Abstract
Control of folate homeostasis is essential to sustain the demand for one-carbon (C1) units that are necessary for major biological functions, including nucleotide synthesis and methylation reactions. In this study, we analyzed the genome-wide and metabolic adaptive response of Arabidopsis (Arabidopsis thaliana) cells to folate depletion induced by the antifolate methotrexate. Drug treatment induced a response typical to xenobiotic stress and important changes in folate content and composition. This resulted in a reduction of cell division and primary energy metabolism that was likely associated with perturbation of nucleotide homeostasis. Through a modification of serine metabolism, folate depletion also induced O-acetylserine accumulation and mimicked sulfur deficiency response. The major adaptive response to folate limitation concerned the composition of the folate pool rather than the intracellular level of cofactors. Thus, no significant change in the expression of genes involved in cofactor synthesis, degradation, or trafficking was observed. However, changes in the distribution of C1 derivative pools and increased expression levels for transcripts coding enzymes manipulating C1 moieties in plastids suggested a reorientation of C1 units toward the synthesis of purine and thymidylate. Also, no genomic or metabolic adaptation was built up to counterbalance the major impairment of the methyl index, which controls the efficiency of methylation reactions in the cell. Together, these data suggested that the metabolic priority of Arabidopsis cells in response to folate limitation was to shuttle the available folate derivatives to the synthesis of nucleotides at the expense of methylation reactions.
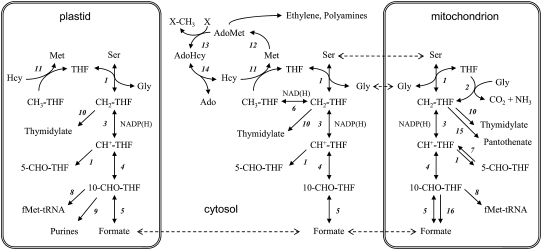
One-carbon (C1) metabolism refers to a complex network in which C1 units carried by folate cofactors are used for essential biosynthetic and regulatory functions. Cellular tetrahydrofolate (THF) derivatives, collectively termed folate(s), differ in the location and oxidation level of the C1 unit they carry and by the length of their (poly)Glu side chain. The 10-formyl derivative of THF is involved in purine and 10-formylmethionine-tRNA synthesis, 5,10-methylene-THF is required for thymidylate and pantothenate synthesis and for Gly to Ser conversion, and 5-methyl-THF is the methyl donor for the synthesis of Met (Fig. 1; Hanson and Roje, 2001). Met is incorporated into protein or converted to S-adenosylmethionine (AdoMet), the universal methyl donor that is involved in dozens methyltransferase reactions and in the biogenesis of ethylene, polyamines, biotin, and nicotianamine (Roje, 2006). C1 metabolism in plants displays unique features as compared to other organisms, in particular regarding its multiple subcellular compartmentation (Fig. 1). Also, plants are able to synthesize THF de novo through a complex biosynthetic route comprising 10 steps located in three subcellular compartments (plastids, cytosol, and mitochondria; Rébeillé et al., 2006).
Figure 1.
Overview of C1 metabolism and its compartmentation in plant cells. 1, SHMT; 2, Gly decarboxylase; 3, 5,10-methylene-THF dehydrogenase; 4, 5,10-methenyl-THF cyclohydrolase; 5, FTHFS; 6, 5,10-methylene-THF reductase; 7, 5-formyl-THF cycloligase; 8, methionyl-tRNA formyltransferase; 9, glycinamide ribonucleotide transformylase and aminoimidazole carboximide ribonucleide transformylase; 10, thymidylate synthase; 11, Met synthase; 12, AdoMet synthetase; 13, AdoMet-dependent methyltransferase; 14, AdoHcy hydrolase; 15, ketopantoate hydroxymethyltransferase; 16, 10-formyl-THF deformylase. Ado, adenosine; THF and its derivatives: CH3- (methyl), CH2- (methylene), CH+- (methenyl), 5-CHO-, and 10-CHO- (formyl).
During plant development, the enzymatic capacity for THF synthesis, the pool of folates, and the demands for C1 units vary importantly (Gambonnet et al., 2001; Basset et al., 2002, 2004a, 2004b; Jabrin et al., 2003). Little is known, however, about how folate homeostasis is controlled to match the supply of C1 units with fluctuating demand and how C1 units are accurately distributed between different anabolic routes. To gain insights into the regulatory processes governing this network, several groups analyzed the consequences of perturbation of the folate pool on C1 metabolism. By exposing Arabidopsis (Arabidopsis thaliana) plants to folate antagonists, Prabhu et al. (1998) determined that a continuous supply of folates was essential to maintain high rates of Ser synthesis during the photorespiratory process. Also, plant cell cultures treated with the folate analog methotrexate (MTX) displayed an important inhibition of their growth (Wu et al., 1993), thus emphasizing the crucial role of folates in nucleotide synthesis and cell division. Last, folate limitation in Arabidopsis cells was associated with important perturbation of the activated methyl cycle, which corresponds to a recycling of Met and AdoMet following methylation reactions (Loizeau et al., 2007). Upon 5-methyl-THF depletion, a delayed restoration of Met and AdoMet homeostasis was observed and correlated with a proteolytic cleavage of the N terminus of the first enzyme involved in de novo Met synthesis (Loizeau et al., 2007). Processing of this enzyme was specifically associated with perturbation of the folate pool, thus demonstrating that a folate-dependent anabolic route can adapt to folate depletion through a posttranslational regulatory process. Drug-induced folate starvation in Arabidopsis cells was also associated with a significant modification of folate composition (distribution of C1 derivatives), suggesting an adaptive response to favor a preferential shuttling of the flux of C1 units to the synthesis of nucleotides over the synthesis of Met (Loizeau et al., 2007).
The aim of this genome-wide and metabolic study was to gain insight into the regulatory process leading to the adaptation of cell physiology, and more particularly of C1 metabolism, upon perturbation of folate homeostasis. We showed that folate limitation due to MTX application was responsible for important metabolic disturbance, including stress response, impairment in cell division, and reduction of energy metabolism. Also, a metabolic priority of Arabidopsis cells was to shuttle the available C1 derivatives to the synthesis of nucleotides at the expense of AdoMet synthesis and methylation reactions.
RESULTS AND DISCUSSION
In this study, we used the antifolate MTX to analyze the genome-wide and metabolic consequences of folate depletion in Arabidopsis cells. MTX is an analog of dihydrofolate that primarily acts on dihydrofolate reductase (DHFR), an enzyme involved in the de novo synthesis of THF and in the regeneration of reduced folate coenzymes from oxidized dihydrofolate species. Previous studies have shown that MTX is useful to manipulate folate pools in plants (Wu et al., 1993; Prabhu et al., 1998; Loizeau et al., 2007). In our study, MTX was used at 100 μm to induce rapid perturbation of folate homeostasis in Arabidopsis cells, and the dynamics of global transcriptomic changes was followed 2, 6, and 24 h following drug addition to the culture medium. Folates were determined by liquid chromatography-electrospray ionization tandem mass spectrometry (LC-MS/MS), and, taking into account experimental bias (De Brouwer et al., 2007), derivatives were grouped into five categories, namely 5-methyl-THF, 5-formyl-THF, THF plus 5,10-methylene-THF, 10-formyl-THF plus 5,10-methenyl-THF, and folic acid. Control Arabidopsis cells contained an average of 12 nmol folates/g of fresh weight (FW) all along the examined period (Supplemental Table S1). In treated cells, MTX uptake was rapid, and the intracellular pool reached a plateau value at about 10 nmol/g FW after 6 h of treatment. Upon MTX treatment, the overall pool of folate decreased by 22%, 56%, and 77% after 2, 6, and 24 h of exposure to the drug, respectively (Supplemental Table S1). As previously described by Loizeau et al. (2007), this treatment also induced an imbalance in C1-substituted cofactors, the pool of 5-methyl-THF being the most severely reduced, whereas other derivatives decreased less rapidly.
Effects of MTX on Genome-Wide Expression in Arabidopsis Cells
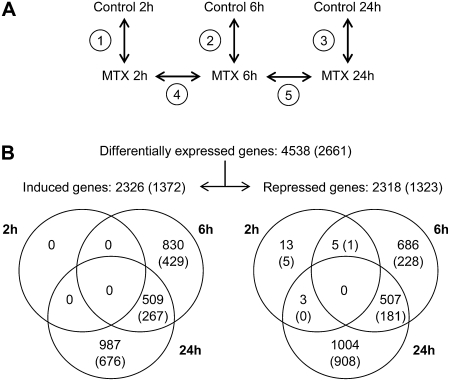
To follow changes in gene expression over the time course of exposure to MTX, we directly compared control and MTX-treated cells at 2, 6, and 24 h (Fig. 2A, comparisons 1–3) using the Complete Arabidopsis Transcriptome MicroArray (CATMA) chips. Statistical analysis using a P-value cutoff of <0.05 after Bonferroni correction (Gagnot et al., 2008) revealed 4,538 genes displaying significant differential expression at some point during drug treatment. Thus, 20% of the 22,089 Arabidopsis nuclear genes represented on the CATMA array displayed changes in mRNA levels in response to MTX treatment. The dynamics of genome-wide response was confirmed through direct comparisons of MTX-treated cells at 2, 6, and 24 h (Fig. 2A, comparisons 4–5). Differentially expressed genes showed an equal distribution between up- and down-regulated groups (2,326 and 2,318 genes, respectively, Fig. 2B). In addition, 106 genes were shared by these two categories as they displayed induction followed by repression, or vice versa, during the time course analysis. The expression profiles of 12 selected genes were analyzed by real-time quantitative RT-PCR (qPCR) and allowed to validate the array experiments (Supplemental Table S2).
Figure 2.
Experimental design for CATMA transcript profiling and distribution of MTX-responsive genes over the time course analysis. A, Schematic representation of the experimental procedure used for transcript profiling of the Arabidopsis cell response to MTX. Five comparisons (1–5) of MTX-treated cells and corresponding controls were performed at three time points (see “Materials and Methods” for details). B, Number of genes regulated in response to MTX treatment. Differentially expressed genes were selected by statistical analysis after Bonferroni correction (P-value cutoff of 0.05). Numbers in parenthesis correspond to highly regulated genes, i.e. genes for which the log2(ratio) value was >1 or <−1 (2-fold changes).
To focus our study on highly responsive transcripts, we have selected differentially expressed genes for which log2(ratio) was >1 or <−1 (corresponding to 2-fold changes). The resulting 2,661 MTX-responsive genes were classified according to their temporal expression, and we could define two principal sets of genes corresponding to an early (6 h) and a late (24 h) response to MTX treatment (Fig. 2B). After 2 h of exposure to MTX, cellular changes due to drug incorporation within cells and the associated modest depletion of the folate pool (22% as compared to control cells) were not sufficient to induce a significant transcriptional response of Arabidopsis cells. The early response to MTX treatment comprised 1,106 highly regulated genes, of which 657 displayed only transient differential expression at 6 h. For 448 genes, the differential expression observed in the early phase of the response to MTX was maintained in the late period. Both the transient and persistent sets of early regulated genes were characterized by a high proportion of up-regulated transcripts (63%). Lastly, genes for whom the differential expression was specific to the late response to MTX accounted for 60% of highly responsive transcripts. Particularly, 908 out of 1,323 down-regulated genes (i.e. 69%) were typical of a late response (Fig. 2B).
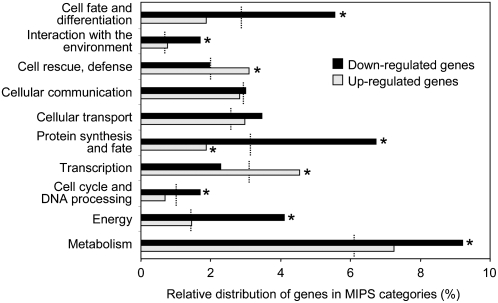
To gain insight into the biological processes in which the highly differentially expressed genes are involved, we performed functional categorization based on the Munich Information Center for Protein Sequences (MIPS) database (Fig. 3; Ruepp et al., 2004). In addition to the large number of genes with no clear function (74% of the Arabidopsis genes and 66% of the MTX-responsive genes), the principal bias of the MIPS classification resides in the important variation in the number of genes in each category. Thus, to highlight biological processes that are significantly affected by the MTX treatment, we compared the proportion (in percent), but not the number, of MTX highly responsive genes assigned in each category to the relative distribution of all the Arabidopsis annotated genes in the different categories (Fig. 3). Using these criteria, the most significantly modified categories among the MTX-repressed genes were metabolism, energy, protein synthesis, cell fate and differentiation, interaction with the environment, and cell cycle. Figure 3 shows that for MTX-induced genes, the MIPS categories principally affected were transcription and cell rescue, whereas protein synthesis was the lowest represented functional group. The function of selected groups of MTX-regulated genes will be analyzed in detail in the next paragraphs.
Figure 3.
Functional categorization of MTX-regulated genes. Highly responsive genes were separated into up-regulated (gray bars) and down-regulated (black bars) sets and classified into functional categories using the MIPS interface (Ruepp et al., 2004). The proportion (in percent) of MTX-responsive genes identified in each category is indicated on the x axis. Dotted lines indicate the distribution (percent) of all the Arabidopsis genes in each MIPS category. Asterisks indicate that the distribution of MTX-responsive genes is significantly different from that of Arabidopsis genes using a chi-square test with an α risk of P < 0.01. Genes of unknown function, which represent 74% of the Arabidopsis genes and 66% of the MTX-responsive genes, were excluded from the figure for clarity.
MTX Treatment Is Associated with a Broad Slowdown of Primary Metabolic and Cellular Processes
In agreement with the antiproliferative properties of MTX, we observed that growth of Arabidopsis cells supplied with the drug was markedly reduced (Supplemental Fig. S1). Cell viability was not affected by MTX, thus indicating that the treatment did not trigger cell death during the examined period of culture. Thus, growth inhibition upon MTX treatment illustrated the crucial role of folates in nucleotide synthesis, DNA synthesis, and cell division. It was not surprising, therefore, to observe that MTX-repressed genes were highly represented in the “cell cycle and DNA processing” MIPS functional category (Fig. 3). More specifically, it appeared that crucial genes involved in the control of the G2/M transition of the cell cycle (Francis, 2007) were down-regulated in MTX-treated cells, mainly in the late phase of the response. Thus, the cyclin-dependent protein kinase B CDKB2, for which the activity normally peaks in mid-to-late G2, the CKS1 and CKS2 proteins that interact with CDKs, and three cyclins A/B, substrates of CDKs, were down-regulated in response to MTX treatment (Supplemental Table S3). A similar behavior was observed for CDC25, which dephosphorylates CDKA/B to allow the fully active kinase to drive cells into mitosis (Francis, 2007). Also, the down-regulation of seven tubulin α- and β-chain coding genes and of the KIS tubulin-folding cofactor gene suggested that the microtubule cytoskeleton, which forms unique arrays during cell division, was markedly affected in MTX-treated cells (Supplemental Table S3). Together, these data suggested that folate depletion probably resulted in perturbation of nucleotide homeostasis and abnormal DNA synthesis during the S phase and induced a subsequent blockage of cell division through a down-regulation of key genes of the G2/M check-point.
As illustrated in Figure 3, the impairment in cell division was accompanied by an important repression of genes involved in protein synthesis and metabolism. Thus, 44 genes coding ribosomal proteins, which are structural constituents of ribosome 40S and 60S, were down-regulated in the early and late phases of MTX response. An important decrease in cellular energy metabolism was also a characteristic of the response induced by MTX. Measurements of respiration rates in control and treated cells indicated an approximately 40% reduction after 24 h of exposure to the antifolate (Supplemental Fig. S1). Photosynthesis, which is not elevated in Arabidopsis cells grown under mixotrophic conditions, was also reduced by about 50% in MTX-treated cells as compared to controls after 24 h of treatment. These measurements fit well with the transcriptomic data showing that key genes of both respiration (e.g. subunits of mitochondrial inner membrane complexes) and photosynthesis (e.g. small subunit of Rubisco and subunits of PSI and PSII) were down-regulated in the late phase of exposure to MTX (Supplemental Fig. S2). Several genes involved in photorespiration, a process that is tightly associated with photosynthesis in C3 plants and depends on folate-dependent steps located in mitochondria, were also down-regulated in response to MTX treatment. These genes encode the principal mitochondrial isoform of Ser hydroxymethyltransferase (SHMT; Voll et al., 2006) and the H, P, and T subunits of the Gly decarboxylase complex (Douce et al., 2001).
In conclusion, the effects of MTX treatment resulted in a broad slowdown of essential physiological functions, including cell division and primary energy metabolism. These changes could be due, to some extent, to perturbations of NTP and dNTP pools following MTX treatment. Indeed, in addition to their essential role as building blocks for nucleic acid synthesis, purine and pyrimidine are also required for energy metabolism and participate in many important biochemical processes (Zrenner et al., 2006). Folate depletion, through the inhibition of both purine and pyrimidine synthesis (Fig. 1), was likely responsible for the observed physiological and genome-wide response of Arabidopsis cells to MTX treatment. To support these assumptions, Kidd et al. (1982) have shown that pea (Pisum sativum) seedlings treated with the antifolate asulam accumulated two intermediates of purine biosynthesis, thus indicating that folate deficiency was associated with an impairment of nucleotide synthesis. Also, growth inhibition of tobacco (Nicotiana tabacum) protoplast-derived cells induced by MTX was prevented by a mixture of Met, adenine, and thymidine (Poll et al., 1984).
MTX Induces a Stress Response Characteristic to Xenobiotic Treatments
According to the MIPS catalogue, 66 putative transcription factors belonging to various families were found to be up-regulated in response to MTX treatment (i.e. 4.5% of the up-regulated genes; Fig. 3). A search in the plant transcription factor database PlantTFDB (Guo et al., 2008) enlarged this list to 146 members. Among them, 86 genes were associated with the early response and 13 displayed log2(ratio) higher than 3 (8-fold induction) after 6 h of exposure to MTX. To analyze whether these genes were specifically associated with MTX treatment and/or perturbation of folate homeostasis, we analyzed their expression profiles using the Genevestigator database and software (Zimmermann et al., 2004). The 13 selected genes were found to be up-regulated to elevated levels by at least one abiotic and biotic stress. Examination of other up-regulated genes coding putative transcription factors suggested that most of these changes were not characteristic for folate depletion but most likely resulted from a stress response induced by the MTX drug. This assumption was strengthened by the observation that MTX-induced genes were highly represented in the MIPS functional category “cell rescue and defense” (Fig. 3). These genes related to stress response include, for example (Supplemental Table S3), the Aox1a and Aox1d genes coding alternative oxidases (Clifton et al., 2006) and genes involved in the synthesis of ethylene and polyamines, two metabolites derived from AdoMet (Fig. 1) and involved in plant stress responses (Fujita et al., 2006; Groppa and Benavides, 2008). Other stress-related genes could be associated more specifically to detoxification of xenobiotics by plants (Coleman et al., 1997). They include several genes coding Cyt P450s, glutathione S-transferases, glycosyl- and sulfotransferases, ATP-binding cassette transporters, and MATE efflux family proteins (Supplemental Table S3). The most important protective phases of the detoxification process involve conjugation of the drug to glutathione or another hydrophilic molecule and sequestration of the conjugates into the vacuole or extrusion outside the cell (Coleman et al., 1997). In animal models, it has been shown that members of the ATP-binding cassette superfamily, including multidrug resistance proteins (MRPs) and breast cancer resistance protein (BCRP), are high-capacity ATP-driven MTX efflux transporters (Assaraf, 2006). Therefore, by actively extruding antifolates, overexpressed MRPs and/or BCRP confer resistance to antifolate chemotherapy. In Arabidopsis, the AtMRP1 and AtMRP4 proteins have been shown to mediate transport of MTX and/or folate (Klein et al., 2004). These two genes were not differentially expressed in MTX-treated cells. However, AtMRP2 and AtMRP3, for which the ability to transport MTX or folate has not yet been tested (Klein et al., 2006), were up-regulated to a limited extent (2- to 3-fold) in response to MTX treatment (Supplemental Table S3). Additional experiments will be necessary to determine whether these MRPs and other MTX-responsive transporters are involved in antifolate detoxification and, eventually, in the transport of physiological folates.
Supplementation with Leucovorin Allows Determining the Specificity of the Response to Folate Depletion
Taken together, the above-presented data indicated that treatment with MTX was responsible for a stress response, in part related to xenobiotic-induced detoxification processes. The main goal of our study was to gain insight into the regulatory process leading to the adaptation of C1 metabolism upon perturbation of folate homeostasis. To discriminate between the response due to the undesired effects of MTX and the one associated with folate depletion, we used leucovorin (LV), a stable folate derivative (5-formyl-THF) that is administered in anticancer chemotherapy after antifolate treatment to rescue normal cells from the cytotoxicity of MTX (Stover and Schirch, 1993; McGuire, 2003). LV is an inhibitor of many folate-dependent enzymes in vitro, but exceptionally high levels of 5-formyl-THF do not much affect fluxes through folate-dependent reaction in vivo in Arabidopsis (Goyer et al., 2005). Thus, this well-tolerated folate derivative was used for supplementation assays. The new set of experiments comprised four culture conditions: cells treated with MTX (100 μm, as for the transcriptomic studies) or LV (500 μm), cells treated with both compounds, and control cells. Cells were collected after 6, 12, and 24 h of exposure to the different compounds.
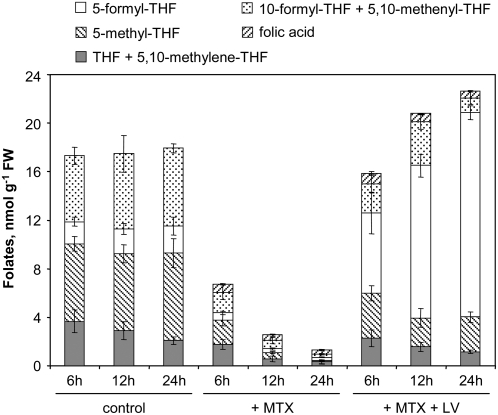
In this series of experiments, control cells contained 17 ± 3 nmol folate/g FW, a pool that is substantially higher than the one measured in control cells used for transcriptomic studies (12 ± 2 nmol/g FW; Supplemental Table S1; Fig. 4). Cell treatment with MTX resulted in an important depletion of folate to 1.3 nmol/g FW, whereas supplementation with LV alone led to intracellular folate accumulation to elevated levels (>50 nmol/g FW; Supplemental Table S1). The simultaneous addition of LV and MTX in the culture medium did not interfere with drug uptake but fully abolished the overall folate depletion induced by MTX (Fig. 4). Indeed, the folate pool was maintained above 16 nmol/g FW throughout the examined period. However, the distribution of C1 derivatives was significantly changed, with an increased ratio of 5-formyl-THF in LV-supplemented cells as compared to control cells (Fig. 4).
Figure 4.
Measurements of folate pools in Arabidopsis cells treated with MTX alone or in combination with LV. Cells were grown in standard conditions or exposed to MTX (100 μm) alone or in combination with LV (5-formyl-THF, 500 μm). The following pools of folates were determined by LC-MS/MS (Zhang et al., 2005): 5-methyl-THF, 5-formyl-THF, THF plus 5,10-methylene-THF, 10-formyl-THF plus 5,10-methenyl-THF, and folic acid. Data are means of three biological replicates and sd. Values are available in Supplemental Table S1.
Analysis of the physiological parameters of cells treated with MTX and LV indicated that supplementation was associated with a partial rescue of cell growth and primary energy metabolism (Supplemental Fig. S1). It seems, therefore, that these cellular processes were not affected by overall folate depletion but rather by changes in individual C1-derivative pools, which were not restored to accurate levels in supplementation assays. The persistent deleterious effects of MTX, albeit LV supplementation, may also be due to the direct inhibition of key enzymes involved in purine and thymidylate synthesis, as demonstrated in animal models (McGuire, 2003). In animals, the folate-dependent enzymes thymidylate synthase, glycinamide ribonucleotide, and aminoimidazole-carboxamide ribonucleotide transformylases (Fig. 1) have been shown to be inhibited by MTX and by dihydrofolates, which accumulate when DHFR, the principal target of MTX, is blocked (McGuire, 2003). The parallel between the animal and plant systems is supported by two observations. First, MTX accumulated to elevated levels in Arabidopsis cells (Supplemental Table S1) so that the activities of the hypothetical secondary targets of MTX should be markedly impaired. Second, folic acid accumulated in Arabidopsis cells treated with MTX alone or in combination with LV (Fig. 4). The increase in folic acid pool was probably due to the inhibition of DHFR enzymes and the accumulation of their substrate, dihydrofolates. These compounds could not be measured directly in MTX-treated cells, because they are very labile and are in part oxidized to folic acid during the analytical procedure used for folate measurements (De Brouwer et al., 2007). Also, it is likely that dihydrofolates were oxidized to folic acid in vivo and potentially catabolized to pteridines and p-aminobenzoylglutamate, as shown in Escherichia coli cells treated with the DHFR inhibitor trimethoprim (Quinlivan et al., 2000).
Together, these results indicated that supplementation of MTX-treated cells with LV prevented overall folate depletion but did not restore a folate composition similar to control cells. Moreover, the subsequent analyses of selected genes and metabolites indicated a delay in the response to folate depletion in the two sets of experiments (transcriptomic and rescue studies), likely because cells initially displayed differences in folate pool size and composition (Supplemental Table S1). Thus, the early response took place after 6 h in cells collected for the transcriptomic studies (Supplemental Table S2), whereas it was delayed to 12 h in cells used for the rescue experiments (Figs. 5 and 7). A detailed analysis of folate pools in the two sets of cultures was done to identify which derivative(s) could be responsible for the observed delay. It appears that most of the changes in gene expression and metabolite levels could be correlated to the cellular level of 5-methyl-THF, with a threshold at about 0.5 nmol/g FW below which cellular responses are induced (Supplemental Table S1). We cannot exclude that another folate derivative can act as a sensor in transcriptional and metabolic controls. For example, the level of 5,10-methylene-THF, which could not been determined because it is converted to THF during the analytical procedure, could play an important role in signaling folate depletion in Arabidopsis cells. In yeast (Saccharomyces cerevisiae), the level of 5,10-methylene-THF in the cytoplasm has a direct role in signaling transcriptional control of several genes involved in C1 metabolism and de novo purine synthesis (Piper et al., 2000; Gelling et al., 2004). As mentioned above, the inhibition of DHFR could have resulted in the accumulation of folate catabolic products in Arabidopsis cells treated with MTX. Although we did not quantify pteridines and p-aminobenzoylglutamate, it is likely that these compounds do not play any important role in the transcriptional and metabolic responses we observed, because LV supplementation abolished or markedly reduced the effects of MTX (see below), albeit folic acid, and potentially folate catabolites, accumulated to similar levels in cells treated with MTX or MTX plus LV (Supplemental Table S1).
Figure 5.
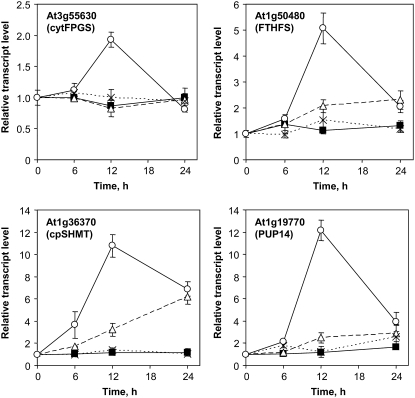
Expression profiles of genes involved in the adaptation of C1 metabolism in response to folate depletion. The relative steady-state transcript levels for At3g55630 (cytFPGS), At1g50480 (FTHFS), At1g36370 (cpSHMT), and At1g19770 (PUP14) were determined by qPCR. Control cells (▪) and cells treated with MTX (○), MTX and LV (△), and LV (×) were analyzed after 0, 6, 12, and 24 h of treatment. The actin2/7 gene (At5g09810) was used to normalize qPCR data. Data are means of three biological replicates and sd.
Figure 7.
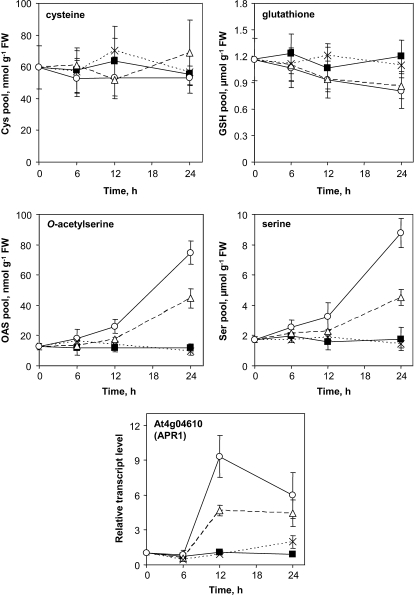
Measurements of key metabolites and gene of S metabolism in folate-depleted cells. Control cells (▪) and cells treated with MTX (○), MTX and LV (△), and LV (×) were analyzed after 0, 6, 12, and 24 h of treatment. The pools of Cys and glutathione, on the one hand, and of OAS and Ser, on the other hand, were determined by RP-HPLC after derivatization with monobromobimane and O-phthaldialdehyde, respectively. The relative steady-state transcript level for At4g04610 (APR1) was determined by qPCR. The actin2/7 gene (At5g09810) was used to normalize qPCR data. Data are means of three biological replicates and sd.
Folate Depletion Is Accompanied by a Limited Response of the Gene Network Controlling the Intracellular Folate Pool
Folate-autotrophs are able to synthesize THF through a complex biosynthetic route comprising 10 reactions (Rébeillé et al., 2006). In plants, the expression level of several enzymes involved in this pathway was correlated with the pool of folate and the demand for C1 units in various physiological situations (Basset et al., 2002, 2004a, 2004b; Jabrin et al., 2003). Our work is, to our knowledge, the first genome-wide analysis of the effect of folate limitation in a folate autotroph, providing the opportunity to study the transcriptional regulation of cofactor biosynthesis. Our transcriptomic data indicate that the expression of only one gene involved in THF synthesis was modified by folate depletion (Supplemental Table S4). This gene encodes the cytosolic isoform of folylpolyglutamate synthetase (cytFPGS), the last enzyme of the biosynthetic route, which is responsible for polyglutamylation of THF and C1-substituted folate coenzymes in the cytosol (Ravanel et al., 2001). The differential expression of the cytFPGS gene (2- to 5-fold induction) was confirmed by qPCR in both cells used for the transcriptomic analysis and the supplementation assays. The cytFPGS expression profiles illustrated the previously mentioned delay between the two series of experiments. In the first experiment, the cytFPGS transcript peaked after 6 h of exposure to MTX (Supplemental Tables S2 and S4), whereas maximal expression level was observed at 12 h in the second series of experiments (Fig. 5). The induction of the cytFPGS gene observed in MTX-treated cells was abolished in cells treated with both MTX and LV, and the steady-state level of the transcript was not affected by supplementation with LV alone (Fig. 5). This suggested that induction of cytFPGS was not a consequence of MTX or LV uptake and accumulation in cells. Rather, this adaptation was due to depletion of a natural folate, possibly 5-methyl-THF and/or 5,10-methylene-THF.
Due to its positioning at the end of THF biosynthetic pathway, cytFPGS is not assumed to change flux toward THF synthesis but rather to influence the extent of folate glutamylation in the cytosol. Under folate-limiting conditions, there may be two advantages in favoring polyglutamylation of folates. First, folylpolyglutamates are the most efficient substrates for most folate-dependent enzymes. For example, the cytosolic and chloroplastic isoforms of Met synthase strictly depend on a polyglutamylated 5-methyl-THF derivative to catalyze their reaction (Ravanel et al., 2004). Second, folylpolyglutamates are less susceptible to catabolism and turnover (Suh et al., 2001). In the absence of transcriptional regulation of Arabidopsis genes involved in folate catabolism or salvage (Supplemental Table S4), one can propose that increasing polyglutamylation in the cytosol is an adaptation to limit folate breakdown in cells treated with MTX.
In folate-auxotroph organisms, the expression of folate transport proteins located on the plasma membrane adapts to extracellular folate levels and thus participates to the regulation of intracellular homeostasis of the cofactor (Matherly and Goldman, 2003). It appears that Arabidopsis cells do not elaborate a coordinated transcriptional regulation of key genes involved in cofactor synthesis, degradation, or trafficking to compensate folate limitation induced by MTX. This finding was unexpected, because the expression of several genes of THF biosynthesis was correlated with folate levels in many physiological situations (Basset et al., 2002, 2004a, 2004b; Jabrin et al., 2003). Also, the overexpression of genes involved in the first steps of THF synthesis led to transgenic plants with increased folate levels (Diaz de la Garza et al., 2007; Storozhenko et al., 2007). Together, our data suggest that folate level does not exert feedback transcriptional control on its own biosynthetic route, a situation that does not exclude regulation through posttranscriptional events.
The Imbalanced Activity of the Methyl Cycle Induced by 5-Methyl-THF Limitation Cannot Be Compensated through Transcriptional Response
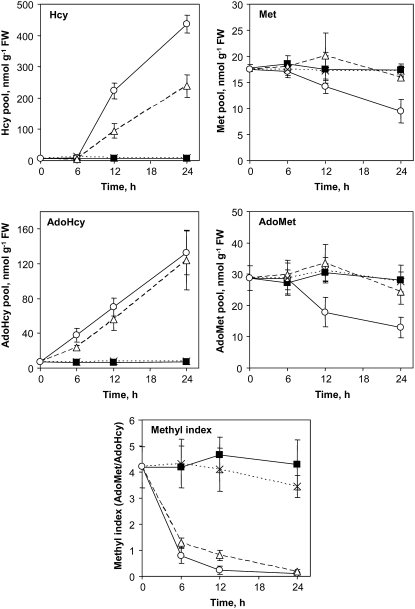
The pool of 5-methyl-THF was rapidly reduced in cells treated with MTX. As a consequence, the homeostasis of the intermediates of the activated methyl cycle was markedly affected, as previously observed by Loizeau et al. (2007). Thus, after 6 h of treatment, 5-methyl-THF was reduced by approximately 70% as compared to controls (Fig. 4) and S-adenosylhomocysteine (AdoHcy), the by-product of AdoMet-methyltransferases (MTases), accumulated by 5-fold (Fig. 6). The rise in the AdoHcy pool could be attributed to the activity of AdoHcy hydrolase, a cytosolic reversible enzyme that catalyzes the hydrolysis of AdoHcy into homo-Cys (Hcy) and adenosine (Fig. 1). The hydrolytic reaction is only favored by removal of reaction products through the enzymes Met synthase and adenosine kinase, respectively (Moffatt and Weretilnyk, 2001). In MTX-treated cells, 5-methyl-THF depletion most probably impaired the reaction catalyzed by Met synthase, thus limiting the removal of Hcy in the cytosol and favoring AdoHcy hydrolase in the direction of AdoHcy synthesis (Fig. 1; Loizeau et al., 2007). In the very early phase (6 h) of the response, the other key intermediates of the activated methyl cycle did not change significantly (Fig. 6), suggesting that the conversion of Hcy to AdoHcy was a metabolic priority, probably to avoid the toxic effect of this compound (Perla-Kajan et al., 2007); and the synthesis of Met and AdoMet was sufficient to follow the demand for anabolic reactions (e.g. protein, ethylene, and polyamine synthesis). To support this last assumption, it is worth noting that the Km value for chloroplastic Met synthase is 4-fold lower than for the cytosolic isoforms (17 versus 60 μm, respectively; Ravanel et al., 2004), suggesting that upon 5-methyl-THF limitation, the de novo synthesis of Met in plastids could be less affected than Met recycling in the cytosol.
Figure 6.
Analysis of key intermediates of the activated methyl cycle in Arabidopsis cells treated with MTX and LV. Control cells (▪) and cells treated with MTX (○), MTX and LV (△), and LV (×) were analyzed after 0, 6, 12, and 24 h of treatment. The pools of Met, Hcy, AdoMet, and AdoHcy were determined by RP-HPLC after derivatization with specific probes as described in “Materials and Methods.” The methyl index was determined as the AdoMet to AdoHcy ratio. Data are means of three biological replicates and sd.
The major consequence of the AdoHcy increase was a 5-fold reduction in the AdoMet to AdoHcy ratio (referred to as the methyl index), which is an indicator of the cell capacity to perform methylation reactions. Indeed, AdoMet-dependent MTases are strongly inhibited by AdoHcy, which behaves as a competitive inhibitor to the substrate AdoMet (Moffatt and Weretilnyk, 2001). Despite these important changes that could have short-term deleterious effects on cell physiology, the key genes involved in the activated methyl cycle were not differentially regulated during the early response to MTX (Supplemental Table S4). Thus, the imbalanced activity of the cycle initiated after 6 h of exposure to MTX propagated until the end of the treatment. As a consequence, AdoHcy and then Hcy accumulated to elevated levels, and the Met and AdoMet pools progressively declined, probably because of an imbalance between fluxes of regeneration and utilization of these metabolites (Fig. 6). Supplementation of MTX-treated cells with LV allowed to maintain the 5-methyl-THF pool at 50% ± 10% of the control levels (Fig. 4), a situation that was sufficient to prevent Met and AdoMet changes (Fig. 6). However, this reduction was sufficient to trigger AdoHcy and Hcy, to a lower extent, accumulation, suggesting again that the cytosolic Met synthases were more sensitive to 5-methyl-THF limitation than their chloroplastic counterpart. In the late phase of the response (24 h), several genes coding enzymes of the activated methyl cycle were down-regulated (Supplemental Table S4), suggesting an attempt to adapt to long-term perturbation of the cycle. In this regard, it has been shown previously that a persistent situation of folate deficiency was associated with a posttranslational modification of the first enzyme specific for de novo Met synthesis and a restoration of Met and AdoMet homeostasis (Loizeau et al., 2007).
After 12 and 24 h of exposure to MTX, the methyl index was reduced by 20- to 40-fold, respectively, suggesting a dramatic impairment in cell ability to perform methylation reactions. Even though, our transcriptomic data indicated that only a limited number of genes coding MTases were differentially expressed under these conditions. Among the 135 genes coding MTases in the Arabidopsis genome, only four were up-regulated and six were down-regulated with at least 2-fold changes in response to MTX treatment (Supplemental Table S4). Three of these differentially expressed MTases are involved in the biogenesis of sterols, which are structural components of membranes and precursors of steroid hormones (He et al., 2003). The potential role of these compounds in the adaptation of Arabidopsis cells to folate depletion is not known. Together, these results indicated that, despite the probable marked limitation of MTases activities, transcriptional regulation of most, if not all, genes coding these enzymes was not dependent on the methyl index.
Folate Depletion Is Associated with a Reorientation of C1 Units toward the Synthesis of Nucleotides in Plastids
The pool of folate is made of a dynamic equilibrium of coenzymes carrying C1 units at different oxidation levels and having different metabolic fates. Several genes coding enzymes involved in the loading of C1 units onto THF and in the conversion of C1 units were differentially expressed in cells treated with MTX (Supplemental Table S4). As previously mentioned, genes coding mitochondrial SHMT and several subunits of the Gly decarboxylase complex were down-regulated in the late phase of the response to MTX treatment. We found that the transcripts coding 10-formyl-THF synthetase (FTHFS) and chloroplastic SHMT (cpSHMT) were up-regulated in the early phase of the response (Fig. 5; Supplemental Tables S2 and S4). The up-regulation of these genes was markedly reduced in cells supplemented with LV, suggesting that theses changes were associated with a limitation in folate availability, possibly 5-methyl-THF and/or 5,10-methylene-THF. Moreover, these results suggested a reorientation of C1 metabolism in plastids to favor the synthesis of 10-formyl-THF and/or 5,10-methylene-THF and thus to allocate C1 units toward the synthesis of nucleotides in this compartment (Fig. 1). First, the ATP-dependent condensation of formate and THF to produce 10-formyl-THF is catalyzed by FTHFS, an enzyme found in the cytosol, mitochondria, and plastids (Hanson and Roje, 2001). Also, the de novo purine biosynthetic pathway, including the 10-formyl-THF-dependent glycinamide ribonucleotide and aminoimidazole-carboxamide ribonucleotide formyltransferases, is located in plastids in Arabidopsis (Zrenner et al., 2006). Second, cpSHMT is involved in the conversion of Ser and THF into Gly and 5,10-methylene-THF (Besson et al., 1995). A bifunctional DHFR/thymidylate synthase is present in plastids (Luo et al., 1997) and could allow the 5,10-methylene-dependent methylation of dUMP to dTMP. The reorientation of C1 metabolism toward the synthesis of nucleotides in plastids was strengthened by the simultaneous up-regulation of the AtPUP14 gene coding a putative purine transporter (Fig. 5). Because this transporter is predicted to be located in chloroplasts (TargetP server; Emanuelsson et al., 2000), one can suggest that it will be involved in the efflux of purine outside of plastids to sustain metabolic processes in the rest of the cell.
Folate Depletion Induces O-Acetylserine Accumulation and Up-Regulates the Sulfate Assimilatory Pathway
The genome-wide analysis of MTX-treated Arabidopsis cells revealed a set of genes involved in the sulfate assimilatory pathway. Most of these genes were up-regulated in the early phase of the response and encode sulfate transporters and key enzymes involved in sulfate activation, reduction, and incorporation into Cys (Supplemental Table S5). The induction of these genes, e.g. APR1 coding adenosine 5′-phosphosulfate reductase (Fig. 7), is characteristic for the adaptive response of plant to sulfur (S) deficiency (Hirai et al., 2003; Nikiforova et al., 2003). Under S-limiting conditions, the pools of the S-containing metabolites Cys and glutathione are also markedly reduced (Hirai et al., 2004; Nikiforova et al., 2005). Our metabolic analyses indicated that folate depletion was not associated with significant changes in the level of Cys and that the pool of glutathione was reduced by about 30% after 24 h of exposure to MTX (Fig. 7). However, we found that O-acetylserine (OAS), a key intermediate of sulfate assimilation, accumulated in folate-depleted cells (Fig. 7). OAS has been shown to act as a regulator of global transcript and metabolites profiles under S-limiting conditions (Hirai et al., 2004; Nikiforova et al., 2005). Thus, OAS accumulation in the early phase of the response to MTX treatment was probably at the origin of the observed transcriptional response. OAS is synthesized from Ser and acetyl-CoA by the enzyme Ser acetyltransferase (Wirtz and Droux, 2005). In MTX-treated cells, the pool of Ser increased gradually, and this could be attributed to an impairment of the conversion of Ser into Gly catalyzed by the folate-dependent enzyme SHMT (Fig. 1; Prabhu et al., 1998; Loizeau et al., 2007). We suggest that the increase in Ser levels in MTX-treated cells was at the origin of the elevation of OAS through an increased activity of Ser acetyltransferase. This assumption is supported by the previous finding that Ser application to Arabidopsis plants increased OAS levels and induced S deficiency response (Ohkama-Ohtsu et al., 2004). Also, in cells treated with MTX and LV, we found that the accumulation of Ser, OAS, and transcripts of the S-responsive gene APR1 was less important and delayed as compared to cells treated with MTX alone (Fig. 7). Together, these data indicated that folate depletion mimicked S deficiency response through the OAS-mediated coordinated transcriptional regulation of key genes of the sulfate assimilatory pathway.
CONCLUSION
The present genome-wide and metabolic analysis revealed several adaptive responses of Arabidopsis cells to MTX treatment and perturbation of folate homeostasis. Transcriptomic data indicated that MTX treatment induced a stress response and suggested that a protective effect builds up rapidly after cellular uptake of the drug. Similar stress-responsive effects have been associated with antifolate application or folate depletion in bacteria, animal models, fly, and yeast (e.g. Huang et al., 1997, 2004; Gelling et al., 2004; Affleck et al., 2006). As a consequence of the multiplicity of cellular functions in which folate derivatives are involved, perturbations to this network had far-reaching consequences. Thus, modification of nucleotide homeostasis resulted in cell division arrest and in reduction of energy metabolism. Also, impairment of Ser catabolism led to OAS accumulation and to a transcriptional response typical of S starvation.
Regarding C1 metabolism, the major adaptive response of Arabidopsis cells to folate depletion concerned the composition of the folate pool rather than the intracellular level of cofactors. Indeed, no significant change in the expression of genes that could have increased the flux toward THF production was observed. However, the distribution of C1 derivatives as well as the expression of several genes coding enzymes manipulating C1 moieties was significantly affected in depleted cells. Because of the predominance of 5-methyl-THF over other folate derivatives in many plant organs, tissues, or cells, previous studies concluded that the synthesis of Met and AdoMet was a metabolic priority in many physiological situations (Hanson and Roje, 2001; Rébeillé et al., 2006). Our study indicates that this situation is not true in Arabidopsis cells for which the pool of folate is limited by the use of MTX. Indeed, no genomic or metabolic adaptation was built up to counterbalance the major impairment of the methyl index, which controls methylation of essential metabolites, proteins, and nucleic acids in the cell. However, folate depletion was associated with increased expression levels for transcripts coding FTHFS, cpSHMT, and a purine transporter, suggesting a reorientation of C1 units toward the synthesis of purine and thymidylate in plastids. To summarize, the metabolic priority of Arabidopsis cells in response to folate limitation was to shuttle the available folate derivatives to the synthesis of nucleotides at the expense of AdoMet synthesis and methylation reactions. Similar conclusions were obtained using human colonic epithelial cells grown in folate-deficient conditions (Hayashi et al., 2007). Moreover, it was proposed that, under certain conditions, the cytoplasmic SHMT acts as a switch to modulate nucleotide synthesis and methylation reactions in animal cells (Herbig et al., 2002). In yeast, a limitation in the cytoplasmic pool of 5,10-methylene-THF was associated with the co-induction of several genes involved in C1 metabolism (referred to as the C1 regulon) as well as genes for enzymes of de novo purine biosynthesis (Gelling et al., 2004). In contrast to the situation existing in plant and animal cells, co-regulation of the C1 regulon and purine synthesis in yeast was not at the expense of Met and AdoMet synthesis. Together, the data obtained in folate-auxotroph and folate-autotroph organisms indicate that C1 metabolism operates as a flexible system whereby folate composition is adapted to maintain the supply of C1 units to anabolic pathways.
MATERIALS AND METHODS
Cells and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia cell suspension cultures were grown under continuous light (40 μE m−2 s−1) at 22°C with rotary agitation at 125 rpm in Gamborg's B5 medium supplemented with 1 μm 2-naphtalene acetic acid and 1.5% (w/v) Suc. Cells were subcultured every 7 d, and chemicals were added to cultures at the beginning of exponential growing phase (3 d after subcloning). At each time point, cells were collected, washed with distilled water, weighed, and frozen in liquid nitrogen. Each experiment was done in triplicate.
Measurements of Respiration, Photosynthesis, and Cell Viability
Measurements of respiration rate were done in 1 mL of Gamborg's B5 medium containing 50 mm sodium bicarbonate, and oxygen consumption was determined in the darkness using an O2 electrode (Hansatech, Eurosep Instruments) at 25°C. Photosynthetic activities were determined in the same conditions using a white light source of 2,000 μE m−2 s−1. Cell viability was determined using the vital dye fluorescein diacetate, as described by Loizeau et al. (2007).
Measurements of Metabolites
Determination of folates was done by LC-MS/MS as described by Zhang et al. (2005). Following extraction, folylpolyglutamates were deconjugated in the presence of rat serum to generate the corresponding monoglutamate derivatives. Folates and MTX eluted from the RP-18 column were detected by electrospray ionization on an Applied Biosystems API4000 tandem quadrupole mass spectrometer. Because the stability and eventual interconversion of folates are affected during the typical sample preparation steps (De Brouwer et al., 2007), the final quantitative data were simplified to five pools of folates: 5-methyl-THF, 5-formyl-THF, THF plus 5,10-methylene-THF, 10-formyl-THF plus 5,10-methenyl-THF, and folic acid.
Soluble amino acids and thiols were extracted in aqueous ethanol buffers and quantified by reverse-phase HPLC after derivatization with O-phthaldialdehyde and monobromobimane, respectively, as described by Loizeau et al. (2007). AdoMet and AdoHcy measurements were done by reverse-phase HPLC after derivatization with chloro-acetaldehyde (Loizeau et al., 2007).
Transcriptome Studies
Microarray analysis was performed with the CATMA array containing 24,576 gene-specific tags corresponding to 22,089 genes from Arabidopsis (Crowe et al, 2003; Hilson et al, 2004). Five comparisons were analyzed in our time-course analysis of Arabidopsis cell response to MTX (Fig. 2). In comparisons 1 to 3, arrays were hybridized with cRNA from control and MTX-treated cells at each time point of the kinetic. Comparisons 4 and 5 corresponded to hybridization with cRNA from MTX-treated cells at 2 and 6 h, and at 6 and 24 h, respectively. For each comparison, we performed a repeat using a second set of cell samples originating from an independent biological repeat. In addition, to avoid dye bias and gene-specific dye bias, a dye-swap experiment was carried out. Therefore, four arrays were used for each comparison. Total RNA was extracted from Arabidopsis cells using the RNeasy Plant Mini kit (Qiagen) following the manufacturer's protocol. RNA integrity, cDNA synthesis, hybridization, and array scanning were performed as described by Lurin et al. (2004).
Statistical Analysis of Microarray Data
Statistical analysis was based on two dye swaps (i.e. four arrays, each containing 24,576 gene-specific tags and 384 controls) as described in Gagnot et al. (2008). To determine differentially expressed genes, we performed a paired t-test on the log ratios, assuming that the variance of the log ratios was the same for all genes. Spots displaying extreme variance (too small or too large) were excluded. The raw P-values were adjusted by the Bonferroni method, which controls the Family Wise Error Rate. We considered as being differentially expressed the genes with a Bonferroni P-value ≤ 0.05, as described in Gagnot et al. (2008). We use the Bonferroni method with a type I error equal to 5% to keep a strong control of the false positives in a multiple-comparison context.
Microarray data from this article were deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/; accession no. GSE10675) and at CATdb (http://urgv.evry.inra.fr/CATdb/; project CEA06-02_Folate) according to the Minimum Information About a Microarray Experiment standards.
qRT-PCR
qRT-PCR experiments were performed using cDNA synthesized from total RNA isolated from control and treated Arabidopsis cells (Reverse-it first strand synthesis kit, ABgene). Specific primer sequences designed for each genes selected for analysis are available in Supplemental Table S6. The real-time PCR reactions were carried out on a Rotor-Gene 3000 instrument (Corbett Research) using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich). Quantification of gene expression was performed using the comparative CT method with the Rotor-Gene 3000 software. The actin2/7 gene (At5g09810) was used to normalize qPCR data.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Physiological parameters of Arabidopsis cells treated with MTX and LV.
Supplemental Figure S2. MapMan-generated overview of MTX-responsive genes involved in respiration and photosynthesis.
Supplemental Table S1. Folate measurements in Arabidopsis cells used for the transcriptomic studies (A) and the rescue experiments (B).
Supplemental Table S2. Validation of the microarray data by qPCR.
Supplemental Table S3. Selected MTX-responsive genes that may be involved in the regulation of cell cycle and response to stress.
Supplemental Table S4. Expression of genes associated with C1 metabolism in Arabidopsis cells treated with MTX.
Supplemental Table S5. Expression of genes involved in the sulfate assimilatory pathway in Arabidopsis cells treated with MTX.
Supplemental Table S6. Sequences of primers used for qPCR.
Supplementary Material
Acknowledgments
We are grateful to Dr. Julie Fiévet for her assistance with the transcriptomic data analysis and to Drs. Claude Alban and Eric Maréchal for helpful discussions and critical reading of the manuscript. We thank Professor Roland Douce for stimulating discussions.
This work was supported by the French Ministry of Research (a Ph.D. fellowship to K.L.). The folate analysis was supported by the Institute for the Promotion of Innovation through Science and Technology in Flanders (IWT Vlaanderen; grant no. GOA 1251204 [Bijzonder Onderzoeksfonds, UGent] to D.V.D.S. and W.L. and a bursary to V.D.B.).
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Stéphane Ravanel (sravanel@cea.fr).
The online version of this article contains Web-only data.
References
- Affleck JG, Neumann K, Wong L, Walker VK (2006) The effects of methotrexate on Drosophila development, female fecundity, and gene expression. Toxicol Sci 89 495–503 [DOI] [PubMed] [Google Scholar]
- Assaraf YG (2006) The role of multidrug resistance efflux transporters in antifolate resistance and folate homeostasis. Drug Resist Updat 9 227–246 [DOI] [PubMed] [Google Scholar]
- Basset G, Quinlivan EP, Ziemak MJ, Diaz De La Garza R, Fischer M, Schiffmann S, Bacher A, Gregory JF III, Hanson AD (2002) Folate synthesis in plants: the first step of the pterin branch is mediated by a unique bimodular GTP cyclohydrolase I. Proc Natl Acad Sci USA 99 12489–12494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basset GJ, Quinlivan EP, Ravanel S, Rebeille F, Nichols BP, Shinozaki K, Seki M, Adams-Phillips LC, Giovannoni JJ, Gregory JF III, et al (2004. a) Folate synthesis in plants: the p-aminobenzoate branch is initiated by a bifunctional PabA-PabB protein that is targeted to plastids. Proc Natl Acad Sci USA 101 1496–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basset GJ, Ravanel S, Quinlivan EP, White R, Giovannoni JJ, Rebeille F, Nichols BP, Shinozaki K, Seki M, Gregory JF III, et al (2004. b) Folate synthesis in plants: the last step of the p-aminobenzoate branch is catalyzed by a plastidial aminodeoxychorismate lyase. Plant J 40 453–461 [DOI] [PubMed] [Google Scholar]
- Besson V, Rebeille F, Neuburger M, Douce R, Cossins EA (1995) Evidence for three serine hydroxymethyltransferases in green leaf cells. Purification and characterisation of the mitochondrial and the chloroplastic isoforms. Plant Physiol Biochem 33 665–673 [Google Scholar]
- Clifton R, Millar AH, Whelan J (2006) Alternative oxidases in Arabidopsis: a comparative analysis of differential expression in the gene family provides new insights into function of non-phosphorylating bypasses. Biochim Biophys Acta 1757 730–741 [DOI] [PubMed] [Google Scholar]
- Coleman JOD, Blake-Kalff MMA, Davies TGE (1997) Detoxification of xenobiotics by plants: chemical modification and vacuolar compartmentation. Trends Plant Sci 2 144–151 [Google Scholar]
- Crowe ML, Serizet C, Thareau V, Aubourg S, Rouze P, Hilson P, Beynon J, Weisbeek P, van Hummelen P, Reymond P, et al (2003) CATMA: a complete Arabidopsis GST database. Nucleic Acids Res 31 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Brouwer V, Zhang GF, Storozhenko S, Straeten DV, Lambert WE (2007) pH stability of individual folates during critical sample preparation steps in prevision of the analysis of plant folates. Phytochem Anal 18 496–508 [DOI] [PubMed] [Google Scholar]
- Diaz de la Garza RI, Gregory JF III, Hanson AD (2007) Folate biofortification of tomato fruit. Proc Natl Acad Sci USA 104 4218–4222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R, Bourguignon J, Neuburger M, Rebeille F (2001) The glycine decarboxylase system: a fascinating complex. Trends Plant Sci 6 167–176 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300 1005–1016 [DOI] [PubMed] [Google Scholar]
- Francis D (2007) The plant cell cycle: 15 years on. New Phytol 174 261–278 [DOI] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K (2006) Crosstalk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol 9 436–442 [DOI] [PubMed] [Google Scholar]
- Gagnot S, Tamby JP, Martin-Magniette ML, Bitton F, Taconnat L, Balzergue S, Aubourg S, Renou JP, Lecharny A, Brunaud V (2008) CATdb: a public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res 36 D986–D990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambonnet B, Jabrin S, Ravanel S, Karan M, Douce R, Rebeille F (2001) Folate distribution during higher plant development. J Sci Food Agric 81 835–841 [Google Scholar]
- Gelling CL, Piper MDW, Hong SP, Kornfeld GD, Dawes IW (2004) Identification of a novel one-carbon metabolism regulon in Saccharomyces cerevisiae. J Biol Chem 279 7072–7081 [DOI] [PubMed] [Google Scholar]
- Goyer A, Collakova E, Diaz de la Garza R, Quinlivan EP, Williamson J, Gregory JF III, Shachar-Hill Y, Hanson AD (2005) 5-Formyltetrahydrofolate is an inhibitory but well tolerated metabolite in Arabidopsis leaves. J Biol Chem 280 26137–26142 [DOI] [PubMed] [Google Scholar]
- Groppa MD, Benavides MP (2008) Polyamines and abiotic stress: recent advances. Amino Acids 34 35–45 [DOI] [PubMed] [Google Scholar]
- Guo AY, Chen X, Gao G, Zhang H, Zhu QH, Liu XC, Zhong YF, Gu X, He K, Luo J (2008) PlantTFDB: a comprehensive plant transcription factor database. Nucleic Acids Res 36 D966–D969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson AD, Roje S (2001) One-carbon metabolism in higher plants. Annu Rev Plant Physiol Plant Mol Biol 52 119–137 [DOI] [PubMed] [Google Scholar]
- Hayashi I, Sohn KJ, Stempak JM, Croxford R, Kim YI (2007) Folate deficiency induces cell-specific changes in the steady-state transcript levels of genes involved in folate metabolism and 1-carbon transfer reactions in human colonic epithelial cells. J Nutr 137 607–613 [DOI] [PubMed] [Google Scholar]
- He JX, Fujioka S, Li TC, Kang SG, Seto H, Takatsuto S, Yoshida S, Jang JC (2003) Sterols regulate development and gene expression in Arabidopsis. Plant Physiol 131 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig K, Chiang EP, Lee LR, Hills J, Shane B, Stover PJ (2002) Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. J Biol Chem 277 38381–38389 [DOI] [PubMed] [Google Scholar]
- Hilson P, Allemeersch J, Altmann T, Aubourg S, Avon A, Beynon J, Bhalerao RP, Bitton F, Caboche M, Cannoot B, et al (2004) Versatile gene-specific sequence tags for Arabidopsis functional genomics: transcript profiling and reverse genetics applications. Genome Res 14 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai MY, Fujiwara T, Awazuhara M, Kimura T, Noji M, Saito K (2003) Global expression profiling of sulfur-starved Arabidopsis by DNA macroarray reveals the role of O-acetyl-l-serine as a general regulator of gene expression in response to sulfur nutrition. Plant J 33 651–663 [DOI] [PubMed] [Google Scholar]
- Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K (2004) Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA 101 10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EY, Mohler AM, Rohlman CE (1997) Protein expression in response to folate stress in Escherichia coli. J Bacteriol 179 5648–5653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q, Jin X, Gaillard ET, Knight BL, Pack FD, Stoltz JH, Jayadev S, Blanchard KT (2004) Gene expression profiling reveals multiple toxicity endpoints induced by hepatotoxicants. Mutat Res 549 147–167 [DOI] [PubMed] [Google Scholar]
- Jabrin S, Ravanel S, Gambonnet B, Douce R, Rebeille F (2003) One-carbon metabolism in plants. Regulation of tetrahydrofolate synthesis during germination and seedling development. Plant Physiol 131 1431–1439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd BR, Stephen NH, Duncan HJ (1982) The effect of asulam on purine biosynthesis. Plant Sci Lett 26 211–217 [Google Scholar]
- Klein M, Burla B, Martinoia E (2006) The multidrug resistance-associated protein (MRP/ABCC) subfamily of ATP-binding cassette transporters in plants. FEBS Lett 580 1112–1122 [DOI] [PubMed] [Google Scholar]
- Klein M, Geisler M, Suh SJ, Kolukisaoglu HU, Azevedo L, Plaza S, Curtis MD, Richter A, Weder B, Schulz B, et al (2004) Disruption of AtMRP4, a guard cell plasma membrane ABCC-type ABC transporter, leads to deregulation of stomatal opening and increased drought susceptibility. Plant J 39 219–236 [DOI] [PubMed] [Google Scholar]
- Loizeau K, Gambonnet B, Zhang GF, Curien G, Jabrin S, Van Der Straeten D, Lambert WE, Rebeille F, Ravanel S (2007) Regulation of one-carbon metabolism in Arabidopsis: the N-terminal regulatory domain of cystathionine gamma-synthase is cleaved in response to folate starvation. Plant Physiol 145 491–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Orsi R, Patrucco E, Pancaldi S, Cella R (1997) Multiple transcription start sites of the carrot dihydrofolate reductase-thymidylate synthase gene, and sub-cellular localization of the bifunctional protein. Plant Mol Biol 33 709–722 [DOI] [PubMed] [Google Scholar]
- Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B, et al (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matherly LH, Goldman DI (2003) Membrane transport of folates. Vitam Horm 66 403–456 [DOI] [PubMed] [Google Scholar]
- McGuire JJ (2003) Anticancer antifolates: current status and future directions. Curr Pharm Des 9 2593–2613 [DOI] [PubMed] [Google Scholar]
- Moffatt BA, Weretilnyk EA (2001) Sustaining S-adenosyl-L-methionine-dependent methyltransferase activity in plant cells. Physiol Plant 113 435–442 [Google Scholar]
- Nikiforova V, Freitag J, Kempa S, Adamik M, Hesse H, Hoefgen R (2003) Transcriptome analysis of sulfur depletion in Arabidopsis thaliana: interlacing of biosynthetic pathways provides response specificity. Plant J 33 633–650 [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R (2005) Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiol 138 304–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkama-Ohtsu N, Kasajima I, Fujiwara T, Naito S (2004) Isolation and characterization of an Arabidopsis mutant that overaccumulates O-acetyl-l-Ser. Plant Physiol 136 3209–3222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perla-Kajan J, Twardowski T, Jakubowski H (2007) Mechanisms of homocysteine toxicity in humans. Amino Acids 32 561–572 [DOI] [PubMed] [Google Scholar]
- Piper MD, Hong SP, Ball GE, Dawes IW (2000) Regulation of the balance of one-carbon metabolism in Saccharomyces cerevisiae. J Biol Chem 275 30987–30995 [DOI] [PubMed] [Google Scholar]
- Poll AM, Missonier C, Grandbastien MA, Caboche M (1984) Growth-inhibition of tobacco protoplast-derived cells by methotrexate: relationships with nitrate assimilation. Plant Sci Lett 36 169–176 [Google Scholar]
- Prabhu V, Chatson KB, Lui H, Abrams GD, King J (1998) Effects of sulfanilamide and methotrexate on 13C fluxes through the glycine decarboxylase/serine hydroxymethyltransferase enzyme system in arabidopsis. Plant Physiol 116 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlivan EP, McPartlin J, Weir DG, Scott J (2000) Mechanism of the antimicrobial drug trimethoprim revisited. FASEB J 14 2519–2524 [DOI] [PubMed] [Google Scholar]
- Ravanel S, Block MA, Rippert P, Jabrin S, Curien G, Rebeille F, Douce R (2004) Methionine metabolism in plants: chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. J Biol Chem 279 22548–22557 [DOI] [PubMed] [Google Scholar]
- Ravanel S, Cherest H, Jabrin S, Grunwald D, Surdin-Kerjan Y, Douce R, Rebeille F (2001) Tetrahydrofolate biosynthesis in plants: molecular and functional characterization of dihydrofolate synthetase and three isoforms of folylpolyglutamate synthetase in Arabidopsis thaliana. Proc Natl Acad Sci USA 98 15360–15365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rébeillé F, Ravanel S, Jabrin S, Douce R, Storozhenko S, Van Der Straeten D (2006) Folates in plants: biosynthesis, distribution, and enhancement. Physiol Plant 126 330–342 [Google Scholar]
- Roje S (2006) S-Adenosyl-L-methionine: beyond the universal methyl group donor. Phytochemistry 67 1686–1698 [DOI] [PubMed] [Google Scholar]
- Ruepp A, Zollner A, Maier D, Albermann K, Hani J, Mokrejs M, Tetko I, Guldener U, Mannhaupt G, Munsterkotter M, et al (2004) The FunCat, a functional annotation scheme for systematic classification of proteins from whole genomes. Nucleic Acids Res 32 5539–5545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storozhenko S, De Brouwer V, Volckaert M, Navarrete O, Blancquaert D, Zhang GF, Lambert W, Van Der Straeten D (2007) Folate fortification of rice by metabolic engineering. Nat Biotechnol 25 1277–1279 [DOI] [PubMed] [Google Scholar]
- Stover P, Schirch V (1993) The metabolic role of leucovorin. Trends Biochem Sci 18 102–106 [DOI] [PubMed] [Google Scholar]
- Suh JR, Herbig AK, Stover PJ (2001) New perspectives on folate catabolism. Annu Rev Nutr 21 255–282 [DOI] [PubMed] [Google Scholar]
- Voll LM, Jamai A, Renne P, Voll H, McClung CR, Weber AP (2006) The photorespiratory Arabidopsis shm1 mutant is deficient in SHM1. Plant Physiol 140 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz M, Droux M (2005) Synthesis of the sulfur amino acids: cysteine and methionine. Photosynth Res 86 345–362 [DOI] [PubMed] [Google Scholar]
- Wu K, Atkinson IJ, Cossins EA, King J (1993) Methotrexate resistance in Datura innoxia (uptake and metabolism of methotrexate in wild-type and resistant cell lines). Plant Physiol 101 477–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang GF, Storozhenko S, Van Der Straeten D, Lambert WE (2005) Investigation of the extraction behavior of the main monoglutamate folates from spinach by liquid chromatography-electrospray ionization tandem mass spectrometry. J Chromatogr A 1078 59–66 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R (2006) Pyrimidine and purine biosynthesis and degradation in plants. Annu Rev Plant Biol 57 805–836 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.