Abstract
Initiation of DNA replication at origins more than once per cell cycle results in rereplication and has been implicated in cancer. Here we use Drosophila to examine the checkpoint responses to rereplication in a developmental context. We find that increased Double-parked (Dup), the Drosophila ortholog of Cdt1, results in rereplication and DNA damage. In most cells, this rereplication triggers caspase activation and apoptotic cell death mediated by both p53-dependent and -independent pathways. Elevated Dup also caused DNA damage in endocycling cells, which switch to a G/S cycle during normal development, indicating that rereplication and the endocycling DNA reduplication program are distinct processes. Unexpectedly, however, endocycling cells do not apoptose regardless of tissue type. Our combined evidence suggests that endocycling apoptosis is repressed in part because proapoptotic gene promoters are silenced. Normal endocycling cells had DNA lesions near heterochromatin, which increased after rereplication, explaining why endocycling cells must constantly repress the genotoxic apoptotic response. Our results reveal a novel regulation of apoptosis in development and new insights into the little-understood endocycle. Similar mechanisms may operate during vertebrate development, with implications for cancer predisposition in certain tissues.
Keywords: DNA replication, DNA damage, endocycle, checkpoint, apoptosis
The timely duplication of the genome during S phase of every cell division cycle requires that DNA replication initiate from thousands of origins. If too few origins initiate, replication forks can collapse, resulting in DNA damage and incomplete replication of the genome. Initiation of DNA replication from origins more than once per cell cycle, however, results in “rereplication” and subsequent DNA damage (Arias and Walter 2007). In recent years, it has become increasingly apparent that problems with DNA replication are common in premalignant cells, with subsequent checkpoint defects leading to genome instability and cancer (Dutta 2007). It remains unclear, however, whether all cells in development are equivalent with respect to their regulation of DNA replication and checkpoint responses. Here, we use Drosophila to investigate the checkpoint responses to rereplication in a developmental context.
Two important steps in the cell cycle regulation of DNA replication are the assembly and activation of a prereplicative complex (pre-RC) (Sivaprasad et al. 2006). The pre-RC assembles onto origins in early G1 and is subsequently activated in S phase. During pre-RC assembly, the hexameric origin recognition complex (ORC) serves as a scaffold for origin association of Cdc6 and Cdt1, which are both required to load the hexameric minichromosome maintenance complex (MCM), the replicative helicase (Randell et al. 2006; Sivaprasad et al. 2006). Once the MCM complex is tightly bound, the origins are considered to be “licensed” and competent to initiate replication (Chong et al. 1995). During the subsequent S phase, cyclin-dependent kinase (CDK2 in multicellular eukaryotes) and another kinase, CDC7, activate the pre-RC, resulting in the association of other proteins with the origin, unwinding of DNA, and establishment of the replication fork (Walter and Araki 2006). Inappropriate rereplication is normally prevented because the pre-RC is disassembled when origins initiate, and reassembly of the pre-RC onto origins is inhibited until after mitotic division. Cyclin/CDKs inhibit pre-RC reassembly in S, G2, and early M phases, although the precise mechanisms can differ depending on organism (Arias and Walter 2007).
Evidence from yeast to humans indicates that rereplication can lead to genotoxic stress, activate cell cycle checkpoints, and promote genome instability in cancer (Dutta 2007; Hook et al. 2007). Tight regulation of Cdt1 is especially important to inhibit rereplication in a variety of organisms. Cdt1 is targeted for ubiquitin-mediated proteolysis at the G1/S transition, or after DNA damage, by at least two ubiquitin ligases (Arias and Walter 2007; Tada 2007). In addition, multicellular eukaryotes express another important inhibitor of pre-RC reassembly, Geminin, which binds Cdt1 and prevents it from reloading the MCM complex after origins initiate (McGarry and Kirschner 1998; Wohlschlegel et al. 2000; Tada 2007). In human cells, increased Cdt1 results in DNA rereplication and DNA damage that activates a G2/M checkpoint arrest, mediated by the Fanconia anemia checkpoint proteins (FANC), or apoptosis mediated by p53 (Melixetian et al. 2004; Zhu et al. 2004; Zhu and Dutta 2006). Cdt1 is up-regulated in several human cancers, and misexpression of Cdt1 in T cells of p53 mutant mice results in aneuploidy and a highly penetrant lymphoma, consistent with other evidence that the DNA damage caused by rereplication can lead to genome instability and oncogenesis if checkpoints fail (Hook et al. 2007). It remains unclear, however, whether Cdt1-mediated rereplication induces breakage at common fragile sites and whether these sites and the rereplication checkpoint responses depend on developmental cell type. These questions are important because a modified sensitivity and checkpoint response to rereplication could underlie cancer predisposition of certain tissues.
A current challenge, therefore, is to understand how cell cycle regulation and genome maintenance are integrated with development. For example, in several tissues in Drosophila, developmental signals induce certain cells to enter a specialized endocycle that is comprised of alternating G and S phases without mitosis, which results in genome polyploidization (Edgar and Orr-Weaver 2001; Lilly and Duronio 2005). Endocycles are controlled by the oscillating activity of the CDK complex, Cyclin E/CDK2, and most of the genome is duplicated exactly once during each endocycle S phase, although some heterochromatic sequences are not duplicated (Gall et al. 1971; Sauer et al. 1995; Lilly and Spradling 1996; Calvi et al. 1998; Parisi et al. 2003). This and other evidence suggest that during a single endocycle S phase, origins initiate no more than once (Su and O’Farrell 1998). A notable exception is the developmental gene amplification that occurs due to origin reinitiation at a few select loci in some endocycling cells (for review, see Calvi 2006). The endocycle program occurs widely in nature, including in humans, yet much remains to be learned about the endocycle, including its core cell cycle oscillator, DNA replication program, checkpoints, and developmental control.
The Drosophila double-parked (dup) gene is the fly ortholog of Cdt1 (Whittaker et al. 2000). We had previously shown that Dup protein is phosphorylated by Cyclin E/CDK2 and targeted for proteolysis at G1/S (Thomer et al. 2004; May et al. 2005). We showed that this proteolysis is important because even small increases in Dup protein levels were sufficient to cause rereplication. Here, we investigate how different cells in development respond to rereplication due to Dup misregulation. We find that an increase in Dup protein causes genotoxic damage, which is frequent in heterochromatin, and that in many cells this triggers a p53-dependent and p53-independent apoptosis. We also show that Dup regulation is important to prevent DNA rereplication and damage during the endocycle, indicating that endoreduplication and rereplication are very different processes. Surprisingly, however, cells in the endocycle do not apoptose. Our data suggest that endocycle apoptosis is repressed by silencing of proapoptotic gene promoters. These results provide evidence for a novel regulation of apoptosis during the little-understood endocycle and underscore the importance of investigating cell cycles in development for understanding the maintenance of genome integrity.
Results
Elevated Dup protein induces rereplication in mitotic cycling and endocycling cells
To study the effects of Dup misregulation in a developmental context, we began by examining the adult ovary. The somatic follicle cells surround the nurse cell/oocyte germline cysts as they exit the germarium to form a stage 1 egg chamber. Egg chambers then move down a structure called the “ovariole” as they mature through stages of oogenesis (stages 1–14) (Spradling 1993). Follicle cells proliferate by a mitotic division cycle up to stage 6, and then switch to the endocycle by stage 7, undergoing three rounds of G/S endoreduplication by stage 10A (Mahowald et al. 1979; Lilly and Spradling 1996; Calvi et al. 1998; Deng et al. 2001; Lopez-Schier and St Johnston 2001). Thereafter, they selectively rereplicate only a few loci, resulting in amplification of gene copy number (Calvi et al. 1998). Similar to mitotic cycles, MCM proteins periodically associate with chromatin during each endocycle G phase, and Dup protein is proteolyzed at the G/S transition, suggesting that Dup regulation may be important for licensing control of origins in the endocycle (Su and O’Farrell 1998; Whittaker et al. 2000; Thomer et al. 2004; Hong et al. 2007). To test this idea, we used a heat-inducible hsp70:Myc:FL-Dup strain to examine the effect of Dup overexpression on mitotic cycle and endocycling follicle cells. Expression was induced by three 30-min heat pulses over 30 h, as we described previously (Thomer et al. 2004). In a subset of both mitotic cycle and endocycling follicle cells, Dup overexpression resulted in enlarged nuclei, an increase in total fluorescent intensity after labeling with the DNA dyes DAPI or TOTO-3, and many incorporated BrdU, suggesting that they were actively rereplicating to a higher DNA copy number (Fig. 1A–D; Table 1). This suggests that Dup protein levels must be tightly regulated for normal origin licensing control during both the mitotic cycle and endocycle.
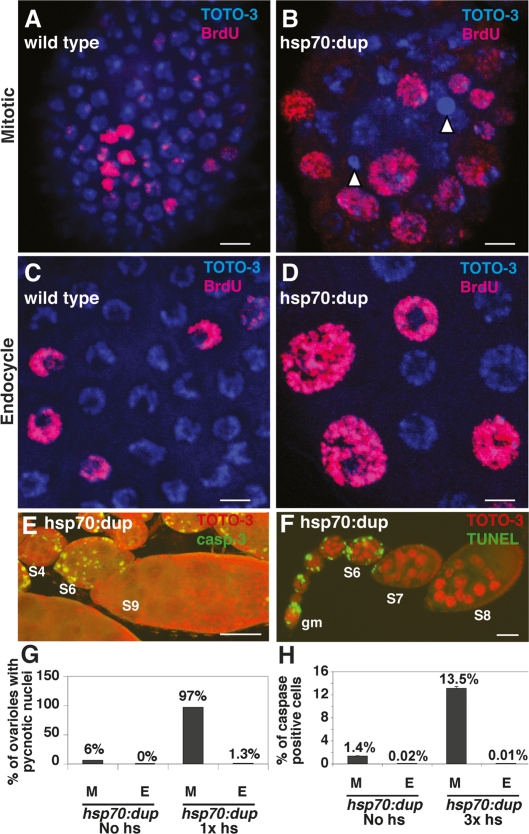
Figure 1.
Dup overexpression causes rereplication in mitotic cycling and endocycling cells, but only mitotic cycling cells apoptose. (A,B) Stage 5 mitotic cycling follicle cell nuclei labeled with TOTO-3 (blue) and BrdU (red) from wild type (A) or hsp70:Myc:FL-Dup overexpressing (B) females. Nuclei of hsp70:Myc:FL-Dup overexpressing follicle cells were enlarged or pycnotic (arrowheads). (C,D) Stage 8 endocycling follicle cells from wildtype (C) or hsp70:Myc:FL-Dup overexpressing (D) females. hsp70:dup overexpression in endocycle follicle cells resulted in enlarged, but not pycnotic, nuclei. (E,F) Dup overexpression activated caspase-3 (green in E) and TUNEL labeling (green in F) in mitotic follicle cells before stage 7 of oogenesis, but not in endocycling follicle cells of later egg chambers, DNA (red). The germarium (gm) and select stages of oogenesis are indicated by numbers. (G) Percent of ovarioles that had five or more pycnotic nuclei in mitotic follicle cells (M) or one or more pycnotic nuclei in endocycle follicle cells (E) from hsp70:Myc:FL-Dup ovaries either without induction or 7 h after one heat treatment (n ≥ 65 ovarioles). (H) Percent of caspase-positive follicle cells from stages 1–5 (M) and stages 9–10 (E) from hsp70:Myc:FL-Dup females without induction or after three heat inductions over 30 h. Cells were counted in a total of 25 ovarioles for each sample. (M vs. E) Heat shock: P < 10−8. Total cells counted: mitotic, n > 10,000; endocycle, n > 5000. Bars: A–D, 5 μm; E,F, 50 μm.
Table 1.
Range of follicle cell nuclear diameters with or without Dup misexpression
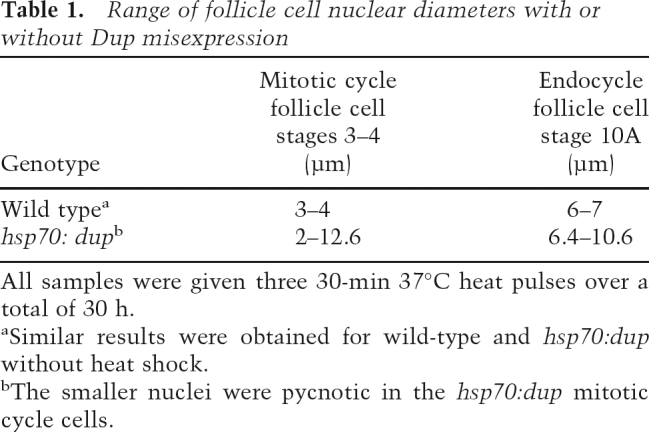
All samples were given three 30-min 37°C heat pulses over a total of 30 h.
aSimilar results were obtained for wild-type and hsp70:dup without heat shock.
bThe smaller nuclei were pycnotic in the hsp70:dup mitotic cycle cells.
Rereplication results in caspase activation and apoptosis in the mitotic cycle but not in the endocycle
We previously noted that after hsp70:Myc:FL-Dup expression, some follicle cells had pycnotic nuclei, a collapsed nuclear phenotype suggestive of apoptotic cell death (Fig. 1B; Thomer et al. 2004). To investigate whether this cell death is indeed apoptosis, we labeled with several markers. Cells with pycnotic nuclei labeled with antibodies against cleaved caspase-3 were permeable to the vital dye acridine orange and had evidence of DNA fragmentation by TUNEL (Fig. 1E,F; Wolff 2000; data not shown). Cell death was inhibited by coexpression of the viral caspase inhibitor p35 from a UAS:p35 transgene (data not shown), further suggesting that these cells were indeed dying by caspase-mediated apoptosis.
We quantified this cell death by counting the total number of ovarioles with dead cells. To subtract out the normal background of developmental cell death that occurs for a small number of polar follicle cells, we counted the number of ovarioles that had greater than five apoptotic follicle cells between stages 1 and 5 (Besse and Pret 2003). Seven hours after one 30-min heat induction of hsp70:Myc:FL-Dup, a large fraction of ovarioles had widespread follicle cell death in the mitotic stages (97% of ovarioles), but many fewer ovarioles had greater than five dead cells in the hsp70:Myc:FL-Dup strain without heat induction (6% of ovarioles) or after heat treatment of control strains that lacked the transgene (0% of ovarioles) (Fig. 1G; data not shown).
In stark contrast to the death of mitotic follicle cells, hsp70:Myc:FL-Dup did not induce apoptosis of follicle cells in the endocycle after stage 6, despite the evidence for rereplication in these cells (Table 2). Labeling with DAPI, TUNEL, acridine orange, and anti-cleaved caspase-3 antibody indicated that only ∼1.3% of ovarioles had any (one or more) apoptotic endocycling follicle cells in stages 9–10 after Dup overexpression, which was similar in frequency to non-heat-shock or wild-type controls (Fig. 1C–H; data not shown). hsp70:Myc:FL-Dup also did not induce follicle cell death in endocycles during developmental amplification that begins in stage 10B (data not shown). Quantifying the number of cells per ovariole that had cleaved caspase-3 confirmed that hsp70:Myc:FL-Dup expression did not induce apoptosis in endocycling cells (Fig. 1H). hsp70:Myc:FL-Dup also induced apoptosis in the germline nurse cells during their mitotic divisions in the germarium, but not after they had entered the endocycle in stage 1 (Fig. 1F; data not shown).
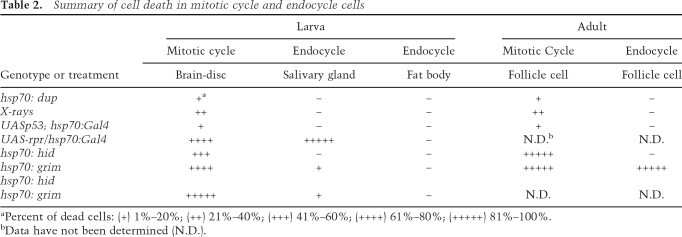
Table 2.
Summary of cell death in mitotic cycle and endocycle cells
aPercent of dead cells: (+) 1%–20%; (++) 21%–40%; (+++) 41%–60%; (++++) 61%–80%; (+++++) 81%–100%.
bData have not been determined (N.D.).
To test if other mitotic cycling and endocycling cells have distinct responses to rereplication, we examined tissues of the developing larva. Imaginal discs are precursors to adult structures and proliferate by a canonical mitotic cell cycle during embryonic and larval development, whereas many other larval tissues are comprised of cells in the endocycle. Expression of hsp70:Myc:FL-Dup in early third-instar larvae induced apoptosis in all imaginal discs as evidenced by pycnotic nuclei and labeling for cleaved caspase-3 and acridine orange (Supplemental Fig. S1A; Table 2; data not shown). Similar results were obtained for mitotic cells of the larval brain (data not shown). In contrast, in the same larvae, there was no evidence of apoptosis in any endocycling cells, such as the salivary glands, fat body, and gut (Supplemental Fig. S1B; Table 2; data not shown). Together, our observations from the larva and adult suggest that the absence of apoptosis in response to DNA rereplication is a common property of endocycling cells.
Rereplication induces genotoxic stress in both mitotic cycling and endocycling cells
It has been shown in several organisms that rereplication results in DNA double-strand breaks (Vaziri et al. 2003; Green and Li 2004; Archambault et al. 2005; Davidson et al. 2006). Therefore, we considered that rereplication may not cause damage in endocycling cells as one possible reason for why these cells do not apoptose. To investigate this, we labeled cells with an antibody against the phosphorylated form of histone H2AV (γ-H2AV). H2AV is phosphorylated on serine 137 by the checkpoint kinase ATM at sites of DNA damage, which can be seen as nuclear repair foci after labeling with anti-γ-H2AV antibodies (Madigan et al. 2002; Mehrotra and McKim 2006). ATM is also the proximal checkpoint kinase that initiates the apoptotic pathway. Induction of hsp70:Myc:FL-Dup expression led to the appearance of tens to hundreds of γ-H2AV foci throughout the nuclei of both mitotic cycling and endocycling follicle cells (Figs. 2A–F). hsp70:Myc:FL-Dup also induced γ-H2AV foci throughout the nucleus of mitotic cycling and endocycling cells of the larva (Fig. 2J–L; data not shown). These repair foci are not induced by apoptotic endonucleases because ∼100% of cells had repair foci, whereas a much smaller fraction activate caspases and label by TUNEL, especially in the endocycle (Fig. 1E–H). This suggests that rereplication causes DNA damage and activates ATM in both mitotic cycling and endocycling cells and therefore does not explain why endocycling cells do not apoptose. Treatment of adult females with 3000 rads of X-ray irradiation also resulted in numerous γ-H2AV repair foci in both mitotic cycle and endocycle cells, but only mitotic cycle cells activated caspases and apoptosed, further supporting that endocycle cells lack a genotoxic apoptotic response downstream from ATM (Table 2; data not shown).
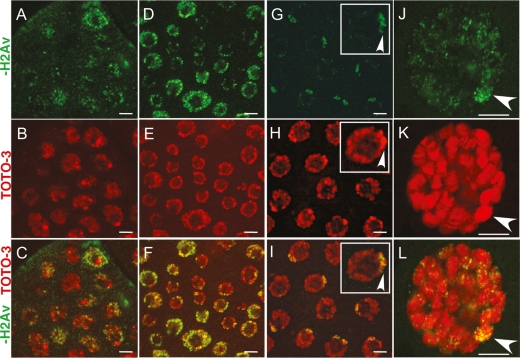
Figure 2.
Dup overexpression causes genotoxic stress in mitotic cycling and endocycling cells Anti-γ-H2AV labeling of repair foci (green) in nuclei (TOTO-3 red) of mitotic cycling or endocycling cells. (A–F) Mitotic follicle cells (A–C) and endocycle follicle cells (D–F) from hsp70:Myc:FL-Dup females 7 h after a single heat pulse. (G–I) Endocycling follicle cells from wild-type females with repair γ-H2AV foci (green) surrounding heterochromatic blocks in the chromocenter (brighter TOTO-3 focus, red, arrowheads). (J–L) A salivary gland polytene nucleus overexpressing Dup with γ-H2AV repair foci (green) throughout the nucleus and numerous repair foci in the heterochromatic chromocenter (arrowheads). Bars: A–I, 5 μm; J–L, 10 μm.
Endocycling follicle cells have DNA repair foci near heterochromatin
The control non-heat-shock or wild-type endocycling follicle cells did not have the hundreds of γ-H2AV repair foci that were seen throughout the nucleus after Dup overexpression. Importantly, this result indicates that DNA rereplication and endoreduplication are distinct processes. These control endocycling follicle cells, however, did consistently label for a smaller number of repair foci clustered in one region of the nucleus (Fig. 2G). Comparison with the DNA stain indicated that these foci were located near the heterochromatic chromocenter, a coalescence of pericentric heterochromatin whose A-T-rich subregions stain brightly with DAPI or TOTO-3 (Fig. 2H,I; Dej and Spradling 1999). Upon closer examination, it appeared that repair foci were located at the border of the A-T-rich TOTO-3 bright regions within the chromocenter (Fig. 2I, inset; Supplemental Fig. S2). Labeling with antibodies that recognize the heterochromatin marker histone H3 trimethylated on lysine 9 confirmed that endocycle cells of the ovary, salivary gland, fat body, and gut normally have repair foci in the chromocenter, which increased in intensity after hsp70:Myc:FL-Dup expression (Supplemental Figs. S2, S3; data not shown). These data suggest that both normal and rereplicating endocycling cells have stalled replication forks or DNA breaks near pericentric heterochromatin that are recognized by an ATM checkpoint pathway. The data further suggest that normal endocycling cells must constantly tolerate genotoxic signaling, explaining their need to suppress apoptosis downstream from active ATM.
A Chk2 and p53-dependent pathway is the major apoptotic response to rereplication
To begin to explore how the response to rereplication differs between mitotic cycling and endocycling cells, we further defined the apoptotic pathway that was engaged in mitotic cycling cells. In Drosophila, DNA damage induced by ionizing radiation activates ATM kinase that phosphorylates CHK2 kinase, which in turn phosphorylates p53, thereby increasing its activity as a transcription factor (Hay and Guo 2006). p53 is required for transcription of several downstream genes involved in apoptosis, DNA repair, and other cellular functions.
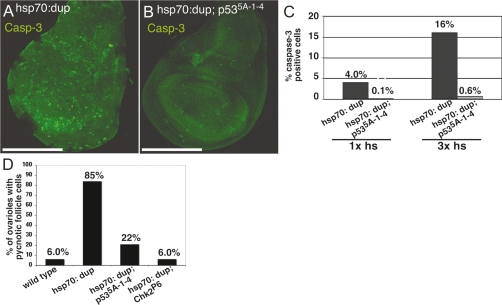
To determine if rereplication induces apoptosis by the canonical genotoxic stress pathway, we induced hsp70:Myc:FL-Dup expression in a p535A-1-4-null deletion mutant background and examined the response of mitotically dividing wing disc cells (Rong et al. 2002). After hsp70:Myc:FL-Dup induction, the fraction of apoptotic cells with cleaved caspase-3 was greatly reduced in the p53 mutant strain compared with wild type (Fig. 3A–C; data not shown). Caspase activation in the p53 mutant was more apparent at 30 h after hsp70:Myc:FL-Dup induction than at 7 h, but the fraction of cells and intensity of cleaved caspase-3 labeling per cell was still greatly reduced relative to wild type. Similar results were obtained for other mitotic cycling cells including those of the adult ovary (Fig. 3D). Analysis of chk2P6-null deletion mutants in the larva and adult ovary showed that Chk2 is also required for most apoptosis after hsp70:Myc:FL-Dup expression (Fig. 3D; Brodsky et al. 2004; data not shown). These results suggest that a response to rereplication in mitotic cycling cells is an induction of apoptosis through the canonical ATM–Chk2–p53 pathway, although a less robust Chk2- and p53-independent apoptotic pathway is also activated.
Figure 3.
p53 and Chk2 are required for most but not all caspase-3 activation in mitotic cycling cells. (A,B) Anti-cleaved caspase-3 labeling (green) in wing discs from hsp70:Myc:FL-Dup (A) and hsp70:Myc:FL-Dup; p535A-1-4 (B) early third-instar larvae after three heat inductions over 30 h. Bar, 50 μm. (C) Quantification of cells with cleaved caspase-3 labeling in hsp70:Myc:FL-Dup and hsp70:Myc:FL-Dup; p535A-1-4 wing discs after one and three heat shocks. hsp70:Myc:FL-Dup versus hsp70:Myc:FL-Dup; p535A-1-4; one heat shock: P < 10−14; three heat shocks: P < 10−18; n > 7000 cells per sample. (D) Number of ovarioles with more than five pycnotic follicle cells in wild type (n = 67), hsp70:Myc:FL-Dup (n = 112), hsp70:Myc:FL-Dup; p535A-1-4 (n = 55), and hsp70:Myc:FL-Dup; chk2P3 (n = 53) after three heat inductions over 30 h. P < 10−4 for hsp70:Myc:FL-Dup versus hsp70:Myc:FL-Dup; p535A-1-4 and hsp70:Myc:FL-Dup versus hsp70:Myc:FL-Dup; chk2P3.
Proapoptotic gene mRNA levels are lower in endocycling cells than mitotic cycling cells
To gain insight into how the rereplication response differs between mitotic cycling and endocycling cells, we analyzed the expression of checkpoint and apoptotic pathway genes. While much of the activation of the checkpoint and apoptotic pathway entails post-transcriptional regulation, a notable exception is the transcriptional activation of proapoptotic genes at the H99 locus. Transcription of three of these proapoptotic genes—sickle (skl), reaper (rpr), and Wrinkled (W also known as Hid)—is dependent on p53, while a fourth gene at the locus, grim, may be regulated by other transcription factors (Hay and Guo 2006). The proteins encoded by all these genes inhibit translation or induce degradation of the Drosophila Inhibitor of Apoptosis Protein (DIAP1) (Yoo et al. 2002; Muro et al. 2006). Since DIAP1 normally restrains activity of initiator caspases, inhibition of DIAP1 by these proapoptotic genes results in cleavage of initiator caspases, which in turn cleave downstream effector caspases (e.g., caspase-3), thereby inducing apoptosis (Hay and Guo 2006). Therefore, the balance of expression between the proapoptotic and DIAP1 genes is crucial in the decision to initiate or prevent apoptosis.
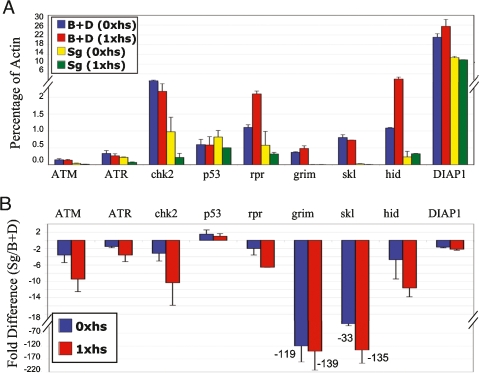
We therefore compared the mRNA levels between mitotic cycling and endocycling cells. To ensure that differences were a property of the cell cycle and not tissue type, we used pooled larval discs and brain tissue as the mitotic cycle sample, and compared it separately with endocycling cells of both larval salivary glands and fat body. The mRNA levels were analyzed by real-time quantitative PCR (RT-QPCR) and were normalized to an Actin 5C control in two biological replicates. We first compared the mRNA levels in unchallenged cells from hsp70:Myc:FL-Dup larvae that were not heat-treated (Fig. 4; Supplemental Fig. S4; Supplemental Tables S1, S2, E/M column). Mitotic cycling and endocycling cells had similar levels of p53 and mei-41 (ATR) mRNA. Endocycling cells had somewhat lower levels of tefu (ATM) mRNA (−3.5-fold), but this is not likely limiting for apoptosis since we saw abundant H2AV phosphorylation in repair foci after rereplication in both salivary gland and fat body. mRNA levels of loki (Chk2) were lower in salivary glands (−3.1-fold), but not in fat body, suggesting Chk2 is also not limiting for apoptosis in endocycling cells. The biggest difference between mitotic cycling and endocycling cells was that the mRNA levels for the proapoptotic genes at the H99 locus were lower in endocycling cells. Salivary glands had lower mRNA levels for all four proapoptotic genes, while fat body had lower levels of three genes with the exception of W (Hid). Most notably, the levels of skl and grim mRNA were tens to hundreds fold lower in endocycling cells, respectively. The anti-apoptotic DIAP1 gene mRNA levels, however, were comparable between mitotic cycling and endocycling cells.
Figure 4.
Endocycling salivary gland cells have low mRNA levels for apoptotic pathway genes. QPCR analysis of mRNA in mitotic cycling brain and disc cells (B + D) and endocycling salivary gland cells (Sg) from hsp70:Myc:FL-Dup early third-instar larvae without heat induction, or 4 h after a single heat shock. (A) mRNA levels as a percentage of Act 5C. Bars represent average and standard deviation from two biological replicates. (B) The endocycling-to-mitotic cycling cell mRNA ratio (from A) converted to fold difference. Bars represent average and standard deviation for endocycle: mitotic fold difference from the two biological replicates.
We next determined if the mRNA levels responded to rereplication. Comparing levels in hsp70:Myc:FL-Dup-expressing versus control mitotic cycling cells revealed that rpr and W expression was induced by rereplication (Fig. 4; Supplemental Fig. S4; Supplemental Tables S1, S2, Mrr/M columns). In salivary glands, only W was induced, while in fat body, mRNA levels for all genes were higher in the rereplicating versus control (Fig. 4; Supplemental Fig. S4; Supplemental Tables S1, S2, Err/E columns). The increase for proapoptotic genes in fat body, however, was relative to an extremely low basal level of these transcripts in unchallenged cells, and therefore represents only a small increase in magnitude. In fact, the mRNA levels for proapoptotic genes at the H99 locus were still lower in the rereplicating endocycling cells even when compared with unchallenged mitotic cycling cells, with the exception of W in the fat body (Fig. 4; Supplemental Fig. S4; Supplemental Tables S1, S2, Err/M columns). The mRNA levels for the anti-apoptotic DIAP1 gene, however, were similar between mitotic cycling and endocycling cells. Analysis of control strains that lacked the hsp70:Myc:FL-Dup transgene confirmed that endocycling cells have lower levels of proapoptotic genes and that these results are not confounded by heat shock (Supplemental Fig. S5). These results suggest that endocycling cells do not apoptose because the balance of expression between the anti-apoptotic DIAP1 and proapoptotic genes at the H99 locus is tipped toward cell survival in the endocycle.
Expression of H99 locus genes from heterologous promoters induces endocycling cell death
To test if the expression of proapoptotic genes may be limiting for endocycling cell death, we determined if overexpression of rpr, grim or hid from heterologous promoters is sufficient to induce apoptosis in mitotic cycling and endocycling cells. Heat induction of hsp70:grim or hsp70:hid transgene expression resulted in rapid and robust cell death of 100% of mitotic cycling follicle cells in the ovary (Table 2; Supplemental Fig. S6; Grether et al. 1995; Chen et al. 1996). During endocycles, expression of hsp70:grim also resulted in the death of 100% of follicle cells, but overexpression of hsp70:hid did not result in endocycling cell death (Table 2; Supplemental Fig. S6).
In early third-instar larvae, heat-induced expression of hsp70:grim, hsp70:hid, or hsp70 Gal4; UAS-rpr/+ strains all caused rapid caspase-3 activation in ∼80%–100% of mitotic cycling cells of the wing disc, eye disc, and brain (Table 2). In endocycling salivary gland cells, however, only rpr overexpression caused rapid and robust cell death in all cells, with complete gland destruction within 30 min (Table 2). Overexpression of grim resulted in only very weak anti-cleaved caspase-3 labeling in a small fraction (0.49%, n = 2905) of salivary gland cells, while hid overexpression did not induce any detectable caspase activation in those cells (Table 2). Coexpression of hid and grim resulted in an increased fraction of salivary gland cells that labeled for activated caspase-3 (7%, n = 900). Expression of the proapoptotic genes alone or hid and grim together did not induce any detectable apoptosis in the fat body (Table 2). The different potencies of these proapoptotic genes in different endocycling cells is consistent with previous evidence that they inhibit DIAP1 by somewhat different mechanisms and that their activities for inducing cell death in mitotic cycling cells are developmentally complex (Hay and Guo 2006). Nevertheless, the ability of the H99 locus genes to induce death of some endocycling cells when expressed from heterologous promoters indicates that these cells are competent to respond to these proapoptotic genes, and is consistent with the interpretation that their transcription may be limiting for endocycling cell apoptosis.
Overexpression of p53 induces apoptosis in mitotic cycling cells but not in endocycling cells
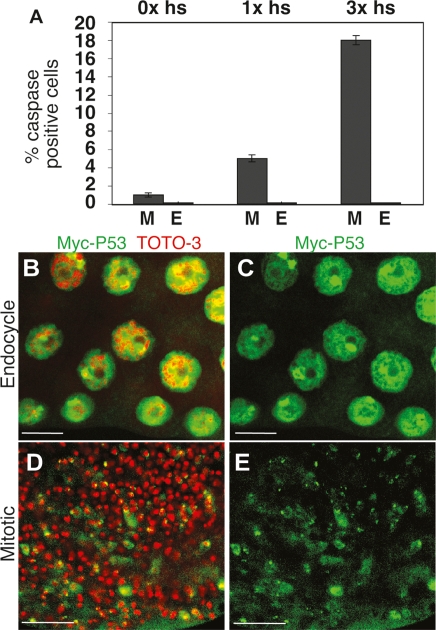
Since endocycling cells labeled for γ-H2AV but not cleaved caspase-3, we knew that the block to apoptosis was upstream of caspase-3 activation, but downstream from ATM. Moreover, QPCR and overexpression analysis suggested that endocycling cell apoptosis is blocked in part by dampening proapoptotic gene expression. Overexpression of p53 is known to be sufficient to induce apoptosis in most cells of Drosophila (Brodsky et al. 2000; Ollmann et al. 2000). Therefore, to test if the block to apoptosis in the endocycle is upstream of or downstream from p53, we induced p53 expression in larvae using a hsp70:GAL4; UAS:p53 strain and monitored apoptosis in mitotic cycling and endocycling tissues (Ollmann et al. 2000). This showed that UAS:p53 overexpression induced apoptosis in up to 18% of mitotic cycling cells of discs, but not in any endocycling cells of salivary glands, fat body, or gut (Fig. 5A; Table 2). Similar results were obtained in the ovary, where overexpression of p53 induced apoptosis in mitotic cycling but not endocycling follicle cells (Supplemental Fig. S7). These results suggest that apoptosis of endocycling cells is blocked at or downstream from p53.
Figure 5.
Overexpressed p53 protein induces apoptosis in mitotic cycling but not endocycling cells. (A) Percent of caspase-positive mitotic cycling cells of wing discs (M) or endocycling cells of salivary gland (E) from UAS:p53/+; hsp70: Gal4/+ early third-instar larvae after zero, one (7 h) or three (30 h) heat inductions. (M vs. E) Zero heat induction, P > 0.05; one heat induction, P < 10−9; three heat inductions, P < 10−12. (B–E) Myc:p53 protein is similarly expressed in mitotic cycling and endocycling cells. Anti-Myc (green) andDNA (TOTO-3, red) labeling of salivary gland (B,C) and wing disc (D,E) cells from P{UAS:6X Myc:p53}/+; hsp70:Gal4/+ early third-instar larvae 2 h after one heat-shock treatment. Bars, 10 μm.
We considered that the inability of p53 to induce apoptosis in the endocycling cells may be because p53 protein translation or stability is compromised in these cells even when overexpressed. To test this idea, we created fly strains with a GAL4-inducible Myc-tagged p53 transgene. We heat-induced expression of this UAS:Myc:p53 transgene using a hsp70:GAL4 driver and compared Myc:p53 protein abundance in mitotic cycling and endocycling cells at different times thereafter by labeling tissues with anti-Myc antibodies. Myc:p53 protein was easily detected in endocycling cells of salivary glands and fat body, as well as in the mitotic cells of discs and brains, but only mitotic cycling cells apoptosed (Fig. 5B–E). Indeed, the intensity of fluorescent labeling suggested that the abundance of Myc:p53 protein was greater in endocycling cells consistent with a much higher genomic DNA copy number in these polyploid cells (up to ∼2048C in salivary glands, and ∼256C in fat body) (Hammond and Laird 1985; Butterworth and Rasch 1986). Similar results were obtained by Western blotting for Myc:p53 (Supplemental Fig. S8). The inability of overexpressed p53 to induce apoptosis during the endocycle, therefore, is not due to compromised p53 protein translation or stability.
Reaper promoter activation is dampened in endocycling cells
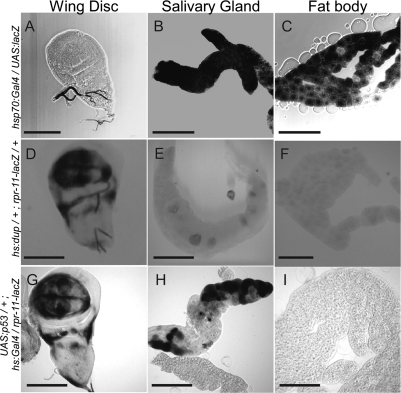
The combined results suggested that proapoptotic gene promoters may be repressed in endocycling cells. To test this idea, we examined the activity of the reaper promoter using a rpr-11-lacZ strain (Nordstrom et al. 1996). This reporter construct contains an 11-kb fragment upstream of the reaper transcription unit, including the p53-binding site, fused to the coding region of the lacZ gene. Since the polyploid cells of fat body and salivary gland have up to hundreds to thousands more copies of the reporter, respectively, we first normalized expression in mitotic cycling versus endocycling cells by analyzing the relative β-gal activity after heat-induced expression of the positive control strain hsp70:GAL4; UAS:lacZ. When the tissue was fixed at 2 h after heat induction, X-gal staining was saturated in salivary gland and fat body cells after only 3 h of X-gal staining reaction, whereas disc cells were not robustly labeled until 15 h of X-gal staining, consistent with the much higher gene copy number in polyploid cells (Fig. 6A–C). Indeed, X-gal labeling was first apparent in salivary glands after only 15 min of the X-gal reaction (data not shown).
Figure 6.
rpr promoter activity is dampened in endocycling cells. X-gal labeling in wing discs (A,D,G), salivary glands (B,E,H), and fat body (C,F,I) of early third-instar larvae. (A–C) Normalization control UAS-lacZ/+; hsp70:Gal4/+. (D–F) hsp70:Myc:FL-Dup/+; rpr11-lacZ/+. (G–I) UAS:p53/+; rpr11-lacZ/hsp70:Gal4. Images depict samples fixed either at 2 h (A–C,G–I) or 7 h (D–F) after heat shock treatment, followed by 3 h of X-Gal staining. Bars, 50 μm.
In contrast, 7 h after expression of Dup in hsp70:Dup/+; rpr-11-lacZ/+, both imaginal discs and brain were robustly labeled after 3 h of X-gal labeling, but only extremely low levels of mosaic staining were observed in a small fraction of salivary gland cells, and all fat body cells were not detectably labeled (Fig. 6D–F). These results suggest that the induction of rpr gene expression after rereplication is dampened in endocycling cells at the level of transcription.
To investigate whether p53 is capable of activating the reaper promoter in endocycling cells, we heat-induced expression of p53 in UAS:p53/+; rpr-11-lacZ/hsp70:GAL4 larvae and labeled with X-gal at different time points. Two hours after heat induction, p53 overexpression induced rpr:lacZ expression in both imaginal discs and salivary gland cells (Fig. 6G,H). Salivary gland labeling was mosaic; some cells were X-gal-positive, whereas others were completely negative. Although the X-gal staining in some salivary gland cells was comparable with that of the disc, the salivary gland cells have ∼1000 more copies of UAS:p53 and rpr-11-lacZ than diploid cells. Therefore, in comparison with the hsp70:GAL4; UAS:lacZ normalization control, this result suggests that p53 activation of rpr-11-lacZ is much less efficient in endocycling salivary gland cells than in mitotic cycling cells. Moreover, there was no detectable X-gal staining in any fat body cells at any time point, indicating that the ∼124 copies of rpr-11-lacZ in those polyploid cells are not detectably induced even when p53 is greatly overexpressed (Fig. 6I). Similar results were obtained when we quantified β-galactosidase activity using a solution assay on tissue extracts (Supplemental Fig. S9). These findings suggest that rpr promoter activation by p53 is blunted in endocycling cells. Moreover, the variegated expression of rpr-lacZ in salivary glands is suggestive of an epigenetic silencing of the rpr promoter that is partially counteracted by p53 overexpression in some endocycling cells but not others.
Discussion
We found that elevated levels of Dup are sufficient to induce DNA rereplication and DNA damage in cycling cells from a variety of tissues. In mitotic cycling cells, this DNA damage resulted in activation of a canonical ATM–Chk2–p53 apoptotic pathway, although a minor apoptotic pathway that is independent of p53 was also activated. In endocycling cells, elevated Dup also induced rereplication, DNA damage, and activated ATM, underscoring that rereplication is a distinct process from regulated endoreduplication. Surprisingly, however, rereplication did not induce apoptosis in endocycling cells from any tissue. Our combined data suggest that the proapoptotic genes are transcriptionally repressed in the endocycle. Both normal and rereplicating endocycling cells had γ-H2AV repair foci at heterochromatin junctions, suggesting that these difficult-to-replicate genomic regions may be fragile sites and that endocycling cells must constantly block apoptosis downstream from active ATM. These results are important because they provide insights into the complexity of genome maintenance and checkpoint signaling in development and reveal a new mechanism for suppressing apoptosis in certain cell types. Similar mechanisms may operate in other cells in flies and may be conserved to humans, with potentially important implications for cancer predisposition of particular cell types.
Rereplication is distinct from the normal process of endoreduplication
Our data suggest that origin licensing control is similar in mitotic cycles and endocycles and underscores that uncontrolled rereplication is not equivalent to the controlled genome endoreduplication of the endocycle. We found that elevated levels of Dup were sufficient to induce rereplication and repair foci in both mitotic cycling and endocycling cells. We had previously shown that Dup is rapidly proteolyzed at the onset of S phase in both mitotic cycles and endocycles (Thomer et al. 2004). The proteolysis of Dup/Cdt1 is now known to be important to prevent rereplication during mitotic cycles of a variety of organisms (Arias and Walter 2007). The proposed importance of Dup regulation for endocycle licensing control is consistent with the observation that MCM proteins periodically associate with chromatin during the Gap phase of each endocycle, and measurements of DNA copy number that showed that most of the genome does not duplicate more than once per endocycle S phase (Lilly and Spradling 1996; Calvi et al. 1998; Su and O’Farrell 1998; A. Kolpakas, S. Maqbool, and B. Calvi, unpubl.). Moreover, previous evidence suggested that the Dup inhibitor Geminin, and periodic inhibition of CDK2, is important for normal endocycle DNA replication (Hong et al. 2007; Narbonne Reveau et al. 2008; Zielke et al. 2008). It is possible, however, that some aspects of the origin licensing mechanism differ between mitotic cycles and endocycles.
Rereplication induces apoptosis in Drosophila
We found that apoptosis is one response of mitotic cycling cells to DNA damage caused by rereplication. Most of this apoptosis was dependent on the ATM–Chk2–p53 pathway; however, a few cells in chk2- and p53-null mutants displayed a lower level of caspase activation after Dup misexpression. This Chk2- and p53-independent cell death pathway is probably similar to the one that mediates a delayed cell death in chk2 and p53 mutants after ionizing radiation, although further experiments will be required to define this pathway (Wichmann et al. 2006). Our results in Drosophila are similar to those from human cells in culture, in which Cdt1 misexpression induces p53-dependent apoptosis, suggesting that the rereplication checkpoint response is conserved (Vaziri et al. 2003; Zhu and Dutta 2006).
Endocycling cells do not apoptose in response to genotoxic stress
A remarkable result is that endocycling cells do not apoptose in response to the genotoxic stress created by rereplication. The absence of an apoptotic response was a common property of all endocycling cells that we examined both in the larva and adult. This difference was not unique to lesions created by rereplication since endocycling cells also did not apoptose in response to DNA double-strand breaks induced by X-irradiation. It appears, therefore, that the endocycle not only represents a fundamental remodeling of the cell cycle oscillator, but also a modified apoptotic checkpoint response to genotoxic stress.
The idea that apoptotic repression is common to the endocycle is consistent with evidence for the normal developmental fate of these cells. All the endocycling cells we studied are destined to undergo developmental programmed cell death. They all do so, however, by mechanisms that differ from standard apoptosis. For example, while proapoptotic genes and caspases become activated during salivary gland death during early pupal life, these cells also activate an autophagic cell death pathway that is required for the normal developmental demise of these tissues (Berry and Baehrecke 2007). In the ovary, nurse cells and follicle cells die in late oogenesis, but nurse cell death does not require rpr, Hid, and skl and also differs in the types and mechanism of caspase activation (McCall 2004). Thus, in the face of apoptotic repression, it appears that endocycling cells have taken alternative paths to developmental cell death.
Endocycle apoptosis is repressed in part by blunting transcriptional activation of proapoptotic genes
Our combined data support that p53 transcriptional activation of the proapoptotic genes at the H99 locus is blunted in endocycle cells and contributes to restraining apoptosis. Endocycling cells had low levels of mRNA for these proapoptotic genes, yet the levels of anti-apoptotic gene DIAP1 was comparable between endocycling and mitotic cycling cells. Analysis of the rpr-11-lacZ enhancer–promoter reporter supports that, at least for this p53 target, the reduced mRNA levels in the endocycling cells reflect repression at the level of transcription. This interpretation is consistent with our findings that overexpressed p53 did not induce apoptosis in any endocycling cells, whereas expression of proapoptotic genes from heterologous promoters did induce endocycling cell apoptosis.
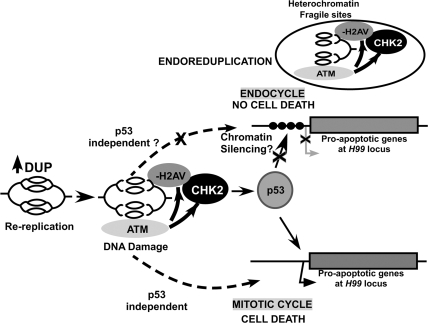
The variegated expression of the rpr:lacZ reporter is suggestive of an epigenetic silencing mechanism. The proapoptotic genes at the H99 locus are separated by large intergenic regions, and the 11 kb of rpr enhancer–promoter in the rpr:lacZ reporter may not contain the full complement of cis sequences that mediate epigenetic silencing at the endogenous H99 locus. Therefore, overexpressed p53 may be able to partially reverse the metastable chromatin silencing of the reporter in some endocycling cells. We did not examine the promoter activities of the other proapoptotic genes at the H99 locus, but their lower mRNA levels in the endocycle suggests that they may also be repressed at the level of transcription. We therefore propose a model wherein epigenetic silencing at the H99 locus blunts their activation by p53, thereby repressing apoptosis in endocycling cells (Fig. 7). This silencing may be reversible since the proapoptotic genes can be activated during developmentally programmed cell death in the salivary gland (Martin and Baehrecke 2004). While this manuscript was in preparation, Zhang et al. (2008) reported that epigenetic silencing at the H99 locus contributes to repressing apoptosis after ionizing radiation in late-stage embryos. It is possible, therefore, that similar epigenetic mechanisms operate to couple developmental entry into the endocycle with the repression of apoptosis. Given that rereplication activated a p53-dependent and p53-independent apoptosis in mitotic cycling cells, both pathways must be inoperative in endocycling cells. It is tempting to speculate that both cell death pathways act through activation of gene expression at the H99 locus, providing a parsimonious explanation for how both pathways could be corepressed by epigenetic silencing in endocycling cells (Fig. 7).
Figure 7.
A model for the response to rereplication in mitotic cycling and endocycling cells. Overexpression of Dup results in rereplication, DNA damage, and activation of ATM, in mitotic cycling and endocycling cells. Active ATM is known to phosphorylate H2AV and also Chk2, which in turn phosphorylates p53, thereby activating its transcriptional activity. In mitotic cycling cells after rereplication, transcription of the proapoptotic genes at the H99 locus is induced by p53, and perhaps also other transcription factors via a p53-independent pathway (dotted arrows). In endocycling cells, proapoptotic gene expression is dampened, perhaps by an epigenetic silencing at the H99 locus (circles depict repressed chromatin). This would blunt the ability of p53 and other transcription factors to induce transcription of these genes in the endocycling cells. (Circle inset above) Unchallenged endocycling cells also have DNA damage at difficult-to-replicate euchromatin–heterochromatin junctions during endoreduplication, explaining their need to block apoptosis downstream from active ATM.
It remains possible, however, that regulation of other steps in the apoptotic pathway also contributes to repression of apoptosis in endocycling cells. Indeed, overexpression of some proapoptotic genes did not result in endocycling cell death. These observations in endocycling cells are consistent with previous evidence that Rpr, Hid, Grim, and Skl proteins inhibit DIAP1 by somewhat different mechanisms, and that the relative activities of these proapoptotic proteins in different mitotic cycling cell types is developmentally complex (Hay and Guo 2006). It is known that some of the proapoptotic genes are repressed by miRNAs, opening the possibility that this regulation also contributes to repressing endocycling cell apoptosis.
Damage at chromatin junctions in both normal and rereplicating endocycling cells
Why is an absence of apoptosis common to the endocycling cells? We noticed that normal, unchallenged endocycling cells from a variety of tissues have γ-H2AV repair foci near pericentric heterochromatin. Some heterochromatic sequences are replicated during late S phase in mitotic cycles, but, because endocycle S phases are shorter, these late-replicating sequences do not get the opportunity to duplicate each endocycle (Hammond and Laird 1985; Lilly and Spradling 1996). The DNA lesions we observe, therefore, likely result from the stalling and collapse of replication forks at the junction between fully replicated euchromatin and underreplicated heterochromatin during truncated endocycle S phases. DNA damage in these regions may be the result of replication forks “catching up” with stalled forks from previous endocycle S phases. These observations and model are consistent with a previous report of heterochromatic repair foci in endocycle cells of the ovary (Hong et al. 2007). In fact, previous analyses had indicated that these chromatin junctions are fragile in the endocycle, and that fragility correlates with the degree of underreplication (Karpen and Spradling 1990; Leach et al. 2000; B. Calvi, unpubl.). The appearance of phosphorylated H2AV in normal endocycling cells indicates that they recognize these sites as genetic lesions and must constantly tolerate genotoxic signaling and suppress apoptosis downstream from ATM. Thus, it is likely that during evolution the developmental entry into the endocycle has been coupled to the suppression of apoptosis so that cells can reap the biosynthetic benefits of euchromatic polyploidization. An important future goal is to understand how developmental pathways couple endocycle entry and apoptosis inhibition.
The appearance of numerous γ-H2AV repair foci in heterochromatin after Dup overexpression suggests that these genomic regions may also be fragile sites during rereplication. This may be due to fork stalling and increased head-to-tail fork collisions in these difficult-to-replicate regions (Davidson et al. 2006). Therefore, the damage created during rereplication may be mechanistically similar to that which normally occurs near heterochromatin during endoreduplication, although confirmation of this model requires future molecular and genomic analyses. This also opens the possibility that other euchromatic/heterochromatic junctions are common rereplication fragile sites and contribute to oncogenesis in vertebrates.
Apoptotic repression, cell proliferation, and cancer
An important question raised by our results is whether silencing of proapoptotic genes occurs in other cell types and is conserved in evolution. The endocycle is phylogenetically widespread, and a number of different cell types enter an endocycle program during vertebrate development (Edgar and Orr-Weaver 2001; Lilly and Duronio 2005). Interestingly, it was previously shown that the giant trophoblast cells of the rodent placenta are resistant to apoptosis, but it is unclear if the mechanism is similar to the one we described here (Soloveva and Linzer 2004). It is possible, however, that epigenetic silencing of proapoptotic genes contributes to repressing apoptosis in these and other vertebrate endocycle cells, and perhaps also in other cells that exit the mitotic cycle and arrest. Apoptosis is a significant barrier to oncogenesis, but repression of apoptosis is less risky in endocycling or arrested cells that have lost their proliferative capacity. If repression of apoptosis becomes uncoupled from proliferation arrest, however, damage caused by rereplication could lead to cell transformation and may explain the predisposition of certain cell types to cancer.
Materials and methods
Details of materials and methods can be found in the Supplemental Material.
Drosophila genetics
Standard techniques were used for culture of Drosophila melanogaster at 25°C. P{w+mC hsp70:Myc:FL-Dup} has been described (Thomer et al. 2004). The y w67c23 strain was used for wild-type controls since it was the transformation host for hsp70:FL-Dup. Fly strains were obtained from the Bloomington Drosophila Stock Center (http://flystocks.bio.indiana.edu), except rpr-11-LacZ (Nordstrom et al. 1996), hsp70:grim (Chen et al. 1996), and hsp70:hid (Grether et al. 1995) were obtained from J.M. Abrams.
Staging of larvae and adult flies
We dissected imaginal discs, brain, salivary gland, and fat body from early third-instar larvae that were collected at 72–75 h after egg deposition at 25°C, a time when these cells are actively cycling, which we confirmed by BrdU incorporation. Conditioning of adult females and ovary analysis was as described previously (Calvi and Lilly 2004).
Immunolabeling and microscopy
Antibody, BrdU, DAPI, and TOTO-3 labeling was done as described previously (Schwed et al. 2002; Calvi and Lilly 2004). We used the following antibodies and concentrations: rabbit polyclonal anti-cleaved caspase-3 antibody 1:50 (Cell Signaling); rabbit polyclonal anti-γ-H2AV (RU018) antibody 1:50 (a gift from Kim McKim); mouse monoclonal anti-Myc 1:50 (clone 9E10; Upstate Biotechnologies); rabbit polyclonal anti-H3K9-trimethyl 1:50 (Upstate Biotechnologies). Secondary antibodies anti-rabbit Alexa 488; anti-rabbit Alexa 568; and anti-mouse Alexa 488 were all used at 1:400. Acridine Orange labeling was as described (Wolff 2000), and TUNEL labeling was as in McCall and Peterson (2004) using the In Situ Cell Death Detection Kit: Fluorescein (Roche).
Real-time QPCR
Total RNA was isolated from 72- to 75-h-old larvae using the RNeasy Micro Kit (Qiagen) and was reverse-transcribed using the QuantiTect Reverse Transcription Kit (Qiagen). QPCR reactions were performed using the iCycler Real-Time PCR system (Bio-Rad) with iQ SYBR Green Supermix (Bio-Rad), following protocols provided by the manufacturer.
Acknowledgments
We thank the following individuals for fly strains and reagents: M. Abrams, B. Hay, K. McKim, Y. Rong, H. Steller, W. Theurkauf, and especially K. Mathews and K. Cook of the Bloomington Drosophila Stock Center. We thank J. Waka and J. Boyd of Leica Microsystems for assistance with image acquisition, R. Raina for use of his QPCR machine, M. Kadaba for assistance with experiments, and S. Kummer and K. McConnell for comments on the manuscript. We declare that we have no conflicts of interest. This research was supported by ACS Research Scholar Grant RSG-04-172-01-CCG and NIH R01 grant GM612290-07 to B.R.C.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1710208.
References
- Archambault V., Ikui A.E., Drapkin B.J., Cross F.R. Disruption of mechanisms that prevent rereplication triggers a DNA damage response. Mol. Cell. Biol. 2005;25:6707–6721. doi: 10.1128/MCB.25.15.6707-6721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias E.E., Walter J.C. Strength in numbers: Preventing rereplication via multiple mechanisms in eukaryotic cells. Genes & Dev. 2007;21:497–518. doi: 10.1101/gad.1508907. [DOI] [PubMed] [Google Scholar]
- Berry D.L., Baehrecke E.H. Growth arrest and autophagy are required for salivary gland cell degradation in Drosophila. Cell. 2007;131:1137–1148. doi: 10.1016/j.cell.2007.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse F., Pret A.M. Apoptosis-mediated cell death within the ovarian polar cell lineage of Drosophila melanogaster. Development. 2003;130:1017–1027. doi: 10.1242/dev.00313. [DOI] [PubMed] [Google Scholar]
- Brodsky M.H., Nordstrom W., Tsang G., Kwan E., Rubin G.M., Abrams J.M. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- Brodsky M.H., Weinert B.T., Tsang G., Rong Y.S., McGinnis N.M., Golic K.G., Rio D.C., Rubin G.M. Drosophila melanogaster MNK/Chk2 and p53 regulate multiple DNA repair and apoptotic pathways following DNA damage. Mol. Cell. Biol. 2004;24:1219–1231. doi: 10.1128/MCB.24.3.1219-1231.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterworth F.M., Rasch E.M. Adipose tissue of Drosophila melanogaster: VII. Distribution of nuclear DNA amounts along the anterior–posterior axis in the larval fat body. J. Exp. Zool. 1986;239:77–85. doi: 10.1002/jez.1402390110. [DOI] [PubMed] [Google Scholar]
- Calvi B.R. Developmental DNA amplification. In: DePamphilis M.L., editor. DNA replication and human disease. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 233–255. [Google Scholar]
- Calvi B.R., Lilly M.A. BrdU labeling and nuclear flow sorting of the Drosophila ovary. In: Henderson D., editor. Drosophila cytogenetics protocols. Humana Press; Totowa, NJ: 2004. pp. 203–213. [DOI] [PubMed] [Google Scholar]
- Calvi B.R., Lilly M.A., Spradling A.C. Cell cycle control of chorion gene amplification. Genes & Dev. 1998;12:734–744. doi: 10.1101/gad.12.5.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Nordstrom W., Gish B., Abrams J.M. grim, a novel cell death gene in Drosophila. Genes & Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- Chong J., Mahbubani H., Khoo C., Blow J. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature. 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- Davidson I.F., Li A., Blow J.J. Deregulated replication licensing causes DNA fragmentation consistent with head-to-tail fork collision. Mol. Cell. 2006;24:433–443. doi: 10.1016/j.molcel.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dej K.J., Spradling A.C. The endocycle controls nurse cell polytene chromosome structure during Drosophila oogenesis. Development. 1999;126:293–303. doi: 10.1242/dev.126.2.293. [DOI] [PubMed] [Google Scholar]
- Deng W.M., Althauser C., Ruohola-Baker H. Notch-Delta signaling induces a transition from mitotic cell cycle to endocycle in Drosophila follicle cells. Development. 2001;128:4737–4746. doi: 10.1242/dev.128.23.4737. [DOI] [PubMed] [Google Scholar]
- Dutta A. Chaotic license for genetic instability and cancer. Nat. Genet. 2007;39:10–11. doi: 10.1038/ng0107-10. [DOI] [PubMed] [Google Scholar]
- Edgar B.A., Orr-Weaver T.L. Endoreplication cell cycles: More for less. Cell. 2001;105:297–306. doi: 10.1016/s0092-8674(01)00334-8. [DOI] [PubMed] [Google Scholar]
- Gall J.G., Cohen E.H., Polan M.L. Repetitive DNA sequences in Drosophila. Chromosoma. 1971;33:319–344. doi: 10.1007/BF00284948. [DOI] [PubMed] [Google Scholar]
- Green B., Li J. Loss of rereplication control in Saccharomyces cerevisiae results in extensive DNA damage. Mol. Cell. Biol. 2004;16:421–432. doi: 10.1091/mbc.E04-09-0833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether M.E., Abrams J.M., Agapite J., White K., Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes & Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- Hammond M.P., Laird C.D. Chromosome structure and DNA replication in nurse and follicle cells of Drosophila melanogaster. Chromosoma. 1985;91:267–278. doi: 10.1007/BF00328222. [DOI] [PubMed] [Google Scholar]
- Hay B.A., Guo M. Caspase-dependent cell death in Drosophila. Annu. Rev. Cell Dev. Biol. 2006;22:623–650. doi: 10.1146/annurev.cellbio.21.012804.093845. [DOI] [PubMed] [Google Scholar]
- Hong A., Narbonne-Reveau K., Riesgo-Escovar J., Fu H., Aladjem M.I., Lilly M.A. The cyclin-dependent kinase inhibitor Dacapo promotes replication licensing during Drosophila endocycles. EMBO J. 2007;26:2071–2082. doi: 10.1038/sj.emboj.7601648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hook S.S., Lin J.J., Dutta A. Mechanisms to control rereplication and implications for cancer. Curr. Opin. Cell Biol. 2007;19:663–671. doi: 10.1016/j.ceb.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpen G.H., Spradling A.C. Reduced DNA polytenization of a minichromosome region undergoing position-effect variegation in Drosophila. Cell. 1990;63:97–107. doi: 10.1016/0092-8674(90)90291-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach T.J., Chotkowski H.L., Wotring M.G., Dilwith R.L., Glaser R.L. Replication of heterochromatin and structure of polytene chromosomes. Mol. Cell. Biol. 2000;20:6308–6316. doi: 10.1128/mcb.20.17.6308-6316.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilly M.A., Duronio R.J. New insights into cell cycle control from the Drosophila endocycle. Oncogene. 2005;24:2765–2775. doi: 10.1038/sj.onc.1208610. [DOI] [PubMed] [Google Scholar]
- Lilly M., Spradling A. The Drosophila endocycle is controlled by Cyclin E and lacks a checkpoint ensuring S-phase completion. Genes & Dev. 1996;10:2514–2526. doi: 10.1101/gad.10.19.2514. [DOI] [PubMed] [Google Scholar]
- Lopez-Schier H., St Johnston D. Delta signaling from the germ line controls the proliferation and differentiation of the somatic follicle cells during Drosophila oogenesis. Genes & Dev. 2001;15:1393–1405. doi: 10.1101/gad.200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan J.P., Chotkowski H.L., Glaser R.L. DNA double-strand break-induced phosphorylation of Drosophila histone variant H2Av helps prevent radiation-induced apoptosis. Nucleic Acids Res. 2002;30:3698–3705. doi: 10.1093/nar/gkf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald A., Caulton J., Edwards M., Floyd A. Loss of centrioles and polyploidization in follicle cells of Drosophila melanogaster. Exp. Cell Res. 1979;118:404–410. doi: 10.1016/0014-4827(79)90167-8. [DOI] [PubMed] [Google Scholar]
- Martin D.N., Baehrecke E.H. Caspases function in autophagic programmed cell death in Drosophila. Development. 2004;131:275–284. doi: 10.1242/dev.00933. [DOI] [PubMed] [Google Scholar]
- May N.R., Thomer M., Murnen K.F., Calvi B.R. Levels of the origin-binding protein Double parked and its inhibitor Geminin increase in response to replication stress. J. Cell Sci. 2005;118:4207–4217. doi: 10.1242/jcs.02534. [DOI] [PubMed] [Google Scholar]
- McCall K. Eggs over easy: Cell death in the Drosophila ovary. Dev. Biol. 2004;274:3–14. doi: 10.1016/j.ydbio.2004.07.017. [DOI] [PubMed] [Google Scholar]
- McCall K., Peterson J.S. Detection of apoptosis in Drosophila. Methods Mol. Biol. 2004;282:191–205. doi: 10.1385/1-59259-812-9:191. [DOI] [PubMed] [Google Scholar]
- McGarry T.J., Kirschner M.W. Geminin, an inhibitor of DNA replication, is degraded during mitosis. Cell. 1998;93:1043–1053. doi: 10.1016/s0092-8674(00)81209-x. [DOI] [PubMed] [Google Scholar]
- Mehrotra S., McKim K.S. Temporal analysis of meiotic DNA double-strand break formation and repair in Drosophila females. PLoS Genet. 2006;2:e200. doi: 10.1371/journal. pgen.0020200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melixetian M., Ballabeni A., Masiero L., Gasparini P., Zamponi R., Bartek J., Lukas J., Helin K. Loss of Geminin induces rereplication in the presence of functional p53. J. Cell Biol. 2004;165:473–482. doi: 10.1083/jcb.200403106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro I., Berry D.L., Huh J.R., Chen C.H., Huang H., Yoo S.J., Guo M., Baehrecke E.H., Hay B.A. The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development. 2006;133:3305–3315. doi: 10.1242/dev.02495. [DOI] [PubMed] [Google Scholar]
- Narbonne Reveau K., Senger S., Pal M., Herr A., Richardson H.E., Asano M., Deak P., Lilly M.A. APC/CFzr/Cdh1 promotes cell cycle progression during the Drosophila endocycle. Development. 2008;135:1451–1461. doi: 10.1242/dev.016295. [DOI] [PubMed] [Google Scholar]
- Nordstrom W., Chen P., Steller H., Abrams J.M. Activation of the reaper gene during ectopic cell killing in Drosophila. Dev. Biol. 1996;180:213–226. doi: 10.1006/dbio.1996.0296. [DOI] [PubMed] [Google Scholar]
- Ollmann M., Young L.M., Di Como C.J., Karim F., Belvin M., Robertson S., Whittaker K., Demsky M., Fisher W.W., Buchman A., et al. Drosophila p53 is a structural and functional homolog of the tumor suppressor p53. Cell. 2000;101:91–101. doi: 10.1016/S0092-8674(00)80626-1. [DOI] [PubMed] [Google Scholar]
- Parisi T., Beck A.R., Rougier N., McNeil T., Lucian L., Werb Z., Amati B. Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. EMBO J. 2003;22:4794–4803. doi: 10.1093/emboj/cdg482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randell J.C., Bowers J.L., Rodriguez H.K., Bell S.P. Sequential ATP hydrolysis by Cdc6 and ORC directs loading of the Mcm2-7 helicase. Mol. Cell. 2006;21:29–39. doi: 10.1016/j.molcel.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Rong Y.S., Titen S.W., Xie H.B., Golic M.M., Bastiani M., Bandyopadhyay P., Olivera B.M., Brodsky M., Rubin G.M., Golic K.G. Targeted mutagenesis by homologous recombination in D. melanogaster. Genes & Dev. 2002;16:1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K., Knoblich J.A., Richardson H., Lehner C.F. Distinct modes of cyclin E/cdc2c kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes & Dev. 1995;9:1327– 1339. doi: 10.1101/gad.9.11.1327. [DOI] [PubMed] [Google Scholar]
- Schwed G., May N., Pechersky Y., Calvi B.R. Drosophila minichromosome maintenance 6 is required for chorion gene amplification and genomic replication. Mol. Biol. Cell. 2002;13:607–620. doi: 10.1091/mbc.01-08-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaprasad U., Dutta A., Bell S.P. Assembly of pre-replication complexes. In: Depamphilis M.L., editor. DNA replication and human disease. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 63–88. [Google Scholar]
- Soloveva V., Linzer D.I. Differentiation of placental trophoblast giant cells requires downregulation of p53 and Rb. Placenta. 2004;25:29–36. doi: 10.1016/S0143-4004(03)00215-7. [DOI] [PubMed] [Google Scholar]
- Spradling A.C. Developmental genetics of oogenesis. In: Bate M., Martinez-Arias A., editors. The development of Drosophila melanogaster. vol. 1. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1993. pp. 1–70. [Google Scholar]
- Su T.T., O’Farrell P.H. Chromosome association of minichromosome maintenance proteins in Drosophila endoreplication cycles. J. Cell Biol. 1998;140:451–460. doi: 10.1083/jcb.140.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada S. Cdt1 and geminin: Role during cell cycle progression and DNA damage in higher eukaryotes. Front. Biosci. 2007;12:1629–1641. doi: 10.2741/2175. [DOI] [PubMed] [Google Scholar]
- Thomer M., May N.R., Aggarwal B.D., Kwok G., Calvi B.R. Drosophila double-parked is sufficient to induce re-replication during development and is regulated by Cyclin E/CDK2. Development. 2004;131:4807–4818. doi: 10.1242/dev.01348. [DOI] [PubMed] [Google Scholar]
- Vaziri C., Saxena S., Jeon Y., Lee C., Murata K., Machida Y., Wagle N., Hwang D.S., Dutta A. A p53-dependent checkpoint pathway prevents rereplication. Mol. Cell. 2003;11:997–1008. doi: 10.1016/s1097-2765(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Walter J.C., Araki H. Activation of pre-replication complexes. In: DePamphilis M.L., editor. DNA replication and human disease. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 89–104. [Google Scholar]
- Whittaker A.J., Royzman I., Orr-Weaver T.L. Drosophila double parked: A conserved, essential replication protein that colocalizes with the origin recognition complex and links DNA replication with mitosis and the downregulation of S phase transcripts. Genes & Dev. 2000;14:1765–1776. [PMC free article] [PubMed] [Google Scholar]
- Wichmann A., Jaklevic B., Su T.T. Ionizing radiation induces caspase-dependent but Chk2- and p53-independent cell death in Drosophila melanogaster. Proc. Natl. Acad. Sci. 2006;103:9952–9957. doi: 10.1073/pnas.0510528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlegel J.A., Dwyer B.T., Dhar S.K., Cvetic C., Walter J.C., Dutta A. Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science. 2000;290:2309–2312. doi: 10.1126/science.290.5500.2309. [DOI] [PubMed] [Google Scholar]
- Wolff T. Histological techniques for the Drosophila eye. Part I: Larva and pupa. In: Sullivan W., et al., editors. Drosophila protocols. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 201–227. [Google Scholar]
- Yoo S.J., Huh J.R., Muro I., Yu H., Wang L., Wang S.L., Feldman R.M., Clem R.J., Muller H.A., Hay B.A. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat. Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lin N., Carroll P.M., Chan G., Guan B., Xiao H., Yao B., Wu S.S., Zhou L. Epigenetic blocking of an enhancer region controls irradiation-induced proapoptotic gene expression in Drosophila embryos. Dev. Cell. 2008;14:481–493. doi: 10.1016/j.devcel.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Dutta A. An ATR- and BRCA1-mediated Fanconi anemia pathway is required for activating the G2/M checkpoint and DNA damage repair upon rereplication. Mol. Cell. Biol. 2006;26:4601–4611. doi: 10.1128/MCB.02141-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W., Chen Y., Dutta A. Rereplication by depletion of geminin is seen regardless of p53 status and activates a G2/M checkpoint. Mol. Cell. Biol. 2004;24:7140–7150. doi: 10.1128/MCB.24.16.7140-7150.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielke N., Querings S., Rottig C., Lehner C., Sprenger F. The anaphase-promoting complex/cyclosome (APC/C) is required for rereplication control in endoreplication cycles. Genes & Dev. 2008;22:1690–1703. doi: 10.1101/gad.469108. [DOI] [PMC free article] [PubMed] [Google Scholar]