Abstract
Loss of the CDK inhibitor p27KIP1 is widely linked with poor prognosis in human cancer. In Wnt10b-expressing mammary tumors, levels of p27KIP1 were extremely low; conversely, Wnt10b-null mammary cells expressed high levels of this protein, suggesting Wnt-dependent regulation of p27KIP1. Interestingly we found that Wnt-induced turnover of p27KIP1 was independent from classical SCFSKP2-mediated degradation in both mouse and human cells. Instead, turnover required Cullin 4A and Cullin 4B, components of an alternative E3 ubiquitin ligase induced in response to active Wnt signaling. We found that CUL4A was a novel Wnt target gene in both mouse and human cells and that CUL4A physically interacted with p27KIP1 in Wnt-responding cells. We further demonstrated that both Cul4A and Cul4B were required for Wnt-induced p27KIP1 degradation and S-phase progression. CUL4A and CUL4B are therefore components of a conserved Wnt-induced proteasome targeting (WIPT) complex that regulates p27KIP1 levels and cell cycle progression in mammalian cells.
Keywords: Basal-like breast cancer, reprogramming, Wnt-induced proteasome targeting (WIPT), mammary progenitor cells, mammary stem cell (MSC), Cdkn1b
Activation of the Wnt/β-catenin pathway is observed in a number of adult stem cell niches and is a common feature of HER2/ERB-B2-negative breast cancers, where nuclear β-catenin is observed in >50% of cases (Cowin et al. 2005; Nusse 2005). Active Wnt signaling is also a feature of colorectal cancer, and a difficult-to-treat subgroup of breast cancers known as basal carcinomas. These cancers are thought to result from proliferation of cells within the stem cell niche. Gene expression profiling of human and animal breast cancers indicate that the oncogenic properties of Wnt signaling directly expand the number of mammary progenitor cells and thus represent a unique challenge for therapeutic intervention (Perou et al. 2000; Li et al. 2003; Sørlie et al. 2003; Cowin et al. 2005).
During development, Wnt10b is the earliest marker of the definitive mammary lineage (Veltmaat et al. 2004). Elevated expression of Wnt10b results in mammary cancer formation in mice, and has been documented in human breast carcinoma cell lines and primary tumor tissue (Bui et al. 1997; Lane and Leder 1997; Sørlie et al. 2003). Multiple cellular phenotypes are observed in both Wnt1-induced (Li et al. 2003) and Wnt10b transgene (Wnt10bTG)-induced mammary tumors (Lane and Leder 1997), a feature consistent with a basal origin. Furthermore, Wnt transgene-induced mammary tumors express high levels of nuclear β-catenin, stem cell antigen 1 (SCA1), keratin 6a (KRT-6A), and other basal/stem cell markers. In the present study, we show that Wnt10b driven tumors express low levels of p27KIP1 (Cdkn1b) and present evidence for post-translational control by a novel Wnt-induced proteasome targeting pathway (WIPT) that is active in Wnt-responding S-phase cells. This represents the first report of direct reprogramming of proteasome functions in Wnt-responding cells.
p27KIP1 is an inhibitor of cyclin-dependent kinases in mammalian cells, and turnover of p27KIP1 is required for S-phase entry in a number of cell types. Previous work has shown that p27KIP1 is regulated by both transcriptional and post-translational mechanisms; however, the major regulatory pathway in cycling cells involves proteasomal degradation of the p27KIP1 protein (Pagano et al. 1995). In quiescent or growth factor-depleted cell lines, p27KIP1 protein is elevated. As cells are released into G1, p27KIP1 is degraded, reaching their lowest levels in late G1 and S (Coats et al. 1996).
The best-understood pathway for p27KIP1 turnover requires polyubiquitination by the SCFSKP2-E3 ligase active in late G1 and early S phase. This process requires phosphorylation of p27KIP1 at Thr187 (T187) by cyclin A/E-CDK2. Phospho-T187 then binds the SKP2 component of SCFSKP2-E3 ubiquitin ligase complex. Subsequent polyubiquitination of p27KIP1 by SCFSKP2 targets the protein for destruction by the 26S proteasome (Sheaff et al. 1997; Carrano et al. 1999; Malek et al. 2001). Substitution of Thr187 (T187) by alanine (T187A) ablates SCFSKP2-mediated p27KIP1 degradation (Montagnoli et al. 1999). In the absence of SKP2, p27KIP1 is stabilized in S-phase cells, and SKP2-deficient cells fail to progress normally through G2/M (Nakayama et al. 2000, 2004).
In the present study, we provide evidence for Wnt-induced turnover of p27KIP1 in mouse and human cells. This turnover is distinguished from previously characterized pathways in that it is independent of CRM1-mediated nuclear export. Furthermore, it does not require T187 phosphorylation or recruitment of the SCFSKP2 complex. Wnt-induced turnover of p27KIP1 was found in mouse mammary tissue in vivo, mammary epithelial cell lines, human fibroblasts, and human breast cancer cells. We found that components of the CUL4-E3 ligase (Cul4A and Cul4B) are expressed in Wnt10b-responding mammary tumors and that both CUL4A and CUL4B are required for Wnt-induced S-phase progression. Cul4A represents a previously unrecognized Wnt target gene, transcriptionally activated in response to recruitment of β-catenin to the Cul4A promoter. Additionally, CUL4A is found in complex with p27KIP1 in Wnt10b-expressing cells. Therefore, CUL4A and CUL4B represent components of a WIPT pathway that may contribute to p27KIP1 turnover in development and disease. Molecular targeting of this pathway may provide a novel therapeutic strategy for inhibiting stem cell-like cancers and for control of Wnt pathway-mediated cell proliferation.
Results
Wnt10b expression correlates inversely with p27KIP1 protein levels in mammary tumors and in mammary epithelial tissue
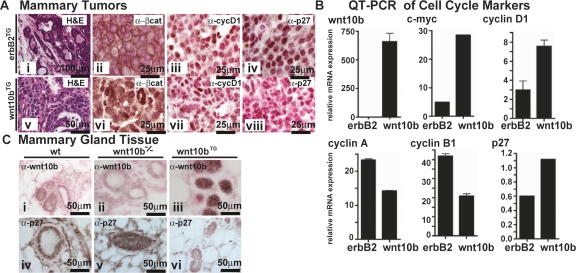
In both male and female transgenic mice, expression of Wnt10b under the regulation of MMTV-LTR (Wnt10bTG) results in precocious ductal formation and the development of stochastic adenocarcinomas (breast cancer) in mammary tissue (Lane and Leder 1997). Wnt10b also promotes the expression of mammary epithelial stem cell markers (Supplemental Figs. S1, S2) and enhances mammary epithelial proliferation. To understand the mechanism by which Wnt10b promotes accelerated cell cycle progression, normal mammary gland and mammary tumors were analyzed for cell cycle markers using immunohistochemistry (IHC) and expression profiling (Supplemental Fig. S2). Wnt10bTG mammary tumors (Lane and Leder 1997) were compared with those from MMTV-ErbB2/c-neu (ErbB2TG) females (Muller et al. 1988). Wnt10bTG tumors demonstrated nuclear localized β-catenin, elevated cyclinD1, and complete loss of p27KIP1 protein (Fig. 1A). In contrast, β-catenin localizes to cellular adhesion junctions in ErbB2TG-expressing tumors and p27KIP1 was expressed at intermediate levels. These results indicate that Wnt10b signals via the canonical Wnt pathway in mammary tissue and suggest that loss of p27KIP1 could be a previously unrecognized target of Wnt-mediated transformation.
Figure 1.
p27KIP1 protein, but not mRNA, is reduced in Wnt10b transgenic mammary tissue and elevated in Wnt10b-null mammary tissue. (A) Five-micrometer paraffin sections from Wnt10bTG (panels i–iv) and MMTV-ErbB2 transgenic (ErbB2TG) (panels v–viii) mammary tumor tissue were stained with H&E (panels i,v) to show morphological features. In parallel sections, protein expression was characterized by IHC (brown): β-catenin (panels ii,vi), cyclin D1 (panels iii,vii), and p27KIP1 (panels iv,viii). Sections were counterstained with nuclear fast red (red) to reveal nuclei. (B) RNA expression profiling (see Supplemental Fig. S2) and QPCR expression analysis was carried out on cDNA from similar tumors. Transcripts were quantified, and values were normalized to 18S rRNA. (C) p27KIP1 (panels i–iii) and WNT10b (panels iv–vi) protein expression was examined in 7-wk-old mammary tissue from wild-type (wt), Wnt10b-null (Wnt10b−/−), and Wnt10bTG mice. Five-micrometer paraffin sections were stained as described above. Bars indicate scale in micrometers.
As a prognostic marker of human breast cancer, nuclear β-catenin correlates significantly with cyclin D1 expression and associates independently with poor prognosis (Lin et al. 2000) and poor survival (Barnes and Gillett 1998). In Wnt10bTG mammary tumors, p27KIP1 protein expression was strongly reduced when compared with ErbB2TG tissue (Fig. 1A, panel iv). In fact, loss of p27KIP1 could be used to reliably distinguish Wnt10TG tumors from ErbB2TG tumors by IHC analysis (Fig. 1A, cf. panels iv and viii). This result indicated that low p27KIP1 (p27KIP1-low) was specifically associated with Wnt10b-mediated tumorgenesis in mammary glands. Furthermore, the levels of cyclin D1 protein were not significantly different for both Wnt10TG tumors and ErbB2TG tumors (Fig. 1A, cf. panels iii and vii). Taken together these data define a mouse mammary tumor model that correlates with nuclear β-catenin, high cyclin D1, and loss of p27KIP1 (ErbB2-negative, nuclear β-catenin-positive, cyclin D1-high, p27-low). This spectrum of markers is a common profile in human mammary and colorectal carcinogenesis and is associated with very poor prognosis (Catzavelos et al. 1997; Barnes and Gillett 1998; Cariou et al. 1998).
The expression of a series of cell cycle regulators was then examined by quantitative PCR (QPCR). As predicted, Wnt10bTG tumors showed elevated expression of several known Wnt target genes, including c-Myc and Cyclin D1 (Fig. 1B). In contrast, ErbB2TG tumors display low levels of c-Myc mRNA. Cyclins A and B1 transcripts were more robustly induced in ErbB2TG tumors (Fig. 1B). Paradoxically, the levels of p27Kip1 mRNA were elevated in the Wnt10bTG tumors when compared with ErbB2TG tumors, suggesting that the decrease in p27KIP1 protein observed by IHC was due to post-translational events. Next we set out to examine the relationship between Wnt10b and p27KIP1 protein in nontransformed mammary tissue.
Wnt10b−/− mice develop normally but have fewer mammary ducts in young adult females (G. Miranda and T.F. Lane, in prep.). In the absence of Wnt10b, we observed stronger nuclear p27KIP1 accumulation in ductal epithelial cells when compared with wild-type controls (Fig. 1C, panel v vs. panel iv). Conversely, Wnt10bTG-derived mammary epithelial cells (precancer) show no detectable p27KIP1 protein expression (Fig. 1C,panel vi). Graded expression of WNT10B protein was also confirmed in these sections using anti-WNT10B antisera (Fig. 1C, panels i–iii). The results of IHC on developing mammary tissue provide evidence that p27KIP1 expression is inversely correlated with WNT10B protein concentrations in vivo, and indicate that Wnt signaling may directly impact p27KIP1 expression during normal development of mammary tissue.
Wnt10b induces proteasome-dependent S-phase turnover of p27KIP1; however, this turnover does not coincide with induction of SKP2 protein in synchronized cells and is independent of CRM1 mediated nuclear export machinery
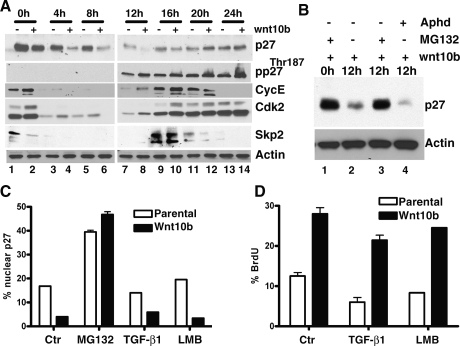
Maximal p27KIP1 protein levels occur in quiescent or prereplicative phases in mammalian cells (Coats et al. 1996). As the cell cycle progresses, p27KIP1 blocks entry into S phase by maintaining Cyclin/CDK complexes in an inactive state. The mechanism of p27KIP1 degradation previously described for cells transitioning to the S phase is dependent on the SCFSKP2 E3 ubiquitin ligase complex and requires phosphorylation of p27KIP1 on Thr187 by Cyclin A/E–CDK2. Protein concentrations of SKP2 and p27KIP1 have been shown previously to be inversely proportional in S-phase cells (Sheaff et al. 1997; Carrano et al. 1999; Nakayama et al. 2000; Malek et al. 2001). To determine the role of SCFSKP2 in Wnt10b-mediated p27KIP1 degradation, we established stable clones of NMuMG (NMG) mammary epithelial cells expressing either pcDNA3 vector alone (NMG) or pcDNA3-Wnt10b (NMG-Wnt10b). NMG and NMG-Wnt10b cells were synchronized in early G1 by maintenance at 100% confluency for 2–3 d. Cells were then released and harvested at various times for protein analysis. Wnt10b-mediated p27KIP1 degradation was maximal at 12 h, the time at which Cyclin E and CDK2 re-expression was first observed (Fig. 2A, lane 7 vs. lane 8). This correlated with enhanced phosphorylation of p27Thr187 but did not coincide with increased SKP2 protein expression levels (Fig. 2A, lanes 7,8). SKP2 protein was absent in 12-h cells (Fig. 2A, lanes 7,8), and maximal SKP2 expression was detected 4 h later (at 16 h) (Fig. 2A, lanes 9,10), down-regulated at 20 h (Fig. 2A, lanes 11,12), and not detected 4 h later as the cells exit mitosis. Paradoxically, high SKP2 was not followed by loss of p27KIP1, suggesting that WNT10B could mediate p27KIP1 turnover through a mechanism other than SCFSKP2 complex in S-phase cells.
Figure 2.
Mammary cell lines expressing Wnt10b show accelerated turnover of p27KIP1 protein in late G1/early S phase, prior to the induction of SKP2. Turnover is blocked by inhibitors of proteasome function but not by inhibitors of CRM-1 nuclear export. NMG and NMG stably expressing Wnt10b (NMG-Wnt10b) cells were synchronized in G1 phase and released by replating. Cell extracts were prepared at the indicated times. (A) Lysates were analyzed by immunoblotting and probed with the indicated antisera. (B) NMG-Wnt10b cells were synchronized at G1 phase. Following release, cells were cultured in the presence or absence of the proteasome inhibitor MG132 (10 μM) or the S-phase blocker aphidicolin (1 μg/mL). Twelve-hour samples were immunoblotted as described above. (C) Nuclear p27KIP1 levels were measured in NMG vector (parental) and NMG-Wnt10b cells following exposure to proteasome blocker MG132 (10 μM), TGFβ (2.5 ng/mL), and LMB (1 μg/mL). Cells were synchronized at G1 phase and released for 12 h, and p27 immunofluorescence was measured quantitatively using an LSC laser cytometer. (D) BrdU incorporation into cells from C was detected by immunofluorescence, and signal was measured quantitatively using an LSC laser cytometer. Experiments were conducted as triplicates. Error bars represent SDM.
The mechanism of Wnt-induced p27KIP1 degradation was examined in NMG-Wnt10b cell lines synchronized at G1 then released for 12 h, as described above. In NMG-Wnt10b, p27KIP1 is high at 0 h but is maximally reduced by 12 h post-release (Fig. 2B, lanes 1,2). In the presence of an inhibitor of proteasomal degradation (10 μM MG132), p27KIP1 turnover was blocked (Fig. 2B, lane 3). In contrast, the levels of p27Kip1 mRNA were consistently elevated in the Wnt10b-expressing cells (Supplemental Fig. S3A), suggesting that loss of p27 protein is due to post-translational events, as previously suggested from the analysis of primary tumor samples from Wnt10bTG mice. To confirm that this degradation was occurring at the G1–S-phase boundary, cells were also treated with aphidicolin (1 μg/mL), an inhibitor of DNA polymerase α, commonly used to block cells in early S phase. Wnt-induced p27KIP1 turnover was not inhibited by aphidicolin treatment (Fig. 2B, lane 4), indicating that S-phase passage was not a prerequisite for degradation.
We quantified the amount of nuclear p27KIP1 in both NMG and NMG-Wnt10b cells 12 h after release in the presence or absence of the proteasomal inhibitor MG132, TGF-β1 or leptomycin B (LMB) (Fig. 2C). LMB has been shown to elevate p27KIP1 by specifically blocking CRM1 function and preventing export of nuclear p27KIP1 to cytosolic proteasomes (Ishida et al. 2002; Connor et al. 2003). MG132 was able to prevent turnover of p27KIP1 in both parental and Wnt10b-expressing cell lines and increased p27-levels approximately ninefold over untreated cells. In contrast, TGF-β1 and LMB were unable to restore p27 in the Wnt-expressing cells. Additionally, we find that LMB treatment of control NMG cells elevates p27KIP1 and that TGF-β1 had no effect on the restoration of p27KIP1.
To determine the effect of either CRM1 inhibition or TGF-β1 treatment on cell cycle progression, we monitored 5-bromo-2-deoxyuridine (BrdU) uptake in treated cells (Fig. 2D). We found that both TGF-β1-mediated and LMB-mediated cell cycle arrest were relieved in Wnt10b-expressing cells. Taken together these data indicate that Wnt10b-mediated turnover of p27KIP1 occurs at the G1–S boundary. Turnover is blocked by standard inhibitors of the 26S proteasome but is insensitive to CRM1 blockade or TGF-β1.
Mutation of p27KIP1 Thr187 to Ala187 does not block Wnt10b-mediated degradation
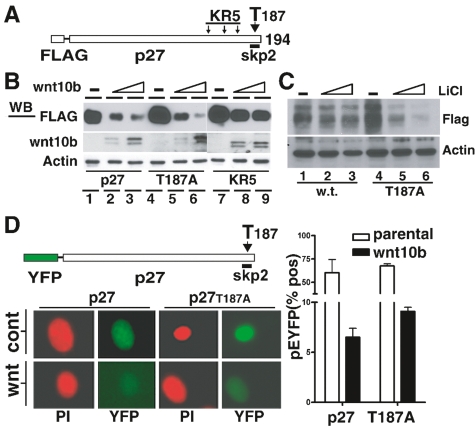
As previously discussed, interaction of p27KIP1 with the SCFSKP2 complex provides an important mechanism for p27KIP1 turnover as cells approach S phase. Interaction of p27KIP1 with the SCFSKP2 requires phosphorylation of Thr187 (T187), and p27KIP1 is stabilized by mutation of T187 to alanine. In addition, SCFSKP2 mediated polyubiquitination requires a cluster of three lysine residues (K134R, K153R and K165R) located near the C terminus of the p27KIP1 protein. When this triad of lysines is substituted with arginine (KR5 mutation), ubiquitination of p27KIP1 is blocked and p27KIP1 protein is stabilized (Shirane et al. 1999). To establish the requirement for T187 and the KR5 domain in Wnt10b-mediated degradation, we used an expression system where human p27KIP1 is tagged with an N-terminal Flag epitope (Ishida et al. 2002). T187A and KR5 mutations were introduced within the backbone of the Flag-human p27KIP1 expression vector (Fig. 3A). Coexpression of Wnt10b with the wild-type human Flag-p27KIP1 resulted in dose-dependent turnover in human embryonic kidney fibroblasts (HEK-293T) by 24 h (Supplemental Fig. S3B). Human HEK-293T cells (arrested in 1% serum for 2 d) were then transfected with Flag-p27wt, Flag-p27T187A, or Flag-p27KR5 plasmids and the indicated amounts of Wnt10b expression plasmid. Immunoblot analysis with anti-Flag antibodies revealed that T187A mutant p27 was degraded when exposed to Wnt10b (Fig. 3B, lanes 1–6). Interestingly, the KR5 mutant was resistant to Wnt10b-mediated degradation (Fig. 3B, lanes 7–9), showing that polyubiquitination at the KR5 site is required in Wnt-induced p27 turnover.
Figure 3.
Wnt-mediated turnover of p27KIP1 is sensitive to mutations that block ubiquitination (KR5) but not to mutations that block SKP2 binding (T187). (A) Schematic illustration of a Flag-tagged human p27KIP1 expression vector with the location of functional mutations KR5 (K134R, K153R, and K165R) or T187 (T187A). (B) Human 293T cells, known to respond to Wnt signals, were synchronized in 1% serum were transfected with Flag-tagged wild-type p27KIP1, or with the indicated mutants in combination with increasing amounts of the pBA-Wnt10b expression vector. Eight hours post-transfection, cells were released with 10% serum and extracts were prepared at 16 h for immunoblotting with the indicated antisera. (C) LiCl (a wnt mimetic) mediates turnover of wild-type and T187A mutated p27KIP1 vectors. Randomly cycling human 293T fibroblasts were transiently transfected with plasmid vectors directing expression of wild-type human p27KIP1 or T187A mutant. Flag epitope vectors (1 μg) alone (lanes 1,4) or with increasing amounts of LiCl (25 mM and 50 mM; lanes 2,3,5,6). Control lanes 1 and 4 had 50 mM of NaCl. Twenty-four hours post-transfection, whole-cell lysates were analyzed by SDS-PAGE and immunoblotted with indicated antisera. (D) Schematic illustration of pYFP-tagged p27 and mutant T187 (T187A). NMG control (cont) or NMG-Wnt10b (wnt) cells, synchronized at G1 phase for 2 d, were then electroporated with pYFP-tagged p27KIP1 or its T187A mutant variant. Cells were released for 12 h and analyzed by immunofluorescence microscopy for YFP expression. Propidium iodide (PI) was used as a counterstain. Cells from D were quantified with an LSC laser cytometer, and the percentage of YFP-positive cells is presented graphically for samples run in triplicate. Error bars represent SDM.
To assess the role of canonical Wnt signaling components in this turnover, we examined the response of cells transiently exposed to LiCl (a Wnt mimetic), known to inhibit GSK3β. We observe that LiCl could induce turnover of both Flag-p27wt and Flag-p27T187A in human HEK-293T fibroblasts (Fig. 3C). These results provide evidence that Wnt-mediated degradation of p27KIP1 is independent of T187 but dependent on the KR5 lysine cluster for ubiquitin conjugation in human fibroblasts. The involvement of the KR5 ubiquitination domain potentially indicates that while SKP2 may not be involved, an alternative E3 complex is likely required for Wnt-mediated p27KIP1 turnover.
To demonstrate that turnover of p27KIP1-T187A also occurred in Wnt-responding mammary epithelial cell lines, we used a vector containing a green fluorescent protein variant (YFP) fused to the N terminus of human p27KIP1 (Connor et al. 2003). Wild-type p27KIP1 was highly unstable in Wnt10b-expressing S-phase cells (Fig. 3D). The YFP-p27T187A variant (lacking the SKP2 recognition motif) was more stable than wild-type p27 when expressed in control NMG cells (Fig. 3D, right panels) but was rapidly degraded in NMG-Wnt10b cells. Quantification of YFP intensity showed comparable loss of wild-type and T187A variants in cells expressing Wnt10b (Fig. 3D, graphical data). These data further support the concept that Wnt10b-mediated turnover of p27KIP1 is independent of T187 phosphorylation but dependent on KR5 polyubiquitination.
Wnt10b promotes the turnover of p27KIP1 in the absence of SKP2 and relieves the G2-to-M progression defect observed in Skp2−/− embryonic fibroblasts
To determine whether Skp2 was required for Wnt-mediated turnover of p27KIP1 in mammary epithelial cells, we used shRNA expression vectors to generate siRNA (Katagiri et al. 2006). Stable Skp2-siRNA-expressing clones were generated in both NMG parental and NMG-Wnt10b cells, and selected clones demonstrating at least 90% reduction in Skp2 mRNA were used for further analysis (data not shown). A control silencing vector (siScrmbl) had no effect on SKP2 protein expression (data not shown).
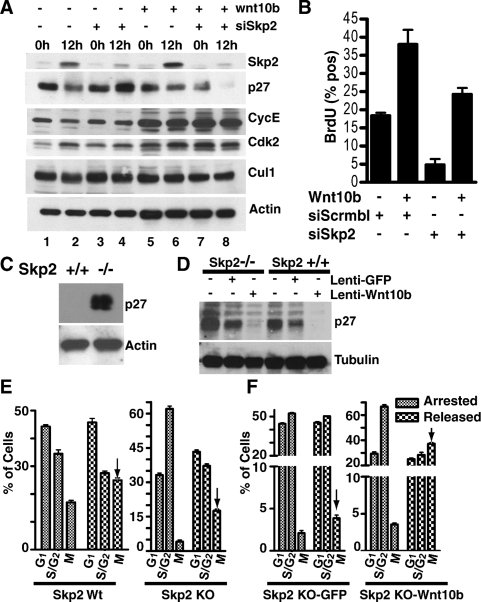
Cells were synchronized in G1 and then released by passage onto fresh plates. To ensure that we could detect SKP2 protein levels at 12 h, we loaded 500 μg of total extract, five times more protein than was analyzed in Figure 2. Immunoblots show that SKP2 protein expression was markedly silenced (>80%) at 12 h in both NMG-siSkp2 and NMG-Wnt10b siSkp2 clones (Fig. 4A, lane 2 vs. lane 4, lane 6 vs. lane 8). Furthermore, the silencing of SKP2 protein expression restored p27KIP1 protein in NMG-siSkp2 control cell lines but not in NMG-Wnt10b-siSkp2 clones (Fig. 4A, lane 4 vs. lane 8). Restoration of cyclin E expression in NMG-siSkp2 (Fig. 4A, lane 2 vs. lane 4) further illustrates that we silenced Skp2 in S-phase cells since it has been reported that SKP2 regulates cyclin E turnover (Nakayama et al. 2000). We observed no change in CULLIN1 (CUL1) and CDK2. Taken together the data provide evidence that Wnt10b is able to mediate the proteolytic degradation of p27KIP1 in mammary epithelial cells engineered to express low levels of SKP2 protein.
Figure 4.
Wnt10b stimulates turnover of p27KIP1 protein in the absence of SKP2 and promotes M-phase entry in Skp2−/− embryonic fibroblasts following synchronization. NMG-control and NMG-Wnt10b cell lines were transfected with pSuppressorNeo vector generating synthetic siRNA for Skp2. (A) Cells were harvested at 0 h and 12 h post-release. Whole-cell lysates were analyzed for expression of SKP2, p27, cyclin E, CDK2, Cullin 1, and β-actin. (B) Cells from A were subsequently pulsed for 15 min with BrdU to label S-phase entry and then fixed at 12 h post-release. BrdU incorporation was assessed by immunofluorescence and quantified by LSC. (C) MEFs from Skp2+/+ and Skp2−/− mice were synchronized in early S phase in the presence of aphidicolin (1 μg/mL). Whole-cell lysates were analyzed for p27KIP1 by immunoblotting. (D) Skp2−/− and Skp2+/+ MEFs were stably transduced with lentivirus expressing GFP or Wnt10b and synchronized with aphidicolin. Whole-cell lysates were immunoblotted to identify p27KIP1 and tubulin. (E) Wild-type and Skp2−/− MEFs from C were arrested in S phase by aphidicolin treatment, released for 24 h and then analyzed for M-phase entry using a nuclear morphology protocol established for the LSC (Luther and Kamentsky 1996). (F) Skp2−/− MEFs cells (±Wnt10b) from D were analyzed for M-phase entry after release. Arrows identify the fraction of M cells 8 h after release into S phase. Experiments were conducted in triplicate, and error bars represent SDM.
We then examined S-phase progression in Skp2-silenced NMG clones using a BrdU incorporation assay. After synchronization, NMG-siSkp2 clones showed a 3.8-fold loss in BrdU labeling when compared with NMG control (18.3% vs. 4.8%) (Fig. 4B). In contrast, NMG-Wnt10b-siSkp2 clones show only minor loss of BrdU-positive cells when compared with NMG-Wnt10b (38% vs. 24.25%). These results provide evidence that NMG, but not NMG-Wnt10b cells, are dependent on SKP2 protein for S-phase progression.
To directly test the role of Skp2 in Wnt-mediated turnover of p27KIP1, we examined p27KIP1 turnover in mouse embryonic fibroblasts (MEFs) derived from Skp2−/−-null mice. As previously shown (Nakayama et al. 2000), p27KIP1 is stabilized in G1–S-arrested (aphidicolin 1 μg/mL treatment for 14 h) Skp2−/− MEFs (Fig. 4C). Skp2−/− and Skp2+/+ MEFs were transduced with lentivirus encoding either hrGFP (GFP control) or Wnt10b. These transduced MEFs were then arrested at the G1–S transition and analyzed for the presence of p27KIP1. The accumulation of p27KIP1 was significantly reduced in the MEFs that expressed Wnt10b (Fig. 4D).
Skp2−/−-null MEFs also display G2-to-M progression defects resulting from p27KIP1 accumulation (Nakayama et al. 2004). We hypothesized that Skp2-independent degradation of p27KIP1 would relieve the defect in M-phase entry observed in Skp2−/− embryonic fibroblasts. Using a nuclear condensation assay developed for the laser-scanning cytometer (LSC) (Luther and Kamentsky 1996), we observed a relief of the G2 arrest when compared with control cells 24 h post-release (Fig. 4E,arrows, indicating the percentage of cells that have entered M phase). Similarly we used the Skp2−/− cells from Figure 4D to test if Wnt10b-expressing cells were able to relieve mitotic accumulation in the absence of SKP2 (Fig. 4F, arrows). The results illustrate that Wnt10b-expressing Skp2−/− MEFs were able to enter M phase fourfold more efficiently than in Skp2−/− control cells. The schematic illustration for the experimental protocol and the scattergram data from one experiment are provided (Supplemental Fig. S4).
Taken together these data provide evidence that Wnt10b signaling can mediate p27KIP1 turnover in the absence of SKP2 protein in S-phase cells. The functional consequences of this pathway include the ability to relieve defective G2-to-M progression in Skp2−/− MEFS.
Wnt10bTG tumors express low SKP2 protein concentrations
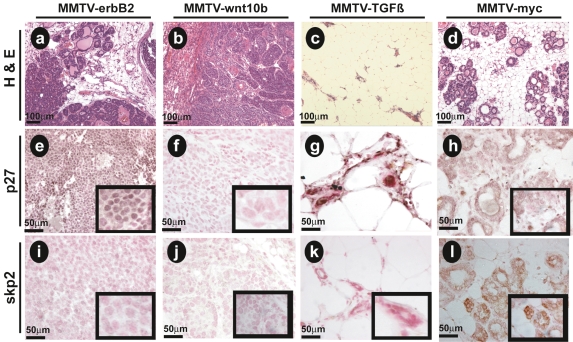
Based on the preceding experiments, we hypothesized that Wnt10bTG tumors would express low p27KIP1 in the context of low SKP2 (SKP2-low), similar to a large group of human breast cancer patients (Traub et al. 2006). We used IHC to analyze the relationship between p27KIP1 (Fig. 5e–h) and SKP2 (Fig. 5i–l) protein expression profiles in transgenic mouse breast specimens. Specifically, we compared expression in tumor sections from MMTV-ErbB2 mammary tumors and MMTV-Wnt10b mammary tumors. Two controls were also included: (1) MMTV-TGF-β1 (TGF-β1TG)-derived mammary tissue (Pierce et al. 1995), modeling low SKP2 protein expression, and (2) MMTV-c-Myc-derived mammary tissue, modeling high expression of SKP2 protein (Stewart et al. 1984).
Figure 5.
p27KIP1-low phenotype of Wnt10bTG mammary tumors correlates with low SKP2 protein. Five-micrometer paraffin sections were prepared from transgenic mouse mammary tumor tissue isolated from ErbB2TG (a,e,i) and Wnt10bTG (b,f,j) animals. Mammary tissue was also isolated from MMTV-TGFß (c,g,k) and MMTV-myc (d,h,l) transgenic mice. H&E (a–d) show morphological features of the tissues (10× objective). IHC analysis with antibodies to p27KIP1 (e–h) and SKP2 (i–l). Immune complexes were visualized by HRP reaction product (brown), and sections were counterstained with nuclear fast red (red) to reveal nuclei. Photomicrographs were collected with 10× (a–d), 40× (e–l), and 100× (inserts) objectives. Five-micrometer paraffin sections were stained as described above. Bars indicate scale in micrometers.
We observed that both p27KIP1 and SKP2 immunoreactivity was absent in Wnt10bTG tumors (Fig. 5f,j). ErbB2TG tumor tissue expressed significant levels of nuclear p27KIP1 but low SKP2 protein (Fig. 5e,i). As expected, TGF-β1TG tissue expressed no SKP2 protein but had the highest levels of nuclear p27KIP1 (Fig. 5g,k). The MMTV-c-Myc tumor tissue displayed intermediate levels of p27KIP1 protein expression (Fig. 5h) when compared with either ErbB2TG or TGF-β1TG. Interestingly, p27KIP1 was diffused throughout the cytoplasm and was not restricted in the nucleus in MMTV-c-Myc tissues (Fig. 5h). Additionally, c-MycTG tumors expressed the highest levels of SKP2 protein (Fig. 5l), consistent with our prediction. A scoring system was established to quantify the results of these immunohistochemical studies, and the data are presented graphically (Supplemental Fig. S5). These results provide strong support for the hypothesis that Wnt-induced tumors display both a p27-low and SKP2-low phenotype.
Cul4A is a direct Wnt target gene necessary for p27KIP1 turnover in mammary epithelial cells expressing Wnt10b, and sufficient for turnover of p27KIP1-T187A
Due to the dispensability of SKP2 in Wnt10b-mediated proteolysis of p27KIP1, we searched for other molecules that might provide mechanistic insight into this SKP2-independent S-phase turnover pathway. Hierarchical analysis of microarray data from Wnt10b-induced tumors revealed that that both Cullin 4A and 4B were elevated in Wnt10bTG tumors (Supplemental Fig. S2A). CUL4A is known to function as a ubiquitin E3 ligase and has been implicated in Dacapo/p27KIP1 turnover in early G1 in Drosophila cells (Bondar et al. 2006; Higa et al. 2006; Li et al. 2006). Cul4-E3 ligase activity has not been implicated previously in CRM1-independent functions, or as a Wnt target gene.
In silico analysis was carried out to identify potential TCF/LEF1-binding sites that could represent Wnt response elements within the Cul4A and Cul4B genes. Both human CUL4A and mouse Cul4A genes were found to contain several potential TCF/LEF1-binding sites based on analysis by multiple algorithms (data not shown).
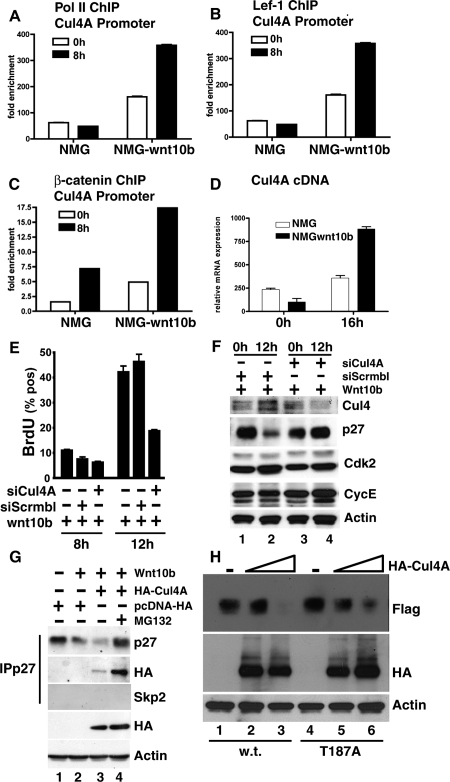
To test the functionality of these putative Lef1 sites, and to examine whether Cul4A represents a previously unrecognized Wnt target gene, we conducted chromatin immunoprecipitation (ChIP) analysis. Initially, we examined Cul4A promoter occupancy by RNA polymerase II (pol II), LEF1, and β-catenin, in Wnt-responding mammary epithelial cells. Figure 6 presents data from synchronized NMG and NMG-Wnt10b cells (at 0 h and 8 h post-release) at the Cul4A proximal promoter. Results illustrate that Wnt10b-expressing cells enhanced the recruitment of RNA pol II to the Cul4A promoter by 2.5-fold at 0 h and by 7.4-fold at 8 h, indicating both cell cycle-independent and cell cycle-dependent activation of the Cul4A promoter (Fig. 6A). In similar extracts, we found Wnt-induced recruitment of both LEF1 (Fig. 6B) and β-catenin (Fig. 6C) to the same region of the Cul4A promoter. Consistent with these events, Cul4A mRNA production was elevated (approximately threefold) at S phase in Wnt ligand-expressing cells (Fig. 6D) and for several other Wnt target genes (Supplemental Fig. S2). The data are consistent with recruitment of canonical Wnt pathway transcriptional mediators and with increased Cul4A mRNA production in S-phase cells. Taken together these data provide evidence that Cul4A is a direct Wnt target gene in mouse mammary cell lines expressing Wnt10b.
Figure 6.
Cul4A is a direct Wnt target gene necessary for p27KIP1 turnover in mammary epithelial cells expressing Wnt10b, and sufficient for turnover of p27KIP1-T187A. (A–C). Wnt-induced activation of Cul4A was demonstrated by ChIP from control NMG and Wnt10b-expressing mammary epithelial clones. Chromatin from growth-arrested (0 h) or released (8 h) cells was precipitated with the indicated antisera. QPCR was used to quantify the amount of Cul4A promoter present in the final precipitates. Values were normalized to both the input template and a nonregulated locus. (D) Induction of Cul4A mRNA was assessed by QPCR on first-strand cDNA prepared at 0 h and 16 h after release. (E) NMG-10b siScrmbl (control) and NMG-10b siCul4A-silenced clones were pulsed with BrdU and analyzed for labeling at 8 h (late G1) and 12 h (early S) by LSC. Experiments were conducted in triplicate, and error bars represent SDM. (F) Whole-cell lysates were analyzed at 0 h or 12 h by immunoblotting and probed with the indicated sera. (G) 293T cells were transiently transfected with pcDNA-Wnt10b, pcDNA-HA-Cul4A, or a control vector pcDNA-HA in the presence or absence of MG132 (10 μM). Immunoprecipitation endogenous p27KIP1 was conducted using a rabbit polyclonal antibody to p27KIP1, and the resulting complexes were immunoblotted with monoclonal antibodies to p27KIP1, SKP2, or HA (12CA5). To control for loading, whole-cell lysates were probed with HA and β-ACTIN. (H) CUL4A is sufficient to accelerate turnover of wild-type or T187A mutant p27KIP1 protein in human cells. Human 293T cells were transiently transfected with Flag-tagged wild-type p27KIP1 or the T187A mutant lacking the SKP2-binding site (1 μg) alone or with increasing amounts of pcDNA-HA-Cul4A (1 μg or 5 μg). Whole-cell extracts were prepared for immunoblotting and analyzed with the indicated antisera.
We hypothesized that CUL4A would regulate p27KIP1 expression and S-phase progression in Wnt10b-expressing cell lines. Cul4A knockout MEF cells are currently unavailable due to early embryonic lethality of Cul4A−/− mice (Li et al. 2002). Therefore, we generated stable clones of NMG-Wnt10b cells expressing shRNA knockdown vectors targeted against Cul4A (siCul4A) or a siScrmbl-control. In NMG-Wnt10b cells, siCul4A reduced Cul4A mRNA production sevenfold, assessed by QPCR (Supplemental Fig. S6C). To examine the functional consequences of Cul4A knockdown, we assessed BrdU uptake at 8 h and 12 h post-release (Fig. 6E). BrdU labeling was reduced by 2.6-fold (47.17% vs. 18.3%) at 12 h, in Cul4A-silenced Wnt10b-expressing cells. No differences were observed at 8 h, prior to S-phase entry, consistent with the notion that CUL4A is required for S-phase progression in Wnt10b-expressing cells.
To determine whether Cul4A-silencing would restore expression of p27KIP1 in NMG-Wnt10b cells, we synchronized cells as previously described and collected extracts at either 0 h or 12 h post-release (Fig. 6F). Importantly, Cul4A silencing restored p27KIP1 expression at 12 h in Wnt10b-expressing clones (Fig. 6F, cf. lanes 2 and 4). Additionally, CDK2 and cyclin E expression were unchanged in Cul4A-silenced clones. This result shows that Cul4A is necessary for turnover of p27KIP1 in Wnt10b-expressing mammary cells.
To test whether CUL4A could interact with endogenous p27KIP1, human HEK-293T cells were transiently transfected with plasmids directing expression of Wnt10b along with HA-tagged CUL4A or a control (pcDNA-HA). Cell lysates were immunoprecipitated with antisera to p27KIP1, and purified complexes were analyzed by SDS-PAGE and immunoblotting (Fig. 6G). We observed that HA-CUL4A protein associated with Flag-p27KIP1 in the presence of a Wnt signal (Fig. 6G, lane 3) and that the complex was stabilized by addition of proteasome inhibitors (Fig. 6G, lane 4).
To test whether CUL4A was sufficient to induce turnover of mutant p27-T187A, HEK-293T cells were transiently transfected with vectors directing the expression of Flag-tagged p27KIP1 and HA-tagged CUL4A. We observed that HA-CUL4A protein initiated turnover of both wild-type (Fig. 6H, lanes 2,3) and T187 mutant p27KIP1 (Fig. 6H, lanes 5,6) in a dose-dependent fashion. These results demonstrate that CUL4A is sufficient to mediate the turnover of both wild-type and mutant T187A p27 in the absence of an identifiable Wnt signal. Therefore, CUL4A represents a genetically critical hub for assembly of this E3 ligase.
Several reports have suggested that CUL4A, CUL4B, and DDB1 are critical structural components of the CUL4-E3 ligase (Lee and Zhou 2007). Interestingly, Cul4B and DDB1 mRNA are also observed in microarray expression assays performed on Wnt10bTG tumor tissue (Supplemental Fig. S2). Analysis of Cul4B promoter suggests that it may also contain Wnt response elements (data not shown). Silencing of Cul4B in Wnt10b-expressing cells also increased endogenous p27KIP1 and reduced S-phase labeling, suggesting that both Cul4A and Cul4B are required for efficient S-phase transit in Wnt-expressing mammary epithelial cells (Supplemental Fig. S6).
These results show that Wnt10b induces Cul4A gene expression in S-phase cells, and indicate a role for CUL4A in a novel, SKP2-independent mechanism of p27KIP1 turnover. Taken together the findings indicate that Wnt10b-mediated proteolysis of p27KIP1 requires a CUL4-dependent ubiquitination system, and that CUL4A is both necessary and sufficient for this function in S-phase cells.
Wnt10b expressed in mammary tumor cell (MCF7) breast cancer cells induces CUL4A and reduces p27KIP1 in S-phase cells
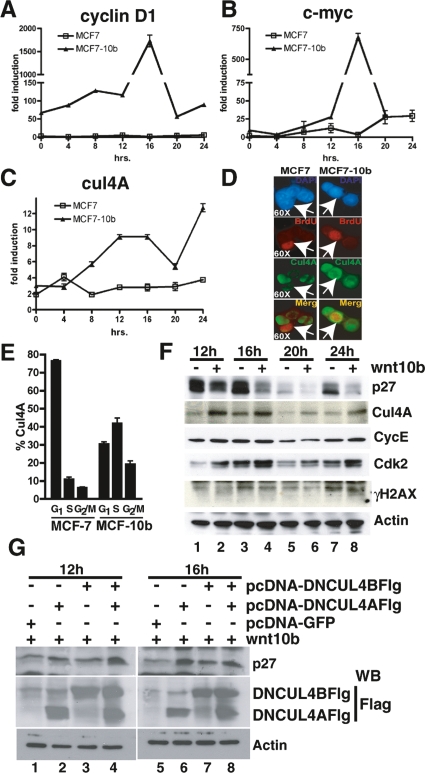
To investigate whether p27KIP1 and CUL4A are regulated similarly in human cells, we generated stable clones in the human breast cancer cell line MCF7 using either pcDNA3 vector alone (MCF7) or pcDNA-Wnt10b (MCF7-10b). We selected clones that expressed modest levels of Wnt10b (twofold over control) to maintain consistency with our mouse cell lines (data not shown). In multiple experiments, we observed that expression of Wnt10b led to stabilization and nuclear accumulation of β-catenin in MCF7 (Supplemental Fig. S7A). Cells were synchronized by contact inhibition for 2–3 d and then released for the indicated times. Wnt10b ligand expression coincided with cell cycle-independent and cell cycle-dependent activation the Wnt target gene cyclin D1 (CCND1) (Fig. 7A) and c-MYC (Fig. 7B). In addition, CUL4A mRNA was highly induced by Wnt10b in these cells (Fig. 7C) but showed slightly earlier induction kinetics. Taken together the data demonstrate that Wnt10b can activate the canonical Wnt signaling pathway in human MCF7 breast cancer cells and elicit a reprogramming that includes elevated expression of Cyclin D1, c-MYC, and CUL4A.
Figure 7.
CUL4A is a Wnt response gene in human MCF7 cells, where CUL4A protein is largely confined to S phase and dominant-negative CUL4 constructs block p27KIP1 turnover. Stable clones of human MCF7 cells were generated carrying either the pcDNA3 vector alone (MCF7) or pcDNA-Wnt10b (MCF7-10b). (A–C) MCF and MFC7-Wnt10b cells were synchronized by contact inhibition and harvested at the indicated times after release. Expression of Wnt response genes was then assessed by QPCR. (A) Cyclin D1 mRNA. (B) c-MYC mRNA. (C) CUL4A mRNA. Experiments were conducted in triplicate and normalized to actin. (D) Sixteen hours after release, BrdU-positive cells (red) were immunostained for CUL4A (green) and counterstained with DAPI (blue). Merged (orange) images of red and green channels are also shown. White arrows illustrate BrdU-positive S-phase cells. Images were collected with a 63× objective. (E) G1, S, and G2/M cells from F were identified by DAPI and BrdU staining and measured quantitatively by an LSC. Data illustrate the reprogramming of CUL4A expression in Wnt10b-expressing cells. Samples were plated in triplicate, and error bars represent SDM. (F) MCF7 and MCF7-10b cells were synchronized in G1 phase and released by replating. Whole-cell extracts were prepared at the indicated times and immunoblotted with the indicated sera. Data indicate stabilization of CUL4A in S-phase cells. The entire time course is presented in S7. (G) MCF7-10b cells were synchronized in G1 phase for 2–3 d and electroporated with pcDNA-GFP or with dominant-negative cullins (−DNCUL4AFlag or −DNCul4BFlag). Cells were released by replating and harvested in S phase. Whole-cell extracts were prepared at the indicated times and immunoblotted with the indicated sera.
The relationship between CUL4A induction in S-phase cells was also examined in single cells by immunofluorescence (Fig. 7D) and quantified by LSC analysis (Fig. 7E). CUL4A (green) was largely confined to G1 in control cells, but expression was dramatically shifted into S phase in Wnt-expressing MCF7 cells. CUL4A-positive S-phase cells were prevalent in Wnt10b-expressing MCF7 clones. Furthermore, we were able to directly quantify the shift of CUL4A protein into S-phase cells using three-color analysis to identify cells within G1, S, or G2/M (Fig. 7D,E). The results show a fourfold increase of CUL4A expression during S phase in Wnt-expressing cells.
To test whether CUL4A protein induction correlates with Wnt-dependent S-phase depletion of p27KIP1 in human cells, we examined synchronized MCF7 cell extracts. Immunoblotting of MCF7 proteins isolated between 12 h and 24 h post-release revealed that CUL4A protein levels are enhanced by approximately fourfold over the control cells at 16 h (corresponding to S phase), and this coincides with augmented turnover of p27KIP1 in MCF7-Wnt10b clones (Fig. 7F). An expanded time course is presented in the Supplemental Material (Supplemental Fig. S7B). At every time point, we observed more CUL4A expressed in the Wnt10b-expressing clones when compared with controls.
To test whether the CUL4A pathway is stimulated by other pathways, we compared induction of CUL4A in MCF-7 cells exposed to estradiol (E2). E2is known to induce proliferation of MCF7 cells but requires SCFSKP2 to initiate turnover of p27KIP1 (Foster et al. 2003). MCF7 cells were synchronized by hormone deprivation for 3 d (Krum et al. 2008) and then released in the presence or absence of LiCl (a Wnt mimetic) or E2. We observed that CUL4A was induced by LiCl, but not by E2 (Supplemental Fig. S7C,E), and that p27KIP1 was turned over by both LiCl and E2 (Supplemental Fig. S7E). These results provide support for the notion that induction of the CUL4A complex is broadly linked to activation of canonical Wnt signaling.
To test whether CUL4A and CUL4B were necessary for p27KIP1 turnover in Wnt-expressing MCF7 cells, we introduced dominant-negative versions of these proteins that have lost their ability to interact with E2 Ub-conjugating enzymes but retain the ability to sequester substrate (Fig. 7G; Jin et al. 2005). We observe that DN-CUL4A (Fig. 7G, lanes 2,6) and DN-CUL4B (Fig. 7G, lanes 3,7) can restore p27KIP1 in S phase of Wnt-expressing MCF7 cells.
Taken together these data indicate that Wnt signaling mediates turnover of p27KIP1 by a novel S-phase process that is independent of SKP2. Cullin 4A, was found to be necessary and sufficient for driving this process in mammalian cells.
Discussion
Wnt signaling regulates cell proliferation, cell fate, and stem cell maintenance in multiple tissues (Cadigan and Nusse 1997). In addition, a clear link has been established between aberrant Wnt signaling and human cancer (Clevers 2004). In the present study, we demonstrate that Wnt10b-mediated transformation of mammary epithelial cells is associated with nuclear localization of β-catenin and enhanced expression of several Wnt target genes and basal cell markers. Novel to this work was the observation that this pattern of gene expression was associated with post-translational loss of p27KIP1 in S-phase cells. This turnover of p27KIP1 was shown to be independent of CRM1 function and to occur in the absence of SKP2. Subsequently, we identified CUL4A as a new Wnt target gene in both human and mouse mammary cells and provide evidence that Wnt10b-mediated turnover of p27KIP1 requires expression of both CUL4A and the related CUL4B. These results indicated that induction of CUL4A and loss of p27KIP1 are unique characteristics of Wnt-responding cells, and may represent features of the basal tumor phenotype in breast cancer. We also found that Wnt10b expression levels correlated inversely with p27KIP1 in a series of mice expressing graded levels of WNT10B protein. This indicates that reduction of p27KIP1 is regulated by a Wnt-dependent mechanism in both normal mammary development and Wnt10b-associated tumor progression.
The data from our experiments demonstrate the dispensability of SKP2 in Wnt10b-mediated p27KIP1 turnover in vitro and a lack of significant SKP2 expression in an in vivo model of Wnt-mediated breast cancer. These results provide evidence that an alternative mechanism of p27KIP1 turnover may exist in Wnt-responsive cells. SKP2 and p27KIP1 have been shown to be important prognostic markers in multivariate analysis correlating with poor survivability in breast cancer patients. In one study, 34% of breast cancers correlated with high levels of SKP2 and low levels of p27KIP1. However, an additional 33.7% of tumors scored negative for both p27KIP1 and SKP2, suggestive of alternative pathways to degrade p27KIP1 protein levels in a large percentage of breast cancer patients (Traub et al. 2006).
We hypothesized that the Wnt-induced degradation of p27KIP1 would alleviate the restriction to enter M in Skp2-null cells (Nakayama et al. 2004). We found that Wnt10b could lessen the accumulation of cells at G2 and reduce the sensitivity of Skp2−/− MEFs responding to serum deprivation. This suggests a link between Wnt signaling and protection from chromosomal abnormalities in situations where SKP2 expression is limiting during development. More importantly, these findings uncover a potential for SCFSKP2-independent p27KIP1 turnover in other stages of the cell cycle.
Since p27 turnover in Wnt10b-expressing cells requires proteasome function and similar p27KIP1 polyubiquitination sites, we searched expression microarray data from Wnt10bTG-induced tumors to identify additional E3-like complexes expressed by these cells. The analysis revealed enhanced expression of Cullin 4A and Cullin 4B. CUL4 genes represent components of a new class of ubiquitin E3 ligases, distinct from the Cullin 1-associated SCFSKP2 complex (Lee and Zhou 2007). CUL4A previously had been found amplified in primary breast cancers and is also amplified and/or overexpressed in a large percentage of breast cancer cell lines (Chen et al. 1998; Schindl et al. 2007). Interestingly, CUL4A has been shown to degrade p27KIP1 during erythropoiesis and is critical for early embryonic development (Li et al. 2006). Cul4A-null embryos die ∼4.5–7.5 d in development (Li et al. 2002). Since Wnt10b is expressed at high levels in 3.5- to 4.5-d blastocysts (Kemp et al. 2005), we hypothesize that CUL4A might be a direct target of Wnt signaling and play a role in Wnt10b-mediated degradation of p27KIP1.
To test this hypothesis, we conducted ChIP assays to identify regulators of Cul4A promoter activity. We observed enhanced recruitment of LEF1 and β-catenin to the Cul4A promoter. RNA pol II holoenzyme was also recruited to the Cul4A promoter and correlated with production of Cul4A mRNA. CUL4A mRNA and protein were similarly enhanced in human MCF7 cells in response to Wnt10b signaling, and CUL4A protein was induced by exposure to LiCl, an activator of canonical Wnt signaling. These results demonstrate that CUL4A is a novel Wnt target gene in mammary epithelial cells.
Using an shRNA strategy to produce siRNA against Cul4A, we were able to directly test if CUL4A was required for the degradation of p27KIP1 and S-phase progression in Wnt10b-expressing cells. No clones were isolated showing complete knockdown of Cul4A mRNA, but clones were recovered with significantly lower levels of Cul4A mRNA expression. Importantly, siCul4A blocked p27KIP1 turnover and restored p27KIP1 protein concentrations in these partially silenced clones. Concurrently, we observed that Cul4A-silenced clones of Wnt10b-expressing cells proliferated at rates ∼2.6-fold slower than control cells. In similar studies, we demonstrated that siCul4B had a similar phenotype as siCul4A, but the effect was less pronounced than in the Cul4A knockdown experiment. In human mammary cells, we found that DN-CUL4A and DN-CUL4B were able to restore endogenous p27KIP1 in S-phase cells.
The loss of CUL4 in Drosophila cells causes G1 accumulation and restoration of the CDK inhibitor Dacapo (p27KIP1), and in human cells silencing of CUL4A restore p27KIP1 in G1 cells (Higa et al. 2006). Previous work has shown that Cul4A–DDB1 cooperates with SCFSKP2, partitioning p27KIP1 degradation between G1/S boundary and S phase (Bondar et al. 2006). Bondar et al. (2006) found that CUL4A–DDB1 plays a role in G1 turnover of p27KIP1, but proposed that SKP2 was still required for S-phase turnover. Our findings represent the first example that illustrates degradation of p27KIP1-mediated by CUL4A in S-phase cells and that demonstrates independence from SKP2 activity. Consequently, we propose that a CUL4-containing E3 ubiquitin ligase complex (potentially including CUL4A–DDB1) functions to reprogram S-phase progression and functionally replace at least some functions of SCFSKP2 in Wnt-responding cells.
In conclusion, we provide evidence that a Wnt-dependent breast cancer model correlates with nuclear β-catenin, cyclin D1-high, p27KIP1-low, and SKP2-low protein expression. The degradation of p27KIP1 in this model appears independent of SKP2 and correlates with induction of a functional CUL4A-E3 ligase complex. These results define a previously unrecognized post-translational component of the Wnt response and define a novel WIPT pathway. The link is significant because a large percentage of human breast cancers have been identified with low p27KIP1 and low SKP2, and their origin is currently unclear. Furthermore, nuclear β-catenin in combination with cycD1high, p27low, and SKP2low may define a subgroup of human breast cancer that are currently not targeted effectively by current therapies. Pharmaceutical suppressors of CUL4A-associated E3 ligase activity could provide a therapeutic target for this subset of cancers, as they would increase p27KIP1 expression and delay proliferation. Such inhibitors could also reduce the genetic instability resulting from SKP2 inhibition. The concept of cancer as a disease of stem or progenitor cells is well illustrated in human mammary carcinogenesis. Mouse tumor models of Wnt-activation should continue to provide insight into the mechanism of basal breast cancer since these tumors are heavily dependent on stem cell progenitors.
Materials and methods
Nomenclature and mice
Mouse genes, RNA, and cDNA are lowercase and italic (e.g., Cul4A, p27Kip1), and human genes are capitalized and italic (e.g., CUL4A, p27KIP1). Mouse and human proteins are listed as capitalized and nonitalic (e.g., WNT10B, CUL4A, or p27KIP1). Naming follows published guidelines for human (http://www.genenames.org/guidelines.html) and mouse (http://www.informatics.jax.org/mgihome/nomen/gene.shtml).
Unless otherwise indicated, all transgenic mice (e.g., Wnt10bTG, ErbB2TG) were derived in the FVB/N strain (Taconic). Wnt10b-null mice were generated in 129/SvEv and backcrossed to FVB/N to create isogenic lines (T. Lane, unpubl.).
Cell culture and synchronization
Mouse mammary epithelial cell lines (NMuMg [NMG] and EpH4), Skp2+/+, and Skp2−/− MEFs, HEK-293T, and human MCF7 were maintained in a humidified atmosphere with 5% CO2 in Dulbecco’s Modified Eagle’s Medium supplemented with 10% fetal calf serum, 100 U/mL penicillin, and 100 μg/mL streptomycin. NMG, EpH4, and MCF7 cell cultures were supplemented with insulin (1 μg/mL). NMG are a nontransformed subclone of NMuMG cells (American Type Culture Collection, CRL-1636). For cell cycle progression analysis, human and mouse mammary epithelial cell lines were synchronized at G1 by maintenance at 100% confluency for 2–3 d (contact inhibition), then released with trypsin and replated for the indicated time. HEK-293T fibroblasts were synchronized at G1 by growth in 1% serum for 2–3 d, then released by replating. Skp2+/+ and Skp2−/− and NMG-Wnt10b were synchronized at the G1–S boundary with aphidicolin (1 mg/mL) for 14 h, as described (Nakayama et. al. 2000). In some experiments, proteasome activity was blocked by pretreatement of cells with MG132 (10 μM), or CRM1-dependent nuclear export machinery was inhibited by pretreatment with LMB (1 μg/mL). Generation of stable cell lines is described in the Supplemental Material.
Transient transfections
Transfection of HEK-293T were carried out using a standard calcium chloride method. NMG and MCF7 cells were electroporated using a Nucleofector (Amaxa) following the manufacturer’s protocol. Plasmids are identified in the text and included pcDNA3.1-Flag-human p27KIP1, p27-KR5, and p27-T187A, which were generously provided by Dr. M. Shirani. pcDNA3.1-YFP-human p27wt was generously provided by Dr. Joyce Slingerland, and pcDNA-HA and pcDNA-Cul4A were provided by Dr. Kristin T. Chun. We purchased pcDNA3-DN-hCUL4A-Flag (15821) and pcDNA3-DN-hCUL4B-Flag (15822) from Addgene. In addition, pBA-Wnt10b, pcDNA3.1-Wnt10b, and pcDNA3.1-YFP-p27-T187A were generated internally and reported here. The inserts of all plasmids were confirmed by direct sequencing. Plasmids used for virus production, gene knockdown, and protein production are described in Supplemental Material.
IHC
Mammary gland tissue or tumors were fixed in 4% paraformaldehyde-PBS and embedded in paraffin. Sections were processed through standard deparafinization protocols followed by 3% H2O2 treatment. Antigen recovery was conducted using the boiling method in 0.1 M sodium citrate (pH 8.5) and 0.1 M citric acid (pH 6.0). The tissue was then incubated in blocking buffer (5% normal goat serum, 2.5% BSA in PBS at pH 7.5) for 30min. Primary antibodies p27KIP1 (C-19, Santa Cruz Biotechnologies), β-Catenin (C2206, Sigma), Cyclin D1 (Ab3, Labvision), SKP2 (Zymed), and WNT10B (Supplemental Material) were incubated overnight at 4°C in a humidified chamber followed by a secondary biotinylated goat anti-mouse or rabbit antibody (Pierce). IHC staining was conducted using Vectastain ABC System (Vector Laboratories) and counterstaining was conducted with Nuclear Fast Red (Vector Laboratories) per the manufacturer’s protocol.
Immunofluorescence and LSC
Cells were fixed with 4% paraformaldehyde-PBS for 30 min, washed, and incubated with blocking buffer (TBS at pH 7.8, 3% BSA, 1% NGS, 1% Triton X-100, 0.01% NaAzide). Primary antisera were purchased against β-catenin (H-102, Santa Cruz Biotechnologies), p27KIP1 (C-19, Santa Cruz Biotechnologies), mouse Cul4 (pan-Cul4A and pan-Cul4B cross-reactive T-15, Santa Cruz Biotechnologies), Sca1 (E13-161.7-FITC, BD-Pharmingen), and human CUL4A (Ab34897, Abcam). Secondary antisera included highly cross-absorbed goat anti-rabbit Alexa-633, goat anti-rabbit Alexa-488, or goat anti-mouse Alexa-633 (Molecular Probes). Cells were counterstained with 6-diamidino-2-phenylindole (DAPI; Molecular Probes) or propidium iodide (PI; Molecular Probes) at a concentration of 10–30 mg/mL to identify DNA and to distinguish cell cycle stages. Immunofluorescence photomicrographs were obtained using 10×, 40×, and 63× objectives on a Leica DM IRBE fluorescent microscope equipped with a Hamamatsu C4742-95 digital camera (Leica Microsystems). Openlab 5.0 software (Improvision) was used for image processing. Cytometric analysis was performed on similarly prepared samples using an LSC (CompuCyte Corp.). The LSC was equipped with UV laser, used to contour DAPI using a minimum cell area of 10 μm3, and Argon and HeNe lasers used to measure immunofluorescence. Cytometric analysis was conducted with WinCyte software (CompuCyte Corp.).
BrdU labeling for cell cycle analysis
Cells were plated in eight-well chamber slides (Nalge NUNC) and treated as indicated. Randomly cycled or released cells were pulsed with 10 mM BrdU (Calbiochem) for 20 min prior to fixation, washed twice with PBS containing 1% BSA, and fixed in place using cold 70% ethanol for 30 min. Chromatin was made accessible by treatment with 2N HCl and 0.5% Triton X-100 for 30 min, followed by 0.1 mM Na2B4O7 ⋅ 10H2O (pH 8.5) at room temperature for 5 min, and blocked in PBS containing 1% BSA for 30 min. BrdU-labeled nuclei were detected with Alexa594-conjugated anti-BrdU (monoclonal-PRB1, Molecular Probes) and counterstained with 30 mg/mL DAPI. Analysis of S-phase labeling was performed using the LSC and immunofluorescence.
Cell extraction and immunoblotting
Cells were lysed in low-salt buffer EBC (20 mM HEPES at pH 7.4, 0.5% Nonidet P-40, 100 mM NaCl) and/or modified RIPA buffer (50 mM Tris-HCl at pH 8.0, 150 mM NaCl, 5mM EDTA, 1% Triton X-100, 0.1% SDS, 1% sodium deoxycholate) supplemented with phosphatase inhibitors (10 mM NaF, 100 mM Na3VO4) and a protease inhibitor mixture (Complete without EDTA; Roche Applied Science) for 30 min on ice. The samples were pelleted at 14,000 rpm for 10 min at 4°C. Protein concentrations were determined using the Bio-Rad Protein Assay (Bio-Rad). Fifty micrograms to 250 μg of protein were loaded per lane and separated by SDS-PAGE 10% gels. After transfer, Immobilon-P (Millipore Corp.) was immunoblotted using the following primary antibodies: β-tubulin (D-10), p27KIP1 (C-19), Cyclin E1 (M-20), and Cullin 4 (T-15) (Santa Cruz Biotechnologies); human CULLIN 4A (Abcam); Cullin 1(Ab1868-500) and Cdk2 (Ab7954-1) (Abcam); Skp2 (32-3400, Zymed) and anti-phospho p27Thr187 (71-7700, Zymed Laboratories); Flag M2 (Sigma); and WNT10B (Supplemental Material).
Immunoprecipitation
Immunoprecipitation reactions were conducted on whole-cell lysates from HEK-293T transiently transfected with pcDNA-HA or pcDNA-HA-Cul4A (Li et al. 2003) in the presence or absence of pBA-Wnt10b or MG132 (10 uM). Immunoprecipitation reactions were conducted with monoclonal p27KIP1 (BD-Pharmigen) on 200 μg of total protein and blotted with previously described antibodies.
RNA, QPCR, and RT–PCR
Isolation of total RNA was performed using TRIzol (Invitrogen) according to the manufacturer’s protocol. RNA was treated with DNA-free kit (Ambion) and converted to cDNA with iScript cDNA Synthesis Kit (Bio-Rad) according to the manufacturers’ protocols. QPCR was performed on cDNA using the Applied Biosystems Power SYBR Green amplification system with the following conditions: 3 min at 95°C, and 40 cycles of 20 sec at 95°C, 30 sec at 55°C, and 20 sec at 72°C. All QPCR reactions were performed with an iCycler thermocycler (Bio-Rad). RT–PCR reactions were carried out using a PTC-200 thermocycler (MJ Research), and products were analyzed on 1.5% agarose/EtBr gels. Reactions included cDNA, 1× Thermopol buffer (New England Biolabs), 0.5 U of Deep Vent (New England Biolabs), 250 uM dNTPs, and 1 μM each oligonucleotide. PCR conditions were as follows: one cycle of 2 min at 95°C; 30 cycles of 20 sec at 95°C, 20 sec at 50°C–62°C, and 30 sec at 72°C; and one cycle of 5 min at 72°C. Primer pairs for each gene are provided in Supplemental Table 1.
ChIP
Cells were grown to confluency for 2–3 d and harvested at 0 h and 8 h post-release. ChIP was performed as described previously (Krum et. al. 2008). Antibodies used included RNA pol II (8WG16, Abcam), LEF1, and β-catenin (N-17 and H-102, Santa Cruz Biotechnologies). Each experiment was repeated at least three times. Primer pairs for each gene are provided (Supplemental Table 1). Additional details are available in the Supplemental Material.
Additional Materials and Methods are detailed in Supplemental Material.
Acknowledgments
We thank the members of the Lane Laboratory for their support and great effort and M. Singer, A. Rizzo, Z. Sanchez-Galvan, and R. Lurvey for technical contributions to this project. We thank Drs. J. Slingerland, S. Kondo, M. Shirani, and M. Kitagawa for reagents and helpful discussions related to p27 and SKP2, and Dr. K.T. Chun for reagents and discussions related to CUL4A. We thank Drs. L. Iruela-Arispe and H. Herschman for comments on the manuscript. The work has been supported by grants from Stop Cancer, the National Cancer Institute (R01-CA107002), and the American Cancer Society (RSG-05-034-01) to T.F.L. G.M.C. is the recipient of fellowships from the CDMRP (DAMD17-02-1-0327) and is currently supported by an NRSA (CA09056). In addition, Q.E.D. is the recipient of an NRSA predoctoral fellowship (GM07185), S.P. is the recipient of an NIH/R03 (HD53888), and S.A.K. is supported by a K12/ BIRCWH grant (HD001400-08).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1692808.
References
- Barnes D.M., Gillett C.E. Cyclin D1 in breast cancer. Breast Cancer Res. Treat. 1998;52:1–15. doi: 10.1023/a:1006103831990. [DOI] [PubMed] [Google Scholar]
- Bondar T., Kalinina A., Khair L., Kopanja D., Nag A., Bagchi S., Raychaudhuri P. Cul4A and DDB1 associate with Skp2 to target p27Kip1 for proteolysis involving the COP9 signalosome. Mol. Cell. Biol. 2006;26:2531–2539. doi: 10.1128/MCB.26.7.2531-2539.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui T.D., Rankin J., Smith K., Huguet E.L., Ruben S., Strachan T., Harris A.L., Lindsay S. A novel human Wnt gene, WNT10B, maps to 12q13 and is expressed in human breast carcinomas. Oncogene. 1997;14:1249–1253. doi: 10.1038/sj.onc.1200936. [DOI] [PubMed] [Google Scholar]
- Cadigan K.M., Nusse R. Wnt signaling: A common theme in animal development. Genes & Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- Cariou S., Catzavelos C., Slingerland J.M. Prognostic implications of expression of the cell cycle inhibitor protein p27Kip1. Breast Cancer Res. Treat. 1998;52:29–41. doi: 10.1023/a:1006154900130. [DOI] [PubMed] [Google Scholar]
- Carrano A.C., Eytan E., Hershko A., Pagano M. SKP2 is required for ubiquitin-mediated degradation of the CDK inhibitor p27. Nat. Cell Biol. 1999;1:193–199. doi: 10.1038/12013. [DOI] [PubMed] [Google Scholar]
- Catzavelos C., Bhattacharya N., Ung Y.C., Wilson J.A., Roncari L., Sandhu C., Shaw P., Yeger H., Morava-Protzner I., Kapusta L., et al. Decreased levels of the cell-cycle inhibitor p27Kip1 protein: Prognostic implications in primary breast cancer. Nat. Med. 1997;3:227–230. doi: 10.1038/nm0297-227. [DOI] [PubMed] [Google Scholar]
- Chen L.C., Manjeshwar S., Lu Y., Moore D., Ljung B.M., Kuo W.L., Dairkee S.H., Wernick M., Collins C., Smith H.S. The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res. 1998;58:3677–3683. [PubMed] [Google Scholar]
- Clevers H. Wnt breakers in colon cancer. Cancer Cell. 2004;5:5–6. doi: 10.1016/s1535-6108(03)00339-8. [DOI] [PubMed] [Google Scholar]
- Coats S., Flanagan W.M., Nourse J., Roberts J.M. Requirement of p27Kip1 for restriction point control of the fibroblast cell cycle. Science. 1996;272:877–880. doi: 10.1126/science.272.5263.877. [DOI] [PubMed] [Google Scholar]
- Connor M.K., Kotchetkov R., Cariou S., Resch A., Lupetti R., Beniston R.G., Melchior F., Hengst L., Slingerland J.M. CRM1/Ran-mediated nuclear export of p27(Kip1) involves a nuclear export signal and links p27 export and proteolysis. Mol. Biol. Cell. 2003;14:201–213. doi: 10.1091/mbc.E02-06-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowin P., Rowlands T.M., Hatsell S.J. Cadherins and catenins in breast cancer. Curr. Opin. Cell Biol. 2005;17:499–508. doi: 10.1016/j.ceb.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Foster J.S., Fernando R.I., Ishida N., Nakayama K.I., Wimalasena J. Estrogens down-regulate p27Kip1 in breast cancer cells through Skp2 and through nuclear export mediated by the ERK pathway. J. Biol. Chem. 2003;278:41355–41366. doi: 10.1074/jbc.M302830200. [DOI] [PubMed] [Google Scholar]
- Higa L.A., Yang X., Zheng J., Banks D., Wu M., Ghosh P., Sun H., Zhang H. Involvement of CUL4A ubiquitin E3 ligases in regulating CDK inhibitors Dacapo/p27Kip1 and cyclin E degradation. Cell Cycle. 2006;5:71–77. doi: 10.4161/cc.5.1.2266. [DOI] [PubMed] [Google Scholar]
- Ishida N., Hara T., Kamura T., Yoshida M., Nakayama K., Nakayama K.I. Phosphorylation of p27Kip1 on serine 10 is required for its binding to CRM1 and nuclear export. J. Biol. Chem. 2002;277:14355–14358. doi: 10.1074/jbc.C100762200. [DOI] [PubMed] [Google Scholar]
- Jin J., Ang X.L., Shirogane T., Harper W.J. Substrates for F-box proteins. Methods Enzymol. 2005;399:287–309. doi: 10.1016/S0076-6879(05)99020-4. [DOI] [PubMed] [Google Scholar]
- Katagiri Y., Hozumi Y., Kondo S. Knockdown of Skp2 by siRNA inhibits melanoma cell growth in vitro and in vivo. J. Dermatol. Sci. 2006;42:215–224. doi: 10.1016/j.jdermsci.2005.12.016. [DOI] [PubMed] [Google Scholar]
- Kemp C., Willems E., Abdo S., Lambiv L., Leyns L. Expression of all Wnt genes and their secreted antagonists during mouse blastocyst and postimplantation development. Dev. Dyn. 2005;233:1064–1075. doi: 10.1002/dvdy.20408. [DOI] [PubMed] [Google Scholar]
- Krum S.A., Miranda-Carboni G.A., Hauschka P.V., Carroll J.S., Lane T.F., Freedman L.P., Brown M. Estrogen protects bone by inducing Fas ligand in osteoblast to regulate osteoclast survival. EMBO J. 2008;27:535–545. doi: 10.1038/sj.emboj.7601984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane T.F., Leder P. Wnt-10b directs hypermorphic development and transformation in mammary glands of male and female mice. Oncogene. 1997;15:2133–2144. doi: 10.1038/sj.onc.1201593. [DOI] [PubMed] [Google Scholar]
- Lee J., Zhou P. DCAFs, the missing link of the CUL4–DDB1 ubiquitin ligase. Mol. Cell. 2007;26:775–780. doi: 10.1016/j.molcel.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Li B., Ruiz J.C., Chun K.T. CUL-4A is critical for early embryonic development. Mol. Cell. Biol. 2002;22:4997–5005. doi: 10.1128/MCB.22.14.4997-5005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Welm B., Podsypanina K., Huang S., Chamorro M., Zhang X., Rowlands T., Egeblad M., Cowin P., Werb Z., et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc. Natl. Acad. Sci. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Jia N., Kapur R., Chun K.T. Cul4A targets p27 for degradation and regulates proliferation, cell cycle exit, and differentiation during erythropoiesis. Blood. 2006;107:4291–4299. doi: 10.1182/blood-2005-08-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S.Y., Xia W., Wang J.C., Kwong K.Y., Spohn B., Wen Y., Pestell R.G., Hung M.C. β-Catenin, a novel prognostic marker for breast cancer: Its roles in cyclin D1 expression and cancer progression. Proc. Natl. Acad. Sci. 2000;97:4262–4266. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luther E., Kamentsky L.A. Resolution of mitotic cells using laser scanning cytometry. Cytometry. 1996;23:272–278. doi: 10.1002/(SICI)1097-0320(19960401)23:4<272::AID-CYTO2>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Malek N.P., Sundberg H., McGrew S., Nakayama K., Kyriakides T.R., Roberts J.M. A mouse knock-in model exposes sequential proteolytic pathways that regulate p27Kip1 in G1 and S phase. Nature. 2001;413:323–327. doi: 10.1038/35095083. [DOI] [PubMed] [Google Scholar]
- Montagnoli A., Fiore F., Eytan E., Carrano A.C., Draetta G.F., Hershko A., Pagano M. Ubiquitination of p27 is regulated by Cdk-dependent phosphorylation and trimeric complex formation. Genes & Dev. 1999;13:1181–1189. doi: 10.1101/gad.13.9.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller W.J., Sinn E., Pattengale P.K., Wallace R., Leder P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 1988;54:105–115. doi: 10.1016/0092-8674(88)90184-5. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Nagahama H., Minamishima Y.A., Matsumoto M., Nakamichi I., Kitagawa K., Shirane M., Tsunematsu R., Tsukiyama T., Ishida N., et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J. 2000;19:2069–2081. doi: 10.1093/emboj/19.9.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama K., Nagahama H., Minamishima Y.A., Miyake S., Ishida N., Hatakeyama S., Kitagawa M., Iemura S., Natsume T., Nakayama K.I. Skp2-mediated degradation of p27 regulates progression into mitosis. Dev. Cell. 2004;6:661–672. doi: 10.1016/s1534-5807(04)00131-5. [DOI] [PubMed] [Google Scholar]
- Nusse R. Wnt signaling in disease and in development. Cell Res. 2005;15:28–32. doi: 10.1038/sj.cr.7290260. [DOI] [PubMed] [Google Scholar]
- Pagano M., Tam S.W., Theodoras A.M., Beer-Romero P., Del Sal G., Chau V., Yew P.R., Draetta G.F., Rolfe M. Role of the ubiquitin–proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- Perou C.M., Sørlie T., Eisen M.B., de van Rijn M., Jeffrey S.S., Rees C.A., Pollack J.R., Ross D.T., Johnsen H., Akslen L.A., et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Pierce D.F., Gorska A.E., Chytil A., Meise K.S., Page D.L., Coffey R.J., Moses H.L. Mammary tumor suppression by transforming growth factor β 1 transgene expression. Proc. Natl. Acad. Sci. 1995;92:4254–4258. doi: 10.1073/pnas.92.10.4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindl M., Gnant M., Schoppmann S.F., Horvat R., Birner P. Overexpression of the human homologue for Caenorhabditis elegans cul-4 gene is associated with poor outcome in node-negative breast cancer. Anticancer Res. 2007;27:949–952. [PubMed] [Google Scholar]
- Sheaff R.J., Groudine M., Gordon M., Roberts J.M., Clurman B.E. Cyclin E–CDK2 is a regulator of p27Kip1. Genes & Dev. 1997;11:1464–1478. doi: 10.1101/gad.11.11.1464. [DOI] [PubMed] [Google Scholar]
- Shirane M., Harumiya Y., Ishida N., Hirai A., Miyamoto C., Hatakeyama S., Nakayama K., Kitagawa M. Down-regulation of p27(Kip1) by two mechanisms, ubiquitin-mediated degradation and proteolytic processing. J. Biol. Chem. 1999;274:13886–13893. doi: 10.1074/jbc.274.20.13886. [DOI] [PubMed] [Google Scholar]
- Sørlie T., Tibshirani R., Parker J., Hastie T., Marron J.S., Nobel A., Deng S., Johnsen H., Pesich R., Geisler S., et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc. Natl. Acad. Sci. 2003;100:8418–8423. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart T.A., Pattengale P.K., Leder P. Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell. 1984;38:627–637. doi: 10.1016/0092-8674(84)90257-5. [DOI] [PubMed] [Google Scholar]
- Traub F., Mengel M., Luck H.J., Kreipe H.H., von Wasielewski R. Prognostic impact of Skp2 and p27 in human breast cancer. Breast Cancer Res. Treat. 2006;99:185–191. doi: 10.1007/s10549-006-9202-3. [DOI] [PubMed] [Google Scholar]
- Veltmaat J.M., Van Veelen W., Thiery J.P., Bellusci S. Identification of the mammary line in mouse by Wnt10b expression. Dev. Dyn. 2004;229:349–356. doi: 10.1002/dvdy.10441. [DOI] [PubMed] [Google Scholar]