Abstract
Although microRNAs (miRNAs) are key regulators of gene expression in normal human physiology and disease, transcriptional regulation of miRNAs is poorly understood, because most miRNA promoters have not yet been characterized. We identified the proximal promoters of 175 human miRNAs by combining nucleosome mapping with chromatin signatures for promoters. We observe that one-third of intronic miRNAs have transcription initiation regions independent from their host promoters and present a list of RNA polymerase II- and III-occupied miRNAs. Nucleosome mapping and linker sequence analyses in miRNA promoters permitted accurate prediction of transcription factors regulating miRNA expression, thus identifying nine miRNAs regulated by the MITF transcription factor/oncoprotein in melanoma cells. Furthermore, DNA sequences encoding mature miRNAs were found to be preferentially occupied by positioned-nucleosomes, and the 3′ end sites of known genes exhibited nucleosome depletion. The high-throughput identification of miRNA promoter and enhancer regulatory elements sheds light on evolution of miRNA transcription and permits rapid identification of transcriptional networks of miRNAs.
Keywords: Transcription, microRNA, promoter, chromatin
MicroRNAs (miRNAs) are small ∼22-nucleotide (nt) noncoding RNAs (Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001) derived from large primary miRNAs (pri-miRNA), which are processed to ∼70-nt precursors (pre-miRNA) and subsequently to the mature form by endonucleases (Lee et al. 2002; Bartel 2004; Cullen 2004; He and Hannon 2004). miRNAs have been discovered in multiple organisms, and many are evolutionarily conserved (Bartel 2004). They regulate various developmental and physiological processes, (Plasterk 2006; Shivdasani 2006) and are implicated in human diseases (Calin and Croce 2006). Examination of miRNA expression patterns revealed that miRNA expression states exhibit spatiotemporal differences (Pasquinelli et al. 2005; Landgraf et al. 2007). These findings highlight the need for a detailed understanding of the mechanisms governing miRNA expression. Evidence exists for important post-transcriptional regulation of miRNA expression, yet a primary level of control is thought to be transcriptional (Cullen 2004). Therefore, identification of genetic and epigenetic elements responsible for transcriptional regulation of miRNA transcription may aid in an understanding of pathways governing miRNA biology.
Because of the labile nature of pri-miRNAs due to their rapid processing, traditional global transcription start site (TSS) mapping strategies relying on RNA analyses are challenging, and, therefore, only a handful of miRNA TSSs have been identified to date (Lee et al. 2002; Bracht et al. 2004; Cai et al. 2004; Houbaviy et al. 2005). These studies indicate that RNA polymerase II (RNAPII) is responsible for miRNA transcription, although an miRNA cluster has recently been shown to be RNA polymerase III (RNAPIII)-transcribed (Borchert et al. 2006). Although RNAPIII function is not as well understood as RNAPII, it is known that the same promoter elements can be used by both polymerases in humans, such as the promoters of globin genes (Carlson and Ross 1983) and c-myc (Chung et al. 1987), and transcription factors (TFs) can also regulate RNAPIII activity (Schramm and Hernandez 2002; Felton-Edkins et al. 2003; Gomez-Roman et al. 2003; Reina et al. 2006; Johnson et al. 2007). It therefore seems plausible that miRNA genes might employ distinct transcriptional machineries for their synthesis, and it is necessary to define the roles of both RNAPs in miRNA transcription in detail.
Recent studies have identified chromatin signatures that can be used for promoter identification in human cells (Guenther et al. 2007; Heintzman et al. 2007; Landgraf et al. 2007; Ozsolak et al. 2007). Several histone modifications, mainly trimethylation of Lys 4 of histone 3 (H3K4me3) and acetylation of Lys 9/14 of histone 3 (H3K9/14Ac), have been established as markers for transcriptionally active promoters (Jenuwein and Allis 2001; Santos-Rosa et al. 2002; Turner 2002; Schneider et al. 2004; Bernstein et al. 2005; Pokholok et al. 2005; Guenther et al. 2007; Li et al. 2007). Heintzman et al. (2007) found distinct histone modification patterns for promoters and enhancers, and demonstrated the feasibility of using them to discover novel regulatory elements with chromatin immunoprecipitation (ChIP)–chip screens to 1- to 2-kb resolution. Furthermore, transcriptionally active genes exhibit nucleosome depletion in the 100- to 130-base-pair (bp) window surrounding their TSS (Mito et al. 2005; Yuan et al. 2005; Ozsolak et al. 2007). We therefore hypothesized that a strategy combining nucleosome positioning patterns with ChIP–chip screens for promoter signatures may identify the transcription initiation regions and upstream regulatory regions of transcriptionally active pri-miRNAs in a high resolution, high-throughput manner. Since a significant fraction of functionally relevant TFs bind in proximity to the TSSs, generally in nucleosome-free regions (Yuan et al. 2005; Ozsolak et al. 2007), the locations of internucleosomal miRNA promoter elements may then be used to rapidly identify functional elements regulating miRNA transcription.
We report here the application of this strategy to three human cancer cell lines, permitting the identification of transcription initiation regions of 175 miRNAs. A short (∼70-bp) nucleosome-depleted region is observed at the core promoter/initiation region, and approximately one-third of the intronic miRNAs were seen to contain transcription initiation sites independent from their host gene’s. We use this information to identify TFs regulating the miRNAs, and focus on those that are regulated by the melanoma TF and oncogene MITF, within the melanocyte lineage. We show that these MITF-binding sites are evolutionarily conserved, and study the evolution of miRNA transcription through comparative genomics. In addition, we define subsets of miRNAs transcribed by RNAPII and/or RNAPIII, and identify distinct chromatin organization patterns surrounding miRNA “coding” regions and 3′ ends of known genes.
Results
High-throughput identification of miRNA promoters
To annotate human pri-miRNAs, we performed nucleosome positioning analyses (Yuan et al. 2005; Ozsolak et al. 2007) and ChIP–chip (Ren et al. 2000; Kim and Ren 2006) screens for H3K4me3, H3K9/14Ac, RNAPII, and RNAPIII in the 20-kb regions upstream of and 1 kb downstream from all miRNAs in two melanoma (UACC62 and MALME) lines and one breast cancer (MCF7) line. Using a scoring function that combined salient features of promoters including nucleosome-depleted regions, high evolutionary conservation, CpG islands, and TF motif occurrence within regions containing H3K4me3, H3K9/14Ac, RNAPII, and/or RNAPIII (described in detail in the Supplemental Material), we determined the transcription initiation regions of 175 miRNAs that are expressed within these cell lines. We demonstrated the power and resolution of this approach by applying it to human genes with available nucleosome positioning data (Ozsolak et al. 2007). The algorithm correctly predicted the transcription initiation regions to a resolution of 150 bp with 81% sensitivity and 95% specificity (Supplemental Material). Furthermore, 75% of our predicted miRNA transcription initiation regions had experimentally observed EST TSSs (University of California at Santa Cruz [UCSC]) or 5′ ends of cDNAs (Kodzius et al. 2006) within 150 bp, supporting the transcriptional initiation capabilities of the identified regions, although additional ESTs or 5′ ends within the upstream 20-kb regions would have precluded identification of start sites purely through use of EST/cDNA datasets.
Among the 175 miRNAs with discovered transcription initiation regions, 87 were intergenic with novel promoters. Eighty-eight miRNA genes were intronic, with 32 containing clearly identifiable promoters distinct from their host’s. We observed poor expression correlations between these 32 intronic miRNAs and their host’s mRNAs (Supplemental Table S1). Similar correlation analyses were previously carried out for 12 of these, with a similar lack of correlation observed in nine of the 12 (Baskerville and Bartel 2005; Liang et al. 2007), suggesting that these miRNAs can be encoded by both the host and their own distinct transcripts. Correlation coefficients were higher for the 56 intronic miRNAs lacking independent TSSs by our data, although it remains possible that novel TSSs may still exist but could not be detected if located beyond the 20-kb distance examined or their relatively low or absent transcriptional activity in the three cell lines studied.
Some miRNAs are organized in clusters (Ambros 2004). We found either one or no promoter upstream of miRNA clusters, suggesting either shared TSS/promoter structure or the presence of additional promoters not identified by our analysis. In agreement with Baskerville and Bartel (2005) as well as Liang et al. (2007), we observed that the expression of miRNAs within clusters are correlated with each other (Supplemental Material). Furthermore, polycistronic transcripts for several miRNA clusters are known to exist (Lee et al. 2002; Aravin et al. 2003). These observations support the possibility that miRNAs in clusters can be encoded by a common pri-miRNA (Baskerville and Bartel 2005; Liang et al. 2007).
Characterization of miRNA core promoters
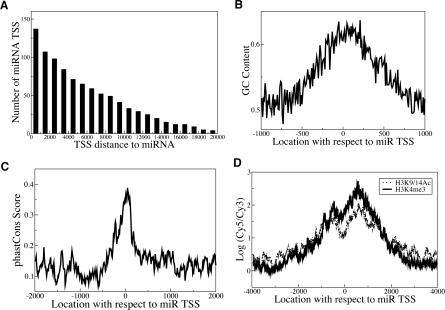
The transcription initiation site distance to the miRNA coding region is variable, ranging from a few hundred bases to 20 kb upstream (Fig. 1A) or longer (Chang et al. 2007). RNAPII-transcribed miRNA promoters are generally similar to known mRNA-encoding promoters. About 64% of miRNA promoters have a CpG island within 500 bp. More precisely, 85% of miRNA promoters corresponding to previously annotated genes have a CpG island, while 51% of novel promoters contain a CpG island. Allowing for 50-bp ambiguity, 25 (19%) of miRNA promoters contain a TATA element, 21% have a TFIIB recognition element (BRE), 64 (47%) have an initiator (Inr), 10 (7%) have a motif 10 element (MTE), 119 (87%) have a downstream promoter element (DPE), and none has a proximal sequence element (PSE).
Figure 1.
miRNA promoters exhibit similarities to known RNAPII promoters. (A) Cumulative histogram showing miRNA promoter distances relative to the corresponding mature miRNA locations in the human genome. The Y-axis represents the number of miRNA promoters with promoter-to-mature miRNA distance greater than or equal to the indicated x-value. (B,C) Average GC content (B) and evolutionary conservation scores (Siepel et al. 2005) (C) are shown at miRNA promoters. The high GC content and evolutionary conservation near transcription initiation regions are similar to the characteristic features found in coding genes (Ozsolak et al. 2007). (D) Composite H3K4me3 and H3K9/14Ac profiles at novel miRNA TSS regions. The X-axis represents the distance relative to the transcription initiation region, the Y-axis the average H3K4me3 or H3K9/14Ac ChIP–chip signal of all miRNA promoters at the indicated x-value. The concentrated H3K4me3 and H3K9/14Ac modifications around transcription initiation regions are similar to the patterns observed for coding gene TSSs (Heintzman et al. 2007).
In comparison, ∼56% of human coding genes have CpG islands. A global search for core elements in promoters from the DBTSS database showed that 22%, 26%, 48%, and 24% of human promoters have TATA, BRE, Inr, and DPE elements, respectively (Gershenzon and Ioshikhes 2005). Therefore, our discovered promoters of miRNAs are similar to those of coding genes in terms of general sequence characteristics. The discrepancy for DPE likely arises from the fact that DPE is a common degenerate 5-mer.
Conservation and GC content increase toward the miRNA promoters (Fig. 1B,C; Fig. Supplemental S1), consistent with mRNA-encoding promoters (Ozsolak et al. 2007). Alignment of H3K4me3 and , with a characteristic dip at the putative TSS (1D), a pattern similar to known genes (Guenther et al. 2007; Koch et al. 2007). The histone modification patterns are indistinguishable between novel miRNA and known coding gene promoters (Supplemental Fig. S2) and are also independent of core promoter elements and CpG islands. RNAPIII’s role in miRNA transcription is discussed below.
Verification of miRNA promoter locations
To test the accuracy of our approach, we first compared our findings with known miRNA TSSs. The method correctly identified the transcription initiation regions for miR-146a and miR-146b (Taganov et al. 2006) in MALME and UACC62, but could not detect any candidate regions in MCF7, as these miRNAs are unexpressed in MCF7. We correctly identified the TSS for miR-155 (Eis et al. 2005) in MALME where it is expressed, but not in UACC62 and MCF7 where it is unexpressed. We could not identify the TSSs for miR-127 (Saito et al. 2006) (miR-127 was unexpressed in the cell lines studied) and miR-34a (Chang et al. 2007) (while miR-34a was expressed in all three lines, its TSS is 30 kb upstream of the mature miR-34a, outside the regions examined). We detected the known TSS of miR-21 in HeLa cells (Cai et al. 2004) and also found a novel transcription initiation region in UACC62, as discussed below. Thus, TSSs for all four expressed, previously annotated miRNAs were correctly identified by this approach.
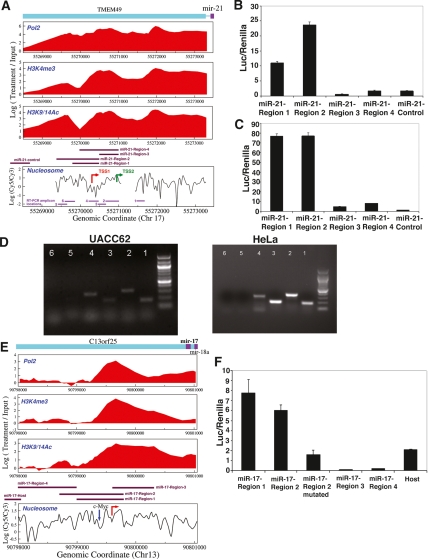
In addition, we experimentally validated six miRNA (miR-17, miR-21, miR-181c/miR-181d cluster, miR-148a, miR-193b/miR-365-1 cluster, and miR-505) transcription initiation regions with RT–PCR and promoter cloning (Fig. 2; Supplemental Fig. S3). miR-17 and miR-21 were chosen because they provide examples of intronic promoters and because of their importance in cancer; the remaining miRNAs were chosen randomly. We first used primers designed to the regions near mature miRNAs for the RT step. We then performed PCR with primer pairs designed to amplify multiple regions between mature miRNAs and the identified promoters as well as regions immediately upstream of the promoters. We detected RNA transcripts extending to the identified transcription initiation regions but not further upstream. Although the initiation regions were reproducible across the cell lines examined here, the approach was also able to identify cell type-specific differences, where they exist, as described below for miR-21. The following section describes Figure 2 and the regulation of miR-17 and miR-21 in detail.
Figure 2.
Identification of miR-21 and miR-17-cluster transcription initiation regions. (A) A nucleosome-depleted area (TSS1) 617 bp upstream of the published position (TSS2) may be the miR-21 TSS. (B,C) To identify regions with promoter function, several segments shown in A were cloned upstream of a luciferase reporter. miR-21-Region-1,2,3,4 and miR-21-control pGL3-basic luciferase reporter constructs (A) (Supplemental Table S6) were transfected into UACC62 (B) and HeLa (C). While miR-21-Region-1 and Region-2 (corresponding to the novel promoter we identified) exhibited promoter activity in both cell lines, we detected weaker activity above negative control levels from region-3 and region-4 (corresponding to the published promoter position) in HeLa, but not in UACC62. Region-4 contained the novel transcription initiation region but not the upstream conserved TF-binding sites. The Y-axis shows the firefly luciferase activity normalized to the control renilla for each sample. (D) RT–PCR was performed using primers designed for the 3′ end of pri-miR-21 construct (Cai et al. 2004) for the RT step. PCR was performed with primer pairs in A (Supplemental Table S7). (E,F) miR-17-cluster genomic regions surrounding the novel (E, red arrow pointing toward right) and host TSSs (Supplemental Table S6) were cloned upstream of luciferase reporter and transfected into UACC62. Unlike negative control constructs (region-3 and region-4), Region-1, Region-2, and host exhibited luciferase activity (F). Mutation of c-Myc site (E, blue down arrow) in miR-17-Region-2 construct (miR-17-Region-2-mutated) resulted in an approximately threefold decrease in luciferase activity relative to the wild-type construct. Data presented are mean ± SEM from three independent experiments.
Six additional transcription initiation regions (miR-205, miR-191, miR-425, miR-423, let-7i, and miR-22) were validated based upon their prior identification in mouse (Fukao et al. 2007). Fukao et al. (2007) found candidate TSSs of 29 miRNAs in mouse by aligning a collection of cDNA sequences from multiple tissues with full-length pre-miRNA sequences. From those 29, 23 had human homologs, and we discovered the transcription initiation regions of 10 homologous miRNAs (miR-205, miR-191, miR-425, miR-423, let-7i, miR-22, miR-339, miR-100, let-7d, and miR-21) in humans. Except for miR-339, we were able to align the TSS regions from human to mouse using the UCSC Batch Coordinate Conversion tool with a minimum of 50% match, and six (miR-205, miR-191, miR-425, miR-423, let-7i, and miR-22) of the mapped TSS regions agreed well with the TSSs found in mouse within 224 ± 182 bp, compared with 569 ± 1182 bp for 500-bp upstream promoter regions of human RefSeq genes aligned to corresponding homologous mouse RefSeq promoters using the UCSC Batch Coordinate Conversion tool with a minimum of 50% match. We could not align the promoter of hsa-miR-339 to the mouse because the flanking regions of hsa-miR-339 were deleted in the mouse. We examined the differences in the three remaining pri-miRNAs (miR-100, let-7d, and miR-21). While we found a promoter region 7 kb upstream of hsa-miR-100, the TSS of an mRNA 130 kb upstream of mmu-miR-100 was found in the mouse. Likewise, while we found an independent intronic transcription initiation region for hsa-miR-21, 3 kb away from mature miR-21, Fukao et al. (2007) found the TSS of the host gene that is almost 100 kb upstream. For hsa-let-7d, we found one long pri-miRNA that encodes hsa-let-7f-1, hsa-let-7d, and hsa-let-7a-1, while a transcript that encodes only mmu-let-7d was found in the mouse. Our transcription initiation region actually corresponds to the TSS of another mouse transcript (AK042687) that encodes all three miRNAs, suggesting that the TSS found by Fukao et al. (2007) may be specific to the mouse and that the transcription initiation region identified here may be utilized in both human and mouse (see Supplemental Fig. S4 for a picture of the genomic locus containing mmu-let-7d).
In summary, we reproduced four previously known miRNA transcription initiation regions and validated six additional intronic and intergenic miRNA promoters experimentally. No false positive predictions were thus found in biochemical validations of 10/10 miRNA promoters. Furthermore, we observed that six additional miRNA transcription initiation regions map to the correct candidate TSSs found in the mouse (Fukao et al. 2007).
Further validation of novel promoters of miRNAs
To further demonstrate the ability of our chromatin-based method to discover novel miRNA promoters, we studied miR-21 and miR-17 in detail. The pri-miR21 transcript was originally cloned from HeLa cells (Cai et al. 2004). The published TSS is in a RNAPII-, H3K4me3-, and H3K9/14Ac-positive region (Fig. 2A). However, based on our data, a nucleosome-depleted region ∼617 bp upstream of the published position was the candidate miR-21 transcription initiation region in UACC62 melanoma cells. To investigate this discrepancy, we cloned constructs covering the two potential TSSs upstream of a luciferase reporter. We detected significant luciferase activity in UACC62 cells from the ∼617-bp upstream region, but not from the previously annotated position (Fig. 2B). In HeLa, luciferase activity was strongest in the upstream region, and the previous TSS also had a detectable but much weaker activity (Fig. 2C). Furthermore, we performed RT–PCR in UACC62 and HeLa cells using primers designed to the 3′ end of the pri-miR-21 transcript for the RT step and PCR primer pairs indicated in Figure 2A. We detected RNA products upstream of the published position, but not upstream of the novel promoter region (Fig. 2D), suggesting that the pri-miR-21 transcript may extend upstream of the published TSS in UACC62 and HeLa (Cai et al. 2004). Our novel promoter region had STAT3-binding activities (Loffler et al. 2007) and CREB-binding activities (data not shown) within 300 bp upstream and also had several other computationally predicted conserved TF-binding sites in the proximal promoter. On the other hand, there were no conserved TF-binding sites between the two TSSs. It could thus be that TF-binding activities favor our novel transcription initiation region, while a HeLa-specific TF may also promote the downstream TSS at a lower level in HeLa.
The intronic miR-17-cluster is a potential “oncomiR” (Cho 2007) regulated by c-Myc binding to the first intron of its host (He et al. 2005; O’Donnell et al. 2005). Previous attempts to map the miR-17 TSS were unsuccessful due to the high GC content of upstream regions (Woods et al. 2007). Our analysis suggested that the miR-17 TSS is ∼2 kb downstream from the host TSS in the three cell lines we studied (Fig. 2E). Luciferase assays showed that both the host and novel transcription initiation regions have promoter activities in UACC62 (Fig. 2F). After confirming c-Myc binding to the published position in UACC62 and MALME (data not shown), we generated reporter constructs including the c-Myc-binding site (wild-type CATGTG or mutated to CATATA) and the novel initiation region. Mutation of the c-Myc site caused luciferase expression to decrease approximately threefold (Fig. 2F). Down-regulation of c-Myc levels by siRNA caused the novel promoter activity to decrease approximately twofold (Supplemental Fig. S5). The miR-17-cluster may thus be encoded by transcripts emanating from both TSSs, and c-Myc regulation of the miR-17 cluster may take place through the intronic promoter.
Predicting TFs that regulate miRNAs: MITF-targeted miRNAs
The ultimate utility of knowing miRNA promoters is in studying the regulation of miRNA expression. Because regulatory proteins preferentially bind nucleosome-free linker DNA (Yuan et al. 2005; Ozsolak et al. 2007), nucleosome occupancy information surrounding the miRNA transcription initiation regions can be used to discover TFs regulating miRNA expression. To demonstrate the power of this approach, we screened the miRNA promoters for E-box elements recognized by MITF, a melanoma oncogene and master transcriptional regulator of melanocyte development (Du et al. 2004). Seventy-three potential binding sites were identified within 1 kb upstream of and 250 bp downstream from 175 miRNA transcription initiation regions. Of these, 50 (68%) were conserved and 19 (26%) showed both conservation and nucleosome depletion (Supplemental Material). Using ChIP, we observed no binding by MITF to 10 randomly chosen nucleosome-occupied E-boxes (out of 54) (data not shown). In contrast, 10 (corresponding to nine miRNAs) of the 19 nucleosome-free E-boxes were occupied by MITF (Table 1).
Table 1.
miRNAs with MITF occupancy near their promoters

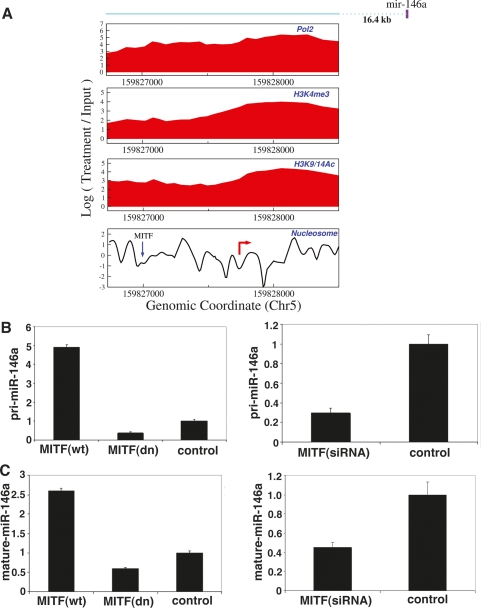
Since the E-box elements may be occupied by TFs other than MITF, we examined published c-Myc ChIP–chip data and found c-Myc to occupy two of the nine non-MITF-bound promoters, including the miR-17 cluster (Fig. 2E). miR-146a (Fig. 3A–C), miR-221/222 (Supplemental Fig. S6) and miR-363 (Supplemental Fig. S7) levels increased in response to MITF overexpression, and decreased in response to MITF down-regulation. Other MITF-targeted miRNAs were also seen to correlate with MITF expression and to be specific to melanoma (Supplemental Material).
Figure 3.
MITF-regulated miRNAs. (A) MITF occupancy (blue down arrow) of an E-box-containing nucleosome-depleted region upstream of miR-146a transcription initiation region (red right-pointing arrow) was shown by ChIP (Supplemental Material) in MALME and UACC62. (B,C) Both pri-miR-146a (B) and mature miR-146a (C) expression levels are increased and decreased by MITF overexpression and down-regulation, respectively, using adenoviruses expressing wild-type (wt) and naturally occurring dominant-negative (dn) MITF (Du et al. 2004) and siRNA against MITF in UACC62. Data presented are mean ± SEM from three independent experiments.
Many of the known or predicted target genes of MITF-targeted miRNAs identified here, such as c-kit (Felli et al. 2005) and p27 (le Sage et al. 2007) by miR-221/222, mcl1 (Mott et al. 2007) by mir-29b and mir-125b, have well-defined roles in pigmentation and survival of melanocytes, potentially linking MITF’s pigmentation/survival roles to miRNA function. This approach can thus rapidly identify TFs likely to modulate target gene expression through identification of their binding motifs within nucleosome-depleted regions.
Evolution of intronic miRNAs and their promoters
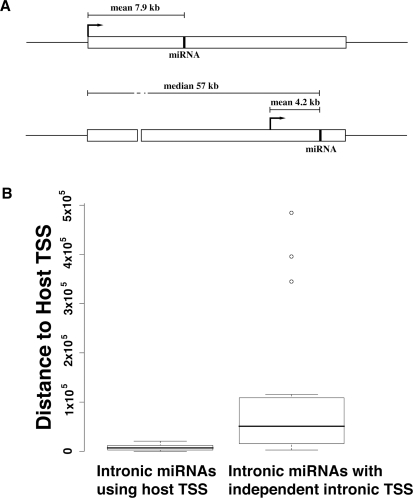
The high-throughput identification of miRNA promoter elements also sheds light on evolution of miRNA transcription mechanisms. Our study revealed that the mean distance between host TSSs and intronic miRNAs utilizing the host TSSs was 7.9 kb ± 6.2 kb, while the mean distance between other intronic and intergenic miRNAs and their independent TSSs was 5.3 kb ± 4.9 kb, significantly shorter than the former (P-value = 1.9 × 10−2, twosided Wicoxon Rank Sum Test). Intronic miRNAs with independent transcription initiation regions had the shortest TSS-to-miRNA distance with mean 4.2 kb ± 3.5 kb; at the same time, these miRNAs were located quite significantly farther (median 57 kb) from their host gene TSSs than the intronic miRNAs utilizing host gene TSSs (P-value = 5.1 × 10−7, two-sided Wilcoxon Rank Sum Test) (Fig. 4A,B). These results suggest that certain intronic miRNAs located far away from their host TSSs may have evolved to adopt nearby independent novel TSSs in order to make the transcription of pri-miRNA faster and more efficient.
Figure 4.
Intronic miRNAs with independent intronic transcription initiation regions. (A) Intronic miRNAs with independent intronic transcription initiation regions are located significantly farther (median distance 57 kb) from host TSSs than those that share the host TSSs. miRNA TSSs are indicated by arrows. (B) Box plots of distance between host TSS and mature miRNA for intronic miRNAs with and without independent transcription initiation regions. There was a significant difference in the two distributions (P-value = 5.1 × 10−7), indicating that intronic miRNAs that have acquired independent promoters for faster transcription are likely located far from host gene TSSs.
We found further differences between the promoters of conserved and nonconserved intronic miRNAs. In particular, intronic miRNAs with their own promoters were more evolutionarily conserved than other intronic miRNAs. To analyze nonconserved miRNAs, we considered a set of 31 intronic miRNAs (1) that have no known homolog in the mouse and (2) <95% of whose pre-miRNAs can be aligned to the mouse genome by using BLAT (Kent 2002). Only six (19%) out of the 31 nonconserved intronic miRNA clusters had their own promoters independent of their host (Supplemental Table S9). On the other hand, out of 43 conserved intronic miRNA clusters with homologues in mouse, 17 (40%) had their own promoters (Supplemental Table S9), a significantly higher proportion compared with nonconserved miRNAs (P-value = 0.05, Fisher’s Exact Test). These observations thus suggest that the “old” intronic miRNAs are more likely to have independent promoters than the relatively recent intronic miRNAs, supporting the hypothesis that as intronic miRNAs arose, they first utilized the host TSSs and may have acquired functional independent promoters later through evolution.
RNAPIII contribution to miRNA transcription
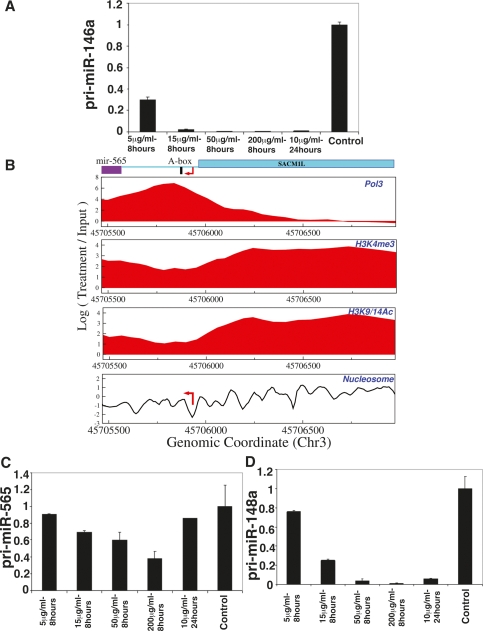
A miRNA cluster was previously found to be transcribed by RNAPIII (Borchert et al. 2006). To find additional RNAPIII-transcribed miRNAs, we performed ChIP–chip for RPC53 (RNAPIII subunit with no paralogs in RNAPI and RNAPII) (Schramm and Hernandez 2002). We identified 11 RPC53-occupied miRNA promoters, which exhibited RNAPIII promoter elements such as A and B boxes (Supplemental Material). As an independent test, we examined α-amanitin sensitivity of several of these miRNAs. α-Amanitin inhibits RNAPII selectively by blocking its translocation along the DNA (Bushnell et al. 2002; Gong et al. 2004), except at high doses where it inhibits both RNAPII and RNAPIII (Lindell et al. 1970). Two RNAPII-occupied miRNAs (miR-146a and let-7d cluster) showed α-amanitin sensitivity at all doses tested (Fig. 5A; Supplemental Fig. S8A). In contrast, the RNAPIII-occupied miRNAs miR-565 (Fig. 5B,C) and mir-128b (Supplemental Fig. S8B) were resistant except at high doses. Eight miRNA promoters exhibited both RNAPII and RNAPIII occupancy. Their α-amanitin sensitivity resembled that of RNAPII-transcribed miRNAs (such as mir-148a in Fig. 5D). We also observed RPC39 and/or RPC62 (two other RNAPIII-specific subunits) localization to these nine promoters (data not shown), suggesting that these are indeed RNAPII- and RNAPIII-occupied, rather than potentially arising through antibody cross-reactivity.
Figure 5.
RNAPIII-occupied miRNAs. (A) α-Amanitin sensitivities of RNAPII-transcribed miR-146a. The level of pri-miR-146a goes down quickly at a low dose of α-amanitin, supporting that it is transcribed by RNAPII and not by RNAPIII. (B) RNAPIII occupancy was observed in proximity to miR-565 transcription initation region. (C,D) α-Amanitin sensitivities of RNAPIII-transcribed miR-565 (C) and RNAPII- and RNAPIII-occupied mir-148a (D) were measured. The Y-axis shows fold change relative to the control sample, normalized to 18S rRNA. Data presented are means ± SEM from three independent experiments.
Distinct chromatin organization surrounding mature miRNA-encoding DNA sequences and transcription end sites
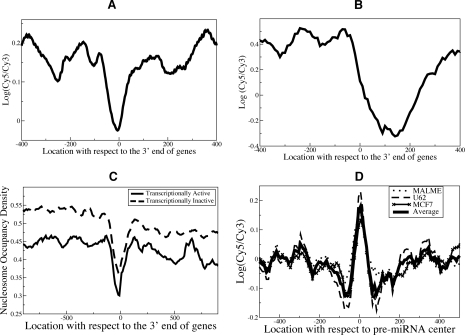
Nucleosomes modulate the accessibility of underlying sequences to regulatory proteins (Anderson and Widom 2000). For instance, TSSs of active genes and TF-binding sites tend to be nucleosome-depleted (Mito et al. 2005; Yuan et al. 2005; Ozsolak et al. 2007). We found that the 3′ ends of genes in humans are also nucleosome-depleted (Fig. 6A). We observed the same trend in yeast (Yuan et al. 2005), although the nucleosome-free nadir was located ∼150 nucleotides 3′ of the translation end site (Fig. 6B). To examine the biological significance of this finding, we analyzed the whole-genome RNAPII ChIP–chip data in MCF7 (Carroll et al. 2006). We observed that the RNAPII (Carroll et al. 2006) occupancy was indeed significantly high downstream from the 3′ ends of genes (Supplemental Figure S9A), suggesting that antisense transcription at the 3′ ends of genes (Kampa et al. 2004; Kapranov et al. 2005) and RNAPII stalling before termination may cause this pattern. While our manuscript was under review, a study reported a similar pattern at the 3′ ends of yeast genes (Mavrich et al. 2008). In that study, Mavrich et al. (2008) found antisense transcripts initiating at the 3′ ends of genes and DNA loops connecting the 5′ and 3′ ends of genes, thus linking the pattern with active transcription. However, we observed nucleosome depletion at the 3′ ends of genes that do not exhibit transcriptional activity (we defined a gene as transcriptionally silent if the gene was unexpressed based on expression profiling and there was no RNAPII binding within the gene and immediate upstream and downstream regions) (Fig. 6C). This suggests additional factors beyond RNAPII pausing and antisense transcription for the observed depletion. For instance, sequence characteristics (such as polyadenylation signaling motifs) of these regions may inherently disfavor nucleosome occupancy.
Figure 6.
Nucleosome positioning in genomic regions surrounding the 3′ end of genes and mature miRNAs-coding sequences exhibits distinct patterns. (A,B) Nucleosome positioning signals at the 3′ end of genes of RefSeq genes in HeLa (A) and translation end sites in yeast (Yuan et al. 2005) (B) were aligned, and average probe signals were calculated. (C) We analyzed the nucleosome positioning maps generated by another group for human CD4+ T cells (Schones et al. 2008) and observed a similar nucleosome depletion at the 3′ ends of transcriptionally active and inactive human genes, further validating our findings. Transcriptionally active genes have less nucleosome occupancy, which may be simply due to nucleosome remodeling/eviction during transcription. (D) Nucleosome positioning signals surrounding all pre-miRNAs were aligned at pre-miRNA centers, and average probe signals were calculated. Preferential occupancy of positioned nucleosomes in pre-miRNA regions (Supplemental Fig. S9C) was observed.
Finally, we observed that the pre-miRNA genomic regions are preferentially occupied by nucleosomes in the three cell lines we studied and CD4+ T cells (Fig. 6D; Supplemental Fig. S9B,C). The observed pattern is independent of the miRNA expression status. The pre-miRNA regions have higher GC content compared with surrounding regions, thus favoring nucleosome formation (Peckham et al. 2007), but exhibit only very weak periodic dinucleotides (Supplemental Material). While the cause and functional significance are not immediately clear, such nucleosome positioning may protect miRNA sequences from external mutagenic pressures or potentially serve as a signal for RNA processing factors.
Discussion
miRNAs play increasingly well understood roles in numerous settings, including normal development and disease pathology (Ambros 2004; Bartel 2004; Alvarez-Garcia and Miska 2005). While mechanisms underlying their post-transcriptional processing and gene silencing effects are yielding important insights, considerably less is known about the pathways governing control of miRNA expression at the transcriptional level. As described here, a high-throughput genomics-based strategy has been applied, first at identification of nucleosome-depleted regions containing TSSs. Then, using high-resolution nucleosome positioning information coupled to ChIP studies, candidate transcriptional regulators could be identified and rapidly tested. The example of MITF was employed here. MITF is a Myc-related basic helix–loop–helix–leucine-zipper family TF whose loss of function results in melanocyte absence (Steingrimsson et al. 2005), and whose oncogenic gene amplification occurs in 10%–20% of human melanomas (Garraway et al. 2005). Among its known transcriptional targets are most (if not all) of the major components of the melanin biosynthesis and processing machinery in melanocytes (Steingrimsson et al. 2004). The identification of candidate MITF-targeted miRNAs through this high-throughput approach significantly broadens the repertoire of biological effects attributable to the TF—a repertoire that can straightforwardly be applied to other TFs as well, with the availability of promoter annotation of miRNAs and functional nucleosome-depleted regions in miRNA promoters. As many of our annotated miRNAs are oncomiRs (e.g., miR-17 and miR-21) or known to play crucial roles in hematopoietic differentiation, lymphoma (miR-10a, miR-155, miR-16, miR-146, and miR-221/222) (Georgantas et al. 2007), and epithelial mesenchymal transition (mir-200 family and miR-205) (Gregory et al. 2008), the ability to identify transcriptional regulators of these miRNAs may provide a useful resource for study of both development and disease pathogenesis.
The method employed here revealed a significant fraction of miRNAs that use their own (novel) transcription initiation regions, including both miRNAs located at intergenic regions and others that are embedded within introns of known coding genes. An additional fraction of intron-embedded miRNAs appear to share TSS use with their host gene, suggesting that alternative post-transcriptional processing plays a key role in determining the fate of each primary transcript. Our study showed that not only are the intronic miRNAs with their own TSSs significantly far from their host TSSs, but they are also more evolutionarily conserved than intronic miRNAs using host TSSs. Our analysis also suggests that while many of the pri-miRNAs are well conserved between human and mouse, there exist important exceptions where novel TSSs—e.g., miR-339—were acquired independently through chromosomal rearrangements or insertions.
It has been shown that certain exons exhibit conserved promoter activities, especially in tissue-specific genes (Carninci et al. 2006). In this work, we found eight intronic miRNA clusters that had independent promoters located in exons, and we thus showed a novel role of exonic promoters for transcribing noncoding RNAs. Carninci et al. (2006) have also observed several bidirectional promoters near CpG islands. Likewise, we found that eight out of 10 miRNAs in divergent bidirectional promoters had a CpG island; e.g., the RNAPIII-transcribed miR-565.
Importantly, the current study has focused on only a limited set of cell types, and possibilities of alternative TSS usage remain possible and indeed are suggested by our findings for miR-21. Given the existence of variable TSS usage as well as alternative TF regulation of specific miRNAs, the use of a high-throughput approach for miRNA proximal promoter annotation (as described here) may significantly facilitate such studies in numerous cell systems, both in normal development and disease. While this approach can only detect transcriptionally active TSS regions in a given cell type and only in nonrepeat regions of the genome that can be represented on microarrays, the use of multiple cell types and emerging technologies such as high-throughput sequencing (Barski et al. 2007; Mikkelsen et al. 2007) may alleviate these limitations.
Several striking molecular features of miRNA gene structure or regulation were observed in the combined nucleosome positioning and ChIP–chip studies reported here. The identification of RNAPIII at multiple miRNA genes suggests a role for this polymerase in addition to RNAPII in miRNA transcription, a finding consistent with earlier reports (Borchert et al. 2006). It remains to be determined whether RNAPII versus RNAPIII occupancy is associated with significant differences in miRNA expression, either in magnitude or regulatory mechanism. A recent study by Listerman et al. (2007) shows that, while U6 snRNA is synthesized by RNAPIII, it is α-amanitin-sensitive. RNAPII is also recruited to U6 snRNA genes and functions to enhance RNAPIII transcription of U6 genes, possibly by facilitating an open chromatin formation for transcription. Although further experiments are needed to determine the precise contributions of RNAPII and RNAPIII to the transcription of the RNAPIII-targeted miRNAs reported here, it is conceivable that similar mechanisms are in place, and both RNAPs cooperate to transcribe these miRNAs. This approach might not detect RNAPIII occupancy in certain repeat-regions transcribed by RNAPIII (e.g., Alu) but masked from the microarray design.
It was also observed that the body of mature miRNA genomic coding regions was strikingly nucleosome-occupied. The functional meaning of this nucleosome patterning is not clear as yet. It could reflect a secondary effect of (as yet) unidentified epigenetic alterations at these genomic locations, and may serve to “mark” miRNA regions for binding by factors that participate in pre-miRNA processing. The pattern of nucleosome occupancy over the miRNA “coding” regions and 3′ ends of known genes observed in this study, as well as previous observations that transcriptionally active TSSs are nucleosome-depleted (Mito et al. 2005; Yuan et al. 2005; Ozsolak et al. 2007) suggest an important interface between chromatin structure and regulation of chromatin function.
Materials and methods
Cell culture and media
MALME and UACC62 were maintained in RPMI with 10% FBS. MCF7 and HeLa were cultured in DMEM with 10% FBS. MALME, HeLa, and UACC62 were obtained from NCI and MCF7 from Myles Brown (Dana-Farber Cancer Institute).
ChIP and ChIP–chip
ChIP and DNA purification of samples were performed as described previously (Ozsolak et al. 2007). The antibodies used were α-H3K4me3 (Upstate Biotechnologies, 07-473), α-H3K9/14Ac (Upstate Biotechnologies, 06-599), α-RNAPII (Abcam, ab5408), MITF (C5), c-MYC (Santa Cruz Biotechnologies, sc-764), and CREB (Upstate Biotechnologies, 06-863). RNAPIII antibodies for RPC53, RPC39, and RPC62 was provided by Robert G. Roeder and have been used previously for ChIP (Borchert et al. 2006). The MITF-binding regions in close proximity to the identified miRNA promoter regions were determined by ChIP (Du et al. 2004; Ozsolak et al. 2007), followed by qPCR analysis using iCycler and SYBR Green iQ SuperMix (Bio-Rad) as described (Heintzman et al. 2007). For ChIP–chip analyses, LM-PCR amplification of samples was performed as described (Ozsolak et al. 2007). Sample labeling and hybridization to synthesized microarrays were performed by Roche NimbleGen, Inc., using standard procedures.
Global nucleosome positioning analysis
Experimental mononucleosome fragments and control DNA samples were obtained under native conditions by partial micrococcal nuclease digestions as described (Ozsolak et al. 2007). Sample labeling, microarray synthesis and hybridization were performed by Roche NimbleGen, Inc., using standard procedures for nucleosome DNA.
miRNA expression analysis
Expression profiles of miRNAs (mirBase version 9.0) in MALME, UACC62, and MCF7 cell lines were performed by LCSciences. Expression data for three cell lines on Affymetrix HGU133 Plus 2.0 arrays were obtained from prior publications (Carroll et al. 2006; Ozsolak et al. 2007) and analyzed as described (Ozsolak et al. 2007). Mature miRNA expression levels were quantified by TaqMan MicroRNA Assays (Applied Biosystems).
Adenovirus infection and RNA precipitation
Cells were infected with adenoviruses to express wild-type MITF, dominant-negative MITF and poly-P control proteins as described (Du et al. 2004). RNA was isolated with mirVana miRNA isolation kit (Ambion) 48 and 72 h after infections.
siRNA transfections
UACC62 cells were transfected with siRNAs against MITF and c-Myc and negative control siRNA (Dharmacon) with Lipofectamine 2000 (Invitrogen) reagent following the manufacturers’ standard protocols. RNA was isolated with the mirVana miRNA isolation kit (Ambion) at 72 and 96 h after transfection.
Cloning
We cloned candidate promoter elements from normal human genomic DNA (BD Biosciences Clontech) using Pfu Polymerase (Stratagene) or with the GC-RICH PCR System (Roche Applied Science) when the GC content of the cloned fragment was high. Primers (synthesized by Operon) were designed with appropriate extensions for cloning into digested pGL3 basic (Promega) plasmid not containing an internal promoter element. Generally, the regions between ∼100 bp downstream from an identified promoter element and 700 bp upstream were cloned. Where indicated, longer fragments were cloned. All clones were sequenced for verification.
Site-directed mutagenesis of the c-Myc-binding site in mir-17 cluster promoter regions was performed using the QuickChange method (Stratagene) according to the supplier’s recommendations. Mutagenesis primers were designed as follows: P1, CCCCTTGTGCGACATATACTGCCGGCCCGG; and P2, CC GGGCCGGCAGTATATGTCGCACAAGGGG.
Luciferase reporter assays
Cells were transfected with 200 ng pGL3-basic vector carrying the identified promoter elements and negative and positive control regions and 1 ng pRL-CMV (Promega) in 24-well format using Lipofectamine 2000 (Invitrogen) reagent following the manufacturer’s recommendations. The cells were assayed with the Dual Luciferase kit (Promega). All measurements were done in triplicate and signals were normalized for transfection efficiency to the internal Renilla control.
Raw data
The nucleosome microarray data are available from Gene Expression Omnibus under accession number GSE9029, http://www.ncbi.nlm.nih.gov/geo.
Acknowledgments
We thank C.S. Chan, R. Cui, V. Igras, A. Kawakami, M. Khaled, A.J. Levine, E. Makino, V. Nanda, and S. Yokoyama for discussions and support. This work was supported by a grant from NIH to D.E.F. D.E.F. is a Distinguished Clinical Scholar of the Doris Duke Medical Foundation. H.L. and X.Z. are employees of Roche NimbleGen, Inc. D.E.F. wishes to disclose consulting relationships with Novartis Pharmaceuticals, Magen BioSciences, and Source MDx.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1706508.
References
- Alvarez-Garcia I., Miska E.A. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Anderson J.D., Widom J. Sequence and position-dependence of the equilibrium accessibility of nucleosomal DNA target sites. J. Mol. Biol. 2000;296:979–987. doi: 10.1006/jmbi.2000.3531. [DOI] [PubMed] [Google Scholar]
- Aravin A.A., Lagos-Quintana M., Yalcin A., Zavolan M., Marks D., Snyder B., Gaasterland T., Meyer J., Tuschl T. The small RNA profile during Drosophila melanogaster development. Dev. Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baskerville S., Bartel D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein B.E., Kamal M., Lindblad-Toh K., Bekiranov S., Bailey D.K., Huebert D.J., McMahon S., Karlsson E.K., Kulbokas E.J., Gingeras T.R., et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Borchert G.M., Lanier W., Davidson B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- Bracht J., Hunter S., Eachus R., Weeks P., Pasquinelli A.E. Trans-splicing and polyadenylation of let-7 microRNA primary transcripts. RNA. 2004;10:1586–1594. doi: 10.1261/rna.7122604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushnell D.A., Cramer P., Kornberg R.D. Structural basis of transcription: α-Amanitin-RNA polymerase II cocrystal at 2.8 Å resolution. Proc. Natl. Acad. Sci. 2002;99:1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Hagedorn C.H., Cullen B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Carlson D.P., Ross J. Human β-globin promoter and coding sequences transcribed by RNA polymerase III. Cell. 1983;34:857–864. doi: 10.1016/0092-8674(83)90543-3. [DOI] [PubMed] [Google Scholar]
- Carninci P., Sandelin A., Lenhard B., Katayama S., Shimokawa K., Ponjavic J., Semple C.A., Taylor M.S., Engstrom P.G., Frith M.C., et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- Carroll J.S., Meyer C.A., Song J., Li W., Geistlinger T.R., Eeckhoute J., Brodsky A.S., Keeton E.K., Fertuck K.C., Hall G.F., et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Chang T.C., Wentzel E.A., Kent O.A., Ramachandran K., Mullendore M., Lee K.H., Feldmann G., Yamakuchi M., Ferlito M., Lowenstein C.J., et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol. Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W.C. OncomiRs: The discovery and progress of microRNAs in cancers. Mol. Cancer. 2007;6:60. doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J., Sussman D.J., Zeller R., Leder P. The c-myc gene encodes superimposed RNA polymerase II and III promoters. Cell. 1987;51:1001–1008. doi: 10.1016/0092-8674(87)90586-1. [DOI] [PubMed] [Google Scholar]
- Cullen B.R. Transcription and processing of human microRNA precursors. Mol. Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Du J., Widlund H.R., Horstmann M.A., Ramaswamy S., Ross K., Huber W.E., Nishimura E.K., Golub T.R., Fisher D.E. Critical role of CDK2 for melanoma growth linked to its melanocyte-specific transcriptional regulation by MITF. Cancer Cell. 2004;6:565–576. doi: 10.1016/j.ccr.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Eis P.S., Tam W., Sun L., Chadburn A., Li Z., Gomez M.F., Lund E., Dahlberg J.E. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc. Natl. Acad. Sci. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felli N., Fontana L., Pelosi E., Botta R., Bonci D., Facchiano F., Liuzzi F., Lulli V., Morsilli O., Santoro S., et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc. Natl. Acad. Sci. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felton-Edkins Z.A., Kenneth N.S., Brown T.R., Daly N.L., Gomez-Roman N., Grandori C., Eisenman R.N., White R.J. Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle. 2003;2:181–184. [PubMed] [Google Scholar]
- Fukao T., Fukuda Y., Kiga K., Sharif J., Hino K., Enomoto Y., Kawamura A., Nakamura K., Takeuchi T., Tanabe M. An evolutionarily conserved mechanism for microRNA-223 expression revealed by microRNA gene profiling. Cell. 2007;129:617–631. doi: 10.1016/j.cell.2007.02.048. [DOI] [PubMed] [Google Scholar]
- Garraway L.A., Widlund H.R., Rubin M.A., Getz G., Berger A.J., Ramaswamy S., Beroukhim R., Milner D.A., Granter S.R., Du J., et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- Georgantas R.W., Hildreth R., Morisot S., Alder J., Liu C.G., Heimfeld S., Calin G.A., Croce C.M., Civin C.I. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: A circuit diagram of differentiation control. Proc. Natl. Acad. Sci. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenzon N.I., Ioshikhes I.P. Synergy of human Pol II core promoter elements revealed by statistical sequence analysis. Bioinformatics. 2005;21:1295–1300. doi: 10.1093/bioinformatics/bti172. [DOI] [PubMed] [Google Scholar]
- Gomez-Roman N., Grandori C., Eisenman R.N., White R.J. Direct activation of RNA polymerase III transcription by c-Myc. Nature. 2003;421:290–294. doi: 10.1038/nature01327. [DOI] [PubMed] [Google Scholar]
- Gong X.Q., Nedialkov Y.A., Burton Z.F. α-Amanitin blocks translocation by human RNA polymerase II. J. Biol. Chem. 2004;279:27422–27427. doi: 10.1074/jbc.M402163200. [DOI] [PubMed] [Google Scholar]
- Gregory P.A., Bert A.G., Paterson E.L., Barry S.C., Tsykin A., Farshid G., Vadas M.A., Khew-Goodall Y., Goodall G.J. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- Guenther M.G., Levine S.S., Boyer L.A., Jaenisch R., Young R.A. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Hannon G.J. MicroRNAs: Small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- He L., Thomson J.M., Hemann M.T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S.W., Hannon G.J., et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman N.D., Stuart R.K., Hon G., Fu Y., Ching C.W., Hawkins R.D., Barrera L.O., Van Calcar S., Qu C., Ching K.A., et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Houbaviy H.B., Dennis L., Jaenisch R., Sharp P.A. Characterization of a highly variable eutherian microRNA gene. RNA. 2005;11:1245–1257. doi: 10.1261/rna.2890305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Johnson S.S., Zhang C., Fromm J., Willis I.M., Johnson D.L. Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol. Cell. 2007;26:367–379. doi: 10.1016/j.molcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Kampa D., Cheng J., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Drenkow J., Cheng J., Long J., Helt G., Dike S., Gingeras T.R. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res. 2005;15:987–997. doi: 10.1101/gr.3455305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent W.J. BLAT—The BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Ren B. Genome-wide analysis of protein–DNA interactions. Annu. Rev. Genomics Hum. Genet. 2006;7:81–102. doi: 10.1146/annurev.genom.7.080505.115634. [DOI] [PubMed] [Google Scholar]
- Koch C.M., Andrews R.M., Flicek P., Dillon S.C., Karaoz U., Clelland G.K., Wilcox S., Beare D.M., Fowler J.C., Couttet P., et al. The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodzius R., Kojima M., Nishiyori H., Nakamura M., Fukuda S., Tagami M., Sasaki D., Imamura K., Kai C., Harbers M., et al. CAGE: Cap analysis of gene expression. Nat. Methods. 2006;3:211–222. doi: 10.1038/nmeth0306-211. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Landgraf P., Rusu M., Sheridan R., Sewer A., Iovino N., Aravin A., Pfeffer S., Rice A., Kamphorst A.O., Landthaler M., et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee Y., Jeon K., Lee J.T., Kim S., Kim V.N. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Sage C., Nagel R., Egan D.A., Schrier M., Mesman E., Mangiola A., Anile C., Maira G., Mercatelli N., Ciafre S.A., et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Carey M., Workman J.L. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Liang Y., Ridzon D., Wong L., Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. doi: 10.1186/1471-2164-8-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindell T.J., Weinberg F., Morris P.W., Roeder R.G., Rutter W.J. Specific inhibition of nuclear RNA polymerase II by α-amanitin. Science. 1970;170:447–449. doi: 10.1126/science.170.3956.447. [DOI] [PubMed] [Google Scholar]
- Listerman I., Bledau A.S., Grishina I., Neugebauer K.M. Extragenic accumulation of RNA polymerase II enhances transcription by RNA polymerase III. PLoS Genet. 2007;3:e212. doi: 10.1371/journal.pgen.0030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffler D., Brocke-Heidrich K., Pfeifer G., Stocsits C., Hackermuller J., Kretzschmar A.K., Burger R., Gramatzki M., Blumert C., Bauer K., et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- Mavrich T.N., Ioshikhes I.P., Venters B.J., Jiang C., Tomsho L.P., Qi J., Schuster S.C., Albert I., Pugh B.F. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen T.S., Ku M., Jaffe D.B., Issac B., Lieberman E., Giannoukos G., Alvarez P., Brockman W., Kim T.K., Koche R.P., et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y., Henikoff J.G., Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat. Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Mott J.L., Kobayashi S., Bronk S.F., Gores G.J. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Ozsolak F., Song J.S., Liu X.S., Fisher D.E. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- Pasquinelli A.E., Hunter S., Bracht J. MicroRNAs: A developing story. Curr. Opin. Genet. Dev. 2005;15:200–205. doi: 10.1016/j.gde.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Peckham H.E., Thurman R.E., Fu Y., Stamatoyannopoulos J.A., Noble W.S., Struhl K., Weng Z. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasterk R.H. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- Pokholok D.K., Harbison C.T., Levine S., Cole M., Hannett N.M., Lee T.I., Bell G.W., Walker K., Rolfe P.A., Herbolsheimer E., et al. Genome-wide map of nucleosome acetylation and methylation in yeast. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Reina J.H., Azzouz T.N., Hernandez N. Maf1, a new player in the regulation of human RNA polymerase III transcription. PLoS One. 2006;1:e134. doi: 10.1371/journal.pone.0000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Saito Y., Liang G., Egger G., Friedman J.M., Chuang J.C., Coetzee G.A., Jones P.A. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H., Schneider R., Bannister A.J., Sherriff J., Bernstein B.E., Emre N.C., Schreiber S.L., Mellor J., Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Schneider R., Bannister A.J., Myers F.A., Thorne A.W., Crane-Robinson C., Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat. Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- Schones D.E., Cui K., Cuddapah S., Roh T.Y., Barski A., Wang Z., Wei G., Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm L., Hernandez N. Recruitment of RNA polymerase III to its target promoters. Genes & Dev. 2002;16:2593–2620. doi: 10.1101/gad.1018902. [DOI] [PubMed] [Google Scholar]
- Shivdasani R.A. MicroRNAs: Regulators of gene expression and cell differentiation. Blood. 2006;108:3646–3653. doi: 10.1182/blood-2006-01-030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S., et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E., Copeland N.G., Jenkins N.A. Melanocytes and the microphthalmia transcription factor network. Annu. Rev. Genet. 2004;38:365–411. doi: 10.1146/annurev.genet.38.072902.092717. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E., Copeland N.G., Jenkins N.A. Melanocyte stem cell maintenance and hair graying. Cell. 2005;121:9–12. doi: 10.1016/j.cell.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Taganov K.D., Boldin M.P., Chang K.J., Baltimore D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner B.M. Cellular memory and the histone code. Cell. 2002;111:285–291. doi: 10.1016/s0092-8674(02)01080-2. [DOI] [PubMed] [Google Scholar]
- Woods K., Thomson J.M., Hammond S.M. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J. Biol. Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- Yuan G.C., Liu Y.J., Dion M.F., Slack M.D., Wu L.F., Altschuler S.J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]