the papaya genome, so that finding no hit in the flanked ancestral space is meaningless. “Believable” for TBLASTN (protein query to a genome translated in six frames) is a score of ≥45, which approximates an E-value of ≤0.001. The 59 thionin genes and 200 defensin (DEFL) genes make fine examples (Table 1). Of the total of 74 nonancestral members of these gene families that were identified (Table 1), only two had believable TBLASTN hits anywhere in papaya. Nor did they have hits in genomes of any species more distantly related than Cp. These families are particularly prone to tandem duplication (25% and 20%, respectively) and have a large number of genes located in “rearrangement-prone” regions (“No Synteny” in Table 1). They are also significantly over-represented among interrupter (I) and near-centromeres (G) genes, as will be shown. The average rapidly evolving gene was covered by overlapping siRNA exact matches at about 20 bp, which is greater than that for the average transcription factor (4 bp) but far less than for transposons.
MIR genes represented a special challenge to detectability. Most MIR genes were either positioned in rearrangement-prone regions or were invalidated (INV) (Table 1). Of the 35 we could analyze by the flanking gene method, 14 were Not Ancestral. Of these, only two were detectable anywhere in Cp. However, even when a MIR gene was not detectable in Cp, it was present (6/14 or 43% of the time) in other dicots more distantly related to At than papaya; this is a signature of high birth-and-death lineages as described in the Introduction. If the frequency of birth-and-death is high enough, new insertions are certainly expected, as with authentic transposons, but the flanking gene method is not useful.
Different categories of genes transpose at different frequencies
Two major results emerged from our flanking gene analyses of the positional stability of several gene families. First, an unexpectedly large percent of genes in almost all detectable families have transposed since the At–Cp split, and second, the distribution of the frequency of transposition varies greatly (0%–93%) among different families of genes. Thus, while families of transcription factors such as GRAS and WRKY are largely positionally static in the rosids, genes in families such as MADS-box, F-box, B3, and NB-LRR are usually found at transposed chromosomal positions (Table 1). The characteristics of each gene family are summarized in Table 2. For convenience, we refer to gene families that are detectable and >80% retained at the same position in At and Cp as “ancestral” families. Families that are >50% transposed are referred to as “gray” families, because their degree of positional stability lies in the gray interval between ancestral genes and authentic transposons.
The ancestral families were picked because we judged them likely to be representative of genes encoding essential phenotypes under continuous purifying selection, which is the case for most transcription factor families or subfamilies. Representatives of ancestral families (named after their protein products) we included in our analyses are: GRAS, WRKY, AS2-LOB, GERMIN, PROTEASOME CORE, DVL, WD40, CaPROTEIN KINASE, PPR, and the parent genes to At retropositioned genes. Among these gene families, the frequency of transposed genes (“Not Ancestral,” Table 1) ranged from 0% to 19%, with a 5% median. In general, these families have average frequencies of retention following the most recent At paleotetraploidy (median 26% compared with a 24% average) and have low frequencies of tandem duplication events (3% compared with a 6.8% average). Also important are the exceptional families. Genes encoding GERMINs, for example, are prone to high levels of tandem duplication, but, exceptionally, are also highly ancestral; however, GERMINS are retained just below average post-paleotetraploidy (23%). Ancestral gene siRNA targeting was low, with an average coverage of 6 bp/gene. Most importantly, all of the 51 nonancestral genes identified by our flanking gene method were detectable in the outgroup.
Data from the Cp outgroup was supported by the more distant Vv outgroup 95% of the time. Examination of the 51 syntenous regions of Vv allowed us to estimate how many, if any, of the putative transpositions into the At lineage actually reflected loss from the Cp outgroup. A total of 39 of these putatively transposed genes from ancestral families could be analyzed unambiguously by the flanking gene method in Vv (grape has had its share of inversions and also has unsequenced regions). Of 39 analyses, 37 found the flanking region but did not find the gene in question. In two cases, the gene was present in the syntenic Vv region. We conclude that the “Not Ancestral” designation based on the At–Cp data organized in Table 1 means that the gene was transposed into At 95% of the time, and was lost in the Cp outgroup 5% (2/39) of the time. These data suggest that Cp makes an excellent outgroup for At positional stability research.
Some gene families in At are rarely in the Brassicales ancestral position, if such a position ever existed. These genes behave like slow transposons. Retropositioned genes (as expected), disease-resistance genes (mainly TIR/CC-NB-LRRs), MADS-box, AB13-Vp1-B3a, many F-box genes, and a large group of genes called “Expressed in V6, Hypotheticals in V5” are the founding members of what we now call “the gray genome.”
Two of the gray families require special explanations because their membership is so large. Genes annotated as encoding an F-box protein are numerous, usually occur in local clusters and are rarely retained post-paleotetraploidy. Preliminary examination also indicated that they were often “Not Ancestral.” We sampled ∼20 F-box genes on each of the five At chromosomes in order to obtain the estimate given in Table 1 of 79% “Not Ancestral,” of which 78% were detectable in Cp (that is, 22% were “rapidly diverged”). Another gene category is not a family at all, but a collection of genes that were annotated as “Hypothetical” in v5, but upgraded to “Unknown” or “Expressed” in v6, usually on the basis of cDNA sequence. These 926 genes were so numerous that we made our criterion for “flanking” as rigorous as possible: We only analyzed genes that were flanked perfectly by their adjacent, orthologous genes in At and Cp based on the best BLASTN hit only (Methods). This stringency inflated greatly the number and proportion of those genes showing “No Synteny.” Because we do not want this proportion compared with the “No Synteny” value of any other family, we include this number in Table 1 only as a footnote. Among those 297 upgraded hypotheticals that were analyzed by the flanking gene method, 68% were “Not Ancestral,” of which 28% were detectable in Cp somewhere, and the remainder were rapidly diverged (like defensins). So, this category is predominantly rapidly diverged, but is also gray. The unverified upgraded V5 category could, in theory, have included unannotated transposons, which would provide a ready explanation for transposability, but this “unannotated transposon” hypothesis will be disproved in the last section of these Results.
Detailed positional analysis of the ancestral At PPR gene family
We wanted to examine one ancestral family in detail. We chose the 364 analyzable genes encoding PPR proteins because they are rarely duplicated in tandem (2.2%) and have a relatively high proportion (15%; 35/230) of transposition for an ancestral family, making statistics meaningful. PPR proteins are thought to function in organellar biogenesis and, sometimes, in fertility and fertility restoration (Saha et al. 2007). Additionally, previous work on these genes indicated that “at least some” PPR genes were nomadic within the family Brassicacea because they did not share syntenic positions between At and its close relative Brassica rapis (Geddy and Brown 2007). We were able to analyze about two-thirds (230) of the PPR genes by using the flanking gene method. Based on analyses using both Cp and Vv outgroups, the 15% of genes that are not ancestral are, with one exception, new transpositions into the At lineage, not losses of the gene in the Cp primary outgroup. Building phylogenetic trees with PPR genes is complicated (Howell et al. 2007); we are fortunate that the Carrington laboratory had completed a tree including almost all PPR genes, and made it available to us. After decorating this tree with our data, it is clear that proved PPR insertions seem to occur at random over the tree (Supplemental Information 3), demonstrating that there is no particular clade that is more likely to have proved transposed members. There is, however, a clade within this tree that cannot be analyzed for being new insertions because they are preferentially located in nonsyntenic (rearrangement-prone) chromosomal regions; these are the nomadic genes described previously (Geddy and Brown 2007). So, even though our flanking gene method could not test it, this detectable clade is likely to be transposed. Interestingly, this clade, which is present in two regions of chromosome 1, is also targeted by a specific set of tasiRNAs derived from MIR173 (Howell et al. 2007).
Among transposed PPR genes, the median number of genes inserted at a new position, counting the initial PPR gene, is one, although insertions including several genes do occur. Figure 2 shows a PPR gene transposed adjacent to three additional transposed genes. Three cases of multiple transposed genes were examined in detail using the CoGeBlast tool and our GEvo Viewer (Methods); these three tiny syntenic groups do not exist in Cp or Vv.
Genes in gene families that are significantly over- and under-represented in G and I chromosomal space
As evidenced in our control experiments, both I-space and G-space are enriched for pseudogenes and transposable elements, perhaps because genes in theses regions are lost via a pseudogene pathway, rather than via deletion. Given this, we predict that any family that has a larger proportion of members that have transposed into new chromosomal positions (based on our flanking gene method) will also have a higher proportion of I-genes and G-genes. This reasoning allows us to estimate indirectly the positional stability of the average At gene whether or not it is detectable in an outgroup.
For the calculations that follow, we use the 26,646 genes/gene-spaces of the minimized TAIR v7 genome. Recall our previous results: Were a gene’s position random, the typical minimized gene has an 8.4% chance of being G and a 2.6% chance of being I.
As a control for gray gene families, we used a collection of genes that that are largely (95%) positioned in the ancestral order. Based on previous work, we focused on genes encoding transcription factors. Of the 1975 genes encoding transcription factors in the 2005 edition of the DATF database, http://datf.cbi.pku.edu.cn/, many of the families had fewer than 20 genes, and five of the smaller families were exceptional in that they tended to be locally duplicated at above the average frequency of about 6%, and to be retained as pairs post-α tetraploidy well below the average frequency of 24%. These families were ARF, AS2 A, and HSF, as well as the two gray families B3 and MADS. Removing these outlier and unpopulated DATF families left a core of 1265 genes encoding transcription factors that we now call “95% ancestral.” Table 3 compares our I and G representation data for genes in Gray families, rapidly diverged families and upgraded hypotheticals (v5 hypotheticals to v7 expressed) to expectations derived from this “95% ancestral” control gene group. These expectations are: I = 1.1%, G = 1.9%. These control values are used to derive the expected numbers of Table 3. The legend of Table 3 indicates exactly which families constitute these three experimental groups of genes.
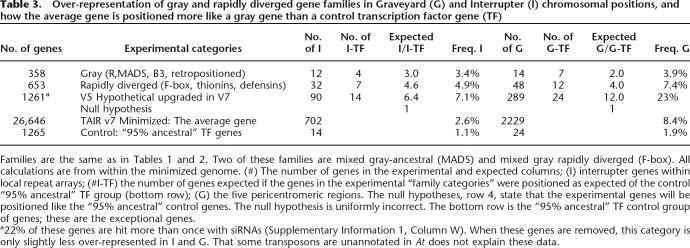
Table 3.
Over-representation of gray and rapidly diverged gene families in Graveyard (G) and Interrupter (I) chromosomal positions, and how the average gene is positioned more like a gray gene than a control transcription factor gene (TF)
Families are the same as in Tables 1 and 2. Two of these families are mixed gray-ancestral (MADS) and mixed gray rapidly diverged (F-box). All calculations are from within the minimized genome. (#) The number of genes in the experimental and expected columns; (I) interrupter genes within local repeat arrays; (#I-TF) the number of genes expected if the genes in the experimental “family categories” were positioned as expected of the control “95% ancestral” TF group (bottom row); (G) the five pericentromeric regions. The null hypotheses, row 4, state that the experimental genes will be positioned like the “95% ancestral” control genes. The null hypothesis is uniformly incorrect. The bottom row is the “95% ancestral” TF control group of genes; these are the exceptional genes.
a22% of these genes are hit more than once with siRNAs (Supplementary Information 1, Column W). When these genes are removed, this category is only slightly less over-represented in I and G. That some transposons are unannotated in At does not explain these data.
The data of Table 3 indicate that all three categories of genes—gray, rapidly diverged and upgraded hypotheticals—are significantly (P < 0.05 by χ2) over-represented in I and G space as compared with expectations derived by the “95% ancestral” control group of genes encoding transcription factors. Gray genes are significantly over-represented in both I and G space. This supports our hypothesis that the increased frequency of insertion of gray genes into syntenic regions is a reflection of overall higher frequency of insertional activity or retention. However, if all genes transposed at about the same rate, and the ones we see transposed are those not removed by purifying selection in I and G, then our control group of genes encoding transcription factors (“95% ancestral”) must have been continuously removed from most regions of chromosome, including I and G space, by some mechanism other than the point mutation/pseudogene pathway, since pseudogenes are vastly over-represented in I and G space.
Most unexpected is the comparative data for the average minimized At gene (Table 3, last row). None of the experimental categories of genes are dramatically atypical except for the huge 23% G representation of upgraded hypothetical genes. What is atypical is the data for the “95% ancestral” control genes themselves, as if the control is far more positionally ancestral than is the average gene. In general, the data summarized in Table 3 paints the average At gene dark gray, behaving more like a slow transposon and less like a static ancestral locus since the split between At and Cp.
Discussion
We have shown that a significant fraction of genes in Arabidopsis (At) have changed location, or transposed, at some point since the divergence of this species from another species in the order Brassicales, papaya (Cp). Although our sampling was certainly biased by the families we chose to examine, the 10 ancestral gene families we chose showed a median transposition frequency of ∼5% (Table 1). This indicates that even in the most positionally conserved families, a substantial fraction of genes have transposed since the At–Cp split. Most of these transpositions are single gene events.
Although a comprehensive analysis of all At genes using the flanking gene method is beyond the scope of this work, we can calculate an approximate minimum transposition frequency. A total of 2.6% of the 26,646 minimized At genes are Interrupter (I) genes, and are certainly insertions, and 8.4% are Graveyard (G) genes. We suggest that many of the G-genes are in fact transposed, given the observation that gene families that are over-represented among I-genes are invariably over-represented among G-genes (Table 3). The five graveyards we sum to be “G” are the largest, but not the only pseudogene/transposon-rich regions in the genome, so we add another 1% of the genes for each chromosome, or 5%. Of the remaining genome, at least 2000 genes are transposed genes in those gray families we have identified. That leaves 20,231 in potentially ancestral families (>80% ancestral), and these average 5% transposed, or at least 1012 genes that escaped from ancestral positions. Based on these estimates, we conservatively estimate that at least 7231 genes, or 27% of the minimized At genome, transposed within the Arabidopsis lineage after the Cp–At divergence. This is a conservative estimate because our data suggests that the average gene is far more likely to be transposed than are members of highly ancestral families of genes (Table 3).
We have also shown that transposition is nonrandom with respect to gene function. Some large gene families are far more likely to have members that have transposed than others. A total of 79% of the F-box genes sampled, for instance, are located in the nonancestral position. Since 78% of these are detectable in the outgroup, most F-box genes are newly transposed. On the other hand, none of the 30 GRAS transcription factor genes were transposed (Table 1). These data clearly indicate a bias with respect the frequency of transposed genes within a given family.
Even though the vast majority of genes we have examined have no defined function, a trend can be clearly discerned. Genes encoding products known to interact specifically with rapidly changing biotic and abiotic extrinsic factors are far more likely to have transposed than genes encoding products involved in relatively stable processes. NB-LRR gene products must rapidly change to meet new pathogenic challenges (Jones and Dangl 2006), as must many plant defensins, which have been implicated in defense against fungal pathogens (Thomma et al. 2002). Similarly, it has been suggested that rapidly evolving and positively selected F-box genes are part of an innate immune system whose function is to degrade various bacterial and viral toxic proteins (Thomas 2006). Although MADS box genes are not required for pathogen response, many of them are involved in floral organ identity and boundary determination (Nam et al. 2003), or flowering time (Dennis and Peacock 2007). Given expected fluctuations in specific pollinator species, flower shape and pollen availability over the at least 50 million year lifespan of the Brassicales, MADS-box genes may well have been particularly exposed to fluctuating selection. In contrast, we hypothesize that ancestral gene families, defined in Tables 1 and 2 as families with >80% genes in the ancestral position, often encode proteins involved in pathways that seem likely to be under continuous selection. Transcription factors that carry out developmental programs, for example, must bind downstream promoter elements that may evolve over time, but these TF genes are unlikely candidates for fluctuating selection. Similarly, PPR genes (85% ancestral) are largely targeted to the mitochondria or chloroplasts and are thought to be involved in RNA processing in these organelles (Small and Peeters 2000; Lurin et al. 2004; Saha et al. 2007). It seems unlikely that these genes would be subjected to strong fluctuating selection (although exceptional PPR genes have been implicated in cytoplasmic incompatibilities/fertility). In summary, it makes sense that the gray and rapidly diverged gene families analyzed so far correlate with selective environments that fluctuate, while ancestral genes may encode functions that are under continuous selection.
Why such dramatic differences in transposition frequency between different categories of genes? There are at least two reasonable and mutually nonexclusive explanations for the transposition bias we observe. The first is that all genes are competent to transpose, and all genes do so at an equal frequency over evolutionary time, but that selection removes transposed copies of some genes or classes of genes more efficiently than others. The other possibility is that some genes or gene families are intrinsically more prone to transpose because a propensity for transposition has a long-term benefit to the organism and has been embedded into the gene’s sequence or into features of its preferred chromosomal positions. The first hypothesis relies purely on direct selection on gene function; the second on what has been called “second order selection,” which can be thought of as a mechanism that increases the propensity to produce new alleles via any distinctive mechanism (Pennisi 1998; Tenaillon et al. 2001). We will briefly discuss each of these possibilities.
The purely selectionist hypothesis suggests that negative (purifying) selection removes transposed copies of members of some gene families preferentially, and/or positive selection favors transposed copies of other gene families. Evidence in favor of negative selection comes from much previous work on the preferential retention of genes encoding transcription factors or other interactive gene products following paleotetraploidy in Arabidopsis, as predicted by the Gene Balance Hypothesis (Birchler and Veitia 2007). In this case, selection operates to prevent imbalanced gene product dosage. For the same reason, selection would be expected to disfavor both tandem duplications and duplicative transpositions of those same genes favored following paleotetraploidy (Freeling and Thomas 2006; Freeling 2008), and this is what we have observed in almost all of the 24 gene categories of Table 1. GRAS transcription factors, for instance have a transposition frequency of 0% (0/29), a tandem duplication frequency of 3% (1/29), and an α duplicate retention frequency of 48%. In contrast, F-box genes have a transposition frequency of 79% (78% detectable), a high tandem duplication frequency of 26%, and low α-duplicate retention frequency of 13%. Cannon and coworkers (Cannon et al. 2004) point out the extremes of this potentially reciprocal relationship, and evidence for it has grown (Freeling and Thomas 2006; Freeling 2008). These observations support the hypothesis that the lack of transposed copies of some genes is a consequence of negative selection against unbalanced gene product levels.
Our data also supports the idea that positive selection favors transposition of genes, in particular gene families. Here, we assume that transposed copies of genes are more likely to be expressed in novel ways, which could be selectively advantageous if the factors with which they interact are constantly changing. If positive selection is strong but acts only periodically, the result could be a constantly shifting population of genes within a family whose positions would vary over time. Clear examples of this are the NB-LRR genes (91% transposed). As new or altered pathogens are encountered, old copies of these genes would become selectively neutral, or even deleterious, and would then be lost. New copies with altered function would continually appear and be selected for. In this case, the intrinsic (and blindly) dynamic nature of the genome may have been harnessed by selection to produce useful variation in the form of a population of genes at various positions. This process is no different in principle from selection on random mutations in coding sequences; the “mutation” in this case being a change in location rather than a change in coding sequence. In contrast, gene families whose basic characteristics are not selected to change, or where negative selection efficiently removes duplicated copies of genes, would not be expected to exhibit a high apparent rate of transposition. Thus, both negative and positive selection could play a role in the frequency that transposed copies of genes are retained or lost.
So what of second order selection? Is it possible that members of some gene families are more prone to transpose than others? Although equivocal, we do have supporting evidence. Our flanking gene method examines regions of the genome that are relatively stable in order to provide evidence of new insertions. However, there are two other independent measures of insertional activity. If a gene is inserted into a tandem array, it is almost certainly transposed. Consistent with this, roughly a quarter of the sequences inserted into tandem arrays are transposons. Among the other genes more likely to have inserted into these arrays are members of gene families, gray gene families, that we had determined to transpose more frequently using the flanking gene method (Table 3). A second independent method for measuring transposition frequency involves examination of the graveyards. These regions of the genome have an excess of transposable elements. Unlike euchromatic regions of the genome, they are also enriched for pseudogenes, suggesting that insertions of all kinds are inefficiently removed from these graveyards. As in the case of interrupter genes, graveyard genes are significantly enriched for gene families that we had determined to transpose more frequently (last row of Table 3). This is true of both potentially functional genes as well as pseudogenes.
Together, these data suggest that the average gene encoding, for instance, a transcription factor (the 95% ancestral control families), transposes into interrupter gene space at a threefold lower rate than the average gene in a gray family and five- to sixfold less than a gene in a “rapidly diverged” family. There are two reasons that this argument is equivocal. First, selection against dosage changes or ectopic expression could act to remove insertions into tandem arrays or the graveyards in the same way that it acts to remove insertions into more stable regions of the genome. However, the presence of large numbers of inactive pseudogenes in the graveyards suggests that insertions into these heterochromatic regions are often lost not by deletion, but by the slow accumulation of point mutations. The absence of significant numbers of transcription-factor pseudogenes in the graveyards is most easily explained if these genes simply transpose into these regions of the genome at a reduced rate. Second, biased gene-loss mechanisms could also account for these data. If our 95% ancestral transcription factor genes were removed from I and G space preferentially by a special deletion mechanism, and did not last long enough to be removed by the pseudogene pathway that obviously operates in I and G space, then these data are also explained. Were this contrived gene-loss mechanism real, this too would be biased and, therefore, of great interest.
Most or all plant lineages have survived repeated paleotetraploidies, and each of these events is a saltation that must have greatly reduced diversity. The Arabidopsis genome has evidence of four paleotetraploidies within its genome, and possibly more that happened too long ago to see clearly. Population-level estimates of NB-LRR gene polymorphism and selection do find diversity (Bakker et al. 2006; Shen et al. 2006; Borevitz et al. 2007), but it is not yet clear whether or not this diversity was necessary in the wild for a plant to have survived pathogens. If the specificity of the plant immune system is actually held at the population level in a great bank of alleles and plus–minus polymorphisms—for example, very many different NB-LRR sequences—then, the early descendants of any polyploid were certainly immune deficient. These tiny populations, beginning with one plant, had no bank of diverse alleles; an allotetraploid has at most four alleles for any one locus. These post-tetraploid populations not only survived, they repeatedly founded major clades of plant life. It is reasonable to entertain the possibility that NB-LRR and similar genes evolved, by second order selection, mechanisms to accelerate diversity.
No matter what mechanism explains why some genes end up transposed more than other genes, gene movement characterizes a large or even the major portion of the Brassicales branch of the Arabidopsis lineage. The new information conferred upon the inserted genes due to their new chromosomal locations has probably had a significant impact on all evolutionary trends and possibilities. But what is the extent of this impact? The ∼3000 species in the family Brassicaceae (crucifers) inhabit all continents except Antarctica, and species exist in virtually all sorts of environments including all edaphic (soil type) environments, in the extremes of these environments known to support plant life, and exhibit most every adaptation known to be possible in plants (Bressan et al. 2001). We need to know whether the large proportion of gray genes characterizing the Arabidopsis and presumably other Brassicaceae genomes describes all other particularly widespread and adaptable clades. Perhaps the gray genome expands along with fractionation of ancient polyploids. Alternatively, perhaps all plants have well-expanded gray genomes like Arabidopsis. We are at such a primitive level of knowledge involving gene expansion by transposition, we can’t begin to estimate how useful our findings are likely to be for understanding trends in eukaryotic evolution. It does seem likely, however, that gene mobility enhances evolvability, especially in intermittently hostile environments. Whether that mobility is a global phenomenon, or one specialized to produce higher rates of movement of particular classes of genes, the intrinsically dynamic nature of genomes has certainly contributed to the mode and tempo of evolution.
Methods
At data acquisition and display
The data acquired or used in this study are listed in an Excel spreadsheet (Supplemental Information 1). Column C of this spreadsheet is one model of every gene in the Arabidopsis genome, version 7, downloaded from The Arabidopsis Information Resource (TAIR) along with a TAIR gene description. Papaya sequence (3×) is version 4 from the Hawaii Papaya Genome Project (Ming et al. 2008). A grape genome was obtained from the French consortium (Jaillon et al. 2007). Both of these shotgun sequence assemblies have regions that are either unassembled or unsequenced; these runs of “n” are color-coded orange in our alignment viewer because our methods require that we keep track of “holes” in the sequence, since missing genes might not be missing, but located in unsequenced chromosome. Small RNAs were downloaded from the Arabidopsis Small RNA Project on 12-2007, (http://asrp.cgrb.oregonstate.edu/db/download.html; 218,928 smRNA sequences). Using these to find exact matches to CDS or, if necessary, mRNA sequence, we report a number of independent hits and total base pair of subject covered by small RNA sequences.
Local repeats and Interrupter genes (I) in At
In order to minimize the genome (Results), to calculate local repeat frequencies (Table 1), and to locate Interrupter genes within tandem arrays (Table 3), we wrote a Perl script that began with the lowest numbered gene on each chromosome and searched for a nearby homologous gene without skipping more than three adjacent, ascending genes. Homology was quantified using BLASTN; the query was a CDS sequence—or RNA if the gene had no CDS—and the subject was the next four CDS/RNAs on the chromosome, using BLASTN at E < 0.0001. We then demanded >50% HSP coverage of the feature. If one of these four genes was hit, skipped genes were labeled “I,” the lowest numbered gene was tagged “parent” arbitrarily, the duplicates were tagged with the locus name of the parent, and the lowest numbered duplicate became the next query in hopes of expanding the array. These tags are noted in Supplemental Information 1 under “Brent’s duplicates.” Most of these arrays are tandem repeats, but “reverse tandems” happen, and a reverse tandem segmental inversion could—in theory—bring unwanted genes into the Interrupter set. We decided to not demand tandem repeats because of the prevalence of single gene inversions (some are visible in Fig. 1). Therefore, our Interrupter gene set is expected to contain some noise.
Establishing candidate At genes for the flanking gene method, and assigning “No Synteny”
The first step in our analysis was to determine whether or not an At gene was located in a syntenic region of At–Cp aligned chromosome. If no syntenic region could be found, then “No Synteny” was recorded in Supplemental Information 1 and on Table 1. To make this syntenic assessment, we constructed a list of all TAIR v7 genes and their descriptions. For each was listed, in separate columns, the best BLASTN hit to Cp at an E < 0.001, its start position on a papaya supercontig, and each of the top five TBLASTN hits, E-values, scores, and start positions more significant than E = 0.1 (which is well within noise). Finally, each gene has, indicated in the last column, a link to GEvo that automatically anchors our alignment viewer on the BLASTN hit. By keeping this list (Supplemental Information 2) sorted on At genes in their actual chromosomal order, it was possible to see whether any particular gene was likely to be (1) present in papaya at the syntenic position, or (2) potentially not in papaya, but surrounded (flanked) by At–Cp orthologs that might provide an accurate definition of the papaya chromosomal region where the At gene might be expected to exist. If the former, “Ancestral” was entered in Table 1 (and “Cp” was entered in Supplemental Information 1). If the latter, we went on to apply the flanking gene method. Even if not flanked by orthologs, sometimes the position of an At gene hit in Cp was at the exact end of a syntenic series of gene positions, thus indicating synteny; for this reason, this initial test for synteny is biased toward finding ancestral genes. To help researchers adverse to “eyeball” methods reproduce our results, we implemented the synteny-finding algorithm DAGchainer (Haas et al. 2004) in an online application called SynMap and now include it in our CoGe suite of genomics databases and tools: http://synteny.cnr.berkeley.edu/CoGe/SynMap.pl. Settings for At–Cp: −g = 200kb; −D = 400kb; −A = 3 generates DAGchainer syntenic pairs as lines of red dots and some reassuring noise. An individual At gene may be found in the graphic using chromosomal position and by mousing over red lines; click any red dot for an anchored At–Cp GEvo alignment. Alternatively, below the SynMap graphic readout are links including “Syntolog File.” Here, every syntenic pair (red dot) between the two genomes is cataloged and each has a GEvo link available by text search.
One gene category used the Supplemental Information 2 prescreen for synteny in an especially rigorous way. The At TAIR v5 hypothetical genes that were upgraded to “expressed” or “unknown” in v7, called “upgraded hypotheticals,” were numerous and mostly not ancestral. We demanded that the nearest upstream and downstream BLASTN hits (E < 0.001) were flanking. This was done in automated fashion. So, even when evidence for synteny was strong, if there was even one intervening nonsyntenic hit within the flanking genes, the gene was called “Not Syntenic,” leading to a comparatively inflated number in this column of Table 1. Therefore, this number in Table 1 is replaced with a footnote to discourage comparisons.
The flanking gene method with a primary and secondary outgroup
We chose one of the GEvo links on a flanking orthologous At–Cp gene very near our gene of interest and examined the regions visually, beginning with a graphic edition of BLASTZ output at our default settings, and choosing the option to color sequences not sequenced (n’s) orange. Figure 2, top and bottom, is a screenshot of such an At–Cp syntenic region surrounding a cluster of four genes, including a PPR gene (the gene under analysis); this cluster of potentially transposed genes is enclosed by the oval in the At panel. The light brown rectangles, BLASTZ At–Cp hits displayed above the Cp (top) and At (bottom) models, show no indication of any of the four genes in the expected Cp region. At this point we rerun the alignment using BLASTN set at an E-value equivalent of a 15/15 exact match—which is just at the noise level—and also TBLASTX (translated protein to translated protein); we found what BLASTZ missed ∼1% of the time. Occasionally, with small genes, we avoided the BLASTN 7-bp nucleation requirement by using Chaos and other alternative alignment algorithms (all available and cited in GEvo.) The light-orange bands in a GEvo graphic denote unsequenced regions of papaya and grape. While there is an unsequenced DNA in the flanked region of Figure 2, it is not big enough to hide any of the test genes, so the genes were denoted “Not Ancestral” in Table 1 (and “new” in Supplemental Information 1). If there had been unsequenced DNA in the flanked region large enough to “hide” half of a test gene at a >5% probability (expert opinion), or if a nearby inversion was judged possibly to have imported such an unsequenced region, the test gene evaluation was terminated, and “Invalidated” was marked in Table 1 (and “INV” was marked in Supplemental Information 1). “Invalidate” is used in this study to indicate invalidation for technical reasons. Each “Not Ancestral” gene was verified for detectability somewhere else in the Cp genome. The E-values/scores of the best TBLASTN hit in Cp was compared with known noise levels of TBLASTN hits in the genome. We set a score of 45—approximating a hit with an E-value of 0.001—as the noise cutoff. Scores above this cutoff were “yes” under the “detectable?” column of Table 1. For MIR genes, BLASTN to Viridiplantae (green plants) at NCBI was performed for each “Not Ancestral” gene. The results were often spotty throughout the plant kingdom, leading to the “high birth-and-death” notation in Tables 1 and 2, as described in the text.
After our At–Cp results were almost complete, we obtained the French grape genome as a second outgroup. We did not use pre-made GEvo links to anchor At:Cp:Vv orthologous chromosomes. Rather, we created our own anchors using a tool in our CoGe platform of comparative genomics databases and tools called “CoGe BLAST”: http://synteny.cnr.berkeley.edu/CoGe/CoGeBlast.pl. For example, the anchor position (yellow exons) of At and Cp was from a pre-made menu of GEvo links, anchor in the middle, grape chromosome derived from CoGe BLAST, where a Cp syntenic group of exons were merged as a BLASTN query to a Vv subject with an E < 0.0001 cutoff. The one to three most likely syntenic Vv regions are made into GEvo links automatically in CoGe BLAST and then evaluated visually. Figure 2 is the result of one such complete At:Cp:Vv analysis. Note that the four-gene region of the “Not Ancestral,” verified PPR gene is not in either Cp nor Vv, although the flanking markers are. Therefore, all four genes are transposed in the Brassicales branch of the At lineage. As should be apparent, the flanking gene method was not automated. We judged that both the choice of candidate syntenous regions and the invalidation by possible inversions nearby were too biologically complicated for automation, but not too complicated to understand if rendered as visual output. The syntenic gene lists and the GEvo multiple sequence alignment viewer and its associated tools in our CoGe platform made it possible for one annotator to generate the gene family data reported here in ∼400 h. A GEvo tutorial designed for the Arabidopsis (rosid) researcher is available in CoGe, and has been summarized (Lyons et al. 2008).
Acknowledgments
We thank all who are part of the multinational Hawaii Papaya Genome Project (http://asgpb.mhpcc.hawaii.edu/papaya/). This research was funded by National Science Foundation research grants DBI-0701871 to M.F. and DBI-0321726 to D.L. Brian C. Thomas was our systems administrator during the early stages of this work. Lakshmi Rapaka provided technical assistance.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.081026.108.
References
- Ameline-Torregrosa C., Wang B.B., O'Bleness M.S., Deshpande S., Zhu H., Roe B., Young N.D., Cannon S.B. Identification and characterization of nucleotide-binding-site leucine-rich-repeat genes in the model plant Medicago truncatula. Plant Physiol. 2008;146:5–21. doi: 10.1104/pp.107.104588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker E.G., Toomajian C., Kreitman M., Bergelson J. A genome-wide survey of R gene polymorphisms in Arabidopsis. Plant Cell. 2006;18:1803–1818. doi: 10.1105/tpc.106.042614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgarten A., Cannon S., Spangler R., May G. Genome-level evolution of resistance genes in Arabidopsis thaliana. Genetics. 2003;165:309–319. doi: 10.1093/genetics/165.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennetzen J.L. Patterns in grass genome evolution. Curr. Opin. Plant Biol. 2007;10:176–181. doi: 10.1016/j.pbi.2007.01.010. [DOI] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A. The gene balance hypothesis: From classical genetics to modern genomics. Plant Cell. 2007;19:395–402. doi: 10.1105/tpc.106.049338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Riddle N.C., Auger D.L., Veitia R.A. Dosage balance in gene regulation: Biological implications. Trends Genet. 2005;21:219–226. doi: 10.1016/j.tig.2005.02.010. [DOI] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz J.O., Hazen S.P., Michael T.P., Morris G.P., Baxter I.R., Hu T.T., Chen H., Werner J.D., Nordborg M., Salt D.E., et al. Genome-wide patterns of single-feature polymorphism in Arabidopsis thaliana. Proc. Natl. Acad. Sci. 2007;104:12057–12062. doi: 10.1073/pnas.0705323104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch N., Caceres M., Cardone M.F., Carreras A., Ballana M.R., Armengol L., Estivill X. Chracterization and evolution of the novel gene family FAM90A in primates originated by multiple duplication and rearrangement events. Hum. Mol. Genet. 2007;16:2572–2582. doi: 10.1093/hmg/ddm209. [DOI] [PubMed] [Google Scholar]
- Bowers J.E., Chapman B.A., Rong J., Paterson A.H. Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature. 2003;422:433–438. doi: 10.1038/nature01521. [DOI] [PubMed] [Google Scholar]
- Bressan R.A., Zhang C., Zhang H., Hasegawa P.M., Bohnert H.J., Zhu J.-K. Lerning from the Arbidopsisexperience. The next gene search pradigm. Plant Physiol. 2001;127:1354–1360. [PMC free article] [PubMed] [Google Scholar]
- Brookfield J.F. The population biology of transposable elements. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1986;312:217–226. doi: 10.1098/rstb.1986.0003. [DOI] [PubMed] [Google Scholar]
- Cannon S.B., Mitra A., Baumgarten A., Young N.D., May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis E.S., Peacock W.J. Epigenetic regulation of flowering. Curr. Opin. Plant Biol. 2007;10:520–527. doi: 10.1016/j.pbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Dietrich C.R., Cui F., Packila M.L., Li J., Ashlock D.A., Nikolau B.J., Schnable P.S. Maize Mu transposons are targeted to the 5′ untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics. 2002;160:697–716. doi: 10.1093/genetics/160.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A., Baum N., Saedler H., Theissen G. Chromosomal mapping of the MADS-box multigene family in Zea mays reveals dispersed distribution of allelic genes as well as transposed copies. Nucleic Acids Res. 1995;23:1901–1911. doi: 10.1093/nar/23.11.1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortna A., Kim Y., MacLaren E., Marshall K., Hahn G., Meltesen L., Brenton M., Hink R., Burgers S., Hernandez-Boussard T., et al. Lineage-specific gene duplication and loss in human and great ape evolution. PLoS Biol. 2004;2:e207. doi: 10.137l/journal.pbio,.0020207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M. The evolutionary position of subfunctionalization, downgraded. Genome Dyn. 2008;4:25–40. doi: 10.1159/000126004. [DOI] [PubMed] [Google Scholar]
- Freeling M., Thomas B.C. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 2006;16:805–814. doi: 10.1101/gr.3681406. [DOI] [PubMed] [Google Scholar]
- Friedman A.R., Baker B.J. The evolution of resistance genes in multi-protein plant resistance systems. Curr. Opin. Genet. Dev. 2007;17:493–499. doi: 10.1016/j.gde.2007.08.014. [DOI] [PubMed] [Google Scholar]
- Gale M.D., Devos K.M. Comparative genetics in the grasses. Proc. Natl. Acad. Sci. 1998;95:1971–1974. doi: 10.1073/pnas.95.5.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geddy R., Brown G.G. Genes encoding pentatricopeptide repeat (PPR) proteins are not conserved in location in plant genomes and may be subject to diversifying selection. BMC Genomics. 2007;8:130. doi: 10.1186/1471-2164-8-130. doi: 10.1186-1471-2164-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon L., Yang S., Tran-Gyamfi M., Baggott D., Christensen M., Hamilton A., Crooijmans R., Groenen M., Lucas S., Ovcharenko I., et al. Comparative analysis of chicken chromosome 28 provides new clues to the evolutionary fragility of gene-rich vertebrate regions. Genome Res. 2007;17:1603–1613. doi: 10.1101/gr.6775107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Delcher A.L., Wortman J.R., Salzberg S.L. DAGchainer: A tool for mining segmental genome duplications and synteny. Bioinformatics. 2004;20:3643–3646. doi: 10.1093/bioinformatics/bth397. [DOI] [PubMed] [Google Scholar]
- Hoen D.R., Park K.C., Elrouby N., Yu Z., Mohabir N., Cowan R.K., Bureau T.E. Transposon-mediated expansion and diversification of a family of ULP-like genes. Mol. Biol. Evol. 2006;23:1254–1268. doi: 10.1093/molbev/msk015. [DOI] [PubMed] [Google Scholar]
- Howell M.D., Fahlgren N., Chapman E.J., Cumbie J.S., Sullivan C.M., Givan S.A., Kasschau K.D., Carrington J.C. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O., Aury J.M., Noel B., Policriti A., Clepet C., Casagrande A., Choisne N., Aubourg S., Vitulo N., Jubin C., et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- Jiang N., Bao Z., Zhang X., Eddy S.R., Wessler S.R. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- Juretic N., Hoen D.R., Huynh M.L., Harrison P.M., Bureau T.E. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 2005;15:1292–1297. doi: 10.1101/gr.4064205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H., Inagaki H., Hosoba E., Kato T., Ohye T., Kogo H., Emanuel B.S. Molecular cloning of a translocation breakpoint hotspot in 22q11. Genome Res. 2007;17:461–469. doi: 10.1101/gr.5769507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance gene. Trends Genet. 2004;20:116–122. doi: 10.1016/j.tig.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Lespinet O., Wolf Y.I., Koonin E.V., Aravind L. The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res. 2002;12:1048–1059. doi: 10.1101/gr.174302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisch D. Pack-MULEs: Theft on a massive scale. BioEssays. 2005;27:353–355. doi: 10.1002/bies.20219. [DOI] [PubMed] [Google Scholar]
- Lurin C., Andres C., Aubourg S., Bellaoui M., Bitton F., Bruyere C., Caboche M., Debast C., Gualberto J., Hoffmann B., et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons E., Freeling M. How to usefully compare homologous plant genes and chromosomes as DNA sequence. Plant J. 2008;53:661–673. doi: 10.1111/j.1365-313X.2007.03326.x. [DOI] [PubMed] [Google Scholar]
- Lyons E., Pedersen B., Kane J., Alam M., Ming R., Tang H., Wang X., Bowers J., Paterson A., Lisch D. Finding and comparing syntenic regions among Arabidopsis and outgroups papaya, poplar and grape: CoGe with Rosids. Plant Physiol. 2008 doi: 10.1104/pp.108.124867. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S., De Bodt S., Raes J., Casneuf T., Van Montagu M., Kuiper M., de Van Peer Y. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino-Ramirez L., Lewis K.C., Landsman D., Jordan I.K. Transposable elements donate lineage-specific regulatory sequences to host genomes. Cytogenet. Genome Res. 2005;110:333–341. doi: 10.1159/000084965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinke D.W., Meinke L.K., Showalter T.C., Schissel A.M., Mueller L.A., Tzafrir I. A sequence-based map of Arabidopsis genes with mutant phenotypes. Plant Physiol. 2003;131:409–418. doi: 10.1104/pp.014134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Shen K.A., Rohani P., Gaut B.S., Michelmore R.W. Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell. 1998;10:1833–1846. doi: 10.1105/tpc.10.11.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers B.C., Lee D.K., Vu T.H., Tej S.S., Edberg S.B., Matvienko M., Tindell L.D. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell. 2003;15:809–834. doi: 10.1105/tpc.009308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming R., Hou S., Feng Y., Yu Q., Dionne-Laporte A., Saw J.H., Senin P., Wang W., Ly B.V., Lewis K.L., et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus) Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgante M., Brunner S., Pea G., Fengler K., Zuccolo A., Rafalski A. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 2005;37:997–1002. doi: 10.1038/ng1615. [DOI] [PubMed] [Google Scholar]
- Nam J., dePamphilis C.W., Ma H., Nei M. Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol. Biol. Evol. 2003;20:1435–1447. doi: 10.1093/molbev/msg152. [DOI] [PubMed] [Google Scholar]
- Nei M. Balanced polymorphism and evolution by the birth and death process in the MHC loci. In: Tsuji K., et al., editors. Proceedings of the 11th histocompatibility workshop and conference. Oxford University Press; Oxford, UK: 1992. pp. 27–38. [Google Scholar]
- Nei M., Rogozin I., Piontkivska H. Purifying selection and birth-and-death evolution in the ubiquitin gene family. Proc. Natl. Acad. Sci. 2000;97:10866–10871. doi: 10.1073/pnas.97.20.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld T.P., Carthew R.W., Rubin G.M. Evolution of gene position: Chromosomal arrangement and sequence comparison of the Drosophila melanogaster and Drosophila virilis sina and Rh4 genes. Proc. Natl. Acad. Sci. 1991;88:10203–10207. doi: 10.1073/pnas.88.22.10203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Li Y., Stein L. Site preferences of insertional mutagenesis agents in Arabidopsis. Plant Physiol. 2005;137:168–175. doi: 10.1104/pp.104.053215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennisi E. How the genome readies itself for evolution. Science. 1998;281:1131–1134. doi: 10.1126/science.281.5380.1131. [DOI] [PubMed] [Google Scholar]
- Petrov D.A., Lozovskaya E.R., Hartl D.L. High intrinsic rate of DNA loss in Drosophila. Nature. 1996;384:346–349. doi: 10.1038/384346a0. [DOI] [PubMed] [Google Scholar]
- Piffanelli P., Droc G., Mieulet D., Lanau N., Bes M., Bourgeois E., Rouviere C., Gavory F., Cruaud C., Ghesquiere A., et al. Large-scale characterization of Tos17 insertion sites in a rice T-DNA mutant library. Plant Mol. Biol. 2007;65:587–601. doi: 10.1007/s11103-007-9222-3. [DOI] [PubMed] [Google Scholar]
- Saha D., Prasad A.M., Srinivasan R. Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol. Biochem. 2007;45:521–534. doi: 10.1016/j.plaphy.2007.03.026. [DOI] [PubMed] [Google Scholar]
- Seoighe C., Gehring C. Genome duplication led to highly selective expansion of the Arabidopsis thaliana proteome. Trends Genet. 2004;20:461–464. doi: 10.1016/j.tig.2004.07.008. [DOI] [PubMed] [Google Scholar]
- Shen J., Araki H., Chen L., Chen J.Q., Tian D. Unique evolutionary mechanism in R-genes under the presence/absence polymorphism in Arabidopsis thaliana. Genetics. 2006;172:1243–1250. doi: 10.1534/genetics.105.047290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small I.D., Peeters N. The PPR motif—A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 2000;25:46–47. doi: 10.1016/s0968-0004(99)01520-0. [DOI] [PubMed] [Google Scholar]
- Tanksley S.D., Ganal M.W., Prince J.P., de Vicente M.C., Bonierbale M.W., Broun P., Fulton T.M., Giovannoni J.J., Grandillo S., Martin G.B., et al. High density molecular linkage maps of the tomato and potato genomes. Genetics. 1992;132:1141–1160. doi: 10.1093/genetics/132.4.1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenaillon O., Taddei F., Radmian M., Matic I. Second-order selection in bacterial evolution: Selection acting on mutation and recombination rates in the course of adaptation. Res. Microbiol. 2001;152:11–16. doi: 10.1016/s0923-2508(00)01163-3. [DOI] [PubMed] [Google Scholar]
- Thomas J.H. Adaptive evolution in two large families of ubiquitin-ligase adapters in nematodes and plants. Genome Res. 2006;16:1017–1030. doi: 10.1101/gr.5089806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.C., Pedersen B., Freeling M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006;16:934–946. doi: 10.1101/gr.4708406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma B.P., Cammue B.P., Thevissen K. Plant defensins. Planta. 2002;216:193–202. doi: 10.1007/s00425-002-0902-6. [DOI] [PubMed] [Google Scholar]
- Tonzetich J., Hayashi S., Grigliatti T.A. Conservation of the sites of tRNA loci among the linkage groups of several Drosophila species. J. Mol. Evol. 1990;30:182–188. doi: 10.1007/BF02099944. [DOI] [PubMed] [Google Scholar]
- Yi S., Charlesworth B. A selective sweep associated with a recent gene transposition in Drosophilamiranda. Genetics. 2000;156:1753–1763. doi: 10.1093/genetics/156.4.1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogeeswaran K., Frary A., York T.L., Amenta A., Lesser A.H., Nasrallah J.B., Tanksley S.D., Nasrallah M.E. Comparative genome analyses of Arabidopsis spp.: Inferring chromosomal rearrangement events in the evolutionary history of A. thaliana. Genome Res. 2005;15:505–515. doi: 10.1101/gr.3436305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wu Y., Liu Y., Han B. Computational identification of 69 retroposons in Arabidopsis. Plant Physiol. 2005;138:935–948. doi: 10.1104/pp.105.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Zhang G., Zhang Y., Xu S., Zhao R., Zhan Z., Xin L., Ding Y., Yang S., Wang W. On the origin of new genes in Drosophila. Genome Res. 2008;18:1446–1455. doi: 10.1101/gr.076588.108. [DOI] [PMC free article] [PubMed] [Google Scholar]