Abstract
Gene expression changes in CD4+Vβ8+ T cells anergized by in vivo exposure to staphylococcal enterotoxin B (SEB) bacterial superantigen compared to CD4+Vβ8+ non-anergic T cells were assessed using DNA microarrays containing 5,184 murine cDNAs. Anergy in splenic T cells of SEB-immunized BALB/c mice was verified by dramatically reduced proliferative capacity and an 8X overexpression of GRAIL mRNA in CD4+Vβ8+ T cells taken from mice 7 d after injection. At an associative t-test threshold of p<0.0005, 96 genes were over-expressed or detected only in anergic T cells, while 256 genes were suppressed or not detected in anergic T cells. Six of eight differential expressions tested using real-time quantitative PCR were validated. Message for B-raf was detected only in non-anergic cells, while expression of the TCR signaling modulator Slap and the TCR ζ-chain specific phosphatase Ptpn3 was enhanced. Modulation of multiple genes suggests down-regulation of Wnt/β-catenin signaling and enhanced Notch signaling in the anergic cells. Consistent with previous reports in a non-superantigen in vivo anergy model, mRNA for CD18 and the transcription factor SATB1 was increased in SEB-anergized T cells. This is the first report of global transcriptional changes in CD4+ T cells made anergic by superantigen exposure.
Keywords: T cell, anergy, superantigen, rodent, microarray
Introduction
Naturally acquired T cell tolerance to self antigens is accounted for by several known mechanisms (reviewed in (1)) including clonal deletion, anergy and immunoregulatory cell control. Anergy is a tolerance mechanism in which lymphocytes are intrinsically functionally inactivated following antigen encounters that produce sub-optimal or altered signaling (2–6). Such cells remain alive for extended periods of time in a hyporesponsive state and are characterized by defective IL-2 production and cell cycle arrest (7, 8). Although these qualities appear to be universal traits of anergic T cells, several other characteristics have been used to subdivide anergic states into two broad and likely non-mutually exclusive categories that differ in several respects, including major modes of biochemical signaling blockade, reversibility by IL-2, shutdown of non-growth-dependent effector functions, and susceptibility of memory versus naïve cells to anergy induction (7).
In BALB/c mice systemically exposed to staphylococcal enterotoxin B (SEB) superantigen, clonal expansion of CD4+Vß8+ T cells precedes death and anergy in sequential stages of the immune response (5) such that, seven or more days post-exposure, remaining non-deleted CD4+Vß8+ T cells are anergic. Studies using this established model have shown defective TCR ζ-chain phosphorylation (9), defective calcium signaling (10, 11) and intact Ras/MAP kinase signaling at very early time points (11), characteristics consistent with an adaptive, or in vivo type of anergy. Reports of the reversibility of SEB-induced anergy by IL-2 (4, 12, 13) and the degree to which non growth-dependent effector functions are impaired or intact in this model (14–16) are conflicting. Interestingly, SEB-induced T cell anergy appears to target primarily cells with a memory phenotype (17, 18), a property usually associated with clonally anergic cells. These studies suggest that the SEB model invokes selective characteristics of both clonal and adaptive anergy, presenting a unique opportunity to appreciate additional factors involved in anergy induction and maintenance.
Initiation of the anergic program requires new protein synthesis that is likely to also be necessary for maintenance of the anergic state (19, 20). In addition, cell fusion studies have shown that the anergic phenotype is dominant (21). Several classes of putative “anergy” factors that may fulfill these roles have been proposed (7) and include inhibitory receptors such as CTLA-4 and PD-1; anti-proliferative factors such as p27kip1; and E3 ubiquitin ligases, including GRAIL, Itch and c-Cbl. While roles for the E3 ubiquitin ligases have been supported by in vivo anergy studies (22, 23), both p27kip1 and CTLA-4 have been shown to be non-essential for anergy induction and/or maintenance (24, 25). Given the increasingly appreciated complexity of the anergic phenotype, the likelihood that additional unknown anergy factors exist has been recently underscored (7, 8).
Towards further defining the anergic T cell state on a molecular basis, this study has demonstrated a number of significant gene expression differences between SEB-anergized CD4+Vβ8+ T cells and similarly isolated non-anergic T cells taken from mice that were not exposed to SEB. This is the first report of differential gene expression in the SEB model. In addition to identifying several expression changes previously associated with T cell anergy, these studies have revealed novel transcriptional programs that are likely to negatively affect vital aspects of TCR signal transduction. Moreover, this is the primary report showing association of the Wnt and Notch signaling pathways with T cell anergy as suggested by differential expression of multiple members of these two fundamental pathways that govern cell fate decisions (26, 27).
Results
Generation of anergic T cells by in vivo exposure to staphylococcal enterotoxin B
The goal of the current study is to determine gene expression profiles of T cells made anergic by exposure to the bacterial superantigen staphylococcal enterotoxin B (SEB). It has previously been established that intraperitoneal immunization of BALB/c mice with a bolus of SEB rapidly activates the vast majority of T cells bearing Vβ8 T cell receptors (5). Following the death of a subset of these cells at approximately 3–4 days post-immunization, surviving SEB-exposed CD4+Vβ8+ T cells are anergic as defined by a loss of proliferative capacity and ability to secrete IL-2 (5, 7). To validate this well-established model in our laboratory, splenic T cells were taken from mice 6 hours after injection with either 50 μg SEB in PBS or with PBS alone and evaluated for activation marker expression by flow cytometry. Six hours post-immunization, over 95% of CD4+Vβ8+ T cells expressed CD69 while more than 90% of CD4+Vβ8+ T cells expressed CD25 at this time point (Figure 1). To evaluate the effectiveness of this immunization protocol for inducing T cell anergy, splenic T cells were taken from mice euthanized seven days after receiving similar injections of SEB or PBS alone and evaluated for in vitro responsiveness to SEB using a standard tritiated thymidine incorporation assay. Figure 2a shows that splenic T cells from SEB-immunized mice responded poorly to SEB while T cells from mice immunized with PBS alone proliferated robustly. Flow cytometry revealed that the fraction of CD4+ T cells expressing Vβ8 in SEB-immunized and PBS-immunized mice seven days post-injection was 21% and 27.5% respectively (data not shown), indicating that deletion of CD4+Vβ8+ T cells in SEB-injected mice was only a minor contributor to the observed abrogation of responsiveness. As expected, splenic T cells from both groups of SEB- and PBS-injected mice responded equally well to the irrelevant Vβ3-binding superantigen staphylococcal enterotoxin A (SEA) (Figure 2b). Further validation of our model is shown in Figure 3 where SEB-exposed CD4+Vβ8+ T cells remain anergic after cell sorting. Sorted CD4+Vβ8+ splenic T cells from BALB/c mice that had been injected 7 d previously with SEB or PBS were rested for 3 days in media + IL-7 then subjected to a [3H]-thymidine proliferation assay. Low [3H]-thymidine incorporation in these sorted cells reflects lack of proliferation in response to restimulation with SEB, indicating anergy. Results from restimulation with SEA reflect the absence of Vβ3+ T cells in the sorted population.
Figure 1. Activation of essentially all CD4+Vβ8+ T cells in mouse spleen two hours after injection of SEB.
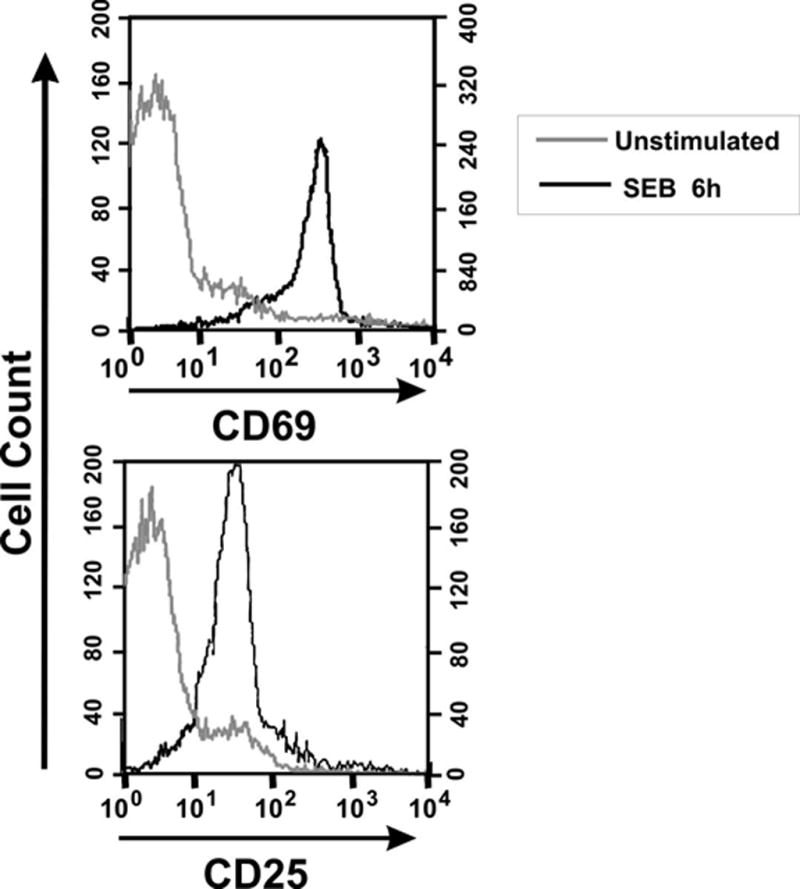
BALB/c mice were injected with 50 μg of SEB in PBS (SEB 6h) or with PBS alone (unstimulated), euthanized 2 h later and their splenocytes evaluated by flow cytometry. Histograms depicting relative expression levels of CD69 and CD25 are gated on CD4+Vβ8+ lymphocytes. Greater than 95% of CD4+Vβ8+ T cells were activated.
Figure 2. Splenic T cells from SEB-immunized mice exhibit defective proliferation to SEB in vitro.
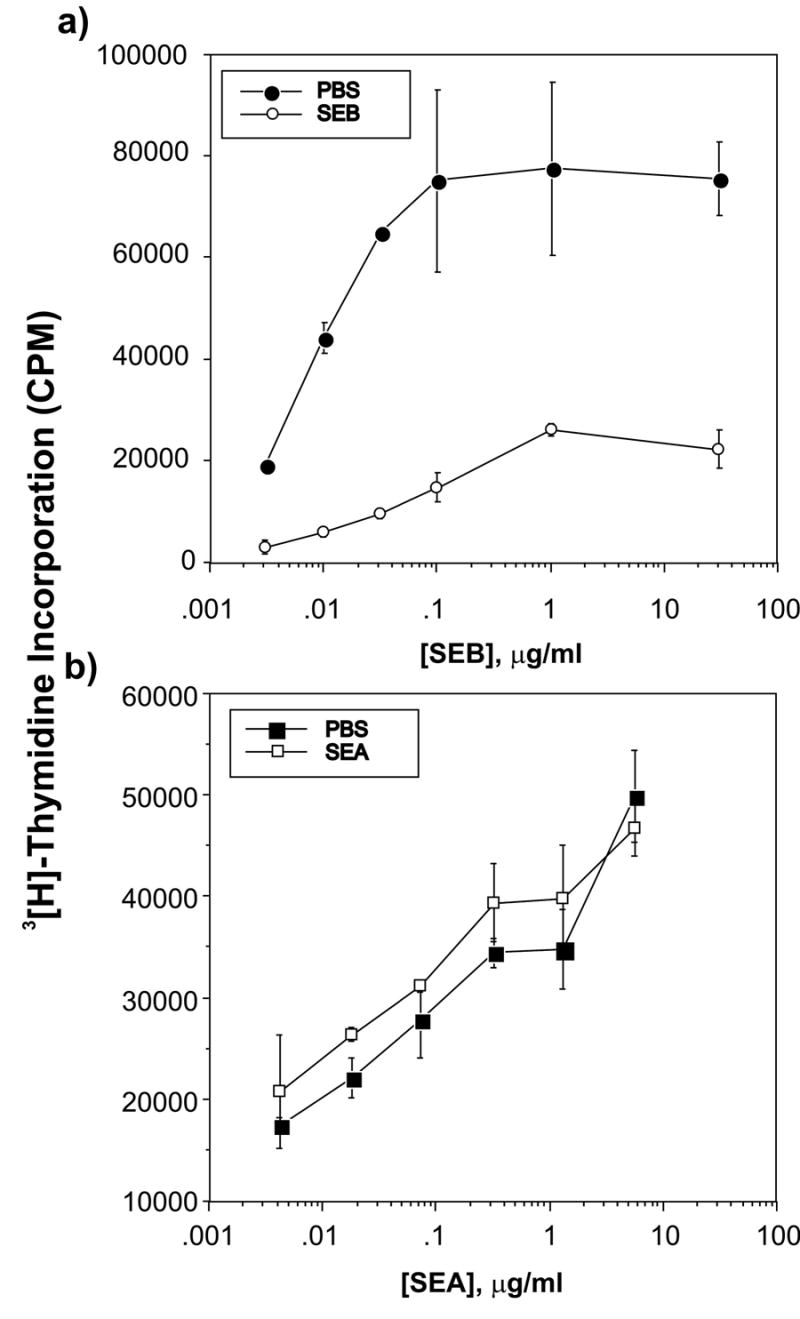
Purified splenic T cells taken from mice injected seven days previously with either 50 μg SEB in PBS (SEB, filled symbols) or with PBS alone (PBS, open symbols) were recalled in vitro with irradiated syngeneic spleen cells taken from uninjected mice and varying concentrations of highly purified SEB (Panel a) the irrelevant non-Vβ8-binding SEA superantigen (Panel b) in triplicate. Results are representative of two independent experiments.
Figure 3. SEB-exposed CD4+Vβ8+ T cells remain anergic after cell sorting.
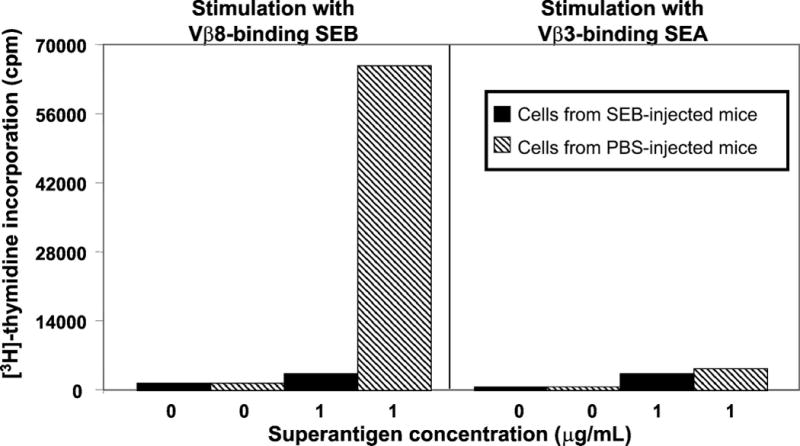
Sorted CD4+Vβ8+ splenic T cells from BALB/c mice that had been injected 7 d previously with SEB (black bars) or PBS (hatched bars) were rested for 3 days in media + IL-7 then subjected to a [3H]-thymidine proliferation assay. Low [3H]-thymidine incorporation reflects lack of proliferation in response to restimulation with SEB, indicating anergy. Results from restimulation with SEA reflect the absence of Vβ3+ T cells in the sorted population.
Evaluation of gene expression profiles using microarray chips containing 5,184 cDNAs
We hypothesized that sustained tolerance in SEB-responsive CD4+ T cells correlates with a distinct genetic program that is likely to be important for maintenance of the anergic state. To test this hypothesis, gene expression profiles of resting SEB-anergized CD4+Vβ8+ T cells and resting non-anergic CD4+Vβ8+ T cells were evaluated using cDNA microarrays containing 5,184 cDNAs and EST’s that were not specifically selected for immunological studies. CD4+Vβ8+ T cells were isolated from groups of mice injected with either SEB in PBS (anergic T cells) or PBS alone (non-anergic T cells) using high speed cell sorting. Post-sort analyses for all sorts revealed cell purities that ranged from 93% to 99%. Only RNA that was judged to be free from degradation was used for experimentation. RNA obtained from four experimentally independent anergic and three experimentally independent non-anergic samples were used for hybridization and statistical analysis. Each independent sample originated from pooled splenocytes of SEB- or mock-immunized mice as described in Materials and Methods and was sorted in independent experiments on different days. All samples gave high quality hybridization signals (not shown).
When the Associative T-test was applied to dramatically reduce false positive determinations, a total of 352 genes and EST’s with expression signals above background as determined by the normalization procedure, were found to be differentially expressed between PBS- and SEB-treated mice at a level of p<0.0005. This analysis and the normalization procedure described in Materials and Methods allows reliable measurement of expression level and fold-expression differences of lowly expressed genes. Of these, 96 genes or ESTs were either detected only in anergic samples or were differentially over-expressed in anergic samples, and 256 genes or ESTs were either detected only in non-anergic samples or were differentially over-expressed in non-anergic samples. Table 1 shows the total number of differentially regulated genes and ESTs categorized by established or proposed function or by cell surface localization as determined using a combination of NCBI databases including Unigene, Locuslink, OMIM and PubMed. Of these differentially expressed genes, approximately 40% are ESTs with no currently assigned function. Due to space limitations, the accession numbers for these sequences are not listed here but may be found in Supplemental Table 1 at the following website: http://microarray.omrf.org/publications/2005/kurella/supplementary_table_1.xls. The functional diversity of the proteins encoded by these genes suggests that complex processes generate and sustain the anergic phenotype in these T cells.
Table 1.
Numbers of differentially expressed genes in Non-Anergic versus Anergic T cells by Functional Category
| Number of Genes expressed in different subgroups * | ||||
|---|---|---|---|---|
| Category | Anergic only | Over expressed in anergic | Non-Anergic only | Over expressed in Non-Anergic |
| Transcription factors | 7 | 4 | 17 | 7 |
| Signal Transduction | 6 | 5 | 13 | 3 |
| Cell cycle/Apoptosis | 2 | 4 | 8 | 5 |
| Cell surface | 4 | 6 | 3 | 1 |
| Others | 14 | 11 | 72 | 17 |
| Total known genes | 33 | 30 | 113 | 32 |
| ESTs | 14 | 19 | 84 | 27 |
| Total differentially regulated genes | 47 | 49 | 197 | 59 |
p<0.0005 by Associative t-test
Numbers of genes within each group are indicated. Placement in "only" categories does not exclude low basal expression in compared samples below the level of detection using this analysis. Identification as a cell surface molecule or transcription factor was given precedence for genes fitting more than one category. Genes listed in the category Others were primarily those of metabolic, structural or unknown function.
Differentially expressed genes encoding signal transduction molecules
Tables 2 through 5 list the identities of differentially expressed genes with known function or activity that are grouped according to functional category (Tables 2–4) or cell surface localization (Table 5). The genes are ranked in order of either expression level in terms of number of SD above background or in terms of fold expression differences as determined by the Associative T-test (all p<0.0005).
Table 2.
Signal Transduction Molecules
| Only Expressed in Anergic Cells
|
Overexpressed in Anergic Cells
|
||||||
|---|---|---|---|---|---|---|---|
| SD Above Background | Accession No. | Gene Symbol | A/N | Non-Anergic SD Above Background | Anergic SD Above Background | Accession No. | Gene Symbol |
| 31.018 | 2.2 | 69.5 | AI323667 | Irg1 | |||
| 2.2 | AI604839 | StmnS | 7.542 | 0.5 | 3.7 | AI451062 | Gucy1a3 |
| 2.0 | AI448261 | Strap | 3.981 | 1.0 | 4.1 | AI451174 | St5 |
| 1.4 | AI465535 | Rab38 | 3.882 | 33.6 | 130.3 | AI451572 | SIa (SIap) |
| 0.9 | AI426390 | Nkd2 | 3.394 | 28.1 | 95.5 | AI323613 | Inpp5d |
| 0.6 | AI4 15054 | PtpnS | |||||
| 0.5 | AI426929 | Hunk | |||||
|
Only Expressed in Non-Anergic Cells
|
Overexpressed in Non-Anergic Cells
|
||||||
| SD Above Background | Accession No. | Gene Symbol | N/A | Non-Anergic SD Above Background | Anergic SD Above Background | Accession No. | Gene Symbol |
| 15.5 | AI448985 | Geft | 7.189 | 256.0 | 35.6 | AI465451 | Lin 7c |
| 6.9 | AI451618 | LCP2/Slp-76 | 4.025 | 22.5 | 5.6 | AI451068 | L fng |
| 3.2 | AI448890 | Ppp1r12a | 3.557 | 6.1 | 1.7 | AI448884 | Asb6 |
| 3.1 | AI893430 | Nle | |||||
| 2.8 | AI448863 | Grem2 | |||||
| 2.8 | AI464368 | Osf2 | |||||
| 2.5 | AI447469 | Braf | |||||
| 2.3 | AI451411 | Tnik | |||||
| 1.9 | AI666784 | Prkcbpl | |||||
| 1.8 | AI429753 | PpmH | |||||
| 0.4 | AI428513 | Traip | |||||
| 0.3 | AI429407 | Mylk | |||||
| 0.3 | AI894311 | Stk23 | |||||
Table 5.
Cell surface molecules
| Only Expressed in Anergic Cells
|
Overexpressed in Anergic Cells
|
||||||
|---|---|---|---|---|---|---|---|
| SD Above Background | Accession No. | Gene Symbol | A/N | Non-Anergic SD Above Background | AnergicSD Above Background | Accession No. | Gene Symbol |
| 2.2 | AI427754 | Gpr83 | 13.733 | 2.7 | 37.0 | AI528633 | Ptpra |
| 1.8 | AI450557 | Gpr114 | 5.197 | 3.2 | 16.5 | AI449406 | Gpr68 |
| 1.1 | AI426162 | EfnbS | 4.514 | 39.3 | 177.5 | AI385637 | FzdS |
| 0.6 | AI413526 | Jag1 | 4.048 | 11.3 | 45.9 | AI465291 | Adora2a |
| 3.808 | 51.1 | 194.6 | AI528527 | Itb2 | |||
| 3.389 | 1.7 | 5.7 | AI450072 | Cdh1 | |||
|
Only Expressed in Non-Anergic Cells
|
Overexpressed in Non-Anergic Cells
|
||||||
| SD Above Background | Accession No. | Gene Symbol | N/A | Non-Anergic SD Above Background | Anergic SD Above Background | Accession No. | Gene Symbol |
| 12.4 | AI661130 | H2-Q1 | 4.565 | 208.7 | 45.7 | AI447815 | Lu |
| 1.7 | AI427644 | Egfr | |||||
| 0.7 | AI323855 | Cd24a | |||||
Table 4.
Cell cycle, proliferation and death
| Only Expressed in Anergic Cells
|
Overexpressed in Anergic Cells
|
||||||
|---|---|---|---|---|---|---|---|
| SD Above Background | Accession No. | Gene Symbol | A/N | Non-Anergic SD Above Background | Anergic SD Above Background | Accession No. | Gene Symbol |
| 3.0 | AI894115 | CcncM (Cyclin D1) | 4.900 | 183.6 | 899.5 | AI448808 | Unc-84a |
| 1.8 | AI451585 | Birdf/NAlP | 4.341 | 2.3 | 9.9 | AI428398 | Mki67 |
| 4.218 | 194.8 | 821.8 | AI323926 | Fau | |||
| 3.034 | 11.0 | 33.3 | AI449265 | Etnk1 | |||
|
Only Expressed in Non-Anergic Cells
|
Overexpressed in Non-Anergic Cells
|
||||||
| SD Above Background | Accession No. | Gene Symbol | N/A | Non-Anergic SD Above Background | Anergic SD Above Background | Accession No. | Gene Symbol |
| 6.3 | AI323649 | P2rx1 | 4.702 | 29.8 | 6.3 | AI452025 | Becn1 |
| 3 7 | AI465262 | Fjx1 | 4 587 | 19 0 | 4 1 | AI413932 | Emd |
| 1.0 | AI426377 | Pcd8 | 4.297 | 5.7 | 1.3 | AI452256 | Kif23 |
| 0.9 | AI324208 | Gel | 4.044 | 44.7 | 11.0 | AI426170 | Igfbp4 |
| 0.7 | AI427986 | Mak3 | |||||
| 0.5 | AI465513 | Top2b | |||||
| 0.5 | AI426979 | Lrdd | |||||
As seen in Table 2, 11 genes participating in signal transduction processes are either over-expressed or expressed only in anergic T cells, while 16 genes are either down-regulated or turned off in the anergic cells. Although the significance of some of these expressions for T cell function can only be speculated, many of the expression changes are likely to critically impact TCR signaling. Of particular interest among these are observations of reduced or undetected levels of mRNA for the src tyrosine kinase SLP-76 and the Ras pathway transducer B-raf. In contrast, enhanced levels of mRNA for the Src-like Adapter Protein (Slap) and the Ptpn3 (also known as PTPH1) phophatase were found in the SEB anergized T cells. Of further potential interest is over-expression of message for the phosphatidyl inositol-3 kinase regulator Inpp5d, or SHIP-1, in the anergic cells. Message for two genes involved in cytoskeletal reorganization downstream of integrin signaling, Protein Phosphatase 1 Regulatory Subunit 12A (Ppp1r12a; also known as Mypt1) and Myosin Light Chain Kinase (Mylk), are down-regulated in anergic T cells. A relatively high level expression of the gene encoding a novel guanine nucleotide exchange factor with controversial specificity, Geft, that has been studied primarily in neuronal cells, was detected only in non-anergic T cells. Two different mRNAs that encode products known to down-regulate the Notch signaling pathway were either turned off (Notchless; Nle) or down-regulated (Lunatic Fringe; Lfng) in anergic cells. Although none of the four known Notch receptors were among the 5,184 genes assessed in this study, detection of mRNA for the Notch-specific transcription factor Hes-5 (Table 3, below) in anergic but not non-anergic T cells suggests active Notch signaling in SEB-anergized T cells, as does overexpression of Jagged-1 (Table 5; see Discussion). Other interesting mRNAs that were overexpressed or detected only in anergic T cells are those encoding the microtubule destabilizing protein Stathmin-like 3 (Stmn3) and the canonical Wnt signaling pathway modulator Naked 2 (Nkd2). Stathmins are rapidly phosphorylated in T cells downstream of both TCR and CD2 (28), and enforced cellular expression of stathmin can cause cellular proliferation defects and cell cycle arrest (29).
Table 3.
Transcription Factors and Modulators
| Only Expressed in Anergic Cells
|
Overexpressed in Anergic Cells
|
||||||
|---|---|---|---|---|---|---|---|
| SD Above Background | Accession No. | Gene Symbol | A/N | Non-Anergic SD Above Background | Anergic SD Above Background | Accession No. | Gene Symbol |
| 3.9 | AI452276 | Nkx6-2 | 17.200 | 5.2 | 89.1 | AI893650 | Usf2 |
| 2.9 | AI427771 | Msc | 5.023 | 0.4 | 1.9 | AI426152 | Smarce1 |
| 2.5 | AI427630 | Ebf3 | 4.926 | 0.9 | 4.3 | AI447611 | Cenpj |
| 2.4 | AI385632 | Foxdl | 4.557 | 26.1 | 119.0 | AI449492 | Nfatc1 |
| 1.4 | AI447744 | Ddx21 | |||||
| 1.0 | AI385524 | Gata4 | |||||
| 0.9 | AI385674 | Hes-5 | |||||
|
Only Expressed in Non-Anergic Cells
|
Overexpressed in Non-Anergic Cells
|
||||||
| SD Above Background | Accession No. | Gene Symbol | N/A | Non-Anergic SD Above Background | Anergic SD Above Background | Accession No. | Gene Symbol |
| 12.2 | AI427147 | Smad4 | 7.260 | 60.9 | 8.4 | AI451958 | PoIr3c |
| 6.9 | AI450635 | Ankra2 | 6.048 | 5.6 | 0.9 | AI447986 | Nfia |
| 4.8 | AI528521 | Satbl | 5.912 | 31.6 | 5.3 | AI431085 | Ski |
| 3.5 | AI447646 | Gtf2a1 | 5.134 | 8.2 | 1.6 | AI426555 | Hdac5 |
| 2.6 | AI385556 | Nab2 | 4.391 | 2.0 | 0.4 | AI324260 | Rbbp4 |
| 2.4 | AI465142 | Hnrpull | 4.326 | 292.2 | 67.5 | AI451430 | Lef1 |
| 2.4 | AI450078 | Epc1 | 3.592 | 85.9 | 23.9 | AI452359 | D11Moh34 |
| 2.2 | AI414482 | Za20d2 | |||||
| 1.4 | AI450810 | Pml | |||||
| 1.2 | AI427022 | Bbx | |||||
| 1.1 | AI429154 | Ewsh | |||||
| 1.1 | AI450125 | Zfp318 | |||||
| 0.8 | AI325529 | Smarcb! | |||||
| 0.8 | AI430751 | Neud4 | |||||
| 0.8 | AI323840 | Ezh2 | |||||
| 0.7 | AI596340 | Pknoxl | |||||
| 0.4 | AI451024 | Ankibl | |||||
Differentially expressed genes encoding transcription factors
The largest functional group of genes modulated between anergic and non-anergic CD4+ T cells are those that are known or strongly suspected to regulate gene transcription (Table 3). In addition to Hes-5, mentioned above, Foxd1 is among transcription factors that are overexpressed or detected only in the anergic cells. This transcription factor is known to be a transcriptional activator of the RI alpha subunit of the cAMP-dependent protein kinase A (PKA) (30), an essential molecule for the induction of cAMP-induced anergy in HIV-1 gp120-exposed CD4+ T cells (31). Potentially important genes that are turned off or down-regulated in the anergic cells include enhancer of zeste 2 (ezh2), retinoblastoma binding protein 4 (rbbp4) and the canonical Wnt pathway transcription factor lef-1 (see Discussion). Message for Smad 4, a transcription factor that mediates TGF-β receptor signal transduction was detected only in non-anergic cells. This, along with the observation that strap, a gene encoding a negative regulator of TGF-β signal transduction is overexpressed in the anergic cells (Table 2), is consistent with observations indicating that the anergic state in SEB-anergized CD4+ T cells is cell-autonomous (32) and thus not subject to the actions of regulatory cytokines. Message for special AT-Rich Binding Protein 1 (Satb1), a matrix attachment region-binding global transcriptional regulator that is known to be important for thymocyte development (33), is detected only in non-anergic T cells, similar to observations made in a non-superantigen-based in vivo model of T cell anergy (34).
Differentially expressed genes encoding regulators of the cell cycle, proliferation and death
In addition to modulated Stmn3 and the pro-proliferative transcription factors noted above, several additional genes known to impact cell cycle, proliferation and death are differentially expressed between anergic and non-anergic T cells (Table 4). Message for two molecules important for cell cycle progression, Emerin (Emd) and Kinesin Factor 23 (Kif23), also known as Mitotic Kinesin-like Protein 1 (MKLP1), were down-regulated in the anergic cells. Other down-regulated mRNAs, as evidenced by detection only in non-anergic populations, included that of two death-promoting genes, programmed cell death 8 (pcd8) and leucine rich and death domain containing (lrdd). Two genes that were either overexpressed or detected only in the anergic cells that may contribute to the viability of SEB-anergized T cells are baculoviral IAP repeat-containing (birc1f, the product of which is also known as Neuronal Apoptosis Inhibitory Protein (NAIP) and Fau. The latter sequence was originally identified in a screen for suppressors of apoptosis in T cells (35). Surprisingly, moderate expression of message cyclin D1 (ccnd1) and the proliferation marker mki67 were detected in the anergic population. This might be explained by a compensatory upregulation of Ccnd1 in response to cell cycle arrest or reductions in the expression of other cyclins that were not measured in this study. This phenomenon has precedence in Cyclin D2 knockout mice, where there was a compensatory upregulation of Cyclin D3, and in anergic B cells (36). The detection of mRNA for Mki67 is more puzzling but might reflect the relatively recent cycling history of the SEB-anergized cells (5).
Differentially expressed genes encoding cell surface molecules
Identification of novel cell surface markers of anergic T cells would represent a significant advance, opening the door to strategies for monitoring the contribution of CD4+ T cell anergy to tolerance in multiple settings. Table 5 lists several novel candidates in this category. Of particular functional interest is the upregulation in anergic cells of mRNA for two cell surface markers impacting β-catenin signaling. One of these, Cadherin 1 (Cdh1, also referred to as E-Cadherin) is an adhesion molecule of the Ig superfamily, the C-terminal domain of which interacts with several catenins including β-catenin. Overexpression of E-cadherin in epithelial and fibroblastoid cells resulted in G1 phase cell growth arrest and was strictly associated with down-regulated β-catenin/LEF activitiy (37), presumably as a result of sequestration of cellular β-catenin. Transcripts for one of the ten known but poorly functionally characterized canonical Wnt receptors, Frizzled-8 (Fzd8) are both highly expressed and over-expressed in the SEB-anergized cells. Detected overexpression of a third cell surface molecule, the adenosine A2a receptor (Adora2a), in the anergic populations is of additional potential functional significance in light of the ability of A2a signaling to stimulate production of cAMP, an intermediate that has been shown to induce T cell anergy in multiple settings by multiple pathways (see Discussion). Interestingly, genes for three other G-protein coupled receptors, gpr83, gpr114 and gpr68, are either overexpressed or detected only in the anergic cells. The product of one of these, Gpr68, has been found to activate p42/p44 MAPKs and inhibit cell proliferation in the Gpr68-transfected human tumor cell line HEK 293 (38). Finally, transcripts for itb2 (CD18) are overrepresented in the SEB-anergized T cells. This was also found by others in a non-superantigen based in vivo anergy model (34), suggesting that upregulation of CD18 could be a universal feature of in vivo T cell anergy.
Differentially expressed genes involved in cell structure, metabolism, or unknown functions
Numerous genes encoding proteins participating in translation, metabolism, maintenance of potassium ion concentration, and cell structure, were reproducibly differentially expressed between the anergic and non-anergic populations. Due to space limitations these are not listed here but may be found in Supplemental Table 2 at the following web site: http://microarray.omrf.org/publications/2005/kurella/supplementary_table_2.xls
Confirmation of differential expression using real-time quantitative RT-PCR
Eight of the differentially expressed genes identified by gene microarray analysis were selected for confirmation by real-time quantitative PCR without regard to expression level or fold-expression differences between the two groups. In addition, this technique was used to determine the relative expression level of the GRAIL gene (not present on the microarrays tested) in anergic and non-anergic samples in order to further validate the data presented herein since GRAIL has been shown to participate in T cell anergy in the SEB model (39). The results of two different sets of experiments evaluating fold-expression levels of mRNA in sorted CD4+Vβ8+ anergic and non-anergic T cells, as normalized on β-actin mRNA expression, are depicted in Figure 4. Significantly, 6 of the 8 microarray-identified expressions were confirmed in both experiments, further establishing the validity of differential expressions described above. Of the two expressions that were not confirmed, myosin light chain kinase (Mylk) was one of the lowest expressed genes (0.3 SD above background) passing the associative T-test criteria. The validity of the microarray observations that myosin light chain kinase (Mylk) and SLP-76 are overexpressed in anergic T cells is not conclusively ruled out by these data since the microarray analysis involved more replicates and may thus be more reliable from a statistical standpoint. Jagged-1, which was detected only in anergic T cells at a level of 0.6 SD above background was confirmed by quantitative PCR in two experiments conducted (values greater than 1). Importantly, GRAIL mRNA was found to be 8X more abundant in anergic T cells compared to non-anergic T cells. Thus, the confirmation of 6 of 8 microarray-identified expressions tested in two separate real-time quantitative PCR experiments conducted and the finding of over-expression of GRAIL mRNA in our SEB-anergized T cells provide a robust validation of the system, techniques, and array results described herein.
Figure 4. Confirmation of microarray data by real-time quantitative PCR analysis of gene expression.
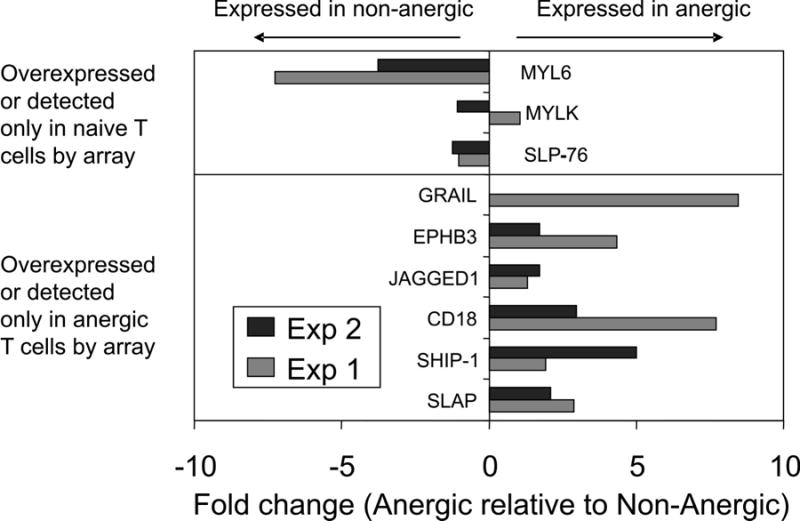
Total RNA prepared from CD4+Vβ8+ T cells sorted from BALB/c mice immunized 7 days previously with SEB in PBS (anergic) or PBS alone (non-anergic) was analyzed by real-time quantitative RT-PCR using the primer pairs detailed in Table 6. Fold changes in expression levels were obtained after normalizing each sample to a house-keeping gene (β-actin). Positive values indicate that the transcript is more abundant in anergic cells, and negative values indicate that the transcript is more abundant in non-anergic cells. Colored bars depict the results of two independent experiments. See the legend to Table 6 for a description of Myl6. Grail expression was confirmed in a single separate experiment.
Enhanced cell surface expression of CD18 on resting anergic T cells
To determine whether CD18 protein is overexpressed on the surface of anergic T cells in the SEB model, its expression was evaluated by flow cytometery. Figure 5 shows the results of a representative experiment establishing that CD18 is indeed over-expressed on the surface of resting anergic T cells. The mean fluorescence intensity (MFI) in anergic CD4+Vβ8+ T cells was 196.64 while the MFI of CD18 expression in non-anergic CD4+Vβ8+ T cells was 165.68. The geometric mean of CD18 expression in anergic T cells was almost twice (113.39) that of non-anergic T cells (60.10). These data confirm that both resting anergic and non-anergic T cells express CD18 and that CD18 is overexpressed on the SEB-anergized cells.
Figure 5. Mean fluorescence intensities of CD18 expression in anergic and non-anergic CD4+Vβ8+ T cells.
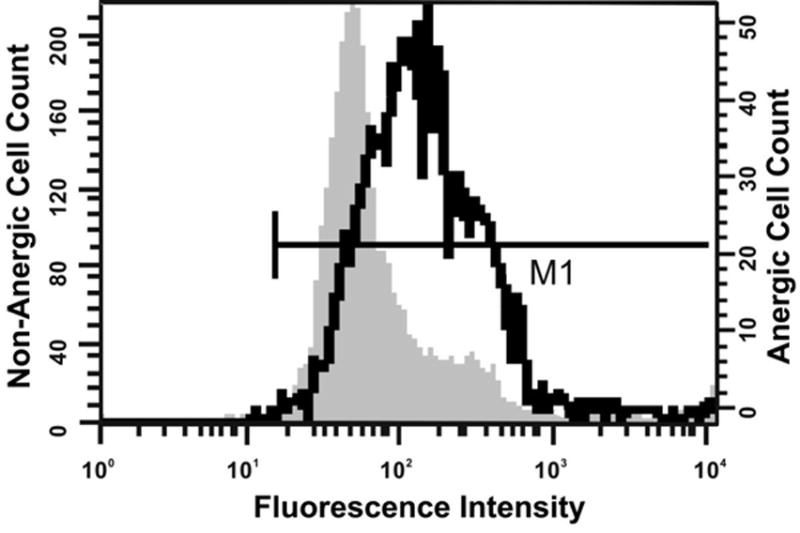
BALB/c mice were injected with 50 μg of SEB in PBS (anergic, heavy line) or with PBS alone (non-anergic; shaded histogram), euthanized 7 d later and their splenocytes evaluated by flow cytometry. Histograms depicting relative expression levels of CD18 are gated on CD4+Vβ8+ lymphocytes. The Y-axis scale at left refers to non-anergic cells, and the Y-axis scale at right refers to anergic cells.
Discussion
This is the first description of the global gene expression profile of T cells anergized by a bacterial superantigen. In the BALB/c SEB anergy system, clonal expansion, death and anergy occur as sequential stages of CD4+ T cell response (5), such that CD4+Vβ8+ T cells isolated 7 or more days post-injection are anergic. This model was validated in our laboratory in three ways. First, we observed the initial activation of nearly all CD4+Vβ8+ T cells, as evidenced by the expression of the very early activation marker CD69 on over 95% of cells within this population 6 hours post-injection. At 7 days post-injection, the proliferative response of CD4+ T cells isolated from SEB-injected mice to SEB in vitro was greatly reduced both in unsorted and sorted populations compared to mock-injected mice, while the proliferative response of the two populations of splenic T cells to the unrelated SEA superantigen were indistinguishable. The detection of only an approximately 6–7% difference in the fraction of CD4+Vβ8+ present in the cultures containing splenic T cells from either SEB- or mock-injected mice compared to a 300–800% difference (depending upon the concentration of SEB present in the cultures) in proliferative capacity between the two populations strongly suggests that clonal deletion was only a minor contributor to the tolerance observed. Residual proliferation of splenic T cells taken from SEB-exposed mice was likely contributed by the presence of CD8+Vβ8+ T cells in the cultures, which have been observed to retain their ability to proliferate to SEB following in vivo SEB exposure (40). Second, the anergic nature of the SEB-responsive CD4+ T cells in this model was further validated by measuring an 8-fold overexpression of GRAIL mRNA in sorted CD4+Vβ8+ T cells taken from SEB-injected mice compared to that sorted from mock-injected mice. GRAIL has been shown to be essential for anergy induction in the SEB model (22). Finally, the utility of the experimental approach used herein for successfully identifying anergy-associated gene expression is further supported by the detection of differential expression of at least three mRNAs, including Satb1, CD18 and Slap, that have been shown by others to be similarly modulated in another, non-superantigen based model of in vivo T cell anergy (34). Thus, under-expressed Satb1 and over-expressed CD18 and Slap mRNA may be universal features of the CD4+ T cell anergic program.
Expression of Slap in Jurkat T cells at physiologic levels was shown to impair TCR signaling, including Ca2+ flux, Ras activation and IL-2 secretion (41) and thus may have a similar impact in SEB-anergized cells. Reproducibly detected expression of Ptpn3/PTPH1 mRNA in the anergic but not non-anergic cells is of particular interest in light of recent work showing that this phosphatase is the predominant cellular protein tyrosine phosphatase capable of specifically and directly dephosphorylating TCR ζ-chains (42). Though the biological role of basal expression of the inositol-5’-phosphatase Inpp5d/SHIP-1 in T cells is still unknown, overexpression of SHIP-1 in anergic T cells could interfere with multiple signaling pathways through its ability to oppose the activity of Phosphatidylinositiol-3-kinase. Signaling cascades that might be influenced by upregulation of SHIP-1 include TCR signaling and LFA-1 dependent adhesion (43).
Expression of the MAP kinase kinase kinase B-raf mRNA was detected only in non-anergic T cells but not in anergic T cells. Interestingly, enforced expression of B-raf in CD4+ T cells of transgenic mice prevented in vitro induction of anergy in these cells by B7-deficient APC by ensuring ERK activation (44). Although these authors did not find appreciable expression of B-raf in unmanipulated primary T cells compared to that of the CD4 promoter-driven B-Raf transgene, Tsukamoto and colleagues demonstrated the presence of B-raf in both human and mouse primary T cells by immunoblotting and intracellular flow cytometry (45). In that study, TCR crosslinking of Jurkat T cells resulted in transient stimulation of Raf-1 activity and sustained stimulation of B-Raf activity. Moreover, expression of dominant negative B-Raf abrogated sustained ERK activation and resulted in reduced NFAT activity and reduced IL-2 production. Thus, down-modulation of B-Raf in SEB anergized T cells may compromise sustained ERK phosphorylation and hence contribute to impaired IL-2 production upon TCR stimulation. Although ERK phosphorylation in SEB-anergized T cells was reported to be normal at very early time points (11), abrogation of extended ERK signaling, which could be mediated by down-regulated B-Raf, has not been investigated in this model. Moreover, that study used SEB-immunized D011.10 TCR transgenic mice as a model system, an approach that has not been reproducible in our laboratory (Yaciuk and Farris, unpublished data). In summary, enhanced expression of Slap, Ptpn3/PTPH1 and possibly SHIP-1 along with reduced expression of B-Raf in SEB-anergized T cells are likely to contribute to the inability of the anergic cells to proliferate and secrete IL-2 upon TCR stimulation.
Cyclic AMP has been shown to induce T cell anergy in Th1 clones (46) and mediate CD4+ T cell anergy induced by HIV-1 envelope glycoprotein gp120 (31). Therefore, the observed overexpression of the seven transmembrane domain (7TM) G protein-coupled receptor adenosine receptor A2a, which upregulates intracellular cAMP upon signaling, in anergic T cells may initiate or reinforce T cell anergy. There is likely to be a surge in extracellular adenosine levels in secondary lymphoid organs during the death phase of the SEB response, thus triggering intracellular cAMP production. Enhanced cell surface expression of the receptor, which responds in the 100–1000 nM range of adenosine, could not only increase this response but might also permit signaling in response to basal adenosine levels which are estimated to be in the 30–300 nM range (47). Several mechanisms whereby cAMP can induce anergy are known and may be categorized based upon their dependency on protein kinase A (PKA). In a PKA-independent pathway, cAMP can directly activate a Rap1 guanine nucleotide exchange factor (GEF) known as Epac (Exchange Protein Activated by cAMP) (48), which in turn is able to inhibit IL-2 production by inhibiting cRaf-1 in the Ras signaling cascade (49). Because Rap1 activates B-Raf, however, suppression of the Ras pathway only occurs in cells that do not express B-Raf. Thus, down-regulation of B-Raf as observed in the present system might be required to prevent activation of IL-2 expression in the presence of active cAMP signaling. In a PKA-dependent pathway, it has been proposed that the inducible cAMP early repressor (ICER) may be synthesized in response to stimulation of unidentified 7TM G protein-coupled receptors leading to the induction and/or maintenance of T cell anergy by inhibition of IL-2 gene transcription (50). ICER is produced as an alternative transcript from the CREM gene but lacks the upstream transactivation domain of CREM and thus acts as a dominant negative inhibitor of CREB- or jun-mediated IL-2 transcription. A second PKA-dependent pathway of cAMP-mediated inhibition of T cell function involves PKA-dependent activation of the COOH-terminal Src kinase (Csk), which then inhibits Lck-mediated TCR ζ-chain phosphorylation and thus hampers proximal TCR signaling (51). Enhanced expression of the PKA transcription factor Foxd1 in the anergic T cells (Table 3) may cooperate with overexpression of the A2a receptor to amplify these latter two pathways.
Overexpression of CD18, the β-chain of LFA-1, in anergic cells was confirmed by quantitative RT-PCR and by protein expression at the cell surface. Also observed in the present study was downregulation of mRNA for at least one molecule that plays an important role in cytoskeletal reorganization that takes place downstream of integrin signaling. Ppp1r12a/Mypt1, is the major regulatory subunit of myosin phosphatase. Downstream of integrin signaling, Mypt1 is phosphorylated by RhoA-activated kinases resulting in inhibition of myosin phosphatase activity that allows cytoskeletal reorganization to take place. Downregulation of Mypt1 expression in anergic cells might thus lead to constitutive activation of the catalytic subunit of myosin phosphatase. In support of this, siRNA knockdown of Mypt1 in HeLa cells was recently shown to inhibit cell migration and adhesion (52). In addition, reduced expression of myosin light chain kinase (Mylk) was observed in anergic T cells by array but not by real-time quantitative PCR. Any reduction in this enzymatic activity would be predicted to contribute to defective cytoskeletal reorganization downstream of LFA-1 signaling. Not only would impaired LFA-1 signaling lead to defects in T cell migration, but such defects might also lead to impairment of signaling molecule reorganization at the T cell/APC synapse during T cell activation. Indeed, disorganized synapse formation was observed in T cells made anergic with ionomycin (23). A final expression difference between anergic and non-anergic T cells that falls into the signaling category and could act downstream of multiple signaling pathways including the integrin pathway, is that of a novel GEF termed Geft. Geft was detected only in non-anergic cells in the present study and was highly expressed in these cells at greater than 15 SD above background. Overexpression of Geft, which was shown to have in vitro specificity for Rac and Cdc42, in NIH 3T3 cells led to actin cytoskeletal reorganization, cell proliferation and migration (53). Thus, down-regulated Geft expression in anergic T cells could also conceivably contribute to defective cytoskeletal reorganization and cell migration.
Although coordinately regulated transcriptional pathways influencing the cell cycle, proliferation and death were not readily apparent from this screen, which examined only a fraction of the mouse genome, expression patterns of potential functional significance were noted. Firstly, differential mRNA expression of at least three genes encoding molecules that influence microtubule dynamics and cell cycle progression was observed and included Emerin, Kif23/MKLP-1 and Stathmin-like 3 (29, 54, 55). The first two of these were at least 4X downregulated in anergic T cells, and Stathmin-like 3 was detected only in anergic T cells at over 2 SD above background. These expression changes are consistent with inhibition of cell cycle progression at different stages. Secondly, the lack of detection of mRNA for the transcriptional repressor Enhancer of Zeste 2 (Ezh2) in the anergic cells is of potential functional significance since Ezh2 is essential for cellular proliferation (56) and since its expression was downregulated in replicatively senescent human fibroblasts (56). Ezh2 by itself does not possess histone methyltransferase activity that might be involved in its repressive activities but Ezh2 is required for such activity by a multiprotein complex that includes both Ezh2 and Rbbp4 (RbAp46/48) (57). Interestingly, Rbbp4 was also found reduced over 4X in the anergic cells. Thirdly, several expressions that would be expected to favor survival of the anergic cells were noted and included detection or enhanced expression of Birc1f/NAIP and ethanolamine kinase, along with failure to detect expression of two genes encoding the death-promoting factors Pcd8 and Lrdd. Overexpression of ethanolamine kinase in NIH 3T3 cells promoted cell survival (58).
Finally, these studies have revealed transcriptional modulation of genes encoding multiple members of the Notch and Wnt signaling pathways in CD4+ anergic T cells. Both of these pathways are of considerable importance in hematopoietic cell development (26, 27), but their roles in mature lymphocyte function are only beginning to be explored. In the present study messages for two regulators of Notch signaling, Notchless (Nle) and Lunatic Fringe (Lfng), were downregulated in the anergic cells (Table 3), while transcripts for the Notch ligand Jagged-1 (Table 5) and the Notch-specific transcription factor Hes-5 (Table 3) were overrepresented in the anergic cells. Taken together, these expression patterns suggest active Notch signaling in the anergic CD4+ T cells. Indeed, active Notch signaling was recently shown to induce increased levels of Jagged-1 ligand in NIH-3T3 cells (59). The results described herein constitute the first report associating Notch signaling with T cell anergy. Although the gene chips used in this study did not permit identification of expressed Notch receptors, Benson and colleagues recently observed that expression of constitutively active Notch-1 in primary human CD4+ T cells prevented cellular proliferation but failed to impair cytokine secretion (60). The expression patterns of several genes encoding important players in a second developmentally important pathway–the Wnt pathway–suggest a global decrease in β-catenin signaling in SEB-anergized T cells. These include upregulation of expression of nkd2 (Table 2) and downregulation of lef-1(Table 3). The former gene encodes an important negative regulator of this pathway, while the latter is one of the major canonical transcription factors of this pathway. In light of recent data demonstrating that mouse Fzd1 acts as an antagonist of the canonical Wnt/β-catenin pathway (61), it will be of interest to determine if Fzd8, which was overexpressed in anergic cells (Table 5), acts in a similar fashion. Upregulation of E-cadherin (Cdh1; Table 5) mRNA in anergic cells predicts further inhibition of this pathway as reported for epithelial and fibroblastoid cells (37). Ephrin B3, a receptor tyrosine kinase whose message was detected only in anergic cells (Table 5) might also be linked to the Wnt signaling pathway via its described ability to bind and phosphorylate a small receptor tyrosine kinase known as Ryk (62). Ryk has been shown to act as a Wnt receptor and to specifically bind the mouse Fzd8 cysteine-rich domain (63). How Ephrins might regulate the Wnt pathway is currently unknown.
In conclusion, analysis of differential gene expression patterns in anergic T cells has proved to be a powerful tool for uncovering the molecular mechanisms of T cell anergy. Although additional studies will be required to establish the functional significance of the gene expression changes identified in this study, multiple novel candidates and pathways that may play central roles in the induction or maintenance of the anergic state have been elucidated. Eventual annotation of the differentially expressed ESTs identified in this study will provide further insight into T cell anergy in the future and may be expected to bring to light the involvement of biologic pathways that remain as yet undiscovered. As additional “anergy factors” emerge, it may become apparent that T cell anergy is not controlled by a single gene or its product but by a balance of signaling events through numerous intermediates in multiple signaling pathways.
Materials and Methods
Immunization model for inducing T cell Anergy
6–8 wk Balb/c female mice (Taconic Farms, Germantown, NY) were immunized intraperitoneally either with 50 μg SEB (Sigma, St. Louis, MO) in 0.2 mL PBS for inducing anergy (5) or 0.2 mL PBS alone for obtaining non-anergic T cells. Mice were euthanized 7 d following immunization, and pooled single cell suspensions were prepared from spleens. The splenocytes were treated with a hypotonic tris ammonium chloride solution to remove RBCs, and T cells were enriched using nylon wool chromatography (64). T cell anergy was verified by standard tritiated-thymidine incorporation of cultures containing enriched T cells, gamma irradiated (2600 rad) whole splenocytes from unimmunized mice, and either SEB or SEA (Toxin Technology, Inc, Sarasota, FL) antigens essentially as described (5). All animal protocols were approved by the IACUC committee of the Oklahoma Medical Research Foundation. All personnel performing studies involving the use of SEB and SEA were approved under the CDC Select Agent Program.
Isolation of CD4+Vβ8+ anergic T cells and flow cytometry
CD4+Vβ8+ T cells were isolated from the splenocytes of mice 7 d following immunization with SEB (anergic) or PBS alone (non-anergic). Nylon wool non-adherent cells were labeled with anti-CD4-PE (GK1.5) and anti-Vβ8-biotin (F23.1), followed by streptavidin-APC. In some analytical experiments unseparated splenocytes were also labeled with anti-CD18-FITC (C71/16), anti-CD69-FITC (H1.2F3) or anti-CD25-FITC (7D4). All mAbs were purchased from BD Biosciences-Pharmingen (San Diego, CA) and were titrated to optimal concentrations before use. Cell suspensions were incubated in the dark on ice for 30 min during primary antibody staining and 15 min during staining with the secondary reagent. They were washed thrice after each staining with PBS supplemented with 0.09% NaN3 and 2% FCS. Highly purified populations of CD4+Vβ8+ were isolated by high speed cell sorting (MoFlo, Dakocytomation, Glostrup, Denmark). Sorting purity was always >93%. Sorted cells were collected into DMEM containing 10% FCS on ice, and sorted cell pellets were used for the isolation of RNA. Analytical data were collected with a FACSCalibur instrument (BD Biosciences) and analyzed using CellQuest software.
Isolation of RNA
RNA was isolated from cells using TRIzol (Invitrogen Life Technologies, City, ST) according to the manufacturer’s protocol with minor modifications. Briefly, the cells were homogenized at 1x106 cells/ml of TRIzol reagent, and then chloroform extraction was performed. After an isopropanol precipitation, the RNA was washed with 70% ethanol and further enriched using the Qiagen RNA Isolation Kit (Valencia, CA). RNA obtained from both anergic and non-anergic samples was analyzed for quantity and quality by spectrophotometry (A260/280 ratio), agarose gel electrophoresis, and capillary gel electrophoresis (Agilent Technologies 2100 Bioanalyzer using RNA nano-chips). RNA samples exhibiting high purity with no detected degradation of 18S or 28S rRNAs were used for gene chip hybridization and preparation of cDNA for real time PCR.
Preparation of cDNA and gene chip hybridization
Synthesis and labeling of cDNA was carried out using the MICROMAXTM TSATM Labeling and Detection Kit (PerkinElmer Life Sciences) according to the manufacturer’s instructions. The samples were hybridized using MICROMAX protocols to microarray chips containing 5,184 mouse cDNAs that were purchased from the microarray core facility at Columbia University (New York; supplied by Dr. Anthony Ferrante) using MICROMAX protocols. The arrays were scanned on an Affymetrix model 428 scanner. Background fluorescence (backgroundFL) was empirically determined from signals generated on a set of randomized negative controls. These probes consist of sequences that are not present in the mouse genome. Only signals that were at least three standard deviations above the thus established background were considered for analysis.
Background determination and normalization procedure
Normalization for differences among experiments was conducted using a procedure described in detail elsewhere (65, 66). The initial step assumes that signals corresponding to mRNAs that are not expressed in the sample (background defined by analysis; backgroundAN) will be normally distributed. Under this assumption, the mean and SD for non-expressed genes were calculated using an iterative nonlinear curve fitting procedure and defined as “backgroundAN”. “Expressed” genes were then defined as having levels outside of the normally distributed homogenous family of background values for that microarray. Data obtained after normalization of each expression profile to its own backgroundAN were subsequently log-transformed, and any negative values were substituted with minimally positive logarithmic numbers.
Normalized expression profiles of genes above backgroundAN were adjusted to each other using a robust regression analysis that has been previously described (65). This analysis is based on the assumption that the majority of genes are equally expressed in compared samples, and that values of each gene are normally distributed around a regression line with a small proportion of differentially expressed “outliers”. The contributions of outliers to the regression analysis were down-weighted in an iterative manner until the residuals were normally distributed as measured by deviations from the regression line calculated against the averaged profile.
The reference group was composed of genes expressed above backgroundAN with normal low variability of expression in control samples as determined by the F-test, and whose residuals approximated a normal distribution based on the Kolmogorov-Smirnov criterion.
Identification of differentially expressed genes
These analyses were performed as previously described (65, 66) and included the assignment of statistically different levels of expression using the Student’s T-test with the commonly accepted significance threshold of p<0.05. However, a significant proportion of genes identified as differentially expressed in this manner are expected to be false positive determinations at this threshold level due to the large number of genes present on microarrays. To address this issue, an Associative T-test was applied, in which the replicated residuals for each gene in the experimental group were compared with the entire set of residuals from the reference group (defined above). The hypothesis that gene expression in the experimental group, presented as replicated residuals (deviations from the averaged control group profile), is distributed similarly to the several thousand members of the normally distributed set of residuals for gene expressions in the reference group is tested in this analysis. The significance threshold was corrected to 1/(number of genes) to make the detection of false positives improbable. Only genes with p-values below the threshold of both the Student T-test and the Associative T-test were defined as differentially expressed. Expression values were then adjusted relative to the normally distributed background (having a mean equal to 0 and a standard deviation equal to 1). Genes expressed distinctively above background in one group and not in another were defined as uniquely expressed genes. A gene was considered to be differentially expressed between groups if there was at least a 1.2-fold difference in the means between the groups and a statistically significant difference with p<0.05 from the normally distributed backgroundAN.
Relative quantitation of gene expression by real-time PCR
Complementary DNA (cDNA) was synthesized from 2 μg total RNA purified as described above using Taqman reverse transcription reagents (Applied Biosystems, Foster City, CA). SYBR Green real time PCR was performed using an ABI Prism 7700 sequence detection system according to the manufacturer’s instructions (Applied Biosystems) with the aid of SYBR Green PCR Master Mix (Applied Biosystems) containing 10X SYBR Green PCR buffer, 3 mM MgCl2, 100mM each dATP, dCTP, dGTP, dUTP, 0.025 U/ml Amplitaq Gold DNA Polymerase, and 0.01 U/ml Amp ERAse UNG. Gene specific forward and reverse primers were designed with one primer spanning an exon-exon junction (to avoid amplification of any contaminating genomic DNA) using Primer Express software (Applied Biosystems) and were synthesized and purified by the Molecular Biology facility, at University of Oklahoma Health Sciences Center. Primers were used at 2 μM in reactions with 1 or 5 ng of cDNA, and β-actin was used as a normalization control. All primer sequences are presented in Table 6. The cycling conditions were: 2 min at 50°C, followed by 10 min at 95°C, followed by 55 cycles of 15 sec at 95°C and 1 min at 60°C. All data points were run in triplicate. Quantitation was done using the comparative CT method as described by Applied Biosystems. All primers were validated for equivalent efficiency to the ß-actin primers by using Universal mouse cDNA as template prior to calculating fold differences in expression between anergic and non-anergic cDNA’s. Dissociation curves of the same were also studied during primer validation and revealed single amplification products with the predicted Tm values for each primer set.
Table 6.
Primers used for real-time quantitative PCR
| Target | Primer | Sequence |
|---|---|---|
| SLP-76 | Forward
Reverse |
5′-TCTTGACCGAGAGCCCTTCAT-3′
5′-TCTTTCGTGTCTGTCCATGGC-3′ |
| SLAP | Forward
Reverse |
5′-GAATCTTCCGTCTTCCCAACAA-3′
5′-CACAGCATAGACCATCAGCCAC -3′ |
| SHIP-1 | Forward
Reverse |
5′-TCTGCAGAGGTTGTTTGACCA-3′
5′-CAATTGGCTGAGTTTGGCAA-3′ |
| CD18 | Forward
Reverse |
5′-GTTTCAGACAGAGGTCGGCAA-3′
5′-GCCAATTTCCTCCGGACAT -3′ |
| Jag1 | Forward
Reverse |
5′-CACTTATTGCTGCGGTTGCA -3′
5′AAGCAACAGACCCAAGCCACT-3′ |
| Epherin B3 | Forward
Reverse |
5′CTGCTGCTGTTAGGTTTTGCG -3′
5′CCGATCTGAGGATAAAGCACGT -3′ |
| Mylk | Forward
Reverse |
5′-AACATCTTTGGCACACCCGA-3′
5′-CGCTCAGGAGGATGTAGGTGAT -3′ |
| Myl6* | Forward
Reverse |
5′-AGGCACCTGCAAAATCCCA-3′
5′-CAGCTCAAAGGCCTCTCTGAAT-3′ |
| GRAIL | Forward
Reverse |
5-GCAGGAAGCAGAGGCAGTTAAA-3′
5′-GCACACAGCACAGCTATCTCCA -3′ |
| β-Actin | Forward
Reverse |
5′-CAACGAGCGGTTCCGATG -3′
5′-GCCACAGGATTCCATACCCA -3′ |
Myl6 (Myosin light chain polypeptide 6; Accession AI450945) is the product of a structural gene that was only expressed in non-anergic cells (1.4SD above background; Supplemental Table 2). All other designations are referred to in the text.
Supplementary Material
Acknowledgments
The authors are grateful to Karen Davis and members of the OMRF Flow Cytometry Core Facility for excellent technical support and to Drs. Craig Cadwell and Jaya Rajaiya for technical advice. The authors thank Drs. Mark Coggeshall and Linda Thompson for helpful discussions and Kathy Bryant for assistance with bioinformatics. Alison Galatian contributed to these studies as an OMRF Fleming Scholar. This work was supported by grants from the Oklahoma Center for the Advancement of Science and Technology (OCAST; HR02-048) and by the National Institutes of Health (AI48097, K02AI51647 and P20 RR016478-05).
References
- 1.Nakken B, Davis KE, Pan ZJ, Bachmann M, Farris AD. T-helper cell tolerance to ubiquitous nuclear antigens. Scand J Immunol. 2003;58:478. doi: 10.1046/j.1365-3083.2003.01323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sloan-Lancaster J, Evavold BD, Allen PM. Th2 cell clonal anergy as a consequence of partial activation. J Exp Med. 1994;180:1195. doi: 10.1084/jem.180.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mirshahidi S, Huang CT, Sadegh-Nasseri S. Anergy in peripheral memory CD4(+) T cells induced by low avidity engagement of T cell receptor. J Exp Med. 2001;194:719. doi: 10.1084/jem.194.6.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rellahan BL, Jones LA, Kruisbeek AM, Fry AM, Matis LA. In vivo induction of anergy in peripheral V beta 8+ T cells by staphylococcal enterotoxin B. J Exp Med. 1990;172:1091. doi: 10.1084/jem.172.4.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MacDonald HR, Baschieri S, Lees RK. Clonal expansion precedes anergy and death of Vβ8+ peripheral T cells responding to staphylococcal enterotoxin B in vivo. Eur J Immunol. 1991;21:1963. doi: 10.1002/eji.1830210827. [DOI] [PubMed] [Google Scholar]
- 6.Powell JD, Ragheb JA, Kitagawa-Sakakida S, Schwartz RH. Molecular regulation of interleukin-2 expression by CD28 co-stimulation and anergy. Immunol Rev. 1998;165:287. doi: 10.1111/j.1600-065x.1998.tb01246.x. [DOI] [PubMed] [Google Scholar]
- 7.Schwartz RH. T Cell Anergy. Annu Rev Immunol. 2003;21:305. doi: 10.1146/annurev.immunol.21.120601.141110. [DOI] [PubMed] [Google Scholar]
- 8.Seroogy CM, Fathman CG. T-cell anergy: from phenotype to genotype and back. Immunol Res. 2003;28:255. doi: 10.1385/IR:28:3:255. [DOI] [PubMed] [Google Scholar]
- 9.Migita K, Eguchi K, Kawabe Y, Tsukada T, Ichinose Y, Nagataki S, Ochi A. Defective TCR-mediated signaling in anergic T cells. J Immunol. 1995;155:5083. [PubMed] [Google Scholar]
- 10.Koide Y, Uchijima M, Yoshida A, Yoshida TO. Effect of staphylococcal enterotoxin B-induced anergy on cytokine gene expression: anergy-sensitive and resistant mRNA expression. J Interferon Cytokine Res. 1996;16:225. doi: 10.1089/jir.1996.16.225. [DOI] [PubMed] [Google Scholar]
- 11.Kimura M, Yamashita M, Kubo M, Iwashima M, Shimizu C, Tokoyoda K, Chiba J, Taniguchi M, Katsumata M, Nakayama T. Impaired Ca/calcineurin pathway in vivo anergized CD4 T cells. Int Immunol. 2000;12:817. doi: 10.1093/intimm/12.6.817. [DOI] [PubMed] [Google Scholar]
- 12.Heeg K, Gaus H, Griese D, Bendigs S, Miethke T, Wagner H. Superantigen-reactive T cells that display an anergic phenotype in vitro appear functional in vivo. Int Immunol. 1995;7:105. doi: 10.1093/intimm/7.1.105. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez-Garcia A, Marchetti P, Castedo M, Zamzami N, Tarazona R, Martinez AC, Kroemer G. Polyethylene glycol-modified IL-2 abrogates superantigen-induced anergy without affecting peripheral clonal deletion in vivo. Clin Immunol Immunopathol. 1996;78:215. doi: 10.1006/clin.1996.0032. [DOI] [PubMed] [Google Scholar]
- 14.Hamel ME, Eynon EE, Savelkoul HF, van Oudenaren A, Kruisbeek AM. Activation and re-activation potential of T cells responding to staphylococcal enterotoxin B. Int Immunol. 1995;7:1065. doi: 10.1093/intimm/7.7.1065. [DOI] [PubMed] [Google Scholar]
- 15.Baschieri S, Lees RK, Lussow AR, MacDonald HR. Clonal anergy to staphylococcal enterotoxin B in vivo: selective effects on T cell subsets and lymphokines. Eur J Immunol. 1993;23:2661. doi: 10.1002/eji.1830231041. [DOI] [PubMed] [Google Scholar]
- 16.Gaus H, Miethke T, Wagner H, Heeg K. Superantigen-induced anergy of V beta 8+ CD4+ T cells induces functional but non-proliferative T cells in vivo. Immunology. 1994;83:333. [PMC free article] [PubMed] [Google Scholar]
- 17.Lee WT, Thrush GR, Vitetta ES. Staphylococcal enterotoxin B induces the expression of activation markers on murine memory T cells in the absence of proliferation or lymphokine secretion. Cell Immunol. 1995;162:26. doi: 10.1006/cimm.1995.1047. [DOI] [PubMed] [Google Scholar]
- 18.Lee WT, Vitetta ES. Memory T cells are anergic to the superantigen staphylococcal enterotoxin B. J Exp Med. 1992;176:575. doi: 10.1084/jem.176.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuh K, Siminovitch KA, Ochi A. T cell anergy is programmed early after exposure to bacterial superantigen in vivo. Int Immunol. 1993;5:1375. doi: 10.1093/intimm/5.11.1375. [DOI] [PubMed] [Google Scholar]
- 20.Quill H, Schwartz RH. Stimulation of normal inducer T cell clones with antigen presented by purified Ia molecules in planar lipid membranes: specific induction of a long-lived state of proliferative nonresponsiveness. J Immunol. 1987;138:3704. [PubMed] [Google Scholar]
- 21.Telander DG, Malvey EN, Mueller DL. Evidence for repression of IL-2 gene activation in anergic T cells. J Immunol. 1999;162:1460. [PubMed] [Google Scholar]
- 22.Seroogy CM, Soares L, Ranheim EA, Su L, Holness C, Bloom D, Fathman CG. The gene related to anergy in lymphocytes, an E3 ubiquitin ligase, is necessary for anergy induction in CD4 T cells. J Immunol. 2004;173:79. doi: 10.4049/jimmunol.173.1.79. [DOI] [PubMed] [Google Scholar]
- 23.Heissmeyer V, Macian F, Im SH, Varma R, Feske S, Venuprasad K, Gu H, Liu YC, Dustin ML, Rao A. Calcineurin imposes T cell unresponsiveness through targeted proteolysis of signaling proteins. Nat Immunol. 2004;5:255. doi: 10.1038/ni1047. [DOI] [PubMed] [Google Scholar]
- 24.Verdoodt B, Blazek T, Rauch P, Schuler G, Steinkasserer A, Lutz MB, Funk JO. The cyclin-dependent kinase inhibitors p27Kip1 and p21Cip1 are not essential in T cell anergy. Eur J Immunol. 2003;33:3154. doi: 10.1002/eji.200323960. [DOI] [PubMed] [Google Scholar]
- 25.Inobe M, Schwartz RH. CTLA-4 engagement acts as a brake on CD4+ T cell proliferation and cytokine production but is not required for tuning T cell reactivity in adaptive tolerance. J Immunol. 2004;173:7239. doi: 10.4049/jimmunol.173.12.7239. [DOI] [PubMed] [Google Scholar]
- 26.Radtke F, Wilson A, Mancini SJ, MacDonald HR. Notch regulation of lymphocyte development and function. Nat Immunol. 2004;5:247. doi: 10.1038/ni1045. [DOI] [PubMed] [Google Scholar]
- 27.Staal FJ, Clevers HC. WNT signalling and haematopoiesis: a WNT-WNT situation. Nat Rev Immunol. 2005;5:21. doi: 10.1038/nri1529. [DOI] [PubMed] [Google Scholar]
- 28.le Gouvello S, Manceau V, Sobel A. Serine 16 of stathmin as a cytosolic target for Ca2+/calmodulin-dependent kinase II after CD2 triggering of human T lymphocytes. J Immunol. 1998;161:1113. [PubMed] [Google Scholar]
- 29.Rubin CI, Atweh GF. The role of stathmin in the regulation of the cell cycle. J Cell Biochem. 2004;93:242. doi: 10.1002/jcb.20187. [DOI] [PubMed] [Google Scholar]
- 30.Dahle MK, Gronning LM, Cederberg A, Blomhoff HK, Miura N, Enerback S, Tasken KA, Tasken K. Mechanisms of FOXC2- and FOXD1-mediated regulation of the RI alpha subunit of cAMP-dependent protein kinase include release of transcriptional repression and activation by protein kinase B alpha and cAMP. J Biol Chem. 2002;277:22902. doi: 10.1074/jbc.M200131200. [DOI] [PubMed] [Google Scholar]
- 31.Masci AM, Galgani M, Cassano S, De Simone S, Gallo A, De Rosa V, Zappacosta S, Racioppi L. HIV-1 gp120 induces anergy in naive T lymphocytes through CD4-independent protein kinase-A-mediated signaling. J Leukoc Biol. 2003;74:1117. doi: 10.1189/jlb.0503239. [DOI] [PubMed] [Google Scholar]
- 32.Attinger A, Acha-Orbea H, MacDonald HR. Cutting edge: cell autonomous rather than environmental factors control bacterial superantigen-induced T cell anergy in vivo. J Immunol. 2000;165:1171. doi: 10.4049/jimmunol.165.3.1171. [DOI] [PubMed] [Google Scholar]
- 33.Yasui D, Miyano M, Cai S, Varga-Weisz P, Kohwi-Shigematsu T. SATB1 targets chromatin remodelling to regulate genes over long distances. Nature. 2002;419:641. doi: 10.1038/nature01084. [DOI] [PubMed] [Google Scholar]
- 34.Lechner O, Lauber J, Franzke A, Sarukhan A, von Boehmer H, Buer J. Fingerprints of anergic T cells. Curr Biol. 2001;11:587. doi: 10.1016/s0960-9822(01)00160-9. [DOI] [PubMed] [Google Scholar]
- 35.Mourtada-Maarabouni M, Kirkham L, Farzaneh F, Williams GT. Regulation of apoptosis by fau revealed by functional expression cloning and antisense expression. Oncogene. 2004;23:9419. doi: 10.1038/sj.onc.1208048. [DOI] [PubMed] [Google Scholar]
- 36.Glynne R, Ghandour G, Rayner J, Mack DH, Goodnow CC. B-lymphocyte quiescence, tolerance and activation as viewed by global gene expression profiling on microarrays. Immunol Rev. 2000;176:216. doi: 10.1034/j.1600-065x.2000.00614.x. [DOI] [PubMed] [Google Scholar]
- 37.Stockinger A, Eger A, Wolf J, Beug H, Foisner R. E-cadherin regulates cell growth by modulating proliferation-dependent beta-catenin transcriptional activity. J Cell Biol. 2001;154:1185. doi: 10.1083/jcb.200104036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y, Zhu K, Hong G, Wu W, Baudhuin LM, Xiao Y, Damron DS. Sphingosylphosphorylcholine is a ligand for ovarian cancer G-protein-coupled receptor 1. Nat Cell Biol. 2000;2:261. doi: 10.1038/35010529. [DOI] [PubMed] [Google Scholar]
- 39.Jeon MS, Atfield A, Venuprasad K, Krawczyk C, Sarao R, Elly C, Yang C, Arya S, Bachmaier K, Su L, Bouchard D, Jones R, Gronski M, Ohashi P, Wada T, Bloom D, Fathman CG, Liu YC, Penninger JM. Essential role of the E3 ubiquitin ligase Cbl-b in T cell anergy induction. Immunity. 2004;21:167. doi: 10.1016/j.immuni.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 40.Sabapathy TK, Hwang LA, Hui KM. Differential effect of staphylococcal enterotoxin B upon the induction of tolerance on peripheral CD4+V beta 8+ and CD8+V beta 8+ T cells. Cell Immunol. 1994;158:83. doi: 10.1006/cimm.1994.1258. [DOI] [PubMed] [Google Scholar]
- 41.Sosinowski T, Killeen N, Weiss A. The Src-like adaptor protein downregulates the T cell receptor on CD4+CD8+ thymocytes and regulates positive selection. Immunity. 2001;15:457. doi: 10.1016/s1074-7613(01)00195-9. [DOI] [PubMed] [Google Scholar]
- 42.Sozio MS, Mathis MA, Young JA, Walchli S, Pitcher LA, Wrage PC, Bartok B, Campbell A, Watts JD, Aebersold R, Van Huijsduijnen RH, van Oers NS. PTPH1 is a predominant protein-tyrosine phosphatase capable of interacting with and dephosphorylating the T cell receptor zeta subunit. J Biol Chem. 2004;279:7760. doi: 10.1074/jbc.M309994200. [DOI] [PubMed] [Google Scholar]
- 43.Mazerolles F, Barbat C, Trucy M, Kolanus W, Fischer A. Molecular events associated with CD4-mediated Down-regulation of LFA-1-dependent adhesion. J Biol Chem. 2002;277:1276. doi: 10.1074/jbc.M110064200. [DOI] [PubMed] [Google Scholar]
- 44.Dillon TJ, Karpitski V, Wetzel SA, Parker DC, Shaw AS, Stork PJ. Ectopic B-Raf expression enhances extracellular signal-regulated kinase (ERK) signaling in T cells and prevents antigen-presenting cell-induced anergy. J Biol Chem. 2003;278:35940. doi: 10.1074/jbc.M301506200. [DOI] [PubMed] [Google Scholar]
- 45.Tsukamoto H, Irie A, Nishimura Y. B-Raf contributes to sustained extracellular signal-regulated kinase activation associated with interleukin-2 production stimulated through the T cell receptor. J Biol Chem. 2004;279:48457. doi: 10.1074/jbc.M403087200. [DOI] [PubMed] [Google Scholar]
- 46.Cone RE, Cochrane R, Lingenheld EG, Clark RB. Elevation of intracellular cyclic AMP induces an anergic-like state in Th1 clones. Cell Immunol. 1996;173:246. doi: 10.1006/cimm.1996.0274. [DOI] [PubMed] [Google Scholar]
- 47.Ballarin M, Fredholm BB, Ambrosio S, Mahy N. Extracellular levels of adenosine and its metabolites in the striatum of awake rats: inhibition of uptake and metabolism. Acta Physiol Scand. 1991;142:97. doi: 10.1111/j.1748-1716.1991.tb09133.x. [DOI] [PubMed] [Google Scholar]
- 48.de Rooij J, Zwartkruis FJ, Verheijen MH, Cool RH, Nijman SM, Wittinghofer A, Bos JL. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396:474. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 49.Boussiotis VA, Freeman GJ, Berezovskaya A, Barber DL, Nadler LM. Maintenance of human T cell anergy: blocking of IL-2 gene transcription by activated Rap1. Science. 1997;278:124. doi: 10.1126/science.278.5335.124. [DOI] [PubMed] [Google Scholar]
- 50.Bodor J, Bodorova J, Gress RE. Suppression of T cell function: a potential role for transcriptional repressor ICER. J Leukoc Biol. 2000;67:774. doi: 10.1002/jlb.67.6.774. [DOI] [PubMed] [Google Scholar]
- 51.Vang T, Torgersen KM, Sundvold V, Saxena M, Levy FO, Skalhegg BS, Hansson V, Mustelin T, Tasken K. Activation of the COOH-terminal Src kinase (Csk) by cAMP-dependent protein kinase inhibits signaling through the T cell receptor. J Exp Med. 2001;193:497. doi: 10.1084/jem.193.4.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xia D, Stull JT, Kamm KE. Myosin phosphatase targeting subunit 1 affects cell migration by regulating myosin phosphorylation and actin assembly. Exp Cell Res. 2005;304:506. doi: 10.1016/j.yexcr.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 53.Guo X, Stafford LJ, Bryan B, Xia C, Ma W, Wu X, Liu D, Songyang Z, Liu M. A Rac/Cdc42-specific exchange factor, GEFT, induces cell proliferation, transformation, and migration. J Biol Chem. 2003;278:13207. doi: 10.1074/jbc.M208896200. [DOI] [PubMed] [Google Scholar]
- 54.Liu J, Lee KK, Segura-Totten M, Neufeld E, Wilson KL, Gruenbaum Y. MAN1 and emerin have overlapping function(s) essential for chromosome segregation and cell division in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2003;100:4598. doi: 10.1073/pnas.0730821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bowerman B. Cell division: timing the machine. Nature. 2004;430:840. doi: 10.1038/430840a. [DOI] [PubMed] [Google Scholar]
- 56.Tang X, Milyavsky M, Shats I, Erez N, Goldfinger N, Rotter V. Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene. 2004;23:5759. doi: 10.1038/sj.onc.1207706. [DOI] [PubMed] [Google Scholar]
- 57.Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Malewicz B, Mukherjee JJ, Crilly KS, Baumann WJ, Kiss Z. Phosphorylation of ethanolamine, methylethanolamine, and dimethylethanolamine by overexpressed ethanolamine kinase in NIH 3T3 cells decreases the co-mitogenic effects of ethanolamines and promotes cell survival. Eur J Biochem. 1998;253:10. doi: 10.1046/j.1432-1327.1998.2530010.x. [DOI] [PubMed] [Google Scholar]
- 59.Ross DA, Kadesch T. Consequences of Notch-mediated induction of Jagged1. Exp Cell Res. 2004;296:173. doi: 10.1016/j.yexcr.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 60.Benson RA, Adamson K, Corsin-Jimenez M, Marley JV, Wahl KA, Lamb JR, Howie SE. Notch1 co-localizes with CD4 on activated T cells and Notch signaling is required for IL-10 production. Eur J Immunol. 2005;35:859. doi: 10.1002/eji.200425562. [DOI] [PubMed] [Google Scholar]
- 61.Roman-Roman S, Shi DL, Stiot V, Hay E, Vayssiere B, Garcia T, Baron R, Rawadi G. Murine Frizzled-1 behaves as an antagonist of the canonical Wnt/beta-catenin signaling. J Biol Chem. 2004;279:5725. doi: 10.1074/jbc.M309233200. [DOI] [PubMed] [Google Scholar]
- 62.Halford MM, Armes J, Buchert M, Meskenaite V, Grail D, Hibbs ML, Wilks AF, Farlie PG, Newgreen DF, Hovens CM, Stacker SA. Ryk-deficient mice exhibit craniofacial defects associated with perturbed Eph receptor crosstalk. Nat Genet. 2000;25:414. doi: 10.1038/78099. [DOI] [PubMed] [Google Scholar]
- 63.Lu W, Yamamoto V, Ortega B, Baltimore D. Mammalian Ryk is a Wnt coreceptor required for stimulation of neurite outgrowth. Cell. 2004;119:97. doi: 10.1016/j.cell.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 64.Julius MH, Simpson E, Herzenberg LA. A rapid method for the isolation of functional thymus-derived murine lymphocytes. Eur J Immunol. 1973;3:645. doi: 10.1002/eji.1830031011. [DOI] [PubMed] [Google Scholar]
- 65.Dozmorov I, Centola M. An associative analysis of gene expression array data. Bioinformatics. 2003;19:204. doi: 10.1093/bioinformatics/19.2.204. [DOI] [PubMed] [Google Scholar]
- 66.Knowlton N, Dozmorov IM, Centola M. Microarray data analysis toolbox (MDAT): for normalization, adjustment and analysis of gene expression data. Bioinformatics. 2004 doi: 10.1093/bioinformatics/bth424. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


