Abstract
In temperate legumes, endosymbiotic nitrogen-fixing rhizobia gain access to inner root tissues via a specialized transcellular apoplastic compartment known as the infection thread (IT). To study IT development in living root hairs, a protocol has been established for Medicago truncatula that allows confocal microscopic observations of the intracellular dynamics associated with IT growth. Fluorescent labeling of both the IT envelope (AtPIP2;1-green fluorescent protein) and the host endoplasmic reticulum (green fluorescent protein-HDEL) has revealed that IT growth is a fundamentally discontinuous process and that the variable rate of root hair invagination is reflected in changes in the host cell cytoarchitecture. The concomitant use of fluorescently labeled Sinorhizobium meliloti has further revealed that a bacteria-free zone is frequently present at the growing tip of the IT, thus indicating that bacterial contact is not essential for thread progression. Finally, these in vivo studies have shown that gaps within the bacterial file are a common feature during the early stages of IT development, and that segments of the file are able to slide collectively down the thread. Taken together, these observations lead us to propose that (1) IT growth involves a host-driven cellular mechanism analogous to that described for intracellular infection by arbuscular mycorrhizal fungi; (2) the non-regular growth of the thread is a consequence of the rate-limiting colonization by the infecting rhizobia; and (3) bacterial colonization involves a combination of bacterial cell division and sliding movement within the extracellular matrix of the apoplastic compartment.
Higher plants are able to establish mutually beneficial endosymbiotic interactions with a variety of microorganisms. Among these, two root endosymbioses are of particular agricultural and ecological importance. Glomeromycota fungi are able to associate with the majority of vascular land plants to form so-called arbuscular mycorrhizas (AMs), whereas root nodulation involving nitrogen-fixing soil bacteria of the Rhizobiaceae family is restricted to the legume family. In these tightly regulated biotrophic associations, host-microbe recognition and initial root entry are crucial steps in the establishment of these two endosymbioses, and, in both cases, outer root penetration by the infecting microsymbiont is transcellular, involving the formation of a specialized host membrane/cell wall interface, which physically separates the microbe from the host cytoplasm (for review, see Brewin, 2004; Genre and Bonfante, 2005; Parniske, 2008). This process, which involves the formation of an apoplastic compartment within the host cell, has been termed accommodation (Parniske, 2000).
For temperate legumes, such as the model legume Medicago truncatula, rhizobia penetrate the host root through epidermal root hairs. The recognition between host and bacteria is based on the synthesis of specific rhizobial lipo-chitooligosaccharides known as Nod factors (NFs). These signal molecules elicit a number of cellular responses in root hairs as a prelude to infection (e.g. Ca2+ oscillations and tip growth reorientation), as well as divisions in inner cortical cells leading to nodule primordium formation (for review, see Oldroyd and Downie, 2008). After recognition, rhizobia attach to root hairs and are subsequently enclosed within an apoplastic space created either by individual root hair tip curling or by the contact between adjacent root hairs. Transcellular apoplastic infection through the root hair then takes place via the progressive formation of a host-derived inwardly growing tubular compartment, known as the infection thread (IT; for review, see Brewin, 2004; Gage, 2004). Rhizobia enter and divide within the IT, which subsequently traverses the entire root hair and outer cortical cells to reach the inner cortex. The path followed by the IT in the cortex is anticipated by the formation of transcellular cytoplasmic bridges known as preinfection threads (van Brussel et al., 1992; Timmers et al., 1999). When ITs reach the nodule primordium, rhizobia are released within membrane-bound compartments into host cells, and subsequent codifferentiation of the two partners leads to the formation of the functional nitrogen-fixing nodule.
Genetic approaches have revealed at least three M. truncatula genes (DMI1, DMI2, and DMI3 [for DOESN'T MAKE INFECTIONS]) whose functions are essential for the establishment of both rhizobial and AM symbioses. The three dmi mutants are totally defective for initial root infection by both endosymbionts (Catoira et al., 2000) and the encoded proteins have been shown to participate in the initial transduction of the rhizobial NF signal in root hairs (for review, see Oldroyd and Downie, 2008). There is also strong evidence suggesting that NFs and DMI genes play key roles during subsequent rhizobial infection (Ardourel et al., 1994; Limpens et al., 2005; Den Herder et al., 2007; Smit et al., 2007). However, IT initiation and elongation also require the presence of other bacterial components, such as rhizobial exopolysaccharides (Cheng and Walker, 1998; Wang et al., 1999). The fact that certain host legume genes required for NF signaling are also necessary for successful mycorrhization suggests the recruitment of host-microbe signaling pathways from the considerably more ancient AM symbiosis (Remy et al., 1994; Parniske, 2008). Because initial root infection for both bacterial and fungal symbionts takes place via transcellular apoplastic compartments, it is possible that the analogy between the rhizobial and AM associations extends to the intracellular mechanisms of host cell infection (Genre and Bonfante, 2007).
Recent in vivo confocal imaging techniques using GFP tagging of cellular components have provided important information about host cell reorganization both prior to and during AM infection (Genre et al., 2005, 2008). These studies have revealed that initial AM infection of non-root hair epidermal cells is preceded by complex intracellular remodeling involving transcellular nuclear migration associated with the formation of a transient cytoplasmic assembly comprising cytoskeletal and endoplasmic reticulum (ER) components. This novel assembly, which has been termed the prepenetration apparatus (PPA), defines the subsequent transcellular path of apoplastic AM infection and is thought to be responsible for synthesizing the perifungal membrane/cell wall interface (for review, see Smith et al., 2006; Parniske, 2008). Furthermore, because dmi mutants are defective in the formation of the PPA, it is probable that a DMI-dependent signaling pathway is required to initiate this major intracellular reorganization (Genre et al., 2005). The PPA-dependent mechanism for intracellular AM infection appears to be conserved between legume and non-legume hosts and also throughout fungal colonization of both the outer and inner root cortex (Genre et al., 2008). In the case of rhizobial infection, cytological studies using fixed tissues have suggested that an array of microtubules is positioned between the nucleus and the inwardly growing IT (Timmers et al., 1999). However, little is currently known about the dynamics of rhizobial infection and, in particular, the subcellular remodeling in the host cell that accompanies IT initiation and development, as well as the coordination with bacterial colonization of the thread.
In this article, we describe experimental techniques that allow direct confocal microscopic observation of fluorescently labeled cellular markers in M. truncatula root hairs to gain insight into the cellular mechanisms involved in IT formation during infection with fluorescently labeled rhizobia. GFP targeting of the host cell ER and the IT membrane has allowed us to investigate the relationships between IT growth, bacterial colonization, and the intracellular dynamics of the root hair. These in vivo data have revealed that IT elongation anticipates bacterial colonization within the thread, indicating that this process is primarily host driven. Furthermore, the growth of the IT appears to be discontinuous, comprising alternating phases of rapid and slower (or temporarily arrested) tip extension. Highly variable IT growth rates are reflected both in the position of the migrating root hair nucleus ahead of the IT tip, as well as in the form of the connecting cytoplasmic bridge. Finally, these observations have revealed that gaps are frequently present within the rhizobial cell file inside the growing IT and have led us to conclude that the colonizing bacteria progress down the newly formed apoplastic compartment by a combination of cell division and collective movement. Based on these findings, we propose a scenario for rhizobial infection of root hairs and compare the cellular mechanism of IT growth with the PPA-based infection process described for intracellular AM infection.
RESULTS
In Vivo Studies of Infection Thread Growth in M. truncatula Root Hairs
To perform confocal microscopy studies of rhizobial infection in M. truncatula root hairs, we have developed an experimental procedure similar to that previously used for AM infection (Genre et al., 2005), which is described in detail in “Materials and Methods.” Briefly, fluorescent (GFP) tags for labeling specific cellular components were introduced into M. truncatula roots using Agrobacterium rhizogenes-mediated transformation (Boisson-Dernier et al., 2001). The composite plants thus generated were grown on a semisolid support and the transgenic roots covered with a gas-permeable film (BioFolie) that allows direct and repeated confocal microscopic observations using water-immersion objectives (Supplemental Fig. S1A). Roots were then inoculated with a stable fluorescently labeled strain of Sinorhizobium meliloti and infection events identified and followed in selected root hairs several days after inoculation. We have validated this experimental system for both the wild-type M. truncatula Jemalong line and the supernodulating mutant sunn (for super numeric nodules; Penmetsa et al., 2003). In both cases, nodulation of composite plants can first be observed under the BioFolie film 4 to 5 d following rhizobial inoculation and the image in Supplemental Figure S1A shows that the roots of composite sunn plants are very efficiently nodulated 2 weeks after inoculation. This shows that the kinetics and efficiency of root nodulation are normal for M. truncatula plants inoculated under these growth conditions.
Because fluorescence labeling of the ER has proved to be extremely useful for visualizing both the cytoplasmic and nuclear dynamics of the host cell during AM infection (Genre et al., 2005), initial confocal experiments were performed using the GFP-HDEL fluorescent marker, which targets the ER compartment (Haseloff et al., 1997). Previous cytological studies had shown that one of the characteristics of growing ITs in legume root hairs is the formation of a broad column (or bridge) of cytoplasm connecting the nucleus to the extending tip of the IT (for review, see Dart, 1974; Gage, 2004). Our initial confocal observations revealed that GFP-HDEL strongly labels this cytoplasmic bridge (see below), thereby allowing the identification of candidate growing ITs in M. truncatula root hairs using this fluorescent tag. As expected, such infected root hairs were considerably more frequent in roots of the supernodulating sunn mutant, and we have estimated that the total number of infection sites per root is at least 10 times higher for sunn plants compared to wild type. Most importantly, microscopic observations performed on over 100 individual infected root hairs revealed that a major proportion of the ITs identified in this way continued their growth down the root hair during the hours following the initial observation. Because similar intracellular dynamics were observed during root hair infection for both wild-type and supernodulator lines, we subsequently focused our studies on sunn because of the significantly enhanced frequency of infection events.
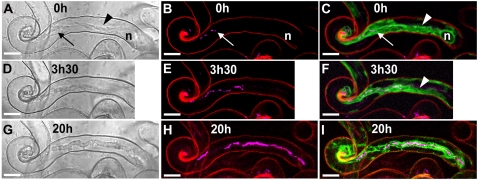
To distinguish the fluorescence labeling of the bacterial partner in the confocal microscope, composite M. truncatula sunn plants expressing 35S:GFP-HDEL were inoculated with an S. meliloti 2011 strain that stably and constitutively expresses the Cerulean version of the cyan fluorescent protein (cCFP; Sm 2011-cCFP, kindly provided by P. Smit, Wageningen, The Netherlands). We have shown that the fluorescence labeling of this strain is stable throughout infection/nodulation (see “Materials and Methods”). The capacity to follow infection in individual root hairs is illustrated in Figure 1, which shows progressive IT growth at three time points over a 20-h period imaged both by bright-field and confocal microscopy. The GFP-HDEL-labeled cytoplasmic bridge located between the migrating nucleus and the growing IT is clearly visible in Figure 1C. Note that the position of the root hair nucleus can be deduced from the strong perinuclear ER labeling (Fig. 1C), and also that the IT is surrounded by ER-rich cytoplasm at all stages of development (Fig. 1, C, F, and I).
Figure 1.
Cytoarchitecture associated with IT development in M. truncatula root hairs. Three successive stages of IT growth in a root hair of the supernodulating M. truncatula mutant sunn expressing the 35S-GFP-HDEL transgene (4 d postinoculation). Bright-field images are shown on the left (A, D, and G). The corresponding confocal images (z axis projections of serial optical sections) showing the cCFP fluorescence of the rhizobia (magenta) are presented in B, E, and H, and combined with the ER-targeted GFP-HDEL (green) in C, F, and I. Cell wall autofluorescence in the confocal images is in red. A to C, At this early stage of IT development, there are only a very few bacteria in the thread, and several gaps are clearly visible within the file (B). The broad ER-rich cytoplasmic bridge (A and C, arrowheads) connecting the nucleus (n) with the elongating IT (arrow) is characteristic of actively growing ITs (C). Note that the IT tip is embedded in the dense, ER-rich cytoplasm and therefore difficult to visualize in the bright-field image. D to F, 3.5 h later, the IT has progressed within the root hair (D), and the rhizobia multiplied within the thread (E). Gaps are again visible within the single file of bacteria. The more mature section of the IT is surrounded by a layer of cytoplasm, whereas the newly synthesized region is embedded in the cytoplasmic bridge (F, arrowhead). Note that the root hair nucleus has moved down the shaft of the root hair and is no longer visible in the image frame. G to I, 20 h later, the IT has grown further down the hair toward the base of the cell. The bright-field image (G) shows the clear outline and uneven surface of the typical mature IT, and the confocal image (I) reveals that this fully differentiated IT is still surrounded by a thin layer of ER-labeled cytoplasm. Thicker stretches of the colonizing bacterial file (H) suggest that there is a doubling in certain regions (H) and, even at this late stage, bacteria-free gaps are still present along the file. Bars = 10 μm.
Having identified growing ITs by this approach, it was then possible to investigate different stages of bacterial colonization in relation to thread development. In mature ITs, which have fully traversed the root hair, the colonizing rhizobia are often in the form of multiple braided files (Supplemental Fig. S1, C and E; Gage, 2002). However, Figure 1, B and E, clearly shows that the bacteria close to the growing tip of the IT are aligned in a single file (Dart, 1974), and indeed we have never observed multiple files of rhizobia in the tip region of an actively growing thread. In addition, the images in Figure 1 show that the file of cCFP-labeled bacteria within the growing IT is often interrupted by gaps of variable length. As discussed in more detail later, these gaps within the bacterial file indicate the absence of bacteria rather than the loss of fluorescence (see also “Materials and Methods”). To our surprise, repeated observations of over 30 growing ITs using the GFP-HDEL marker revealed significant variations in growth rate, the shape and diameter of the cytoplasmic bridge, as well as the distance between the IT and the migrating nucleus. To further investigate this variability, and in particular the dynamics of IT elongation and associated bacterial colonization, it was first necessary to identify a fluorescent marker for the newly synthesized tip of the IT compartment. This was of crucial importance because the precise localization of the growing tip region of the IT in either bright-field or confocal images of GFP-HDEL-labeled root hairs is extremely difficult because this region is usually surrounded by dense, ER-rich cytoplasm (e.g. Fig. 1, A and C; Dart, 1974).
A Plasma Membrane Aquaporin-GFP Fusion Labels the Apoplastic Interface of the IT
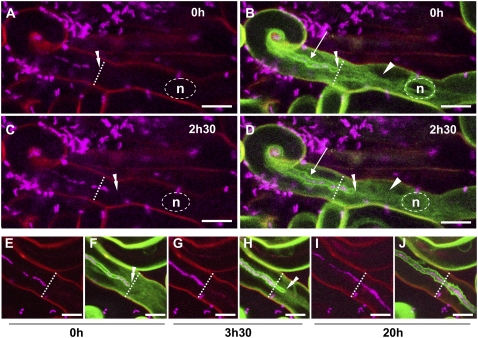
As stated earlier, the apoplastic compartment within the IT is separated from the cell cytoplasm by a membrane-extracellular matrix interface, with the matrix lining the inner surface of the thread. The IT membrane is contiguous with the plasma membrane (PM) and is thought to be initiated from within the curled root hair containing rhizobia via the invagination of the PM (Brewin, 2004; Gage, 2004). In an attempt to identify a PM marker that would also label the IT membrane interface, an Arabidopsis (Arabidopsis thaliana) aquaporin AtPIP2;1-GFP fusion (kindly provided by D.-T. Luu, Montpellier, France; Boursiac et al., 2005) was introduced into M. truncatula roots by A. rhizogenes-mediated transformation. Figure 2 shows that AtPIP2;1-GFP fluorescence labels both the M. truncatula root hair PM as well as the tubular membrane invagination that surrounds the file of colonizing bacteria, including the tip region. In addition to strong fluorescence in the PM and IT membranes, weak cytoplasmic fluorescence can also be seen within the infected root hair. We presume that this is due to the presence of the fusion protein in the ER/secretory pathway, as already reported for other PM-resident proteins (Bhat et al., 2005). Thus, in addition to membrane labeling and although weaker than the GFP-HDEL marker, AtPIP2;1-GFP has also proved useful in certain experiments for visualizing the root hair cytoarchitecture.
Figure 2.
Labeling of the IT interface with GFP-tagged AtPIP2;1 aquaporin. Growing ITs in two different root hairs of sunn plants expressing the AtPIP2;1-GFP fusion. Confocal images A to J (z axis projections of serial optical sections) show the cCFP fluorescence labeling the rhizobia (magenta) and images B, D, F, H, and J the additional fluorescence of the AtPIP2;1-GFP fusion (green). Cell wall autofluorescence in the confocal images is shown in red. The transverse dashed line indicates the initial position of the IT tip (0 h time point) in each image. A to D, Recently initiated IT (4 d postinoculation) observed at two successive time points (0 h [A and B] and 2.5 h [C and D]). In addition to the plant cell membrane, the entire IT membrane (arrows) is labeled by the aquaporin-GFP fusion (B and D). As in Figure 1, the single file of aligned rhizobia within the IT is interrupted by a number of gaps (A and C). In addition, the aquaporin-GFP label reveals that there is a space between the leading bacteria in the file and the growing tip of the IT (double arrowheads) for both time points (B and D). The weak GFP labeling surrounding the nucleus (n) and within the cytoplasmic bridge (arrowheads) is probably indicative of an intracellular pool of the aquaporin fusion protein. E to J, Progressive growth and rhizobial colonization of an IT (5 d postinoculation) observed at three successive time points in a root hair (0 h [E and F], 3.5 h [G and H], and 20 h [I and J]). Initially, the leading bacterium of the discontinuous file is very close to the tip of the IT (E and F); 3.5 h later the tip has grown several micrometers further down the hair, but the leading bacterium has not moved from its original position (G and H), thus creating a space behind the tip. At the same time, most of the initial gaps within the bacterial file have been filled. Finally, the initial smooth appearance of the IT interface (F) has started to become uneven at the 3.5-h time point (H). Twenty hours later (I and J), the IT has progressed down the shaft of the root hair, and displays the characteristic uneven surface of a mature IT. Note that, as in Figure 1, gaps are still present within the bacterial file of the mature IT, and that certain stretches of the file appear to have doubled. The nucleus of the cell is not visible in any of these images, but in all cases was positioned ahead of the growing thread (data not shown). Bars = 10 μm.
Growth of the IT within the root hair is shown in detail in Figure 2, A to D, where the tip has moved approximately 6 μm down the hair shank during the 2.5-h period between the two observations. The labeling of the IT interface reveals that there is a space between the leading cell of the bacterial file and the tip of the growing IT (Fig. 2, B and D). A second infection event is illustrated in Figure 2, E to J, monitored at three time points over a 20-h period. In this case, the leading end of the bacterial file is initially close to the IT tip (Fig. 2, E and F), whereas 3.5 h later, a significant space has been created in front of the bacterial file resulting from IT tip elongation (Fig. 2, G and H). This intriguing observation is dealt with in more detail in the following sections. Whereas the surface of the IT close to the advancing tip has a relatively smooth appearance (Fig. 2, F and H), the same segment of the IT observed 20 h later (Fig. 2J) now possesses a very uneven AtPIP2;1-GFP labeling. In fact, this irregular appearance of the fluorescent labeling is already visible in the older segment of the growing IT at the 3.5-h time point (Fig. 2H), suggesting that this modification occurs at a relatively early stage of IT development. We presume that the irregular contour of the IT interface reflects developmental changes in the underlying extracellular matrix.
In conclusion, the AtPIP2;1-GFP tag is an excellent tool for visualizing both IT elongation and developmental changes of the IT interface throughout root hair infection, as well as for studying the relationship between IT elongation and both the intracellular dynamics of the host cell and the colonization of the apoplastic compartment by the microsymbiont. In the following sections, the kinetics of IT development and the dynamic interplay between IT growth and bacterial colonization will be examined in more detail.
IT Development Is Discontinuous
In the second infection event illustrated in Figure 2, E to J, the mean growth rate of the IT had been estimated as 12 μm h−1 over the 3.5-h period preceding the initial stage depicted in Figure 2F (data not shown). During the following 3.5-h period (Fig. 2, F and H), the mean extension rate of the IT then dropped to only 2 μm h−1. In spite of this significant reduction in average elongation rate, the IT nevertheless successfully reached the base of the root hair the following day (Fig. 2J). To examine the kinetics of IT tip progression in more detail, two independent experiments were performed using roots of composite sunn plants expressing AtPIP2;1-GFP. Based on a total of eight individual growing ITs observed over periods between 2 and 6 h, we calculated an average tip elongation rate of 4.0 ± 2.5 μm h−1 (n = 19). A similar rate of 5.0 ± 2.0 μm h−1 (n = 9) was estimated for a smaller sample of growing ITs observed in a transgenic wild-type line expressing the GFP-HDEL construction (see “Materials and Methods”). However, it is important to underline that, as in the case of the infection event illustrated in Figure 2, E to J, average growth rates for individual root hairs over 2- to 3-h periods ranged from 1 to 12 μm h−1, and frequently changed during IT elongation. These observations indicate that the rate of IT extension can be highly variable throughout the growth of an individual IT.
We therefore asked whether the variability in growth rate of an individual IT could be related to the accompanying intracellular dynamics of the infected root hair and, in particular, to the position of the nucleus relative to the growing IT and the form of the cytoplasmic bridge linking the nucleus to the IT. An analysis of 30 images of root hairs with growing ITs revealed that the distance between the IT and the nucleus can vary from 0 to 40 μm, with an average of 20 ± 10 μm. The rapid changes in nuclear position that can occur during the growth of an individual IT are well illustrated in the time series presented in Supplemental Figure S2. Initially, the nucleus is at a typical distance (approximately 30 μm) from the growing IT and the connecting cytoplasmic bridge is relatively broad (Supplemental Fig. S2C). However, 3 h later, the nucleus has moved away from the growing IT with the cytoplasmic bridge becoming longer and much narrower (Supplemental Fig. S2F). This cytoarchitecture then reverses during the following 3.5 h period with a shortening of the IT-to-nuclear distance and a broadening of the bridge (Supplemental Fig. S2I). Interestingly, the progression of the colonizing bacterial file indicates that IT growth is particularly slow during the initial 3-h period. Although these observations do not allow a precise correlation to be drawn between the IT-to-nuclear distance and the rate of IT extension, it is nevertheless tempting to link the slow progression of the IT over the initial 3-h period to the movement of the nucleus away from the growing invagination and the associated narrowing of the cytoplasmic bridge.
In conclusion, confocal observations of growing ITs using fluorescent markers for both the ER and the IT interface indicate that the construction of the apoplastic compartment is a discontinuous process, most probably involving phases of rapid elongation alternating with slow (or pausing) IT extension, and that these phases are likely correlated with changes in intracellular dynamics.
Elongation of the IT Precedes Bacterial Colonization
Our in vivo studies of IT growth in M. truncatula root hairs have revealed the presence of bacteria-free spaces of variable length between the IT tip and the leading cell of the bacterial file (Fig. 2). To understand the significance of this in relation to discontinuous IT growth, we examined eight growing ITs at several time points in two independent experiments using sunn composite plants expressing the AtPIP2;1-GFP tag. In approximately 65% of the images (n = 27), we observed a bacteria-free space behind the IT tip ranging in length from 2 to 10 μm, and with an average size of 4.0 ± 2.0 μm (n = 17). In certain cases, as in Figure 2, F and H, large variations in the distance between the IT tip and the bacterial file were observed for the same growing thread at different time points. We therefore deduce that extension of the invagination precedes bacterial colonization of the apoplastic compartment and does not require direct physical contact of the IT tip with the rhizobia. In addition, the discontinuous growth of ITs also appears to be reflected in the variability of the distance between the IT tip and the bacterial file, suggesting that these two parameters may be interrelated.
Colonization of the IT Combines Bacterial Cell Division with Coordinated File Movement
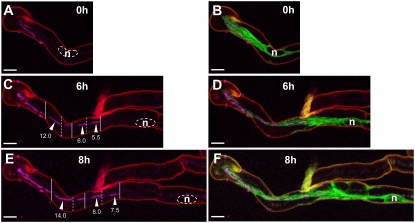
Gaps within the bacterial file were observed in approximately two-thirds of the elongating ITs in root hairs of both sunn and wild-type plants. In the case of the two infections shown in Figures 1 and 2, E to J, multiple gaps are present within the respective files at the first time point (Figs. 1B and 2E). When the two infection events were observed 3.5 h later, bacteria had multiplied within the IT and several of these gaps had disappeared (Figs. 1E and 2G). Finally, as infection continued down the root hair, new gaps appeared within this upper section of the IT (20-h time point; Figs. 1H and 2I). These observations suggest that the bacteria are able to physically move down the thread and also that cell multiplication can occur elsewhere than at the leading end of the bacterial file behind the IT tip. This is particularly well illustrated in Figure 3, which shows infection within a root hair of a wild-type M. truncatula plant expressing GFP-HDEL. This time series reveals that, 6 h after initial imaging (Fig. 3C), several short bacterial files of variable length and separated by gaps of variable sizes are present within the IT. Two hours later (Fig. 3E), these files and the accompanying gaps have moved together down the IT and the files have extended their length, presumably resulting from a round of bacterial cell divisions. The high stability of the pHC60-derived plasmid carrying the cCFP marker, illustrated by the recovery of 100% fluorescent S. meliloti colonies from crushed nodules (“Materials and Methods”), argues that these gaps are not composed of rhizobia that have lost their fluorescence. This is supported by the fact that internal bacterial file segments frequently extend to fill preexisting gaps during IT progression (e.g. Fig. 2, E and G). Similar gap formation and filling within the rhizobial file was also observed when we replaced Sm 2011-cCFP by the same strain labeled with GFP (Limpens et al., 2003; “Materials and Methods”). This is particularly well illustrated in the 2-h-long time-lapse sequence presented in Supplemental Movie S1, which shows progressive colonization of a recently initiated IT by a file of GFP-labeled Sm 2011. This series of confocal images reveals a combination of concerted rhizobial movement, cell elongation, and cell division, resulting in both gap filling and modifications in gap size within the file. As stated earlier and clearly seen in these time-lapse images, colonization of the growing IT is always associated with a single bacterial file in the IT tip region. On the other hand, stretches with parallel files of variable length are often present in more mature IT regions (e.g. Figs. 1H and 2I), and even more frequently in completed ITs (Supplemental Fig. S1, C and E), and we assume that this is due to subsequent bacterial cell division occurring within the IT.
Figure 3.
Bacteria colonize ITs by division and movement. Three successive stages (0, 6, 8 h) of the growth of an IT in a root hair of a wild-type M. truncatula transgenic line expressing the ER-targeted GFP-HDEL (2 d postinoculation). Confocal images A to F (z axis projections of serial optical sections) show the cCFP fluorescence labeling the rhizobia (magenta) and images B, D, and F the additional fluorescence of the GFP-HDEL fusion (green). Cell wall autofluorescence is shown in red. A and B, Early infection stage (0 h). Two short ITs have initiated in one curled root hair. C and D, One of the ITs has elongated and the second has aborted (6 h). Note the bacteria-free spaces separating short files of bacteria within the growing IT. The alternating continuous and dashed vertical lines (C) indicate the positions of the leading bacteria of five contiguous bacterial files separated by spaces. The numbers indicate the length (μm) of three of these bacterial files (arrowheads). E and F, IT has further elongated within the root hair (8 h). The vertical continuous and dashed lines indicate the new positions of the leading bacteria of each of the five bacterial files marked in C. The bacteria have progressed down the thread mainly due to the collective movement of the files. Cell divisions have also taken place in the files, as indicated by their increased length (μm). Bars = 10 μm. n, Nucleus.
In conclusion, bacterial colonization of growing ITs in M. truncatula root hairs involves both bacterial division and the collective movement of bacteria down the thread, and we observe that these events can occur simultaneously and over a significant portion of the IT. The interplay between discontinuous IT growth and the mechanism of rhizobial colonization will be discussed below.
DISCUSSION
The remarkable cellular process involving the initiation and growth of the tubular intracellular structure known as the IT, which allows the controlled entry of nitrogen-fixing rhizobia into the host legume root, has been the subject of microscopic studies over the last half-century (e.g. Fåhraeus, 1957; Nutman, 1959; Dart, 1974; Kijne, 1992; Brewin, 2004; Gage, 2004). With the development of confocal microscopy, it is now possible to examine the intracellular dynamics associated with the elaboration of this specialized apoplastic compartment. In this article, we describe an experimental protocol for performing in vivo confocal studies of rhizobial infection within root hairs of the model legume M. truncatula and the identification of appropriate fluorescent markers, which allow concomitant visualization of the growing IT, the infecting microsymbiont, and various cellular components that play an active role in IT development. The results that have come out of these studies have revealed new information about the process of transcellular IT growth, the complex coordination of IT development with rhizobial colonization, and the mechanism of rhizobial colonization within the IT.
Coordination of IT Growth with Rhizobial Colonization
The possibility of following the growth of individual ITs over time and, in particular, the concomitant monitoring of both IT development and rhizobial colonization using fluorescent markers, has revealed a number of important features of this complex process. One of the most striking is the apparent irregularity of IT progression, illustrated most clearly by the highly variable rate of IT growth (ranging from 1–12 μm h−1). This irregularity is reflected both in the variable distance between the nucleus and the growing thread (0–40 μm) and the form of the connecting cytoplasmic bridge (Fig. 3; and Supplemental Fig. S2). These variations have been observed not only between different infection events, but also throughout the growth of individual ITs. It is likely that the IT elongation rate is directly related to the cytoarchitecture of the infected root hair, and we strongly suspect, based on our observations, that a closely positioned nucleus and a broad cytoplasmic bridge are associated with periods of rapid thread extension (Supplemental Fig. S2). Indeed, it is probable that a broad cytoplasmic bridge is necessary for supplying the growing IT tip with sufficient quantities of exo- and endocytotic vesicles required for the assembly of the matrix/membrane components of the thread, similar to the situation described recently for AM infection (Genre et al., 2008). Finally, it should be noted that a positive correlation between IT growth and the proximity of the nucleus was previously reported in the remarkable studies performed by Nutman, Dart, and colleagues, who used light microscopy to follow IT development in root hairs of Trifolium species (Nutman, 1959; Nutman et al., 1973; Dart, 1974).
The aquaporin AtPIP2;1-GFP fusion labels the IT membrane within the root hair and thereby enables visualization of the elongating tip of the IT in the confocal microscope (Fig. 2). This is important because the apex of the IT is usually embedded in dense cytoplasm and hence difficult to observe in the light microscope (e.g. Fig. 1A). Concomitant fluorescent labeling of the colonizing bacteria has revealed that the leading rhizobia are generally located at a distance of up to 10 μm from the growing tip (Fig. 2, B, D, and H). This strongly argues that IT growth is a host-driven process that does not require permanent physical contact with the bacteria within the thread. Taken together, our data suggest that IT growth is intermittent, with phases of active tip extension alternating with periods of slow or paused growth. To comprehend the mechanisms underlying this irregularity, it is necessary to first consider the process of bacterial colonization within the progressively extending apoplastic compartment.
We have calculated from our in vivo experiments that the average growth rate for the IT in M. truncatula root hairs is 4.0 ± 2.5 μm h−1. This is similar to the IT growth rate previously estimated for clover (5–8 μm h−1; Fåhraeus, 1957; Nutman et al., 1973; Dart, 1974) and alfalfa (Medicago sativa; approximately 8 μm h−1; Gage et al., 1996) root hair infection. Assuming that dividing bacteria (approximately 1–2 μm in length) double at a maximal rate of 4 h within the thread (Gage, 2002), then this implies that at least 10 bacteria need to be actively dividing at any one time during IT development. Our confocal studies have revealed that gaps are often present within the bacterial file in growing ITs (Figs. 1 and 2; Supplemental Movie S1), that these gaps are subsequently filled via active division of bacteria (Fig. 2, F and H; Supplemental Movie S1), and furthermore that segments of the bacterial file can move in unison within the IT (Fig. 3). In this scenario, this sliding movement (discussed in more detail below) is primarily responsible for both creating the gaps and moving bacteria toward the region behind the tip. We therefore propose that rhizobial colonization combines physical movement down the thread, multiple gap creation between short files of bacteria, and concomitant cell division, which then fills the gaps. In this context, it is interesting to note that mixed inoculation experiments performed by Gage (2002) on alfalfa plants using red and green fluorescent rhizobia occasionally resulted in mature ITs containing both types of fluorescent bacteria organized in alternating red/green sectors of variable length. The author concluded from this sectoring pattern that there must be some form of collective movement of the bacteria down the thread and proposed a model in which concomitant bacterial division occurs within a 60-μm-long region proximal to the growing IT tip (Gage, 2002, 2004). These observations and predictions are perfectly coherent with our findings based on in vivo studies in M. truncatula.
Because rhizobia lack flagella when inside the thread, Gage and Margolin (2000) have proposed that such collective movement could be related to so-called sliding motility, a process well documented for a number of bacteria, including mycobacteria (Martinez et al., 1999). Sliding motility is characterized by the collective movement of the bacterial population, often arranged as head-to-tail pseudofilaments and, in the case of mycobacteria, results from reduced surface friction due to the presence of amphiphilic glycopeptidolipids on the outer cell envelope (Recht et al., 2000). Because rhizobia synthesize a variety of exopolysaccharides (EPS), which are important for both initiation and propagation of ITs (for review, see Jones et al., 2007), it is tempting to speculate that one of the roles of these extracellular polysaccharides is to facilitate movement within the thread. Electron microscopy has shown that rhizobia are surrounded by a capsule of EPS within the IT (Rathbun et al., 2002), and that the IT lumen contains secreted plant extracellular glycoproteins (Rae et al., 1992). Based on this, Brewin (2004) has suggested that the EPS-coated rhizobia are maintained as a form of emulsion within the secreted host matrix and that the colonization capacity of the bacteria depends upon the fluidity of this matrix. Further developing this idea, Gucciardo et al. (2005) have proposed that the lumen of the IT contains a rod-like glycoprotein with potential lubricant properties resulting from end-to-end Tyr cross-linking. The anisotropic properties of such a matrix material could thus control the orientation and sliding motility of the rhizobia. Finally, Gucciardo et al. (2005) have pointed out that alternative oxidative cross-linking of these glycoproteins in more mature regions of the thread could subsequently lead to a fluid-to-solid phase transition and hence prevent further movement of the bacterial cells.
A Model for Discontinuous IT Growth
Despite the combination of sliding movement and cell division, bacterial colonization of the IT is likely to be a limiting step in IT growth rate. Indeed, it is striking that bacteria are particularly sparse in recently initiated ITs and in the tip region, in general (e.g. Figs. 1B, and 2, A and F). Furthermore, the average IT growth rate of 4 μm h−1 is only 30% of the maximum extension rate (12 μm h−1) observed in our experiments. We therefore propose that the discontinuity in IT growth within the root hair results from the disparity between the potential tip elongation rate and the limiting speed of bacterial colonization. In such a scenario, IT progression would comprise periods of rapid host-driven tip extension, which creates space in front of the leading bacterial file, alternating with pauses during which the colonizing bacteria fill the space by combining sliding movement and division (see model in Fig. 4). Presumably, the lumen of this bacteria-free space is composed of secreted matrix material of host origin (Rathbun et al., 2002). Significantly, although the distance between the IT tip and the leading bacteria fluctuates considerably throughout growth, this distance never exceeded 10 μm in the growing ITs that we have observed. This suggests that the progression of the IT tip requires that colonizing bacteria are within a certain distance, and therefore that some form of bacterial/host signaling is responsible for regulating the coordinated growth of the infected thread. One obvious candidate for such signaling would be the rhizobial lipo-chitooligosaccharide NFs, believed to play a role in successful IT development (Limpens et al., 2005; Den Herder et al., 2007; Smit et al., 2007), and which can function as diffusible signals. It remains to be shown that such signaling does indeed occur within the IT, and of course it is possible that other factors may be involved in regulating the concerted growth of the two symbiotic organisms.
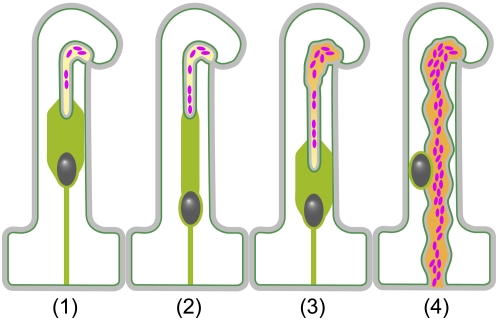
Figure 4.
Schematic illustration of IT growth in M. truncatula root hairs. Representation of consecutive stages of IT growth in the root hair deduced from the in vivo confocal time-lapse studies described in this article. Note that, for simplification, the IT within the curled root hair is represented by an open structure, although of course IT initiation always takes place within the closed chambers formed within three-dimensional curls. Recently initiated ITs are generally sparsely populated with bacteria and the surface of the IT has a smooth appearance (1). Host-driven polar growth of the apoplastic envelope creates a space in front of the leading bacteria within the IT (1, 3). Collective sliding movement of bacteria toward the IT tip, coupled with concomitant cell divisions at different positions within the IT, contributes to colonization of the thread (2, 3). A broad cytoplasmic bridge generally connects the IT tip to the migrating nucleus (1, 3), but at certain stages of development the nucleus-to-IT distance can increase significantly with associated narrowing of the cytoplasmic bridge (2). In addition to the cytoplasmic bridge linking the nucleus to the IT, there is always a thin cytoplasmic strand connecting the nucleus to the basal part of the root hair. The more mature section of the thread progressively develops an uneven appearance (3), probably reflecting developmental changes to the underlying extracellular matrix. This is often accompanied by secondary radial multiplication of the bacterial file. When the mature IT (4) reaches the base of the root hair, the surface has now become very uneven and the nucleus is no longer positioned at the growing end of the IT. Multiple files of bacteria are commonly found within the thickened mature thread. Dark gray, Nucleus; light green, main endoplasmic ER accumulations; dark green, PM; light gray, cell wall; yellow/orange, color transition indicates proposed maturation of the IT matrix; magenta, bacteria. The cortical cytoplasm is not indicated.
Conserved Host Cellular Mechanisms in Endosymbiotic Infection
To what extent can analogies be drawn between the cellular mechanisms involved in the creation of the apoplastic compartments during AM and rhizobial infection? In the case of the AM association, primary root infection by the endosymbiotic fungus involves the assembly of an ER-rich cytoplasmic bridge called the PPA between the site of fungal adhesion on the epidermal cell surface and the transcellular migrating host nucleus (Genre et al., 2005). Similar nuclear-directed cytoplasmic bridges have also been identified preceding and directing AM fungal colonization in outer and inner cortical root tissues of both M. truncatula and carrot (Daucus carota), suggesting that this could be a general mechanism for intracellular infection by these obligate AM fungi (Genre et al., 2008). Obvious similarities with rhizobial infection of root hairs include the transcellular migration of the host nucleus, which prefigures the path of progressive IT development as well as the formation of the broad ER-rich cytoplasmic bridge (e.g. Fig. 1C), which presumably provides both a cytoskeletal scaffold (Timmers et al., 1999) and the cell machinery for IT growth. Because of the relatively short distance (approximately 20 μm) across the root epidermis, the PPA appears as a transient cytoplasmic assembly prior to AM fungal infection (Genre et al., 2005). However, the much longer distances required to traverse root hairs (100–200 μm) implies that equivalent tip elongation machinery would have to move progressively down the hair and this is indeed what is observed during IT growth (Fig. 3; Supplemental Fig. S2). Finally, the rate of transcellular AM infection across the M. truncatula epidermal cell layer (estimated as 15–20 μm h−1; Genre et al., 2005) is only marginally higher than the maximal rate (12 μm h−1) observed for IT growth in root hairs. In conclusion, the intracellular dynamics associated with IT growth, combining directed nuclear migration and the progressive formation of an ER-rich cytoplasmic bridge prefiguring IT formation, clearly resemble the PPA-dependent mechanism described for intracellular AM infection. However, additional studies are now needed to describe in more detail the intracellular dynamics of endosymbiotic infection, and to compare the mechanisms of polarized cell invagination and interface formation with other well-characterized tip-growing processes in plants such as root hair or pollen tube growth.
MATERIALS AND METHODS
Biological Materials
In this study, we have primarily used the Medicago truncatula sunn-2 mutant, kindly provided by E.-P. Journet (Toulouse, France; Schnabel et al., 2005). Experiments on wild-type M. truncatula plants were performed using either the genotype Jemalong A17 or a regenerated transgenic line (referred to as A2) expressing the 35S-GFP-HDEL construct (Chabaud et al., 2003).
Sinorhizobium meliloti 2011 strains expressing either GFP (Sm 2011-GFP) or the cerulean version of the CFP (Sm 2011-cCFP) were kindly provided by P. Smit (Wageningen, The Netherlands) and propagated on selective TY medium supplemented with 10 μg/mL tetracycline. The Sm 2011-GFP strain (Limpens et al., 2003) carries the pHC60 (tetR) plasmid described by Cheng and Walker (1998), which constitutively expresses GFP and possesses the stabilization region of the broad-host-range plasmid RK2. Strain Sm 2011-cCFP carries the identical plasmid, but with the cCFP fluorescent label replacing GFP. To evaluate the stability of the fluorescent labeling during in vivo infection experiments, we repeated the experiment originally performed by Cheng and Walker (1998) for the Sm 1021 strain expressing the pHC60 plasmid. Nodules formed on M. truncatula roots after inoculation with either the Sm 2011-GFP or -cCFP strains were surface sterilized and then crushed to release the symbiotic bacteria. Following dilution and plating on TY medium without tetracycline, 100% of the bacterial colonies were found to be fluorescent, thus confirming the remarkable stability of these plasmids throughout infection/nodulation.
GFP-Labeled Intracellular Markers
For ER labeling in M. truncatula root cells, we used the 35S-GFP-HDEL construct, also known as mgfp4-ER (Haseloff et al., 1997) and kindly provided by J. Haseloff (Cambridge, UK). For PM labeling, we exploited an AtPIP2;1 aquaporin C-terminal fusion to GFP (AtPIP2;1-GFP; Boursiac et al., 2005) under the control of a double cauliflower mosaic virus 35S promoter in the pGreen vector 0179 and kindly provided by D.-T. Luu (Montpellier, France). These cellular markers were introduced into sunn and wild-type A17 plants using the Agrobacterium rhizogenes-mediated transformation technique described by Boisson-Dernier et al. (2001), and rhizobial inoculation experiments were performed on the roots of the resulting composite plants (see below).
In Vivo Microscopic Observation of Rhizobial Infection in M. truncatula Roots
Surface-sterilized seeds of all M. truncatula lines were germinated on inverted agar plates for 3 d at 8°C in the dark. In the case of A. rhizogenes-mediated transformation of both sunn and A17 lines, seedlings were transferred to agar-Fåhraeus plates (Boisson-Dernier et al., 2001) supplemented with 200 mg L−1 Augmentin (amoxicillin:clavulanic acid [5:1]; GlaxoSmithKline) 10 d after A. rhizogenes inoculation to limit the extent of A. rhizogenes multiplication. Six to 12 d later, plants with transformed roots displaying strong and uniform fluorescence were selected for rhizobial inoculation experiments. For transgenic A2 seedlings, the tip was removed to stimulate lateral root emergence and the plantlets grown for about 1 week on agar-Fåhraeus medium. All plants were grown in a culture room at 25°C, with a 16-h photoperiod and a light intensity of 70 μE s−1 m−2.
For in vivo microscopy studies, an experimental setup previously used for monitoring root hair growth for A. rhizogenes-transformed composite plants of M. truncatula (Sieberer et al., 2005; Timmers et al., 2007) was modified to make it compatible with rhizobial inoculation and subsequent infection/nodulation. Selected composite or A2 plants were transferred to 12- × 12-cm petri dishes containing modified Fåhraeus medium (MgSO4 concentration increased to 3 mm) and 0.5% Phytagel (Sigma), supplemented with 50 nm 2-amino ethoxyvinyl Gly (AVG). The AVG is included to limit ethylene production, which could inhibit the nodulation process (Guinel and Geil, 2002, and refs. therein). For microscopic observation, roots were covered with a sterile, gas-permeable and transparent plastic film (BioFolie 25; Sartorius AG, Vivascience), which has the same optical refractive index as water. This allows the use of water-immersion objectives (see below), limits water evaporation from the plates during observation, and reduces the risk of contamination of the roots. In addition, and very importantly, the roots that grow in the thin water layer between the plate surface and plastic film display very low levels of endogenous fluorescence compared to roots growing in air (Genre et al., 2005). Plants were grown vertically in the culture room with the plates slightly tilted to favor the growth of roots along the plastic film and with the roots protected from light using black plastic bags. Inoculation with S. meliloti was performed by pipetting an aqueous suspension of exponentially growing bacteria (approximately 107 bacteria in 1 mL) between the plastic foil and the semisolid medium. Potential infection sites on inoculated roots were identified using both epifluorescence and bright-field illumination several days after rhizobial addition. In the case of bright-field illumination, low light and a green filter were used to minimize light-induced stress. Plants were returned to the culture room between observations.
Confocal Microscopy
Rhizobial infection sites were imaged using a Leica TCS SP2 AOBS confocal laser-scanning microscope equipped with a long-distance ×40 water-immersion objective (HCX Apo L 0.80). The argon laser bands of 458 and 488 nm were used alternatively to excite CFP and GFP, respectively, and a 561-nm diode to observe cell wall autofluorescence. Specific emission windows of 460 to 480 nm, 500 to 540 nm, and 600 to 670 nm were used for CFP, GFP, and autofluorescence signals, which were false-colored in magenta, green and red, respectively. During scanning, the GFP signal and the combined CFP plus autofluorescence signals were acquired alternatively for each line, using the sequential mode. The images shown are maximal projections of selected planes of a z-stack. Images were acquired and projected using Leica confocal software and processed using the Leica CS, ImageJ (http://rsb.info.nih.gov/ij), and Image Pro Plus (Media Cybernetics) software. Distance measurements were carried out using the Leica CS.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Experimental setup for in vivo confocal microscopic observation of rhizobial infection in M. truncatula root hairs.
Supplemental Figure S2. Conformational changes of the cytoplasmic bridge linking the nucleus to the infection thread tip during growth in a M. truncatula root hair.
Supplemental Movie S1. Dynamics of S. meliloti colonization during early infection of a M. truncatula root hair.
Supplementary Material
Acknowledgments
We are grateful to P. Smit (Wageningen, The Netherlands) for providing the S. meliloti 2011 strains expressing cCFP and GFP, to D.-T. Luu (Montpellier, France) for providing the AtPIP2;1-GFP fusion for membrane labeling, and to A. Genre (Torino, Italy) for frequent discussions and critical reading of the manuscript.
This work was supported by an international program for scientific cooperation, titled “Cellular mechanisms of plant root infection by endosymbiotic soil microbes,” of the Centre National de la Recherche Scientifique and by the Institut National de la Recherche Agronomique (postdoctoral grant to B.J.S.).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.or) is: Joëlle Fournier (joelle.fournier@toulouse.inra.fr).
The online version of this article contains Web-only data.
Open access articles can be viewed online without a subscription.
References
- Ardourel M, Demont N, Debelle F, Maillet F, de Billy F, Prome JC, Denarie J, Truchet G (1994) Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell 6 1357–1374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat RA, Miklis M, Schmelzer E, Schulze-Lefert P, Panstruga R (2005) Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc Natl Acad Sci USA 102 3135–3140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisson-Dernier A, Chabaud M, Garcia F, Becard G, Rosenberg C, Barker DG (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact 14 695–700 [DOI] [PubMed] [Google Scholar]
- Boursiac Y, Chen S, Luu DT, Sorieul M, van den Dries N, Maurel C (2005) Early effects of salinity on water transport in Arabidopsis roots. Molecular and cellular features of aquaporin expression. Plant Physiol 139 790–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewin NJ (2004) Plant cell wall remodelling in the Rhizobium-legume symbiosis. Crit Rev Plant Sci 23 293–316 [Google Scholar]
- Catoira R, Galera C, de Billy F, Penmetsa RV, Journet E-P, Maillet F, Rosenberg C, Cook D, Gough C, Denarie J (2000) Four genes of Medicago truncatula controlling components of a Nod factor transduction pathway. Plant Cell 12 1647–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabaud M, de Carvalho-Niebel F, Barker DG (2003) Efficient transformation of Medicago truncatula cv. Jemalong using the hypervirulent Agrobacterium tumefaciens strain AGL1. Plant Cell Rep 22 46–51 [DOI] [PubMed] [Google Scholar]
- Cheng HP, Walker GC (1998) Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol 180 5183–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dart PJ (1974) The infection process. In A Quispel, ed, The Biology of Nitrogen Fixation. North-Holland Publishing Company, Amsterdam, pp 381–429
- Den Herder J, Vanhee C, De Rycke R, Corich V, Holsters M, Goormachtig S (2007) Nod factor perception during infection thread growth fine-tunes nodulation. Mol Plant Microbe Interact 20 129–137 [DOI] [PubMed] [Google Scholar]
- Fåhraeus G (1957) The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J Gen Microbiol 16 374–381 [DOI] [PubMed] [Google Scholar]
- Gage DJ (2002) Analysis of infection thread development using Gfp- and DsRed-expressing Sinorhizobium meliloti. J Bacteriol 184 7042–7046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ (2004) Infection and invasion of roots by symbiotic, nitrogen-fixing Rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev 68 280–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ, Bobo T, Long SR (1996) Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa). J Bacteriol 178 7159–7166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage DJ, Margolin W (2000) Hanging by a thread: invasion of legume plants by rhizobia. Curr Opin Microbiol 3 613–617 [DOI] [PubMed] [Google Scholar]
- Genre A, Bonfante P (2005) Building a mycorrhizal cell: how to reach compatibility between plants and arbuscular mycorrhizal fungi. J Plant Interact 1 3–13 [Google Scholar]
- Genre A, Bonfante P (2007) Check-in procedures for plant cell entry by biotrophic microbes. Mol Plant Microbe Interact 20 1023–1030 [DOI] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Faccio A, Barker DG, Bonfante P (2008) Prepenetration apparatus assembly precedes and predicts the colonization patterns of arbuscular mycorrhizal fungi within the root cortex of both Medicago truncatula and Daucus carota. Plant Cell 20 1407–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genre A, Chabaud M, Timmers T, Bonfante P, Barker DG (2005) Arbuscular mycorrhizal fungi elicit a novel intracellular apparatus in Medicago truncatula root epidermal cells before infection. Plant Cell 17 3489–3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gucciardo S, Rathbun EA, Shanks M, Jenkyns S, Mak L, Durrant MC, Brewin NJ (2005) Epitope tagging of legume root nodule extensin modifies protein structure and cross-linking in cell walls of transformed tobacco leaves. Mol Plant Microbe Interact 18 24–32 [DOI] [PubMed] [Google Scholar]
- Guinel FC, Geil RD (2002) A model for the development of the rhizobial and arbuscular mycorrhizal symbioses in legumes and its use to understand the roles of ethylene in the establishment of these two symbioses. Can J Bot 80 695–720 [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc Natl Acad Sci USA 94 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC (2007) How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol 5 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijne JW (1992) The rhizobium infection process. In G Stacey, RH Burris, HJ Evans, eds, Biological Nitrogen Fixation. Chapman and Hall, New York, pp 349–398
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R (2003) LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 24 630–633 [DOI] [PubMed] [Google Scholar]
- Limpens E, Mirabella R, Fedorova E, Franken C, Franssen H, Bisseling T, Geurts R (2005) Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc Natl Acad Sci USA 102 10375–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A, Torello S, Kolter R (1999) Sliding motility in mycobacteria. J Bacteriol 181 7331–7338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nutman PS (1959) Some observations on root-hair infection by nodule bacteria. J Exp Bot 10 250–263 [Google Scholar]
- Nutman PS, Doncaster CC, Dart PJ (1973) Infection of Clover by Root Nodule Bacteria. Black and white, 16-mm optical sound track film. The British Film Institute, London
- Oldroyd GED, Downie JA (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol 59 519–546 [DOI] [PubMed] [Google Scholar]
- Parniske M (2000) Intracellular accommodation of microbes by plants: a common developmental program for symbiosis and disease? Curr Opin Plant Biol 3 320–328 [DOI] [PubMed] [Google Scholar]
- Parniske M (2008) Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat Rev Microbiol 6 763–775 [DOI] [PubMed] [Google Scholar]
- Penmetsa RV, Frugoli JA, Smith LS, Long SR, Cook DR (2003) Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol 131 998–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae AL, Bonfante-Fasolo P, Brewin NJ (1992) Structure and growth of infection threads in the legume symbiosis with Rhizobium leguminosarum. Plant J 2 385–395 [Google Scholar]
- Rathbun EA, Naldrett MJ, Brewin NJ (2002) Identification of a family of extensin-like glycoproteins in the lumen of Rhizobium-induced infection threads in pea root nodules. Mol Plant Microbe Interact 15 350–359 [DOI] [PubMed] [Google Scholar]
- Recht J, Martinez A, Torello S, Kolter R (2000) Genetic analysis of sliding motility in Mycobacterium smegmatis. J Bacteriol 182 4348–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy W, Taylor TN, Hass H, Kerp H (1994) Four hundred-million-year-old vesicular arbuscular mycorrhizae. Proc Natl Acad Sci USA 91 11841–11843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnabel E, Journet EP, de Carvalho-Niebel F, Duc G, Frugoli J (2005) The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol Biol 58 809–822 [DOI] [PubMed] [Google Scholar]
- Sieberer BJ, Timmers ACJ, Emons AMC (2005) Nod factors alter the microtubule cytoskeleton in Medicago truncatula root hairs to allow root hair reorientation. Mol Plant Microbe Interact 18 1195–1204 [DOI] [PubMed] [Google Scholar]
- Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SE, Barker SJ, Zhu YG (2006) Fast moves in arbuscular mycorrhizal symbiotic signalling. Trends Plant Sci 11 369–371 [DOI] [PubMed] [Google Scholar]
- Timmers A, Auriac M, Truchet G (1999) Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126 3617–3628 [DOI] [PubMed] [Google Scholar]
- Timmers ACJ, Vallotton P, Heym C, Menzel D (2007) Microtubule dynamics in root hairs of Medicago truncatula. Eur J Cell Biol 86 69–83 [DOI] [PubMed] [Google Scholar]
- van Brussel AAN, Bakhuizen R, van Spronsen PC, Spaink HP, Tak T, Lugtenberg BJJ, Kijne JW (1992) Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science 257 70–72 [DOI] [PubMed] [Google Scholar]
- Wang LX, Wang Y, Pellock B, Walker GC (1999) Structural characterization of the symbiotically important low-molecular-weight succinoglycan of Sinorhizobium meliloti. J Bacteriol 181 6788–6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.