Abstract
The SAGA complex is a conserved multifunctional coactivator known to play broad roles in eukaryotic transcription. To gain new insights into its functions, we performed biochemical and genetic analyses of SAGA in the fission yeast, Schizosaccharomyces pombe. Purification of the S. pombe SAGA complex showed that its subunit composition is identical to that of Saccharomyces cerevisiae. Analysis of S. pombe SAGA mutants revealed that SAGA has two opposing roles regulating sexual differentiation. First, in nutrient-rich conditions, the SAGA histone acetyltransferase Gcn5 represses ste11+, which encodes the master regulator of the mating pathway. In contrast, the SAGA subunit Spt8 is required for the induction of ste11+ upon nutrient starvation. Chromatin immunoprecipitation experiments suggest that these regulatory effects are direct, as SAGA is physically associated with the ste11+ promoter independent of nutrient levels. Genetic tests suggest that nutrient levels do cause a switch in SAGA function, as spt8Δ suppresses gcn5Δ with respect to ste11+ derepression in rich medium, whereas the opposite relationship, gcn5Δ suppression of spt8Δ, occurs during starvation. Thus, SAGA plays distinct roles in the control of the switch from proliferation to differentiation in S. pombe through the dynamic and opposing activities of Gcn5 and Spt8.
Keywords: S. pombe, transcription, differentiation, SAGA, Gcn5, Ste11
Transcription initiation is highly regulated and plays critical roles in the growth and differentiation of cells. In eukaryotes, its regulation involves several different classes of factors, including gene-specific regulators, general factors, and coactivators (Martinez 2002). Coactivators are multiprotein complexes that can possess different functions, including histone modification and nucleosome remodeling activities, as well as interactions with gene-specific activators and general transcription factors (Naar et al. 2001). While studies have clearly demonstrated that some coactivator complexes contain multiple activities, there is little understanding of how these activities are coordinated in transcriptional regulation.
One well-studied, multifunctional coactivator is the SAGA complex (Spt–Ada–Gcn5 acetyltransferase). SAGA was initially discovered in Saccharomyces cerevisiae (Grant et al. 1997), where it has been extensively characterized and shown to be composed of 19 distinct subunits (Daniel and Grant 2007). Three sets of SAGA components are known to play direct roles in transcriptional regulation. First, Gcn5 is a histone acetyltransferase (HAT) and, along with two other SAGA components, Ada2 and Ada3 (Ngg1), forms a functional module within SAGA that acetylates histone H3 on four residues—K9, K14, K18, and K23—within nucleosomal substrates (Grant et al. 1999; Balasubramanian et al. 2002). Second, Spt3 and Spt8 recruit the general factor TATA-binding protein (TBP) to certain promoters (Dudley et al. 1999; Bhaumik and Green 2001, 2002; Larschan and Winston 2001). Spt3 and Spt8 also have been shown to negatively regulate the recruitment of TBP (Belotserkovskaya et al. 2000; Yu et al. 2003; Warfield et al. 2004). Third, Ubp8 is a histone deubiquitylase for histone H2B at K123 (Henry et al. 2003; K.K. Lee et al. 2005). Ubp8 forms a functional module with other SAGA components—Sgf11, Sgf73, and Sus1 (Ingvarsdottir et al. 2005; K.K. Lee et al. 2005; Shukla et al. 2006; Kohler et al. 2008). A combination of genetic, microarray, and structural studies strongly suggests that these three sets of SAGA factors function in distinct and independent fashions within SAGA (Grant et al. 1997; Roberts and Winston 1997; Sterner et al. 1999; Lee et al. 2000; Ingvarsdottir et al. 2005). Intriguingly, one study reported that different SAGA activities, Gcn5 and Spt3, have opposing roles in transcriptional regulation of the HO gene in S. cerevisiae (Yu et al. 2003). Finally, other SAGA components have been shown to have other transcriptional roles, including interactions with transcriptional activators and other general transcription factors, mRNA export, transcription elongation, and structural integrity of SAGA (Daniel and Grant 2007).
SAGA is highly conserved, as orthologous complexes with compositions similar to S. cerevisiae SAGA have been identified in larger eukaryotes, including human and Drosophila (Lee and Workman 2007; Nagy and Tora 2007). In addition, electron microscopic studies have shown a high degree of structural conservation between the human and yeast complexes (Brand et al. 1999; Wu et al. 2004). Several studies have revealed a variety of important roles for SAGA in metazoans, such as disease, development, DNA repair, and gene regulation (Lee and Workman 2007; Nagy and Tora 2007). Thus, SAGA plays broad and important regulatory roles from yeast to human.
We initiated analysis of SAGA from the fission yeast Schizosaccharomyces pombe. S. pombe is highly divergent from S. cerevisiae (Wood et al. 2002), and numerous studies have demonstrated that, in many ways, S. pombe chromatin structure and transcriptional regulation are more closely related to mammalian cells than to S. cerevisiae (Grewal and Jia 2007). Therefore, study of SAGA in S. pombe provides an opportunity to elucidate the functions of this coactivator in a highly divergent organism, but with the availability of all the tools of yeast research. Some studies have already identified and initiated characterization of putative components of S. pombe SAGA. Among these, S. pombe Gcn5 was identified previously by sequence homology (Mitsuzawa et al. 2001) and, together with Ada2, was shown to contribute to local meiotic recombination through histone H3-K9 and K14 acetylation at the ade6-M26 hotspot (Yamada et al. 2004; Hirota et al. 2008). In addition, microarray analyses have shown that Gcn5 is involved in the regulation of a subset of genes induced during salt stress (Johnsson et al. 2006). Other S. pombe orthologs of S. cerevisiae SAGA components have also been identified by sequence homology, including Spt3 (Madison and Winston 1998), Taf5 (Yamamoto et al. 1997; Mitsuzawa et al. 2001), Taf6 (Mitsuzawa and Ishihama 2002), and Tra1 (Hayashi et al. 2007). Finally, sep9+, which was identified in a screen for genes required for cytokinesis and sexual differentiation, was suggested to be the ortholog of S. cerevisiae SPT8 (Grallert et al. 1999; Sipiczki et al. 1999).
Here, we characterized the S. pombe SAGA complex using both biochemical and genetic approaches. We purified S. pombe SAGA and found that its subunit composition is identical to that of S. cerevisiae, emphasizing the high degree of conservation of this complex. Genetic analysis of S. pombe SAGA mutants has revealed that SAGA plays at least two key roles during the sexual differentiation pathway. This pathway, from mating to meiosis, is specifically activated in G1 in response to nutrient starvation, and it serves as an excellent model to study how cells respond to external cues to initiate differentiation (Davey 1998; Harigaya and Yamamoto 2007). The switch from proliferation to differentiation is driven by an extensive gene expression program, controlled by the master regulator Ste11, a high-mobility-group box-containing transcription factor (Sugimoto et al. 1991; Mata and Bähler 2006; Mata et al. 2007). Among the direct targets of Ste11 activation is mei2+, which functions as the trigger of meiosis. Our studies show that Gcn5 is required to repress transcription of ste11+ and mei2+ in nutrient-rich conditions, where these genes are normally repressed. In contrast, Spt8 is required for the induction of these genes that normally occurs upon a shift to starvation medium. Chromatin immunoprecipitation (ChIP) analyses suggested that these opposing regulatory roles are direct. Finally, genetic evidence suggests that Gcn5 and Spt8 play antagonistic and dynamic roles to allow SAGA to function both as a negative and a positive regulator, depending on nutrient conditions. Taken together, our results reveal that the control of a master regulator of cell fate decision, S. pombe Ste11, depends on different activities of the SAGA complex for the switch from mitotic growth to sexual differentiation.
Results
The composition of the SAGA complex is identical between S. pombe and S. cerevisiae
To study the SAGA coactivator complex from S. pombe, we first wanted to identify all the components of the complex. Homology searches identified the putative S. pombe orthologs of the S. cerevisiae SPT7 and ADA1 (HFI1) genes, which encode two core SAGA subunits. To purify S. pombe SAGA, a tandem affinity purification (TAP) sequence was fused onto the 3′ end of each of these two S. pombe genes. Both spt7∷TAP and ada1∷TAP alleles appeared functional, as these strains did not exhibit the poor growth phenotype observed for strains in which these genes were deleted (our unpublished observations). Each TAP-tagged protein was then affinity-purified using three salt concentrations (150 mM NaCl, 350 mM NaCl, and 600 mM NaCl), and the copurifying proteins were analyzed by mass spectrometry. By this analysis, purifications of Spt7 and Ada1 each identified an identical complex of 19 proteins at all three salt concentrations (Supplemental Table 1). The composition of S. pombe SAGA is highly conserved, as each member of the complex was identified as the corresponding ortholog of each subunit of S. cerevisiae SAGA (Supplemental Table 1). We also separated the purified SAGA complex on a gradient SDS-PAGE and visualized it by silver staining, revealing several specific bands at the expected sizes (Fig. 1). A few other proteins were also recovered but are likely nonspecific, as all corresponded to highly abundant ribosomal proteins, translation factors, and metabolic enzymes. We did not detect additional subunits that have been identified as part of related SAGA complexes, such as Chd1, Rtg2, and Ahc1 in S. cerevisiae, or Atac1 in Drosophila melanogaster (Eberharter et al. 1999; Pray-Grant et al. 2002, 2005; Guelman et al. 2006). BLAST searches failed to identify proteins orthologous to S. cerevisiae Rtg2 and Ahc1 or to D. melanogaster Atac1 in S. pombe. We thus purified the SAGA coactivator complex from S. pombe and showed that its subunit composition is identical to that of S. cerevisiae and mammalian SAGA complexes (Supplemental Table 2). This result fits well with the high degree of conservation in the composition and structure of SAGA suggested by previous studies (Wu et al. 2004; Lee and Workman 2007).
Figure 1.
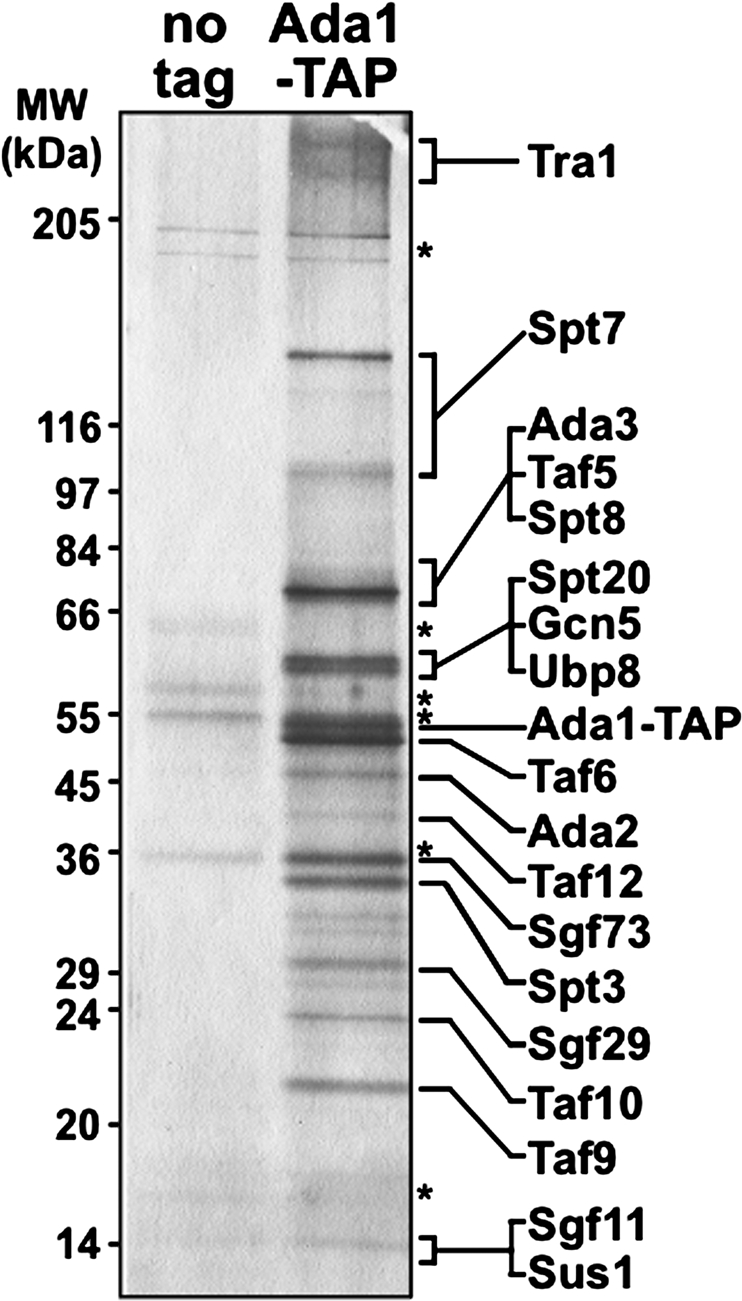
Purification of the SAGA coactivator complex from S. pombe. Proteins copurifying with Ada1-TAP were separated on an SDS 5%–20% gradient polyacrylamide gel and visualized by silver nitrate staining. An untagged control strain is shown (“no tag”), and corresponding nonspecific bands are marked with asterisks. Each putative SAGA subunit was assigned to a specific band on the gel based on predicted molecular weight and isoelectric point (Supplemental Table 2) and, in some cases, Western blot analysis. The positions of molecular weight markers (MW) are shown on the left.
Transcriptome analysis of the SAGA HAT module suggests a role in mating and meiosis
SAGA has been previously shown to regulate transcription using many different activities (Daniel and Grant 2007). We focused on the HAT function, which has been studied extensively both in S. cerevisiae and in mammalian systems (Nagy and Tora 2007). Gcn5 possesses the catalytic HAT activity of the SAGA complex (Grant et al. 1997), and two other SAGA components, Ada2 and Ada3, are required for Gcn5 to acetylate nucleosomal histone H3 (Grant et al. 1999). To identify the genes whose expression is regulated by SAGA HAT activity, we constructed gcn5Δ, ada2Δ, and ada3Δ mutants and performed microarray analyses, comparing each mutant with wild-type. The expression profiles of all three mutants were highly similar and, surprisingly, the expression of only a small number of genes was affected more than twofold (∼55 genes; 1.1% of total) (Fig. 2A; Supplemental Tables 3–5). In S. cerevisiae, gcn5Δ causes a greater effect, as mutant cells show a modest, global diminution of transcript levels (Huisinga and Pugh 2004) with the expression of ∼4% of all genes being affected more than twofold (Lee et al. 2000). To test for such a global effect in our data set, the expression data were renormalized using bacterial spikes added in equal quantities to the RNA samples. This approach allows detection of global changes that would otherwise be lost after classical microarray normalization procedures (K.M. Lee et al. 2005). Interestingly, we were unable to detect any global decrease in transcript levels in gcn5Δ, ada2Δ, or ada3Δ mutants by this analysis (data not shown). Our results, then, suggest that the HAT module of SAGA regulates a limited number of genes or it is functionally redundant with other regulators in S. pombe. The high overlap between Gcn5-, Ada2- and Ada3-regulated genes in S. pombe is consistent with previous data showing that these three subunits form a distinct functional module in S. cerevisiae.
Figure 2.
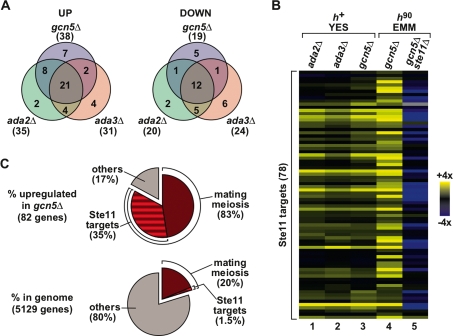
Transcriptome analysis of SAGA HAT mutants. (A) Venn diagrams show the high degree of overlap of genes with increased (“UP,” left panel) or reduced (“DOWN,” right panel) mRNA levels in heterothallic h+ gcn5Δ, ada2Δ and ada3Δ deletion mutants, grown in complete rich media (YES). The total number of genes affected more than twofold for each data set is indicated in parentheses. (B) Hierarchical clustering of the 78 Ste11 target genes, as defined previously (Mata and Bähler 2006), shows the increased level of mRNAs for the Ste11-dependent targets in gcn5Δ, ada2Δ, and ada3Δ deletion mutants. The dendrogram shows Ste11 target genes in rows with their expression changes in h+ gcn5Δ, ada2Δ, and ada3Δ mutants grown in YES (columns 1–3) or in h90 gcn5Δ and gcn5Δ ste11Δ mutants grown in minimal rich medium (EMM) (columns 4,5). The data are presented as a mutant/wild-type ratio of hybridization signals and are color-coded as indicated in the key. (C) Pie charts showing the enrichment of functional categories related to mating and meiosis in the set of genes with increased mRNA levels in homothallic h90 gcn5Δ mutants grown in EMM (top chart), compared with their representation in the genome (bottom chart). The percentage value of a category in the genome corresponds to the percentage observed if that category was found randomly in the data set. The category “mating/meiosis” (dark red, 958 genes) includes all genes whose expression is up-regulated during pat1-114-driven meiosis (as defined in Mata et al. 2002), including the set of “Ste11 targets” (dark- and light-red stripes, 78 genes, defined in A). The category “others” contains all genes not present in either of the two categories mentioned above.
To determine whether any particular biological process was affected in gcn5Δ, ada2Δ, and ada3Δ mutants, genes whose expression was significantly altered in all three mutants were analyzed for enrichment of Gene Ontology (GO) categories (Aslett and Wood 2006) or transcriptional modules (Tanay et al. 2005). Among the genes with decreased mRNA levels, there was no significant enrichment for any category. However, the set of genes with increased mRNA levels was significantly enriched for genes involved in mating or meiosis and sporulation (in gcn5Δ mutants, P = 3.2 × 10−5 and 9.8 × 10−6 for mating and meiosis-sporulation genes, respectively). This observation prompted us to look specifically at genes known to be targets of the transcription factor Ste11, the master regulator of the sexual differentiation pathway, which directs cells from mitotic growth to mating and meiosis in S. pombe (Sugimoto et al. 1991; Mata and Bähler 2006). We found that Ste11 target genes were significantly enriched and represented ∼20% of genes with elevated mRNA levels in gcn5Δ, ada2Δ, and ada3Δ mutants (Fig. 2B, columns 1–3). This set included ste11+ itself, whose expression is autoregulated (Kunitomo et al. 2000).
These expression profiles were obtained from a heterothallic strain, which contains cells of only one mating type and is thus unable to express all the genes required for sexual differentiation. Therefore, we recreated the gcn5Δ mutant strain in a mating-competent homothallic h90 background and measured mRNA levels by microarrays. In the h90 gcn5Δ strain grown under rich conditions, we found that 82 genes, including ste11+, had increased mRNA levels, and 36 genes had decreased levels greater than twofold (Supplemental Tables 6, 7). Analysis of GO categories in the set of genes with increased mRNA levels revealed an even more striking representation of genes involved in all aspects of sexual differentiation (P = 2.4 × 10−19 for mating genes; P = 3.7 × 10−38 for meiosis-sporulation genes; P = 5.3 × 10−30 for Ste11 target genes). Of the 82 genes with increased mRNA levels in gcn5Δ mutants, 68 (83%) are involved in mating and/or meiosis, including 29 (35%) direct Ste11 target genes (Fig. 2C). These percentages represent a highly significant enrichment over their proportion in the S. pombe genome (20%, P = 3.6 × 10−37 for mating/meiosis; 1.5%, P = 2.4 × 10−136 for Ste11 targets). To test the role of Ste11 in the derepression of mating and meiosis genes observed in the gcn5Δ mutant, an h90 gcn5Δ ste11Δ double mutant was constructed and analyzed by microarrays. In this case, we did not observe enrichment of any mating- or meiosis-related GO categories within the set of genes with increased mRNA levels (Supplemental Tables 6, 8). In fact, Ste11 targets appeared to have decreased mRNA levels in h90 gcn5Δ ste11Δ double mutants (Fig. 2B, cf. columns 4 and 5). A similar observation has been made in h90 ste11Δ fus1Δ double mutants (Mata and Bähler 2006) and is likely attributable to a basal activity of Ste11 even in nutrient-rich conditions. Together, these results suggest that the widespread derepression of mating and meiosis genes in the absence of Gcn5 is the consequence of increased Ste11 activity, suggesting that Gcn5 negatively regulates mating in nutrient-rich conditions by repressing Ste11 expression and/or activity.
Regulation of ste11+ is specific for the HAT module of SAGA
Given the results of our microarray experiments, we wanted to characterize further the transcriptional effects caused by gcn5Δ, ada2Δ, and ada3Δ mutations, as well as to determine whether these changes are specific for the HAT function of SAGA. First, we used Northern hybridization analysis to measure the mRNA levels of two Ste11 targets, ste11+ and mei2+. Our results show that in gcn5Δ, ada2Δ, and ada3Δ mutants, ste11+ and mei2+ mRNA levels are increased approximately fourfold and 10-fold, respectively, in the three mutant strains, compared with wild type (Fig. 3A, lanes 1–4). To examine the genetic relationship between gcn5+, ada2+, and ada3+ in this regulation, we analyzed gcn5Δ ada2Δ and gcn5Δ ada3Δ double mutants, as well as a gcn5Δ ada2Δ ada3Δ triple mutant. Northern hybridization analysis of these strains showed very similar effects to the single mutants, as ste11+ and mei2+ mRNA levels were again increased approximately fourfold and 10-fold, respectively (Fig. 3A, lanes 5–7). To test whether the Gcn5 acetylation activity is required for ste11+ regulation, we tested a Gcn5-E191Q mutant specifically impaired for this activity. Our results (Supplemental Fig. 1A) show that, similar to gcn5Δ, this mutant also has increased ste11+ and mei2+ mRNA levels. These results strongly suggest that all three genes regulate ste11+ and mei2+ by a common mechanism, likely Gcn5-dependent acetylation.
Figure 3.
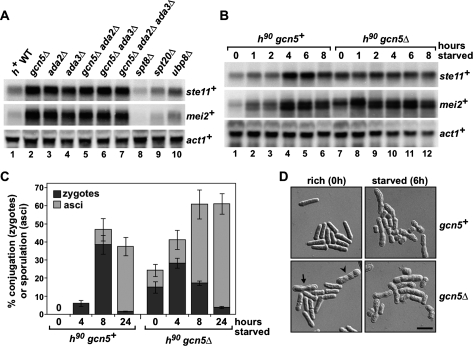
Gcn5 negatively regulates sexual differentiation and expression of mating genes. (A) Northern blot analysis of ste11+ and mei2+ mRNA levels in nutrient-rich conditions, showing that depression of transcription is specific to strains with gcn5Δ, ada2Δ, and ada3Δ mutations. RNA was prepared from h+ heterothallic strains grown in rich medium. act1+ served as a loading control. (B) Northern blot analysis of a time course of ste11+ and mei2+ induction upon nutrient starvation. RNA was prepared from h90 homothallic prototrophic wild-type and gcn5Δ strains. Cells were grown to mid-log phase in minimal rich medium and then shifted for the indicated time (in hours) to starvation medium. act1+ served as a loading control. Each Northern blot is representative of four independent experiments. (C,D) Cells were processed as described in B and collected at the indicated time points. Cells were examined under direct light microscopy (C) or differential interference contrast microscopy (D). Conjugation or sporulation rates (%) were calculated by dividing the number of zygotes (one zygote counted as two cells) or asci (one ascus counted as two cells) by the number of total cells (at least 300 cells). The numbers of zygotes (dark gray) or asci (light gray) are shown as stacked columns. Thus, each column corresponds to the total percentage of cells undergoing sexual differentiation. Each column represents the mean value ± SEM (n = 3–6). Representative photomicrographs are shown in D. The arrow indicates a zygote and the arrowhead indicates an ascus in an h90 gcn5Δ mutant grown in rich medium. Bar, 10 μm.
To characterize the gcn5Δ mutant phenotype during the induction of the sexual differentiation pathway, we also performed a time-course analysis of ste11+ and mei2+ expression upon nutrient starvation of h90 wild-type and gcn5Δ cells (see the Materials and Methods). In wild type, as expected, both ste11+ and mei2+ mRNA levels increased during starvation, peaking 4 h after the shift to starvation medium (Fig. 3B, lanes 1–6). In contrast, in gcn5Δ mutants, ste11+ mRNA levels were almost maximally derepressed in rich medium, and they remained constant during starvation. mei2+ mRNA levels, already elevated in gcn5Δ mutants in rich medium, were further induced during starvation (Fig. 3B, lanes 7–12). Notably, starvation-induced induction of mei2+ was faster and greater in gcn5Δ mutants than in wild-type strains and affected the upper, slower migrating band (Fig. 3B, cf. lanes 2 and 8). Starvation-regulated expression of multiple mei2+ transcripts has been previously described (Shimoda et al. 1987). Together, these results show that Gcn5, Ada2, and Ada3 act in a common pathway to specifically repress ste11+ and mei2+ expression, suggesting that the HAT module of the SAGA complex negatively regulates mating gene expression in nutrient-rich conditions.
Finally, to test whether the regulation of ste11+ and mei2+ is restricted to the HAT module of SAGA, we constructed spt8Δ, ubp8Δ, and spt20Δ deletion mutants and performed similar Northern experiments. Spt8 and Ubp8 represent SAGA activities that are distinct and independent from Gcn5, and Spt20 is a core component of SAGA, contributing to SAGA integrity (Grant et al. 1997; Wu and Winston 2002). We found that neither ste11+ nor mei2+ mRNA levels were significantly increased in any of these mutants (Fig. 3A, lanes 8–10). Thus, SAGA-mediated repression of ste11+ and mei2+ appears to be dependent specifically on its HAT module. As described below, Spt8 is required for induction of ste11+ and mei2+ transcription upon induction of the sexual differentiation pathway.
Gcn5 negatively regulates sexual differentiation in nutrient-rich conditions
In wild-type S. pombe strains, no mating occurs unless the cells are shifted to the appropriate starvation medium. Overexpression of ste11+, however, has been shown to be sufficient to induce mating independently of nutrient levels (Sugimoto et al. 1991). The constitutive expression of ste11+ in gcn5Δ mutants prompted us to examine the mating behavior of these mutants. Microscopic examination of liquid cultures of homothallic gcn5Δ mutants revealed that, in nutrient-rich conditions, 15% of cells had undergone conjugation and formed zygotes and, in addition, 9.2% of cells had sporulated and formed asci, whereas no zygotes or asci were observed in a wild-type control culture (Fig. 3C,D). During nutrient starvation, the gcn5Δ mutants were still able to sense nutrient depletion as the level of zygotes and asci increased, reaching the same level that was eventually achieved by wild-type cultures (Fig. 3C). This increase in gcn5Δ mutants is consistent with the fact that expression of mei2+ is further induced during starvation in gcn5Δ mutants (Fig. 3B). Overall, these results demonstrate that the elevated levels of Ste11 targets in gcn5Δ mutants are sufficient to induce mating and meiosis under normally nonpermissive conditions.
Evidence that Gcn5 directly represses the ste11+ and mei2+ promoters as part of the SAGA complex
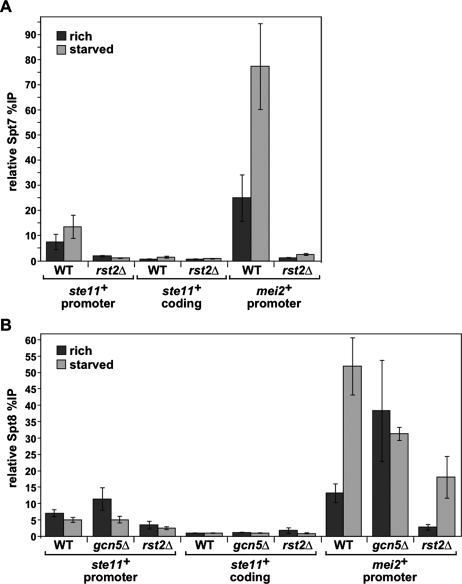
To test whether Gcn5 directly regulates mating genes, we determined its physical association with the ste11+ and mei2+ regulatory regions in rich medium, when these genes are repressed, and under starvation conditions, when they are induced. For this analysis, we performed ChIP experiments using an h90 prototrophic strain in which Gcn5 was tagged with 13 copies of a myc epitope (Materials and Methods). In rich medium, our results showed a significant level of Gcn5 association at both the ste11+ and mei2+ regulatory regions (Fig. 4A). To test the specificity of this association, we also examined the entire ste11+ gene and found that Gcn5 is specifically recruited to the proximal region of the ste11+ promoter (Fig. 4A; Supplemental Fig. 2). If Gcn5 serves as a repressor of ste11+ and mei2+, then one would expect that when these genes are induced upon nutrient starvation, the level of Gcn5 association would decrease. In contrast to this expectation, we observed approximately the same level of Gcn5 association at the ste11+ promoter upon a shift to starvation medium (Fig. 4A). Furthermore, we observed a fivefold increase of Gcn5 association at the mei2+ promoter upon nutrient starvation (Fig. 4A). This specific increase of Gcn5 occupancy at the mei2+ promoter is consistent with the fact that, during starvation, mei2+ expression is induced more dramatically than ste11+ (Figs. 3B, 6, below). These results show that Gcn5 is physically associated with the ste11+ and mei2+ promoters in both rich and starved conditions. Thus, the binding profile of Gcn5 at these promoters is not consistent with a role for Gcn5 as a repressor.
Figure 4.
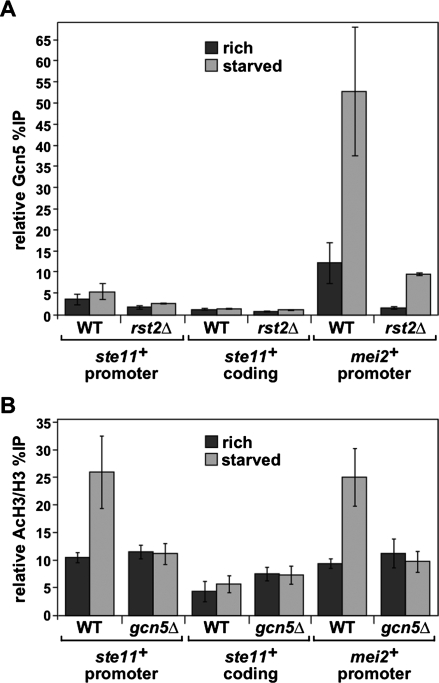
Gcn5 is physically associated with the ste11+ and mei2+ promoters in nutrient-rich and starved conditions. (A) Gcn5 occupancy was assessed by ChIP analysis of chromatin extracts from strains in which Gcn5 was myc-tagged either in a wild-type or in an rst2Δ background. Cells were grown to mid-log phase in minimal rich medium and then shifted for 4 h to either rich medium (dark gray) or starvation medium (light gray). Occupancy levels were quantified by real-time PCR of anti-myc immunoprecipitation over input DNA and normalized to background levels detected at a nontranscribed region (mat3), which are set at 1. The promoter and coding regions within ste11+ that were tested are depicted in Supplemental Figure 2 (promoter, primer pair “B”; coding, primer pair “D”). The background percentage immunoprecipitation values obtained from ChIP analysis of the corresponding “no tag” strains are shown in Supplemental Figure 6. Each column represents the mean percnetage immunoprecipitation value ± SEM (n = 4–10). (B) Culture conditions and analyses are as described in A, except using wild-type or gcn5Δ mutant strains. ChIPs were performed using antibodies specific for acetylated H3-K9/K14 residues (AcH3) or against unmodified histone H3 (H3). Individual AcH3, H3, and “no antibody” percentage immunoprecipitation values are presented in Supplemental Figure 3A–C. Each column represents the mean percentage immunoprecipitation value ± SEM (n = 4).
Figure 6.
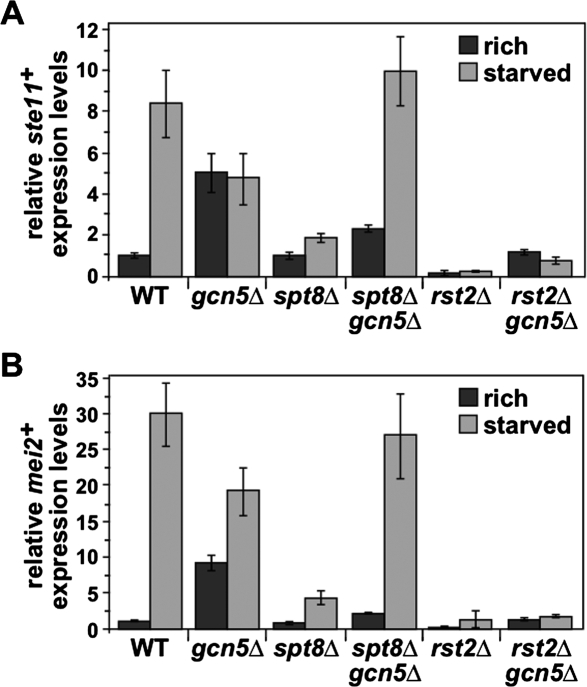
Opposing genetic interactions between Gcn5 and Spt8 in the regulation of ste11+ and mei2+ transcription. Expression analysis of ste11+ (A) and mei2+ (B) by quantitative RT–PCR, showing an antagonistic genetic interaction between Gcn5 and Spt8 that differs, depending on nutrient levels. RNA was prepared from h90 homothallic strains. Cells were grown to mid-log phase in minimal rich medium and then shifted for 4 h to either rich medium (dark gray) or starvation medium (light gray). act1+ served as a control for normalization across RT–PCR samples. Normalized ste11+ or mei2+ mRNA levels in a wild-type strain grown in rich medium were set at 1 to allow comparisons across culture conditions and mutant strains. Each column represents the mean value ± SEM (n = 4).
To test whether Gcn5 affects the level of histone H3 acetylation, we performed additional ChIP experiments using antibodies specific for H3-K9/K14 acetylation. Our results show that the level of acetylation of histone H3-K9/K14 (normalized to the level of total histone H3) increases ∼2.5-fold at the ste11+ and mei2+ promoters upon induction of these genes by starvation (Fig. 4B; Supplemental Fig. 3). In gcn5Δ mutants, the starvation-induced H3-K9/K14 hyperacetylation was absent at both promoters, although the basal level of H3-K9/K14 acetylation remained similar to that seen in wild-type strains (Fig. 4B). As an additional control, we measured H3-K9/K14 acetylation at the act1+ promoter, which is not affected by nutrient starvation, and found no Gcn5-dependent acetylation (data not shown). Western blot analysis showed a reduction of H3-K9/K14 acetylation levels in gcn5Δ and gcn5-191 (encoding E191Q) mutants compared with wild type (Supplemental Fig. 1B), suggesting that most H3-K9/K14 acetylation is Gcn5-dependent. Thus, Gcn5 is required for hyperacetylation of the ste11+ and mei2+ promoters upon their induction. However, the hyperacetylation of H3 at ste11+ and mei2+ does not correlate with a role for Gcn5 as a repressor, suggesting that Gcn5 represses these genes by a different mechanism than histone acetylation.
Previous studies in S. cerevisiae have established that Gcn5 is present in other complexes besides SAGA, including two SAGA-related complexes named ADA and SLIK/SALSA (Eberharter et al. 1999; Pray-Grant et al. 2002; Sterner et al. 2002; Wu and Winston 2002). To gain evidence that it is SAGA, rather than another Gcn5-related complex, that is physically present at the ste11+ and mei2+ promoters, we performed additional ChIP experiments to test the association of two other SAGA components, Spt7 and Spt8. In S. cerevisiae, Spt7 is in both SAGA and SLIK/SALSA, but not ADA, while Spt8 is only in SAGA. Our results show that, like Gcn5, both Spt7 and Spt8 are associated at the ste11+ promoter regardless of nutrient levels and of its mRNA levels. At the mei2+ promoter, we observed a significant level of Spt7 and Spt8 association in rich medium and a threefold to fivefold increase of their recruitment upon a shift to starvation medium (Fig. 5). Similar ChIP results were obtained for another SAGA core component, Ada1 (data not shown). These results suggest that Gcn5 plays a direct role in repression of ste11+ and mei2+ during growth in rich medium as part of SAGA. Finally, we performed ChIP analysis of Spt8 in a gcn5Δ mutant, in which ste11+ and mei2+ are induced regardless of nutrient levels. We found that Spt8 association at the mei2+ promoter was significantly increased in gcn5Δ mutant cells grown in rich medium, as compared with wild-type cells (Fig. 5B). Thus, the binding profile of SAGA positively correlates with the levels of expression of mei2+, suggesting that SAGA plays an additional role in the regulation of some Ste11 target genes and contributes to their induction.
Figure 5.
Spt7 and Spt8 are physically associated with the ste11+ and mei2+ promoters in nutrient-rich and starved conditions. Spt7 (A) or Spt8 (B) occupancy was assessed by ChIP analysis of chromatin extracts from strains in which either Spt7 or Spt8 was myc-tagged in a wild-type, gcn5Δ, or rst2Δ background. Culture conditions and analyses are as described in the legend for Figure 4. Each column represents the mean percentage immunopreciptation value ± SEM (n = 3–5).
Our ChIP results show that Gcn5 is specifically associated over the UASst and TR1 regulatory sites in the ste11+ promoter (Fig. 4A; Supplemental Fig. 2). Previous work showed that these sites are bound by the two main activators of ste11+ transcription, Rst2 and Ste11 itself (Sugimoto et al. 1991; Kunitomo et al. 2000). Furthermore, deletion of rst2+ completely abolishes ste11+ (Kunitomo et al. 2000; Higuchi et al. 2002) and mei2+ transcription regardless of nutrient levels (Fig. 6A,B). To test if Rst2 is required for SAGA recruitment, we performed additional ChIP analysis of Gcn5, Spt7, and Spt8 in an rst2Δ mutant. For all three SAGA components tested, their association at the ste11+ and mei2+ promoters was greatly reduced in the rst2Δ mutant when cells were grown in either rich or starvation medium (Figs. 4A, 5). As a control, Western blot analysis showed that steady-state levels of Gcn5, Spt7, and Spt8 were not affected in rst2Δ mutants compared with wild-type cells (Supplemental Fig. 4). These results show that Rst2 is required for recruitment of SAGA at ste11+ and mei2+ promoters in both nutrient conditions.
The SAGA subunits Spt8 and Spt20 positively regulate mating in S. pombe
The transcriptional changes in gcn5Δ mutants, along with our ChIP results, suggest that Gcn5 directly represses ste11+ and mei2+ transcription. However, the binding profile of the SAGA complex at the ste11+ and mei2+ promoters suggested that SAGA may play a second role in the regulation of these two genes—their activation upon nutrient starvation. Given the results described above, such an activation role would not be dependent on Gcn5, Ada2, or Ada3, but rather, on other SAGA components. To examine this possibility, we tested mating in deletion mutants for three other SAGA components—Ubp8, Spt8, and Spt20. As shown above, Ubp8, Spt8, and Spt20 are not required for repression of ste11+ and mei2+ transcription in rich medium (Fig. 3A). We found that heterothallic spt8Δ and spt20Δ strains were sterile and unable to mate even when a wild-type mating partner was provided, whereas heterothallic ubp8Δ showed normal mating, comparable with that of wild type (data not shown). These results suggest that the SAGA subunits Spt8 and Spt20 have a positive role in mating, in contrast with the negative role of Gcn5, Ada2, and Ada3.
To measure mating efficiency more quantitatively, we constructed spt8Δ and spt20Δ mutants in an h90 (homothallic) background and performed quantitative mating assays in liquid culture. To compare these SAGA mutants with previously characterized mutants that are also defective for mating, we included ste11Δ and rst2Δ mutants in our analysis. Our results confirmed that both spt8Δ and spt20Δ mutants are unable to mate upon starvation, even after 24 h, behaving similarly to the ste11Δ and rst2Δ mutants (Supplemental Table 9). To determine the epistasis relationship between the nonmating phenotype of spt8Δ and spt20Δ mutants with the constitutive mating phenotype of a gcn5Δ mutant, we constructed and assayed spt8Δ gcn5Δ and spt20Δ gcn5Δ double mutants. These experiments showed that the double mutants are unable to mate; that is, spt8Δ and spt20Δ each suppresses the gcn5Δ mutant mating phenotype. Similar observations were made for the ste11Δ gcn5Δ and rst2Δ gcn5Δ double mutants. These results show that Spt8 and Spt20 are required for the positive regulation of S. pombe sexual differentiation. Furthermore, the constitutive mating phenotype observed in gcn5Δ mutants is dependent on these functions, as well as on the transcription activators Ste11 and Rst2. Thus, different SAGA subunits, although part of the same coactivator complex, have opposite roles in the regulation of mating in S. pombe.
Genetic interactions between Spt8 and Gcn5 in regulation of mating gene expression
The spt8Δ sterility phenotype and the association of SAGA with the ste11+ and mei2+ promoters raised the possibility that Spt8 is required for the induction of ste11+ and mei2+ transcription upon starvation. To address this possibility, we performed quantitative RT–PCR to measure ste11+ and mei2+ mRNA levels in an spt8Δ strain compared with the levels in wild-type and gcn5Δ strains. This analysis revealed that spt8Δ mutants are severely impaired for induction of both genes (Fig. 6). In wild type, ste11+ and mei2+ mRNA levels increased 11-fold and 36-fold upon nutrient starvation, respectively. However, while spt8Δ mutants had similar ste11+ and mei2+ mRNA levels in rich medium, the levels increased only 1.7-fold and fivefold upon nutrient starvation (Fig. 6). Our results also provided a quantitative measurement of the derepression of ste11+ and mei2+ mRNA levels in a gcn5Δ mutant, showing that ste11+ and mei2+ mRNA levels are increased by 5.3-fold and 9.9-fold, respectively, compared with wild-type cells in nutrient-rich conditions. Upon starvation, ste11+ was not further induced, while mei2+ mRNA levels increased by 2.5-fold in gcn5Δ mutants (Fig. 6A,B). These RT–PCR experiments, then, provide evidence that Spt8 is required for activation of ste11+ and mei2+ in starvation medium, while Gcn5 is required for repression of ste11+ and mei2+ in rich medium.
Given the opposite regulatory roles of Gcn5 and Spt8, we wanted to determine which one acts more directly in regulation of ste11+ and mei2+ transcription. To do this, we constructed an spt8Δ gcn5Δ double mutant and determined its effect on ste11+ and mei2+ mRNA levels under both rich and starvation conditions. In rich medium, the spt8Δ gcn5Δ double mutant has a low level of ste11+ and mei2+ mRNA, far below the levels of a gcn5Δ single mutant and more similar to the levels in spt8Δ and wild-type strains (Fig. 6A,B). Thus, in rich medium, the spt8Δ mutation suppresses the aberrant activation caused by the gcn5Δ mutation. However, during starvation, the opposite result was obtained, as the spt8Δ gcn5Δ double mutant had a high level of ste11+ and mei2+ mRNA, similar to the levels in gcn5Δ and wild-type strains (Fig. 6A,B). During starvation, then, gcn5Δ suppresses spt8Δ. Therefore, the induction of ste11+ and mei2+ upon starvation does not require Spt8 if Gcn5 is absent. We then compared this behavior of spt8Δ with another mutation that impairs ste11+ activation, rst2Δ. In marked contrast with what we observed in an spt8Δ gcn5Δ double mutant, in an rst2Δ gcn5Δ double mutant, both ste11+ and mei2+ mRNA levels are greatly decreased, regardless of nutrient levels (Fig. 6A,B). Taken together, these results suggest that nutrient levels modulate the roles of Gcn5 and Spt8 to control whether SAGA acts to repress or activate ste11+ and mei2+ transcription.
Discussion
In this study, we provided the first characterization of the S. pombe SAGA coactivator complex, and we identified key roles for this complex in regulation of the S. pombe sexual differentiation pathway. Previous studies had found a high level of conservation for SAGA complexes between S. cerevisiae, D. melanogaster, and mammals (Lee and Workman 2007), and our studies showed that S. pombe SAGA is also highly conserved, thus establishing S. pombe as a relevant system for the study of SAGA function in vivo. Our results showed that, depending on nutrient levels, SAGA uses at least two of its multiple activities to perform both negative and positive regulatory roles in the regulation of the S. pombe sexual differentiation pathway. First, during growth in rich medium, Gcn5 is a negative regulator of genes required for mating and meiosis, including ste11+, which encodes the master regulator of sexual differentiation in S. pombe. In contrast, upon starvation, another SAGA subunit, Spt8, is required for induction of ste11+ and mei2+. ChIP experiments indicate that Gcn5 and Spt8 perform these regulatory roles as part of SAGA and that SAGA is recruited to the promoters of Ste11 target genes by the transcriptional activator Rst2.
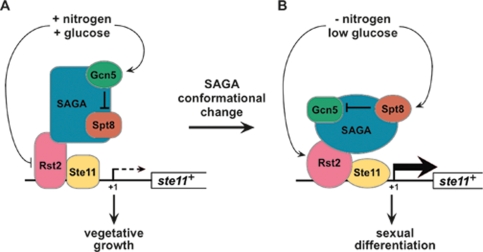
Our results suggest a model in which SAGA responds to a change in nutrient levels in a dynamic fashion to allow it to switch from a negative to a positive regulator of Ste11 target genes (Fig. 7). According to this model, in nutrient-rich conditions, SAGA is recruited to Ste11 target gene promoters, where Gcn5 acts to inhibit Spt8 activity (Fig. 7A). This aspect of the model is supported by our result that, in gcn5Δ mutants, Spt8 is required for the aberrant activation of Ste11 target genes in rich medium. Then, upon nutrient starvation, the opposite occurs, and Gcn5 is inhibited by Spt8 in order for SAGA to activate Ste11 target genes, thereby committing cells to the sexual differentiation pathway (Fig. 7B). This aspect of the model is supported by our finding that the defect in activation of the pathway caused by spt8Δ is suppressed by gcn5Δ. In response to changes in nutrient levels, SAGA may undergo a change in either conformation or in composition specifically at Ste11 target gene promoters in order to regulate the antagonistic interaction between Gcn5 and Spt8. Thus, we suggest that different modules of the same coactivator complex can regulate each other at one promoter, allowing cells to rapidly modulate transcriptional changes upon an environmental shift. In addition to that mechanism, nutrient starvation leads to activation of the transcription factors Ste11 and Rst2 (Harigaya and Yamamoto 2007), which would recruit an active form of SAGA, and presumably other coactivators, at the promoter of some Ste11 target genes, such as mei2+. We note that ste11+ mRNA levels do not reach wild-type levels in starved gcn5Δ mutants (Figs. 3B, 6A); raising the possibility that Gcn5 also contributes to the expression of this gene upon nutrient starvation. In S. cerevisiae, Gcn5 can act as both an activator and a repressor of ARG1 expression, depending on nutrient conditions (Ricci et al. 2002).
Figure 7.
A model for SAGA-dependent regulation of a Ste11 target gene. In this model, the SAGA coactivator complex undergoes a conformational change in response to nutrient levels at the promoters of Ste11 target genes. This conformational change controls the opposing roles for two of its subunits, Gcn5 and Spt8. (A) In rich conditions, the transcription activator Rst2, and possibly Ste11, recruits SAGA to the promoter of a Ste11 target gene, such as ste11+ itself. In these conditions, Rst2 is not activated and allows only basal levels of transcription that do not require SAGA. Rather, SAGA is in a conformation where Gcn5 prevents SAGA-dependent activation by inhibiting Spt8 activity. (B) Upon nutrient starvation, Rst2 and Ste11 become activated and allow the full induction of expression of the Ste11 target gene. In addition, SAGA undergoes a conformational change, thereby allowing it to function as a positive regulator, which contributes to the full induction of transcription. Under these conditions, the repressive activity of Gcn5 is inhibited in SAGA by Spt8.
An extensive network of factors has been shown to regulate ste11+ expression and other Ste11 target genes, including the cAMP–PKA and stress-responsive MAPK pathways (Harigaya and Yamamoto 2007). Therefore, repression of ste11+ by Gcn5 might occur indirectly, by regulation of a factor acting upstream in one of these pathways. However, we think that this possibility is unlikely. First, based on our microarray results, there are no significant effects of a gcn5Δ mutation on the mRNA levels of the known regulators that act in these pathways. Second, we asked whether gcn5Δ might impair the cAMP–PKA pathway, as previous work has shown that decreased levels of cAMP can derepress ste11+ expression (Maeda et al. 1990, 1994; Isshiki et al. 1992). To test this possibility, we measured mRNA levels for fbp1+, another gene repressed by this pathway. Our results show that fbp1+ transcription is still tightly repressed in gcn5Δ mutants (data not shown), demonstrating that the cAMP–PKA pathway is still functional. Furthermore, genetic analyses showed that Gcn5 negatively regulates mating independently of the cAMP and stress-responsive MAPK pathways (data not shown). Third, we considered whether Gcn5 might control Tor2 levels as inactivation of Tor2 causes derepression of ste11+ and entry into the sexual differentiation pathway in rich medium, phenotypes similar to those of gcn5Δ mutants (Alvarez and Moreno 2006; Uritani et al. 2006; Matsuo et al. 2007; Weisman et al. 2007). However, there are clear differences between tor2 and gcn5Δ mutants, as tor2 mutants fully mimic nitrogen starvation, including decreased cell size and altered expression of most nitrogen-responsive genes (Alvarez and Moreno 2006; Matsuo et al. 2007). In contrast, gcn5Δ mutants do not have decreased cell size and cause a more limited effect on gene expression, primarily limited to Ste11 targets. Finally, we tested whether Gcn5 regulates cell cycle progression, as mating is restricted to the G1 phase in S. pombe, and a G1 delay can induce ectopic mating (Obara-Ishihara and Okayama 1994; Kjaerulff et al. 2007). However, flow cytometry analysis of gcn5Δ mutants revealed no defect in G1 progression (data not shown). Overall, the simplest interpretation of our results is that Gcn5, as part of SAGA, functions directly at the ste11+ and mei2+ promoters, and likely at other Ste11 target promoters, to directly regulate the sexual differentiation pathway.
What, then, is the mechanism by which Gcn5 represses Ste11 target genes during growth in rich medium? Previous studies have shown that in mammalian cells, Gcn5 and its homolog PCAF can regulate transcription, both positively and negatively, by direct acetylation of transcription factors (Sterner and Berger 2000; Nagy and Tora 2007). In particular, Gcn5 has been shown to inhibit through acetylation the transcriptional activity of PGC-1α, which is the master regulator of hepatic gluconeogenesis in response to hormone and nutrient levels in mice (Lerin et al. 2006). Similarly, we propose that Gcn5 repression of Ste11 target genes occurs by acetylation and the consequent inactivation of a factor normally required for Ste11 activation. The simplest possibility, based on our model, is that Gcn5 acetylates, and thereby inactivates, Spt8. However, our mass spectrometry data strongly suggest that Spt8 is not acetylated (data not shown), making it an unlikely target. We observed that other SAGA components are acetylated. Possibly, Gcn5-dependent acetylation of one of these components induces a conformational or compositional change within SAGA that in turn inhibits Spt8. Additional mass spectrometry analysis and extensive mutagenesis will be required to test this possibility. Another possible target for Gcn5 is Ste11. We attempted to test this possibility by purification of Ste11 in nutrient-rich conditions followed by mass spectrometry analysis. Unfortunately, we could not obtain a conclusive result due to low Ste11 levels and severe instability during purification. Finally, we believe that this regulation does not involve acetylation of histones. Our results showed that Gcn5-dependent acetylation of histone H3 increases at the ste11+ and mei2+ promoters upon induction of these genes, inconsistent with a role for H3 acetylation in repression. The regulation of Gcn5 activity by nutrient levels also remains an open question; this event likely occurs post-transcriptionally, as gcn5+ mRNA levels remain constant upon nutrient starvation (Supplemental Fig. 5).
One unexpected result from our studies is that Spt8 is required for activation of Ste11 targets upon nutrient starvation, yet Spt8 does not act directly in this activation. Rather, based on our finding that the spt8Δ activation defect is suppressed by gcn5Δ, Spt8 appears to inhibit the repressive activity of Gcn5. Previous studies in S. cerevisiae have implicated Spt8 in transcriptional activation by recruitment of TBP (Eisenmann et al. 1994; Belotserkovskaya et al. 2000; Bhaumik and Green 2002; Warfield et al. 2004; Sermwittayawong and Tan 2006; Laprade et al. 2007). It is not clear how such an activity fits with our results and raises the possibility of other mechanisms by which Spt8 can control transcription. We note that, although gcn5Δ spt8Δ double mutants appear to regulate ste11+ and mei2+ similarly to wild-type cells in response to nutrient levels, these double mutants are incapable of mating (Supplemental Table 9), suggesting additional regulatory roles for Gcn5 and/or Spt8 in this pathway. Additional studies are required to address these roles and to determine whether other SAGA components also participate in this regulation. In conclusion, our studies revealed important roles for S. pombe SAGA in choosing between a proliferative state and a differentiation pathway. These roles are accomplished by the dynamic utilization of distinct activities of SAGA in response to changes in environmental conditions. Further investigations will determine whether SAGA coordinates these or other combinations of its activities, in yeast or larger eukaryotes, to control additional cell fate decisions.
Materials and methods
S. pombe strains and genetic procedures
The S. pombe strains used in this study are listed in Supplemental Table 10. Standard S. pombe culture media (Yeast Extract Supplemented [YES] or Edinburgh Minimal Medium [EMM]) and genetic manipulations were used, as described previously (Forsburg and Rhind 2006). The media and methods are described in detail in the Supplemental Material.
TAP and mass spectrometry analysis
The S. pombe SAGA complex was purified by the TAP method (Supplemental Material).
Microarray experiments
Microarray experiments were done using total RNA as described in the Supplemental Material. The data have been submitted to ArrayExpress; the number is E-MTAB-53.
Northern blot and RT–PCR analysis
Total RNA was extracted using hot acidic phenol as described (Lyne et al. 2003). Northern blot analysis was performed as described (Hickman and Winston 2007). Details on these methods and quantitative RT–PCR are provided in the Supplemental Material.
Mating assays
Mating assays were performed using homothallic strains as described in the Supplemental Material.
ChIP
ChIP experiments were performed as described previously (Hickman and Winston 2007), with minor modifications. The method is described in detail in the Supplemental Material.
Acknowledgments
We thank Mark Hickman, Erica Larschan, and Brendon Monahan for critical reading of the manuscript. We are grateful to Brad Cairns, Charlie Hoffman, Nick Rhind, Chikashi Shimoda, and Masayuki Yamamoto for providing antibodies, plasmids, and strains. We thank David Drubin and Pamela Silver for use of their microscope. This work was supported by NIH grants GM45720 to F.W. and HG3456 to S.G., and CRUK grant C9546/A6517 to J.B. S.M. was supported by a fellowship for Advanced Researchers from the Swiss National Science Foundation, and D.H. was supported by a Long-Term Post-Doctoral Research Fellowship from the Human Frontier Science Program (HFSP).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1719908.
References
- Alvarez B., Moreno S. Fission yeast Tor2 promotes cell growth and represses cell differentiation. J. Cell Sci. 2006;119:4475–4485. doi: 10.1242/jcs.03241. [DOI] [PubMed] [Google Scholar]
- Aslett M., Wood V. Gene Ontology annotation status of the fission yeast genome: Preliminary coverage approaches 100% Yeast. 2006;23:913–919. doi: 10.1002/yea.1420. [DOI] [PubMed] [Google Scholar]
- Balasubramanian R., Pray-Grant M.G., Selleck W., Grant P.A., Tan S. Role of the Ada2 and Ada3 transcriptional coactivators in histone acetylation. J. Biol. Chem. 2002;277:7989–7995. doi: 10.1074/jbc.M110849200. [DOI] [PubMed] [Google Scholar]
- Belotserkovskaya R., Sterner D.E., Deng M., Sayre M.H., Lieberman P.M., Berger S.L. Inhibition of TATA-binding protein function by SAGA subunits Spt3 and Spt8 at Gcn4-activated promoters. Mol. Cell. Biol. 2000;20:634–647. doi: 10.1128/mcb.20.2.634-647.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S.R., Green M.R. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes & Dev. 2001;15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhaumik S.R., Green M.R. Differential requirement of SAGA components for recruitment of TATA-box-binding protein to promoters in vivo. Mol. Cell. Biol. 2002;22:7365–7371. doi: 10.1128/MCB.22.21.7365-7371.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand M., Leurent C., Mallouh V., Tora L., Schultz P. Three-dimensional structures of the TAFII-containing complexes TFIID and TFTC. Science. 1999;286:2151–2153. doi: 10.1126/science.286.5447.2151. [DOI] [PubMed] [Google Scholar]
- Daniel J.A., Grant P.A. Multi-tasking on chromatin with the SAGA coactivator complexes. Mutat. Res. 2007;618:135–148. doi: 10.1016/j.mrfmmm.2006.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey J. Fusion of a fission yeast. Yeast. 1998;14:1529–1566. doi: 10.1002/(SICI)1097-0061(199812)14:16<1529::AID-YEA357>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Dudley A.M., Rougeulle C., Winston F. The Spt components of SAGA facilitate TBP binding to a promoter at a post-activator-binding step in vivo. Genes & Dev. 1999;13:2940–2945. doi: 10.1101/gad.13.22.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A., Sterner D.E., Schieltz D., Hassan A., Yates J.R., Berger S.L., Workman J.L. The ADA complex is a distinct histone acetyltransferase complex in Saccharomyces cerevisiae. Mol. Cell. Biol. 1999;19:6621–6631. doi: 10.1128/mcb.19.10.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann D.M., Chapon C., Roberts S.M., Dollard C., Winston F. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics. 1994;137:647–657. doi: 10.1093/genetics/137.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S.L., Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A., Grallert B., Zilahi E., Szilagyi Z., Sipiczki M. Eleven novel sep genes of Schizosaccharomyces pombe required for efficient cell separation and sexual differentiation. Yeast. 1999;15:669–686. doi: 10.1002/(SICI)1097-0061(19990615)15:8<669::AID-YEA411>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Duggan L., Côté J., Roberts S.M., Brownell J.E., Candau R., Ohba R., Owen-Hughes T., Allis C.D., Winston F., et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Grant P.A., Eberharter A., John S., Cook R.G., Turner B.M., Workman J.L. Expanded lysine acetylation specificity of Gcn5 in native complexes. J. Biol. Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- Grewal S.I., Jia S. Heterochromatin revisited. Nat. Rev. Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- Guelman S., Suganuma T., Florens L., Swanson S.K., Kiesecker C.L., Kusch T., Anderson S., Yates J.R., Washburn M.P., Abmayr S.M., et al. Host cell factor and an uncharacterized SANT domain protein are stable components of ATAC, a novel dAda2A/dGcn5-containing histone acetyltransferase complex in Drosophila. Mol. Cell. Biol. 2006;26:871–882. doi: 10.1128/MCB.26.3.871-882.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harigaya Y., Yamamoto M. Molecular mechanisms underlying the mitosis–meiosis decision. Chromosome Res. 2007;15:523–537. doi: 10.1007/s10577-007-1151-0. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Hatanaka M., Nagao K., Nakaseko Y., Kanoh J., Kokubu A., Ebe M., Yanagida M. Rapamycin sensitivity of the Schizosaccharomyces pombe tor2 mutant and organization of two highly phosphorylated TOR complexes by specific and common subunits. Genes Cells. 2007;12:1357–1370. doi: 10.1111/j.1365-2443.2007.01141.x. [DOI] [PubMed] [Google Scholar]
- Henry K.W., Wyce A., Lo W.S., Duggan L.J., Emre N.C., Kao C.F., Pillus L., Shilatifard A., Osley M.A., Berger S.L. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes & Dev. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickman M.J., Winston F. Heme levels switch the function of Hap1 of Saccharomyces cerevisiae between transcriptional activator and transcriptional repressor. Mol. Cell. Biol. 2007;27:7414–7424. doi: 10.1128/MCB.00887-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi T., Watanabe Y., Yamamoto M. Protein kinase A regulates sexual development and gluconeogenesis through phosphorylation of the Zn finger transcriptional activator Rst2p in fission yeast. Mol. Cell. Biol. 2002;22:1–11. doi: 10.1128/MCB.22.1.1-11.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota K., Mizuno K.I., Shibata T., Ohta K. Distinct chromatin modulators regulate the formation of accessible and repressive chromatin at the fission yeast recombination hotspot ade6-M26. Mol. Biol. Cell. 2008;19:1162–1173. doi: 10.1091/mbc.E07-04-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisinga K.L., Pugh B.F. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- Ingvarsdottir K., Krogan N.J., Emre N.C., Wyce A., Thompson N.J., Emili A., Hughes T.R., Greenblatt J.F., Berger S.L. H2B ubiquitin protease Ubp8 and Sgf11 constitute a discrete functional module within the Saccharomyces cerevisiae SAGA complex. Mol. Cell. Biol. 2005;25:1162–1172. doi: 10.1128/MCB.25.3.1162-1172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isshiki T., Mochizuki N., Maeda T., Yamamoto M. Characterization of a fission yeast gene, gpa2, that encodes a Gα subunit involved in the monitoring of nutrition. Genes & Dev. 1992;6:2455–2462. doi: 10.1101/gad.6.12b.2455. [DOI] [PubMed] [Google Scholar]
- Johnsson A., Xue-Franzen Y., Lundin M., Wright A.P. Stress-specific role of fission yeast Gcn5 histone acetyltransferase in programming a subset of stress response genes. Eukaryot. Cell. 2006;5:1337–1346. doi: 10.1128/EC.00101-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaerulff S., Andersen N.R., Borup M.T., Nielsen O. Cdk phosphorylation of the Ste11 transcription factor constrains differentiation-specific transcription to G1. Genes & Dev. 2007;21:347–359. doi: 10.1101/gad.407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler A., Schneider M., Cabal G.G., Nehrbass U., Hurt E. Yeast Ataxin-7 links histone deubiquitination with gene gating and mRNA export. Nat. Cell Biol. 2008;10:707–715. doi: 10.1038/ncb1733. [DOI] [PubMed] [Google Scholar]
- Kunitomo H., Higuchi T., Iino Y., Yamamoto M. A zinc-finger protein, Rst2p, regulates transcription of the fission yeast ste11(+) gene, which encodes a pivotal transcription factor for sexual development. Mol. Biol. Cell. 2000;11:3205–3217. doi: 10.1091/mbc.11.9.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laprade L., Rose D., Winston F. Characterization of new Spt3 and TATA-binding protein mutants of Saccharomyces cerevisiae: Spt3 TBP allele-specific interactions and bypass of Spt8. Genetics. 2007;177:2007–2017. doi: 10.1534/genetics.107.081976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E., Winston F. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes & Dev. 2001;15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.K., Workman J.L. Histone acetyltransferase complexes: One size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- Lee T.I., Causton H.C., Holstege F.C., Shen W.C., Hannett N., Jennings E.G., Winston F., Green M.R., Young R.A. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 2000;405:701–704. doi: 10.1038/35015104. [DOI] [PubMed] [Google Scholar]
- Lee K.K., Florens L., Swanson S.K., Washburn M.P., Workman J.L. The deubiquitylation activity of Ubp8 is dependent upon Sgf11 and its association with the SAGA complex. Mol. Cell. Biol. 2005;25:1173–1182. doi: 10.1128/MCB.25.3.1173-1182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.M., Miklos I., Du H., Watt S., Szilagyi Z., Saiz J.E., Madabhushi R., Penkett C.J., Sipiczki M., Bähler J., et al. Impairment of the TFIIH-associated CDK-activating kinase selectively affects cell cycle-regulated gene expression in fission yeast. Mol. Biol. Cell. 2005;16:2734–2745. doi: 10.1091/mbc.E04-11-0982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerin C., Rodgers J.T., Kalume D.E., Kim S.H., Pandey A., Puigserver P. GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metab. 2006;3:429–438. doi: 10.1016/j.cmet.2006.04.013. [DOI] [PubMed] [Google Scholar]
- Lyne R., Burns G., Mata J., Penkett C.J., Rustici G., Chen D., Langford C., Vetrie D., Bähler J. Whole-genome microarrays of fission yeast: Characteristics, accuracy, reproducibility, and processing of array data. BMC Genomics. 2003;4:27. doi: 10.1186/1471-2164-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison J.M., Winston F. Identification and analysis of homologues of Saccharomyces cerevisiae Spt3 suggest conserved functional domains. Yeast. 1998;14:409–417. doi: 10.1002/(SICI)1097-0061(19980330)14:5<409::AID-YEA237>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Maeda T., Mochizuki N., Yamamoto M. Adenylyl cyclase is dispensable for vegetative cell growth in the fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. 1990;87:7814–7818. doi: 10.1073/pnas.87.20.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T., Watanabe Y., Kunitomo H., Yamamoto M. Cloning of the pka1 gene encoding the catalytic subunit of the cAMP-dependent protein kinase in Schizosaccharomyces pombe. J. Biol. Chem. 1994;269:9632–9637. [PubMed] [Google Scholar]
- Martinez E. Multi-protein complexes in eukaryotic gene transcription. Plant Mol. Biol. 2002;50:925–947. doi: 10.1023/a:1021258713850. [DOI] [PubMed] [Google Scholar]
- Mata J., Bähler J. Global roles of Ste11p, cell type, and pheromone in the control of gene expression during early sexual differentiation in fission yeast. Proc. Natl. Acad. Sci. 2006;103:15517–15522. doi: 10.1073/pnas.0603403103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J., Lyne R., Burns G., Bähler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat. Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- Mata J., Wilbrey A., Bähler J. Transcriptional regulatory network for sexual differentiation in fission yeast. Genome Biol. 2007;8:R217. doi: 10.1186/gb-2007-8-10-r217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo T., Otsubo Y., Urano J., Tamanoi F., Yamamoto M. Loss of the TOR kinase Tor2 mimics nitrogen starvation and activates the sexual development pathway in fission yeast. Mol. Cell. Biol. 2007;27:3154–3164. doi: 10.1128/MCB.01039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuzawa H., Ishihama A. Identification of histone H4-like TAF in Schizosaccharomyces pombe as a protein that interacts with WD repeat-containing TAF. Nucleic Acids Res. 2002;30:1952–1958. doi: 10.1093/nar/30.9.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuzawa H., Seino H., Yamao F., Ishihama A. Two WD repeat-containing TATA-binding protein-associated factors in fission yeast that suppress defects in the anaphase-promoting complex. J. Biol. Chem. 2001;276:17117–17124. doi: 10.1074/jbc.M100248200. [DOI] [PubMed] [Google Scholar]
- Naar A.M., Lemon B.D., Tjian R. Transcriptional coactivator complexes. Annu. Rev. Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- Nagy Z., Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- Obara-Ishihara T., Okayama H. A B-type cyclin negatively regulates conjugation via interacting with cell cycle ‘start’ genes in fission yeast. EMBO J. 1994;13:1863–1872. doi: 10.1002/j.1460-2075.1994.tb06455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant M.G., Schieltz D., McMahon S.J., Wood J.M., Kennedy E.L., Cook R.G., Workman J.L., Yates J.R., Grant P.A. The novel SLIK histone acetyltransferase complex functions in the yeast retrograde response pathway. Mol. Cell. Biol. 2002;22:8774–8786. doi: 10.1128/MCB.22.24.8774-8786.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pray-Grant M.G., Daniel J.A., Schieltz D., Yates J.R., Grant P.A. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature. 2005;433:434–438. doi: 10.1038/nature03242. [DOI] [PubMed] [Google Scholar]
- Ricci A.R., Genereaux J., Brandl C.J. Components of the SAGA histone acetyltransferase complex are required for repressed transcription of ARG1 in rich medium. Mol. Cell. Biol. 2002;22:4033–4042. doi: 10.1128/MCB.22.12.4033-4042.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts S.M., Winston F. Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada, and Gcn5 proteins, with the Snf/Swi and Srb/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sermwittayawong D., Tan S. SAGA binds TBP via its Spt8 subunit in competition with DNA: Implications for TBP recruitment. EMBO J. 2006;25:3791–3800. doi: 10.1038/sj.emboj.7601265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoda C., Uehira M., Kishida M., Fujioka H., Iino Y., Watanabe Y., Yamamoto M. Cloning and analysis of transcription of the mei2 gene responsible for initiation of meiosis in the fission yeast Schizosaccharomyces pombe. J. Bacteriol. 1987;169:93–96. doi: 10.1128/jb.169.1.93-96.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla A., Stanojevic N., Duan Z., Sen P., Bhaumik S.R. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol. Cell. Biol. 2006;26:3339–3352. doi: 10.1128/MCB.26.9.3339-3352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipiczki M., Grallert A., Miklos I., Zilahi E., Bozsik A., Szilagyi Z. Genetics, physiology and cytology of yeast-mycelial dimorphism in fission yeasts. Acta Microbiol. Immunol. Hung. 1999;46:297–302. doi: 10.1556/AMicr.46.1999.2-3.21. [DOI] [PubMed] [Google Scholar]
- Sterner D.E., Berger S.L. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E., Grant P.A., Roberts S.M., Duggan L.J., Belotserkovskaya R., Pacella L.A., Winston F., Workman J.L., Berger S.L. Functional organization of the yeast SAGA complex: Distinct components involved in structural integrity, nucleosome acetylation, and TATA-binding protein interaction. Mol. Cell. Biol. 1999;19:86–98. doi: 10.1128/mcb.19.1.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterner D.E., Belotserkovskaya R., Berger S.L. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proc. Natl. Acad. Sci. 2002;99:11622–11627. doi: 10.1073/pnas.182021199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto A., Iino Y., Maeda T., Watanabe Y., Yamamoto M. Schizosaccharomyces pombe ste11+ encodes a transcription factor with an HMG motif that is a critical regulator of sexual development. Genes & Dev. 1991;5:1990–1999. doi: 10.1101/gad.5.11.1990. [DOI] [PubMed] [Google Scholar]
- Tanay A., Regev A., Shamir R. Conservation and evolvability in regulatory networks: The evolution of ribosomal regulation in yeast. Proc. Natl. Acad. Sci. 2005;102:7203–7208. doi: 10.1073/pnas.0502521102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uritani M., Hidaka H., Hotta Y., Ueno M., Ushimaru T., Toda T. Fission yeast Tor2 links nitrogen signals to cell proliferation and acts downstream of the Rheb GTPase. Genes Cells. 2006;11:1367–1379. doi: 10.1111/j.1365-2443.2006.01025.x. [DOI] [PubMed] [Google Scholar]
- Warfield L., Ranish J.A., Hahn S. Positive and negative functions of the SAGA complex mediated through interaction of Spt8 with TBP and the N-terminal domain of TFIIA. Genes & Dev. 2004;18:1022–1034. doi: 10.1101/gad.1192204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman R., Roitburg I., Schonbrun M., Harari R., Kupiec M. Opposite effects of tor1 and tor2 on nitrogen starvation responses in fission yeast. Genetics. 2007;175:1153–1162. doi: 10.1534/genetics.106.064170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V., Gwilliam R., Rajandream M.A., Lyne M., Lyne R., Stewart A., Sgouros J., Peat N., Hayles J., Baker S., et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Wu P.Y., Winston F. Analysis of Spt7 function in the Saccharomyces cerevisiae SAGA coactivator complex. Mol. Cell. Biol. 2002;22:5367–5379. doi: 10.1128/MCB.22.15.5367-5379.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P.Y., Ruhlmann C., Winston F., Schultz P. Molecular architecture of the S. cerevisiae SAGA complex. Mol. Cell. 2004;15:199–208. doi: 10.1016/j.molcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Yamada T., Mizuno K., Hirota K., Kon N., Wahls W.P., Hartsuiker E., Murofushi H., Shibata T., Ohta K. Roles of histone acetylation and chromatin remodeling factor in a meiotic recombination hotspot. EMBO J. 2004;23:1792–1803. doi: 10.1038/sj.emboj.7600138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T., Poon D., Weil P.A., Horikoshi M. Molecular genetic elucidation of the tripartite structure of the Schizosaccharomyces pombe 72 kDa TFIID subunit which contains a WD40 structural motif. Genes Cells. 1997;2:245–254. doi: 10.1046/j.1365-2443.1997.1180316.x. [DOI] [PubMed] [Google Scholar]
- Yu Y., Eriksson P., Bhoite L.T., Stillman D.J. Regulation of TATA-binding protein binding by the SAGA complex and the Nhp6 high-mobility group protein. Mol. Cell. Biol. 2003;23:1910–1921. doi: 10.1128/MCB.23.6.1910-1921.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]