Abstract
Although the role of liganded nuclear receptors in mediating coactivator/corepressor exchange is well-established, little is known about the potential regulation of chromosomal organization in the 3-dimensional space of the nucleus in achieving integrated transcriptional responses to diverse signaling events. Here, we report that ligand induces rapid interchromosomal interactions among specific subsets of estrogen receptor α-bound transcription units, with a dramatic reorganization of nuclear territories, which depends on the actions of nuclear actin/myosin-I machinery and dynein light chain 1. The histone lysine demethylase, LSD1, is required for these ligand-induced interactive loci to associate with distinct interchromatin granules, long thought to serve as “storage” sites for the splicing machinery, some critical transcription elongation factors, and various chromatin remodeling complexes. We demonstrate that this 2-step nuclear rearrangement is essential for achieving enhanced, coordinated transcription of nuclear receptor target genes.
Keywords: enhancement of gene expression, nuclear architecture, chromosomal movement, interchromosomal interactions, SC35 domains
The molecular mechanism used by nuclear receptors to mediate coactivator/corepressor exchanges in gene activation has been well elucidated (1, 2). However, recent genome-wide analyses of DNA binding sites for transcription factors, including estrogen receptor (ERα), revealed numerous intergenic sites, only a few of which have clearly been established to function as enhancers in vivo (3–5), raising intriguing questions about whether and how some of these remote binding sites might communicate with their putative target genes via long-distance intrachromosomal and (potentially) interchromosomal interactions.
Although the architectural organization of the nucleus is still poorly understood (6, 7), repartitioning of active genes from interior to the periphery of nuclear territories has been suggested for Hox genes in ES cells (8), IgH in β lymphocytes (9), c-maf in T cells (10), Mash1 in neuronal cells (11), and Cftr in adenocarcinoma cells (12). Many recent studies have documented interchromosomal interactions to provide novel control mechanisms for regulated gene expression in interphase nuclei (13–20).
Here, we report rapid and 17β-estradiol (E2)-induced interactions between gene loci located in different chromosomes, suggesting a potentially dynamic system that operates to provide coordinated control for regulated gene expression in mammalian cells. We found that the interacting loci exhibit LSD1-dependent interactions with interchromatin granules that harbor key factors for transcriptional elongation and pre-mRNA splicing, suggesting at least 2 steps for nuclear reorganization and gene repartitioning critical for enhanced gene expression. These findings provide a model for coordinated regulation of specific gene transcription in the nucleus, and we suggest that the strategy may be widely used for signal-induced gene expression programs.
Results
Identification of Estrogen-Induced Interchromosomal Interactions.
We devised an initial, open-ended approach to detecting long-distance genomic interactions by coupling the chromosome conformation capture (3C) assay (21) with the ChIP-DSL strategy that we recently developed for large-scale promoter array and tiling array analyses (22), a method we refer to as deconvolution of DNA interaction by DSL or the 3D assay (Fig. 1A). In pilot experiments on MCF7 cells treated with E2 for 60min, we isolated DNA after in situ restriction digestion followed by ligation under an extreme dilution condition according to the established 3C protocol, and after sonication of the DNA, we used a biotinylated oligonucleotides to capture DNA fragments that contain the enhancer of the well-studied E2-regulated TFF1 gene. To detect DNA fragments that were linked to the TFF1 enhancer during 3C, we annealed a set of DSL oligonucleotide pairs targeting individual genomic blocks in a ≈1.4 Mb tiled path surrounding the TFF1 gene in chromosome 21 (Fig. S1A). After selection, ligation, amplification, and hybridization on the corresponding tiling array according to the DSL protocol, we identified a series of specific intrachromosomal interactions that frequently involve other ERα-bound genomic loci, which were confirmed by the conventional 3C assay (Fig. S1B).
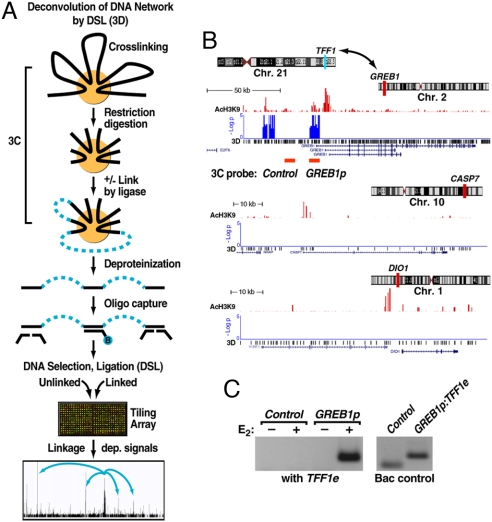
Fig. 1.
Identification of long-range, estrogen induced chromosomal interactions by 3D. (A) Diagram of the 3D technology. The initial steps are identical to the established 3C technology. A key extension is DNA capturing by using a specific biotinylated oligonucleotide followed by DNA selection and ligation to detect cocaptured DNA fragments in a high-throughput and unbiased fashion. Specific signals were identified based on relative enrichment of DNA fragments linked by ligase compared with those from the parallel minus ligase control under an extensive dilution condition. (B) Specific interchromosomal interactions predicted by 3D. Three of the 6 assessed chromosomal intervals are shown, plotting the location of AcH3K9, an activation mark (red), and signal enrichment in 3D assay (blue) at the GREB1 locus (chromosome 2). Two negative controls (CASP7 in chromosome 10 and DIO1 in chromosome 1) show significant levels of AcH3K9 but no 3D signal. (C) 3C validation of the detected interchromosomal interaction predicted by 3D between TFF1 enhancer (chromosome 21) and GREB1 promoters (chromosome 2) in mock-treated and E2-induced (60 min) HMECs in which only the sample treated with E2 shows a strong interaction. (Right) Shows the primer efficiency obtained on randomly ligated BAC controls. The location of the 3C primers for GREB1 is indicted below the 3D signal track in B.
We included in the 3D experiment a set of tiled intervals on 6 different chromosomes, one of which encompassed GREB1, a well-characterized ERα-inducible gene located in chromosome 2 (22, 23). Based on selection of TFF1 interactants, 3D capture revealed 2 clusters of significant signals coincident with an enhancer and promoter in the GREB1 gene (Fig. 1B), whereas the other 5 tiled genomic regions on other chromosomes showed no signals, two of which are illustrated (Fig. 1B). This finding suggested E2-inducible interchromosomal interactions between TFF1 and GREB1, which was validated by the conventional 3C assay (Fig. 1C).
To ensure that the interaction between the TFF1 and GREB1 genes, which reside in chromosome 21 and chromosome 2, respectively, occurred in both tumor and normal cells, we performed FISH analysis (24) on MCF7 and primary cultures of human mammary epithelial cells (HMECs), using a standardized protocol, including 4 days under serum deprivation and temporal synchronization of interphase nuclei. In each case approximately half of the cells exhibited monoallelic interactions, whereas the other half exhibited biallelic interactions (Fig. 2A), with the precise percentage differing for different lots of MCF7 cells or HMECs. Monoallelic interactions appear prevalent in other studies of interchromosomal interactions (14, 20, 25, 26). As controls, Dio1 and Casp7 loci exhibited no E2-induced interaction with TFF1 above FISH assay background (Fig. S2A).
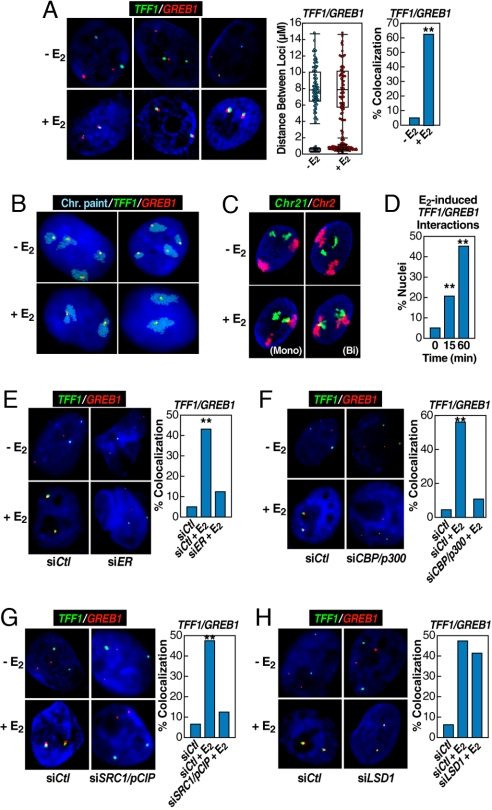
Fig. 2.
Rapid induction of interchromosomal interactions by nuclear hormone signaling. (A) 3D-FISH confirmation of E2-induced (60 min) TFF1:GREB1 interchromosomal interactions in HMECs with the distribution of loci distances measured (box plot with scatter plot) and quantification of colocalization (bar graph) before and after E2 treatment. Cells exhibiting mono- or biallelic interactions were combined for comparison with cells showing no colocalization; statistical significance in the bar graph was determined by χ2 test (**, P < 0.001). (B) 2D FISH confirmation of the interchromosomal interactions in MCF7 cells by combining chromosome paint (aqua) and specific DNA probes (green and red). (Upper) Illustrates two examples of mock-treated cells. (Lower) Shows the biallelic interactions/nuclear reorganization after E2 treatment for 60 min, exhibiting kissing events between chromosome 21 and chromosome 2. (C) Similar analysis on HMECs, but in this case using 3D FISH to paint chromosome 2 (red) and chromosome 21 (green), showing E2-induced chromosome 2–chromosome 21 interaction. Both assays revealed neither chromosome 21–chromosome 21 nor chromosome 2–chromosome 2 interactions in response to E2. (D) Temporal kinetics of GREB1:TFF1 interactions by 3D FISH in HMECs (**, P < 0.001 by χ2). (E–G) Nuclear microinjection of siRNA against ERα, CBP/p300, or SRC1/pCIP prevented E2-induced interchromosomal interactions, counting both mono- and biallelic interactions (**, P < 0.001 by χ2). The injection of siER and siDLC1 were done in the same experiment, sharing the same control group. (H) Nuclear microinjection of siRNA against LSD1, which was shown to be required for estrogen-induced gene expression (22), did not block E2-induced interchromosomal interactions. The injection of siLSD1 and SRC1/pCIP were done in a single experiment, sharing the same control group.
To determine whether the detected interchromosomal interactions resulted only from long-distance DNA looping or were also accompanied by chromosomal movements/interactions, we performed chromosome painting in the presence or absence of FISH, using TFF1 and GREB1 probes, finding that, although chromosome 21 and chromosome 2 were independently localized in the nucleus before E2 treatment, the two chromosomes became intimately associated in many cells after hormone treatment (Fig. 2 B and C), suggesting the possibility that estrogen treatment exerted dramatic effects on nuclear architecture. Although we reproducibly detected >5-fold difference in chromosome 21–chromosome 2 pairing between mock-treated and E2-treated cells, the frequency of interacting chromosomes was lower than E2-induced TFF1:GREB1 interactions detected with specific DNA probes, suggesting that chromosomal movement, in addition to long-distance DNA looping, occurs in response to E2. Interestingly, neither chromosome 21–chromosome 21 nor chromosome 2–chromosome 2 interactions were observed in response to E2. A time-course with FISH analysis further revealed that the interchromosomal interactions had already occurred at the initial time point determined (15 min) after E2 treatment (Fig. 2D). These results establish rapid nuclear reorganization in interphase cells in response to ligand.
Requirements for ERα-Dependent Interchromosomal Interactions.
Interactions between TFF1 and GREB1 were apparently dependent on ERα binding, because a specific siRNA, proven to effectively knockdown ERα (5, 27), effectively blocked the E2-induced GREB1:TFF1 interactions (Fig. 2E). We next performed single cell nuclear microinjection, using specific siRNAs or blocking antibodies against several specific coactivators, including CBP/p300 (Fig. 2F) and the p160 coactivator SRC1/pCIP (Fig. 2G), as established (5, 27), finding that inactivation of these coactivators for ERα also abolished the E2-dependent TFF1:GREB1 interactions. We also examined the histone lysine demethylase 1 (LSD1), which was recently shown to be essential for E2-dependent gene activation (5). Unexpectedly, we observed that LSD1 knockdown had little effect on the E2-dependent interchromosomal interactions between the two genes (Fig. 2H). This finding implies that E2-induced interchromosomal interactions precede gene activation.
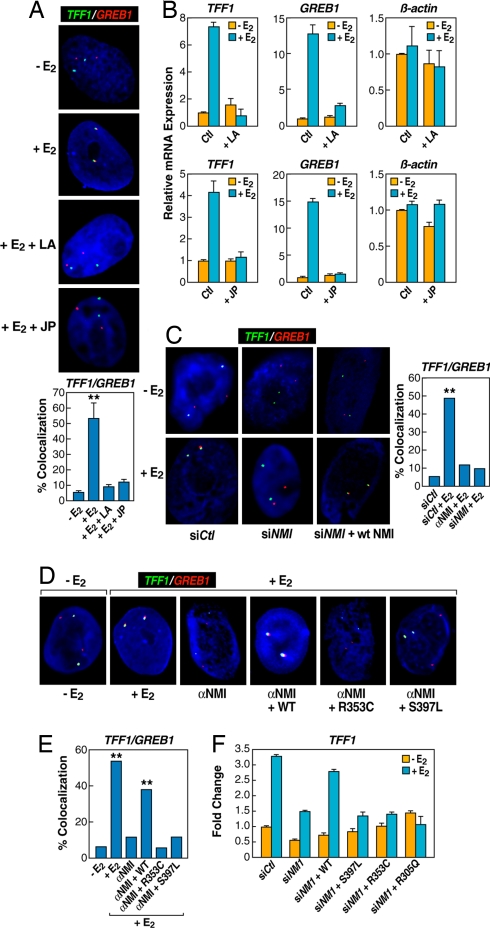
Although there is no filamentous actin in the nucleus, nuclear actin is present in many transcriptional complexes and reported to play an important role in transcriptional activation in yeast (28, 29). We could also detect oligomerized g-actin with a specific monoclonal antibody (30) in the nucleus of normal breast epithelial cells (Fig. S3A). Treatment of E2-stimulated breast epithelial cells with either latrunculin, a well-characterized drug that blocks actin polymerization (31), or Jasplakinolide, which inhibits depolymerization of actin networks (32), caused a complete loss of E2-induced interchromosomal interactions (Fig. 3A and Fig. S2B). These treatments also inhibited E2-induced expression of TFF1 and GREB1, but not constitutive genes (e.g., β-actin) in MCF7 cells (Fig. 3B).
Fig. 3.
The nuclear actin/myosin machinery is required for long-distance chromosomal interactions. (A) Chemical disruption of actin polymerization with latrunculin (LA) (10 μM, 2 h), and prevention of actin depolymerization by jasplakinolide (JP) (10 μM, 2 h) impaired E2-induced interchromosomal interactions. The bar graph shows the percentage of cells ± SEM that showed colocalization under individual conditions (**, P < 0.001 by t test). (B) qPCR analysis of gene expression affected by JP and LA treatment. (C) Nuclear microinjection of siRNA or antibody against nuclear myosin I (NMI) abolished E2-induced TFF1:GREB1 interchromosomal interactions (**, P < 0.001 by χ2). (D and E) The requirement for the actin binding and ATPase activity of nuclear myosin I in mediating E2-induced interchromosomal interactions. Cells injected with antibody against NMI were coinjected with the plasmid expressing either WT or mutant NMI containing specific mutations in the myosin “head,” which were shown to be critical for actin binding (R353C) and ATPase activity (S497L) of the motor protein (**, P < 0.001 by χ2). (F) Rescue of E2-induced expression of TFF1 by expression of WT, but not mutant, NMI. Results are the average of triplicates ± SD differing by <10%; similar results were observed in duplicate experiments.
To further investigate the possibility that interchromosomal interactions depend on dynamic nuclear reorganization, we performed single cell nuclear microinjection assay to determine the potential requirement for Nuclear Myosin-I (NMI) (33), finding that a specific siRNA against NMI abolished E2-induced TFF1:GREB1 interactions (Fig. 3C). We also found that nuclear injection of antibodies against NMI blocked E2-induced TFF1:GREB1 interactions (Fig. 3C Right and Fig. S3 B–D), as did similar single cell nuclear microinjection of the monoclonal antibody (2G2) against g-actin (Fig. S3 B–D). Immunohistochemical analysis confirmed nuclear localization of injected IgG against NMI and g-actin (Fig. S3 B and D), strongly suggesting a direct functional requirement for NMI in the nucleus, rather than an indirect effect caused by a disrupted cytoskeleton. To document the involvement of NMI-based nuclear motors in mediating E2-induced interchromosomal interactions, we preformed rescue experiments, using NMI mutants that impair actin binding (e.g., R353C) or the ATPase activity (e.g., S397L) in the nuclear myosin-I “head” (34, 35). We found that the interchromosomal interactions abolished with anti-NMI IgG could be restored using the expression vector expressing WT, but not mutant NMI defective in actin binding or lacking ATPase activity (Fig. 3 D and E). This finding agrees with a recent report showing that NMI is required for transcription activation-induced DNA looping out of the nuclear territory (36). We also documented that WT, but not functionally inactive NMI mutants, rescued the expression of TFF1 (Fig. 3F) and GREB1 (data not shown) after knockdown of endogenous NMI in MCF7.
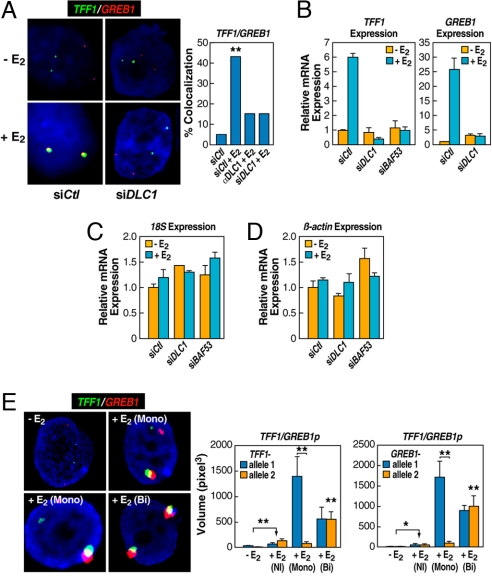
Based on a recent report that the dynein light chain-1 (DLC1) directly interacts with liganded ERα (37), we examined the effect of DLC1 inactivation by siRNA or injection of a blocking antibody, both of which effectively abolished GREB1:TFF1 interactions in E2-treated primary breast epithelial cells (Fig. 4A and Fig. S4) and inhibited E2-induced TFF1 and GREB1 expression, but not expression of constitutive genes examined (Fig. 4 B–D and Fig. S5); similar data were observed with siRNA against the chromatin remodeling factor BAF53 (Fig. 4 B–D), consistent with the multifactorial orchestration of interchromosomal interactions. Disruption of NMI or DLC1 by siRNA did not cause aberrant recruitment of ERα or its coactivators, such as CBP/p300, to the TFF1 promoter (Fig. S6), which suggests that these components of the nuclear motor system act after initial ERα binding and coactivator recruitment events.
Fig. 4.
Enhanced gene expression resulting from interchromosomal interactions and the requirement for a key nuclear motor component. (A) Effects of nuclear microinjection of siRNA or antibody against DLCI on inhibiting E2-induced TFF1:GREB1 interactions (**, P < 0.001 by χ2). (B–D) Cells were treated with siRNAs against DLCI and/or BAF53, and the expression of specific genes as indicated, were quantified by RT-qPCR. Mean ± SD of triplicate determinations. (E) RNA FISH demonstrates the requirement for interchromosomal interactions to achieve enhanced, E2-induced gene expression. Quantification of expression from noninteracting (NI) and interacting allelic regions was based on the diameter of individual signals, which was then converted to volume (M ± SEM). Plot shows the significant increase of expression even in the absence of colocalization of either allele; and the comparison of interacting: noninteracting alleles (*, P < 0.01; **, P < 0.001 by t test).
Functional Consequences of Induced Interchromosomal Interactions.
Both the siRNA and the antibody microinjection experiments described above established a correlation between hormone-induced interchromosomal interactions and gene activation. To directly demonstrate the functional requirement of E2-induced interchromosomal interactions for gene activation, we took advantage of the observation that ≈50% of cells exhibited monoallelic interactions between chromosome 2 and chromosome 21 to ask whether the transcription of gene loci engaged in the interchromosomal interactions was enhanced compared with noninteracting alleles in the same cells. We performed RNA FISH using probes that span exon-exon junctions and confirmed the specificity of the probes in detecting RNA by showing that RNase A treatment (14) abolished hybridization signals (data not shown). By determining the volume of the RNA signals for each transcript for colocalized and noncolocalized signals, we were able to quantify the level of transcription associated with interacting and noninteracting alleles before and after the E2 treatment (Fig. 4E). We found that E2 induced modest activation of TFF1 and GREB1 from “noninteracting” alleles, suggesting that both alleles may be equally competent in transcriptional activation. Remarkably, expression of these ERα target genes exhibited enhancement when engaged in specific interchromosomal interactions, therefore demonstrating the functional significance of interchromosomal interactions in enhancement of gene activation; indeed, this was actually more robust in cells with monoallelic, as opposed to biallelic, interactions (Fig. 4E). Three color DNA:RNA serial FISH experiments, using a TFF1 DNA FISH probe confirmed the colocalization of the genomic locus, and the two colocalized RNA transcripts (data not shown).
Interchromatin Granules: Hubs for Interchromosomal Interactions?
Having established nuclear actin/myosin-mediated gene networking in the nucleus, we next determined potential nuclear domains that permit or underlie such functional interchromosomal interactions. We suspected a possible spatial relationship with nuclear speckles, formally known as interchromatin granules that are enriched with several key transcriptional elongation factors, chromatin remodeling complexes, and essentially all factors required for pre-mRNA splicing (7, 38). To test this hypothesis, we determined colocalization between FISH probes and the splicing factor SC35, a marker for interchromatin granules (39). In mock-treated primary breast epithelial cells, the position of the TFF1 and GREB1 foci were entirely distinct from SC35-positive speckles. Upon E2 treatment, however, the interacting TFF1/GREB1 foci became intimately associated with two of the SC35-positive speckles. Intriguingly, in cells exhibiting monoallelic interactions, the interacting loci, but not noninteracting loci, were observed to associate with interchromatin granules (Fig. 5A). In cells with biallelic interactions, both were present in interchromatin granules (Fig. 5B). Blocking actin oligomerization with latrunculin or actin depolymerization with jasplakinolide impaired the association (Fig. S5C). Likewise, siRNAs against DLC1 or BAF53 all similarly blocked the colocalization of the FISH probes with interchromatin granules (Fig. S5 B and C and data not shown). These findings suggest that interchromatin granules may function as hubs for gene networking in the nucleus.
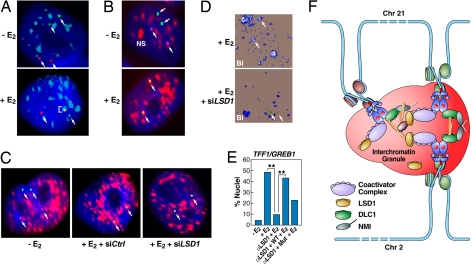
Fig. 5.
Interchromatin granules are hubs for interchromosomal interactions. (A) 2D-FISH shows selective association of interacting loci with interchromatin granules. Only the interacting (TFF1:GREB1) alleles intermingle within interchromatin granules (ICGs) stained with αSC35 (pseudocolored blue), whereas the remaining noncolocalized alleles show no colocalization with ICGs. (B) 2D-FISH of biallelic TFF1/GREB1 interactions (purple/green as indicated by arrows) coincident with the IGCs. (C) LSD1 is required for the association of the interacting TFF1:GREB1 loci with IGCs (pseudocolored red). Microinjection of siRNA against LSD1 abolished the colocalization between the interacting TFF1:GREB1 loci and ICGs. (D) 3D-FISH demonstrating the requirement of LSD1 for association of the interacting TFF1:GREB1 loci (arrows) with IGCs. (E) Percentage of cells exhibiting IGC association in response to control and specific siRNA against LSD1 (**, P < 0.001 by χ2). The rescure experiments indicate that the enzymatic activity of LSD1 is at least partially required for mediating the association of the interacting gene loci with ICGs. (F) Proposed model of E2-induced, actin/myosin1/DLC1-mediated chromosomal movement and LSD1-dependent interactions with interchromatin granules, creating a 3-dimensional enhancer hub in the nucleus.
Because LSD1 siRNA was able to block E2-dependent transcription of TFF1 and GREB1, but not their interchromosomal interactions, we investigated whether there might be an effect on their coalescence with nuclear speckles. Intriguingly, depletion of LSD1 by specific siRNA prevented the TFF1/GREB1 loci from interacting with interchromatin granules as determined by both 2D and 3D FISH with wild-type, but not enzymatically inactive LSD1, fully rescuing hub: interchromatin granule interaction (Fig. 5 C–E). In concert with our previous observation that LSD1 siRNA decreased, but did not abolish ERα recruitment (5), we detected only a modest reduction in the recruitment of coactivators, such as CBP/p300, to ERα target genes in response to E2 (Fig. S7). Together, these findings reveal an unexpected role of LSD1 in exerting a key regulatory function in linking transcriptional initiation to full gene activation by promoting the association of initial interacting loci to nuclear domains enriched with critical factors for transcription and cotranscriptional processing.
Discussion
Our findings reveal a previously unappreciated role of liganded nuclear receptors in initiating specific interchromosomal interactions. The formation of such gene networks now proves to be of functional importance for ligand-dependent enhancement of gene transcription. Our data also connect nuclear receptor-mediated recruitment of coactivators to the actions of the nuclear motor machinery in establishing ligand-induced intra- and interchromosomal interactions and the association of interacting gene loci with interchromatin granules, which constitutes a more complex program for hormone-induced gene expression than previously suspected. Our finding that NMI plays a key role in E2-induced gene activation program is consistent with its documented role in gene expression in other experimental systems (28, 29, 32, 40–42).
These data suggest a general model in which dynamic nuclear architecture permits rapid association of specific interacting genomic loci with potential “enhancer hubs” for higher level interactions of DNA-bound transcription factors with their cofactors (Fig. 5F). These events appear to represent at least a “two-step” program, with a specific histone lysine demethylase (LSD1) serving as a critical mediator to link initial induced interacting gene loci to interchromatin granules. Our findings of rapid, regulated nuclear reorganization is in accord with the documented role of nuclear actin in mediating the assembly of Pol II and coactivator complexes (29, 43), and with specific interchromosomal interaction events observed in other signal-induced transcription systems (20, 44–47). These findings underscore the chromosome mobility in interphase cells, which has been appreciated only recently (36, 48, 49).
In contrast to yeast, higher eukaryotic cells seem to have partitioned their nucleus into various subdomains (50). Strikingly, the dynamic E2-dependent, ERα-mediated interchromosomal interactions has proven to coincide with interchromatin granules, and surprisingly, LSD1 is required for the association of initial interacting gene loci with these interchromosomal hubs, which have long been considered as “storage” sites for splicing factors (51). Because interchromatin granules are also enriched for phosphorylated Pol II, several transcriptional elongation factors, such as p-TEFb, and key chromatin remodeling complexes, such as SWI/SNF (reviewed in ref. 7), we may now consider this nuclear domain as a specialized “nuclear factory.” For hormone-induced genes, the detected interchromosomal interactions in interchromatin granules may play an important role in coordinated and enhanced regulation of gene expression by permitting efficient coupling of transcriptional initiation, elongation, and RNA processing events.
Materials and Methods
Detailed protocols for cell culture, signal cell nuclear microinjection, and pharmacological treatment of cells are described in the SI Methods.
ChIP-DSL, 3D, and 3C assays.
Genomic tiling by ChIP-DSL was described (5, 22). Two anti-ERα antibodies (HC-20 and H-184; Santa Cruz Biotechnology) were combined for ChIP analyses. The 3D assay began with the conventional 3C assay after restriction digestion with BamH1 and BglII, using the procedure identical to that described for mammalian cells (52). Details of these assays are presented in the SI Methods. Oligonucleotides used for 3C validation are listed in Table S1.
DNA and RNA FISH.
The cells were processed for FISH essentially as described in ref. 24 except that oligonucleotide probes labeled with specific haptens were used as listed in Table S2. Both 2D and 3D FISH were performed. For triple-labeled FISH, probes to promoter regions were labeled at the 5′ position with digoxigenin (DIG) and probes to enhancer regions were labeled with either Biotin (Bio) or Fluorescein (FITC). For double-labeled FISH, promoters were labeled with Bio and enhancers with FITC. After hybridization, specific probes were detected by using a mix of quantum dot (Q-dot)-conjugated antibodies in 1:200 dilution (sheep anti-DIG Fab fragment primary antibody-conjugated with Qdot 655, streptavidin-conjugated with Q-dot 605, and goat anti-FITC whole IgG primary antibody-conjugated with Q-dot 525, all from Invitrogen). For a complete list of antibodies used in this study, see Table S3. Q-dots were mildly sonicated before use.
Single chromosome paint probes were commercially acquired from Applied Spectral Imaging (Vista). Each probe was custom-labeled with different fluorophores: chromosome 1 (1-585-605), chromosome 2 (1-585-606) and chromosome 21 (1–585-649) in aqua, red and green, respectively. Hybridization and detection protocols were performed as recommended by the manufacturer. Data acquisition and analysis are described in the SI Methods.
RNA FISH was performed with modification of published techniques (CSH Protocols; 2007–prot4763). All reagents were RNase-free. Cells were prepared as described in ref. 24 with the addition of a dehydration step by means of an ethanol series to 100% ethanol. Sequence of the specific oligo probes used is provided in Table S2B. Slides were washed and signal was detected with Q-dot antibodies as previously described. Slides were treated with RNase A before signal detection as a control.
Supplementary Material
Acknowledgments.
We thank V. Malhotra and S. Emr for insightful suggestions on nuclear motor experiments, P. Kannanganat and D. Spector for expert help in performing initial DNA FISH to independently confirm E2-induced interchromosomal interactions, J. Feramisco and C. Murre for invaluable assistance in microscopy studies, Trey Ideker for advice on statistical analysis, Begem Lee for technical assistance, C. Nelson for cell culture, M-J. Jin and L-X Duan of Aviva Systems Biology for assistance in performing ChIP-DSL and 3D assays, J. Hightower and M. Fisher for assistance with figure and manuscript preparation, and S. Cattaert of Santa Curz Biotechnology for advice on antibody reagents. Q.H was supported by the Cancer Research Institute; M.D.C. was supported by an American-Italian Cancer Foundation Postdoctoral Research Fellowship. M.G.R. is a Howard Hughes Medical Institute Investigator. This work was supported by National Institutes of Health Grants CA52599 and HL088129 (to G.K.C.), GM049369 and CA114184 (to X.-D.F.), and CA97134, DK39949, NS034934 HL65445, DK018477; DOD W81XWH-07-PCRP-IDA and W81XWH-08-1-0665, PCF (to M.G.R.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810634105/DCSupplemental.
References
- 1.Dennis AP, O'Malley BW. Rush hour at the promoter: How the ubiquitin proteasome pathway polices the traffic flow of nuclear receptor-dependent transcription. J Steroid Biochem Mol Biol. 2005;93:139–151. doi: 10.1016/j.jsbmb.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 2.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20:1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 3.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 5.Garcia-Bassets I, et al. Histone methylation dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell. 2007;128:505–518. doi: 10.1016/j.cell.2006.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Handwerger KE, Gall JG. Subnuclear organelles: New insights into form and function. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Lamond AI, Spector DL. Nuclear speckles: A model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 8.Chambeyron S, Bickmore WA. Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev. 2004;18:1119–1130. doi: 10.1101/gad.292104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kosak ST, et al. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 10.Hewitt SL, High FA, Reiner SL, Fisher AG, Merkenschlager M. Nuclear repositioning marks the selective exclusion of lineage-inappropriate transcription factor loci during T helper cell differentiation. Eur J Immunol. 2004;34:3604–3613. doi: 10.1002/eji.200425469. [DOI] [PubMed] [Google Scholar]
- 11.Williams RR, et al. Neural induction promotes large-scale chromatin reorganization of the Mash1 locus. J Cell Sci. 2006;119:132–140. doi: 10.1242/jcs.02727. [DOI] [PubMed] [Google Scholar]
- 12.Zink D, et al. Transcription-dependent spatial arrangements of CFTR and adjacent genes in human cell nuclei. J Cell Biol. 2004;166:815–825. doi: 10.1083/jcb.200404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Interchromosomal associations between alternatively expressed loci. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 14.Lomvardas S, et al. Interchromosomal interactions and olfactory receptor choice. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 15.Murrell A, Heeson S, Reik W. Interaction between differentially methylated regions partitions the imprinted genes Igf2 and H19 into parent-specific chromatin loops. Nat Genet. 2004;36:889–893. doi: 10.1038/ng1402. [DOI] [PubMed] [Google Scholar]
- 16.Zhao Z, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra and interchromosomal interactions. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 17.Ling JQ, et al. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science. 2006;312:269–272. doi: 10.1126/science.1123191. [DOI] [PubMed] [Google Scholar]
- 18.Sexton T, Schober H, Fraser P, Gasser SM. Gene regulation through nuclear organization. Nat Struc Mol Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 19.Mitchell JA, Fraser P. Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev. 2008;22:20–25. doi: 10.1101/gad.454008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apostolou E, Thanos D. Virus infection induces NF-κB-dependent interchromosomal associations mediating monoallelic IFN-β gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 21.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 22.Kwon YS, et al. Sensitive ChIP-DSL technology reveals an extensive estrogen receptor alpha-binding program on human gene promoters. Proc Natl Acad Sci USA. 2007;104:4852–4857. doi: 10.1073/pnas.0700715104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh MG, Thompson DA, Weigel RJ. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 2000;60:6367–6375. [PubMed] [Google Scholar]
- 24.Cai S, Kohwi-Shigematsu T. Intranuclear relocalization of matrix binding sites during T cell activation detected by amplified fluorescence in situ hybridization. Methods. 1999;19:394–402. doi: 10.1006/meth.1999.0875. [DOI] [PubMed] [Google Scholar]
- 25.Cook P. Duplicating a tangled genome. Science. 1998;281:1466–1467. doi: 10.1126/science.281.5382.1466. [DOI] [PubMed] [Google Scholar]
- 26.Paixao T, Carvalho TP, Calado DP, Carneiro J. Quantitative insights into stochastic monoallelic expression of cytokine genes. Immunol Cell Biol. 2007;85:315–322. doi: 10.1038/sj.icb.7100057. [DOI] [PubMed] [Google Scholar]
- 27.Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG. A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell. 2004;116:511–526. doi: 10.1016/s0092-8674(04)00133-3. [DOI] [PubMed] [Google Scholar]
- 28.Hofmann WA, de Lanerolle P. Nuclear actin: To polymerize or not to polymerize. J Cell Biol. 2006;172:495–496. doi: 10.1083/jcb.200601095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Percipalle P, Visa N. Molecular functions of nuclear actin in transcription. J Cell Biol. 2006;172:967–971. doi: 10.1083/jcb.200512083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gonsior SM, et al. Conformational difference between nuclear and cytoplasmic actin as detected by a monoclonal antibody. J Cell Sci. 1999;112:797–809. doi: 10.1242/jcs.112.6.797. [DOI] [PubMed] [Google Scholar]
- 31.Rizk RS, Walczak CE. Chromosome dynamics: Actin's gone fishing. Curr Biol. 2005;15:R841–842. doi: 10.1016/j.cub.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 32.Holzinger A. Jasplakinolide. An actin-specific reagent that promotes actin polymerization. Methods Mol Biol. 2001;161:109–120. doi: 10.1385/1-59259-051-9:109. [DOI] [PubMed] [Google Scholar]
- 33.Hofmann WA, Johnson T, Klapczynski M, Fan JL, de Lanerolle P. From transcription to transport: Emerging roles for nuclear myosin I. Biochem Cell Biol. 2006;84:418–426. doi: 10.1139/o06-069. [DOI] [PubMed] [Google Scholar]
- 34.Croft JA, et al. Differences in the localization and morphology of chromosomes in the human nucleus. J Cell Biol. 1999;145:1119–1131. doi: 10.1083/jcb.145.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q, Moncman CL, Winkelmann DA. Mutations in the motor domain modulate myosin activity and myofibril organization. J Cell Sci. 2003;116:4227–4238. doi: 10.1242/jcs.00709. [DOI] [PubMed] [Google Scholar]
- 36.Chuang C, et al. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 37.Rayala SK, et al. Functional regulation of oestrogen receptor pathway by the dynein light chain 1. EMBO Rep. 2005;6:538–544. doi: 10.1038/sj.embor.7400417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saitoh N, et al. Proteomic analysis of interchromatin granule clusters. Mol Biol Cell. 2004;15:3876–3890. doi: 10.1091/mbc.E04-03-0253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 40.Pestic-Dragovich L, et al. A myosin I isoform in the nucleus. Science. 2000;290:337–341. doi: 10.1126/science.290.5490.337. [DOI] [PubMed] [Google Scholar]
- 41.Percipalle P, Farrants AK. Chromatin remodeling and transcription: Be-WICHed by nuclear myosin 1. Curr Opin Cell Biol. 2006;18:267–274. doi: 10.1016/j.ceb.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 42.Kahle M, Pridalva J, Spacek M, Dzijak R, Hozak P. Nuclear myosin is ubiquitously expressed and evolutionary conserved in vertebrates. Hostochem Cell Biol. 2007;127:139–148. doi: 10.1007/s00418-006-0231-0. [DOI] [PubMed] [Google Scholar]
- 43.Bettinger BT, Gilbert DM, Amberg DC. Actin up in the nucleus. Nat Rev Mol Cell Biol. 2004;5:410–415. doi: 10.1038/nrm1370. [DOI] [PubMed] [Google Scholar]
- 44.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Long-range chromatin regulatory interactions in vivo. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 45.Osborne CS, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 46.Branco MR, Pombo An intermingling of chromosome territories in interphase suggests role in translocations and transcription-dependent associations. PLoS Biol. 2006;4:e138. doi: 10.1371/journal.pbio.0040138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active β-globin locus. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 48.Dillon N. Gene regulation and large-scale chromatin organization in the nucleus. Chromosome Res. 2006;14:117–126. doi: 10.1007/s10577-006-1027-8. [DOI] [PubMed] [Google Scholar]
- 49.Gasser SM. Visualizing chromatin dynamics in interphase nuclei. Science. 2002;296:1412–1416. doi: 10.1126/science.1067703. [DOI] [PubMed] [Google Scholar]
- 50.Spector DL. Macromolecular domains within the cell nucleus. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 51.Singer RH, Green MR. Compartmentalization of eukaryotic gene expression: Causes and effects. Cell. 1997;91:291–294. doi: 10.1016/s0092-8674(00)80411-0. [DOI] [PubMed] [Google Scholar]
- 52.Vakoc CR, et al. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.