Abstract
RNA-enveloped viruses bud from infected cells by exploiting the multivesicular body (MVB) pathway. In this context, ubiquitination of structural viral proteins and their direct interaction with cellular factors involved in the MVB biogenesis through short proline rich regions, named late domains (L-domains), are crucial mechanisms. Here we report that, in contrast with the human immunodeficiency virus (HIV), the feline immunodeficiency virus (FIV), a non-primate lentivirus, is strictly dependent for its budding on a “PSAP”-type L-domain, mapping in the carboxy-terminal region of Gag, irrespective of a functional viral protease. Moreover, we provide evidence that FIV egress is related to Gag ubiquitination, that is linked to the presence of an active L-domain. Finally, although FIV Gag does not contain a PPxY motif, we show that the Nedd4-2s ubiquitin ligase enhances FIV Gag ubiquitination and it is capable to rescue viral mutants lacking a functional L-domain. In conclusion, our data bring to light peculiar aspects of FIV egress, but we also demonstrate that a non-primate lentivirus shares with HIV-1 a novel mechanism of connection to the cellular budding machinery.
Keywords: FIV, ubiquitin, Nedd4-2s, MVBs
Introduction
Feline immunodeficiency virus (FIV), a non primate lentivirus, causes an immunodeficiency syndrome in domestic cats that is striking similar to AIDS in humans (Pedersen et al., 1989; Bendinelli et al., 1995). FIV genome is simpler than that of primate lentiviruses, as the immunodeficiency virus (HIV), being characterised by three (rev, vif, orf2) rather than six accessory genes (Elder and Phillips, 1993; Poeschla et al., 1998). However, FIV is also similar to HIV in many molecular and biochemical properties, thus representing an attractive model for AIDS research (Elder et al., 1998). Indeed, FIV and its natural host, have already provided important insights into different aspects of lentiviral pathogenesis (Burkhard and Dean, 2003; Power et al., 2004). In addition, FIV has been successfully employed for studying viral resistance to chemotherapy and genetic variations that leads to drug resistance and to host immune response evasion (Dias et al., 2006). Indeed, the use of animal model is an essential step in the development of antiviral drugs and combination therapies. Tacking into account that even the best animal model is not completely predictive of human responses, studies carried on employing the FIV system could contribute information about pharmakokinetic and pharmakodynamic of new anti-HIV drugs, and about their potential toxicity as well as antiviral activity and could help to optimize clinical protocol. In addition, the FIV model could provide sound conceptual grounds on which to design candidate anti-HIV vaccines, allowing to study a virus that closely resembles HIV in its natural host (Dunham, 2006; Uhl et al., 2008). Finally, since FIV shares with HIV-1 the ability to infect dividing and non-dividing human cells, FIV-based vectors could be the basis for the development of safe systems aimed to target specific cells in different organs and tissues (Saenz and Poeschla, 2004).
In order to fully take advantage of these important potentialities and considering that our present understanding of FIV lags behind knowledge accumulated on HIV, different aspects of FIV biology need to be further investigated. Under this respect, due to its impact in the development of new therapeutic and vaccination approaches, as well as in the design and optimization of gene-transfer vectors, a deep dissection of the major structural protein of FIV, Gag, could be extremely useful. As for all the other retroviruses, FIV Gag can assemble and bud from cells in the absence of any other viral factors (Gottlinger, 2001). All the conserved Gag domains, matrix, capsid and nucleocapsid, are involved in driving the different steps of viral particle maturation and egress. Moreover, studies carried on by mutagenesis of the carboxy-terminal region of HIV-1 Gag, p6, have led to the identification of short proline rich motifs named late domains (L-domains), in that their functional knock out results in viral assembly arrest at late stages (Gottlinger et al., 1991). Early reports on HIV-1 had demonstrated that mutational inactivation of the viral protease could reverse defects in the L-domain region, suggesting a functional linkage between p6 and the proteolytic processing of the Gag precursor protein, during the budding of progeny virions (Huang et al., 1995). Up to date, three different classes of L-domains have been well characterised (PT/SAP, PPxY or YPxL), and it has been established that these motifs are not an unique feature of retroviruses, but are present in both positive- and negative-strand RNA enveloped-viruses (Demirov and Freed, 2004). Moreover, it has been demonstrated that L-domains are docking sites for different cellular proteins that are essential in the biogenesis of a cellular organelle, the multivesicular body (MVB). The MVB is involved in the degradation of post-Golgi integral membrane proteins in all eukaryotic cell. During the biogenesis of this organelle a process takes place that is topologically identical to the budding of viruses away from the cytosol (Piper and Katzamann, 2007). Thus, structural viral proteins, carrying L-domains, hijack the MVB biogenesis machinery to execute their exit from infected cells. Interestingly, due to their nature of docking site for cellular factors, L-domains constitute autonomous modules that are not dependent on a particular position within Gag and that are transferable between unrelated viruses (Parent et al., 1995). Moreover, Gag is ubiquitinated and the level of ubiquitination depends on the L-domain that is present, suggesting a role for ubiquitin in retroviral release (Martin-Serrano et al., 2007). While ubiquitination of PPxY containing Gag is clearly linked to the ability of such a domain to specifically interact with ubiquitin ligases (Harty et al., 2000; Martin-Serrano et al., 2005), HIV-1 and other lentiviruses, which do not present such an amino acid motif within their Gag, are not expected to exploit ubiquitin ligases for their budding. However, two recent reports have linked HIV-1 egress to Nedd4-2s, a member of the Nedd4 ubiquitin ligase family (Chung et al., 2008; Usami et al., 2008), suggesting that either i) Gag ubiquitination may facilitate its interaction with ubiquitin-binding proteins of the MVB pathway, or that ii) MVB factor ubiquitination may influence their ability to recruit Gag. In any case, the involvement of Nedd4-2s represents a novel mechanism by which primate lentiviruses can be connected to the cellular budding machinery (Chung et al., 2008; Usami et al., 2008).
FIV Gag is characterised by extensive variability in the carboxy-terminal peptide (p2), which correspond to HIV-1 p6. However, a PSAP L-domain within this region appears to be highly conserved (Manrique et al., 2004). It has been recently reported that mutagenesis of this motif inhibits FIV release in the context of an active viral protease (Luttge et al., 2008). In addition, the involvement of the MVB biogenesis pathway in FIV assembly and egress has been demonstrated (Luttge et al., 2008).
In the present study we report a further dissection of the L-domain region of FIV Gag, in particular in the context of the lack of an active protease, pointing out some peculiar features of FIV budding that should be taken into account when this lentivirus is employed as a model for studying HIV-1 pathogenesis and therapy. Moreover, we demostrate that a non-primate lentivirus respond to Nedd4-2s ubiquitin ligase, sharing with HIV-1 a novel mechanism of connection to the MVB pathway.
Materials and Methods
Cell lines
Human (293T, ATCC® Number: CRL-11268™) and feline (CrFK, ATCC® Number: CCL-94™) kidney cells were grown in Dulbecco's modified Eagle's medium (DMEM) with addition of 10 % heat inactivated fetal calf serum (complete medium).
Viral constructs
pΔenv1, plasmid kindly provided by Prof. Mauro Pistello (University of Pisa, Italy), contains Gag/Pol and Rev encoding sequences of the feline immunodeficiency virus (FIV), p34TF10 strain (NC_001482), (Pistello et al., 2007). Starting from this construct, a series of mutant in the Gag carboxy-terminal region were generated as following. A portion of the Gag-Pol encoding sequence, from nucleotide 1183 to nucleotide 2542, considering nucleotide 1 the first of the Gag coding sequence, was amplified by PCR and cloned in the pCR®2.1-Topo® plasmid (Invitrogen). Mutations of the L-domain region was performed by employing the QuickChange® II Site-directed mutagenesis kit (Stratagene). The fragments carrying the desired L-domain mutations were back-inserted into the pΔenv1 plasmid, replacing the wild-type (wt) counterpart.
Mammalian expression vectors
The pBJ5-Vps4 plasmid encodes the human Vps4-A protein with a carboxy-terminal FLAG epitope, while the pBJ5-Vps4E228Q construct encodes FLAG-tagged version of Vps4-A characterised by the E228Q mutation. The pBJ5-HA-Ub construct expresses a hemagglutinin (HA)-tagged version of wt ubiquitin. The pBJ5-Nedd4-2 plasmid contains the entire coding sequence for the “ancestral” isoform II of Nedd4-2 with an intact N-terminal C2 domain preceded by a FLAG tag. The pBJ5-Nedd4-2s construct contains the coding sequence for the native Nedd4-2s (residues 122–955 of Nedd4-2 isoform II) with a N-terminal FLAG tag, while the pBJ5-Nedd4-2s C801S construct encodes a FLAG-tagged Nedd4-2s version characterised by the mutation of the active site cysteine (801) in the HETC domain to serine. The empty vectors pBJ5 was employed in place of the correspondent expression plasmids when required. All the above constructs have been previously described (Strack et al. 2002, Strack et al. 2003, Usami et al., 2008).
Analysis of Gag processing and virus-like particle (VLP) release
CrFK cells (1.5x 106) or 293T cells (1.5x 106) were seeded into 25-cm2 tissue culture flasks. Twenty four h later the cells were transfected by calcium-phosphate method with either 8 μg (CrFK cells) or 1.25 μg (293T cells) of the appropriate viral construct. In co-transfection experiments, along with the viral construct, different mammalian expression vectors were employed in the following amount: pBJ5-Vps4, pBJ5-Vps4E228Q, pBJ5-HA-Ub 2 μg; pBJ5-Nedd4-2, pBJ5-Nedd4-2s, pBJ5-Nedd4-2s C801S 1 μg. In all the conditions, the total amount of transfected DNA was brought to either 10 μg (CrFK cells) or to 3.25 μg (293T cells) with carrier DNA (pBluescriptKS+, Stratagene). pBluescriptKS+ was also employed, when required, as transfection negative control. Twenty four h post-transfection, the culture supernatants were collected and the cells were lysed in radioimmunoprecipitation assay (RIPA) buffer [140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1 % Nonidet P-40, 0.5 % sodium deoxycholate, 0.05 % sodium dodecyl sulfate (SDS)]. Supernatants were clarified by low-speed centrifugation and passaged through 0.45-μm-pore-size filters. Released VLPs were spun through 20 % sucrose cushions for 2 h at 4 °C and 27,000 rpm in a Beckman SW41 rotor. Pelleted VLPs were lysed in RIPA buffer, and viral proteins were analysed by SDS-polyacrylamide gel electrophoresis (PAGE), followed by western blotting. Cell lysates were subjected to the same procedure in order to examine the intracellular Gag expression levels.
Western blotting analysis
For immunoblot analysis, aliquots of lysed VLPs and of the relative cell lysates were resolved by SDS-PAGE and electroblotted onto a Hybond-C Extra membrane (Amersham Pharmacia). The membranes were incubated with the appropriate antibody, namely: a monoclonal anti-FIV capsid antiserum (anti FIV-p24 Gag, Serotec) or a mouse monoclonal anti-HA antibody (Covance), followed by a peroxidase-conjugated anti-mouse IgG antibody (GE Healthcare). The blots were developed with enhanced chemiluminescence reagents (Amersham Pharmacia), as described (Strack et al., 2000).
Results
FIV requires the PSAP motif for budding from transfected cells, independently from the functionality of the viral protease
It has been recently reported by Luttge and coworkers that FIV release and replication requires an intact PSAP motif, present in the carboxy-terminal region of the structural viral Gag protein (Luttge et al., 2008). It is well known that Gag is the only viral protein required for assembly and budding of virus-like particles (VLPs) (Gottlinger, 1991) and that, in the case of HIV-1, a functional linkage between p6 and the proteolytic processing of the Gag precursor protein exists during release of progeny virions (Huang et al., 1995). In particular, in different cell types such as 293T cells and HeLa cells, HIV-1 constructs lacking a functional L-domain and defective in the protease were not impaired in viral release when compared to the wild type (Huang et al., 1995; Martin Serrano et al., 2004).
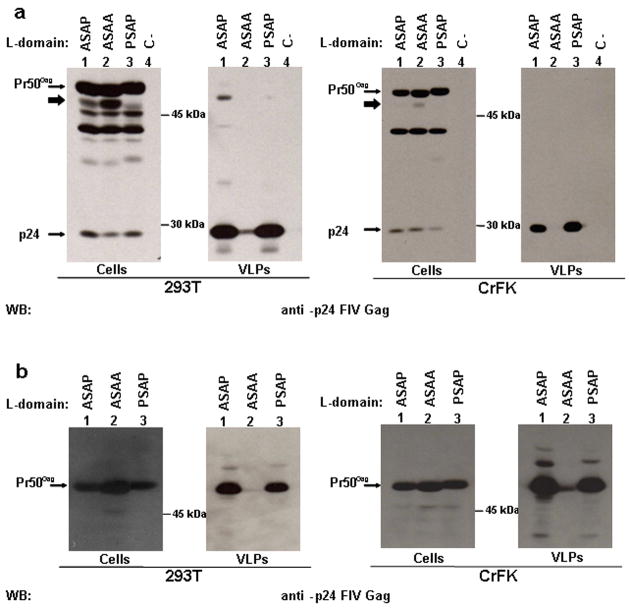
In order to contribute to the characterisation of the carboxy-terminal region of FIV Gag and to analyse the effect of Gag processing on particle release, starting from a plasmid encoding the FIV Gag/Pol region (pΔenv1, herein indicated as WT), we obtained, by site specific mutagenesis, four different constructs, characterised by the substitution of one or both prolines of the PSAP motif with alanines, in combination or not with the introduction of a STOP codon in place of aminoacid 10 of the protease (Table 1). The recombinant DNAs were transfected in CrFK cells and the released particles were purified and analysed by western blotting. Moreover we employed 293T cells as a control, since it is known that this cell line can be used to produce FIV particles upon transfection (Pistello et al., 2007) and that HIV-1 constructs lacking a functional L-domain and viral protease are fully competent for particle release in this cellular context (Martin Serrano et al., 2004). Our data confirm that the PSAP motif is essential for particle egress and that the substitution of both prolines with alanines significantly inactivates the L-domain function, independently from the cell line employed (Fig. 1a). Interestingly, in contrast with HIV-1, the mutation of the first proline is not sufficient to impair particle release (Fig. 1a). Moreover, the PSAP inactivation has a strong impact on virus like particles egress also in the absence of viral protease (Fig. 1b).
Table 1.
Schematic representation of the mutagenesis steps of the FIV L-domain a
| 438 | 439 | 440 | 441 | 442 | 443 | 444 | 445 | ||
|---|---|---|---|---|---|---|---|---|---|
| WT | P | S | A | P | P | M | E | E | |
|
|
|||||||||
| WT-Stop | P | S | A | P | P | M | E | E | + STOP |
|
|
|||||||||
| ASAP | A | S | A | P | P | M | E | E | |
|
|
|||||||||
| ASAA | A | S | A | A | P | M | E | E | |
The FIV wt L-domain (PSAP) was mutated starting from the WT construct, obtaining the ASAP construct, where Pro438 is replaced by Ala and the ASAA construct, where both Pro438 and Pro441 are replaced by Ala. The same mutations were introduced in the WT-Stop construct, which is characterised by the introduction of a STOP codon in place of aminoacid 10 of viral protease, obtaining ASAP-Stop and ASAA-Stop respectively.
Fig. 1.
PSAP motif is essential for FIV particle egress, independently from an active viral protease. 293T (left panel) or CrFK (right panel) cells were transfected with different plasmids encoding FIV-Gag/Pol either wt (a) or characterised by a STOP codon in the Pol reading frame (b) and carrying the wt PSAP motif or a mutated L-domain, as indicated. Twenty-four h later, cell lysates (Cells) and virus like particles (VLPs) were analysed by western blotting; C-: transfection negative control. The bold arrows point to a partially processed form of Gag, most likely the MA-CA-p1-NC precursor.
It has been recently shown that FIV Gag exploits the MVB biogenesis machinery to egress from infected cells (Luttge et al., 2008). In order to support our observation that the PSAP function is essential also in the context of unprocessed Gag, we blocked the MVB pathway by the over-expression of a dominant negative form of the cellular AAA-ATPase Vps4 (Vps4E228Q, Strack et al., 2003). In agreement with our hypothesis, under this condition, a significant reduction of VLP release was observed also in the absence of viral protease (Fig. 2, lane 1-3). Thus it can be concluded that, in the case of FIV, the MVB machinery is required for budding, regardless of viral protease.
Fig. 2.
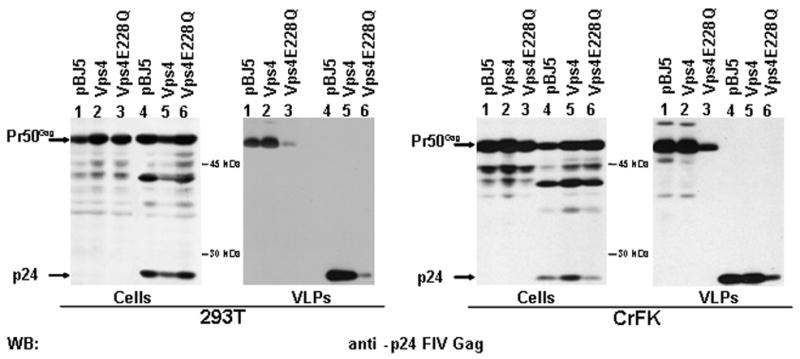
MVB functionality is essential for FIV budding from transfected cells, also in the absence of a viral protease. The MVB biogenesis was blocked by over-expressing a dominant negative form of the AAA-ATPase Vps4 (Vps4E228Q). The cells were co-transfected either with WT-Stop or WT construct in both 293T (left panel) or CrFK (right panel) cells. Twenty-four h later, cell lysates (Cells) and purified virus like particles (VLPs) were analysed by western blotting.
Ebola L-domain is working also in the context of full length FIV Gag, but does not lead to a significant enhancement in particle production
FIV has promising potentialities for the development of safe and efficient gene transfer vectors and for the generation of VLPs to be tested in the animal model for vaccination purposes (Bendinelli et al., 1995). One of the limiting steps in lentiviral vector and VLP production is the amount of viral structural proteins produced by the systems currently used (Lesch et al., 2008). Since our data suggested that the L-domain region is crucial for FIV Gag release, even in the absence of an active protease, we wanted to analyse whether it could be possible to increase the amount of processed and unprocessed VLPs released in the culture supernatants by manipulating this region. Thus, we sequentially mutagenised the sequence spanning from aminoacid 438 to aminoacid 444 of Gag protein in the WT background, in order to obtain constructs where the natural FIV L-domain was transformed in the PTAPPEY motif, characteristic of the Ebola virus (WT-Eb). Indeed, in specific context, like in HIV-1 based minimal Gag constructs engineered to contain this motif (Strack et al., 2002), the PTAPPEY domain is particularly efficient in mediating particle release. It has been previously reported that the substitution of the two glutamic acids in position 444 and 445, without affecting the overlapping Pol reading frame, led to the release of un-processed VLPs (Manrique et al., 2004). In order to obtain the PTAPPEY motif we needed to replace E444 with a Y. Thus, we firstly investigated the impact of the single E444 to A mutation on Gag processing and VLP release (MUT1). We were able to show that transfection of MUT1 in both 293T and CrFK cells led to the release of mature VLPs (Fig. 3, lane 2). Thus the single E444 to A mutation does not impair the ability of FIV Gag-Pol to produce mature viral particles. The same phenotype result was obtained when the A444 was exchanged in Y (MUT2, Fig. 3, lane 3) and when a T was inserted in place of S439 (MUT3, Fig. 3, lane 4). Finally, by combining the latest two mutations a construct bearing the L-domain characteristic Ebola virus (WT-Eb) was obtained. All the constructs are reported in Table 2. When the amount of particles produced by MUT1, MUT2, MUT3 and WT-Eb constructs were quantified by densitometry and normalized for the intracellular Gag content, we did not notice a significant increase in particle production with respect to WT FIV (data not shown). Thus, our data seem to indicate that, even though the Ebola L-domain is working also in the context of full length FIV Gag, it does not lead to a significant enhancement in particle production.
Fig. 3.
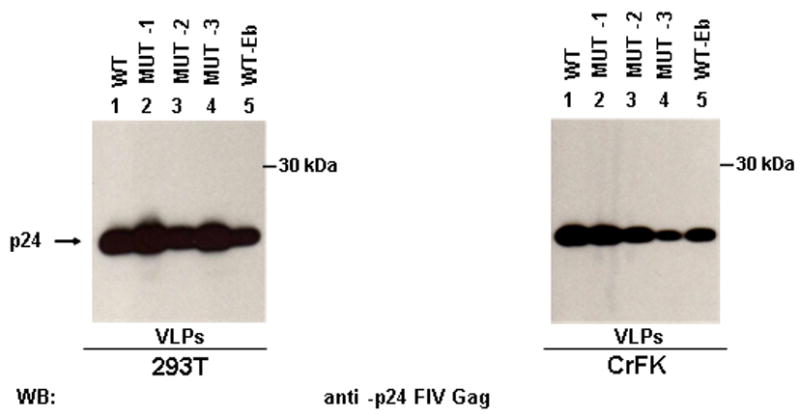
Effect of the Ebola L-domain on FIV particle release. 293T (left panel) and CrFK (right panel) cells were transfected with FIV Gag/Pol constructs, bearing different mutations at the level of the carboxy-terminal region of Gag, as described in Table 2. Twenty four h post-transfection, VLPs were spun down by ultracentrifugation and analysed by western blotting.
Table 2.
Mutational analysis of the carboxy-terminal region of FIV-Gag proteina
| 438 | 439 | 440 | 441 | 442 | 443 | 444 | Gag processing | |
|---|---|---|---|---|---|---|---|---|
| WT | P | S | A | P | P | M | E | + |
|
|
||||||||
| MUT-1 | P | S | A | P | P | M | A | + |
|
|
||||||||
| MUT-2 | P | S | A | P | P | M | Y | + |
|
|
||||||||
| MUT-3 | P | T | A | P | P | M | Y | + |
|
|
||||||||
| WT-Eb | P | T | A | P | P | E | Y | + |
The table shows the amminoacid sequence relative to the carboxy-terminal region of different constructs obtained by site specific mutagenesis, starting from the WT construct. The effect on Gag processing is reported. The mutated nucleotides are underlined.
The L-domain-induced bands are ubiquitin conjugates Gag forms
We have previously reported the appearance of modified Gag products in VLPs of different retroviruses, related to the L-domain function (Strack et al., 2000). Here we observed that higher molecular Gag bands are present also in FIV VLPs, obtained by transfecting both 293T and CrFK cells with FIV-Stop (Fig. 4a). These bands are particularly evident in the case of FIV-Stop construct with the wt L-domain replaced by the one characteristic of Ebola virus (Ebola-Stop, Fig. 4a). Since it is known that this latter L-domain leads to strong ubiquitination, we hypothesized that these bands may be ubiquitin-conjugated Gag forms. Thus, we co-transfected WT-Stop and Ebola-Stop plasmids along with a vector expressing an HA-tagged version of ubiquitin. VLPs were spun down, and subjected to western blotting analysis directed against the HA-tag. The results clearly showed that the modified forms of Gag were indeed ubiquitin-conjugates (Fig. 4b). Moreover, the ubiquitination seemed to be L-domain induced, since, as expected in this case, it appeared to be stronger in the context of the Ebola-Stop Gag (Fig. 4b). In order to further support this conclusion and to analyse whether there was a correlation between L-domain function and ubiquitination, we co-transfected 293T cells and CrfK cells with either ASAA-Stop or WT-Stop construct, along with the HA-ubiquitin expressing plasmid. Released VLPs were then purified and the samples were normalized for p24 content, prior to SDS-gel analysis and western blotting. We were able to confirm that the ubiquitin-conjugated bands were indeed L-domain-induced and disappeared in the presence of mutations that affect L-domain functionality (Fig. 4b). In conclusion, our data suggest a correlation between FIV Gag ubiquitination and late domain function.
Fig. 4.
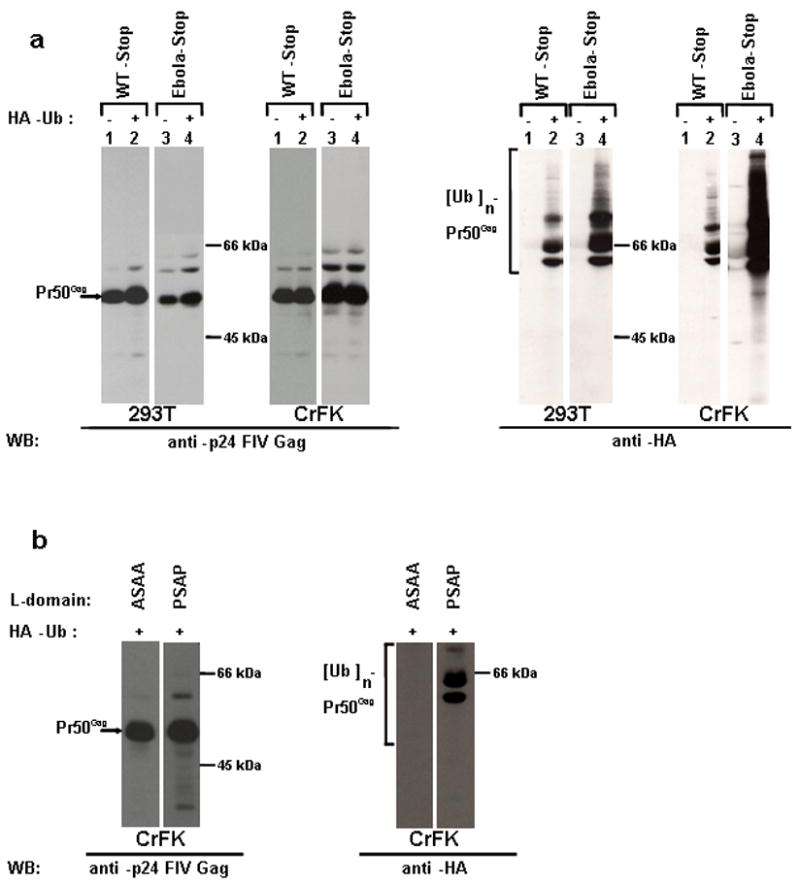
A functional L-domain induces Gag ubiquitination. (a) 293T cells or CrFK cells were transfected with WT-Stop or Ebola-Stop constructs along with either the empty vector pBJ5 (lane 1 and 3) or an HA-tagged ubiquitin expressing plasmid (lane 2 and 4). Purified VLPs were subjected to SDS-PAGE, followed by western blotting analysis, as indicated. (b) CrFK cells were transfected with either ASAA-Stop or WT-Stop construct along with an HA-tagged ubiquitin expressing plasmid. VLPs were purified and p24 levels were normalized prior to SDS-PAGE. A western blotting analysis was performed, as indicated.
Nedd4-2s ubiquitin ligase is involved in FIV Gag egress
It is known that the PPxY motif leads to strong ubiquitination of structural viral proteins and facilitate viral egress by recruiting a subset of ubiquitin ligases, belonging to the Nedd4 family (Martin-Serrano et al., 2005). However, it is less clear how this phenomenon can take place in the case of Gag proteins lacking such a motif. This is particularly true for FIV Gag which appears to be ubiquitinated. It has been recently shown that the budding of various HIV-1 L-domain mutants is dramatically enhanced by ectopic Nedd4-2s, a native isoform with a truncated C2 domain (Chung et al., 2008; Usami et al., 2008). In order to analyse whether this ubiquitin ligase was involved in FIV Gag ubiquitination and budding, we over-expressed WT-Stop with Nedd4-2s in the presence of HA-tagged ubiquitin. We were able to show that, under these conditions, the Gag ubiquitination is enhanced (Fig. 5a). Moreover, when we co-transfected ASAA construct along with Nedd4-2s we were able to show a significant rescue of viral budding (Fig. 5b). Significantly, the ASAA mutant was not rescued when a Nedd4-2s C801S, a form mutated at the level of the active site, (Usami et al., 2008) was co-expressed (Fig. 5b), indicating that the ability of the cellular protein to form thioester bond plays an important role in this context. Moreover, a rescue was observed also when the full-length version of Nedd4-2 was expressed (Fig. 5b), even though to a less extend when compared to the Nedd4-2s effect. This latter finding is in agreement with the data reported in the case of HIV-1 and with a possible inhibitory effect of the C2 domain on Nedd4-2 activity (Usami et al., 2008).
Fig. 5.
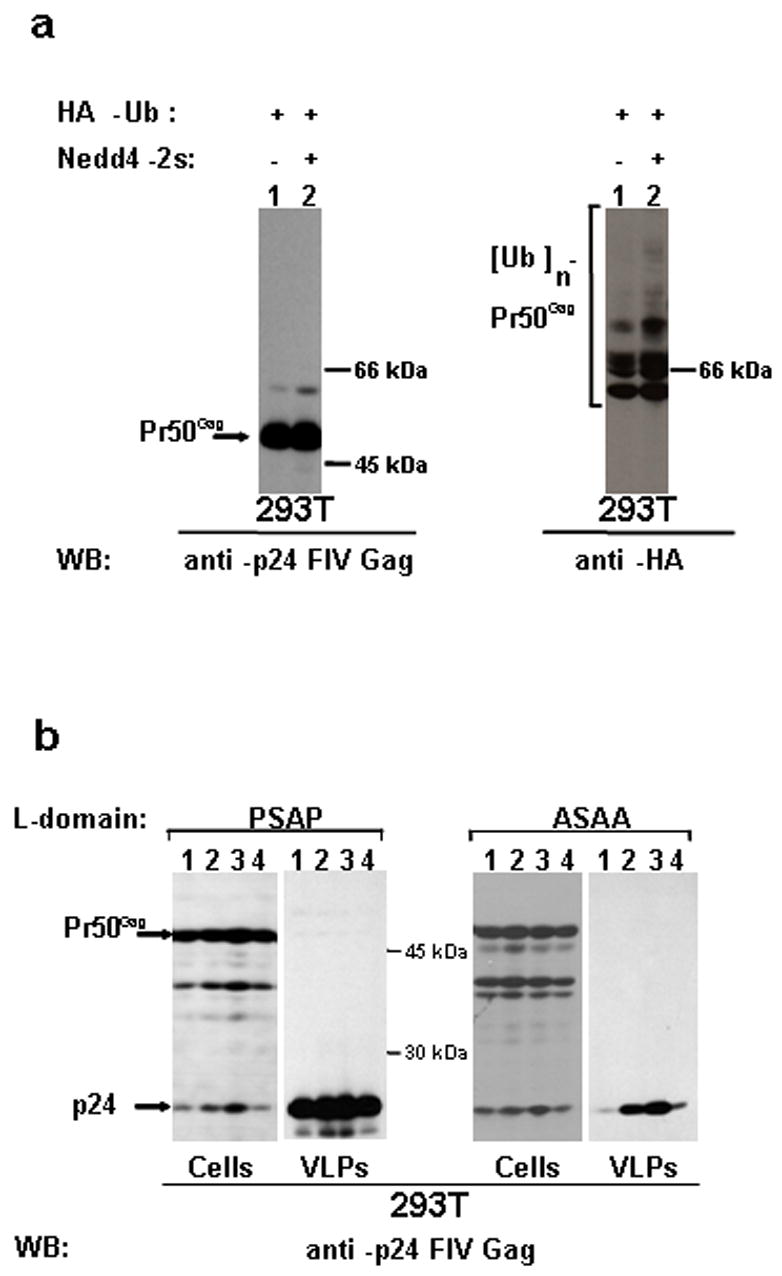
Nedd4-2s ubiquitin ligase leads to an enhancement of FIV-Gag ubiquitination and to the rescue of constructs lacking a functional late domain. (a) 293T cells were transfected with either WT-Stop in combination with an HA-tagged ubiquitin expressing plasmid alone (lane 1) or in combination with a construct encoding Nedd4-2s (lane 2). Purified VLPs were normalised for p24 content and analysed by western blotting, as indicated. (b). 293T cells were transfected with either WT or ASAA constructs, as indicated, alone (lane 4) or in combination with constructs expressing either Nedd4-2s C801S (lane 1), or Nedd4-2 (lane 2), or Nedd4-2s (lane 3). Twenty-four h later, cell lysates (Cells) and purified virus like particles (VLPs) were analysed by western blotting.
Discussion
Many biological and pathogenetic aspects render FIV an attractive model for AIDS research. Indeed, FIV and is natural host, the domestic cat, have already provided important insights into relevant aspect of HIV infection and pathogenesis (Burkhard and Dean, 2003; Power et al., 2004). Moreover, this model has been proposed for testing new specific HIV drugs, for the analysis of resistance mechanisms and for the design of candidate HIV vaccine. The fact that efficacious anti-FIV therapeutic approach would be advantageous also in veterinary medicine, adds value to the system. Finally, the development of safe gene-transfer vectors based on FIV is another interesting application of this non-primate lentivirus.
Despite this great potentiality, some of the experiments carried on in order to test the efficacy of specific HIV-1 drugs and to analyse viral resistance mechanisms (North and LaCasse, 1995), employing FIV as a model, have clearly brought to light that in many areas our present understanding of FIV lags behind knowledge accumulated on HIV. In this context, a further and deep dissection of FIV structural protein Gag functions could have a significant impact on clarifying different aspects of viral pathogenesis, as well as on drug development and vaccination strategy design.
It has been recently reported that FIV, like other enveloped viruses, exploits the multivesicular body (MVB) biogenesis pathway for its budding (Luttge et al., 2008). In particular, a four aminoacidic motif (PSAP), present in the carboxy-terminal region of Gag, appears to be crucial in this context (Luttge et al., 2008). In the present work we intended to contribute to the dissection of the carboxy-terminal region of the FIV Gag protein, in particular focusing on its contribution to VLPs production. Thus, we obtained four different constructs, characterised by the substitution of one or both prolines of the PSAP motif with alanines, in combination or not with the introduction of a STOP codon in place of aminoacid 10 of the protease. Early reports on HIV-1 had demonstrated that mutational inactivation of the viral protease could reverse defects in the L-domain region, suggesting a functional linkage between p6 and the proteolytic processing of the Gag precursor protein during the budding of progeny virions (Huang et al., 1995). Here we show that, in contrast with HIV-1, a FIV Gag characterised by an inactive L-domain is impaired in the release from transfected cells also in the absence of the viral protease. In agreement with this finding, we demonstrate that a block of the MVB pathway strongly impairs unprocessed FIV Gag from budding. Interestingly, our data do not rule out the functional linkage between FIV L-domain and protease activity. Indeed, when the L-domain is inactive, in the context of an active viral protease, we do notice the accumulation of intracellular partially processed Gag (Fig. 1a, lane 2 “Cells” panel). In particular the MA-CA-p1-NC intermediate seems to accumulate. It is known that in the case of HIV-1, defects in Gag processing, such as the p25 to p24 increased ratio, are typically associated with p6 mutants (Huang et al., 1995). Thus, even though our data suggest that also in the case of FIV the inactivation of the L-domain is linked to protease activity, we still see a strong impairment in the unprocessed Gag ability to bud, when the L-domain is inactivated. The fact that the carboxy-terminal region of FIV Gag does not overlap with the open reading frame of the viral protease, may contribute to explain this difference between the two lentiviruses. Moreover, the specific properties of the feline protease, that have been identified (Schnölzer et al., 1996; Lin et al., 2003), like, for instance, the different order of Gag processing (Lin et al., 2006), may also contribute to this phenomenon. It is worth to note that an other difference between HIV-1 and FIV emerged form our study. Indeed, we demonstrate that the first proline of the PSAP motif is not involved in the late domain function, while it appears to be essential in the case of HIV-1 (Huang et al., 1995). It is known that the PSAP finds in the cellular protein Tsg101 the connection with the MVB pathway (Garrus et al., 2001). Moreover it has been recently reported that knocking down Tsg101 a significant reduction in FIV budding is achieved (Luttge et al., 2008). Since Sundquist’s group has reported that the first proline of the HIV-1 PT/SAP makes critical contacts with the UEV domain of Tsg101 (Pornillos et al., 2002) it would be interesting to analyse whether Tsg101 is incorporated in FIV VLPs and to determine whether the FIV ASAP mutant is still able to interact with Tsg101. Moreover, it would also be useful to know whether Tsg101 depletion affects the release of the ASAP mutant.
As mentioned above, FIV may find interesting application in the development of gene-transfer vectors. However, the production of replication-defective lentiviral vectors for a large-scale clinical use is challenging (Lesch et al., 2008). One of the limiting steps in the process is represented by the low vector titer obtained with the packaging system currently adopted (Segura et al., 2007). Since our data demonstrated the impact of the L-domain region in FIV VLP production and since it has been reported that L-domains are transferable between unrelated viruses (Parent et al., 1995), we wanted to investigate the particle release efficiency of a FIV Gag chimera characterised by the Ebola virus L-domain replacing the PSAP motif (WT-Eb). Indeed, it has been shown that HIV-1 based minimal Gag constructs that have been engineered to contain the PTAPPEY motif, which is the one characteristic of Ebola virus, are released very efficiently from transfected cells (Strack et al., 2002). When we quantified the amount of particles produced by the constructs containing a more or less optimised Ebola virus L-domain (WT-Eb, MUT1, MUT2 and MUT3), we did not notice a significant increase in particle production, with respect to FIV WT. Thus, even though Ebola L-domain is working also in the context of full length FIV Gag, it does not lead to a significant enhancement in particle production. However, our data do not rule out the possibility that carefully manipulating the carboxy-terminal region of FIV Gag the amount of VLP released could be positively influenced. It is worth to mention that by few steps of site specific mutagenesis it is possible to modify the sequence downstream the PSAP motif in order to obtain a Gag construct containing the Ebola virus L-domain combined with the YPxL domain. Thus, it could be interesting to test the impact of this combination between different L-domains on the particle release efficiency.
An other finding of our work is that unprocessed wt FIV Gag is modified by ubiquitination. This is again a peculiar feature of FIV Gag, when compared to HIV-1, which appears to be much less ubiquitinated (Strack et al., 2000; Martin Serrano et al., 2004), but it is supported by similar findings published in the case of other lentiviruses, such as SIVmac (Strack et al., 2000). Moreover, Gag ubiquitination appears to be linked to the functionality of the L-domain. Thus, our data suggest that, in the case of FIV Gag, ubiquitination may be directly linked to budding. We and others have reported that a functional L-domain leads to retroviral Gag ubiquitination and that ubiquitin residues involved in endocytosis are crucial for budding (Strack et al., 2002; Martin-Serrano et al., 2004; Gottwein et al., 2006). However, the role of ubiquitin in the egress of retroviruses, and in particular of lentiviruses, is still under debate. Indeed, even though several findings argue for a functional contribution of the ubiquitin modification to virus release, there are studies suggesting that ubiquitination could be merely a bystander effect (Martin-Serrano, 2007). In addition, in agreement with our present findings, the enhancement of ubiquitination, does not always correlate with an enhancement in particle release (Strack et al., 2000; Martin Serrano, 2007). Finally, while ubiquitination of PPxY-containing Gag is easily explained by the direct interaction of the viral structural protein with Nedd4 family ubiquitin ligases (Harty et al., 2000, Martin-Serrano et al., 2005), the involvement of ubiquitin in the egress of viruses lacking such a motif is less clear. However, two recent studies (Chung et al., 2008; Usami et al., 2008) have shown that Nedd4-2s, a member of Nedd4-ubiquitin ligases, plays a role in HIV-1 budding. The authors’ conclusion is that Nedd4-2s, even in the absence of a functional PPxY motif, may cooperate with the other L-domains to enhance viral budding, maybe leading to the ubiquitination and consequent activation of MVB components or facilitating Gag interaction with ubiquitin binding protein of the pathway. Moreover, the authors clearly demonstrated that Nedd4-2s is the only ubiquitin ligase showing the ability to rescue L-domain deficient HIV-1 constructs (Usami et al., 2008). In order to confirm and extend this conclusion to non-primate lentiviruses and since our data suggested that ubiquitin could play a role in FIV egress, we decided to analyse whether Nedd4-2s was involved in this process. We were able to show that indeed Nedd4-2s is involved in FIV Gag ubiquitination and it is capable of rescuing FIV Gag lacking a functional L-domain in its ability to bud from transfected cells. This finding is interesting because it demonstrates that lentiviruses in general respond to Nedd4-2s in the absence of a PPxY motif, in contrast to other retroviruses, like the Mason-Pfizer monkey virus (Usami et al., 2008).
Overall our data bring to light peculiar aspects of FIV Gag carboxy-terminal region, that may have an impact when FIV is employed as a model for studying HIV pathogenesis and therapy, or for biotechnological applications of this lentivirus. On the other hand, we extend the recent observation that ubiquitin ligases may cooperate with other late domains to enhance the efficiency of viral release to a non-primate lentivirus, demonstrating that this novel mechanism of connection to the cellular budding machinery is shared between different members of this viral family.
Acknowledgments
This work was supported by grants from Regione Veneto, MIUR (PRIN-2005), Istituto Superiore di Sanità (Rome-AIDS Projects GRANT no. 30G.24 and 40F.57) and University of Padova (Ex 60% and ATENEO 2006) to A.C., G.P. and C.P. and by NIH grant number AI29873 to H.G. We thank Paola Sette and Elena Sartori for help and technical support. Marta Celegato is a PhD student in the Virology and Microbial Biotechnologies program at the University of Padova. Michele Celestino is a PhD student in the Biomedicine program at the University of Padova. Cristiano Salata is supported by a Post-Doctoral fellowship of the Azienda Ospedaliera of Padova, Padova Hospital.
Literature Cited
- Bendinelli M, Pistello M, Lombardi S, Pli A, Garzelli C, Matteucci D, Ceccherini-Nelli L, Malvalia G, Zozzini F. Feline immunodeficiency virus: an interesting model for AIDS studies and an important cat pathogen. Clin Microbiol Rev. 1995;8:87–112. doi: 10.1128/cmr.8.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhard MJ, Dean GA. Transmission and immunopathogenesis of FIV in cats as a model for HIV. Curr HIV Res. 2003;1:15–29. doi: 10.2174/1570162033352101. [DOI] [PubMed] [Google Scholar]
- Chung HY, Morita E, von Schwedler U, Müller B, Kräusslich HG, Sundquist WI. NEDD4L Overexpression rescues release and infectivity of HIV-1 constructs lacking PTAP and YPXL late domains. J Virol. 2008;82:4884–4897. doi: 10.1128/JVI.02667-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirov DG, Freed EO. Retrovirus budding. Virus Res. 2004;106:87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Dias AS, Bester MJ, Britz RF, Apostolides Z. Animal models used for the evaluation of antiretroviral therapies. Curr HIV Res. 2006;4:431–446. doi: 10.2174/157016206778560045. [DOI] [PubMed] [Google Scholar]
- Dunham S. Lesson from the cat: development of vaccines against lentivirus. Vet Immunol Immunopathol. 2006;112:67–77. doi: 10.1016/j.vetimm.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Elder JH, Phillips TR. Molecular properties of feline immunodeficiency virus (FIV) Infect Agents Dis. 1993;2:361–374. [PubMed] [Google Scholar]
- Elder JH, Dean GA, Hoover EA, Hoxie JA, Malim MH, Mathes L, Neil JC, North TW, Sparger E, Tompkins MB, Tompkins WA, Yamamoto J, Yuhki N, Pedersen NC, Miller RH. Lessons from the cat: feline immunodeficiency virus as a tool to develop intervention strategies against human immunodeficiency virus type 1. AIDS Res Hum Retroviruses. 1998;14:797–801. doi: 10.1089/aid.1998.14.797. [DOI] [PubMed] [Google Scholar]
- Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Cote M, Rich RL, Myszka DG, Sundquist WI. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001;107:55–65. doi: 10.1016/s0092-8674(01)00506-2. [DOI] [PubMed] [Google Scholar]
- Göttlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Göttlinger HG. The HIV-1 assembly machine. AIDS. 2001;15(Suppl 5):S13–20. doi: 10.1097/00002030-200100005-00003. [DOI] [PubMed] [Google Scholar]
- Gottwein E, Jäger S, Habermann A, Kräusslich HG. Cumulative mutations of ubiquitin acceptor sites in human immunodeficiency virus type 1 gag cause a late budding defect. J Virol. 2006;80:6267–6275. doi: 10.1128/JVI.02177-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty RN, Brown ME, Wang G, Huibregtse J, Hayes FP. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc Natl Acad Sci USA. 2000;97:13871–13876. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesch HP, Turpeinen S, Niskanen EA, Mähönen AJ, Airenne KJ, Ylä-Herttuala S. Generation of lentivirus vectors using recombinant baculoviruses. Gene Ther. 2008 doi: 10.1038/gt.2008.76. [DOI] [PubMed] [Google Scholar]
- Lin YC, Beck Z, Morris GM, Olson AJ, Elder JH. Structural basis for distinctions between substrate and inhibitor specificities for feline immunodeficiency virus and human immunodeficiency virus proteases. J Virol. 2003;77:6589–6600. doi: 10.1128/JVI.77.12.6589-6600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Brik A, de Parseval A, Tam K, Torbett BE, Wong CH, Elder JH. Altered gag polyprotein cleavage specificity of feline immunodeficiency virus/human immunodeficiency virus mutant proteases as demonstrated in a cell-based expression system. J Virol. 2006;80:7832–7843. doi: 10.1128/JVI.00374-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttge BG, Shehu-Xhilaga M, Demirov DG, Adamson CS, Soheilian F, Nagashima K, Stephen AG, Fisher RJ, Freed EO. Molecular characterization of feline immunodeficiency virus budding. J Virol. 2008;82:2106–2119. doi: 10.1128/JVI.02337-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrique ML, Rauddi ML, Gonzalez SA, Affranchino JL. Functional domains in the feline immunodeficiency virus nucleocapsid protein. Virology. 2004;327:83–92. doi: 10.1016/j.virol.2004.06.019. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Perez-Caballero D, Bieniasz PD. Context-dependent effects of L domains and ubiquitination on viral budding. J Virol. 2004;78:5554–5563. doi: 10.1128/JVI.78.11.5554-5563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol. 2005;168:89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J. The role of ubiquitin in retroviral egress. Traffic. 2007;8:1297–1303. doi: 10.1111/j.1600-0854.2007.00609.x. [DOI] [PubMed] [Google Scholar]
- North TW, LaCasse RA. Testing anti-HIV drugs in the FIV model. Nat Med. 1995;1:410–411. doi: 10.1038/nm0595-410. [DOI] [PubMed] [Google Scholar]
- Parent LJ, Bennett RP, Craven RC, Nelle TD, Krishna NK, Bowzard JB, Wilson CB, Puffer BA, Montelaro RC, Wills JW. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen NC, Yamamoto JK, Ishida T, Hansen H. Feline immunodeficiency virus infection. Vet Immunol Immunopathol. 1989;21:111–29. doi: 10.1016/0165-2427(89)90134-7. [DOI] [PubMed] [Google Scholar]
- Piper RC, Katzmann DJ. Biogenesis and function of multivesicular bodies. Annu Rev Cell Dev Biol. 2007;23:519–547. doi: 10.1146/annurev.cellbio.23.090506.123319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistello M, Vannucci L, Ravani A, Bonci F, Chiuppesi F, Del Santo B, Freer G, Bendinelli M. Streamlined design of a self-inactivating feline immunodeficiency virus vector for transducing ex vivo dendritic cells and T lymphocytes. Genet Vaccines Ther. 2007;5:8. doi: 10.1186/1479-0556-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poeschla E, Wong-Staal F, Looney D. Efficient transduction of nondividing cells by feline immunodeficiency virus lentiviral vectors. Nat Med. 1998;4:354–357. doi: 10.1038/nm0398-354. [DOI] [PubMed] [Google Scholar]
- Pornillos O, Alam SL, Davis DR, Sundquist WI. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat Struct Biol. 2002;9:812–817. doi: 10.1038/nsb856. [DOI] [PubMed] [Google Scholar]
- Power C, Zhang K, van Marle G. Comparative neurovirulence in lentiviral infections: The roles of viral molecular diversity and select proteases. J Neurovirol. 2004;10(Suppl 1):113–117. doi: 10.1080/753312762. [DOI] [PubMed] [Google Scholar]
- Saenz DT, Poeschla EM. FIV: from lentivirus to lentivector. J Gene Med. 2004;6(Suppl 1):S95–104. doi: 10.1002/jgm.500. [DOI] [PubMed] [Google Scholar]
- Schnölzer M, Rackwitz HR, Gustchina A, Laco GS, Wlodawer A, Elder JH, Kent SB. Comparative properties of feline immunodeficiency virus (FIV) and human immunodeficiency virus type 1 (HIV-1) proteinases prepared by total chemical synthesis. Virology. 1996;224:268–275. doi: 10.1006/viro.1996.0528. [DOI] [PubMed] [Google Scholar]
- Segura MM, Garnier A, Durocher Y, Coelho H, Kamen A. Production of lentiviral vectors by large-scale transient transfection of suspension cultures and affinity chromatography purification. Biotechnol Bioeng. 2007;98:789–799. doi: 10.1002/bit.21467. [DOI] [PubMed] [Google Scholar]
- Strack B, Calistri A, Accola MA, Palù G, Göttlinger HG. A role for ubiquitin ligase recruitment in retrovirus release. Proc Natl Acad Sci USA. 2000;97:13063–13068. doi: 10.1073/pnas.97.24.13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Göttlinger HG. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J Virol. 2002;76:5472–5479. doi: 10.1128/JVI.76.11.5472-5479.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Göttlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Uhl EW, Martin M, Coleman JK, Yamamoto JK. Advances in FIV vaccine technology. Vet Immunol Immunopathol. 2008;123:65–80. doi: 10.1016/j.vetimm.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usami Y, Popov S, Popova E, Göttlinger HG. Efficient and specific rescue of human immunodeficiency virus type 1 budding defects by a Nedd4-like ubiquitin ligase. J Virol. 2008;82:4898–907. doi: 10.1128/JVI.02675-07. [DOI] [PMC free article] [PubMed] [Google Scholar]