Abstract
In plants, myoinositol signaling pathways have been associated with several stress, developmental, and physiological processes, but the regulation of these pathways is largely unknown. In our efforts to better understand myoinositol signaling pathways in plants, we have found that the WD40 repeat region of a myoinositol polyphosphate 5-phosphatase (5PTase13; At1g05630) interacts with the sucrose nonfermenting-1-related kinase (SnRK1.1) in the yeast two-hybrid system and in vitro. Plant SnRK1 proteins (also known as AKIN10/11) have been described as central integrators of sugar, metabolic, stress, and developmental signals. Using mutants defective in 5PTase13, we show that 5PTase13 can act as a regulator of SnRK1 activity and that regulation differs with different nutrient availability. Specifically, we show that under low-nutrient or -sugar conditions, 5PTase13 acts as a positive regulator of SnRK1 activity. In contrast, under severe starvation conditions, 5PTase13 acts as a negative regulator of SnRK1 activity. To delineate the regulatory interaction that occurs between 5PTase13 and SnRK1.1, we used a cell-free degradation assay and found that 5PTase13 is required to reduce the amount of SnRK1.1 targeted for proteasomal destruction under low-nutrient conditions. This regulation most likely involves a 5PTase13-SnRK1.1 interaction within the nucleus, as a 5PTase13:green fluorescent protein was localized to the nucleus. We also show that a loss of function in 5PTase13 leads to nutrient level-dependent reduction of root growth, along with abscisic acid (ABA) and sugar insensitivity. 5ptase13 mutants accumulate less inositol 1,4,5-trisphosphate in response to sugar stress and have alterations in ABA-regulated gene expression, both of which are consistent with the known role of inositol 1,4,5-trisphosphate in ABA-mediated signaling. We propose that by forming a protein complex with SnRK1.1 protein, 5PTase13 plays a regulatory role linking inositol, sugar, and stress signaling.
Myoinositol (inositol) signaling pathways are important for many different developmental and physiological processes in eukaryotes (Boss et al., 2006; Berridge, 2007). In plants, inositol signaling is used in the response to abscisic acid (ABA; Sanchez and Chua, 2001; Xiong et al., 2001; Burnette et al., 2003; Gunesekera et al., 2007; Lee et al., 2007), salt stress (DeWald et al., 2001; Takahashi et al., 2001), gravity (Perera et al., 2001, 2006), and pathogens (Ortega and Perez, 2001; Andersson et al., 2006). The inositol signaling pathway makes use of an inositol 1,4,5-trisphosphate (InsP3) second messenger that triggers intracellular Ca2+ release from various sources within the cell (Berridge, 1993; Tang et al., 2007), although the precise mechanism in plants is not yet known (Krinke et al., 2007). To terminate signaling events driven by InsP3, cells utilize the myoinositol polyphosphate 5-phosphatases (5PTases; EC 3.1.3.56) to remove the 5-phosphate, thus initiating second messenger breakdown (Astle et al., 2007). Eukaryotes have a family of diverse 5PTases, with the Arabidopsis (Arabidopsis thaliana) genome containing 15 genes predicted to encode conserved 5PTases (Berdy et al., 2001). Of the 15 Arabidopsis 5PTase genes, four encode 5PTase enzymes with a large, N-terminal extension containing five to six WD40 repeat regions (Zhong and Ye, 2004). WD40 repeats are found in a number of eukaryotic regulatory proteins (Li and Roberts, 2001), where they are speculated to serve as a stable propeller-like platform to which proteins can bind either stably or reversibly. In some cases, sequences outside of the WD40 repeats may confer a new functional specificity to the protein even as conserved protein interactions are maintained through the WD40 repeats, potentially adapting basic cellular mechanisms to organism-specific processes. WD40-containing 5PTases are unique to plants and certain fungi, as no other genomes contain candidate genes for these proteins (Zhong and Ye, 2004).
By examining plants containing either a gain or loss of function in specific 5PTases, investigators have established that 5PTases are critical in plant development and in ABA signaling. In particular, mutations in the CVP2 (5PTase6; At1g05470) and 5PTase13 (At1g05630) genes have been linked to altered cotyledon vascular patterning and/or blue light responses and phototropin 1 signaling (Carland and Nelson, 2004; Lin et al., 2005; Chen et al., 2008). In contrast, fra3 (At1g65580) and mrh3 (5PTase5; At5g65090) mutants are altered in fiber cell development (Zhong et al., 2004) and root hair initiation (Jones et al., 2006), respectively. With regard to stress signaling, ectopic expression of either 5PTase1 or 5PTase2 has been shown to decrease ABA signaling by increasing hydrolysis of InsP3 (Sanchez and Chua, 2001; Burnette et al., 2003). Loss-of-function mutants in these same genes result in ABA hypersensitivity in germinating seeds and increased seedling hypocotyl elongation in the dark, which is accompanied by an increase in InsP3 levels (Gunesekera et al., 2007).
5PTases have been reported to play a role in Glc sensing/metabolism in animal cells (Wada et al., 2001; Sasaoka et al., 2004; Kagawa et al., 2008). Overexpression of an SH2-containing inositol phosphatase 2 (SHIP2) inhibited insulin-induced signaling leading to Glc uptake and glycogen synthesis via hydrolysis of phosphatidylinositol 3,4,5-trisphosphate (PtdIns-P3) in 3T3-L1 adipocytes and L6 myotubes (Sasaoka et al., 2001; Wada et al., 2001). On the other hand, loss of SHIP2 in mice resulted in increased sensitivity to insulin, which is characterized by deregulated expression of genes involved in gluconeogenesis and perinatal death (Clement et al., 2001). Although there are recent reports delineating some relationship between inositol signaling and sugar sensing/metabolism in plant cells (Im et al., 2007; Lou et al., 2007), a specific role of 5PTases in sugar signaling has not yet been determined. The mechanisms by which plants sense sugars and regulate carbohydrate metabolism are complex and often facilitated by protein complexes. Therefore, we sought to identify 5PTase-interacting proteins involved in signaling and metabolic events.
We report here that the WD40 repeat region of the 5PTase13 gene interacts specifically with a Suc nonfermenting-1-related kinase (SnRK1.1, AKIN10; At3g01090), which functions as a sensor of energy and stress in plants (Baena-Gonzalez et al., 2007; Hardie, 2007; Hue and Rider, 2007). Using T-DNA insertion mutants in the 5PTase13 gene, we present evidence that 5PTase13 is a nuclear protein that can act as a regulator of SnRK1.1 under low-nutrient conditions by decreasing the amount of SnRK1.1 degraded by the proteasome.
RESULTS
5PTase13 Is a Member of a Unique Group of WD40-Containing Proteins
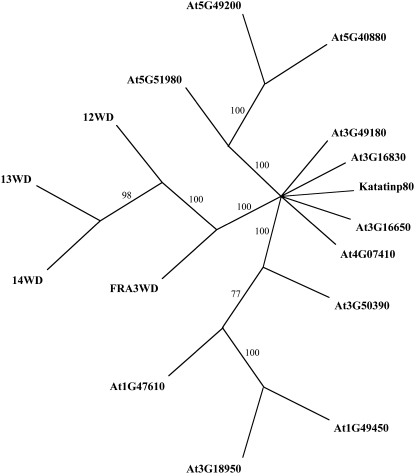
Besides containing a conserved inositol polyphosphate 5-phosphatase catalytic domain, 5PTase13 also contains five WD40 repeat regions in the N terminus that may allow for unique protein interactions. Three other genes in the Arabidopsis genome, 5PTase12 (At2g43900), 5PTase14 (At2g31830), and FRA3 (At1g65580), encode similar proteins (Berdy et al., 2001; Zhong and Ye, 2004), and the amino acid identity of the WD40 repeat region varies from 45.5% to 75.3% among these proteins (Supplemental Table S1). We used the amino acid sequences corresponding to these WD40 repeat regions in BLASTp searches and obtained 12 related sequences with e-values less than 1. ClustalW and PAUP4.0 were used to generate phylogenetic trees using parsimony (Fig. 1), which showed that the 5PTase WD40 repeat regions are contained on a separate branch, thus indicating that they are more similar to one another than to other WD40 repeat regions.
Figure 1.
Phylogenetic tree of the WD40 regions from 5PTases and other WD40-containing Arabidopsis proteins. ClustalW and PAUP4.0 were used to generate a tree. Bootstrapping (1,000 times) yielded the confidence level indicated at each branch. The results show that the WD40 region of 5PTase14 (14WD) is the closest relative to 5PTase13 (13WD). Furthermore, the tree indicates that the 5PTase WD40 regions (12WD, 13WD, 14WD, and FRA3WD) are more closely related to each other than to other WD40-containing proteins.
The similarity of the 5PTases (Zhong and Ye, 2004) suggests that these proteins may function in a redundant manner. Microarray data from GENEVESTIGATOR (Zimmermann et al., 2004) indicates that FRA3 is broadly and abundantly expressed compared with 5PTase12, 5PTase13, and 5PTase14 (Supplemental Fig. S1A). We conclude that the WD40-containing 5PTases are a small group of proteins with the potential to form protein complexes and that functional redundancy may be present.
The WD40 Repeat Region of 5PTase13 Interacts with SnRK1.1
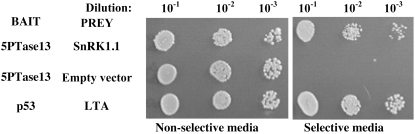
To investigate the ability of the WD40 regions of 5PTase13 to participate in protein complexes, we used the yeast two-hybrid system. The 533 N-terminal amino acids from 5PTase13 containing the WD40 repeats were used as bait in a yeast two-hybrid screen of an Arabidopsis 3-d-old etiolated seedling cDNA library. We screened over 1 million yeast transformants and obtained a positive clone containing the C-terminal domain of the SnRK1.1 gene (At3g01090; also known as AKIN10). We retransformed this positive clone into yeast and verified the interaction (Fig. 2). Negative controls, including the empty DNA binding domain and activation domain vectors, established that SnRK1.1 binds to the WD40 repeat region of the 5PTase13 protein in yeast.
Figure 2.
Yeast two-hybrid screen. An Arabidopsis cDNA prey library was screened using the WD40 region of 5PTase13 as a bait in the yeast two-hybrid system. Over 1 million yeast transformants were screened for reporter gene expression, which resulted in the isolation of a prey plasmid expressing the C-terminal domain of SnRK1.1. Serially diluted yeast strains were spotted onto agar plates that are either nonselective or selective for expression of the two-hybrid reporter genes.
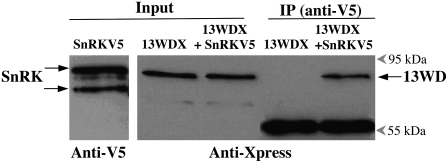
To examine the interaction between 5PTase13 and SnRK1.1 in vitro, we fused the sequence encoding the Xpress epitope tag to the C terminus of the WD40 repeat region of 5PTase13 (13WDX) and the V5 epitope tag sequence to the C terminus of SnRK1.1 (SnRKV5) and expressed both proteins in Escherichia coli. The SnRKV5 construct directs the expression of a 61.7-kD protein detected by an anti-V5 monoclonal antibody, and in most cases we detected two SnRKV5 bands, perhaps as a result of phosphorylation or proteolytic cleavage (Fig. 3, lane 1). The 13WDX construct directs the expression of a 65.9-kD protein detected by an anti-Xpress monoclonal antibody (Fig. 3, lane 2). These interactions are specific, as SnRKV5 is not detected by the anti-Xpress antibody and 13WDX is not detected by the anti-V5 antibody (data not shown). To determine whether the SnRK1.1 recombinant protein can “pull down” 13WDX, immunoprecipitations using anti-V5:protein A-Sepharose beads were performed, and the resulting complex was then analyzed by western blotting with an anti-Xpress antibody (Fig. 3, IP lanes). As shown in Figure 3, the WD40 repeat region from 5PTase13 (13WDX) is only detected in this pull-down assay when it has been incubated in the presence of SnRKV5. We conclude that the WD40 repeat region from 5PTase13 and SnRK1.1 can form a protein complex in vitro.
Figure 3.
13WD40-Xpress protein interacts with SnRK1.1-V5 in vitro. Immunoprecipitation (IP) was carried out using an anti-V5 antibody bound to a protein A-Sepharose (PAS). Purified and dialyzed proteins (13WDX and SnRKV5) were incubated for 2 h at room temperature in immunoprecipitation buffer before transferring to the antiV5:PAS complex. The resulting immunoprecipitation fractions were analyzed using protein gel blotting. Lane 1, Input fraction of SnRKV5 probed with an anti-V5 antibody; lane 2, input fraction of 13WDX; lane 3, input fraction of 13WDX in combination with SnRKV5; lane 4, bound fraction of 13WDX alone; lane 5, bound fraction of 13WDX in combination with SnRKV5. Lanes 2 through 5 were probed with an anti-Xpress antibody. The 55-kD band is the immunoglobin heavy chain component of the anti-V5 antibody. The blot images are representative of four independent experiments.
SnRK1.1, along with its closely related gene family member SnRK1.2, encodes a Suc nonfermenting-1-related kinase implicated as a central integrator of energy signaling and metabolic regulation in yeast, plants, and animals. The interaction of 5PTase13 and SnRK1.1 is novel and is perhaps unique to plants in that yeast and animal 5PTases do not contain WD40 regions. This interaction may indicate that InsP3 signal termination via 5PTase13 function affects the SnRK1 “energy sensor” in plants. Data obtained from the GENEVESTIGATOR database indicate that SnRK1.1, SnRK1.2, and 5PTase13 are detected in most plant tissues examined, although 5PTase13 expression levels are very low compared with SnRK1 gene expression (Supplemental Fig. S1B).
5PTase13 Mutant Identification
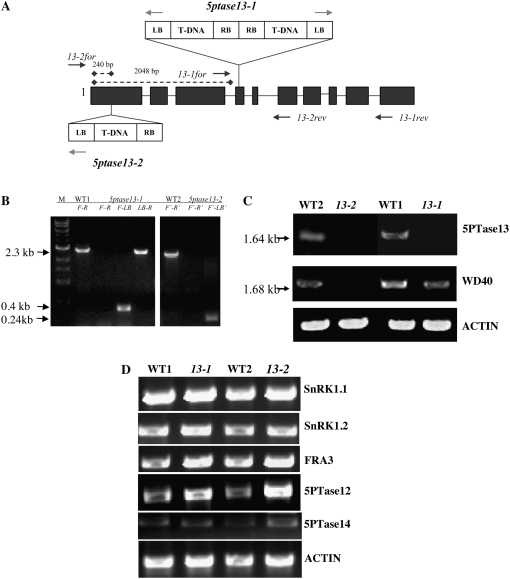
To further explore the link between InsP3 signaling and the energy sensor, SnRK1.1, we isolated two independent T-DNA insertion mutants in the 5PTase13 gene. Two potential mutants were identified in the SALK T-DNA database and named 5ptase13-1 (SAIL_350_FO1) and 5ptase13-2 (SALK_081991) and were compared with their corresponding wild-type accessions (Fig. 4A). The presence of the T-DNA insertion was verified by diagnostic PCR in each mutant using genomic DNA and primers specific for the T-DNA left border (LB) and gene-specific primers that flank the T-DNA insertion (Fig. 4, A and B). The resulting LB gene-specific fragments were sequenced, indicating that in 5ptase13-1 mutants, a second T-DNA insertion is found in tandem in the fourth exon (Fig. 4, A and B, LB-R band). This is in contrast to the previously reported analysis by Lin et al. (2005) showing that the 5ptase13-1 mutant contains a single T-DNA insertion.
Figure 4.
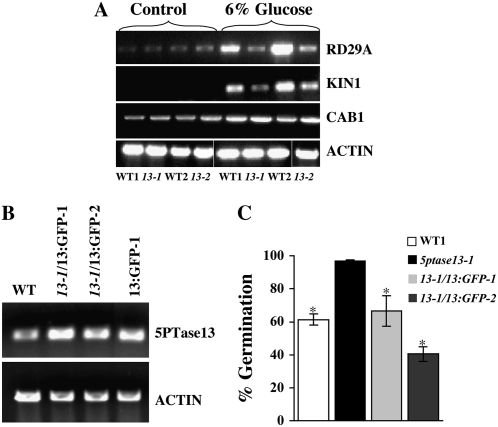
T-DNA insertions and gene expression in 5ptase13 mutant lines. A, T-DNA insertions in the 5PTase13 gene. Exons are shown as dark gray boxes; light gray arrows indicate primers used to amplify the LB of the T-DNA; dark gray arrows indicate positions of gene-specific primers. RB, Right border. B, PCR performed with gene-specific and LB primers confirms that both lines are homozygous. Gene-specific primers that flank the T-DNA insertion (primers 13-1for [F] and 13-1rev [R]) in conjunction with the T-DNA LB primer amplify 0.4- and 2.3-kb fragments in 5ptase13-1 mutants, indicating the presence of two LB sequences in proximity to the 5PTase13 gene. C and D, Expression of genes in 5ptase13 and wild-type seedlings. Total RNA (1–2 μg) was isolated, and semiquantitative RT-PCR was performed with the indicated primers (Supplemental Table S4). C, Verification of the loss of 5PTase13 expression in 2-week-old leaves from soil-grown plants. D, SnRK1 and 5PTase expression from 7-d-old wild-type and 5ptase13 (13-1 and 13-2) seedlings grown on 0.5× MS salts and 0.8% agar in the dark. The experiment was independently repeated two times.
Using primers specific for the 3′ end of 5PTase13, we detected a PCR product in both wild-type lines used (CS60000 and CS908) but not in 5ptase13-1 and 5ptase13-2 mutants (5PTase13 in Fig. 4C). Using primers that amplify the 5′ end, we detected a 1.68-kb product in both wild-type lines and in the 5ptase13-1 mutant, but not in 5ptase13-2 (Fig. 4C). We conclude that the 5ptase13-2 mutant is totally lacking 5PTase13 expression and that both 5ptase13-1 and 5ptase13-2 mutant lines do not express transcripts capable of encoding a full-length 5PTase13 protein.
To examine how the loss of 5PTase13 affects the expression of its binding partner SnRK1.1, we examined the expression of SnRK1.1 and its closely related isoform, SnRK1.2, in 7-d-old dark-grown 5ptase13 and wild-type seedlings. The results reveal that there are no large changes in SnRK1.1 and SnRK1.2 in 5ptase13 mutants (Fig. 4D). As shown in Figure 4D, we found that FRA3 expression remains unchanged in 5ptase13 mutants, and 5PTase14 expression is barely detectable but also unchanged. In contrast, the expression of 5PTase12 is increased in both 5ptase13 mutant lines, revealing a possible means of compensation for the loss of 5PTase13 function (Fig. 4D).
Under standard laboratory conditions, 5ptase13 mutants did not exhibit any abnormalities in plant growth or development. Since Lin et al. (2005) previously reported that 5ptase13-1 mutants are altered in cotyledon vein development, we carefully examined both 5ptase13-1 and 5ptase13-2 mutants and their corresponding wild-type accessions for cotyledon vein development, as described by Carland and Nelson (2004). Using 7-d-old light-grown seedlings of wild-type and 5ptase13 mutant soil-grown plants, we found no evidence for a cotyledon vein phenotype (Supplemental Tables S2 and S3). Our analysis revealed that 33.3% of total WT1 cotyledons and 26.18% of total 5ptase13-1 cotyledons have abnormal patterns and that only 15.9% of total WT2 cotyledons and 19.4% of total 5ptase13-2 cotyledons can be classified as abnormal (Supplemental Table S3). Thus, there is variation in cotyledon vein development within different wild-type accessions, but not between 5ptase13 mutants and their matched wild-type accessions.
SnRK1 Activity Is Altered in 5ptase13 Mutants and Varies with Nutrient Conditions
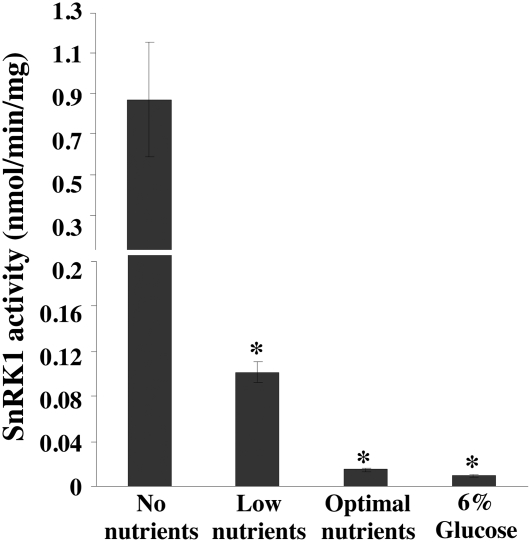
To determine whether 5PTase13 affects SnRK1 function, we measured the activity of SnRK1 in 5ptase13 mutants and wild-type seedlings grown under various nutrient conditions. It is well documented that SnRK1 regulates multiple transcription cascades in response to sugar or energy deprivation (Baena-Gonzalez et al., 2007); however, whether in planta SnRK1 activity changes under various nutrient conditions is not known. To address this, we measured SnRK1 activity from 7-d-old wild-type seedlings grown under low light (40 μE) with different added nutrients to establish various “low-energy” conditions: no nutrients (agar and water alone), low nutrients (0.5× Murashige and Skoog [MS] salts and agar), and optimal nutrients (0.5× MS salts, agar, and 3% Suc). In addition, we examined a stressful level of added sugar (0.5× MS salts, agar, and 6% Glc). For this work, we used a well-established SnRK1 assay (Radchuk et al., 2006) and incubated a sucrose phosphate synthase (SPS) substrate peptide (Huang and Huber, 2001), radiolabeled [γ-32P]ATP, and 5 μg of plant protein extract for 30 min. Validation that these conditions are in the linear range of product accumulation is shown in Supplemental Figure S2.
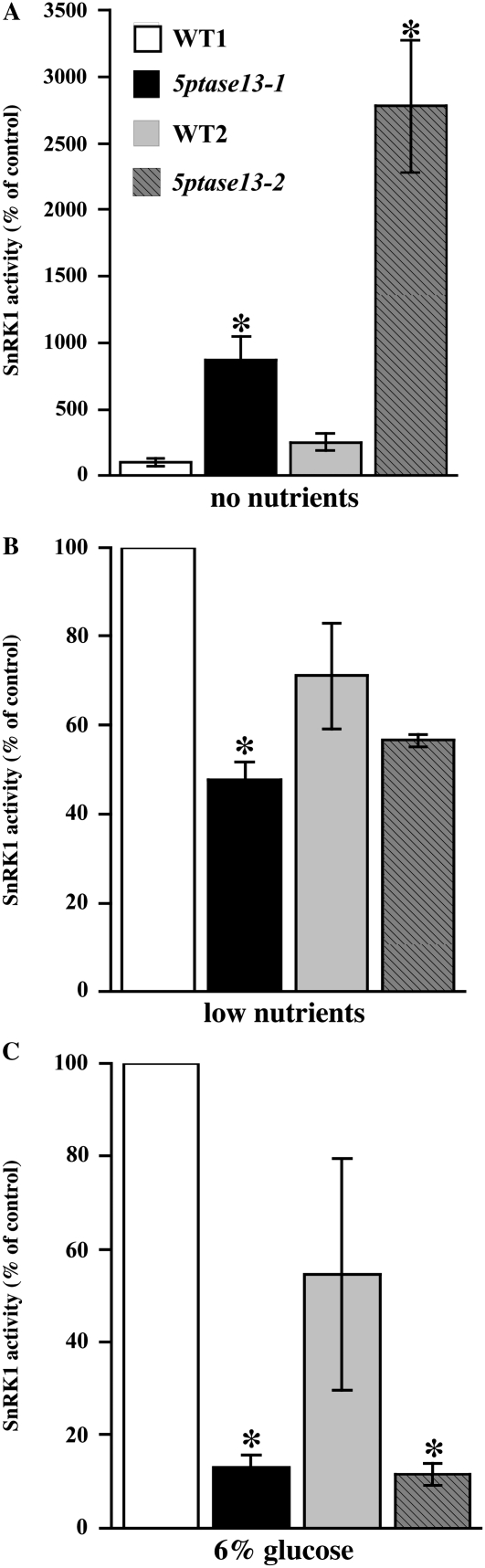
As expected from its role as a low-energy sensor, SnRK1 activity is higher in seedlings grown on low nutrients compared with extracts prepared from seedlings grown on optimal nutrients or 6% Glc (Fig. 5). Figure 6A shows that the activity of SnRK1 significantly increases in 5ptase13-1 and 5ptase13-2 mutants when seedlings are grown with no nutrients (8.1- and 11.3-fold increases, respectively). In contrast, SnRK1 activity decreases in 5ptase13-1 and 5ptase13-2 seedlings compared with wild-type seedlings when grown with low nutrients (Fig. 6B). Furthermore, the decline in SnRK1 activity is even more dramatic when 6% Glc is added (7.1- and 3.3-fold reduction, respectively; Fig. 6C). We conclude that a loss of 5PTase13 function affects SnRK1 activity, and the impact differs depending on nutrient availability. Our data support a role for 5PTase13 as a negative regulator of SnRK1 activity in the absence of nutrients and as a positive regulator when either low nutrients or 6% Glc is present.
Figure 5.
SnRK1 activity varies with different nutrient conditions. Seeds from wild-type (WT2-CS60000) plants were grown on 0.8% agar, no nutrients, low nutrients (0.5× MS salts), optimal nutrients (0.5× MS salts, 3% Suc), or 6% Glc (0.5× MS salts, 6% Glc) under low light for 7 d. The seeds grown in 6% Glc were allowed to germinate in 0.5× MS medium for 4 d and then transferred to 6% Glc medium for 3 d. SnRK1 activity with an SPS peptide was measured from crude plant extracts precipitated with ammonium sulfate according to the method described (Radchuk et al., 2006). Bars represent means ± se of three replicates. The experiment was independently repeated two times. *, P < 0.05 compared with no nutrients.
Figure 6.
SnRK1 activity in 5ptase13 mutants. SnRK1 activity was measured in crude plant extracts from 7-d-old seedlings precipitated with ammonium sulfate according to the method described (Radchuk et al., 2006) using an SPS peptide. Values for SnRK1 activity (nmol inorganic phosphate min−1 mg−1 protein) are normalized to WT1. Extracts are from seedlings grown in no nutrients (A), low nutrients (0.5× MS salts; B), or 6% Glc (0.5× MS salts, 6% Glc; C). Bars represent means ± se of three replicates. The experiment was independently repeated two times. *, P < 0.05 compared with the wild type.
5PTase13 Affects SnRK1.1 Stability
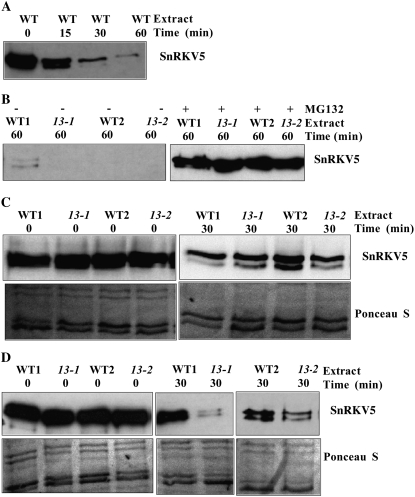
To explore whether 5PTase13 regulates SnRK1.1 stability, we used a similar approach as Lee et al. (2008) to examine the stability of recombinant V5-tagged SnRK1.1 (SnRKV5) in a cell-free degradation assay with wild-type or 5ptase13 extracts. The time course of SnRKV5 degradation with wild-type seedling extracts indicated that SnRKV5 is mostly degraded within 60 min (Fig. 7A). MG132, a proteasome-specific inhibitor, blocked SnRKV5 degradation in both 5ptase13 and wild-type extracts (Fig. 7B), indicating that SnRKV5 is destroyed by the 26S proteasome pathway in these assays.
Figure 7.
Degradation of V5-tagged SnRK1.1 in cell extracts from wild-type (WT) and 5ptase13 seedlings grown under different nutrient conditions. SnRK1.1-V5 protein (SnRKV5; 500 ng) was incubated in extracts (30 μg) prepared from 7-d-old light-grown wild-type and 5ptase13 seedlings for the indicated times at 30°C and was analyzed by western blotting with an anti-V5 antibody. A and B, Extracts from seedlings grown on low nutrients were incubated in the presence or absence of 10 μm MG132. C, SnRKV5 protein degradation in extracts prepared from seedlings grown with no nutrients. D, SnRKV5 protein degradation in extracts prepared from seedlings grown with low nutrients (0.5× MS salts). Ponceau S-stained filters are shown as controls for equivalent loading. The blot images are representative of two independent experiments.
To test the hypothesis that 5PTase13 destabilizes SnRK1.1 under the no-nutrient conditions, we examined a 30-min time point for analysis as a midpoint to total SnRKV5 degradation. The data indicate no difference in SnRKV5 stability in wild-type and 5ptase13 extracts under the no-nutrient conditions (Fig. 7C). In contrast, when extracts are prepared from seedlings grown on low nutrients, we find less SnRKV5 accumulation in 5ptase13 extracts compared with wild-type extracts (Fig. 7D). We conclude that under the low-nutrient conditions, 5PTase13 is required to stabilize SnRKV5 protein and slow its degradation by the 26S proteasome pathway. Furthermore, the increased degradation of SnRKV5 seen with the low-nutrient conditions correlates well with the lower SnRK1 activity levels measured in 5ptase13 mutants under these same nutrient conditions (Fig. 6B). In contrast, 5PTase13 is not required to stabilize SnRKV5 when seedlings are grown with no nutrients, and this is consistent with the switch in SnRK1 activity levels we found in 5ptase13 mutants grown with no nutrients (Fig. 6A). However, since we did not observe an increase in SnRKV5 stability in 5ptase13 extracts prepared from the no-nutrient conditions, we speculate that there is an additional mechanism that influences the elevated SnRK1 activity in 5ptase13 mutants under these conditions.
Role of 5PTase13 in Root Growth under Different Nutrient Conditions
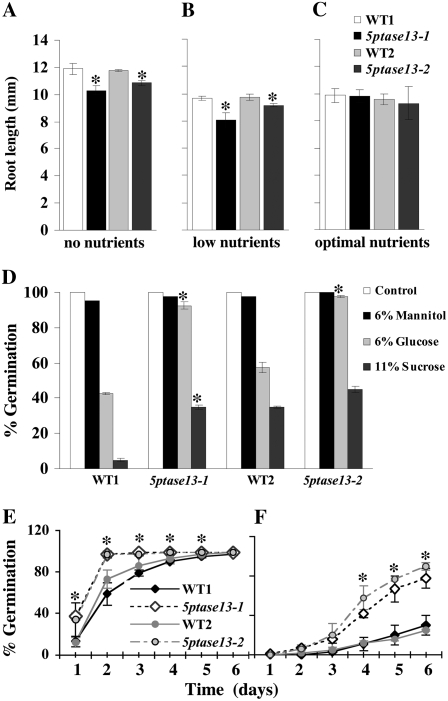
We compared 5ptase13 mutants with SnRK1.1 mutants, which have a small reduction in root growth under the low-nutrient conditions and exhibit enhanced root growth with 1% to 3% Suc (Baena-Gonzalez et al., 2007). 5ptase13-1 and 5ptase13-2 mutants exhibited reduced root growth under no-nutrient and low-nutrient conditions and no change in root growth when exogenously supplied with 3% Suc (Fig. 8, A–C). A similar trend in nutrient-dependent root growth reduction of 5ptase13 mutants was also found when seedlings were grown in the dark (data not shown). This indicates that along with a decrease in SnRK1 activity (Fig. 6B), 5ptase13 mutants grown on the low-nutrient conditions have the same root phenotype as the SnRK1.1 mutants (Baena-Gonzalez et al., 2007), suggesting that the decrease in root growth may be an outcome of decreased SnRK1 activity. However, since the 5ptase13 mutants grown with no nutrients have elevated SnRK1 activity (Fig. 6A), we conclude that under these conditions the decrease in 5ptase13 mutant root growth cannot be a function of lower SnRK1 activity and most likely involves another mechanism.
Figure 8.
Phenotypes of 5ptase13 mutants. A to C, Roles of 5PTase13 in root development under different nutrient conditions. Wild-type and 5ptase13 mutant (13-1 and 13-2) seeds were grown in low light for 6 d on 0.8% agar plates and no nutrients (A), low nutrients (0.5× MS salts; B), or optimal nutrients (0.5× MS salts, 3% Suc; C). The root length for each group of seedlings was measured on days 2, 4, and 6. The results represent the root length measured after 4 d. Values are means ± se (n ≥ 40). The experiment was independently repeated two times. D, Comparison of germination of wild-type and 5ptase13 seeds grown on 0.5× MS salts and 0.8% agar that were untreated (control) or treated with 6% Glc, 6% mannitol, or 11% Suc after 7 d in the dark. Values are means ± se (n = 50). The data are representative of three independent experiments. E and F, Wild-type and 5ptase13 mutant seeds were germinated in the light for 6 d on 0.5× MS salts, 0.8% agar, and either 1 μm ABA (E) or 3 μm ABA (F). The germination rate was scored for each group of seeds starting from day 1. Values are means ± se (n = 50). Significant differences from wild-type germination were noted at days 1 to 5 for 5ptase13-1 (1 μm ABA) and days 4 to 6 for both mutants (3 μm ABA). The experiment was independently repeated two times. *, P < 0.05 compared with the wild type.
5PTase13 Alterations Change Plant Sensitivity to Sugars and ABA
To test whether 5PTase13 is required for sugar and ABA responses, we analyzed age-matched seeds for germination in the presence of 0%, 1%, 3%, or 6% Glc or 0%, 1%, 2%, 3%, 6%, or 11% Suc. At low concentrations of sugar, there were no differences in the germination of wild-type and 5ptase13 seeds in the dark or light (data not shown). However, at a high exogenous sugar concentration (6% Glc and 11% Suc) in the dark, we found that 5ptase13 mutants were significantly less sensitive to sugar (Fig. 8D; Supplemental Figs. S3 and S4). We saw the same trend in sugar insensitivity when 5ptase13 mutant seeds were germinated in the presence of 6% Glc in the light, although the sugar insensitivity was less apparent (Supplemental Fig. S3). This sugar insensitivity was also noted for 5ptase13-1 and 5ptase13-2 mutant seeds germinated in the presence of 11% Suc, in which maximal increases of 2.5- and 2.2-fold for 5ptase13-1 and 5ptase13-2, respectively, were noted compared with wild-type seeds (Supplemental Fig. S4). Germination in the presence of mannitol, a nonmetabolizable sugar, was not altered, indicating that the sugar insensitivity of 5ptase13 mutants is not due to a general osmotic stress tolerance (Fig. 8D).
We also germinated 5ptase13 mutant seeds in the presence of 0, 1, 2, and 3 μm ABA and measured the impact on germination during a 6-d period. As expected, germination of wild-type seeds was delayed by ABA in a concentration-dependent manner during the 6-d period under light conditions (Fig. 8, E and F). In contrast, 5ptase13-1 and 5ptase13-2 mutant seeds were ABA insensitive, reaching 100% germination on day 2 in the presence of 1 μm ABA and 78% to 90% on day 6 in the presence of 3 μm ABA (Fig. 8, E and F). Since we did not find reduced seed dormancy in our mutants (data not shown), this ABA insensitivity of 5ptase13 mutants most likely does not correlate with changes in de novo synthesis of ABA (Gubler et al., 2005) and is in accordance with the previously reported results for ABA insensitivity of the 5ptase13-1 mutant (Lin et al., 2005).
Expression of Glc- and ABA-Regulated Genes Is Altered in 5ptase13 Mutants
To determine whether there are differences in the expression patterns of Glc- and/or ABA-regulated genes (RD29A, KIN1, and CAB1 genes; Price et al., 2004) in 5ptase13 mutants, we examined expression in 4-d-old dark-grown wild-type and 5ptase13 seedlings exposed to 6% Glc for 3 d (Fig. 9A). Treatment with 6% Glc strongly induced the expression of RD29A and KIN1 in wild-type seedlings. In contrast, 5ptase13-1 and 5ptase13-2 mutants grown in 6% Glc contained much smaller increases in RD29A and KIN1 transcript levels (Fig. 9A), indicating that induction of these genes is diminished, but not abolished, in 5ptase13 mutants. The regulation of CAB1, on the other hand, was not significantly altered in 5ptase13 mutants.
Figure 9.
A, Expression of Glc- and ABA-inducible genes from dark-grown 4-d-old wild-type and 5ptase13 seedlings that were either untreated (control; 0.5× MS salts, 0.8% agar) or treated with 6% Glc (0.5× MS salts, 0.8% agar) in the dark for 3 d. The experiment was independently repeated two times. B, Verification of overexpression of 5PTase13 in complemented lines and a 5PTase13:GFP line. Total RNA (1–2 μg) was isolated, and semiquantitative RT-PCR was performed with the indicated primers (Supplemental Table S4). C, The 5PTase13:GFP gene complements the ABA-insensitive phenotype of 5ptase13-1 mutants. Seeds from WT1, 5ptase13-1, and 5ptase13-1 containing a 5PTase13:GFP transgene were germinated and grown in the light for 6 d on 0.5× MS salts, 0.8% agar, and 3 μm ABA. The germination rate was scored for each group of seeds at day 3. Values are means ± se (n = 50). The experiment was independently repeated two times. *, P < 0.05 compared with the 5ptase13-1 mutant.
Complementation of the 5ptase13-1 Mutant
To ensure that the alterations noted in 5ptase13 mutants are due to loss of 5PTase13 expression, we expressed a 5PTase13:GFP fusion under the control of the 35S cauliflower mosaic virus promoter in wild-type and 5ptase13-1 plants (13:GFP and 13-1/13:GFP plants, respectively). We examined two lines of 13-1/13:GFP plants with good expression of the transgene (Fig. 9B) along with wild-type and 5ptase13-1 plants in ABA-sensitivity assays. Both 13-1/13:GFP lines exhibited more ABA sensitivity in germination assays compared with 5ptase13 mutants (Fig. 9C). Since 5ptase13-1 mutants contain decreased SnRK1 activity under low-nutrient conditions compared with wild-type plants (47%; Fig. 6B), we also measured SnRK1 activity in the 13-1/13:GFP-1 and 13-1/13:GFP-2 lines and found, as expected, similar or increased SnRK1 activity compared with wild-type plants (140% ± 10% for 13-1/13:GFP-1 and 102% ± 5% for 13-1/13:GFP-2). We conclude that expression of the 5PTase13:GFP transgene complements the 5ptase13-1 mutant.
Glc-Stimulated InsP3 Levels Are Altered in 5ptase13 Mutant Seedlings
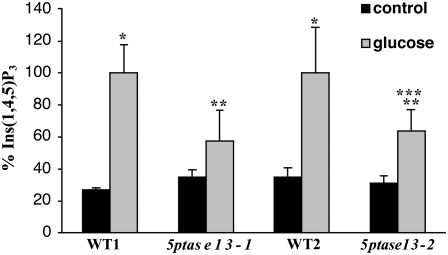
We examined whether the Glc insensitivity of 5ptase13 mutants is accompanied by alterations in mass InsP3 levels by measuring mass InsP3 levels. The results in Figure 10 indicate that neither 5ptase13-1 nor 5ptase13-2 mutant seedlings differ significantly from wild-type seedlings in their InsP3 mass levels under control conditions. When wild-type seedlings are exposed to 6% Glc for 3 d, mass InsP3 levels increase 2.8- to 3.7-fold, which is a statistically significant elevation. However, when 5ptase13 mutants are grown for 3 d in the presence of 6% Glc, mass InsP3 level changes are smaller, with an increase of 1.6- to 2-fold over basal levels, and statistically significant only in the 5ptase13-2 mutant. More importantly, the Glc-stimulated InsP3 levels in both 5ptase13 mutants differ significantly from the Glc-stimulated InsP3 levels in wild-type seedlings. We conclude that 5ptase13 mutants are impaired in their ability to accumulate InsP3 in response to Glc and that this correlates with the sugar and ABA insensitivity noted in the germination assays.
Figure 10.
Comparison of mass InsP3 levels in wild-type and 5ptase13 seedlings. Dark-grown 4-d-old wild-type and 5ptase13 seedlings were untreated (control; 0.5× MS salts, 0.8% agar) or treated with 6% Glc and 0.5× MS salts for 3 d. The seedlings were frozen in liquid nitrogen, ground, and analyzed for mass InsP3 levels as described in “Materials and Methods.” Values for InsP3 (pmol g−1) were normalized to the matched wild-type Glc-treated sample; raw values are found in “Materials and Methods.” Bars represent means ± se of three to five replicates. The experiment was repeated three times. *, P < 0.05 compared with the control, untreated sample; **, P < 0.10 compared with the matched wild-type Glc sample; ***, P < 0.10 compared with the control, untreated sample.
Subcellular Localization of the 5PTase13:GFP Fusion Protein
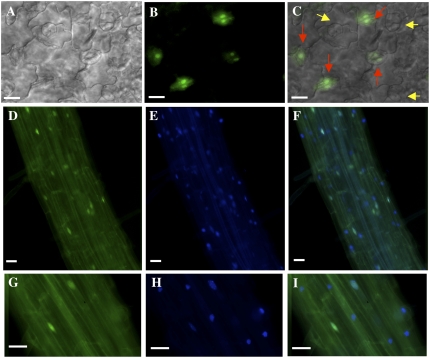
To investigate the subcellular location of the 5PTase13 protein, we performed imaging experiments with 5ptase13-1 mutants complemented with the 5PTase13:GFP construct (13-1/13:GFP plants) and wild-type plants containing the same 5PTase13:GFP construct. As the 5PTase13:GFP construct we used allowed for complementation of the ABA and sugar insensitivity phenotype in 5ptase13-1 mutants (Fig. 9, B and C), it is likely that this fusion protein undergoes the same posttranslational modifications and subcellular localization as the native 5PTase13 protein. We analyzed T2 progeny from two independent 13-1/13:GFP lines with fluorescence deconvolution microscopy and found a similar pattern in both lines. GFP fluorescence was associated with the nucleus in many, but not all, cells in cotyledon epidermis (Fig. 11, B and C), hypocotyls (data not shown), and roots (Fig. 11, D–I). Nuclei from some but not all guard cells contained the 13:GFP protein (Fig. 11C). To confirm the nuclear localization, we stained 13-1/13:GFP seedlings with the nuclear dye 4′,6-diamidino-2-phenylindole (DAPI) and imaged GFP and DAPI fluorescence simultaneously (Fig. 11, D–I). Once again, we found 13:GFP fluorescence in only a portion of root nuclei, while DAPI fluorescence was present in all nuclei. We conclude that 5PTase13 protein is located in the nucleus of seedlings and that its presence in the nucleus is restricted in some cells.
Figure 11.
Subcellular localization of GFP-tagged 5PTase13 in Arabidopsis seedlings. GFP-tagged 5PTase13 (13:GFP) was expressed in the 5ptase13-1 mutant background, and the subcellular location in cotyledons (A–C) and roots (D–I) from 7-d-old seedlings was examined with fluorescence deconvolution microscopy. A, Differential interference contrast. B, D, and G, GFP fluorescence. C, Overlay of A and B. E and H, Seedlings were stained with DAPI and examined with a standard UV fluorescence filter set. F and I, Overlay of GFP and DAPI fluorescence. Note that red arrows in C indicate nuclei from guard cells expressing 13:GFP, while yellow arrows correspond to nuclei lacking expression. Bars = 20 μm.
DISCUSSION
Since control of second messengers is critical for signaling, there is interest in determining how the plant cell regulates levels of second messengers such as InsP3. We identified a potential regulator of InsP3 signaling by isolating protein interactors of 5PTase13. We focused on 5PTase13 because it contains several conserved WD40 repeats (for review, see van Nocker and Ludwig, 2003) and the presence of WD40 repeats within 5PTase genes is unique to plants and certain fungi (Zhong and Ye, 2004). To date, the WD40 repeats of 5PTases have not been functionally characterized, and no potential protein partners have been identified. We show here an important and novel interaction between the WD40 repeat region of 5PTase13 and the central metabolic regulator, SnRK1.1. SnRK1.1 and its orthologues in yeast and mammals is one subunit of a complex that modulates cellular metabolism in response to nutrient and environmental conditions (Baena-Gonzalez et al., 2007; Hardie, 2007; Hue and Rider, 2007). Recent work in Arabidopsis has elegantly delineated the role of SnRK 1.1 in the transcriptional activation of genes from catabolic pathways that provide routes for alternative energy sources and coordinate repression of a large number of anabolic genes (Baena-Gonzalez et al., 2007). Our discovery of a 5PTase13:SnRK1.1 complex points to the novel interaction of this metabolic modulator and InsP3 signaling.
It has been shown previously that a loss of function in the 5PTase13 gene leads to defects in auxin-regulated development (Lin et al., 2005) and in blue light responses (Chen et al., 2008). Because of the 5PTase13 interaction with SnRK1.1, we investigated the role of 5PTase13 in nutrient sensing and stress responses. Recent studies have shown that Arabidopsis plants overexpressing SnRK1.1 have greater Suc sensitivity and that mutants lacking SnRK1.1 use exogenous sugars more efficiently (Baena-Gonzalez et al., 2007). When virus-induced gene silencing is used to silence both SnRK1.1 and SnRK1.2, growth is severely affected, indicating the importance of the SnRK1 modulator to overall growth (Baena-Gonzalez et al., 2007). Repressing a pea (Pisum sativum) homolog of SnRK1 with antisense RNA results in seed maturation effects resembling ABA insensitivity (Radchuk et al., 2006), while a loss of function in SnRK1 in rice (Oryza sativa) delays seed germination and seedling growth (Lu et al., 2007). We report here that 5ptase13 mutants exhibit reduced root growth under limited nutrient conditions, but not when 3% Suc is supplied. The variability of the root growth phenotype under different nutrient conditions is thus similar, although not identical, to growth responses of SnRK1.1 mutants. In addition, we documented that 5ptase13 mutants are altered in their responses to ABA and sugar stress (Figs. 8–10). When 5ptase13 mutants are challenged with a sugar stress, alterations in germination, gene expression, and InsP3 accumulation occur, indicating that 5ptase13 mutants are sugar insensitive. This sugar insensitivity is similar to that noted for the regulator of G-protein signaling1 mutant (Chen et al., 2006b) but differs from the Glc-insensitive (Zhou et al., 1998; Arenas-Huertero et al., 2000; Moore et al., 2003; Lin et al., 2007) and sugar-insensitive (Laby et al., 2000; Gibson et al., 2001) mutants that are also insensitive to the developmental arrest induced by sugar that occurs over a period of 21 d. Since there are other WD40-containing 5PTases, we speculate that the sensitivity to developmental arrest noted in 5ptase13 mutants grown on high sugar may be due to this redundancy. Together, the data on nutrient and sugar stress responses of 5ptase13 mutants are consistent with the existence of a 5PTase13:SnRK1.1 complex that modulates seedling responses to nutrient conditions and stress.
To understand how a 5PTase13:SnRK1.1 complex might regulate nutrient and stress signaling, we measured SnRK1 activity in 5ptase13 mutants and wild-type seedlings grown under different nutrient conditions. Our data show that the presence of 5PTase13 affects SnRK1 activity and that nutrient availability is an important switch in 5PTase13 regulation of SnRK1.1. Specifically, 5PTase13 is required to maintain wild-type levels of SnRK1 activity when low nutrient or high sugar is present (Fig. 6, B and C). In contrast, under the greatest starvation conditions of no added nutrients, 5PTase13 appears to be a negative regulator of SnRK1, as 5ptase13 mutants contain significantly elevated SnRK1 activity (Fig. 6A). To further delineate the mechanism of 5PTase13 regulation of SnRK1.1, we used a cell-free degradation assay and showed that the decrease in SnRK1 activity in 5ptase13 mutants correlates with increased degradation of SnRK1.1 by the proteasome (Fig. 7D).
Interestingly, another WD40 repeat-containing protein called PRL1 (for PLEITROPIC REGULATOR LOCUS1) also interacts with SnRK1.1 and is able to inhibit the activity of SnRK1.1 and SnRK1.2 (Bhalerao et al., 1999; Farras et al., 2001). Recent evidence shows that there is a decrease in SnRK1 degradation in prl1 mutant extracts, and additional evidence suggests that PRL1 acts in delivering SnRK1 to the CUL4-DDB1 complex for proteasomal degradation (Lee et al., 2008). Thus, we predict that 5PTase13 and PRL1 have opposing functions regarding SnRK1 stability under low-nutrient and sugar-stress conditions. The fact that prl1 mutants are ABA and sugar hypersensitive (Bhalerao et al., 1999), which is the opposite of 5ptase13 mutants, also supports opposing roles for 5PTase13 and PRL1 in regulating SnRK1 degradation.
Our data reported here, along with data from other groups, support the model of SnRK1 as a sensor of low nutrient status and cellular stress. Under low-nutrient or sugar-stress conditions, 5PTase13 acts as a positive regulator of SnRK1.1 activity by reducing the amount of SnRK1 targeted for proteasomal destruction. This regulation most likely involves a 5PTase13-SnRK1.1 interaction within the nucleus, as that is where we find 5PTase13:GFP (Fig. 11), and SnRK1 is most likely nuclear as well (Pierre et al., 2007; Thelander et al., 2007). A loss of 5PTase function, therefore, results in a lack of response, or insensitivity, to stress conditions. In contrast, nuclear PRL1 acts as a negative regulator of SnRK1.1 and facilitates its proteasomal destruction. Thus, the increased SnRK1 levels found in prl1 mutants could potentiate low-nutrient and/or stress signaling, resulting in the hypersensitivity to sugar and ABA found in this mutant. Nemeth et al. (1998) showed that PRL1 is located in the nucleus, as are CUL4 and other SnRK1-interacting proteasomal components such as SKP/ASK1 and a4/PAD1 (Farras et al., 2001; Chen et al., 2006a).
While data support this model, we currently do not know how second messenger InsP3 affects the SnRK1 pathway. We have shown that InsP3 levels increase when seedlings are given a sugar stress, implicating InsP3 as a second messenger under high-sugar conditions (Fig. 10). Given that the 5PTase13 enzyme has been shown to hydrolyze InsP3 in vitro (Zhong and Ye, 2004), we expected that a loss of 5PTase13 function would elevate InsP3. However, 5ptase13 mutants have, instead, lower InsP3 levels upon sugar stress (Fig. 10). The lack of increase in Glc-stimulated InsP3 accumulation in 5ptase13 mutants may indicate a more complex role for InsP3 in 5PTase13 function and/or could be the result of compensatory actions of the other 5PTase enzymes. Elevated InsP3 levels have been found in other 5ptase mutants, and this increase in InsP3 is sometimes associated with ABA hypersensitivity (Carland and Nelson, 2004; Gunesekera et al., 2007). It should be noted that 5ptase13 mutant seeds are also ABA insensitive, a trait they share with other sugar-insensitive mutants. It is currently thought that the sugar-induced delay of germination involves the action of ABA, as ABA synthesis mutants are also resistant to the effects of sugars (Price et al., 2003; Rognoni et al., 2007).
There have been previous reports that inositol signaling and sugar sensing/metabolism are linked. Im et al. (2007) found that expressing a human phosphatidylinositol 4-phosphate 5-kinase (PIP5K) gene in tissue culture cells dramatically elevated phosphatidylinositol 4,5-bisphosphate (PtdInsP2) and InsP3 and resulted in increased sugar use and oxygen uptake. This work importantly defined the PIP5K enzyme as the flux-limiting enzyme for PtdInsP2 production and InsP3 signaling in plant cells. In separate studies, Lou et al. (2007) found that one member of the PIP5K gene family, At3g09920, negatively regulates a cytosolic invertase by direct protein interactions. These investigators found that a gain of function in this same PIP5K gene results in less invertase activity and a resulting sugar insensitivity in transgenic seedlings. Additional evidence supporting the importance of PtdInsP2 in sugar sensing/metabolism comes from transcriptional studies. A rigorous correlative analysis of gene expression shows that a different PIP5K gene (At4g17080) is one of 278 genes coactivated by SnRK1.1 and sugar starvation and down-regulated by sugar treatment (Baena-Gonzalez et al., 2007). Together, these studies indicate that PIP5K-catalyzed production of PtdInsP2 may play a critical role in mediating the response of plant cells to sugar. In addition to transcriptional changes in PIP5K, protoplasts overexpressing SnRK1.1 repress two genes required for inositol synthesis (At4g39800 and At3g02870) and activate three genes encoding other 5PTases (5PTase1, 5PTase12, and FRA3; Baena-Gonzalez et al., 2007). These alterations indicate the likelihood that the SnRK1 modulator decreases the energy-consuming synthesis of inositol but increases the ability of the plant cell to hydrolyze inositol-containing second messengers such as InsP3.
Together, the data presented here support a unique role for 5PTase13 functioning as a binding partner and regulator of the SnRK1.1 modulator of energy and stress signaling. Given the previously established role of 5PTase13 in blue light signaling (Chen et al., 2008), this work identifies a new connection between inositol signaling and the sensing of cellular metabolism, energy, light, and stress.
MATERIALS AND METHODS
Phylogenetic Tree Construction
BLASTp was used to identify 12 related WD40 regions. ClustalW and PAUP4.0 were used to generate phylogenetic trees using parsimony. One thousand bootstraps were performed.
Yeast Two-Hybrid Screen
The Matchmaker Two-Hybrid System 3 was used (BD Biosciences Clontech). The cDNA corresponding to amino acids 1 to 533 of 5PTase13 was amplified by PCR and ligated into pGBKT7 bait vector (BD Biosciences Clontech), verified by DNA sequencing, and transformed into yeast strain AH109. The yeast strain AH109 containing the 5PTase13 WD40 repeat domain was transformed with an Arabidopsis (Arabidopsis thaliana) 3-d-old etiolated seedling cDNA library (Thelogis). Screening for interactors was performed with SD/-Ade-His-Leu-Trp, 10 mm 3-amino-1,2,4-triazole, and 20 μg mL−1 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside plates. Candidate clones that grew were rescued from yeast and retested in the original bait and control strains. Prey plasmids that passed all tests were sequenced to identify the Arabidopsis gene insert.
Mutant Isolation
Arabidopsis ecotype Columbia was used for all experiments. Growth conditions of soil-grown plants have been described (Berdy et al., 2001). Regular-light conditions were 100 μE for 16-h days, while low-light conditions were 40 μE for 16-h days. 5ptase13-1 (At1g05630, SAIL_350_F1) and 5ptase13-2 (SALK_081991) mutants were identified from the SIGnAL database (Alonso et al., 2003). Methods for PCR screening of mutants has been described (Ercetin et al., 2008) and utilized SALK and SAIL LB primers and 5PTase13 gene-specific primers (13-1for, 13-1rev, 13-2for, and 13-2rev; Supplemental Table S4). The resulting PCR fragments were sequenced to map the T-DNA insertions.
Reverse Transcription-PCR
Conditions for reverse transcription (RT)-PCR have been described previously (Ercetin et al., 2008) and used the primers listed in Supplemental Table S4. Amplification of the WD40 region of 5PTase13 was performed using primers 13WD40for and 13WD40rev. Conditions for actin (Berdy et al., 2001), KIN1 (Knight et al., 1998), and RD29A (Sanchez and Chua, 2001) amplification have been described and generate 428-, 342-, and 715-bp products, respectively. Each RT-PCR experiment was independently repeated at least two times to verify the observed changes in expression.
Seedling Growth and Seed Germination Assays
Age-matched seeds used for assays were harvested from plants grown in parallel on the same shelf in a growth room, and seeds were harvested on the same day and ripened for 6 weeks at room temperature. Seeds were surface sterilized and plated on no salts or 0.5× MS salts solution (pH 5.7) containing 0.8% agar. Seeds were stratified on plates at 4°C for 3 d and germinated at 23°C in the light or dark. For seed germination and root growth assays, seeds were plated on medium containing 0%, 1%, 3%, or 6% Glc or mannitol (Sigma-Aldrich) or 0%, 1%, 2%, 3%, 6%, or 11% Suc (Sigma-Aldrich) in the light or dark at 23°C. Germination was scored as positive when the radicle protruded through the seed coat. For ABA sensitivity experiments, ABA (Sigma-Aldrich) was dissolved in 100% ethanol and added to cooled, sterile medium at a final concentration of 1, 2, or 3 μm ABA. The germination assay was performed as before. Hormone/sugar treatment experiments were repeated two or three times.
Immunoprecipitations
The entire open reading frame (encoding 535 amino acids) of At3g01090 was amplified with gene-specific primers SnRK1.1pFor and SnRK1.1pRev (Supplemental Table S4) and ligated to pCRT7/CT-TOPO vector. This resulted in pSnRKV5, which directed expression of a 61.7-kD SnRK1.1 protein with a C-terminal 6xHis tag and V5 epitope tag (Invitrogen). The WD40 repeat region of 5PTase13 was amplified with gene-specific primers WD40For and WD40Rev (Supplemental Table S4) and ligated to pCRT7/NT-TOPO vector. This resulted in p13WDX, which directed expression of a 65.9-kD protein that corresponds to the WD40 region of 5PTase13 and an N-terminal 6xHis tag and Xpress epitope (Invitrogen). For expression, pSnRKV5 and p13WDX were transformed in Escherichia coli BL21(DE3) pLysS cells (Invitrogen), and the resulting recombinant proteins 13WDX and SnRKV5 were purified using nickel-nitrilotriacetic acid agarose columns (Invitrogen). SnRKV5 protein appeared on the SDS protein gel as two separate bands. These bands represented an active SnRKV5 recombinant protein as determined by the SnRK1 activity assay.
For the immunoprecipitation, between 40 and 90 ng of the purified and dialyzed proteins was used. The immunoprecipitation reactions were carried out as described by Ercetin et al. (2008). Western-blot conditions were as described previously (Burnette et al., 2003). For detection of either the V5 or the Xpress epitope tag, a 1:5,000 dilution of the mouse anti-V5 or anti-Xpress monoclonal antibody, followed by a 1:20,000 dilution of goat anti-mouse horseradish peroxidase-conjugated antibody (Amersham), was used. Purified positope protein (175 ng; Invitrogen) was loaded on the same gel to estimate the amount of recombinant V5- or Xpress-tagged proteins.
SnRK1 Activity Assay
Whole 7-d-old seedlings were ground in liquid nitrogen and resuspended in extraction buffer (50 mm Tris-HCl, pH 7.8, 1 mm EGTA, 1 mm EDTA, 2 mm dithiothreitol [DTT], 0.05% Nonidet P-40, 0.5 mm phenylmethylsulfonyl fluoride, 1 mm benzamide, 3 mm glycerophosphate, and plant protease cocktail; Sigma). After centrifugation at 13.2 rpm for 15 min at 4°C, ammonium sulfate was slowly added to the supernatant to 40% saturation while stirring for 10 min at 4°C and centrifuged for 15 min at 13.2 rpm and 4°C. Precipitated protein was resuspended in 50 μL of fractionation buffer (50 mm Tris-HCl, pH 7.8, 1 mm EGTA, 1 mm EDTA, 2 mm DTT, 0.05% Nonidet P-40, 0.5 mm phenylmethylsulfonyl fluoride, 1 mm benzamide, 3 mm glycerophosphate, 10% glycerol, and plant protease cocktail; Sigma). Protein concentration was determined according to the Bradford method (Bradford, 1976), and equal amounts of protein were added to each assay. SnRK1 activity assay was performed as described by Radchuk et al. (2006) with slight modifications. Protein extract (5 μg in 5 μL) was mixed with 5 μL of kinase buffer (50 mm Tris-HCl and 1 mm DTT, pH 7.0), 5 μL of sterile water, 5 μL of SPS peptide stock solution (peptide sequence RDHMPRIRSEMQIWSED; 200 μm), and 5 μL of labeled ATP stock solution (5 mm [γ-32P]ATP, 5 mm unlabeled ATP, and 125 mm magnesium chloride). The samples were incubated for 30 min at 30°C, and 10-μL aliquots were spotted twice onto P81 paper. The pieces were washed three times in 125 mm phosphoric acid for 20 min each and transferred to scintillation vials for counting. Two different control reactions were examined along with each reaction assay. The first reaction control contained no SPS peptide, and the second control contained no protein extract. Activity was expressed as nanomoles of phosphate incorporated into peptide per minute per milligram of protein. The assay was performed using two independently prepared extracts and three replicates of each extract.
Cell-Free Degradation Assay
Conditions described by Lee et al. (2008) were followed. Briefly, 7-d-old light-grown seedlings were ground in liquid nitrogen, resuspended in buffer (25 mm Tris, pH 7.5, 10 mm MgCl2, 5 mm DTT, and 10 mm NaCl), and centrifuged at 13.2 rpm for 10 min at 4°C. Purified recombinant SnRK1.1-V5 (SnRKV5; 500 ng) and total cell extracts (30 μg) were mixed in a reaction buffer (25 mm Tris, pH 7.5, 10 mm MgCl2, 5 mm DTT, 10 mm NaCl, and 10 mm ATP) and incubated at 30°C for the indicated times. Reactions were analyzed by SDS-PAGE, followed by western blotting with a 1:10,000 dilution of goat anti-mouse horseradish peroxidase-conjugated anti-V5 antibody (Amersham). Ponceau S staining was performed to ensure that equivalent amounts of extracts were analyzed.
Mutant Complementation and GFP Imaging
The 3,408-bp coding region of 5PTase13 minus the stop codon was amplified by high-fidelity PCR, confirmed by sequencing, cloned into the pENTR/D-TOPO vector (Invitrogen), and recombined via the Gateway system (Invitrogen) using the manufacturer's instructions into pK7FWG2. The resulting 35S cauliflower mosaic virus promoter:5PTase13:GFP construct was transformed into Agrobacterium tumefaciens by cold shock and was used in the transformation of 5ptase13-1 and wild-type plants as described (Bechtold et al., 1993). 13-1/13:GFP seedlings were identified on kanamycin plates and by screening for GFP production using a Zeiss Axioimager microscope equipped with fluorescence optics. Two independent complemented lines with detectable GFP expression (13-1/13:GFP-1 and 13-1/13:GFP-2) were used in growth assays and for subcellular localization. Seven-day-old seedlings were used for imaging utilizing Axiovision software (Zeiss). Z-stack series of 15 to 20 1-μm sections were collected, and deconvolution with an iterative algorithm was applied. The resulting deconvolved images were reconstituted into a single image using the maximal intensity projection function of Axiovision. To visualize nuclei, seedlings were stained with 1 μg mL−1 DAPI (Molecular Probes) solution for 5 min, excess liquid was removed, and the seedlings were mounted in water. Photographs were taken with a Zeiss MC100 camera. GFP was imaged with a filter set consisting of an excitation filter of 540 to 580 nm, a dichroic mirror of 595 nm, and a barrier filter of 600 to 660 nm. DAPI staining was visualized with a standard UV fluorescence filter set.
Extraction and Measurement of Mass InsP3
Filters containing whole 4-d-old dark-grown seedlings were floated on a 6% Glc solution (0.5× MS salts, pH 5.7) or on a control solution (0.5× MS salts, pH 5.7, only) in the dark for 3 d and then frozen in liquid nitrogen at the end of the treatment. Tissues were harvested and mass InsP3 measurements were made as described previously (Gunesekera et al., 2007; Ercetin et al., 2008). The assays were performed in triplicate, and the experiment was repeated two times. Raw values for InsP3 in Glc-treated samples are as follows: WT1, 1,313 ± 203 pmol g−1; WT2, 1,313 ± 372 pmol g−1.
Histological Characterization
The histological analysis was performed as described (Carland et al., 1999) with slight modifications. Seven-day-old wild-type and mutant cotyledons of soil-grown seedlings in a growth room were fixed in ethanol:chloroform:acetic acid solution (6:3:1) overnight at 4°C and cleared sequentially in 80% ethanol, 95% ethanol overnight at 4°C, and 10% NaOH for 1 h at 42°C. Cotyledons then were stained briefly (30 s to 2 min) in 0.002% safranin-O (Sigma). Specimens were mounted on slides in 50% glycerol and visualized with differential interference contrast on a Zeiss Axioimager microscope and Spot digital camera (Zeiss). A total of 50 cotyledons per variant were used.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: 5PTase13, At1g05630, NP_172054; SnRK1.1, At3g01090, NP_001118546; 5PTase12, At2g43900, NP_181918; 5PTase14, At2g31830, NP_180742; FRA3, At1g65580, NP_176736.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. GENEVESTIGATOR expression analysis.
Supplemental Figure S2. Validation of the SnRK1 activity assay.
Supplemental Figure S3. 5ptase13 mutants are less sensitive to 6% Glc under light.
Supplemental Figure S4. 5ptase13 mutants are less sensitive to 11% Suc under light.
Supplemental Table S1. Identity of the WD40 regions of 5PTases in Arabidopsis.
Supplemental Table S2. Normal cotyledon vein development patterns in wild-type and 5ptase13 plants.
Supplemental Table S3. Abnormal cotyledon vein development patterns in wild-type and 5ptase13 plants.
Supplemental Table S4. Primer sequences.
Supplementary Material
Acknowledgments
This paper is dedicated to the memory of Emily Jane Hilscher. We are grateful to SIGnAL and the Arabidopsis Biological Resource Center for supplying mutant seeds. We also thank Dr. P.J. Kennelly (Virginia Tech) for advice on the SnRK1 activity assay, Dr. Jae-Hoon Lee and Dr. Xing Wang Deng (Yale University) for advice on the cell-free degradation assay, and Janet Donahue (Virginia Tech) for assistance with InsP3 measurements.
This work was supported by the National Science Foundation (grant no. MCB–0641954 to G.E.G.) and the U.S. Department of Agriculture (grant no. 2003–35318–13690 to G.E.G.) and by Hatch program funds (grant no. VA–135583).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Glenda E. Gillaspy (gillaspy@vt.edu).
The online version of this article contains Web-only data.
Open Access articles can be viewed online without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Andersson MX, Kourtchenko O, Dangl JL, Mackey D, Ellerstrom M (2006) Phospholipase-dependent signalling during the AvrRpm1- and AvrRpt2-induced disease resistance responses in Arabidopsis thaliana. Plant J 47 947–959 [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, Leon P (2000) Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes Dev 14 2085–2096 [PMC free article] [PubMed] [Google Scholar]
- Astle MV, Horan KA, Ooms LM, Mitchell CA (2007) The inositol polyphosphate 5-phosphatases: traffic controllers, waistline watchers and tumour suppressors? Biochem Soc Symp 74 161–181 [DOI] [PubMed] [Google Scholar]
- Baena-Gonzalez E, Rolland F, Thevelein JM, Sheen J (2007) A central integrator of transcription networks in plant stress and energy signalling. Nature 448 938–942 [DOI] [PubMed] [Google Scholar]
- Bechtold N, Ellis J, Pelletier G (1993) In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C R Acad Sci Ser III Sci Vie 316 1194–1199 [Google Scholar]
- Berdy S, Kudla J, Gruissem W, Gillaspy G (2001) Molecular characterization of At5PTase1, an inositol phosphatase capable of terminating IP3 signaling. Plant Physiol 126 801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ (1993) Inositol trisphosphate and calcium signaling. Nature 361 315–325 [DOI] [PubMed] [Google Scholar]
- Berridge MJ (2007) Inositol trisphosphate and calcium oscillations. Biochem Soc Symp 74 1–7 [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Salchert K, Bako L, Okresz L, Szabados L, Muranaka T, Machida Y, Schell J, Koncz C (1999) Regulatory interaction of PRL1 WD protein with Arabidopsis SNF1-like protein kinases. Proc Natl Acad Sci USA 96 5322–5327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss WF, Davis AJ, Im YJ, Galvão RM, Perera IY (2006) Phosphoinositide metabolism: towards an understanding of subcellular signaling. Subcell Biochem 39 181–205 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72 248–254 [DOI] [PubMed] [Google Scholar]
- Burnette RN, Gunesekera BM, Gillaspy GE (2003) An Arabidopsis inositol 5-phosphatase gain-of-function alters abscisic acid signaling. Plant Physiol 132 1011–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland FM, Berg BL, FitzGerald JN, Jinamornphongs S, Nelson T, Keith B (1999) Genetic regulation of vascular tissue patterning in Arabidopsis. Plant Cell 11 2123–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carland FM, Nelson T (2004) Cotyledon vascular pattern2-mediated inositol (1,4,5) triphosphate signal transduction is essential for closed venation patterns of Arabidopsis foliar organs. Plant Cell 16 1263–1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Shena Y, Tang X, Yb Y, Wang J, Guoa L, Zhang Y, Zhang H, Feng S, Strickland E, et al (2006. a) Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell 18 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Lin WH, Wang Y, Luan S, Xue HW (2008) An inositol polyphosphate 5-phosphatase functions in PHOTOTROPIN1 signaling in Arabidopsis by altering cytosolic Ca2+. Plant Cell 20 353–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Ji F, Xie H, Liang J, Zhang J (2006. b) The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination. Plant Physiol 140 302–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement S, Krause U, Desmedt F, Tanti JF, Behrends J, Pesesse X, Sasaki T, Penninger J, Doherty M, Malaisse W, et al (2001) The lipid phosphatase SHIP2 controls insulin sensitivity. Nature 409 92–97 [DOI] [PubMed] [Google Scholar]
- DeWald DB, Torabinejad J, Jones CA, Shope JC, Cangelosi AR, Thompson JE, Prestwich GD, Hama H (2001) Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed Arabidopsis. Plant Physiol 126 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercetin M, Ananieva E, Safaee N, Robinson J, Gillaspy G (2008) Phosphoinositide-specific myo-inositol polyphosphate 5-phosphatase required for seedling growth. Plant Mol Biol 67 375–388 [DOI] [PubMed] [Google Scholar]
- Farras R, Ferrando A, Jasik J, Kleinow T, Okresz L, Tiburcio A, Salchert K, del Pozo C, Schell J, Koncz C (2001) SKP1-SnRK protein kinase interactions mediate proteasomal binding of a plant SCF ubiquitin ligase. EMBO J 20 2742–2756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson SI, Laby RJ, Kim D (2001) The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochem Biophys Res Commun 280 196–203 [DOI] [PubMed] [Google Scholar]
- Gubler F, Millar AA, Jacobsen JV (2005) Dormancy release, ABA and pre-harvest sprouting. Curr Opin Plant Biol 8 183–187 [DOI] [PubMed] [Google Scholar]
- Gunesekera B, Torabinejad J, Robinson J, Gillaspy GE (2007) Inositol polyphosphate 5-phosphatases 1 and 2 are required for regulating seedling growth. Plant Physiol 143 1408–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG (2007) AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol 8 774–785 [DOI] [PubMed] [Google Scholar]
- Huang JZ, Huber SC (2001) Phosphorylation of synthetic peptides by a CDPK and plant SNF1-related protein kinase: influence of proline and basic amino acid residues at selected positions. Plant Cell Physiol 42 1079–1087 [DOI] [PubMed] [Google Scholar]
- Hue L, Rider MH (2007) The AMP-activated protein kinase: more than an energy sensor. Essays Biochem 43 121–137 [DOI] [PubMed] [Google Scholar]
- Im YJ, Perera IY, Brglez I, Davis AJ, Stevenson-Paulik J, Phillippy BQ, Johannes E, Allen NS, Boss WF (2007) Increasing plasma membrane phosphatidylinositol(4,5)bisphosphate biosynthesis increases phosphoinositide metabolism in Nicotiana tabacum. Plant Cell 19 1603–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Smirnoff N (2006) Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J 45 83–100 [DOI] [PubMed] [Google Scholar]
- Kagawa S, Soeda Y, Ishihara H, Oya T, Sasahara M, Yaguchi S, Oshita R, Wada T, Tsuneki H, Sasaoka T (2008) Impact of transgenic overexpression of SH2-containing inositol 5′-phosphatase 2 on glucose metabolism and insulin signaling in mice. Endocrinology 149 642–650 [DOI] [PubMed] [Google Scholar]
- Knight H, Brandt S, Knight MR (1998) A history of stress alters drought calcium signalling pathways in Arabidopsis. Plant J 16 681–687 [DOI] [PubMed] [Google Scholar]
- Krinke O, Novotna Z, Valentova O, Martinec J (2007) Inositol trisphosphate receptor in higher plants: is it real? J Exp Bot 58 361–376 [DOI] [PubMed] [Google Scholar]
- Laby RJ, Kincaid MS, Kim D, Gibson SI (2000) The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. Plant J 23 587–596 [DOI] [PubMed] [Google Scholar]
- Lee JH, Terzaghi W, Gusmaroli G, Charron JB, Yoon HJ, Chen H, He YJ, Xiong Y, Deng XW (2008) Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 20 152–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim YW, Jeon BW, Park KY, Suh SJ, Seo J, Kwak JM, Martinoia E, Hwang I (2007) Phosphatidylinositol 4,5-bisphosphate is important for stomatal opening. Plant J 5 803–816 [DOI] [PubMed] [Google Scholar]
- Li D, Roberts R (2001) WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci 58 2085–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PC, Hwang SG, Endo A, Okamoto M, Koshiba T, Cheng WH (2007) Ectopic expression of ABSCISIC ACID2/GLUCOSE INSENSITIVE1 in Arabidopsis promotes seed dormancy and stress tolerance. Plant Physiol 143 745–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WH, Wang Y, Mueller-Roeber B, Brearley CA, Xu ZH, Xue HW (2005) At5PTase13 modulates cotyledon vein development through regulating auxin homeostasis. Plant Physiol 139 1677–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y, Gou JY, Xue HW (2007) PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar-mediated root growth. Plant Cell 19 163–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu CA, Lin CC, Lee KW, Chen JL, Huang LF, Ho SL, Liu HJ, Hsing YI, Yu SM (2007) The SnRK1A protein kinase plays a key role in sugar signaling during germination and seedling growth of rice. Plant Cell 19 2484–2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300 332–336 [DOI] [PubMed] [Google Scholar]
- Nemeth K, Salchert K, Putnoky P, Bhalerao R, Koncz-Kalman Z, Stankovic-Stangeland B, Bako L, Mathur J, Okresz L, Stabel S, et al (1998) Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev 12 3059–3073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega X, Perez LM (2001) Participation of the phosphoinositide metabolism in the hypersensitive response of Citrus limon against Alternaria alternata. Biol Res 34 43–50 [DOI] [PubMed] [Google Scholar]
- Perera IY, Heilmann I, Chang SC, Boss WF, Kaufman PB (2001) A role for inositol 1,4,5-trisphosphate in gravitropic signaling and the retention of cold-perceived gravistimulation of oat shoot pulvini. Plant Physiol 125 1499–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera IY, Hung CY, Brady S, Muday GK, Boss WF (2006) A universal role for inositol 1,4,5-trisphosphate-mediated signaling in plant gravitropism. Plant Physiol 140 746–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre M, Traverso JA, Boisson B, Domenichini S, Bouchez D, Giglione C, Meinnel T (2007) N-Myristoylation regulates the SnRK1 pathway in Arabidopsis. Plant Cell 19 2804–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Laxmi A, St Martin SK, Jang JC (2004) Global transcription profiling reveals multiple sugar signal transduction mechanisms in Arabidopsis. Plant Cell 16 2128–2150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J, Li TC, Kang SG, Na JK, Jang JC (2003) Mechanisms of glucose signaling during germination of Arabidopsis. Plant Physiol 132 1424–1438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radchuk R, Radchuk V, Weschke W, Borisjuk L, Weber H (2006) Repressing the expression of the SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE gene in pea embryo causes pleiotropic defects of maturation similar to an abscisic acid-insensitive phenotype. Plant Physiol 140 263–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rognoni S, Teng S, Arrus L, Smeekens S, Perata P (2007) Sugar effects on early seedling development in Arabidopsis. Plant Growth Regul 52 217–228 [Google Scholar]
- Sanchez JP, Chua NH (2001) Arabidopsis plc1 is required for secondary responses to abscisic acid signals. Plant Cell 13 1143–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaoka T, Hori H, Wada T, Ishiki M, Haruta T, Ishihara H, Kobayashi M (2001) SH2-containing inositol phosphatase 2 negatively regulates insulin-induced glycogen synthesis in L6 myotubes. Diabetologia 44 1258–1267 [DOI] [PubMed] [Google Scholar]
- Sasaoka T, Wada T, Fukui K, Murakami S, Ishihara H, Suzuki R, Tobe K, Kadowaki T, Kobayashi M (2004) SH2-containing inositol phosphatase 2 predominantly regulates Akt2, and not Akt1, phosphorylation at the plasma membrane in response to insulin in 3T3-L1 adipocytes. J Biol Chem 279 14835–14843 [DOI] [PubMed] [Google Scholar]
- Takahashi S, Katagiri T, Hirayama T, Yamaguchi-Shinozaki K, Shinozaki K (2001) Hyperosmotic stress induces a rapid and transient increase in inositol 1,4,5-trisphosphate independent of abscisic acid in Arabidopsis cell culture. Plant Cell Physiol 42 214–222 [DOI] [PubMed] [Google Scholar]
- Tang RH, Han S, Zheng H, Cook CW, Choi CS, Woerner TE, Jackson RB, Pei ZM (2007) Coupling diurnal cytosolic Ca2+ oscillations to the CAS-IP3 pathway in Arabidopsis. Science 315 1423–1426 [DOI] [PubMed] [Google Scholar]
- Thelander M, Nilsson A, Olsson T, Johansson M, Girod P-A, Schaefer DG, Zryd J-P, Ronne H (2007) The moss genes PpSKI1 and PpSKI2 encode nuclear SnRK1 interacting proteins with homologues in vascular plants. Plant Mol Biol 64 559–573 [DOI] [PubMed] [Google Scholar]
- van Nocker S, Ludwig P (2003) The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics 4 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Sasaoka T, Funaki M, Hori H, Murakami S, Ishiki M, Haruta T, Asano T, Ogawa W, Ishihara H, et al (2001) Overexpression of SH2-containing inositol phosphatase 2 results in negative regulation of insulin-induced metabolic actions in 3T3-L1 adipocytes via its 5′-phosphatase catalytic activity. Mol Cell Biol 21 1633–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong L, Lee B, Ishitani M, Lee H, Zhang C, Zhu JK (2001) FIERY1 encoding an inositol polyphosphate 1-phosphatase is a negative regulator of abscisic acid and stress signaling in Arabidopsis. Genes Dev 15 1971–1984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Burk DH, Morrison WH III, Ye ZH (2004) FRAGILE FIBER3, an Arabidopsis gene encoding a type II inositol polyphosphate 5-phosphatase, is required for secondary wall synthesis and actin organization in fiber cells. Plant Cell 16 3242–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH (2004) Molecular and biochemical characterization of three WD-repeat-domain-containing inositol polyphosphate 5-phosphatases in Arabidopsis thaliana. Plant Cell Physiol 45 1720–1728 [DOI] [PubMed] [Google Scholar]
- Zhou L, Jang JC, Jones TL, Sheen J (1998) Glucose and ethylene signal transduction crosstalk revealed by an Arabidopsis glucose-insensitive mutant. Proc Natl Acad Sci USA 95 10294–10299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.