Abstract
The Tuberous Sclerosis Complex component, TSC1, functions as a tumor suppressor via its regulation of diverse cellular processes, particularly cell growth. TSC1 exists in a complex with TSC2 and functions primarily as a key negative regulator of mammalian target of rapamycin complex 1 (mTORC1) signaling and protein synthesis, although the TSC1/TSC2 complex also shows mTORC1-independent outputs to other pathways. Here, we explored the role of TSC1 in various aspects of stem cell biology and dissected the extent to which TSC1 functions are executed via mTORC1-dependent versus mTORC1-independent pathways. Using hematopoietic stem cells (HSCs) as a model system, we demonstrate that somatic deletion of TSC1 produces striking stem cell and derivative effector cell phenotypes characterized by increased HSC cell cycling, mobilization, marked progressive depletion, defective long-term repopulating potential, and hematopoietic lineage developmental aberrations. On the mechanistic level, we further establish that TSC1 regulation of HSC quiescence and long-term repopulating potential and hematopoietic lineage development is mediated through mTORC1 signaling. In contrast, TSC1 regulation of HSC mobilization is effected in an mTORC1-independent manner, and gene profiling and functional analyses reveals the actin-bundling protein FSCN1 as a key TSC1/TSC2 target in the regulation of HSC mobilization. Thus, TSC1 is a critical regulator of HSC self-renewal, mobilization, and multilineage development and executes these actions via both mTORC1-dependent and -independent pathways.
Keywords: FSCN1, hematopoietic stem cell, Tuberous Sclerosis Complex
Germ-line mutation of either TSC1 and TSC2 genes causes Tuberous Sclerosis Complex (TSC), a rare condition manifesting as hamartoma formation in a wide range of tissues (1). TSC1 and TSC2 form a stable complex and function as the GTPase activating protein of the small GTPase, Rheb, which is a positive upstream regulator of mammalian target of rapamycin complex 1 (mTORC1). The TSC1/TSC2 complex thus inhibits mTORC1 activity by stimulating Rheb GTP hydrolysis (2). mTORC1 functions as the key regulator of cell growth (increase of cell size), which is a prerequisite for cell cycle progression (3). TSC1/TSC2–mTORC1 signaling activities and its capacity to regulate cell growth and macromolecular synthesis involves its ability to control mRNA translation, ribosome synthesis, metabolism-related gene expression, and autophagy (4).
TSC1/TSC2 signaling can also impact the regulation of cell proliferation and cell adhesion/migration via interaction with many other signaling pathways, including B-Raf, β-catenin, ERM proteins, small GTPase Rho, and mTORC2-Akt signaling (5–8). mTORC1 pharmacological inhibition and mutagenesis approaches have indicated that some of these functions are mTORC1-independent (6, 8). The extent to which TSC1/TSC2 complex operates via mTORC1 or additional pathways in the regulation of tissue homeostasis in vivo remains an area of active investigation.
Given the multitude of TSC1/TSC2 downstream targets and the context-specific actions of TSC1/TSC2 complex interactions with other signaling components, we elected to assess TSC1 function via genetic means in a well-defined model system that is subject to stringent regulation of cell growth, proliferation, and differentiation. Hematopoietic stem cell (HSC) affords such a biological system. Adult HSCs sustain all blood lineages throughout life via a highly orchestrated process involving a balance between self-renewal and differentiation. HSCs exist in a relatively quiescent state in the bone marrow microenvironment and can be activated to rapidly enter cell cycle to regulate hematopoiesis as physiological demands dictate (9). The maintenance of HSC reserves therefore demands strict control over quiescence, renewal, and differentiation processes in the context of various intrinsic and extrinsic cues (9). In contrast to considerable information supporting the key roles of components governing cell cycle entry in the regulation of HSC biology (10), much less is known about the cell growth regulatory circuits governing HSC homeostasis and its effector lineages.
In this study designed to assess the impact of somatic deletion of TSC1 in the adult hematopoietic system, an integrated genetic, transcriptomic, functional validation and pharmacological analysis establishes that both mTORC1-dependent and -independent TSC1 activities regulate HSC homeostasis and that the actin-bundling protein FSCN1 serves as a key mTORC1-independent mediator of TSC1 in the regulation of HSC mobilization.
Results
Somatic Deletion of TSC1 Leads to Fatal Bone Marrow Failure and Multiple Lineage Defects.
To dissect TSC1 in vivo function in the adult mouse and circumvent the embryonic lethality associated with germ-line nullizygosity (11, 12), we adopted a conditional somatic KO strategy that uses the established inducible deletor allele Rosa26-CreERT2 (13) and the conditional KO allele TSC1L (11). Treatment of adult mice with tamoxifen resulted in complete deletion of TSC1 in hematopoietic organs (Fig. S1 A and B). Within 4–7 weeks after tamoxifen treatment, all TSC1 KO mice exhibited constitutional signs of illness, including weight loss (Fig. S1C), lethargy, scruffy fur, hunched posture, and ultimately death; in contrast, TSC1 WT mice remained viable and healthy (Fig. 1A).
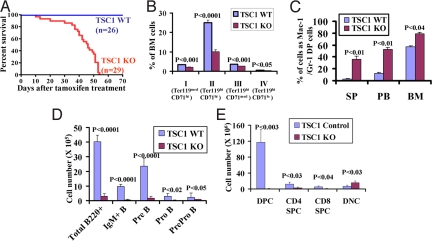
Fig. 1.
Somatic deletion of TSC1 leads to fatal bone marrow failure and multiple lineage defects. (A) Kaplan–Meier overall survival analysis of Rosa26-CreERT2+, TSC1L/L (TSC1 KO) and Rosa26-CreERT2-, TSC1L/L or Rosa26-CreERT2+, TSC1+/+ (TSC1 WT) after tamoxifen treatment. (B) Bar graph showing decreased percentages of erythroid cells at different developmental stages, including proerythroblasts (population I, Ter119medCD71hi), basophilic erythroblasts (population II, Ter119hiCD71hi), late erythroblasts (populations III-IV, Ter119hiCD71med and Ter119hiCD71lo), in TSC1 KO bone marrow cells. (C) Bar graph showing increased percentages of Mac-1/Gr-1 double positive (DP) cells from spleen (SP), peripheral blood (PB), and bone marrow (BM) in TSC1 KO mice. (D) Bar graph showing decreased number of various B cell lineages in TSC1 KO bone marrow cells. (E) Bar graph showing absolute number of various subpopulations of thymic T cells, including CD4/CD8 double-positive cells (DPC), CD4 single-positive cells (CD4 SPC), CD8 single-positive cells (CD8 SPC), and CD4/CD8 double-negative cells (DNC). n > 3 for each genotyping. P values are shown in the bar graphs (B–E).
Extensive analysis revealed severe multilineage defects in the hematopoietic systems of TSC1 KO mice. In addition, a subset of TSC1 KO mice developed polycystic kidney disease (data not shown), in line with the previous report in TSC2 mutant rat model (14). TSC1 deletion results in severe anemia as reflected by dramatic decline in red blood cells, hemoglobin, and hematocrit counts that reached lethal levels by 40–50 days post completing tamoxifen injection (DPI) only in the TSC1 KO mice (Fig. S1D). Detailed analysis of erythroid differentiation indicated severe reductions in total Ter119+ cells in TSC1 KO bone marrow (Fig. S1E) and erythroid progenitors of various developmental stages (Fig. 1B), a profile consistent with decreased survival in TSC1 KO Ter119+ population (Fig. S1F) and a significantly reduced erythroid burst-forming units (BFU-E) and colony-forming units-erythroid (CFU-E) activities of TSC1 KO bone marrow cells (Fig. S1G). Analysis of the myeloid progenitor compartment (Lin−, Sca-1−, c-Kit+) revealed that TSC1 deletion leads to increased number of the granulocyte–monocyte progenitor (GMP) and decreased number of the megakaryocyte–erythrocyte progenitor (MEP) subpopulation without any obvious changes in the common myeloid progenitor (CMP) compartment (Fig. S1H). Furthermore, TSC1 KO mice also show reduced platelet counts (Fig. S1I). Together, decreased erythroid survival and selective suppression of the MEP compartment likely contribute to the anemia phenotypes observed in TSC1 KO mice.
Within 30 DPI, the TSC1 KO mice also exhibited myeloproliferative disease characterized by a significantly higher percentage of Gr-1+/Mac-1+ cells in bone marrow, spleen, peripheral blood (Fig. 1C and Fig. S2A), and abundant chloroacetate-esterase-positive myeloid cells in spleen and liver (Fig. S2B). TSC1 KO animals also developed extensive extramedullary haematopoiesis with splenomegaly (Fig. S2C), increased spleen cellularity (Fig. S2D), and effaced splenic architecture caused by expanded numbers of myeloid cells and megakaryocytes (data not shown). Furthermore, CFU-C assays showed dramatic increased representation of myeloid colony formation from splenocytes in the TSC1 KO samples compared with WT controls (Fig. S2E)
Severe progressive B and T lymphoid defects were also evident in the TSC1 KO mice. By 30 DPI, TSC1 KO mice showed dramatic reduction of peripheral blood lymphocytes (Fig. S3 A and B) and total and various subpopulation of bone marrow B220+ cells (Fig. 1D). Correspondingly, there was significant increase of apoptosis and cell death in TSC1 KO B220+IgM− cells (Fig. S3C) and decreased pre-B colony-forming activity in vitro only in TSC1 KO bone marrow cells (Fig. S3D). Finally, TSC1 KO mice also exhibited a progressive reduction in thymus weight (Fig. S3E) and cellularity (Fig. S3F) and a dramatic decrease in thymic CD4+/CD8+ cells (DPC) (Fig. 1E and Fig. S3G). In summary, TSC1 deficiency causes severe multilineage defects characterized as anemia, myeloid expansion, and suppression of lymphoid lineage development.
TSC1 Deficiency Causes Short-Term Expansion, but Provokes Long-Term Reduction of HSC Reserves and Repopulating Potential.
The severe multilineage defects in TSC1 KO mice prompted detailed examination of HSC and hematopoietic progenitor populations. Quantitative RT-PCR from various sorted hematopoietic stem and progenitor populations showed that TSC1 is expressed in most of the hematopoietic cells tested, but most highly expressed in the Lin−, Sca-1+, c-Kit+ (LSK) CD34− HSCs (Fig. S4A). Consistent with TSC1 function in the regulation of protein synthesis and cell size, TSC1 deletion is associated with an ≈10% increased average cell size of the LSK compartment (Fig. S4 B and C).
The LSK cells are comprised of a heterogeneous mixture of long-term HSCs (LT-HSCs; LSK CD34− Flt-3−), short-term HSCs (LSK CD34+ Flt-3−), and multipotent progenitors (LSK CD34+ Flt-3+) (15, 16). Correspondingly, multiparameter flow cytometry revealed that documented complete TSC1 deletion (Fig. S4A) results in marked acute increase in both percentage and absolute number of HSC-enriched LSK cells in bone marrow immediately after completion of tamoxifen treatment (0, 1, and 3 DPI; Fig. 2B and Fig. S4D), an effect observed in all subpopulations of LSK cells, particularly the LT-HSCs in the TSC1 KO mice at 3 DPI (Fig. S4E). However, such expansion of the LSK population in TSC1 KO bone marrow was transient, because serial analysis shows decreasing LSK cell numbers in the TSC1 KO mice beyond 10 DPI (Fig. 2 A and B).
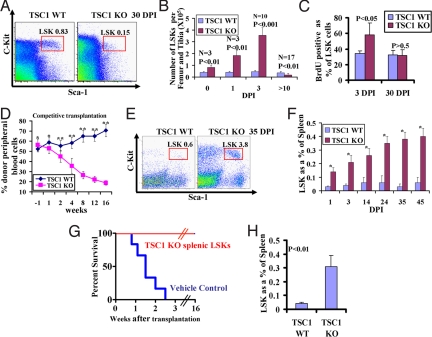
Fig. 2.
TSC1 regulates HSC quiescence and mobilization. (A) Flow cytometry analysis of LSK cell from bone marrow at 30 DPI from representative TSC1 KO and WT mice. The averaged percentage of LSK population from the Lin− population is also indicated. (B) Bar graph showing the absolute numbers of LSK cells per femur and tibia at various DPIs in TSC1 KO and WT mice. P value and the number of mice of each genotype at each time point are indicated. (C) Bar graph showing the percentage of BrdU-positive LSK cells in bone marrow in TSC1 KO and WT mice at 3 and 30 DPI. n = 3 for each genotype. P value for each day is indicated. (D) Recipient mice from competitive transplantation were analyzed by CD45 staining to examine the contribution of donor-derived cells in peripheral blood at various time points before or after tamoxifen treatment. Fifteen recipients for each genotype were used in competitive transplantation. *, P > 0.1; **, P < 0.01. (E) Flow cytometry analysis of LSK cell from spleen at 35 DPI from representative TSC1 KO and WT mice. The averaged percentage of LSK population from Lin− population is also indicated. (F) Bar graph showing the percentage of LSK cells from spleen in TSC1 KO and WT mice at various DPIs as indicated. n > 3 for each genotype at each time point. *, P < 0.01. (G) Kaplan–Meier overall survival analysis of lethally irradiated hosts transplanted with TSC1 KO splenic LSKs or vehicle control. n = 6 for each genotyping. (H) Bar graph showing the percentage of LSK cells from spleen in TSC1 KO and WT transplants [tamoxifen-treated WT recipient mice reconstituted with Rosa26-CreERT2+, TSC1L/L (TSC1 KO) and Rosa26-CreERT2−, TSC1L/L (TSC1 WT) BM cells]. n = 6 for each genotyping.
This pattern of acute and transient expansion of LSK population followed by its contraction is suggestive of TSC1 deficiency leading to enhanced HSC exit from quiescence without self-renewal maintenance leading to exhaustion of its reserve. Accordingly, 7-amino-actinomycin D (7-AAD)/Annexin-V staining of the LSK compartment showed no obvious difference in apoptosis between TSC1 KO and WT controls at different time points we examined (1, 3, 14, and 30 DPI; data not shown). In contrast, BrdU labeling revealed significantly higher percentage of BrdU-positive cells in TSC1 KO LSK cells relative to WT LSK controls at 3 DPI, but not at 30 DPI (Fig. 2C).
Next, competitive and noncompetitive transplantation assays were performed to assess the impact of TSC1 status on HSC repopulating ability in vivo. Bone marrow cells from TSC1L/L, Rosa26-CreERT2 or littermate WT mice were transplanted into lethally irradiated mice, followed by tamoxifen treatment 5–6 weeks posttransplantation to delete TSC1 in hematopoietic systems in the recipient hosts (Fig. S5A). Before tamoxifen treatment, flow cytometry analysis of peripheral blood showed that TSC1 KO and WT bone marrow cells possessed similar reconstitution activity in both noncompetitive and competitive settings. However, with tamoxifin treatment, TSC1 KO transplants exhibited progressively diminished repopulating potential relative to WT controls, commencing at 2 weeks posttamoxifen treatment and thereafter (Fig. 2D and Fig. S5B). At 16 weeks posttamoxifen treatment, bone marrow analysis of TSC1 KO transplants showed reductions in all hematopoietic lineages examined (Fig. S5 C and D). Together, the data indicate that TSC1 is essential for proper HSC cell cycle control and the maintenance of repopulating potential, prompting us to propose that a failure to maintain quiescence in TSC1 KO mice results in transient expansion, yet long-term decline, in the bone marrow LSK population and hematopoiesis support of the periphery.
Deletion of TSC1 Results in HSC Mobilization.
Adult HSCs normally reside in the bone marrow microenvironment and are minimally present in extramedullary sites (17). In the TSC1 KO mice, we observed that the decrease in LSK cells in the bone marrow coincided with a dramatic increase in LSK cells in the extramedullary sites, including peripheral blood (data not shown) and spleen (Fig. 2E). Increased LSK cell numbers in TSC1 KO spleens were first evident as early as 1 DPI and rose steadilyy thereafter to a 7- to 10-fold increase of the percentage of LSK cells by 30 DPI (Fig. 2F). This fold increase is likely an underestimate given the significantly increased cellularity of the TSC1 KO spleens (Fig. S2D). Furthermore, these TSC1 KO splenic LSK cells could support lethally irradiated mice via transplantation for at least 16 weeks (Fig. 2G), suggesting that these HSCs possess some degree of functional repopulating potential. Additionally, the TSC1 KO transplants described above (Fig. 2D) also displayed the HSC mobilization phenotype (Fig. 2H). Collectively, these data point to an HSC intrinsic role for TSC1 in the maintenance of a productive interaction between the HSC and its bone marrow niche that serves to constrain HSC mobilization and extramedullary hematopoiesis.
TSC1 Operates via both mTORC1-Dependent and -Independent Mechanisms to Regulate HSC Biology.
To begin to understand the role of mTORC1-dependent and -independent signaling in TSC1-directed HSC biology, we assessed the impact of mTORC1 pharmacological inhibition on TSC1 KO HSC self-renewal, mobilization, and lineage development. We administered daily rapamycin or vehicle control to TSC1 KO and WT mice from 30 DPI thereafter, a time frame during which severe hematopoietic defects are well-established (see above) (Fig. S6A). Importantly, we first documented that rapamycin treatment indeed reversed the hyperactivation of the mTORC1-S6K signaling axis in TSC1 KO mice; note the restoration of S6 phosphorylation to levels detected in TSC1 WT mice treated with vehicle (Fig. S6 B and C).
Within several days of rapamycin treatment, the TSC1 KO mice were noticeably more active and healthier than vehicle-treated TSC1 KO controls, which experienced progressive illness and death 7–15 days into the treatment protocol (i.e., 37–45 DPI). Rapamycin treatment produced a prominent rescue of myeloid expansion in spleen (Fig. S6E), restored bone marrow B220+ B cells and thymic CD4+/CD8+ cells (Fig. 3A), and reversed anemia (Fig. S6D) with normalization of bone marrow Ter119+ cells (Fig. 3A). Thus, TSC1 regulation of lineage development is mediated largely through the mTORC1 pathway.
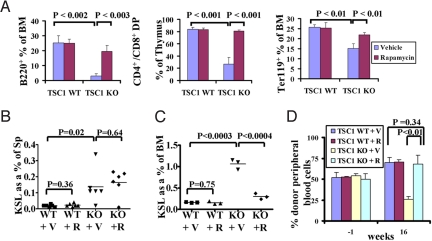
Fig. 3.
Rapamycin treatment reveals both mTORC1-dependent and -independent TSC1-deficient phenotypes in HSC. (A) Bar graph showing the percentage of B220+ B cells, Ter119+ erythroid cells in bone marrow and CD4+/CD8+ T cells in thymus from the mice shown in Fig. S6A. n = 3 for each genotype. (B and C) Scatter plots showing the percentage of LSK cells from spleen (B) and bone marrow (C) of the mice at 4 DPI as shown in Fig. S6F. (D) Bar graph showing the percentage of donor-derived cells in peripheral blood from 1 week before and 16 weeks after tamoxifen treatment. n = 3 for each genotyping. See SI Text for a detailed description of rapamycin treatment of TSC1 KO transplants.
Notably, certain TSC1 KO phenotypes were unaffected by the rapamycin treatment. Specifically, rapamycin-treated TSC1 KO mice continued to exhibit increased LSK cells in the spleen (data not shown). One explanation may be that HSCs have already been mobilized to the spleen at the time rapamycin treatment was initiated at 30 DPI. To address this, we also subjected mice to prophylactic rapamycin administration. To that end, at the time of tamoxifen treatment, we initiated daily rapamycin or vehicle protocols to TSC1 KO and WT mice. The continuously treated mice were analyzed at 4 DPI (Fig. S6F), revealing minimal impact of rapamycin treatment on LSK splenic mobilization in TSC1 KO mice (Fig. 3B), but clear reversal of LSK expansion in the BM (Fig. 3C). Continuous rapamycin treatment up to 14 DPI still had very limited effect on splenic mobilization in TSC1 KO mice (Fig. S3G). Finally, rapamycin treatment blocked the long-term exhaustion of HSC reserve (data not shown) and restored the defective long-term repopulating capability in TSC1 KO transplants (Fig. 3D) Taken together, TSC1 acts through mTORC1 to regulate HSC quiescence and long-term repopulating capability, but acts through an mTORC1-independent mechanism to regulate HSC mobilization to the periphery.
FSCN1 Plays a Key Role in Mediating TSC1 Regulation of HSC Mobilization.
To gain insight into the mechanistic basis for the mTORC1-independent regulation of HSC mobilization by TSC1, we performed comparative transcriptome analysis of sorted LSK cells from TSC1 WT and KO bone marrows at 3 DPI (Fig. 4A). Note at 3 DPI, there is documented TSC1 deletion in TSC1 KO LSK cells (Fig. S4A), yet no discernable bone marrow failure phenotype (see above). It is worth noting that, although the TSC1/TSC2 complex mainly functions to regulate protein synthesis through mTOR, transcriptomic network analysis can readily identify targets/pathways modulated by TSC1 via its engagement of other signaling outputs.
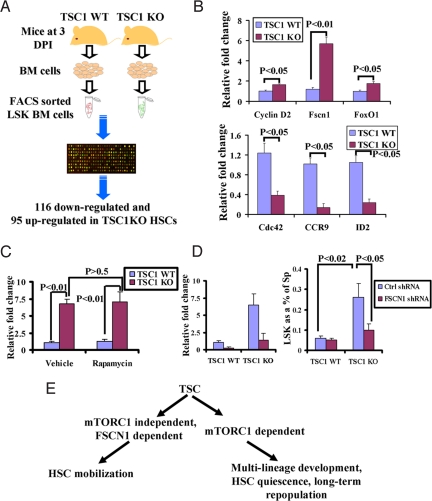
Fig. 4.
FSCN1 plays a key role in mediating TSC1 regulation of HSC mobilization. (A) Schematic representation of transcriptome analysis. See SI Text for a detailed description. (B) Bar graph showing relative expression fold change of selected genes between TSC1 WT and KO LSKs. n = 3 for each genotyping. (C) Bar graph showing relative expression fold change of FSCN1 between TSC1 WT and KO LSKs treated with rapamycin or vehicle. n = 3 for each genotyping. (D) Bar graph showing relative expression fold change of FSCN1 in spleen LSKs (Left) and percentage of LSK cells (Right) from spleen in TSC1 WT and KO transplants infected with control shRNA or FSCN1 shRNA-expressing lentivirus. (E) The working model showing that TSC1 regulation of HSC quiescence, long-term repopulating potential and lineage development is mediated through mTORC1 signaling, whereas TSC1 regulates HSC mobilization in an mTORC1-independent, but FSCN1-dependent manner.
The TSC1 KO transcriptome contains ≈200 significantly differentially expressed genes, 116 down-regulated, 95 up-regulated (Table S1). Knowledge-based pathway analysis of these genes showed significant enrichment of genes involved in the amino acid biosynthesis/metabolism pathway (Table S2), consistent with TSC1's major functions in protein synthesis and metabolism. In addition, there was enrichment for known regulators of cell migration/adhesion and homing, including CCR9, Cdc42, FSCN1, ID2, and Racgap1 (Rac GTPase activating protein 1) (Table S2). Differential expression was validated by quantitative RT-PCR in the TSC1 KO HSCs (Fig. 4B).
To test whether these cell migratory genes might mediate TSC1 regulation of HSC mobilization, we focused on FSCN1, an actin-bundling protein with known functions in cell mobilization (18). The selection of FSCN1 was based on the observations that (i) it is 1 of the most significantly up-regulated genes upon TSC1 deletion, and (ii) rapamycin treatment has no impact on FSCN1 expression in TSC1 KO HSCs (Fig. 4C), whereas rapamycin treatment restores the expression of Cdc42, CCR9, and Racgap1 (data not shown). Because our data in the transplantation experiments indicated intrinsic regulation of HSC mobilization by TSC1, we used the transplantation approach to further examine the role of FSCN1 in TSC1-mediated HSC mobilization. In the transplant studies, TSC1 WT and KO bone marrow cells were infected with scramble or FSCN1 shRNA-expressing lentivirus and then transplanted into lethally irradiated mice. FSCN1 shRNA causes 80% expression knockdown in TSC1 KO HSCs, i.e., FSCN1 expression levels similar to those in TSC1 WT HSCs (Fig. 4D Left). Remarkably, FSCN1 knockdown in TSC1 KO transplants significantly blocked the HSC mobilization phenotype (Fig. 4D Right). Thus, FSCN1 is a validated key mediator of TSC1 in the regulation of HSC mobilization.
Discussion
Here, we establish a role for TSC1 as an essential regulator of HSC homeostasis. Consistent with the major function of TSC1 as the negative regulator of mTORC1, rapamycin treatment shows that the TSC–mTORC1 axis controls HSC quiescence maintenance, long-term repopulating potential, and lineage development, but that HSC mobilization is controlled primarily by a TSC–mTORC1-independent pathway. Integrated transcriptomic and functional analyses identified the cell mobility protein, FSCN1, as at least 1 key mediator of this TSC1-dependent, mTORC1-independent process (Fig. 4E).
FSCN1 is an actin-cross-linking protein and plays an important role in the regulation of cytoskeleton reorganization and cell motility. FSCN1 has been shown to be up-regulated in a number of human cancers and functions to promote cancer cell migration and invasion (18). Interestingly, FSCN1 has been identified as a transcriptional target of β-catenin/T cell factor (TCF) signaling (19), and this signaling can be inhibited by the TSC1/TSC2 complex (7). Whether TSC1/TSC2 regulates FSCN1 via β-catenin/TCF signaling awaits further investigation. Because abnormal stem cell mobilization often correlates with neoplastic transformation and metastasis (17), our findings prompt speculation that TSC1 regulation of FSCN1 might play a role in the dissemination of hematopoeitic cells in the cancer setting.
The phenotypes from our TSC1-deficient mice share many similarities with those of previously reported PTEN KO mice (20, 21), including increased proliferation, but not aberrant apoptosis, leading to short-term expansion, but long-term depletion of HSCs, HSC mobilization to extramedullary sites, lineage development defects with blockade of lymphocyte development, and expansion of myeloid development. Our study thus provides direct genetic evidence that the TSC–mTORC1 pathway is an important downstream target of the PI3K–PTEN–Akt pathway to regulate HSC biology. At the same time, it is important to note that the phenotypic impacts of TSC1 versus PTEN deletion are not identical. For example, the myeloproliferative disease phenotype in TSC1 KO mice is much less severe than in the PTEN KO model, which progresses to leukemia. Nevertheless, the TSC1/TSC2 complex may still be an important target of PTEN′s actions as evidenced by TSC1 deletion in an experimental model of thymic lymphoma (22). At the same time, the PTEN versus TSC1 phenotypic differences provide genetic evidence that the actions of PTEN extend beyond its regulation of the TSC1/TSC2 complex. Along these lines, our recent studies (23) and those of others (24) show that the FoxO transcriptional factors can serve as important regulators of HSC homeostasis. Specifically, conditional deletion of FoxOs in adult murine hematopoietic system results in a phenotype with similarities to that of PTEN-deficient mice (23). Thus, the findings of this TSC1 study, together with the PTEN and FoxO studies (20, 21, 23, 24), support the view that the TSC–mTORC1 and FoxO pathways collectively serve as 2 key downstream effector arms of PI3K–PTEN–Akt signaling in the regulation of HSC self-renewal. The extent to which the TSC1/TSC2 and FoxO signaling arms are interconnected will require further investigation as current models of feedback regulation do not adequately rationalize the concordance of HSC phenotypes.
Materials and Methods
Generation and Analysis of Mice.
Rosa26CreERT2+ deletor mice (13) were crossed with TSC1 L/L mice (11) to generate TSC1 L/+, Rosa26CreERT2+ mice. TSC1 L/+, Rosa26CreERT2+ mice were backcrossed 5 generations into C57BL/Ka-CD45.2:Thy-1.1 background, which were then intercrossed to generate mice of the desired genotypes. Recipients in transplantation assays were adult C57BL/Ka-CD45.1:Thy-1.2 mice (Jackson Laboratory). The protocols for tamoxifen administration and rapamycin administration are described in SI Text.
Flow Cytometric Analysis.
LSK, GMP, MEP, and CMP populations were analyzed and sorted by using FACSAria (Becton Dickinson) as reported (23). Apoptosis was determined by staining freshly harvested bone marrow mononuclear cells with lineage, stem, and progenitor markers, followed by Annexin V and 7-AAD staining. Cell cycle analysis was carried out as reported (25). Additional analyses were carried out on a 4-color FACSCalibur cytometer (Becton Dickinson) with a minimum of 10,000 events acquired. The data were analyzed by FlowJo and CellQuest softwares. Additional details are provided in SI Text.
In Vitro Colony Assays.
The details for myeloid and pre-B colony-plating assays and CFU-E and BFU-E assays are provided in SI Text.
Microarray Analysis and Quantitative Real-Time PCR Analysis.
Microarray and quantitiative real-time PCR were performed as described (26). Details of the microarray and quantitative real-time PCR experiments are provided in SI Text.
Western Blot Analysis.
Tissues and sorted cells were lysed with RIPA buffer [20 mM Tris (pH 7.5), 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 1 mM EDTA, 0.1% SDS] containing complete miniprotease inhibitors (Roche). Western blot analysis was performed by using 20–50 μg of lysate protein, incubated with the antibodies S6 and Ser-240/244 phospho-S6 (Cell Signaling Technology).
Supplementary Material
Acknowledgments.
We thank Anton Berns (Netherlands Cancer Institute, Amsterdam) for the Rosa26-CreERT2 mouse strain; Shan (Julia) Zhou for assistance in the animal facility; Carol Lim and Stewart Borden Reed for the assistance with genotyping; Suzan Lazo-Kallanian, John Daley, and Peter Schow for assistance with flow cytometry; Liansheng Zheng, Giovanni Tonon, and David J. Hiller for bioinformatics analysis of gene profiling; Gerry Chu for helpful suggestion on histology; the Dana–Farber Cancer Institute–Broad RNAi Consortium for lentiviral shRNA vectors; and Herald von Boehmer and Yaoqi A. Wang for helpful discussion. This research was supported by National Cancer Institute Grant R21CA135057 (to R.A.D.). B. G. is the Research Fellow of the Leukemia and Lymphoma Society. K.L.S. is the Research Fellow of American Cancer Society (Grant PF-07-039-01-CSM) and National Institutes of Health grant DK62757 (to D.A.W.). R.A.D. is the American Cancer Society Research Professor and an Ellison Medical Foundation Senior Scholar and is supported by the Robert A. and Renee E. Belfer Foundation Institute for Innovative Cancer Science.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810584105/DCSupplemental.
References
- 1.Kwiatkowski DJ, Manning BD. Tuberous sclerosis: A GAP at the crossroads of multiple signaling pathways. Hum Mol Genet. 2005;14:R251–R258. doi: 10.1093/hmg/ddi260. [DOI] [PubMed] [Google Scholar]
- 2.Huang J, Manning BD. The TSC1–TSC2 complex: A molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Conlon I, Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- 4.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006;124:471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 5.Lamb RF, et al. The TSC1 tumor suppressor hamartin regulates cell adhesion through ERM proteins and the GTPase Rho. Nat Cell Biol. 2000;2:281–287. doi: 10.1038/35010550. [DOI] [PubMed] [Google Scholar]
- 6.Karbowniczek M, et al. Regulation of B-Raf kinase activity by tuberin and Rheb is mammalian target of rapamycin (mTOR)-independent. J Biol Chem. 2004;279:29930–29937. doi: 10.1074/jbc.M402591200. [DOI] [PubMed] [Google Scholar]
- 7.Mak BC, Takemaru K, Kenerson HL, Moon RT, Yeung RS. The tuberin–hamartin complex negatively regulates β-catenin signaling activity. J Biol Chem. 2003;278:5947–5951. doi: 10.1074/jbc.C200473200. [DOI] [PubMed] [Google Scholar]
- 8.Huang J, Dibble CC, Matsuzaki M, Manning BD. The TSC1–TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol. 2008;28:4104–4115. doi: 10.1128/MCB.00289-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Orkin SH, Zon LI. Hematopoiesis: An evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orford KW, Scadden DT. Deconstructing stem cell self-renewal: Genetic insights into cell-cycle regulation. Nat Rev Genet. 2008;9:115–128. doi: 10.1038/nrg2269. [DOI] [PubMed] [Google Scholar]
- 11.Kwiatkowski DJ, et al. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas and up-regulation of p70S6 kinase activity in Tsc1 null cells. Hum Mol Genet. 2002;11:525–534. doi: 10.1093/hmg/11.5.525. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi T, et al. A germ-line Tsc1 mutation causes tumor development and embryonic lethality that are similar, but not identical to, those caused by Tsc2 mutation in mice. Proc Natl Acad Sci USA. 2001;98:8762–8767. doi: 10.1073/pnas.151033798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vooijs M, Jonkers J, Berns A. A highly efficient ligand-regulated Cre recombinase mouse line shows that LoxP recombination is position dependent. EMBO Rep. 2001;2:292–297. doi: 10.1093/embo-reports/kve064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleymenova E, et al. Tuberin-dependent membrane localization of polycystin-1: A functional link between polycystic kidney disease and the TSC2 tumor suppressor gene. Mol Cell. 2001;7:823–832. doi: 10.1016/s1097-2765(01)00226-x. [DOI] [PubMed] [Google Scholar]
- 15.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 16.Adolfsson J, et al. Dentification of Flt3+ lympho-myeloid stem cells lacking erythro-megakaryocytic potential: A revised road map for adult blood lineage commitment. Cell. 2005;121:295–306. doi: 10.1016/j.cell.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 17.Laird DJ, von Andrian UH, Wagers AJ. Stem cell trafficking in tissue development, growth, and disease. Cell. 2008;132:612–630. doi: 10.1016/j.cell.2008.01.041. [DOI] [PubMed] [Google Scholar]
- 18.Hashimoto Y, Skacel M, Adams JC. Roles of fascin in human carcinoma motility and signaling: Prospects for a novel biomarker? Int J Biochem Cell Biol. 2005;37:1787–1804. doi: 10.1016/j.biocel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 19.Vignjevic D, et al. Fascin, a novel target of β-catenin-TCF signaling, is expressed at the invasive front of human colon cancer. Cancer Res. 2007;67:6844–6853. doi: 10.1158/0008-5472.CAN-07-0929. [DOI] [PubMed] [Google Scholar]
- 20.Zhang J, et al. PTEN maintains hematopoietic stem cells and acts in lineage choice and leukemia prevention. Nature. 2006;441:518–522. doi: 10.1038/nature04747. [DOI] [PubMed] [Google Scholar]
- 21.Yilmaz OH, et al. Pten dependence distinguishes hematopoietic stem cells from leukemia-initiating cells. Nature. 2006;441:475–482. doi: 10.1038/nature04703. [DOI] [PubMed] [Google Scholar]
- 22.Maser RS, et al. Chromosomally unstable mouse tumors have genomic alterations similar to diverse human cancers. Nature. 2007;447:966–971. doi: 10.1038/nature05886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tothova Z, et al. FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell. 2007;128:325–339. doi: 10.1016/j.cell.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 24.Miyamoto K, et al. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1:101–112. doi: 10.1016/j.stem.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 25.Hock H, et al. Gfi-1 restricts proliferation and preserves functional integrity of hematopoietic stem cells. Nature. 2004;431:1002–1007. doi: 10.1038/nature02994. [DOI] [PubMed] [Google Scholar]
- 26.Paik JH, et al. FoxOs are lineage-restricted redundant tumor suppressors and regulate endothelial cell homeostasis. Cell. 2007;128:309–323. doi: 10.1016/j.cell.2006.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.